
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 09 March 2023
Sec. Molecular and Cellular Pathology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1120984
This article is part of the Research TopicThe 6th Israeli Zebrafish Meeting: Zebrafish as a Model for Biomedical Research In IsraelView all 5 articles
Research on learning and memory formation at the level of neural networks, as well as at the molecular level, is challenging due to the immense complexity of the brain. The zebrafish as a genetically tractable model organism can overcome many of the current challenges of studying molecular mechanisms of learning and memory formation. Zebrafish have a translucent, smaller and more accessible brain than that of mammals, allowing imaging of the entire brain during behavioral manipulations. Recent years have seen an extensive increase in published brain research describing the use of zebrafish for the study of learning and memory. Nevertheless, due to the complexity of the brain comprising many neural cell types that are difficult to isolate, it has been difficult to elucidate neural networks and molecular mechanisms involved in memory formation in an unbiased manner, even in zebrafish larvae. Therefore, data regarding the identity, location, and intensity of nascent proteins during memory formation is still sparse and our understanding of the molecular networks remains limited, indicating a need for new techniques. Here, we review recent progress in establishing learning paradigms for zebrafish and the development of methods to elucidate neural and molecular networks of learning. We describe various types of learning and highlight directions for future studies, focusing on molecular mechanisms of long-term memory formation and promising state-of-the-art techniques such as cell-type-specific metabolic labeling.
Learning and memory are an integral part of our daily lives. Cognitive and behavioral alterations during learning and memory are mediated by changes at the molecular level, most of which seem to be remarkably conserved across species (Kandel et al., 2014). Although research to understand the molecular mechanisms of learning and memory has made great progress, and several protein factors important for plasticity have been identified (Kelleher et al., 2004; Sutton and Schuman, 2006), the exact underlying molecular pathways are yet to be fully characterized (Evans et al., 2021).
A major obstacle to research focused on the neural networks and molecular pathways involved in learning and memory formation is the immense complexity of the brain. Therefore, efforts have been made to develop learning paradigms in model organisms with useful behavioral repertoires, including invertebrates with simple nervous systems and vertebrates in which overall regions of the central nervous system have been conserved. So far, most research on learning and memory has been performed in two highly related rodent model organisms, rats and mice (Figure 1A). The ability to form memories and recall previous events and thereby alter future behaviors provides such a strong evolutionary advantage that it is well conserved in both invertebrates and vertebrates including zebrafish (Farley and Alkon, 1985).
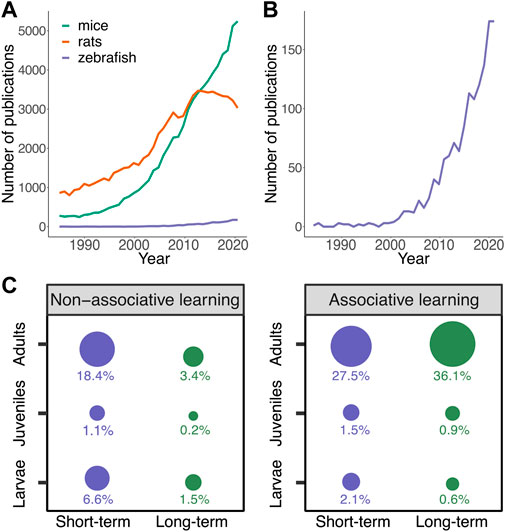
FIGURE 1. Studies of learning and memory formation in zebrafish. PubMed results for search terms (species; rats, mice or zebrafish) AND (learning OR memory). (A) PubMed results (1985–2021) for rats, mice and zebrafish indicating the dominance of rodents for studying learning and memory. (B) Focus on PubMed results for zebrafish indicating the increase in publications in zebrafish for studying learning and memory. (C) PubMed results (date of search 16/10/2022) for zebrafish were screened (N = 1,307) and 433 papers that we could relate to learning and memory formation (e.g., not including reviews or behavioral studies that we could not categorize as learning) were included and their data were classified for associative or non-associative learning, short- or long-term memory. Larvae were defined as younger than 13 dpf, juveniles as 13 dpf to 2 months old, and adults as 2 months or older. Size of the circle represents the percentage out of total included experiments. Data were analyzed and visualized using R.
The zebrafish is a relative newcomer as a model organism in behavioral neuroscience, with an observed sharp increase in published studies on learning and memory in the recent decade (Figure 1B). Characteristics of zebrafish are beneficial to study the conserved and general principles of vertebrate nervous system functions, including learning and memory, making zebrafish a powerful model organism to study memory consolidation. Zebrafish are genetically tractable, translucent vertebrates with a small and relatively accessible brain that possesses numerous features of brain organization conserved across vertebrates. In addition, zebrafish exhibit complex behaviors, including learning and memory behaviors. Combining these characteristics with their suitability to high-throughput studies, zebrafish provides an excellent complementary vertebrate model for studying the molecular and neural basis of learning and memory (Gerlai, 2020).
Here, we provide a brief review of recent progress in learning paradigms for larval, juvenile, and adult zebrafish, as well as experimental methods to elucidate neural and molecular networks of learning and memory. We describe various types of learning and highlight directions for future studies, focusing on molecular mechanisms of long-term memory formation and promising state-of-the-art techniques, such as cell-type-specific metabolic labeling.
Zebrafish has long been a favorite model organism for developmental biologists and geneticists, mainly due to their suitability for genetic manipulation and translucent nature from oocyte to developed larvae. Over the last 2 decades, researchers have begun to appreciate the organism’s usefulness for behavioral studies, including for social behavior and, more recently, for studying mechanisms of learning and memory formation. Here, we discuss 1) behaviors related to learning and memory formation, and 2) research paradigms used to study learning and memory in zebrafish.
Many basic neural systems mediating learning and memory have been evolutionarily conserved. Mammalian brain regions known to be key for memory, including the hippocampus and amygdala, have functional homologous structures in the zebrafish brain. The dorsomedial telencephalic region is involved in place preference conditioning as was demonstrated by activity of the immediately early gene c-fos (von Trotha et al., 2014). Further analyses of specific markers involved in emotional behaviors proposed this region to be homologous to the mammalian amygdala (von Trotha et al., 2014; Porter and Mueller, 2020; Lal et al., 2018). Likewise, the lateral pallium is involved in spatial learning and fulfills hippocampus-like functions (Broglio et al., 2010; Mueller, 2012; Mazzitelli-Fuentes et al., 2022).
In addition to studying established regions for memory consolidation, unbiased approaches of studying molecular mechanisms of learning and memory could identify additional conserved brain regions and networks important for the consolidation of memory, as discussed in the section “Zebrafish for the study of molecular mechanisms of learning and memory.”
Zebrafish exhibit a broad range of behaviors depending on their developmental stage (Nelson and Granato, 2022). These behaviors can be manipulated and studied in the context of learning and memory experiments. Within the first day post-fertilization (dpf), larvae exhibit spontaneous coiling (Granato and Nüsslein-Volhard, 1996; Saint-Amant and Drapeau, 1998). At three dpf, larvae react to touch by tail beating and backward movement and exhibit escape behavior when exposed to touch, acoustic, or electrical stimuli (Nelson and Granato, 2022; Santistevan et al., 2022). From three dpf, larval zebrafish avoid both hot and cold temperatures (Prober et al., 2008; Gau et al., 2013; Haesemeyer et al., 2015). At four dpf, larvae perform spontaneous swimming and start to exhibit habituation learning following repeated stimuli (Best et al., 2007). At five dpf, larvae can feed on their own, and at 6–7 dpf they actively hunt live prey, as well as keep a certain distance with respect to conspecifics (Marques et al., 2017). At 6–8 dpf, larvae show preference to a light over dark environment and actively swim to the light environment (Hinz et al., 2013). This light-dark preference will change to dark preference in adults, but the exact ontology is not yet known. Social preference starts 1 week post-fertilization and becomes robust at 3 weeks (Hinz et al., 2013; Dreosti et al., 2015). In accordance to the behavioral repertoire during development, numerous learning paradigms have been reported for zebrafish across ages (Kenney, 2020; R; Gerlai, 2016; Mu et al., 2020) (Figure 1C).
Larval zebrafish have been predominantly used to study habituation learning (Figure 2A) (paragraph 2. b.1; (Roberts et al., 2013), which has been demonstrated as early as 4 dpf (Eaton et al., 1977). In addition to this relatively simple form of learning, it was reported that 6–8 dpf larvae can perform in an associative place-preference paradigm (Hinz et al., 2013). At eight dpf, partially mounted larvae performed Relief of Aversive Stimulus by Turn (ROAST) in operant-conditioning task where the larvae could avoid an aversive heat stimulus by moving the tail (Figure 2B) (Lin et al., 2019). Simultaneous Ca2+ imaging revealed functional connectivity changes between the cerebellum and habenula, which correlated with decision outcomes. Other studies showed that ten dpf larvae are able to perform a visual lateralization novel object recognition task (Andersson et al., 2015). Classical and operant conditioning paradigms using electroshocks showed that learning improves during development from seven dpf, starts to be robust at around week three and reaches adult performance at week six (Valente et al., 2012). Another study demonstrated that a spatial alternation task based on a food reward was successfully performed by young adult (6–8 weeks) and adult fish (>8 weeks), while 3-4-week-old juveniles did not learn the task (Williams et al., 2002).
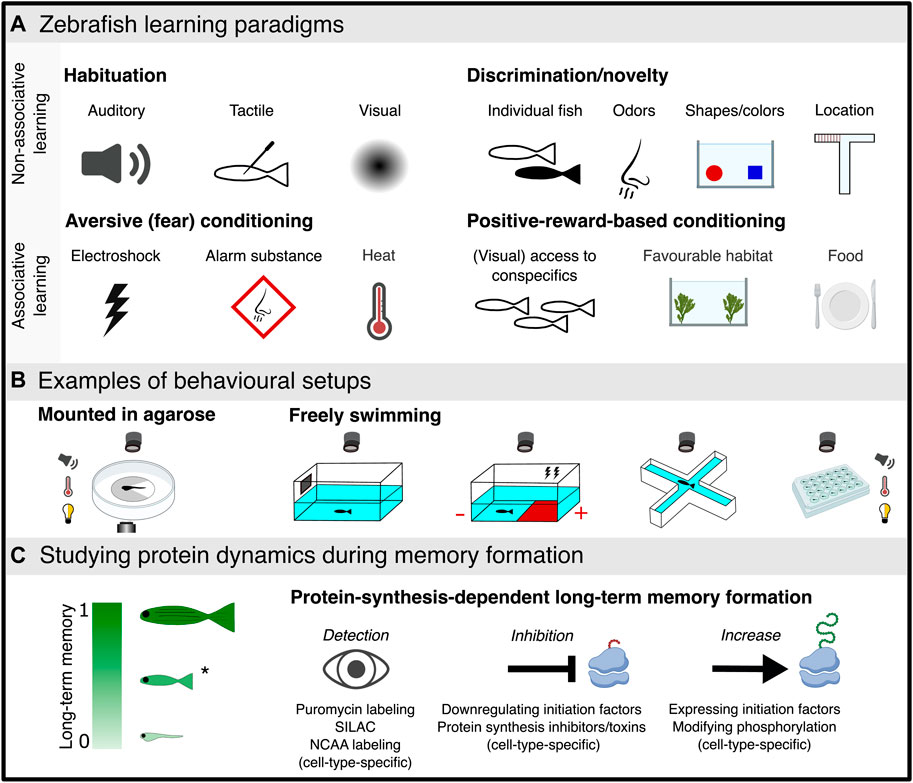
FIGURE 2. Schematic overview of studying learning in zebrafish. (A) Schematic representation of learning and memory paradigms used in zebrafish including forms of non-associative learning (habituation, recognition and novelty learning) and associative learning (aversive and positive-reward based learning). (B) Behavioural setups for zebrafish learning paradigms (partially mounted and freely swimming) can include recording of behaviour, neural activity combined with various stimuli as presented in (A), or light-dark environment (white-yellow light bulb). (C) Protein dynamics during long-term memory formation. Left, the ability to form long-term memory increases with age (larvae, juvenile and adult from bottom to top, respectively) with only some evidence in larvae while most studies have used adult fish (Figure 1C), highlighting the rationale to further examine long-term memory consolidation in juvenile zebrafish (*) that possess a more accessible brain than adults. Right, a suite of methodologies allowing for the detection and manipulation of protein dynamics during protein-synthesis-dependent long-term memory formation. The figure was created with BioRender and Inkscape.
Adult zebrafish are able to perform complex learning tasks, which depends on their ability to discriminate between different sensory stimuli (Figure 2A). They can discriminate between visual stimuli such as shapes and colors (Colwill et al., 2005; Risner et al., 2005; Gatto et al., 2020; Santacà et al., 2021; Santaca et al., 2022), different odors (Braubach et al., 2008; Namekawa et al., 2018), and individual fish (Madeira and Oliveira, 2017). These cues have been used to train fish during complex spatial learning tasks (Williams et al., 2002; Levin et al., 2003; Xu et al., 2006; Baratti et al., 2019; Baratti et al., 2021). For example, fish can use the geometry of an arena to orient themselves in order to find the exit and gain a reward (Baratti et al., 2021).
Forms of avoidance and fear learning paradigms have also been widely used with zebrafish (Figure 2A) (Pradel et al., 2000; Xu et al., 2006; Castro et al., 2009; Baker and Wong, 2019). For example, adult zebrafish can form memories of a natural olfactory alarm cue using a contextual fear learning paradigm that depends on stress-coping styles of the zebrafish (Baker and Wong, 2019). Most associative learning paradigms resulting in long-term memory formation have been demonstrated in adult zebrafish (Figure 1C). A recent study used electroshock fear conditioning in juveniles and found that the dorsolateral habenula is required for updating learned behaviors (Palumbo et al., 2020). In addition to negative reinforcers or punishment, such as electric shocks and natural olfactory alarm cues, positive reinforcers have been studied for classical conditioning paradigms, including food (Bilotta et al., 2005; Colwill et al., 2005; Sison and Gerlai, 2009; Manabe et al., 2013a) and visual access to conspecifics (Al-Imari and Gerlai, 2008; Sison and Gerlai, 2011; Hinz et al., 2013; Fernandes et al., 2016). For example, adult zebrafish have been successfully trained to perform well during visual discrimination and amodal completion tasks, using both food and conspecifics as a reward (Sovrano et al., 2022).
Learning can be divided into two main forms: non-associative and associative (Figure 2A). Non-associative learning is a simple yet fundamental form of learning, not requiring stimuli association or pairing. It means that a response to a single event or stimulus, an animal can change their behavior (Ioannou and Anastassiou-Hadjicharalambous, 2021). Examples of non-associative learning include habituation, sensitization, perceptual learning, priming and recognition memory (Pereira and Kooy, 2013; Ioannou and Anastassiou-Hadjicharalambous, 2021). Habituation and sensitization learning are implicit or procedural forms of learning that respectively attenuates or augments (sensitizes) an animal’s sensory percept or behavioral response to a sensory stimulus upon repeated or continual presentation of the stimulus (Harris, 1943; Thompson and Spencer, 1966; Poon and Schmid, 2012). The altered response to the repeated stimuli of fixed intensity is not due to sensory adaptation, fatigue, or injury. Habituation has been commonly used in zebrafish paradigms based on a rapid startle response that decreases over time upon repeated exposure to a sensory stimulus (auditory, visual, or tactile) (Eaton et al., 1977; Best et al., 2007; Wolman et al., 2011). The light preference of larval zebrafish has been used to show dynamic learning including sensitization and habituation during a dark-avoidance task (Xu et al., 2021). The use of virtual reality paradigms with larval zebrafish embedded in agarose combined with light sheet microscopy allows for whole brain imaging during short-term motor learning (Kawashima et al., 2016). Regarding recognition learning, both short- and long-term memory formation have been demonstrated using novel object recognition and location paradigms in zebrafish (Lucon-Xiccato and Dadda, 2014; Oliveira et al., 2014; Andersson et al., 2015; May et al., 2015; Gaspary et al., 2018). Additionally, Y- and T-mazes have been primarily used to study working and short-term memory of a previously explored arm (Cleal et al., 2020; Fontana et al., 2021; Brinza et al., 2022).
Associative learning involves establishing a relationship (association) between at least two separate stimuli. A basic form of associative learning is classical conditioning (Pavlov, 1951; Sokolov, 1963; Rehman et al., 2022). Here, animals learn how to associate a neutral stimulus (conditioned stimulus, CS) with a reinforcing stimulus which can be either positive or negative (unconditioned stimulus, US) (Figure 2B). As the result of the paired delivery of a CS and US, the animal learns that the CS predicts the occurrence of the US. Consequently, the response to the CS becomes similar to its initial response to the US. Classical conditioning has been reported in both larval (Aizenberg and Schuman, 2011; Valente et al., 2012; Hinz et al., 2013) and adult zebrafish (Braubach et al., 2008; Agetsuma et al., 2010; Karnik and Gerlai, 2012; Aoki et al., 2013) using both positive (Colwill et al., 2005; Al-Imari and Gerlai, 2008; Gómez-Laplaza and Gerlai, 2009; Sison and Gerlai, 2009; Sison and Gerlai, 2011; Manabe et al., 2013b) and negative (Xu et al., 2006; Castro et al., 2009; Lee et al., 2010; Pradel et al., 2000; Levin and Chen, 2004; Baker and Wong, 2019) reinforcing stimuli. The use of classical conditioning in adult zebrafish is dominant in the literature (Figure 1C), yet, recent studies have adapted long-term memory associative learning-based paradigms for juveniles e.g., (Palumbo et al., 2020).
Zebrafish can breed throughout the year and have progenies of hundreds of eggs, which allows for studying memory formation in large numbers or while testing numerous conditions in a high-throughput manner. There have been developments in designing learning paradigms that are more appropriate for high-throughput screens (Stewart et al., 2015). For example, automated systems for imaging, tracking, and analyzing dozens of larvae simultaneously (Figure 1B) (Ahmed et al., 2010; Wolman et al., 2011; Doyle et al., 2016; Randlett et al., 2019; Barreiros et al., 2021); paradigms that can be performed with multiple adult fish at the same time (Kareklas et al., 2018; Samaras and Pavlidis, 2020; Barreiros et al., 2021); and relatively short paradigms that do not last for more than 2 or 3 days (Hinz et al., 2013; Lucon-Xiccato and Dadda, 2014). Efforts should be made to ensure that fish are habituated properly to the paradigm setup and show no signs of stress or anxiety, especially for paradigms that use individual zebrafish (Gerlai, 2016).
The zebrafish model is advantageous due its nervous system complexity and practical accessibility. Zebrafish are evolutionary ancient vertebrates but still possess numerous conserved features across multiple levels of biological organization in the brain. Zebrafish larvae are the only vertebrates with a translucent brain, allowing for imaging of the entire brain at a (sub)-cellular level, even while the fish is alive and during learning tasks described above (Naumann et al., 2016). Using optogenetic tools, it is possible to manipulate neuronal activity in a specific and reversible manner. Light-gated channels (Szobota et al., 2007; Douglass et al., 2008; Arrenberg et al., 2009; Bundschuh et al., 2012; Fajardo et al., 2013) can be used to either excite or inhibit neurons, which consequently can be imaged in the intact behaving fish (Wyart and Del Bene, 2011; Del Bene and Wyart, 2012; Portugues et al., 2013). The transparency of the larval zebrafish allows for non-invasive optogenetic detection and modulation of neural activity, and pharmacological tools and genetic lines exist that increase this transparency into adulthood (Karlsson et al., 2001; Bergmann et al., 2018). Indeed, there have been successful reports on the use of optogenetics to study both larval (Harmon et al., 2017) and adult zebrafish behavior (Douglass et al., 2008; Ahrens et al., 2012; Del Bene and Wyart, 2012; Portugues et al., 2013). Still, application during learning and memory paradigms is challenging, mainly due to the need for restraining the fish during the experimental procedure (Wyart et al., 2009).
Both associative and non-associative learning can lead to short- or long-term memory formation. Short-term memory lasts from seconds to minutes and its formation mostly relies on biochemical changes to existing proteins (Kandel et al., 2014). Long-term memory lasts from hours to years and its formation is protein synthesis dependent (Flexner et al., 1962; Josefa et al., 1963; Nee et al., 2008). This has been demonstrated in different organisms and using various learning paradigms (Agranoff et al., 1966; Gal-Ben-Ari and Rosenblum, 2011; Gal-Ben-Ari et al., 2012; Kandel et al., 2014).
In zebrafish, both long-term habituation (Wolman et al., 2011; Roberts et al., 2013; Roberts et al., 2016) and classical conditioning leading to long term memory formation (Pradel et al., 1998; Pradel et al., 2000; Blank et al., 2009) depend on the ability to synthesize new proteins. For example, an essential role for the synthesis of cell adhesion molecules in memory consolidation and recall in adult zebrafish was discovered using an active avoidance paradigm (Pradel et al., 2000). A conditioned place preference paradigm in zebrafish larvae reported protein-synthesis-dependent long-term memory formation and a role for NMDA-receptor activation in this process (Hinz et al., 2013). The place preference for an environment with visual access to conspecifics develops with age and becomes more robust 2–3 weeks post fertilization (Dreosti et al., 2015), suggesting that this promising paradigm may be more robust in juvenile zebrafish (Figure 2C). A recent study measured brain protein dynamics following adaptation of zebrafish to water currents induced by magnetic stirrers and detected 57 regulated proteins in larvae exposed to the water vortex (Langebeck-Jensen et al., 2019). However, when measuring total protein content, newly synthesized (nascent) proteins can be masked by already existing proteins, thus hindering their detection. In addition, the use of whole tissue proteomics hinders detection of cell-type-specific protein alterations. This calls for methodologies that allow for the detection of newly synthesized proteins, preferably in cell types of interest.
Due to the complexity of the brain, comprising many cell types including neurons that possess long processes and are entangled in the respective tissue, it is difficult to reveal the newly synthesized proteome during learning and memory formation in an unbiased manner. In this section, we will review evidence for the role of protein synthesis during long-term memory formation and discuss novel methods that allow for the labelling of nascent proteins in cell types of interest (Figure 1C). See also reviews on de novo proteomic methods in relation to memory consolidation (Evans et al., 2021; Ross et al., 2021).
The pioneering studies that first showed the need for protein synthesis during long-term memory formation used protein synthesis inhibitors such as puromycin, delivered non-specifically (Flexner et al., 1963). Delivering protein synthesis inhibitors can done by injection to a brain region or for zebrafish, by adding it to the water bath of the fish resulting in universal inhibition (Hinz et al., 2012; Shahar and Schuman, 2020). However, within the complex structure of the brain, different brain regions and cell types enable the formation, consolidation, and recall of memory (Camina and Güell, 2017). Although inhibitors can be delivered directly to specific brain regions at different stages of long-term memory formation, they cannot be restricted to specific cell types and therefore cannot distinguish between the role of neuronal subtypes or glial cells in memory consolidation. Furthermore, although memory research has predominantly focused on hippocampal neurons, other brain cells including glia have been shown to play a role too (Yoo et al., 2021). Recently, tools that enable cell-type-specific drug-inducible inhibition of protein synthesis have been developed including a toxin from Maize that can be expressed in a cell-type-specific manner (Heumüller et al., 2019) which has so far only been demonstrated in vitro. Another approach enables rapid and reversible phosphorylation of eukaryotic initiation factor 2α, leading to inhibition of general translation (Shrestha et al., 2020). Such tools have the potential to increase the spatiotemporal resolution in which protein synthesis can be detected during learning and memory adaptations. Similarly, protein synthesis has been artificially increased in specific cell types through overexpression of initiation factors (Shrestha et al., 2020; Xu et al., 2020). Another way to modulate translation of memory consolidation related proteins and in a cell-type-specific manner is by modifying the kinase activity of elf2a, which affects memory consolidation in mice (Gould et al., 2020; Sharma et al., 2020). Future studies in zebrafish could provide additional information on conserved vertebrate brain regions and cell types involved in different stages of memory formation.
Given the evidence that protein synthesis is required for long-term memory formation, identifying which proteins are synthesized, and their specific roles, is fundamental for understanding the complexity of the underpinning molecular mechanisms (Flexner et al., 1963; Sutton and Schuman, 2006; Costa-Mattioli et al., 2008; Gal-Ben-Ari et al., 2012; Hinz et al., 2013). Manipulation of protein synthesis provides useful information about the importance of protein synthesis in certain cell types and brain regions during memory consolidation, but it does not identify which proteins are being synthesized. Therefore, efforts have been made to identify newly synthesized proteins in a given brain region in a cell-type-specific manner. One widely used technique is Stable Isotope Labelling with Amino Acids in cell culture (SILAC) (Koren et al., 2019). Isotope tagging of proteins leads to a shift in the molecular mass of the labelled peptide, thus enabling identification via mass spectrometry (Engmann et al., 2010; Chen et al., 2015). However, although this technique labels the newly synthesized proteins, it does not enrich for them, therefore, newly synthesized proteins in low abundance may be missed due to masking by highly abundant existing proteins. Additionally, although the technique has been adapted for its use in animal models (Price et al., 2010; Rauniyar et al., 2013), it cannot be directed to specific cell types, since it uses endogenous amino acids to label newly synthesized proteins.
A method that purifies only nascent proteins uses the general protein synthesis inhibitor puromycin (Nemoto et al., 1999). Puromycin is molecularly similar to aminoacyl-tRNA and uses the endogenous translational machinery to integrate itself into proteins as they are being synthesized (Dieck et al., 2015). Puromycin-tagged proteins can be labeled with anti-puromycin antibodies and subsequently visualized via immunohistochemistry, or purified and identified via mass spectrometry. Because of the rapid integration of puromycin into the newly synthesized amino-acid chain, this method can be used to examine local protein synthesis (Hafner et al., 2019). This is important because both in neurons and glial cells, it has been demonstrated that protein synthesis occurs locally, which likely plays an important role in synaptic plasticity (Sutton and Schuman, 2006; Sakers et al., 2017; Spaulding and Burgess, 2017). Click-chemistry compatible analogs of puromycin have been developed (Liu et al., 2012) and used to label neuronal nascent proteins (Holt et al., 2019). A disadvantage of puromycilation is its interference with the translation machinery and the resulting truncated peptides (Schmidt et al., 2009) and recent evidence suggests that puromycilation may not be a good indicator of nascent proteins (Enam et al., 2020; Hobson et al., 2020). Puromycin-independent techniques have been developed as well.
Non-canonical amino acid tagging (NCAT) has emerged as a strategy for identifying nascent proteins without terminating translation. Bio-Orthogonal NCAT (BONCAT) and Fluorescent NCAT (FUNCAT) methodologies tag newly synthesized proteins with either azide- or alkyne-bearing non-canonical amino acids (NCAAs) (Kiick et al., 2002; Link et al., 2004; Yang et al., 2018). Using click chemistry, the azide or alkyne group of the NCAA can be clicked to biotin for purification and mass spectrometry analysis (BONCAT), or a fluorophore for imaging (FUNCAT) (Hinz et al., 2012; Ngo et al., 2013; Ullrich et al., 2014; Lehner et al., 2017). As a result of the low toxicity, higher concentrations of NCAAs and longer labelling periods can be used in vivo, increasing proteome coverage (Koren et al., 2019). Moreover, since NCAAs do not affect the rate or efficiency of protein translation (Calve et al., 2016), this method is much more suitable for examining protein synthesis during long-term memory formation.
Modifications to NCAAs allow for cell-type-specific tagging of newly synthesized proteins. For example, the NCAA azidonorleucine (ANL) is not recognized by the endogenous methionine tRNA synthetase and therefore does not integrate into proteins in wild-type cells. ANL incorporates only into newly synthesized proteins in cells expressing mutant methionyl-tRNA synthetase (MetRS) in which, in zebrafish, Leucine 270 is replaced with Glycine (MetRSL270G) (Shahar and Schuman, 2020). In mice, this technology has been successfully used to label nascent proteins in hippocampal excitatory principal neurons and cerebellar Purkinje neurons, to discover differentially regulated proteins in mice exposed to an enriched environment (Alvarez-Castelao et al., 2017) and to identify 156 proteins in hippocampal excitatory neurons during an aversive cue learning paradigm (Evans et al., 2020). We have used both BONCAT and FUNCAT in zebrafish larvae expressing cell-type-specific MetRSL270G in a pan-neuronal manner to label neuron-specific nascent protein, which revealed elevated levels of neuronal newly synthesized proteins following induced neuronal activity (Shahar and Schuman, 2020). FUNCAT provides spatial information and indicates the intensity of newly synthesized proteins. Moreover, a Proximity Ligation Assay (PLA) can be used in combination with FUNCAT (PLA-FUNCAT) to reveal the cellular location of nascent proteins-of-interest (Dieck et al., 2015; Evans et al., 2019). The PLA detects the spatial colocalization of two antibodies: one that identifies the newly synthesized protein by using click chemistry to the azide or alkyne group of the NCAA, and another that identifies a specific epitope in a protein of interest. Only when the two antibodies are in proximity, will a ligation amplification circle reaction occur, resulting in fluorescent signal (Dieck et al., 2015). The relative ease of creating transgenic zebrafish lines expressing mutant MetRS under glial and neuronal subtype promotors enables the exciting opportunity to identify cell-type-specific protein synthesis during long-term memory formation in vertebrates.
Thus, NCAT-based methods seem very promising for the analysis of protein-synthesis-dependent memory formation. It is important, however, to realize that strategies for NCAA delivery and labeling durations can affect experimental outcomes, which likely differs between species. While longer labelling periods enable the labeling of a large number of newly synthesized proteins, shorter labelling periods may be preferable for examining memory-phase-specific protein synthesis. In addition, labeling tools for nascent proteins have a bias towards proteins with a high turnover, because they are synthesized more regularly and thus more likely integrate the NCAA or puromycin (Schanzenbacher et al., 2016). Lastly, while these methods are able to identify the nascent proteome, it does not reveal their functionality. Their identity can be used for functional prediction, which can be further examined as next steps.
The characteristics of zebrafish, including their 1) ability to learn, 2) relative ease of genetic manipulation, 3) suitability for high-throughput studies, and 4) translucent brain in young ages, make it an excellent vertebrate model to study the molecular underpinnings of learning and memory. The collective data thus far suggest that larval stages are easier to work with due to the more accessible brain and translucency, and that complex learning such as long-term memory formation work better in juveniles, starting at about two to 3 weeks post-fertilization and becoming more robust in adults. The fact that the brain of young juveniles (2–4 weeks post fertilization) is smaller and more accessible than that of adult zebrafish, poises them as a promising tool for future research. Combining young juveniles with advanced, novel techniques, in particular labeling with NCAAs in a cell-type-specific manner, is relatively unexplored but starting to be used for examining the nascent proteome during various forms of learning and memory. We envision a rise in the use and further refinement of the zebrafish model, including applications of these novel techniques, which will hopefully increase the understanding of conserved mechanisms of long-term memory formation in vertebrates at the molecular level.
KR performed the literature screening, prepared the figures and wrote the manuscript. HS performed the literature screening; OS designed the study, edited the figures and wrote the manuscript.
MIGAL internal funds and startup package.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agetsuma, M., Aizawa, H., Aoki, T., Nakayama, R., Takahoko, M., Goto, M., et al. (2010). “The habenula is crucial for experience-dependent modification of fear responses in zebrafish”. Nat. Neurosci. 13 (11), 1354–1356. doi:10.1038/nn.2654
Agranoff, B. W., Davis, R. E., and Brink, J. J. (1966). “Chemical studies on memory fixation in goldfish.”. Brain Res. 1 (3), 303–309. doi:10.1016/0006-8993(66)90095-3
Ahmed, O., Seguin, D., and Gerlai, R. (2010). “An automated predator avoidance task in zebrafish”. Behav. Brain Res. 216 (1), 166–171. doi:10.1016/j.bbr.2010.07.028
Ahrens, M. B., Li, J. M., Orger, M. B., Robson, D. N., Schier, A. F., Engert, F., et al. (2012). “Brain-wide neuronal dynamics during motor adaptation in zebrafish”. Nature 485 (7399), 471–477. doi:10.1038/nature11057
Aizenberg, M., and Schuman, E. M. (2011). “Cerebellar-Dependent learning in larval zebrafish.”. J. Neurosci. 31 (24), 8708–8712. doi:10.1523/JNEUROSCI.6565-10.2011
Al-Imari, Lina, and Gerlai, Robert (2008). “Sight of conspecifics as reward in associative learning in zebrafish (Danio rerio).”. Behav. Brain Res. 189 (1), 216–219. doi:10.1016/j.bbr.2007.12.007
Andersson, M. Å., Ek, F., and Olsson, R. (2015). “Using visual lateralization to model learning and memory in zebrafish larvae.”. Sci. Rep. 5, 8667. doi:10.1038/SREP08667
Aoki, T., Kinoshita, M., Aoki, R., Agetsuma, M., Aizawa, H., Yamazaki, M., et al. (2013). Imaging of neural ensemble for the retrieval of a learned behavioral program. Neuron 78 (5), 881–894. doi:10.1016/j.neuron.2013.04.009
Arrenberg, A. B., Del Bene, F., and Baier, H. (2009). “Optical control of zebrafish behavior with halorhodopsin”. Proc. Natl. Acad. Sci. U. S. A. 106 (42), 17968–17973. doi:10.1073/pnas.0906252106
Baker, M. R., and Wong, R. Y. (2019). “Contextual fear learning and memory differ between stress coping styles in zebrafish.”. Sci. Rep. 9 (1), 9935. doi:10.1038/S41598-019-46319-0
Baratti, G., Potrich, D., and Sovrano, V. A. (2019). “The environmental geometry in spatial learning by zebrafish (Danio rerio)”. Zebrafish 17 (2), 131–138. doi:10.1089/zeb.2019.1845
Baratti, G., Rizzo, A., Elena Miletto Petrazzini, M., and Sovrano, V. A. (2021). “Learning by doing: The use of distance, corners and length in rewarded geometric tasks by zebrafish (Danio rerio).”. Animals 11 (7), 2001. doi:10.3390/ANI11072001
Barreiros, M. D. O., Gomes Barbosa, F., Dantas, D. D. O., Santos, D. d. M. L. d., Ribeiro, S., and Barros, A. K. (2021). “Zebrafish automatic monitoring system for conditioning and behavioral analysis.”. Sci. Rep. 11 (1), 9330. doi:10.1038/S41598-021-87502-6
Alvarez-Castelao, B., Schanzenbächer, C. T., Hanus, C., Glock, C., Dieck, S. T., Dörrbaum, A. R., et al. (2017). “Cell-Type-Specific metabolic labeling of nascent proteomes in vivo.”. Nat. Biotechnol. 35 (12), 1196–1201. doi:10.1038/nbt.4016
Bergmann, K., Santoscoy, P. M., Lygdas, K., Nikolaeva, Y., MacDonald, R. B., Cunliffe, V. T., et al. (2018). “Imaging neuronal activity in the optic tectum of late stage larval zebrafish.”. J. Dev. Biol. 6 (1), 6. doi:10.3390/JDB6010006
Best, J. D., Berghmans, S., Hunt, J. F. G., Clarke, S., Fleming, A., Goldsmith, P., et al. (2007). “Non-Associative learning in larval zebrafish.”. Neuropsychopharmacology 33 (5), 1206–1215. doi:10.1038/sj.npp.1301489
Bilotta, J., Risner, M. L., Davis, E. C., and Haggbloom, S. J. (2005). “Assessing appetitive choice discrimination learning in zebrafish”. Zebrafish 2 (4), 259–268. doi:10.1089/zeb.2005.2.259
Blank, M., Guerim, L. D., Cordeiro, R. F., and Vianna, M. R. M. (2009). “A one-trial inhibitory avoidance task to zebrafish: Rapid acquisition of an NMDA-dependent long-term memory”. Neurobiol. Learn. Mem. 92 (4), 529–534. doi:10.1016/j.nlm.2009.07.001
Braubach, O. R., Wood, H-D., Gadbois, S., Fine, A., and Croll, R. P. (2008). “Olfactory conditioning in the zebrafish (Danio rerio)”. Behav. Brain Res. 198 (1), 190–198. doi:10.1016/j.bbr.2008.10.044
Brinza, I., Eldahshan, O. A., Raey, M. A. E., Hritcu, L., El-Kashak, W., Eldahshan, O. A, et al. (2022), “Sweroside ameliorated memory deficits in scopolamine-induced zebrafish (Danio rerio) model: Involvement of cholinergic system and brain oxidative stress.” Molecules Vol. 27, (18): 5901. doi:10.3390/molecules27185901
Broglio, C., Rodríguez, F., Gómez, A., Arias, J. L., and Salas, C. (2010). “Selective involvement of the goldfish lateral pallium in spatial memory”. Behav. Brain Res. 210 (2), 191–201. doi:10.1016/j.bbr.2010.02.031
Bundschuh, S. T., Zhu, P., Zhang Schärer, Y. P., and Friedrich, R. W. (2012). “Dopaminergic modulation of mitral cells and odor responses in the zebrafish olfactory bulb.”. J. Neurosci. 32 (20), 6830–6840. doi:10.1523/JNEUROSCI.6026-11.2012
Calve, S., Witten, A. J., Ocken, A. R., and Kinzer-Ursem, T. L. (2016). “Incorporation of non-canonical amino acids into the developing murine proteome.”. Sci. Rep. 6 (1), 32377. doi:10.1038/srep32377
Camina, E., and Güell, F. (2017). “The neuroanatomical, neurophysiological and psychological basis of memory: Current models and their origins.”. Front. Pharmacol. 8, 438. doi:10.3389/fphar.2017.00438
Castro, M. R., Ventura Lima, J., Paula Salomão de Freitas, D., Valente, R. D. S., Seus Dummer, N., Borba de Aguiar, R., et al. (2009). “Behavioral and neurotoxic effects of arsenic exposure in zebrafish (Danio rerio, teleostei: Cyprinidae).”. Comp. Biochem. Physiology - C Toxicol. Pharmacol. 150 (3), 337–342. doi:10.1016/J.CBPC
Chen, X., Wei, S., Ji, Y., Guo, X., and Yang, F. (2015). “Quantitative proteomics using SILAC: Principles, applications, and developments”. proteomics 15 (18), 3175–3192. doi:10.1002/pmic.201500108
Cleal, M., Fontana, B. D., Ranson, D. C., McBride, S. D., Swinny, J. D., Redhead, E. S., et al. (2020). “The free-movement pattern Y-maze: A cross-species measure of working memory and executive function.”. Behav. Res. Methods 53 (2), 536–557. doi:10.3758/s13428-020-01452-x
Colwill, R. M., Raymond, M. P., Ferreira, L., and Escudero, H. (2005). “Visual discrimination learning in zebrafish (Danio rerio)”. Behav. Process. 70 (1), 19–31. doi:10.1016/j.beproc.2005.03.001
Costa-Mattioli, M., Sossin, W. S., Klann, E., and Sonenberg, N. (2008). “Translational control of long-lasting synaptic plasticity and memory.”. Neuron 61 (1), 10–26. doi:10.1016/j.neuron.2008.10.055
Del Bene, F., and Wyart, C. (2012). “Optogenetics: A new enlightenment age for zebrafish neurobiology”. Dev. Neurobiol. 72 (3), 404–414. doi:10.1002/dneu.20914
Dieck, S. T., Kochen, L., Hanus, C., Heumüller, M., Bartnik, I., Nassim-Assir, B., et al. (2015). “Direct visualization of newly synthesized target proteins in situ”. Nat. Methods 12 (5), 411–414. doi:10.1038/nmeth.3319
Douglass, A. D., Kraves, S., Deisseroth, K., Schier, A. F., and Engert, F. (2008). “Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons”. Curr. Biol. CB 18 (15), 1133–1137. doi:10.1016/j.cub.2008.06.077
Doyle, J. M., Merovitch, N., Wyeth, R. C., Stoyek, M. R., Schmidt, M., Wilfart, F., et al. (2016). “A simple automated system for appetitive conditioning of zebrafish in their home tanks”. Behav. Brain Res. 317, 444–452. doi:10.1016/j.bbr.2016.09.044
Dreosti, E., Wilson, S. W., Lopes, G., and Kampff, A. R. (2015). “Development of social behavior in young zebrafish.”. Front. Neural Circuits 9, 39–9. doi:10.3389/fncir.2015.00039
Eaton, R. C., Farley, R. D., Kimmel, C. B., and Schabtach, E. (1977). “Functional development in the mauthner cell system of embryos and larvae of the zebra fish.”. J. Neurobiol. 8 (2), 151–172. doi:10.1002/neu.480080207
Enam, S. U., Zinshteyn, B., Goldman, D. H., Cassani, M., Livingston, N. M., Seydoux, G., et al. (2020). “Puromycin reactivity does not accurately localize translation at the subcellular level.”. ELife 9, 1–34. doi:10.7554/ELIFE.60303
Engmann, O., Campbell, J., Ward, M., Peter Giese, K., and Thompson, A. J. (2010). “Comparison of a protein-level and peptide-level labeling strategy for quantitative proteomics of synaptosomes using isobaric tags.”. J. Proteome Res. 9 (5), 2725–2733. doi:10.1021/pr900627e_FILE/PR900627E_SI_004.PDF
Evans, H. T., Benetatos, J., Roijen, M. V., Bodea, L-G., and Götz, J. (2019). “Decreased synthesis of ribosomal proteins in tauopathy revealed by non-canonical amino acid labelling.”. EMBO J. 38 (13), 101174. doi:10.15252/EMBJ.2018101174
Evans, H. T., Gabriel Bodea, L., and Götz, J. (2020). “Cell-Specific non-canonical amino acid labelling identifies changes in the de Novo proteome during memory formation.”. ELife 9, 1–19. doi:10.7554/eLife.52990
Evans, T. H., Blackmore, D., Götz, J., and Bodea, L. G. (2021). “De novo proteomic methods for examining the molecular mechanisms underpinning long-term memory.”. Brain Res. Bull. 169, 94–103. doi:10.1016/j.brainresbull.2020.12.015
Fajardo, O., Zhu, P., and Friedrich, R. W. (2013). “Control of a specific motor program by a small brain area in zebrafish.”. Front. Neural Circuits 7, 67. doi:10.3389/FNCIR.2013.00067
Farley, J., and Alkon, D. L. (1985). “Cellular mechanisms of learning, memory, and information storage.”. Annu. Rev. Psychol. 36, 419–494. doi:10.1146/annurev.ps.36.020185.002223
Fernandes, Y. M., Rampersad, M., Luchiari, A. C., and Gerlai, R. (2016). “Associative learning in the multichamber tank: A new learning paradigm for zebrafish”. Behav. Brain Res. 312, 279–284. doi:10.1016/j.bbr.2016.06.038
Flexner, J. B., Flexner, L. B., and Stellar, E. (1963). “Memory in mice as affected by intracerebral puromycin.”. Science 141 (3575), 57–59. doi:10.1126/science.141.3575.57
Flexner, J. B., Flexner, L. B., Stellar, E., and de la Haba, G. (1962). “Inhibition of protein synthesis in brain and learning and memory following puromycin*.”. J. Neurochem. 9 (6), 595–605. doi:10.1111/j.1471-4159.1962.tb04216.x
Fontana, B. D., Gibbon, A. J., Cleal, M., Sudwarts, A., Pritchett, D., Elena Miletto Petrazzini, M., et al. (2021). “Moderate early life stress improves adult zebrafish (Danio rerio) working memory but does not affect social and anxiety-like responses”. Dev. Psychobiol. 63 (1), 54–64. doi:10.1002/dev.21986
Gal-Ben-Ari, S., Kenney, J. W., Ounalla-Saad, H., Taha, E., David, O., Levitan, D., et al. (2012). “Consolidation and translation regulation.”. Learn. Mem. 19 (9), 410. doi:10.1101/LM.026849.112
Gal-Ben-Ari, S., and Rosenblum, K. (2011). “Molecular mechanisms underlying memory consolidation of taste information in the cortex.”. Front. Behav. Neurosci. 5, 87. doi:10.3389/FNBEH
Gaspary, K. V., Kellermann Reolon, G., Gusso, D., and Bonan, C. D. (2018). “Novel object recognition and object location tasks in zebrafish: Influence of habituation and NMDA receptor antagonism.”. Neurobiol. Learn. Mem. 155, 249–260. doi:10.1016/j.nlm.2018.08.005
Gatto, E., Lucon-Xiccato, T., Bisazza, A., Manabe, K., and Dadda, M. (2020). “The devil is in the detail: Zebrafish learn to discriminate visual stimuli only if salient.”. Behav. Process. 179, 104215. doi:10.1016/J.BEPROC.2020.104215
Gau, P., Poon, J., Ufret-Vincenty, C., Snelson, C. D., Gordon, S. E., Raible, D. W., et al. (2013). “The zebrafish ortholog of TRPV1 is required for heat-induced locomotion.”. J. Neurosci. 33 (12), 5249–5260. doi:10.1523/JNEUROSCI.5403-12.2013
Gerlai, R. (2020). Evolutionary conservation, translational relevance and cognitive function: The future of zebrafish in behavioral neuroscience. Neurosci. Biobehav. Rev. 116, 426–435. doi:10.1016/j.neubiorev.2020.07.009
Gerlai, R. (2016). Learning and memory in zebrafish (Danio rerio). Methods Cell Biol. 134, 551. doi:10.1016/bs.mcb.2016.02.005
Gómez-Laplaza, L. M., and Gerlai, R. (2009). Latent learning in zebrafish (Danio rerio). Behav. Brain Res. 208 (2), 509–515. doi:10.1016/j.bbr.2009.12.031
Gould, N. L., Sharma, V., Hleihil, M., Kolatt Chandran, S., David, O., Edry, E., et al. (2020). “Dopamine-Dependent QR2 pathway activation in CA1 interneurons enhances novel memory formation.”. J. Neurosci. 40 (45), 8698–8714. doi:10.1523/JNEUROSCI.1243-20.2020
Granato, M., and Nüsslein-Volhard, C. (1996). “Fishing for genes controlling development.”. Curr. Opin. Genet. Dev. 6 (4), 461–468. doi:10.1016/s0959-437x(96)80068-2
Hafner, A-S., Donlin-Asp, P. G., Leitch, B., Herzog, E., and Schuman, E. M. (2019). “Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments.” Science 364(6441)3644. doi:10.1126/SCIENCE.AAU3644
Haesemeyer, M., Robson, D. N., Li, J. M., Schier, A. F., and Engert, F. (2015). “The structure and timescales of heat perception in larval zebrafish.”. Cell Syst. 1 (5), 338–348. doi:10.1016/j.cels.2015.10.010
Harmon, T. C., Magaram, U., McLean, D. L., and Raman, I. M. (2017). “Distinct responses of Purkinje neurons and roles of simple spikes during associative motor learning in larval zebrafish.”. ELife 6, 22537. doi:10.7554/ELIFE.22537
Harris, J. D. (1943). “Habituatory response decrement in the intact organism.”. Psychol. Bull. 40 (6), 385–422. doi:10.1037/h0053918
Heumüller, M., Glock, C., Rangaraju, V., Biever, A., and Schuman, E. M. (2019). “A genetically encodable cell-type-specific protein synthesis inhibitor.”. Nat. Methods 16 (8), 699–702. doi:10.1038/s41592-019-0468-x
Hinz, F. I., Aizenberg, M., Tushev, G. E., and Schuman, E. M. (2013). “Protein synthesis-dependent associative long-term memory in larval zebrafish.”. J. Neurosci. 33 (39), 15382–15387. doi:10.1523/JNEUROSCI.0560-13.2013
Hinz, F. I., Dieterich, D. C., Tirrell, D. A., and Schuman, E. M. (2012). Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem. Neurosci. 3 (1), 40–49. doi:10.1021/cn2000876
Hobson, B. D., Kong, L., Hartwick, E. W., Gonzalez, R. L., and Sims, P. A. (2020). “Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes.”. ELife 9, 1–19. doi:10.7554/eLife.60048
Holt, C. E., Martin, K. C., and Schuman, E. M. (2019). “Local translation in neurons: Visualization and function.”. Nat. Struct. Mol. Biol. 26 (7), 557–566. doi:10.1038/s41594-019-0263-5
Ioannou, A., and Anastassiou-Hadjicharalambous, X. (2021). “Non-Associative learning.”. Encycl. Evol. Psychol. Sci., 5419–5432. doi:10.1007/978-3-319-19650-3_1027
Kandel, E. R., Dudai, Y., and Mayford, M. R. (2014). The molecular and systems biology of memory. Cell 157 (1), 163–186. doi:10.1016/j.cell.2014.03.001
Kareklas, K., Elwood, R. W., and Holland, R. A. (2018). “Fish learn collectively, but groups with differing personalities are slower to decide and more likely to split.” Biol. Open 7(5)033613. doi:10.1242/BIO.033613
Karlsson, J., Von Hofsten, J., and Olsson, P. E. (2001). “Generating transparent zebrafish: A refined method to improve detection of gene expression during embryonic development.”. Mar. Biotechnol. 3 (6), 522–527. doi:10.1007/s1012601-0053-4
Karnik, I., and Gerlai, R. (2012). “Can zebrafish learn spatial tasks? An empirical analysis of place and single CS-US associative learning.”. Behav. Brain Res. 233 (2), 415–421. doi:10.1016/j.bbr.2012.05.024
Kawashima, T., Ahrens, M. B., Maarten, F. Z., Yang, C-T., Mensh, B. D., and Ahrens, M. B. (2016). “The serotonergic system tracks the outcomes of actions to mediate short-term motor learning”. Cell 167 (4), 933–946. doi:10.1016/j.cell.2016.09.055
Kelleher, R. J., Govindarajan, A., Yoon Jung, H., Kang, H., and Tonegawa, S. (2004). “Translational control by MAPK signaling in long-term synaptic plasticity and memory.”. Cell 116 (3), 467–479. doi:10.1016/s0092-8674(04)00115-1
Kenney, J. W. (2020). ““Associative and nonassociative learning in adult zebrafish.”,” in Behavioral and neural genetics of zebrafish (Elsevier), 187–204. doi:10.1016/B978-0-12-817528-6.00012-7
Kiick, K. L., Saxon, E., Tirrell, D. A., and Bertozzi, C. R. (2002). “Incorporation of azides into recombinant proteins for chemoselective modification by the staudinger ligation.”. Proc. Natl. Acad. Sci. U. S. A. 99 (1), 19–24. doi:10.1073/pnas.012583299
Koren, S. A., Gillett, D. A., D’alton, S. V., Hamm, M. J., and Abisambra, J. F. (2019). “Proteomic techniques to examine neuronal translational dynamics.” Int. J. Mol. Sci. 3524 20(14): 3524. doi:10.3390/ijms20143524
Lal, P., Tanabe, H., Maximiliano, L. S., Ailani, D., Kotani, Y., Muto, A., et al. (2018). “Identification of a neuronal population in the telencephalon essential for fear conditioning in zebrafish.”. BMC Biol. 16 (1), 45. doi:10.1186/S12915-018-0502-Y
Langebeck-Jensen, K., Shahar, O. D., Schuman, E. M., Langer, J. D., and Ryu, S. (2019). “Larval zebrafish proteome regulation in response to an environmental challenge.”. PROTEOMICS 19 (14), 1900028. doi:10.1002/pmic.201900028
Lee, A., Korzh, V., Penney, T. B., Ajay, S. M., Teh, C., Kibat, C., et al. (2010). “The habenula prevents helpless behavior in larval zebrafish.”. Curr. Biol. CB 20 (24), 2211–2216. doi:10.1016/j.cub.2010.11.025
Lehner, F., Kudlinzki, D., Richter, C., Müller-Werkmeister, H. M., Eberl, K. B., Bredenbeck, J., et al. (2017). “Impact of azidohomoalanine incorporation on protein structure and ligand binding.”. ChemBioChem 18 (23), 2340–2350. doi:10.1002/cbic.201700437
Levin, E. D., and Chen, E. (2004). Nicotinic involvement in memory function in zebrafish. Neurotoxicology Teratol. 26 (6), 731–735. doi:10.1016/j.ntt.2004.06.010
Levin, E. D., Chrysanthis, E., Yacisin, K., and Linney, E. (2003). “Chlorpyrifos exposure of developing zebrafish: Effects on survival and long-term effects on response latency and spatial discrimination.”. Neurotoxicology Teratol. 25 (1), 51–57. doi:10.1016/s0892-0362(02)00322-7
Lin, Q., Robson, D. N., Manley, J., Helmreich, M., Schlumm, F., Li, J. M., et al. (2019). “Cerebellar neurodynamics predict decision timing and outcome on the single-trial level”. Cell 180 (3), 536–551. doi:10.1016/j.cell.2019.12.018
Link, A. J., Vink, M. K. S., and Tirrell, D. A. (2004). “Presentation and detection of azide functionality in bacterial cell surface proteins.”. J. Am. Chem. Soc. 126 (34), 10598–10602. doi:10.1021/ja047629c
Liu, J., Xu, Y., Stoleru, D., and Salic, A. (2012). “Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin.”. Proc. Natl. Acad. Sci. U. S. A. 109 (2), 413–418. doi:10.1073/pnas.1111561108
Lucon-Xiccato, T., and Dadda, M. (2014). “Assessing memory in zebrafish using the one-trial test”. Behav. Process. 106, 1–4. doi:10.1016/j.beproc.2014.03.010
Madeira, N., and Oliveira, R. F. (2017). “Long-Term social recognition memory in zebrafish”. Zebrafish 14 (4), 305–310. doi:10.1089/zeb.2017.1430
Manabe, K., Dooling, R. J., and Takaku, S. (2013b). “An automated device for appetitive conditioning in zebrafish (Danio rerio).”. Zebrafish 10 (4), 518–523. doi:10.1089/ZEB.2012.0776
Manabe, K., Dooling, R. J., and Takaku, S. (2013a). “Differential reinforcement of an approach response in zebrafish (Danio rerio)”. Behav. Process. 98, 106–111. doi:10.1016/j.beproc.2013.05.013
Marques, J. C., Lackner, S., Félix, R., and Orger, M. B. (2017). Structure of the zebrafish locomotor repertoire revealed with unsupervised behavioral clustering. Curr. Biol. CB 28 (2), 181–195. doi:10.1016/j.cub.2017.12.002
May, Z., Morrill, A., Holcombe, A., Johnston, T., Gallup, J., Fouad, K., et al. (2015). Object recognition memory in zebrafish. Behav. Brain Res. 296, 199–210. doi:10.1016/j.bbr.2015.09.016
Mazzitelli-Fuentes, L. S., Román, F. R., Julio, R., Elías, C., Deleglise, E. B., and Mongiat, L. A. (2022). “Spatial learning promotes adult neurogenesis in specific regions of the zebrafish pallium.”. Front. Cell Dev. Biol. 10, 1. doi:10.3389/FCELL.2022.840964
Mu, Y., Ahrens, M. B., Narayan, S., and Mensh, B. D. (2020). Brain-wide, scale-wide physiology underlying behavioral flexibility in zebrafish. Curr. Opin. Neurobiol. 64, 151–160. doi:10.1016/j.conb.2020.08.013
Mueller, T. (2012). “What is the thalamus in zebrafish?”. Front. Neurosci. 6, 64. doi:10.3389/fnins.2012.00064
Namekawa, I., Moenig, N. R., and Friedrich, R. W. (2018). “Rapid olfactory discrimination learning in adult zebrafish.”. Exp. Brain Res. 236 (11), 2959–2969. doi:10.1007/s00221-018-5352-x
Naumann, E. A., Fitzgerald, J. E., Dunn, T. W., Rihel, J., Sompolinsky, H., and Engert, F. (2016). From whole-brain data to functional circuit models: The zebrafish optomotor response. Cell 167 (4), 947–960. doi:10.1016/j.cell.2016.10.019
Nee, D. E., Berman, M. G., Sledge Moore, K., and Jonides, J. (2008). “Neuroscientific evidence about the distinction between short- and long-term memory.”. Curr. Dir. Psychol. Sci. 17 (2), 102–106. doi:10.1111/j.1467-8721.2008.00557.x
Nelson, J. C., and Granato, M. (2022). “Zebrafish behavior as a gateway to nervous system assembly and plasticity.”. Development 149 (9), 177998. doi:10.1242/DEV.177998/275367
Nemoto, N., Miyamoto-Sato, E., and Yanagawa, H. (1999). “Fluorescence labeling of the C-terminus of proteins with a puromycin analogue in cell-free translation systems.”. FEBS Lett. 462 (1–2), 43–46. doi:10.1016/s0014-5793(99)01474-x
Ngo, J. T., Schuman, E. M., and Tirrell, D. A. (2013). “Mutant methionyl-TRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells.”. Proc. Natl. Acad. Sci. U. S. A. 110 (13), 4992–4997. doi:10.1073/pnas.1216375110_FILE/SD01.PDF
Oliveira, J., Silveira, M., Chacon, D., and Luchiari, A. (2014). “The zebrafish world of colors and shapes: Preference and discrimination”. Zebrafish 12 (2), 166–173. doi:10.1089/zeb.2014.1019
Palumbo, F., Serneels, B., Pelgrims, R., and Yaksi, E. (2020). “The zebrafish dorsolateral habenula is required for updating learned behaviors.”. Cell Rep. 32 (8), 108054. doi:10.1016/J.CELREP
Pavlov, I. P. (1951). “Conditioned reflex.”. Feldsher Akusherka 10, 6–12. https://pubmed.ncbi.nlm.nih.gov/14887723/.
Pereira, S., and Kooy, D. V. D. (2013). “Entwined engrams: The evolution of associative and non-associative learning”. Worm 2 (2), e22725. doi:10.4161/worm.22725
Poon, C-S., and Schmid, S. (2012). “Nonassociative learning.”. Encycl. Sci. Learn., 2475–2477. doi:10.1007/978-1-4419-1428-6_1849
Porter, B. A., and Mueller, T (2020). “The zebrafish amygdaloid complex – functional ground plan, molecular delineation, and everted topology.”. Front. Neurosci. 14, 00608. doi:10.3389/FNINS.2020.00608
Portugues, R., Severi, K. E., Wyart, C., and Ahrens, M. B. (2013). “Optogenetics in a transparent animal: Circuit function in the larval zebrafish.”. Curr. Opin. Neurobiol. 23 (1), 119–126. doi:10.1016/j.conb.2012.11.001
Pradel, G., Schmidt, R., and Schachner, M. (2000). “Involvement of L1.1 in memory consolidation after active avoidance conditioning in zebrafish.”. J. Neurobiol. 43 (4), 389–403. doi:10.1002/1097-4695(20000615)43:4<389::aid-neu7>3.0.co;2-x
Pradel, G., Schachner, M., and Schmidt, R. (1998). “Inhibition of memory consolidation by antibodies against cell adhesion molecules after active avoidance conditioning in zebrafish.”. J. Neurobiol. 39 (2), 197–206. https://onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291097-4695%28199905%2939%3A2%3C197%3A%3AAID-NEU4%3E3.0.CO%3B2-9#references-section.
Price, J. C., Guan, S., Burlingame, A., Prusiner, S. B., and Ghaemmaghami, S. (2010). “Analysis of proteome dynamics in the mouse brain.”. Proc. Natl. Acad. Sci. U. S. A. 107 (32), 14508–14513. doi:10.1073/pnas.1006551107
Prober, D. A., Zimmerman, S., Myers, B. R., McDermott, B. M., Hyung Kim, S., Sophie Caron, J. R., et al. (2008). “Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function.”. J. Neurosci. 28 (40), 10102–10110. doi:10.1523/JNEUROSCI.2740-08.2008
Randlett, O., Schier, A. F., Engert, F., Haesemeyer, M., Forkin, G., Shoenhard, H., et al. (2019). “Distributed plasticity drives visual habituation learning in larval zebrafish.”. Curr. Biol. CB 29 (8), 1337–1345. doi:10.1016/j.cub.2019.02.039
Rauniyar, N., McClatchy, D. B., and Yates, J. R. (2013). Stable isotope labeling of mammals (SILAM) for in vivo quantitative proteomic analysis”. Methods 61 (3), 260–268. doi:10.1016/j.ymeth.2013.03.008
Rehman, I., Mahabadi, N., Sanvictores, T., and Rehman, C. I. (2022). “Classical conditioning.”. Encycl. Hum. Behav., 484–491. Second Edition, August. doi:10.1016/B978-0-12-375000-6.00090-2
Risner, M. L., Lemerise, E., Vukmanic, E. V., and Moore, A. (2005). “Behavioral spectral sensitivity of the zebrafish (Danio rerio).”. Vis. Res. 46 (17), 2625–2635. doi:10.1016/j.visres.2005.12.014
Roberts, A. C., Bill, B. R., and Glanzman, D. L. (2013). “Learning and memory in zebrafish larvae.”. Front. Neural Circuits 7, 126. doi:10.3389/fncir.2013.00126
Roberts, A. C., Pearce, K. C., Choe, R. C., Alzagatiti, J. B., Yeung, A. K., Bill, B. R, et al. (2016). “Long-Term habituation of the C-start escape response in zebrafish larvae.” Neurobiol. Learn. Mem. 134 Pt B (Pt B): 360–368. doi:10.1016/J.NLM.2016.08.014
Ross, A. B., David Langer, J., and Jovanovic, M. (2021). “Proteome turnover in the spotlight: Approaches, applications, and perspectives.”. Mol. Cell. Proteomics 20, 100016. doi:10.1074/MCP.R120.002190
Saint-Amant, L., and Drapeau, P. (1998). “Time course of the development of motor behaviors in the zebrafish embryo”. J. Neurobiol. 37 (4), 622–632. doi:10.1002/(sici)1097-4695(199812)37:4<622::aid-neu10>3.0.co;2-s
Sakers, K., Dani, A., Lake, A. M., Khazanchi, R., Ouwenga, R., Vasek, M. J., et al. (2017). “Astrocytes locally translate transcripts in their peripheral processes.”. Proc. Natl. Acad. Sci. U. S. A. 114 (19), E3830–E3838. doi:10.1073/pnas.1617782114
Samaras, A., and Pavlidis, M. (2020). “Behavioural and physiological responses to a conditioning protocol for adult zebrafish, Danio rerio, held in groups.”. Behav. Process. 179, 104201. doi:10.1016/J.BEPROC.2020.104201
Santacà, M., Dadda, M., Dalla Valle, L., Fontana, C., Gjinaj, G., and Bisazza, A. (2022). “Learning and visual discrimination in newly hatched zebrafish.”. IScience 25 (5), 104283. doi:10.1016/J.ISCI.2022.104283
Santacà, M., Dadda, M., Elena Miletto Petrazzini, M., and Bisazza, A. (2021). “Learning bias and visual discrimination in zebrafish (Danio rerio)”. Behav. Process. 192, 104499. doi:10.1016/j.beproc.2021.104499
Santistevan, N. J., Nelson, J. C., Ortiz, E. A., Miller, A. H., Kenj Halabi, D., Sippl, Z. A., et al. (2022). “Cacna2d3, a voltage-gated calcium channel subunit, functions in vertebrate habituation learning and the startle sensitivity threshold.”. PloS One 17 (7), 0270903. doi:10.1371/JOURNAL.PONE.0270903
Schanzenbächer, C. T., Sambandan, S., Langer, J. D., and Schuman, E. M. (2016). “Nascent proteome remodeling following homeostatic scaling at hippocampal synapses”. Neuron 92 (2), 358–371. doi:10.1016/j.neuron.2016.09.058
Schmidt, E. K., Clavarino, G., Ceppi, M., and Pierre, P. (2009). “SUnSET, a nonradioactive method to monitor protein synthesis”. Nat. Methods 6 (4), 275–277. doi:10.1038/nmeth.1314
Shahar, O. D., and Schuman, E. M. (2020). “Large-Scale cell-type-specific imaging of protein synthesis in a vertebrate brain.”. ELife 9, 1–14. doi:10.7554/eLife.50564
Sharma, V., Sood, R., Khlaifia, A., Javad Eslamizade, M., Yu Hung, T., Lou, D., et al. (2020). “EIF2α controls memory consolidation via excitatory and somatostatin neurons.”. Nature 586 (7829), 412–416. doi:10.1038/s41586-020-2805-8
Shrestha, P., LeDoux, J. E., Ayata, P., Herrero-Vidal, P., Longo, F., Gastone, A., et al. (2020). “Cell-Type-Specific drug-inducible protein synthesis inhibition demonstrates that memory consolidation requires rapid neuronal translation.”. Nat. Neurosci. 23 (2), 281–292. doi:10.1038/s41593-019-0568-z
Sison, M., and Gerlai, R. (2009). “Associative learning in zebrafish (Danio rerio) in the plus maze”. Behav. Brain Res. 207 (1), 99–104. doi:10.1016/j.bbr.2009.09.043
Sison, M., and Gerlai, R. (2011). “Associative learning performance is impaired in zebrafish (Danio rerio) by the NMDA-R antagonist MK-801.”. Neurobiol. Learn. Mem. 96 (2), 230–237. doi:10.1016/j.nlm.2011.04.016
Sokolov, E. N. (1963). “Higher nervous functions; the orienting reflex”. Annu. Rev. Physiology 25, 545–580. doi:10.1146/annurev.ph.25.030163.002553
Sovrano, V. A., Vicidomini, S., Potrich, D., Elena Miletto Petrazzini, M., Baratti, G., and Rosa-Salva, O. (2022). “Visual discrimination and amodal completion in zebrafish.”. PloS One 17 (3). doi:10.1371/JOURNAL.PONE.0264127
Spaulding, E. L., and Burgess, R. W. (2017). “Accumulating evidence for axonal translation in neuronal homeostasis”. Front. Neurosci. 11, 312. doi:10.3389/fnins.2017.00312
Stewart, A. M., Gerlai, R., and Kalueff, A. V. (2015). “Developing HighER-throughput zebrafish screens for in-vivo CNS drug discovery.”. Front. Behav. Neurosci. 9, 00014. doi:10.3389/FNBEH.2015.00014
Sutton, M. A., and Schuman, E. M. (2006). “Dendritic protein synthesis, synaptic plasticity”. Cell 127 (1), 49–58. doi:10.1016/j.cell.2006.09.014
Szobota, S., Wyart, C., Fortin, D. L., Kolstad, K. D., Gorostiza, P., Del Bene, F., et al. (2007). “Remote control of neuronal activity with a light-gated glutamate receptor”. Neuron 54 (4), 535–545. doi:10.1016/j.neuron.2007.05.010
Thompson, R. F., and Spencer, W. A. (1966). “Habituation: A model phenomenon for the study of neuronal substrates of behavior”. Psychol. Rev. 73 (1), 16–43. doi:10.1037/h0022681
Ullrich, M., Liang, V., Lian Chew, Y., Banister, S., Song, X., Zaw, T., et al. (2014). “Bio-Orthogonal labeling as a tool to visualize and identify newly synthesized proteins in Caenorhabditis elegans.”. Nat. Protoc. 9 (9), 2237–2255. doi:10.1038/nprot.2014.150
Valente, A., Huang, K-H., Portugues, R., and Engert, F. (2012). “Ontogeny of classical and operant learning behaviors in zebrafish.”. Learn. Mem. 19 (4), 170–177. doi:10.1101/lm.025668.112
von Trotha, J. W., Vernier, P., and Bally-Cuif, L. (2014). “Emotions and motivated behavior converge on an amygdala-like structure in the zebrafish.”. Eur. J. Neurosci. 40 (9), 3302–3315. doi:10.1111/ejn.12692
Williams, F. E., White, D., and Messer, W. S. (2002). “A simple spatial alternation task for assessing memory function in zebrafish.”. Behav. Process. 58 (3), 125–132. doi:10.1016/s0376-6357(02)00025-6
Wolman, M. A., Jain, R. A., Liss, L., and Granato, M. (2011). “Chemical modulation of memory formation in larval zebrafish.”. Proc. Natl. Acad. Sci. U. S. A. 108 (37), 15468–15473. doi:10.1073/pnas.1107156108
Wyart, C., Baier, H., Del Bene, F., Warp, E., Scott, E. K., Trauner, D., et al. (2009). “Optogenetic dissection of a behavioural module in the vertebrate spinal cord.”. Nature 461 (7262), 407–410. doi:10.1038/nature08323
Wyart, C., and Del Bene, F. (2011). “Let there Be light: Zebrafish neurobiology and the optogenetic revolution.”. Rev. Neurosci. 22 (1), 121–130. doi:10.1515/RNS.2011.013
Xu, Z-X., Kim, G. H., Tan, J-W., Riso, A. E., Sun, Y., Xu, E. Y., et al. (2020). “Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations.”. Nat. Commun. 11 (1), 1–17. doi:10.1038/s41467-020-15530-3
Xu, J., Casanave, R., and Guo, S. (2021). “Larval zebrafish display dynamic learning of aversive stimuli in a constant visual surrounding.”. Learn. Mem. 28 (7), 228–238. doi:10.1101/lm.053425.121
Xu, X., Scott-Scheiern, T., Kempker, L., and Simons, K. (2006). “Active avoidance conditioning in zebrafish (Danio rerio).”. Neurobiol. Learn. Mem. 87 (1), 72–77. doi:10.1016/j.nlm.2006.06.002
Yang, A. C., Du Bois, H., Olsson, N., Gate, D., Lehallier, B., Berdnik, D., et al. (2018). “Multiple click-selective TRNA synthetases expand mammalian cell-specific proteomics.”. J. Am. Chem. Soc. 140 (23), 7046–7051. doi:10.1021/jacs.8b03074
Keywords: zebrafish, memory consolidation, protein synthesis, memory, long-term memory, learning, translation
Citation: Reemst K, Shahin H and Shahar OD (2023) Learning and memory formation in zebrafish: Protein dynamics and molecular tools. Front. Cell Dev. Biol. 11:1120984. doi: 10.3389/fcell.2023.1120984
Received: 10 December 2022; Accepted: 20 February 2023;
Published: 09 March 2023.
Edited by:
Yaniv M. Elkouby, Hebrew University of Jerusalem, IsraelReviewed by:
Thomas Mueller, Kansas State University, United StatesCopyright © 2023 Reemst, Shahin and Shahar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Or David Shahar, b3JAbWlnYWwub3JnLmls
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.