- 1Department of Systems Biochemistry, Institute for Biochemistry and Pathobiochemistry, Faculty of Medicine, Ruhr University Bochum, Bochum, Germany
- 2Department of Medical Biochemistry and Molecular Biology, Saarland University, Homburg, Germany
- 3Department Molecular Cell Biology, Institute of Biochemistry and Pathobiochemistry, Faculty of Medicine, Ruhr University Bochum, Bochum, Germany
PEX19 binding sites are essential parts of the targeting signals of peroxisomal membrane proteins (mPTS). In this study, we characterized PEX19 binding sites of PEX11, the most abundant peroxisomal and glycosomal membrane protein from Trypanosoma brucei and Saccharomyces cerevisiae. TbPEX11 contains two PEX19 binding sites, one close to the N-terminus (BS1) and a second in proximity to the first transmembrane domain (BS2). The N-terminal BS1 is highly conserved across different organisms and is required for maintenance of the steady-state concentration and efficient targeting to peroxisomes and glycosomes in both baker’s yeast and Trypanosoma brucei. The second PEX19 binding site in TbPEX11 is essential for its glycosomal localization. Deletion or mutations of the PEX19 binding sites in TbPEX11 or ScPEX11 results in mislocalization of the proteins to mitochondria. Bioinformatic analysis indicates that the N-terminal region of TbPEX11 contains an amphiphilic helix and several putative TOM20 recognition motifs. We show that the extreme N-terminal region of TbPEX11 contains a cryptic N-terminal signal that directs PEX11 to the mitochondrion if its glycosomal transport is blocked.
1 Introduction
Peroxisomes are single membrane bound organelles performing a wide range of functions (Rhodin, 1954; De Duve and Baudhuin, 1966). Glyoxysomes in plants, Woronin bodies in fungi, and glycosomes in trypanosomatid parasites are specialized forms of peroxisomes (Reichle and Alexander, 1965; Breidenbach and Beevers, 1967; Opperdoes and Borst, 1977). Peroxisomes can multiply by growth and division, or they can form de novo from pre-peroxisomal vesicles that are supposed to bud from the endoplasmic reticulum (Hoepfner et al., 2005; Motley and Hettema, 2007). Peroxisomes import matrix as well as membrane proteins post-translationally (Goldman Blobel, 1978; Lazarow and Fujiki, 1985; Sacksteder et al., 2000; Jones et al., 2004). The import depends on a machinery of Peroxins (PEX proteins) and requires the presence of peroxisomal targeting signals in the cargo proteins (Gould et al., 1989; Swinkels et al., 1991; Faber et al., 1995). Biogenesis of peroxisomes requires two distinct machineries for protein targeting: The first is responsible for the formation of the peroxisomal membrane by the targeting and insertion of peroxisomal membrane proteins (PMPs), and the second machinery is responsible for the import of peroxisomal matrix proteins (reviewed in (Agrawal and Subramani, 2016)). The trafficking of proteins destined for the peroxisome matrix has been well studied. A striking feature is that peroxisomes can import folded, even oligomeric proteins (McNew andGoodman, 1994; Leon et al., 2006). Peroxisomal matrix proteins contain type 1 or type 2 peroxisomal targeting signals (PTS1/PTS2) at the extreme C-terminus or close to the N-terminus, respectively (Gould et al., 1989; Swinkels et al., 1991; Faber et al., 1995). Some proteins contain internal targeting signals (Galland et al., 2010) and some are transported by piggy-backing onto a PTS-containing protein (Islinger et al., 2009). The import of peroxisomal matrix proteins depends on cycling receptors that recognize peroxisomal proteins via their PTS in the cytosol and target them to a docking complex at the peroxisomal membrane. Import takes place through a transient pore or hydrogel-filled pore in an unknown fashion (Erdmann and Schliebs, 2005; Meinecke et al., 2016; Gao et al., 2022). The cargo-unloaded receptors are mono-ubiquitinated and released to the cytosol for another round of import in an ATP-dependent manner by the peroxisomal exportomer (Platta et al., 2004; Platta et al., 2005). The machinery that is responsible for the topogenesis of membrane proteins is distinct from the import machinery for matrix proteins (Reviewed in (Hasan et al., 2013; Mayerhofer, 2016)). Only three peroxins with a direct role in PMP targeting have been identified, namely, PEX3 (Hettema et al., 2000), PEX16 in mammals (South and Gould, 1999; Sacksteder et al., 2000), and PEX19 (Sacksteder et al., 2000) (Also reviewed in (Kalel and Erdmann, 2018)). In cells lacking any of these proteins, PMPs are either degraded or mistargeted to other subcellular compartments such as mitochondria, endoplasmic reticulum (ER), and membranes of unknown origin (Ghaedi et al., 2000; Hettema et al., 2000; Sacksteder et al., 2000). PMPs contain multiple binding sites (BSs) for the cytosolic receptor and chaperone PEX19 (Jones et al., 2004). These binding sites are essential for targeting of the PMPs to the peroxisomal membrane, as they can function as mPTS i.e., membrane peroxisome targeting signal (Halbach et al., 2005). The mPTS often comprises part of the transmembrane domains and a short adjacent sequence, which contains either a cluster of basic residues or a mixture of basic and hydrophobic amino acids (Marshall et al., 1996) (Reviewed in (Baerends et al., 2000; Murphy et al., 2003; Van Ael and Fransen, 2006)). Rottensteiner et al., developed PEX19 binding site prediction methodology using peptide arrays (Rottensteiner et al., 2004). Unlike PTS1 and PTS2 signals, which can be predicted more reliably (Kamoshita et al., 2022; Kunze, 2023), PEX19 BSs are comparatively degenerate and can be present multiple times in a PMP. Therefore, an efficient PEX19BS predictor is still needed. Nonetheless, PEX19 binding sites (BSs) have been identified in various yeast, human and parasite PMPs, which shows evolutionary conservation across eukaryotes (Rottensteiner et al., 2004; Saveria et al., 2007). In most eukaryotes, PEX19 harbors a farnesylation motif (CaaX box), and farnesylation has been shown to increase the binding efficiency of PMPs (Rucktaschel et al., 2009). However, trypanosomatid parasite PEX19 proteins lack such a CaaX motif (Banerjee et al., 2005).
PEX11 is an integral peroxisomal membrane protein with at least two predicted alpha-helical transmembrane domains and both termini facing the cytosol (Abe et al., 1998b; Lorenz et al., 1998; Anton et al., 2000; Bonekamp et al., 2013). In the yeast Saccharomyces cerevisiae, Pex11p, Pex25p, and Pex27p are the three members of the PEX11 protein family (Erdmann and Blobel, 1995; Rottensteiner et al., 2003; Tam et al., 2003). Similarly, mammals also encode three PEX11-family proteins namely, PEX11α, PEX11β, and PEX11γ (Li and Gould, 2002; Koch et al., 2010). In plants, there are five PEX11 homologs, for e. g., in Arabidopsis thaliana AtPEX11a, -b, -c, -d, and -e (Lingard and Trelease, 2006). PEX11 family proteins are involved in the proliferation of peroxisome in yeasts, plants, and mammals (Erdmann and Blobel, 1995; Abe and Fujiki, 1998a; Schrader et al., 1998; Orth et al., 2007; Koch et al., 2010). Deletion of PEX11 in yeast has an effect on the β-oxidation of fatty acids, which can be due to defects in the transport of metabolites across the peroxisomal membrane (Sulter et al., 1993). PEX11β is widely expressed in mammalian tissues and it has a well-recognized function in the initial phase of peroxisomal fission when it remodels and elongates peroxisomal membranes (Delille et al., 2010; Yoshida et al., 2015; Schrader et al., 2016). The functions of PEX11α and PEX11γ are less clear (Schrader et al., 2016). Of the PEX11 proteins in mammals, only PEX11β deficiency was associated with the pathology of peroxisome biogenesis disorders (PBDs) (Li and Gould, 2002; Thoms and Gartner, 2012; Schrader et al., 2016).
In trypanosomes, three PEX11 family proteins are known, namely, PEX11, GIM5A and GIM5B (Lorenz et al., 1998; Maier et al., 2001; Voncken et al., 2003). Like in mammals, yeast, and plants, both N- and C-termini of TbPEX11 are exposed to the cytosol (Lorenz et al., 1998). Overexpression of TbPEX11 induces growth inhibition and transforms the globular glycosomes into long tubule clusters that occupy a large portion of the cytoplasm (Lorenz et al., 1998). Accordingly, TbPEX11 appears to play a role in the proliferation of glycosomes in trypanosomes like its homologs in yeast and mammalian cells (van Roermund et al., 2000) (Reviewed in (Moyersoen et al., 2004)). PEX11 and both GIM5 proteins are essential for the survival of parasites (Lorenz et al., 1998; Voncken et al., 2003). At primary sequence level, PEX11 family proteins contain several conserved helices particularly in the N-terminal region (Lorenz et al., 1998; Opalinski et al., 2018). PEX19 binding sites have been identified in various glycosomal membrane proteins (Saveria et al., 2007), but not in TbPEX11. Therefore, in this study, we characterized PEX19 binding sites of PEX11 from Trypanosoma brucei and Saccharomyces cerevisiae. TbPEX11 contains two PEX19 binding sites, the N-terminal PEX19 binding site (BS1) in PEX11 is highly conserved across different organisms and is required for maintenance of the steady-state concentration as well as efficient targeting to peroxisomes and glycosomes in both baker’s yeast and T. brucei. Deletion or mutations of the PEX19 binding site in TbPEX11 (second PEX19 BS, i.e., BS2) or ScPEX11 (single PEX19 BS) results in a mislocalization of the proteins to mitochondria. Trypanosomes contain multiple small glycosomes, but harbor a single mitochondrion (Tyler et al., 2001). We show that the extreme N-terminal region of TbPEX11 contains a cryptic N-terminal signal that directs PEX11 to the mitochondrion if its glycosomal transport is blocked.
2 Materials and methods
2.1 Cloning
Escherichia coli, yeast, and Trypanosoma expression plasmid constructs and cloning strategies are listed in Table 1, and oligonucleotide sequences are listed in Table 2. Point mutations in ScPEX11, TbPEX111-89aa, and the gene fragment deletions (TbPEX111-76aa and TbPEX11-GFP constructs) were generated by overlap extension PCR. Sequences of the constructs, mutations, and gene fragment deletions were verified for all constructs by automated Sanger sequencing.
2.2 Cell culture
2.2.1 Trypanosoma
Trypanosoma procyclic form (PCF) 29–13 cell line (co-expressing T7 RNAP and TetR) was used in this study. PCF cells were grown in SDM-79 medium supplemented with 10% FBS at 28°C (Brun and Schonenberger, 1979; Krishna et al., 2023). PCF cultures were maintained at 1 × 106–30 × 106 cells/mL. Transfections were performed with NotI-linearized plasmid constructs (pGN1-TbPEX11 constructs), which was genomically integrated into the rRNA locus in the genome of cell line 29–13. Clones were selected using Blasticidin (10 μg/mL) as described previously (Kalel et al., 2015).
2.2.2 Yeast
S. cerevisiae wild-type strain BY4742 (for microscopy) and strain PCY2 (WT or Δpex19 for yeast two-hybrid assay) were grown in double dropout SD synthetic media as described in section 2.3 and 2.4.2. Yeast cells were transformed by the traditional Lithium-acetate method (Gietz and Woods, 2002).
2.2.3 Escherichia coli
Escherichia coli strain TOP10 was used for all plasmid amplifications and BL21 (DE3) strain was used for heterologous expression of recombinant GST-PEX19 fusion proteins. Liquid E. coli cultures were grown at 37°C under continuous shaking in LB medium containing the appropriate selective antibiotic (100 μg/mL Ampicillin).
2.3 Yeast two-hybrid analysis (Y2H)
Y2H studies were performed based on the Yeast protocols handbook (Clontech, Protocol No. PT3024-1, Version No. PR742227). Full length or various truncations of Trypanosoma or Human PEX11 were cloned in pPC97 vector containing GAL4-DNA Binding Domain (BD) and full-length Trypanosoma or human PEX19 were cloned in pPC86 vector containing GAL4-Activation domain (AD), as described in Table 1. Co-transformation of various two-hybrid plasmids i.e., BD and AD constructs were performed in WT PCY2 or Δpex19 PCY2 strain in case of HsPEX19-HsPEX11 constructs. The clones were selected on SD synthetic medium without tryptophan and leucine. A filter-based β-galactosidase assay and liquid culture assay using ONPG were performed in three replicates as described in the Yeast protocols handbook (Clontech).
2.4 Microscopy
2.4.1 Trypanosoma
Trypanosoma stable cell lines (Procyclic 29:13) encoding various tetracycline inducible PEX11-GFP constructs (full-length and mutants) were induced with 1 μg/mL tetracycline or treated with DMSO alone as negative control. Cells were sedimented and fixed by resuspension in 4% paraformaldehyde in PBS (phosphate-buffered saline, pH 7.4) at 4°C for 15 min. Fixed Trypanosoma cells were washed two times with PBS and stored at 4°C in a dark box. For imaging, fixed cells were immobilized on a glass slide (StarFrost 76 × 26 mm, Knittel Glass) pre-coated with 10% (v/v) of poly-L-lysine (Sigma-Aldrich) in water for 1 h at room temperature (RT). Further, the cell membranes were permeabilized with PBS containing 0.125% Triton X-100 and incubated for 10–15 min, followed by blocking with PBS containing 3% BSA, 0.25% Tween-20 for 1 h at RT. Rabbit α-Aldolase antibody (1:500 in blocking buffer) was used as glycosomal marker and incubated at RT for 1 h. Following 5 washes with PBS, anti-rabbit Alexa fluor 594 secondary antibodies (1:200 dilution) in PBS was applied and incubated for 30 min at RT in the dark. Further, the stained samples were washed, dried, and layered with anti-fading mounting medium, i.e., Mowiol with DAPI (4′,6-diamidino-2-phenylindole).
For mitochondrial staining, tetracycline-induced or uninduced (DMSO-treated) Trypanosoma cells were harvested and resuspended in the culture medium containing 75 nM MitoTracker® Deep Red and incubated for 5 min at 28°C. Following incubation, cells were washed with PBS twice and resuspended in the culture medium and further incubated for 30 min at 28°C. Subsequently, cells were fixed with 4% paraformaldehyde in PBS, and samples were prepared for microscopy as mentioned above.
Glycosome- or mitochondrion-stained cells were visualized and imaged with a Zeiss Elyra microscope and were analyzed using Zeiss Zen 3.2 software (blue edition). Both aldolase and MitoTracker which are markers for glycosome and mitochondrion respectively, are pseudo-colored to magenta for visualization.
2.4.2 Yeast
BY4742 yeast strain co-transformed with the plasmids encoding ScPEX11-GFP (WT (Boutouja et al., 2019) and L35 to P mutant, with the endogenous promoter) and DsRed-SKL (as a peroxisomal reporter (Kuravi et al., 2006)) were grown overnight (16 h) with shaking in an SD synthetic medium without uracil and histidine. Next day the precultures were diluted to 0.1 OD600/mL and were incubated under shaking until the cell density reached 0.6–0.8 OD600/mL. After incubation, 1–2 mL cultures were harvested and washed with water. For mitochondrial staining, 5 mL of yeast cells, grown to a density of 0.6–0.8 OD600/mL, expressing the ScPEX11 constructs were stained with 150 nM MitoTracker™ Orange CMTMRos (Invitrogen) for 30 min with shaking in dark. Following incubation, 1–2 mL cultures were harvested and washed with water. All incubation steps were performed at 30°C. Yeast cells expressing various fluorescent proteins were directly visualized microscopically without fixation. Microscopy was performed with Carl Zeiss Microscope, using the Axiovision 4.6.3 software, and images were analyzed using Zen 3.2 (blue edition), a Carl Zeiss software. The DsRed-SKL, a peroxisomal reporter is pseudo-colored to magenta for visualization.
2.5 Peptide array
The immobilized peptides of 15-amino acids length, sequentially overlapping by 13 residues (2aa shift), representing the entire sequence of TbPEX11 or the N-terminal domains of three human PEX11 isoforms were synthesized on a cellulose membrane as described previously (Hilpert et al., 2007; Neuhaus et al., 2014). The peptide array was first washed with ethanol for 10 min with gentle shaking followed by three washes with TBS (50 mM Tris, 137 mM NaCl, 2.7 mM KCl, adjusted to pH 8) for 10 min each. Further, the peptide array was incubated with a blocking buffer (TBS +3% BSA +0.05% Tween-20) for 2 h at room temperature (RT). The purified recombinant proteins GST-TbPEX19, GST-HsPEX19, or GST alone (10 mL of 1 µM solution prepared in blocking buffer) were incubated with the arrays for 1 h at 4°C. Then, the arrays were washed three times for 10 min at RT with TBS, and subsequently incubated with the anti-GST monoclonal antibody (Sigma, 1:2000) at RT for 1 h. Followed by three washes with TBS (10 min each), a secondary antibody (Horseradish peroxidase-coupled anti-mouse IgGs, 1:5,000 in blocking buffer) was applied, and the array was further incubated for 1 h at RT. After three washes with TBS, the array was scanned with chemiluminescence substrate (WesternBright Sirius) using Azure sapphire biomolecular imager.
2.6 Protein expression and purification, in vitro pull-downs and AlphaScreen binding assay
2.6.1 Protein expression and purification
The expression plasmids pGEX4T2, pGEX4T2-TbPEX19 or pGEX4T1-HsPEX19, encoding for GST, GST-TbPEX19 or GST-HsPEX19, respectively, were transformed into BL21 (DE3) E. coli strain. Single colonies were inoculated in LB medium containing ampicillin and incubated overnight with shaking at 37°C. On the following day, the cultures were reinoculated with 0.1 OD600/mL and further incubated at 37°C with shaking, until the cell density reached 0.6 OD600/mL. Protein expression was induced with 1 mM IPTG for 4 h at 30°C. Harvested cell pellets were stored at −20°C before use. For protein purification, E. coli cell pellets were resuspended in PBS with protease inhibitors (5 μg/mL Antipain, 2 μg/mL Aprotinin, 0.35 μg/mL Bestatin, 6 μg/mL Chymostatin, 2.5 μg/mL Leupeptin, 1 μg/mL Pepstatin, 0.1 mM PMSF, 25 μg/mL DNAse and 1 mM DTT). Cells were disrupted using EmulsiFlex and unbroken cells were removed by centrifugation at 4,500 rpm for 15 min (rotor SX4400, Beckman Coulter). The resulting supernatant (SN1) was subjected to a high-speed centrifugation at 14,000 rpm for 1 h (rotor SS-34, Thermo Scientific), which yielded supernatant 2 (SN2), a soluble fraction that included overexpressed proteins. Proteins were purified by affinity chromatography using Glutathione Agarose 4B beads (Macherey-Nagel). To this end, SN2 was incubated with the pre-equilibrated glutathione agarose beads for 2 h in a tube rotator. After collection of the flow-through, using a gravity flow column, the protein-bound beads were washed five times with PBS. Proteins were eluted with 10 mM reduced glutathione in 50 mM Tris-Cl (pH 8). The buffer of the eluted protein was exchanged to PBS using Amicon centrifugation tubes with molecular weight cut-off (MWCO) 10 kDa. The concentration of the proteins was determined by the Bradford method (Thermo, Coomassie Plus assay kit), and protein aliquots were stored at—80°C. All the purification steps were performed at 4°C.
2.6.2 In vitro pull-down
20 µL bed volume Glutathione Agarose 4B beads (Macherey-Nagel) were incubated with 200 µg of recombinantly purified GST and GST-TbPEX19 proteins in separate tubes for 2 h at 4°C with gentle rotation. Following incubation, beads were washed with PBS to remove unbound proteins. Subsequently, 25 µg of C-terminally His6-tagged synthetic peptides of crude grade, containing the corresponding TbPEX19 binding regions in TbPEX11 (BS1-BS3) were loaded to the respective tubes and were incubated for 2 h at 4°C with gentle rotation to allow binding of the peptides to GST-TbPEX19 or control GST. Following washes with PBS, the bound proteins/peptides were eluted with 50 µL 10 mM reduced glutathione in 50 mM Tris (pH 8). The eluted samples were analyzed by SDS-PAGE followed by Coomassie staining and immunoblotting. The sequences of the TbPEX11 peptides used for the pull-down are as follows, BS1: QTDGRDKILKAFSGVFKALGSLD-GS-His6, BS2: CRAKGKVNMKEVLKFLRVLCNFL-GS-His6 and BS3: VLDVVALYGALQKRASDPATS-GS-His6.
2.6.3 AlphaScreen binding assay
N-terminal GST tagged TbPEX19 and TbPEX11 peptides with C-/N-terminal His6 were used for the interaction study with the AlphaScreen system. The final reaction volume used for the study was 25 μL, which consist of 5 μL of each protein solution (30 nM for PEX19 and 300 nM of PEX11 peptides), 5 μL of buffer, and 5 μL of solution for each of the donor and acceptor beads (5 μg/mL). The above solutions were prepared in reaction buffer [0.5% BSA v/v, PBS (pH7.4)] on the day of the assay. Compounds were incubated with the proteins for 30 min at room temperature (RT). 5 μL of AlphaScreen Nickel-chelate acceptor beads (cat. no. 6760619C, PerkinElmer®) were added to the above mixture following 15 min incubation at RT. 5 μL AlphaScreen Glutathione donor beads (cat. no. 6765300, PerkinElmer®) were added to the mixture. The complete 25 μL reaction solutions were incubated for 45 min at RT in the dark, and Alpha signals were captured with Cytation 5 plate reader (BioTek®) with the gain value set at 180. All above concentrations mentioned for the AlphaScreen assays were final concentrations unless otherwise stated. The sequences of the TbPEX11 peptides used for the AlphaScreen assay are as follows, BS1: QTDGRDKILKAFSGVFKALGSLD-GS-His6, His6-GS-QTDGRDKILKAFSGVFKALGSLD and BS2: CRAKGKVNMKEVLKFLRVLCNFL-GS-His6. The binding assay were performed in three biological replicates, with 3 technical replicates each.
2.7 Immunoblotting
Proteins separated by SDS-PAGE were transferred on a nitrocellulose membrane with a pore size of 0.45 μm (Amersham Biosciences). Blotting was performed by using the MiniProtean III cell (BioRad) with blot transfer buffer (Dunn carbonate buffer) for 1 h with a constant current of 300 mA per chamber. Further, the membrane was blocked for 1 h at room temperature (RT), under constant swirling with 3% BSA in blot washing buffer (TBS with 0.05% Tween-20) to avoid nonspecific binding of antibodies. Then, the membrane was washed three times for 5 min at RT, and subsequently incubated with the primary antibodies in blot washing buffer at 4°C overnight. Following primary antibodies were used in this study: mouse anti-GFP (Sigma, 1:2,000), anti-GAL4 AD/-BD (Santa Cruz Biotechnology, 1:1,000) or anti-His6 (Invitrogen, 1:2,000); rabbit anti-Trypanosoma Aldolase (1:20,000) or Enolase (1:20,000), and anti-Porin (S. cerevisiae, 1:10,000). After three washing steps, the corresponding secondary antibodies i.e., goat anti-rabbit IRDye 680 or goat anti-mouse IRDye 800CW (LI-COR Biosciences, both 1:15,000 in blot wash buffer) were applied, and the membrane was further incubated for 30 min at RT in the dark. Following three washes, immunoblots were scanned using the Li-Cor Odyssey 9120 Infrared Imaging System.
2.8 Statistical analysis
Microscopic data was collected from two independent S. cerevisiae or T. brucei cultures. Images were quantified using Pearson’s correlation coefficient, which was calculated with colocalization tool of Zen 3.6 pro (blue edition). Statistical significances for colocalization studies were calculated using a one-way ANOVA (mixed) by Dunnett’s multiple comparisons test (comparison with WT control) with each row representing matched or repeated measures. Statistical analysis for AlphaScreen results was done using two-way ANOVA with Bonferroni’s multiple comparison test (comparison with respective controls) with the values obtained from three independent biological replicates, each with three technical replicates.
3 Results
3.1 Identification and validation of PEX19 binding sites in Trypanosoma brucei PEX11
PEX19 acts as a cytosolic chaperone and receptor for the import of newly synthesized class 1 peroxisomal membrane proteins (PMPs), except the class II PMP i.e., PEX3, which can be imported independent of PEX19 (Sacksteder et al., 2000; Jones et al., 2004). PMPs contain multiple PEX19 binding sites, which are well characterized in yeast and humans. This includes the most abundant yeast PMP Pex11p and related PEX11-family proteins Pex25p and Pex27p (Rottensteiner et al., 2004; Halbach et al., 2005). In trypanosomatid parasites, PEX19 binding sites have been identified in various glycosomal membrane proteins as well as parasite specific PEX11 family proteins GIM5A/B (Saveria et al., 2007). However, the PEX19-binding sites (-BS) in parasite PEX11 remained uncharacterized. To identify these binding sites, we obtained synthetic peptide arrays, containing consecutive 15-amino acid peptides with two amino acid shifts, representing the entire sequence of TbPEX11. Affinity purified recombinant GST-TbPEX19 (Purification profile described in Supplementary Figure S1), or GST alone were incubated with the arrays and bound proteins were immuno-detected using monoclonal anti-GST antibodies. Immunodetection of at least three consecutive spots were considered as potential PEX19 binding sites (Rottensteiner et al., 2004). Comparison of control and test peptide arrays revealed the presence of three potential PEX19 binding sites (BS1-BS3) in TbPEX11 (Figure 1). The topological prediction of transmembrane domains (TMDs) using Phobius webserver (Kall et al., 2007) indicates that TbPEX11 contains four TMDs and an N-terminal extension of about 90 amino acids to the cytosol (Supplementary Figure S2). The first PEX19 binding site (BS1) is present close to the N-terminus of TbPEX11 between amino acid (aa) residues 13–35, the second and third PEX19 binding sites are located between aa77-99 and aa139-159, respectively, in proximity of the first and second predicted transmembrane domains (Figure 1C). Both N- and C-termini of TbPEX11 face the cytosol (Lorenz et al., 1998), which implies that the BS1 would remain exposed to the cytosol even after targeting and insertion of TbPEX11 into the glycosomal membrane.
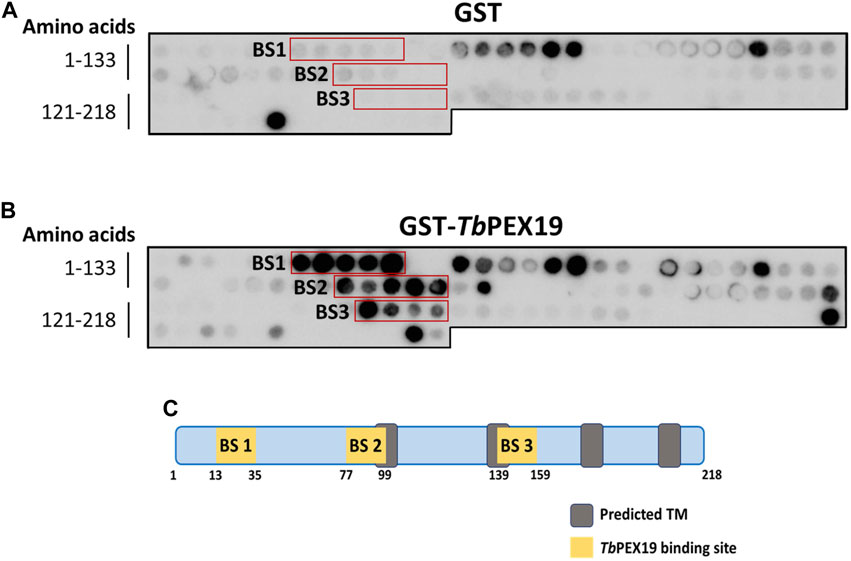
FIGURE 1. Identification of PEX19 binding sites in Trypanosoma PEX11 using synthetic peptide arrays. Synthetic 15-mer peptides with 2-amino acids shifts corresponding to the complete TbPEX11 protein sequence were synthesized on cellulose membrane and probed with GST as negative control (A) or GST-TbPEX19 (B). Bound analyte was immuno-detected using primary antibodies against GST and horseradish peroxidase coupled secondary antibodies, followed by the signal detection using chemiluminescence. Three regions in TbPEX11 showed clear and specific interaction with TbPEX19 as compared to the GST control (red boxes, marked BS1-BS3). (C) Scheme of TbPEX11 showing the identified binding regions in relation to transmembrane segments predicted using Phobius webtool (https://phobius.sbc.su.se/) (Supplementary Figure S2).
PEX19 binding motifs are conserved between peroxisomal proteins of yeast or mammals and trypanosomal glycosomal proteins (Saveria et al., 2007). Probing of the TbPEX11 peptide array with GST-tagged recombinant human PEX19 also revealed a similar binding pattern (Supplementary Figure S3) as observed with TbPEX19 (Figure 1). This further demonstrates the conservation of PEX19-BSs, which can be recognized by PEX19 from different organisms.
Binding of TbPEX19 to the newly identified regions in TbPEX11 were further investigated by yeast two-hybrid (Y2H) analysis and in vitro binding assays using pull-down and AlphaScreen. For the Y2H assay, GAL4-AD fusion of TbPEX19 was used since the corresponding GAL4-BD fusions showed autoactivation (not shown). In addition to this, the corresponding GAL4-BD fused PEX11 constructs did not result in the auto activation when tested with GAL4-AD alone (not shown). Various TbPEX11 constructs fused to GAL4-BD were tested for interaction with TbPEX19 (Figure 2A). Full length TbPEX11 showed only a very weak interaction with TbPEX19. This could be due to the presence of several predicted transmembrane domains in TbPEX11, which may hinder the translocation into the nucleus and activation of GAL-promoter. However, the N-terminal fragment of TbPEX111-89aa that has been predicted to be soluble (Supplementary Figure S2) showed a clear and strong interaction with the full length TbPEX19, in both plate-based (sensitive) and liquid Y2H assays (quantitative) (Figure 2A). Construct that lacks the N-terminal domain but contains BS3 (TbPEX1190-218aa) did not interact with TbPEX19. TbPEX111-89 contains BS1 as well as partial BS2. We also tested a shorter construct that contains only BS1 (TbPEX111-76), which still showed a strong interaction with TbPEX19. Immunoblotting confirmed that all constructs are expressed in yeast at correct molecular weights (Figure 2B). Furthermore, we opted to introduce two mutations in Trypanosoma PEX11. The first mutation replaced serine 25, which is in the TbPEX19 BS1 region, by aspartate to mimic phosphorylation (based on the post-translational modifications database). For the second mutation, we referred to a study in baker’s yeast, which showed that replacing leucine at position 35 by proline results in the loss of interaction (Rottensteiner et al., 2004). We aligned the sequence with ScPEX11 and chose the closest leucine residue (position 31) for replacement by proline. Mutation of serine25 to aspartate i.e., phospho-mimicking did not affect the interaction, whereas mutation of serine25 or leucine31 individually and together to proline within BS1 in TbPEX111-89 led to a reduced or complete abolishment of the interaction with TbPEX19 (Supplementary Figure S4). Based on the peptide blot and the Y2H studies, it can be concluded that BS1 is a bona fide PEX19 binding motif.
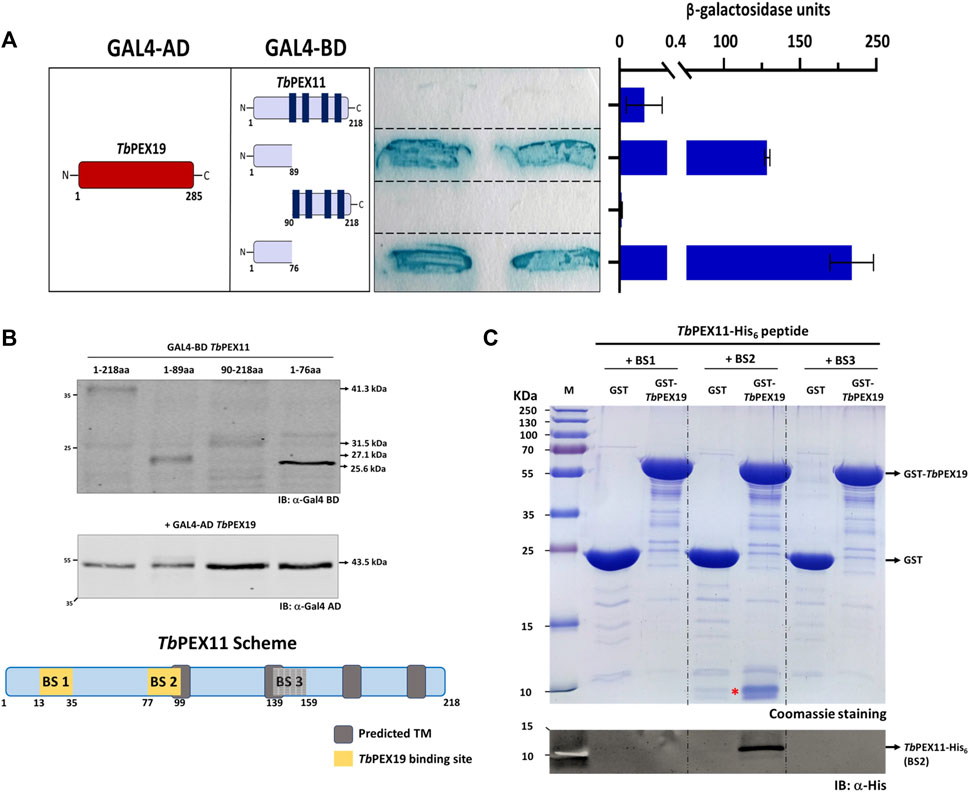
FIGURE 2. Validation of TbPEX19 binding sites in TbPEX11 (A) Yeast two-hybrid assay (Y2H): Full length TbPEX19 and various TbPEX11 constructs fused to the GAL4-activation domain (AD) or -binding domain (BD) as indicated (left panel) were co-transformed into PCY2 yeast strain and analyzed by Colony-lift filter assay (middle panel) and liquid ONPG assay (right panel). Both assays were done in three replicates and the β-galactosidase activity units shown are an average of the technical replicates with three different clones. Error bars represent mean with standard deviations. The soluble N-terminal fragments of TbPEX11 comprising BS1 and partially BS2 (1–89) as well as a shorter fragment containing only BS1 (1–76) showed a clear interaction with full-length TbPEX19. Full length TbPEX11 showed a weaker interaction, while the PEX11-fragment (90–218), lacking the N-terminal PEX19 BSs, did not interact with TbPEX19. The GAL4-AD fusion of TbPEX19 and the various GAL4-BD fusions of TbPEX11 were tested for autoactivation and no coloration of the filter was seen (not shown). (B) Expression of the GAL4-AD and -BD fusion proteins in the yeast two-hybrid (Y2H) assay was confirmed by immunoblotting with monoclonal antibodies against GAL4-AD and -BD as indicated. Arrow marks on the right correspond to the predicted molecular weights of the fusion proteins. The scheme at the bottom shows the modular structure of TbPEX11, highlighting the identified putative TbPEX19 binding regions. (C) In vitro pull-down of TbPEX19 with His6-tagged synthetic peptides of TbPEX11 that correspond to the binding regions highlighted in the PEX11 scheme. Recombinant GST-TbPEX19 or GST as negative control were pre-incubated with Glutathione agarose beads, followed by incubation with the C-terminally His6-tagged synthetic peptides of TbPEX11. After thorough washing, bound proteins were eluted with reduced glutathione and analyzed by SDS-PAGE and staining with Coomassie brilliant blue (upper panel). The peptide corresponding to BS2 (migrating at ∼10 kDa and marked with a red asterisk) was pulled down with GST-TbPEX19, which was also confirmed by immunoblotting using an anti-His monoclonal antibody (lower panel). The putative BS3 could not be validated by either of the assays, therefore it was not considered further and is shown as grey box in the scheme in (B). AD: Activation domain, BD: Binding domain, ONPG: ortho-Nitrophenyl-ß-galactoside.
As an alternative, we obtained C-terminally His6-tagged synthetic peptides of TbPEX11 corresponding to the three putative PEX19 binding sites. Affinity pull-down was performed with GST-TbPEX19 or GST alone as negative control, which were bound to the glutathione affinity beads. Glutathione eluates of the in vitro pull-downs were analyzed by Coomassie staining as well as immunoblotting using anti-His6 tag antibodies (Figure 2C; full profile of the pull-downs is shown in Supplementary Figure S5). The analysis shows that the synthetic peptide corresponding to BS2 (running at ∼10 kDa) is efficiently retained with TbPEX19 but not with GST alone. Similar binding of the BS2 representing peptide of TbPEX11 was also observed with recombinant human GST-PEX19 (not shown). The synthetic peptides corresponding to BS1 and BS3 (both running at ∼10 kDa) did not bind to recombinant GST-TbPEX19 in this assay. The third putative PEX19 binding site (BS3) in TbPEX11 that was identified in the peptide array analysis (Figure 1) could not be further validated by the methods employed here and was not considered further.
Finally, we analyzed the interaction of BS1 and BS2 with TbPEX19 with the more sensitive AlphaScreen assay. This assay was performed using C-terminally His6-tagged peptides that represent the TbPEX11 binding sites with GST-TbPEX19 or GST as negative control (Supplementary Figure S6). Again, the BS2 showed a clear interaction with TbPEX19, while the BS1 did not interact. As the interacting BS1 containing region was N-terminally tagged in the Y2H assay (Figure 2A), we considered that the orientation of the tag might have an influence and therefore analyzed the interaction of an N-terminally tagged BS1-peptide, which indeed showed a significant interaction with TbPEX19 (Supplementary Figure S6). Taken together this study identified two PEX19 binding sites in TbPEX11 (BS1 and BS2).
3.2 Role of PEX19 binding sites in glycosomal targeting of TbPEX11
We performed immunofluorescence microscopy analysis to assess the relevance of the newly identified PEX19 binding sites for the topogenesis of TbPEX11. Tetracycline inducible stable cell lines of Trypanosoma were generated, which express C-terminally GFP-tagged full-length TbPEX11 and variants lacking either BS1 or BS2. Glycosomal localization of the constructs was investigated by analysis of colocalization of the fluorescent GFP-fusions of TbPEX11 with the glycosomal marker enzyme aldolase, which was monitored by immunofluorescence microscopy. Overexpressed TbPEX11WT-GFP colocalized with the glycosomal marker, indicative for its glycosomal localization (Figure 3A, upper panel). However, frequently glycosomes appeared to cluster, confirming an earlier study reporting that overexpression of TbPEX11 results in clustering of glycosomes in bloodstream form of T. brucei (Lorenz et al., 1998). The GFP fluorescence of cells expressing both truncated TbPEX11 variants was much weaker in comparison to the wild-type protein and clustering of glycosomes was not seen. This is explained by the decreased steady-state concentration of both truncated proteins, which is much lower in comparison to the full-length TbPEX11 as indicated by the corresponding immunoblots. (Figure 3D). However, the fluorescence was bright enough to allow investigation of their subcellular localization. TbPEX11 lacking BS1 (TbPEX11△BS1-GFP) still showed a partial glycosomal localization (Figure 3A, Middle panel), while the TbPEX11 variant lacking BS2 (TbPEX11△BS2-GFP) was mislocalized, as indicated by the lacking colocalization with the glycosomal marker (Figure 3A, lower panel). Taken together this result demonstrated that deficiency in either BS1 or BS2 affects the steady-state concentration of TbPEX11. Thus, binding of PEX19 to either of these sites might stabilize the protein. This is in agreement with studies in yeast, which showed that various PMPs, including PEX11, are unstable and their steady state levels are significantly reduced in PEX19-or PEX3-deficient cells (Hettema et al., 2000). In the absence of BS1, the remaining small amount of TbPEX11 is still directed to glycosomes, while in the absence of BS2, PEX11 is mistargeted, indicating that BS2 is essential for glycosomal targeting of TbPEX11.
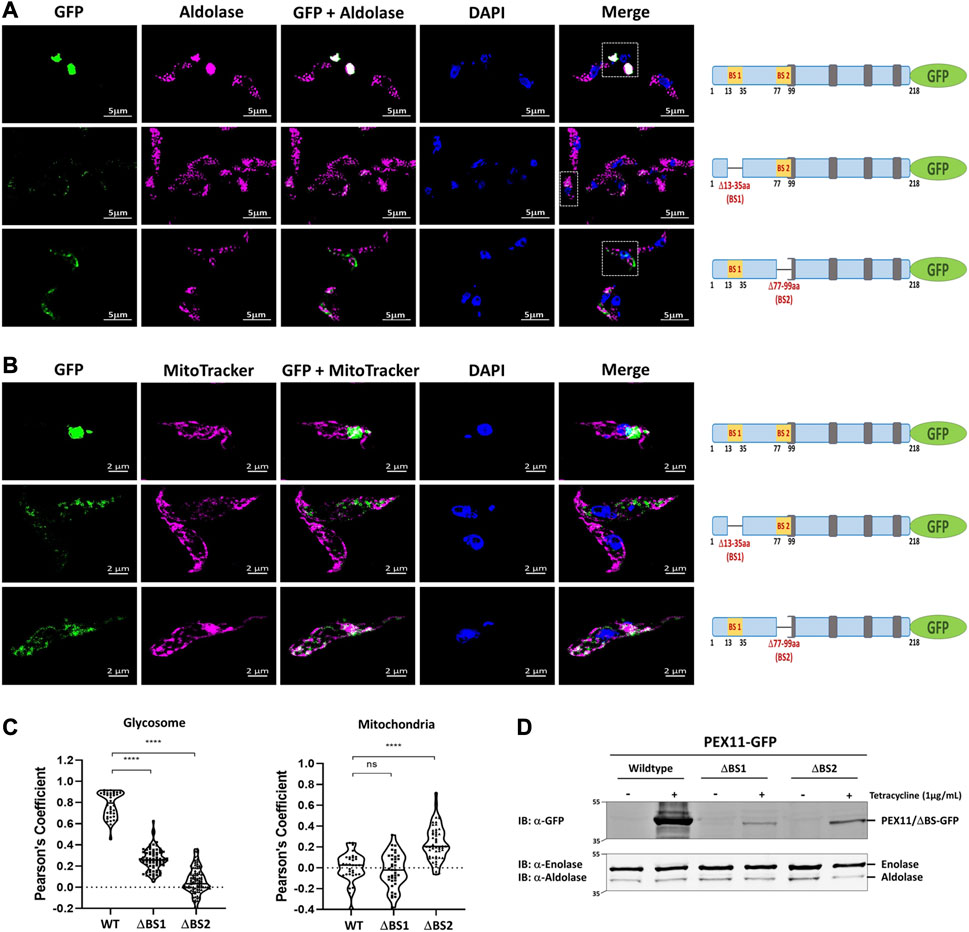
FIGURE 3. PEX19 binding sites are required for maintenance and essential for glycosomal targeting of TbPEX11. (A) Binding site 2 (BS2) is required for glycosomal targeting of TbPEX11. Trypanosoma brucei parasites (procyclic form) expressing tetracycline-induced and C-terminally GFP tagged TbPEX11 constructs (wildtype or mutant proteins lacking PEX19 binding sites) were analyzed for localization of the GFP fusion proteins, the glycosomal marker aldolase, as well as the DAPI-stained nucleus and kinetoplast by fluorescence- or immunofluorescence microscopy. The GFP fusion of wildtype TbPEX11 (upper panel) did colocalize with the glycosome marker aldolase (pseudo-colored to magenta). It is also evident that the overexpression of the full-length TbPEX11 results in the clustering of glycosomes as previously reported (Lorenz et al., 1998). The mutant lacking the first PEX19 binding site (middle panel) partially colocalized with the glycosome marker aldolase. In this case, a clustering of glycosomes was not seen, most likely as the steady-state concentration of the truncated protein was much lower than the corresponding full-length TbPEX11 (see below). PEX11-GFP harboring deletion of BS2 (Δ77-99aa) did not colocalize with the glycosomal marker aldolase (lower panel), but instead showed mislocalization to mitochondrion as demonstrated by colocalization with the mitochondrial marker MitoTracker (pseudo-colored to magenta) (B). Scale bar—5 μm and −2 μm. Schematic representation of the various PEX11-GFP constructs is shown on the right. (C) Quantification of the colocalization to glycosomes (left) or mitochondrion (right). The Pearson’s coefficient of colocalization to respective organelle is shown. Dots within the violin plot indicates individual Pearson correlation coefficient data points and the central line represents the median. Statistical significance were calculated by one-way ANOVA, with Dunnett’s multiple comparisons test (n ≥ 35 cells). ****, p < 0.0001; ns, not significant. (D) Analysis of the expression levels of PEX11-GFP (wildtype and mutants) upon tetracycline induction (+/−) by immunoblotting with anti-GFP antibodies. Cytosolic marker enolase and glycosomal marker aldolase served as the loading controls (lower panel). Wildtype TbPEX11 expression was highly induced, resulting in a high steady-state concentration, while the steady-state concentration of both truncation mutants of TbPEX11 were very low in comparison to the wildtype TbPEX11, most likely due to an instability of the TbPEX11 constructs lacking either of the PEX19 binding sites.
In yeast, PEX11 mislocalizes to mitochondria in cells lacking peroxisomal membranes (Hettema et al., 2000; Mattiazzi Usaj et al., 2015). To assess whether mislocalized TbPEX11 is targeted to mitochondria also in trypanosomes, mitochondrial staining was performed. Indeed, colocalization of the truncated TbPEX11 with the MitoTracker indicated that TbPEX11 lacking BS2 is mistargeted to the mitochondrion (Figure 3B, lower panel).
Multiple sequence alignment of the N-terminal region comprising BS1 of Trypanosoma, yeast, human, and plant PEX11 family proteins or isoforms indicates a high degree of sequence conservation, suggesting that the region corresponding to trypanosomal BS1 is conserved among PEX11 species (Figure 4A). To investigate the capacity of this region of human PEX11 proteins for PEX19 binding, we obtained synthetic peptide array of N-terminal soluble domains of all three human PEX11 isoforms (15mer peptides with 2-amino acids shifts). The arrays were probed with GST alone as a negative control, which showed little or no background (Figure 4B, upper panel). Probing the array with GST-HsPEX19 revealed that the peptides from PEX11γ did not bind PEX19, while PEX11α and PEX11β do contain potential PEX19-BS (Figure 4B, lower panel). To validate the interactions, Y2H analysis was performed to investigate the interaction of N-terminal domains of PEX11 isoforms and human PEX19 (Figure 4C). An interaction was seen only with PEX11β, in both the plate-based and the liquid assay. Immunoblot analysis of lysates of yeast cell used in Y2H shows that PEX11α and β were expressed, but not PEX11γ (Figure 4D). As PEX11γ was not expressed, no conclusion can be drawn from the negative result of the two-hybrid study. However, the results are clear in that PEX11β indeed does contain a PEX19 binding site in the region that corresponds to trypanosomal BS1.
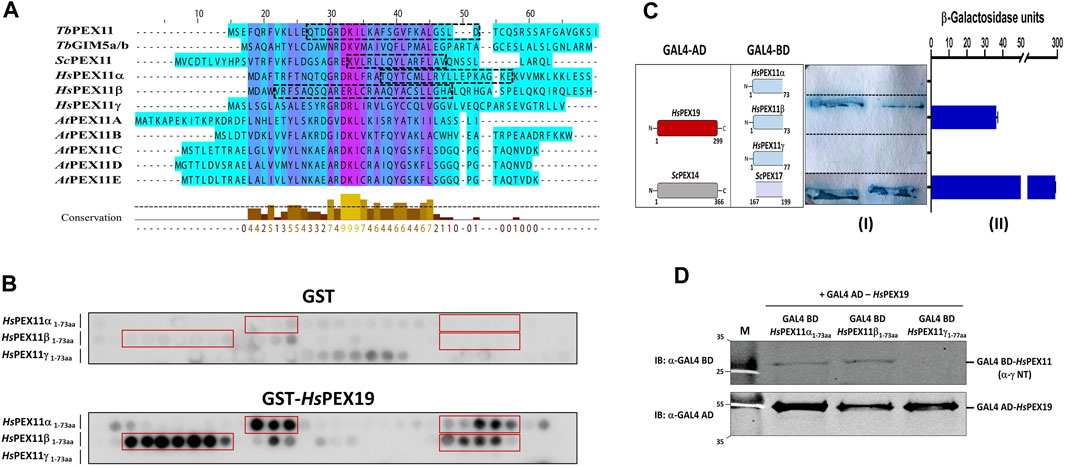
FIGURE 4. The N-terminal PEX19 binding site in PEX11 is conserved among species. (A) Multiple sequence alignment of N-terminal sequences of PEX11 proteins from parasites, baker’s yeast, humans, and plants shows a high degree of conservation. The N-terminal PEX19 binding site (BS1) identified in Trypanosoma brucei is indicated by a black dotted box. The known PEX19 binding site from yeast and the newly identified PEX19 binding sites in humans are also indicated by black dotted boxes. (B) Identification of N-terminal PEX19 binding site in the human members of the PEX11 family. Synthetic 15-mer peptides with 2-amino acids shift of the N-terminal protein sequence of HsPEX11 (α, β and γ) were synthesized on cellulose membrane and probed with GST as a negative control (upper panel) or GST-HsPEX19 (lower panel). The bound analyte was immunodetected using primary antibodies against GST and horseradish peroxidase coupled secondary antibody. The signal was monitored using chemiluminescence. Binding regions in HsPEX11 (α and β) showed clear interaction with HsPEX19 (red boxes). (C) Validation of identified binding sites by Yeast two-hybrid analysis using Δpex19 PCY2 strain. Scheme of the cotransformed HsPEX19 and HsPEX11 (α, β and γ) N-terminal constructs is shown on the left. Colony lift filter assay (I) and liquid ONPG assay (II) were performed using full-length HsPEX19 and different constructs of HsPEX11 fused to GAL4-AD and -BD as indicated. The interaction of yeast PEX14-PEX17 served as a positive control. Both assays were performed in three replicates and the β-galactosidase activity units shown are an average of the technical replicates with three different clones. Error bars represents mean with standard deviations. Of the three members of the PEX11-family only the N-terminal fragment of HsPEX11β showed a clear interaction with full-length HsPEX19. (D) Expression of the GAL4-AD and -BD fused proteins were tested by immunoblotting using GAL4-AD and -BD with monoclonal antibodies. No expression of Gal BD-HsPEX11γ N-terminal fragment was detected.
In yeast, ScPEX11 contains only one PEX19 binding site, spanning amino acids 27–41 (Rottensteiner et al., 2004), that is homologous to trypanosomal BS1. Mutational analysis of this binding site indicated that the L35P mutation completely abolished interaction with ScPEX19 (Rottensteiner et al., 2004). Here we introduced this mutation into the full-length sequence of PEX11 fused to GFP and analyzed its subcellular localization in comparison to wild-type PEX11 by fluorescence microscopy (Figure 5A). As expected, the full-length PEX11-GFP is targeted to peroxisomes as indicated by its co-localization with the peroxisomal marker DsRed-SKL (Figure 5A, middle panel). The PEX11-GFP fusion harboring the L35P exchange, however, was partly mislocalized to tubular structures (Figure 5A, lower panel). We further performed staining of yeast cells that express PEX11L35P-GFP together with a mitochondrial marker (MitoTracker) (Figure 5B, lower panel), confirming that the L35P mutant is mislocalized to mitochondria. Immunoblot analysis of cells shown in Figure 5A show that GFP-tagged wild-type PEX11 is stable, but the steady-state concentration of L35P mutant protein that cannot bind to PEX19 is very low in comparison (Figure 5D).
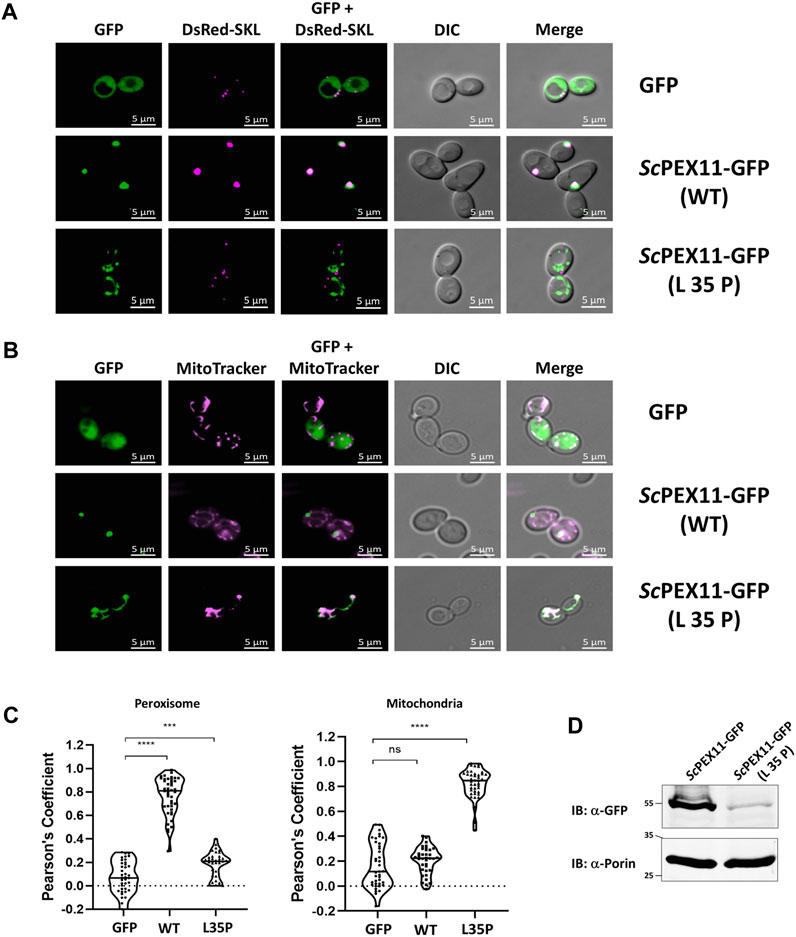
FIGURE 5. The N-terminal binding site for PEX19 is essential for peroxisomal localization of ScPEX11. Plasmids expressing ScPEX11-GFP (wildtype and L35P-mutant) or the peroxisomal marker protein DsRed-SKL were cotransformed in the BY4742 yeast strain. (A) Clones expressing GFP fusion proteins and the peroxisomal marker DsRed-SKL were grown on plates and visualized by fluorescence microscopy. Merged images reveal peroxisomal colocalization of ScPEX11-GFP (wildtype) with DsRed-SKL (pseudo-colored to magenta), indicative for its peroxisomal localization. In contrast, the L35P exchange that is known to block PEX19 binding site result in mislocalization of ScPEX11. (B) ScPEX11-GFP with mutation L35P mislocalizes to mitochondria as demonstrated by its colocalization with MitoTracker (pseudo-colored to magenta). DIC–Differential Interference Contrast, Scale bar—5 μm. (C) Quantification of the colocalization of ScPEX11-GFP to peroxisomes (left) or mitochondria (right). The Pearson’s coefficient of colocalization to respective organelle is shown. Dots within the violin plot indicates individual Pearson correlation coefficient data points and the central line represents the median. Statistical significances were calculated by one-way ANOVA, with Dunnett’s multiple comparisons test (n ≥ 35 cells). ****, p < 0.0001; ***, p = 0.0003; ns, not significant. (D) Expression of ScPEX11-GFP (wildtype and L35P mutant) was tested by immunoblotting with anti-GFP antibody, which revealed that the steady-state concentration of the L35P is much lower than that of the corresponding wild-type protein. Porin served as the loading control.
The data show that PEX11 from Trypanosoma and yeast as well as PEX11β from humans contain a conserved N-terminal region that can bind PEX19. This region, corresponding to BS1 in Trypanosoma, is required to maintain the steady-state concentration of PEX11 in all studied species, and at least for yeast, it is shown that it is also required for efficient targeting of PEX11 to peroxisomes.
3.3 Cryptic N-terminal targeting signal of trypanosomal PEX11
In the absence of PEX19, PMPs are mislocalized to the cytosol and rapidly degraded, or mislocalized to other membranes. For example, PEX3 localizes to the endoplasmic reticulum (Hoepfner et al., 2005) but many PMPs, including yeast PEX11 and PEX13, accumulate in mitochondria when peroxisomes are absent in the cell (Nuebel et al., 2021). PEX13 that is mislocalized to mitochondria can recruit functional docking and import peroxin complexes to mitochondria and also some peroxisomal matrix proteins (Nuebel et al., 2021). Peroxins also accumulate in mitochondria of Zellweger patient-derived cells leading to mitochondrial dysfunction (Nuebel et al., 2021). This can be rescued by overexpressing mitochondrial quality control ATPase ATAD1. We showed that TbPEX11-GFP lacking BS2 is mislocalized to mitochondrion in trypanosomes (Figure 3B, lower panel). However, glycosomal targeting also requires the presence of a transmembrane segment for correct targeting. Accordingly, the TbPEX11 construct containing the first 90 amino acids fused to GFP that lacks the transmembrane domain (TMD) is also mistargeted to mitochondrion. This construct is used here as a control to investigate the requirement for the targeting of TbPEX11 to mitochondria (Figure 6C, upper panel). Normally, mitochondrial proteins are targeted via N-terminal or internal mitochondrial targeting signals (Backes et al., 2018; Bykov et al., 2022), After mitochondrial import, the N-terminal targeting presequences of proteins are removed by mitochondrial processing peptidase (MPP) to allow the proper folding of the imported protein (Kunova et al., 2022). Here, we applied the Mitofates webtool to predict putative mitochondrial targeting signals in the N-terminal region of TbPEX11 (Fukasawa et al., 2015). Although the tool does not identify the TbPEX11-NTD as a classical mitochondrial presequence, it predicts the presence of two tandem TOM20 recognition motifs and a positively charged amphiphilic region with mitochondrial processing peptidase (MPP) cleavage site (Figure 6A, upper panel). The putative TOM20 recognition motifs are in the N-terminal region of TbPEX11 (4–31 aa), partially overlapping with the identified BS1-binding region for TbPEX19 (13–35aa) (Figure 6A, upper panel). Further, we looked for the TOM20 motifs by performing multiple sequence alignment of N-terminal region of PEX11 across organisms (Figure 6A, lower panel), which also contains PEX19 binding site (BS) of yeast (Rottensteiner et al., 2004) and the identified PEX19-BS in T. brucei and human (this study). This alignment indicates the conservation of TOM20 motifs in the N-terminal region of PEX11, pointing to its role in mitochondrial mislocalization. To test the putative signal sequences for functionality, we analyzed the role of this region for mitochondrial and glycosomal targeting by fluorescence microscopy (Figure 6B). To this end, GFP tagged N-terminal 90 amino acid region of TbPEX11, with or without BS1 (Δ13-35aa) was analyzed for co-localization with glycosomal marker aldolase (Figure 6B, upper and middle panel). Both fusion proteins were expressed and not targeted to glycosomes but mislocalized to mitochondrion as evident from co-staining with mitochondrial marker (Figure 6C). The expression of these constructs was confirmed by immunoblotting using α-GFP monoclonal antibody (Supplementary Figure S7). Finally, TbPEX11NTD(1-90aa)-GFP lacking amino acid residues 2–11, which were predicted to contain TOM20 motifs and positively charged amphiphilicity was analyzed. Expression of this construct did result in a diffuse cytosolic labelling (Figures 6B,C, lower panels), indicating that deletion of this extreme N-terminal region prevented mitochondrial targeting of the fusion protein.
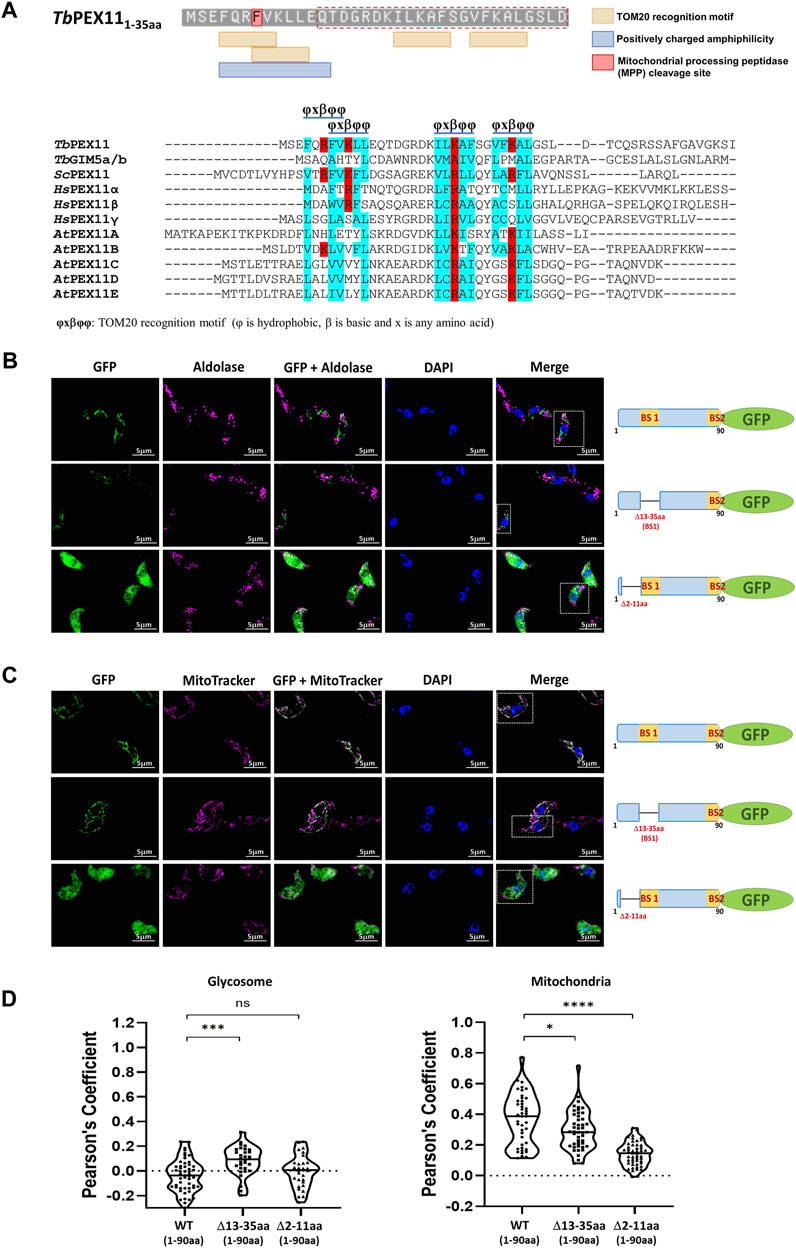
FIGURE 6. Cryptic signal at the N-terminus causes mitochondrial mislocalization of TbPEX11. (A) Upper panel: Prediction of two overlapping putative TOM20 motifs with positively charged amphiphilicity within the N-terminal region TbPEX11 and partially overlapping with the PEX19 binding site region (red dotted box). The residue highlighted in red indicates the presence of a mitochondrial processing peptidase (MPP) cleavage site. The presence of a predicted TOM20 recognition motif and positively charged amphiphilic region with MPP cleavage site were determined by Mitofates webtool (https://mitf.cbrc.pj.aist.go.jp/MitoFates/cgi-bin/top.cgi). Lower panel: Multiple sequence alignment of N-terminal sequences of PEX11 proteins from parasites, yeast, humans, and plants indicates the presence of conserved TOM20 motifs with positively charged (red) residues encased by hydrophobic (blue) amino acid residues. (B) Subcellular localization of the N-terminal domain of TbPEX11 (1–90aa) fused to GFP with and without deletion of the PEX19-binding site 1 (Δ13-35aa) or deletion of a N-terminal putative TOM20 binding motif (Δ2-11aa) by fluorescence and immunofluorescence microscopy. The non-truncated fusion with and without PEX19 binding site 1 (BS1) did not co-localize with the glycosomal marker aldolase but was targeted to mitochondrion as shown below (upper and middle panel). The TbPEX11-GFP lacking the TOM20 motif did mislocalize to the cytosol as indicated by the overall cell labelling (lower panel). (C) The non-truncated fusion with and without PEX19 binding site 1 (BS1) localized to the mitochondrion, demonstrated by their colocalization with MitoTracker (upper and middle panel, respectively). The glycosomal marker aldolase (pseudo-colored to magenta) was labelled with the corresponding antibody, nuclei and kinetoplasts were stained with DAPI, and mitochondrion was visualized by MitoTracker (pseudo-colored to magenta). Scale bar—5 μm. (D) Quantification of the colocalization to glycosomes (left) or the mitochondrion (right). The Pearson’s coefficient of colocalization to respective organelle is shown. Dots within the violin plot indicate individual Pearson correlation coefficient data points and the central line represents the median. Statistical significances were calculated by one-way ANOVA, with Dunnett’s multiple comparisons test (n ≥ 35 cells). ****, p < 0.0001; ***, p = 0.0001; *, p = 0.0262; ns, not significant.
4 Discussion
Here we show that the glycosomal membrane protein TbPEX11 contains two PEX19 binding sites in its N-terminal region, as shown by peptide array analysis, yeast two-hybrid studies, pull-down experiments, and AlphaScreen assays. PEX19 is a peroxisomal membrane protein (PMP) receptor and chaperone that stabilizes its cargo proteins and targets them to peroxisomes (Hettema et al., 2000; Sacksteder et al., 2000; Jones et al., 2004). PEX19 binding sites are distributed across the length of cargo proteins, including the C-terminus in case of tail-anchored (TA) proteins (Halbach et al., 2006). Apart from yeast Pex11p (aa 27–41), PEX19 binding sites close to the N-terminus were also found in yeast Pex3p (aa 28–42) and Pxa1p (aa 33–47) (Rottensteiner et al., 2004). Along this line, the N-terminal 95 residues of Pxa1p have recently been shown to be sufficient for targeting a reporter protein to peroxisomes. Interestingly, truncated Pxa1p lacking residues 1–95 still localized to peroxisomes but its targeting depended on the presence of its interaction partner Pxa2 (Jansen et al., 2023).
Eukaryotic organisms contain multiple proteins belonging to the PEX11 family. In yeast, Pex11p contains a single PEX19-BS near its N-terminus, while in the other ScPEX11-family members, Pex25p and Pex27p, binding of PEX19 occurs far distal from the N-terminus (Rottensteiner et al., 2004). A recent study identified a classical PEX19-BS near the N-terminus of Pichia pastoris PEX11 (Zientara-Rytter et al., 2022). However, the study also showed that amphipathic helix 4 (H4) located in the C-terminal region of PpPEX11, functions as a second, PEX19-independent mPTS, which is preserved among PEX11-family proteins (Zientara-Rytter et al., 2022). Thus, unlike most PMPs, PEX11 of Pichia pastoris can use two mechanisms of transport to peroxisomes, where only one of them depends on its direct interaction with PEX19, but the other does not. The presence of such a PEX19-independent targeting signal is not confirmed in our studies for PEX11 from T. brucei and S. cerevisiae, as N-terminal mutations or truncations of PEX19-binding sites BS1 or BS2 but not of the amphiphilic helix 4 did prevent efficient glycosomal or peroxisomal targeting of the proteins.
Our data indicate that the N-terminal binding site (BS1) for PEX19 is conserved among TbPEX11 orthologues but not in PEX11-family member GIM5/B of Trypanosoma or Leishmania (Saveria et al., 2007). Our results also indicate differences within PEX11 isoforms in humans. All three isoforms, PEX11α, PEX11β, and PEX11γ show sequence similarities to the established PEX19 binding site BS1 in yeast and T. brucei, but only HsPEX11β did bind human PEX19 (Figures 4B,C). PEX11β is a key factor in the regulation of peroxisome abundance in mammals (reviewed in (Schrader et al., 2016)). It functions as a membrane-remodeling protein that can deform and elongate the peroxisome membrane prior to fission (Delille et al., 2010; Yoshida et al., 2015). Accordingly, PEX11β is the functional counterpart of yeast and T. brucei PEX11 that are targeted to their destination in a PEX19-dependent manner and contribute to the morphogenesis of the peroxisomal membrane, which is required for subsequent fission. Overall, the N-terminal PEX11-binding sites for PEX19 are conserved among species. In this study, this is highlighted by the peptide array analysis of TbPEX11, which revealed that human PEX19 binds to the same regions as trypanosomal PEX19 (Supplementary Figure S3).
In the absence of peroxisomes, many PMPs are unstable and degraded or mistargeted to other organelles such as ER and mitochondria. This is seen in yeast as well as in human cells derived from patients suffering from a Peroxisome Biogenesis Disorder (PBD) (Hettema et al., 2000; Nuebel et al., 2021). In yeast, PEX3 localizes to the ER (Toro et al., 2009), while several peroxins/PMPs including PEX13, PEX14, PEX17 (peroxisomal docking complex) as well as PEX11 and PEX25 accumulated on mitochondria (Nuebel et al., 2021). Here we show that mislocalization to mitochondria is seen for TbPEX11 lacking its second PEX19-BS and ScPEX11 harboring mutation of its sole PEX19-binding site, (Figure 3B; 5B). Mitochondria have been described as emergency landing places for abandoned peroxins, which results in a partial reconstitution of the peroxisomal import machinery and routing of a substantial part of the peroxisomal proteome to mitochondria (Nuebel et al., 2021; Vogtle and Meisinger, 2021). The mislocalization of PMPs, especially peroxins, to mitochondria are supposed to be the cause for mitochondrial dysfunction in PBD patients (Hettema et al., 2000; Nuebel et al., 2021). Therefore, it is of interest to gain insight into why some PMPs mislocalize to this particular organelle. Mitochondrial targeting signals (MTS) have been extensively characterized across different organisms and consensus motifs can be predicted. Bioinformatic prediction indicated the presence of positively charged amphiphilicity in the extreme N-terminal Helix 1 and detected four TOM20 recognition motifs in NTD of TbPEX11, out of which two tandem motifs coincide with the N-terminal PEX19-BS (Figure 6A). There is no obvious structural similarity between PEX19 and TOM20. The TOM20 recognizes motifs in cargo proteins via its TPR (tetratricopeptide repeat) domain, which is not present in PEX19. In our in vitro studies, PEX19 can directly interact with the PEX19-BSs, without the requirement of an additional cofactor. Surprisingly, the iMTS-Ls predictor also recognizes PEX19-BSs in Tb and ScPEX11 as having Internal Matrix Targeting Signal-like Sequences (iMTSL) propensity (Supplementary Figure S8) (Schneider et al., 2021). How these signals that have primary sequence and potential structural similarities, are faithfully recognized by the correct receptor, and targeted to the correct location requires further investigation. The expression of TbPEX11-NTD lacking N-terminal Helix 1 did not any more mistarget to the mitochondrion. This demonstrates that the N-terminal amphipathic helix at the extreme N-terminus of TbPEX11 is essential for the mitochondrial mislocalization. This result indicates that TbPEX11 harbors a cryptic mitochondrial targeting signal. Whether this is also true for human PEX11 and other PMPs, and involvement of TOM complex machinery requires further investigation.
4.1 What could be the role of a mitochondrial targeting signal of PEX11?
In mature glycosomes/peroxisomes of the wild type cells, TbPEX11-NTD is exposed to the cytosol. In this case, the cryptic N-terminal signals may be masked by the oligomerization of PEX11. However, in newly formed glycosomes, which are importing PEX11, these signals may be still accessible to interact with the mitochondrial TOM machinery, and this may mediate glycosome-mitochondrion membrane contact site (MCS). Accordingly, association of ScPEX11 with the mitochondrial TOM complex has been seen in two studies, i) 37-fold enrichment of ScPEX11 in the SILAC based interactome of yeast TOM22 (Opalinski et al., 2018), and ii) interaction of ScPEX11 with TOM22 in split-ubiquitin assay (Eckert and Johnsson, 2003). Recently, a nuclear membrane protein Cnm1 (Contact nucleus mitochondria 1) was shown to interact with TOM70, a component of the mitochondrial TOM (translocase of outer membrane) complex (Eisenberg-Bord et al., 2021). This interaction establishes nuclear-mitochondrial contact sites, which are regulated by phosphatidylcholine metabolism. Interestingly, Cnm1 harbors two predicted transmembrane domains close to the N-terminus, while C-terminal end contains internal mitochondrial targeting signal-like (iMTS-L) sequences, which are known to directly bind to TOM70 (Backes et al., 2018). Similarly, PEX11 localized to the glycosomal membrane could still associate with the mitochondrial preprotein import machinery to establish glycosome-mitochondrial contact. Interestingly, PEX11 of parasite Entamoeba histolytica shows dual localization to peroxisomes and mitosomes (Verner et al., 2021). In baker’s yeast, PEX11 interacts with Mdm34, a component of the ER–mitochondria encounter structure (ERMES), and act as a peroxisome–mitochondria tether (Mattiazzi Usaj et al., 2015). It has been shown that a mutant form of Mdm34, a component of the ERMES, which impairs ERMES formation and diminishes its association with the peroxisomal membrane protein PEX11, also causes defects in pexophagy (Liu et al., 2018). Along this line, a role for ERMES complex proteins on regulating peroxisome abundance has been reported (Esposito et al., 2019).
We do not yet know whether the newly identified cryptic mitochondrial targeting signal of TbPEX11 is of functional relevance. However, peroxisomes are not only metabolically linked to mitochondria but also share components of their division machinery (Schrader et al., 2015). These include the tail-anchored adaptor proteins FIS1 and MFF, which are dually targeted to both peroxisomes and mitochondria, where they recruit the fission GTPase DRP1 (also known as DNML1) to the organelle membrane (Schrader et al., 2022). In this context, it is interesting to note that targeting of PEX11β to mitochondria induces mitochondrial division in human cells. Accordingly, like PEX11β also TbPEX11 might have the potential to modulate mitochondrial dynamics.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
CK, VK, and RE conceived and planned the experiments. CK carried out the experiments. NS and KW contributed to the super-resolution fluorescence microscopy. BT performed the AlphaScreen assay. MJ synthesized the peptide array. CK, VK, and RE wrote the manuscript with support from WS. VK and RE supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This project work has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 812968—PerICo (to CK, and RE). The work was supported by DFG grant FOR1905 to RE.
Acknowledgments
We thank Paul Michels for kindly providing various antibodies and Wolfgang Girzalsky for critical reading of the manuscript. We acknowledge support by the Open Access Publication Funds of the Ruhr-Universität Bochum.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1213761/full#supplementary-material
References
Abe, I., and Fujiki, Y. (1998a). cDNA cloning and characterization of a constitutively expressed isoform of the human peroxin Pex11p. Biochem. Biophys. Res. Commun. 252 (2), 529–533. doi:10.1006/bbrc.1998.9684
Abe, I., Okumoto, K., Tamura, S., and Fujiki, Y. (1998b). Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett. 431 (3), 468–472. doi:10.1016/s0014-5793(98)00815-1
Agrawal, G., and Subramani, S. (2016). De novo peroxisome biogenesis: evolving concepts and conundrums. Biochim. Biophys. Acta 1863 (5), 892–901. doi:10.1016/j.bbamcr.2015.09.014
Albertini, M., Rehling, P., Erdmann, R., Girzalsky, W., Kiel, J. A., Veenhuis, M., et al. (1997). Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89 (1), 83–92. doi:10.1016/s0092-8674(00)80185-3
Anton, M., Passreiter, M., Lay, D., Thai, T. P., Gorgas, K., and Just, W. W. (2000). ARF- and coatomer-mediated peroxisomal vesiculation. Cell Biochem. Biophys. 32 (1), 27–36. doi:10.1385/cbb:32:1-3:27
Breidenbach, R. W., and Beevers, H. (1967). Association of the glyoxylate cycle enzymes in a novel subcellular particle from castor bean endosperm. Biochem. Biophys. Res. Commun. 27 (4), 462–469. doi:10.1016/s0006-291x(67)80007-x
Backes, S., Hess, S., Boos, F., Woellhaf, M. W., Godel, S., Jung, M., et al. (2018). Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 217 (4), 1369–1382. doi:10.1083/jcb.201708044
Baerends, R. J., Faber, K. N., Kiel, J. A., van der Klei, I. J., Harder, W., and Veenhuis, M. (2000). Sorting and function of peroxisomal membrane proteins. FEMS Microbiol. Rev. 24 (3), 291–301. doi:10.1111/j.1574-6976.2000.tb00543.x
Banerjee, S. K., Kessler, P. S., Saveria, T., and Parsons, M. (2005). Identification of trypanosomatid PEX19: functional characterization reveals impact on cell growth and glycosome size and number. Mol. Biochem. Parasitol. 142 (1), 47–55. doi:10.1016/j.molbiopara.2005.03.008
Bonekamp, N. A., Grille, S., Cardoso, M. J., Almeida, M., Aroso, M., Gomes, S., et al. (2013). Self-interaction of human Pex11pβ during peroxisomal growth and division. PLoS One 8 (1), e53424. doi:10.1371/journal.pone.0053424
Boutouja, F., Stiehm, C. M., Mastalski, T., Brinkmeier, R., Reidick, C., El Magraoui, F., et al. (2019). Vps10-mediated targeting of Pep4 determines the activity of the vacuole in a substrate-dependent manner. Sci. Rep. 9 (1), 10557. doi:10.1038/s41598-019-47184-7
Brun, R., and Schonenberger, (1979). Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 36 (3), 289–292.
Bykov, Y. S., Flohr, T., Boos, F., Zung, N., Herrmann, J. M., and Schuldiner, M. (2022). Widespread use of unconventional targeting signals in mitochondrial ribosome proteins. EMBO J. 41 (1), e109519. doi:10.15252/embj.2021109519
De Duve, C., and Baudhuin, P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46 (2), 323–357. doi:10.1152/physrev.1966.46.2.323
Delille, H. K., Agricola, B., Guimaraes, S. C., Borta, H., Luers, G. H., Fransen, M., et al. (2010). Pex11pbeta-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J. Cell Sci. 123 (16), 2750–2762. doi:10.1242/jcs.062109
Eckert, J. H., and Johnsson, N. (2003). Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J. Cell Sci. 116 (17), 3623–3634. doi:10.1242/jcs.00678
Eisenberg-Bord, M., Zung, N., Collado, J., Drwesh, L., Fenech, E. J., Fadel, A., et al. (2021). Cnm1 mediates nucleus-mitochondria contact site formation in response to phospholipid levels. J. Cell Biol. 220 (11), e202104100. doi:10.1083/jcb.202104100
Erdmann, R., and Blobel, G. (1995). Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128 (4), 509–523. doi:10.1083/jcb.128.4.509
Erdmann, R., and Schliebs, W. (2005). Peroxisomal matrix protein import: the transient pore model. Nat. Rev. Mol. Cell Biol. 6 (9), 738–742. doi:10.1038/nrm1710
Esposito, M., Hermann-Le Denmat, S., and Delahodde, A. (2019). Contribution of ERMES subunits to mature peroxisome abundance. PLoS One 14 (3), e0214287. doi:10.1371/journal.pone.0214287
Faber, K. N., Keizer-Gunnink, I., Pluim, D., Harder, W., Ab, G., and Veenhuis, M. (1995). The N-terminus of amine oxidase of Hansenula polymorpha contains a peroxisomal targeting signal. FEBS Lett. 357 (2), 115–120. doi:10.1016/0014-5793(94)01317-t
Fukasawa, Y., Tsuji, J., Fu, S. C., Tomii, K., Horton, P., and Imai, K. (2015). MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell Proteomics 14 (4), 1113–1126. doi:10.1074/mcp.M114.043083
Galland, N., de Walque, S., Voncken, F. G., Verlinde, C. L., and Michels, P. A. (2010). An internal sequence targets Trypanosoma brucei triosephosphate isomerase to glycosomes. Mol. Biochem. Parasitol. 171 (1), 45–49. doi:10.1016/j.molbiopara.2010.01.002
Gao, Y., Skowyra, M. L., Feng, P., and Rapoport, T. A. (2022). Protein import into peroxisomes occurs through a nuclear pore-like phase. Science 378 (6625), eadf3971. doi:10.1126/science.adf3971
Ghaedi, K., Tamura, S., Okumoto, K., Matsuzono, Y., and Fujiki, Y. (2000). The peroxin pex3p initiates membrane assembly in peroxisome biogenesis. Mol. Biol. Cell 11 (6), 2085–2102. doi:10.1091/mbc.11.6.2085
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi:10.1016/s0076-6879(02)50957-5
Girzalsky, W., Hoffmann, L. S., Schemenewitz, A., Nolte, A., Kunau, W. H., and Erdmann, R. (2006). Pex19p-dependent targeting of Pex17p, a peripheral component of the peroxisomal protein import machinery. J. Biol. Chem. 281 (28), 19417–19425. doi:10.1074/jbc.M603344200
Goldman, B. M., and Blobel, G. (1978). Biogenesis of peroxisomes: intracellular site of synthesis of catalase and uricase. Proc. Natl. Acad. Sci. U. S. A. 75 (10), 5066–5070. doi:10.1073/pnas.75.10.5066
Gould, S. J., Keller, G. A., Hosken, N., Wilkinson, J., and Subramani, S. (1989). A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108 (5), 1657–1664. doi:10.1083/jcb.108.5.1657
Halbach, A., Landgraf, C., Lorenzen, S., Rosenkranz, K., Volkmer-Engert, R., Erdmann, R., et al. (2006). Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119 (12), 2508–2517. doi:10.1242/jcs.02979
Halbach, A., Lorenzen, S., Landgraf, C., Volkmer-Engert, R., Erdmann, R., and Rottensteiner, H. (2005). Function of the PEX19-binding site of human adrenoleukodystrophy protein as targeting motif in man and yeast. PMP targeting is evolutionarily conserved. J. Biol. Chem. 280 (22), 21176–21182. doi:10.1074/jbc.M501750200
Hasan, S., Platta, H. W., and Erdmann, R. (2013). Import of proteins into the peroxisomal matrix. Front. Physiol. 4, 261. doi:10.3389/fphys.2013.00261
Hettema, E. H., Girzalsky, W., van Den Berg, M., Erdmann, R., and Distel, B. (2000). Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19 (2), 223–233. doi:10.1093/emboj/19.2.223
Hilpert, K., Winkler, D. F., and Hancock, R. E. (2007). Peptide arrays on cellulose support: sPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2 (6), 1333–1349. doi:10.1038/nprot.2007.160
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122 (1), 85–95. doi:10.1016/j.cell.2005.04.025
Islinger, M., Li, K. W., Seitz, J., Volkl, A., and Luers, G. H. (2009). Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes--evidence for a natural piggyback import mechanism in mammals. Traffic 10 (11), 1711–1721. doi:10.1111/j.1600-0854.2009.00966.x
Jansen, R. L. M., van den Noort, M., Krikken, A. M., Bibi, C., Bohm, A., Schuldiner, M., et al. (2023). Novel targeting assay uncovers targeting information within peroxisomal ABC transporter Pxa1. Biochim. Biophys. Acta Mol. Cell Res. 1870 (5), 119471. doi:10.1016/j.bbamcr.2023.119471
Jones, J. M., Morrell, J. C., and Gould, S. J. (2004). PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164 (1), 57–67. doi:10.1083/jcb.200304111
Kalel, V. C., and Erdmann, R. (2018). Unraveling of the structure and function of peroxisomal protein import machineries. Subcell. Biochem. 89, 299–321. doi:10.1007/978-981-13-2233-4_13
Kalel, V. C., Schliebs, W., and Erdmann, R. (2015). Identification and functional characterization of Trypanosoma brucei peroxin 16. Biochim. Biophys. Acta 1853 (10), 2326–2337. doi:10.1016/j.bbamcr.2015.05.024
Kall, L., Krogh, A., and Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 35, W429–W432. Web Server issue). doi:10.1093/nar/gkm256
Kamoshita, M., Kumar, R., Anteghini, M., Kunze, M., Islinger, M., Martins Dos Santos, V., et al. (2022). Insights into the peroxisomal protein inventory of zebrafish. Front. Physiol. 13, 822509. doi:10.3389/fphys.2022.822509
Koch, J., Pranjic, K., Huber, A., Ellinger, A., Hartig, A., Kragler, F., et al. (2010). PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123 (19), 3389–3400. doi:10.1242/jcs.064907
Krishna, C. K., Franke, L., Erdmann, R., and Kalel, V. C. (2023). “Isolation of glycosomes from trypanosoma brucei,” in Peroxisomes: Methods and protocols. Editor M. Schrader (New York, NY: Springer US), 33–45.
Kunova, N., Havalova, H., Ondrovicova, G., Stojkovicova, B., Bauer, J. A., Bauerova-Hlinkova, V., et al. (2022). Mitochondrial processing peptidases-structure, function and the role in human diseases. Int. J. Mol. Sci. 23 (3), 1297. doi:10.3390/ijms23031297
Kunze, M. (2023). Computational evaluation of peroxisomal targeting signals in metazoa. Methods Mol. Biol. 2643, 391–404. doi:10.1007/978-1-0716-3048-8_28
Kuravi, K., Nagotu, S., Krikken, A. M., Sjollema, K., Deckers, M., Erdmann, R., et al. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119 (19), 3994–4001. doi:10.1242/jcs.03166
Lazarow, P. B., and Fujiki, Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530. doi:10.1146/annurev.cb.01.110185.002421
Leon, S., Goodman, J. M., and Subramani, S. (2006). Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta 1763 (12), 1552–1564. doi:10.1016/j.bbamcr.2006.08.037
Li, X., and Gould, S. J. (2002). PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 156 (4), 643–651. doi:10.1083/jcb.200112028
Lingard, M. J., and Trelease, R. N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119 (9), 1961–1972. doi:10.1242/jcs.02904
Liu, X., Wen, X., and Klionsky, D. J. (2018). Endoplasmic reticulum–mitochondria contacts are required for pexophagy in Saccharomyces cerevisiae. Contact 2, 2158. Thousand Oaks) 2. doi:10.1177/2515256418821584
Lorenz, P., Maier, A. G., Baumgart, E., Erdmann, R., and Clayton, C. (1998). Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 17 (13), 3542–3555. doi:10.1093/emboj/17.13.3542
Maier, A., Lorenz, P., Voncken, F., and Clayton, C. (2001). An essential dimeric membrane protein of trypanosome glycosomes. Mol. Microbiol. 39 (6), 1443–1451. doi:10.1046/j.1365-2958.2001.02333.x
Marshall, P. A., Dyer, J. M., Quick, M. E., and Goodman, J. M. (1996). Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135 (1), 123–137. doi:10.1083/jcb.135.1.123
Mattiazzi Usaj, M., Brloznik, M., Kaferle, P., Zitnik, M., Wolinski, H., Leitner, F., et al. (2015). Genome-wide localization study of yeast Pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J. Mol. Biol. 427 (11), 2072–2087. doi:10.1016/j.jmb.2015.03.004
Mayerhofer, P. U. (2016). Targeting and insertion of peroxisomal membrane proteins: eR trafficking versus direct delivery to peroxisomes. Biochim. Biophys. Acta 1863 (5), 870–880. doi:10.1016/j.bbamcr.2015.09.021
McNew, J. A., and Goodman, J. M. (1994). An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127 (5), 1245–1257. doi:10.1083/jcb.127.5.1245
Meinecke, M., Bartsch, P., and Wagner, R. (2016). Peroxisomal protein import pores. Biochim. Biophys. Acta 1863 (5), 821–827. doi:10.1016/j.bbamcr.2015.10.013
Motley, A. M., and Hettema, E. H. (2007). Yeast peroxisomes multiply by growth and division. J. Cell Biol. 178 (3), 399–410. doi:10.1083/jcb.200702167
Moyersoen, J., Choe, J., Fan, E., Hol, W. G., and Michels, P. A. (2004). Biogenesis of peroxisomes and glycosomes: trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 28 (5), 603–643. doi:10.1016/j.femsre.2004.06.004
Murphy, M. A., Phillipson, B. A., Baker, A., and Mullen, R. T. (2003). Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133 (2), 813–828. doi:10.1104/pp.103.027870
Neuhaus, A., Kooshapur, H., Wolf, J., Meyer, N. H., Madl, T., Saidowsky, J., et al. (2014). A novel Pex14 protein-interacting site of human Pex5 is critical for matrix protein import into peroxisomes. J. Biol. Chem. 289 (1), 437–448. doi:10.1074/jbc.M113.499707
Nuebel, E., Morgan, J. T., Fogarty, S., Winter, J. M., Lettlova, S., Berg, J. A., et al. (2021). The biochemical basis of mitochondrial dysfunction in Zellweger Spectrum Disorder. EMBO Rep. 22 (10), e51991. doi:10.15252/embr.202051991
Opalinski, L., Song, J., Priesnitz, C., Wenz, L. S., Oeljeklaus, S., Warscheid, B., et al. (2018). Recruitment of cytosolic J-proteins by TOM receptors promotes mitochondrial protein biogenesis. Cell Rep. 25 (8), 2036–2043.e5. doi:10.1016/j.celrep.2018.10.083
Opperdoes, F. R., and Borst, P. (1977). Localization of nine glycolytic enzymes in a microbody-like organelle in trypanosoma brucei: the glycosome. FEBS Lett. 80 (2), 360–364. doi:10.1016/0014-5793(77)80476-6
Orth, T., Reumann, S., Zhang, X., Fan, J., Wenzel, D., Quan, S., et al. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19 (1), 333–350. doi:10.1105/tpc.106.045831
Platta, H. W., Girzalsky, W., and Erdmann, R. (2004). Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 384 (1), 37–45. doi:10.1042/BJ20040572
Platta, H. W., Grunau, S., Rosenkranz, K., Girzalsky, W., and Erdmann, R. (2005). Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7 (8), 817–822. doi:10.1038/ncb1281
Reichle, R. E., and Alexander, J. V. (1965). Multiperforate septations, Woronin bodies, and septal plugs in Fusarium. J. Cell Biol. 24 (3), 489–496. doi:10.1083/jcb.24.3.489
Rhodin, J. (1954). “Correlation of ultrastructural organization: and function in normal and experimentally changed proximal convoluted tubule cells of the mouse kidney: an electron microscopic study,” (Sweden: Dept. of Anatomy, Karolinska Institutet). PhD Thesis.
Rottensteiner, H., Kramer, A., Lorenzen, S., Stein, K., Landgraf, C., Volkmer-Engert, R., et al. (2004). Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol. Biol. Cell 15 (7), 3406–3417. doi:10.1091/mbc.e04-03-0188
Rottensteiner, H., Stein, K., Sonnenhol, E., and Erdmann, R. (2003). Conserved function of pex11p and the novel pex25p and pex27p in peroxisome biogenesis. Mol. Biol. Cell 14 (10), 4316–4328. doi:10.1091/mbc.e03-03-0153
Rucktaschel, R., Thoms, S., Sidorovitch, V., Halbach, A., Pechlivanis, M., Volkmer, R., et al. (2009). Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis. J. Biol. Chem. 284 (31), 20885–20896. doi:10.1074/jbc.M109.016584
Sacksteder, K. A., Jones, J. M., South, S. T., Li, X., Liu, Y., and Gould, S. J. (2000). PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148 (5), 931–944. doi:10.1083/jcb.148.5.931
Saveria, T., Halbach, A., Erdmann, R., Volkmer-Engert, R., Landgraf, C., Rottensteiner, H., et al. (2007). Conservation of PEX19-binding motifs required for protein targeting to mammalian peroxisomal and trypanosome glycosomal membranes. Eukaryot. Cell 6 (8), 1439–1449. doi:10.1128/EC.00084-07
Schneider, K., Zimmer, D., Nielsen, H., Herrmann, J. M., and Muhlhaus, T. (2021). iMLP, a predictor for internal matrix targeting-like sequences in mitochondrial proteins. Biol. Chem. 402 (8), 937–943. doi:10.1515/hsz-2021-0185
Schrader, M., Costello, J., Godinho, L. F., and Islinger, M. (2015). Peroxisome-mitochondria interplay and disease. J. Inherit. Metab. Dis. 38 (4), 681–702. doi:10.1007/s10545-015-9819-7
Schrader, M., Costello, J. L., Godinho, L. F., Azadi, A. S., and Islinger, M. (2016). Proliferation and fission of peroxisomes - an update. Biochim. Biophys. Acta 1863 (5), 971–983. doi:10.1016/j.bbamcr.2015.09.024
Schrader, M., Reuber, B. E., Morrell, J. C., Jimenez-Sanchez, G., Obie, C., Stroh, T. A., et al. (1998). Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273 (45), 29607–29614. doi:10.1074/jbc.273.45.29607
Schrader, T. A., Carmichael, R. E., Islinger, M., Costello, J. L., Hacker, C., Bonekamp, N. A., et al. (2022). PEX11β and FIS1 cooperate in peroxisome division independently of mitochondrial fission factor. J. Cell Sci. 135 (13), jcs259924. doi:10.1242/jcs.259924
South, S. T., and Gould, S. J. (1999). Peroxisome synthesis in the absence of preexisting peroxisomes. J. Cell Biol. 144 (2), 255–266. doi:10.1083/jcb.144.2.255
Sulter, G. J., Verheyden, K., Mannaerts, G., Harder, W., and Veenhuis, M. (1993). The in vitro permeability of yeast peroxisomal membranes is caused by a 31 kDa integral membrane protein. Yeast 9 (7), 733–742. doi:10.1002/yea.320090707
Swinkels, B. W., Gould, S. J., Bodnar, A. G., Rachubinski, R. A., and Subramani, S. (1991). A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10 (11), 3255–3262. doi:10.1002/j.1460-2075.1991.tb04889.x
Tam, Y. Y., Torres-Guzman, J. C., Vizeacoumar, F. J., Smith, J. J., Marelli, M., Aitchison, J. D., et al. (2003). Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14 (10), 4089–4102. doi:10.1091/mbc.e03-03-0150
Thoms, S., and Gartner, J. (2012). First PEX11β patient extends spectrum of peroxisomal biogenesis disorder phenotypes. J. Med. Genet. 49 (5), 314–316. doi:10.1136/jmedgenet-2012-100899
Toro, A. A., Araya, C. A., Cordova, G. J., Arredondo, C. A., Cardenas, H. G., Moreno, R. E., et al. (2009). Pex3p-dependent peroxisomal biogenesis initiates in the endoplasmic reticulum of human fibroblasts. J. Cell Biochem. 107 (6), 1083–1096. doi:10.1002/jcb.22210
Tyler, K. M., Matthews, K. R., and Gull, K. (2001). Anisomorphic cell division by African trypanosomes. Protist 152 (4), 367–378. doi:10.1078/1434-4610-00074
Van Ael, E., and Fransen, M. (2006). Targeting signals in peroxisomal membrane proteins. Biochim. Biophys. Acta 1763 (12), 1629–1638. doi:10.1016/j.bbamcr.2006.08.020
van Roermund, C. W., Tabak, H. F., van Den Berg, M., Wanders, R. J., and Hettema, E. H. (2000). Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150 (3), 489–498. doi:10.1083/jcb.150.3.489
Verner, Z., Zarsky, V., Le, T., Narayanasamy, R. K., Rada, P., Rozbesky, D., et al. (2021). Anaerobic peroxisomes in Entamoeba histolytica metabolize myo-inositol. PLoS Pathog. 17 (11), e1010041. doi:10.1371/journal.ppat.1010041
Vogtle, F. N., and Meisinger, C. (2021). Mitochondria as emergency landing for abandoned peroxins. EMBO Rep. 22 (10), e53790. doi:10.15252/embr.202153790
Voncken, F., van Hellemond, J. J., Pfisterer, I., Maier, A., Hillmer, S., and Clayton, C. (2003). Depletion of GIM5 causes cellular fragility, a decreased glycosome number, and reduced levels of ether-linked phospholipids in trypanosomes. J. Biol. Chem. 278 (37), 35299–35310. doi:10.1074/jbc.M301811200
Yoshida, Y., Niwa, H., Honsho, M., Itoyama, A., and Fujiki, Y. (2015). Pex11mediates peroxisomal proliferation by promoting deformation of the lipid membrane. Biol. Open 4 (6), 710–721. doi:10.1242/bio.201410801
Keywords: peroxisome, glycosome, peroxin, PEX19, PMP, mPTS, MTS
Citation: Krishna CK, Schmidt N, Tippler BG, Schliebs W, Jung M, Winklhofer KF, Erdmann R and Kalel VC (2023) Molecular basis of the glycosomal targeting of PEX11 and its mislocalization to mitochondrion in trypanosomes. Front. Cell Dev. Biol. 11:1213761. doi: 10.3389/fcell.2023.1213761
Received: 28 April 2023; Accepted: 03 August 2023;
Published: 17 August 2023.
Edited by:
Nathan Alder, University of Connecticut, United StatesReviewed by:
Dejana Mokranjac, Ludwig Maximilian University of Munich, GermanyRichard Rachubinski, University of Alberta, Canada
Copyright © 2023 Krishna, Schmidt, Tippler, Schliebs, Jung, Winklhofer, Erdmann and Kalel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Erdmann, cmFsZi5lcmRtYW5uQHJ1Yi5kZQ==; Vishal C. Kalel, dmlzaGFsLmthbGVsQHJ1Yi5kZQ==
 Chethan K. Krishna
Chethan K. Krishna Nadine Schmidt
Nadine Schmidt Bettina G. Tippler
Bettina G. Tippler Wolfgang Schliebs
Wolfgang Schliebs Martin Jung
Martin Jung Konstanze F. Winklhofer
Konstanze F. Winklhofer Ralf Erdmann
Ralf Erdmann Vishal C. Kalel
Vishal C. Kalel