
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 03 February 2023
Sec. Membrane Traffic and Organelle Dynamics
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1112954
This article is part of the Research Topic The Evolving Role of Lipid Droplets: Advancements and Future Directions View all 8 articles
Cells store excess energy in the form of lipid droplets (LDs), a specialized sub-compartment of the endoplasmic reticulum (ER) network. The lipodystrophy protein seipin is a key player in LD biogenesis and ER-LD contact site maintenance. Recent structural and in silico studies have started to shed light on the molecular function of seipin as a LD nucleator in early LD biogenesis, whilst new cell biological work implies a role for seipin in ER-mitochondria contact sites and calcium metabolism. In this minireview, I discuss recent insights into the molecular function of seipin.
Seipin mutations have been linked to severe congenital lipodystrophy (BSCL2), motor neuronal disorders and congenital encephalopathy (Magré et al., 2001; Windpassinger et al., 2004; Ito and Suzuki, 2009; Guillén-Navarro et al., 2013). BSCL2 arises via loss-of seipin function, whereas neuronal disorders may be related to accumulation of toxic seipin oligomers (Rao and Goodman, 2021). Mechanistically, seipin localizes to ER-LD contact sites and is crucial for LD formation across the evolutionary tree (Chapman et al., 2019). Seipin is also required for adipogenesis and adipocyte maintenance, and it is an important open question whether the as-yet-unresolved molecular function of seipin at LDs is the culprit for defective adipogenesis.
As there are several excellent recent reviews on this topic (e.g. (Gao et al., 2019; Renne et al., 2020; Rao and Goodman, 2021; Schneiter and Choudhary, 2022), I will focus on most recent insights and emerging themes in the ongoing voyage to uncover the molecular function of seipin, a “mysterious” protein (Agarwal and Garg, 2004).
During LD biogenesis, neutral lipids (NLs, triglycerides, TAG, and sterol esters, SE) accumulate within the ER bilayer, with rising concentrations leading to the formation of nanoscale lenses (Thiam and Ikonen, 2020). Lenses have been predicted by simulations (Khandelia et al., 2010; Zoni et al., 2021a; Kim et al., 2022b) and 40–60 nm-in-size lenses have been observed in yeast by electron microscopy (EM) (Choudhary et al., 2015). Upon further growth lenses bud out to the cytoplasmic side of the ER as nascent LDs, which remain in contact with the ER. It is accepted that virtually all LDs remain in contact with the ER in yeast (Jacquier et al., 2011). In mammalian systems it is often stated that LDs detach from the ER after their formation, but there is no clear evidence supporting this. Instead, LDs likely remain in contact with the ER (Salo et al., 2016), and this connection may only be severed in specialized cases, such as during LD engulfment to degradatory compartments (Schulze et al., 2020) or in milk-secreting epithelial cells of the mammary gland (Monks et al., 2020).
Nascent LD formation is influenced by biophysical factors, including local ER lipid composition, membrane shape and surface tension (Ben M’barek et al., 2017; Choudhary et al., 2018; Chorlay et al., 2019; Santinho et al., 2020; Zoni et al., 2021a). Specific proteins also impact LD assembly, including FIT2, DFCP1, LDAF1, ACSL3, perilipins, ER curvature proteins and the yeast lipin orthologue Pah1 (Adeyo et al., 2011; Kassan et al., 2014; Choudhary et al., 2015; Gao et al., 2017; Li D. et al., 2019; Chung et al., 2019; Chen et al., 2021). An elegant recent study ordered the recruitment of many of these proteins during LD formation in yeast, providing clear evidence for a stepwise assembly of LDs (Choudhary et al., 2020). In this LD assembly cascade, seipin plays a pivotal role in both LD nucleation and controlling subsequent LD growth at ER-LD contacts.
By live cell microscopy in human cells, partially immobilized seipin complexes preclude the recruitment of many LD markers, such as ACSL3, Perilipin-3, Bodipy, LD540 or LiveDrop (Chung et al., 2019; Salo et al., 2019). MCTP2 was also recently shown to mark nascent LD formation sites in the ER (Joshi et al., 2018; 2021), MCTP2-sites appeared more immobile than seipin-defined sites. DFCP1 (Li D. et al., 2019) and FIT2, Reep5 and Rtn4A (Chen et al., 2021) were also recently shown to transiently accumulate at ER sites prior to LiveDrop appearance. As these proteins likely harbor membrane-shaping capabilities, these data further imply that ER curvature is vital for early LDs (Santinho et al., 2020). It will be crucial in the future to investigate the temporal relationship of these players in respect to seipin.
Seipin homo-oligomers consist of N- and C-terminal cytosolic regions and a conserved luminal domain flanked by two transmembrane domains (TMDs) (Lundin et al., 2006; Binns et al., 2010). The cryo-EM structure of human and Drosophila seipin luminal domains indicates that seipin forms ER luminal disks of 11–12 subunits, with a diameter of ∼15 nm (Sui et al., 2018; Yan et al., 2018) (Figure 1A). The outer layer of this ring is composed of closely interacting beta-sandwich folds with structural similarity to lipid-binding domains, binding anionic phospholipids in vitro (Yan et al., 2018). These putative phospholipid binding domains are especially interesting in the context that seipin has been implicated in localized control of phosphatidic acid (PA) metabolism (Fei et al., 2011; Sim et al., 2012; 2020; Jiang et al., 2014; Han et al., 2015; Talukder et al., 2015; Wolinski et al., 2015; Pagac et al., 2016; Soltysik et al., 2020). Indeed, one model for seipin function is that it prevents ectopic accumulation of PA in the ER, which could lead to both aberrant LD formation and disturb the adipogenic programme by interfering with the key transcription factor PPAR-gamma (Rao and Goodman, 2021).
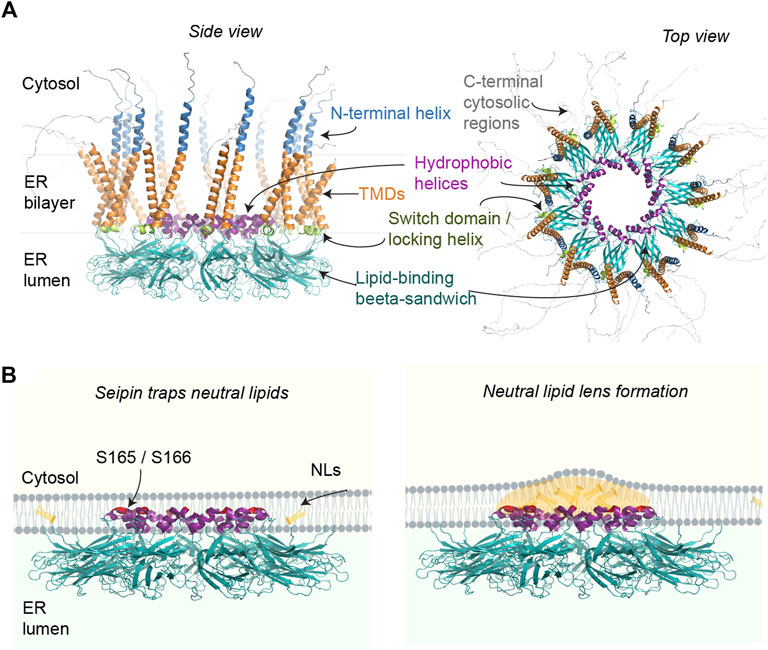
FIGURE 1. (A) Putative structural elements of the human seipin oligomer (isoform 1), predicted using Alphafold Multimer (Evans et al., 2022) and visualized in PyMol. Only the luminal regions containing the hydrophobic helices and lipid-binding beta sandwich of human seipin have been solved by cryo-EM. The rest of the annotations are based on published yeast structures and functional analyses. Alphafold predictions suggest cytosolic N- and C-terminal regions (only partially shown) may be mostly disordered in vivo. (B) Working model for how seipin may catalyze LD formation by facilitating NL nucleation in the ER. For clarity, only select ER luminal regions of seipin are depicted, with the putative NL-interacting residues highlighted in red.
Besides PA, a recent study indicated that PI3P, another anionic phospholipid, accumulates in ER foci in seipin KO (Lukmantara et al., 2022). Remarkably, decreasing PI3P was sufficient to partially rescue the LD phenotype of seipin KO. Increased PI3P via seipin KO was shown to impair the activity of DFCP1 in facilitating early LD formation. Seipin and DFCP1 have been mechanistically linked previously (Li D. et al., 2019) and although endogenous proteins do not directly interact (Lukmantara et al., 2022) it will be crucial to further decipher their interplay in LD formation. As DFCP1 harbors NTPase activity (Ismail et al., 2022; Nähse et al., 2022) facilitating omegasome constriction (Nähse et al., 2022) it could be directly involved in membrane deformation during LD budding.
Seipin also contains hydrophobic helices (HHs) in the center of the ring, partially embedded in the ER bilayer. Molecular simulations supported by mutational analysis indicate that the HHs may induce nanoscale clustering of TAG molecules (Zoni et al., 2021b; Klug et al., 2021; Prasanna et al., 2021; Kim et al., 2022a; Renne et al., 2022) (Figure 1B). Importantly, seipin induces this TAG lensing at a lower concentration than TAG alone, providing a plausible explanation why LDs form at sites marked by seipin and ER TAG is increased in seipin KO (Gao et al., 2017; Salo et al., 2019; Choudhary et al., 2020).
Biochemically, TAG has been found to co-purify with seipin, but only in the presence of its important interactor LDAF1, a likely homolog of yeast ldo proteins (Eisenberg-Bord et al., 2018; Teixeira et al., 2018; Castro et al., 2019). Indeed, analysis using acute seipin removal with the auxin-inducible degradation system indicated that LDAF1 may serve to protect seipin-TAG lenses (Prasanna et al., 2021). It is plausible that even in TAG-deprived situations minute amounts of TAGs may become stably clustered within the seipin disk, with a high number of seipin-LDAF1 complexes always primed for LD formation. In the absence of LDAF1, LDs are fewer in number but larger (Chung et al., 2019; Chartschenko et al., 2021), which could be a consequence of defectively primed (“TAGless”) seipins needing higher ER TAG concentrations before LD formation can begin.
The structure of yeast seipin was recently resolved by cryo-EM, and shows an overall similar fold to its mammalian counterparts, albeit with 10 subunits (Klug et al., 2021; Arlt et al., 2022). In yeast, seipin function is carried out by two interacting proteins, Sei1 and Ldb16, which form a stable complex. However, density could not be resolved for Ldb16, indicating structural flexibility. Nevertheless, the structures of Sei1 and supporting cross-linking analysis revealed that in yeast the HHs important for LD nucleation are likely provided by Ldb16 (Klug et al., 2021).
The structures also resolved seipin TMDs and one of the structures indicated that the TMDs of seipin exist in two separate conformations (Arlt et al., 2022). It was speculated that during LD formation the seipin ring may thus open up via a conformational switch of the TMDs as additional TAGs are accommodated. In line with this, a conserved, luminal region near the TMDs, called switch domain or locking helix, appears to be important to control TMD orientation and flexibility (Klug et al., 2021; Arlt et al., 2022). Indeed, simulations indicate that the TMDs and nearby residues are important for seipin function in both attracting TAGs in the ER bilayer (Zoni et al., 2021b) and facilitating the conversion of a flat TAG-lens into a budding LD (Kim et al., 2022a).
It is well documented that many seipin foci (likely corresponding to individual seipin oligomers although this has not been formally shown) localize stably to ER-LD contact sites in yeast, Drosophila, C. elegans and human cell models (Szymanski et al., 2007; Fei et al., 2008; Salo et al., 2016; Wang et al., 2016; Cao et al., 2019) (Figure 2). In human non-adipocyte cell models, there is typically one seipin-mediated contact per LD, which displays direct membrane continuity, possibly as a consequence of LD budding taking place through the seipin disk (Salo et al., 2020). At ER-LD contacts, seipin appears to be required for continuous LD growth via the ER-LD contact (Salo et al., 2019) and protein trafficking between the ER and LDs (Grippa et al., 2015; Salo et al., 2016; Cottier and Schneiter, 2022). The aforementioned TAG-attracting propensity of seipin may be important for this (Zoni et al., 2021b; Prasanna et al., 2021). Seipin may physically stabilize the contact site, which is supported by the notion that by electron tomography seipin-mediated ∼15 nm ER-LD necks display uniform membrane architecture and size (Salo et al., 2019). The TMDs of seipin, likely exhibiting a degree of flexibility, may also effect phospholipid and/or protein diffusion at the ER-LD neck. Recent in vitro studies in protein-free systems have begun to shed light on the unique properties of ER-LD membrane continuities (Chorlay et al., 2021; Puza et al., 2022), paving the way to understand the contribution of protein factors such as seipin in ER-LD cargo exchange.
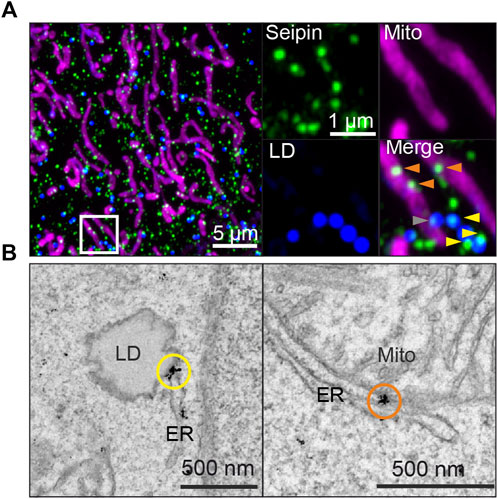
FIGURE 2. Endogenously tagged seipin localizes to both ER-LD and ER-mitochondria contact sites in human A431 cells, as shown by (A) Airyscan microscopy of fixed cells and (B) immuno-EM gold labeling. Yellow arrowheads and circles: seipin localization at ER-LD contact sites, orange arrowheads and circles: seipin localization at ER-mitochondria contact sites. Gray arrowheads indicate seipins near both sites simultaneously. For detailed methodology, see (Combot et al., 2022) from wherein image is adapted from.
In addition to seipin-mediated ER-LD contact sites, there are also other types of ER-LD contacts, including wider ER-LD membrane proximities resembling canonical membrane contact sites (Salo and Ikonen, 2019; Bohnert, 2020). These may be regulated by ER-LD tethering factors such as Rab18, Snx14, Vps13 and MOSPD2 (Kumar et al., 2018; Xu et al., 2018; Datta et al., 2019; Zouiouich et al., 2022) although the division of labor between such contact sites and direct membrane continuities is unclear.
Recently, in a tour de force study Drosophila cells, a specific set of membrane-fusion machinery was proposed to generate seipin-independent ER-LD membrane bridges to enable specialized protein cargo, such as GPAT4, to traffic to a subset of LDs upon their maturation (Song et al., 2022). Using FRAP, GPAT4 was found to traffic to LDs via the ER at sites not directly harboring a seipin complex (Song et al., 2022). However, the resolution of light microscopy in the crowded ER-LD network makes interpretation of such data challenging. An alternative possibility is that the described protein machinery acts on matured LDs and directly alters their surface properties, such as membrane tension (Gong et al., 2011; Thiam et al., 2013), thus allowing specific protein cargo to travel to the LD via the existing seipin-mediated ER-LD neck. In any case, it will be interesting to compare the membrane architecture and nanoscale dimensions of these seipin-dependent and -independent contact sites and how this correlates with their functionality.
To date, most reports of seipin in LD formation have focused on TAG-LDs. However, recent work has begun to shed light on the role of seipin specifically in SE-LDs. SE levels were unaltered in BSCL2 patient lymphoid cells (Boutet et al., 2009), decreased in seipin KO A431 cells (Salo et al., 2016) and slightly increased in seipin KO yeast (Wang et al., 2014; Grippa et al., 2015). As seipin has been documented to reside at ER-LD contacts of SE-LDs in both yeast and human cells (Santinho et al., 2020; Molenaar et al., 2021), seipin may also play a role in SE-LD formation. However, the assembly of SE-LDs may be quite different to TAG-LDs due to different physical chemistry (Thiam and Ikonen, 2020). Indeed, in contrast to TAG-LDs, seipin could not determine SE-LD biogenesis site in yeast (Molenaar et al., 2021) suggesting seipin-independent pathways for SE-LD formation. Seipin could also not determine the biogenesis site for retinyl ester-LDs, although it did facilitate their packaging into pre-existing TAG-LDs (Molenaar et al., 2021).
In a recent study, careful quantification of LD sizes in yeast seipin mutants engineered to produce SE- or retinyl ester -only LDs revealed marked alterations in their sizes (Renne et al., 2022). Simulations also showed that the HHs in the seipin ring could cluster SEs similarly as TAGs, suggesting seipin could facilitate SE-LD nucleation (Renne et al., 2022). These data suggest seipin can act as a universal NL-clustering machine in the ER, although some of the observed differences may also be caused by defective ER-LD lipid flux rather than defective initial LD formation. Very recently seipin was also shown to be essential for SE-LD maintenance and formation in vivo, in steroidogenic tissues of mice (Shen et al., 2022).
Interestingly, emerging work suggests that LDs can undergo phase transition, with liquid crystalline lattices detected by in situ cryo-electron tomography and linked to high SE/TAG ratios (Czabany et al., 2008; Mahamid et al., 2019; Shimobayashi and Ohsaki, 2019). Driven by lipolysis of TAGs or other metabolic cues (Suzuki et al., 2012; Mahamid et al., 2019; Rogers et al., 2022), rising SE concentrations thus lead to the formation of lattices at the periphery of LDs, whilst LD cores remain amorphous with TAGs. How such lattices may impact the nanoscale architecture of seipin-mediated ER-LD contact sites remains to be investigated, but proteomic changes in SE vs. TAG LDs (Hsieh et al., 2012; Khor et al., 2014; Rogers et al., 2022) could be related to altered functionality of seipin-mediated ER-LD necks.
In addition to its role in LD assembly and PA metabolism, several studies have linked seipin function to calcium fluxes. Seipin was found to physically interact with the ER calcium pump SERCA in Drosophila, acting as a positive SERCA regulator (Bi et al., 2014). In seipin KO flies, ER calcium was reduced and fat body lipogenesis defects could be partially rescued by disturbing ER-to-cytosol calcium efflux. In a follow-up study, it was found that the defective ER calcium stores led to decreased mitochondrial calcium and concomitant defects in TCA cycle and mitochondrial function (Ding et al., 2018). Indeed, several studies have since uncovered altered calcium homeostasis in seipin knockdown systems (Li Q. et al., 2019; Wu et al., 2021).
We recently investigated seipin links to mitochondria and calcium fluxes using human and mouse cells as well as inducibly seipin depleted mice. We found that within hours of acute seipin removal, calcium flux to mitochondria was reduced, whilst ER calcium stores remained initially intact (Combot et al., 2022). Reduced mitochondrial calcium flux was accompanied by mitochondrial defects in multiple cell types including BSCL2 patient cells. Mechanistically, a subset of seipins were found to localize at ER-mitochondria contact sites (MsAMs) (Figure 2) in a nutritionally regulated manner, in close proximity to calcium regulators SERCA and IP3R, suggesting seipin may be important in stabilizing MAMs for calcium flux into mitochondria.
Another recent study also reported that a subset of seipin localized to MAMs in human cells (Guyard et al., 2022). They found that an Orp5/Orp8 complex is important for LD formation and localizes to newly described ER-MAM-LD tripartite contact sites, wherein LDs were proposed to emerge. Importantly, Orp5 was necessary to recruit seipin to MAMs, which were also found to be enriched in PA. Interestingly, whilst (Guyard et al., 2022) linked MAM-seipins to LD formation, (Combot et al., 2022), found MAM-seipin localization to decrease in lipogenic conditions and increase in starvation. These discrepancies could be related to differences between cell-types. Altogether, nutritional status of the cell may effect MAM lipid environment, leading to seipin recruitment via Orp5, wherein seipin could be important for stabilizing calcium-regulating ER subdomains.
These studies highlight the emerging concept that seipin may be involved in multiple ER contact sites and/or organelle assembly platforms (LDs and mitochondria in mammals and LDs and nascent peroxisomes in yeast (Joshi et al., 2018; Wang et al., 2018)). Interestingly, two seipin isoforms in Arabidopsis were recently shown to physically interact with VAP during LD formation (Greer et al., 2020). Whilst mammalian seipin contains no obvious VAP-interacting FFAT motif, pulldowns of endogenously tagged mouse seipin did contain VAPA (Combot et al., 2022), prompting further study. The dynamic mobility of most MAM-associated seipins (Combot et al., 2022) appears similar to the dynamic VAPB MAM domains recently described by single molecule imaging (Obara et al., 2022).
At present, the two main, mutually non-exclusive hypothesis of seipin function (control of phospholipid metabolism and as a LD nucleator) are both supported by strong experimental clues. There appear to be at least three, partially overlapping populations of seipins: ER, ER-LD and ER-mitochondrial seipins. These may exert different functions due to altered conformations or binding partners. For further cataloguing of seipin subcellular behavior and localization, it would be important to investigate endogenous-level seipin, since overexpression may alter or mask its true localization (Lak et al., 2021). Furthermore, care should be exercised when assessing seipin LD phenotypes; LD markers of sufficient sensitivity are required to fully characterize the typical seipin KO phenotype of tiny and supersized LDs (Salo et al., 2020).
In regards to seipin-TAG lenses, direct observations of them have not been reported and the previously reported lenses in yeast cells seem too large to be contained within the seipin disk (Choudhary et al., 2015). Going forward, reconstitution of seipin in TAG-enriched model membranes would help to tackle its role in LDs, aided by recent advances in mimicking LD nucleation in protein-free systems (Santinho et al., 2020; Hegaard et al., 2022). High resolution in situ cryo-EM could be another avenue to directly visualize seipin-mediated early LDs. Further structural work on seipin in complex with its myriad interactors will also be crucial to decipher the function of seipin at different membrane contact sites.
The author confirms being the sole contributor of this work and has approved it for publication.
This work was supported by Marie Skłodowska-Curie Actions (101028297), the Biomedicum Helsinki Foundation and the Orion Foundation.
The author acknowledges Dr. Rasmus K. Jensen for help with AlphaFold multimer. Figure 1B was generated with the help of Biorender.com.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adeyo, O., Horn, P. J., Lee, S., Binns, D. D., Chandrahas, A., Chapman, K. D., et al. (2011). The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055. doi:10.1083/jcb.201010111
Agarwal, A. K., and Garg, A. (2004). Seipin: A mysterious protein. Trends Mol. Med. 10, 440–444. doi:10.1016/j.molmed.2004.07.009
Arlt, H., Sui, X., Folger, B., Adams, C., Chen, X., Remme, R., et al. (2022). Seipin forms a flexible cage at lipid droplet formation sites. Nat. Struct. Mol. Biol. 29, 194–202. doi:10.1038/s41594-021-00718-y
Ben M’barek, K., Ajjaji, D., Chorlay, A., Vanni, S., Forêt, L., and Thiam, A. R. (2017). ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604. doi:10.1016/j.devcel.2017.05.012
Bi, J., Wang, W., Liu, Z., Hueng, X., Jiang, Q., Liu, G., et al. (2014). Seipin promotes adipose tissue fat storage through the ER Ca²⁺-ATPase SERCA. Cell Metab. 19, 861–871. doi:10.1016/j.cmet.2014.03.028
Binns, D., Lee, S., Hilton, C. L., Jiang, Q.-X., and Goodman, J. M. (2010). Seipin is a discrete homooligomer. Biochemistry 49, 10747–10755. doi:10.1021/bi1013003
Bohnert, M. (2020). Tethering fat: Tethers in lipid droplet contact sites. Contact 3, 251525642090814. doi:10.1177/2515256420908142
Boutet, E., El Mourabit, H., Prot, M., Nemani, M., Khallouf, E., Colard, O., et al. (2009). Seipin deficiency alters fatty acid Delta9 desaturation and lipid droplet formation in Berardinelli-Seip congenital lipodystrophy. Biochimie 91, 796–803. doi:10.1016/j.biochi.2009.01.011
Cao, Z., Hao, Y., Lee, Y. Y., Wang, P., Li, X., Xie, K., et al. (2019). Dietary fatty acids promote lipid droplet diversity through seipin enrichment in an ER subdomain. Nat. Commun. 10, 2902. doi:10.1038/s41467-019-10835-4
Castro, I. G., Eisenberg-Bord, M., Persiani, E., Rochford, J. J., Schuldiner, M., and Bohnert, M. (2019). Promethin is a conserved seipin partner protein. Cells 8, 268. doi:10.3390/cells8030268
Chapman, K. D., Aziz, M., Dyer, J. M., and Mullen, R. T. (2019). Mechanisms of lipid droplet biogenesis. Biochem. J. 476, 1929–1942. doi:10.1042/bcj20180021
Chartschenko, E., Hugenroth, M., Akhtar, I., Droste, A., Kolkhof, P., Bohnert, M., et al. (2021). CG32803 is the fly homolog of LDAF1 and influences lipid storage in vivo. Insect biochem. Mol. Biol. 133, 103512. doi:10.1016/j.ibmb.2020.103512
Chen, F., Yan, B., Ren, J., Lyu, R., Wu, Y., Guo, Y., et al. (2021). FIT2 organizes lipid droplet biogenesis with ER tubule-forming proteins and septins. J. Cell Biol. 220, e201907183. doi:10.1083/jcb.201907183
Chorlay, A., Forêt, L., and Thiam, A. R. (2021). Origin of gradients in lipid density and surface tension between connected lipid droplet and bilayer. Biophys. J. 120, 5491–5503. doi:10.1016/j.bpj.2021.11.022
Chorlay, A., Monticelli, L., Veríssimo Ferreira, J., Ben M’barek, K., Ajjaji, D., Wang, S., et al. (2019). Membrane asymmetry imposes directionality on lipid droplet emergence from the ER. Dev. Cell 50, 25–42. doi:10.1016/j.devcel.2019.05.003
Choudhary, V., El Atab, O., Mizzon, G., Prinz, W. A., and Schneiter, R. (2020). Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J. Cell Biol. 219, e201910177. doi:10.1083/jcb.201910177
Choudhary, V., Golani, G., Joshi, A. S., Cottier, S., Schneiter, R., Prinz, W. A., et al. (2018). Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr. Biol. 28, 915–926. doi:10.1016/j.cub.2018.02.020
Choudhary, V., Ojha, N., Golden, A., and Prinz, W. A. (2015). A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211, 261–271. doi:10.1083/jcb.201505067
Chung, J., Wu, X., Lambert, T. J., Lai, Z. W., Walther, T. C., Farese, R. V., et al. (2019). LDAF1 and seipin form a lipid droplet assembly complex. Dev. Cell 51, 1–13. doi:10.1016/j.devcel.2019.10.006
Combot, Y., Salo, V. T., Chadeuf, G., Hölttä, M., Ven, K., Pulli, I., et al. (2022). Seipin localizes at endoplasmic-reticulum-mitochondria contact sites to control mitochondrial calcium import and metabolism in adipocytes. Cell Rep. 38, 110213. doi:10.1016/j.celrep.2021.110213
Cottier, S., and Schneiter, R. (2022). Lipid droplets form a network interconnected by the endoplasmic reticulum through which their proteins equilibrate. J. Cell Sci. 135, jcs258819. doi:10.1242/jcs.258819
Czabany, T., Wagner, A., Zweytick, D., Lohner, K., Leitner, E., Ingolic, E., et al. (2008). Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283, 17065–17074. doi:10.1074/jbc.M800401200
Datta, S., Liu, Y., Hariri, H., Bowerman, J., and Henne, W. M. (2019). Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. 218, 1335–1351. doi:10.1083/jcb.201808133
Ding, L., Yang, X., Tian, H., Liang, J., Zhang, F., Wang, G., et al. (2018). Seipin regulates lipid homeostasis by ensuring calcium-dependent mitochondrial metabolism. EMBO J. 37, 1–17. doi:10.15252/embj.201797572
Eisenberg-Bord, M., Mari, M., Weill, U., Rosenfeld-Gur, E., Moldavski, O., Castro, I. G., et al. (2018). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217, 269–282. doi:10.1083/jcb.201704122
Evans, R., O’Neill, M., Pritzel, A., Antropova, N., Senior, A., Green, T., et al. (2022). “Protein complex prediction with AlphaFold-Multimer,”. bioRxiv.
Fei, W., Shui, G., Gaeta, B., Du, X., Kuerschner, L., Li, P., et al. (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482. doi:10.1083/jcb.200711136
Fei, W., Shui, G., Zhang, Y., Krahmer, N., Ferguson, C., Kapterian, T. S., et al. (2011). A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201. doi:10.1371/journal.pgen.1002201
Gao, M., Huang, X., Song, B.-L., and Yang, H. (2019). The biogenesis of lipid droplets: Lipids take center stage. Prog. Lipid Res. 75, 100989. doi:10.1016/j.plipres.2019.100989
Gao, Q., Binns, D. D., Kinch, L. N., Grishin, N. V., Ortiz, N., Chen, X., et al. (2017). Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J. Cell Biol. 216, 3199–3217. doi:10.1083/jcb.201610013
Gong, J., Sun, Z., Wu, L., Xu, W., Schieber, N., Xu, D., et al. (2011). Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 195, 953–963. doi:10.1083/jcb.201104142
Greer, M. S., Cai, Y., Gidda, S. K., Esnay, N., Kretzschmar, F. K., Seay, D., et al. (2020). SEIPIN isoforms interact with the membrane-tethering protein VAP27-1 for lipid droplet formation. Plant Cell 32, 2932–2950. doi:10.1105/tpc.19.00771
Grippa, A., Buxó, L., Mora, G., Funaya, C., Idrissi, F.-Z., Mancuso, F., et al. (2015). The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–844. doi:10.1083/jcb.201502070
Guillén-Navarro, E., Sánchez-Iglesias, S., Domingo-Jiménez, R., Victoria, B., Ruiz-Riquelme, A., Rábano, A., et al. (2013). A new seipin-associated neurodegenerative syndrome. J. Med. Genet. 50, 401–409. doi:10.1136/jmedgenet-2013-101525
Guyard, V., Monteiro-Cardoso, V. F., Omrane, M., Sauvanet, C., Houcine, A., Boulogne, C., et al. (2022). ORP5 and ORP8 orchestrate lipid droplet biogenesis and maintenance at ER–mitochondria contact sites. J. Cell Biol. 221, e202112107. doi:10.1083/jcb.202112107
Han, S., Binns, D. D., Chang, Y.-F., and Goodman, J. M. (2015). Dissecting seipin function: The localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1pΔNterm only in combination with Ldb16p. BMC Cell Biol. 16, 29. doi:10.1186/s12860-015-0075-3
Hegaard, F. V., Klenow, M. B., and Simonsen, A. C. (2022). Lens nucleation and droplet budding in a membrane model for lipid droplet biogenesis. Langmuir 38, 9247–9256. doi:10.1021/acs.langmuir.2c01014
Hsieh, K., Lee, Y. K., Londos, C., Raaka, B. M., Dalen, K. T., and Kimmel, A. R. (2012). Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-esterspecific intracellular lipid storage droplets. J. Cell Sci. 125, 4067–4076. doi:10.1242/jcs.104943
Ismail, V. A., Naismith, T., and Kast, D. J. (2022). The NTPase Activity of the Double FYVE Domain Containing Protein 1 Regulates Lipid Droplet Metabolism. J. Biol. Chem., 299. doi:10.1016/j.jbc.2022.102830
Ito, D., and Suzuki, N. (2009). Seipinopathy: A novel endoplasmic reticulum stress-associated disease. Brain 132, 8–15. doi:10.1093/brain/awn216
Jacquier, N., Choudhary, V., Mari, M., Toulmay, A., Reggiori, F., and Schneiter, R. (2011). Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124, 2424–2437. doi:10.1242/jcs.076836
Jiang, M., Gao, M., Wu, C., He, H., Guo, X., Zhou, Z., et al. (2014). Lack of testicular seipin causes teratozoospermia syndrome in men. Proc. Natl. Acad. Sci. 111, 7054–7059. doi:10.1073/pnas.1324025111
Joshi, A. S., Nebenfuehr, B., Choudhary, V., Satpute-Krishnan, P., Levine, T. P., Golden, A., et al. (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9, 2940. doi:10.1038/s41467-018-05277-3
Joshi, A. S., Ragusa, J. V., Prinz, W. A., and Cohen, S. (2021). Multiple C2 domain-containing transmembrane proteins promote lipid droplet biogenesis and growth at specialized endoplasmic reticulum subdomains. Mol. Biol. Cell 32, 1147–1157. doi:10.1091/MBC.E20-09-0590
Kassan, A., Herms, A., Fernández-Vidal, A., Bosch, M., Schieber, N. L., Reddy, B. J. N., et al. (2013). Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Sci. 203, 985–1001. doi:10.1083/jcb.201305142
Khandelia, H., Duelund, L., Pakkanen, K. I., and Ipsen, J. H. (2010). Triglyceride blisters in lipid bilayers: Implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS One 5, e12811–e12818. doi:10.1371/journal.pone.0012811
Khor, V. K., Ahrends, R., Lin, Y., Shen, W. J., Adams, C. M., Roseman, A. N., et al. (2014). The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One 9, 1–11. doi:10.1371/journal.pone.0105047
Kim, S., Chung, J., Arlt, H., Pak, A. J., Farese Jnr, R. V., Walther, T. C., et al. (2022a). Seipin transmembrane segments critically function in triglyceride nucleation and lipid droplet budding from the membrane. Elife 11, e75808–e75820. doi:10.7554/eLife.75808
Kim, S., Li, C., Farese, R. V., Walther, T. C., and Voth, G. A. (2022b). Key factors governing initial stages of lipid droplet formation. J. Phys. Chem. B 126, 453–462. doi:10.1021/acs.jpcb.1c09683
Klug, Y. A., Deme, J. C., Corey, R. A., Renne, M. F., Stansfeld, P. J., Lea, S. M., et al. (2021). Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Nat. Commun. 12, 5892. doi:10.1038/s41467-021-26162-6
Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F. A., Li, P., Lees, J. A., et al. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639. doi:10.1083/jcb.201807019
Lak, B., Li, S., Belevich, I., Sree, S., Butkovic, R., Ikonen, E., et al. (2021). Specific subdomain localization of ER resident proteins and membrane contact sites resolved by electron microscopy. Eur. J. Cell Biol. 100, 151180. doi:10.1016/j.ejcb.2021.151180
Li, D., Zhao, Y. G., Li, D., Zhao, H., Huang, J., Miao, G., et al. (2019a). The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep. 27, 343–358. doi:10.1016/j.celrep.2019.03.025
Li, Q., Li, Y., Zhang, Z., Kang, H., Zhang, L., Zhang, Y., et al. (2019b). SEIPIN overexpression in the liver may alleviate hepatic steatosis by influencing the intracellular calcium level. Mol. Cell. Endocrinol. 488, 70–78. doi:10.1016/j.mce.2019.03.005
Lukmantara, I., Chen, F., Mak, H. Y., Zadoorian, A., Du, X., Xiao, F. N., et al. (2022). PI(3)P and DFCP1 regulate the biogenesis of lipid droplets. Mol. Biol. Cell. 33, ar131. doi:10.1091/mbc.E22-07-0279
Lundin, C., Nordström, R., Wagner, K., Windpassinger, C., Andersson, H., von Heijne, G., et al. (2006). Membrane topology of the human seipin protein. FEBS Lett. 580, 2281–2284. doi:10.1016/j.febslet.2006.03.040
Magré, J., Delépine, M., Khallouf, E., Gedde-Dahl, T., Van Maldergem, L., Sobel, E., et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365–370. doi:10.1038/ng585
Mahamid, J., Tegunov, D., Maiser, A., Arnold, J., Leonhardt, H., Plitzko, J. M., et al. (2019). Liquid-crystalline phase transitions in lipid droplets are related to cellular states and specific organelle association. Proc. Natl. Acad. Sci. 116, 16866–16871. doi:10.1073/pnas.1903642116
Molenaar, M. R., Yadav, K. K., Toulmay, A., Wassenaar, T. A., Mari, M. C., Caillon, L., et al. (2021). Retinyl esters form lipid droplets independently of triacylglycerol and seipin. J. Cell Biol. 220, e202011071. doi:10.1083/jcb.202011071
Monks, J., Ladinsky, M. S., and McManaman, J. L. (2020). Organellar contacts of milk lipid droplets. Contact 3, 251525641989722. doi:10.1177/2515256419897226
Nähse, V., Raiborg, C., Wee Tan, K., Mørk, S., Lyngaas Torgersen, M., Wenzel, M., et al. (2022). “ATPase activity of DFCP1 controls selective autophagy,”. bioRxiv, 2022.02.24.481614.
Obara, C. J., Nixon-abell, J., Moore, A. S., Hoffman, D. P., Shtengel, G., Xu, C. S., et al. (2022). “Motion of single molecular tethers reveals dynamic subdomains at ER-mitochondria contact sites,”. J. bioRxiv.
Pagac, M., Cooper, D. E., Qi, Y., Lukmantara, I. E., Mak, H. Y., Wu, Z., et al. (2016). SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17, 1546–1559. doi:10.1016/j.celrep.2016.10.037
Prasanna, X., Salo, V. T., Li, S., Ven, K., Vihinen, H., Jokitalo, E., et al. (2021). Seipin traps triacylglycerols to facilitate their nanoscale clustering in the endoplasmic reticulum membrane. PLoS Biol. 19, e3000998. doi:10.1371/journal.pbio.3000998
Puza, S., Caesar, S., Poojari, C., Jung, M., Seemann, R., Hub, J. S., et al. (2022). Lipid droplets embedded in a model cell membrane create a phospholipid diffusion barrier. Small 18, e2106524. doi:10.1002/smll.202106524
Rao, M. J., and Goodman, J. M. (2021). Seipin: Harvesting fat and keeping adipocytes healthy. Trends Cell Biol. 31, 912–923. doi:10.1016/j.tcb.2021.06.003
Renne, M. F., Corey, R. A., Ferreira, J. V., Stansfeld, P. J., and Carvalho, P. (2022). Seipin concentrates distinct neutral lipids via interactions with their acyl chain carboxyl esters. J. Cell Biol. 221, e202112068. doi:10.1083/jcb.202112068
Renne, M. F., Klug, Y. A., and Carvalho, P. (2020). Lipid droplet biogenesis: A mystery “ unmixing. Semin. Cell Dev. Biol. 108, 14–23. doi:10.1016/j.semcdb.2020.03.001
Rogers, S., Gui, L., Kovalenko, A., Zoni, V., Carpentier, M., Ramji, K., et al. (2022). Triglyceride lipolysis triggers liquid crystalline phases in lipid droplets and alters the LD proteome. J. Cell Biol. 221, e202205053. doi:10.1083/jcb.202205053
Salo, V. T., Belevich, I., Li, S., Karhinen, L., Vihinen, H., Vigouroux, C., et al. (2016). Seipin regulates ER–lipid droplet contacts and cargo delivery. EMBO J. 24, 2699–2716. doi:10.15252/embj.201695170
Salo, V. T., Hölttä-Vuori, M., and Ikonen, E. (2020). Seipin-mediated contacts as gatekeepers of lipid flux at the endoplasmic reticulum – lipid droplet nexus. Contact 3, 251525642094582. doi:10.1177/2515256420945820
Salo, V. T., and Ikonen, E. (2019). Moving out but keeping in touch: Contacts between endoplasmic reticulum and lipid droplets. Curr. Opin. Cell Biol. 57, 64–70. doi:10.1016/j.ceb.2018.11.002
Salo, V. T., Li, S., Vihinen, H., Hölttä-Vuori, M., Szkalisity, A., Horvath, P., et al. (2019). Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev. Cell 50, 478–493. doi:10.1016/j.devcel.2019.05.016
Santinho, A., Salo, V. T., Chorlay, A., Li, S., Zhou, X., Omrane, M., et al. (2020). Membrane curvature catalyzes lipid droplet assembly. Curr. Biol. 30, 1–14. doi:10.1016/j.cub.2020.04.066
Schneiter, R., and Choudhary, V. (2022). Seipin collaborates with the ER membrane to control the sites of lipid droplet formation. Curr. Opin. Cell Biol. 75, 102070. doi:10.1016/j.ceb.2022.02.004
Schulze, R. J., Krueger, E. W., Weller, S. G., Johnson, K. M., Casey, C. A., Schott, M. B., et al. (2020). Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc. Natl. Acad. Sci. 117, 32443–32452. doi:10.1073/pnas.2011442117
Shen, W.-J., Cortez, Y., Singh, A., Chen, W., Azhar, S., and Kraemer, F. B. (2022). Mice deficient in ER protein seipin have reduced adrenal cholesteryl ester lipid droplet formation and utilization. J. Lipid Res. 100309, 100309. doi:10.1016/j.jlr.2022.100309
Shimobayashi, S. F., and Ohsaki, Y. (2019). Universal phase behaviors of intracellular lipid droplets. Proc. Natl. Acad. Sci. 116, 741264. doi:10.1101/741264
Sim, M. F. M., Dennis, R. J., Aubry, E. M., Ramanathan, N., Sembongi, H., Saudek, V., et al. (2012). The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol. Metab. 2, 38–46. doi:10.1016/j.molmet.2012.11.002
Sim, M. F. M., Persiani, E., Talukder, M. M. U., Mcilroy, G. D., Roumane, A., Edwardson, J. M., et al. (2020). Oligomers of the lipodystrophy protein seipin may co-ordinate GPAT3 and AGPAT2 enzymes to facilitate adipocyte differentiation. Sci. Rep. 10, 1–14. doi:10.1038/s41598-020-59982-5
Soltysik, K., Ohsaki, Y., Tatematsu, T., Cheng, J., Maeda, A., Morita, S., et al. (2020). Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220, e202005026. doi:10.1083/jcb.202005026
Song, J., Mizrak, A., Lee, C. W., Cicconet, M., Lai, Z. W., Tang, W. C., et al. (2022). Identification of two pathways mediating protein targeting from ER to lipid droplets. Nat. Cell Biol. 24, 1364–1377. doi:10.1038/s41556-022-00974-0
Sui, X., Arlt, H., Kelly, B., Zong, L., Frank, D., Debora, M., et al. (2018). Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 217, 4080–4091. doi:10.1083/jcb.201809067
Suzuki, M., Ohsaki, Y., Tatematsu, T., Shinohara, Y., Maeda, T., Cheng, J., et al. (2012). Translation inhibitors induce formation of cholesterol ester-rich lipid droplets. PLoS One 7, e42379–9. doi:10.1371/journal.pone.0042379
Szymanski, K. M., Binns, D., Bartz, R., Grishin, N. V., Li, W., Agarwal, A. K., et al. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A. 104, 20890–20895. doi:10.1073/pnas.0704154104
Talukder, M. M. U., Sim, M. F. M., O’Rahilly, S., Edwardson, J. M., and Rochford, J. J. (2015). Seipin oligomers can interact directly with AGPAT2 and lipin 1, physically scaffolding critical regulators of adipogenesis. Mol. Metab. 4, 199–209. doi:10.1016/j.molmet.2014.12.013
Teixeira, V., Johnsen, L., Martínez-Montañés, F., Grippa, A., Buxó, L., Idrissi, F. Z., et al. (2018). Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217, 127–138. doi:10.1083/jcb.201704115
Thiam, A. R., Farese, R. V., and Walther, T. C. (2013). The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786. doi:10.1038/nrm3699
Thiam, A. R., and Ikonen, E. (2020). Lipid droplet nucleation. Trends Cell Biol. 1, 108–118. doi:10.1016/j.tcb.2020.11.006
Wang, C.-W., Miao, Y.-H., and Chang, Y.-S. (2014). Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127, 1214–1228. doi:10.1242/jcs.137737
Wang, H., Becuwe, M., Housden, B. E., Chitraju, C., Porras, A. J., Graham, M. M., et al. (2016). Seipin is required for converting nascent to mature lipid droplets. Elife 5, 133–150. doi:10.7554/eLife.16582
Wang, S., Idrissi, F. Z., Hermansson, M., Grippa, A., Ejsing, C. S., and Carvalho, P. (2018). Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9, 1–12. doi:10.1038/s41467-018-05278-2
Windpassinger, C., Auer-Grumbach, M., Irobi, J., Patel, H., Petek, E., Hörl, G., et al. (2004). Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat. Genet. 36, 271–276. doi:10.1038/ng1313
Wolinski, H., Hofbauer, H. F., Hellauer, K., Cristobal-Sarramian, A., Kolb, D., Radulovic, M., et al. (2015). Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1851, 1450–1464. doi:10.1016/j.bbalip.2015.08.003
Wu, X., Liu, X., Wang, H., Zhou, Z., Yang, C., Li, Z., et al. (2021). Seipin deficiency accelerates heart failure due to calcium handling abnormalities and endoplasmic reticulum stress in mice. Front. Cardiovasc. Med. 8, 1–14. doi:10.3389/fcvm.2021.644128
Xu, D., Li, Y., Wu, L., Li, Y., Zhao, D., Yu, J., et al. (2018). Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995. doi:10.1083/jcb.201704184
Yan, R., Qian, H., Lukmantara, I., Gao, M., Du, X., Yan, N., et al. (2018). Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256. doi:10.1016/j.devcel.2018.09.010
Zoni, V., Khaddaj, R., Campomanes, P., Thiam, A. R., Schneiter, R., and Vanni, S. (2021a). Pre-existing bilayer stresses modulate triglyceride accumulation in the er versus lipid droplets. Elife 10, 1–24. doi:10.7554/eLife.62886
Zoni, V., Shinoda, W., and Vanni, S. (2021b). Seipin accumulates and traps diacylglycerols and triglycerides in its ring-like structure. Proc. Natl. Acad. Sci. 118, e2017205118. doi:10.1101/2020.10.27.357079
Keywords: seipin, lipid droplet, lipid droplet-ER contact sites, membrane contact site, endoplasmic reticulum, mitochondria-ER contact sites
Citation: Salo VT (2023) Seipin—still a mysterious protein?. Front. Cell Dev. Biol. 11:1112954. doi: 10.3389/fcell.2023.1112954
Received: 30 November 2022; Accepted: 17 January 2023;
Published: 03 February 2023.
Edited by:
Joel M. Goodman, University of Texas Southwestern Medical Center, United StatesReviewed by:
Pedro Carvalho, University of Oxford, United KingdomCopyright © 2023 Salo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veijo T. Salo, dmVpam8uc2Fsb0BlbWJsLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.