
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 07 November 2023
Sec. Embryonic Development
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1273006
Neuropeptide Y receptor Y2 (npy2r) is an important receptor gene involved in anxiety and feeding regulation in mammals. Since NPY receptors have different receptor gene deletions in mammals and teleost fish, it is not clear whether npy2r has the similar function in fish as in mammals. In this study, we used the CRISPR/Cas9 system to establish npy2r-deficient medaka (Oryzias latipes). Unexpectedly, the deletion of npy2r resulted in the npy2r+/− medaka were all-male, therefore, npy2r homozygous mutant lines could not be established. The deletion of npy2r increased the food intake in medaka, and the expression levels of appetite stimulating genes (agrp, npy) increased significantly, while the expression levels of anorexia factors (cck, pomc) decreased significantly. Moreover, the absence of npy2r significantly increased the total length and body weight of medaka. The mirror test and open field test showed that npy2r+/− medaka improved sociability and reduced anxiety-like behavior, qRT-PCR analysis showed that the expression levels of anxiety related genes (th1, th2, gr1, gr2, and mr) in npy2r+/− medaka were significantly decreased. So far, this is the first npy2r gene knockout model established in fish and demonstrates that npy2r plays an important role in the regulation of reproduction, feeding and anxiety in fish.
As an important neurotransmitter, Neuropeptide Y (NPY) was first identified in the pig brain (Tatemoto et al., 1982), and it is widely distributed in the central nervous system (Loh et al., 2015) and peripheral nervous system (Kuo et al., 2007) of vertebrates. By binding to different subtype receptors, NPY activates signal transduction pathways, which can regulate feeding (Matsuda et al., 2012), energy balance (Loh et al., 2015), anxiety (Kask et al., 2002; Shiozaki et a., 2020), circadian rhythm (Harrington et al., 2007) and other physiological processes. NPY receptors (NPYRs) belong to the G protein-coupled receptor family (Gehlert, 2004), at present, five receptor gene subtypes have been identified in mammals, namely npy1r, npy2r, npy4r, npy5r, and npy6r (Wraith et al., 2000). Based on pharmacological data of human and mouse cardiomyocytes, it is speculated that npy3r may exist, but it has not been cloned and characterized in any vertebrate, so it cannot exist as an independent gene (Lee and Miller, 1998; Larhammar et al., 2001; Pedragosa-Badia et al., 2013). In addition to the five known receptors, npy7r and npy8r were also identified in teleost fishes, while npy7r may have been lost in mammals (Larhammar and Salaneck, 2004; Sundström et al., 2013; Zhou et al., 2013; Wang et al., 2019). In particular, npy8r has two subtypes, npy8ar and npy8br respectively, which has been reported in zebrafish (Sundström et al., 2013), orange spotted grouper (Wang et al., 2014) and Chinese perch (Zhang et al., 2021).
According to amino acid sequence homology and functional characteristics, NPYRs can be divided into Y1 subfamily (NPY1R, NPY4R, NPY6R, and NPY8R), Y2 subfamily (NPY2R, NPY7R) and Y5 subfamily (NPY5R) (Larhammar and Salaneck, 2004). Different receptors regulate different functions, in npy receptor knockout mice, npy1r and npy5r knockout mice exhibited decreased food intake and body weight. npy4r knockout mice showed that npy4r promotes obesity induced by high fat diet (Wang et al., 2023). Deletion of npy6r resulted in body weight loss and late-life obesity in mice (Yulyaningsih et al., 2014). Injection of siRNA-npy8br into Chinese perch’s ventricle indicates that npy8br plays an important role in appetite regulation (Zhang et al., 2021).
At present, npy2r has been cloned and characterized in mammals including human (Larhammar et al., 1992; Gerald et al., 1995), mouse (Nakamura et al., 1996), pig (Wraith et al., 2000) and some teleost fishes such as rainbow trout (Larsson et al., 2006), orange-spotted grouper (Wang et al., 2014) and large yellow croaker (Wang et al., 2019). Immunohistochemical localization revealed that NPY2R is presynaptic receptor and is expressed in many regions of the amygdala in mouse (Stanić et al., 2011). In fish, it has been confirmed that zebrafish NPY2R is homologous to mammals and is most similar in pharmacology to chicken (Wang et al., 2019). Studies have shown that npy2r knockout mice gain weight, increase food intake, and increase fat deposition (Naveilhan et al., 1999). Through maze experiment, open field and light/dark test, it has been demonstrated that npy2r plays an important role in anxiety and stress-related behavior in mice (Tschenett et al., 2003). It has been shown that loss of npy2r in the hippocampus of mice decreases episodic fear memory, but improves working memory and spatial memory (Tschenett et al., 2003; Hörmer et al., 2018). Studies have shown that npy2r is highly expressed in the brain, liver and gonads of fish (Larsson et al., 2006; Wang et al., 2014; Wang et al., 2019). However, functional studies of npy2r are mainly focused on mammals such as human and mice, and only a few teleost fish have been preliminarily explored based on pharmacological characteristics, while the physiological function of npy2r in fish has not been investigated.
In order to explore the physiological function of npy2r in fish, in particular, whether fish npy2r also plays an important role in anxious behavior and feeding activity, it is necessary to establish gene knockout models. Japanese medaka (Oryzias latipes) is an important model organism, it has the advantages of transparent embryo, fast growth rate and easy cultivation. In this study, we used the CRISPR/Cas9 technology to knockout the npy2r gene in Japanese medaka, in order to further explore the physiological function of npy2r gene.
The male and female wild-type (WT) Japanese medaka HdrR (orange-red strain) were obtained from the Institute of Hydrobiology, Chinese Academy of Sciences (Wuhan, China). They were bred in the circulating water system of the Research Center of Mandarin Fish in Huazhong Agricultural University with 27°C of room temperature and 14 h of light and 10 h of darkness. Embryos were collected in the morning and incubated at 28°C incubator. The male WT, male npy2r+/− medaka and female npy2r+/+ medaka used in this study were 5 months old.
NCBI (https://www.ncbi.nlm.nih.gov/) and Ensembl (https://asia.ensembl.org/index.html) were used to search the NPY receptors genes and amino acid sequences of Japanese medaka, Human (Homo sapiens), Mouse (Mus musculus), Seabass (Dicentrarchus labrax), Swamp eel (Monopterus albus), Spotted gar (Lepisosteus oculatus), Nile tilapia (Oreochromis niloticus), Zebrafish (Danio rerio), Rainbow trout (Oncorhynchus mykiss), Atlantic cod (Gadus morhua), Chinese perch (Siniperca chuatsi), Japanese flounder (Paralichthys olivaceus), African clawed frog (Xenopus laevis), and Torafugu (Takifugu rubripes). The accession numbers are shown in Supplementary Table S1. Multiple amino acid alignment of npy2r among medaka and other Vertebrate were performed by using Clustal X, and the protein genealogies of NPY receptors were assessed by the Neighbor-Joining using Mega11.
The CCTOP web (https://cctop.cos.uni-heidelberg.de/) was used to design medaka single guide RNA (sgRNA). Two targets on the second exon of medaka npy2r gene were designed. The sgRNAs were then cloned into PMD-19T vector and synthesized using TranscriptAid T7 High Yield Transcription kit (Thermo, Scientific). Medaka embryos at the single-cell stage were microinjected with 2 nL of a mixed solution consisting of 1 μL of sgRNAs (50 ng/μL), 1 μL of Cas9 mRNA (300 ng/μL), 0.5 μL of phenol red indicator, and 2.5 μL of DEPC water. After 24 h of microinjection, genomic DNA was extracted from medaka embryos, and the effectiveness of sgRNAs were tested by PCR and sequencing. The microinjected embryos were cultured to adulthood. The caudal fin genome DNA was extracted, and the F0 mutant line was obtained by PCR amplification and agarose gel electrophoresis. After that, the F0 mutant (male) and WT (female) medaka were crossed to generate F1 heterozygous mutants. The F2 homozygous mutant medaka was obtained by crossing F1 of the same mutant type. The sgRNAs and detection primers of npy2r are shown in Table 1.
In order to identify the genotypic sex of adult medaka, we used DNA extraction kit to extract DNA from the tail fin of medaka according to the manufacturer’s instructions. Sex-specific primers dmrt1bY (5′-AGAGGAGGAGCTTGGGATTTGTAG-3′) and dmrt1a (5′-CAGACGCTTCCTCGCCGTAA-3′) were then used for PCR amplification (Patil and Hinze et al., 2008).
The male WT and male npy2r deficient male medaka were anesthetized with MS222, placed on ice and dissected immediately, and then the removed gonads were placed in fixative for 24 h. The gonads were dehydrated and embedded in paraffin. Paraffin sections were made using a sectioning mechanism and stained with hematoxylin and eosin (H&E).
The male WT, female npy2r+/+ and male npy2r+/− medaka were randomly selected and placed under the same experimental conditions to determine the food intake. Before the experiment, the medaka were starved for 24 h. Before feeding, the medaka were weighed and placed individually in 1-L tank. The medaka were fed an excessive amount of brine shrimp (Artemia nauplii) and allowed to eat freely for 1 h, then the weight of medaka were measured again (Shi et al., 2020). The difference before and after feeding is the food intake of medaka.
The mirror test, or mirror approaching behavior, is a good model for measuring social interaction and social anxiety (Ansai et al., 2016; Lucon-Xiccato et al., 2022). Mirror test was performed with reference to the method already reported (Shi et al., 2020; Shiozaki et al., 2020). In this experiment, a non-transparent rectangular tank (20 × 10 × 10 cm) filled with water was used, and put a mirror on one side. The male WT, female npy2r+/+ and male npy2r+/− medaka were randomly selected, one fish at a time, and slowly placed into the tank for 2 min to adapt to the environment. After that, they were allowed to explore freely without being disturbed (n = 7). The movement trajectory for 10 min was recorded, EthoVision XT software was used to analyze the biting time and swimming distance.
The Open-field test can be used to assess medaka anxiety levels and locomotor activity of medaka (Lucon-Xiccato et al., 2022). In this experiment, a non-transparent rectangular tank (20 × 10 × 10 cm) filled with water was used, and a camera was placed above the tank to record the movement trajectory of medaka. The male WT, female npy2r+/+ and male npy2r+/− medaka were randomly selected, one fish at a time, and slowly placed into the tank for 2 min to adapt to the environment. After that, they were allowed to explore freely without being disturbed (n = 7). The movement trajectory for 10 min was recorded, and EthoVision XT software was used to analyze the swimming distance, movement time and total freezing time.
The expression levels of anxiety and appetite genes were analyzed by real-time quantitative PCR (qRT-PCR) using medaka brain cDNA as template. The eye, brain, gill, heart, kidney, ovary, testis, liver and spleen from medaka were sampled for tissue expression analysis of npy2r. The total RNA was extracted by TRIzol Reagent (Takara, Japan), and then 1 μg RNA was reverse transcribed into cDNA using reverse transcription kit (Vazyme, China). SYBR (Vazyme, China) and th1 (tryptophan hydroxylase1), th2, agrp (agouti-related protein), cck (cholecystokinin), pomc (pro-opiomelanocortin), npy (neuropeptide Y), gr1 (glucocorticoid receptor 1), gr2, mr (mineralocorticoid receptor) genes specific primers were used for qRT-PCR. Table 2 shows the specific primers sequences of npy2r, th1, th2, agrp, cck, pomc, npy, gr1, gr2, and mr genes. The 20 μL RT-qPCR reaction system included 1 μL cDNA, 0.5 μL forward and reserve primers (10 mmol/μL), 10 μL SYBR, and 8 μL double distilled water (ddH2O). The conditions for PCR were as follows: 95°C for 3 min initially, followed by 40 cycles at 95°C for 10 s, 58°C for 30 s and 72°C for 30 s, and melting curve assay from 65°C gradually increasing 0.5°C s−1°C to 95°C, with acquisition data at every 6 s. The mRNA expression levels of target genes were quantified relative to the expression of β-actin using the optimized comparative Ct (2−ΔΔCt) value method (Livak and Schmittgen, 2001).
The SPSS 25.0 software was used for statistical analysis. All data were expressed as means ± S.E.M, and analyzed by independent-samples t-test. GraphPad Prism 8.0.2 used to make data analysis diagram, p < 0.05 and p < 0.01 were considered statistically significant.
Amino acid sequence alignment in vertebrates revealed low conservation of npy2r. The npy2r amino acid sequence of medaka showed 79.2% homology with seabass, 74.9% homology with swamp eel, 55.7% homology with human and 49.2% homology with mouse, respectively (Supplementary Figure S1). Comparing the medaka, npy2r adjacent genes with other vertebrates, synteny analysis showed that the npy2r adjacent genes were highly conserved among the medaka, Chinese perch, Torafugu, Atlantic cod, Rainbow trout and Nile tilapia, but showed different synteny from human and mouse (Figure 1A). The phylogenetic tree showed that medaka npy2r is homologous to human and mouse npy2r, moreover, medaka is more closely related to Torafugu, Chinese perch and Nile tilapia (Figure 1B).
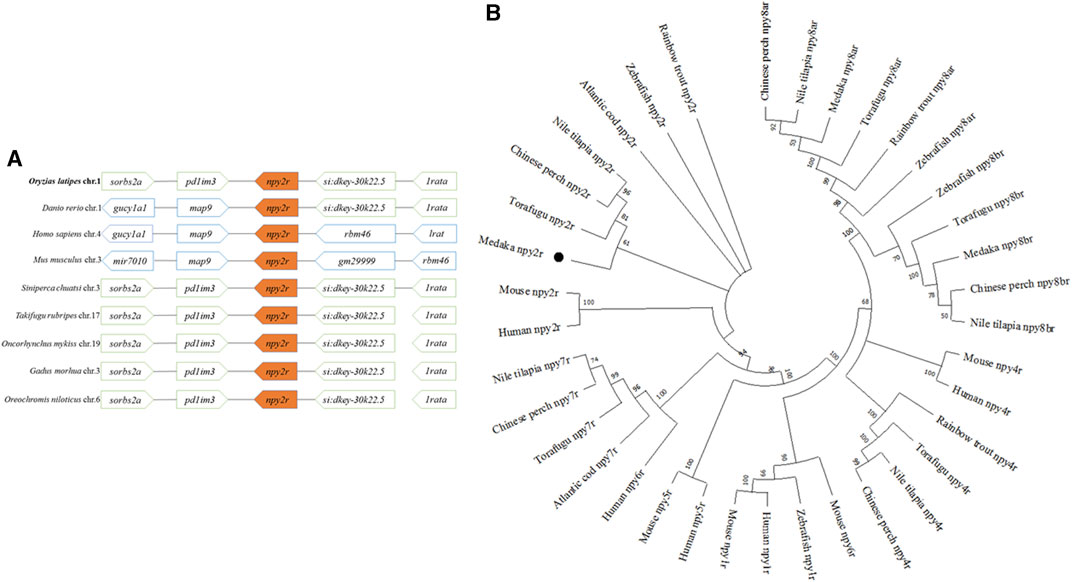
FIGURE 1. Amino Acid Sequence analysis of npy2r. (A): Synteny analyses of npy2r in medaka, zebrafish, human, mouse, Chinese perch, Torafugu, Rainbow trout, Atlantic cod and Nile tilapia. (B): Phylogenetic analysis of npy2r in vertebrates.
The genome target was located in the second exon, and the F0 mutant was obtained by microinjection (Figure 2A). The F1 heterozygote (npy2r+/−) was obtained by PCR and sequencing after hybridization of FO and WT medaka, namely 297 bp deletion, 5 bp addition and 1 bp mutation (Figure 2B). Agarose gel electrophoresis showed that npy2r+/+ had only one bright band at approximately 1,000 bp, while npy2r+/− had three bands (Figure 2C). The additional insertion, deletion, and mutation of the bases resulted in premature termination of npy2r gene translation compared with WT. As a result, the protein only had 44 amino acids and the conserved 7 transmembrane domains of the npy2r protein were completely lost (Figure 2D). The mRNA level of npy2r was significantly decreased in npy2r+/− medaka (Figure 2E).
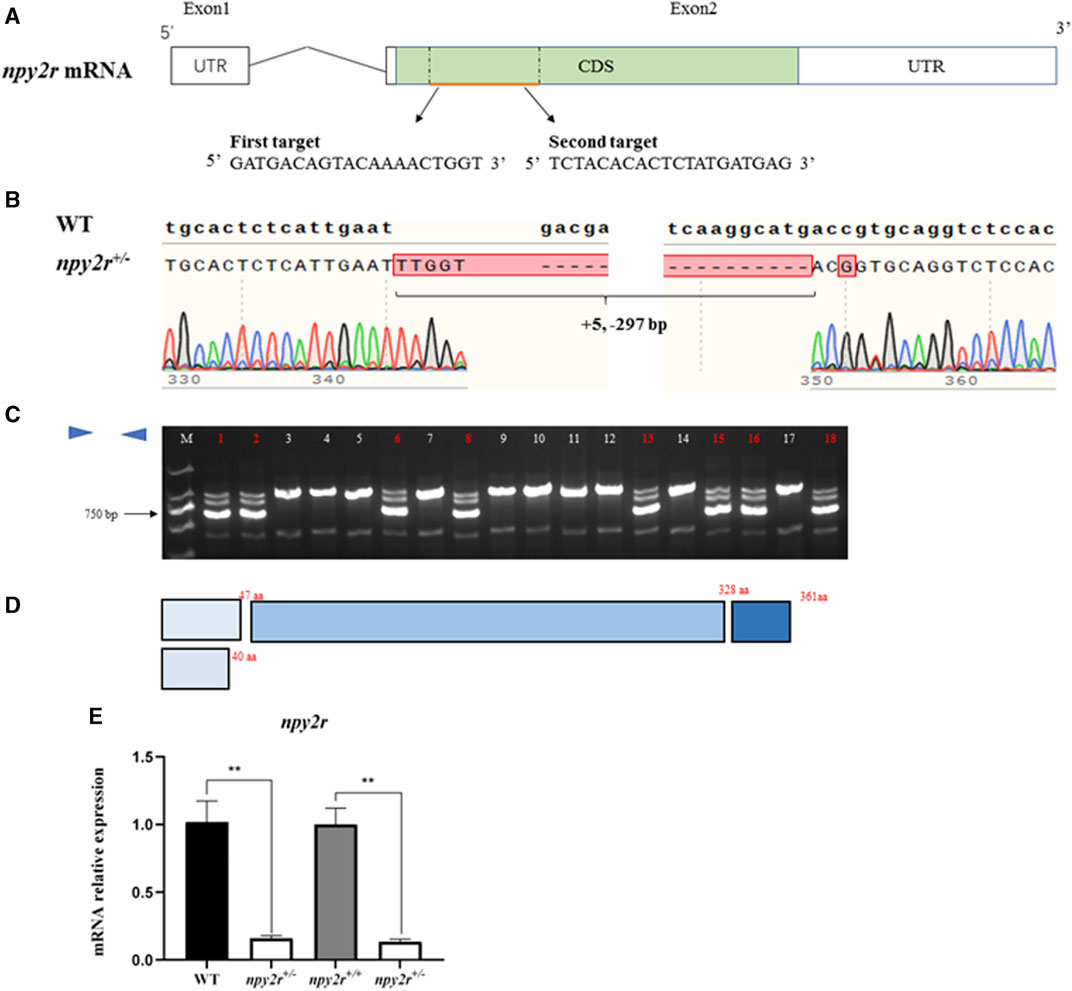
FIGURE 2. Establishment of npy2r mutant line medaka. (A): The npy2r gene structure was composed of two exons (white box) and an intron (black line), in which the mRNA coding sequence (CDS) (green box) is entirely present in exon2. The genomic target of CRISPR/Cas9 is located in the CDS region. (B): The result of WT and npy2r+/− DNA sequencing showed that the mutant had 297 bp deletion, 5 bp addition and 1bp mutation. (C): Agarose gel electrophoresis. M:marker; The lanes marked by red numbers are npy2r+/− and the white numbers are npy2r+/+. (D): Comparison of npy2r+/− mutant with npy2r+/+ protein structure. (E): mRNA expression level of npy2r+/− medaka. * represents significant differences exist between WT and npy2r+/− medaka or npy2r+/+ and npy2r+/− medaka. Data were presented as means ± S.E.M. (n = 6). *p < 0.05, * *p < 0.01.
In the process of creating homozygous mutant, based on the shape of the dorsal fin and anal fin, it was found that the npy2r+/− medaka in F1 generation were exclusively male, while the npy2r+/+ medaka were exclusively female. Therefore, we conducted genetic sex identification to determine whether there was sex reversal in the F1 mutant. The results showed that the genetic sex of F1 medaka were consistent with the physiological sex, that is, there was no sex reversal (Figures 3A, C). Our statistics showed that npy2r+/− medaka accounted for 56.96% and npy2r+/+ medaka accounted for 43.04% in the F1 medaka (Figure 3B). H&E sections of the gonad revealed that spermatogonia, spermatogonia, spermatocytes and spermatozoa developed normally in WT and npy2r+/− medaka (Figure 3D). Tissue expression analysis showed that npy2r was expressed in the gonad of medaka (Figure 3E).
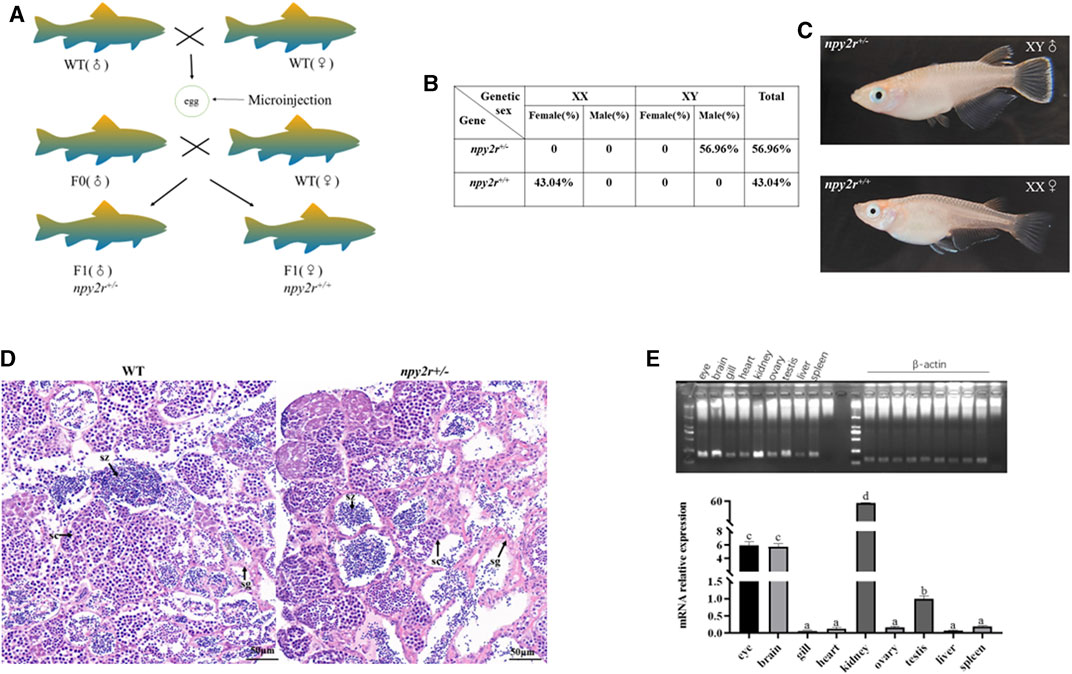
FIGURE 3. npy2r mutant line medaka had sex development disorder. (A) Genotypes and sex types of medaka. (B) The proportion of genetic sex in the F1 generation of medaka, n = 200. (C) Sexual phenotype of F1 generation of medaka. (D): Gonad histology of WT and npy2r+/− medaka. Scale bar: 50 μm. sg, spermatogonia; sc, spermatocytes; sz, spermatozoa. (E): npy2r tissue expression differences were analyzed by polymerase chain reaction (PCR) and qRT-PCR. Data were presented as means ± S.E.M. (n = 6). Different small letters above the bars indicate significant differences at p < 0.05.
The medaka was fed freely for 1 h, the food intake statistics showed that the food intake of npy2r+/− medaka was higher than WT and npy2r+/+ medaka, and there were significantly different between npy2r+/− medaka and both WT and npy2r+/+ medaka (p < 0.05) (Figure 4B). By measuring the growth performance of WT, npy2r+/+ and npy2r+/− medaka, we found that the total length and body weight of npy2r+/− medaka increased significantly (p < 0.01) (Figures 4A, C, D). Compared with npy2r+/+ medaka, the total length of npy2r+/− medaka was significantly increased (p < 0.01), but the body weight was not significantly different between npy2r+/+ and npy2r+/− medaka (p > 0.05) (Figures 4C, D).

FIGURE 4. Growth performance of npy2r deficient medaka. (A):Appearance of WT, npy2r+/+ and npy2r+/− medaka. (B): Food intake of WT, npy2r+/+ and npy2r+/− medaka (WT, n = 9; npy2r+/−, n = 10; npy2r+/+, n = 10). (C): Total length of WT, npy2r+/+ and npy2r+/− medaka. (D): Body weight of WT, npy2r+/+ and npy2r+/− medaka. (n = 15). Data were presented as means ± S.E.M. *p < 0.05, * *p < 0.01, n.s, not significant.
In the mirror test, the medaka will infer the presence of another fish based on the mirror and then display either touching or biting the mirror. Therefore, the social behavior of medaka can be evaluated by the mirror test. In this study, according to the WT, npy2r+/+ and npy2r+/− medaka three-dimensional trajectory graph can clearly see npy2r+/− medaka swam back and forth in one side of the mirror, however, WT and npy2r+/+ medaka swam all over the region (Figures 5A–C). Compared with WT and npy2r+/+ medaka, npy2r+/− medaka significantly increased the total distance travelled and contact time with the mirror during the whole behavior test (p < 0.05) (Figures 5D, E).
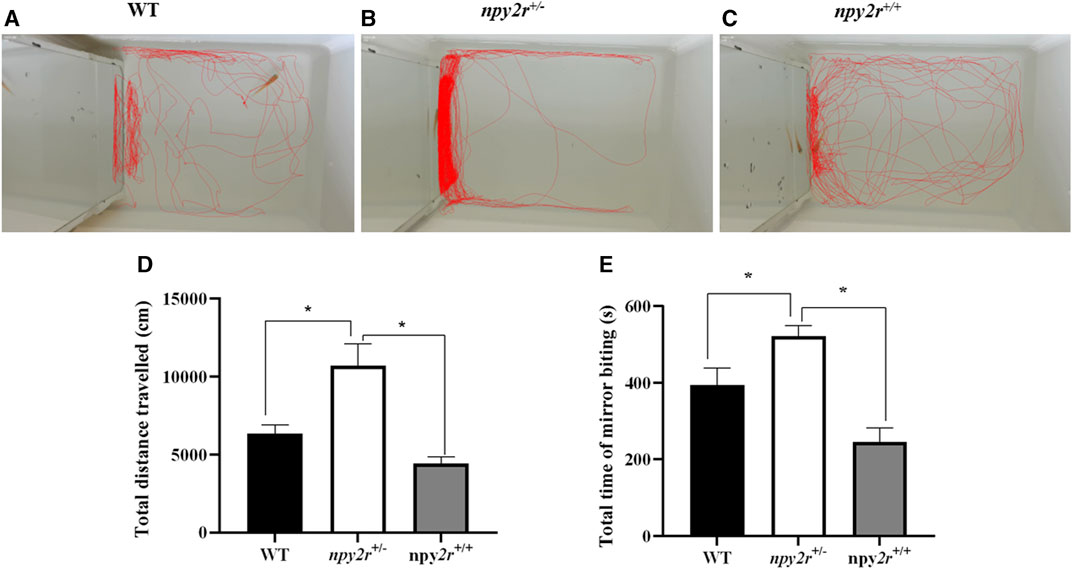
FIGURE 5. Mirror test analysis of npy2r deficient medaka. (A): WT medaka swimming trajectories. (B): npy2r+/+ medaka swimming trajectories. (C): npy2r+/− medaka swimming trajectories. (D): Total distance travelled. (E): Total time of mirror biting. Data were presented as means ± S.E.M. (n = 7). * represents significant differences exist between WT and npy2r+/− medaka or npy2r+/+ and npy2r+/− medaka. p < 0.05.
When placed in a new environment, the medaka display exploratory behavior in both marginal and central areas, thus, it is possible to assess the anxiety of medaka by their locomotor behavior. In open field test, the movement trajectories of WT, npy2r+/+ and npy2r+/− medaka are shown in Figure 4. WT and npy2r+/− medaka are more inclined to explore the edges (Figures 6A–C). Compared with WT and npy2r+/+ medaka, the total distance travelled and movement time of npy2r+/− medaka decreased significantly (p < 0.05) (Figures 6D, E), while the total freezing time was significantly increased (p < 0.05) (Figure 6F).
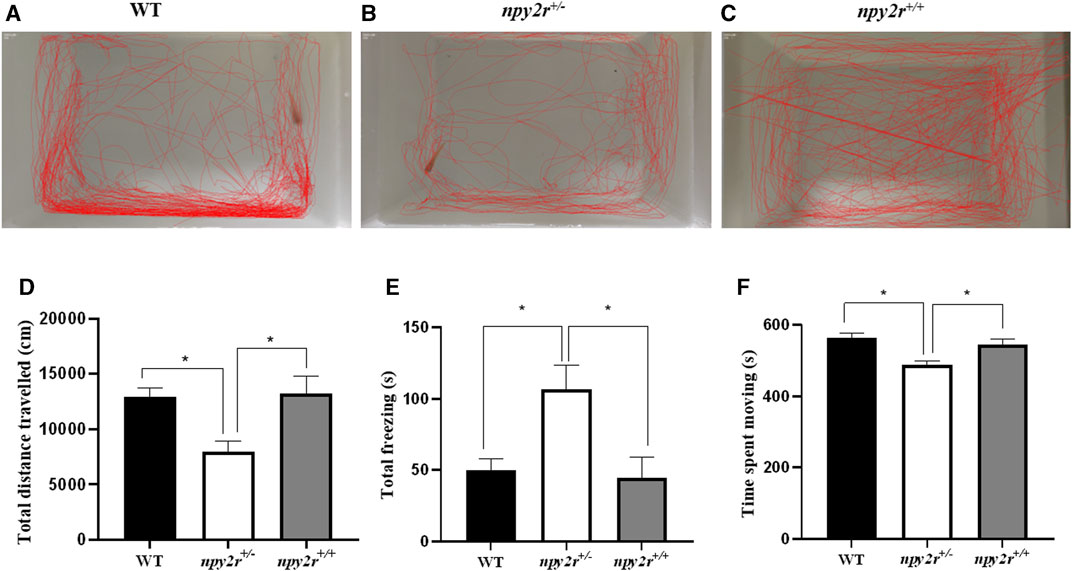
FIGURE 6. Open-field test behavior analysis of npy2r deficient medaka. (A): WT medaka swimming trajectories. (B): npy2r+/− medaka swimming trajectories. (C): npy2r+/+medaka swimming trajectories. (D): Total distance travelled. (E): Total freezing. (F): Time spent moving. Data were presented as means ± S.E.M. (n = 7). * represents significant differences exist between WT and npy2r+/− medaka or npy2r+/+ and npy2r+/− medaka. p < 0.05.
In order to compare the expression patterns of anxiety and appetite related genes of WT, npy2r+/+ and npy2r+/− medaka, we measured the mRNA expression levels of genes associated with anxiety behavior (th1, th2, gr1, gr2, mr), appetitive genes (agrp, npy), and anorexigenic factors (pomc, cck) by qRT-PCR. The results showed that compared with WT, the expression levels of th2, gr1, gr2, cck gene in npy2r+/− medaka were significantly decreased (p < 0.05) (Figures 7B–D, G), agrp, npy expression levels of npy2r+/− medaka were significantly increased (p < 0.05) (Figures 7F, I), and th1, mr, and pomc were not significantly different between WT and npy2r+/− medaka (p > 0.05) (Figures 7A, E, H). Compared with npy2r+/+, the expression levels of th1, th2, gr1, cck and pomc in npy2r+/− medaka were significantly decreased (p < 0.05) (Figures 7A–C, G, H), agrp, npy expression levels of npy2r+/− medaka were significantly increased (p < 0.05) (Figures 7F, I), and gr2, mr and npy were not significantly different between npy2r+/+ and npy2r+/ medaka (p > 0.05) (Figures 7D, E, I).
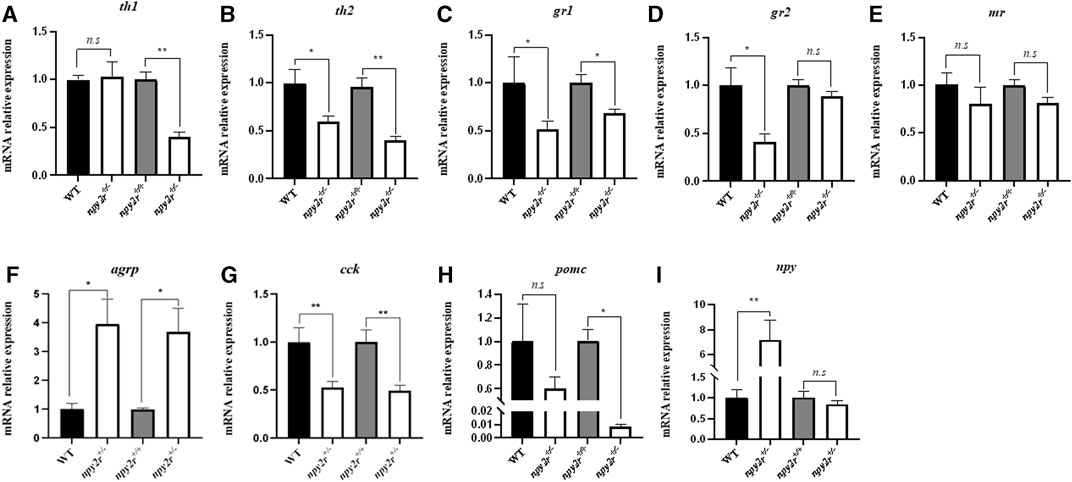
FIGURE 7. The mRNA levels of th1 (A), th2 (B), gr1 (C), gr2 (D), mr (E), agrp (F), cck (G), pomc (H), npy (I) in WT, npy2r+/+ and npy2r+/− medaka. The mRNA levels in npy2r+/− medaka is shown as a relative value to that in WT or npy2r+/+ medaka in each gene analysis. Data were presented as means ± S.E.M. (n = 6). *p < 0.05, **p < 0.01, n.s, not significant.
NPY is composed of 36 highly conserved amino acids and forms the NPY family together with peptide YY and pancreatic peptides (Cerdá-Reverter et al., 2000; Hansel et al., 2001; Conlon, 2002). NPY and its receptors have been studied extensively in the past few decades. The NPY receptor family eventually produced seven receptor subtypes named NPY1R, NPY2R, NPY4R, NPY5R, NPY6R, NPY7R, and NPY8R through two whole-genome duplications in the early stage of vertebrate evolution (Larsson et al., 2009; Sundström et al., 2013; Xu et al., 2015). NPY receptors have different receptor gene deletions in mammals and teleost fish, in this study, the phylogenetic tree showed that medaka npy2r is homologous to human and mouse npy2r.
At present, the establishment of npy2r knockout mice model revealed that npy2r plays an important role in anxiety behavior, feeding activity and learning (Naveilhan et al., 1999). However, it is not clear whether npy2r has the similar function in fish as in mammals, especially the knowledge related to anxiety and feeding. Based on this, we established npy2r mutant medaka by CRISPR/Cas9 system, namely 297 bp deletion, 5 bp addition and 1 bp mutation. The additional insertion, deletion, and mutation of the bases resulted in premature termination of npy2r gene translation compared with WT. As a result, the protein only had 44 amino acids and the conserved 7 transmembrane domains of the npy2r protein were completely lost. Meanwhile, the mRNA level of npy2r was significantly reduced in npy2r+/− medaka. Unexpectedly, during the establishment of npy2r homozygous mutant medaka, we found that npy2r+/− medaka were all-male. Therefore, npy2r+/− medaka could not be produced by self-fertilization of npy2r+/− medaka. H&E sections of gonad showed that the spermatogonia, spermatocytes and spermatozoa of npy2r+/− medaka developed normally and had normal fertility. Tissue expression analysis showed that npy2r was highly expressed in the testis and ovary of medaka. Studies have shown that npy2r may be involved in ovarian metabolism based on the npy2r expression profile and ligand binding characteristics of orange-spotted grouper (Wang et al., 2014). Consistently, tissue expression analysis of npy2r in rainbow trout suggested that npy2r may play a role in reproductive regulation (Larsson et al., 2006). However, no relevant reports were reported in npy2r knockout mice models. Therefore, npy2r knockout may affect the ovarian metabolism in medaka, resulting in the npy2r+/− medaka being all-male.
We investigated the effect of npy2r on food intake and the result showed that the food intake of npy2r+/− medaka increased significantly. Moreover, the total length and body weight were significantly increased compared with WT. The growth pattern of fish is different from higher vertebrates, which continue to grow until they reach a critical size. Studies have shown that fish are one of the animals with the highest efficiency in converting food into body tissue (Talbot, 1993). Therefore, the increase in food intake of npy2r+/− medaka will ultimately be reflected in growth indicators. We detected the expression of appetite genes by qRT-PCR, and the results showed that the knockout of npy2r in medaka significantly increased the expression of appetite stimulating genes (agrp, npy), and significantly decreased the expression of anorexia factors (cck, pomc), which further indicated that npy2r play an important role in feeding regulation of medaka. Previous studies have shown that npy2r knockout mice body weight and food intake were significantly increased (Naveilhan et al., 1999), and our study is consistent with previous research. Based on the exploration of npy2r gene in orange-spotted grouper, it is speculated that npy2r may play an important role in metabolism and energy balance (Wang et al., 2014). Studies have shown that npy2r is widely expressed in the brain, stomach and intestines of large yellow croaker, so npy2r may be involved in food intake and body weight regulation (Wang et al., 2019). Therefore, our research shows that npy2r plays an important role in fish feeding regulation.
Elevated mazes, open fields, light/dark tests are commonly used to evaluate anxiety-like behaviors in rodents such as mice (Bourin et al., 2007; Hurst and West, 2010). Similarly, there are many behavioral tests in fish. Since most animals, such as birds and fish, do not recognize their own mirror images, the mirror test is often used to assess sociability (Cattelan et al., 2017). Studies have shown that the mirror test is a good model for measuring social interaction and social anxiety (Ansai et al., 2016; Lucon-Xiccato et al., 2022). The medaka uses the visual stimulus formed by the mirror response to approach the mirror or bite the mirror. Thus, the sociability of the species can be assessed by the number of times they bite the mirror or the amount of time they spend in the area near the mirror (Lucon-Xiccato et al., 2022). In the present study, as shown by mirror test, npy2r knockout resulted in medaka significantly increased the total distance travelled and contact time with the mirror during the whole behavior test. Therefore, npy2r may play an important role in the social preference and sociability of the species.
The open-field test can be used to reflect the anxiety level and locomotor activity in medaka (Lucon-Xiccato et al., 2022). Therefore, we used the open field to evaluate the anxious behavior of npy2r deficient medaka. The results showed that npy2r knockout caused the total distance travelled and movement time of npy2r+/− medaka decreased, while the total freezing time increased. Previous studies have demonstrated that npy2r plays an important role in anxiety and stress-related behavior in mice using elevated maze, open field, and light/dark tests (Tschenett et al., 2003), and our study is consistent with previous research. Studies have shown that the medaka exhibit an anxious response when placed in a new environment, that is, it may first explore the entire region before showing a preference for edges (Hong and Zha, 2019; Lucon-Xiccato et al., 2022). Medaka will attempt to escape open filed in anxious situations, resulting in vigorous swimming (Lucon-Xiccato et al., 2020). We detected the expression levels of anxiety-related genes (th1, th2, gr1, gr2, mr) by qRT-PCR, the results showed that the expression levels of th1, th2, gr1, gr2, and mr gene in npy2r+/− medaka decreased significantly. Since th1 and th2 catalyze the production of dopamine, excessive exercise may lead to significantly increased expression of th1 and th2 genes in the brain (Otsuka et al., 2022). Studies have shown that the expression levels of th1, th2, gr and mr gene are significantly increased when zebrafish are in a state of anxiety or stress (Shiozaki et al., 2020). Therefore, the present study suggests that npy2r is involved in the regulation of anxiety behavior in medaka.
In conclusion, in this study, we established the npy2r-deficient medaka. It was found that the deletion of npy2r resulted in the npy2r+/− medaka were all-male, therefore, npy2r homozygous mutant lines could not be established. The study of npy2r+/− medaka showed that the absence of npy2r can improve social interaction and reduce anxiety behavior. In addition, the deletion of npy2r gene can promote the feeding and significantly increase the total length and weight in medaka. So far, this is the first npy2r gene knockout model established in fish and demonstrates that npy2r plays an important role in the regulation of reproduction, feeding and anxiety in fish.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author.
The animal study was approved by the Institutional Animal Care and Use Ethics Committee of Huazhong Agricultural University (HZAUFI-2020-0024). The study was conducted in accordance with the local legislation and institutional requirements.
KL: Conceptualization, Data curation, Methodology, Writing–original draft. XJ: Conceptualization, Data curation, Methodology, Writing–original draft. JW: Data curation, Writing–review and editing. QW: Data curation, Writing–review and editing. X-FL: Writing–review and editing, Conceptualization, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Research & Development Program of Hubei Province (2022BBA0051) and the National Natural Science Foundation of China (31972809).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1273006/full#supplementary-material
Ansai, S., Hosokawa, H., Maegawa, S., and Kinoshita, M. (2016). Chronic fluoxetine treatment induces anxiolytic responses and altered social behaviors in medaka, Oryzias latipes. Behav. Brain Res. 303, 126–136. doi:10.1016/j.bbr.2016.01.050
Bourin, M., Petit-Demoulière, B., Nic Dhonnchadha, B., and Hascöet, M. (2007). Animal models of anxiety in mice. Fundam. Clin. Pharmacol. 21 (6), 567–574. doi:10.1111/j.1472-8206.2007.00526.x
Cattelan, S., Lucon-Xiccato, T., Pilastro, A., and Griggio, M. (2017). Is the mirror test a valid measure of fish sociability? Anim. Behav. 127, 109–116. doi:10.1016/j.anbehav.2017.03.009
Cerdá-Reverter, J. M., Martınez-Rodrıguez, G., Zanuy, S., Carrillo, M., and Larhammar, D. (2000). Molecular evolution of the neuropeptide Y (NPY) family of peptides: cloning of three NPY-related peptides from the sea bass (Dicentrarchus labrax). Regul. Pept. 95 (1-3), 25–34. doi:10.1016/s0167-0115(00)00132-4
Conlon, J. M. (2002). The origin and evolution of peptide YY (PYY) and pancreatic polypeptide (PP). Peptides 23 (2), 269–278. doi:10.1016/s0196-9781(01)00608-8
Gehlert, D. R. (2004). Introduction to the reviews on neuropeptide Y. Y. Neuropeptides. 38 (4), 135–140. doi:10.1016/j.npep.2004.07.002
Gerald, C., Walker, M. W., Vaysse, P. J. J., He, C., Branchek, T. A., and Weinshank, R. L. (1995). Expression cloning and pharmacological characterization of a human hippocampal neuropeptide Y/peptide YY Y2 receptor subtype. J. Biol. Chem. 270 (45), 26758–26761. doi:10.1074/jbc.270.45.26758
Harrington, M., Molyneux, P., Soscia, S., Prabakar, C., McKinley-Brewer, J., Lall, G., et al. (2007). Behavioral and neurochemical sources of variability of circadian period and phase: studies of circadian rhythms of npy–/– mice. American Journal of Physiology-Regulatory. Integrative and Comparative Physiology 292 (03), R1306–R1314. doi:10.1152/ajpregu.00383.2006
Hansel, D. E., Eipper, B. A., and Ronnett, G. V. (2001). Neuropeptide Y functions as a neuroproliferative factor. Nature 410 (6831), 940–944. doi:10.1038/35073601
Hong, X., and Zha, J. (2019). Fish behavior: a promising model for aquatic toxicology research. Sci. total Environ. 686, 311–321. doi:10.1016/j.scitotenv.2019.06.028
Hörmer, B. A., Verma, D., Gasser, E., Wieselthaler-Hölzl, A., Herzog, H., and Tasan, R. O. (2018). Hippocampal NPY Y2 receptors modulate memory depending on emotional valence and time. Neuropharmacology 143, 20–28. doi:10.1016/j.neuropharm.2018.09.018
Hurst, J. L., and West, R. S. (2010). Taming anxiety in laboratory mice. Nat. methods 7 (10), 825–826. doi:10.1038/nmeth.1500
Kask, A., Harro, J., von Hörsten, S., Redrobe, J. P., Dumont, Y., and Quirion, R. (2002). The neurocircuitry and receptor subtypes mediating anxiolytic-like effects of neuropeptide Y. Neurosci. Biobehav. Rev. 26 (3), 259–283. doi:10.1016/s0149-7634(01)00066-5
Kuo, L. E., Kitlinska, J. B., Tilan, J. U., Li, L., Baker, S. B., Johnson, M. D., et al. (2007). Neuropeptide Y acts directly in the periphery on fat tissue and mediates stress-induced obesity and metabolic syndrome. Nat. Med. 13 (7), 803–811. doi:10.1038/nm1611
Larhammar, D., Blomqvist, A. G., Yee, F., Jazin, E., Yoo, H., and Wahlested, C. (1992). Cloning and functional expression of a human neuropeptide Y/peptide YY receptor of the Y1 type. J. Biol. Chem. 267 (16), 10935–10938. doi:10.1016/s0021-9258(19)49854-2
Larhammar, D., and Salaneck, E. (2004). Molecular evolution of NPY receptor subtypes. Neuropeptides 38 (4), 141–151. doi:10.1016/j.npep.2004.06.002
Larhammar, D., Wraith, A., Berglund, M. M., Holmberg, S. K., and Lundell, I. (2001). Origins of the many NPY-family receptors in mammals. Peptides 22 (3), 295–307. doi:10.1016/s0196-9781(01)00331-x
Larsson, T. A., Larson, E. T., Fredriksson, R., Conlon, J. M., and Larhammar, D. (2006). Characterization of NPY receptor subtypes Y2 and Y7 in rainbow trout Oncorhynchus mykiss. Peptides 27 (6), 1320–1327. doi:10.1016/j.peptides.2005.10.008
Larsson, T. A., Tay, B. H., Sundström, G., Fredriksson, R., Brenner, S., Larhammar, D., et al. (2009). Neuropeptide Y-family peptides and receptors in the elephant shark, Callorhinchus milii confirm gene duplications before the gnathostome radiation. Genomics 93 (3), 254–260. doi:10.1016/j.ygeno.2008.10.001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lee, C. C., and Miller, R. J. (1998). Is there really an NPY Y3 receptor? Regul. Pept. 75, 71–78. doi:10.1016/s0167-0115(98)00054-8
Loh, K., Herzog, H., and Shi, Y. C. (2015). Regulation of energy homeostasis by the NPY system. Trends Endocrinol. Metabolism 26 (3), 125–135. doi:10.1016/j.tem.2015.01.003
Lucon-Xiccato, T., Conti, F., Loosli, F., Foulkes, N. S., and Bertolucci, C. (2020). Development of open-field behaviour in the medaka, Oryzias latipes. Biology 9 (11), 389. doi:10.3390/biology9110389
Lucon-Xiccato, T., Loosli, F., Conti, F., Foulkes, N. S., and Bertolucci, C. (2022). Comparison of anxiety-like and social behaviour in medaka and zebrafish. Sci. Rep. 12 (1), 10926. doi:10.1038/s41598-022-14978-1
Matsuda, K., Sakashita, A., Yokobori, E., and Azuma, M. (2012). Neuroendocrine control of feeding behavior and psychomotor activity by neuropeptide Y in fish. Neuropeptides 46 (6), 275–283. doi:10.1016/j.npep.2012.09.006
Nakamura, M., Aoki, Y., and Hirano, D. (1996). Cloning and functional expression of a cDNA encoding a mouse type 2 neuropeptide Y receptor. Biochimica Biophysica Acta (BBA)-Biomembranes. 1284 (2), 134–137. doi:10.1016/s0005-2736(96)00166-6
Naveilhan, P., Hassani, H., Canals, J. M., Ekstrand, A. J., Larefalk, A., Chhajlani, V., et al. (1999). Normal feeding behavior, body weight and leptin response require the neuropeptide Y Y2 receptor. Nat. Med. 5 (10), 1188–1193. doi:10.1038/13514
Otsuka, A., Shimomura, Y., Sakikubo, H., Miura, K., and Kagawa, N. (2022). Effects of single and repeated heat stress on anxiety-like behavior and locomotor activity in medaka fish. Fish. Sci. 88, 45–54. doi:10.1007/s12562-021-01561-2
Patil, J. G., and Hinze, S. J. (2008). Simplex PCR assay for positive identification of genetic sex in the Japanese medaka, Oryzias latipes. Mar. Biotechnol. 10 (6), 641–644. doi:10.1007/s10126-008-9106-9
Pedragosa-Badia, X., Stichel, J., and Beck-Sickinger, A. G. (2013). Neuropeptide Y receptors: how to get subtype selectivity. Front. Endocrinol. 4, 5. doi:10.3389/fendo.2013.00005
Shi, C., Lu, Y., Zhai, G., Huang, J., Shang, G., Lou, Q., et al. (2020). Hyperandrogenism in POMCa-deficient zebrafish enhances somatic growth without increasing adiposity. J. Mol. Cell Biol. 12 (4), 291–304. doi:10.1093/jmcb/mjz053
Shiozaki, K., Kawabe, M., Karasuyama, K., Kurachi, T., Hayashi, A., Ataka, K., et al. (2020). Neuropeptide Y deficiency induces anxiety-like behaviours in zebrafish (Danio rerio). Sci. Rep. 10 (1), 5913. doi:10.1038/s41598-020-62699-0
Stanić, D., Mulder, J., Watanabe, M., and Hökfelt, T. (2011). Characterization of NPY Y2 receptor protein expression in the mouse brain. II. Coexistence with NPY, the Y1 receptor, and other neurotransmitter-related molecules. J. Comp. Neurology 519 (7), 1219–1257. doi:10.1002/cne.22608
Sundström, G., Larsson, T. A., Xu, B., Heldin, J., and Larhammar, D. (2013). Interactions of zebrafish peptide YYb with the neuropeptide Y-family receptors Y4, Y7, Y8a, and Y8b. Front. Neurosci. 7, 29. doi:10.3389/fnins.2013.00029
Talbot, C. (1993). Some aspects of the biology of feeding and growth in fish. Proc. Nutr. Soc. 52 (3), 403–416. doi:10.1079/pns19930081
Tatemoto, K., Carlquist, M., and Mutt, V. (1982). Neuropeptide Y—a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature 296, 659–660. doi:10.1038/296659a0
Tschenett, A., Singewald, N., Carli, M., Balducci, C., Salchner, P., Vezzani, A., et al. (2003). Reduced anxiety and improved stress coping ability in mice lacking NPY-Y2 receptors. Eur. J. Neurosci. 18 (1), 143–148. doi:10.1046/j.1460-9568.2003.02725.x
Wang, F., Chen, W., Lin, H., and Li, W. (2014). Cloning, expression, and ligand-binding characterization of two neuropeptide Y receptor subtypes in orange-spotted grouper, Epinephelus coioides. Fish physiology Biochem. 40, 1693–1707. doi:10.1007/s10695-014-9960-5
Wang, L., Zeng, F., Huang, R. F., Lin, S., Zhang, Z. H., and Li, M. D. (2023). Adipocyte reconstitution of npy4r gene in npy4r silenced mice promotes diet-induced obesity. Hereditas 45 (02), 144–155. doi:10.16288/j.yczz.22-302
Wang, T., Liang, J., Xiang, X., Chen, X., Zhang, B., Zhou, N., et al. (2019). Pharmacological characterization, cellular localization and expression profile of NPY receptor subtypes Y2 and Y7 in large yellow croaker, Larimichthys crocea. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 238, 110347. doi:10.1016/j.cbpb.2019.110347
Wraith, A., Törnsten, A., Chardon, P., Harbitz, I., Chowdhary, B. P., Andersson, L., et al. (2000). Evolution of the neuropeptide Y receptor family: gene and chromosome duplications deduced from the cloning and mapping of the five receptor subtype genes in pig. Genome Res. 10 (3), 302–310. doi:10.1101/gr.10.3.302
Xu, B., Lagman, D., Sundström, G., and Larhammar, D. (2015). Neuropeptide Y family receptors Y1 and Y2 from sea lamprey, Petromyzon marinus. General Comp. Endocrinol. 222, 106–115. doi:10.1016/j.ygcen.2015.08.005
Yulyaningsih, E., Loh, K., Lin, S., Lau, J., Zhang, L., Shi, Y., et al. (2014). Pancreatic polypeptide controls energy homeostasis via Npy6r signaling in the suprachiasmatic nucleus in mice. Cell metab. 19 (1), 58–72. doi:10.1016/j.cmet.2013.11.019
Zhang, Y., Zhang, Z., Liang, X. F., He, S., Xu, J., Smyth, S. S., et al. (2021). Role of NPY receptor 8 in regulating of food intake in Chinese perch (Siniperca chuatsi). Aquac. Int. 29, 2619–2623. doi:10.1182/bloodadvances.2020003041
Keywords: NPY2R, Japanese medaka, knockout, food intake, anxiety
Citation: Lu K, Jia X, Wu J, Wang Q and Liang X-F (2023) Neuropeptide Y receptor Y2 (npy2r) deficiency reduces anxiety and increases food intake in Japanese medaka (Oryzias latipes). Front. Cell Dev. Biol. 11:1273006. doi: 10.3389/fcell.2023.1273006
Received: 05 August 2023; Accepted: 23 October 2023;
Published: 07 November 2023.
Edited by:
Michael Schubert, UMR7009 Laboratoire de Biologie du Développement de Villefranche sur Mer, FranceReviewed by:
Satoshi Ogawa, Monash University Malaysia, MalaysiaCopyright © 2023 Lu, Jia, Wu, Wang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu-Fang Liang, eHVmYW5nX2xpYW5nQGhvdG1haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.