
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol., 07 December 2023
Sec. Evolutionary Developmental Biology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1323378
This article is part of the Research TopicTunicates in Evolutionary Developmental BiologyView all 12 articles
 Haiyang Feng1,2*
Haiyang Feng1,2* Eric M. Thompson2,3*
Eric M. Thompson2,3*A single Aurora kinase found in non-vertebrate deuterostomes is assumed to represent the ancestor of vertebrate Auroras A/B/C. However, the tunicate Oikopleura dioica, a member of the sister group to vertebrates, possesses two Aurora kinases (Aurora1 and Aurora2) that are expressed in proliferative cells and reproductive organs. Previously, we have shown that Aurora kinases relocate from organizing centers to meiotic nuclei and were enriched on centromeric regions as meiosis proceeds to metaphase I. Here, we assessed their respective functions in oogenic meiosis using dsRNA interferences. We found that Aurora1 (Aur1) was involved in meiotic spindle organization and chromosome congression, probably through the regulation of microtubule dynamics, whereas Aurora2 (Aur2) was crucial for chromosome condensation and meiotic spindle assembly. In vitro kinase assays showed that Aur1 and Aur2 had comparable levels of kinase activities. Using yeast two-hybrid library screening, we identified a few novel interaction proteins for Aur1, including c-Jun-amino-terminal kinase-interacting protein 4, cohesin loader Scc2, and mitochondrial carrier homolog 2, suggesting that Aur1 may have an altered interaction network and participate in the regulation of microtubule motors and cohesin complexes in O. dioica.
The Aurora kinases are a group of evolutionarily conserved serine/threonine protein kinases that regulate diverse cellular events during mitosis and meiosis (Hochegger et al., 2013). Aurora originated as a single gene in the last eukaryotic common ancestor (LECA) and evolved independently in plants, fungi, protists, and metazoans (Brown et al., 2004). However, the Aurora kinase sequences of invertebrates are highly divergent from those of vertebrates and often exhibit long-branch attraction artifacts in phylogenetic analyses (Brown et al., 2004). The single, bifunctional Aurora present in the echinoderm starfish and tunicate ascidians are believed to represent the ancestral kinase that gave rise to Auroras A and B in vertebrates (Abe et al., 2010; Hebras and McDougall, 2012). Whether this scenario is generally applicable in chordates requires stringent functional evidence across closely related phyla, in order to better understand the selection pressures that restrain the concerted functions of Aurora homologs.
In species with a single Aurora, it functions at the spindle poles, chromosomes, and central spindle in mitosis (Li et al., 2008; Abe et al., 2010; Hebras and McDougall, 2012). In metazoans possessing two Aurora kinases, functional studies consistently reveal the division of labor between A- and B-type Aurora kinases at the spindle poles and equator (Hochegger et al., 2013). In contrast, reports on the meiotic functions of Aurora kinases in a few model organisms are sparse and less consistent (Nguyen and Schindler, 2017). Aurora A has a common role in regulating microtubule assembly and maintenance in the meiotic spindle, but this task is carried out at different sites. Aurora A associates with acentriolar microtubule organizing centers (aMTOCs) at the spindle poles in mouse oocytes and induces the fragmentation of aMTOCs through the phosphorylation of polo-like kinase 1 (PLK1) (Blengini et al., 2021), while active Aurora A/AIR-1 is concentrated on chromosomes during prometaphase I and on interchromosomal microtubules during anaphase I in C. elegans oocytes (Sumiyoshi et al., 2015). The chromosomal passenger complex (CPC) kinase, Aurora B, is involved in multiple aspects of meiotic division, such as chromosome condensation, cohesion, and error correction in kinetochore-microtubule (KT-MT) attachments. In C. elegans oocytes, Aurora B/AIR-2 is localized on the equatorial axis of holocentric bivalents where it provides spatial cues to load condensin I, promotes ring complex (RC) assembly, and releases interhomolog cohesins at the onset of anaphase I (Rogers et al., 2002; Collette et al., 2011; Divekar et al., 2021). In Drosophila oocytes, Aurora B/ial interacts with non-centromeric chromosomes and regulates spindle bipolarity and chromosome biorientation at the central spindle ring (Radford et al., 2012). In mouse oocytes, Aurora C replaces Aurora B at the interchromatid axis (ICA) and centromeres and also localizes on aMTOCs. Aurora C regulates the loading of condensin, chromosome alignment, correction of erroneous KT-MT attachments, and aMTOC clustering (Balboula and Schindler, 2014; Nguyen et al., 2014; Balboula et al., 2016). Subtle differences in the localizations and catalytic activities of Aurora kinases are imposed by their interacting proteins (Carmena et al., 2009). It remains unclear to what extent the distinct requirements of Aurora kinases in meiotic progression reflect the diversity of their interaction proteins and substrates in different organisms. This is of interest in the context of gene gains and losses during metazoan evolution.
The tunicate, Oikopleura dioica, in the sister group to vertebrates, is an emerging model used to study variants in cell cycle regulation (Delsuc et al., 2006; Campsteijn et al., 2012; Ma et al., 2022). It has a compact genome of 70 Mb, accompanied by massive gene losses and lineage-specific gene duplications (Denoeud et al., 2010; Ferrández-Roldán et al., 2021). In contrast to ascidians, appendicularians possess two Aurora kinase homologs. In this study, we assessed the distinct roles of two Aurora kinases during oogenesis in O. dioica. We found that Aurora1 regulated microtubule dynamics during meiotic spindle assembly, and Aurora2 was essential for chromosome condensation and meiotic spindle assembly. We also identified some novel interaction proteins for Aurora1, suggesting new avenues for further investigation of the evolutionary trajectory of Aurora kinases in chordates.
Oikopleura dioica culture was carried out as before (Bouquet et al., 2009). dsRNA was synthesized using the T7 RiboMAX™ Express RNAi System (Promega) from a 300–400-bp DNA fragment within the CDS of the target gene as templates. At day 4, females were placed in artificial seawater, removed from their houses, anesthetized in ethyl 3-aminobenzoate methanesulfonate salt (MS-222, 0.125 mg/mL, Sigma), and injected with 400 ng/μL of dsRNA solutions. Shortly before spawning, the injected mature females were individually transferred to artificial seawater in six-well plates coated with 0.1% gelatin. After spawning, oocytes were collected for RT–qPCR or used for phenotypic analyses. Capped Aur1-GFP mRNA was synthesized from a linearized pANZ-OdAur1-GFP fusion construct (IDT) using the mMESSAGE mMACHINE T7 Transcription Kit (Ambion) and was injected into the 1-cell embryos at 400 ng/μL. The injected embryos were collected at 2–8 cell stages for immunostaining using an anti-GFP antibody (AMS Biotechnology, TP401) to detect the localizations of Aurora1.
Primary antibodies include anti-Histone H3-pS10 (Upstate, 06–570), anti-H3-pS28 (Abcam, ab10543), anti-Aurora A (pT288)/Aurora B (pT232)/Aurora C (pT198) (Cell Signaling Technology, D13A11), anti-α-Tubulin (Abcam, ab6161), anti-GFP (AMS Biotechnology, TP401), and anti-His tag (Santa Cruz, sc-8036). Secondary antibodies against rabbit, mouse, or rat IgG (conjugated Alexa Fluor 488, 568 or HRP) were supplied by molecular probes. Primary antibodies were used at 1:100 dilutions, and secondary antibodies were used at 1:300 dilutions for immunofluorescence. Primary and secondary antibodies were used at 1:2000 and 1:5000 dilutions, respectively, in Western blots.
Following immunostaining, nuclei were counterstained with 1 µM TO-PRO-3 iodide (Molecular Probes). Fluorescence images were acquired using a Leica SP5 Confocal with 40x/NA1.25 objective. Brightfield images were acquired using a Nikon TE2000-S microscope with 20x/NA0.45 objective. All images were processed using ImageJ.
The coding sequences of OdAur1 or OdAur2 and INbox sequences (residues 493–572) of OdINCENPa were cloned into the first and second cassettes of a bi-cistronic vector pHTvAMP1-SGC (gift from Dr. Jonathan M. Elkins) (Abdul Azeez et al., 2019). The resulting constructs expressed as the complexes of his-tagged Aurora (N-terminal fusion residues: MGSSHHHHHHSQDPENLYFQGANSSSARLQ for OdAur1 and MGSSHHHHHHSQDPENLYFQGANS for OdAur2) and INboxes with an additional N-terminal methionine. The constructs were transformed into E. coli BL21 (DE3)-R3-pRARE2. The resulting colonies were inoculated into the 5-mL SOB media and cultured overnight. These overnight cultures were inoculated into the 1-L SOB media and were grown to OD600 of 0.5 at 37°C. A final concentration of 0.5 mM IPTG was added to induce protein expression, and afterward, the cultures were grown at 20°C overnight. The next day, the bacterial cells were harvested by centrifugation, resuspended in binding buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 20 mM imidazole, 5% glycerol, and protease inhibitor cocktail), and lysed by sonication. After high-speed centrifugation, the supernatant was recovered and passed through a 0.45-µm syringe filter before loading to a column packed with 1 mL of Ni Sepharose resin (GE Healthcare). The resin was washed with an increasing gradient (20 mM–100 mM) of imidazole. The purified proteins were eluted with three bed volumes of 250 mM imidazole and concentrated using an Amicon Ultra-4 10K centrifugal filter device (Millipore). Protein concentrations were determined by the Bradford assay (Thermo Fisher).
Equal amounts of purified proteins were used in kinase assays with the CycLex Aurora Family Kinase Assay Kit (MBL|Medical & Biological Laboratories), following the manufacturer’s instructions. In brief, 10 µL of protein sample and 90 µL of kinase reaction buffer containing 0.125 mM ATP were added to the well that was pre-coated with “Aurora-substrate-1” and were incubated for 30 min at 30°C. After washing the well, 100 µL of HRP-conjugated antibody, recognizing the phosphorylated substrate, was added and incubated for 30 min at 25°C. After another round of washing, 100 µL of chromogenic reagent was added and incubated for 15 min at 25°C. Finally, 100 µL of stop solution was added to terminate the color development reaction. Absorbance was measured at 450 nm with a TECAN microplate reader.
Total RNA from mixed samples containing oocytes and early embryos was extracted using the RNAqueous™-Micro Total RNA Isolation Kit (Ambion). A cDNA library was generated and incorporated into the prey vector pGADT7-pla using the all-direct method (Shanghai Biogen Biotechnology Corporation) and then transformed into a yeast strain Y187 following the LiAc transformation procedure (Clontech) and, hereafter, named the prey library as pGADT7-Od-pla. A bait vector pGBKCg containing Aur1 (pGBKCg-Aur1) was transformed into AH109. The autoactivation of the bait vector was tested and confirmed that the bait vector did not activate HIS3, ADE2, and LacZ reporters. Next, prey library cells were incubated with the overnight culture of bait cells for 24 h at 30°C. One aliquot of zygotes was gradient-diluted and plated on SD-TL plates to calculate mating efficiency. The rest were plated on SD-TLH +5 mM 3AT plates and maintained at 30°C for 3–4 days. Positive colonies were further inoculated on SD-TLHA plates to test the ADE2 reporter. Plasmids were rescued from the colonies that passed both HIS3 and ADE2 tests and sequenced. Sequences were blasted against NCBI and OikoBase (http://oikoarrays.biology.uiowa.edu/Oiko/) databases to retrieve their protein identities and mRNA expression profiles at different developmental stages of O. dioica.
Appendicularians, also known as larvaceans, include three families: Oikopleuridae, Fritillaridae, and Kowalevskiidae. In the available genome and transcriptome databases of appendicularians, we ran a local tBLASTn search using Aurora ortholog sequences from both invertebrates and vertebrates as queries. We consistently found two Aurora homologs with very significant E-values (2e-066–7e-144) in Oikopleura dioica, Oikopleura albicans, and Fritillaria borealis. Among the vertebrates, phylogenetic trees clearly showed that vertebrate Aurora A formed one orthologous clade. Auroras B and C in mammals and Aurora B in amphibians formed another clade. However, the nodes leading to other invertebrate deuterostomes were not well resolved with high-bootstrap values. The single Aurora observed in amphioxus, hemichordates, and echinoderms was weakly clustered together. Auroras in ascidians have occupied a separate branch. Auroras in appendicularians underwent rapid evolution as witnessed by their long-branch lengths, and they also underwent an independent gene duplication event which generated two lineages: Aurora1 and Aurora2 in O. dioica, O. albicans, and F. borealis (Figure 1A; Supplementary Data S1). OdAur1-capped mRNA with a C-terminal GFP fusion was injected into 1-cell embryos, and the localizations of OdAur1-GFP fusion protein during early embryonic mitoses were analyzed. Aurora1 was localized on centrosomes throughout mitosis. Using a commercial phospho-Aurora antibody that can recognize the phosphorylated forms of both Aurora1 and Aurora2 in O. dioica, we assessed the combined signals of the two Aurora kinases. As expected, during early embryonic cleavages, signals were found on centrosomes and chromosomes in prophase and metaphase, and on centrosomes and the central spindle in anaphase (Figure 1B). As such, Aurora2 would occupy the chromosome-central spindle axis. This suggests that Aurora1 and Aurora2 in O. dioica represent the polar and equatorial forms of Aurora kinases, respectively, and they are most likely to be functional equivalents of vertebrate Auroras A and B during mitosis.
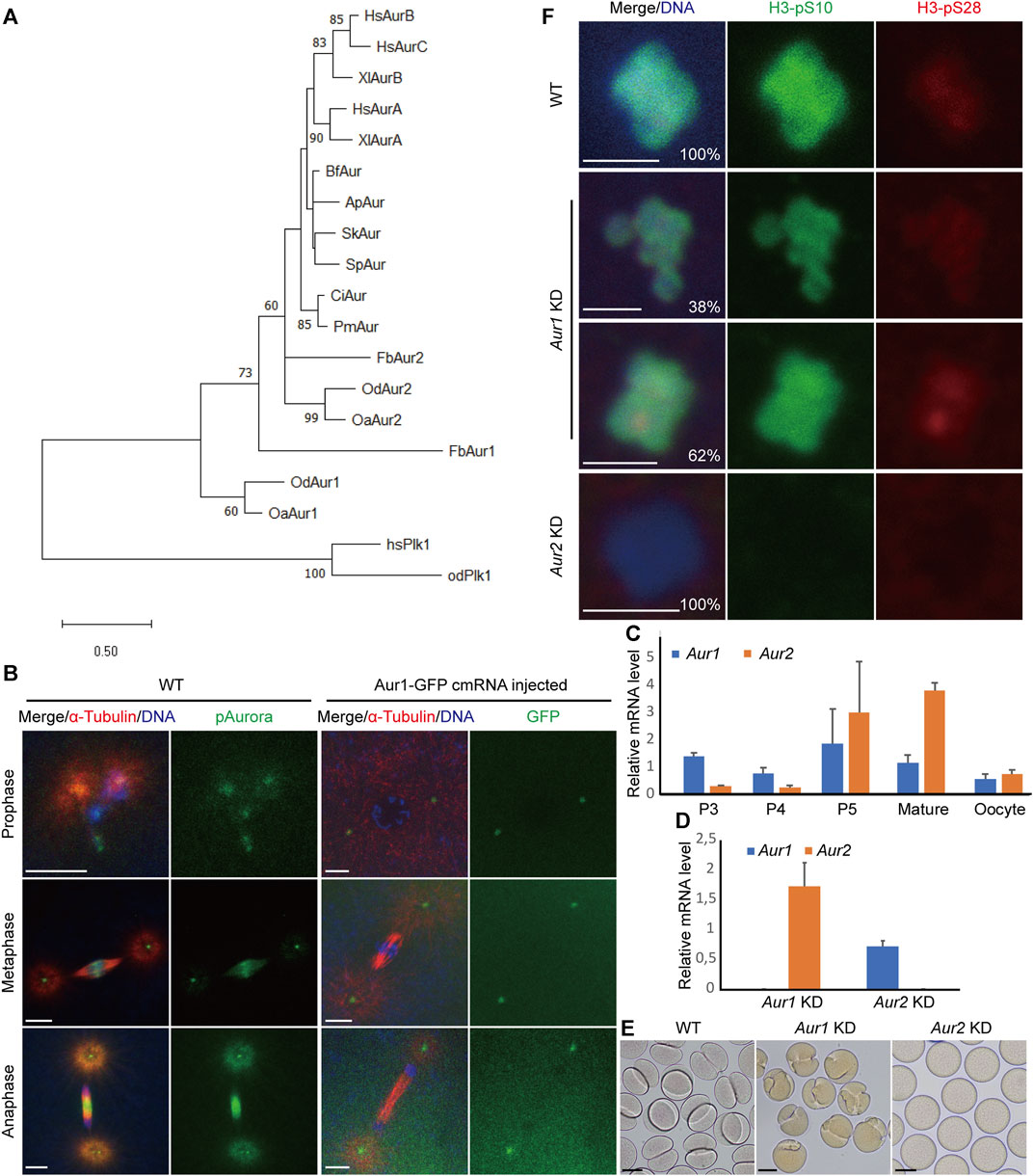
FIGURE 1. Identification, expression profiles, and distinct functions of Aurora1 and Aurora2 kinases during oogenic meiosis in Oikopleura dioica. (A) Phylogenetic relationships of Aurora kinases of three larvaceans in deuterostomes. The tree was constructed in MEGA11 using the maximum likelihood method based on the alignment of the catalytic domains of Aurora kinases (Supplementary Data S1) (Tamura et al., 2021). An LG + G amino acid substitution model was used in a 1,000-iteration bootstrap test. Only bootstrap percentages >50% are shown on the nodes. Tree was drawn to scale, with branch lengths measured in the expected number of substitutions per site. Plk1 kinases were used as an outgroup to root the tree. The abbreviations of the selected species are as follows: Hs, Homo sapiens; Xl, Xenopus laevis; Od, Oikopleura dioica; Oa, Oikopleura albicans; Fb, Fritillaria borealis; Ci, Ciona intestinalis; Pm, Phallusia mammillata; Bf, Branchiostoma floridae; Sk, Saccoglossus kowalevskii; Ap, Asterina pectinifera; and Sp, Strongylocentrotus purpuratus. (B) Localizations of combined Aurora signals and Aurora1 during embryonic mitoses. Scale bars: 5 µm. (C) Relative mRNA levels of Aur1 and Aur2 during O. dioica oogenesis. Coenocytic oogenesis is subdivided into five phases, P1 to P5 (Ganot et al., 2007). P3 corresponds to pachytene and diplotene, during which the coenocyst grows rapidly. In P4, a subset of pro-oocytes increases in size by cytoplasmic transfer through ring canals and meiotic chromatin condenses. During P5, oocytes attained their full size, and nurse and excess meiotic nuclei underwent apoptosis. Subsequently, oocyte maturation is completed, and spawning occurs via the rupture of the gonad wall. Mean values normalized to EF1 transcripts are shown with standard error bars (n = 3). (D) Knockdown efficiencies of target genes in spawned oocytes were obtained using RT–qPCR after dsRNA-mediated knockdowns of Aur1 and Aur2, respectively. Fold changes of mRNA levels relative to those in wild type are shown with standard error bars (n = 3). (E) At 30 min after exposure to wild-type sperm, WT embryos underwent first cleavage (left). Aur1 KD embryos showed “boxing glove”-like shapes (middle), whereas Aur2 KD oocytes were non-viable (right). Scale bars: 50 µm. (F) Knockdowns of Aur1 and Aur2 had distinct effects on histone H3-S10 and H3-S28 phosphorylation on oocyte chromosomes. In WT (top), the H3-pS10 signal was present along the entire chromosomes, and H3-pS28 was enriched at centromeres. In Aur1 KD (n = 52, middle), although H3-pS10 appeared normal, 38% of oocytes showed faint signals of H3-pS28 spreading along the entire chromosomes, and 62% of oocytes showed enriched signals of H3-pS28 at centromeres. In all Aur2 KD oocytes (n = 36, bottom), both H3-pS10 and H3-pS28 signals were absent on decondensed chromosomes. Scale bars: 2 µm.
The expression profiles of the two Aurora kinases during oogenesis were analyzed by RT–qPCR. Their transcripts co-existed during oogenesis, with maximum expression levels at P5 and maturation phases during which nurse nuclei exported RNAs and proteins to rapidly growing pro-oocytes (Figure 1C). Our previous studies have shown that combined Aurora signals were present in meiotic nuclei and organizing centers (OCs) during P3, relocated to selected meiotic nuclei during P4, and enriched toward centromeric regions during prometaphase I. This coincided with the changes of histone H3-S10 and H3-S28 phosphorylation from P3 to P4 in selected meiotic nuclei and acentrosomal spindle assembly during prometaphase I (Øvrebø et al., 2015; Feng and Thompson, 2018).
To explore their potential roles in the aforementioned meiotic features, dsRNA knockdowns (Aur1 KD or Aur2 KD) were carried out by gonad injection. Knockdown efficiencies were calculated by RT–qPCR, showing that the mRNAs of target genes dropped to extremely low levels, whereas paralogous genes were not significantly affected (Figure 1D). After exposure to wild-type sperm, some Aur1 KD oocytes seemed to extrude two polar bodies with normal timing. However, when the first mitotic division started, cleavage furrows emerged from one side of most zygotes and abnormally progressed inwards, appearing as “boxing gloves” at 30 min post-fertilization (Figure 1E). Thereafter, some furrows regressed, whereas others formed one or more furrows from other locations, resulting in two or three cells with unequal sizes or disrupted division axes (Supplementary Figure S1). In contrast, Aur2 KD oocytes were non-viable, and polar body extrusion was not observed (Figure 1E). We suspect that defective mitotic cleavages in Aur1 KD might be caused by defects in meiotic progression. We have previously shown that H3-S10 and H3-S28 phosphorylation can indicate the progression of meiosis I. Therefore, we assessed these two histone modifications in Aur1 KD and Aur2 KD oocytes (Figure 1F). H3-pS10 was present on the entire chromosomes in all the Aur1 KD oocytes that have been analyzed, similar to WT, but H3-pS28 was either spread along entire chromosomes in 38% of oocytes or enriched on centromeres in 62% of oocytes. This suggests that Aurora1 is involved in promoting the progression of prometaphase I. In all the Aur2 KD oocytes, both H3-pS10 and H3-pS28 were absent, and chromosomes were decondensed, reminiscent of INCENPa knockdown phenotypes (Feng et al., 2019). This confirms that Aurora2 and INCENPa, as the kinase and scaffold proteins, respectively, in the CPC, are essential for H3-S10 and -S28 phosphorylation and chromosome condensation during oogenic meiosis of O. dioica.
The first meiotic spindle is organized without centrosomes in O. dioica, potentially through a chromatin-dependent pathway mainly driven by Aurora kinases. The enrichment of Aurora signals on centromeric regions was concomitant with the establishment of robust microtubules around the chromosomes (Figure 2A). Intriguingly, a variety of defects in meiotic spindle assembly in Aur1 KD oocytes were observed and classified into 10 categories based on spindle morphology and karyotype (Figure 2B). Among 315 oocytes from six Aur1 KD females, only 6% had normal-looking spindles. An additional 36% of spindles appeared to be elongated, compact, multi-polar, or rudimentary, with diffuse Aurora signals on most chromosomes. Spindles were absent in the remaining majority, with few, sporadic microtubules in contact with chromosomes. Interestingly, in 6% of oocytes, we also observed homologous chromosomes detached from a partially organized spindle. To assess the extent that these defects can affect chromosome segregation, Aur1 KD oocytes were fertilized with WT sperm and collected within 5 min post-fertilization to analyze the progression of anaphase I. In WT, Aurora signals relocated to the central spindle and overlapped with fine and robust microtubule fibers which emerged between segregating homologous chromosomes, when thick microtubule fibers at the two poles were degraded (Figure 2A). In contrast, among 245 oocytes from six Aur1 KD females, 93% of defective spindles were not capable of segregating chromosomes. In rare cases when Aurora signals were enriched on the central spindle and chromosomes managed to segregate, 5% showed less robust microtubule staining in the central spindle, and 2% showed lagging chromosomes along the central spindle (Figure 2C). In all the Aur2 KD oocytes observed in seven females, neither Aurora signals nor microtubules were detected around decondensed chromosomes (Figure 2D).
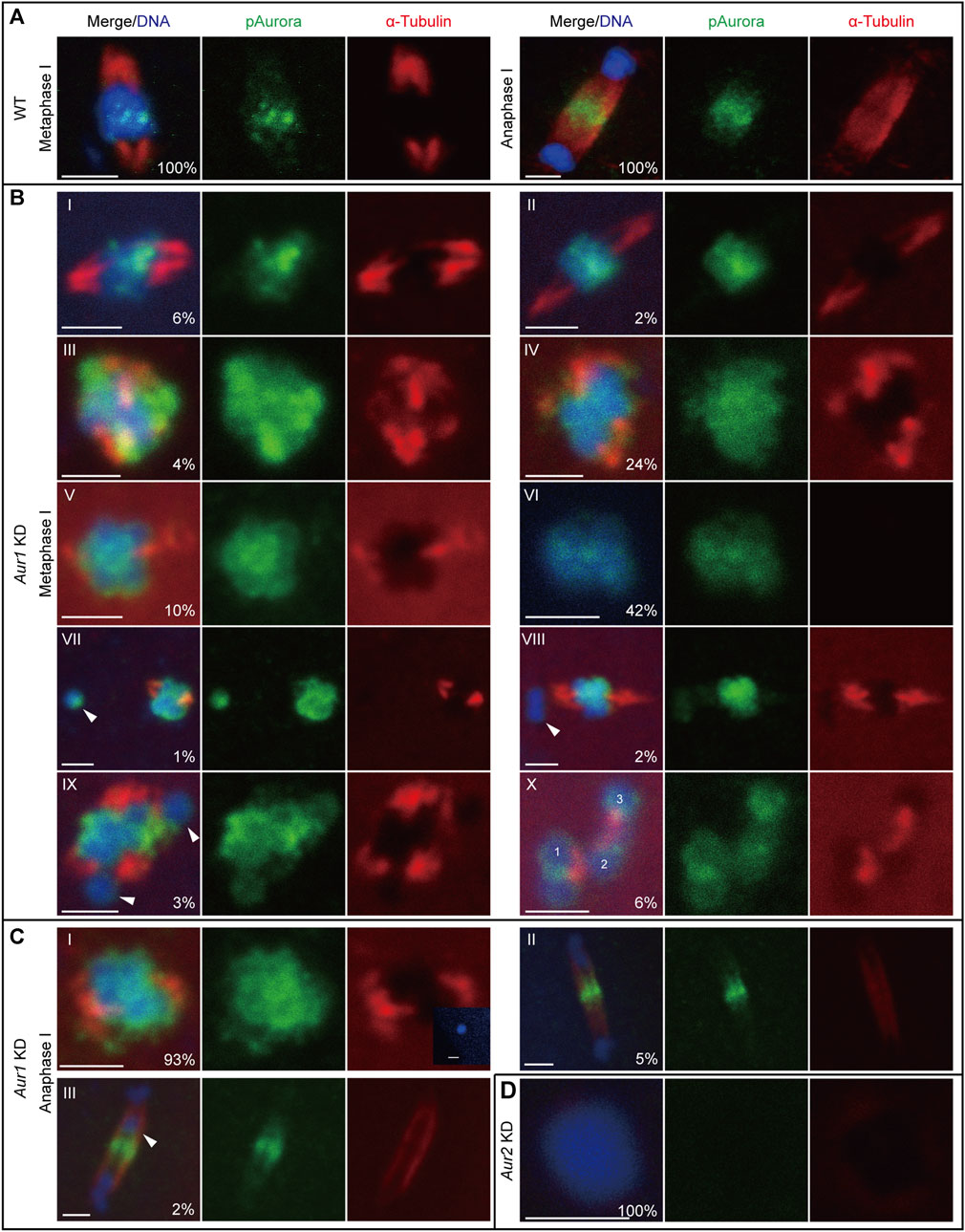
FIGURE 2. Differential requirements of Aurora1 and Aurora2 in meiotic spindle assembly. (A) In WT oocytes, phospho-Aurora signals were enriched on centromeres and robust microtubule fibers radiated from opposite poles to form a bipolar spindle at metaphase I (left). During anaphase I (right), phospho-Aurora signals accumulated at the central spindle, and interchromosomal microtubules formed between the two masses of segregating homologous chromosomes. (B) Aur1 KD caused a range of defects in meiotic spindle assembly at metaphase I. These defects were categorized, with proportions labeled in the merge panel (n = 315). (I) 6% showed normal spindles with lengths of 4–5 μm; (II) 2% showed elongated spindles with lengths >6 μm; (III) 4% showed compact spindles with lengths <4 μm, exhibiting delocalized Aurora signals surrounding chromosomes; (IV) 24% showed thick microtubules radiating from opposite poles to form rudimentary spindles; (V) 10% lacked spindles with few microtubules attached to chromosomes; and (VI) 42% lacked spindles and showed no detectable microtubule staining. In categories IV to VI, diffuse Aurora signals were spread along chromosomes. In addition, small proportions of defects in homolog chromosome congression were also observed and distributed in the following categories: (VII) 1% showed one homologous chromosome (arrowhead) detached from the spindle; (VIII) 2% showed one bivalent (a pair of homologous chromosomes, arrowhead) detached from the spindle; (IX) 3% showed two separate chromosomes (arrowhead) detached from the spindle, and the spindle seemed to be multi-polar; and (X) 6% showed three separate bivalents with microtubules between them. (C) Within 5 min after exposure to WT sperm, the progression of anaphase I in Aur1 KD oocytes was analyzed (n = 245). (I) 93% showed defective spindles. Insert panel showed a sperm nucleus inside an Aur1 KD oocyte. (II) 5% showed normal segregating chromosomes with enriched Aurora signals on the central spindle and less robust interchromosomal microtubule fibers. (III) 2% showed lagging chromosomes (arrowhead) along the central spindle. (D) In all Aur2 KD oocytes (n = 26), chromosomes were decondensed, and phospho-Aurora signals and microtubule staining were absent. Scale bars: 2 µm.
In summary, Aurora1 was involved in the enrichment of Aurora2 on centromeres and the regulation of microtubule dynamics to promote acentrosomal spindle assembly that was particularly driven by centromeric Aurora2. The deregulation of Aurora1 can lead to defective chromosome congression during metaphase I and subsequent mis-segregation during anaphase I.
It was observed that Aurora1 can assist Aurora2 in regulating meiotic progression. Then, we assessed whether Aurora1 has any intrinsic enzymatic activity. To make a comparison, the His-tagged full-length proteins of Aurora1 and Aurora2 were expressed and purified in complexes with the INbox motif of INCENPa using a bi-cistronic vector pHTvAMP1-SGC in E. coli. Western blot analyses confirmed that the concentrations of purified protein complexes were similar before carrying out the Aurora kinase assay (Figure 3A). Interestingly, in vitro, INCENPa activated Aurora1 to similar levels as it activated Aurora2 when provided with a synthetic substrate that contained an Aurora consensus phosphorylation site (Figure 3B). This implies that O. dioica Aurora1 is a bona fide Aurora kinase and can be activated by a known interaction partner.
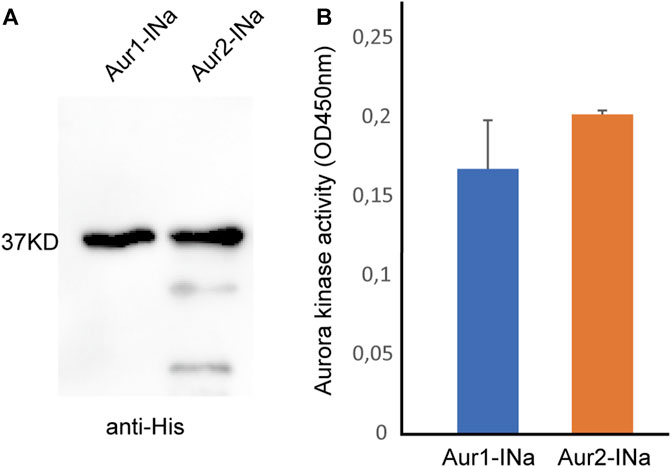
FIGURE 3. Kinase activity assay of Aurora1 or Aurora2 complexed with the INbox motif of INCENPa. (A) Western blot of purified Aurora1 and Aurora2 complexed with the INbox of INCENPa (INa) using an anti-His tag antibody. (B) INbox motif can stimulate the kinase activities of Aurora1 and Aurora2. Mean values with standard errors were calculated from four independent experiments.
Aurora kinases are organized in regulatory kinase cascades by their interaction/scaffolding proteins that are involved in the activation and spatiotemporal targeting of kinases. Our initial genome-wide search failed to detect TPX2, a microtubule-binding protein that normally localizes Aurora A toward the spindle and the mid-body (Bayliss et al., 2003), implying that the Aurora1-scaffold complement might have changed in O. dioica. Considering new interaction proteins of Aurora1, a cDNA prey library (number of independent clones: 9.60×106) generated from oocytes and early embryos of O. dioica was screened with Aurora1 as bait using a yeast two-hybrid library screening approach. This approach identified eight possible interaction proteins (Table 1). To our knowledge, none of them have been previously reported as Aurora A interaction proteins.
Given the loss of canonical interaction proteins of Aurora A, such as TPX2 in O. dioica, it is perhaps not surprising that several new binding partners of Aurora1 may have developed. c-Jun-amino-terminal kinase-interacting protein 4 (JIP4) is the scaffolding protein in the JNK/mitogen-activated protein kinases (MAPK) signaling pathway (Nguyen et al., 2005). JIP4 is involved in the bidirectional transportation of cargos along the microtubules through interaction with kinesin-1 and dynactin/dynein motors (Montagnac et al., 2009). The interface between JIP4 and kinesin-1 lies in the tetratricopeptide repeat (TPR) motif of kinesin-1 light chain (KLC1). Interestingly, we also identified a TPR-containing protein (Ttc36) as an Aurora1 interaction protein. Although Ttc36 is less studied, the TPR motif comprises 3–16 tandem-repeats of 34 amino acid residues and forms an all-helical structure, often mediating protein–protein interactions and cell cycle regulation (D’Andrea and Regan, 2003). Thus, it is tempting to speculate that JIP4 brings Aurora1 in close proximity to microtubule motors, allowing Aurora1 to phosphorylate kinesin-1 or dynactin/dynein subunits and regulate microtubule dynamics. Aurora A phosphorylates dynactin’s largest subunit p150glued on its N-terminal microtubule-binding domain (MBD), which regulates the disassociation of the dynactin/dynein complex from the spindle poles after NEBD in Drosophila S2 cells (Romé et al., 2010). This phosphorylation is also involved in central spindle assembly at anaphase in human cells (Reboutier et al., 2013). It would be of great interest to test whether Aurora1 regulates microtubule dynamics through the interaction with JIP4.
Scc2/Nipbl regulates the loading of cohesin onto chromatin before the S phase (Mattingly et al., 2022). In addition to cohesin loading, Scc2 can move between cohesin complexes on chromosomes after DNA replication (Rhodes et al., 2017). Its interaction with Aurora1 suggests that Aurora1 might make transient contact with cohesin complexes at crossover sites, where homologous chromosomes are held together, and on centromeres between sister chromatids in meiosis I. Indeed, we observed one or two homologous chromosomes as being detached from the main chromosome mass in a minor proportion of Aur1 KD oocytes. Further investigation is needed to test whether Aurora1 is involved in the maintenance of cohesion at metaphase I.
Although the function of Aurora in mitochondria is outside the scope of this study, we did identify mitochondrial carrier homolog 2 (MTCH2), a regulator of mitochondrial fusion, as an Aurora1 interaction protein (Bahat et al., 2018). Human Aurora A is transported into mitochondria in the interphase and induces mitochondrial fragmentation (Bertolin et al., 2018). This transport relies on its N-terminal 36 amino acids serving as an atypical mitochondrial targeting sequence (MTS). Once inside the mitochondria, MTS is cleaved off, generating two forms of truncated Aurora A. The N terminal of O. dioica Aurora1 is approximately 90 aa shorter than human Aurora A and does not have an MTS motif (Supplementary Data S1). It is possible that MTCH2 might serve as a transporter to import Aurora1 into the mitochondria in O. dioica.
It would be of interest to develop specific antibodies for the aforementioned interaction proteins, in order to assess these interactions in vivo by Co-IP and colocalization assays. Targeted perturbations would be informative to dissect their roles in the regulation of Aurora1 in more detail.
Considering their distinct roles in meiotic spindle organization, we argue that the two Aurora kinases co-exist in meiotic nuclei. Aurora1 has opportunities to contact microtubules that are synthesized through an Aurora2-dependent pathway on chromosomes. This proposed pattern is distinct from other invertebrate and vertebrate models reported thus far. Aurora1 in O. dioica, or in appendicularians in general, may have evolved to regulate microtubule dynamics during meiotic spindle assembly. The role of O. dioica Aurora2 in regulating chromosome condensation and acentrosomal spindle assembly is consistent with the roles in vertebrates. However, a key residue (Gly198 in human Aurora A or Asn142 in human Aurora B) in the catalytic domain that determines the interaction partners, subcellular localizations, and functions of the respective Aurora kinases in vertebrates has been altered to Gly in the two O. dioica Aurora kinases (Gly105 in Aurora1 or Gly91 in Aurora2, Supplementary Data S1) (Fu et al., 2009). The two Aurora kinases in O. dioica share this feature with the single ancestral Aurora which possesses Gly at the equivalent residue in most invertebrate deuterostomes, social amoeba, and yeast.
A question raised for many years is why independent evolution of Aurora kinases in a few invertebrates has led to a similar functional separation in spindle and chromosome regulations. Although our study cannot definitively respond to this question, we can provide some new insights. As shown in this study, O. dioica Aurora1 has substantially changed its interaction proteins and regulated meiotic spindle organization from distinct locations. There is also cross-talk between Aurora1 and Aurora2 in promoting their respective localizations, along with other signaling cascades that are brought together by novel interaction/scaffolding proteins.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, SCLG00000000.1 https://www.ncbi.nlm.nih.gov/genbank/, SDII00000000.1.
This manuscript presents research on animals that do not require ethical approval for their study.
HF: conceptualization, data curation, formal analysis, investigation, methodology, resources, validation, writing–original draft, and writing–review and editing. ET: conceptualization, formal analysis, funding acquisition, project administration, resources, supervision, and writing–review and editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the SARS Centre core budget (EMT).
The authors thank Anne E. Aasjord for providing animals from the appendicularian facility and the Chourrout Group at the Michael Sars Centre for access to sequence data from hermaphroditic appendicularian species. They also thank Dr. Jonathan M. Elkins at the University of Oxford for providing the bi-cistronic vector pHTvAMP1-SGC. Yeast two-hybrid library screening was carried out by the Shanghai Biogen Biotechnology Corporation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1323378/full#supplementary-material
Abdul Azeez, K. R., Chatterjee, S., Yu, C., Golub, T. R., Sobott, F., and Elkins, J. M. (2019). Structural mechanism of synergistic activation of Aurora kinase B/C by phosphorylated INCENP. Nat. Commun. 10, 3166. doi:10.1038/s41467-019-11085-0
Abe, Y., Okumura, E., Hosoya, T., Hirota, T., and Kishimoto, T. (2010). A single starfish Aurora kinase performs the combined functions of Aurora-A and Aurora-B in human cells. J. Cell Sci. 123, 3978–3988. doi:10.1242/jcs.076315
Bahat, A., Goldman, A., Zaltsman, Y., Khan, D. H., Halperin, C., Amzallag, E., et al. (2018). MTCH2-mediated mitochondrial fusion drives exit from naïve pluripotency in embryonic stem cells. Nat. Commun. 9, 5132. doi:10.1038/s41467-018-07519-w
Balboula, A. Z., Nguyen, A. L., Gentilello, A. S., Quartuccio, S. M., Drutovic, D., Solc, P., et al. (2016). Haspin kinase regulates microtubule-organizing center clustering and stability through Aurora kinase C in mouse oocytes. J. Cell Sci. 129, 3648–3660. doi:10.1242/jcs.189340
Balboula, A. Z., and Schindler, K. (2014). Selective disruption of Aurora C kinase reveals distinct functions from Aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 10, e1004194. doi:10.1371/journal.pgen.1004194
Bayliss, R., Sardon, T., Vernos, I., and Conti, E. (2003). Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862. doi:10.1016/S1097-2765(03)00392-7
Bertolin, G., Bulteau, A.-L., Alves-Guerra, M.-C., Burel, A., Lavault, M.-T., Gavard, O., et al. (2018). Aurora kinase A localises to mitochondria to control organelle dynamics and energy production. Elife 7, e38111. doi:10.7554/eLife.38111
Blengini, C. S., Ibrahimian, P., Vaskovicova, M., Drutovic, D., Solc, P., and Schindler, K. (2021). Aurora kinase A is essential for meiosis in mouse oocytes. PLoS Genet. 17, e1009327. doi:10.1371/journal.pgen.1009327
Bouquet, J.-M., Spriet, E., Troedsson, C., Otterå, H., Chourrout, D., and Thompson, E. M. (2009). Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J. Plankton Res. 31, 359–370. doi:10.1093/plankt/fbn132
Brown, J. R., Koretke, K. K., Birkeland, M. L., Sanseau, P., and Patrick, D. R. (2004). Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol. Biol. 4, 39. doi:10.1186/1471-2148-4-39
Campsteijn, C., Øvrebø, J. I., Karlsen, B. O., and Thompson, E. M. (2012). Expansion of cyclin D and CDK1 paralogs in Oikopleura dioica, a chordate employing diverse cell cycle variants. Mol. Biol. Evol. 29, 487–502. doi:10.1093/molbev/msr136
Carmena, M., Ruchaud, S., and Earnshaw, W. C. (2009). Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21, 796–805. doi:10.1016/j.ceb.2009.09.008
D’Andrea, L. D., and Regan, L. (2003). TPR proteins: the versatile helix. Trends biochem. Sci. 28, 655–662. doi:10.1016/j.tibs.2003.10.007
Delsuc, F., Brinkmann, H., Chourrout, D., and Philippe, H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968. doi:10.1038/nature04336
Denoeud, F., Henriet, S., Mungpakdee, S., Aury, J.-M., Da Silva, C., Brinkmann, H., et al. (2010). Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–1385. doi:10.1126/science.1194167
Divekar, N. S., Davis-Roca, A. C., Zhang, L., Dernburg, A. F., and Wignall, S. M. (2021). A degron-based strategy reveals new insights into Aurora B function in C. elegans. PLoS Genet. 17, e1009567. doi:10.1371/journal.pgen.1009567
Feng, H., Raasholm, M., Moosmann, A., Campsteijn, C., and Thompson, E. M. (2019). Switching of INCENP paralogs controls transitions in mitotic chromosomal passenger complex functions. Cell Cycle 18, 2006–2025. doi:10.1080/15384101.2019.1634954
Feng, H., and Thompson, E. M. (2018). Specialization of CDK1 and cyclin B paralog functions in a coenocystic mode of oogenic meiosis. Cell Cycle 17, 1425–1444. doi:10.1080/15384101.2018.1486167
Ferrández-Roldán, A., Fabregà-Torrus, M., Sánchez-Serna, G., Duran-Bello, E., Joaquín-Lluís, M., Bujosa, P., et al. (2021). Cardiopharyngeal deconstruction and ancestral tunicate sessility. Nature 599, 431–435. doi:10.1038/s41586-021-04041-w
Fu, J., Bian, M., Liu, J., Jiang, Q., and Zhang, C. (2009). A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc. Natl. Acad. Sci. U. S. A. 106, 6939–6944. doi:10.1073/pnas.0900833106
Ganot, P., Kallesøe, T., and Thompson, E. M. (2007). The cytoskeleton organizes germ nuclei with divergent fates and asynchronous cycles in a common cytoplasm during oogenesis in the chordate Oikopleura. Dev. Biol. 302, 577–590. doi:10.1016/j.ydbio.2006.10.022
Hebras, C., and McDougall, A. (2012). Urochordate ascidians possess a single isoform of Aurora kinase that localizes to the midbody via TPX2 in eggs and cleavage stage embryos. PLoS One 7, e45431. doi:10.1371/journal.pone.0045431
Hochegger, H., Hégarat, N., and Pereira-Leal, J. B. (2013). Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 3, 120185. doi:10.1098/rsob.120185
Li, H., Chen, Q., Kaller, M., Nellen, W., Gräf, R., and De Lozanne, A. (2008). Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot. Cell 7, 894–905. doi:10.1128/EC.00422-07
Ma, X., Øvrebø, J. I., and Thompson, E. M. (2022). Evolution of CDK1 paralog specializations in a lineage with fast developing planktonic embryos. Front. Cell Dev. Biol. 9, 770939. doi:10.3389/fcell.2021.770939
Mattingly, M., Seidel, C., Muñoz, S., Hao, Y., Zhang, Y., Wen, Z., et al. (2022). Mediator recruits the cohesin loader Scc2 to RNA Pol II-transcribed genes and promotes sister chromatid cohesion. Curr. Biol. 32, 2884–2896.e6. doi:10.1016/j.cub.2022.05.019
Montagnac, G., Sibarita, J.-B., Loubéry, S., Daviet, L., Romao, M., Raposo, G., et al. (2009). ARF6 interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19, 184–195. doi:10.1016/j.cub.2008.12.043
Nguyen, A. L., Gentilello, A. S., Balboula, A. Z., Shrivastava, V., Ohring, J., and Schindler, K. (2014). Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes. J. Cell Sci. 127, 5066–5078. doi:10.1242/jcs.158840
Nguyen, A. L., and Schindler, K. (2017). Specialize and divide (twice): functions of three Aurora kinase homologs in mammalian oocyte meiotic maturation. Trends Genet. 33, 349–363. doi:10.1016/j.tig.2017.03.005
Nguyen, Q., Lee, C. M., Le, A., and Reddy, E. P. (2005). JLP associates with kinesin light chain 1 through a novel leucine zipper-like domain. J. Biol. Chem. 280, 30185–30191. doi:10.1074/jbc.M505499200
Øvrebø, J. I., Campsteijn, C., Kourtesis, I., Hausen, H., Raasholm, M., and Thompson, E. M. (2015). Functional specialization of chordate CDK1 paralogs during oogenic meiosis. Cell Cycle 14, 880–893. doi:10.1080/15384101.2015.1006000
Radford, S. J., Jang, J. K., and McKim, K. S. (2012). The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192, 417–429. doi:10.1534/genetics.112.143495
Reboutier, D., Troadec, M.-B., Cremet, J.-Y., Chauvin, L., Guen, V., Salaun, P., et al. (2013). Aurora A is involved in central spindle assembly through phosphorylation of Ser 19 in P150Glued. J. Cell Biol. 201, 65–79. doi:10.1083/jcb.201210060
Rhodes, J., Mazza, D., Nasmyth, K., and Uphoff, S. (2017). Scc2/Nipbl hops between chromosomal cohesin rings after loading. Elife 6, e30000. doi:10.7554/eLife.30000
Romé, P., Montembault, E., Franck, N., Pascal, A., Glover, D. M., and Giet, R. (2010). Aurora A contributes to p150glued phosphorylation and function during mitosis. J. Cell Biol. 189, 651–659. doi:10.1083/jcb.201001144
Sumiyoshi, E., Fukata, Y., Namai, S., and Sugimoto, A. (2015). Caenorhabditis elegans Aurora A kinase is required for the formation of spindle microtubules in female meiosis. Mol. Biol. Cell 26, 4187–4196. doi:10.1091/mbc.E15-05-0258
Keywords: tunicate, Aurora kinase, INCENP, interaction/scaffolding proteins, oogenic meiosis, molecular evolution
Citation: Feng H and Thompson EM (2023) Functional specialization of Aurora kinase homologs during oogenic meiosis in the tunicate Oikopleura dioica. Front. Cell Dev. Biol. 11:1323378. doi: 10.3389/fcell.2023.1323378
Received: 17 October 2023; Accepted: 21 November 2023;
Published: 07 December 2023.
Edited by:
Lucia Manni, University of Padua, ItalyReviewed by:
Alfonso Ferrández Roldán, University of Barcelona, SpainCopyright © 2023 Feng and Thompson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyang Feng, aHlmZW5nQGxpdmUuY24=; Eric M. Thompson, ZXJpYy50aG9tcHNvbkB1aWIubm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.