
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 23 May 2023
Sec. Stem Cell Research
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1166934
This article is part of the Research TopicSingle Cell Analysis to Study Cell Differentiation in Human DiseasesView all 6 articles
Introduction: Porphyromonas gingivalis and Enterococcus faecalis promote the development of pulpitis and periapical periodontitis. These bacteria are difficult to eliminate from the root canal systems, leading to persistent infection and poor treatment outcomes. We explored the response of human dental pulp stem cells (hDPSCs) to bacterial invasion and the mechanisms underlying the impact of residual bacteria on dental pulp regeneration.
Methods: Single-cell sequencing was used to categorize the hDPSCs into clusters based on their response to P. gingivalis and E. faecalis. We depicted a single-cell transcriptome atlas of hDPSCs stimulated by P. gingivalis or E. faecalis.
Results: The most differentially expressed genes in the Pg samples were THBS1, COL1A2, CRIM1, and STC1, which are related to matrix formation and mineralization, and HILPDA and PLIN2, which are related to the cellular response to hypoxia. A cell cluster characterized by high expression levels of THBS1 and PTGS2 was increased after P. gingivalis stimulation. Further signaling pathway analysis showed that hDPSCs prevented P. gingivalis infection by regulating the TGF-β/SMAD, NF-κB, and MAPK/ERK signaling pathways. Differentiation potency and pseudotime trajectory analyses showed that hDPSCs infected by P. gingivalis undergo multidirectional differentiation, particularly to the mineralization-related cell lineage. Furthermore, P. gingivalis can create a hypoxia environment to effect cell differentiation. The Ef samples were characterized by the expression of CCL2, which is related to leukocyte chemotaxis, and ACTA2, which is related to actin. There was an increased proportion of a cell cluster that was similar to myofibroblasts and exhibited significant ACTA2 expression. The presence of E. faecalis promoted the differentiation of hDPSCs into fibroblast-like cells, which highlights the role of fibroblast-like cells and myofibroblasts in tissue repair.
Discussion: hDPSCs do not maintain their stem cell status in the presence of P. gingivalis and E. faecalis. They differentiate into mineralization-related cells in the presence of P. gingivalis and into fibroblast-like cells in the presence of E. faecalis. We identified the mechanism underlying the infection of hDPSCs by P. gingivalis and E. faecalis. Our results will improve understanding of the pathogenesis of pulpitis and periapical periodontitis. Furthermore, the presence of residual bacteria can have adverse effects on the outcomes of regenerative endodontic treatment.
Dental pulpitis and periapical periodontitis are common infections of the oral cavity (Tibúrcio-Machado et al., 2021; León-López et al., 2022). Several bacteria can participate these diseases, including those from the phyla Firmicutes, Actinomycetes, Fusobacteria, Spirochaetes, and Bacteroides (Siqueira & Roca., 2022). Culture and identification techniques have shown that Firmicutes and Bacteroides are closely associated with the development of irreversible pulpitis and periapical periodontitis (Rôças et al., 2016; Sánchez-Sanhueza et al., 2018; de Brito et al., 2020; Zahran et al., 2021; Buonavoglia et al., 2023).
Enterococcus faecalis is a Gram-positive facultative anaerobe from the Firmicutes phylum. This bacterium is highly resilient (Love, 2001) and typically infects the dental pulp, leading to persistent root canal infection (Hancock et al., 2001; Kayaoglu AND Orstavik, 2004; Stuart et al., 2006; Zhu et al., 2010). The effects of Enterococcus faecalis on the pulp and periapical tissue are unclear. The bacteria releases byproducts such as lysase, gelatinase, hyaluronidase, and cytolysin, which cause tissue damage (Kayaoglu AND Orstavik, 2004) or modulate the immune responses of pulp cells, leading to further tissue damage (Sipert et al., 2010; Fransson et al., 2014). Furthermore, this bacteria inhibits osteoblast differentiation (Karygianni et al., 2012; Park et al., 2015) and upregulates the expression of osteogenic genes (RUNX2, ALP, COL1A1, and ALP) in human dental pulp stem cells (hDPSCs) (Lee et al., 2022), which might affect the repair of the pulp and periapical lesions. However, the response of hDPSCs to E. faecalis is incompletely understood.
Porphyromonas gingivalis is a Gram-negative anaerobe from the Bacteroides phylum and is the primary bacterial pathogen involved in periodontal disease. This microbe invades the dental pulp through the channels within the periodontal pocket, leading to retrograde pulpitis. P. gingivalis has been detected in the pulp and adjacent deep periodontal pocket of infected patients (Li et al., 2014), and is typically found in the apical region of the root canal. It is a dominant bacteria found in persistent periapical lesions (Zakaria et al., 2015). P. gingivalis and its byproduct, lipopolysaccharide (LPS), can affect dental pulp cells (Yang et al., 2003; Sipert et al., 2013; Biedermann et al., 2014; Liu et al., 2015). When hDPSCs are exposed to the LPS produced by P. gingivalis (Pg-LPS), the expression of genes involved in mineralization (DSPP and OCD) is downregulated (Yamagishi et al., 2011). Although P. gingivalis facilitates immune evasion in periodontal tissue (Zheng et al., 2021), its role in dental pulp tissue needs further investigation.
In addition to causing infections, bacteria negatively impact treatment outcomes. Regenerative endodontic treatment (RET) involves biological processes that aim to replace damaged tooth structures and cells in the pulp-dentine complex (Murray et al., 2007; Hargreaves et al., 2013). However, histological studies have shown that the new tissue that forms in the canal space after pulp RET is more like bone, cementum, and periodontal ligament than like pulp or dentin (Altaii et al., 2017). Residual bacteria may negatively impact the outcomes of RET (Siqueira & Rocas., 2008; Verma et al., 2017; Zaky et al., 2020). The current root canal disinfection methods cannot completely remove bacteria, their biofilms and byproducts. Active E. faecalis and its metabolic products have been detected in the apical region after disinfection (Baca et al., 2011; Pinheiro et al., 2015; Zhang et al., 2015; Vishwanat et al., 2017). The presence of residual bacteria in root canals can affect the biological properties of endogenous and exogenous stem cells during pulp regeneration treatment, as well as the regenerated tissues (Fouad, 2011; Nosrat et al., 2015; Fouad, 2017). A previous study showed that the quantity of dentin-associated mineralized tissue was significantly less in teeth with vs. without residual bacteria (E. faecalis) (Siqueira & Rocas., 2008; Verma et al., 2017). Furthermore, Pg-LPS inhibits odontoblast differentiation (Yamagishi et al., 2011; Vishwanat et al., 2017), which inhibits pulp regeneration. However, the mechanisms underlying these effects are unclear.
Some studies have investigated the impact of two common bacteria, E. faecalis and P. gingivalis, on dental pulp tissue. These bacteria interact with cells during invasion of dental pulp tissue through various mechanisms, such as adhesion and internalization (Biedermann et al., 2014). Furthermore, bacteria possess multiple virulence factors, such as lysase, gelatinase, hyaluronidase, and cytolysin. A single virulence factor (Sipert et al., 2013) and related cytokines and genes alone will not reflect the overall cell response (Yamagishi et al., 2011; Lee et al., 2022). In vitro coculture models are widely used to explore the interactions among microorganisms and a host. This approach can be used to evaluate bacterial adhesion and invasion, and cellular immune and inflammatory reactions (Puschhof et al., 2021). Coculture models of the interactions between dental stem cells and P. gingivalis have demonstrated the occurrence of cell adhesion and internalization (Biedermann et al., 2014). Such models comprehensively reflect the interaction between cells and microorganisms.
We used single-cell sequencing to create a hDPSCs atlas following infection with E. faecalis or P. gingivalis. We analyzed the differentiation trends and pathway enrichment of clusters of infected and noninfected hDPSCs, constructed an interaction network of multiple cells, proposed their possible evolutionary trajectories, and explored the effects of bacterial invasion on hDPSCs differentiation and proliferation. Based on our results, we proposed some mechanisms underlying the impact of residual bacteria in root canal systems on dental pulp regeneration.
hDPSCs were isolated as described previously (Liu et al., 2015). Impacted third molars were obtained from healthy individuals (n = 6; age: 18–23 years) at the Oral and Maxillofacial Surgery Department. The study protocol was approved by the Ethics Committee of Peking University School and Hospital of Stomatology (PKUSSIRB-202163047). Immediately after extraction, pulp tissues were minced into small pieces (0.1 × 0.1 × 0.1 cm3) and digested with 3 mg/mL collagenase type I (Worthington Biochemical Corp., Lakewood, NJ, United States) for 30 min at 37°C. The digested tissue and single cells were seeded into six-well plates with minimal essential medium-α (Gibco, Grand Island, NY, United States) supplemented with 10% fetal bovine serum (Kang Yuan Biology) and 1% penicillin-streptomycin (Gibco) under 5% CO2 at 37°C. The resulting heterogeneous populations of adherent, clonogenic dental stem/progenitor cells were analyzed for their cell surface marker expression by flow cytometry (positive for STRO-1, CD146, CD90, and CD105, and negative for CD45). The cells used in the present study were from passage 4 or 5.
E. faecalis ATCC 29212 and P. gingivalis ATCC 33277 were gifted by the Central Laboratory of Peking University School of Stomatology. E. faecalis were grown aerobically (5% CO2, 37°C) in brain heart infusion medium, whereas P. gingivalis were grown anaerobically in brain heart infusion medium supplemented with 1% hemin and 0.5% vitamin K.
For coculture, hDPSCs from six donors were mixed to reduce the impact of individual differences on the results. The cells were seeded at a density of 5 × 104 cells/well in a 24-well plate. The final concentration of the bacterial suspension was measured based on the optical density at 630 nm. The microbial concentration was measured at optical density (630 nm) = 0.1 as 1 × 109 CFU/mL, and the solution was diluted to achieve the desired multiplicity of infection. After cell adhesion, the cells were infected with live bacteria at a multiplicity of infection of 1:1 and incubated for 4 h at 5% CO2 and 37°C. Before harvesting, cells were incubated with 200 µM 4sU for 2 h in each group. 4sU is incorporated into new RNA during transcription and converted to a cytosine analogue using iodoacetamide (IAA) before RNA sequencing (Eramo et al., 2018). We used 4sU as a metabolic marker and quantified the newly synthesized RNA by labeling.
The quality and concentration of the single-cell preparations were evaluated using a hemocytometer and an inverted microscope. Single-cell suspensions were prepared in phosphate-buffered saline (HyClone, Logan, UT, United States) at a concentration of 1 × 105 cells/mL and loaded onto microfluidic devices. ScRNA-seq libraries were constructed using GEXSCOPE® Single-Cell RNA Library Kit (Singleron Biotechnologies, Nanjing, China) according to the manufacturer’s protocol. Individual libraries were diluted to 4 nM and pooled for sequencing using Illumina novaseq 6,000 with 150 bp paired end reads.
Raw reads were processed to generate gene expression matrixes using CeleScope (https://github.com/singleron-RD/CeleScope) v1.9.0 pipeline. Briefly, low-quality reads were removed using CeleScope; then Cutadapt v1.17 (Martin, 2011) was used to trim the poly-A tail and adapter sequences. The cell barcode and unique molecular identifiers were extracted. Thereafter, we used STAR v2.6.1a (Dobin et al., 2013) to map the reads to the reference genome GRCh38 (ensembl version 92 annotation). The unique molecular identifier and gene counts of each cell were acquired using featureCounts v2.0.1 (Liao et al., 2014), and were used to generate expression matrix files, including total, new, and old RNA matrices, for subsequent analysis.
We input the filtered feature barcode matrices using the function “Read10X” and “CreateSeuratObject.” Quality control, normalization, and downstream analysis were performed using the R package Seurat (ver. 4.0.2; https://github.com/satijalab/seurat). We extracted cells that expressed 200–4,000 genes and had < 15% mitochondrial genes to exclude low-quality cells and those with artifacts. Data were normalized to transform the gene expression matrices. The integrated data and correct batch of each group were analyzed by integrating data using “FindIntegrationAnchors” and “IntegrateData.”
Sequenced cells were assigned a cell cycle phase “score” and scaled to reduce cell cycle heterogeneity. The first 50 PCs and the parameter resolution to 1 were used to cluster data into cell types. Cluster-specific genes were identified using “FindAllMarkers,” and cell clusters were manually annotated based on these genes.
Gene set over-representation of significantly upregulated cluster-defining genes was analyzed using the gsfisher R package (ver. 0.2; https://github.com/sansomlab/gsfisher/) and the corresponding gene sets were obtained from the gene ontology database.
The differentiation potency score was calculated using the “AddModuleScore” function. The gene sets were used to rank the cells in the osteogenic, chondrogenic, adipogenic, endothelial, neurogenic, and vascular endothelial growth factor sets (Lee et al., 2022). Wilcoxon signed-rank tests were used to compare the groups.
The CellChat R package was used to visualize cell–cell interactions among immune cells, followed by the standard workflow and preprocessing steps. Then the potential interactions between ligands and receptors were explored based on the number of interactions and interaction strength.
Single-cell pseudotime trajectories were analyzed using the Monocle3 R package (ver. 1.2.0; https://cole-trapnell-lab.github.io/monocle3/). The “learn_graph” function was used to analyze the trajectories. Then the cells were organized in pseudotime using the “order_cells” function with a selected node that represents the hDPSCs to identify the pseudotime root node. RNA velocities were evaluated using the veloctyo.py v0.17.16 and scVelo algorithms (0.2.5.dev5+g1805ab4 [python 3.8.0]) (Bergen et al., 2020). The “Scv.tl.velocity” function was used to calculate the velocity. Velocity plots were visualized using the “scvelo.tl.velocity_embedding_stream (basis = ‘umap’)” function.
The top 200 genes with the highest multiple difference of new and old RNA rates between the infection and control groups were selected to analyze pathway enrichment using the “Enrichr” tool (Kuleshov et al., 2016). Then we focused on the KEGG 2021 human pathway databases.
Statistical analyses were performed using Origin (2020b) and R (version 4.0.3) software. Statistical significance was assessed using an unpaired two-tailed Student’s t test, non-parametric Wilcoxon rank sum test, and analysis of variance. p values < 0.05 were considered indicative of statistical significance.
We used single-cell profiling to explore the cellular and molecular characteristics of hDPSCs infected with P. gingivalis or E. faecalis (Figure 1A). After removal of low-quality cells, 72,486 cells remained, including 21,736 non-infected cells (control sample), 30,599 cells infected with P. gingivalis (Pg sample), and 20,151 cells infected with E. faecalis (Ef sample). Supplementary Figure S1C is the bar graph of the cell populations.
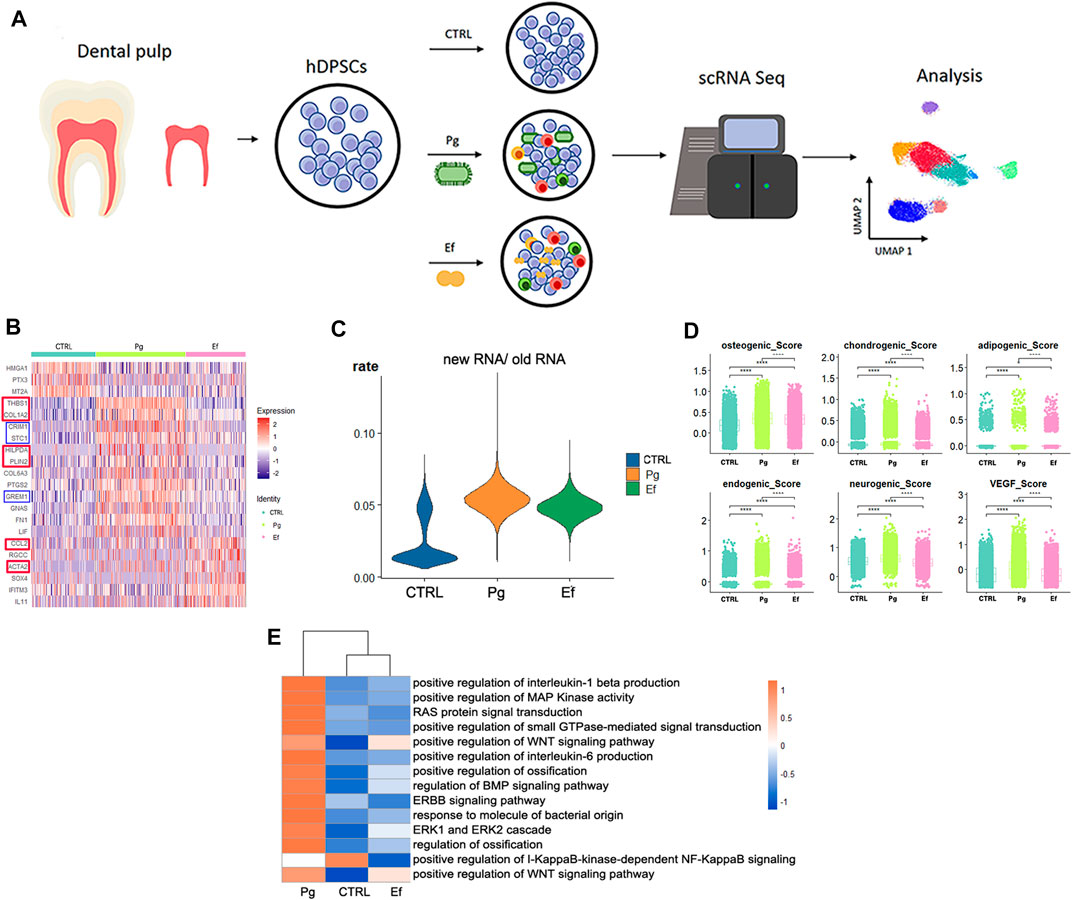
FIGURE 1. Study design and differential gene analysis. (A) Schematic graph showing the experimental design for single-cell RNA sequencing and analysis. (B) Heatmaps of the most differentially expressed genes among control, Pg, and Ef samples. Red and blue square frames indicated genes mentioned in the article. (C) Violin plots of the new RNA/old RNA rate of hDPSCs among each sample. (D) hDPSCs differentiation scores among three samples, including osteogenic, chondrogenic, adipogenic, endogenic, neurogenic, and VEGF scores. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. (E) Heatmaps of enriched pathways among the three groups.
We analyzed the differentially expressed genes (DEGs) among control, Pg, and Ef samples. The control cells expressed HMGA1 and MT2A, which are involved in cell cycle regulation. The DEGs in the Pg sample were THBS1, COL1A2, CRIM1, and STC1, which are related to matrix formation and mineralization, and HILPDA and PLIN2, which are related to the cellular response to hypoxia. The Ef sample was characterized by CCL2 expression, which is related to leukocyte chemotaxis, and ACTA2, which is related to actin (Figure 1B). The proportion of newly synthesized RNA in cells was increased after bacterial stimulation in the Pg and Ef samples (Figure 1C).
We conducted differentiation potential analysis of the three cell groups. The hDPSCs of the Pg sample showed the highest scores (p < 0.05), including the osteogenic, chondrogenic, adipogenic, endothelial, neurogenic, and VEGF scores (Figure 1D). These results suggest that hDPSCs could not maintain their stem cell status, but had multidirectional differentiation trends under conditions of P. gingivalis stimulation.
Figure 1E shows the differences in the expression of signaling pathways among the three cell groups. Certain signaling pathways were upregulated after P. gingivalis or E. faecalis infection, including the interleukin-1 beta, MAPK, WNT, ossification, BMP, extracellular signal-regulated kinase 1 (ERK1), and ERK2 pathways. The expression of the WNT signaling pathway is significantly upregulated by P. gingivalis or E. faecalis infection. The Pg sample showed significant upregulation of the BMP signaling pathway and ossification. The ERBB signaling pathway, RAS protein signal transduction, and small GTPase-mediated signal transduction were upregulated in Pg samples and downregulated in Ef samples. The I-KappaB-kinase-dependent NF-KappaB signaling pathway was significantly downregulated in the Pg and Ef samples (Figure 1E).
We used unsupervised graph clustering to partition the cells into 20 clusters and visualized the clusters via uniform manifold approximation and projection (UMAP) (Figure 2A; Supplementary Figure S2). Subsequently, we used marker genes to identify four types of hDPSCs (Figure 2A), including classical mesenchymal stem cells (MSCs; CD105, CD90, and CD73), fibroblast-like hDPSCs (DCN, COL1A1, COL3A1, LUM, S100A4, FAP, PDPN, and TPM1), monocyte-like hDPSCs (IFITM3, IFIT1, OASL, and MX1), and perivascular-like hDPSCs (ACTA2, MCAM, TAGLN, and PDGFRβ) (Supplementary Figure S3). The four cell types were present in different proportions in each sample. The number of fibroblast-like hDPSCs was higher in the Ef sample than in control and Pg samples (49%, 28%, and 28%, respectively) (Figure 2B).
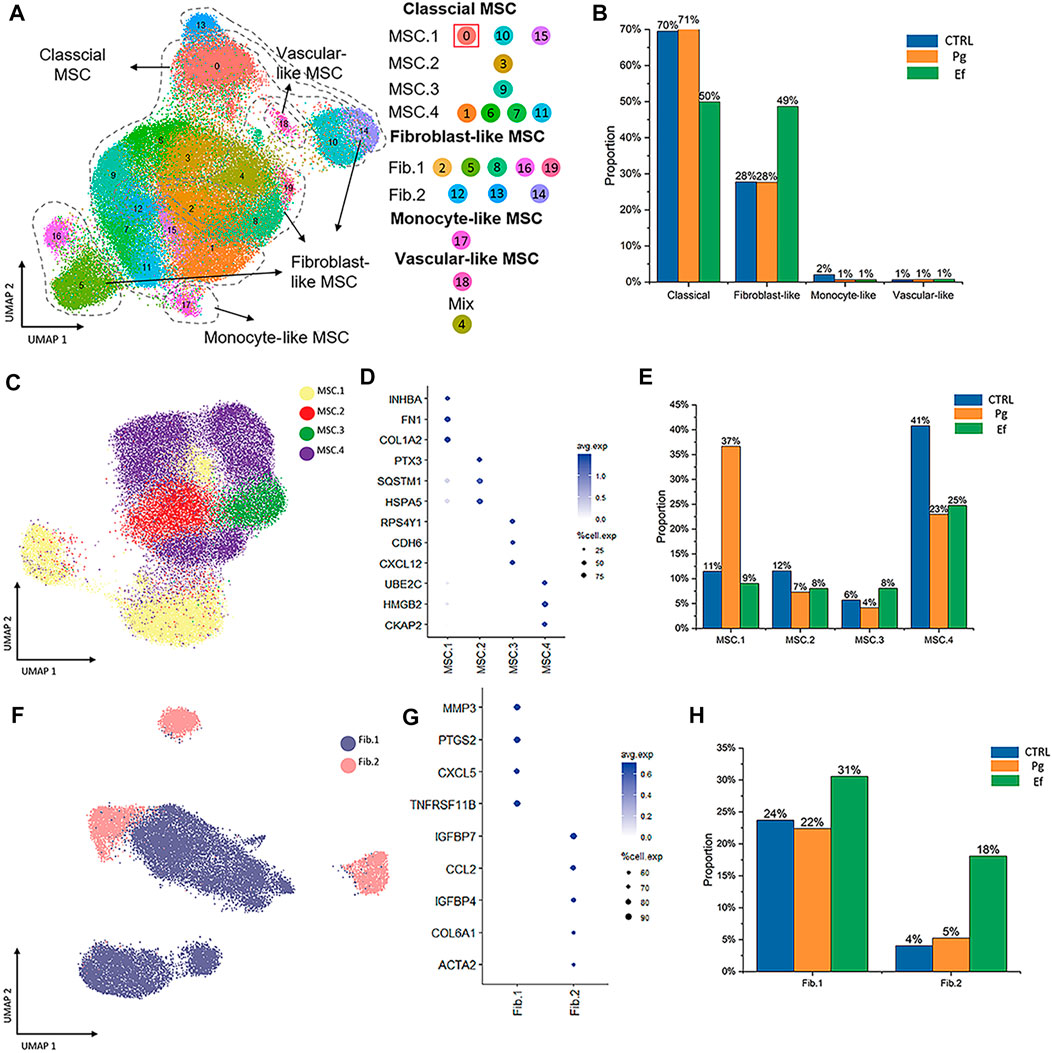
FIGURE 2. Cell types and clustering in the control, Ef, and Pg samples. (A) UMAP plot of the distribution of the 20 clusters. Clusters 0, 1, 3, 6, 7, 9, 10, 11, 15, and 20 were identified as MSC.1 (clusters 0, 10, and 15), MSC.2 (cluster 3), MSC.3 (cluster 9), and MSC.4 (clusters 1, 6, 7, and 11). Clusters 2, 5, 8, 12–14, 16, and 19 were fibroblast-like hDPSCs and included Fib.1 (clusters 2, 5, 8, 16, and 19) and Fib.2 (clusters 12–14). Cluster 17 included monocyte-like hDPSCs. Cluster 18 included perivascular-like hDPSCs. (B) Classical MSCs (46,902 cells), fibroblast-like hDPSCs (24,292 cells), monocyte-like hDPSCs (771 cells), and perivascular-like hDPSCs (160 cells) are shown. Proportion plots showing the percentage of four cell subclusters from each sample. (C) UMAP plot of classical MSC.1 (mineralization and ossification; 15,530 cells), MSC.2 (stress and inflammatory response; 6,367 cells), MSC.3 (osteoclastic reaction; 4,128 cells), and MSC.4 (cell cycle; 20,877 cells) populations. (D) Dot plots showing the expression of cluster-defining genes and percentage of cells expressing each gene in the four subtypes of classical MSC. The expression values are normalized and scaled averages. (E) Proportion plots showing the percentage of four subcluster types from MSCs. (F) UMAP plot of Fib.1 (inflammatory properties; 18,164 cells) and Fib.2 (myofibroblast-like hDPSCs; 6,128 cells) populations. (G) Dot plots showing the expression of cluster-defining genes and percentage of cells expressing each gene of the two subtypes of fibroblast-like hDPSCs. Expression values are normalized and scaled averages. (H) Proportion plots showing the percentage of two subcluster types from fibroblast-like hDPSCs.
Next, we identified four subtypes of classical MSCs and two subtypes of fibroblast-like hDPSCs based on their marker genes and biological characteristics (Figure 2). Cluster 0, 10, and 15, which were identified as MSC.1, were linked to mineralization and ossification with expression of INHBA, FN1, and COL1A2 (Figure 2D); their number was significantly higher in the Pg sample than in control and Ef samples (37%, 11%, and 9%, respectively) (Figure 2E). MSC.2 (cluster 3) was related to stress and the inflammatory response with expression of PTX3, SQSTM1, and HSPA5 (Figure 2D). Because cluster 9 was associated with a unique gene ontology pathway, i.e., negative regulation of osteoblast differentiation and ossification, it was classified as MSC.3. MSC.4 included clusters 1, 6, 7, and 11 (Figure 2A).
Fib.1 included clusters 2, 5, 8, 16, and 19, and Fib.2 included clusters 12–14 (Figures 2A, F, G). The proportions of Fib.1 and Fib.2 were higher in Ef samples than control and Pg samples (Fib.1: 31%, 24%, and 22%, respectively; Fib.2: 18%, 4%, and 5%, respectively) (Figure 2H).
The above data showed that the distribution of hDPSCs under P. gingivalis or E. faecalis stimulation was obviously different (Figure 2).
Characteristics of and communication among classical MSC, fibroblast-like, monocyte-like, and perivascular-like hDPSCs
The cell types had unique characteristics. Differential gene heatmaps of the cell clusters are shown in Supplementary Figure S4. MSC.1 was associated with mineralization and ossification and the expression of INHBA, FN1, and COL1A2 (Figure 2D); there were also present at a significantly higher proportion in Pg samples than in other samples (Figure 2E), indicating significant odontogenic and osteogenic differentiation of hDPSCs after stimulation by P. gingivalis. The DEGs of MSC.1 were enriched in the cellular respiration and ion transport pathway in control samples. MSC.1 was associated with odontogenesis and response to the hypoxia pathway in Pg samples, and neutrophil chemotaxis and osteoblast differentiation pathways in Ef samples (Figure 3A).
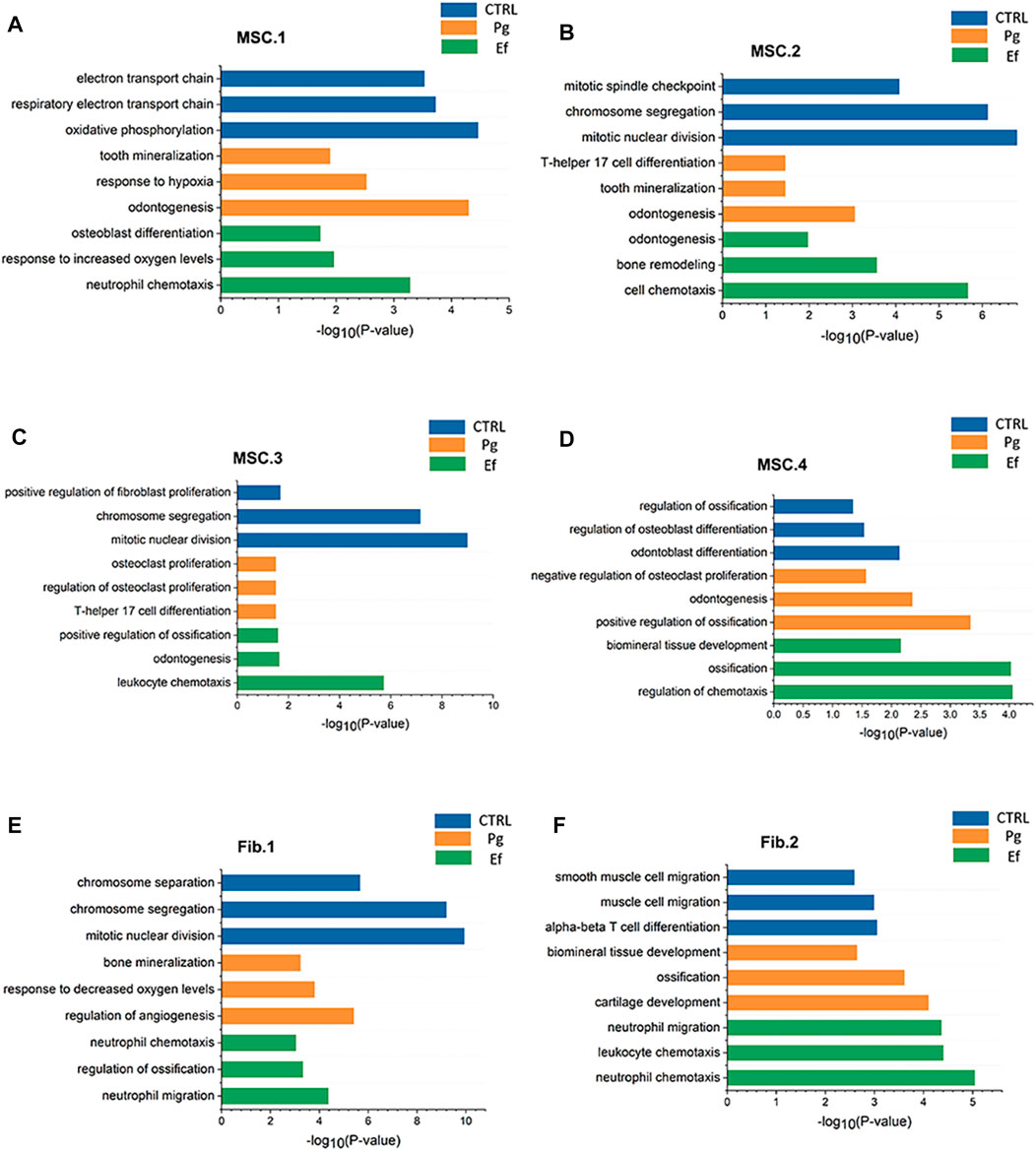
FIGURE 3. Pathway atlas for the characterization of classical MSCs and fibroblast-like hDPSCs. Bar charts of the most typical pathways enriched in MSC.1 (A), MSC.2 (B), MSC.3 (C), MSC.4 (D), Fib.1 (E), and Fib.2 (F) subtypes from each sample. The length of the bar graph indicate the number of genes from each pathway.
MSC.2 (cluster 3) was related to the stress and inflammatory response associated with the expression of PTX3, SQSTM1, and HSPA5 (Figure 2D). MSC.2 was associated with cell cycle function in control samples, odontogenesis, and the T-helper 17 cell differentiation pathway in Pg samples, and cell chemotaxis and bone remodeling in Ef samples (Figure 3B).
MSC.3 expressed genes related to osteoclasts (RPS4Y1, CDH6, and CXCL12) (Figure 2D). MSC.3 cluster genes were related to the regulation of fibroblast proliferation in control samples, osteoclast proliferation in Pg samples, and odontogenesis and positive regulation of ossification in Ef samples (Figure 3C).
MSC.4 expressed genes involved in the regulation of the cell cycle (UBE2C, HMGB2, and CKAP2) (Figure 2D). The MSC.4 genes were upregulated to a higher level in control samples than in Pg and Ef samples (Figure 2E). We speculated that, when hDPSCs are infected with E. faecalis and P. gingivalis, the cell cycle is affected and the stem cells differentiate into other cell types. MSC.4 was involved in the regulation of osteoblast differentiation and differentiation in control samples, positive regulation of ossification and negative regulation of osteoclast proliferation in Pg samples, and ossification and regulation of chemotaxis in Ef samples (Figure 3D). The increase in the odontogenic and osteogenic pathways in control samples indicated that hDPSCs can differentiate into odontoblasts and promote odontogenesis and osteogenesis without exogenous stimulation.
Fib.1 consisted of clusters 2, 5, 8, 16, and 19, and was associated with genes related to inflammatory and hypoxia-related gene expression (MMP3, PTGS2, CXCL5, and TNFRSF11B) (Figure 2G). The Fib.1 genes were associated with the cell cycle regulation in control samples, bone mineralization in Pg samples, and cell chemotaxis and ossification in Ef samples (Figure 3E).
Fib.2 are the characteristic effector cells stimulated by E. faecalis infection. The proportion of Fib.2 cells was significantly higher in Ef samples than in the other two samples, which had few Fib.2 cells (Figure 2H). Similar to myofibroblasts, these cells showed upregulated ACTA2 expression (Figure 2G) and enrichment of the muscle cell migration signaling pathways (Figure 3F).
Monocyte-like hDPSCs exhibited upregulation of genes involved in immune responses. Perivascular-like hDPSCs showed similar characteristics to perivascular cells, and a similar pattern of gene expression across the control, Pg, and Ef samples as for monocyte-like hDPSCs (Supplementary Figure S4). The samples had similar proportions of monocyte-like and perivascular-like hDPSCs.
Next, we performed cell differentiation potential analyses of the cell clusters. MSC.4 had the highest differentiation potential, whereas MSC.1 had the lowest, indicating that MSC.1 was relatively active in the late stages of differentiation (Figure 4A). Perivascular-like hDPSCs had a higher endogenic score (Figure 4B). The osteogenic score of MSC.1 was higher than that of other MSC clusters (Figure 4C).
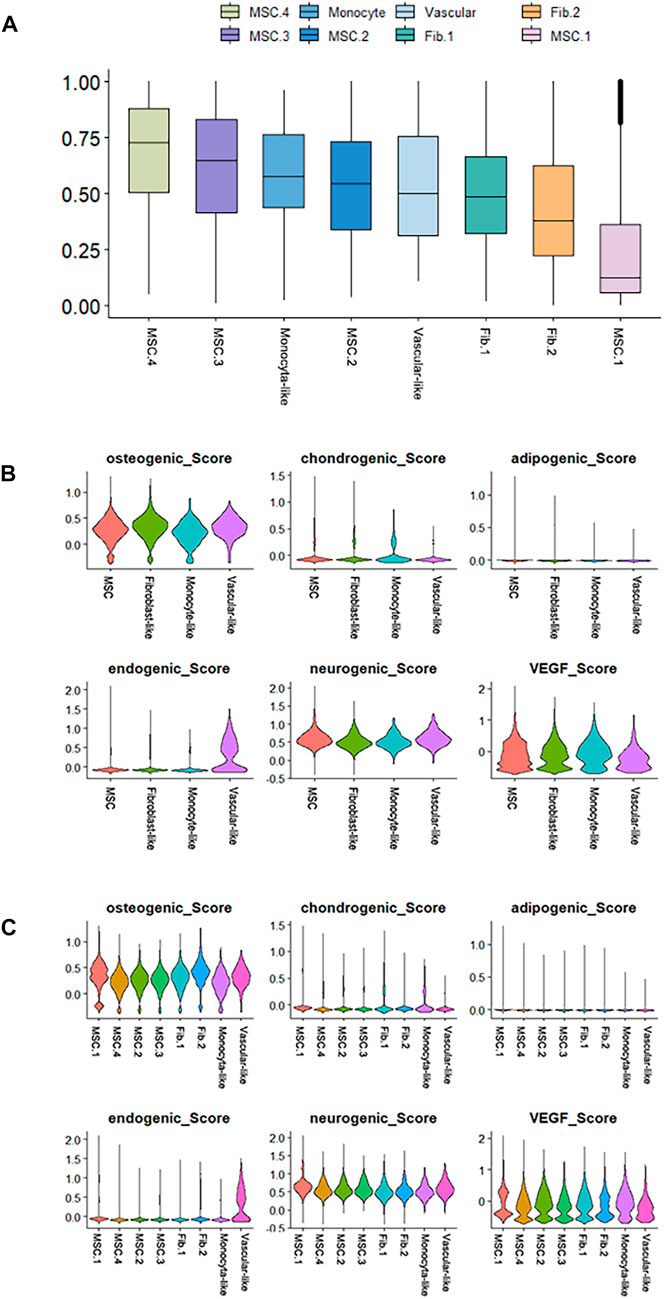
FIGURE 4. Differentiation potency scores. (A) Box diagrams showing the overall differentiation potential of various cell types. (B) Violin plots showing the hDPSCs differentiation scores among the four cell types. (C) Violin plots showing the hDPSCs differentiation scores among the subtypes.
We used the Monocle3 R package and scVelo to reconstruct the fate decisions and pseudotime trajectories of hDPSCs and explore the gene-specific transcriptional dynamics of hDPSCs over time.
Three hDPSCs samples were analyzed (Figure 5). We selected classical MSC.4 (cell cycle) as the root principal point because of its role in the cell cycle. The analysis of cell differentiation potential showed that MSC.4 was relatively active in the early stages of differentiation. The UMAP plot exhibited different patterns for each sample. In the control samples, fibroblast-like hDPSCs were ordered by later pseudotime. In Pg samples, hDPSCs related to the cell cycle evolved into classical MSC.1 (mineralization and ossification), whereas in Ef samples, MSC.4 developed into fibroblast-like hDPSCs (Figure 5).
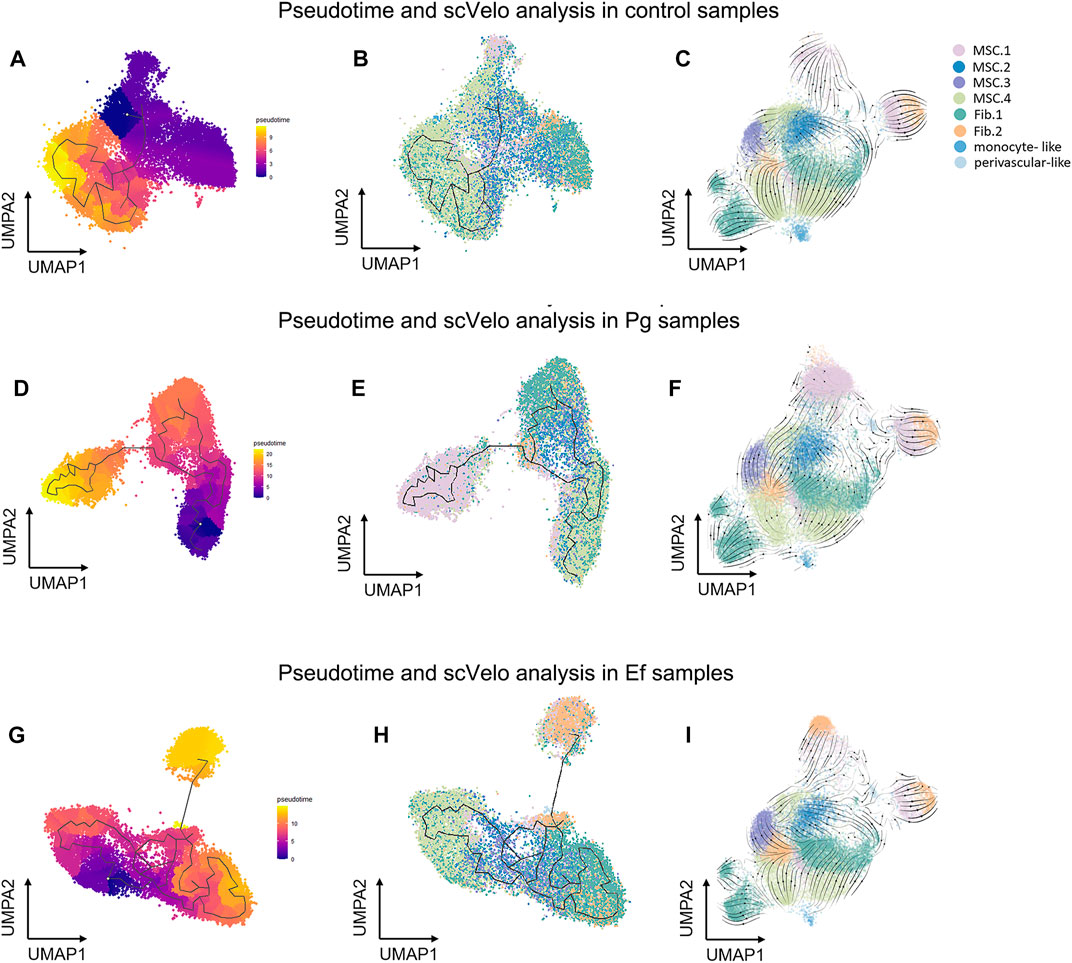
FIGURE 5. Pseudotime and scVelo analysis of hDPSCs subtypes. (A , D, G) UMAP plots showing branched cell trajectories (black lines) and cell type in control (A), Pg (D), and Ef (G) samples. (B, E, H) UMAP plots showing hDPSCs colored by pseudotime in control (B), Pg (E), and Ef (H) samples. (C, F , I) UMAP plots showing the velocity trajectory across hDPSCs subtypes in control (C), Pg (F), and Ef (I) samples. The arrows indicate the predicted future cell states.
The cellular communications of hDPSCs are involved in multiple metabolic processes and biological functions. Using the CellChat R package, we analyzed the mutual relationships of eight cell subtypes via ligand–receptor interactions, which regulate hDPSCs differentiation and development. Bacterial stimulation promotes communication among cells, particularly between MSC.2 (stress and inflammatory response) and monocyte-like hDPSCs, indicating activation of the immune inflammatory response. Fib.2 myofibroblasts and perivascular-like hDPSCs were relatively independent of each other (Figure 6; Supplementary Figure S6), indicating that they represented late stages of differentiation.
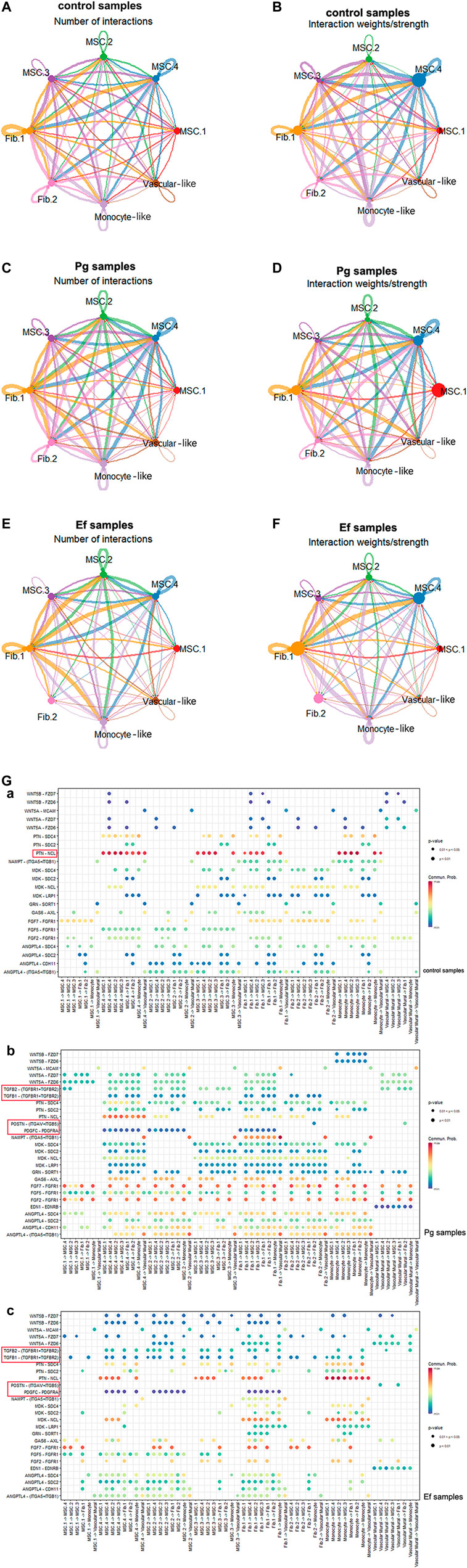
FIGURE 6. Cell–cell interactions among different subtypes. (A , C , E) Circle plots showing the number of cell–cell interactions in control (A), Pg (C), and Ef (E) samples. The edge width is proportional to the indicated number of ligand–receptor pairs. (B , D , F) Circle plots showing the weight/strength of cell–cell interactions in control (B), Pg (D), and Ef (F) samples. The edge width is proportional to the indicated weight/strength of ligand–receptor pairs. (G) Dot plots showing the ligand–receptor expression among various cell types. (a), (b), (c).
Analyses of ligand–receptor interactions that regulate hDPSC differentiation and development showed similar results. In control samples, there were significant cellular interactions among cell groups, as well as the presence of ligand receptors among different groups (Figure 6G). PTN-NCL was highly expressed in control samples but significantly downregulated in Pg samples (Figure 6H). The TGF-related ligand receptors, POSTN-(ITGAV + ITGB5) and PDGFC-PDGFRA were activated in response to P. gingivalis and E. faecalis infection. These ligand receptor pairs may be related to cell migration and adhesion. Additionally, we observed enhanced intercellular interactions between FGF and ANGPTL, which are related to angiogenesis.
BMP5-(ACVR1+ACVR2A), BMP7-(ACVR1+ACVR2A), and BTLA-TNFRSF14 were only detected in control samples. In the Pg sample, we observed interactions between ADM and CALCRL, which is related to periodontitis; EDA and EDA2R, which is related to ectodermal development; and SPP1 and CD44 and SPP1 and (ITGA4+ITGB1), which promotes phosphoprotein secretion and regulates bone salinization. The Ef samples also showed interactions between C3 and C3AR1 and HC and C5AR1, which are associated with complement activation; and SST and SSTR2, which negatively regulates cell proliferation (Supplementary Table S1).
We analyzed 20 small clusters of cells, and found different trends among different samples for each cluster (Figure 7). Interestingly, the cell percentage of cluster 0 was nearly 10-fold higher in Pg samples than in control and Ef samples. The control and Ef samples had a similar number of cells in cluster 0 (Figure 7A), indicating that cluster 0 cells are the effector cells against P. gingivalis stimulation. DEGs in Cluster 0 were related to the positive regulation of biomineralization and response to the hypoxia pathway in the Pg samples (Figure 7B). This cell cluster exhibited increased levels of new RNA and upregulated expression of HILPDA and DDIT4, which are closely related to the cellular response to hypoxia; and COL1A2, which is closely related to cell mineralization (Figures 7C, D).
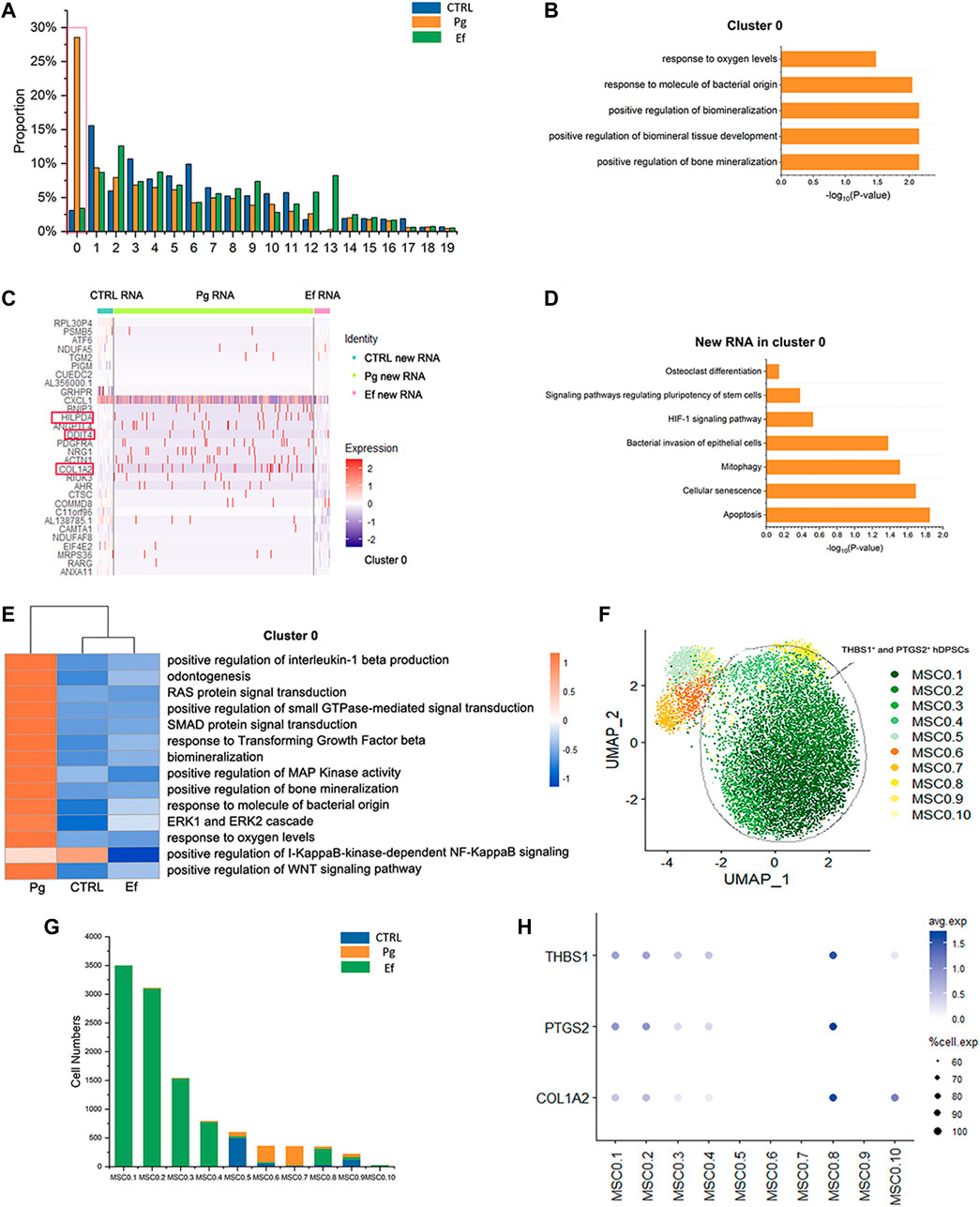
FIGURE 7. Responses of hDPSCs to P. gingivalis infection. (A) Proportion plots showing the percentage of 20 subclusters from each sample. Red line used to mark the cluster 0, the vast majority of cells in the 0 clusters originate from Sample Pg. (B) Bar chart of pathways enriched in cluster 0. The length of the bar graph indicates the number of genes from each pathway. (C) Heatmap of the most differentially expressed genes of new RNA in cluster 0 from each sample. (D) Bar chart of new RNA pathways enriched in Cluster 0. The length of the bar graph indicates the number of new genes from each pathway. (E) Heatmap of pathways enriched in cluster 0 among the three groups. (F) UMAP plot depicting cluster 0.1–0.10 populations in cluster 0. (G) Number of cluster 0.1–0.10 cells. (H) Dot plots showing the expression of THBS1, PTGS2, and COL1A2 among clusters 0.1–0.10.
The pathways enriched in cluster 0 of hDPSCs included the hypoxia response, mineralization, stem cell maintenance, and osteoclasts (Figures 7B, D). Changes were detected in several signal transduction factors, including positive regulation of pathway-restricted SMAD protein phosphorylation, positive regulation of Ras protein signal transduction, ERK1 and ERK2 cascades, and the TGF-beta signaling pathway, as well as negative regulation of the I-KappaB-kinase-dependent NF-KappaB signaling pathway (Figure 7E). These findings suggest the involvement of the classical signaling transduction pathway and represent the response of hDPSCs to P. gingivalis invasion.
We further analyzed cluster 0 cells, divided them into 10 new clusters (Figure 7F), and screened cells resistant to P. gingivalis infection. MSC0.1, MSC0.2, MSC0.3, MSC0.4, and MSC0.8 were present in Pg samples (Figure 7G) but not in control or Ef samples. These results suggest that the cells produced by P. gingivalis stimulation are characterized by high expression levels of THBS1, PTGS2, and COL1A2 (Figure 7H). These cells were categorized as THBS1+ and PTGS2+ hDPSCs.
The Ef samples were characterized by expression of CCL2, which is related to leukocyte chemotaxis, and ACTA2, which is an actin-related gene (Figure 1B).
Fib.1 and Fib.2 had higher proportions in Ef samples than in Pg and control samples (Fib.1: 31%, 22%, and 24%, respectively; Fib.2: 18%, 5%, and 4%, respectively) (Figure 2H). These results indicate that hDPSCs differentiate into fibroblast-like cells after E. faecalis infection. Fibroblasts and myofibroblasts play a key role in the defense of dental pulp against invasion by E. faecalis. Pseudotime analysis also showed that hDPSCs differentiated into fibroblasts and myofibroblasts.
We evaluated the effects of P. gingivalis and E. faecalis on hDPSCs in a bacteria–cell coculture model using single-cell sequencing. The hDPSCs had different responses to the two types of bacteria in terms of the cell clusters mobilized and their differentiation. After P. gingivalis stimulation, hDPSCs showed matrix mineralization-related characteristics, whereas E. faecalis caused the cells to differentiate into fibroblast-like cells. These findings provide insight into the mechanisms underlying the responses of hDPSCs to invasion by distinct bacterial species and the effects of residual bacteria on RET outcomes. Figure 8 summarized the highlights of the findings.
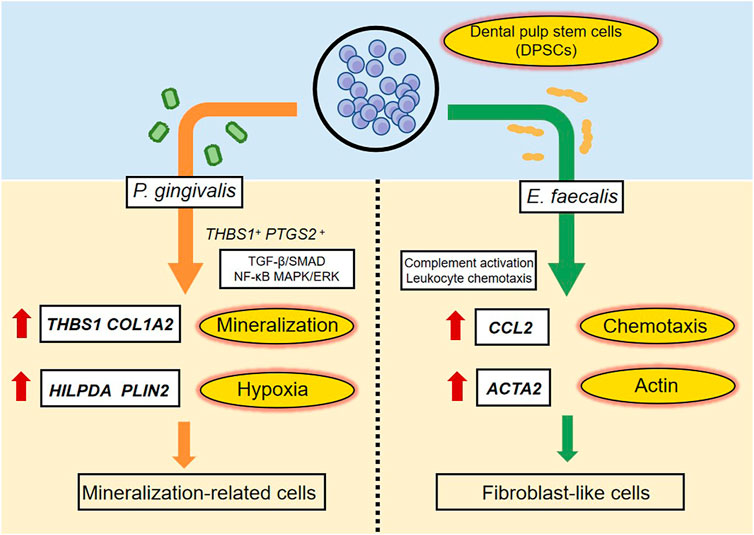
FIGURE 8. A schematic diagram recapitulating the response of hDPSCs to P. gingivalis or E. faecalis. When hDPSCs infected by P. gingivalis, THBS1 and COL1A2, which are related to matrix formation and mineralization, and HILPDA and PLIN2, which are related to the cellular response to hypoxia were the most differentially expressed genes. THBS1+ PTGS2+ hDPSCs prevented P. gingivalis infection by regulating the TGF-β/SMAD, NF-κB, and MAPK/ERK signaling pathways. hDPSCs infected by P. gingivalis undergo multidirectional differentiation, particularly to the mineralization-related cell lineage. The Ef samples were characterized by the expression of CCL2, which is related to leukocyte chemotaxis, and ACTA2, which is related to actin. hDPSCs mediated complement activation and leukocyte chemotaxis pathways. The presence of E. faecalis promoted the differentiation of hDPSCs into fibroblast-like cells.
In response to P. gingivalis stimulation, a cluster of cells characterized by high expression levels of thrombospondin 1 (THBS1) and prostaglandin-endoperoxide synthase 2 (PTGS2) (i.e., THBS1+ and PTGS2+ hDPSCs) were identified. In a previous study, the level of TSP1 protein encoded by THBS1 was increased in gingival tissues of patients with periodontitis and were upregulated by Pg-LPS (Gokyu et al., 2014). The TSP1 protein is highly expressed in and secreted by odontoblasts; it is involved in biomineralization and odontogenesis (Ueno et al., 1998). PTGS2, also known as cyclooxygenase, is responsible for the prostanoid biosynthesis involved in inflammation and mitogenesis. P. gingivalis is associated with increased PTGS2 expression, PGE2 secretion (Symmank et al., 2021), and the development of pulpitis. In the present study, the high expression levels of THBS1 and PTGS2 in the Pg samples suggest that hDPSCs stimulate the immune inflammatory response against bacteria and prevent P. gingivalis invasion through mineralization and repair.
In cluster 0, the TGF-β and WNT signaling pathways were upregulated (Figure 7E). These conservative signaling pathways regulate multiple cellular functions, including cell growth, adhesion, migration, cell-fate determination, differentiation, and apoptosis (Kubiczkova et al., 2012). These pathways play a key role in the synthesis of dentin proteins and proteases (Niwa et al., 2018). Additionally, positive regulation of pathway-restricted SMAD protein phosphorylation was found to be enriched. Hwang et al. (2008) reported that TGF-β is released during reparative dentin formation and activates the Smad proteins to control TGF-β cell signaling. In Pg samples, the enrichment of TGF β and SMAD may be related to the formation of repair tissue. The regulation of the ERK1 and ERK2 cascades and small RAS GTPase were enriched in Cluster 0. The ERK1 and ERK2 proteins belong to a family of structurally related kinases called mitogen-activated protein kinases (MAPKs), and are essential components of the TGF-β signaling pathway and mitogenic signals largely channeled by small RAS GTPases. This pathway is an important therapeutic target for inflammatory diseases and can induce osteogenic gene expression (Lavoie et al., 2020). The MAPK/ERK pathways are activated to regulate inflammation through downstream RAS GTPase.
The signaling pathway of inhibitor of nuclear factor kappa-B (IκB) was suppressed (Figure 7E), suggesting upregulation of nuclear factor kappa-B (NF-κB) expression. NF-κB is associated with multiple biological reactions, cytokines, chemokines, cell adhesion molecules, growth factors, and immune receptors (such as IL-1β and PTGS2, which were detected in our study in Figures 7E, H) (Paudel et al., 2014). NF-κB and MAPK pathways are activated by the LPS in dental pulp cells (Kim et al., 2012; Lee et al., 2022). These results suggest that hDPSCs resist P. gingivalis invasion by regulating the TGF-β/SMAD, NF-κB, and MAPK/ERK pathways.
The differentiation potency scores, including osteogenic, chondrogenic, adipogenic, endothelial, neurogenic, and VEGF factors, showed the multi-directional differentiation potential of hDPSCs of the Pg samples (Figure 1D). However, after P. gingivalis stimulation, the cells differentiated into terminal cells (MSC.1) related to mineralization expression. In Pg samples, the number of MSC.1 cells was increased (Figure 2E), and pseudotime analysis showed differentiation of hDPSCs into MSC.1 cells (Figure 5). The genes that were significantly overexpressed in Pg samples (STC1, CRIM1, and GREM1) (Figure 1B, blue square frame marked) were involved in the regulation of matrix mineralization and bone formation (Yoshiko et al., 2002; Liu et al., 2021).
Interestingly, the presence of bacteria was associated with altered expression of hypoxia-related genes, although the coculture was not performed in a hypoxic environment. The highly expressed genes HILPDA, DDIT4, and HIF1A are closely related to the cellular response to hypoxia (Figure 1B). Previous studies have shown that Pg-LPS and hypoxia promote periodontal tissue destruction (Gölz et al., 2015). Hypoxia also enhances the odontogenic and osteogenic properties of hDPSCs through the HIF1α-Wnt/β-catenin signaling pathway (Orikasa et al., 2022). Based on our classification, the enrichment of hypoxic and hyperoxic pathways in Fib.1 and MSC.1 (Figure 3) indicates that these cell types respond to changes in the oxygen level.
Multiple cell chemotaxis-related and complement-activated signaling pathways were detected in the Ef samples, whereas the corresponding pathways were not significantly enriched in the Pg samples (Figure 3). These findings indicate an immune escape effect of P. gingivalis. P. gingivalis is internalized into cells through MAPK activation to avoid extracellular clearance, inhibit IL-8 production by serine dephosphorylation of the p65 subunit of NF-κB, and reduce neutrophil migration and killing. P. gingivalis cleaves the complement component C3 into C3a and C3b, and degrades them to disrupt the complement cascade reaction and inhibit T cell activation to avoid being killed by the host’s immune response (Zheng et al., 2021). However, previous studies have mainly focused on periodontal tissue. The present study found that immune evasion can also occur during dental pulp inflammation and pulp regeneration.
The numbers of fibroblast-like cells were significantly increased after E. faecalis infection, including typical myofibroblasts (Fib.2) with high ACTA2 expression (Figure 2). In contrast to the changes in Ef samples, hDPSCs after P. gingivalis infection did not differentiate into fibroblast-like cells (Figure 2B). These differences may be due to the different characteristics of the bacteria. The hDPSCs that were exposed to E. faecalis differentiated into fibroblast-like cells via chemotaxis of leukocytes and phagocytes, and activation of the complement system. Dental pulp fibroblasts play pivotal roles in the production of the complement system proteins involved in defense processes, control of inflammation, dentin-pulp regeneration, and dental pulp wound healing (Tsai et al., 2022). The high activity of the leukocyte migration and complement pathways in the Ef samples in our study are consistent with these findings (Figure 3). Myofibroblasts in the dental pulp originate from MSCs (Dimitrova-Nakov et al., 2014) and promote extracellular matrix remodeling in damaged dental pulp (Edanami et al., 2017) and tissue repair through collagen synthesis. Some myofibroblasts produce newly differentiated odontoblast-like cells that synthesize restorative dentin (Álvarez-Vásquez & Castañeda-Alvarado, 2022). Our study showed that myofibroblasts play a significant role in tissue repair after E. faecalis invasion.
The different responses to the two types of bacteria may be related to their different characteristics, in terms of aspects such as oxygen demand and virulence. LPS and lipoteichoicacid (LTA) have different molecular structures and antigens, as well as recognition patterns, leading to different responses against bacteria (Colombini-Ishikiriama et al., 2020). The upregulation of the TLR/MyD88/NF-κB pathway of LPS-treated hDPSCs is associated with increased interleukin production and odontoblastic differentiation (Brodzikowska et al., 2022). LTA-stimulated hDPSCs release IL-6 and IL-8 in a dose-dependent manner and promote cell proliferation, cell migration, and the local inflammatory response through cytokine release; however, LTA does not influence osteogenic or odontoblastic differentiation (Shayegan et al., 2021). In our study, the effects of the two virulence factors on the cells were consistent with those of the two bacteria on hDPSCs.
Based on previous studies, the tissues that formed in the canals of revascularized/revitalized teeth were identified as fibrous connective tissue, similar to that found in the periodontal ligament and cementum-like or bone-like tissue. Fibrous connective tissue, which is mainly characterized by fibroblasts and collagen fibers, filled the radiographically empty canal space; no odontoblast-like cells were observed, particularly in the area of severe inflammation (Becerra et al., 2014; Nosrat et al., 2015). The hDPSCs could differentiate into odontoblast-like cells and express dentin sialophosphoprotein (DSPP) and osteocalcin (OCN), which are used to regenerate dentin (Alongi et al., 2010; Liao et al., 2011). However, the corresponding genes were not detected in our study, which may be related to the presence of bacteria. Some studies have shown that P. gingivalis suppresses the expression of DSPP and OCN in hDPSCs (Yamagishi et al., 2011), suggesting a negative effect on pulp regeneration. We did not detect cementogen-related genes (CDGF, CAP, and CEMP1), which may be due to the limited stimulation time, as we only observed early reactions.
In conclusion, hDPSCs do not maintain their stem cell status in the presence of P. gingivalis and E. faecalis (Figure 4A). The proportion of MSC.4 with the highest differentiation potential score (Figure 4A) decreased after bacterial invasion and differentiated into MSC.1or Fib.2 (Figure 5), whose differentiation potential are lower than MSC.4. They differentiate into mineralization-related cells in the presence of P. gingivalis and into fibroblast-like cells in the presence of E. faecalis. Additionally, we identified certain hDPSC clusters and signaling pathways that respond to each type of bacteria. These findings enhance our understanding of the molecular mechanisms underlying the responses of dental pulp to bacterial invasion. Future studies should create a microenvironment that is conducive to the survival and directional differentiation of stem cells during RET.
Although scRNA-seq is useful for determining the mechanisms underlying the responses of hDPSCs to various bacterial infections, our findings need to be validated in future studies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author FC (Y2hlbmZlbmcyMDExQGhzYy5wa3UuZWR1LmNu).
The studies involving human participants were reviewed and approved by Ethics Committee of Peking University School and Hospital of Stomatology (PKUSSIRB-202163047). The patients/participants provided their written informed consent to participate in this study.
FC, XZ, and LY conceived and designed this study; TX and WZ analyzed and interpreted the data; YL, WZ, and YZ recruited the study participants and conducted the experiments; and WZ and TX wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The study was supported by grants-in-aid from the National Science Foundation China (No. 81991501) and National Key R&D Program of China (No. 2022YFE0118300).
We thank the study participants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1166934/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Quality control of samples (A) Violin plots showing the number of features, RNA counts, and percent mitochondrial transcripts in the control, Pg, and Ef samples prior to quality control. (B) Violin plots showing the number of features, RNA counts, and percent mitochondrial transcripts following quality control. (C) Bar graphs showing the number of cells before (preQC) and after (postQC) quality control in each sample.
SUPPLEMENTARY FIGURE S2 | Differentially expressed genes for the 20 cell clusters. Heatmap plot showing the average expression of the top differentially regulated genes for the 20 cell clusters.
SUPPLEMENTARY FIGURE S3 | Identification of cell types. Feature plots (UMAP) displaying the main signatures for each subtype of classical MSC (A), fibroblast-like hDPSCs (B), perivascular-like hDPSCs (C), and monocyte-like hDPSCs (D).
SUPPLEMENTARY FIGURE S4 | Genes expressed in MSC.1, MSC.2, MSC.3, MSC.4, Fib.1, Fib.2. Heatmap of the expression of the most differentially expressed genes among the three samples in MSC.1(A) , MSC.2 (B), MSC.3 (C), MSC.4 (D), Fib.1 (E) and Fib.2 (F).
SUPPLEMENTARY FIGURE S5 | Genes expressed in monocyte-like and perivascular-like hDPSCs. Heatmaps showing the expression of the most differentially expressed genes between each cluster compared to the other clusters in monocyte-like hDPSCs (A) and perivascular-like hDPSCs (B).
SUPPLEMENTARY FIGURE S6 | Cell–cell interactions between different subtypes. (A, C, E) Circle plots showing the number of cell–cell interactions in control (A), Pg (C), and Ef (E) samples. The edge width is proportional to the indicated number of ligand–receptor pairs. (B, D, F) Circle plots showing the weight/strength of cell–cell interactions in control (B), Pg (D), and Ef (F) samples. The edge width is proportional to the indicated weight/strength of ligand–receptor pairs.
SUPPLEMENTARY TABLE S1 | Ligand-receptors pairs in control, Pg, and Ef samples.
Alongi, D. J., Yamaza, T., Song, Y., Fouad, A. F., Romberg, E. E., Shi, S., et al. (2010). Stem/progenitor cells from inflamed human dental pulp retain tissue regeneration potential. Regen. Med. 5 (4), 617–631. doi:10.2217/rme.10.30
Altaii, M., Richards, L., and Rossi-Fedele, G. (2017). Histological assessment of regenerative endodontic treatment in animal studies with different scaffolds: A systematic review. Dent. Traumatology Official Publ. Int. Assoc. Dent. Traumatology 33 (4), 235–244. doi:10.1111/edt.12338
Álvarez-Vásquez, J. L., and Castañeda-Alvarado, C. P. (2022). Dental pulp fibroblast: A star cell. J. Endod. S0099-2399 (22), 00341–00347. doi:10.1016/j.joen.2022.05.004
American Association of Endodontists (Aae), Clinical considerations for a regenerative procedure. Revised 2021[R/ OL]. (2021). Available At: https://f3f142zs0k2w1g84k5p9i1o-wpengine.netdna-ssl.com/specialty/wp-content/uploads/sites/.
Baca, P., Junco, P., Arias-Moliz, M. T., González-Rodríguez, M. P., and Ferrer-Luque, C. M. (2011). Residual and antimicrobial activity of final irrigation protocols on Enterococcus faecalis biofilm in dentin. J. Endod. 37 (3), 363–366. doi:10.1016/j.joen.2010.11.036
Becerra, P., Ricucci, D., Loghin, S., Gibbs, J. L., and Lin, L. M. (2014). Histologic study of a human immature permanent premolar with chronic apical abscess after revascularization/revitalization. J. Endod. 40 (1), 133–139. doi:10.1016/j.joen.2013.07.017
Bergen, V., Lange, M., Peidli, S., Wolf, F. A., and Theis, F. J. (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38 (12), 1408–1414. doi:10.1038/s41587-020-0591-3
Biedermann, A., Kriebel, K., Kreikemeyer, B., and Lang, H. (2014). Interactions of anaerobic bacteria with dental stem cells: An in vitro study. PloS one 9 (11), e110616. doi:10.1371/journal.pone.0110616
Brodzikowska, A., Ciechanowska, M., Kopka, M., Stachura, A., and Włodarski, P. K. (2022). Role of lipopolysaccharide, derived from various bacterial species, in pulpitis: A systematic review. Biomolecules 12 (1), 138. doi:10.3390/biom12010138
Buonavoglia, A., Zamparini, F., Lanave, G., Pellegrini, F., Diakoudi, G., Spinelli, A., et al. (2023). Endodontic microbial communities in apical periodontitis. J. Endod. 49 (2), 178–189. doi:10.1016/j.joen.2022.11.015
Burns, L. E., Kim, J., Wu, Y., Alzwaideh, R., McGowan, R., and Sigurdsson, A. (2022). Outcomes of primary root canal therapy: An updated systematic review of longitudinal clinical studies published between 2003 and 2020. Int. Endod. J. 55 (7), 714–731. doi:10.1111/iej.13736
Caetano, A. J., Yianni, V., Volponi, A., Booth, V., D’Agostino, E. M., and Sharpe, P. (2021). Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. eLife 10, e62810. doi:10.7554/eLife.62810
Chrepa, V., Pitcher, B., Henry, M. A., and Diogenes, A. (2017). Survival of the apical papilla and its resident stem cells in a case of advanced pulpal necrosis and apical periodontitis. J. Endod. 43 (4), 561–567. doi:10.1016/j.joen.2016.09.024
Colombini-Ishikiriama, B. L., Dionisio, T. J., Garbieri, T. F., da Silva, R. A., Machado, M. A. A. M., de Oliveira, S. H. P., et al. (2020). What is the response profile of deciduous pulp fibroblasts stimulated with E. coli LPS and E. faecalis LTA? BMC Immunol. 21 (1), 38. doi:10.1186/s12865-020-00367-8
Cui, Y., Ji, W., Gao, Y., Xiao, Y., Liu, H., and Chen, Z. (2021). Single-cell characterization of monolayer cultured human dental pulp stem cells with enhanced differentiation capacity. Int. J. Oral Sci. 13 (1), 44. doi:10.1038/s41368-021-00140-6
de Brito, L. C. N., Doolittle-Hall, J., Lee, C. T., Moss, K., Bambirra Júnior, W., Tavares, W. L. F., et al. (2020). The apical root canal system microbial communities determined by next-generation sequencing. Sci. Rep. 10 (1), 10932. doi:10.1038/s41598-020-67828-3
Diogenes, A., and Hargreaves, K. M. (2017). Microbial modulation of stem cells and future directions in regenerative endodontics. J. Endod. 43 (9), S95–S101. doi:10.1016/j.joen.2017.07.012
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). Star: Ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 29 (1), 15–21. doi:10.1093/bioinformatics/bts635
Edanami, N., Yoshiba, N., Ohkura, N., Takeuchi, R., Tohma, A., Noiri, Y., et al. (2017). Characterization of dental pulp myofibroblasts in rat molars after pulpotomy. J. Endod. 43 (7), 1116–1121. doi:10.1016/j.joen.2017.02.018
El Karim, I. A., Linden, G. J., Irwin, C. R., and Lundy, F. T. (2009). Neuropeptides regulate expression of angiogenic growth factors in human dental pulp fibroblasts. J. Endod. 35 (6), 829–833. doi:10.1016/j.joen.2009.03.005
Eramo, S., Natali, A., Pinna, R., and Milia, E. (2018). Dental pulp regeneration via cell homing. Int. Endod. J. 51 (4), 405–419. doi:10.1111/iej.12868
Fouad, A. F. (2017). Microbial factors and antimicrobial strategies in dental pulp regeneration. J. Endod. 43 (9S), S46–S50. doi:10.1016/j.joen.2017.06.010
Fouad, A. F. (2011). The microbial challenge to pulp regeneration. Adv. Dent. Res. 23 (3), 285–289. doi:10.1177/0022034511405388
Fransson, H., Petersson, K., and Davies, J. R. (2014). Effects of bacterial products on the activity of odontoblast-like cells and their formation of type 1 collagen. Int. Endod. J. 47 (4), 397–404. doi:10.1111/iej.12160
Gokyu, M., Kobayashi, H., Nanbara, H., Sudo, T., Ikeda, Y., Suda, T., et al. (2014). Thrombospondin-1 production is enhanced by Porphyromonas gingivalis lipopolysaccharide in THP-1 cells. PloS One 9 (12), e115107. doi:10.1371/journal.pone.0115107
Gölz, L., Memmert, S., Rath-Deschner, B., Jäger, A., Appel, T., Baumgarten, G., et al. (2015). Hypoxia and P. gingivalis synergistically induce HIF-1 and NF-κB activation in PDL cells and periodontal diseases. Mediat. Inflamm., 2015, 438085. doi:10.1155/2015/438085
Hancock, H. .H., Sigurdsson, A., Trope, M., and Moiseiwitsch, J. (2001). Bacteria isolated after unsuccessful endodontic treatment in a North American population. Oral Surg. Oral Med. Oral Pathol. 91, 579–586. doi:10.1067/moe.2001.113587
Hargreaves, K. .M., Diogenes, A., and Teixeira, F. .B. (2013). Treatment options: Biological basis of regenerative endodontic procedures. J. Endod. 39, S30–S43. doi:10.1016/j.joen.2012.11.025
Huang, G. T. (2009). Apexification: The beginning of its end. Int. Endod. J. 42 (10), 855–866. doi:10.1111/j.1365-2591.2009.01577.x
Huang, G. T., Gronthos, S., and Shi, S. (2009). Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 88 (9), 792–806. doi:10.1177/0022034509340867
Huang, G. T., Yamaza, T., Shea, L. D., Djouad, F., Kuhn, N. Z., Tuan, R. S., et al. (2010). Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng. Part A 16 (2), 605–615. doi:10.1089/ten.TEA.2009.0518
Hwang, Y. C., Hwang, I. N., Oh, W. M., Park, J. C., Lee, D. S., and Son, H. H. (2008). Influence of TGF-beta1 on the expression of BSP, DSP, TGF-beta1 receptor I and Smad proteins during reparative dentinogenesis. J. Mol. Histology 39 (2), 153–160. doi:10.1007/s10735-007-9148-8
Iwaya, S. I., Ikawa, M., and Kubota, M. (2001). Revascularization of an immature permanent tooth with apical periodontitis and sinus tract. Dent. Traumatology Official Publ. Int. Assoc. Dent. Traumatology 17 (4), 185–187. doi:10.1034/j.1600-9657.2001.017004185.x
Karygianni, L., Wiedmann-Al-Ahmad, M., Finkenzeller, G., Sauerbier, S., Wolkewitz, M., Hellwig, E., et al. (2012). Enterococcus faecalis affects the proliferation and differentiation of ovine osteoblast-like cells. Clin. Oral Investig. 16 (3), 879–887. doi:10.1007/s00784-011-0563-6
Kayaoglu, G., and Ørstavik, D. (2004). Virulence factors of Enterococcus faecalis: Relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 15 (5), 308–320. doi:10.1177/154411130401500506
Kim, J. .C., Lee, Y. .H., Yu, M. .K., Lee, N. H., Park, J. D., Bhattarai, G., et al. (2012). Anti-inflammatory mechanism of PPARγ on LPS-induced pulp cells: Role of the ROS removal activity. Arch. Oral Biol. 57, 392–400. doi:10.1016/j.archoralbio.2011.09.009
Kim, S. G., Malek, M., Sigurdsson, A., Lin, L. M., and Kahler, B. (2018). Regenerative endodontics: A comprehensive review. Int. Endod. J. 51 (12), 1367–1388. doi:10.1111/iej.12954
Kubiczkova, L., Sedlarikova, L., Hajek, R., and Sevcikova, S. (2012). TGF-β: An excellent servant but a bad master. J. Transl. Med. 10, 183. doi:10.1186/1479-5876-10-183
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. doi:10.1093/nar/gkw377
Lavoie, H., Gagnon, J., and Therrien, M. (2020). ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632. doi:10.1038/s41580-020-0255-7
Lee, S., Chen, D., Park, M., Kim, S., Choi, Y. J., Moon, S. J., et al. (2022). Single-cell RNA sequencing analysis of human dental pulp stem cell and human periodontal ligament stem cell. J. Endod. 48 (2), 240–248. doi:10.1016/j.joen.2021.11.005
León-López, M., Cabanillas-Balsera, D., Martín-González, J., Montero-Miralles, P., Saúco-Márquez, J. J., and Segura-Egea, J. J. (2022). Prevalence of root canal treatment worldwide: A systematic review and meta-analysis. Int. Endod. J. 55 (11), 1105–1127. doi:10.1111/iej.13822
Li, H., Guan, R., Sun, J., and Hou, B. (2014). Bacteria community study of combined periodontal-endodontic lesions using denaturing gradient gel electrophoresis and sequencing analysis. J. Periodontology 85 (10), 1442–1449. doi:10.1902/jop.2014.130572
Liao, J., Al Shahrani, M., Al-Habib, M., Tanaka, T., and Huang, G. T. (2011). Cells isolated from inflamed periapical tissue express mesenchymal stem cell markers and are highly osteogenic. J. Endod. 37 (9), 1217–1224. doi:10.1016/j.joen.2011.05.022
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinforma. Oxf. Engl. 30 (7), 923–930. doi:10.1093/bioinformatics/btt656
Lin, X., Chi, D., Meng, Q., Gong, Q., and Tong, Z. (2022). Single-cell sequencing unveils the heterogeneity of nonimmune cells in chronic apical periodontitis. Front. Cell Dev. Biol. 9, 820274. doi:10.3389/fcell.2021.820274
Liu, H., Han, X., Yang, H., Cao, Y., Zhang, C., Du, J., et al. (2021). GREM1 inhibits osteogenic differentiation, senescence and BMP transcription of adipose-derived stem cells. Connect. Tissue Res. 62 (3), 325–336. doi:10.1080/03008207.2020.1736054
Liu, J. Y., Chen, X., Yue, L., Huang, G. T., and Zou, X. Y. (2015). CXC chemokine receptor 4 is expressed paravascularly in apical papilla and coordinates with stromal cell-derived factor-1α during transmigration of stem cells from Apical Papilla. J. Endod. 41 (9), 1430–1436. doi:10.1016/j.joen.2015.04.006
Love, R. M. (2001). Enterococcus faecalis: A mechanism for its role in endodontic failure. Int. Endod. J. 34 (5), 399–405. doi:10.1046/j.1365-2591.2001.00437.x
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi:10.14806/ej.17.1.200
Murray, P. E., Garcia-Godoy, F., and Hargreaves, K. M. (2007). Regenerative endodontics: A review of current status and a call for action. J. Endod. 33 (4), 377–390. doi:10.1016/j.joen.2006.09.013
Niwa, T., Yamakoshi, Y., Yamazaki, H., Karakida, T., Chiba, R., Hu, J. C., et al. (2018). The dynamics of TGF-β in dental pulp, odontoblasts and dentin. Sci. Rep. 8 (1), 4450. doi:10.1038/s41598-018-22823-7
Nosrat, A., Kolahdouzan, A., Hosseini, F., Mehrizi, E. A., Verma, P., and Torabinejad, M. (2015). Histologic outcomes of uninfected human immature teeth treated with regenerative endodontics: 2 case reports. J. Endod. 41 (10), 1725–1729. doi:10.1016/j.joen.2015.05.004
Orikasa, S., Kawashima, N., Tazawa, K., Hashimoto, K., Sunada-Nara, K., Noda, S., et al. (2022). Hypoxia-inducible factor 1α induces osteo/odontoblast differentiation of human dental pulp stem cells via Wnt/β-catenin transcriptional cofactor BCL9. Sci. Rep. 12 (1), 682. doi:10.1038/s41598-021-04453-8
Park, O. J., Kim, J., Yang, J., Yun, C. H., and Han, S. H. (2015). Enterococcus faecalis inhibits osteoblast differentiation and induces chemokine expression. J. Endod. 41 (9), 1480–1485. doi:10.1016/j.joen.2015.04.025
Paudel, U., Lee, Y. H., Kwon, T. H., Park, N. H., Yun, B. S., Hwang, P. H., et al. (2014). Eckols reduce dental pulp inflammation through the ERK1/2 pathway independent of COX-2 inhibition. Oral Dis. 20 (8), 827–832. doi:10.1111/odi.12266
Pinheiro, E. T., Candeiro, G. T., Teixeira, S. R., Shin, R. C., Prado, L. C., Gavini, G., et al. (2015). RNA-based assay demonstrated Enterococcus faecalis metabolic activity after chemomechanical procedures. J. Endod. 41 (9), 1441–1444. doi:10.1016/j.joen.2015.04.020
Puschhof, J., Pleguezuelos-Manzano, C., and Clevers, H. (2021). Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 29 (6), 867–878. doi:10.1016/j.chom.2021.04.002
Qian, S. J., Huang, Q. R., Chen, R. Y., Mo, J. J., Zhou, L. Y., Zhao, Y., et al. (2021). Single-cell RNA sequencing identifies new inflammation-promoting cell subsets in Asian patients with chronic periodontitis. Front. Immunol. 12, 711337. doi:10.3389/fimmu.2021.711337
Rôças, I. N., Alves, F. R., Rachid, C. T., Lima, K. C., Assunção, I. V., Gomes, P. N., et al. (2016). Microbiome of deep dentinal caries lesions in teeth with symptomatic irreversible pulpitis. PloS One 11 (5), e0154653. doi:10.1371/journal.pone.0154653
Sánchez-Sanhueza, G., Bello-Toledo, H., González-Rocha, G., Gonçalves, A. T., Valenzuela, V., and Gallardo-Escárate, C. (2018). Metagenomic study of bacterial microbiota in persistent endodontic infections using next-generation sequencing. Int. Endod. J. 51 (12), 1336–1348. doi:10.1111/iej.12953
Shayegan, A., Zucchi, A., De Swert, K., Balau, B., Truyens, C., and Nicaise, C. (2021). Lipoteichoic acid stimulates the proliferation, migration and cytokine production of adult dental pulp stem cells without affecting osteogenic differentiation. Int. Endod. J. 54 (4), 585–600. doi:10.1111/iej.13448
Sipert, C. R., Moraes, I. G., Bernardinelli, N., Garcia, R. B., Bramante, C. M., Gasparoto, T. H., et al. (2010). Heat-killed Enterococcus faecalis alters nitric oxide and CXCL12 production but not CXCL8 and CCL3 production by cultured human dental pulp fibroblasts. J. Endod. 36 (1), 91–94. doi:10.1016/j.joen.2009.10.014
Sipert, C. R., Morandini, A. C., Modena, K. C., Dionísio, T. J., Machado, M. A., Oliveira, S. H., et al. (2013). CCL3 and CXCL12 production in vitro by dental pulp fibroblasts from permanent and deciduous teeth stimulated by Porphyromonas gingivalis LPS. J. Appl. Oral Sci. Revista FOB 21 (2), 99–105. doi:10.1590/1678-7757201300004
Siqueira, J. F., and Rôças, I. N. (2008). Clinical implications and microbiology of bacterial persistence after treatment procedures. J. Endod. 34 (11), 1291–1301. doi:10.1016/j.joen.2008.07.028
Siqueira, J. F., and Rôças, I. N. (2022). Present status and future directions: Microbiology of endodontic infections. Int. Endod. J. 55 (3), 512–530. doi:10.1111/iej.13677
Stuart, C. H., Schwartz, S. A., Beeson, T. J., and Owatz, C. B. (2006). Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J. Endod. 32 (2), 93–98. doi:10.1016/j.joen.2005.10.049
Symmank, J., Appel, S., Bastian, J. A., Knaup, I., Marciniak, J., Hennig, C. L., et al. (2021). Hyperlipidemic conditions impact force-induced inflammatory response of human periodontal ligament fibroblasts concomitantly challenged with P. gingivalis-LPS. Int. J. Mol. Sci. 22 (11), 6069. doi:10.3390/ijms22116069
Tibúrcio-Machado, C. S., Michelon, C., Zanatta, F. B., Gomes, M. S., Marin, J. A., and Bier, C. A. (2021). The global prevalence of apical periodontitis: A systematic review and meta-analysis. Int. Endod. J. 54 (5), 712–735. doi:10.1111/iej.13467
Tsai, C. L., Hung, S. L., Lee, Y. Y., Ho, Y. C., and Yang, S. F. (2022). The role of fibroblasts in the modulation of dental pulp inflammation. J. Formos. Med. Assoc. = Taiwan yi zhi 121 (2), 342–349. doi:10.1016/j.jfma.2021.05.007
Ueno, A., Yamashita, K., Nagata, T., Tsurumi, C., Miwa, Y., Kitamura, S., et al. (1998). cDNA cloning of bovine thrombospondin 1 and its expression in odontoblasts and predentin. Biochimica Biophysica Acta 1382 (1), 17–22. doi:10.1016/s0167-4838(97)00188-x
Verma, P., Nosrat, A., Kim, J. R., Price, J. B., Wang, P., Bair, E., et al. (2017). Effect of residual bacteria on the outcome of pulp regeneration in vivo. J. Dent. Res. 96 (1), 100–106. doi:10.1177/0022034516671499
Vishwanat, L., Duong, R., Takimoto, K., Phillips, L., Espitia, C. O., Diogenes, A., et al. (2017). Effect of bacterial biofilm on the osteogenic differentiation of stem cells of apical papilla. J. Endod. 43 (6), 916–922. doi:10.1016/j.joen.2017.01.023
Yamagishi, V. T., Torneck, C. D., Friedman, S., Huang, G. T., and Glogauer, M. (2011). Blockade of TLR2 inhibits Porphyromonas gingivalis suppression of mineralized matrix formation by human dental pulp stem cells. J. Endod. 37 (6), 812–818. doi:10.1016/j.joen.2011.03.013
Yang, L. C., Huang, F. M., Lin, C. S., Liu, C. M., Lai, C. C., and Chang, Y. C. (2003). Induction of interleukin-8 gene expression by black-pigmented Bacteroides in human pulp fibroblasts and osteoblasts. Int. Endod. J. 36 (11), 774–779. doi:10.1046/j.1365-2591.2003.00740.x
Yoshiko, Y., Aubin, J. E., and Maeda, N. (2002). Stanniocalcin 1 (STC1) protein and mRNA are developmentally regulated during embryonic mouse osteogenesis: The potential of stc1 as an autocrine/paracrine factor for osteoblast development and bone formation. J. Histochem. Cytochem. Official J. Histochem. Soc. 50 (4), 483–492. doi:10.1177/002215540205000405
Zahran, S., Witherden, E., Mannocci, F., and Koller, G. (2021). Characterization of root canal microbiota in teeth diagnosed with irreversible pulpitis. J. Endod. 47 (3), 415–423. doi:10.1016/j.joen.2020.12.009
Zakaria, M. N., Takeshita, T., Shibata, Y., Maeda, H., Wada, N., Akamine, A., et al. (2015). Microbial community in persistent apical periodontitis: A 16S rRNA gene clone library analysis. Int. Endod. J. 48 (8), 717–728. doi:10.1111/iej.12361
Zaky, S. H., AlQahtani, Q., Chen, J., Patil, A., Taboas, J., Beniash, E., et al. (2020). Effect of the periapical “inflammatory plug” on dental pulp regeneration: A histologic in vivo study. J. Endod. 46 (1), 51–56. doi:10.1016/j.joen.2019.10.006
Zhang, C., Du, J., and Peng, Z. (2015). Correlation between Enterococcus faecalis and persistent intraradicular infection compared with primary intraradicular infection: A systematic review. J. Endod. 41 (8), 1207–1213. doi:10.1016/j.joen.2015.04.008
Zheng, S., Yu, S., Fan, X., Zhang, Y., Sun, Y., Lin, L., et al. (2021). Porphyromonas gingivalis survival skills: Immune evasion. J. Periodontal Res. 56 (6), 1007–1018. doi:10.1111/jre.12915
Keywords: regenerative endodontic treatment, human dental pulp stem cell, single-cell RNA sequencing, Porphyromonas gingivalis, Enterococcus faecalis, cell-cell interactions and development
Citation: Zhang W, Xu T, Li X, Zhang Y, Zou X, Chen F and Yue L (2023) Single-cell atlas of dental pulp stem cells exposed to the oral bacteria Porphyromonas gingivalis and Enterococcus faecalis. Front. Cell Dev. Biol. 11:1166934. doi: 10.3389/fcell.2023.1166934
Received: 15 February 2023; Accepted: 09 May 2023;
Published: 23 May 2023.
Edited by:
Jue Fan, Singleron Biotechnologies, ChinaReviewed by:
W. Scott Goebel, Indiana University School of Medicine, United StatesCopyright © 2023 Zhang, Xu, Li, Zhang, Zou, Chen and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoying Zou, em91eGlhb3lpbmcxMTI1QDE2My5jb20=; Feng Chen, Y2hlbmZlbmcyMDExQGhzYy5wa3UuZWR1LmNu; Lin Yue, a3FsaW55dWVAYmptdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.