- 1Department of Biology, Integrated Sciences Complex, University of Massachusetts Boston, Boston, MA, United States
- 2NYU Langone Medical Center, New York, NY, United States
- 3Department of Biology, University of Fribourg, Fribourg, Switzerland
- 4Arizona College of Osteopathic Medicine, Midwestern University, Glendale, AZ, United States
- 5Department of Pharmacology, College of Graduate Studies, Midwestern University, Glendale, AZ, United States
During terminal differentiation of the mammalian retina, transcription factors control binary cell fate decisions that generate functionally distinct subtypes of photoreceptor neurons. For instance, Otx2 and RORβ activate the expression of the transcriptional repressor Blimp-1/PRDM1 that represses bipolar interneuron fate and promotes rod photoreceptor fate. Moreover, Otx2 and Crx promote expression of the nuclear receptor Nrl that promotes rod photoreceptor fate and represses cone photoreceptor fate. Mutations in these four transcription factors cause severe eye diseases such as retinitis pigmentosa. Here, we show that a post-mitotic binary fate decision in Drosophila color photoreceptor subtype specification requires ecdysone signaling and involves orthologs of these transcription factors: Drosophila Blimp-1/PRDM1 and Hr3/RORβ promote blue-sensitive (Rh5) photoreceptor fate and repress green-sensitive (Rh6) photoreceptor fate through the transcriptional repression of warts/LATS, the nexus of the phylogenetically conserved Hippo tumor suppressor pathway. Moreover, we identify a novel interaction between Blimp-1 and warts, whereby Blimp-1 represses a warts intronic enhancer in blue-sensitive photoreceptors and thereby gives rise to specific expression of warts in green-sensitive photoreceptors. Together, these results reveal that conserved transcriptional regulators play key roles in terminal cell fate decisions in both the Drosophila and the mammalian retina, and the mechanistic insights further deepen our understanding of how Hippo pathway signaling is repurposed to control photoreceptor fates for Drosophila color vision.
1 Introduction
Color vision requires the expression of light-sensing pigments with different wavelength-sensitivities in different photoreceptor (PR) subtypes (Rister and Desplan, 2011). For instance, human color vision is based on three cone PR subtypes that express short-, medium-, or long wavelength-sensitive pigments (Nathans, 1999; Hofer et al., 2005). Likewise, the Drosophila melanogaster retina expresses five color-sensing Rhodopsin (Rh) pigments in distinct PR subtypes (Poupault et al., 2021). The “outer” PRs R1-R6 express blue/green-sensitive Rh1 and mediate dim light vision equivalent to human rods (O'Tousa et al., 1985; Zuker et al., 1985). The two ‘inner’ PRs R7/R8, which are arranged in tandem, each occur in two subtypes (Figure 1A) that are sensitive to different wavelengths and mediate color vision equivalent to human cones (Roorda and Williams, 1999). 35% of R7 PRs express the short UV-sensitive Rh3 and are coupled to proximally located R8 PRs that express the blue-sensitive Rh5 (Figure 1A, left), while the other 65% of R7 PRs express the long UV-sensitive Rh4 coupled with R8 PRs that express the green-sensitive Rh6 (Figure 1A, right) (Fortini and Rubin, 1990; Chou et al., 1996; Papatsenko et al., 1997; Chou et al., 1999).
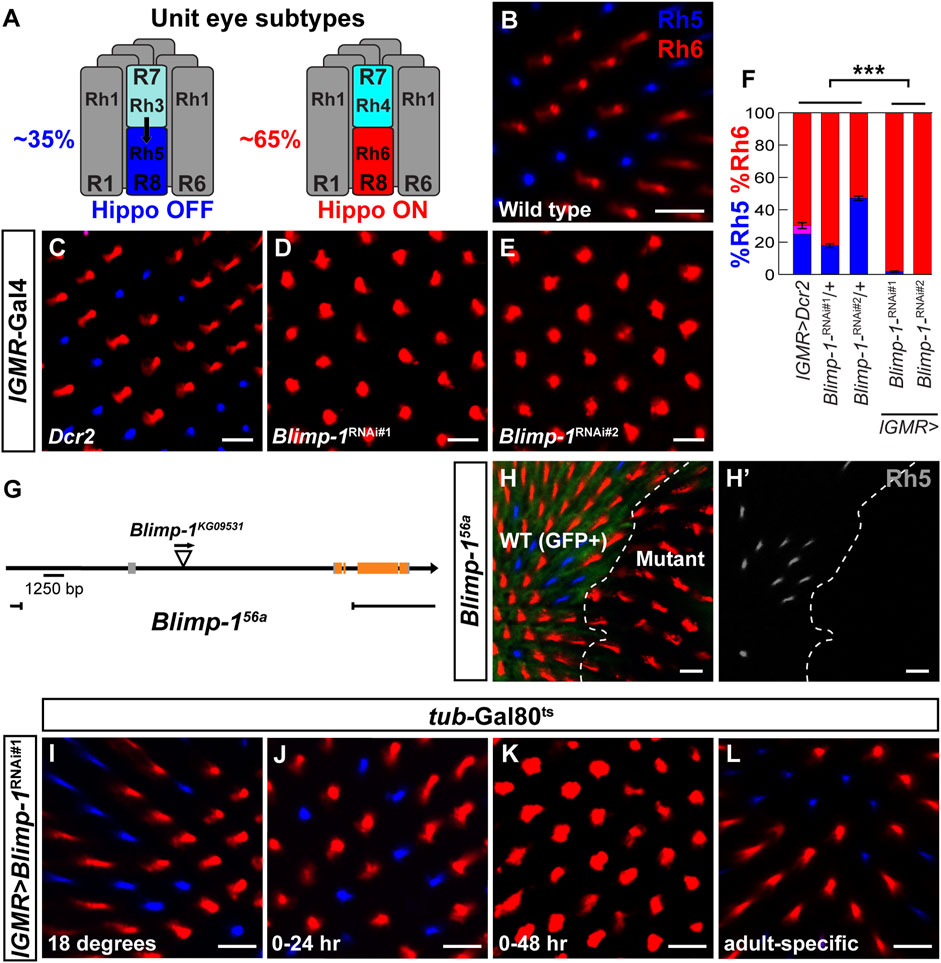
FIGURE 1. Blimp-1 is required to promote Rh5 and to repress Rh6. (A) Schematic side view of the two different unit eye subtypes in the Drosophila retina. Black arrow represents the Activin signal from Rh3-expressing R7 photoreceptors (35%, left) that induces Rh5 fate (Hippo OFF) in the proximally located R8 photoreceptors. Rh4-expressing R7 photoreceptors (65%, right) do not send the Activin signal and the corresponding R8 photoreceptors express Rh6 (Hippo ON) by default. (B–E) Confocal images of whole mounted adult retinas stained with antibodies for Rh5 (blue) and Rh6 (red). (B) A wild-type Canton S retina has an Rh5:Rh6 ratio of ∼35:65. (C) Expression of Dcr2 with the pan-photoreceptor driver lGMR-Gal4 does not affect the Rh5:Rh6 ratio. (D and E) Expression of two independent Blimp-1 RNAi constructs, Blimp-1-RNAikk (Blimp-1RNAi#1) and Blimp-1-RNAiTRiP #36634 (Blimp-1RNAi#2), in combination with Dcr2 causes a dramatic loss of Rh5 and a corresponding gain of Rh6. (F) Quantification of R8 subtypes in controls and Blimp-1 knockdowns. Graph shows the percentage of R8 photoreceptors that express exclusively Rh5 (blue), exclusively Rh6 (red), or co-express Rh5 and Rh6 (magenta). Mean %Rh6 was compared among genotypes with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 7-8 retinas were scored for each genotype. (G) Schematic showing the location of the Blimp-156a deletion that was generated through imprecise P-element (triangle) excision. (H) Confocal image showing wild type R8 photoreceptors (GFP positive, left) exhibiting a normal Rh5:Rh6 ratio and Blimp-156a null mutant clones (GFP negative, right) that exclusively express Rh6. The white dashed line denotes the boundary between wild-type and Blimp-1 mutant tissue. (H′) Rh5 channel (grayscale) shows the loss of Rh5 in the Blimp-1 mutant tissue. (I–L) Confocal images of temporally restricted lGMR > Blimp-1RNAi#1 knockdown with tub-Gal80ts. (I) Control flies raised throughout development and adulthood at 18°C, i.e., no Blimp-1 knockdown, have a normal Rh5:Rh6 ratio. (J) Pupal Blimp-1 knockdown from 0 to 24 h after puparium formation at 29°C does not affect the Rh5:Rh6 ratio. (K) Pupal Blimp-1 knockdown from 0 to 48 h after puparium formation at 29°C causes a loss of Rh5 and gain of Rh6. (L) Adult-specific Blimp-1 knockdown for 7 days at 29°C following eclosion does not affect the Rh5:Rh6 ratio. All scale bars, 10 µm.
The binary cell-fate decision to express either Rh5 or Rh6 in R8 PRs (Figure 1B) requires an Activin/TGFβ signal from the distally located R7 PRs (arrow in Figure 1A) (Wells et al., 2017) and differential regulation of the Hippo tumor suppressor pathway in R8 PRs (Mikeladze-Dvali et al., 2005; Jukam and Desplan, 2011; Jukam et al., 2013). In the canonical and conserved role of the Hippo pathway (Zeng and Hong, 2008; Zhao et al., 2008; Zhang et al., 2009; Chan et al., 2011; Halder and Johnson, 2011; Harvey and Hariharan, 2012; Halder and Camargo, 2013), the kinase Warts (Wts) restricts tissue growth by phosphorylating the transcriptional co-activator and oncogene Yorkie (Yki), which prevents Yki from entering the nucleus (Huang et al., 2005; Oh and Irvine, 2008). When Yki is not phosphorylated, it enters the nucleus and associates with transcription factors such as Scalloped (Sd) to activate genes that function in promoting growth, inhibiting apoptosis, and negative feedback regulation (Hamaratoglu et al., 2006; Neto-Silva et al., 2010).
In terminally differentiating Drosophila PRs, the Hippo pathway is repurposed to promote green-sensitive Rh6 PR fate, while its inactivation promotes blue-sensitive Rh5 PR fate (Figure 2A) (Mikeladze-Dvali et al., 2005; Jukam and Desplan, 2011; Jukam et al., 2013; Pojer et al., 2021). In this post-mitotic context, Yki/Sd promote Rh5 fate and inactivate the Hippo pathway by repressing wts at the transcriptional level (Mikeladze-Dvali et al., 2005; Jukam et al., 2013; Xie et al., 2019). Wts repression results in the activation of melted (melt), which encodes a Pleckstrin Homology domain protein (Mikeladze-Dvali et al., 2005; Teleman et al., 2005). For wts repression and melt activation, Yki/Sd require the transcription factors Orthodenticle (Otd) and Traffic Jam (Tj), which act in a coherent feedforward loop (Vandendries et al., 1996; Mears et al., 2001; Tahayato et al., 2003; Montana et al., 2011; Fichelson et al., 2012; Hao et al., 2012; Jukam et al., 2013). Conversely, when wts is expressed, it represses Yki; this leads to the transcriptional repression of melt, which allows the Hippo pathway to remain active and to promote Rh6 fate (Figure 2A). Thus, the Hippo pathway functions as a bi-stable switch in post-mitotic PRs: wts expression (Hippo ON) and melt repression specify Rh6 fate, while melt expression and wts repression (Hippo OFF) specify Rh5 fate (Figure 2A). The transcriptional regulation of wts in this context remains poorly understood.
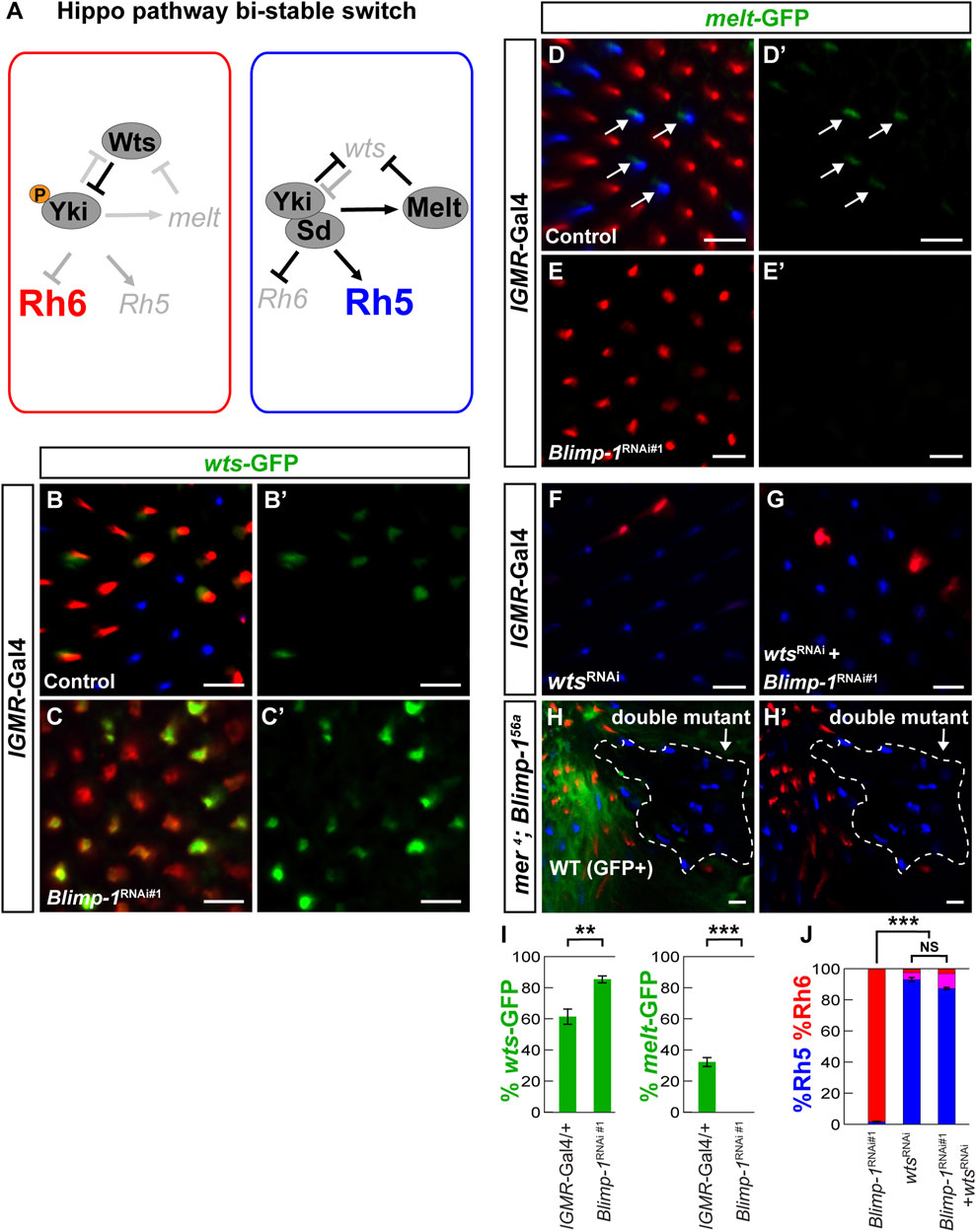
FIGURE 2. Blimp-1 is required for warts repression and melted expression. (A) Schematic of the repurposed Hippo pathway that controls a binary Rh5 vs. Rh6 cell fate decision in post-mitotic R8 photoreceptors. In Rh6 expressing photoreceptors (left), Wts represses Yki by phosphorylation, which prevents the expression of melt and Rh5 and permits Rh6 expression. In Rh5 expressing photoreceptors (right), Yki binds to Sd, activates melt and Rh5, and represses wts and Rh6. (B) In the lGMR-Gal4/+ driver control, the wts-GFP reporter is expressed in most Rh6 photoreceptors and absent in all Rh5 photoreceptors. (C) In lGMR > Blimp-1RNAi#1 retinas, wts-GFP is de-repressed with Rh6. (B′,C′) GFP channel. (D) In the lGMR-Gal4/+ driver control, the melt-GFP reporter is expressed in most Rh5 photoreceptors but absent in Rh6 photoreceptors. White arrows indicate melt-GFP expression. (E) In lGMR > Blimp-1RNAi#1 retinas, melt-GFP expression is completely lost. (D′, E′) GFP channels. (F) Knockdown of wts with lGMR-Gal4 causes a gain of Rh5 and loss of Rh6. (G) Knockdown of Blimp-1 in addition to wts does not modify the wts knockdown phenotype. (H) mer4; Blimp-156a double mutant clones (encircled by the white dashed line) exclusively express Rh5, which is the opposite of Blimp-156a mutant clones that exclusively express Rh6. All scale bars, 10 µm. (I) Quantification of the wts-GFP-expressing R8 photoreceptors (left graph) and the melt-GFP expressing R8 photoreceptors (right graph) in the driver control and Blimp-1 knockdown. Mean %wts-GFP (left) or %melt-GFP reporter (right) expression in driver control vs. knockdown was compared with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 6 retinas were analyzed for each genotype. (J) Quantification of R8 subtypes, mean %Rh5 was compared among genotypes with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001.5-8 retinas were scored for each genotype.
Here, we show that Drosophila color PR subtype specification requires ecdysone signaling as well as the orthologs of transcription factors that promote rod PR fate in the mammalian retina: B lymphocyte-induced maturation protein-1/PR domain containing 1 (Blimp-1/PRDM1) and Hr3 (RAR-related orphan receptor β/RORβ) repress the Hippo pathway in post-mitotic Drosophila PRs, thereby promoting blue-sensitive Rh5 PR fate and repressing green-sensitive Rh6 PR fate. Moreover, we identify a novel mechanism of wts regulation in which Blimp-1 represses a wts enhancer to give rise to Rh6 PR subtype-specific wts expression. However, we find that this regulation of the Hippo pathway by Blimp-1 is context-specific, as Blimp-1 is required for wing growth independently of Wts. In summary, these results show that conserved transcriptional regulators play key roles in terminal PR differentiation in both Drosophila and mammals. Moreover, these insights deepen our understanding of the mechanisms that allow the Hippo pathway to control a binary cell-fate decision in terminally differentiating color PRs.
2 Materials and methods
2.1 Stocks and maintenance
The following Drosophila stocks were used in this study: mer4 FRT19A (Fehon et al., 1997), ey-FLP, P{ubiGFP} FRT19A (from Jessica Treisman, NYU), ey-FLP, P{ubiGFP} FRT80 (from Claude Desplan, NYU), lGMR-Gal4 (Wernet et al., 2006), UAS-Babo* (Wells et al., 2017), sens-Gal4 (from Graeme Mardon, Baylor College of Medicine), and ex-LacZ (Yu and Pan, 2018). From Bloomington Drosophila Stock Center, we obtained: MS1096-Gal4; UAS-Dcr2 (#25706), UAS-Dcr2; nubbin-Gal4 (#25754), tubP-Gal80ts (#7017), en2.4-Gal4 (#30564), UAS-Dcr2; en2.4-Gal4, UAS-2XEGFP (#25752), UAS-Hr3-RNAi #1 (TRiP.JF02542, #27253), UAS-Hr3-RNAi #2 (TRiP.JF02543, #27254), UAS-wts-RNAi (TRiP HMS00026) (#34064), UAS-EcR-RNAi (TRiP.HMJ22371, #58286), UAS-EcR-A-dsRNA (#9328), UAS-EcR-b1-dsRNA (#9329), UAS-Blimp-1-RNAi #2 (TRiP.GL00594, #36634), and Blimp-1KG09531 (#15195). From Vienna Drosophila Resource Center, we obtained UAS-Blimp-1-RNAi #1 (KK #108374).
All stocks were maintained on standard lab food at 25°C, 50% humidity, and under a 12 h/12 h light/dark cycle. All experiments and controls were conducted at 25°C with 2–5 day-old female flies, unless stated otherwise. For RNAi experiments with the sens-Gal4 driver, third instar larvae of the experimental and control crosses were shifted from a 25°C incubator to a 29°C incubator.
For staging of pupae, newly formed white prepupae—corresponding to 0 h after puparium formation—were circled with a marker pen on the food vial (Hsiao et al., 2012). At the desired time point after puparium formation, pupae were gently removed from the vial using forceps. For Gal80ts experiments, white prepupae were shifted from a 25°C incubator to a 29°C incubator and then shifted at the desired time point (see Results) from the 29°C incubator to an 18°C incubator.
2.2 Generation of the Blimp-1 null mutant
We used the P-element insertion Blimp-1KG09531 to generate the imprecise excision allele Blimp-156a, following standard procedures (Engels et al., 1990). The deletion breakpoints of the imprecise excision allele were initially determined by Next-Generation Sequencing of the 3L chromosome from predominantly homozygous Blimp-156a mutant first instar larvae that had reduced or no GFP expression from the TM3, P{GAL4-Kr.C}DC2, P{UAS-GFP.S65T}DC10, Sb1 balancer chromosome. Initial breakpoints were determined by using the bioinformatics tool BreakDancer (Fan et al., 2014) to evaluate the chromosome for large structural variants. BreakDancer predicted a deletion from bases 3L:5624169.5645326, lining up with an area of low sequence coverage noticeable in the alignment. Subsequent Sanger sequencing surrounding this region—using the forward primer TTTTCCAGGTCATCGTTTCC and the reverse primer ATCGTCGTCTCAGGATCCAC—revealed that the chromosomal region that was deleted is 3L:5624036.5645326 (Figure 1G), based off the D. melanogaster genome release 6.45.
2.3 Generation of warts and melted reporters
The first melt intron was digested with KpnI and NotI to obtain a 2 kb enhancer fragment from its 3′ region (Jukam et al., 2013); the second wts intron (in TOPO plasmid/ThermoFisher, a gift from David Jukam) was digested with BamHI, BglII, and XhoI to obtain a 2.6 kb enhancer fragment from its 3’ region. The obtained enhancer sequences were confirmed by Sanger sequencing. The following primers were used to mutate the two Blimp-1 motifs in the wts intron (mutated bases are in bold) with the Q5® Site-Directed Mutagenesis Kit (New England Biolabs).
wts-ΔBlimp-1 motif 1 forward: acacgctAATCCACTTAAGCTCCTCTG
wts-ΔBlimp-1 motif 1 reverse: atgccacGGCAATTCTACGGTTCGTG
wts-ΔBlimp-1 motif 2 forward: acgcCAATGGGCTCACTAAATC
wts-ΔBlimp-1 motif 2 reverse: tagcaAATTTGCCGTAAATTGGC
We inserted the wild type or mutant enhancer constructs into a transformation plasmid containing an egfp reporter gene, a mini-white+ transformation marker, an hsp70 minimal promoter, and an attB site for phiC31-mediated transgenesis (Rister et al., 2015). The constructs were inserted in the second chromosomal landing site attP40 or the third chromosomal landing site J36 (ZH-attP-86Fb) (Bischof et al., 2007).
2.4 Luciferase reporter assay
The wts-hsp70 enhancer-promoter construct (see above) was amplified using the Phusion® High-Fidelity PCR Kit (New England Biolabs) and fused to a firefly luciferase reporter gene (wts-hsp70-luc), digested with BglII and KpnI, and subcloned into the pGL3-basic vector (Promega) with the NEBuilder® HiFi DNA Assembly master mix (New England Biolabs). The following primers were used to amplify wts-hsp70 (pGL3 overhangs are in lowercase).
wts-hsp70 forward: atttctctatcgataggtacCTGACATATTTGGTGCTACACATGTAATCCC
wts-hsp70 reverse: ccaagcttacttagatcgcaGGTGGCGACCGGTGGATC
To generate the Blimp-1 expression construct (Ac5-Blimp-1), Blimp-1-RA cDNA was amplified from a UAS-Blimp-1-RA plasmid (Drosophila Genomics Resource Center #1634032) and inserted into the pAc5.1/V5-His A vector (Invitrogen) by digest with XhoI and AgeI and using the NEBuilder® HiFi DNA Assembly master mix. The following primers were used to amplify the Blimp-1-RA insert (pAc5.1/V5-His overhangs are in lowercase).
Blimp-1RA forward: tccagcacagtggcggccgcAGTTTCCCGTAAGCAACAAAAC
Blimp-1RA reverse: aatggtgatggtgatgatgatACGTGCATTCGATGATCATG
To generate the Hr3 expression construct (Ac5-Hr3), we ordered the Hr3-RA coding sequence inserted into the p-UCIDT-AMP vector from Integrated DNA Technologies (IDT). From that plasmid we amplified Hr3-RA and inserted it into the pAc5.1/V5-His A vector (Invitrogen) by digest with NotI and EcoRI and using the NEBuilder® HiFi DNA Assembly master mix. The following primers were used to amplify the Hr3-RA insert (pAc5.1/V5-His overhangs are in lowercase).
Hr3-1RA forward: ctactagtccagtgtggtggATGTATACGCAACGTATGTTTG
Hr3-1RA reverse: agggccctctagactcgagcTTATGTCAGGTCCTGCTG
Luciferase reporter assays were performed using the Dual-Luciferase® Reporter Assay System (Promega). For all experiments and controls, 100 ng of wts-hsp70-luc were co-transfected with 100 ng of a Renilla luciferase gene fused to a TK promoter (pRL-TK) (Promega) that served as a control reporter. For controls, wts-hsp70-luc and pRL-TK were co-transfected with 100 ng of pAc5.1/V5-His A empty vector; for experiments, wts-hsp70-luc and pRL-TK were co-transfected with 100 ng of either Ac5-Blimp-1 or Ac5-Hr3. Drosophila S2 cells (Gibco) were maintained in Schneider’s Medium with 10% fetal bovine serum (Gibco) at room temperature. 1 × 10^6 cells were plated in 6-well tissue culture dishes (Corning) 24 h prior to transfection with 7.5 μL Effectene Transfection Reagent (Qiagen). Samples were transfected in triplicate for each experiment, and each experiment was performed at least three independent times. Background luminescence was determined using non-transfected cells and subtracted from control and experimental luminescence readings. Firefly luminescence results were normalized to Renilla luminescence results and presented as Relative Luminescence Units (RLU). Error bars for luciferase data represent ±S.E.M.
2.5 Wing size assay
Adult wings of female flies raised at 25°C were removed with forceps and mounted with mounting medium–lactic acid/CMCP-10 high viscosity mountant (Polysciences) (1:3, v/v)—on a glass slide. 5 μL of isopropanol (Sigma-Aldrich) was added to the wings and allowed to evaporate without drying out the wings. Next, 10 µL of mounting medium was added to the wings on the slide, wings were oriented, and air bubbles were removed. To determine relative wing sizes, wing areas were calculated using ImageJ. At least five wings were measured per genotype, and average wing areas were normalized to the average wing area of the MS1096-Gal4/+ driver control. Error bars indicate the standard error of the mean (S.E.M.).
2.6 Imaging the Drosophila eye
Adult flies were embedded in an agarose gel that was prepared in a 500 mL Erlenmeyer flask by mixing 2 g of UltraPure Agarose (Invitrogen) with 100 mL of distilled water. The mixture was microwaved until bubbles were seen and the agarose was fully dissolved. Next, the Erlenmeyer flask with the dissolved agarose was transferred to a 60°C water bath (Thermo Scientific) to cool the agarose gel but maintaining it liquid. Flies were anesthetized with CO2 and transferred to a 60 mm Petri dish (Falcon) filled approximately halfway with the liquid agarose gel. Wings and legs were submerged and the head was oriented with forceps such that one compound eye faced the microscope lens. Next, the Petri dish was placed on ice to allow the gel to solidify and then positioned under a Stemi 508 Trinoc microscope (model #4350649030, Zeiss); the eye was imaged using an Axiocam 208 HD/4k color camera (model #4265709000) set to auto exposure and auto white balance. Image processing was performed using Fiji (https://imagej.net/software/fiji/), Adobe Photoshop 2020, and Adobe Illustrator 2020 software.
2.7 Retina whole-mounts, immunohistochemistry, and confocal microscopy
We performed adult retina dissections and immunohistochemistry as previously described (Hsiao et al., 2012). We dissected adult retinas in cold phosphate-buffered saline (PBS) and fixed them in 3.7% formaldehyde solution (in PBS) for 15 min at room temperature. After two washes with PBS and one with PBST (PBS +0.2% Triton-X, Sigma), we removed the laminas. Next, we incubated the retinas overnight in the primary antibodies that were diluted in PBST (sheep anti-GFP—from AbD Serotec—1:100; mouse anti-Rh3—from Steve Britt/University of Texas—1:10; mouse anti-Rh5—from Steve Britt—1:400, guinea pig anti-Rh4—from Claude Desplan/NYU—1:1,000; or rabbit anti-Rh6—from Claude Desplan—1:1,000). After three PBST washes, we incubated the retinas overnight at room temperature in the secondary antibodies (Alexa Fluor 488-, 555-, or 647-conjugated raised in donkey; Molecular Probes) that were diluted 1:800 in PBST. Alexa Fluor (488 or 555)-coupled Phalloidin (1:150, Invitrogen) was used to visualize the photoreceptor rhabdomeres. After three PBST washes, we mounted the retinas on bridge slides with SlowFade (Molecular Probes).
We performed pupal retina dissections as previously described but with some modifications (Hsiao et al., 2012; Wang et al., 2022). We circled white prepupae, which were raised at 25°C, for staged pupal dissections. At the desired stage (after 24, 48, or 72 h, respectively), pupae were taped to a dissecting plate with double-sided tape and removed from the pupal case with forceps. The pupa was then submerged in ice-cold PBS and the head was removed using microdissection scissors. Next, the retina-brain complexes were removed using forceps and fixed in 3.7% formaldehyde solution (in PBS) for 15 min at room temperature. After two washes with PBS and one wash with PBST, the retina-brain complexes were incubated at 4°C overnight in primary antibodies (guinea pig anti-Blimp-1—from Sudipto Roy/National University of Singapore—1:400; rat anti-Elav—from DHSB—1:50) diluted in PBST and 5% normal donkey serum. After three PBST washes, we incubated the retinas overnight at 4°C for three hours in the secondary antibodies (see above). After three PBST washes, we performed a secondary fixation step by submerging retina-brain complexes in 3.7% formaldehyde solution (in PBS) for 20 min at room temperature. After washing three times with PBST, we removed the retina from the brain in PBS and mounted the retina on a slide using Slowfade. We imaged both adult and pupal retina whole mounts with a Leica SP5 or a Zeiss LSM 8 confocal microscope. We processed the confocal images with Fiji, Adobe Photoshop 2020, and Adobe Illustrator 2020 software.
2.8 Quantification of rhodopsin and reporter expression patterns
As previously described (Poupault et al., 2021), we manually scored the number of rhabdomeres that expressed the markers Rh5, Rh6, or a GFP reporter with the count tool in Adobe Photoshop 2020. The percentage of R8 PRs that expressed the respective marker was calculated for each retina as well as the mean percentage of all retinas within a genotype. Statistical comparisons across genotypes were performed using the Mann-Whitney U Test; significance levels are given as p values and error bars indicate the standard error of the mean (S.E.M.).
2.9 Conservation analysis of Blimp-1 motifs
To analyze the evolutionary conservation of Blimp-1 motifs, we obtained alignments of the wts second intron sequences from ten Drosophila species (Clark et al., 2007) from the UCSC genome browser (https://genome.ucsc.edu/).
3 Results
3.1 Blimp-1 is required for blue-sensitive photoreceptor subtype specification
We performed a candidate RNAi screen to identify sequence-specific transcription factors that are required for the wild type 35/65 ratio of the Rh5/Rh6-expressing R8 PR subtypes and found that the knockdown of B lymphocyte-induced maturation protein-1 (Blimp-1) with the pan-PR driver lGMR-Gal4 (Wernet et al., 2006) caused a dramatic gain of Rh6-expressing PRs and a loss of Rh5-expressing PRs (Figures 1C–F). This loss of blue-sensitive PR fate and gain of green-sensitive PR fate was observed with two independent Blimp-1-RNAi constructs that targeted different parts of the Blimp-1 transcript (Figures 1D,E). To validate the RNAi results, we generated a Blimp-1 null mutant through imprecise P-element excision (see Materials and methods), which resulted in a genomic deletion that spans from 6.8 kb upstream of the Blimp-1 transcription start site to the third coding exon (Blimp-156a, Figure 1G). Since the Blimp-156a null allele was embryonic lethal, we used eyeless > Flp to generate mutant clones in the eye with the FLP/FRT recombination system (Newsome et al., 2000). Consistent with the RNAi results, Blimp-156a null mutant clones exclusively expressed Rh6 at the expense of Rh5, confirming that Blimp-1 is required for expression of Rh5 and repression of Rh6 (Figures 1H,H’). Taken together, Blimp-1 is required for the terminal differentiation of the blue-sensitive, Rh5-expressing R8 PR subtype.
3.2 Blimp-1 promotes blue-sensitive photoreceptor subtype fate in mid-pupal photoreceptors
Our next goal was to determine the time window during which Blimp-1 affects R8 PR subtype specification. We detected weak Blimp-1 expression (Ng et al., 2006) in all PRs at 24 h after puparium formation (APF) and stronger expression at 48 h APF (Supplementary Figures S1A, A’, B, B’), but no expression at 72 h APF and in adult eyes (Supplementary Figures S1C, C′, D, D′). Moreover, RNAi-mediated knockdown of Blimp-1 abolished Blimp-1 expression (Figures S1E–F′). This suggests that Blimp-1 expression is transiently increased in PRs from early-to mid-pupal development, when R8 PR subtypes are distinguished (Jukam and Desplan, 2011), and is turned off in late pupal development. Interestingly, most of the Blimp-1 signal was not restricted to the nucleus, but surrounded it: at 48 h APF, we observed a partial overlap of Blimp-1 with the Elav nuclear marker in wild-type and occasional strong Blimp-1 PR nuclear localization in the driver control, suggesting that Blimp-1’s nuclear localization is regulated and transient. We therefore propose that Blimp-1’s nuclear entry and/or export is regulated to restrict its activity during pupal development.
Since Blimp-1 expression and nuclear localization is lost after 48 h APF, we hypothesized that Blimp-1 is required for the specification of Rh5 fate but not for its maintenance at later stages. To further investigate this transient requirement of Blimp-1 for R8 subtype specification, we used a temperature sensitive mutant of the Gal4 inhibitor Gal80 (Gal80ts) to temporally restrict the Blimp-1 knockdown (McGuire et al., 2004). Permissive temperature (Figure 1I), RNAi-mediated Blimp-1 knockdown from 0 to 24 h APF (Figure 1J), or Blimp-1 knockdown for seven days after eclosion (Figure 1L) did not affect the Rh5:Rh6 ratio. In contrast, Blimp-1 knockdown from 0 to 48 h APF (Figure 1K) caused a dramatic loss of Rh5 and gain of Rh6. Taken together, these data suggest that Blimp-1 is required in early to mid-pupal PRs for the specification of Rh5 fate.
We next asked if Blimp-1 acts cell autonomously in R8 PRs to promote Rh5 fate. To this end, we performed an RNAi-mediated knockdown of Blimp-1 using the R8 driver sens-Gal4 and two copies of UAS-Blimp-1-RNAi. This again led to a dramatic gain of Rh6 and loss of Rh5 (Supplementary Figures S1G–J), suggesting that Blimp-1 is required cell autonomously in R8 PRs to promote Rh5 PR fate and to repress Rh6 PR fate. In a complementary approach, we took advantage of the fact that the specification of Rh5 fate requires an Activin signal from a subset of R7 PRs (Figure 1A) that activates the type I receptor Baboon (Babo) in the proximally located subset of R8 PRs (Wells et al., 2017). If Blimp-1 acts cell autonomously in R8 PRs to promote Rh5 fate, then Blimp-1 should act downstream of the Activin signal. Indeed, when we expressed a constitutively active form of Babo (Babo*) (Wells et al., 2017) in combination with an RNAi-mediated knockdown of Blimp-1, Babo activation was no longer able to specify Rh5 fate and we observed the Blimp-1 mutant phenotype (loss of Rh5 and gain of Rh6; Supplementary Figures S1K, L). In summary, these data suggest that Blimp-1 specifies Rh5 fate cell autonomously in early to mid-pupal R8 PRs downstream of the Activin signal and the Babo receptor.
3.3 Blimp-1 is required for the activation of melted and the repression of warts
Because the Hippo pathway is inactivated in the Rh5 PRs that require Blimp-1 for their specification (Mikeladze-Dvali et al., 2005; Jukam et al., 2013) and Blimp-1 is a transcriptional repressor (Keller and Maniatis, 1991; Yu et al., 2000; Agawa et al., 2007; Ozturk-Colak et al., 2018), we asked whether Blimp-1 represses the Hippo pathway in R8 PRs. A candidate target for Hippo pathway repression is its nexus wts, which is transcriptionally repressed in Rh5 PRs (Figure 2A) (Mikeladze-Dvali et al., 2005; Jukam et al., 2013; Pojer et al., 2021). We identified an enhancer in the second wts intron (Figure 3A; see Materials and methods) that was sufficient to recapitulate PR subtype-specific wts expression in the Rh6 PRs when fused to an egfp reporter gene (wts-GFP); this allowed us to test whether Blimp-1 is required to repress wts transcription in Rh5 PRs. Indeed, wts-GFP was de-repressed with Rh6 upon Blimp-1 knockdown (Figures 2B–C′, I; Supplementary Figures S2A–C), suggesting that Blimp-1 is required for the transcriptional repression of wts. Since melt is expressed in Rh5 PRs and repressed by Wts in Rh6 PRs, we tested if Blimp-1 is also required for melt activation in Rh5 PRs. Indeed, melt transcriptional reporter expression (Figures 2D, D′) was lost when Blimp-1 was knocked down (Figures 2E, E’, 2I). Taken together, these data suggest that Blimp-1 is required for both the repression of wts and the expression of melt.
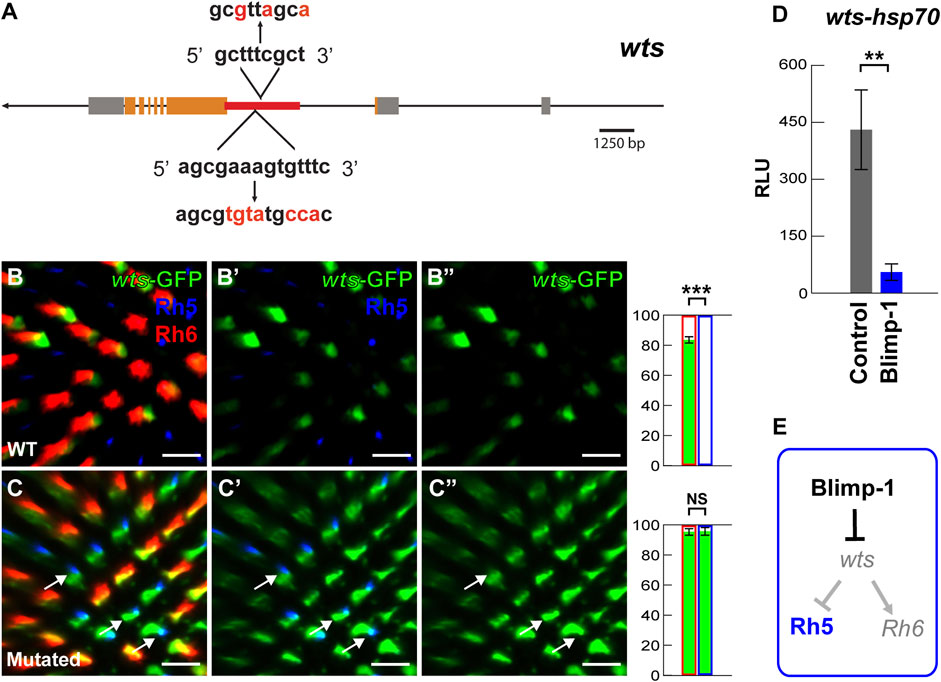
FIGURE 3. Blimp-1 represses the warts intronic enhancer. (A) Schematic of the wts locus. The red line indicates the location of the 2.6 kb wts enhancer in the second intron that contains two conserved Blimp-1 motifs. The red letters indicate introduced mutations. (B–B’) The 2.6 kb wts enhancer fused to an hsp70 promoter drives GFP (hereafter referred to as wts-GFP reporter) in the majority of Rh6 photoreceptors. Right: percentage of Rh6 photoreceptors (red outline) or Rh5 photoreceptors (blue outline) that co-express GFP (green bars), respectively. %Rh6/GFP co-expression vs. %Rh5/GFP co-expression was compared with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. N = 5 retinas. (C–C’) Mutation of the two conserved Blimp-1 motifs causes wts-GFP reporter de-repression in Rh5 photoreceptors. White arrows indicate examples of wts-GFP de-repression. Right: percentage of Rh6 photoreceptors or Rh5 photoreceptors that co-express GFP, respectively. N = 5 retinas. All scale bars, 10 µm. (D) Dual luciferase reporter assay. The intronic wts reporter (wts-hsp70-luc) is active in S2 cells, but Blimp-1 strongly represses it (N = 3). Y-axis: Relative Luminescence Units (RLU) of wts-hsp70-luc normalized to the Renilla luciferase control reporter (see Materials and methods). (E) Schematic of Blimp-1 function in color photoreceptor specification: Blimp-1 represses wts/Rh6 fate to promote Rh5 fate.
The wts and melt reporter results suggest that Blimp-1 acts genetically upstream of wts. To test this hypothesis, we performed epistasis experiments. Indeed, RNAi-mediated knockdown of wts caused a gain of Rh5 and loss of Rh6 (Figure 2F) even with concomitant knockdown of Blimp-1 (Figures 2G,J). Since wts knockdown reverses the Blimp-1 knockdown phenotype (gain of Rh6 and loss of Rh5), this provides further support that Blimp-1 acts upstream of wts. To corroborate this result, we analyzed merlin (mer) mutant clones. Mer is a FERM domain-containing protein that is required for Wts activity in R8 PRs, and mer mutant clones exclusively express Rh5 (Jukam and Desplan, 2011). Likewise, we found that mer; Blimp-1 double mutants also exclusively expressed Rh5 (Figure 2H, H’). In summary, the perturbation of Wts activity, either through RNAi-mediated knockdown of wts or mutation of mer, reverses the Blimp-1 mutant phenotype. These data show that Blimp-1 acts genetically upstream of wts.
Lastly, we investigated the possibility that Blimp-1 directly represses wts transcription. We identified two conserved motifs (Supplementary Figure S3A) in the wts intronic enhancer that match the Blimp-1 consensus motif AGNGAAAG (Kuo and Calame, 2004; Ancelin et al., 2006; Katoh et al., 2010) as well as the Drosophila Blimp-1 Position Weight Matrix (Zhu et al., 2011) (Figure 3A) (see Materials and methods). Strikingly, the mutation of the conserved Blimp-1 motifs caused wts-GFP reporter de-repression in Rh5 PRs (compare Figures 3B-B”, C-C”), suggesting that the Blimp-1 motifs are required for wts repression in Rh5 PRs. Consistent with these in vivo data, Blimp-1 dramatically reduced (∼9-fold) wts-hsp70-luc reporter expression in Drosophila S2 cells (Figure 3D) (see Material and methods). Taken all the data together, we propose that Blimp-1 represses Rh6 fate through repression of wts, the nexus of the Hippo pathway, and thereby promotes Rh5 fate (Figure 3E).
3.4 The role of Blimp-1 in wing growth is independent of the hippo pathway
Since the canonical role of the Hippo pathway is to regulate organ growth (Halder and Camargo, 2013) and we found that Blimp-1 represses the core component wts in the post-mitotic PR context, we asked whether Blimp-1 regulates the Hippo pathway in mitotically active tissue. While Blimp-1 knockdown caused a glossy eye phenotype (Wang et al., 2022) (Supplementary Figures S4A, B), it did not decrease the size of the adult eye (Supplementary Figures S4A, B), indicating that Blimp-1 does not regulate tissue growth in the developing eye. Moreover, knockdown of wts did not rescue the glossy eye phenotype (Supplementary Figure S4C), indicating that this Blimp-1 mutant phenotype is not related to the Hippo pathway. In addition, Blimp-1 knockdown in the developing wing caused a dramatic decrease in adult wing size with two different wing disc drivers (Figures 4A, B; Supplementary Figures S4D, E). However, in contrast to the terminal PR differentiation context, and consistent with the glossy eye phenotype, the wing size defect could not be modified by concomitant knockdown of wts (Figures 4C–E). Moreover, knockdown of Blimp-1 in the posterior half of the wing disc with engrailed-Gal4 did not have any obvious effects on the size of the third instar larval wing disc in comparison to the anterior half (Figure 4F), suggesting that Blimp-1 likely regulates wing development during pupal stages, similar to its pupal role in post-mitotic PRs. Again, in contrast to post-mitotic PRs, this other role of Blimp-1 does not appear to involve regulation of Hippo pathway activity: expanded-LacZ (Yu and Pan, 2018), a transcriptional reporter of Yki activity, was unaffected when Blimp-1 was knocked down in the posterior half of the developing wing disc (Figures 4G, G’). Furthermore, the intronic wts enhancer that is repressed by Blimp-1 in post-mitotic PRs was not detectable in the wing disc (Supplementary Figure S4F). Together, these data suggest that Blimp-1 is required for proper wing growth, but this function appears to be independent of the Hippo pathway.
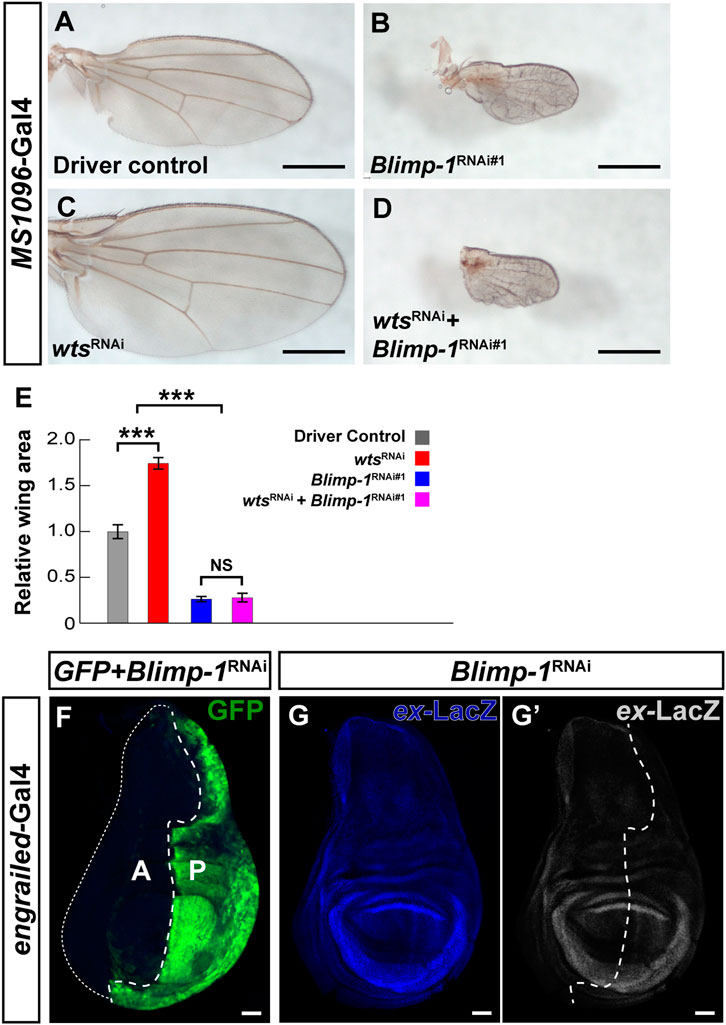
FIGURE 4. Blimp-1 plays a role in wing development independent of the Hippo pathway. (A–D) Wings of 2–4 days-old female flies raised at 25°C. Scale bars, 500 µm. (A) Wing disc driver control MS1096-Gal4/+. (B) Blimp-1 knockdown causes a dramatic reduction in wing size. (C) wts knockdown causes an increase in wing size. (D) Knockdown of both Blimp-1 and wts resembles the Blimp-1 knockdown. (E) Quantification of wing area for each genotype normalized to the MS1096-Gal4 driver control. Relative wing areas were compared with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 6-8 wings were scored for each genotype. (F) Confocal image of a third instar larval wing disc with engrailed-Gal4 driving GFP and Blimp-1RNAi#1 in the posterior half. Blimp-1 knockdown does not appear to affect the size of the posterior half of the wing disc (marked by GFP) compared to the anterior half of the wing disc (GFP negative). White dashed line encircles the anterior half of the wing disc. Anterior is labeled with “A”, and posterior is labeled with “P”. (G) Blimp-1 knockdown does not affect expanded-lacZ reporter expression in the posterior wing disc. (G′) Expanded-LacZ staining in grayscale. White dotted line indicates the approximate boundary between anterior and posterior halves based on the GFP staining in (F). All scale bars for larval wing discs, 50 µm.
3.5 Ecdysone signaling is cell autonomously required for blue-sensitive photoreceptor fate
Blimp-1 is activated by the steroid hormone ecdysone (Agawa et al., 2007; Akagi and Ueda, 2011; Akagi et al., 2016; Ozturk-Colak et al., 2018) and Blimp-1 expression in the pupal retina requires the ecdysone receptor (EcR) (Wang et al., 2022). Therefore, we analyzed whether ecdysone signaling is required to specify Rh5 fate by performing RNAi-mediated knockdown of EcR with lGMR-Gal4. Closely resembling the Blimp-1 mutant phenotype, EcR knockdown with an RNAi construct that targets all EcR isoforms caused a complete loss of Rh5 PRs and a gain of Rh6 PRs (Figures 5A,B). Since there are three EcR isoforms (EcR-A, EcR-b1, and EcR-b2) that have identical DNA binding domains but differ in their N-terminal A/B domains that allow them to elicit differential transcriptional responses (Mouillet et al., 2001; Schubiger et al., 2003), we additionally performed isoform-specific knockdowns. The knockdown of EcR-A and EcR-b1 (an RNAi line specifically targeting EcR-b2 was not available) each caused a loss of Rh5 PRs and gain of Rh6 PRs (Figures 5C, D), respectively, suggesting that both isoforms are non-redundantly required to specify Rh5 fate. Similarly, EcR knockdown using the R8 PR driver sens-Gal4 also caused a loss of Rh5 PRs and gain of Rh6 PRs (Figures 5E, F). Taken together (Figure 5G), ecdysone signaling is cell autonomously required in R8 PRs to specify Rh5 fate and to repress Rh6 fate.
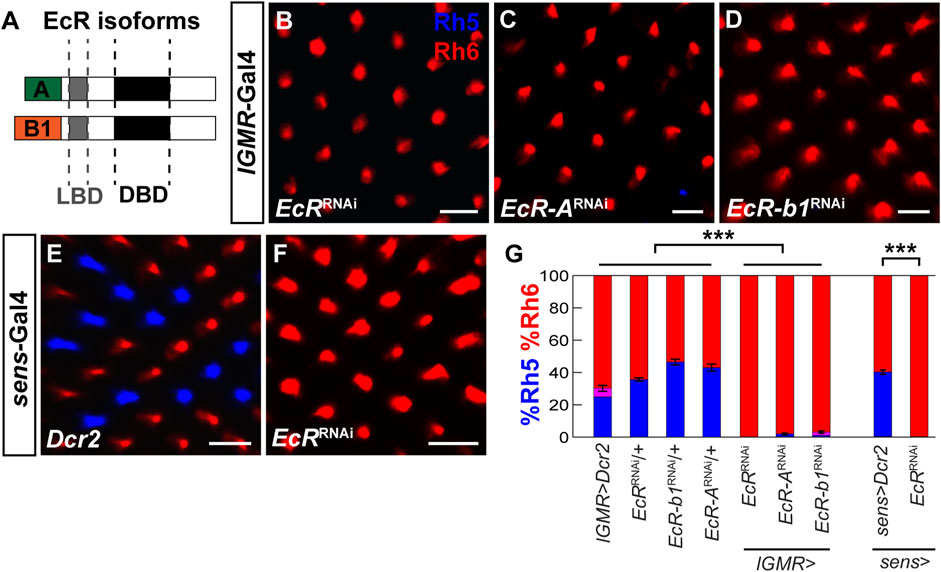
FIGURE 5. EcR is required cell autonomously to specify Rh5 fate. (A) Schematic of two EcR isoforms, EcR-A and EcR-b1, which contain identical DNA binding domains (DBD) and ligand binding domains (LBD) but differ in their N-terminal A/B domains. (B) Pan-photoreceptor knockdown of all EcR isoforms causes a loss of Rh5 and a gain of Rh6. (C, D) Pan-photoreceptor knockdown of the individual EcR isoforms EcR-A (C) and EcR-b1 (D) causes a loss of Rh5 and gain of Rh6. (E) Expression of Dcr2 with the R8 driver sens-Gal4 at 29°C does not affect the Rh5:Rh6 ratio. (F) Expression of EcRRNAi and Dcr2 with sens-Gal4 at 29°C causes a loss of Rh5 and gain of Rh6. All scale bars, 10 µm. (G) Quantification of R8 subtypes in controls vs. EcR knockdowns. Mean %Rh6 was compared among genotypes with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 5-8 retinas were scored for each genotype.
3.6 Hr3 acts cell autonomously in R8 photoreceptors to promote blue-sensitive photoreceptor fate
In the developing mouse retina, the nuclear receptor RORβ and the transcription factor Otx2 activate Blimp-1 in retinal progenitor cells to repress bipolar interneuron fate and promote rod PR fate (Jia et al., 2009; Brzezinski et al., 2010; Katoh et al., 2010; Brzezinski et al., 2013; Wang et al., 2014; Goodson et al., 2020). Because the Drosophila ortholog of Otx2, Otd, is required for Rh5 fate (McDonald et al., 2010; Jukam et al., 2013), we asked whether the Drosophila ortholog of RORβ, the ecdysone-responsive Hormone Receptor 3 (Hr3) (Kageyama et al., 1997; Lam et al., 1999) is also required for Rh5 fate. Indeed, RNAi-mediated knockdown of Hr3 with two different RNAi lines combined with the lGMR-Gal4 driver (Figures 6A, B) or the sens-Gal4 driver (Figures 6C, D) caused a nearly complete loss of Rh5 PRs and gain of Rh6 PRs. Next, we hypothesized that Hr3 represses the Hippo pathway in Rh5 PRs. Consistent with this hypothesis, Hr3 knockdown caused a significant de-repression of the wts-GFP reporter (Figures 6E–F’) and concomitant knockdown of wts reversed the Hr3 knockdown phenotype (Figure 6G). As expected from the gain of wts, Hr3 knockdown also caused a loss of melt-GFP reporter expression (Supplementary Figures S5A–C). Moreover, Hr3 significantly reduced wts-hsp70-luc reporter expression in Drosophila S2 cells, albeit to a lesser extent (∼3 fold) than Blimp-1 (Supplementary Figure SD). In summary (Figures 6H, I), these data suggest that both Blimp-1 and Hr3 are cell autonomously required to specify Rh5 fate in R8 PRs by repressing wts (Figure 6J).
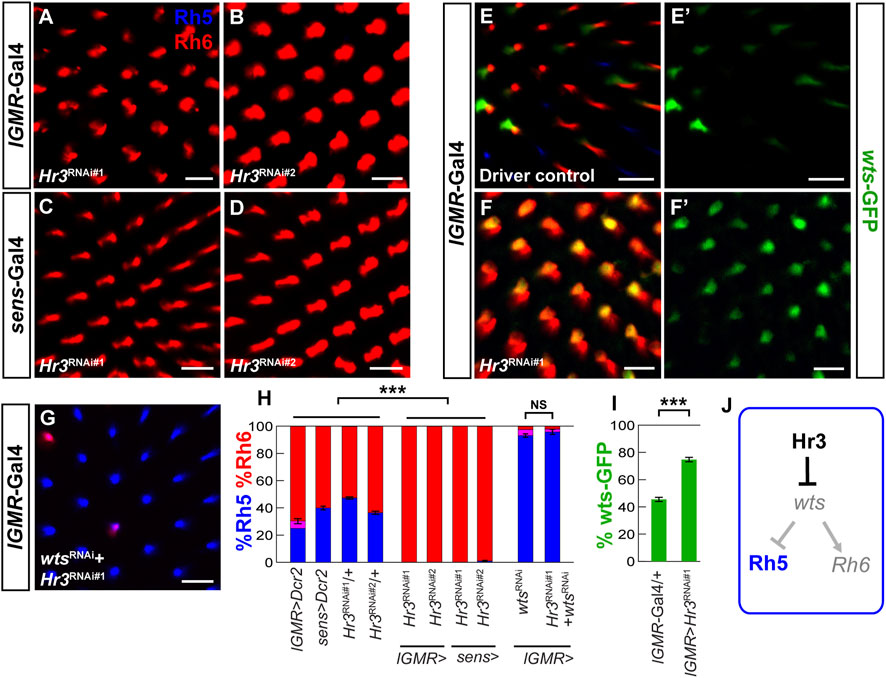
FIGURE 6. Hr3/RORβ is required to repress warts and to specify Rh5 fate.(A,B) Pan-photoreceptor knockdown of Hr3 using two separate RNAi constructs, TRiP#27253 (Hr3RNAi#1) and TRiP#27254 (Hr3RNAi#2) causes a complete loss of Rh5 and gain of Rh6. (C,D) R8 photoreceptor knockdown with two separate RNAi constructs also causes a dramatic loss of Rh5 and gain of Rh6. (E,E′) In the heterozygous lGMR-Gal4/+ driver control, the wts-GFP transcriptional reporter is expressed in most Rh6 photoreceptors. (F, F′) Pan-photoreceptor knockdown of Hr3 causes a de-repression of the wts-GFP reporter together with Rh6. (G) Pan-photoreceptor knockdown of both Hr3 and wts causes a gain of Rh5 and loss of Rh6, resembling the wts knockdown phenotype. All scale bars, 10 µm. (H) Quantification of R8 subtypes in controls and Hr3 knockdowns. Mean %Rh6 was compared among genotypes with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 5-8 retinas were scored for each genotype. (I) Quantification of the GFP-expressing R8 photoreceptors in the driver control and Hr3 knockdown. Mean %wts-GFP reporter expression in driver control vs. knockdown was compared with an ANOVA and a post hoc Tukey HSD Test; ***p < 0.0001. 6 retinas were analyzed for each genotype. (J) Schematic of Hr3 function in Rh5 photoreceptors: Hr3 represses wts and thereby promotes Rh5 fate.
We next asked whether Hr3 is required for Blimp-1 expression. To this end, we performed RNAi-mediated knockdown of Hr3 and assessed the expression of Blimp-1 in pupal PRs. While Blimp-1 knockdown abolished Blimp-1 expression, Hr3 knockdown did not affect Blimp-1 expression compared to the driver control (Supplementary Figures S5E–G’). Therefore, Hr3 is not required for Blimp-1 expression in pupal PRs. Taken together, these data suggest that Hr3 promotes Rh5 fate by acting in parallel with Blimp-1 to repress wts (Figure 7B).
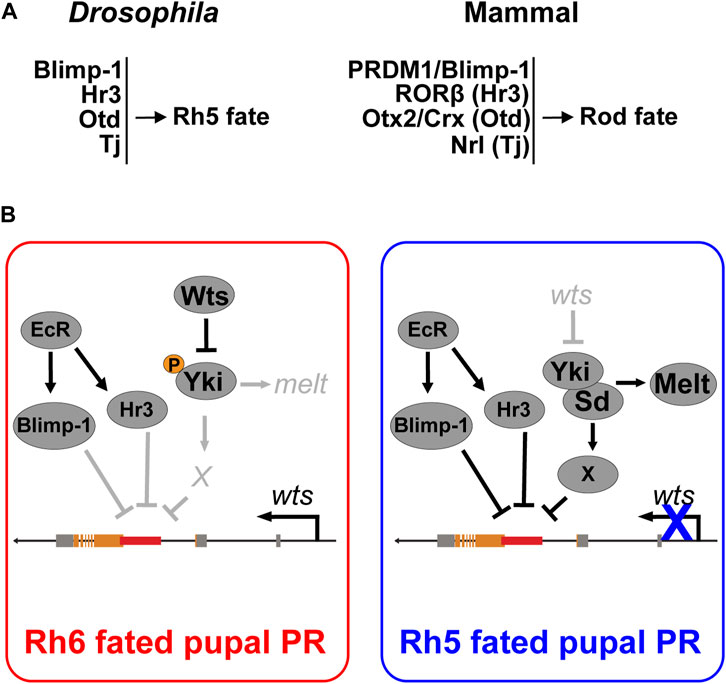
FIGURE 7. Cell fate decisions in the Drosophila and mammalian retina. (A) Summary of conserved transcription factors that are necessary to specify Rh5 photoreceptor fate in Drosophila and rod photoreceptor fate in mammals. (B) Model for Blimp-1’s function as a permissive factor that represses wts at the transcriptional level in parallel with the ecdysone-responsive Hr3. Blimp-1 is expressed in pupal PRs in an EcR-dependent manner, but when Yki is not activated, Blimp-1 is not sufficient to repress wts and thereby gives rise to Rh6 fate. However, when Yki is transiently activated, possibly in response to the TGFβ signal from the distal R7 photoreceptor, Yki/Sd activate an unknown Rh5 subtype-specific transcription factor “X” that acts in combination with Blimp-1 and Hr3 to repress the wts enhancer, thereby silencing wts transcription and giving rise to robust Yki activation as well as Rh5 fate.
4 Discussion
4.1 Conserved transcription factors control binary cell fate decisions in the mammalian and the Drosophila retina
Here, we analyzed the regulatory mechanisms that specify two related Drosophila PR subtypes that express different color-sensing pigments (Rh5 or Rh6) in a mutually exclusive manner. We discovered that the Drosophila orthologs of the mammalian rod PR fate determinants Blimp-1/PRDM1 (Brzezinski et al., 2010; Katoh et al., 2010; Brzezinski et al., 2013; Goodson et al., 2020) and Hr3/RORβ (Wang et al., 2014) also play a role in terminal Drosophila PR specification, but in the cone-equivalent “inner” R8 PRs rather than the rod-equivalent “outer” R1-R6 PRs. In the binary R8 PR subtype decision, Blimp-1 and Hr3 promote the blue-sensitive/Rh5 PR fate and repress the green-sensitive/Rh6 PR fate by repressing wts and activating melt.
A previous study had unraveled that Otd, the ortholog of the mammalian PR fate determinants Otx2 and Crx (McDonald et al., 2010), and Tj, the ortholog of the mammalian rod fate determinant Nrl, are also required to specify Rh5 fate (Jukam et al., 2013). Otd and Tj form a coherent feedforward loop that allows Yki/Sd to activate melt and to repress wts (Jukam et al., 2013). In the mammalian retina, the Otd and Tj orthologs Otx2/Crx and Nrl, respectively, promote rod PR fate: Otx2 and Crx are both necessary for the expression of Nrl (Montana et al., 2011; Roger et al., 2014), which is necessary and sufficient for rod PR fate (Mears et al., 2001; Oh et al., 2007). NRL mutations have been associated with retinitis pigmentosa (Bessant et al., 1999) and mutations in Blimp-1, RORβ, Otx2, Crx, or Nrl are associated with a loss of rod PRs in mammals (Mears et al., 2001; Nishida et al., 2003; Daniele et al., 2005; Koike et al., 2007; Jia et al., 2009; Katoh et al., 2010). Likewise, loss of Blimp-1, Hr3, Otd, or Tj are each associated with a loss of blue-sensing PR fate (this study) (Jukam et al., 2013). Although the Drosophila eye and the mammalian eye seem to use different mechanisms for eye and PR development (Rister and Desplan, 2011; Cepko, 2015; Eldred et al., 2018), our current study and previous results (Jukam et al., 2013) suggest that a conserved set of transcription factors is used in both animal groups for specific binary PR fate decisions (Figure 7A).
While Blimp-1/PRDM1 and Hr3/RORβ promote rod fate in mammals and blue-sensitive/Rh5 PR fate in Drosophila respectively, the mechanisms by which they regulate these cell fate decisions differ: in mammals, Hr3/RORβ is required to activate Blimp-1/PRDM1 to repress bipolar fate and specify rod fate (Wang et al., 2014). However, in developing Drosophila PRs, Hr3 does not regulate Blimp-1 expression (Supplementary Figures SE–G’) but rather acts in parallel with Blimp-1 to repress wts. Given that conserved Blimp-1 motifs are required to repress wts in blue-sensitive/Rh5 PRs and Blimp-1 represses a wts-hsp70-luc reporter in vitro (Figures 3B–D), it is likely that Blimp-1 represses the wts enhancer directly. Since we did not find conserved Hr3 motifs in the wts enhancer, future studies will have to analyze the in vivo relevance of the Hr3-mediated wts repression that we found in cultured cells.
4.2 Blimp-1 acts as a permissive factor to promote blue-sensitive photoreceptor fate
Blimp-1/PRDM1 controls cell fate decisions in diverse developmental contexts (Bikoff et al., 2009). In pupal Drosophila PRs, Blimp-1 expression is ecdysone-dependent and regulates the terminal differentiation of the eye non-autonomously in non-neuronal cells (Wang et al., 2022). In the current study, we revealed a novel cell autonomous role of Blimp-1 in the terminal differentiation of color-sensing PR neurons. Blue-sensitive PR fate requires ecdysone signaling (EcR) and the ecdysone-responsive regulators Blimp-1 and Hr3, which both act to promote Rh5 PR fate. In contrast, in the larval fat body, Blimp-1 and Hr3 play antagonistic roles in regulating the regulatory gene ftz-f1 to control pupation timing: Blimp1 represses ftz-f1, while Hr3 activates ftz-f1 (Kageyama et al., 1997; Lam et al., 1999; Agawa et al., 2007).
The R8 PR specification network involves several permissive transcription factors that are not restricted to one of the two subtypes. Blimp-1 is transiently expressed during the early differentiation of both R8 PR subtypes, where it on the one hand represses wts in Rh5-fated PRs but on the other hand permits wts expression in Rh6-fated PRs. Likewise, Otd and Tj are expressed in both Rh5- and Rh6-fated PRs and act as permissive factors for Rh5 fate. Yki/Sd are unable to activate melt in the absence of Otd or Tj, or to activate Rh5 in the absence of Otd (Jukam et al., 2013). However, Tj and Otd are not sufficient to activate melt in the absence of Yki or Sd, and Otd is not sufficient to activate Rh5 (Jukam et al., 2013). Therefore, the conserved PR fate specification module is required to establish a post-mitotic context wherein melt and wts can function as a bi-stable switch to rewire the Hippo pathway. The proposed context-specificity is consistent with the finding that Blimp-1 represses wts in the post-mitotic PR context, but not in the wing growth context, and that the intronic wts enhancer drives expression in Rh6 PRs but not in the wing disc.
Since Blimp-1 acts as a permissive factor that represses wts in Rh5-fated PRs, a possible regulatory scenario is that an unknown transcription factor is specifically expressed in the Rh5-fated PRs and acts in combination with Blimp-1 to repress wts, analogous to how Yki/Sd activate melt in combination with Otd and Tj. Since Yki is active in Rh5 PRs but not in Rh6 PRs (Jukam et al., 2013), another possibility is that Yki is transiently activated in Rh5-fated pupal PRs, and then acts in combination with Blimp-1 to repress wts transcriptionally, permanently inactivating the Hippo pathway to give rise to robust Yki activation and Rh5 fate. Alternatively, we propose a scenario that contains an intermediate step (Figure 7B) in which Yki/Sd activate an additional transcription factor that acts with Blimp-1 to repress wts in a combinatorial manner.
In conclusion, the Drosophila color PR subtype specification is an excellent model to study how terminal cell fate decisions mediate the differential expression of sensory receptor proteins in related subsets of sensory neurons. The analysis of the underlying mechanisms gives insights into how conserved transcription factors generate sensory neuron diversity and potentially inform treatments for diseases that affect specific sensory neuron types.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JR and JB conceived the experiments, analyzed the data, and wrote the article. JR obtained financial support. JB, JR, SB, AP, MB, PB, RD, EN, GC, MT, and SS collected the data. All authors read and approved the final article.
Funding
This work was supported by an R00/Pathway to Independence Award (R00EY023995) to JR from the NEI/NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding to SGS was provided by the Swiss National Science foundation grant 310030_188471.
Acknowledgments
We thank Steve Britt, Claude Desplan, and Sudipto Roy for antibodies, as well as Johannes Bischof and Konrad Basler (UZH) for the J36 landing site stock.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1058961/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Blimp-1 acts in mid-pupal R8 photoreceptors. (A-F’) Confocal images of whole mounted retinas from different pupal timepoints stained with antibodies for Elav (red, photoreceptor nuclei) and Blimp-1 (blue). (A,A’) Blimp-1 is weakly expressed in photoreceptors at 24 h after puparium formation. Note the partial overlap with the Elav nuclear marker (white arrow). Inset shows ommatidium of the arrowed photoreceptor nucleus. (B,B’) Blimp-1 is strongly expressed at 48 h after puparium formation. White arrow indicates partial Blimp-1/Elav overlap; inset shows ommatidium of the arrowed photoreceptor nucleus. (C,C’) Blimp-1 is absent in R8 photoreceptors at 72 h after puparium formation. (D,D’) Blimp-1 is absent in adult R8 photoreceptors. (E,E’) Blimp-1 expression at 48 h after puparium formation in the heterozygous lGMR-Gal4; UAS-Dcr2 driver control. Note individual strong Blimp-1-positive nuclei (one example is indicated by white arrow). (F,F’) Pan-photoreceptor knockdown of Blimp-1 causes a loss of Blimp-1 staining. (G–I,K,L) Confocal images of whole mounted retinas from adult males raised at 29°C. (G) In driver control males with hemizygous UAS-Dcr2 on the X chromosome and heterozygous sens-Gal4 on the third chromosome, the ratio of Rh5:Rh6 is normal. (H) Blimp-1 knockdown with two copies of UAS-Blimp-1RNAi#1, one copy of sens-Gal4, and hemizygous UAS-Dcr2 causes a loss of Rh5 and gain of Rh6. (I) The Rh5:Rh6 ratio is normal in the homozygous UAS-Blimp-1RNAi#1 control. (J) Quantification of R8 subtypes in controls and Blimp-1 knockdown. Graph shows %R8 photoreceptors that express Rh5 exclusively (blue), Rh6 exclusively (red), or co-express Rh5 and Rh6 (magenta). Mean %Rh6 was compared among genotypes with an ANOVA and a post-hoc Tukey HSD Test; *** p<0.0001. 8 retinas were scored for each genotype. (K) Pan-photoreceptor expression of a constitutively active form of Babo (Babo*) causes a gain of Rh5 and loss of Rh6. (L) Pan-photoreceptor Blimp-1 knockdown causes a loss of Rh5 and gain of Rh6 even with concomitant ectopic Babo* expression. All scale bars, 10 µm.
SUPPLEMENTARY FIGURE S2 | Blimp-1 knockdown de-represses wts-GFP. (A) Longitudinal section of the heterozygous lGMR-Gal4 driver control with a wts-GFP transcriptional reporter. Note that wts-GFP overlaps with Rh6 and not with Rh5; white arrow indicates an Rh6-expressing photoreceptor that does not express wts-GFP, suggesting that the heterozygous wts reporter is not strong enough to label all Rh6 photoreceptors. (A’) Rh5 channel. (A’’) Rh6 channel. (A’’’) wts-GFP channel (grayscale). (B) Longitudinal section of Blimp-1RNAi#1 expressed with lGMR-Gal4 in the presence of a wts-GFP transcriptional reporter. There is a loss of Rh5 and gain of Rh6, and wts-GFP expands with Rh6. Morphological defects caused by Blimp-1 knockdown made it difficult to observe the wts-GFP reporter; longitudinal sections circumvented this issue. Similar to the driver control, there were occasional instances of Rh6-expressing photoreceptors that did not express wts-GFP (white arrow). (B’) Rh5 channel. (B’’) Rh6 channel. (B’’’) wts-GFP channel (grayscale). (C) Quantification of the percentage of Rh6 photoreceptors (red outline) that co-express GFP (green bars) in the lGMR-Gal4 driver control and Blimp-1 knockdown, respectively. %wts-GFP/Rh6 co-expression was compared for the two genotypes with an ANOVA and a post-hoc Tukey HSD Test; *** p<0.0001. N = 6 retinas.
SUPPLEMENTARY FIGURE S3 | Conservation analysis of Blimp-1 motifs in the wts intron. (A) Alignments of the two Blimp-1 motif regions located in the intronic wts enhancer of ten Drosophila species. Blue indicates parts of the Blimp-1 motifs matching the consensus AGNGAAAG.
SUPPLEMENTARY FIGURE S4 | Blimp-1 does not regulate wts in a growth context. (A,B) Knockdown of Blimp-1 using the eye driver GMR-Gal4 did not affect adult eye size compared to the heterozygous driver control. Note the glossy eye phenotype caused by Blimp-1 knockdown due to its role in corneal lens formation (Wang et al., 2022). (C) Concomitant knockdown of wts with Blimp-1 using GMR-Gal4 did not affect the glossy eye phenotype that is caused by Blimp-1 knockdown. (D,E) Knockdown of Blimp-1 using the wing disc driver nubbin-Gal4 causes a dramatic decrease in wing size compared to the heterozygous driver control. Scale bars, 500 µm (E) The intronic wts-GFP reporter that is expressed in post-mitotic photoreceptors is not active in the third instar larval wing disc. White dashed line indicates the edge of the wing disc. Scale bar, 50 µm.
Supplementary Figure S5 | Hr3 regulates melt and wts but not Blimp-1. (A) In the heterozygous lGMR-Gal4 driver control, melt-GFP is expressed in most Rh5 PRs and absent from Rh6 PRs. (A’) GFP channel. White arrows indicate examples of Rh5/melt-GFP co-expression. (B) Knockdown of Hr3 using lGMR-Gal4 causes a loss of melt-GFP. (B’) GFP channel. All scale bars, 10 µm. (C) Quantification of the GFP-expressing R8 photoreceptors in the driver control and Hr3 knockdown. Mean %melt-GFP reporter expression in driver control vs. knockdown was compared with an ANOVA and a post-hoc Tukey HSD Test; *** p<0.0001. 6 retinas were analyzed for each genotype. (D) Dual luciferase reporter assay. The intronic wts reporter (wts-hsp70-luc) is active in S2 cells, but Hr3 represses it (N = 3). Y-axis: Relative Luminescence Units (RLU) of wts-hsp70-luc normalized to the Renilla luciferase control reporter (see Materials and methods). (E-G’) Pupal retinas with lGMR-Gal4 at 48 h after puparium formation stained for Blimp-1 (blue) and the nuclear marker Elav (red). (E) The heterozygous lGMR-Gal4; UAS-Dcr2 driver control shows Blimp-1 expression with occasional strong nuclear localization. White arrow indicates nuclear Blimp-1. (E’) Blimp-1 channel. (F) Pan-photoreceptor knockdown of Blimp-1 abolishes Blimp-1 signal. (F’) Blimp-1 channel. (G) Pan-photoreceptor knockdown of Hr3 does not affect Blimp-1 expression. Moreover, there was occasional strong nuclear localization, similar to the driver control, suggesting that Hr3 is neither required for Blimp-1 expression, nor its nuclear localization. White arrow indicates nuclear Blimp-1. (G’) Blimp-1 channel.
References
Agawa, Y., Sarhan, M., Kageyama, Y., Akagi, K., Takai, M., Hashiyama, K., et al. (2007). Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol. Cell Biol. 27 (24), 8739–8747. doi:10.1128/MCB.01304-07
Akagi, K., Sarhan, M., Sultan, A. R., Nishida, H., Koie, A., Nakayama, T., et al. (2016). A biological timer in the fat body comprising Blimp-1, βFtz-f1 and Shade regulates pupation timing in Drosophila melanogaster. Development 143 (13), 2410–2416. doi:10.1242/dev.133595
Akagi, K., and Ueda, H. (2011). Regulatory mechanisms of ecdysone-inducible Blimp-1 encoding a transcriptional repressor that is important for the prepupal development in Drosophila. Dev. Growth Differ. 53 (5), 697–703. doi:10.1111/j.1440-169X.2011.01276.x
Ancelin, K., Lange, U. C., Hajkova, P., Schneider, R., Bannister, A. J., Kouzarides, T., et al. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8 (6), 623–630. doi:10.1038/ncb1413
Bessant, D. A., Payne, A. M., Mitton, K. P., Wang, Q. L., Swain, P. K., Plant, C., et al. (1999). A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat. Genet. 21 (4), 355–356. doi:10.1038/7678
Bikoff, E. K., Morgan, M. A., and Robertson, E. J. (2009). An expanding job description for Blimp-1/PRDM1. Curr. Opin. Genet. Dev. 19 (4), 379–385. doi:10.1016/j.gde.2009.05.005
Bischof, J., Maeda, R. K., Hediger, M., Karch, F., and Basler, K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 104 (9), 3312–3317. doi:10.1073/pnas.0611511104
Brzezinski, J. A. t., Lamba, D. A., and Reh, T. A. (2010). Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137 (4), 619–629. doi:10.1242/dev.043968
Brzezinski, J. A. t., Uoon Park, K., and Reh, T. A. (2013). Blimp1 (Prdm1) prevents re-specification of photoreceptors into retinal bipolar cells by restricting competence. Dev. Biol. 384 (2), 194–204. doi:10.1016/j.ydbio.2013.10.006
Cepko, C. L. (2015). The determination of rod and cone photoreceptor fate. Annu. Rev. Vis. Sci. 1, 211–234. doi:10.1146/annurev-vision-090814-121657
Chan, S. W., Lim, C. J., Chen, L., Chong, Y. F., Huang, C., Song, H., et al. (2011). The Hippo pathway in biological control and cancer development. J. Cell Physiol. 226 (4), 928–939. doi:10.1002/jcp.22435
Chou, W. H., Hall, K. J., Wilson, D. B., Wideman, C. L., Townson, S. M., Chadwell, L. V., et al. (1996). Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17 (6), 1101–1115. doi:10.1016/s0896-6273(00)80243-3
Chou, W. H., Huber, A., Bentrop, J., Schulz, S., Schwab, K., Chadwell, L. V., et al. (1999). Patterning of the R7 and R8 photoreceptor cells of Drosophila: Evidence for induced and default cell-fate specification. Development 126 (4), 607–616. doi:10.1242/dev.126.4.607
Clark, A. G., Eisen, M. B., Smith, D. R., Bergman, C. M., Oliver, B., Markow, T. A., et al. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 (7167), 203–218. doi:10.1038/nature06341
Daniele, L. L., Lillo, C., Lyubarsky, A. L., Nikonov, S. S., Philp, N., Mears, A. J., et al. (2005). Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest. Ophthalmol. Vis. Sci. 46 (6), 2156–2167. doi:10.1167/iovs.04-1427
Eldred, K. C., Hadyniak, S. E., Hussey, K. A., Brenerman, B., Zhang, P. W., Chamling, X., et al. (2018). Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348. doi:10.1126/science.aau6348
Engels, W. R., Johnson-Schlitz, D. M., Eggleston, W. B., and Sved, J. (1990). High-frequency P element loss in Drosophila is homolog dependent. Cell 62 (3), 515–525. doi:10.1016/0092-8674(90)90016-8
Fan, X., Abbott, T. E., Larson, D., and Chen, K. (2014). BreakDancer: Identification of genomic structural variation from paired-end read mapping. Curr. Protoc. Bioinforma. 45, 1–11. doi:10.1002/0471250953.bi1506s45
Fehon, R. G., Oren, T., LaJeunesse, D. R., Melby, T. E., and McCartney, B. M. (1997). Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics 146 (1), 245–252. doi:10.1093/genetics/146.1.245
Fichelson, P., Brigui, A., and Pichaud, F. (2012). Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc. Natl. Acad. Sci. U. S. A. 109, 7893–7898. doi:10.1073/pnas.1120276109
Fortini, M. E., and Rubin, G. M. (1990). Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 4 (3), 444–463. doi:10.1101/gad.4.3.444
Goodson, N. B., Park, K. U., Silver, J. S., Chiodo, V. A., Hauswirth, W. W., and Brzezinski, J. A. (2020). Prdm1 overexpression causes a photoreceptor fate-shift in nascent, but not mature, bipolar cells. Dev. Biol. 464 (2), 111–123. doi:10.1016/j.ydbio.2020.06.003
Halder, G., and Camargo, F. D. (2013). The hippo tumor suppressor network: From organ size control to stem cells and cancer. Cancer Res. 73 (21), 6389–6392. doi:10.1158/0008-5472.CAN-13-2392
Halder, G., and Johnson, R. L. (2011). Hippo signaling: Growth control and beyond. Development 138 (1), 9–22. doi:10.1242/dev.045500
Hamaratoglu, F., Willecke, M., Kango-Singh, M., Nolo, R., Hyun, E., Tao, C., et al. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8 (1), 27–36. doi:10.1038/ncb1339
Hao, H., Kim, D. S., Klocke, B., Johnson, K. R., Cui, K., Gotoh, N., et al. (2012). Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 8 (4), e1002649. doi:10.1371/journal.pgen.1002649
Harvey, K. F., and Hariharan, I. K. (2012). The hippo pathway. Cold Spring Harb. Perspect. Biol. 4, a011288. doi:10.1101/cshperspect.a011288
Hofer, H., Carroll, J., Neitz, J., Neitz, M., and Williams, D. R. (2005). Organization of the human trichromatic cone mosaic. J. Neurosci. 25 (42), 9669–9679. doi:10.1523/JNEUROSCI.2414-05.2005
Hsiao, H. Y., Johnston, R. J., Jukam, D., Vasiliauskas, D., Desplan, C., and Rister, J. (2012). Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J. Vis. Exp., 4347. doi:10.3791/4347
Huang, J., Wu, S., Barrera, J., Matthews, K., and Pan, D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122 (3), 421–434. doi:10.1016/j.cell.2005.06.007
Jia, L., Oh, E. C., Ng, L., Srinivas, M., Brooks, M., Swaroop, A., et al. (2009). Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc. Natl. Acad. Sci. U. S. A. 106 (41), 17534–17539. doi:10.1073/pnas.0902425106
Jukam, D., and Desplan, C. (2011). Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev. Cell 21 (5), 874–887. doi:10.1016/j.devcel.2011.10.004
Jukam, D., Xie, B., Rister, J., Terrell, D., Charlton-Perkins, M., Pistillo, D., et al. (2013). Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342 (6155), 1238016. doi:10.1126/science.1238016
Kageyama, Y., Masuda, S., Hirose, S., and Ueda, H. (1997). Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes cells. 2 (9), 559–569. doi:10.1046/j.1365-2443.1997.1460344.x
Katoh, K., Omori, Y., Onishi, A., Sato, S., Kondo, M., and Furukawa, T. (2010). Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci. 30 (19), 6515–6526. doi:10.1523/JNEUROSCI.0771-10.2010
Keller, A. D., and Maniatis, T. (1991). Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5 (5), 868–879. doi:10.1101/gad.5.5.868
Koike, C., Nishida, A., Ueno, S., Saito, H., Sanuki, R., Sato, S., et al. (2007). Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol. Cell Biol. 27 (23), 8318–8329. doi:10.1128/MCB.01209-07
Kuo, T. C., and Calame, K. L. (2004). B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 173 (9), 5556–5563. doi:10.4049/jimmunol.173.9.5556
Lam, G., Hall, B. L., Bender, M., and Thummel, C. S. (1999). DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev. Biol. 212 (1), 204–216. doi:10.1006/dbio.1999.9343
McDonald, E. C., Xie, B., Workman, M., Charlton-Perkins, M., Terrell, D. A., Reischl, J., et al. (2010). Separable transcriptional regulatory domains within Otd control photoreceptor terminal differentiation events. Dev. Biol. 347 (1), 122–132. doi:10.1016/j.ydbio.2010.08.016
McGuire, S. E., Roman, G., and Davis, R. L. (2004). Gene expression systems in Drosophila: A synthesis of time and space. Trends Genet. 20 (8), 384–391. doi:10.1016/j.tig.2004.06.012
Mears, A. J., Kondo, M., Swain, P. K., Takada, Y., Bush, R. A., Saunders, T. L., et al. (2001). Nrl is required for rod photoreceptor development. Nat. Genet. 29 (4), 447–452. doi:10.1038/ng774
Mikeladze-Dvali, T., Wernet, M. F., Pistillo, D., Mazzoni, E. O., Teleman, A. A., Chen, Y. W., et al. (2005). The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122 (5), 775–787. doi:10.1016/j.cell.2005.07.026
Montana, C. L., Lawrence, K. A., Williams, N. L., Tran, N. M., Peng, G. H., Chen, S., et al. (2011). Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. J. Biol. Chem. 286 (42), 36921–36931. doi:10.1074/jbc.M111.279026
Mouillet, J. F., Henrich, V. C., Lezzi, M., and Vogtli, M. (2001). Differential control of gene activity by isoforms A, B1 and B2 of the Drosophila ecdysone receptor. Eur. J. Biochem. 268 (6), 1811–1819. doi:10.1046/j.1432-1327.2001.02051.x
Nathans, J. (1999). The evolution and physiology of human color vision: Insights from molecular genetic studies of visual pigments. Neuron 24 (2), 299–312. doi:10.1016/s0896-6273(00)80845-4
Neto-Silva, R. M., de Beco, S., and Johnston, L. A. (2010). Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell 19 (4), 507–520. doi:10.1016/j.devcel.2010.09.009
Newsome, T. P., Asling, B., and Dickson, B. J. (2000). Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127 (4), 851–860. doi:10.1242/dev.127.4.851
Ng, T., Yu, F., and Roy, S. (2006). A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Dev. Genes Evol. 216 (5), 243–252. doi:10.1007/s00427-005-0044-5
Nishida, A., Furukawa, A., Koike, C., Tano, Y., Aizawa, S., Matsuo, I., et al. (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6 (12), 1255–1263. doi:10.1038/nn1155
O'Tousa, J. E., Baehr, W., Martin, R. L., Hirsh, J., Pak, W. L., and Applebury, M. L. (1985). The Drosophila ninaE gene encodes an opsin. Cell 40 (4), 839–850. doi:10.1016/0092-8674(85)90343-5
Oh, E. C., Khan, N., Novelli, E., Khanna, H., Strettoi, E., and Swaroop, A. (2007). Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc. Natl. Acad. Sci. U. S. A. 104 (5), 1679–1684. doi:10.1073/pnas.0605934104
Oh, H., and Irvine, K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135 (6), 1081–1088. doi:10.1242/dev.015255
Ozturk-Colak, A., Stephan-Otto Attolini, C., Casanova, J., and Araujo, S. J. (2018). Blimp-1 mediates tracheal lumen maturation in Drosophila melanogaster. Genetics 210 (2), 653–663. doi:10.1534/genetics.118.301444
Papatsenko, D., Sheng, G., and Desplan, C. (1997). A new rhodopsin in R8 photoreceptors of Drosophila: Evidence for coordinate expression with Rh3 in R7 cells. Development 124 (9), 1665–1673. doi:10.1242/dev.124.9.1665
Pojer, J. M., Manning, S. A., Kroeger, B., Kondo, S., and Harvey, K. F. (2021). The Hippo pathway uses different machinery to control cell fate and organ size. iScience 24 (8), 102830. doi:10.1016/j.isci.2021.102830
Poupault, C., Choi, D., Lam-Kamath, K., Dewett, D., Razzaq, A., Bunker, J., et al. (2021). A combinatorial cis-regulatory logic restricts color-sensing Rhodopsins to specific photoreceptor subsets in Drosophila. PLoS Genet. 17 (6), e1009613. doi:10.1371/journal.pgen.1009613
Rister, J., and Desplan, C. (2011). The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 71 (12), 1212–1226. doi:10.1002/dneu.20905
Rister, J., Razzaq, A., Boodram, P., Desai, N., Tsanis, C., Chen, H., et al. (2015). Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science 350 (6265), 1258–1261. doi:10.1126/science.aab3417
Roger, J. E., Hiriyanna, A., Gotoh, N., Hao, H., Cheng, D. F., Ratnapriya, R., et al. (2014). OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J. Clin. Invest. 124 (2), 631–643. doi:10.1172/JCI72722
Roorda, A., and Williams, D. R. (1999). The arrangement of the three cone classes in the living human eye. Nature 397 (6719), 520–522. doi:10.1038/17383
Schubiger, M., Tomita, S., Sung, C., Robinow, S., and Truman, J. W. (2003). Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech. Dev. 120 (8), 909–918. doi:10.1016/s0925-4773(03)00134-5
Tahayato, A., Sonneville, R., Pichaud, F., Wernet, M. F., Papatsenko, D., Beaufils, P., et al. (2003). Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev. Cell 5 (3), 391–402. doi:10.1016/s1534-5807(03)00239-9
Teleman, A. A., Chen, Y. W., and Cohen, S. M. (2005). Drosophila Melted modulates FOXO and TOR activity. Dev. Cell 9 (2), 271–281. doi:10.1016/j.devcel.2005.07.004
Vandendries, E. R., Johnson, D., and Reinke, R. (1996). Orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev. Biol. 173 (1), 243–255. doi:10.1006/dbio.1996.0020
Wang, H., Morrison, C. A., Ghosh, N., Tea, J. S., Call, G. B., and Treisman, J. E. (2022). The Blimp-1 transcription factor acts in non-neuronal cells to regulate terminal differentiation of the Drosophila eye. Development 149 (7). doi:10.1242/dev.200217
Wang, S., Sengel, C., Emerson, M. M., and Cepko, C. L. (2014). A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev. Cell 30 (5), 513–527. doi:10.1016/j.devcel.2014.07.018
Wells, B. S., Pistillo, D., Barnhart, E., and Desplan, C. (2017). Parallel Activin and BMP signaling coordinates R7/R8 photoreceptor subtype pairing in the stochastic Drosophila retina. Elife 6, e25301. doi:10.7554/eLife.25301
Wernet, M. F., Mazzoni, E. O., Celik, A., Duncan, D. M., Duncan, I., and Desplan, C. (2006). Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440 (7081), 174–180. doi:10.1038/nature04615
Xie, B., Morton, D. B., and Cook, T. A. (2019). Opposing transcriptional and post-transcriptional roles for Scalloped in binary Hippo-dependent neural fate decisions. Dev. Biol. 455 (1), 51–59. doi:10.1016/j.ydbio.2019.06.022
Yu, J., Angelin-Duclos, C., Greenwood, J., Liao, J., and Calame, K. (2000). Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell Biol. 20 (7), 2592–2603. doi:10.1128/mcb.20.7.2592-2603.2000
Yu, J., and Pan, D. (2018). Validating upstream regulators of Yorkie activity in Hippo signaling through scalloped-based genetic epistasis. Development 145 (4). doi:10.1242/dev.157545
Zeng, Q., and Hong, W. (2008). The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13 (3), 188–192. doi:10.1016/j.ccr.2008.02.011
Zhang, L., Yue, T., and Jiang, J. (2009). Hippo signaling pathway and organ size control. Fly. (Austin) 3 (1), 68–73. doi:10.4161/fly.3.1.7788
Zhao, B., Lei, Q. Y., and Guan, K. L. (2008). The hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20 (6), 638–646. doi:10.1016/j.ceb.2008.10.001
Zhu, L. J., Christensen, R. G., Kazemian, M., Hull, C. J., Enuameh, M. S., Basciotta, M. D., et al. (2011). FlyFactorSurvey: A database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39, D111–D117. doi:10.1093/nar/gkq858
Keywords: rhodopsin, retina, photoreceptor, ecdysone, BLIMP-1, rorβ, color vision, hippo pathway
Citation: Bunker J, Bashir M, Bailey S, Boodram P, Perry A, Delaney R, Tsachaki M, Sprecher SG, Nelson E, Call GB and Rister J (2023) Blimp-1/PRDM1 and Hr3/RORβ specify the blue-sensitive photoreceptor subtype in Drosophila by repressing the hippo pathway . Front. Cell Dev. Biol. 11:1058961. doi: 10.3389/fcell.2023.1058961
Received: 30 September 2022; Accepted: 20 February 2023;
Published: 07 March 2023.
Edited by:
Rajalekshmy Shyam, Indiana University Bloomington, United StatesReviewed by:
Sudershana Nair, New York University, United StatesDeepika Vasudevan, University of Pittsburgh, United States
Copyright © 2023 Bunker, Bashir, Bailey, Boodram, Perry, Delaney, Tsachaki, Sprecher, Nelson, Call and Rister. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Rister, amVucy5yaXN0ZXJAdW1iLmVkdQ==
†Present addresses: Pamela Boodram, NYU Langone Medical Center, New York, NY, United States
 Joseph Bunker
Joseph Bunker Mhamed Bashir
Mhamed Bashir Sydney Bailey
Sydney Bailey Pamela Boodram1,2†
Pamela Boodram1,2† Simon G. Sprecher
Simon G. Sprecher Jens Rister
Jens Rister