- 1Department of Physiology and Immunology, College of Medicine and Health Sciences, Khalifa University, Abu Dhabi, United Arab Emirates
- 2Department of Pediatrics, College of Medicine and Health Sciences, Khalifa University, Abu Dhabi, United Arab Emirates
- 3Healthcare Engineering Innovation Center (HEIC), Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates
- 4Department of Biology, College of Arts and Science, Khalifa University, Abu Dhabi, United Arab Emirates
- 5Department of Comparative Medicine, King Faisal Specialist Hospital and Research Centre, Riyadh, Saudi Arabia
The recent COVID-19 pandemic led to many drastic changes in not only society, law, economics, but also in science and medicine, marking for the first time when drug regulatory authorities cleared for use mRNA-based vaccines in the fight against this outbreak. However, while indeed representing a novel application of such technology in the context of vaccination medicine, introducing RNA into cells to produce resultant molecules (proteins, antibodies, etc.) is not a novel principle. It has been common practice to introduce/inject mRNA into oocytes and embryos to inhibit, induce, and identify several factors in a research context, while such aspects have also been proposed as potential therapeutic and diagnostic applications to combat infertility in humans. Herein, we describe key areas where mRNA-based platforms have thus far represented potential areas of clinical applications, describing the advantages and limitations of such applications. Finally, we also discuss how recent advances in mRNA-based platforms, driven by the recent pandemic, may stand to benefit the treatment of infertility in humans. We also present brief future directions as to how we could utilise recent and current advancements to enhance RNA therapeutics within reproductive biology, specifically with relation to oocyte and embryo delivery.
Introduction
mRNA and its therapeutic potential
Messenger RNA (mRNA)-based therapies revolve around the concept of translating exogenous mRNA into functional proteins. Exogenous mRNAs are synthesized by in vitro transcription, and a cap analogue is attached to their 5′end for cellular recognition. Since mRNA is largely unstable, targeted delivery requires a form of delivery vehicle which encapsulates the mRNA, such as lipid nanoparticles (LNPs), polyplexes and polymeric nanoparticles, lipopolyplexes (LPPs), and cationic polypeptides. In 1990, the first trial to introduce exogenous mRNA successfully was performed by Wolff et al. (1990), while Martinon et al. (1993) used the concept of transcript mRNA of influenza nucleoprotein to produce a vaccine. The mRNA was encapsulated into liposomes and injected into mice, observing the production of virus-specific cytotoxic T lymphocytes. However, the first clinical application of such an approach was the utilisation of an mRNA-based strategy as a novel rabies vaccine in 2013 (NCT02241135), successfully yielding a functional antibody response targeting viral rabies antigens (Alberer et al., 2017). The most recent example of such technology was also observed in 2020, with the advent of mRNA-based vaccine against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (Kim, 2022). Three such vaccines, Pfizer-BioNTech (BNT162b2), Moderna (mRNA-1273), and CureVac101–103 were the fastest vaccines to be developed in medical history, with the Pfizer-BioNTech iteration being the first vaccine to be approved by the FDA for commercialization and use in children 5–11 years old (Yaqinuddin et al., 2021; Fang et al., 2022), with subsequent vaccines also being approved (albeit with some concerns) (Kashir et al., 2022a). Indeed, numerous other applications are also attributed to mRNA platforms within Biomedicine (Table 1).
The causative factor underlying the global coronavirus disease-19 (COVID-19) pandemic was of course sever acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which enters host cells via the surface S protein - comprising S1 and S2. S1 consists of the receptor-binding domain (RBD), while S2 oversees viral-cell membrane fusion and cellular entry through its cognate receptor - the angiotensin-converting enzyme 2 (ACE2) receptor, which seems to be the case with all current variants of the virus (Kashir et al., 2021; Shafqat et al., 2022). The main antigen target in the case of the COVID vaccine was the S antigen, designed following a 2P mutation, and S1/S2 cleavage site, strategies. In the 2P mutation strategy, two amino acids at the top of the helical position of the S2 subunit center were substituted with prolines (K986P and V987P), to enhance the stability of S protein (Corbett et al., 2020; Wrapp et al., 2020; Zhang et al., 2020). For the S1/S2 cleavage site strategy, part of Q677TNSPRRARYSV687 sequence in wild-type SARS-CoV-2 protein S was deleted to Q677TILRYSV683, causing a change in the amino acid sequence from (RRAR to GGSG), preventing S protein degradation in the host cell. It is cleaved into two subunits, S1 and S2 by the enzyme, Furin, and transmembrane serine protease 2 (TMPRSS2) (Hoffmann et al., 2020a; Hoffmann et al., 2020b). The role of these subunits is to interact with cellular angiotensin-converting enzyme 2 (ACE2), mediating viral fusion to the host cell membrane, resulting in post-fusion confirmation (Fang et al., 2022).
Due to its large molecular weight (104–106 Da), negative charge, and proneness to degradation by nucleases, mRNA cannot pass through the phospholipid bilayer membrane of the host cell. Thus, delivery vesicles such as nano-scale vesicle were developed, composed of structures such as ionizable lipids, helper phospholipids, cholesterol, and polyethylene glycosylated (PEGylated) lipids. These components facilitate the endosomal escape of mRNA, determine the specificity of target organs, and reduce aggregation (Malone et al., 1989; Kauffman et al., 2015; Chen et al., 2016; Cheng & Lee, 2016; Cullis & Hope, 2017; Ball et al., 2018; Sabnis et al., 2018; Hassett et al., 2019; Lokugamage et al., 2019; Buschmann et al., 2021; Fang et al., 2022).
Perhaps most importantly, mRNA vaccine strategies (both ssRNA and dsRNA) exhibit ‘self-adjuvant’ effects, while also inducing antibody production, and adequate immune responses (Alexopoulou et al., 2001; Diebold et al., 2004; Verbeke et al., 2019; Linares-Fernández et al., 2020; Xu et al., 2020). However, regardless of the synthesis/delivery strategy, the concept is that the host cell translates the mRNA sequence by cellular ribosomes to express the antigen of interest, which is degraded into small peptides that will be presented by MHC class I to CD8+ cytotoxic T Cells as endogenous antigens. These antigens can be secreted to the extracellular membrane as exogenous antigens and presented by MHC class II to CD4+ T Cells (Cagigi & Loré, 2021), which in turn secrete cytokines and activate B Cells for humoral immune response. Upon infection, the immune system recognizes the S antigen on the surface of the virus and triggers humoral and cellular responses (Fang et al., 2022).
RNA/mRNA processing in oocytes/embryos
RNAs are frequently kept in membrane-free compartments that arise naturally when proteins and/or nucleic acids spontaneously phase separate. Earlier research found many membrane-less compartment types, including P-granules in Caenorhabditis elegans (Brangwynne et al., 2009; Seydoux, 2018) and polar granules in Drosophila (Malone et al., 1989; Trcek & Lehmann, 2019; Bose et al., 2022), that store mRNAs in non-mammalian oocytes (Flemr et al., 2010; Cheng et al., 2022). Mammalian oocytes actively transcribe a large number of mRNAs. During the last stages of oocyte maturation, transcription arrests until after fertilization when the embryonic genome is activated. The mRNAs that the oocyte/embryo can employ to create new proteins during this time are those that have been stored. Hence, for meiosis to transform oocytes into embryos following fertilization, proper maternal mRNA storage is essential. Cheng et al. (2022) collectively suggested that oocytes of at least mammals, maternal mRNAs, and RNA-binding proteins are primarily deposited around mitochondria.
Interactions between membrane-bound and membrane-less compartments play key roles in cellular architecture and function, according to recent studies. Oocytes undergo a lengthy growth phase during which their mitochondria remain largely inactive. In addition to supplying energy for early embryonic development and oocyte meiotic maturation, maternal mitochondria can produce reactive oxygen species (ROS), which can jeopardize the integrity of the genetic material in the mitochondria and the nucleus (Sasaki et al., 2019; Rodríguez-Nuevo et al., 2022). Fewer ROS are produced, which leads to less DNA oxidative damage (Cheng et al., 2022). This helps to maintain the genetic material’s stability in oocytes. Only fully developed oocytes with surrounded nucleoli (SN oocytes) have completely polarized mitochondria, which provide the necessary energy for oocyte meiotic maturation and subsequent embryogenesis (Cheng et al., 2022).
Jansova et al. (2018) used fluorescent oligo (dT) probes to detect the localization of RNA in the oocyte and two-cell embryos, indicating that whereas the cytoplasm contained comparable amounts of polyA RNA, the embryo’s nucleus contained significantly more than the oocyte’s. To visualize the whole cellular transcriptome, Rolling Circle Amplification (RCA) (Larsson et al., 2010; Lee J. H. et al., 2015) was adopted, and results were similar to RNA FISH; the intensity of fluorescence in the nucleus and cytoplasm was equal in the general transcriptome, while in the 2-celled embryo, the RNA intensity was significantly lower in the cytoplasm. Oocytes store and transcribe mRNA in its growth phase until it reaches its full size, which is approximately 3 mm in cattle, where transcription is decreased. The mRNA’s stability has been proven to depend on the poly(A) tail extension at the 3′ end, whereby the tail’s length is correlated with its capability of successful development; the shorter the poly (A) tail, the lower its competence (Wrenzycki et al., 2007).
Understanding the control of mRNA stability in mammalian oocytes and zygotes has lately made significant advances. In reproductive and developmental biology, the precise processes by which maternal mRNAs are degraded during the maternal-to-zygotic transition (MZT) have long been a matter of debate. The zinc finger protein 36-like 2 (ZFP36L2) protein and CNOT6L, a catalytic component of CCR4-NOT deadenylase, are crucial for mRNA decay that occurs in conjunction with oocyte meiotic maturation. All animal species undergo the MZT, the first stage of early development (Sha et al., 2020), during which maternal gene transcripts are degraded, and the zygotic genome activated. The dynamics of such mRNA and the mechanisms that control the stepwise maternal mRNA clearance during MZT in humans are still unknown, despite such findings in model animals of lower-level species.
Genetic and high-throughput sequencing studies on model systems, including Drosophila, zebrafish, and Xenopus, have shown that the elimination of maternal transcripts is accomplished by two sequential pathways. The first is entirely mediated by maternal factors accumulated in the mature oocytes and is referred to as maternal (M)-decay; the second depends on de novo zygotic transcription products after fertilization and is referred to as zygotic (Z)-decay (Sha et al., 2020). Maternal (M)-decay, entirely mediated by maternal factors accumulated in the mature oocytes, is the first pathway, while zygotic (Z)-decay, which depends on de novo zygotic transcription products after fertilization, is the second.
The oocyte-specific adaptor protein of CCR4-NOT, B Cell translocation gene-4 (BTG4), was discovered to be an MZT-licensing factor in mice that mediated mRNA clearance before ZGA9-11 (Morgan et al., 2017; Horvat et al., 2018). Terminal uridine transferase-4/(Tut4/Tut7) and potentially additional early zygotic genes encoding unidentified mRNA destabilizers are transcriptionally activated by the maternal transcriptional coactivator YAP1 and its co-transcription factor TEAD4 (Yagi et al., 2007; Yu et al., 2016; Morgan et al., 2017; Chang et al., 2018). These processes are also important elements of the murine Z-decay system (Sha et al., 2020).
Thus, the RNA landscape in oocytes and embryos is extremely dynamic, and understanding such mechanisms would further allow the development of specific targets using such strategies to be used either therapeutically or diagnostically in the context of specific fertility applications within the clinic.
mRNA technology for oocyte activation
PLCζ/oocyte activation therapeutics
At fertilization, the pivotal signal for oocyte activation in every animal species studied to date is a rise in intracellular calcium concentration (Ca2+). In mammalian oocytes, the initial Ca2+ rise is followed by repetitive Ca2+ transients or ‘oscillations’, triggered by a soluble factor introduced into the ooplasm at gamete fusion by the fertilizing sperm. These oscillations are also observed following intracytoplasmic sperm injection (ICSI; direct injection of a single sperm into the ooplasm) in human and mouse oocytes (Kashir et al., 2010; Kashir et al., 2012a; Nomikos et al., 2017; Kashir et al., 2018). The sperm-specific phospholipase C (PLC), PLCzeta (PLCζ) exhibits the expected properties of the oocyte activating factor (Cox et al., 2002; Saunders et al., 2002), and is considered the strongest candidate for the sperm factor, although several other candidates have also been proposed but without much further evidence or independent support (Parrington et al., 1996; Sette et al., 1997; Sette et al., 1998; Sette et al., 2002; Wu et al., 2007; Kashir et al., 2013b; Aarabi et al., 2014; Nomikos et al., 2015a; Kashir et al., 2015). Interestingly, injection of recombinant mouse and human PLCζ RNA into mouse oocytes not only evoked Ca2+ oscillations similar to those induced by sperm, but also promoted subsequent embryo development until the blastocyst stage (Cox et al., 2002; Saunders et al., 2002; Rogers et al., 2004), with a similar response elicited following by microinjection of recombinant mouse PLCζ protein into mouse oocytes (Kouchi et al., 2004; Yoda et al., 2004).
The clinical application of ICSI has been of major importance in human-assisted reproductive medicine in the treatment of male infertility. However, in a small percentage (2%–3%) of such patients, oocytes fail to activate following ICSI and this appears to be due to a deficiency in oocyte activation, specifically due to sperm defects in PLCζ (either reduced/absent levels or abnormal localisation), in even sperm with normal morphology (Yoon et al., 2008; Heytens et al., 2009; Kashir et al., 2014; Saleh et al., 2020). One circumstance where fertilization following ICSI appears to be severely diminished is in globozoospermia, a rare disorder affecting ∼0.1% of infertile men and characterized by round-headed, acrosome-less sperm cells (Liu et al., 1995; Heindryckx et al., 2005; Dam et al., 2006; Fesahat et al., 2019). Such infertile, males also tend to exhibit mutations in the PLCζ gene (Heytens et al., 2009; Kashir et al., 2011b), with numerous such mutations now identified multiple independent studies and correlated with oocyte activation failure in humans (Abdulsamad et al., 2023; Jones et al., 2022; Kashir, 2020). Indeed, PLCζ also seems to be an important indicator of sperm health, correlating with all examined sperm parameters used within the clinic (Kashir et al., 2010; Kashir et al., 2011a; Kashir et al., 2011b; Kashir et al., 2012b; Kashir et al., 2012c; Kashir et al., 2012d; Kashir et al., 2012e; Ramadan et al., 2012; Yelumalai et al., 2012; Kashir et al., 2013a; Nomikos et al., 2013a; Kashir et al., 2013b; Kashir et al., 2014; Yelumalai et al., 2015; Kashir et al., 2016; Kashir, 2020; Kashir et al., 2020; Kashir et al., 2022b; Kashir et al., 2023).
Most interestingly, co-injection of such sperm with PLCζ mRNA in mouse oocytes was able to ‘rescue’ the induction of Ca2+ oscillations, enabling oocyte activation and fertilisation to complete (Yoon et al., 2008; Heytens et al., 2009), the idea of course being that the oocytes active transcriptional machinery converts the RNA to protein, leading to activity. Conversely, injecting mutant, inactive, PLCζ RNA in mouse oocytes failed to elicit suitable Ca2+ release, resulting in failed activation in mouse oocytes (reviewed by Kashir, 2020). Collectively, such studies suggest that recombinant PLCζ injection represents perhaps an immensely important therapeutic avenue to rescue cases of ICSI failure (Figure 1). Indeed, this has predominantly been pursued in the form of protein injection (Swann et al., 2012; Nomikos et al., 2013b; Nomikos et al., 2014; Nomikos et al., 2015b; Sanusi et al., 2015), driven predominantly by problems associated with RNA injection into oocytes (discussed later herein). However, this is not to say that injection of such protein is not challenging. Indeed, numerous issues also persist in such applications (reviewed further by Kashir, 2020) leaving some room for improved RNA technology-driven injection of PLCζ.
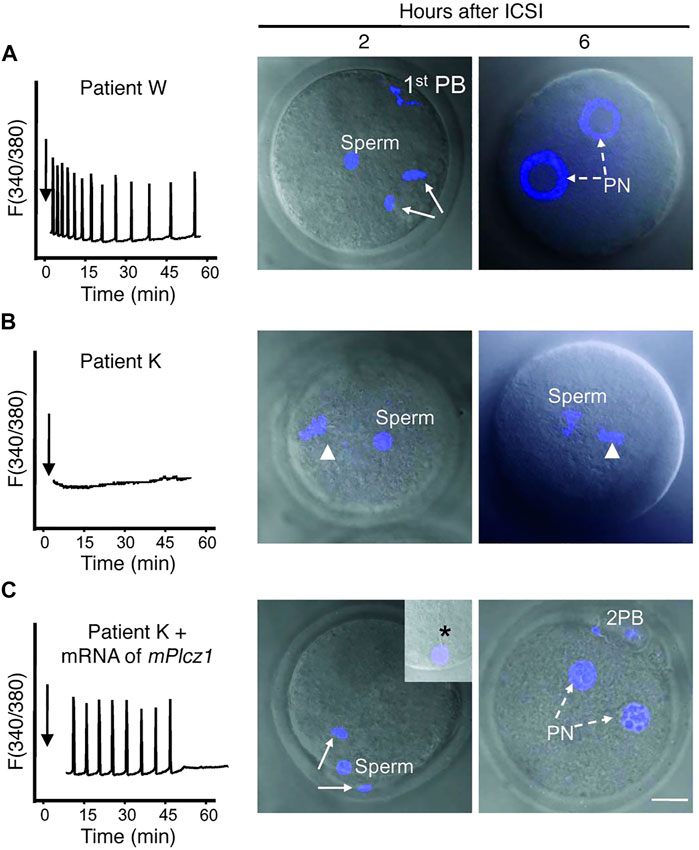
FIGURE 1. Co-injection of mouse PLCζ mRNA alongside infertile humans sperm in mouse oocytes can rescue Ca2+ oscillatory ability. (A) Injection of fertile sperm into mouse oocytes exhibited normal Ca2+ release patterns and oocyte activation and pronuclear (PN) formation (broken arrows). (B) Injection of sperm from an infertile patient (unable to result in fertilisation) was unable to initiate Ca2+ release and failed to induce oocyte activation (arrowhead denotes MII chromatin). (C) However, co-injection of sperm from the same patient alongside mouse PLCζ mRNA initiated Ca2+ oscillations comparable to fertile controls, and enabled resumption of meiosis and PN formation. 1st PB: first polar body; 2 PB: second polar body. Asterisk in inset points to the persistence of the human sperm tail in mouse eggs. Scale bar: 10 μm. Figure adapted from Yoon et al. (2008) with permission.
Another approach could be to target aspects of oocyte activation involved downstream of the action of PLCζ-induced Ca2+ release–specifically factors involved in maintaining cell cycle arrest. A major function of oocyte activation (via Ca2+ release) is to alleviate arrest through the proteolysis of cyclin B1 by ubiquitin or proteasome activation (Miyazaki and Ito, 2006; Kashir et al., 2022b). Ca2+–calmodulin interactions then further repetitively activates calmodulin-dependent kinase II (CaMKII) coincident with each Ca2+ peak during fertilization (at least in mouse oocytes), polyubiquitinating cyclin B1 by the anaphase promoting complex/cyclosome (APC/C), a E3 ubiquitin ligase (Swann and Lai, 2016). This entire pathway is inhibited by cytostatic factor (CSF) (Hyslop et al., 2004; Jones, 2004; Jones, 2005; Miyazaki and Ito, 2006), which is inhibited by CaMKII (Hyslop et al., 2004). Finally, Ca2+ oscillations also contribute to pronuclear formation by reducing mitogen-associated protein kinase activity (Ducibella et al., 2002; Miyazaki and Ito, 2006).
Ca2+ also activates protein kinase C (PKC) to phosphorylate myristoylated alanine-rich C kinase substrate (MARCKS), causing its disassociation from F-actin and leading to actin breakdown in the oocyte cortex, facilitating cortical granule exocytosis. Application of protein synthesis or kinase inhibitors to block the synthesis of cyclin B or CDK1 activity respectively (Heindryckx et al., 2010; Kashir & Swann, 2018), and targeting Emi2 activity (inhibiting cyclin B activity and MPF activity) (Suzuki et al., 2010; Lee K. et al., 2015) are methods that show promise in a supplementary role to Ca2+ (Ducibella and Fissore, 2008; Heindryckx et al., 2010; Kashir and Swann, 2018). To this degree, perhaps it would be worthwhile to consider RNA-mediated therapeutic targeting of such factors/processes as an additional/alternative measure to PLCζ activity.
Ca2+ imaging diagnostics
Ca2+ imaging is important in oocytes to confirm Ca2+ oscillations and oocyte activation. Multiple tools have been used in the past to image changes in intracellular Ca2+, including the photoprotein aequorin in medaka fish, Ca2+ sensitive fluorescent dyes such as Fura-2 in sea urchin and hamsters, and calcium green-1-dextran in Xenopus (Derrick et al., 2016). The mouse oocyte calcium analysis (MOCA) test quantitatively measures free cytosolic Ca2+ spikes in the oocyte using fluorescent probes, and analyzed based on the frequency of Ca2+spikes and classified into 4 categories, similar to the mouse oocyte activation test (MOAT), which is used for more severe abnormalities in Ca2+ release (Heindryckx et al., 2005; Vanden Meerschaut et al., 2013; Ferrer-Buitrago et al., 2018; Cardona Barberán et al., 2020). On the level of neuronal cells, other kinds of dyes have been used, including Oregon Green BAPTA (OGB)-1 or fluo-4 (Grienberger and Konnerth, 2012; Oh et al., 2019).
However, such methods hold major ethical concerns, with sensitivities of oocytes to Ca2+ stimuli perhaps masking specific outcomes Cardona Barberán et al., 2020 Kashir, 2020). If such concerns were to be alleviated, however, there are also other concerns pertaining to Ca2+ imaging using fluorescents dyes–namely, that the constant imaging and light exposure exert lethality upon the developing embryo and are thus not used in clinical human IVF and ICSI (Swann et al., 2012). Furthermore, the delivery method of some dyes involves cell permeabilization by a whole-cell patch clamp or with an acute bulk loading protocol that may damage the cell (Oh et al., 2019).
A proposed alternative are genetically encoded calcium indicators (GECIs), which bind to Ca2+ ions, based on the Ca2+-binding with calmodulin/calmodulin-binding proteins, emitting fluorescence based upon fluorescence resonance energy transfer (FRET), which use a combined form of two fluorescent proteins. Attachment of Ca2+ reduces the distance between two proteins to <10 nm, facilitating energy transfer between donor and acceptor, and thus fluorescence emission (Jares-Erijman & Jovin, 2003; Oh et al., 2019). Single-fluorophore based GECI methods are mediated through conformational changes mostly through calmodulin (CaM) and induced chromophore deprotonation leading to elevated fluorescence emission (Nakamura et al., 1999; Akerboom et al., 2009; Oh et al., 2019).
Such methods were have been further modified to examine Ca2+ release specifically from acidic organelles, as is the case in oocytes (via the endoplasmic reticulum; ER)—termed genetically encoded Ca2+ indicators for optical imaging (GECOs) (Suzuki et al., 2016), which would potentially minimise light-induced damage due to exposure to shorter wavelengths of light, and minimization of reactive oxygen species (ROS) generation (which are generally harmful for cells in large quantities) (Nasr-Esfahani et al., 1990; Squirrell et al., 1999). Satouh et al. (2017) utilized such GECOs, via injection of RNA encoding for various GECOs based upon CaM, to successfully image Ca2+ release dynamics without interfering with the efficacy of embryogenesis or birth rates of pups in comparison with controls, whilst also being able to visualize specific Ca2+-dependent events such as cortical granule exocytosis. Perhaps such methods would be preferable to using Ca2+ dyes, but would of course be subject to limitations associated with RNA use as discussed later. Another aspect to consider is that some GECOs may interfere with Ca2+ signalling pathways and it may alter Ca2+ oscillations in the oocyte. However, Satouh et al. (2017) and others using similar applications (Cappa et al., 2018; Morita et al., 2021) did not seem to experience such drawbacks.
Embryogenic targets
During mammalian oocyte maturation, the female genome is transcriptionally and translationally active, generating a large amount of maternal proteins (De Leon et al., 1983) which are required due to the arrest of transcription prior to ovulation until the 2-cell stage. This MZT process heavily requires large stores of such maternal components to initiate development and activate the embryonic genome (Tadros and Lipshitz, 2009; Li et al., 2013; Zhang et al., 2022). Microarray and proteomic analyses indicate that maternal effect genes affect multiple processes ranging from pronuclear formation, embryogenic cell divisions and gene transcription (Tong et al., 2000; Payer et al., 2003; Tang et al., 2007; Li et al., 2008; Philipps et al., 2008; Kashir et al., 2012b; Li et al., 2013).
The initiation of embryonic genome transcription, also known as zygotic genome activation (ZGA) varies among species, ranging from the 1-cell mouse zygote, the 4-8 cell stage in humans and pigs, to the 8–16 cell stage in cows and sheep (Davis, 1985; Braude et al., 1988; Crosby et al., 1988; Frei et al., 1989; Latham et al., 1991; Ram and Schultz, 1993; Bouniol et al., 1995; Li et al., 2013). Large-scale global gene expression profiles seem significantly dynamic, occuring in ‘waves’ during the MZT and preimplantation development in various mammals, during which maternal transcripts are degraded and zygotic genes are activated (Hamatani et al., 2004; Wang et al., 2004; Zeng et al., 2004; Sirard et al., 2005; Whitworth et al., 2005; Zeng and Schultz, 2005; Misirlioglu et al., 2006; Vallée et al., 2008; Li et al., 2013) (Figure 2). For successful embryogenesis, maternal micro RNAs (miRNAs) require degradation concurrent to de novo miRNA synthesis at the 2-cell stage (Giraldez et al., 2006; Tang et al., 2007; Bushati et al., 2008; Lund et al., 2009; Ma et al., 2010; Suh et al., 2010), which may be regulated by various short interfering RNA (siRNA) pathways (Tam et al., 2008; Watanabe et al., 2008; Ma et al., 2010).
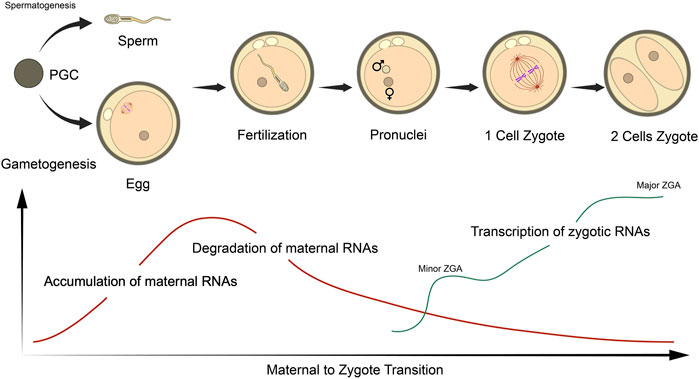
FIGURE 2. Schematic representation of the maternal to zygotic (MZT) and zygotic genome activation (ZGA) in relation to RNA status in mouse embryos. The MZT initiates following sperm/oocyte fusion at fertilization, which undergoes the various stages of embryogenesis. Oocytes accumulate a large pool of maternal RNA throughout ovulation that are essential for these processes. Post-fertilization, maternal RNAs are gradually degraded, while transcription of embryonic transcripts are initiated at the late zygote stage (minor ZGA) and robustly activated at 2- and 4-cell stages (major ZGA). Figure adapted from Li et al. (2013) with permission.
During the MZT, maternal mRNAs are translated concurrent to specific developmental events that occur before ZGA, dependent upon cis-regulatory elements within transcripts. Such transcripts seem associated with cellular homeostasis and protein biosynthesis (Potireddy et al., 2006; Li et al., 2013). ZGA is also crucial for embryogenesis regulated by multiple regulatory mechanisms involving maternal effect genes, chromatin remodelling and DNA replication (Schultz, 1993; Minami et al., 2007; Li et al., 2013). To this degree, the control of mRNA degradation/translation status is major regulatory step, maintained both spatially and temporally, with specific regulatory factors exerting a critical role during early embryogenesis. Thus, such factors may also serve as important potential targets for RNA-based therapies (knockdown/expression as appropriate) (Teixeira and Lehmann, 2019; Alhajeri et al., 2022; Zhang et al., 2022). Further to maternal factors, Yuan et al. (2023) reported that the paternal genome also exerts a significant influence during maternal RNA degradation (MRD) and ZGA in human early embryos, and mused whether the infertility of some patients may be attributable to defects of paternal contributors of human ZGA.
Some factors that could potentially represent targets for mRNA-based interventions include October 4 (also known as POU5F1), a transcription factor in human embryos, and expressed as maternal transcript and protein in mouse oocytes, linked to cellular pluripotency (Tan et al., 2013; Wu and Schöler, 2014; Cui et al., 2019). Zar1/Zar2 play a critical rule in oocyte meiotic maturation, as well as a major role in inducing 2-cell stage arrest, bind mRNAs, and regulate the stability of the maternal transcriptome and MZT, and trigger mRNA clearance during MZT by interacting with other RNA-binding proteins (Rong et al., 2019). Mater is another essential protein expressed exclusively in oocyte cytoplasms but exert significant effect throughout embryogenesis, remaining present throughout the late blastocyst stages (Tong et al., 2000). Finally, another major player, Nanog, is highly expressed and localized to epiblast, deficiencies in which underlie a failure in appropriate implantation. Importantly, Nanog interacts with numerous other factors (such as CDX2, a Mediator Complex Protein–MED, and Oct4) to regulate status of various RNA transcripts and overall embryonic health (Wu and Schöler, 2014; Cui et al., 2019) (more factors exhaustively reviewed by (Jiang et al., 2023).
All such factors underlying these essential events could represent therapeutic targets for embryos, targeting as appropriate to regulates key events during embryogenesis to perhaps enhance chances of successful pregnancy within the clinic. Another more recent development, however, also would perhaps allow for effective diagnostic analysis of such factors, both in terms of quantification and localisation, within embryos. Jansova et al. (2021) developed a technique further to previous methods (Tanenbaum et al., 2014), using a combination of RNA-FISH and the puromycilation proximity ligation assay, to examine localized translation of non-coding and mRNAs in mouse oocytes and embryos, establishing simultaneous visualization of mRNA and in situ translation at the subcellular level, allowing quantitative spatio-temporal analysis. Using such methods, analyses of such RNA also represent significant diagnostic targets.
Issues associated with RNA-therapies/diagnostics in oocytes and embryos
mRNA therapeutics have given researchers great hope in combating widespread incurable diseases. However, while such technology is associated with certain advantages, such new mRNA technology also exhibit significant drawbacks such as mRNA efficiency, safety, stability, immunogenicity, and enhancement of delivery systems (Table 2; (Almalki et al., 2021; Liu et al., 2021; Pardi et al., 2018; Polack et al., 2020; Sahin et al., 2020; Weng et al., 2020; Zhang et al., 2019). It must be considered, however, that numerous issues pertaining to RNA delivery in the body may not necessarily apply to direct delivery in a much smaller oocyte cell–such as thermodynamic stability. Another example is that while injection of large volumes of RNA solution into the body will result in several hundred thousand–fold dilutions, this issue will not be as profound for oocyte delivery (Guo et al., 2012).
mRNA is a very large unstable molecule, prone to degradation by nucleases (Weng et al., 2020). Indeed, the half-life of mRNA transcribed in vitro is a crucial factor hindering application of mRNA-based therapeutics, while natural RNA is extremely sensitive to RNase degradation in the body or within serum (Guo et al., 2012). Rapid progress in recent years have developed chemical base modifications; phosphate linkage modifications; alteration of the 2′carbon (Watts et al., 2008; Singh et al., 2010); use of polycarbamate nucleic acids (Madhuri and Kumar, 2010), locked/bridged nucleic acids (Mathé and Périgaud, 2008); and 5′- and 3′-end capping (Jemielity et al., 2003; Patra and Richert, 2009; Li and Kiledjian, 2010; Ziemniak et al., 2013; Weng et al., 2020), all serving to increase RNase resistance in vitro and in vivo. However, such modifications may further affect folding and functional properties of the RNA (some more than others) (Hong et al., 2010; Liu et al., 2011; Guo et al., 2012; Weng et al., 2020).
Other chemical modifications include modifying the coding region or the poly(A) tail - plays an important role in regulating the stability and efficiency of the translation of mRNA in union with the 5′cap, the entry site of the internal ribosome, and other factors (Gallie, 1991). Modification of the poly(A) tail could be used to optimize efficiency of mRNA use (Sahin et al., 2014), whereby replacing rare codons with synonymous but frequent codons can improve the translational yield (Gustafsson et al., 2004) via reuse of the same tRNA accelerated translation which occurs due to amino-acylation of tRNAs in the locality of the ribosomes (Cannarozzi et al., 2010). However, a limitation of this method is that each RNA preparation contains a mixture of RNA species that differ in the length of the poly(A) tail (Sahin et al., 2014).
In addition to genetic diseases and cancer, uncontrolled mRNA expression can also result from exposure to environmental toxins or drugs that disrupt normal gene expression patterns. For instance, exposure to heavy metals such as lead can interfere with mRNA processing and transport, leading to an altered mRNA expression pattern. Similarly, certain drugs can affect mRNA synthesis, processing, and stability, leading to unintended consequences for protein production. Another potential drawback suggests that external mRNA introduction could elicit an immunogenic response via Toll-like receptors (Weng et al., 2020). However, such potential issues would perhaps not apply to direct oocyte injection of RNA transcripts.
Most problematic, however, is that data increasingly suggest that mouse zygotes possess a retrotransposon-encoded reverse transcriptase activity, triggered a few hours after fertilization and lasting up to at least the two-cell stage, with products absent in zygotes treated with reverse transcriptase inhibitors (Pittoggi et al., 2003; Sciamanna et al., 2011). This ‘reverse transcription wave’ could generate cDNA products, capable of retention and possibly integration, in zygotic pronuclei and embryonic nuclei. Indeed, it would seem that such activity may even be an essential process driving murine preimplantation development (Sciamanna et al., 2011; Kohlrausch et al., 2022). Indeed, telomere reverse transcriptase (Tert) is highly expressed in oocytes, although this decreased with reproductive age (Blackburn, 2001; Yamada-Fukunaga et al., 2013). Furthermore, expression of long interspersed elements 1 (LINE-1 or L1) - the most common autonomous retrotransposons in humans–is required by mouse embryos alongside activity of endogenous reverse transcriptase for embryogenic development to occur (Beraldi et al., 2006; Kohlrausch et al., 2022).
Future directions
Rather astoundingly, Ostermeier et al. (2004) revealed that sperm-based mRNAs are transferred to the oocyte. The mRNAs that gain access are degraded, but they play a role in the zygote: for example, clusterin has several roles including, but not limited to, enhancing fertility rate, transport of lipids, and controlling apoptosis. The functions listed, amongst others, are crucial in the early zygote and embryonic development, but not in the oocyte. Yet, as part of the complex paternal contribution, spermatozoa mRNA also supplies vital genomic organelle (the centriole), and male-specific proteomic components (Krawetz, 2005).
Considering that sperm is already contributing an RNA-load to the oocyte/embryo, it may be also worth considering using the fertilising sperm as a delivery vehicle to delivery therapeutic/diagnostic RNA. Indeed, Barkalina et al. (2014) showed that spherical mesoporous silica nanoparticles (MSNPs) with hexagonal pore symmetry, loaded common types of cargo (nucleic acids/protein), could form strong associations with porcine sperm following in vitro incubation without exerting negative effect upon sperm health (Figure 3), which was preserved following introduction of a cell-penetrating peptide (C105Y) (Barkalina et al., 2015). This was also further applied to mammalian cell (HEK2983T)-derived exosomes, which interacted with boar sperm without affecting parameters of sperm function (Vilanova-Perez et al., 2020) indicating that RNA delivery using such methods could potentially present a less invasive method of introducing RNA to oocytes without injection.
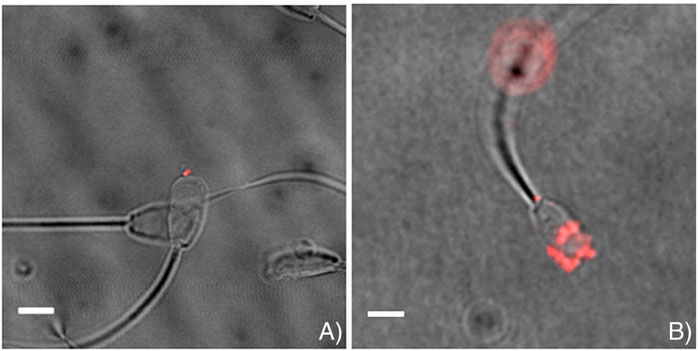
FIGURE 3. Representative images indicating successful association of loaded mesoporous silica nanoparticles (MSNPs) with sperm. MSNPs were loaded with (A) Lamin A/C siRNA, and (B) mCherry fluorescent protein. Scalebar = 5 μm. Figure adapted from (Barkalina et al., 2014) with permission.
Indeed, numerous other methods also exist to compartmentalize RNA with various encapsulations, including liposome complexes/nanoparticles (Pollard et al., 2013; Pardi et al., 2015; Pardi et al., 2019; Liu et al., 2021; Liu et al., 2021), lipid and other materials-based nanoparticles (Wang et al., 2020; Bahmani et al., 2021; Swetha et al., 2023), and various polymer-based strategies (Zhao et al., 2016; Yang et al., 2023). Another approach worth investigating further is the use of hydrogels, which can be easily modified engineered to deliver RNA in a specific and controlled spatiotemporal manner (Zhong et al., 2023). Such approaches involve a relatively easy mechanism of loading for naked/single stranded RNA (Zhong et al., 2023), and may perhaps minimise potential reverse transcriptase activity. However, the efficacy of such a system needs further evaluation in the context of gametes/embryos.
Conclusion
The use of mRNA therapeutics in the context of specific diseases has matured rapidly in recent years, accelerated further by the devastating COVID pandemic, maturing into a range of improved modifications and delivery systems that have proven to be extremely effective in generating effective responses against specific viral infections. Such strategies present significant promise for other non-viral infectious diseases, as well as numerous types of cancers.
Herein, we discuss another potential area wherein mRNA therapeutics could represent a significantly powerful tool to perhaps enhance quality and success of fertility treatments in vitro–specifically within the context of delivery and expression within oocytes and embryos (Figure 4). Indeed, RNA has been traditionally a powerful tool in an extensive repertoire of methods used to study reproductive biology. However, such applications have been traditionally bereft with limitations and safety considerations, which have prevented their application in a clinical format both as a therapeutic and potential diagnostic. As we discuss, however, given the recent advances and improvements in mRNA delivery, stability, and efficiency, perhaps it is time to reconsider.
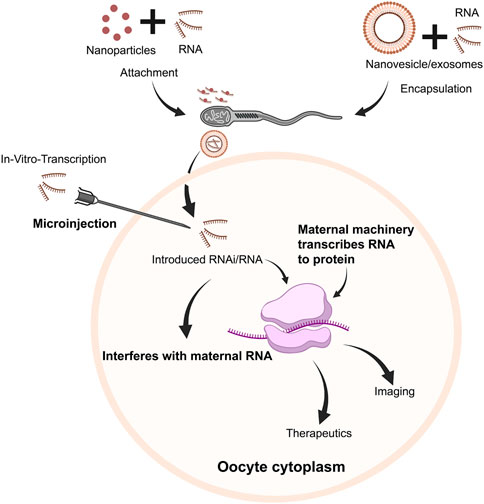
FIGURE 4. Schematic overview of potential pathways of RNA use within oocytes/embryos. Following in vitro transcription, RNA transcripts (mRNA/RNAi, etc.) could be injected into oocytes and embryos. Alternatively, RNA transcripts could either be attached with nanoparticles, or encapsulated in nanovesicles/exosomes and attached/associated with sperm, which could be delivered to the oocyte along with RNA cargo. RNAi could downregulate specific pathways to regulate embryogenesis, targeting specific genes/transcripts. Concurrently, mRNA would be translated to protein, serving therapeutic or diagnostic purposes.
Author contributions
JK and HH were responsible for the manuscript scope and design, along with WB, HA, SMA-Q, RYE, RTE, AA, SA, and ZD who contributed to the literature search and information collection. All authors contributed to the article and approved the submitted version.
Funding
JK is supported by a faculty startup grant (FSU-2023-15) and HH is supported by a faculty startup grant (FSU-2022-002) awarded by Khalifa University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aarabi, M., Balakier, H., Bashar, S., Moskovtsev, S. I., Sutovsky, P., Librach, C. L., et al. (2014). Sperm-derived WW domain-binding protein, PAWP, elicits calcium oscillations and oocyte activation in humans and mice. Faseb J. 28 (10), 4434–4440. doi:10.1096/fj.14-256495
Abdulsamad, H. M. R., Murtaza, Z. F., AlMuhairi, H. M., Bafleh, W. S., AlMansoori, S. A., AlQubaisi, S. A., et al. (2023). The therapeutic and diagnostic potential of phospholipase C zeta, oocyte activation, and calcium in treating human infertility. Pharmaceuticals 16 (3), 441. doi:10.3390/ph16030441
Akerboom, J., Rivera, J. D. V., Guilbe, M. M. R., Malavé, E. C. A., Hernandez, H. H., Tian, L., et al. (2009). Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J. Biol. Chem. 284 (10), 6455–6464. doi:10.1074/jbc.M807657200
Alberer, M., Gnad-Vogt, U., Hong, H. S., Mehr, K. T., Backert, L., Finak, G., et al. (2017). Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 390 (10101), 1511–1520. doi:10.1016/s0140-6736(17)31665-3
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413 (6857), 732–738. doi:10.1038/35099560
Alhajeri, M. M., Alkhanjari, R. R., Hodeify, R., Khraibi, A., and Hamdan, H. (2022). Neurotransmitters, neuropeptides and calcium in oocyte maturation and early development. Front. Cell Dev. Biol. 10, 980219. doi:10.3389/fcell.2022.980219
Almalki, W. H., Alghamdi, S., Alzahrani, A., and Zhang, W. (2021). Emerging paradigms in treating cerebral infarction with nanotheranostics: Opportunities and clinical challenges. Drug Discov. Today 26 (3), 826–835. doi:10.1016/j.drudis.2020.12.018
Anttila, V., Saraste, A., Knuuti, J., Jaakkola, P., Hedman, M., Svedlund, S., et al. (2020). Synthetic mRNA encoding VEGF-A in patients undergoing coronary artery bypass grafting: Design of a phase 2a clinical trial. Mol. Ther. Methods Clin. Dev. 18, 464–472. doi:10.1016/j.omtm.2020.05.030
Bahmani, B., Gong, H., Luk, B. T., Haushalter, K. J., DeTeresa, E., Previti, M., et al. (2021). Intratumoral immunotherapy using platelet-cloaked nanoparticles enhances antitumor immunity in solid tumors. Nat. Commun. 12 (1), 1999. doi:10.1038/s41467-021-22311-z
Ball, R. L., Hajj, K. A., Vizelman, J., Bajaj, P., and Whitehead, K. A. (2018). Lipid nanoparticle formulations for enhanced Co-delivery of siRNA and mRNA. Nano Lett. 18 (6), 3814–3822. doi:10.1021/acs.nanolett.8b01101
Bareche, Y., Venet, D., Ignatiadis, M., Aftimos, P., Piccart, M., Rothe, F., et al. (2018). Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 29 (4), 895–902. doi:10.1093/annonc/mdy024
Barkalina, N., Jones, C., Kashir, J., Coote, S., Huang, X., Morrison, R., et al. (2014). Effects of mesoporous silica nanoparticles upon the function of mammalian sperm in vitro. Nanomedicine 10 (4), 859–870. doi:10.1016/j.nano.2013.10.011
Barkalina, N., Jones, C., Townley, H., and Coward, K. (2015). Functionalization of mesoporous silica nanoparticles with a cell-penetrating peptide to target mammalian sperm in vitro. Nanomedicine 10 (10), 1539–1553. doi:10.2217/nnm.14.235
Beraldi, R., Pittoggi, C., Sciamanna, I., Mattei, E., and Spadafora, C. (2006). Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol. Reprod. Dev. 73 (3), 279–287. doi:10.1002/mrd.20423
Blackburn, E. H. (2001). Switching and signaling at the telomere. Cell 106 (6), 661–673. doi:10.1016/s0092-8674(01)00492-5
Bose, M., Lampe, M., Mahamid, J., and Ephrussi, A. (2022). Liquid-to-solid phase transition of oskar ribonucleoprotein granules is essential for their function in Drosophila embryonic development. Cell 185 (8), 1308–1324.e23. doi:10.1016/j.cell.2022.02.022
Bouniol, C., Nguyen, E., and Debey, P. (1995). Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp. Cell Res. 218 (1), 57–62. doi:10.1006/excr.1995.1130
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324 (5935), 1729–1732. doi:10.1126/science.1172046
Braude, P., Bolton, V., and Moore, S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332 (6163), 459–461. doi:10.1038/332459a0
Burris, H. A., Patel, M. R., Cho, D. C., Clarke, J. M., Gutierrez, M., Zaks, T. Z., et al. (2019). A phase I multicenter study to assess the safety, tolerability, and immunogenicity of mRNA-4157 alone in patients with resected solid tumors and in combination with pembrolizumab in patients with unresectable solid tumors. J. Clin. Oncol. 37 (15), 2523. doi:10.1200/JCO.2019.37.15_suppl.2523
Buschmann, M. D., Carrasco, M. J., Alishetty, S., Paige, M., Alameh, M. G., and Weissman, D. (2021). Nanomaterial delivery systems for mRNA vaccines. Vaccines 9 (1), 65. doi:10.3390/vaccines9010065
Bushati, N., Stark, A., Brennecke, J., and Cohen, S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18 (7), 501–506. doi:10.1016/j.cub.2008.02.081
Cagigi, A., and Loré, K. (2021). Immune responses induced by mRNA vaccination in mice, monkeys and humans. Vaccines 9 (1), 61. doi:10.3390/vaccines9010061
Cannarozzi, G., Schraudolph, N. N., Faty, M., von Rohr, P., Friberg, M. T., Roth, A. C., et al. (2010). A role for codon order in translation dynamics. Cell 141 (2), 355–367. doi:10.1016/j.cell.2010.02.036
Cappa, A. I., de Paola, M., Wetten, P., De Blas, G. A., and Michaut, M. A. (2018). Live imaging of cortical granule exocytosis reveals that in vitro matured mouse oocytes are not fully competent to secrete their content. Biol. Open 7 (12), bio031872. doi:10.1242/bio.031872
Cardona Barberán, A., Boel, A., Vanden Meerschaut, F., Stoop, D., and Heindryckx, B. (2020). Diagnosis and treatment of male infertility-related fertilization failure. J. Clin. Med. 9 (12), 3899. doi:10.3390/jcm9123899
Carlsson, L., Clarke, J. C., Yen, C., Gregoire, F., Albery, T., Billger, M., et al. (2018). Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 Week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 9, 330–346. doi:10.1016/j.omtm.2018.04.003
Chang, H., Yeo, J., Kim, J.-g., Kim, H., Lim, J., Lee, M., et al. (2018). Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Mol. Cell 70 (1), 72–82. doi:10.1016/j.molcel.2018.03.004
Chen, S., Tam, Y. Y. C., Lin, P. J. C., Sung, M. M. H., Tam, Y. K., and Cullis, P. R. (2016). Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release 235, 236–244. doi:10.1016/j.jconrel.2016.05.059
Cheng, S., Altmeppen, G., So, C., Welp, L. M., Penir, S., Ruhwedel, T., et al. (2022). Mammalian oocytes store mRNAs in a mitochondria-associated membraneless compartment. Science 378 (6617), eabq4835. doi:10.1126/science.abq4835
Cheng, X., and Lee, R. J. (2016). The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 99, 129–137. doi:10.1016/j.addr.2016.01.022
Corbett, K. S., Flynn, B., Foulds, K. E., Francica, J. R., Boyoglu-Barnum, S., Werner, A. P., et al. (2020). Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 383 (16), 1544–1555. doi:10.1056/NEJMoa2024671
Cox, L. J., Larman, M. G., Saunders, C. M., Hashimoto, K., Swann, K., and Lai, F. A. (2002). Sperm phospholipase Czeta from humans and cynomolgus monkeys triggers Ca2+ oscillations, activation and development of mouse oocytes. Reproduction 124, 611–623. doi:10.1530/rep.0.1240611
Crosby, I. M., Gandolfi, F., and Moor, R. M. (1988). Control of protein synthesis during early cleavage of sheep embryos. J. Reprod. Fertil. 82 (2), 769–775. doi:10.1530/jrf.0.0820769
Cui, W., Marcho, C., Wang, Y., Degani, R., Golan, M., Tremblay, K. D., et al. (2019). MED20 is essential for early embryogenesis and regulates NANOG expression. Reproduction 157 (3), 215–222. doi:10.1530/rep-18-0508
Cullis, P. R., and Hope, M. J. (2017). Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25 (7), 1467–1475. doi:10.1016/j.ymthe.2017.03.013
Dam, A. H. D. M., Feenstra, I., Westphal, J. R., Ramos, L., van Golde, R. J. T., and Kremer, J. A. M. (2006). Globozoospermia revisited. Hum. Reprod. Update 13 (1), 63–75. doi:10.1093/humupd/dml047
De Leon, V., Johnson, A., and Bachvarova, R. (1983). Half-lives and relative amounts of stored and polysomal ribosomes and poly(A) + RNA in mouse oocytes. Dev. Biol. 98 (2), 400–408. doi:10.1016/0012-1606(83)90369-x
Derrick, C. J., York-Andersen, A. H., and Weil, T. T. (2016). Imaging calcium in Drosophila at egg activation. JoVE 114, e54311. doi:10.3791/54311
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis e Sousa, C. (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303 (5663), 1529–1531. doi:10.1126/science.1093616
Ducibella, T., and Fissore, R. (2008). The roles of Ca2+, downstream protein kinases, and oscillatory signaling in regulating fertilization and the activation of development. Dev. Biol. 315 (2), 257–279. doi:10.1016/j.ydbio.2007.12.012
Ducibella, T., Huneau, D., Angelichio, E., Xu, Z., Schultz, R. M., Kopf, G. S., et al. (2002). Egg-to-Embryo transition is driven by differential responses to Ca2+ oscillation number. Dev. Biol. 250 (2), 280–291. doi:10.1006/dbio.2002.0788
Fang, E., Liu, X., Li, M., Zhang, Z., Song, L., Zhu, B., et al. (2022). Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 7 (1), 94. doi:10.1038/s41392-022-00950-y
Ferrer-Buitrago, M., Dhaenens, L., Lu, Y., Bonte, D., Vanden Meerschaut, F., De Sutter, P., et al. (2018). Human oocyte calcium analysis predicts the response to assisted oocyte activation in patients experiencing fertilization failure after ICSI. Hum. Reprod. 33 (3), 416–425. doi:10.1093/humrep/dex376
Fesahat, F., Henkel, R., and Agarwal, A. (2019). Globozoospermia syndrome: An update. Andrologia 52 (2), e13459. doi:10.1111/and.13459
Flemr, M., Ma, J., Schultz, R. M., and Svoboda, P. (2010). P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol. Reprod. 82 (5), 1008–1017. doi:10.1095/biolreprod.109.082057
Frei, R. E., Schultz, G. A., and Church, R. B. (1989). Qualitative and quantitative changes in protein synthesis occur at the 8-16-cell stage of embryogenesis in the cow. J. Reprod. Fertil. 86 (2), 637–641. doi:10.1530/jrf.0.0860637
Gallie, D. R. (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes. Dev. 5 (11), 2108–2116. doi:10.1101/gad.5.11.2108
Gan, L. M., Lagerström-Fermér, M., Carlsson, L. G., Arfvidsson, C., Egnell, A. C., Rudvik, A., et al. (2019). Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 10 (1), 871. doi:10.1038/s41467-019-08852-4
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312 (5770), 75–79. doi:10.1126/science.1122689
Gote, V., Bolla, P. K., Kommineni, N., Butreddy, A., Nukala, P. K., Palakurthi, S. S., et al. (2023). A comprehensive Review of mRNA vaccines. Int. J. Mol. Sci. 24 (3), 2700. doi:10.3390/ijms24032700
Grienberger, C., and Konnerth, A. (2012). Imaging calcium in neurons. Neuron 73 (5), 862–885. doi:10.1016/j.neuron.2012.02.011
Guo, P., Haque, F., Hallahan, B., Reif, R., and Li, H. (2012). Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid. Ther. 22 (4), 226–245. doi:10.1089/nat.2012.0350
Gustafsson, C., Govindarajan, S., and Minshull, J. (2004). Codon bias and heterologous protein expression. Trends Biotechnol. 22 (7), 346–353. doi:10.1016/j.tibtech.2004.04.006
Hamatani, T., Carter, M. G., Sharov, A. A., and Ko, M. S. (2004). Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6 (1), 117–131. doi:10.1016/s1534-5807(03)00373-3
Hassett, K. J., Benenato, K. E., Jacquinet, E., Lee, A., Woods, A., Yuzhakov, O., et al. (2019). Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. - Nucleic Acids 15, 1–11. doi:10.1016/j.omtn.2019.01.013
Heindryckx, B., De Sutter, P., and Gerris, J. (2010). “Somatic nuclear transfer to in vitro-matured human germinal vesicle oocytes,” in Stem cells in human reproduction: Basic science and therapeutic potential. Editors C Simón, and A Pellicer. 2nd ed. (Boca Raton: CRC Press).
Heindryckx, B., Van der Elst, J., De Sutter, P., and Dhont, M. (2005). Treatment option for sperm- or oocyte-related fertilization failure: Assisted oocyte activation following diagnostic heterologous ICSI. Hum. Reprod. 20 (8), 2237–2241. doi:10.1093/humrep/dei029
Heytens, E., Parrington, J., Coward, K., Young, C., Lambrecht, S., Yoon, S. Y., et al. (2009). Reduced amounts and abnormal forms of phospholipase C zeta (PLCzeta) in spermatozoa from infertile men. Hum. Reprod. 24 (10), 2417–2428. doi:10.1093/humrep/dep207
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78 (4), 779–784. doi:10.1016/j.molcel.2020.04.022
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181 (2), 271–280. doi:10.1016/j.cell.2020.02.052
Hong, J., Huang, Y., Li, J., Yi, F., Zheng, J., Huang, H., et al. (2010). Comprehensive analysis of sequence-specific stability of siRNA. Faseb J. 24 (12), 4844–4855. doi:10.1096/fj.09-142398
Horvat, F., Fulka, H., Jankele, R., Malik, R., Jun, M., Solcova, K., et al. (2018). Role of Cnot6l in maternal mRNA turnover. Life Sci. Alliance 1 (4), e201800084. doi:10.26508/lsa.201800084
Hyslop, L. A., Nixon, V. L., Levasseur, M., Chapman, F., Chiba, K., McDougall, A., et al. (2004). Ca(2+)-promoted cyclin B1 degradation in mouse oocytes requires the establishment of a metaphase arrest. Dev. Biol. 269 (1), 206–219. doi:10.1016/j.ydbio.2004.01.030
Jabulowsky, R. A., Loquai, C., Derhovanessian, E., Mitzel-Rink, H., Utikal, J., Hassel, J., et al. (2018). A first-in-human phase I/II clinical trial assessing novel mRNA-lipoplex nanoparticles encoding shared tumor antigens for immunotherapy of malignant melanoma. Ann. Oncol. 29, viii439. doi:10.1093/annonc/mdy288.109
Jansova, D., Aleshkina, D., Jindrova, A., Iyyappan, R., An, Q., Fan, G., et al. (2021). Single molecule RNA localization and translation in the mammalian oocyte and embryo. J. Mol. Biol. 433 (19), 167166. doi:10.1016/j.jmb.2021.167166
Jansova, D., Tetkova, A., Koncicka, M., Kubelka, M., and Susor, A. (2018). Localization of RNA and translation in the mammalian oocyte and embryo. PLOS ONE 13 (3), e0192544. doi:10.1371/journal.pone.0192544
Jares-Erijman, E. A., and Jovin, T. M. (2003). FRET imaging. Nat. Biotechnol. 21 (11), 1387–1395. doi:10.1038/nbt896
Jemielity, J., Fowler, T., Zuberek, J., Stepinski, J., Lewdorowicz, M., Niedzwiecka, A., et al. (2003). Novel “anti-reverse” cap analogs with superior translational properties. Rna 9 (9), 1108–1122. doi:10.1261/rna.5430403
Jiang, Y., Adhikari, D., Li, C., and Zhou, X. (2023). Spatiotemporal regulation of maternal mRNAs during vertebrate oocyte meiotic maturation. Biol. Rev. 98, 900–930. doi:10.1111/brv.12937
John, S., Yuzhakov, O., Woods, A., Deterling, J., Hassett, K., Shaw, C. A., et al. (2018). Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine 36 (12), 1689–1699. doi:10.1016/j.vaccine.2018.01.029
Jones, C., Meng, X., and Coward, K. (2022). Sperm Factors and Egg Activation: Phospholipase C zeta (PLCZ1) and the clinical diagnosis of oocyte activation deficiency. Reproduction 164 (1), F53–F66. doi:10.1530/REP-21-0458
Jones, K. T. (2005). Mammalian egg activation: From Ca2+ spiking to cell cycle progression. Reproduction 130 (6), 813–823. doi:10.1530/rep.1.00710
Jones, K. T. (2004). Turning it on and off: M-Phase promoting factor during meiotic maturation and fertilization. Mol. Hum. Reprod. 10 (1), 1–5. doi:10.1093/molehr/gah009
Kashir, J., AlKattan, K., and Yaqinuddin, A. (2021). COVID-19: Cross-immunity of viral epitopes may influence severity of infection and immune response. Signal Transduct. Target. Ther. 6 (1), 102. doi:10.1038/s41392-021-00490-x
Kashir, J., Ambia, A. R., Shafqat, A., Sajid, M. R., AlKattan, K., and Yaqinuddin, A. (2022a). Scientific premise for the involvement of neutrophil extracellular traps (NETs) in vaccine-induced thrombotic thrombocytopenia (VITT). J. Leukoc. Biol. 111 (3), 725–734. doi:10.1002/JLB.5COVR0621-320RR
Kashir, J., Buntwal, L., Nomikos, M., Calver, B. L., Stamatiadis, P., Ashley, P., et al. (2016). Antigen unmasking enhances visualization efficacy of the oocyte activation factor, phospholipase C zeta, in mammalian sperm. Mol. Hum. Reprod. 23 (1), 54–67. doi:10.1093/molehr/gaw073
Kashir, J., Deguchi, R., Jones, C., Coward, K., and Stricker, S. A. (2013a). Comparative biology of sperm factors and fertilization-induced calcium signals across the animal kingdom. Mol. Reprod. Dev. 80 (10), 787–815. doi:10.1002/mrd.22222
Kashir, J., Ganesh, D., Jones, C., and Coward, K. (2022b). Oocyte activation deficiency and assisted oocyte activation: Mechanisms, obstacles and prospects for clinical application. Hum. Reprod. Open 2022 (2), hoac003. doi:10.1093/hropen/hoac003
Kashir, J., Heindryckx, B., Jones, C., De Sutter, P., Parrington, J., and Coward, K. (2010). Oocyte activation, phospholipase C zeta and human infertility. Hum. Reprod. Update 16 (6), 690–703. doi:10.1093/humupd/dmq018
Kashir, J., Heynen, A., Jones, C., Durrans, C., Craig, J., Gadea, J., et al. (2011a). Effects of cryopreservation and density-gradient washing on phospholipase C zeta concentrations in human spermatozoa. Reprod. Biomed. Online 23 (2), 263–267. doi:10.1016/j.rbmo.2011.04.006
Kashir, J. (2020). Increasing associations between defects in phospholipase C zeta and conditions of male infertility: Not just ICSI failure? J. Assist. Reprod. Genet. 37 (6), 1273–1293. doi:10.1007/s10815-020-01748-z
Kashir, J., Jones, C., Child, T., Williams, S. A., and Coward, K. (2012a). Viability assessment for artificial gametes: The need for biomarkers of functional competency. Biol. Reproduction 87 (5), 114. doi:10.1095/biolreprod.112.103853
Kashir, J., Jones, C., and Coward, K. (2012b). Calcium oscillations, oocyte activation, and phospholipase C zeta. Adv. Exp. Med. Biol. 740, 1095–1121. doi:10.1007/978-94-007-2888-2_50
Kashir, J., Jones, C., Lee, H. C., Rietdorf, K., Nikiforaki, D., Durrans, C., et al. (2011b). Loss of activity mutations in phospholipase C zeta (PLCζ) abolishes calcium oscillatory ability of human recombinant protein in mouse oocytes. Hum. Reprod. 26 (12), 3372–3387. doi:10.1093/humrep/der336
Kashir, J., Jones, C., Mounce, G., Ramadan, W. M., Lemmon, B., Heindryckx, B., et al. (2013b). Variance in total levels of phospholipase C zeta (PLC-ζ) in human sperm may limit the applicability of quantitative immunofluorescent analysis as a diagnostic indicator of oocyte activation capability. Fertil. Steril. 99 (1), 107–117. doi:10.1016/j.fertnstert.2012.09.001
Kashir, J., Konstantinidis, M., Jones, C., Heindryckx, B., De Sutter, P., Parrington, J., et al. (2012c). Characterization of two heterozygous mutations of the oocyte activation factor phospholipase C zeta (PLCζ) from an infertile man by use of minisequencing of individual sperm and expression in somatic cells. Fertil. Steril. 98 (2), 423–431. doi:10.1016/j.fertnstert.2012.05.002
Kashir, J., Konstantinidis, M., Jones, C., Lemmon, B., Lee, H. C., Hamer, R., et al. (2012d). A maternally inherited autosomal point mutation in human phospholipase C zeta (PLCζ) leads to male infertility. Hum. Reprod. Oxf. Engl. 27 (1), 222–231. doi:10.1093/humrep/der384
Kashir, J., Mistry, B. V., BuSaleh, L., Abu-Dawas, R., Nomikos, M., Ajlan, A., et al. (2020). Phospholipase C zeta profiles are indicative of optimal sperm parameters and fertilisation success in patients undergoing fertility treatment. Andrology 8, 1143–1159. doi:10.1111/andr.12796
Kashir, J., Mistry, B. V., BuSaleh, L., Nomikos, M., Almuqayyil, S., Abu-Dawud, R., et al. (2023). Antigen unmasking is required to clinically assess levels and localisation patterns of phospholipase C zeta in human sperm. Pharmaceuticals 16 (2), 198. doi:10.3390/ph16020198
Kashir, J., Nomikos, M., and Lai, F. A. (2018). Phospholipase C zeta and calcium oscillations at fertilisation: The evidence, applications, and further questions. Adv. Biol. Regul. 67, 148–162. doi:10.1016/j.jbior.2017.10.012
Kashir, J., Nomikos, M., Lai, F. A., and Swann, K. (2014). Sperm-induced Ca2+ release during egg activation in mammals. Biochem. Biophysical Res. Commun. 450 (3), 1204–1211. doi:10.1016/j.bbrc.2014.04.078
Kashir, J., Nomikos, M., Swann, K., and Lai, F. A. (2015). PLCζ or PAWP: Revisiting the putative mammalian sperm factor that triggers egg activation and embryogenesis. Mol. Hum. Reprod. 21 (5), 383–388. doi:10.1093/molehr/gav009
Kashir, J., Sermondade, N., Sifer, C., Oo, S. L., Jones, C., Mounce, G., et al. (2012e). Motile sperm organelle morphology evaluation-selected globozoospermic human sperm with an acrosomal bud exhibits novel patterns and higher levels of phospholipase C zeta. Hum. Reprod. 27 (11), 3150–3160. doi:10.1093/humrep/des312
Kashir, J., and Swann, K. (2018). “Assisted oocyte activation: Current understanding, practice, and future perspectives,” in Textbook of assisted reproductive techniques: Volume 1: Laboratory perspectives. Editors A Weissman, CM Howles, and Z Shoham. 5th ed. (Boca Raton: CRC Press).
Kauffman, K. J., Dorkin, J. R., Yang, J. H., Heartlein, M. W., DeRosa, F., Mir, F. F., et al. (2015). Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 15 (11), 7300–7306. doi:10.1021/acs.nanolett.5b02497
Kim, Y.-K. (2022). RNA therapy: Rich history, various applications and unlimited future prospects. Exp. Mol. Med. 54 (4), 455–465. doi:10.1038/s12276-022-00757-5
Kohlrausch, F. B., Berteli, T. S., Wang, F., Navarro, P. A., and Keefe, D. L. (2022). Control of LINE-1 expression maintains genome integrity in germline and early embryo development. Reprod. Sci. 29 (2), 328–340. doi:10.1007/s43032-021-00461-1
Kouchi, Z., Fukami, K., Shikano, T., Oda, S., Nakamura, Y., Takenawa, T., et al. (2004). Recombinant phospholipase Czeta has high Ca2+ sensitivity and induces Ca2+ oscillations in mouse eggs. J. Biol. Chem. 279 (11), 10408–10412. doi:10.1074/jbc.m313801200
Kranz, L. M., Diken, M., Haas, H., Kreiter, S., Loquai, C., Reuter, K. C., et al. (2016). Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534 (7607), 396–401. doi:10.1038/nature18300
Krawetz, S. A. (2005). Paternal contribution: New insights and future challenges. Nat. Rev. Genet. 6 (8), 633–642. doi:10.1038/nrg1654
Larsson, C., Grundberg, I., Söderberg, O., and Nilsson, M. (2010). In situ detection and genotyping of individual mRNA molecules. Nat. Methods 7 (5), 395–397. doi:10.1038/nmeth.1448
Latham, K. E., Garrels, J. I., Chang, C., and Solter, D. (1991). Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development 112 (4), 921–932. doi:10.1242/dev.112.4.921
Le, T., Sun, C., Chang, J., Zhang, G., and Yin, X. (2022). mRNA vaccine development for emerging animal and zoonotic diseases. Viruses 14 (2), 401. doi:10.3390/v14020401
Lee, J. H., Daugharthy, E. R., Scheiman, J., Kalhor, R., Ferrante, T. C., Terry, R., et al. (2015a). Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10 (3), 442–458. doi:10.1038/nprot.2014.191
Lee, K., Davis, A., Zhang, L., Ryu, J., Spate, L. D., Park, K. W., et al. (2015b). Pig oocyte activation using a Zn²⁺ chelator, TPEN. Theriogenology 84 (6), 1024–1032. doi:10.1016/j.theriogenology.2015.05.036
Li, L., Baibakov, B., and Dean, J. (2008). A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell 15 (3), 416–425. doi:10.1016/j.devcel.2008.07.010
Li, L., Lu, X., and Dean, J. (2013). The maternal to zygotic transition in mammals. Mol. Asp. Med. 34 (5), 919–938. doi:10.1016/j.mam.2013.01.003
Li, Y., and Kiledjian, M. (2010). Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA 1 (2), 253–265. doi:10.1002/wrna.15
Linares-Fernández, S., Lacroix, C., Exposito, J.-Y., and Verrier, B. (2020). Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol. Med. 26 (3), 311–323. doi:10.1016/j.molmed.2019.10.002
Liu, J., Guo, S., Cinier, M., Shlyakhtenko, L. S., Shu, Y., Chen, C., et al. (2011). Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano 5 (1), 237–246. doi:10.1021/nn1024658
Liu, J., Nagy, Z., Joris, H., Tournaye, H., Devroey, P., and Van Steirteghem, A. (1995). Andrology: Successful fertilization and establishment of pregnancies after intracytoplasmic sperm injection in patients with globozoospermia. Hum. Reprod. 10 (3), 626–629. doi:10.1093/oxfordjournals.humrep.a136000
Liu, T., Liang, Y., and Huang, L. (2021). Development and delivery systems of mRNA vaccines. Front. Bioeng. Biotechnol. 9, 718753. doi:10.3389/fbioe.2021.718753
Lokugamage, M. P., Sago, C. D., Gan, Z., Krupczak, B. R., and Dahlman, J. E. (2019). Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv. Mater. 31 (41), 1902251. doi:10.1002/adma.201902251
Lund, E., Liu, M., Hartley, R. S., Sheets, M. D., and Dahlberg, J. E. (2009). Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna 15 (12), 2351–2363. doi:10.1261/rna.1882009
Ma, J., Flemr, M., Stein, P., Berninger, P., Malik, R., Zavolan, M., et al. (2010). MicroRNA activity is suppressed in mouse oocytes. Curr. Biol. 20 (3), 265–270. doi:10.1016/j.cub.2009.12.042
Madhuri, V., and Kumar, V. A. (2010). Design, synthesis and DNA/RNA binding studies of nucleic acids comprising stereoregular and acyclic polycarbamate backbone: Polycarbamate nucleic acids (PCNA). Org. Biomol. Chem. 8 (16), 3734–3741. doi:10.1039/c003405n
Malone, R. W., Felgner, P. L., and Verma, I. M. (1989). Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. 86 (16), 6077–6081. doi:10.1073/pnas.86.16.6077
Martinon, F., Krishnan, S., Lenzen, G., Magné, R., Gomard, E., Guillet, J.-G., et al. (1993). Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 23 (7), 1719–1722. doi:10.1002/eji.1830230749
Mathé, C., and Périgaud, C. (2008). Recent approaches in the synthesis of conformationally restricted nucleoside analogues. Eur. J. Org. Chem. 2008 (9), 1489–1505. doi:10.1002/ejoc.200700946
Minami, N., Suzuki, T., and Tsukamoto, S. (2007). Zygotic gene activation and maternal factors in mammals. J. Reprod. Dev. 53 (4), 707–715. doi:10.1262/jrd.19029
Misirlioglu, M., Page, G. P., Sagirkaya, H., Kaya, A., Parrish, J. J., First, N. L., et al. (2006). Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl. Acad. Sci. U. S. A. 103 (50), 18905–18910. doi:10.1073/pnas.0608247103
Miyazaki, S., and Ito, M. (2006). Calcium signals for egg activation in mammals. J. Pharmacol. Sci. 100 (5), 545–552. doi:10.1254/jphs.cpj06003x
Morgan, M., Much, C., DiGiacomo, M., Azzi, C., Ivanova, I., Vitsios, D. M., et al. (2017). mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 548 (7667), 347–351. doi:10.1038/nature23318
Morita, A., Satouh, Y., Kosako, H., Kobayashi, H., Iwase, A., and Sato, K. (2021). Clathrin-mediated endocytosis is essential for the selective degradation of maternal membrane proteins and preimplantation development. Development 148 (14), dev199461. doi:10.1242/dev.199461
Nakamura, T., Barbara, J. G., Nakamura, K., and Ross, W. N. (1999). Synergistic release of Ca2+ from IP3-sensitive stores evoked by synaptic activation of mGluRs paired with backpropagating action potentials. Neuron 24 (3), 727–737. doi:10.1016/s0896-6273(00)81125-3
Nasr-Esfahani, M. H., Aitken, J. R., and Johnson, M. H. (1990). Hydrogen peroxide levels in mouse oocytes and early cleavage stage embryos developed in vitro or in vivo. Development 109 (2), 501–507. doi:10.1242/dev.109.2.501
Nomikos, M., Kashir, J., and Lai, F. A. (2017). The role and mechanism of action of sperm PLC-zeta in mammalian fertilisation. Biochem. J. 474 (21), 3659–3673. doi:10.1042/bcj20160521
Nomikos, M., Kashir, J., Swann, K., and Lai, F. A. (2013a). Sperm PLCζ: From structure to Ca2+oscillations, egg activation and therapeutic potential. FEBS Lett. 587 (22), 3609–3616. doi:10.1016/j.febslet.2013.10.008
Nomikos, M., Sanders, J. R., Kashir, J., Sanusi, R., Buntwal, L., Love, D., et al. (2015a). Functional disparity between human PAWP and PLCζ in the generation of Ca2+oscillations for oocyte activation. Mol. Hum. Reprod. 21 (9), 702–710. doi:10.1093/molehr/gav034
Nomikos, M., Sanders, J. R., Parthimos, D., Buntwal, L., Calver, B. L., Stamatiadis, P., et al. (2015b). Essential role of the EF-hand domain in targeting sperm phospholipase cζ to membrane phosphatidylinositol 4,5-bisphosphate (PIP2). J. Biol. Chem. 290 (49), 29519–29530. doi:10.1074/jbc.m115.658443
Nomikos, M., Theodoridou, M., Elgmati, K., Parthimos, D., Calver, B. L., Buntwal, L., et al. (2014). Human PLC exhibits superior fertilization potency over mouse PLC in triggering the Ca2+ oscillations required for mammalian oocyte activation. Mol. Hum. Reprod. 20 (6), 489–498. doi:10.1093/molehr/gau011
Nomikos, M., Yu, Y., Elgmati, K., Theodoridou, M., Campbell, K., Vassilakopoulou, V., et al. (2013b). Phospholipase Cζ rescues failed oocyte activation in a prototype of male factor infertility. Fertil. Steril. 99 (1), 76–85. doi:10.1016/j.fertnstert.2012.08.035
Oh, J., Lee, C., and Kaang, B.-K. (2019). Imaging and analysis of genetically encoded calcium indicators linking neural circuits and behaviors. kjpp 23 (4), 237–249. doi:10.4196/kjpp.2019.23.4.237
Ostermeier, G. C., Miller, D., Huntriss, J. D., Diamond, M. P., and Krawetz, S. A. (2004). Reproductive biology: Delivering spermatozoan RNA to the oocyte. Nature 429 (6988), 154. doi:10.1038/429154a
Pardi, N., Hogan, M. J., Porter, F. W., and Weissman, D. (2018). mRNA vaccines — A new era in vaccinology. Nat. Rev. Drug Discov. 17 (4), 261–279. doi:10.1038/nrd.2017.243
Pardi, N., LaBranche, C. C., Ferrari, G., Cain, D. W., Tombácz, I., Parks, R. J., et al. (2019). Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol. Therapy-Nucleic Acids 15, 36–47. doi:10.1016/j.omtn.2019.03.003
Pardi, N., Tuyishime, S., Muramatsu, H., Kariko, K., Mui, B. L., Tam, Y. K., et al. (2015). Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 217, 345–351. doi:10.1016/j.jconrel.2015.08.007
Parrington, J., Swann, K., Shevchenko, V. I., Sesay, A. K., and Lai, F. A. (1996). Calcium oscillations in mammalian eggs triggered by a soluble sperm protein. Nature 379 (6563), 364–368. doi:10.1038/379364a0
Patel, A. K., Kaczmarek, J. C., Bose, S., Kauffman, K. J., Mir, F., Heartlein, M. W., et al. (2019). Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 31 (8), 1805116. doi:10.1002/adma.201805116
Patra, A., and Richert, C. (2009). High fidelity base pairing at the 3'-terminus. J. Am. Chem. Soc. 131 (35), 12671–12681. doi:10.1021/ja9033654
Payer, B., Saitou, M., Barton, S. C., Thresher, R., Dixon, J. P., Zahn, D., et al. (2003). Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13 (23), 2110–2117. doi:10.1016/j.cub.2003.11.026
Philipps, D. L., Wigglesworth, K., Hartford, S. A., Sun, F., Pattabiraman, S., Schimenti, K., et al. (2008). The dual bromodomain and WD repeat-containing mouse protein BRWD1 is required for normal spermiogenesis and the oocyte-embryo transition. Dev. Biol. 317 (1), 72–82. doi:10.1016/j.ydbio.2008.02.018
Pittoggi, C., Sciamanna, I., Mattei, E., Beraldi, R., Lobascio, A. M., Mai, A., et al. (2003). Role of endogenous reverse transcriptase in murine early embryo development. Mol. Reprod. Dev. 66 (3), 225–236. doi:10.1002/mrd.10349
Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383 (27), 2603–2615. doi:10.1056/NEJMoa2034577
Pollard, C., Rejman, J., De Haes, W., Verrier, B., Van Gulck, E., Naessens, T., et al. (2013). Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 21 (1), 251–259. doi:10.1038/mt.2012.202
Potireddy, S., Vassena, R., Patel, B. G., and Latham, K. E. (2006). Analysis of polysomal mRNA populations of mouse oocytes and zygotes: Dynamic changes in maternal mRNA utilization and function. Dev. Biol. 298 (1), 155–166. doi:10.1016/j.ydbio.2006.06.024
Ram, P. T., and Schultz, R. M. (1993). Reporter gene expression in G2 of the 1-cell mouse embryo. Dev. Biol. 156 (2), 552–556. doi:10.1006/dbio.1993.1101
Ramadan, W. M., Kashir, J., Jones, C., and Coward, K. (2012). Oocyte activation and phospholipase C zeta (PLCζ): Diagnostic and therapeutic implications for assisted reproductive technology. Cell Commun. Signal. CCS 10 (1), 12. doi:10.1186/1478-811X-10-12
Richner, J. M., Himansu, S., Dowd, K. A., Butler, S. L., Salazar, V., Fox, J. M., et al. (2017). Modified mRNA vaccines protect against zika virus infection. Cell 168 (6), 176. doi:10.1016/j.cell.2017.03.016
Rodríguez-Nuevo, A., Torres-Sanchez, A., Duran, J. M., De Guirior, C., Martínez-Zamora, M. A., and Böke, E. (2022). Oocytes maintain ROS-free mitochondrial metabolism by suppressing complex I. Nature 607 (7920), 756–761. doi:10.1038/s41586-022-04979-5
Rogers, N. T., Hobson, E., Pickering, S., Lai, F. A., Braude, P., and Swann, K. (2004). Phospholipase Czeta causes Ca2+ oscillations and parthenogenetic activation of human oocytes. Reproduction 128 (6), 697–702. doi:10.1530/rep.1.00484
Rong, Y., Ji, S.-Y., Zhu, Y.-Z., Wu, Y.-W., Shen, L., and Fan, H.-Y. (2019). ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res. 47 (21), 11387–11402. doi:10.1093/nar/gkz863
Sabnis, S., Kumarasinghe, E. S., Salerno, T., Mihai, C., Ketova, T., Senn, J. J., et al. (2018). A novel amino lipid series for mRNA delivery: Improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26 (6), 1509–1519. doi:10.1016/j.ymthe.2018.03.010
Sahin, U., Karikó, K., and Türeci, Ö. (2014). mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 13 (10), 759–780. doi:10.1038/nrd4278
Sahin, U., Oehm, P., Derhovanessian, E., Jabulowsky, R. A., Vormehr, M., Gold, M., et al. (2020). An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585 (7823), 107–112. doi:10.1038/s41586-020-2537-9
Saifullah, , Motohashi, N., Tsukahara, T., and Aoki, Y. (2022). Development of therapeutic RNA manipulation for muscular dystrophy. Front. Genome Ed. 4, 863651. doi:10.3389/fgeed.2022.863651
Saleh, A., Kashir, J., Thanassoulas, A., Safieh-Garabedian, B., Lai, F. A., and Nomikos, M. (2020). Essential role of sperm-specific PLC-zeta in egg activation and male factor infertility: An update. Front. Cell Dev. Biol. 8, 28. doi:10.3389/fcell.2020.00028
Sanusi, R., Yu, Y., Nomikos, M., Lai, F. A., and Swann, K. (2015). Rescue of failed oocyte activation after ICSI in a mouse model of male factor infertility by recombinant phospholipase Cζ. Mol. Hum. Reprod. 21 (10), 783–791. doi:10.1093/molehr/gav042
Sarhadi, V. K., and Armengol, G. (2022). Molecular biomarkers in cancer. Biomolecules 12 (8), 1021. doi:10.3390/biom12081021
Sasaki, H., Hamatani, T., Kamijo, S., Iwai, M., Kobanawa, M., Ogawa, S., et al. (2019). Impact of oxidative stress on age-associated decline in oocyte developmental competence. Front. Endocrinol. (Lausanne) 10, 811. doi:10.3389/fendo.2019.00811
Satouh, Y., Nozawa, K., Yamagata, K., Fujimoto, T., and Ikawa, M. (2017). Viable offspring after imaging of Ca2+ oscillations and visualization of the cortical reaction in mouse eggs. Biol. Reproduction 96 (3), 563–575. doi:10.1093/biolre/iox002
Saunders, C. M., Larman, M. G., Parrington, J., Cox, L. J., Royse, J., Blayney, L. M., et al. (2002). PLC zeta: A sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 129 (15), 3533–3544. doi:10.1242/dev.129.15.3533
Schultz, R. M. (1993). Regulation of zygotic gene activation in the mouse. Bioessays 15 (8), 531–538. doi:10.1002/bies.950150806
Sciamanna, I., Vitullo, P., Curatolo, A., and Spadafora, C. (2011). A reverse transcriptase-dependent mechanism is essential for murine preimplantation development. Genes. (Basel) 2 (2), 360–373. doi:10.3390/genes2020360
Sette, C., Bevilacqua, A., Bianchini, A., Mangia, F., Geremia, R., and Rossi, P. (1997). Parthenogenetic activation of mouse eggs by microinjection of a truncated c-kit tyrosine kinase present in spermatozoa. Development 124 (11), 2267–2274. doi:10.1242/dev.124.11.2267
Sette, C., Bevilacqua, A., Geremia, R., and Rossi, P. (1998). Involvement of phospholipase Cgamma1 in mouse egg activation induced by a truncated form of the C-kit tyrosine kinase present in spermatozoa. J. Cell. Biol. 142 (4), 1063–1074. doi:10.1083/jcb.142.4.1063
Sette, C., Paronetto, M. P., Barchi, M., Bevilacqua, A., Geremia, R., and Rossi, P. (2002). Tr-kit-induced resumption of the cell cycle in mouse eggs requires activation of a Src-like kinase. EMBO J. 21 (20), 5386–5395. doi:10.1093/emboj/cdf553
Seydoux, G. (2018). The P granules of C. elegans: A genetic model for the study of RNA-protein condensates. J. Mol. Biol. 430 (23), 4702–4710. doi:10.1016/j.jmb.2018.08.007
Sha, Q.-Q., Zheng, W., Wu, Y.-W., Li, S., Guo, L., Zhang, S., et al. (2020). Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 11 (1), 4917. doi:10.1038/s41467-020-18680-6
Shafqat, A., Shafqat, S., Salameh, S. A., Kashir, J., Alkattan, K., and Yaqinuddin, A. (2022). Mechanistic insights into the immune pathophysiology of COVID-19; an in-depth Review. Front. Immunol. 13, 835104. doi:10.3389/fimmu.2022.835104
Shinawi, T., Nasser, K. K., Moradi, F. A., Mujalli, A., Albaqami, W. F., Almukadi, H. S., et al. (2022). A comparative mRNA- and miRNA transcriptomics reveals novel molecular signatures associated with metastatic prostate cancers. Front. Genet. 13, 1066118. doi:10.3389/fgene.2022.1066118
Singh, Y., Murat, P., and Defrancq, E. (2010). Recent developments in oligonucleotide conjugation. Chem. Soc. Rev. 39 (6), 2054–2070. doi:10.1039/b911431a
Sirard, M. A., Dufort, I., Vallée, M., Massicotte, L., Gravel, C., Reghenas, H., et al. (2005). Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: Preliminary analysis using a bovine embryonic array. Reprod. Fertil. Dev. 17 (1-2), 47–57. doi:10.1071/rd04113
Squirrell, J. M., Wokosin, D. L., White, J. G., and Bavister, B. D. (1999). Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat. Biotechnol. 17 (8), 763–767. doi:10.1038/11698
Stitz, L., Vogel, A., Schnee, M., Voss, D., Rauch, S., Mutzke, T., et al. (2017). A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 11 (12), e0006108. doi:10.1371/journal.pntd.0006108
Suh, N., Baehner, L., Moltzahn, F., Melton, C., Shenoy, A., Chen, J., et al. (2010). MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 20 (3), 271–277. doi:10.1016/j.cub.2009.12.044
Sun, H., Zhang, Y., Wang, G., Yang, W., and Xu, Y. (2023). mRNA-based therapeutics in cancer treatment. Pharmaceutics 15 (2), 622. doi:10.3390/pharmaceutics15020622
Suzuki, J., Kanemaru, K., and Iino, M. (2016). Genetically encoded fluorescent indicators for organellar calcium imaging. Biophys. J. 111 (6), 1119–1131. doi:10.1016/j.bpj.2016.04.054
Suzuki, T., Yoshida, N., Suzuki, E., Okuda, E., and Perry, A. C. (2010). Full-term mouse development by abolishing Zn2+-dependent metaphase II arrest without Ca2+ release. Development 137 (16), 2659–2669. doi:10.1242/dev.049791
Swann, K., and Lai, F. A. (2016). The sperm phospholipase C-ζ and Ca2+ signalling at fertilization in mammals. Biochem. Soc. Trans. 44 (1), 267–272. doi:10.1042/bst20150221
Swann, K., Windsor, S., Campbell, K., Elgmati, K., Nomikos, M., Zernicka-Goetz, M., et al. (2012). Phospholipase C-ζ-induced Ca2+ oscillations cause coincident cytoplasmic movements in human oocytes that failed to fertilize after intracytoplasmic sperm injection. Fertil. Steril. 97 (3), 742–747. doi:10.1016/j.fertnstert.2011.12.013
Swetha, K., Kotla, N. G., Tunki, L., Jayaraj, A., Bhargava, S. K., Hu, H., et al. (2023). Recent advances in the lipid nanoparticle-mediated delivery of mRNA vaccines. Vaccines 11 (3), 658. doi:10.3390/vaccines11030658
Tadros, W., and Lipshitz, H. D. (2009). The maternal-to-zygotic transition: A play in two acts. Development 136 (18), 3033–3042. doi:10.1242/dev.033183
Tam, O. H., Aravin, A. A., Stein, P., Girard, A., Murchison, E. P., Cheloufi, S., et al. (2008). Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453 (7194), 534–538. doi:10.1038/nature06904
Tan, M. H., Au, K. F., Leong, D. E., Foygel, K., Wong, W. H., and Yao, M. W. (2013). An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Syst. Biol. 9 (1), 632. doi:10.1038/msb.2012.65
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S., and Vale, R. D. (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159 (3), 635–646. doi:10.1016/j.cell.2014.09.039
Tang, F., Kaneda, M., O'Carroll, D., Hajkova, P., Barton, S. C., Sun, Y. A., et al. (2007). Maternal microRNAs are essential for mouse zygotic development. Genes. Dev. 21 (6), 644–648. doi:10.1101/gad.418707
Teixeira, F. K., and Lehmann, R. (2019). Translational control during developmental transitions. Cold Spring Harb. Perspect. Biol. 11 (6), a032987. doi:10.1101/cshperspect.a032987
Tong, Z. B., Gold, L., Pfeifer, K. E., Dorward, H., Lee, E., Bondy, C. A., et al. (2000). Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 26 (3), 267–268. doi:10.1038/81547
Trcek, T., and Lehmann, R. (2019). Germ granules in Drosophila. Traffic 20 (9), 650–660. doi:10.1111/tra.12674
Vallée, M., Aiba, K., Piao, Y., Palin, M. F., Ko, M. S., and Sirard, M. A. (2008). Comparative analysis of oocyte transcript profiles reveals a high degree of conservation among species. Reproduction 135 (4), 439–448. doi:10.1530/rep-07-0342
Vanden Meerschaut, F., Leybaert, L., Nikiforaki, D., Qian, C., Heindryckx, B., and De Sutter, P. (2013). Diagnostic and prognostic value of calcium oscillatory pattern analysis for patients with ICSI fertilization failure. Hum. Reprod. 28 (1), 87–98. doi:10.1093/humrep/des368
Verbeke, R., Lentacker, I., De Smedt, S. C., and Dewitte, H. (2019). Three decades of messenger RNA vaccine development. Nano Today 28, 100766. doi:10.1016/j.nantod.2019.100766
Vilanova-Perez, T., Jones, C., Balint, S., Dragovic, R., Dustin, M. L., Yeste, M., et al. (2020). Exosomes derived from HEK293T cells interact in an efficient and noninvasive manner with mammalian sperm in vitro. Nanomedicine (Lond) 15 (20), 1965–1980. doi:10.2217/nnm-2020-0056
Wang, Q. T., Piotrowska, K., Ciemerych, M. A., Milenkovic, L., Scott, M. P., Davis, R. W., et al. (2004). A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell 6 (1), 133–144. doi:10.1016/s1534-5807(03)00404-0
Wang, W., Zhou, X., Bian, Y., Wang, S., Chai, Q., Guo, Z., et al. (2020). Dual-targeting nanoparticle vaccine elicits a therapeutic antibody response against chronic Hepatitis B. Nat. Nanotechnol. 15 (5), 406–416. doi:10.1038/s41565-020-0648-y
Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., et al. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453 (7194), 539–543. doi:10.1038/nature06908
Watts, J. K., Deleavey, G. F., and Damha, M. J. (2008). Chemically modified siRNA: Tools and applications. Drug Discov. Today 13 (19-20), 842–855. doi:10.1016/j.drudis.2008.05.007
Weng, Y., Li, C., Yang, T., Hu, B., Zhang, M., Guo, S., et al. (2020). The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 40, 107534. doi:10.1016/j.biotechadv.2020.107534
Whitworth, K. M., Agca, C., Kim, J. G., Patel, R. V., Springer, G. K., Bivens, N. J., et al. (2005). Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol. Reprod. 72 (6), 1437–1451. doi:10.1095/biolreprod.104.037952
Wolff, J. A., Malone, R. W., Williams, P., Chong, W., Acsadi, G., Jani, A., et al. (1990). Direct gene transfer into mouse muscle in vivo. Science 247 (4949), 1465–1468. doi:10.1126/science.1690918
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367 (6483), 1260–1263. doi:10.1126/science.abb2507
Wrenzycki, C., Herrmann, D., and Niemann, H. (2007). Messenger RNA in oocytes and embryos in relation to embryo viability. Theriogenology 68 (1), S77–S83. doi:10.1016/j.theriogenology.2007.04.028
Wu, A. T. H., Sutovsky, P., Manandhar, G., Xu, W., Katayama, M., Day, B. N., et al. (2007). PAWP, a sperm-specific WW domain-binding protein, promotes meiotic resumption and pronuclear development during fertilization. J. Biol. Chem. 282 (16), 12164–12175. doi:10.1074/jbc.m609132200
Wu, G., and Schöler, H. R. (2014). Role of Oct4 in the early embryo development. Cell Regen. 3 (1), 7. doi:10.1186/2045-9769-3-7
Xu, S., Yang, K., Li, R., and Zhang, L. (2020). mRNA vaccine era—mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 21 (18), 6582. doi:10.3390/ijms21186582
Yagi, R., Kohn, M. J., Karavanova, I., Kaneko, K. J., Vullhorst, D., DePamphilis, M. L., et al. (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134 (21), 3827–3836. doi:10.1242/dev.010223
Yamada-Fukunaga, T., Yamada, M., Hamatani, T., Chikazawa, N., Ogawa, S., Akutsu, H., et al. (2013). Age-associated telomere shortening in mouse oocytes. Reproductive Biol. Endocrinol. 11 (1), 108. doi:10.1186/1477-7827-11-108
Yang, T., Xia, L., Li, G., Zhao, J., Li, J., Ge, J., et al. (2023). Novel bionic inspired nanosystem construction for precise delivery of mRNA. Front. Bioeng. Biotechnol. 11, 1160509. doi:10.3389/fbioe.2023.1160509
Yaqinuddin, A., Shafqat, A., Kashir, J., and Alkattan, K. (2021). Effect of SARS-CoV-2 mutations on the efficacy of antibody therapy and response to vaccines. Vaccines 9 (8), 914. doi:10.3390/vaccines9080914
Yelumalai, S., Kashir, J., Jones, C., Bagheri, H., Oo, S. L., McLaren, L., et al. (2012). Clinician-induced (iatrogenic) damage incurred during human infertility treatment: Detrimental effects of sperm selection methods and cryopreservation upon the viability, DNA integrity, and function of human sperm. Asian Pac. J. Reproduction 1 (1), 69–75. doi:10.1016/S2305-0500(13)60052-4
Yelumalai, S., Yeste, M., Jones, C., Amdani, S. N., Kashir, J., Mounce, G., et al. (2015). Total levels, localization patterns, and proportions of sperm exhibiting phospholipase C zeta are significantly correlated with fertilization rates after intracytoplasmic sperm injection. Fertil. Steril. 104 (3), 561–568. doi:10.1016/j.fertnstert.2015.05.018
Yoda, A., Oda, S., Shikano, T., Kouchi, Z., Awaji, T., Shirakawa, H., et al. (2004). Ca2+ oscillation-inducing phospholipase C zeta expressed in mouse eggs is accumulated to the pronucleus during egg activation. Dev. Biol. 268 (2), 245–257. doi:10.1016/j.ydbio.2003.12.028
Yoon, S.-Y., Jellerette, T., Salicioni, A. M., Lee, H. C., Yoo, M.-S., Coward, K., et al. (2008). Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J. Clin. investigation 118 (11), 3671–3681. doi:10.1172/JCI36942
Yu, C., Ji, S.-Y., Dang, Y.-J., Sha, Q.-Q., Yuan, Y.-F., Zhou, J.-J., et al. (2016). Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 26 (3), 275–287. doi:10.1038/cr.2016.20
Yuan, S., Zhan, J., Zhang, J., Liu, Z., Hou, Z., Zhang, C., et al. (2023). Human zygotic genome activation is initiated from paternal genome. Cell Discov. 9 (1), 13. doi:10.1038/s41421-022-00494-z
Zeng, F., Baldwin, D. A., and Schultz, R. M. (2004). Transcript profiling during preimplantation mouse development. Dev. Biol. 272 (2), 483–496. doi:10.1016/j.ydbio.2004.05.018
Zeng, F., and Schultz, R. M. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283 (1), 40–57. doi:10.1016/j.ydbio.2005.03.038
Zhang, C., Maruggi, G., Shan, H., and Li, J. (2019). Advances in mRNA vaccines for infectious diseases. Front. Immunol. 10, 594. doi:10.3389/fimmu.2019.00594
Zhang, C., Wang, M., Li, Y., and Zhang, Y. (2022). Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv. 8 (5), eabj3967. doi:10.1126/sciadv.abj3967
Zhang, N.-N., Li, X.-F., Deng, Y.-Q., Zhao, H., Huang, Y.-J., Yang, G., et al. (2020). A thermostable mRNA vaccine against COVID-19. Cell 182 (5), 1271–1283. doi:10.1016/j.cell.2020.07.024
Zhao, M., Li, M., Zhang, Z., Gong, T., and Sun, X. (2016). Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 23 (7), 2596–2607. doi:10.3109/10717544.2015.1038856
Zhong, R., Talebian, S., Mendes, B. B., Wallace, G., Langer, R., Conde, J., et al. (2023). Hydrogels for RNA delivery. Nat. Mater. doi:10.1038/s41563-023-01472-w
Keywords: oocyte, sperm, embryo, RNA, oocyte activation, calcium, therapeutics
Citation: Bafleh WS, Abdulsamad HMR, Al-Qaraghuli SM, El Khatib RY, Elbahrawi RT, Abdukadir AM, Alsawae SM, Dimassi Z, Hamdan H and Kashir J (2023) Applications of advances in mRNA-based platforms as therapeutics and diagnostics in reproductive technologies. Front. Cell Dev. Biol. 11:1198848. doi: 10.3389/fcell.2023.1198848
Received: 03 April 2023; Accepted: 18 May 2023;
Published: 26 May 2023.
Edited by:
Matteo Avella, Sidra Medicine, QatarReviewed by:
Nady El Hajj, Hamad bin Khalifa University, QatarTalluri Thirumala Rao, Indian Council of Agricultural Research (ICAR), India
Copyright © 2023 Bafleh, Abdulsamad, Al-Qaraghuli, El Khatib, Elbahrawi, Abdukadir, Alsawae, Dimassi, Hamdan and Kashir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junaid Kashir, anVuYWlka2FzaGlyQGhvdG1haWwuY29t
 Wjdan S. Bafleh1
Wjdan S. Bafleh1 Sally M. Al-Qaraghuli
Sally M. Al-Qaraghuli Hamdan Hamdan
Hamdan Hamdan Junaid Kashir
Junaid Kashir