
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 17 May 2023
Sec. Signaling
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1205540
This article is part of the Research TopicProtein-RNA Interplay-Regulated Signaling in Stem Cells and CancerView all 10 articles
Enhancers are a class of cis-regulatory elements in the genome that instruct the spatiotemporal transcriptional program. Last decade has witnessed an exploration of non-coding transcripts pervasively transcribed from active enhancers in diverse contexts, referred to as enhancer RNAs (eRNAs). Emerging evidence unequivocally suggests eRNAs are an important layer in transcriptional regulation. In this mini-review, we summarize the well-established regulatory models for eRNA actions and highlight the recent insights into the structure and chemical modifications of eRNAs underlying their functions. We also explore the potential roles of eRNAs in transcriptional condensates.
Enhancers are distal cis-regulatory elements in the genome that direct spatiotemporal transcription programs in response to diverse cues (Andersson et al., 2014; Long et al., 2016). An estimate of over 400,000 putative enhancers in human genome, plus the identification of disease-associated traits within enhancers, underscores the essence of exploring the regulatory grammar encrypted within these elements (Consortium, 2012; Long et al., 2016; Sur and Taipale, 2016; Furlong and Levine, 2018; Schoenfelder and Fraser, 2019).
The advent of state-of-art genomic approaches unveils that the human genome is pervasively transcribed, yielding a plethora of non-coding RNA (ncRNA) species (Hangauer et al., 2013). Among them, RNA transcripts emanating from enhancers, dubbed enhancer RNA (eRNAs), have attracted a particular interest considering their potential roles in enhancer regulation (De Santa et al., 2010; Kim et al., 2010; Lam et al., 2014; Li et al., 2016; Sartorelli and Lauberth, 2020; Harrison and Bose, 2022). It is noteworthy that distinct terms, e.g., eRNAs and enhancer-associated lncRNAs (elncRNAs), appear in the literature to represent transcripts from enhancer regions (Orom et al., 2010; Marques et al., 2013; Andersson et al., 2014; Li et al., 2016; Hon et al., 2017; Statello et al., 2021; Mattick et al., 2023). Strictly, eRNAs are short, bidirectional ones, which are generally non-polyadenylated and unstable, whereas elncRNAs are usually polyadenylated and have higher stability (Li et al., 2016; Statello et al., 2021; Mattick et al., 2023). However, concerning gene-activating mechanisms, elncRNAs and eRNAs share some common themes (Orom et al., 2010; Wang et al., 2011; Lai et al., 2013; Grossi et al., 2020) and we do not distinguish these different terms in this mini-review. As an integral component of active enhancers, eRNA transcription generally correlates with enhancer activation and can serve as an independent marker of active enhancers (Carullo et al., 2020). Although there isn’t yet a consensus regarding whether the functions come from the transcription process or eRNA transcripts per se, accumulating evidence has shown a subset of eRNAs are pivotal for the transcription of cognate targets and coined several well-appreciated themes for eRNA actions.
In this mini-review, we outline current models for eRNA actions in transcriptional regulation. In addition, we highlight recent findings concerning eRNAs secondary structure and post-transcriptional modifications in bestowing diverse functional features of eRNAs. Finally, we discuss an emerging paradigm of transcriptional condensates wherein eRNAs partaken and contribute.
The well-appreciated model for enhancer action is that chromatin loops form between enhancers and cognate promoters bringing these two elements into physically close proximity, which involves the participation of cohesin complex, and transcriptional coactivator Mediator complex (Kagey et al., 2010). Li et al. provided the first piece of evidence that estrogen-induced eRNAs bind with SMC3 and RAD21, components of cohesin complex. Depletion of eRNAs abrogates cohesin increment to enhancers, thus abolishing enhancer-promoter interactions and target genes activation (Li et al., 2013). Similarly, Lai et al. (2013) revealed that ncRNA-a interacts with Mediator subunits and is involved in chromatin looping between ncRNA-a loci and their regulated promoters. Since then, further studies identify the direct interactions between eRNAs and chromatin looping factors [e.g., hnRNPU (Jiao et al., 2018), CTCF (Xiang et al., 2014), MED1 (Hsieh et al., 2014), MED12 (Tan et al., 2019)], suggesting modulating chromatin looping is one common theme underlying eRNA functions (Figure 1A).
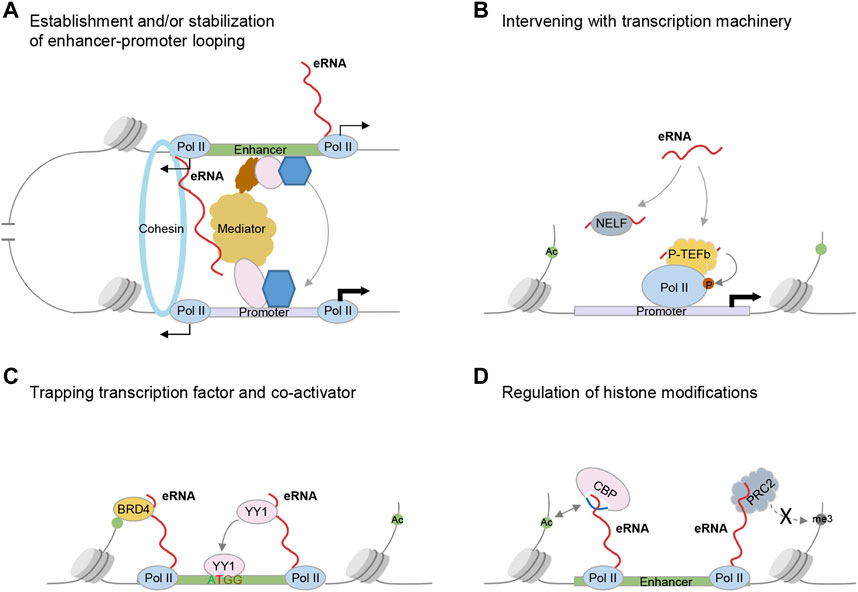
FIGURE 1. Established mechanisms underlying eRNA functions in transcriptional regulation. (A) Regulating chromatin looping. eRNAs interact with Cohesin complex or Mediator to establish and/or stabilize enhancer-promoter looping. (B) Intervening with the transcription machinery. eRNAs promote RNAP II pause release into productive elongation stage via acting as decoy for NELF and interacting with the P-TEFb. eRNAs also stimulate transcription through the intermediate hnRNPL. (C) Trapping transcription factors or transcription coactivators. eRNAs enhance the enhancer binding of TF YY1 and transcription coactivator BRD4 through direct interaction with them. (D) Modulating enhancer chromatin environment. eRNAs interact with CBP, stimulate its catalytic activity, and increase the deposition of histone acetylation on enhancers. eRNAs also inhibit the catalytic activity of PRC2 by binding the EZH2 subunit and inhibit repressive H3K27me3 deposition.
In addition to regulating chromatin looping, eRNAs can directly intervene with transcription machinery (Figure 1B). Lines of evidence suggest eRNAs can modulate RNA polymerase II (Pol II) pause release (Schaukowitch et al., 2014; Zhao et al., 2016; Shi et al., 2018). Schaukowitch et al. found eRNAs bind to NELF-E, and decoy this negative elongation factor (NELF) complex away from immediate early genes, thus promoting Pol II pause release into the productive elongation stage (Schaukowitch et al., 2014). In another study, Zhao et al. uncovered that PSA eRNA stimulates transcription through forming a complex with the positive elongation factor (P-TEFb) (Zhao et al., 2016). Congruent with these works, the following studies added more examples demonstrating interactions between eRNAs and NELF or P-TEFb (Shi et al., 2018). Besides these direct interactions, our group identified eRNAs interact with hnRNPL via a CAAA tract and modulate the appropriate loading of hnRNPL to the target locus (Zhao et al., 2019). hnRNPL has been shown to interact with KMT3A to regulate H3K36me3 enrichment (Yuan et al., 2009) and impinge on transcription elongation via interacting with P-TEFb components, CDK9 and CCNT1 (Giraud et al., 2014). In this scenario, hnRNPL acts as an intermediate to bridge the interaction between eRNAs and transcription machinery.
Another important paradigm of eRNA functions is that eRNAs can trap transcription factors and transcription co-activators, and enhance their binding to local chromatin (Figure 1C). Sigova et al. showed nascent RNAs transcribed from enhancers and promoters, through interactions with transcription factor (TF) YY1, increase YY1 binding to these regulatory elements (Sigova et al., 2015). One recent study reinforces this idea, showing a broad scope of TFs bind to RNA through arginine-rich motif (ARM)-like domains and such interactions contribute to TF association with chromatin (Oksuz et al., 2022). Deletion of ARM-like domains skews TF nuclear dynamics: it reduces the immobile and subdiffusive fractions of TFs while enhancing the diffusing molecules. A positive-feedback loop is thus proposed that nascent RNA produced from enhancer (eRNA) or promoter regions can trap dissociating TFs through RNA-mediated weak interactions, which facilitates TFs rebind to these regulatory elements and augments the transcription outputs. Similarly, eRNAs interact directly with BRD4 via its bromodomains and promote BRD4 binding to acetylated histones, which in turn maintains enhancers in an active state (Rahnamoun et al., 2018).
Lastly, eRNAs can modulate chromatin state. Depletion of eRNAs has been shown to decrease chromatin accessibility at enhancers and cognate promoters (Mousavi et al., 2013; Tsai et al., 2018). Besides these, eRNAs can directly interact with chromatin modifiers that deposit histone acetylation or methylation marks (Figure 1D). Specifically, Bose et al. demonstrated eRNAs interact with histone acetyltransferase CBP via its RNA binding region within the activation loop of HAT domain (Bose et al., 2017). Such interaction displaces the activation loop from the catalytic site and enhances CBP binding to its histone substrate. Similarly, eRNAs can also stimulate p300 catalytic activity and increase H3K27 acetylation at enhancers (Hou and Kraus, 2022). In addition to promoting histone acetylation, eRNAs also repel the PRC2-mediated deposition of the repressive histone modification H3K27me3 (Ounzain et al., 2015). Consistently, PRC2 binds to nascent RNA promiscuously at nearly all active genes, which antagonizes its binding to chromatin and thus alleviates the deposition of the repressive H3K27me3 mark (Beltran et al., 2016; Wang et al., 2017).
As mentioned above, caution needs to be taken to discern whether eRNAs function in a transcript-dependent or -independent manner. Engreitz et al. (2016) provided compelling evidence to show regulatory roles of many lncRNA loci stem from DNA elements or transcription processes, instead of their specific transcripts. Similar findings have been reported in other works (Kaikkonen et al., 2013; Anderson et al., 2016; Paralkar et al., 2016; Winkler et al., 2022). Thus, more rigorous methodologies are warranted in future studies to distinguish this point (Engreitz et al., 2016; Joung et al., 2017).
Despite the substantial advances concerning eRNA functions and mechanisms, their regulatory roles instructed by eRNA structures are poorly studied. As mentioned above, eRNA can interact with and activate P-TEFb. Such interaction requires a TAR RNA-like (TAR-L) motif, whose secondary structure is akin to the 3′ end of the small nuclear RNA 7SK. AR-eRNA, through competitive binding with P-TEFb, can help release P-TEFb from the inhibitory complex (7SK snRNP) and promotes effective transcription elongation (Zhao et al., 2016). On the contrary, interactions between eRNAs and NELF may not depend on structural motifs. Instead, adequate length (>200 nt) and the presence of unpaired guanosines are indispensable, which enables simultaneous and allosteric interactions between eRNAs and NELF subunits -A and -E (Gorbovytska et al., 2022).
The DRReRNA (also known as MUNC) is a well-studied pro-myogenic eRNA, which is transcribed from an enhancer region of the myogenic master TF, MyoD (Mousavi et al., 2013; Mueller et al., 2015). DRReRNA functions in trans to activate Myogenin transcription through directing cohesin loading at Myogenin locus (Cichewicz et al., 2018; Tsai et al., 2018). A recent study employed SHAPE-MaP (2′-hydroxyl acylation analyzed by primer extension coupled with mutational profiling) chemical probing approach to decode the secondary structure of DRReRNA and unraveled multiple structural domains that confer distinct features of DRReRNA for cohesion binding, genomic interaction, and gene expression regulation (Przanowska et al., 2022).
In addition to structured features embedded in eRNAs themselves, accumulating evidence underpins the regulatory code underlying intermolecular interactions. A prominent example comes from MALAT1, which interacts with many pre-mRNAs at active gene loci indirectly through RNA binding protein (RBP) intermediates (Engreitz et al., 2014; West et al., 2014). Recently, Cai et al. (2020) developed a novel approach termed RIC-seq (RNA in situ conformation sequencing), which can map RNA-RNA interactions in situ in an unbiased manner, and discovered MALAT1 interaction with highly transcribed nascent RNAs. Similarly, in this study, researchers also revealed extensive interactions between eRNAs and promoter upstream antisense RNAs (uaRNAs), which can be leveraged to infer enhancer-promoter connections. Intriguingly, modulating such interaction between eRNAs and uaRNAs influences chromatin looping (Figure 2A). Specifically, depletion of the super-enhancer-derived lncRNA CCAT1-5L markedly attenuates the chromatin looping between its parental CCAT1 locus and MYC locus, and weakens Pol II deposition at MYC promoter. In this specific scenario, the interaction relies on the RBP hnRNPK, which can physically interact with Pol II and form a homodimer. Thus, hnRNPK-mediated interaction between eRNA-uaRNA pairs may serve as modulator for enhancer-promoter chromatin interactions and Pol II delivery from enhancer regions to target promoter regions.
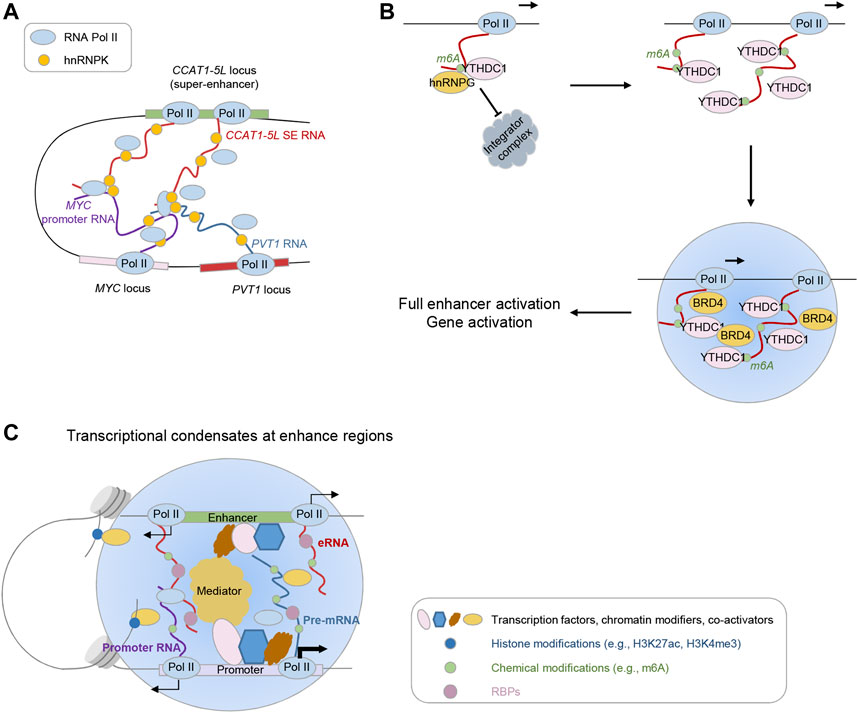
FIGURE 2. Expanding themes contributing to eRNA function. (A) Inter-molecular eRNA-uaRNA pair modulates enhancer-promoter chromatin interactions. (B) m6A modifications on eRNAs feed back on transcription. (C) eRNA-mediated regulation in transcriptional condensate formation.
Besides the RNA structures through RNA-RNA interactions, eRNAs can form DNA/RNA hybrid structure co-transcriptionally, termed R-loops. Competing evidence about R-loop functions comes from individual studies. Watts et al. (2022) found the enhancer RNA AANCR transcription leads to R-loops formation and in the R-loops eRNA is enzymatically modified to bear abasic sites, which helps stabilize R-loops, thus resulting in RNA Pol II pausing. Upon hypertonic stress, the R-loops are resolved and eRNA is fully transcribed to activate the target APOE activation. On the contrary, Tan-Wong et al. (2019) demonstrate that R-loops, often found at promoters, enhancers, and terminators, promote antisense transcription in these regions. More recently, local R-loops formation between an antisense eRNA PEARL and HS5-1 enhancer region facilitates chromatin looping between distal enhancers and target promoters (Zhou et al., 2021).
N6-methyladenosine (m6A) methylation, the most abundant RNA internal modification, has been shown to deposit on chromatin associated RNAs, including eRNAs (Louloupi et al., 2018; Xiao S. et al., 2019; Liu et al., 2020; Xu et al., 2021). Notably, the distribution of m6A methylation on these transcripts is not restricted to the 3′ end and is proven to regulate chromatin state and transcription directly (Louloupi et al., 2018; Xiao S. et al., 2019; Liu et al., 2020; Liu et al., 2021; Xu et al., 2021). Liu et al. found m6A-marked eRNAs, recognized by the nuclear reader YTHDC1, are subject to subsequent nuclear degradation by the nuclear exosome targeting (NEXT) complex. Knockout of the m6A writer Mettl3 increases carRNAs abundance and promotes downstream transcription in mouse embryonic stem cells (mESCs). Mechanistically, m6A erasure upon Mettl3 knockout stabilizes the carRNAs, rendering the following recruitment of active TFs (e.g., YY1 and CBP/EP300) and repelling of repressive factors (e.g., PRC2), thus tunes the nearby active chromatin state and stimulates downstream transcription.
The effects of m6A methylation on nuclear nascent transcripts and the transcription process could vary depending on different cell contexts. The recent two findings, on the contrary, show that m6A modification protects eRNAs from nuclear degradation, enhances the recruitment of m6A machinery components on enhancers and promoters, and stimulates effective transcription progress (Figure 2B) (Lee et al., 2021; Xu et al., 2022). In one study, Xu et al. (2022) revealed the chromatin binding of m6A methyltransferase complex (MTC) components METTL3/METTL14/WTAP locates at active enhancers and in turn decorates m6A modification on the 5′ end of nascent RNAs, neighboring to MTC chromatin binding sites. METTL3 depletion results in a loss of nascent RNAs emanating from enhancers at the TSS (transcription start site) proximal regions. Mechanistically, m6A modification recruits m6A reader/binder proteins such as hnRNPG and YTHDC1 to the nascent RNAs (including eRNAs), which protects these transcripts from cleavage by the Integrator complex. Loss of MTC would otherwise promote the recruitment of INS11 (the endonuclease subunit of the Integrator complex), leading to premature transcription termination. Of note, MTC recruitment to the promoter is augmented by active transcription elongation (Akhtar et al., 2021). Thus, m6A modification along with m6A reader proteins shields nascent eRNAs from premature termination, and the productive elongation in turn fosters MTC recruitment, establishing a positive feedback control over the transcription process.
In the other study, Lee et al. (2021) employed a high-sensitive method dubbed methylation-inscribed nascent transcripts sequencing (MINT-seq) to capture m6A methylome directly on nascent RNAs. They uncovered m6A is pervasively decorated with enrichment in the middle of eRNA transcripts and m6A modification positively correlates with eRNA length and abundance. In agreement with the canonical “RRACH” motif identified on mRNAs, “GGACT” motif sequences are identified with eRNA m6A peaks. Functionally, m6A-modified eRNAs can stimulate enhancer activation through reader protein YTHDC1 recruitment. Targeted m6A erasure, genetic and chemical perturbation of m6A writer and reader impair the enhancer activation, eRNA transcription, and subsequent target gene activation. Mechanistically, YTHDC1 can phase-separate into liquid-like condensates and co-assemble into BRD4 transcriptional condensates, while m6A-eRNAs presence augments the size of condensates. Concordantly, either perturbation of YTHDC1 levels or its condensate formation ability attenuates BRD4 recruitment to enhancers and BRD4 condensate formation.
In addition to m6A modification, enrichment of 5-methylcytosine (m5C) marked eRNAs were found at a set of enhancers upon metabolic stress (Aguilo et al., 2016). Under this circumstance, the interaction between PGC-1a and the NOP2/Sun RNA methyltransferase 7 (NSUN7) is essential in instructing m5C deposition on eRNAs.
Recent studies have shown liquid-liquid phase separation (LLPS) occurs at super-enhancers, which compartmentalizes crowded transcription regulators (e.g., TFs, transcriptional co-activators, RNA Pol II, and RNA) and promotes the formation of transcriptional condensates (Hnisz et al., 2017; Boija et al., 2018; Cho et al., 2018; Sabari et al., 2018; Shrinivas et al., 2019). Considering the established multivalent interactions between eRNAs with a myriad of factors (e.g., TFs, chromatin modifiers, DNA, RNA), eRNAs could potentially play a broad role in the formation of transcriptional condensates at enhancers (Roden and Gladfelter, 2021). Nair et al. (2019) recently reported an indispensable role of eRNA in controlling the assembly of MegaTrans complex at the ligand-activated enhancers, which exhibit properties of phase-separated components. Intriguingly, the complex components include several transcription factors (e.g., GATA3, ERα, RARA, FOXA1, AP2γ), which harbor intrinsically disordered regions (IDRs). The authors demonstrated two of them, GATA3 and ERα, are capable of liquid phase condensation at enhancers. Depletion of eRNA affects the diffusion properties of MegaTrans components, thus abolishing the full assembly of MegaTrans at the cognate enhancer. Notably, chronic enhancer activation alters the physicochemical properties of this enhancer RNA-dependent ribonucleoprotein (eRNP) complex to a more gel-like state. This study provides compelling evidence showing eRNAs directly contribute to the formation of phase-separated condensates and enhancer activation.
Based on current findings, we can extrapolate eRNAs play a broad role in controlling the formation, dissociation, and dynamics of transcriptional condensates at enhancers and/or cognate promoters via scaffolding multivalent interactions between condensate components (Maharana et al., 2018; Henninger et al., 2021; Quinodoz et al., 2021; Roden and Gladfelter, 2021). First, eRNAs may have a role in contributing to the formation of transcriptional condensates. Many eRNAs-interacting protein partners, as mentioned above, harbor IDRs that are essential in the induction of phase separation. For example, eRNAs interact with MED1 and BRD4, the IDRs of which have been demonstrated to foster super-enhancer formation through phase separation (Cho et al., 2018; Sabari et al., 2018). eRNAs also interact with P-TEFb and the recent finding supports the promoting role of CCNT1, a component of P-TEFb, in phase separation via its histidine-rich domain, which subsequently compartmentalizes RNA Pol II C-terminal domain (CTD) into CCNT1 droplets to ensure CTD hyperphosphorylation and transcription elongation (Lu et al., 2018). In addition to TFs and co-activators, increasing evidence has shown RBPs pervasively bind to regulatory elements and mediate the phase separation (Xiao et al., 2019a; Shao et al., 2022). Shao et al. discovered one RBP PSPC1 exhibits liquid-like properties and the presence of RNA augments the PSPC1-mediated transcriptional condensates that compartmentalize the CTD for enhanced phosphorylation. The low-complexity sequences (LCS) and RNA recognition motifs (RRMs) of PSPC1 are the prerequisites for the synergistic interplay between PSPC1 and RNA, the resultant PSPC1 chromatin binding and phase separation. Remarkably, the discovery that chrRBPs tend to co-occupy at regulatory regions, such as super-enhancers and promoters, provides a chance that diverse RBPs act collaboratively, in synergy with RNAs from these regulatory elements, in promoting the formation of transcriptional condensates. Second, eRNAs may not only engage in the formation but also regulate the dissociation and composition of transcriptional condensates. Maharana et al. proposed that RNA concentration determines distinct phase separation behaviors: higher RNA concentration impedes phase separation of RBPs in the nucleus, while lower RNA concentration facilitates aggregation (Maharana et al., 2018). Consistently, Henninger et al. (2021) recently reported low levels of RNA generated due to transcription initiation at regulatory elements, including eRNAs, promote condensate formation, whereas high production of RNAs during transcription elongation results in condensate dissolution. Considering the majority of eRNAs are short, unstable, and lowly expressed, eRNAs more likely partaken in the formation of transcriptional condensates. Interestingly, a recent work revealed nascent RNAs primarily impede the association of diverse categories of proteins with chromatin, including transcriptional regulators and chromatin modifiers (Skalska et al., 2021). RNA directly binds to these factors, and in turn blocks their binding to nucleosomes, suggesting an antagonistic relationship between their RNA- and chromosome-binding. Whether these proteins contribute to the formation of phase-separated condensates awaits further investigation. In addition, the phosphorylation status of RNAPII CTD affects the compartmentalization of RNAPII into distinct condensates (Guo et al., 2019). As eRNAs are proven to interact with protein components from different condensates (e.g., Mediator complex and RBPs involved in RNA processing and splicing), they are likely to influence the dynamic exchange of RNAPII between different condensates.
Despite the substantial progress of eRNA studies, much more efforts are warranted in delineating their functions and underlying mechanisms, considering the heterogenous nature in terms of their expression, length, secondary structures, and post-transcriptional modifications. For instance, our understanding of eRNA structures is poorly explored. To tackle this situation, more structural studies (e.g., SHAPE-MaP) are anticipated to uncover intramolecular secondary structures crucial for distinct properties of eRNAs. Equally important is the cataloging a more comprehensive list of eRNA binding partners (e.g., RBPs). Analyses of such data can provide insights into how eRNAs interact, and whether common sequence motifs or structural features of eRNAs exist conferring the interaction specificity. It will be also important to further explore the recently identified intermolecular RNA-RNA interactions and RNA-chromatin interactions, which present an intriguing possibility that eRNAs potentially participate in nuclear compartmentalization (Cai et al., 2020; Quinodoz et al., 2021). Besides, the existence and functions of epitranscriptomic modifications on eRNAs, such as m6A, m5C, hydroxymethyl cytosine (5hmC), and methyl-1-adenosine (m1A), need to be further explored. One trending direction is to demystify the regulatory feedback from these chemical modifications on nascent RNAs (including eRNAs) to chromatin and transcription. As mentioned above, pieces of evidence point to the involvement of eRNAs in phase-separated transcriptional condensates. Multivalent interactions mediated by eRNAs, e.g., RNA-RNA, RNA-protein, RNA-DNA interactions, render them great potential in mediating phase separation (Figure 2C). Several important questions need to be addressed in the future. How do eRNAs contribute to the formation of transcriptional condensates and what features are important (e.g., length, motifs, secondary structures, intermolecular RNA-RNA interactions, and post-transcriptional modifications)? Do eRNAs regulate the transition from transcription initiation into elongation condensates? In addition, the linkage is largely unclear between eRNA-involved transcriptional condensate formation and higher-order 3D genome organization. Finally, functional investigations are required to delineate the roles of eRNAs in disease entities (Zhang et al., 2019; Chen and Liang, 2020) and to dissect how the altered eRNA features favor disease development. Answers to these questions will provide deeper insights not only into eRNA functions and regulatory mechanisms but also into eRNA-centric therapeutic strategies.
YZ and QC conceived the project; QC, YZ, JK, MH, NL, KS, and YZ performed the investigation; QC and YZ wrote the paper. YZ created figures. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the Natural Science Foundation of Guangdong Province, China (2021A1515012058 to YZ); National Natural Science Foundation of China (NSFC) (Grant No: 32270587, 32100673 to YZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilo, F., Li, S., Balasubramaniyan, N., Sancho, A., Benko, S., Zhang, F., et al. (2016). Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell. Rep. 14, 479–492. doi:10.1016/j.celrep.2015.12.043
Akhtar, J., Renaud, Y., Albrecht, S., Ghavi-Helm, Y., Roignant, J. Y., Silies, M., et al. (2021). m(6)A RNA methylation regulates promoter-proximal pausing of RNA polymerase II. Mol. Cell. 81, 3356–3367.e6. doi:10.1016/j.molcel.2021.06.023
Anderson, K. M., Anderson, D. M., Mcanally, J. R., Shelton, J. M., Bassel-Duby, R., and Olson, E. N. (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. doi:10.1038/nature20128
Andersson, R., Gebhard, C., Miguel-Escalada, I., Hoof, I., Bornholdt, J., Boyd, M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. doi:10.1038/nature12787
Beltran, M., Yates, C. M., Skalska, L., Dawson, M., Reis, F. P., Viiri, K., et al. (2016). The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 26, 896–907. doi:10.1101/gr.197632.115
Boija, A., Klein, I. A., Sabari, B. R., Dall'agnese, A., Coffey, E. L., Zamudio, A. V., et al. (2018). Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 175, 1842–1855. doi:10.1016/j.cell.2018.10.042
Bose, D. A., Donahue, G., Reinberg, D., Shiekhattar, R., Bonasio, R., and Berger, S. L. (2017). RNA binding to CBP stimulates histone acetylation and transcription. Cell. 168, 135–149. doi:10.1016/j.cell.2016.12.020
Cai, Z., Cao, C., Ji, L., Ye, R., Wang, D., Xia, C., et al. (2020). RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 582, 432–437. doi:10.1038/s41586-020-2249-1
Carullo, N. V. N., Phillips, R. A., Simon, R. C., Soto, S. a. R., Hinds, J. E., Salisbury, A. J., et al. (2020). Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res. 48, 9550–9570. doi:10.1093/nar/gkaa671
Chen, H., and Liang, H. (2020). A high-resolution map of human enhancer RNA loci characterizes super-enhancer activities in cancer. Cancer Cell. 38, 701–715. doi:10.1016/j.ccell.2020.08.020
Cho, W. K., Spille, J. H., Hecht, M., Lee, C., Li, C., Grube, V., et al. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. doi:10.1126/science.aar4199
Cichewicz, M. A., Kiran, M., Przanowska, R. K., Sobierajska, E., Shibata, Y., and Dutta, A. (2018). MUNC, an enhancer RNA upstream from the MYOD gene, induces a subgroup of myogenic transcripts in trans independently of MyoD. Mol. Cell. Biol. 38, e00655-17. doi:10.1128/MCB.00655-17
Consortium, E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi:10.1038/nature11247
De Santa, F., Barozzi, I., Mietton, F., Ghisletti, S., Polletti, S., Tusi, B. K., et al. (2010). A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384. doi:10.1371/journal.pbio.1000384
Engreitz, J. M., Sirokman, K., Mcdonel, P., Shishkin, A. A., Surka, C., Russell, P., et al. (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 159, 188–199. doi:10.1016/j.cell.2014.08.018
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi:10.1038/nature20149
Furlong, E. E. M., and Levine, M. (2018). Developmental enhancers and chromosome topology. Science 361, 1341–1345. doi:10.1126/science.aau0320
Giraud, M., Jmari, N., Du, L., Carallis, F., Nieland, T. J., Perez-Campo, F. M., et al. (2014). An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc. Natl. Acad. Sci. U. S. A. 111, 1491–1496. doi:10.1073/pnas.1323535111
Gorbovytska, V., Kim, S. K., Kuybu, F., Gotze, M., Um, D., Kang, K., et al. (2022). Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat. Commun. 13, 2429. doi:10.1038/s41467-022-29934-w
Grossi, E., Raimondi, I., Goni, E., Gonzalez, J., Marchese, F. P., Chapaprieta, V., et al. (2020). A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 11, 936. doi:10.1038/s41467-020-14623-3
Guo, Y. E., Manteiga, J. C., Henninger, J. E., Sabari, B. R., Dall'agnese, A., Hannett, N. M., et al. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548. doi:10.1038/s41586-019-1464-0
Hangauer, M. J., Vaughn, I. W., and Mcmanus, M. T. (2013). Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9, e1003569. doi:10.1371/journal.pgen.1003569
Harrison, L. J., and Bose, D. (2022). Enhancer RNAs step forward: New insights into enhancer function. Development 149, dev200398. doi:10.1242/dev.200398
Henninger, J. E., Oksuz, O., Shrinivas, K., Sagi, I., Leroy, G., Zheng, M. M., et al. (2021). RNA-mediated feedback control of transcriptional condensates. Cell. 184, 207–225.e24. doi:10.1016/j.cell.2020.11.030
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K., and Sharp, P. A. (2017). A phase separation model for transcriptional control. Cell. 169, 13–23. doi:10.1016/j.cell.2017.02.007
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5' ends. Nature 543, 199–204. doi:10.1038/nature21374
Hou, T. Y., and Kraus, W. L. (2022). Analysis of estrogen-regulated enhancer RNAs identifies a functional motif required for enhancer assembly and gene expression. Cell. Rep. 39, 110944. doi:10.1016/j.celrep.2022.110944
Hsieh, C. L., Fei, T., Chen, Y., Li, T., Gao, Y., Wang, X., et al. (2014). Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U. S. A. 111, 7319–7324. doi:10.1073/pnas.1324151111
Jiao, W., Chen, Y., Song, H., Li, D., Mei, H., Yang, F., et al. (2018). HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene 37, 2728–2745. doi:10.1038/s41388-018-0128-0
Joung, J., Engreitz, J. M., Konermann, S., Abudayyeh, O. O., Verdine, V. K., Aguet, F., et al. (2017). Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346. doi:10.1038/nature23451
Kagey, M. H., Newman, J. J., Bilodeau, S., Zhan, Y., Orlando, D. A., Van Berkum, N. L., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. doi:10.1038/nature09380
Kaikkonen, M. U., Spann, N. J., Heinz, S., Romanoski, C. E., Allison, K. A., Stender, J. D., et al. (2013). Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 51, 310–325. doi:10.1016/j.molcel.2013.07.010
Kim, T. K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. doi:10.1038/nature09033
Lai, F., Orom, U. A., Cesaroni, M., Beringer, M., Taatjes, D. J., Blobel, G. A., et al. (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. doi:10.1038/nature11884
Lam, M. T., Li, W., Rosenfeld, M. G., and Glass, C. K. (2014). Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 39, 170–182. doi:10.1016/j.tibs.2014.02.007
Lee, J. H., Wang, R., Xiong, F., Krakowiak, J., Liao, Z., Nguyen, P. T., et al. (2021). Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell. 81, 3368–3385.e9. doi:10.1016/j.molcel.2021.07.024
Li, W., Notani, D., Ma, Q., Tanasa, B., Nunez, E., Chen, A. Y., et al. (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520. doi:10.1038/nature12210
Li, W., Notani, D., and Rosenfeld, M. G. (2016). Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223. doi:10.1038/nrg.2016.4
Liu, J., Dou, X., Chen, C., Chen, C., Liu, C., Xu, M. M., et al. (2020). N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586. doi:10.1126/science.aay6018
Liu, J., Gao, M., He, J., Wu, K., Lin, S., Jin, L., et al. (2021). The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature 591, 322–326. doi:10.1038/s41586-021-03313-9
Long, H. K., Prescott, S. L., and Wysocka, J. (2016). Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell. 167, 1170–1187. doi:10.1016/j.cell.2016.09.018
Louloupi, A., Ntini, E., Conrad, T., and Orom, U. a. V. (2018). Transient N-6-Methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell. Rep. 23, 3429–3437. doi:10.1016/j.celrep.2018.05.077
Lu, H., Yu, D., Hansen, A. S., Ganguly, S., Liu, R., Heckert, A., et al. (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. doi:10.1038/s41586-018-0174-3
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. doi:10.1126/science.aar7366
Marques, A. C., Hughes, J., Graham, B., Kowalczyk, M. S., Higgs, D. R., and Ponting, C. P. (2013). Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 14, R131. doi:10.1186/gb-2013-14-11-r131
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. Online ahead of print. doi:10.1038/s41580-022-00566-8
Mousavi, K., Zare, H., Dell'orso, S., Grontved, L., Gutierrez-Cruz, G., Derfoul, A., et al. (2013). eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 51, 606–617. doi:10.1016/j.molcel.2013.07.022
Mueller, A. C., Cichewicz, M. A., Dey, B. K., Layer, R., Reon, B. J., Gagan, J. R., et al. (2015). MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell. Biol. 35, 498–513. doi:10.1128/MCB.01079-14
Nair, S. J., Yang, L., Meluzzi, D., Oh, S., Yang, F., Friedman, M. J., et al. (2019). Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 26, 193–203. doi:10.1038/s41594-019-0190-5
Oksuz, O., Henninger, J. E., Warneford-Thomson, R., Zheng, M. M., Erb, H., Overholt, K. J., et al. (2022). Transcription factors interact with RNA to regulate genes. bioRxiv. New York, NY: Cold Spring Harbor Laboratory. doi:10.1101/2022.09.27.509776
Orom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell. 143, 46–58. doi:10.1016/j.cell.2010.09.001
Ounzain, S., Micheletti, R., Arnan, C., Plaisance, I., Cecchi, D., Schroen, B., et al. (2015). CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 89, 98–112. doi:10.1016/j.yjmcc.2015.09.016
Paralkar, V. R., Taborda, C. C., Huang, P., Yao, Y., Kossenkov, A. V., Prasad, R., et al. (2016). Unlinking an lncRNA from its associated cis element. Mol. Cell. 62, 104–110. doi:10.1016/j.molcel.2016.02.029
Przanowska, R. K., Weidmann, C. A., Saha, S., Cichewicz, M. A., Jensen, K. N., Przanowski, P., et al. (2022). Distinct MUNC lncRNA structural domains regulate transcription of different promyogenic factors. Cell. Rep. 38, 110361. doi:10.1016/j.celrep.2022.110361
Quinodoz, S. A., Jachowicz, J. W., Bhat, P., Ollikainen, N., Banerjee, A. K., Goronzy, I. N., et al. (2021). RNA promotes the formation of spatial compartments in the nucleus. Cell. 184, 5775–5790.e30. doi:10.1016/j.cell.2021.10.014
Rahnamoun, H., Lee, J., Sun, Z., Lu, H., Ramsey, K. M., Komives, E. A., et al. (2018). RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 25, 687–697. doi:10.1038/s41594-018-0102-0
Roden, C., and Gladfelter, A. S. (2021). RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell. Biol. 22, 183–195. doi:10.1038/s41580-020-0264-6
Sabari, B. R., Dall'agnese, A., Boija, A., Klein, I. A., Coffey, E. L., Shrinivas, K., et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. doi:10.1126/science.aar3958
Sartorelli, V., and Lauberth, S. M. (2020). Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 27, 521–528. doi:10.1038/s41594-020-0446-0
Schaukowitch, K., Joo, J. Y., Liu, X., Watts, J. K., Martinez, C., and Kim, T. K. (2014). Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 56, 29–42. doi:10.1016/j.molcel.2014.08.023
Schoenfelder, S., and Fraser, P. (2019). Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455. doi:10.1038/s41576-019-0128-0
Shao, W., Bi, X., Pan, Y., Gao, B., Wu, J., Yin, Y., et al. (2022). Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nat. Chem. Biol. 18, 70–80. doi:10.1038/s41589-021-00904-5
Shi, L., Li, S., Maurer, K., Zhang, Z., Petri, M., and Sullivan, K. E. (2018). Enhancer RNA and NFκB-dependent P300 regulation of ADAMDEC1. Mol. Immunol. 103, 312–321. doi:10.1016/j.molimm.2018.09.019
Shrinivas, K., Sabari, B. R., Coffey, E. L., Klein, I. A., Boija, A., Zamudio, A. V., et al. (2019). Enhancer features that drive formation of transcriptional condensates. Mol. Cell. 75, 549–561. doi:10.1016/j.molcel.2019.07.009
Sigova, A. A., Abraham, B. J., Ji, X., Molinie, B., Hannett, N. M., Guo, Y. E., et al. (2015). Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981. doi:10.1126/science.aad3346
Skalska, L., Begley, V., Beltran, M., Lukauskas, S., Khandelwal, G., Faull, P., et al. (2021). Nascent RNA antagonizes the interaction of a set of regulatory proteins with chromatin. Mol. Cell. 81, 2944–2959.e10. doi:10.1016/j.molcel.2021.05.026
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sur, I., and Taipale, J. (2016). The role of enhancers in cancer. Nat. Rev. Cancer 16, 483–493. doi:10.1038/nrc.2016.62
Tan, S. H., Leong, W. Z., Ngoc, P. C. T., Tan, T. K., Bertulfo, F. C., Lim, M. C., et al. (2019). The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood 134, 239–251. doi:10.1182/blood.2018874503
Tan-Wong, S. M., Dhir, S., and Proudfoot, N. J. (2019). R-loops promote antisense transcription across the mammalian genome. Mol. Cell. 76, 600–616. doi:10.1016/j.molcel.2019.10.002
Tsai, P. F., Dell'orso, S., Rodriguez, J., Vivanco, K. O., Ko, K. D., Jiang, K., et al. (2018). A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol. Cell. 71, 129–141. doi:10.1016/j.molcel.2018.06.008
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi:10.1038/nature09819
Wang, X., Paucek, R. D., Gooding, A. R., Brown, Z. Z., Ge, E. J., Muir, T. W., et al. (2017). Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 24, 1028–1038. doi:10.1038/nsmb.3487
Watts, J. A., Grunseich, C., Rodriguez, Y., Liu, Y., Li, D., Burdick, J. T., et al. (2022). A common transcriptional mechanism involving R-loop and RNA abasic site regulates an enhancer RNA of APOE. Nucleic Acids Res. 50, 12497–12514. doi:10.1093/nar/gkac1107
West, J. A., Davis, C. P., Sunwoo, H., Simon, M. D., Sadreyev, R. I., Wang, P. I., et al. (2014). The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 55, 791–802. doi:10.1016/j.molcel.2014.07.012
Winkler, L., Jimenez, M., Zimmer, J. T., Williams, A., Simon, M. D., and Dimitrova, N. (2022). Functional elements of the cis-regulatory lincRNA-p21. Cell. Rep. 39, 110687. doi:10.1016/j.celrep.2022.110687
Xiang, J. F., Yin, Q. F., Chen, T., Zhang, Y., Zhang, X. O., Wu, Z., et al. (2014). Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell. Res. 24, 513–531. doi:10.1038/cr.2014.35
Xiao, R., Chen, J. Y., Liang, Z., Luo, D., Chen, G., Lu, Z. J., et al. (2019a). Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell. 178, 107–121. doi:10.1016/j.cell.2019.06.001
Xiao, S., Cao, S., Huang, Q., Xia, L., Deng, M., Yang, M., et al. (2019b). The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell. Biol. 21, 651–661. doi:10.1038/s41556-019-0315-4
Xu, W., Li, J., He, C., Wen, J., Ma, H., Rong, B., et al. (2021). METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 591, 317–321. doi:10.1038/s41586-021-03210-1
Xu, W., He, C., Kaye, E. G., Li, J., Mu, M., Nelson, G. M., et al. (2022). Dynamic control of chromatin-associated m(6)A methylation regulates nascent RNA synthesis. Mol. Cell. 82, 1156–1168.e7. doi:10.1016/j.molcel.2022.02.006
Yuan, W., Xie, J., Long, C., Erdjument-Bromage, H., Ding, X., Zheng, Y., et al. (2009). Heterogeneous nuclear ribonucleoprotein L Is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J. Biol. Chem. 284, 15701–15707. doi:10.1074/jbc.M808431200
Zhang, Z., Lee, J. H., Ruan, H., Ye, Y., Krakowiak, J., Hu, Q., et al. (2019). Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 10, 4562. doi:10.1038/s41467-019-12543-5
Zhao, Y., Wang, L., Ren, S., Wang, L., Blackburn, P. R., Mcnulty, M. S., et al. (2016). Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell. Rep. 15, 599–610. doi:10.1016/j.celrep.2016.03.038
Zhao, Y., Zhou, J., He, L., Li, Y., Yuan, J., Sun, K., et al. (2019). MyoD induced enhancer RNA interacts with hnRNPL to activate target gene transcription during myogenic differentiation. Nat. Commun. 10, 5787. doi:10.1038/s41467-019-13598-0
Keywords: eRNA, transcriptional regulation, chromatin looping, phase separation, epitranscriptome
Citation: Chen Q, Zeng Y, Kang J, Hu M, Li N, Sun K and Zhao Y (2023) Enhancer RNAs in transcriptional regulation: recent insights. Front. Cell Dev. Biol. 11:1205540. doi: 10.3389/fcell.2023.1205540
Received: 14 April 2023; Accepted: 09 May 2023;
Published: 17 May 2023.
Edited by:
Liang Zhou, Southern Medical University, ChinaReviewed by:
Krishna Shrinivas, Harvard University, United StatesCopyright © 2023 Chen, Zeng, Kang, Hu, Li, Sun and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhao, emhhb3l1MjVAbWFpbC5zeXN1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.