- Cancer Metabolism and Microenvironment Program, Sanford Burnham Prebys, La Jolla, CA, United States
Phosphoinositides serve as essential players in numerous biological activities and are critical for overall cellular function. Due to their complex chemical structures, localization, and low abundance, current challenges in the phosphoinositide field include the accurate measurement and identification of specific variants, particularly those with acyl chains. Researchers are intensively developing innovative techniques and approaches to address these challenges and advance our understanding of the impact of phosphoinositide signaling on cellular biology. This article provides an overview of recent advances in the study of phosphoinositides, including mass spectrometry, lipid biosensors, and real-time activity assays using fluorometric sensors. These methodologies have proven instrumental for a comprehensive exploration of the cellular distribution and dynamics of phosphoinositides and have shed light on the growing significance of these lipids in human health and various pathological processes, including cancer. To illustrate the importance of phosphoinositide signaling in disease, this perspective also highlights the role of a family of lipid kinases named phosphatidylinositol 5-phosphate 4-kinases (PI5P4Ks), which have recently emerged as exciting therapeutic targets for cancer treatment. The ongoing exploration of phosphoinositide signaling not only deepens our understanding of cellular biology but also holds promise for novel interventions in cancer therapy.
1 Introduction
The phosphoinositide family of phospholipids plays pivotal roles in nearly all aspects of cellular function. These phospholipids are one of the most functionally versatile membrane lipid families involved in human health and disease (Di Paolo et al., 2006). The base structure of all phosphoinositides contains phosphatidylinositol (PI), which is made up of an inositol head group and two long-chain fatty acids linked to a glycerol backbone. Combinatorial phosphorylation of residues in the PI head group gives rise to seven other PI classes, namely, PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3 (Figure 1A). These lipids spatiotemporally control the activities of numerous proteins possessing phosphoinositide-binding motifs and, importantly, these motifs can bind to various PI species with differing affinities to regulate physiological processes in cells.
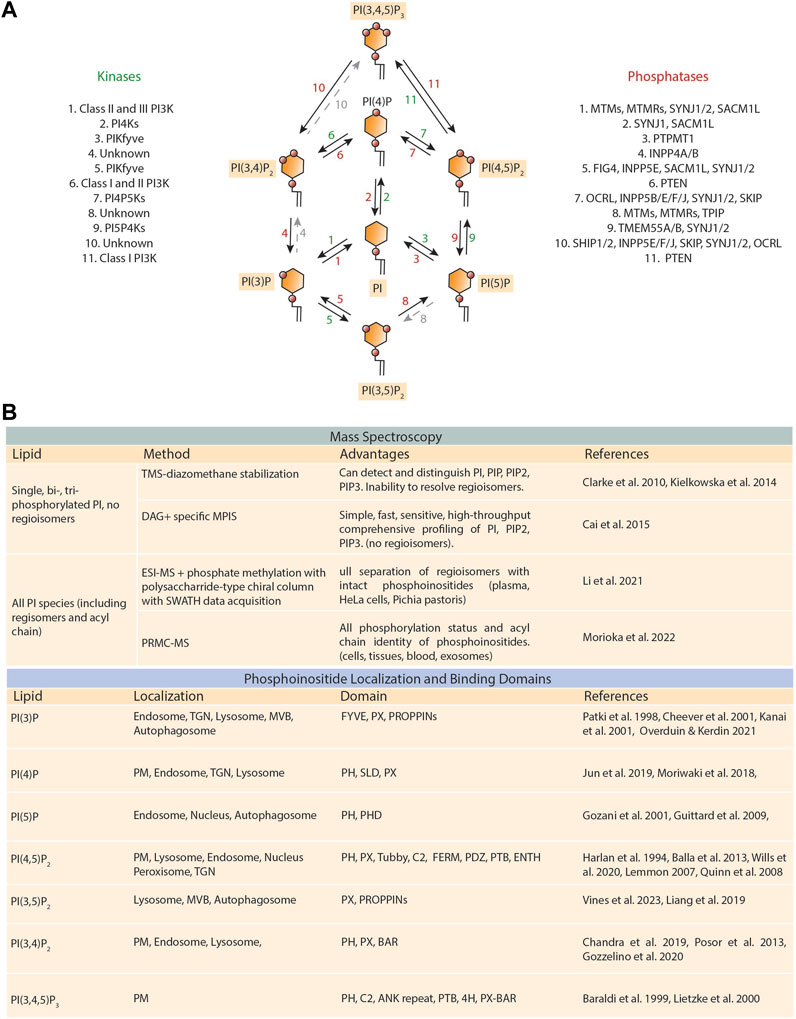
FIGURE 1. Lipid enzymes that regulate phosphoinositide metabolism and techniques for phosphoinositide measurement and identification. (A) Schematic representation displaying the lipid kinases (shown in green) and phosphatases (depicted in red) involved in the generation of the seven phosphoinositide species. Phosphoinositides are formed through the phosphorylation of the third, fourth, and fifth hydroxyl groups on the inositol ring of phosphatidylinositol (PI). The green and red numbers correspond to the lipid kinases and phosphatases that allow each reaction. (B) (top) Table of phosphoinositide relevant mass spectrometry advancements. TMS-diazomethane, trimethylsilyl diazomethane; DAG + specific MPIS, charged diacylglycerol fragment ion-specific multiple precursor ion scanning; ESI-MS, electrospray ionization mass spectrometry; SWATH, sequential window acquisition of all theoretical mass spectra; PRMC-MS, phosphoinositide regioisomer measurement by chiral column chromatography and mass spectrometry. (bottom) Localization and binding domains of the seven unique phosphoinositide species. TGN, trans-Golgi network; MVB, multivesicular body; PM, plasma membrane; FYVE, Fab1, YOTB, Vac1, and EEA1; PX, phox homology; PROPPIN, β-propellers that bind phosphoinositides; PH, pleckstrin homology; PHD, plant homeobox domain; SLD, simple-like domain; C2, protein kinase C conserved region 2; FERM, Four-point-one, ezrin, radixin, moesin; PDZ, PSD95, DlgA, Zo-1; PTB, phosphotyrosine binding; ENTH, epsin NH2-terminal homology; BAR, Bin-amphiphysin-Rvs.
In addition to the lipids themselves, the importance of phosphoinositide signaling is underscored by the mutations and/or dysregulation of the of PI-metabolizing enzymes, such as kinases, phosphatases, lipases, and acyltransferases, which have been implicated in the pathogenesis of various diseases, including cancer (Burke et al., 2023). Current challenges in the phosphoinositide field include accurately measuring the lipid substrate and reaction products of these important PI enzymes, as well as assessing specific phosphoinositide acyl variants. Here, we will attempt to summarize current techniques to study phosphoinositides and focus on an underappreciated phosphoinositide enzyme family that is emerging as a promising target for several cancer subtypes.
2 Advances in phosphoinositide detection techniques
Measuring cellular levels of phospholipids is a continuously developing field, as technical limitations to localization, polarity, and cellular concentrations prove difficult for accurate measurements. Lipidomics, the practice of measuring the different lipid classes in parallel, has proven successful for broad classification and identification of lipid concentrations of several different species. However, the acidic nature of phosphoinositides makes them challenging to measure using these methods. Despite their difficult nature, critical advances in mass spectrometry (MS), lipid biosensors, and sensor-based activity assays have provided an intriguing new outlook for the future of measuring phosphoinositides.
2.1 Elucidating regioisomers and acyl chain specificities of phosphoinositides
There are several challenges in investigating the distribution of phosphoinositide species using MS techniques. For a historical review of the utility of MS-based detection of phosphoinositides, we point the reader to an excellent review (Kim et al., 2010). The traditional protocol utilizing extraction, separation, and autoradiograph analysis was sufficient for comparing the relative levels of the eight phosphoinositide classes. Major advancements in absolute quantification of phosphoinositide species (Figure 1B) were initially sparked by the methylation of phosphoinositides using TMS-diazomethane to stabilize the phosphate group and allow for subsequent measuring of the species PI, PIP2, and PIP3. However, this method did not yet delineate regioisomers (Clark et al., 2011; Kielkowska et al., 2014). Using a similar stabilization method with charged diacylglycerol fragment ion-specific multiple precursor ion scanning (DAG-specific MPIS) allowed for rapid identification of PI, PIP2, and PIP3 species in tissue extracts and human cell lines with additional information on fatty acyl chain variations, but nevertheless, the limitation of identifying the remaining regioisomers of phosphoinositides still remained (Cai et al., 2015; Cai et al., 2016). Recent advancements combining electrospray ionization MS (ESI-MS) and sequential window acquisition of all theoretical fragment ion mass spectra (SWATH) technology enabled the full picture of phosphoinositide regioisomers in human plasma and cultured HeLa cells (Li and Lammerhofer, 2021). This elegant use of advanced technical deconvolution of MS datapoints using SWATH, coupled with strategic mapping of column elution time of regioisomers, allowed for the untangling of miniscule differences between phosphoinositide regioisomers. Similarly, a newly developed technique to measure regioisomers and acyl chain length termed phosphoinositide regioisomer measurement by chiral column chromatography MS (PRMC-MS) was successful in deciphering phosphoinositides and acyl chain length in cells, blood, and cancerous tissue (Morioka et al., 2022). Intriguingly, this method revealed oncogene induced acyl chain signatures and extracellular phosphoinositide mobilization (Morioka et al., 2022). In addition to furthering our knowledge of the cellular phosphoinositide milieu, the use of mass spectrometry imaging (MSI) can provide a powerful tool for uncovering phosphoinositide distribution in tissue-based applications (Buchberger et al., 2018). For instance, high-resolution matrix-assisted laser desorption/ionization imaging MS (MALDI-MS) has been used to identify several phosphoinositide species accumulating in breast cancer tissues (Kawashima et al., 2013). The application for MSI allows for both uncovering the precise spatial distribution of phosphoinositides in breast cancer tissues compared to normal tissue, as well as identifying altered acyl chain distributions of phosphoinositides that could serve as future biomarkers (Kawashima et al., 2013). Further, recent work utilizing MALDI-MS on histological samples from breast cancer patients revealed distinct differences in phosphoinositide acyl chain distribution in invasive cancer cells compared to normal tissues, which when coupled with gene expression analysis, showed an association with PD-1-related immune checkpoint pathway (Kawashima et al., 2020). This can also be extended to in vivo metastasis studies, where metastatic brain lesions, originated from orthotopically implanted human MDA-MB-435 breast cancer cells, were found to contain specific altered phosphoinositide populations compared to surrounding tissues (Roux et al., 2023). Taken together, the rigorous identification of phosphoinositide species, including their variations in acyl chain length, may bring the field forward to identifying these alterations as biomarkers in the cancer setting.
2.2 Advances in domain-based lipid biosensor detection of phosphoinositides
Phosphoinositides are critically important for cell signaling processes, as their local concentration at both the plasma membrane and intracellular membranes directs recruitment of effector proteins. To date, the subcellular mapping of phosphoinositides present at each membrane is well-defined and can be found in many excellent reviews (Balla et al., 2009; Falkenburger et al., 2010; Balla, 2013; Burke, 2018; Dickson and Hille, 2019). Here, we will focus on the developing progress of fluorescently encoded biosensors based on phosphoinositide interacting domains. The best characterized domains are the PH-, FYVE-, and PX-domains (Lee et al., 2005; Chandra et al., 2019). The largest family of lipid-binding domains is the pleckstrin homology (PH) domain, although a continuously developing list of lipid-binding domains recognizing phosphoinositides now enables the possibility of identifying all species (Figure 1B) (Varnai and Balla, 1998). Intriguingly, the identification of these domains has resulted in the development of genetically encoded biosensors facilitating the study of phosphoinositides in their native cellular context (Greenwald et al., 2018; Hertel et al., 2020; Hammond et al., 2022; Posor et al., 2022). Successful development of these probes requires high selectivity of target lipid (due to low abundance) and the target lipid alone, sans the presence of helper proteins, must drive localization of the protein domain to the site of activity (Hammond and Balla, 2015; Wills et al., 2018).
The drawback to genetically encoded biosensors is the requirement for expression in the cell of interest, thus complicating the accuracy of quantitation, as the expression level of these proteins can be highly variable. To resolve this issue, new biosensor development encompassing Fluorescence Resonance Energy Transfer (FRET)-based and dimerization-dependent fluorescent protein-based biosensors utilizing PH domains enable density-based measurements of localization with the tagged proteins in live cells (van der Wal et al., 2001; Sato et al., 2003; Hertel et al., 2020). Using the dimerization-dependent biosensor strategy, the generation of a series of phosphoinositide reporters that preserve the native cellular environment and importantly, are spatially targetable, enabled subcellular location-specific monitoring of phosphoinositide dynamics at areas away from the plasma membrane (Hertel et al., 2020). Recent work has combined activity assays and a predictive algorithm to accurately predict full-length protein containing-PH domain binding to phosphoinositides (Singh et al., 2021). A similar investigative effort to characterize the binding profile of the 49 known PX-domain containing proteins has led to a new grouping classification system for proteins containing these domains, which bind to not only PI(3)P, but to other phosphorylated phosphoinositides as well (Chandra et al., 2019). In future studies, fine-tuning the ability to predict protein binding and localization based on their domain interactions with phosphoinositides could provide an enhanced toolbox for drug discovery. Recently, several groups have shown the utility of phosphoinositide binding antibodies, although the stabilization of lipids and permeabilization techniques must be highly considered to preserve the native lipid structure after fixation. Nonetheless, these antibodies provide a powerful visualization tool without the requirement of engineered cell lines. Discovery of new phosphoinositide related functions in the nuclear compartment demonstrate the need for nuclear visualization of phosphoinositides (Shah et al., 2013). Antibody labeling in direct comparison to overexpressed domains shows a separate and distinctive patterning that was only replicated by preparing purified domains fused with eGFP (Irino et al., 2012; Kalasova et al., 2016). As our knowledge of protein-binding domains specific to phosphoinositides grows, so too will our capabilities of using new biosensors to delineate phosphoinositide dynamics and localization spatially and temporally to subcellular compartments.
2.3 Functional assays for phosphoinositide drug discovery
With our advancing knowledge of measuring phosphoinositide species through MS and using fluorescent probes to further compartmentalize phosphoinositide dynamics at subcellular membranes, quantitatively measuring enzymatic reactions that interconvert these lipids is equally important for a comprehensive view of phosphoinositide relevance in cellular function. Historically, phosphoinositide kinase activity assays were performed using radioactivity-based assays, thus preventing direct measurement of this process in the native cellular context. The development of solvatochromatic fluorophores in conjunction with protein-based lipid sensors allows for quantification of metabolically linked signaling lipids, such as PI(4,5)P2 and PI(3,4,5)P3. However, this technology is amenable to any combination of signaling molecules (Sharma et al., 2020). Recently, the development of real-time activity assays using fluorometric sensors allows for quantitative analysis of enzyme kinetics in response to small molecule modulators (Sun et al., 2020). This assay has for the first time provided a modular system, capable of rapidly screening small molecule inhibitors to lipid kinases. Indeed, as the landscape of phosphoinositide localization throughout the cell becomes better defined, and we improve domain binding algorithms, targeting lipid-protein interactions may prove to be an attractive target (Saliba et al., 2015; Singaram et al., 2023).
3 Integrating lipid biology knowledge into novel approaches for cancer treatment
The techniques discussed above have collectively shed light on the critical significance of maintaining a precise balance of phosphoinositide levels to ensure optimal cellular functionality. Notably, numerous studies have demonstrated a correlation between alterations in the composition of cellular phosphoinositides and the development and progression of several cancer types (Bunney and Katan, 2010). Central to these alterations are lipid kinases, phosphatases, and phospholipases, making these lipid-modifying enzymes promising candidates for targeted cancer therapy. Here, we will highlight the role of phosphatidylinositol 5-phosphate 4-kinases (PI5P4Ks) in cancer as an example of what we believe to be exciting novel therapeutic targets for cancer treatment.
3.1 Role of PI5P4Ks in tumor progression
PI5P4Ks phosphorylate PI(5)P to generate PI(4,5)P2. Within healthy cells, PI(4,5)P2 serves as a platform to activate various signaling pathways that regulate crucial cellular processes, such as cell growth, proliferation, migration, and apoptosis (Llorente et al., 2023). In the context of cancer, variations in PI(5)P and PI(4,5)P2 levels disrupt normal cellular signaling, fostering cancer cell survival and contributing to tumor progression and metastasis (Arora et al., 2022). Deregulated expression of PI5P4Ks has been reported in several cancer types, including leukemia, glioblastoma, soft tissue sarcoma, and prostate and breast cancers (Emerling et al., 2013; Jude et al., 2015; Shin et al., 2019; Ravi et al., 2021; Triscott et al., 2023).
3.1.1 Localization and impact
The PI5P4K family of lipid kinases include three members: PI5P4Kα, PI5P4Kβ, and PI5P4Kγ, with differences in their catalytic activity and localization within the cell. While all three PI5P4K isoforms are primarily found within intracellular membranes, their precise subcellular distributions vary. PI5P4Kα is distributed across lysosomes, autophagosomes, and peroxisomes; PI5P4Kβ primarily resides within the nucleus but may also be detected in autophagosomes, and PI5P4Kγ can be observed in autophagosomes, endomembrane compartments, and the Golgi apparatus (Clarke et al., 2008; Bultsma et al., 2010; Vicinanza et al., 2015; Hu et al., 2018; Lundquist et al., 2018). Additionally, their localization can be influenced by their capacity to heterodimerize (Bultsma et al., 2010; Wang et al., 2010). Interestingly, recent data has shown that PI5P4Ks can also be recruited to the plasma membrane by PI(4,5)P2 to inhibit phosphatidylinositol-4-phosphate 5-kinases (PI4P5Ks) as a homeostasis mechanism (Wills et al., 2023). By regulating the balance between PI(5)P and PI(4,5)P2, PI5P4Ks modulate cellular signaling, protein activation at specific subcellular locations and protein transport. Furthermore, growing evidence suggests that these kinases possess catalytic-independent roles, which adds a layer of complexity to their functional repertoire (Llorente et al., 2023). Notably, in vitro assays have shown a markedly higher kinase activity for PI5P4Kα when compared to PI5P4Kβ, while PI5P4Kγ exhibits the lowest activity (Clarke et al., 2008; Bultsma et al., 2010; Wang et al., 2010). The substantial difference in kinase activity, with PI5P4Kγ being several orders of magnitude lower than the other two kinases, suggests that it potentially plays a more significant role as a scaffolding protein.
3.1.2 Metabolic adaptations through the action of PI5P4Ks in tumor cells
During tumorigenesis, cancer cells typically rely on metabolic reprogramming to adapt to energy and oxidative stresses. The synthetic lethal interaction involving p53, PI5P4Kα, and PI5P4Kβ is a very compelling illustration of this phenomenon (Emerling et al., 2013). The frequent loss of p53 in cancer renders cells more susceptible to oxidative stress. In this context, PI5P4Kα and PI5P4Kβ play critical roles in regulating oxygen consumption, ROS generation, glucose metabolism and AKT signaling to manage such stress, allowing cancer cell survival despite challenging conditions. Other studies support the role of PI5P4Ks in metabolic homeostasis through modulation of insulin signaling, PI3K, AKT, and mTORC pathways, and oxidative stress (Carricaburu et al., 2003; Gupta et al., 2013; Mackey et al., 2014; Lundquist et al., 2018; Wang et al., 2019). It is worth noting that PI5P4Kβ preferentially uses GTP over ATP for PI(4,5)P2 synthesis, acting as an intracellular GTP sensor. The GTP-sensing ability of PI5P4Kβ plays a crucial role in both metabolic adaptation and tumor development (Sumita et al., 2016).
PI5P4Ks have also been implicated in autophagy and organelle communication. Loss of PI5P4Ks expression increases autophagosome biogenesis and results in a defect in autophagosome-lysosome fusion and subsequent accumulation of autophagic vesicles (Vicinanza et al., 2015; Lundquist et al., 2018). In addition, PI5P4Kα regulates PI(4,5)P2 levels at the peroxisomal membrane to facilitate the lysosome-peroxisome membrane contacts necessary for proper intracellular cholesterol transport (Hu et al., 2018). Moreover, regulation of the peroxisomal PI(4,5)P2 pool by the action of PI5P4Kα and PI5P4Kβ is also required for the traffic and peroxisomal oxidation of very long chain fatty acids and consequently for ensuring proper mitochondrial metabolism (Ravi et al., 2021). Consistently, inhibition of these kinases negatively impacts mitochondrial ATP production, disrupting cell energy metabolism (Chen et al., 2021). Together, all these studies demonstrate the ability of PI5P4Ks to maintain the metabolic homeostasis required for cancer cell survival.
Deregulation of stress response pathways, such as autophagy, frequently contributes to drug resistance and tumor progression. Given the implication of PI5P4Ks in energy stress responses and their reported involvement in cellular trafficking (Kamalesh et al., 2017), including membrane receptor recycling (Zheng and Conner, 2018), we speculate that in the context of cancer, PI5P4Ks might play a role in fostering resistance to targeted therapies, especially those involving membrane receptors. Exploring the interplay between PI5P4Ks and resistance pathways would provide valuable insights for enhancing the durability and effectiveness of targeted cancer treatments.
3.1.3 Immunomodulatory roles of PI5P4Ks
Beyond their influence on cancer cell intrinsic processes, PI5P4Ks may also contribute to immune modulation within the tumor microenvironment. Single nucleotide polymorphisms (SNPs) near PIP4K2C (the gene coding for PI5P4Kγ) have been associated with susceptibility to autoimmune diseases (Raychaudhuri et al., 2008; Fung et al., 2009). Interestingly, mice lacking PI5P4Kγ exhibit increased T helper (Th) and decreased regulatory T (Treg) cell populations, along with heightened proinflammatory cytokine levels, resulting in immune hyperactivation (Shim et al., 2016). Furthermore, there is evidence indicating the necessity of PI5P4Kβ and PI5P4Kγ for Treg-mediated immune suppression (Poli et al., 2021). The activity of these PI5P4K isoforms impacts the PI3K, mTORC1 and MAPK signaling pathways, leading to consequential changes in FOXP3 expression that govern Treg reprogramming and functionality. Collectively, these data reveal the significant role of these kinases in immune regulation, underscoring the potential of targeting PI5P4Kγ as a therapeutic strategy to enhance cancer immunotherapy and open new possibilities for cancer treatment.
3.2 PI5P4Ks as targets for cancer therapy
Alterations in phosphoinositide metabolism play a critical role in various human diseases, offering numerous opportunities for therapeutic modulation of the enzymes involved in this process. Currently, significant research efforts have resulted in the development of several compounds targeting lipid kinases and phosphatases in multiple cancer subtypes, with some PI3K inhibitors already having received clinical approval (Figure 2). Further, an increase in the understanding of PI5P4Ks functions have brought to the surface their potential to serve as therapeutic targets for cancer treatment. Here, we are going to provide a short overview of PI5P4K targeting agents that have been developed until now. For an extensive review on the preclinical and clinical development of PI5P4K and other phosphoinositide kinase inhibitors we refer the reader to (Burke et al., 2023).
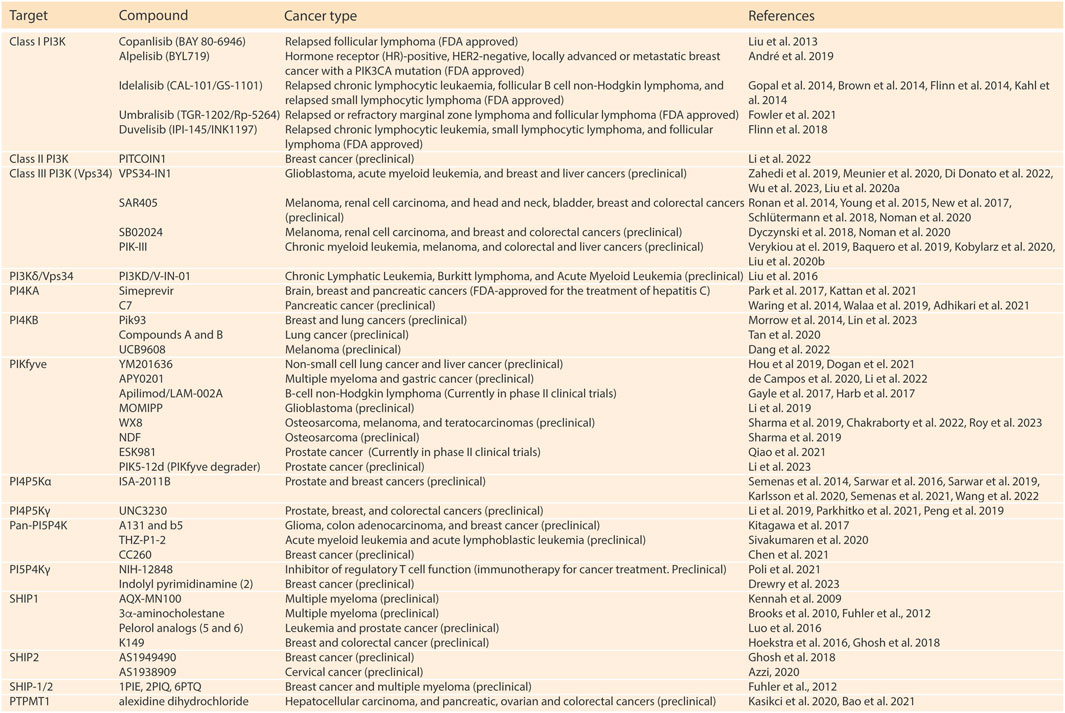
FIGURE 2. Relevant compounds targeting phosphoinositide kinases and phosphatases for the treatment of cancer. (Harlan et al., 1994; Patki et al., 1998; Baraldi et al., 1999; Lietzke et al., 2000; Cheever et al., 2001; Kanai et al., 2001; Gozani et al., 2003; Quinn, Behe, and Tinker, 2008; Guittard et al., 2009; Kennah et al., 2009; Brooks et al., 2010; Fuhler et al., 2012; Liu et al., 2013; Brown et al., 2014; Flinn et al., 2014; Gopal et al., 2014; Kahl et al., 2014; Morrow et al., 2014; Ronan et al., 2014; Semenas et al., 2014; Waring et al., 2014; Young et al., 2015; Hoekstra et al., 2016; Liu et al., 2016; Luo et al., 2016; Sarwar et al., 2016; Gayle et al., 2017; Harb et al., 2017; Kitagawa et al., 2017; New et al., 2017; Park et al., 2017; Dyczynski et al., 2018; Flinn et al., 2018; Ghosh et al., 2018; Moriwaki et al., 2018; Schlütermann et al., 2018; Li L. et al., 2019; André et al., 2019; Baquero et al., 2019; Li Z. et al., 2019; Chandra et al., 2019; Hou et al., 2019; Jun et al., 2019; Liang et al., 2019; Peng et al., 2019; Sarwar et al., 2019; Sharma et al., 2019; Verykiou et al., 2019; Walaa et al., 2019; Zahedi et al., 2019; Liu F. et al., 2020; Azzi, 2020; Liu et al., 2020b; de Campos et al., 2020; Gozzelino et al., 2020; Karlsson et al., 2020; Kasikci et al., 2020; Kobylarz et al., 2020; Meunier et al., 2020; Noman et al., 2020; Sivakumaren et al., 2020; Tan et al., 2020; Adhikari et al., 2021; Bao et al., 2021; Chen et al., 2021; DoĞan et al., 2021; Fowler et al., 2021; Kattan et al., 2021; Overduin and Kervin, 2021; Parkhitko et al., 2021; Poli et al., 2021; Qiao et al., 2021; Semenas et al., 2021; Li et al., 2022a; Li et al., 2022b; Chakraborty et al., 2022; Dang et al., 2022; Di Donato et al., 2022; Wang et al., 2022; Drewry et al., 2023; Li et al., 2023; Lin et al., 2023; Roy et al., 2023; Wu et al., 2023)
3.2.1 Small molecule inhibitors
Developing small molecule inhibitors against PI5P4Ks emerges as a promising avenue, allowing precise modulation of PI(5)P and PI(4,5)P2 dynamics and its downstream effects. Efforts have been made to develop pan-PI5P4K inhibitors (Kitagawa et al., 2017; Manz et al., 2020; Sivakumaren et al., 2020; Chen et al., 2021; Lima et al., 2022), as well as isoform-specific inhibitors that selectively target PI5P4Kα (Davis et al., 2013; Wortmann et al., 2021; Willems et al., 2023), PI5P4Kβ (Voss et al., 2014), and PI5P4Kγ (Clarke et al., 2015; Al-Ramahi et al., 2017; Boffey et al., 2022; Drewry et al., 2023; Rooney et al., 2023). Despite a dedicated endeavor to enhance the potency and selectivity of these compounds, PI5P4K inhibitors have not yet advanced to the clinical stage.
3.2.2 PI5P4Ks Proteolysis Targeting Chimeras (PROTACs)
Exploring the potential of PI5P4Ks degraders offers a distinct approach, allowing complete abrogation of protein function, including both kinase-dependent and independent functions. Currently, there is active work directed towards the development of PROTACS targeting PI5P4Kα and PI5P4Kβ, while highly potent and selective PI5P4Kγ degraders have already been documented (Ji et al., 2023; Teng et al., 2023). Further, progress in investigating protein-protein interactions, facilitated by techniques like proximity-dependent biotin identification (BioID) (Roux et al., 2012), hold promise in shedding light on the lesser-understood functions of PI5P4Ks that do not rely on their kinase activity, as well as on their currently less-known upstream regulators. Indeed, the efficacy of this approach to identify lipid kinases relevant in the context of cancer has already been demonstrated. In the exploration of KRAS interactions through BioID, researchers successfully identified the type I lipid kinase PIP5K1A (Adhikari and Counter, 2018). Using proximity-based labeling to identify PI5P4K interactors could uncover significant functions beyond phosphoinositide signaling, providing a rationale for the development of molecules that target kinase-dependent and independent functions and shifting the balance from small molecule inhibitors to PROTACs.
4 Discussion
Altogether, with our ever-increasing toolbox for phosphoinositide quantification, localization, and turnover, the combination of these technologies in clinically relevant models can potentially provide proof of concept for advancing drug targeting studies of phosphoinositides and their metabolizing enzymes. Further, enhancing the specificity of genetically encoded biosensors will unveil previously undefined pools of phosphoinositides at areas outside the defined phosphoinositide map that is widely accepted, leading to previously unknown protein localizations and involvement in cellular signaling cascades.
Finally, with the wide breadth of phosphoinositide involvement in cellular functions, it is likely to be understated due to our developing, yet limited ability to fully understand their involvement in these processes through measurement. However, the union of these novel technologies paints a bright future for discovery in the field of phosphoinositide biology and, importantly, for the feasibility of targeting phosphoinositide enzymes in human diseases. This is evident with the rising drug discovery efforts of the PI5P4K family of enzymes for cancer treatment, as highlighted here in this perspective.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
AL: Writing–original draft. RL: Writing–original draft, Writing–review and editing. BE: Writing–original draft, Writing–review and editing.
Funding
This work is supported by grants from the NCI (R01 CA237536), NIH (R01 GM143583), and ACS (RSG-20-064-01-TBE) to BME, NCI Training Fellowship (T32CA211036) to RML, and The Conrad Prebys Foundation to AL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adhikari, H., and Counter, C. M. (2018). 'Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nat. Commun. 9, 3646. doi:10.1038/s41467-018-05692-6
Adhikari, H., Kattan, W. E., Kumar, S., Zhou, P., Hancock, J. F., and Counter, C. M. (2021). 'Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Nat. Commun. 12, 5248. doi:10.1038/s41467-021-25523-5
Al-Ramahi, I., Giridharan, S. S. P., Chen, Y. C., Patnaik, S., Safren, N., Hasegawa, J., et al. (2017). 'Inhibition of PIP4Kgamma ameliorates the pathological effects of mutant huntingtin protein. Elife 6. doi:10.7554/eLife.29123
André, F., Ciruelos, E., Rubovszky, G., Campone, M., Loibl, S., Rugo, H. S., et al. (2019). 'Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940. doi:10.1056/NEJMoa1813904
Arora, G. K., Palamiuc, L., and Emerling, B. M. (2022). Expanding role of PI5P4Ks in cancer: a promising druggable target. FEBS Lett. 596, 3–16. doi:10.1002/1873-3468.14237
Azzi, A. (2020). 'SHIP2 inhibition alters redox-induced PI3K/AKT and MAP kinase pathways via PTEN over-activation in cervical cancer cells. FEBS Open Bio 10, 2191–2205. doi:10.1002/2211-5463.12967
Balla, T. (2013). 'Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. doi:10.1152/physrev.00028.2012
Balla, T., Szentpetery, Z., and Kim, Y. J. (2009). 'Phosphoinositide signaling: new tools and insights. Physiol. (Bethesda) 24, 231–244. doi:10.1152/physiol.00014.2009
Bao, M. H., Yang, C., Tse, A. P., Wei, L., Lee, D., Zhang, M. S., et al. (2021). 'Genome-wide CRISPR-Cas9 knockout library screening identified PTPMT1 in cardiolipin synthesis is crucial to survival in hypoxia in liver cancer. Cell. Rep. 34, 108676. doi:10.1016/j.celrep.2020.108676
Baquero, P., Dawson, A., Mukhopadhyay, A., Kuntz, E. M., Mitchell, R., Olivares, O., et al. (2019). 'Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia 33, 981–994. doi:10.1038/s41375-018-0252-4
Baraldi, E., Djinovic Carugo, K., Hyvonen, M., Surdo, P. L., Riley, A. M., Potter, B. V., et al. (1999). 'Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure 7, 449–460. doi:10.1016/s0969-2126(99)80057-4
Boffey, H. K., Rooney, T. P. C., Willems, H. M. G., Edwards, S., Green, C., Howard, T., et al. (2022). 'Development of selective phosphatidylinositol 5-phosphate 4-kinase gamma inhibitors with a non-ATP-competitive, allosteric binding mode. J. Med. Chem. 65, 3359–3370. doi:10.1021/acs.jmedchem.1c01819
Brooks, R., Fuhler, G. M., Iyer, S., Smith, M. J., Park, M. Y., Paraiso, K. H., et al. (2010). 'SHIP1 inhibition increases immunoregulatory capacity and triggers apoptosis of hematopoietic cancer cells. J. Immunol. 184, 3582–3589. doi:10.4049/jimmunol.0902844
Brown, J. R., Byrd, J. C., Coutre, S. E., Benson, D. M., Flinn, I. W., Wagner-Johnston, N. D., et al. (2014). 'Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood 123, 3390–3397. doi:10.1182/blood-2013-11-535047
Buchberger, A. R., DeLaney, K., Johnson, J., and Li, L. (2018). 'Mass spectrometry imaging: a review of emerging advancements and future insights. Anal. Chem. 90, 240–265. doi:10.1021/acs.analchem.7b04733
Bultsma, Y., Keune, W. J., and Divecha, N. (2010). 'PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem. J. 430, 223–235. doi:10.1042/BJ20100341
Bunney, T. D., and Katan., M. (2010). 'Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer 10, 342–352. doi:10.1038/nrc2842
Burke, J. E. (2018). 'Structural basis for regulation of phosphoinositide kinases and their involvement in human disease. Mol. Cell. 71, 653–673. doi:10.1016/j.molcel.2018.08.005
Burke, J. E., Triscott, J., Emerling, B. M., and Hammond, G. R. V. (2023). 'Beyond PI3Ks: targeting phosphoinositide kinases in disease. Nat. Rev. Drug Discov. 22, 357–386. doi:10.1038/s41573-022-00582-5
Cai, T., Shu, Q., Hou, J., Liu, P., Niu, L., Guo, X., et al. (2015). 'Profiling and relative quantitation of phosphoinositides by multiple precursor ion scanning based on phosphate methylation and isotopic labeling. Anal. Chem. 87, 513–521. doi:10.1021/ac503224j
Cai, T., Shu, Q., Liu, P., Niu, L., Guo, X., Ding, X., et al. (2016). 'Characterization and relative quantification of phospholipids based on methylation and stable isotopic labeling. J. Lipid Res. 57, 388–397. doi:10.1194/jlr.M063024
Carricaburu, V., Lamia, K. A., Lo, E., Favereaux, L., Payrastre, B., Cantley, L. C., et al. (2003). 'The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. U. S. A. 100, 9867–9872. doi:10.1073/pnas.1734038100
Chakraborty, A. R., Vassilev, A., Jaiswal, S. K., O'Connell, C. E., Ahrens, J. F., Mallon, B. S., et al. (2022). 'Selective elimination of pluripotent stem cells by PIKfyve specific inhibitors. Stem Cell. Rep. 17, 397–412. doi:10.1016/j.stemcr.2021.12.013
Chandra, M., Chin, Y. K., Mas, C., Feathers, J. R., Paul, B., Datta, S., et al. (2019). 'Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 10, 1528. doi:10.1038/s41467-019-09355-y
Cheever, M. L., Sato, T. K., de Beer, T., Kutateladze, T. G., Emr, S. D., and Overduin, M. (2001). 'Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell. Biol. 3, 613–618. doi:10.1038/35083000
Chen, S., Chandra Tjin, C., Gao, X., Xue, Y., Jiao, H., Zhang, R., et al. (2021). 'Pharmacological inhibition of PI5P4Kalpha/beta disrupts cell energy metabolism and selectively kills p53-null tumor cells'. Proc. Natl. Acad. Sci. U. S. A. 118. doi:10.1073/pnas.2002486118
Clark, J., Anderson, K. E., Juvin, V., Smith, T. S., Karpe, F., Wakelam, M. J., et al. (2011). 'Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 8, 267–272. doi:10.1038/nmeth.1564
Clarke, J. H., Emson, P. C., and Irvine, R. F. (2008). 'Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am. J. Physiol. Ren. Physiol. 295, F1422–F1430. doi:10.1152/ajprenal.90310.2008
Clarke, J. H., Giudici, M. L., Burke, J. E., Williams, R. L., Maloney, D. J., Marugan, J., et al. (2015). The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem. J. 466, 359–367. doi:10.1042/BJ20141333
Dang, N., Waer, M., Sprangers, B., and Lin, Y. (2022). 'Establishment of operational tolerance to sustain antitumor immunotherapy. J. Heart Lung Transpl. 41, 568–577. doi:10.1016/j.healun.2022.01.019
Davis, M. I., Sasaki, A. T., Shen, M., Emerling, B. M., Thorne, N., Michael, S., et al. (2013). 'A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS One 8, e54127. doi:10.1371/journal.pone.0054127
de Campos, C. B., Zhu, Y. X., Sepetov, N., Romanov, S., Bruins, L. A., Shi, C. X., et al. (2020). 'Identification of PIKfyve kinase as a target in multiple myeloma. Haematologica 105, 1641–1649. doi:10.3324/haematol.2019.222729
Di Donato, M., Giovannelli, P., Migliaccio, A., and Bilancio, A. (2022). Inhibition of Vps34 and p110δ PI3K impairs migration, invasion and three-dimensional spheroid growth in breast cancer cells. Int. J. Mol. Sci. 23, 9008. doi:10.3390/ijms23169008
Di Paolo, H., Gilbert, L., and De Camilli, P. (2006). 'Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657. doi:10.1038/nature05185
Dickson, E. J., and Hille, B. (2019). 'Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 476, 1–23. doi:10.1042/BCJ20180022
DoĞan, E., DüzgÜn, Z., Yildirim, Z., Özdİl, B., AktuĞ, H., and Bozok ÇetİntaŞ, V. (2021). The effects of PIKfyve inhibitor YM201636 on claudins and malignancy potential of nonsmall cell cancer cells. Turk J. Biol. 45, 26–34. doi:10.3906/biy-2010-32
Drewry, D. H., Potjewyd, F. M., Axtman, A. D., and Axtman, A. D. (2023). 'Identification of a chemical probe for lipid kinase phosphatidylinositol-5-phosphate 4-kinase gamma (PI5P4Kγ). Curr. Res. Chem. Biol. 3, 100036. doi:10.1016/j.crchbi.2022.100036
Dyczynski, M., Yu, Y., Otrocka, M., Parpal, S., Braga, T., Henley, A. B., et al. (2018). 'Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 435, 32–43. doi:10.1016/j.canlet.2018.07.028
Emerling, B. M., Hurov, J. B., Poulogiannis, G., Tsukazawa, K. S., Choo-Wing, R., Wulf, G. M., et al. (2013). 'Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 155, 844–857. doi:10.1016/j.cell.2013.09.057
Falkenburger, B. H., Jensen, J. B., Dickson, E. J., Suh, B. C., and Hille, B. (2010). 'Phosphoinositides: lipid regulators of membrane proteins. J. Physiol. 588, 3179–3185. doi:10.1113/jphysiol.2010.192153
Flinn, I. W., Kahl, B. S., Leonard, J. P., Furman, R. R., Brown, J. R., Byrd, J. C., et al. (2014). 'Idelalisib, a selective inhibitor of phosphatidylinositol 3-kinase-δ, as therapy for previously treated indolent non-Hodgkin lymphoma. Blood 123, 3406–3413. doi:10.1182/blood-2013-11-538546
Flinn, I. W., O'Brien, S., Kahl, B., Patel, M., Oki, Y., Foss, F. F., et al. (2018). 'Duvelisib, a novel oral dual inhibitor of PI3K-δ,γ, is clinically active in advanced hematologic malignancies. Blood 131, 877–887. doi:10.1182/blood-2017-05-786566
Fowler, N. H., Samaniego, F., Jurczak, W., Ghosh, N., Derenzini, E., Reeves, J. A., et al. (2021). 'Umbralisib, a dual PI3Kδ/CK1ε inhibitor in patients with relapsed or refractory indolent lymphoma. J. Clin. Oncol. 39, 1609–1618. doi:10.1200/JCO.20.03433
Fuhler, G. M., Brooks, R., Toms, B., Iyer, S., Gengo, E. A., Park, M. Y., et al. (2012). 'Therapeutic potential of SH2 domain-containing inositol-5'-phosphatase 1 (SHIP1) and SHIP2 inhibition in cancer. Mol. Med. 18, 65–75. doi:10.2119/molmed.2011.00178
Fung, E. Y., Smyth, D. J., Howson, J. M., Cooper, J. D., Walker, N. M., Stevens, H., et al. (2009). 'Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes. Immun. 10, 188–191. doi:10.1038/gene.2008.99
Gayle, S., Landrette, S., Beeharry, N., Conrad, C., Hernandez, M., Beckett, P., et al. (2017). 'Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood 129, 1768–1778. doi:10.1182/blood-2016-09-736892
Ghosh, S., Scozzaro, S., Delcambre, S., Chevalier, C., Pavel Krejci, H., and Erneux, C. (2018). Inhibition of SHIP2 activity inhibits cell migration and could prevent metastasis in breast cancer cells. J. Cell. Sci. 131, jcs216408. doi:10.1242/jcs.216408
Gopal, A. K., Kahl, B. S., de Vos, S., Wagner-Johnston, N. D., Schuster, S. J., Jurczak, W. J., et al. (2014). PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N. Engl. J. Med. 370, 1008–1018. doi:10.1056/NEJMoa1314583
Gozani, O., Karuman, P., Jones, D. R., Ivanov, D., Cha, J., Lugovskoy, A. A., et al. (2003). The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 114, 99–111. doi:10.1016/s0092-8674(03)00480-x
Gozzelino, L., De Santis, M. C., Gulluni, F., Hirsch, E., and Martini, M. (2020). 'PI(3,4)P2 signaling in cancer and metabolism. Front. Oncol. 10, 360. doi:10.3389/fonc.2020.00360
Greenwald, E. C., Mehta, S., and Zhang, J. (2018). 'Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794. doi:10.1021/acs.chemrev.8b00333
Guittard, G., Gerard, A., Dupuis-Coronas, S., Tronchere, H., Mortier, E., Favre, C., et al. (2009). Cutting edge: dok-1 and Dok-2 adaptor molecules are regulated by phosphatidylinositol 5-phosphate production in T cells. J. Immunol. 182, 3974–3978. doi:10.4049/jimmunol.0804172
Gupta, A., Toscano, S., Trivedi, D., Jones, D. R., Mathre, S., Clarke, J. H., et al. (2013). 'Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc. Natl. Acad. Sci. U. S. A. 110, 5963–5968. doi:10.1073/pnas.1219333110
Hammond, G. R., and Balla, T. (2015). 'Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim. Biophys. Acta 1851, 746–758. doi:10.1016/j.bbalip.2015.02.013
Hammond, G. R. V., Ricci, M. M. C., Weckerly, C. C., and Wills, R. C. (2022). 'An update on genetically encoded lipid biosensors. Mol. Biol. Cell. 33, tp2. doi:10.1091/mbc.E21-07-0363
Harb, W. A., Diefenbach, C. S., Lakhani, N., Rutherford, S. C., Schreeder, M. T., Ansell, S. M., et al. (2017). 'Phase 1 clinical safety, pharmacokinetics (PK), and activity of apilimod dimesylate (LAM-002A), a first-in-class inhibitor of phosphatidylinositol-3-phosphate 5-kinase (PIKfyve), in patients with relapsed or refractory B-cell malignancies. Blood 130, 4119. doi:10.1182/blood.V130.Suppl_1.4119.4119
Harlan, J. E., Hajduk, P. J., Yoon, H. S., and Fesik, S. W. (1994). 'Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature 371, 168–170. doi:10.1038/371168a0
Hertel, F., Li, S., Chen, M., Pott, L., Mehta, S., and Zhang, J. (2020). 'Fluorescent biosensors for multiplexed imaging of phosphoinositide dynamics. ACS Chem. Biol. 15, 33–38. doi:10.1021/acschembio.9b00691
Hoekstra, E., Das, A. M., Willemsen, M., Swets, M., Kuppen, P. J., van der Woude, C. J., et al. (2016). Lipid phosphatase SHIP2 functions as oncogene in colorectal cancer by regulating PKB activation. Oncotarget 7, 73525–73540. doi:10.18632/oncotarget.12321
Hou, J. Z., Xi, Z. Q., Niu, J., Li, W., Wang, X., Liang, C., et al. (2019). 'Inhibition of PIKfyve using YM201636 suppresses the growth of liver cancer via the induction of autophagy. Oncol. Rep. 41, 1971–1979. doi:10.3892/or.2018.6928
Hu, A., Zhao, X. T., Tu, H., Xiao, T., Fu, T., Wang, Y., et al. (2018). 'PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P(2) homeostasis. J. Lipid Res. 59, 507–514. doi:10.1194/jlr.M082149
Irino, Y., Tokuda, E., Hasegawa, J., Itoh, T., and Takenawa, T. (2012). 'Quantification and visualization of phosphoinositides by quantum dot-labeled specific binding-domain probes. J. Lipid Res. 53, 810–819. doi:10.1194/jlr.D019547
Ji, W., Wang, E. S., Manz, T. D., Jiang, J., Donovan, K. A., Abulaiti, X., et al. (2023). Development of potent and selective degraders of PI5P4Kγ. Eur. J. Med. Chem. 247, 115027. doi:10.1016/j.ejmech.2022.115027
Jude, J. G., Spencer, G. J., Huang, X., Somerville, T. D. D., Jones, D. R., Divecha, N., et al. (2015). 'A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene 34, 1253–1262. doi:10.1038/onc.2014.77
Jun, Y. W., Lee, J. A., and Jang, D. J. (2019). 'Novel GFP-fused protein probes for detecting phosphatidylinositol-4-phosphate in the plasma membrane. Anim. Cells Syst. Seoul. 23, 164–169. doi:10.1080/19768354.2019.1599424
Kahl, B. S., Spurgeon, S. E., Furman, R. R., Flinn, I. W., Coutre, S. E., Brown, J. R., et al. (2014). 'A phase 1 study of the PI3Kδ inhibitor idelalisib in patients with relapsed/refractory mantle cell lymphoma (MCL). Blood 123, 3398–3405. doi:10.1182/blood-2013-11-537555
Kalasova, I., Faberova, V., Kalendova, A., Yildirim, S., Ulicna, L., Venit, T., et al. (2016). 'Tools for visualization of phosphoinositides in the cell nucleus. Histochem Cell. Biol. 145, 485–496. doi:10.1007/s00418-016-1409-8
Kamalesh, K., Trivedi, D., Toscano, S., Sharma, S., Kolay, S., and Raghu, P. (2017). 'Phosphatidylinositol 5-phosphate 4-kinase regulates early endosomal dynamics during clathrin-mediated endocytosis. J. Cell. Sci. 130, 2119–2133. doi:10.1242/jcs.202259
Kanai, F., Liu, H., Field, S. J., Akbary, H., Matsuo, T., Brown, G. E., et al. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell. Biol. 3, 675–678. doi:10.1038/35083070
Karlsson, R., Larsson, P., Miftakhova, R., Syed Khaja, A. S., Sarwar, M., Semenas, J., et al. (2020). 'Establishment of prostate tumor growth and metastasis is supported by bone marrow cells and is mediated by PIP5K1α lipid kinase. Basel): Cancers, 12.
Kasikci, E., Aydemir, E., Yogurtcu, B. M., Sahin, F., and Bayrak, O. F. (2020). 'Repurposing of alexidine dihydrochloride as an apoptosis initiator and cell cycle inhibitor in human pancreatic cancer. Anticancer Agents Med. Chem. 20, 1956–1965. doi:10.2174/1871520620666200508085439
Kattan, W. E., Liu, J., Montufar-Solis, D., Liang, H., Brahmendra Barathi, B., van der Hoeven, R., et al. (2021). 'Components of the phosphatidylserine endoplasmic reticulum to plasma membrane transport mechanism as targets for KRAS inhibition in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A. 118. doi:10.1073/pnas.2114126118
Kawashima, M., Iwamoto, N., Kawaguchi-Sakita, N., Sugimoto, M., Ueno, T., Mikami, Y., et al. (2013). 'High-resolution imaging mass spectrometry reveals detailed spatial distribution of phosphatidylinositols in human breast cancer. Cancer Sci. 104, 1372–1379. doi:10.1111/cas.12229
Kawashima, M., Tokiwa, M., Nishimura, T., Kawata, Y., Sugimoto, M., Kataoka, T. R., et al. (2020). 'High-resolution imaging mass spectrometry combined with transcriptomic analysis identified a link between fatty acid composition of phosphatidylinositols and the immune checkpoint pathway at the primary tumour site of breast cancer. Br. J. Cancer 122, 245–257. doi:10.1038/s41416-019-0662-8
Kennah, M., Yau, T. Y., Nodwell, M., Krystal, G., Andersen, R. J., Ong, C. J., et al. (2009). 'Activation of SHIP via a small molecule agonist kills multiple myeloma cells. Exp. Hematol. 37, 1274–1283. doi:10.1016/j.exphem.2009.08.001
Kielkowska, A., Niewczas, I., Anderson, K. E., Durrant, T. N., Clark, J., Stephens, L. R., et al. (2014). 'A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv. Biol. Regul. 54, 131–141. doi:10.1016/j.jbior.2013.09.001
Kim, Y., Shanta, S. R., Zhou, L. H., and Kim, K. P. (2010). 'Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: a brief review. Exp. Mol. Med. 42, 1–11. doi:10.3858/emm.2010.42.1.001
Kitagawa, M., Liao, P. J., Lee, K. H., Wong, J., Shang, S. C., Minami, N., et al. (2017). 'Dual blockade of the lipid kinase PIP4Ks and mitotic pathways leads to cancer-selective lethality. Nat. Commun. 8, 2200. doi:10.1038/s41467-017-02287-5
Kobylarz, M. J., Goodwin, J. M., Kang, Z. B., Annand, J. W., Hevi, S., O'Mahony, E., et al. (2020). An iron-dependent metabolic vulnerability underlies VPS34-dependence in RKO cancer cells. PLoS One 15, e0235551. doi:10.1371/journal.pone.0235551
Lee, S. A., Eyeson, R., Cheever, M. L., Geng, J., Verkhusha, V. V., Burd, C., et al. (2005). 'Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. U. S. A. 102, 13052–13057. doi:10.1073/pnas.0503900102
Li, C., Qiao, Y., Jiang, X., Liu, L., Zheng, Y., Qiu, Y., et al. (2023). 'Discovery of a first-in-class degrader for the lipid kinase PIKfyve. J. Med. Chem. 66, 12432–12445. doi:10.1021/acs.jmedchem.3c00912
Li, H., Jin, X., Zhang, S., Li, B., Zeng, L., He, Y., et al. (2022a). 'APY0201 represses tumor growth through inhibiting autophagy in gastric cancer cells. J. Oncol. 2022, 7104592. doi:10.1155/2022/7104592
Li, H., Prever, L., Hsu, M. Y., Lo, W. T., Margaria, J. P., De Santis, M. C., et al. (2022b). 'Phosphoinositide conversion inactivates R-RAS and drives metastases in breast cancer. Adv. Sci. (Weinh) 9, e2103249. doi:10.1002/advs.202103249
Li, L., Kołodziej, T., Jafari, N., Chen, J., Zhu, H., Rajfur, Z., et al. (2019a). 'Cdk5-mediated phosphorylation regulates phosphatidylinositol 4-phosphate 5-kinase type I γ 90 activity and cell invasion. Faseb J. 33, 631–642. doi:10.1096/fj.201800296R
Li, P., and Lammerhofer, M. (2021). 'Isomer selective comprehensive lipidomics analysis of phosphoinositides in biological samples by liquid chromatography with data independent acquisition tandem mass spectrometry. Anal. Chem. 93, 9583–9592. doi:10.1021/acs.analchem.1c01751
Li, Z., Mbah, N. E., Overmeyer, J. H., Sarver, J. G., George, S., Trabbic, C. J., et al. (2019b). 'The JNK signaling pathway plays a key role in methuosis (non-apoptotic cell death) induced by MOMIPP in glioblastoma. BMC Cancer 19, 77. doi:10.1186/s12885-019-5288-y
Liang, R., Ren, J., Zhang, Y., and Feng, W. (2019). 'Structural conservation of the two phosphoinositide-binding sites in WIPI proteins. J. Mol. Biol. 431, 1494–1505. doi:10.1016/j.jmb.2019.02.019
Lietzke, S. E., Bose, S., Cronin, T., Klarlund, J., Chawla, A., Czech, M. P., et al. (2000). 'Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 6, 385–394. doi:10.1016/s1097-2765(00)00038-1
Lima, K., Pereira-Martins, D. A., de Miranda, L. B. L., Coelho-Silva, J. L., Leandro, G. D. S., Weinhauser, I., et al. (2022). 'The PIP4K2 inhibitor THZ-P1-2 exhibits antileukemia activity by disruption of mitochondrial homeostasis and autophagy. Blood Cancer J. 12, 151. doi:10.1038/s41408-022-00747-w
Lin, C. Y., Huang, K. Y., Kao, S. H., Lin, M. S., Lin, C. C., Yang, S. C., et al. (2023). 'Small-molecule PIK-93 modulates the tumor microenvironment to improve immune checkpoint blockade response. Sci. Adv. 9, eade9944. doi:10.1126/sciadv.ade9944
Liu, F., Wu, X., Qian, Y., Jiang, X., Wang, Y., and Gao, J. (2020b). 'PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. Cell. Death Dis. 11, 427. doi:10.1038/s41419-020-2631-9
Liu, F., Wu, X., Qian, Y., Jiang, X., Wang, Y., and Gao, J. (2020a). 'PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. Cell. Death Dis. 11, 427. doi:10.1038/s41419-020-2631-9
Liu, N., Rowley, B. R., Bull, C. O., Schneider, C., Haegebarth, A., Schatz, C. A., et al. (2013). BAY 80-6946 is a highly selective intravenous PI3K inhibitor with potent p110α and p110δ activities in tumor cell lines and xenograft models. Mol. Cancer Ther. 12, 2319–2330. doi:10.1158/1535-7163.MCT-12-0993-T
Liu, X., Wang, A., Liang, X., Liu, J., Zou, F., Chen, C., et al. (2016). 'Simultaneous inhibition of Vps34 kinase would enhance PI3Kδ inhibitor cytotoxicity in the B-cell malignancies. Oncotarget 7, 53515–53525. doi:10.18632/oncotarget.10650
Llorente, A., Arora, G. K., Grenier, S. F., and Emerling, B. M. (2023). PIP kinases: a versatile family that demands further therapeutic attention. Adv. Biol. Regul. 87, 100939. doi:10.1016/j.jbior.2022.100939
Lundquist, M. R., Goncalves, M. D., Loughran, R. M., Possik, E., Vijayaraghavan, T., Yang, A., et al. (2018). 'Phosphatidylinositol-5-Phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol. Cell. 70, 531–544. doi:10.1016/j.molcel.2018.03.037
Luo, Y., Chen, H., Weng, J., and Lu, G. (2016). 'Synthesis of pelorol and its analogs and their inhibitory effects on phosphatidylinositol 3-kinase'. Mar. Drugs 14. doi:10.3390/md14060118
Mackey, A. M., Sarkes, D. A., Bettencourt, I., Asara, J. M., and Rameh, L. E. (2014). PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci. Signal 7, ra104. doi:10.1126/scisignal.2005191
Manz, T. D., Sivakumaren, S. C., Ferguson, F. M., Zhang, T., Yasgar, A., Seo, H. S., et al. (2020). Discovery and structure-activity relationship study of (Z)-5-Methylenethiazolidin-4-one derivatives as potent and selective pan-phosphatidylinositol 5-phosphate 4-kinase inhibitors. J. Med. Chem. 63, 4880–4895. doi:10.1021/acs.jmedchem.0c00227
Meunier, G., Birsen, R., Cazelles, C., Belhadj, M., Cantero-Aguilar, L., Kosmider, O., et al. (2020). 'Antileukemic activity of the VPS34-IN1 inhibitor in acute myeloid leukemia. Oncogenesis 9, 94. doi:10.1038/s41389-020-00278-8
Morioka, S., Nakanishi, H., Yamamoto, T., Hasegawa, J., Tokuda, E., Hikita, T., et al. (2022). 'A mass spectrometric method for in-depth profiling of phosphoinositide regioisomers and their disease-associated regulation. Nat. Commun. 13, 83. doi:10.1038/s41467-021-27648-z
Moriwaki, Y., Ohno, Y., Ishii, T., Takamura, Y., Kita, Y., Watabe, K., et al. (2018). 'SIMPLE binds specifically to PI4P through SIMPLE-like domain and participates in protein trafficking in the trans-Golgi network and/or recycling endosomes. PLoS One 13, e0199829. doi:10.1371/journal.pone.0199829
Morrow, A. A., Bridges, D., Yao, Z., Saltiel, A. R., and Lee, J. M. (2014). The lipid kinase PI4KIIIβ is highly expressed in breast tumors and activates akt in cooperation with Rab11a. Mol. Cancer Res. 12, 1492–1508. doi:10.1158/1541-7786.MCR-13-0604
New, J., Arnold, L., Ananth, M., Alvi, S., Thornton, M., Werner, L., et al. (2017). 'Secretory autophagy in cancer-associated fibroblasts promotes head and neck cancer progression and offers a novel therapeutic target. Cancer Res. 77, 6679–6691. doi:10.1158/0008-5472.CAN-17-1077
Noman, M. Z., Parpal, S., Van Moer, K., Xiao, M., Yu, Y., Viklund, J., et al. (2020). Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 6, eaax7881. doi:10.1126/sciadv.aax7881
Overduin, M., and Kervin, T. A. (2021). 'The phosphoinositide code is read by a plethora of protein domains. Expert Rev. Proteomics 18, 483–502. doi:10.1080/14789450.2021.1962302
Park, Y., Park, J. M., Kim, D. H., Kwon, J., and Kim, I. A. (2017). Inhibition of PI4K IIIα radiosensitizes in human tumor xenograft and immune-competent syngeneic murine tumor model. Oncotarget 8, 110392–110405. doi:10.18632/oncotarget.22778
Parkhitko, A. A., Singh, A., Hsieh, S., Hu, Y., Binari, R., Lord, C. J., et al. (2021). Cross-species identification of PIP5K1-splicing- and ubiquitin-related pathways as potential targets for RB1-deficient cells. PLoS Genet. 17, e1009354. doi:10.1371/journal.pgen.1009354
Patki, V., Lawe, D. C., Corvera, S., Virbasius, J. V., and Chawla, A. (1998). 'A functional PtdIns(3)P-binding motif. Nature 394, 433–434. doi:10.1038/28771
Peng, W., Huang, W., Ge, X., Xue, L., Zhao, W., and Xue, J. (2019). 'Type Iγ phosphatidylinositol phosphate kinase promotes tumor growth by facilitating Warburg effect in colorectal cancer. EBioMedicine 44, 375–386. doi:10.1016/j.ebiom.2019.05.015
Poli, A., Abdul-Hamid, S., Zaurito, A. E., Campagnoli, F., Bevilacqua, V., Sheth, B., et al. (2021). 'PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity'. Proc. Natl. Acad. Sci. U. S. A. 118. doi:10.1073/pnas.2010053118
Posor, Y., Jang, W., and Haucke, V. (2022). 'Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell. Biol. 23, 797–816. doi:10.1038/s41580-022-00490-x
Qiao, Y., Choi, J. E., Tien, J. C., Simko, S. A., Rajendiran, T., Vo, J. N., et al. (2021). Autophagy inhibition by targeting PIKfyve potentiates response to immune checkpoint blockade in prostate cancer. Nat. Cancer 2, 978–993. doi:10.1038/s43018-021-00237-1
Quinn, K. V., Behe, P., and Tinker, A. (2008). 'Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 586, 2855–2871. doi:10.1113/jphysiol.2008.153791
Ravi, A., Palamiuc, L., Loughran, R. M., Triscott, J., Arora, G. K., Kumar, A., et al. (2021). 'PI5P4Ks drive metabolic homeostasis through peroxisome-mitochondria interplay. Dev. Cell. 56, 1661–1676.e10. doi:10.1016/j.devcel.2021.04.019
Raychaudhuri, S., Remmers, E. F., Lee, A. T., Hackett, R., Guiducci, C., Burtt, N. P., et al. (2008). 'Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 40, 1216–1223. doi:10.1038/ng.233
Ronan, B., Flamand, O., Vescovi, L., Dureuil, C., Durand, L., Fassy, F., et al. (2014). 'A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019. doi:10.1038/nchembio.1681
Rooney, T. P. C., Aldred, G. G., Boffey, H. K., Willems, H. M. G., Edwards, S., Chawner, S. J., et al. (2023). The identification of potent, selective, and brain penetrant PI5P4Kγ inhibitors as in vivo-Ready tool molecules. J. Med. Chem. 66, 804–821. doi:10.1021/acs.jmedchem.2c01693
Roux, A., Winnard, P. T., Van Voss, M. H., Muller, L., Jackson, S. N., Hoffer, B., et al. (2023). 'MALDI-MSI of lipids in a model of breast cancer brain metastasis provides a surrogate measure of ischemia/hypoxia. Mol. Cell. Biochem. doi:10.1007/s11010-023-04685-4
Roux, K. J., Kim, D. I., Raida, M., and Burke, B. (2012). 'A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell. Biol. 196, 801–810. doi:10.1083/jcb.201112098
Roy, A., Chakraborty, A. R., Nomanbhoy, T., and DePamphilis, M. L. (2023). 'PIP5K1C phosphoinositide kinase deficiency distinguishes PIKFYVE-dependent cancer cells from non-malignant cells. Autophagy 19, 2464–2484. doi:10.1080/15548627.2023.2182594
Saliba, A. E., Vonkova, I., and Gavin, A. C. (2015). 'The systematic analysis of protein-lipid interactions comes of age. Nat. Rev. Mol. Cell. Biol. 16, 753–761. doi:10.1038/nrm4080
Sarwar, M., Aleskandarany, M., Karlsson, R., Althobiti, M., Ødum, N., Mongan, N. P., et al. (2019). The role of PIP5K1α/pAKT and targeted inhibition of growth of subtypes of breast cancer using PIP5K1α inhibitor. Oncogene 38, 375–389. doi:10.1038/s41388-018-0438-2
Sarwar, M., Semenas, J., Miftakhova, R., Simoulis, A., Robinson, B., Gjörloff Wingren, A., et al. (2016). Targeted suppression of AR-V7 using PIP5K1α inhibitor overcomes enzalutamide resistance in prostate cancer cells. Oncotarget 7, 63065–63081. doi:10.18632/oncotarget.11757
Sato, M., Ueda, Y., Takagi, T., and Umezawa, Y. (2003). 'Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell. Biol. 5, 1016–1022. doi:10.1038/ncb1054
Schlütermann, D., Skowron, M. A., Berleth, N., Böhler, P., Deitersen, J., Stuhldreier, F., et al. (2018). 'Targeting urothelial carcinoma cells by combining cisplatin with a specific inhibitor of the autophagy-inducing class III PtdIns3K complex. Urol. Oncol. 36, 160.e1–160. doi:10.1016/j.urolonc.2017.11.021
Semenas, J., Hedblom, A., Miftakhova, R. R., Sarwar, M., Larsson, R., Shcherbina, L., et al. (2014). The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 111, E3689–E3698. doi:10.1073/pnas.1405801111
Semenas, J., Wang, T., Sajid Syed Khaja, A., Firoj Mahmud, A., Simoulis, A., Grundström, T., et al. (2021). 'Targeted inhibition of ERα signaling and PIP5K1α/Akt pathways in castration-resistant prostate cancer. Mol. Oncol. 15, 968–986. doi:10.1002/1878-0261.12873
Shah, Z. H., Jones, D. R., Sommer, L., Foulger, R., Bultsma, Y., D'Santos, C., et al. (2013). 'Nuclear phosphoinositides and their impact on nuclear functions. FEBS J. 280, 6295–6310. doi:10.1111/febs.12543
Sharma, A., Sun, J., Singaram, I., Ralko, A., Lee, D., and Cho, W. (2020). 'Photostable and orthogonal solvatochromic fluorophores for simultaneous in situ quantification of multiple cellular signaling molecules. ACS Chem. Biol. 15, 1913–1920. doi:10.1021/acschembio.0c00241
Sharma, G., Guardia, C. M., Roy, A., Vassilev, A., Saric, A., Griner, L. N., et al. (2019). 'A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy 15, 1694–1718. doi:10.1080/15548627.2019.1586257
Shim, H., Wu, C., Ramsamooj, S., Bosch, K. N., Chen, Z., Emerling, B. M., et al. (2016). 'Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl. Acad. Sci. U. S. A. 113, 7596–7601. doi:10.1073/pnas.1600934113
Shin, Y. J., Sa, J. K., Lee, Y., Kim, D., Chang, N., Cho, H. J., et al. (2019). PIP4K2A as a negative regulator of PI3K in PTEN-deficient glioblastoma. J. Exp. Med. 216, 1120–1134. doi:10.1084/jem.20172170
Singaram, I., Sharma, A., Pant, S., Lihan, M., Park, M. J., Pergande, M., et al. (2023). 'Targeting lipid-protein interaction to treat Syk-mediated acute myeloid leukemia. Nat. Chem. Biol. 19, 239–250. doi:10.1038/s41589-022-01150-z
Singh, N., Reyes-Ordonez, A., Compagnone, M. A., Moreno, J. F., Leslie, B. J., Ha, T., et al. (2021). 'Redefining the specificity of phosphoinositide-binding by human PH domain-containing proteins. Nat. Commun. 12, 4339. doi:10.1038/s41467-021-24639-y
Sivakumaren, S. C., Shim, H., Zhang, T., Ferguson, F. M., Lundquist, M. R., Browne, C. M., et al. (2020). Targeting the PI5P4K lipid kinase family in cancer using covalent inhibitors. Cell. Chem. Biol. 27, 525–537. doi:10.1016/j.chembiol.2020.02.003
Sumita, K., Lo, Y. H., Takeuchi, K., Senda, M., Kofuji, S., Ikeda, Y., et al. (2016). The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis. Mol. Cell. 61, 187–198. doi:10.1016/j.molcel.2015.12.011
Sun, J., Singaram, I., Soflaee, M. H., and Cho, W. (2020). 'A direct fluorometric activity assay for lipid kinases and phosphatases. J. Lipid Res. 61, 945–952. doi:10.1194/jlr.D120000794
Tan, X., Banerjee, P., Pham, E. A., Rutaganira, F. U. N., Basu, K., Bota-Rabassedas, N., et al. (2020). 'PI4KIIIβ is a therapeutic target in chromosome 1q-amplified lung adenocarcinoma. Sci. Transl. Med. 12, eaax3772. doi:10.1126/scitranslmed.aax3772
Teng, M., Jiang, J., Wang, E. S., Geng, Q., Toenjes, S. T., Donovan, K. A., et al. (2023). 'Targeting the dark lipid kinase PIP4K2C with a potent and selective binder and degrader. Angew. Chem. Int. Ed. Engl. 62, e202302364. doi:10.1002/anie.202302364
Triscott, J., Reist, M., Kung, L., Moselle, F. C., Lehner, M., Gallon, J., et al. (2023). PI5P4Kα supports prostate cancer metabolism and exposes a survival vulnerability during androgen receptor inhibition. Sci. Adv. 9, eade8641. doi:10.1126/sciadv.ade8641
van der Wal, J., Habets, R., Varnai, P., Balla, T., and Jalink, K. (2001). 'Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J. Biol. Chem. 276, 15337–15344. doi:10.1074/jbc.M007194200
Varnai, P., and Balla, T. (1998). 'Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell. Biol. 143, 501–510. doi:10.1083/jcb.143.2.501
Verykiou, S., Alexander, M., Edwards, N., Plummer, R., Chaudhry, B., Lovat, P. E., et al. (2019). 'Harnessing autophagy to overcome mitogen-activated protein kinase kinase inhibitor-induced resistance in metastatic melanoma. Br. J. Dermatol 180, 346–356. doi:10.1111/bjd.17333
Vicinanza, M., Korolchuk, V. I., Ashkenazi, A., Puri, C., Menzies, F. M., Clarke, J. H., et al. (2015). 'PI(5)P regulates autophagosome biogenesis. Mol. Cell. 57, 219–234. doi:10.1016/j.molcel.2014.12.007
Voss, M. D., Czechtizky, W., Li, Z., Rudolph, C., Petry, S., Brummerhop, H., et al. (2014). Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem. Biophys. Res. Commun. 449, 327–331. doi:10.1016/j.bbrc.2014.05.024
Walaa, E. K., Chen, W., Ma, X., Hung, L. T., Hoeven Dharini van der, , Ransome van der, H., et al. (2019). 'Targeting plasma membrane phosphatidylserine content to inhibit oncogenic KRAS function. Life Sci. Alliance 2, e201900431. doi:10.26508/lsa.201900431
Wang, D. G., Paddock, M. N., Lundquist, M. R., Sun, J. Y., Mashadova, O., Amadiume, S., et al. (2019). 'PIP4Ks suppress insulin signaling through a catalytic-independent mechanism. Cell. Rep. 27, 1991–2001. doi:10.1016/j.celrep.2019.04.070
Wang, M., Bond, N. J., Letcher, A. J., Richardson, J. P., Lilley, K. S., Irvine, R. F., et al. (2010). 'Genomic tagging reveals a random association of endogenous PtdIns5P 4-kinases IIalpha and IIbeta and a partial nuclear localization of the IIalpha isoform. Biochem. J. 430, 215–221. doi:10.1042/BJ20100340
Wang, T., Sarwar, M., Whitchurch, J. B., Collins, H. M., Green, T., Semenas, J., et al. (2022). 'PIP5K1α is required for promoting tumor progression in castration-resistant prostate cancer. Front. Cell. Dev. Biol. 10, 798590. doi:10.3389/fcell.2022.798590
Waring, M. J., Andrews, D. M., Faulder, P. F., Flemington, V., McKelvie, J. C., Maman, S., et al. (2014). Potent, selective small molecule inhibitors of type III phosphatidylinositol-4-kinase α- but not β-inhibit the phosphatidylinositol signaling cascade and cancer cell proliferation. Chem. Commun. (Camb) 50, 5388–5390. doi:10.1039/c3cc48391f
Willems, H. M. G., Edwards, S., Boffey, H. K., Chawner, S. J., Green, C., Romero, T., et al. (2023). Identification of ARUK2002821 as an isoform-selective PI5P4Kα inhibitor. RSC Med. Chem. 14, 934–946. doi:10.1039/d3md00039g
Wills, R. C., Doyle, C. P., Zewe, J. P., Pacheco, J., Hansen, S. D., and Hammond, G. R. V. (2023). 'A novel homeostatic mechanism tunes PI(4,5)P2-dependent signaling at the plasma membrane. J. Cell. Sci. 136, jcs261494. doi:10.1242/jcs.261494
Wills, R. C., Goulden, B. D., and Hammond, G. R. V. (2018). 'Genetically encoded lipid biosensors. Mol. Biol. Cell. 29, 1526–1532. doi:10.1091/mbc.E17-12-0738
Wortmann, L., Brauer, N., Holton, S. J., Irlbacher, H., Weiske, J., Lechner, C., et al. (2021). 'Discovery and characterization of the potent and highly selective 1,7-naphthyridine-based inhibitors BAY-091 and BAY-297 of the kinase PIP4K2A. J. Med. Chem. 64, 15883–15911. doi:10.1021/acs.jmedchem.1c01245
Wu, Q., Zhou, D., Shen, Z., Chen, B., Wang, G., Wu, L., et al. (2023). 'VPS34-IN1 induces apoptosis of ER+ breast cancer cells via activating PERK/ATF4/CHOP pathway. Biochem. Pharmacol. 214, 115634. doi:10.1016/j.bcp.2023.115634
Young, C. D., Arteaga, C. L., and Cook, R. S. (2015). 'Dual inhibition of Type I and Type III PI3 kinases increases tumor cell apoptosis in HER2+ breast cancers. Breast Cancer Res. 17, 148. doi:10.1186/s13058-015-0656-2
Zahedi, S., Fitzwalter, B. E., Morin, A., Grob, S., Desmarais, M., Nellan, A., et al. (2019). 'Effect of early-stage autophagy inhibition in BRAF(V600E) autophagy-dependent brain tumor cells. Cell. Death Dis. 10, 679. doi:10.1038/s41419-019-1880-y
Keywords: phosphoinositide, lipid, metabolism, cancer, signaling, PI5P4K, lipidomics, mass spectometry
Citation: Llorente A, Loughran RM and Emerling BM (2023) Targeting phosphoinositide signaling in cancer: relevant techniques to study lipids and novel avenues for therapeutic intervention. Front. Cell Dev. Biol. 11:1297355. doi: 10.3389/fcell.2023.1297355
Received: 19 September 2023; Accepted: 12 October 2023;
Published: 25 October 2023.
Edited by:
Banafshe Larijani, University of Bath, United KingdomReviewed by:
Wenping Zhou, Yale University, United StatesCopyright © 2023 Llorente, Loughran and Emerling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brooke M. Emerling, YmVtZXJsaW5nQHNicGRpc2NvdmVyeS5vcmc=
†These authors have contributed equally to this work
 Alicia Llorente
Alicia Llorente Ryan M. Loughran
Ryan M. Loughran Brooke M. Emerling
Brooke M. Emerling