
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 26 September 2023
Sec. Cancer Cell Biology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1237530
Deubiquitination is an important form of post-translational modification that regulates protein homeostasis. Ovarian tumor domain-containing proteins (OTUDs) subfamily member OTUD3 was identified as a deubiquitinating enzyme involved in the regulation of various physiological processes such as immunity and inflammation. Disturbances in these physiological processes trigger diseases in humans and animals, such as cancer, neurodegenerative diseases, diabetes, mastitis, etc. OTUD3 is aberrantly expressed in tumors and is a double-edged sword, exerting tumor-promoting or anti-tumor effects in different types of tumors affecting cancer cell proliferation, metastasis, and metabolism. OTUD3 is regulated at the transcriptional level by a number of MicroRNAs, such as miR-520h, miR-32, and miR101-3p. In addition, OTUD3 is regulated by a number of post-translational modifications, such as acetylation and ubiquitination. Therefore, understanding the regulatory mechanisms of OTUD3 expression can help provide insight into its function in human immunity and disease, offering the possibility of its use as a therapeutic target to diagnose or treat disease.
Ubiquitination which is involved in the regulation of many eukaryotic signaling pathways is an important post-translational modification of proteins (Swatek and Komander, 2016). The process of ubiquitination modification is achieved by three enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (Scheffner et al., 1995; Koegl et al., 1999; Rotin and Kumar, 2009). The ubiquitin molecule itself has eight ubiquitination modification sites, namely, seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and the N-terminal methionine residue (Met1) (Kulathu and Komander, 2012; Kwon and Ciechanover, 2017). The ubiquitination process can also be regulated by deubiquitinating enzymes (DUB), which cleave ubiquitin from substrate proteins and thus regulate the dynamic balance of ubiquitination and deubiquitination in the organism.
The human genome encodes about 100 DUBs, which are usually divided into two groups, the majority cysteine proteases, and metalloproteases. In the past, cysteine proteases were classified into four categories which are ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), ovarian protease (OTU), and Machado-Joseph disease protease (MJD) (Nijman et al., 2005). In recent studies, two other categories MINDY (Abdul Rehman et al., 2016) and ZUFSP (Kwasna et al., 2018) were added to it. The JAB1/MPN/Mov34 metalloenzymes (JAMMs) with zinc-dependent properties are the only members of the metalloproteases family (Pan et al., 2022). Unlike USP family members that are non-specific for ubiquitin chain selection, OTU family members are linkage specific for different types of ubiquitin chains (Faesen Alex et al., 2011; Mevissen Tycho ET et al., 2013). Growing evidence that OTU subfamily members are associated with many human diseases such as cancer (Lin et al., 2019), immunity (Zhang et al., 2018), inflammation (Zhao et al., 2020), DNA damage repair (Kato et al., 2014), etc. The dysfunction that occurs with abnormal OTUD3 expression can also lead to the development of tumors, diabetes, and neurodegenerative diseases. In this paper, we first review the progress of research on the structure and molecular mechanisms and functions of OTUD3 in human and animal diseases and then describe the regulatory mechanisms of OTUD3 activity and expression.
The OTU structural domain was first identified in the Drosophila ovarian tumor gene from Drosophila melanogaster (Makarova et al., 2000). OTU-type DUBs can be divided into four subclasses based on the catalytic and other structural domains, the Otubains subfamily (OTUB1 and OTUB2), the OTUD subfamily (OTUD1, OTUD2, OTUD3, OTUD4, OTUD5, OTUD6A, OTUD6B, ALG13) (Figure 1), the A20-like OTUs subfamily (A20, OTUD7A, OTUD7B, Trabid, VCPIP1), and the OTULIN subfamily (Du et al., 2020).
In the OTU family, OTU domain-containing protein 3 (OTUD3) is described as a deubiquitinase that cleaves mainly Lys6 and Lys63 junctions and, to a lesser extent, Lys11 and Lys 48. OTUD3 is highly conserved evolutionarily and its protein structure consists of the catalytic OTU structural domain and the ubiquitin-associated (UBA) structural domain. The OTU structural domain contains a cysteine protease catalytic triad consisting of cysteine, aspartate, and histidine residues (Balakirev et al., 2003). The UBA structural domain is a small structural domain that consists of 40 residues with ubiquitin-binding functions (Buchberger, 2002). OTUD3 is localized on chromosome 1p36.13 and maintains its catalytic activity dependent on Cys76 (Du et al., 2019).
OTUD3 is expressed in all cells but at higher levels in neuron-derived tissues. And it is mainly distributed in the cytoplasmic interstitial space bound to microtubules in the cytoplasm and less in the nucleus. OTUD3 entry into the nucleus may depend on the interaction between itself and microtubule network dynamics. The substrate of OTUD3, p53, is present in the nucleus. P53 is associated with cellular microtubules and translocates to the nucleus in a kinesin-dependent manner (Marchenko et al., 2010). OTUD3 may enter the nucleus to mediate the process of deubiquitination of p53. Since a small fraction of OTUD3 is localized in the nucleus of non-stressed cells, it may also participate in the DNA repair pathway by counteracting Lys6 ubiquitination when it is not required. These are of course speculations and the function of OTUD3 into the nucleus needs to be further investigated (Job et al., 2022).
OTUD3 has many biological functions as a deubiquitinating enzyme that deubiquitinates a variety of substrates or interacts with related proteins (Table 1). Several recent studies have shown that OTUD3 is associated with many human and animal diseases, such as innate immunity, inflammation, tumors, diabetes, and clinical mastitis in cows.
OTUD3 is a deubiquitinase dependent on its Lys129-acetylation, and acetylated OTUD3 hydrolyzes polyubiquitination of Lys63 on MAVS during resting state. During viral infection, SIRT1 deacetylates OTUD3, thereby rapidly inactivating OTUD3 and providing effective innate anti-RNA virus immunity (Zhang et al., 2020a). OTUD3 can also reduce the binding of RIG-I and MDA5 to viral RNA and MAVS by binding RIG-I and MDA5 and removing k63-linked ubiquitination, thereby inhibiting the innate antiviral response triggered by RNA virus (Figure 2A). Interestingly, in contrast to RNA virus, OTUD3 promotes DNA virus-triggered innate immunity. OTUD3 binds cGAS and targets the Lys279 site for deubiquitination of k48- and k27-linked ubiquitination, thereby enhancing cGAS protein stability and DNA binding capacity (Cai et al., 2022). OTUD3 can also promote the DNA binding ability and dimerization of cGAS, thereby enhancing the enzymatic activity of cGAS (Lyu et al., 2023) (Figure 2B).
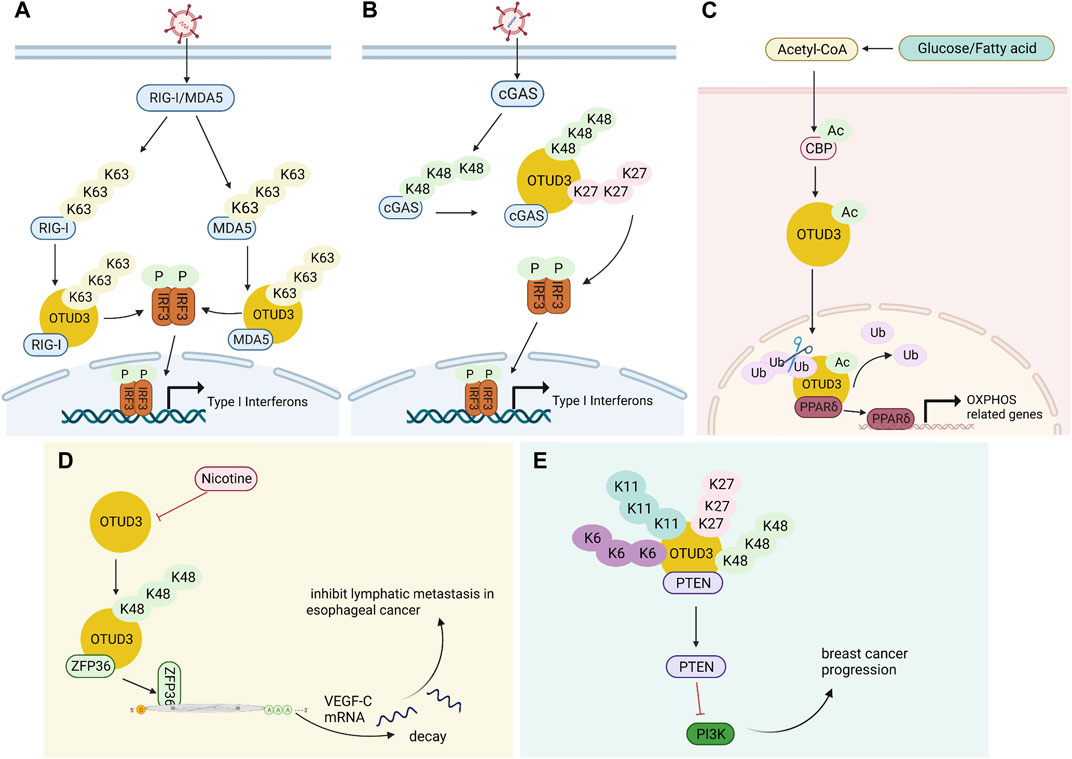
FIGURE 2. Multiple functions of OTUD3. (A) OTUD3 remove k63-linked ubiquitination of RIG-I and MDA5, thereby inhibiting the innate antiviral response triggered by RNA virus. (B) OTUD3 promotes DNA virus-triggered innate immunity. It binds cGAS and targets the Lys279 site for deubiquitination of k48- and k27-linked ubiquitination, thereby enhancing cGAS protein stability and DNA binding capacity. (C) The CBP promotes the acetylation of OTUD3 to its nuclear translocation, and OTUD3 regulates OXPHOS processes by stabilizing PPARδ. (D) OTUD3 stabilizes the k48-linked ubiquitinated ZFP36, and ZFP36 recruits RNA degradation complexes that mediate the decay of VEGF-C mRNA. (E) OTUD3-PTEN-PI3K axis may be the molecular mechanism of OTUD3 tumor suppression in breast cancer.
OTUD3 not only participates in innate antiviral immunity, but also triggers inflammatory immunity. The expression of lipopolysaccharide-binding protein (LBP) and cyclooxygenase 2 (COX2) on the classical pathway of nigrostriatal NF-κB and the expression of C-C motif chemokine 19 (CCL19) and cell adhesion factor 1 (ICAM1) on the non-classical pathway were both upregulated after OTUD3 knockdown in mice. All of these suggest that OTUD3 may be involved in the neuroinflammatory response (Gao et al., 2021).
OTUD3 and USP21 are deubiquitinating enzymes that antagonize ZNF598-mediated 40S ubiquitination and limit RQC activation. Cells lacking USP21 or OTUD3 alter the activity of RQC, delaying the deubiquitination of eS10 causing the ribosome to retain its ubiquitin tag longer and more likely to stall and fail to continue reading mRNA (Garshott et al., 2020).
OTUD3 interacts with PTEN in the cytoplasm, deubiquitinates and stabilizes PTEN, ultimately inhibiting tumor growth and metastasis in breast cancer (Yuan et al., 2015). PTEN is a key negative regulator of the PI3K/p-AKT/mTOR pathway (Lee et al., 2018). Inhibition of PI3K/p-AKT/mTOR pathway activation thereby inhibits breast cancer progression with increased drug sensitivity (Song et al., 2012; Marcotte et al., 2016) (Figure 2E). The expression of OTUD3 is regulated by small non-coding RNAs. MiR-520h binds to the 3′-untranslated region (3′-UTR) of OTUD3 and inhibits OTUD3 protein expression, which in turn inhibits PTEN expression and ultimately activates the downstream pathway AKT. Activation of AKT pathway enhances proliferation, invasiveness and resistance to paclitaxel in breast cells (Geng et al., 2020). Similarly, MicroRNA 32 (miR-32) can directly target the 3′-untranslated region (3′-UTR) of OTUD3 to inhibit OTUD3 expression, which can promote proliferation and motility of colon cancer cells and inhibits apoptosis (Jin et al., 2019). OTUD3 inhibits BC cell proliferation and makes BC cells more sensitive to chemotherapeutic drug-induced apoptosis by directly interacting with p53 through the amino-terminal OTU region to deubiquitinate it from murine double minute 2 (Mdm2)-mediated ubiquitination and degradation (Pu et al., 2020).
Carboxyl terminus of Hsc70-interacting protein CHIP is the ubiquitin ligase of OTUD3. OTUD3 is stabilized in lung cancer cell lines by the deletion of CHIP, and the level of intracellular GRP78 increases, thus promoting lung cancer cell invasion and tumor metastasis (Zhang et al., 2020b). A small molecule OTUD3 inhibitor named OTUDin3 has been reported for the treatment of non-small cell lung cancer. In NSCLC, OTUDin3 exhibited significant anti-proliferative and pro-apoptotic effects by inhibiting the deubiquitination activity of OTUD3 (Zhang et al., 2023). OTUD3 mediates Alpha-actinin 4 ACTN4 deubiquitination in HCC to stabilize it and promote hepatocellular carcinoma cell growth and metastasis (Xie et al., 2021). In addition, ACTN4 promotes metastasis to lSymph nodes in colorectal cancer as well as induces epithelial mesenchymal transition and tumorigenesis in cervical cancer, and it can be inferred that OTUD3 is also involved in these processes by regulating ACTN4 (Honda et al., 2005; An et al., 2016). OTUD3 also deubiquitinates k48-linked ubiquitination to stabilize ZFP36 ring finger protein (ZFP36), which ZFP36 recruits RNA degradation complexes to mediate the decay of VEGF-C mRNA. Downregulation of OTUD3 expression by Nicotine in esophageal cancer leads to downregulation of ZFP36, which induces VEGF-C production and promotes lymphatic metastasis in esophageal cancer (Wang et al., 2021) (Figure 2D). TRIM56 acts as a ubiquitin ligase to increase the ubiquitination level of lung adenocarcinoma cancer-promoting factor OTUD3 and promote its ubiquitin-proteasome pathway degradation. OTUD3 also feedback regulates TRIM56 and is able to remove the Lys48-linked polyubiquitin chain on TRIM56 and stabilize its protein expression. TRIM56 acts as a tumor suppressor, inhibiting the migration and invasion of lung adenocarcinoma (Lu, 2021).
OTUD3 is highly expressed in human lung cancer tissues and is associated with poor patient survival. OTUD3 interacts with the glucose-regulated protein GRP78 to deubiquitinate and stabilize GRP78, thereby inhibiting lung cancer cell growth and migration (Du et al., 2019). OTUD3 increases SOAT1 protein stability by specifically removing the polyubiquitination modification of sterol O-acyltransferase 1 (SOAT1), dependent on its deubiquitinating enzyme activity (Gai et al., 2021). SOAT1 affects the growth and migration of hepatocellular carcinoma cells by promoting the synthesis of cholesterol and consequently (Jiang et al., 2019).
OTUD3 has been elucidated to exert pro- or oncogenic functions in tumors by deubiquitinating and stabilizing the expression of related proteins. However, the expression and mechanism of action of OTUD3 in other tumors remain unknown. PTEN and OTUD3 protein expression was significantly lower in C6 cells compared to primary astrocytes, and survival time was longer in patients with high OTUD3-expressing gliomas than in patients with low-expressing gliomas. It was concluded only that the low expression of OTUD3 in glioma cells may be involved in gliomagenesis, but the mechanism behind this is unclear (Liu et al., 2020). Similarly, upon genomic analysis of human cancers, OTUD3 locus deletion may be responsible for the development of multiple squamous cell carcinomas (SCCs). However, the molecular basis of OTUD3 in SCC as a tumor suppressor has not been fully elucidated. It is speculated that OTUD3 function may be impaired in SCCs, destabilizing the tumor suppressor PTEN and thus activating the AKT pathway to cause cancer (Gatti et al., 2020).
OTUD3 appears to have both pro- and anti-cancer effects in cancer. Whether it exerts a cancer-promoting or cancer-suppressing effect may depend on its substrate protein. If the substrate protein is a tumor suppressor, OTUD3 stabilizes the expression of the substrate protein by deubiquitination and is also a tumor suppressor. However, more clinical evidence and mechanistic studies are still needed to further elucidate the role of OTUD3 in cancer in order to target it for antitumor therapy.
In recent years there have been many studies on the factors involved in the degeneration of dopaminergic neurons in PD, and iron deposition has received much attention (Moreau et al., 2018). Iron is essential for cellular biosynthesis, metabolic activity, and the balance of iron uptake, efflux and storage in cells is regulated by iron regulatory proteins regulating IRPs (Hentze et al., 2010). Intracellular IRP2 is degraded via the ubiquitin-proteasome pathway. OTUD3 acts as a deubiquitinating enzyme for IRP2, promoting the removal of k48-linked polyubiquitination of IRP2 in the cytoplasm and stabilizing IRP2 protein in an iron-independent manner. Thus OTUD3 deficiency leads to disruption of iron metabolism (Jia et al., 2022). OTUD3 deficiency also leads to dopaminergic neuronal damage, which may be another reason for the appearance of Parkinson’s symptoms. OTUD3 regulates the level of fortilin ubiquitination, affects endoplasmic reticulum stress, and inhibits dopaminergic neuronal apoptosis (Chen et al., 2023).
The OTUD3 c.863G>A (rs78466831) mutation was identified in the obesity and diabetes family, a mutation that reduces the stability and catalytic activity of OTUD3. Otud3−/− mice show a significant impairment of energy metabolism and are susceptible to obesity and metabolic disorders. To gain more insight into the function of OTUD3, RNA-seq analysis was performed on the mutant and wild type. GSEA analysis (Subramanian et al., 2005) showed that OTUD3 loss-of-function-associated genes scored high in diabetes, PPAR signaling pathway, glycolipid metabolism, and fatty acid degradation. Glucose and fatty acid stimulation leads to increased levels of acetyl coenzyme a and increased levels of acetylation of CREB-binding protein (CBP) (Wellen et al., 2009; Cordero-Herrera et al., 2017). The acetyltransferase CBP promotes OTUD3 acetylation to its nuclear translocation, where OTUD3 regulates glycolipid metabolism and oxidative phosphorylation (OXPHOS) processes through the stabilization of the metabolism-associated factor peroxisome-proliferator-activated receptor delta (PPARδ) (Zhou et al., 2022) (Figure 2C). In addition, a comparison of the prevalence of rs78466831 in patients with type 2 diabetes mellitus T2DM in different regions revealed that the rs78466831 mutation was strongly associated with T2DM in a province in eastern China and had a higher risk of developing diabetes retinopathy DR (Liu et al., 2022). It is possible that mutations in specific amino acids in OTUD3 lead to changes in protein conformation (Kar et al., 2017), which in turn leads to a much higher risk of developing diabetes.
Defective OTUD3 gene triggers tissue inflammation in humans and animals. OTUD3 is a susceptibility gene for Crohn’s disease and ulcerative colitis (Jung et al., 2023). The three RNF186-OTUD3-PLA2G2E genes are located on chromosome 1p36, and 1p36 contains two different ulcerative colitis risk SNPs (rs1317209 and rs6426833). Previous studies have shown that OTUD3 transcripts are higher in immune tissues and lymphocytes than in CD14-positive cells, suggesting that OTUD3 regulates the immune response (McGovern et al., 2010), and that mutations in it may be responsible for developing ulcerative colitis. Further studies found that the approximately 8 kb SNP rs4654903, located upstream of OTUD3, is closer to the OTUD3 position and has a stronger signal than the two previous SNPs, similar to Caucasian and Asian (Yang et al., 2013). A 200 bp deletion in the Copy number variant regions (CNVRs) of OTUD3 (BTA2) in Holstein-Friesian cows was positively correlated with the incidence of clinical mastitis (CM) (Szyda et al., 2019).
Long-stranded noncoding RNA SNHG6 and OTUD3 are upregulated and miR-101-3p is downregulated in recurrent spontaneous abortion (RSA) patients. In RSA, SNHG6 binds to and negatively regulates miR101-3p expression and OTUD3 is a miR101-3p target. SNHG6 reduces trophoblast cell viability and migration capacity by regulating miR101-3p/OTUD3 and increases trophoblast apoptosis. SNHG6/miR-101-3p/OTUD3 could be a potential target for the prevention or treatment of RSA (Zhao et al., 2022).
OTUD3 is involved in the regulation of iron metabolism and glucose metabolism through deubiquitination of metabolism-related proteins. When its deficiency occurs, it induces metabolism-related diseases. Also, OTUD3 is a susceptibility gene for inflammatory diseases. Therefore, uncovering more information about OTUD3 can help to understand the pathogenesis of these diseases and to treat them.
As with most deubiquitinating enzymes, the activity and expression of OTUD3 is regulated by mechanisms of action at the transcriptional and post-translational levels (Table 2).
OTUD3 is regulated at the transcriptional level by a number of microRNAs which target the 3′-UTR region of OTUD3 mRNA and are involved in the regulation of cell proliferation and apoptosis. For example, miR-520h binds to OTUD3 to inhibit the expression of OTUD3 protein in breast cancer cells, which in turn enhances the resistance of breast cancer cells to paclitaxel (Geng et al., 2020). MiR-32 also inhibits the expression of OTUD3 and promotes cell proliferation and motility and inhibits apoptosis in colon cancer cells (Jin et al., 2019). In addition, miR101-3p restored the viability and migration ability of trophoblast cells by inhibiting OTUD3 expression, while reducing trophoblast cell apoptosis (Zhao et al., 2022). In Bronchopulmonary dysplasia (BPD) mouse, miR-34c-5p negatively targeted OTUD3 which enhanced proliferation, angiogenesis, and migration in human pulmonary microvascular endothelial cells (HPMECs) (He et al., 2023) (Figure 3A).
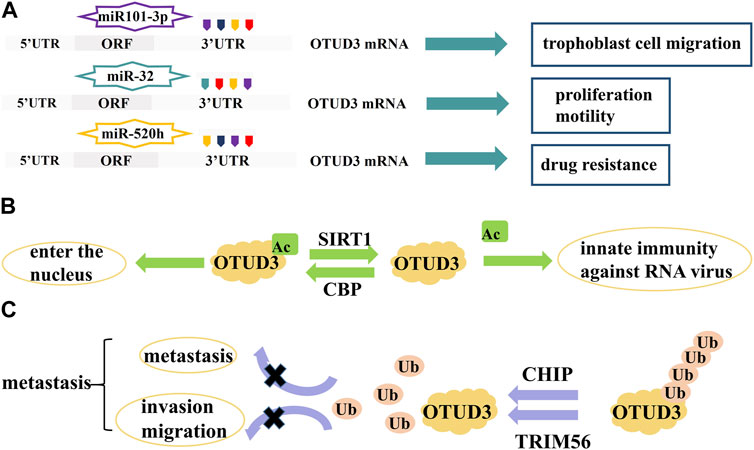
FIGURE 3. Proteins and RNAs regulating OTUD3 activity and expression. (A) MicroRNAs regulate the transcription of OTUD3. (B) SIRTI and CBP regulate the acetylation of OTUD3. (C) CHIP and TRIM56 regulate the ubiquitination of OTUD3.
OTUD3 is regulated by a number of post-translational modifications, such as acetylation and ubiquitination.
OTUD3 activity is dependent on its Lys129-acetylation. During the onset of innate anti-RNA virus immunity, OTUD3 is regulated by SIRT1 deacetylation and activity is lost, resulting in effective resistance to RNA viruses (Zhang et al., 2020a). OTUD3 acetylation can also affect its cellular localization. Normally, OTUD3 is localized in the cytoplasm and can enter the nucleus after acetylation. For example, the acetyltransferase CBP promotes OTUD3 acetylation for its nuclear translocation, allowing OTUD3 to deubiquitinate and stabilize the intranuclear metabolism-associated transcription factor PPARδ thereby affecting gene expression related to glycolipid metabolism and oxidative phosphorylation (OXPHOS) (Zhou et al., 2022) (Figure 3B).
The ubiquitin ligase CHIP ubiquitinates OTUD3 to degrade it, and inhibition of the OTUD3-GRP78 signaling axis reduces tumor metastasis in lung cancer (Zhang et al., 2020b). OTUD3 can also be ubiquitinated by TRIM56, thereby inhibiting invasion and migration of lung adenocarcinoma (Marcotte et al., 2016) (Figure 3C).
This paper reviews the physiopathological roles of OTUD3 in human diseases such as innate immunity, inflammatory response, tumorigenesis, diabetes, and neurodegenerative diseases, and explores in depth the regulatory mechanisms of its activity and expression. Interestingly, OTUD3 is a double-edged sword for tumors, playing a pro- or oncogenic role in different types of tumors, yet the regulatory mechanisms of OTUD3 expression, localization and function in these tumors are still not fully explored. Understanding these mechanisms can help develop drugs targeting OTUD3 to inhibit cancer progression and provide new ideas for cancer therapy.
OTUD3 mRNA levels were significantly lower in breast cancer tissues compared to paraneoplastic tissues, while prognostic risk assessment models for OTUD3 showed patients with strong OTUD3 mRNA expression had a better prognosis (Qi et al., 2019). This could serve as evidence that OTUD3 acts as a tumor suppressor in breast cancer. During tumorigenesis, the PI3K/p-AKT/mTOR pathway is regulated by OTUD3, and PTEN is a key regulatory protein of the PI3K/p-AKT/mTOR pathway. Since OTUD3 is a specific cytoplasmic deubiquitinase of PTEN, the OTUD3-PTEN-PI3K axis may also be the molecular mechanism of OTUD3 tumor suppression in breast cancer, glioma and SCC. In addition to exerting cancer-inhibiting effects, OTUD3 also exerts cancer-promoting functions in liver and lung cancers, promoting the growth of liver and lung cancer cells. OTUD3 is like a double-sided person, which side it exhibits may depend on its partner substrate proteins. If the substrate protein is a tumor suppressor, OTUD3 stabilizes the expression of the substrate protein by deubiquitination and also functions as a cancer suppressor. In antiviral immune responses, OTUD3 is similarly two-sided in that it inhibits the innate antiviral immune response to RNA viruses but promotes the innate antiviral immune response to DNA viruses. Most of the proteins identified to date are consistent in regulating innate antiviral immunity in response to DNA and RNA viruses, either promoting or inhibiting, with few opposing effects found (Zhang et al., 2020c). Deubiquitinases that remove K63 or K48-linked polyubiquitin chains exerting opposite effects, like OTUD3, have rarely been reported. The opposite role of OTUD3 in the innate antiviral immune response may be consistent with its opposite mechanism of action in cancer, both depending on the function of its substrate.
OTUD3 activity is influenced by post-translational modifications and its activity is dependent on Lys129-acetylation. As a deubiquitinating enzyme, OTUD3 itself can be degraded by ubiquitination mediated by two E3 ubiquitin ligases, CHIP and TRIM56. In addition to acetylation and ubiquitination modifications, other post-translational modifications such as phosphorylation, hydroxylation and hypusination (Kar et al., 2021; Park et al., 2022; Ziegler et al., 2022) were not reported.
We still have not found the commonality of OTUD3 substrate proteins and the real reason for OTUD3 entry into the nucleus. Therefore, it is necessary to further explore the molecular mechanisms of OTUD3 in diseases and its unique functions. In the future, structural modeling analysis of OTUD3 protein can be performed using proteomics technology and bioinformatics to predict its substrate proteins and discover their structural commonalities, which can help to elucidate the regulatory mechanisms of OTUD3 in human diseases. As kinases and phosphatases are main drug targets for various cancers (Nemec et al., 2019), researchers attempted to find kinases that are up- or downregulated in the OTUD3 pathway, and unfortunately did not find any. However, a small molecule inhibitor, OTUDin3, which directly targets OTUD3, has been found. Given the oncogenic role of OTUD3 in many cancers, if more small molecule inhibitors of OTUD3 for cancer therapy are fortunately screened and identified to inhibit its expression or activity, it is expected to be a promising therapeutic strategy for the treatment of a range of human tumors.
Conceptualization: HJ and XD; Supervision, project administration and funding acquisition: XD, LH, SZ, and HJ; Manuscript writing: QX; Writing—review and editing: HJ. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant Nos. 82103184, 81672632, 81972312, and 32000622), the Science and Technology Innovation Program of Hunan Province of China (Grant No. 2022RC1165), the Natural Science Foundation of Hunan Province of China (Grant Nos. 2021JJ40720, 2021JJ30912, 2021JJ40052, and 2022JJ40579), and the Research Project of Health Commission of Hunan Province of China (Grant No. 202202085572).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdul Rehman, S. A., Kristariyanto, Y. A., Choi, S. Y., Nkosi, P. J., Weidlich, S., Labib, K., et al. (2016). MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 63 (1), 146–155. doi:10.1016/j.molcel.2016.05.009
An, H. T., Yoo, S., and Ko, J. (2016). α-Actinin-4 induces the epithelial-to-mesenchymal transition and tumorigenesis via regulation of Snail expression and β-catenin stabilization in cervical cancer. Oncogene 35 (45), 5893–5904. doi:10.1038/onc.2016.117
Balakirev, M. Y., Tcherniuk, S. O., Jaquinod, M., and Chroboczek, J. (2003). Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4 (5), 517–522. doi:10.1038/sj.embor.embor824
Buchberger, A. (2002). From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell. Biol. 12 (5), 216–221. doi:10.1016/s0962-8924(02)02269-9
Cai, X., Zhou, Z., Zhu, J., Liu, X., Ouyang, G., Wang, J., et al. (2022). Opposing effects of deubiquitinase OTUD3 in innate immunity against RNA and DNA viruses. Cell. Rep. 39 (10), 110920. doi:10.1016/j.celrep.2022.110920
Chen, L., Huan, X., Jia, F., Zhang, Z., Bi, M., Fu, L., et al. (2023). Deubiquitylase OTUD3 mediates endoplasmic reticulum stress through regulating fortilin stability to restrain dopaminergic neurons apoptosis. Antioxidants (Basel, Switz. 12 (4), 809. doi:10.3390/antiox12040809
Cordero-Herrera, I., Chen, X., Ramos, S., and Devaraj, S. (2017). (-)-Epicatechin attenuates high-glucose-induced inflammation by epigenetic modulation in human monocytes. Eur. J. Nutr. 56 (3), 1369–1373. doi:10.1007/s00394-015-1136-2
Du, T., Li, H., Fan, Y., Yuan, L., Guo, X., Zhu, Q., et al. (2019). The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat. Commun. 10 (1), 2914. doi:10.1038/s41467-019-10824-7
Du, J., Fu, L., Sui, Y., and Zhang, L. (2020). The function and regulation of OTU deubiquitinases. Front. Med. 14 (5), 542–563. doi:10.1007/s11684-019-0734-4
Faesen Alex, C., Luna-Vargas Mark, P. A., Geurink Paul, P., Clerici, M., Merkx, R., van Dijk Willem, J., et al. (2011). The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18 (12), 1550–1561. doi:10.1016/j.chembiol.2011.10.017
Gai, W. B., Bi, M. X., and Jiang, H. (2021). Mechanism of deubiquitylase OTUD3 in regulating the protein stability of sterol O-acyltransferase-1. J. Qingdao Univ. Sci. 57 (05), 637–641.
Gao, Y. J., Jia, F. J., Jiao, Q., Chen, X., Du, X. X., and Jiang, H. (2021). Changes in the expression of nuclear factor-Kappa B-related inflammatory factors in the substantia nigra of mice with ovarian tumor domain-containing protein 3 knockout and related mechanisms. J. Precis. Med. 36 (02), 95–98.
Garshott, D. M., Sundaramoorthy, E., Leonard, M., and Bennett, E. J. (2020). Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. Elife 9, e54023. doi:10.7554/eLife.54023
Gatti, V., Bernassola, F., Talora, C., Melino, G., and Peschiaroli, A. (2020). The impact of the ubiquitin system in the pathogenesis of squamous cell carcinomas. Cancers (Basel) 12 (6), 1595. doi:10.3390/cancers12061595
Geng, W., Song, H., Zhao, Q., Dong, K., Pu, Q., Gao, H., et al. (2020). miR-520h stimulates drug resistance to paclitaxel by targeting the OTUD3-PTEN Axis in breast cancer. Biomed. Res. Int. 2020, 9512793. doi:10.1155/2020/9512793
He, X., Kuang, J., Wang, Y., Lan, G., and Shi, X. (2023). Bone marrow stromal cell-secreted extracellular vesicles containing miR-34c-5p alleviate lung injury and inflammation in bronchopulmonary dysplasia through promotion of PTEN degradation by targeting OTUD3. Immunol. Investig. 52, 681–702. doi:10.1080/08820139.2023.2217854
Hentze, M. W., Muckenthaler, M. U., Galy, B., and Camaschella, C. (2010). Two to tango: regulation of mammalian iron metabolism. Cell. 142 (1), 24–38. doi:10.1016/j.cell.2010.06.028
Honda, K., Yamada, T., Hayashida, Y., Idogawa, M., Sato, S., Hasegawa, F., et al. (2005). Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 128 (1), 51–62. doi:10.1053/j.gastro.2004.10.004
Jia, F., Li, H., Jiao, Q., Li, C., Fu, L., Cui, C., et al. (2022). Deubiquitylase OTUD3 prevents Parkinson's disease through stabilizing iron regulatory protein 2. Cell. Death Dis. 13 (4), 418. doi:10.1038/s41419-022-04704-0
Jiang, Y., Sun, A., Zhao, Y., Ying, W., Sun, H., Yang, X., et al. (2019). Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567 (7747), 257–261. doi:10.1038/s41586-019-0987-8
Jin, Y., Cheng, H., Cao, J., and Shen, W. (2019). MicroRNA 32 promotes cell proliferation, migration, and suppresses apoptosis in colon cancer cells by targeting OTU domain containing 3. J. Cell. Biochem. 120 (11), 18629–18639. doi:10.1002/jcb.28874
Job, F., Mai, C., Villavicencio-Lorini, P., Herfurth, J., Neuhaus, H., Hoffmann, K., et al. (2022). OTUD3: A Lys6 and Lys63 specific deubiquitinase in early vertebrate development. Biochim. Biophys. Acta Gene Regul. Mech. 1866 (1), 194901. doi:10.1016/j.bbagrm.2022.194901
Jung, S., Kim, Y., Park, D., Lee, Y., Park, S., Baek, J., et al. (2023). Case-case genome-wide association analysis identifying genetic loci with divergent effects on Crohn's disease and ulcerative colitis. Hum. Mol. Genet. 32 (4), 677–684. doi:10.1093/hmg/ddac241
Kar, R. K., Kharerin, H., Padinhateeri, R., and Bhat, P. J. (2017). Multiple conformations of Gal3 protein drive the galactose-induced allosteric activation of the GAL genetic switch of Saccharomyces cerevisiae. J. Mol. Biol. 429 (1), 158–176. doi:10.1016/j.jmb.2016.11.005
Kar, R. K., Hanner, A. S., Starost, M. F., Springer, D., Mastracci, T. L., Mirmira, R. G., et al. (2021). Neuron-specific ablation of eIF5A or deoxyhypusine synthase leads to impairments in growth, viability, neurodevelopment, and cognitive functions in mice. J. Biol. Chem. 297 (5), 101333. doi:10.1016/j.jbc.2021.101333
Kato, K., Nakajima, K., Ui, A., Muto-Terao, Y., Ogiwara, H., and Nakada, S. (2014). Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell. 53 (4), 617–630. doi:10.1016/j.molcel.2014.01.030
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 96 (5), 635–644. doi:10.1016/s0092-8674(00)80574-7
Kulathu, Y., and Komander, D. (2012). Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell. Biol. 13 (8), 508–523. doi:10.1038/nrm3394
Kwasna, D., Abdul Rehman, S. A., Natarajan, J., Matthews, S., Madden, R., De Cesare, V., et al. (2018). Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell. 70 (1), 150–164. doi:10.1016/j.molcel.2018.02.023
Kwon, Y. T., and Ciechanover, A. (2017). The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42 (11), 873–886. doi:10.1016/j.tibs.2017.09.002
Lee, Y. R., Chen, M., and Pandolfi, P. P. (2018). The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell. Biol. 19 (9), 547–562. doi:10.1038/s41580-018-0015-0
Lin, D. D., Shen, Y., Qiao, S., Liu, W. W., Zheng, L., Wang, Y. N., et al. (2019). Upregulation of OTUD7B (cezanne) promotes tumor progression via AKT/VEGF pathway in lung squamous carcinoma and adenocarcinoma. Front. Oncol. 9, 862. doi:10.3389/fonc.2019.00862
Liu, Y. Z., Du, X. X., Zhao, Q. Q., Jiao, Q., and Jiang, H. (2020). The expression change of OTUD3-PTEN signaling axis in glioma cells. Ann. Transl. Med. 8 (7), 490. doi:10.21037/atm.2020.03.51
Liu, J. P., Yang, A. P., Lei, G., Yu, M., Peng, Y., and Le, A. P. (2022). Prevalence and clinical characteristics of T2DM patients with OTUD3 gene rs78466831 SNP at a single academic center in China. Front. Endocrinol. (Lausanne) 13, 1059641. doi:10.3389/fendo.2022.1059641
Lu, K. (2021). Function and regulation mechanism of TRIM56 in lung adenocarcinoma. Qingdao University. doi:10.27262/d.cnki.gqdau.2021.000385
Lyu, H., Sun, L., Guan, Z., Li, J., Yin, C., Zhang, Y., et al. (2023). Proximity labeling reveals OTUD3 as a DNA-binding deubiquitinase of cGAS. Cell. Rep. 42 (4), 112309. doi:10.1016/j.celrep.2023.112309
Makarova, K. S., Aravind, L., and Koonin, E. V. (2000). A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25 (2), 50–52. doi:10.1016/s0968-0004(99)01530-3
Marchenko, N. D., Hanel, W., Li, D., Becker, K., Reich, N., and Moll, U. M. (2010). Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell. Death Differ. 17 (2), 255–267. doi:10.1038/cdd.2009.173
Marcotte, R., Sayad, A., Brown, K. R., Sanchez-Garcia, F., Reimand, J., Haider, M., et al. (2016). Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 164 (1-2), 293–309. doi:10.1016/j.cell.2015.11.062
McGovern, D. P., Gardet, A., Torkvist, L., Goyette, P., Essers, J., Taylor, K. D., et al. (2010). Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42 (4), 332–337. doi:10.1038/ng.549
Mevissen Tycho Et, Hospenthal Manuela, K., Geurink Paul, P., Elliott Paul, R., Akutsu, M., Arnaudo, N., Ekkebus, R., et al. (2013). OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 154 (1), 169–184. doi:10.1016/j.cell.2013.05.046
Moreau, C., Duce, J. A., Rascol, O., Devedjian, J. C., Berg, D., Dexter, D., et al. (2018). Iron as a therapeutic target for Parkinson's disease. Mov. Disord. official J. Mov. Disord. Soc. 33 (4), 568–574. doi:10.1002/mds.27275
Nemec, C. M., Singh, A. K., Ali, A., Tseng, S. C., Syal, K., Ringelberg, K. J., et al. (2019). Noncanonical CTD kinases regulate RNA polymerase II in a gene-class-specific manner. Nat. Chem. Biol. 15 (2), 123–131. doi:10.1038/s41589-018-0194-1
Nijman, S. M., Luna-Vargas, M. P., Velds, A., Brummelkamp, T. R., Dirac, A. M., Sixma, T. K., et al. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell. 123 (5), 773–786. doi:10.1016/j.cell.2005.11.007
Pan, X., Wu, S., Wei, W., Chen, Z., Wu, Y., and Gong, K. (2022). Structural and functional basis of JAMM deubiquitinating enzymes in disease. Biomolecules 12 (7), 910. doi:10.3390/biom12070910
Park, M. H., Kar, R. K., Banka, S., Ziegler, A., and Chung, W. K. (2022). Post-translational formation of hypusine in eIF5A: implications in human neurodevelopment. Amino acids 54 (4), 485–499. doi:10.1007/s00726-021-03023-6
Pu, Q., Lv, Y. R., Dong, K., Geng, W. W., and Gao, H. D. (2020). Tumor suppressor OTUD3 induces growth inhibition and apoptosis by directly deubiquitinating and stabilizing p53 in invasive breast carcinoma cells. BMC Cancer 20 (1), 583. doi:10.1186/s12885-020-07069-9
Qi, L., Yao, Y., Zhang, T., Feng, F., Zhou, C., Xu, X., et al. (2019). A four-mRNA model to improve the prediction of breast cancer prognosis. Gene 721, 144100. doi:10.1016/j.gene.2019.144100
Rotin, D., and Kumar, S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 10 (6), 398–409. doi:10.1038/nrm2690
Scheffner, M., Nuber, U., and Huibregtse, J. M. (1995). Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373 (6509), 81–83. doi:10.1038/373081a0
Song, M. S., Salmena, L., and Pandolfi, P. P. (2012). The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell. Biol. 13 (5), 283–296. doi:10.1038/nrm3330
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Swatek, K. N., and Komander, D. (2016). Ubiquitin modifications. Cell. Res. 26 (4), 399–422. doi:10.1038/cr.2016.39
Szyda, J., Mielczarek, M., Fraszczak, M., Minozzi, G., Williams, J. L., and Wojdak-Maksymiec, K. (2019). The genetic background of clinical mastitis in Holstein-Friesian cattle. Animal 13 (10), 2156–2163. doi:10.1017/S1751731119000338
Wang, M., Li, Y., Xiao, Y., Yang, M., Chen, J., Jian, Y., et al. (2021). Nicotine-mediated OTUD3 downregulation inhibits VEGF-C mRNA decay to promote lymphatic metastasis of human esophageal cancer. Nat. Commun. 12 (1), 7006. doi:10.1038/s41467-021-27348-8
Wellen, K. E., Hatzivassiliou, G., Sachdeva, U. M., Bui, T. V., Cross, J. R., and Thompson, C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Sci. (New York, NY) 324 (5930), 1076–1080. doi:10.1126/science.1164097
Xie, P., Chen, Y., Zhang, H., Zhou, G., Chao, Q., Wang, J., et al. (2021). The deubiquitinase OTUD3 stabilizes ACTN4 to drive growth and metastasis of hepatocellular carcinoma. Aging (Albany NY) 13 (15), 19317–19338. doi:10.18632/aging.203293
Yang, S. K., Hong, M., Zhao, W., Jung, Y., Tayebi, N., Ye, B. D., et al. (2013). Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm. Bowel Dis. 19 (5), 954–966. doi:10.1097/MIB.0b013e3182802ab6
Yuan, L., Lv, Y., Li, H., Gao, H., Song, S., Zhang, Y., et al. (2015). Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat. Cell. Biol. 17 (9), 1169–1181. doi:10.1038/ncb3218
Zhang, L., Liu, J., Qian, L., Feng, Q., Wang, X., Yuan, Y., et al. (2018). Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathog. 14 (5), e1007067. doi:10.1371/journal.ppat.1007067
Zhang, Z., Fang, X., Wu, X., Ling, L., Chu, F., Li, J., et al. (2020a). Acetylation-dependent deubiquitinase OTUD3 controls MAVS activation in innate antiviral immunity. Mol. Cell. 79 (2), 304–319. doi:10.1016/j.molcel.2020.06.020
Zhang, P., Li, C., Li, H., Yuan, L., Dai, H., Peng, Z., et al. (2020b). Ubiquitin ligase CHIP regulates OTUD3 stability and suppresses tumour metastasis in lung cancer. Cell. Death Differ. 27 (11), 3177–3195. doi:10.1038/s41418-020-0571-7
Zhang, Z-D., Xiong, T-C., Yao, S-Q., Wei, M-C., Chen, M., Lin, D., et al. (2020c). RNF115 plays dual roles in innate antiviral responses by catalyzing distinct ubiquitination of MAVS and MITA. Nat. Commun. 11 (1), 5536. doi:10.1038/s41467-020-19318-3
Zhang, Y., Du, T., Liu, N., Wang, J., Zhang, L., Cui, C-P., et al. (2023). Discovery of an OTUD3 inhibitor for the treatment of non-small cell lung cancer. Cell. Death Dis. 14 (6), 378. doi:10.1038/s41419-023-05900-2
Zhao, M., Song, K., Hao, W., Wang, L., Patil, G., Li, Q., et al. (2020). Non-proteolytic ubiquitination of OTULIN regulates NF-κB signaling pathway. J. Mol. Cell. Biol. 12 (3), 163–175. doi:10.1093/jmcb/mjz081
Zhao, L., Liu, X., Ren, C., Zhang, H., and Gao, L. (2022). Silencing of LncRNA SNHG6 protects trophoblast cells through regulating miR-101-3p/OTUD3 axis in unexplained recurrent spontaneous abortion. J. Mol. Histol. 53 (6), 871–882. doi:10.1007/s10735-022-10102-9
Zhou, N., Qi, H., Liu, J., Zhang, G., Liu, J., Liu, N., et al. (2022). Deubiquitinase OTUD3 regulates metabolism homeostasis in response to nutritional stresses. Cell. Metab. 34 (7), 1023–1041.e8. doi:10.1016/j.cmet.2022.05.005
Keywords: deubiquitinase, OTUD3, cancer, immunity, disease
Citation: Xu Q, He L, Zhang S, Di X and Jiang H (2023) Deubiquitinase OTUD3: a double-edged sword in immunity and disease. Front. Cell Dev. Biol. 11:1237530. doi: 10.3389/fcell.2023.1237530
Received: 09 June 2023; Accepted: 28 August 2023;
Published: 26 September 2023.
Edited by:
Anjan Kumar Pradhan, Virginia Commonwealth University, United StatesReviewed by:
Balarko Chakraborty, VCU School of Dentistry, United StatesCopyright © 2023 Xu, He, Zhang, Di and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotang Di, ZGl4aWFvdGFuZ0Bjc3UuZWR1LmNu; Hao Jiang, amlhbmdoYW8xMjA5QGNzdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.