
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 21 March 2023
Sec. Molecular and Cellular Reproduction
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1121231
This article is part of the Research Topic Challenges in Fertilization and Implantation Success View all 9 articles
Worldwide increase in life expectancy has boosted research on aging. Overcoming the concept of chronological age, higher attention has been addressed to biological age, which reflects a person’s real health state, and which may be the resulting combination of both intrinsic and environmental factors. As epigenetics may exert a pivotal role in the biological aging, epigenetic clocks were developed. They are based on mathematical models aimed at identifying DNA methylation patterns that can define the biological age and that can be adopted for different clinical scopes (i.e., estimation of the risks of developing age-related disorders or predicting lifespan). Recently, epigenetic clocks have gained a peculiar attention in the fertility research field, in particular in the female counterpart. The insight into the possible relations between epigenetic aging and women’s infertility might glean additional information about certain conditions that are still not completely understood. Moreover, they could disclose significant implications for health promotion programs in infertile women. Of relevance here is that the impact of biological age and epigenetics may not be limited to fertility status but could translate into pregnancy issues. Indeed, epigenetic alterations of the mother may transfer into the offspring, and pregnancy itself as well as related complications could contribute to epigenetic modifications in both the mother and newborn. However, even if the growing interest has culminated in the conspicuous production of studies on these topics, a global overview and the availability of validated instruments for diagnosis is still missing. The present narrative review aims to explore the possible bonds between epigenetic aging and fertility timeline. In the “infertility” section, we will discuss the advances on epigenetic clocks focusing on the different tissues examined (endometrium, peripheral blood, ovaries). In the “pregnancy” section, we will discuss the results obtained from placenta, umbilical cord and peripheral blood. The possible role of epigenetic aging on infertility mechanisms and pregnancy outcomes represents a question that may configure epigenetic clock as a bond between two apparently opposite worlds: infertility and pregnancy.
The female reproductive timeline represents a shadowy and attractive topic of research. Albeit with individual variations and peculiarities, four milestones can be identified in the female reproductive lifespan: 1) menarche, corresponding to the beginning of fertile phase; 2) gestation, the fulfilment of the potential fertility; 3) the end of the fertile period occurring at about 10 years prior to menopause, and 4) menopause itself, marking the end of the ovarian hormonal activity and the onset of the senior age (te Velde and Pearson, 2002). The improved life expectancy and the social trend of postponing pregnancy have determined an increase in the average age of pregnant women, but also an increase in the rate of age-related infertility, boosting the interest on this topic (Mills et al., 2011; ESHRE Capri workshop group, 2017).
In the aging research field, the concept of biological age has recently gained major attention since it may better reflect the real physiological status and function of an individual compared to chronological age (Lowsky et al., 2014). Epigenetics, defined as genome modifications that do not involve changes in DNA sequence but rather modifications of chromatin, is a crucial determinant of biological age (Bell et al., 2019; López-Otín et al., 2013). The growing awareness of the role of epigenetics in aging has encouraged the introduction of several estimators of biological age. Among them, epigenetic clocks have been heralded as the most accurate. It is possible that these clocks may glean additional information regarding fertility timeline, in which aging exerts a pivotal part. The possible impact of epigenetic aging on infertility mechanisms, but also the epigenetic modifications during gestation and the impact of pregnancy on a woman’s biological age are intriguing queries. Within an evolutionary framework, aging and reproduction are indeed intrinsically linked (Levine et al., 2018).
The present narrative review considers the interaction between epigenetic clocks and fertility timeline. We mainly aimed to provide an overview of the knowledge gained on the role of epigenetic clocks in two apparent opposite faces of fertility, that is infertility and pregnancy. Table 1 summarizes the main features of the studies discussed in the present review.
Epigenome has ushered a new era of molecular research in the field of aging (Erema et al., 2022) as DNA methylation (DNAm) has emerged as one of the most powerful strategies to predict chronological age (Horvath and Raj, 2018; Oblak et al., 2021; Simpson and Chandra, 2021). The precision of this instrument is astonishing, to the point to be used for forensic purposes to estimate chronological age in burned and defaced corpses or in biological specimens (Freire-Aradas et al., 2022). The availability of a precise predictive tool of chronological age can also allow to identify subjects whose chronological and biological age diverge and investigate the health consequences of this variation.
DNA methylation refers to the addition of a methyl group to the 5’ end of DNA under the action of DNA methyltransferase (DNMT), thereby affecting the regulation of gene expression. Usually, this modification occurs on cytosine (C)—phosphate (p)—guanine G) (CpG). A section of DNA rich in CpG in the genome is called a CpG island, with a length of 300–3,000 bp. CpG islands are usually located in the promoter region of the gene, so the CpG island of the gene promoter is a common location where methylation occurs. Approximately 70%–80% of the CpG in the human genome is methylated but its status can vary according to the aging process: CpGs undergo hypermethylation in the promoter regions, while they become hypomethylated in others (Saul and Kosinsky, 2021; Duan et al., 2022). DNAm can be measured by different methods: microarrays (Horvath et al., 2014), pyrosequencing (Zbieć-Piekarska et al., 2015), quantitative PCR and next-generation sequencing methods (Lu et al., 2018). Advances in microarray technology, biostatistics development and the emergence of a new open-access scientific mentality finally allowed the development of epigenetic clocks, the new metrics of biological age based on DNAm patterns (Horvath and Raj, 2018). Epigenetic clocks derive from machine learning models that automatically select the most informative CpGs by regressing (i.e., penalized, cox, elastic net regression model) a transformed version of chronological age on a set of CpGs (Horvath and Raj, 2018; Rutledge et al., 2022). Even if the large number of CpG included may be a guarantee of higher accurateness, a moderate number of CpGs could be robust enough, as epigenetic clocks depict comprehensive properties of the methylome (Duan et al., 2022). This notion has led to the development of simplified and less expensive epigenetic clocks.
Epigenetic clocks may be based on one tissue (single-tissue clock) so that their accurateness is permitted only if applied on the tissue type on which they were trained (Hannum et al., 2013) or they could apply to all tissues and cell types across the entire duration of the human lifespan (multi-tissue clock) (Horvath et al., 2014; Levine et al., 2018; Lu et al., 2019). Huge differences in biological age exist among different tissues. The first multi-tissue predictor of age (Horvath Clock) was devised using n = 8,000 samples from 82 Illumina DNA methylation array datasets and encompassing n = 51 healthy tissues and cell types (Horvath et al., 2014). Since 2012, multiple models have spread throughout the world (Oblak et al., 2021; Shahal et al., 2022). While the first-generation clock goal was to be accurate in chronological age prediction (Horvath et al., 2014), the second ones aimed to investigate a possible causative role of DNAm in the ageing process and onset of age-related disorders (i.e., GrimAge, PhenoAge) (Levine et al., 2018; Lu et al., 2019). While GrimAge has been adopted for lifespan prediction, the PhenoAge clock could estimate cancer, or Alzheimer’s disease risk. A multitude of other clocks have spread for different clinical purposes (Levine et al., 2018; Lu et al., 2019; Johnstone et al., 2022). The possible difference between a person’s chronological age and the biological one, provided by epigenetic clocks, is defined as “age acceleration or deceleration”. If predicted DNAm age is greater than chronological age, age acceleration will be positive, if the predicted biological age is less than chronological one, it will be negative (decelerated age). While genetic and inner process could cause “intrinsic” age acceleration (Horvath and Raj, 2018), lifestyle and environmental factors may be detrimental for “extrinsic” one (Franzago et al., 2022). Not only epigenetic clocks may provide a clue on the impact of modifiable factors on the natural process of aging, but also they may give a glimpse of possible effective strategies to make the clock tick backwards (Levine et al., 2018). In fact, the growing awareness of the modulation role of nutrients and other food components on DNAm patterns has paved the way for new studies on the relationship between lifestyle interventions and epigenetic aging (Mathers et al., 2010; Horvath et al., 2014; Zannas et al., 2015; Quach et al., 2017; Franzago et al., 2022). Intriguingly, folates, a standard supplementation both in the pre-conception period and during early pregnancy, have been recently investigated in the aging field (Sae-Lee et al., 2018; Monasso et al., 2021; Nwanaji-Enwerem et al., 2021a). Chanachai Sae-Lest et al. observed that folic acid plus vitamin B12 intake can provide a different effect depending on MTHFR genotype, proving a decreased epigenetic age in women with the more common 677CC genotype (Sae-Lee et al., 2018). Owing to the relevance of the topic and its practical implications, these preliminary data corroborate the need for solid evidence. A description of the main features of most common clocks is provided in Figure 1.
The worldwide trend toward delayed parenthood to the late thirties and even beyond has boosted the focus of reproductive medicine on the possible impact of aging on conceiving success (De Geyter et al., 2020). Women’s natural fertility sharply declines after 35 years and terminates at a median age of 41–42 years (Eijkemans et al., 2014). The subsequent 10 years until the advent of menopause are still characterized by ovulatory cycles, but the quality of the recruited oocytes is incompatible with live births (te Velde and Pearson, 2002). The lost of fertility is not due to a reduction of the remnant ovarian reserve but, conversely, to the deterioration of oocytes quality. Concepts like diminished ovarian reserve and ovarian aging have become “daily bread” for experts dealing with infertility issues. However, clarifying the real meaning of ovarian aging represents a worthwhile endeavor. In fact, despite the increasing number of studies on this topic, the exact biological changes occurring within ovaries overtime are not completely known. The current social trend for couples to delay childbearing has given more urgency to the need of investigating age rhythm and ‘ageing oocyte’ (Ge et al., 2015). Increasing literature has suggested that female reproductive timeline could be somehow punctuated by epigenetic mechanisms and clocks rhythm (Das and Destouni, 2022).
One of the most stimulating theories supports a new concept of age-related infertility as the first sign of an “accelerated aging disease” (Llarena and Hine, 2021; Loose et al., 2022). In other words, ovaries could be the first compartment to show signs of a general process of accelerated aging. The result would be subfertility or even infertility. Thus, infertility could be remarked as part of a more general systemic illness that is defined by accelerated or premature aging process. Proving that infertility could be associated with age acceleration of a person’s biological clock may revolutionize reproductive medicine. Fertility status would no longer concern only couples desirous of offspring, but it could become a matter of global health, also involving the general practitioner who faces a young woman not yet seeking pregnancy. Prompt identification of women at higher risk of earlier exhaustion of fertility could provide them with a double opportunity: on one hand if it is a contemplated wish, they may have time enough for scheduling their family building plan; on the other one, they could prevent the raise of age-related disorders such as cardiovascular or bone health problems. A prompt family building plan could consist of two different chances: anticipating their offspring search time or overcoming the ageing process by freezing eggs or ovarian tissue.
Not at last, biological aging biomarkers could glean additional information to traditional procreative testing (Morin et al., 2018; Monseur et al., 2020; Li Piani et al., 2022). Moreover, as at current state no consensus exists on whether the available traditional ovarian markers could be adopted for a large-scale screening of reproductive senescence, they could be evaluated for this purpose too. Noteworthy, the advantage of the availability of a measurement of ovarian biological age is not limited to a diagnostic purpose. It may also become a precious tool to predict the success of Assisted Reproductive Techniques (ART). Women with predicted compromised oocytes could indeed avoid embarking in the expensive, burdensome, and risky ART journey. They could be straightly advised for egg donation. Apart from these appealing potential clinical applications, studies on the topic are yet initial and insufficient. Available evidence is from here on reviewed, taking into consideration separately the endometrial, peripheral, and ovarian districts.
Endometrium is a unique dynamic tissue that cyclically sheds, rejuvenates, and endures complex modifications aimed to promote embryo implantation. However, uterine ageing is often left out of the picture of reproductive female timeline, so that few and contrasting data are available on age-related functional decline in the uterine tissues (Deryabin and Borodkina, 2022). Epigenetic mechanisms are however emerging as key players in regulating cyclic modulation of endometrium (Retis-Resendiz AM et al., 2021). The increase of the global acetylation of some histones (i.e., H3K9ac, H2AK5ac, H3K14ac, and H4K8ac) during the early proliferative phase may be supportive of the regeneration of the endometrial functional layer (Munro et al., 2010), while genes associated with transcription regulation (i.e., ID2, NFAM1, RUNX3, ZNF57) are more methylated in the secretory phase (Saare et al., 2016). In addition, gynecological diseases such as endometriosis may be linked to DNA methylation alterations (Leap et al., 2022). There is evidence of a perturbed biological age of the endometrium in affected women (Guo, 2009; Houshdaran et al., 2016; Saare et al., 2016) and altered speed of epigenetic clocks may predict the progression of some endometrial pathologies, including recurrent implantation failure and endometriosis (Deryabin and Borodkina, 2022).
In 2013, Horvath reported that endometrial epigenetic age badly correlated with chronological one (cor = 0.55), (Horvath et al., 2014). In fact, when the multi-tissue Horvath clock was applied to healthy endometrium samples in the test data, the authors found an high error rate in the chronological age prediction, the endometrium being much younger based on epigenetic data (Horvath et al., 2014). However, Olesen et al. who specifically focused on the endometrium did not confirm this evidence (Olesen et al., 2018). Considering endometrial biopsies sampled at the same point in two consecutive cycles, these authors also described a good correlation between endometrial epigenetic and chronological age, as well as a modest inter-cycle variability. Endometrium was characterized by higher epigenetic acceleration (4.4 years higher than chronological age, p < 0.012) and no difference was detected across the different cycles. The authors also postulated that endometrial DNA methylation profile might change according to the phase of menstrual cycle (Olesen et al., 2018). In fact, the bad correlation found in Horvath’s work could derive from the insufficient attention given to the phase of the cycle for the assessment. However, the small sample size of the study of Olesen et al. (n = 9) hampers robust conclusions (Olesen et al., 2018). The cyclic variation of endometrium, its hormone-related nature, and the easy availability of samples all encourage deeper investigations to expand current understanding of the relation between DNA methylation and endometrial aging and function. The uterine factor still represents a neglected topic that should be part in the comprehension of age-related female fertility decline (Deryabin and Borodkina, 2022).
Most common models have been validated on peripheral blood cells, and in particular in leukocytes. This font of DNAm was used to test the possibility that biological age, more specifically age acceleration, could better explain fertility decline variability than chronological age alone.
In a pilot single-center cohort study, Monseur et al. evaluated the epigenetic age in the peripheral blood cells of 39 women undergoing ART. They used a modified Hovarth clock (they added some intergenic sites) to investigate whether age acceleration could be associated to fertility biomarkers (Monseur et al., 2020). They observed that not only chronological but also biological age negatively correlated with remnant ovarian reserve and oocytes yield. Women with age acceleration showed lower levels of anti-mullerian hormone (AMH) (1.29 vs. 2.29 ng/mL, p = 0.05), lower antral follicular count (AFC) (8 vs. 14.5, p = 0.05), lower number of oocytes retrieved (5.5 vs. 14.5, p = 0.002) and lower number of two pronuclear zygotes (3 vs. 7, p = 0.007), compared to the women without age acceleration. However, the design of this pilot study did not allow the authors to compare the predictive capacity of age acceleration with that of common biomarkers of ART success. Moreover, they could not disentangle whether age acceleration may provide independent information on the quality of the oocytes (Monseur et al., 2020). Apparently in contrast, Christensen et al. did not yield any indication of accelerated aging in n = 55 young women with idiopathic early ovarian aging (defined as: minimum two cycles with ≤5 oocytes obtained by ovarian stimulation with at least 250 IU recombinant FSH), when adjusting for chronological age (Christensen et al., 2021). However, the small sample size and the lack of adjustments for other variables (i.e., BMI, smoking) limit the value of the study.
Another possible hypothesis postulates that the information provided by epigenetic clocks may go beyond ovarian reserve. In our latest project, we adopted Zbieć-Piekarska epigenetic model, that is set on five genes and based on pyrosequencing technique (Li Piani et al., 2022). In a group of n = 181 infertile patients aged 37–39 years, we could confirm that women who subsequently conceived and delivered after ART were epigenetically younger than those who did not (respectively: 36.1 ± 4.2 and 37.3 ± 3.3 years, p < 0.04). In contrast with Monseur et al. and Lee et al. (Monseur et al., 2020; Lee et al., 2022), we verified that adjusting for the variables that differed between the two groups (AFC, FSH, number of oocytes retrieved) the difference remained statistically significant (p = 0.028). Although these findings should be considered with caution for some aspects (restricted patients’ age, choice of the epigenetic model), they suggest that the contribution of epigenetic clocks may not be limited to reflect ovarian reserve. They may capture other aspects of reproductive health (Li Piani et al., 2022).
In 2022, the role of age acceleration was investigated in a large population of pregnant women and using five different epigenetic aging clocks (DunedinPoAm, PhenoAge, DNAmTL, Hannum clock, Horvath Clock) (Lee et al., 2022). The authors considered a much greater sample size, comparing pregnant women conceiving naturally (n = 1,000) with those who conceived after ART treatment (n = 995 patients) (Lee et al., 2022). They noted that ART mothers aged 0.021 years faster than non-ART ones (p < 0.03), when the DunedinPoAm clock was adopted. They also found that the etiology of infertility might interplay with epigenetic clock behavior: women with tubal, anovulatory, or idiopathic infertility factor were epigenetically older than the non-ART counterparts (Lee et al., 2022). According to the authors, DunedinPoAm could surpass the other biomarkers in the field of reproductive medicine for its nature of “pace of aging” estimator. It is the only one that reflects aging pace from the time of sample to 10–15 years back and it is more sensitive to weigh environmental exposure burden. This large study stands out from the others as it has included fertile women as control group, it has recruited pregnant women, and it has pioneered novel epigenetic model. It has however some limitations. The authors did not report any information regarding the ovarian reserve, and, by study design, they excluded women with severe infertility that could not be overcome by ART. Finally, data could not be generalized to unpregnant women (Lee et al., 2022).
Ovaries could be renamed as “women’s heart”, from a reproductive point of view. They are a time-limited forge of hormones and substances that are crucial for the general wellbeing. Follicular granulosa cells are responsible for the declining of ovarian steroid production associated with reproductive age and they are engaged in an intense crosstalk with the surrounding oocytes. Granulosa cells are crucial for supporting oocytes during follicular growth, maturation, and estrogen production. While epigenetic clocks have been widely validated on easily accessible tissues, their application to several other body regions including the ovaries has been quite limited. Intriguingly, Horvath clock performance seemed lower when tested on hormone-dependent tissues, such as endometrial and breast tissues (Horvath et al., 2014). More specifically, they resulted epigenetically older. Thus, it is not surprising that ovarian microenviroment has been attractive for studying epigenetic clock performance.
The first study that investigated ovarian epigenetic age in infertile women was published in 2018 (Morin et al., 2018). The authors applied the Horvath model to leukocytes and cumulus cells of women who underwent ART. They divided the enrolled subjects in four groups according to chronological age and ovarian response (A: young good responder; B: young poor responder; C: old good responder; D: old poor responder). They showed that the compartment of cumulus granulosa cells followed a different methylation trajectory with age compared to leukocytes: these cells resulted extremely younger, without difference among the groups for either chronological age or ovarian response. Not at last, they noted that this result was in line with a longer length of telomeres, which represents another valid aging biomarker. Overall, the results suggested a potentially distinct epigenetic timeline in the ovarian compartment. Inferences should however be drawn with caution since the sample size was not large (n = 77) and the authors could not include a control group of fertile women (Morin et al., 2018).
Two years later, the same group sought to verify their results in a larger sample size, recruiting n = 175 women who were undergoing ovarian stimulation and divided into two groups (good responders: >5 oocytes retrieved, poor responders: <5 oocytes retrieved) (Hanson et al., 2020). To note, the definition of ovarian reserve was different compared to their previous work (Morin et al., 2018). The authors confirmed their previous observation: cumulus granulosa cells remained epigenetically younger than the chronological age (average 8.6 ± 2.1 vs. 35.3 ± 4.1 years) when Horvath model was applied. Going further, they attempted to generate an epigenetic model based on cumulus granulosa cell DNA methylation signature, but it turned out to be unsuccessful (Hanson et al., 2020).
All these data also convey the idea that epigenetic aging patterns may be distinct in hormonally responsive tissues. As aforementioned before, Horvath model had a bad performance in case of endometrial tissues, that resulted epigenetically older (Horvath et al., 2014) or it could vary according to the intermenstrual period (Olesen et al., 2018). Cumulus granulosa cells resulted conversely epigenetically extremely younger, and the evidence of a higher telomere length, that is another common aging biomarker, comforted this finding (Morin et al., 2018; Hanson et al., 2020).
Granulosa cells may reflect a different ageing process than other tissues: they are silent for decades, and then rapidly proliferate from 10–15 quiescent granulosa cells in primordial follicles to 60 million in the ovulatory phase. Given these premises, Olsen et al. postulated that ovarian compartment has a distinct epigenetic heritage than other somatic tissues (Olsen et al., 2018). The authors investigated the process of aging in mural granulosa cells, which are part of the follicular compartment (cumulus granulosa cells being those cells in close contact with the oocytes and mural granulosa cells being the main component of the follicular wall). Data from n = 118 women providing peripheral leukocytes and n = 59 women providing mural granulosa cells were tested using genome-wide methylation analyses (Olsen et al., 2018). Along with Horvath clock and Skin and Blood clock (Horvath et al., 2014; Horvath and Raj, 2018), a new epigenetic model (Granulosa Cell clock), developed by the same authors, was tested on mural granulosa cells (Olsen et al., 2018). As an upgrading of the popular Skin & Blood clock, the Granulosa Cell clock consisted of 296 CpG sites and yielded an improved correlation with chronological age of 0.85 (p = 4.3 × 10−34) when applied to leukocytes (Olsen et al., 2018). Mural granulosa cells were confirmed to be quite younger (an average of 2.7 years based on Horvath clock) (Olsen et al., 2018). The findings are mainly in line with those emerged from the study by Morin et al. (Morin et al., 2018). These latter authors also reported on cumulus granulosa cells and showed an average of 6.8 years based on Horvath clock and an average of 2.7 years based on Skin and Blood clock (Morin et al., 2018). In both studies, cumulus cells and mural granulosa cells epigenetic age did not reflect chronological age (Morin et al., 2018; Olsen et al., 2018). Besides, in mural granulosa cells, age acceleration difference was negative and the gap with chronological age was larger along with older age. To note, the rate of epimutations in terms of accumulation of DNA methylation errors was higher in advanced maternal age, but its entity was much more considerable in mural granulosa cells than in the corresponding peripheral leukocytes (p < 0.003) (Olsen et al., 2018).
Olsen et al. applied the Granulosa Cell and Skin & Blood clocks to verify whether epigenetic age could also depict the magnitude of a woman’s ovarian reserve (Olsen et al., 2018). A higher number of epimutations in terms of greater DNA methylation variability characterized mural granulosa cells of women with diminished ovarian reserve. More specifically, variability in epimutations involved AMH and IGF2 genes, both coding for proteins that are involved in folliculogenesis. These epimutations were also higher in advanced maternal age, suggesting that women with diminished ovarian reserve are similar to older women (Olsen et al., 2018).
Symmetrically, Knight et al. proved a significant association between epigenetic age derived from granulosa cells and traditional ovarian reserve markers (Knight et al., 2022). The Grim Age clock was chosen as independent from chronological age, so that it could provide a unique perspective to evaluate oocyte competence. In a group of n = 70 women undergoing ART treatment, the GrimAge acceleration was negatively associated with AMH levels (p = 0.003), AFC (p = 0.0001), number of mature oocytes retrieved (p = 0.0003) and number of fertilized oocytes (p = 0.008) (Knight et al., 2022).
These findings, however, deserve confirmation. Indirect findings from studies not designed to address this issue are less trenchant (Li Piani et al., 2022). In addition, one must weigh the clinical relevance.
Overall, increasing literature has focused its attention on the application of epigenetic clocks to the different tissues of female reproductive organs. However, solid knowledge is not available yet and deeper investigations should be encouraged.
It is not surprising that the world of pregnancy has been attractive for potential applications of epigenetic hallmarks too (Dieckmann et al., 2021). As a progress of the earlier fetal origin of adult diseases (FOAD) hypothesis (Barker, 2007), the Developmental Origins of Health and Diseases (DOHaD) theory establishes that the interaction at an early stage (from fertilization to early childhood) between genome and external factors (i.e., nutrition, stress, endocrine-disrupting chemicals) may be detrimental for non-communicable disease risk later in life (Arima and Fukuoka, 2020). These theories are based on the ‘developmental plasticity’ assumption, whereby organisms exhibit ‘plasticity’ to master an optimal fit between phenotype and environment, throughout critical developmental periods (Bateson et al., 2004). As this is a time-limited phenomenon, intrauterine “programming” has been the focus of several studies, where epigenetics has emerged as one of the most exciting themes (Bird, 2002; Hochberg et al., 2011). Prenatal and early postnatal periods represent windows of “epigenetic susceptibility” (Coussons-Read, 2013; Reimann et al., 2022).
All these premises underscore the utility of epigenetic clocks to investigate “epigenetic programming” during the early developmental period, as epigenetic clocks are reliable epigenetic footprint. In case of advanced maternal age, epigenetic indices could capture unique signatures that go beyond the known higher rate of adverse newborns outcomes risk. Not only, in case of ART, the possible interference with the extensive epigenetic reprogramming in the early embryo could be disclosed by a change in the epigenetic gestational age acceleration (GAA) (Carlsen et al., 2022). Finally, the search of a solid marker for gestational age (GA) definition has become a research priority, as GA is crucial to forecast fetal maturity. Compared to traditional variables (i.e., last menstrual period (LMP), crown-rump length (CRL), or embryo transfer date (EDT) if conceptions are obtained with ART), epigenetic gestational aging could reveal a higher performance.
Despite the interest that this area deserves, studies are underrepresented and what is lacking is a state of the evidence to date. In the following sections, we will discuss the results gathered according to the tissue included (placenta, cord-blood, peripheral blood) and finally the possible effects of pregnancy on mother’s epigenetic rhythm.
Figure 2 shows a summary of findings regarding epigenetic patterns in case of the two mirrors of fertility time: infertility and pregnancy.
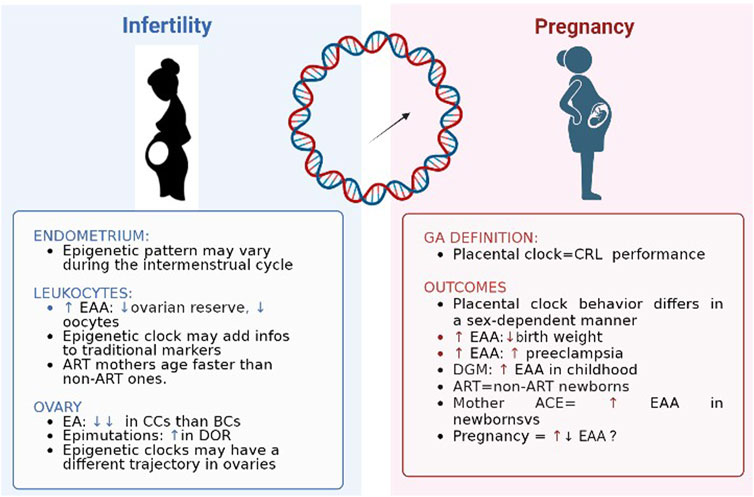
FIGURE 2. Epigenetic clocks and two mirrors of female fertility time. EAA: epigenetic age acceleration; ART: assisted reproductive technique; EA: epigenetic age; CC: cumulus cells; BC: blood cell; DOR: diminished ovarian reserve; CRL: crown-rump length; DGM: gestational diabetes.
Placenta is a highly specialized organ that orchestrates the whole course of pregnancy thanks to its multiple functions (i.e., gas and nutrients exchange, immunity transfer, hormone secretion), allowing the normal growth and development of the fetus. Conversely, placental pathology or dysfunction may be the origin of some high-risk pregnancies or fetal adverse events (Gude et al., 2004). It is reasonable that placental DNA methylation profile could provide information about how epigenetic variations ‘work under the skin’ during pregnancy to influence obstetric outcomes (Clark et al., 2022).
Mayne et al. investigated the precise changes in DNA methylation that occur in the placenta across gestation and determined whether DNA methylation data could be used to predict the GA of a placenta (Mayne et al., 2017). Including placental tissue samples from 12 different datasets of only healthy singleton pregnancies (n = 387), the authors developed their placental GA calculator (DNAm GA) based on 62 CpG sites. The performance of DNAm GA sorted out excellent, with high correlation with the chronological age (p < 0.001) (Mayne et al., 2017). Starting from the results provided by Mayne et al., Lee et al. used a 6 times larger placental training set (n = 1,102) to create a new more robust placental clock, that could be well suited for GA estimation (Lee et al., 2019). Intriguingly, they also proved that placenta is quite distinct from other tissues regarding DNAm estimators’ application, observing no correlation between their placental GA model and other common epigenetic clocks (Lee et al., 2019). The reliability of these two models was also tested in a following sample of n = 427 placentas, founding an overall satisfactory accurateness and confirming the superiority of the model from Lee et al. (Lee et al., 2019) on the one by Mayne and coworkers (Mayne et al., 2017) (Dieckmann et al., 2021).
Whether on one hand placental epigenetic clocks may be intriguing as a possible marker for GA estimation, on the other they could glean additional information in case of pregnancy-related complications. Indeed, it is feasible to speculate that a state of either maternal or fetal biological disorders may lead to disrupted placental epigenetic ageing, since it is the primary interface and balancing system between the maternal-fetal compartment in pregnancy.
While Lee et al. proved that the DNAm GA performance was not impaired by common adverse conditions, the clock by Mayne et al. under/overestimated GA according to pregnancy conditions (Mayne et al., 2017; Lee et al., 2019). When comparing n = 70 placentas from preeclamptic pregnancies with n = 62 placentas from uncomplicated ones, Mayne et al. showed different methylation pattern in 741 CpG sites. They also underlined that only in case of early-onset preeclampsia, placentas were characterized by accelerated placental aging (p < 0.001) (Mayne et al., 2017). Systematic deviations of this clock might reveal interesting biological effects (Mayne et al., 2017). In fact, their result regarding preeclampsia is in line with the most supported theory according to which early preeclampsia may reflect a pathological distressed placenta, while late preeclampsia could be seen as the effect of a maternal status (Hales and Barker, 2001; Valensise et al., 2008; Staff, 2019). This result could be strengthened by an apparent opposite finding provided by Workalemahu et al. (Workalemahu et al., 2021) In n = 312 placentas, the authors found an association between placental epigenetic age deceleration with a lower cardiometabolic profile of the mother (pre-pregnancy BMI, gain of weight, maternal blood pressure). In multivariable-adjusted models, each 1 kg/week increase in their weight was significantly associated with 1.46 (95%CI: −2.61, −0.31), 1.70 (−3.00, −0.40), and 1.71 (−3.11, −0.32) week lower placental epigenetic age acceleration, for each trimester respectively. It could be indeed postulated that the placental immaturity, usually connected to maternal risk factors, may be reflected by an epigenetic deceleration. According to this vision, only early-onset preeclampsia would be characterized by placental age acceleration, and not the late-onset one, that is usually the consequence of maternal adverse profile and would be characterized by the opposite effect, such as age deceleration (Workalemahu et al., 2021).
Moving forward, also sex-specific fetal growth differences and consequent early origins of neonatal and long-term health disparities have been explored also from an epigenetic point of view. Placental acceleration aging behavior differed in a sex-dependent manner (Dieckmann et al., 2021; Workalemahu et al., 2021), proving to be detrimental for fetal weight in males rather than in females (Mayne et al., 2017). Two different explanations could be plausible. First, it could be due to the higher in-utero energy demand in male fetuses. Alternatively, female fetuses may display a more efficient risk averse response against the ensuing accelerated aging (Mayne et al., 2017). It is also easily comprehensible that this relation is more manifest at later gestational periods that are typically distinguished by greater fetal oxygen and nutrients demand as well as higher oxidative stress (Tekola-Ayele et al., 2019). Considering n = 408 placentas from the Extremely Low Gestational Age Newborns cohort, Clark et al. revealed differences according to maternal race. In fact, among infants born to black mothers, smoking during pregnancy was associated with higher placental GA acceleration than in other ethnic groups (+0.89 weeks, 95% CI: 0.38, 1.40) and with the duration of admission in neonatal intensive care units (NICU) (−0.08 days/weeks-GA, 95% CI: 0.12, −0.05) (Clark et al., 2022).
All these data advocate the appropriateness of this novel approach to “old issues” like pregnancy disorders (Blair et al., 2013; Leavey et al., 2017). Nevertheless, some limits should be considered in on-going research. Attention should be addressed to the trimester considered. The DNAm GA proposed by Mayne et al. was based on I-II trimester healthy pregnancy, that could conceal later adverse events in III trimester (Mayne et al., 2017) while on the opposite way, the clock by Lee et al. may encompass some biases due to the training set based on III trimester (Lee et al., 2022). Not at last, as studies in twin pregnancies GA acceleration showed that heritability reached only 57%, environmental factors should be taken into account in the data analysis (Tekola-Ayele et al., 2019).
Cord blood could be a suitable model for DNA methylation study, as it contains stem cells, and changes in DNA methylation profile of stem cells are known to likely correlate with GA (Bocker et al., 2011).
In 2016, Knight et al. adopted umbilical cord blood samples derived from n = 1,135 newborns to build an accurate neonatal predictor of GA (Knight et al., 2016). In the same year, from the Norwegian Mother and Child Birth Cohort (MoBa) study, Bohlin et al. confirmed the correlation between their prediction model derived by DNA extracted from n = 1,753 cord blood samples and GA based on CRL or LMP (Bohlin et al., 2016). In both studies, DNAm GA correlated more strongly with clinical GA estimates based on CRL than those based exclusively on LMP (Bohlin et al., 2016; Knight et al., 2016). However, this is expected since ultrasound is generally regarded as the most reliable method for GA estimation amongst practitioners. Not only, cord-blood epigenetic clocks proved to be more precise than those provided on placenta samples (Dieckmann et al., 2021). In fact, they observed higher correlation in DNAm GA provided by the cord-blood clocks (r = 0.77, p < 0.001) than by placental ones (r = 0.44, p < 0.001) (Dieckmann et al., 2021) and a superiority of the clock by Bohlin et al. on that by Knight’s group (Dieckmann et al., 2021; Daredia et al., 2022).
In a prospective setting, Michaeli et al. designed a new robust epigenetic clock, based on n = 41 cord-blood samples, that showed high concordance with GA determined by common parameters (r = 0.77, p < 0.001) (Falick Michaeli et al., 2019). The groundbreaking feature of the work was the choice of reduced representation bisulfite sequencing (RRBS) protocol for DNAm: it provides comprehensive genomic CpG coverage, it requires small amount of DNA and it guarantees a more cost-effectiveness than the 450 K array (Falick Michaeli et al., 2019).
Haftorn et al. also developed a new epigenetic clock (EPIC GA) that outperformed both the previous clocks in terms of GA prediction accuracy (Haftorn et al., 2021). In fact, EPIC clock showed high precision in terms of prediction of GA (R2 = 0.724) with a median difference (MAD) of 3.24 days. Performance of clocks from Bohlin and Knight’s groups was lower (respectively: R2 = 0.610 MAD = 6.69 days; and R2 = 0.406 MAD = 4.55) (Bohlin et al., 2016; Knight et al., 2016).
Besides, Haftorn et al. focused on the possible role of conception mode, comparing n = 200 newborns from natural conception with n = 838 ART ones. Although ART has been postulated to impair epigenetic signature, Haftorn et al. found no sharp different accuracy between epigenetic clocks set on CRL or ETD (Haftorn et al., 2021). Indeed, not only ART newborns were comparable to non-ART ones in terms of age acceleration, but a closer inspection in the specific ART procedure (ICSI vs. conventional IVF, fresh transfer vs. frozen transfer) did not show any significant variation (Haftorn et al., 2021).
In n = 814 mother-child pairs, adopting clock by Knight et al. to cord-blood samples, Girchenko et al. found a correlation between newborns’ GA with some adverse maternal or obstetrics variables: maternal age of above 40 years at delivery, preeclampsia or fetal demise in a previous pregnancy, more than two pregnancy risk factors for preeclampsia or intra uterine growth restriction (IUGR). They showed that adverse maternal characteristics influence the newborn developmental stage and fetal organ and tissue maturation, determining variations in the offspring’s DNAm GA at birth (Girchenko et al., 2017). Conversely, the association of maternal age with newborn’s GA was not confirmed by Daredia et al. (Daredia et al., 2022). Indeed, the authors postulated that also negative acceleration (deceleration) of epigenetic gestational age could reflect either prenatal adverse conditions or developmental immaturity (Daredia et al., 2022), as aforementioned before (Workalemahu et al., 2021).
All these findings are in line with the aforementioned DOHaD theory, according to which specific prenatal epigenetic perturbations of the “developmental programming” stage may have consequences during early life and cause accelerated aging (Simpkin et al., 2015; Reimann et al., 2022). Thus, cord blood DNAm GA could be a useful proxy for assessing developmental maturity (Javed et al., 2016). GA acceleration significantly predicted birthweight (p = 0.033) in the study of Knight et al. (Knight et al., 2016), consistent with the idea that DNAm GA may reflect general fetal maturity.
Overall, placenta and cord blood seem quite distinct from other tissues regarding the development and application of DNAm-based age estimators. In clinical practice, one could envisage a plethora of applications: its usefulness for GA prediction could be especially foreseen in women who seek prenatal care late in their pregnancy or are unsure of LMP. As a proxy of fetal maturity, placental or cord-blood GA models could allow easier identification of the right treatment in case of preterm birth. Besides, it could be fruitful as a complemental clinical tool to recognize newborns that could benefit from additional monitoring and care.
Nevertheless, cord-blood and placenta samples are not routinary collected before delivery. This aspect is not a matter of minority, as it would make the device currently unsuitable from a clinical point of view except after birth or confined to research purposes only. However, the presence of fetal cells in the maternal circulation is currently ascertained and may open the opportunity for new models based on cell-free fetal DNA (cffDNA), making GA estimate potentially extremely accurate simply by collecting maternal blood samples at any time during pregnancy. Future studies should also focus on amniocytes obtained with amniocentesis because this is a more accessible and safer modality to obtain fetal cells.
The bond between pregnancy and DNA methylation profile has been deeply investigated in mother and newborns’ peripheral blood. As fetal experience might permanently influence adult health through “developmental plasticity” (Hales and Baker, 2001; Wadhwa et al., 2009), it is not surprising that fetal birthweight has deserved a great attention. While at first it has been seen as an index of maternal nutrition, later, birthweight has gained the role of general intrauterine health proxy, due to the awareness that multiple factors can influence it (Rondó et al., 2003; Grote et al., 2010). Ross et al. were the first to describe an inverse association between second trimester maternal GrimAge acceleration and gestational length or birthweight (Ross et al., 2020). As part of a longitudinal study designed to test the impact of antenatal maternal mood on pregnancy and postpartum outcome (Healthy Babies Before Birth HB3), Ross et al. described the association between GrimAge profile of n = 77 women at their II trimester and newborns’ birthweight and length. The significant correlation between an increase of mothers’ GrimAge acceleration and lower newborns’ birthweight (p = 0.001) highlights the potential role of maternal biological aging on birth outcomes. The authors themselves however underline the possible selection bias due to the recruitment of low-risk pregnancies as well as the lack of validation of epigenetic indices in pregnancy (Ross et al., 2020).
Madden et al. corroborated their results, presenting a significant relation between low birthweight and GrimAge acceleration (Madden et al., 2021). Based on a Scottish family-based cohort, the authors conducted an epigenome-wide study of birthweight using whole blood DNA derived from n = 1,757 adult samples. Among the five adopted epigenetic models (Horvath clock, Hannum, PhenoAge, GrimAge, DNAmTL), only GrimAge and DNAmTL revealed a significant increase of epigenetic age along with low birthweight (Madden et al., 2021). The authors postulated that while DNAmTL could reflect aging phenomenon at a cellular level, GrimAge could describe this aspect at a whole-body level. Finally, they could state that GrimAge outperforms the other models in the framework of pregnancy (Madden et al., 2021). To note, results by Madden et al. regarding DNAmTL are in contrast with previous works that excluded a role of telomere system in birth outcome (Akkad et al., 2006) and with data from Ross et al., where DNAmTL seemed independent from birthweight (Ross et al., 2020).
Symmetrically, extremely low birth weight (ELBW≤1,000 g) could provide a model from nature for how early adversity may shape adult phenotypes (Saigal et al., 2020). Adopting Horvath clock to n = 45 ELBW and n = 47 normal birthweight (NBW) adult survivors, Mathewson et al. observed that ELBW survivors had 2.16 epigenetic years more than their NBW peers for each additional adult risk factor (i.e., BMI, respiratory sinus arrhythmia, blood pressure) (Mathewson et al., 2021). The association between epigenetic age and cumulative risks was characterized by an increasing degree over time, reaching higher significance at their 40s (Mathewson et al., 2021). Their numbers supported the idea that extreme perinatal adversity, such as ELBW, may cause perturbations in the “epigenetic maintaining system” and influencing the “developmental plasticity” in a negative way. However, further considerations are limited for some reasons: the reduced sample size, the single time-point nature of the study that does not allow assessment of dynamic changes in epigenetic aging, the missed choice of second-generation models that may be more appropriate for the purpose (Mathewson et al., 2021).
In other projects, epigenetic rhythm disturbances have been exploited as a potential mechanism linking maternal gestational diabetes (GDM) with offspring’s metabolic risk in later life. In a study set on mother-child pairs (n = 1,156), Shiau et al. adopted Horvath and Hannum clock to verify whether offspring exposed to GDM exhibited accelerated epigenetic ageing in their early childhood, compared to those not exposed (Shiau et al., 2021). Even after adjustment for potential confounders, offspring age acceleration remained higher in the GDM group than in the non-GDM group (according to Horvath clock: +4.96 months higher, p = 0.0002; Hannum clock: +11.2 months, p < 0.0001) (Shiau et al., 2021). Similarly, even if in a smaller sample, Kanney et al. examined the association of epigenetic acceleration with five metabolic biomarkers of interest and GDM (Kanney et al., 2022). In a total of n = 137 mother-child dyads in the Health After Pregnancy - Intergenerational Transmission of Obesity (HAPi) study, the authors compared the result provided by Hannum and Horvath clock in mothers with a prior GDM pregnancy and those without. They could show not only a higher mean epigenetic age in GDM group than in the non-GDM one, but also that among the biomarkers evaluated (leptin, HOMA IR, HDL C, fasting glucose), a statistically significant association was registered between HDL-C and epigenetic age acceleration residuals for the Hannum clock (p = 0.0063) in GDM mothers (Kanney et al., 2022). Both these studies showed the effect of a suboptimal prenatal developmental milieu of the offspring and go along with other data that supported a possible impact of hyperglycemia on developmentally crucial genes (i.e., energy metabolism, metabolic regulation), causing persistent epigenetic changes (Bouchard et al., 2012; Reichetzeder et al., 2016). Nevertheless, additional investigation across the life course using longitudinal samples is warranted (Shiau et al., 2021).
Moving further and going beyond DOHaD theory, the starting point of ageing process could be placed even before intrauterine lifetime and it could be postulated that the “epigenetic programming and plasticity” could be shaped by maternal experience, even before becoming mother. Adverse child events (ACE) refer to negative psychosocial experiences that individuals experience during the first 18 years of life, that can span from abuse, household dysfunction and neglect (Felitti et al., 1998). There is consistent evidence that ACE could cause important consequences in later health life and a consequent hypothesis formulates a possible intergenerational effect, in part due to epigenetic inheritance (Deschênes et al., 2021). In other words, it is possible that events in the mother’s childhood could have effect during pregnancy and more specifically on her future newborns’ outcomes. Nwanaji-Enwerem et al. verified whether preconception maternal ACE could cause stress-related physiologic reprogramming that might result in child health changes, causing intrinsic epigenetic acceleration age, due to intergenerational effects (Nwanaji-Enwerem et al., 2021b) (Figure 3). Describing ACEs experienced by mothers during their childhood and epigenetic aging of their children at 7, 9, and 14 years, Nwanaji-Enwerem et al. showed that children exposed to adverse events were 0.76-year older than children of mothers who did not report these events, in terms of Horvath epigenetic age acceleration (p = 0.004) (Nwanaji-Enwerem et al., 2021a). These findings are the first to exploit the relation between early mother life and epigenetic consequences on their children, and they can provide a molecular basis for the perpetuation of health disparities over time in marginalized populations (Nwanaji-Enwerem et al., 2021b). These data agree and indeed go beyond the results provided by George et al., who showed an influence of social disadvantage exposure in childhood on the biological ageing during adulthood (George et al., 2021). In a cohort of 53-year-old n = 1,376 adults, George et al. exhibited that disadvantaged childhood and lower adult socioeconomic positions were characterized by an accelerated aging process according to GrimAge estimator (p < 0.001) (George et al., 2021).
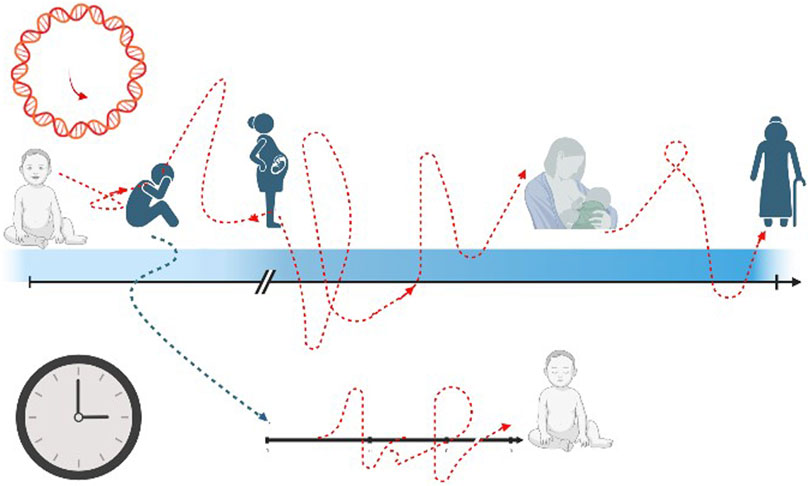
FIGURE 3. The transgenerational effect of epigenetic pattern. The black arrow and black clock represent the chronological time rhythm, that is linear and straight. The red arrow and red clock stand for epigenetic age with a non-linear way of proceeding, with acceleration and decelerated slopes. The blue arrow figures the possible impact of mother’s past adverse events (even in childhood) on the future newborn’s health.
However, another explanation to the data published by Nwanaji-Enwerem et al. may be provided by Herrero-Roldán et al. (Herrero-Roldán et al., 2021). Comparing n = 87 mothers with n = 50 women characterized by “neglect attitude” towards their newborns (neglect group NG), the authors observed that the NG was typically composed by women who have had a disadvantaged background: a higher number of pregnancies in particular at younger age, lower education level, greater childhood maltreatment, propensity for psychiatric disorder, and more economic difficulties. Besides, the authors observed that NG was 2 years epigenetically older than the control group, applying PhenoAge estimator to the collected buccal samples (Herrero-Roldán et al., 2021).
Thus, data from Nwanaji-Enwerem et al. could be justified by two different hypotheses. On one hand, as women with ACE are characterized by epigenetic accelerated age (Herrero-Roldán et al., 2021), their newborns could inherit this disadvantaged epigenetic profile. On the other one, it is also likely that women with ACE experiences are at higher risk of adopting neglect attitude towards their newborns (Herrero-Roldán et al., 2021), causing a prenatal or neonatal adverse environment that would be detrimental for the newborns’ developmental maturity and the cause of their epigenetic acceleration profile.
To sum up, epigenetic clocks further underscore the urgence of providing timely and effective mental health support to vulnerable populations, especially during pregnancy. These last papers illustrate how the mother’s mental health state, that has been mostly “neglected”, can influence the quality of the prenatal environment and consequent multiple aspects of child development (McGill et al., 2022).
As epigenetic clocks have been exploited to detect the impact of pregnancy and maternal characteristic on obstetric and newborns’ outcomes, the possibility that pregnancy may have consequences on maternal health status has raised the interest in epigenetic age behavior, too. In a very preliminary work (n = 33 women), Carroll et al. investigated the effect of sleep deprivation in the postpartum phase and on the biological clock (Carroll et al., 2021). Applying Horvath clock and PhenoAge estimator to peripheral blood samples, they observed that insufficient sleep duration during the early postpartum period was associated with accelerated biological aging (p = 0.03 and p = 0.001 with Horvath and PhenoAge) (Carroll et al., 2021). It is not hard to believe a possible trade-off between reproductive effort and life expectancy (Bolund et al., 2016). Reproductive effort could change brain structures involved in the sociability required for parenting, likely influencing both mAge acceleration and lifespan (Nishitani et al., 2022). Based on a cohort of n = 51 mothers aged 27–46 years, Nishitani et al. explored the relation between epigenetic age and the regional gray matter volumes measured by magnetic resonance imaging (MRI). They noted that multiparity (less than four newborns) encompasses a process of mAge deceleration (p = 0.02) and that precuneus volume increases as mAge deceleration occurred. However, their findings should be considered in the context of some limitations. Saliva samples did not let the adoption of second-generation epigenetic clock, and nulliparous women should be included in future studies to confirm their results (Nishitani et al., 2022). On the other hand, this study opens new fascinating scenarios. This proof could represent the molecular description of the dynamic structural and functional phenomena during pregnancy, linked to “maternalization” of the brain (Hoekzema et al., 2017; Luders et al., 2020) and could give the clarification of longer life expectancy in child-rearing mothers (less than four children) according to demographic studies.
Evidence is however not univocal. In a cohort of n = 397 20–22-year-old women, Ryan et al. also described a raise of DNAmAge acceleration with parity (p = 0.005), consistent with the idea that reproduction comes at a cost of ‘maintenance’ and that epigenetic biomarkers could reflect this “stress” (Ryan et al., 2018). However, they failed to detect an increase of DNAmAge in case of low socio-economic status, a robust “stressor”, even if they justified it as a plausible effect of young age compensation. The narrow age range chosen could indeed introduce some biases (Ryan et al., 2018) that may explain at least in part the difference with previous works (Nishitani et al., 2022).
Gestational epigenetic age clocks are still in their nascent stages. Given that pregnancy emphasizes unique and dynamic physiological adaptations across physiological systems, epigenetic clocks should be validated in pregnant samples. Besides, even if the set cohort homogeneity provides robustness to epigenetic indices, further validation is mandatory in larger and more heterogenous samples to guarantee its generalizability.
In this review, we mainly discussed the possible role of epigenetic clocks in the female reproductive health, starting with physiological fertility issues but then inevitably encompassing the linked topics of infertility and pregnancy. The role and mechanisms of aging in female reproductive lifespan has just recently gain scientific attention but multiple aspects are still neglected.
Epigenetic clocks represent a promising research tool for the coming years and may bring us closer than ever to understanding the complexities of reproductive function. It is reasonable that exploiting epigenetic mechanisms may help advance inquiry into the relation between fertility time and aging. In case of infertility, clinical practice could take advantage of epigenetic biomarkers. Biological age could become a precious information in the diagnostic fertility work-up. Women affected by an “accelerated aging” process could be promptly addressed to adequate health promotion programs and provided with the awareness of “family plan” urgency, if it is a desired wish (Figure 4). Similarly, oocyte or ovarian tissue freezing may not only relegated as “social freezing”, but this choice could be part of health promotion and prevention medicine programs, even in those countries where elective egg freezing is limited to small subgroups of women at high risk of ovarian reserve exhaustion such as those scheduled for radio or chemotherapy.
Regarding pregnancy, epigenetic clocks have the merit of opening a new vision to old arguments. Pregnancy may be not confined to a restricted time window for a woman’s life, but may deserve higher consideration for the whole life. The possibility that environmental exposure could deeply impact on epigenetic footprints and determine the epigenetic rhythm of ageing could renovate the way of dealing with pregnancy issues too. Epigenetic clocks may serve as screening tool for high-risk pregnancies, and they may foster improvement of pregnancy outcomes and postpartum wellbeing. Not only, but it could represent a new tool for dealing with social disparities and help the support of “vulnerable” pregnant populations. Then the future development of GA estimator based on fetal free DNA in maternal blood could completely revolutionize even the prenatal medicine. Whether these epigenetic patterns are stable or shift over the course of reproductive female life represents another key point in the knowledge that may unveil pragmatic therapeutic approach to “turn back the clock” in (a very) distant future. Last, but not least, there is the need to explore more in depth the possible impact of a history of infertility and ART on the epigenetics of newborns.
Future research should explore the unique epigenetic mechanisms underlying aging, that may play a role in the infertility and the obstetric world, with all the possible consequent implications. Finally, it is important to emphasize that pregnancy and infertility may not be seen as two different “worlds” but could represent “chapters” of a same story, whose red thread is the epigenetic rhythm.
LP designed the review and wrote the first draft of the manuscript; LP drew the figures and tables; PV and ES revised and corrected the manuscript. All the authors revised the final version of the manuscript.
This study was (partially) funded by Italian Ministry of Health- Current research IRCCS.
Figures created with BioRender.com. Figure 1. adapted from “Gut-Brain Axis regulators” by BioRender.com (2022) Retrieved from https://app.biorender.com/biorender-templates.
ES received personal honoraria to give talks at international meetings from Theramex and Merck-Serono, received a donation (US machine) from Merck-Serono for the ART unit and handled grants of research from Ferring and Theramex.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akkad, A., Hastings, R., Konje, J. C., Bell, S. C., Thurston, H., and Williams, B. (2006). Telomere length in small-for-gestational-age babies. BJOG : Int. J. obstetrics Gynaecol. 113 (3), 318–323. doi:10.1111/j.1471-0528.2005.00839.x
Arima, Y., and Fukuoka, H. (2020). Developmental origins of health and disease theory in cardiology. J. Cardiol. 76 (1), 14–17. doi:10.1016/j.jjcc.2020.02.003
Barker, D. J. (2007). The origins of the developmental origins theory. J. Intern. Med. 261 (5), 412–417. doi:10.1111/j.1365-2796.2007.01809.x
Bateson, P., Barker, D., Clutton-Brock, T., Deb, D., D'Udine, B., Foley, R. A., et al. (2004). Developmental plasticity and human health. Nature 430 (6998), 419–421. doi:10.1038/nature02725
Bell, C. G., Lowe, R., Adams, P. D., Baccarelli, A. A., Beck, S., Bell, J. T., et al. (2019). DNA methylation aging clocks: Challenges and recommendations. Genome. Biol. 21 (1), 249. doi:10.1186/s13059-019-1824-y
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes & Dev. 16 (1), 6–21. doi:10.1101/gad.947102
Blair, J. D., Yuen, R. K., Lim, B. K., McFadden, D. E., von Dadelszen, P., and Robinson, W. P. (2013). Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol. Hum. Reprod. 19 (10), 697–708. doi:10.1093/molehr/gat044
Bocker, M. T., Hellwig, I., Breiling, A., Eckstein, V., Ho, A. D., and Lyko, F. (2011). Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood 117 (19), e182–e189. doi:10.1182/blood-2011-01-331926
Bohlin, J., Håberg, S. E., Magnus, P., Reese, S. E., Gjessing, H. K., Magnus, M. C., et al. (2016). Prediction of gestational age based on genome-wide differentially methylated regions. Genome Biol. 17 (1), 207. doi:10.1186/s13059-016-1063-4
Bolund, E., Lummaa, V., Smith, K. R., Hanson, H. A., and Maklakov, A. A. (2016). Reduced costs of reproduction in females mediate a shift from a male-biased to a female-biased lifespan in humans. Sci. Rep. 6, 24672. doi:10.1038/srep24672
Bouchard, L., Hivert, M. F., Guay, S. P., St-Pierre, J., Perron, P., and Brisson, D. (2012). Placental adiponectin gene DNA methylation levels are associated with mothers' blood glucose concentration. Diabetes 61 (5), 1272–1280. doi:10.2337/db11-1160
Carlsen, E. Ø., Lee, Y., Magnus, P., Jugessur, A., Page, C. M., Nustad, H. E., et al. (2022). An examination of mediation by DNA methylation on birthweight differences induced by assisted reproductive technologies. Clin. epigenetics 14 (1), 151. doi:10.1186/s13148-022-01381-w
Carroll, J. E., Ross, K. M., Horvath, S., Okun, M., Hobel, C., Rentscher, K. E., et al. (2021). Postpartum sleep loss and accelerated epigenetic aging. Sleep. health 7 (3), 362–367. doi:10.1016/j.sleh.2021.02.002
Christensen, M. W., Keefe, D. L., Wang, F., Hansen, C. S., Chamani, I. J., Sommer, C., et al. (2021). Idiopathic early ovarian aging: Is there a relation with premenopausal accelerated biological aging in young women with diminished response to ART? J. assisted reproduction Genet. 38 (11), 3027–3038. doi:10.1007/s10815-021-02326-7
Clark, J., Bulka, C. M., Martin, C. L., Roell, K., Santos, H. P., O'Shea, T. M., et al. (2022). Placental epigenetic gestational aging in relation to maternal sociodemographic factors and smoking among infants born extremely preterm: A descriptive study. Epigenetics 17 (13), 2389–2403. doi:10.1080/15592294.2022.2125717
Coussons-Read, M. E. (2013). Effects of prenatal stress on pregnancy and human development: Mechanisms and pathways. Obstet. Med. 6 (2), 52–57. doi:10.1177/1753495X12473751
Daredia, S., Huen, K., Van Der Laan, L., Collender, P. A., Nwanaji-Enwerem, J. C., Harley, K., et al. (2022). Prenatal and birth associations of epigenetic gestational age acceleration in the Center for the Health Assessment of Mothers and Children of Salinas (CHAMACOS) cohort. Epigenetics 17 (13), 2006–2021. doi:10.1080/15592294.2022.2102846
Das, A., and Destouni, A. (2022). Novel insights into reproductive ageing and menopause from genomics. Hum. Reprod. Oxf. Engl. 38, 195–203. Advance online publication. doi:10.1093/humrep/deac256
De Geyter, C., Calhaz-Jorge, C., Kupka, M. S., Wyns, C., Mocanu, E., Motrenko, T., et al. (2020). ART in europe, 2015: Results generated from European registries by ESHRE. Hum. Reprod. open 2020 (1), hoz038. doi:10.1093/hropen/hoz038
Deryabin, P. I., and Borodkina, A. V. (2022). Epigenetic clocks provide clues to the mystery of uterine ageing. Hum. Reprod. update, dmac042. Advance online publication. doi:10.1093/humupd/dmac042
Deschênes, S. S., Kivimaki, M., and Schmitz, N. (2021). Adverse childhood experiences and the risk of coronary heart disease in adulthood: Examining potential psychological, biological, and behavioral mediators in the whitehall II cohort study. J. Am. Heart Assoc. 10 (10), e019013. doi:10.1161/JAHA.120.019013
Dieckmann, L., Lahti-Pulkkinen, M., Kvist, T., Lahti, J., DeWitt, P. E., Cruceanu, C., et al. (2021). Characteristics of epigenetic aging across gestational and perinatal tissues. Clin. epigenetics 13 (1), 97. doi:10.1186/s13148-021-01080-y
Duan, R., Fu, Q., Sun, Y., and Li, Q. (2022). Epigenetic clock: A promising biomarker and practical tool in aging. Ageing Res. Rev. 81, 101743. doi:10.1016/j.arr.2022.101743
Eijkemans, M. J. C., van Poppel, F., Habbema, D. F., Smith, K. R., Leridon, H., and te Velde, E. R. (2014). Too old to have children? Lessons from natural fertility populations. Hum. Reprod. 29, 1304–1312. doi:10.1093/humrep/deu056
Erema, V. V., Yakovchik, A. Y., Kashtanova, D. A., Bochkaeva, Z. V., Ivanov, M. V., Sosin, D. V., et al. (2022). Biological age predictors: The status quo and future trends. Int. J. Mol. Sci. 23 (23), 15103. doi:10.3390/ijms232315103
ESHRE Capri Workshop Group (2017). A prognosis-based approach to infertility: Understanding the role of time. Hum. Reprod. 32 (8), 1556–1559. doi:10.1093/humrep/dex214
Falick Michaeli, T., Spiro, A., Sabag, O., Karavani, G., Yagel, S., Eventov-Friedman, S., et al. (2019). Determining gestational age using genome methylation profile: A novel approach for fetal medicine. Prenat. Diagn. 39 (11), 1005–1010. doi:10.1002/pd.5535
Felitti, V. J., Anda, R. F., Nordenberg, D., Williamson, D. F., Spitz, A. M., Edwards, V., et al. (1998). Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. The Adverse Childhood Experiences (ACE) Study. Am. J. Prev. Med. 14 (4), 245–258. doi:10.1016/s0749-3797(98)00017-8
Franzago, M., Pilenzi, L., Di Rado, S., Vitacolonna, E., and Stuppia, L. (2022). The epigenetic aging, obesity, and lifestyle. Front. cell Dev. Biol. 10, 985274. doi:10.3389/fcell.2022.985274
Freire-Aradas, A., Girón-Santamaría, L., Mosquera-Miguel, A., Ambroa-Conde, A., Phillips, C., Casares de Cal, M., et al. (2022). A common epigenetic clock from childhood to old age. Forensic Sci. Int. Genet. 60, 102743. doi:10.1016/j.fsigen.2022.102743
Ge, Z. J., Schatten, H., Zhang, C. L., and Sun, Q. Y. (2015). Oocyte ageing and epigenetics. Reprod. Camb. Engl. 149 (3), R103–R114. doi:10.1530/REP-14-0242
George, A., Hardy, R., Castillo Fernandez, J., Kelly, Y., and Maddock, J. (2021). Life course socioeconomic position and DNA methylation age acceleration in mid-life. J. Epidemiol. community health 75 (11), 1084–1090. doi:10.1136/jech-2020-215608
Girchenko, P., Lahti, J., Czamara, D., Knight, A. K., Jones, M. J., Suarez, A., et al. (2017). Associations between maternal risk factors of adverse pregnancy and birth outcomes and the offspring epigenetic clock of gestational age at birth. Clin. epigenetics 9, 49. doi:10.1186/s13148-017-0349-z
Grote, N. K., Bridge, J. A., Gavin, A. R., Melville, J. L., Iyengar, S., and Katon, W. J. (2010). A meta-analysis of depression during pregnancy and the risk of preterm birth, low birth weight, and intrauterine growth restriction. Archives general psychiatry 67 (10), 1012–1024. doi:10.1001/archgenpsychiatry.2010.111
Gude, N. M., Roberts, C. T., Kalionis, B., and King, R. G. (2004). Growth and function of the normal human placenta. Thrombosis Res. 114 (5-6), 397–407. doi:10.1016/j.thromres.2004.06.038
Guo, S. W. (2009). Epigenetics of endometriosis. Mol. Hum. Reprod. 15 (10), 587–607. doi:10.1093/molehr/gap064
Haftorn, K. L., Lee, Y., Denault, W. R. P., Page, C. M., Nustad, H. E., Lyle, R., et al. (2021). An EPIC predictor of gestational age and its application to newborns conceived by assisted reproductive technologies. Clin. epigenetics 13 (1), 82. doi:10.1186/s13148-021-01055-z
Hales, C. N., and Barker, D. J. (2001). The thrifty phenotype hypothesis. Br. Med. Bull. 60, 5–20. doi:10.1093/bmb/60.1.5
Hannum, G., Guinney, J., Zhao, L., Zhang, L., Hughes, G., Sadda, S., et al. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. cell 49 (2), 359–367. doi:10.1016/j.molcel.2012.10.016
Hanson, B. M., Tao, X., Zhan, Y., Jenkins, T. G., Morin, S. J., Scott, R. T., et al. (2020). Young women with poor ovarian response exhibit epigenetic age acceleration based on evaluation of white blood cells using a DNA methylation-derived age prediction model. Hum. Reprod. Oxf. Engl. 35 (11), 2579–2588. doi:10.1093/humrep/deaa206
Herrero-Roldán, S., Rodrigo, M. J., Hernández-Cabrera, J. A., Mitchell, C., López, M., Alcoba-Florez, J., et al. (2021). Reduction in epigenetic age acceleration is related to empathy in mothers with neglectful caregiving. Brain Sci. 11 (11), 1376. doi:10.3390/brainsci11111376
Hochberg, Z., Feil, R., Constancia, M., Fraga, M., Junien, C., Carel, J. C., et al. (2011). Child health, developmental plasticity, and epigenetic programming. Endocr. Rev. 32 (2), 159–224. doi:10.1210/er.2009-0039
Hoekzema, E., Barba-Müller, E., Pozzobon, C., Picado, M., Lucco, F., García-García, D., et al. (2017). Pregnancy leads to long-lasting changes in human brain structure. Nat. Neurosci. 20 (2), 287–296. doi:10.1038/nn.4458
Horvath, S., Erhart, W., Brosch, M., Ammerpohl, O., von Schönfels, W., Ahrens, M., et al. (2014). Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. United States of America 111 (43), 15538–15543. doi:10.1073/pnas.1412759111
Horvath, S., and Raj, K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19 (6), 371–384. doi:10.1038/s41576-018-0004-3
Houshdaran, S., Nezhat, C. R., Vo, K. C., Zelenko, Z., Irwin, J. C., and Giudice, L. C. (2016). Aberrant endometrial DNA methylome and associated gene expression in women with endometriosis. Biol. reproduction 95 (5), 93. doi:10.1095/biolreprod.116.140434
Javed, R., Chen, W., and Liang, H. (2016). Infant's DNA methylation age at birth and epigenetic aging accelerators. BioMed Res. Int. 2016, 4515928. doi:10.1155/2016/4515928
Johnstone, S. E., Gladyshev, V. N., Aryee, M. J., and Bernstein, B. E. (2022). Epigenetic clocks, aging, and cancer. Sci. (New York, N.Y.) 378 (6626), 1276–1277. doi:10.1126/science.abn4009
Kanney, N., Patki, A., Chandler-Laney, P., Garvey, W. T., and Hidalgo, B. A. (2022). Epigenetic age acceleration in mothers and offspring 4-10 Years after a pregnancy complicated by gestational diabetes and obesity. Metabolites 12 (12), 1226. doi:10.3390/metabo12121226
Knight, A. K., Craig, J. M., Theda, C., Bækvad-Hansen, M., Bybjerg-Grauholm, J., Hansen, C. S., et al. (2016). An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 17 (1), 206. doi:10.1186/s13059-016-1068-z
Knight, A. K., Hipp, H. S., Abhari, S., Gerkowicz, S. A., Katler, Q. S., McKenzie, L. J., et al. (2022). Markers of ovarian reserve are associated with reproductive age acceleration in granulosa cells from IVF patients. Hum. Reprod. Oxf. Engl. 37 (10), 2438–2445. doi:10.1093/humrep/deac178
Kovalenko, T. F., Morozova, K. V., Pavlyukov, M. S., Anufrieva, K. S., Bobrov, M. Y., Gamisoniya, A. M., et al. (2021). Methylation of the PTENP1 pseudogene as potential epigenetic marker of age-related changes in human endometrium. PloS one 16 (1), e0243093. doi:10.1371/journal.pone.0243093
Leap, K., Yotova, I., Horvath, S., and Martinez-Agosto, J. A. (2022). Epigenetic age provides insight into tissue origin in endometriosis. Sci. Rep. 12 (1), 21281. doi:10.1038/s41598-022-25416-7
Leavey, K., Benton, S. J., Grynspan, D., Bainbridge, S. A., Morgen, E. K., and Cox, B. J. (2017). Gene markers of normal villous maturation and their expression in placentas with maturational pathology. Placenta 58, 52–59. doi:10.1016/j.placenta.2017.08.005
Lee, Y., Bohlin, J., Page, C. M., Nustad, H. E., Harris, J. R., Magnus, P., et al. (2022). Associations between epigenetic age acceleration and infertility. Hum. Reprod. Oxf. Engl. 37 (9), 2063–2074. doi:10.1093/humrep/deac147
Lee, Y., Choufani, S., Weksberg, R., Wilson, S. L., Yuan, V., Burt, A., et al. (2019). Placental epigenetic clocks: Estimating gestational age using placental DNA methylation levels. Aging 11 (12), 4238–4253. doi:10.18632/aging.102049
Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S., et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging 10 (4), 573–591. doi:10.18632/aging.101414
Li Piani, L., Reschini, M., Somigliana, E., Ferrari, S., Busnelli, A., Viganò, P., et al. (2022). Peripheral mitochondrial DNA, telomere length and DNA methylation as predictors of live birth in in vitro fertilization cycles. PLoS One 17 (1), e0261591. doi:10.1371/journal.pone.0261591
Llarena, N., and Hine, C. (2021). Reproductive longevity and aging: Geroscience approaches to maintain long-term ovarian fitness. J. Geronto. Ser. A, Biol. Sci. Med. Sci. 76 (9), 1551–1560. doi:10.1093/gerona/glaa204
Loose, J. A., Amrit, F. R. G., Patil, T., Yanowitz, J. L., and Ghazi, A. (2022). Meiotic dysfunction accelerates somatic aging in Caenorhabditis elegans. Aging cell 21 (11), e13716. doi:10.1111/acel.13716
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153 (6), 1194–1217. doi:10.1016/j.cell.2013.05.039
Lowsky, D. J., Olshansky, S. J., Bhattacharya, J., and Goldman, D. P. (2014). Heterogeneity in healthy aging. J. Gerontol. A Biol. Sci. Med. Sci. 69 (6), 640–649. doi:10.1093/gerona/glt162
Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11 (2), 303–327. doi:10.18632/aging.101684
Lu, A. T., Xue, L., Salfati, E. L., Chen, B. H., Ferrucci, L., Levy, D., et al. (2018). GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat. Commun. 9 (1), 387. doi:10.1038/s41467-017-02697-5
Luders, E., Kurth, F., Gingnell, M., Engman, J., Yong, E. L., Poromaa, I. S., et al. (2020). From baby brain to mommy brain: Widespread gray matter gain after giving birth. Cortex; a J. devoted study Nerv. Syst. Behav. 126, 334–342. doi:10.1016/j.cortex.2019.12.029
Madden, R. A., McCartney, D. L., Walker, R. M., Hillary, R. F., Bermingham, M. L., Rawlik, K., et al. (2021). Birth weight associations with DNA methylation differences in an adult population. Epigenetics 16 (7), 783–796. doi:10.1080/15592294.2020.1827713
Mathers, J. C., Strathdee, G., and Relton, C. L. (2010). Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet. 71, 3–39. doi:10.1016/B978-0-12-380864-6.00001-8
Mathewson, K. J., McGowan, P. O., de Vega, W. C., Morrison, K. M., Saigal, S., Van Lieshout, R. J., et al. (2021). Cumulative risks predict epigenetic age in adult survivors of extremely low birth weight. Dev. Psychobiol. 63 (1), e22222. doi:10.1002/dev.22222
Mayne, B. T., Leemaqz, S. Y., Smith, A. K., Breen, J., Roberts, C. T., and Bianco-Miotto, T. (2017). Accelerated placental aging in early onset preeclampsia pregnancies identified by DNA methylation. Epigenomics 9 (3), 279–289. doi:10.2217/epi-2016-0103
McGill, M. G., Pokhvisneva, I., Clappison, A. S., McEwen, L. M., Beijers, R., Tollenaar, M. S., et al. (2022). Maternal prenatal anxiety and the fetal origins of epigenetic aging. Biol. psychiatry 91 (3), 303–312. doi:10.1016/j.biopsych.2021.07.025
Mills, M., Rindfuss, R. R., McDonald, P., and te Velde, E.ESHRE Reproduction and Society Task Force (2011). Why do people postpone parenthood? Reasons and social policy incentives. Hum. Reprod. update 17 (6), 848–860. doi:10.1093/humupd/dmr026
Monasso, G. S., Küpers, L. K., Jaddoe, V. W. V., Heil, S. G., and Felix, J. F. (2021). Associations of circulating folate, vitamin B12 and homocysteine concentrations in early pregnancy and cord blood with epigenetic gestational age: The generation R study. Clin. epigenetics 13 (1), 95. doi:10.1186/s13148-021-01065-x
Monseur, B., Murugappan, G., Bentley, J., Teng, N., and Westphal, L. (2020). Epigenetic clock measuring age acceleration via DNA methylation levels in blood is associated with decreased oocyte yield. J. assisted reproduction Genet. 37 (5), 1097–1103. doi:10.1007/s10815-020-01763-0
Morin, S. J., Tao, X., Marin, D., Zhan, Y., Landis, J., Bedard, J., et al. (2018). DNA methylation-based age prediction and telomere length in white blood cells and cumulus cells of infertile women with normal or poor response to ovarian stimulation. Aging 10 (12), 3761–3773. doi:10.18632/aging.101670
Munro, S. K., Farquhar, C. M., Mitchell, M. D., and Ponnampalam, A. P. (2010). Epigenetic regulation of endometrium during the menstrual cycle. Mol. Hum. Reprod. 16 (5), 297–310. doi:10.1093/molehr/gaq010
Nishitani, S., Kasaba, R., Hiraoka, D., Shimada, K., Fujisawa, T. X., Okazawa, H., et al. (2022). Epigenetic clock deceleration and maternal reproductive efforts: Associations with increasing gray matter volume of the precuneus. Front. Genet. 13, 803584. doi:10.3389/fgene.2022.803584
Nwanaji-Enwerem, J. C., Colicino, E., Gao, X., Wang, C., Vokonas, P., Boyer, E. W., et al. (2021b). Associations of plasma folate and vitamin B6 with blood DNA methylation age: An analysis of one-carbon metabolites in the VA normative aging study. J. Geronto. Ser. A, Biol. Sci. Med. Sci. 76 (5), 760–769. doi:10.1093/gerona/glaa257
Nwanaji-Enwerem, J. C., Van Der Laan, L., Kogut, K., Eskenazi, B., Holland, N., Deardorff, J., et al. (2021a). Maternal adverse childhood experiences before pregnancy are associated with epigenetic aging changes in their children. Aging 13 (24), 25653–25669. doi:10.18632/aging.203776
Oblak, L., van der Zaag, J., Higgins-Chen, A. T., Levine, M. E., and Boks, M. P. (2021). A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res. Rev. 69, 101348. doi:10.1016/j.arr.2021.101348
Olesen, M. S., Starnawska, A., Bybjerg-Grauholm, J., Bielfeld, A. P., Agerholm, I., Forman, A., et al. (2018). Biological age of the endometrium using DNA methylation. Reprod. Camb. Engl. 155 (2), 167–172. doi:10.1530/REP-17-0601
Quach, A., Levine, M. E., Tanaka, T., Lu, A. T., Chen, B. H., Ferrucci, L., et al. (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 9 (2), 419–446. doi:10.18632/aging.101168
Reichetzeder, C., Dwi Putra, S. E., Pfab, T., Slowinski, T., Neuber, C., Kleuser, B., et al. (2016). Increased global placental DNA methylation levels are associated with gestational diabetes. Clin. epigenetics 8, 82. doi:10.1186/s13148-016-0247-9
Reimann, B., Martens, D. S., Wang, C., Ghantous, A., Herceg, Z., Plusquin, M., et al. (2022). Interrelationships and determinants of aging biomarkers in cord blood. J. Transl. Med. 20 (1), 353. doi:10.1186/s12967-022-03541-1
Retis-Resendiz, A. M., González-García, I. N., León-Juárez, M., Camacho-Arroyo, I., Cerbón, M., and Vázquez-Martínez, E. R. (2021). The role of epigenetic mechanisms in the regulation of gene expression in the cyclical endometrium. Clin. epigenetics 13 (1), 116. doi:10.1186/s13148-021-01103-8
Rondó, P. H., Ferreira, R. F., Nogueira, F., Ribeiro, M. C., Lobert, H., and Artes, R. (2003). Maternal psychological stress and distress as predictors of low birth weight, prematurity and intrauterine growth retardation. Eur. J. Clin. Nutr. 57 (2), 266–272. doi:10.1038/sj.ejcn.1601526
Ross, K. M., Carroll, J. E., Horvath, S., Hobel, C. J., Coussons-Read, M. E., and Dunkel Schetter, C. (2020). Epigenetic age and pregnancy outcomes: GrimAge acceleration is associated with shorter gestational length and lower birthweight. Clin. Epigenetics 12 (1), 120. doi:10.1186/s13148-020-00909-2
Rutledge, J., Oh, H., and Wyss-Coray, T. (2022). Measuring biological age using omics data. Nat. Rev. Genet. 23 (12), 715–727. doi:10.1038/s41576-022-00511-7
Ryan, C. P., Hayes, M. G., Lee, N. R., McDade, T. W., Jones, M. J., Kobor, M. S., et al. (2018). Reproduction predicts shorter telomeres and epigenetic age acceleration among young adult women. Sci. Rep. 8 (1), 11100. doi:10.1038/s41598-018-29486-4
Saare, M., Modhukur, V., Suhorutshenko, M., Rajashekar, B., Rekker, K., Sõritsa, D., et al. (2016). The influence of menstrual cycle and endometriosis on endometrial methylome. Clin. epigenetics 8, 2. doi:10.1186/s13148-015-0168-z
Sae-Lee, C., Corsi, S., Barrow, T. M., Kuhnle, G. G. C., Bollati, V., Mathers, J. C., et al. (2018). Dietary intervention modifies DNA methylation age assessed by the epigenetic clock. Mol. Nutr. food Res. 62 (23), e1800092. doi:10.1002/mnfr.201800092
Saigal, S., Morrison, K., and Schmidt, L. A. (2020). Health, wealth and achievements of former very premature infants in adult life. Seminars fetal & neonatal Med. 25 (3), 101107. doi:10.1016/j.siny.2020.101107
Saul, D., and Kosinsky, R. L. (2021). Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. 22 (1), 401. doi:10.3390/ijms22010401
Shahal, T., Segev, E., Konstantinovsky, T., Marcus, Y., Shefer, G., Pasmanik-Chor, M., et al. (2022). Deconvolution of the epigenetic age discloses distinct inter-personal variability in epigenetic aging patterns. Epigenetics chromatin 15 (1), 9. doi:10.1186/s13072-022-00441-y
Shiau, S., Wang, L., Liu, H., Zheng, Y., Drong, A., Joyce, B. T., et al. (2021). Prenatal gestational diabetes mellitus exposure and accelerated offspring DNA methylation age in early childhood. Epigenetics 16 (2), 186–195. doi:10.1080/15592294.2020.1790924
Simpkin, A. J., Suderman, M., Gaunt, T. R., Lyttleton, O., McArdle, W. L., Ring, S. M., et al. (2015). Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum. Mol. Genet. 24 (13), 3752–3763. doi:10.1093/hmg/ddv119
Simpson, D. J., and Chandra, T. (2021). Epigenetic age prediction. Aging cell 20 (9), e13452. doi:10.1111/acel.13452
Staff, A. C. (2019). The two-stage placental model of preeclampsia: An update. J. reproductive Immunol. 134, 1–10. doi:10.1016/j.jri.2019.07.004
te Velde, E. R., and Pearson, P. L. (2002). The variability of female reproductive ageing. Hum. Reprod. update 8 (2), 141–154. doi:10.1093/humupd/8.2.141
Tekola-Ayele, F., Workalemahu, T., Gorfu, G., Shrestha, D., Tycko, B., Wapner, R., et al. (2019). Sex differences in the associations of placental epigenetic aging with fetal growth. Aging 11 (15), 5412–5432. doi:10.18632/aging.102124
Valensise, H., Vasapollo, B., Gagliardi, G., and Novelli, G. P. (2008). Early and late preeclampsia: Two different maternal hemodynamic states in the latent phase of the disease. Hypertension 52 (5), 873–880. doi:10.1161/HYPERTENSIONAHA.108.117358
Wadhwa, P. D., Buss, C., Entringer, S., and Swanson, J. M. (2009). Developmental origins of health and disease: Brief history of the approach and current focus on epigenetic mechanisms. Seminars reproductive Med. 27 (5), 358–368. doi:10.1055/s-0029-1237424
Workalemahu, T., Shrestha, D., Tajuddin, S. M., and Tekola-Ayele, F. (2021). Maternal cardiometabolic factors and genetic ancestry influence epigenetic aging of the placenta. J. Dev. Orig. health Dis. 12 (1), 34–41. doi:10.1017/S2040174419000801
Zannas, A. S., Arloth, J., Carrillo-Roa, T., Iurato, S., Röh, S., Ressler, K. J., et al. (2015). Lifetime stress accelerates epigenetic aging in an urban, african American cohort: Relevance of glucocorticoid signaling. Genome Biol. 16, 266. doi:10.1186/s13059-015-0828-5
Keywords: epigenetic, aging, infertility, pregnancy, epigenetic clock, newborn
Citation: Li Piani L, Vigano' P and Somigliana E (2023) Epigenetic clocks and female fertility timeline: A new approach to an old issue?. Front. Cell Dev. Biol. 11:1121231. doi: 10.3389/fcell.2023.1121231
Received: 11 December 2022; Accepted: 03 March 2023;
Published: 21 March 2023.
Edited by:
Claudia Massarotti, University of Genoa, ItalyReviewed by:
Barbara Barboni, University of Teramo, ItalyCopyright © 2023 Li Piani, Vigano' and Somigliana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Letizia Li Piani, bGV0aXppYS5saXBpYW5pQHVuaW1pLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.