- 1Whitney Laboratory for Marine Biosciences, University of Florida, St. Augustine, FL, United States
- 2Departments of Neuroscience and McKnight Brain Institute, University of Florida, Gainesville, FL, United States
Transposable elements (TE) could serve as sources of new transcription factors (TFs) in plants and some other model species, but such evidence is lacking for most animal lineages. Here, we discovered multiple independent co-options of TEs to generate 788 TFs across Metazoa, including all early-branching animal lineages. Six of ten superfamilies of DNA transposon-derived conserved TF families (ZBED, CENPB, FHY3, HTH-Psq, THAP, and FLYWCH) were identified across nine phyla encompassing the entire metazoan phylogeny. The most extensive convergent domestication of potentially TE-derived TFs occurred in the hydroid polyps, polychaete worms, cephalopods, oysters, and sea slugs. Phylogenetic reconstructions showed species-specific clustering and lineage-specific expansion; none of the identified TE-derived TFs revealed homologs in their closest neighbors. Together, our study established a framework for categorizing TE-derived TFs and informing the origins of novel genes across phyla.
1 Introduction
Transposable elements (TEs) or transposons identified by Barbara McClintock during the 1940-the 50s are now recognized as pivotal regulatory elements (Biemont and Vieira, 2006) controlling roughly 25% of the human genes (Jordan et al., 2003). TEs are also major constituents of all eukaryotic genomes, frequently occupying from 20% to more than 70% of genomes. The inherent ability of TEs to self-replicate, move and mutate transformed the initial assessment of TEs as “selfish gene” parasites and “junk DNA” into powerful evolutionary forces (Miller et al., 1999). The process of genomic integration of TE and thus generating or expanding cis-regulatory elements, genes, and other elements such as micro (microRNAs) or non-coding RNAs (ncRNAs) followed by suppression of parasitic self-propagation properties is called molecular domestication or exaptation (Gould and Vrba, 1982; Miller et al., 1999; Volff, 2006).
A domesticated TE-derived gene regulator can benefit the host and be an adaptive advantage (Miller et al., 1999; Biemont and Vieira, 2006; Volff, 2006; Feschotte and Pritham, 2007). The TE-associated domestication events can be sources of novel genes (Miller et al., 1999), ncRNAs, microRNAs, etc., (Borchert et al., 2011; Li et al., 2011; Chuong et al., 2013; Henaff et al., 2014; Zhang et al., 2016). There are multiple examples of such beneficial domestication events, and the scope of this process is expanding with sequenced genomes (Miller et al., 1999; Jordan et al., 2003; Volff, 2006; Feschotte and Pritham, 2007; Koonin et al., 2020; Sundaram and Wysocka, 2020). There are also examples of convergent domestication, reflecting TE’s nature (Casola et al., 2008; Mateo and Gonzalez, 2014). For example, the emergence of the placenta from the TE-derived Syncytin gene in mammals and lizards occurred through two independent occurrences of TE domestication; it is portrayed as a classic example of convergent evolution (Miller et al., 1999; Lavialle et al., 2013; Cornelis et al., 2017).
Perhaps, the most critical domestication episodes associated with the rise of biological novelties are the recruitments of TEs in the evolution of transcription factors (TFs). TFs are known to be master regulators of gene expression across Metazoa (Lewis, 1978; Gehring, 1996), including body patterning (Pearson et al., 2005; Peter and Davidson, 2011) and cell fate commitment (Lin et al., 2010; Vervoort and Ledent, 2001). Mechanisms of the origins and lineage-specific TF gene expansion are primarily unknown. A classical hypothesis implies ancestral TF gene duplication, followed by the divergence of the duplicated gene (Ohno et al., 1968). However, this scenario does not apply to the TFs that are solely organism-specific and have no bona fide one-to-one orthologs in closest relatives.
The complementary scenario is the origin of TFs and the novel TF-binding sites with the contribution of TEs. DNA-binding properties of TEs, in particular the evidence that TEs contain TF-binding sites, perfectly match structural genome constraints as a potential “pre-adaptation” and sources to form novel cis-regulatory elements and TFs. Thus, incorporating non-coding and new TF genes into existing transcriptional networks (Sundaram and Wysocka, 2020) can also lead to the origins of new functions and transformative biological innovations, as well as the diversification of both genes and forms.
The most notable examples of TE-derived TFs came from plants (Lin et al., 2007; Henaff et al., 2014) and such model animal species as insects, e.g., Drosophila (Miller et al., 1999; Casola et al., 2007; Mateo and Gonzalez, 2014) or vertebrates (Hammer et al., 2005; Cayrol et al., 2007; Balakrishnan et al., 2009; Markljung et al., 2009; Hayward et al., 2013; Majumdar et al., 2013). However, the broad comparative scope of these events is less explored, with little knowledge about the majority of animal phyla.
Practically nothing is known about the most diverse bilaterian lineage–Lophotrochozoa. This clade consists of more than a dozen phyla (Kocot et al., 2017), including Mollusca—the second most species-rich phylum and one of the most diverse groups of animals (Ponder and Linderg, 2008). The evidence of TE domestication events outside Bilateria in four other basal metazoan lineages (Ctenophora, Porifera, Placozoa, and Cnidaria) is also lacking.
Here, we generated a catalog of potentially TE-derived TFs across Metazoa and proposed independent co-option of six out of ten superfamilies of TEs to create hundreds of TFs in all early-branching animal lineages.
2 Results and discussion
1. Mosaic distribution and parallel evolution of transposon-derived transcription factors across metazoans
Using tblastn searches against target genomes we first identified and curated a complete dataset of transcription factors (TFs) encoded in representatives of four animal phyla with the sequenced genomes, including two bilaterians (Aplysia californica and Octopus bimaculoides), one ctenophore (Pleurobrachia bachei), a sponge (Amphimedon queenslandica), and a placozoan (Trichoplax adhaerens). As a query, we used the most completed, annotated, and published dataset of 1,600 TFs encoded in the human genome to represent the deuterostomes clade (Lambert et al., 2018) and 755 predicted sequence-specific TFs in Drosophila, the model representative of the Ecdysozoa clade, as the initial queries for the tblastn searches (Shokri et al., 2019). Utilizing these complete and initial datasets, we identified that the sea slug Aplysia genome encodes 824 transcription factors. Similarly, using all Aplysia, Drosophila, and human TFs as queries in tblastn searches against their genomes, we identified the complete repertoire of TFs encoded in the Octopus bimaculoides, and the other three (Trichoplax, Amphimedon, Pleurobrachia) basal metazoan genomes.
Next, we identified TF families in these five animal phyla that have undergone lineage-specific TFs gene expansions, including the ones that have originated through tandem duplications. To our surprise, we found that the full-length TFs that derived from the class II DNA transposable elements (TEs) were primarily associated with species-specific TFs family gene expansion (Figure 1). Within this framework, Cosby et al. (Cosby et al., 2021) not only described the tendency of class II TE for being domesticated as TFs in mammals but also study mechanisms and proposed a model for this process, taking into count the binding sites of transposases. There are ten superfamilies of Class II TEs that are known to use the “cut-and-paste” mechanism for transposition from one position in the genome to another (Feschotte and Pritham, 2007; Zattera and Bruschi, 2022). Representatives of each of these subfamilies TE encoded full-length TF proteins were used as a query to screen for potentially TE-derived TFs across nine metazoan phyla (Figure 1; Supplementary Table S1). We determined that six of these TEs superfamilies could be independently recruited into the metazoan TFs: ZBED, CENPB, FHY3, HTH-Psq, THAP, and FLYWCH (Figure 1). Phylogenetic reconstruction suggested independent recruitment due to the absence of a “one-to-one” homolog in the closest species (Figure 2). The domain organization of newly identified potentially TE-derived metazoan TFs (summarized in Figure 3) also revealed the presence of transposon-like components within the protein-coding open reading frames (ORFs). The occurrence of TEs components within the TFs was further supported by sequence similarity searches against the de novo assembled transcriptome (RNA-Seq) dataset (https://neurobase.rc.ufl.edu).
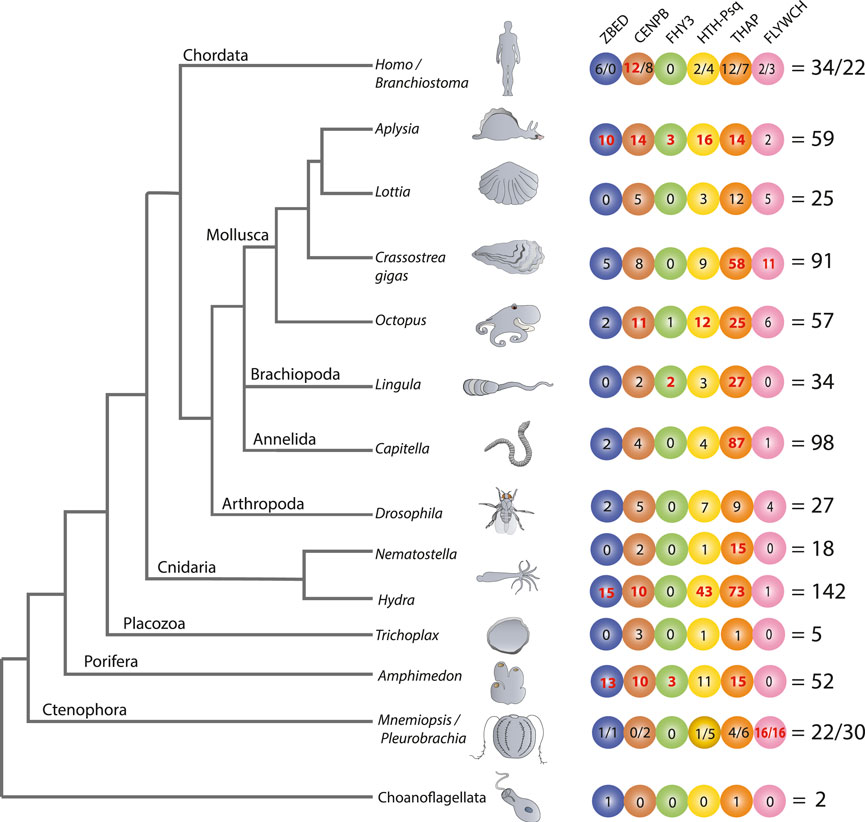
FIGURE 1. Transposon-derived transcription factors across metazoans. The diagram shows lineage-specific expansion and mosaic distributions of six families of transposon-derived transcription factors (TFs) across metazoans. All TFs depicted in the tree are lineage-specific genes that have no homolog in other classes or phyla. Each colored circle represents one of the six potentially TE-derived TF gene families: ZBED, CNPB, FHY3, HTH-Psq, THAP, and FLYWCH. Figures within circles indicate several independent species-specific events of the domestication of a particular TF family. The total numbers of transposon-derived TFs identified in each reference species are shown on the right. We observed the most extensive expansion of transposon-derived TFs in four bilaterian lineages led to the hydrozoan polyp—Hydra (142), the oligochaete—Capitella (98), the sea slug—Aplysia (59), and the bivalve—Crassostrea (91). Of note, a significant expansion of the THAP gene family occurred in Capitella (87), Hydra (73), and Crassostrea (58). Independent species-specific expansions of the FLYWCH gene family occurred in ctenophores Mnemiopsis (16) and Pleurobrachia (16). The “/” symbol is used to differentiate the numbers identified under both species, such as in Homo/Branchostoma and Mnemiopsis/Pleurobrachia, etc., The bold red letter indicates when the values are significantly higher in numbers compared to other species.
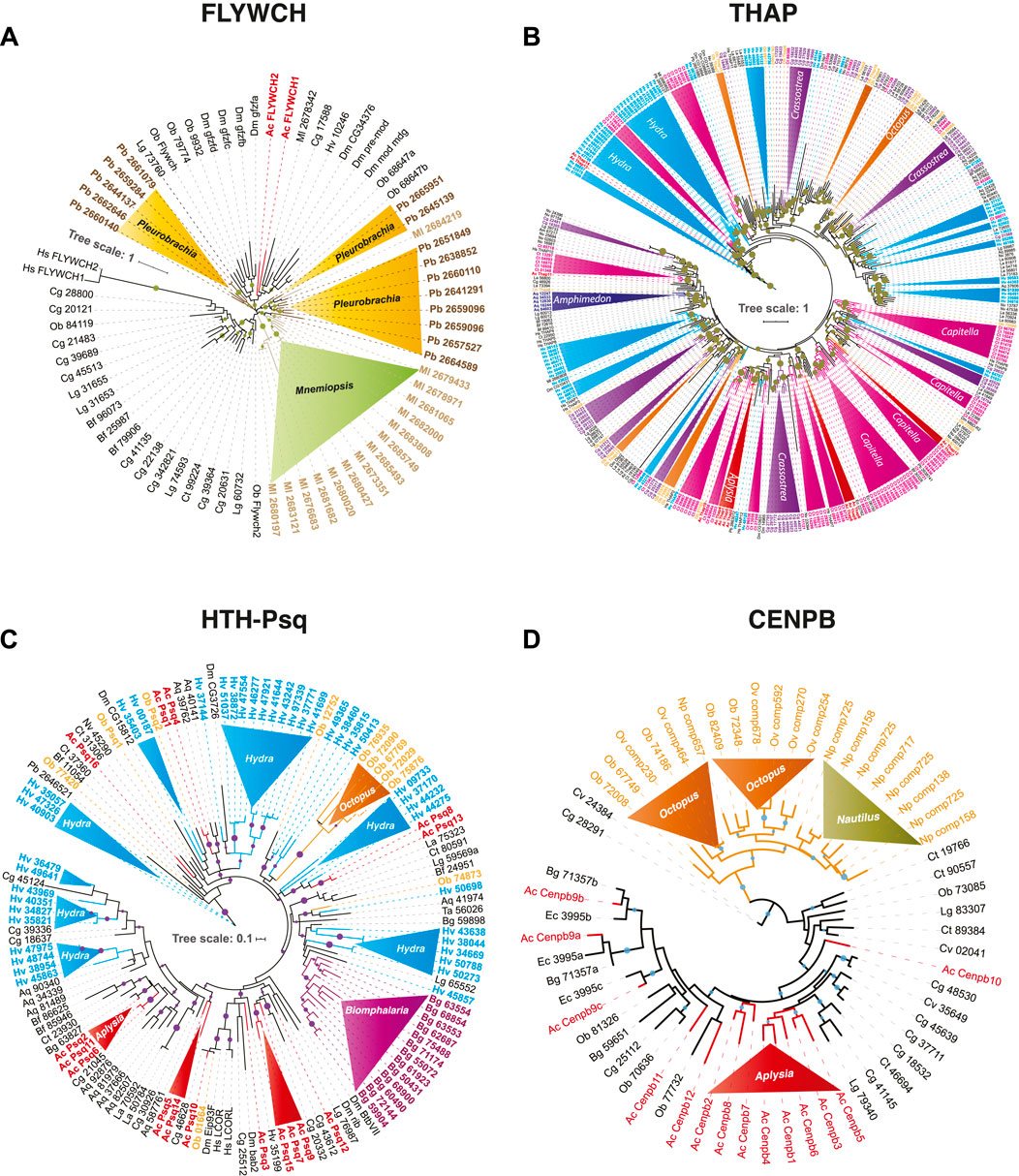
FIGURE 2. Independent expansion and convergent evolution of transposon-derived transcription factors in Metazoa. The phylogenetic tree represents the independent expansion and evolution of transposon-derived transcription factors protein families across metazoans. Each solid-color triangle represents species-specific expansion that has no homologs in related species. We used the following DNA binding domains–FLYWCH (A), THAP (B), HTH-Psq (C), and CENPB (D)—as illustrative examples to build the maximum likelihood (ML) tree. The trees show independent FLYWCH gene expansion in the ctenophores Mnemiopsis and Pleurobrachia (A). Similarly, independent THAP genes expansion in Capitella, Octopus, Crassostrea, Hydra (B), HTH-Psq expansion in Hydra, Biomphalaria, Aplysia, and Octopus (C), and Independent convergent domestication of CENPB genes in Octopus, Nautilus, and Aplysia (A). High-resolution images of each of these trees are presented in Supplementary Figures.S1–S4.
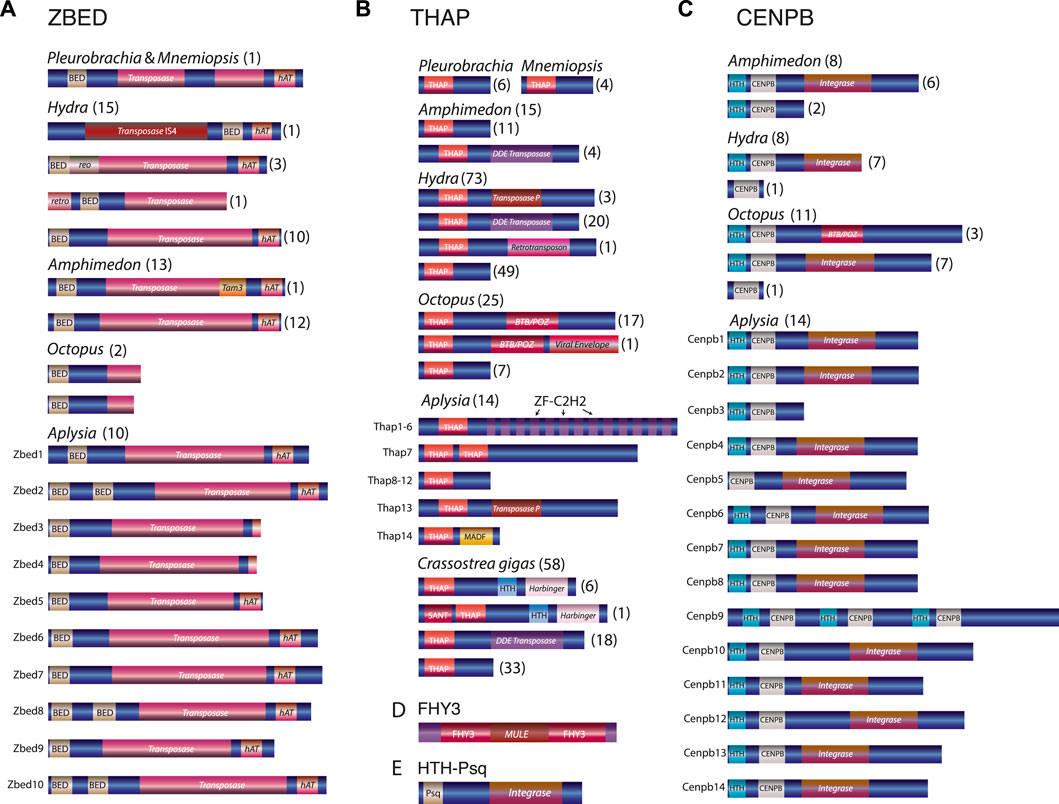
FIGURE 3. Domain organizations of the transposon-derived transcription factors across metazoans (A–E). Transposon insertions domains are shown in shaded red color and labeled as integrase, transposase, Harbinger, BTB/POZ, etc., Note that the same transcription factor protein families have different transposon components. For example, Octopus CENPB and THAP proteins have derived mostly from BTB/Poxvirus BTB (Godt et al., 1993)/POZ (Bardwell and Treisman, 1994) transposable elements, whereas, in other species, the same TFs have originated from multiple different transposable elements. Similarly, Hydra ZBED genes could have derived from at least three transposon sources such as retrotransposon, reoviruses, and transposon IS4, whereas all Aplysia ZBED genes seem to have derived from Ac transposon (Supplementary Figures S5, S6). Numbers within parentheses indicate the number of genes identified with a similar domain organization.
All predicted TE-derived TF families identified in our analysis showed low ( <1; Z-test p < 0.05) non-synonymous substitutions versus synonymous substitution (Ka/Ks) ratios (Supplementary Excel File S2, S3), indicating negative or purifying selection acting to maintain evolutionarily conserved sets of amino acid sequences. Similarly, the low Ka/Ks ratio of predicted TE-derived TFs suggests stationary domesticated genes (Gao et al., 2020). Furthermore, maintaining low Ka/Ks also suggest that their transposition ability can be maintained (Dazeniere et al., 2022). In addition to the Z test, Fast Unbiased Bayesian Approximation (FUBAR) (Murrell et al., 2013) estimation of the dN/dS ratio also confirmed negative or purifying selection pressure acting on these TFs (Figure 4). The total number of the proposed transposon-derived TFs is 788 (Supplementary Excel File S1). Supplementary Table S3 includes species such as the sea slug, Elysia chlorotica, the hemipteran insect Myzus persicae, and the rainbow trout Oncorhynchus mykiss (Supplementary Excel File S1).
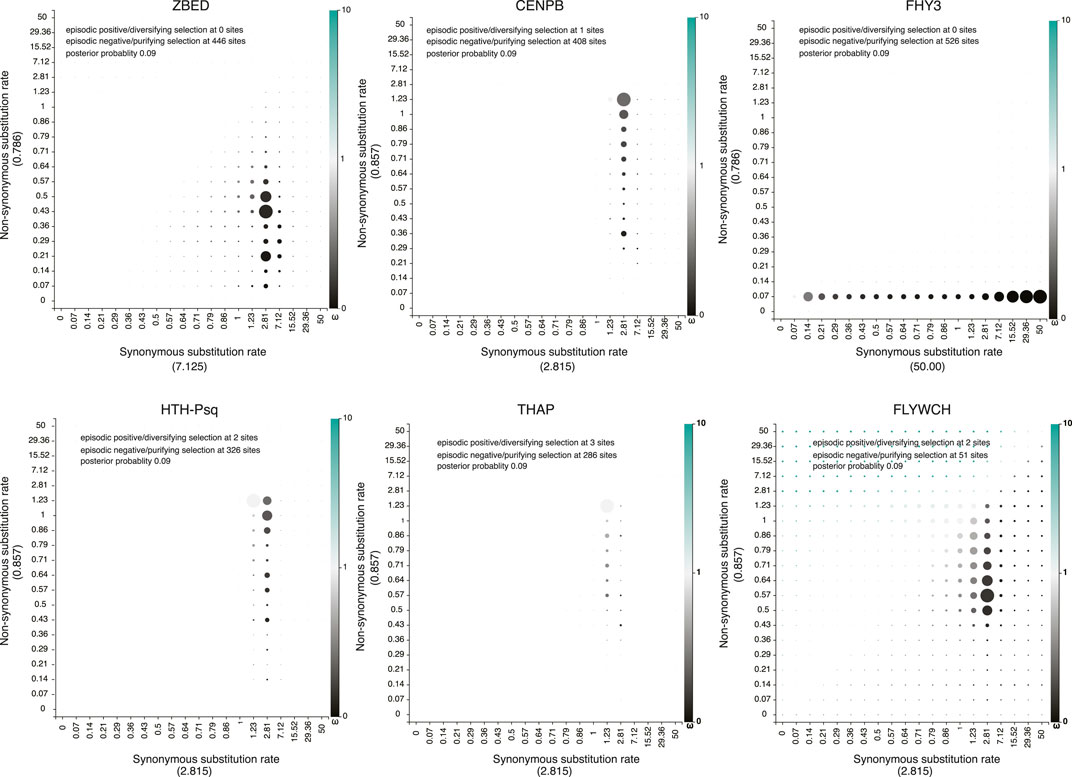
FIGURE 4. Non-synonymous (dN) versus synonymous substitution (dS) ratio show transposon-derived transcription factors evolving under purifying selection pressure. Non-synonymous versus synonymous substitutions were calculated across all potentially TE-derived TF families using the Fast Unbiased Bayesian Approximation (FUBAR) approach (Murrell et al., 2013). Synonymous substitutions (dS) rates calculated under each family are shown in X-axis inside the parentheses. Similarly, Non-synonymous substitutions (dN) rates calculated under each family showed in the Y axis inside the parentheses. Gray to intense black color-coding dots signifies negative or purifying (dN/dS < 1) selection, while light green to intense green represents sites under diversifying or positive (dN/dS > 1) selection.
Figure 1 illuminates the mosaic-type distribution in the recruitments of transposon-derived TF subfamilies across major metazoan lineages studied here. In the sister group to all Metazoa—Choanoflagellata—we found only two genes likely encoding transposon-derived TFs from ZBED and THAP superfamilies, respectively.
Ctenophores are often viewed as the earliest branching lineage of animals, sister to the rest of Metazoa (Ryan et al., 2013; Moroz et al., 2014; Whelan et al., 2015; Whelan et al., 2017), although the reconstruction of the basal metazoan phylogeny is still a highly debated topic (Kapli and Telford, 2020; Li et al., 2021; Redmond and McLysaght, 2021), and might not be convincingly resolved. Unlike other studied metazoans, both the ctenophores Mnemiopsis and Pleurobrachia showed tremendous expansions of the FLYWCH transcription factor gene family (Figure 2A). FLYWCH (Dorn and Krauss, 2003; Ow et al., 2008), which is a distinct DNA-binding zinc finger domain-containing protein family known to have originated from the Mutator transposase (Marquez a Pritham, 2010). FLYWCH domains are evolutionary conserved but relatively rarely occur in animals. They were initially identified in Drosophila (Dai et al., 2004) and then in C. elegans, where it plays regulatory roles during embryogenesis by repressing microRNAs (Ow et al., 2008). The most recent evidence suggests that FLYWCH, in complex with β-catenin, repressed specific genes of the Wnt pathways and, therefore, can control cell polarity, migration, and metastasis (Muhammad et al., 2018). Surprisingly, none of the newly identified FLYWCH domain-containing genes have homologs in each other ctenophore species (Figure 2A; Supplementary Figure S1). Unfortunately, there are no functional studies of these genes, and the roles of these TFs in ctenophores will be subjects of future studies.
There are three species with the broadest overall domestication of TEs: the hydroid polyp—Hydra (142 TFs), the polychaete annelid—Capitella (98 TFs), and the gastropod mollusk, Aplysia (59 TFs). In these animals, the identified domestication events are both species-specific and TF-type-specific. In other words, for each animal studied, we noticed an independent expansion of one or more families of potentially TE-derived TFs (Figure 1). The most notable examples of predicted TE exaptation we found in Hydra and the ctenophore Pleurobrachia (5 out of 6 superfamilies), Aplysia (6 out of 6 superfamilies), and the sponge Amphimedon (5 out of 6 superfamilies). Surprisingly, the lineage that led to the sponges also revealed multiple examples of independent domestication and expansion of potentially TE-derived TFs compared to other non-bilaterian metazoans (except Hydra), which correlate to astonishing diversification within the phylum Porifera in general.
In contrast, the placozoan Trichoplax—the simplest known free-living animal (Grell and Ruthmann, 1991; Srivastava et al., 2008; Romanova et al., 2021; 2022), had the smallest number (5) of predicted TE-derived TFs, which might reflect the observed morphological simplicity of these disk-shaped benthic animals with only three layers of cells gliding on algal substrates (Srivastava et al., 2008; Smith et al., 2014; Eitel et al., 2018).
Likewise, the anthozoan Nematostella also had a modest representation of potentially TE-derived TFs, mostly related to just one superfamily; there are 15 Thanatos and associated protein (THAP) domain-containing genes. THAP genes were found in Drosophila, and they are known to have originated from P element transposes (Roussigne et al., 2003). Our analysis support events of the independent diversification of THAP genes in Hydra (73), Capitella (87), Crassostrea (58) (see details in the next section and Figure 2B; Supplementary Figure S2); and at a lesser degree in a living fossil—the brachiopod, Lingula (27) and Octopus (25).
In summary, THAP genes represent the largest class of potentially TE-derived TFs identified in this study, including the basally branched chordate amphioxus (Branchiostoma) and humans. THAP- TF functions in invertebrates are primarily unknown (Nicholas et al., 2008). On the other hand, THAP TFs in humans were implicated in epigenetic regulation, maintenance of pluripotency, transposition, cancers, and other disorders like hemophilia. For example, THAP0 is a member of the apoptotic cascade induced by IFN-γ (Lin et al., 2002). THAP1, with RRM1, regulates cell proliferation (Cayrol et al., 2007). THAP5 acts as a cell cycle inhibitor (Balakrishnan et al., 2009). THAP9 is an active transposase in humans (Majumdar et al., 2013). The THAP11 homolog in mice is essential for embryogenesis (Dejosez et al., 2008).
Two other groups presently identified TE-derived TFs are also prominent in humans and Branchiostoma: ZBED and CENPB (Figure 1; Supplementary Figures S5–S7).
BED zinc fingers or ZBED genes reported having derived from the hAT (hobo, Ac, Tam3) superfamily of DNA transposon (Aravind, 2000), and members of this superfamily regulate an extensive array of functions in vertebrates. For example, ZBED6 affects development, cell proliferation, wound healing, and muscle growth (Markljung et al., 2009). ZBEDs are present in mammals, birds, reptiles, and fish; however, they are absent from jawless fishes. Based on these findings, it was proposed that ZBED genes in vertebrates originated due to at least two independent hAT DNA transposon domestication events in primitive jawed-vertebrate ancestors (Hayward et al., 2013). Our searches against the Branchiostoma belcheri genome uncovered a full-length ZBED gene, which was surprisingly absent from the Branchiostoma floridae genome, further suggesting species-specific and mosaic exaptation of TE-encoded genes.
Also, using both the DNA binding BED domain and known full-length ZBED genes, we find that ZBED genes form a monophyletic cluster in three mollusks (Aplysia, Biomphalaria, Crassostrea), the sponge Amphimedon, and Hydra (Supplementary Figures S5–S6).
Centromere-binding proteins-B (CENPB) transcription factor (Lein et al., 2007) involved in chromosome segregation maintenance and genome stability (Morozov et al., 2017) recurrently domesticated from pogo-like transposons (Casola et al., 2008; Mateo and Gonzalez, 2014) across Metazoa (Supplementary Figure S7). CENPB homologs were found in mammals (Sullivan and Glass, 1991) but not in other vertebrates. Nevertheless, we identified CENPB TFs from both Branchiostoma belcheri and B. floridae genomes, indicating their presence before the divergence of vertebrates. Thus, this finding suggests either loss of CENPBs in most of the extant lineages of vertebrates or their independent domestication in mammalian species, which is a more likely scenario (Casola et al., 2008). There is also a remarkable diversification and independent expansion of the CENPB superfamily in Mollusca (Supplementary Figure S7), which we will discuss in the following section.
The most stunning example of mosaic recruitment of TEs can be illustrated using Mule transposons. Mule transposon-derived transcription factor far-red elongated hypocotyls 3 (FHY3) group are critical for far-red (near-infrared) light signaling and survival of chloroplast in plants (Lin et al., 2007; Chang et al., 2015). Here for the first time, we identified FHY3 in animals (Figures 1, 3D). Our cross-species comparison across metazoans showed that FHY3 was present in three copies, both in the demosponge Amphimedon and the sea slug Aplysia genomes. There are two copies in the brachiopod Lingula and one in Octopus genomes (Figure 1). However, we did not find FHY3 in the sequenced ctenophores (Pleurobrachia and Mnemiopsis), placozoan (Trichoplax), and cnidarian (Nematostella and Hydra) and human genomes. Thus, FHY3 can be absent or present in a mosaic fashion without a recognized taxonomical specification. Our phylogenetic analysis (Supplementary Excel File S1) showed that FHY3 had been repeatedly domesticated over 550 + million years of animal evolution (see Supplementary Figure 8S), including examples from selected molluscs (e.g., the algae-eating sea slugs Aplysia californica, Elysia chlorotica, and the oyster—Crassostrea), some arthropods (Myzus persicae and Limulus polyphemus) and chordates (Branchiostoma).
In conclusion, we obtained evidence that the majority of TFs are the results of the species-specific convergent domestication events across animal phyla tested here. Figure 2; Supplementary Figures S1–S8 illustrate these cases. Of note, although some of the studied species show a predominant exaptation of just one or two categories of genes, many domesticated events occurred independently, even within the same superfamily of potentially TE-derived TFs (Figure 2; Supplementary Figures S1–S8). This situation is summarized below, focusing on the Lophotrochozoan lineage.
2. Transposon-derived TFs showed independent species-specific expansion and evolution in Molluscs.
Lophotrochozoa or Spiralia, including the phylum Mollusca, is the most morphologically and biochemically diverse animal clade (Kocot et al., 2017). None of the predicted TE-derived TFs were previously reported in Lophotrochozoa (Table 1). The phylum Mollusca in our analysis is represented by seven species (Aplysia, Biomphalaria, Elysia, Lottia, Crassostrea, Octopus, and Nautilus), with Aplysia showing the most remarkable expansion of potentially TE-derived TFs (Figure 1). First, we systematically scanned the complete set of the TFs encoded in the Aplysia californica genome a prominent neuroscience model (Kandel, 2001; Moroz et al., 2006; Moroz, 2011), resulting in the identification of 824 transcription factors.
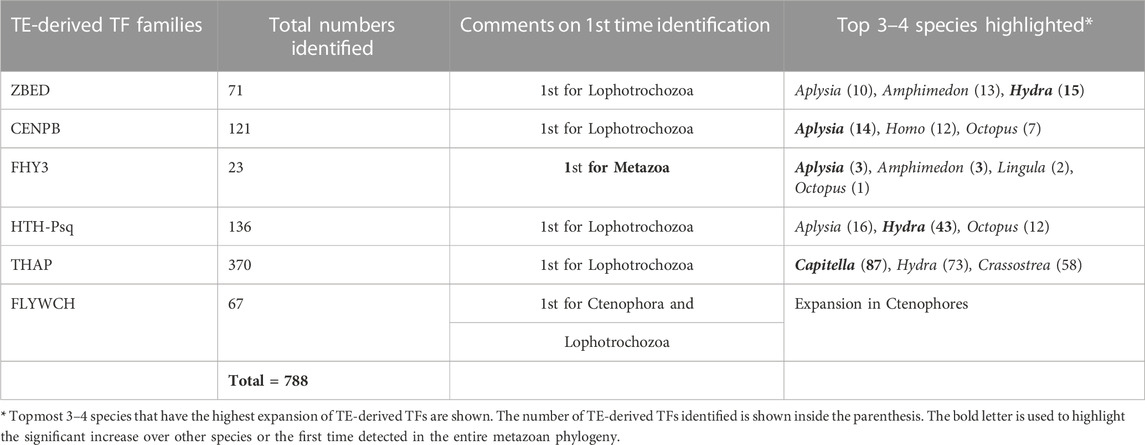
TABLE 1. The total number of potentially TE-derived TFs identified in this study. (See Figure 1; Supplementary Table S1 for details).
Then, we identified 59 novel (∼7%) transposon-derived TFs that have no homolog in closely related species such as in Biomphalaria the freshwater pulmonated snail (Adema et al., 2017) or the limpet Lottia (Simakov et al., 2013). This finding indicates that these TFs did not originate from canonical gene duplication events (Supplementary Excel File S1); they do not follow the canonical subfunctionalization (Stoltzfus, 1999) and neofunctionalization (Force et al., 1999) characteristics. Of these 59 Aplysia lineage-specific TFs, 42 were coupled with the transposase (TPase) domain (Figure 3), confirming the hypothesis that these genes, including their DNA-binding domain, may have originated by unique mechanisms involving “cut-and-paste” DNA transposons.
In molluscs, we also revealed that the lineage-specific TFs, even those belonging to identical TF families, originated both from similar and different transposon sources: the majority of potentially TE-derived TF domestication events were not detected from related species. Thus, the most likely parsimonious scenario is a broad scope of independent domestication events leading to the convergent evolution of TE-derived TFs within animal lineages studied here. Figure 2; Supplementary Figures S1–S8 illustrates bursts of parallel expansions of transposon-derived TFs subfamilies. Three examples are outlined below.
(1) There are convergent domestications of pogo-derived CENPB sequences in Aplysia, cephalopods, and other Lophotrochozoan species, such as in Crassostrea (Figure 2D). Within the cephalopod lineage, we identified two distinct events of pogo domestication—one, in the lineage leading to Nautilus and another event occurring in the lineage leading to Octopus (Figure 2D).
(2) Helix-turn-helix motif of pipsqueak (HTH-Psq) proteins form a family of transcription factors known to have derived from Drosophila pogo transposase (Siegmund and Lehmann, 2002). We find the Aplysia genome encodes 16 HTH-Psq subfamily transcription factors while the Biomphalaria genome encodes 15. Surprisingly none of these Biomphalaria TFs has direct homologs in the Aplysia genome and vice versa (Figure 2C; Supplementary Figure S3), indicating species-specific expansion event. Similarly, both Hydra and Octopus showed independent species-specific expansions of transposon-derived HTH-Psq genes. Thus, independent domestication of Psq genes might occur at least five times in Aplysia, Biomphalaria, Octopus, and the Hydra and Amphimedon genomes (Figure 2C).
(3) Myb-SANT, like in Adf (MADF) domain-containing genes initially identified in Drosophila known to have originated from the P instability factor or PIF superfamily of DNA transposon (Lin et al., 2007). We find that MADF genes were expanded in Amphimedon, Drosophila, and, most of all, Aplysia with at least six predicted independent domestication events. Although MADF genes are likely derived from the PIF superfamily of DNA transposon, we have excluded MADF genes from this analysis owing to the growing concern that these genes do not harbor a recognized transposon-derived transposase domain within the protein-coding gene.
Altogether our results suggest a substantial lineage-specific diversification and independent evolution of new genes originating from a modular diversity of cut-and-paste DNA transposons, as outlined in the next section.
3 Domain analysis revealed the presence of transposons derived components within the protein-coding TFs
All subfamilies of transposon-derived TFs predicted in this analysis have a modular domain architecture (Figure 3). Within each subfamily, most TFs encode recognizable transposon-derived components within exons of these protein-coding genes. For example, transposon-derived ZBED TFs, besides encoding the canonical DNA-binding BED zinc finger motif, also encoded a transposon-derived transposase domain and an hAT dimerization domain (Figure 3A). Strikingly, we find that ZBED genes across metazoans derived from diverse transposable element components (Supplementary Figures S5, S6). For instance, Homo ZBED5 is known to have derived from Buster DNA transposon (Hayward et al., 2013), which, in our analysis, forms a robust clade with one of the Octopus ZBED genes indicating its Buster transposon origin (Supplementary Figures S5, S6). In contrast, the second Octopus ZBED gene forms a robust cluster with the Hydra retrotransposon-derived ZBED gene (Supplementary Figures S5, S6). The two truncated ZBED genes from the Octopus bimaculoides genome lack an intact transposase and an hAT dimerization domain. In addition, we could not recover the full-length transposase domain and the hAT dimerization domain from the Octopus bimaculoides genome associated with them. This result indicates that the two Octopus ZBED genes may have evolved from two independent transposon components.
Similarly, the Hydra retrotransposon-derived ZBED gene encodes an intron that separates the N-terminal reverse transcriptase (RT) domain against the C-terminal BED finger and the transposase domain. This result suggests that the Hydra BED and the transposase domains are no longer part of the retrotransposon component. In addition, Hydra ZBED genes contained at least three transposon components, such as retrotransposons, reoviruses, and transposon IS4 (Figure 3A; Supplementary Figures S5, S6). Likewise, while Octopus THAP genes are mostly derived from BTB (Godt et al., 1993) (Broad- Complex, Tramtrack, Bric a Brac) or POZ (Bardwell and Treisman, 1994) (poxvirus and zinc finger) transposon sources—the Hydra THAP genes, however, found to be derived from versatile transposon sources such as Transposase P element, DDE transposase (DDE_Tnp_4) and retrotransposon. In contrast, some Crassostrea gigas THAP genes contained sequences associated with the Harbinger-derived transposon domain (Figure. 3B).
Also, while most of the Octopus CENPB TFs were associated with the transposon-derived BTB/POZ domain, none of the genes from another mollusc, Aplysia, contained this domain (Figure 3C).
Both CENPB and HTH-Psq genes had a signature of the viral rve superfamily of the retroviral integrase domain (Figure 3C, E). Integrase is the retroviral enzyme that catalyzes the integration of virally derived DNA into the host cell’s nuclear DNA, forming a provirus that can be activated to produce viral proteins (Delelis et al., 2008). In the same way, FHY3 genes share remarkable sequence similarities with MURA (Hudson et al., 2003), the transposable element encoded by the Mutator element of maize, and the predicted transposase of the maize mobile element Jittery (Xu et al., 2004). Both transposons are a member of the Mutator-like elements (MULE) (Lisch, 2002) (Figure 3D).
These results, for the first time, indicate that even within the same subfamily of transposon-derived TFs—similar domains have derived from multiple transposon components across the animal kingdom. Together our phylogenetic analysis and the revealed domain organizations suggest that similar domain architecture originated in parallel from numerous transposon resources across phyla.
4 Conclusion
By systematic analysis of about seven thousand animal TFs, we have predicted a total of 788 ( >10%) novel DNA transposons-derived TFs across metazoans (Figure 1; Supplementary Excel File S1). Our study was limited to 6 previously known TE-derived TF families used as a query to search for the new domestication events. Although predictably derived from the TE components, we had to exclude the MADF genes from the current analysis owing to the absence of a potential transposase domain.
The Aplysia genome encodes 41 MADF genes, and a many of them expressed in developmental stages as well as in specific neuronal populations, suggesting their involvement in the control of cell-specific phenotypes (data not shown) as well as contributing to the very origin of neuronal organizations and diversification events (Erwin, 2009; Mustafin and Khusnutdinova, 2020; Moroz and Romanova, 2021). Homologs of these Aplysia MADF genes are missing in the sequenced Biomphalaria genome a related gastropod species (Adema et al., 2017; Kocot et al., 2011), which encodes only three of these MADF genes. Thus, careful systematic analysis is needed to identify novel domestication events in the evolution of TE-derived TFs within molluscs.
Overall, predicted TE-derived TFs show mosaic patterns in their distribution with extreme heterogeneity and with a ‘sudden’ appearance in one lineage and, at the same time, found to be ‘missing’ in more closely related species.
Although most studied species predict a predominant exaptation of just one category of genes, many domesticated events might occur independently in evolution, even within the same superfamily of potentially TE-derived TFs (Figure 2).
Our results suggest a substantial lineage-specific diversification and independent origins of new TF genes originated from a broad array and a modular diversity of cut-and-paste DNA transposons and related viroid-like elements. Many described TFs preserved the original modular gene organization (Figure 3) and could act as highly dynamic modules shaping the genome-wide reorganization within Metazoa.
5 Materials and Methods
5.1 Identification of potentially TE-derived TFs
We used representatives of published and confirmed domesticated transposable element-derived TFs protein families from plants and animals as a query (Supplementary Table S2). Both PSI-BLAST, as well as Tblastn searches, were performed using both the command-line version at the NCBI standalone BLAST (version 2.2.18) (Camacho et al., 2009) as well as at the online BLAST web interface (Boratyn et al., 2013; Shi et al., 2018) using default e-value cut off for the online version and 10−5 to 10−10 cut off for the stand-alone blast to identify all potential homologs. Homologs were detected not solely based on e-value cut-off but other criteria such as coverage statistics, bit score, etc., were considered. Protein sequences recovered from one round of TBLASTN or PSI-BLAST searches were recursively used as queries until no further sequences were detected. Each protein blast hit was manually inspected following multiple sequence alignment (MSA) and validated utilizing several databases including the NCBI conserved domain database (CDD) (Marchler-Bauer et al., 2011), Hmmer (Finn et al., 2011), Pfam (Punta et al., 2011), and SMART (Letunic and Bork, 2018). In the case of the non-availability of the gene model (exome), genome sequences surrounding the coding region were excised, and homology-based gene prediction based on hidden Markov models (HMMs) was performed in FGENESH+ (www.softberry.com) to identify the complete open reading frame. Finally, TE insertions within the TFs were further validated by similarity searches against the de novo assembled RNA-Seq (transcriptome) datasets obtained in Moroz lab (https://neurobase.rc.ufl.edu).
5.2 Multiple sequence alignment and protein domain identification
Protein functional domains were identified by sequence search of the NCBI conserved domain databases (Marchler-Bauer et al., 2011; Marchler-Bauer et al., 2017). Results were verified via sequence searches of the SMART (Letunic and Bork, 2018) and Pfam database (Punta et al., 2011). Also, sequences were aligned in MUSCLE (Edgar, 2004a; Edgar, 2004b) and displayed in clustalX (Larkin et al., 2007) and manually confirmed the domain architecture by examining the sequences based on protein secondary structure analysis and profile alignments. Multiple sequence alignment (MSA) obtained through MUSCLE was used to build the HMMER v3.1b2 (Finn et al., 2011) position-specific scoring matrix (PSM) to search against the reference proteome datasets.
5.3 Phylogeny reconstruction
Maximum-likelihood (ML) trees were inferred using PhyML v3.0 (Guindon and Gascuel, 2003; Guindon et al., 2010), with the best-fit evolutionary model identified using the AIC criterion estimated by ProtTest (Abascal et al., 2005). ML phylogenies were performed using the JTT model of rate heterogeneity, estimated proportion of invariable sites, four rate categories, and estimated alpha distribution parameter. Tree topology searches were optimized using the best of both NNI (nearest-neighbor interchanges) and SPR (subtree pruning and regrafting) moves (Hordijk and Gascuel, 2005). Clade support was calculated using the SH-like approximate likelihood ratio test (Anisimova et al., 2011). Unless otherwise mentioned, all phylogenetic trees presented throughout the manuscript show SH-support of 80 or greater. The resulting phylogenetic trees were viewed and edited with iTol version 2.0 (Letunic and Bork, 2007).
5.4 Estimation of codon substitution pattern and inference of selective pressure
Protein sequences of potentially TE-derived transcription factors under each family were aligned using MUSCLE (Edgar, 2004a), and the conversion of protein alignments to corresponding nucleotide coding sequences was obtained using PAL2NAL webserver (Suyama et al., 2006). Codon-based tests of neutrality and negative or purifying selection were conducted using MEGA with a Z test by calculating the substitution ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) versus synonymous substitution per synonymous sites (Ks) using the Nei-Gojobori method (Nei and Gojobori, 1986). Orthologous sequences with a Ka/Ks value of <1 (Z-test, p < 0.05) were defined as having been under purifying selection shown with yellow color (Supplementary Excel files S3, S4).
Of note that the extended methods section is summarized in the Supplementary Method section online.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KM and LM: Conceptualization; Writing an original draft, Writing-review and editing, Data obtaining, and curation. KM: Formal computational analysis, Investigation, Methodology, Software, Validation, Data Visualization, LM: Funding Acquisition, Project Administration, Resources, and Supervision.
Funding
This work was supported by the Human Frontiers Science Program (RGP0060/2017), National Science Foundation (Grants 1146575, 1557923, 1548121, and 1645219), National Institute of Health (R01 NS114491) to LM.
Acknowledgments
The authors would like to thank Drs. Caleb Bostwick, Peter Williams, and Andrea Kohn for the generation of RNA-seq libraries and initial annotations. Thanks to Gayle Prevatt for the initial drawing of the animal sketches.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1113046/full#supplementary-material
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi:10.1093/bioinformatics/bti263
Adema, C. M., Hillier, L. W., Jones, C. S., Loker, E. S., Knight, M., Minx, P., et al. (2017). Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 8, 15451. doi:10.1038/ncomms15451
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi:10.1093/sysbio/syr041
Aravind, L. (2000). The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25, 421–423. doi:10.1016/s0968-0004(00)01620-0
Balakrishnan, M. P., Cilenti, L., Mashak, Z., Popat, P., Alnemri, E. S., and Zervos, A. S. (2009). THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am. J. Physiol. Heart Circ. Physiol. 297, H643–H653. doi:10.1152/ajpheart.00234.2009
Bardwell, V. J., and Treisman, R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes. Dev. 8, 1664–1677. doi:10.1101/gad.8.14.1664
Biemont, C., and Vieira, C. (2006). Genetics: Junk DNA as an evolutionary force. Nature 443, 521–524. doi:10.1038/443521a
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). Blast: A more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi:10.1093/nar/gkt282
Borchert, G. M., Holton, N. W., Williams, J. D., Hernan, W. L., Bishop, I. P., Dembosky, J. A., et al. (2011). Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins. Mob. Genet. Elem. 1, 8–17. doi:10.4161/mge.1.1.15766
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinforma. 10, 421. doi:10.1186/1471-2105-10-421
Casola, C., Hucks, D., and Feschotte, C. (2008). Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 25, 29–41. doi:10.1093/molbev/msm221
Casola, C., Lawing, A. M., Betran, E., and Feschotte, C. (2007). PIF-like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol. 24, 1872–1888. doi:10.1093/molbev/msm116
Cayrol, C., Lacroix, C., Mathe, C., Ecochard, V., Ceribelli, M., Loreau, E., et al. (2007). The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109, 584–594. doi:10.1182/blood-2006-03-012013
Chang, N., Gao, Y., Zhao, L., Liu, X., and Gao, H. (2015). Arabidopsis FHY3/CPD45 regulates far-red light signaling and chloroplast division in parallel. Sci. Rep. 5, 9612. doi:10.1038/srep09612
Chuong, E. B., Rumi, M. A., Soares, M. J., and Baker, J. C. (2013). Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329. doi:10.1038/ng.2553
Cornelis, G., Funk, M., Vernochet, C., Leal, F., Tarazona, O. A., Meurice, G., et al. (2017). An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. U. S. A. 114, E10991–E11000. doi:10.1073/pnas.1714590114
Cosby, R. L., Judd, J., Zhang, R., Zhong, A., Garry, N., Pritham, E. J., et al. (2021). Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371, eabc6405. doi:10.1126/science.abc6405
Dai, M. S., Sun, X. X., Qin, J., Smolik, S. M., and Lu, H. (2004). Identification and characterization of a novel Drosophila melanogaster glutathione S-transferase-containing FLYWCH zinc finger protein. Gene 342, 49–56. doi:10.1016/j.gene.2004.07.043
Dazeniere, J., Bousios, A., and Eyre-Walker, A. (2022). Patterns of selection in the evolution of a transposable element. G3 (Bethesda) 12, jkac056. doi:10.1093/g3journal/jkac056
Dejosez, M., Krumenacker, J. S., Zitur, L. J., Passeri, M., Chu, L. F., Songyang, Z., et al. (2008). Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 133, 1162–1174. doi:10.1016/j.cell.2008.05.047
Delelis, O., Carayon, K., Saib, A., Deprez, E., and Mouscadet, J. F. (2008). Integrase and integration: Biochemical activities of HIV-1 integrase. Retrovirology 5, 114. doi:10.1186/1742-4690-5-114
Dorn, R., and Krauss, V. (2003). The modifier of mdg4 locus in Drosophila: Functional complexity is resolved by trans splicing. Genetica 117, 165–177. doi:10.1023/a:1022983810016
Edgar, R. C. (2004a). Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 5, 113. doi:10.1186/1471-2105-5-113
Edgar, R. C. (2004b). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Eitel, M., Francis, W. R., Varoqueaux, F., Daraspe, J., Osigus, H. J., Krebs, S., et al. (2018). Comparative genomics and the nature of placozoan species. PLoS Biol. 16, e2005359. doi:10.1371/journal.pbio.2005359
Erwin, D. H. (2009). Early origin of the bilaterian developmental toolkit. Philos. Trans. R. Soc. Lond B Biol. Sci. 364, 2253–2261. doi:10.1098/rstb.2009.0038
Feschotte, C., and Pritham, E. J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368. doi:10.1146/annurev.genet.40.110405.090448
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. doi:10.1093/genetics/151.4.1531
Gao, B., Wang, Y., Diaby, M., Zong, W., Shen, D., Wang, S., et al. (2020). Evolution of pogo, a separate superfamily of IS630-Tc1-mariner transposons, revealing recurrent domestication events in vertebrates. Mob. DNA 11, 25. doi:10.1186/s13100-020-00220-0
Gehring, W. J. (1996). The master control gene for morphogenesis and evolution of the eye. Genes. cells. 1, 11–15.
Godt, D., Couderc, J. L., Cramton, S. E., and Laski, F. A. (1993). Pattern formation in the limbs of Drosophila: Bric a brac is expressed in both a gradient and a wave-like pattern and is required for specification and proper segmentation of the tarsus. Development 119, 799–812.
Gould, S. J., and Vrba, E. S. (1982). Exaptation—A missing term in the science of form. Paleobiology 8, 4–15. doi:10.1017/S0094837300004310
Grell, K. G., and Ruthmann, A. (1991). “Placozoa,” in Microscopic anatomy of invertebrates. Editor F. W. Harrison (New York: Wiley-Liss), 13–27.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi:10.1080/10635150390235520
Hammer, S. E., Strehl, S., and Hagemann, S. (2005). Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol. Biol. Evol. 22, 833–844. doi:10.1093/molbev/msi068
Hayward, A., Ghazal, A., Andersson, G., Andersson, L., and Jern, P. (2013). ZBED evolution: Repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One 8, e59940. doi:10.1371/journal.pone.0059940
Henaff, E., Vives, C., Desvoyes, B., Chaurasia, A., Payet, J., Gutierrez, C., et al. (2014). Extensive amplification of the E2F transcription factor binding sites by transposons during evolution of Brassica species. Plant J. 77, 852–862. doi:10.1111/tpj.12434
Hordijk, W., and Gascuel, O. (2005). Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21, 4338–4347. doi:10.1093/bioinformatics/bti713
Hudson, M. E., Lisch, D. R., and Quail, P. H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34, 453–471. doi:10.1046/j.1365-313x.2003.01741.x
Jordan, I. K., Rogozin, I. B., Glazko, G. V., and Koonin, E. V. (2003). Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72. doi:10.1016/s0168-9525(02)00006-9
Kandel, E. R. (2001). The molecular biology of memory storage: A dialogue between genes and synapses. Science 2 (294), 1030–1028. doi:10.1126/science.1067020
Kapli, P., and Telford, M. J. (2020). Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 6, eabc5162. doi:10.1126/sciadv.abc5162
Kocot, K. M., Cannon, J. T., Todt, C., Citarella, M. R., Kohn, A. B., Meyer, A., et al. (2011). Phylogenomics reveals deep molluscan relationships. Nature 4 (477), 452–456. doi:10.1038/nature10382
Kocot, K. M., Struck, T. H., Merkel, J., Waits, D. S., Todt, C., Brannock, P. M., et al. (2017). Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256–282. doi:10.1093/sysbio/syw079
Koonin, E. V., Makarova, K. S., Wolf, Y. I., and Krupovic, M. (2020). Evolutionary entanglement of mobile genetic elements and host defence systems: Guns for hire. Nat. Rev. Genet. 21, 119–131. doi:10.1038/s41576-019-0172-9
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell. 175, 598–599. doi:10.1016/j.cell.2018.09.045
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi:10.1093/bioinformatics/btm404
Lavialle, C., Cornelis, G., Dupressoir, A., Esnault, C., Heidmann, O., Vernochet, C., et al. (2013). Paleovirology of 'syncytins', retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond B Biol. Sci. 368, 20120507. doi:10.1098/rstb.2012.0507
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. doi:10.1038/nature05453
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi:10.1093/nar/gkx922
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi:10.1093/bioinformatics/btl529
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi:10.1038/276565a0
Li, Y., Li, C., Xia, J., and Jin, Y. (2011). Domestication of transposable elements into MicroRNA genes in plants. PLoS One 6, e19212. doi:10.1371/journal.pone.0019212
Li, Y., Shen, X. X., Evans, B., Dunn, C. W., and Rokas, A. (2021). Rooting the animal tree of life. Mol. Biol. Evol. 38, 4322–4333. doi:10.1093/molbev/msab170
Lin, R., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305. doi:10.1126/science.1146281
Lin, Y. C., Jhunjhunwala, S., Benner, C., Heinz, S., Welinder, E., Mansson, R., et al. (2010). A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11, 635–643. doi:10.1038/ni.1891
Lin, Y., Khokhlatchev, A., Figeys, D., and Avruch, J. (2002). Death-associated protein 4 binds MST1 and augments MST1-induced apoptosis. J. Biol. Chem. 277, 47991–48001. doi:10.1074/jbc.M202630200
Lisch, D. (2002). Mutator transposons. Trends Plant Sci. 7, 498–504. doi:10.1016/s1360-1385(02)02347-6
Majumdar, S., Singh, A., and Rio, D. C. (2013). The human THAP9 gene encodes an active P-element DNA transposase. Science 339, 446–448. doi:10.1126/science.1231789
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi:10.1093/nar/gkw1129
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2011). Cdd: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225gkq1189–229. doi:10.1093/nar/gkq1189
Markljung, E., Jiang, L., Jaffe, J. D., Mikkelsen, T. S., Wallerman, O., Larhammar, M., et al. (2009). ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 7, e1000256. doi:10.1371/journal.pbio.1000256
Marquez, C. P., and Pritham, E. J. (2010). Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics 185, 1507–1517. doi:10.1534/genetics.110.116673
Mateo, L., and Gonzalez, J. (2014). Pogo-like transposases have been repeatedly domesticated into CENP-B-related proteins. Genome Biol. Evol. 6, 2008–2016. doi:10.1093/gbe/evu153
Miller, W. J., McDonald, J. F., Nouaud, D., and Anxolabehere, D. (1999). Molecular domestication--more than a sporadic episode in evolution. Genetica, 107, 197–207.
Moroz, L. L., Edwards, J. R., Puthanveettil, S. V., Kohn, A. B., Ha, T., Heyland, A., et al. (2006). Neuronal transcriptome of Aplysia: Neuronal compartments and circuitry. Cell 29 (127), 1453–1467. doi:10.1016/j.cell.2006.09.052
Moroz, L. L., Kocot, K. M., Citarella, M. R., Dosung, S., Norekian, T. P., Povolotskaya, I. S., et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. doi:10.1038/nature13400
Moroz, L. L., and Romanova, D. Y. (2021). Selective advantages of synapses in evolution. Front. Cell. Dev. Biol. 9, 726563. doi:10.3389/fcell.2021.726563
Morozov, V. M., Giovinazzi, S., and Ishov, A. M. (2017). CENP-B protects centromere chromatin integrity by facilitating histone deposition via the H3.3-specific chaperone Daxx. Epigenetics Chromatin 10, 63. doi:10.1186/s13072-017-0164-y
Muhammad, B. A., Almozyan, S., Babaei-Jadidi, R., Onyido, E. K., Saadeddin, A., Kashfi, S. H., et al. (2018). FLYWCH1, a novel suppressor of nuclear beta-catenin, regulates migration and morphology in colorectal cancer. Mol. Cancer Res. 16, 1977–1990. doi:10.1158/1541-7786.MCR-18-0262
Murrell, B., Moola, S., Mabona, A., Weighill, T., Sheward, D., Kosakovsky Pond, S. L., et al. (2013). Fubar: A fast, unconstrained bayesian approximation for inferring selection. Mol. Biol. Evol. 30, 1196–1205. doi:10.1093/molbev/mst030
Mustafin, R. N., and Khusnutdinova, E. K. (2020). Involvement of transposable elements in neurogenesis. Vavilovskii Zhurnal Genet. Sel. 24, 209–218. doi:10.18699/VJ20.613
Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. doi:10.1093/oxfordjournals.molbev.a040410
Nicholas, H. R., Lowry, J. A., Wu, T., and Crossley, M. (2008). The Caenorhabditis elegans protein CTBP-1 defines a new group of THAP domain-containing CtBP corepressors. J. Mol. Biol. 375, 1–11. doi:10.1016/j.jmb.2007.10.041
Ohno, S., Wolf, U., and Atkin, N. B. (1968). Evolution from fish to mammals by gene duplication. Hereditas 59, 169–187. doi:10.1111/j.1601-5223.1968.tb02169.x
Ow, M. C., Martinez, N. J., Olsen, P. H., Silverman, H. S., Barrasa, M. I., Conradt, B., et al. (2008). The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes. Dev. 22, 2520–2534. doi:10.1101/gad.1678808
Pearson, J. C., Lemons, D., and McGinnis, W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904. doi:10.1038/nrg1726
Peter, I. S., and Davidson, E. H. (2011). Evolution of gene regulatory networks controlling body plan development. Cell. 144, 970–985. doi:10.1016/j.cell.2011.02.017
Ponder, W. F., and Linderg, D. R. (2008). Molluscan Evolution and Phylogeny: An introduction. Berkeley (CA): Univ of California Press.
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2011). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi:10.1093/nar/gkr1065
Redmond, A. K., and McLysaght, A. (2021). Evidence for sponges as sister to all other animals from partitioned phylogenomics with mixture models and recoding. Nat. Commun. 12, 1783. doi:10.1038/s41467-021-22074-7
Romanova, D. Y., Nikitin, M. A., Shchenkov, S. V., and Moroz, L. L. (2022). Expanding of Life Strategies in Placozoa: Insights From Long-Term Culturing of Trichoplax and Hoilungia. Front. Cell Dev. Biol. 10, 823283. doi:10.3389/fcell.2022.823283
Romanova, D. Y., Varoqueaux, F., Daraspe, J., Nikitin, M. A., Eitel, M., Fasshauer, D., et al. (2021). Hidden cell diversity in Placozoa: Ultrastructural insights from Hoilungia hongkongensis. Cell Tissue Res. 385, 623–637. doi:10.1007/s00441-021-03459-y
Roussigne, M., Kossida, S., Lavigne, A. C., Clouaire, T., Ecochard, V., Glories, A., et al. (2003). The THAP domain: A novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 28, 66–69. doi:10.1016/S0968-0004(02)00013-0
Ryan, J. F., Pang, K., Schnitzler, C. E., Nguyen, A. D., Moreland, R. T., Simmons, D. K., et al. (2013). The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592. doi:10.1126/science.1242592
Shi, M., Lin, X. D., Chen, X., Tian, J. H., Chen, L. J., Li, K., et al. (2018). The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202. doi:10.1038/s41586-018-0012-7
Shokri, L., Inukai, S., Hafner, A., Weinand, K., Hens, K., Vedenko, A., et al. (2019). A comprehensive Drosophila melanogaster transcription factor interactome. Cell. Rep. 27, 955–970.e7. doi:10.1016/j.celrep.2019.03.071
Siegmund, T., and Lehmann, M. (2002). The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev. Genes. Evol. 212, 152–157. doi:10.1007/s00427-002-0219-2
Simakov, O., Marletaz, F., Cho, S. J., Edsinger-Gonzales, E., Havlak, P., Hellsten, U., et al. (2013). Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531. doi:10.1038/nature11696
Smith, C. L., Varoqueaux, F., Kittelmann, M., Azzam, R. N., Cooper, B., Winters, C. A., et al. (2014). Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24, 1565–1572. doi:10.1016/j.cub.2014.05.046
Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. doi:10.1038/nature07191
Stoltzfus, A. (1999). On the possibility of constructive neutral evolution. J. Mol. Evol. 49, 169–181. doi:10.1007/pl00006540
Sullivan, K. F., and Glass, C. A. (1991). CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma 100, 360–370. doi:10.1007/BF00337514
Sundaram, V., and Wysocka, J. (2020). Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos. Trans. R. Soc. Lond B Biol. Sci. 375, 20190347. doi:10.1098/rstb.2019.0347
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi:10.1093/nar/gkl315
Vervoort, M., and Ledent, V. (2001). The evolution of the neural basic Helix-Loop-Helix proteins. ScientificWorldJournal 1, 396–426. doi:10.1100/tsw.2001.68
Volff, J. N. (2006). Turning junk into gold: Domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28, 913–922. doi:10.1002/bies.20452
Whelan, N. V., Kocot, K. M., Moroz, L. L., and Halanych, K. M. (2015). Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5773–5778. doi:10.1073/pnas.1503453112
Whelan, N. V., Kocot, K. M., Moroz, T. P., Mukherjee, K., Williams, P., Paulay, G., et al. (2017). Ctenophore relationships and their placement as the sister group to all other animals. Nat. Ecol. Evol. 1, 1737–1746. doi:10.1038/s41559-017-0331-3
Xu, Z., Yan, X., Maurais, S., Fu, H., O'Brien, D. G., Mottinger, J., et al. (2004). Jittery, a mutator distant relative with a paradoxical mobile behavior: Excision without reinsertion. Plant Cell. 16, 1105–1114. doi:10.1105/tpc.019802
Zattera, M. L., and Bruschi, D. P. (2022). Transposable elements as a source of novel repetitive DNA in the eukaryote genome. Cells 11, 3373. doi:10.3390/cells11213373
Keywords: placozoa, ctenophora, porifera, cnidaria, mollusca, convergent domestication, transcription factors, class II DNA transposons
Citation: Mukherjee K and Moroz LL (2023) Transposon-derived transcription factors across metazoans. Front. Cell Dev. Biol. 11:1113046. doi: 10.3389/fcell.2023.1113046
Received: 01 December 2022; Accepted: 09 February 2023;
Published: 07 March 2023.
Edited by:
Pedro Martinez, University of Barcelona, SpainReviewed by:
Stephane Boissinot, New York University Abu Dhabi, United Arab EmiratesKirill Ustyantsev, University Medical Center Groningen, Netherlands
Manuel Fernández Moreno, Center for Genomic Regulation (CRG), Spain
Copyright © 2023 Mukherjee and Moroz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonid L. Moroz, bW9yb3pAd2hpdG5leS51ZmwuZWR1; Krishanu Mukherjee, a3Jpc2hhbnVAdWZsLmVkdQ==
 Krishanu Mukherjee
Krishanu Mukherjee Leonid L. Moroz
Leonid L. Moroz