- 1Laboratory of Research in Parkinson’s Disease and Related Disorders, Health Sciences Institute, China Medical University, Shenyang, China
- 2Key Laboratory of Biomedical Engineering of Hainan Province, School of Biomedical Engineering, Hainan University, Haikou, China
- 3Faculty of Medicine, Dynamics and Mechanics of Epithelia Group, CNRS, Institute of Genetics and Development of Rennes (IGDR), University Rennes, Rennes, France
- 4Laboratory of Molecular Oncology and Innovative Therapies, Military Institute of Medicine (WIM-PIB), Warsaw, Poland
- 5State Key Laboratory of Animal Biotech Breeding, College of Biological Sciences, China Agricultural University, Beijing, China
- 6Sanya Institute of China Agricultural University, Sanya, China
- 7Key Laboratory of Bioresource Research and Development of Liaoning Province, College of Life and Health Sciences, Northeastern University, Shenyang, China
Editorial on the Research Topic
Applying large animals for developmental study and disease modeling
Introduction
We have a great pleasure to organize the Research Topic “Applying large Animals for Developmental Study and Disease Modeling”. It is well known that large animals show similarities in many physical and pathological characteristics with human beings and have numerous advantages in developmental and medical research. In recent years, an important breakthrough has been achieved in gene editing technique, including ZFN, TALEN, CRISPR/Cas9, and so on. Especially for the emergence of CRISPR/Cas9 mediated gene modification strategies, various types of human diseases have been recapitulated in large animal models. It has to be mentioned that the progress in establishing embryonic stem cells and inducible pluripotent stem cells is also critical for the application in diseases modeling and therapies. In our Research Topic more than ten papers have been collected, and most of them focus on pig, monkey, rabbit and horse (Figure 1).
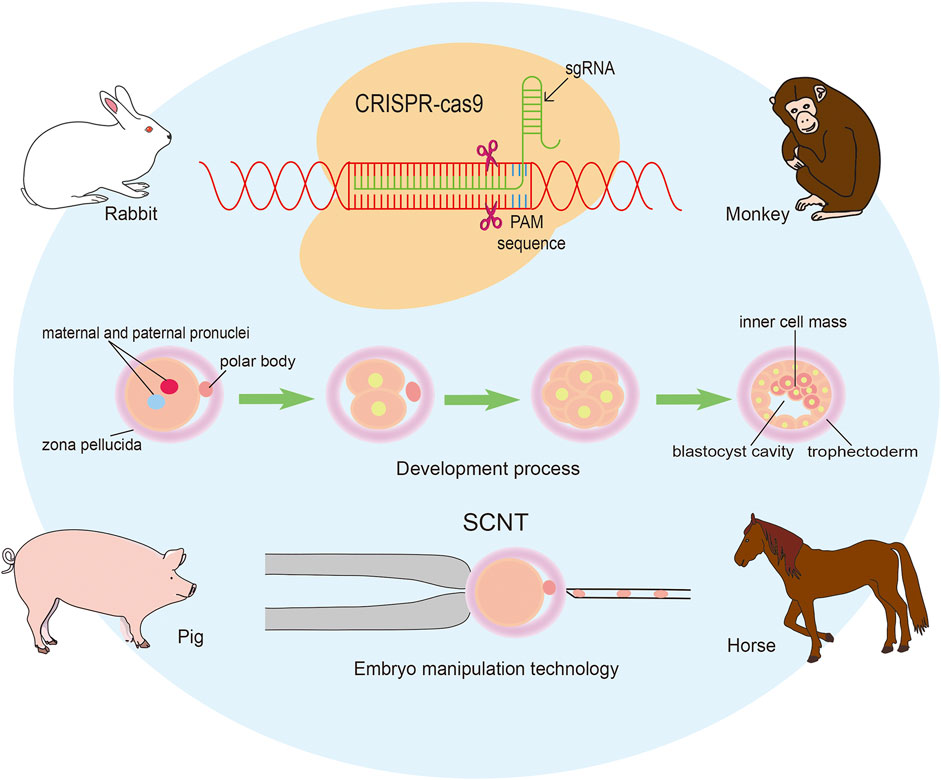
FIGURE 1. Applying genomic editing technology and somatic cell nuclear transfer to generate large animal models.
Pig
The pig, as one of the most common livestock, exhibits several advantages in biomedical research. These are: comparable human and pig body sizes, similar anatomical and physiological characteristics, genomic composition and ordinary diets (Hou et al., 2022). It makes the pig a promising alternative animal model for humans. Rapid growth rate, early sexual maturity, short generation intervals, high number of offspring per litter, and standardized breeding techniques, also make easier the application of pigs in the study on human diseases. In recent decades, the advances in technologies contribute to the development of genetically engineered pig models of human diseases. The emergence of somatic cell nuclear transfer (SCNT) and reliable genome editing techniques make possible for the generation of porcine models of human diseases. The efficacy of porcine SCNT has been largely improved and several kinds of small molecular inhibitors have been applied to enhance the developmental competence of SCNT embryos (Ouyang et al., 2021; Hou et al., 2022). CRISPR/Cas9-mediated gene editing strategies are widely used in the establishment of porcine models of human diseases. Various genomic genetically engineered pigs have been generated, including chimeric gene knock in, point mutations, large genome fragment deletions, multifunctional live cell sensors, etc. One of the most promising potential application of pigs in the biomedical field is xenotransplantation. In 7 January 2022, the medical school of the University of Maryland in the United States conducted the first-ever life-saving cardiac xenotransplantation and it was successful in extending the patient’s life for about 8 weeks (Rothblatt, 2022). The breakthrough sheds light on xenotransplantation using porcine organs as donors. Actually, corneas, lungs, nerve cells, kidneys, livers, and islets of pigs are also potential candidates for xenotransplantation. In the Research Topic, Junliang Li et al. evaluated the methylation status of IG-DMR and gene expression profile in the DLK1-DIO3 region (Li et al.); Lin et al. revealed that intravenous injection of AAV9-GFP could result in widespread expression of transgene in various porcine organs (Lin et al.); Hilansi Rawat et al. found that porcine expanded pluripotent stem cells could differentiate into cardiovascular progenitor cells, functional cardiomyocytes, epicardial cells and epicardial-derived cells, and they established an enhanced system for whole-embryo culture allowing ex utero development of porcine post-implantation embryos from ED14 up to ED17 (Hilansi et al.); and many papers review the progress of applying pigs in xenotransplantation and biomedical research.
Nonhuman primates
Nonhuman primates (NHP) emerge increasingly as excellent models for translational research, due to even closer proximity to human beings in terms of physiology, biochemistry, immunology, pathology and genetic evolution. For example, NHP models are widely used in studies on Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). In fact, NHP models of HIV, ZIKV or Ebola virus infection mirror closely the pathogenesis in human patients. SCNT monkeys have been successfully produced using fetal fibroblasts as donor cells in recent years (Liu et al., 2018). Furthermore, BMAL1 gene-edited monkeys via the CRISPR/Cas9 technique were generated with both intracytoplasmic sperm injection and SCNT method (Liu et al., 2019; Qiu et al., 2019). Cynomolgus monkey blastoids resembling blastocysts in morphology and transcriptomics using naive ESC develop to embryonic disk with the structures of yolk sac, amnion cavity, chorionic cavity and primitive streak via prolonged in vitro culture (Li et al., 2023), making it possible to investigate primate embryonic development without the same ethical concerns associated humans. Various NHP models of human diseases have been established in recent years, including bilaterally delivering synthetic Aβ oligomers into the cerebral parenchyma of cynomolgus monkeys to drive early pathological progression of Alzheimer’s disease (Yue et al., 2021). However, in the near future, researchers might face monkey shortage crisis (Grimm, 2023). In the Research Topic, Liang et al. summarizes the recent progress in using genomic editing technology in the establishment of NHP models and discusses the factors limiting the wide application of NHP models of human diseases (Liang et al.).
Rabbit
Rabbit is another commonly used experimental model for investigating equivalents of human diseases. With the emergence of microinjecting, SCNT and genomic editing technology, a large number of rabbit models of human diseases were established. The pronuclear microinjection is still the most common method in the generation of transgenic rabbit models. Novel genomic editing technologies, such as CRISPR/Cas9, remarkably promote precision in rabbit genome manipulation (Song et al., 2020). In previous studies, CRISPR/Cas9 technique was applied to introduce mutation of αA-Crystallin or GJA8 gene to induce congenital cataracts in Rabbits (Yuan et al., 2016; Yuan et al., 2017). Recently, SpRY-ABEmax mediated base substitution has been used to generate YIPF5 (p.W218R) mutation to generate rabbit primary microcephaly model, which precisely recapitulate the typical symptoms of human primary microcephaly (Liu et al., 2023). With recent technological innovations in genomic editing techniques, rabbit models will certainly play a much more important role in the study on human diseases. In our Research Topic, a carrageenan-induced abdominal aortic adventitial inflammatory model in hypercholesterolemic rabbits is described (Chen et al.). It determines the role of MMP-12 secreted from adventitial macrophages in the pathogenesis of this diseases (Chen et al.).
Horse
Horses are commonly used as animal models for studying several human diseases, including neurodegenerative diseases, mental and behavioral disorders, neuropsychiatric disorders and spontaneous sepsis. Using SCNT and genomic editing technology to generate horse models of human diseases seems to be less attractive than porcine and rabbit models. However, the rapid development in horse models have been achieved in recent years. A recent report has revealed that a total of 12 SCNT foals were born (Cortez et al., 2023), and gene-edited horse embryos have been generated by CRISPR/Cas9 technology (Maniego et al., 2022). In our Research Topic, Neil Marr et al. utilized immunolabelling for CD146 to determine horse tendon cell population, providing the intrinsic evidences for the relationships between local interfascicular matrix vascular and basement membrane constituents (Marr et al.).
In conclusion, the Research Topic “Applying large Animals for Developmental Study and Disease Modeling” delivers a whole plethora of excellent examples of the fascinating progress made recently in this field of biomedical research.
Author contributions
LY and YH drafted the manuscript, YH revised the draft, FY, JK and SW made substantial contributions to the work through in-depth discussion. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (Nos. 82201594 and 81502582). Funding was also provided by the Fundamental Research Funds for the Central Universities (N182004002 and N2220002), Natural Science Foundation of Liaoning Province (2021-MS-104, 2022-YGJC-39 and 2022-MS-228), Fundamental Scientific Research Fund of Liaoning Provincial Education Department (LJKQZ2021002), Key Laboratory of Bioresource Research and Development of Liaoning Province (2022JH13/10200026), Hainan Key Research and Development Project (ZDYF2021SHFZ049), and JK was supported by an internal grant nr. 612/2023 from Wojskowy Instytut Medyczny-Panstwowy Instytut Badawczy, Warsaw, Poland.
Acknowledgments
We would like to thank Wanlu Zhang in Northeastern University for assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cortez, J. V., Hardwicke, K., Cuervo-Arango, J., and Grupen, C. G. (2023). Cloning horses by somatic cell nuclear transfer: Effects of oocyte source on development to foaling. Theriogenology 203, 99–108. Epub 2023/04/04PubMed PMID: 37011429. doi:10.1016/j.theriogenology.2023.03.018
Grimm, D. U. S. (2023). U.S., European researchers face monkey shortage crisis. Science 380 (6645), 567–568. Epub 2023/05/11PubMed PMID: 37167380. doi:10.1126/science.adi6512
Hou, N., Du, X., and Wu, S. (2022). Advances in pig models of human diseases. Anim. Model Exp. Med. 5 (2), 141–152. Epub 2022/03/29PubMed PMID: 35343091; PubMed Central PMCID: PMCPMC9043727. doi:10.1002/ame2.12223
Li, J., Zhu, Q., Cao, J., Liu, Y., Lu, Y., Sun, Y., et al. (2023). Cynomolgus monkey embryo model captures gastrulation and early pregnancy. Cell Stem Cell 30 (4), 362–377.e7. Epub 2023/04/08PubMed PMID: 37028403. doi:10.1016/j.stem.2023.03.009
Liu, X., Yang, J., Li, Z., Liu, R., Wu, X., Zhang, Z., et al. (2023). YIPF5 (p.W218R) mutation induced primary microcephaly in rabbits. Neurobiol. Dis. 182, 106135. Epub 2023/05/05PubMed PMID: 37142085. doi:10.1016/j.nbd.2023.106135
Liu, Z., Cai, Y., Liao, Z., Xu, Y., Wang, Y., Wang, Z., et al. (2019). Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl. Sci. Rev. 6 (1), 101–108. Epub 2019/01/01PubMed PMID: 34691835; PubMed Central PMCID: PMCPMC8291622. doi:10.1093/nsr/nwz003
Liu, Z., Cai, Y., Wang, Y., Nie, Y., Zhang, C., Xu, Y., et al. (2018). Cloning of macaque monkeys by somatic cell nuclear transfer. Cell 174 (1), 245. Epub 2018/06/30PubMed PMID: 29958110. doi:10.1016/j.cell.2018.01.036
Maniego, J., Pesko, B., Habershon-Butcher, J., Hincks, P., Taylor, P., Tozaki, T., et al. (2022). Use of mitochondrial sequencing to detect gene doping in horses via gene editing and somatic cell nuclear transfer. Drug Test. Anal. 14 (8), 1429–1437. Epub 2022/04/02PubMed PMID: 35362263. doi:10.1002/dta.3267
Ouyang, H., Han, J., and Huang, Y. (2021). Pig cloning using somatic cell nuclear transfer. Methods Mol. Biol. 2239, 1–18. Epub 2020/11/24PubMed PMID: 33226609. doi:10.1007/978-1-0716-1084-8_1
Qiu, P., Jiang, J., Liu, Z., Cai, Y., Huang, T., Wang, Y., et al. (2019). BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl. Sci. Rev. 6 (1), 87–100. Epub 2019/01/01PubMed PMID: 34691834; PubMed Central PMCID: PMCPMC8291534. doi:10.1093/nsr/nwz002
Rothblatt, M. (2022). Commentary on achievement of first life-saving xenoheart transplant. Xenotransplantation 29 (3), e12746. Epub 2022/04/27PubMed PMID: 35471736. doi:10.1111/xen.12746
Song, J., Zhang, J., Xu, J., Garcia-Barrio, M., Chen, Y. E., and Yang, D. (2020). Genome engineering technologies in rabbits. J. Biomed. Res. 35 (2), 135–147. Epub 2020/09/17PubMed PMID: 32934190; PubMed Central PMCID: PMCPMC8038526. doi:10.7555/JBR.34.20190133
Yuan, L., Sui, T., Chen, M., Deng, J., Huang, Y., Zeng, J., et al. (2016). CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci. Rep. 6, 22024. Epub 2016/02/26PubMed PMID: 26912477; PubMed Central PMCID: PMCPMC4766569. doi:10.1038/srep22024
Yuan, L., Yao, H., Xu, Y., Chen, M., Deng, J., Song, Y., et al. (2017). CRISPR/Cas9-Mediated mutation of αa-crystallin gene induces congenital cataracts in rabbits. Invest. Ophthalmol. Vis. Sci. 58 (6), BIO34–BIO41. Epub 2017/05/06PubMed PMID: 28475701. doi:10.1167/iovs.16-21287
Yue, F., Feng, S., Lu, C., Zhang, T., Tao, G., Liu, J., et al. (2021). Synthetic amyloid-beta oligomers drive early pathological progression of Alzheimer's disease in nonhuman primates. iScience 24 (10), 103207. Epub 2021/10/28PubMed PMID: 34704001; PubMed Central PMCID: PMCPMC8524197. doi:10.1016/j.isci.2021.103207
Keywords: gene editing, CRISPR/Cas9, embryo, development, pig, rabbit, monkey
Citation: Yuan L, Yue F, Kubiak JZ, Wu S and Huang Y (2023) Editorial: Applying large animals for developmental study and disease modeling. Front. Cell Dev. Biol. 11:1225060. doi: 10.3389/fcell.2023.1225060
Received: 18 May 2023; Accepted: 24 May 2023;
Published: 30 May 2023.
Edited and reviewed by:
Valerie Kouskoff, The University of Manchester, United KingdomCopyright © 2023 Yuan, Yue, Kubiak, Wu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Yuan, bHl1YW5AY211LmVkdS5jbg==; Feng Yue, Znl1ZWVAaG90bWFpbC5jb20=; Jacek Z. Kubiak, amFjZWsua3ViaWFrQHVuaXYtcmVubmVzMS5mcg==; Sen Wu, c3d1QGNhdS5lZHUuY24=; Yongye Huang, aHVhbmd5b25neWU4OEAxNjMuY29t
 Lin Yuan
Lin Yuan Feng Yue
Feng Yue Jacek Z. Kubiak
Jacek Z. Kubiak Sen Wu
Sen Wu Yongye Huang
Yongye Huang