- 1Department of Dentistry—Orthodontics and Craniofacial Biology, Radboud Institute of Molecular Life Sciences (RIMLS), Radboud University Medical Center, Nijmegen, Netherlands
- 2Department of Animal Ecology and Physiology, Radboud Institute for Biological and Environmental Sciences (RIBES), Radboud University, Nijmegen, Netherlands
- 3Department of Human Genetics, Radboud University Medical Center, Nijmegen, Netherlands
Introduction: Mutations in the FOXE1 gene are implicated in cleft palate and thyroid dysgenesis in humans.
Methods: To investigate whether zebrafish could provide meaningful insights into the etiology of developmental defects in humans related to FOXE1, we generated a zebrafish mutant that has a disruption in the nuclear localization signal in the foxe1 gene, thereby restraining nuclear access of the transcription factor. We characterized skeletal development and thyroidogenesis in these mutants, focusing on embryonic and larval stages.
Results: Mutant larvae showed aberrant skeletal phenotypes in the ceratohyal cartilage and had reduced whole body levels of Ca, Mg and P, indicating a critical role for foxe1 in early skeletal development. Markers of bone and cartilage (precursor) cells were differentially expressed in mutants in post-migratory cranial neural crest cells in the pharyngeal arch at 1 dpf, at induction of chondrogenesis at 3 dpf and at the start of endochondral bone formation at 6 dpf. Foxe1 protein was detected in differentiated thyroid follicles, suggesting a role for the transcription factor in thyroidogenesis, but thyroid follicle morphology or differentiation were unaffected in mutants.
Discussion: Taken together, our findings highlight the conserved role of Foxe1 in skeletal development and thyroidogenesis, and show differential signaling of osteogenic and chondrogenic genes related to foxe1 mutation.
Introduction
Disruption of sonic hedgehog signaling during development can result in varying craniofacial malformations, as observed in clinical examples and confirmed in animal models (Wada et al., 2005; Xavier et al., 2016). Forkhead box transcription factor E 1 (FOXE1, also known as TTF-2) is a transcriptional regulator associated with the SHH pathway, that uses a forkhead DNA-binding domain to regulate transcription of downstream targets (Clifton-Bligh et al., 1998; Brancaccio et al., 2004; Eichberger et al., 2004). FOXE1 can be activated by GLI2 and regulates the transcription of genes involved in craniofacial development, including a transcription factor (MSX1) and growth factors (TGFβ3, WNT5A) and regulates thyroid differentiation factors (TPO, NIS, TG) (Venza et al., 2011; Lopez-Marquez et al., 2019).
FOX-family proteins and their characteristic DNA-binding domains are highly conserved among vertebrates (Figure 1). FOXE1 is a single exon gene containing two functional regions; the forkhead DNA-binding domain and a 16–19 residue polyalanine stretch (Zannini et al., 1996). Cytoplasmic FOXE1 gains entry to the nucleus by two identical FHD-flanking nuclear localization signals. These localization signals facilitate nuclear entry of FOXE1 by binding to importin-α upon which it is transported by importin-β (McLane and Corbett 2009). FOXE1 is also retained in the nucleus by NLS activity, thus the NLSs are paramount for transcriptional regulation by FOXE1 (Romanelli et al., 2003; Castanet and Polak 2010).

FIGURE 1. Foxe1 protein structure in human, mouse and zebrafish. Multiple sequence alignment of full-length human, mouse and zebrafish FOXE1 protein. The protein contains three characterized domains which are annotated with colored lines. The similarity of the Forkhead domain (FHD) is 97% and 100%, respectively, between zebrafish and humans and between mouse and human. The sequence similarity of the whole protein between human and zebrafish is 54%. NLS is 100% identical between all species and the poly alanine stretch (PAS) is only present in humans and mouse. Red boxes mark the putative DNA-binding residues and asterisks (*) mark locations of missense mutations that were reported in individuals with Bamforth–Lazarus syndrome (Clifton-Bligh et al., 1998; Castanet et al., 2002; Baris et al., 2006; Castanet and Polak 2010; Carre et al., 2014; Sarma et al., 2022). NLS: nuclear localization signal, FHD: forkhead box domain PAS: poly alanine stretch in human and mouse FOXE1. Alignment made using Clustal Omega.
Homozygous loss- and gain-of-function missense mutations in the forkhead domain of FOXE1 cause Bamforth–Lazarus syndrome, which is characterized by thyroid dysgenesis, cleft palate, spiky hair, as well as, in some cases, choanal atresia and bifid epiglottis (Bamforth et al., 1989; Castanet and Polak 2010; Carre et al., 2014). Very recently, a novel homozygous protein-truncating frameshift mutation in FOXE1 outside the FHD (Leu29Profs*75) was also reported to cause Bamforth–Lazarus syndrome (Sarma et al., 2022). Bamforth–Lazarus syndrome is an extremely rare condition with an incidence of <1:1.000.000. However, variants in the FOXE1 locus have also been shown to contribute significantly to non-syndromic orofacial clefts in multiple case-control and GWAS studies. Indeed, the gene is thought to contribute substantially to both CP and CLP cases (Moreno et al., 2009; Dixon et al., 2011; Lammer et al., 2016; Leslie et al., 2017).
How FOXE1 is involved in palate formation is largely unknown, and etiological mechanisms for FOXE1-related orofacial clefts remain therefore elusive. This prompts the need for research models to investigate genotype-phenotype associations. Foxe1−/− mice feature severe cleft palate and the migration of the thyroid primordium is blocked in these mutants, resulting in thyroid dysgenesis (De Felice et al., 1998). Indeed, these phenotypes partly recapitulate the symptoms of Bamforth-Lazarus syndrome, yet Foxe1−/− mice die upon birth due to the extent of the clefts. This limits the use of this model for post-embryonic research and there is thus a need for alternative models that can provide insight into the role of FOXE1 in pathogenesis.
Zebrafish allow the modeling of human congenital craniofacial disorders since developmental processes are well-conserved during evolution (Swartz et al., 2011; Mork and Crump 2015). Cartilage and bone elements of zebrafish and other vertebrates’ craniofacial skeleton originate primarily from cranial neural crest cells. Induction in the neural plate, epithelial-mesenchymal transition, and subsequent migration of CNCC to the pharyngeal arches of this vertebrate-specific cell population occurs in zebrafish within the first 24 h after fertilization (Schilling and Kimmel 1994; Kague et al., 2012). The craniofacial cartilages subsequently develop from the pharyngeal arches, and then function as a template for bone development. In vertebrate skeletal development, three modes of bone formation are described, which all occur in zebrafish. First, endochondral ossification describes the formation of bone from within a cartilage template, starting at a primary ossification center in the cartilage structure. Second, intramembranous or dermal ossification describes direct bone formation without a cartilage template. Third, perichondral ossification refers to the covering of a cartilage template with bone in a sheath-like manner (Tonelli et al., 2020; Valenti et al., 2020; Dietrich et al., 2021).
A zebrafish study using morpholino-mediated knock-down of foxe1 reported defects in chondrogenesis, specifically shortening of the Meckel’s cartilage, and inverted or shortened ceratohyal cartilages (Nakada et al., 2009). In the same study, fgfr2 upregulation was indicated as the main mediator of the detrimental effects on the cranial skeleton. In addition, the role of foxe1 in thyroid development in zebrafish was questioned, as no differences in the expression of thyroid markers were observed upon knock-down of foxe1 (Nakada et al., 2009). Today, stable mutant lines are considered in most instances superior over morphants, due to non-specific binding artifacts and p53-mediated apoptosis in morphants (Schulte-Merker and Stainier 2014; Kok et al., 2015; Duncan et al., 2017; El-Brolosy and Stainier 2017). Importantly, morpholinos induce transient knock-downs only, of which effects wear off before the onset of mineralization, while stable mutant lines allow the study of effects during the entire lifespan.
To investigate whether a zebrafish foxe1 mutant model could provide meaningful insights into human developmental defects related to FOXE1, the present study aimed to characterize the function of Foxe1 in craniofacial development and thyroidogenesis in zebrafish. We generated a foxe1 mutant and observed skeletal mineralization defects and CNCC dysregulation. We also found that the expression of genes involved in osteogenesis and chondrogenesis in early development are modulated by foxe1 disruption.
Results
Foxe1 is expressed in multiple larval tissues, including the oral epithelium, ethmoid plate, and the thyroid follicles
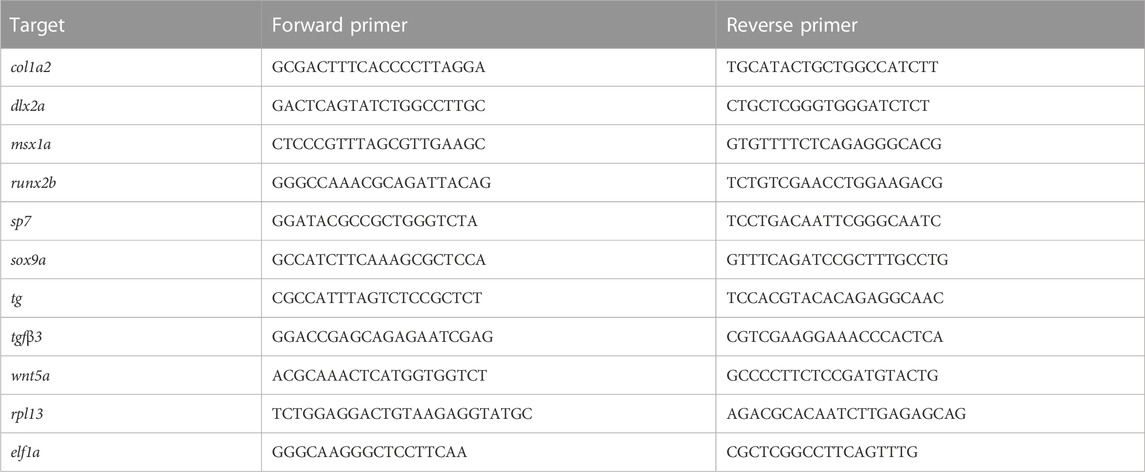
We first assessed spatiotemporal foxe1 expression at both the transcriptional and the translational level in wild type zebrafish. Foxe1 was diffusely detected by in situ hybridization at 12.5 hpf (Figure 2A) until 24 hpf in the head and notochord (Figures 2B–D, arrowheads in Figure 2C). A diffuse foxe1 expression pattern was observed until 48 hpf with increased intensity in the thyroid primordium (Figure 2E); after that, foxe1 continued to be expressed throughout the larvae and was specifically intense in the (sub)pharyngeal area (Figure 2F). At 96 hpf, foxe1 expression was most abundant and was also observed in the cartilages of the viscerocranium (Figures 2G, I). Contrary to previous reports of foxe1 spatiotemporal expression in the developing zebrafish larvae (Nakada et al., 2009; Lidral et al., 2015), in the present study foxe1 expression was observed in the viscerocranial structures at 120 hpf. However, this expression was lower than that at 96 hpf (Figures 2H, I). Normalized gene expression levels reflected the patterns of foxe1 expression throughout early development and indeed showed a sharp increase between 48 and 72 hpf as thyroid morphogenesis and craniofacial cartilage formation commenced (Figure 2I).
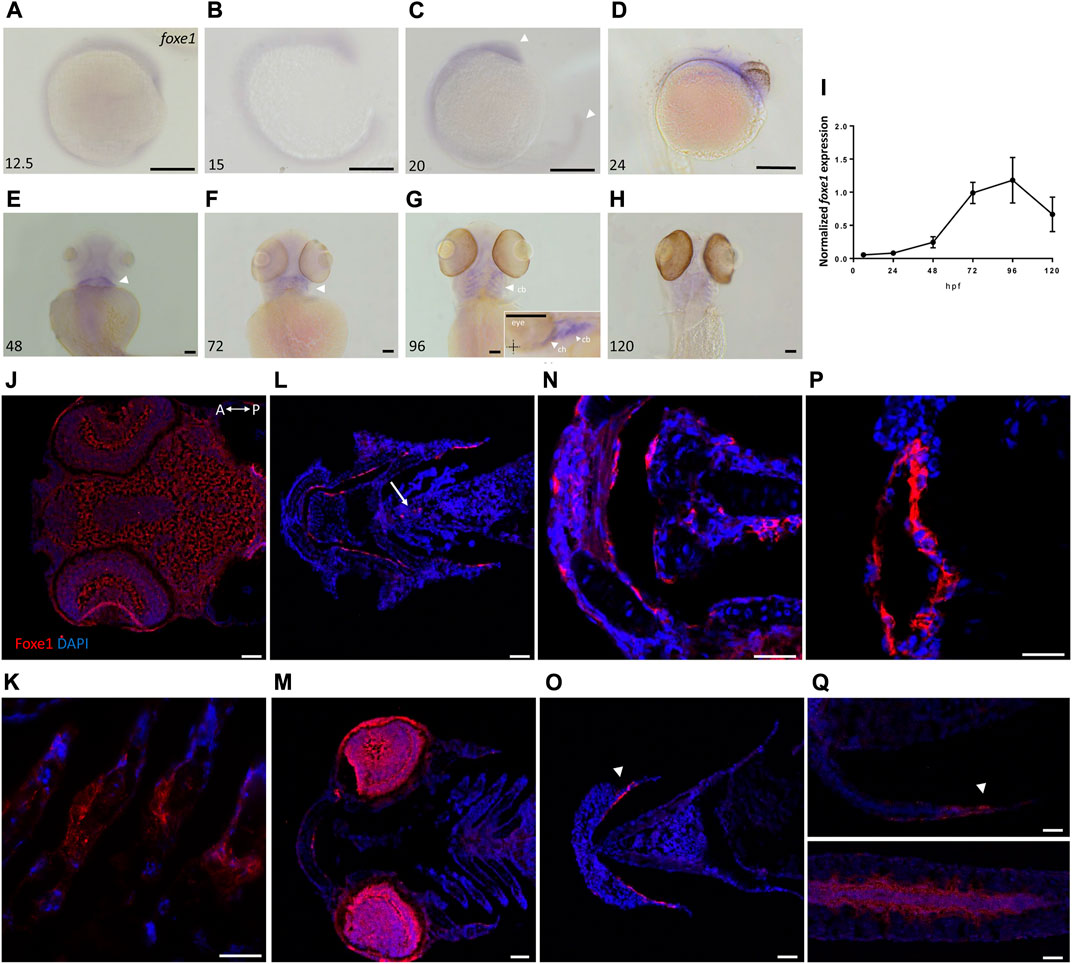
FIGURE 2. Foxe1 is expressed in a range of larval tissues, including the oral epithelium, ethmoid plate, and the brain. (A–H) foxe1 transcripts were detected diffusely throughout the embryo from early development at 12.5 hpf (A) which continued through early larval stages (B–D) where transcripts also seemed specifically upregulated in the subpharyngeal area (E, F) before being enhanced in the ceratobranchials and ceratohyal ((G), insert lateral view). (A-D) lateral view and (E-H) ventral view. (I) Normalized relative expression of foxe1 during embryonic and larval stages as determined by qPCR. Error bars indicate standard deviations. (J-Q) Detailed protein localization at 96 hpf showed Foxe1-positive cells in the brain (J), the ceratobranchials (K), in the oral epithelium and in a linear cluster in the subpharyngeal area (white arrow) (L), the eyes (M), on the ethmoid plate (N), on the lining of the ceratohyal (O), in the mouth opening (P) and to some extent in the fin tips (Q), upper image) and in the notochordal sheath ((Q), lower image). Posterior-anterior axis in J applies to all images. cb: ceratobranchals, ch: ceratohyal. A-H scalebar 100 μm, (J-Q) scalebar 50 µm. Antibody validation in Supplementary Figure S1.
Foxe1 was most abundantly expressed at 96 hpf, and a more detailed localization of the protein at this timepoint was examined by immunohistochemistry. Throughout the head we found Foxe1-positive cells, many of which would have been too scattered to be distinguished by whole mount in situ hybridization. Foxe1 was localized in the brain (Figure 2J), ceratobranchials (Figure 2K), oral epithelium (Figure 2L), eyes (Figure 2M), ethmoid plate (Figure 2N), lining of the ceratohyal cartilage (Figure 2O), mouth opening (Figure 2P), and, to some extent, in the tips of the fins and the notochordal sheath (Figure 2Q).
Foxe1-positive cells were also consistently observed in linear clusters in the subpharyngeal area at 96 hpf (indicated by the white arrow in Figure 2L). As Foxe1 is important in thyroid development in many vertebrates, we set out to establish whether this signal originated from the developing thyroid follicles. The zebrafish thyroid is not organized as a discrete gland, but as individual follicles scattered along the ventral aorta (Alt et al., 2006). These follicles appear from 72 to 96 hpf and can be visualized using thyroid hormone (T4) antibody staining (Zhang et al., 2021). Figure 3 shows two sequential sections of the same individual stained with Foxe1-and T4-antibodies. Thyroid hormone was observed in the colloid lumen of the thyroid follicle, where it is produced from its precursor thyroglobulin. Colocalization of the T4 signal with Foxe1 on follicular level was clearly detected and strongly supports a role for Foxe1 in the developing thyroid.
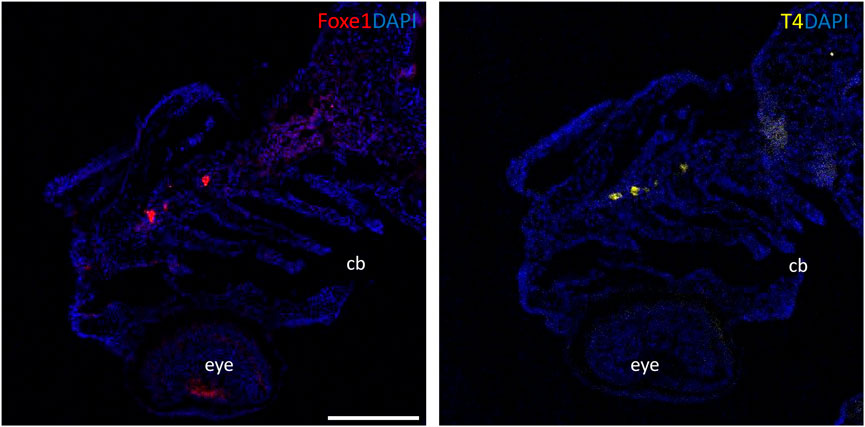
FIGURE 3. Foxe1 and T4 are colocalized in the subpharyngeal area. Foxe1 and T4 staining on adjacent coronal sections of the subpharyngeal area of the larval head. T4 and Foxe1 staining in the developing thyroid follicles at 96 hpf. Nuclear counterstain with DAPI. cb; ceratobranchials, e; eye. Scale bar 200 µm.
Foxe1 mutants show enhanced Foxe1 expression, normal growth rates, and normal thyroid follicle morphology
Although the role of Foxe1 in zebrafish thyroid follicle development was previously questioned, we here show that Foxe1 is expressed in the developing follicles (Nakada et al., 2009). To better understand the role of Foxe1 in craniofacial development and thyroidogenesis, we generated a foxe1 mutant zebrafish line, using CRISPR-Cas9 targeting 5’ end of the single foxe1 exon. Whilst screening the F1 generation, in-frame indels were identified with a 6-bp deletion and 3-bp insertion that we outcrossed to obtain a stable mutant line (which we termed foxe1^rdb2). These indels lead to an arginine deletion and arginine to leucine substitution in the NLS domain, resulting in the following amino acid sequence change: “RRRKR” to “RKKR” (Figures 4A, B). According to 3D structural predictions by homology modeling of the winged helix domain of Foxe1 in YASARA, this amino acid change alters the structure and function of the NLS domain in this mutant, affecting nuclear translocation and retention of Foxe1 (Supplementary Figure S2). Indeed, subcellular localization of Foxe1 shows that in wild types the protein is present inside the cell nucleus, while in mutants the disrupted NLS failed to translocate and retain Foxe1 into the nucleus and it was found to be localized mostly in the cytoplasm (Figure 4C).
FIGURE 4. The foxe1rdb2 mutant. (A) Wild type and foxe1rdb2 mutant DNA sequence at (C) 84–105. The protospacer adjacent motif is underlined (full gRNA sequence: 5′-GCCGCAAAGAGGCCGTCGGAGG-3′). The deletion is highlighted in red and the insertion in green. (B) Schematic representation of the zebrafish Foxe1 protein and the position of the amino acid changes as indicated by the red bars. (C) Nuclear localization of wild type Foxe1 and cytoplasmatic localization of mutant Foxe1 in zebrafish keratinocytes on scale. Scale bar 50 µm. (D) Representative images of wild types and foxe1 mutants at 6 dpf and 2 months post fertilization. Scale bars 1 mm.
To assess the overall fitness of the homozygous foxe1rdb2 mutants (referred to as mutant or −/− in the rest of the manuscript), survival, general morphology and growth rate analyses up to juvenile stages were performed in the F3 generation, according to previously established protocols (Parichy et al., 2009; Beekhuijzen et al., 2015). General morphology analyses included tracking of tail formation, pericardial development, and swim bladder formation. No significant differences between wild type, heterozygote, and homozygous mutant fish were observed in growth, development or survival (data not shown). Both the heterozygous and homozygous mutants were viable up to adulthood and fertile (Figure 4D). The immuno-localization of Foxe1 in different tissues was not altered by the in-frame mutation, but a more widespread abundance in the mutants was found (Figure 5). Overexpression of a mutated allele is a common feature of genetic mutants and can indicate loss-of-function (El-Brolosy et al., 2019).
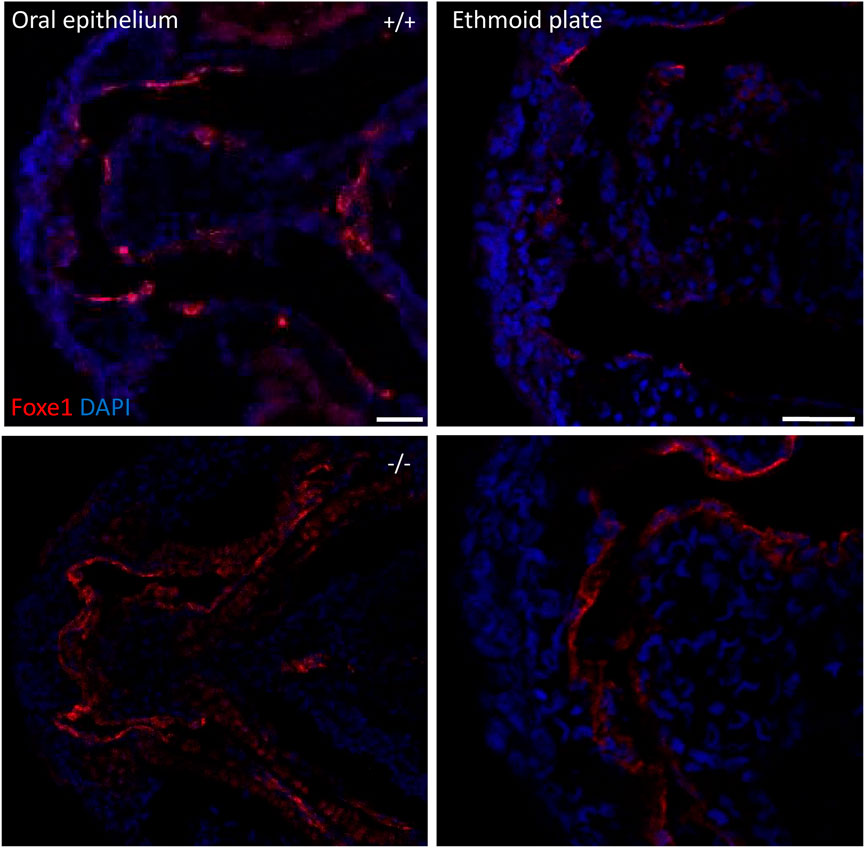
FIGURE 5. foxe1 mutants show upregulated Foxe1 expression. Foxe1 expression in the oral epithelium and the ethmoid plate of wild type and foxe1 mutants at 96 hpf. Foxe1 was more abundantly expressed in the mutants compared to the wild types, but the tissue localization of the protein remained unaffected. Scale bar 50 µm.
The effect of the mutation on the development of the thyroid follicles was assessed by gene expression of thyroglobulin (tg) and morphological analysis of the thyroid follicles at 6 dpf. The foxe1 mutants showed no statistically significant gene expression differences in tg at 3 dpf compared to wild types (Figure 6A). tg expression reflects the function of thyroid follicles (De Felice et al., 1998). Follicle morphology at 6 dpf by T4 antibody staining showed that there were no differences in average number nor size (by volume) of differentiated follicles per individual between mutants and wild types (Figures 6B–D).
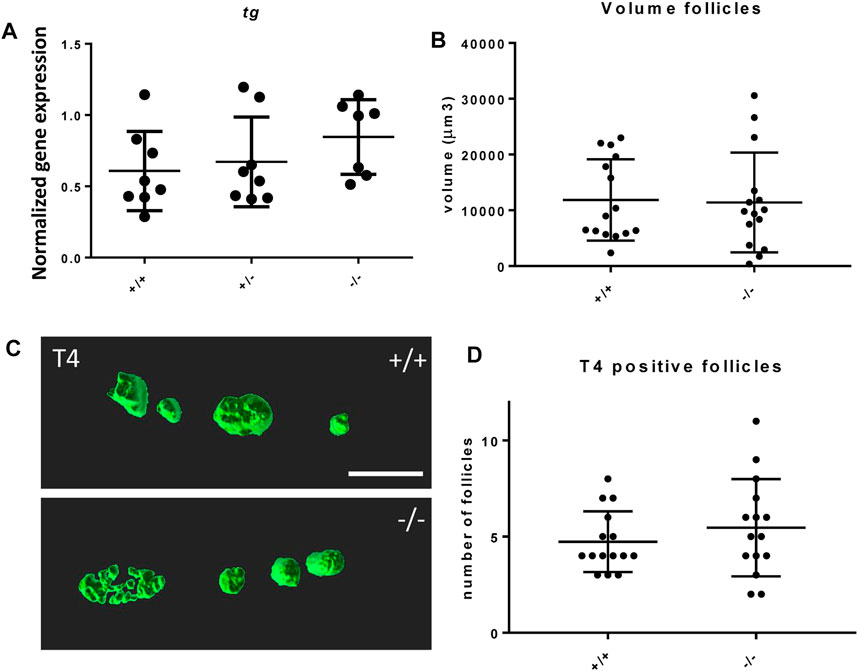
FIGURE 6. Thyroid follicle development in foxe1 mutants. (A) Normalized gene-expression of thyroid marker thyroglobulin during early development (3 dpf). (B, D) Analysis of T4 positive follicles by whole mount immunohistochemistry in 6 dpf larvae showed no difference in mean follicle count or volume. (C) Representative images of wild type and mutant thyroid follicles. Larvae were imaged from the ventral side and T4-positive follicle surfaces were rendered from z-stack images using Imaris 9.0 as previously described (Zhang et al., 2021). Scale bar 200 µm. (D) Data were assessed for normality with the D’Agostino-Pearson normality test. Normally distributed data were analyzed for statistical differences using a one-way ANOVA and post hoc Tukey test or unpaired t-test. Non-parametric data were compared with a Kruskal–Wallis test with post hoc Dunn’s Multiple comparison test or Mann-Whitney test. Error bars indicate standard deviation.
Bone and cartilage formation are adversely affected in the foxe1 mutants
We next studied the effects of the foxe1 mutation on craniofacial development. In 19% and 36% percent of heterozygote and homozygote mutants, respectively, a kink in one or both ceratohyal cartilages was observed upon bone and cartilage staining at 8 dpf (Figures 7A, B). We observed this specific collapse after staining, but not during live-imaging studies of 5 dpf zebrafish larvae carrying the foxe1 mutation homozygously in a transgenic col2a1a:mCherry background (Figure 7B’). The morphology of other bone and cartilage structures in the larval head was unaffected. Measurements of head skeletal structures showed no differences between mutants and wild types (Supplementary Figure S3). The larvae’s total body length and the number of mineralized vertebrae at 8 dpf were unaffected (Figures 7C, D). An analysis of the total molar content of essential elements revealed a reduction in the levels of calcium, magnesium, and phosphorus in the foxe1 mutants at 8 dpf, indicating a more fragile bone structure in mutants (Figures 7E–G).
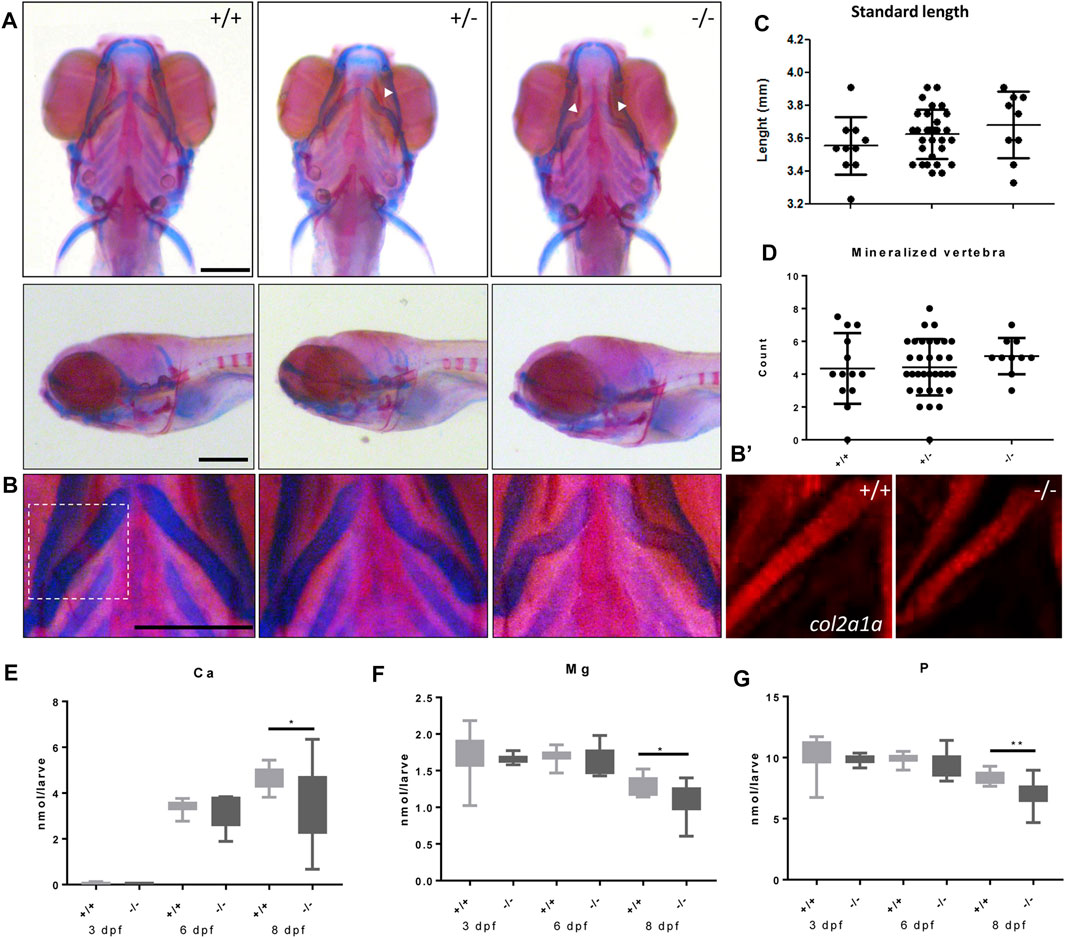
FIGURE 7. Malformed ceratohyal cartilage and reduced content of Ca, Mg and P in foxe1 mutants. (A) Representative images of wild types and foxe1 heterozygous- and homozygous mutants, ventral and lateral view. (B) High magnification images of ceratohyal phenotype. (B′) Col2a1a positive cells in the in ceratohyal. (C) Standard length at 8 dpf. (D) Count of mineralized vertebrae in foxe1 mutants versus wild types at 8 dpf. (E–G) Molar calcium, magnesium and phosphorus content in foxe1 mutants versus wild type larvae during early skeletal development three to 8 dpf. n = 10–33. Scale bar 200 µm. Data were assessed for normality with the D’Agostino-Pearson normality test. Normally distributed data were analyzed for statistical differences using a one-way ANOVA and post-hoc Tukey test or unpaired t-test. Non-parametric data were compared with a Kruskal–Wallis test with post-hoc Dunn’s Multiple comparison test or Mann-Whitney test. Error bars indicate standard deviation.
To better understand the effects of the foxe1 mutation on head skeleton development, the relative expression of direct target genes of Foxe1 was determined. Tgfb3 was consistently upregulated in mutants during embryonic and larval development, suggesting a function for Foxe1 as a transcriptional repressor of this gene, as it is for some other genes (Figure 8A) (Perrone et al., 2000; Venza et al., 2011; Lane et al., 2014). Wtn5a, reported to be regulated by Foxe1, was also consistently upregulated in mutants (Figure 8B).
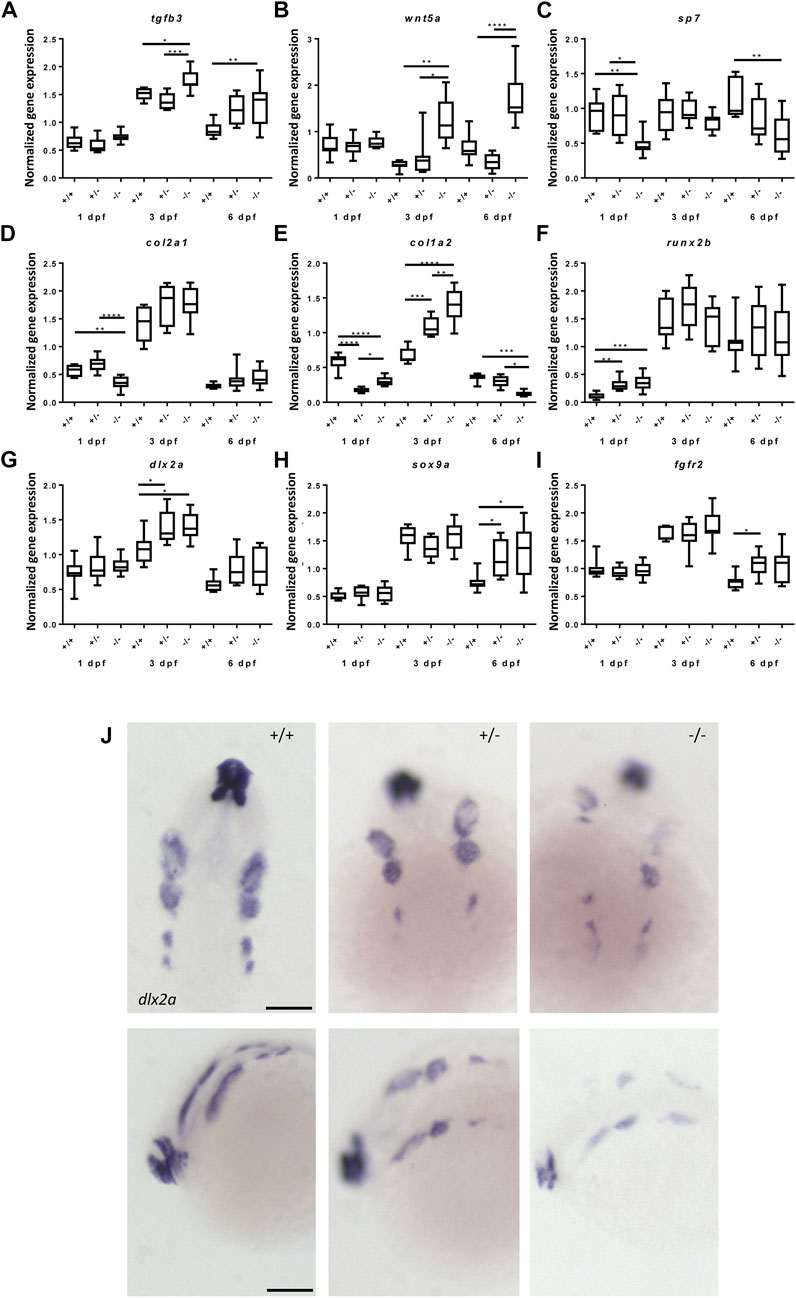
FIGURE 8. Gene expression of cartilage and bone (precursor) cells is affected in foxe1 mutants. (A-I) Relative gene expression of tgfβ3, wnt5a, sp7, col2a1, col1a2, runx2b, dlx2a, sox9a and fgfr2 in mutants versus wild types. Axis description from (A) applies to all graphs. (J) dlx2a positive post migratory neural crest cells at 24 hpf (Prim-5) in wild types, 20% of the heterozygous- and 60% of the homozygous mutants. Asterisks indicate the level of significance: * = p< 0.05, ** = p< 0.01, *** = p< 0.001, **** = p< 0.0001 (n = 10–15). Data were assessed for normality with the D’Agostino-Pearson normality test. Normally distributed data were analyzed using a one-way ANOVA and post-hoc Tukey test. Non-parametric data were compared with a Kruskal–Wallis test with post-hoc Dunn’s Multiple comparison test. Error bars indicate standard deviation. Scale bar 100 µm.
Because we observed apparent abnormalities in mineralization in the mutants, we assessed the gene expression of important markers of cartilage and bone development in mutants and wild types. Gene expression levels of sp7, col2a1 and col1a2 were downregulated at 1 dpf while runx2b was upregulated in foxe1 mutants (Figures 8C–F). At 3 dpf, col1a2 and dlx2a were upregulated in mutants, suggesting that both bone and cartilage development are (indirectly) affected by Foxe1 (Figures 8D,G). Sox9a was upregulated at 6 dpf, when it is expressed in both cartilage cells and developing osteoblasts in foxe1 mutant zebrafish (Figure 8H) (Eames et al., 2012). Col1a2 was differentially expressed in mutants over time but opposing effects on transcription of col1a2 were observed at three and 6 dpf (Figure 8E). During the latter timepoint, col1a2 is specifically expressed in the ceratohyal perichondrium (Eames et al., 2012). The osteoblast-specific transcription factor sp7 (Osterix) was significantly downregulated throughout development of foxe1 mutant zebrafish. We hypothesize that this is causally associated with the reduced amount of Ca, P, and Mg in mutants, as sp7 functions as a major regulator of bone development. Indeed, mineralization deficiencies were previously found in sp7 mutants (Kague et al., 2016; Niu et al., 2017; Chen et al., 2019).
Craniofacial malformations often originate from disrupted regulation of the CNCCs. To investigate whether the foxe1 mutation had effects on the CNCC cell populations, the spatiotemporal and quantitative expression of the CNCC marker dl2xa was assessed. Dlx2a relative gene expression by qPCR did not differ significantly between mutants and wild types at 24 hpf (Figure 8G). However, in situ hybridization showed that dlx2a positive CNCCs populations were affected in 20% of the heterozygous mutants and 60% of the homozygous mutants, this was not observed in wild types (Figure 8J). The various streams of CNCCs showed depletion of dlx2a-positive cells and appeared asymmetrically organized or populations were even entirely absent.
Discussion
The function of FOXE1 in most tissues is as yet not fully understood. Our study shows that foxe1 is involved in chondrogenesis and osteogenesis in early developmental stages. For the first time we show the localization of Foxe1 protein in developing zebrafish and demonstrate that it is widely expressed; specifically, Foxe1 was expressed in the eyes, the thyroid follicles, the brain, the oral epithelium, the notochordal sheath, and the fin tips. Staining intensity in in situ hybridization images did not equal signal strength upon antibody staining in our study. Previously, two reports described the spatiotemporal distribution of foxe1 transcripts from 11 to 96 hpf in the notochord, the thyroid primordium and the pharyngeal skeleton and oral epithelium (Nakada et al., 2009; Lidral et al., 2015). Nakada et al. reported Foxe1 transcripts in multiple adult zebrafish tissues, most strongly in the eyes, the brain, the gills, and the heart. In human embryos, FOXE1 was detected in the oropharyngeal epithelium and thymus, while proteomic expression data (www.ebi.ac.uk) suggest that it is much more widely expressed in adults (Trueba et al., 2005; Wang et al., 2019). In mice, expression of FOXE1 was specifically found in the thyroid, pharyngeal wall and arches, and in the tongue, the palate, the epiglottis, the pharynx, the esophagus, and the whiskers (Dathan et al., 2002). Together, these data indicate that the FOXE1 expression patterns are evolutionarily conserved across vertebrate species and ontogeny.
To better understand the function of FOXE1, we generated and characterized a zebrafish foxe1 mutant. We did not obtain frame-shift mutants, which may indicate these were early lethal. It is noteworthy that, as opposed to the previously used mouse and zebrafish models, Bamforth–Lazarus syndrome mutations typically impair FHD DNA-binding, but do not affect the C-terminal protein regions (Castanet and Polak 2010). Therefore, the previously described mouse knockout model may also not be fully representative for the clinical syndrome (De Felice et al., 1998). As a strategy to compromise Foxe1 function, we disrupted the N-terminal NLS sequence of foxe1. The N-terminal NLS is involved in nuclear retention while the C-terminal NLS is important for nuclear translocation (Romanelli et al., 2003). We observed a compromised nuclear localization and differential expression of genes downstream of Foxe1 in the NLS-disrupted mutants. Deregulation of NLS-dependent protein import into the nucleus has been linked to cancer and several developmental disorders (McLane and Corbett 2009). For instance, changes in the NLS sequence of the SRY gene cause Swyer syndrome because of reduced nuclear translocation of the protein (Harley et al., 2003).
Strict regulation of FOXE1 activity appears essential for normal development, as dysregulation has detrimental effects. In humans, two homozygous FOXE1 probands have been described with the mutations S57 N and A65 V (Clifton-Bligh et al., 1998; Castanet et al., 2002). These mutations in highly conserved FHD amino acids impair FOXE1 DNA binding capacity partly and completely, respectively (Castanet et al., 2002). Based on the phenotypes in these individuals, features of the Bamforth–Lazarus syndrome were deemed more extensive, as the capacity of FOXE1 DNA binding decreased (Castanet et al., 2002). However, a homozygous gain-of-function mutation (R73 S), which increased DNA binding capacity, led to the same Bamforth–Lazarus phenotypes as loss-of-function variants (Carre et al., 2014). Similar to these loss- and gain-of-function mutations, we show that in our zebrafish model the transcriptional activity of Foxe1 is compromised, in our case due to disrupted nuclear localization.
A typical symptom of Bamforth-Lazarus syndrome is cleft palate, which refers to a defective fusion of the two palatal shelves (Castanet and Polak 2010; Sarma et al., 2022). The ethmoid plate of the zebrafish is analogous to the human hard palate, and cleft phenotypes have been described in multiple zebrafish mutants (Mork and Crump 2015). These phenotypes include rough anterior edges of the ethmoid plate at varying degrees of severity, or a lack of cells in the medial ethmoid plate (Swartz et al., 2011; Mork and Crump 2015; Duncan et al., 2017). Although we did not observe obvious effects of the foxe1 mutation on the morphology of the ethmoid plate, we did find differential expression of sox9, tgfβ3 and col2a1. These are important factors for the development of the ethmoid plate, thus more subtle defects in chondrogenesis of this structure cannot be ruled out.
In our foxe1 mutant, we observed a kink of the ceratohyal cartilage, reminiscent of previously reported kinks in the ceratohyals of lrp5 mutants (also after bone and cartilage staining), which aimed to mimic osteoporotic phenotypes (Bek et al., 2021). This structure is the only head skeleton cartilage that mineralizes via endochondral ossification at this timepoint (Tonelli et al., 2020). As the kink is observed at the middle of the ceratohyal, which is the primary ossification center, a decreased ossification may have led to a collapse of the structure after the staining procedure which expands soft tissues slightly. Nakada et al. showed specific upregulation of fgfr2 surrounding the ceratohyal in Foxe1 morphants, which is in line with the increased fgfr2 transcripts levels we observed in our mutants (Nakada et al., 2009). FGFR2 gain- and loss-of-function mutations have been associated with multiple skeletal syndromes such as bent bone dysplasia, in which long bones are bent and contain smaller hypertrophic chondrocytes in the growth plate and a thickened periosteum (Merrill et al., 2012). It is thus tempting to speculate that the foxe1 mutation has led to a less rigid structure of the ceratohyal bone which led to a collapse upon staining.
It was proposed by Xu and others (2018) that a Fox code for craniofacial patterning may exist. Fox-F and Fox-C-proteins were found to be individually redundant for cartilage morphogenesis in zebrafish. However, there were severe additive effects in case of multiple Fox-mutations. Xu and others described reduced or absent lower jaw cartilage elements caused by Fox-F mutations and malformed upper facial cartilages associated with Fox-C mutations (Xu et al., 2018). Interestingly, this study reported specifically that the ceratohyal cartilages were not affected by any of the mutations. Together with our data, this indicates there may be a specific role for Foxe1 in ceratohyal patterning within the suggested Fox code (Xu et al., 2018).
Another clinical symptom reported in two infants with Bamforth–Lazarus syndrome, is delayed growth. This was specified in one individual as a severe delay in bone maturation with absent ossification centers in the tibial epiphyses (Castanet et al., 2002). Our model may have subtly recapitulated this clinical symptom associated with a FOXE1 mutation at 8 dpf in the ceratohyals.
We further observed a reduced Ca, Mg and P content in mutant larvae at 8 dpf compared to wild type siblings. This is in line with the reduced rigidity of the ceratohyals and with the observed downregulation of sp7. Sp7 functions as a master gene for osteoblast differentiation from sox9 and runx2 positive progenitors (Akiyama et al., 2005; Flores et al., 2006). Sp7−/− mice develop cartilage normally, but no osteoblasts or bone are formed, while the mutation is embryonically lethal (Nakashima et al., 2002; Sinha and Zhou 2013). Cartilage formation was also normal in zebrafish with truncated sp7, but bone growth and mineralization defects were observed as well as the absence of teeth (Kague et al., 2016; Niu et al., 2017; Chen et al., 2019). Altered expression of other Fox-family proteins in CNCCs inhibited bone development and expansion of cartilages, which may be reflected in the upregulation of chondrocyte markers and sp7 downregulation in our mutants (Xu et al., 2018). Col1a2 was also downregulated in our mutants at 6 dpf. Mutation of this gene causes osteogenesis imperfecta, characterized by low bone mass and high bone fragility, also previously modeled in zebrafish (Gistelinck et al., 2016).
An essential process in CNCCs is EMT, in which morphological and behavioral characteristics change to promote migration (Theveneau and Mayor 2012). Loss- and gain-of-function studies of FOXE1 in thyroid cancer cells show that FOXE1 may be an EMT modulator (Morillo-Bernal, Fernandez, and Santisteban 2020). Previously, Nakada and others suggested that CNCC migration and specification following EMT is not affected by foxe1 knock-down. In our study, we observed no differences in total expression of dlx2a; however, the spatial distribution of dlx2a at 24 hpf was severely altered in a subset of heterozygous and homozygous mutants. These differences may be mediated through tgfb3, which was consistently upregulated in our mutants, likely due to dysregulation of Foxe1. At 24 hpf, tgfb3 is expressed in post-migratory CNCCs in the pharyngeal arches (Perrone et al., 2000; Cheah et al., 2010; Venza et al., 2011). Both overexpression and knock-down of tgfb3 resulted in reduced amounts of post-migratory dlx2a-positive CNCCs in a study by Cheah and others with similar distorted stream morphology to foxe1 mutants. Despite the distorted dlx2a expression in CNCCs, the craniofacial cartilages developed normally, as was the case for most cartilage structures in our foxe1 mutants (Cheah et al., 2010). Studies on knockdown and knockout of dlx2a in zebrafish give conflicting evidence on the importance of dlx2a on CNCC survival and craniofacial malformations. Knockdown resulted in increased apoptotic CNCC death and lead to defects in the ceratobranchials (Sperber et al., 2008). However, stable dlx2a−/− mutants showed no apparent malformations up to adulthood (Yu et al., 2021). Although the effects of disrupted dlx2a expression are unclear, our findings strongly suggest that Foxe1 is involved in neural crest cell processes.
In the present study we found consistent Foxe1 expression and colocalization with T4 in the developing thyroid follicles of zebrafish. This suggests a role for this transcription factor in thyroidogenesis in zebrafish, although this was previously debated (Nakada et al., 2009). However, Lidral and others characterized the expression of the zebrafish foxe1 enhancer region hsCNE-67.7 and reported that it also colocalizes with T4 at 96 hpf, which supports our findings (Lidral et al., 2015). Although the presence of Foxe1 in the developing follicles suggests a developmental function, its exact role requires further study. In regard of Bamforth–Lazarus characteristics, there is clearly a role for the FOXE1 FHD in thyroid development, but the C-terminal PAS region of the protein is also important. Shorter PAS length in humans is associated with susceptibility to thyroid dysgenesis. However, it does not affect the binding of FOXE1 to the promoters of the TGFβ3 and MSX1 genes, which are important for skeletal development (Carre et al., 2007; Venza et al., 2011; Pimentel et al., 2017). The absence of the PAS region in the zebrafish Foxe1 protein may partially explain why, up to now, no function of Foxe1 in thyroid development was found.
Taken together, this study shows that the developmental functions of Foxe1 are at least partly conserved between humans and zebrafish. We reported widespread expression of foxe1 during development and showed that small amino acid changes in the NLS alters its function in craniofacial bone development. We describe novel factors associated with Foxe1 that guide CNCC development and bone formation. Our model provides critical new insights into how foxe1 mutations affect craniofacial development and may aid in elucidating FOXE1-related disease etiologies, such as in Bamforth-Lazarus syndrome. Our observations that only in a subset of the foxe1 mutants a phenotype is obvious, indicates that a set of unknown additional factors determines whether or not an aberrant phenotype develops. Gene-gene and gene-environment interactions have been broadly reported to play a significant role in craniofacial malformations in general (Raterman et al., 2020). Some of these interactions have also been specifically associated with the SHH pathway, and our future studies will focus on these interactions and the persistence of skeletal defects in adults. The fact that the foxe1 gene in our model system is compromised but not completely abolished as it would be in a knockout, makes this a powerful model system to separate the relative influence of genetic and environmental risk factors.
Methods
Animals
Animal procedures were carried out in accordance with the Dutch Animals Act and European laws. Ethical approval for the experiments was granted by Radboud University’s Institutional Animal Care and Use Committee (AVD10300202115245). Zebrafish (Danio rerio) were raised and kept under standard husbandry conditions (28 °C under a 14 h light/10 h dark cycle) in the Radboud University Zebrafish Facility. For breeding, males and females were separated overnight by a transparent divider in breeding tanks. The next day, at the start of the light period, the divider was removed and breeding water was added to induce spawning. Eggs were collected and transferred to Petri dishes with E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 0.00001% Methylene Blue). Larvae were raised in an incubator at 28.5 °C with a 14 h light/10 h dark cycle and medium was refreshed daily.
CRISPR/Cas9 mutant generation
Using CHOPCHOP (https://chopchop.cbu.uib.no) an efficient single guide (sg) RNA was selected at the 5′ end of the target exon. sgRNAs were generated using templates for in vitro transcription as previously described (Schellens et al., 2021). Briefly, a 20 nt (GGCCGCAAAGAGGCCGTCGG) target specific oligonucleotide with a T7 promotor sequence (5′-TAATACGACTCACTATA-3′) and a complementary region (5′-GTTTTAGAGCTAGAAATAGCAAG-3′) to a constant oligonucleotide encoding the reverse complement of the tracrRNA tail were annealed. Using Phusion™ High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, United States #M0530 L) the ssDNA overhang was filled. Next, the template was purified using the GenElute™ PCR cleanup kit. In-vitro transcription was performed using T7 MEGAshortscript kit. The transcripts were purified using MEGAclear transcription clean up kit.
Before injection, the sgRNA/Cas9 ribonucleoproteincomplex was formed by incubating a mix of 80 ng/μL sgRNA, 800 ng/μL Cas9 protein (Intergrated DNA Technologies), 0.2 M KCl and 0.05% phenol red at 37 °C for 5 min. Injection needles (World Precision Instruments, Friedberg, Germany, #TW120F-3) were prepared using a micropipette puller (Sutter Instrument Company, Novato, CA, United States, Model P-97). Wild type AB strain zebrafish embryos were collected and injected at the single cell stage with 1 nL sgRNA/Cas9 mixture using a Pneumatic PicoPump pv280 (World Precision Instruments, Friedberg, Germany). Subsequently, embryos were raised at 28.5 °C in E3 medium. At 24 hpf, a sample of the injected embryos (8 pools of three embryos) was analyzed for the presence of indels. Therefore, genomic DNA was extracted from injected and control embryos. The target site was amplified in a PCR reaction (fw, 5′-CCCTCTGAAACCACTCTTCCAG-3’; rev, 5′-TGTATGGTGGTTTGCCTCGC-3′) and products were analyzed for indel induction by HRM as described previously (Thomas et al., 2014). Sanger sequencing was used for confirmation of indels. In the F1 generation, one indel was selected and outcrossed. After outcrossing with wild types the F3 generation was used for experiments.
Immunohistochemistry
Immunohistochemistry was performed as described earlier (Schellens et al., 2021). Larvae were euthanized and fixated in 4% PFA overnight and embedded in Optimal Cutting Temperature (Agar Scientific) for cryo-sectioning. 7 µm horizontal sections were mounted on a glass slide and washed in PBS before incubation in PBS with 0.01% Tween (Sigma) for 20 min. After PBS rinse, blocking was performed using 10% NGS (Thermo Fisher) 2% BSA (Sigma) before overnight primary antibody incubation in blocking buffer at 4 °C. The polyclonal antibody for zebrafish Foxe1 was custom-made at Boster Bio (DZ41149, using C-terminal peptide AVGSSGAYIRHPAYSG as the immunogen) and it was applied at a dilution of 1:500. The T4 antibody (MP Biomedicals rabbit anti-T4) was diluted to 1:1,000. Secondary antibodies (Alexa Fluor 488/568 Thermo Fisher) were applied at 1:800 in blocking buffer. After washing, a short incubation with 300 nM DAPI was performed before post-fixation in 4% PFA. Sections were then sealed in FluorSave (Merck Millipore). Signals were imaged using an SP8x confocal laser scanning microscope (Leica-microsystems) with Leica Application Suite X software at magnifications x20 or 100x.
Whole mount immunohistochemistry was performed exactly as described previously (Hammond-Weinberger and ZeRuth 2020) using T4 antibody (MP Biomedicals rabbit anti-T4) diluted at 1:1,000 for whole embryos and Foxe1 Boster Bio (DZ41149) at 1:500 on scales. Z-stack images of the T4 staining were taken on an SPX8 confocal microscope at magnification ×20 and were processed by surface rendering using Imaris 9.0 software to calculate volumes as previously described (Zhang et al., 2021).
In situ hybridization
Whole mount in situ hybridization was performed as previously reported (Thisse and Thisse 2008) (Gebuijs et al., 2019). Probes were generated using respective primers: dlx2a fw, 5′-AGTGTGCTTTTGCGGTATGA-3′, rev, 5′-AATATGGTCCCGGCGCTAAC-3′ and foxe1 fw, 5′-ATGCCTGTGGTTAAAGTGGAGAGT-3′, rev, 5′-TGGCCCATAATGCTGAGTGCT-3’. In brief, sequences were amplified by PCR and cloned in pGEM®-T Vector (Promega). Sanger sequencing was used to confirm positive clones. Plasmid linearization was performed using Notl and Ncol (Thermo Fisher Scientific), before purification with GeneJET PCR purification kit (Thermo Fisher Scientific). Digoxigenin (DIG) labeling of the probe was performed using the DIG labeling Sp6/T7 Kit (Roche). Samples were fixed in 4% PFA for 12 h. Digestion with Proteinase K (Sigma-Aldrich) was applied to samples >2 dpf for 30 min at RT. Samples were pre-hybridized at 70°C for 2 h in hybridization mix (HM) (50% formamide (VWR), five x saline-sodium citrate buffer (SSC), 50 μg/ml heparin (Sigma-Aldrich), 500 μg/ml tRNA (Sigma-Aldrich), 0.1% Tween20 and 9.2 mM citric acid). Then, hybridization with DIG-labeled anti-sense probes was performed overnight at 70°C in HM (∼100 ng/ml probe). After hybridization, samples were washed in gradients of HM (without tRNA and heparin) and SSC to PBT and then samples were pre-incubated with 2% NCS (HyClone) in PBT. Next, samples were incubated with anti-DIG AP fragments (Roche) (1:2000) in 2% NCS overnight at 4°C. After washes, labeling was performed using Nitro Blue Tetrazolium (NBT)/5-Bromo-4-chloro-3-indolyl phosphate (BCIP) (Roche) in alkaline phosphatase buffer (100 mM Tris (pH 9.5), 50 mM MgCl2, 100 mM NaCl and 0.1% Tween 20). For imaging, larvae were cleared in a 2:1 mixture of benzyl benzoate (Merck) and benzyl alcohol (Merck). Images were acquired with a binocular microscope (Leica DMRE) using Leica Application Suite (LAS 3.3, Leica).
Quantitative polymerase chain reaction
Total larval RNA was extracted in 400 μL Trizol reagent (Invitrogen, Carlsbad, United States of America) following the manufacturer’s protocol and quantified using a NANODROP 1000 spectrophotometer. For cDNA synthesis, the iScript cDNA Synthesis Kit (Bio-Rad, Cressier, Switzerland) was applied. Used primers are listed in Table 1. The qPCR mix contained 10 μL SYBR green mix (2X) (Bio-Rad, Hercules, United States of America) and qPCR was performed using a CFX 96 (Bio-Rad) machine. Relative expression was calculated based on normalization against two reference genes: elongation factor alpha (elf1a) and ribosomal protein L13 (rpl13) (Vandesompele et al., 2002).
Bone and cartilage staining
Staining was performed as described by Walker and Kimmel with small modifications (Walker and Kimmel 2007). Larvae were euthanized on ice and fixed for 1 h in 2% PFA. After washing with 100 mM Tris/40 mM MgCl2 (pH 7.5) larvae were incubated in 0.04% Alcian Blue (Sigma-Aldrich)/40 mM MgCl2 for 2.5 h. After gradual rehydration, bleaching was performed using 3% H2O2/0.5% KOH. After two short incubations in 25% glycerol/0.1% KOH bone staining with 0.03% Alizarin Red (pH 7.5, Sigma-Aldrich) for 1 h. Finally, larvae were imaged in 3% methylcellulose using Leica Application Suite (LAS 3.3, Leica).
Live imaging
Wild type zebrafish and Foxe1 mutants in a transgenic col2a1a:mCherry background were anaesthetized at 5 dpf and embedded in agarose before short term live imaging on an SPX8 confocal microscope at magnification ×20.
Inductively Coupled Plasma - Optical Emission Spectroscopy
Larvae were euthanized on ice and rinsed in ultrapure water, transferred to 2 ml Eppendorf tubes and any excess liquid was removed. 100 µL of 65% nitric acid (HNO3) was added and the next day, the nitric acid containing the dissolved samples was added to 6 ml ultrapure water. Inductively Coupled Plasma - Optical Emission Spectroscopy (ICP-OES) was used to determine the molar contents of calcium, phosphorus, and magnesium in the samples. Samples were measured on an ARCOSMV (Spectro, Kleve, Germany) on axial view, with 1400 W plasma power. Samples were nebulized with a seaspray nebulizer combined with a cyclone chamber and an argon flow of 0.7 L/min.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Radboud University’s Institutional Animal Care and Use Committee (AVD10300202115245).
Author contributions
Conceptualization: SR, JVDH, JM, and FW; Methodology: SR, JVDH, JM, FW, JZ, EDV, SD, TTM, CDV, and SB; Formal analysis: SR, JZ, SD, TTM, and CDV; Writing—original draft: SR; Writing—review and editing: SR, JVDH, JM, and FW; Supervision: JVDH, JM, and FW.
Funding
This work was funded by the Radboudumc and by the Dr. Vaillant Foundation.
Acknowledgments
We would like to thank Hanka Venselaar for help with 3D protein structure predictions. We thank Antoon van der Horst and Jeroen Boerrigter for animal care at the Radboud University Zebrafish Facility. Paul van de Ven and Sebastian Krosse are gratefully acknowledged for the elemental analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1143844/full#supplementary-material
Abbreviations
BSA; Bovine serum albumin, CNCC; Cranial neural crest cells, COL1A2; Collagen type I alpha 2 chain, COL2A1; Collagen type II alpha 1 chain, CP/CLP; Cleft palate/Cleft lip and palate, CRISPR; Clustered Regularly Interspaced Short Palindromic Repeats, DAPI; 4′,6-diamidino-2-phenylindole, DPF; Days post fertilization, EMT; Epithelial mesenchymal transition, FGFR2; Fibroblast growth factor receptor 2, FHD; Forkhead domain, FOXE1; Forkhead transcription factor E1/TTF-2, GLI2; GLI Family Zinc Finger 2, GWAS; Genome wide association study, HPF; Hours post fertilization, HRM; High resolution meltcurve, LPR5; Low-density lipoprotein receptor-related protein 5, MPF; Months post fertilization, MSX1; Msh Homeobox 1, NGS; Normal goat serum, NIS; Sodium/iodide cotransporter, NLS; Nuclear localization signal, PAS; Polyalanine stretch, RUNX2B; RUNX family transcription factor 2 b, SHH; Sonic hedgehog, SOX9A; SRY-box transcription factor 9a, SP7; Sp7 Transcription Factor, T4; Thyroid hormone, TG; Thyroglobulin, TGFB3; Transforming Growth Factor Beta 3, TPO; Thyroid peroxidase, TTF-2; Thyroid transcription factor 2/FOXE1, WNT5A; Wnt Family Member 5 A.
References
Akiyama, H., Kim, J. E., Nakashima, K., Balmes, G., Iwai, N., Deng, J. M., et al. (2005). 'Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A. 102, 14665–14670. doi:10.1073/pnas.0504750102
Alt, B., Reibe, S., Feitosa, N. M., Elsalini, O. A., Wendl, T., and Rohr, K. B. (2006). 'Analysis of origin and growth of the thyroid gland in zebrafish. Dev. Dyn. 235, 1872–1883. doi:10.1002/dvdy.20831
Bamforth, J. S., Hughes, I. A., Lazarus, J. H., Weaver, C. M., and Harper, P. S. (1989). 'Congenital hypothyroidism, spiky hair, and cleft palate. J. Med. Genet. 26, 49–51. doi:10.1136/jmg.26.1.49
Baris, I., Arisoy, A. E., Smith, A., Agostini, M., Mitchell, C. S., Park, S. M., et al. (2006). 'A novel missense mutation in human TTF-2 (FKHL15) gene associated with congenital hypothyroidism but not athyreosis. J. Clin. Endocrinol. Metab. 91, 4183–4187. doi:10.1210/jc.2006-0405
Beekhuijzen, M., de Koning, C., Flores-Guillen, M. E., de Vries-Buitenweg, S., Tobor-Kaplon, M., van de Waart, B., et al. (2015). 'From cutting edge to guideline: A first step in harmonization of the zebrafish embryotoxicity test (zet) by describing the most optimal test conditions and morphology scoring system. Reprod. Toxicol. 56, 64–76. doi:10.1016/j.reprotox.2015.06.050
Bek, J. W., Shochat, C., De Clercq, A., De Saffel, H., Boel, A., Metz, J., et al. (2021). 'Lrp5 mutant and crispant zebrafish faithfully model human osteoporosis, establishing the zebrafish as a platform for CRISPR-based functional screening of osteoporosis candidate genes. J. Bone Mineral Res. 36, 1749–1764. doi:10.1002/jbmr.4327
Brancaccio, A., Minichiello, A., Grachtchouk, M., Antonini, D., Sheng, H., Parlato, R., et al. (2004). 'Requirement of the forkhead gene Foxe1, a target of sonic hedgehog signaling, in hair follicle morphogenesis. Hum. Mol. Genet. 13, 2595–2606. doi:10.1093/hmg/ddh292
Carre, A., Castanet, M., Sura-Trueba, S., Szinnai, G., Van Vliet, G., Trochet, D., et al. (2007). 'Polymorphic length of FOXE1 alanine stretch: Evidence for genetic susceptibility to thyroid dysgenesis. Hum. Genet. 122, 467–476. doi:10.1007/s00439-007-0420-5
Carre, A., Hamza, R. T., Kariyawasam, D., Guillot, L., Teissier, R., Tron, E., et al. (2014). A novel FOXE1 mutation (R73S) in Bamforth-Lazarus syndrome causing increased thyroidal gene expression. Thyroid 24, 649–654. doi:10.1089/thy.2013.0417
Castanet, M., Park, S. M., Smith, A., Bost, M., Leger, J., Lyonnet, S., et al. (2002). 'A novel loss-of-function mutation in TTF-2 is associated with congenital hypothyroidism, thyroid agenesis and cleft palate. Hum. Mol. Genet. 11, 2051–2059. doi:10.1093/hmg/11.17.2051
Castanet, M., and Polak, M. (2010). 'Spectrum of human foxe1/TTF2 mutations. Hormone Res. Paediatr. 73, 423–429. doi:10.1159/000281438
Cheah, F. S., Winkler, C., Jabs, E. W., and Chong, S. S. (2010). 'Tgfbeta3 regulation of chondrogenesis and osteogenesis in zebrafish is mediated through formation and survival of a subpopulation of the cranial neural crest. Mech. Dev. 127, 329–344. doi:10.1016/j.mod.2010.04.003
Chen, Z. J., Song, Z. Y., Yang, J. J., Huang, J., and Jiang, H. B. (2019). 'Sp7/osterix positively regulates dlx2b and bglap to affect tooth development and bone mineralization in zebrafish larvae. J. Biosci. 44, 127. doi:10.1007/s12038-019-9948-5
Clifton-Bligh, R. J., Wentworth, J. M., Heinz, P., Crisp, M. S., John, R., Lazarus, J. H., et al. (1998). 'Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet. 19, 399–401. doi:10.1038/1294
Dathan, N., Parlato, R., Rosica, A., De Felice, M., and Di Lauro, R. (2002). 'Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev. Dyn. 224, 450–456. doi:10.1002/dvdy.10118
De Felice, M., Ovitt, C., Biffali, E., Rodriguez-Mallon, A., Arra, C., Anastassiadis, K., et al. (1998). 'A mouse model for hereditary thyroid dysgenesis and cleft palate. Nat. Genet. 19, 395–398. doi:10.1038/1289
Dietrich, K., Fiedler, I. A., Kurzyukova, A., Lopez-Delgado, A. C., McGowan, L. M., Geurtzen, K., et al. (2021). Skeletal Biology and disease modeling in zebrafish. J. Bone Min. Res. 36, 436–458. doi:10.1002/jbmr.4256
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). 'Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167–178. doi:10.1038/nrg2933
Duncan, K. M., Mukherjee, K., Cornell, R. A., and Liao, E. C. (2017). 'Zebrafish models of orofacial clefts. Dev. Dyn. 246, 897–914. doi:10.1002/dvdy.24566
Eames, B. F., Amores, A., Yan, Y. L., and Postlethwait, J. H. (2012). Evolution of the osteoblast: Skeletogenesis in gar and zebrafish. Bmc Evol. Biol. 12, 27. doi:10.1186/1471-2148-12-27
Eichberger, T., Regl, G., Ikram, M. S., Neill, G. W., Philpott, M. P., Aberger, F., et al. (2004). 'FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J. Investigative Dermatology 122, 1180–1187. doi:10.1111/j.0022-202X.2004.22505.x
El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Gunther, S., Fukuda, N., et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. doi:10.1038/s41586-019-1064-z
El-Brolosy, M. A., and Stainier, D. Y. R. (2017). Genetic compensation: A phenomenon in search of mechanisms. Plos Genet. 13, e1006780. doi:10.1371/journal.pgen.1006780
Flores, M. V., Lam, E. Y. N., Crosier, P., and Crosier, K. (2006). A hierarchy of Runx transcription factors modulate the onset of chondrogenesis in craniofacial endochondral bones in zebrafish. Dev. Dyn. 235, 3166–3176. doi:10.1002/dvdy.20957
Gebuijs, I. G. E., Raterman, S. T., Metz, J. R., Swanenberg, L., Zethof, J., Van den Bos, R., et al. (2019). 'Fgf8a mutation affects craniofacial development and skeletal gene expression in zebrafish larvae. Biol. Open 8, bio039834. doi:10.1242/bio.039834
Gistelinck, C., Gioia, R., Gagliardi, A., Tonelli, F., Marchese, L., Bianchi, L., et al. (2016). 'Zebrafish collagen type I: Molecular and biochemical characterization of the major structural protein in bone and skin. Sci. Rep. 6, 21540. doi:10.1038/srep21540
Hammond-Weinberger, D. R., and ZeRuth, G. T. (2020). 'whole mount immunohistochemistry in zebrafish embryos and larvae. Jove-Journal of Visualized Experiments.
Harley, V. R., Layfield, S., Mitchell, C. L., Forwood, J. K., John, A. P., Briggs, L. J., et al. (2003). 'Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. U. S. A. 100, 7045–7050. doi:10.1073/pnas.1137864100
Kague, E., Gallagher, M., Burke, S., Parsons, M., Franz-Odendaal, T., and Fisher, S. (2012). 'Skeletogenic fate of zebrafish cranial and trunk neural crest. Plos One 7, e47394. doi:10.1371/journal.pone.0047394
Kague, E., Roy, P., Asselin, G., Hu, G., Simonet, J., Stanley, A., et al. (2016). 'Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev. Biol. 413, 160–172. doi:10.1016/j.ydbio.2016.03.011
Kok, F. O., Shin, M., Ni, C. W., Gupta, A., Grosse, A. S., van Impel, A., et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108. doi:10.1016/j.devcel.2014.11.018
Lammer, E. J., Mohammed, N., Iovannisci, D. M., Ma, C., Lidral, A. C., and Shaw, G. M. (2016). 'Genetic variation of FOXE1 and risk for orofacial clefts in a California population. Am. J. Med. Genet. Part A 170, 2770–2776. doi:10.1002/ajmg.a.37871
Lane, J., Yumoto, K., Pisano, J., Azhar, M., Thomas, P. S., and Kaartinen, V. (2014). 'Control elements targeting Tgfb3 expression to the palatal epithelium are located intergenically and in introns of the upstream Ift43 gene. Front. Physiology 5, 258. doi:10.3389/fphys.2014.00258
Leslie, E. J., Carlson, J. C., Shaffer, J. R., Butali, A., Buxo, C. J., Castilla, E. E., et al. (2017). 'Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 136, 275–286. doi:10.1007/s00439-016-1754-7
Lidral, A. C., Liu, H., Bullard, S. A., Bonde, G., Machida, J., Visel, A., et al. (2015). 'A single nucleotide polymorphism associated with isolated cleft lip and palate, thyroid cancer and hypothyroidism alters the activity of an oral epithelium and thyroid enhancer near FOXE1. Hum. Mol. Genet. 24, 3895–3907. doi:10.1093/hmg/ddv047
Lopez-Marquez, A., Fernandez-Mendez, C., Recacha, P., and Santisteban, P. (2019). Regulation of foxe1 by thyrotropin and transforming growth factor beta depends on the interplay between thyroid-specific, CREB and SMAD transcription factors. Thyroid 29, 714–725. doi:10.1089/thy.2018.0136
McLane, L. M., and Corbett, A. H. (2009). Nuclear localization signals and human disease. Iubmb Life 61, 697–706. doi:10.1002/iub.194
Merrill, A. E., Sarukhanov, A., Krejci, P., Idoni, B., Camacho, N., Estrada, K. D., et al. (2012). 'Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am. J. Hum. Genet. 90, 550–557. doi:10.1016/j.ajhg.2012.02.005
Moreno, L. M., Mansilla, M. A., Bullard, S. A., Cooper, M. E., Busch, T. D., Machida, J., et al. (2009). 'FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet. 18, 4879–4896. doi:10.1093/hmg/ddp444
Morillo-Bernal, J., Fernandez, L. P., and Santisteban, P. (2020). 'FOXE1 regulates migration and invasion in thyroid cancer cells and targets ZEB1. Endocrine-Related Cancer 27, 137–151. doi:10.1530/ERC-19-0156
Mork, L., and Crump, G. (2015). Zebrafish craniofacial development: A window into early patterning. Curr. Top. Dev. Biol. 115, 235–269. doi:10.1016/bs.ctdb.2015.07.001
Nakada, C., Iida, A., Tabata, Y., and Watanabe, S. (2009). Forkhead transcription factor foxe1 regulates chondrogenesis in zebrafish. J. Exp. Zool. B Mol. Dev. Evol. 312, 827–840. doi:10.1002/jez.b.21298
Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z. P., Deng, J. M., Behringer, R. R., et al. (2002). The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29. doi:10.1016/s0092-8674(01)00622-5
Niu, P. F., Zhong, Z. M., Wang, M. Y., Huang, G. D., Xu, S. H., Hou, Y., et al. (2017). Zinc finger transcription factor Sp7/Osterix acts on bone formation and regulates col10a1a expression in zebrafish. Sci. Bull. 62, 174–184. doi:10.1016/j.scib.2017.01.009
Parichy, D. M., Elizondo, M. R., Mills, M. G., Gordon, T. N., and Engeszer, R. E. (2009). Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 238, 2975–3015. doi:10.1002/dvdy.22113
Perrone, L., di Magliano, M. P., Zannini, M., and Di Lauro, R. (2000). The thyroid transcription factor 2 (TTF-2) is a promoter-specific DNA-binding independent transcriptional repressor. Biochem. Biophysical Res. Commun. 275, 203–208. doi:10.1006/bbrc.2000.3232
Pimentel, C. P., Cortinhas-Alves, E. A., de Oliveira, E. H. C., and Santana-da-Silva, L. C. (2017). Does the polymorphism in the length of the polyalanine tract of FOXE1 gene influence the risk of thyroid dysgenesis occurrence? J. Thyroid. Res. 2017, 2793205. doi:10.1155/2017/2793205
Raterman, S. T., Metz, J. R., Wagener, F. A. D. T. G., and Von den Hoff, J. W. (2020). 'Zebrafish models of craniofacial malformations: Interactions of environmental factors. Front. Cell Dev. Biol. 8, 600926. doi:10.3389/fcell.2020.600926
Romanelli, M. G., Tato', L., Lorenzi, P., and Morandi, C. (2003). Nuclear localization domains in human thyroid transcription factor 2. Biochimica Biophysica Acta-Molecular Cell Res. 1643, 55–64. doi:10.1016/j.bbamcr.2003.09.002
Sarma, A. S., Banda, L., Rao Vupputuri, M., Desai, A., and Dalal, A. (2022). 'A new FOXE1 homozygous frameshift variant expands the genotypic and phenotypic spectrum of Bamforth-Lazarus syndrome. Eur. J. Med. Genet. 65, 104591. doi:10.1016/j.ejmg.2022.104591
Schellens, R., de Vrieze, E., Graave, P., Broekman, S., Nagel-Wolfrum, K., Peters, T., et al. (2021). 'Zebrafish as a model to evaluate a CRISPR/Cas9-Based exon excision approach as a future treatment option for EYS-associated retinitis pigmentosa. Int. J. Mol. Sci. 22, 9154. doi:10.3390/ijms22179154
Schilling, T. F., and Kimmel, C. B. (1994). 'Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483–494. doi:10.1242/dev.120.3.483
Schulte-Merker, S., and Stainier, D. Y. (2014). Out with the old, in with the new: Reassessing morpholino knockdowns in light of genome editing technology. Development 141, 3103–3104. doi:10.1242/dev.112003
Sinha, K. M., and Zhou, X. (2013). 'Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 114, 975–984. doi:10.1002/jcb.24439
Sperber, S. M., Saxena, V., Hatch, G., and Ekker, M. (2008). 'Zebrafish dlx2a contributes to hindbrain neural crest survival, is necessary for differentiation of sensory ganglia and functions with dlx1a in maturation of the arch cartilage elements. Dev. Biol. 314, 59–70. doi:10.1016/j.ydbio.2007.11.005
Swartz, M. E., Sheehan-Rooney, K., Dixon, M. J., and Eberhart, J. K. (2011). Examination of a palatogenic gene program in zebrafish. Dev. Dyn. 240, 2204–2220. doi:10.1002/dvdy.22713
Theveneau, E., and Mayor, R. (2012). 'Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 366, 34–54. doi:10.1016/j.ydbio.2011.12.041
Thisse, C., and Thisse, B. (2008). 'High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi:10.1038/nprot.2007.514
Thomas, H. R., Percival, S. M., Yoder, B. K., and Parant, J. M. (2014). High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS ONE 9 (12), e114632. doi:10.1371/journal.pone.0114632
Tonelli, F., Bek, J. W., Besio, R., De Clercq, A., Leoni, L., Salmon, P., et al. (2020). 'Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. (Lausanne) 11, 489. doi:10.3389/fendo.2020.00489
Trueba, S. S., Auge, J., Mattei, G., Etchevers, H., Martinovic, J., Czernichow, P., et al. (2005). 'PAX8, TITF1, and FOXE1 gene expression patterns during human development: New insights into human thyroid development and thyroid dysgenesis-associated malformations. J. Clin. Endocrinol. Metabolism 90, 455–462. doi:10.1210/jc.2004-1358
Valenti, M. T., Marchetto, G., Mottes, M., and Dalle Carbonare, L. (2020). 'Zebrafish: A suitable tool for the study of cell signaling in bone. Cells 9, 1911. doi:10.3390/cells9081911
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). 'Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. doi:10.1186/gb-2002-3-7-research0034
Venza, I., Visalli, M., Parrillo, L., De Felice, M., Teti, D., and Venza, M. (2011). MSX1 and TGF-beta3 are novel target genes functionally regulated by FOXE1. Hum. Mol. Genet. 20, 1016–1025. doi:10.1093/hmg/ddq547
Wada, N., Javidan, Y., Nelson, S., Carney, T. J., Kelsh, R. N., and Schilling, T. F. (2005). 'Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977–3988. doi:10.1242/dev.01943
Walker, M. B., and Kimmel, C. B. (2007). 'A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28. doi:10.1080/10520290701333558
Wang, D. X., Eraslan, B., Wieland, T., Hallstrom, B., Hopf, T., Zolg, D. P., et al. (2019). 'A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 15, e8503. doi:10.15252/msb.20188503
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). 'SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi:10.1093/nar/gky427
Xavier, G. M., Seppala, M., Barrell, W., Birjandi, A. A., Geoghegan, F., and Cobourne, M. T. (2016). 'Hedgehog receptor function during craniofacial development. Dev. Biol. 415, 198–215. doi:10.1016/j.ydbio.2016.02.009
Xu, P. F., Balczerski, B., Ciozda, A., Louie, K., Oralova, V., Huysseune, A., et al. (2018). Fox proteins are modular competency factors for facial cartilage and tooth specification. Development, 145.
Yu, E. P. Y., Perin, S., Saxena, V., and Ekker, M. (2021). 'Novel cross-regulation interactions between dlx genes in larval zebrafish. Gene 801, 145848. doi:10.1016/j.gene.2021.145848
Zannini, M., Acebron, A., DeFelice, M., Arnone, M. I., MartinPerez, J., Santisteban, P., et al. (1996). Mapping and functional role of phosphorylation sites in the thyroid transcription factor-1 (TTF-1). J. Biol. Chem. 271, 2249–2254. doi:10.1074/jbc.271.4.2249
Keywords: zebrafish, FOXE1, bamforth-lazarus syndrome, cleft palate, craniofacial malformations, skeletal development, thyroidogenesis
Citation: Raterman ST, Von Den Hoff JW, Dijkstra S, De Vriend C, Te Morsche T, Broekman S, Zethof J, De Vrieze E, Wagener FADTG and Metz JR (2023) Disruption of the foxe1 gene in zebrafish reveals conserved functions in development of the craniofacial skeleton and the thyroid. Front. Cell Dev. Biol. 11:1143844. doi: 10.3389/fcell.2023.1143844
Received: 13 January 2023; Accepted: 28 February 2023;
Published: 13 March 2023.
Edited by:
Jakub Konrad Famulski, University of Kentucky, United StatesReviewed by:
Natércia Conceição, University of Algarve, PortugalBen Lovely, University of Louisville, United States
Copyright © 2023 Raterman, Von Den Hoff, Dijkstra, De Vriend, Te Morsche, Broekman, Zethof, De Vrieze, Wagener and Metz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie T. Raterman, c29waGllLnJhdGVybWFuQHJhZGJvdWR1bWMubmw=
 Sophie T. Raterman
Sophie T. Raterman Johannes W. Von Den Hoff
Johannes W. Von Den Hoff Sietske Dijkstra2
Sietske Dijkstra2 Cheyenne De Vriend
Cheyenne De Vriend Erik De Vrieze
Erik De Vrieze Frank A. D. T. G. Wagener
Frank A. D. T. G. Wagener Juriaan R. Metz
Juriaan R. Metz