
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Cell Dev. Biol. , 23 October 2023
Sec. Signaling
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1045260
This article is a correction to:
Single-Cell RNA Sequencing Reveals the Role of Phosphorylation-Related Genes in Hepatocellular Carcinoma Stem Cells
 Fuwen Yao1,2†
Fuwen Yao1,2† Yongqiang Zhan1†
Yongqiang Zhan1† Changzheng Li3†
Changzheng Li3† Ying Lu2
Ying Lu2 Jiao Chen2
Jiao Chen2 Jing Deng2
Jing Deng2 Zijing Wu2
Zijing Wu2 Qi Li2
Qi Li2 Yi’an Song2
Yi’an Song2 Binhua Chen1
Binhua Chen1 Jinjun Chen1
Jinjun Chen1 Kuifeng Tian1
Kuifeng Tian1 Zuhui Pu4*
Zuhui Pu4* Yong Ni1*
Yong Ni1* Lisha Mou1,2*
Lisha Mou1,2*A Corrigendum on
Single-cell RNA sequencing reveals the role of phosphorylation-related genes in hepatocellular carcinoma stem cells
by Yao F, Zhan Y, Li C, Lu Y, Chen J, Deng J, Wu Z, Li Q, Song Y, Chen B, Chen J, Tian K, Pu Z, Ni Y and Mou L (2022). Front. Cell Dev. Biol. 9:734287. doi: 10.3389/fcell.2021.734287
In the published article, there was an error in Figure 7C as published. The red label and black label were reversed. The corrected Figure 7C and its caption is as follows: “(C) Hep3B and Huh7 cells were treated with gambogenic acid (EZH2 inhibitor, 2 μM) or alisertib (AURKA inhibitor, 10 μM) for 0, 12, 24, 32, 48, and 60 h, and cell viability was determined by Calcein-AM/PI staining assays.”
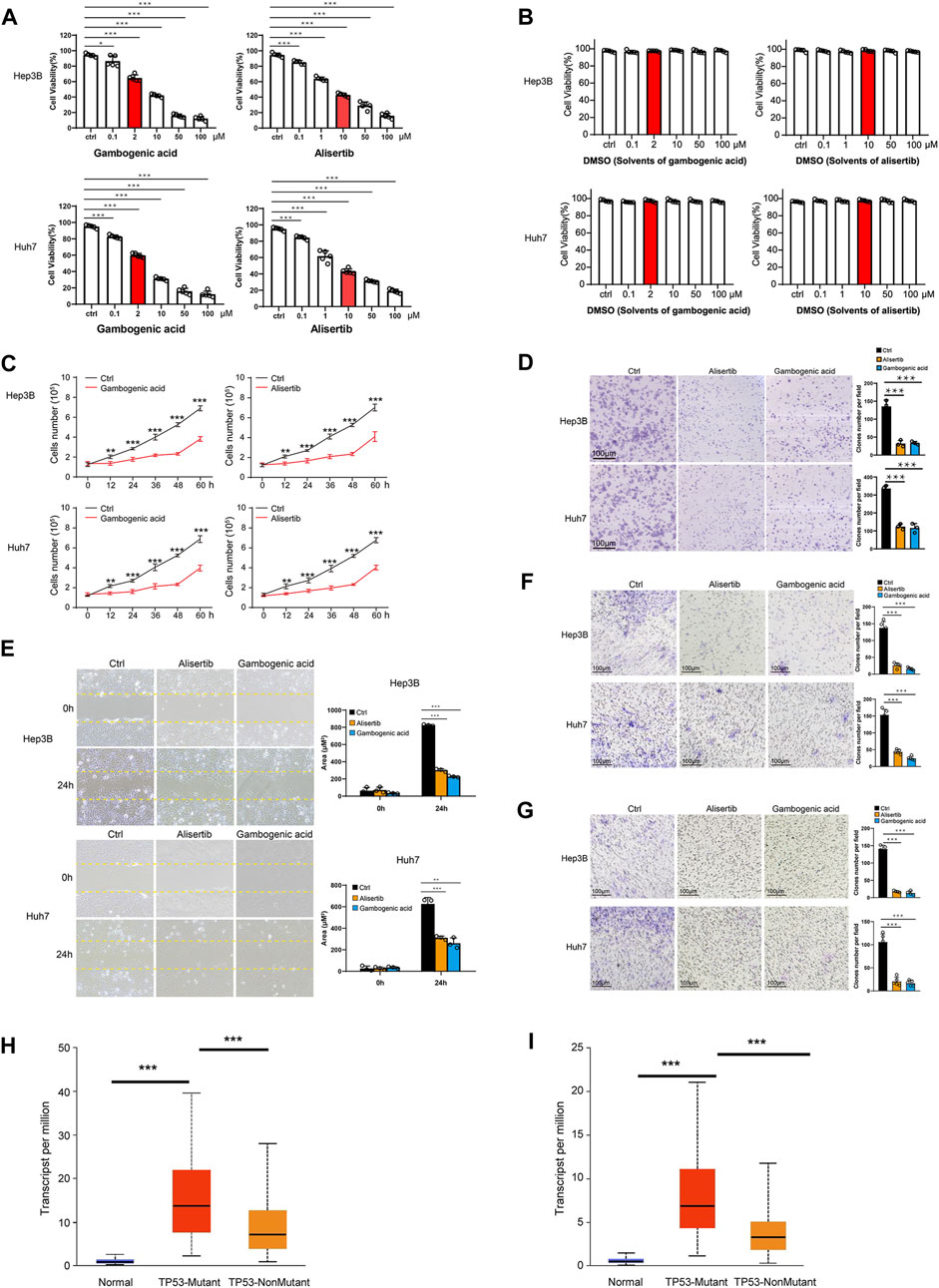
FIGURE 7. Gambogenic acid (EZH2 inhibitor) and alisertib (AURKA inhibitor) inhibit HCC cell proliferation, migration, and invasion. (A) Hep3B and Huh7 cells were treated with gambogenic acid (EZH2 inhibitor) or alisertib (AURKA inhibitor) at different concentrations (0–100 μM) for 48 h, and cell viability was determined by Calcein–AM/PI staining assays. (B) Hep3B and Huh7 cells were treated with DMSO (solvents of gambogenic acid and alisertib) at different concentrations (0–100 μM) for 48 h, and cell viability was determined by Calcein–AM/PI staining assays. (C) Hep3B and Huh7 cells were treated with gambogenic acid (EZH2 inhibitor, 2 μM) or alisertib (AURKA inhibitor, 10 μM) for 0, 12, 24, 32, 48, and 60 h, and cell viability was determined by Calcein–AM/PI staining assays. (D) Colony formation assays were conducted to analyze Hep3B and Huh7 cell proliferation with gambogenic acid (2 μM) or alisertib (10 μM) treatment. (E, F) Wound healing assays (E) and Transwell assays (F) were performed to detect the cell migratory abilities of Hep3B and Huh7 cells treated with gambogenic acid (2 μM) or alisertib (10 μM). (G) Transwell assays were performed to detect the cell invasion abilities of Hep3B and Huh7 cells treated with gambogenic acid (2 μM) or alisertib (10 μM). Data are expressed as the means ± s.d. Differences were considered statistically significant if p < 0.05. ns, no significance, *p < 0.05, **p < 0.01, ***p < 0.001. (H, I) Expression of AURKA (H) and EZH2 (I) in TCGA–LIHC based on TP3 mutation status.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: AURKA, EZH2, tyrosine kinase inhibitors, TKI, protein kinases, cell cycle, single-cell RNA sequencing, hepatocellular carcinoma
Citation: Yao F, Zhan Y, Li C, Lu Y, Chen J, Deng J, Wu Z, Li Q, Song Y, Chen B, Chen J, Tian K, Pu Z, Ni Y and Mou L (2023) Corrigendum: Single-cell RNA sequencing reveals the role of phosphorylation-related genes in hepatocellular carcinoma stem cells. Front. Cell Dev. Biol. 11:1045260. doi: 10.3389/fcell.2023.1045260
Received: 15 September 2022; Accepted: 11 October 2023;
Published: 23 October 2023.
Edited and reviewed by:
Lei Dong, Beijing Institute of Technology, ChinaCopyright © 2023 Yao, Zhan, Li, Lu, Chen, Deng, Wu, Li, Song, Chen, Chen, Tian, Pu, Ni and Mou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zuhui Pu, cHVwZXRlcjE5MEAxNjMuY29t; Yong Ni, c3puaXlvbmdAc2luYS5jb20=; Lisha Mou, bGlzaGFtb3VAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.