- 1Department of Dermatology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China
- 2General Practice Department, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China
Tumor Necrosis Factor-alpha (TNF-α) is ubiquitous in the human body and plays a significant role in various physiological and pathological processes. However, TNF-α-induced diseases remain poorly understood with limited efficacy due to the intricate nature of their mechanisms. N6-methyladenosine (m6A) methylation, a prevalent type of epigenetic modification of mRNA, primarily occurs at the post-transcriptional level and is involved in intranuclear and extranuclear mRNA metabolism. Evidence suggests that m6A methylation participates in TNF-α-induced diseases and signaling pathways associated with TNF-α. This review summarizes the involvement of TNF-α and m6A methylation regulators in various diseases, investigates the impact of m6A methylation on TNF-α-induced diseases, and puts forth potential therapeutic targets for treating TNF-α-induced diseases.
Introduction
Tumor Necrosis Factor-alpha (TNF-α), a cytokine ubiquitous in the human body, is pivotal in the pathogenesis of various diseases, particularly immune and inflammatory disorders. The intricate mechanisms and aggressive therapeutic interventions necessitate a further investigation of the underlying pathogenesis and potential therapeutic targets. The widespread involvement of m6A methylation in physiopathological processes is a well-established phenomenon. Recent studies have demonstrated the existence of m6A-induced gene expression regulation in various TNF-α-induced diseases. This regulation involves the participation of the writer, eraser, and reader and plays a crucial role in multiple biological processes, including RNA stability, degradation, and variable cleavage. This study aims to summarize the TNF-α-induced diseases and elucidate the relationship between signaling pathways and m6A methylation. This endeavor provides a promising opportunity to gain further insights into the role of m6A methylation in TNF-α-induced diseases and identify potential therapeutic targets.
TNF-α
TNF-α, or cachectin and TNFSF1A, is a constituent of the tumor necrosis factor superfamily and comprises a 157 amino acid homotrimeric protein (Campanati A et al., 2019). Various immune cells, including macrophages, monocytes, neutrophils, CD4+ T cells, and Natural killer cells (NK cells), secrete TNF-α in response to endotoxin stimulation (Chadwick et al., 2008). TNF-α exits in two biologically active forms: transmembrane TNF-alpha (tmTNF-α) and secretory TNF-alpha (sTNF-α). The former predominantly exists in a membrane-bound configuration and is converted into sTNF-α by the TNF-α converting enzyme TACE. The systemic circulation is the primary site of action for sTNF-α, imparting an endocrine role to TNF-α (Yang et al., 2018).
The cellular receptors for TNF-α are classified into TNFR1 and TNFR2. TNFR1 is expressed ubiquitously on almost all cells and can be activated by sTNF-α and tmTNF-α. It interacts with TNFR1-associated death domain (TRADD) adaptor proteins (Pobezinskaya and Liu, 2012). Conversely, TNFR2 expression is restricted to immune cells, endothelial cells, and neurons. Furthermore, sTNF signaling via TNFR1 primarily initiates pro-inflammatory pathways, while tmTNF binds to TNFR2 for immune regulation and tissue regeneration (Zhang et al., 2017).
Role of TNF-α in autoimmune/inflammatory diseases
TNF-α plays an important role in physiology and pathology. Physiologically, TNF-α is a critical component of normal immune responses. TNF-α is regulated by immune system activation and has various biological activities, such as regulating inflammation, apoptosis, chemokine production, and metabolism (Brenner et al., 2015). However, abnormal or excessive TNF-α activation can lead to or aggravate chronic inflammation and neoplastic diseases (Idriss and Naismith, 2000; van Horssen et al., 2006). Figure 1 illustrates the role of TNF-α in TNF-α-induced diseases.
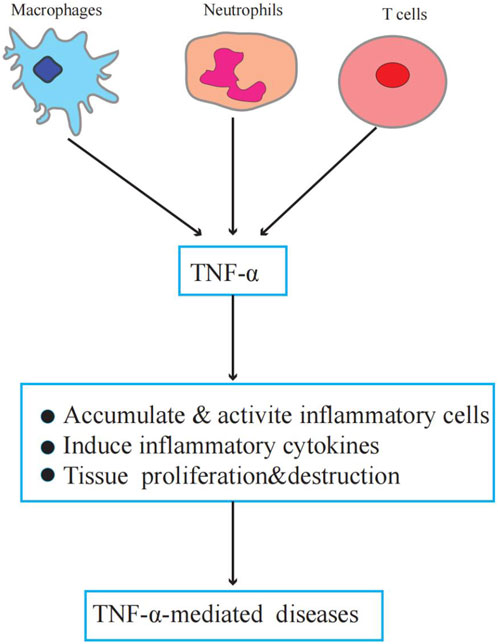
FIGURE 1. The role of TNF-α in TNF-α-induced diseases. Macrophages, neutrophils, and T cells secrete TNF-α. TNF-α promotes inflammatory cell activation and accumulation induces inflammatory factor production, forms a local inflammatory environment, and leads to tissue proliferation and destruction.
Autoimmune/inflammatory diseases are a series of tissue injuries caused by systemic or organ-specific inflammation (Wang L et al., 2015). Epidemiology suggests that autoimmune/inflammatory diseases affect 3%–5% of the population (Surace and Hedrich, 2019). Autoimmune/inflammatory diseases are monogenic or polygenic, with autoantibodies and/or reactive lymphocyte populations being induced. They are influenced by individuals and the environment, ultimately leading to disease development (Wang X et al., 2015; He et al., 2018). TNF-α is widely involved in autoimmune/inflammatory diseases, such as rheumatoid arthritis (RA), inflammatory bowel disease (IBD), ankylosing spondylitis (AS) arteriosclerosis, psoriasis (PS), and noninfectious uveitis (NIU). TNF-α expression is elevated in tissues and/or blood and is associated with disease activity in TNF-α-induced autoimmune/inflammatory diseases (Arican et al., 2005; Wehkamp et al., 2016; Bullock et al., 2018; Ebrahimiadib et al., 2021). Meanwhile, TNF-α inhibitors have proven therapeutic in treating the above diseases (Udalova et al., 2016). These studies suggest an important role for TNF-α in autoimmune/inflammatory diseases.
Macrophages and Th1 cells secrete TNF-α in RA, activate synovial fibroblasts, and produce excessive cathepsins and MMPs, destroying cartilage and bone and causing joint erosion. TNF-α promotes osteoclast generation and induces synovial hyperplasia and angiogenesis. TNF-α can also produce other inflammatory cytokines and promote an inflammatory milieu in the synovium, manifesting clinically as pain and joint swelling (Koch et al., 1994; Brennan and McInnes, 2008; Lin et al., 2020).
Stressed keratinocytes secret TNF-α and other cytokines in PS, activating DCs macrophages and neutrophil accumulation, which is pathologically characterized by parakeratosis, acanthosis, and Munro microabscesses. TNF-α also induces keratinocyte proliferation and anti-apoptosis via the NF-κB/MAPK signaling pathway (Jang et al., 2021).
IBD primarily comprises Crohn’s disease (CD) and ulcerative colitis (UC). TNF-α induces pro-inflammatory effects, including macrophages and T cell activation, chemotaxis of neutrophils and monocytes, increased endothelial cell adhesion molecules (EcAMs) expression, and activation of coagulation and fibrinolytic responses, resulting in epithelial injury, endothelial activation, and vascular destruction. TNF-α also promotes intestinal fibrosis caused by intestinal fibroblasts, leading to intestinal strictures formation (Liou and Storz, 2010; Bounder et al., 2020).
Role of TNF-α in tumors
TNF-α is a double-edged sword in cancer. When TNF-α is secreted in excess, it stimulates tumor cells to release pro-inflammatory mediators, inhibits the anti-tumor activity of immune cells, suppresses mucosal healing, and disrupts mucosal barrier function, inducing cancer cell proliferation, immune escape, invasion, angiogenesis, and metastasis (Havell et al., 1988). The mechanisms may be related to reactive oxygen species (ROS) and reactive nitrogen species (RNS) generation (Adegbola et al., 2018). High TNF-α concentrations may also play an anti-tumor role (Levin et al., 2016). Additionally, the role of TNF-α depends on its expression site in the tumor (Landskron et al., 2014).
m6A methylation
m6A refers to the adenine methylation at position N6 and is a rich and extensive RNA modification, accounting for approximately 80% of all RNA methylation modifications (Dominissini et al., 2012; Yue et al., 2015). m6A modifications are widespread in mammals, plants, and viruses and are highly conserved among species (Horowitz et al., 1984; Fu et al., 2014; Liu et al., 2014).
Mammalian m6A methylation sites near the mRNA stop codon and in the 3′untranslated region (3′UTR) (Fu et al., 2014), non-coding RNAs, such as long non-coding RNAs (lncRNAs) and microRNAs (miRNA), also have m6A modification sites (Alarcón et al., 2015a; Jacob et al., 2017). m6A transferases, demethylases, and binding proteins regulate involvement in m6A modification and play essential roles in biological processes, such as mRNA stability, degradation, alternative splicing, nuclear export, and translation (Wang et al., 2014; Zaccara et al., 2019; Wei and He, 2021). A recent study of m6A methylation has demonstrated a rapid increase trend with the rapid update of new technologies. m6A modification plays an essential role in the body’s primary metabolism and basic physiological functions (Dominissini et al., 2012; Frye et al., 2018; Shi et al., 2019). Methylation of m6A has been studied in various diseases, such as tumors, cardiovascular diseases, neurological diseases, viral infections, and autoimmune inflammation (Zhao et al., 2014; Geula et al., 2015; Sun et al., 2019). The m6A binding protein binds to the corresponding m6A modification site to achieve their respective biological functions and can also change RNA secondary structure and play a regulatory role (Zhang C et al., 2019; Chai et al., 2021).
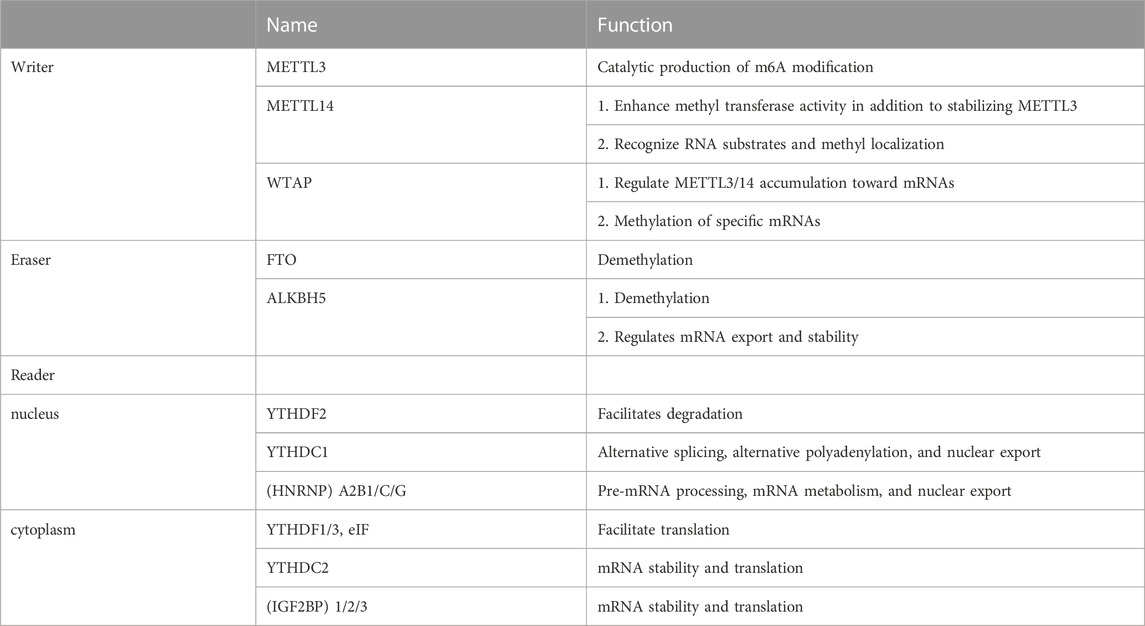
Enzymes involved in catalyzing and recognizing m6A modifications can be divided into writer, eraser, and reader based on their function. Table 1 presents their functions. The writer, including major components, such as methyltransferase-like 3 (METTL3), methyltransferase-like14 (METTL14), and Wilms’ tumor 1-associated protein (WTAP), is primarily responsible for catalyzing m6A modifications and writing methyl groups in RNA. METTL3 is a prominent constituent member of this complex. METTL3 localizes to nuclear patch areas of the nucleus and forms a stable heterodimer with METTL14 (Liu et al., 2020; Zeng et al., 2020). METTL3 recognizes S-adenosyl methionine (SAM) during catalysis. It exhibits catalytic activity (Liu et al., 2014), whereas METTL14 recognizes RNA substrates and methyl localization, enhancing methyltransferase activity besides stabilizing METTL3 (Wang et al., 2016; Zhang et al., 2020). WTAP regulates the METTL3/14 accumulation on mRNAs and the specific mRNA methylation as its primary function (Fan et al., 2022). Other components of the m6A methyltransferase complex include KIAA1429, RNA-binding motif protein 15 (RBM15), and Zinc finger CCCH-type containing 13 (ZC3H13) (Hu et al., 2016; Huang et al., 2021; Jiang et al., 2021; Zhang, et al., 2022a).
m6A demethylase catalyzes the reversible m6A modification process. Demethylases, primarily Fat mass and obesity-associated protein (FTO) and Alkylation repair homolog protein 5 (ALKBH5), play a role in catalyzing demethylation (Ito et al., 2011). FTO belongs to the iron/α-ketoglutarate–dependent AlkB protein family and is tightly linked to obesity (Dina et al., 2007; Frayling et al., 2007; Gerken et al., 2007). However, the substrate-bound by FTO does not appear to be m6A alone. It has been claimed that FTO impacts mRNA stability and translation efficiency by regulating m6A.m. levels (Mauer et al., 2017). Moreover, FTO localization varies in different cells, with FTO mediating the m6A demethylation in the nucleus and mediating the demethylation of m6A.m. and m6A in the cytoplasm (Wei et al., 2018). Except for demethylation, ALKBH5 regulates mRNA export and stability by affecting the splicing and mRNA production rates and participating in functions, such as mRNA transport, metabolic processing, and degradation (Zheng et al., 2013; Li et al., 2020).
The “writer” and “eraser” of m6A act on intranuclear mRNAs, making m6A modifications dynamic and reversible. Furthermore, downstream regulation at the mRNA translation level by the “reader” is required to achieve biological function. Some m6A readers have biological roles in the nucleus. Nuclear m6A readers include nuclear ribonucleoprotein (HNRNP) A2B1/C/G, YT521-B homology (YTH) domain-containing heterogeneous proteins, such as YTHDC1 (Zhang et al., 2021). HNRNPs are RNA-binding proteins associated with the precursor mRNA (pre-mRNA) maturation in the nucleus and affect processes, such as pre-mRNA processing, mRNA metabolism, and nuclear export (Das, et al., 2019). HNRNPA2B1 initially promotes the primary miRNA processing and maturation by binding transcription of m6A methylation (Alarcon et al., 2015b). YTHDC1 mediates alternative splicing, alternative polyadenylation, and nuclear export (Xiao et al., 2016; Chen et al., 2021). A portion of the reader plays a biological role in the cytoplasm. Extranuclear m6A reader primarily includes YTHDF1/2/3, YTHDC2, and Insulin-like growth factor 2 mRNA binding protein (IGF2BP) 1/2/3 (Xiao et al., 2016). YTHDF1/3 and eIF can act on mRNA translation initiation factors and contribute to mRNA translation (WANG X et al., 2015). YTHDF2 binds to mRNA and promotes its degradation, affecting stability and translation status (Wang and Liu, 2021). IGF2BP1/2/3 promotes m6A-induced mRNA stability and translation (Liu et al., 2015).
TNF-α and m6A methylation
Many studies have revealed that m6A methylation is involved in TNF-α-induced disease progression. It mainly involves changes in global m6A methylation, enzyme levels and activities, and TNF-α-induced pathways. Figure 2 shows the role of m6A in TNF-α-medicated diseases.

FIGURE 2. The role of m6A in TNF-α-medicated diseases. m6A regulator levels (writers, erasers, and readers) change upon TNF-α stimulation. Simultaneously, TNF-α levels change with altered m6A regulator expressions, implying that TNF-α and m6A regulators interact with each other and jointly impact the disease condition. However, the causal relationship between TNF-α and m6A regulators is currently unknown.
Li et al. (2021) discovered that differentially expressed m6A methylation sites between lncRNAs and circRNAs were primarily enriched in TNF signaling pathways after mapping m6A methylation profiles in bladder cancer. These results suggest that TNF signaling may differ in global m6A methylation levels in some diseases.
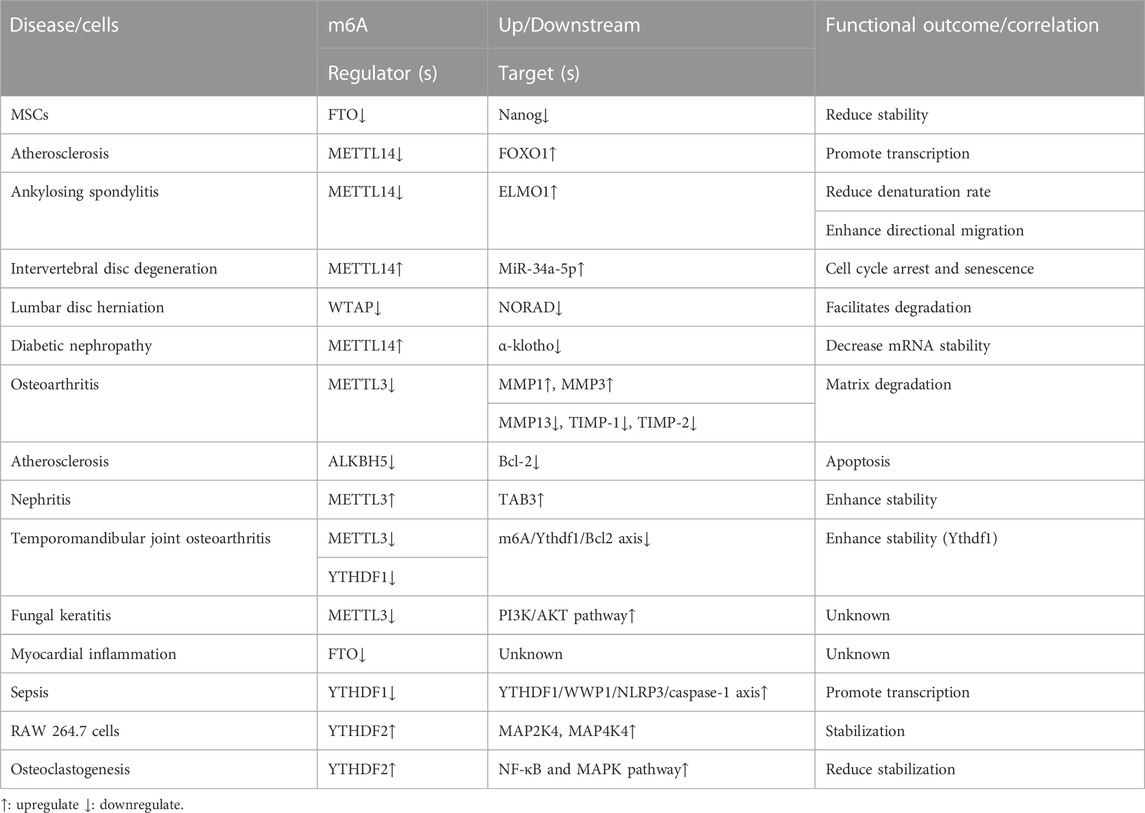
m6A regulator levels (writers, erasers, and readers) change upon TNF-α stimulation, ultimately impacting the disease. These results suggest that m6A methylation regulators may be involved in TNF-α-induced disease progression. Wang, et al. (2019) discovered that TNF-α stimulation decreased FTO expression in mesenchymal stromal cells (MSCs), shortened the half-life of Nanog mRNA, led to decreased mRNA and protein expression, and ultimately significantly inhibited the ability of MSCs to differentiate into sweat gland cells. Jian D et al. (2020) discovered that when endothelial cells are stimulated with TNF-α, METTL14 expression increases, acts directly on the promoter regions of adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM1), increases the FOXO1 gene m6A methylation level and promotes its transcription, enhances expression, ultimately leads to atherosclerotic plaque formation. METTL14 binds to the engulfment and cell motility 1 (ELMO1) mRNA 3 ‘UTR and promotes mRNA degeneration in Ankylosing spondylitis. TNF-α stimulation decreased METTL14 expression and decreased the ELMO1 degeneration rate, thereby increasing ELMO1 expression, leading to enhanced directional migration of AS-MSC in vivo and in vitro, ultimately aggravating the disease (Xie et al., 2021). Zhu et al. (2021) revealed that METTL14 expression positively correlated with TNF-α concentration in abnormal nucleus pulposus cells. METTL14 was involved in TNF-α-induced m6A modification of miR-34a-5p and interacted with DGCR8 to promote cell cycle arrest and aberrant degeneration in nucleus pulposus (NP) cells from intervertebral disc degeneration (IVDD) patients. In lumbar disc herniation (LDH), WTAP can bind to METTL3 and METTL14 as well as NORAD to promote its m6A methylation. Following TNF-α stimulation of nucleus pulposus cells, WTAP expression was decreased.RNA immunoprecipitation (RIP) revealed that YTHDF2 was the predominant reader protein, promoting NORAD degradation and ultimately exacerbating disease (Li et al., 2022).
Altered m6A methylation regulator levels can also significantly affect TNF-α levels and/or related signaling pathway expressions, ultimately affecting the disease. Li et al. (2020) indicated that m6A RNA modification is essential for Th cell differentiation and proliferation in vivo. METTL3 knockdown mice significantly decreased the proportion of naive T cells differentiating into Th1 and Th17 cells and a significant increase in Th2 cells. METTL14 expression is significantly increased in diabetic nephropathy, and METTL14 overexpression leads to increased inflammatory factor secretion, such as TNF-α in serum. Mechanistically, METTL14 downregulated α-klotho expression in an m6A-dependent manner, leading to glomerular endothelial cell apoptosis in the kidneys of diabetic nephropathy (DN) patients (Li M.et al., 2021). Sang et al. (2021) exposed that METTL3 expression was decreased in osteoarthritis (OA), and METTL3 overexpression decreased inflammatory factor levels, such as TNF-α. METTL3 overexpression adjusts to the balance between TIMPs and MMPs, possibly leading to extracellular matrix degradation in OA. ALKBH5 expression has been decreased in TNF-α-treated human umbilical vein endothelial cells (HUVECs). ALKBH5 inhibits TNF-α-induced endothelial cell apoptosis via Bcl-2, thereby reducing atherosclerosis (Zhang T. et al., 2022). Studies have revealed that METTL3 is overexpressed in acute kidney injury (AKI) models, and METTL3 deficiency reduces renal cell inflammation and programmed cell death induced by TNF-α. METTL3 promoted m6A modifications of TAB3 and enhanced stability via IGF2BP2-dependent mechanisms (Wang B et al., 2022). METTL3 expression is reduced in temporomandibular joint osteoarthritis (TMJ OA). Further investigation indicated that TNF-α-induced osteoclast autophagy and apoptosis are inhibited via the m6A/Ythdf1/Bcl2 signaling axis (He et al., 2022). METTL3 expression was increased in mouse fungal keratitis (FK). METTL3 deficiency reduced TNF-α expression and decreased PI3K/AKT pathway protein expression, ultimately inhibiting the inflammatory response (Huang et al., 2022). FTO expression was reduced in LPS-treated cardiomyocytes, and FTO knockdown in cardiomyocytes may lead to inflammatory cytokine overexpression, such as TNF-α. The m6A-RNA immunoprecipitation (MeRIP) assay revealed that FTO knockdown would lead to TNF-α mRNA transcript hypermethylation (Dubey et al., 2022). YTHDF1 was downregulated and acts as YTHDF1/WWP1/NLRP3/caspase-1 axis in sepsis. Overexpressed YTHDF1 stabilizes the TNF-α expression in CLP-induced mice (Zhang et al., 2022c). The YTHDF2 expression was increased in lipopolysaccharide (LPS)-induced RAW 264.7 cells, and the YTHDF2 knockdown significantly increased the TNF-α, MAP2K4, MAP4K4, and activated downstream signaling pathway expressions, such as NF-κB and MAPK (Yu H et al., 2019). However, YTHDF2 expression was elevated in LPS-induced osteoclastogenesis cells, and the YTHDF2 knockdown significantly increased the inflammatory factor expression levels, such as TNF-α and activated downstream pathways, including NF-κB and MAPK signaling (Fang et al., 2021).
Influencing the activity of m6A methylation regulators can similarly influence TNF-α levels and disease progression. LuHui derivative (LHD), an FTO inhibitor, binds explicitly the pocket containing the regulatory sites of RNA methylation in LHD protein, leads to loss of FTO function, inhibits palmitate-induced cardiac inflammation, reduces CD36 expression by reducing the stability of CD36 mRNA, and inhibits inflammatory factor expressions, such as TNF-α and IL-6, thereby relieving hyperlipidemic cardiomyopathy (Yu et al., 2021). Emodin decreases inflammation, such as TNF-α expression in LPS-treated 1321N1 cells, by inactivating METTL3-induced NLRP3 expression (Wang J et al., 2022). Further, the important role of m6A methylation in TNF-α-induced diseases is illustrated.
In summary, m6A methylation is widely involved in TNF-α-induced diseases involving various diseases and cells. Table 2 presents the role of m6A methylation in TNF-α-medicated diseases. However, whether TNF-α acts directly on m6A methylation regulators and how m6A methylation regulators are altered in TNF-α-induced diseases still needs further investigation.
TNF-α signaling pathways
TNF-α exerts its biological function primarily via nuclear factor-kappaB (NF-κB) and mitogen-activated protein kinases (MAPK) signaling pathways. Studies have revealed that the above signaling pathways are also involved in m6A methylation in other diseases.
NF-κB is a eukaryotic transcription factor family with five subunits: p65 (RelA), RelB, c-Rel, p105/p50, and p100/p52. It involves various processes, such as immune response, inflammation, apoptosis, growth, and development (Lawrence, 2009; Wertz and Dixit, 2010; Kumar et al., 2015). NF-κB signaling pathway contains canonical and non-canonical signaling pathways. Classical signaling pathways initiate transcription by degrading IκB and releasing NF-κB dimers, translocating to the nucleus and activating target genes rapidly but transiently (Chen and Greene, 2004; Ruland, 2011). A few TNF superfamily receptors activate non-canonical signaling pathways. The central event of the non-canonical NF-κB signaling pathway is processed for p100 to obtain p52. Proteolytic processing of p100 leads to the p52 production and RelB release, forming a RelB/p52 dimer and translocation to the nucleus, thereby activating signaling pathways (Dejardin, 2006; Hayden and Ghosh, 2008; Sun, 2011).
The m6A methylation is involved in the NF-κB pathway in various diseases and impacts the disease. METTL3 has been overexpressed in RA, positively associated with disease activity. Furthermore, METTL3 inhibited LPS-induced macrophages in inflammation via NF-κB (Wang et al., 2019). Specific METTL14 deletion leads to colon stem cell apoptosis in IBD, mucosal barrier dysfunction, and severe colitis. Mechanistically, METTL14 mitigates colonic epithelial cell death by controlling Nfkbia mRNA stability and the NF-κB signaling pathway (Zhang et al., 2022d). Mice with loss of METTL3 in myeloid cells develop less age-associated incidence of non-alcoholic fatty liver disease (NAFLD) and obesity. In macrophages, METTL3 deficiency may cause significantly increased DNA Damage Inducible Transcript 4 (DDIT4) expression and activates the mTORC1/NF-κB signaling pathway, finally improving NAFLD (Qin et al., 2021). The research discovered that IL-6 cooperates with CUDR, causing METTL3 to interact with SUV39h1 mRNA3 ‘UTR and promote SUV39h1 expression. SUV39h1 enhances the NF-κB expression and phosphorylation, leading to malignant transformation of the human embryonic hepatocyte-like stem cells (Zheng Q et al., 2016). ALKBH5 had higher expression in cancer stem cells than in normal tissue. TLR4 increases ALKBH5 expression via activating the NF-κB pathway. The upregulated ALKBH5, whose m6A modification gene is NANOG, promotes ovarian cancer cell aggressiveness (Jiang et al., 2020).
MAPK is a signal-regulated protein kinase important in cell proliferation, differentiation, autophagy, apoptosis, immune response, and other physiological and pathological conditions (Arthur and Ley, 2013). MAPKs act simultaneously as signaling messengers, transducing extracellular signals into the cell and regulating the various cytokine and chemokine expressions (Zhao et al., 2018). MAPK signaling pathway comprises four subfamilies: ERK1/2, JNK1/2/3, p38 MAPK, and ERK5 (Uddin et al., 2021). This signaling pathway activates the MAPKs in a three-tier kinase activity from MAPKKK, MAPKK to MAPK (Burotto et al., 2014). The m6A methylation is widely involved in regulating the MAPK signaling pathway. METTL3 expression is decreased in tumor-infiltrating NK cells. SHP-2, an m6A-modified gene whose protein expression is reduced in METTL3-deficient NK cells, may render NK cells hyporesponsive to IL-15 via the MAPK signaling pathway (Song et al., 2021). YTHDF2 interacts with mRNAs encoding proteins, epithelial-to-mesenchymal transition, and increased translation rates in MYC-driven breast cancer to inhibit cell death via the MAPK signaling pathway (Einstein et al., 2021). Highly expressed WTAP is associated with shorter overall survival and lymph node metastasis in ovarian cancer cells. WTAP downregulation decreased several MAPK protein expressions, such as p-ERK, ERK, p-JNK, and JNK, suggesting a possible correlation between WTAP and MAPK signaling pathways (Yu R et al., 2019). METTL3, whose m6A target modification is miR-1246, is upregulated in colorectal cancer and promotes cell migration and invasion. SPRED2 is a downstream target of miR-1246, and its downregulation promotes MAPK signaling pathway expression (Peng et al., 2019).
Conclusion and future perspectives
This study reviews TNF-α and TNF-α-induced diseases and summarizes the role of m6A methylation in TNF-α-induced diseases and the main signaling pathways involved in TNF-α. Moreover, an in-depth study of m6A methylation may elucidate the pathogenesis of TNF-α-induced diseases and provide more possibilities to identify potential therapeutic targets.
If an m6A methylation regulator is aberrantly expressed and changes with TNF-α levels in TNF-α-induced diseases, we can search the binding sites of m6A methylation regulators to differentially expressed genes via methylated RNA immunoprecipitation sequencing (MeRIP-seq) and transcriptomic RNA sequencing (RNA-seq).
Studies have demonstrated that the external environment influences the m6A methylation level. Cigarette smoke condensate (CSC) induces hypomethylation of the METTL3 promoter, resulting in aberrant METTL3 overexpression. The miR-25–3p promotes the pancreatic ductal adenocarcinoma (PDAC) development in smokers by inhibiting PH domain leucine-rich repeat protein phosphatase 2 (PHLPP2) and activating AKT-p70S6K (Zhang J et al., 2019). Chronic exposure of human bronchial epithelial (HBE) cells to arsenite sodium (NaAsO2) increases m6A methylation levels significantly and induces a malignant phenotype. Silencing METTL3 reversed these changes (Gu et al., 2018). Given the potential link between TNF-α and m6A methylation, whether TNF-α affects RNA functions, such as secondary structure, decoration affinities, and protein binding partners, by affecting m6A methylation regulators remains to be investigated.
Thus, m6A modification is a complex process working as a double-edged sword for TNF-α-medicated diseases. Does the relationship between TNF-α and m6A methylation regulators are causal or associations? Therefore, unknown regulators exist between TNF-α and m6A methylation. Furthermore, TNF-α has a dual role of cytokines and inflammatory mediators. It is also worth investigating whether TNF-α and m6A methylation regulators can influence each other between different cells. For example, in TNF-α-induced diseases that concern various immune cells, can changes in the m6A methylation regulator expression levels in one cell influence the m6A methylation regulator levels and TNF-α in another immune cell? Further exploration is needed.
Modulators of m6A methylation provide novel potential therapeutic targets. If altered levels of m6A methylation regulators improve disease conditions, inhibitors/activators targeting m6A methylation regulators may provide potential therapeutic possibilities. The newly identified m6A modifiers will open novel therapeutic avenues to understand the m6A methylation-associated diseases.
Author contributions
YW drafted the manuscript. YW and JL designed the review, wrote, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adegbola, S. O., Sahnan, K., Warusavitarne, J., Hart, A., and Tozer, P. (2018). Anti-TNF therapy in crohn's disease. Int. J. Mol. Sci. 19 (8), 2244. doi:10.3390/ijms19082244
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell. 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Arican, O., Aral, M., Sasmaz, S., and Ciragil, P. (2005). Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediat. M. 2005 (5), 273–279. doi:10.1155/MI.2005.273
Arthur, J. S., and Ley, S. C. (2013). Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13 (9), 679–692. doi:10.1038/nri3495
Bounder, G., Jouimyi, M. R., Boura, H., Touati, E., Michel, V., Badre, W., et al. (2020). Associations of the -238(G/A) and -308(G/A) TNF-α promoter polymorphisms and TNF-α serum levels with the susceptibility to gastric precancerous lesions and gastric cancer related to Helicobacter pylori infection in a Moroccan population. Pac J. Cancer Prev. 21 (6), 1623–1629. doi:10.31557/APJCP.2020.21.6.1623
Brennan, F. M., and McInnes, I. B. (2008). Evidence that cytokines play a role in rheumatoid arthritis. J. Clin. Invest. 118 (11), 3537–3545. doi:10.1172/JCI36389
Brenner, D., Blaser, H., and Mak, T. W. (2015). Regulation of tumour necrosis factor signalling: Live or let die. Nat. Rev. Immunol. 15 (6), 362–374. doi:10.1038/nri3834
Bullock, J., Rizvi, S. A. A., Saleh, A. M., Ahmed, S. S., Do, D. P., Ansari, R. A., et al. (2018). Rheumatoid arthritis: A brief overview of the treatment. Med. Pract. 27 (6):501–507. doi:10.1159/000493390
Burotto, M., Chiou, V. L., Lee, J. M., and Kohn, E. C. (2014). The MAPK pathway across different malignancies: A new perspective. Cancer 120 (22), 3446–3456. doi:10.1002/cncr.28864
Campanati, A., Paolinelli, M., Diotallevi, F., Martina, E., Molinelli, E., and Offidani, A. (2019). Pharmacodynamics of TNF-α inhibitors for the treatment of psoriasis. Expert Opin. Drug Metab. Toxicol. 15 (11), 913–925. doi:10.1080/17425255.2019.1681969
Chadwick, W., Magnus, T., Martin, B., Keselman, A., Mattson, M. P., and Maudsley, S. (2008). Targeting TNF-alpha receptors for neurotherapeutics. Trends Neurosci. 31 (10), 504–511. doi:10.1016/j.tins2008.07.005
Chai, R. C., Chang, Y. Z., Chang, X., Pang, B., An, S. Y., Zhang, K. N., et al. (2021). YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J. Hematol. Oncol. 14 (1), 109. doi:10.1186/s13045-021-01124-z
Chen, L. F., and Greene, W. C. (2004). Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell. Biol. 5 (5), 392–401. doi:10.1038/nrm1368
Chen, C., Liu, W., Guo, J., Liu, Y., Liu, X., Liu, J., et al. (2021). Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell. 12 (6), 455–474. doi:10.1007/s13238-021-00837-8
Das, U., Nguyen, H., and Xie, J. (2019). Transcriptome protection by the expanded family of hnRNPs. RNA Biol. 16 (2), 155–159. doi:10.1080/15476286.2018.1564617
Dejardin, E. (2006). The alternative NF-kappaB pathway from biochemistry to biology: Pitfalls and promises for future drug development. Biochem. Pharmacol. 72 (9), 1161–1179. doi:10.1016/j.bcp.2006.08.007
Dina, C., Meyre, D., Gallina, S., Durand, E., Körner, A., Jacobson, P., et al. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 39 (6), 724–726. doi:10.1038/ng2048
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dubey, P. K., Patil, M., Singh, S., Dubey, S., Ahuja, P., Verma, S. K., et al. (2022). Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol. Cell. Biochem. 477 (1), 129–141. doi:10.1007/s11010-021-04267-2
Ebrahimiadib, N., Berijani, S., Ghahari, M., and Pahlaviani, F. G. (2021). Ankylosing spondylitis. J. Ophthalmic Vis. Res. 16 (3), 462–469. doi:10.18502/jovr.v16i3.9440
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell. 81 (15), 3048–3064.e9. doi:10.1016/j.molcel.2021.06.014
Fan, Y., Li, X., Sun, H., Gao, Z., Zhu, Z., and Yuan, K. (2022). Role of WTAP in cancer: From mechanisms to the therapeutic potential. Biomolecules 12 (9), 1224. doi:10.3390/biom12091224
Fang, C., He, M., Li, D., and Xu, Q. (2021). YTHDF2 mediates LPS-induced osteoclastogenesis and inflammatory response via the NF-κB and MAPK signaling pathways. Cell. Signal 85, 110060. doi:10.1016/j.cellsig.2021.110060
Frayling, T. M., Timpson, N. J., Weedon, M. N., Zeggini, E., Freathy, R. M., Lindgren, C. M., et al. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316 (5826), 889–894. doi:10.1126/science.1141634
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Sci. (New York, N.Y.) 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. J. Sci. 318 (5855), 1469–1472. doi:10.1126/science.1151710
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347 (6225), 1002–1006. doi:10.1126/science.1261417
Gu, S., Sun, D., Dai, H., and Zhang, Z. (2018). N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol. Lett. 292, 1–11. doi:10.1016/j.toxlet.2018.04.018
Havell, E. A., Fiers, W., and North, R. J. (1988). The antitumor function of tumor necrosis factor (TNF), I. Therapeutic action of TNF against an established murine sarcoma is indirect,immunologically dependent, and limited by severe toxicity. J. Exp. Med. 167 (3), 1067–1085. doi:10.1084/jem.167.3.1067
Hayden, M. S., and Ghosh, S. (2008). Shared principles in NF-kappaB signaling. Cell. 132 (3), 344–362. doi:10.1016/j.cell.2008.01.020
He, H., Hu, Z., Xiao, H., Zhou, F., and Yang, B. (2018). The tale of histone modifications and its role in multiple sclerosis. Hum. Genomics 12 (1), 31. doi:10.1186/s40246-018-0163-5
He, Y., Wang, W., Xu, X., Yang, B., Yu, X., Wu, Y., et al. (2022). Mettl3 inhibits the apoptosis and autophagy of chondrocytes in inflammation through mediating Bcl2 stability via Ythdf1-mediated m6A modification. Bone 154, 116182. doi:10.1016/j.bone.2021.116182
Horowitz, S., Horowitz, A., Nilsen, T. W., Munns, T. W., and Rottman, F. M. (1984). Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci. U. S. A. 81 (18), 5667–5671. doi:10.1073/pnas.81.18.5667
Hu, M., Yang, Y., Ji, Z., and Luo, J. (2016). RBM15 functions in blood diseases. Curr. cancer drug targets 16 (7), 579–585. doi:10.2174/1568009616666160112105706
Huang, J., Shao, Y., and Gu, W. (2021). Function and clinical significance of N6-methyladenosine in digestive system tumours. Exp. Hematol. Oncol. 10 (1), 40. doi:10.1186/s40164-021-00234-1
Huang, L., Tang, H., and Hu, J. (2022). METTL3 attenuates inflammation in Fusarium solani-induced keratitis via the PI3K/AKT signaling pathway. Invest. Ophthalmol. Vis. Sci. 63 (10), 20. doi:10.1167/iovs.63.10.20
Idriss, H. T., and Naismith, J. H. (2000). TNF alpha and the TNF receptor superfamily: Structure-function relationship(s). Microsc. Res. Tech. 50 (3), 184–195. doi:10.1002/1097-0029(20000801)50:3<184::AID-JEMT2>3.0.CO;2-H
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333 (6047), 1300–1303. doi:10.1126/science.1210597
Jacob, R., Zander, S., and Gutschner, T. (2017). The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 18 (11), 2387. doi:10.3390/ijms18112387
Jang, D. I., Lee, A. H., Shin, H. Y., Song, H. R., Park, J. H., Kang, T. B., et al. (2021). The role of tumor necrosis factor alpha (TNF-α) in autoimmune disease and current TNF-α inhibitors in therapeutics. Int. J. Mol. Sci. 22 (5), 2719. doi:10.3390/ijms22052719
Jian, D., Wang, Y., Jian, L., Tang, H., Rao, L., Chen, K., et al. (2020). METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics 10 (20), 8939–8956. doi:10.7150/thno.45178
Jiang, Y., Wan, Y., Gong, M., Zhou, S., Qiu, J., and Cheng, W. (2020). RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway. J. Cell. Mol. Med. 24 (11), 6137–6148. doi:10.1111/jcmm.15228
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6 (1), 74. doi:10.1038/s41392-020-00450-x
Koch, A. E., Harlow, L. A., Haines, G. K., Amento, E. P., Unemori, E. N., Wong, W. L., et al. (1994). Vascular endothelial growth factor. A cytokine modulating endothelial function in rheumatoid arthritis. J. Immunol. 152 (8), 4149–4156. doi:10.4049/jimmunol.152.8.4149
Kumar, D., Singla, S. K., Puri, V., and Puri, S. (2015). The restrained expression of NF-kB in renal tissue ameliorates folic acid-induced acute kidney injury in mice. PLoS One 10 (1), e115947. doi:10.1371/journal.pone.0115947
Landskron, G., De la Fuente, M., Thuwajit, P., Thuwajit, C., and Hermoso, M. A. (2014). Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014:149185. doi:10.1155/2014/149185
Lawrence, T. (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1 (6), a001651. doi:10.1101/cshperspect.a001651
Levin, A. D., Wildenberg, M. E., and van den Brink, G. R. (2016). Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J. Crohns Colitis 10 (8), 989–997. doi:10.1093/ecco-jcc/jjw053
Li, A., Gan, Y., Cao, C., Ma, B., Zhang, Q., Zhang, Q., et al. (2021). Transcriptome-wide map of N6-methyladenosine methylome profiling in human bladder cancer. Front. Oncol. 11, 717622. doi:10.3389/fonc.2021.717622
Li, H. B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017). m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 (7667), 338–342. doi:10.1038/nature23450
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Li, G., Ma, L., He, S., Luo, R., Wang, B., Zhang, W., et al. (2022). WTAP-mediated m6A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat. Commun. 13 (1), 1469. doi:10.1038/s41467-022-28990-6
Li, M., Deng, L., and Xu, G. (2021). METTL14 promotes glomerular endothelial cell injury and diabetic nephropathy via m6A modification of α-klotho. Mol. Med. 27 (1), 106. doi:10.1186/s10020-021-00365-5
Lin, Y. J., Anzaghe, M., and Schülke, S. (2020). Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells 9 (4), 880. doi:10.3390/cells9040880
Liou, G. Y., and Storz, P. (2010). Reactive oxygen species in cancer. Free Radic. Res. 44 (5), 479–496. doi:10.3109/10715761003667554
Liu, J., and Jia, G. (2014). Methylation modifications in eukaryotic messenger RNA. J. Genet. Genomics 41 (1), 21–33. doi:10.1016/j.jgg.2013.10.002
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, S., Zhuo, L., Wang, J., Zhang, Q., Li, Q., Li, G., et al. (2020). METTL3 plays multiple functions in biological processes. Am. J. cancer Res. 10 (6), 1631–1646.
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
Peng, W., Li, J., Chen, R., Gu, Q., Yang, P., Qian, W., et al. (2019). Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 38 (1), 393. doi:10.1186/s13046-019-1408-4
Pobezinskaya, Y. L., and Liu, Z. (2012). The role of TRADD in death receptor signaling. Cell. Cycle 11 (5), 871–876. doi:10.4161/cc.11.5.19300
Qin, Y., Li, B., Arumugam, S., Lu, Q., Mankash, S. M., Li, J., et al. (2021). m6A mRNA methylation-directed myeloid cell activation controls progression of NAFLD and obesity. Cell. Rep. 37 (6):109968. doi:10.1016/j.celrep.2021.109968
Ruland, J. (2011). Return to homeostasis: Downregulation of NF-κB responses. Nat. Immunol. 12 (8), 709–714. doi:10.1038/ni.2055
Sang, W., Xue, S., Jiang, Y., Lu, H., Zhu, L., Wang, C., et al. (2021). METTL3 involves the progression of osteoarthritis probably by affecting ECM degradation and regulating the inflammatory response. Life Sci. 278, 119528. doi:10.1016/j.lfs.2021.119528
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Song, H., Song, J., Cheng, M., Zheng, M., Wang, T., Tian, S., et al. (2021). METTL3-mediated m6A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat. Commun. 12 (1), 5522. doi:10.1038/s41467-021-25803-0
Sun, T., Wu, R., and Ming, L. (2019). The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 112, 108613. doi:10.1016/j.biopha.2019.108613
Sun, S. C. (2011). Non-canonical NF-κB signaling pathway. Cell. Res. 21 (1), 71–85. doi:10.1038/cr.2010.177
Surace, A. E. A., and Hedrich, C. M. (2019). The role of epigenetics in autoimmune/inflammatory disease. Front. Immunol. 10, 1525. doi:10.3389/fimmu.2019.01525
Udalova, I., Monaco, C., Nanchahal, J., and Feldmann, M. (2016). Anti-TNF therapy. Microbiol. Horizonium 4 (4). doi:10.1128/microbiolspec.mchd-0022-2015
Uddin, M. B., Wang, Z., and Yang, C. (2021). The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20 (1), 61. doi:10.1186/s12943-021-01356-0
van Horssen, R., Ten Hagen, T. L., and Eggermont, A. M. (2006). TNF-Alpha in cancer treatment: Molecular insights, antitumor effects, and clinical utility. Oncologist 11 (4), 397–408. doi:10.1634/theoncologist.11-4-397
Wang, J. Y., and Lu, A. Q. (2021).The biological function of m6A reader YTHDF2 and its role in human disease. Cancer Cell. Int. 21, 109. doi:10.1186/s12935-021-01807-0
Wang, B., Liu, Y., Jiang, R., Liu, Z., Gao, H., Chen, F., et al. (2022). Emodin relieves the inflammation and pyroptosis of lipopolysaccharide-treated 1321N1 cells by regulating methyltransferase-like 3 -mediated NLR family pyrin domain containing 3 expression. Bioengineered 13 (3), 6740–6749. doi:10.1080/21655979.2022.2045836
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, J., Yan, S., Lu, H., Wang, S., and Xu, D. (2019). METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-κB signaling pathway. Mediat. Inflamm. M. 2019, 3120391. doi:10.1155/2019/3120391
Wang, J., Wang, F., Ke, J., Li, Z., Xu, C. H., Yang, Q., et al. (2022). Inhibition of METTL3 attenuates renal injury and inflammation by alleviating TAB3 m6A modifications via IGF2BP2-dependent mechanisms. Sci. Transl. Med. 14 (640), eabk2709. doi:10.1126/scitranslmed.abk2709
Wang, L., Wang, F. S., and Gershwin, M. E. (2015). Human autoimmune diseases: A comprehensive update. J. Intern Med. 278 (4), 369–395. doi:10.1111/joim.12395
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Wang, R., Yao, B., Hu, T., Li, Z., Liu, Y., et al. (2019). TNF-α suppresses sweat gland differentiation of MSCs by reducing FTO-mediated m6A-demethylation of Nanog mRNA. Sci. China Life Sci. 62, 80–91. doi:10.1007/s11427-019-9826-7
Wehkamp, J., Götz, M., Herrlinger, K., Steurer, W., and Stange, E. F. (2016). Inflammatory bowel disease. Dtsch. Arztebl Int. 113 (5), 72–82. doi:10.3238/arztebl.2016.0072
Wei, J., and He, C. (2021). Chromatin and transcriptional regulation by reversible RNA methylation. Curr. Opin. Cell. Biol. 70, 109–115. doi:10.1016/j.ceb.2020.11.005
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 71 (6), 973–985.e5. doi:10.1016/j.molcel.2018.08.011
Wertz, I. E., and Dixit, V. M. (2010). Signaling to NF-kappaB: Regulation by ubiquitination. Cold Spring Harb. Perspect. Biol. 2 (3), a003350. doi:10.1101/cshperspect.a003350
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m (6) A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xie, Z., Yu, W., Zheng, G., Li, J., Cen, S., Ye, G., et al. (2021). TNF-α-mediated m6A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat. Commun. 12 (1), 5373. doi:10.1038/s41467-021-25710-4
Yang, S., Wang, J., Brand, D. D., and Zheng, S. G. (2018). Role of TNF-TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 9, 784. doi:10.3389/fimmu.2018.00784
Yu, Y., Pan, Y., Fan, Z., Xu, S., Gao, Z., Ren, Z., et al. (2021). LuHui derivative, A novel compound that inhibits the Fat mass and obesity-associated (FTO), alleviates the inflammatory response and injury in hyperlipidemia-induced cardiomyopathy. Front. Cell. Dev. Biol. 9, 731365. doi:10.3389/fcell.2021.731365
Yu, H., Ma, X. D., Tong, J. F., Li, J. Q., Guan, X. J., Yang, J. H., et al. (2019). WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther. 12, 6191–6201. doi:10.2147/OTT.S205730
Yu, R., Li, Q., Feng, Z., Cai, L., and Xu, Q. (2019). m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int. J. Mol. Sci. 20 (6), 1323. doi:10.3390/ijms20061323
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes. Dev. 29 (13), 1343–1355. doi:10.1101/gad.262766.115
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell. Biol. 20 (10), 608–624. doi:10.1038/s41580-019-0168-5
Zeng, C., Huang, W., Li, Y., and Weng, H. (2020). Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 13 (1), 117. doi:10.1186/s13045-020-00951-w
Zhang, C., Fu, J., and Zhou, Y. (2019). A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 10, 922. doi:10.3389/fimmu.2019.00922
Zhang, M., Wang, J., Jia, L., Huang, J., He, C., Hu, F., et al. (2017). Transmembrane TNF-α promotes activation-induced cell death by forward and reverse signaling. Oncotarget 8 (38), 63799–63812. doi:10.18632/oncotarget.19124
Zhang, B., Jiang, H., Dong, Z., Sun, A., and Ge, J. (2020). The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases. Genes. Dis. 8 (6), 746–758. doi:10.1016/j.gendis.2020.07.011
Zhang, N., Zuo, Y., Peng, Y., and Zuo, L. (2021). Function of N6-methyladenosine modification in tumors. J. Oncol. 2021, 6461552. doi:10.1155/2021/6461552
Zhang, S., Guan, X., Liu, W., Zhu, Z., Jin, H., Zhu, Y., et al. (2022a). YTHDF1 alleviates sepsis by upregulating WWP1 to induce NLRP3 ubiquitination and inhibit caspase-1-dependent pyroptosis. Cell. Discov. 8 (1), 244. caspase-4. doi:10.1038/s41420-022-00872-2
Zhang, T., Ding, C., Chen, H., Zhao, J., Chen, Z., Chen, B., et al. (2022b). m6A mRNA modification maintains colonic epithelial cell homeostasis via NF-κB-mediated antiapoptotic pathway. Sci. Adv. 8 (12), eabl5723. doi:10.1126/sciadv.abl5723
Zhang, X., Deng, S., Peng, Y., Wei, H., and Tian, Z. (2022c). ALKBH5 inhibits TNF-α-induced apoptosis of HUVECs through Bcl-2 pathway. Open Med. (Wars) 17 (1), 1092–1099. doi:10.1515/med-2022-0484
Zhang, X., Li, M. J., Xia, L., and Zhang, H. (2022d). The biological function of m6A methyltransferase KIAA1429 and its role in human disease. PeerJ 10, e14334. doi:10.7717/peerj.14334
Zhang, J., Bai, R., Li, M., Ye, H., Wu, C., Wang, C., et al. (2019). Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 10 (1), 1858. doi:10.1038/s41467-019-09712-x
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell. Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, W., Xiao, S., Li, H., Zheng, T., Huang, J., Hu, R., et al. (2018). MAPK phosphatase-1 deficiency exacerbates the severity of imiquimod-induced psoriasiform skin disease. Front. Immunol. 9, 569. doi:10.3389/fimmu.2018.00569
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, Q., Lin, Z., Li, X., Xin, X., Wu, M., An, J., et al. (2016). Inflammatory cytokine IL6 cooperates with CUDR to aggravate hepatocyte-like stem cells malignant transformation through NF-κB signaling. Sci. Rep. 6, 36843. doi:10.1038/srep36843
Keywords: N6-methyladenosine (m6A), TNF-α, signaling pathway, immunity, autoimmune disease
Citation: Wang Y, Liu J and Wang Y (2023) Role of TNF-α-induced m6A RNA methylation in diseases: a comprehensive review. Front. Cell Dev. Biol. 11:1166308. doi: 10.3389/fcell.2023.1166308
Received: 15 February 2023; Accepted: 13 July 2023;
Published: 24 July 2023.
Edited by:
Suresh Kumar, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Praveen K. Dubey, University of Alabama at Birmingham, United StatesJiangbo Wei, The University of Chicago, United States
Copyright © 2023 Wang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchen Wang, eW9uZ2NoZW53YW5nQDE2My5jb20=
†These authors have contributed equally to this work
 Youlin Wang
Youlin Wang Jing Liu1†
Jing Liu1†