- 1School of Life Sciences, Arizona State University, Tempe, AZ, United States
- 2Biodesign Institute, Center for Immunotherapy, Vaccines and Virotherapy (CIVV), Arizona State University, Tempe, AZ, United States
Insights into the effect of the microbiome’s composition on immune cell function have recently been discerned and further characterized. Microbiome dysbiosis can result in functional alterations across immune cells, including those required for innate and adaptive immune responses to malignancies and immunotherapy treatment. Dysbiosis can yield changes in or elimination of metabolite secretions, such as short-chain fatty acids (SCFAs), from certain bacterial species that are believed to impact proper immune cell function. Such alterations within the tumor microenvironment (TME) can significantly affect T cell function and survival necessary for eliminating cancerous cells. Understanding these effects is essential to improve the immune system’s ability to fight malignancies and the subsequent efficacy of immunotherapies that rely on T cells. In this review, we assess typical T cell response to malignancies, classify the known impact of the microbiome and particular metabolites on T cells, discuss how dysbiosis can affect their function in the TME then further describe the impact of the microbiome on T cell-based immunotherapy treatment, with an emphasis on recent developments in the field. Understanding the impact of dysbiosis on T cell function within the TME can carry substantial implications for the design of immunotherapy treatments and further our understanding of factors that could impact how the immune system combats malignancies.
Introduction
Microbial species inhabit nearly every organ of the human body; their significance has recently been established in proper health and immune cell function, with potential impacts in the tumor microenvironment (TME) through the presence or absence of microbial-derived metabolites such as short-chain fatty acids (SCFAs), that can impact T cell functioning. The human microbiome comprises a complex network of various organisms, including those of bacterial, archaeal, fungal, viral, and protozoan populations, many of which are capable of symbiotic or pathogenic manifestations on the host, particularly when oscillations in microbial composition occur (Riiser, 2015). In a healthy host, microbial populations typically outnumber human cells; current studies have estimated the number of bacteria alone in the human body is roughly the same as that of human cells (Sender et al., 2016). Technological advancements such as metagenomic sequencing and sophisticated data analysis have allowed scientists to characterize the abundance and diversity of the human microbiome, classifications that have allowed researchers to elucidate the potential mechanisms by which these species impact health and disease (Freilich et al., 2009; Qin et al., 2010; Methé et al., 2012). Studies support the necessity of a diverse, stable, and balanced microbiome to maintain general health and proper immunity to disease, with negative impacts during microbiome dysbiosis (Tuddenham and Sears, 2015). Microbial dysbiosis is an imbalance in the composition of microbial communities within a host resulting in perturbations from normal cellular and organ function, illness, or reduced treatment efficacy for an infection or disease (Petersen and Round, 2014). The etiology of dysbiosis is diverse and includes pathologies resulting from inflammation, infection, diet, genetics, and antibiotic administration (Willing et al., 2010; Claesson et al., 2012; Pham and Lawley, 2014). Current studies have also demonstrated the impact of host-microbiome interactions during dysbiosis on cancer progression and treatment, including disease presence and chronic inflammation (Czesnikiewicz-Guzik and Müller, 2018). Dysbiosis is believed to impact the regular functioning of immune cells caused by modifications in microbial-produced metabolites needed for proper performance (Arpaia et al., 2013; Luu et al., 2019). Such alterations in immune cells can impact not only how we respond to pathogenic infections but also the immune response to neoplastic events.
The degree of T cell tumor site infiltration and proper function within tumor sites can significantly affect tumor progression or regression (Al-Shibli et al., 2008). Therefore, an alteration in T cell function can cause a massive change in the efficacy of how T cells respond within the TME and, ultimately, their ability to clear malignancies. Understanding the mechanistic impact of microbial composition on T cells and their function within the TME is essential if we hope to understand and improve treatment strategies for malignancies. Here, we review canonical T-cell responses to malignancies, the impacts of the microbiome in the context of its typical and dysbiotic state on T-cell signaling and further how this can change T-cell function within the TME, recent discoveries in the field, and the potential that research stemming from these investigations can have on how we design and administer cancer therapies.
Conventional anti-tumoral activity of T-cells
Adaptive immune cells, such as T cells, play a considerable role in the antitumor immune response, with the presence of tumor-infiltrating T lymphocytes exhibiting positive prognostic value across a wide range of cancers (Zhang et al., 2003; Taylor et al., 2007). To understand how microbiome dysbiosis can affect the ability of T cells to react to malignancies, we must first understand conventional anti-tumoral T-cell mechanisms.
Typical CD4 T cell signaling and function in response to malignancies
CD4 T cells are directly and peripherally involved in the antitumor immune response through effects on innate and adaptive immune cells such as macrophages and T cells. Individually, CD4 T cells can be cytotoxic to incipient and progressive malignancies, acting directly on unhealthy cells that present neoepitopes to eliminate them through differentiation into Th1 cells and ensuing perforin/granzyme B-dependent killing or target cell elimination via ligation of Fas/FasL (or other TNF/TNFR family) death receptors (Figure 1A) (Lundin et al., 2004). Peripherally, CD4 T cells can also secrete IL-2 and IFN-γ, which have been found to elicit M1 macrophages to inhibit tumor propagation via secretion of nitric oxide synthase (iNOS), reactive oxygen species (ROS), and IL-12, resulting in direct and indirect clearance of tumor cells. Additionally, M1 macrophages can engulf malignant cells to eliminate them (Figure 1B) (Zhou et al., 2021). These M1 mechanisms are productive during MHC class II negativity, which can result from the loss of MHC Class II trans-activator (CIITA) expression during carcinogenesis (Haabeth et al., 2014). Additionally, CD4 T cells are necessary to support continued CD8 memory T cell survival and function (Janssen et al., 2003). They secrete cytokines, such as INFγ and IL-2, that can act on CD8 T cells to improve the anticancer immune response (Figure 1C) (Ossendorp et al., 1998).
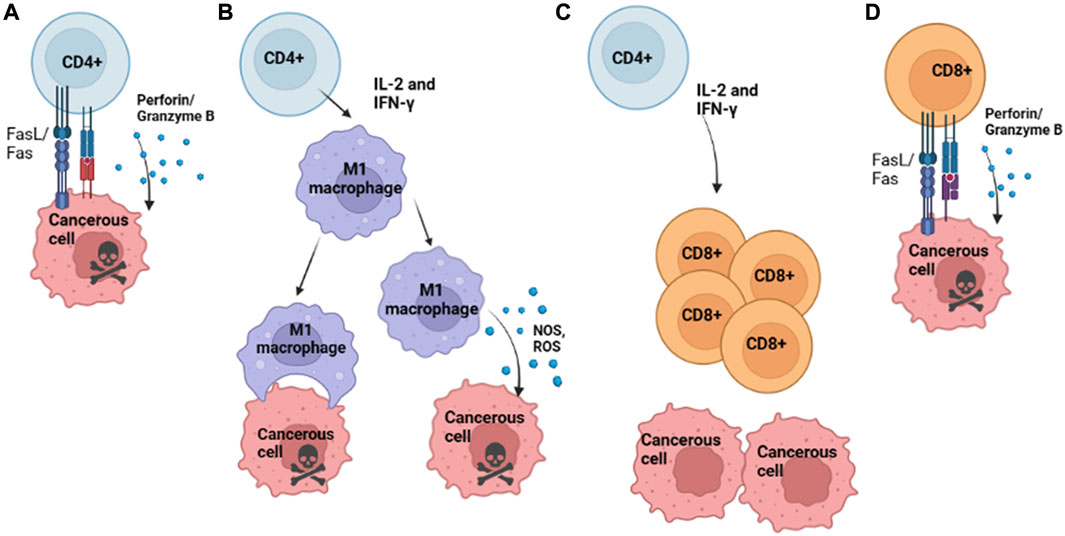
FIGURE 1. Conventional Mechanisms of T Cell Anti-Tumoral Activity. (A) Th1 differentiated CD4 T cell killing of cancerous cells that present neoepitopes on MHC II via secretion of perforin and granzyme B. (B) CD4 T cells release cytokines IL-2 and IFN-γ to recruit M1 macrophages; activated M1 Macrophages can kill cancerous cells either by direct phagocytosis or release of NOS or ROS, which results in tumor cell death and continue to secrete IL-12, among other cytokines which recruit T cells to the site of the tumor. (C) Activated CD4 T cell release of cytokines such as IL-2 and IFN-γ, which recruits CD8 T cells to the site of tumor growth. (D) CD8 T cells directly kill cancerous cells that present neoepitopes via MHC I via the Fas/FasL and the release of perforin/ granzyme B, resulting in tumor cell killing. Image Created with BioRender.com.
Typical CD8 T Cell signaling and function in response to malignancies
The presence of CD8 T cells within the TME has been associated with improved tumor clearance and overall prognosis, often characterized by the concomitant presence of pro-inflammatory cytokines such as Type I IFN (Trujillo et al., 2018). The mechanisms by which CD8 T cells within the TME are believed to eliminate cancerous cells are via ligation of the Fas ligand (Fas L) on T cells with the Fas receptor (FasR) on target cells or by the perforin/granzyme B pathway, with a preference for FasL/FasR pathway for cancerous cells (Figure 1D) (Chávez-Galán et al., 2009). Though these mechanisms can aid in the clearance of tumors, tumor-infiltrating lymphocytes often display upregulation of inhibitory markers, such as PD-1 and CTLA-4, which bind PD-L1 and CD80/CD86, respectively; these can halt anti-tumoral effector functions and result in reduced effector cytokine production (Ahmadzadeh et al., 2009).
Molecular impact of the microbiome on anti-tumoral activity of T-cells
There is evidence that microbial species within the mucosal tissue of a host and within the tumor microbiome contribute to patient tumor immune response variations, with correlations between metabolic functions of microbes present within the TME and clinical patient presentation (Nejman et al., 2020). These trends have been documented for several pancreatic, bone, and breast cancers (Riquelme et al., 2019; Nejman et al., 2020; Banerjee et al., 2021). Alteration of the host microbiome can change host cell function, including that of innate and adaptive immune cells (Russo et al., 2016; Thaiss et al., 2016). The mechanisms underlying these cellular changes are still being investigated but have been better characterized in recent studies and are believed to be primarily associated with alterations in metabolite secretions by microbial species that subsequently impact immune cell function (Luu et al., 2019; Yang et al., 2020). In particular, microbes produce fermentation products known as short-chain fatty acids (SCFAs); these free fatty acids contain short aliphatic carbon chains and are composed of less than six carbons. Typically when referring to SCFAs, the following are included: formic acid (C1), acetic acid (C2), propionic acid (C3), butyric acid (C4), and valeric acid (C5) (Tan et al., 2014). SCFAs are predominantly water-soluble, and therefore easily transported throughout the body, and are believed to play a significant role in the differentiation, function, and regulation of T cells and other immune cells through the promotion of pro- or anti-inflammatory cytokines needed for particular effector functions (Arpaia et al., 2013; Ryu et al., 2022). Thus, alterations in microbiome composition and consequent changes in microbial metabolite secretion may disrupt T cells’ conventional effector functions against malignancies.
Tumor-specific microbiome associations with malignancies
Microbial composition within the TME varies across cancers and further differs from adjacent healthy tissues, even in solid tumors with no direct contact with the external environment (Nejman et al., 2020). Several cancers, including breast, lung, ovarian, colorectal, melanoma, brain, prostate, and bone, have exhibited the presence of specific microbial species contributing to a dysbiotic state within tumor tissue (Sfanos et al., 2008; Apostolou et al., 2011; Castellarin et al., 2012; Thompson et al., 2017; Costantini et al., 2018; Greathouse et al., 2018; Nejman et al., 2020). These tumor-specific microbial populations can sometimes vary for different cancer types within the same organ systems (Banerjee et al., 2018). Considering the known impact of microbial changes on immune cell function, changes within the TME regarding tissue-specific microbial populations may initiate changes in T cell function within neoplasms (Nejman et al., 2020). Intriguingly, Rotter-Maskowitz et al. recently identified correlations between intratumoral bacterial presence and predicted clinical presentation and response to anticancer treatment (Nejman et al., 2020). Similar concepts can be traced back to William Coley in the late 19th century, who showed that injecting killed bacterial species into tumor tissue resulted in tumor regression, which we now believe is due to adjuvant effects via activation of innate immune receptors and engagement of subsequent immune responses within the tumor (Kopenhaver et al., 2020). Further investigations are needed to determine if bacterial presence in cancer impacts cancer progression via changes in immune cell function or if metabolites preferred by certain bacteria that are present as a result of cancer progression provide a niche for those found in specific tumor tissues, the presence of particular bacterial species in tumor tissue compared to healthy tissue indicates a potential avenue for understanding better what factors impact cancer progression within the TME (Thompson et al., 2017).
Gut microbiome associations with malignancies
Investigations during cancer progression and treatment suggest certain gut microbial presence outside the TME correlate with systemic inflammatory processes that affect the TME, including the upregulation of pro-inflammatory cytokines such as tumor necrosis factor (TNF) caused by increased immune cell responses, resulting in more significant tumor regression (Iida et al., 2013). These systemic alterations could be associated with secreted metabolites from specific bacteria, noting that metagenomic studies concluded that the enrichment of anabolic pathways resulting from cellular metabolism and differences in pro-inflammatory cytokines caused by some bacteria’s presence affect tumor response (Gopalakrishnan et al., 2018). Additionally, several studies suggest that gut microbiome composition plays a role in cancer progression at mucosal sites and in tumors not confined to mucosal tissue, which may be partly due to metabolites produced by microbes in the gut (Zhuang et al., 2019; Sánchez-Alcoholado et al., 2020).
Transport of microbial-derived metabolites to T immune cells
Metabolites can be produced by gut-specific or organ-specific bacteria and passively or actively transported to other locations impacting organ and cellular function at local and peripheral sites (Cummings et al., 1987; Kamp and Hamilton, 2006). For this reason changes in microbiome composition will also impact metabolite presence. For instance, during dysbioisis, certain SCFAs or other metabolites may be reduced in quantity if the bacteria that produce them are no longer present. Active diffusion of metabolites in T cells can occur via membrane transporters, including MCT1 (monocarboxylate transporter-1/Slc16a1) and SMCT1 (sodium-coupled monocarboxylate transporter-1/Slc5 a8) (Park et al., 2014). Once inside the cell, SCFAs are known to act via G-protein-coupled receptor (GPCR) signaling, inhibition of histone deacetylase (HDAC), production of acetyl-CoA, and further changes in the metabolism of the cell resulting in increased or decreased functionality (Kim, 2021). Evidence shows that SCFA presence activates mTOR, and STAT3 in T cells via GPCR41, GPCR43, GPCR109a resulting in Blimp-1 expression, which triggers the expression of many downstream signaling cascades (Zhao et al., 2018). Studies in germ-free mice suggest that changes in microbiota can directly impact the expression of toll-like receptors (TLRs) (Lundin et al., 2008) and can affect antigen-presenting cell presence, T cell differentiation, and systemic immunity (Rangan and Mondino, 2022).
Molecular effects of dysbiosis on CD4 T cell signaling and function
Recent studies showed C2, C3, and C4 metabolites, which can be produced by microbiota, could exhibit immunomodulatory functions by altering CD4 T cell differentiation in a concentration-dependent manner, as high concentrations of C2 and C3 drove expression of IL-17A, IL-17F, RORα, RORγt, T-bet, and IFN-γ, cytokines associated with Th17 and Th1 profiles (Figure 2A). (Park et al., 2014). More specifically, the natural killer group 2, member D (NKG2D) ligand system is a central immunomodulatory system in which immune cells recognize NKG2DL on stressed or infected cells through NKG2DR, present on immune cells such as CD4 T cells, CD8 T cells, and NK cells, activating effector functions (Groh et al., 2003). C3 produced by propionibacteria during cellular metabolism can induce the expression of NKG2D ligands MICA/B on both activated T lymphocytes and cancer cells in an intracellular calcium-dependent manner, allowing for proper immune effector function and potential prophylaxis of malignant cells (Figure 2B). (Andresen et al., 2009). This finding also implies the absence of C3 during the elimination of microbial populations that produce it can limit the degree of NKG2D ligands and, therefore, limit subsequent NKG2D/NKG2D interactions, resulting in a reduced overall T cell effector function within the TME. Further, C4 presence was shown to drive Treg differentiation in a concentration-dependent manner both in vitro and in vivo (Figure 2C). (Furusawa et al., 2013; Kespohl et al., 2017). Additional studies have indicated issues with the development of regulatory T cells (Tregs) in antibiotic-treated mice, indicating a requirement for healthy intestinal microbiota to achieve even normal Treg development (Han et al., 2021).
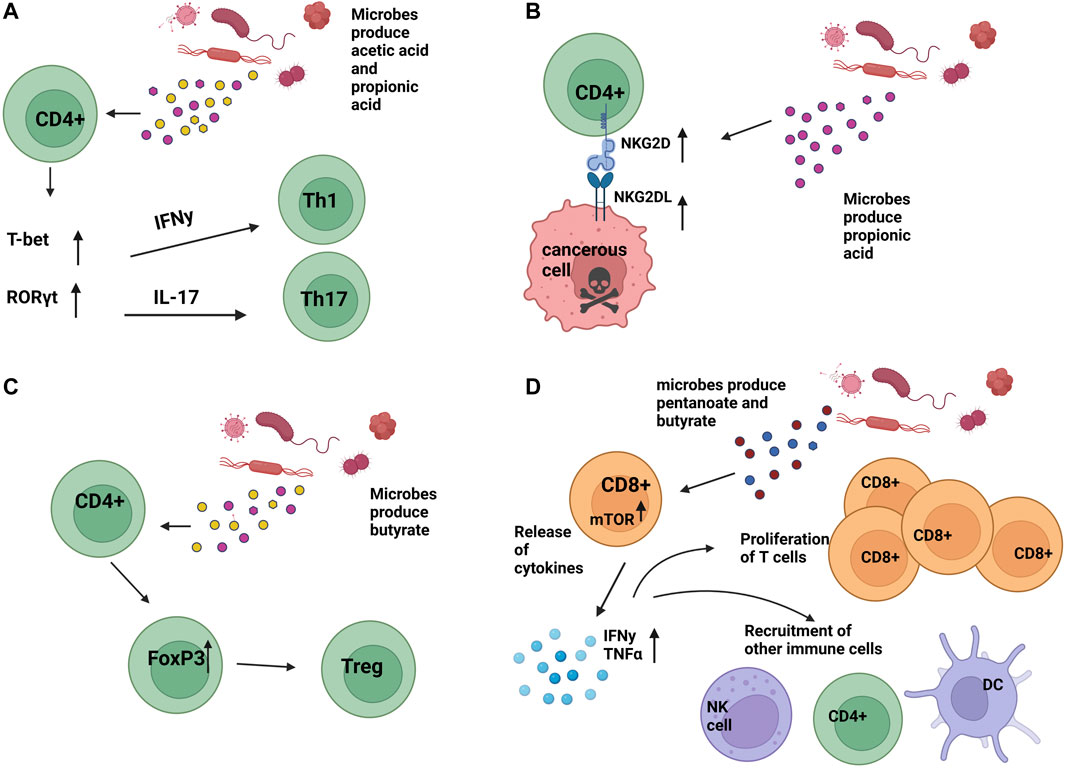
FIGURE 2. Impact of microbial secreted metabolites on T cell function (A). Bacterial-produced acetic acid (C2) and propionic acid (C3) can result in upregulation of T-bet and RORγt, causing upregulation of IFNy and IL-17 and subsequent differentiation into Th1 and Th17 T cells, respectively. (B). Microbial produced propionic acid (C3) has resulted in the upregulation of NKG2D and NKG2DL on CD4 T cells, CD8 T cells, and NK cells, contributing to an anti-tumoral environment and death of cancer cells. (C). Microbial-produced butyrate has been shown to cause the differentiation of T-regulatory cells (Tregs) in a concentration-dependent manner. (D). Microbial-produced pentanoate and butyrate have been shown to contribute to increased CD8 T cell effector functions. Image created with BioRender.com.
Molecular impact of dysbiosis on CD8 T cell signaling and function
In pre-clinical mouse models, microbial dysbiosis induced by maternal antibiotic treatment found that the offspring had altered CD8+ T Cell receptor signaling apparent by an inability to sustain interferon-gamma (IFN-γ) production in vivo after vaccination and in vitro upon T cell receptor (TCR) stimulation. Resultantly, these cells did not maintain protein tyrosine phosphorylation and Erk1/2 activation, which are necessary for the proper functioning of CD8+ T Cells (Brownlie and Zamoyska, 2013). Further, SCFA presence was shown to enhance the function of cytotoxic T lymphocytes through an increased function of mTOR post-treatment of T cells with pentanoate and C4 and driving supplementary inhibition of class I histone deacetylase activity. mTOR typically drives differentiation into Th1, Th2, and Th17 but is also a critical regulator in CD8 T cell differentiation through the regulation of cytolytic effector molecules (Finlay et al., 2012; Liu et al., 2015). Effector molecules such as CD25, IFN-γ, and TNF-α were then elevated in these treated cells, demonstrating enhanced cytotoxic activity and potential for pentanoate and C4 as supplements to cancer treatment for some malignancies (Figure 2D). (Schiweck et al., 2022).
Elimination of microbial species by antibiotics and the resulting effect on T cell populations
Considering the impact of microbial species on T-cell function, clinicians should be cognizant of the immunopharmacological behavior of antibiotics regarding microbial species-specific elimination based on antibiotic type. For instance, neomycin, which predominantly eradicates facultative gram-negative species, and vancomycin which predominantly eradicates gram-positive species, have both been associated with the reduced expansion of T cells in mouse models during antibiotic administration (Duan et al., 2010; Cheng et al., 2017). Cocktails of ampicillin, vancomycin, neomycin and metronidazole (AVNM) have been associated with lower immune function and decreased concentrations of bacterial metabolites C3 and C4 in mice (Ubeda and Pamer, 2012). γδ T cells are a part of the Th17 subset and are a source of the pro-inflammatory cytokine IL-17. Antibiotics have also been shown to modulate γδ T cell populations, with variations depending on the antibiotic type used and species of bacteria eliminated (Duan et al., 2010). Understanding which antibiotics eliminate microbial populations may be critical when designing patient treatment regimens to reduce the chances of anomalous immune cell function.
Interactions of dysbiosis on T cell-based immunotherapy in the TME
Both CD4 and CD8 T cells play an essential role in the clearance of malignancies. The impact of microbiome dysbiosis on immune cell function generates a challenging dilemma, considering that many immunotherapies in pre-clinical and clinical use, such as immune checkpoint blockade and adoptive cellular therapies, are T-cell-based.
Impact of dysbiosis on T cell dependent immune checkpoint blockade
It has been documented that specific microbes in the gut can impact ICB immunotherapy approaches across several cancers (McCulloch et al., 2022). For instance, anti-CTLA-4 treatment for melanoma relies on the presence of Bacteroides species; additionally, the treatment showed no effect on germ-free and antibiotic-treated mice (Vétizou et al., 2015). Bacteroides species can be propionogenic, having the capacity to generate the SCFA C3. Therefore, the absence of C3 can alter the proper function of T cells, which anti-CTLA-4 treatment relies on (Louis et al., 2014). In a separate study, the introduction and restoration of propionogenic bacteria during antibiotic-induced dysbiosis resulted in the restoration of C3 levels, indicating that the re-establishment of propionogenic bacteria could counteract decreases in SCFAs required for the efficacy of particular immunotherapy (El Hage et al., 2019). In mice and humans, high C4 concentrations in the blood were associated with resistance to anti-CTLA-4 therapies, evidenced by restrained upregulation of B7 on T cells (Coutzac et al., 2020). Similar results of reliance of immunotherapy efficacy on bacterial presence were seen in anti-PD-L1 treatment for melanoma, which depended on the presence of Bifidobacterium (Sivan et al., 2015). Bifidobacterium species produce SCFAs C2, C3, and C4 SCFAs that contribute to immune cell function (Louis et al., 2014). Strikingly, more recent investigations have shown fecal transplants from ICB responders to non-responders for melanoma treatment saw that more than one-third of human patients previously unresponsive to treatment become responsive after transplants (Davar et al., 2021).
Impact of dysbiosis on adoptive T-cell therapy
Recent studies in mice have further shown that differences in gut microbiome composition and dysbiosis due to antibiotic administration could alter the efficacy of adoptive T-cell cancer treatments. However, these changes in response to treatment are likely species-specific since they vary based on the type of antibiotic administered. In fact, some mice treated with vancomycin displayed an increase in CD8α+ dendritic cells (DCs) with supplemental decreases in tumor burden in an IL-12-dependent manner. At the same time, alternative antibiotics did not produce the same effect (Uribe-Herranz et al., 2018). Additional studies have indicated that severe cytokine release syndrome during CAR- T cell therapy is associated with particular microbiome alterations and a higher abundance of Bifidobacterium, Leuconostoc, Stenotrophomonas, and Staphylococcus and that desired responses may require specific gut microbial presence (Hu et al., 2022; Smith et al., 2022). These results demonstrate that microbiome composition can also impact T-cell therapies whose efficacy relies on proper immune cell function.
The impact of diet on T cell based cancer therapies
It is well known that diet can influence the establishment of microbial communities within a host (David et al., 2013). Since we now know that microbial community composition can impact T cell function, investigations regarding how diet can impact cancer treatments that rely on T cell function have been recently investigated. In mice, the western diet of high fat, high carbohydrate, and low fiber diet can decrease downstream production of short-chain fatty acids (SCFA), which originate from microbiota; this could impact T cell function and efficacy of treatments that rely on T cells. (Statovci et al., 2017). Other findings have shown that ketogenic diets can increase antitumor immunosurveillance by reducing PD-L1 expression on tumor cells in a malignant glioma model. (Lussier et al., 2016). Additionally, when placed on a ketogenic diet, mice have displayed enhancement of the anticancer effects of PD-1 blockade. (Ferrere et al., 2021). More recent studies have shown that when fucoidan, a polysaccharide naturally derived from brown algae, was co-administered with ICB treatment, it significantly improved the antitumoral activity of PD-1 antibodies in a murine melanoma model in vivo through consistent activation of tumor-infiltrating CD8+ T cells. (Yang et al., 2021). Recent studies also indicate that calorie restriction can increase the antitumoral ability of T cells (Pietrocola and Kroemer, 2019), an approach that, when applied to a murine triple-negative breast cancer model, augmented radiation efficacy (Saleh et al., 2013). Further, clinical trials analyzing melanoma patients showed that patients who consumed a high-fiber diet were five times more likely to respond to PD-1 therapy (Spencer et al., 2019). Modulation of diet serves as a potential interventional strategy that can be used to enhance T cell-based immunotherapies therapies in the future.
Discussion
With the onset of technological advancements such as molecular sequencing and sophisticated Metabolic-network modeling, insights into how microbiome composition impacts health, disease, immune cell function disease, cancer, and immune cell function have been recently better illuminated. Further insights into cohesion between microbiome composition and proper immune cell function may be a helpful resource that will eventually allow scientists to regulate the immune response to and clearance of malignancies. Further, broadening our understanding of T cell function in the TME concerning microbiome composition may improve our understanding of how best to administer current antineoplastic drugs and therapies.
There also exists potential to exploit the dysbiotic microbiome’s impact on immune cell function as an augmentation to anti-tumoral therapies for some cancers originating in common lymphoid progenitors (CLP). For instance, patients with cutaneous T-cell lymphoma (CTCL) reported decreased overall tumor burden when treated with an aggressive antibiotic regimen. Upon immunohistochemistry analysis, samples displayed a decrease in interleukin-2 high-affinity receptors in T cells at these sites, indicating a decline in mechanisms that allow for T cell proliferation (Lindahl et al., 2019). Similar regimens have also been pursued in Mucosal Associated Lymphoid Tissue (MALT) lymphomas, improving 5- year survival rates (Ferreri et al., 2018). These incidents demonstrate the importance of identifying how antibiotics can affect immune cells and the specific malignant cells clinicians might target during treatment.
Though the composition of the microbiome and microbial dysbiosis can impact immune cell function and subsequent immune response to malignancy, research gaps still exist that must be pursued to mitigate the adverse effects of microbial dysbiosis and immune system dysfunction. Further research is needed into what comprises the “optimal” microbiome and whether this composition differs for particular diseases and malignancies. Current data indicate a requirement for the presence of specific microbial species’ therapeutic efficiency (Sivan et al., 2015), but this also tends to vary from cancer to cancer, making it difficult to classify what an optimal microbiome for patients might look like. For this reason, a more significant effort is needed to determine what microbes may offer benefits during cancer treatment and what causes these microbiomes to be beneficial or harmful in particular organ systems. Since both gut and tumor-specific microbial composition can potentially permute disease progression, identifying what comprises microbial populations both locally and systemically across specific cancers may serve as a helpful resource for future diagnostic approaches. Several microbial signatures between blood and tissues have already been identified across cancers, indicating a potential diagnostic approach to cancer treatment that may soon be available (Poore et al., 2020).
Further research into how the dysbiotic state of the microbiome can impact T cell function can lead to potentially better treatments for cancer patients through the modulation of microbes present in the host. Knowledge pertaining to which bacterial communities and their associated mechanisms are needed for proper immune cell function would allow physicians to be aware of the implications associated with specific antibiotic use and subsequent species-specific elimination of bacterial communities in conjunction with ICB treatments. Overall, expanding our knowledge about the microbiomes’ interconnection with the immune system and T-cell function during cancer progression and treatment can improve our knowledge of how to design best and administer cancer treatments and ultimately improve patient outcomes.
Author contributions
MD conceived, designed, and wrote the first draft of this manuscript. MD and JB read and critically revised the manuscript.
Funding
Funding for this work has been supported by the Sarcoma Foundation of America (SFA) and Jeff Gordon Children’s Foundation (JGCF). Funding for this article was also provided in-part by the Graduate and Professional Student Association (GPSA) at Arizona State University (ASU) in conjunction with the Graduate College at ASU.
Acknowledgments
All figure images in this article were created using BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadzadeh, M., Johnson, L. A., Heemskerk, B., Wunderlich, J. R., Dudley, M. E., White, D. E., et al. (2009). Tumor antigen–specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114, 1537–1544. doi:10.1182/BLOOD-2008-12-195792
Al-Shibli, K. I., Donnem, T., Al-Saad, S., Persson, M., Bremnes, R. M., and Busund, L. T. (2008). Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin. Cancer Res. 14, 5220–5227. doi:10.1158/1078-0432.CCR-08-0133
Andresen, L., Hansen, K. A., Jensen, H., Pedersen, S. F., Stougaard, P., Hansen, H. R., et al. (2009). Propionic acid secreted from propionibacteria induces NKG2D ligand expression on human-activated T lymphocytes and cancer cells. J. Immunol. 183, 897–906. doi:10.4049/JIMMUNOL.0803014
Apostolou, P., Tsantsaridou, A., Papasotiriou, I., Toloudi, M., Chatziioannou, M., and Giamouzis, G. (2011). Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 6, 137. doi:10.1186/1749-8090-6-137
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., Van Der Veeken, J., Deroos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nat 504, 451–455. doi:10.1038/nature12726
Banerjee, S., Tian, T., Wei, Z., Shih, N., Feldman, M. D., Peck, K. N., et al. (2018). Distinct microbial signatures associated with different breast cancer types. Front. Microbiol. 9, 951. doi:10.3389/fmicb.2018.00951
Banerjee, S., Wei, Z., Tian, T., Bose, D., Shih, N. N. C., Feldman, M. D., et al. (2021). Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 129 (12), 831. doi:10.1038/s41419-021-04092-x
Brownlie, R. J., and Zamoyska, R. (2013). T cell receptor signalling networks: Branched, diversified and bounded. Nat. Rev. Immunol. 13, 257–269. doi:10.1038/nri3403
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi:10.1101/GR.126516.111
Chávez-Galán, L., Arenas-Del Angel, M. C., Zenteno, E., Chávez, R., and Lascurain, R. (2009). Cell death mechanisms induced by cytotoxic lymphocytes. Cell. Mol. Immunol. 6, 15–25. doi:10.1038/cmi.2009.3
Cheng, R. Y., Li, M., Li, S. S., He, M., Yu, X. H., Shi, L., et al. (2017). Vancomycin and ceftriaxone can damage intestinal microbiota and affect the development of the intestinal tract and immune system to different degrees in neonatal mice. Pathog. Dis. 75, 104. doi:10.1093/FEMSPD/FTX104
Claesson, M. J., Jeffery, I. B., Conde, S., Power, S. E., O’connor, E. M., Cusack, S., et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nat 488, 178–184. doi:10.1038/nature11319
Costantini, L., Magno, S., Albanese, D., Donati, C., Molinari, R., Filippone, A., et al. (2018). Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 8, 16893–16899. doi:10.1038/s41598-018-35329-z
Coutzac, C., Jouniaux, J. M., Paci, A., Schmidt, J., Mallardo, D., Seck, A., et al. (2020). Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 11, 2168. doi:10.1038/s41467-020-16079-x
Cummings, J. H., Pomare, E. W., Branch, H. W. J., Naylor, C. P. E., and MacFarlane, G. T. (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227. doi:10.1136/GUT.28.10.1221
Czesnikiewicz-Guzik, M., and Müller, D. N. (2018). Scientists on the Spot: Salt, the microbiome, and cardiovascular diseases. Cardiovasc. Res. 114, e72–e73. doi:10.1093/cvr/cvy171
Davar, D., Dzutsev, A. K., McCulloch, J. A., Rodrigues, R. R., Chauvin, J. M., Morrison, R. M., et al. (2021). Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602. doi:10.1126/science.abf3363
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2013). Diet rapidly and reproducibly alters the human gut microbiome. Nat 505, 559–563. doi:10.1038/nature12820
Duan, J., Chung, H., Troy, E., and Kasper, D. L. (2010). Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe 7, 140–150. doi:10.1016/j.chom.2010.01.005
El Hage, R., Hernandez-Sanabria, E., Calatayud Arroyo, M., Props, R., and Van De Wiele, T. (2019). Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front. Microbiol. 10, 1206. doi:10.3389/fmicb.2019.01206
Ferrere, G., Alou, M. T., Liu, P., Goubet, A. G., Fidelle, M., Kepp, O., et al. (2021). Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. JCI Insight 6, e145207. doi:10.1172/JCI.INSIGHT.145207
Ferreri, A. J. M., Cecchetti, C., Kiesewetter, B., Sassone, M., Calimeri, T., Perrone, S., et al. (2018). Clarithromycin as a “repurposing drug” against MALT lymphoma. Br. J. Haematol. 182, 913–915. doi:10.1111/BJH.14878
Finlay, D. K., Rosenzweig, E., Sinclair, L. V., Carmen, F. C., Hukelmann, J. L., Rolf, J., et al. (2012). PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 209, 2441–2453. doi:10.1084/JEM.20112607
Freilich, S., Kreimer, A., Borenstein, E., Yosef, N., Sharan, R., Gophna, U., et al. (2009). Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 10, R61–R68. doi:10.1186/gb-2009-10-6-r61
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nat 504, 446–450. doi:10.1038/nature12721
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. doi:10.1126/science.aan4236
Greathouse, K. L., White, J. R., Vargas, A. J., Bliskovsky, V. V., Beck, J. A., von Muhlinen, N., et al. (2018). Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123. doi:10.1186/s13059-018-1501-6
Groh, V., Brühl, A., El-Gabalawy, H., Nelson, J. L., and Spies, T. (2003). Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. 100, 9452–9457. doi:10.1073/PNAS.1632807100
Haabeth, O. A. W., Tveita, A. A., Fauskanger, M., Schjesvold, F., Lorvik, K. B., Hofgaard, P. O., et al. (2014). How do CD4+ T cells detect and eliminate tumor cells that either lack or express MHC class II molecules? Front. Immunol. 5, 174. doi:10.3389/fimmu.2014.00174
Han, H., Yan, H., and King, K. Y. (2021). Broad-spectrum antibiotics deplete bone marrow regulatory T cells. Cells 10, 1–9. doi:10.3390/CELLS10020277
Hu, Y., Li, J., Ni, F., Yang, Z., Gui, X., Bao, Z., et al. (2022). CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat. Commun. 13, 5313–5314. doi:10.1038/s41467-022-32960-3
Iida, N., Dzutsev, A., Stewart, C. A., Smith, L., Bouladoux, N., Weingarten, R. A., et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967. doi:10.1126/science.1240527
Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., Von Herrath, M. G., and Schoenberger, S. P. (2003). CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nat 421, 852–856. doi:10.1038/nature01441
Kamp, F., and Hamilton, J. A. (2006). How fatty acids of different chain length enter and leave cells by free diffusion. Prostagl. Leukot. Essent. Fat. Acids 75, 149–159. doi:10.1016/J.PLEFA.2006.05.003
Kespohl, M., Vachharajani, N., Luu, M., Harb, H., Pautz, S., Wolff, S., et al. (2017). The microbial metabolite butyrate induces expression of Th1- associated factors in cD4+ T cells. Front. Immunol. 8, 1036. doi:10.3389/fimmu.2017.01036
Kim, C. H. (2021). Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 185 (18), 1161–1171. doi:10.1038/s41423-020-00625-0
Kopenhaver, J., Carlson, R. D., and Snook, A. E. (2020). Mobilizing toxins for cancer treatment: Historical perspectives and current strategies. Toxins 12, 416. doi:10.3390/TOXINS12060416
Lindahl, L. M., Willerslev-Olsen, A., Gjerdrum, L. M. R., Nielsen, P. R., Blümel, E., Rittig, A. H., et al. (2019). Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 134, 1072–1083. doi:10.1182/BLOOD.2018888107
Liu, C., Chapman, N. M., Karmaus, P. W. F., Zeng, H., and Chi, H. (2015). mTOR and metabolic regulation of conventional and regulatory T cells. J. Leukoc. Biol. 97, 837–847. doi:10.1189/JLB.2RI0814-408R
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 1210 (12), 661–672. doi:10.1038/nrmicro3344
Lundin, A., Bok, C. M., Aronsson, L., Björkholm, B., Gustafsson, J. Å., Pott, S., et al. (2008). Gut flora, toll-like receptors and nuclear receptors: A tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 10, 1093–1103. doi:10.1111/J.1462-5822.2007.01108.X
Lundin, K. U., Screpanti, V., Omholt, H., Hofgaard, P. O., Yagita, H., Grandien, A., et al. (2004). CD4+ T cells kill Id+ B-lymphoma cells: FasLigand-fas interaction is dominant in vitro but is redundant in vivo. Cancer Immunol. Immunother. 53, 1135–1145. doi:10.1007/s00262-004-0538-4
Lussier, D. M., Woolf, E. C., Johnson, J. L., Brooks, K. S., Blattman, J. N., and Scheck, A. C. (2016). Enhanced immunity in a mouse model of malignant glioma is mediated by a therapeutic ketogenic diet. BMC Cancer 16, 310–10. doi:10.1186/s12885-016-2337-7
Luu, M., Pautz, S., Kohl, V., Singh, R., Romero, R., Lucas, S., et al. (2019). The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat. Commun. 10, 760. doi:10.1038/s41467-019-08711-2
McCulloch, J. A., Davar, D., Rodrigues, R. R., Badger, J. H., Fang, J. R., Cole, A. M., et al. (2022). Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 28, 545–556. doi:10.1038/s41591-022-01698-2
Methé, B. A., Nelson, K. E., Pop, M., Creasy, H. H., Giglio, M. G., Huttenhower, C., et al. (2012). A framework for human microbiome research. Nat 486, 215–221. doi:10.1038/nature11209
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. doi:10.1126/science.aay9189
Ossendorp, F., Mengedé, E., Camps, M., Filius, R., and Melief, C. J. M. (1998). Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J. Exp. Med. 187, 693–702. doi:10.1084/JEM.187.5.693
Park, J., Kim, M., Kang, S. G., Jannasch, A. H., Cooper, B., Patterson, J., et al. (2014). Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR–S6K pathway. Mucosal Immunol. 81 (8), 80–93. doi:10.1038/mi.2014.44
Petersen, C., and Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16, 1024–1033. doi:10.1111/CMI.12308
Pham, T. A. N., and Lawley, T. D. (2014). Emerging insights on intestinal dysbiosis during bacterial infections. Curr. Opin. Microbiol. 17, 67–74. doi:10.1016/J.MIB.2013.12.002
Pietrocola, F., and Kroemer, G. (2019). Caloric restriction promotes the stemness and antitumor activity of T lymphocytes. Oncoimmunology 8, e1616153. doi:10.1080/2162402X.2019.1616153
Poore, G. D., Kopylova, E., Zhu, Q., Carpenter, C., Fraraccio, S., Wandro, S., et al. (2020). Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nat 579, 567–574. doi:10.1038/s41586-020-2095-1
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nat 464, 59–65. doi:10.1038/nature08821
Rangan, P., and Mondino, A. (2022). Microbial short-chain fatty acids: A strategy to tune adoptive T cell therapy. J. Immunother. Cancer 10, e004147. doi:10.1136/JITC-2021-004147
Riiser, A. (2015). The human microbiome, asthma, and allergy. Allergy, Asthma Clin. Immunol. 11, 35–37. doi:10.1186/S13223-015-0102-0
Riquelme, E., Zhang, Y., Zhang, L., Montiel, M., Zoltan, M., Dong, W., et al. (2019). Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806. doi:10.1016/J.CELL.2019.07.008
Russo, E., Taddei, A., Ringressi, M. N., Ricci, F., and Amedei, A. (2016). The interplay between the microbiome and the adaptive immune response in cancer development. Ther. Adv. Gastroenterol. 9, 594–605. doi:10.1177/1756283X16635082
Ryu, T. Y., Kim, K., Han, T. S., Lee, M. O., Lee, J., Choi, J., et al. (2022). Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 165 (16), 1205–1221. doi:10.1038/s41396-021-01119-1
Saleh, A. D., Simone, B. A., Palazzo, J., Savage, J. E., Sano, Y., Dan, T., et al. (2013). Caloric restriction augments radiation efficacy in breast cancer. Cell Cycle 12, 1955–1963. doi:10.4161/CC.25016
Sánchez-Alcoholado, L., Ramos-Molina, B., Otero, A., Laborda-Illanes, A., Ordóñez, R., Medina, J. A., et al. (2020). The role of the gut microbiome in colorectal cancer development and therapy response. Cancers 12, 1406. doi:10.3390/CANCERS12061406
Schiweck, C., Edwin Thanarajah, S., Aichholzer, M., Matura, S., Reif, A., Vrieze, E., et al. (2022). Regulation of CD4+ and CD8+ T cell Biology by short-chain fatty acids and its relevance for autoimmune pathology. Int. J. Mol. Sci. 23, 8272. doi:10.3390/IJMS23158272
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 14, e1002533. doi:10.1371/JOURNAL.PBIO.1002533
Sfanos, K. S., Sauvageot, J., Fedor, H. L., Dick, J. D., De Marzo, A. M., and Isaacs, W. B. (2008). A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 68, 306–320. doi:10.1002/PROS.20680
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi:10.1126/science.aac4255
Smith, M., Dai, A., Ghilardi, G., Amelsberg, K. V., Devlin, S. M., Pajarillo, R., et al. (2022). Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 28, 713–723. doi:10.1038/s41591-022-01702-9
Spencer, C. N., Gopalakrishnan, V., McQuade, J., Andrews, M. C., Helmink, B., Khan, M. A. W., et al. (2019). Abstract 2838: The gut microbiome (GM) and immunotherapy response are influenced by host lifestyle factors. Cancer Res. 79, 2838. doi:10.1158/1538-7445.AM2019-2838
Statovci, D., Aguilera, M., MacSharry, J., and Melgar, S. (2017). The impact of Western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front. Immunol. 8, 838. doi:10.3389/fimmu.2017.00838
Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., and Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119. doi:10.1016/B978-0-12-800100-4.00003-9
Taylor, R. C., Patel, A., Panageas, K. S., Busam, K. J., and Brady, M. S. (2007). Tumor-infiltrating lymphocytes predict sentinel lymph node positivity in patients with cutaneous melanoma. J. Clin. Oncol. 25, 869–875. doi:10.1200/JCO.2006.08.9755
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nat 535, 65–74. doi:10.1038/nature18847
Thompson, K. J., Ingle, J. N., Tang, X., Chia, N., Jeraldo, P. R., Walther-Antonio, M. R., et al. (2017). A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS One 12, e0188873. doi:10.1371/JOURNAL.PONE.0188873
Trujillo, J. A., Sweis, R. F., Bao, R., and Luke, J. J. (2018). T cell–inflamed versus non-T cell–inflamed tumors: A conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol. Res. 6, 990–1000. doi:10.1158/2326-6066.CIR-18-0277
Tuddenham, S., and Sears, C. L. (2015). The intestinal microbiome and health. Curr. Opin. Infect. Dis. 28, 464–470. doi:10.1097/QCO.0000000000000196
Ubeda, C., and Pamer, E. G. (2012). Antibiotics, microbiota, and immune defense. Trends Immunol. 33, 459–466. doi:10.1016/J.IT.2012.05.003
Uribe-Herranz, M., Bittinger, K., Rafail, S., Guedan, S., Pierini, S., Tanes, C., et al. (2018). Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI insight 3, e94952. doi:10.1172/JCI.INSIGHT.94952
Vétizou, M., Pitt, J. M., Daillère, R., Lepage, P., Waldschmitt, N., Flament, C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi:10.1126/science.aad1329
Willing, B. P., Dicksved, J., Halfvarson, J., Andersson, A. F., Lucio, M., Zheng, Z., et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854. doi:10.1053/J.GASTRO.2010.08.049
Yang, J., Yang, X., Pan, W., Wang, M., Lu, Y., Zhang, J., et al. (2021). Fucoidan-supplemented diet potentiates immune checkpoint blockage by enhancing antitumor immunity. Front. Cell Dev. Biol. 9, 2198. doi:10.3389/fcell.2021.733246
Yang, W., Yu, T., Huang, X., Bilotta, A. J., Xu, L., Lu, Y., et al. (2020). Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 11, 4457. doi:10.1038/s41467-020-18262-6
Zhang, L., Conejo-Garcia, J. R., Katsaros, D., Gimotty, P. A., Massobrio, M., Regnani, G., et al. (2003). Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 348, 203–213. doi:10.1056/NEJMoa020177
Zhao, Y., Chen, F., Wu, W., Sun, M., Bilotta, A. J., Yao, S., et al. (2018). GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 11, 752–762. doi:10.1038/MI.2017.118
Zhou, X., Liu, X., Huang, L., Zhou, X., Huang, L., and Liu, X. (2021). Macrophage-Mediated tumor cell phagocytosis: Opportunity for nanomedicine intervention. Adv. Funct. Mater. 31, 2006220. doi:10.1002/ADFM.202006220
Keywords: microbiome, T cell, tumor microenvironment (TME), dysbiosis, metabolites, T cell signaling, short chain fatty acids (SCFAs), immunotherapy
Citation: DiPalma MP and Blattman JN (2023) The impact of microbiome dysbiosis on T cell function within the tumor microenvironment (TME). Front. Cell Dev. Biol. 11:1141215. doi: 10.3389/fcell.2023.1141215
Received: 10 January 2023; Accepted: 06 March 2023;
Published: 17 March 2023.
Edited by:
Kristian Michael Hargadon, Hampden–Sydney College, United StatesReviewed by:
Manoj Chelvanambi, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2023 DiPalma and Blattman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph N. Blattman, am9zZXBoLmJsYXR0bWFuQGFzdS5lZHU=
 Michelle P. DiPalma
Michelle P. DiPalma Joseph N. Blattman
Joseph N. Blattman