
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 22 June 2023
Sec. Molecular and Cellular Reproduction
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1174211
This article is part of the Research Topic Omics for Infertility and Contraception: Two Sides of Same Coin View all 5 articles
 Oladele A. Oluwayiose1
Oladele A. Oluwayiose1 Emily Houle1
Emily Houle1 Brian W. Whitcomb2
Brian W. Whitcomb2 Alexander Suvorov3
Alexander Suvorov3 Tayyab Rahil4
Tayyab Rahil4 Cynthia K. Sites4
Cynthia K. Sites4 Stephen A. Krawetz1,5
Stephen A. Krawetz1,5 Pablo E. Visconti6
Pablo E. Visconti6 J. Richard Pilsner1,7*
J. Richard Pilsner1,7*Background: Infertility remains a global health problem with male-factor infertility accounting for around 50% of cases. Understanding the molecular markers for the male contribution of live birth success has been limited. Here, we evaluated the expression levels of seminal plasma extracellular vesicle (spEV) non-coding RNAs (ncRNAs) in men of couples in relation with those with and without a successful live birth after infertility treatment.
Method: Sperm-free spEV small RNA profiles were generated from 91 semen samples collected from male participants of couples undergoing assisted reproductive technology (ART) treatment. Couples were classified into two groups based on successful live birth (yes, n = 28) and (no, n = 63). Mapping of reads to human transcriptomes followed the order: miRNA > tRNA > piRNA > rRNA> “other” RNA > circRNA > lncRNA. Differential expression analysis of biotype-specific normalized read counts between groups were assessed using EdgeR (FDR<0.05).
Result: We found a total of 12 differentially expressed spEV ncRNAs which included 10 circRNAs and two piRNAs between the live birth groups. Most (n = 8) of the identified circRNAs were downregulated in the no live birth group and targeted genes related to ontology terms such as negative reproductive system and head development, tissue morphogenesis, embryo development ending in birth or egg hatching, and vesicle-mediated transport. The differentially upregulated piRNAs overlapped with genomic regions including coding PID1 genes previously known to play a role in mitochondrion morphogenesis, signal transduction and cellular proliferation.
Conclusion: This study identified novel ncRNAs profiles of spEVs differentiating men of couples with and without live birth and emphasizes the role of the male partner for ART success.
Infertility is the inability to conceive within 1 year of regular unprotected sexual intercourse. It remains a global reproductive problem affecting 15% of heterosexual couples and nearly 190 million individuals with the desire to conceive (Rutstein and Shah, 2004; Mascarenhas et al., 2012). The diagnosis of couple infertility involves the evaluation of both male and female factors (Smith et al., 2003; Wright et al., 2003; Campbell et al., 2021). However, approximately 30% of cases are unexplained with unknown etiology when the results of standard infertility evaluations are normal (Quaas and Dokras, 2008).
Due to increased access and delayed parenthood (Kocourkova et al., 2014; van Loendersloot et al., 2014), there is a steady rise in the use of ART including in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) (Sunderam et al., 2017). However, despite that ART has existed for several decades, the global overall success rate, as measured by live birth outcomes following a single embryo transfer, is approximately 30% (Smith et al., 2015; Gleicher et al., 2019; Khalife et al., 2020). This may prompt multiple attempts, constituting physical, financial and/or emotional burden among couples (Domar et al., 1992). Thus, understanding the molecular markers that are associated with live birth outcomes prior to the initiation of the ART treatment cycle may provide a pathway to enhance clinical care.
As such, profiling seminal plasma to identify novel markers of reproductive success has been proposed (Crawford et al., 2015; Ayaz et al., 2021; Rodriguez-Martinez et al., 2021). Seminal plasma is enriched in biologically active molecules including coding and non-coding RNAs (Vojtech et al., 2014; Ayaz et al., 2021; Rodriguez-Martinez et al., 2021; Oluwayiose et al., 2022), which are either found freely or encapsulated in extracellular vesicles (EVs). Importantly, there is a growing interest in unraveling the reproductive functions of ncRNAs retained in EVs found in seminal plasma. Findings suggest that injection of ncRNA cargo of seminal plasma EV (spEV) can modulate gene expression, potentially regulating biological processes involved in embryonic developments (Chen et al., 2016; Chen et al., 2021).
Recent findings from our group (Oluwayiose et al., 2022) complement prior evidence of ncRNA enrichment in spEVs (Vojtech et al., 2014; Joshi et al., 2022) and showed these ncRNAs were differentially expressed in men with poor semen quality, suggesting the relevance of spEV ncRNAs in reproductive outcomes. However, whether these spEV ncRNAs also serve as markers of live birth outcomes, the major goal of infertility treatment in clinical setting, is unknown. Thus, this study evaluated the expression of spEV ncRNAs in men of couples undergoing ART treatment in relation to those with and without live birth.
Study population. The study population includes men (n = 91) of couples receiving fertility treatment at Baystate Medical Center located in Springfield, Massachusetts who agreed to participate in the Sperm Environmental Epigenetics and Developments Study (SEEDS). SEEDS is prospective observational cohort study designed to investigate the associations of male preconception endocrine disrupting chemical exposure with sperm epigenetics and subsequent early-life development. The inclusion criteria were male partners of at least 18 years old without vasectomy and fresh ejaculate sperm used for ART treatment while females were included if 40 years or younger. Relevant demographics (race, age, height, and weight), lifestyle factors (current and past alcohol and cigarette use), and medical history (diagnoses of infertility) data were collected by clinic personnel during the ART cycle for both partners. Male fertility diagnoses were based on semen quality parameters according to World Health Organization (WHO) reference values (Cooper et al., 2010) while female diagnoses were based on CDC criteria including polycystic ovarian syndrome, anovulation, tubal factors, diminished ovarian reserve and endometriosis (Wright et al., 2003). Females received ovarian stimulation protocols based on infertility diagnoses and medical history. Egg retrieval was carried out about 36 h after the leading follicle reached 18–20 mm diameter Written informed consent was obtained from eligible males who showed voluntary interest in participating in the study by the attending physicians. The study is approved by the institutional review boards at Baystate Medical Center and at the University of Massachusetts Amherst (IRB number: BH-12-190).
Isolation of EVs from seminal plasma. Following a 2–3 days abstinence period, freshly collected ejaculates were processed using a two-step (40% and 80%) gradient fractionation method (Wu et al., 2015) and the resulting seminal plasma was stored in −80°C until subsequent analysis. To isolate EVs and total RNA, frozen seminal plasma samples were thawed for 20 min at room temperature, followed by extraction of EV RNA isolation using Qiagen exoRNAeasy Midi Kit (Cat. #77144) with slight modification to manufacturer’s protocol (Enderle et al., 2015). Briefly, equal volume of PBS and seminal plasma were mixed, centrifuged at 12,000 g for 45 min at 4°C to pellet sperm and other debris, and the resulting supernatant carefully separated and pre-filtered through a 0.45 μm syringe (Whatman PURADISC 25, Cat.#6780-2505). This was mixed with one volume of DNA binding buffer (XBP) and centrifuged at 500 g for 3 min in a spin column. The column-bound EVs were then purified using XWP buffer and lysed with QIAzol prior to phase-separation with chloroform for total RNA isolation. The aqueous phase containing RNA was mixed with 2 volumes of 100% ethanol and transferred onto a spin column. The column-bound RNA was purified, washed in 80% ethanol and eluted in nuclease free water following by total RNA quantification and quality assessment using nanodrop and fragment analyzer respectively.
Small RNA library preparation and sequencing. End repair of the isolated total RNA was performed using T4 Polynucleotide kinase (NEB, Cat #M0201) according to manufacturer’s instructions. All steps including polyadenylated RNA library preparation, reverse transcription cDNA synthesis, and 6 cycles of PCR amplification followed modified SMARTer smRNA-Seq protocol (Clontech). SPRI beads were used for library cleanup, and size selection and quality assessment were performed using AMPure X beads and fragment analyzer, respectively. Multiplexed samples were loaded onto the flow cell using Illumina barcodes (1—24) and sequencing of 50-bp single-end reads was performed on an Illumina HiSeq 4000. Libraries construction and sequencing were conducted at the UMass Medical Genomics Core Facility.
small RNA sequencing analysis. The quality of the raw reads was assessed using FastQC (Andrews, 2010). Trimming of the first three nucleotides and polyA sequences as well as filtering of reads with low quality (PHRED score < 20) and reads shorter than 16 nt was performed using cutadapt (Martin, 2011). Sequencing contaminants were eliminated by alignment to UniVec database using Bowtie2 with default parameters (Langmead and Salzberg, 2012). Sequential alignment to human transcriptome databases for ncRNA biotypes (Rozowsky et al., 2019; Oluwayiose et al., 2022) followed the order: miRNA > piRNA > tRNA > rRNA>“other ncRNAs”>circRNA > lncRNA using STAR (v2.7.6a) (Dobin et al., 2013). Multi-mapping reads were assigned based on the best mapping quality. We allowed incremental mismatch: no mismatches in the reads ≤25 bases and 1 mismatch in 26–50 bases. Samtools (v1.9) (Li et al., 2009) was used for BAM/SAM data manipulation and counting of mapped reads. All raw and processed sequences were deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE203494. Details of the count output were reported in our previous publication (Oluwayiose et al., 2022).
Data analysis. Biotype-specific counts were normalized using weighted trimmed mean of M-values TMM (Robinson and Oshlack, 2010) and species with normalized count ≤1 RPM in all samples were filtered. Male participants fertility diagnosis was defined based on WHO 5th centile reference cutoff points (Cooper et al., 2010) and were categorized based on couple live birth status (Yes/No, referent = Yes). Live birth was defined as clinically confirmed delivery of a living infant from the current ART treatment cycle. Differential expression analysis between men with and without successful live birth was performed using EdgeR (Robinson et al., 2010). ncRNA species with absolute value of log2 fold change (FC) ≥ 0.585 and FDR<0.05 was considered significant. Student’s t-test or the nonparametric Mann–Whitney U test was used to compare participants’ demographic and clinical characteristics between groups while proportions were compared using chi-square statistics (p < 0.05). All analyses were performed in R (v4.1.0) software.
Gene target determination and functional annotation. BEDTools (Quinlan and Hall, 2010) was first used to annotate the differentially expressed piRNAs to non-repeat genomic region obtained from piRBase and total human genome (https://www.gencodegenes.org/human/release_38lift37.html). Based on evidence suggesting shared gene-regulating mechanisms between piRNA and miRNA (Robine et al., 2009; Gainetdinov et al., 2022; Kamenova et al., 2022), we utilized miranda algorithm (Enright et al., 2003) to predict potential interactions between our differentially expressed piRNAs and 3′UTR mRNA using alignment score >160 and ∆G ≤ −10 kcal/mol. For differentially expressed circRNA, top 5% of predicted circRNA-miRNA pairs curated either in miranda or targetscan database were identified (Zhou et al., 2020). This was followed by the identification of conserved miRNA-gene pairs which were predicted in both miranda and targetscan. Ontology and pathway analyses of target genes were performed using metascape (https://metascape.org/gp/index.html#/main/step1) (Zhou et al., 2019).
The overall demographic and clinical characteristics of the participants (n = 91) are presented in Table1. The participants were mostly non-Hispanic white (81.3%) with an average age (years ±SD) and BMI (kg/m2) of 36.2 ± 6.0 and 29.9 ± 5.5, respectively. Table 1 also compares the demographic and semen characteristics of participants stratified based on couple live birth status (Live birth n = 28; no live birth, n = 63). There was no statistical difference (p > 0.05) in the couple demographics, infertility diagnoses and ART outcomes such as the number of oocytes retrieved and fertilized eggs between the two groups. As expected, the total counts of day 3 high quality embryos were significantly higher (p < 0.05) in the group with livebirth relative to the group without successful livebirth. While similar observation was also observed in day 5 high quality embryos, it did not achieve statistical significance (p > 0.05).
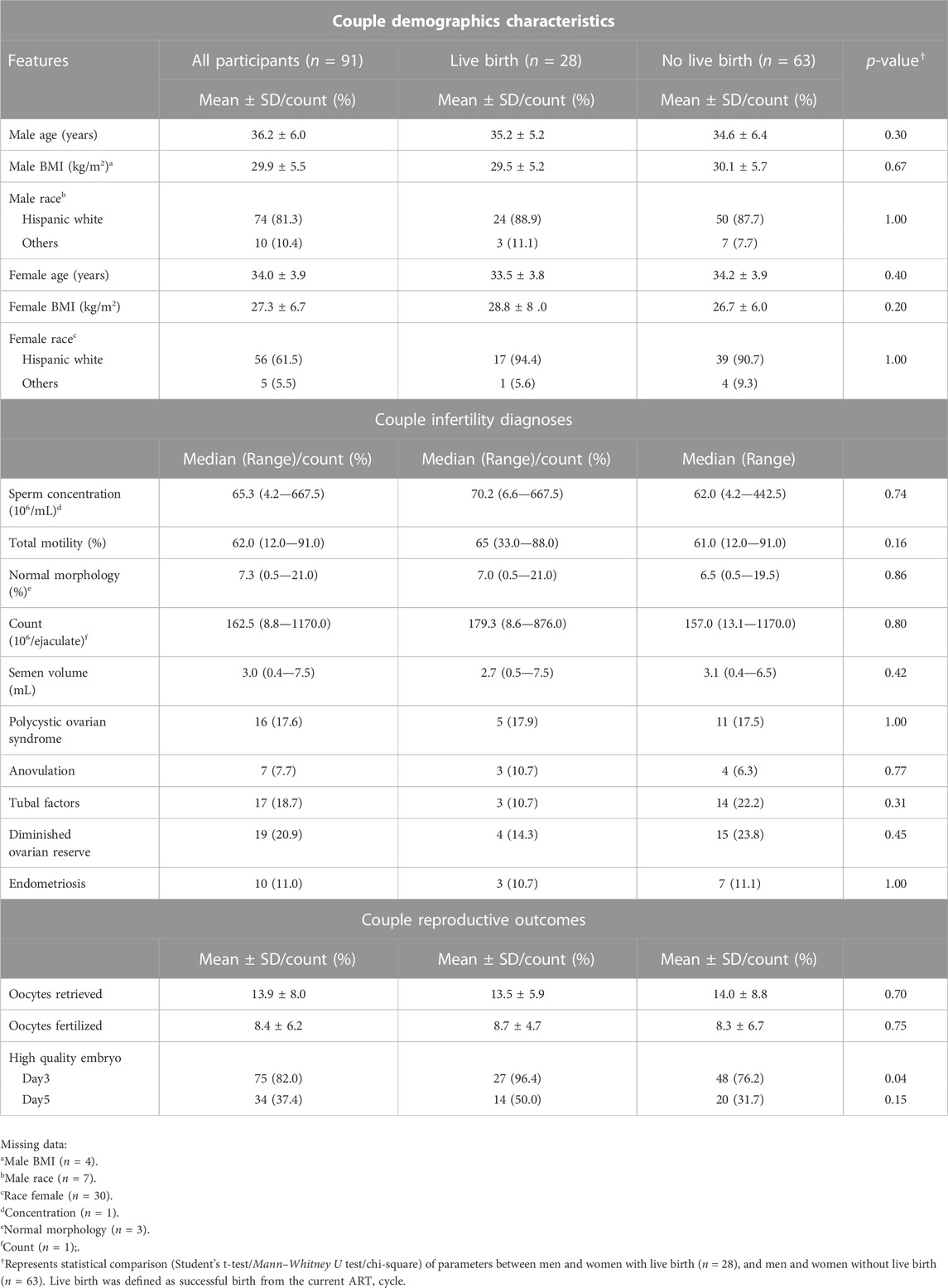
TABLE 1. Demographic and clinical features of SEEDS participants (n = 91) and stratification by live birth status.
First, we sought to identify spEV derived differentially expressed ncRNAs in men without successful live births compared to men with live births. We found a total of 12 ncRNAs were differentially expressed in men without live birth (Figures 1A, B; Supplementary Table S1). The most prominent differentially expressed ncRNA species were annotated to circRNAs (n = 10, 83%), of which the majority (80%) had lower expression in spEVs among men of couples without successful live birth compared to those with live birth. The remaining two differentially expressed spEV ncRNAs were annotated as piRNAs (piR-28478 and piR-1077), in which both were over-expressed in men without live birth compared to men of couples with a successful live birth.
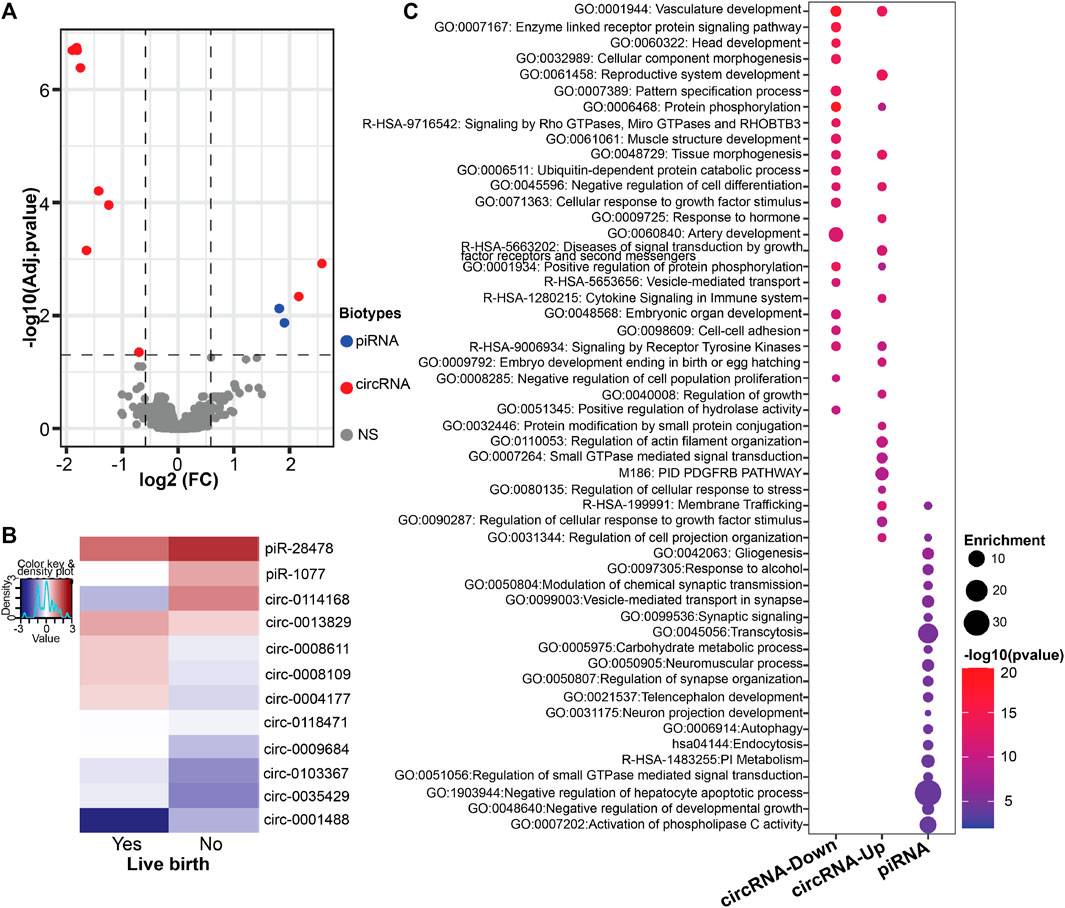
FIGURE 1. spEV ncRNA expression levels by live birth status (Yes/No, referent = Yes). (A) Volcano plot of differential expression of all identified spEV ncRNA biotypes. Dashed lines represent cutoff points [horizontal line = FDR of 0.05; vertical lines = absolute value of log2 fold change (FC) ≥ 0.585]. Colored and grayed datapoints are statistically significant and non-significant (NS) differentially expressed ncRNAs, respectively. (B) Heatmaps of all differentially expressed spEV ncRNAs (C) Dotplot of ontology analysis of differentially expressed spEV circRNA and piRNA gene targets.
Next, we performed gene ontology analyses to understand the biological significance of the differentially expressed circRNAs and piRNAs (Figure 1C). Potential gene target analyses revealed that the 10 downregulated circRNAs were associated with 69 miRNAs, which in turn, targeted 815 unique genes (Supplementary Tables S2, S3). The downregulated differentially expressed circRNAs gene targets were enriched in several developmental related ontology terms including head development, cell component morphogenesis, pattern specification process, muscle structure and artery development, vesicle-mediated transport and embryonic organ development. For the two upregulated circRNAs, 60 miRNAs were identified as potential target which in-turn resulted in 731 unique gene targets (Supplementary Tables S2, S3). Similar to those of the downregulated circRNAs, gene targets of the upregulated circRNAs showed enrichment in biological functions such as reproductive system development, membrane trafficking, embryo development ending in birth or egg hatching and regulation of growth. The two differentially expressed piRNAs (piR-28478 and piR-1077) showed alignment to protein coding gene (PID1) and processed pseudogene (RP11-1315F8), respectively. The piRNAs were also predicted to target 470 unique genes (Supplementary Table S4) with enrichment in ontology terms such as gliogenesis, membrane trafficking, vesicle-mediated transport, synaptic signaling, telencephalon and neuron projection development (Figure 1C).
Finally, we were interested to examine the overlap of differentially expressed spEV derived circRNAs identified in the present study with those identified in our previous study among men with poor, compared to normal, semen quality (Oluwayiose et al., 2022). Out of the 12 and 34 spEV circRNAs identified in the current and previous study, respectively, four (circ-0103367, circ-0009684, circ-0114168, circ-0001488) overlapped between the two studies (Supplementary Table S5). Of these, two (circ-0103367, circ-0009684) showed a consistent downregulation pattern and targeted genes that were enriched in important ontology terms including mitotic cell cycle, blood vessel morphogenesis, brain development, vesicle-mediated transport and growth (Supplementary Table S1).
To our knowledge, our study is the first to investigate the potential role of spEV-derived ncRNA and the subsequent ART outcome of successful live birth. In doing so, we leveraged our prospective cohort study, SEEDS, which recruited couples prior to undergoing infertility treatment and who were followed through their ART cycle that resulted with or without a live birth. Data for spEV ncRNA expression were obtained from banked seminal plasma samples from the same semen sample used for ART treatment. Sequencing of ncRNA libraries identified 12 spEV ncRNAs (10 circRNAs and 2 piRNAs) that were differentially expressed (q < 0.05) between live birth groups, in which predominantly (66.7%) were downregulated among men of couples without successful live birth. Finally, gene targets of the identified circRNAs and piRNAs were enriched in functional terms related to cellular communications, signaling, organ morphogenesis, and early embryonic development and birth.
Infertility treatment for couples has become prevalent especially in high income countries. However, the success of a live birth from one ART cycle has not improved over decades and results in multiple attempts that are accompanied by significant financial and emotional costs (Farley Ordovensky Staniec and Webb, 2007; Smith et al., 2015; Gleicher et al., 2019). In attempts to understand significant factors associated with pregnancy success prior to conception, the majority of previous studies have utilized machine learning algorithms to predict live birth outcomes based only on female characteristics (Metello et al., 2019; Liu et al., 2021). However, this resulted in low prediction accuracies as it failed to account for other effective predictors (Liu et al., 2021) like male factors, such as sperm DNA integrity (Nicopoullos et al., 2019), which has been reported to play critical role in embryo quality (Colaco and Sakkas, 2018). For example, our group has observed that, irrespective of female measurements, male chronological age (Oluwayiose et al., 2021) and preconception exposure to environmental phthalates (Wu et al., 2017a) were negatively associated with ART outcomes including fertilization, embryo quality and live birth. These associations were mediated by sperm DNA methylation at genomic loci related to embryo development (Wu et al., 2017b; Oluwayiose et al., 2021) and highlights the importance of male sperm epigenome in ART outcomes.
Other epigenetic mechanisms, such as EV-derived ncRNAs, are implicated in remodeling sperm RNAs during epididymal maturation and ejaculation (Du et al., 2016; Sharma et al., 2016; Jodar, 2019) and have also been observed to influence embryo development in mice and human (Chen et al., 2016; Sharma et al., 2018; Chen et al., 2021). In the present study, we found the expression of 12 spEVs ncRNAs were significantly altered among men of couples without live birth. Interestingly, four of the ten circRNAs identified in this study were also found to be differentially expressed in men with poor semen quality (Oluwayiose et al., 2022), in which two (circ-0103367, circ-0009684) showed a consistent downregulation pattern among men with poor semen quality and those without a successful live birth. The networks generated in enrichment analyses for the predicted gene targets of these two circRNAs revealed several functions related to signaling, vesicular transport, growth and development. This suggests spEV circRNAs may serve as important markers of both poor semen quality and couple reproductive outcomes.
circRNAs are single-stranded RNAs produced from alternative backsplicing events of corresponding messenger RNA (mRNA) resulting in a covalently closed-loop structures without 5′caps and 3′poly tails (Quan and Li, 2018). Despite the unique features of circRNAs, which include resistance to exonucleases, evolutionary conservation, high abundance, and participation in competing endogenous RNA network system, few studies have explored their roles in reproductive outcomes (Quan and Li, 2018). Advanced sequencing technologies have identified diverse species of circRNAs at different stages of pre-implantation embryos of human (Dang et al., 2016) and animal models (Fan et al., 2015; Kuang et al., 2020) with ontology terms related to organ and tissue morphogenesis, an observation consistent with our current findings.
The biological role of EV piRNAs in human body fluids in disease outcomes has been well documented (Goh et al., 2022). However, little is known about the contribution of EV piRNAs from seminal plasma with respect to reproductive outcomes. In the present study, two spEV piRNAs (piR-28478, piR-1077) were upregulated in men without live birth and predicted to regulate genes that are important in signaling, vesicle-mediated transport and neuron development. These piRNAs were also aligned within the protein coding region of PID1 and the pseudogene, RP11-1315F8, respectively. PID1, also known as NYGGF4, encodes for a protein that contains a phosphotyrosine-interacting domain containing 1 and plays a role in diverse physiological processes including mitochondrion morphogenesis, signal transduction and glucose uptake (Zhao et al., 2010; Wu et al., 2011). The function of PID1 has been suggested to differ based on cellular context, and its silencing in cell lines resulted in reduced proliferation rate (Monteleone et al., 2016). While the biological role of these two spEV piRNAs in relation to live birth is unknown and warrants further investigation, our observations suggest altered piRNAs in spEVs of men without live birth may be important in early development.
This study has notable strengths. We were the first to utilize high throughput small RNA sequencing to identify spEV ncRNAs that distinguished couples undergoing ART treatment that resulted with and without live birth. Additionally, the seminal plasma utilized in our study was collected from the same semen sample used for IVF treatment; thus, our prospective study design allowed us to connect spEV RNA cargo obtained during fertility treatment with subsequent live birth outcomes. We also acknowledge that this study has several limitations. First, spEV ncRNAs were compared between men of couples seeking infertility treatment; thus selection bias and the generalizability of our findings should be noted. Also, given there were no differences in couple demographic and male infertility status between the two live birth groups, we did not include these covariates in our statistical models. However, we cannot rule out the potential for other unmeasured confounders such as differences in nutrition, environmental exposure patterns or underlying health conditions. While the adaptor-ligation free protocol used for generating our small RNA libraries exhibits less sensitivity to modifications to RNA nucleotides resulting in more diverse libraries (Dard-Dascot et al., 2018), distinguishing naturally occurring and library-generated adenine bases at the 3’ end of libraries is a shortcoming. Lastly, our study did not investigate potential mechanisms by which spEV ncRNAs impact live birth outcomes. While our results suggest that select spEV ncRNAs provide prognostic value for predicting successful births, future studies should also focus on their association with early outcomes such as fertilization rates and embryonic development.
In conclusion, through the use of small RNA-sequencing, our study is the first, to our knowledge, to identify spEV ncRNA cargo that distinguished men of couples with and without successful live birth during infertility treatment. These findings highlight that spEVs, derived from several accessory sex glands, contain important molecular cargo that may impact the trajectory of early-life development and subsequent reproductive success, and moreover, the importance of the male partner for reproductive success.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by The University of Massachusetts Amherst and Baystate Medical Center. The patients/participants provided their written informed consent to participate in this study.
OO: Data acquisition, analyses and interpretation; drafting and revising of manuscript. EH: Data acquisition, interpretation and revising of manuscript. BW: Data interpretation and revising of manuscript. AS: Data interpretation and revisin of manuscript. TR: Revising of manuscript. CS: Revising of manuscript. SK: Revising of manuscript. PV: Revising of manuscript. JP: Conception and design, data acquisition, interpretation and revision of manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1174211/full#supplementary-material
Ayaz, A., Houle, E., and Pilsner, J. R. (2021). Extracellular vesicle cargo of the male reproductive tract and the paternal preconception environment. Syst. Biol. Reprod. Med. 67, 103–111. doi:10.1080/19396368.2020.1867665
Campbell, M. J., Lotti, F., Baldi, E., Schlatt, S., Festin, M. P. R., Björndahl, L., et al. (2021). Distribution of semen examination results 2020 – a follow up of data collated for the WHO semen analysis manual 2010. Andrology 9, 817–822. doi:10.1111/andr.12983
Chen, Q., Yan, M., Cao, Z., Li, X., Zhang, Y., Shi, J., et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. SCIENCE 351, 397–400. doi:10.1126/science.aad7977
Chen, X., Sun, Q., Zheng, Y., Liu, Z., Meng, X., Zeng, W., et al. (2021). Human sperm tsRNA as potential biomarker and therapy target for male fertility. Reproduction 161, 111–122. doi:10.1530/REP-20-0415
Colaco, S., and Sakkas, D. (2018). Paternal factors contributing to embryo quality. J. Assisted Reproduction Genet. 35, 1953–1968. doi:10.1007/s10815-018-1304-4
Cooper, T. G., Noonan, E., Von Eckardstein, S., Auger, J., Baker, H. W., Behre, H. M., et al. (2010). World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 16, 231–245. doi:10.1093/humupd/dmp048
Crawford, G., Ray, A., Gudi, A., Shah, A., and Homburg, R. (2015). The role of seminal plasma for improved outcomes during in vitro fertilization treatment: Review of the literature and meta-analysis. Hum. Reprod. Update 21, 275–284. doi:10.1093/humupd/dmu052
Dang, Y., Yan, L., Hu, B., Fan, X., Ren, Y., Li, R., et al. (2016). Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 17, 130. doi:10.1186/s13059-016-0991-3
Dard-Dascot, C., Naquin, D., D’Aubenton-Carafa, Y., Alix, K., Thermes, C., and Van Dijk, E. (2018). Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 19, 118. doi:10.1186/s12864-018-4491-6
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). Star: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
Domar, A. D., Broome, A., Zuttermeister, P. C., Seibel, M., and Friedman, R. (1991). The prevalence and predictability of depression in infertile women*†. Fertil. Steril., 58, 1158–1163. doi:10.1016/s0015-0282(16)55562-9
Du, J., Shen, J., Wang, Y., Pan, C., Pang, W., Diao, H., et al. (2016). Boar seminal plasma exosomes maintain sperm function by infiltrating into the sperm membrane. Oncotarget 7, 58832–58847. doi:10.18632/oncotarget.11315
Enderle, D., Spiel, A., Coticchia, C. M., Berghoff, E., Mueller, R., Schlumpberger, M., et al. (2015). Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLOS ONE 10, e0136133. doi:10.1371/journal.pone.0136133
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5, R1. doi:10.1186/gb-2003-5-1-r1
Fan, X., Zhang, X., Wu, X., Guo, H., Hu, Y., Tang, F., et al. (2015). Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148. doi:10.1186/s13059-015-0706-1
Farley Ordovensky Staniec, J., and Webb, N. J. (2007). Utilization of infertility services: How much does money matter? Health Serv. Res. 42, 971–989. doi:10.1111/j.1475-6773.2006.00640.x
Gainetdinov, I., Cecchini, K., Vega-Badillo, J., Bagci, A., Colpan, C., Arif, A., et al. (2022). "Relaxed targeting rules allow PIWI-clade Argonaute proteins to silence ever-mutating transposons. bioRxiv. doi:10.1101/2022.08.04.502788
Gleicher, N., Kushnir, V. A., and Barad, D. H. 2019. Worldwide decline of IVF birth rates and its probable causes. Hum Reprod Open, 2019, hoz017.
Goh, T. X., Tan, S. L., Roebuck, M. M., Teo, S. H., and Kamarul, T. (2022). A systematic review of extracellular vesicle-derived piwi-interacting RNA in human body fluid and its role in disease progression. Tissue Eng. Part C Methods 28, 511–528. doi:10.1089/ten.TEC.2022.0092
Jodar, M. (2019). Sperm and seminal plasma RNAs: What roles do they play beyond fertilization? Reproduction 158, R113–R123. doi:10.1530/REP-18-0639
Joshi, M., Andrabi, S. W., Yadav, R. K., Sankhwar, S. N., Gupta, G., and Rajender, S. (2022). Qualitative and quantitative assessment of sperm miRNAs identifies hsa-miR-9-3p, hsa-miR-30b-5p and hsa-miR-122-5p as potential biomarkers of male infertility and sperm quality. Reproductive Biol. Endocrinol. 20, 122. doi:10.1186/s12958-022-00990-7
Kamenova, S., Sharapkhanova, A., Akimniyazova, A., Kuzhybayeva, K., Kondybayeva, A., Rakhmetullina, A., et al. (2022). piRNA and miRNA can suppress the expression of multiple sclerosis candidate genes. Nanomaterials 13, 22. doi:10.3390/nano13010022
Khalife, D., Nassar, A., Khalil, A., Awwad, J., Abu Musa, A., Hannoun, A., et al. (2020). Cumulative live-birth rates by maternal age after one or multiple in vitro fertilization cycles: An institutional experience. Int. J. Fertil. Steril. 14, 34–40. doi:10.22074/ijfs.2020.5855
Kocourkova, J., Burcin, B., and Kucera, T. (2014). Demographic relevancy of increased use of assisted reproduction in European countries. Reprod. Health 11, 37. doi:10.1186/1742-4755-11-37
Kuang, L., Lei, M., Li, C., Guo, Z., Ren, Y., Zhang, X., et al. (2020). Whole transcriptome sequencing reveals that non-coding RNAs are related to embryo morphogenesis and development in rabbits. Genomics 112, 2203–2212. doi:10.1016/j.ygeno.2019.12.016
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Liu, R., Bai, S., Jiang, X., Luo, L., Tong, X., Zheng, S., et al. (2021). Multifactor prediction of embryo transfer outcomes based on a machine learning algorithm. Front. Endocrinol. 12, 745039. doi:10.3389/fendo.2021.745039
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10. doi:10.14806/ej.17.1.200
Mascarenhas, M. N., Flaxman, S. R., Boerma, T., Vanderpoel, S., and Stevens, G. A. (2012). National, regional, and global trends in infertility prevalence since 1990: A systematic analysis of 277 health surveys. PLoS Med. 9, e1001356. doi:10.1371/journal.pmed.1001356
Metello, J. L., Tomás, C., and Ferreira, P. (2019). Can we predict the IVF/ICSI live birth rate? JBRA Assist. Reprod. 23, 402–407. doi:10.5935/1518-0557.20190043
Monteleone, F., Vitale, M., Caratù, G., D’Ambrosio, C., Di Giovanni, S., Gorrese, M., et al. (2016). Inhibition of PID1/NYGGF4/PCLI1 gene expression highlights its role in the early events of the cell cycle in NIH3T3 fibroblasts. J. Enzyme Inhibition Med. Chem. 31, 45–53. doi:10.1080/14756366.2016.1217855
Nicopoullos, J., Vicens-Morton, A., Lewis, S. E. M., Lee, K., Larsen, P., Ramsay, J., et al. (2019). Novel use of COMET parameters of sperm DNA damage may increase its utility to diagnose male infertility and predict live births following both IVF and ICSI. Hum. Reprod. 34, 1915–1923. doi:10.1093/humrep/dez151
Oluwayiose, O. A., Houle, E., Whitcomb, B. W., Suvorov, A., Rahil, T., Sites, C. K., et al. (2022). Altered non-coding RNA profiles of seminal plasma extracellular vesicles of men with poor semen quality undergoing in vitro fertilization treatment. Andrology 11, 677–686. doi:10.1111/andr.13295
Oluwayiose, O. A., Wu, H., Saddiki, H., Whitcomb, B. W., Balzer, L. B., Brandon, N., et al. (2021). Sperm DNA methylation mediates the association of male age on reproductive outcomes among couples undergoing infertility treatment. Sci. Rep. 11, 3216. doi:10.1038/s41598-020-80857-2
Quaas, A., and Dokras, A. (2008). Diagnosis and treatment of unexplained infertility. Rev. Obstet. Gynecol. 1, 69–76. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2505167/
Quan, G., and Li, J. (2018). Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 11, 9. doi:10.1186/s13048-018-0381-4
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi:10.1093/bioinformatics/btq033
Robine, N., Lau, N. C., Balla, S., Jin, Z., Okamura, K., Kuramochi-Miyagawa, S., et al. (2009). A broadly conserved pathway generates 3′utr-directed primary piRNAs. Curr. Biol. 19, 2066–2076. doi:10.1016/j.cub.2009.11.064
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi:10.1093/bioinformatics/btp616
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. doi:10.1186/gb-2010-11-3-r25
Rodriguez-Martinez, H., Martinez, E. A., Calvete, J. J., Peña Vega, F. J., and Roca, J. (2021). Seminal plasma: Relevant for fertility? Int. J. Mol. Sci. 22, 4368. doi:10.3390/ijms22094368
Rozowsky, J., Kitchen, R. R., Park, J. J., Galeev, T. R., Diao, J., Warrell, J., et al. (2019). exceRpt: A comprehensive analytic platform for extracellular RNA profiling. Cell Syst. 8, 352–357. doi:10.1016/j.cels.2019.03.004
Rutstein, S. O., and Shah, I. H. (2004). Infecundity infertility and childlessness in developing countries. Geneva: World Health Organization, 249–251.
Sharma, U., Conine, C. C., Shea, J. M., Boskovic, A., Derr, A. G., Bing, X. Y., et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. doi:10.1126/science.aad6780
Sharma, U., Sun, F., Conine, C. C., Reichholf, B., Kukreja, S., Herzog, V. A., et al. (2018). Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46, 481–494. doi:10.1016/j.devcel.2018.06.023
Smith, A. D. A. C., Tilling, K., Nelson, S. M., and Lawlor, D. A. (2015). Live-birth rate associated with repeat in vitro fertilization treatment cycles. JAMA 314, 2654–2662. doi:10.1001/jama.2015.17296
Smith, S., Pfeifer, S. M., and Collins, J. A. (2003). Diagnosis and management of female infertility. JAMA 290, 1767–1770. doi:10.1001/jama.290.13.1767
Sunderam, S., Kissin, D. M., Crawford, S. B., Folger, S. G., Jamieson, D. J., Warner, L., et al. (2017). Assisted reproductive technology surveillance — United States, 2014. MMWR. Surveill. Summ. 66, 1–24. doi:10.15585/mmwr.ss6606a1
Van Loendersloot, L., Repping, S., Bossuyt, P. M., Van Der Veen, F., and Van Wely, M. (2014). Prediction models in in vitro fertilization; where are we? A mini review. J. Adv. Res. 5, 295–301. doi:10.1016/j.jare.2013.05.002
Vojtech, L., Woo, S., Hughes, S., Levy, C., Ballweber, L., Sauteraud, R. P., et al. (2014). Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 42, 7290–7304. doi:10.1093/nar/gku347
Wright, V. C., Schieve, L. A., Reynolds, M. A., and Jeng, G. (2003). Assisted reproductive technology surveillance–United States, 2000. MMWR Surveill. Summ. 52, 1–16. doi:10.15585/mmwr.ss6703a1
Wu, H., Ashcraft, L., Whitcomb, B. W., Rahil, T., Tougias, E., Sites, C. K., et al. (2017a). Parental contributions to early embryo development: Influences of urinary phthalate and phthalate alternatives among couples undergoing IVF treatment. Hum. Reprod. 32, 65–75. doi:10.1093/humrep/dew301
Wu, H., De Gannes, M. K., Luchetti, G., and Pilsner, J. R. (2015). Rapid method for the isolation of mammalian sperm DNA. BioTechniques 58, 293–300. doi:10.2144/000114280
Wu, H., Estill, M. S., Shershebnev, A., Suvorov, A., Krawetz, S. A., Whitcomb, B. W., et al. (2017b). Preconception urinary phthalate concentrations and sperm DNA methylation profiles among men undergoing IVF treatment: A cross-sectional study. Hum. Reprod. 32, 2159–2169. doi:10.1093/humrep/dex283
Wu, W. L., Gan, W. H., Tong, M. L., Li, X. L., Dai, J. Z., Zhang, C. M., et al. (2011). Over-expression of NYGGF4 (PID1) inhibits glucose transport in skeletal myotubes by blocking the IRS1/PI3K/AKT insulin pathway. Mol. Genet. Metab. 102, 374–377. doi:10.1016/j.ymgme.2010.11.165
Zhao, Y., Zhang, C., Chen, X., Gao, C., Ji, C., Chen, F., et al. (2010). Overexpression of NYGGF4 (PID1) induces mitochondrial impairment in 3T3-L1 adipocytes. Mol. Cell Biochem. 340, 41–48. doi:10.1007/s11010-010-0398-5
Zhou, W.-Y., Cai, Z.-R., Liu, J., Wang, D.-S., Ju, H.-Q., and Xu, R.-H. (2020). Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 19, 172. doi:10.1186/s12943-020-01286-3
Keywords: seminal plasma, extracellular vesicles, small non-coding RNA, live birth, infertility, art
Citation: Oluwayiose OA, Houle E, Whitcomb BW, Suvorov A, Rahil T, Sites CK, Krawetz SA, Visconti PE and Pilsner JR (2023) Non-coding RNAs from seminal plasma extracellular vesicles and success of live birth among couples undergoing fertility treatment. Front. Cell Dev. Biol. 11:1174211. doi: 10.3389/fcell.2023.1174211
Received: 26 February 2023; Accepted: 12 June 2023;
Published: 22 June 2023.
Edited by:
Kavindra Kumar Kesari, Aalto University, FinlandReviewed by:
Singh Rajender, Central Drug Research Institute (CSIR), IndiaCopyright © 2023 Oluwayiose, Houle, Whitcomb, Suvorov, Rahil, Sites, Krawetz, Visconti and Pilsner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Richard Pilsner, cnBpbHNuZXJAd2F5bmUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.