- 1National Translational Science Center for Molecular Medicine, Department of Cell Biology, School of Basic Medicine, The Fourth Military Medical University, Xi’an, China
- 2Department of Oncology, Tangdu Hospital, The Fourth Military Medical University, Xi’an, China
- 3Unit 94710 of the PLA, Wuxi, China
- 4Department of Urology, Xijing Hospital, The Fourth Military Medical University, Xi’an, China
Background: Kidney renal clear cell carcinoma (KIRC) is an immunogenic tumor, and immune infiltrates are relevant to patients’ therapeutic response and prognosis. NDUFS1, the core subunit of mitochondrial complex I, has been reported to be associated with KIRC patients’ prognosis. However, the upstream regulator for NDUFS1 and their correlations with immune infiltration remain unclear.
Methods: The expression of NDUFS genes in KIRC and their influences on patients’ survival were investigated by UALCAN, ENCORI, Oncomine, TIMER as well as Kaplan-Meier Plotter. miRNAs regulating NDUFS1 were predicted and analyzed by TargetScan and ENCORI. The correlations between NDUFS1 expression and immune cell infiltration or gene marker sets of immune infiltrates were analyzed via TIMER. The overall survival in high/low NDUFS1 or hsa-miR-320b expressed KIRC patients with or without immune infiltrates were analyzed via Kaplan-Meier Plotter. The combined NDUFS1 expression and/or CD4+ T cell infiltration on KIRC patients’ overall survival were validated by multiplexed immunofluorescence (mIF) staining in tissue microarray (TMA). Furthermore, the influences of NDUFS1 expression on the chemotaxis of CD4+ T cells to KIRC cells were performed by transwell migration assays.
Results: We found that the low expression of NDUFS1 mRNA and protein in KIRC was correlated with unfavorable patients’ survival and poor infiltration of CD4+ T cells. In patients with decreased CD4+ T cell infiltration whose pathological grade less than III, TMA mIF staining showed that low expression of NDUFS1 had significantly poor OS than that with high expression of NDUFS1 did. Furthermore, hsa-miR-320b, a possible negative regulator of NDUFS1, was highly expressed in KIRC. And, low NDUFS1 or high hsa-miR-320b consistently correlated to unfavorable outcomes in KIRC patients with decreased CD4+ T cell infiltration. In vitro, NDUFS1 overexpression significantly increased the chemotaxis of CD4+ T cell to KIRC cells.
Conclusion: Together, NDUFS1, upregulated by decreased hsa-miR-320b expression in KIRC patients, might act as a biomarker for CD4+ T cell infiltration. And, the combination of NDUFS1 with CD4+ T cell infiltration predicts favorable prognosis in KIRC.
1 Introduction
Kidney renal clear cell carcinoma (KIRC), characterized by biallelic loss of function of the von Hippel Lindau (VHL) tumor suppressor gene, is the adenocarcinoma derived from the renal tubular epithelial cell. The amount of new KIRC cases is ranking seventh in all cancer types around the world (Siegel et al., 2022). As a deadly or fatal disease, one-third of patients have advanced or metastatic disease at diagnosis; and 30%–40% of patients have tumor recurrence after surgical resection (Toth and Cho, 2020; Singh, 2021). As an important strategy for KIRC, the systemic therapy has successively experienced the 4 stages of development: the precytokine era, the cytokine era, the molecularly targeted era (2005–2014), and the immune checkpoint blockade era (2015 to present), with 2 stages relating to immunotherapy (Hasanov et al., 2020).
In contrast to limited efficacy and serious toxicity of cytokine therapy that adopts high dose of IL-2 and IFN-α, immune checkpoint inhibitors (ICIs) targeting programmed death (PD)-1 or CTLA-4 have improved efficacy and safety greatly. However, a large percentage of KIRC patients fail to obtain complete responses and durable remissions, whether by using ICIs single-agent or the combination of ICIs with tyrosine kinase inhibitors (TKIs) (Hasanov et al., 2020; Wang et al., 2021). The reasons for the resistant mechanisms may include absence of antigenic proteins or defects in antigen presentation, decreased T cell activity, the existence of other inhibitory checkpoints such as VISTA, LAG-3, and TIM-3, and having other immunosuppressive cells like tumor-associated macrophages, or regulatory T cells (Hasanov et al., 2020). Besides these, immune infiltration has been considered as a significant determinant that helps to select optimal patients or to predict therapeutic response and prognosis. For example, it is reported that the recurrence of KIRC was related to lower T cells, while a higher T effector (Teff)/Treg ratio would reduce recurrence rate (Ghatalia et al., 2019). In COMPARZ phase III trial of first-line TKIs sunitinib or pazopanib, patients’ comparably therapeutic effect was sufficiently affected by the immune infiltration level (Laurell et al., 2017). Thus, biomarkers identifying immune cell infiltration are urgently required.
Mitochondrial metabolism is essential for macromolecular synthesis, cell proliferation, cell migration/invasion, cell division or cell differentiation, etc (Vasan et al., 2020). By adopting general features of immune-based groupings and incorporating transcriptomic and proteomic features, Clark DJ et al. divided clear cell renal cell carcinoma (ccRCC) cohort into four subtypes: CD8+ infiamed, CD8− infiamed, VEGF immune desert and metabolic immune desert (Clark et al., 2019). Metabolic immune desert tumors display low immune scores but elevated mitochondrial, oxidative phosphorylation (OXPHOS) and glycolysis protein expression, indicating that metabolism is tightly associated with immune microenvironment in ccRCC. These findings suggest that cell metabolism is connected to immune signatures, and mitochondrial or OXPHOS protein that associated with immune cell infiltration would be a potential novel biomarker for patient selection or prognosis monitoring.
Mitochondrial complex I (NADH dehydrogenase) is the first and largest mitochondrial enzyme complex of OXPHOS. In many cancers including KIRC (or ccRCC), the dysfunction of complex I resulted in activating Akt pathway and cancer progression (Santidrian et al., 2013; Ellinger et al., 2017). Complex I is composed of 45 subunits in mammals, and 8 of which form a chain of iron-sulfur (Fe/S) clusters that encoded by NDUFS [(NADH dehydrogenase (ubiquinone) Fe-S protein)] genes. Their encoded protein, namely, NDUFSs, are responsible to transfer electron from NADH to coenzyme Q or ubiquinone. Although the expression profiles and prognosis values of NDUFS genes in KIRC have been reported recently, the relationship between NDUFS1 and immune cell infiltration in cancer have not been explored. Besides, the detailed role, mechanism and regulation of NDUFS genes in cancer progression remain unknown.
In this study, through digging into public online databases, we firstly screened the key player in NDUFS genes family based on expression levels and patients’ survival in KIRC; then we explored whether NDUFS genes were correlated with immune infiltration and whether their correlation could combinedly predict the patients’ prognosis; Finally we probed the upstream regulator for NDUFS1 and validated their individual correlations with immune cell infiltration and/or with KIRC patients’ prognosis. Our findings highlight that NDUFS1, downregulated by hsa-miR-320b, might act as a biomarker for CD4+ T cell infiltration and predict favorable prognosis in KIRC patients. Our understanding of NDUFS genes in predicting KIRC immune cell infiltration would help to identify patients that benefited from ICIs and to develop the rational combinations of mitochondrial inhibitors with ICIs for future novel and efficacious anti-cancer treatments.
2 Materials and methods
2.1 Study design and data collection
As described in the flow chart (Supplementary Figure S1), we screened differentially expressed genes (DEGs) in NDUFS gene family and overall survival (OS) analysis of NDUFS genes in KIRC by intersecting three online databases, respectively. We further explored NDUFS1 protein expression, its prognostic value and NDUFS1-correlated immune infiltrates in KIRC by digging into UALCAN and conducting TMA multiplexed immunofluorescence (mIF) validation. Meanwhile, possible miRNAs targeting NDUFS1 and NDUFS1-related cuproptosis genes were predicted and analyzed via online databases. Moreover, to analyze the combined prognostic effect of NDUFS1 with miRNAs or cuproptosis-regulated gene in KIRC, we collected mRNA and miRNA sequencing data of TCGA database from Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov).
2.2 UALCAN database analysis
UALCAN (RRID:SCR_015827) (http://ualcan.path.uab.edu/index.html) is a comprehensive and interactive web resource which is designed to analyze cancer OMICS data including TCGA and CPTAC database, and to evaluate epigenetic regulation of gene expression (Chandrashekar et al., 2017). We used UALCAN for investigating the NDUFS genes’ expression profiles in KIRC and the relationship of NDUFS1 expression with tumor grades, individual cancer stages or KIRC subtypes. Besides, the protein expression and promoter methylation of NDUFS1 in KIRC were also analyzed.
2.3 ENCORI database analysis
ENCORI (RRID:SCR_016303) (http://starbase.sysu.edu.cn/index.php) is an open-source platform for studying miRNA-target interactions, of which 2.5 million miRNA-mRNA interactions are included. ENCORI also provides platforms for analyzing the differential expression and survival of miRNAs and the targeted mRNAs (Li et al., 2014). Herein on this platform, the expression profiles and overall survival of NDUFS genes, the predicted miRNAs, and their correlation with patients’ survival time in KIRC were explored.
2.4 GEPIA database analysis
GEPIA 2.0 (Gene Expression Profiling Interactive Analysis, RRID:SCR_018294) (http://gepia2.cancer-pku.cn/) is an interactive web server for analyzing the RNA sequencing expression data from the TCGA and the GTEx projects, which provides differential expression analysis, patient survival analysis, etc (Tang et al., 2019). Thus, we acquired the NDUFS genes’ expression profiles between normal and cancer tissues and the overall survival data on this platform.
2.5 Kaplan-Meier Plotter database analysis
The Kaplan Meier plotter (RRID:SCR_018753) (https://kmplot.com/analysis/) is an online database which is capable to assess the effect of 54,000 genes (mRNA, miRNA, protein) on patients’ survival in 21 cancer types from GEO, EGA, and TCGA (Nagy et al., 2021). We used this database to predict the OS and disease-free survival (DFS) of KIRC patients according to the expression of NDUFS genes or hsa-miR-320b and the association of their expression with clinical outcome at different levels of immune infiltrates.
2.6 Oncomine database analysis
Oncomine (RRID:SCR_007834) (https://www.oncomine.org/resource/main.html) datasets are composed of microarray data measuring either mRNA expression or DNA copy number in primary tumors, cell lines or xenografts, usually from published research (Rhodes et al., 2007). We use Oncomine database to compare the expression profiles of NDUFS1 between normal and tumor tissues in different cancer types.
2.7 TIMER database analysis
TIMER 2.0 (RRID:SCR_018737) (http://timer.cistrome.org/) is a comprehensive web server for estimating the abundance of immune infiltrates across 32 cancer types from The Cancer Genome Atlas (TCGA) (Li et al., 2020). We applied TIMER for analyzing the following three aspects: the NDUFS1 expression in different types of cancers and its correlation with the abundance of immune infiltrates including NK, neutrophils, CD4+ T, macrophage M1 and NKT cells were explored via gene modules; the correlation of NDUFS1 expression with the gene markers of tumor-infiltrating immune cells was analyzed via correlation modules; and the association between immune infiltrates and clinical outcomes was investigated via outcome modules.
2.8 CProSite database analysis
The cProSite (Cancer Proteogenomic Data Analysis Site) (https://cprosite.ccr.cancer.gov/) is a web-based interactive platform providing visualization of proteomic analysis from the datasets of the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) and National Cancer Institute’s International Cancer Proteogenome Consortium (ICPC). We used cProSite to analyze correlation between the level of NDUFS1 protein and its mRNA or FDX1 protein in KIRC.
2.9 TargetScan database analysis
TargetScan (RRID:SCR_010845) (http://www.targetscan.org) can predict biological targets of miRNAs by searching for the presence of conserved 8mer and 7mer sites that match the seed region of each miRNA (Lewis et al., 2005). We used TargetScan to predict the possibly conserved miRNAs that regulate NDUFS1.
2.10 Tissue microarray (TMA) and multiplexed immunofluorescence (mIF) staining
Human KIRC TMA (Cat.HKidE180Su03, OUTDO BIOTECH, China) were constructed with tumor and paired adjacent tissues from a cohort including 90 primary KIRC patients who were operated between 2006.10 and 2008.2 and were followed for 7–9 years. Patient information including age, gender and follow-up information such as survival times were recorded. For mIF staining, TMA slides were firstly incubated with rabbit monoclonal anti-NDUFS1 antibody (1:200, Cat.ab169540, Abcam, United Kingdom), mouse monoclonal anti-CK antibody (1:400, Cat.PA125, Abcarta, China) and rabbit monoclonal anti-CD4 antibody (1:200, Cat.PA285, Abcarta, China), followed by incubation with goat anti-mouse (1:250, Cat.35502, ThermoFisher Scientific, United States) or goat anti-rabbit (1:500, Cat.A23320, Abbkine, United States) secondary antibodies. The samples were finally counterstained with DAPI and the images were analyzed by TissueFAXS Viewer software. The percentage of NDUFS1 and CK double positive cells in each tissue was described as the expression level of NDUFS1 in KIRC and adjacent normal tissues, and the percentage of CD4 positive cells in each tumor tissues was described as the infiltration level of CD4+ T cell in KIRC. For the survival analysis, patients were divided into low or high expression groups which is cut off by mean value of NDUFS1 or CD4 expression in KIRC.
2.11 Cell culture and transfection
The human kidney renal clear cell lines ACHN and 786-O, and immortal human CD4 T lymphocytes Jurkat T were purchased from the Shanghai Institutes for Biological Sciences (Shanghai, China). Cells were cultured in DMEM (Cat.11965092, Invitrogen) containing 10% fetal bovine serum (Cat.10100147, Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Cat.15240062, Invitrogen). The pcDNA3.1-NDUFS1 plasmid and pcDNA3.1 vector control plasmid were obtained from Tsingke Co., Ltd. (Beijing, China). The transfection was performed by Lipofectamine 2000 (Cat.11668500, Invitrogen).
2.12 Quantitative real-time PCR
Total RNA was isolated from KIRC cells using the total RNA kit II (Omega, CA, United States) and was reverse transcribed into complementary DNA by PrimeScriptTM RT reagent kit (TaKaRa Biotechnology, Otsu, Japan). Single stranded complementary DNA was amplified by quantitative real-time PCR (qRT-PCR) using the SYBR Premix ExTaq kit (TaKaRa Biotechnology) in the Mx3005P Real-Time PCR system (Agilent Technologies, Germany). The following primers were used for qRT-PCR analysis: NDUFS1 forward 5′-TTAGCAAATCACCCATTGGACTG-3′, reverse: 5′-CCCCTCTAAAAATCGGCTCCTA-3′; β-actin forward: 5′-AGCGAGCATCCCCCAAAGTT-3′, reverse:5′- GGGCACGAAGGCT CATCATT-3′.
2.13 Western blot analysis
Western blot analysis was performed according to the standard protocol. Briefly, the proteins were extracted by RIPA (10% PMSF) from KIRC cells, and then were electrophoresed by SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked in 5% skimmed milk and incubated with anti-NDUFS1 (1:10,000, Cat.Ab169540, Abcam, United Kingdom) and anti-β-actin (1:20,000, Cat.66009-1-Ig, Proteintech, China) at 4°C overnight, and then labeled with anti-mouse IgG (Pierce, IL, United States) or anti-rabbit IgG (Pierce) at room temperature for 1 h, respectively. The protein signal was visualized using a Western-Light chemiluminescent detection system (Image Station 4000 MM Pro, MA, United States of America).
2.14 Chemotaxis assays in vitro
Chemotaxis assays in vitro were performed using transwell chambers (Cat.3413, Corning, United States). A total of 5 × 105 Jurkat T cells in 100 μl DMEM culture medium were placed in the upper chambers, 1 × 105 NDUFS1-overexpressed or control KIRC cells in 300 μl DMEM culture medium were seeded in the lower wells and the chambers were incubated for 24 h at 37°C. The migrated Jurkat T cells on the inserts were stained with crystal violet. Chemotaxis ability was determined by counting the stained cells on the bottom surface of each membrane in 4 random fields, and images were captured at 200×magnification.
2.15 Statistical analysis
Pearson or Spearman’s correlation coefficients (R or Rho) and p-values were used to evaluate the correlations of NDUFS1 with miRNAs or immune infiltrates. OS and DFS generated by Kaplan-Meier plots were used to evaluate the prognostic impacts of NDUFS genes or miRNAs and their combination with immune infiltrates, and the results are displayed with p-values based on a log-rank test and a hazard ratio (HR). Two-tailed Student’s t-tests were used to test the significance of differences between two groups in TMA analysis and chemotaxis assays in vivo. p-values less than 0.05 were considered statistically significant.
3 Results
3.1 The expression and survival analysis of NDUFS genes in KIRC
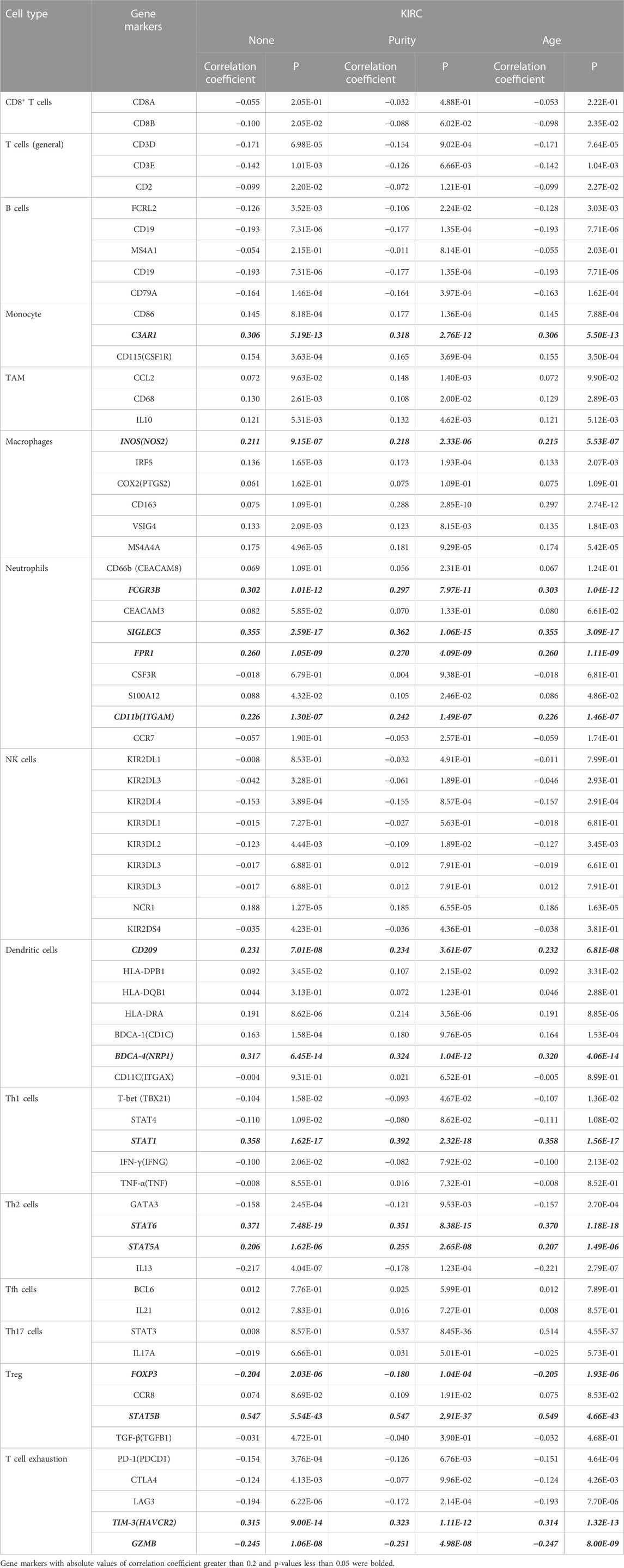
To figure out the expression of NDUFS genes in KIRC patients, we firstly investigated three online databases (Supplementary Figure S2). In UALCAN, NDUFS5 mRNA level was significantly increased while all other seven NDUFS mRNA levels were significantly decreased. In ENCORI, we discovered that NDUFS5 mRNA level again was significantly increased (p = 0.00074), but all other six NDUFS mRNA levels except NDUFS4 were significantly decreased in KIRC tissues as compared to those in normal tissues. When it comes to GEPIA, only NDUFS1 mRNA level was significantly decreased in KIRC tissues comparing with that in normal tissues. Then, we drew Venn diagrams to summarize the above online databases results. As shown in Figure 1A, NDUFS1 was the only overlapped NDUFS gene that differentially expressed between tumor and normal tissues in KIRC patients among UALCAN, ENCORI and GEPIA databases.
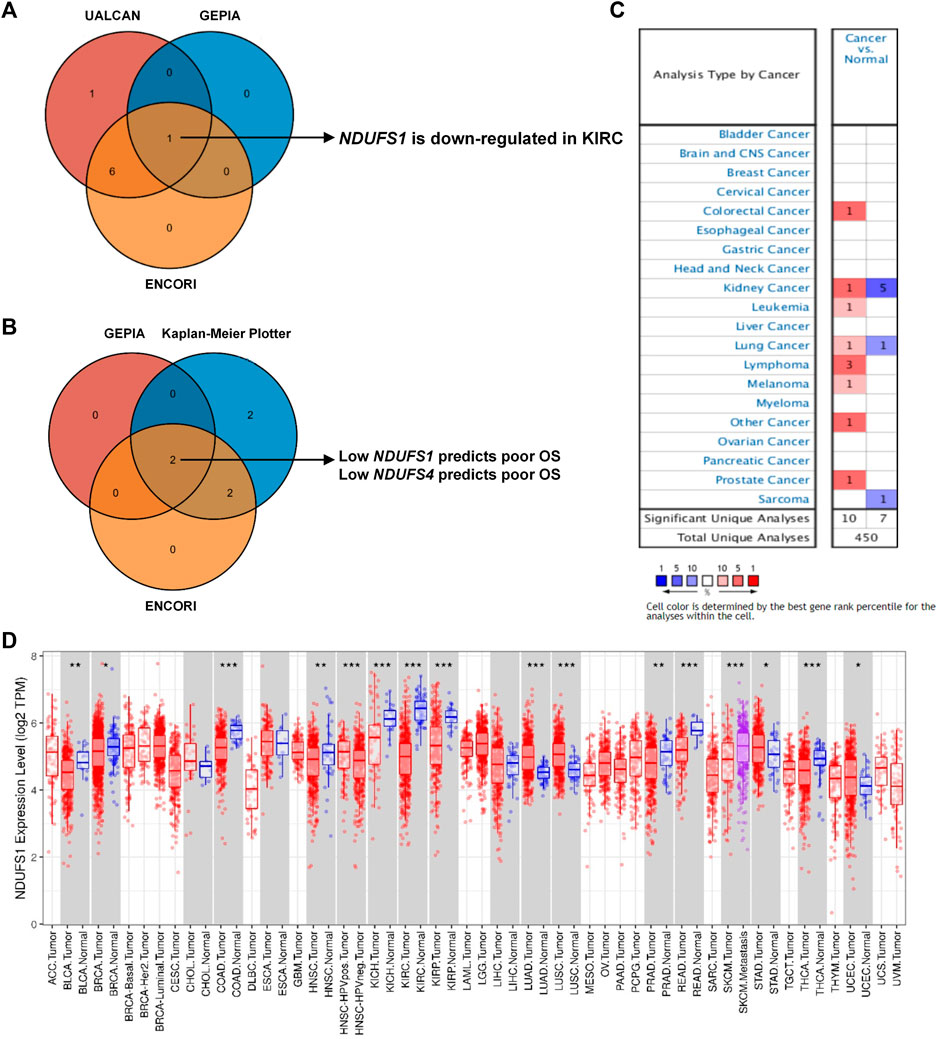
FIGURE 1. Comprehensive NDUFS1 mRNA expression profiles in pan-cancer. (A) Venn diagram comparison of NDUFS genes’ expression among UALCAN, ENCORI and GEPIA database. (B) Venn diagram comparison of the association of NDUFS genes with patient’s survival among GEPIA, Kaplan-Meier Plotter and ENCORI database. (C) NDUFS1 mRNA expression levels in various cancer tissues comparing with normal tissues were analyzed by the Oncomine database. (D) NDUFS1 expression levels in pan-cancers were analyzed by TIMER database. (*p < 0.05, **p < 0.01, ***p < 0.001).
To further explore the association of NDUFS genes’ expression with patients’ survival in KIRC, we investigated the OS in patients with high or low expression of NDUFS genes (Supplementary Figure S3). From GEPIA, we found that KIRC patients with low expression of NDUFS1 or NDUFS4 had significantly poor OS than the patients with high expression did. Meanwhile, Kaplan-Meier Plotter displayed a significantly unfavorable OS in KIRC patients who have lower expression of NDUFS1 or NDUFS4, while a significantly favorable OS in KIRC patients with lower expression of NDUFS3, NDUFS5, NDUFS6, NDUFS7 or NDUFS8. Analysis from ENCORI demonstrated that low expression of NDUFS1 or NDUFS4 was associated with patients’ shorter OS; in contrast, the high level of NDUFS6 or NDUFS8 was related to longer OS in KIRC patients. Moreover, GEPIA database analysis showed that the KIRC patients with low level of NDUFS1, NDUFS2 or NDUFS4 had poor DFS than those with high level did (Supplementary Figure S4). Together, the above three databases consistently demonstrated that the expression of NDUFS1 or NDUFS4 has a significantly positive correlation with favorable survival in KIRC patients (Figure 1B). Combined with the expression profile of NDUFS genes, NDUFS1 is downregulated in KIRC and low expression of NDUFS1 is associated with poor prognosis but high expression associated with favorable prognosis.
3.2 NDUFS1 expression is negatively correlated with clinicopathological features in KIRC
To extend our analysis of NDUFS1 mRNA level in KIRC to the universal expression profiles in other cancer types, we further investigated the Oncomine and TIMER databases. Oncomine database revealed that NDUFS1 mRNA expression was increased significantly in colorectal cancer, leukemia, lymphoma, melanoma and prostate cancer while was decreased significantly in sarcoma (Figure 1C). As for kidney cancer, NDUFS1 was reported to be increased in 1 study while decreased in 5 studies. In TIMER database, NDUFS1 was upregulated significantly in LUAD, LUSC, STAD and UCEC but was downregulated significantly in BLCA, BRCA, COAD, HNSC, PRAD, READ, THCA, and KICH, KIRC, KIRP (Figure 1D). These two databases consistently displayed that NDUFS1 expression was significantly downregulated in the majority of tumor tissues, especially decreased in kidney cancers, including KIRC, KIRP and KICH.
As for the protein level, the expression of NDUFS1 in primary KIRC tumor was also significantly decreased as compared to that in normal tissues, and patients with high NDUFS1 expression had significantly favorable survival than those with low expression did (Figures 2A,B). And, 5-year survivals in high or low NDUFS1 expressed patients were 76% or 51% respectively. Besides, we investigated the correlation between protein and mRNA levels of NDUFS1 in KIRC via cProSite, and we found that the correlation coefficient in adjacent normal tissue was 0.58 while in tumor was only 0.23 (Supplementary Figure S5A), indicating that NDUFS1 mRNA in tumor may be further epigenetically modified at post-transcriptional level. In pan-cancer analysis, the protein level of NDUFS1 was significantly decreased in 6 cancers of breast, colon, pancreas, head and neck, glioblastoma and liver, with decreasing most greatly in ccRCC (KIRC) (Figure 2C). Furthermore, in mIF staining of KIRC TMA, we found that the protein level of NDUFS1 was significantly decreased in KIRC tissues as compared to that in normal tissues (p < 0.0001, Figures 2D,E). And KIRC patients with low NDUFS1 expression had significantly poor overall survival times than those with high NDUFS1 expression did (p = 0.017, Figure 2F). Together, all these data suggested that the expression of both NDUFS1 mRNA and protein were significantly decreased in KIRC patients and correlated with poor patients’ survival.
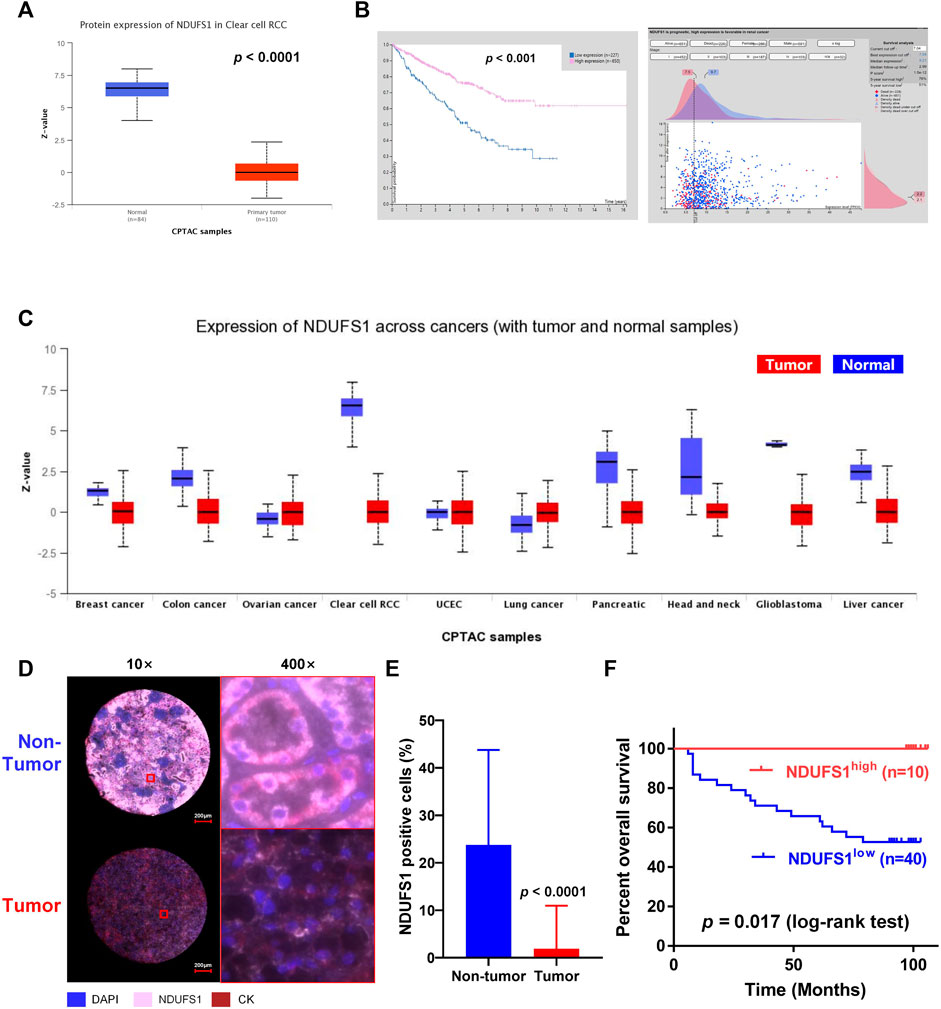
FIGURE 2. The NDUFS1 protein expression and patient’s overall survival analysis in KIRC. (A) The protein expression of NDUFS1 were analyzed by UALCAN database. (B) The percent of 5-year survivals in patients with high or low NDUFS1 protein expression were analyzed by Human Protein Atlas database. (C) NDUFS1 expression of pan-cancer with tumor and non-tumor samples were analyzed by UALCAN database. (D) Representative immunofluorescence images of NDUFS1 positive cells in non-tumor and KIRC tissues. Blue, DAPI; pink, NDUFS1; red, CK. (E) The quantitative analysis of NDUFS1 expression in tumor tissues (n = 50) and non-tumor tissues (n = 82) of KIRC TMA. (F) The overall survival analysis of KIRC patients with high or low NDUFS1 expression.
Moreover, we analyzed the NDUFS1 expression based on different tumor grades, patients’ stages and subtypes through digging into UALCAN database. We found that the expression of NDUFS1 was decreased gradually when the tumor grade was increasing or the individual cancer stage was enhancing, with the lowest NDUFS1 expression occurring in Grade 4 and Stage 4 (Supplementary Figure S5B, C). As far as subtypes were concerned, ccB subtype with poor risk had much lower level of NDUFS1 protein expression than ccA subtype with good risk did (Supplementary Figure S5D). Therefore, lower NDUFS1 expression was correlated with advanced tumor grades, patients’ stages and worse subtype in KIRC.
3.3 The low NDUFS1 expression and the associated poor CD4+ T cell infiltration combinedly predict unfavorable prognosis in KIRC
To figure out whether the low NDUFS1 expression associated poor OS in KIRC patients was related to the altered immune cell infiltration, we then searched the TIMER database. As shown in Figure 3A, NDUFS1 expression was significantly correlated with the infiltration of NK, neutrophil, CD4+ T, macrophage M1 and NKT cells. Judging by the correlation coefficient, however, the NDUFS1 expression has a moderately positive correlation with the infiltration of neutrophil cells (r = 0.555) and CD4+ T cells (r = 0.571), while has a moderately negative correlation with NKT cell infiltration (r = −0.608).
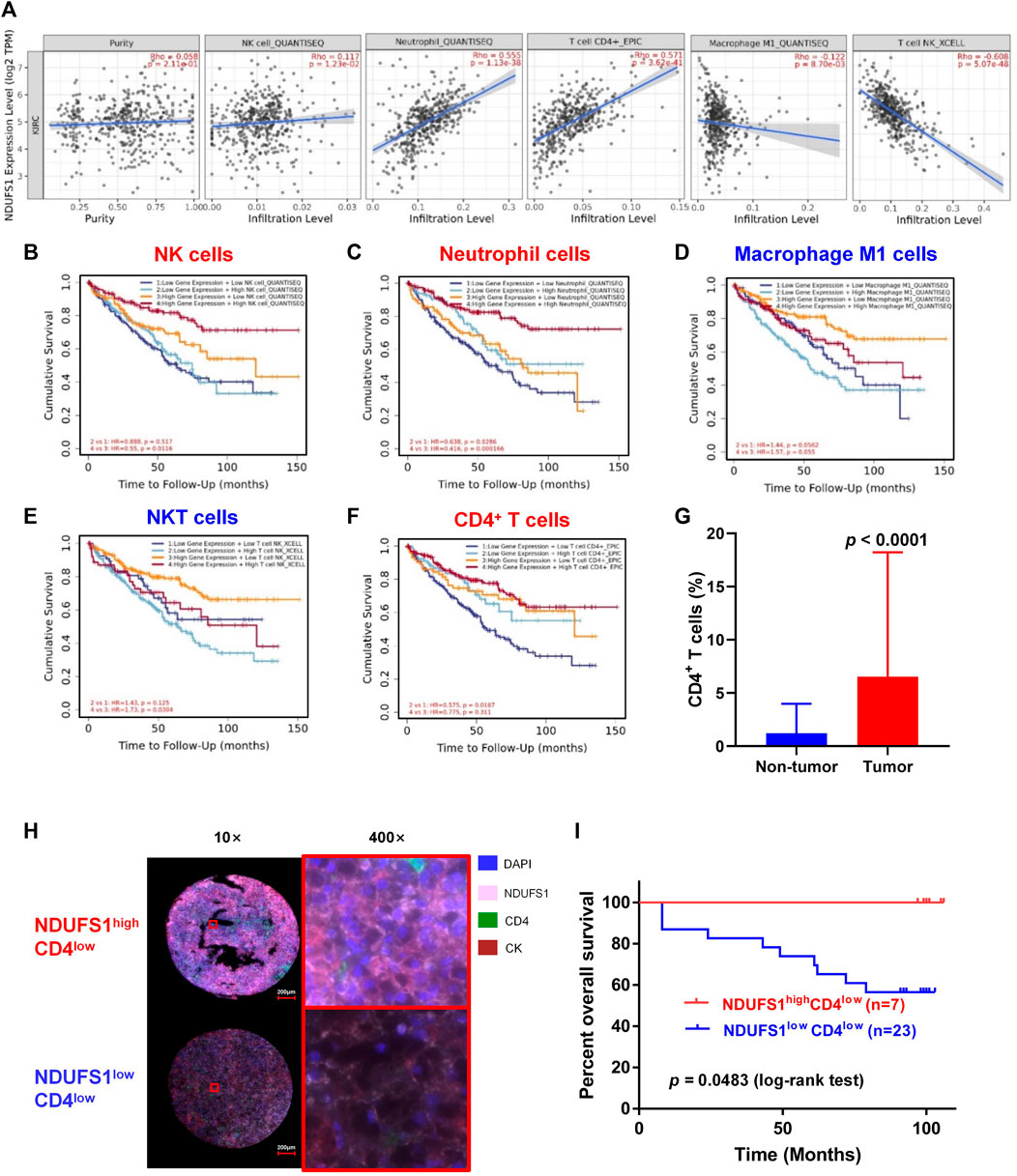
FIGURE 3. The low NDUFS1 expression and associated poor CD4+ T cell infiltration combinedly predict unfavorable survival in KIRC patients. (A) Correlation analysis of NDUFS1 expression with immune infiltration were done by TIMER database. (B–F) Survival curves combining high or low NDUFS1 expression levels with high or low infiltration of immune cells were analyzed by TIMER. (G) The quantitative analysis of CD4 expression in tumor tissues (n = 50) and non-tumor tissues (n = 82) by mIF staining in KIRC TMA. (H) Representative immunofluorescence images of NDUFS1high CD4low and NDUFS1lowCD4low cells in KIRC TMA. Blue, DAPI; pink, NDUFS1; green, CD4; red, CK. (I) The overall survival analysis in NDUFS1high CD4low and NDUFS1lowCD4low KIRC patients whose pathological grade less than Ⅲ.
We further explored the expression correlation of NDUFS1 with marker genes of immune cells (Table 1). Firstly, we found that NDUFS1 was positively correlated with 4 marker genes in neutrophil cells that including FCGR3B (r = 0.302), SIGLEC5 (r = 0.355), FPR1 (r = 0.260) and CD11b (r = 0.226). Then, a positive correlation between the expression of NDUFS1 and 5 marker genes in CD4+ T cell subtypes was seen, for example, STAT1 in Th1 cells (r = 0.358), STAT6 (r = 0.371) and STAT5A (r = 0.206) in Th2 cells, as well as FOXP3 (r = −0.204) and STAT5B (r = 0.547) in Treg cells. Also, the expression of NDUFS1 was positively correlated with TIM-3 (r = 0.315) while negatively correlated with GZMB (r = −0.245) in T cell exhaustion. In addition, NDUFS1 was positively correlated with C3AR1 (r = 0.306) in monocyte, INOS (r = 0.211) in macrophages, as well as CD209 (r = 0.231) and BDCA-4 (r = 0.317) in dendritic cells. And, all above correlations were not affected by tumor purity or patients’ age. Taken together, these results suggested that NDUFS1 highly expressed KIRC had enriched infiltration of immune cells, for example, neutrophil and CD4+ T cells, and that the unfavorable prognosis in NDUFS1 lowly expressed KIRC might be related to the poor infiltration of these immune cells.
We further investigated the influence of combined NDUFS1 expression and immune cell infiltration on KIRC patients’ prognosis by analyzing the OS. We found that in NDUFS1 highly expressed KIRC patients, poor NK cells (HR = 0.55, p = 0.0116, Figure 3B) or neutrophil cells (HR = 0.416, p = 0.000166, Figure 3C) was connected to unfavorable survival; while poor macrophage M1 cells (HR = 1.57, p = 0.055, Figure 3D) or NKT cells (HR = 0.173, p = 0.0304, Figure 3E) was associated with favorable survival. When it comes to the low NDUFS1 expression groups, KIRC patients with poor neutrophil cells (HR = 0.638, p = 0.0286) or CD4+ T cells (HR = 0.575, p = 0.0187, Figure 3F) had a shorter survival time; while KIRC patients with poor Macrophage M1 cells (HR = 1.44, p = 0.0562) had a longer survival time. Combined together, for CD4+ T cells that have moderately positive correlation with NDUFS1 expression, lower immune infiltration may further exacerbate poor survival in lower NDUFS1 expressed patients. For neutrophils cells, the unfavorable prognosis of neutrophils infiltration in KIRC was independent of NDUFS1 expression; however, low NDUFS1 expression and low neutrophils infiltration would have superimposed effects on patients’ survival. Therefore, the combination of NDUFS1 with CD4+ T cell infiltration would improve the efficacy in predicting KIRC patients’ prognosis.
We then validated the above results by conducting mIF staining in KIRC TMA, we found that CD4 expression was significantly increased in KIRC tissues as compared to that in normal tissues (p < 0.0001, Figure 3G). More importantly, in patients with decreased CD4+ T cell infiltration whose pathological grade less than III, low expression of NDUFS1 had significantly poor OS than that with high expression of NDUFS1 did (p = 0.0483, Figures 3H,I), which was consistent to the results of bioinformatic analysis. To reveal the association of NDUFS1 expression with clinicopathological features in KIRC patients, we performed univariate and multivariate Cox proportional hazard analysis of independent predictors for KIRC patients’ overall survival based on clinicopathological features of TMA (Tables 2, 3). We found that advanced age, high AJCC stage, low expression of NDUFS1 and the combination of low NDUFS1 with low CD4 were risk factors for KIRC patients in univariate Cox proportional hazard analysis. After eliminating the influence of other factors by multivariate Cox proportional hazard analysis, we found that only advanced age, low NDUFS1, and the combination of low NDUFS1 with low CD4 were independent risk factors for KIRC patients’ worse prognosis.
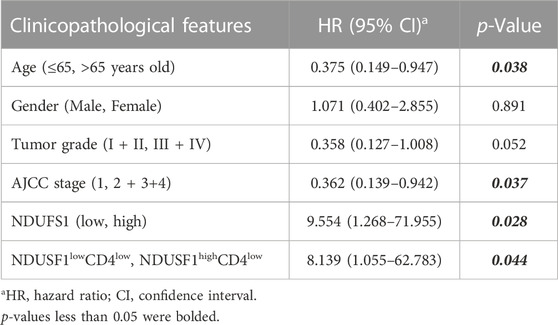
TABLE 2. The univariate Cox proportional hazard analysis of independent predicators for overall survival of KIRC patients.
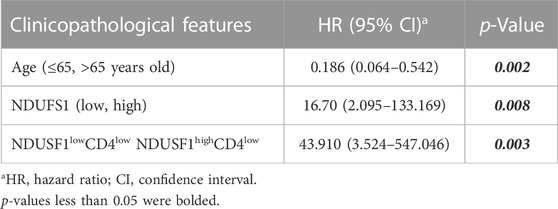
TABLE 3. The multivariate Cox proportional hazard analysis of independent predicators for overall survival of KIRC patients.
Moreover, we performed in vitro transwell migration assays to analyze the influences of NDUFS1 expression on T cell chemotaxis to KIRC cells. We found that the overexpression of NDUFS1 significantly promoted the migration of CD4+ Jurkat T cells to ACHN and 786-O KIRC cells (Figures 4A–C).
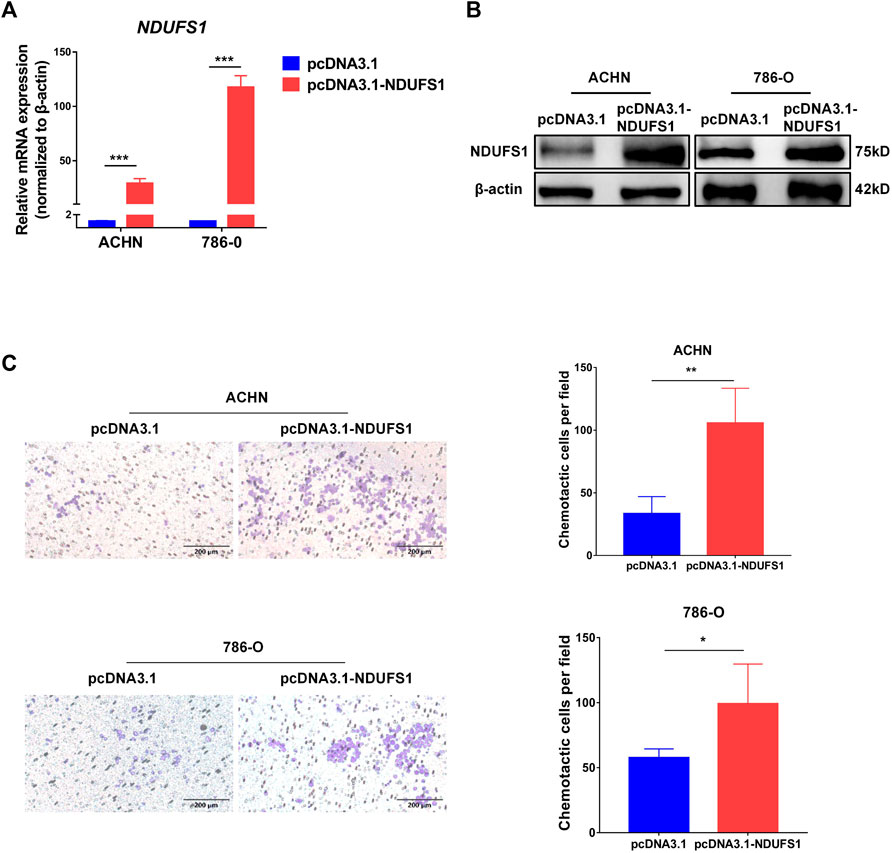
FIGURE 4. NDUFS1 overexpression promotes T cell chemotaxis to KIRC cells. (A, B) qRT-PCR and Western blot analysis of NDUFS1 mRNA (A) and protein (B) expression in KIRC cells that transfected with NDUFS1 cDNA. (C) The effect of NDUFS1 overexpression on CD4+ Jurkat T cell chemotaxis to KIRC cells (*p < 0.05, **p < 0.01, ***p < 0.001).
3.4 Low expression of both NDUFS1 and FDX1 is associated with worse outcome in KIRC patients
In order to further elucidate potential cellular mechanism of NDUFS1 in affecting KIRC progression, we focused on the key regulator of cuproptosis, namely, FDX1, which is reported to be regulated by mitochondrial respiration. From GEPIA and TIMER database, we found that FDX1 was significantly correlated with NDUFS1 on the mRNA levels in KIRC respectively (R = 0.55, p = 0, Figure 5A; R = 0.619, p = 1.18e-57; Figure 5B). As for protein level, NDUFS1 and FDX1 was also positively correlative with the correlation coefficient in tumor was only 0.40 while in adjacent normal tissues was 0.82 (Supplementary Figure S5E), which was consisting with the trend between the difference in NDUFS1 mRNA and protein.
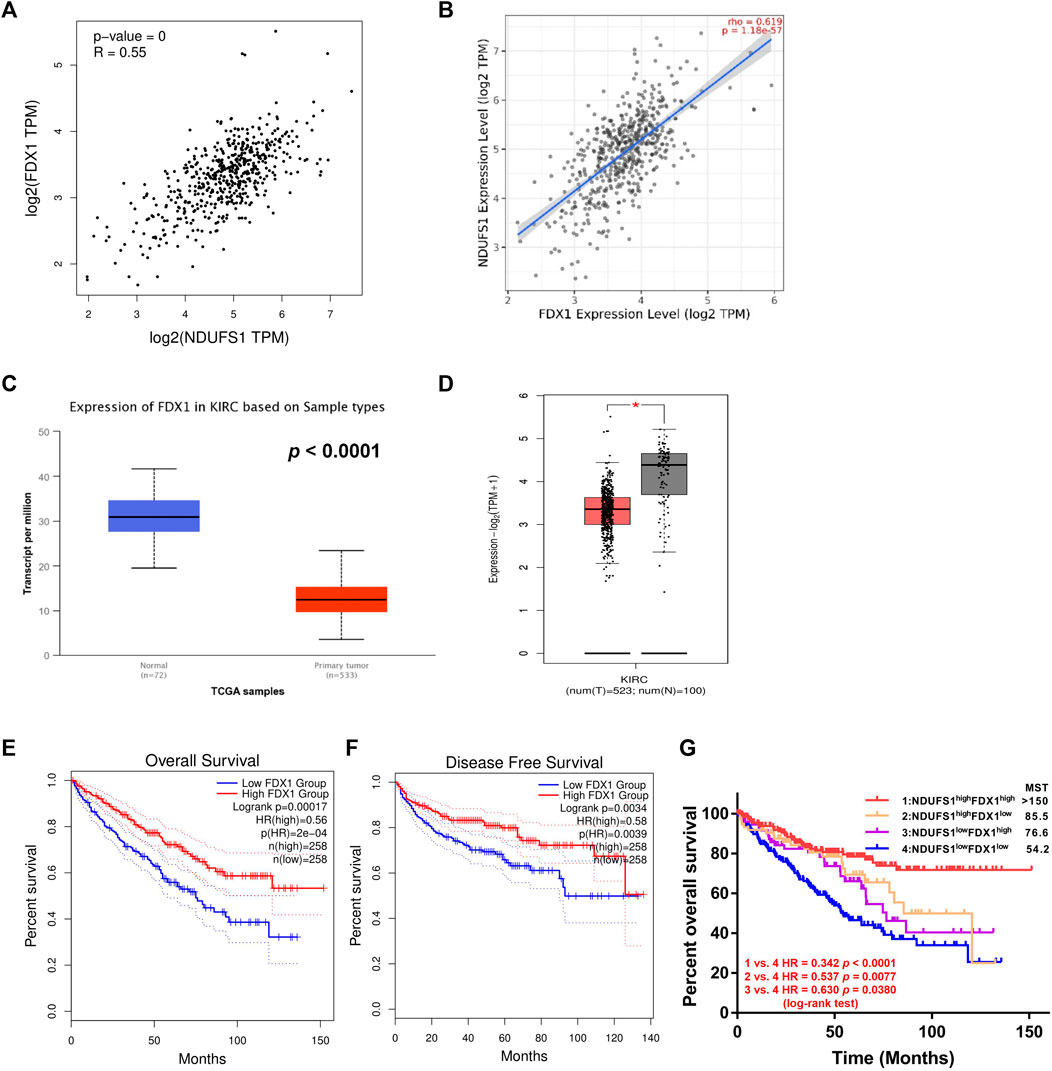
FIGURE 5. Low expression of both NDUFS1 and FDX1 are associated with worse outcomes in KIRC patients. (A, B) The correlation between NDUFS1 and FDX1 mRNA expression was analyzed by GEPIA (A) and TIMER (B) database respectively. (C, D) FDX1 mRNA expression was analyzed in UALCAN (C) and GEPIA (D) database. (E, F) The overall survival (E) and disease free survival (F) analysis in KIRC patients with high or low FDX1 expression was conducted in GEPIA database. (G) The overall survival analysis in KIRC patients with combined expression of NDUFS1 and FDX1. MST, median survival time.
Moreover, we found that FDX1 in KIRC tissues was considerably downregulated as compared to that in normal tissues through digging into UALCAN and GEPIA database respectively (P < 1E-12, Figure 5C; p < 0.05; Figure 5D). Further analysis from GEPIA database displayed that KIRC patients with low FDX1 had significantly poor OS (p = 0.00017, Figure 5E) and DFS (p = 0.0034, Figure 5F) than patients with high expression did. By analyzing the downloaded gene expression profiles and corresponding clinical information from TCGA database, we found that patients in NDUFS1lowFDX1low group had significantly worse prognosis than those in NDUFS1lowFDXhigh group (HR = 0.630, p = 0.0380, Figure 5G) or those in NDUFS1highFDXlow group (HR = 0.537, p = 0.0077, Figure 5G). In comparison with using either NDUFS1 or FDX1 as a single marker, the combination of low FDX1 and low NDUFS1 predict significantly worse outcome. Together, bioinformatic analysis in a series of online databases have revealed that FDX1, highly related to NDUFS1, was downregulated in KIRC; and low expression of FDX1 was associated with poor prognosis in a similar trend consistent with NDUFS1.
3.5 NDUFS1 expression is probably downregulated by hsa-miR-320b in KIRC
Considering the low correlation between NDUFS1 protein and mRNA levels in tumor, as well as the fact that the decreased promoter methylation of NDUFS1 was not consistent to the low expression of NDUFS1 mRNA in KIRC (Supplementary Figure S6), we speculated that the expression level of NDUFS1 was probably regulated at the post-transcriptional level. As recent studies showed that mitochondrial biogenesis was in part modified by miRNA silence, resulting in decreased mtDNA expression (Zhang and Xu, 2016). Thus, we tried to explore the potential miRNAs for down-regulating the expression of NDUFS1 in KIRC by digging into TargetScan database. There were 6 conserved miRNAs, including hsa-miR-599, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d and hsa-miR-4429, were predicted to bind to the 3’ UTR region of NDUFS1 mRNA (Figure 6A). Among them, the level of hsa-miR-599 was positively correlated (r = 0.176, p = 5.55e-05) while hsa-miR-320b (r = −0.259, p = 2.27e-09), hsa-miR-320a (r = −0.219, p = 4.95e-07), hsa-miR-320c (r = −0.165, p = 1.66e-04) and hsa-miR-320d (r = −0.140, p = 1.36e-03) were negatively correlated with the level of NDUFS1 mRNA in KIRC (Figure 6B). The differential expression analysis demonstrated that hsa-miR-599 (p = 6.0e-6), hsa-miR-320a (p = 0.003), hsa-miR-320b (p = 0.00021), hsa-miR-320c (p = 3.6e-7) and hsa-miR-320d (p = 1.2e-7) were significantly upregulated in KIRC tissues as compared with those in normal tissues (Figure 6C).

FIGURE 6. The expression of miRNAs and their correlation with NDUFS1 in KIRC. (A) miRNAs involving in regulating NDUFS1 were predicted by TargetScan database. (B) The correlations between the expression of miRNAs and NDUFS1 were analyzed by ENCORI. (C) The expression levels of miRNAs in KIRC were analyzed by ENCORI.
Next, the association of these 6 miRNAs with KIRC patients’ OS were analyzed. Kaplan-Meier Plotter showed that the patients with high level of has-miR-320b had unfavorable OS (p = 2.9e-05, Figure 7A) in contrast to the favorable prognosis in KIRC patients with high expression of hsa-miR-599 (p = 0.00072), hsa-miR-320d (p = 0.024), and hsa-miR-4429 (P = 3e-10). In ENCORI, only high expression of has-miR-320b (p = 0.00035) was associated with shorter OS in KIRC whereas other miRNAs had no statistical correlation with KIRC patients’ OS (Figure 7B). Moreover, by analyzing mRNA and miRNA expression profiles and the corresponding clinical information in TCGA database, we found that patients in NDUFSlowhsa-miR-320bhigh group had significantly shorter median survival times than those in NDUFS1highhsa-miR-320bhigh group did (53.3 months vs. >150 months, HR = 0.405, p < 0.0001, Figure 8A) but not those in NDUFS1lowhsa-miR-320blow group did. In comparison with using hsa-miR-320b as a single marker, the combination of low NDUFS1 and high hsa-miR-320b predicted significantly worse outcome. To summarize, although 6 miRNAs were predicted to probably affect the transcription of NDUFS1, only hsa-miR-320b was the one that was expressed at high levels, correlated to poor OS and had consistent prognostic values with those of low NDUFS1 expression in KIRC patients. Therefore, hsa-miR-320b was possibly served as an upstream regulator for NDUFS1 in KIRC.
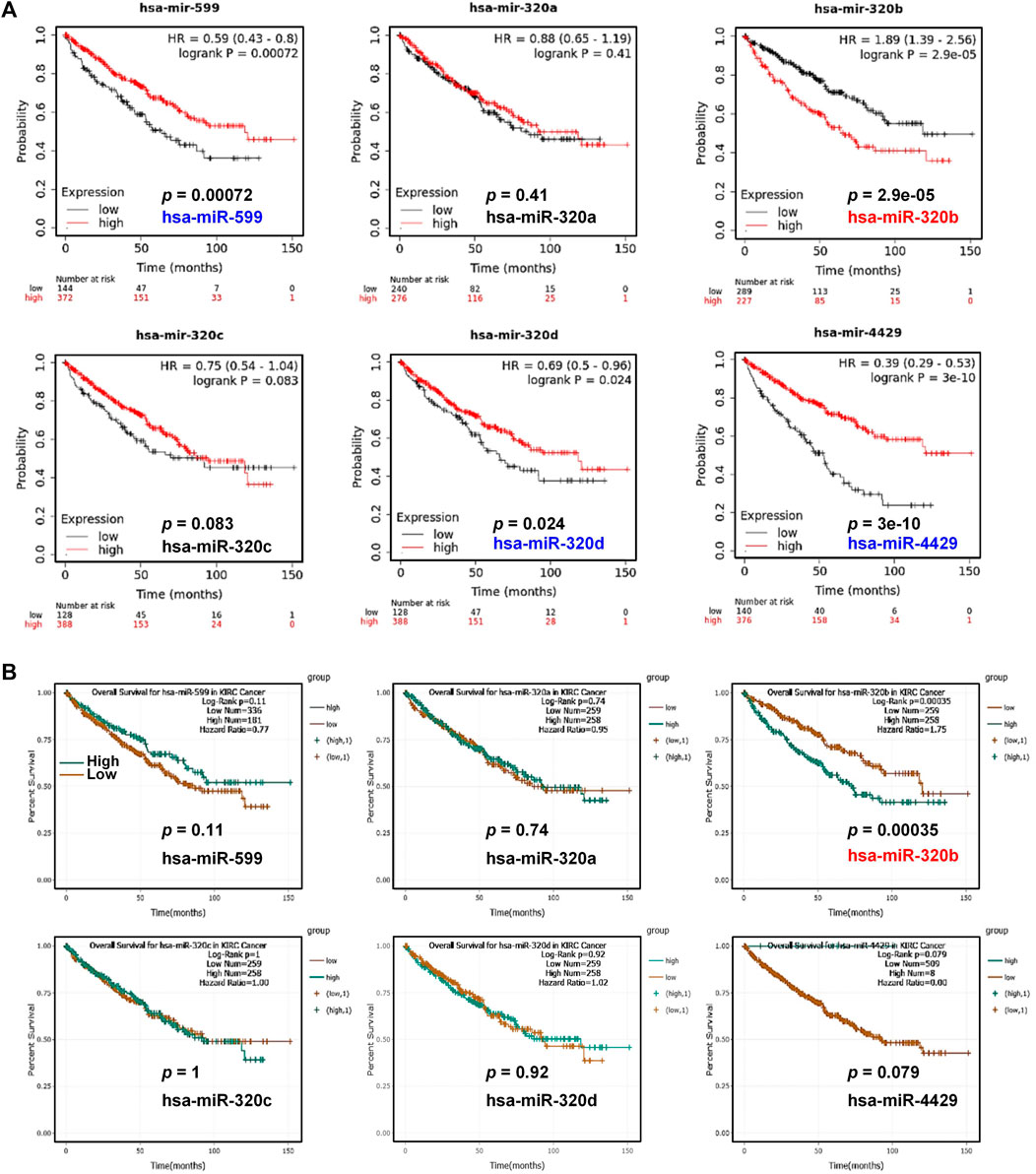
FIGURE 7. The overall survival analysis of miRNAs in KIRC patients. (A, B) The overall survival analysis in KIRC patients with high or low expression of predicted miRNAs were conducted by Kaplan-Meier Plotter (A) and ENCORI (B).
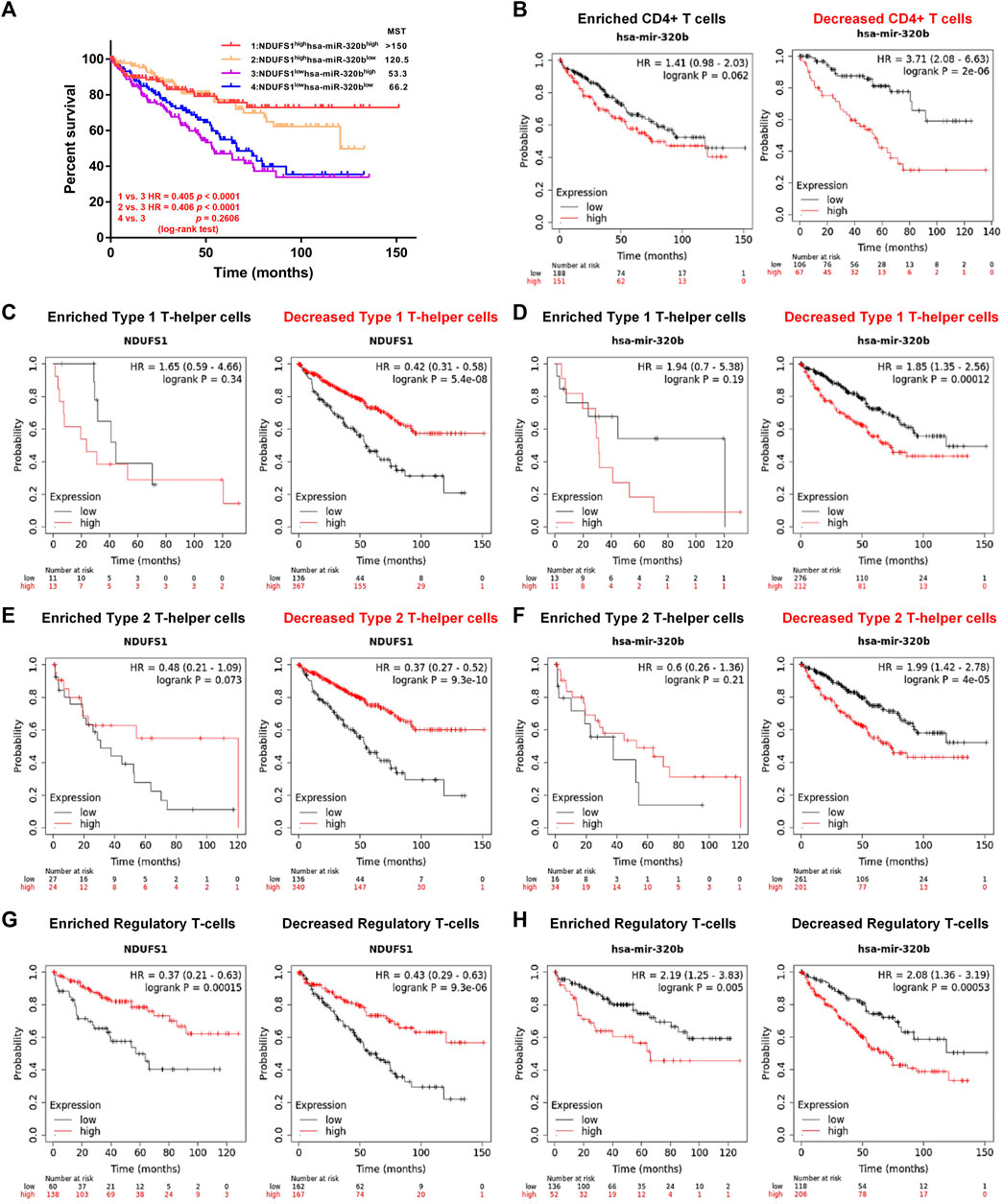
FIGURE 8. Low NDUFS1 or high hsa-miR-320b is correlated to poor clinical outcomes in KIRC patients with decreased CD4+ T cells infiltration. (A) The overall survival analysis in KIRC patients with combined expression of hsa-miR-320b and NDUFS1. (B) The overall survival analysis in high or low has-miR-320b expressed KIRC patients were analyzed by Kaplan-Meier Plotter based on CD4+T cell infiltration. (C, D) The overall survival curves in high/low NDUFS1 (C) or high/low has-miR-320b (D) expressed KIRC patients based on Th1 cell infiltration. (E, F) The overall survival cures in high/low NDUFS1 (E) or high/low has-miR-320b (F) expressed KIRC patients based on Th2 cell infiltration. (G, H) The overall survival curves in high/low NDUFS1 (G) or high/low has-miR-320b (H) expressed KIRC patients based on regulatory T-cell infiltration.
3.6 Low NDUFS1 or high hsa-miR-320b is correlated to poor outcomes in KIRC patients with decreased CD4+ T cell infiltration
We further explored the correlation of hsa-miR-320b expression with patients’ survival based on immune cell infiltration in KIRC. We found that in KIRC patients with decreased CD4+ T cells, high hsa-miR-320b was correlated with poor OS (P = 2e-06, Figure 8B), whose prognostic roles in KIRC was consistent to low NDUFS1. When it comes to the subgroup of CD4+ T cells, low NDUFS1 (p = 5.4e-08, Figure 8C; p = 9.3e-10; Figure 8E) or high hsa-miR-320b (p = 0.00012, Figure 8D; P = 4e-05; Figure 8F) both were associated with poor OS in KIRC patients with decreased Th1 and Th2 cells, respectively. However, in KIRC patients whether with enriched or with decreased Treg cells, low NDUFS1 or high hsa-miR-320b was all associated with poor OS (Figures 8G,H), suggesting that the prognostic value of NDUFS1 or hsa-miR-320b was not connected to Treg cell infiltration, but might be related to Th1 and Th2 cell infiltration. To sum up, the expression of low NDUFS1 or high hsa-miR-320b was connected to poor prognosis in KIRC patients with decreased CD4+ T cell infiltration, especially in those with decreased Th1 and Th2 cell infiltration.
4 Discussion
In this study, we have explored the potential biomarker associated immune cell infiltration in a view of mitochondrial metabolism, and our findings indicated that NDUFS1, a core subunit of mitochondrial complex I whose expression was probably downregulated by hsa-miR-320b and correlated with cuproptosis, might act as a biomarker for CD4+ T cell infiltration in KIRC. And, NDUFS1 expression promoted CD4+ T cell in vitro chemotaxis to KIRC cells. Moreover, the combined low NDUFS1 expression with poor CD4+ T cell infiltration predicts unfavorable prognosis. Together, we speculated that NDUFS1 may reprogram the tumor immune microenvironment via affecting cell metabolism.
The above findings were obtained by bioinformatics digging and biological experiment validation. For NDUFS1 mRNA or protein expression and its influence on patients’ survival, both online database and TMA mIF analysis consistently supported that NDUFS1 was downregulated in KIRC and low NDUFS1 was associated with unfavorable survival. As regards to relationship between the combined NDUFS1 expression with CD4+ T cell infiltration and patients’ survival, bioinformatics analysis revealed that low NDUFS1 mRNA expression correlated poor CD4+ T cell infiltration was related to unfavorable survival in all KIRC patients; while TMA quantitative data revealed that low NDUFS1 protein expression correlated decreased CD4+ T cell infiltration predicted unfavorable prognosis only in KIRC patients whose pathological stage less than III. Based on TMA mIF analysis in the protein level, we hypothesized that NDUFS1 might serve as a prognostic biomarker that predicts better outcome for CD4+ T cell infiltration in KIRC patients whose pathological stage less than III. However, due to limited sample size, further larger sample size of KIRC cohorts is required.
A few studies have demonstrated the expression and corresponding function of NDUFS genes in the tumor. As prognostic factors, patients with high expression of NDUFS8 in NSCLC or acute myeloid leukemia has poor overall survival (Su et al., 2016; Wei et al., 2020). However, the opposite prognostic trends are reported for NDUFS3 and NDUFS5. In serous ovarian adenocarcinoma, the downregulation of NDUFS3 is related to advanced tumor stage and shorter overall survival (Wang et al., 2013); but in invasive breast carcinoma whose upregulation is positively aggressiveness-correlated biomarker signature (Suhane et al., 2011). Besides, the elevated level of NDUFS5 is associated with good survival in lung adenocarcinoma, but related to significantly reduced time to first progression in gastric cancer (Sotgia and Lisanti, 2017). In the view of drug targets, NDUFS4 or NDUFS7 is reported as the targeted gene by casticin or nebivolol for its role in inhibiting cell migration and invasion in mouse melanoma B16F10 cells (Shih et al., 2017), or in blocking complex I activity related colon and breast tumor growth (Nuevo-Tapioles et al., 2020), respectively. As an interacting partner with mitochondrial OSMR or CD147, NDUFS1/2 or NDUFS6 confers their individually roles in mediating oxidative phosphorylation related IR resistance of brain tumor stem cells (Sharanek et al., 2020), or in regulating complex I activity and apoptosis in human melanoma (Luo et al., 2014), respectively. As far as the molecular function is concerned, NDUFS2 silencing inhibits mitochondrial complex I activity and dramatically decreases tumor growth and metastasis rates in lung cancer (Liu L. et al., 2019), and NDUFS4 knockdown in melanoma cells results in decreased oxidative metabolism, significant decreased CD8+ T cell numbers but with superior functional, and an increased response to anti-PD-1 therapy (Najjar et al., 2019). However, the detailed function or mechanism of action for NDUFS1, and its relationship with KIRC immunotherapy or immune infiltrates, remain unclear. Although there were similar articles indicating the possible prognostic role of NDUFS1 in KIRC (Ellinger et al., 2017; Stein et al., 2019), our research for the first time suggested the connection of NDUFS1 with immune cell infiltrates. Moreover, their combination could predict the patient’s clinical outcomes. Due to the fact that high NDUFS1 expression predicts favorable prognosis in KIRC, we speculate that the NDUFS1 associated favorable prognosis may be correlated with immune cell infiltration.
Currently, the possible interventions for NDUFS1 include gene expression intervention such as overexpression, RNA interference or CRISPR/Cas9-mediated knockout, and small molecule inhibitors. For exploring NDUFS1’s function using gain-of-function strategy, the overexpression of NDUFS1 were used to investigate its role in mediating the radiosensitization of colorectal cancer (Shi et al., 2021) or in miR-3130-5p mediated invasiveness of lung adenocarcinoma (Zhan et al., 2021). In addition, loss-of-function strategy using siRNA or shRNA mediated RNA interference were used to reveal NDUFS1-mediated colorectal cancer cell proliferation and tumorigenesis (Ren et al., 2023), or to study the MDM2-binding associated ROS production (Elkholi et al., 2019), respectively. Besides, sgRNA mediated knockout strategy were utilized to validate its necessity in maintaining cancer cell survival under low pH (Michl et al., 2022). For NDUFS1 as diagnostic or therapeutic target, a near-infrared (NIR) small-molecule fluorophore dye IR-34, directly cleaves NDUFS1 and disrupts electron transporting in the respiratory chain, could be a potentially useful multifunctional theranostic agent for cancer cell targeting, NIR imaging, and therapeutic ER stress inducing (Wang et al., 2018). More interestingly, by exploring the global gene expression and docking profiling of COVID-19 infection, Jabeen, A et al. found that herbal drugs (apigenin, quercetin, and resveratrol) has the potentials to bind and inhibit NDUFS1 (Jabeen et al., 2022), which suggesting these drugs could be possible inhibitors for NDUFS1.
At present, potential companion diagnostic biomarkers involving gene expression are approved in clinics to identify optimal patients or to screen individual treatment. Besides the biomarkers on tumor cells, the future of prognostic biomarker is likely to rely on the components of the tumor microenvironment. As the main cellular components in tumor microenvironment, the cellular composition and functional state of tumor-infiltrating immune cells vary considerably across tumors. In KIRC that contains a large number of immune infiltrates, consisting of NK, DC, T cells, macrophages, etc., the type or the status of immune infiltrates is varied and needed to be predicted. Consistent with facts that NDUFS4 mediated oxidative metabolism acts as a barrier to the response of PD-1 blockade in melanoma (Najjar et al., 2019), and more and more studies convey the concept that tumor cell metabolites epigenetically regulate the immune cell phenotype (Jiang et al., 2020). Therefore, we thus focused on the prediction of immune infiltrates by mitochondrial metabolism. And, our analysis in this paper revealed that NDUFS1 could be a metabolic indicator for CD4+ T and neutrophil cell infiltrations in KIRC, and the low NDUFS1 correlated decreased CD4+ T predicted unfavorable prognosis. At present, researches on the detailed influences of NDUFS1 on the infiltration of CD4+ T especially Th1 and Th2 cells in KIRC were ongoing.
In KIRC, the significance of miRNA modifications and their prognostic prediction have been reported in bioinformatic analysis (Christinat and Krek, 2015; Liu S. et al., 2019). Moreover, some miRNAs engaging in the occurrence and development of KIRC are identified (Wang et al., 2019; Xue et al., 2019). In this paper, we found that hsa-miR-320b was negatively correlated with NDUFS1 and predicted poor prognosis of KIRC patients when expressed at high level. And, we further revealed that low NDUFS1 or high hsa-miR-320b consistently correlated to unfavorable outcomes in KIRC patients with decreased CD4+ T cell infiltration. Although our analysis in this paper suggested that hsa-miR-320b could be a potential upstream down-regulator for NDUFS1, further validated experiment is still needed. Interestingly, hsa-miR-320b has been recently reported to be a potential biomarker for predicting the efficacy of immunotherapy in advanced NSCLCs (Peng et al., 2020), which partly supporting our finding that hsa-miR-320b is associated with immune infiltrates. Therefore, hsa-miR-320b alone or in combination with its potential target NDUFS1 could be a miRNA-mitochondrial signature for KIRC immunotherapy.
Recently, a study released on Science has revealed that mitochondrial respiration regulates cuproptosis in the process of which FDX1 served as the key regulator (Tsvetkov et al., 2022). As for targeting tumor cells, Jiang et al. (Jiang et al., 2022) has reviewed three typical copper induced tumor cell death mechanisms, including oxidative stress, proteasome inhibition and antiangiogenesis, from which oxidative stress produced by mitochondria was considered the most critical and effective tumor-killing method. Additionally, Yao, et al. (Yao et al., 2023) revealed that FDX1 may serve as an independent factor affecting the prognosis of KIRC, and was correlated with CD4+ T cell infiltration. Hence, we tried to explore possible relationship between NDUFS1 and cuproptosis in KIRC. We found that FDX1, decreased in KIRC, was positively related with NDUFS1 and predicted poor prognosis when expressed at low level. Therefore, our findings indicated NDUFS1 may correlate to KIRC cuproptosis, which may be served as a possible mechanism of NDUFS1 in regulating KIRC cells.
From the view of immune-epigenetic-metabolism, we have obtained a preliminary understanding of the possible functions and upstream regulator of NDUFS1 in KIRC immune infiltration. Nevertheless, our data are mostly derived from dynamically updated online databases, thus more or less leading to possibly unstable results. And, the sample size in TMA with qualified mIF images and available survival information was small. Moreover, the regulation of hsa-miR-320b on NDUFS1 or NDUFS1 on cuproptosis are still needed to be further investigated via in vitro and in vivo biological experiments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conception and design: JQ and LL. Administrative support: JQ and LL. Provision of study materials or patients: DW, LH, ZX, and RF-T. Collection and assembly of data: DW, XY-F, and RF-T. Data analysis and interpretation: JF and JA. Manuscript writing: DW, LH, ZX, and RF-T. Final approval of manuscript: JQ and LL. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by National Natural Science Foundation of China (#31571469, #81872349 and #82130084).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1168462/full#supplementary-material
References
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). Ualcan: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19 (8), 649–658. doi:10.1016/j.neo.2017.05.002
Christinat, Y., and Krek, W. (2015). Integrated genomic analysis identifies subclasses and prognosis signatures of kidney cancer. Oncotarget 6 (12), 10521–10531. doi:10.18632/oncotarget.3294
Clark, D. J., Dhanasekaran, S. M., Petralia, F., Pan, J., Song, X., Hu, Y., et al. (2019). Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179 (4), 964–983.e31. doi:10.1016/j.cell.2019.10.007
Elkholi, R., Abraham-Enachescu, I., Trotta, A. P., Rubio-Patiño, C., Mohammed, J. N., Luna-Vargas, M. P. A., et al. (2019). MDM2 integrates cellular respiration and apoptotic signaling through NDUFS1 and the mitochondrial network. Mol. Cell 74 (3), 452–465.e7. doi:10.1016/j.molcel.2019.02.012
Ellinger, J., Poss, M., Brüggemann, M., Gromes, A., Schmidt, D., Ellinger, N., et al. (2017). Systematic expression analysis of mitochondrial complex I identifies NDUFS1 as a biomarker in clear-cell renal-cell carcinoma. Clin. Genitourin. Cancer 15 (4), e551–e562. doi:10.1016/j.clgc.2016.11.010
Ghatalia, P., Gordetsky, J., Kuo, F., Dulaimi, E., Cai, K. Q., Devarajan, K., et al. (2019). Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J. Immunother. Cancer 7 (1), 139. doi:10.1186/s40425-019-0621-1
Hasanov, E., Gao, J., and Tannir, N. M. (2020). The immunotherapy revolution in kidney cancer treatment: Scientific rationale and first-Generation results. Cancer J. 26 (5), 419–431. doi:10.1097/PPO.0000000000000471
Jabeen, A., Ahmad, N., and Raza, K. (2022). Global gene expression and docking profiling of COVID-19 infection. Front. Genet. 13, 870836. doi:10.3389/fgene.2022.870836
Jiang, Y., Huo, Z., Qi, X., Zuo, T., and Wu, Z. (2022). Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine (Lond). 17 (5), 303–324. doi:10.2217/nnm-2021-0374
Jiang, Z., Hsu, J. L., Li, Y., Hortobagyi, G. N., and Hung, M. C. (2020). Cancer cell metabolism bolsters immunotherapy resistance by promoting an immunosuppressive tumor microenvironment. Front. Oncol. 10, 1197. doi:10.3389/fonc.2020.01197
Laurell, A., Lönnemark, M., Brekkan, E., Magnusson, A., Tolf, A., Wallgren, A. C., et al. (2017). Intratumorally injected pro-inflammatory allogeneic dendritic cells as immune enhancers: A first-in-human study in unfavourable risk patients with metastatic renal cell carcinoma. J. Immunother. Cancer 5, 52. doi:10.1186/s40425-017-0255-0
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 (1), 15–20. doi:10.1016/j.cell.2004.12.035
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97. doi:10.1093/nar/gkt1248
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48 (W1), W509–W514. doi:10.1093/nar/gkaa407
Liu, L., Qi, L., Knifley, T., Piecoro, D. W., Rychahou, P., Liu, J., et al. (2019a). S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J. Biol. Chem. 294 (18), 7516–7527. doi:10.1074/jbc.RA118.004365
Liu, S., Deng, X., and Zhang, J. (2019b). Identification of dysregulated serum miR-508-3p and miR-885-5p as potential diagnostic biomarkers of clear cell renal carcinoma. Mol. Med. Rep. 20 (6), 5075–5083. doi:10.3892/mmr.2019.10762
Luo, Z., Zeng, W., Tang, W., Long, T., Zhang, J., Xie, X., et al. (2014). CD147 interacts with NDUFS6 in regulating mitochondrial complex I activity and the mitochondrial apoptotic pathway in human malignant melanoma cells. Curr. Mol. Med. 14 (10), 1252–1264. doi:10.2174/1566524014666141202144601
Michl, J., Wang, Y., Monterisi, S., Blaszczak, W., Beveridge, R., Bridges, E. M., et al. (2022). CRISPR-Cas9 screen identifies oxidative phosphorylation as essential for cancer cell survival at low extracellular pH. Cell Rep. 38 (10), 110493. doi:10.1016/j.celrep.2022.110493
Nagy, Á., Munkácsy, G., and Győrffy, B. (2021). Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11 (1), 6047. doi:10.1038/s41598-021-84787-5
Najjar, Y. G., Menk, A. V., Sander, C., Rao, U., Karunamurthy, A., Bhatia, R., et al. (2019). Tumor cell oxidative metabolism as a barrier to PD-1 blockade immunotherapy in melanoma. JCI Insight 4 (5), e124989. doi:10.1172/jci.insight.124989
Nuevo-Tapioles, C., Santacatterina, F., Stamatakis, K., Núñez de Arenas, C., Gómez de Cedrón, M., Formentini, L., et al. (2020). Coordinate β-adrenergic inhibition of mitochondrial activity and angiogenesis arrest tumor growth. Nat. Commun. 11 (1), 3606. doi:10.1038/s41467-020-17384-1
Peng, X. X., Yu, R., Wu, X., Wu, S. Y., Pi, C., Chen, Z. H., et al. (2020). Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 8 (1), e000376. doi:10.1136/jitc-2019-000376
Ren, L., Meng, L., Gao, J., Lu, M., Guo, C., Li, Y., et al. (2023). PHB2 promotes colorectal cancer cell proliferation and tumorigenesis through NDUFS1-mediated oxidative phosphorylation. Cell death Dis. 14 (1), 44. doi:10.1038/s41419-023-05575-9
Rhodes, D. R., Kalyana-Sundaram, S., Mahavisno, V., Varambally, R., Yu, J., Briggs, B. B., et al. (2007). Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9 (2), 166–180. doi:10.1593/neo.07112
Santidrian, A. F., Matsuno-Yagi, A., Ritland, M., Seo, B. B., LeBoeuf, S. E., Gay, L. J., et al. (2013). Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J. Clin. Invest. 123 (3), 1068–1081. doi:10.1172/JCI64264
Sharanek, A., Burban, A., Laaper, M., Heckel, E., Joyal, J. S., Soleimani, V. D., et al. (2020). OSMR controls glioma stem cell respiration and confers resistance of glioblastoma to ionizing radiation. Nat. Commun. 11 (1), 4116. doi:10.1038/s41467-020-17885-z
Shi, Y., Wang, Y., Jiang, H., Sun, X., Xu, H., Wei, X., et al. (2021). Mitochondrial dysfunction induces radioresistance in colorectal cancer by activating [Ca(2+)](m)-PDP1-PDH-histone acetylation retrograde signaling. Cell death Dis. 12 (9), 837. doi:10.1038/s41419-021-03984-2
Shih, Y. L., Chou, H. M., Chou, H. C., Lu, H. F., Chu, Y. L., Shang, H. S., et al. (2017). Casticin impairs cell migration and invasion of mouse melanoma B16F10 cells via PI3K/AKT and NF-κB signaling pathways. Environ. Toxicol. 32 (9), 2097–2112. doi:10.1002/tox.22417
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Singh, D. (2021). Current updates and future perspectives on the management of renal cell carcinoma. Life Sci. 264, 118632. doi:10.1016/j.lfs.2020.118632
Sotgia, F., and Lisanti, M. P. (2017). Mitochondrial biomarkers predict tumor progression and poor overall survival in gastric cancers: Companion diagnostics for personalized medicine. Oncotarget 8 (40), 67117–67128. doi:10.18632/oncotarget.19962
Stein, J., Tenbrock, J., Kristiansen, G., Müller, S. C., and Ellinger, J. (2019). Systematic expression analysis of the mitochondrial respiratory chain protein subunits identifies COX5B as a prognostic marker in clear cell renal cell carcinoma. Int. J. Urol. 26 (9), 910–916. doi:10.1111/iju.14040
Su, C. Y., Chang, Y. C., Yang, C. J., Huang, M. S., and Hsiao, M. (2016). The opposite prognostic effect of NDUFS1 and NDUFS8 in lung cancer reflects the oncojanus role of mitochondrial complex I. Sci. Rep. 6, 31357. doi:10.1038/srep31357
Suhane, S., Berel, D., and Ramanujan, V. K. (2011). Biomarker signatures of mitochondrial NDUFS3 in invasive breast carcinoma. Biochem. Biophys. Res. Commun. 412 (4), 590–595. doi:10.1016/j.bbrc.2011.08.003
Tang, Z., Kang, B., Li, C., Chen, T., and Zhang, Z. (2019). GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47 (W1), W556–W560. doi:10.1093/nar/gkz430
Toth, A. T., and Cho, D. C. (2020). Emerging therapies for advanced clear cell renal cell carcinoma. J. Kidney Cancer VHL 7 (4), 17–26. doi:10.15586/jkcvhl.2020.156
Tsvetkov, P., Coy, S., Petrova, B., Dreishpoon, M., Verma, A., Abdusamad, M., et al. (2022). Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375 (6586), 1254–1261. doi:10.1126/science.abf0529
Vasan, K., Werner, M., and Chandel, N. S. (2020). Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 32 (3), 341–352. doi:10.1016/j.cmet.2020.06.019
Wang, L., Yang, G., Zhao, D., Wang, J., Bai, Y., Peng, Q., et al. (2019). CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Mol. Cancer 18 (1), 86. doi:10.1186/s12943-019-0997-z
Wang, P., Cheng, X., Fu, Z., Zhou, C., Lu, W., and Xie, X. (2013). Reduced expression of NDUFS3 and its clinical significance in serous ovarian cancer. Int. J. Gynecol. Cancer 23 (4), 622–629. doi:10.1097/IGC.0b013e318287a90d
Wang, X., Lopez, R., Luchtel, R. A., Hafizi, S., Gartrell, B., and Shenoy, N. (2021). Immune evasion in renal cell carcinoma: Biology, clinical translation, future directions. Kidney Int. 99 (1), 75–85. doi:10.1016/j.kint.2020.08.028
Wang, Y., Luo, S., Zhang, C., Liao, X., Liu, T., Jiang, Z., et al. (2018). An NIR-fluorophore-based therapeutic endoplasmic reticulum stress inducer. Adv. Mater. 30, e1800475. doi:10.1002/adma.201800475
Wei, J., Xie, Q., Liu, X., Wan, C., Wu, W., Fang, K., et al. (2020). Identification the prognostic value of glutathione peroxidases expression levels in acute myeloid leukemia. Ann. Transl. Med. 8 (11), 678. doi:10.21037/atm-20-3296
Xue, D., Wang, H., Chen, Y., Shen, D., Lu, J., Wang, M., et al. (2019). Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol. Cancer 18 (1), 151. doi:10.1186/s12943-019-1072-5
Yao, Y., Chen, H., Lou, M., and Chen, T. (2023). Cuproptosis-related gene FDX1 as a prognostic biomarker for kidney renal clear cell carcinoma correlates with immune checkpoints and immune cell infiltration. Front. Genet. 14, 1071694. doi:10.3389/fgene.2023.1071694
Zhan, J., Sun, S., Chen, Y., Xu, C., Chen, Q., Li, M., et al. (2021). MiR-3130-5p is an intermediate modulator of 2q33 and influences the invasiveness of lung adenocarcinoma by targeting NDUFS1. Cancer Med. 10 (11), 3700–3714. doi:10.1002/cam4.3885
Keywords: kidney renal clear cell carcinoma, NDUFS1, survival, hsa-miR-320b, CD4+ T cell infiltration
Citation: Wu D, He L, Xu Z, Tian R-F, Fan X-Y, Fan J, Ai J, Bian H-J, Qin W-J, Qin J and Li L (2023) The combination of NDUFS1 with CD4+ T cell infiltration predicts favorable prognosis in kidney renal clear cell carcinoma. Front. Cell Dev. Biol. 11:1168462. doi: 10.3389/fcell.2023.1168462
Received: 17 February 2023; Accepted: 21 June 2023;
Published: 04 July 2023.
Edited by:
Brajendra Tripathi, National Institutes of Health (NIH), United StatesReviewed by:
Dunrui Wang, National Cancer Institute (NIH), United StatesRizaldy Taslim Pinzon, Duta Wacana Christian University, Indonesia
Tianhe Huang, Wuhan University, China
Copyright © 2023 Wu, He, Xu, Tian, Fan, Fan, Ai, Bian, Qin, Qin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Jun Qin, qinweij@fmmu.edu.cn; Jun Qin, 292632954@qq.com; Ling Li, liling25@fmmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Dong Wu
Dong Wu