
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 13 September 2022
Sec. Epigenomics and Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.985274
This article is part of the Research TopicDNA Methylation: The Ageing ClockView all 5 articles
The prevalence of obesity has dramatically increased worldwide over the past decades. Aging-related chronic conditions, such as type 2 diabetes and cardiovascular disease, are more prevalent in individuals with obesity, thus reducing their lifespan. Epigenetic clocks, the new metrics of biological age based on DNA methylation patterns, could be considered a reflection of the state of one’s health. Several environmental exposures and lifestyle factors can induce epigenetic aging accelerations, including obesity, thus leading to an increased risk of age-related diseases. The insight into the complex link between obesity and aging might have significant implications for the promotion of health and the mitigation of future disease risk. The present narrative review takes into account the interaction between epigenetic aging and obesity, suggesting that epigenome may be an intriguing target for age-related physiological changes and that its modification could influence aging and prolong a healthy lifespan. Therefore, we have focused on DNA methylation age as a clinical biomarker, as well as on the potential reversal of epigenetic age using a personalized diet- and lifestyle-based intervention.
Nowadays, the aging of the population represents a major social, economic, and public health challenge. Since aging, defined as a progressive loss of physiological integrity, is a key risk factor for many non-communicable diseases (NCD), including diabetes, cardiovascular, and metabolic diseases (Correa-Burrows et al., 2022). People of the same age may show differences in age-related functional decline as well as in their susceptibility to disease (Fransquet et al., 2019). Among the nine hallmarks which are considered the common denominators of aging at the molecular and cellular level, epigenetic changes, that are modifications of DNA or chromatin not altering the primary nucleotide sequence, can impact significantly on the aging process (López-Otín et al., 2013).
In this view, epigenetic marks, including DNA methylation, also known as “epigenetic clocks”, are emerging as a molecular tool for measuring biological aging (Tekola-Ayele, 2021) and, in this light, they could be considered as special “indicators” of the state of one’s health (Raj, 2018). In detail, the average of DNA methylation levels at multiple CpG sites (CpGs) is used to calculate the “epigenetic age”.
Many genes and pathways have been correlated with longevity regulation which could be possible epigenetic biomarkers of aging. The DNA epigenetic pattern changes during life. ELOVL2 (fatty acid elongase 2) gene is the most extreme example of age-related hypermethylation, representing a bridge between the early stages of development and the aging process (Garagnani et al., 2012; Bacalini et al., 2017). This gene encodes for an enzyme that catalyzes one of the rate-limiting steps in the synthesis of long chain polyunsaturated fatty acids, which plays an important role in development (Gregory et al., 2011). ELOVL2 is unmethylated in newborns, whereas its methylation levels significantly increase with age in different tissues, suggesting it as a kind of rheostat for aging-the more the gene is methylated, the older is the subject (Garagnani et al., 2012). Other possible genes proposed as biomarkers of aging include tumor suppressors (COX7A1, LOX, RUNX3, TIG1, p16INK4A, RASSF1, DUSP22) and genes involved in growth and development (IGF2, cFos), cell-cell adhesion (CDH1), metabolism (SLC38A4, SLC22A18, MGC3207, ECRG4, ATP13A4, AGPAT2, LEP), DNA repair (MLH1) and the control of signal transmission (FZD1, FZD7) (Christensen et al., 2009; Taormina and Mirisola, 2015). In addition, transcription factor-binding sites and promoters of genes involved in the regulation of gene expression, senescence, apoptosis and tumorigenesis are hypermethylated and silenced in a tissue-specific manner (Waki et al., 2003; Grönniger et al., 2010; Hernandez et al., 2011; Salminen et al., 2011). For example, the silencing of the p16INK4a gene (Keyes et al., 2007), which is a tumor suppressor and an aging-associated gene, is induced by hypermethylation of the transcription factor E2F-1 binding site within the gene promoter (Li and Tollefsbol, 2011).
Horvath et al. (Horvath, 2013) suggested that DNA methylation age (DNAmage) can evaluate the cumulative effect of an epigenetic maintenance system, identifying tissues that show evidence of accelerated age due to disease (Horvath, 2013). Epigenetic-age acceleration, defined as the discrepancy between the biological age measure and chronological age, have been used to examine the associations between epigenetic aging and several traits (Nannini et al., 2019). Overall, negative values of epigenetic-age acceleration (i.e., slower aging) are correlated with lifestyle factors, while positive values are associated with age-related health outcomes such as frailty, cancer, lung function and physical and cognitive fitness (Marioni et al., 2015; Breitling et al., 2016; Perna et al., 2016; Zheng et al., 2016).
Recently, it has been demonstrated that obesity accelerates epigenetic aging of metabolically active tissues including visceral adipose tissue and liver (Horvath et al., 2014; de Toro-Martín et al., 2019). Therefore, epigenetic-age acceleration has been correlated not only with obesity (Nevalainen et al., 2017), but also with physical fitness (Marioni et al., 2015) and stress (Zannas et al., 2015), albeit little is known about the link between epigenetic aging rates and lifestyle factors, including diet, alcohol abuse, and physical activity. The present narrative review considers the interaction between epigenetic clocks and age-related diseases, with particular emphasis on obesity. Some authors suggested the epigenome as an intriguing target for age-related physiological changes and its modification could prolong a healthy lifespan (López-Otín et al., 2013). Therefore, we have focused on DNA methylation age as clinical biomarker and on potential reversal of epigenetic age using a diet and lifestyle intervention.
The prevalence of obesity has increased dramatically over the past decades. Obesity is associated with a higher risk of several age-related conditions and diseases, including cardiovascular disease (CVD), hypertension, type 2 diabetes mellitus (T2DM), and cancer (Pi-Sunyer, 2009). As a consequence, this condition shortens life expectancy by up to 20 years with the risk of premature death by 1.45 to 2.76-fold (Fontaine et al., 2003; Reuser et al., 2008). The mortality is lowest at a BMI of 22.5–25 kg/m2 range, with each 5 kg/m2 increase in the BMI associated with a 30% higher mortality, confirming the important role of BMI in the determination of lifespan (Prospective Studies Collaboration Whitlock et al., 2009). It is possible to suggest that obesity can accelerate aging, thus having a profound impact on human health. However, the relationship between aging and obesity is complex, involving cellular and molecular phenotypic signatures. Tam et al. (Tam et al., 2020) defined obesity and aging as “two sides of the same coin” since the pathology of the first condition parallels those of aging at multiple levels (Nannini et al., 2019).
In particular, the metabolic dysregulation correlated with obesity is similar to the one observed in aging and inflammation, oxidative stress, and impaired immune functions seem to be important mediators of this similarity (Santos and Sinha, 2021) (Figure 1). For example, the increased level of pro-inflammatory molecules is one of the major hallmarks of aging, in turn plays an important role in the adipose tissue enlargement. Obesity disrupts homeostatic resilience mechanisms which preserve physiological integrity, and the adipose tissue of obese subjects shows several of the hallmarks of aging, such as mitochondrial dysfunction, increased apoptosis, cellular senescence, insufficient autophagy, and chronic inflammation ((Pérez et al., 2016; Correa-Burrows et al., 2022). The excessive reactive oxygen species (ROS) formation triggers by chronic inflammatory cellular environment leading in oxidative stress and mitochondrial dysfunction, which results in further ROS formation and exacerbation of inflammatory processes. Overnutrition and obesity decrease autophagic flux resulting in excessive accumulation of malfunctioning organelles and misfolded proteins, which are common features of ageing. ROS formation compromises the mitochondrial DNA (mtDNA) integrity affecting the function of multiple tissues (Taylor and Turnbull, 2005), while the upregulation of pro-apoptotic genes and the caspase activation cascades resulting in the proteolytic cleavage of major structural proteins and the subsequent apoptosis (Yuzefovych et al., 2013). Moreover, DNA damage also triggers the upregulation of p21, p16ink4a playing important roles in DNA damage response. On the other hand, the chronic activation of DNA damage response and p53 induce senescence growth arrest (Freund et al., 2012). Chromatin reorganization triggers by the subsequent lamin B1 depletion in senescent cells, causes upregulation of senescence-associated secretory phenotypes (SASP) genes as well as packaging and silencing of proliferative genes promoting cellular senescence (Freund et al., 2012). Taken together, excessive ROS formation in obesity results in a broad range of cellular events which are key contributors to the onset of age-associated diseases. Moreover, obesity appears to influence telomere length as well as accelerate epigenetic clocks associated with aging (Buxton et al., 2011; Horvath, 2013; Clemente et al., 2019) as reviewed below.
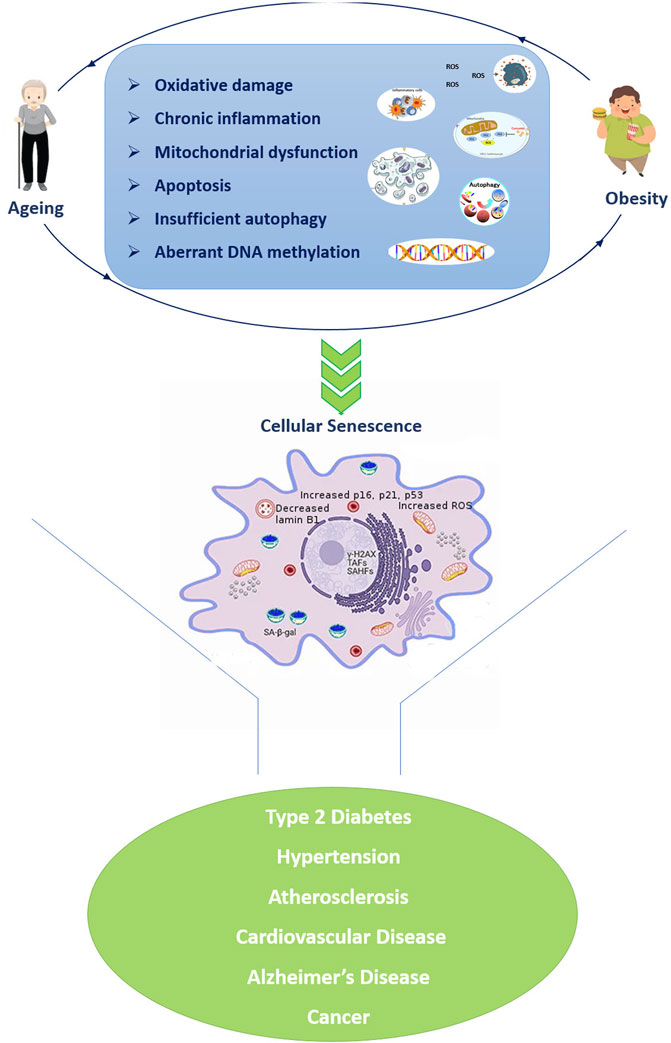
FIGURE 1. Similarities in cellular and molecular alterations of both ageing and obesity resulting in a vicious propagatory cycle which triggers the development of age-related diseases. The senescent cell was adapted from Narasimhan et al., 2021. Abbreviations: TAFs: telomere-associated DNA damage foci; SAHF: Senescence-associated heterochromatin foci; SA-β-gal: senescence-associated β-galactosidase.
Epigenetic mechanisms, such as DNA methylation, histone modification and microRNA (miRNA) regulation, can modulate gene expression without changing the DNA sequence. Environmental and lifestyle stressors disturb these epigenetic mechanisms, inducing long-term changes and potentially leading to disease in later life or even in the offspring, with a transgenerational effect, according to the developmental origin of health and disease (DOHaD) hypothesis (Stuppia et al., 2015). The aging process involves mainly DNA methylation, occurring at cytosines in a CpG dinucleotide (Lewis and Tollefsbol, 2017; Unnikrishnan et al., 2019) catalyzed by DNA methyltransferases (DNMTs). DNMTs family is responsible for establishing (DNMT3a and DNMT3b) and maintaining (DNMT1) the process of DNA methylation. Currently, there is a variability of reported findings showing the effects of age on the expression of the DNMTs. The life span of the heterozygous Dnmt1 knockout mice, which show a 33% decrease in DNMT1 expression, was similar to wild-type mice (Ray et al., 2006). On the other hand, the expression of the DNA methyltransferase Dnmt3a2 in the hippocampus were found to decrease with age (Cui and Xu, 2018) and the rescue of Dnmt3a2 levels restored cognitive functions (Oliveira et al., 2012). The expression of DNMT1 and DNMT3a were significantly higher in young dwarf mice compared with young wild-type mice; however, this difference was disappeared with age (Armstrong et al., 2014). Based on the results mentioned above, further studies are required to elucidate the expressional changes of DNMTs based on the aging.
A relationship between epigenetics and aging was first observed about 40 years ago in studies carried out in pink salmon and rat, which showed an overall decrease in DNA methylation with increasing age (Berdyshev et al., 1967; Vanyushin et al., 1973) This decrease has been subsequently also reported in humans. Drinkwater et al (Drinkwater et al., 1989) comparing DNAm in blood of young (average age 25 years) and older individuals (average age 75 years), showed that the elderly group had significantly reduced levels of 5 mC (Drinkwater et al., 1989). Several approaches for quantifying biological aging have been used over time, including telomere length and most recently epigenetic aging clocks. Telomeres, nucleoprotein structures containing tandem repeats of DNA (5′-TTAGGG-3′), are situated at the end of the chromosomes ((Biron-Shental et al., 2010)) and maintain their integrity and the stability of the genome, while preventing end-to-end chromosomal fusions as well (Lin et al., 2013). Shelterin, a complex of six telomere-specific proteins (TRF1, TRF2, TIN2, POT1, TPP1, and RAP1), is responsible for the protection of the genetic material, forming the so-called “t-loop” structure and together with the ribonucleoprotein reverse transcriptase, known as telomerase, modulate telomere length (de Lange, 2005). A proper function of the complex is required to prevent unwanted repair events and the trigger of the DNA damage response (Grun et al., 2018).
In the majority of somatic cells, telomeres become progressively shorter with each cell division, a process increased by oxidative stress and inflammation. As a result of telomere shortening, cell division slows down over the years, leading to accelerated cell senescence and a higher risk for developing age-related diseases such as CVD, T2DM, insulin resistance (Gardner et al., 2005), hypertension (Yang et al., 2009) and increased mortality (Levine et al., 2018). The association of leucocyte telomere length (LTL) and obesity has been also evidenced. The impact of high‐fat diet induced obesity and advancing age on the shelterin complex, and telomerase is unclear. Obesity and age influence the expression telomere shelterin complex and telomerase gene expression, altering telomere function in adipose tissue and thereby increasing inflammation and the risk of chronic disease (Bloom et al., 2020). Grun et al. (Grun et al., 2018) investigated TL from peripheral blood mononuclear cells in obese subjects analyzing the expression of shelterin genes and the plasma redox state. The authors showed dysregulation of the shelterin components that was explained by TRF1 upregulation, suggesting TRF1 as a major contributor for telomeres uncapping in the context of obesity (Grun et al., 2018). Some studies have suggested that weight loss via a structured multidisciplinary, personalized lifestyle intervention program of healthy diet and physical exercise might increase LTL, which, in turn, could potentially predict a better weight loss response to an intervention program in overweight and obese adolescents (Garcia-Calzon et al., 2014; Paltoglou et al., 2021).
Nowadays, epigenetic clocks play a crucial role in determining chronological age based on evaluation of hypo- and hyper-methylation changes in many regions of the genome (Fraga and Esteller, 2007; Zheng et al., 2016). The first-generation clocks used algorithms, based on DNAm as biomarker of aging (the Horvath and Hannum clocks (Hannum et al., 2013; Horvath, 2013), and were similar to a machine learning method that regresses chronological age on its selected set of the most informative CpG sites (CpGs) for age prediction. Thus, these models convert the methylation status of the selected CpGs into units of years, known as “DNAm age” (Horvath et al., 2014; Horvath and Raj, 2018). The Hannum estimator has been tested on blood-derived DNA and is based on DNA methylation at 71 CpGs selected from the Illumina 450k array, while the Horvath estimator (17) has been constructed across multiple tissues, including the blood data from Hannum et al., and comprises 353 CpGs that were present on the earlier generation Illumina 27k array. In recent years, ‘second generation clock’ models, such as DNAmGrimAge and the PhenoAge, have outperformed the ‘first-generation clocks’ in predicting longevity and the onset of many age-related pathological conditions and diseases (Levine et al., 2018). The DNAm-age estimator of Levine et al., defined PhenoAge clock, consists of 513 CpGs and is based on a measure of biological age comprising age and nine clinical biomarkers, including lymphocyte percentage, albumin, and glucose levels (Levine et al., 2018). Another multifactorial clock, DNAmGrimA, combines seven sets of CpGs, estimating the concentration of a different plasma protein, chronological age, sex and smoking habits (Lu et al., 2019).
More recently, a third-generation epigenetic clock, known as the Dunedin Pace of Aging methylation clock (DunedinPoAm) takes into account longitudinal change over time in 18 biomarkers (i.e. BMI, LTL and HDL cholesterol) of organ-system integrity among individuals who are all the same chronological age, recording how fast the subject is aging (Belsky et al., 2020) (Table1).
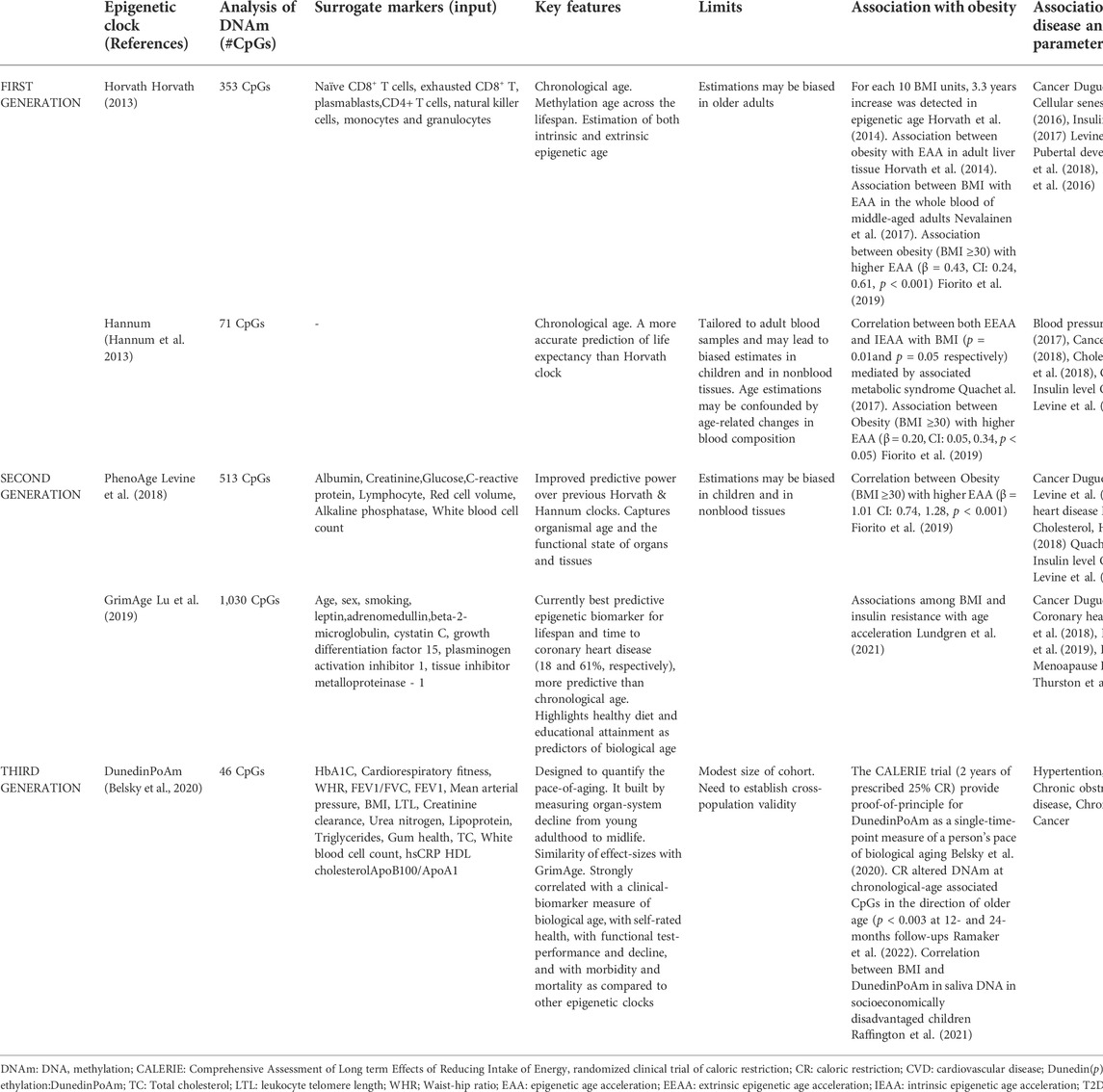
TABLE 1. Summary of first-generation (Hannum and Horvath), second (DNAm PhenoAge, GrimAge) and third-generation (DunedinPoAm) epigenetic clocks.
Epigenetic age acceleration (EAA) has been defined as the discrepancy between epigenetic age, estimated using the models described above, and chronological age. EAA can be divided into intrinsic acceleration (IEAA), which captures intrinsic cellular properties of the aging process and extrinsic acceleration (EEAA) which measures immune system aging (18). These measures have been widely used to evaluate the associations between epigenetic aging and numerous clinical traits and it has been evidenced that EEAA is affected by lifestyle (Nannini et al., 2019).
Research on epigenetic biomarkers described more than a hundred of genes distributed over all chromosomes and associated with obesity and adiposity (Samblas et al., 2019).
BMI and obesity are associated with alterations of DNA methylation levels at several CpGs within metabolic genes including HIF3A, IGFBP3, SREBF1, TNF, TRIM3 and UBASH3A as well as global methylation levels of Alu–elements genes (Houde et al., 2015; Samblas et al., 2019). However, some studies demonstrated that these obesity-related DNA methylation changes do not overlap those seen in aging-associated modifications (Marttila et al., 2015; Kananen et al., 2016), whereas other studies identified sites with a common effect in obesity and aging. In particular, it has been identified, by performing genome wide methylation analysis, 10 sites with an interaction effect between obesity and aging. In eight (ADCY1, CXADR, KCNS2, LMX1B, FNDC4, NAT8L, AQPEP, and FBLIM1) of the ten sites, the obese individuals displayed decreased level of methylation with age when compared to their lean counterparts (Almén et al., 2014). In contrast, the remaining two sites (RNH1 and NNAT) were hypermethylated with aging in the obese group compared to lean subjects (Almén et al., 2014). Leukocyte DNA methylation levels of several CpGs located at genes involved in longevity-regulating pathways have been evaluated in obesity, suggesting a role of DNA methylation in aging-related metabolic alterations. In details, the methylation levels of 58 CpGs located at genes involved in longevity-regulating pathways were associated with BMI. Fifteen of them were differentially methylated between younger and older individuals that exhibited at least one metabolic alteration. Of note, six of these CpGs, within MTOR, ULK1, ADCY6, IGF1R, CREB5, and RELA, were common to the metabolic traits, and CREB5, RELA, and ULK1 were statistically correlated with age (Salas-Pérez et al., 2019).
Although the effects of DNA methylation on adipocyte aging process are yet to be fully elucidated, it has been demonstrated that some environmental factors and aging lead to modifications in methylation patterns in adipocytes. In particular, the senescence relevant genes hypomethylation such as p21, p16 as well as the telomerase reverse transcriptase (TERT)’s hypermethylation accelerate adipose progenitor cellular senescence and exhaustion (Qimuge et al., 2019; Pan et al., 2020). Moreover, an increase in methylation levels of the regulator regions in Ppar-γ ADIPOQ and LEP (Melzner et al., 2002; Fujiki et al., 2009; Kim et al., 2015) has been associated in white adipocyte glucose and lipid metabolism alterations, which further worsen insulin resistance, obesity, and inflammation. In addition, hypermethylation of PRDM16 and the enhancer region of UCP1 during cellular senescence, triggers to loss of beige adipose tissue and thermogenic properties of brown adipocytes (Yang et al., 2016; Foti and Brunetti, 2017). Of note, instead of focusing on single CpGs hypo- or hypermethylation patterns in single genes, the use of an epigenetic clock to study the relationship between obesity and the DNA methylation ages is based on the evaluation of the methylation status in many regions of the genome (Table 1). For example, Hovarth et al. (Horvath et al., 2014) showed 279 genes was underexpressed in older liver samples that are highly enriched with nuclear mitochondrial genes playing a role in oxidative phosphorylation and electron transport.
Although epigenetic clocks are recent metrics of biological age correlating with mortality risk (Marioni et al., 2018), it remains unclear whether epigenetic aging rates are associated with lifestyle factors such as diet, alcohol consumption, and BMI. For instance, the association between body composition and obesity with epigenetic clocks is not well understood. To date, it is unclear whether obesity is a driver or a consequence of epigenetic age acceleration (Tekola-Ayele, 2021). Some studies have investigated the obesity-related epigenetic aging in metabolically active tissues, confirming obesity-related epigenetic age acceleration in blood, liver, adipose, and buccal tissues (Horvath et al., 2014; Simpkin et al., 2017; de Toro-Martín et al., 2019).
Physical activity seems to attenuate the link between obesity and acceleration of the epigenetic clock: lower abilities in physical and mental fitness, measured by several markers (i.e. lung function, walking speed, grip strength and cognitive ability) have been associated with accelerated epigenetic aging (Marioni et al., 2015). Positive associations of body composition and physical activity level with multiple measures of epigenetic age acceleration have been recently found in blood from 2,758 non-Hispanic White women (Kresovich et al., 2021). Another previous study included data in blood tissue of more 4,100 postmenopausal female participants from the Women’s Health Initiative, as well as 402 male and female participants from the Italian cohort study. Interestingly, the research shows that diet, education, physical activity, low BMI, but not smoking habits, are associated with EEAA.
It is noteworthy that these findings confirmed the conventional benefits of eating a high plant diet with lean meats, moderate alcohol consumption, physical activity, and education, as well as the health risks of obesity and metabolic syndrome (Quach et al., 2017).
To characterize the significance of obesity in epigenetic aging, Nevalainen et al. (Nevalainen et al., 2017) studied the association between BMI and epigenetic age in three age groups: young adults, middle-aged, and nonagenarian individuals. Interestingly, the study confirmed the correlation between increased BMI and accelerated epigenetic aging in the blood cells of the middle-aged subjects, and this result was showed even when the BMI increased in adulthood (Nevalainen et al., 2017). More recent reporting (Mozhui et al., 2022) carried out in mouse models found that more rapid age-dependent changes in methylation in which higher weight gains at younger age are associated with higher epigenetic age acceleration later in life. In addition, the study showed that the effects of diet on epigenetic age acceleration was mediated by the changes in metabolic traits and weight, particularly, the latter had a stronger age-accelerating impact.
While the results described above confirm the link between accelerated epigenetic aging and obesity in adults, such evidence appears limited as far as it concerns children. Epigenetic age acceleration at birth and childhood has been associated with maternal habits, including smoking and alcohol consumption during the prenatal period (Simpkin et al., 2016). The link between epigenetic age acceleration with growth and development in early life remains unclear but the trajectory of the blood DNA methylome aging rate appears largely set before adulthood, remaining remarkably stable thereafter (Etzel et al., 2022). Recently, the association between accelerated epigenetic aging and BMI in children using multiple epigenetic clocks has been demonstrated (Gentilini et al., 2013). If the correlation between obesity and accelerated epigenetic aging begins in early life, epigenetic programming may offer potential and novel opportunity of intervention with implications for the promotion of health and the mitigation of future disease risk. In this context, since DNA methylation status changes with age, thus contributing to the development of age-related diseases, the study of centenarians’ offspring seems to be a valid approach to understand the role of epigenetics in the modulation of healthy aging. In this context, an age-related decrease in global DNA methylation and a delay of this process in centenarians’ offspring has been observed (Horvath et al., 2015). Moreover, a similar trend was reported by Horvath showing a lower intrinsic epigenetic ageing rate in peripheral blood mononuclear cells from Italian semi-supercentenarians and their offspring (Sae-Lee et al., 2018). Thus, the offspring of semi supercentenarians may be very informative when it comes to identifying epigenetic determinants of healthy aging and future relevant studies might be able to detect how to extend the benefits of successful aging.
There is a growing interest to identifying therapeutic interventions modulating the mechanisms of aging or at least have the onset of age-related diseases slowed (Schork et al., 2022).
Among therapies targeting that slow the aging rate and mitigate damage to the body, have been proposed several approaches: 1) the seek to mimic the beneficial molecular effects of caloric restriction (Mahmoudi et al., 2019), 2) attempt to clear out senescent cells and the deleterious age-related debris that they secrete (Baker et al., 2016; Simpson et al., 2021), 3) the identification of circulating factors associated with healthy youth that could be infuse into older individuals (Conboy et al., 2013), 4) take advantage insights into stem cell biology and cellular rejuvenation (Olova et al., 2019; Browder et al., 2022).
On the other hand, the alteration of the environment and drug administration have been proposed as new approaches that could be utilized to extend life span due to their impact on epigenetic mechanisms and related metabolic activity (Longo et al., 2015). Some drugs, known as “epigenetic drugs” or “epi-drugs”, are chemical modifiers of epigenetic enzymes, specifically target DNMTs and are mainly used to treat cancer (Bates, 2020). On the other hand, based on a many similar mechanisms epigenetic regulation between aging and cancer (i.e., decline of heterochromatin DNA methylation, inhibitory histone modifications, increased DNA methylation in growth-related and tumor suppressor gene promoters), it could also be envisaged to apply epigenetic anticancer drugs to age-related diseases. This attractive strategy is being tested in ongoing clinical trials but demonstrate great promise about rejuvenation (Johnson et al., 2012) and may propose new possibilities to manage diseases.
Many studies support the hypothesis that dietary factors can reverse epigenetic alterations influencing the aging process: this newer approach could also offer compelling opportunities for the design of anti-aging treatments. Among dietary compounds, vitamins and polyphenols contribute to the production of the methyl donor S-adenosyl-l-methionine (SAM) and to the inhibition of DNA methyltransferases (DNMT) activity, respectively. By using publicly available Illumina Infinium 450K methylation datasets, it has been proven that dietary supplementation with folic acid + vitamin B12 as a methyl donor and flavanols as DNMT inhibitors can decrease epigenetic age and appear to be gender and MTHFR genotype-specific (Sae-Lee et al., 2018). It has been showed that calorie restriction and dietary rapamycin can slow molecular changes associated with an epigenetic clock in mice livers (Wang et al., 2017). Of note, caloric restriction decreased the age-related decline in DNMT1, DNMT3B, TET1, and TET3 gene expression in colon mucosa isolated from mice (Unnikrishnan et al., 2019). Previously, also Chouliaras et al. (Chouliaras et al., 2012) showed that the age-related increase in the level of DNMT3a protein in the hippocampus can be attenuated by dietary restriction.
However, although these results related to the reversal of epigenetic age might be exciting, it should be noted that while exercise can induce genome-wide changes in DNA methylation, the DNAm age of the adipose tissue seems not to be influenced (Rönn et al., 2013). These observations agree with previous studies showing that the bariatric surgery with significant weight loss, improved metabolic abnormalities but did not reverse epigenetic age acceleration in the liver of subjects within a 9-month period (Horvath et al., 2014). A 24- month randomized factorial intervention trial carried out in healthy postmenopausal women provided strong evidence of a causal relationship between lifestyle and healthy aging-related epigenetic mechanisms. The effect of a dietary intervention reflected a slowing of the DNAmGrimAge clock, and the increase of physical activity led to a reduction of stochastic epigenetic mutations, which have been proposed as a complementary DNAm-based biomarker of healthy aging (Fiorito et al., 2021). Taken together, the key issues remain whether: 1) these tissue-specific effects are reflected epigenetic aging rates in the same way as for other tissues; 2) the reversal of epigenetic ageing is effectively possible through lifestyle improvement.
Some findings shed light on the potential utility of epigenetic clocks for studying key traits and changes during critically important period of life. In women menopause represents a crucial turning point, because it not only marks the end of the fertile age, but also accelerates the general aging processes, with significant repercussions on health as a whole. Addressing sexual aging adequately –(particularly with the healthy lifestyle) can prevent several issues. Aging and reproduction are intrinsically linked although future work examining the relationship between epigenetic clocks and reproductive health is warranted. The effect of reproduction on epigenetic clocks may be tied to modifications in immune cell composition of blood and to hormonal changes (Ryan, 2021). Postmenopausal women with a late onset of menopause are epigenetically younger than women with an early onset of menopause (Levine et al., 2016). In addition, women who have undergone surgical menopause (bilateral oophorectomy) showed accelerated epigenetic age, appearing to be decelerated with menopausal hormone therapies (Levine et al., 2016). Similarly, severe vasomotor symptoms (hot flashes) among older women were associated with higher DNAm PhenoAge as well as late-occurring hot flashes were linked to accelerated DNAm PhenoAge and DNAm GrimAge (Thurston et al., 2020).
The rapid rise of obesity causes for concern, and it is attributed mainly to the lifestyle and diet adopted recently. However, both genetic variants and epigenetic changes may affect some subjects who are more susceptible to this obesogenic environment, leading to obesity and its complications (Franzago et al., 2020). Since obesity is linked to aging, epigenetic processes might play a role in healthy human aging. There is great interest for epigenetic markers with regard to their future beneficial application in the promotion for healt. Although lifestyle factors such as diet, alcohol consumption, and physical activity are connected to health-related outcomes, the literature on the direct influence of these factors on molecular aging rates is only at its early stage. Clarifying the molecular determinants of healthy aging and longevity would provide a major contribution to current research. The promising ultimate tools for investigating the relationship between lifestyle and aging are the molecular biomarkers known as the “epigenetic clocks”, which are based on the quantification of DNA methylation levels, apart from LTL.
The interest of the epigenetic mechanisms is due to their reversibility as well as their possible transgenerational effects (Franzago et al., 2019), which can be detected by the predictor of epigenetic age. The above has the potential to elucidate epigenetic inheritance and the effects caused by the environment and lifestyle that can slow down or accelerate epigenetic aging. In particular, the recent attention for epigenetic clocks as a biomarker of ageing is due to a 1) their application to several human tissues, 2) their accurate measurement of chronological age (Gibbs, 2014), 3) their independent predictive value for all-cause mortality in later life (Marioni et al., 2015), 4) their association with cognitive and physical factors in the elderly (Marioni et al., 2015) and 5) their usefulness in identifying accelerated ageing effects due to obesity and other conditions (Lowe et al., 2016; Binder et al., 2018; Dugué et al., 2018; Fiorito et al., 2019; Lundgren et al., 2021; Narasimhan et al., 2021; Ramaker et al., 2022)
Overall, epigenetic clocks are considered accurate biomarkers of the wider process of epigenetic remodeling occurring in different tissues during aging and might provide optimal strategies to improve the health of the population, especially if targeting specific groups, such as obese subjects (Figure 2). Recent literature supported that the modifications in epigenetic acceleration are correlated with change in BMI, suggesting that epigenetic aging may respond to changes in lifestyle, at least with respect to change in obesity (Quach et al., 2017). This perspective leads the way towards considering epigenetic biomarkers in dietary intervention studies in the future. Further research is warranted in order to gain further insight into the physiological relevance of epigenetic aging and to its role in the onset of lifestyle-related diseases such as obesity.
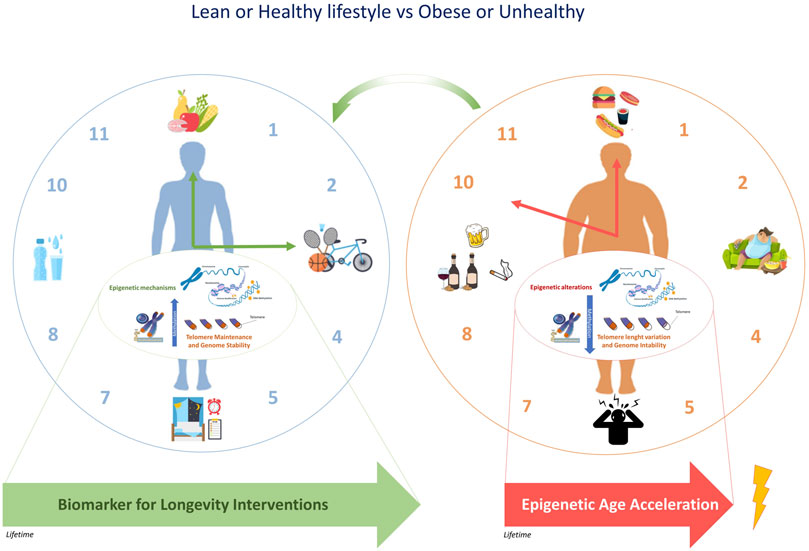
FIGURE 2. The epigenetic clock and telomere length are associated with chronological age. The epigenetic modifications induced by an unhealthy lifestyle can accelerate epigenetic aging. Due to the potential reversal of epigenetic aging, some targeted population groups, such as obese subjects might respond to changes in lifestyle.
MF and EV conceived this manuscript. MF, SD, and LP carried out the search of literature about epigenetic aging, obesity and lifestyle. The manuscript was drafted by MF. EV and LS contributed to the editing the manuscript. All authors read and approved the final version of the manuscript.
MF was supported by the Fondazione Umberto Veronesi.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Almén, M. S., Nilsson, E. K., Jacobsson, J. A., Kalnina, I., Klovins, J., Fredriksson, R., et al. (2014). Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene 548 (1), 61–67. doi:10.1016/j.gene.2014.07.009
Armstrong, V. L., Rakoczy, S., Rojanathammanee, L., and Brown-Borg, H. M. (2014). Expression of DNA methyltransferases is influenced by growth hormone in the long-living Ames dwarf mouse in vivo and in vitro. J. Gerontol. A Biol. Sci. Med. Sci. 69 (8), 923–933. doi:10.1093/gerona/glt133
Bacalini, M. G., Deelen, J., Pirazzini, C., De Cecco, M., Giuliani, C., Lanzarini, C., et al. (2017). Systemic age-associated DNA hypermethylation of ELOVL2 gene: In vivo and in vitro evidences of a cell replication process. J. Gerontol. A Biol. Sci. Med. Sci. 72 (8), 1015–1023. doi:10.1093/gerona/glw185
Baker, D. J., Childs, B. G., Durik, M., Wijers, M. E., Sieben, C. J., Zhong, J., et al. (2016). Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530 (7589), 184–189. doi:10.1038/nature16932
Bates, S. E. (2020). Epigenetic therapies for cancer. N. Engl. J. Med. 383 (7), 650–663. doi:10.1056/NEJMra1805035
Belsky, D. W., Caspi, A., Arseneault, L., Baccarelli, A., Corcoran, D. L., Gao, X., et al. (2020). Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife 9, e54870. doi:10.7554/eLife.54870
Berdyshev, G. D., Korotaev, G. K., Boiarskikh, G. V., and Vaniushin, B. F. (1967). [Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning]. Biokhimiia Mosc. Russ. 32 (5), 988–993.
Binder, A. M., Corvalan, C., Mericq, V., Pereira, A., Santos, J. L., Horvath, S., et al. (2018). Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics 13 (1), 85–94. doi:10.1080/15592294.2017.1414127
Biron-Shental, T., Sukenik Halevy, R., Goldberg-Bittman, L., Kidron, D., Fejgin, M. D., and Amiel, A. (2010). Telomeres are shorter in placental trophoblasts of pregnancies complicated with intrauterine growth restriction (IUGR). Early Hum. Dev. 86 (7), 451–456. doi:10.1016/j.earlhumdev.2010.06.002
Bloom, S. I., Tuluca, A., Ives, S. J., and Reynolds, T. H. (2020). High-fat diet induced obesity and age influence the telomere shelterin complex and telomerase gene expression in mouse adipose tissue. Physiol. Rep. 8 (11), e14461. doi:10.14814/phy2.14461
Breitling, L. P., Saum, K. U., Perna, L., Schöttker, B., Holleczek, B., and Brenner, H. (2016). Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenetics 8, 21. doi:10.1186/s13148-016-0186-5
Browder, K. C., Reddy, P., Yamamoto, M., Haghani, A., Guillen, I. G., Sahu, S., et al. (2022). In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat. Aging 2, 243–253. doi:10.1038/s43587-022-00183-2
Buxton, J. L., Walters, R. G., Visvikis-Siest, S., Meyre, D., Froguel, P., and Blakemore, A. I. (2011). Childhood obesity is associated with shorter leukocyte telomere length. J. Clin. Endocrinol. Metab. 96 (5), 1500–1505. doi:10.1210/jc.2010-2924
Chouliaras, L., van den Hove, D. L., Kenis, G., Keitel, S., Hof, P. R., van Os, J., et al. (2012). Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiol. Aging 33 (8), 1672–1681. doi:10.1016/j.neurobiolaging.2011.06.003
Christensen, B. C., Houseman, E. A., Marsit, C. J., Zheng, S., Wrensch, M. R., Wiemels, J. L., et al. (2009). Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 5 (8), e1000602. doi:10.1371/journal.pgen.1000602
Clemente, D., Maitre, L., Bustamante, M., Chatzi, L., Roumeliotaki, T., Fossati, S., et al. (2019). Obesity is associated with shorter telomeres in 8 year-old children. Sci. Rep. 9 (1), 18739. doi:10.1038/s41598-019-55283-8
Conboy, M. J., Conboy, I. M., and Rando, T. A. (2013). Heterochronic parabiosis: Historical perspective and methodological considerations for studies of aging and longevity. Aging Cell 12 (3), 525–530. doi:10.1111/acel.12065
Correa-Burrows, P., Burrows, R., Albala, C., Court, F. A., Salech, F., Sanhueza, G., et al. (2022). Multiple events case-control study in a prospective cohort to identify systemic, cellular, and molecular biomarkers of obesity-induced accelerated aging in 30-years-olds: The ObAGE study protocol. BMC Geriatr. 22 (1), 387. doi:10.1186/s12877-022-03032-4
Cui, D., and Xu, X. (2018). DNA methyltransferases, DNA methylation, and age-associated cognitive function. Int. J. Mol. Sci. 19 (5), 1315. doi:10.3390/ijms19051315
de Lange, T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19 (18), 2100–2110. doi:10.1101/gad.1346005
de Toro-Martín, J., Guénard, F., Tchernof, A., Hould, F. S., Lebel, S., Julien, F., et al. (2019). Body mass index is associated with epigenetic age acceleration in the visceral adipose tissue of subjects with severe obesity. Clin. Epigenetics 11 (1), 172. doi:10.1186/s13148-019-0754-6
Drinkwater, R. D., Blake, T. J., Morley, A. A., and Turner, D. R. (1989). Human lymphocytes aged in vivo have reduced levels of methylation in transcriptionally active and inactive DNA. Mutat. Res. 219 (1), 29–37. doi:10.1016/0921-8734(89)90038-6
Dugué, P. A., Bassett, J. K., Joo, J. E., Jung, C. H., Ming Wong, E., Moreno-Betancur, M., et al. (2018). DNA methylation-based biological aging and cancer risk and survival: Pooled analysis of seven prospective studies. Int. J. Cancer 142 (8), 1611–1619. doi:10.1002/ijc.31189
Etzel, L., Hastings, W. J., Hall, M. A., Heim, C. M., Meaney, M. J., Noll, J. G., et al. (2022). Obesity and accelerated epigenetic aging in a high-risk cohort of children. Sci. Rep. 12 (1), 8328. doi:10.1038/s41598-022-11562-5
Fiorito, G., Caini, S., Palli, D., Bendinelli, B., Saieva, C., Ermini, I., et al. (2021). DNA methylation-based biomarkers of aging were slowed down in a two-year diet and physical activity intervention trial: The DAMA study. Aging Cell 20 (10), e13439. doi:10.1111/acel.13439
Fiorito, G., McCrory, C., Robinson, O., Carmeli, C., Ochoa-Rosales, C., Zhang, Y., Colicino, E., Dugué, P. A., Artaud, F., McKay, G. J., Jeong, A., Mishra, P. P., Nøst, T. H., Krogh, V., Panico, S., Sacerdote, C., Tumino, R., Palli, D., Matullo, G., Guarrera, S., et al. Lifepath consortium (2019). Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: A multi-cohort analysis. Aging 11 (7), 2045–2070. doi:10.18632/aging.101900
Fontaine, K. R., Redden, D. T., Wang, C., Westfall, A. O., and Allison, D. B. (2003). Years of life lost due to obesity. JAMA 289 (2), 187–193. doi:10.1001/jama.289.2.187
Foti, D. P., and Brunetti, A. (2017). Editorial: "Linking hypoxia to obesity. Front. Endocrinol. 8, 34. doi:10.3389/fendo.2017.00034
Fraga, M. F., and Esteller, M. (2007). Epigenetics and aging: The targets and the marks. Trends Genet. 23 (8), 413–418. doi:10.1016/j.tig.2007.05.008
Fransquet, P. D., Wrigglesworth, J., Woods, R. L., Ernst, M. E., and Ryan, J. (2019). The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin. Epigenetics 11 (1), 62. doi:10.1186/s13148-019-0656-7
Franzago, M., Fraticelli, F., Stuppia, L., and Vitacolonna, E. (2019). Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 14 (3), 215–235. doi:10.1080/15592294.2019.1582277
Franzago, M., Santurbano, D., Vitacolonna, E., and Stuppia, L. (2020). Genes and diet in the prevention of chronic diseases in future generations. Int. J. Mol. Sci. 21 (7), 2633. doi:10.3390/ijms21072633
Freund, A., Laberge, R. M., Demaria, M., and Campisi, J. (2012). Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23 (11), 2066–2075. doi:10.1091/mbc.E11-10-0884
Fujiki, K., Kano, F., Shiota, K., and Murata, M. (2009). Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 7, 38. doi:10.1186/1741-7007-7-38
Garagnani, P., Bacalini, M. G., Pirazzini, C., Gori, D., Giuliani, C., Mari, D., et al. (2012). Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 11 (6), 1132–1134. doi:10.1111/acel.12005
Garcia-Calzon, S., Moleres, A., Marcos, A., Campoy, C., Moreno, L. A., Azcona-Sanjulián, M. C., et al. EVASYON Study Group (2014). Telomere length as a biomarker for adiposity changes after a multidisciplinary intervention in overweight/obese adolescents: The EVASYON study. PloS one 9 (2), e89828. doi:10.1371/journal.pone.0089828
Gardner, J. P., Li, S., Srinivasan, S. R., Chen, W., Kimura, M., Lu, X., et al. (2005). Rise in insulin resistance is associated with escalated telomere attrition. Circulation 111 (17), 2171–2177. doi:10.1161/01.CIR.0000163550.70487.0B
Gentilini, D., Mari, D., Castaldi, D., Remondini, D., Ogliari, G., Ostan, R., et al. (2013). Role of epigenetics in human aging and longevity: Genome-wide DNA methylation profile in centenarians and centenarians' offspring. Age Dordr. Neth. 35 (5), 1961–1973. doi:10.1007/s11357-012-9463-1
Gibbs, W. W. (2014). Biomarkers and ageing: The clock-watcher. Nature 508 (7495), 168–170. doi:10.1038/508168a
Gregory, M. K., Gibson, R. A., Cook-Johnson, R. J., Cleland, L. G., and James, M. J. (2011). Elongase reactions as control points in long-chain polyunsaturated fatty acid synthesis. PloS one 6 (12), e29662. doi:10.1371/journal.pone.0029662
Grönniger, E., Weber, B., Heil, O., Peters, N., Stäb, F., Wenck, H., et al. (2010). Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 6 (5), e1000971. doi:10.1371/journal.pgen.1000971
Grun, L. K., Teixeira, N., Mengden, L. V., de Bastiani, M. A., Parisi, M. M., Bortolin, R., et al. (2018). TRF1 as a major contributor for telomeres' shortening in the context of obesity. Free Radic. Biol. Med. 129, 286–295. doi:10.1016/j.freeradbiomed.2018.09.039
Hannum, G., GuinneyJ, ZhaoL., Zhao, L., Zhang, L., Hughes, G., Sadda, S., et al. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. doi:10.1016/j.molcel.2012.10.016
Hernandez, D. G., Nalls, M. A., Gibbs, J. R., Arepalli, S., van der Brug, M., Chong, S., et al. (2011). Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum. Mol. Genet. 20 (6), 1164–1172. doi:10.1093/hmg/ddq561
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14 (10), R115. doi:10.1186/gb-2013-14-10-r115
Horvath, S., Erhart, W., Brosch, M., Ammerpohl, O., von Schönfels, W., Ahrens, M., et al. (2014). Obesity accelerates epigenetic aging of human liver. Proc. Natl. Acad. Sci. U. S. A. 111 (43), 15538–15543. doi:10.1073/pnas.1412759111
Horvath, S., Pirazzini, C., Bacalini, M. G., Gentilini, D., Di Blasio, A. M., Delledonne, M., et al. (2015). Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 7 (12), 1159–1170. doi:10.18632/aging.100861
Horvath, S., and Raj, K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19 (6), 371–384. doi:10.1038/s41576-018-0004-3
Houde, A. A., Légaré, C., Biron, S., Lescelleur, O., Biertho, L., Marceau, S., et al. (2015). Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL-cholesterol levels in severely obese men and women. BMC Med. Genet. 16, 29. doi:10.1186/s12881-015-0174-1
Johnson, A. A., Akman, K., Calimport, S. R., Wuttke, D., Stolzing, A., and de Magalhães, J. P. (2012). The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res. 15 (5), 483–494. doi:10.1089/rej.2012.1324
Kananen, L., Marttila, S., Nevalainen, T., Kummola, L., Junttila, I., Mononen, N., et al. (2016). The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: Evidence from two longitudinal studies. Age Dordr. Neth. 38 (3), 65. doi:10.1007/s11357-016-9927-9
Keyes, M. K., Jang, H., Mason, J. B., Liu, Z., Crott, J. W., Smith, D. E., et al. (2007). Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. J. Nutr. 137 (7), 1713–1717. doi:10.1093/jn/137.7.1713
Kim, A. Y., Park, Y. J., Pan, X., Shin, K. C., Kwak, S. H., Bassas, A. F., et al. (2015). Obesity-induced DNA hypermethylation of the adiponectin gene mediates insulin resistance. Nat. Commun. 6, 7585. doi:10.1038/ncomms8585
Kresovich, J. K., Garval, E. L., Martinez Lopez, A. M., Xu, Z., Niehoff, N. M., White, A. J., et al. (2021). Associations of body composition and physical activity level with multiple measures of epigenetic age acceleration. Am. J. Epidemiol. 190 (6), 984–993. doi:10.1093/aje/kwaa251
Levine, M. E., Lu, A. T., Chen, B. H., Hernandez, D. G., Singleton, A. B., Ferrucci, L., et al. (2016). Menopause accelerates biological aging. Proc. Natl. Acad. Sci. U. S. A. 113 (33), 9327–9332. doi:10.1073/pnas.1604558113
Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S., et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging 10 (4), 573–591. doi:10.18632/aging.101414
Lewis, K. A., and Tollefsbol, T. O. (2017). The influence of an epigenetics diet on the cancer epigenome. Epigenomics 9 (9), 1153–1155. doi:10.2217/epi-2017-0077
Li, Y., and Tollefsbol, T. O. (2011). p16(INK4a) suppression by glucose restriction contributes to human cellular lifespan extension through SIRT1-mediated epigenetic and genetic mechanisms. PloS one 6 (2), e17421. doi:10.1371/journal.pone.0017421
Lin, S., Huo, X., Zhang, Q., Fan, X., Du, L., Xu, X., et al. (2013). Short placental telomere was associated with cadmium pollution in an electronic waste recycling town in China. PloS one 8 (4), e60815. doi:10.1371/journal.pone.0060815
Longo, V. D., Antebi, A., Bartke, A., Barzilai, N., Brown-Borg, H. M., Caruso, C., et al. (2015). Interventions to slow aging in humans: Are we ready? Aging Cell 14 (4), 497–510. doi:10.1111/acel.12338
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153 (6), 1194–1217. doi:10.1016/j.cell.2013.05.039
Lowe, D., Horvath, S., and Raj, K. (2016). Epigenetic clock analyses of cellular senescence and ageing. Oncotarget 7 (8), 8524–8531. doi:10.18632/oncotarget.7383
Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11 (2), 303–327. doi:10.18632/aging.101684
Lundgren, J. R., Janus, C., Jensen, S., Juhl, C. R., Olsen, L. M., Christensen, R. M., et al. (2021). Healthy weight loss maintenance with exercise, liraglutide, or both combined. N. Engl. J. Med. 384 (18), 1719–1730. doi:10.1056/NEJMoa2028198
Mahmoudi, S., Xu, L., and Brunet, A. (2019). Turning back time with emerging rejuvenation strategies. Nat. Cell Biol. 21 (1), 32–43. doi:10.1038/s41556-018-0206-0
Marioni, R. E., Harris, S. E., Shah, S., McRae, A. F., von Zglinicki, T., Martin-Ruiz, C., et al. (2018). The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int. J. Epidemiol. 45 (2), 424–432. doi:10.1093/ije/dyw041
Marioni, R. E., Shah, S., McRae, A. F., Ritchie, S. J., Muniz-Terrera, G., Harris, S. E., et al. (2015). The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 44 (4), 1388–1396. doi:10.1093/ije/dyu277
Marttila, S., Kananen, L., Häyrynen, S., Jylhävä, J., Nevalainen, T., Hervonen, A., et al. (2015). Ageing-associated changes in the human DNA methylome: Genomic locations and effects on gene expression. BMC genomics 16 (1), 179. doi:10.1186/s12864-015-1381-z
Melzner, I., Scott, V., Dorsch, K., Fischer, P., Wabitsch, M., Brüderlein, S., et al. 2002). Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J. Biol. Chem. 277 (47), 45420–45427. doi:10.1074/jbc.M208511200
Mozhui, K., Lu, A. T., Li, C. Z., Haghani, A., Sandoval-Sierra, J. V., Wu, Y., et al. (2022). Genetic loci and metabolic states associated with murine epigenetic aging. eLife 11, e75244. doi:10.7554/eLife.75244
Nannini, D. R., Joyce, B. T., Zheng, Y., Gao, T., Liu, L., Yoon, G., et al. (2019). Epigenetic age acceleration and metabolic syndrome in the coronary artery risk development in young adults study. Clin. Epigenetics 11 (1), 160. doi:10.1186/s13148-019-0767-1
Narasimhan, A., Flores, R. R., Robbins, P. D., and Niedernhofer, L. J. (2021). Role of cellular senescence in type II diabetes. Endocrinology 162, bqab136. doi:10.1210/endocr/bqab136
Nevalainen, T., Kananen, L., Marttila, S., Jylhävä, J., Mononen, N., Kähönen, M., et al. (2017). Obesity accelerates epigenetic aging in middle-aged but not in elderly individuals. Clin. Epigenetics 9, 20. doi:10.1186/s13148-016-0301-7
Oliveira, A. M., Hemstedt, T. J., and Bading, H. (2012). Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15 (8), 1111–1113. doi:10.1038/nn.3151
Olova, N., Simpson, D. J., Marioni, R. E., and Chandra, T. (2019). Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell 18 (1), e12877. doi:10.1111/acel.12877
Paltoglou, G., Raftopoulou, C., Nicolaides, N. C., Genitsaridi, S. M., Karampatsou, S. I., Papadopoulou, M., et al. (2021). A comprehensive, multidisciplinary, personalized, lifestyle intervention program is associated with increased leukocyte telomere length in children and adolescents with overweight and obesity. Nutrients 13 (8), 2682. doi:10.3390/nu13082682
Pan, X., Yan, W., Qiu, B., Liao, Y., Liao, Y., Wu, S., et al. (2020). Aberrant DNA methylation of Cyclind-CDK4-p21 is associated with chronic fluoride poisoning. Chem. Biol. Interact. 315, 108875. doi:10.1016/j.cbi.2019.108875
Pérez, L. M., Pareja-Galeano, H., Sanchis-Gomar, F., Emanuele, E., Lucia, A., and Gálvez, B. G. (2016). Adipaging': Ageing and obesity share biological hallmarks related to a dysfunctional adipose tissue. J. Physiol. 594 (12), 3187–3207. doi:10.1113/JP271691
Perna, L., Zhang, Y., Mons, U., Holleczek, B., Saum, K. U., and Brenner, H. (2016). Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenetics 8, 64. doi:10.1186/s13148-016-0228-z
Pi-Sunyer, X. (2009). The medical risks of obesity. Postgrad. Med. 121 (6), 21–33. doi:10.3810/pgm.2009.11.2074
Qimuge, N., He, Z., Qin, J., Sun, Y., Wang, X., Yu, T., et al. (2019). Overexpression of DNMT3A promotes proliferation and inhibits differentiation of porcine intramuscular preadipocytes by methylating p21 and PPARg promoters. Gene 696, 54–62. doi:10.1016/j.gene.2019.02.029
Quach, A., Levine, M. E., Tanaka, T., Lu, A. T., Chen, B. H., Ferrucci, L., et al. (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 9 (2), 419–446. doi:10.18632/aging.101168
Raffington, L., Belsky, D. W., Kothari, M., Malanchini, M., Tucker-Drob, E. M., and Harden, K. P. (2021). Socioeconomic disadvantage and the pace of biological aging in children. Pediatrics 147 (6), e2020024406. doi:10.1542/peds.2020-024406
Raj, K. (2018). “The epigenetic clock and aging,” in Epigenetics of aging and longevity (Academic Press), 95–118.
Ramaker, M. E., Corcoran, D. L., Apsley, A. T., Kobor, M. S., Kraus, V. B., Kraus, W. E., et al. (2022). Epigenome-wide association study analysis of calorie restriction in humans, CALERIE TM Trial analysis. J. Gerontol. A Biol. Sci. Med. Sci. 168, glac168. doi:10.1093/gerona/glac168
Ray, D., Wu, A., Wilkinson, J. E., Murphy, H. S., Lu, Q., Kluve-Beckerman, B., et al. (2006). Aging in heterozygous dnmt1-deficient mice: Effects on survival, the DNA methylation genes, and the development of amyloidosis. J. Gerontol. A Biol. Sci. Med. Sci. 61 (2), 115–124. doi:10.1093/gerona/61.2.115
Reuser, M., Bonneux, L., and Willekens, F. (2008). The burden of mortality of obesity at middle and old age is small. A life table analysis of the US Health and Retirement Survey. Eur. J. Epidemiol. 23 (9), 601–607. doi:10.1007/s10654-008-9269-8
Rönn, T., Volkov, P., Davegårdh, C., Dayeh, T., Hall, E., Olsson, A. H., et al. (2013). A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 9 (6), e1003572. doi:10.1371/journal.pgen.1003572
Ryan, C. P. (2021). Epigenetic clocks": Theory and applications in human biology. Am. J. Hum. Biol. 33 (3), e23488. doi:10.1002/ajhb.23488
Sae-Lee, C., Corsi, S., Barrow, T. M., Kuhnle, G., Bollati, V., Mathers, J. C., et al. (2018). Dietary intervention modifies DNA methylation age assessed by the epigenetic clock. Mol. Nutr. Food Res. 62 (23), e1800092. doi:10.1002/mnfr.201800092
Salas-Pérez, F., Ramos-Lopez, O., Mansego, M. L., Milagro, F. I., Santos, J. L., Riezu-Boj, J. I., et al. (2019). DNA methylation in genes of longevity-regulating pathways: Association with obesity and metabolic complications. Aging 11 (6), 1874–1899. doi:10.18632/aging.101882
Salminen, A., Ojala, J., and Kaarniranta, K. (2011). Apoptosis and aging: Increased resistance to apoptosis enhances the aging process. Cell. Mol. Life Sci. 68 (6), 1021–1031. doi:10.1007/s00018-010-0597-y
Samblas, M., Milagro, F. I., and Martínez, A. (2019). DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics 14 (5), 421–444. doi:10.1080/15592294.2019.1595297
Santos, A. L., and Sinha, S. (2021). Obesity and aging: Molecular mechanisms and therapeutic approaches. Ageing Res. Rev. 67, 101268. doi:10.1016/j.arr.2021.101268
Schork, N. J., Beaulieu-Jones, B., Liang, W., Smalley, S., and Goetz, L. H. (2022). Does modulation of an epigenetic clock define a geroprotector? Adv. Geriatr. Med. Res. 4 (1), e220002. doi:10.20900/agmr20220002
Simpkin, A. J., Cooper, R., Howe, L. D., Relton, C. L., Davey Smith, G., Teschendorff, A., et al. (2017). Are objective measures of physical capability related to accelerated epigenetic age? Findings from a British birth cohort. BMJ open 7 (10), e016708. doi:10.1136/bmjopen-2017-016708
Simpkin, A. J., Hemani, G., Suderman, M., Gaunt, T. R., Lyttleton, O., Mcardle, W. L., et al. (2016). Prenatal and early life influences on epigenetic age in children: A study of mother-offspring pairs from two cohort studies. Hum. Mol. Genet. 25 (1), 191–201. doi:10.1093/hmg/ddv456
Simpson, D. J., Olova, N. N., and Chandra, T. (2021). Cellular reprogramming and epigenetic rejuvenation. Clin. Epigenetics 13 (1), 170. doi:10.1186/s13148-021-01158-7
Stuppia, L., Franzago, M., Ballerini, P., Gatta, V., and Antonucci, I. (2015). Epigenetics and male reproduction: The consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin. Epigenetics 7 (1), 1–5. doi:10.1186/s13148-015-0155-4
Tam, B. T., Morais, J. A., and Santosa, S. (2020). Obesity and ageing: Two sides of the same coin. Obes. Rev. 21 (4), e12991. doi:10.1111/obr.12991
Taormina, G., and Mirisola, M. G. (2015). Longevity: Epigenetic and biomolecular aspects. Biomol. Concepts 6 (2), 105–117. doi:10.1515/bmc-2014-0038
Taylor, R. W., and Turnbull, D. M. (2005). Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6 (5), 389–402. doi:10.1038/nrg1606
Tekola-Ayele, F. (2021). Invited commentary: Epigenetic clocks and obesity-towards the next frontier using integrative approaches and early-life models. Am. J. Epidemiol. 190 (6), 994–997. doi:10.1093/aje/kwaa252
Thurston, R. C., Carroll, J. E., Levine, M., Chang, Y., Crandall, C., Manson, J. E., et al. (2020). Vasomotor symptoms and accelerated epigenetic aging in the Women's health initiative (WHI). J. Clin. Endocrinol. Metab. 105 (4), dgaa081–1227. doi:10.1210/clinem/dgaa081
Unnikrishnan, A., Freeman, W. M., Jackson, J., Wren, J. D., Porter, H., and Richardson, A. (2019). The role of DNA methylation in epigenetics of aging. Pharmacol. Ther. 195, 172–185. doi:10.1016/j.pharmthera.2018.11.001
Vanyushin, B. F., Nemirovsky, L. E., Klimenko, V. V., Vasiliev, V. K., and Belozersky, A. N. (1973). The 5-methylcytosine in DNA of rats. Gerontology 19 (3), 138–152. doi:10.1159/000211967
Waki, T., Tamura, G., Sato, M., and Motoyama, T. (2003). Age-related methylation of tumor suppressor and tumor-related genes: An analysis of autopsy samples. Oncogene 22 (26), 4128–4133. doi:10.1038/sj.onc.1206651
Wang, T., Tsui, B., Kreisberg, J. F., Robertson, N. A., Gross, A. M., Yu, M. K., et al. (2017). Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18 (1), 57. doi:10.1186/s13059-017-1186-2
Prospective Studies Collaboration Whitlock, G., Lewington, S., Sherliker, P., Clarke, R., Emberson, J., Halsey, J., et al. (2009). Body-mass index and cause-specific mortality in 900 000 adults: Collaborative analyses of 57 prospective studies. Lancet (London, Engl. 373 (9669), 1083–1096. doi:10.1016/S0140-6736(09)60318-4
Yang, Q., Liang, X., Sun, X., Zhang, L., Fu, X., Rogers, C. J., et al. (2016). AMPK/α-Ketoglutarate Axis dynamically mediates DNA demethylation in the Prdm16 promoter and Brown adipogenesis. Cell Metab. 24 (4), 542–554. doi:10.1016/j.cmet.2016.08.010
Yang, Z., Huang, X., Jiang, H., Zhang, Y., Liu, H., Qin, C., et al. (2009). Short telomeres and prognosis of hypertension in a Chinese population. Hypertension 53 (4), 639–645. doi:10.1161/HYPERTENSIONAHA.108.123752
Yuzefovych, L. V., Musiyenko, S. I., Wilson, G. L., and Rachek, L. I. (2013). Mitochondrial DNA damage and dysfunction, and oxidative stress are associated with endoplasmic reticulum stress, protein degradation and apoptosis in high fat diet-induced insulin resistance mice. PloS one 8 (1), e54059. doi:10.1371/journal.pone.0054059
Zannas, A. S., Arloth, J., Carrillo-Roa, T., Iurato, S., Röh, S., Ressler, K. J., et al. (2015). Lifetime stress accelerates epigenetic aging in an urban, african American cohort: Relevance of glucocorticoid signaling. Genome Biol. 16, 266. doi:10.1186/s13059-015-0828-5
Keywords: DNA methylation, DNAm age, epigenetics, obesity, lifestyle, BMI
Citation: Franzago M, Pilenzi L, Di Rado S, Vitacolonna E and Stuppia L (2022) The epigenetic aging, obesity, and lifestyle. Front. Cell Dev. Biol. 10:985274. doi: 10.3389/fcell.2022.985274
Received: 03 July 2022; Accepted: 22 August 2022;
Published: 13 September 2022.
Edited by:
Maria Rosaria Matarazzo, National Research Council (CNR), ItalyReviewed by:
Apiwat Mutirangura, Chulalongkorn University, ThailandCopyright © 2022 Franzago, Pilenzi, Di Rado, Vitacolonna and Stuppia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liborio Stuppia, c3R1cHBpYUB1bmljaC5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.