
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 27 March 2023
Sec. Stem Cell Research
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1084068
This article is part of the Research Topic Cellular Heterogeneity in Physiological and Pathological Myogenesis View all 5 articles
 Alexander B. Andre1,2
Alexander B. Andre1,2 Katherina P. Rees1,2
Katherina P. Rees1,2 Samantha O’Connor3,4
Samantha O’Connor3,4 Grant W. Severson1,2
Grant W. Severson1,2 Jason M. Newbern1
Jason M. Newbern1 Jeanne Wilson-Rawls1
Jeanne Wilson-Rawls1 Christopher L. Plaisier3
Christopher L. Plaisier3 Alan Rawls1*
Alan Rawls1*Background: The expression of proinflammatory signals at the site of muscle injury are essential for efficient tissue repair and their dysregulation can lead to inflammatory myopathies. Macrophages, neutrophils, and fibroadipogenic progenitor cells residing in the muscle are significant sources of proinflammatory cytokines and chemokines. However, the inducibility of the myogenic satellite cell population and their contribution to proinflammatory signaling is less understood.
Methods: Mouse satellite cells were isolated and exposed to lipopolysaccharide (LPS) to mimic sterile skeletal muscle injury and changes in the expression of proinflammatory genes was examined by RT-qPCR and single cell RNA sequencing. Expression patterns were validated in skeletal muscle injured with cardiotoxin by RT-qPCR and immunofluorescence.
Results: Satellite cells in culture were able to express Tnfa, Ccl2, and Il6, within 2 h of treatment with LPS. Single cell RNA-Seq revealed seven cell clusters representing the continuum from activation to differentiation. LPS treatment led to a heterogeneous pattern of induction of C-C and C-X-C chemokines (e.g., Ccl2, Ccl5, and Cxcl0) and cytokines (e.g., Tgfb1, Bmp2, Il18, and Il33) associated with innate immune cell recruitment and satellite cell proliferation. One cell cluster was enriched for expression of the antiviral interferon pathway genes under control conditions and LPS treatment. Activation of this pathway in satellite cells was also detectable at the site of cardiotoxin induced muscle injury.
Conclusion: These data demonstrate that satellite cells respond to inflammatory signals and secrete chemokines and cytokines. Further, we identified a previously unrecognized subset of satellite cells that may act as sensors for muscle infection or injury using the antiviral interferon pathway.
Skeletal muscle has the intrinsic capacity to repair itself in response to acute injury through crosstalk between the muscle and the innate immune system (reviewed in Tidball, 2017; Howard et al., 2020). Inflammatory signals, such as cytokines and chemokines, expressed at the site of injury, serve to activate and recruit monocytes and neutrophils responsible for phagocytosis of debris and necrotic fibers. These signals also promote a promyogenic signaling cascade directing the activation, proliferation, and differentiation of satellite cells (MuSCs), which are necessary to regenerate muscle fibers. Inhibition, or prolonged expression, of these factors diminishes muscle regeneration and promotes fibrosis (Dogra et al., 2006; Dogra et al., 2007; Munoz-Canoves et al., 2013). Despite the importance of cytokines and chemokines to this process, the contribution of muscle cells to immune cell recruitment and the proinflammatory environment remains poorly understood.
In response to acute injury, a proinflammatory environment is established at the site of injury that is necessary for the clearance of necrotic muscle fibers and cell debris (Yang and Hu, 2018). This is initiated immediately after tissue damage by resident neutrophils and degranulating mast cells releasing proinflammatory cytokines, including tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ) and interleukin-1β (IL-1β) (Gordon and Galli, 1990; Fielding et al., 1993). Subsequently, tissue resident macrophages responding to damage-associated molecular patterns (DAMPs) secrete TNF-α, IFN-γ, IL-1β, and IL-6, as well as chemoattractants, such as C-X-C-chemokine ligand 1 (CXCL1) and C-C-chemokine ligand 2 (CCL2) (Brigitte et al., 2010; Venereau et al., 2012; Yang and Hu, 2018). Collectively, these signals promote the recruitment of additional neutrophils and proinflammatory (M1) macrophages that produce reactive oxygen species necessary for the degradation of the damaged muscle fibers (Collins and Grounds, 2001; Teixeira et al., 2003). Single-cell transcriptomic analyses of regenerating muscle have demonstrated that macrophages, neutrophils, and fibroadipogenic progenitor cells (FAPs) residing in the endomysial space of muscle are the primary source of proinflammatory signals (Oprescu et al., 2020; McKellar et al., 2021). The importance of these signals to successful muscle repair has been demonstrated in mice that were deficient for Tnfα, Ccl2, or their respective receptors (Collins and Grounds, 2001; Warren et al., 2002; Chen et al., 2005; Lu et al., 2011). These mutations resulted in reduced muscle inflammation, as measured by decreased infiltration of neutrophils and M1 macrophages after injury, concomitant with a decrease in the clearance of necrotic fibers and diminished muscle fiber regeneration.
Proinflammatory cytokines also promote muscle repair by direct regulation of the myogenic progenitor cell population. Quiescent satellite cells are positioned between the basement and plasma membrane of muscle (Chargé and Rudnicki, 2004). They are distinguished by Pax7 expression, but not the myogenic transcription factors, Myod1, Myf5, or myogenin (Myog). Upon injury, MuSCs start expressing Myod1 and Myf5 and pass through a period of rapid cell population expansion. Daughter cells undergoing differentiation to contribute to muscle fiber repair decrease Pax7 expression and increase expression of a third myogenic transcription factor, Myog. IL-6 and IFN-γ secreted at the site of injury contribute to the activation, proliferation, and migration of the MuSC population (Munoz-Canoves et al., 2013; Fu et al., 2015). TNF-α differentially regulates MuSCs based on its level of expression, at high levels it promotes proliferation and migration and inhibits differentiation. As the inflammation is resolved, TNF-α levels decrease, and MuSCs begin to differentiate and form new muscle fibers (Li, 2003; Torrente et al., 2003; Langen et al., 2004; Dogra et al., 2006; Fu et al., 2015). C-C and C-X-C chemokines have been implicated in the migration, proliferation, and differentiation of satellite cells during muscle repair (Griffin et al., 2010; Ge et al., 2013; Zhu et al., 2016; Hogan et al., 2018). Thus, proinflammatory cytokines tightly coordinate the activity of both myeloid and myogenic cell lineages during muscle repair.
Proinflammatory factors also contribute to idiopathic inflammatory myopathies, including dermatomyositis (DM) and polymyositis (PM) (Kuru et al., 2003). The affected muscle expresses proteins associated with the activation of the type I interferon pathway (Boehm et al., 1997; Greenberg et al., 2005; Salajegheh et al., 2010; Suárez-Calvet et al., 2014). These observations and several in vitro studies raise the possibility that skeletal muscle can also contribute to activation of the inflammatory response associated with healthy muscle repair in response to injury. For example, the well-studied inflammatory activator, bacterial lipopolysaccharide (LPS), induced C2C12 and L6 myoblast cell lines to express cytokines (Frost et al., 2003). LPS treatment is used to stimulate sterile inflammation in culture (Frost et al., 2002) by acting on Toll Like Receptor 4 (TLR4), a cellular membrane sensor of damage and infection that also triggers the caspase-11 non-canonical inflammasome (Yang et al., 2015; Cieslelska et al., 2021). LPS treatment induced Tnfα, Il6, Ccl2, and Cxcl1 expression within 2 h in these cell lines (Frost et al., 2002; Frost et al., 2003; Bivona et al., 2021). Primary human MuSCs have also been shown to express a broad set of cytokines and chemokines associated with muscle repair, including Tnfa, Il6, Ccl2, and Ccl5 when treated with TNF-α or IFN-γ (De Rossi et al., 2000).
In this study, we wanted to explore whether MuSCs contributed to the proinflammatory response during healthy muscle repair post-injury. We observed that primary mouse MuSC cultures were comprised of cells along the continuum from activation through differentiation and they upregulated cytokine expression in response to LPS treatment. Single cell RNA sequencing (scRNA-Seq) revealed that a broad set of cytokines and chemokines were differentially expressed in response to LPS. For example, Ccl2 and Cxcl1 were broadly expressed throughout the culture. In contrast, a small subset of MuSCs expressed Ccl5 and genes belonging to the antiviral IFN pathway. These cells were identifiable in PBS treated control cells, suggesting this is a stable cell population. Further, CCL5 positive MuSCs were detectable in cardiotoxin (CTX) injured muscle suggesting that chemokine and cytokine expression by these cells has a distinct function in muscle repair.
B6129SF1/J mice were purchased from Jax Labs (Bar Harbor, ME) and bred and housed in a vivarium at Arizona State University (ASU). These mice were kept on a 10 h light: 14 h dark schedule with ad libitum access to food and water. ASU is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC). All procedures were carried out in compliance with the ASU institutional animal care and use committee and AALAC under an approved research protocol.
Primary muscle satellite cell cultures (MuSCs) were established from cells isolated from five 12 week old B6129SF1/J mice as previously described (Palade et al., 2019). Briefly, hind limb quadriceps femoris muscles were excised, trimmed of fat and connective tissue, and finely minced. The muscle tissue was digested with 1.25 mg of protease XIV (Sigma-Aldrich, St. Louis, MO) for 1 h at 37°C. The cell suspension was filtered, differentially centrifuged and pre-plated in DMEM (Corning, Corning NY), containing 2% donor horse serum (HS) (Atlanta Biologicals, Flowery Branch, GA), and 100 μg/mL Primocin (Invivogen, San Diego, CA). Fibroblasts were removed by three pre-plating rounds of 3 h. Satellite cells were grown in a humidified chamber at 37°C with 5% CO2, on Matrigel Matrix Basement Membrane (BD Biosciences, Bedford, MA) coated plates, in growth medium, Hams F-10 (Corning), containing 20% FBS (Atlanta Biologicals), 10 ng/mL bFGF (BD Biosciences) and 100 μg/mL Primocin (Invivogen).
MuSCs were seeded onto Matrigel (BD Biosciences) coated 6 well plates at a density of 1 × 105 cells/well in growth medium. Cells were then treated with LPS (1 mg/mL; Sigma-Aldrich) or phosphate buffered saline (PBS) for 1, 2, 4, 6, or 8 h. Cells were lysed in TRIzol (ThermoFisher Scientific, Waltham, MA) for RNA isolation, per the manufacturer’s protocol, and 2 μg of total RNA was used to synthesize cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and random hexamer primers.
Quantitative Real Time PCR (RT-qPCR) was carried out using SYBRgreen (Eurogentec, Fremont, CA) on an ABI 7900 HT thermocycler using a 384 well format in 10 μL reactions. All samples were normalized to the Gapdh transcript and relative gene expression was calculated using ΔΔCt analysis (Haimes and Kelley, 2010). Primer sequences are listed in Supplementary Table S1. All data are the result of triplicates from 3 biological samples.
MuSCs were treated with LPS as above for 2 h. MuSCs were then trypsinized (0.05% trypsin 0.53 M EDTA, Mediatech, Manassas VA) to create a single cell suspension. Cells were then counted on an automated cell counter and RNA library preparation was performed by the ASU Genomics Core facility. Samples were processed using a 10X Chromium single cell 3′ GEM, Library and Gel Bead kit V3 with a Chromium single cell B chip. The quality of each library was determined using Agilent TapeStation automated electrophoresis. Samples were sequenced at a read depth of ∼31,000 reads per cell (BioFrontiers Sequencing Core, University of Colorado, Boulder). The 10X Genomics CellRanger v6.0.1 was used to align to the Mus musculus reference genome mm10-2020-A (GRCm39), quantify, and provide basic quality control metrics for the scRNA-seq data. Data was then analyzed utilizing the Seurat R package. Cells with < 2000 reads or >80,000 reads were discarded as well as cells with < 0.9% or >20% mitochondrial genes. Clusters of cells with similar expression patters were identified using the ‘FindClusters’ function which used a shared nearest neighbor modularity optimization-based clustering approach.
Acute muscle injury was generated in the quadriceps of 3 month old mice by intramuscular injection of 50 μL of 10 μM cardiotoxin (CTX) (Sigma-Aldrich) solution in DMSO (Garry et al., 2000). At 24 h post-injection, Evan’s blue dye (Sigma-Aldrich) was injected intraperitoneally at 1 mg/10 g body weight to allow for visualization of damaged cellular membranes. Mice were sacrificed at designated time points and tissue was harvested for analysis. The uninjured quadriceps muscle from the contralateral leg was also harvested for use as control tissue. Total RNA was isolated from muscle biopsies using TRIzol (ThermoFisher Scientific) as described in Majumdar et al. (2012).
Mice (n = 3) were injected with CTX, quadriceps were harvested, and fixed in 4% paraformaldehyde in PBS overnight at 4°C. Tissue was cryopreserved in sucrose prior to embedding in Tissue-Tek OCT. Sections were then permeabilized and blocked in a buffer containing 0.1% Triton-X 100% and 5% normal donkey serum (NDS, Sigma-Aldrich) in PBS. Samples were then incubated in primary antibody solution containing antibodies recognizing a muscle marker, M-cadherin (Santa Cruz #sc-81471 1:250) and CCL5 or CCL2 (CCL5, R&D Systems AF478 1:300, CCL2 Novus Biologicals NBP 1-07035 1:500). Sections were then rinsed and incubated in secondary antibody solutions containing Alexa-Fluor conjugated antibodies against mouse (Thermofisher, A-21202, 1:1000), rabbit (Thermofisher, A-31573, 1:1000) or goat IgG (Thermofisher, A-21447, 1:1000) and DAPI (Sigma Aldrich 10,236,276,001, 1:1000) diluted in blocking solution. Tissue was rinsed in PBS and cover-slipped for microscopic analysis. Images were collected on a Zeiss LSM800 laser scanning confocal microscope and optimized for brightness and contrast. Blinded evaluators quantified M-cadherin positive (MCAD+ve) and MCAD+ve/CCL5+ve cells in random 20X fields. Data were averaged for both CTX injured (n = 418 MCAD+ve cells) and contralateral control quadriceps (n = 416 MCAD+ve cells). Student’s t-test was used to determine statistical significance.
MuSCs treated with LPS (1 mg/mL) or PBS control for 2 h, were prepared for immunofluorescence (IF) by washing twice with cold PBS followed by fixation in 4% PFA for 20 min. Cells were then washed 3 times with PBS before antibody staining. Monoclonal mouse anti-MYOD (NB100-56511, Bio-techne Minneapolis, MN, 1:200) and polyclonal goat anti-CCL5 (AF478-SP, R&D Systems, 1:200) were used to detect cellular expression in culture. Anti-mouse-Alexa Fluor 488 (1:1000), anti-goat Alexa Fluor 647 (1:1000) and DAPI (1:1000) were then used to visualize MYOD, CCL5, and nuclei, respectively. Images were collected on a Zeiss LSM800 laser scanning confocal microscope at 20X magnification. Cells from 50 fields were then counted to determine the percent of total cells positive for these markers.
To assess the significance of the expression of cultured MuSCs post-treatment a one-way ANOVA was used. The ‘FindMarkers’ function in the Seurat package was used to determine the significance of the differential expression of marker genes from each cluster in the scRNA-Seq data. Marker genes were determined as having an adjusted p-value less than 0.05 and an average log 2 fold-change greater than 0.25.
Proinflammatory cytokines promote neutrophil and M1 macrophage infiltration to the damaged muscle via diapedesis and promote the activation and proliferation of MuSCs necessary for muscle regeneration (Howard et al., 2020). There are several studies that indicate the potential for MuSCs to contribute to the proinflammatory environment. For example, the pathogen-associated molecular pattern molecule (PAMP) LPS induced the expression of Tnfa, Il6, Ccl2, and Ccl5 in C2C12 and L6 muscle cell lines and human MuSCs (De Rossi et al., 2000; Frost et al., 2002; Frost et al., 2003). Further, expression of genes in the antiviral IFN pathway was elevated in the muscle of DM patients, predicting that muscle plays a role in the onset of inflammation (Moneta et al., 2019; Bivona et al., 2021). To examine the potential induction of an inflammatory response by MuSCs, we examined primary cell cultures derived from MuSCs isolated from the quadriceps of 3 month old B6129SF1/J mice treated with LPS (1 mg/mL) or a PBS control for 1, 2, 4, 6, or 8 h. Total RNA was isolated and transcription of Tnfa, Il6, and Ccl2 was assessed by RT-qPCR. We found that Tnfa was rapidly induced and reached its maximum at 1 h post-treatment (p < 0.0001) with a significant drop off between 2 and 4 h. In contrast, Il-6 and Ccl2 transcription were induced to maximal levels at 2 h (p < 0.0001) (Figure 1).
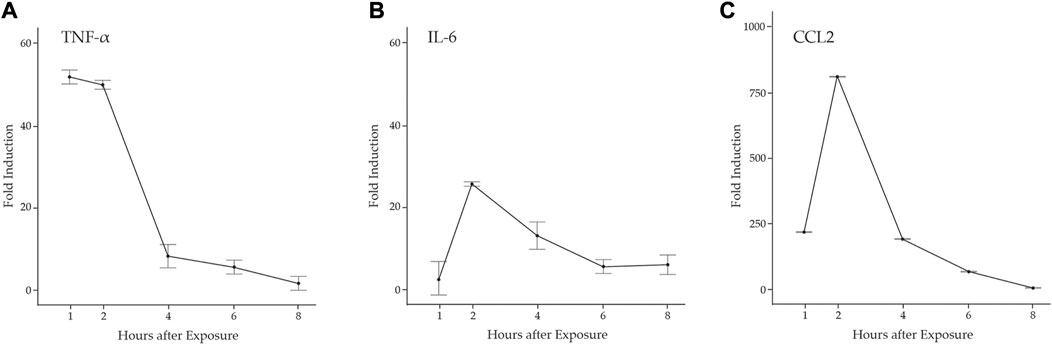
FIGURE 1. LPS induction of proinflammatory cytokines in primary mouse MuSCs. Transcription of (A) Tnfα (B) Il6, and (C) Ccl2 from MuSCs treated with LPS (1 mg/mL) for 1, 2, 4, 6, and 8 h was quantified by RT-qPCR. RNA transcript levels are express as the fold induction over control MuSCs treated with PBS. At 1, 2, and 4 h, p < 0.001.
It is becoming increasingly clear that MuSCs are heterogeneous based on the expression of specific markers, cell function, and their ability to contribute to muscle regeneration (Biressi and Rando, 2010; Rodriguez-Outeiriño et al., 2021). Thus, it is likely that the chemokine and cytokine profiles of individual MuSCs will vary across different subpopulations. To examine this, scRNA-Seq was performed on actively proliferating MuSCs treated with LPS or PBS. Based on the results in Figure 1, cells were exposed to LPS for 2 h for maximal induction of chemokines and cytokines. Single cells were isolated and profiled by scRNA-Seq. Following quality control for mitochondrial RNA content and reads per cell, this resulted in a PBS-treated library of 2,751 cells with 20,235 mean reads/cell and an LPS-treated library of 2,802 cells with 19,330 mean reads/cell.
Unsupervised clustering and UMAP embedding was performed using the Seurat R package and identified seven unique clusters in PBS-treated MuSCs based on differentially expressed genes (Figures 2A,B; Supplementary Table S2). All clusters were determined to consist of myogenic lineage cells based on the expression of one or more of the muscle-specific transcription factors, Pax7, Myod1, Myog, and Myf5 (Figure 2C), known to have dynamic expression in satellite cells as they progress along the continuum from quiescent satellite cell through activation, proliferation, and differentiation (Figure 3A). The pattern of transcription of these four factors and other differentially expressed genes was used to identify the cell clusters within the PBS-treated MuSC culture (Figure 3B). Activated satellite cells (ASC) were identified based on transcription of Pax7 and the satellite cell activation and proliferation markers H19, EIF2S3Y, and HGF (Sheehan et al., 2000; Li et al., 2016; Martinet et al., 2016). The vast majority of the cells sorted into 3 cell clusters, PSC1—PSC3, with a similar pattern of reduced Pax7, and increased Myod1, transcription. PSC1 had a marked increase in ribosomal protein expression (Rspa, Rps2, Rps26, and Rps29), necessary for ribosome biogenesis and post-transcriptional regulation. Expression of these genes is associated with the increase in protein production needed for the transition of ASCs to proliferating myoblasts (Chaillou et al., 2017; Gayraud-Morel et al., 2018). The second and largest cell cluster, PSC2, was enriched for markers of proliferating myoblasts, including Peg3, Gas6, and C1qtnf3 (Correra et al., 2018; Mervis et al., 2020; Otani et al., 2020). The third PSC cluster (PSC3) had a higher expression level of glutathione peroxidase 3 (Gpx3) and the RNA helicase Mtrex, implying that these cells were responding to local environmental stressors that may not be related to the normal progression of satellite cells (El Haddad et al., 2012). There was a small cell population related to the PSC cell clusters, except for reduced transcription of Myod1 and Myog and enrichment for genes in the antiviral IFN pathway and downstream interferon stimulated genes (ISG), including Oasl1, Ifit1, Ifit3, and Rsad2 (Figure 3B). Thus, we have designated this cluster, IFN stimulated cells or ISCs. Finally, two clusters of differentiating myocytes (DMC1 and 2) were present in the culture, based on the expression of Myog and Myod1, as well as sarcomeric proteins including Myl1, Acta1, Tnnc2, and Myh1. Thus, the PBS treated primary satellite cell culture was comprised of cells along the continuum from activated satellite cells to differentiating myocytes with several subsets of myoblasts with distinct patterns of gene transcription.
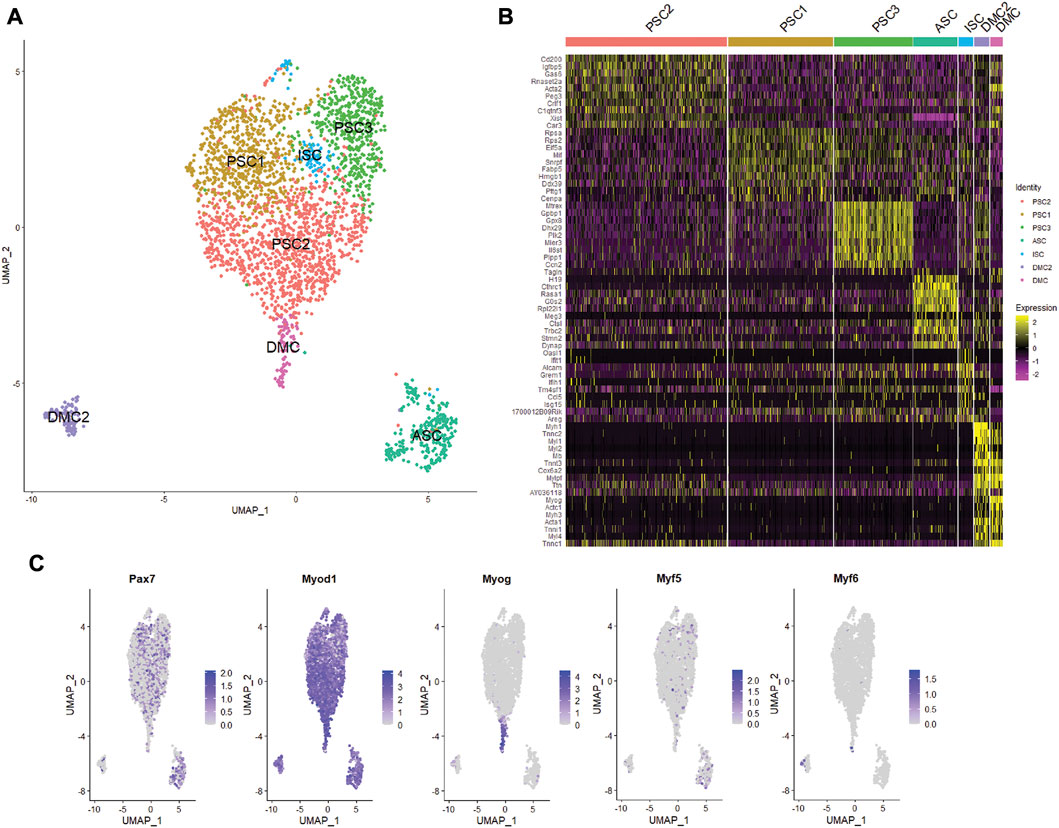
FIGURE 2. Single cell RNA-seq analysis of primary mouse MuSC culture. (A) UMAP clustering of the PBS-treated MuSCs. Cells clustered into seven groups based on differential gene transcription. (B) Heatmap depicting top 10 differentially expressed genes for each cluster. (C) Feature plots show the distribution of the myogenic transcription factors of Pax7, Myod1, Myog, Myf5, and Myf6 in the MuSCs.
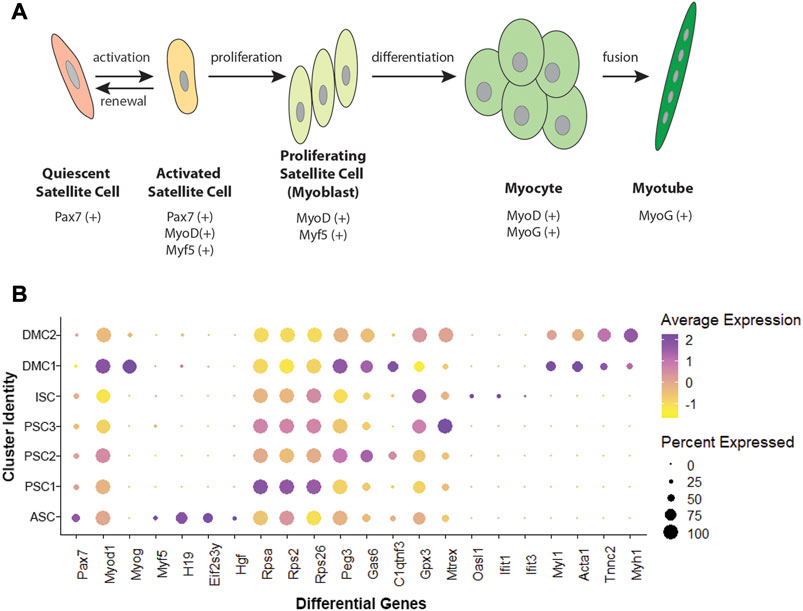
FIGURE 3. Myogenic transcription factors and differentially expressed genes in MuSCs. (A) Schematic of satellite cell progression to differentiated myotube, including the myogenic transcription factor profile for each stage. (B) Dotplot showing average and percent expression of genes that differentiate the 7 cell clusters in PBS-treated MuSCs.
To extend our evaluation of LPS induction of cytokines and chemokines in muscle (Figure 1), MuSC cultures were treated with LPS (1 mg/mL) for 2 h and profiled by scRNA-Seq. Unsupervised clustering and UMAP embedding was performed using the Seurat R package under the same conditions as the control culture (Figure 4, Supplementary Table S3). A similar set of cell clusters, as described above, were identified based on differential gene expression. However, there were exceptions, including a fourth PSC cluster (PSC4) and the absence of the second DMC cluster. The PSC4 cluster was distinguished by decreased expression of more than 20 ribosomal proteins. The significance of this reduction of the translational machinery is not clear, though there is evidence of LPS reducing protein synthesis in muscle tissue (Gordon et al., 2013).
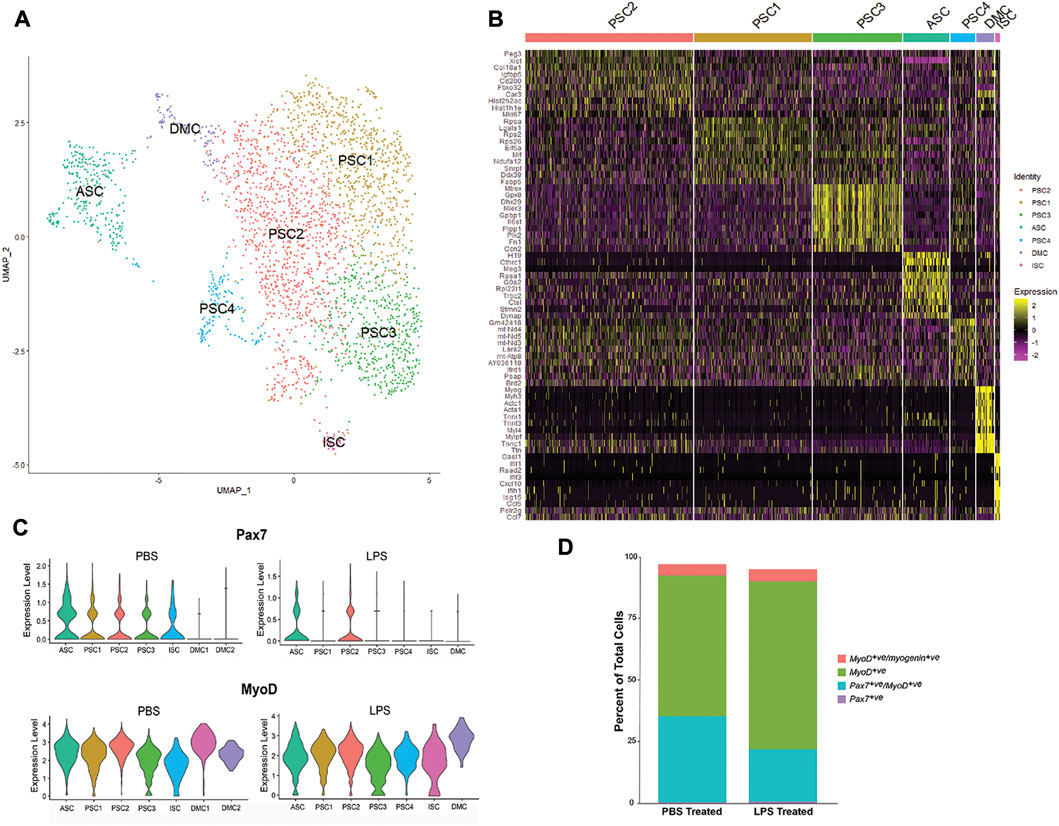
FIGURE 4. Single-cell RNA-SEQ analysis of LPS-treated primary mouse MuSCs. (A) UMAP clustering of the LPS-treated MuSC. Cells cluster into seven groups based on differential gene transcription. (B) Heatmap depicting top 10 differentially expressed genes for each cluster. (C) Violin plots of Pax7 and Myod1 transcription in PBS and LPS treated MuSCs. (D) The distribution of satellite cells (Pax7+ve only), activated satellite cells (Pax7+ve/Myod1+ve), proliferating myoblasts (Myod1+ve only) and differentiating myocytes (Myod1+ve/Myog+ve) in PBS and LPS treated cultures. Cell populations are presented as percent of the total population.
The level of transcription of the myogenic markers, Pax7 and Myod1, was examined in the cell clusters of PBS and LPS treated cells (Figure 4C). Pax7 transcription was reduced in PSC1, PSC3 and ISC clusters and reduced in the ASC cluster. In contrast, Myod1 transcription levels are unaffected by LPS treatment. Alternatively, the distribution of cells along the satellite cell continuum was evaluated independent of cell clusters by comparing the number of cells co-expressing the myogenic markers, Pax7, Myod1, and Myog (Figure 4D). While the number of Pax7+ve-only satellite cells and Myod1+ve/Myog+ve differentiating myocytes remained relatively constant, there was an expansion of the Myod1+ve myoblasts cells at the cost of the Pax7+ve/Myod1+ve activated satellite cells. This is consistent with the reduction of Pax7 transcription after LPS treatment. It remains to be determined if this is due to direct regulation by LPS signaling, or the indirect result of promoting proliferation of myoblasts through cytokine production.
An evaluation of chemokine production in response to LPS by scRNA-Seq showed that 13 members of the C-C and C-X-C subfamilies (Ccl2, Ccl5, Ccl7, Ccl17, Ccl20, Ccl27a, Cxcl1, Cxcl2, Cxcl4, Cxcl10, Cxcl12, Cxcl16, and Cx3cl1) were expressed in MuSCs. A comparison of transcription of these factors across the cell clusters in the LPS and PBS control conditions revealed two distinct patterns (Figure 5). The first pattern included chemokines not expressed in the control cells and broadly transcribed after treatment with LPS (e.g., Ccl2, Cxcl1, Ccl7, and Cx3c1). This is in contrast to chemokines that were induced in a single cell cluster. This was observed for Ccl5 and Cxcl10, which were induced in the ISC cluster, and Ccl17 and Cxcl16 in the ASC cluster (Figure 5B). Ccl5 and Cxcl10 are induced by type I IFNs (Kelly-Scumpia et al., 2010; Nakano et al., 2012) consistent with our designation of this cluster as IFN stimulated.
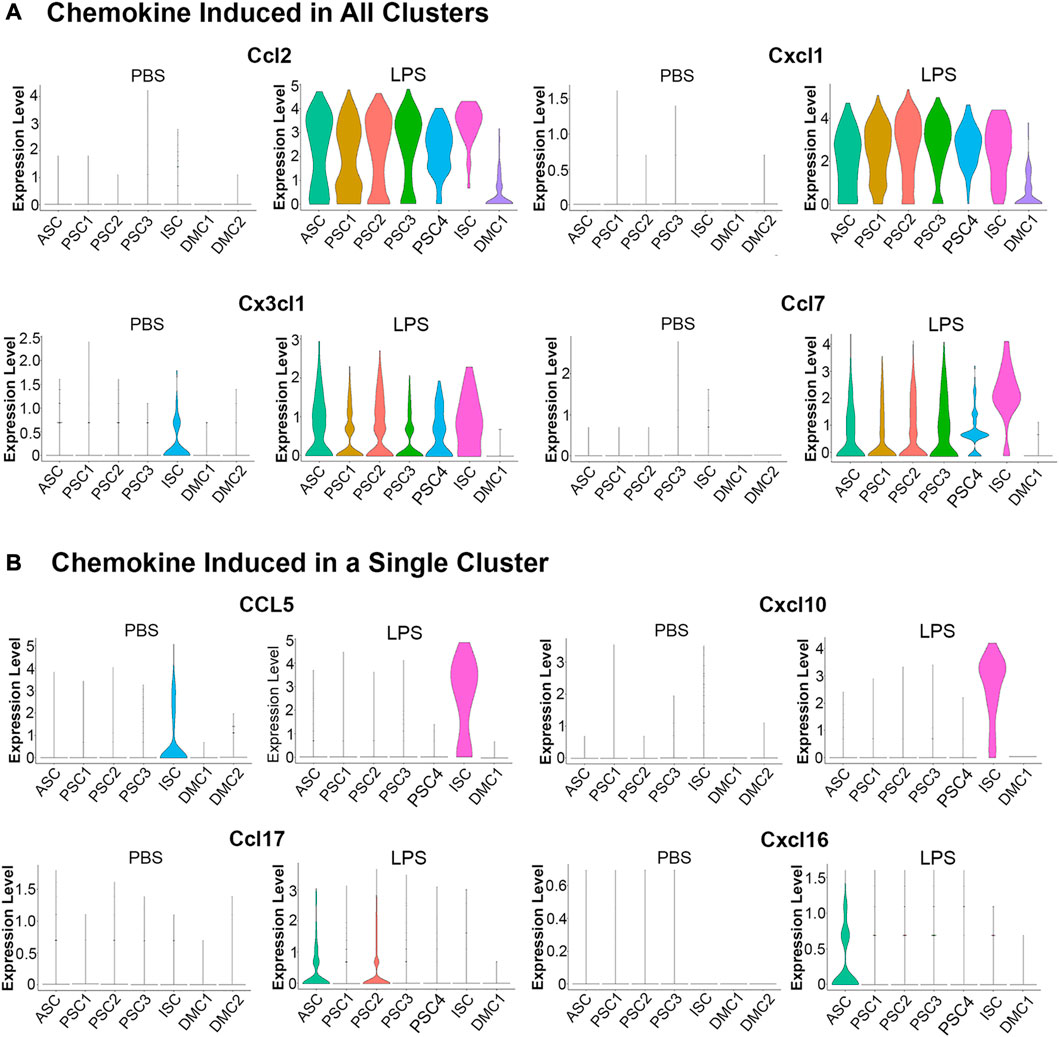
FIGURE 5. Violin plots of chemokine transcription in PBS and LPS treated MuSCs. (A) The transcription of chemokines Ccl2, Cxcl1, Cx3cl1 and Ccl7 were upregulated in all cluster after LPS treatment. (B) The transcription of chemokines Ccl5, Cxcl10, Ccl17 and Cxcl16 were upregulated in a singular cluster post-LPS treatment. Expression is depicted as Log 2 fold change above average expression across all cells.
LPS induction of chemokines was validated in independently isolated primary MuSCs by RT-qPCR of total RNA using gene specific primers. Transcripts for the chemokines Ccl2, Ccl5, Ccl7, Cxcl1, and Cxcl10 were significantly higher in LPS-treated cells as compared to the PBS-treated controls (Supplementary Table S4). Overall, these studies revealed that MuSCs can be induced to express many C-C and C-X-C chemokines with LPS and they are differentially expressed across the cell clusters. ASC and ISC cell clusters express separate sets of chemokines, raising the possibility that they serve unique functions during muscle inflammation response.
An examination of proinflammatory cytokine transcription in response to LPS treatment revealed a modest induction of a limited number of factors when compared to the robust induction of chemokines (Figure 5). Among the TGFβ superfamily, known to positively regulate cells of the innate immune system (Chen and ten Dijke, 2016), Tgfb1 was constitutively transcribed at low levels in the ASC, PSC2, PSC3, and ISC clusters. With LPS treatment, Tgfb1 was induced in PSC1 cells (Figure 6A). Similarly, Tgfb2 was induced in PSC1 and PSC2 cells, but constitutively expressed in DMCs (data not shown). In contrast, Bmp2, a known antiviral ISG in bone marrow macrophages (Liu et al., 2012), was induced selectively in the ISC cluster (Figure 6A). Interestingly, Il18 and Il33, members of the IL-1 family of proinflammatory cytokines (Chan et al., 2019; Martinon et al., 2009), were weakly induced in ISCs (Figure 6B). Overall, the MuSCs expressed cytokines and chemokines in a heterogeneous pattern in response to inflammatory signals.
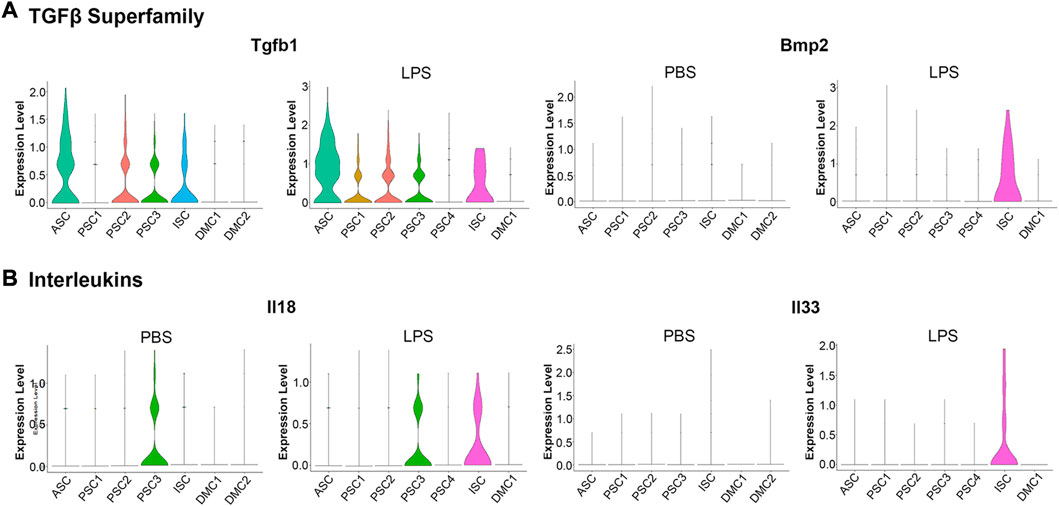
FIGURE 6. Violin plots of cytokine transcription in PBS and LPS treated MuSCs. (A) The transcription pattern of the TGFβ superfamily genes Tgfb1 and Bmp2. (B) The transcription pattern of the Interleukins superfamily genes Il18 and Il33. Expression is depicted as Log 2 FC above average expression across all cells.
Single cell sequencing of the PBS control cells revealed a low level expression of components of the IFN signaling pathway in the ISC cluster (Figure 2). LPS induced the expression of Ccl5, Cxcl10, Il18, and Il33 in these cells (Figures 5, 6), supporting a proinflammatory role for this cluster. Since LPS acts through the TLR4 receptor to induce IFN α and γ expression (Richez et al., 2010; Wang et al., 2017), transcription of members of the antiviral IFN pathway were examined (Table 1). Of note, two cytosolic sensors of viral DNA and RNA, Ifih1/MDA-5 and Ddx58/Rig-1 (Loo and Gale, 2011), as well as their downstream transcription factor, Irf7, were selectively induced in the ISC cells. Further, twelve other IFN induced signaling factors associated with antiviral activity were also induced (Table 1). It should be noted that IFN-α, -β, or -γ transcription was not detected, raising the possibility that the antiviral IFN pathway was activated through the cytosolic receptors independent of the IFNs.
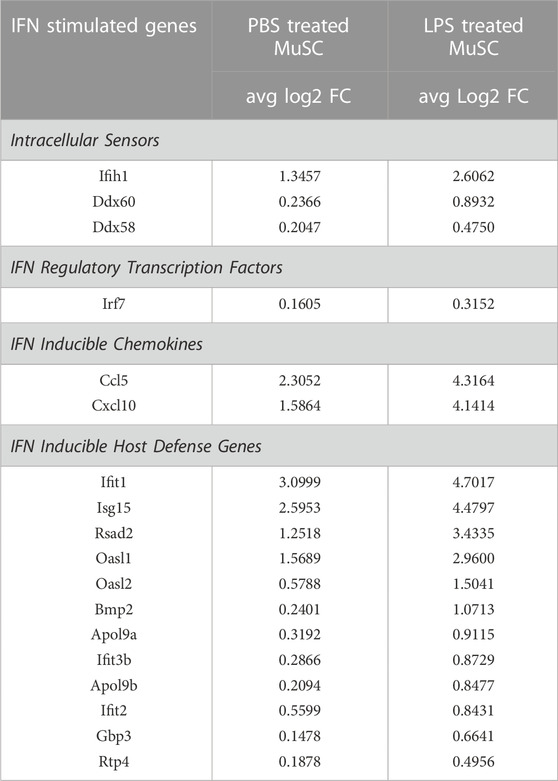
TABLE 1. Induction of genes in the antiviral IFN pathway in the ISC cluster of PBS and LPS treated MuSCs. Transcription of genes reported to participate in the antiviral IFN pathway or be downstream induced genes were quantified by scRNA-Seq and expressed as Log2 fold change above average expression across all cells in the PBS- or LPS-treated MuSC library.
The expression of the IFN pathway marker, CCL5, in a small subset of MuSCs was confirmed by IF done using antibodies specific to both MYOD and CCL5. CCL5+ve/MYOD+ve cells were detected in newly isolated MuSCs from the quadriceps femoris of 3 month old mice treated with LPS or PBS (Figure 7). Double positive cells represented 2.3% of the LPS treated MuSCs, which was comparable to the distribution (2.08%) of Myod1 positive cells expressing ISG genes identified in the scRNA-Seq analysis (Table 2). These genes were also detectable in PBS treated control cultures (Figure 7), indicating that activation of the ISG gene expression was independent of LPS activation, but the expression level was increased in response to this inflammatory signal.
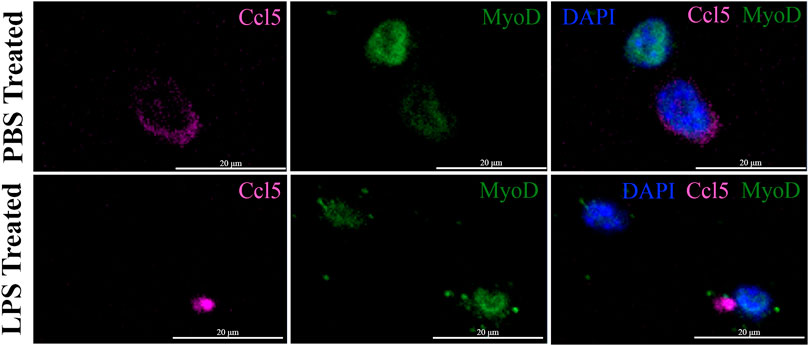
FIGURE 7. Co-expression of CCL5 and MYOD in proliferating MuSCs treated with LPS and PBS. The expression of CCL5 (magenta) and MYOD (green) was detected by indirect IF in MuSCs treated with LPS (1 mg/mL) or control PBS, for 2 h or control PBS treatment. CCL5 can be detected in the cytoplasm and MYOD in the nucleus. The nuclei were visualized with DAPI (blue). Images were taken at ×20 magnification. Scalebar = 20 µm.
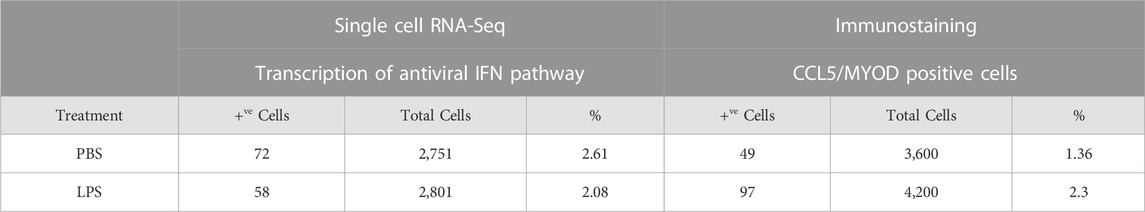
TABLE 2. Distribution of MuSCs with the activated antiviral IFN pathway in PBS- and LPS-treated cells. Representation of the ISC cluster within the control and LPS treated MuSCs was evaluated by co-expression of Isg15, Ifih1, Oasl, Ifit1, and Myod1 as measured by scRNA-Seq. ISC cells were independently assess in culture by co-expression of CCL5 and MYOD by IF.
Activation of the antiviral IFN pathway is associated with the chronic muscle inflammation noted in DM patients and IFN-γ has been linked to muscle regeneration (Greenberg et al., 2005; Cheng et al., 2008; Suárez-Calvet et al., 2014). Since we observed that the IFN pathway was activated in a small percentage of MuSCs isolated from healthy mice, it was possible that it has a role in skeletal muscle repair as well. We further examined if we could detect activation of a subset of the ISG in response to acute injury. CTX was injected into the quadriceps femoris of Bl10 mice and total RNA isolated from the site of injury at 1, 2, 3, and 5 days post-injury (DPI) and from the uninjured contralateral leg. RT-qPCR was done using gene specific primers. When compared to the uninjured muscle, both Ifih1 and Isg15 transcription were rapidly induced at 1 DPI followed by a gradual decline. In contrast, expression of the IFN-regulated chemokines, Ccl5 and Cxcl10, peaked at 3 DPI before sharply declining (Figure 8). This is distinctly different from the broadly transcribed chemokines, Ccl2 and Cxcl1, that peaked earlier at 1 DPI. Thus, these data demonstrate that components of the antiviral IFN pathway originally observed with LPS treatment are expressed in vivo in response to injury also.
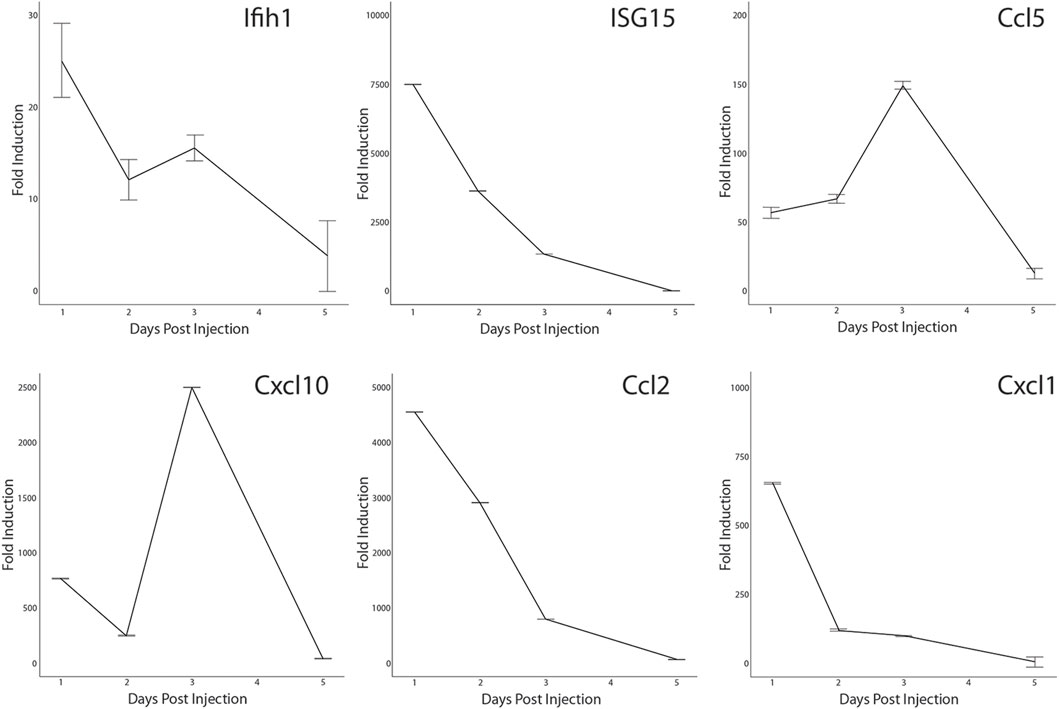
FIGURE 8. Transcription of antiviral interferon pathway genes in CTX-injured quadriceps. CTX-injured quadriceps had total RNA isolated at 1, 2, 3, and 5 DPI. Transcripts of Ifih1, Isg15, Ccl5, Cxcl10, Ccl2, and Cxcl1 were quantified by RT-qPCR. RNA transcript levels are expressed as the fold induction over RNA isolated from the uninjured contralateral leg. Significance was determined by Student’s t-test, all timepoints p < 0.0001 except for Cxcl1, Ifih1, and Ccl5 at 5 DPI.
The distribution of satellite cells that had the antiviral IFN pathway activated in muscle was evaluated by IF with antibodies specific for CCL5 and M-Cadherin (MCAD), a cell adhesion protein expressed on satellite cells in their niche and proliferating myoblasts. MCAD expression is used to distinguish the mononuclear satellite cells from the adjacent muscle fiber (Yin et al., 2013). IF was performed on sections of CTX injured and contralateral control quadriceps. Analysis demonstrated that CCL5+ve/MCAD+ve cells represented 10.18% of total MCAD+ve satellite cells of uninjured muscle and 10.52% of the total MCAD+ve cells in CTX injured muscle at 3 DPI (Figure 9A). In comparison, we found CCL2, which was transcribed at high levels in LPS-induced MuSCs, was broadly expressed in both satellite cells and intact muscle fibers at 1 DPI (Figure 9B).
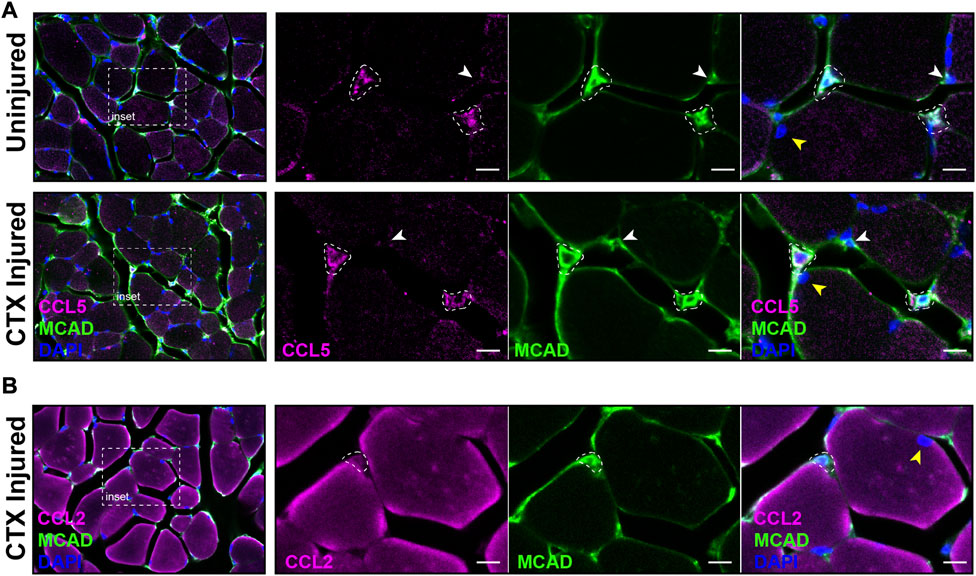
FIGURE 9. Co-expression of CCL2, CCL5 and MCAD in CTX-injured skeletal muscle. (A) The expression of CCL5 (magenta) and MCAD (green) were visualized in DAPI-labeled (blue) CTX-injured and uninjured quadriceps at 3 DPI by IF. White dotted outlines mark CCL5/MCAD co-expressing satellite cells, the white arrowhead indicates a MCAD-only expressing satellite cell, and the yellow arrowhead marks a muscle fiber nucleus. (B) CCL2 (magenta) and MCAD (green) co-expressing satellite cells (white dotted outline) were detected in CTX-injured quadriceps at 1 DPI. The nuclei were labeled with DAPI (blue). In contrast to CCL5, significant expression of CCL2 was detected in muscle fibers. The yellow arrowhead denotes a muscle fiber nucleus. Scalebar = 10 µm.
In this study, we re-evaluated the muscle-immune paradigm during muscle damage and repair. The transcription of immune effector genes has been reported in muscle through a MYF6-dependent pathway (Barruet et al., 2020; Lazure et al., 2020). However, their capacity to be induced to express pro-inflammatory genes in response to infection or injury remains less clear. Here we leveraged primary satellite cell cultures and single cell RNA sequencing technology to examine the capacity of satellite cells and myoblasts to express cytokines and chemokines. Here, we demonstrated that non-sterile PAMPs (LPS) induced a heterogeneous pattern of expression of proinflammatory cytokines and chemokines (Figures 3–6). Single-cell RNA-SEQ revealed that proliferating MuSCs in culture were comprised of distinct clusters that ranged from activated satellite cells, proliferating myoblasts, to differentiating myocytes (Figures 2, 3). Though many of the chemokines and cytokines were broadly expressed across these clusters, there were examples of selective transcription in ISC, ASC, and PSC1. Thus, the responsiveness of the MuSCs to inflammatory signals can vary dependent on a cell’s myogenic status on the pathway towards differentiation. Of particular interest are the ISCs that appear to have primed the antiviral IFN pathway which then can be activated by LPS to selectively increase the expression of Ccl5, Cxcl10, Il18, and Il33. There are descriptions of satellite cell subsets predicted to have an adaptive response for injury and stress (GAlert) (Rodgers et al., 2014; Scaramozza et al., 2019; De Micheli et al., 2020). Our data is the first indication that a subgroup of satellite cells that could be acting as sensors for infection and injury in the muscle.
An evaluation of the chemokines and cytokines expressed in response to LPS predicts that MuSCs can promote the proinflammatory environment in muscle. Many of these factors have been reported to recruit neutrophils, monocytes, and T cells to the site of muscle injury, including CCL2, CCL5, CCL7, CXCL16, and CX3CL3 (Figure 5; Zhang et al., 2009; Martinez et al., 2010; Crescioli et al., 2012; Kohno et al., 2011; Strömberg et al., 2016). Further, our finding that CCL2 and CCL5 were expressed in satellite cells and muscle fibers at the site of CTX-induced injury, indicated that induction occurs in vivo and this response is not restricted to LPS. Several of the chemokines expressed in LPS-treated MuSC are predicted to participate in the expansion of the satellite cell population during skeletal muscle repair. CCL2 and CXCL16 promote satellite cell proliferation (Yahiaoui et al., 2008; Zhang et al., 2009), while CXCL1, CXCL10, and CCL17, negatively regulate myoblast differentiation, preventing premature exit from the cell cycle (Ge et al., 2013; Hogan et al., 2018). Thus, the pattern of expression is consistent with a dual role for MuSCs during regeneration, recruiting the innate immune cells and promoting myoblast expansion.
An unexpected outcome of the scRNA-Seq analysis was the identification of a small population of MuSCs with the antiviral IFN pathway activated in the absence of LPS. Approximately, 2% of the proliferating MuSCs in culture and 10% of satellite cells in muscle tissue expressed genes associated with the IFN-independent early antiviral components of the pathway that sense cytosolic viral DNA and double stranded RNA. Among the IFN genes expressed were the cytosolic sensors, Ddx58 and Ifih1, which bind the triphosphate group at the 5’ end of viral RNA (Wu and Chen, 2014). This interaction induces expression of type I IFNs and a subset ISG proteins mediated by the transcription factors nuclear factor κ-light-chain enhancer of activated B cells (NF-κB), and the IFN regulatory factors IRF3 and IRF7 (Grandvaux et al., 2002). Among the antiviral pathway genes detected at high levels in uninduced MuSC ISCs, Oasl, OasII, Isg15, Rsad2, and Irf7, are positive regulators of IFIH1/DDX58-dependent signaling (Kim et al., 2008; MacMicking et al., 2012; Hee and Cresswell, 2017; Li et al., 2017; Liu et al., 2021). Transcription of these genes in the PBS treated controls suggests these cells are maintained in a primed state. How the antiviral pathway is activated at low levels in the absence of a viral infection is unclear. Further, the presence of satellite cells with activated antiviral IFN pathway 3 days after a sterile muscle injury reinforces the presence of a mechanism of activation that is virus independent. One possible explanation comes from the observation that the endoribonuclease, RNase L, and the 2′,5′-oligoadenylate synthetase, OAS, can produce small self-RNAs from cellular mRNA that is recognizable by IFIH1 and DDX58 (Malathi et al., 2007; Stok et al., 2020). This suggests that ISCs are capable of self-priming or possibly detecting RNA from adjacent damaged cells post-injury in the absence of infection.
Expression of the IFN signaling pathway in satellite cells has been previously described in patients with dermatomyositis (DM) and polymyositis, demonstrating the capacity of muscle to regulate this pathway (Greenberg et al., 2005; Salajegheh et al., 2010; Suárez-Calvet et al., 2014). This suggests a mechanism for the autoimmune pathology of DM patients through overstimulation of IFIH1/DDX58 sensors by self-RNA. Consistent with this, muscle from these DM patients express type I IFN and upregulate Isg15 and other ISG genes at levels approaching 100 fold higher than unaffected muscle (Baechler et al., 2011). Interestingly, activation of the IFN pathway also was detected in uninjured human vastus lateralis satellite cells by scRNA-Seq (Barruet et al., 2020). They identified a small population of Pax7+ve cells that expressed many of the IFN genes described in our study. It should be noted that the chemokines, CCL5 and CXCL10, were not observed in the human satellite cells, whereas our data demonstrated expression of Ccl5 and Cxcl10 transcripts in cultured satellite cells, and CCL5 protein in muscle. As our current studies demonstrate that IFN pathway and chemokine expressing MuSCs are present and expression of these genes can be upregulated by LPS, we have an opportunity for further study of the role of Ccl5 and IFN pathway genes in muscle inflammation.
Heterogeneity of the adult satellite cell population has been well studied as it relates to tissue origin, cell cycle, and function in skeletal muscle repair (Biressi and Rando, 2010). In this study, we expand our view of heterogeneity to include the capacity of MuSCs to produce chemokines and cytokines that act on the myeloid and myogenic cells present at the site of injury. Pax7+ve satellite cells (ASC) selectively expressed Ccl17 and Cxcl16 that promoted cell proliferation in myogenic cells, as well recruiting neutrophils and macrophages. Perhaps more surprising, was the identification of ISC population, present in both treated and control MuSC cultures and in the satellite cells of injured muscle. Their presence in the absence of injury or inflammatory signals suggests that they serve as sentinel cells. The cells are specifically predisposed to activate the IFIH1/DDX58-dependent pathway. This is underscored by the results of our LPS treatment of MuSCs. LPS induces a broad set of proinflammatory cytokines and chemokines in immune and non-immune cells through a TLR4-dependent manner, including the IFIH1/DDX58-dependent pathway (Lu et al., 2008; Imaizumi et al., 2013). While LPS induced transcription of Ccl2, Ccl7, and Cxcl1 in all cell clusters, induction of Ifih1, Ddx58, Ccl5 and Cxcl10 transcription was restricted to the ISC cluster. The nature of the signals regulating the heterogeneity of MuSC response to LPS or other immunogens remains to determined.
Overall, our studies demonstrate that satellite cells and proliferating myoblasts are competent to respond to PAMPs and DAMPs by expressing of chemokines and cytokines. Functionally, this is likely a combination of amplifying the signals at the site of injury that recruit and activate proinflammatory immune cells and autocrine signaling promoting the rapid expansion of the satellite cells prior to differentiation into muscle fibers. These studies suggest that the antiviral IFN pathway plays a greater role in sterile inflammation in response to muscle damage than previously considered. Additional studies are needed to understand its contribution to the regulation of the innate immune response and satellite cell proliferation and differentiation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE216848.
The animal study was reviewed and approved by Arizona State University Institutional Animal Care and Use Committee.
AA as the first author to lead on the execution of all aspects of the research. KR and JN provided expertise with indirect immunofluorescence and confocal imaging of chemokine expression in culture and injured muscle. SO’C and CP provide expertise on the scRNA-Seq analysis of chemokine and cytokine induction in cell culture. GS contributed to the validation of chemokine transcription by RT-qPCR. AR and JW-R contributed to the design and interpretation of the experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1084068/full#supplementary-material
Baechler, E. C., Bilgic, H., and Reed, A. M. (2011). Type I interferon pathway in adult and juvenile dermatomyositis. Arthritis Res. Ther. 13 249, 249. doi:10.1186/ar3531
Barruet, E., Garcia, S. M., Striedinger, K., Wu, J., Lee, S., Byrnes, L., et al. (2020). Functionally heterogeneous human satellite cells identified by single cell RNA sequencing. eLife 9, e51576. doi:10.7554/eLife.51576
Biressi, S., and Rando, T. A. (2010). Heterogeneity in the muscle satellite cell population. Semin. Cell Dev. Biol. 21, 845–854. doi:10.1016/j.semcdb.2010.09.003
Bivona, J. J., Crymble, H. M., Guigni, B. A., Stapleton, R. D., Files, D. C., Toth, M. J., et al. (2021). Macrophages augment the skeletal muscle proinflammatory response through TNFα following LPS-induced acute lung injury. FASEB J. 35, e21462. doi:10.1096/fj.202002275RR
Boehm, U., Klamp, T., Groot, M., and Howard, J. C. (1997). Cellular responses to interferon-gamma.Annu. Rev. Immunol. 15, 749–795. doi:10.1146/annurev.immunol.15.1.749
Brigitte, M., Schilte, C., Plonquet, A., Baba-Amer, Y., Henri, A., Charlier, C., et al. (2010). Muscle resident macrophages control the immune cell reaction in a mouse model of notexin-induced myoinjury. Arthritis Rheum. 62, 268–279. doi:10.1002/art.27183
Chaillou, T., Kirby, T. J., and McCarthy, J. J. (2017). Ribosome biogenesis: Emerging evidence for a central role in the regulation of skeletal muscle mass. J. Cell. Physiol. 229, 1584–1594. doi:10.1002/jcp.24604
Chan, B. C. L., Lam, C. W. K., Tam, L.-S., and Wong, C. K. (2019). IL33: Roles in allergic inflammation and therapeutic perspectives. Front. Immunol. 10, 364. doi:10.3389/fimmu.2019.00364
Chargé, S. B., and Rudnicki, M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238. doi:10.1152/physrev.00019.2003
Chen, S. E., Gerken, E., Zhang, Y., Zhan, M., Mohan, R. K., Li, A. S., et al. (2005). Role of TNF-{alpha} signaling in regeneration of cardiotoxin-injured muscle. Am. J. Physiol. Cell Physiol. 289, C1179–C1187. doi:10.1152/ajpcell.00062.2005
Chen, W., and ten Dijke, P. (2016). Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immuno 16, 723–740. doi:10.1038/nri.2016.112
Cheng, M., Nguyen, M.-H., and Koh, T. J. (2008). Endogenous interferon-gamma is required for efficient skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 294, C1183–C1191. doi:10.1152/ajpcell.00568.2007
Ciesielska, A., Matyjek, M., and Kwiatkowska, K. (2021). TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol. Life Sci. 78, 1233–1261. doi:10.1007/s00018-020-03656-y
Collins, R. A., and Grounds, M. D. (2001). The role of tumor necrosis factor-α (TNFα) in skeletal muscle regeneration. Studies in TNFα −/− and TNFα −/−/LTα −/− mice. J. Histochem Cytochem 49, 989–1001. doi:10.1177/002215540104900807
Correra, R. M., Ollitrault, D., Valente, M., Mazzola, A., Adalsteinsson, B. T., Ferguson-Smith, A. C., et al. (2018). The imprinted gene Pw1/Peg3 regulates skeletal muscle growth, satellite cell metabolic state, and self-renewal. Sci. Rep. 8, 14649. doi:10.1038/s41598-018-32941-x
Crescioli, C., Sottili, M., Bonini, P., Cosmi, L., Chiarugi, P., Romagnani, P., et al. (2012). Inflammatory response in human skeletal muscle cells: CXCL10 as a potential therapeutic target. Eur. J. Cell Biol. 91, 139–149. doi:10.1016/j.ejcb.2011.09.011
De Michelli, A. J., Spector, J. A., Elemento, O., and Cosgrove, B. D. (2020). A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skelet. Muscle 10, 19. doi:10.1186/s13395-020-00236-3
De Rossi, M., Bernasconi, P., Baggi, F., de Waal Malefyt, R., and Mantegazza, R. (2000). Cytokines and chemokines are both expressed by human myoblasts: Possible relevance for the immune pathogenesis of muscle inflammation. Int. Immunol. 12, 1329–1335. doi:10.1093/intimm/12.9.1329
Dogra, C., Changotra, H., Mohan, S., and Kumar, A. (2006). Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-kappaB and degradation of MyoD protein. J. Biol. Chem. 281, 10327–10336. doi:10.1074/jbc.m511131200
Dogra, C., Changotra, H., Wedhas, N., Qin, X., Wergedal, J. E., and Kumar, A. (2007). TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 21, 1857–1869. doi:10.1096/fj.06-7537com
El Haddad, M., Jean, E., Turki, A., Hugon, G., Vernus, B., Bonnieu, A., et al. (2012). Glutathione peroxidase 3, a new retinoid target gene, is crucial for human skeletal muscle precursor cell survival. J. Cell Sci. 125, 6147–6156. doi:10.1242/jcs.115220
Fielding, R. A., Manfredi, T. J., Ding, W. J., Fiatarone, M. A., Evans, W. J., and Cannon, J. G. (1993). Acute-phase response in exercise. III. Neutrophil and IL-1 beta accumulation in skeletal muscle. Am. J. Physiol. 265, R166–R172. doi:10.1152/ajpregu.1993.265.1.R166
Frost, R. A., Nystrom, G. J., and Lang, C. H. (2003). Lipopolysaccharide and proinflammatory cytokines stimulateinterleukin-6 expression in C2C12 myoblasts: Role of the jun NH2-terminal kinase. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285, R1153–R1164. doi:10.1152/ajpregu.00164.2003
Frost, R. A., Nystrom, G. J., and Lang, C. H. (2002). Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283, R698–R709. doi:10.1152/ajpregu.00039.2002
Fu, X., Xiao, J., Wei, Y., Li, S., Liu, Y., Yin, J., et al. (2015). Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 25, 655–673. doi:10.1038/cr.2015.58
Garry, D. J., Meeson, A., Elterman, J., Zhao, Y., Yang, P., Bassel-Duby, R., et al. (2000). Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. U.S.A. 97, 5416–5421. doi:10.1073/pnas.100501197
Gayraud-Morel, B., Le Bouteiller, M., Commere, P.-H., Cohen-Tannoudji, M., and Tajbakhsh, S. (2018). Notchless defines a stage-specific requirement for ribosome biogenesis during lineage progression in adult skeletal myogenesis. Development 145, dev162636–12. doi:10.1242/dev.162636
Ge, Y., Waldemer, R. J., Nalluri, R., Nuzzi, P. D., and Chen, J. (2013). RNAi screen reveals potentially novel roles of cytokines in myoblast differentiation. PLoS One 8, e68068. doi:10.1371/journal.pone.0068068
Gordon, B. S., Kelleher, A. R., and Kimball, S. R. (2013). Regulation of muscle protein synthesis and the effects of catabolic states. Int. J. Biochem. Cell Biol. 45, 2147–2157. doi:10.1016/j.biocel.2013.05.039
Gordon, J. R., and Galli, S. J. (1990). Mast cells as a source of both preformed and immunologically inducible TNF-alpha/cachectin. Nature 346, 274–276. doi:10.1038/346274a0
Grandvaux, N., Servant, M. J., ten Oever, B., Sen, G. C., Balachandran, S., Barber, G. N., et al. (2002). Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76, 5532–5539. doi:10.1128/JVI.76.11.5532-5539.2002
Greenberg, S. A., Pinkus, J. L., Pinkus, G. S., Burleson, T., Sanoudou, D., Tawil, R., et al. (2005). Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann. Neurol. 57, 664–678. doi:10.1002/ana.20464
Griffin, C. A., Apponi, L. H., Long, K. K., and Pavlath, G. K. (2010). Chemokine expression and control of muscle cell migration during myogenesis. J. Cell Sci. 123, 3052–3060. doi:10.1242/jcs.066241
Haimes, J., and Kelley, M. (2010). Demonstration of a ΔΔCq calculation method to compute relative gene expression from qPCR data. Thermo. Sci. Technol. Note, 1–4.
Hee, J. S., and Cresswell, P. (2017). Viperin interaction with mitochondrial antiviral signaling protein (MAVS) limits viperin-mediated inhibition of the interferon response in macrophages. PLoS One 12, e0172236. doi:10.1371/journal.pone.0172236
Hogan, K. A., Cho, D. S., Arneson, P. C., Samani, A., Palines, P., Yang, Y., et al. (2018). Tumor-derived cytokines impair myogenesis and alter the skeletal muscle immune microenvironment. Cytokine107 107, 9–17. doi:10.1016/j.cyto.2017.11.006
Howard, E. E., Pasiakos, S. M., Blesso, C. N., Fussell, M. A., and Rodriquez, N. R. (2020). Divergent roles of inflammation in skeletal muscle recovery from injury. Front. Physiology 11, 87–13. doi:10.3389/fphys.2020.00087
Imaizumi, T., Aizawa-Yashiro, T., Watanabe, S, Matsumiya, T., Yoshida, H., Tatsuta, T., et al. (2013). TLR4 signaling induces retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5 in mesangial cells. J. Nephrol. 26, 886–893. doi:10.5301/jn.5000254
Kelly-Scumpia, K. M., Scumpia, P. O., Delano, M. J., Weinstein, J. S., Cuenca, A. G., Wynn, J. L., et al. (2010). Type I interferon signaling in hematopoietic cells is required for survival in mouse polymicrobial sepsis by regulating CXCL10. J. Exp. Med. 207, 319–326. doi:10.1084/jem.20091959
Kim, M.-J., Hwang, S. Y., Imaizumi, T., and Yoo, J. Y. (2008). Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J. Virol. 82, 1474–1483. doi:10.1128/JVI.01650-07
Kohno, S., Ueji, T., Abe, T., Nakao, R., Hirasaka, K., Oarada, M., et al. (2011). Rantes secreted from macrophages disturbs skeletal muscle regeneration after cardiotoxin injection in Cbl-b-deficient mice. Muscle Nerve 43, 223–229. doi:10.1002/mus.21829
Kuru, S., Inukai, A., Kato, T., Lian, T., Kimura, S., and Sobue, G. (2003). Expression of tumor necrosis factor-α in regenerating muscle fibers in inflammatory and non-inflammatory myopathies. Acta Neuropathol. 105, 217–224. doi:10.1007/s00401-002-0635-4
Langen, R. C., Van Der Velden, J. L., Schols, A. M., Kelders, M. C., Wouters, E. F., and Janssen-Heininger, Y. M. (2004). Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 18, 227–237. doi:10.1096/fj.03-0251com
Lazure, F., Blackburn, D. M., Corchado, A. H., Sahinyan, K., Karam, N., Sharanek, A., et al. (2020). Myf6/MRF4 is a myogenic niche regulator required for the maintenance of the muscle stem cell pool. EMBO Rep. 21, e49499. doi:10.15252/embr.201949499
Li, L.-F., Yu, J., Zhang, Y., Yang, Q., Li, Y., Zhang, L., et al. (2017). Interferon-Inducible oligoadenylate synthetase-like protein acts as an antiviral effector against classical swine fever virus via the MDA5-mediated type I interferon-signaling pathway. J. Virol. 91, e01514. doi:10.1128/JVI.01514-16
Li, N., Mu, H., Zheng, L., Li, B., Wu, C., Niu, B., et al. (2016). EIF2S3Y suppresses the pluripotency state and promotes the proliferation of mouse embryonic stem cells. Oncotarget 7, 11321–11331. doi:10.18632/oncotarget.7187
Li, Y. P. (2003). TNF-alpha is a mitogen in skeletal muscle. Am. J. Physiol. Cell Physiol. 285, C370–C376. doi:10.1152/ajpcell.00453.2002
Liu, G. Q., Lee, J.-H., Parker, Z. M., Acharya, D., Chiang, J. J., van Gent, M., et al. (2021). ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 6, 467–478. doi:10.1038/s41564-021-00884-1
Liu, S.-Y., Sanchez, D. J., Aliyari, R., Lu, S., and Cheng, G. (2012). Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 109, 4239–4244. doi:10.1073/pnas.1114981109
Loo, Y.-M., and Gale, M. (2011). Immune signaling by RIG-I-like receptors. Immunity 34, 680–692. doi:10.1016/j.immuni.2011.05.003
Lu, H., Huang, D., Ransohoff, R. M., and Zhou, L. (2011). Acute skeletal muscle injury: CCL2 expression by both monocytes and injured muscle is required for repair. FASEB J. 25, 3344–3355. doi:10.1096/fj.10-178939
Lu, Y.-C., Yeh, W.-C., and Ohashi, P. S. (2008). LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151. doi:10.1016/j.cyto.2008.01.006
MacMicking, J. D. (2012). Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382. doi:10.1038/nri3210
Majumdar, G., Vera, S., Elam, M. B., and Raghow, R. (2015). A streamlined protocol for extracting RNA and genomic DNA from archived human blood and muscle. Anal. Biochem. 474, 25–27. doi:10.1016/j.ab.2014.12.021
Malathi, K., Dong, B., Gale, M., and Silverman, R. H. (2007). Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819. doi:10.1038/nature06042
Martinet, C., Monnier, P., Louault, Y., Benard, M., Gabory, A., and Dandolo, L. (2016). H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 143, 962–971. doi:10.1242/dev.131771
Martinez, C. O., McHale, M. J., Wells, J. T., Ochoa, O., Michalek, J. E., McManus, L. M., et al. (2010). Regulation of skeletal muscle regeneration by CCR2-activating chemokines is directly related tomacrophage recruitment. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R832–R842. –R842. doi:10.1152/ajpregu.00797.2009
Martinon, F., Mayor, A., and Tschopp, J. (2009). The inflammasomes: Guardians of the body. Annu. Rev. Immunol. 27, 229–265. doi:10.1146/annurev.immunol.021908.132715
McKellar, D. W., Walter, L. D., Song, L. T., Mantri, M., Wang, M. F. Z., De Vlaminck, I., et al. (2021). Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun. Biol. 4, 1280. doi:10.1038/s42003-021-02810-x
Mervis, M. J., Matsumura, M. S., Olsen, Z. E., Hirschi-Budge, K. M., Reynolds, P. R., Arroyo, J. A., et al. (2020). The effect of gas6-axl double knockout on satellite cell proliferation and skeletal muscle regeneration after injury. FASEB J. 34, 1. doi:10.1096/fasebj.2020.34.s1.09814
Moneta, G. M., Marafon, D. P., Marasco, E., Rosina, S., Verardo, M., Fiorillo, C., et al. (2019). Muscle expression of type I and type II interferons is increased in juvenile dermatomyositis and related to clinical and histologic features. Arthritis Rheumatol. 71, 1011–1021. doi:10.1002/art.40800
Munoz-Canoves, P., Scheele, C., Pedersen, B. K., and Serrano, A. L. (2013). Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 280, 4131–4148. doi:10.1111/febs.12338
Nakano, M., Fujii, T., Hashimoto, M., Yukawa, N., Yoshifuji, H., Ohmura, K., et al. (2012). Type I interferon induces CX3CL1 (fractalkine) and CCL5 (RANTES) production in human pulmonary vascular endothelial cells. Clin. Exp. Immunol. 170, 94–100. doi:10.1111/j.1365-2249.2012.04638.x
Oprescu, S. N., Yue, F., Qiu, J., Brito, L. F., and Kuang, S. (2020). Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration. iScience 23, 100993. doi:10.1016/j.isci.2020.100993
Otani, M., Furukawa, S., Wakisaka, S., and Maeda, T. (2020). A novel adipokine C1q/TNF-related protein 3 is expressed in developing skeletal muscle and controls myoblast proliferation and differentiation. Mol. Cell. Biochem. 409, 271–282. doi:10.1007/s11010-015-2531-y
Palade, J., Pal, A., Rawls, A., Stabenfeldt, S., and Wilson-Rawls, J. (2019). Molecular analysis of muscle progenitor cells on extracellular matrix coatings and hydrogels. Acta Biomater. 97, 296–309. doi:10.1016/j.actbio.2019.08.019
Richez, C., Yasuda, K., Watkins, A. A., Akira, S., Lafyatis, R., Van Seventer, J., et al. (2010). TLR4 ligands induce IFN-alpha production by mouse conventional dendritic cells and human monocytes after IFN-beta priming. J. Immunol. 182, 820–828. doi:10.4049/jimmunol.182.2.820
Rodgers, J. T., King, K. Y., Brett, J. O., Cromie, M. J., Charville, G. W., Maguire, K. K., et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396. doi:10.1038/nature13255
Rodriguez-Outeiriño, L., Hernandez-Torres, F., Ramírez-de Acuña, F., Matías-Valiente, L., Sanchez-Fernandez, C., Franco, D., et al. (2021). Muscle satellite cell heterogeneity: Does embryonic origin matter? Front. Cell Dev. Biol. 15 (9), 750534. doi:10.3389/fcell.2021.750534
Salajegheh, M., Kong, S. W., Pinkus, J. L., Walsh, R. J., Liao, A., Nazareno, R., et al. (2010). Interferon-stimulated gene 15 (ISG15) conjugates proteins in dermatomyositis muscle with perifascicular atrophy. Ann. Neurol. 67, 53–63. doi:10.1002/ana.21805
Scaramozza, A., Park, D., Kollu, S., Beerman, I., Sun, X., Rossi, D. J., et al. (2019). Lineage tracing reveals a subset of reserve muscle stem cells capable of clonal expansion under stress. Cell Stem Cell 24, 944–957.e5. doi:10.1016/j.stem.2019.03.020
Sheehan, S. M., Tatsumi, R., Temm-Grove, C. J., and Allen, R. E. (2000). HGF is an autocrine growth factor for skeletal muscle satellite cells in vitro. Muscle and Nerve 23, 239–245. doi:10.1002/(sici)1097-4598(200002)23:2<239:aid-mus15>3.0.co;2-u
Stok, J. E., Quiroz, M. E. V., and van der Veen, A. G. (2020). Self RNA sensing by RIG-I-like receptors in viral infection and sterile inflammation. J. Immunol205 205, 883–891. doi:10.4049/jimmunol.2000488
Strömberg, A., Olsson, K., Dijksterhuis, J. P., Rullman, E., Schulte, G., and Gustafsson, T. (2016). CX3CL1--a macrophage chemoattractant induced by a single bout of exercise in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R297–R304. doi:10.1152/ajpregu.00236.2015
Suárez-Calvet, X., Gallardo, E., Nogales-Gadea, G., Querol, L., Navas, M., Diaz-Manera, J., et al. (2014). Altered RIG-I/DDX58-mediated innate immunity in dermatomyositis. J. Pathol. 233, 258–268. doi:10.1002/path.4346
Teixeira, C. F., Zamuner, S. R., Zuliani, J. P., Fernandes, C. M., Cruz-Hofling, M. A., Fernandes, I., et al. (2003). Neutrophils do not contribute to local tissue damage, but play a key role in skeletal muscle regeneration, in mice injected with Bothrops asper snake venom. Muscle Nerve 28, 449–459. doi:10.1002/mus.10453
Tidball, J. G. (2017). Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 17 (3), 165–178. doi:10.1038/nri.2016.150
Torrente, Y., El Fahime, E., Caron, N. J., Bo, R. D., Belicchi, M., Pisati, F., et al. (2003). Tumor necrosis factor-alpha (TNF-alpha) stimulates chemotactic response in mouse myogenic cells. Cell Transpl. 12, 91–100. doi:10.3727/000000003783985115
Venereau, E., Casalgrandi, M., Schiraldi, M., Antoine, D. J., Cattaneo, A., De Marchis, F., et al. (2012). Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 209, 1519–1528. doi:10.1084/jem.20120189
Wang, P., Han, X., Mo, B., Huang, G., and Wang, C. (2017). LPS enhances TLR4 expression and IFN-γ production via the TLR4/IRAK/NF-κB signaling pathway in rat pulmonary arterial smooth muscle cells. Mol. Med. Rep. 16, 3111–3116. doi:10.3892/mmr.2017.6983
Warren, G. L., Hulderman, T., Jensen, N., McKinstry, M., Mishra, M., Luster, M. I., et al. (2002). Physiological role of tumor necrosis factor-α in traumatic muscle injury. FASEB J. 16, 1630–1632. doi:10.1096/fj.02-0187fje
Yahiaoui, L., Gvozdic, D., Danialou, G., Mack, M., and Petrof, B. J. (2008). CC family chemokines directly regulate myoblast responses to skeletal muscle injury. J. Physiol. 586, 3991–4004. doi:10.1113/jphysiol.2008.152090
Yang, J., Zhao, Y., and Shao, F. (2015). Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 32, 78–83. doi:10.1016/j.coi.2015.01.007
Yang, W., and Hu, P. (2018). Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Transl. 13, 25–32. doi:10.1016/j.jot.2018.01.002
Yin, H., Price, F., and Rudnicki, M. A. (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67. doi:10.1152/physrev.00043.2011
Zhang, L., Ran, L., Garcia, G. E., Wang, X. H., Han, S., Du, J., et al. (2009). Chemokine CXCL16 regulates neutrophil and macrophage infiltration into injured muscle, promoting muscle regeneration. Am. J. Pathol. 175, 2518–2527. doi:10.2353/ajpath.2009.090275
Keywords: satellite cells, chemokines, scRNA-seq, interferon, inflammation, muscle repair
Citation: Andre AB, Rees KP, O’Connor S, Severson GW, Newbern JM, Wilson-Rawls J, Plaisier CL and Rawls A (2023) Single cell analysis reveals satellite cell heterogeneity for proinflammatory chemokine expression. Front. Cell Dev. Biol. 11:1084068. doi: 10.3389/fcell.2023.1084068
Received: 29 October 2022; Accepted: 16 March 2023;
Published: 27 March 2023.
Edited by:
Monica Dentice, University of Naples Federico II, ItalyReviewed by:
Guglielmo Sorci, University of Perugia, ItalyCopyright © 2023 Andre, Rees, O’Connor, Severson, Newbern, Wilson-Rawls, Plaisier and Rawls. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan Rawls, YWxhbi5yYXdsc0Bhc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.