- Department of Stem Cell and Regenerative Biotechnology, Institute of Advanced Regenerative Science, Konkuk University, Seoul, South Korea
Epigenetic mechanisms are mandatory for endothelial called lymphangioblasts during cardiovascular development. Dot1l-mediated gene transcription in mice is essential for the development and function of lymphatic ECs (LECs). The role of Dot1l in the development and function of blood ECs blood endothelial cells is unclear. RNA-seq datasets from Dot1l-depleted or -overexpressing BECs and LECs were used to comprehensively analyze regulatory networks of gene transcription and pathways. Dot1l depletion in BECs changed the expression of genes involved in cell-to-cell adhesion and immunity-related biological processes. Dot1l overexpression modified the expression of genes involved in different types of cell-to-cell adhesion and angiogenesis-related biological processes. Genes involved in specific tissue development-related biological pathways were altered in Dot1l-depleted BECs and LECs. Dot1l overexpression altered ion transportation-related genes in BECs and immune response regulation-related genes in LECs. Importantly, Dot1l overexpression in BECs led to the expression of genes related to the angiogenesis and increased expression of MAPK signaling pathways related was found in both Dot1l-overexpressing BECs and LECs. Therefore, our integrated analyses of transcriptomics in Dot1l-depleted and Dot1l-overexpressed ECs demonstrate the unique transcriptomic program of ECs and the differential functions of Dot1l in the regulation of gene transcription in BECs and LECs.
1 Introduction
The blood endothelial cells (BECs) line the inner cavity of the vascular system and provide a physical barrier, transportation of molecules and oxygen, and immune responses (Galley and Webster, 2004; McCarron et al., 2017). Although, in general, BECs originate from mesodermal lineages, ample evidence suggests that multiple developmental stages- and/or organ-type-specific sub-mesoderm origins of BECs exist. The molecular, functional, and structural heterogeneity in different types of ECs has been identified (Aird, 2007a; Aird, 2007b; Aird, 2012; Jambusaria et al., 2020; Becker et al., 2022; Hou et al., 2022).
Early studies proposed that a specialized mesoderm-origin cell, called a hemangioblast, differentiates into hematopoietic stem cells and angioblasts after gastrulation. Subsequently, angioblasts generate BEC progenitors (Chung et al., 2002). In mice, embryonic vasculatures arise simultaneously in embryonic and extraembryonic tissues via vasculogenesis, a process of de novo blood vessel formation (Drake and Fleming, 2000; Chong et al., 2011). After that, BECs respond to angiogenic growth factors, sprouting from pre-existing vessels by angiogenesis (Majesky, 2018) and undergo specialization into venous or arterial BECs before the formation of mature vasculature by diverse factors, such as hemodynamic forces and transcriptional programs (Jain, 2003; Heil et al., 2006; Lin, 2007) After arterial or venous BEC fate is determined, each BEC expresses distinct sets of transcriptomes and displays different branching capacities (Jain, 2003; Torres-Vazquez et al., 2003). Following the maturation of blood vessels, some of the venous ECs in the anterior part of the cardinal vein (CV) express Prox1, Lyve1, and Sox18, and differentiation to lymphatic EC (LECs) begins at approximately embryonic day (E) 9–10 in mice (Tammela and Alitalo, 2010; Potente and Makinen, 2017). Accumulating evidence suggests that LECs of developing mouse organs have heterogeneity in both cellular origin and function (Klotz et al., 2015; Martinez-Corral et al., 2015; Pichol-Thievend et al., 2018; Maruyama et al., 2019; Lioux et al., 2020; Aquino et al., 2021). Furthermore, Prox1 (+) LECs in the CV are differentiated from Pax3 (+) paraxial mesoderm-derived LEC progenitor cells (called lymphangioblasts) in mouse (Stone and Stainier, 2019; Lupu et al., 2022).
During the development of BECs and LECs, various epigenetic mechanisms, including DNA methylation and histone modifications are involved in EC development and functions (Gutierrez et al., 2021; Yu et al., 2021). Furthermore, there are epigenetic differences in ECs depending on organotypic and developmental origin factors (Bronneke et al., 2012; Nakato et al., 2019; Gutierrez et al., 2021; Tacconi et al., 2021). However, epigenetic modifiers establishing vessel type-specific epigenetic landscapes that contribute to the development of unique transcriptomic signatures during development remain elusive.
Dot1l is the only known histone H3K79 methyltransferase that produces mono-, di-, tri-methyl H3K79 (H3K79me1-3) (Feng et al., 2002). Dot1l has diverse roles in biological processes, including DNA repair (Huyen et al., 2004), transcription elongation (Mueller et al., 2007), telomeric silencing (Jones et al., 2008), regulation of the cell cycle (Jones et al., 2008), pluripotent stem cell differentiation (Barry et al., 2009), and immune responses (Kealy et al., 2020). Dot1l is essential for embryonic blood vessel, heart, and lymphatic vessel development and functions via transcriptional regulation of lymphatic genes (Jones et al., 2008; Nguyen et al., 2011; Yoo et al., 2020). Dot1l loss in BECs leads to decreased tube formation and sprouting in vitro and vessel network formation in vivo (Duan et al., 2016). Our group previously showed that Dot1l loss in both BECs and LECs causes severe defects in LEC development and function. In particular, we revealed functional heterogeneity of Dot1l in LEC defects depending on the developmental origin and organotypes of LECs (Yoo et al., 2020). However, the regulatory mechanism by which Dot1l functions differentially in BECs vs. LECs at the molecular level remains unclear.
Using genome-scale transcriptomic analysis, this study showed that mouse Dot1l regulates common or distinct transcriptomic programs in an EC type-dependent manner, providing background information for identifying factors orchestrating the transcriptional change together with Dot1l and conditional target specificity of Dot1l. Furthermore, since BECs and LECs branch off from the same source (Wigle and Oliver, 1999; Srinivasan et al., 2007; Francois et al., 2012) and have distinguishable phenotypes, this study may provide insights into the cell fate decision mechanism and the function gaining process of these cell types.
This study found that Dot1l loss changes the expression of genes involved in lipid metabolism in BECs and the chemotaxis in LECs. Meanwhile, Dot1l overexpression alters the expression of genes involved in ion transportation and immune response regulation in both BECs and LECs. Taken together, these findings reveal a distinct Dot1l function in developing BECs and LECs and the heterogeneity of the transcriptomic program in the ECs.
2 Materials and methods
2.1 Animals and isolation of skin BECs
The endothelial-specific Dot1l conditional knockout mouse has been described previously (Yoo et al., 2020). Briefly, E15.5 Tie2-Cre(+); Dot1l2f/2f (cKO), and Tie2-Cre(−); Dot1l2f/2f (Cont) embryo skins were harvested and dissociated in media containing type II and IV collagenase and DNase I (LS006333; Worthington Biochemical Corp.) for 20 min at 37°C. Cells that passed through 40-µm cell strainers were incubated with anti-CD45 and anti-F4/80 antibodies (13–0451 and 13–4801, eBioscience) for 1 h at room temperature to deplete macrophages. F4/80 (−)/CD45 (−) cells were collected and incubated with Lyve1 antibody (13–0443, eBioscience) to collect LECs. Together with the LECs, isolated F4/80 (−)/CD45 (−)/Lyve1 (−) BECs were subjected to RT-qPCR or RNA-seq analysis. All animal studies were reviewed and approved by the Institute of Animal Care and Use Committee (IACUC) of Konkuk University (IACUC#KU18027).
2.2 Lentivirus-mediated Dot1l overexpression in BECs and LECs
The overexpression of Dot1l has been described in a previous study (Yoo et al., 2020). Briefly, guide RNAs targeting the Dot1l promoter were designed and subcloned into the BbsI sites of the vectors (cat. #53186, #53187, #53188, and #53189, Addgene). The vectors were then cloned into a lentivirus vector (59791, Addgene) containing catalytically dead Cas9 (dCas9) fused with VP64, guide RNAs targeting Dot1l promoter regions and enhanced green fluorescent protein genes (Figure 3A). The Dot1l overexpression vector with lentiviral packaging vectors [psPAX2 (cat. #12260, Addgene) and pMD2.G (cat. #12259, Addgene) vectors] was transfected into HEK293T cells to produce lentiviral particles. Mouse embryonic dermal LECs (C57-6064L, Cell Biologics) and embryonic yolk sac BECs (CRL-2581, ATCC) were used for viral transduction. Each cell line was maintained according to the manufacturer’s instructions. Once ECs reached approximately 50% confluency, viral particles were added to BECs and LECs for Dot1l overexpression. ECs transduced with empty lentivirus were used as controls. EGFP(+) cells were sorted using FACS Aria (BD Biosciences) and used for RT-qPCR and RNA-sequencing analysis.
2.3 RNA isolation and quantitative RT-PCR (RT-qPCR)
Dot1l cKO ECs from E15.5 skins and cultured Dot1l overexpression ECs were used for RT-qPCR. Total RNAs were extracted using RNeasy Plus Mini Kit (Qiagen). SMARTer Pico PCR cDNA synthesis kit (Takara) along with Advantage 2 PCR Kit (Takara) and TOPscript RT DryMix cDNA synthesis kit (Enzynomics) were used for cDNA synthesis of Dot1l cKO ECs and Dot1l overexpression ECs, respectively. RT-qPCR was performed on StepOnePlusTM platform (Thermo Fisher Scientific) using Fast SYBR® Green Master Mix (Thermo Fisher Scientific). Gene expression in each sample was normalized to the expression of Gapdh. The sequences for primers used for RT-qPCR are as follows: Gapdh; 5′-CATGGCCTTCCGTGTTCCTA-3′, 5′-GCCTGCTTCACCACCTTCTT-3′, Dot1l; 5′-TGGCAAGCCTGTCTCCTACT-3′, 5′-CTGCTCCTCCCTGAGTTTTG-3′, Mmp3: 5′- ACATGGAGACTTTGTCCCTTTTG-3′, 5′-TTGGCTGAGTGGTAGAGTCCC-3′ and Cldn1 5′-GGGGACAACATCGTGACCG-3′, 5′-AGGAGTCGAAGACTTTGCACT-3′.
2.4 RNA-seq analysis
For the RNA-sequencing (RNA-seq) analysis in the Cont and cKO BECs, pooled total RNAs isolated from BECs of 2–3 E15.5 pup skins, with RNA integrity number (RIN) greater than 7, were used to make a library for each genotype. The BEC RNAs were from the same embryos which the previously described LEC RNAs were extracted from (Yoo et al., 2020). For control and Dot1l overexpression BECs or LECs, cells from 2 to 3 T25 flask cultures were harvested to make libraries for each genotype. For RNA extraction, RNeasy Plus Mini Kit (Qiagen) was used according to the manufacturer’s instructions and total RNA isolated from BECs and LECs were used to prepare RNA-sequencing (RNA-seq) libraries using the ScriptSeq v2 kit (Illumina) according to the manufacturer’s instructions. Sequencing of the library was performed to produce single raw data for each genotype and cell type combination. RNA-seq data generated in the E15.5 Tie2-Cre(+); Dot1l2f/2f and Tie2-Cre(−); Dot1l2f/2f embryonic skin LECs (GSE104811) were downloaded and analyzed.
For post-sequencing data analysis, adapter sequences in the raw reads were trimmed with skewer (v0.2.2) (Jiang et al., 2014) with -L 75 and -e options specified. The skewer output reads were aligned to the mm10 UCSC mouse reference genome using a STAR read aligner (v2.7.9a) (Dobin et al., 2013). We quantified the mapped reads using VERSE (v0.1.5) (Zhu et al., 2016) with the -S option. Samples were normalized with the median of ratios method using the DESeq2 (v1.36.0) (Love et al., 2014) package in R (v4.2.0). Genes with normalized expression values ≥ 5 and values exceeding three times the value of the sample in comparison were defined as differentially expressed genes (DEGs) in Dot1l cKO samples, and those with normalized expression values ≥ 3 and values exceeding three times the value of the sample in comparison were defined as DEGs in OE samples. Genes with normalized expression values ≥ 5 and under three times the value of the sample in comparison were defined as unchanged genes (none-DEGs). DEG lists were used as inputs for GO enrichment analysis conducted on DAVID for GO terms in the biological pathway subset or KEGG pathways (v2022q2) (Huang da et al., 2009; Sherman et al., 2022). 10 GO terms or KEGG pathways with the lowest p-value were presented from each analysis. In Gene Set Enrichment Analysis (GSEA), “weighted” was selected for enrichment statistics and “log2 ratio of classes” was selected for metric of ranking genes (Subramanian et al., 2005). Gene expression change was presented using heatmap2 function of gplots R package (Warnes et al., 2022). Integrative genomic viewer (IGV) was used to verify the expression of the representative genes (Robinson et al., 2011).
2.5 ChIP-seq analysis
For chromatin immunoprecipitation-sequencing (ChIP-seq) analysis of H3K79me2 in LEC, the data was downloaded from Gene Expression Omnibus (GEO) under the accession code GSE104811. Adapters were trimmed from the raw reads using skewer (v0.2.2) (Jiang et al., 2014) using -L and -e options, and the reads were aligned to mm10 UCSC mouse genome using bowtie2 (v2.3.4.2) (Langmead and Salzberg, 2012). BedGraph files were constructed using MACS3 (v3.0.0b1) (Zhang et al., 2008) from the output of read alignment. The file was used to create H3K79me2 profile on the gene bodies using deepTools (v3.5.1) (Ramirez et al., 2016).
3 Results
3.1 Depletion of Dot1l transcripts in BECs leads to global transcriptomic changes
To analyze Dot1l function in BECs, we performed RNA-sequencing (RNA-seq) analysis of Dot1l-depleted BECs isolated from E15.5 Tie2-Cre(+); Dot1l2f/2f (conditional knockout, cKO) embryonic skin (Figure 1A). BECs isolated from E15.5 littermate Tie2-Cre(−); Dot1l2f/2f embryonic skin were used as the control (Cont). As shown in Figure 1B, 794 genes were upregulated and 369 genes were downregulated in cKO BECs. Dot1l cKO was validated using RT-qPCR (Figure 1C). Gene ontology (GO) analysis of these DEGs in cKO BECs revealed associations between the downregulated genes and lipid catabolic process, cell adhesion, and immune response, and between the upregulation of genes and peptide cross-linking, positive regulation of gene expression, and immune response (Figure 1D). Pathway analysis of the DEGs showed that the genes involved in the tumor necrosis factor (TNF) signal-, peroxisome proliferator-activated receptor signal-, and interleukin (IL)17-related pathways were downregulated, and genes involved in drug metabolism and cytokine/receptor-related pathways were upregulated (Figure 1E).
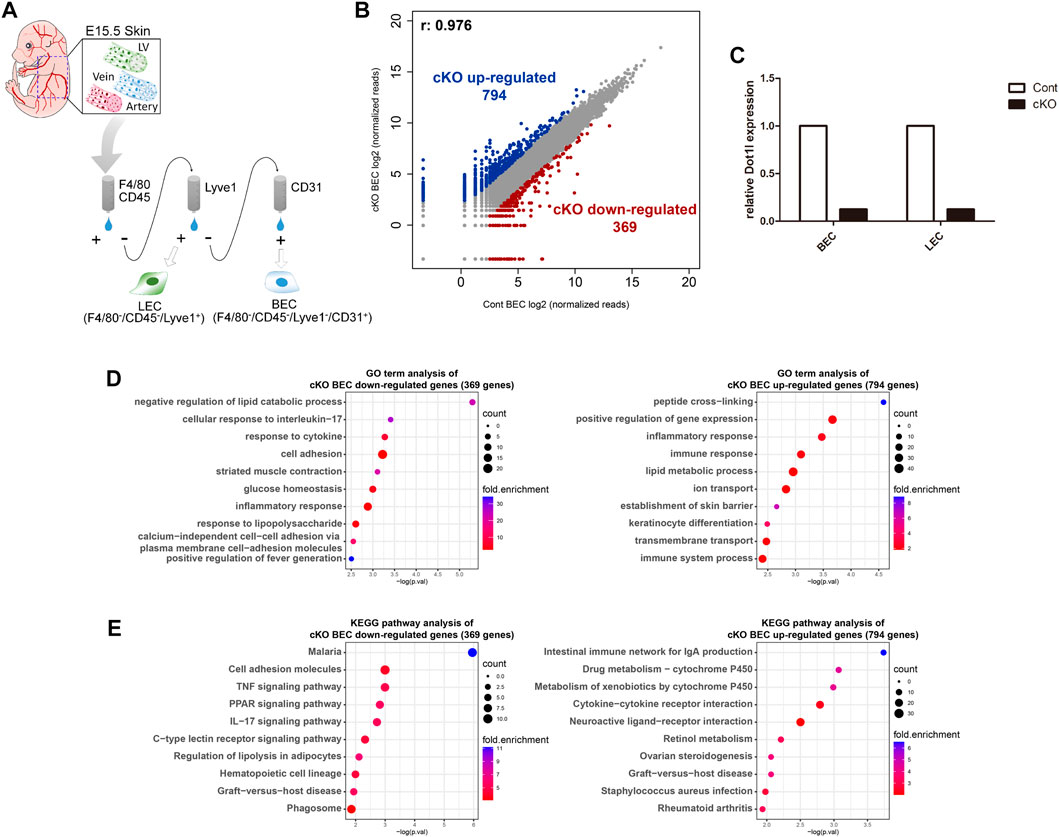
FIGURE 1. RNA-seq analysis of Dot1l cKO BECs. (A) Schematic diagram of the isolation of BECs from E15.5 cKO or control embryos. Cell surface markers used for BEC isolation were macrophage markers (F4/80/CD45)-negative, LEC marker (Lyve1)-negative, and BEC marker (CD31)-positive. (B) Scatterplot of differentially expressed genes (DEGs) according to a cutoff value, ≥5 normalized read counts, and fold change ≥3. (C) RT-qPCR results for validation of Tie2-Cre mediated Dot1l cKO. Expression of Dot1l is normalized to the expression value of Gapdh. (n = 1) (D) GO term analysis of down- and upregulated DEGs. (E) KEGG pathway analysis with down- and upregulated DEGs. Abbreviations: cKO, conditional knockout; LV, lymphatic vessel; LEC, lymphatic endothelial cell and BEC, blood endothelial cell.
3.2 Dot1l regulates common or distinct transcriptome in BECs and LECs
Next, we analyzed the regulatory function of Dot1l in the transcription of BECs and LECs. We previously showed that Dot1l loss represses genes involved in cell adhesion and angiogenesis in E15.5 skin LECs (Yoo et al., 2020). In this study, we compared the transcriptomic changes induced by Dot1l cKO in BECs and LECs (Figures 2A,B). The gene set repressed uniquely in the cKO LECs included genes related to cell adhesion, nervous system development, and angiogenesis (out of 2,134 genes) (Figure 2C). Meanwhile, genes related to immune response were enriched in cKO BECs repressed gene set (24 out of 227 genes, 10.6%; Figure 2C, Supplementary Figure S1A). The cKO LEC GO analysis of uniquely enhanced gene set also exhibited enrichment of immune response-related genes (375 out of 2,401 genes, 15.6%; Figure 2D). We further investigated expression pattern change of immunity-related genes using Gene Set Enrichment Analysis to identify which category of the immune system was up- or downregulated in each case (Subramanian et al., 2005). 18 child terms of the term “immune response” was obtained from the Amigo2 database (Carbon et al., 2009). Out of the 18 child terms, all 18 terms were significantly more upregulated in the Dot1l cKO LECs, while cKO BECs exhibited downregulation of “innate immune response in mucosa”, “organ or tissue specific immune response”, “immunological memory process” and “type 2 immune response” (Figure 2E; Supplementary Figure S2). This result attributes to decreased expression of genes such as Il6, Arg2, and Irf1 in the cKO BECs (Supplementary Figure S1A).
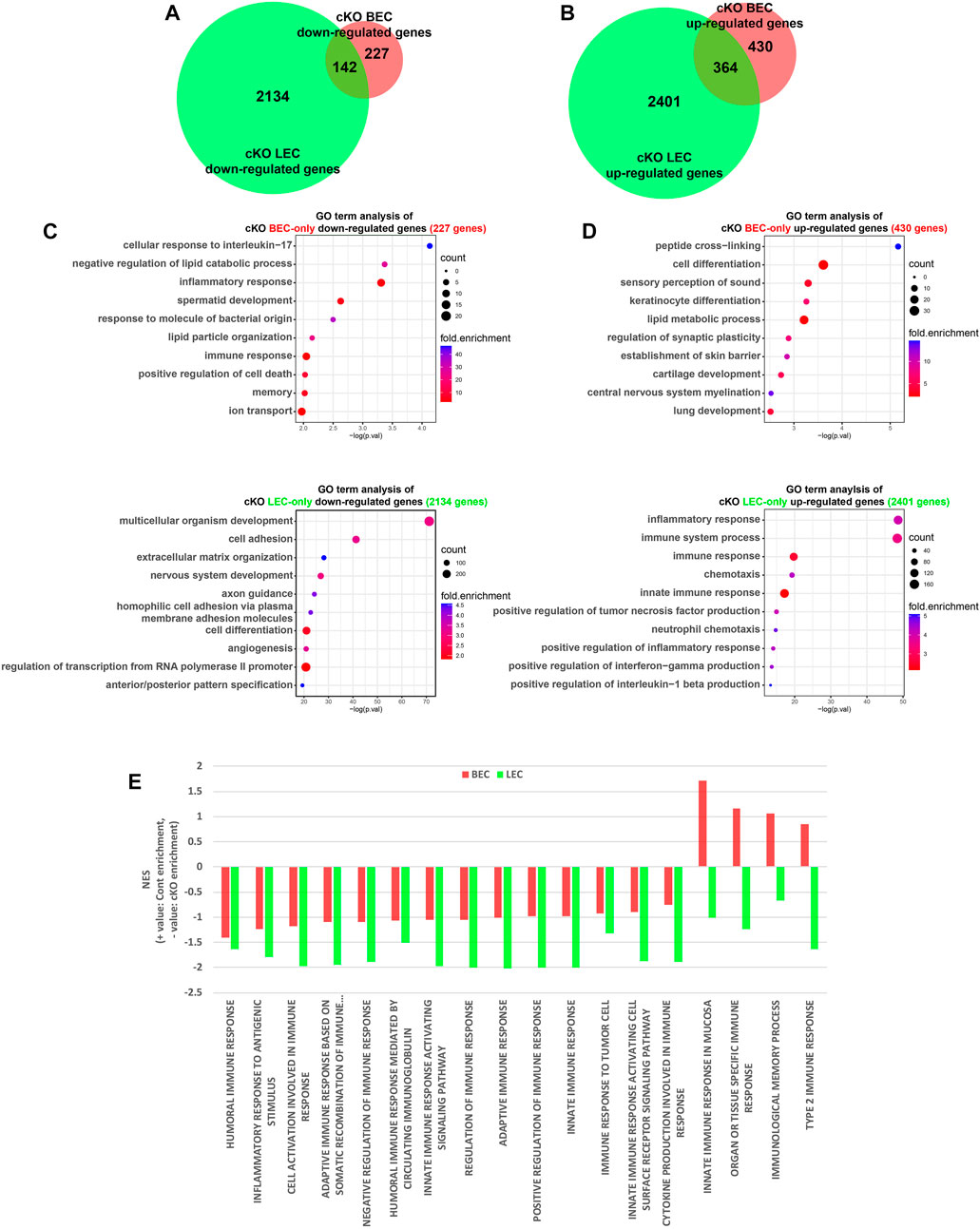
FIGURE 2. Comparison of DEGs between cKO BECs and LECs. (A) Venn diagram showing numbers of common and BEC- and LEC-only downregulated genes in cKO cells. (B) Venn diagram showing numbers of common and BEC- and LEC-only upregulated genes in the cKO. (C) GO terms identified for BEC- and LEC-only downregulated genes. (D) GO terms identified for BEC- and LEC-only upregulated genes. (E) Bar graph of normalized enrichment score (NES) of 18 child terms of the term “immune response”. Positive NES indicates enrichment in the control ECs and negative NES indicates enrichment in the Dot1l cKO ECs.
To investigate whether the DEGs in Dot1l cKO ECs are a result of differential H3K79 methylation, LEC H3K79me2 ChIP data from our previous study was reanalyzed (Yoo et al., 2020). Reduction of H3K79me2 levels upon Dot1l inhibition by EPZ-5676 (also known as pinometostat) was observed in the gene bodies of commonly unchanged (none-DEGs) and downregulated DEGs but not in the commonly upregulated DEGs (Supplementary Figure S1C). The result provides additional evidence for association with the change of Dot1l-mediated H3K79 methylation and transcriptional activation. At the same time, the decrease of methylation level in the gene bodies of unchanged genes suggests that Dot1l is involved in transcription-independent cellular and molecular mechanisms.
The analysis of overlapping DEGs revealed 142 and 364 commonly repressed and elevated genes, respectively, in both ECs as a result of Dot1l depletion (Figures 2A,B). Analysis of GO terms revealed that downregulated genes were associated with glucose metabolism and upregulated genes were associated with cell-to-cell adhesion, and immunity-related terms (immune system process, inflammatory response, and others) (Supplementary Figure S1B). Representative genes visualized in the IGV tracks included Cldn11 (downregulated in BECs and LECs), CD84 (upregulated in BECs and LECs), Socs3 (downregulated in BECs), Spink5 (upregulated in BECs), Cdh5 (downregulated in LECs), and Tnf (upregulated in LECs) (Supplementary Figure S1D).
3.3 Forced Overexpression of Dot1l transcripts changes distinct gene sets in BECs or LECs
Next, we induced Dot1l overexpression in BECs (OE-BECs) or LECs (OE-LECs) to investigate its role in the regulation of gene transcription (Figure 3A). Overexpression of Dot1l and expression alteration of Mmp3 and Cldn1 in both types of cells were confirmed by RT-qPCR and in the IGV (Figures 3B,C; Supplementary Figures S3A, B). RNA-seq analysis identified 1,175 upregulated and 723 downregulated genes in OE-BECs, and 384 upregulated and 289 downregulated genes in OE-LECs (Figure 3D). Analysis of GO terms revealed OE-BECs had enhanced expression of genes associated with cell adhesion, development, and positive regulation of angiogenesis, and repressed expression of genes associated with positive regulation of angiogenesis, extracellular matrix (ECM) organization, cell adhesion, and cell proliferation. OE-LECs displayed enhanced transcriptional activity of genes associated with cellular response to interferon-beta and response to virus and immune system processes, and repressed activity of genes associated with ECM organization, cell adhesion, extracellular signal-regulated kinase (ERK)1 and ERK2 cascade, and establishment of the endothelial barrier (Figures 3F,G).
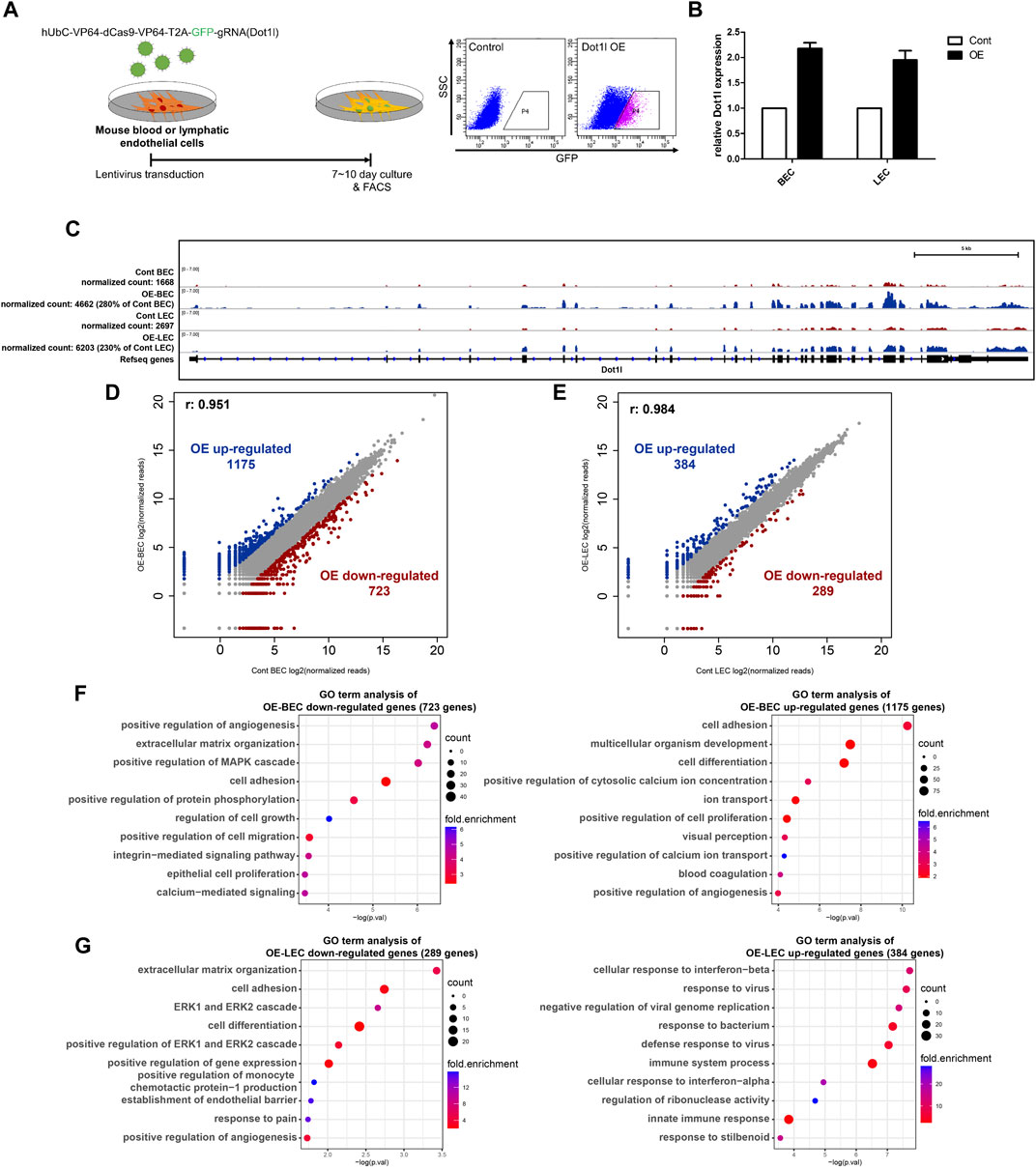
FIGURE 3. RNA-seq analysis in the Dot1l-overexpression (OE) BECs and LECs. (A) Schematic diagram showing lentivirus-mediated OE in either LECs or BECs via hUbC-VP64-dCas9-dCas9-T2A-GFP-gRNA (for Dot1l) vector and fluorescence-activated cell sorting of transduced cells (green fluorescent protein-positive [GFP+] cells). (B) RT-qPCR results for validation of lentiviral transduction mediated Dot1l OE. Dot1l expression is normalized to the expression value of Gapdh. (n = 2) (C) IGV track confirming Dot1l overexpression in LECs or BECs. (D) Scatter plot showing DEGs between control and OE-BECs. (E) Scatter plot showing DEGs between control and OE-LECs. Analysis of GO terms with down- or upregulated DEGs in OE-BECs (F) and OE-LECs (G).
Next, we sought to identify genes that are highly dependent on Dot1l for their expression by comparing gene expression change between the cKO and OE samples. To that end, genes with normalized expression values that were either more than three times higher or less than one-third of the expression value in the control were selected. In total, there were 16 genes for BECs and 22 genes for LECs (Supplementary Figure S3C). Notably, GO term analysis in the cKO vs. OE DEGs of LECs revealed an enrichment of genes related to cell adhesion and blood vessel remodeling among the selected genes (Supplementary Figure S3D). The genes Stab2, Fat2, Emp2, and Sulf1 were associated with the term cell adhesion, and Sema3c and Axl were associated with the term blood vessel remodeling.
3.4 Comparison of transcriptomic alterations between OE-BECs and OE-LECs
We then compared genes that were up- or downregulated in OE-BECs and OE-LECs (Figures 4A,B). As shown in Figure 4A, out of a total of 76 genes, genes associated with ECM organization, differentiation, EC proliferation involved in wound healing, and cell adhesion were commonly downregulated in OE-BECs and OE-LECs (Figure 4A). Of a total of 86 genes, those associated with positive regulation of cell migration, chemotaxis, and inflammatory response were commonly upregulated in OE-BECs and OE-LECs (Figure 4B). Interestingly, common and related terms, such as positive regulation of angiogenesis, extracellular matrix organization, cell adhesion, positive regulation of MAPK cascade, and ERK1 and ERK2 cascade, were found in the most significantly enriched terms in downregulated DEG sets of both types of OE-ECs (BEC: 38 out of 723 genes, 5.3%; LEC: 28 out of 289 genes, 9.7%; Figures 3E,F). The term positive regulation of ERK1 and ERK2 cascade were also enriched in GO term analysis results of the commonly upregulated gene set of both Dot1l OE-ECs (5 out of 86 genes, 5.8%; Figures 4B,C). This implies a close connection between Dot1l-mediated transcriptional regulation and MAPK/ERK signaling pathway during EC development. In the analysis result of upregulated genes, OE-BEC upregulated genes are enriched in GO terms such as cell adhesion and ion transport, while OE-LEC upregulated genes are enriched in immunity related GO terms (Figures 3F,G). GO term analysis of the overlapping DEGs from each comparison presented enrichment in both calcium ion transport and inflammatory response terms (Figures 4B,D).
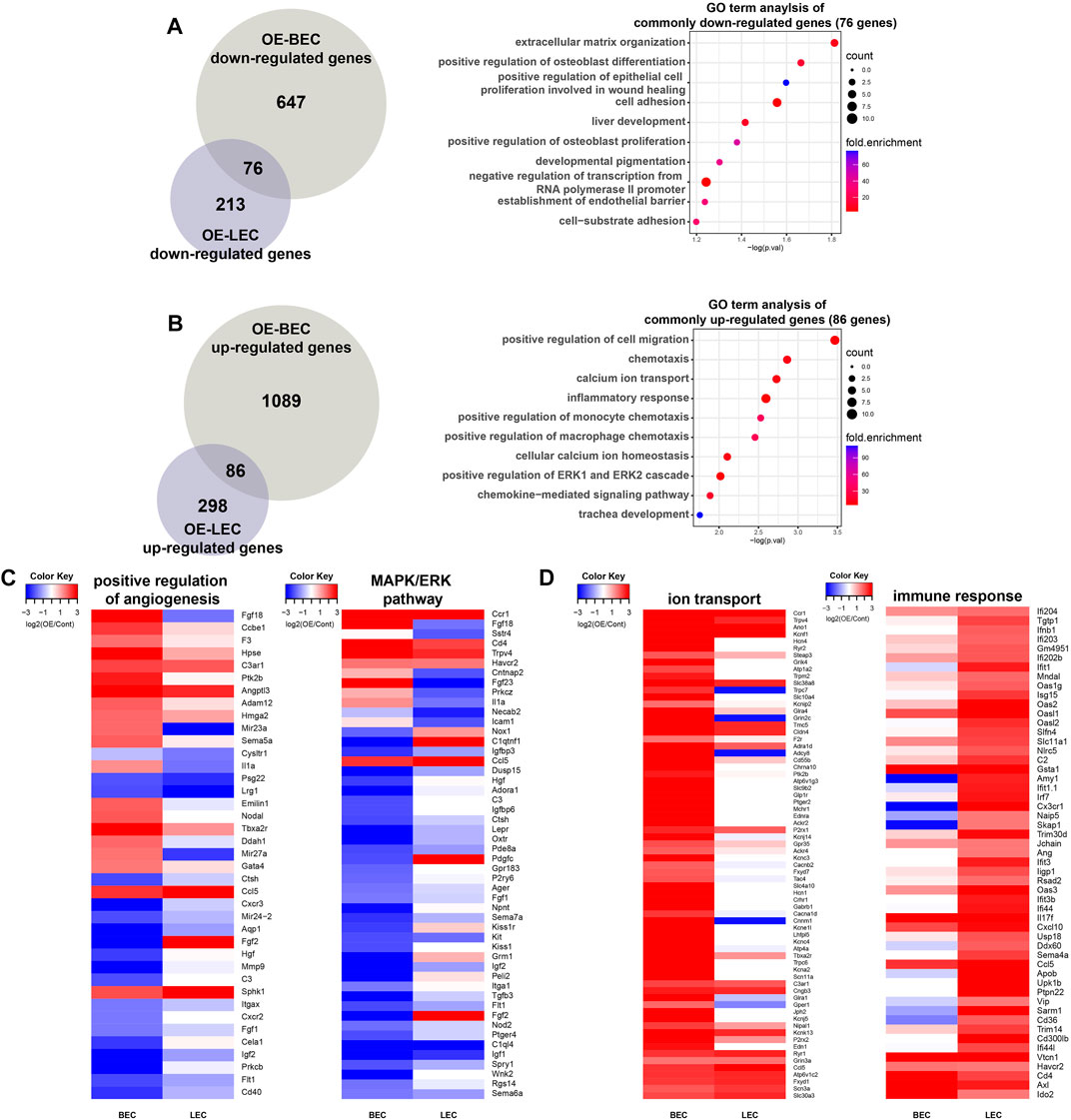
FIGURE 4. Comparison of DEGs between OE-BECs and OE-LECs. (A) Venn diagram showing numbers of common and BEC- and LEC-only downregulated genes in an analysis of GO terms. (B) Venn diagram showing numbers of common and BEC- and LEC-only upregulated genes in an analysis of GO terms. (C) Heatmap representing expression change of genes related to positive regulation of angiogenesis and MAPK/ERK pathway. (D) Heatmap representing expression change of genes related to ion transport and immune response.
4 Discussion
Dot1l has critical roles in cardiovascular development and function. Dot1l loss leads to defects in heart development and function in vivo (Nguyen et al., 2011), LEC development and function in vivo (Yoo et al., 2020), and BEC function in vitro (Duan et al., 2016). Using genetic and epigenomic approaches, we previously provided evidence that Dot1l is essential for LEC differentiation from the hemogenic endothelium and takes part in the transcriptional regulation process. Consistent with our previous study, another study showed that Dot1l is a critical factor that regulates human LEC migration (Williams et al., 2017). A genome-wide study showed that LECs require multiple-layered signaling pathways systemically regulated by each other. However, the function of Dot1l in BECs, especially function in transcriptional regulation, remains unclear. Furthermore, to the best of our knowledge, no study has directly compared the transcriptomes of BECs and LECs in which epigenetic factors are depleted. However, it has been clearly shown that epigenetic mechanisms involved in establishing, diversifying, and maintaining endothelial function depend on their location in different organs, their function, and cell type (BEC vs. LEC) (Bronneke et al., 2012; Yan and Marsden, 2015; Schlereth et al., 2018; Nakato et al., 2019; Gutierrez et al., 2021; Tacconi et al., 2021).
A well-known regulatory function of Dot1l in gene transcription is mediated through H3K79 methylation. The H3K79 methylation promotes the euchromatin state and consequent transcription factor recruitment and transcription initiation (Steger et al., 2008; Wang et al., 2008; Sarno et al., 2020). Using the previous H3K79me2 ChIP-seq analysis in cKO LECs after EPZ-5676 treatment, we found decrease in methylation level of gene bodies of Dot1l cKO common downregulated DEGs (Supplementary Figure S1C). H3K79 di-/tri-methylation is strongly correlated with active gene transcription (Steger et al., 2008) and other active histone markers in mammalian cells. Dot1l knockdown impaired the differentiation potential of KIND1 and HES3 cells by reducing NKX2.5 expression (Pursani et al., 2018) and Dot1l cKO caused lymph vessel development failure by reducing Sox18, Vegfr3, Ramp2, Foxc2, Efnb2, and Eph4 expression. On the other hand, Dot1l-mediated transcriptional repression has also been reported in a number of studies. H3K79me3 has been demonstrated to localize more at silent promoters than active promoters except for narrow region surrounding TSS (Barski et al., 2007). Dot1l-depleted hematopoietic progenitor cells (HPCs) exhibited increased expression of genes involved in HPC differentiation induction (Borosha et al., 2022). Non-cardiomyocyte related genes were increased in Dot1l KO neonatal cardiomyocytes (Cattaneo et al., 2022). GO term analysis of SGC0946 Dot1l inhibitor treatment or Dot1l KO upregulated genes found enrichment of inflammatory response and related terms in macrophages (Willemsen et al., 2022). Consistently, our analysis suggests repressive activity of Dot1l by demonstrating that Dot1l cKO can induce increased gene expression and Dot1l OE can induce reduced gene expression. Specifically, genes associated with immune responses were increased in Dot1l cKO BECs and LECs and genes associated with cell adhesion was repressed in OE-BECs and OE-LECs (Supplementary Figure S1B; Figure 4A). Furthermore, the decrease of H3K79 methylation level in the gene bodies of common none-DEGs suggests that Dot1l could be involved in transcription independent functions such as DNA repair (Huyen et al., 2004) and increased H3K79me2 level does not necessarily result in increased gene transcription (Supplementary Figure S1C). Further research is required to identify factors working in conjunction with Dot1l or Dot1l-mediated H3K79 methylation in transcription regulation.
Neighboring ECs are tightly held together via the expression of various types of adhesion molecules by the cells. These include tight junction species, including junctional adhesion molecules, EC-selective adhesion molecules, and claudins, as well as adherens junction species that include vascular endothelial cadherin (Dejana et al., 2009; Giannotta et al., 2013; Reglero-Real et al., 2016). Furthermore, cell adhesion molecules, such as claudin 1 (CLDN1) and CLDN11, are differentially expressed in an organotypic manner (Nakato et al., 2019). It is interesting to note that CLDN11 expression was repressed in both cKO BECs and cKO LECs. These DEGs are most likely mediated by differential epigenetic marks and/or priming (Matouk and Marsden, 2008; Turgeon et al., 2020). Enrichment of GO term hemophilic cell adhesion via plasma membrane adhesion molecules was also highlighted in our analysis of genes diminished in both cKO EC types. Protocadherin beta 2 (Pcdhb2), Pcdhb18, Pcdhb, Pcdhb3, Pcdhb12, Pcdhb11, and Pcdhb10 genes were found to be involved (Supplementary Figure S1A). The PCDH family is a member of the cadherin superfamily and is essential for neural development (Sano et al., 1993; Hayashi and Takeichi, 2015). PCDHs can be divided into two subgroups: clustered and non-clustered PCDHs (Colas-Algora and Millan, 2019). The genes encoding clustered PCDHs include the PCDHα, β, and γ families and they are located closely to each other within a small region in the genome, while the genes encoding non-clustered PCDHs are dispersed throughout the genome. In our analysis, the expression change was prominent in the PCDH β family. The function of PCDHB genes should be further explored, as no study has clearly provided their function in EC development.
Owing to their location in vessels, ECs are continuously exposed to pathogens, immunogenic signals, or stimuli (Amersfoort et al., 2022). Therefore, it is important that ECs promptly respond to these signals via gene transcription. Evidence suggests that a subset of ECs acts as immune cells by modulating immunity (Pober and Sessa, 2007; Shao et al., 2020). Furthermore, some ECs display typical features of immune cells, in which ECs express co-inhibitory and co-stimulatory receptors (Carman and Martinelli, 2015), induce apoptosis of tumor cells (Motz et al., 2014), secrete cytokines, and act as antigen-presenting cells (Daar et al., 1984a; Daar et al., 1984b). Endothelial hepatitis A virus cellular receptor 2 that is upregulated in both cKO ECs enhances defense mechanism against Rickettsia heilongjiangensis (Yang et al., 2016).
In our analyses, cell migration was ranked as the most enriched GO term in the analysis of commonly upregulated genes in OE BECs and LECs. C-X-C motif chemokine ligand 10, one of the upregulated genes in OE-Dot1l BECs and LECs, promotes the migration of cardiac microvascular ECs (CMEC), likely by regulating the p38/focal adhesion kinase pathways without changing CMEC proliferation and viability (Xia et al., 2016). A recent study showed that LETR1, an LEC-dominant long noncoding RNA, modulates the expression of semaphorin 3C, one of the upregulated genes in OE-Dot1l BECs and LECs, and promotes the growth and migration of LECs by changing chromatin structure (Ducoli et al., 2021). Williams et al. have performed a series of siRNA screening assays by targeting individual protein-coding genes and identified 111 genes critical for BEC and/or LEC migration (Williams et al., 2017). We have found that the mouse homologs of their screening results were present in our DEG list. Among the overlapping genes, Cwc22 has been identified to be downregulated in both cKO ECs but not in the OE ECs, and Hpdl and Ugt1a9 were upregulated in both OE ECs and not in the cKO ECs. On the other hand, Abcc3 and Gcgr were upregulated in both cKO ECs and not in the OE ECs. Cwc22, Hpdl and Ugt1a9 represent the transcriptional activation, and Abcc3 and Gcgr represents the transcriptional repression mediated by Dot1l in genes related to cell migration (data not shown). Furthermore, using the mouse LEC H3K79me2 ChIP data from our previous research, we have confirmed that H3K79me2 levels in their gene bodies reduce when Dot1l function is inhibited by EPZ-5676, providing evidence for Dot1l regulated transcription (data not shown). Genes which do not show a Dot1l-dependent expression pattern are expected to be regulated by Dot1l-independent mechanism or represent the difference between the mouse and human ECs.
Positive regulation of the ERK1/ERK2 cascade was identified as the one of the most significantly enriched GO term among the uniquely downregulated genes of OE-LECs (Figure 3F). VEGFR3 signaling activates ERK1/2 signaling and induces LEC proliferation (Qin et al., 2021). Interestingly, OE-Dot1l in BECs selectively led to repression of genes related to positive regulation of angiogenesis, whereas OE-Dot1l in the LECs induced a set of genes involved in the function of cell adhesion. In contrast, cellular response to interferon-beta related genes were enriched in OE-LECs upregulated gene set. The comprehensive analysis demonstrates EC type-dependent transcriptional regulation of angiogenesis-related genes by Dot1l. Histone modifications are significant contributors to the regulation of angiogenic gene expression. A recent study clearly showed common (or core) and diverse gene signatures and epigenetic landscapes across developmental time and organotypic space in developing mouse and adult BECs (Gutierrez et al., 2021). Our study further demonstrates that a single epigenetic factor can differentially function and build-up unique chromatin structure to regulate gene transcription even in the developmentally close cells. Recently, Kanki et al. (2022) showed that the genes of immediate-early angiogenic transcription factors exclusively acquire protein regulator of cytokinesis (PRC)1.3/PRC2-mediated bivalent histone markers (both H3K4me3-and H3K27me3-positive) after VEGF stimulus. In addition, Ezh2, a component of the PRC2 complex, inhibits the endothelial expression of Fosl1, Creb3l1, Klf5, and Mmp9 to maintain vascular integrity during embryonic development (Delgado-Olguin et al., 2014). Lysine demethylase 3A (Kdm3a) expression is induced in BECs by hypoxia. Its elevated expression accelerates matrix metalloproteinase 12 expression via histone H3K9 demethylation and facilitates trophoblast invasion and uterine vascular remodeling (Chakraborty et al., 2016). The Jmjd6 histone arginine demethylase regulates the expression and splicing of the angiogenic factor Flt1 and controls the angiogenic sprouting of BECs (Boeckel et al., 2011).
Gene OE experiments frequently result in suboptimal physiological protein levels, potentially enabling the formation of transcriptional complexes that would not typically occur (Moriya, 2015). Nevertheless, ECs in our OE experiments exhibited 2∼3-fold upregulation of Dot1l expression, suggesting that it falls within the physiological range (Figure 3B). Therefore, the result of our study can be used for understanding the Dot1l-mediated transcription regulation. As for Dot1l expression in disease condition, it was shown to potentially contribute to the development of multiple cancers including leukemia (Okada et al., 2005; Bernt et al., 2011), breast cancer (Cho et al., 2015; Nassa et al., 2019), ovarian cancer (Zhang et al., 2017), prostate cancer (Vatapalli et al., 2020), and other cancers (Jacinto et al., 2009; Campbell et al., 2016; Zhu et al., 2018; Liu et al., 2020). Given that tumor metastasis is positively or negatively mediated by lymphatics (Dieterich et al., 2022; La et al., 2022), the identified genes regulated by Dot1l can be targets for the detection or prevention of cancer progression. Indeed, studies suggest that Dot1l itself can serve as a prognostic biomarker for ovarian cancer and gastric cancer (Zhang et al., 2017; Song et al., 2020), and pharmacological inhibition of its activities with EPZ-5676 has a beneficial effect in patients with mixed lineage leukemia (Stein et al., 2018).
In the present study, we directly compared the capability of mouse Dot1l in the regulation of gene transcription in BECs and LECs. Common and distinct sets of genes regulated by Dot1l were identified depending on EC types and H3K79me2 levels in LEC has also provided insight into the relationship between epigenetic modification and transcriptional regulation. To the best of our knowledge, this is the first study to delineate the regulatory functions of a specific epigenetic factor in two different ECs, in which its function is abolished or strengthened. Nevertheless, analysis of H3K79 methylation in BECs and investigation of histone methylation-independent transcriptional change in aberrant Dot1l expression could provide more thorough understanding of molecular mechanism governing EC type-specific functions. Additionally, follow-up studies on changes and differences in transcriptomic and epigenetic landscapes in developing and adult ECs in multiple organs would be helpful in precisely understanding the cellular heterogeneity of ECs.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: [GSE214945]. Publicly available datasets were analyzed in this study. This data can be found here: [GSE104811]. Links to the repositories are as follows GSE214945: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214945, GSE104811: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104811.
Ethics statement
The animal study was reviewed and approved by Institute of Animal Care and Use Committee of Konkuk University (IACUC#KU18027).
Author contributions
KH conceptualized the idea and KH, HY, and HL (2nd author) participated in the designing of the manuscript. HY and HL (2nd author), CP, SY, HL (5th author), HS, JTD, YC, and KH conducted the experiments and validated the results. HY and HL (2nd author), CP, SY, HL (5th author), HS, JTD, YC, and KH performed RNA-seq and downstream bioinformatics analysis. HY and HL (2nd author), CP, SY, HL (5th author), HS, JTD, YC, and KH conducted investigation on previous research. HY and HL (2nd author), CP, SY, HL (5th author), HS, JTD, YC, and KH prepared the original draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Konkuk University (2022).
Acknowledgments
The authors are indebted to all the members of the KH lab for helpful discussion.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1176115/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | (A) Heatmap representing expression change of genes under the GO term immune response, glucose metabolism and cell-to-cell adhesion. (B) GO terms identified for commonly up- or down-regulated genes. (C) H3K79me2 profiles on DMSO and EPZ-5676 treated LEC. The gene body profiles of none-DEGs, down- and up-regulated DEGs are shown. (D) IGV visualization of Cont and cKO EC RNA-seq for representative genes.
SUPPLEMENTARY FIGURE S2 | Results of Cont and cKO EC comparison in 18 child terms of the term “immune response” shown in GSEA enrichment plot.
SUPPLEMENTARY FIGURE S3 | (A) RT-qPCR results of Mmp3 and Cldn1. Gene expression is normalized to the expression value of Gapdh in Cont and OE EC. (n=2) (B) IGV track visualization of Cont and OE EC RNA-seq for Mmp3 and Cldn1. (C) Heatmap representing expression change of Dot1l dependent genes. (D) Upper panel shows the GO term analysis results for the Dot1l dependent genes in the BECs and the lower panel shows the GO term analysis results for the LECs.
References
Aird, W. C. (2007a). Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 100 (2), 158–173. doi:10.1161/01.RES.0000255691.76142.4a
Aird, W. C. (2007b). Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ. Res. 100 (2), 174–190. doi:10.1161/01.RES.0000255690.03436.ae
Aird, W. C. (2012). Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2 (1), a006429. doi:10.1101/cshperspect.a006429
Amersfoort, J., Eelen, G., and Carmeliet, P. (2022). Immunomodulation by endothelial cells - partnering up with the immune system? Nat. Rev. Immunol. 22 (9), 576–588. doi:10.1038/s41577-022-00694-4
Aquino, J. B., Sierra, R., and Montaldo, L. A. (2021). Diverse cellular origins of adult blood vascular endothelial cells. Dev. Biol. 477, 117–132. doi:10.1016/j.ydbio.2021.05.010
Barry, E. R., Krueger, W., Jakuba, C. M., Veilleux, E., Ambrosi, D. J., Nelson, C. E., et al. (2009). ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 27 (7), 1538–1547. doi:10.1002/stem.86
Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell. 129 (4), 823–837. doi:10.1016/j.cell.2007.05.009
Becker, L. M., Chen, S. H., Rodor, J., de Rooij, L., Baker, A. H., and Carmeliet, P. (2022). Deciphering endothelial heterogeneity in health and disease at single cell resolution: Progress and perspectives. Cardiovasc Res. 119, 6–27. doi:10.1093/cvr/cvac018
Bernt, K. M., Zhu, N., Sinha, A. U., Vempati, S., Faber, J., Krivtsov, A. V., et al. (2011). MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 20 (1), 66–78. doi:10.1016/j.ccr.2011.06.010
Boeckel, J. N., Guarani, V., Koyanagi, M., Roexe, T., Lengeling, A., Schermuly, R. T., et al. (2011). Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U. S. A. 108 (8), 3276–3281. doi:10.1073/pnas.1008098108
Borosha, S., Ratri, A., Ghosh, S., Malcom, C. A., Chakravarthi, V. P., Vivian, J. L., et al. (2022). DOT1L mediated gene repression in extensively self-renewing erythroblasts. Front. Genet. 13, 828086. doi:10.3389/fgene.2022.828086
Bronneke, S., Bruckner, B., Peters, N., Bosch, T. C., Stab, F., Wenck, H., et al. (2012). DNA methylation regulates lineage-specifying genes in primary lymphatic and blood endothelial cells. Angiogenesis 15 (2), 317–329. doi:10.1007/s10456-012-9264-2
Campbell, J. D., Alexandrov, A., Kim, J., Wala, J., Berger, A. H., Pedamallu, C. S., et al. (2016). Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48 (6), 607–616. doi:10.1038/ng.3564
Carbon, S., Ireland, A., Mungall, C. J., Shu, S., Marshall, B., Lewis, S., et al. (2009). AmiGO: Online access to ontology and annotation data. Bioinformatics 25 (2), 288–289. doi:10.1093/bioinformatics/btn615
Carman, C. V., and Martinelli, R. (2015). T lymphocyte-endothelial interactions: Emerging understanding of trafficking and antigen-specific immunity. Front. Immunol. 6, 603. doi:10.3389/fimmu.2015.00603
Cattaneo, P., Hayes, M. G. B., Baumgarten, N., Hecker, D., Peruzzo, S., Aslan, G. S., et al. (2022). DOT1L regulates chamber-specific transcriptional networks during cardiogenesis and mediates postnatal cell cycle withdrawal. Nat. Commun. 13 (1), 7444. doi:10.1038/s41467-022-35070-2
Chakraborty, D., Cui, W., Rosario, G. X., Scott, R. L., Dhakal, P., Renaud, S. J., et al. (2016). HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. U. S. A. 113 (46), E7212–E7221. doi:10.1073/pnas.1612626113
Cho, M. H., Park, J. H., Choi, H. J., Park, M. K., Won, H. Y., Park, Y. J., et al. (2015). DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat. Commun. 6, 7821. doi:10.1038/ncomms8821
Chong, D. C., Koo, Y., Xu, K., Fu, S., and Cleaver, O. (2011). Stepwise arteriovenous fate acquisition during mammalian vasculogenesis. Dev. Dyn. 240 (9), 2153–2165. doi:10.1002/dvdy.22706
Chung, Y. S., Zhang, W. J., Arentson, E., Kingsley, P. D., Palis, J., and Choi, K. (2002). Lineage analysis of the hemangioblast as defined by FLK1 and SCL expression. Development 129 (23), 5511–5520. doi:10.1242/dev.00149
Colas-Algora, N., and Millan, J. (2019). How many cadherins do human endothelial cells express? Cell. Mol. Life Sci. 76 (7), 1299–1317. doi:10.1007/s00018-018-2991-9
Daar, A. S., Fuggle, S. V., Fabre, J. W., Ting, A., and Morris, P. J. (1984a). The detailed distribution of HLA-A, B, C antigens in normal human organs. Transplantation 38 (3), 287–292. doi:10.1097/00007890-198409000-00018
Daar, A. S., Fuggle, S. V., Fabre, J. W., Ting, A., and Morris, P. J. (1984b). The detailed distribution of MHC Class II antigens in normal human organs. Transplantation 38 (3), 293–298. doi:10.1097/00007890-198409000-00019
Dejana, E., Tournier-Lasserve, E., and Weinstein, B. M. (2009). The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell. 16 (2), 209–221. doi:10.1016/j.devcel.2009.01.004
Delgado-Olguin, P., Dang, L. T., He, D., Thomas, S., Chi, L., Sukonnik, T., et al. (2014). Ezh2-mediated repression of a transcriptional pathway upstream of Mmp9 maintains integrity of the developing vasculature. Development 141 (23), 4610–4617. doi:10.1242/dev.112607
Dieterich, L. C., Tacconi, C., Ducoli, L., and Detmar, M. (2022). Lymphatic vessels in cancer. Physiol. Rev. 102 (4), 1837–1879. doi:10.1152/physrev.00039.2021
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). Star: Ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi:10.1093/bioinformatics/bts635
Drake, C. J., and Fleming, P. A. (2000). Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood 95 (5), 1671–1679. doi:10.1182/blood.v95.5.1671.005k39_1671_1679
Duan, Y., Wu, X., Zhao, Q., Gao, J., Huo, D., Liu, X., et al. (2016). DOT1L promotes angiogenesis through cooperative regulation of VEGFR2 with ETS-1. Oncotarget 7 (43), 69674–69687. doi:10.18632/oncotarget.11939
Ducoli, L., Agrawal, S., Sibler, E., Kouno, T., Tacconi, C., Hon, C. C., et al. (2021). LETR1 is a lymphatic endothelial-specific lncRNA governing cell proliferation and migration through KLF4 and SEMA3C. Nat. Commun. 12 (1), 925. doi:10.1038/s41467-021-21217-0
Feng, Q., Wang, H., Ng, H. H., Erdjument-Bromage, H., Tempst, P., Struhl, K., et al. (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12 (12), 1052–1058. doi:10.1016/s0960-9822(02)00901-6
Francois, M., Short, K., Secker, G. A., Combes, A., Schwarz, Q., Davidson, T. L., et al. (2012). Segmental territories along the cardinal veins generate lymph sacs via a ballooning mechanism during embryonic lymphangiogenesis in mice. Dev. Biol. 364 (2), 89–98. doi:10.1016/j.ydbio.2011.12.032
Galley, H. F., and Webster, N. R. (2004). Physiology of the endothelium. Br. J. Anaesth. 93 (1), 105–113. doi:10.1093/bja/aeh163
Giannotta, M., Trani, M., and Dejana, E. (2013). VE-Cadherin and endothelial adherens junctions: Active guardians of vascular integrity. Dev. Cell. 26 (5), 441–454. doi:10.1016/j.devcel.2013.08.020
Gutierrez, M. E. C., Hill, M. C., Largoza, G., Martin, J. F., and Wythe, J. D. (2021). Defining the transcriptional and epigenetic basis of organotypic endothelial diversity in the developing and adult mouse. bioRxiv. doi:10.1101/2021.11.15.468651
Hayashi, S., and Takeichi, M. (2015). Emerging roles of protocadherins: From self-avoidance to enhancement of motility. J. Cell. Sci. 128 (8), 1455–1464. doi:10.1242/jcs.166306
Heil, M., Eitenmuller, I., Schmitz-Rixen, T., and Schaper, W. (2006). Arteriogenesis versus angiogenesis: Similarities and differences. J. Cell. Mol. Med. 10 (1), 45–55. doi:10.1111/j.1582-4934.2006.tb00290.x
Hou, S., Li, Z., Dong, J., Gao, Y., Chang, Z., Ding, X., et al. (2022). Heterogeneity in endothelial cells and widespread venous arterialization during early vascular development in mammals. Cell. Res. 32 (4), 333–348. doi:10.1038/s41422-022-00615-z
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 (1), 44–57. doi:10.1038/nprot.2008.211
Huyen, Y., Zgheib, O., Ditullio, R. A., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., et al. (2004). Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432 (7015), 406–411. doi:10.1038/nature03114
Jacinto, F. V., Ballestar, E., and Esteller, M. (2009). Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene 28 (47), 4212–4224. doi:10.1038/onc.2009.267
Jain, R. K. (2003). Molecular regulation of vessel maturation. Nat. Med. 9 (6), 685–693. doi:10.1038/nm0603-685
Jambusaria, A., Hong, Z., Zhang, L., Srivastava, S., Jana, A., Toth, P. T., et al. (2020). Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife 9, e51413. doi:10.7554/eLife.51413
Jiang, H., Lei, R., Ding, S. W., and Zhu, S. (2014). Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinforma. 15, 182. doi:10.1186/1471-2105-15-182
Jones, B., Su, H., Bhat, A., Lei, H., Bajko, J., Hevi, S., et al. (2008). The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4 (9), e1000190. doi:10.1371/journal.pgen.1000190
Kanki, Y., Muramatsu, M., Miyamura, Y., Kikuchi, K., Higashijima, Y., Nakaki, R., et al. (2022). Bivalent-histone-marked immediate-early gene regulation is vital for VEGF-responsive angiogenesis. Cell. Rep. 38 (6), 110332. doi:10.1016/j.celrep.2022.110332
Kealy, L., Di Pietro, A., Hailes, L., Scheer, S., Dalit, L., Groom, J. R., et al. (2020). The histone methyltransferase DOT1L is essential for humoral immune responses. Cell. Rep. 33 (11), 108504. doi:10.1016/j.celrep.2020.108504
Klotz, L., Norman, S., Vieira, J. M., Masters, M., Rohling, M., Dube, K. N., et al. (2015). Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 522 (7554), 62–67. doi:10.1038/nature14483
La, H., Yoo, H., Park, Y. B., Thang, N. X., Park, C., Yoo, S., et al. (2022). Role of transcriptional and epigenetic regulation in lymphatic endothelial cell development. Cells 11 (10), 1692. doi:10.3390/cells11101692
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Lin, J. D. (2007). Papillary thyroid carcinoma with lymph node metastases. Growth factors. 25 (1), 41–49. doi:10.1080/08977190601000095
Lioux, G., Liu, X., Temino, S., Oxendine, M., Ayala, E., Ortega, S., et al. (2020). A second heart field-derived vasculogenic niche contributes to cardiac lymphatics. Dev. Cell. 52 (3), 350–363. doi:10.1016/j.devcel.2019.12.006
Liu, C., Yang, Q., Zhu, Q., Lu, X., Li, M., Hou, T., et al. (2020). CBP mediated DOT1L acetylation confers DOT1L stability and promotes cancer metastasis. Theranostics 10 (4), 1758–1776. doi:10.7150/thno.39013
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lupu, I.-E., Kirschnick, N., Weischer, S., Martinez-Corral, I., Forrow, A., Lahmann, I., et al. (2022). Direct specification of lymphatic endothelium from non-venous angioblasts. bioRxiv. doi:10.1101/2022.05.11.491403
Majesky, M. W. (2018). Vascular development. Arterioscler. Thromb. Vasc. Biol. 38 (3), e17–e24. doi:10.1161/ATVBAHA.118.310223
Martinez-Corral, I., Ulvmar, M. H., Stanczuk, L., Tatin, F., Kizhatil, K., John, S. W., et al. (2015). Nonvenous origin of dermal lymphatic vasculature. Circ. Res. 116 (10), 1649–1654. doi:10.1161/CIRCRESAHA.116.306170
Maruyama, K., Miyagawa-Tomita, S., Mizukami, K., Matsuzaki, F., and Kurihara, H. (2019). Isl1-expressing non-venous cell lineage contributes to cardiac lymphatic vessel development. Dev. Biol. 452 (2), 134–143. doi:10.1016/j.ydbio.2019.05.002
Matouk, C. C., and Marsden, P. A. (2008). Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 102 (8), 873–887. doi:10.1161/CIRCRESAHA.107.171025
McCarron, J. G., Lee, M. D., and Wilson, C. (2017). The endothelium solves problems that endothelial cells do not know exist. Trends Pharmacol. Sci. 38 (4), 322–338. doi:10.1016/j.tips.2017.01.008
Moriya, H. (2015). Quantitative nature of overexpression experiments. Mol. Biol. Cell. 26 (22), 3932–3939. doi:10.1091/mbc.E15-07-0512
Motz, G. T., Santoro, S. P., Wang, L. P., Garrabrant, T., Lastra, R. R., Hagemann, I. S., et al. (2014). Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 20 (6), 607–615. doi:10.1038/nm.3541
Mueller, D., Bach, C., Zeisig, D., Garcia-Cuellar, M. P., Monroe, S., Sreekumar, A., et al. (2007). A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110 (13), 4445–4454. doi:10.1182/blood-2007-05-090514
Nakato, R., Wada, Y., Nakaki, R., Nagae, G., Katou, Y., Tsutsumi, S., et al. (2019). Comprehensive epigenome characterization reveals diverse transcriptional regulation across human vascular endothelial cells. Epigenetics Chromatin 12 (1), 77. doi:10.1186/s13072-019-0319-0
Nassa, G., Salvati, A., Tarallo, R., Gigantino, V., Alexandrova, E., Memoli, D., et al. (2019). Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells. Sci. Adv. 5 (2), eaav5590. doi:10.1126/sciadv.aav5590
Nguyen, A. T., Xiao, B., Neppl, R. L., Kallin, E. M., Li, J., Chen, T., et al. (2011). DOT1L regulates dystrophin expression and is critical for cardiac function. Genes. Dev. 25 (3), 263–274. doi:10.1101/gad.2018511
Okada, Y., Feng, Q., Lin, Y., Jiang, Q., Li, Y., Coffield, V. M., et al. (2005). hDOT1L links histone methylation to leukemogenesis. Cell. 121 (2), 167–178. doi:10.1016/j.cell.2005.02.020
Pichol-Thievend, C., Betterman, K. L., Liu, X., Ma, W., Skoczylas, R., Lesieur, E., et al. (2018). A blood capillary plexus-derived population of progenitor cells contributes to Genesis of the dermal lymphatic vasculature during embryonic development. Development 145 (10), dev160184. doi:10.1242/dev.160184
Pober, J. S., and Sessa, W. C. (2007). Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7 (10), 803–815. doi:10.1038/nri2171
Potente, M., and Makinen, T. (2017). Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell. Biol. 18 (8), 477–494. doi:10.1038/nrm.2017.36
Pursani, V., Bhartiya, D., Tanavde, V., Bashir, M., and Sampath, P. (2018). Transcriptional activator DOT1L putatively regulates human embryonic stem cell differentiation into the cardiac lineage. Stem Cell. Res. Ther. 9 (1), 97. doi:10.1186/s13287-018-0810-8
Qin, L., Zhang, H., Li, B., Jiang, Q., Lopez, F., Min, W., et al. (2021). CCM3 loss-induced lymphatic defect is mediated by the augmented VEGFR3-ERK1/2 signaling. Arterioscler. Thromb. Vasc. Biol. 41 (12), 2943–2960. doi:10.1161/ATVBAHA.121.316707
Ramirez, F., Ryan, D. P., Gruning, B., Bhardwaj, V., Kilpert, F., Richter, A. S., et al. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165. doi:10.1093/nar/gkw257
Reglero-Real, N., Colom, B., Bodkin, J. V., and Nourshargh, S. (2016). Endothelial cell junctional adhesion molecules: Role and regulation of expression in inflammation. Arterioscler. Thromb. Vasc. Biol. 36 (10), 2048–2057. doi:10.1161/ATVBAHA.116.307610
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29 (1), 24–26. doi:10.1038/nbt.1754
Sano, K., Tanihara, H., Heimark, R. L., Obata, S., Davidson, M., St John, T., et al. (1993). Protocadherins: A large family of cadherin-related molecules in central nervous system. EMBO J. 12 (6), 2249–2256. doi:10.1002/j.1460-2075.1993.tb05878.x
Sarno, F., Nebbioso, A., and Altucci, L. (2020). DOT1L: A key target in normal chromatin remodelling and in mixed-lineage leukaemia treatment. Epigenetics 15 (5), 439–453. doi:10.1080/15592294.2019.1699991
Schlereth, K., Weichenhan, D., Bauer, T., Heumann, T., Giannakouri, E., Lipka, D., et al. (2018). The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. Elife 7, e34423. doi:10.7554/eLife.34423
Shao, Y., Saredy, J., Yang, W. Y., Sun, Y., Lu, Y., Saaoud, F., et al. (2020). Vascular endothelial cells and innate immunity. Arterioscler. Thromb. Vasc. Biol. 40 (6), e138–e152. doi:10.1161/ATVBAHA.120.314330
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). David: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Song, Z., Wei, Z., Wang, Q., Zhang, X., Tao, X., Wu, N., et al. (2020). The role of DOT1L in the proliferation and prognosis of gastric cancer. Biosci. Rep. 40 (1). doi:10.1042/BSR20193515
Srinivasan, R. S., Dillard, M. E., Lagutin, O. V., Lin, F. J., Tsai, S., Tsai, M. J., et al. (2007). Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes. Dev. 21 (19), 2422–2432. doi:10.1101/gad.1588407
Steger, D. J., Lefterova, M. I., Ying, L., Stonestrom, A. J., Schupp, M., Zhuo, D., et al. (2008). DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28 (8), 2825–2839. doi:10.1128/MCB.02076-07
Stein, E. M., Garcia-Manero, G., Rizzieri, D. A., Tibes, R., Berdeja, J. G., Savona, M. R., et al. (2018). The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131 (24), 2661–2669. doi:10.1182/blood-2017-12-818948
Stone, O. A., and Stainier, D. Y. R. (2019). Paraxial mesoderm is the major source of lymphatic endothelium. Dev. Cell. 50 (2), 247–255. doi:10.1016/j.devcel.2019.04.034
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Tacconi, C., He, Y., Ducoli, L., and Detmar, M. (2021). Epigenetic regulation of the lineage specificity of primary human dermal lymphatic and blood vascular endothelial cells. Angiogenesis 24 (1), 67–82. doi:10.1007/s10456-020-09743-9
Tammela, T., and Alitalo, K. (2010). Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 140 (4), 460–476. doi:10.1016/j.cell.2010.01.045
Torres-Vazquez, J., Kamei, M., and Weinstein, B. M. (2003). Molecular distinction between arteries and veins. Cell. Tissue Res. 314 (1), 43–59. doi:10.1007/s00441-003-0771-8
Turgeon, P. J., Chan, G. C., Chen, L., Jamal, A. N., Yan, M. S., Ho, J. J. D., et al. (2020). Epigenetic heterogeneity and mitotic heritability prime endothelial cell gene induction. J. Immunol. 204 (5), 1173–1187. doi:10.4049/jimmunol.1900744
Vatapalli, R., Sagar, V., Rodriguez, Y., Zhao, J. C., Unno, K., Pamarthy, S., et al. (2020). Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer. Nat. Commun. 11 (1), 4153. doi:10.1038/s41467-020-18013-7
Wang, Z., Zang, C., Rosenfeld, J. A., Schones, D. E., Barski, A., Cuddapah, S., et al. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40 (7), 897–903. doi:10.1038/ng.154
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., et al. (2022). gplots: Various R programming tools for plotting data. R package version 3.1.3 ed.
Wigle, J. T., and Oliver, G. (1999). Prox1 function is required for the development of the murine lymphatic system. Cell. 98 (6), 769–778. doi:10.1016/s0092-8674(00)81511-1
Willemsen, L., Prange, K. H. M., Neele, A. E., van Roomen, C., Gijbels, M., Griffith, G. R., et al. (2022). DOT1L regulates lipid biosynthesis and inflammatory responses in macrophages and promotes atherosclerotic plaque stability. Cell. Rep. 41 (8), 111703. doi:10.1016/j.celrep.2022.111703
Williams, S. P., Odell, A. F., Karnezis, T., Farnsworth, R. H., Gould, C. M., Li, J., et al. (2017). Genome-wide functional analysis reveals central signaling regulators of lymphatic endothelial cell migration and remodeling. Sci. Signal 10 (499), eaal2987. doi:10.1126/scisignal.aal2987
Xia, J. B., Mao, C. Z., Chen, Z. Y., Liu, G. H., Wu, H. Y., Zhou, D. C., et al. (2016). The CXCL10/CXCR3 axis promotes cardiac microvascular endothelial cell migration via the p38/FAK pathway in a proliferation-independent manner. Exp. Mol. Pathol. 100 (2), 257–265. doi:10.1016/j.yexmp.2016.01.010
Yan, M. S., and Marsden, P. A. (2015). Epigenetics in the vascular endothelium: Looking from a different perspective in the epigenomics era. Arterioscler. Thromb. Vasc. Biol. 35 (11), 2297–2306. doi:10.1161/ATVBAHA.115.305043
Yang, X., Jiao, J., Han, G., Gong, W., Wang, P., Xiong, X., et al. (2016). Enhanced expression of T-cell immunoglobulin and mucin domain protein 3 in endothelial cells facilitates intracellular killing of Rickettsia heilongjiangensis. J. Infect. Dis. 213 (1), 71–79. doi:10.1093/infdis/jiv463
Yoo, H., Lee, Y. J., Park, C., Son, D., Choi, D. Y., Park, J. H., et al. (2020). Epigenetic priming by Dot1l in lymphatic endothelial progenitors ensures normal lymphatic development and function. Cell. Death Dis. 11 (1), 14. doi:10.1038/s41419-019-2201-1
Yu, X., Liu, Y., Liu, X., Xiong, H., and He, A. (2021). Single-cell epigenomic tracing of lifelong endothelial cell plasticity across mouse organs. bioRxiv. doi:10.1101/2021.05.12.443777
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9 (9), R137. doi:10.1186/gb-2008-9-9-r137
Zhang, X., Liu, D., Li, M., Cao, C., Wan, D., Xi, B., et al. (2017). Prognostic and therapeutic value of disruptor of telomeric silencing-1-like (DOT1L) expression in patients with ovarian cancer. J. Hematol. Oncol. 10 (1), 29. doi:10.1186/s13045-017-0400-8
Zhu, Q. F. S., Shallcross, J., and Kim, J. (2016). Verse: A versatile and efficient RNA-seq read counting tool. bioRxiv. doi:10.1101/053306
Keywords: blood endothelial cell, lymphatic endothelial cell, epigenetics, DOT1l, transcriptional regulation
Citation: Yoo H, La H, Park C, Yoo S, Lee H, Song H, Do JT, Choi Y and Hong K (2023) Common and distinct functions of mouse Dot1l in the regulation of endothelial transcriptome. Front. Cell Dev. Biol. 11:1176115. doi: 10.3389/fcell.2023.1176115
Received: 28 February 2023; Accepted: 06 June 2023;
Published: 15 June 2023.
Edited by:
Mellissa Mann, University of Pittsburgh, United StatesReviewed by:
Coral Wille, University of Wisconsin-Madison, United StatesChunsik Lee, Sun Yat-sen University, China
Copyright © 2023 Yoo, La, Park, Yoo, Lee, Song, Do, Choi and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwonho Hong, hongk@konkuk.ac.kr
†These authors have contributed equally to this work
 Hyunjin Yoo†
Hyunjin Yoo†