
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 15 November 2023
Sec. Epigenomics and Epigenetics
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1294541
Introduction: miR-21 is a critical microRNA for the regulation of various processes in oocytes and granulosa cells. It is involved in the modulation of apoptosis and can influence other epigenetic mechanisms. Among these mechanisms, DNA methylation holds significant importance, particularly during female gametogenesis. Evidence has demonstrated that microRNAs, including miR-21, can regulate DNA methylation. Bisphenol A (BPA) is a widespread chemical that disrupts oocyte maturation and granulosa cell function. Recent findings suggested that BPA can act through epigenetic pathways, including DNA methylation and microRNAs.
Methods: This study uses anti-miR-21 LNAs to explore the involvement of miR-21 in the regulation of DNA methylation in bovine Cumulus-Oocyte-Complexes (COCs) and granulosa cells, in the presence and absence of BPA. This study investigated 5 mC/5hmC levels as well as gene expression of various methylation enzymes using qPCR and western blotting.
Results and discussion: Results reveal that BPA reduces 5mC levels in granulosa cells but not in COCs, which can be attributed to a decrease in the methylating enzymes DNMT1 and DNMT3A, and an increase in the demethylating enzyme TET2. We observed a significant increase in the protein levels of DNMT1, DNMT3A, and TET2 upon inhibition of miR-21 in both COCs and granulosa cells. These findings directly imply a strong correlation between miR-21 signaling and the regulation of DNA methylation in bovine COCs and granulosa cells under BPA exposure.
Oocyte competence, essential for embryo development, includes equipping the oocyte with the critical RNA, proteins, and nutrients required for appropriate embryo development (Coticchio et al., 2015). Whether an oocyte is developmentally competent, is heavily dependent on intricate molecular pathways governed by both genetic and epigenetic mechanisms. Epigenetics, the study of molecular pathways capable of altering gene expression without altering the DNA sequence, consists of DNA methylation, histone modifications, and non-coding RNA (ncRNA) (Felsenfeld, 2014). The disruption of any of these pathways during the critical window of oocyte maturation can have detrimental effects on subsequent fertilization, early embryo development, and the birth of a live offspring (Jacobs et al., 2017). The endocrine disrupting compound bisphenol A (BPA) is capable of interfering with all three epigenetic pathways (Jacobs et al., 2017). It is shown that these pathways govern thousands of regulatory processes essential for adequate cellular function. There is also increasing evidence to suggest that these pathways can control each other in a complex regulatory network that remains to be uncovered.
The focus of this study is on a specific microRNA, miR-21, and its ability to orchestrate DNA methylation in bovine COCs and granulosa cells. The highly complex nature of miRNA synthesis indicates that multiple routes of disruption are possible. Estrogen and androgen signaling are examples of hormonal pathways that are involved in regulating miRNA expression in different tissues (Gulyaeva and Kushlinskiy, 2016). Environmental factors including endocrine-disrupting compounds, such as bisphenols, are capable of interfering with gene expression via miRNA activity (Huumonen et al., 2014; Gulyaeva and Kushlinskiy, 2016).
miR-21 is an important miRNA conserved in several species such as rats, bovine, and humans (Yerushalmi et al., 2018). It is critical for mammalian reproduction and abundantly expressed in bovine, rats, pigs, and human oocytes and granulosa cells (Yerushalmi et al., 2018). Previous research by our group reports that BPA significantly increases miR-21 expression in bovine granulosa cells and Cumulus-Oocyte-Complexes (COCs) (Sabry et al., 2021). miR-21 is important for cell survival by regulating apoptotic genes such as PDCD4, PRC1, and CDC25a (Zi et al., 2017; Yerushalmi et al., 2018). miR-21 knockdown studies in pigs and mice report disrupted meiotic maturation and increased apoptosis of cumulus cells and embryonic arrest at the 4 - 8 cell stage due to the inhibition (Han et al., 2017). The DNA region that encodes miR-21 contains an estrogen response element (ER) in its promoter; therefore, it has the potential to be regulated by estrogen and estrogen-mimic compounds, such as bisphenols (Klinge, 2009).
Another crucial epigenetic pathway is DNA methylation, which involves the addition by specialized enzymes of a methyl group on specific sites within the genome, known as CpG islands (Jin et al., 2011). These methyl marks most commonly repress transcription and thereby govern which genes are turned “on or off” in which cells and during which developmental stage. DNA methylation functions to maintain genomic integrity, imprint on specific genes, and inactivate the X chromosome (Fedoriw et al., 2012; Toranõ et al., 2016). DNA methyltransferases (DNMTs) are responsible for the methyl addition within the genome to either maintain methylation (DNMT1) or to establish de novo methylation patterns (DNMT3s). Other genes involved in DNA demethylation, such as TETs and TDG, also play critical roles in regulating genome wide demethylation to activate gene expression. Decreased levels of DNMT3 enzymes are linked to embryonic lethality (Liao et al., 2015), several cancers (Ramassone et al., 2018), and EDC exposure, including BPA (Chao et al., 2012). Similar to miRNAs, methylation pathways and DNMT activities can also be affected by BPA. In development, BPA exposure resulted in abnormal methylation on the X chromosome, decreased DNMT activity, inhibition of meiotic development, and genome-wide methylation errors (Chao et al., 2012; Kim et al., 2013; Trapphoff et al., 2013; Kahlon, 2016; Wang et al., 2016). Previous research by our group reported that BPA exposure was correlated with significant increases and decreases in DNMT3A mRNA and protein expression, respectively, in bovine granulosa cells (Sabry, 2019).
Recently, researchers have begun to investigate the interactions between miRNAs and DNA methylation: an intricate regulatory network exists between these epigenetic modulators to control gene expression. By altering the promoter regions of miRNA genes on the genome, DNA methylation can modulate the biosynthesis of miRNAs (Lujambio et al., 2008). miR-34b and miR-34c, which are tumour suppressor miRNAs that target a number of oncogenes, are tightly regulated by the methylation status of their primary gene promoter region in cancer cells (Suzuki et al., 2010).
On the other hand, miRNAs can regulate DNA methylation by targeting and modulating genes that encode for methylation enzymes and other factors that comprise the DNA methylation machinery. miR-29b is found to target DNMT3A and 3B, thereby regulating de novo methylation of several target genes (Fabbri et al., 2007). Furthermore, miR-148a has been shown to target DNMT1, responsible for the maintenance DNA methylation (Shimizu et al., 2020). Additionally, miR-21 was shown to be able to modulate the expression of TET2, which is responsible for the demethylation of DNA (Cao et al., 2019). Furthermore, miR-21 can regulate the expression of DNMT3A, and thereby control the activation of the promoter regions that promote hepatocellular carcinoma (HCC) cell growth and proliferation (Lin et al., 2023). Additionally, it was reported that treatment with a miR-21 inhibitor increased DNMT3A expression, increased promoter methylation patterns, and arrested the growth of these HCC cells (Lin et al., 2023).
A significant increase and decrease of miR-21 expression and the methylation regulator, DNA methyltransferase 3A (DNMT3A), respectively, after BPA treatment suggests a plausible link between these two genes in the context of female reproductive toxicity. DNMT3A is one of the predicted downstream targets of miR-21 in in vitro bovine embryos (Mondou et al., 2012). These studies present interesting correlations suggesting miR-21 regulation of DNA methylation. Knocking down miR-21 in bovine cumulus cells using locked nucleic acid (LNA) technology is a simple, precise, and accurate method to answer whether this interplay between epigenetic regulators exists in bovine oocytes and granulosa cells during in vitro oocyte maturation and in vitro granulosa cell culture. Any alteration to DNMT levels most likely contributes to downstream disruptions of maintenance and de novo DNA methylation of crucial genes within the oocyte, which leads to altered gene expression, and ultimately disrupted oocyte maturation.
miRNAs and DNA methylation are essential epigenetic mechanisms that regulate gene expression in various cell types. These vital mechanisms interact through complex regulatory processes co-orchestrating the precise tuning of gene expression and cellular functions. Their interaction carries severe biological implications on a broad range of physiological operations, including development, differentiation, and disease progression. This research is focused on exploring miR-21’s functional capacity to regulate the DNA methylation process. By analyzing their intertwined partnership, we aim to obtain insights into the convoluted regulatory mechanisms governing these cellular functions in both healthy and environmentally-disrupted reproductive functions.
miR-21 was inhibited in bovine COCs over the course of in vitro maturation. The transfection and knockdown efficiencies were previously confirmed in in vitro cultured granulosa cells (Sabry et al., 2022) and therefore, miR-21 inhibition will be confirmed only in COCs. Figure 1 depicts miR-21 expression in bovine COCs with both the scramble and inhibitor LNAs in the presence and absence of Lipofectamine 3,000. The results show a significant reduction in miR-21 expression when the inhibitor was used alone at 0.5 µM with no changes in the scramble group at that same concentration [F (5,12) = 16.7, p = 0.0000486477]. The difference between the control and the miR-21 LNA-only groups is an 80% (±1.8%) reduction in expression values indicating effective inhibition of miR-21 activity. This concentration was used for further transfection of COCs followed by treatments with either ethanol alone or ethanol with BPA at 0.05 mg/mL.
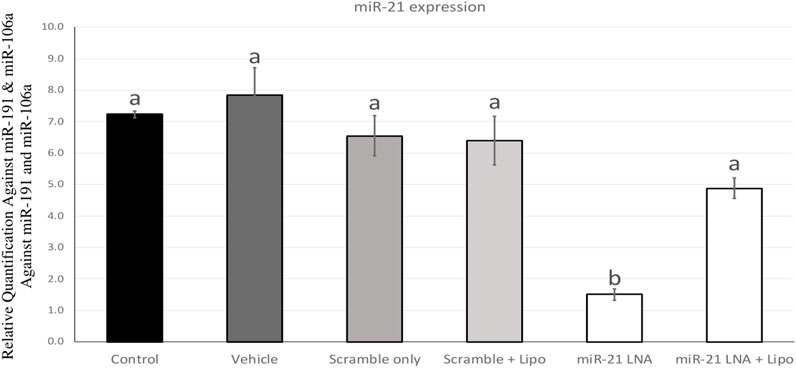
FIGURE 1. miR-21 expression in in vitro matured bovine COCs. Bovine COCs were matured in in vitro S-IVM media and treated with either an LNA scramble or LNA inhibitor in the presence and absence of Lipofectamine 3,000 at 0.1 µM and 0.5 µM, respectively. Results show that treatment with the miR-21 inhibitor alone resulted in an 80% knockdown of miR-21 expression. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05. Bars represent the mean ± SEM.
24 h after in vitro maturation, the COCs from all 9 groups were imaged and are represented in Figure 2. As shown through qualitative assessment, treatment of COCs with BPA halfway through maturation negatively impacts the observable qualities of the COCs. The BPA-treated COCs exhibited incomplete cumulus expansion and the cytoplasm of the cumulus cells appeared darker and more fragmented than the control. This appeared to be the case for all BPA-treated COCs regardless of transfection conditions indicating miR-21 inhibition has no observable effect on COC phenotypes.
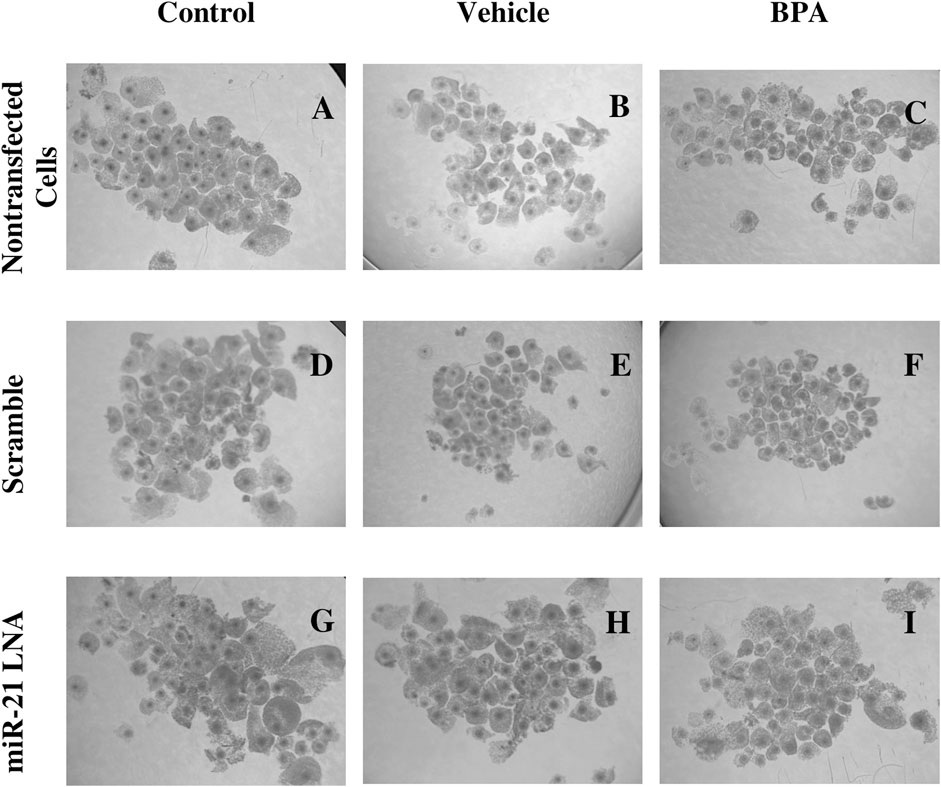
FIGURE 2. Oocyte Morphology after 24 h in vitro maturation with anti-miR-21 LNAs and BPA. Bovine COCs were placed into maturation media alone (A–C), in the presence of a scramble (D–F), or in the presence of anti-miR-21 LNA inhibitors (G–I) at 0.5 µM. 12 h into maturation, COCs were also treated with either a vehicle (B,E,H) or BPA (C,F,I) at 0.05 mg/mL for another 12 h. BPA-treated COCs appear poorer in quality with less cumulus cell expansion and increased heterogeneity among COC phenotypes. Scale bar = 500 µm.
Immunofluorescence of 5 mC and 5 hmC provided initial information on the effects of BPA on global methylation patterns and whether these effects are reliant on miR-21-mediated pathways. Figures 3–6 display the results of 5 mC and 5 hmC staining in oocytes, which displayed that BPA had no significant effects on global methylation patterns as shown by confocal microscopy (Figures 3, 5) and analyzed by ImageJ (Figures 4, 6).
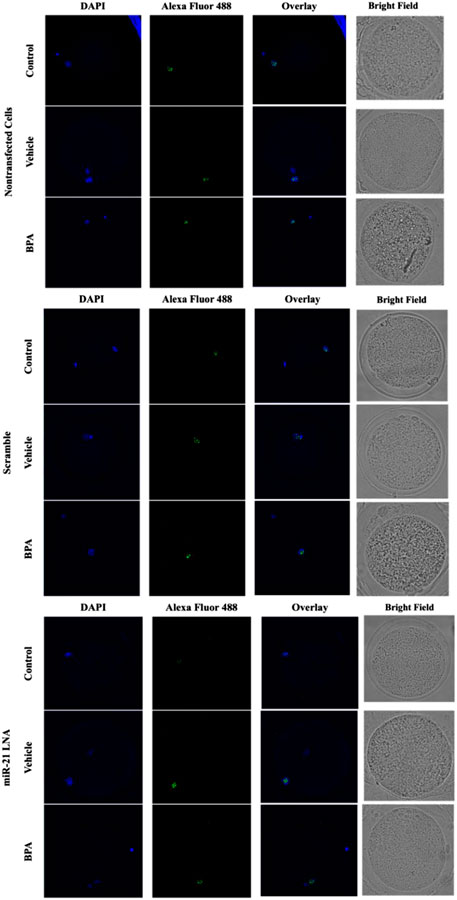
FIGURE 3. Confocal Microscopy of 5′methylcytosine in Bovine Oocytes. Transfected and treated COCs were fixed and permeabilized to allow for immunostaining of 5 mC. Immunodetection of 5 mC in COCs was achieved by utilizing an Alexa Fluor 488 conjugated secondary antibody coupled with confocal microscopy. Confocal Images are represented for all 9 groups.
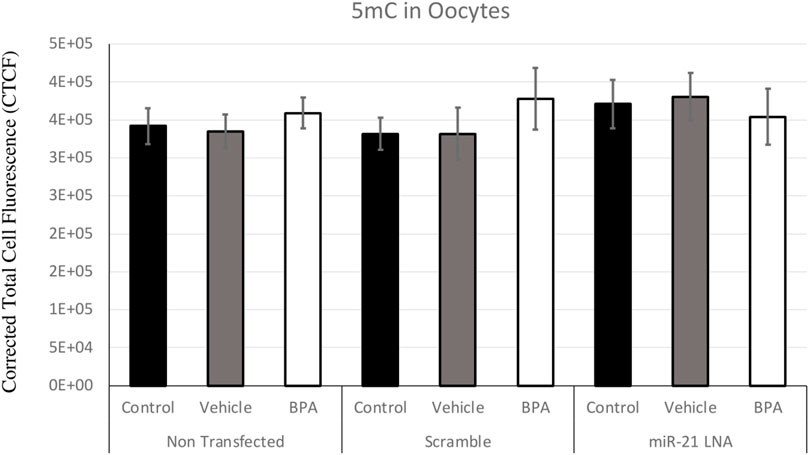
FIGURE 4. Corrected Total Cell Fluorescence of 5′methylcytosine in Bovine Oocytes. Transfected and treated COCs were fixed and permeabilized to allow for immunostaining of 5 mC. The corrected total cell fluorescence was calculated using ImageJ on a minimum of 15 COCs per group for a total of 140 COCs analyzed. Bars represent the mean ± SEM at p < 0.05.
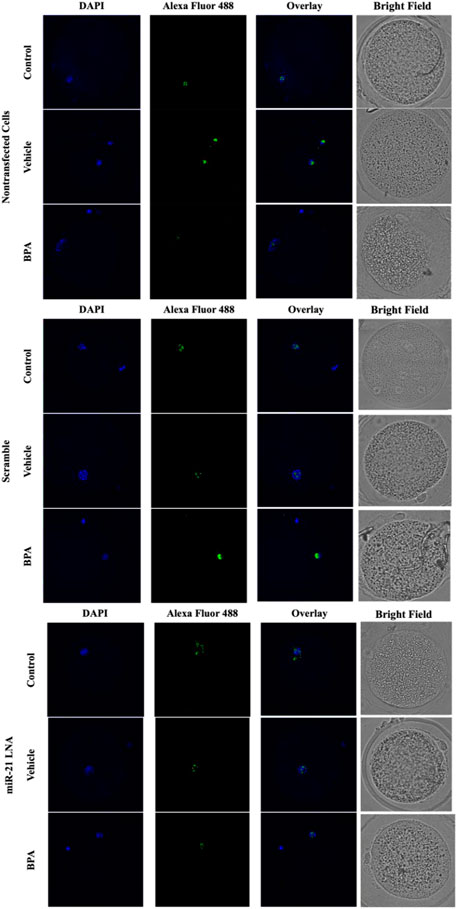
FIGURE 5. Confocal Microscopy of 5′hydroxymethylcytosine in Bovine Oocytes. Transfected and treated COCs were fixed and permeabilized to allow for immunostaining of 5 hmC. Immunodetection of 5 hmC in COCs was also achieved by utilizing an Alexa Fluor 488 conjugated secondary antibody coupled with confocal microscopy. Confocal Images are represented for all 9 groups.
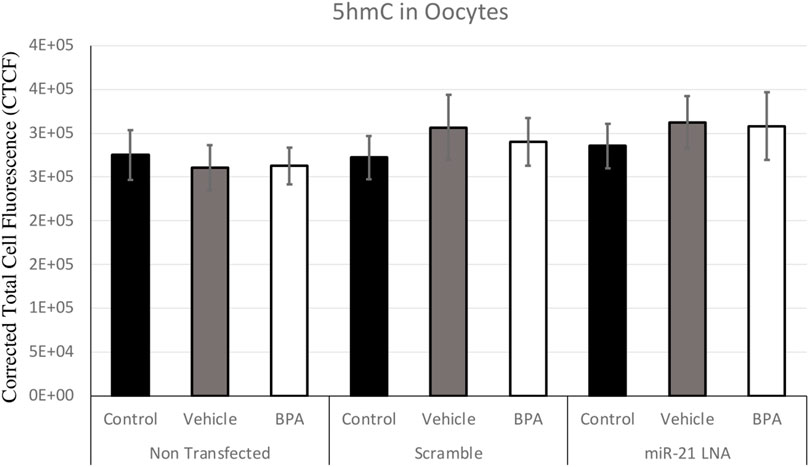
FIGURE 6. Corrected Total Cell Fluorescence of 5′hydroxymethylcytosine in Bovine Oocytes. Transfected and treated COCs were fixed and permeabilized to allow for immunostaining of 5 hmC. The corrected total cell fluorescence was calculated using ImageJ on a minimum of 15 COCs per group for a total of 140 COCs analyzed. Bars represent the mean ± SEM at p < 0.05.
Figure 7, 8 display the results of 5 mC and 5 hmC staining in GCs, which interestingly revealed opposite findings than the COCs. BPA significantly decreased 5 mC staining in GCs as shown by Flow cytometry (Supplementary Figure S2). Analysis using FlowJo showed that BPA reduces 5 mC staining in nontransfected GCs and in scramble GCs [H (8) = 23.4562, p = 0.002825] (Figure 7). Additionally, miR-21 inhibition in GCs reversed the effects of BPA on 5 mC staining in GCs where there are no significant changes between the inhibited controls and treated groups (Figure 7). Similar to the COCs, miR-21 inhibition and BPA did not alter 5 hmC staining in GCs either (Figure 8).
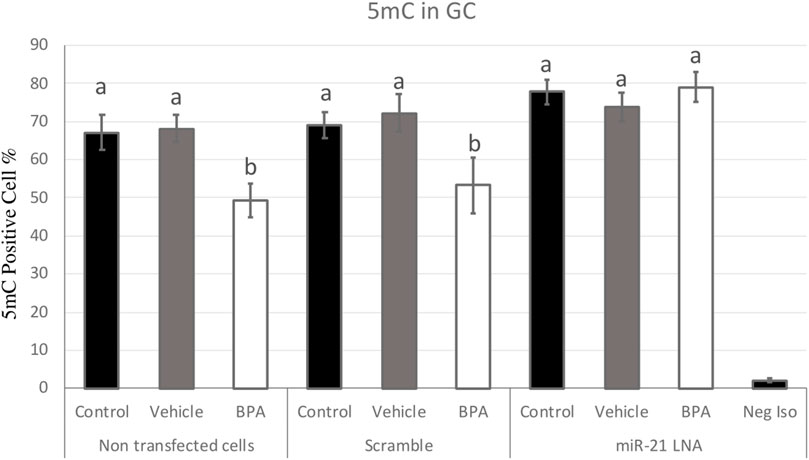
FIGURE 7. Quantification of 5′methylcytosine (5 mC) in Bovine Granulosa Cells. Analysis of 5 mC levels was done on FlowJo on a minimum of three biological replicates. The last column represents the negative control where a negative isotype was used as the primary antibody. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05. Bars represent the mean ± SEM and n = 7.
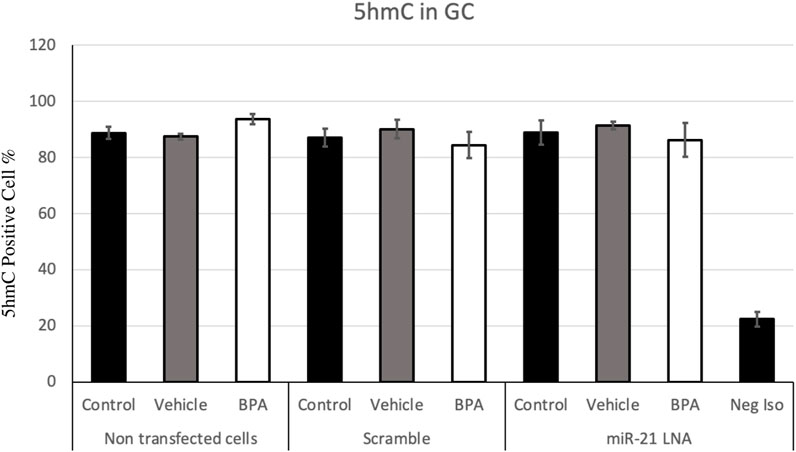
FIGURE 8. Quantification of 5′hydroxymethylcytosine (5 hmC) in Bovine Granulosa Cells. The analysis was done on FlowJo on a minimum of three biological replicates. The last column represents the negative control where a negative isotype was used as the primary antibody. Bars represent the mean ± SEM at p < 0.05and n = 4.
To further uncover the source of aberrant methylation patterns due to BPA exposure, the expression of methylation and demethylation transcripts were quantified using qPCR. Methylators (DNMT1, 3A, and 3B) and demethylators (TET1, 2, 3, and TDG) were quantified and normalized to housekeeping genes (GAPDH, B-actin, and YWHAZ). Figures 9, 10 display the results in COCs and GCs, respectively. In COCs, all transcripts except for DNMT3B were significantly increased after BPA exposure regardless of transfection conditions (p < 0.05) (Figure 9). In GCs, the results are similar to the COCs for the methylators investigated; DNMT1 and DNMT3A were significantly increased after BPA exposure, while DNMT3B was unaffected (Figures 10A–C).
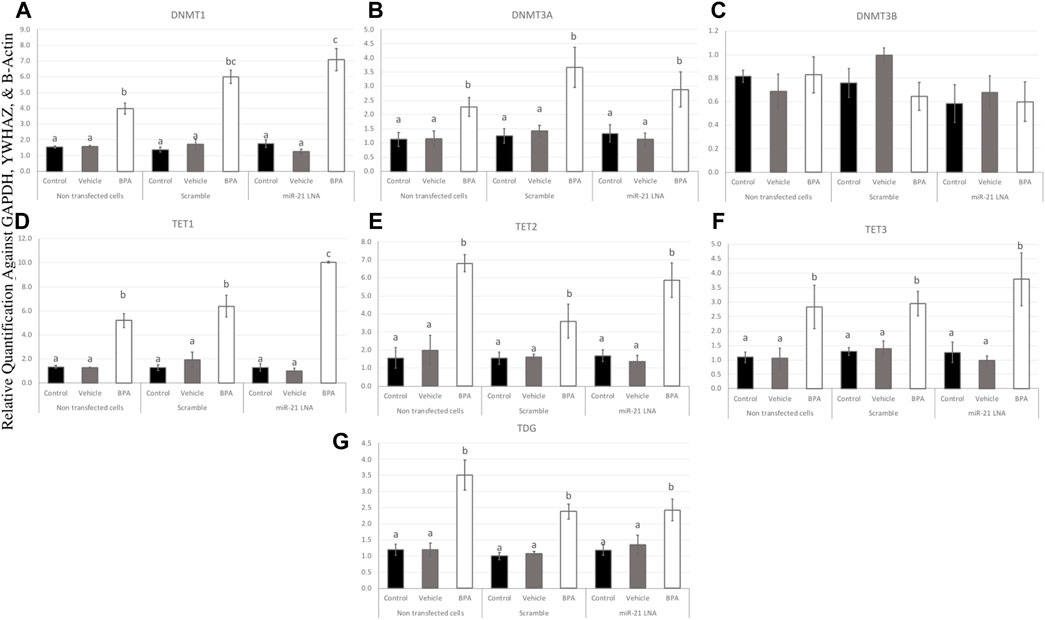
FIGURE 9. Expression of methylating and demethylating mRNAs after miR-21 inhibition and BPA treatment in Bovine COCs. COCs were transfected with LNA inhibitor probes at 0.5 μM then treated with BPA (0.05 mg/mL). All transcripts including DNMT1 (A), DNMT3A (B), TET1—3 (D–F), and TDG (G) were significantly increased after BPA exposure in all transfection groups apart from DNMT3B (C). Quantification is normalized to reference targets GAPDH, YWHAZ, and B-Actin. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05, c indicating a significantly different mean than a and b, and d indicating a significantly different mean than a, b, and c at p < 0.05. ab indicates no differences between a or b and bc indicates no differences between b or c. Bars represent the mean ± SEM.
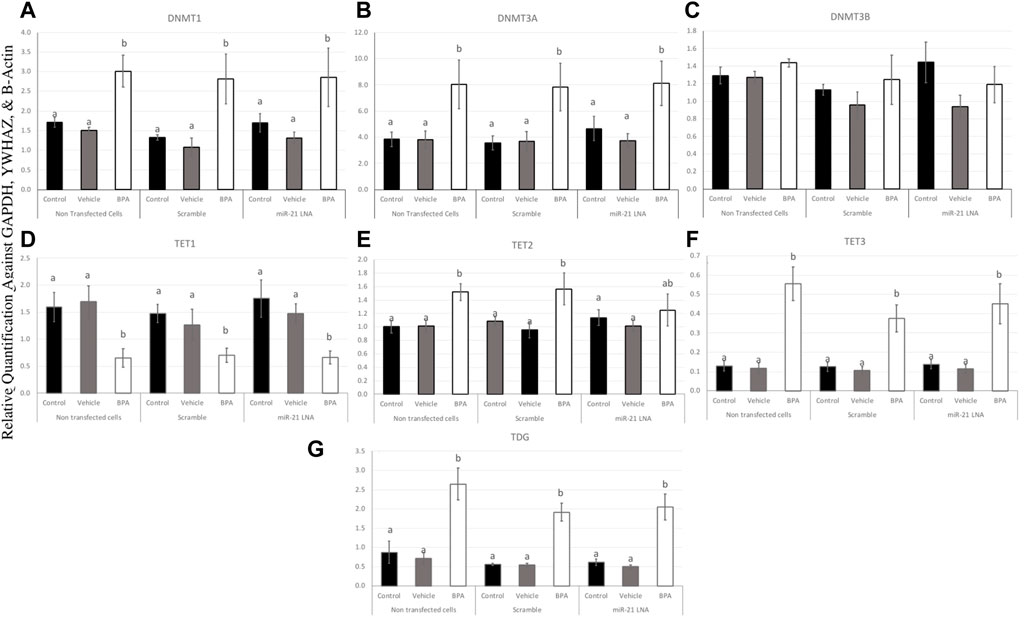
FIGURE 10. Expression of methylating and demethylating mRNAs after miR-21 inhibition and BPA treatment in Bovine GCs. Cells were transfected with LNA inhibitor probes at 0.5 μM for 12 h and then treated with BPA (0.05 mg/mL) for another 12 h. Most transcripts including DNMT1 (A), DNMT3A (B), TET3 (F), and TDG (G) were significantly increased after BPA exposure in all transfection groups apart from TET1 (D) that was decreased, DNMT3B (C) that was unaffected, and TET2 (E) that was unaffected in the inhibited group. Quantification is normalized to reference targets GAPDH, YWHAZ, and B-Actin. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05. Bars represent the mean ± SEM.
In terms of the demethylators, TET3 and TDG were also increased in the BPA-treated cells regardless of transfection conditions (p < 0.05) (Figures 10F, G). TET2 was also significantly increased after BPA treatment, but only in the nontransfected and scramble groups (p < 0.05), whereas the miR-21 inhibited cells that were also treated with BPA exhibited a non-significant increase in TET2 (Figure 10E). Finally, TET1 was the only transcript in GCs that was significantly decreased as a result of BPA exposure and this was independent of miR-21 inhibition (p < 0.05) (Figure 10D). This was the opposite of the significant increase of this gene observed in BPA-treated COCs (Figure 9D). The individual statistic H, F, and p values for each target can be found in Supplementary Table S2.
To interpret the effects of miR-21 inhibition and BPA on key methylation genes, it is important to quantify the protein levels to demonstrate if these effects are translating. In COCs, DNMT1, DNMT3A, and TET2 were quantified at the protein level (Figure 11A). DNMT1 was unaffected by BPA but interestingly, miR-21 inhibition alone significantly increased DNMT1 protein expression in the control groups (p = 0.021) and in the vehicle groups (p = 0.005) (Figure 11B). DNMT3A, on the other hand, was affected by BPA exposure but not by miR-21 inhibition. DNMT3A was significantly decreased after BPA exposure in the nontransfected group (p = 0.046), in the scramble group (p = 0.011), and in the inhibited group (p = 0.014). Unlike DNMT1, there is no significant increase of DNMT3A after miR-21 inhibition (Figure 11C).
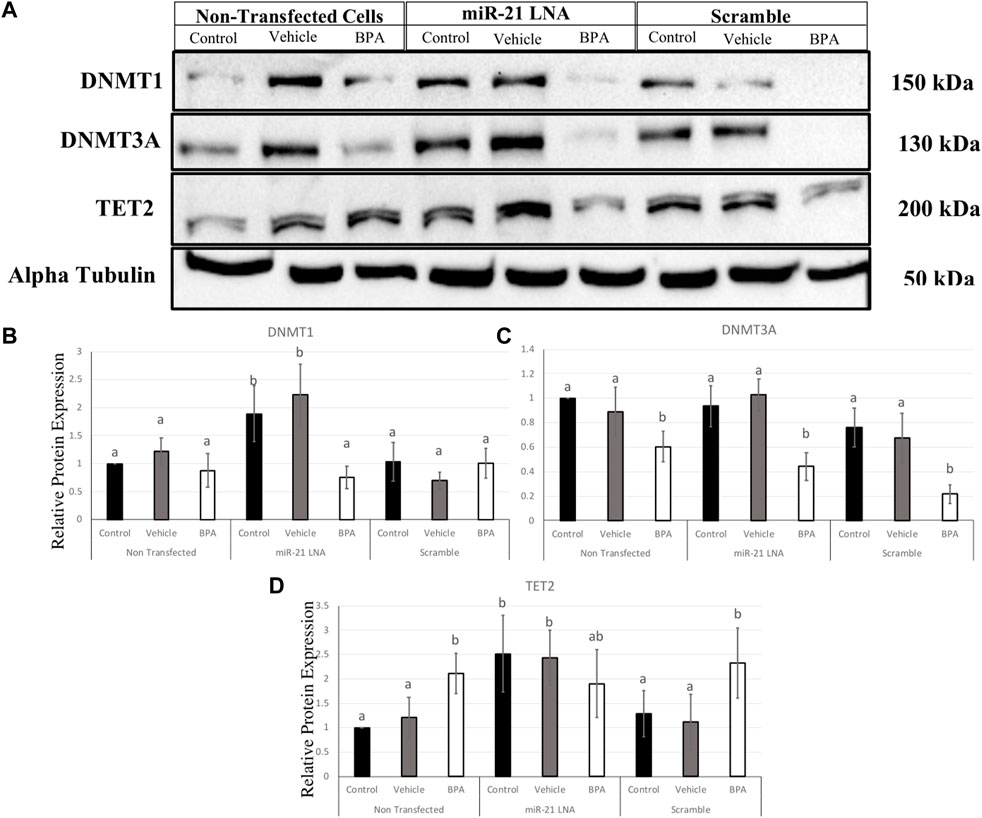
FIGURE 11. Relative protein expression of DNMT1, DNMT3A, and TET2 after miR-21 inhibition and BPA treatment in Bovine COCs. Transfections were done with LNA inhibitor probes at 0.5 μM for 12 h followed by BPA treatment for another 12 h. Western blots (A) and graphical representations of DNMT1 (B), DNMT3A (C), and TET2 (D) revealed that BPA decreased and increased DNMT3A and TET2 proteins, respectively. miR-21 Inhibition also induced protein expression in the control-only groups for DNMT1 and TET2. Densitometric analysis was performed relative to the loading control, α-tubulin. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05. Bars represent the mean ± SEM. n = 6 for DNMT1; n = 9 for DNMT3A; n = 6 for TET2.
BPA had the opposite effect on TET2 protein with a significant increase in protein levels in the nontransfected group (p = 0.015) and in the scramble group (p = 0.049), but not in the inhibited group (Figure 11D), this can be attributed to a significant increase in TET2 protein in the control and vehicle groups with miR-21 inhibition. Just like the DNMTs, TET2 was shown to be significantly upregulated when miR-21 was inhibited in the control group (p = 0.028) and in the vehicle group (p = 0.048) indicating it is implicated in miR-21 signaling.
In GCs, DNMT1, DNMT3A, and TET2 were also quantified at the protein level (Figure 12A). DNMT1 was significantly decreased after BPA exposure in the nontransfected group (p = 0.049), in the scramble group (p = 0.049), and in the inhibited group (p = 0.038) (Figure 12B). Interestingly, miR-21 inhibition alone significantly increased DNMT1 protein expression in the control groups (p = 0.004), the vehicle groups (p = 0.048), and there was a significant recovery of DNMT1 after BPA exposure in the miR-21 inhibited group (p = 0.024) (Figure 12B).
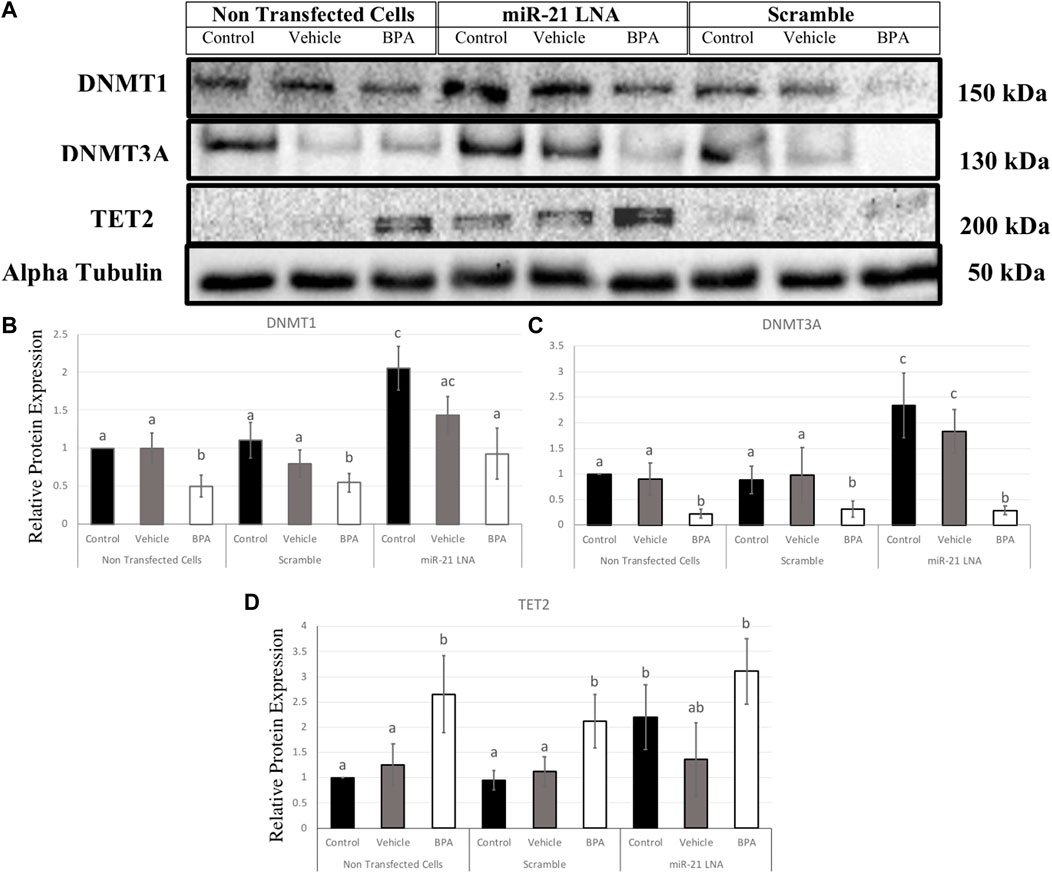
FIGURE 12. Relative protein expression of DNMT1, DNMT3A, and TET2 after miR-21 inhibition and BPA treatment in Bovine GCs. Transfections were done with LNA inhibitor probes at 0.5 μM for 12 h followed by BPA treatment for another 12 h. Western blots (A) and graphical representations of DNMT1 (B), DNMT3A (C), and TET2 (D) revealed that BPA decreased and increased DNMTs and TET2 proteins, respectively. miR-21 Inhibition also induced all protein expression in the control-only groups. Densitometric analysis was performed relative to the loading control, α-tubulin. Different letters indicate significant differences, with b indicating a significantly different mean than a at p < 0.05. Bars represent the mean ± SEM. n = 9 for DNMT1, n = 6 for DNMT3A, n = 6 for TET2.
Similarly, DNMT3A exhibited similar results in terms of BPA exposure and miR-21 inhibition. DNMT3A was significantly decreased after BPA exposure in the nontransfected group (p = 0.025), in the scramble group (p = 0.02), and in the inhibited group (p = 0.005). Furthermore, miR-21 inhibition alone significantly increased DNMT3A protein expression, just like DNMT1, in the control groups (p = 0.011) and in the vehicle groups (p = 0.039). Unlike DNMT1, there is no significant recovery of DNMT3A after BPA exposure in the miR-21-inhibited group (Figure 12C).
BPA had the opposite effect on TET2 protein with a significant increase in protein levels regardless of transfection conditions. This was seen in the nontransfected group (p = 0.036), in the scramble group (p = 0.024), and in the inhibited group (p = 0.029) (Figure 12D). Just like the DNMTs, TET2 was shown to be significantly upregulated when miR-21 was inhibited in the control groups (p = 0.037) indicating it is implicated as a miR-21 target.
Uncovering the pathways utilized by microRNAs in oocyte maturation is key due to their crucial involvement in regulating an array of biological processes, such as cell proliferation, differentiation and apoptosis (Toms et al., 2018). In particular, research has delved into the role that individual microRNAs play in oocytes over the course of oocyte maturation; studies have demonstrated specific involvement with meiosis regulation, spindle assembly and cytoskeleton organization (Wang et al., 2017). A comprehensive analysis regarding each unique function served by these epigenetic players may provide insights into molecular mechanisms responsible for proper oocyte development, thus enhancing assisted reproductive technologies. miR-21 is one excellent example of a microRNA worth further investigations. It is upregulated during oocyte maturation in several species, including murine, porcine, bovine, and human (Tscherner et al., 2018; Dehghan et al., 2021). The functions miR-21 play in the oocyte and its surrounding granulosa cells are numerous.
miR-21 expression is crucial in granulosa cells surrounding the oocyte (Aldakheel et al., 2021). In accordance with Bartolucci et al. (Bartolucci et al., 2020), miR-21 expression is positively correlated with oocyte developmental competence; furthermore, mimicking miR-21 in granulosa cells led to improved developmental competence by way of promoting mitochondrial function while combating oxidative stress (Bartolucci et al., 2020). It is also noted that miR-21 is capable of regulating other epigenetic pathways including DNA methylation which is essential for proper oocyte maturation (Pan et al., 2010; Cao et al., 2019). It is linked to various aspects of oocyte maturations including spindle formation and maternal mRNA stability (Dehghan et al., 2021). Methylation of the granulosa cell genome has also been linked to granulosa cell function and subsequently oocyte competence. For example, adequate expression of DNMTs in granulosa cells is associated with proper methylation and normal cumulus cell expansion and steroidogenesis (Sagvekar et al., 2019).
Understanding the regulation of DNA methylation in these oocytes and granulosa cells is important since alterations in DNA methylation patterns can lead to developmental defects and infertility. This is carried out in this study where the role of miR-21 in regulating DNA methylation in bovine COCs and granulosa cells is investigated in a model of BPA toxicity.
This study showed that in bovine granulosa cells BPA induced hypomethylation, which was attenuated by inhibition of miR-21. The observation of hypomethylation is supported in literature where BPA treatment was associated with decreased 5 mC staining in various cell types including mouse (Khaghani et al., 2021) and porcine oocytes (Wang et al., 2016) that also reported differently methylated genes, decreased maturation rates, increased oxidative stress, and increased apoptosis. The importance of stable 5 mC presence in the oocyte has been proven in numerous studies. Ivanova et al. (Ivanova et al., 2020) found that 5 mC levels increased during meiotic progression, particularly in the pericentric heterochromatin regions, and that reducing 5 mC levels impaired meiotic progression and spindle assembly in pig oocytes. The study highlights that 5 mC dynamics play an important role in regulating oocyte quality in pigs.
The levels of 5 hmC in the oocyte and granulosa cells are equally as crucial for regulating gene expression for proper development (Sakashita et al., 2014). 5 hmC levels also significantly increase as oocytes grow and mature and this is linked with active transcription; this is reasonable considering the maturing oocyte requires fine tuning of gene expression for its specific unique needs (Sakashita et al., 2014). Regardless of this, there were no changes in 5 hmC levels due to BPA or miR-21 inhibition. This finding is supported in literature with no changes in 5 hmC levels due to BPA in mice blood (Kochmanski et al., 2018). However, it is worth noting that these effects are dependent on several factors including cell type, dose of BPA, and timing of exposure as other studies have reported BPA induced changes in 5 hmC levels (Senyildiz et al., 2017; Zheng et al., 2017).
The interesting finding at the level of global methylation patterns is the attenuation of BPA induced hypomethylation in the miR-21 inhibited granulosa cells. This suggests that BPA utilizes a mechanism likely dependent on miR-21 signalling. miR-21 expression is significantly increased due to BPA (Sabry et al., 2021). Therefore, increased miR-21 expression could be linked to hypomethylation of the genome. This speculation is supported by increased methylation (higher 5 mC staining) in the BPA treated granulosa cells that were also treated with a miR-21 inhibitor. To further characterize these pathways, this study investigated crucial methylation genes at the transcript and protein levels as microRNAs exert their functions by acting on transcripts and interrupting protein synthesis (Bhaskaran and Mohan, 2014).
At the mRNA level, BPA increased the amount of all transcripts analyzed except for DNMT3B in both COCs and in granulosa cells and TET1 in granulosa cells. TET1 mRNA was increased in COCs, but decreased in granulosa cells, whereas DNMT3B was unaffected. These effects occurred regardless of transfection conditions with another exception of TET2 mRNA being rescued after BPA treatment in the miR-21 inhibited granulosa cells. All these genes are known to play crucial roles in methylation within the oocyte (Sendžikaitė and Kelsey, 2019). DNMT3B, however, plays a more dominant role in early embryonic development as opposed to de novo methylation in the oocyte (Gao et al., 2020). DNMT3A is another de novo methylator for early embryogenesis, but has been proven to be critical for imprinting during oogenesis (Gao et al., 2020). This could explain why DNMT3B was the only transcript to be unaffected during oocyte maturation.
The significant disruptions of transcripts at the mRNA level indicate a disturbance in the bioavailability of methylation machinery within these cell types and this must contribute to aberrant DNA methylation as a result of BPA. These results are supported in literature with several studies showing that BPA increased the expression of DNMTs at the mRNA levels. BPA was reported to increase mRNA expression of DNMT1, TET2, and TET3 in MCF-7 cells with no changes on DNMT3A and DNMT3B (Awada et al., 2019). In addition, BPA induced an increase in DNMT1 and DNMT3A in zebrafish ovaries (Santangeli et al., 2016). These overall effects are shown to be tissue specific and dependent on experimental conditions. Bhandari et al. (Bhandari et al., 2019) reported that BPA affected DNMT mRNA expression levels differently depending on the dose of BPA used on mesenchymal cells.
Interestingly, the effects of BPA on TET2 mRNA expression in granulosa cells were reversed when miR-21 was inhibited, which suggests a link between these two genes. miR-21 has previously been shown to regulate TETs in hepatocellular carcinomas (Cao et al., 2019). They reported significant downregulation of all TET proteins when they treated cells with a miR-21 mimic (Cao et al., 2019). They suggested a direct link where miR-21 was able to directly bind to the 3′UTR sequences of TET1, TET2, and TET3. In this study, only TET2 was affected at the mRNA level in granulosa cells, supporting a novel report of miR-21 regulation of TET2 in bovine granulosa cells that are also treated with BPA. The rescue of TET2 at the mRNA level could explain the rescue that is observed in methylation patterns when miR-21 was inhibited in granulosa cells. To adequately understand the ultimate effect of BPA and miR-21 inhibition on these genes, it is crucial to quantify the functional proteins. This is also essential to confirm a functional link between miR-21 and any of these downstream methylating enzymes.
In COCs, BPA decreased and increased DNMT3A and TET2 protein levels, respectively. In granulosa cells, BPA was shown to decrease the protein levels of both DNMT1 and DNMT3A while TET2 proteins were significantly increased. A decrease in methylating enzymes and an increase in demethylating enzymes directly explain the overall hypomethylation observed in granulosa cells that were treated with BPA. The opposing finding of DNMT transcript and protein levels are not surprising since these levels are not always directly proportional (Liu et al., 2016). This also suggests the involvement of microRNAs, which are post transcriptional regulators that can interfere with protein levels without necessarily interfering with transcript levels (Bhaskaran and Mohan, 2014). The increase in transcripts can actually be a result of decreased proteins whereby the cellular response to low translation is to compensate by increasing transcription (Bhaskaran and Mohan, 2014).
In the case of TET2, the effects do appear to be directly proportional with both increased transcripts and proteins. The increase in TET2 is shown to be a direct pathway that is utilized by BPA to induce DNA hypomethylation. Li et al. (Li et al., 2020) reported that activation of the estrogen receptor by BPA modulated DNA hydroxymethylation followed by demethylation that was primarily regulated by TET2 in breast cancer cells. This effect was attenuated when TET2 was silenced (Li et al., 2020). This is supported in numerous other studies that found BPA reduced global 5 mC levels coupled with an increase in TET enzyme expression in mouse Leydig cells (Zhou et al., 2023) and mouse brains (Malloy et al., 2019).
DNMT1, DNMT3A, and TET2 were all significantly increased at the protein level in the miR-21 inhibited granulosa cells in the absence of BPA treatment. In COCs, only DNMT1 and TET2 were significantly increased as a result of miR-21 inhibition. Several recent studies characterize the ability of miRNAs to also regulate DNA methylation by targeting the transcripts for DNMTs or TETs and thereby modulating protein levels (Wang et al., 2017). miR-29b is known to target DNMTs and TETs in porcine embryos which tightly regulates DNA methylation patterns (Zhang et al., 2018). The miR-21 findings in this study are the first of its kind within this specific model; here it is reported that these three genes can be regulated by miR-21 signaling in bovine oocytes and granulosa cells. This regulation is speculated to be a direct interaction with the 3′UTR of these genes as reported in literature of other species and other cell types (Li et al., 2020).
In a study by Pan et al. (Pan et al., 2010), miR-21 and miR-148a were investigated to characterize their role in DNA hypomethylation in T cells from patients with lupus. The authors treated cells with a miR-21 mimic and reported compatible findings with significant decreases of DNMT1 at the protein level (Pan et al., 2010). As previously described, TET2 was shown to be a direct target of miR-21 in liver cancer cells (Cao et al., 2019). Furthermore, DNMT3A was predicted to be a target of miR-21 in the bovine genome as shown by Mondou et al. (Mondou et al., 2012) that investigated potential targets within the bovine embryo. Correlation analysis also revealed that miR-21 had a positive relationship with DNMT1 and DNMT3A with no significant relationship with DNMT3B in hepatocellular carcinomas (Lin et al., 2023). To the best of our knowledge, this is the first study to support these findings within bovine granulosa cells, thus rendering this relationship crucial in reproductive research. By targeting DNMT1, DNMT3A and TET2, miR-21 exhibits a significant contribution to maintaining DNA methylation patterns in bovine COCs and granulosa cells.
Despite the findings of miR-21-dependant expression of these proteins, BPA effects on these genes were not reversed when miR-21 was inhibited. DNMTs and TET2 were decreased and increased, respectively, after BPA treatment regardless of transfection conditions. This suggests that BPA effects are potent enough to mask the miR-21 regulation observed in the absence of BPA. Other microRNAs could compensate for a miR-21 knockdown and perform its function, as miRNAs are known to have high amounts of redundancies in their tasks (Fischer et al., 2015). miR-29 is another miRNA that is significantly increased after BPA treatment (Sabry et al., 2021) and it can also regulate DNMTs and TETs (Zhang et al., 2018). However, this study did observe a rescue in BPA induced hypomethylation in granulosa cells after miR-21 inhibition. Therefore, an alternative pathway by which BPA induces an increase in miR-21 expression that results in subsequent reduction of 5 mC levels in granulosa cells might exist. It is possible that there are recoveries in the other genes investigated, such as TET1 or TET3. A limitation of this study is the quantification of only TET2 at the protein level due to the commercial availability of specific antibodies that are able to detect the TET1 and TET3 isoforms within the bovine species. Antibodies for humans, mice, and rats were purchased and unsuccessfully tested on the bovine species alongside species-specific positive controls.
The discrepancies in the findings between granulosa cells and COCs can be attributed to the presence of the oocyte or likely due to differences in experimental manipulation. COCs are matured in the presence of IVM hormones (LH, FSH, and estradiol) where cumulus granulosa cells maintain communication with the oocyte; thus, COCs that are examined experience changes associated with the maturation period. The granulosa cells that were cultured separately were removed from the oocyte prior to maturation and likely represent an alternate stage of cumulus granulosa cell development with pre-maturation expression patterns. This is taken into account using a control group that allows comparisons between groups but not amongst sample types. For this reason, this study does not aim to compare granulosa cells and COCs, but rather, reports the findings of two separate experimental models and accounts for these combined observations when interpreting the data.
Concisely, the significance of these finding consists in the understanding of the role of a key microRNA in early female reproduction, miR-21, and how it is affected by Endocrine Disrupting Compounds, such as Bisphenol A (BPA), ultimately affecting fertility. BPA has been proven to have deleterious effects in early reproduction as it is widely spread in the environment, eliciting its action at low doses of exposure. Although extensively studied, BPA mechanism of action and impacts on molecular aspects of granulosa cells viability remains elusive, especially at the non-genomic and epigenetic level. miR-21 is a well-studied microRNA that is proven as a crucial miRNA in regulating DNA methylation; it is also the most documented miRNA to be affected by BPA. This study investigates the involvement of miR-21 in BPA-induced aberrant DNA methylation and characterizes its potential molecular mechanism of action. This significantly contributes to the field of molecular biosciences particularly focused on epigenetic modulation of a specific microRNA by environmental toxicants. Countless miRNAs are continuously being identified on lengthy lists as differentially expressed in different physiological and pathological conditions. It is crucial to unpack these findings and work up with a single miRNA, tracking its mechanism to better understand its function. This may ultimately help us to further regulate BPA exposure and to better understand its role in female infertility.
The anti-miR-21 Locked Nucleic Acid (LNA) and the nonspecific scrambled control LNA were purchased from Qiagen (Toronto, ON, Canada). The LNA sequence was complementary to miR-21: 5′-CAACATCAGTCTGATAAGCT-3′, and the scrambled control was a random mix of nucleotides: 5′-TAACACGTCTATACGCCCA-3′.
Bovine ovaries (Bos taurus) were collected from a local abattoir (Cargill Meat Solutions, Guelph, ON, Canada). Ovaries were transported at a temperature between 34–36°C and washed with sterile saline solution. COCs were aspirated from follicles using a vacuum pump and collected into a tube containing 1 mL of oocyte collection media comprised of 1 M HEPES-buffered Ham’s F-10 media (Sigma-Aldrich) supplemented with 2% Fetal Bovine Serum (Gibco), Heparin (0.2 IU/mL) (Fresenius Kabi Canada Ltd.), Sodium Bicarbonate (Sigma), and Penicillin/Streptomycin (1%) (Gibco). COCs were divided into 6 groups of 20 COCs per group into in vitro HEPES buffered TCM199 (Sigma-Aldrich) maturation media (S-IVM) containing 20% steer serum and sodium pyruvate (Sigma-Aldrich) and supplemented with 1 μg/mL LH (NIH), 0.5 μg/mL FSH (Follitropin V), 1 μg/mL Estradiol (Sigma-Aldrich).
This initial pilot experiment was to optimize miR-21 knockdown conditions in COCs and compare transfection efficiencies at different concentrations in the presence and absence of a transfection reagent (Lipofectamine 3,000; ThermoFisher). Transfection reagents are commonly used and are necessary to deliver the LNAs into most cell types but are cytotoxic and can introduce confounding results. The inhibitors purchased in this study have the potential to enter cells without the use of a transfection reagent if used at higher concentrations. To determine the most optimal conditions for our cell type and our experimental setting, it is crucial to assess the efficiency of knockdown with and without lipofectamine. The 6 groups were prepared as follows: Control group with S-IVM only, Lipofectamine alone in S-IVM (Vehicle), anti-miR-21 LNA in Lipofectamine in S-IVM (0.1 μM), Scramble LNA in Lipofectamine in S-IVM (0.1 μM), anti-miR-21 LNA alone in S-IVM (0.5 μM), and Scramble LNA alone in S-IVM (0.5 μM). Higher concentrations are needed for unassisted uptake while lower concentrations were used with lipofectamine as per the manufacturer’s recommendations. Samples were matured for 24 h at 38.5°C and 5% CO2. After maturation, COCs were snap frozen in liquid nitrogen for miR-21 quantification using qPCR.
Once the transfection conditions were determined using qPCR, COCs were collected again, but this time divided into three groups of 60 COCs per group. The 60 COCs were placed into 4-well dishes with 20 COCs per well in 3 wells for a total of 180 COCs across the three groups. The three groups were the nontransfected COCs (S-IVM only), Scramble COCs (0.5 μM of scramble LNA in S-IVM), and the miR-21 knockdown group (0.5 μM of the miR-21 inhibitor LNA in S-IVM). COCs were incubated at 38.5°C and 5% CO2. Halfway through maturation at 12 h, each 4-well dish was further divided into 3 groups labelled “Control, Vehicle, and BPA” to result in 9 groups altogether. The following treatments were prepared as followed and were then added into their respective wells: S-IVM only (Control), 0.1% ethanol in S-IVM (Vehicle), and 0.05 mg/mL of BPA (Sigma-Aldrich) dissolved in 0.1% ethanol in S-IVM. COCs were put back into the incubator to complete maturation for another 12 h in a humidified atmosphere at 38.5°C and 5% CO2. After 24 h, 20 COCs per group for 9 groups were imaged to observe qualitative effects alone. The COCs were then frozen in liquid nitrogen and stored at −80°C for downstream RNA and protein analysis.
COCs were aspirated from follicles using the methods described above. Approximately 100–200 COCs were stripped of their granulosa cells using mechanical disruptions via a micropipette. Granulosa cells were placed in 15 mL conical tubes containing 8 mL of 1X Dulbecco’s Modified Eagle Medium (DMEM) (Gibco), glutamine (2 mM) (Sigma-Aldrich), and penicillin/streptomycin (1%). Cells were resuspended in DMEM supplemented with 20% FBS and cultured at 38.5°C in 5% CO2 for 6–7 days with media replacement every 48 h until no empty patches were observed. At 100% confluency, the cells were passaged twice, split at passage 2 into 9 different groups in DMEM containing serum (10% FBS) in 6 well plates at 1 × 105 cells/mL. After 24 h, cells were serum restricted using OptiMEM for another 24 h before being transfected with the LNAs. Optimal GC transfection with these LNAs have been previously optimized as described in Sabry et al. (2022). Briefly, cells were transfected with 0.5 μM of both LNAs for 12 h. Transfected cells were then treated with a vehicle (0.1% ethanol) or BPA (0.05 mg/mL in 0.1% ethanol) in OptiMEM for 12 h then frozen in liquid nitrogen and stored at −80°C for RNA and protein analysis.
Global methylation patterns were assessed by quantifying 5mC and 5 hmC using immunofluorescence followed by confocal microscopy and flow cytometry for COCs and granulosa cells (GCs), respectively. In both cases, COCs and GCs were matured and cultured under the conditions previously described. However, instead of snap freezing in liquid nitrogen, the cells were fixed in 4% paraformaledhyde (PFA) for 30 min at 37°C for GCs and for 1 h at Room Temp (RT) for COCs. Cells were then washed 2X in PBS before being permeabilized in 0.1% Triton X-100 with 5% BSA for 1 h at RT. Primary antibodies were prepared in the permeabilization buffer at 1:100 for both 5 mC (ab10805) and 5 hmC (ab214728) antibodies. Cells were then incubated in the primary antibodies for 1 h at RT and then washed 3X in PBS to remove unbound primary antibodies, followed by incubations in the alexa-fluor 488 conjugated secondary antibodies [Anti-Mouse for 5 mC (ab150113) and Anti-Rabbit for 5 hmC (ab150077)] for 45 min at RT in the dark. Negative controls were included for each antibody and each sample type by replacing the primary antibody with an Isotype control. The Mouse IgG1 Isotype (ab170190) was used for 5 mC and the Rabbit IgG Isotype (ab172730) was used for 5 hmC as negative controls. Following incubation in the secondary antibodies, samples were washed 3X in PBS.
GCs were then resuspended in PBS with 5% BSA, strained through 40 μM cell strainers, and run through the BD Accuri C6 Flow Cytometer to quantify the fluorescence, which was analyzed using FlowJo V10 on a minimum of 3 biological replicates. COCs were mounted onto slides and mixed with Dakocytomation. Slides were then sealed and visualized using an Olympus FV1200 Confocal Microscope. Images were then analyzed using ImageJ and corrected total cell fluorescence was calculated according to the following formula: [CTCF = Integrated Density—(Area of Selected cell X Mean Area of Background Fluorescence)]. This analysis was performed on 15 individual COCs per group for 9 groups resulting in 135 COCs analyzed for 5mC and 135 COCs for 5 hmC.
Total RNA was isolated using the miRNeasy Micro Kit (Qiagen, Toronto, ON, Canada) according to the manufacturer’s instructions from a minimum of three biological replicates for both COCs and GCs. RNA concentration and quality were measured using the Nanodrop 2000c (ThermoFisher). 1 μg of mRNAs and 0.2 μg of miRNAs were reverse transcribed using qScript complementary DNA (cDNA) Supermix (Quantabio, Beverly Hills, MA, United States) and miRCURY LNA RT kit (Qiagen), respectively, in a T100 Thermal Cycler (BioRad, Mississauga, ON, Canada). cDNA was diluted with RNase-free water to a concentration of 5 ng/μL for mRNAs and the miRNAs were diluted 1:60 as per manufacturer guidelines for qPCR amplification.
mRNA and miRNAs expression levels of a minimum of three biological replicates were quantified via quantitative real-time PCR (qPCR) using a CFX96 Touch Real-Time PCR Detection System (BioRad). mRNA was amplified using the SsoFast EvaGreen Supermix (BioRad), while miRNAs were amplified using the miRCURY LNA SYBR Green PCR Kit (Qiagen). Primers for mRNAs were purchased from Sigma-Aldrich and primers for miRNAs were purchased from Qiagen as predesigned primers from the miRCURY LNA miRNA PCR Assays. All primers were tested using standard curves with efficiencies accepted only with values between 90% and 110%. Gene expression was calculated using the efficiency-corrected method (ΔΔCt). Primer sequences and efficiencies can be found in Table 1. mRNA expression was normalized to housekeeping genes Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), and Beta-actin (ACTB), as they were determined to be the most stable reference genes according to a GeNorm Analysis using the CFX Maestro Software 2.3 (Sabry et al., 2022). miRNA expression was normalized to miR-191 and miR-106a, as they are stable reference targets across treatments. Quantification was run on at least three biological replicates in technical triplicates.
miRNA PCR signal acquisition was carried out using the following two-step PCR cycling protocol: 95°C for 2 min followed by 40 cycles of 95°C for 10 s and 56°C for 60 s, ending with melt curve acquisition from 60°C to 95°C. mRNA PCR signal acquisition was carried out using the following two-step PCR cycling protocol: 95°C for 2 min followed by 44 cycles of 95°C for 10 s, 60°C for 30 s, ending with melt curve acquisition from 60°C to 95°C.
All Western blotting buffers and reagents were made in-house unless otherwise specified.
Quantification of DNMT1, DNMT3A, and TET2 proteins were performed by Western blotting on a minimum of 3 biological replicates in both GCs and COCs. Samples were lysed in 50 μL radioimmunoprecipitation assay (RIPA) buffer and 1% (v/v) protease inhibitors (Biotool, Jupiter, FL, USA), followed by freeze–thaw cycles in liquid nitrogen. Samples were then sheared using 0.33 mm (29 G) syringes (BD Biosciences) to break down the clumping of genomic DNA, placed in a water bath sonicator for 30 min followed by centrifugation at 12,000 × g RPM at 4°C for 10 min. Protein concentrations were quantified using the Bio-Rad DC protein assay (BioRad) and 40 and 30 μg of proteins for GCs and COCs, respectively, were loaded onto gels. Equal volumes of 3 ×reducing buffer with β-mercaptoethanol (Sigma-Aldrich) were added to each sample. Polyacrylamide gels (8%) were prepared using Bio-Rad standard gel recipes.
Proteins were heated for denaturation, separated on the 8% gels in an Invitrogen wet transfer Western blot apparatus (Invitrogen, Burlington, ON, Canada) at 125 V for 2 h and then transferred (40 V for 2 h) onto nitrocellulose membranes (Biorad) using a transfer buffer of Tris, Glycine, and water. Nitrocellulose blots were washed in Tris-buffered saline pH 7.6 with 0.1% Tween 20 (Thermo Fisher Scientific, Whitby, ON, Canada) (TBST), blocked for 1 h in 5% Bovine Serum Albumin (BSA) (Sigma-Aldrich) for TET2 and 5% Skim milk for DNMT1 and DNMT3A in TBST, to limit nonspecific binding. Blots were then incubated with each target primary antibody at 4°C overnight: DNMT1 at 1:500 (Cell Signaling D63A6, mAb#5032), DNMT3A at 1:1000 (ab228691), and TET2 at 1:500 (Cell Signaling, #45010).
After TBST washes, blots were incubated with the anti-rabbit IgG HRP-linked secondary antibody (Cell signaling Technology; 70,745) at 1:2000 dilution for DNMT1, 1:5000 for DNMT3A, and 1:1000 for TET2. All secondary antibodies were left on blots for 1 h at room temp and incubated with Clarity Western ECL Blotting Substrate (Bio-Rad) for 2–3 min. Blots were imaged on a ChemiDoc XRS + Imaging System (Bio-Rad). α-tubulin (Cell Signalling Technology) was used as a loading control and densitometric analysis was performed using the Bio-Rad Image Lab software and quantified as a ratio to α-tubulin expression.
GraphPad Prism 6 software was used to analyze the statistical difference among the treatment groups. Each data set was tested for normality using the Shapiro Wilk test for Normality. Normally distributed data sets were analyzed using a one-way Analysis of Variance (ANOVA) and non-parametric distributed data sets were analyzed using the Kruskal–Wallis test. Differences at a two-tailed p-value ≤0.05 were considered statistically significant. Parametric tests, specifically a one-way ANOVA, were suitable for normally distributed data as this test assumes homogeneity of variances and as such is sensitive to deviations from normality. When these assumptions are met, parametric tests offer greater statistical power and precision. Furthermore, a normally distributed data set that was found to have significant differences between the means of our tested variables was then subjected to a Tukeys Post-hoc test to establish which specific group differences are statistically significant. It helps identify which pairs of groups have means that differ significantly from each other after considering all the groups simultaneously. These differences are reported in the results section and in the graphs are denoted as different letters.
Non-parametric tests, like the Kruskal–Wallis test, are used for non-normally distributed data or data that does not meet the assumptions of parametric tests. They do not rely on the same distributional assumptions, which makes them more robust for skewed data and can also be useful when working with small sample sizes. Any data set that did not comply with parametric assumptions and found to have significant differences between the mean were also subjected to Dunn’s Multiple comparison tests which serves to perform post hoc pairwise comparisons between multiple groups in the context of non-parametric data. The data shown represent the mean ± standard error of the mean (SEM) for the biological replicates and statistical differences were determined at a two-tailed p-value ≤0.05, therefore, any differences with p ≤ 0.05 were considered significant.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because This study uses granulosa cells and COCs derived from ovaries obtained from Cargill Meat Solutions (Guelph, ON). The Animal Care Committee, which reviews the ethical implications of animal research taking place at the University of Guelph, did not require the study to be reviewed or approved by an ethics committee because these materials are a by-product of routine slaughterhouse procedures. Ethics approval is only required for live animal research that falls under categories B-E according to the Canadian Council on Animal Care “Categories of Invasiveness in Animal Experiments.” The current study includes the use of isolated live cells (Category A) which does not require ethics approval.
RS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing–original draft, Writing–review and editing. DM: Data curation, Investigation, Writing–review and editing. LF: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) (Grant #401511 to LF) and a General Purpose Grant (Grant #072049 to LF), the Ontario Veterinary College (OVC) and the Ontario Graduate Scholarships (OGS) to RS in the Department of Biomedical Sciences at the University of Guelph.
The authors would like to acknowledge all members of the Reproductive Health and Biotechnology laboratory in the Department of Biomedical Sciences at the University of Guelph for all their support and stimulating discussions. Special mention to the support of the Lab Manager, Dr. Monica Antenos, and research technicians Liz St. John and Allison MacKay. The authors would also like to thank Cargill Meat Solutions in Guelph, ON, Canada for supplying the bovine ovaries from which the bovine oocytes and granulosa cells are obtained to perform this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1294541/full#supplementary-material
Aldakheel, F. M., Abuderman, A. A., Alduraywish, S. A., Xiao, Y., and Guo, W. W. (2021). MicroRNA-21 inhibits ovarian granulosa cell proliferation by targeting SNHG7 in premature ovarian failure with polycystic ovary syndrome. J. Reprod. Immunol. 146, 103328. doi:10.1016/J.JRI.2021.103328
Awada, Z., Nasr, R., Akika, R., Cahais, V., Cuenin, C., Zhivagui, M., et al. (2019). DNA methylome-wide alterations associated with estrogen receptor-dependent effects of bisphenols in breast cancer. Clin. Epigenetics 11, 138–217. doi:10.1186/s13148-019-0725-y
Bartolucci, A. F., Uliasz, T., and Peluso, J. J. (2020). MicroRNA-21 as a regulator of human cumulus cell viability and its potential influence on the developmental potential of the oocyte. Biol. Reprod. 103, 94–103. doi:10.1093/BIOLRE/IOAA058
Bhandari, R. K., Taylor, J. A., Sommerfeld-Sager, J., Tillitt, D. E., Ricke, W. A., and Vom Saal, F. S. (2019). Estrogen receptor 1 expression and methylation of Esr1 promoter in mouse fetal prostate mesenchymal cells induced by gestational exposure to bisphenol A or ethinylestradiol. Environ. Epigenet 5, dvz012–8. doi:10.1093/EEP/DVZ012
Bhaskaran, M., and Mohan, M. (2014). MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 51, 759–774. doi:10.1177/0300985813502820
Cao, L. Q., Yang, X. W., Chen, Y. B., Zhang, D. W., Jiang, X. F., and Xue, P. (2019). Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer 18, 148–214. doi:10.1186/s12943-019-1075-2
Chao, H. H., Zhang, X. F., Chen, B., Pan, B., Zhang, L. J., Li, L., et al. (2012). Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem Cell. Biol. 137, 249–259. doi:10.1007/S00418-011-0894-Z
Coticchio, G., Dal Canto, M., Renzini, M. M., Guglielmo, M. C., Brambillasca, F., Turchi, D., et al. (2015). Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum. Reprod. Update 21, 427–454. doi:10.1093/HUMUPD/DMV011
Dehghan, Z., Mohammadi-Yeganeh, S., Rezaee, D., and Salehi, M. (2021). MicroRNA-21 is involved in oocyte maturation, blastocyst formation, and pre-implantation embryo development. Dev. Biol. 480, 69–77. doi:10.1016/J.YDBIO.2021.08.008
Fabbri, M., Garzon, R., Cimmino, A., Liu, Z., Zanesi, N., Callegari, E., et al. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104, 15805–15810. doi:10.1073/PNAS.0707628104
Fedoriw, A., Mugford, J., and Magnuson, T. (2012). Genomic imprinting and epigenetic control of development. Cold Spring Harb. Perspect. Biol. 4, 0081366–a8215. doi:10.1101/CSHPERSPECT.A008136
Felsenfeld, G. (2014). A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 6, a018200. doi:10.1101/CSHPERSPECT.A018200
Ferris, J., Mahboubi, K., MacLusky, N., King, W. A., and Favetta, L. A. (2016). BPA exposure during in vitro oocyte maturation results in dose-dependent alterations to embryo development rates, apoptosis rate, sex ratio and gene expression. Reprod. Toxicol. 59, 128–138. doi:10.1016/J.REPROTOX.2015.12.002
Fischer, S., Handrick, R., Aschrafi, A., and Otte, K. (2015). Unveiling the principle of microRNA-mediated redundancy in cellular pathway regulation. RNA Biol. 12, 238–247. doi:10.1080/15476286.2015.1017238
Gao, L., Emperle, M., Guo, Y., Grimm, S. A., Ren, W., Adam, S., et al. (2020). Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 11, 3355. doi:10.1038/s41467-020-17109-4
Gulyaeva, L. F., and Kushlinskiy, N. E. (2016). Regulatory mechanisms of microRNA expression. J. Transl. Med. 14, 143. doi:10.1186/S12967-016-0893-X
Han, X., Xue, R., Yuan, H. J., Wang, T. Y., Lin, J., Zhang, J., et al. (2017). MicroRNA-21 plays a pivotal role in the oocyte-secreted factor-induced suppression of cumulus cell apoptosis. Biol. Reprod. 96, 1167–1180. doi:10.1093/BIOLRE/IOX044
Huumonen, K., Korkalainen, M., Viluksela, M., Lahtinen, T., Naarala, J., and Juutilainen, J. (2014). Role of microRNAs and DNA methyltransferases in transmitting induced genomic instability between cell generations. Front. Public Health 2, 139. doi:10.3389/fpubh.2014.00139
Ivanova, E., Canovas, S., Garcia-Martínez, S., Romar, R., Lopes, J. S., Rizos, D., et al. (2020). DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 12, 64. doi:10.1186/S13148-020-00857-X
Jacobs, M. N., Marczylo, E. L., Guerrero-Bosagna, C., and Rüegg, J. (2017). Marked for life: epigenetic effects of endocrine disrupting chemicals. Annu. Rev. Environ. Resour. 42, 105–160. doi:10.1146/ANNUREV-ENVIRON-102016-061111
Jin, B., Li, Y., and Robertson, K. D. (2011). DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes. Cancer 2, 607–617. doi:10.1177/1947601910393957
Kahlon, N. (2016). The effects of oocyte exposure to bisphenol A on early bovine embryo development. Thesis. Guelph: University of Guelph.
Khaghani, A. J., Farrokh, P., and Zavareh, S. (2021). Epigenetic effects of Bisphenol A on granulosa cells of mouse follicles during in vitro culture: an experimental study. Int. J. Reprod. Biomed. 19, 129–136. doi:10.18502/IJRM.V19I2.8471
Kim, J. H., Rozek, L. S., Soliman, A. S., Sartor, M. A., Hablas, A., Seifeldin, I. A., et al. (2013). Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah, Egypt. Environ. Health 12, 33. doi:10.1186/1476-069X-12-33
Klinge, C. (2009). Estrogen regulation of MicroRNA expression. Curr. Genomics 10, 169–183. doi:10.2174/138920209788185289
Kochmanski, J. J., Marchlewicz, E. H., Cavalcante, R. G., Perera, B. P. U., Sartor, M. A., and Dolinoy, D. C. (2018). Longitudinal effects of developmental bisphenol A exposure on epigenome-wide DNA hydroxymethylation at imprinted loci in mouse blood. Environ. Health Perspect. 126, 077006. doi:10.1289/EHP3441
Li, Z., Lyu, C., Ren, Y., and Wang, H. (2020). Role of TET dioxygenases and DNA hydroxymethylation in bisphenols-stimulated proliferation of breast cancer cells. Environ. Health Perspect. 128, 27008. doi:10.1289/EHP5862
Liao, J., Karnik, R., Gu, H., Ziller, M. J., Clement, K., Tsankov, A. M., et al. (2015). Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 47, 469–478. doi:10.1038/ng.3258
Lin, X. H., Li, D. P., Liu, Z. Y., Zhang, S., Tang, W., Xin, C. R., et al. (2023). Six immune-related promising biomarkers may promote hepatocellular carcinoma prognosis: a bioinformatics analysis and experimental validation. Cancer Cell. Int. 23, 52–20. doi:10.1186/s12935-023-02888-9
Liu, Y., Beyer, A., and Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell. 165, 535–550. doi:10.1016/J.CELL.2016.03.014
Lujambio, A., Calin, G. A., Villanueva, A., Ropero, S., Sánchez-Céspedes, M., Blanco, D., et al. (2008). A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U. S. A. 105, 13556–13561. doi:10.1073/PNAS.0803055105
Malloy, M. A., Kochmanski, J. J., Jones, T. R., Colacino, J. A., Goodrich, J. M., Dolinoy, D. C., et al. (2019). Perinatal bisphenol A exposure and reprogramming of imprinted gene expression in the adult mouse brain. Front. Genet. 10, 951. doi:10.3389/fgene.2019.00951
Misirlioglu, M., Page, G. P., Sagirkaya, H., Kaya, A., Parrish, J. J., First, N. L., et al. (2006). Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl. Acad. Sci. U. S. A. 103, 18905–18910. doi:10.1073/PNAS.0608247103
Mondou, E., Dufort, I., Gohin, M., Fournier, E., and Sirard, M. A. (2012). Analysis of microRNAs and their precursors in bovine early embryonic development. Mol. Hum. Reprod. 18, 425–434. doi:10.1093/MOLEHR/GAS015
O’doherty, A. M., O’shea, L. C., and Fair, T. (2012). Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins. Biol. Reprod. 86, 67. doi:10.1095/BIOLREPROD.111.094946
Pan, W., Zhu, S., Yuan, M., Cui, H., Wang, L., Luo, X., et al. (2010). MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 184, 6773–6781. doi:10.4049/JIMMUNOL.0904060
Ramassone, A., Pagotto, S., Veronese, A., and Visone, R. (2018). Epigenetics and MicroRNAs in cancer. Int. J. Mol. Sci. 19, 459. doi:10.3390/IJMS19020459
Sabry, R. (2019). The effects of BPA and BPS on microRNAs during bovine oocyte maturation and early embryo development. Thesis. Guelph: University of Guelph.
Sabry, R., Saleh, A. C., Stalker, L., LaMarre, J., and Favetta, L. A. (2021). Effects of bisphenol A and bisphenol S on microRNA expression during bovine (Bos taurus) oocyte maturation and early embryo development. Reprod. Toxicol. 99, 96–108. doi:10.1016/J.REPROTOX.2020.12.001
Sabry, R., Williams, M., Werry, N., LaMarre, J., and Favetta, L. A. (2022). BPA decreases PDCD4 in bovine granulosa cells independently of miR-21 inhibition. Int. J. Mol. Sci. 23, 8276. doi:10.3390/IJMS23158276
Sagvekar, P., Kumar, P., Mangoli, V., Desai, S., and Mukherjee, S. (2019). DNA methylome profiling of granulosa cells reveals altered methylation in genes regulating vital ovarian functions in polycystic ovary syndrome. Clin. Epigenetics 11, 61. doi:10.1186/S13148-019-0657-6
Sakashita, A., Kobayashi, H., Wakai, T., Sotomaru, Y., Hata, K., and Kono, T. (2014). Dynamics of genomic 5-hydroxymethylcytosine during mouse oocyte growth. Genes. Cells 19, 629–636. doi:10.1111/GTC.12164
Santangeli, S., Maradonna, F., Gioacchini, G., Cobellis, G., Piccinetti, C. C., Dalla Valle, L., et al. (2016). BPA-induced deregulation of epigenetic patterns: effects on female zebrafish reproduction. Sci. Rep. 6, 21982. doi:10.1038/srep21982
Sendžikaitė, G., and Kelsey, G. (2019). The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 63, 691–705. doi:10.1042/EBC20190043
Senyildiz, M., Karaman, E. F., Bas, S. S., Pirincci, P. A., and Ozden, S. (2017). Effects of BPA on global DNA methylation and global histone 3 lysine modifications in SH-SY5Y cells: an epigenetic mechanism linking the regulation of chromatin modifiying genes. Toxicol. Vitro 44, 313–321. doi:10.1016/J.TIV.2017.07.028
Sharma, J., and Madan, P. (2022). Differential regulation of Hippo signaling pathway components between 8-cell and blastocyst stages of bovine preimplantation embryogenesis. Mol. Reprod. Dev. 89, 146–161. doi:10.1002/MRD.23564
Shimizu, T., Sohn, Y., Choi, E., Petersen, C. P., Prasad, N., and Goldenring, J. R. (2020). Decrease in MiR-148a expression during initiation of chief cell transdifferentiation. Cell. Mol. Gastroenterol. Hepatol. 9, 61–78. doi:10.1016/J.JCMGH.2019.08.008
Suzuki, H., Yamamoto, E., Nojima, M., Kai, M., Yamano, H. O., Yoshikawa, K., et al. (2010). Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 31, 2066–2073. doi:10.1093/CARCIN/BGQ203
Toms, D., Pan, B., and Li, J. (2018). Endocrine regulation in the ovary by MicroRNA during the estrous cycle. Front. Endocrinol. (Lausanne) 8, 8. doi:10.3389/fendo.2017.00378
Toranõ, E. G., Garciá, M. G., Fernández-Morera, J. L., Ninõ-Garciá, P., and Fernández, A. F. (2016). The impact of external factors on the epigenome: in utero and over lifetime. Biomed. Res. Int. 2016, 2568635. doi:10.1155/2016/2568635
Trapphoff, T., Heiligentag, M., El Hajj, N., Haaf, T., and Eichenlaub-Ritter, U. (2013). Chronic exposure to a low concentration of bisphenol A during follicle culture affects the epigenetic status of germinal vesicles and metaphase II oocytes. Fertil. Steril. 100, 1758–1767. doi:10.1016/J.FERTNSTERT.2013.08.021
Tscherner, A., Brown, A. C., Stalker, L., Kao, J., Dufort, I., Sirard, M. A., et al. (2018). STAT3 signaling stimulates miR-21 expression in bovine cumulus cells during in vitro oocyte maturation. Sci. Rep. 8 (8), 11527. doi:10.1038/s41598-018-29874-w
Wang, S., Wu, W., and Claret, F. X. (2017). Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 12, 187–197. doi:10.1080/15592294.2016.1273308
Wang, T., Han, J., Duan, X., Xiong, B., Cui, X. S., Kim, N. H., et al. (2016). The toxic effects and possible mechanisms of Bisphenol A on oocyte maturation of porcine in vitro. Oncotarget 7, 32554–32565. doi:10.18632/oncotarget.8689
Yerushalmi, G. M., Salmon-Divon, M., Ophir, L., Yung, Y., Baum, M., Coticchio, G., et al. (2018). Characterization of the miRNA regulators of the human ovulatory cascade. Sci. Rep. 8, 15605. doi:10.1038/S41598-018-33807-Y
Zhang, J., Hao, L., Wei, Q., Zhang, S., Cheng, H., Zhai, Y., et al. (2020). TET3 overexpression facilitates DNA reprogramming and early development of bovine SCNT embryos. Reproduction 160, 379–391. doi:10.1530/REP-20-0021
Zhang, Z., Cao, Y., Zhai, Y., Ma, X., An, X., Zhang, S., et al. (2018). MicroRNA-29b regulates DNA methylation by targeting Dnmt3a/3b and Tet1/2/3 in porcine early embryo development. Dev. Growth Differ. 60, 197–204. doi:10.1111/DGD.12537
Zheng, H., Zhou, X., Li, D. K., Yang, F., Pan, H., Li, T., et al. (2017). Genome-wide alteration in DNA hydroxymethylation in the sperm from bisphenol A-exposed men. PLoS One 12, e0178535. doi:10.1371/JOURNAL.PONE.0178535
Zhou, S., bo, Y. W., Zhi, L. J., Qiang, C. H., Zeng, Y., Wang, N., et al. (2023). TET1 involved in bisphenol A induced TM3 Leydig cell toxicity by regulating Cav3.3 hydroxymethylation. Chemosphere 312, 137171. doi:10.1016/J.CHEMOSPHERE.2022.137171
Keywords: miR-21, DNA methylation, bisphenol A, oocytes, granulosa cells
Citation: Sabry R, May DR and Favetta LA (2023) The relationship between miR-21, DNA methylation, and bisphenol a in bovine COCs and granulosa cells. Front. Cell Dev. Biol. 11:1294541. doi: 10.3389/fcell.2023.1294541
Received: 14 September 2023; Accepted: 31 October 2023;
Published: 15 November 2023.
Edited by:
Giovanni Nassa, University of Salerno, ItalyReviewed by:
Fahiel Casillas, Metropolitan Autonomous University, MexicoCopyright © 2023 Sabry, May and Favetta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura A. Favetta, bGZhdmV0dGFAdW9ndWVscGguY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.