
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 14 July 2023
Sec. Stem Cell Research
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1172385
This article is part of the Research Topic Methods in Induced Pluripotent Stem Cells - Neuroscience View all 7 articles
 Shuo Cao1,2†
Shuo Cao1,2† Xinyue Gao1,2†
Xinyue Gao1,2† Fangyuan Liu1,2
Fangyuan Liu1,2 Yanglin Chen1,2
Yanglin Chen1,2 Qin Na1,2,3
Qin Na1,2,3 Qiaoqiao Meng1,2
Qiaoqiao Meng1,2 Peng Shao1,2
Peng Shao1,2 Chen Chen1,2
Chen Chen1,2 Yongli Song1,2
Yongli Song1,2 Baojiang Wu1,2
Baojiang Wu1,2 Xihe Li1,2,4*
Xihe Li1,2,4* Siqin Bao1,2*
Siqin Bao1,2*The emergence and development of induced pluripotent stem cells (iPSCs) provides an approach to understand the regulatory mechanisms of cell pluripotency and demonstrates the great potential of iPSCs in disease modeling. Acute myelitis defines a group of inflammatory diseases that cause acute nerve damage in the spinal cord; however, its pathophysiology remains to be elusive. In this study, we derived skin fibroblasts from a patient with acute myelitis (P-HAF) and then reprogrammed P-HAF cells to iPSCs using eight exogenous factors (namely, OCT4, SOX2, c-MYC, KLF4, NANOG, LIN28, RARG, and LRH1). We performed transcriptomic analysis of the P-HAF and compared the biological characteristics of the iPSCs derived from the patient (P-iPSCs) with those derived from normal individuals in terms of pluripotency, transcriptomic characteristics, and differentiation ability toward the ectoderm. Compared to the control iPSCs, the P-iPSCs displayed similar features of pluripotency and comparable capability of ectoderm differentiation in the specified culture. However, when tested in the common medium, the P-iPSCs showed attenuated potential for ectoderm differentiation. The transcriptomic analysis revealed that pathways enriched in P-iPSCs included those involved in Wnt signaling. To this end, we treated iPSCs and P-iPSCs with the Wnt signaling pathway inhibitor IWR1 during the differentiation process and found that the expression of the ectoderm marker Sox1 was increased significantly in P-iPSCs. This study provides a novel approach to investigating the pathogenesis of acute myelitis.
Human embryonic stem cells (hESCs) were first established by Thomson et al. (1998). Takahashi et al. (2007) derived the first human-induced pluripotent stem cells (hiPSCs). Human pluripotent stem cells (hPSCs), including hESCs and hiPSCs, have played an important role in regenerative medicine and disease research. hPSCs are characterized by the potential for self-renewal and multilineage differentiation and have since been established and studied in many laboratories (Loser et al., 2010; Fraga et al., 2011). Gao et al. (2019) established the expanded pluripotent human stem cell line (hEPSC) derived from iPSCs. Recently, Guo et al. (2021) found that hESCs differed from those of mice and showed that hESCs have the potential to differentiate directly into trophoblast cells.
iPSC technology has been extensively utilized for the development of cell therapies, drug discoveries, and disease modeling in humans. Researchers use differentiated cell populations derived from hESCs or hiPSCs to screen for effective compounds or new therapeutic targets affecting disease-related signaling pathways. Thus far, using iPSC technology, researchers have made significant progress in modeling diseases such as heart disease (Sasaki et al., 2016; McNally and Mestroni, 2017; Li et al., 2018), eye disease (Guo et al., 2018; Yan et al., 2019; Zhou et al., 2020; Klipfel et al., 2021), muscle disease (Choi et al., 2016; Ueki et al., 2017), and diabetes (Braverman-Gross et al., 2018; Yabe et al., 2019).
Acute myelitis, also known as acute transverse myelitis (ATM), is a rare inflammatory demyelinating disease. The incidence of ATM is estimated three cases per 100,000 population per year (0.003%). However, ATM can have devastating effects on the nervous system, with up to two-thirds of patients having moderate-to-severe disability (West et al., 2012). Symptoms of ATM include impaired sensory and autonomic function, which is easily recognized clinically. ATM is an acute or subacute condition with gradual deterioration within 4 h–21 days of onset (de Seze et al., 2005). ATM symptoms peak on average within 2–4 days, but not more than after 7 days, when urinary retention usually occurs. Thus, patients require catheterization, and in the acute phase, up to 89% of patients are unable to walk or need ventilator assistance for breathing (Kahn, 2020). Currently, acute inflammation in ATM is usually treated with high doses of intravenous corticosteroids, but also by plasmapheresis or intravenous immunoglobulin. Few studies have been conducted evaluating the pathogenesis of ATM.
Although iPSC disease models have played an important role in many studies, the current application of human stem cells in myelitis is mainly focused on hematopoietic stem cell transplantation for the treatment of optic neuromyelitis (Ceglie et al., 2020), and there have been no reports of the establishment of iPSCs in patients with acute myelitis. In this study, human embryonic fibroblasts and skin fibroblasts derived from a patient with acute myelitis were transfected with eight exogenous factors through the PiggyBac transposition method described by Gao et al. (2019) and reprogrammed into iPSCs. Here, we compared the biological characteristics of the patient iPSCs with normal iPSCs in terms of pluripotency, transcriptomic characteristics, and the ability to differentiate toward the ectoderm. This study provides an approach to explore the pathological mechanisms underlying acute myelitis.
The human embryonic stem cell line W24 was obtained from the Wellcome Trust/Cancer Research UK Gurdon Institute. The normal human embryonic fibroblasts (HEFs) were received from Nanjing Medical University, and patient fibroblasts (P-HAF) derived from patients with acute myelitis were donated by volunteers at the hospital.
A DNA mixture consisting of 3.0 μg PB-Tre-h4F, 1.0 μg PB-Tre-hRL, 1.0 μg PB-Tre-P2F, 1.0 μg EF1α, and 1.0 μg Pbase was prepared for transfection into cells. The transfected cells (1.0 × 106) were seeded on STO (10-cm dish) in the M15 medium supplemented with doxycycline (Dox) (1.0 μg/mL). Dox was removed at day 14, and the medium was switched to the mTeSR (StemCell, 85,850) medium and expanded to stable iPSC lines.
iPSCs/P-iPSCs were maintained on an STO feeder cultured in the M15+Dox medium, while iPSCs/P-iPSCs were maintained feeder-free and cultured in the mTeSR (STEMCELL, 85,850) medium. The colonies were dissociated by Versen for 3 min and removed of Versen, and then cells were suspended by mTeSR medium. Cell suspensions were placed a new well at a ratio of 1:5 and the mTeSR medium was replaced regularly every day. All culture plates used were coated with VTN (Gibco A14700) for at least 0.5 h prior to use.
Before staining, the cells were washed briefly with PBS and fixed in 4% paraformaldehyde at room temperature for 30 min, before adding the alkaline phosphatase (AP) staining solution. The AP staining solution was prepared as follows: 50 µL sodium nitrite solution was gently combined with 50 µL FRV alkaline solution, and the mixture was placed at 37°C for 3 min. Then, 2.25 mL of H2O and 50 µL naphthol-As-BI alkaline solution were added to the mixture.
A solution of 0.2 μg/mL colchicine was added to the cells in good growth conditions and treated at 37°C for 2.5 h. The cells were exposed to Versen for 3 min, and detached cells were centrifuged at 1,300 rpm for 3 min. The cells were placed in 0.075 mol/L KCL for hypotonic treatment for 50 min (stationary liquid: methanol: glacial acetic acid = 3:1). The cells were then centrifuged at 1,000 rpm for 10 min, and the cell pellets were placed in 8 mL of stationary medium at 37°C for 10 min. This step was repeated twice. Next, 300 μL ice-cold stationary liquid was added to the cell pellet and introduced dropwise onto glass slides. The glass slides were placed in a 70°C oven for 1 h. After drying, the cells were stained with Giemsa stain (Sigma) for 10 min at 37°C. Finally, the slides were placed under a microscope, and the chromosomes were arranged and analyzed using the LUCIA Cytogenetics platform.
RNA was achieved using an RNA extraction kit (Qiagen, cat. no. 74104), and cDNA was prepared using Promega’s reverse transcription kit (cat. no. A5001). The primers used are listed in Supplementary Table S1.
Cells were washed briefly with PBS and fixed in 4% paraformaldehyde at room temperature for 30 min. Immunofluorescence (IF) buffer was added to permeabilize cells for 30 min at room temperature (0.1% Triton X-100 [Sigma] and 1% BSA in PBS). The primary antibody was prepared according to the antibody dilution ratio, and 150 μL of the primary antibody was added to each well before incubation at 4°C overnight. The next day, the cells were washed with IF buffer three times for 5 min each, and the secondary antibody was added at room temperature for 1 h in the dark. In addition, Hoechst 33,342 was added, and the nuclei were stained for 5 min at room temperature. The cells were analyzed using confocal microscopy. The antibodies used are listed in Supplementary Table S1.
Genomic DNA was extracted using the Genomic DNA Kit (TIANGEN Biotech, Beijing, China) and treated with the EZ DNA Methylation-Gold™ Kit (Zymo Research, Irvine, CA, United States). Then, the bisulfite-modified DNA was amplified by PCR using the following primers: Oct4 sense-GTTAGAGGTTAAGGTTAGTGGGTG and antisense-AAACCTTAAAAACTTAACCAAATCC. The amplified PCR products were randomly selected and cloned into the Pmd19T Cloning vector ((Takara, Otsu, Shiga, Japan) and sequenced.
Pre-differentiated iPSC/P-iPSCs were detached using Versen and then seeded in 24-well plates at a density of 1.25 × 104 cells in 1 mL of the M10 medium (DMEM/F12 supplemented with 10% FBS). After 12 days of culture, the cells were collected for analysis.
In this study, HEF cells and iPSCs were used as controls. Differential gene analysis was performed using the transcriptome data of HEF and P-HAF cells, as well as of iPSC and P-iPSC cells. Gene expression was first normalized, and differential analysis was conducted to select differentially expressed genes (DEGs) from the transcriptome sequencing results of HEF, P-HAF, iPSC and P-iPSC. Log2 Fold change >1 and p-value <0.05 were selected as screening criteria for differential genes. Gene ontology (GO) term enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, principal component analysis (PCA), and Gene Set Enrichment Analysis (GSEA) were performed using OmicStudio and OmicShare tools, online platforms for data analysis. The PPI analysis was performed using String tools.
The hESCs, iPSCs, and P-iPSCs were dissociated with 0.25% trypsin/EDTA and seeded in gelatin-coated well plates. Differentiation to neural crest (NC) cells: the cells were cultured in the BM medium supplemented with 1.0 ng/mL BMP4 (R&D systems, 314-BP-010), 10 μM SB431542 (Selleck, S1067), and 600 nM CHIR99021 (Miltenyi Biotech, 130-103-926). Two days later, the cells were cultured in the BM medium supplemented with 10 μM SB431542 and 1.5 μM CHIR99021. The cells were collected on day 12 for analysis. Differentiation to cranial placode (CP) cells: the cells were cultured in the BM medium supplemented with 5 ng/mL of BMP4 and 10 μM SB431542. Two days later, the cells were cultured in the BM medium supplemented with 10 μM SB431542 and 50 ng/mL bFGF (R&D systems, 233-FB-025). The cells were collected on day 12 for analysis. The BM medium (500 mL) was prepared as follows: 245 mL DMEM/F12 (Gibco, 11320-033), 245 mL Neurobasal Medium (Gibco, 21103-049), 2.5 mL N2 supplement (Gibco, 17502-048), 5.0 mL B27 supplement (Gibco, 17504-044), 5.0 mL 100 × GlutaMax (Gibco, 35050-061), and 0.1 mM 2-mercaptoethanol (Sigma, M3148).
Total protein was extracted from cells by cell lysis buffer (Solarbio, R1010) (PMSF and phosphatase inhibitor were added to lysis buffer immediately before use). The concentrations of extracted proteins were measured using the CoomassiePlus (Bradford) Assay (Thermo Scientific). Equal amounts of protein (30 μg) were separated by SDS-PAGE, and proteins were transferred to PVDF membranes (Amersham). The blots were blocked in 5% non-fat powdered milk in TBST at room temperature for 1 h and then incubated with the primary antibodies overnight at 4°C. The next day, secondary antibodies were incubated at room temperature for 1 h. The expression of protein was detected by ECL (Thermo Scientific).
ATM is a rare inflammatory demyelinating disease characterized by relatively acute motor, sensory, and autonomic dysfunction. ATM involves multiple components of the central nervous system, including oligodendrocytes, neurons, axons, and myelin, and is a mixed inflammatory disease rather than a pure demyelinating disease (Kerr and Ayetey, 2002; Krishnan et al., 2004; Weng et al., 2017b).
In order to establish iPSCs derived from the patient with ATM, we first established human embryonic fibroblasts (HEFs) and P-HAF derived from the patient. We show that HEF and P-HAF exhibited normal spindle-shaped symmetry (Figure 1A). To gain further insight into the underlying molecular mechanisms characterizing HEF and P-HAF, we performed RNA sequencing (RNA-seq) and analyzed DEGs using the DESeq2 R package. There were 3,059 DEGs in P-HAF compared to HEF cells, of which 1,472 genes were significantly downregulated and 1,587 genes were significantly upregulated (Figure 1B; Supplementary Table S2). To identify the functions of DEGs, we performed GO enrichment analysis. Comparing the GO enrichment results of HEF and P-HAF, we found that some terms involved in the positive regulation of nervous system development, axon development, and regulation of neuron projection development were enriched in DEGs (Figure 1C; Supplementary Table S3). KEGG pathway enrichment analysis revealed that enriched pathways were involved in the PI3K-AKT signaling pathway, the Hippo signaling pathway, the JAK-STAT signaling pathway, and the Wnt signaling pathway (Figure 1D; Supplementary Table S4). In addition, the GSEA showed enrichment in the NF-kB signaling pathway and in the IL-17 signaling pathway (Figure 1E). Furthermore, we observed that the expression of genes from the PI3K-AKT signaling pathway showed the most significant differences between HEF and P-HAF cells (Figure 1F). We used protein–protein interaction (PPI) to examine the top 12 proteins. The network revealed that ITGA2 and COL4A1 may play a key role in the PI3K-AKT signaling pathway (Figure 1G). Taken together, gene sets related to inflammation, Hippo, JAK-STAT, and Wnt signaling pathways are enriched in P-HAF cells.
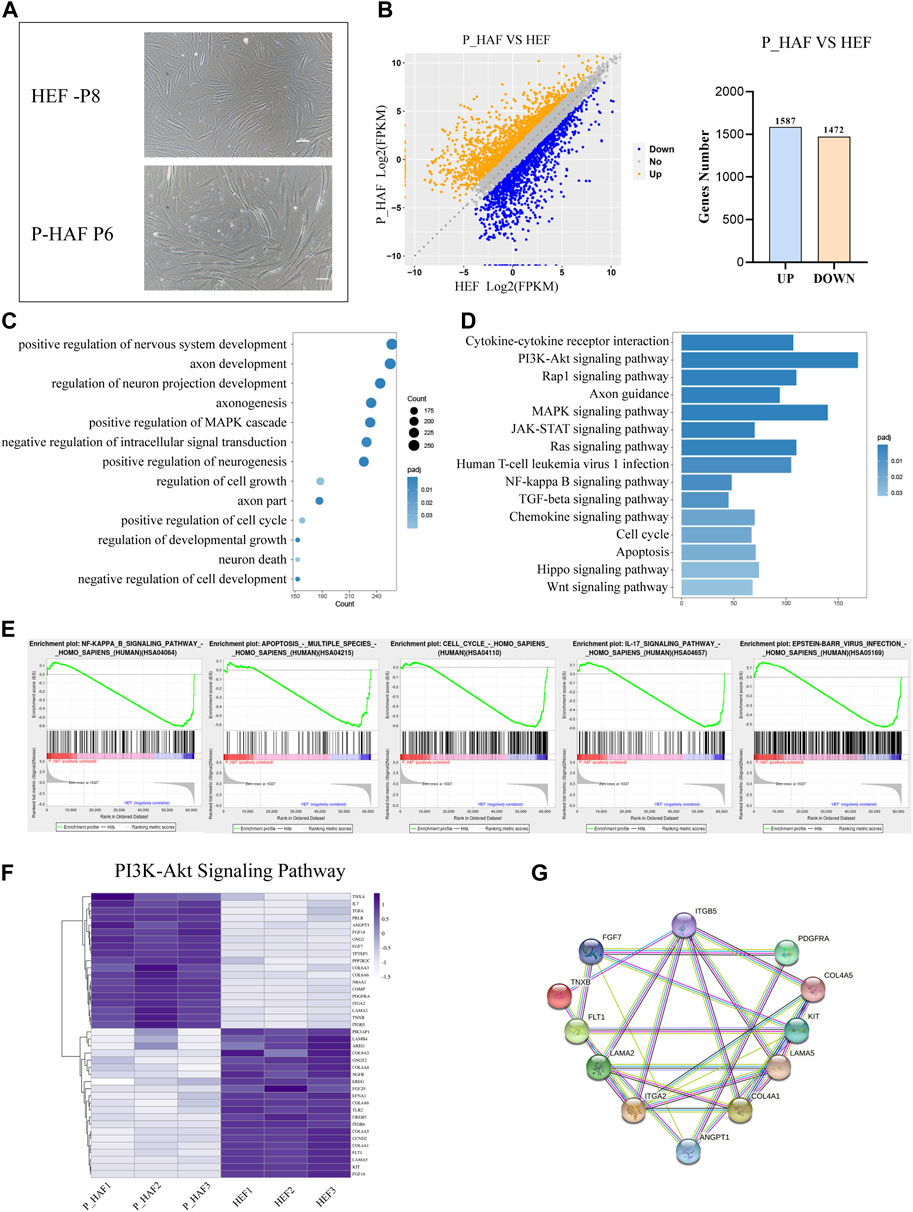
FIGURE 1. Transcriptomic features of HEF and P-HAF cells. (A) Morphology of HEF and P-HAF (scale bars, 100 μm). (B) DEGs analysis of HEF and P-HAF cells. (C) GO analysis of the DEGs between HEF and P-HAF. (D) KEGG pathway analysis of DEGs between HEF and P-HAF. (E) GSEA showing enriched GO-BP terms for upregulated DEGs and downregulated DEGs, respectively. (F) DEGs (|log2 (fold change)| > 1, FDR< 0.05) related to the PI3K-Akt signaling pathway are shown in the heatmap. (G) Network of candidate targets in the PI3K-Akt signaling pathway.
In this study, iPSCs were established by reprogramming normal HEFs and skin fibroblasts derived from a patient with ATM, using piggyBac transposition and doxycycline (Dox) inducible expression of eight exogenous factors (namely, OCT4, SOX2, c-Myc, KLF4, NANOG, LIN28, RARG, and LRH1) (Figure 2A). The Dox induction system allowed HEF and P-HAF cells to form primary colonies, which were selected from day 7 to day 14. The selected colonies were passaged in a serum-containing medium (MEM+15% FCS: M15) on STO feeder cells (Figure 2B). Based on the mechanical passaging method, the medium was changed to mTeSR, and the cells were passaged into cell culture dishes (the first passage was named P1), which were not feeder-or Dox-containing cultures (Figure 2B). We named the normal embryonic fibroblast-induced pluripotency stem cell as iPSCs and the patient (with ATM) skin fibroblast-derived iPSC cell line was named P-iPSC cells.
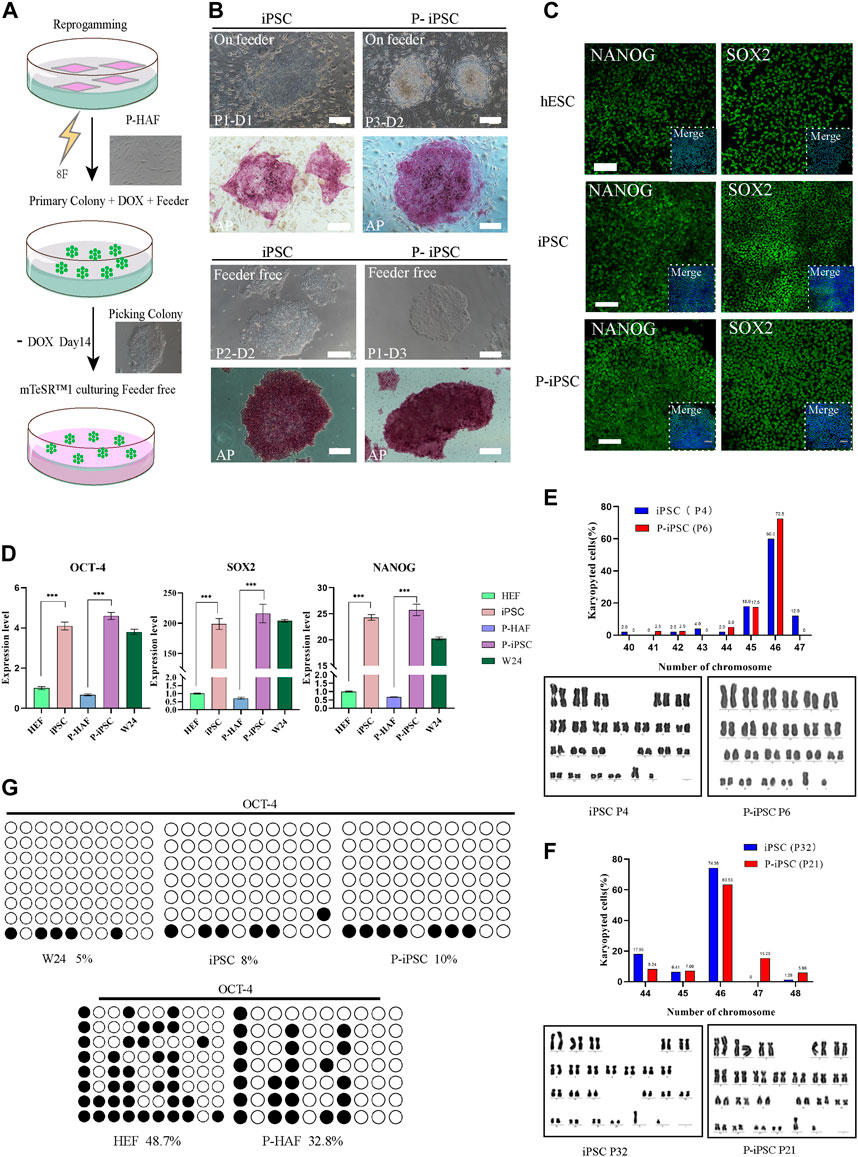
FIGURE 2. Reprogramming HEF and P-HAF cells to iPSCs, P-iPSCs, and the features of two types of iPSCs. (A) Schedule of reprogramming HEF and P-HAF to iPSCs and P-iPSCs. (B) Morphology and AP staining of iPSCs and P-iPSCs on feeder cells or feeder-free cultures (scale bars, 200 μm). (C) Immunostaining of NANOG and SOX2 in hESCs, iPSCs, and P-iPSCs (scale bars, 100 μm). (D) Real-time PCR analysis of pluripotency-associated gene expression in the hESCs, iPSCs, and P-iPSCs. HEF and P-HAF cells were used as the control. Data were obtained in triplicate and presented as the mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.01, and ***p < 0.001. (E) Karyotyping analysis of iPSCs (passage 4) and P-iPSCs (passage 6). (F) Karyotyping analysis of iPSCs (passage 32) and P-iPSCs (passage 21). (G) Analysis of the DNA CpG methylation analysis of the OCT4 promoter loci via bisulfite sequencing in HEF, P-HAF, hESCs, iPSCs, and P-iPSCs.
A remarkable feature of iPSCs and P-iPSCs was that they remained undifferentiated colonies under feeder-free conditions and without Dox for more than 30 passages, during which the cells proliferated robustly. Both iPSC and P-iPSC colonies were AP positive. Then, we determined the protein expression of SOX2 and NANOG, which were shown to be expressed at similar levels in hESCs (W24), iPSCs, and P-iPSCs (Figure 2C). The expression of pluripotency genes (OCT4, SOX2, and NANOG) in hESC (W24), iPSC, and P-iPSC was significantly higher than that of HEF and P-HAF cells (Figure 2D). Furthermore, iPSCs and P-iPSCs were genetically stable and exhibited a normal karyotype (Figures 2E, F). Furthermore, we found that the degree of DNA methylation of the OCT4 promoter loci in HEF and P-HAF was higher than that in hESC, iPSC, and P-iPSC (Figure 2G), which is consistent with previous findings (Guan et al., 2022).
Taken together, we successfully established human iPSCs from normal embryonic and patient-derived fibroblasts using eight exogenous factors. We showed that the iPSCs and P-iPSCs display no obvious differences in the clonal morphology, both were positive for alkaline phosphatase activity and harbored normal karyotypes and activation of endogenous pluripotent gene expression in both lines.
To gain further insight into the molecular mechanisms of P-iPSCs, we performed transcriptomic analyses by RNA sequencing of the HEF, P-HAF, hESCs, iPSCs, and P-iPSCs. PCA showed that iPSCs and P-iPSCs were closely related to hESCs and were distinct from HEF and P-HAF cells (Figure 3A). To examine the differences between iPSCs and P-iPSCs, we compared the DEGs. Overall, there were 1,014 DEGs in P-iPSC compared to normal iPSCs, of which 458 genes were significantly downregulated and 556 genes were significantly upregulated (Figure 3B; Supplementary Table S5). GO analysis revealed enrichment of the regulation of cell cycle phase transition, negative regulation of the Wnt signaling pathway, negative regulation of the canonical Wnt signaling pathway, mechanisms involving the mitochondrial inner membrane, metabolic processes involving ATP, and nuclear-transcribed catabolic processes of mRNA (Figure 3C; Supplementary Table S6). The KEGG pathway enrichment analysis showed the enriched pathways (Figure 3D; Supplementary Table S7), including the Hippo signaling pathway, the Wnt signaling pathway, the cell cycle, and the JAK-STAT signaling pathway. Notably, we observed that the expression of cell cycle genes was significantly different between normal iPSCs and P-iPSCs (Figure 3E). The expression of fibroblast-related genes and pluripotency-related genes in HEF, P-HAF, iPSCs, P-iPSCs, and hESCs were then analyzed. We found that fibroblast-related genes, such as ANPEP, JUN, FOSL1, and RFX2, are highly expressed in HEF and P-HAF but not expressed in iPSC, P-iPSC, and hESC (Figure 3F). Pluripotency-related genes, such as GPC4, JARID2, SOX2, and NANOG, were highly expressed in iPSCs, P-iPSCs, and hESCs but not in HEF and P-HAF (Figure 3F). Furthermore, the GSEA showed enrichment of the Hippo signaling pathway and the IL-17 signaling pathway (Figure 3G). In particular, we observed significant expression of the Wnt signaling pathway genes between normal iPSCs and P-iPSCs (Figure 3H). We constructed PPI networks to examine the top 10 proteins. The PPI network revealed that WNT5A and FZD2 may play a key role in the Wnt signaling pathway (Figure 3I).
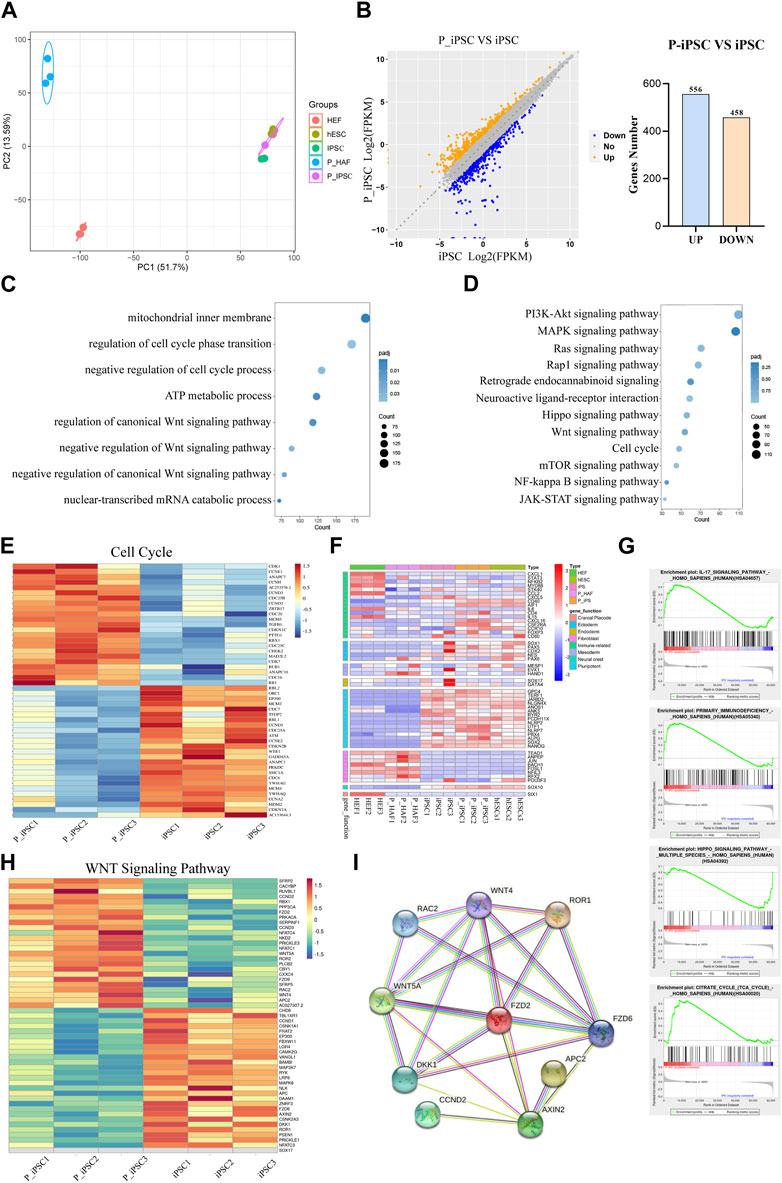
FIGURE 3. Transcriptome changes between iPSCs and P-iPSCs. (A) Principal component analysis of HEF, P-HAF, hESCs, iPSCs, and P-iPSCs. (B) DEGs of iPSCs and P-iPSCs. (C) GO analysis of the DEGs comparing iPSCs and P-iPSCs. (D) KEGG pathway analysis comparing iPSCs and P-iPSCs. (E) DEGs (|log2 (fold change)| > 1, FDR< 0.05) related to the cell cycle are shown in the heatmap. (F) Heatmap showing the expression of marker genes in HEF, P-HAF, iPSCs, P-iPSCs, and hESCs. (G) GSEA showing enriched GO-BP terms for up- and downregulated DEGs, respectively. (H) DEGs (|log2 (fold change)| > 1, FDR< 0.05) related to the Wnt signaling pathway are shown in the heatmap. (I) Network of candidate targets in the Wnt signaling pathway.
These results indicate that iPSCs, P-iPSCs, and hESCs are pluripotent and display molecular features of pluripotency. In addition, our data indicate that P-iPSCs exhibit unique molecular features compared to iPSCs.
To identify the differentiation property of P-iPSCs, we differentiated both P-iPSCs and iPSCs to the ectoderm. RT-qPCR analysis showed that the expression of ectoderm and neuron markers were significantly different between in iPSCs and P-iPSCs (Figure 4A). The ectoderm consists of four major ectoderm lineages, namely, the neuroectoderm, neural crest (NC), cranial placode (CP), and non-neural ectoderm (Tchieu et al., 2017). The neuroectoderm is the main structure that forms the central nervous system, and the NC and CP are involved in the formation of the peripheral nervous system (Breau and Schneider-Maunoury, 2014; Ji et al., 2019). To determine the capabilities of the iPSCs and P-iPSCs to differentiate into neurons, we used the human embryonic stem cell line (hESCs), iPSCs, and P-iPSCs to induce the formation of the NC and CP. According to the cell morphology and AP staining results, the NC and CP presented different cell morphologies, and the cell morphology of NC was similar to that of nerve cells, while the cellular morphology of CP resembled that of fibroblasts (Figure 4B). NC cells induced from hESCs, iPSCs, and P-iPSCs express the NC-specific marker SOX10 at the protein level and the ectodermal marker NESTIN and PAX6. CP cells induced from hESCs, iPSCs, and P-iPSCs express the CP-specific marker SIX1, as well as the ectoderm markers NESTIN and PAX6, while SOX2 is weakly expressed (Figure 4C). These data indicate that hESCs, iPSCs, and P-iPSCs were all capable of differentiating to NC and CP cells by our differentiation model, and these NC- and CP-induced cells showed comparable cell morphology and AP staining.
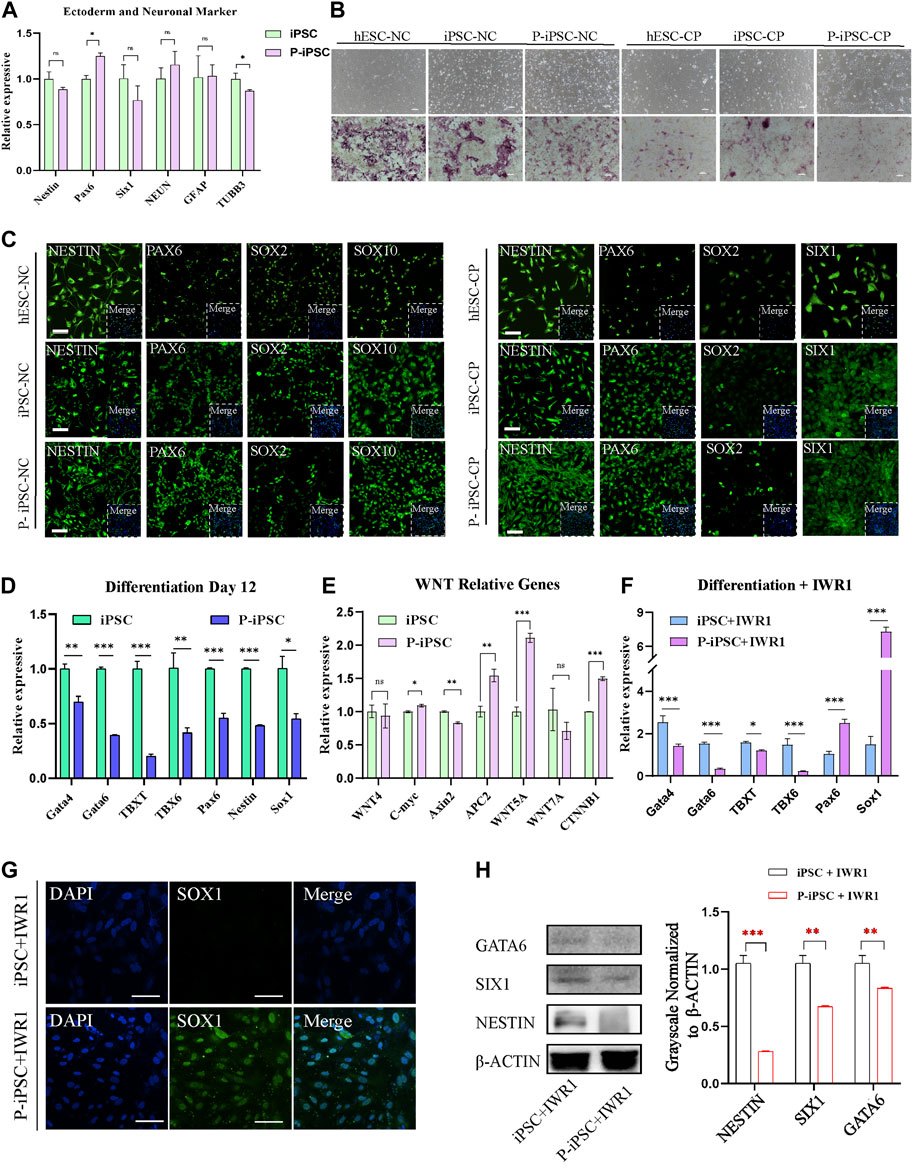
FIGURE 4. Differentiation of iPSCs and P-iPSCs toward the ectoderm. (A) Real-time PCR analysis of ectoderm- and neuronal-associated gene expression in the iPSCs and P-iPSCs; iPSCs were used as the control. Data were obtained in triplicate and presented as the mean ± SD. p-values were calculated by two-way ANOVA, *p < 0.05, **p < 0.01, and ***p < 0.001. (B) Cell morphology and AP staining of hESCs, iPSCs, and P-iPSCs differentiated to the ectoderm (scale bars, 200 μm). (C) Immunofluorescence for ectoderm-oriented cell markers derived from hESCs, iPSCs, and P-iPSCs (scale bars, 100 μm). (D) Real-time PCR analysis of endoderm-, mesoderm-, and ectoderm-related gene expression in iPSCs and P-iPSCs differentiated on day 12; iPSCs were used as the control. (E) Real-time PCR analysis of the Wnt signaling pathway-associated gene expression in the iPSCs and P-iPSCs; iPSCs were used as the control. (F) Real-time PCR analysis of endoderm-, mesoderm-, and ectoderm-related gene expression in iPSCs and P-iPSCs differentiated on day 12 of IWR1 treatment; iPSCs were used as the control. (G) Immunostaining of SOX1 in iPSCs and P-iPSCs differentiated on day 12 of IWR1 treatment (scale bars, 100 μm). (H) Western blotting analysis for GATA6, SIX1, and NESTIN in iPSCs and P-iPSCs differentiated on day 12 of IWR1 treatment, and the band intensity of Western blotting was quantified in the control.
Interestingly, when we tested the ability of iPSCs and P-iPSCs to differentiate in vitro by the common method in the M10 basic medium for 12 days, we observed that the expression of all mesoderm, endoderm, and ectoderm marker genes was significantly decreased in P-iPSCs compared to iPSCs as analyzed by pPCR (Figure 4D).
Since our RNA-seq analysis showed enrichment of the Wnt signaling pathway in P-iPSCs when compared to iPSCs, we performed RT-qPCR on key genes, and the data were consistent with the RNA-Seq data (Figure 4E). We then treated iPSCs and P-iPSCs with 2 μM of the Wnt signaling pathway inhibitor IWR1 during the differentiation process and found that the expression of ectoderm markers Pax6 and Sox1 significantly increased in P-iPSCs (Figure 4F). These findings were confirmed by immunofluorescence staining against SOX1 (Figure 4G). Furthermore, the protein level of endoderm marker GATA6 and ectoderm markers SIX1 and NESTIN were downregulated in P-iPSCs compared with iPSCs differentiated on day 12 of IWR1 treatment (Figure 4H). These data indicated that after inhibiting the Wnt signaling pathway, the ectoderm marker Sox1 was very sensitive in P-iPSCs. Previous studies have also shown that the Wnt signaling pathway plays an important role in the regulation of mESC (Ying et al., 2008) and in hESC pluripotency (Neagu et al., 2020; Bayerl et al., 2021). Recently, Liu et al. (2021) treated hPSCs with the Wnt activator (CHIR-99021) and leukemia inhibitory factor (LIF) in a chemically defined medium (N2B27) to induce neural stem cells (NSCs). Together, these results provide further support that the Wnt signaling pathway may play important roles in the differentiation of the ectoderm.
In this study, we successfully established human iPSCs from normal and patient fibroblasts. RNA sequencing data showed DEGs in iPSCs and P-iPSCs were enriched in the Wnt pathway, PI3K-AKT pathway, and JAK-STAT pathway. In particular, the NC and CP were successfully induced from iPSCs and P-iPSCs. Importantly, P-iPSCs were more sensitive to the Wnt pathway, and Wnt signaling inhibition resulted in an increased expression of SOX1, an ectoderm marker.
In 2008, iPSC technology was used to generate disease-specific iPSCs from somatic cells from patients with Parkinson’s, Huntington’s, and Gaucher’s disease (Park et al., 2008). Here, we used the PiggyBac eight-factor system and successfully established human iPSCs from the fibroblasts of normal individual and patient with ATM. A recent study also showed that using a similar eight-factor bovine protocol could establish expanded potential bovine stem cells (Zhao et al., 2021). iPSCs undergo epigenetic reprogramming (Takikawa et al., 2013; Li et al., 2019; Yagi et al., 2019), and some human iPSCs in culture displayed an increased chance of chromosomal abnormalities (Aasen et al., 2008). Using our approach, both iPSC and P-iPSC exhibited normal karyotypes (Figures 2E, F).
GSEA on HEF and P-HAF, as well as iPSCs and P-iPSCs, showed that the most significantly enriched pathways were related to inflammation. These findings indicate that although reprogramming will reduce the number of DEGs, some genes, such as those associated with inflammation-related pathways, are conserved in these processes. Thus, this process of establishing iPSCs using the eight-factor system is very stable. P-iPSCs can, therefore, be used as a model for further studies on ATM.
At present, research on ATM has focused more on clinical imaging analysis, treatment, and prognosis and is based generally on case reports. There are very few studies investigating the underlying pathologic mechanisms of ATM. In a study involving 42 patients with ATM, the levels of thyroid-stimulating hormone (TSH) and free triiodothyronine (FT3) in patients were lower than those in the healthy control group (Weng et al., 2017a). ATM, which involves multiple components of the central nervous system, including oligodendrocytes, neurons, axons, and the myelin sheath, is a mixed inflammatory disease (Kerr and Ayetey, 2002). In addition, histopathological studies have shown that ATM is associated with the activation of astrocytes and microglia (Krishnan et al., 2004).
The clinical features of ATM include a relatively acute onset of motor, sensory, and autonomic dysfunction, although peripheral nervous system lesions or damage have also been reported in patients with optic neuromyelitis and acute disseminated encephalomyelitis (Tavasoli and Tabrizi, 2018). To date, very little is known about the pathophysiology of the disease. Our study demonstrates that the PiggyBac eight-factor system is a successful approach for reprograming the patient fibroblasts to P-iPSCs, and it would be interesting to apply this in reprogramming thyrocytes, astrocytes, and microglia. These studies will provide powerful tools for understanding the pathogenesis of ATM.
Interestingly, we found that the expression of the ectoderm marker Sox1 significantly increased in P-iPSCs following exposure to Wnt signaling inhibitor IWR1. The SOX1 and Wnt signaling pathways may be a clue to understand the underlying molecular developmental defects in ATM pathogenesis. Kan et al. (2004) reported that SOX1 binds to β-catenin, potentially attenuating the Wnt signaling pathway to regulate neurogenesis. Furthermore, Venere et al. (2012) found that SOX1 is expressed in a subset of astrocytes, and the subpopulation of SOX1-marked cells has long-term neurogenic potential. It remains to be investigated how SOX1 and Wnt signaling pathways play their roles in ectoderm differentiation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All animal experiments were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Inner Mongolia University, China. The study approval number is “NMGDX (Wu) 2022-0,002.” Informed consent was obtained from all subjects involved in the study.
Conceptualization: SB and XL; methodology: SC, XG, FL, YC, QM, QN, and PS; writing—original draft preparation: SC and SB; writing—review and editing: SC, SB, XL, BW, and YS; and bioinformatic analyses: FL. All authors contributed to the article and approved the submitted version.
This work was supported by grants National Key R&D Program of China (2022YFD1302202/3); Program of Higher-Level Talents of Inner Mongolia University (10000-21311201/058); Inner Mongolia Autonomous Region Natural Science Foundation (2021MS03003); Program of Advanced Foreign Experts Plan of Inner Mongolia Autonomous Region (21400-5213401).
The authors are grateful to Yu Tian for RNA-seq analyses. The authors thank Juan Li for reviewing the manuscript, Ren Bu for providing material, and Yunxia Li, Qing Qing, and Ya Ji for their generous cooperation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1172385/full#supplementary-material
Aasen, T., Raya, A., Barrero, M. J., Garreta, E., Consiglio, A., Gonzalez, F., et al. (2008). Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284. doi:10.1038/nbt.1503
Bayerl, J., Ayyash, M., Shani, T., Manor, Y. S., Gafni, O., Massarwa, R., et al. (2021). Principles of signaling pathway modulation for enhancing human naive pluripotency induction. Cell. Stem Cell. 28, 1549–1565.e12. doi:10.1016/j.stem.2021.04.001
Braverman-Gross, C., Nudel, N., Ronen, D., Beer, N. L., McCarty, M. I., and Benvenisty, N. (2018). Derivation and molecular characterization of pancreatic differentiated MODY1-iPSCs. Stem Cell. Res. 31, 16–26. doi:10.1016/j.scr.2018.06.013
Breau, M. A., and Schneider-Maunoury, S. (2014). Mechanisms of cranial placode assembly. Int. J. Dev. Biol. 58, 9–19. doi:10.1387/ijdb.130351mb
Ceglie, G., Papetti, L., Valeriani, M., and Merli, P. (2020). Hematopoietic stem cell transplantation in neuromyelitis optica-spectrum disorders (NMO-SD): State-of-the-Art and future perspectives. Int. J. Mol. Sci. 21, 5304. doi:10.3390/ijms21155304
Choi, I. Y., Lim, H., Estrellas, K., Mula, J., Cohen, T. V., Zhang, Y., et al. (2016). Concordant but varied phenotypes among Duchenne muscular dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell. Rep. 15, 2301–2312. doi:10.1016/j.celrep.2016.05.016
de Seze, J., Lanctin, C., Lebrun, C., Malikova, I., Papeix, C., Wiertlewski, S., et al. (2005). Idiopathic acute transverse myelitis: Application of the recent diagnostic criteria. Neurology 65, 1950–1953. doi:10.1212/01.wnl.0000188896.48308.26
Fraga, A. M., de Araujo, E. S. S., Stabellini, R., Vergani, N., and Pereira, L. V. (2011). A survey of parameters involved in the establishment of new lines of human embryonic stem cells. Stem Cell. Rev. Rep. 7, 775–781. doi:10.1007/s12015-011-9250-x
Gao, X. F., Nowak-Imialek, M., Chen, X., Chen, D. S., Herrmann, D., Ruan, D. G., et al. (2019). Establishment of porcine and human expanded potential stem cells. Nat. Cell. Biol. 21, 687–699. doi:10.1038/s41556-019-0333-2
Guan, J. Y., Wang, G., Wang, J. L., Zhang, Z. Y., Fu, Y., Cheng, L., et al. (2022). Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature 605, 325–331. doi:10.1038/s41586-022-04593-5
Guo, G., Stirparo, G. G., Strawbridge, S. E., Spindlow, D., Yang, J., Clarke, J., et al. (2021). Human naive epiblast cells possess unrestricted lineage potential. Cell. Stem Cell. 28, 1040–1056.e6. doi:10.1016/j.stem.2021.02.025
Guo, Y. L., Zeng, Q. L., Liu, S. W., Yu, Q., Wang, P. Y., Ma, H. J., et al. (2018). Generation of an iPS cell line via a non-integrative method using urine-derived cells from a patient with USH2A-associated retinitis pigmentosa. Stem Cell. Res. 29, 139–142. doi:10.1016/j.scr.2018.03.022
Ji, Y., Hao, H. Y., Reynolds, K., McMahon, M., and Zhou, C. J. J. (2019). Wnt signaling in neural crest ontogenesis and oncogenesis. Cells-Basel 8, 1173. doi:10.3390/cells8101173
Kahn, I. (2020). Acute transverse myelitis and acute disseminated encephalomyelitis. Pediatr. Rev. 41, 313–320. doi:10.1542/pir.2019-0179
Kan, L., Israsena, N., Zhang, Z., Hu, M., Zhao, L. R., Jalali, A., et al. (2004). Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 269, 580–594. doi:10.1016/j.ydbio.2004.02.005
Kerr, D. A., and Ayetey, H. (2002). Immunopathogenesis of acute transverse myelitis. Curr. Opin. Neurol. 15, 339–347. doi:10.1097/00019052-200206000-00019
Klipfel, L., Cordonnier, M., Thiebault, L., Clerin, E., Blond, F., Millet-Puel, G., et al. (2021). A splice variant in SLC16A8 gene leads to lactate transport deficit in human iPS cell-derived retinal pigment epithelial cells. Cells-Basel 10, 179. doi:10.3390/cells10010179
Krishnan, C., Kaplin, A. I., Deshpande, D. M., Pardo, C. A., and Kerr, D. A. (2004). Transverse myelitis: Pathogenesis, diagnosis and treatment. Front. Biosci-Landmrk 9, 1483–1499. doi:10.2741/1351
Li, S. S., Pan, H. Y., Tan, C., Sun, Y. P., Song, Y. R., Zhang, X., et al. (2018). Mitochondrial dysfunctions contribute to hypertrophic cardiomyopathy in patient iPSC-derived cardiomyocytes with MT-RNR2 mutation. Stem Cell. Rep. 10, 808–821. doi:10.1016/j.stemcr.2018.01.013
Li, X. J., Li, M. J., Yang, Y., and Bai, Y. (2019). Effects of reprogramming on genomic imprinting and the application of pluripotent stem cells. Stem Cell. Res. 41, 101655. doi:10.1016/j.scr.2019.101655
Liu, B. C., Liu, F. Y., Gao, X. Y., Chen, Y. L., Meng, Q. Q., Song, Y. L., et al. (2021). Global transcriptional analyses of the wnt-induced development of neural stem cells from human pluripotent stem cells. Int. J. Mol. Sci. 22, 7473. doi:10.3390/ijms22147473
Loser, P., Schirm, J., Guhr, A., Wobus, A. M., and Kurtz, A. (2010). Human embryonic stem cell lines and their use in international research. Stem Cells 28, 240–246. doi:10.1002/stem.286
McNally, E. M., and Mestroni, L. (2017). Dilated cardiomyopathy genetic determinants and mechanisms. Circ. Res. 121, 731–748. doi:10.1161/CIRCRESAHA.116.309396
Neagu, A., van Genderen, E., Escudero, I., Verwegen, L., Kurek, D., Lehmann, J., et al. (2020). In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states. Nat. Cell. Biol. 22, 534–545. doi:10.1038/s41556-020-0508-x
Park, I. H., Arora, N., Huo, H., Maherali, N., Ahfeldt, T., Shimamura, A., et al. (2008). Disease-specific induced pluripotent stem cells. Cell. 134, 877–886. doi:10.1016/j.cell.2008.07.041
Sasaki, K., Makiyama, T., Yoshida, Y., Wuriyanghai, Y., Kamakura, T., Nishiuchi, S., et al. (2016). Patient-specific human induced pluripotent stem cell model assessed with electrical pacing validates S107 as a potential therapeutic agent for catecholaminergic polymorphic ventricular tachycardia. Plos One 11, e0164795. doi:10.1371/journal.pone.0164795
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131, 861–872. doi:10.1016/j.cell.2007.11.019
Takikawa, S., Ray, C., Wang, X., Shamis, Y., Wu, T. Y., and Li, X. J. (2013). Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell. Res. 11, 861–873. doi:10.1016/j.scr.2013.05.011
Tavasoli, A., and Tabrizi, A. (2018). Acute transverse myelitis in children, literature Review. Iran. J. Child. Neurol. 12, 7–16.
Tchieu, J., Zimmer, B., Fattahi, F., Amin, S., Zeltner, N., Chen, S. B., et al. (2017). A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell. Stem Cell. 21, 399–410.e7. doi:10.1016/j.stem.2017.08.015
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. doi:10.1126/science.282.5391.1145
Ueki, J., Nakamori, M., Nakamura, M., Nishikawa, M., Yoshida, Y., Tanaka, A., et al. (2017). Myotonic dystrophy type 1 patient-derived iPSCs for the investigation of CTG repeat instability. Sci. Rep. 7, 42522. doi:10.1038/srep42522
Venere, M., Han, Y. G., Bell, R., Song, J. S., Alvarez-Buylla, A., and Blelloch, R. (2012). Sox1 marks an activated neural stem/progenitor cell in the hippocampus. Development 139, 3938–3949. doi:10.1242/dev.081133
Weng, Y., Lin, H., Ye, X., Xie, D., Chen, Z., Zheng, J., et al. (2017a). Thyroid hormone level is associated with the frequency and severity of acute transverse myelitis. Neuroreport 28, 292–297. doi:10.1097/WNR.0000000000000753
West, T. W., Hess, C., and Cree, B. A. (2012). Acute transverse myelitis: Demyelinating, inflammatory, and infectious myelopathies. Seminars neurology 32, 97–113. doi:10.1055/s-0032-1322586
Yabe, S. G., Nishida, J., Fukuda, S., Takeda, F., Nasiro, K., Yasuda, K., et al. (2019). Expression of mutant mRNA and protein in pancreatic cells derived from MODY3-iPS cells. Plos One 14, e0217110. doi:10.1371/journal.pone.0217110
Yagi, M., Kabata, M., Ukai, T., Ohta, S., Tanaka, A., Shimada, Y., et al. (2019). De novo DNA methylation at imprinted loci during reprogramming into naive and primed pluripotency. Stem Cell. Rep. 12, 1113–1128. doi:10.1016/j.stemcr.2019.04.008
Yan, X., Guo, Y. L., Chen, J., Cui, Z. K., Gu, J. N., Wang, Y. N., et al. (2019). Establishment of CSUASOi001-A, a non-integrated induced pluripotent stem cell line from urine-derived cells of a Chinese patient carrying RS1 gene mutation. Stem Cell. Res. 38, 101466. doi:10.1016/j.scr.2019.101466
Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. doi:10.1038/nature06968
Zhao, L. X., Gao, X. F., Zheng, Y. X., Wang, Z. X., Zhao, G. P., Ren, J., et al. (2021). Establishment of bovine expanded potential stem cells. P Natl. Acad. Sci. U. S. A. 118, e2018505118. doi:10.1073/pnas.2018505118
Zhou, Y. L., Ding, C. C., Xia, S. T., Jing, Y. T., Mao, S. R., Liu, J., et al. (2020). Establishment of induced pluripotent stem cell line CSUASOi003-A from an autosomal recessive retinitis pigmentosa patient carrying compound heterozygous mutations in CRB1 gene. Stem Cell. Res. 44, 101742. doi:10.1016/j.scr.2020.101742
Keywords: induced pluripotent stem cells, acute myelitis, reprogramming, ectoderm, differentiation
Citation: Cao S, Gao X, Liu F, Chen Y, Na Q, Meng Q, Shao P, Chen C, Song Y, Wu B, Li X and Bao S (2023) Derivation and characteristics of induced pluripotent stem cells from a patient with acute myelitis. Front. Cell Dev. Biol. 11:1172385. doi: 10.3389/fcell.2023.1172385
Received: 23 February 2023; Accepted: 03 July 2023;
Published: 14 July 2023.
Edited by:
Qiang Wu, Macau University of Science and Technology, Macao SAR, ChinaReviewed by:
Min Huang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Cao, Gao, Liu, Chen, Na, Meng, Shao, Chen, Song, Wu, Li and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siqin Bao, YmFvc3FAaW11LmVkdS5jbg==, Xihe Li, bGl4aEBpbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.