
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 07 September 2023
Sec. Stem Cell Research
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1112069
This article is part of the Research TopicFrom Single Stem Cells to Organoids, Organ Repair, and Public Health Volume IIView all 6 articles
Mutations in the transcription factor gene grainyhead-like 2 (GRHL2) are associated with progressive non-syndromic sensorineural deafness autosomal dominant type 28 (DFNA28) in humans. Since complete loss of Grhl2 is lethal in mouse embryos, we studied its role during inner ear pathology and hearing loss in vitro. To this end, we generated different homozygous deletions to knockout Grhl2 in mouse embryonic stem cells (Grhl2-KO ESCs), including some mimicking naturally occurring truncations in the dimerisation domain related to human DFNA28. Under naïve culture conditions, Grhl2-KO cells in suspension were more heterogenous in size and larger than wild-type controls. Adherent Grhl2-KO cells were also larger, with a less uniform shape, flattened, less circular morphology, forming loose monolayer colonies with poorly defined edges. These changes correlated with lower expression of epithelial cadherin Cdh1 but no changes in tight junction markers (Ocln, Tjp2) or other Grhl isoforms (Grhl1, Grhl3). Clonogenicity from single cells, proliferation rates of cell populations and proliferation markers were reduced in Grhl2-KO ESCs. We next induced stepwise directed differentiation of Grhl2-KO ESCs along an otic pathway, giving rise to three-dimensional inner ear-like organoids (IELOs). Quantitative morphometry revealed that Grhl2-KO cells initially formed larger IELOs with a less compacted structure, more eccentric shape and increased surface area. These morphological changes persisted for up to one week. They were partially rescued by forced cell aggregation and fully restored by stably overexpressing exogenous Grhl2 in Grhl2-KO ESCs, indicating that Grhl2 alters cell-cell interactions. On day 8, aggregates were transferred into minimal maturation medium to allow self-guided organogenesis for another two weeks. During this period, Grhl2-KO cells and wild-type controls developed similarly, expressing neural, neuronal and sensory hair cell markers, while maintaining their initial differences in size and shape. In summary, Grhl2 is required for morphological maintenance of ESCs and orderly formation of IELOs, consistent with an essential role in organising epithelial integrity during inner ear development. Our findings validate quantitative morphometry as a useful, non-invasive screening method for molecular phenotyping of candidate mutations during organoid development.
Hearing loss is the most common sensory disability, affecting over 5% of the world’s population and estimated to double by 2050 due to an ageing population (Exeter et al., 2015; WHO, 2023). Inherited dominant hearing loss accounts for approximately 40% of non-syndromic hearing loss and there are currently 68 genetic loci associated with the autosomal dominant form of hearing loss (Van Camp and Smith, 2023). Understanding the complex genetic basis of non-syndromic age-related hearing loss is essential for developing restorative or preventative measures but is complicated by genetic and clinical heterogeneity (Astuto et al., 2000; Keats, 2005). The auditory system requires 26 different cell types for functional hearing (Dror and Avraham, 2010) and 124 genes have been associated with non-syndromic hearing loss alone (Van Camp and Smith, 2023).
Grainyhead-like 2 (GRHL2) is associated with progressive, non-syndromic sensorineural deafness autosomal dominant type 28 (DFNA28) in humans. The gene encodes a highly conserved transcription factor (Bray and Kafatos, 1991) that contains an N-terminal transactivation domain, a unique CP2 DNA-binding domain and a C-terminal dimerisation domain (Ming et al., 2018). DFNA28 was first identified in a multi-generational family with affected individuals displaying bilateral hearing loss. A single nucleotide polymorphism (SNP) in exon 13 (c.1609dupC), resulting in a frameshift and a premature stop codon in exon 14, was associated as the causative mutation (Peters et al., 2002). Variants in GRHL2 rarely associate with autosomal dominant hearing loss, with only eight heterozygous, pathogenic variants known (Trebusak Podkrajsek et al., 2021). Half of these are located in the DNA-binding domain (Iwasa et al., 2016; Vona et al., 2013; Wu et al., 2020; Ji et al., 2014) and the other half in the dimerisation domain (Peters et al., 2002; Trebusak Podkrajsek et al., 2021; Iwasa et al., 2016); so far, none have been reported to affect the transactivation domain. No homozygous GRHL2 mutations have been described in human DFNA28.
GRHL2 is expressed exclusively in ectodermal and endodermal epithelia and their derivatives, suggesting a role in epithelial development, maintenance and repair (Auden et al., 2006; Werth et al., 2010; Senga et al., 2012; Mehrazarin et al., 2015; Walentin et al., 2015; Ray and Niswander, 2016). Homozygous Grhl2 mutations in mouse are embryonic-lethal with defects in neural tube closure (Rifat et al., 2010). Consequently, analysing Grhl2 function in inner ear development has been restricted to examining otocyst formation (Rifat et al., 2010) and Grhl2 expression up to mid-gestation (Auden et al., 2006). Wild-type and heterozygous Grhl2 KO mice show high expression of Grhl2 in both the otic cup and otocyst at embryonic day 8.5–11.5. Subsequently, Grhl2 expression is high in the vestibular and cochlear duct (Auden et al., 2006) as well as in the hair cells, support cells and stria vascularis (Peters et al., 2002). No neural tube defects were observed in heterozygous Grhl2 KO mice which are viable, fertile and live a normal life-span (Rifat et al., 2010).
Apical junctional complexes are essential for epithelial remodelling in the developing inner ear (Bryant et al., 2002; Barald and Kelley, 2004). Formation of the otic vesicle involves tight junctions for determining cell polarity and adherens junctions for invaginating the placode to form the otic cup (Torres and Giraldez, 1998), as well as for maintaining sensory hair and non-sensory support cells in the sensory epithelium (Hibino and Kurachi, 2006; Nakajima, 2015). Loss of the tight junction protein claudin 14 (Cldn14) has been associated with autosomal recessive deafness DFNB29 due to degeneration of the hair cells after birth (Wilcox et al., 2001; Ben-Yosef et al., 2003). Cdh1 regulates cell-cell adhesion in many epithelia, including the inner ear. Loss of Cdh1 in otic tissue impaired hearing function through failure to determine cell polarity (Desai et al., 2009) and establish a functional epithelial basal layer within the stria vascularis (Trowe et al., 2011).
During embryogenesis, GRHL2 activates genes involved in establishing functional epithelia via apical junction complexes (Werth et al., 2010; Walentin et al., 2015; Walentin et al., 2016). Specifically, Grhl2 binds to the Cldn4 and Cdh1 promoters to upregulate their expression, which is critical for forming tight and adherens junctions, respectively (Senga et al., 2012). In Grhl2 KO mice, Cldn4 and Cdh1 were reduced and compensatory Cdh2 mRNA expression was increased, allowing formation of intact adherens junctions (Werth et al., 2010). Loss of Grhl2 dysregulated epithelial morphogenesis and impaired epithelial barrier function (Chen et al., 2010; Varma et al., 2012; Gao et al., 2013; Petrof et al., 2014; Hinze et al., 2018; Kersbergen et al., 2018).
Since Grhl2 KO mice die at mid-gestation, cell culture models have been used to better define the Grhl2 loss-of-function phenotype during ear development. Embryonic stem cells (ESCs) can give rise to all somatic cell types (“pluripotency”) and self-renew indefinitely from single cells (“clonogenicity”). This allows for long-term culture of ESC-derived three-dimensional (3D) cultures to assay in vitro differentiation potential of genetically engineered cells. Organoid differentiation from ESCs was initiated by forming floating embryoid bodies (EBs), derestricting cytomechanic forces, tissue movement and growth, to create miniaturised organ-like aggregates. These organoids offer an alternative in vitro model system, which mimics aspects of in vivo development, to study human disorders, and regenerative therapeutics (Artegiani and Clevers, 2018). This is especially relevant for modelling GRHL2, which is involved in cell-cell and cell-extracellular matrix (ECM) interactions during epithelial morphogenesis.
ESCs have been differentiated into inner ear-like organoids (IELOs) using the self-organising stem cell culture technique known as SFEBq (serum-free floating culture of EB-like aggregates with quick re-aggregation) (Eiraku et al., 2008). The resulting 3D-organoids resemble the saccules of the vestibular apparatus and recapitulate aspects of inner ear formation in vitro (Koehler et al., 2013; Koehler and Hashino, 2014; DeJonge et al., 2016; Liu et al., 2016; Nie et al., 2017; Tang et al., 2019). To elucidate the role of Grhl2 during inner ear biogenesis, we engineered different homozygous deletions of Grhl2 in ESCs. This included truncations in the three different functional domains of Grhl2, two of which are associated with human DFNA28. We demonstrate that all of these mutations cause pathological features in naïve ESCs and IELOs that are consistent with an essential role of Grhl2 in organising epithelial integrity during inner ear development.
Chemicals were supplied by Sigma-Aldrich (Auckland, New Zealand), unless indicated otherwise.
Parental v6.5 ESCs were cultured on 0.1% gelatin-coated tissue culture dishes at 37°C in 2iL medium, comprising of PD0325901 (0.4 μM), CHIR99021 (3 μM), and recombinant human LIF (20 ng/mL) in Hepes-buffered DMEM (hDMEM) with N2 and mixed 1:1 with Neurobasal medium, supplemented with B27 and 1 mM L-glutamine (“N2B27”) (Ying et al., 2008). Cells were passaged using 0.25% trypsin.
For knocking out (KO) two or three functional domains of Ghrl2, wild-type (WT) ESCs were transfected with pools of two different gRNA/Cas9 expression vectors from Santa Cruz Biotechnology (SCBT, #sc-434250), targeting Cas9 to exons 2 and 3 and co-expressing 2a-linked green fluorescent protein (GFP). Homology-directed repair (HDR) plasmids from a genome-scale CRISPR knock-out (GeCKO) v2 library (Sanjana et al., 2014) were used for inserting red fluorescent protein (RFP) and a 2a-linked puromycin resistance gene into exons 2 and 3 (#sc-434250 HDR). For targeting the dimerisation domain in exon 13, two gRNAs were cloned into the pSpCas9(BB) 2A puro (PX459) plasmid containing a puromycin antibiotic selection marker (Ran et al., 2013). Sequences for genome editors are shown in Supplementary Table S1. To isolate correctly targeted clones, cells were selected with 2 µg/mL puromycin 7 days after transfection with lipofectamine® LTX. Resulting puromycin-resistant clones were picked approximately 10 days later and expanded in puromycin-free medium. Picked clones were analysed for correct targeting by endpoint polymerase chain reaction (PCR) of genomic DNA using the KAPA2G Fast HotStart PCR kit under standard conditions (Merck, New Zealand). Exons 2 and 3 of the non-targeted wild-type locus were detected with primer pairs 2 and 3, respectively. Successful targeting at exon 2 and 3 was detected with exon primers paired with forward or reverse primers targeting the transgene insert (FHDR and RHDR). Integration of the HDR-RFP_puro and gRNA/Cas9_GFP plasmids was tested using primer pairs FPlasmid/RHDR and FPlasmid/RGFP, respectively. Primer sequences are shown in Supplementary Table S2. Excised bands were verified by Sanger sequencing (Massey Genome Service, New Zealand).
Full-length mouse Ghrl1 and Ghrl2 open reading frames were synthesised by GeneArt (ThermoFisher, New Zealand) and ligated into the EGFP N1 expression vector (Clontech) under control of the CMV promoter. Above described Grhl2-KO and wild-type ESC strains were transfected with lipofectamine LTX, placed under 1 mg/mL G418 antibiotic selection and GFP expression confirmed. Selected overexpression strains were subjected to genomic and RT-PCR to validate integration and expression of Ghrl1 and Ghrl2, respectively, with appropriate primer pairs (Supplementary Table S2).
All Grhl2-KO and overexpression ESC strains described above were subjected to IELO differentiation. Culture was carried out as described, including centrifugation at 200 × g for 3 min to promote EB-like formation via SFEBq-spin (Liu et al., 2016; Longworth-Mills et al., 2016), and Wnt signalling modulation through CHIR 99021 treatment on Day 8 (DeJonge et al., 2016). ESCs were cultured in ectodermal differentiation (ED) medium (95.5 mL GMEM [Gibco, ThermoFisher, #11710 035], 1.5 mL KO serum replacement [Gibco, ThermoFisher, #10828 010], 100 μL sodium pyruvate [1 M], 1 mL MEM-non-essential amino acids [100x], 180 μL β-mercaptoethanol). Cells were dissociated with trypsin, washed in PBS and GMEM and resuspended in 1 mL of ED medium. Single cell suspensions were passed through a 40 μm cell strainer (Corning, Sigma #352350), washed with ED medium, diluted to 3 × 104 cell/mL and 100 µL (3,000 cells per well) seeded in ultra-low attachment 96-well plates overnight (Corning Costar®, Sigma CLS3474). On day 1, 2% (v/v) matrigel was added to promote formation of a basal membrane (Corning, Sigma #354230). On day 3, non-neural ectoderm was induced and mesendoderm formation inhibited with 10 ng/mL BMP4 (ProSpec, United States, #cyt 361) and 1 μM SB 431542 (Stemgent, United States, #04 0010 05), respectively. On day 4.5, pre-placodal ectoderm was induced with 25 ng/mL FGF2 (R&D systems, United States, #233 fb) and 1 μM LDN 193189 (Reagents Direct, United States, #36 f52). On day 8, aggregates were removed from their wells, pooled by cell strain and treatment, washed with PBS and resuspended in maturation medium (Advanced DMEM/F12, N2 supplement, 1% Matrigel) and 3 μM CHIR 99021. Single aggregates were plated in a 96-well low-attachment plate and incubated for 48 h. On day 10, aggregates transitioned to self-guided IELO culture in maturation medium with daily half medium changes for the next 10–20 days. For each experiment, 16 wells served as controls by removing either substrate (matrigel), cytokines (BMP4, FGF2) or small molecules (SB 431542, LDN 193819) from the ED medium. Additionally, 3,000 ESCs were cultured in 2iL to ensure normal growth prior to differentiation.
For complementation, KO- and WT-ESCs were prepared as described above for IELO differentiation (trypsinised, washed, size-selected) and seeded together, each at 1.5 × 104 cell/mL per 96-well. Images of mixed colonies were acquired after 1 day in co-culture.
For EB-like formation in suspension, ESCs were trypsinised into single cells, washed, and seeded in 2iL medium at 1.7 × 105 cells/cm2 per Petri dish. After 6 days in suspension culture, with occasional agitation, colonies were sequentially filtered through 70 μm and 100 μm cell strainers and collected in a Petri dish. Size-selected EB-like aggregates were placed into wells of a 96-well low-attachment plate with ED medium and incubated for 24 h before imaging.
Cells were either processed fresh or snap-frozen in liquid nitrogen. Following lysis in RNAGEM™ Tissue PLUS (microGEM, Custom Science, New Zealand), cDNA was synthesised as described using Superscript IV (Bray and Kafatos, 1991). Reverse transcriptase was omitted in one sample, each time a batch was processed for cDNA synthesis (‘-RT’). Primers were designed using NCBI/Primer-BLAST, spanning introns, when possible (Supplementary Tables S2, S3), and synthesised by Integrated DNA Technologies (IDT, IA, United States).
For RT-PCR and RT-qPCR, a Mic PCR instrument (BioMolecular Systems, United States) was used. All experiments were performed with the Takara Bio SYBR® Ex Taq qPCR master mix (Norrie Biotech, New Zealand). The master mix consisted of 1.0 µL of each primer (10 µM), 10.0 µL master mix, 0.4 µL ROX dye, 7.44 µL DEPC water and 1.0 µL cDNA template. The following four-segment program was used: 1) denaturation (20 s at 95°C); 2) amplification and quantification (60 s at 95°C, 20 s at 60°C, followed by a single fluorescent measurement repeated 40 times); 3) melting curve (95°C for 5 min, final extension at 70°C for 1 min, heating at 0.003°C/s to 95°C while continuously measuring fluorescence); and 4) cooling to 4°C. Product identity was confirmed by gel electrophoresis and melting curve analysis. Assays were optimised to ensure i) a single melting peak corresponding to the correct PCR product size, ii) sufficient dynamic range over four orders of magnitude by running a 4-fold dilution series and iii) reduced or absent primer-dimer formation. Only the primer pairs that passed these quality controls were used for analysis. Relative quantification was carried out as described (Ruijter et al., 2009), taking reaction efficiency into account by amplification curve analysis. The mean reaction efficiencies for each assay are listed (Supplementary Table S3). When amplicons were not detected (as indicated by failure to amplify and/or absence of a single specific melting peak) an arbitrary baseline was set for that sample with a Ct of 35, which was normalised on the geometric mean of three or four reference genes.
AP activity was determined using a Leukocyte AP Kit (#86R), based on α-naphtholum coupled with diazonium salt, according to the manufacturer’s protocol. Cells were counterstained with Hoechst 33,342 and mounted in DAKO. Qualitative and quantitative comparisons were made using brightfield and fluorescence microscopy, respectively.
Proteins were extracted with RIPA buffer (0.025 mL 1 M Tris/HCl pH 7.4, 0.3 mL 1 M NaCl, 0.05 mL 1 M EDTA, 0.5 mL 1 M Triton-X 100, 0.5 mL 10% SDS, 48 mL H2O and 1 pill cOmplete™ mini EDTA-free [Roche, New Zealand, #040693116001]). Protein extracts were separated by NuPage 4%–12% SDS PAGE, transferred onto a PVDF membrane (Amersham Life Science) and probed with anti-proliferating cell nuclear antigen (PCNA, Abcam, #ab29) and histone 3 (H3, Abcam, #ab1791) for 4°C overnight. Following incubation with a secondary antibody for 30 min at room temperature, bands were visualised with enhanced chemiluminescence. Signal intensity was normalised for the H3 signal and quantified using a Bio Rad image Quant Analyser and Image Lab 6.0 (Bio-Rad Laboratories Inc., New Zealand) (Supplementary Tables S4, S5). After imaging, the blot was stained with Ponceau, rinsed briefly in tap water to remove excess stain and place into a zip lock bag to image on the photocopier. For re-probing, the blot was stripped using 20 mM TCEP in 1% SDS solution for 2 h at 55°C and washed thoroughly in tap water before proceeding with washes, blocking and antibody incubation as above.
IELOs were fixed in 4% (w/v) paraformaldehyde/4% (w/v) sucrose at room temperature for 20 min (Day 1–10 samples) or 1 h (Day 11–30 samples), washed in PBS and incubated in increasing concentrations of sucrose (10, 20, 30%) until they sank to the bottom. IELOs were embedded in Tissue-Tek O.C.T. Compound, frozen in an isopentane/dry ice slurry and stored at −80°C. Serial 7 μm sections were cut on a Leica CM1850 UV cryostat at −14°C and collected on Polysine® slides, coated in 2 g/L gelatin.
Fixed sections were quenched in NH4Cl for 10 min, permeabilised in 0.1% (v/v) Triton X-100 in PBS for 10 min at room temperature, blocked in 5% goat or donkey serum for at least 1 h and incubated with the primary antibody at 4°C overnight. Samples were rinsed in PBS and incubated with the secondary antibody and 5 µg/mL Hoechst stain, diluted in blocking solution, for 30 min at 37°C (Supplementary Tables S4, S5). Samples were washed thrice in PBS and once in H2O before mounting in Dako (Dako, Australia) and sealed with nail polish. Images were taken on an epifluorescence microscope (AX-70, Olympus, New Zealand) equipped with a Spot RT-KE slider CCD camera (Diagnostics Instruments Inc., Sterling Heights, MI, United States) and analysed with ImageJ 1.45S.
For ESC morphometry, single cells and group cultures were seeded onto flat-bottomed 96-well plates and 8-well chambers, respectively, with images taken every day (AMG, EVOS FL digital microscope, United States). Clonogenicity was defined as the proportion of colonies formed per single cells seeded after 12 days. For all other measures, cells were fixed in 4% paraformaldehyde for 15 min at room temperature, stained with Eosin/Hoechst 33,342 and images acquired under transmission or fluorescence settings. Images were loaded into OrganoSeg software (Borten et al., 2018) and cells or colonies segmented to produce binary images. Binary images, along with the raw brightfield and fluorescent images, were imported into CellProfiler 3.1.5 software (McQuin et al., 2018) to create masks for cells, colonies and backgrounds of each image type. The mean background intensity was subtracted from each corresponding image for normalisation before measuring area, cell density and circularity. All masked images, raw and calculated measurements were exported as a .csv file and analysed in R Studio (R Core Team, 2023). Each experimental run was treated as a technical replicate with cell strains (clonogenicity, area) or clonal colonies (density, circularity) as biological replicates. Nuclei were manually counted in ImageJ ((Schneider et al., 2012) Version: 1.4.3.67). For IELOs, images were taken daily for 9 days after seeding and processed as for naïve ESCs using OrganoSeg and CellProfiler software to measure area, perimeter, circularity and compactness. Each experimental run was treated as a technical replicate with clonal colonies as biological replicates.
Proliferation rate was measured from clonal colonies and group cultures, seeding either single cells onto 96-well plates or 1.2 × 104 cells/well onto 12-well plates, respectively. After 5 days of culture, all cells were lifted from each well, dispersed with trypsin, and single cells counted with a haemocytometer to determine the population doubling (PD) time.
Real-time changes in cell number, viability, and morphology were quantified as a cell index (CI) using an RTCA-SP xCELLigence™ system (Roche, New Zealand). Cells were seeded in 100 μL 2iL+2% FBS medium at 3,000 or 5,000 cells per well onto 0.1% gelatin-coated 96-well E-plates (ACEA Biosciences, New Zealand, #5232368001). Each ESC strain (N = 6, N = 4 and N = 3 for WT, 2KO/3KO, and 1KO strains, respectively) and negative control (medium only) was run in 6-8 technical replicates per run. CI readings were taken every 15 min for 12 days. For RTCA data analysis, normalised CI values were calculated for each well by normalising the CI value 30 min after media changes. Normalised CI values were automatically calculated by RTCA Software 2.0 (Roche) and used threshold data for baseline corrections. Once normalised and baseline-corrected, a sigmoid curve was fitted using the Growthcurver package in R (Sprouffske and Wagner, 2016). Outlier wells were removed before processing.
Variables were analysed using ANOVA (aov in R) by genotype or experimental conditions (R Core Team, 2023). Means were compared using TukeyHSD (multcompLetters4 in R). Values are presented as box-whisker plots, including individual data points, mean and 95% confidence interval for the median, unless indicated otherwise. For RT-qPCR gene expression analysis, log transformed relative expression was analysed by linear mixed-effects model fit by REML (Pinheiro et al., 2007). A Ct value of 35 was given for missing values for computation of the baseline relative expression.
ESCs were transfected with pools of three different gRNA/Cas9 expression vectors and HDR plasmids, either targeting insertion of RFP-puromycin into exons 2 and 3 of the murine wild-type Grhl2 gene or introducing indels into exon 13 (Figure 1A). These edits were designed to either create a complete KO by disrupting at least two functional protein domains or mimicking the SNP associated with human DFNA28 by only targeting the putative dimerisation domain (Figure 1B). We identified 16 RFP-positive, puromycin-resistant clones with targeted HDR events. These included biallelic cell strains with loss of all three functional domains (3KO) or homozygous disruption of the DNA-binding and dimerisation domains (2KO-1, -2) (Figure 1C). Following NHEJ-mediated editing, we isolated three biallelic ESC strains with random deletions that introduced a premature stop codon in exon 13, similar to the human SNP. These DFNA28-like (1KO) strains (1KO-1, -2, -3) carried compound heterozygous frameshift mutations disrupting the putative dimerisation domain and were selected for further characterisation (Figure 1C). Wild-type and targeted ESC strains were screened for integration of the HDR-RFP_puro and gRNA/Cas9_GFP plasmids using primer pairs FPlasmid/RHDR and FPlasmid/RGFP, respectively, and strains carrying plasmid integration were excluded (Supplementary Figure S1A, B). Disruption of wild-type exons 2 and/or 3 via HDR-mediated transgene integration was verified by genomic PCR and sequencing of the excised bands (Supplementary Figures S1C–E). Disruption of exon 13 via NHEJ-mediated indels was confirmed by Sanger sequencing (Supplementary Figure S1F). ESC strains were also verified using PCR to detect the presence of wild-type length mRNA transcripts (Supplementary Figure S1G). For 2KO and 3KO strains, the HDR transgene insert included an SV40 polyA transcription termination sequence leading to premature transcriptional arrest of mRNA resulting in the failure to amplify with mRNA primers spanning either exons 2-3 or exons 10-16. For 1KO strains, agarose gel electrophoresis was not able to resolve 1-4 bp differences introduced by NHEJ-mediated indels.
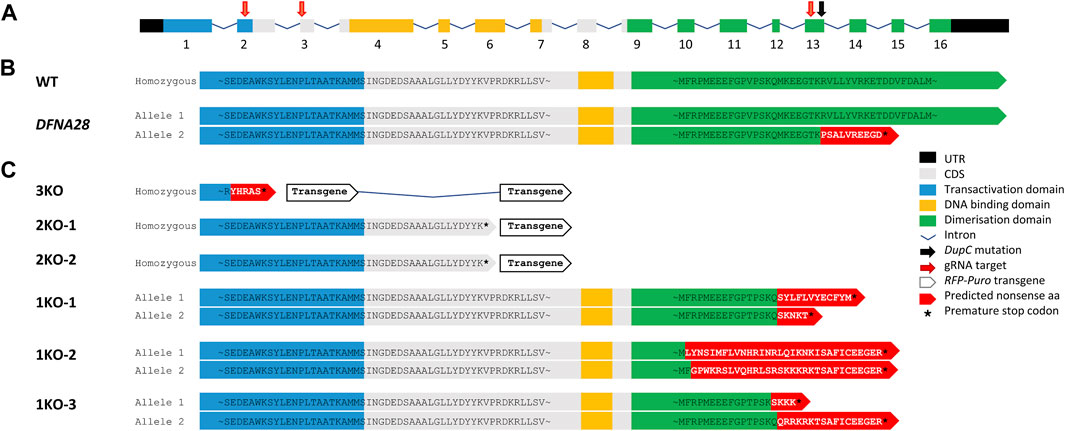
FIGURE 1. gRNA/Cas9 genome editing of murine Grhl2. (A) Murine Grhl2 wild-type (WT) gene structure with untranslated regions (black), numbered exons of coding sequence (CDS, grey), including functional domains for transactivation (blue), DNA-binding (yellow) and dimerisation (green). Mapped to the sequence are the gRNA targets (red arrows) and the c.1609dupC mutation in human DFNA28 (black arrow). (B) Predicted amino acid (aa) sequences for murine WT and human DFNA28 heterozygous genotype. (C) Predicted amino acid sequences for gene edited ESC strains with loss of three or two functional domains after homology-directed insertion of red fluorescent protein (RFP) and puromycin (Puro) transgene (3KO or 2KO-1, -2, respectively). Compound heterozygous DFNA28-like frameshift mutations after non-homologous end joining only affect the dimerisation domain (1KO-1, -2, -3).
We first evaluated the effect of Grhl2 disruption on ESC morphology under naïve culture conditions. Qualitatively, adherent Grhl2-KO ESC colonies were flatter than the uniformly dome-shaped wild-type controls, instead forming heterogenous, loose monolayers with poorly defined edges, which showed alkaline phosphatase activity (Figure 2A). Grhl2-KO ESCs were less than half as clonogenic as wild-type controls (Figure 2B), whose colony formation rate was within the normal range for ESC strains in 2iL medium (Carey et al., 2015; Mulas et al., 2019). Quantitative morphometry showed that individual Grhl2-KO ESCs in suspension were 1.3-fold larger on average, but more variable, while their size in colonies was about 2.6-fold larger. They formed less compact colonies with a lower cell packaging density and reduced circularity (Figure 2B).
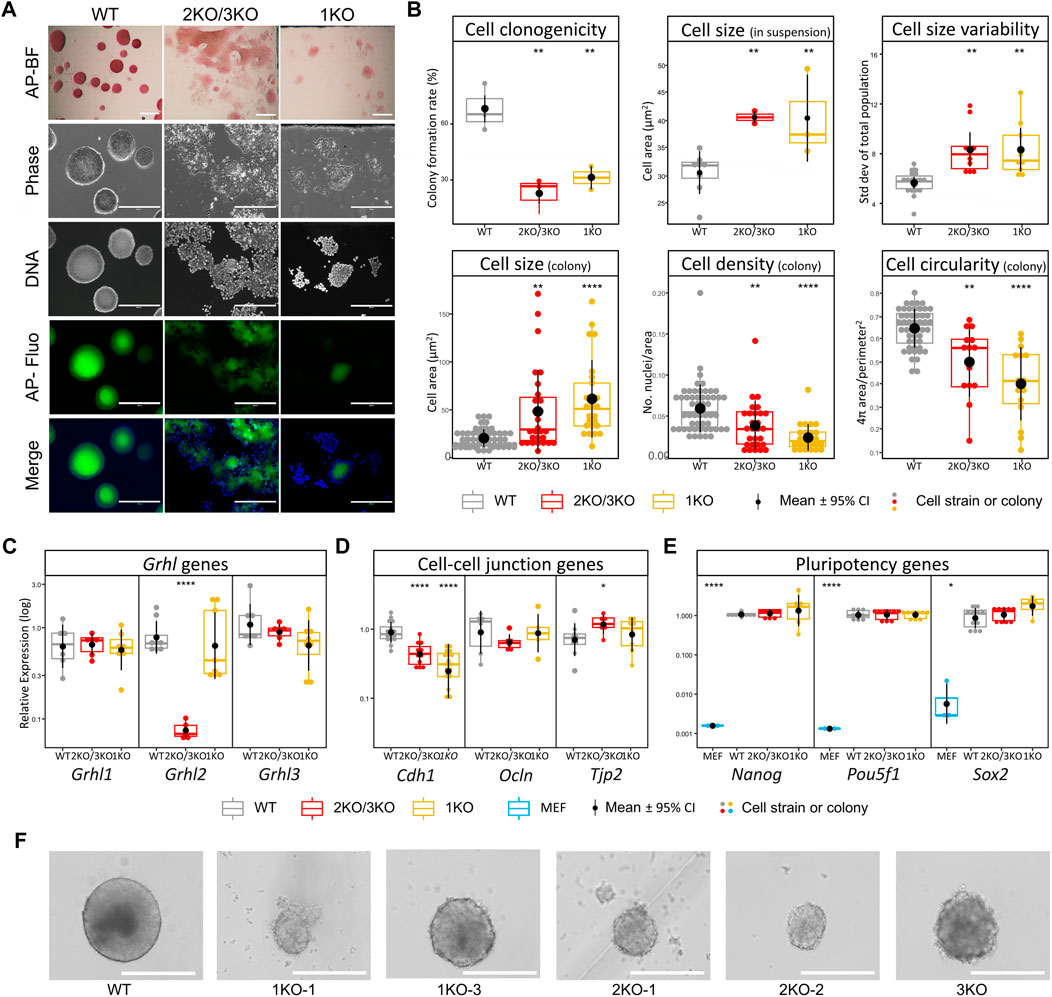
FIGURE 2. Loss of Grhl2 alters ESC morphology. (A) Qualitative morphology of representative Grhl2-KO grown in group cultures, compared to wild-type (WT) ESCs. Alkaline phosphatase (AP) activity was stained with a colorimetric assay and visualised with brightfield (AP-BF), phase contrast (Phase) or epifluorescence in the FITC channel (AP-Fluo). DNA was visualised with Hoechst 33,342. Scale bar = 400 µm. (B) Quantitative morphometry of single ESCs and ESC colonies. Colony formation assay from single cells (clonogenicity) was determined in several strains for each genotype (N = 3, N = 3 and N = 2 for WT, 2KO/3KO and 1KO, respectively). Single cells were sized in suspension with several strains analysed for each genotype (N = 6, N = 4 and N = 3 for WT, 2KO/3KO and 1KO, respectively). To determine cell size variability, each strain was analysed in 3 replicates (N = 1,122, N = 708 and N = 568 cells, respectively). Colony morphology and composition (cell size within colonies, density and circularity) was calculated using colonies from the colony formation assay (N = 49, N = 14, N = 13 colonies for WT, 2KO/3KO and 1KO, respectively). All measurements were taken 12 days after plating single ESCs. (C–E) Gene expression changes for Grhl transcription factors (C), cell-cell junctional markers (D) and pluripotency markers (E). Expression was normalised on the geometric mean of three reference genes. *, **** = p < 0.05, p < 0.0001, respectively. (F) Representative images of EB-like aggregates formed during hanging drop differentiation for several days. Scale bar = 400 µm.
Grhl2 expression was barely detectable in 2KO/3KO cell strains, but not affected in 1KO strains, consistent with the endpoint PCR results reported above. Closely related Grhl1 and Grhl3 transcripts were not significantly altered (Figure 2C). Given the observed morphological changes, we investigated the expression of genes encoding candidate junctional proteins (Figure 2D). Epithelial cadherin (Cdh1) expression was 2-3-fold decreased in Grhl2-KO strains compared to wild-type, while Ocln and Tjp2 expression, both encoding tight junction-associated proteins, showed small or no changes. To determine the state of ESCs, gene expression of canonical pluripotency markers was carried out (Nanog, Sox2 and Pou5f1). Expression of these markers was compared between wild-type and edited ESC strains (3KO, 2KO-1, -2, and 1KO-1, -2, -3) and the data pooled for each knockout (3KO/2KO and 1KO). The results show abundant expression of pluripotency markers, with no significant differences between non-edited vs edited ESCs (Figure 2E). Further, morphologically normal KO-aggregates were formed after prolonged ESC culture in suspension and subsequent differentiation into EB-like structures (Figure 2F). Collectively, this indicates that Grhl2 editing did not affect ESC differentiation per se.
We next determined the proliferation behaviour of Grhl2-KO ESCs. Based on cell counts of group cultures, population doubling times in wild-type ESCs was about 18 h, close to the range of the expected generation time (10–16 h) for naïve ESCs grown in 2iL medium (Carey et al., 2015). This was nearly doubled in both single and group cultures of Grhl2–KO ESCs with a mean generation time of 32 and 30 h, respectively (Supplementary Figure S2A). Using xCELLigence real-time kinetic profiling, both Grhl2-KO ESC genotypes behaved very similar, showing a flattened growth curve with lower peak and endpoint cell indices than wild-type controls (n = 4 biological replicates, Supplementary Figure S2B). Their growth rate, based on the RTCA cell index changes in group culture, was significantly reduced against wild-type cells (Supplementary Figure S2C). Reduced proliferation rates correlated with a 2.4-fold reduced abundance of PCNA, a cofactor of DNA polymerase and marker of cells synthesising DNA (Supplementary Figures S2D, E).
To elucidate the role of Grhl2 during differentiation, we generated 3D-organoids using the SFEBq technique. On day 1 after seeding, wild-type ESCs aggregated into uniform, spherical, tightly packed aggregates with a smooth surface. By contrast, both 2KO/3KO and DFNA28L-KO ESCs formed multiple heterogeneous, disorganised aggregates (Figure 3A). An additional centrifugation step (“SFEBq-spin”) partially rescued the KO phenotype and reduced the number of aggregates (Figure 3A). Quantitative morphometry showed that Grhl2-KO ESCs had a larger surface area and perimeter, were more eccentrically shaped and had a less compact structure (Figure 3B). These differences were still visible, but not as pronounced, in the SFEBq-spin group, indicating that centrifugation could partially rescue Grhl2-dependent cell-to-cell interactions during initial EB-like formation. The centrifugation resulted in smaller aggregates for both wild-type vs mutated ESCs and a reduced surface area in Grhl2-KO cells. KO aggregates had a more homogenous shape, as well as producing a single aggregate more often than smaller, discrete aggregates that very loosely clumped together. Although the KO EB-like morphology was improved by centrifugation (SFEBq-spin group), differences between wild-type and KO remained significant for area, perimeter, circularity and compactness, compared to SFEBq alone. Accordingly, Cdh1 expression was about 4-fold decreased in Grhl2-KO strains compared to wild-type. By contrast to naïve ESCs, abundance of tight junction-related genes Cldn12, Ocln, and Tjp2 was also about 4-fold reduced (Figure 3C).
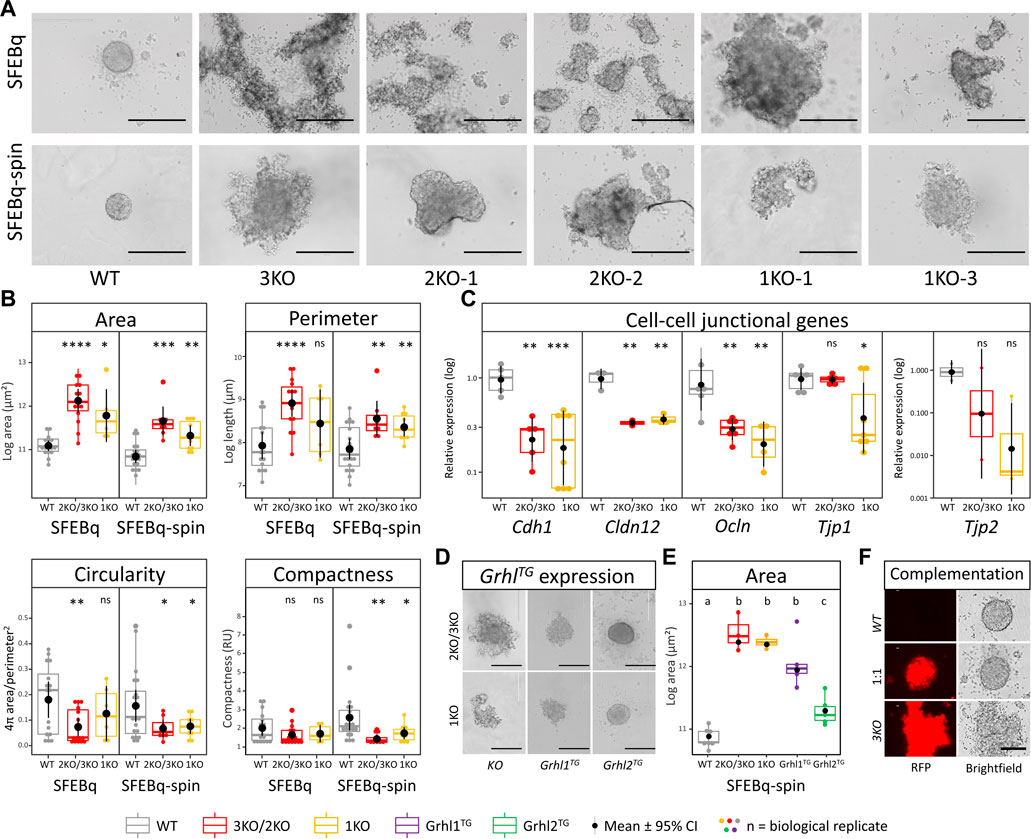
FIGURE 3. Loss of Grhl2 disrupts early IELO integrity. (A) SFEBq-spin partially restores IELO morphology; scale bar = 400 μm (B) Quantitative morphometry of Grhl2-KO IELOs. Morphology (area, perimeter, circularity and compactness) was determined after 1 day in culture. ns = non-significant. *, **, *** = p < 0.05, p < 0.01 and p < 0.001, respectively. (C) Gene expression changes for cell-cell junctional markers, normalised on the geometric mean of four reference genes. (D) Qualitative and (E) quantitative restoration of area by overexpressing Grhl1, or -2 transgenes (Grhl-TG). ab = letters differ by p < 0.05; scale bar = 400 μm. (F) Complementation of 3KO-ESCs carrying a red fluorescent protein (RFP) transgene with WT-ESC (1:1). Non-aggregated 3KO and WT ESCs provide negative and positive controls, respectively. Scale bars = 200 µm.
EB-like formation by SFEBq-spin was also carried out with KO ESCs that carried randomly integrated Grhl1 or Grhl2 overexpression constructs. This resulted in a qualitative rescue of the phenotype for Grhl2, but not Grhl1, overexpressing lines, producing spherical and tightly packed colonies with a smooth surface (Figure 3D). Consequently, the surface area of Grhl2, but not Grhl1, stably overexpressing lines differed significantly from their parental KO but not from wild-type EB-like aggregates (Figure 3E). Rescue of morphology was also achieved by complementing RFP-positive Grhl2-KO ESCs with equal numbers of wild-type ESCs, indicating non-cell-autonomous actions of GRHL2 (Figure 3F). All transgenic overexpression strains were verified by genomic PCR (Supplementary Figure S3A) and Sanger sequencing of the excised bands (Supplementary Figure S3B).
Development of ESC-derived IELOs is defined by the addition of different small molecules and cytokines that initiate formation of defined developmental milestones. This process was tracked over time by imaging aggregates daily for up to three weeks. The gradual size increase was common for both genotypes and no qualitative differences in morphology were seen between the two methods used to form EB-like aggregates (Supplementary Figure S4A). Size and shape variations seen in the KO lines using SFEBq methods, carried through to subsequent days of intermediate differentiation and produced larger, less circular and less compacted aggregates (Supplementary Figure S4B).
On day 8, aggregates were transferred into minimal maturation medium to allow self-guided organogenesis for another two weeks. Grhl2 was not required for late IELO differentiation as both KO- and WT-derived IELOs developed presumptive otic vesicles after day 17 (Figure 4A).
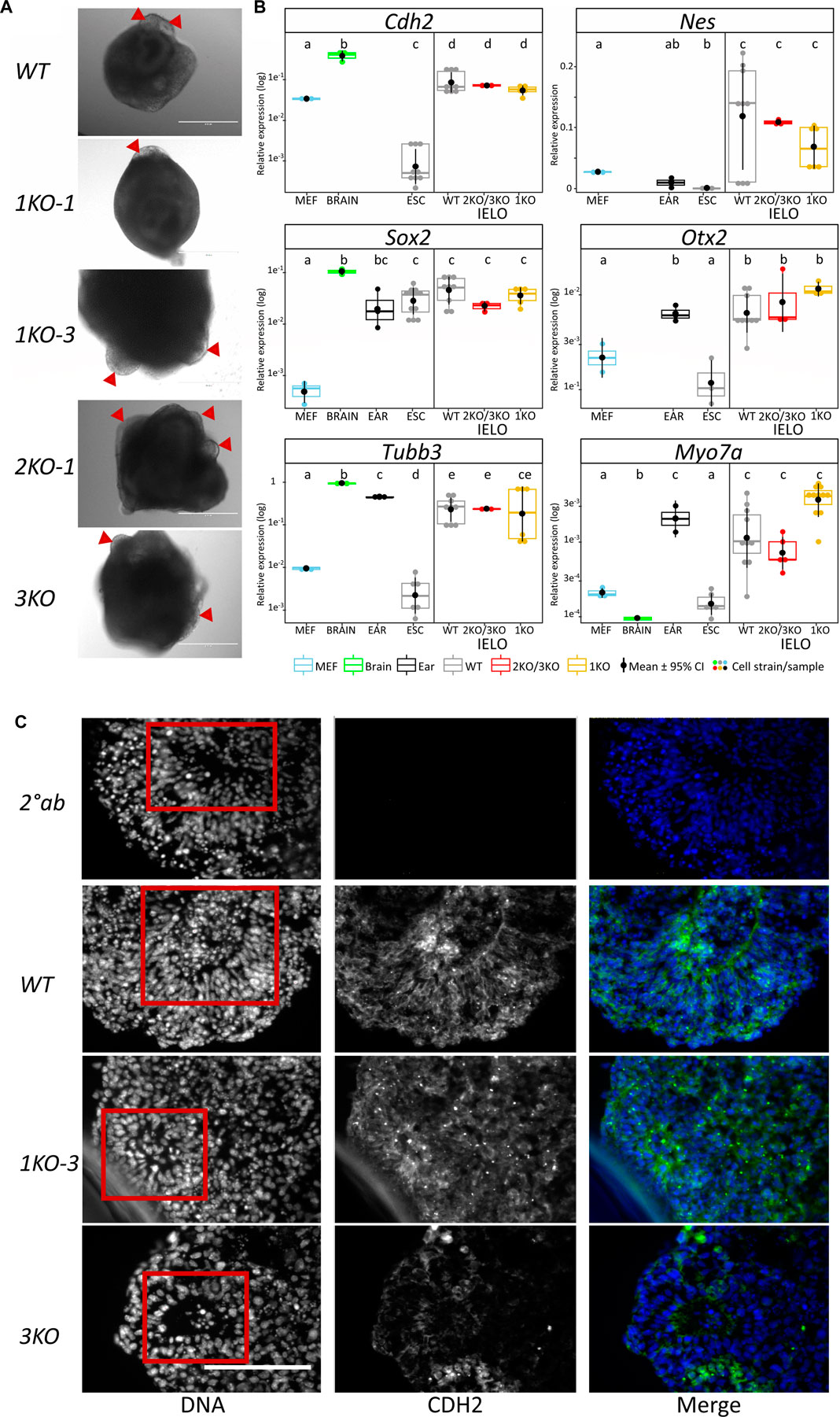
FIGURE 4. Grhl2 is not required for late IELO differentiation. (A) Brightfield images of IELOs showing presumptive otic vesicles on day 17–21 (red triangles). (B) Gene expression changes for neural (Cdh2, Nes, Otx2, Sox2), neuronal (Tubb3) and hair cell specific (Myo7a) markers in day 26 IELOs. Gene expression relative to Gapdh. abcde = samples with different letters differ by p < 0.05. (C) IELOs at day 21 contained neural rosettes (red box) with cells expressing CDH2 (green). Negative control was only incubated with the secondary antibody (2° Ab). DNA was counterstained with Hoechst 33,342 (blue). Scale bars = 400 µm.
To assess differentiation, we quantified gene expression of neural markers (Cdh2, Nes, Otx2, Sox2), neuronal Tubb3, and hair cell-specific Myo7a in edited vs wild-type IELOs after 26 days in culture (Figure 4B). ESCs, MEFs and mouse adult ear or brain tissue served as negative and positive controls, respectively. There was significant expression, relative to undifferentiated ESCs and/or MEFs, of Cdh2, Nes, Otx2, Sox2 and neuronal Tubb3 in all D26 IELOs, indicating neural differentiation. Sensory hair marker Myo7a was equally expressed in both WT- and KO-genotypes and not significantly different from ear tissue. Protein expression of Cdh2 was present in both WT- and KO-IELOs, confirming neural differentiation around distinct rosette structures (Figure 4C). In summary, disruption of Grhl2, either by completely knocking out most functional domains or by mimicking the DFNA28 mutation, altered early IEOL differentiation in ways that are consistent with Grhl2 organising epithelial integrity during development, but did not affect their ability to give rise to end-point IELO containing neuronal and sensory cells.
We have shown that mutagenising three, two or one of the Grhl2 functional domains, including one truncation in the dimerisation domain associated with human DFNA28, causes loss of epithelial integrity in naïve ESCs and early IELOs. These findings are consistent with an essential role of GRHL2 during inner ear development and validate quantitative morphometry as a non-invasive system for screening phenotypes of candidate pathogenic mutations in organoids.
All homozygous Grhl2-KO mice are embryonic-lethal, despite encompassing different mutations and genetic backgrounds, and none have reported a hearing loss phenotype (Werth et al., 2010; Menke et al., 2015; Walentin et al., 2015). Many inbred mouse strains display impaired hearing (Zheng et al., 1999; Noben-Trauth et al., 2003; Noben-Trauth and Johnson, 2009), including those from which the v6.5 ESC line derives (F1-C57BL/6 × 129/Sv). As both parental strains carry Cdh23 mutations, which are associated with age-related hearing loss (Noben-Trauth et al., 2003), we used isogenic wild-type ESC controls to account for any potential phenotype caused by the background genetics. The Grhl2 gene was disrupted in exons 2 or 3 by HDR-mediated transgene insertion or in exon 13 by NHEJ-mediated introduction of premature stop codons. The latter strategy was devised after an HDR-mediated approach failed to introduce the dupC mutation associated with DFNA28, possibly because the HDR template was complementary to the C57BL/6 reference sequence, which differs from the corresponding 129/Sv sequence by two SNPs and a 3 bp deletion. The NHEJ-introduced indels were not an exact match for the c.1609dupC mutation, resulting in 10 non-sense codons and a premature stop codon in exon 14, but caused a similar C-terminal truncation in the dimerisation domain. We also did not succeed in creating heterozygous DFNA28-like mutations. In the future, the sequence variations in exon 13 between C57BL/6 and 129/Sv could be exploited to introduce the +1 frameshift mutation to only one copy of the gene. Ideally, the human GRHL2 sequence could be knocked into the mouse locus, before introducing any of the exact heterozygous pathogenic variants, either in the DNA-binding (Vona et al., 2013; Ji et al., 2014; Iwasa et al., 2016; Wu et al., 2020) or dimerisation domain (Peters et al., 2002; Iwasa et al., 2016; Trebusak Podkrajsek et al., 2021).
In keratinocytes and cancer cell lines, GRHL2 expression levels correlated with increased and decreased PCNA expression after overexpression and knockdown (Coronado et al., 2013), respectively. This is consistent with our results and with Grh proteins binding the PCNA promoter and enhancing expression (Hayashi et al., 1999). Proliferation promotes pluripotency reprogramming, whereas cell cycle arrest inhibits it and induces differentiation (Araki et al., 2010; Smith et al., 2010; Ruiz et al., 2011). Even though proliferation was reduced in Grhl2-KO compared to naïve ESCs, there were no signs of premature differentiation. It remains to be shown whether the anti-proliferative effect of Grhl2 in ESCs was due to lengthening or blocking individual cell cycle phases, decreasing the proliferative cell fraction, inducing cell death or indirect effects. The assays we used, cell counts and RTCA, are both effected by the strength of cell-ECM or cell-cell interactions. Loss of Grhl2 might weaken these interactions, increasing cell loss during media changes and overestimating the anti-proliferative effect. By contrast, wild-type ESCs grow as multi-layered colonies with a relatively small substrate footprint compared to the larger Grhl2-KO ESC monolayers, which will likely reduce their cell index. Uncoupling Grhl2 effects on proliferation from substrate or cell adhesion will require different methods to measure self-renewal, such as flow cytometry of cells in suspension.
Pluripotency progresses from an early naïve to a late primed phase, represented by ESCs and epiblast-derived stem cells (EpiSCs), respectively (Nichols and Smith, 2009). ESCs form compact, dome-shaped 3D colonies, while EpiSCs form flattened 2D colonies and epiblast-like cells (EpiLCs) form a transitory intermediate with a heterogenous population of cells that are enriched in formative phase cells (Kalkan et al., 2017). Adherent ESCs exiting the naïve ground state show early morphological changes, involving cell movement and flattening, as they develop apicobasal polarity and epithelialize (Hayashi et al., 2011; Kinoshita et al., 2021). These in vitro events precede lineage priming and mirror cell biological changes in vivo when epithelial integrity consolidates during development from the pre- to the post-implantation epiblast (Smith, 2017).
Cdh1 is essential for maintaining tightly packed colonies through cell-cell interactions. In EpiLCs or differentiating cells, Cdh1 is downregulated or delocalised and cell-ECM interactions are increased. This results in a flattened elongated morphology, reduced proliferation and reduced clonogenicity. Loss or reduction of Cdh1 pushes ESCs towards a flattened EpiLC morphology and impairs differentiation (Bao et al., 2009; Tian et al., 2011; Murray et al., 2013). Mimicking this phenotype, loss of Cdh1, ESC morphology, proliferation and self-renewal was also observed in Grhl2-KO cells.
Grhl2-KO ESCs were independently generated by mutagenesis of exon 2 (Chen et al., 2018). The authors observed that during the transition from wild-type ESCs to EpiLCs, Grhl2 expression increased ten-fold, activating enhancers that maintained expression of genes involved in epithelialisation and cell adhesion (Chen et al., 2018). Grhl2-KO EpiLCs no longer formed compact colonies, increased mesenchymal Cdh2 and decreased epithelial markers (Cdh1, Tjp2). This phenotype is reminiscent of Grhl2-KO IELOs. In contrast to our results, which documented larger, more heterogenous ESCs and flatter colonies, this phenotypic change was only described in EpiLCs. These differences may relate to lower levels of Grhl2 expression, due to variations in culture conditions and pluripotency status. Both studies used the parental v6.5 ESCs and 2iL supplementation, which results in a homogenous population shielded from differentiation (Tamm et al., 2013). However, our culture did not include 15% KO serum, which can introduce variable cell morphology, alter gene expression and lower chimaerism rates, all indicative of increased differentiation (Tamm et al., 2013; Martin Gonzalez et al., 2016). We also included N2/B27 supplemented Neurobasal/hDMEM, rather than KO DMEM base medium. The culture substrate differed between both studies, either comprising inactivated mouse embryonic fibroblasts (MEFs) or gelatin, which can impact colony morphology. Naïve ESCs reduce cell-ECM interactions by downregulating focal adhesion and integrins, forming tightly packed, rounded colonies on gelatin-coated plates (Taleahmad et al., 2017). Such weakly adherent substrate can maintain self-renewal and pluripotency without LIF or serum (Murray et al., 2013). By contrast, strongly adhesive surfaces (MEFs, fibronectin, laminin or collagen) can promote attachment and downregulation of pluripotency markers (Murray et al., 2013; Taleahmad et al., 2017). The pluripotent state is a continuum and these differences may collectively contribute to the observed differences in Grhl2-KO phenotype.
The effect in ESCs fits with the role of GRHL2 during epithelial-to-mesenchymal transitions (EMT) and the reverse process (MET) where it also suppresses mesenchymal proteins (Chung et al., 2019). During EMT, cell-cell adhesion and polarity are lost as cells delaminate and migrate, while MET decreases cell motility, increases polarity and increases cell-cell interactions, leading to a more mesenchymal phenotype. GRHL2 regulates EMT/MET plasticity through epigenetic methylation of CpG sites and nucleosome remodelling (Cieply et al., 2013; Frisch et al., 2017; Chen et al., 2018). A key event here is again the loss or reduction of CDH1 expression, which weakens cell-cell contacts.
To better understand the effect of Grhl2 KO on pluripotency will require a more in-depth analysis of the molecular and functional features that are diagnostic of naïve vs formative vs primed stem cells. This would require comprehensive profiling of their karyotype, transcriptional, epigenetic, metabolic and cell signalling properties, as well as their ability to form germ cells and pre- or post-implantation chimaeras, ideally at the single cell level (Kalkan et al., 2017).
The initial differentiation steps are critical for understanding the end-point in organoid production systems, which is often less reproducible (Huch et al., 2017). We observed reproducible changes during EB-like formation, the first step of generating IELOs. Under SFEBq conditions, Grhl2-KO ESCs formed large, loose, heterogenous aggregates. Brief centrifugation partially rescued aggregation and EB-like morphology, consistent with centrifugal force enhancing cell-cell attachment via induced cell shape change (Kinoshita et al., 2020). These force-dependent cellular events involve accumulation of F-actin at the apical cell surface and tight junction components (Tjp1 or ZO-1) at the junction, enhancing cell-cell contact. Mechanical force can trigger Erk2 activation, remodelling cell-cell adhesion and tight junctions. It is also been reported that ZO-1 conformation changes in a tension-dependent manner (Spadaro et al., 2017). We have not studied the mechanisms underlying partial physical rescue of the Grhl2-KO phenotype by centrifugation. We speculate that the centrifugal force may have enhanced bridging of cell-cell junctions, possibly through accumulating junctional proteins, which would compensate for the loss of Grhl2 and associated downregulation of junctional markers (Cdh1 Cldn12, Ocln, and Tjp2). However, spinning did not completely rescue KO morphology, indicating that the reduction of molecular mediators of cell adhesion, e.g., Cdh1, could not be overcome by mechanically enforcing cell-cell interactions. Instead, prolonging EB-like formation by a few days allowed the formation of more homogenous and compact aggregates.
Despite the initial differences in EB-like size and shape, and the carry-on effect seen up until about Day 7 of differentiation, KO cells developed along similar pathways to the wild-type cells. This indicates a change in developmental timing, rather than a complete loss of developmental ability.
Self-guided organogenesis within organoid differentiation protocols can be highly variable (Fatehullah et al., 2016; Huch et al., 2017). The IELO differentiation method used here relies on a homogenous, naïve population of ESCs forming a uniform, spherical aggregate composed of ∼3,000 cells. These were treated at specific time points to sequentially induce neural and non-neural ectoderm, eventually forming the otic placode and otic vesicles, respectively (Koehler et al., 2013; Koehler and Hashino, 2014). The differentiation protocol entails about 3 weeks of culture and results in heterogenous aggregates of <1 mm in diameter that contain multiple structures and cell types. The ability to accurately reproduce the IELO end-points relies on many factors: a homogenous ESC starting population; the specific ESC lines and strains used; complete incorporation of these cells into aggregates; uniform aggregate size and shape; batch-batch variation in reagents; timing for treatment with small molecules and proteins, which varies within a 24–48 h window for different cell lines (Eiraku et al., 2008; Eiraku and Sasai, 2011; Koehler et al., 2013). While differentiation of Grhl2-KO EB-like aggregates yielded similar structures to those produced by wild-type, the variability of the resulting organoids made quantification difficult. When a phenotype has already been described in other systems, for example, in vivo mouse studies, the IELO protocol can provide additional valuable insights. This was exemplified by analysing cellular phenotypes from mutations in the type II transmembrane protease 3 gene and its effect on hair cell apoptosis (Tang et al., 2019).
During bronchosphere organoid culture, loss of GRHL2 after formation of apical junction complexes delocalised TJP2, and decreased levels of CDH1 and CLDN4 (Gao et al., 2013). This induced morphological changes towards less closely adherent cells with a more extended morphology. Single cells seeded into an ECM and cultured for 3 weeks had reduced clonogenicity, produced smaller spheres and lacked ciliated cells (Gao et al., 2013). Conditional GRHL2 deletion of exons 1 or 3 was further analysed in mouse tracheal cells in vivo and in 3D tracheospheres (Gao et al., 2015). Again, GRHL2 KO cells were less clonogenic and produced smaller organoids, indicative of reduced proliferation or decreased luminal spaces. The luminal cells within the spheres were flatter, more squamous with shorter basolateral membrane domain and fewer ciliated cells than wild-type controls. Wild-type cells compensated for the loss of conditionally deleted GRHL2 in basal tracheal cells, maintaining epithelial barrier function and demonstrating the non-cell-autonomous actions of GRHL2. We observed a similar rescue of EB-like morphology in Grhl2-KO ESCs complemented with equal numbers of wild-type ESCs. By stimulating proliferation and organising epithelial integrity, GRHL2 appears to play similar roles during airway and inner ear development.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The animal study was approved by the Ruakura Animal Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
All authors contributed to conception and design of the study. BF-G performed all of the experiments. BO wrote the first draft of the manuscript. BF-G and BO contributed to manuscript revision. All authors read the article and approved the submitted version.
BF-G was funded by fellowships from the Health Research Council of New Zealand, the University of Waikato, AgResearch Ltd., the Māori Education Trust, the Todd Foundation, the National Foundation for the Deaf, the Ministry of Health, Ngāti Ranginui Iwi and the Tauranga Moana Māori Trust Board.
We thank Dr Rudolf Jaenisch for kindly donating the v6.5 ESC line, as well as Dr Harold Henderson for help with statistical analyses.
BF-G and BO were employed by AgResearch Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1112069/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Validating gRNA/Cas9 genome edits of murine Grhl2. A commercial Grhl2 gRNA/Cas9 KO HDR kit (SCBT) was used, consisting of pooled plasmids: (A) Grhl2 gRNA/Cas9 KO Plasmid (sc-434250), targeting exons 2 and 3 (red arrows—(C), and (B) two HDR plasmids (sc-434250-HDR) with homology arms (pink) targeting exons 2 and 3. This plasmid combination was used to produce clonal ESC strains 3KO and 2KO. Gels show screening for integration of HDR (FPlasmid/RHDR) and KO (FPlasmid/402R) plasmid. (C) Murine Grhl2 wild-type (WT) genomic DNA (gDNA) with untranslated regions (black) and coding sequence (CDS, grey) and complementary 5’ and 3’ homology arms (pink). Shown are binding sites for primers against exon 2 (primer pair: 2F/2R) and exon 3 (3F/3R). (D) Detection of edited exon 2 (2F/RHDR and FHDR/2R) after homology-directed (HDR) insertion of red fluorescent protein (RFP) and puromycin (Puro) transgene. (E) Edited exon 3 (3F/RHDR and FHDR/3R). (F) Edited exon 13 (primer pair: 13F/13R) after non-homologous end joining (NHEJ) showing Sanger sequencing. (G) Presence of Grhl2 RNA with primers against exons 2-4 (2FmRNA/3RmRNA) and 10-16 (10FmRNA/16RmRNA). 3/2/1-KO = knockout of transactivation (blue), DNA-binding (yellow) and/or dimerisation (green) domains, respectively; M = marker; ntc = no template controls (water); Null = other clonal ESC strains that were not characterised further; RT- = no reverse transcriptase control.
SUPPLEMENTARY FIGURE S2 | Reduced proliferation in Grhl2–KO ESCs and rescue of EB-like formation. (A) Proliferation rate of 2KO/3KO and 1KO compared to wild-type (WT) ESCs. Cells within colonies were counted five days after plating single ESCs (clonal culture) and populations of ESCs in standard 96-well and 96-well E-plates (group culture). (B) xCelligence real-time kinetic profiling. Normalised mean cell indices and standard deviation (light grey) were determined in 15 min intervals for 12 days. Sigmoid curves were fitted using the Growthcurver package in R (n = 2 biological replicates, N = 48 technical replicates per genotype). (C) Cell index quantification from xCelligence growth curve. (D) Representative immunoblot for proliferating cell nuclear antigen (PCNA) and reference histone 3 (H3) from group colonies. (E) Results represent normalised abundance for each genotype. *, **, *** = p < 0.05, p < 0.01, and p < 0.001, respectively, abc = groups with different letters differ by P < 0.01.
SUPPLEMENTARY FIGURE S3 | Validating transgenic overexpression lines. (A) PCR from genomic DNA of Grhl2-KO-Grhl1-TG and Grhl2-KO-Grhl2-TG was amplified using primers specific for the Grhl ORFs. (B) 3KO-Grhl1-TG and 3KO-Grhl2-TG sequence confirmation of integration. ORF nucleotide sequences (3’ end only) with 100% identity to the reference sequence. Exon 16 (grey) of Grhl1 and Grhl2 ending in a histidine tag (HIS) and Not1 restriction enzyme (RE) cut site. Generated using Geneious Prime® 2019.1.1.
SUPPLEMENTARY FIGURE S4 | Grhl2 is required for early IELO differentiation. (A) Representative images of single IELOs of different genotypes (WT, KO) tracked for eight days of differentiation (day 1–8). Scale bars = 400 µm. (B) Quantitative morphometry of area, perimeter, circularity and compactness during IELO differentiation. Data from SFEBq and SFEBq-spin culture conditions were pooled.
Araki, R., Jincho, Y., Hoki, Y., Nakamura, M., Tamura, C., Ando, S., et al. (2010). Conversion of ancestral fibroblasts to induced pluripotent stem cells. Stem Cells 28, 213–220. doi:10.1002/stem.282
Artegiani, B., and Clevers, H. (2018). Use and application of 3D-organoid technology. Hum. Mol. Genet. 27, R99–R107. doi:10.1093/hmg/ddy187
Astuto, L. M., Weston, M. D., Carney, C. A., Hoover, D. M., Cremers, C. W., Wagenaar, M., et al. (2000). Genetic heterogeneity of usher syndrome: analysis of 151 families with usher type I. Am. J. Hum. Genet. 67, 1569–1574. doi:10.1086/316889
Auden, A., Caddy, J., Wilanowski, T., Ting, S. B., Cunningham, J. M., and Jane, S. M. (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr. Patterns 6, 964–970. doi:10.1016/j.modgep.2006.03.011
Bao, S., Tang, F., Li, X., Hayashi, K., Gillich, A., Lao, K., et al. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461, 1292–1295. doi:10.1038/nature08534
Barald, K. F., and Kelley, M. W. (2004). From placode to polarization: new tunes in inner ear development. Development 131, 4119–4130. doi:10.1242/dev.01339
Ben-Yosef, T., Belyantseva, I. A., Saunders, T. L., Hughes, E. D., Kawamoto, K., Van Itallie, C. M., et al. (2003). Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 12, 2049–2061. doi:10.1093/hmg/ddg210
Borten, M. A., Bajikar, S. S., Sasaki, N., Clevers, H., and Janes, K. A. (2018). Automated brightfield morphometry of 3D organoid populations by OrganoSeg. Sci. Rep. 8, 5319. doi:10.1038/s41598-017-18815-8
Bray, S. J., and Kafatos, F. C. (1991). Developmental function of elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes and Dev. 5, 1672–1683. doi:10.1101/gad.5.9.1672
Bryant, J., Goodyear, R. J., and Richardson, G. P. (2002). Sensory organ development in the inner ear: molecular and cellular mechanisms. Br. Med. Bull. 63, 39–57. doi:10.1093/bmb/63.1.39
Carey, B. W., Finley, L. W., Cross, J. R., Allis, C. D., and Thompson, C. B. (2015). Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416. doi:10.1038/nature13981
Chen, A. F., Liu, A. J., Krishnakumar, R., Freimer, J. W., DeVeale, B., and Blelloch, R. (2018a). GRHL2-Dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell 23, 226–238. doi:10.1016/j.stem.2018.06.005
Chen, W., Dong, Q., Shin, K. H., Kim, R. H., Oh, J. E., Park, N. H., et al. (2010). Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5'-CpG island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863. doi:10.1074/jbc.M110.103812
Chen, W., Kang, K. L., Alshaikh, A., Varma, S., Lin, Y. L., Shin, K. H., et al. (2018b). Grainyhead-like 2 (GRHL2) knockout abolishes oral cancer development through reciprocal regulation of the MAP kinase and TGF-beta signaling pathways. Oncogenesis 7, 38. doi:10.1038/s41389-018-0047-5
Chung, V. Y., Tan, T. Z., Ye, J., Huang, R. L., Lai, H. C., Kappei, D., et al. (2019). The role of GRHL2 and epigenetic remodeling in epithelial-mesenchymal plasticity in ovarian cancer cells. Commun. Biol. 2, 272. doi:10.1038/s42003-019-0506-3
Cieply, B., Farris, J., Denvir, J., Ford, H. L., and Frisch, S. M. (2013). Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 73, 6299–6309. doi:10.1158/0008-5472.CAN-12-4082
Coronado, D., Godet, M., Bourillot, P. Y., Tapponnier, Y., Bernat, A., Petit, M., et al. (2013). A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 10, 118–131. doi:10.1016/j.scr.2012.10.004
DeJonge, R. E., Liu, X. P., Deig, C. R., Heller, S., Koehler, K. R., and Hashino, E. (2016). Modulation of Wnt signaling enhances inner ear organoid development in 3D culture. PLoS One 11, e0162508. doi:10.1371/journal.pone.0162508
Desai, R. A., Gao, L., Raghavan, S., Liu, W. F., and Chen, C. S. (2009). Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 122, 905–911. doi:10.1242/jcs.028183
Dror, A. A., and Avraham, K. B. (2010). Hearing impairment: A panoply of genes and functions. Neuron 68, 293–308. doi:10.1016/j.neuron.2010.10.011
Eiraku, M., and Sasai, Y. (2011). Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 7, 69–79. doi:10.1038/nprot.2011.429
Eiraku, M., Watanabe, K., Matsuo-Takasaki, M., Kawada, M., Yonemura, S., Matsumura, M., et al. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. doi:10.1016/j.stem.2008.09.002
Exeter, D. J., Wu, B., Lee, A. C., and Searchfield, G. D. (2015). The projected burden of hearing loss in New Zealand (2011-2061) and the implications for the hearing health workforce. N. Z. Med. J. 128 (1419), 12–21.
Fatehullah, A., Tan, S. H., and Barker, N. (2016). Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254. doi:10.1038/ncb3312
Frisch, S. M., Farris, J. C., and Pifer, P. M. (2017). Roles of Grainyhead-like transcription factors in cancer. Oncogene 36, 6067–6073. doi:10.1038/onc.2017.178
Gao, X., Bali, A. S., Randell, S. H., and Hogan, B. L. (2015). GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 211, 669–682. doi:10.1083/jcb.201506014
Gao, X., Vockley, C. M., Pauli, F., Newberry, K. M., Xue, Y., Randell, S. H., et al. (2013). Evidence for multiple roles for grainyhead-like 2 in the establishment and maintenance of human mucociliary airway epithelium.[corrected]. Proc. Natl. Acad. Sci. U. S. A. 110, 9356–9361. doi:10.1073/pnas.1307589110
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S., and Saitou, M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532. doi:10.1016/j.cell.2011.06.052
Hayashi, Y., Yamagishi, M., Nishimoto, Y., Taguchi, O., Matsukage, A., and Yamaguchi, M. (1999). A binding site for the transcription factor Grainyhead/Nuclear transcription factor-1 contributes to regulation of the Drosophila proliferating cell nuclear antigen gene promoter. J. Biol. Chem. 274, 35080–35088. doi:10.1074/jbc.274.49.35080
Hibino, H., and Kurachi, Y. (2006). Molecular and physiological bases of the K+ circulation in the mammalian inner ear. Physiol. (Bethesda) 21, 336–345. doi:10.1152/physiol.00023.2006
Hinze, C., Ruffert, J., Walentin, K., Himmerkus, N., Nikpey, E., Tenstad, O., et al. (2018). GRHL2 is required for collecting duct epithelial barrier function and renal osmoregulation. J. Am. Soc. Nephrol. 29, 857–868. doi:10.1681/ASN.2017030353
Huch, M., Knoblich, J. A., Lutolf, M. P., and Martinez-Arias, A. (2017). The hope and the hype of organoid research. Development 144, 938–941. doi:10.1242/dev.150201
Iwasa, Y. I., Nishio, S. Y., and Usami, S. I. (2016). Comprehensive genetic analysis of Japanese autosomal dominant sensorineural hearing loss patients. PLoS One 11, e0166781. doi:10.1371/journal.pone.0166781
Ji, H., Lu, J., Wang, J., Li, H., and Lin, X. (2014). Combined examination of sequence and copy number variations in human deafness genes improves diagnosis for cases of genetic deafness. BMC Ear Nose Throat Disord. 14, 9. doi:10.1186/1472-6815-14-9
Kalkan, T., Olova, N., Roode, M., Mulas, C., Lee, H. J., Nett, I., et al. (2017). Tracking the embryonic stem cell transition from ground state pluripotency. Development 144, 1221–1234. doi:10.1242/dev.142711
Keats, B. (2005). Genetics and hearing loss. ASHA Lead. 10, 6–18. doi:10.1044/leader.ftr1.10122005.6
Kersbergen, A., Best, S. A., Dworkin, S., Ah-Cann, C., de Vries, M. E., Asselin-Labat, M. L., et al. (2018). Lung morphogenesis is orchestrated through Grainyhead-like 2 (Grhl2) transcriptional programs. Dev. Biol. 443, 1–9. doi:10.1016/j.ydbio.2018.09.002
Kinoshita, M., Barber, M., Mansfield, W., Cui, Y., Spindlow, D., Stirparo, G. G., et al. (2021). Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell 28, 2180–2471.e8. doi:10.1016/j.stem.2021.11.002
Kinoshita, N., Hashimoto, Y., Yasue, N., Suzuki, M., Cristea, I. M., and Ueno, N. (2020). Mechanical stress regulates epithelial tissue integrity and stiffness through the FGFR/Erk2 signaling pathway during embryogenesis. Cell Rep. 30, 3875–3888. doi:10.1016/j.celrep.2020.02.074
Koehler, K. R., and Hashino, E. (2014). 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat. Protoc. 9, 1229–1244. doi:10.1038/nprot.2014.100
Koehler, K. R., Mikosz, A. M., Molosh, A. I., Patel, D., and Hashino, E. (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221. doi:10.1038/nature12298
Liu, X. P., Koehler, K. R., Mikosz, A. M., Hashino, E., and Holt, J. R. (2016). Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat. Commun. 7, 11508. doi:10.1038/ncomms11508
Longworth-Mills, E., Koehler, K. R., and Hashino, E. (2016). Generating inner ear organoids from mouse embryonic stem cells. Methods Mol. Biol. 1341, 391–406. doi:10.1007/7651_2015_215
McQuin, C., Goodman, A., Chernyshev, V., Kamentsky, L., Cimini, B. A., Karhohs, K. W., et al. (2018). CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970. doi:10.1371/journal.pbio.2005970
Martin Gonzalez, J., Morgani, S. M., Bone, R. A., Bonderup, K., Abelchian, S., Brakebusch, C., et al. (2016). Embryonic stem cell culture conditions support distinct states associated with different developmental stages and potency. Stem Cell Rep. 7, 177–191. doi:10.1016/j.stemcr.2016.07.009
Mehrazarin, S., Chen, W., Oh, J. E., Liu, Z. X., Kang, K. L., Yi, J. K., et al. (2015). The p63 gene is regulated by grainyhead-like 2 (GRHL2) through reciprocal feedback and determines the epithelial phenotype in human keratinocytes. J. Biol. Chem. 290, 19999–20008. doi:10.1074/jbc.M115.659144
Menke, C., Cionni, M., Siggers, T., Bulyk, M. L., Beier, D. R., and Stottmann, R. W. (2015). Grhl2 is required in nonneural tissues for neural progenitor survival and forebrain development. Genesis 53, 573–582. doi:10.1002/dvg.22875
Ming, Q., Roske, Y., Schuetz, A., Walentin, K., Ibraimi, I., Schmidt-Ott, K. M., et al. (2018). Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 46, 2082–2095. doi:10.1093/nar/gkx1299
Mulas, C., Kalkan, T., von Meyenn, F., Leitch, H. G., Nichols, J., and Smith, A. (2019). Defined conditions for propagation and manipulation of mouse embryonic stem cells. Development 146, dev173146. doi:10.1242/dev.173146
Murray, P., Prewitz, M., Hopp, I., Wells, N., Zhang, H., Cooper, A., et al. (2013). The self-renewal of mouse embryonic stem cells is regulated by cell-substratum adhesion and cell spreading. Int. J. Biochem. Cell Biol. 45, 2698–2705. doi:10.1016/j.biocel.2013.07.001
Nakajima, Y. (2015). Signaling regulating inner ear development: cell fate determination, patterning, morphogenesis, and defects. Congenit. Anomalies 55, 17–25. doi:10.1111/cga.12072
Nichols, J., and Smith, A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. doi:10.1016/j.stem.2009.05.015
Nie, J., Koehler, K. R., and Hashino, E. (2017). Directed differentiation of mouse embryonic stem cells into inner ear sensory epithelia in 3D culture. Methods Mol. Biol. 1597, 67–83. doi:10.1007/978-1-4939-6949-4_6
Noben-Trauth, K., and Johnson, K. R. (2009). Inheritance patterns of progressive hearing loss in laboratory strains of mice. Brain Res. 1277, 42–51. doi:10.1016/j.brainres.2009.02.012
Noben-Trauth, K., Zheng, Q. Y., and Johnson, K. R. (2003). Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 35, 21–23. doi:10.1038/ng1226
Peters, L. M., Anderson, D. W., Griffith, A. J., Grundfast, K. M., San Agustin, T. B., Madeo, A. C., et al. (2002). Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. . Hum. Mol. Genet. 11, 2877–2885. doi:10.1093/hmg/11.23.2877
Petrof, G., Nanda, A., Howden, J., Takeichi, T., McMillan, J. R., Aristodemou, S., et al. (2014). Mutations in GRHL2 result in an autosomal-recessive ectodermal Dysplasia syndrome. Am. J. Hum. Genet. 95, 308–314. doi:10.1016/j.ajhg.2014.08.001
Pinheiro, J., Bates, D., DebRoy, S., and Sarkar, D. (2007). Nlme: linear and nonlinear mixed effects models. R. Dev. Core Team, 1–97.
R Core Team, R. (2023). in A language and environment for statistical computing (Vienna, Austria: R.F.f.S. Computing).
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi:10.1038/nprot.2013.143
Ray, H. J., and Niswander, L. A. (2016). Grainyhead-like 2 downstream targets act to suppress epithelial-to-mesenchymal transition during neural tube closure. Development 143, 1192–1204. doi:10.1242/dev.129825
Rifat, Y., Parekh, V., Wilanowski, T., Hislop, N. R., Auden, A., Ting, S. B., et al. (2010). Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345, 237–245. doi:10.1016/j.ydbio.2010.07.017
Ruijter, J. M., Ramakers, C., Hoogaars, W. M. H., Karlen, Y., Bakker, O., van den Hoff, M. J. B., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45. doi:10.1093/nar/gkp045
Ruiz, S., Panopoulos, A. D., Herrerias, A., Bissig, K. D., Lutz, M., Berggren, W. T., et al. (2011). A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 21, 45–52. doi:10.1016/j.cub.2010.11.049
Sanjana, N. E., Shalem, O., and Zhang, F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. doi:10.1038/nmeth.3047
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Senga, K., Mostov, K. E., Mitaka, T., Miyajima, A., and Tanimizu, N. (2012). Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol. Biol. Cell 23, 2845–2855. doi:10.1091/mbc.E12-02-0097
Smith, A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373. doi:10.1242/dev.142679
Smith, Z. D., Nachman, I., Regev, A., and Meissner, A. (2010). Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 28, 521–526. doi:10.1038/nbt.1632
Spadaro, D., Le, S., Laroche, T., Mean, I., Jond, L., Yan, J., et al. (2017). Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr. Biol. 27, 3783–3795. doi:10.1016/j.cub.2017.11.014
Sprouffske, K., and Wagner, A. (2016). Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinforma. 17, 172. doi:10.1186/s12859-016-1016-7
Taleahmad, S., Mirzaei, M., Samadian, A., Hassani, S. N., Haynes, P. A., Salekdeh, G. H., et al. (2017). Low focal adhesion signaling promotes ground state pluripotency of mouse embryonic stem cells. J. Proteome Res. 16, 3585–3595. doi:10.1021/acs.jproteome.7b00322
Tamm, C., Pijuan Galito, S., and Anneren, C. (2013). A comparative study of protocols for mouse embryonic stem cell culturing. PLoS One 8, e81156. doi:10.1371/journal.pone.0081156
Tang, P. C., Alex, A. L., Nie, J., Lee, J., Roth, A. A., Booth, K. T., et al. (2019). Defective tmprss3-associated hair cell degeneration in inner ear organoids. Stem Cell Rep. 13, 147–162. doi:10.1016/j.stemcr.2019.05.014
Tian, X., Liu, Z., Niu, B., Zhang, J., Tan, T. K., Lee, S. R., et al. (2011). E-cadherin/β-catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, 567305. doi:10.1155/2011/567305
Torres, M., and Giraldez, F. (1998). The development of the vertebrate inner ear. Mech. Dev. 71, 5–21. doi:10.1016/s0925-4773(97)00155-x
Trebusak Podkrajsek, K., Tesovnik, T., Bozanic Urbancic, N., and Battelino, S. (2021). Novel GRHL2 gene variant associated with hearing loss: A case report and review of the literature. Genes (Basel) 12, 484. doi:10.3390/genes12040484
Trowe, M. O., Maier, H., Petry, M., Schweizer, M., Schuster-Gossler, K., and Kispert, A. (2011). Impaired stria vascularis integrity upon loss of E-cadherin in basal cells. Dev. Biol. 359, 95–107. doi:10.1016/j.ydbio.2011.08.030
Van Camp, G., and Smith, R. J. H. (2023). Hereditary hearing loss homepage. Available at: https://hereditaryhearingloss.org.
Varma, S., Cao, Y., Tagne, J. B., Lakshminarayanan, M., Li, J., Friedman, T. B., et al. (2012). The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J. Biol. Chem. 287, 37282–37295. doi:10.1074/jbc.M112.408401
Vona, B., Nanda, I., Neuner, C., Muller, T., and Haaf, T. (2013). Confirmation of GRHL2 as the gene for the DFNA28 locus. Am. J. Med. Genet. A 161A, 2060–2065. doi:10.1002/ajmg.a.36017
Walentin, K., Hinze, C., and Schmidt-Ott, K. M. (2016). The basal chorionic trophoblast cell layer: an emerging coordinator of placenta development. Bioessays 38, 254–265. doi:10.1002/bies.201500087
Walentin, K., Hinze, C., Werth, M., Haase, N., Varma, S., Morell, R., et al. (2015). A Grhl2-dependent gene network controls trophoblast branching morphogenesis. Development 142, 1125–1136. doi:10.1242/dev.113829
Werth, M., Walentin, K., Aue, A., Schonheit, J., Wuebken, A., Pode-Shakked, N., et al. (2010). The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137, 3835–3845. doi:10.1242/dev.055483
WHO (2023). Deafness and hearing loss. Available at: https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss.
Wilcox, E. R., Burton, Q. L., Naz, S., Riazuddin, S., Smith, T. N., Ploplis, B., et al. (2001). Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104, 165–172. doi:10.1016/s0092-8674(01)00200-8
Wu, D., Huang, W., Xu, Z., Li, S., Zhang, J., Chen, X., et al. (2020). Clinical and genetic study of 12 Chinese Han families with nonsyndromic deafness. Mol. Genet. Genomic Med. 8, e1177. doi:10.1002/mgg3.1177
Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., et al. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. doi:10.1038/nature06968
Keywords: autosomal-dominant hearing loss, inner ear, embryonic stem cells, organoids, grainyhead, quantitative morphometry, cell adhesion, cadherin
Citation: Forrester-Gauntlett B, Peters L and Oback B (2023) Grainyhead-like 2 is required for morphological integrity of mouse embryonic stem cells and orderly formation of inner ear-like organoids. Front. Cell Dev. Biol. 11:1112069. doi: 10.3389/fcell.2023.1112069
Received: 30 November 2022; Accepted: 21 August 2023;
Published: 07 September 2023.
Edited by:
Lon J. Van Winkle, Rocky Vista University, United StatesReviewed by:
Sally Ann Moody, George Washington University, United StatesCopyright © 2023 Forrester-Gauntlett, Peters and Oback. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Björn Oback, Ympvcm4ub2JhY2tAYWdyZXNlYXJjaC5jby5ueg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.