
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 15 December 2023
Sec. Cancer Cell Biology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1295555
This article is part of the Research TopicCRISPR Screening in Metabolic Stress and CancerView all 4 articles
Lung cancer is an extremely aggressive and highly prevalent disease worldwide, and it is one of the leading causes of cancer death. Deciphering intrinsic genetic mechanism, finding new targets, and overcoming drug resistance are the key to lung cancer treatment. High-throughput CRISPR screening has been extensively used to obtain the genes related to cancers including lung cancer. This review describes CRISPR/Cas9 or CRISPR/dCas9-based technologies for high-throughput screening. We summarize the applications of CRISPR screening technology in exploring the mechanism of lung cancer development in vivo or in vitro, overcoming drug resistance, improving the effect of immunotherapy, and discovering new therapeutic targets. This review highlights the potential of CRISPR screening in combination with tumor barcoding and high-throughput sequencing (Tuba-seq) to precisely quantify the impact of alterations in many tumor suppressor genes on lung cancer.
Lung cancer, an extremely aggressive and highly prevalent disease worldwide, is the most frequently occurring cancer and the first leading cause of cancer death in men, and the second leading cause of cancer death in women next to breast cancer (Sung et al., 2021). Lung cancer can be divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), of which NSCLC accounts for the vast majority of all cases, and NSCLC can be divided into adenocarcinoma (40%), squamous cell carcinoma (25%), and others (Leiter et al., 2023). Traditional therapies such as surgery, chemotherapy, and radiotherapy are the first choice, but drug resistance and systemic toxicity cannot be avoided in chemotherapy. Emerging therapies, such as targeted therapy and immunotherapy, have relatively low toxicity, but their application is limited, and they are also prone to induce drug resistance (Zheng X. et al., 2023). Therefore, overcoming drug resistance or finding new targets is the key to the treatment of lung cancer.
Prokaryotic CRISPR (clustered regularly interspaced short palindromic repeats)/Cas adaptive immune system can facilitate RNA-guided site-specific DNA cleavage. Under the guidance of short RNAs, Cas9 nuclease can also induce precise cleavage of endogenous genomic loci in human and mouse cells (Cong et al., 2013). The CRISPR/Cas system is not only useful for single-gene perturbations, but can also be used for high-throughput screening such as pooled screens, which has been widely used to obtain the genes related to cancer (Zhou et al., 2023). Unlike other gene editing tools, CRISPR-Cas9 has the multiple advantages such as wider editing site, lower off-target effects, and lower cost (Zheng R. et al., 2023).
This review introduces CRISPR/Cas9 and CRISPR/dCas9-based technologies including CRISPR knockout (CRISPRko), CRISPR interference (CRISPRi), and CRISPR activation (CRISPRa) for high-throughput screening and summarizes their application in revealing the mechanisms of lung cancer development in vivo or in vitro, overcoming the drug resistance, improving the effect of immunotherapy, and discovering more new therapeutic targets.
Traditionally, RNA interference (RNAi) technology is employed for genome-wide screening in mammalian cells. Compared to CRISPR screening, RNAi exhibits some limitations such as single function and off-target effects (Shalem et al., 2014). Currently, there are three CRISPR-based genome perturbation technologies (CRISPRko, CRISPRi, and CRISPRa) for screening (Figure 1). By designing sgRNAs and modifying Cas proteins, CRISPR technology enables flexible gene editing and screening (Shalem et al., 2014; Przybyla and Gilbert, 2022; Wang and Doudna, 2023).
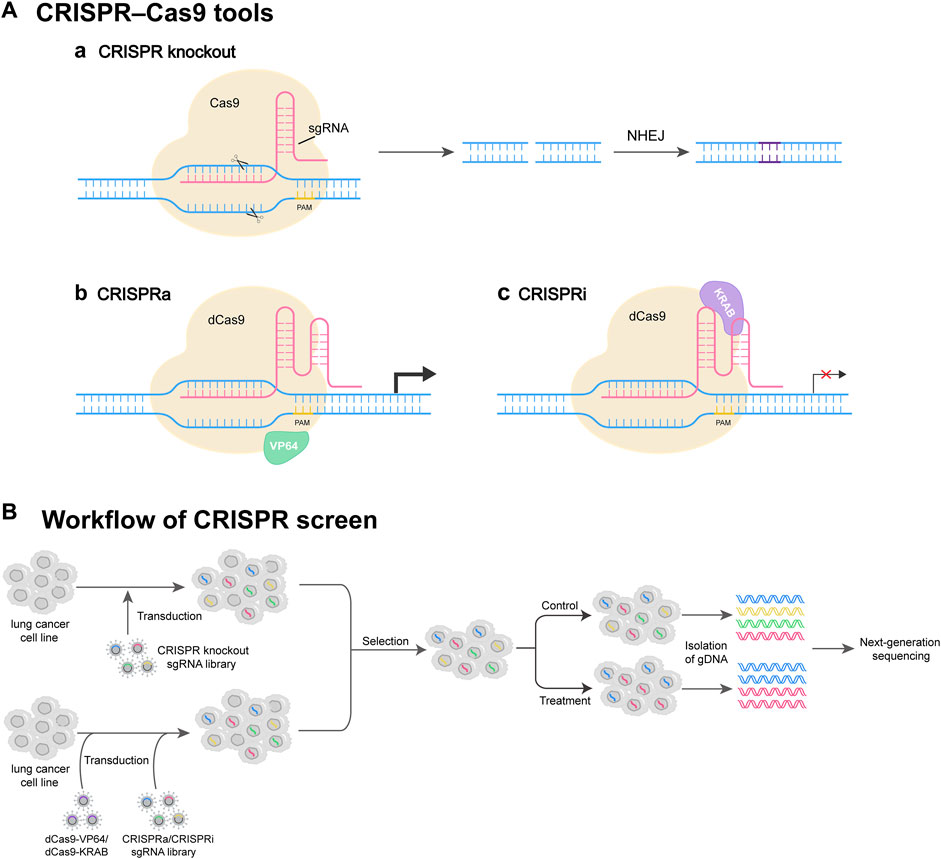
FIGURE 1. CRISPR screen tools and workflow. (A) Commonly used CRISPR perturbation methods. (Aa) CRISPR-Cas9 system consists of sgRNA and Cas9. sgRNA targets the gene of interest, and Cas9 induces a DNA double-strand break (DSB) at the target site. The DSB can be repaired by NHEJ, resulting in gene knockout. (Ab) CRISPRa system activates target gene expression through dCas9 fused with transcriptional activator VP64. (Ac) CRISPRi system represses target gene expression through dCas9 fused with transcription repressor protein KRAB. (B) Workflow of CRISPR screen. The lung cancer cells are transduced by a lentivirus-based CRISPR knockout sgRNA library, or by dCas9-VP64/dCas9-KRAB virus and CRISPRa/CRISPRi sgRNA library. Then, the cells successfully infected are selected and further propagated under control or treatment conditions. At appropriate times, gDNAs of cells are isolated, followed by next-generation DNA sequencing (NGS).
CRISPR RNA (crRNA) can hybridize with a small transactivating CRISPR RNA (tracrRNA), which is further recognized and bound by Cas9 to generate a ribonucleoprotein (RNP) complex. The RNP complex can bind to the genome to recognize the sequences matching the spacer region on the crRNA (Chavez et al., 2023). If there is a homologous sequence, Cas9 acts as a nuclease to cleave approximately 3 bp bases upstream the protospacer adjacent motif (PAM), causing a double-stranded DNA break (Hille et al., 2018; Liu et al., 2023). To simplify the above system, crRNA and tracrRNA are fused into a single guide RNA (sgRNA), which guides Cas9 to a specific gene locus to generate a double-stranded DNA break, highly possibly resulting in a error-prone repair via nonhomologous end joining (NHEJ) (Chavez et al., 2023; Zhou et al., 2023). NHEJ leads to insertions or deletions of bases at the cleavage site, thus achieving loss-of-function (LOF) mutations eventually resulting in disruption of the target gene (Shalem et al., 2015; Zhou et al., 2023).
Cas9 system has found various applications such as genome editing and large-scale screen (Chong et al., 2020). Ophir et al. (Shalem et al., 2014). First performed a genome-wide CRISPR/cas9 screen in human cells, confirming the feasibility of this technique. Genome-wide CRISPR screen in CD8 T cell-transferred mice in cancer immunotherapy setting identifies the regulators of tumor infiltration and degranulation and robustly reidentifies the regulators of canonical immune responses such as PD-1 and Tim-3. As a therapeutic target, a new gene Dhx37 was discovered in T cells (Dong et al., 2019). Not only can the whole genome library be used for CRISPR screening, but also a specific sub-library can be constructed for screening according to the scientific questions to be studied. To screen RNA-binding proteins (RBPs) critical for macrophage activation, a lentiviral mini-library targeting 782 genes encoding known canonical RBPs in the mouse genome was constructed, and METTL9 was identified as a positive regulator of macrophage innate responses via CRISPR-Cas9 screen (Tong et al., 2021).
The dCas9, a nuclease-dead version of the protein, is generated from the removal of the catalytic activity of Cas9, and dCas9 can be used as a platform for transcriptional regulation (Chavez et al., 2023). The dCas9 can change transcription of any gene without genetically altering the target sequence (Qi et al., 2013). The dCas9 is bound with sequence-specific sgRNA to form the dCas9-sgRNA complex, and this complex can interfere with transcription elongation and hinder transcription initiation by disrupting transcription factor binding (Dominguez et al., 2016). The transcription repression function of dCas9 can be enhanced by linking the transcriptional repression domain Krüppel-associated box (KRAB) to dCas9 (Bendixen et al., 2023; Chavez et al., 2023).
CRISPR interference (CRISPRi) can inhibit the expression of pseudogenes without directly interfering with the transcription of parental genes. To investigate pseudogene function, the researchers designed the first genome-wide CRISPRi single-guide RNA (sgRNA) library targeting human pseudogene promoter-proximal regions for CRISPRi screening in breast cancer cells, and CRISPRi screening identified many pseudogenes that affect the fitness of breast cancer cells (Sun et al., 2021). Induced indels in open reading frames can disrupt translation of functional proteins, but the function of most noncoding genes is not affected, and large genomic deletions may affect nearby or overlapping genes (Goyal et al., 2017). In one previous study, CRISPR interference (CRISPRi) screen was used to identify MYC-regulated lncRNAs based on a library targeting sequences surrounding the transcription start sites (TSS) of non-coding genes, and CRISPRi screen successfully identified 320 non-coding sites that are vital for cell growth (Raffeiner et al., 2020).
In the past, genome-wide acquisition screening was performed using cDNA overexpression library, but the complexity of transcription isoform variation, long cDNA sequence, and high cost limit its application, and thus it is necessary to seek new techniques for genome-scale gain-of-function perturbation (Konermann et al., 2015). CRISPRa is one promising therapeutic gene editing strategy (Zhang et al., 2021). By fusing four copies of the transcriptional activator VP16 or one copy of the p65 activation domain (AD) to dCas9, the obtained dCas9-VP64 and dCas9-p65AD can effectively activate reporter gene expression (Gilbert et al., 2013). Compared with VP64, the second-generation activators (VPR, SAM, and Suntag) showed higher activation levels, mainly through the recruitment of multiple activation domains to the dCas9 complex (Chavez et al., 2016; Joung et al., 2017).
In addition to being used for the activation of individual genes, CRISPR activation can also be used for high-throughput screening. To evaluate lncRNA function, a CRISPR-activated lncRNA (CaLR) strategy was developed, and a library targeting lncRNA genes was designed to perform CRISPRa screening, resulting in the identification of new cell cycle regulators, survival/apoptosis and cancer signaling genes (Bester et al., 2018). A genome-scale CRISPRa screen was used to identify candidate genes related to the drug-resistance phenotype from PC9 cell lines under osimertinib treatment, and the identified genes could be new targets (Müller et al., 2023).
In addition to using a CRISPR screening method alone, multiple methods are combined for cancer treatment research. To systematically understand the regulators that coordinate T cell activation with gene perturbations, regulators responsible for cytokine production were discovered using genome-wide pooled CRISPRa and CRISPRi screens (Schmidt et al., 2022).
In addition to traditional CRISPR-Cas9 mediated genome editing methods, two novel techniques, namely, base editing and prime editing, are gaining momentum in the field of genome editing. Base editing (BE) is a highly efficient and permanent technique for introducing single nucleotide variants (SNVs) into the DNA of living cells (Porto et al., 2020). BE does not require dependence on DSB, HDR, or a donor DNA template and it catalyzes the deamination of cytidine (C) or adenosine (A), which leads to the conversion of the bases to thymine (T) or guanine (G), respectively (Zhao et al., 2023). Base editor screening is a promising strategy for examining the functional effects of point mutations, which will contribute to genetic disease diagnosis and the identification of potential treatment strategies. Prime editing (PE) can create any kind of DNA substitutions, small insertions, and deletions at targeted sites within the genome of living cells, without relying on DSBs (Chen and Liu, 2023). PE consists of nCas9 (H840A) coupled to an engineered reverse transcriptase (RT) and prime editing guide RNA (pegRNA), which both matches the target site and encodes the required editing (Zhao et al., 2023). By sequencing integrated pegRNAs to identify genetic alterations, the prime editing system may also enable high-throughput pooled screens eventually (Chen and Liu, 2023).
The CRISPR/Cas system is simple and efficient, and can modify multiple genes simultaneously, making it suitable for high-throughput genetic screening (Wang and Doudna, 2023). The above CRISPR screening methods can be used to explore the mechanism of the occurrence and development of lung cancer, overcome the drug resistance, improve the effect of immunotherapy, and discover more new therapeutic targets, and others (Figure 2) (Table 1).
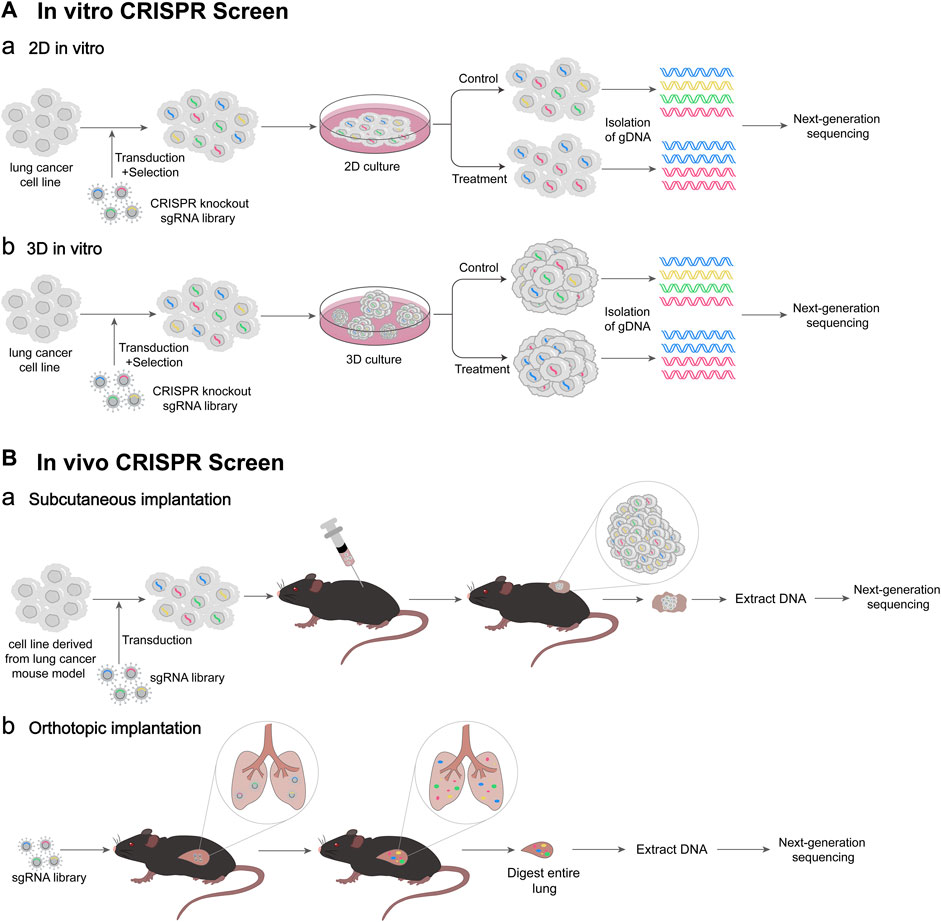
FIGURE 2. Schematic diagram of CRISPR screen. (A) Workflow of in vitro CRISPR screen. (Aa) 2D culture-based in vitro CRISPR screen. Lentivirus-based CRISPR knockout sgRNA library is transfected into lung cancer cells, and the cells successfully infected are selected. Then, 2D culture of cells is performed under control or treatment conditions. At appropriate times gDNAs of cells are isolated, followed by next-generation DNA sequencing (NGS). (Ab) 3D culture-based in vitro CRISPR screen. 3D culture of cells is performed under control or treatment condition. Then, gDNAs of 3D spheroids are isolated. (B) Workflow of in vivo CRISPR screen. (Ba) Cancer cell subcutaneous implantation. A lentivirus-based sgRNA library is transfected into lung cancer cells derived from lung cancer mouse model. The transfected cells are transplanted into mice subcutaneously. Tumor-derived DNA was extracted, followed by NGS. (Bb) Virus orthotopic inoculation. The lentivirus-based sgRNA library is delivered to mice lung intratracheally. After the entire mouse lung is digested, DNA is extracted, followed by NGS.
Since genetic changes induced by drug treatment allow cells to acquire resistance or susceptibility, it is possible to screen relevant targets through CRISPRa-mediated gain-of-function and CRISPRi- and CRISPRko-mediated loss-of-function (Bodapati et al., 2020). Resistance to EGFR inhibitors (EGFRi) is a significant barrier to NSCLC treatment. CRISPR-based functional genomic screen is a new method to discover possible resistance indicators (Gogleva et al., 2022). In PC9 cells treated with osimertinib, a new potential driver of EGFR inhibitor resistance, WWTR1, was discovered by a genome-wide CRISPRa, and WWTR1 knockdown markedly decreased the development of drug-resistant cells (Müller et al., 2023). A kinome-wide CRISPR/Cas9 screen was performed in FGFR1-amplified lung cancer cells treated with FGFR inhibitors, identifying PLK1 as a potent synthetic lethal target for this cancer type (Yang et al., 2021).
A genome-scale CRISPR/Cas9 deletion screen in DDP- and VP16-treated and untreated resistant SCLC cells results in the identification of the serine/threonine kinase cell division cycle 7 (CDC7) as a promising target for chemotherapeutic synergy. The combined use of CDC7 inhibitors and other drugs helps to overcome chemoresistance in the treatment of SCLC and improves patient prognosis (Deng et al., 2023). A human-genome-wide library was employed to screen the genes necessary for CHK1i sensitivity in lung cancer cells, and several MMB-FOXM1 complex components were discovered (Branigan et al., 2021). Many models are naturally resistant to MCL1 inhibitor. A whole-genome CRISPR screen based on flow cytometry identified three components of the cullin-RING ubiquitin ligase complex CRL5 (CUL5, RNF7 and UBE2F), all of which resensitized cells to MCL1 inhibition (Kabir et al., 2019).
Radiotherapy is the cornerstone of cancer treatment, but radioresistance has always been a major obstacle, and the response rate of advanced NSCLC to radiotherapy remains unsatisfactory (Kim et al., 2020). Poly-ADP-ribose polymerase (PARP) inhibitors have been shown to radiosensitize SCLC tumors, and radiotherapy in combination with PARP inhibitor may be a promising therapy to overcome radioresistance (Laird et al., 2018; Cheng et al., 2021). CRISPR has been used to screen genes affecting radioresistance in glioblastoma (GBM) (Zhu et al., 2021), nasopharyngeal carcinoma (NPC) (Shen et al., 2021), and pancreatic cancer (PC) (Lan et al., 2022), providing new solutions for cancer treatment.
CRISPR screens have also been successfully applied in lung cancer treatment to discover genes responsible for radioresistance. A genome-scale CRISPR screen determined plakophilin 2 (PKP2) as a key driver of radioresistance in lung cancer cell line A549 cells by comparing sgRNA abundance (Cheng et al., 2021).
A CRISPR/Cas9 screen was used in a mouse lung cancer model to assess the immunomodulatory potential of genes related to cytotoxicity alteration in human cancers. The results showed that the cancer testis antigen Adam2 was an immunomodulator and also an oncogene, and it could inhibit interferon and TNF cytokine signaling and reduce the presentation of tumor-associated antigens (Dervovic et al., 2023). Based on a mouse KP lung cancer model, an in vitro and in vivo CRISPR screening platform was constructed to thoroughly assess intracellular regulators of antitumor immunity. Tsc2 was identified, and its deletion improved the response of tumors to ICB treatment (Huang et al., 2022). A genome-wide CRISPR screen based on fluorescence-activated cell sorting (FACS) identified the regulators of PD-L1 gene in the human NSCLC cell line NCI-H358, and PD-L1 gene was found to be efficiently induced when the heme biosynthetic pathway was disrupted (Suresh et al., 2020). A study investigated the roles of epigenetic genes in anti-tumor immunity using an epigenetic-focused CRISPR screen in the LUAD model, and discovered anti-silencing function protein 1 homolog A (Asf1a) as a possible therapeutic target (Li et al., 2020).
CRISPR screen can be used to explore novel targets. In order to determine the impacts of loss of a single target on cell proliferation of LUAD, the researchers performed a CRISPR screening in eight LUAD cell lines. They found that CASP8AP2 was a crucial activity factor regulating tumor cell death (Myacheva et al., 2023). Two genome-scale CRISPR/Cas9 screenings were conducted in the wild-type TP53 and RTK-containing cell lines with BEAS-2B cells serving as a control. The results showed that MDM2 was a potential therapeutic target (Wang et al., 2022). A genome-scale screen was used to identify tumor suppressor genes through knockout by CRISPR/Cas9 system. ZNF24 was found to be a powerful tumor suppressor in mouse xenograft and autologous lung cancer models as well as lung cell lines (Liu et al., 2022).
A library targeting potential druggable genes was used to conduct genetic CRISPR screening in tumor cell lines derived from mouse models. The results showed that the de novo pyrimidine biosynthesis pathway was critical target of SCLC. In several in vivo SCLC models, pharmacological inhibition of DHODH, one of the enzymes in this system, reduced tumor growth (Li et al., 2019). One previous study identified protein neddylation through a genome-wide CRISPR/Cas9 screen in five SCLC-derived cell lines from genetic animal models and mouse embryonic fibroblasts (MEFs) (Norton et al., 2021).
CRISPR screen has also been used to explore the mechanism of lung cancer development. A genome-wide CRISPR/Cas9 screen identified Cul5 as a crucial gene regulating SCLC metastasis in a SCLC spontaneous metastatic mouse model. Deletion of Cul5 dramatically increased SCLC development by preventing integrin β1 degradation (Zhao et al., 2019). Additionally, CRISPR screens identified IGF1R and ERBB3 as key mediators of KEAP1-mutant cell growth (Vartanian et al., 2019). CRISPR-Cas9 is also used to screen potential tumor suppressors in an early-stage SCLC cell line. One of the top hits is MAX, which is mutated in human SCLC. MAX has been found to be an essential heterodimerization partner for MYC family proteins. Max deletion significantly speeds up SCLC progression by enhancing cell proliferation and transformation. In contrast, Max deletion stops tumor development in SCLC that overexpresses MYCL (Augert et al., 2020). A CRISPR screen reveals that histone demethylase UTX deficiency significantly promotes lung cancer progression (Wu et al., 2018).
Deletion of synthetic lethality target genes aggravates ROS-induced cell death in NRF2-hyperactivated NSCLC cells. CRISPR/Cas9 screens identified mitochondrial superoxide dismutase 2 (SOD2) as antioxidant enzymes genes in KEAP1-mutated NSCLC cell lines A549, and loss of SOD2 enhanced the efficacy of β-Lapafenone (Jiang et al., 2022). Domain-focused CRISPR screen is a method to identify key transcriptional regulators in cancer. A large-scale CRISPR interference (CRISPRi) screen in combination with epithelial-mesenchymal transition (EMT) and fate mapping by genetic tracing was employed to identify the genes involved in RTK signaling and epigenome regulation, resulting in the discovery of a large number of chromatin regulators and a limited number of kinome genes as regulators of EMT interconversion (Serresi et al., 2021).
Although many tumor suppressors have been identified by cancer genome sequencing, the strategies to test their in vivo functions in a quantitative manner are limited (Rogers et al., 2017). The tumor barcode and high-throughput sequencing enables high-resolution quantification of tumor growth in mouse models. Tumor barcode sequencing (Tuba-seq) and CRISPR technology when combined can utilize their unique strengths. The former provides precise measurement of tumor size whereas the latter modifies the expression of specific genes (Rogers et al., 2018). Two barcodes placed on the lentiviral vector, (one tracking TSG-targeting guide RNA (sgRNA), and the other tracking individual tumor), are used to measure tumor cell number and tumor size (Kim and Minna, 2018). The schematic diagram of Tuba-seq technology is shown in the figure (Figure 3).
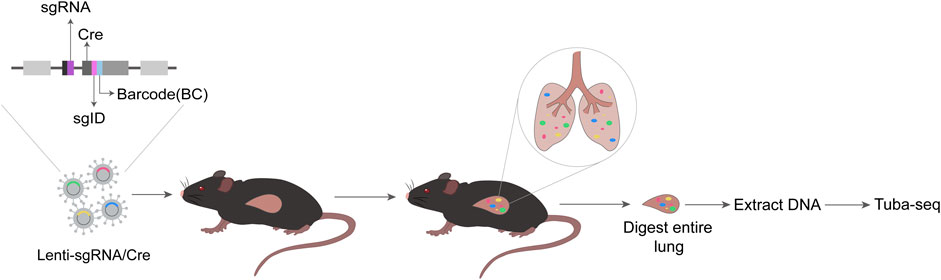
FIGURE 3. Schematic diagram of Tuba-seq. Each vector in the Lenti-sgRNA/Cre vector library contains a sgRNA, a Cre, and a two-component barcode sgID-BC consisting of an 8-nt ‘sgID’ corresponding to each sgRNA and a random 15-nt barcode (BC). The Lenti-sgRNA/Cre is delivered to mice intratracheally. At appropriate times, entire lung of mice is digested, and DNA is extracted. Then, Tuba-seq is performed, followed by data analysis.
On the one hand, the combination of Tuba-seq and CRISPR technology can more accurately characterize the effects of genetic alterations on tumor growth. Using CRISPR/Tuba-seq, a study of 48 TSGs characterizes the functional landscape of tumor suppression in an KRAS-driven lung cancer model, revealing novel genetics of KRAS-driven lung cancer initiation, overall and abnormal growth (Cai et al., 2021). A modified CRISPR/Tuba-seq method was used to quantify TSG function in EGFR-driven lung tumors and to identify key TSGs that limit the growth of EGFR-mutant tumors (Foggetti et al., 2021). Furthermore, multiple candidate KRAS-interacting proteins are studied for their effects on lung cancer growth in a genetically engineered mouse model using multiplex somatic CRISPR/Cas9 screenig combined with Tuba-seq. The results indicate that NRAS and HRAS act as tumor suppressor genes (Tang et al., 2023). Tuba-seq is suitable for the investigation of lentiviral transduction and condition-dependent regulation of oncogenic programs in vivo models, but it is not applicable to study the mechanisms underlying the natural development of SCLC. Considering this, a Lenti-Cre-based barcoded SCLC mouse model was developed to track tumors and investigate candidate regulators controlling tumor initiation and growth in their natural environment (Lee et al., 2023).
On the other hand, CRISPR/Tuba-seq allows for the analysis of how various genetic modifications affect therapeutic response, finding the therapeutic vulnerabilities, ultimately aiding in clinical drug guidance. The use of CRISPR/Tuba-seq indicated that KEAP1 inactivation considerably diminished the sensitivity of EGFR-driven lung adenocarcinoma to the EGFR inhibitor osimertinib in a TSGs pool (Foggetti et al., 2021). This study showed KEAP1 inactivation as a prominent resistance mechanism of osimertinib in lung adenocarcinoma. Another research employing CRISPR/Tuba-seq mapped a landscape of genotype-specific treatment responses and established over 20% of potential relationships between genotypes and treatment responses showing resistance or sensitivity (Li et al., 2021). Furthermore, CRISPR/Tuba-seq could also be used to find therapeutic vulnerabilities in lung cancer patients with no targetable proto-oncogenic mutations. Recent research through CRISPR/Tuba-seq have demonstrated that the inactivation of RAS and PI3K suppressors drives the development of lung tumor without oncogenic mutations and uncovered their therapeutic vulnerabilities (Yousefi et al., 2022). CRISPR/Tuba-seq contributes to the development of precision therapy by propelling the analysis of tumor genotypes and potential target identifications.
Loss-of-function CRISPR screens (CRISPRko and CRISPRi) and gain-of-function CRISPR screens (CRISPRa) are powerful tools that have facilitated comprehensive investigations of the genomic landscape of lung cancer. These approaches can be used to decipher the mechanism underlying the occurrence and development of lung cancer, overcome the cancer treatment resistance, improve the effect of immunotherapy, and discover more new potential therapeutic targets.
However, there are certain limitations with these approaches, such as off-target effects (Goyal et al., 2017), continuous technology innovation are expected to reduce the likelihood of off-target issues. Additionally, each CRISPR screen method has its own disadvantages. For instance, CRISPRko screen may not effectively identify genes critical for cell viability or those with functional redundancy (Chong et al., 2020). Hence, multiplexed screens are suggested to enhance the reliability and validity of research work.
In summary, the CRISPR/Cas9 system provides a potent approach for high-throughput screening. However, the limitations of this system should also be considered in order to obtain reliable results. To avoid false positives of CRISPR-based experiments, orthogonal techniques can be used for validation. In the future, CRISPR screening will play a prominent role in lung cancer research.
WS: Writing–original draft, Writing–review and editing. FH: Methodology, Visualization, Writing–review and editing. PL: Funding acquisition, Methodology, Resources, Supervision, Writing–review and editing. YT: Funding acquisition, Methodology, Resources, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PL is supported by the Science and Technology Research Project of Hubei Education Department (No. B2019110); YT is supported by the Hubei Province medical leading talents training project (No. 201947).
We are grateful to members of the Yao Lab for the discussion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Augert, A., Mathsyaraja, H., Ibrahim, A. H., Freie, B., Geuenich, M. J., Cheng, P. F., et al. (2020). MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38 (1), 97–114. doi:10.1016/j.ccell.2020.04.016
Bendixen, L., Jensen, T. I., and Bak, R. O. (2023). CRISPR-Cas-mediated transcriptional modulation: the therapeutic promises of CRISPRa and CRISPRi. Mol. Ther. 31 (7), 1920–1937. doi:10.1016/j.ymthe.2023.03.024
Bester, A. C., Lee, J. D., Chavez, A., Lee, Y. R., Nachmani, D., Vora, S., et al. (2018). An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 173 (3), 649–664. doi:10.1016/j.cell.2018.03.052
Bodapati, S., Daley, T. P., Lin, X., Zou, J., and Qi, L. S. (2020). A benchmark of algorithms for the analysis of pooled CRISPR screens. Genome Biol. 21 (1), 62. doi:10.1186/s13059-020-01972-x
Branigan, T. B., Kozono, D., Schade, A. E., Deraska, P., Rivas, H. G., Sambel, L., et al. (2021). MMB-FOXM1-driven premature mitosis is required for CHK1 inhibitor sensitivity. Cell Rep. 34 (9), 108808. doi:10.1016/j.celrep.2021.108808
Cai, H., Chew, S. K., Li, C., Tsai, M. K., Andrejka, L., Murray, C. W., et al. (2021). A functional taxonomy of tumor suppression in oncogenic KRAS-driven lung cancer. Cancer Discov. 11 (7), 1754–1773. doi:10.1158/2159-8290.Cd-20-1325
Chavez, A., Tuttle, M., Pruitt, B. W., Ewen-Campen, B., Chari, R., Ter-Ovanesyan, D., et al. (2016). Comparison of Cas9 activators in multiple species. Nat. Methods 13 (7), 563–567. doi:10.1038/nmeth.3871
Chavez, M., Chen, X., Finn, P. B., and Qi, L. S. (2023). Advances in CRISPR therapeutics. Nat. Rev. Nephrol. 19 (1), 9–22. doi:10.1038/s41581-022-00636-2
Chen, P. J., and Liu, D. R. (2023). Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 24 (3), 161–177. doi:10.1038/s41576-022-00541-1
Cheng, C., Pei, X., Li, S. W., Yang, J., Li, C., Tang, J., et al. (2021). CRISPR/Cas9 library screening uncovered methylated PKP2 as a critical driver of lung cancer radioresistance by stabilizing β-catenin. Oncogene 40 (16), 2842–2857. doi:10.1038/s41388-021-01692-x
Chong, Z. S., Wright, G. J., and Sharma, S. (2020). Investigating cellular recognition using CRISPR/Cas9 genetic screening. Trends Cell Biol. 30 (8), 619–627. doi:10.1016/j.tcb.2020.05.005
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi:10.1126/science.1231143
Deng, L., Yang, L., Zhu, S., Li, M., Wang, Y., Cao, X., et al. (2023). Identifying CDC7 as a synergistic target of chemotherapy in resistant small-cell lung cancer via CRISPR/Cas9 screening. Cell Death Discov. 9 (1), 40. doi:10.1038/s41420-023-01315-2
Dervovic, D., Malik, A. A., Chen, E. L. Y., Narimatsu, M., Adler, N., Afiuni-Zadeh, S., et al. (2023). In vivo CRISPR screens reveal Serpinb9 and Adam2 as regulators of immune therapy response in lung cancer. Nat. Commun. 14 (1), 3150. doi:10.1038/s41467-023-38841-7
Dominguez, A. A., Lim, W. A., and Qi, L. S. (2016). Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 17 (1), 5–15. doi:10.1038/nrm.2015.2
Dong, M. B., Wang, G., Chow, R. D., Ye, L., Zhu, L., Dai, X., et al. (2019). Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178 (5), 1189–1204. doi:10.1016/j.cell.2019.07.044
Foggetti, G., Li, C., Cai, H., Hellyer, J. A., Lin, W. Y., Ayeni, D., et al. (2021). Genetic determinants of EGFR-driven lung cancer growth and therapeutic response in vivo. Cancer Discov. 11 (7), 1736–1753. doi:10.1158/2159-8290.Cd-20-1385
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 (2), 442–451. doi:10.1016/j.cell.2013.06.044
Gogleva, A., Polychronopoulos, D., Pfeifer, M., Poroshin, V., Ughetto, M., Martin, M. J., et al. (2022). Knowledge graph-based recommendation framework identifies drivers of resistance in EGFR mutant non-small cell lung cancer. Nat. Commun. 13 (1), 1667. doi:10.1038/s41467-022-29292-7
Goyal, A., Myacheva, K., Groß, M., Klingenberg, M., Duran Arqué, B., and Diederichs, S. (2017). Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 45 (3), e12. doi:10.1093/nar/gkw883
Hille, F., Richter, H., Wong, S. P., Bratovič, M., Ressel, S., and Charpentier, E. (2018). The Biology of CRISPR-cas: backward and forward. Cell 172 (6), 1239–1259. doi:10.1016/j.cell.2017.11.032
Huang, Q., Li, F., Hu, H., Fang, Z., Gao, Z., Xia, G., et al. (2022). Loss of TSC1/TSC2 sensitizes immune checkpoint blockade in non-small cell lung cancer. Sci. Adv. 8 (5), eabi9533. doi:10.1126/sciadv.abi9533
Jiang, C., Ward, N. P., Prieto-Farigua, N., Kang, Y. P., Thalakola, A., Teng, M., et al. (2022). A CRISPR screen identifies redox vulnerabilities for KEAP1/NRF2 mutant non-small cell lung cancer. Redox Biol. 54, 102358. doi:10.1016/j.redox.2022.102358
Joung, J., Konermann, S., Gootenberg, J. S., Abudayyeh, O. O., Platt, R. J., Brigham, M. D., et al. (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12 (4), 828–863. doi:10.1038/nprot.2017.016
Kabir, S., Cidado, J., Andersen, C., Dick, C., Lin, P. C., Mitros, T., et al. (2019). The CUL5 ubiquitin ligase complex mediates resistance to CDK9 and MCL1 inhibitors in lung cancer cells. Elife 8, e44288. doi:10.7554/eLife.44288
Kim, B. H., Kim, Y. J., Kim, M. H., Na, Y. R., Jung, D., Seok, S. H., et al. (2020). Identification of FES as a novel radiosensitizing target in human cancers. Clin. Cancer Res. 26 (1), 265–273. doi:10.1158/1078-0432.Ccr-19-0610
Kim, J., and Minna, J. D. (2018). Evaluating tumor-suppressor gene combinations. Nat. Genet. 50 (4), 480–482. doi:10.1038/s41588-018-0095-y
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 (7536), 583–588. doi:10.1038/nature14136
Laird, J. H., Lok, B. H., Ma, J., Bell, A., de Stanchina, E., Poirier, J. T., et al. (2018). Talazoparib is a potent radiosensitizer in small cell lung cancer cell lines and xenografts. Clin. Cancer Res. 24 (20), 5143–5152. doi:10.1158/1078-0432.Ccr-18-0401
Lan, B., Zeng, S., Zhang, S., Ren, X., Xing, Y., Kutschick, I., et al. (2022). CRISPR-Cas9 screen identifies DYRK1A as a target for radiotherapy sensitization in pancreatic cancer. Cancers (Basel) 14 (2), 326. doi:10.3390/cancers14020326
Lee, M. C., Cai, H., Murray, C. W., Li, C., Shue, Y. T., Andrejka, L., et al. (2023). A multiplexed in vivo approach to identify driver genes in small cell lung cancer. Cell Rep. 42 (1), 111990. doi:10.1016/j.celrep.2023.111990
Leiter, A., Veluswamy, R. R., and Wisnivesky, J. P. (2023). The global burden of lung cancer: current status and future trends. Nat. Rev. Clin. Oncol. 20 (9), 624–639. doi:10.1038/s41571-023-00798-3
Li, C., Lin, W. Y., Rizvi, H., Cai, H., McFarland, C. D., Rogers, Z. N., et al. (2021). Quantitative in vivo analyses reveal a complex pharmacogenomic landscape in lung adenocarcinoma. Cancer Res. 81 (17), 4570–4580. doi:10.1158/0008-5472.Can-21-0716
Li, F., Huang, Q., Luster, T. A., Hu, H., Zhang, H., Ng, W. L., et al. (2020). In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in kras-mutant lung adenocarcinoma. Cancer Discov. 10 (2), 270–287. doi:10.1158/2159-8290.Cd-19-0780
Li, L., Ng, S. R., Colón, C. I., Drapkin, B. J., Hsu, P. P., Li, Z., et al. (2019). Identification of DHODH as a therapeutic target in small cell lung cancer. Sci. Transl. Med. 11 (517), eaaw7852. doi:10.1126/scitranslmed.aaw7852
Liu, L., Lei, Y., Chen, W., Zhou, Q., Zheng, Z., Zeng, G., et al. (2022). In vivo genome-wide CRISPR screening identifies ZNF24 as a negative NF-κB modulator in lung cancer. Cell Biosci. 12 (1), 193. doi:10.1186/s13578-022-00933-0
Liu, Z., Shi, M., Ren, Y., Xu, H., Weng, S., Ning, W., et al. (2023). Recent advances and applications of CRISPR-Cas9 in cancer immunotherapy. Mol. Cancer 22 (1), 35. doi:10.1186/s12943-023-01738-6
Müller, N., Lorenz, C., Ostendorp, J., Heisel, F. S., Friese, U. P., Cartolano, M., et al. (2023). Characterizing evolutionary dynamics reveals strategies to exhaust the spectrum of subclonal resistance in EGFR-mutant lung cancer. Cancer Res. 83 (15), 2471–2479. doi:10.1158/0008-5472.Can-22-2605
Myacheva, K., Walsh, A., Riester, M., Pelos, G., Carl, J., and Diederichs, S. (2023). CRISPRi screening identifies CASP8AP2 as an essential viability factor in lung cancer controlling tumor cell death via the AP-1 pathway. Cancer Lett. 552, 215958. doi:10.1016/j.canlet.2022.215958
Norton, J. P., Augert, A., Eastwood, E., Basom, R., Rudin, C. M., and MacPherson, D. (2021). Protein neddylation as a therapeutic target in pulmonary and extrapulmonary small cell carcinomas. Genes Dev. 35 (11-12), 870–887. doi:10.1101/gad.348316.121
Porto, E. M., Komor, A. C., Slaymaker, I. M., and Yeo, G. W. (2020). Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19 (12), 839–859. doi:10.1038/s41573-020-0084-6
Przybyla, L., and Gilbert, L. A. (2022). A new era in functional genomics screens. Nat. Rev. Genet. 23 (2), 89–103. doi:10.1038/s41576-021-00409-w
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 (5), 1173–1183. doi:10.1016/j.cell.2013.02.022
Raffeiner, P., Hart, J. R., García-Caballero, D., Bar-Peled, L., Weinberg, M. S., and Vogt, P. K. (2020). An MXD1-derived repressor peptide identifies noncoding mediators of MYC-driven cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 117 (12), 6571–6579. doi:10.1073/pnas.1921786117
Rogers, Z. N., McFarland, C. D., Winters, I. P., Naranjo, S., Chuang, C. H., Petrov, D., et al. (2017). A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat. Methods 14 (7), 737–742. doi:10.1038/nmeth.4297
Rogers, Z. N., McFarland, C. D., Winters, I. P., Seoane, J. A., Brady, J. J., Yoon, S., et al. (2018). Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat. Genet. 50 (4), 483–486. doi:10.1038/s41588-018-0083-2
Romero, R., Sánchez-Rivera, F. J., Westcott, P. M. K., Mercer, K. L., Bhutkar, A., Muir, A., et al. (2020). Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nat. Cancer 1 (6), 589–602. doi:10.1038/s43018-020-0071-1
Schmidt, R., Steinhart, Z., Layeghi, M., Freimer, J. W., Bueno, R., Nguyen, V. Q., et al. (2022). CRISPR activation and interference screens decode stimulation responses in primary human T cells. Science 375 (6580), eabj4008. doi:10.1126/science.abj4008
Serresi, M., Kertalli, S., Li, L., Schmitt, M. J., Dramaretska, Y., Wierikx, J., et al. (2021). Functional antagonism of chromatin modulators regulates epithelial-mesenchymal transition. Sci. Adv. 7 (9), eabd7974. doi:10.1126/sciadv.abd7974
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelson, T., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343 (6166), 84–87. doi:10.1126/science.1247005
Shalem, O., Sanjana, N. E., and Zhang, F. (2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16 (5), 299–311. doi:10.1038/nrg3899
Shen, L., Li, C., Chen, F., Shen, L., Li, Z., and Li, N. (2021). CRISPR/Cas9 genome-wide screening identifies LUC7L2 that promotes radioresistance via autophagy in nasopharyngeal carcinoma cells. Cell Death Discov. 7 (1), 392. doi:10.1038/s41420-021-00783-8
Sun, M., Wang, Y., Zheng, C., Wei, Y., Hou, J., Zhang, P., et al. (2021). Systematic functional interrogation of human pseudogenes using CRISPRi. Genome Biol. 22 (1), 240. doi:10.1186/s13059-021-02464-2
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Suresh, S., Chen, B., Zhu, J., Golden, R. J., Lu, C., Evers, B. M., et al. (2020). eIF5B drives integrated stress response-dependent translation of PD-L1 in lung cancer. Nat. Cancer 1 (5), 533–545. doi:10.1038/s43018-020-0056-0
Tang, R., Shuldiner, E. G., Kelly, M., Murray, C. W., Hebert, J. D., Andrejka, L., et al. (2023). Multiplexed screens identify RAS paralogues HRAS and NRAS as suppressors of KRAS-driven lung cancer growth. Nat. Cell Biol. 25 (1), 159–169. doi:10.1038/s41556-022-01049-w
Tong, J., Wang, X., Liu, Y., Ren, X., Wang, A., Chen, Z., et al. (2021). Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci. Adv. 7 (18), eabd4742. doi:10.1126/sciadv.abd4742
Vartanian, S., Lee, J., Klijn, C., Gnad, F., Bagniewska, M., Schaefer, G., et al. (2019). ERBB3 and IGF1R signaling are required for nrf2-dependent growth in KEAP1-mutant lung cancer. Cancer Res. 79 (19), 4828–4839. doi:10.1158/0008-5472.CAN-18-2086
Wang, J. Y., and Doudna, J. A. (2023). CRISPR technology: a decade of genome editing is only the beginning. Science 379 (6629), eadd8643. doi:10.1126/science.add8643
Wang, Q., Li, J., Zhu, J., Mao, J., Duan, C., Liang, X., et al. (2022). Genome-wide CRISPR/Cas9 screening for therapeutic targets in NSCLC carrying wild-type TP53 and receptor tyrosine kinase genes. Clin. Transl. Med. 12 (6), e882. doi:10.1002/ctm2.882
Wu, Q., Tian, Y., Zhang, J., Tong, X., Huang, H., Li, S., et al. (2018). In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 115 (17), E3978-E3986–E3986. doi:10.1073/pnas.1716589115
Yang, Z., Liang, S. Q., Yang, H., Xu, D., Bruggmann, R., Gao, Y., et al. (2021). CRISPR-mediated kinome editing prioritizes a synergistic combination therapy for FGFR1-amplified lung cancer. Cancer Res. 81 (11), 3121–3133. doi:10.1158/0008-5472.Can-20-2276
Yousefi, M., Boross, G., Weiss, C., Murray, C. W., Hebert, J. D., Cai, H., et al. (2022). Combinatorial inactivation of tumor suppressors efficiently initiates lung adenocarcinoma with therapeutic vulnerabilities. Cancer Res. 82 (8), 1589–1602. doi:10.1158/0008-5472.Can-22-0059
Zhang, X., Lv, S., Luo, Z., Hu, Y., Peng, X., Lv, J., et al. (2021). MiniCAFE, a CRISPR/Cas9-based compact and potent transcriptional activator, elicits gene expression in vivo. Nucleic Acids Res. 49 (7), 4171–4185. doi:10.1093/nar/gkab174
Zhao, G., Gong, L., Su, D., Jin, Y., Guo, C., Yue, M., et al. (2019). Cullin5 deficiency promotes small-cell lung cancer metastasis by stabilizing integrin β1. J. Clin. Invest. 129 (3), 972–987. doi:10.1172/JCI122779
Zhao, Z., Shang, P., Mohanraju, P., and Geijsen, N. (2023). Prime editing: advances and therapeutic applications. Trends Biotechnol. 41 (8), 1000–1012. doi:10.1016/j.tibtech.2023.03.004
Zheng, R., Zhang, L., Parvin, R., Su, L., Chi, J., Shi, K., et al. (2023a). Progress and perspective of CRISPR-cas9 technology in translational medicine. Adv. Sci. (Weinh) 10, e2300195. doi:10.1002/advs.202300195
Zheng, X., Wu, Y., Zuo, H., Chen, W., and Wang, K. (2023b). Metal nanoparticles as novel agents for lung cancer diagnosis and therapy. Small 19 (18), e2206624. doi:10.1002/smll.202206624
Zhou, X., Renauer, P. A., Zhou, L., Fang, S. Y., and Chen, S. (2023). Applications of CRISPR technology in cellular immunotherapy. Immunol. Rev. 2023, 13241. doi:10.1111/imr.13241
Keywords: CRISPR screening, lung cancer, treatment, drug resistance, Tuba-seq
Citation: Shen W, Hu F, Lei P and Tang Y (2023) Applications of CRISPR screening to lung cancer treatment. Front. Cell Dev. Biol. 11:1295555. doi: 10.3389/fcell.2023.1295555
Received: 16 September 2023; Accepted: 14 November 2023;
Published: 15 December 2023.
Edited by:
Subhadip Mukhopadhyay, New York University, United StatesReviewed by:
Kajal Biswas, National Cancer Institute, United StatesCopyright © 2023 Shen, Hu, Lei and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pan Lei, bGVpcGFuQHRhaWhlaG9zcGl0YWwuY29t; Yijun Tang, dHlqXzc5OUB0YWloZWhvc3BpdGFsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.