- Department of Cancer Biology, Mayo Clinic, Jacksonville, FL, United States
Cadherin-catenin complexes are integral components of the adherens junctions crucial for cell-cell adhesion and tissue homeostasis. Dysregulation of these complexes is linked to cancer development via alteration of cell-autonomous oncogenic signaling pathways and extrinsic tumor microenvironment. Advances in multiomics have uncovered key signaling events in multiple cancer types, creating a need for a better understanding of the crosstalk between cadherin-catenin complexes and oncogenic pathways. In this review, we focus on the biological functions of classical cadherins and associated catenins, describe how their dysregulation influences major cancer pathways, and discuss feedback regulation mechanisms between cadherin complexes and cellular signaling. We discuss evidence of cross regulation in the following contexts: Hippo-Yap/Taz and receptor tyrosine kinase signaling, key pathways involved in cell proliferation and growth; Wnt, Notch, and hedgehog signaling, key developmental pathways involved in human cancer; as well as TGFβ and the epithelial-to-mesenchymal transition program, an important process for cancer cell plasticity. Moreover, we briefly explore the role of cadherins and catenins in mechanotransduction and the immune tumor microenvironment.
Introduction
The integrity of the epithelial monolayer is critical for tissue morphogenesis and depends on dynamic interactions between cells to maintain tissue homeostasis. A group of transmembrane proteins termed cadherins and their associated catenins play a critical role in these interactions at areas of cell-cell contact. Cadherin-catenin complexes (CCC) coalesce to form adherens junctions (AJs), which in addition to their adhesive function, are dynamic structures and hubs for intracellular signaling conveying signals related to cohesion, tension, proliferation, and inflammation. The ultimate goal of this signaling is maintenance of epithelial barrier function and tissue homeostasis.
Not surprisingly, dysfunction of the CCC is associated with loss of epithelial architecture, increased cell proliferation and survival, as well as enhanced cell migration and invasion. In line with these altered cellular behaviors, CCC components are frequently deregulated in human cancer and implicated in cancer progression. For example, loss of epithelial cadherin (E-cadherin, encoded by CDH1) is considered as the most prominent cancer driving event in invasive lobular carcinoma of the breast (ILC) (Ciriello et al., 2015), as well as hereditary and a subset of sporadic diffuse gastric cancer (Wang et al., 2014; Cho et al., 2017). The mechanism by which E-cadherin loss promotes cancer has been studied in several animal models [reviewed in (Bruner and Derksen, 2018)]. Examples of catenin misregulation include recurrent homozygous deletion of α-catenin (encoded by CTNNA1) in basal-like breast cancer (Ding et al., 2010). Genomic alterations of p120-catenin (encoded by CTNND1) are relatively rare in human cancer (cBioPortal). Epigenetic downregulation of p120-catenin expression promotes E-cadherin degradation and cancer progression in non-small-cell lung cancer (Mortazavi et al., 2010). Conversely, high p120-catenin expression is thought to be a key event in the progression of inflammatory breast cancer (Silvera et al., 2009).
Advances in sequencing technologies and multi-omics approaches are unraveling the oncogenic landscape of human tumors, including the elucidation of key oncogenic signaling pathways that play major roles in tumor initiation and progression. In this review, after a brief overview of CCC organization and function, we discuss recent advances in our understanding of the relationship between cadherins/catenins and oncogenic pathways, including Hippo-YAP/TAZ, receptor tyrosine kinase (RTK), WNT, Notch, hedgehog (HH), and TGFβ/epithelial-mesenchymal transition (EMT) pathways. In each section, we cover the introduction of an oncogenic pathway of interest, its involvement in particular cancer types according to omics data, and its bidirectional interplay with cadherins and catenins. As most human cancer is epithelial in origin, our focus will be E-cadherin and its related catenins, with occasional discussion on roles of other classical cadherins, such as neural cadherin (N-cadherin, encoded by CDH2) and vascular cadherin (VE-cadherin, encoded by CDH5). The role of cadherins/catenins in mechanotransduction and their crosstalk with the tumor microenvironment (TME) will also be discussed.
Cadherin-catenin complex
E-cadherin is the prototypical member of the classical cadherin family. It is a transmembrane adhesion receptor that contains five extracellular repeats, a single-span transmembrane region, and a cytoplasmic tail (Figure 1). E-cadherin ectodomains (ECs) mediate calcium-dependent homophilic interactions with E-cadherins in the same (cis interactions via EC1 in one cadherin and EC2 and EC3 of an adjacent cadherin) and apposing cells (trans interactions via EC1 from both cadherin molecules from each cell, existing in weak X-dimer and strong strand-swap dimer conformations) (Yagi and Takeichi, 2000; Wu et al., 2010; Harrison et al., 2011; Wu et al., 2011; Brasch et al., 2012; Fichtner et al., 2014) that cooperatively strengthen each other’s stability (Zhang et al., 2009; Thompson et al., 2021). The structural and dynamic cis/trans interactions of E-cadherin molecules are beyond the scope of this review and have been reviewed in detail elsewhere (Brasch et al., 2012; Troyanovsky, 2022). On the other hand, the extracellular adhesive contact is further enforced by interaction of E-cadherin’s cytoplasmic tail with various catenins (inside-out signaling) (Nagafuchi and Takeichi, 1988; Ozawa et al., 1989; Thoreson et al., 2000; Gottardi and Gumbiner, 2001; Petrova et al., 2012; Shashikanth et al., 2015; Maiden et al., 2016; Koirala et al., 2021) (Figure 1).
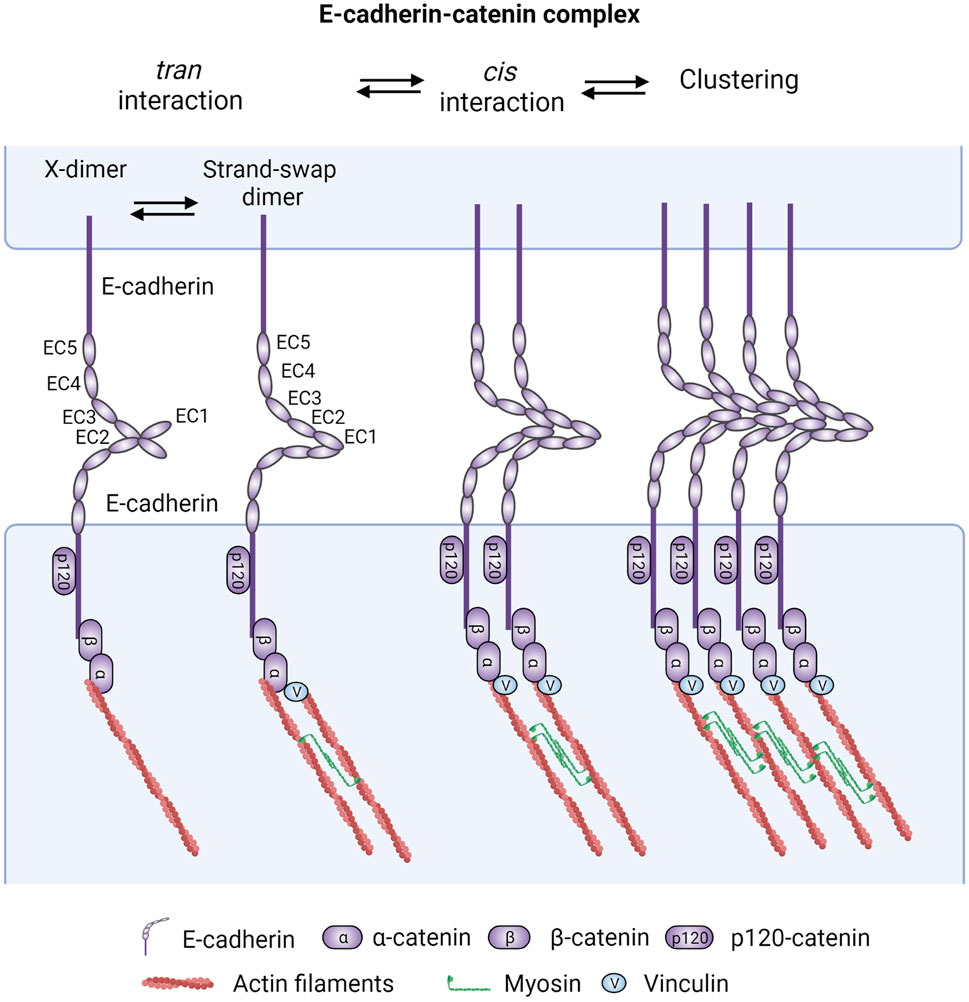
FIGURE 1. The core epithelial cadherin-catenin cell adhesion system. Epithelial cadherin-catenin complexes are the main building blocks of intercellular adhesions and are comprised of at least membrane-bound E-cadherin associated with p120-catenin (p120), β-catenin (β-cat), and α-catenin (α-cat). Fundamentally, these adhesions are mediated by homophilic E-cadherin adhesive (trans dimerization forming the X-dimer and strand-swap dimer) interactions between the ectodomains (EC) of cadherins on opposite cells and lateral (cis) binding between cadherin molecules on the same cell. These cis/trans interactions reciprocally strengthen each other’s stability, building up cadherin clusters. The clustering dynamics and stability can be further fine tuned by cellular and external biochemical or mechanical inputs. Of note, p120 binding to E-cadherin has also been shown to strengthen E-cadherin cis dimerization and trans interaction Additionally, intracellular actomyosin-mediated pulling forces, which are linked to cadherin molecules through α-catenin binding to β-catenin, further potentiate the stability of adherens junctions through a tension-mediated recruitment of vinculin (v) and its interaction with actin filaments. Figure created with BioRender.com.
It is noteworthy that cadherins associate with a large number of proteins that collectively form the cadherin adhesome (also known as cadhesome) and structurally support and/or regulate the dynamics of cadherin junctions (Guo et al., 2014; Van Itallie et al., 2014; Bertocchi et al., 2017; Li et al., 2019; Shafraz et al., 2020). These associations are thought to occur at distinct microclusters during the process of CCC clustering [reviewed in (Yap et al., 2015; Troyanovsky, 2022)]. While many of these interacting proteins are likely to have roles in cancer, this review will focus primarily on catenins, the most studied and better understood cadherin interacting partners that mediate linkage to the cytoskeleton and regulate adhesion-induced signaling. β-catenin (encoded by CTNNB1) binds directly to the C-terminal “catenin-binding domain” of the cadherin cytoplasmic tail (Nagafuchi and Takeichi, 1988; Ozawa et al., 1989; Ozawa et al., 1990; McCrea and Gumbiner, 1991; Stappert and Kemler, 1994; Thoreson et al., 2000; Gottardi and Gumbiner, 2001; Huber and Weis, 2001) and recruits alpha epithelial catenin (αE-catenin) to bridge E-cad adhesions to the actin cytoskeleton (Rimm et al., 1995; Drees et al., 2005; Desai et al., 2013). αE-Catenin also functions as allosteric regulator of AJ remodeling in a force-dependent manner, acting as a tension transducer via its homolog vinculin (Peng et al., 2012; Thomas et al., 2013; Yao et al., 2014; Ishiyama et al., 2018; Seddiki et al., 2018). Actomyosin tension changes the conformation of αE-catenin from a closed to an open state, allowing it to recruit vinculin to further reinforce cadherin adhesive forces (Figure 1). p120-catenin (encoded by CTNND1) binds to the “juxtamembrane domain” of the cadherin cytoplasmic tail to support cadherin stability, cis lateral clustering, and E-cadherin trans affinity (Yap et al., 1998; Thoreson et al., 2000; Ireton et al., 2002; Davis et al., 2003; Xiao et al., 2003; Davis and Reynolds, 2006; Kourtidis et al., 2013; Vu et al., 2021). Mature AJs form at apical regions of polarized epithelia, at the zonula adherens (ZA) (Nishimura and Takeichi, 2009). E-cadherin is considered a master regulator of the epithelial phenotype, due in part to its role in associating the ZA with a circumferential actomyosin ring that stabilizes the epithelial architecture (Miyoshi and Takai, 2008). Additionally, p120-catenin regulates the cytoskeleton via RhoGTPases (Anastasiadis, 2007), while the ZA interacts with microtubules either via β-catenin and dynein (Ligon et al., 2001) or via p120-catenin (Chen et al., 2003; Yanagisawa et al., 2004) and its interacting partner PLEKHA7 (Meng et al., 2008; Paschoud et al., 2014).
Effects of deregulated cadherin-catenin signaling on major oncogenic pathways
Contact inhibition and Hippo-YAP/TAZ signaling
In multicellular organisms, cell proliferation is tightly controlled by signals in the microenvironment and adjoining cells. Normal diploid cells in culture grow and divide in response to nutrients and growth factor signaling until they reach confluence. As cell contact increases in a density-dependent manner, cell proliferation slows down and is eventually halted in a process referred to as contact inhibition of proliferation (CIP) (Levine et al., 1965; Eagle and Levine, 1967; Hanahan and Weinberg, 2000; Motti et al., 2005; Hanahan and Weinberg, 2011). CIP is crucial for tissue morphogenesis and organ development. Loss of CIP allows cells to overgrow in epithelial monolayers, altering tissue architecture and leading to tumor formation (Levine et al., 1965). Hence, loss of CIP is one of the hallmarks of cancer and can deregulate signaling pathways that are normally suppressed by cell-cell interactions (Hanahan and Weinberg, 2000; Hanahan and Weinberg, 2011).
In an early study to identify molecular mechanisms underlying CIP, Whittenberger and Glaser observed that membrane isolates from cell monolayers were able to decrease proliferation of cells at sub-confluence (Whittenberger and Glaser, 1977). Since cadherins localize to the plasma membrane and mediate cell-cell interactions, they are prime candidates to initiate CIP. In agreement, re-expression of E-cadherin in cancer cells lacking endogenous expression suppressed cell proliferation via upregulation of cyclin-dependent kinase inhibitor p27 and reduction of cyclin E-cdk2 activity (St Croix et al., 1998; Motti et al., 2005). Addition of E-cadherin-neutralizing antibodies to these E-cadherin transfected cells repressed E-cadherin-mediated growth inhibition (St Croix et al., 1998). Moreover, re-expression of α-catenin in a lung adenocarcinoma cell line (PC9) lacking endogenous α-catenin rescued E-cadherin-mediated growth inhibition (Watabe et al., 1994). Another study reported that β-catenin binding to E-cadherin and E-cadherin engagement in subconfluent cells are sufficient to suppress growth in control or EGF treated cells (Perrais et al., 2007). Collectively these studies argue that E-cadherin-based adhesions contribute to CIP.
As cell proliferation requires transcriptional regulation of cell cycle related genes, one immediate question regarding CIP is how cadherins and AJs at the plasma membrane transduce growth signals to the nuclear transcriptional machinery. Recent studies indicate a key role for the Hippo-YAP/TAZ signaling pathways. Nuclear accumulation of the transcriptional cofactors YAP and/or TAZ promotes cell cycle progression while Hippo signaling suppresses YAP/TAZ nuclear accumulation and inhibits cell growth (Zhao et al., 2008; Piccolo et al., 2014). In mammalian cells, the Hippo pathway includes the serine threonine protein kinases Mst1/2 (homologs of Drosophila Hippo) and Lats1/2 (homologs of Drosophila Warts) and their associated regulatory scaffold proteins Sav and Mob (Figure 2). Activation of Lats kinases by Mst results in inhibitory phosphorylation of YAP/TAZ (homologs of Drosophila Yorkie and WWTR1, respectively) (Meng et al., 2016; Fu et al., 2017). Once phosphorylated, YAP/TAZ undergo cytoplasmic retention via 14-3-3 interaction or proteasome-mediated degradation. When the Hippo kinases are suppressed, YAP/TAZ become active and can interact with transcription factors (TFs), such as TEAD proteins, following nuclear translocation, to transactivate a variety of pro-proliferative and anti-apoptosis genes. Consistent with a role for this pathway in CIP, YAP phosphorylation is increased with cell density (Zhao et al., 2007). Moreover, overexpression of YAP de-represses cell proliferation in high density culture and expression of a dominant negative YAP mutant rescues the CIP phenotype in cells with Hippo pathway deficiency (Zhao et al., 2007).
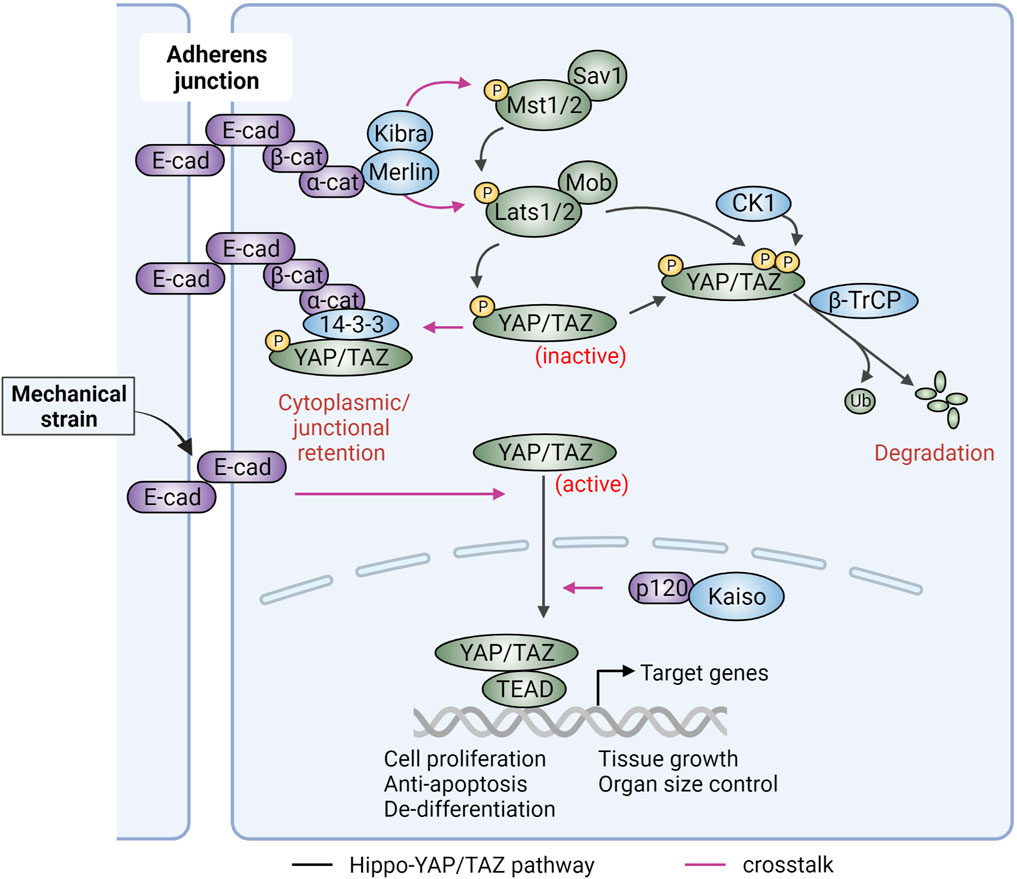
FIGURE 2. Hippo-YAP/TAZ signaling and its regulation by adherens junctions. Hippo-YAP/TAZ signaling includes the upstream Hippo components, which suppress activation of the downstream YAP/TAZ transcriptional coactivators. The core components of Hippo signaling include Mst1/2 and Lats1/2 kinases in complex with Sav1 and Mob scaffold proteins. Activated Lats1/2 promotes YAP/TAZ serine inhibitory phosphorylation, resulting in their cytoplasm/junctional retention (via 14-3-3) or β-TrCP mediated proteolytic degradation [if YAP/TAZ are further phosphorylated by casein kinase (CK1)]. Active, unphosphorylated YAP/TAZ can shuttle into the nucleus, where they cooperate with TEAD transcription factors to promote tumorigenic gene expression. This activation can be achieved by Hippo kinase inactivation, activation of nuclear p120-catenin (p120)/Kaiso signaling, or mechanical strain exerted at E-cadherin (E-cad) cell-cell junctions. In the absence of force, cadherin-mediated junctions can activate Hippo pathway kinases via β-catenin (β-cat) and α-catenin (α-cat) to inhibit YAP/TAZ-induced pro-tumorigenic signaling. Alpha-catenin can interact with YAP/TAZ and 14-3-3 preventing YAP/TAZ nuclear translocation. Merlin, which is linked to adherens junctions via α-cat, functions in a complex with the Kibra tumor suppressor to promote contact inhibition of proliferation by regulating Hippo-YAP/TAZ signaling. Figure created with BioRender.com.
Given their roles in CIP, molecular alterations promoting activation of YAP/TAZ or inhibition of Hippo kinases would be expected in human cancers. While only a few somatic or germline mutations in the Hippo-YAP/TAZ pathway have been identified (Harvey et al., 2013; Huyghe et al., 2019), homozygous deletions and inactivating mutations of the Hippo kinase LATS2 were reported in malignant mesothelioma (Murakami et al., 2011). Merlin (encoded by NF2), which activates Hippo kinases (Hamaratoglu et al., 2006; Zhang et al., 2010; Yin et al., 2013), is also frequently mutated in neurofibromatosis (Asthagiri et al., 2009) and malignant mesothelioma (Murakami et al., 2011). DNA amplification and upregulation of YAP/TAZ was seen in various cancers, such as squamous cancers and pancreatic cancer (Yu et al., 2015). In line with these findings, preclinical studies largely support the notion that deregulation of Hippo-YAP/TAZ signaling promotes cancer development [reviewed in (Harvey et al., 2013)]. Moreover, upregulation of a YAP/TAZ gene signature was found to be associated with worse clinical outcomes and chemotherapy resistance in cancer (Rozengurt et al., 2018; Nguyen and Yi, 2019).
A direct relationship between E-cadherin-mediated CIP and YAP/TAZ signaling was revealed using cells where E-cadherin engagement and CIP were triggered by E-cadherin-coated beads. Under these conditions, knockdown of Hippo core kinases or overexpression of YAP (wild-type and non-phosphorylatable active mutant) were both able to reverse E-cadherin mediated CIP (Kim et al., 2011). Conversely, expression of E-cadherin in cells lacking endogenous E-cadherin promoted cytoplasmic retention of YAP (Kim et al., 2011). Furthermore, α- and β-catenin, but not p120-catenin, were required for E-cadherin’s ability to retain YAP in the cytoplasm under conditions of high density (Kim et al., 2011). Mechanistically, an association between phosphorylated YAP and α-catenin via 14-3-3 in a tripartite complex was reported in epidermal keratinocytes (Schlegelmilch et al., 2011) (Figure 2). In this case, α-catenin functions as a negative upstream regulator of YAP. Upon calcium-mediated cell adhesion, YAP is associated with α-catenin at cellular junctions and knockdown of α-catenin leads to reduced phosphorylation and increased nuclear localization of YAP. Animal experiments with α-catenin gene targeting further argue that the tumor suppressing effects of α-catenin can be attributed to dysregulation of YAP activity (Silvis et al., 2011). This hypothesis is also supported by an inverse correlation between the levels of α-catenin and nuclear YAP in squamous cell carcinoma (Silvis et al., 2011). A role of α-catenin in suppressing YAP activity was also reported in endothelial cells, modulated by the actin processing protein EPS8. In cells with nascent junctions, YAP was active and able to shuttle into the nucleus as its binding to α-catenin was prevented by EPS8. Conversely, in mature junctions, YAP was phosphorylated by AKT downstream of VE-cadherin clustering and recruited to endothelial junctions via 14-3-3 and α-catenin (Giampietro et al., 2015). The data argue that α-catenin is a negative regulator of YAP signaling in both E-cadherin and VE-cadherin based cell-cell junctions.
Of note, α-catenin can bind directly to Merlin, a membrane-associated scaffold protein that functions as a tumor suppressor. This interaction is critical for AJ maturation (Gladden et al., 2010). Several studies have shown that Merlin in association with Kibra acts upstream of Hippo kinases (Hamaratoglu et al., 2006; Yin et al., 2013) (Figure 2). The biological impact of the connection between cadherins/catenins and Merlin was highlighted by a recent study in human mesothelioma cells showing that E-cadherin adhesions activate Merlin-Hippo signaling to resist ferroptosis, a form of cell death regulated by cellular metabolism and cellular iron (Wu et al., 2019). As several cancer-targeted agents induce ferroptosis (Lei et al., 2022), molecular alterations in cadherin/catenin and/or Merlin-YAP signaling could serve as biomarkers predicting cancer cell responsiveness to ferroptosis-inducing therapies.
As with α-catenin, β-catenin at junctions can induce YAP phosphorylation and cytoplasmic retention (Kim et al., 2011). However, the relationship between Hippo-YAP/TAZ signaling and β-catenin is more complex as β-catenin can shuttle between different subcellular pools (i.e., the plasma membrane, cytoplasm, and nucleus) in response to cellular and environmental cues (Henderson and Fagotto, 2002; Bienz, 2005; Krieghoff et al., 2006; Wu et al., 2008; Phelps et al., 2009). The most studied scenario is how WNT and Hippo-YAP/TAZ signaling regulate each other via β-catenin (see WNT section). Moreover, the impact of mechanical forces across E-cadherin junctions on β-catenin and YAP interaction has been examined in quiescent epithelial monolayers experiencing varying degrees of mechanical strain. In the absence of external stretching, YAP1 is localized to the cell cortex and cytoplasm and β-catenin is restricted at cell junctions. In response to mechanical strain that triggers E-cadherin engagement, both YAP1 and β-catenin translocate into the nucleus, with YAP1 driving cell cycle re-entry and β-catenin mediating cell transition into the S phase for DNA replication (Benham-Pyle et al., 2015) (Figure 2). Additionally, the association of nuclear p120-catenin with TF Kaiso, suppresses Kaiso’s transcriptional repressing activity resulting in the nuclear accumulation of YAP/TAZ through a mechanism that involves suppression of YAP/TAZ phosphorylation (Zhu et al., 2012) (Figure 2).
The effect of cadherin-catenin complexes on Hippo-YAP/TAZ signaling also extends to non-epithelial cells. Mesenchymal stem cells (MSCs) are pluripotent adult stromal cells that can differentiate into distinct cell lineages upon different mechanical cues and this process is known to be heavily regulated by YAP/TAZ. Interestingly, these cells can form “mechanical memory” by “remembering” the mechanical forces applied to them. Cells that have experienced stiff environment for some time (∼10 days) maintain nuclear accumulation of YAP/TAZ, even when replated onto more compliant substrates (Yang et al., 2014; Totaro et al., 2018). Using a hydrogel-based system capable of mimicking N-cadherin based cell-cell adhesions (using HAVDI peptide derived from the EC1 repeat of N-cadherin) and integrin-mediated cell-extracellular matrix (ECM) interactions (using RGD peptides), cell-ECM adhesions were found to mediate the perception and retention of mechanical memory via nuclear YAP. This feature was reversed by N-cadherin ligation and junctional β-catenin, which promoted re-localization of nuclear YAP to the cytoplasm (Zhang et al., 2021).
In summary, available data suggest that CIP is at least in part mediated by CCC-induced downregulation of YAP/TAZ signaling. E-cadherin can directly restrain YAP1 nuclear translocation via its extracellular engagement (Benham-Pyle et al., 2015), and also promote Hippo kinase activity to repress YAP/TAZ mediated transcription through its cytoplasmic domain associated catenins (Kim et al., 2011). Further, CCCs can function as a nexus to transduce mechanical cues to Hippo-YAP/TAZ signaling. As the majority of these studies utilized simple cell line models, studies in model organisms, patient-derived organoids [reviewed in (Kim et al., 2020)], or tumor-on-a-chip models [reviewed in (Sontheimer-Phelps et al., 2019; Liu et al., 2021)] could provide further insight into the interplay between CCC and Hippo-YAP/TAZ signaling and its role in cancer.
Receptor tyrosine kinase signaling pathways
RTKs and their downstream signaling pathways are crucial for cell proliferation, differentiation, and survival during development (Schlessinger, 2014) (Figure 3A). Activation of RTK signaling is common in cancer, and often attributed to amplifications, mutations, rearrangements, and overexpression of pathway components. For example, activating mutations and amplifications of epithelial growth factor receptors (EGFRs) are frequently identified in glioma (Wong et al., 1992; Cancer Genome Atlas Research Network, 2008) and lung adenocarcinoma (Cancer Genome Atlas Research Network, 2014b). ERBB2 (also known as HER2) is a commonly amplified and overexpressed proto-oncogene in a subset of breast cancer (Slamon et al., 1987; Slamon et al., 1989; Cancer Genome Atlas Network, 2012). Aberrations of fibroblast growth factor receptors (FGFRs), c-Met, and insulin-like growth factor 1 receptor (IGF1R) are frequently found in urothelial carcinoma (Cancer Genome Atlas Research Network, 2014a; Helsten et al., 2016), hepatocellular carcinoma (HCC) (Park et al., 1999; Cancer Genome Atlas Research Network, 2017) and breast cancer (Cancer Genome Atlas Network, 2012; Farabaugh et al., 2015), respectively. In addition to these growth factor signaling pathways that are activated in cancer cells, vascular endothelial growth factor receptor (VEGFR) signaling is frequently activated in endothelial cells by vascular endothelial growth factor (VEGF) released from tumor cells and stroma to promote neo-angiogenesis to facilitate tumor growth (Apte et al., 2019).
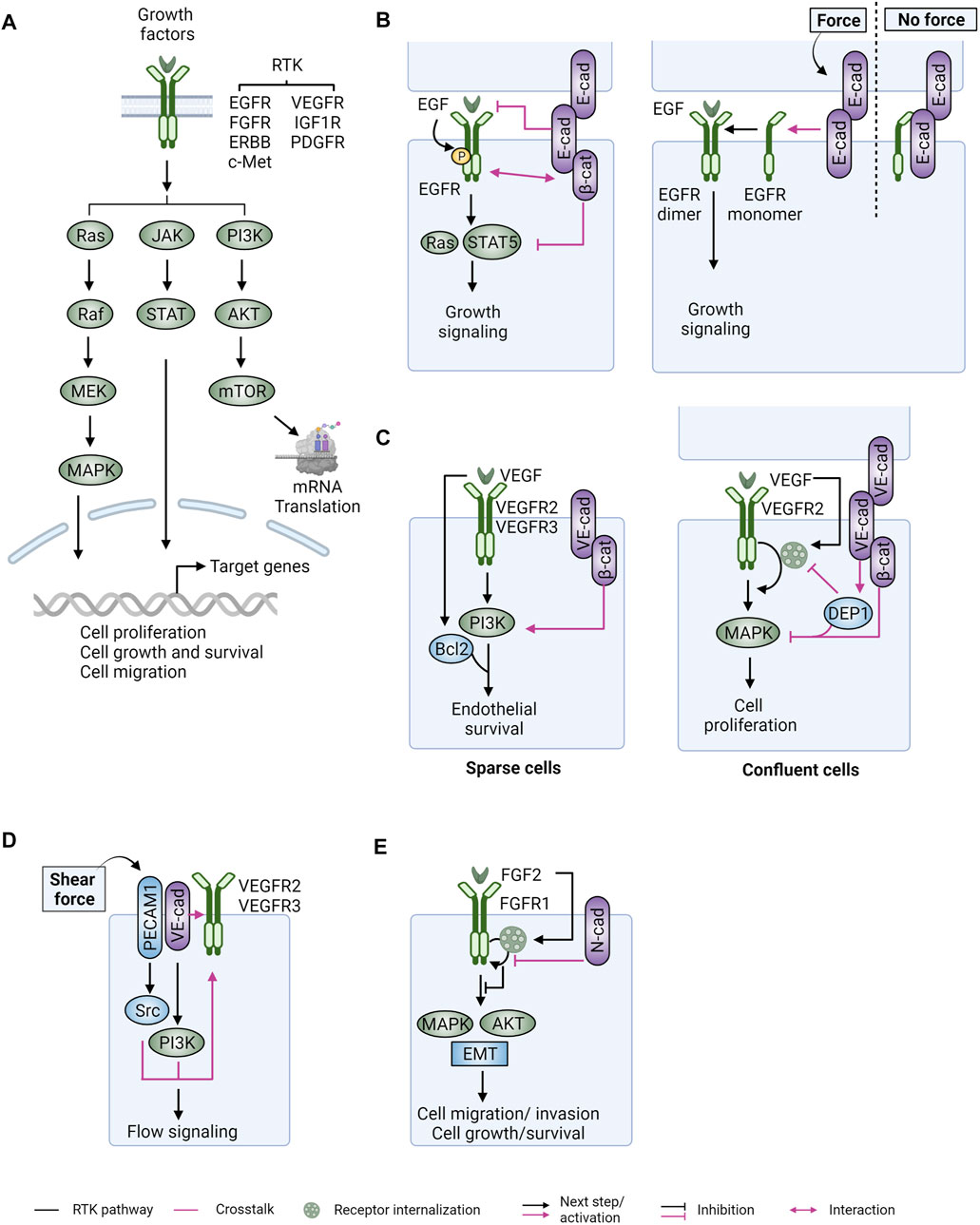
FIGURE 3. Crosstalk between receptor tyrosine kinases and cadherin-catenin complexes. (A) RTK binding to their cognate ligands (i.e., growth factors) and subsequent receptor dimerization activates three major downstream pathways (RAS-RAF-MEK-MAPK, JAK-STAT, and PI3K-AKT-mTOR) that regulate gene expression and various cellular processes. (B) (Left) In epithelial cells, E-cadherin (E-cad) can associate with EGFR through its lateral ectodomains and/or intracellular β-catenin (β-cat). E-cadherin ligation and its interaction with β-catenin are required for inhibition of EGF/EGFR-mediated cell growth via STAT5 and RAS. In general, E-cadherin junctions suppress EGFR activation and the conveyance of signal transduction to downstream effectors, while EGFR activation can promote the disassembly of E-cadherin junctions. (Right) EGFR is sequestered by E-cadherin in the absence of force, whereas it is released to set off growth signaling when external force is applied to E-cadherin junctions. (C) (Left) In endothelial cells with sparse cell contacts (sparse), VEGF-A ligand activated VEGFR2 associates with vascular adherens junctions via direct interaction with VE-cadherin (VE-cad) or through a linkage between PI3K and β-cat, resulting in increased endothelial proliferation (via VEGF-A induced Bcl2 upregulation), mechanotransduction, and angiogenesis. (Right) In confluent cells, junctional phosphatase DEP1 is recruited to VE-cad/β-cat junctions to inhibit VEGFR signaling by suppressing its receptor internalization and transduction to MAPK. (D) VEGFR2/3 can be activated upon shear stress in the absence of VEGF ligands. In this scenario, a tripartite complex of PECAM1, VE-cad, and VEGFR is formed. Activation of Src and PI3K kinases, downstream of PECAM1 and VE-cad, respectively, potentiates VEGFR signaling, collaboratively contributing to flow signaling. (E) In tumor cells, interaction of N-cadherin (N-cad) and fibroblast growth factor receptors (FGFRs) upon ligand stimulation sustains RTK signaling to promote cell motility and invasion. Figure created with BioRender.com.
Support for crosstalk between CCCs and RTKs was initially provided by observations that catenins are phosphorylated in response to growth factors (Hoschuetzky et al., 1994; Shibamoto et al., 1994; Hazan and Norton, 1998). Different modes of physical interaction between CCCs and EGFR have been reported (Figure 3B). The EGFR can associate with CCCs through interaction with β-catenin (Hoschuetzky et al., 1994; Perrais et al., 2007). E-cadherin can also interact with EGFR via ECs without binding to β-catenin (Qian et al., 2004). The interaction between these two signaling hubs allows a reciprocal regulation, by which EGFR promotes cell junction disassembly in part due to increased endocytosis of CCCs (Hazan and Norton, 1998; Fujita et al., 2002), whereas E-cadherin adhesions modulate activation and cellular signaling of EGFR in a cell-cell contact and/or cell density dependent manner (Pece and Gutkind, 2000; Qian et al., 2004; Perrais et al., 2007). In confluent epithelial cells, E-cadherin junctions are generally thought to suppress EGFR activation and downstream RAS signaling by sequestering EGFR away from apical cell domains, thus suppressing EGF binding to EGFR (Takahashi and Suzuki, 1996; Qian et al., 2004) (Figure 3B, left). Consistent with its ability to promote E-cadherin stability, p120-catenin through its interaction with E-cadherin suppresses overall tyrosine phosphorylation levels in epithelial cells and inhibits RAS signaling (Soto et al., 2008). Moreover E-cadherin binding to β-catenin, despite not affecting autophosphorylation of EGFR, suppresses EGFR-mediated STAT5 activation (Perrais et al., 2007) (Figure 3B, left). In response to mechanical stress at E-cadherin junctions, EGFR growth signaling becomes active followed by its dissociation with E-cadherin (Sullivan et al., 2022) (Figure 3B, right). EGFR further functions as a positive regulator for force-mediated E-cadherin signaling that increases cell stiffness through the activation of PI3K and Abl kinases (Sehgal et al., 2018). Interestingly, the basolateral localization and apical exclusion of EGFR in polarized epithelial cells may exclude EGF, but promote signaling by alternate ligands, like amphiregulin or epiregulin, which localize exclusively to the basolateral domain (Damstrup et al., 1999; Singh et al., 2013). Finally, E-cadherin complexes also associate with and are reciprocally regulated by other RTKs such as c-Met, HER2, and IGF1R (Gamallo et al., 1993; Moll et al., 1993; Shibamoto et al., 1994; Hiscox and Jiang, 1999; Kamei et al., 1999; Fujita et al., 2002; Cozzolino et al., 2003; Yanagisawa et al., 2004; Nagle et al., 2018). Collectively, these studies indicate the presence of intricate crosstalk between CCCs and RTK signaling that is likely critical for cell growth and tissue homeostasis (Kim et al., 2009).
This reciprocal relationship is clearly evident in the Drosophila gut where healthy enterocytes suppress stem cell division through the repression of EGF secretion by E-cadherin. Conversely, dying cells activate EGFR signaling by losing E-cadherin to increase cell division. This coordinated feedback regulation ensures that the total number of cells in the organ remain constant and thus prevents hyperplasia or atrophy (Liang et al., 2017; Ngo et al., 2020). The interaction of CCCs with Merlin provides an additional mechanism of EGFR pathway regulation. Upon cell-cell contact, the adaptor protein Na+/H+ exchanger regulatory factor 1 (NHERF1) links EGFR to Merlin, which sequesters the receptor at the cortical membrane to prevent its internalization and activation (Curto et al., 2007; Cole et al., 2008). At the plasma membrane-cytoskeleton interface, lateral mobility and recycling of EGFR within the membrane is restrained by a coordinated effort involving Merlin, cortical actomyosin cytoskeleton, and the composition of membrane lipids (Chiasson-MacKenzie et al., 2015; Chiasson-MacKenzie et al., 2018). Perturbation of this inhibitory effect of Merlin on EGFR signaling promotes tumorigenesis in the mouse liver (Benhamouche et al., 2010).
Recently, a new E-cadherin variant was found to be overexpressed in glioblastoma and to affect EGFR signaling in an unprecedented manner. This variant is encoded by E-cadherin circular RNA (circRNA) and exhibits distinct functions from the common E-cadherin encoded by linear RNA (Gao et al., 2021). Circular RNAs (circRNAs) encompass one or multiple exons, are more stable than linear RNAs, and are often considered as miRNA sponges that shield mRNAs from miRNA-dependent degradation (Kristensen et al., 2019). While full-length E-cadherin is low or absent in brain tumor cells, expression of variant E-cadherin from circRNAs was high, was secreted by glioma stem cells, and acted as an autocrine ligand to amplify EGFR signaling independent of EGF, resulting in sustained glioblastoma stemness (Gao et al., 2021).
Other cadherins can also modulate the function of growth factor receptors (GFRs). In endothelial cells, VEGF and its cognate receptors (VEGFRs) control cell survival, proliferation, and angiogenesis [reviewed in (Koch and Claesson-Welsh, 2012)]. Upon VEGF binding, VEGFR-2 (also known as FLK1, encoded by KDR) can form physical interactions with VE-cadherin through β-catenin to modulate downstream signaling events in a cell density-dependent manner (Carmeliet et al., 1999; Grazia Lampugnani et al., 2003). In cells with reduced cell contacts, this complex (consisting of VEGFR-2, VE-cadherin, and β-catenin) recruits PI3K to activate Akt and involves Bcl-2 to drive endothelial survival and proliferation (Carmeliet et al., 1999) (Figure 3C, left). In confluent cells, junctional phosphatase CD148/DEP-1 is recruited to this complex upon VE-cadherin engagement to suppress VEGFR-2 phosphorylation and internalization, thus inhibiting endothelial proliferation (Grazia Lampugnani et al., 2003; Lampugnani et al., 2006) (Figure 3C, right).
On the other hand, VEGFRs can be activated by shear flow in a ligand-independent manner (Jin et al., 2003). Shear force in the vasculature is generated by the blood flowing through the vessels, imparting physical forces on endothelial cells which comprise vessel walls. Under this condition, the complexes of junctional VE-cadherin with VEGFRs (VEGFR-2 and VEGFR-3) function as a crucial node that transmits flow signals, and converts them into biochemical responses (Shay-Salit et al., 2002; Tzima et al., 2005; Coon et al., 2015; Conway et al., 2017). The VE-cadherin-VEGFR2 associated complex can also be coupled with another adhesion receptor PECAM-1 to transmit shear stress signals, leading to the activation of Src kinases and the PI3K pathway (Shay-Salit et al., 2002; Tzima et al., 2005; Conway et al., 2013) (Figure 3D). Interestingly, the mechanotransduction mediated by local VE-cadherin adhesions and the downstream intercellular junctional remodeling can be propagated to junctions distal to where the force is applied (Barry et al., 2015).
VEGF and its cognate receptors are crucial for neo-angiogenesis. Interestingly, in ovarian cancer, VE-cadherin on endothelial cells can form a direct heterophilic interaction with a form of soluble E-cadherin (sE-cadherin) localized on the surface of exosomes. sE-cadherin is the cleaved-off ectodomain of E-cadherin and is highly expressed in malignant ascites as well as tumor cells with high metastasis capabilities. This unique interaction between endothelial cells and exosomes leads to angiogenesis via VEGF-independent activation of Wnt/β-catenin and NF-κB signaling (Tang et al., 2018). Due to the close crosstalk between VE-cadherin and VEGFR-2, an interesting question to ask is whether sE-cadherin/VE-cadherin interactions promote angiogenesis by affecting VEGFR-2 signaling.
In tumor cells, N-cadherin is frequently upregulated, enhancing migratory and invasive cellular behaviors (reviewed in (Derycke and Bracke, 2004). Inspired by the synergistic activity of N-cadherin and FGFR in neurite outgrowth (Williams et al., 1994), the relationship between the two in cell motility and invasion was explored. Molecularly, FGFR signaling is sustained in the presence of N-cadherin as FGF-2 stimulation brings N-cadherin and FGFR1 together to inhibit ligand-mediated receptor internalization, resulting in increased stability of FGFR1 and cell motility/invasiveness (Suyama et al., 2002; Hulit et al., 2007) (Figure 3E). In line with these findings, expression of N-cadherin in mouse cancer models supports the notion that N-cadherin and FGFR synergistically potentiate downstream MAPK and AKT signaling with EMT to promote cancer metastasis (Shintani et al., 2006; Hulit et al., 2007; Su et al., 2012; Qian et al., 2014). Furthermore, β-catenin association with NHERF2 induced the interaction of N-cadherin with platelet derived growth factor receptor beta (PDGFR-β) to promote cell motility (Theisen et al., 2007).
Cadherin-associated catenins have also been shown to play a vital role in growth factor signaling which involves signal transduction from the upstream GFRs to the downstream Ras-MAPK pathway. In mouse skin with α-catenin ablation, a sustained upregulation of Ras-MAPK signaling was found in keratinocytes. Detailed analyses showed that there is no upregulation of RTK levels and/or activity in these knockout cells despite an increased sensitivity to insulin and IGF-1 and activation of Ras-MAPK. Upon insulin stimulation of α-catenin null cells, a physical association between the E-cadherin/β-catenin complex and phosphorylated insulin receptor substrate 1 (IRS-1) is formed. This potentiates the propagation of IGF1R signaling to Ras-MAPK, promoting epidermal hyperplasia (Vasioukhin et al., 2001). In early stage hepatocellular carcinoma (HCC), the AJ-associated β-catenin is reported to increase EGFR stability, promoting tumor growth (Kim et al., 2019). p120-catenin, on the other hand, inhibits Ras when binding to E-cadherin but promotes Ras-MAPK activation and tumor cell growth when associated with mesenchymal cadherins upon E-cadherin loss (Soto et al., 2008). p120-catenin also plays key roles in EGF- and HGF-mediated cell migration/invasion and scattering via its ability to modulate the activities of Rac and Rho small GTPases (Cozzolino et al., 2003; Yanagisawa et al., 2008). Moreover, a study in Drosophila showed that nuclear p120-catenin can induce expression of the EGF maturation factor rhomboid (Liang et al., 2017), whose mammalian paralog RHBDD1 promotes colorectal cancer tumorigenesis via EGFR-RAS-MAPK signaling (Song et al., 2015).
From an evolutionary standpoint, it is notable that the transition to multicellularity is marked by the simultaneous addition of adhesion proteins, including cadherins, and RTKs in choanoflagellates (King et al., 2003; Abedin and King, 2008). The reciprocal crosstalk between these two receptor classes fine tunes both adhesion and growth mediated responses through mechanisms that can be dependent or independent of cell contacts and/or GFR ligands and is commonly deregulated in cancer. RTK alterations are one of the most prominent driving events in human cancer (Sanchez-Vega et al., 2018) and prime targets for cancer therapy [reviewed in (Yamaoka et al., 2018; Pottier et al., 2020)]. Unfortunately, crosstalk between different RTKs often mitigate tumor cell response to a given RTK inhibitor. As CCCs collectively regulate the function of multiple RTKs, a better understanding of their crosstalk could provide novel mechanistic and therapeutic insights for the treatment of cancer.
Oncogenic Wnt/β-catenin signaling
The Wnt signaling pathway is essential for tissue homeostasis, embryonic development, and stem cell regeneration and maintenance, and commonly categorized into canonical and non-canonical forms based on β-catenin dependence [Grumolato et al., 2010; Zhan et al., 2017)]. Here, we focus on β-catenin dependent, canonical Wnt signaling (as known as Wnt/β-catenin signaling), which is one of the key cancer driver pathways (Sanchez-Vega et al., 2018). For readers with a particular interest in non-canonical Wnt, several comprehensive reviews are available (Kohn and Moon, 2005; Wang, 2009; Akoumianakis et al., 2022). Notably, non-canonical Wnt signaling is suppressed by the transcriptional repressor Kaiso, and this suppression is relieved by association of Kaiso with p120 catenin (Kim et al., 2004).
In canonical Wnt signaling, β-catenin is a key downstream effector to promote transcription of Wnt target genes (Figure 4). In the absence of Wnt ligands (Wnt OFF), the levels of cytosolic β-catenin are kept low by the Destruction Complex, which contains the tumor suppressors Axin and adenomatous polyposis coli (APC), and the serine/threonine kinases glycogen synthase kinase 3β (GSK-3β) and casein kinase 1 (CK1) (Zhan et al., 2017). In this complex APC serves as a platform, allowing β-catenin to interact with other proteins. GSK3β and CK1 phosphorylate β-catenin to prime its recognition by the E3-ubiquitin ligase β-TrCP, marking β-catenin for degradation by the proteasome. In Wnt/β-catenin signaling (Wnt ON), trafficking and secretion of Wnt ligands are enabled by the Porcupine acyltransferase and Wntless membrane protein (Takebe et al., 2015). Secreted Wnt ligands then bind to Frizzled (FZD) family receptors and associated low-density lipoprotein receptor-related protein 5/6 (LRP5/6) co-receptors on neighboring cells to initiate Wnt signaling. Ligand-bound FZD receptors then recruit and activate dishevelled (Dvl) at the plasma membrane. Activated Dvl, in turn, recruits and inhibits the β-catenin Destruction Complex, leading to accumulation of β-catenin in the cytoplasm. Stabilized β-catenin translocates to the nucleus where it interacts with the T cell factor (TCF)/lymphocyte enhance factor (LEF) family of TFs to drive gene expression. The E3 ubiquitin ligases zinc and ring finger 3 (ZNRF3) and its homologue RNF43, which are Wnt target genes, in turn form a negative feedback circuit to target FZD receptors for lysosomal degradation (de Lau et al., 2011; Koo et al., 2012). R-spondin ligands, on the other hand, form a complex with the G-protein-coupled receptors (GPCRs) Lgr4/5/6 to maintain Wnt signaling via inhibition of ZNRF3/RNF43 (Hao et al., 2012; de Lau et al., 2014). In cancer, alterations of these molecules can lead to activation of the Wnt pathway, promoting cancer initiation and progression.
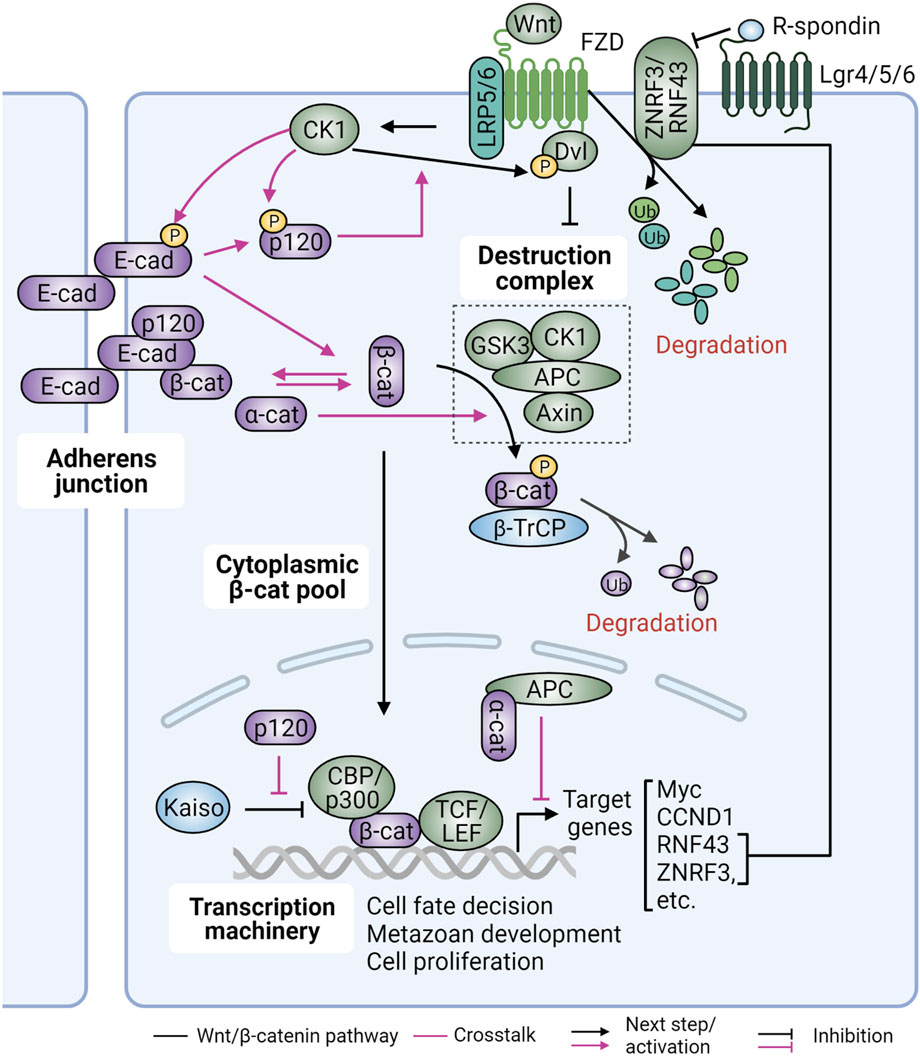
FIGURE 4. Interrelationship between Wnt/β-catenin signaling and cadherins/catenins. Canonical Wnt signaling is a crucial regulator of cell fate determination, cell proliferation and embryonic development. It is kept inert when cytoplasmic β-catenin (β-cat) levels are low by the action of the multiprotein Destruction Complex and β-TrCP-mediated proteasomal degradation. Wnt ligand binding to Frizzled (FZD) receptors and low-density lipoprotein receptor-related protein 5/6 (LRP5/6) co-receptors results in dishevelled (Dvl) recruitment and activation by casein kinase (CK1), leading to β-cat release from the Destruction Complex. The freed cytoplasmic β-cat is then translocated into the nucleus where it interacts with transcription (co)factors TCF/LEF and CBP/p300 to drive gene expression. Wnt signaling is also negatively regulated by RNF43 and ZNRF3 ubiquitin E3 ligases, which degrade FZD and LRP receptors. Conversely, R-spondin and Dvl positively regulate Wnt signaling by binding to Lgr4/5/6 receptors to remove RNF43/ZNRF3 from the plasma membrane, or by inhibiting the β-cat Destruction Complex, respectively. Upon Wnt stimulation, E-cadherin (E-cad) is phosphorylated by CK1 resulting in dissociation of β-cat, which can then participate in Wnt signaling or be degraded. This process can be induced by cytoplasmic α-catenin (α-cat) via increased β-cat binding to adenomatous polyposis coli (APC). Additionally, nuclear α-cat can recruit APC to the β-cat-TCF/LEF complex to suppress expression of Wnt target genes. P120-catenin (p120), on the other hand, is also phosphorylated by CK1, leading to its dissociation from E-cadherin and increased formation of the Wnt signalosome via interaction with LRP5/6. Finally, p120 in the nucleus can de-repress Kaiso’s action on β-cat/TCF-TEF activity. Figure created with BioRender.com.
The primary mechanism leading to hyper-activation of Wnt signaling in cancer is loss of function (LoF) in APC, an event that is commonly observed in hereditary (familial adenomatous polyposis) and sporadic colorectal carcinomas (CRC) (Nishisho et al., 1991; Cottrell et al., 1992; Powell et al., 1992; Rowan et al., 2000). Dysregulation of APC negates its normal function to restrict β-catenin levels through the Destruction Complex (Rubinfeld et al., 1993; Morin et al., 1997). Activating mutations of CTNNB1 (β-catenin) are also found in CRC and are mutually exclusive with APC alterations (Morin et al., 1997; Sparks et al., 1998). In endometrial carcinomas, mutations of CTNNB1 (β-catenin) are the predominant mechanism that triggers Wnt/β-catenin signaling (Fukuchi et al., 1998; Cancer Genome Atlas Research Network et al., 2013). These mutational events often occur in the NH2-terminal regulatory domain (exon 3, in particular) of β-catenin to prevent its phosphorylation and subsequent ubiquitination and degradation (Gao et al., 2018). Intriguingly, accumulation of β-catenin and thus activation of Wnt signaling are also implicated in triple-negative breast cancer (TNBC) despite no evidence of CTNNB1 mutations (Khramtsov et al., 2010; Geyer et al., 2011). Moreover, the R-spondin-ZNRF3/RNF43 module is often deregulated to activate Wnt signaling in cancer. R-spondin gene fusions can drive colon tumor formation and progression (Seshagiri et al., 2012; Han et al., 2017), whereas inactivating mutations of RNF43 confer sensitivity to porcupine inhibitors in Wnt-dependent CRC and endometrial tumor organoids (Giannakis et al., 2014; van de Wetering et al., 2015).
As β-catenin is an essential component of both AJs and canonical Wnt signaling, crosstalk between the two has long been suspected. Initial studies in Xenopus and Drosophila showed that cadherin overexpression can antagonize Wnt signaling by sequestering β-catenin to the plasma membrane (Heasman et al., 1994; Sanson et al., 1996) (Figure 4). Similar observations were also reported in cell culture systems overexpressing cadherins (Sadot et al., 1998; Gottardi et al., 2001; Stockinger et al., 2001). The competition between junctional and nuclear β-catenin can be explained by the observation that the interaction domains of β-catenin for cadherin, APC, and TCF overlap and thus interactions are mutually exclusive. Functionally, this implied that loss of E-cadherin during cancer progression would be sufficient to promote canonical Wnt signaling. However, the lack of upregulation of β-catenin signaling upon cadherin loss in several models argues that β-catenin released from cadherin complexes is rapidly degraded by the Destruction Complex, and additional events including Wnt activation and/or reduced degradation are required for the induction of β-catenin signaling (Mendonsa et al., 2018). Under these conditions, loss of E-cadherin could potentiate canonical Wnt signaling and contribute to cancer progression.
Recently, Wnt signaling was shown to interconnect with Hippo-YAP/TAZ signaling via β-catenin. Briefly, nuclear β-catenin forms a complex with active YAP to potentiate the transcriptional programs of both WNT/β-catenin and YAP pathways (Heallen et al., 2011; Rosenbluh et al., 2012). Moreover, cytoplasmic YAP/TAZ are recruited to the Destruction Complex by β-catenin, ensuring the recruitment of the β-TrCP E3 ubiquitin ligase for protein degradation (Azzolin et al., 2012; Azzolin et al., 2014). Consequently, both β-catenin and TAZ are maintained at low levels, preventing overactivation of WNT and YAP/TAZ pathways (Azzolin et al., 2012; Azzolin et al., 2014). These findings highlight multiple mechanisms that link Hippo-YAP/TAZ and Wnt/β-catenin pathways and suggest potential crosstalk with cadherin catenin complexes.
Finally, α- and p120-catenins also influence transcription of Wnt target genes (Figure 4). Cytoplasmic α-catenin physically associates with APC to promote ubiquitination and subsequent degradation of β-catenin. Upon Wnt3A stimulation, nuclear α-catenin recruits APC to canonical Wnt response elements to regulate β-catenin turnover (Choi et al., 2013). Independent of the destruction complex, nuclear α-catenin can also block the interaction of the β-catenin/TCF transcriptional complex with DNA, thus suppressing TCF-dependent transcription (Giannini et al., 2000). Cadherin-associated p120-catenin is phosphorylated by CK1ε in response to Wnt3a, which promotes recruitment of CK1 and Dvl2 to the Wnt co-receptors LRP5/6 (Casagolda et al., 2010). By promoting the formation of the Wnt signalosome, p120-catenin promotes β-catenin stability and transcriptional activity. Another mechanism by which p120-catenin promotes β-catenin signaling is by de-repressing transcription of Wnt target genes by promoting the dissociation of the transcriptional repressor Kaiso from the promoters of canonical Wnt target genes (Park et al., 2005).
In summary, in the absence of activating events (Wnt ligand, APC mutations, etc.) loss of E-cadherin alone is insufficient to promote Wnt/β-catenin signaling. However, in the presence of activating events, cadherins and catenins are able to modulate canonical Wnt signaling by acting at different subcellular locations and with various mechanisms of action to regulate signalosome formation, β-catenin degradation, or transcriptional activation.
Notch and Hedgehog developmental and stemness pathways
Similar to Wnt signaling, Notch and HH play crucial roles in tissue development and homeostasis (Takebe et al., 2015). Extensive crosstalk between these three pathways collectively governs self-renewal and cell-fate decisions (Takebe et al., 2011; Takebe et al., 2015). The Notch signaling pathway begins with the interaction between transmembrane-bound Notch ligands and receptors on neighboring cells (Figure 5A). Five Notch ligands, Delta-like ligand 1 (DLL1), DLL3, DLL4, Jagged 1 (Jag1), and Jag2, and four Notch receptors (Notch1-4), are involved in this pathway. Different ligands and receptors are expressed in different tissues and tumor types [reviewed in (Takebe et al., 2011; Takebe et al., 2015)]. Upon ligand-receptor association, Notch receptors undergo a two-step proteolytic cleavage by the a-disintegrin and metalloproteinase (ADAM) enzymes (ADAM10 or ADAM17) and γ-secretase, releasing the Notch intracellular domain (NICD) (Gordon et al., 2007). Released NICD then translocates to the nucleus to interact with the DNA-binding transcription regulator RBPJ and transcriptional coactivators to activate or repress gene expression. The best-known Notch target genes include members of the Hes/Hey family of basic helix-loop-helix (βHLH) TFs, CCND1 (encodes cyclin D1), CDKN1A (encodes p21) and Myc (Bailey and Posakony, 1995; Jarriault et al., 1998; Rangarajan et al., 2001; Ronchini and Capobianco, 2001; Iso et al., 2003; Klinakis et al., 2006; Weng et al., 2006). This signaling can be terminated by degradation of NICD by the E3 ubiquitin ligase F-box and WD repeat domain containing 7 (FBXW7) (Tsunematsu et al., 2004).
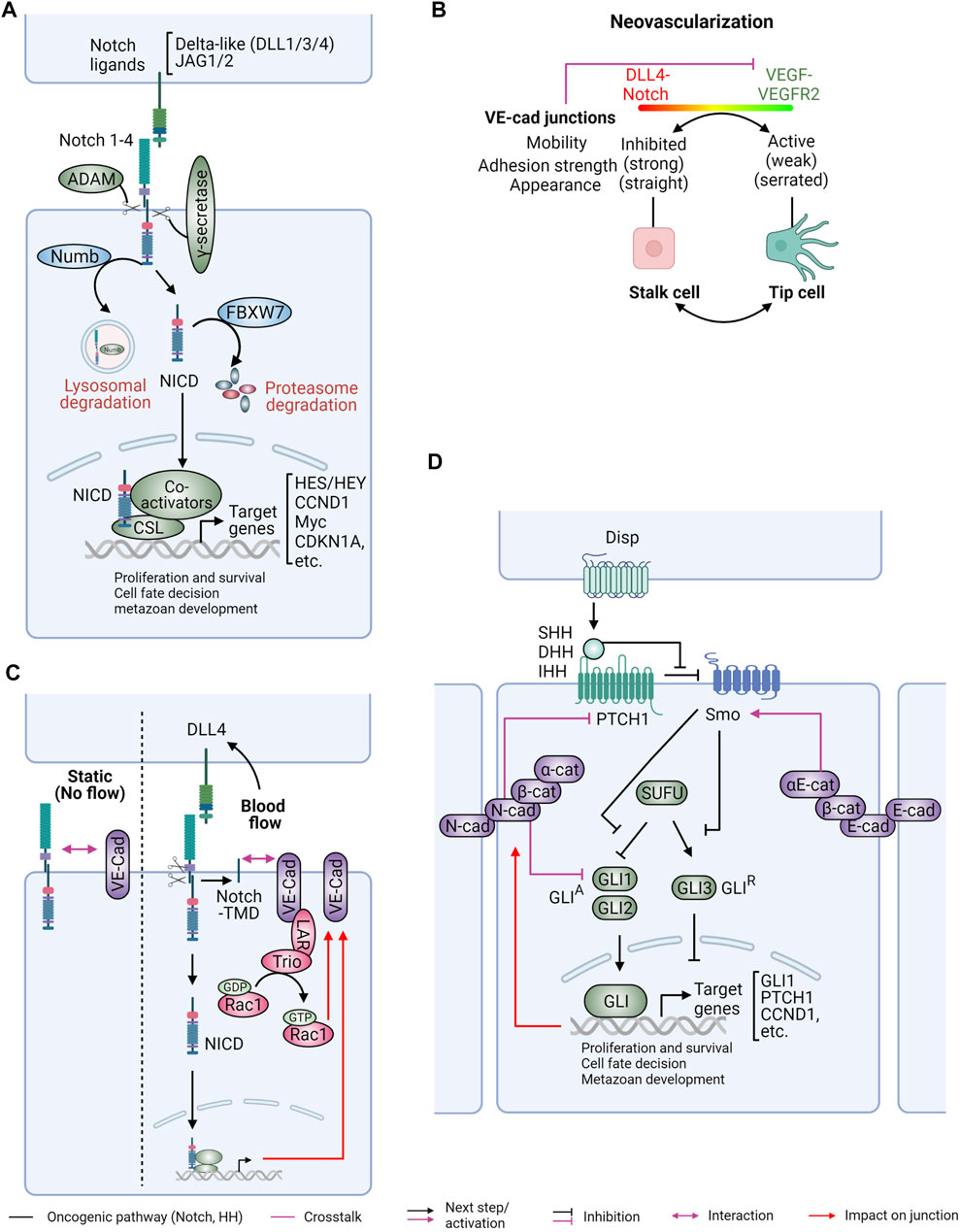
FIGURE 5. Interplay between Notch and Hedgehog signaling with cadherin/catenin complexes. Notch and hedgehog (HH) are key pathways for tissue homeostasis, embryonal development, and cancer stemness. (A) Notch signaling commences when transmembrane Notch ligands bind to the corresponding receptors on neighboring cells. This interaction results in proteolytic cleavage of Notch receptors by ADAMs and γ-secretase enzymes, releasing the Notch intracellular domain (NICD). The NICD translocates into the nucleus, where it functions as a transcriptional regulator to promote gene expression. Notch signaling can be negatively regulated by the E3 ubiquitin ligase specificity factor FBXW7 and the endocytosis regulator Numb. (B) During neovascularization, Dll4/Notch and vascular endothelial growth factor (VEGF)/VEGF receptor 2 (VGFR2) signaling cooperatively determine the stability and dynamics of junctional VE-cadherin (VE-cad) to drive the rearrangement of tip cells and stalk cells during angiogenic sprouts. Endothelial cells become stalk cells when they encounter high level of Notch signaling with low VEGFR2 activity. These cells exhibit reduced VE-cadherin junctions with stronger adhesions and straight appearance. In contrast, low Notch activity with high VEGFR2 signaling promotes the transformation of endothelial cells to tip cells. As a feedback mechanism, VE-cad association with VEGFR2 inhibits VEGFR2 signaling. (C) The interplay between VE-cad and Notch signaling can be modulated by mechanical signals. (Left) Under no flow static condition, VE-cad can associate with full-length Notch1. (Right) Shear stress, on the other hand, has been shown to induce both conventional transcription-dependent and unconventional transcription-independent Notch signaling with induced Dll4 expression. Increased expression of Notch target genes upon hemodynamic forces is critical for the maintenance of VE-cad junctions. Additionally, Notch cleavage can lead to release of its transmembrane domain (TMD), which forms a direct association with VE-cad to promote endothelial barrier function via the LAR phosphatase/Trio guanine-nucleotide exchange factor/Rac1 signaling axis. (D) In the absence of HH ligands, the activity of Smo is inhibited by the Patched 1 (PTCH1) receptor. Once HH ligands are released by the transporter Dispatched (Disp) at neighboring cells and binding to PTCH1, Smo becomes active and then inhibits Suppressor of Fused (SUFU), resulting in nuclear translocation of glioma-associated oncogene homolog (Gli) proteins. Gli proteins regulate expression of HH target genes with Gli1 and Gli2 being transcriptional activators (GLIA) and Gli3 acting as a transcriptional repressor (GLIR). Cadherin (E-cadherin, N-cadherin) adhesions restrict activation of HH signaling, in part through α-catenin (α-cat), although the underlying molecular mechanisms have not been fully elucidated. Activated HH signaling, in turn, promotes assembly of N-cad cell junctions. Figure created with BioRender.com.
The first evidence linking Notch signaling to cancer was a chromosomal translocation targeting Notch1 in T-cell acute lymphoblastic leukemia (T-ALL) (Ellisen et al., 1991). Later studies showed that activating mutations of Notch 1 and inactivating mutations of the tumor suppressor gene (TSG) FBXW7 were common in T-ALL (Weng et al., 2004; Malyukova et al., 2007; Maser et al., 2007; Thompson et al., 2007). These molecular alterations lead to constitutive activation of Notch signaling and drive neoplastic transformation. A pro-tumorigenic role for Notch signaling has also been reported in other hematological malignancies and solid tumors (Ranganathan et al., 2011; Misiorek et al., 2021). Activation of Notch signaling in these diseases can be also attributed to overexpression of Notch receptors and ligands and inactivating mutations of the negative regulators FBXW7 and Numb. However, the role of Notch signaling in cancer is nuanced by observations that in some contexts it can act as a tumor suppressor. For example, Notch signaling promotes cell differentiation and inhibits proliferation in skin keratinocytes (Lowell et al., 2000; Rangarajan et al., 2001; Nicolas et al., 2003). A similar inhibitory effect on cell growth was also reported in small cell lung cancer (SCLC) (Sriuranpong et al., 2001). Two genome-wide association studies (GWAS) delineating the molecular mechanisms underlying head and neck squamous cell carcinoma (HNSCC) have identified inactivating mutations in Notch1, suggesting a tumor-suppressing function of Notch in this disease (Agrawal et al., 2011; Stransky et al., 2011). In contrast, activation of Notch target genes HES1/HEY1 has also been reported in HNSCC, suggesting a tumor promoting function (Sun et al., 2014). Moreover, Notch 1 and Notch 2 appear to have opposite functions in embryonic brain tumors (Fan et al., 2004) and pancreatic ductal adenocarcinoma (PADC) (Hanlon et al., 2010; Mazur et al., 2010), adding another layer of complexity on the role of Notch signaling in tumor progression.
A relationship between cadherin and Notch signaling was suggested by the observation that Notch activity promotes AJ remodeling and cell morphogenesis via the regulation of E-cadherin spatial expression during Drosophila oogenesis (Grammont, 2007). E-cadherin, in normal transit-amplifying prostatic cells, can also affect Notch 1 signaling in a calcium-dependent manner. Reduced E-cadherin adhesion under conditions of low calcium promotes constitutive activation of Notch 1 and cell survival. Conversely, when these cells are grown in high calcium, Notch signaling is only active upon ligand-binding, and can be strengthened by E-cadherin-mediated adhesion (Dalrymple et al., 2005).
In endothelial cells, DLL4/Notch signaling cooperates with VEGF/VEGFR2 signaling to control the turnover of VE-cadherin and cell adhesion during vascular morphogenesis (Bentley et al., 2014) (Figure 5B). During angiogenesis, vascular sprouting from pre-existing vessels is regulated by the dynamic interaction of leading tip cells and stalk cells within each sprout. Cells encountering high Notch signals contain straight junctions and are static due to reduced junctional turnover of VE-cadherin, whereas cells encountering high VEGF exhibit serrated junctions and are highly motile, moving toward the leading tip. The impact of Notch on VE-cadherin junctions is also extended to endothelial barrier function (Mack et al., 2017; Polacheck et al., 2017) (Figure 5C). Using a microfabrication system to study endothelial responses to shear forces induced by fluid flow, Notch 1 was found to be associated with VE-cadherin under control conditions. However, under shear force Notch 1 was cleaved releasing its transmembrane domain (TMD), which recruited the receptor protein tyrosine phosphatase LAR, activated Rac1 signaling, and reinforced the AJs (Figure 5C). This adaptive response reinforced the barrier function of the endothelial monolayer to shear stress. Notably, this effect was independent of Notch transcriptional signaling but instead attributed to the physical interaction of VE-cadherin with the Notch TMD (Polacheck et al., 2017).
Bidirectional regulation between Notch2 and N-cadherin was reported in the context of chronic lymphocytic leukemia (CLL) (Mangolini et al., 2018). N-cadherin homotypic interactions between CLL cells and bone marrow-derived MSCs allow tumor cells to induce Notch2 activation in stromal cells. Active stromal Notch2, in turn, transcriptionally increases expression of N-cadherin, which surprisingly potentiates β-catenin stabilization and promotes Wnt signaling in CLL cells (Mangolini et al., 2018).
A close relationship between Notch and different cellular pools of β-catenin has also been suggested. Membrane-bound Notch physically interacts with dephosphorylated, active β-catenin, promoting its degradation (Hayward et al., 2005; Jin et al., 2009). This interaction is also observed in mouse embryonic stem cells where active β-catenin is downregulated by Notch in a ligand-independent manner. This Notch-dependent β-catenin degradation is surprisingly independent of the GSK3β/APC-mediated Destruction Complex and thus offers an alternative means to restrict the levels of active β-catenin and Wnt signaling in cells (Kwon et al., 2011). However, it is worth noting that ligand-induced Notch signaling in primary melanoma cells can upregulate β-catenin levels to promote tumor progression (Balint et al., 2005). Collectively, the data argue that Notch is a key regulator of β-catenin activity, but its effects are highly dependent on cellular context and subcellular localization. Conversely, β-catenin can also affect Notch signaling. Activation of Wnt by nuclear β-catenin can increase Notch signaling via the transcriptional activation of Jag1, leading to increased tumorigenesis (Rodilla et al., 2009; Kode et al., 2014).
Unlike Notch signaling, the HH pathway can promote both short-range and long-range signaling via HH ligands which are released by the transporter-like protein Dispatched (DISP1) (Burke et al., 1999; Zeng et al., 2001; Ma et al., 2002). HH ligands comprise sonic hedgehog (SHH), Indian hedgehog (IHH), and desert hedgehog (DHH) (Figure 5D). Other critical components in this pathway include the membrane receptors Patched (encoded by PTCH1) and Smoothened (encoded by SMO), the glioma-associated oncogene (GLI) TFs, and the adaptor protein suppressor of fused (SUFU). The function of HH signaling was initially revealed by studies on primary cilia, microtubule-enriched sensory organelles emanated from the plasma membrane of quiescent cells. In the absence of HH ligands, PTCH1 binds and prevents SMO translocation to the primary cilia (Gong et al., 2018). This inhibitory effect is released upon HH ligand binding to PTCH1, permitting cilia localization and activation of SMO. Activated SMO induces the release of GLI proteins from SUFU, which normally retains GLIs in the cytoplasm in the absence of HH ligands. Three GLI TFs (GLI1, GLI2, and GLI3) are the ultimate effectors of this pathway by regulating the expression of HH target genes Interestingly, while all three GLI proteins contain a C-terminal activation domain, GLI2 and GLI3 also possess repressor domains in their N-terminus and therefore can repress gene expression when their C-terminus is removed (Wu et al., 2017). GLI1 and GLI2 are widely considered as transcriptional activators (GLIA) in response to HH stimulation, whereas GLI3 mainly functions as a transcriptional repressor (GLIR) to inhibit HH target gene expression in the absence of HH ligands (Hui and Angers, 2011).
The oncogenic function of HH was initially suggested by the observation that loss of heterozygosity (LOH) and germline inactivating mutations in PTCH1 are associated with the Gorlin syndrome, a hereditary disorder that predisposes patients to basal cell carcinomas (BCCs), medulloblastomas, and rhabdomyosarcomas (Hahn et al., 1996; Johnson et al., 1996). Similarly, SUFU, the negative regulator of HH signaling, is also commonly mutated in Gorlin syndrome-related medulloblastomas (Taylor et al., 2002; Smith et al., 2014). In the setting of sporadic cancers, somatic deleterious mutations and deletions of PTCH1 and SUFU have been observed in a subset of BCCs (Teh et al., 2005; Jayaraman et al., 2014) and medulloblastomas (Raffel et al., 1997; Taylor et al., 2002). Activating mutations and overexpression of the oncogenic HH signaling components, such as SMO, are also linked to tumorigenesis in these cancers (Xie et al., 1998; Drummond et al., 2018; Tan et al., 2018). Enrichment of HH signaling is currently used as a biomarker in the clinic to guide targeted therapy for BCCs and to subcategorize medulloblastoma.
Similar to Notch, a bidirectional relationship exists between the HH pathway and AJs. Deletion of αE-catenin in neural progenitor cells causes disruption of apical junctions and loss of cell polarity, resulting in ectopic activation of HH signaling in the developing brain cortex (Lien et al., 2006) (Figure 5D). This, in turn, shortens the cell cycle and decreases cell death, leading to cortical hyperplasia and the formation of invasive tissue masses that resemble medulloblastoma and other brain tumors. In the zebrafish dorsal neural tube, impaired cell adhesion due to loss of functional N-cadherin also results in hyperactivation of HH signaling and hyperproliferation (Chalasani and Brewster, 2011) (Figure 5D). Activation of HH, in turn, promotes N-cadherin mediated AJ assembly as a negative feedback mechanism to restrict its own activity (Jarov et al., 2003; Fournier-Thibault et al., 2009; Chalasani and Brewster, 2011) (Figure 5D). Collectively, these studies indicated that AJs exert an inhibitory effect on HH signaling during neurulation.
Interestingly, Wnt/β-catenin signaling is critically involved in HH-mediated tumorigenesis (Yang et al., 2008). Moreover, the nuclear translocation of β-catenin induced by HH signaling can be suppressed by E-cadherin (Li et al., 2007; Inaguma et al., 2011). To circumvent this inhibitory effect of E-cadherin, HH signaling can either repress the expression or disrupt the junctional localization of E-cadherin. To repress E-cadherin expression HH promotes the expression of Snail or Maf TFs (Li et al., 2007; Inaguma et al., 2011; Lai et al., 2017). Additionally, mucin MUC5AC, a direct SHH target in pancreatic adenocarcinoma cells, is activated and its localization to intercellular junctions destabilizes E-cadherin adhesions (Inaguma et al., 2011). Overall, the data are consistent with a model whereby cadherin-mediated AJs suppress HH oncogenic signaling, whereas HH activation suppresses AJ function and promotes Wnt/β-catenin signaling.
The bilateral interactions between VE-cadherin and Notch modulated by hemodynamic stress as well as between N-cadherin and HH in the context of cell polarity have been nicely delineated in endothelial cells and central nervous system (CNS) precursor cells, respectively. However, our current understanding of the interplay between CCC, Notch, and HH signaling in cancer is far from complete. As Notch signaling can be tumor suppressing or promoting depending on cellular context, the role of CCCs in these different disease settings is unclear and warrants further investigation. Little is also known about the crosstalk between CCCs and HH signaling in human cancer despite strong evidence of negative feedback regulation between AJs and HH signaling during development, which is generally supported by cancer studies described in this review.
Epithelial-mesenchymal transition and TGF-beta signaling
Metastasis involves a series of events from cell invasion and dissemination from the primary tumor mass to survival in circulation, extravasation and colonization of distant organs (Nieto et al., 2016). EMT, an important developmental process that regulates cell fate decisions and tissue specification, is often co-opted during metastasis to promote cell dissemination, followed by its reverse process mesenchymal-epithelial transition (MET) to re-establish the tumor at distant sites. Instead of a bimodal switch, EMT is now considered a dynamic and plastic program that includes a spectrum of EMT phenotypes involving intermediate partial and full EMTs (Nieto et al., 2016). Full EMT is characterized by loss of epithelial features accompanied by gain of mesenchymal phenotypes, endowing cells with invasive and migratory properties, whereas partial EMT states present varying degrees of epithelial and mesenchymal characteristics.
A potent inducer of EMT is transforming growth factor beta (TGFβ), which functions in SMAD-dependent and independent manners (Lamouille et al., 2014) (Figure 6). The TGFβ family of ligands include TGFβs and bone morphogenetic proteins (BMPs), and they form homodimers or heterodimers to bind and activate TGFβ receptors. Activated receptors phosphorylate receptor-regulated SMADs (R-SMADs), which form a complex with SMAD4 to activate or repress gene expression following nuclear translocation. These nuclear SMAD complexes directly or indirectly promote the expression and transcriptional activity of EMT-TFs, including SNAIL1/2, ZEB1/2, and TWIST. These EMT-TFs function individually or collaboratively with each other or other TFs to repress expression of epithelial markers such as E-cadherin and increase the levels of mesenchymal markers such as N-cadherin, vimentin and fibronectin. This “cadherin switch” leads to altered cell-cell adhesion and potentiates EMT phenotypes. Independent of SMAD-mediated gene expression, TGFβ can also induce EMT phenotypes via the E3 ubiquitin ligase SMAD ubiquitination regulator factor 1 (SMURF1). SMURF1 is recruited to TGFβ receptors and targets localized RhoA for degradation, leading to dissociation of cortical actin (Ozdamar et al., 2005). Additionally, p120-catenin is mono-ubiquitinated by SMURF1 following TGFβ induced MAPK activation, leading to AJ and then tight junction (TJ) disassembly and ultimately lung metastasis of murine breast cancer (Wu et al., 2020) (Figure 6).
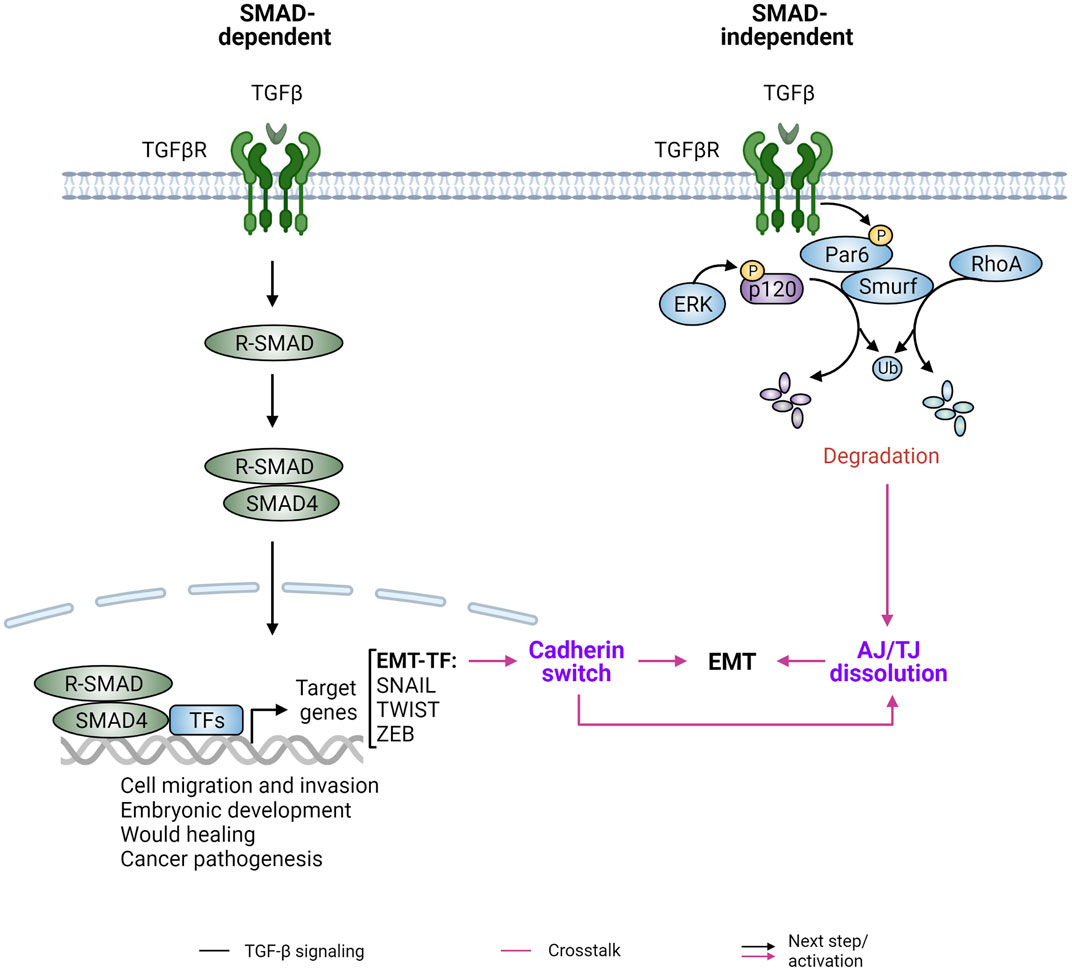
FIGURE 6. TGFβ signaling and cadherin/catenin plasticity. TGFβ signaling is a major inducer of epithelial-mesenchymal transition (EMT) and is mediated by SMAD-dependent and SMAD-independent downstream pathways. (Left) The canonical TGF-β signaling pathway uses Smad proteins to transduce its biological signals and effects. Upon TGFβ binding and the subsequent formation of the TGFβ-TGFβ receptor (TGFβRs) hetero-oligomeric signaling complex, receptor-activated SMADs (R-SMAD) are released from the TGFβ-TGFβR complex and form a heterotrimeric complex with SMAD4. The R-SMAD/SMAD4 complex then translocates into the nucleus and interacts with other transcription factors (TFs) and co-factors to drive expression of TGFβ target genes, such as the EMT master regulators SNAIL, TWIST, and ZEB. This leads to downregulation of E-cadherin and upregulation of N-cadherin, a process referred to as “cadherin switch.” (Right) The epithelial polarity protein partitioning-defective protein 6 (Par6) is one of the intracellular factors involved in SMAD-independent, non-canonical TGFβ signaling. Specifically, activated TGFβRII phosphorylates Par6, leading to the recruitment of the E3 ubiquitin ligase Smurf1 and localized degradation of RhoA and p120-catenin (p120) to affect downstream signaling. Figure created with BioRender.com.
EMT can endow individual cells with decreased adhesion to the main tumor mass and with increased cell migration and invasion properties (Lamouille et al., 2014). This “identity crisis” is thought to allow tumor cells to disseminate, and to be critically involved in cancer metastasis. A vast body of evidence concurs that EMT promotes cell migration and invasion; however, its role in cancer metastasis remains under debate [reviewed in (Lamouille et al., 2014; Brabletz et al., 2018; Yang et al., 2020)]. Conditions at the leading edges of tumors favor a transient EMT that contributes to metastatic dissemination (Tsai et al., 2012; Tran et al., 2014; Puram et al., 2017). However, metastasis often involves groups of E-cadherin expressing cells that collectively migrate and invade tissues (Shamir et al., 2014; Westcott et al., 2015), and consistent with this, tumor cells with inhibited EMT still retain metastatic potential (Fischer et al., 2015; Zheng et al., 2015). For example, studies in mouse genetically engineered models (GEMM) suggested that EMT-mediated gene expression is dispensable for tumor cell invasion and metastasis, but rather promotes chemoresistance (Fischer et al., 2015; Zheng et al., 2015). Moreover, circulating tumor cell (CTC) clusters that contain a group of cells with intact epithelial junctions are more prevalent than single CTCs lacking E-cadherin expression in the blood of cancer patients (Friedl et al., 2012; Aceto et al., 2014), suggesting that retention of E-cadherin is common during key stages of metastatic spread.
The role of E-cadherin loss in cancer metastasis is also under debate, despite strong evidence that E-cadherin suppresses tumor cell migration, invasion, and anchorage-independent growth. Two recent studies used breast cancer mouse models to elucidate the role of E-cadherin in cancer metastasis (Padmanaban et al., 2019; Na et al., 2020). In one study, E-cadherin was conditionally knocked out in order to evaluate the role of E-cadherin expression in the metastasis cascade (Padmanaban et al., 2019), whereas the other used E-cadherin activating antibodies to elucidate the role of E-cadherin adhesive function in this process (Na et al., 2020). Both studies supported an inhibitory role of E-cadherin in cell invasion but showed opposite functions of E-cadherin in the formation of CTCs and gross metastasis. In the knockout study, E-cadherin loss decreased CTCs in the bloodstream and suppressed metastasis due to reduced cell proliferation and increased apoptosis (Padmanaban et al., 2019), while increased E-cadherin engagement in the second study resulted in suppression of cancer metastasis (Na et al., 2020). The discrepancies in E-cadherin effects could be related to cellular context and specifically the status of p53 and/or the apoptotic machinery. p120-catenin, α-catenin and E-cadherin are all thought to act as haploinsufficient tumor suppressors, whereby complete biallelic loss is only allowed under permissive conditions, like prior loss of p53 function (Shibata et al., 2007; Short et al., 2017). Conversely, epithelial cells expressing E-cadherin undergo cell death by anoikis upon anchorage independent conditions similar to those of CTCs in circulation, which may account for the increased apoptosis of CTCs and reduced metastasis of tumor cells treated with E-cadherin activating antibodies.
Our group reported that E-cadherin expression suppresses Rac1 and Src activation by mesenchymal cadherins resulting in reduced cell migration, invasion, and anchorage-independent growth (Yanagisawa and Anastasiadis, 2006; Soto et al., 2008). More recently we delineated two distinct E-cadherin containing complexes, one apical at the ZA containing PLEKHA7, and one basolateral lacking PLEKHA7 and characterized by increased Rac1 and Src activities (Kourtidis et al., 2015). Furthermore, the apical complex potently suppressed the pro-tumorigenic activity of the basolateral complex by recruiting the RNAi machinery to the ZA and regulating the translation of key pro-tumorigenic markers through a junction associated RNA induced silencing complex (RISC). The data argued that the tumor suppressive function of E-cadherin is specifically associated with the integrity of the apical ZA, and that in the absence of apical junctions, basolateral E-cadherin can promote tumor progression.
Despite contextual differences on the role of EMT and E-cadherin in cancer progression and metastasis, EMT remains an attractive therapeutic target to suppress metastasis and/or to overcome chemoresistance. While EMT does not only impact cell-cell adhesion, the switch in cadherin expression is a key part of the process. A deeper understanding of each tumor’s genomic and transcriptomic landscape, combined with the status of E-cadherin and the apical junctions, and elucidation of factors regulating dynamic EMT states may finally reconcile contextual differences and uncover the role of cadherin complexes in metastatic spread.
Cadherin and catenin effects on the tumor immune microenvironment
NF-κB and inflammation
The TME is crucial for tumor progression and comprises the ECM, stromal fibroblasts, endothelial cells, and tissue resident or infiltrating immune cells. These immune cells include lymphocytes and dendritic cells (DCs), which mediate adaptive immunity, and macrophages, natural killer (NK) cells, etc., which are involved in innate immunity. Through direct cell-cell interactions or communication via soluble factors, such as cytokines, the crosstalk between different cell types in the TME modulates biochemical pathways and cellular responses to suppress or promote tumor progression [reviewed in (Binnewies et al., 2018; Labani-Motlagh et al., 2020; Bejarano et al., 2021)].
The most studied role for cadherins and catenins in the TME is their effect on inflammatory responses via nuclear factor kappa B (NF-κB) signaling. NF-κB promotes inflammation, cell proliferation, angiogenesis, and cell migration and invasion and is frequently upregulated in cancer (Liu et al., 2017). In the absence of stimuli, NF-κB is sequestered in the cytoplasm by the inhibitor of κBs (IκBs) (Figure 7). In response to stimuli, such as cytokines, mitogens, or cellular stresses, IκB is phosphorylated and degraded, allowing nuclear translocation of NF-κB and expression of NF-κB target genes, driving inflammation.
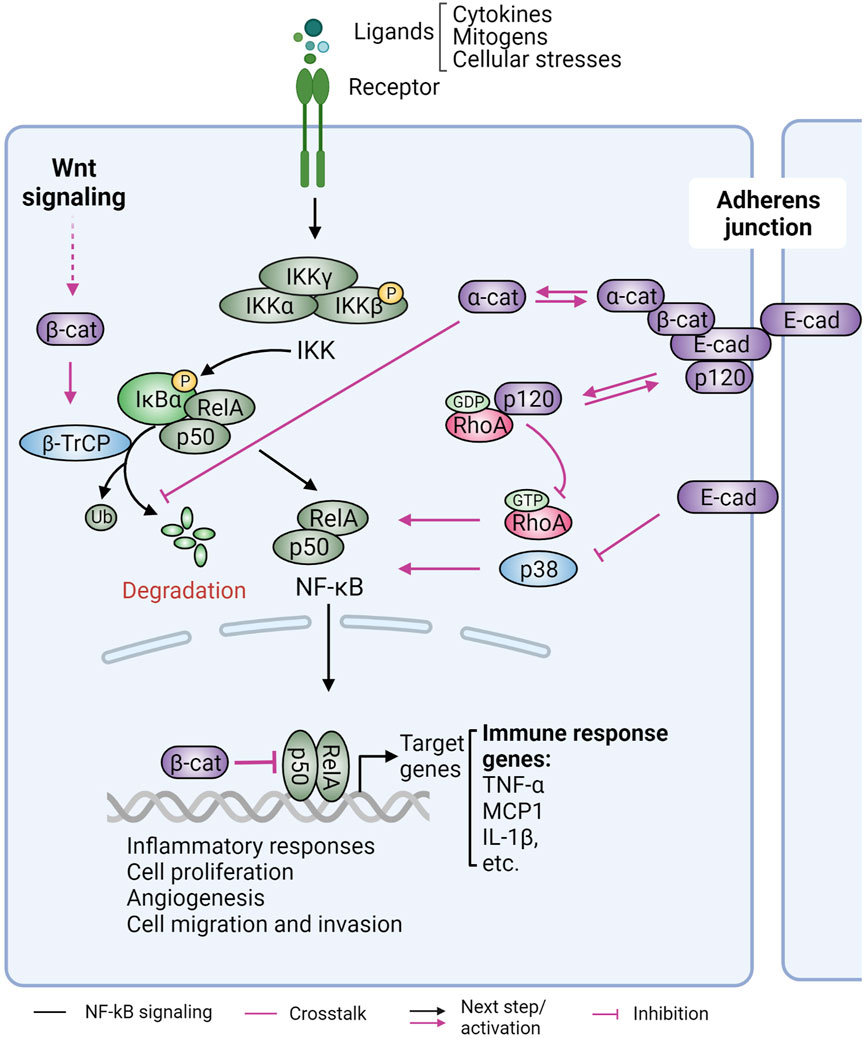
FIGURE 7. Role of cadherins and catenins in NF-κB signaling. Nuclear factor kappa B (NF-κB) signaling is a critical regulator of inflammation, an immune response that is linked to cancer formation and progression. Under normal conditions, NF-κB transcription factors (the most prominent dimer pair RelA-p50 shown) are retained in the cytoplasm by the inhibitor of κB (IκB). Various stimuli induce activation of the IκB kinase (IKK), which phosphorylates and inhibits IκB, resulting in the release and nuclear translocation of NF-κB and pro-inflammatory gene expression. Adherens junction proteins inhibit NF-κB signaling. E-cadherin (E-cad) loss in malignant melanoma cells blocks p38-mediated NF-kB activation. E-cadherin unbound p120-catenin (p120) functions as a guanine-nucleotide dissociation inhibitor to suppress RhoA activity, leading to NF-κB activation. Alpha-catenin (α-cat), independent of its binding to E-cadherin, interacts with IκBα to prevent IκBα ubiquitination and subsequent proteolytic degradation. Alpha-cat can also suppress NF-κB nuclear translocation. Beta-catenin (β-cat), on the other hand, can directly interact with NF-κB and repress its DNA-binding and transactivation activity. In response to canonical Wnt signaling, β-cat can stabilize β−TrCP1 mRNA. This results in elevated β−TrCP1 protein expression, which can increase NF-κB activation via downregulation of IκB. Figure created with BioRender.com.
Hermiston and Gordon initially reported that transgenic expression of a dominant negative N-cadherin in mouse small intestine epithelial cells results in inflammatory bowel disease (IBD) and epithelial dysplasia that leads to adenoma (Hermiston and Gordon, 1995). Loss of E-cadherin was then shown to induce activation of NF-κB signaling in malignant melanoma cells (Kuphal et al., 2004) (Figure 7). Furthermore, conditional knockout of p120-catenin in the mouse epidermis resulted in sustained inflammation, hyperproliferation, and skin cancer formation (Perez-Moreno et al., 2006; Perez-Moreno et al., 2008). Interestingly, the hyperplasia phenotype was linked to chronic inflammation and was independent of AJ instability, suggesting an involvement of soluble components. Indeed, a large array of pro-inflammatory cytokines (e.g., tumor necrosis factor alpha), interleukins (IL1β, IL6, IL13, and IL15), and chemokines [e.g., macrophage chemotactic protein (MCP1)] were substantially increased in CTNND1 (p120-catenin) null epidermis and induced infiltration of immune cells. Mechanistically, cadherin-uncoupled p120-catenin inhibits guanine-nucleotide exchange to restrict RhoA activity (Anastasiadis et al., 2000). Loss of p120-catenin therefore results in the upregulation of RhoA-ROCK signaling, which leads to nuclear localization and activation of NF-κB (Figure 7). These inflammatory responses and the resulting tumor development were similarly observed when p120-catenin was knocked out in the intestine, squamous oral cavity, esophagus, forestomach, and a TP53 null noninvasive breast cancer model (Smalley-Freed et al., 2010; Smalley-Freed et al., 2011; Stairs et al., 2011; Schackmann et al., 2013).
Alpha-catenin also has an anti-inflammatory role through the modulation of NF-κB signaling (Figure 7). Similar to p120-catenin ablation (Perez-Moreno et al., 2006), depletion of α-catenin results in NF-κB activation (Kobielak and Fuchs, 2006). In basal-like breast cancer cells, loss of α-catenin promotes NF-κB activation in an E-cadherin independent manner. A direct association of α-catenin with IκBα reportedly sustains IκBα stability and thus increases cytoplasmic retention of NF-kB (RelA) (Piao et al., 2014). β-catenin, on the other hand, exhibits a complicated relationship with NF-κB signaling by either suppressing or enhancing its activity in a context-dependent manner [reviewed in (Ma and Hottiger, 2016)]. In colon, liver, and breast cancer cells, a physical association of β-catenin with NF-κB is thought to decrease NF-κB’s DNA binding and transactivation activity (Deng et al., 2002; Du et al., 2009) (Figure 7). In prostate cancer cells, β-catenin also forms a transcription repressing complex to suppress NF-κB, resulting in downregulation of metastasis suppressor KAI1, a NFκB target gene (Kim et al., 2005) (Figure 7). Conversely, upregulation or increased stabilization of β-catenin by Wnt signaling resulted in NF-κB activation through upregulation of the β−TrCP E3 ubiquitin ligase, which mediates the degradation of NF-κB inhibitor protein IκB (Winston et al., 1999; Spiegelman et al., 2000; Noubissi et al., 2006) (Figure 7).
Overall, despite different degrees of involvement and underlying mechanisms, loss of cadherins (E-cadherin and N-cadherin) or catenins (p120-and α-catenin) results in NF-κB hyperactivation and production of pro-inflammatory signals that globally regulate the TME, induce immune infiltration and desmoplasia, and promote tumor progression.
Immune regulation
In addition to their role in inflammation, increasing evidence supports a direct role of cadherin-catenin complexes in immune regulation. While not a focus of this review, compelling evidence exists supporting an immune suppressive role for Wnt/β-catenin signaling that also extends to resistance to treatment with immune checkpoint inhibitors (ICIs) (Ruiz de Galarreta et al., 2019; Du et al., 2020). Surprisingly, E-cadherin is expressed in certain immune cells and is critical for their function. In DCs, disruption of E-cadherin-mediated DC-DC adhesions triggers DC maturation via activation of β-catenin/TCF signaling. Intriguingly, different from the DCs that undergo the typical pathogen-induced maturation process, these DCs are linked to immune tolerance rather than immunity initiation (Jiang et al., 2007). Further, E-cadherin homophilic ligation links multiple myeloma cells and plasmacytoid DCs (pDCs), a subset of DCs known for their role in anti-virus innate immunity, and unexpectedly allows tumor cells to condition pDCs to promote tumor growth (Bi et al., 2018). Moreover, E-cadherin on epithelial cells can form heterotypic interactions with integrin αEβ7 on lymphocytes, recruiting these immune cells into epithelial tumors (Cepek et al., 1994; Le Floc’h et al., 2007). These studies highlight the largely unexplored roles of cadherins and catenins beyond the epithelium, and their potential in modulating the immune TME to regulate tumor development.
Conclusion and perspectives
The diverse cellular functions regulated by cadherins and catenins highlight the importance of adhesion signaling in tumorigenesis and cancer progression. Extensive research has uncovered several molecular mechanisms by which cadherins/catenins regulate major oncogenic pathways involved in human cancer, and conversely, elucidated how oncogenic pathways regulate cadherin turnover and adhesive behaviors to promote cancer initiation and progression. The interplay between the two (cadherins/catenins and cancer pathways) is very dynamic and sensitive to their surrounding cells and the microenvironment in the tumor ecosystem.
This complexity stresses the importance of using physiological and disease relevant models to study cadherin/catenin biology and function in cancer. Human cancers are molecularly and cellularly complex comprised of sophisticated tissue architectures. Models that maintain this cellular diversity and overall architecture will be invaluable in elucidating the complex roles of CCCs in cancer. Significant questions remain related to the function of CCCs in cancer initiation, the TME, cancer progression, and metastasis. Addressing these questions will not only advance our understanding of signaling crosstalk between cadherin/catenin signaling and cancer driving events, but may also identify proper patient populations for given cancer therapies.
Notably, despite a wealth of information from TCGA, GWAS and functional studies regarding the expression, molecular alterations, and role of cadherins and catenins in particular cancer types, no directed treatment strategies are currently available to target cadherin/catenin dysfunction. Fortunately, several large-scale drug screening platforms have revealed potential lead compounds to target cadherin-dissociated β-catenin in Wnt signaling [reviewed in (Cui et al., 2018)]. Moreover, the introduction of novel gene therapy methods [reviewed in (Gregory and Copple, 2023)], epigenetic drugs, etc., may provide additional tools to target the multifaceted upregulation of oncogenic pathways induced by CCC dysfunction.
Author contributions
W-HL and PZA contributed to the planning, writing, and editing of this review article. LC is involved in the editing of this work. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedin, M., and King, N. (2008). The premetazoan ancestry of cadherins. Science 319, 946–948. doi:10.1126/science.1151084
Aceto, N., Bardia, A., Miyamoto, D. T., Donaldson, M. C., Wittner, B. S., Spencer, J. A., et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122. doi:10.1016/j.cell.2014.07.013
Agrawal, N., Frederick, M. J., Pickering, C. R., Bettegowda, C., Chang, K., Li, R. J., et al. (2011). Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333, 1154–1157. doi:10.1126/science.1206923
Akoumianakis, I., Polkinghorne, M., and Antoniades, C. (2022). Non-canonical WNT signalling in cardiovascular disease: Mechanisms and therapeutic implications. Nat. Rev. Cardiol. 19, 783–797. doi:10.1038/s41569-022-00718-5
Anastasiadis, P. Z., Moon, S. Y., Thoreson, M. A., Mariner, D. J., Crawford, H. C., Zheng, Y., et al. (2000). Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2, 637–644. doi:10.1038/35023588
Anastasiadis, P. Z. (2007). p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta 1773, 34–46. doi:10.1016/j.bbamcr.2006.08.040
Apte, R. S., Chen, D. S., and Ferrara, N. (2019). VEGF in signaling and disease: Beyond discovery and development. Cell 176, 1248–1264. doi:10.1016/j.cell.2019.01.021
Asthagiri, A. R., Parry, D. M., Butman, J. A., Kim, H. J., Tsilou, E. T., Zhuang, Z., et al. (2009). Neurofibromatosis type 2. Lancet 373, 1974–1986. doi:10.1016/S0140-6736(09)60259-2
Azzolin, L., Zanconato, F., Bresolin, S., Forcato, M., Basso, G., Bicciato, S., et al. (2012). Role of TAZ as mediator of Wnt signaling. Cell 151, 1443–1456. doi:10.1016/j.cell.2012.11.027
Azzolin, L., Panciera, T., Soligo, S., Enzo, E., Bicciato, S., Dupont, S., et al. (2014). YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170. doi:10.1016/j.cell.2014.06.013
Bailey, A. M., and Posakony, J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609–2622. doi:10.1101/gad.9.21.2609
Balint, K., Xiao, M., Pinnix, C. C., Soma, A., Veres, I., Juhasz, I., et al. (2005). Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J. Clin. Invest. 115, 3166–3176. doi:10.1172/JCI25001
Barry, A. K., Wang, N., and Leckband, D. E. (2015). Local VE-cadherin mechanotransduction triggers long-ranged remodeling of endothelial monolayers. J. Cell Sci. 128, 1341–1351. doi:10.1242/jcs.159954
Bejarano, L., Jordao, M. J. C., and Joyce, J. A. (2021). Therapeutic targeting of the tumor microenvironment. Cancer Discov. 11, 933–959. doi:10.1158/2159-8290.CD-20-1808
Benhamouche, S., Curto, M., Saotome, I., Gladden, A. B., Liu, C. H., Giovannini, M., et al. (2010). Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 24, 1718–1730. doi:10.1101/gad.1938710
Benham-Pyle, B. W., Pruitt, B. L., and Nelson, W. J. (2015). Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science 348, 1024–1027. doi:10.1126/science.aaa4559
Bentley, K., Franco, C. A., Philippides, A., Blanco, R., Dierkes, M., Gebala, V., et al. (2014). The role of differential VE-cadherin dynamics in cell rearrangement during angiogenesis. Nat. Cell Biol. 16, 309–321. doi:10.1038/ncb2926
Bertocchi, C., Wang, Y., Ravasio, A., Hara, Y., Wu, Y., Sailov, T., et al. (2017). Nanoscale architecture of cadherin-based cell adhesions. Nat. Cell Biol. 19, 28–37. doi:10.1038/ncb3456
Bi, E., Li, R., Bover, L. C., Li, H., Su, P., Ma, X., et al. (2018). E-cadherin expression on multiple myeloma cells activates tumor-promoting properties in plasmacytoid DCs. J. Clin. Invest. 128, 4821–4831. doi:10.1172/JCI121421
Bienz, M. (2005). beta-Catenin: a pivot between cell adhesion and Wnt signalling. Curr. Biol. 15, R64–R67. doi:10.1016/j.cub.2004.12.058
Binnewies, M., Roberts, E. W., Kersten, K., Chan, V., Fearon, D. F., Merad, M., et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 24, 541–550. doi:10.1038/s41591-018-0014-x
Brabletz, T., Kalluri, R., Nieto, M. A., and Weinberg, R. A. (2018). EMT in cancer. Nat. Rev. Cancer 18, 128–134. doi:10.1038/nrc.2017.118
Brasch, J., Harrison, O. J., Honig, B., and Shapiro, L. (2012). Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol. 22, 299–310. doi:10.1016/j.tcb.2012.03.004
Bruner, H. C., and Derksen, P. W. B. (2018). Loss of E-cadherin-dependent cell-cell adhesion and the development and progression of cancer. Cold Spring Harb. Perspect. Biol. 10, a029330. doi:10.1101/cshperspect.a029330
Burke, R., Nellen, D., Bellotto, M., Hafen, E., Senti, K. A., Dickson, B. J., et al. (1999). Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803–815. doi:10.1016/s0092-8674(00)81677-3
Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. doi:10.1038/nature11412
Cancer Genome Atlas Research Network Kandoth, C., Schultz, N., Cherniack, A. D., Akbani, R., Liu, Y., Shen, H., et al. (2013). Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73. doi:10.1038/nature12113
Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. doi:10.1038/nature07385
Cancer Genome Atlas Research Network (2014a). Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322. doi:10.1038/nature12965
Cancer Genome Atlas Research Network (2014b). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. doi:10.1038/nature13385
Cancer Genome Atlas Research Network (2017). Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23. doi:10.1016/j.cell.2017.05.046
Carmeliet, P., Lampugnani, M. G., Moons, L., Breviario, F., Compernolle, V., Bono, F., et al. (1999). Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell 98, 147–157. doi:10.1016/s0092-8674(00)81010-7
Casagolda, D., Del Valle-Perez, B., Valls, G., Lugilde, E., Vinyoles, M., Casado-Vela, J., et al. (2010). A p120-catenin-CK1epsilon complex regulates Wnt signaling. J. Cell Sci. 123, 2621–2631. doi:10.1242/jcs.067512
Cepek, K. L., Shaw, S. K., Parker, C. M., Russell, G. J., Morrow, J. S., Rimm, D. L., et al. (1994). Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature 372, 190–193. doi:10.1038/372190a0
Chalasani, K., and Brewster, R. M. (2011). N-cadherin-mediated cell adhesion restricts cell proliferation in the dorsal neural tube. Mol. Biol. Cell 22, 1505–1515. doi:10.1091/mbc.E10-08-0675
Chen, X., Kojima, S., Borisy, G. G., and Green, K. J. (2003). p120 catenin associates with kinesin and facilitates the transport of cadherin-catenin complexes to intercellular junctions. J. Cell Biol. 163, 547–557. doi:10.1083/jcb.200305137
Chiasson-Mackenzie, C., Morris, Z. S., Baca, Q., Morris, B., Coker, J. K., Mirchev, R., et al. (2015). NF2/Merlin mediates contact-dependent inhibition of EGFR mobility and internalization via cortical actomyosin. J. Cell Biol. 211, 391–405. doi:10.1083/jcb.201503081
Chiasson-Mackenzie, C., Morris, Z. S., Liu, C. H., Bradford, W. B., Koorman, T., and Mcclatchey, A. I. (2018). Merlin/ERM proteins regulate growth factor-induced macropinocytosis and receptor recycling by organizing the plasma membrane:cytoskeleton interface. Genes Dev. 32, 1201–1214. doi:10.1101/gad.317354.118
Cho, S. Y., Park, J. W., Liu, Y., Park, Y. S., Kim, J. H., Yang, H., et al. (2017). Sporadic early-onset diffuse gastric cancers have high frequency of somatic CDH1 alterations, but low frequency of somatic RHOA mutations compared with late-onset cancers. Gastroenterology 153, 536–549. doi:10.1053/j.gastro.2017.05.012
Choi, S. H., Estaras, C., Moresco, J. J., Yates, J. R., and Jones, K. A. (2013). α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev. 27, 2473–2488. doi:10.1101/gad.229062.113
Ciriello, G., Gatza, M. L., Beck, A. H., Wilkerson, M. D., Rhie, S. K., Pastore, A., et al. (2015). Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519. doi:10.1016/j.cell.2015.09.033
Cole, B. K., Curto, M., Chan, A. W., and Mcclatchey, A. I. (2008). Localization to the cortical cytoskeleton is necessary for Nf2/merlin-dependent epidermal growth factor receptor silencing. Mol. Cell Biol. 28, 1274–1284. doi:10.1128/MCB.01139-07
Conway, D. E., Breckenridge, M. T., Hinde, E., Gratton, E., Chen, C. S., and Schwartz, M. A. (2013). Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr. Biol. 23, 1024–1030. doi:10.1016/j.cub.2013.04.049
Conway, D. E., Coon, B. G., Budatha, M., Arsenovic, P. T., Orsenigo, F., Wessel, F., et al. (2017). VE-cadherin phosphorylation regulates endothelial fluid shear stress responses through the polarity protein LGN. Curr. Biol. 27, 2219–2225. doi:10.1016/j.cub.2017.06.020
Coon, B. G., Baeyens, N., Han, J., Budatha, M., Ross, T. D., Fang, J. S., et al. (2015). Intramembrane binding of VE-cadherin to VEGFR2 and VEGFR3 assembles the endothelial mechanosensory complex. J. Cell Biol. 208, 975–986. doi:10.1083/jcb.201408103
Cottrell, S., Bicknell, D., Kaklamanis, L., and Bodmer, W. F. (1992). Molecular analysis of APC mutations in familial adenomatous polyposis and sporadic colon carcinomas. Lancet 340, 626–630. doi:10.1016/0140-6736(92)92169-g
Cozzolino, M., Stagni, V., Spinardi, L., Campioni, N., Fiorentini, C., Salvati, E., et al. (2003). p120 Catenin is required for growth factor-dependent cell motility and scattering in epithelial cells. Mol. Biol. Cell 14, 1964–1977. doi:10.1091/mbc.e02-08-0469
Cui, C., Zhou, X., Zhang, W., Qu, Y., and Ke, X. (2018). Is beta-catenin a druggable target for cancer therapy? Trends Biochem. Sci. 43, 623–634. doi:10.1016/j.tibs.2018.06.003
Curto, M., Cole, B. K., Lallemand, D., Liu, C. H., and Mcclatchey, A. I. (2007). Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 177, 893–903. doi:10.1083/jcb.200703010
Dalrymple, S., Antony, L., Xu, Y., Uzgare, A. R., Arnold, J. T., Savaugeot, J., et al. (2005). Role of notch-1 and E-cadherin in the differential response to calcium in culturing normal versus malignant prostate cells. Cancer Res. 65, 9269–9279. doi:10.1158/0008-5472.CAN-04-3989
Damstrup, L., Kuwada, S. K., Dempsey, P. J., Brown, C. L., Hawkey, C. J., Poulsen, H. S., et al. (1999). Amphiregulin acts as an autocrine growth factor in two human polarizing colon cancer lines that exhibit domain selective EGF receptor mitogenesis. Br. J. Cancer 80, 1012–1019. doi:10.1038/sj.bjc.6690456
Davis, M. A., and Reynolds, A. B. (2006). Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell 10, 21–31. doi:10.1016/j.devcel.2005.12.004
Davis, M. A., Ireton, R. C., and Reynolds, A. B. (2003). A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163, 525–534. doi:10.1083/jcb.200307111
De Lau, W., Barker, N., Low, T. Y., Koo, B. K., Li, V. S., Teunissen, H., et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297. doi:10.1038/nature10337
De Lau, W., Peng, W. C., Gros, P., and Clevers, H. (2014). The R-spondin/lgr5/rnf43 module: Regulator of Wnt signal strength. Genes Dev. 28, 305–316. doi:10.1101/gad.235473.113
Deng, J., Miller, S. A., Wang, H. Y., Xia, W., Wen, Y., Zhou, B. P., et al. (2002). beta-catenin interacts with and inhibits NF-kappa B in human colon and breast cancer. Cancer Cell 2, 323–334. doi:10.1016/s1535-6108(02)00154-x
Derycke, L. D., and Bracke, M. E. (2004). N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int. J. Dev. Biol. 48, 463–476. doi:10.1387/ijdb.041793ld
Desai, R., Sarpal, R., Ishiyama, N., Pellikka, M., Ikura, M., and Tepass, U. (2013). Monomeric alpha-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15, 261–273. doi:10.1038/ncb2685
Ding, L., Ellis, M. J., Li, S., Larson, D. E., Chen, K., Wallis, J. W., et al. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005. doi:10.1038/nature08989
Drees, F., Pokutta, S., Yamada, S., Nelson, W. J., and Weis, W. I. (2005). Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell 123, 903–915. doi:10.1016/j.cell.2005.09.021
Drummond, C. J., Hanna, J. A., Garcia, M. R., Devine, D. J., Heyrana, A. J., Finkelstein, D., et al. (2018). Hedgehog pathway drives fusion-negative rhabdomyosarcoma initiated from non-myogenic endothelial progenitors. Cancer Cell 33, 108–124. doi:10.1016/j.ccell.2017.12.001
Du, Q., Zhang, X., Cardinal, J., Cao, Z., Guo, Z., Shao, L., et al. (2009). Wnt/beta-catenin signaling regulates cytokine-induced human inducible nitric oxide synthase expression by inhibiting nuclear factor-kappaB activation in cancer cells. Cancer Res. 69, 3764–3771. doi:10.1158/0008-5472.CAN-09-0014
Du, L., Lee, J. H., Jiang, H., Wang, C., Wang, S., Zheng, Z., et al. (2020). β-Catenin induces transcriptional expression of PD-L1 to promote glioblastoma immune evasion. J. Exp. Med. 217, e20191115. doi:10.1084/jem.20191115
Eagle, H., and Levine, E. M. (1967). Growth regulatory effects of cellular interaction. Nature 213, 1102–1106. doi:10.1038/2131102a0
Ellisen, L. W., Bird, J., West, D. C., Soreng, A. L., Reynolds, T. C., Smith, S. D., et al. (1991). TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661. doi:10.1016/0092-8674(91)90111-b
Fan, X., Mikolaenko, I., Elhassan, I., Ni, X., Wang, Y., Ball, D., et al. (2004). Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 64, 7787–7793. doi:10.1158/0008-5472.CAN-04-1446
Farabaugh, S. M., Boone, D. N., and Lee, A. V. (2015). Role of IGF1R in breast cancer subtypes, stemness, and lineage differentiation. Front. Endocrinol. (Lausanne) 6, 59. doi:10.3389/fendo.2015.00059
Fichtner, D., Lorenz, B., Engin, S., Deichmann, C., Oelkers, M., Janshoff, A., et al. (2014). Covalent and density-controlled surface immobilization of E-cadherin for adhesion force spectroscopy. PLoS One 9, e93123. doi:10.1371/journal.pone.0093123
Fischer, K. R., Durrans, A., Lee, S., Sheng, J., Li, F., Wong, S. T., et al. (2015). Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476. doi:10.1038/nature15748
Fournier-Thibault, C., Blavet, C., Jarov, A., Bajanca, F., Thorsteinsdottir, S., and Duband, J. L. (2009). Sonic hedgehog regulates integrin activity, cadherin contacts, and cell polarity to orchestrate neural tube morphogenesis. J. Neurosci. 29, 12506–12520. doi:10.1523/JNEUROSCI.2003-09.2009
Friedl, P., Locker, J., Sahai, E., and Segall, J. E. (2012). Classifying collective cancer cell invasion. Nat. Cell Biol. 14, 777–783. doi:10.1038/ncb2548
Fu, V., Plouffe, S. W., and Guan, K. L. (2017). The Hippo pathway in organ development, homeostasis, and regeneration. Curr. Opin. Cell Biol. 49, 99–107. doi:10.1016/j.ceb.2017.12.012
Fujita, Y., Krause, G., Scheffner, M., Zechner, D., Leddy, H. E., Behrens, J., et al. (2002). Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 4, 222–231. doi:10.1038/ncb758
Fukuchi, T., Sakamoto, M., Tsuda, H., Maruyama, K., Nozawa, S., and Hirohashi, S. (1998). Beta-catenin mutation in carcinoma of the uterine endometrium. Cancer Res. 58, 3526–3528.
Gamallo, C., Palacios, J., Suarez, A., Pizarro, A., Navarro, P., Quintanilla, M., et al. (1993). Correlation of E-cadherin expression with differentiation grade and histological type in breast carcinoma. Am. J. Pathol. 142, 987–993.
Gao, C., Wang, Y., Broaddus, R., Sun, L., Xue, F., and Zhang, W. (2018). Exon 3 mutations of CTNNB1 drive tumorigenesis: A review. Oncotarget 9, 5492–5508. doi:10.18632/oncotarget.23695
Gao, X., Xia, X., Li, F., Zhang, M., Zhou, H., Wu, X., et al. (2021). Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 23, 278–291. doi:10.1038/s41556-021-00639-4
Geyer, F. C., Lacroix-Triki, M., Savage, K., Arnedos, M., Lambros, M. B., Mackay, A., et al. (2011). β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol. 24, 209–231. doi:10.1038/modpathol.2010.205
Giampietro, C., Disanza, A., Bravi, L., Barrios-Rodiles, M., Corada, M., Frittoli, E., et al. (2015). The actin-binding protein EPS8 binds VE-cadherin and modulates YAP localization and signaling. J. Cell Biol. 211, 1177–1192. doi:10.1083/jcb.201501089
Giannakis, M., Hodis, E., Jasmine Mu, X., Yamauchi, M., Rosenbluh, J., Cibulskis, K., et al. (2014). RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46, 1264–1266. doi:10.1038/ng.3127
Giannini, A. L., Vivanco, M., and Kypta, R. M. (2000). alpha-catenin inhibits beta-catenin signaling by preventing formation of a beta-catenin*T-cell factor*DNA complex. J. Biol. Chem. 275, 21883–21888. doi:10.1074/jbc.M001929200
Gladden, A. B., Hebert, A. M., Schneeberger, E. E., and Mcclatchey, A. I. (2010). The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev. Cell 19, 727–739. doi:10.1016/j.devcel.2010.10.008
Gong, X., Qian, H., Cao, P., Zhao, X., Zhou, Q., Lei, J., et al. (2018). Structural basis for the recognition of sonic hedgehog by human Patched1. Science 361, eaas8935. doi:10.1126/science.aas8935
Gordon, W. R., Vardar-Ulu, D., Histen, G., Sanchez-Irizarry, C., Aster, J. C., and Blacklow, S. C. (2007). Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14, 295–300. doi:10.1038/nsmb1227
Gottardi, C. J., and Gumbiner, B. M. (2001). Adhesion signaling: How beta-catenin interacts with its partners. Curr. Biol. 11, R792–R794. doi:10.1016/s0960-9822(01)00473-0
Gottardi, C. J., Wong, E., and Gumbiner, B. M. (2001). E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153, 1049–1060. doi:10.1083/jcb.153.5.1049
Grammont, M. (2007). Adherens junction remodeling by the Notch pathway in Drosophila melanogaster oogenesis. J. Cell Biol. 177, 139–150. doi:10.1083/jcb.200609079
Grazia Lampugnani, M., Zanetti, A., Corada, M., Takahashi, T., Balconi, G., Breviario, F., et al. (2003). Contact inhibition of VEGF-induced proliferation requires vascular endothelial cadherin, beta-catenin, and the phosphatase DEP-1/CD148. J. Cell Biol. 161, 793–804. doi:10.1083/jcb.200209019
Gregory, G. L., and Copple, I. M. (2023). Modulating the expression of tumor suppressor genes using activating oligonucleotide technologies as a therapeutic approach in cancer. Mol. Ther. Nucleic Acids 31, 211–223. doi:10.1016/j.omtn.2022.12.016
Grumolato, L., Liu, G., Mong, P., Mudbhary, R., Biswas, R., Arroyave, R., et al. (2010). Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 24, 2517–2530. doi:10.1101/gad.1957710
Guo, Z., Neilson, L. J., Zhong, H., Murray, P. S., Zanivan, S., and Zaidel-Bar, R. (2014). E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal 7, rs7. doi:10.1126/scisignal.2005473
Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., et al. (1996). Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851. doi:10.1016/s0092-8674(00)81268-4
Hamaratoglu, F., Willecke, M., Kango-Singh, M., Nolo, R., Hyun, E., Tao, C., et al. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36. doi:10.1038/ncb1339
Han, T., Schatoff, E. M., Murphy, C., Zafra, M. P., Wilkinson, J. E., Elemento, O., et al. (2017). R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 8, 15945. doi:10.1038/ncomms15945
Hanahan, D., and Weinberg, R. A. (2000). The hallmarks of cancer. Cell 100, 57–70. doi:10.1016/s0092-8674(00)81683-9
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144, 646–674. doi:10.1016/j.cell.2011.02.013
Hanlon, L., Avila, J. L., Demarest, R. M., Troutman, S., Allen, M., Ratti, F., et al. (2010). Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 70, 4280–4286. doi:10.1158/0008-5472.CAN-09-4645
Hao, H. X., Xie, Y., Zhang, Y., Charlat, O., Oster, E., Avello, M., et al. (2012). ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200. doi:10.1038/nature11019
Harrison, O. J., Jin, X., Hong, S., Bahna, F., Ahlsen, G., Brasch, J., et al. (2011). The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 19, 244–256. doi:10.1016/j.str.2010.11.016
Harvey, K. F., Zhang, X., and Thomas, D. M. (2013). The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257. doi:10.1038/nrc3458
Hayward, P., Brennan, K., Sanders, P., Balayo, T., Dasgupta, R., Perrimon, N., et al. (2005). Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development 132, 1819–1830. doi:10.1242/dev.01724
Hazan, R. B., and Norton, L. (1998). The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton. J. Biol. Chem. 273, 9078–9084. doi:10.1074/jbc.273.15.9078
Heallen, T., Zhang, M., Wang, J., Bonilla-Claudio, M., Klysik, E., Johnson, R. L., et al. (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461. doi:10.1126/science.1199010
Heasman, J., Crawford, A., Goldstone, K., Garner-Hamrick, P., Gumbiner, B., McCrea, P., et al. (1994). Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell 79, 791–803. doi:10.1016/0092-8674(94)90069-8
Helsten, T., Elkin, S., Arthur, E., Tomson, B. N., Carter, J., and Kurzrock, R. (2016). The FGFR landscape in cancer: Analysis of 4,853 tumors by next-generation sequencing. Clin. Cancer Res. 22, 259–267. doi:10.1158/1078-0432.CCR-14-3212
Henderson, B. R., and Fagotto, F. (2002). The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep. 3, 834–839. doi:10.1093/embo-reports/kvf181
Hermiston, M. L., and Gordon, J. I. (1995). Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science 270, 1203–1207. doi:10.1126/science.270.5239.1203
Hiscox, S., and Jiang, W. G. (1999). Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells. Biochem. Biophys. Res. Commun. 261, 406–411. doi:10.1006/bbrc.1999.1002
Hoschuetzky, H., Aberle, H., and Kemler, R. (1994). Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J. Cell Biol. 127, 1375–1380. doi:10.1083/jcb.127.5.1375
Huber, A. H., and Weis, W. I. (2001). The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105, 391–402. doi:10.1016/s0092-8674(01)00330-0
Hui, C. C., and Angers, S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537. doi:10.1146/annurev-cellbio-092910-154048
Hulit, J., Suyama, K., Chung, S., Keren, R., Agiostratidou, G., Shan, W., et al. (2007). N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 67, 3106–3116. doi:10.1158/0008-5472.CAN-06-3401
Huyghe, J. R., Bien, S. A., Harrison, T. A., Kang, H. M., Chen, S., Schmit, S. L., et al. (2019). Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 51, 76–87. doi:10.1038/s41588-018-0286-6
Inaguma, S., Kasai, K., and Ikeda, H. (2011). GLI1 facilitates the migration and invasion of pancreatic cancer cells through MUC5AC-mediated attenuation of E-cadherin. Oncogene 30, 714–723. doi:10.1038/onc.2010.459
Ireton, R. C., Davis, M. A., Van Hengel, J., Mariner, D. J., Barnes, K., Thoreson, M. A., et al. (2002). A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476. doi:10.1083/jcb.200205115
Ishiyama, N., Sarpal, R., Wood, M. N., Barrick, S. K., Nishikawa, T., Hayashi, H., et al. (2018). Force-dependent allostery of the alpha-catenin actin-binding domain controls adherens junction dynamics and functions. Nat. Commun. 9, 5121. doi:10.1038/s41467-018-07481-7
Iso, T., Kedes, L., and Hamamori, Y. (2003). HES and HERP families: Multiple effectors of the notch signaling pathway. J. Cell Physiol. 194, 237–255. doi:10.1002/jcp.10208
Jarov, A., Williams, K. P., Ling, L. E., Koteliansky, V. E., Duband, J. L., and Fournier-Thibault, C. (2003). A dual role for Sonic hedgehog in regulating adhesion and differentiation of neuroepithelial cells. Dev. Biol. 261, 520–536. doi:10.1016/s0012-1606(03)00351-8
Jarriault, S., Le Bail, O., Hirsinger, E., Pourquie, O., Logeat, F., Strong, C. F., et al. (1998). Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell Biol. 18, 7423–7431. doi:10.1128/mcb.18.12.7423
Jayaraman, S. S., Rayhan, D. J., Hazany, S., and Kolodney, M. S. (2014). Mutational landscape of basal cell carcinomas by whole-exome sequencing. J. Invest. Dermatol 134, 213–220. doi:10.1038/jid.2013.276
Jiang, A., Bloom, O., Ono, S., Cui, W., Unternaehrer, J., Jiang, S., et al. (2007). Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity 27, 610–624. doi:10.1016/j.immuni.2007.08.015
Jin, Z. G., Ueba, H., Tanimoto, T., Lungu, A. O., Frame, M. D., and Berk, B. C. (2003). Ligand-independent activation of vascular endothelial growth factor receptor 2 by fluid shear stress regulates activation of endothelial nitric oxide synthase. Circ. Res. 93, 354–363. doi:10.1161/01.RES.0000089257.94002.96
Jin, Y. H., Kim, H., Ki, H., Yang, I., Yang, N., Lee, K. Y., et al. (2009). Beta-catenin modulates the level and transcriptional activity of Notch1/NICD through its direct interaction. Biochim. Biophys. Acta 1793, 290–299. doi:10.1016/j.bbamcr.2008.10.002
Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., et al. (1996). Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671. doi:10.1126/science.272.5268.1668
Kamei, T., Matozaki, T., Sakisaka, T., Kodama, A., Yokoyama, S., Peng, Y. F., et al. (1999). Coendocytosis of cadherin and c-Met coupled to disruption of cell-cell adhesion in MDCK cells--regulation by Rho, Rac and Rab small G proteins. Oncogene 18, 6776–6784. doi:10.1038/sj.onc.1203114
Khramtsov, A. I., Khramtsova, G. F., Tretiakova, M., Huo, D., Olopade, O. I., and Goss, K. H. (2010). Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 176, 2911–2920. doi:10.2353/ajpath.2010.091125
Kim, S. W., Park, J. I., Spring, C. M., Sater, A. K., Ji, H., Otchere, A. A., et al. (2004). Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat. Cell Biol. 6, 1212–1220. doi:10.1038/ncb1191
Kim, J. H., Kim, B., Cai, L., Choi, H. J., Ohgi, K. A., Tran, C., et al. (2005). Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 434, 921–926. doi:10.1038/nature03452
Kim, J. H., Kushiro, K., Graham, N. A., and Asthagiri, A. R. (2009). Tunable interplay between epidermal growth factor and cell-cell contact governs the spatial dynamics of epithelial growth. Proc. Natl. Acad. Sci. U. S. A. 106, 11149–11153. doi:10.1073/pnas.0812651106
Kim, N. G., Koh, E., Chen, X., and Gumbiner, B. M. (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U. S. A. 108, 11930–11935. doi:10.1073/pnas.1103345108
Kim, E., Lisby, A., Ma, C., Lo, N., Ehmer, U., Hayer, K. E., et al. (2019). Promotion of growth factor signaling as a critical function of beta-catenin during HCC progression. Nat. Commun. 10, 1909. doi:10.1038/s41467-019-09780-z
Kim, J., Koo, B. K., and Knoblich, J. A. (2020). Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584. doi:10.1038/s41580-020-0259-3
King, N., Hittinger, C. T., and Carroll, S. B. (2003). Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301, 361–363. doi:10.1126/science.1083853
Klinakis, A., Szabolcs, M., Politi, K., Kiaris, H., Artavanis-Tsakonas, S., and Efstratiadis, A. (2006). Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. U. S. A. 103, 9262–9267. doi:10.1073/pnas.0603371103
Kobielak, A., and Fuchs, E. (2006). Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc. Natl. Acad. Sci. U. S. A. 103, 2322–2327. doi:10.1073/pnas.0510422103
Koch, S., and Claesson-Welsh, L. (2012). Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb. Perspect. Med. 2, a006502. doi:10.1101/cshperspect.a006502
Kode, A., Manavalan, J. S., Mosialou, I., Bhagat, G., Rathinam, C. V., Luo, N., et al. (2014). Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature 506, 240–244. doi:10.1038/nature12883
Kohn, A. D., and Moon, R. T. (2005). Wnt and calcium signaling: Beta-catenin-independent pathways. Cell Calcium 38, 439–446. doi:10.1016/j.ceca.2005.06.022
Koirala, R., Priest, A. V., Yen, C. F., Cheah, J. S., Pannekoek, W. J., Gloerich, M., et al. (2021). Inside-out regulation of E-cadherin conformation and adhesion. Proc. Natl. Acad. Sci. U. S. A. 118, e2104090118. doi:10.1073/pnas.2104090118
Koo, B. K., Spit, M., Jordens, I., Low, T. Y., Stange, D. E., Van De Wetering, M., et al. (2012). Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669. doi:10.1038/nature11308
Kourtidis, A., Ngok, S. P., and Anastasiadis, P. Z. (2013). p120 catenin: an essential regulator of cadherin stability, adhesion-induced signaling, and cancer progression. Prog. Mol. Biol. Transl. Sci. 116, 409–432. doi:10.1016/B978-0-12-394311-8.00018-2
Kourtidis, A., Ngok, S. P., Pulimeno, P., Feathers, R. W., Carpio, L. R., Baker, T. R., et al. (2015). Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat. Cell Biol. 17, 1145–1157. doi:10.1038/ncb3227
Krieghoff, E., Behrens, J., and Mayr, B. (2006). Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J. Cell Sci. 119, 1453–1463. doi:10.1242/jcs.02864
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Kuphal, S., Poser, I., Jobin, C., Hellerbrand, C., and Bosserhoff, A. K. (2004). Loss of E-cadherin leads to upregulation of NFkappaB activity in malignant melanoma. Oncogene 23, 8509–8519. doi:10.1038/sj.onc.1207831
Kwon, C., Cheng, P., King, I. N., Andersen, P., Shenje, L., Nigam, V., et al. (2011). Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nat. Cell Biol. 13, 1244–1251. doi:10.1038/ncb2313
Labani-Motlagh, A., Ashja-Mahdavi, M., and Loskog, A. (2020). The tumor microenvironment: A milieu hindering and obstructing antitumor immune responses. Front. Immunol. 11, 940. doi:10.3389/fimmu.2020.00940
Lai, C. M., Lin, K. Y., Kao, S. H., Chen, Y. N., Huang, F., and Hsu, H. J. (2017). Hedgehog signaling establishes precursors for germline stem cell niches by regulating cell adhesion. J. Cell Biol. 216, 1439–1453. doi:10.1083/jcb.201610063
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. doi:10.1038/nrm3758
Lampugnani, M. G., Orsenigo, F., Gagliani, M. C., Tacchetti, C., and Dejana, E. (2006). Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 174, 593–604. doi:10.1083/jcb.200602080
Le Floc'h, A., Jalil, A., Vergnon, I., Le Maux Chansac, B., Lazar, V., Bismuth, G., et al. (2007). Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J. Exp. Med. 204, 559–570. doi:10.1084/jem.20061524
Lei, G., Zhuang, L., and Gan, B. (2022). Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 22, 381–396. doi:10.1038/s41568-022-00459-0
Levine, E. M., Becker, Y., Boone, C. W., and Eagle, H. (1965). Contact inhibition, macromolecular synthesis, and polyribosomes in cultured human diploid fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 53, 350–356. doi:10.1073/pnas.53.2.350
Li, X., Deng, W., Lobo-Ruppert, S. M., and Ruppert, J. M. (2007). Gli1 acts through Snail and E-cadherin to promote nuclear signaling by beta-catenin. Oncogene 26, 4489–4498. doi:10.1038/sj.onc.1210241
Li, Y., Merkel, C. D., Zeng, X., Heier, J. A., Cantrell, P. S., Sun, M., et al. (2019). The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J. Cell Sci. 132, jcs221606. doi:10.1242/jcs.221606
Liang, J., Balachandra, S., Ngo, S., and O'brien, L. E. (2017). Feedback regulation of steady-state epithelial turnover and organ size. Nature 548, 588–591. doi:10.1038/nature23678
Lien, W. H., Klezovitch, O., Fernandez, T. E., Delrow, J., and Vasioukhin, V. (2006). alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science 311, 1609–1612. doi:10.1126/science.1121449
Ligon, L. A., Karki, S., Tokito, M., and Holzbaur, E. L. (2001). Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol. 3, 913–917. doi:10.1038/ncb1001-913
Liu, T., Zhang, L., Joo, D., and Sun, S. C. (2017). NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2, 17023-. doi:10.1038/sigtrans.2017.23
Liu, X., Fang, J., Huang, S., Wu, X., Xie, X., Wang, J., et al. (2021). Tumor-on-a-chip: From bioinspired design to biomedical application. Microsyst. Nanoeng. 7, 50. doi:10.1038/s41378-021-00277-8
Lowell, S., Jones, P., Le Roux, I., Dunne, J., and Watt, F. M. (2000). Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10, 491–500. doi:10.1016/s0960-9822(00)00451-6
Ma, B., and Hottiger, M. O. (2016). Crosstalk between wnt/β-catenin and NF-κB signaling pathway during inflammation. Front. Immunol. 7, 378. doi:10.3389/fimmu.2016.00378
Ma, Y., Erkner, A., Gong, R., Yao, S., Taipale, J., Basler, K., et al. (2002). Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63–75. doi:10.1016/s0092-8674(02)00977-7
Mack, J. J., Mosqueiro, T. S., Archer, B. J., Jones, W. M., Sunshine, H., Faas, G. C., et al. (2017). NOTCH1 is a mechanosensor in adult arteries. Nat. Commun. 8, 1620. doi:10.1038/s41467-017-01741-8
Maiden, S. L., Petrova, Y. I., and Gumbiner, B. M. (2016). Microtubules inhibit E-cadherin adhesive activity by maintaining phosphorylated p120-catenin in a colon carcinoma cell model. PLoS One 11, e0148574. doi:10.1371/journal.pone.0148574
Malyukova, A., Dohda, T., Von Der Lehr, N., Akhoondi, S., Corcoran, M., Heyman, M., et al. (2007). The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 67, 5611–5616. doi:10.1158/0008-5472.CAN-06-4381
Mangolini, M., Gotte, F., Moore, A., Ammon, T., Oelsner, M., Lutzny-Geier, G., et al. (2018). Notch2 controls non-autonomous Wnt-signalling in chronic lymphocytic leukaemia. Nat. Commun. 9, 3839. doi:10.1038/s41467-018-06069-5
Maser, R. S., Choudhury, B., Campbell, P. J., Feng, B., Wong, K. K., Protopopov, A., et al. (2007). Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 447, 966–971. doi:10.1038/nature05886
Mazur, P. K., Einwachter, H., Lee, M., Sipos, B., Nakhai, H., Rad, R., et al. (2010). Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A. 107, 13438–13443. doi:10.1073/pnas.1002423107
McCrea, P. D., and Gumbiner, B. M. (1991). Purification of a 92-kDa cytoplasmic protein tightly associated with the cell-cell adhesion molecule E-cadherin (uvomorulin). J. Biol. Chem. 266, 4514–4520. doi:10.1016/s0021-9258(20)64353-8
Mendonsa, A. M., Na, T. Y., and Gumbiner, B. M. (2018). E-cadherin in contact inhibition and cancer. Oncogene 37, 4769–4780. doi:10.1038/s41388-018-0304-2
Meng, W., Mushika, Y., Ichii, T., and Takeichi, M. (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell 135, 948–959. doi:10.1016/j.cell.2008.09.040
Meng, Z., Moroishi, T., and Guan, K. L. (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17. doi:10.1101/gad.274027.115
Misiorek, J. O., Przybyszewska-Podstawka, A., Kalafut, J., Paziewska, B., Rolle, K., Rivero-Muller, A., et al. (2021). Context matters: NOTCH signatures and pathway in cancer progression and metastasis. Cells 10, 94. doi:10.3390/cells10010094
Miyoshi, J., and Takai, Y. (2008). Structural and functional associations of apical junctions with cytoskeleton. Biochim. Biophys. Acta 1778, 670–691. doi:10.1016/j.bbamem.2007.12.014
Moll, R., Mitze, M., Frixen, U. H., and Birchmeier, W. (1993). Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am. J. Pathol. 143, 1731–1742.
Morin, P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B., et al. (1997). Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275, 1787–1790. doi:10.1126/science.275.5307.1787
Mortazavi, F., An, J., Dubinett, S., and Rettig, M. (2010). p120-catenin is transcriptionally downregulated by FOXC2 in non-small cell lung cancer cells. Mol. Cancer Res. 8, 762–774. doi:10.1158/1541-7786.MCR-10-0004
Motti, M. L., Califano, D., Baldassarre, G., Celetti, A., Merolla, F., Forzati, F., et al. (2005). Reduced E-cadherin expression contributes to the loss of p27kip1-mediated mechanism of contact inhibition in thyroid anaplastic carcinomas. Carcinogenesis 26, 1021–1034. doi:10.1093/carcin/bgi050
Murakami, H., Mizuno, T., Taniguchi, T., Fujii, M., Ishiguro, F., Fukui, T., et al. (2011). LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 71, 873–883. doi:10.1158/0008-5472.CAN-10-2164
Na, T. Y., Schecterson, L., Mendonsa, A. M., and Gumbiner, B. M. (2020). The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc. Natl. Acad. Sci. U. S. A. 117, 5931–5937. doi:10.1073/pnas.1918167117
Nagafuchi, A., and Takeichi, M. (1988). Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 7, 3679–3684. doi:10.1002/j.1460-2075.1988.tb03249.x
Nagle, A. M., Levine, K. M., Tasdemir, N., Scott, J. A., Burlbaugh, K., Kehm, J., et al. (2018). Loss of E-cadherin enhances IGF1-IGF1R pathway activation and sensitizes breast cancers to anti-IGF1R/InsR inhibitors. Clin. Cancer Res. 24, 5165–5177. doi:10.1158/1078-0432.CCR-18-0279
Ngo, S., Liang, J., Su, Y. H., and O'brien, L. E. (2020). Disruption of EGF feedback by intestinal tumors and neighboring cells in Drosophila. Curr. Biol. 30, 1537–1546. doi:10.1016/j.cub.2020.01.082
Nguyen, C. D. K., and Yi, C. (2019). YAP/TAZ signaling and resistance to cancer therapy. Trends Cancer 5, 283–296. doi:10.1016/j.trecan.2019.02.010
Nicolas, M., Wolfer, A., Raj, K., Kummer, J. A., Mill, P., Van Noort, M., et al. (2003). Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 33, 416–421. doi:10.1038/ng1099
Nieto, M. A., Huang, R. Y., Jackson, R. A., and Thiery, J. P. (2016). Emt: 2016. Cell 166, 21–45. doi:10.1016/j.cell.2016.06.028
Nishimura, T., and Takeichi, M. (2009). Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 89, 33–54. doi:10.1016/S0070-2153(09)89002-9
Nishisho, I., Nakamura, Y., Miyoshi, Y., Miki, Y., Ando, H., Horii, A., et al. (1991). Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253, 665–669. doi:10.1126/science.1651563
Noubissi, F. K., Elcheva, I., Bhatia, N., Shakoori, A., Ougolkov, A., Liu, J., et al. (2006). CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature 441, 898–901. doi:10.1038/nature04839
Ozawa, M., Baribault, H., and Kemler, R. (1989). The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 8, 1711–1717. doi:10.1002/j.1460-2075.1989.tb03563.x
Ozawa, M., Ringwald, M., and Kemler, R. (1990). Uvomorulin-catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc. Natl. Acad. Sci. U. S. A. 87, 4246–4250. doi:10.1073/pnas.87.11.4246
Ozdamar, B., Bose, R., Barrios-Rodiles, M., Wang, H. R., Zhang, Y., and Wrana, J. L. (2005). Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307, 1603–1609. doi:10.1126/science.1105718
Padmanaban, V., Krol, I., Suhail, Y., Szczerba, B. M., Aceto, N., Bader, J. S., et al. (2019). E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444. doi:10.1038/s41586-019-1526-3
Park, W. S., Dong, S. M., Kim, S. Y., Na, E. Y., Shin, M. S., Pi, J. H., et al. (1999). Somatic mutations in the kinase domain of the Met/hepatocyte growth factor receptor gene in childhood hepatocellular carcinomas. Cancer Res. 59, 307–310.
Park, J. I., Kim, S. W., Lyons, J. P., Ji, H., Nguyen, T. T., Cho, K., et al. (2005). Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell 8, 843–854. doi:10.1016/j.devcel.2005.04.010
Paschoud, S., Jond, L., Guerrera, D., and Citi, S. (2014). PLEKHA7 modulates epithelial tight junction barrier function. Tissue Barriers 2, e28755. doi:10.4161/tisb.28755
Pece, S., and Gutkind, J. S. (2000). Signaling from E-cadherins to the MAPK pathway by the recruitment and activation of epidermal growth factor receptors upon cell-cell contact formation. J. Biol. Chem. 275, 41227–41233. doi:10.1074/jbc.M006578200
Peng, X., Maiers, J. L., Choudhury, D., Craig, S. W., and Demali, K. A. (2012). α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 287, 7728–7737. doi:10.1074/jbc.M111.297481
Perez-Moreno, M., Davis, M. A., Wong, E., Pasolli, H. A., Reynolds, A. B., and Fuchs, E. (2006). p120-catenin mediates inflammatory responses in the skin. Cell 124, 631–644. doi:10.1016/j.cell.2005.11.043
Perez-Moreno, M., Song, W., Pasolli, H. A., Williams, S. E., and Fuchs, E. (2008). Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc. Natl. Acad. Sci. U. S. A. 105, 15399–15404. doi:10.1073/pnas.0807301105
Perrais, M., Chen, X., Perez-Moreno, M., and Gumbiner, B. M. (2007). E-cadherin homophilic ligation inhibits cell growth and epidermal growth factor receptor signaling independently of other cell interactions. Mol. Biol. Cell 18, 2013–2025. doi:10.1091/mbc.e06-04-0348
Petrova, Y. I., Spano, M. M., and Gumbiner, B. M. (2012). Conformational epitopes at cadherin calcium-binding sites and p120-catenin phosphorylation regulate cell adhesion. Mol. Biol. Cell 23, 2092–2108. doi:10.1091/mbc.E11-12-1060
Phelps, R. A., Chidester, S., Dehghanizadeh, S., Phelps, J., Sandoval, I. T., Rai, K., et al. (2009). A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 137, 623–634. doi:10.1016/j.cell.2009.02.037
Piao, H. L., Yuan, Y., Wang, M., Sun, Y., Liang, H., and Ma, L. (2014). α-catenin acts as a tumour suppressor in E-cadherin-negative basal-like breast cancer by inhibiting NF-κB signalling. Nat. Cell Biol. 16, 245–254. doi:10.1038/ncb2909
Piccolo, S., Dupont, S., and Cordenonsi, M. (2014). The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 94, 1287–1312. doi:10.1152/physrev.00005.2014
Polacheck, W. J., Kutys, M. L., Yang, J., Eyckmans, J., Wu, Y., Vasavada, H., et al. (2017). A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature 552, 258–262. doi:10.1038/nature24998
Pottier, C., Fresnais, M., Gilon, M., Jerusalem, G., Longuespee, R., and Sounni, N. E. (2020). Tyrosine kinase inhibitors in cancer: Breakthrough and challenges of targeted therapy. Cancers (Basel) 12, 731. doi:10.3390/cancers12030731
Powell, S. M., Zilz, N., Beazer-Barclay, Y., Bryan, T. M., Hamilton, S. R., Thibodeau, S. N., et al. (1992). APC mutations occur early during colorectal tumorigenesis. Nature 359, 235–237. doi:10.1038/359235a0
Puram, S. V., Tirosh, I., Parikh, A. S., Patel, A. P., Yizhak, K., Gillespie, S., et al. (2017). Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624. doi:10.1016/j.cell.2017.10.044
Qian, X., Karpova, T., Sheppard, A. M., Mcnally, J., and Lowy, D. R. (2004). E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 23, 1739–1748. doi:10.1038/sj.emboj.7600136
Qian, X., Anzovino, A., Kim, S., Suyama, K., Yao, J., Hulit, J., et al. (2014). N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene 33, 3411–3421. doi:10.1038/onc.2013.310
Raffel, C., Jenkins, R. B., Frederick, L., Hebrink, D., Alderete, B., Fults, D. W., et al. (1997). Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 57, 842–845.
Ranganathan, P., Weaver, K. L., and Capobianco, A. J. (2011). Notch signalling in solid tumours: A little bit of everything but not all the time. Nat. Rev. Cancer 11, 338–351. doi:10.1038/nrc3035
Rangarajan, A., Talora, C., Okuyama, R., Nicolas, M., Mammucari, C., Oh, H., et al. (2001). Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436. doi:10.1093/emboj/20.13.3427
Rimm, D. L., Koslov, E. R., Kebriaei, P., Cianci, C. D., and Morrow, J. S. (1995). Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. U. S. A. 92, 8813–8817. doi:10.1073/pnas.92.19.8813
Rodilla, V., Villanueva, A., Obrador-Hevia, A., Robert-Moreno, A., Fernandez-Majada, V., Grilli, A., et al. (2009). Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 106, 6315–6320. doi:10.1073/pnas.0813221106
Ronchini, C., and Capobianco, A. J. (2001). Induction of cyclin D1 transcription and CDK2 activity by notch(ic): Implication for cell cycle disruption in transformation by notch(ic). Mol. Cell Biol. 21, 5925–5934. doi:10.1128/mcb.21.17.5925-5934.2001
Rosenbluh, J., Nijhawan, D., Cox, A. G., Li, X., Neal, J. T., Schafer, E. J., et al. (2012). β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473. doi:10.1016/j.cell.2012.11.026
Rowan, A. J., Lamlum, H., Ilyas, M., Wheeler, J., Straub, J., Papadopoulou, A., et al. (2000). APC mutations in sporadic colorectal tumors: A mutational "hotspot" and interdependence of the "two hits. Proc. Natl. Acad. Sci. U. S. A. 97, 3352–3357. doi:10.1073/pnas.97.7.3352
Rozengurt, E., Sinnett-Smith, J., and Eibl, G. (2018). Yes-associated protein (YAP) in pancreatic cancer: At the epicenter of a targetable signaling network associated with patient survival. Signal Transduct. Target Ther. 3, 11. doi:10.1038/s41392-017-0005-2
Rubinfeld, B., Souza, B., Albert, I., Muller, O., Chamberlain, S. H., Masiarz, F. R., et al. (1993). Association of the APC gene product with beta-catenin. Science 262, 1731–1734. doi:10.1126/science.8259518
Ruiz De Galarreta, M., Bresnahan, E., Molina-Sanchez, P., Lindblad, K. E., Maier, B., Sia, D., et al. (2019). β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 9, 1124–1141. doi:10.1158/2159-8290.CD-19-0074
Sadot, E., Simcha, I., Shtutman, M., Ben-Ze'ev, A., and Geiger, B. (1998). Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc. Natl. Acad. Sci. U. S. A. 95, 15339–15344. doi:10.1073/pnas.95.26.15339
Sanchez-Vega, F., Mina, M., Armenia, J., Chatila, W. K., Luna, A., La, K. C., et al. (2018). Oncogenic signaling pathways in the cancer genome Atlas. Cell 173, 321–337.e10. doi:10.1016/j.cell.2018.03.035
Sanson, B., White, P., and Vincent, J. P. (1996). Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature 383, 627–630. doi:10.1038/383627a0
Schackmann, R. C., Klarenbeek, S., Vlug, E. J., Stelloo, S., Van Amersfoort, M., Tenhagen, M., et al. (2013). Loss of p120-catenin induces metastatic progression of breast cancer by inducing anoikis resistance and augmenting growth factor receptor signaling. Cancer Res. 73, 4937–4949. doi:10.1158/0008-5472.CAN-13-0180
Schlegelmilch, K., Mohseni, M., Kirak, O., Pruszak, J., Rodriguez, J. R., Zhou, D., et al. (2011). Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782–795. doi:10.1016/j.cell.2011.02.031
Schlessinger, J. (2014). Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 6, a008912. doi:10.1101/cshperspect.a008912
Seddiki, R., Narayana, G., Strale, P. O., Balcioglu, H. E., Peyret, G., Yao, M., et al. (2018). Force-dependent binding of vinculin to alpha-catenin regulates cell-cell contact stability and collective cell behavior. Mol. Biol. Cell 29, 380–388. doi:10.1091/mbc.E17-04-0231
Sehgal, P., Kong, X., Wu, J., Sunyer, R., Trepat, X., and Leckband, D. (2018). Epidermal growth factor receptor and integrins control force-dependent vinculin recruitment to E-cadherin junctions. J. Cell Sci. 131, jcs206656. doi:10.1242/jcs.206656
Seshagiri, S., Stawiski, E. W., Durinck, S., Modrusan, Z., Storm, E. E., Conboy, C. B., et al. (2012). Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664. doi:10.1038/nature11282
Shafraz, O., Xie, B., Yamada, S., and Sivasankar, S. (2020). Mapping transmembrane binding partners for E-cadherin ectodomains. Proc. Natl. Acad. Sci. U. S. A. 117, 31157–31165. doi:10.1073/pnas.2010209117
Shamir, E. R., Pappalardo, E., Jorgens, D. M., Coutinho, K., Tsai, W. T., Aziz, K., et al. (2014). Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol. 204, 839–856. doi:10.1083/jcb.201306088
Shashikanth, N., Petrova, Y. I., Park, S., Chekan, J., Maiden, S., Spano, M., et al. (2015). Allosteric regulation of E-cadherin adhesion. J. Biol. Chem. 290, 21749–21761. doi:10.1074/jbc.M115.657098
Shay-Salit, A., Shushy, M., Wolfovitz, E., Yahav, H., Breviario, F., Dejana, E., et al. (2002). VEGF receptor 2 and the adherens junction as a mechanical transducer in vascular endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 99, 9462–9467. doi:10.1073/pnas.142224299
Shibamoto, S., Hayakawa, M., Takeuchi, K., Hori, T., Oku, N., Miyazawa, K., et al. (1994). Tyrosine phosphorylation of beta-catenin and plakoglobin enhanced by hepatocyte growth factor and epidermal growth factor in human carcinoma cells. Cell Adhes. Commun. 1, 295–305. doi:10.3109/15419069409097261
Shibata, H., Takano, H., Ito, M., Shioya, H., Hirota, M., Matsumoto, H., et al. (2007). Alpha-catenin is essential in intestinal adenoma formation. Proc. Natl. Acad. Sci. U. S. A. 104, 18199–18204. doi:10.1073/pnas.0705730104
Shintani, Y., Hollingsworth, M. A., Wheelock, M. J., and Johnson, K. R. (2006). Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH(2)-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res. 66, 11745–11753. doi:10.1158/0008-5472.CAN-06-2322
Short, S. P., Kondo, J., Smalley-Freed, W. G., Takeda, H., Dohn, M. R., Powell, A. E., et al. (2017). p120-Catenin is an obligate haploinsufficient tumor suppressor in intestinal neoplasia. J. Clin. Invest. 127, 4462–4476. doi:10.1172/JCI77217
Silvera, D., Arju, R., Darvishian, F., Levine, P. H., Zolfaghari, L., Goldberg, J., et al. (2009). Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell Biol. 11, 903–908. doi:10.1038/ncb1900
Silvis, M. R., Kreger, B. T., Lien, W. H., Klezovitch, O., Rudakova, G. M., Camargo, F. D., et al. (2011). α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal 4, ra33. doi:10.1126/scisignal.2001823
Singh, B., Bogatcheva, G., Washington, M. K., and Coffey, R. J. (2013). Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proc. Natl. Acad. Sci. U. S. A. 110, 8960–8965. doi:10.1073/pnas.1305508110
Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., and Mcguire, W. L. (1987). Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182. doi:10.1126/science.3798106
Slamon, D. J., Godolphin, W., Jones, L. A., Holt, J. A., Wong, S. G., Keith, D. E., et al. (1989). Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244, 707–712. doi:10.1126/science.2470152
Smalley-Freed, W. G., Efimov, A., Burnett, P. E., Short, S. P., Davis, M. A., Gumucio, D. L., et al. (2010). p120-catenin is essential for maintenance of barrier function and intestinal homeostasis in mice. J. Clin. Invest. 120, 1824–1835. doi:10.1172/JCI41414
Smalley-Freed, W. G., Efimov, A., Short, S. P., Jia, P., Zhao, Z., Washington, M. K., et al. (2011). Adenoma formation following limited ablation of p120-catenin in the mouse intestine. PLoS One 6, e19880. doi:10.1371/journal.pone.0019880
Smith, M. J., Beetz, C., Williams, S. G., Bhaskar, S. S., O'sullivan, J., Anderson, B., et al. (2014). Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J. Clin. Oncol. 32, 4155–4161. doi:10.1200/JCO.2014.58.2569
Song, W., Liu, W., Zhao, H., Li, S., Guan, X., Ying, J., et al. (2015). Rhomboid domain containing 1 promotes colorectal cancer growth through activation of the EGFR signalling pathway. Nat. Commun. 6, 8022. doi:10.1038/ncomms9022
Sontheimer-Phelps, A., Hassell, B. A., and Ingber, D. E. (2019). Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 19, 65–81. doi:10.1038/s41568-018-0104-6
Soto, E., Yanagisawa, M., Marlow, L. A., Copland, J. A., Perez, E. A., and Anastasiadis, P. Z. (2008). p120 catenin induces opposing effects on tumor cell growth depending on E-cadherin expression. J. Cell Biol. 183, 737–749. doi:10.1083/jcb.200805113
Sparks, A. B., Morin, P. J., Vogelstein, B., and Kinzler, K. W. (1998). Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 58, 1130–1134.
Spiegelman, V. S., Slaga, T. J., Pagano, M., Minamoto, T., Ronai, Z., and Fuchs, S. Y. (2000). Wnt/beta-catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol. Cell 5, 877–882. doi:10.1016/s1097-2765(00)80327-5
Sriuranpong, V., Borges, M. W., Ravi, R. K., Arnold, D. R., Nelkin, B. D., Baylin, S. B., et al. (2001). Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 61, 3200–3205.
St Croix, B., Sheehan, C., Rak, J. W., Florenes, V. A., Slingerland, J. M., and Kerbel, R. S. (1998). E-Cadherin-dependent growth suppression is mediated by the cyclin-dependent kinase inhibitor p27(KIP1). J. Cell Biol. 142, 557–571. doi:10.1083/jcb.142.2.557
Stairs, D. B., Bayne, L. J., Rhoades, B., Vega, M. E., Waldron, T. J., Kalabis, J., et al. (2011). Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell 19, 470–483. doi:10.1016/j.ccr.2011.02.007
Stappert, J., and Kemler, R. (1994). A short core region of E-cadherin is essential for catenin binding and is highly phosphorylated. Cell Adhes. Commun. 2, 319–327. doi:10.3109/15419069409014207
Stockinger, A., Eger, A., Wolf, J., Beug, H., and Foisner, R. (2001). E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol. 154, 1185–1196. doi:10.1083/jcb.200104036
Stransky, N., Egloff, A. M., Tward, A. D., Kostic, A. D., Cibulskis, K., Sivachenko, A., et al. (2011). The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160. doi:10.1126/science.1208130
Su, Y., Li, J., Witkiewicz, A. K., Brennan, D., Neill, T., Talarico, J., et al. (2012). N-cadherin haploinsufficiency increases survival in a mouse model of pancreatic cancer. Oncogene 31, 4484–4489. doi:10.1038/onc.2011.574
Sullivan, B., Light, T., Vu, V., Kapustka, A., Hristova, K., and Leckband, D. (2022). Mechanical disruption of E-cadherin complexes with epidermal growth factor receptor actuates growth factor-dependent signaling. Proc. Natl. Acad. Sci. U. S. A. 119, e2100679119. doi:10.1073/pnas.2100679119
Sun, W., Gaykalova, D. A., Ochs, M. F., Mambo, E., Arnaoutakis, D., Liu, Y., et al. (2014). Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 74, 1091–1104. doi:10.1158/0008-5472.CAN-13-1259
Suyama, K., Shapiro, I., Guttman, M., and Hazan, R. B. (2002). A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2, 301–314. doi:10.1016/s1535-6108(02)00150-2
Takahashi, K., and Suzuki, K. (1996). Density-dependent inhibition of growth involves prevention of EGF receptor activation by E-cadherin-mediated cell-cell adhesion. Exp. Cell Res. 226, 214–222. doi:10.1006/excr.1996.0221
Takebe, N., Harris, P. J., Warren, R. Q., and Ivy, S. P. (2011). Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 8, 97–106. doi:10.1038/nrclinonc.2010.196
Takebe, N., Miele, L., Harris, P. J., Jeong, W., Bando, H., Kahn, M., et al. (2015). Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 12, 445–464. doi:10.1038/nrclinonc.2015.61
Tan, I. L., Wojcinski, A., Rallapalli, H., Lao, Z., Sanghrajka, R. M., Stephen, D., et al. (2018). Lateral cerebellum is preferentially sensitive to high sonic hedgehog signaling and medulloblastoma formation. Proc. Natl. Acad. Sci. U. S. A. 115, 3392–3397. doi:10.1073/pnas.1717815115
Tang, M. K. S., Yue, P. Y. K., Ip, P. P., Huang, R. L., Lai, H. C., Cheung, A. N. Y., et al. (2018). Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat. Commun. 9, 2270. doi:10.1038/s41467-018-04695-7
Taylor, M. D., Liu, L., Raffel, C., Hui, C. C., Mainprize, T. G., Zhang, X., et al. (2002). Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306–310. doi:10.1038/ng916
Teh, M. T., Blaydon, D., Chaplin, T., Foot, N. J., Skoulakis, S., Raghavan, M., et al. (2005). Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 65, 8597–8603. doi:10.1158/0008-5472.CAN-05-0842
Theisen, C. S., Wahl, J. K., Johnson, K. R., and Wheelock, M. J. (2007). NHERF links the N-cadherin/catenin complex to the platelet-derived growth factor receptor to modulate the actin cytoskeleton and regulate cell motility. Mol. Biol. Cell 18, 1220–1232. doi:10.1091/mbc.e06-10-0960
Thomas, W. A., Boscher, C., Chu, Y. S., Cuvelier, D., Martinez-Rico, C., Seddiki, R., et al. (2013). α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem. 288, 4957–4969. doi:10.1074/jbc.M112.403774
Thompson, B. J., Buonamici, S., Sulis, M. L., Palomero, T., Vilimas, T., Basso, G., et al. (2007). The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204, 1825–1835. doi:10.1084/jem.20070872
Thompson, C. J., Vu, V. H., Leckband, D. E., and Schwartz, D. K. (2021). Cadherin cis and trans interactions are mutually cooperative. Proc. Natl. Acad. Sci. U. S. A. 118, e2019845118. doi:10.1073/pnas.2019845118
Thoreson, M. A., Anastasiadis, P. Z., Daniel, J. M., Ireton, R. C., Wheelock, M. J., Johnson, K. R., et al. (2000). Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148, 189–202. doi:10.1083/jcb.148.1.189
Totaro, A., Panciera, T., and Piccolo, S. (2018). YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20, 888–899. doi:10.1038/s41556-018-0142-z
Tran, H. D., Luitel, K., Kim, M., Zhang, K., Longmore, G. D., and Tran, D. D. (2014). Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 74, 6330–6340. doi:10.1158/0008-5472.CAN-14-0923
Troyanovsky, S. M. (2022). Adherens junction: The ensemble of specialized cadherin clusters. Trends Cell Biol. 33, 374–387. doi:10.1016/j.tcb.2022.08.007
Tsai, J. H., Donaher, J. L., Murphy, D. A., Chau, S., and Yang, J. (2012). Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736. doi:10.1016/j.ccr.2012.09.022
Tsunematsu, R., Nakayama, K., Oike, Y., Nishiyama, M., Ishida, N., Hatakeyama, S., et al. (2004). Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423. doi:10.1074/jbc.M312337200
Tzima, E., Irani-Tehrani, M., Kiosses, W. B., Dejana, E., Schultz, D. A., Engelhardt, B., et al. (2005). A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature 437, 426–431. doi:10.1038/nature03952
Van De Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi:10.1016/j.cell.2015.03.053
Van Itallie, C. M., Tietgens, A. J., Aponte, A., Fredriksson, K., Fanning, A. S., Gucek, M., et al. (2014). Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J. Cell Sci. 127, 885–895. doi:10.1242/jcs.140475
Vasioukhin, V., Bauer, C., Degenstein, L., Wise, B., and Fuchs, E. (2001). Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell 104, 605–617. doi:10.1016/s0092-8674(01)00246-x
Vu, V., Light, T., Sullivan, B., Greiner, D., Hristova, K., and Leckband, D. (2021). P120 catenin potentiates constitutive E-cadherin dimerization at the plasma membrane and regulates trans binding. Curr. Biol. 31, 3017–3027.e7. doi:10.1016/j.cub.2021.04.061
Wang, K., Yuen, S. T., Xu, J., Lee, S. P., Yan, H. H., Shi, S. T., et al. (2014). Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 46, 573–582. doi:10.1038/ng.2983
Wang, Y. (2009). Wnt/planar cell polarity signaling: A new paradigm for cancer therapy. Mol. Cancer Ther. 8, 2103–2109. doi:10.1158/1535-7163.MCT-09-0282
Watabe, M., Nagafuchi, A., Tsukita, S., and Takeichi, M. (1994). Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 127, 247–256. doi:10.1083/jcb.127.1.247
Weng, A. P., Ferrando, A. A., Lee, W., Morris, J. P. T., Silverman, L. B., Sanchez-Irizarry, C., et al. (2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271. doi:10.1126/science.1102160
Weng, A. P., Millholland, J. M., Yashiro-Ohtani, Y., Arcangeli, M. L., Lau, A., Wai, C., et al. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109. doi:10.1101/gad.1450406
Westcott, J. M., Prechtl, A. M., Maine, E. A., Dang, T. T., Esparza, M. A., Sun, H., et al. (2015). An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Invest. 125, 1927–1943. doi:10.1172/JCI77767
Whittenberger, B., and Glaser, L. (1977). Inhibition of DNA synthesis in cultures of 3T3 cells by isolated surface membranes. Proc. Natl. Acad. Sci. U. S. A. 74, 2251–2255. doi:10.1073/pnas.74.6.2251
Williams, E. J., Furness, J., Walsh, F. S., and Doherty, P. (1994). Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 13, 583–594. doi:10.1016/0896-6273(94)90027-2
Winston, J. T., Strack, P., Beer-Romero, P., Chu, C. Y., Elledge, S. J., and Harper, J. W. (1999). The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13, 270–283. doi:10.1101/gad.13.3.270
Wong, A. J., Ruppert, J. M., Bigner, S. H., Grzeschik, C. H., Humphrey, P. A., Bigner, D. S., et al. (1992). Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Natl. Acad. Sci. U. S. A. 89, 2965–2969. doi:10.1073/pnas.89.7.2965
Wu, X., Tu, X., Joeng, K. S., Hilton, M. J., Williams, D. A., and Long, F. (2008). Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell 133, 340–353. doi:10.1016/j.cell.2008.01.052
Wu, Y., Jin, X., Harrison, O., Shapiro, L., Honig, B. H., and Ben-Shaul, A. (2010). Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc. Natl. Acad. Sci. U. S. A. 107, 17592–17597. doi:10.1073/pnas.1011247107
Wu, Y., Vendome, J., Shapiro, L., Ben-Shaul, A., and Honig, B. (2011). Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 475, 510–513. doi:10.1038/nature10183
Wu, F., Zhang, Y., Sun, B., Mcmahon, A. P., and Wang, Y. (2017). Hedgehog signaling: From basic biology to cancer therapy. Cell Chem. Biol. 24, 252–280. doi:10.1016/j.chembiol.2017.02.010
Wu, J., Minikes, A. M., Gao, M., Bian, H., Li, Y., Stockwell, B. R., et al. (2019). Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 572, 402–406. doi:10.1038/s41586-019-1426-6
Wu, Q., Li, G., Wen, C., Zeng, T., Fan, Y., Liu, C., et al. (2020). Monoubiquitination of p120-catenin is essential for TGFβ-induced epithelial-mesenchymal transition and tumor metastasis. Sci. Adv. 6, eaay9819. doi:10.1126/sciadv.aay9819
Xiao, K., Allison, D. F., Buckley, K. M., Kottke, M. D., Vincent, P. A., Faundez, V., et al. (2003). Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J. Cell Biol. 163, 535–545. doi:10.1083/jcb.200306001
Xie, J., Murone, M., Luoh, S. M., Ryan, A., Gu, Q., Zhang, C., et al. (1998). Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92. doi:10.1038/34201
Yagi, T., and Takeichi, M. (2000). Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes Dev. 14, 1169–1180. doi:10.1101/gad.14.10.1169
Yamaoka, T., Kusumoto, S., Ando, K., Ohba, M., and Ohmori, T. (2018). Receptor tyrosine kinase-targeted cancer therapy. Int. J. Mol. Sci. 19, 3491. doi:10.3390/ijms19113491
Yanagisawa, M., and Anastasiadis, P. Z. (2006). p120 catenin is essential for mesenchymal cadherin-mediated regulation of cell motility and invasiveness. J. Cell Biol. 174, 1087–1096. doi:10.1083/jcb.200605022
Yanagisawa, M., Kaverina, I. N., Wang, A., Fujita, Y., Reynolds, A. B., and Anastasiadis, P. Z. (2004). A novel interaction between kinesin and p120 modulates p120 localization and function. J. Biol. Chem. 279, 9512–9521. doi:10.1074/jbc.M310895200
Yanagisawa, M., Huveldt, D., Kreinest, P., Lohse, C. M., Cheville, J. C., Parker, A. S., et al. (2008). A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J. Biol. Chem. 283, 18344–18354. doi:10.1074/jbc.M801192200
Yang, S. H., Andl, T., Grachtchouk, V., Wang, A., Liu, J., Syu, L. J., et al. (2008). Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/beta3-catenin signaling. Nat. Genet. 40, 1130–1135. doi:10.1038/ng.192
Yang, C., Tibbitt, M. W., Basta, L., and Anseth, K. S. (2014). Mechanical memory and dosing influence stem cell fate. Nat. Mater 13, 645–652. doi:10.1038/nmat3889
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., et al. (2020). Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352. doi:10.1038/s41580-020-0237-9
Yao, M., Qiu, W., Liu, R., Efremov, A. K., Cong, P., Seddiki, R., et al. (2014). Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 5, 4525. doi:10.1038/ncomms5525
Yap, A. S., Niessen, C. M., and Gumbiner, B. M. (1998). The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 141, 779–789. doi:10.1083/jcb.141.3.779
Yap, A. S., Gomez, G. A., and Parton, R. G. (2015). Adherens junctions revisualized: Organizing cadherins as nanoassemblies. Dev. Cell 35, 12–20. doi:10.1016/j.devcel.2015.09.012
Yin, F., Yu, J., Zheng, Y., Chen, Q., Zhang, N., and Pan, D. (2013). Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154, 1342–1355. doi:10.1016/j.cell.2013.08.025
Yu, F. X., Zhao, B., and Guan, K. L. (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163, 811–828. doi:10.1016/j.cell.2015.10.044
Zeng, X., Goetz, J. A., Suber, L. M., Scott, W. J., Schreiner, C. M., and Robbins, D. J. (2001). A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 411, 716–720. doi:10.1038/35079648
Zhan, T., Rindtorff, N., and Boutros, M. (2017). Wnt signaling in cancer. Oncogene 36, 1461–1473. doi:10.1038/onc.2016.304
Zhang, Y., Sivasankar, S., Nelson, W. J., and Chu, S. (2009). Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc. Natl. Acad. Sci. U. S. A. 106, 109–114. doi:10.1073/pnas.0811350106
Zhang, N., Bai, H., David, K. K., Dong, J., Zheng, Y., Cai, J., et al. (2010). The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27–38. doi:10.1016/j.devcel.2010.06.015
Zhang, C., Zhu, H., Ren, X., Gao, B., Cheng, B., Liu, S., et al. (2021). Mechanics-driven nuclear localization of YAP can be reversed by N-cadherin ligation in mesenchymal stem cells. Nat. Commun. 12, 6229. doi:10.1038/s41467-021-26454-x
Zhao, B., Wei, X., Li, W., Udan, R. S., Yang, Q., Kim, J., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761. doi:10.1101/gad.1602907
Zhao, B., Lei, Q. Y., and Guan, K. L. (2008). The hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646. doi:10.1016/j.ceb.2008.10.001
Zheng, X., Carstens, J. L., Kim, J., Scheible, M., Kaye, J., Sugimoto, H., et al. (2015). Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530. doi:10.1038/nature16064
Keywords: cadherins, Yap/Taz, growth factor receptors, WNT, Notch, hedgehog, epithelial-mesenchymal transition, immune responses
Citation: Lin W-H, Cooper LM and Anastasiadis PZ (2023) Cadherins and catenins in cancer: connecting cancer pathways and tumor microenvironment. Front. Cell Dev. Biol. 11:1137013. doi: 10.3389/fcell.2023.1137013
Received: 03 January 2023; Accepted: 03 May 2023;
Published: 15 May 2023.
Edited by:
Antonis Kourtidis, Medical University of South Carolina, United StatesReviewed by:
Maria Elena Fernandez Sanchez, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceBlair Benham-Pyle, Baylor College of Medicine, United States
Cristina Bertocchi, Pontificia Universidad Católica de Chile, Chile
Copyright © 2023 Lin, Cooper and Anastasiadis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Panos Z. Anastasiadis, QW5hc3Rhc2lhZGlzLnBhbm9zQG1heW8uZWR1
 Wan-Hsin Lin
Wan-Hsin Lin Panos Z. Anastasiadis
Panos Z. Anastasiadis