- 1Department of Biology, Drexel University, Philadelphia, PA, United States
- 2Department of Neurobiology and Anatomy, Drexel University, Philadelphia, PA, United States
- 3School of Biomedical Engineering, Science and Health Systems, Drexel University, Philadelphia, PA, United States
The past 15–20 years has seen a remarkable shift in our understanding of astrocyte contributions to central nervous system (CNS) function. Astrocytes have emerged from the shadows of neuroscience and are now recognized as key elements in a broad array of CNS functions. Astrocytes comprise a substantial fraction of cells in the human CNS. Nevertheless, fundamental questions surrounding their basic biology remain poorly understood. While recent studies have revealed a diversity of essential roles in CNS function, from synapse formation and function to blood brain barrier maintenance, fundamental mechanisms of astrocyte development, including their expansion, migration, and maturation, remain to be elucidated. The coincident development of astrocytes and synapses highlights the need to better understand astrocyte development and will facilitate novel strategies for addressing neurodevelopmental and neurological dysfunction. In this review, we provide an overview of the current understanding of astrocyte development, focusing primarily on mammalian astrocytes and highlight outstanding questions that remain to be addressed. We also include an overview of Drosophila glial development, emphasizing astrocyte-like glia given their close anatomical and functional association with synapses. Drosophila offer an array of sophisticated molecular genetic tools and they remain a powerful model for elucidating fundamental cellular and molecular mechanisms governing astrocyte development. Understanding the parallels and distinctions between astrocyte development in Drosophila and vertebrates will enable investigators to leverage the strengths of each model system to gain new insights into astrocyte function.
Introduction
Astrocytes comprise a diverse population of cells responsible for a broad array of functions in the nervous system. Nevertheless, our understanding of fundamental principles of astrocyte biology, including their development, remains far behind that of other glial cell populations in the nervous system, including oligodendrocytes and Schwann cells (Jessen and Mirsky, 2005). Astrocyte progenitors are specified during embryonic development. However, their production, migration, and maturation occur predominantly during the first three to four weeks of postnatal development, coincident with synapse formation (Sauvageot and Stiles, 2002; Farhy-Tselnicker and Allen, 2018). Astrocytes are required for synapse formation and a growing number of studies further demonstrate the role of astrocytes in modulating synaptic function and behavior across several invertebrate and vertebrate species (for review, see Freeman, 2015; Nagai et al., 2021). Understanding the mechanisms governing the production, migration, and maturation of astrocytes will not only facilitate novel insight into the establishment of neural circuits underlying behavior but will further enable the development of novel therapeutic strategies for treating neurological dysfunction arising from neurodevelopmental disorders or injury.
Much of our understanding about astrocyte development is derived from classic neuroanatomical studies of the mammalian neocortex. At birth, radial glia begin retracting their processes and transform into protoplasmic astrocytes (Figure 1). Immature astrocytes then undergo morphological and functional maturation over the first few weeks of postnatal development. Mature astrocytes exhibit diverse morphologies, as well as differential expression of ion channels (Kelley et al., 2018), transporters (Lehre et al., 1995), transcription factors (Garcia et al., 2010), signaling molecules and cell surface markers (John Lin et al., 2017; Blanco-Suarez et al., 2018; Batiuk et al., 2020; Bayraktar et al., 2020), among other genes (Westergard and Rothstein, 2020), raising the question of functional and molecular heterogeneity within this large population of cells in the CNS. Indeed, a growing number of RNA sequencing studies from many labs have identified multiple populations of mature astrocytes and their progenitors with defined transcriptional signatures throughout the rodent CNS (Chai et al., 2017; Boisvert et al., 2018; Loo et al., 2019; Cheng et al., 2022; Endo et al., 2022), among many others). How such diversity might arise remains poorly understood. While local, neuronally derived cues have been shown to regulate expression of certain genes during postnatal development (Farmer et al., 2016; Hill et al., 2019; Bayraktar et al., 2020), there is evidence that lineage may also confer specific transcriptional signatures (Ohayon et al., 2021). Whether and how this sets the stage for interpreting local signals is not known.
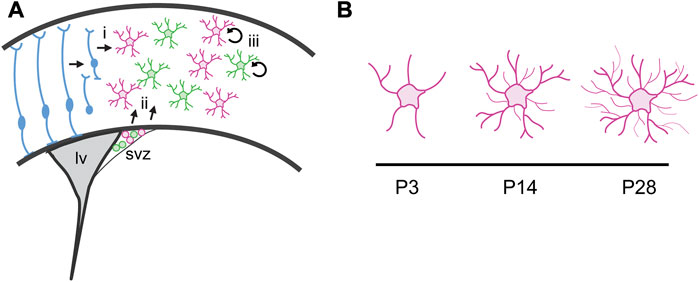
FIGURE 1. Astrocyte production and maturation during postnatal development. (A) Cortical astrocytes are derived from two progenitor cell sources. (i) Radial glia transform into cortical astrocytes and also serve as scaffolding for migration of immature astrocyte progenitor cells. (ii) A second pool of astrocyte progenitor cells reside in the dorsolateral corner of the subventricular zone (svz) at birth. The pool includes two molecularly distinct populations defined by Shh signaling. These cells migrate into the cortex during the first week of postnatal development and continue to proliferate locally, expanding the astrocyte population. (iii) Local proliferation of mature, differentiated astrocytes in the cortex expands the astrocyte population. lv, lateral ventricle. (B) Astrocytes undergo dramatic morphogenesis during the first few weeks after birth, elaborating a complex network of fine processes that interact with synapses.
Several fundamental principles of astrocyte development remain to be explored and defined. Among these are the identity of intermediate astrocyte progenitor cells that rapidly expand the astrocyte population, similar to transient amplifying cells or oligodendrocyte precursor cells that expand the neuronal and oligodendrocyte populations, respectively. What is the molecular identity of these cells and what are the underlying mechanisms regulating their cellular behaviors? What are the cues that regulate astrocyte migration to their final positions and are these cues intrinsic or environmentally derived? How do intrinsic and extrinsic cues coordinate to confer astrocyte identity and how fixed or plastic is this identity? In this review, we provide a broad overview of key molecular and cellular mechanisms that have been identified underlying various developmental events during astrocytogenesis. Because much of the focus on astrocyte development has historically centered on molecular and genetic programs regulating acquisition of astrocyte cell fate, our understanding of these mechanisms is much more advanced than that of later stages of development. For a comprehensive review of molecular regulation of astrocyte specification in the vertebrate CNS, we refer the reader to several excellent reviews, including (Rowitch and Kriegstein, 2010; Kanski et al., 2014; Takouda et al., 2017). Here, we aim to highlight later developmental events, such as migration, morphogenesis, and diversity where many opportunities remain for closing existing fundamental gaps in knowledge.
While much of our understanding of astrocyte development is derived primarily from rodent studies, there is a growing recognition and appreciation for glial cells in the Drosophila CNS whose functional properties suggest their analogous relationship to vertebrate astrocytes. Although the precise evolutionary relationship of Drosophila glia to mammalian astrocytes remains to be clearly defined (Freeman and Rowitch, 2013; Losada-Perez, 2018), similarities across functional, morphological, and molecular axes suggest analogous, if not homologous, relationships between these cells (Freeman and Rowitch, 2013; Yang and Jackson, 2019). Most notably, astrocyte-like glia (ALG) and ensheathing glia (EG) share several key phenotypical and functional properties with mammalian astrocytes, including complex morphologies, regulation of synapse formation and function, and phagocytosis of debris following injury (Awasaki et al., 2008; Freeman, 2015). Given the striking similarities between these cells in the Drosophila CNS and mammalian astrocytes, a greater understanding of fundamental principles underlying their development and maturation may yield important insights into mammalian astrocyte development. Here, we provide an overview of key principles of Drosophila glial development, focusing primarily on ALG. Although early events, such as the stereotyped production and migration of cells during embryonic and larval development differ substantially from vertebrate development, later events, such as morphogenesis and synaptic contact, have direct parallels with astrocyte development in rodents. We focus our discussion on these later events, and aim to highlight open questions and exciting areas for future investigation. For a more comprehensive discussion of Drosophila glia and their development, we refer the reader to several excellent reviews (Altenhein, 2015; Freeman, 2015).
Astrocyte specification: Embryonic beginnings
Much of the history of the study of astrocyte development has focused largely on the specification of astrocyte progenitors from multipotent neural precursor cells in the developing embryo. In vertebrates, radial glia undergo a gliogenic switch and shift from producing neurons to glia. This gliogenic switch is orchestrated by a combination of both intrinsic and extrinsic molecular cues. Neural precursor cells isolated from the cortex of embryonic day 10 (E10) mice and cultured in clonal conditions initially produce neurons. Glia are not observed in the cultures until 10 days later (Qian et al., 2000), demonstrating that individual cells in the embryonic CNS possess intrinsic molecular programs that drive developmentally-dependent competence to generate neurons first, then glia. However, extrinsic molecular cues also play an instructive role in cell fate acquisition. Cortical progenitor cells isolated from E15 mouse brains produce astrocytes when they are grown on cortical slices from postnatal day 15 (P15) brains. However, when the cells are grown on E18 slices, the cells instead produce neurons (Morrow et al., 2001). Thus, the gliogenic competence of neural progenitor cells is a coordinated effort between intrinsic and extrinsic programs that include mechanisms to repress neuron production while promoting glial production.
A major challenge in understanding astrocyte development is the paucity of molecular identifiers of astrocytes and their progenitors in both vertebrates and invertebrates. Historically, vertebrate studies have relied heavily on expression of glial acidic fibrillary protein (GFAP) to identify the acquisition of an astrocytic fate by neural precursors. Although GFAP expression is low in most astrocytes in the healthy adult brain, fate mapping studies show that nearly all astrocytes in the mouse cortex and throughout the forebrain are derived from a GFAP-expressing progenitor cell (Tatsumi et al., 2018; Hill et al., 2019), supporting its utility as a tool for identifying astrocyte progenitors. Several key molecular signaling programs have been identified that regulate the switch from neurogenesis to gliogenesis. Among these is Jak/STAT signaling, which is stimulated by activation of ciliary neurotrophic factor-leukemia inhibitory factor (CNTF/LIF) receptors in progenitor cells (Bonni et al., 1997). Activation of Jak/STAT signaling leads to repression of proneuronal genes neurogenin-1 and neurogenin-2 and promotes GFAP expression (Bonni et al., 1997; Sun et al., 2001). As neural development proceeds, newborn neurons express the cytokine, cardiotrophin-1 (CT-1), which signals back to neural precursors to generate astrocytes (Barnabé-Heider et al., 2005). In addition, BMP signaling promotes the acquisition of astrocyte characteristics, including GFAP expression (Gross et al., 1996; Bonaguidi et al., 2005). This is accomplished by the generation of a molecular complex composed of SMAD/p300 and STAT which promotes the transcription of astrocytic genes including GFAP and S100β (Nakashima et al., 1999). In vivo conditional knockout of STAT3 using nestin-cre in the spinal cord and the cortex show a decrease of GFAP expression and an increase in proliferation of stem cells (Hong and Song, 2014; Su et al., 2020). Another key signaling pathway in astrocyte specification is Notch signaling. In vertebrates, activation of Notch upregulates GFAP expression (Ge et al., 2012). Notch activation promotes demethylation of the GFAP promoter (Namihira et al., 2009).
Much of the molecular signaling regulating the gliogenic switch was identified through studies in vitro. Studies in vivo have identified key transcription factors that coordinate the repression of neurogenic genes while promoting the acquisition of astrocyte cell fate. The first such transcription factor identified was NFIA (Deneen et al., 2006). Its expression in vivo is initially detected at embryonic day 11.5 in the mouse spinal cord, and coincides with expression of the glutamate transporter, GLAST, expressed in astrocyte progenitors (Shibata et al., 1997). NFIA interacts with Notch signaling to inhibit neurogenesis and is necessary and sufficient for gliogenesis (Deneen et al., 2006). Expression of another transcription factor, Sox9, precedes NFIA induction, and its deletion in the CNS severely impairs the production of astrocytes in the mouse spinal cord (Stolt et al., 2003). Later studies identified Sox9 as a regulator of NFIA expression in the mouse and chick spinal cord (Kang et al., 2012). Despite the essential requirement of NFIA and Sox9 expression for astrocyte production, neither are exclusive for astrocyte progenitors, suggesting that additional regulatory mechanisms are necessary to specify astrocytic identity. In the mouse neocortex, the transcriptional repressor, Zbtb20, cooperates with Sox9 and NFIA to drive gliogenesis. Overexpression of Zbtb20 by electroporation increases astrocyte production at the expense of neuron production in the mouse cortex, suggesting that it plays a role in the suppression of neuronal genes (Nagao et al., 2016). The critical role of NFIA in astrogliogenesis was further confirmed in a recent study deploying transcriptomic and epigenomic approaches to identify specific transcriptional programs and epigenetic states during astrogliogenesis (Tiwari et al., 2018). NFIA, together with ATF3 and RUNX2, acts at regulatory elements of genes associated with astrogliogenesis. Electroporation of each of these transcription factors alone, or in combination, in the mouse cortex, was sufficient to enhance astrocyte production at the expense of neurons.
How does the brain achieve the proper number and distribution of astrocytes?
Much of the emphasis on astrocyte development has focused largely on glial specification events during embryonic development. However, astrocyte production, migration, and maturation occur largely during postembryonic development, peaking early in the perinatal period (Sauvageot and Stiles, 2002). Whereas the mechanisms governing the gliogenic switch during embryonic development are well characterized, the cellular and molecular mechanisms governing developmental events occurring during postembryonic stages, such as expansion of the progenitor population, migration of immature cells to their final positions, and maturation into fully differentiated astrocytes, are considerably less well understood.
Classic neuroanatomical studies from Rakic and colleagues demonstrated the gradual disappearance of radial glia together with the concomitant appearance of differentiated astrocytes in the primate neocortex (Schmechel and Rakic, 1979; Levitt and Rakic, 1980; Cameron and Rakic, 2004), suggesting that cortical astrocytes are derived from the transformation of radial glia (Figure 1). In the rodent, this process begins around birth and is largely complete by about P7 (Kriegstein and Alvarez-Buylla, 2009).
In addition to radial glia, the early rodent postnatal cortex also harbors a pool of glial progenitor cells residing in the dorsolateral corner of the subventricular zone (dlSVZ; Figure 1). These glial progenitors contribute both astrocytes and oligodendrocytes to the overlying cortex and white matter (Levison and Goldman, 1993; Zerlin et al., 1995) and can be identified by expression of BLBP and Sox9. These dlSVZ progenitors are thought to be derived from radial glia (Kriegstein and Alvarez-Buylla, 2009). Whether astrocytes derived directly from radial glial transformation or from dlSVZ progenitors represent molecularly or functionally distinct populations is not known. Addressing these questions will require developing approaches to molecularly dissect these astrocyte progenitor populations. Finally, a third pool of progenitor cells responsible for expanding the cortical astrocyte population has been reported (Ge et al., 2012). These “pioneer” cells are derived from radial glia and dlSVZ progenitors that colonize the cortex and acquire mature, differentiated characteristics before proliferating locally (Figure 1).
Astrocytes comprise a substantial number of cells in the mature brain. How expansion of astrocyte progenitors is accomplished to achieve the appropriate number of mature astrocytes is not well understood. Expansion of neuronal and oligodendrocyte populations is achieved through intermediate progenitor populations which each exhibit defined molecular signatures at distinct developmental stages (Bergles and Richardson, 2015; Hevner, 2019). Oligodendrocyte precursors and their progeny, for example, express Sox10 and Olig2, premyelinating oligodendrocyte progenitor cells express PDGFRa and NG2, and mature myelinating oligodendrocytes express myelin basic protein (MBP), myelin associated glycoprotein (MAG) and carbonic anhydrase 1 (CA1). In contrast, efforts to identify a defined pool of intermediate progenitor cells for vertebrate astrocytes are hindered by the inability to distinguish these cells molecularly from their neural precursor or their daughter cells. Temporal profiling of developing astrocytes in the spinal cord across several mid-embryonic to early postnatal time points identified the stage-specific gene expression profiles (Molofsky et al., 2013; Chaboub et al., 2016). It would be interesting to examine whether these genes identify specific astrocyte progenitor populations or play a role in regulating progenitor cell behaviors. As discussed above, local proliferation of “pioneer” astrocytes in the cortex has been suggested as one source of intermediate progenitors. However, their mature gene expression and morphological characteristics preclude their prospective identification and fail to distinguish them from their post-mitotic daughter cells. In the mouse spinal cord, a population of Aldh1L1 positive intermediate progenitor cells undergo transit amplification and are differentiated from radial glia progenitor cells (Tien et al., 2012). However, mature astrocytes also express Aldh1L1, leaving open the question of how to define the intermediate astrocyte progenitor pool and distinguish them from earlier gliogenic precursors and later mature astrocytes.
The molecular and cellular mechanisms that govern the precise distribution and localization of astrocytes are poorly defined. However, emerging evidence suggests that unlike neurons and oligodendrocytes which can migrate long distances to their final position, astrocytes appear to have considerably more limited migration potential. Sparse labeling of embryonic cortical precursors demonstrates that cortical astrocytes are distributed within a radially oriented column, mimicking the columnar organization of cortical neurons (Magavi et al., 2012). Indeed, fate mapping from regionally defined progenitor zones in the forebrain and spinal cord shows that astrocyte migration is limited to the radial territory directly overlying their progenitor domain (Hochstim et al., 2008; Tsai et al., 2012). While radial migration of astrocyte progenitors has been well documented, advanced 3D serial microscopy imaging approaches of large volumes combined with multiclonal lineage tracing demonstrated that individual clones also undergo wide dispersion in the cortex that is not strictly radial (Clavreul et al., 2019). This distribution is thought to arise from stochastic “seeding” of the cortex by astrocyte progenitor cells, followed by local proliferation and expansion. Because earlier studies analyzed individual tissue sections, it is possible that such large scale dispersion could not be easily detected. Indeed, imaging individual sections labeled by the same multiclonal lineage approach also produces clones with a columnar distribution (Loulier et al., 2014). Taken together, these studies demonstrate that cortical astrocytes use different strategies to migrate to their final positions. Because the methods used in these studies to mark progenitor cells relied on expression and recombination in non-specific astrocyte progenitor cells, whether these different migration strategies can be adopted by cells within a given lineage or whether this reflects an intrinsic heterogeneity between specific astroctyte progenitor cells is not known. Such heterogeneity may arise from underlying genetic programs or from temporal cues orchestrated by molecular signaling programs that remain to be identified.
While many clones contained cells with shared morphologies, some clones were comprised of both protoplasmic and pial astrocytes, cells with distinct morphological characteristics. These mixed clones suggest that local cues may drive the adoption of mature phenotypes. Consistent with this is the observation that cortical astrocytes exhibit distinct molecular signatures based on their position in upper or deep layers (Bayraktar et al., 2020). However, in mutants with cortical neuron layer defects, these defined astrocyte layer molecular signatures are disrupted, suggesting that neurons provide molecular cues that instruct molecular or positional characteristics. As discussed below, Sonic hedgehog (Shh) signaling has been identified in the cerebellum as one such cue. Elucidating additional cues and their underlying mechanisms will be important for advancing our understanding of astrocyte development.
How do astrocytes mature?
The production and migration of immature astrocytes occurs predominantly during the first week of postnatal development, followed by the acquisition of mature functional and morphological characteristics that develops over the second—fourth postnatal weeks (Stogsdill et al., 2017). By comparing the transcriptional signatures of immature and mature human and mouse astrocytes, Li et al., observed that astrocyte-astrocyte contact inhibits proliferation and promotes the expression of genes associated with mature cells (Li et al., 2019). Notably, astrocyte conditioned media failed to produce the same degree of transcriptional maturation, suggesting that the signals promoting astrocyte maturation are not secreted but instead require direct contact between cells. Further experiments showed that inhibiting EGFR signaling promotes astrocyte maturation. Consistent with this, heparin-binding epidermal growth factor-like growth factor (HBEGF), a ligand for EGFR, inhibits astrocyte maturation whereas BMP signaling downregulates EGFR and promotes astrocyte maturation (Scholze et al., 2014). RNA-seq identified genes associated with cilium assembly as highly enriched and upregulated following HBEGF removal or EGFR inhibition. Sonic hedgehog signaling is transduced in primary cilia (Goetz et al., 2009). Thus, these observations are consistent with the identification of Shh activity in astrocyte progenitor cells (Gingrich et al., 2022) and suggests a role for Shh signaling in astrocyte maturation.
Mature astrocytes exhibit complex morphologies that mediate their intimate relationships with synapses. Understanding how these morphologies develop has been limited by a paucity of available tools to study morphogenesis. GFAP expression is limited to larger processes, exposing only the primary cytoarchitecture while the small branchlets associated with synapses remain concealed. Further, the full complexity of their morphological characteristics is lost when dissociated, and astrocytes in culture instead take on flat morphologies that lack the fine branchlets and leaflets associated with synapses and blood vessels. This limits the utility of in vitro approaches for investigating the mechanisms driving their morphogenesis. However, in vivo studies are beginning to illuminate such mechanisms. The development of technologies including intracellular dye injections or fluorescent reporters targeting the membrane or filling the cell, which can be delivered by transgenes or viral vectors, now enable full visualization of astrocyte morphologies in vivo. Application of these approaches reveals that astrocytes begin to exhibit numerous processes and complex morphologies by the end of the first week of postnatal development, with further elaboration of fine processes that produce their spongiform morphology between P14 to P28 (Figure 1; Bushong et al., 2004; Morel et al., 2014; Holt et al., 2019). The temporal coincidence between astrocyte maturation and the major period of synaptogenesis in the rodent brain, during the second—fourth weeks of postnatal development, together with the observations that astrocytes respond dynamically to neuronal activity (Cornell-Bell et al., 1990a; Cornell-Bell et al., 1990b; Hirrlinger et al., 2004; Genoud et al., 2006), have long led to speculation that synaptogenesis instructively cooperates with astrocyte morphogenesis. Consistent with this, loss of glutamatergic signaling through mGluR5 reduces the domain size of individual astrocytes, leading to a reduction in the number of synapses ensheathed by an astrocyte process (Morel et al., 2014). However, synaptic activity alone is not sufficient for astrocyte morphogenesis. Complete blockade of synaptic activity with TTX treatment does not impair the extension of astrocyte processes (Stogsdill et al., 2017), pointing to contact-mediated mechanisms underlying astrocyte morphogenesis. Astrocytes interact with neurons through neuroligins which bind to neurexins found on presynaptic terminals. Perturbation of this interaction in vitro and in vivo reduces astrocyte complexity and volume of processes infiltrating the neuropil (Stogsdill et al., 2017).
Growth factor signaling also plays a role in astrocyte morphogenesis. The growth factor, brain derived neurotropic factor (BDNF) regulates astrocyte morphogenesis in vitro and in vivo (Holt et al., 2019). A truncated isoform of the BDNF receptor, TrkB.T1 lacking the tyrosine kinase domain but containing an additional exon, is found predominantly in astrocytes. Loss of TrkB.T1 expression decreases astrocyte volume and morphological complexity in vitro and in vivo.
In zebrafish, astrocytes fail to elaborate complex morphologies in the absence of fgfr3 and fgfr4 (Chen et al., 2020). As discussed below, FGFR signaling is required for morphogenesis of Drosophila ALG, suggesting that the requirement for FGF signaling in astrocyte morphogenesis is conserved in vertebrates. This is supported by in vitro studies from rodent cells where addition of FGFs or overexpression of fgfr3 promotes enhanced branch number and complexity (Perraud et al., 1988; Kang et al., 2014). Indeed, several members of the fgf family and their receptors are expressed in astrocytes (Miyake et al., 1996; Oh et al., 2003). Identifying the precise members of the fgf family and their mechanism of action on astrocyte morphogenesis would advance our understanding of their development and may shed new light on their interactions with synapses.
A key feature of astrocyte morphology is the complete tiling of the neuropil in a non-overlapping manner, ensuring synaptic coverage. While the precise function of this cellular behavior is not well understood, it is observed in both invertebrate and vertebrate astrocytes (Bushong et al., 2004; Halassa et al., 2007; Stork et al., 2014; Chen et al., 2020; Pogodalla et al., 2022) and severe injury or disease of the nervous system perturbs the distinct territories established by individual astrocytes (Sofroniew, 2014). Astrocytes express a large complement of cell adhesion molecules (Zhang et al., 2014; Hillen et al., 2018) and growing evidence shows that these molecules mediate homophilic and heterophilic interactions between other astrocytes and neurons that regulate astrocyte morphogenesis. Co-culturing astrocytes with neurons enhances morphological complexity (Holt et al., 2019). This effect requires physical contact with neurons as the addition of neuron-conditioned media fails to induce this complexity (Stogsdill et al., 2017). The hepatocyte cell adhesion molecule, hepaCAM, was recently identified as a molecular mediator of astrocyte-astrocyte interactions that regulates their complexity and territory volume (Baldwin et al., 2021). Selective perturbation of hepaCAM expression from a sparse population of cortical astrocytes increases territory overlap between neighboring cells with differential hepaCAM expression.
Drosophila glia
Drosophila offer powerful genetic and molecular tools, together with several connectome datasets (Zheng et al., 2018; Scheffer et al., 2020; Phelps et al., 2021) and well-characterized behaviors that can be exploited for advancing our understanding of astrocyte development and its role in neural circuits and behavior. Drosophila possess glial cells that, like mammalian astrocytes, play key roles in synapse formation and function, blood brain barrier function, phagocytosis of debris, and response to neural injury [Table 1; (Doherty et al., 2009; Limmer et al., 2014; Freeman, 2015)]. Interestingly, whereas these diverse functional roles are assigned to astrocytes in the mammalian CNS, glial cells in Drosophila performing these roles are assigned to several distinct glial cell classes. Surface glia, comprised of perineural and subperineural glia, cover the entire surface of the brain and perform blood-brain barrier functions (Figure 2; Awasaki et al., 2008; Stork et al., 2014). Cortex glia are found in the cell cortex of the CNS, ensheath neuronal soma, elicit transient calcium signaling, and provide phagocytic support during CNS development (Figure 2; Melom and Littleton, 2013; Coutinho-Budd et al., 2017; Nakano et al., 2019). Cortex glia form a dense meshwork, with each cell surrounding up to 100 individual neuronal soma (Kremer et al., 2017). Interestingly, mammalian astrocytes also surround neuronal soma, though at a considerably lower ratio of an average of 4 neurons per astrocyte (Halassa et al., 2007). Despite these intriguing morphological and functional properties, cortex glia are considerably understudied relative to other classes of Drosophila glia, and will not be discussed further here. Surrounding the neuropil are ensheathing glia (EG) and astrocyte-like glia (ALG) which together are known as neuropil glia (Figure 2; Pereanu et al., 2005). EG are found on the border of the neuropil where they define subcompartments in the CNS. Their processes interact with axon tracts and neuronal cell bodies and, EG participate in the phagocytosis of neuronal debris in the healthy and injured brain (Doherty et al., 2009). ALG are also found surrounding the neuropil. However, whereas, EG processes are largely excluded from the neuropil, ALG processes extend deep into the neuropil where they interact intimately with synapses and play key roles in their formation and modulating their function (Awasaki et al., 2008; Stork et al., 2014). Drosophila ALG maintain the balance of excitatory and inhibitory neurotransmitters and regulate synapse formation (Liu et al., 2014). ALG exhibit complex morphologies that tile the neuropil with little overlap in their processes, much like vertebrate astrocytes (Stork et al., 2014). Notably, some of the molecular signatures of ALG in larva and adult CNS are shared with vertebrate astrocytes, including the glutamate transporter, Eaat1, the GABA transporter, gat, and glutamine synthetase (Huang et al., 2015; Ng et al., 2016). As in the mammalian CNS, the development of ALG and synaptogenesis occur in parallel, highlighting their essential roles in synaptogenesis and synapse function (Muthukumar et al., 2014).
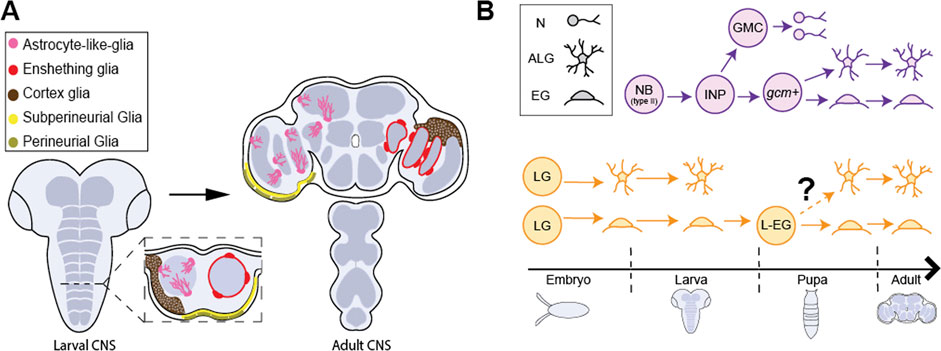
FIGURE 2. Glial cell types in Drosophila CNS. (A) The larval CNS (left) and adult CNS (right) with the 5 primary classes of glial cells, astrocyte-like glia (pink), ensheathing glia (red), cortex glia (brown), subperineural glia (yellow) and perineural glia (gold). (B) The adult CNS harbors neuropil glia (ALG and EG) arising from different progenitor populations at different stages in development. (Top, purple) Type II neuroblasts (Type II NB) generate bipotent intermediate neural progenitors (INPs) postembryonically. INPs generate ganglion mother cells (GMC) and gcm+ glial precursors which are responsible for generating a large fraction of ALG and EG after expansion. (Bottom, yellow) Embryonic glioblasts (not shown) generate longitudinal glia (LG) that are committed to generating ALG or EG cells in larva. It has been proposed that larval ALG are ultimately eliminated and surviving larval, EG (L-EG) dedifferentiate in pupa and become bipotent progenitors that can generate both ALG and EG for the adult CNS.
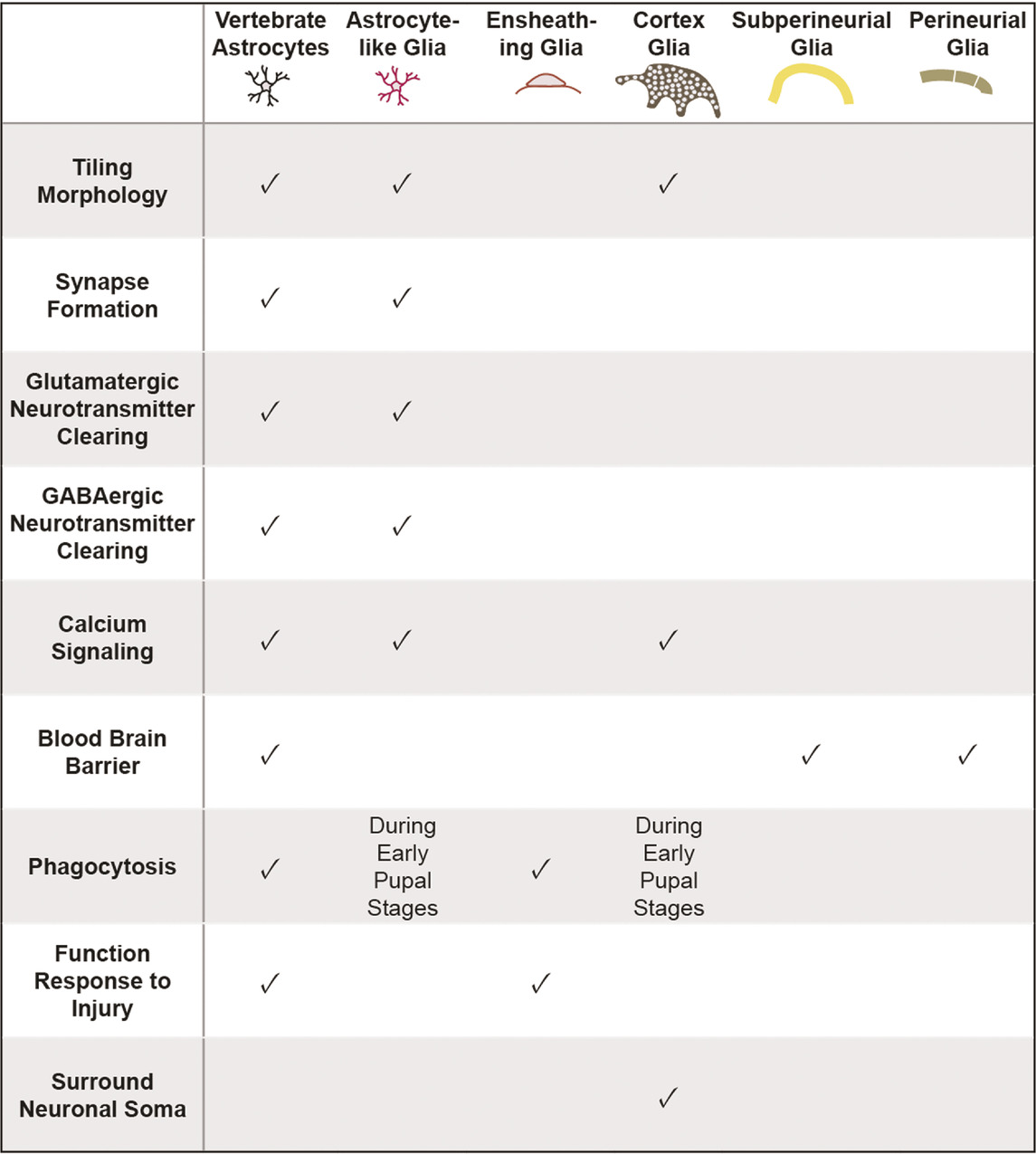
TABLE 1. Functional characteristics shared between vertebrate astrocytes and various classes of Drosophila glia. Neuropil glia, encompassing astrocyte-like and ensheathing glia, share many morphological and functional properties with vertebrate astrocytes.
As in the rodent CNS, glial progenitors in Drosophila are also specified during embryonic development. Neuroblasts (NB) delaminate from the epithelium and generate neurons and glia of the larval CNS through neuron or glia-restricted neuroblasts or glioblasts, respectively, or bipotent neuroglioblasts, generating both neurons and glia (Hartenstein, 2011). A key determinant of glial cell fate is glial cells missing (gcm), a major effector for Drosophila glial specification. Loss of function of gcm perturbs differentiation of embryonically and postembryonically derived glia, while ectopic expression of gcm produces excess glia at the expense of neurons (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). As in vertebrates, Drosophila glial cell fate is established through suppression of pro-neuronal genes and upregulation of pro-glial genes. In Drosophila, this is mediated through transcription factors repo, pointed and tramtrack that act downstream of gcm (Halter et al., 1995; Giesen et al., 1997; Badenhorst, 2001; Yuasa et al., 2003). Interestingly, the vertebrate homologs, gcm1 and gcm2, are not required for glial specification in the mouse nervous system. Whereas these genes are easily detected in non-neural tissues of the developing mouse embryo, their expression in neural tissues is low (Kim et al., 1998). Nevertheless, expression of vertebrate gcm1 or gcm2 in Drosophila embryos is sufficient to promote glial fate and rescues Drosophila gcm mutants (Kim et al., 1998), highlighting the key role of this gene in Drosophila gliogenesis.
Notch is a conserved protein and similar to rodents, plays key roles in Drosophila glial specification and production. Notch signaling regulates the expression of gcm (Umesono et al., 2002). Mutants lacking Notch activity show a loss of glial cells (Ren et al., 2018) whereas ectopic activation of Notch activity produces an overabundance of glial cells (Udolph et al., 2001).
During metamorphosis, the Drosophila larval CNS is dramatically transformed through the combined actions of cell elimination, rewiring of connections and the production of new cells to generate the adult nervous system. Whereas the cellular and molecular mechanisms mediating neuron production are well characterized (Doe, 2017; Miyares and Lee, 2019), how glia of the adult CNS arise is not as well established. During late embryonic stages, NB that do not undergo apoptosis become quiescent but then resume proliferation in the larval stage, creating a second wave of neurogenesis responsible for 90% of neurons in the adult CNS (Truman and Bate, 1988; Homem and Knoblich, 2012; Lacin and Truman, 2016; Walsh and Doe, 2017; Harding and White, 2018). Similarly, glia progress through a second, post-embryonic stage of production and expansion to generate glia for the adult CNS (Hartenstein, 2011). This begins during the larval stage and is achieved through the proliferation of glial precursors, postembryonic NB, and differentiated glia in the larva (Pereanu et al., 2005; Omoto et al., 2015; Enriquez et al., 2018; Ren et al., 2018).
Drosophila astrocyte-like glia (ALG) generation, migration, and morphogenesis
ALG generation—Who am I?
While the key roles for gcm and Notch in glial cell fate specification are well established, the molecular determinants underlying specification of ALG and other classes of Drosophila glia remain poorly understood. Unlike vertebrate CNS development, the cellular lineage and final position of individual NBs is highly stereotyped and the NB lineages responsible for generating various classes of glial cells have been mapped (Beckervordersandforth et al., 2008; Altenhein et al., 2016). Neuropil glia arise from the longitudinal glioblast that generates longitudinal glia which differentiate into six ALG and three EG for each larval ventral nerve cord hemisegment (Jacobs et al., 1989; Stacey et al., 2007; Beckervordersandforth et al., 2008; Peco et al., 2016). Notch, in addition to its role in glial specification, drives embryonic ALG differentiation by regulating the expression of the homeodomain transcription factor prospero (pros) (Peco et al., 2016). Overexpression of a Notch repressor or knock down of pros leads to the loss of ALG markers including Eaat1, Gat, and Ebony and increase in the EG marker Eaat2, and ALG fail to infiltrate the neuropil.
While longitudinal glia have been identified as the embryonic precursors of larval neuropil glia, the precise source of ALG and EG found in the adult brain remains under debate. At the onset of metamorphosis, larval ALG begin to express draper, a gene necessary for phagocytosis, and adopt a spherical morphology similar to that of EG (Kato et al., 2020). It is currently hypothesized that larval ALG undergo apoptosis after performing phagocytosis of the degenerating larval nervous system (Tasdemir-Yilmaz and Freeman, 2014; Omoto et al., 2015; Pinto-Teixeira et al., 2016). This is supported by lineage tracing studies demonstrating that embryonically generated ALG soma are observed during early stages of pupal development, but are rare by 72 h after pupal formation (APF) and completely absent at 96 h APF (Omoto et al., 2015), suggesting these cells are eliminated before eclosion. TUNEL staining of GAT-positive cells demonstrates that ALG undergo programmed cell death (Omoto et al., 2015). Using an intersectional lineage tracing strategy, postembryonic Type II NB were identified as a source of adult ALG (Figure 2; Omoto et al., 2015; Enriquez et al., 2018; Ren et al., 2018). Type II NB produce intermediate neural progenitors (INPs) and rapidly expand the population of neurons and glia. Type II NBs can be multipotent, generating neurons, ALG, and EG (Ren et al., 2018). In addition to Type II NBs as a source of adult ALG, recent evidence demonstrates that some larval EG dedifferentiate to then generate adult ALG and EG during metamorphosis (Kato et al., 2020). However, to date, the exact contribution of embryonic and larval NBs to the full complement of adult ALG has not been definitively established. It is possible that different regions use differing strategies for generating the adult ALG population. In support of this, lineage tracing of Type II-derived ALG are found predominantly in the core of the brain whereas peripheral brain regions remain largely unlabeled (Ren et al., 2018). In addition, molecular signatures for lineage-specific identification of ALG precursors have not been well established. Because larval and adult flies exhibit distinct motor behaviors and underlying circuits, understanding how ALG in each of these distinct stages are generated and incorporate into neural circuits will be an important step towards understanding the development and function of neural circuits driving behavior.
Type II neuroblasts produce postembryonic INPs capable of generating neurons or glia (Urbach and Technau, 2003; Viktorin et al., 2011). Like vertebrate neural progenitor cells, multipotent Type II NBs generate glia following neuron production (Ren et al., 2018). Notch regulates gliogenesis from INPs which generate glial precursors that then undergo an expansion period during pupal stages before differentiating into adult ALG (Viktorin et al., 2011; Omoto et al., 2015). The mechanisms for INP expansion and neuropil glia differentiation are not well established, however, recent studies have again pinpointed Notch signaling as a major regulator of expansion (Ren et al., 2018).
ALG migration—How did I get here?
How ALG localize to their positions surrounding the neuropil is also incompletely understood. In contrast to vertebrate astrocytes, the migration and position of neuropil glia precursors in Drosophila embryos is highly stereotyped. Glial precursors migrate to precise positions within the ventral nerve cord and brain preceding their differentiation into larval ALG and EG (Ito et al., 1995; Hartenstein et al., 1998; Beckervordersandforth et al., 2008; Omoto et al., 2015). Adult ALG also appear to have distinct migration patterns linked to their type II neuroblast origins, often incorporating with the neural cell types generated by the same neuroblast (Viktorin et al., 2011; Omoto et al., 2015; Ren et al., 2018). The mechanisms underlying migration patterns and the existence of location dependent molecular signatures remain relatively unexplored. The highly stereotyped manner by which ALG precursors migrate to their final positions suggest intrinsic molecular cues that instruct this process. The identification of such cues may provide insight into how vertebrate astrocytes find their way to their final positions in the CNS and how this process is coordinated with local environmental cues.
ALG morphogenesis—How do I look?
A distinguishing characteristic of both Drosophila ALG and vertebrate astrocytes is their complex, bushy-like morphology comprised of fine processes that interact intimately with synapses. These processes tile the neuropil, establishing discrete territories of synapses within reach of an individual cell (Freeman, 2015). How these elaborate morphologies are established and maintained is only beginning to be understood. Similar to mammalian astrocytes, ALG morphogenesis coincides with synaptogenesis and the refinement of developing neural circuits. ALG processes are observed infiltrating the neuropil as early as late stage embryos (Stork et al., 2014). Morphogenesis of larval ALG cells is strongly regulated by the FGF receptor Heartless (Htl) (Stork et al., 2014). Loss of Htl does not disturb ALG specification, but results in failure of ALG invasion into the neuropil. Two key FGFs, Pyramus (Pyr) and Thisbe (Ths), activate Htl and promote morphogenesis and neuropil invasion. In conjunction with FGF signaling, lapsyn, a member of the extracellular leucine-rich repeat superfamily, is involved in ALG neuropil invasion and branch morphogenesis (Richier et al., 2017). Although ALG processes are observed into late larval stages and at puparium formation, they are absent from the neuropil by 48 h APF (Tasdemir-Yilmaz and Freeman, 2014). However, by 60 h APF, ALG processes are once again observed in the neuropil, and continue to elaborate their processes until 84 h APF, a time course coincident with synapse formation (Muthukumar et al., 2014). The signals that initiate invasion of ALG processes into the neuropil in the embryo or during pupal stages are not known. In addition, whether the seemingly distinct processes of neuropil invasion and branch morphogenesis are differentially regulated remain to be explored. Whereas the highly stereotyped manner by which Drosophila ALG are generated differs markedly from vertebrate astrocyte production, the remarkable parallels between Drosophila and vertebrates in astrocyte morphogenesis and its coordination with synaptogenesis offer an exciting opportunity to leverage discoveries from Drosophila for advancing fundamental principles of astrocyte development. Indeed, the identification of FGF receptor signaling as an important regulator of astrocyte morphogenesis in both Drosophila and vertebrates suggests that additional molecular cues may be conserved (Table 2).
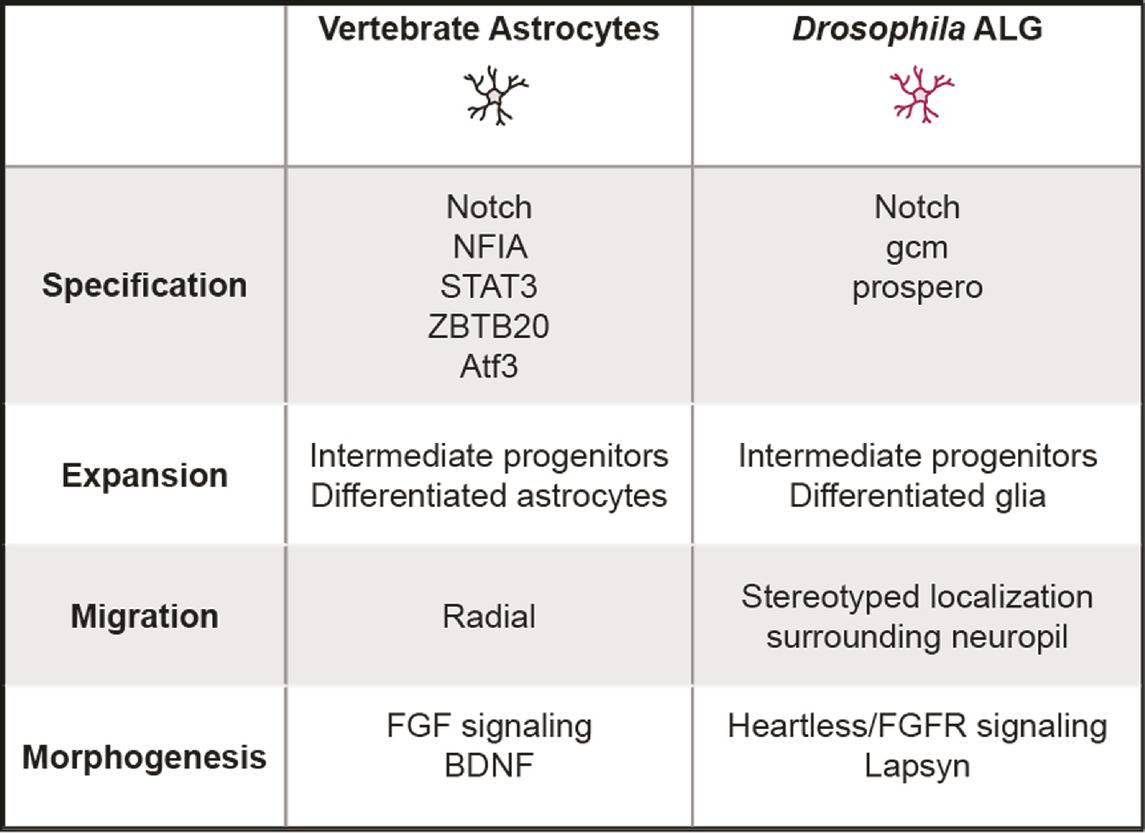
TABLE 2. Key molecular and cellular mechanisms across various stages in vertebrate and Drosophila astrocyte-like glia development.
How does astrocyte diversity arise?
Historically, astrocytes have been divided into two primary morphological classes, protoplasmic astrocytes found in gray matter and fibrous astrocytes in white matter. Yet the abundance of these cells in the CNS, together with the recognition of their diverse functional roles, has prompted great interest in their molecular and functional diversity. The emergence of RNA sequencing and bioinformatic analysis has enabled the discovery of transcriptionally distinct populations of astrocytes throughout the CNS. Such approaches have been applied most fervently in mouse tissues. However, emerging studies in Drosophila have also revealed previously unappreciated diversity of glia with defined transcriptional signatures. The nature of this heterogeneity and how it arises remain poorly understood.
In the mouse, a remarkable degree of molecular diversity has been demonstrated among astrocytes between regions, as well as within a region (Lanjakornsiripan et al., 2018; Hill et al., 2019; Batiuk et al., 2020; Bayraktar et al., 2020). In depth discussion of astrocyte heterogeneity in vertebrates is available from several excellent recent reviews and we refer the reader to these (Ben Haim and Rowitch, 2017; Khakh and Deneen, 2019; Pestana et al., 2020). An important question that arises from the discovery of these genetically defined populations is the role of intrinsic versus externally derived drivers of these molecular signatures.
Several studies demonstrate that neuronally derived cues regulate astrocyte gene expression. In the cerebellum, Sonic hedgehog (Shh) derived from Purkinje neurons regulates expression of genes in nearby Bergmann glia, such as GluA1, GluA4, GLAST and Kir4.1 (Farmer et al., 2016). Ectopic activation of Shh signaling in velate astrocytes, a morphologically and regionally distinct astrocyte population in the cerebellum that expresses low levels of these genes, increases expression of these and other genes associated with Bergmann glia. In the forebrain, mutants with aberrant patterning of cortical layers disrupts gene expression and distribution of morphologically defined astrocytes in a manner that follows the perturbed neuronal layers (Lanjakornsiripan et al., 2018; Bayraktar et al., 2020). These studies demonstrate remarkable plasticity in astrocyte phenotype and suggest that local, neuronally derived cues can drive molecular phenotypic diversity.
However, fate mapping studies provide evidence that developmental origin also plays a role in generating diversity. In the spinal cord, white matter astrocytes exhibit combinatorial expression of Reelin and Slit1 (Hochstim et al., 2008). These cells are derived from distinct progenitor cell domains in the ventricular zone defined by Pax6 and Nkx6.1. Similarly, protoplasmic astrocytes in the forebrain can be fate mapped from ventral, intermediate and dorsal progenitor domains expressing Nkx2.1, Dbx, and Emx1, respectively (Tsai et al., 2012). In both the spinal cord and forebrain, astrocytes derived from these progenitors occupy specific territories directly overlying these domains, in a manner suggesting that astrocytes undergo radial, but not tangential, migration. Notably, ablation of cells from one domain failed to recruit astrocytes from a different domain (Tsai et al., 2012), suggesting that lineage confers regional identities that are fixed. While these studies demonstrate regional specialization of astrocytes, evidence is emerging for diversity within a region.
In the spinal cord, a subpopulation of gray matter astrocytes express Olig2 (Ohayon et al., 2019). These cells are derived from a subpopulation of Olig2-expressing precursor cells in the pMN domain that are regulated by expression of the heparin sulfate modification enzyme, Sulf2. Olig2-expressing astrocytes are intermixed among nonOlig2 astrocytes in the gray matter of the spinal cord, resulting in a heterogeneous mix of cells derived from a molecularly defined lineage. RNASeq identified specific molecular signatures of the Olig2 and nonOlig2 astrocyte populations, including enrichment of inka2 and kcnip3 in Olig2 astrocytes (Ohayon et al., 2021). In the mouse forebrain, recent studies demonstrated that activity of the Sonic hedgehog (Shh) signaling pathway occurs in a subpopulation of dlSVZ progenitors (Tong et al., 2015; Gingrich et al., 2022). These cells are identified by expression of the transcription factor, Gli1, a readout of Shh signaling. Using fate mapping approaches, Gingrich et al., demonstrated that these cells migrate out of the SVZ and proliferate locally in the cortex, undergoing rapid expansion early in the first postnatal week. Whether these cells correspond to “pioneer” cells with mature characteristics is not known. Astrocytes within the Gli1 lineage comprise nearly half the total cortical astrocyte population and are found throughout all layers, suggesting that the cortex harbors astrocytes from two molecularly defined astrocyte progenitor lineages. Consistent with this, single cell RNA sequencing (scRNA-seq) identified two molecularly distinct populations of astrocyte progenitor cells in the rodent forebrain (Liu et al., 2022). And using clonal analysis, homogenous clones of protoplasmic and fibrous astrocytes were observed in the cortex and corpus callosum, respectively, suggesting distinct lineages for morphologically defined astrocyte populations (Garcia-Marques and Lopez-Mascaraque, 2013). Whether and how these molecularly and morphologically defined lineages intersect remains to be discovered.
Taken together, these initial studies suggest that both lineage and local cues work in concert to define astrocyte characteristics. It is likely that molecular signaling programs initiate a transcriptional landscape that confers competence to interpret local cues in specific ways. What is the identity of these molecular signaling programs and how do they instruct cell identity? How plastic or fixed is astrocyte identity and how do these identities correspond to functional specialization? Early studies have begun to address these questions, but further efforts are needed to elucidate the nature of astrocyte heterogeneity and how it emerges.
There is also emerging evidence for greater heterogeneity among Drosophila glia than is currently appreciated. Although Drosophila glia have been classically organized into five classes defined by their distinct morphological and anatomical characteristics, recent transcriptomic atlases generated by scRNA-seq identified 19 transcriptionally distinct clusters of glial cells in the optic lobes of pupae (Kurmangaliyev et al., 2020). Remarkably, an independent study similarly identified 19 glial clusters from the adult optic lobe (Ozel et al., 2021). The convergence of 19 glial clusters from two independent studies across two life-cycle stages suggests that glia diversity may arise as a function of lineage. Indeed, Drosophila ALG demonstrate region specific morphological differences, particularly in the optic lobes of the adult brain (Edwards et al., 2012). Studies are needed to determine whether these morphological phenotypes correspond to specific transcriptional signatures. Type II neuroblasts reliably generate defined, molecularly and morphologically distinct neuronal cell types (Bayraktar and Doe, 2013; Ren et al., 2018), however, neuroblast specific signatures in ALG have yet to be uncovered. Alternatively, the clusters may reflect different physiological states of cells within a given class. ALG establish territories that are highly stereotyped across flies (Ren et al., 2018), but are still able to compensate for the loss of a lineage in both neuropil coverage and final cell counts (Stork et al., 2014; Richier et al., 2017), highlighting that environmental cues may also play a substantial role in within-class heterogeneity. Lineage tracing and functional studies are needed to distinguish between these possibilities.
Future directions
It is remarkable to consider how far behind we are in our understanding of fundamental principles of astrocyte development, relative to what is known about development of myelinating cells and neurons. Across each stage of development lie many unanswered, yet basic, questions. What is the identity of intermediate astrocyte progenitor cells and how can they be distinguished from their more primitive parent cells? What are the cues that instruct the localization and distribution of astrocytes? How do intrinsic and extrinsic signals coordinate to drive the maturation and adoption of specific phenotypic and functional identities of astrocytes?
To date, rodent models have been predominant in the study of astrocytes. However, the emerging recognition of cells with key structural and functional properties of astrocytes in other model organisms, including zebrafish (Chen et al., 2020) and Drosophila (Stork et al., 2014), offers an opportunity to leverage the unique advantages of different models for complementary and parallel studies to advance our understanding of this historically understudied cell. Drosophila, in particular, due to their relatively inexpensive maintenance and short generation times, together with an abundance of existing highly sophisticated genetic tools including RNAi and intersectional tools for single cell manipulations, can be a workhorse for mechanistic discoveries.
While interrogating and manipulating astrocyte progenitor cells remains challenging due to insufficient tools and markers, the advanced molecular, genetic and imaging tools now available nevertheless afford opportunities for building on the existing foundations established by early neuroanatomical and molecular studies. In particular, the emergence of sequencing technologies opens the door to such discoveries and a growing number of transcriptional profiles of astrocytes from both Drosophila and mouse brains are now available for interrogation. Transcriptional profiling identified over 3,000 genes enriched in larval ALG (Huang et al., 2015). Notably, among these genes are those involved in cell morphogenesis and synapse development and function. Comparison with a comparable mouse study identified mouse homologs for nearly half of the larval-enriched genes, a majority of which are enriched in astrocytes (Huang et al., 2015). Together with the more recently published single cell transcriptional atlases of pupal development (Kurmangaliyev et al., 2020), and adult brains (Ozel et al., 2021), it is now possible to identify and interrogate the transcriptional signatures of glial cells across development. Identifying and mapping the in vivo expression of these genes may illuminate molecular and functional diversity that remains poorly understood. Single cell labeling using multi-color flip-out approaches and gene deletion approaches, for example, could facilitate analysis of morphological phenotypes and illuminate functional significance of these genes. Because many neural circuits and their behavioral outputs are well characterized in Drosophila, such targeted analyses provide an opportunity for dissecting how astrocytes modulate circuit function and behavior. There are also a growing number of transcriptomic atlases of the mouse brain, astrocyte-specific or including other cell types, across developmental stages and under various environmental conditions (Zhang et al., 2014; Hrvatin et al., 2018; Saunders et al., 2018, among many others). While many of these are restricted to cortical regions, there is nevertheless an unprecedented opportunity to identify novel molecular candidates that are conserved between species. Accompanied by lineage tracing and functional analysis, such studies will yield important novel insights into astrocyte development.
Author contributions
KM, JCS, and JS performed literature review and contributed writing; CV and DG performed literature review and contributed writing and editing of the manuscript.
Funding
DG is funded by National Institute of Neurological Disorders and Stroke R21 NS116664 and R01 NS096100 which support research on astrocyte neuron interactions in synaptic plasticity and astrocyte development. CV is funded by National Institute of Neurological Disorders and Stroke R01 NS118562 and the National Science Foundation IOS-1921065 which support research on neural circuit function in Drosophila. DG and CV also receive funding from the PA Department of Health for work on the role of astrocyte glia in neural circuit function in the Drosophila visual system.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altenhein, B., Cattenoz, P. B., and Giangrande, A. (2016). The early life of a fly glial cell. Wiley Interdiscip. Rev. Dev. Biol. 5, 67–84. doi:10.1002/wdev.200
Altenhein, B. (2015). Glial cell progenitors in the Drosophila embryo. Glia 63, 1291–1302. doi:10.1002/glia.22820
Awasaki, T., Lai, S. L., Ito, K., and Lee, T. (2008). Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 28, 13742–13753. doi:10.1523/JNEUROSCI.4844-08.2008
Badenhorst, P. (2001). Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development 128, 4093–4101. doi:10.1242/dev.128.20.4093
Baldwin, K. T., Tan, C. X., Strader, S. T., Jiang, C., Savage, J. T., Elorza-Vidal, X., et al. (2021). HepaCAM controls astrocyte self-organization and coupling. Neuron 109, 2427–2442 e10. doi:10.1016/j.neuron.2021.05.025
Barnabé-Heider, F., Wasylnka, J. A., Fernandes, K. J., Porsche, C., Sendtner, M., Kaplan, D. R., et al. (2005). Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron 48, 253–265. doi:10.1016/j.neuron.2005.08.037
Batiuk, M. Y., Martirosyan, A., Wahis, J., de Vin, F., Marneffe, C., Kusserow, C., et al. (2020). Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220. doi:10.1038/s41467-019-14198-8
Bayraktar, O. A., and Doe, C. Q. (2013). Combinatorial temporal patterning in progenitors expands neural diversity. Nature 498, 449–455. doi:10.1038/nature12266
Bayraktar, O. A., Bartels, T., Holmqvist, S., Kleshchevnikov, V., Martirosyan, A., Polioudakis, D., et al. (2020). Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nat. Neurosci. 23, 500–509. doi:10.1038/s41593-020-0602-1
Beckervordersandforth, R. M., Rickert, C., Altenhein, B., and Technau, G. M. (2008). Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech. Dev. 125, 542–557. doi:10.1016/j.mod.2007.12.004
Ben Haim, L., and Rowitch, D. H. (2017). Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 18, 31–41. doi:10.1038/nrn.2016.159
Bergles, D. E., and Richardson, W. D. (2015). Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 8, a020453. doi:10.1101/cshperspect.a020453
Blanco-Suarez, E., Liu, T. F., Kopelevich, A., and Allen, N. J. (2018). Astrocyte-secreted chordin-like 1 drives synapse maturation and limits plasticity by increasing synaptic GluA2 AMPA receptors. Neuron 100, 1116–1132 e13. doi:10.1016/j.neuron.2018.09.043
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N., and Allen, N. J. (2018). The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285. doi:10.1016/j.celrep.2017.12.039
Bonaguidi, M. A., Mcguire, T., Hu, M., Kan, L., Samanta, J., and Kessler, J. A. (2005). LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development 132, 5503–5514. doi:10.1242/dev.02166
Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., et al. (1997). Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278, 477–483. doi:10.1126/science.278.5337.477
Bushong, E. A., Martone, M. E., and Ellisman, M. H. (2004). Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int. J. Dev. Neurosci. 22, 73–86. doi:10.1016/j.ijdevneu.2003.12.008
Cameron, R. S., and Rakic, P. (2004). Glial cell lineage in the cerebral cortex: A review and synthesis. Glia 4, 124–137. doi:10.1002/glia.440040204
Chaboub, L. S., Manalo, J. M., Lee, H. K., Glasgow, S. M., Chen, F., Kawasaki, Y., et al. (2016). Temporal profiling of astrocyte precursors reveals parallel roles for asef during development and after injury. J. Neurosci. 36, 11904–11917. doi:10.1523/JNEUROSCI.1658-16.2016
Chai, H., Diaz-Castro, B., Shigetomi, E., Monte, E., Octeau, J. C., Yu, X., et al. (2017). Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron 95, 531–549 e9. doi:10.1016/j.neuron.2017.06.029
Chen, J., Poskanzer, K. E., Freeman, M. R., and Monk, K. R. (2020). Live-imaging of astrocyte morphogenesis and function in zebrafish neural circuits. Nat. Neurosci. 23, 1297–1306. doi:10.1038/s41593-020-0703-x
Cheng, S., Butrus, S., Tan, L., Xu, R., Sagireddy, S., Trachtenberg, J. T., et al. (2022). Vision-dependent specification of cell types and function in the developing cortex. Cell 185, 311–327 e24. doi:10.1016/j.cell.2021.12.022
Clavreul, S., Abdeladim, L., Hernandez-Garzon, E., Niculescu, D., Durand, J., Ieng, S. H., et al. (2019). Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nat. Commun. 10, 4884. doi:10.1038/s41467-019-12791-5
Cornell-Bell, A. H., Finkbeiner, S. M., Cooper, M. S., and Smith, S. J. (1990a). Glutamate induces calcium waves in cultured astrocytes: Long-range glial signaling. Science 247, 470–473. doi:10.1126/science.1967852
Cornell-Bell, A. H., Thomas, P. G., and Smith, S. J. (1990b). The excitatory neurotransmitter glutamate causes filopodia formation in cultured hippocampal astrocytes. Glia 3, 322–334. doi:10.1002/glia.440030503
Coutinho-Budd, J. C., Sheehan, A. E., and Freeman, M. R. (2017). The secreted neurotrophin Spatzle 3 promotes glial morphogenesis and supports neuronal survival and function. Genes Dev. 31, 2023–2038. doi:10.1101/gad.305888.117
Deneen, B., Ho, R., Lukaszewicz, A., Hochstim, C. J., Gronostajski, R. M., and Anderson, D. J. (2006). The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968. doi:10.1016/j.neuron.2006.11.019
Doe, C. Q. (2017). Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240. doi:10.1146/annurev-cellbio-111315-125210
Doherty, J., Logan, M. A., Tasdemir, O. E., and Freeman, M. R. (2009). Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768–4781. doi:10.1523/JNEUROSCI.5951-08.2009
Edwards, T. N., Nuschke, A. C., Nern, A., and Meinertzhagen, I. A. (2012). Organization and metamorphosis of glia in the Drosophila visual system. J. Comp. Neurol. 520, 2067–2085. doi:10.1002/cne.23071
Endo, F., Kasai, A., Soto, J. S., Yu, X., Qu, Z., Hashimoto, H., et al. (2022). Molecular basis of astrocyte diversity and morphology across the CNS in health and disease. Science 378, eadc9020. doi:10.1126/science.adc9020
Enriquez, J., Rio, L. Q., Blazeski, R., Bellemin, S., Godement, P., Mason, C., et al. (2018). Differing strategies despite shared lineages of motor neurons and glia to achieve robust development of an adult neuropil in Drosophila. Neuron 97, 538–554 e5. doi:10.1016/j.neuron.2018.01.007
Farhy-Tselnicker, I., and Allen, N. J. (2018). Astrocytes, neurons, synapses: A tripartite view on cortical circuit development. Neural Dev. 13, 7. doi:10.1186/s13064-018-0104-y
Farmer, W. T., Abrahamsson, T., Chierzi, S., Lui, C., Zaelzer, C., Jones, E. V., et al. (2016). Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 351, 849–854. doi:10.1126/science.aab3103
Freeman, M. R., and Rowitch, D. H. (2013). Evolving concepts of gliogenesis: A look way back and ahead to the next 25 years. Neuron 80, 613–623. doi:10.1016/j.neuron.2013.10.034
Freeman, M. R. (2015). Drosophila central nervous system glia. Cold Spring Harb. Perspect. Biol. 7, a020552. doi:10.1101/cshperspect.a020552
Garcia, A. D., Petrova, R., Eng, L., and Joyner, A. L. (2010). Sonic hedgehog regulates discrete populations of astrocytes in the adult mouse forebrain. J. Neurosci. 30, 13597–13608. doi:10.1523/JNEUROSCI.0830-10.2010
Garcia-Marques, J., and Lopez-Mascaraque, L. (2013). Clonal identity determines astrocyte cortical heterogeneity. Cereb. Cortex 23, 1463–1472. doi:10.1093/cercor/bhs134
Ge, W. P., Miyawaki, A., Gage, F. H., Jan, Y. N., and Jan, L. Y. (2012). Local generation of glia is a major astrocyte source in postnatal cortex. Nature 484, 376–380. doi:10.1038/nature10959
Genoud, C., Quairiaux, C., Steiner, P., Hirling, H., Welker, E., and Knott, G. W. (2006). Plasticity of astrocytic coverage and glutamate transporter expression in adult mouse cortex. PLoS Biol. 4, e343. doi:10.1371/journal.pbio.0040343
Giesen, K., Hummel, T., Stollewerk, A., Harrison, S., Travers, A., and Klambt, C. (1997). Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 124, 2307–2316. doi:10.1242/dev.124.12.2307
Gingrich, E. C., Case, K., and Garcia, A. D. R. (2022). A subpopulation of astrocyte progenitors defined by Sonic hedgehog signaling. Neural Dev. 17, 2. doi:10.1186/s13064-021-00158-w
Goetz, S. C., Ocbina, P. J., and Anderson, K. V. (2009). The primary cilium as a Hedgehog signal transduction machine. Methods Cell Biol. 94, 199–222. doi:10.1016/S0091-679X(08)94010-3
Gross, R. E., Mehler, M. F., Mabie, P. C., Zang, Z., Santschi, L., and Kessler, J. A. (1996). Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 17, 595–606. doi:10.1016/s0896-6273(00)80193-2
Halassa, M. M., Fellin, T., Takano, H., Dong, J. H., and Haydon, P. G. (2007). Synaptic islands defined by the territory of a single astrocyte. J. Neurosci. 27, 6473–6477. doi:10.1523/JNEUROSCI.1419-07.2007
Halter, D. A., Urban, J., Rickert, C., Ner, S. S., Ito, K., Travers, A. A., et al. (1995). The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development 121, 317–332. doi:10.1242/dev.121.2.317
Harding, K., and White, K. (2018). Drosophila as a model for developmental biology: Stem cell-fate decisions in the developing nervous system. J. Dev. Biol. 6, 25. doi:10.3390/jdb6040025
Hartenstein, V., Nassif, C., and Lekven, A. (1998). Embryonic development of the Drosophila brain. II. Pattern of glial cells. J. Comp. Neurol. 402, 32–47. doi:10.1002/(sici)1096-9861(19981207)402:1<32::aid-cne3>3.0.co;2-v
Hartenstein, V. (2011). Morphological diversity and development of glia in Drosophila. Glia 59, 1237–1252. doi:10.1002/glia.21162
Hevner, R. F. (2019). Intermediate progenitors and Tbr2 in cortical development. J. Anat. 235, 616–625. doi:10.1111/joa.12939
Hill, S. A., Blaeser, A. S., Coley, A. A., Xie, Y., Shepard, K. A., Harwell, C. C., et al. (2019). Sonic hedgehog signaling in astrocytes mediates cell type-specific synaptic organization. Elife 8, e45545. doi:10.7554/eLife.45545
Hillen, A. E. J., Burbach, J. P. H., and Hol, E. M. (2018). Cell adhesion and matricellular support by astrocytes of the tripartite synapse. Prog. Neurobiol. 165-167, 66–86. doi:10.1016/j.pneurobio.2018.02.002
Hirrlinger, J., Hulsmann, S., and Kirchhoff, F. (2004). Astroglial processes show spontaneous motility at active synaptic terminals in situ. Eur. J. Neurosci. 20, 2235–2239. doi:10.1111/j.1460-9568.2004.03689.x
Hochstim, C., Deneen, B., Lukaszewicz, A., Zhou, Q., and Anderson, D. J. (2008). Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 133, 510–522. doi:10.1016/j.cell.2008.02.046
Holt, L. M., Hernandez, R. D., Pacheco, N. L., Torres Ceja, B., Hossain, M., and Olsen, M. L. (2019). Astrocyte morphogenesis is dependent on BDNF signaling via astrocytic TrkB.T1. Elife 8, e44667. doi:10.7554/eLife.44667
Homem, C. C., and Knoblich, J. A. (2012). Drosophila neuroblasts: A model for stem cell biology. Development 139, 4297–4310. doi:10.1242/dev.080515
Hong, S., and Song, M. R. (2014). STAT3 but not STAT1 is required for astrocyte differentiation. PLoS One 9, e86851. doi:10.1371/journal.pone.0086851
Hosoya, T., Takizawa, K., Nitta, K., and Hotta, Y. (1995). Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82, 1025–1036. doi:10.1016/0092-8674(95)90281-3
Hrvatin, S., Hochbaum, D. R., Nagy, M. A., Cicconet, M., Robertson, K., Cheadle, L., et al. (2018). Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129. doi:10.1038/s41593-017-0029-5
Huang, Y., Ng, F. S., and Jackson, F. R. (2015). Comparison of larval and adult Drosophila astrocytes reveals stage-specific gene expression profiles. G3 (Bethesda) 5, 551–558. doi:10.1534/g3.114.016162
Ito, K., Urban, J., and Technau, G. M. (1995). Distribution, classification, and development ofDrosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch. Dev. Biol. 204, 284–307. doi:10.1007/BF02179499
Jacobs, J. R., Hiromi, Y., Patel, N. H., and Goodman, C. S. (1989). Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron 2, 1625–1631. doi:10.1016/0896-6273(89)90051-2
Jessen, K. R., and Mirsky, R. (2005). The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682. doi:10.1038/nrn1746
John Lin, C. C., Yu, K., Hatcher, A., Huang, T. W., Lee, H. K., Carlson, J., et al. (2017). Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405. doi:10.1038/nn.4493
Jones, B. W., Fetter, R. D., Tear, G., and Goodman, C. S. (1995). Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82, 1013–1023. doi:10.1016/0092-8674(95)90280-5
Kang, P., Lee, H. K., Glasgow, S. M., Finley, M., Donti, T., Gaber, Z. B., et al. (2012). Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74, 79–94. doi:10.1016/j.neuron.2012.01.024
Kang, K., Lee, S. W., Han, J. E., Choi, J. W., and Song, M. R. (2014). The complex morphology of reactive astrocytes controlled by fibroblast growth factor signaling. Glia 62, 1328–1344. doi:10.1002/glia.22684
Kanski, R., van Strien, M. E., van Tijn, P., and Hol, E. M. (2014). A star is born: New insights into the mechanism of astrogenesis. Cell Mol. Life Sci. 71, 433–447. doi:10.1007/s00018-013-1435-9
Kato, K., Orihara-Ono, M., and Awasaki, T. (2020). Multiple lineages enable robust development of the neuropil-glia architecture in adult Drosophila. Development 147, dev184085. doi:10.1242/dev.184085
Kelley, K. W., Ben Haim, L., Schirmer, L., Tyzack, G. E., Tolman, M., Miller, J. G., et al. (2018). Kir4.1-Dependent astrocyte-fast motor neuron interactions are required for peak strength. Neuron 98, 306–319 e7. doi:10.1016/j.neuron.2018.03.010
Khakh, B. S., and Deneen, B. (2019). The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 42, 187–207. doi:10.1146/annurev-neuro-070918-050443
Kim, J., Jones, B. W., Zock, C., Chen, Z., Wang, H., Goodman, C. S., et al. (1998). Isolation and characterization of mammalian homologs of the Drosophila gene glial cells missing. Proc. Natl. Acad. Sci. U. S. A. 95, 12364–12369. doi:10.1073/pnas.95.21.12364
Kremer, M. C., Jung, C., Batelli, S., Rubin, G. M., and Gaul, U. (2017). The glia of the adult Drosophila nervous system. Glia 65, 606–638. doi:10.1002/glia.23115
Kriegstein, A., and Alvarez-Buylla, A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184. doi:10.1146/annurev.neuro.051508.135600
Kurmangaliyev, Y. Z., Yoo, J., Valdes-Aleman, J., Sanfilippo, P., and Zipursky, S. L. (2020). Transcriptional programs of circuit assembly in the Drosophila visual system. Neuron 108, 1045–1057 e6. doi:10.1016/j.neuron.2020.10.006
Lacin, H., and Truman, J. W. (2016). Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. Elife 5, e13399. doi:10.7554/eLife.13399
Lanjakornsiripan, D., Pior, B.-J., Kawaguchi, D., Furutachi, S., Tahara, T., Katsuyama, Y., et al. (2018). Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 9, 1623–1715. doi:10.1038/s41467-018-03940-3
Lehre, K. P., Levy, L. M., Ottersen, O. P., Storm-Mathisen, J., and Danbolt, N. C. (1995). Differential expression of two glial glutamate transporters in the rat brain: Quantitative and immunocytochemical observations. J. Neurosci. 15, 1835–1853. doi:10.1523/JNEUROSCI.15-03-01835.1995
Levison, S. W., and Goldman, J. E. (1993). Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron 10, 201–212. doi:10.1016/0896-6273(93)90311-e
Levitt, P., and Rakic, P. (1980). Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J. Comp. Neurol. 193, 815–840. doi:10.1002/cne.901930316
Li, J., Khankan, R. R., Caneda, C., Godoy, M. I., Haney, M. S., Krawczyk, M. C., et al. (2019). Astrocyte-to-astrocyte contact and a positive feedback loop of growth factor signaling regulate astrocyte maturation. Glia 67, 1571–1597. doi:10.1002/glia.23630
Limmer, S., Weiler, A., Volkenhoff, A., Babatz, F., and Klambt, C. (2014). The Drosophila blood-brain barrier: Development and function of a glial endothelium. Front. Neurosci. 8, 365. doi:10.3389/fnins.2014.00365
Liu, H., Zhou, B., Yan, W., Lei, Z., Zhao, X., Zhang, K., et al. (2014). Astrocyte-like glial cells physiologically regulate olfactory processing through the modification of ORN-PN synaptic strength in Drosophila. Eur. J. Neurosci. 40, 2744–2754. doi:10.1111/ejn.12646
Liu, J., Wu, X., and Lu, Q. (2022). Molecular divergence of mammalian astrocyte progenitor cells at early gliogenesis. Development 149, dev199985. doi:10.1242/dev.199985
Loo, L., Simon, J. M., Xing, L., Mccoy, E. S., Niehaus, J. K., Guo, J., et al. (2019). Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134. doi:10.1038/s41467-018-08079-9
Losada-Perez, M. (2018). Glia: From 'just glue' to essential players in complex nervous systems: A comparative view from flies to mammals. J. Neurogenet. 32, 78–91. doi:10.1080/01677063.2018.1464568
Loulier, K., Barry, R., Mahou, P., Le Franc, Y., Supatto, W., Matho, K. S., et al. (2014). Multiplex cell and lineage tracking with combinatorial labels. Neuron 81, 505–520. doi:10.1016/j.neuron.2013.12.016
Magavi, S., Friedmann, D., Banks, G., Stolfi, A., and Lois, C. (2012). Coincident generation of pyramidal neurons and protoplasmic astrocytes in neocortical columns. J. Neurosci. 32, 4762–4772. doi:10.1523/JNEUROSCI.3560-11.2012
Melom, J. E., and Littleton, J. T. (2013). Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J. Neurosci. 33, 1169–1178. doi:10.1523/JNEUROSCI.3920-12.2013
Miyake, A., Hattori, Y., Ohta, M., and Itoh, N. (1996). Rat oligodendrocytes and astrocytes preferentially express fibroblast growth factor receptor-2 and -3 mRNAs. J. Neurosci. Res. 45, 534–541. doi:10.1002/(SICI)1097-4547(19960901)45:5<534::AID-JNR3>3.0.CO;2-D
Miyares, R. L., and Lee, T. (2019). Temporal control of Drosophila central nervous system development. Curr. Opin. Neurobiol. 56, 24–32. doi:10.1016/j.conb.2018.10.016
Molofsky, A. V., Glasgow, S. M., Chaboub, L. S., Tsai, H. H., Murnen, A. T., Kelley, K. W., et al. (2013). Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia 61, 1518–1532. doi:10.1002/glia.22538
Morel, L., Higashimori, H., Tolman, M., and Yang, Y. (2014). VGluT1+ neuronal glutamatergic signaling regulates postnatal developmental maturation of cortical protoplasmic astroglia. J. Neurosci. 34, 10950–10962. doi:10.1523/JNEUROSCI.1167-14.2014
Morrow, T., Song, M. R., and Ghosh, A. (2001). Sequential specification of neurons and glia by developmentally regulated extracellular factors. Development 128, 3585–3594. doi:10.1242/dev.128.18.3585
Muthukumar, A. K., Stork, T., and Freeman, M. R. (2014). Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat. Neurosci. 17, 1340–1350. doi:10.1038/nn.3791
Nagai, J., Yu, X., Papouin, T., Cheong, E., Freeman, M. R., Monk, K. R., et al. (2021). Behaviorally consequential astrocytic regulation of neural circuits. Neuron 109, 576–596. doi:10.1016/j.neuron.2020.12.008
Nagao, M., Ogata, T., Sawada, Y., and Gotoh, Y. (2016). Zbtb20 promotes astrocytogenesis during neocortical development. Nat. Commun. 7, 11102. doi:10.1038/ncomms11102
Nakano, R., Iwamura, M., Obikawa, A., Togane, Y., Hara, Y., Fukuhara, T., et al. (2019). Cortex glia clear dead young neurons via Drpr/dCed-6/Shark and Crk/Mbc/dCed-12 signaling pathways in the developing Drosophila optic lobe. Dev. Biol. 453, 68–85. doi:10.1016/j.ydbio.2019.05.003
Nakashima, K., Wiese, S., Yanagisawa, M., Arakawa, H., Kimura, N., Hisatsune, T., et al. (1999). Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J. Neurosci. 19, 5429–5434. doi:10.1523/JNEUROSCI.19-13-05429.1999
Namihira, M., Kohyama, J., Semi, K., Sanosaka, T., Deneen, B., Taga, T., et al. (2009). Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245–255. doi:10.1016/j.devcel.2008.12.014
Ng, F. S., Sengupta, S., Huang, Y., Yu, A. M., You, S., Roberts, M. A., et al. (2016). TRAP-Seq profiling and RNAi-based genetic screens identify conserved glial genes required for adult Drosophila behavior. Front. Mol. Neurosci. 9, 146. doi:10.3389/fnmol.2016.00146
Oh, L. Y., Denninger, A., Colvin, J. S., Vyas, A., Tole, S., Ornitz, D. M., et al. (2003). Fibroblast growth factor receptor 3 signaling regulates the onset of oligodendrocyte terminal differentiation. J. Neurosci. 23, 883–894. doi:10.1523/JNEUROSCI.23-03-00883.2003
Ohayon, D., Escalas, N., Cochard, P., Glise, B., Danesin, C., and Soula, C. (2019). Sulfatase 2 promotes generation of a spinal cord astrocyte subtype that stands out through the expression of Olig2. Glia 67, 1478–1495. doi:10.1002/glia.23621
Ohayon, D., Aguirrebengoa, M., Escalas, N., Jungas, T., and Soula, C. (2021). Transcriptome profiling of the Olig2-expressing astrocyte subtype reveals their unique molecular signature. iScience 24, 102806. doi:10.1016/j.isci.2021.102806
Omoto, J. J., Yogi, P., and Hartenstein, V. (2015). Origin and development of neuropil glia of the Drosophila larval and adult brain: Two distinct glial populations derived from separate progenitors. Dev. Biol. 404, 2–20. doi:10.1016/j.ydbio.2015.03.004
Ozel, M. N., Simon, F., Jafari, S., Holguera, I., Chen, Y. C., Benhra, N., et al. (2021). Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95. doi:10.1038/s41586-020-2879-3
Peco, E., Davla, S., Camp, D., Stacey, S. M., Landgraf, M., and van Meyel, D. J. (2016). Drosophila astrocytes cover specific territories of the CNS neuropil and are instructed to differentiate by Prospero, a key effector of Notch. Development 143, 1170–1181. doi:10.1242/dev.133165
Pereanu, W., Shy, D., and Hartenstein, V. (2005). Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 283, 191–203. doi:10.1016/j.ydbio.2005.04.024
Perraud, F., Labourdette, G., Miehe, M., Loret, C., and Sensenbrenner, M. (1988). Comparison of the morphological effects of acidic and basic fibroblast growth factors on rat astroblasts in culture. J. Neurosci. Res. 20, 1–11. doi:10.1002/jnr.490200102
Pestana, F., Edwards-Faret, G., Belgard, T. G., Martirosyan, A., and Holt, M. G. (2020). No longer underappreciated: The emerging concept of astrocyte heterogeneity in neuroscience. Brain Sci. 10, 168. doi:10.3390/brainsci10030168
Phelps, J. S., Hildebrand, D. G. C., Graham, B. J., Kuan, A. T., Thomas, L. A., Nguyen, T. M., et al. (2021). Reconstruction of motor control circuits in adult Drosophila using automated transmission electron microscopy. Cell 184, 759–774 e18. doi:10.1016/j.cell.2020.12.013
Pinto-Teixeira, F., Konstantinides, N., and Desplan, C. (2016). Programmed cell death acts at different stages of Drosophila neurodevelopment to shape the central nervous system. FEBS Lett. 590, 2435–2453. doi:10.1002/1873-3468.12298
Pogodalla, N., Winkler, B., and Klambt, C. (2022). Glial tiling in the insect nervous system. Front. Cell Neurosci. 16, 825695. doi:10.3389/fncel.2022.825695
Qian, X., Shen, Q., Goderie, S. K., He, W., Capela, A., Davis, A. A., et al. (2000). Timing of CNS cell generation: A programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80. doi:10.1016/s0896-6273(00)00086-6
Ren, Q., Awasaki, T., Wang, Y. C., Huang, Y. F., and Lee, T. (2018). Lineage-guided Notch-dependent gliogenesis by Drosophila multi-potent progenitors. Development 145, dev160127. doi:10.1242/dev.160127
Richier, B., Vijandi, C. M., Mackensen, S., and Salecker, I. (2017). Lapsyn controls branch extension and positioning of astrocyte-like glia in the Drosophila optic lobe. Nat. Commun. 8, 317. doi:10.1038/s41467-017-00384-z
Rowitch, D. H., and Kriegstein, A. R. (2010). Developmental genetics of vertebrate glial-cell specification. Nature 468, 214–222. doi:10.1038/nature09611
Saunders, A., Macosko, E. Z., Wysoker, A., Goldman, M., Krienen, F. M., de Rivera, H., et al. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 e16. doi:10.1016/j.cell.2018.07.028
Sauvageot, C. M., and Stiles, C. D. (2002). Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 12, 244–249. doi:10.1016/s0959-4388(02)00322-7
Scheffer, L. K., Xu, C. S., Januszewski, M., Lu, Z., Takemura, S. Y., Hayworth, K. J., et al. (2020). A connectome and analysis of the adult Drosophila central brain. Elife 9, e57443. doi:10.7554/eLife.57443
Schmechel, D. E., and Rakic, P. (1979). Arrested proliferation of radial glial cells during midgestation in rhesus monkey. Nature 277, 303–305. doi:10.1038/277303a0
Scholze, A. R., Foo, L. C., Mulinyawe, S., and Barres, B. A. (2014). BMP signaling in astrocytes downregulates EGFR to modulate survival and maturation. PLoS One 9, e110668. doi:10.1371/journal.pone.0110668
Shibata, T., Yamada, K., Watanabe, M., Ikenaka, K., Wada, K., Tanaka, K., et al. (1997). Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J. Neurosci. 17, 9212–9219. doi:10.1523/JNEUROSCI.17-23-09212.1997
Sofroniew, M. V. (2014). Astrogliosis. Cold Spring Harb. Perspect. Biol. 7, a020420. doi:10.1101/cshperspect.a020420
Stacey, S. M., Thomas, G. B., Labbe, A., and van Meyel, D. J. (2007). Longitudinal glia in the fly CNS: Pushing the envelope on glial diversity and neuron-glial interactions. Neuron Glia Biol. 3, 27–33. doi:10.1017/S1740925X07000506
Stogsdill, J. A., Ramirez, J., Liu, D., Kim, Y. H., Baldwin, K. T., Enustun, E., et al. (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. doi:10.1038/nature24638
Stolt, C. C., Lommes, P., Sock, E., Chaboissier, M. C., Schedl, A., and Wegner, M. (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689. doi:10.1101/gad.259003
Stork, T., Sheehan, A., Tasdemir-Yilmaz, O. E., and Freeman, M. R. (2014). Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 83, 388–403. doi:10.1016/j.neuron.2014.06.026
Su, Y., Zhang, W., Patro, C. P. K., Zhao, J., Mu, T., Ma, Z., et al. (2020). STAT3 regulates mouse neural progenitor proliferation and differentiation by promoting mitochondrial metabolism. Front. Cell Dev. Biol. 8, 362. doi:10.3389/fcell.2020.00362
Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M. Z., Zubiaga, A., Hua, X., et al. (2001). Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. doi:10.1016/s0092-8674(01)00224-0
Takouda, J., Katada, S., and Nakashima, K. (2017). Emerging mechanisms underlying astrogenesis in the developing mammalian brain. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 386–398. doi:10.2183/pjab.93.024
Tasdemir-Yilmaz, O. E., and Freeman, M. R. (2014). Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28, 20–33. doi:10.1101/gad.229518.113
Tatsumi, K., Isonishi, A., Yamasaki, M., Kawabe, Y., Morita-Takemura, S., Nakahara, K., et al. (2018). Olig2-Lineage astrocytes: A distinct subtype of astrocytes that differs from GFAP astrocytes. Front. Neuroanat. 12, 8. doi:10.3389/fnana.2018.00008
Tien, A. C., Tsai, H. H., Molofsky, A. V., Mcmahon, M., Foo, L. C., Kaul, A., et al. (2012). Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development 139, 2477–2487. doi:10.1242/dev.077214
Tiwari, N., Pataskar, A., Peron, S., Thakurela, S., Sahu, S. K., Figueres-Onate, M., et al. (2018). Stage-specific transcription factors drive astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell 23, 557–571 e8. doi:10.1016/j.stem.2018.09.008
Tong, C. K., Fuentealba, L. C., Shah, J. K., Lindquist, R. A., Ihrie, R. A., Guinto, C. D., et al. (2015). A dorsal SHH-dependent domain in the V-SVZ produces large numbers of oligodendroglial lineage cells in the postnatal brain. Stem Cell Rep. 5, 461–470. doi:10.1016/j.stemcr.2015.08.013
Truman, J. W., and Bate, M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125, 145–157. doi:10.1016/0012-1606(88)90067-x
Tsai, H. H., Li, H., Fuentealba, L. C., Molofsky, A. V., Taveira-Marques, R., Zhuang, H., et al. (2012). Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337, 358–362. doi:10.1126/science.1222381
Udolph, G., Rath, P., and Chia, W. (2001). A requirement for Notch in the Genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development 128, 1457–1466. doi:10.1242/dev.128.8.1457
Umesono, Y., Hiromi, Y., and Hotta, Y. (2002). Context-dependent utilization of Notch activity in Drosophila glial determination. Development 129, 2391–2399. doi:10.1242/dev.129.10.2391
Urbach, R., and Technau, G. M. (2003). Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130, 3621–3637. doi:10.1242/dev.00533
Viktorin, G., Riebli, N., Popkova, A., Giangrande, A., and Reichert, H. (2011). Multipotent neural stem cells generate glial cells of the central complex through transit amplifying intermediate progenitors in Drosophila brain development. Dev. Biol. 356, 553–565. doi:10.1016/j.ydbio.2011.06.013
Vincent, S., Vonesch, J. L., and Giangrande, A. (1996). Glide directs glial fate commitment and cell fate switch between neurones and glia. Development 122, 131–139. doi:10.1242/dev.122.1.131
Walsh, K. T., and Doe, C. Q. (2017). Drosophila embryonic type II neuroblasts: Origin, temporal patterning, and contribution to the adult central complex. Development 144, 4552–4562. doi:10.1242/dev.157826
Westergard, T., and Rothstein, J. D. (2020). Astrocyte diversity: Current insights and future directions. Neurochem. Res. 45, 1298–1305. doi:10.1007/s11064-020-02959-7
Yang, Y., and Jackson, R. (2019). Astrocyte identity: Evolutionary perspectives on astrocyte functions and heterogeneity. Curr. Opin. Neurobiol. 56, 40–46. doi:10.1016/j.conb.2018.11.006
Yuasa, Y., Okabe, M., Yoshikawa, S., Tabuchi, K., Xiong, W. C., Hiromi, Y., et al. (2003). Drosophila homeodomain protein REPO controls glial differentiation by cooperating with ETS and BTB transcription factors. Development 130, 2419–2428. doi:10.1242/dev.00468
Zerlin, M., Levison, S. W., and Goldman, J. E. (1995). Early patterns of migration, morphogenesis, and intermediate filament expression of subventricular zone cells in the postnatal rat forebrain. J. Neurosci. 15, 7238–7249. doi:10.1523/JNEUROSCI.15-11-07238.1995
Zhang, Y., Chen, K., Sloan, S. A., Bennett, M. L., Scholze, A. R., O'Keeffe, S., et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947. doi:10.1523/JNEUROSCI.1860-14.2014
Keywords: astrocyte, glia, development, Drosophila, astrocyte-like glia, neuropil glia, last modified: 10/7/22 12:36 p.m. p. 2
Citation: Markey KM, Saunders JC, Smuts J, von Reyn CR and Garcia ADR (2023) Astrocyte development—More questions than answers. Front. Cell Dev. Biol. 11:1063843. doi: 10.3389/fcell.2023.1063843
Received: 07 October 2022; Accepted: 14 March 2023;
Published: 27 March 2023.
Edited by:
Matthew Holt, Universidade do Porto, PortugalReviewed by:
Benjamin Deneen, Baylor College of Medicine, United StatesIsabella Farhy-Tselnicker, Texas A&M University, United States
Copyright © 2023 Markey, Saunders, Smuts, von Reyn and Garcia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Denise R. Garcia, YWRnODJAZHJleGVsLmVkdQ==
 Kathryn M. Markey1
Kathryn M. Markey1 Jana Smuts
Jana Smuts Catherine R. von Reyn
Catherine R. von Reyn A. Denise R. Garcia
A. Denise R. Garcia