
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 02 June 2023
Sec. Cancer Cell Biology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1198949
 Bingxin Zhang1
Bingxin Zhang1 Quanqiang Wang1
Quanqiang Wang1 Zhili Lin1
Zhili Lin1 Ziwei Zheng1
Ziwei Zheng1 Shujuan Zhou1
Shujuan Zhou1 Tianyu Zhang1
Tianyu Zhang1 Dong Zheng1
Dong Zheng1 Zixing Chen2
Zixing Chen2 Sisi Zheng1
Sisi Zheng1 Yu Zhang1
Yu Zhang1 Xuanru Lin1
Xuanru Lin1 Rujiao Dong1
Rujiao Dong1 Jingjing Chen1
Jingjing Chen1 Honglan Qian1
Honglan Qian1 Xudong Hu1
Xudong Hu1 Yan Zhuang1
Yan Zhuang1 Qianying Zhang1
Qianying Zhang1 Zhouxiang Jin2*
Zhouxiang Jin2* Songfu Jiang1*
Songfu Jiang1* Yongyong Ma1,3,4*
Yongyong Ma1,3,4*Background: Metabolic reprogramming is an important hallmark of cancer. Glycolysis provides the conditions on which multiple myeloma (MM) thrives. Due to MM’s great heterogeneity and incurability, risk assessment and treatment choices are still difficult.
Method: We constructed a glycolysis-related prognostic model by Least absolute shrinkage and selection operator (LASSO) Cox regression analysis. It was validated in two independent external cohorts, cell lines, and our clinical specimens. The model was also explored for its biological properties, immune microenvironment, and therapeutic response including immunotherapy. Finally, multiple metrics were combined to construct a nomogram to assist in personalized prediction of survival outcomes.
Results: A wide range of variants and heterogeneous expression profiles of glycolysis-related genes were observed in MM. The prognostic model behaved well in differentiating between populations with various prognoses and proved to be an independent prognostic factor. This prognostic signature closely coordinated with multiple malignant features such as high-risk clinical features, immune dysfunction, stem cell-like features, cancer-related pathways, which was associated with the survival outcomes of MM. In terms of treatment, the high-risk group showed resistance to conventional drugs such as bortezomib, doxorubicin and immunotherapy. The joint scores generated by the nomogram showed higher clinical benefit than other clinical indicators. The in vitro experiments with cell lines and clinical subjects further provided convincing evidence for our study.
Conclusion: We developed and validated the utility of the MM glycolysis-related prognostic model, which provides a new direction for prognosis assessment, treatment options for MM patients.
Multiple myeloma (MM) is the second most common hematological malignancy and is attributed to bone marrow infiltration of monoclonal plasma cells (Laubach et al., 2011; Siegel et al., 2021). It is characterized by hypercalcemia, renal damage, anemia, and bone lesions (Röllig et al., 2015; Kumar et al., 2017). MM is a highly heterogenous disease, that exits on a continuous disease spectrum ranging from precancerous states monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM) driven by the accumulating genetic changes and immune escape (Kyle et al., 2002; Kyle et al., 2007; Landgren et al., 2009).
In the past decade, novel treatments have increased MM’s survival rates, such as proteasome inhibitor bortezomib (BTZ) and immunomodulatory drug (IMiD) thalidomide (Kumar et al., 2008; Bianchi et al., 2015; Moreau et al., 2015). Despite the improvements in overall survival, relapse frequently occurs (Krishnan et al., 2016; Wallington-Beddoe et al., 2018). Meanwhile, MM is still an incurable disease, and the prevalence continues to rise owing to the aging population (Cowan et al., 2018). The Revised International Staging System (R-ISS) combines cytogenetic abnormalities (CAs), serum lactate dehydrogenase (LDH), and ISS traits and is the most widely recognized risk-stratification tool for newly diagnosed multiple myeloma (NDMM) patients (Palumbo et al., 2015). However, a large number of people with heterogeneous risk factors were concentrated in R-ISS stage II (Cho et al., 2017; Jung et al., 2018). New approaches are needed to better risk stratify MM patients to inform prognosis and treatment options for better individualized management.
The most crucial hallmark of cancer is metabolic reprogramming (Hanahan, 2022). Cancer cells preferentially employ glycolysis, even when there is abundant oxygen. This trait of cancer is known as “Warburg effect” (Warburg, 1956). Consistently, a typical feature of MM is the presence of an increased glycolytic gene profile and sensitivity to glycolysis inhibitors (D'Souza and Bhattacharya, 2019). Several key enzymes in glycolysis were highly upregulated in MM, including hexokinase 2 (HK2) and lactate dehydrogenase A (LDHA) (He et al., 2015; Ikeda et al., 2020). Furthermore, compared with NDMM patients, these enzymes were upregulated more significantly in relapsed and bortezomib-refractory MM patients (Mulligan et al., 2007; Maiso et al., 2015). Drugs targeting glycolysis have increasingly become a promising direction for cancer treatment. HK2 inhibitor, 3-bromopyruvate (3-BP), in addition to disrupting ATP production, significantly impaired autophagy and induced apoptosis, which was demonstrated in MM cells (Ikeda et al., 2020). Likewise, the GLUT4 inhibitor ritonavir has been shown to reduce the proliferation and viability of myeloma and improve chemotherapy sensitivity (McBrayer et al., 2012). Dichloroacetic acid is thought to achieve the apoptosis and proliferation inhibition of MM cells by activating the pyruvate dehydrogenase complex, and can increase the sensitivity of MM cell lines to bortezomib (Sanchez et al., 2013). Given the significance of glucose metabolism reprogramming, it is crucial to investigate the prognostic signatures and novel treatments of MM from the perspective of glycolysis.
In this study, we constructed a risk scoring model based on glycolysis-related genes (GRGs) to predict the prognosis and guide clinical treatment of MM. We further performed therapeutic response prediction and comparative analyses of biological function and tumor microenvironment (TME) to explore the underlying mechanisms. Then, a nomogram combing the gene signature and clinical manifestations was developed to improve the predictive power and clinical applicability. Finally, the expression of the selected genes was further verified by quantitative real-time PCR (qRT-PCR).
From the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), expression profiles were downloaded for 4 MM datasets, including GSE136337, GSE24080, GSE4204, and GSE6477. The datasets were log2 transformed after normalising with a microarray muite 5.0 (MAS5.0) method. Detailed clinicopathological data and survival data were also extracted from GEO (Table 1). The GSE136337 was used as the training, while the GSE24080 and GSE4204 datasets were used for validation. And we used the GSE6477 to reveal the evolution of GRGs in normal individuals, MGUS, SMM, and active MM groups. Glycolysis-related genomes “c2.cp.biocarta.v7.2.symbols”, “c2.cp.hallmark.v5.0.symbols”, “c2.cp.reactome.v7.5.symbols”, “c2.cp.kegg.v7.2.symbols”, “c2.cp.wikipathways.v7.2.symbols” in Gene Set Enrichment Analysis (GSEA) (http://www.gsea-msigdb.org/gsea/msigdb) were the source of the GRGs. After taking intersections with the three GEO datasets (GEO136337, GSE24080 and GSE4204), 293 genes were finally used for further study (Supplementary Table S1).
The prognostic glycolytic genes identified by univariable Cox regression analysis (p < 0.001) were then subjected to the Least absolute shrinkage and selection operator (LASSO) Cox regression analysis. Using the R package “glmnet”, the penalty parameter λ was set as 0.09 to determine the best weighting coefficient of glycolytic genes.
The co-expression correlation matrix of the 12 genes was constructed by “ggcorrplot” R package. The protein-protein interaction (PPI) network of the 12 genes was acquired from the STRING database (version 11.5) (https://www.string-db.org/) (Szklarczyk et al., 2011). The genetic mutation landscape of the GRGs in MM was explored with The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) with the “maftools” package and cBioPortal for Cancer Genomics (http://www.cbioportal.org/). We also used the Cancer Cell Line Encyclopedia database (CCLE, https://portals.broadinstitute.org/ccle) to further validate these prognostic genes’ expression.
To further investigate the heterogeneity between subgroups, we compared clinical characteristics and drug sensitivity among the subtypes. As for cytogenetics, based on the existing data in the training dataset and the previous findings, we defined high-risk cytogenetic abnormalities (HRCAs) as follows: del17p, amp1q, t (4; 14), t (14; 20), t (14; 16) or MYC aberrations determined by fluorescent in situ hybridization (FISH) or conventional cytogenetics (Chng et al., 2014; Sonneveld et al., 2016; Abdallah et al., 2020; Schmidt et al., 2021; Wallington-Beddoe and Mynott, 2021). Patients with at least one HRCA were defined as the high-risk cohort. The low-risk subgroup included those with other abnormalities [del13q, del16q, del1p, del1q, t (11; 14), t (12; 14)]. The rest belonged to the non-mutation cohort.
The “pRRophetic” package was used to assess the drug susceptibility between the low- and high-scoring group (Geeleher et al., 2014a; Geeleher et al., 2014b). This method builds the statistical model based on the data from the Cancer Genome Project (CGP) (Garnett et al., 2012), consisting of baseline gene expression microarray data and sensitivity to 138 drugs in a panel of almost 700 cell lines.
To reveal the underlying mechanism, we employed the weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008, 2012). Also, we conducted an association study between modules and clinical aspects and identified critical genes linked to the model. Further use of the Metascape platform (https://metascape.org/gp/index.html) was made to implement the Gene Ontology (GO) analysis of the important genes. GSEA (GSEA v4.2.2 software, http://software.broadinstitute.org/gsea/login.jsp) was also conducted to uncover the differences in biological functions with the Kyoto Encyclopedia of Genes and Genomes (KEGG). Statistical significance was defined as p < 0.05 and q < 0.25.
We employed five techniques to measure the immune microenvironment of subgroups, including EPIC, MCPCounter (Becht et al., 2016), quanTIseq (Plattner et al., 2020), the single-sample gene set enrichment analysis (ssGSEA) and xCell (Aran, 2020), to remove variances across different algorithms. The correlation analysis of the GRGs with immune-related genes and functional status from Thorsson et al. was performed. (Thorsson et al., 2018). mRNAsi is a tool for evaluating how closely cancer cells resemble stem cells (Malta et al., 2018). The sensitivity to immune checkpoint inhibitor (ICI) was evaluated using the immunophenotype score (IPS) (Charoentong et al., 2017) and the tumor immune dysfunction and exclusion (TIDE) (Jiang et al., 2018).
A nomogram combing age, ISS stage and glycolytic risk score was constructed. For the nomogram’s self-verification, the calibration curve was conducted. Based on the merged scores and other clinical parameters, time-dependent receiver operating characteristic (time-ROC) curves were computed for 1-, 3-, and 5-year survival. With the “ggDCA” package, survival net benefits of each clinical feature and the risk score were estimated with decision curve analysis (DCA). DCA curves focus on evaluating the clinical benefit of the model, predicting which threshold probability range treatment for patients in the model will achieve a higher clinical benefit, meeting the practical needs of clinical decision-making. For the prediction model, the net benefit (NB) is a composite indicator that incorporates both true positives and false positives. In the DCA curve, there are two reference lines, one with no intervention for anyone, called treat_none, and the other with intervention for everyone, called treat_all. The model has real value only if its NB is higher than both treat_all and treat_none at some threshold probability (Vickers and Elkin, 2006).
MM cell lines including LP-1, RPMI8226, NCI-H929, MM1.R, I9.2, and U266 cells were collected from Fenghui Biotechnology Co., Ltd. (Hunan, China). Cells were grown in an incubator set at 37 °C with a humid environment that contained 5% CO2. In addition to 10% fetal bovine serum, the cells were cultured in RPMI-1640 medium (Gibco, Shanghai, China) with 100 IU/mL penicillin and 100 mg/mL streptomycin.
35 MM patients were subjected to our experiment, of whom four were relapsed MM (RMM). As controls, normal bone marrow samples were taken from 21 healthy volunteers for PCR on cell lines and patient samples. Table 2 displayed the clinical traits of the patients. The study was authorized by the First Affiliated Hospital of Wenzhou Medical University’s ethics committee, and all operations were conducted according to the informed consent and Helsinki Declaration.
Using the Righton DNA&RNA Blood and Tissue Kit (Righton Bio, Shanghai, China), total RNA was isolated from total bone marrow-derived mononuclear cells as directed by the manufacturer. The cDNA synthesis kit (Vazyme, Nanjing, China) was used to execute reverse transcription in order to produce cDNAs. With the help of the Taq Pro universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), qRT-PCR was used to find the levels of glycolysis-related gene expression, with β-ACTIN acting as an internal reference. The primers used was provided in Table 3.
The statistical analysis was performed by SPSS version 24.0 (SPSS Inc., Chicago, IL, United States), R software version 4.1.1 (R Foundation for Statistical Computing, Vienna, Austria), and GraphPad Prism version 9.0.0 (GraphPad-Software Inc., San Diego, CA, United States). The Student’s t test was used to compare two groups for quantitative variables with a normal distribution. The Chi-square test was for the categorical variables. The Mann-Whitney U test was employed for the abnormal distributional variables. Multiple groups were compared using one-way analysis of variance (ANOVA) and the Kruskal-Wallis test. LSD test is used for multiple comparisons after ANOVA. Pearson’s correlation test was for the correlation evaluation between variables with normal distributions, and the Spearman’s correlation test was for variables with aberrant distributions. Statistics were judged significant at p < 0.05.
The glycolytic model was developed using the GSE136337. The GSE24080 and GSE4204 were used for model validation. Survival data were available for 1,514 subjects in the three datasets (GSE136337, n = 424; GSE24080, n = 556; GSE4204, n = 534). Clinicopathological features in sufficient samples allowed for subsequent Cox regression analysis (GSE136337, n = 415; GSE24080, n = 556) (Table 1). Moreover, the GSE6477 (n = 162) was used to reveal the evolution of GRGs in normal individuals, MGUS, SMM, and active MM groups.
In the GSE136337 (n = 424), 22 genes related to glycolysis were associated with prognosis (p < 0.001) (Figure 1A). Then, the glycolytic model was constructed using 12 genes (λ = 0.09) (Figure 1B). The formula was as follows: glycolytic risk score = (0.0803 × LDHB) + (0.0365 × SOD1) + (0.2087 × MDH2) - (0.1617 × PAM) + (0.1063 × MPC2) + (0.0182 × PRPS1) - (0.0262 × GMPPB) + (0.0217 × CYB5A) + (0.0593 × POLR3K) + (0.0721 × PAXIP1) + (0.1125 × AURKA) + (0.0507 × NSDHL). The median risk score was used to divide the subjects into high-risk and low-risk subgroups.
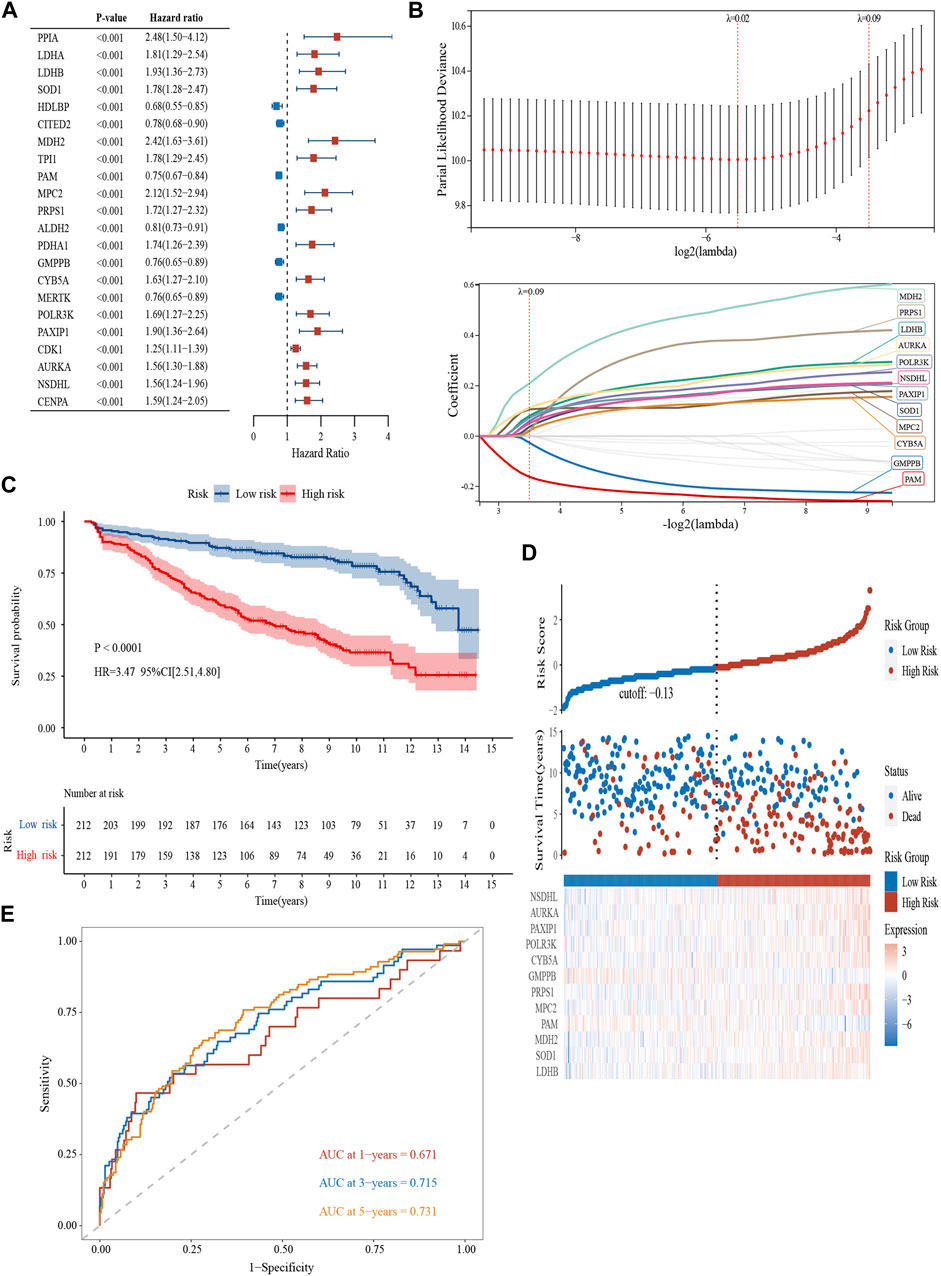
FIGURE 1. Construction and validation of a glycolytic prognostic model (GSE136337, n = 424). (A) Forest plot of hazard ratios manifesting the prognostic values of glycolysis-related genes. (B) LASSO Cox regression analysis for variable selection. (C) Kaplan-Meier curves of patients in the high- and low-risk group (p < 0.0001). (D) The AUC of the model assessed by time-dependent ROC curves. (E) The distribution of the survival outcomes and the expression of the prognostic genes among the subgroups.
Kaplan–Meier curves revealed survival differences among the subtype in the training (Figure 1C) and the two validation datasets (Supplementary Figures S1A, B). The high-scoring population had worse survival (HR = 3.47, 95% CI = 2.51–4.80, p < 0.0001; HR = 2.46, 95% CI = 1.78–3.41, p < 0.0001; HR = 2.09, 95% CI = 1.35–3.22, p < 0.001). The heterogeneity of survival outcomes was further validated in Figure 1D; Supplementary Figures S1C, D. Heat maps were also plotted to compare the expression of the 12 glycolytic genes. In general, the high-risk groups had lower levels of GMPPB and PAM expression, whereas other genes exhibit the opposite pattern (Figure 1D; Supplementary Figures S1C, D). AUCs of the 1-, 3-, and 5-year survival were 0.671, 0.715, and 0.731 in the GSE136337 (n = 424) to assess the sensitivity and specificity (Figure 1E). The results of the validation datasets were exhibited in Supplementary Figure S1E (GSE24080, n = 556) and 1F (GSE4204, n = 534).
Depending on the TCGA data, we explored the mutation landscape of 293 genes in the candidate glycolytic gene set in MM. Figure 2A showed the top 20 genes with the highest mutation rate. Among them, missense mutations accounted for the major part and the single nucleotide polymorphisms (SNPs) occurred more frequently than insertions or deletions. Furthermore, we used cBioPortal to reveal the single nucleotide variants (SNVs) and copy number variations (CNVs) of the 12 genes (Figure 2B). The alterations of LDHB, SOD1, MDH2, MPC2, PRPS1, AURKA, PAXIP1 and NSDHL in cancer cell lines were 6%, 2.1%, 5%, 3%, 1.4%, 10%, 10%, and 4% separately. Amplification was the most common change. In contrast, both PAM and GMPPB were altered at a frequency of 5%, and their most common change was deletion (Figure 2B). These results were in general agreement with our data.
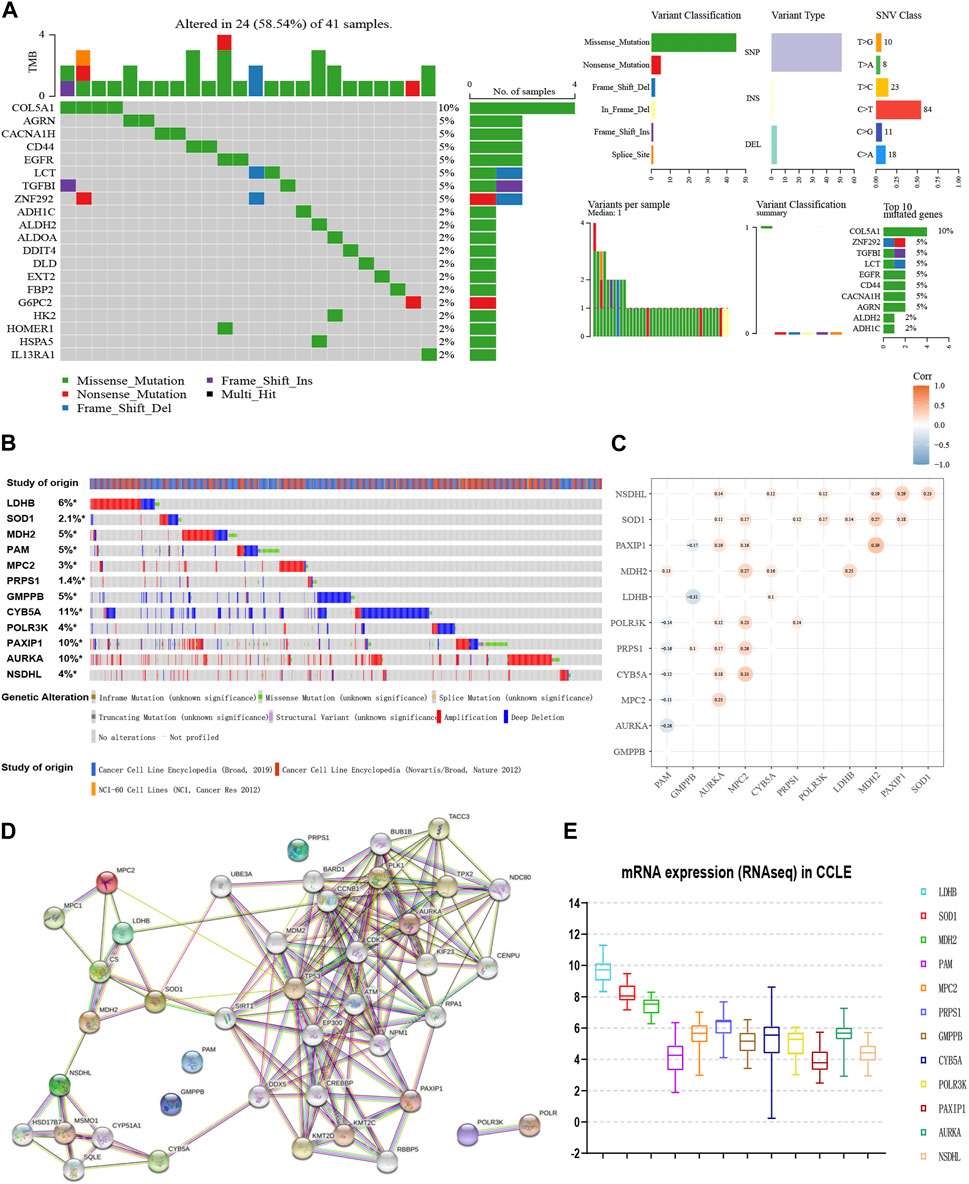
FIGURE 2. Interaction network and genetic variation map of glycolysis-related genes. (A) The genetic mutation landscape of the candidate genes from TCGA-MM. (B) The SNVs and CNVs of the 12 genes with cBioPortal for Cancer Genomics. (C) Co-expression analyses of 12 glycolytic gene signatures (GSE136337, n = 424). (D) Protein-protein interaction network of the 12 genes and other closely related proteins. (E) The external validation of the expression levels of the 12 genes using CCLE.
Moreover, the co-expression matrix and the PPI network showed a close relationship between the 12 genes (Figures 2C, D). In the CCLE database, LDHB, SOD1, MDH2, MPC2, PRPS1, CYB5A, POLR3K, and AURKA were over-expressed at the cellular level in MM, while PAM and GMPPB were under-expressed (Figure 2E), corresponding to the model equation above.
The glycolytic risk score was shown to be independently associated with survival (GSE136337, n = 415, HR = 3.191 (2.295, 4.436), p < 0.001; GSE24080, n = 556, HR = 1.662 (1.214, 2.275), p = 0.020) (Table 4). Subsequently, we analyzed the relationship between the 12-gene signature and clinicopathological traits in the GSE136337 (n = 415). The risk scores of the higher LDH and the high-risk cytogenetics groups were higher than those counterparts (p < 0.001) (Figure 3A). We observed the gradually increasing tendency of the risk score with the increasing ISS or R-ISS stage (p < 0.05) (Figure 3A). The distribution of the patient’s characteristics and survival outcomes were illustrated in Figure 3B.
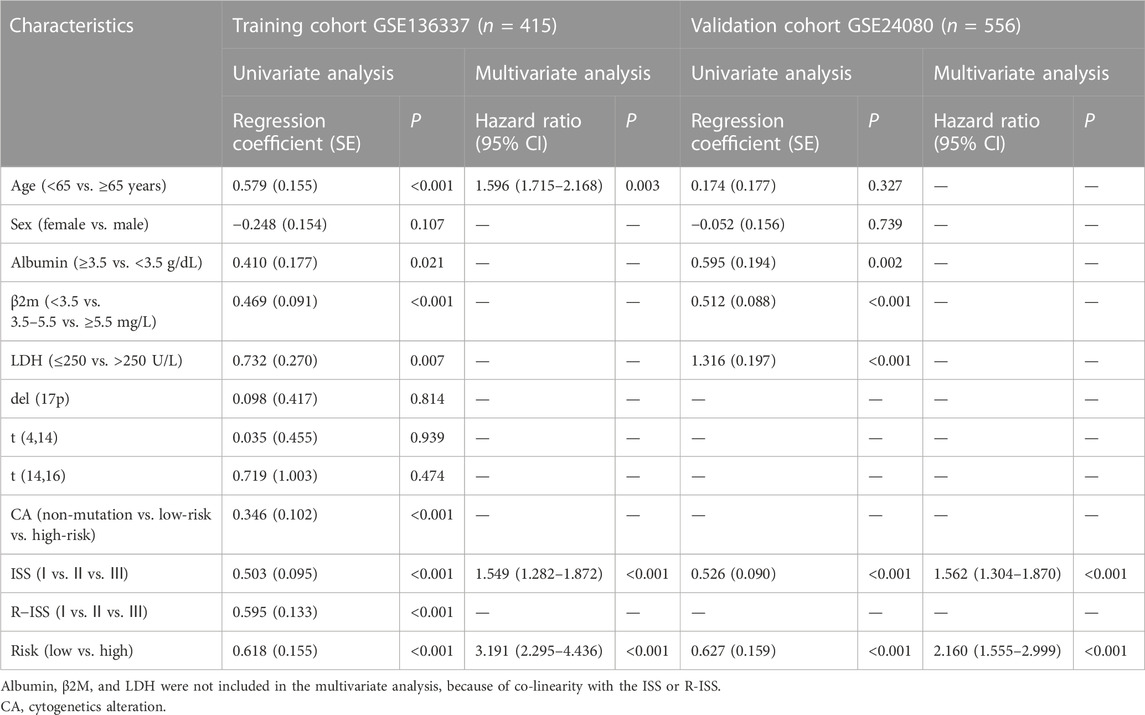
TABLE 4. Univariate and multivariate Cox regression analyses of survival in the training and validation cohorts.
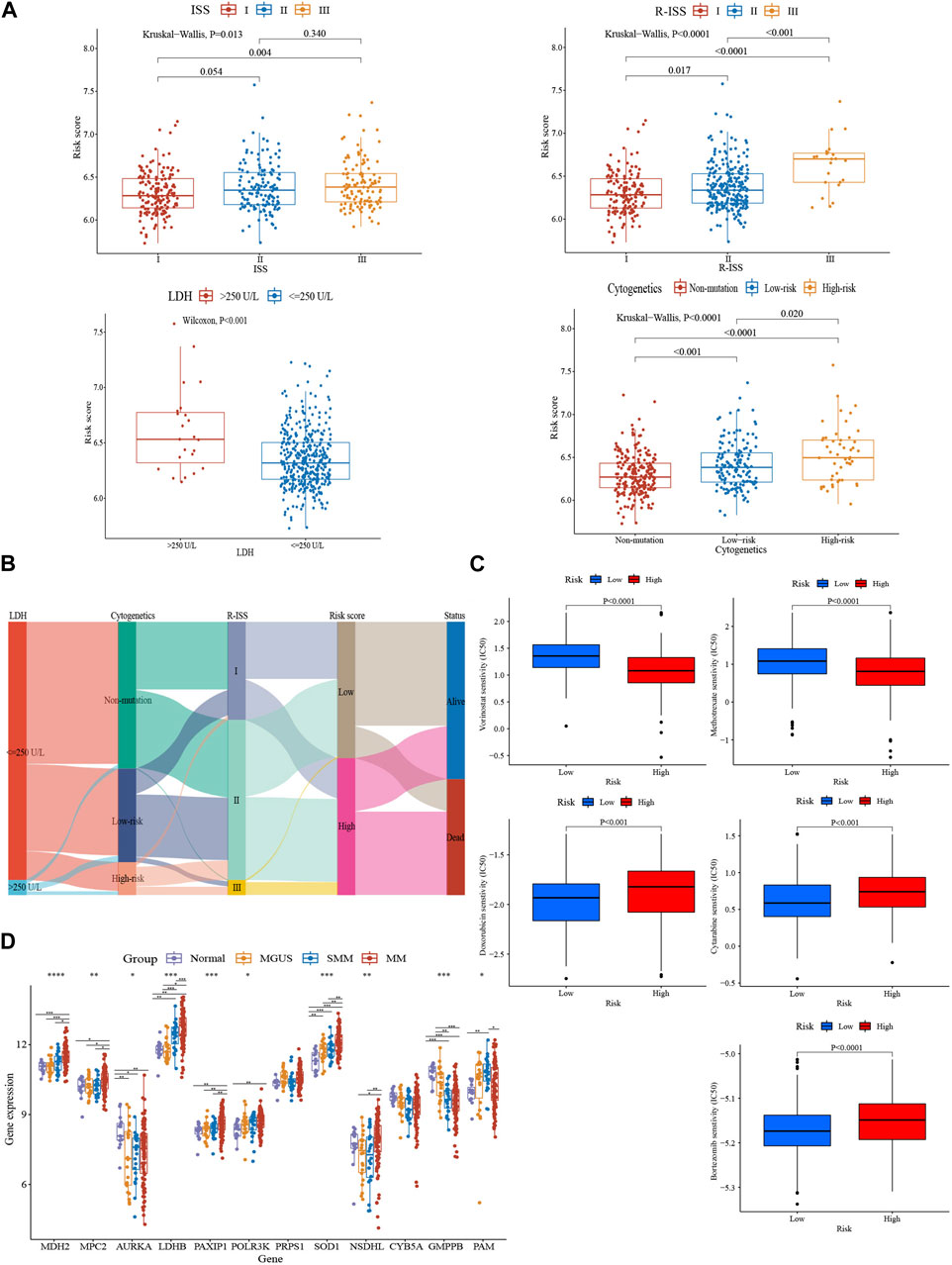
FIGURE 3. Comparative analysis of clinical characteristics and treatment responsiveness between subgroups. (A) Relationship between risk score and distinct clinical traits (GSE136337, n = 415). (B) A sankey diagram exhibiting the changes of LDH level, R-ISS stage, cytogenetic variation risk, glycolytic risk score, and prognosis (GSE136337, n = 415). (C) Evaluations of the drug susceptibility among the subtypes (GSE136337, n = 424). (D) Variations in these 12 genes’ expression levels as MM progressed (GSE6477, n = 162). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
According to the estimated IC50 values of the chemotherapeutic agents, the high-risk cohort showed the resistance to bortezomib, doxorubicin, and cytarabine (p < 0.001), but more sensitive to methotrexate and vorinostat (p < 0.0001), compared to the low-risk group (Figure 3C). Additionally, glycolysis-related gene expression changes were also present in the evolving disease spectrum of myeloma (GSE6477, n = 162) (Figure 3D). The expression levels of LDHB and SOD1 exhibited an increasing trend as the disease progressed. In addition, MDH2, MPC2, PAXIP1, and NSDHL were upregulated in myeloma patients. GMPPB, however, revealed a pattern of decline.
We conducted an investigation of biological functions among subgroups to uncover the mechanism underlying the glycolytic signature. WGCNA was implemented to build a weighted gene co-expression network. To ensure proper connectivity and sustain the network nearly scale-free, the soft threshold was set at four (Figure 4A). 19 gene modules were produced after grouping related modules (Figure 4B). The red module and the risk score had the strongest association (r = 0.58, p < 0.001) (Figure 4C). The red module’s 221 genes were analyzed for GO enrichment using the online database Metascape. Important processes in tumors, including the mitotic cell cycle, cell cycle checkpoints, DNA metabolism, and the retinoblastoma gene in cancer, were the main sites of gene module concentration (Figure 4D).
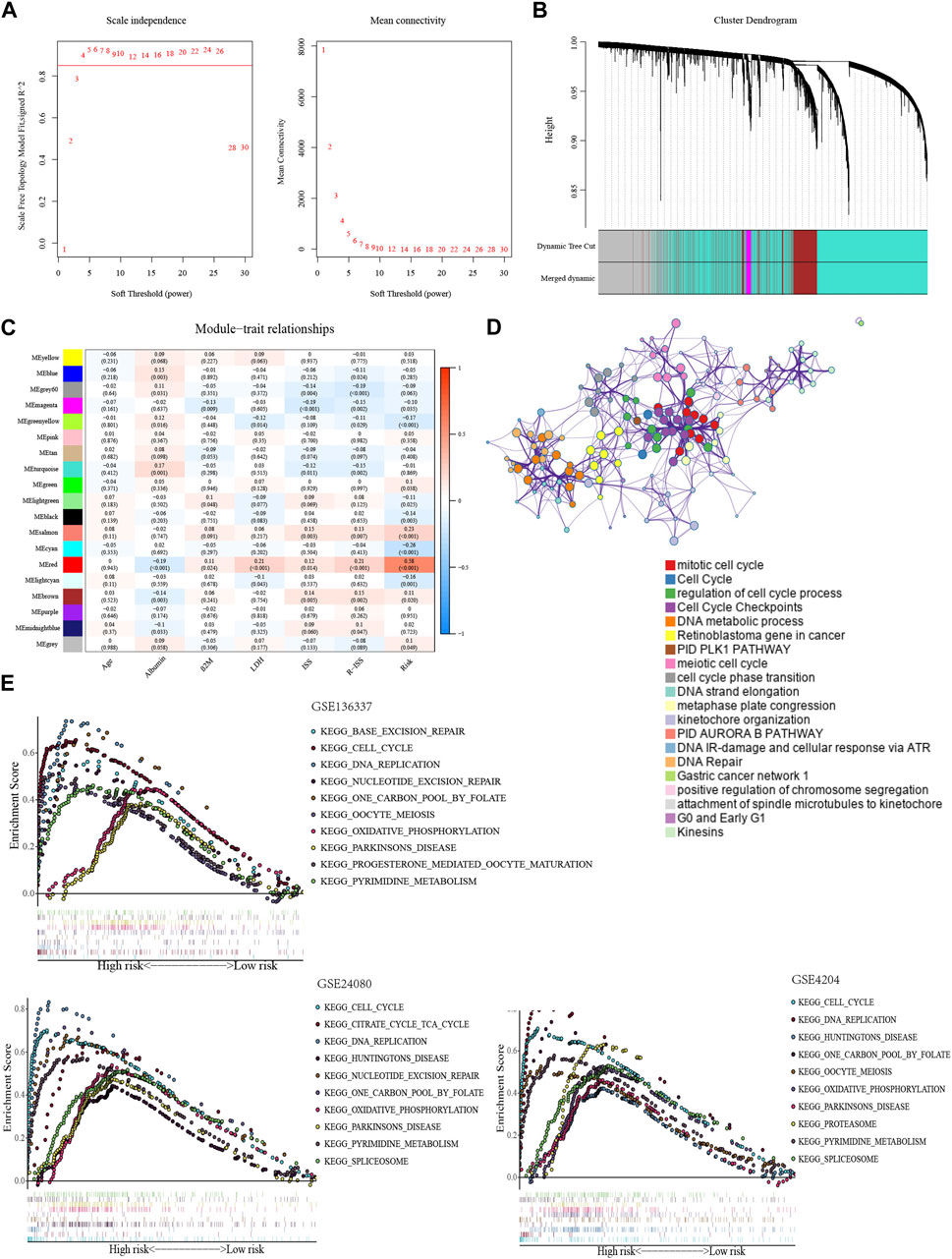
FIGURE 4. Exploration of biological functions based on the glycolytic signature. (A) Selection of optimal soft thresholds in WGCNA (GSE136337, n = 424). (B) Hierarchical clustering dendrogram of genes using WGCNA (GSE136337, n = 424). (C) Associations of gene module with risk model and clinical features (GSE136337, n = 415). (D) Enrichment clustering network using Metascape database (GSE136337, n = 424). (E) The top 10 enriched pathways in the training dataset and validation datasets (GSE136337, n = 424; GSE24080, n = 556; GSE4204, n = 534).
In GSEA, the high-scoring population was home to the considerably enriched pathways, which were mostly linked to glycolysis and included pyrimidine metabolism, one carbon pool via folate, and oxidative phosphorylation. Consistent with previous results, the pathways of cell cycle, DNA repair, and nucleotide excision repair can also be found (Figure 4E).
The immune microenvironment is incredibly important to the growth of MM. Based on several different algorithms, the heat map demonstrated the heterogeneity in the immune landscape among the subgroups (Figure 5A). Immune cell infiltration levels were higher in the low-scoring cohort (p < 0.05), such as active B cells, effector memory CD8+ T cells, immature B cells, NK cells, plasmacytoid dendritic cells, and Th2, while stromal cell abundance was higher in the high-scoring population (p < 0.01) (Figures 5B, C). Moreover, we performed a correlation analysis of the GRGs with immune-related genes and functional status. The expression of most prognostic genes was negatively correlated with the immune-related genes (Figure 5D). For example, LDHB, SOD1, MDH2, POLR3K, and PAXIP1 were negatively correlated with immune-related genes in terms of antigen presentation and cell adhesion, while GMPPB was positively correlated (Figure 5D).
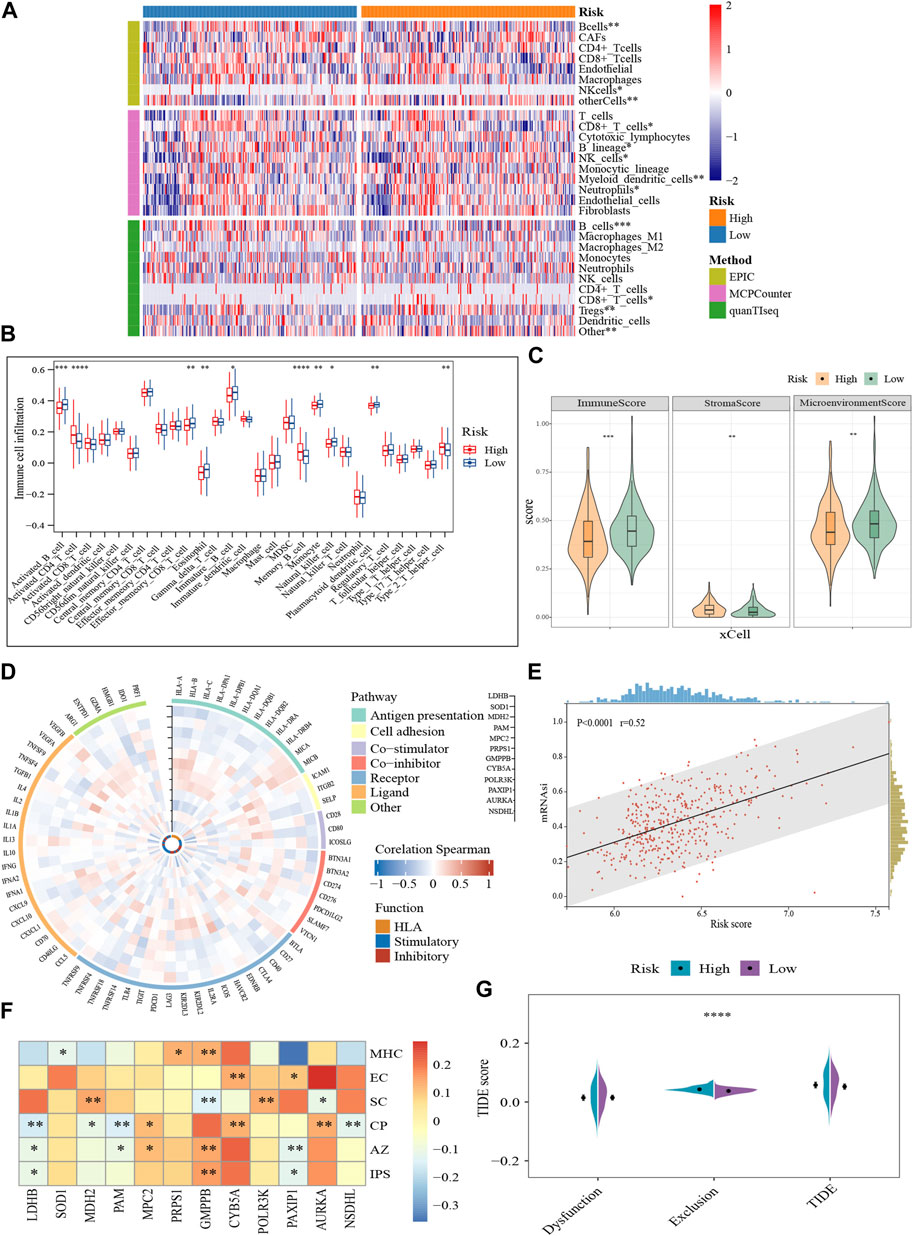
FIGURE 5. Characterization of the tumor microenvironment and immune treatment sensitivity of the glycolytic model (GSE136337, n = 424). (A) Visualization of differences in immune cell abundance based on the various algorithms. (B) Comparison of 28 immune cell infiltration levels between high- and low-scoring cohorts (C). Immune-related scores calculated by xCell. (D) Correlation of prognostic genes with immune-related pathways and functions. The vertical axes with black arrows indicate different prognostic genes. (E) Correlation between risk score and stemness index. (F) Relationships of prognostic genes with distinct immune phenotypes. (G) Assessment of T-cell function and infiltration levels. IPS, immunophenotype score; MHC, antigen presentation; EC, effector cells; SC, suppressor cells; CP, checkpoint marker; z-score, AZ; TIDE, tumor immune dysfunction and exclusion. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Meanwhile, higher risk scores were observed to be strongly associated with the stemness index (Figure 5E). IPS characterizes the immune phenotype of the cells from four perspectives (“antigen-presenting, AP; effector cells, EC; suppressor cells, SC; checkpoints, CP”) (Charoentong et al., 2017). And a total score (z-score, AZ) is finally generated after normalization. A higher z-score corresponds to higher ICI responsiveness. Consistent to our previous results, GMPPB was positively correlated with antigen presentations (p < 0.01). Compared to GMPPB, MDH2, and POLR3K were linked to higher concentrations of immunosuppressive cells (p < 0.01). Furthermore, high expression of GMPPB was often accompanied by greater sensitivity to ICI (p < 0.01), while LDHB and PAXIP1 showed the opposite pattern (p < 0.05) (Figure 5F). The quantity and functional status of T cells is important mediators in immune-targeted therapy. The T-cell exclusion was more prone to occur in the TME of the high-scoring cohort (p < 0.0001), foreshadowing their possible resistance to ICI (Figure 5G). In conclusion, glycolysis-related markers may be a useful tool for determining the immune status of MM and for gauging the effectiveness of immunotherapy.
For improving the survival prediction efficiency of the model, a combined nomogram model was constructed with age, ISS stage, and glycolytic risk score (GSE136337, n = 415) (Figure 6A). Considering the clinical information of the validation cohort, we did not include cytogenetics. The nomogram showed a strong predictive value, as shown by a C-index of 0.728. An ideal concordance between the forecast and observation was visible on the survival probability calibrations plot (Figure 6B). The 1-, 3- and 5-year AUCs were 0.75, 0.75, and 0.78, respectively, higher than the AUCs of the ISS and R-ISS (Figure 6C). The same was in the validation set GSE24080 (n = 556) (Figure 6D).
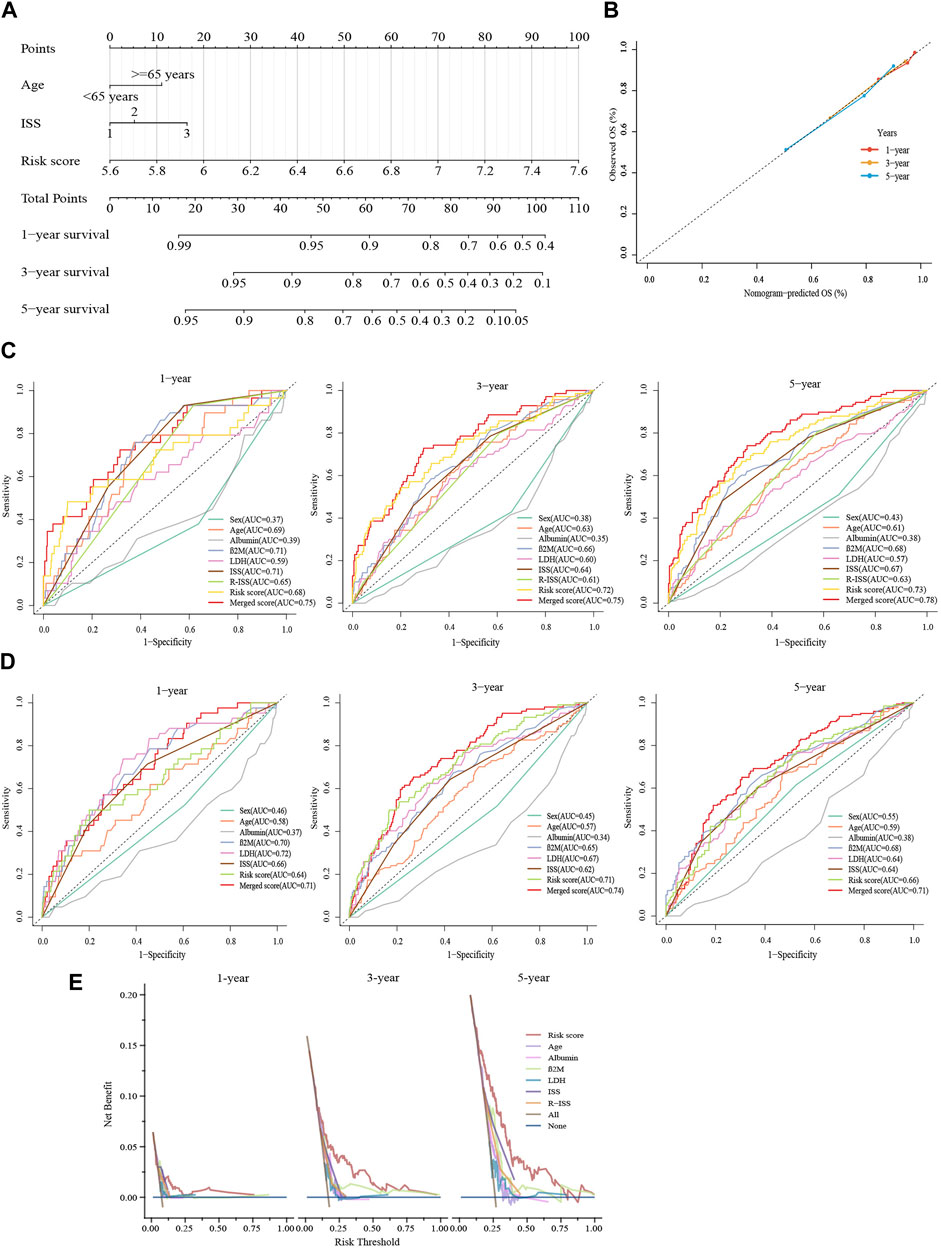
FIGURE 6. Constructing a predictive nomogram to assess clinical applicability. (A) The nomogram based on age, ISS phase, and glycolytic risk score in the training cohort. (B) Calibration plots were made to validate the accuracy in predicting 1-, 3- and 5-year survivals. (C,D) Time-dependent ROC analyses at 1-, 3- and 5-year with the merged score and other clinical covariates. (E) Survival net benefits of each clinical feature and the risk score were estimated with decision curve analysis (DCA). C shows GSE136337 (n = 415); D displays GSE24080 (n = 556).
Moreover, graphical representations provided by DCA showed that the glycolytic risk score outperformed other metrics, such as the R-ISS, in terms of the net benefit to survival (GSE136337, n = 415) (1-year, threshold:0.125–0.250; 3-year, threshold:0.125–0.563; 5-year, threshold:0.125–0.750) (Figure 6E).
MM cell lines and patient samples were subjected to PCR to further validate our predictive model. In these six cell lines (LP-1, RPMI8226, NCI-H929, MM1.R, I9.2, and U266), AURKA, CYB5A, LDHB, MDH2, MPC2, NSDHL, PAXIP1, POLR3K, PRPS1, and SOD1 exhibited primarily upregulation (p < 0.01). GMPPB and PAM, on the other hand, underwent downregulation (p < 0.05) (Figure 7). Correspondingly, the experimental results from the patients were consistent with the cell lines (p < 0.0001) (Figure 8A). We then further investigated the changes in gene expression profiles related to glycolysis among the RMM patients. Figure 8B showed that the expression of GMPPB and PAM was significantly lower than that of controls (p < 0.001), while the opposite was true for other genes (p < 0.01).
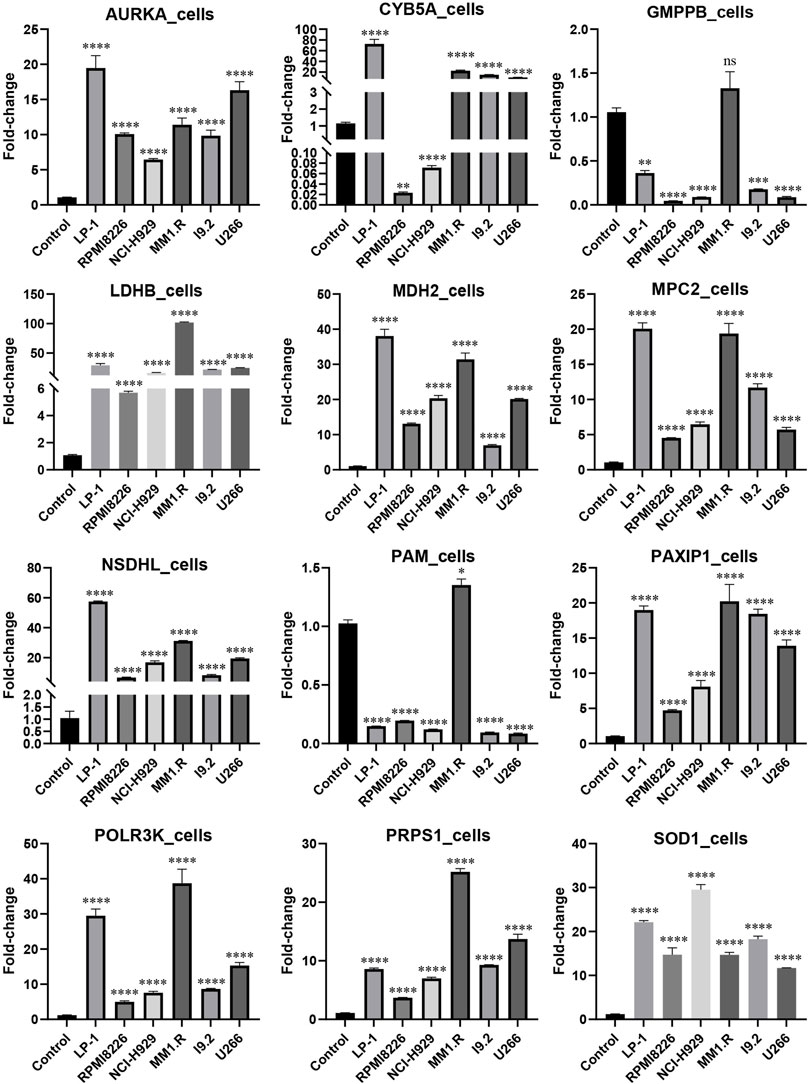
FIGURE 7. External validation in MM cell lines using qRT-PCR (Mean ± SEM). ns, no statistical significance; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
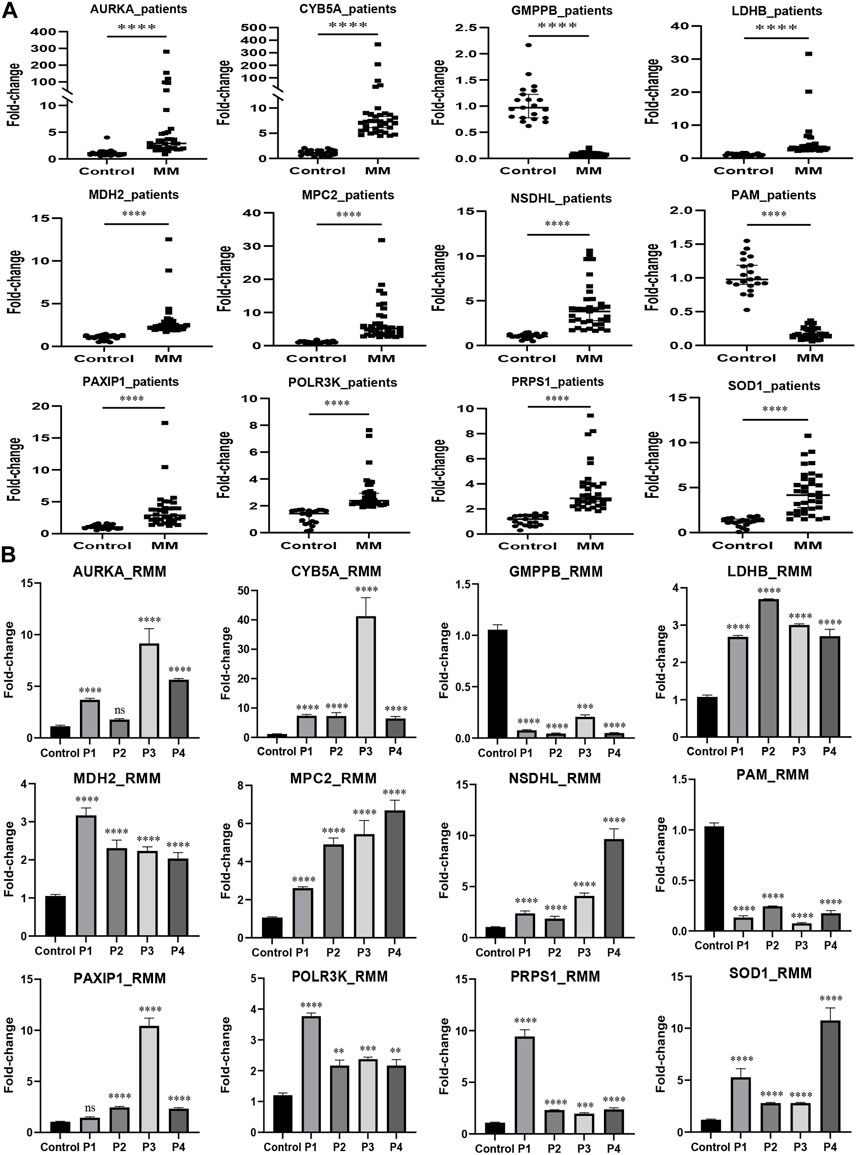
FIGURE 8. External validation in MM patients using qRT-PCR. (A) The expression levels of the prognostic genes in MM patients (n = 35). (B) The expression levels of the prognostic genes in RMM patients (n = 4) (Mean ± SEM). Control, n = 21; ns, no statistical significance; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Metabolic reprogramming, as a defining feature of cancer, has been extensively studied in recent years (Hanahan, 2022). Warburg effect is a manifestation of glucose metabolism reprogramming and is considered an adaptive mechanism to support the biosynthetic demands of uncontrolled proliferation of cancer cells (Warburg, 1956). MM is thought to be a glycolytic tumor because of an elevated glycolytic gene profile associated with disease progression (Evans et al., 2022; Misund et al., 2022) and its sensitivity to glycolysis inhibitors, such as GLUTs and key enzymes (D'Souza and Bhattacharya, 2019; Okabe et al., 2022; Yao et al., 2023). Some key enzymes in the glycolysis process are over-expressed in MM, and more highly expressed in relapsed and refractory patients, such as HK2 and LDHA (Mulligan et al., 2007; Maiso et al., 2015). At the same time, glycolysis as a new therapeutic target has attracted extensive attention. Hypoxia-inducible HK2 has been shown to induce an anti-apoptotic phenotype in myeloma cells through autophagy activation (Ikeda et al., 2020). The combination of phloretin (GLUT1 inhibitor) and daunorubicin (a chemotherapeutic drug) enhances the effect of the latter under hypoxia (Cao et al., 2007; Zub et al., 2015). The production of large amounts of lactate in glycolysis leads to acidification of the tumor microenvironment, which in turn impairs T cells’ proliferation and damages NK cells’ function. Buffering the pH can improve the efficacy of immunotherapy and can be useful in MM (Beckermann et al., 2017; Kouidhi et al., 2018; Wu et al., 2020).
Recently, advances in genomic technology have led to a better understanding of the underlying genetic abnormalities in myeloma. Gene expression profiles (GEP) are the most important tumor-related prognostic biomarkers (Chng et al., 2014). In recent years, GEP has been extensively studied as a potential tool to assess the risk of MM, and several GEP classifiers have been developed (Shaughnessy et al., 2007; Kuiper et al., 2012; Kuiper et al., 2015). However, research on biomarkers of glycolysis-related genes in MM is still limited.
In our study, we constructed a prognostic model featuring 12 GRGs using LASSO regression analysis and validated it in two independent external cohorts. The high-risk group was identified as having poor prognoses. The prognostic accuracy of the model was further assessed by the time-ROC analysis. Furthermore, the multivariate Cox analysis demonstrated that the glycolytic model was an independent prognostic factor and truly served as a powerful prognostic signature of MM.
Clinical traits and drug sensitivity varied between the subtypes. The higher-scoring group with a worse prognosis had more high-risk factors, such as higher LDH levels, ISS, RISS staging, and higher-risk cytogenetic markers. Most of the risk genes in the model were upregulated in myeloma patients, and this upregulation tendency progressed as MM developed. Furthermore, the high-risk cohort exhibited resistance to bortezomib, doxorubicin, and cytarabine. These results were consistent with previous analyses of prognosis. We found that the high-risk subgroup exhibited more responsiveness to vorinostat. As a histone deacetylases (HDAC) inhibitor, vorinostat causes cell cycle arrest and cell death, lowers angiogenesis, and modifies immunological responses in cancer cells (McClure et al., 2018). Several clinical trials have demonstrated the enormous promise of vorinostat in the combination treatment of MM (Richardson et al., 2008; Badros et al., 2009; Weber et al., 2012; Vesole et al., 2015). Chemosensitization allows it to increase the efficiency with which IMiDs or conventional chemotherapeutic drugs kill MM cells (Mitsiades et al., 2004). Panobinostat, also an HDAC inhibitor, has been approved by the U.S. Food and Drug Administration for the treatment of relapsed and refractory multiple myeloma.
Furthermore, we used the WGCNA and GSEA to explore the underlying mechanism of glycolytic prognostic gene markers. Functional analysis revealed many pathways enriched in the high-risk subgroup, including those closely related to metabolism and tumor progression. These findings suggested the strong link between poor prognosis in high-risk cohort and activation of tumor metabolic reprogramming, providing a potential molecular mechanism to elucidate the relationship between the signature and MM progression.
More and more data point to the important role of the evolution of tumor cells and their surroundings in the emergence of MM. Immune evasion and the progression of MM are made possible by the TME. Signature components such as BMSC in TME reduce immune surveillance and promote migration, proliferation, and drug resistance of malignant plasma cells (Garcia-Ortiz et al., 2021; Holthof et al., 2021). The further factors in MM cells avoiding cytotoxic T lymphocyte killing are defective antigen presentation and immunoreactive T cells during bone marrow tumorigenesis (Cohen et al., 2020; Dhatchinamoorthy et al., 2021; Samur et al., 2021). Moreover, immune escape and immunotherapy resistance have increasingly been linked to mutations in genes relevant to antigen presentation (Lee et al., 2020; Jhunjhunwala et al., 2021). And it is believed that one of the crucial mechanisms for the invasion and metastasis of cancer cells is detective intercellular adhesion (Cavallaro and Christofori, 2004; Sousa et al., 2019). In our study, the high-risk group displayed dysregulation of immune function. It had a lesser number of immune cells and more stromal cell infiltration. Additionally, a higher rejection of T cells was noted. The expression of genes related to antigen presentation and cell adhesion was negatively correlated with most risk genes, whereas the opposite was true for the protective gene GMPPB. Biological processes similar to those present in stem cells were also more prevalent in the high-scoring subtype. Furthermore, the high-risk group had a higher degree of tumor dedifferentiation and a subsequent higher likelihood of metastatic and recurrence (Sampieri and Fodde, 2012; Hsu et al., 2018). All of these findings point to the promise of glycolysis-related models in characterizing the immune microenvironment, predicting responsiveness to immune-targeted therapies, and assessing prognosis.
The host factors, tumor-related factors, tumor stage are considered as crucial predictors of survival outcomes by the International Myeloma Working Group (IMWG). The most significant host factor is age, the most significant tumor-related factors are genetic aberration and GEP (Chng et al., 2014). Our glycolysis-related gene signature risk model belongs to tumor-related prognostic factors. MM is highly heterogeneous, and the prognosis of patients varies greatly among individuals. A single prognostic biomarker does not meet the requirements for accurate prognosis prediction. Therefore, combining the information from the training and validation datasets, we integrated multiple prognostic factors to construct a nomogram, including age, risk score, and ISS stage. The nomogram’s ability to predict survival at various time points was the most reliable and potent of any other single variable.
In our prognostic model, AURKA, SOD1, MPC2, LDHB, PAXIP1, MDH2, PRPS1, CYB5A, POLR3K, and NSDHL were identified as risk genes, while PAM and GMPPB were identified as protective genes. The majority of them are reportedly closely related to the occurrence and progression of cancer. AURKA is a serine/threonine kinase that regulates chromosome arrangement, centrosome amplification, and mitotic spindle formation (Hirota et al., 2003; Fu et al., 2007). Overexpression of AURKA has been linked to oncogenic transformation, including chromosomal instability and disruption of multiple oncoprotein regulatory pathways and tumor suppressors (Zhou et al., 1998; Lens et al., 2010; Nikonova et al., 2013). Through AURKA-mediated phosphorylation of LDHB, glycolysis and biosynthesis were effectively promoted, thereby promoting tumor progression (Cheng et al., 2019). AURKA participates in the control of NF-κB and Wnt/β-catenin pathways, associated with the resistance and progression of MM (Dutta-Simmons et al., 2009; Mazzera et al., 2013; Zhou et al., 2013; Huynh et al., 2018). Both in vitro and in vivo, inhibition of AURKA can induce apoptosis and cell death of MM (Shi et al., 2007; Gorgun et al., 2010). AURKA inhibitor can synergize with BTZ to kill t (4,14)-positive MM cells (Jiang et al., 2022). Several AURKA inhibitors are currently being studied in clinical trials in MM or other cancers (Dees et al., 2012; Caputo et al., 2014; Kelly et al., 2014; Long et al., 2015). In the phase I clinical trial studies of RMM, AURKA had shown potential efficacy (Gorgun et al., 2010). Superoxide dismutase (SOD) catalyzes the disproportionation of superoxide free radicals into hydrogen peroxide and molecular oxygen. SOD1 (CuZnSOD) is the main SOD in mammals (Crapo et al., 1992; Tsang et al., 2014). SOD1 overexpression is associated with MM progression and poor prognosis, as well as bortezomib resistance (Salem et al., 2015; Wang et al., 2020; Du et al., 2021). Dysregulation of the intrinsic oxidative environment is an important feature of cancer cells, including MM cells (Aykin-Burns et al., 2009). As an antioxidant, SOD1 had been shown to counteract the cytotoxic effects induced by proteasome inhibitors, which relied mainly on the production of ROS (McConkey and Zhu, 2008; Salem et al., 2015). MPC2 and LDHB were found to be associated with the carfilzomib-related cardiotoxicity in MM (Tantawy et al., 2021). And they were linked to the reorganization of BTZ metabolism in MM cells, which results in resistance to BTZ and poor prognosis (Findlay et al., 2023). MPC2 is one of the rate-limiting proteins involved in glycolytic metabolism. Inhibition of MPC complexes impaired myeloma cell core bioenergetics to increase proteasome inhibitor-induced anti-MM effects (Findlay et al., 2023). Silencing LDHB selectively inhibited basal autophagy and cell proliferation in cancer cells and induced cell death (Brisson et al., 2016). For humoral immunity (Su et al., 2017) and the reorganization of B lymphocyte class switches (Daniel et al., 2010), PAXIP1 is necessary. It is reported that PAXIP1 was necessary to encourage osteoclast differentiation to maintain bone marrow niche structure (Das et al., 2018). Upregulation of PAXIP1 promoted cell proliferation and inhibited apoptosis (Ma and Zheng, 2021). MDH2 is one of the major players in the malate-aspartate shuttle (Minarik et al., 2002). Through increased cell viability and reduced apoptosis, MDH2 contributed to the doxorubicin resistance (Lo et al., 2015). The above may explain the resistance to bortezomib and doxorubicin in the high glycolysis scoring group. PRPS1 is a crucial enzyme in nucleotide synthesis. Mutations in PRPS increase the metabolic vulnerability of the patients with acute lymphoblastic leukemia (ALL), thereby reducing relapse and progression (Srivastava et al., 2021; Song et al., 2023), which is also seen in colorectal and hepatocellular carcinomas (Li et al., 2016; Jing et al., 2019). As a membrane-bound cytochrome, CYB5A carries electron for several membrane-bound oxygenases (Hegesh et al., 1986; Kurian et al., 2006). Studies have shown that CYB5A is upregulated in recurrent ALL (Bartsch et al., 2022). POLR3K is responsible for synthesizing transfer and small ribosomal RNAs in eukaryotes. POLR3K may contribute to the proliferation and angiogenesis of cancer cells by inducing NF-B signaling. (Ablasser et al., 2009; Chiu et al., 2009; Taniguchi and Karin, 2018). The cholesterol-metabolizing enzyme NSDHL is a potential metastatic driver in breast cancer (Yoon et al., 2020; Chen et al., 2021). PAM plays an auxiliary role in amidation and regulates the activity of peptides in the adrenal medulla and pheochromocytoma cells. Oligonucleotide hybridization and immunohistochemical staining showed that PAM was downregulated in neuroendocrine tumors, which was associated with the malignant behavior of the tumors (Thouennon et al., 2007; Horton et al., 2020). GMPPB was negatively associated with poor outcomes in endometrial cancer (Wang et al., 2019; Liu et al., 2020). The relationship between some genes and MM still needs further study.
However, our study has several limitations that need to be addressed. First, the construction and validation of our model relied on the retrospective data from the public database and our clinical samples, so the prognostic robustness and clinical utility of the glycolysis-associated gene signature need to be further verified in larger prospective studies. Secondly, the validation datasets we used lacked complete clinical information, such as detailed cytogenetic information. Finally, the specific role of each gene in MM is unclear, and additional studies in vivo and in vitro are further needed.
In summary, our research offers a fresh perspective for comprehending glycolysis’ function in MM. Different glycolysis-related patterns exhibited heterogeneity in terms of clinical traits and the sensitivity of chemotherapeutic drugs and immunotherapy. This prognostic signature was highly coordinated with multiple malignant features such as immune dysfunction, stem cell-like features, and cancer-related pathways, and was associated with survival outcomes in MM patients. Its clinical effectiveness had been further validated, showing promise in prognostic assessment and treatment options for MM patients.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by The First Affiliated Hospital of Wenzhou Medical University’s ethics committee. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
BZ created the study’s layout, finished data analysis and manuscript writing. The conclusion of the qRT-PCR was aided by QW. All authors helped with gathering clinical samples. The manuscript received significant revisions from YM. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation (grant no. 82270212), the Natural Science Foundation of Zhejiang province (grant no. LY20H080003) and the Wenzhou Municipal Science and Technology Bureau (grant no. Y20220716).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1198949/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Validation of the prognostic model. (A,B) Kaplan-Meier curves for the validation datasets between the high- and low-scoring subtypes. (C,D) The disparities in the 12 genes’ expressions and survival outcomes. (E,F) Time-dependent ROC analysis was used to evaluate the model’s sensitivity and specificity. (A,C,E) display GSE24080 (n = 556); (B,D,F) display GSE4204 (n = 534).
Abdallah, N., Baughn, L. B., Rajkumar, S. V., Kapoor, P., Gertz, M. A., Dispenzieri, A., et al. (2020). Implications of MYC rearrangements in newly diagnosed multiple myeloma. Clin. Cancer Res. 26 (24), 6581–6588. doi:10.1158/1078-0432.CCR-20-2283
Ablasser, A., Bauernfeind, F., Hartmann, G., Latz, E., Fitzgerald, K. A., and Hornung, V. (2009). RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10 (10), 1065–1072. doi:10.1038/ni.1779
Aran, D. (2020). Cell-Type enrichment analysis of bulk transcriptomes using xCell. Methods Mol. Biol. 2120, 263–276. doi:10.1007/978-1-0716-0327-7_19
Aykin-Burns, N., Ahmad, I. M., Zhu, Y., Oberley, L. W., and Spitz, D. R. (2009). Increased levels of superoxide and H2O2 mediate the differential susceptibility of cancer cells versus normal cells to glucose deprivation. Biochem. J. 418 (1), 29–37. doi:10.1042/BJ20081258
Badros, A., Burger, A. M., Philip, S., Niesvizky, R., Kolla, S. S., Goloubeva, O., et al. (2009). Phase I study of vorinostat in combination with bortezomib for relapsed and refractory multiple myeloma. Clin. Cancer Res. 15 (16), 5250–5257. doi:10.1158/1078-0432.CCR-08-2850
Bartsch, L., Schroeder, M. P., Hanzelmann, S., Bastian, L., Lazaro-Navarro, J., Schlee, C., et al. (2022). An alternative CYB5A transcript is expressed in aneuploid ALL and enriched in relapse. BMC Genom Data 23 (1), 30. doi:10.1186/s12863-022-01041-1
Becht, E., Giraldo, N. A., Lacroix, L., Buttard, B., Elarouci, N., Petitprez, F., et al. (2016). Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17 (1), 218. doi:10.1186/s13059-016-1070-5
Beckermann, K. E., Dudzinski, S. O., and Rathmell, J. C. (2017). Dysfunctional T cell metabolism in the tumor microenvironment. Cytokine Growth Factor Rev. 35, 7–14. doi:10.1016/j.cytogfr.2017.04.003
Bianchi, G., Richardson, P. G., and Anderson, K. C. (2015). Promising therapies in multiple myeloma. Blood 126 (3), 300–310. doi:10.1182/blood-2015-03-575365
Brisson, L., Banski, P., Sboarina, M., Dethier, C., Danhier, P., Fontenille, M. J., et al. (2016). Lactate dehydrogenase B controls lysosome activity and autophagy in cancer. Cancer Cell 30 (3), 418–431. doi:10.1016/j.ccell.2016.08.005
Cao, X., Fang, L., Gibbs, S., Huang, Y., Dai, Z., Wen, P., et al. (2007). Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia. Cancer Chemother. Pharmacol. 59 (4), 495–505. doi:10.1007/s00280-006-0291-9
Caputo, E., Miceli, R., Motti, M. L., Tate, R., Fratangelo, F., Botti, G., et al. (2014). AurkA inhibitors enhance the effects of B-RAF and MEK inhibitors in melanoma treatment. J. Transl. Med. 12, 216. doi:10.1186/s12967-014-0216-z
Cavallaro, U., and Christofori, G. (2004). Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 4 (2), 118–132. doi:10.1038/nrc1276
Charoentong, P., Finotello, F., Angelova, M., Mayer, C., Efremova, M., Rieder, D., et al. (2017). Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18 (1), 248–262. doi:10.1016/j.celrep.2016.12.019
Chen, M., Zhao, Y., Yang, X., Zhao, Y., Liu, Q., Liu, Y., et al. (2021). NSDHL promotes triple-negative breast cancer metastasis through the TGFβ signaling pathway and cholesterol biosynthesis. Breast Cancer Res. Treat. 187 (2), 349–362. doi:10.1007/s10549-021-06213-8
Cheng, A., Zhang, P., Wang, B., Yang, D., Duan, X., Jiang, Y., et al. (2019). Aurora-A mediated phosphorylation of LDHB promotes glycolysis and tumor progression by relieving the substrate-inhibition effect. Nat. Commun. 10 (1), 5566. doi:10.1038/s41467-019-13485-8
Chiu, Y. H., Macmillan, J. B., and Chen, Z. J. (2009). RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138 (3), 576–591. doi:10.1016/j.cell.2009.06.015
Chng, W. J., Dispenzieri, A., Chim, C. S., Fonseca, R., Goldschmidt, H., Lentzsch, S., et al. (2014). IMWG consensus on risk stratification in multiple myeloma. Leukemia 28 (2), 269–277. doi:10.1038/leu.2013.247
Cho, H., Yoon, D. H., Lee, J. B., Kim, S. Y., Moon, J. H., Do, Y. R., et al. (2017). Comprehensive evaluation of the revised international staging system in multiple myeloma patients treated with novel agents as a primary therapy. Am. J. Hematol. 92 (12), 1280–1286. doi:10.1002/ajh.24891
Cohen, A. D., Raje, N., Fowler, J. A., Mezzi, K., Scott, E. C., and Dhodapkar, M. V. (2020). How to train your T cells: Overcoming immune dysfunction in multiple myeloma. Clin. Cancer Res. 26 (7), 1541–1554. doi:10.1158/1078-0432.CCR-19-2111
Cowan, A. J., Allen, C., Barac, A., Basaleem, H., Bensenor, I., Curado, M. P., et al. (2018). Global burden of multiple myeloma: A systematic analysis for the global burden of disease study 2016. JAMA Oncol. 4 (9), 1221–1227. doi:10.1001/jamaoncol.2018.2128
Crapo, J. D., Oury, T., Rabouille, C., Slot, J. W., and Chang, L. Y. (1992). Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. U. S. A. 89 (21), 10405–10409. doi:10.1073/pnas.89.21.10405
D'Souza, L., and Bhattacharya, D. (2019). Plasma cells: You are what you eat. Immunol. Rev. 288 (1), 161–177. doi:10.1111/imr.12732
Daniel, J. A., Santos, M. A., Wang, Z., Zang, C., Schwab, K. R., Jankovic, M., et al. (2010). PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science 329 (5994), 917–923. doi:10.1126/science.1187942
Das, P., Veazey, K. J., Van, H. T., Kaushik, S., Lin, K., Lu, Y., et al. (2018). Histone methylation regulator PTIP is required to maintain normal and leukemic bone marrow niches. Proc. Natl. Acad. Sci. U. S. A. 115 (43), E10137–E10146. doi:10.1073/pnas.1806019115
Dees, E. C., Cohen, R. B., von Mehren, M., Stinchcombe, T. E., Liu, H., Venkatakrishnan, K., et al. (2012). Phase I study of aurora A kinase inhibitor MLN8237 in advanced solid tumors: Safety, pharmacokinetics, pharmacodynamics, and bioavailability of two oral formulations. Clin. Cancer Res. 18 (17), 4775–4784. doi:10.1158/1078-0432.CCR-12-0589
Dhatchinamoorthy, K., Colbert, J. D., and Rock, K. L. (2021). Cancer immune evasion through loss of MHC class I antigen presentation. Front. Immunol. 12, 636568. doi:10.3389/fimmu.2021.636568
Du, T., Song, Y., Ray, A., Chauhan, D., and Anderson, K. C. (2021). Proteomic analysis identifies mechanism(s) of overcoming bortezomib resistance via targeting ubiquitin receptor Rpn13. Leukemia 35 (2), 550–561. doi:10.1038/s41375-020-0865-2
Dutta-Simmons, J., Zhang, Y., Gorgun, G., Gatt, M., Mani, M., Hideshima, T., et al. (2009). Aurora kinase A is a target of Wnt/beta-catenin involved in multiple myeloma disease progression. Blood 114 (13), 2699–2708. doi:10.1182/blood-2008-12-194290
Evans, L. A., Anderson, E. A., Jessen, E., Nandakumar, B., Atilgan, E., Jevremovic, D., et al. (2022). Overexpression of the energy metabolism transcriptome within clonal plasma cells is associated with the pathogenesis and outcomes of patients with multiple myeloma. Am. J. Hematol. 97 (7), 895–902. doi:10.1002/ajh.26577
Findlay, S., Nair, R., Merrill, R. A., Kaiser, Z., Cajelot, A., Aryanpour, Z., et al. (2023). The mitochondrial pyruvate carrier complex potentiates the efficacy of proteasome inhibitors in multiple myeloma. Blood Adv. 2023, 2022008345. doi:10.1182/bloodadvances.2022008345
Fu, J., Bian, M., Jiang, Q., and Zhang, C. (2007). Roles of Aurora kinases in mitosis and tumorigenesis. Mol. Cancer Res. 5 (1), 1–10. doi:10.1158/1541-7786.MCR-06-0208
Garcia-Ortiz, A., Rodriguez-Garcia, Y., Encinas, J., Maroto-Martin, E., Castellano, E., Teixido, J., et al. (2021). The role of tumor microenvironment in multiple myeloma development and progression. Cancers (Basel) 13 (2), 217. doi:10.3390/cancers13020217
Garnett, M. J., Edelman, E. J., Heidorn, S. J., Greenman, C. D., Dastur, A., Lau, K. W., et al. (2012). Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483 (7391), 570–575. doi:10.1038/nature11005
Geeleher, P., Cox, N., and Huang, R. S. (2014a). pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 9 (9), e107468. doi:10.1371/journal.pone.0107468
Geeleher, P., Cox, N. J., and Huang, R. S. (2014b). Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 15 (3), R47. doi:10.1186/gb-2014-15-3-r47
Gorgun, G., Calabrese, E., Hideshima, T., Ecsedy, J., Perrone, G., Mani, M., et al. (2010). A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood 115 (25), 5202–5213. doi:10.1182/blood-2009-12-259523
Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discov. 12 (1), 31–46. doi:10.1158/2159-8290.CD-21-1059
He, Y., Wang, Y., Liu, H., Xu, X., He, S., Tang, J., et al. (2015). Pyruvate kinase isoform M2 (PKM2) participates in multiple myeloma cell proliferation, adhesion and chemoresistance. Leuk. Res. 39 (12), 1428–1436. doi:10.1016/j.leukres.2015.09.019
Hegesh, E., Hegesh, J., and Kaftory, A. (1986). Congenital methemoglobinemia with a deficiency of cytochrome b5. N. Engl. J. Med. 314 (12), 757–761. doi:10.1056/NEJM198603203141206
Hirota, T., Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Nitta, M., et al. (2003). Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114 (5), 585–598. doi:10.1016/s0092-8674(03)00642-1
Holthof, L. C., van der Schans, J. J., Katsarou, A., Poels, R., Gelderloos, A. T., Drent, E., et al. (2021). Bone marrow mesenchymal stromal cells can render multiple myeloma cells resistant to cytotoxic machinery of CAR T cells through inhibition of apoptosis. Clin. Cancer Res. 27 (13), 3793–3803. doi:10.1158/1078-0432.CCR-20-2188
Horton, T. M., Sundaram, V., Lee, C. H., Hornbacker, K., Van Vleck, A., Benjamin, K. N., et al. (2020). PAM staining intensity of primary neuroendocrine neoplasms is a potential prognostic biomarker. Sci. Rep. 10 (1), 10943. doi:10.1038/s41598-020-68071-6
Hsu, J. M., Xia, W., Hsu, Y. H., Chan, L. C., Yu, W. H., Cha, J. H., et al. (2018). STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 9 (1), 1908. doi:10.1038/s41467-018-04313-6
Huynh, M., Pak, C., Markovina, S., Callander, N. S., Chng, K. S., Wuerzberger-Davis, S. M., et al. (2018). Hyaluronan and proteoglycan link protein 1 (HAPLN1) activates bortezomib-resistant NF-κB activity and increases drug resistance in multiple myeloma. J. Biol. Chem. 293 (7), 2452–2465. doi:10.1074/jbc.RA117.000667
Ikeda, S., Abe, F., Matsuda, Y., Kitadate, A., Takahashi, N., and Tagawa, H. (2020). Hypoxia-inducible hexokinase-2 enhances anti-apoptotic function via activating autophagy in multiple myeloma. Cancer Sci. 111 (11), 4088–4101. doi:10.1111/cas.14614
Jhunjhunwala, S., Hammer, C., and Delamarre, L. (2021). Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 21 (5), 298–312. doi:10.1038/s41568-021-00339-z
Jiang, H., Wang, Y., Wang, J., Wang, Y., Wang, S., He, E., et al. (2022). Posttranslational modification of Aurora A-NSD2 loop contributes to drug resistance in t(4;14) multiple myeloma. Clin. Transl. Med. 12 (4), e744. doi:10.1002/ctm2.744
Jiang, P., Gu, S., Pan, D., Fu, J., Sahu, A., Hu, X., et al. (2018). Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24 (10), 1550–1558. doi:10.1038/s41591-018-0136-1
Jing, X., Wang, X. J., Zhang, T., Zhu, W., Fang, Y., Wu, H., et al. (2019). Cell-cycle-dependent phosphorylation of PRPS1 fuels nucleotide synthesis and promotes tumorigenesis. Cancer Res. 79 (18), 4650–4664. doi:10.1158/0008-5472.CAN-18-2486
Jung, S. H., Kim, K., Kim, J. S., Kim, S. J., Cheong, J. W., Kim, S. J., et al. (2018). A prognostic scoring system for patients with multiple myeloma classified as stage II with the Revised International Staging System. Br. J. Haematol. 181 (5), 707–710. doi:10.1111/bjh.14701
Kelly, K. R., Shea, T. C., Goy, A., Berdeja, J. G., Reeder, C. B., McDonagh, K. T., et al. (2014). Phase I study of MLN8237--investigational Aurora A kinase inhibitor--in relapsed/refractory multiple myeloma, non-Hodgkin lymphoma and chronic lymphocytic leukemia. Invest. New Drugs 32 (3), 489–499. doi:10.1007/s10637-013-0050-9
Kouidhi, S., Ben Ayed, F., and Benammar Elgaaied, A. (2018). Targeting tumor metabolism: A new challenge to improve immunotherapy. Front. Immunol. 9, 353. doi:10.3389/fimmu.2018.00353
Krishnan, S. R., Jaiswal, R., Brown, R. D., Luk, F., and Bebawy, M. (2016). Multiple myeloma and persistence of drug resistance in the age of novel drugs (Review). Int. J. Oncol. 49 (1), 33–50. doi:10.3892/ijo.2016.3516
Kuiper, R., Broyl, A., de Knegt, Y., van Vliet, M. H., van Beers, E. H., van der Holt, B., et al. (2012). A gene expression signature for high-risk multiple myeloma. Leukemia 26 (11), 2406–2413. doi:10.1038/leu.2012.127
Kuiper, R., van Duin, M., van Vliet, M. H., Broijl, A., van der Holt, B., El Jarari, L., et al. (2015). Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood 126 (17), 1996–2004. doi:10.1182/blood-2015-05-644039
Kumar, S. K., Rajkumar, S. V., Dispenzieri, A., Lacy, M. Q., Hayman, S. R., Buadi, F. K., et al. (2008). Improved survival in multiple myeloma and the impact of novel therapies. Blood 111 (5), 2516–2520. doi:10.1182/blood-2007-10-116129
Kumar, S. K., Rajkumar, V., Kyle, R. A., van Duin, M., Sonneveld, P., Mateos, M. V., et al. (2017). Multiple myeloma. Nat. Rev. Dis. Prim. 3, 17046. doi:10.1038/nrdp.2017.46
Kurian, J. R., Chin, N. A., Longlais, B. J., Hayes, K. L., and Trepanier, L. A. (2006). Reductive detoxification of arylhydroxylamine carcinogens by human NADH cytochrome b5 reductase and cytochrome b5. Chem. Res. Toxicol. 19 (10), 1366–1373. doi:10.1021/tx060106t
Kyle, R. A., Remstein, E. D., Therneau, T. M., Dispenzieri, A., Kurtin, P. J., Hodnefield, J. M., et al. (2007). Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N. Engl. J. Med. 356 (25), 2582–2590. doi:10.1056/NEJMoa070389
Kyle, R. A., Therneau, T. M., Rajkumar, S. V., Offord, J. R., Larson, D. R., Plevak, M. F., et al. (2002). A long-term study of prognosis in monoclonal gammopathy of undetermined significance. N. Engl. J. Med. 346 (8), 564–569. doi:10.1056/NEJMoa01133202
Landgren, O., Kyle, R. A., Pfeiffer, R. M., Katzmann, J. A., Caporaso, N. E., Hayes, R. B., et al. (2009). Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: A prospective study. Blood 113 (22), 5412–5417. doi:10.1182/blood-2008-12-194241
Langfelder, P., and Horvath, S. (2012). Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 46 (11), i11. doi:10.18637/jss.v046.i11
Langfelder, P., and Horvath, S. (2008). Wgcna: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559. doi:10.1186/1471-2105-9-559
Laubach, J., Richardson, P., and Anderson, K. (2011). Multiple myeloma. Annu. Rev. Med. 62, 249–264. doi:10.1146/annurev-med-070209-175325
Lee, M. Y., Jeon, J. W., Sievers, C., and Allen, C. T. (2020). Antigen processing and presentation in cancer immunotherapy. J. Immunother. Cancer 8 (2), e001111. doi:10.1136/jitc-2020-001111
Lens, S. M., Voest, E. E., and Medema, R. H. (2010). Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer 10 (12), 825–841. doi:10.1038/nrc2964
Li, X., Qian, X., Peng, L. X., Jiang, Y., Hawke, D. H., Zheng, Y., et al. (2016). A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat. Cell Biol. 18 (5), 561–571. doi:10.1038/ncb3338
Liu, J., Li, S., Feng, G., Meng, H., Nie, S., Sun, R., et al. (2020). Nine glycolysis-related gene signature predicting the survival of patients with endometrial adenocarcinoma. Cancer Cell Int. 20, 183. doi:10.1186/s12935-020-01264-1
Lo, Y. W., Lin, S. T., Chang, S. J., Chan, C. H., Lyu, K. W., Chang, J. F., et al. (2015). Mitochondrial proteomics with siRNA knockdown to reveal ACAT1 and MDH2 in the development of doxorubicin-resistant uterine cancer. J. Cell Mol. Med. 19 (4), 744–759. doi:10.1111/jcmm.12388
Long, Z. J., Wang, L. X., Zheng, F. M., Chen, J. J., Luo, Y., Tu, X. X., et al. (2015). A novel compound against oncogenic Aurora kinase A overcomes imatinib resistance in chronic myeloid leukemia cells. Int. J. Oncol. 46 (6), 2488–2496. doi:10.3892/ijo.2015.2960
Ma, Y., and Zheng, W. (2021). H3K27ac-induced lncRNA PAXIP1-AS1 promotes cell proliferation, migration, EMT and apoptosis in ovarian cancer by targeting miR-6744-5p/PCBP2 axis. J. Ovarian Res. 14 (1), 76. doi:10.1186/s13048-021-00822-z
Maiso, P., Huynh, D., Moschetta, M., Sacco, A., Aljawai, Y., Mishima, Y., et al. (2015). Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res. 75 (10), 2071–2082. doi:10.1158/0008-5472.CAN-14-3400
Malta, T. M., Sokolov, A., Gentles, A. J., Burzykowski, T., Poisson, L., Weinstein, J. N., et al. (2018). Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173 (2), 338–354.e15. doi:10.1016/j.cell.2018.03.034
Mazzera, L., Lombardi, G., Abeltino, M., Ricca, M., Donofrio, G., Giuliani, N., et al. (2013). Aurora and IKK kinases cooperatively interact to protect multiple myeloma cells from Apo2L/TRAIL. Blood 122 (15), 2641–2653. doi:10.1182/blood-2013-02-482356
McBrayer, S. K., Cheng, J. C., Singhal, S., Krett, N. L., Rosen, S. T., and Shanmugam, M. (2012). Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: Implications for glucose transporter-directed therapy. Blood 119 (20), 4686–4697. doi:10.1182/blood-2011-09-377846
McClure, J. J., Li, X., and Chou, C. J. (2018). Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv. Cancer Res. 138, 183–211. doi:10.1016/bs.acr.2018.02.006
McConkey, D. J., and Zhu, K. (2008). Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist Updat 11 (4-5), 164–179. doi:10.1016/j.drup.2008.08.002
Minarik, P., Tomaskova, N., Kollarova, M., and Antalik, M. (2002). Malate dehydrogenases--structure and function. Gen. Physiol. Biophys. 21 (3), 257–265.
Misund, K., Hofste Op Bruinink, D., Coward, E., Hoogenboezem, R. M., Rustad, E. H., Sanders, M. A., et al. (2022). Clonal evolution after treatment pressure in multiple myeloma: Heterogenous genomic aberrations and transcriptomic convergence. Leukemia 36 (7), 1887–1897. doi:10.1038/s41375-022-01597-y
Mitsiades, C. S., Mitsiades, N. S., McMullan, C. J., Poulaki, V., Shringarpure, R., Hideshima, T., et al. (2004). Transcriptional signature of histone deacetylase inhibition in multiple myeloma: Biological and clinical implications. Proc. Natl. Acad. Sci. U. S. A. 101 (2), 540–545. doi:10.1073/pnas.2536759100
Moreau, P., Attal, M., and Facon, T. (2015). Frontline therapy of multiple myeloma. Blood 125 (20), 3076–3084. doi:10.1182/blood-2014-09-568915
Mulligan, G., Mitsiades, C., Bryant, B., Zhan, F., Chng, W. J., Roels, S., et al. (2007). Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood 109 (8), 3177–3188. doi:10.1182/blood-2006-09-044974
Nikonova, A. S., Astsaturov, I., Serebriiskii, I. G., Dunbrack, R. L., and Golemis, E. A. (2013). Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol. Life Sci. 70 (4), 661–687. doi:10.1007/s00018-012-1073-7
Okabe, S., Tanaka, Y., and Gotoh, A. (2022). Therapeutic targeting of PFKFB3 and PFKFB4 in multiple myeloma cells under hypoxic conditions. Biomark. Res. 10 (1), 31. doi:10.1186/s40364-022-00376-2
Palumbo, A., Avet-Loiseau, H., Oliva, S., Lokhorst, H. M., Goldschmidt, H., Rosinol, L., et al. (2015). Revised international staging system for multiple myeloma: A report from international myeloma working group. J. Clin. Oncol. 33 (26), 2863–2869. doi:10.1200/JCO.2015.61.2267
Plattner, C., Finotello, F., and Rieder, D. (2020). Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol. 636, 261–285. doi:10.1016/bs.mie.2019.05.056
Richardson, P., Mitsiades, C., Colson, K., Reilly, E., McBride, L., Chiao, J., et al. (2008). Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leuk. Lymphoma 49 (3), 502–507. doi:10.1080/10428190701817258
Röllig, C., Knop, S., and Bornhäuser, M. (2015). Multiple myeloma. Lancet 385 (9983), 2197–2208. doi:10.1016/s0140-6736(14)60493-1
Salem, K., McCormick, M. L., Wendlandt, E., Zhan, F., and Goel, A. (2015). Copper-zinc superoxide dismutase-mediated redox regulation of bortezomib resistance in multiple myeloma. Redox Biol. 4, 23–33. doi:10.1016/j.redox.2014.11.002
Sampieri, K., and Fodde, R. (2012). Cancer stem cells and metastasis. Semin. Cancer Biol. 22 (3), 187–193. doi:10.1016/j.semcancer.2012.03.002
Samur, M. K., Fulciniti, M., Aktas Samur, A., Bazarbachi, A. H., Tai, Y. T., Prabhala, R., et al. (2021). Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat. Commun. 12 (1), 868. doi:10.1038/s41467-021-21177-5
Sanchez, W. Y., McGee, S. L., Connor, T., Mottram, B., Wilkinson, A., Whitehead, J. P., et al. (2013). Dichloroacetate inhibits aerobic glycolysis in multiple myeloma cells and increases sensitivity to bortezomib. Br. J. Cancer 108 (8), 1624–1633. doi:10.1038/bjc.2013.120
Schmidt, T. M., Fonseca, R., and Usmani, S. Z. (2021). Chromosome 1q21 abnormalities in multiple myeloma. Blood Cancer J. 11 (4), 83. doi:10.1038/s41408-021-00474-8
Shaughnessy, J. D., Zhan, F., Burington, B. E., Huang, Y., Colla, S., Hanamura, I., et al. (2007). A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109 (6), 2276–2284. doi:10.1182/blood-2006-07-038430
Shi, Y., Reiman, T., Li, W., Maxwell, C. A., Sen, S., Pilarski, L., et al. (2007). Targeting aurora kinases as therapy in multiple myeloma. Blood 109 (9), 3915–3921. doi:10.1182/blood-2006-07-037671
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71 (1), 7–33. doi:10.3322/caac.21654
Song, L., Li, P., Sun, H., Ding, L., Wang, J., Li, B., et al. (2023). PRPS2 mutations drive acute lymphoblastic leukemia relapse through influencing PRPS1/2 hexamer stability. Blood Sci. 5 (1), 39–50. doi:10.1097/BS9.0000000000000139
Sonneveld, P., Avet-Loiseau, H., Lonial, S., Usmani, S., Siegel, D., Anderson, K. C., et al. (2016). Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the international myeloma working group. Blood 127 (24), 2955–2962. doi:10.1182/blood-2016-01-631200
Sousa, B., Pereira, J., and Paredes, J. (2019). The crosstalk between cell adhesion and cancer metabolism. Int. J. Mol. Sci. 20 (8), 1933. doi:10.3390/ijms20081933
Srivastava, S., Sahu, U., Zhou, Y., Hogan, A. K., Sathyan, K. M., Bodner, J., et al. (2021). NOTCH1-driven UBR7 stimulates nucleotide biosynthesis to promote T cell acute lymphoblastic leukemia. Sci. Adv. 7 (5), eabc9781. doi:10.1126/sciadv.abc9781
Su, D., Vanhee, S., Soria, R., Gyllenback, E. J., Starnes, L. M., Hojfeldt, M. K., et al. (2017). PTIP chromatin regulator controls development and activation of B cell subsets to license humoral immunity in mice. Proc. Natl. Acad. Sci. U. S. A. 114 (44), E9328–E9337. doi:10.1073/pnas.1707938114
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth, A., Minguez, P., et al. (2011). The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568. doi:10.1093/nar/gkq973
Taniguchi, K., and Karin, M. (2018). NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 18 (5), 309–324. doi:10.1038/nri.2017.142
Tantawy, M., Chekka, L. M., Huang, Y., Garrett, T. J., Singh, S., Shah, C. P., et al. (2021). Lactate dehydrogenase B and pyruvate oxidation pathway associated with carfilzomib-related cardiotoxicity in multiple myeloma patients: Result of a multi-omics integrative analysis. Front. Cardiovasc Med. 8, 645122. doi:10.3389/fcvm.2021.645122
Thorsson, V., Gibbs, D. L., Brown, S. D., Wolf, D., Bortone, D. S., Ou Yang, T. H., et al. (2018). The immune landscape of cancer. Immunity 48 (4), 812–830.e14. doi:10.1016/j.immuni.2018.03.023
Thouennon, E., Elkahloun, A. G., Guillemot, J., Gimenez-Roqueplo, A. P., Bertherat, J., Pierre, A., et al. (2007). Identification of potential gene markers and insights into the pathophysiology of pheochromocytoma malignancy. J. Clin. Endocrinol. Metab. 92 (12), 4865–4872. doi:10.1210/jc.2007-1253
Tsang, C. K., Liu, Y., Thomas, J., Zhang, Y., and Zheng, X. F. (2014). Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5, 3446. doi:10.1038/ncomms4446
Vesole, D. H., Bilotti, E., Richter, J. R., McNeill, A., McBride, L., Raucci, L., et al. (2015). Phase I study of carfilzomib, lenalidomide, vorinostat, and dexamethasone in patients with relapsed and/or refractory multiple myeloma. Br. J. Haematol. 171 (1), 52–59. doi:10.1111/bjh.13517
Vickers, A. J., and Elkin, E. B. (2006). Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 26 (6), 565–574. doi:10.1177/0272989X06295361
Wallington-Beddoe, C. T., and Mynott, R. L. (2021). Prognostic and predictive biomarker developments in multiple myeloma. J. Hematol. Oncol. 14 (1), 151. doi:10.1186/s13045-021-01162-7
Wallington-Beddoe, C. T., Sobieraj-Teague, M., Kuss, B. J., and Pitson, S. M. (2018). Resistance to proteasome inhibitors and other targeted therapies in myeloma. Br. J. Haematol. 182 (1), 11–28. doi:10.1111/bjh.15210
Wang, Q., Zhao, D., Xian, M., Wang, Z., Bi, E., Su, P., et al. (2020). MIF as a biomarker and therapeutic target for overcoming resistance to proteasome inhibitors in human myeloma. Blood 136 (22), 2557–2573. doi:10.1182/blood.2020005795
Wang, Z. H., Zhang, Y. Z., Wang, Y. S., and Ma, X. X. (2019). Identification of novel cell glycolysis related gene signature predicting survival in patients with endometrial cancer. Cancer Cell Int. 19, 296. doi:10.1186/s12935-019-1001-0
Warburg, O. (1956). On the origin of cancer cells. Science 123 (3191), 309–314. doi:10.1126/science.123.3191.309
Weber, D. M., Graef, T., Hussein, M., Sobecks, R. M., Schiller, G. J., Lupinacci, L., et al. (2012). Phase I trial of vorinostat combined with bortezomib for the treatment of relapsing and/or refractory multiple myeloma. Clin. Lymphoma Myeloma Leuk. 12 (5), 319–324. doi:10.1016/j.clml.2012.07.007
Wu, S., Kuang, H., Ke, J., Pi, M., and Yang, D. H. (2020). Metabolic reprogramming induces immune cell dysfunction in the tumor microenvironment of multiple myeloma. Front. Oncol. 10, 591342. doi:10.3389/fonc.2020.591342
Yao, Y., Fong Ng, J., Park, W. D., Samur, M. K., Morelli, E., Encinas, J., et al. (2023). CDK7 controls E2F- and MYC-driven proliferative and metabolic vulnerabilities in multiple myeloma. Blood 2023, 2022018885. doi:10.1182/blood.2022018885
Yoon, S. H., Kim, H. S., Kim, R. N., Jung, S. Y., Hong, B. S., Kang, E. J., et al. (2020). NAD(P)-dependent steroid dehydrogenase-like is involved in breast cancer cell growth and metastasis. BMC Cancer 20 (1), 375. doi:10.1186/s12885-020-06840-2
Zhou, H., Kuang, J., Zhong, L., Kuo, W. L., Gray, J. W., Sahin, A., et al. (1998). Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20 (2), 189–193. doi:10.1038/2496
Zhou, W., Yang, Y., Xia, J., Wang, H., Salama, M. E., Xiong, W., et al. (2013). NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell 23 (1), 48–62. doi:10.1016/j.ccr.2012.12.001
Zub, K. A., Sousa, M. M., Sarno, A., Sharma, A., Demirovic, A., Rao, S., et al. (2015). Modulation of cell metabolic pathways and oxidative stress signaling contribute to acquired melphalan resistance in multiple myeloma cells. PLoS One 10 (3), e0119857. doi:10.1371/journal.pone.0119857
Keywords: multiple myeloma, glycolysis, prognostic signature, tumor microenvironment, risk stratification, therapeutic targets
Citation: Zhang B, Wang Q, Lin Z, Zheng Z, Zhou S, Zhang T, Zheng D, Chen Z, Zheng S, Zhang Y, Lin X, Dong R, Chen J, Qian H, Hu X, Zhuang Y, Zhang Q, Jin Z, Jiang S and Ma Y (2023) A novel glycolysis-related gene signature for predicting the prognosis of multiple myeloma. Front. Cell Dev. Biol. 11:1198949. doi: 10.3389/fcell.2023.1198949
Received: 02 April 2023; Accepted: 25 May 2023;
Published: 02 June 2023.
Edited by:
Si Zhang, Fudan University, ChinaReviewed by:
Pan Chen, Central South University, ChinaCopyright © 2023 Zhang, Wang, Lin, Zheng, Zhou, Zhang, Zheng, Chen, Zheng, Zhang, Lin, Dong, Chen, Qian, Hu, Zhuang, Zhang, Jin, Jiang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhouxiang Jin, d3pqaW56eEAxNjMuY29t; Songfu Jiang, amlhbmdzb25nZnVAMTg5LmNu; Yongyong Ma, bWF5eUB3bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.