
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 31 May 2023
Sec. Molecular and Cellular Reproduction
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1166517
 Bo Yuan1
Bo Yuan1 Yi Yang1
Yi Yang1 Zhichao Yan1
Zhichao Yan1 Chun He1
Chun He1 Yu H. Sun2
Yu H. Sun2 Fei Wang1
Fei Wang1 Beibei Wang1
Beibei Wang1 Jiamin Shi1
Jiamin Shi1 Shan Xiao1
Shan Xiao1 Fang Wang1
Fang Wang1 Qi Fang1
Qi Fang1 Fei Li1
Fei Li1 Xinhai Ye1,3,4*
Xinhai Ye1,3,4* Gongyin Ye1
Gongyin Ye1The linker histone H1 binds to the nucleosome core particle at the site where DNA enters and exits, and facilitates folding of the nucleosomes into a higher-order chromatin structure in eukaryotes. Additionally, some variant H1s promote specialized chromatin functions in cellular processes. Germline-specific H1 variants have been reported in some model species with diverse roles in chromatin structure changes during gametogenesis. In insects, the current understanding of germline-specific H1 variants comes mainly from the studies in Drosophila melanogaster, and the information on this set of genes in other non-model insects remains largely unknown. Here, we identify two H1 variants (PpH1V1 and PpH1V2) that are specifically predominantly expressed in the testis of the parasitoid wasp Pteromalus puparum. Evolutionary analyses suggest that these H1 variant genes evolve rapidly, and are generally maintained as a single copy in Hymenoptera. Disruption of PpH1V1 function in the late larval stage male by RNA interference experiments has no phenotype on spermatogenesis in the pupal testis, but results in abnormal chromatin structure and low sperm fertility in the adult seminal vesicle. In addition, PpH1V2 knockdown has no detectable effect on spermatogenesis or male fertility. Collectively, our discovery indicates distinct functions of male germline-enriched H1 variants between parasitoid wasp Pteromalus and Drosophila, providing new insights into the role of insect H1 variants in gametogenesis. This study also highlights the functional complexity of germline-specific H1s in animals.
Eukaryotic genomes are packed in the form of chromatin within the cell nucleus, and the linker histone H1 protein is one of the main components of eukaryotic chromatin, which binds to adjacent nucleosomes and linker-DNA, facilitating the forming of the higher-order chromatin structure known as the chromatin fiber (Fan et al., 2005; Harshman et al., 2013; Fyodorov et al., 2018). The H1 gene family has evolved faster than other core histone families (H2A, H2B, H3, and H4), which has contributed to the emergence of many H1 variants with redundant or specific functions (Izzo et al., 2008; Ponte et al., 2017). Among the diverse H1 variants, a group exhibits a highly-germline-enriched expression pattern, and plays key roles in changing the chromatin structure during gametogenesis and early embryogenesis (Pérez-Montero et al., 2016). In mammalians, a total of eleven H1 variants are identified, seven of which are somatic subtypes (H1.0, H1.1 to H1.5, and H1x), and the additional four are germline-specific subtypes: H1t, H1T2, HILS1 in the testis (Kumaroo and Irvin, 1980; Seyedin and Kistler, 1980; Kremer and Kistler, 1991; Drabent et al., 1993; Yan et al., 2003; Martianov et al., 2005; Tanaka et al., 2005), and H1oo in the oocyte (Tanaka et al., 2001). In general, germline-specific H1s are strictly regulated during gametogenesis and they may function in different stages. For example, H1t is the first variant to be expressed in meiotic spermatocytes and compacts chromatin to a lesser extent and binds DNA with lower affinity, which may facilitate meiotic recombination (De Lucia et al., 1994; Khadake and Rao, 1995; Talaszt et al., 1999). Later, H1T2 and HILS1 express and gradually replace H1t, which contributes to the compaction of sperm chromatin during spermatid stages (Iguchi et al., 2003; Yan et al., 2003; Martianov et al., 2005; Tanaka et al., 2005). Other germline-specific H1s were also identified in some model species in other taxa, such as zebrafish (H1M) (Müller et al., 2002), clawed frog (H1fx and B4/H1M) (Smith et al., 1988; Shechter et al., 2009), sea urchin (SpH1 and Cs-H1) (Strickland et al., 1980; Brandt et al., 1997; Mandl et al., 1997), and nematode (H1.1/HIS-24) (Vanfleteren et al., 1988). These studies showed that, although these variants are largely restricted to the germline, they may have distinct expression pattern and different functions (Pérez-Montero et al., 2016), suggesting the high complexity of germline-specific H1s in animals.
Insects comprise extremely diverse species and are important models for understanding the functional diversification of genes, such as chemosensory-related gene families (Eyun et al., 2017; Robertson, 2019). To date, our ideas of insect germline-specific H1s are solely from studying the fruit fly Drosophila melanogaster. A germline-specific H1 variant dBigH1 has been reported (Pérez-Montero et al., 2013), which is expressed in both the female and male germlines, and early embryos. dBigH1 is abundant during early embryogenesis and regulates zygotic genome activation (Pérez-Montero et al., 2013). In male, dBigH1 is essential for germ stem cell (GSC) maintenance and spermatocyte differentiation (Carbonell et al., 2017). Another H1-like linker protein Mst77F is detected from late canoe stage spermatids to mature sperm overlapping with protamines. And Mst77F mutants show low fertility and morphologically abnormal spermatids (Kimura and Loppin, 2016). Although the studies on model organism Drosophila greatly advance our knowledge of germline-specific H1 variants in insects, the relevant information of this group of genes in other non-model insects, including their sequences, origins, evolutionary trajectories and their biological functions, is poorly understood.
In this study, we identify two male germline-enriched histone H1 variants (PpH1V1 and PpH1V2) in the model parasitoid wasp, Pteromalus puparum (from Hymenoptera). Our evolutionary analyses show that these two H1 variant genes generally keep as a single copy in all hymenopterans we examined, although they evolve rapidly. We knockdown PpH1V1 and PpH1V2 using RNA interference (RNAi), and demonstrate that PpH1V1 may play a critical role in the chromatin organization of sperm in the seminal vesicle, and is associated with male fertility. However, PpH1V2 knockdown has no detectable effect on spermatogenesis or male fertility. This study provides new insights into the role of H1 variants in gametogenesis, and contributes to expanding our understanding of insect H1 variants in non-model species.
The P. puparum genome sequencing project identified 18 histone H1 genes in the official gene set (OGS) by an automatic genome annotation pipeline which integrated the evidences from homology mapping, de novo prediction and gene expression (Ye et al., 2020). In this study, to obtain a full list of H1 genes in P. puparum, we performed a comprehensive homology-based search using the previously identified H1 sequences. We further determined the presence of the functional domain (Pfam id: PF00538, Linker_histone) and checked the complete open reading frame for each candidate. In total, we confirmed the accurate gene annotation of H1 genes in OGS, and did not find additional genes which were miss-annotated in our previous genome annotation. Among these 18 genes, two were further assigned as putative H1 variants, according to the HistoneDB 2.0 online service (Draizen et al., 2016). We thus named them as P. puparum H1 variant 1 (PpH1V1, OGS id: PPU09681) and P. puparum H1 variant 2 (PpH1V2, OGS id: PPU12916). Gene structure analysis showed that both putative H1 variants contain introns (Figure 1A), which is distinctly different from generic histone H1 (such as PpH1, OGS id: PPU08399) that lack introns (Hentschel and Birnstiel, 1981). In addition, previous studies have shown that the insertion of an intron into a histone gene could alter the histone 3′end formation and thus result in the formation of polyadenylated mRNA (Pandey et al., 1990). As expected, we used rapid amplification of cDNA ends (RACE) to amplify the full-length mRNAs and confirmed the existence of poly(A) tails in the mRNAs of these H1 variants (Supplementary Table S1). Sequence analysis of the PpH1V1 full length mRNA (1,093 base pair, bp) revealed a 651 bp open reading frame encoding a 216 amino acid protein of predicted molecular mass 24.24 kDa and the PpH1V2 full length mRNA (698 bp) revealed a 444 bp open reading frame encoding a 147 amino acid protein of predicted molecular mass 16.04 kDa (Supplementary Figure S1).
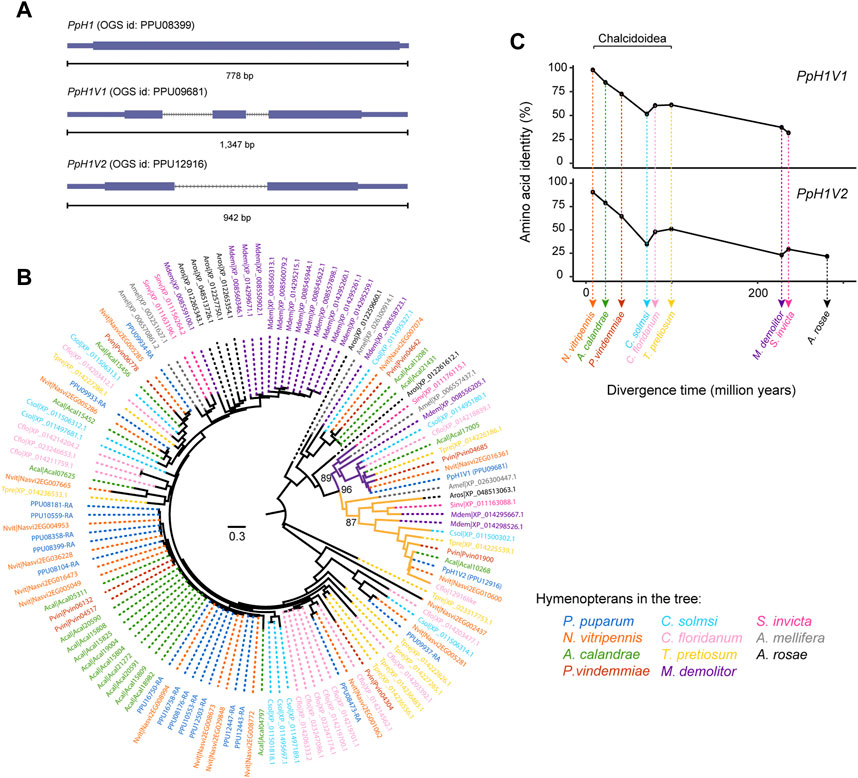
FIGURE 1. Two rapidly evolving histone H1 variants in P. pupraum genome. (A) Comparison of the gene structures of a generic histone H1 gene and two histone H1 variant genes (PpH1V1 and PpH1V2). The coding exons are shown as blocks connected by horizontal lines representing introns, and the 5′and 3′untranslated regions (UTRs) are shown as thinner blocks. (B) A maximum-likelihood protein phylogeny of the histone H1 sequences from 11 representative hymenopteran species. The clade containing PpH1V1 was highlighted in purple, and the clade with PpH1V2 was in yellow. Statistical support for the phylogeny was assessed by ultrafast bootstrap analysis using 1,000 replicates, and bootstrap values at selected nodes were indicated. (C) Pairwise amino acid identity analysis showing comparisons of PpH1V1 or PpH1V2 versus the hymenopteran ortholog of each variant. Percent identities were plotted against species divergence time. The species divergence times were obtained from previous studies (Peters et al., 2017; Ye et al., 2022).
To understand the evolutionary history of H1 genes in Hymenoptera, we identified the H1 genes in other ten hymenopteran species with high-quality genomes, representing parasitoid wasp, ant, bee, and sawfly lineages. Then, we performed maximum likelihood phylogenetic analysis with protein sequences. Our analysis revealed that PpH1V1 and PpH1V2 belonged to two distinct clades, and both clades were overall broadly distributed among hymenopterans (Figure 1B). The genes in these clades generally kept as a single copy in all hymenopterans we examined, except Microplitis demolitor. In addition, the two H1 variant clades were further clustered together with high bootstrap support (96%), suggestive of the evolutionary origin of one of the H1 variants by an ancient gene duplication event. Ortholog inference based on reciprocal mapping, phylogenetic tree and synteny could find an unambiguous ortholog of PpH1V2 in all hymenopterans we examined; however, we failed to infer the unambiguous ortholog of PpH1V1 in Athalia rosae (early branch of Hymenoptera), because the candidate gene (XP_012261612.1) was not clustered with other orthologs of PpH1V1, and there was no synteny signal. We next investigated the protein divergence rates of these H1 variants in Hymenoptera spanning 281 million years of evolution. To this end, we measured the pairwise identity of each H1 variant protein to its ortholog of P. puparum and plotted it with the species divergence time. We showed that the pairwise amino acid identity decreased rapidly over the divergence time (from 97% to 32% for PpH1V1, and from 92% to 22% for PpH1V2), reflecting the rapid evolution of these H1 variant proteins (Figure 1C).
We next sought to investigate the expression of the two H1 variants in P. puparum. First, we performed quantitative real-time PCR (qRT-PCR) for each variant on different developmental stages (totally seven time points from early larva to adult) of both sexes. The results showed that these two H1 variants were lowly expressed (34–161 and 38–110 copies per ng total RNA, respectively) in females, and there were no significant expression differences during the female development. By contrast, the expressions of these H1 variants were much higher (38–29,198 and 26–7,450 copies per ng total RNA, respectively) in males (Figures 2A, B). Importantly, we found that, from the late larval stage to the late yellow pupal stage, these H1 variants steadily increased their expressions with statistically significant differences (Figures 2A, B, p < 0.0001, Tukey’s multiple comparisons test), resulting in the highest expression level at the late yellow pupal stage. Then, the mRNA levels of these H1 variants decreased gradually from the late yellow pupal stage to the adult stage. These male-biased, and developmental-stage-biased gene expression patterns prompted us to test whether these H1 variants are highly specific to the germline and may be related to spermatogenesis, because the expression patterns are highly consistent with the spermatogenesis process in parasitoid wasps (Ferree et al., 2019). Next, we measured the expressions of these H1 variants on a representative set of tissues (head, thorax, testis, male accessory gland and body remnant) from the late-stage yellow pupal males. As expected, although the expressions were detectable in other tissues, we observed the significantly higher expressions of these H1 variants in the testis (Figures 2C, D, p < 0.0001, Tukey’s multiple comparisons test). To investigate the immunolocalization of PpH1V1 and PpH1V2 in the germ cells, we raised polyclonal antibodies against them, separately. We observed that spermatids with elongating nuclei were specifically stained by both antibodies (Supplementary Figures S2A, B). Taken together, our results indicate that both H1 variants showed the male-biased, and testis-specific high expression patterns, implying their potential roles in male fertility.
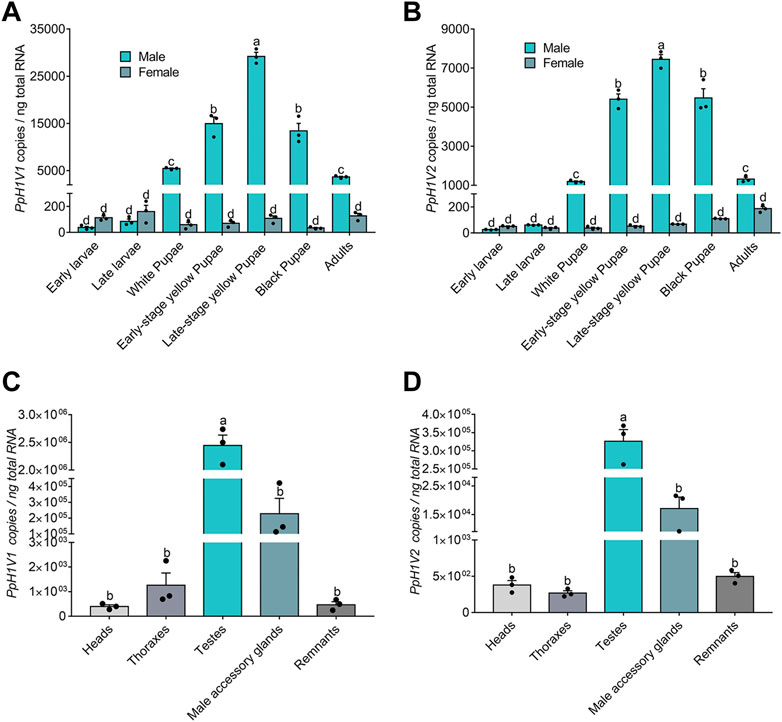
FIGURE 2. Expression patterns of PpH1V1 and PpH1V2 mRNA in different developmental stages and different tissues. (A–B) Expression levels of PpH1V1 gene (A) and PpH1V2 gene (B) in different developmental stages (from the larval stage to the adult stage, in both sexes). (C–D) Expression levels of PpH1V1 gene (C) and PpH1V2 gene (D) in different tissues of the late-stage yellow pupal males. For each measurement, three biological replicates were done. The qRT-PCR data were presented as mean ± standard error. Differences between groups were analyzed by one-way ANOVA with Tukey’s multiple comparisons test, and bars annotated with the same letters are not significantly different (p > 0.05).
Next, we knocked down the two H1 variants by injecting dsRNA into the male larvae. To reduce the likelihood of false positives due to sequence-specific off-target effects, we used two independent, non-overlapping dsRNAs against PpH1V1 and PpH1V2. At the late yellow pupal stage (5 days post injection), the transcript abundance of dsPpH1V1 and dsPpH1V2 treated wasps were significantly decreased compared to the negative controls (over 95% reductions), which were treated with ddH2O and dsEGFP (EGFP: enhanced green fluorescent protein gene), respectively (Figures 3A, B, p < 0.0001 and p < 0.001, Tukey’s multiple comparisons test). RNAi effects were also evaluated at the protein levels, and the immunofluorescence staining analysis revealed strong reductions of PpH1V1 (Figures 4A–C) and PpH1V2 (Figures 4D–F) protein contents in the dsRNA treated wasps, respectively. Then, the dsRNA and negative control treated wasps were kept for phenotype observation until emergence, and we did not observe significant differences in either pupation proportion or emergence proportion between the dsRNA treated groups and the negative controls (Figures 3C, D, p = 0.8094 and p = 0.7464, Tukey’s multiple comparisons test). To test whether these two H1 variants play important roles in male fertility, we paired the dsRNA treated or negative control male adults with virgin female adults, and allowed them to mate for 48 h, then put females individually in glass tubes with one newly-pupated host. During the following 5 days, each host encountered by the female wasp was individually transferred into a new glass tube every day, and the fresh pupa and food were provided and replaced. According to the special haplodiploidy sex-determination system of parasitoid wasps, males develop from unfertilized eggs and are haploid, and females develop from fertilized eggs and are diploid (Cockburn, 1993; Heimpel and De Boer, 2008). Therefore, theoretically, mating with the male wasps that are infertile would result in fertilization failure, and thus induce more male offspring and lower offspring sex ratio (number of female offspring/total number of offspring). Indeed, our results revealed that the dsRNA treatments had no influence on the offspring number (Figure 3E), but knockdown of PpH1V1 in males induced a significantly lower offspring sex ratio (27%) than those in the control groups (87% for ddH2O and 87% for dsEGFP) (Figure 3F, p < 0.0001, Tukey’s multiple comparisons test), suggesting the crucial role of PpH1V1 in male fertility. However, we did not find any significant effects on offspring sex ratio in dsPpH1V2 treatment compared to the controls (Figure 3F).
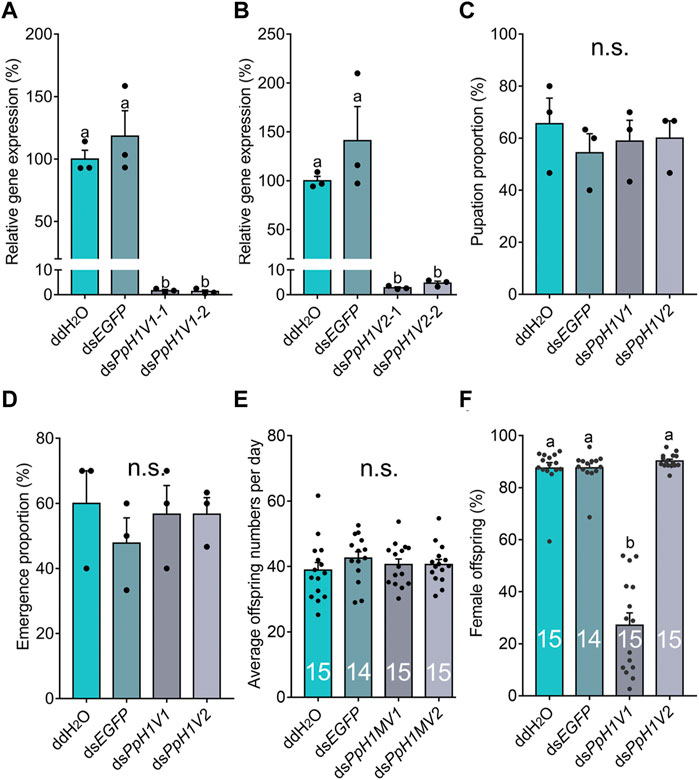
FIGURE 3. Knockdown of PpH1V1 in males results in significantly lower sex ratio in offspring. (A–B) Expressions of PpH1V1 (A) and PpH1V2 (B) were successfully knocked down in pupal males, three biological replicates. (C–D) Knockdown of PpH1V1 or PpH1V2 in male caused no effect on pupation proportion (C) or emergence proportion (D), sample size = 10, three biological replicates. (E) Knockdown of PpH1V1 or PpH1V2 in male resulted in no effect on average offspring number after mating with one female, sample size = 14 or 15. (F) Knockdown of PpH1V1 in male leaded to the significantly decrease of the sex ratio (female/male) of offspring, sample size = 14 or 15. All values were presented as mean ± standard error. Differences between groups were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. Bars annotated with the same letters are not significantly different (p > 0.05); n.s. not significantly different.
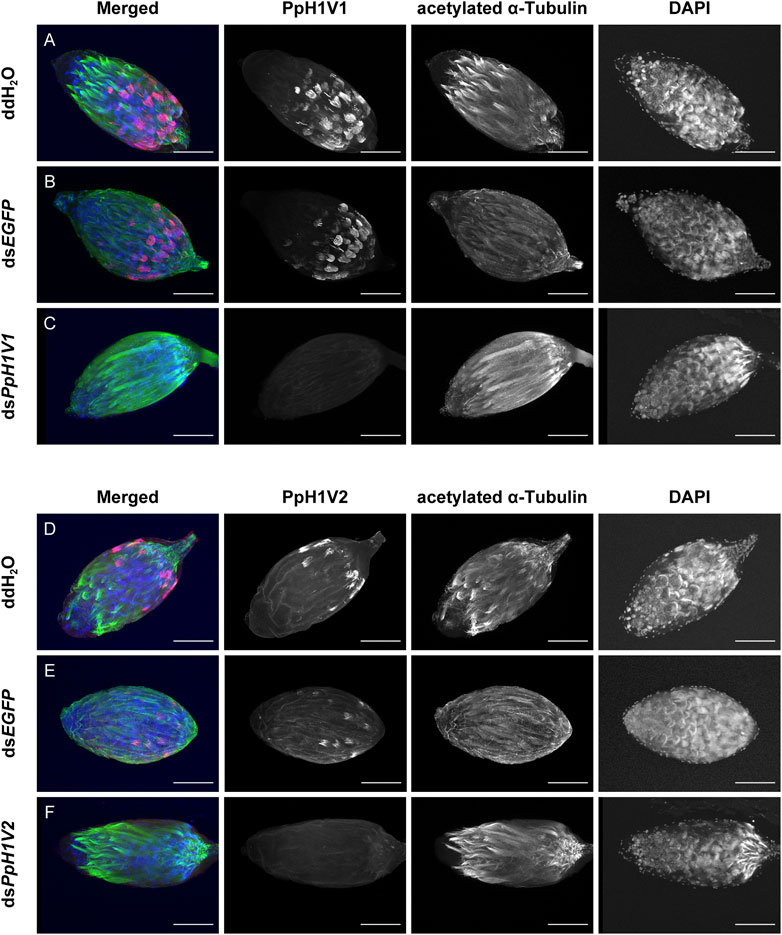
FIGURE 4. RNA interference (RNAi) silencing effects on PpH1V1 and PpH1V2 at the protein levels. Immunofluorescence staining showing the testes of control and knockdown RNAi yellow-black pupal males. PpH1V1 (A–C) or PpH1V2 (D–F) was detected using a rabbit anti-PpH1V1 or a rabbit anti-PpH1V2 polyclonal antibody and goat anti-rabbit IgG (H + L) with Alexa Fluor™ 488 secondary antibody (shown in red). Flagellum was stained with acetylated α Tubulin Alexa Fluor® 647 antibody (shown in green). Nuclei was stained with DAPI (shown in blue). Images are shown in greyscale for single channels. Three biological replicates were performed for each measurement. Scale bars correspond to 100 μm.
We next tested whether PpH1V1 was involved in the spermatogenesis of P. puparum males. For this purpose, we first examined the spermatogenesis process in the testes of RNAi-treated late-stage yellow pupal males. At this developmental stage, we observed several cells at distinct status, for example, germ cells at interphase (Figure 5A’), germ cells with condensed nuclei (Figure 5A’’) which entry into spermiogenesis (Supplementary Figure S3), and germ cells reorganized into a hemispherical shape (cup-like) (Figure 5A’’’). Compared to ddH2O and dsEGFP treatments (Figures 5A, B), we did not find obvious disorders of spermatogenesis in the dsPpH1V1-treated wasps, nor in the dsPpH1V2-treated groups (Figures 5C, D). The results from transmission electron microscopy (TEM) indicated that no phenotype during early spermatogenesis process in the testes of RNAi-treated wasps (Supplementary Figure S4A). Moreover, we did not find significant expression changes of a conserved spermatogenesis-related marker gene Vasa between RNAi-treated wasps and controls (Supplementary Figure S4B). In the older testes (black pupae), bundles of elongating spermatid nuclei or fully elongated sperm nuclei were detected in controls, dsPpH1V1- and dsPpH1V2-treated wasps (Figures 5E–H), suggesting that knockdown of these two H1 variants does not affect the spermiogenesis in testis.
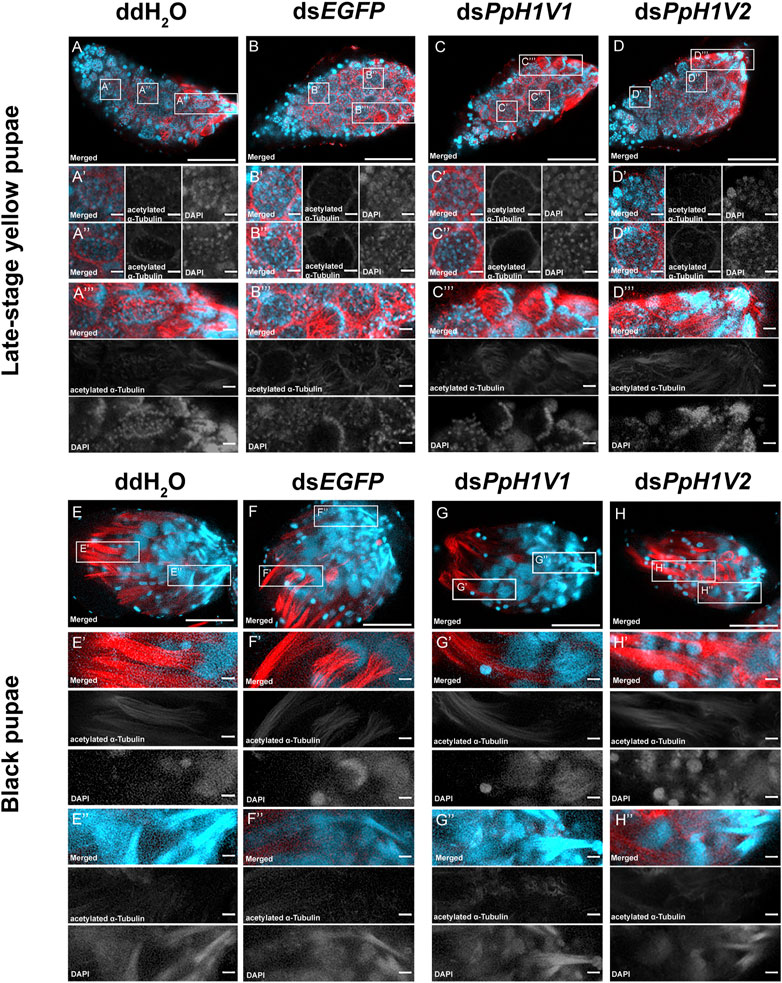
FIGURE 5. PpH1V1 knockdown does not affect the spermatogenesis in the testis. (A–D) Immunofluorescence staining showing the testes of late-stage yellow pupal males under four different treatments. (A′–D′) Small cyst of germ cells at the apical end of the testis. (A′′–D′′) Germ cells in a more posterior-located cyst have entered into spermiogenesis, as indicated by their condensed nuclei. (A‴–D‴) Bundles of spermatids with elongating tails. (E–H) Immunofluorescence staining showing the testes of black pupal males under four different treatments. (E′–H′) Bundles of spermatids with elongating nucleus. (E′′–H′′) Bundles of sperm with fully elongated sperm nucleus. Flagellum was stained with acetylated α Tubulin Alexa Fluor® 488 antibody (showed in red). Nucleus was stained with DAPI (shown in light blue). Images are shown in greyscale for single channels. Three biological replicates were performed for each measurement. Scale bars correspond to 100 μm in (A-H) and 10 μm in all others.
The above results can not explain why PpH1V1 knockdown could influence male fertility and result in the change of sex ratio of offspring. We then asked whether PpH1V1 was required for maintaining the chromatin organization of sperm in the seminal vesicle, a downstream organ of testis for sperm storage. In contrast to the well-organized needle-shaped nuclei of sperm in the seminal vesicles of adult wasps from the control treatments (Figures 6A, B, E, F), the nuclei of sperm in the seminal vesicles of dsPpH1V1-treated wasps were in disarray (Figures 6C, G), indicating potential disorder in the sperm chromatin. In addition, we investigated the nuclear morphology of sperm in seminal vesicles of RNAi-treated wasps and controls. We observed that the sperm nuclei of the control treatments were needle-like, while the sperm nuclei of the dsPpH1V1-treated wasps were clearly deformed and irregular in shape, with short but bent or crumpled nuclei (Supplementary Figure S5). In contrast, we did not find such abnormal chromatin status in the sperm from dsPpH1V2-treated wasps (Figures 6D, H; Supplementary Figure S5). Using transmission electron microscopy (TEM), we did not observe a significant reduction in sperm density in the seminal vesicles of dsPpH1V1-treated wasps compared to the control treatments (Supplementary Figure S6).
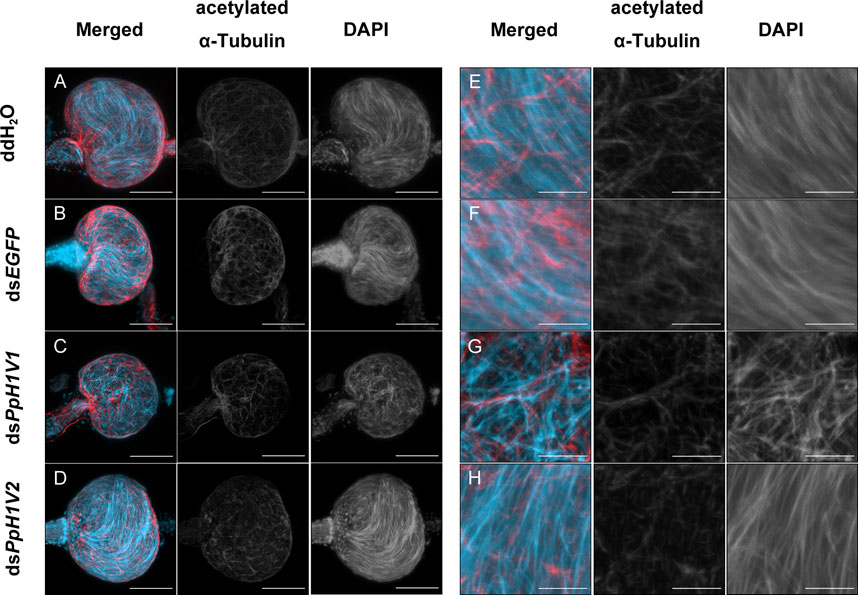
FIGURE 6. Knockdown of PpH1V1 resulted in obvious abnormal sperm nuclei in the seminal vesicles of adult males. (A–D) Immunofluorescence staining showing the seminal vesicles of adult males under four different treatments. (E–H) The local enlarged images of (A–D). Flagellum was stained with acetylated α Tubulin Alexa Fluor® 488 antibody (showed in red). Nucleus was stained with DAPI (shown in light blue). Images are shown in greyscale for single channels. Three biological replicates were performed for each measurement. Scale bars correspond to 50 μm in (A–D) and 10 μm in (E–H).
We also performed a sperm viability assay to test whether PpH1V1 knockdown induced more dead sperm than controls. Indeed, the results revealed that knockdown of PpH1V1 in males induced significantly lower sperm viability (54%) than those in control groups (82% for ddH2O and 80% for dsEGFP) (Figures 7A, B, p < 0.0001, Tukey’s multiple comparisons test). And PpH1V2 knockdown did not affect the sperm viability compared to controls. Altogether, our results suggest that PpH1V1 is likely to be involved in the chromatin organization of sperm, and thus plays a critical role in male fertility in P. puparum.
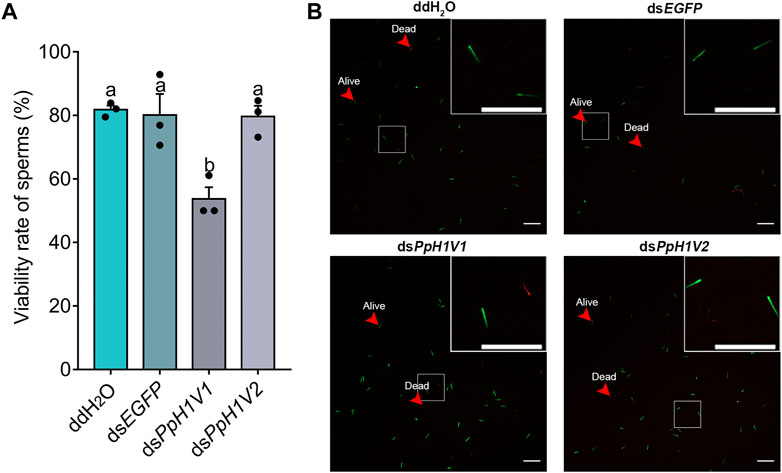
FIGURE 7. Sperm viability was significantly reduced in PpH1V1 knockdown males. (A) Viability rate of sperm in the seminal vesicles of adult males. Three biological replicates were performed for each measurement. All values were presented as mean ± standard error. Differences between groups were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. Bars annotated with the same letters are not significantly different (p > 0.05). (B) Microscopic image of sperm. Live and dead sperm were shown in green and red, respectively. Red arrows indicate examples of sperm that were alive or dead. Scale bars correspond to 100 μm.
Germline-enriched histone H1 variants have been reported in many metazoan species, and studies on the model species of some key taxa have revealed that the germline-enriched H1 variants play crucial roles in gametogenesis (Pérez-Montero et al., 2016). In insects, one of the most diverse groups of animals on Earth, the knowledge of germline-enriched H1 variants was limited to Drosophila species (Carbonell et al., 2017; Climent-Cantó et al., 2021), and very little is known about these variants in other insects. In this study, we reported two testis-specific highly expressed H1 variant genes (PpH1V1 and PpH1V2) in the parasitoid wasp P. puparum, and functional studies suggested that PpH1V1 is associated with male fertility in the wasp.
In Drosophila, a germline-enriched H1 variant (dBigH1) and three types of sperm nuclear basic proteins have been so far identified (Mst35Ba/b, Mst77F and Prtl99C), with different expression patterns and roles in spermatogenesis (Tirmarche et al., 2014; Eren-Ghiani et al., 2015; Kimura and Loppin, 2016; Carbonell et al., 2017). In particular, the expression of dBigH1 variant is largely restricted to spermatocytes, and it interplays with bag-of-marbles (Bam) to form a repressor loop that is essential for male germ stem cell differentiation (Carbonell et al., 2017). The loss of dBigH1 in germ cells of testes prevents spermatogonia from proliferation to differentiation, resulting in the accumulation of mitotic spermatogonia that fails to differentiate into spermatocytes and mature sperm (Carbonell et al., 2017). While Mst77F, which shows weak similarity to HILS1, expresses in elongating spermatids and is involve in shaping the nuclear of sperm (Jayaramaiah Raja and Renkawitz-Pohl, 2005; Rathke et al., 2010). Mst77F mutants show low fertility and morphologically abnormal spermatids (Jayaramaiah Raja and Renkawitz-Pohl, 2005; Kimura and Loppin, 2016). In this study, we showed that knockdown of PpH1V1 resulted in abnormal chromatin patterns and low fertility sperm in the seminal vesicles, and this may be the cause of the observation of the significantly reduced male fertility. And we did not find obvious spermatogenesis interruption in testis. This result suggests that, like the Mst77F in Drosophila, PpH1V1 may play a key role in the late process of spermatogenesis. However, although PpH1V1 knockdown induced obviously disordered chromatin organization of sperm, mature sperm were still present in the seminal vesicles. This phenotype differs from that of Mst77F mutants in Drosophila, which had no mature sperm in the seminal vesicles (Kimura and Loppin, 2016). This finding suggests that, although PpH1V1 and Mst77F may involve in the late process of spermatogenesis, they have distinct functions. Additionally, PpH1V1 showed very weak protein sequence identity with Mst77F (24%), and Blastp against Drosophila proteins fails to find any hits, indicating no strong evidence of the orthologous relationship between two proteins. The functions of PpH1V2 variant remain still unclear, such as the knockdown revealed no detectable fertility defects or spermatogenesis interruption, although it carried a testis-specific expression pattern. A similar phenomenon was also found in mice, in which the loss of germline H1 do not reduce male fertility (Drabent et al., 2000; Lin et al., 2000; Fantz et al., 2001). Overall, our findings clearly revealed distinct functions of male germline-enriched H1 variants in parasitoid wasp and Drosophila (Kimura and Loppin, 2016; Carbonell et al., 2017), implying the highly diverged roles of this kind of H1 genes in insects.
As we mentioned above, male germline-enriched H1 variants are often detectable in a specific cell type or process during spermatogenesis. For example, in mammals, H1t generally expresses in meiotic spermatocytes and is also detected during spermatid differentiation, depending on the species (Seyedin and Kistler, 1980; Drabent et al., 1996; 1998; Steger et al., 1998), while HILS1 and H1T2 are restricted to spermatids (Iguchi et al., 2003; Yan et al., 2003; Martianov et al., 2005; Tanaka et al., 2005). In Drosophila, dBigH1 expression is tightly turned on in male germ stem cells and spermatocytes in the early stages of spermatogenesis (Carbonell et al., 2017), whereas Mst77F is detected together with protamines from late canoe stage spermatids to mature sperm (Rathke et al., 2010; Kimura and Loppin, 2016). Here our qPCR experiments revealed that, in the parasitoid wasp P. puparum, the two H1 variants were expressed predominantly in the testis. And their protein expression in the germ cells was demonstrated by immunolocalization. Moreover, we found that these proteins were detected in spermatids with elongating nuclei, but not in mature sperm with elongated nuclei, suggesting that these H1 variants are probably involved in restructuring of the chromatin of elongating spermatids and are subsequently replaced in mature sperm.
We found the two H1 variant genes generally kept as a single copy in all hymenopterans we examined. This discovery suggests H1V1 gene could thus conservatively work as a key factor for male fertility in Hymenoptera. However, further evidence is needed to test this hypothesis by expanded sampling and functional studies. Protein sequence analyses showed rapid evolution of these H1 variants, consistent with the finding that H1 is the most divergent and heterogeneous group of histones (Zhang et al., 2012). Given this fast-evolving pattern, it is challenging to determine the origin and evolutionary trajectory of these two variant genes. Our gene scanning failed to detect putative orthologs of the two H1 variants outside Hymenoptera, thus we conclude that they either arose only in Hymenoptera, or there are not enough distinguishing sequence features for us to unambiguously identify their orthologs outside Hymenoptera. Phylogenomic analysis identified distinct H1V1 and H1V2 variant clades in Hymenoptera, and these two clades were clustered together, suggesting that one of the variants is likely raised by an ancient gene duplication.
In conclusion, we present two rapidly evolving male germline-enriched histone H1 variants in a parasitoid wasp, apparently representing the first view of germline-enriched H1 variants of non-model insects. We then demonstrate that one of the variants, PpH1V1 is associated with male fertility. The roles of these variants in wasps are likely to be different from the H1 variants reported in Drosophila, suggesting the functional diversification of male germline-enriched H1 variants in insects. Thus, our study provides new insights into the role of H1 variants in gametogenesis, and paves the way for future functional and evolutionary studies of H1 variants in insects.
The laboratory cultures of P. puparum adult wasps were fed with 20% honey solution (v/v, honey/deionized distilled water) and maintained at 25°C, 75% relative humidity and a 14:10 h (light: dark) photoperiod. The wasp host, Pieris rapae, was reared on fresh cabbage leaves under the same conditions. Newly pupated P. rapae were used as hosts for the wasps to maintain wasp generation in the laboratory (Wang et al., 2017).
We first searched candidate histone H1 genes in the eleven hymenopteran species (Supplementary Table S2) with high-quality genomes using Blastp (E-value ≤ 1e-5), then these sequences were predicted by searching the sequences against the HMM profiles of the Pfam database (Mistry et al., 2021) and SMART (Letunic et al., 2021) (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/) (E-value ≤ 1e-5), using the default parameters. We also checked the complete open reading frame for each candidate. The H1 sequences for Blast search were downloaded from the database HistoneDB 2.0 (Draizen et al., 2016). Finally, each candidate H1 gene was manually inspected and used for analyses.
The protein sequences of histone H1 genes were aligned by MAFFT v 7.487 (Katoh and Standley, 2013) and filtered by trimAl v1.2 (Capella-Gutiérrez et al., 2009) with the default parameters. The best model for phylogenetic analysis was determined by ModelFinder (Kalyaanamoorthy et al., 2017), and the gene tree was inferred in IQ-Tree v2.0 (Minh et al., 2020) with 1,000 ultrafast bootstrap approximation replicates. We used full-length protein sequences of all H1 variants to calculate pairwise identities between representative hymenopteran orthologs. The species divergence times were obtained from previous studies(Peters et al., 2017; Ye et al., 2022).
Total RNA was extracted from male yellow pupae of P. puparum using RNAiso Plus (Takara Bio, Otsu, Japan) and the RNA concentration of each sample was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Single-strand cDNA was synthesized from the total RNA (1 μg per sample) using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China).
The synthesized cDNA was used as a template for polymerase chain reaction (PCR). PCR primers were designed using Primer3Plus (https://www.primer3plus.com) and specific amplification of coding sequences was performed using the KOD One™ PCR Master Mix (Toyobo, Osaka, Japan). The 5′-and 3′-flanking regions of the H1 variant genes were confirmed by 5′ and 3′ rapid amplification of cDNA ends (RACE) using the SMART RACE cDNA amplification kit (Clontech, California, USA). All amplified PCR products were cloned into the pCE2 TA/Blunt-Zero vector (Vazyme, Nanjing, China) and positive cloning was verified by DNA sequencing. Information about the primers used in this study is provided (Supplementary Table S3).
To detect the developmental expression patterns at different stages of the histone H1 variant genes in P. puparum, larvae, pupae, and adult wasps were collected and transferred into RNAiso Plus (Takara Bio). Thirty early larvae or five late larvae/pupae/adult wasps were pooled as one biological replicate and each life stage was performed in triplicate. The total RNA extraction and the single-strand cDNA synthesis of different samples were conducted as described above. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to quantify the expression level of H1 variant genes using specific primers. An absolute standard curve was constructed from a plasmid clone of each gene using specific primers. PCR products were cloned into pCE2 TA/Blunt-Zero vectors (Vazyme) and then sequenced. Standard curves were generated by determination of copy numbers (103–108 copies) of standard plasmid. qRT-PCR was performed using the Bio-Rad CFX 96 Real-Time Detection System (Bio-Rad, Hercules, CA, United States) with SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio). Thermal cycling conditions were 94°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. Three biological replicates for each group were performed. The equation of y = −3.5095x + 39.678 (y = Ct value; x = the logarithm of plasmid copy number to base 10; R2 = 0.9965) was used to calculate the copy number of PpH1V1. The equation of y = −3.6725x + 40.492 (y = Ct value; x = the logarithm of plasmid copy number to base 10; R2 = 0.9986) was used to calculate the copy number of PpH1V2.
The sex and development stage with the highest copy number of both genes was selected to detect the expression in different tissues. Late-stage yellow pupal males were used for dissections. Heads, thoraxes, testes, male accessory glands or the leftover remnants were dissected using a dissecting microscope (Leica, Wetzlar, Germany) then pooled as one biological replicate and performed in triplicate. The extraction of total RNA, the synthesis of single-strand cDNA, and qRT-PCR were conducted for each sample as described above.
To investigate the functional significance of the H1 variants on P. puparum, RNAi knockdown of the genes were utilized to reduce the abundance of the H1 variants, and then the effect of these knockdown on male fecundity were evaluated. RNAi effects on the gene expressions were evaluated at the transcript/mRNA levels.
We used two different, non-overlapping, double-stranded RNA (dsRNA) fragments to exclude off-target effects. The H1 variant genes specific primers and primers targeting enhanced green fluorescent protein gene (EGFP; negative control) were designed with added T7 promoter adaptors. All amplified PCR products (150–500 bp) were cloned into pCE2 TA/Blunt-Zero vector (Vazyme) and sequenced. The correct PCR products were used as templates for dsRNA synthesis with the T7 High Yield RNA Transcription Kit (Vazyme), according to the manufacturer’s instructions. Synthesized dsRNA was purified by phenol/chloroform extraction and isopropanol precipitation, dissolved in diethylpyrocarbonate-treated water, and quantified using a NanoDrop 2000 Spectrophotometer (Thermo Scientific) at 260 nm.
46.4 nL of dsRNA (4 × 103 ng/μL) was injected into each larval male using Drummond Nanoject II™ Auto-Nanoliter Injector (Drummond Scientific Company, Broomall, United States). The expression level of the genes in the late-stage yellow pupae developed from injected larvae were quantified by qRT-PCR. 15 uninjected female wasps that had mated with injected male wasps for 48 h were selected to investigate the offspring number and sex ratio.
Major biological parameters including the pupation proportion, adult emergence proportion, offspring number and sex ratio of P. puparum were compared among the male RNAi wasps and calculated as described previously (Wang et al., 2017). Reproductive performance of mated females was evaluated in pairings of RNAi or control males with virgin females.
We designed the primers (Supplementary Table S3) by the coding sequence of PpH1V1 or PpH1V2 with 18 bp extension homologous to the vector ends and amplified the two genes by PCR using the pCE2 TA/Blunt-Zero vector with the full-length cDNA sequences inserted as the templates. Linear pET-32a(+) vectors were generated by FastDigest BamHI and HindIII (Thermo Scientific) restrictions. Then the pET-32a(+)-PpH1V1 and pET-32a(+)-PpH1V2 plasmids were constructed by seamless cloning using MonClone™ Hi-Fusion Cloning Mix V2 (Monad Biotech, Suzhou, China). The recombinant plasmids were transformed into Escherichia coli bacterial strain BL21(DE3) (TransGen Biotech). A single positive clone containing the inserts of PpH1V1 or PpH1V2 was selected and incubated in Luria-Bertani (LB) liquid medium (supplemented with 100 μg/mL ampicillin) in a MaxQ 4000 rotating incubator (Thermo Scientific) at 37°C with 200 rpm rotation until the OD600 of the culture reached 0.6. Isopropyl β-D-1-thiogalactopyranoside (Sangon Biotech, Shanghai, China) was added at 1 mmol/L to induce protein expression, then the bacterial culture was incubated at 16°C with 120 rpm rotation for 16 h. Bacterial cells were collected by centrifugation and disrupted with BugBuster Master Mix (Novagen, San Diego, CA, United States) using standard procedures. The insoluble recombinant His-tagged PpH1V1 protein (43.47 kDa) and PpH1V2 protein (35.24 kDa) were purified using High Affinity Ni-NTA Resin (GenScript, Nanjing, China). To confirm the identity of the recombinant proteins, proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (PVDF, Millipore Corporation, Billerica, MA, United States) by an eBlot L1 fast wet transfer system (GenScript), and detected with an anti-His monoclonal antibody (GenScript). Signals were visualized with an enhanced chemiluminescence detection system (Super Signal West Pico Chemiluminescent Substrate; Pierce, Rockford, IL, United States). The recombinant plasmids were submitted to Wuhan Daian Biotechnology Company (Wuhan, Hubei Province, China) for production of purified proteins, which were then used as antigens for immunization of Japanese white rabbits. The obtained polyclonal antibodies to PpH1V1 and PpH1V2 protein were purified from antiserum by the company.
For immunofluorescence (IF), testes and seminal vesicles from P. puparum late-stage yellow pupae, yellow-black pupae, black pupae or adult wasps were dissected, washed, and handled, as described previously (Wu and Luo, 2006). The primary antibodies were rabbit anti-PpH1V1 and rabbit anti-PpH1V2, diluted 1:200 in 0.1 M, pH 7.0 phosphate-buffered saline (PBS; Wisent Biotech, Nanjing, Jiangsu Province, China), containing 0.3% Triton X-100 (Solarbio, Beijing, China) and 5% goat serum (Sangon Biotech). The secondary antibody was Alexa Fluor™ 488 conjugated Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed (Invitrogen, Carlsbad, CA, United States), diluted 1:200 in 0.1 M, pH 7.0 PBS (Wisent Biotech). Antibodies for marking sperm flagellum were acetylated α Tubulin Alexa Fluor® 647 antibody (Santa Cruz Biotechnology, Dallas, Texas, United States) and acetylated α Tubulin Alexa Fluor® 488 antibody (Santa Cruz Biotechnology), diluted 1:200 in 0.1 M, pH 7.0 PBS (Wisent Biotech), containing 0.3% Triton X-100 (Solarbio) and 5% goat serum (Sangon Biotech). Cell nuclei were stained with 1 μg/mL 4′,6-diamidino-2-phenylindole (YEASEN Biotech, Shanghai, China). Tissues were analyzed mounted onto slides using SlowFade Gold Antifade Mountant (Invitrogen) and images recorded with a Zeiss LSM 880 confocal microscope (Carl Zeiss SAS, Jena, Germany) and a Zeiss LSM 800 confocal microscope (Carl Zeiss SAS). A stack of consecutive confocal optical sections (Z-stacks) was recorded at 8-bit resolution. Images were merged and scale bars were added using LSM ZEN 3.2 software (Carl Zeiss SAS).
Testes of P. puparum were dissected from late-stage yellow pupal males using a dissecting microscope (Leica). Seminal vesicles were dissected from 2-day-old adult males. The dissected samples were pre-fixed overnight at 4°C with 2.5% glutaraldehyde (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China) in PBS (0.1 M, pH 7.0). After three washes in PBS for 15 min each, the samples were embedded in 2% agarose diluted in PBS and then fixed with 1% osmium tetroxide (OsO4; SPI, West Chester, PA, United States) in PBS for 1–2 h and washed again for three times. The samples were first dehydrated by a graded series of ethanol (30%, 50%, 70%, 80% for 15 min each), then dehydrated by a graded series of acetone (90%, 95% for 15 min each). In the end, the samples were dehydrated twice by absolute acetone for 20 min each. Samples were infiltrated with a 1:1 mixture of absolute acetone and the final Spurr’s resin (SPI Supplies, West Chester, PA, United States) mixture for 1 h at room temperature, transferred to a 1:3 mixture of absolute acetone and the final resin mixture for 3 h, and then placed into a final Spurr’s resin mixture overnight. Finally, each specimen was placed in an Eppendorf tube containing Spurr’s resin, incubated at 70°C for more than 9 h, and then sectioned using an EM UC7 ultramicrotome (Leica). Ultrathin sections were double-stained with uranyl acetate (SPI Supplies) for 5 min and alkaline lead citrate (Electron Microscopy Sciences) for 10 min and then were observed using an H-7650 TEM (Hitachi, Tokyo, Japan) at an accelerating voltage of 80 kV.
The sperm viability was determined using the Live/Dead sperm viability kit (Invitrogen) as described previously (Zhang et al., 2018). Two fluorescence probes that bind to DNA were used. The first probe (SYBR-14) emits green fluorescence and is actively incorporated by living cells, and the second probe (propidium iodide, PI) emits red fluorescence and can only enter cells with damaged membranes. Seminal vesicles from individual males at 2–3 days post emergence were dissected, washed three times and sperm were collected in 5 µL HEPES solution (10 mM HEPES, 150 mM NaCl, pH 7.4). First, 5 µL SYBR-14 working solution (2 µL SYBR-14 stock in 98 µL HEPES solution) was added to the semen solution, followed by incubation for 10 min in the dark. Then, the solution was incubated with 2 µL PI for another 7 min. Images were recorded with a Zeiss LSM 800 confocal microscope (Carl Zeiss SAS). Sperm viability was obtained for each sample by calculating the percentage of live sperm in the total number of sperm counted. Number of seminal vesicle replications for examining sperm viability is three. A stack of consecutive confocal optical sections (Z-stacks) was recorded at 8-bit resolution. Images were merged, and scale bars were added using LSM ZEN 3.2 software (Carl Zeiss SAS).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
GY and XY conceived and supervised the project. BY performed experiments. XY, YY, ZY, and CH conducted gene identification and evolutionary analyses. FeW, BW, and JS assisted in performing partial experiments. SX, FaW, and QF participated in discussions. BY and XY wrote the draft manuscript. YS, YY, CH, QF, FL, and GY revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Key Program of National Natural Science Foundation of China (NSFC) (Grant no. 31830074 to GY), the Program of NSFC (Grant no. 32202376 to XY), the China Postdoctoral Science Foundation (Grant no. 2021M700125 to XY), and the Young Elite Scientists Sponsorship Program by China Association for Science and Technology (CAST) (Grant no. 2022QNRC001 to XY).
We thank Yunshen Cao (Institute of Insect Sciences, Zhejiang University) for assistance in material preparation. We thank Jiao Zhang, Lei Guo, Cong Dang, Wenxi Jia, Bin Wan, Shijiao Xiong, Xiaomu Qiao and Xuyang Ni (Institute of Insect Sciences, Zhejiang University) for technical guidance and assistance. We thank Jiqiang Song and Kaili Yu (Institute of Insect Sciences, Zhejiang University) for helpful advice during revision of the manuscript. We thank Yunqin Li (Bio-ultrastructure Analysis Laboratory, Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University) and Jianhua Huang (Institute of Insect Sciences, Zhejiang University) for confocal microscopy support. We thank Li Xie, Junying Li, Jin Niu, Weilan Wang and Huanmin Qian (Bio-ultrastructure Analysis Laboratory, Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University) for transmission electron microscopy (TEM) support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1166517/full#supplementary-material
Brandt, W. F., Schwager, S. U. L., Rodrigues, J. A., and Busslinger, M. (1997). Isolation and amino acid sequence analysis reveal an ancient evolutionary origin of the cleavage stage (CS) histones of the sea urchin. Eur. J. Biochem. 247, 784–791. doi:10.1111/j.1432-1033.1997.00784.x
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi:10.1093/bioinformatics/btp348
Carbonell, A., Pérez-Montero, S., Climent-Cantó, P., Reina, O., and Azorín, F. (2017). The germline linker histone dBigH1 and the translational regulator bam form a repressor loop essential for male germ stem cell differentiation. Cell Rep. 21, 3178–3189. doi:10.1016/j.celrep.2017.11.060
Climent-Cantó, P., Carbonell, A., Tamirisa, S., Henn, L., Pérez-Montero, S., Boros, I. M., et al. (2021). The tumour suppressor brain tumour (Brat) regulates linker histone dBigH1 expression in the Drosophila female germline and the early embryo. Open Biol. 11, 200408. doi:10.1098/rsob.200408
Cockburn, A. (1993). Sex determination in the hymenoptera: A review of models and evidence. Hered. (Edinb). 71, 421–435.
De Lucia, F., Faraone-Mennella, M. R., D’Erme, M., Quesada, P., Caiafa, P., and Farina, B. (1994). Histone-induced condensation of rat testis chromatin: Testis-specific H1t versus somatic H1 variants. Biochem. Biophys. Res. Commun. 198, 32–39. doi:10.1006/bbrc.1994.1005
Drabent, B., Bode, C., Bramlage, B., and Doenecke, D. (1996). Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 106, 247–251. doi:10.1007/BF02484408
Drabent, B., Bode, C., and Doenecke, D. (1993). Structure and expression of the mouse testicular H1 histone gene (H1t). BBA - Gene Struct. Expr. 1216, 311–313. doi:10.1016/0167-4781(93)90162-7
Drabent, B., Bode, C., Miosge, N., Herken, R., and Doenecke, D. (1998). Expression of the mouse histone gene H1t begins at premeiotic stages of spermatogenesis. Cell Tissue Res. 291, 127–132. doi:10.1007/s004410050986
Drabent, B., Saftig, P., Bode, C., and Doenecke, D. (2000). Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem. Cell Biol. 113, 433–442. doi:10.1007/s004180000146
Draizen, E. J., Shaytan, A. K., Mariño-Ramírez, L., Talbert, P. B., Landsman, D., and Panchenko, A. R. (2016). HistoneDB 2.0: A histone database with variants - an integrated resource to explore histones and their variants. Database 2016, 1–10. doi:10.1093/database/baw014
Eren-Ghiani, Z., Rathke, C., Theofel, I., and Renkawitz-Pohl, R. (2015). Prtl99C acts together with protamines and safeguards male fertility in Drosophila. Cell Rep. 13, 2327–2335. doi:10.1016/j.celrep.2015.11.023
Eyun, S. Il, Soh, H. Y., Posavi, M., Munro, J. B., Hughes, D. S. T., Murali, S. C., et al. (2017). Evolutionary history of chemosensory-related gene families across the arthropoda. Mol. Biol. Evol. 34, 1838–1862. doi:10.1093/molbev/msx147
Fan, Y., Nikitina, T., Zhao, J., Fleury, T. J., Bhattacharyya, R., Bouhassira, E. E., et al. (2005). Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212. doi:10.1016/j.cell.2005.10.028
Fantz, D. A., Hatfield, W. R., Horvath, G., Kistler, M. K., and Kistler, W. S. (2001). Mice with a targeted disruption of the H1t gene are fertile and undergo normal changes in structural chromosomal proteins during spermiogenesis. Biol. Reprod. 64, 425–431. doi:10.1095/biolreprod64.2.425
Ferree, P. M., Aldrich, J. C., Jing, X. A., Norwood, C. T., Van Schaick, M. R., Cheema, M. S., et al. (2019). Spermatogenesis in haploid males of the jewel wasp Nasonia vitripennis. Sci. Rep. 9, 1–13. doi:10.1038/s41598-019-48332-9
Fyodorov, D. V., Zhou, B. R., Skoultchi, A. I., and Bai, Y. (2018). Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206. doi:10.1038/nrm.2017.94
Harshman, S. W., Young, N. L., Parthun, M. R., and Freitas, M. A. (2013). H1 histones: Current perspectives and challenges. Nucleic Acids Res. 41, 9593–9609. doi:10.1093/nar/gkt700
Heimpel, G. E., and De Boer, J. G. (2008). Sex determination in the hymenoptera. Annu. Rev. Entomol. 53, 209–230. doi:10.1146/annurev.ento.53.103106.093441
Hentschel, C. C., and Birnstiel, M. L. (1981). The organization and expression of histone gene families. Cell 25, 301–313. doi:10.1016/0092-8674(81)90048-9
Iguchi, N., Tanaka, H., Yomogida, K., and Nishimune, Y. (2003). Isolation and characterization of a novel cDNa encoding a DNA-binding protein (Hils1) specifically expressed in testicular haploid germ cells. Int. J. Androl. 26, 354–365. doi:10.1046/j.0105-6263.2003.00449.x
Izzo, A., Kamieniarz, K., and Schneider, R. (2008). The histone H1 family: Specific members, specific functions? Biol. Chem. 389, 333–343. doi:10.1515/BC.2008.037
Jayaramaiah Raja, S., and Renkawitz-Pohl, R. (2005). Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol. Cell. Biol. 26, 3682. doi:10.1128/mcb.26.9.3682.2006
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi:10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi:10.1093/molbev/mst010
Khadake, J. R., and Rao, M. R. (1995). DNA- and chromatin-condensing properties of rat testes H1a and H1t compared to those of rat liver H1bdec; H1t is a poor condenser of chromatin. Biochemistry 34, 15792–15801. doi:10.1021/bi00048a025
Kimura, S., and Loppin, B. (2016). The Drosophila chromosomal protein Mst77F is processed to generate an essential component of mature sperm chromatin. Open Biol. 6, 160207. doi:10.1098/rsob.160207
Kremer, E. J., and Kistler, W. S. (1991). Localization of mRNA for testis-specific histone H1t by in situ hybridization. Exp. Cell Res. 197, 330–332. doi:10.1016/0014-4827(91)90442-W
Kumaroo, K. K., and Irvin, J. L. (1980). Isolation of histone TH1-xB from rat testis. Biochem. Biophys. Res. Commun. 94, 49–54. doi:10.1016/S0006-291X(80)80185-9
Letunic, I., Khedkar, S., and Bork, P. (2021). Smart: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi:10.1093/nar/gkaa937
Lin, Q., Sirotkin, A., and Skoultchi, A. I. (2000). Normal spermatogenesis in mice lacking the testis-specific linker histone H1t. Mol. Cell. Biol. 20, 2122–2128. doi:10.1128/mcb.20.6.2122-2128.2000
Mandl, B., Brandt, W. F., Superti-Furga, G., Graninger, P. G., Birnstiel, M. L., and Busslinger, M. (1997). The five cleavage-stage (CS) histones of the sea urchin are encoded by a maternally expressed family of replacement histone genes: Functional equivalence of the CS H1 and frog H1M (B4) proteins. Mol. Cell. Biol. 17, 1189–1200. doi:10.1128/mcb.17.3.1189
Martianov, I., Brancorsini, S., Catena, R., Gansmuller, A., Kotaja, N., Parvinen, M., et al. (2005). Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl. Acad. Sci. U. S. A. 102, 2808–2813. doi:10.1073/pnas.0406060102
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi:10.1093/molbev/msaa015
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi:10.1093/nar/gkaa913
Müller, K., Thisse, C., Thisse, B., and Raz, E. (2002). Expression of a linker histone-like gene in the primordial germ cells in zebrafish. Mech. Dev. 117, 253–257. doi:10.1016/S0925-4773(02)00174-0
Pandey, N. B., Chodchoy, N., Liu, T. J., and Marzluff, W. F. (1990). Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res. 18, 3161–3170. doi:10.1093/nar/18.11.3161
Pérez-Montero, S., Carbonell, A., and Azorín, F. (2016). Germline-specific H1 variants: The “sexy” linker histones. Chromosoma 125, 1–13. doi:10.1007/s00412-015-0517-x
Pérez-Montero, S., Carbonell, A., Morán, T., Vaquero, A., and Azorín, F. (2013). The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 26, 578–590. doi:10.1016/j.devcel.2013.08.011
Peters, R. S., Krogmann, L., Mayer, C., Donath, A., Gunkel, S., Meusemann, K., et al. (2017). Evolutionary history of the hymenoptera. Curr. Biol. 27, 1013–1018. doi:10.1016/j.cub.2017.01.027
Ponte, I., Romero, D., Yero, D., Suau, P., and Roque, A. (2017). Complex evolutionary history of the mammalian histone H1.1-H1.5 gene family. Mol. Biol. Evol. 34, 545–558. doi:10.1093/molbev/msw241
Rathke, C., Barckmann, B., Burkhard, S., Jayaramaiah-Raja, S., Roote, J., and Renkawitz-Pohl, R. (2010). Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur. J. Cell Biol. 89, 326–338. doi:10.1016/j.ejcb.2009.09.001
Robertson, H. M. (2019). Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64, 227–242. doi:10.1146/annurev-ento-020117-043322
Seyedin, S. M., and Kistler, W. S. (1980). Isolation and characterization of rat testis H1t. An H1 histone variant associated with spermatogenesis. J. Biol. Chem. 255, 5949–5954. doi:10.1016/s0021-9258(19)70722-4
Shechter, D., Nicklay, J. J., Chitta, R. K., Shabanowitz, J., Hunt, D. F., and David Allis, C. (2009). Analysis of histones in Xenopus laevis I. a distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J. Biol. Chem. 284, 1064–1074. doi:10.1074/jbc.M807273200
Smith, R. C., Dworkin-Rastl, E., and Dworkin, M. B. (1988). Expression of a histone H1-like protein is restricted to early Xenopus development. Genes Dev. 2, 1284–1295. doi:10.1101/gad.2.10.1284
Steger, K., Klonisch, T., Gavenis, K., Drabent, B., Doenecke, D., and Bergmann, M. (1998). Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Mol. Hum. Reprod. 4, 939–945. doi:10.1093/molehr/4.10.939
Strickland, W. N., Strickland, M., Brandt, W. F., Von Holt, C., Lehmann, A., and Wittmann-Liebold, B. (1980). The primary structure of histone H1 from sperm of the sea urchin Parechinus angulosus 2. Sequence of the C-terminal CNBr peptide and the entire primary structure. Eur. J. Biochem. 104, 567–578. doi:10.1111/j.1432-1033.1980.tb04460.x
Talaszt, H., Sapojnikova, N., Helliger, W., Lindner, H., and Puschendorf, B. (1999). In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J. Biol. Chem. 273, 32236–32243. doi:10.1074/jbc.273.48.32236
Tanaka, H., Iguchi, N., Isotani, A., Kitamura, K., Toyama, Y., Matsuoka, Y., et al. (2005). HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Mol. Cell. Biol. 25, 7107–7119. doi:10.1128/mcb.25.16.7107-7119.2005
Tanaka, M., Hennebold, J. D., Macfarlane, J., and Adashi, E. Y. (2001). A mammalian oocyte-specific linker histone gene H1oo: Homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 128, 655–664. doi:10.1242/dev.128.5.655
Tirmarche, S., Kimura, S., Sapey-Triomphe, L., Sullivan, W., Landmann, F., and Loppin, B. (2014). Drosophila protamine-like Mst35Ba and Mst35Bb are required for proper sperm nuclear morphology but are dispensable for male fertility. G3 Genes, Genomes, Genet. 4, 2241–2245. doi:10.1534/g3.114.012724
Vanfleteren, J. R., Van Bun, S. M., and Van Beeumen, J. J. (1988). The primary structure of the major isoform (H1.1) of histone H1 from the nematode Caenorhabditis elegans. Biochem. J. 255, 647–652.
Wang, F., Fang, Q., Wang, B., Yan, Z., Hong, J., Bao, Y., et al. (2017). A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 13, 1–27. doi:10.1371/journal.ppat.1006201
Wu, J. S., and Luo, L. (2006). A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 1, 2110–2115. doi:10.1038/nprot.2006.336
Yan, W., Ma, L., Burns, K. H., and Matzuk, M. M. (2003). HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc. Natl. Acad. Sci. U. S. A. 100, 10546–10551. doi:10.1073/pnas.1837812100
Ye, X., Yan, Z., Yang, Y., Xiao, S., Chen, L., Wang, J., et al. (2020). A chromosome-level genome assembly of the parasitoid wasp Pteromalus puparum. Mol. Ecol. Resour. 20, 1384–1402. doi:10.1111/1755-0998.13206
Ye, X., Yang, Y., Zhao, C., Xiao, S., Sun, Y. H., He, C., et al. (2022). Genomic signatures associated with maintenance of genome stability and venom turnover in two parasitoid wasps. Nat. Commun. 13, 34202. doi:10.1038/s41467-022-34202-y
Zhang, J. L., Yuan, X. B., Chen, S. J., Chen, H. H., Xu, N., Xue, W. H., et al. (2018). The histone deacetylase NlHDAC1 regulates both female and male fertility in the Brown planthopper, Nilaparvata lugens. Open Biol. 8, 180158. doi:10.1098/rsob.180158
Keywords: parasitoid wasp, Pteromalus puparum, histone H1 variant, chromatin, spermatogenesis
Citation: Yuan B, Yang Y, Yan Z, He C, Sun YH, Wang F, Wang B, Shi J, Xiao S, Wang F, Fang Q, Li F, Ye X and Ye G (2023) A rapidly evolving single copy histone H1 variant is associated with male fertility in a parasitoid wasp. Front. Cell Dev. Biol. 11:1166517. doi: 10.3389/fcell.2023.1166517
Received: 15 February 2023; Accepted: 22 May 2023;
Published: 31 May 2023.
Edited by:
Judith Yanowitz, Magee-Womens Research Institute, United StatesReviewed by:
Giovanni Messina, Sapienza University of Rome, ItalyCopyright © 2023 Yuan, Yang, Yan, He, Sun, Wang, Wang, Shi, Xiao, Wang, Fang, Li, Ye and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhai Ye, eWV4aW5oYWlAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.