- 1School of Life Sciences, Gibbet Hill Campus, University of Warwick, Coventry, United Kingdom
- 2Randall Centre for Cell and Molecular Biophysics, New Hunt’s House, Guy’s Campus, King’s College London, London, United Kingdom
- 3Warwick Medical School, Gibbet Hill Campus, University of Warwick, Coventry, United Kingdom
- 4Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
The T-box family transcription factor Eomesodermin (Eomes) is present in all vertebrates, with many key roles in the developing mammalian embryo and immune system. Homozygous Eomes mutant mouse embryos exhibit early lethality due to defects in both the embryonic mesendoderm and the extraembryonic trophoblast cell lineage. In contrast, zebrafish lacking the predominant Eomes homologue A (Eomesa) do not suffer complete lethality and can be maintained. This suggests fundamental differences in either the molecular function of Eomes orthologues or the molecular configuration of processes in which they participate. To explore these hypotheses we initially analysed the expression of distinct Eomes isoforms in various mouse cell types. Next we compared the functional capabilities of these murine isoforms to zebrafish Eomesa. These experiments provided no evidence for functional divergence. Next we examined the functions of zebrafish Eomesa and other T-box family members expressed in early development, as well as its paralogue Eomesb. Though Eomes is a member of the Tbr1 subfamily we found evidence for functional redundancy with the Tbx6 subfamily member Tbx16, known to be absent from eutherians. However, Tbx16 does not appear to synergise with Eomesa cofactors Mixl1 and Gata5. Finally, we analysed the ability of Eomesa and other T-box factors to induce zebrafish left-right organiser progenitors (known as dorsal forerunner cells) known to be positively regulated by vgll4l, a gene we had previously shown to be repressed by Eomesa. Here we demonstrate that Eomesa indirectly upregulates vgll4l expression via interlocking feedforward loops, suggesting a role in establishment of left-right asymmetry. Conversely, other T-box factors could not similarly induce left-right organiser progenitors. Overall these findings demonstrate conservation of Eomes molecular function and participation in similar processes, but differential requirements across evolution due to additional co-expressed T-box factors in teleosts, albeit with markedly different molecular capabilities. Our analyses also provide insights into the role of Eomesa in left-right organiser formation in zebrafish.
Introduction
T-box transcription factors (TFs) are an ancient family of transcriptional regulators with diverse roles in development and disease (Papaioannou, 2014). Eomesodermin (Eomes) belongs to the Tbr1 subfamily of T-box TFs, consisting of similarly sized N- and C-terminal domains (NTD and CTD) flanking a central DNA binding domain known as the T-box. Amongst the species where Eomes is best studied are mouse and zebrafish (Probst and Arnold, 2017). Mice have a single copy of Eomes, whereas zebrafish owing to the whole genome duplication in the teleost lineage have two paralogous genes, eomesa and eomesb (Glasauer and Neuhauss, 2014). During mouse embryogenesis Eomes plays essential roles in trophectoderm (Russ et al., 2000; Strumpf et al., 2005), in the primitive streak for epithelial-to-mesenchymal transition, mesoderm migration and specification of definitive endoderm and cardiac mesoderm during gastrulation (Arnold et al., 2008a; Costello et al., 2011). Additionally Eomes acts in the visceral endoderm to control anterior-posterior axis identity (Nowotschin et al., 2013) and later has key functions in cortical neuron progenitors (Arnold et al., 2008b). It is also expressed in progenitors of the left-right organiser known in mammals as the node, and is required for correct formation of the node suggesting a potential role in establishing left-right asymmetry (Arnold et al., 2008a; Costello et al., 2011). In zebrafish Eomesa also plays multiple roles in mesendoderm formation. It acts in conjunction with Hwa to control expression of Nodal pathway ligands ndr1/2, leading to mesendoderm induction (Xing et al., 2022). Eomesa can also induce ectopic endoderm if overexpressed with essential interacting factors (Bjornson et al., 2005), and is sufficient to induce dorsal mesoderm markers and represses ectoderm gene expression in early development (Bruce et al., 2003; Nelson et al., 2014). Furthermore, Eomesa is sufficient to induce progenitors of the left-right organiser, known as dorsal forerunner cells (DFCs) in zebrafish (Bjornson et al., 2005). However, we previously found that Eomesa represses expression of the transcriptional cofactor vgll4l (Nelson et al., 2014), a key positive regulator of DFC proliferation, survival and function (Fillatre et al., 2019). Here we further investigate these paradoxical findings.
Mouse Eomes and zebrafish eomesa display similar expression domains during early development (Russ et al., 2000; Mione et al., 2001; Pearce et al., 2003; Takizawa et al., 2007; Du et al., 2012; Takizawa et al., 2014). However, surprisingly endoderm, cardiac mesoderm and axial patterning proceed normally in eomesa loss-of-function mutants (Du et al., 2012). This observation cannot be explained simply by rescue by eomesb, which is not co-expressed with eomesa in early development, nor is it induced in eomesa mutant embryos (Vesterlund et al., 2011; Nelson et al., 2014). The extent to which Eomes functional activities are conserved between zebrafish and mouse remains unknown.
One possibility is that these distinct loss-of-function phenotypes could potentially be due to functional diversification during evolution. The process of alternative splicing (AS) allows a single gene to give rise to multiple isoforms with different functional characteristics. The prevalence of AS has expanded across evolutionary time, allowing increased proteome diversity out of proportion with gene number (Keren et al., 2010). For example, only ∼25% of nematode genes have alternative isoforms compared to >90% in human (Wang et al., 2008; Ramani et al., 2011). AS leading to functional diversification may account for altered functions of Eomes between species. However, it is also possible that differential requirement for Eomes is due to functional redundancy owing to altered complements of T-box factors in different vertebrate evolutionary lineages. The most ancient T-box factor Brachyury (otherwise known as Tbxt) is present in several non-metazoan lineages, however, the T-box family is considerably expanded in Metazoa, reflecting its developmental importance (Sebe-Pedros et al., 2013). Additionally, the complement of T-box factors has varied across vertebrate evolution, with gain or loss of individual factors in certain lineages. For example, the Tbx6 subfamily member tbx16 is present in fish, frogs, birds, marsupials and monotremes but lost in placental mammals (Ahn et al., 2012). The T-box domain itself directly binds DNA in a sequence-specific manner. Genome-wide profiling of multiple T-box factors including Eomes, Tbx16, Tbx6 and Brachyury in zebrafish, mice, Xenopus and human has revealed they bind most frequently to variants of an eight to nine base pair core consensus of (T)TVRCACHT, interchangeably allowing occupancy of different T-box factors at the same genomic sites e.g. (Morley et al., 2009; Teo et al., 2011; Nelson et al., 2012; Gentsch et al., 2013; Lolas et al., 2014; Nelson et al., 2014; Faial et al., 2015; Tsankov et al., 2015; Windner et al., 2015; Gentsch et al., 2017; Nelson et al., 2017). T-box factors therefore often exhibit redundancy through regulation of the same target genes through the same cis-regulatory modules.
We therefore sought to answer three key questions: 1. Are zebrafish and mouse Eomes genes functionally equivalent? 2. What is the basis for the observed differences in severity of loss-of-function phenotypes between mouse and zebrafish? and; 3. How can Eomesa promote DFC gene expression while repressing the key DFC regulator vgll4l?
Our analyses suggest that the molecular function of Eomes is highly conserved throughout vertebrate evolution. Our data also reveal that while alternative splicing of mouse Eomes transcript occurs at exon 6, functionally the encoded proteins were virtually indistinguishable. We found that Eomesa and Tbx16 share overlapping functions and capabilities in the presumptive endoderm, suggesting that phenotypic rescue by Tbx16 may explain eomesa mutant viability. Finally, we found that Eomesa acts within interlocking feedforward loops to both repress vgll4l and activate it indirectly via the essential SOX family transcription factor Sox32. Our results therefore advance our understanding of T-box factor functional conservation during early vertebrate embryogenesis, and regulatory networks controlling left-right organiser progenitor gene expression.
Materials and methods
Zebrafish strains
AB and mutant zebrafish were reared as described (Westerfield, 2000). For eomesa mutant experiments eggs from homozygous eomesafh105/fh105 females were in vitro fertilized with eomesa+/fh105 sperm yielding a mixture of Meomesa and MZeomesa mutant embryos. Since previous studies have revealed no differences in endodermal or mesodermal expression between Meomesa and MZeomesa mutant embryos we did not distinguish between them in this study (Du et al., 2012; Xu et al., 2014). All zebrafish studies complied fully with the United Kingdom Animals (Scientific Procedures) Act 1986 as implemented by King’s College London, The University of Warwick, or were in accordance with the policies of the University of Toronto Animal Care Committee.
Cloning for in vitro production of mRNAs and mammalian expression vectors
Full length tbx16 and eomesb open reading frames were cloned with C-terminal myc tags into XhoI and XbaI sites in pCS2+ by PCR from zebrafish cDNA using the following primers: tbx16-myc CATACTCGAGATGCAGGCTATCAGAGACCT and CGCGTCTAGACTACAGATCCTCTTCTGAGATGAGTTTTTGTTCCCAGCACGAGTATGAGAAAA; eomesb-myc ATATCTCGAGATGCCCGGAGAAGGATCCAG and GCGCTCTAGACTACAGATCCTCTTCTGAGATGAGTTTTTGTTCGCTGCTGGTGTAGAAGGCGTA. Full length gata5 cDNA with a C-terminal HA tag was similarly cloned into pCS2+ EcoRI and XhoI sites using primers CGCCGAATTCATGTATTCGAGCCTGGCTTT and AATGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTACGCTTGAGACAGAGCACACC. Eomes cloning into pCS2+ was performed between EcoRI and XhoI sites. pCS2+eomesaN320K was produced by PCR mutagenesis of the wild type construct using AAACTGAAGCTAACCAACAAGAAAGGAGCAAATAACAACAAT and TCCGAAAGATATTTCTTGTCT followed by recircularization. Eomesa ∆CTD was similarly produced using the following primers TAAGAACTGCTTTTCAAGATCCTTTATCAATCC and CGAATCATAATTGTCCCTGAA. The ∆NTD mutation was produced by removing the EcoRI/BstEII fragment from pCS2+eomesa and replacing with the EcoRI/BstEII fragment produced by PCR from pCS2+eomesa with primer pair GCCCTCGAATTCACAGTTAAGAATGGCGCGGGCGC and CCCGCAGGTCACCCACTTTCCGCCCTGAAATCTCCA.
mRNA, morpholinos and microinjections
All capped mRNA were synthesized from plasmids encoding proteins of interest in pCS2+ NotI linearization followed by SP6 transcription as described (Bruce et al., 2003), with the exception tbxta which was produced from pSP64T as described (Marcellini et al., 2003). mRNA quantities for T-box factors were scaled in order to inject equimolar amounts of each mRNA per embryo. One-cell stage embryos were injected with the following quantities: eomesa – 400pg; Eomes∆VR – 410 pg; EomesFL – 420 pg; eomesa∆NTD – 308 pg; eomesa∆CTD – 285 pg; eomesaN320K – 400 pg; eomesb-myc – 286 pg, tbx16-myc – 217 pg; tbxta – 223 pg; gata5-HA – 140 pg; mixl1 – 200 pg. For Tbx16 knockdown one-cell stage embryos were injected with, 0.5 pmol of a previously characterized tbx16 morpholino (GeneTools) (Bisgrove et al., 2005).
In vitro protein production
Unlabelled in vitro translated protein was produced using rabbit reticulocyte lysates according to manufacturer’s protocol (Promega).
Northern blot
Total RNA was extracted from specified cell types using Rneasy Mini Kits (QIAGEN), and polyA selected using Oligotex mRNA Mini Kits (QIAGEN). 500ng polyA + RNA per lane was size fractionated on a 1.5% agarose/MOPS gel, transferred onto Hybond N membranes (GE Healthcare), and probed with 32P-random-primed 1 kb XmaI-EcoRV cDNA fragment corresponding to the exon 1–4T-box region.
Western blot
Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer, subjected to SDS–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Membranes were blocked with 5% milk powder in Tris-buffered saline with Tween 20, incubated in primary antibodies overnight including rabbit anti-Eomes CTD (Abcam, ab23345, 1:2,000), rabbit anti-Eomes NTD (Santa Cruz, sc-98555, 1:1,000) and rat anti-Eomes (eBioscience, 14–4,876, 1:1,000). Secondary antibodies were donkey anti-rabbit horseradish peroxidase (GE Healthcare NA934, 1:2,000) and goat anti-rat horseradish peroxidase (GE Healthcare NA935, 1:2,000). Blots were developed by chemiluminescence using Amersham ECL Prime Detection Reagent (GE Healthcare).
Embryonic stem cell differentiation
Wild type (+), Eomesnull/null (null), feeder-depleted ESCs were cultured in DMEM (ThermoFisher) with 15% FCS, 1% non-essential amino acids, 0.1 mM β-mercaptoethanol and 1,000 U/ml recombinant leukaemia inhibitory factor (Millipore). For differentiation ESCs were resuspended at 1×104 cells/ml in DMEM (ThermoFisher) with 15% FCS, 1% non-essential amino acids, 0.1 mM β-mercaptoethanol in hanging drops (10 μL) plated on the inside lids of bacteriological dishes. After 48 h embryoid bodies were transferred in 10 ml medium to 10 cm bacteriological dishes and RNA extracted at the appropriate timepoints.
P19Cl6 cell culture and differentiation
P19Cl6 embryonal carcinoma cells were cultured in α-MEM (ThermoFisher) supplemented with 10% FCS. To induce differentiation, cells were seeded at 3.7×105 cells/6 cm dish in media containing 1% DMSO (Sigma) and RNA harvested after 72 h.
Cytotoxic T-cell lymphocyte, neomycin- and hygromycin B-resistant STO fibroblasts (SNH) and HeLa cell culture
CTLL cells derived from the ATCC TIB-214 line were maintained at 104–105 cells per ml in complete T cell medium supplemented with IL-2. SNH fibroblasts and HeLa cells were maintained on 0.1% gelatin coated dishes in DMEM supplemented with 10% bovine calf serum (Hyclone).
Reverse transcription–polymerase chain reaction
Cytoplasmic RNA was produced as previously described (Eggermont and Proudfoot, 1993). Total RNA was produced using Rneasy Mini Kits according to manufacturers protocol (QIAGEN). RT-PCR was performed using OneStep RT-PCR Kit (QIAGEN) using the following primers: Total Eomes–TGTTTTCGTGGAAGTGGTTCTGGC and AGGTCTGAGTCTTGGAAGGTTCATTC; Eomes exon 4-6 to distinguish FL and ∆VR isoforms ATCGTGGAAGTGACAGAGGACG and CGGGAAGAAGTTTTGAACGCC; Gapdh–TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG; Eomes start codon to ∆CTD 3′ UTR–ATATCTCGAGATGCAGTTGGGAGAGCAGCTC and TGGGCTCGAAGATGAAACTC; HBB exon 2 to Eomes exon 6 – GCACGTGGATCCTGAGAACT and CGGGAAGAAGTTTTGAACGCC. For nested PCR to test exon 5-6 splicing association with the long Eomes 3’ UTR the initial primer pair used was ATCGTGGAAGTGACAGAGGACG and CAAGTACGGAGGCAGCTGAG.
Whole-mount embryo staining
Whole-mount in situ hybridization (WISH) of zebrafish embryos were performed as described (Jowett and Lettice, 1994). Anti-sense riboprobes for noto (Talbot et al., 1995), chrd (Miller-Bertoglio et al., 1997), vgll4l (Nelson et al., 2014), zic3 (Grinblat and Sive, 2001), mixl1 (Alexander et al., 1999) and sox32 (Dickmeis et al., 2001) were produced as described. Blinding and randomisation was performed prior to categorical scoring of WISH embryos to prevent bias.
Cloning and mutagenesis to test eomes exon 6 splice sequences
Clones to test the splicing efficiency at Eomes exon 6 were generated by cloning PCR products using primers ACGGCAATTGGCCTCGAACATTCTTGCTTC and CCAGCCATCACTTTGGTCAAAGGTGGAAGGCAAAAG into MfeI-BstXI sites of the human HBB gene (GenBank accession no. U01317) within a previously described TAT-inducible expression vector (Ashe et al.,1997). Mutation of splicing sequences for the ∆VR and FL Eomes isoforms were introduced by PCR using primers: ∆VR - CATGTACACGGCTTCAGAAAACGACAGGTTAACGCCAAGTCCGACGGATTCCCCTCGATCCCATCAGATTGTCCCTGG and CTACAATATAAAGAGAGACACTTAAAAATAAAAAACAACCCTCACGTTGTCCCCAAACAAGCTGCCTCCCAGAAGC; FL–CATGTACACGGCTTCAGAAAATGACAGGTTAACTCCA TCTCCCACGGATTCCCC and GGACATTATATACACCGCCTCTTATATTTTTACACCAACCCTCACGTTGTCCCCAAACAAGCTGCCTCCCAGAAGC followed by recircularization of the resulting PCR products. Deletion of the VR was similarly performed using primers ATCCCATCAGATTGTCCCTGGA and CTACAATATAAAGAGAGACACTTAAAAATAAAAAACAACCCTCACGTTGTCCCCAAACAAGC TGCCTCCCAGAAGC. HBB plasmids were co-transfected with a plasmid expressing the HIV transactivator protein TAT (Adams et al., 1988), into HeLa cells using Lipotectamine 2000 according to manufacturers protocol (ThermoFisher).
Conservation analysis
The Tbr1 subfamily Gene Tree was generated by Ensembl (Yates et al., 2016). Eomes conservation measurements (phyloP) across 60 vertebrate species were visualized in UCSC Genome Browser (http://genome.ucsc.edu/) (Karolchik et al., 2004; Speir et al., 2016). Sequence logos of the Eomes variant region in placental mammals, other tetrapods and teleosts were based on alignment of the same 60 vertebrate species and visualized using WebLogo (Crooks et al., 2004).
Full-length protein alignments were performed using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011; McWilliam et al., 2013) and visualized using JalView (Waterhouse et al., 2009). BLOSUM62 average distance gene tree was also produced using Jalview.
Single-cell ribonucleic acid sequencing analysis
Single-cell (sc) RNA sequencing count data of zebrafish embryonic cells from Wagner et al (2018) was downloaded from GEO (Barrett et al., 2013; Wagner et al., 2018). Raw UMI-filtered count data in CSV format from 6 h post fertilisation (h.p.f.) embryos (GSM3067190) was imported in to R v3.6.2 and analysed using Seurat v3.0.2 (Stuart et al., 2019). Cells with less than 200 features, and features detected in <3 cells were discarded. The remaining count data were then normalised via SCTransform v0.2.1 (Hafemeister and Satija, 2019) with mitochondrial genes passed as a regression variable. Genes were clustered using UMAP utilising the R package uwot v0.1.5 (Melville et al., 2020), with the following parameters: dims = 1:30, n.neighbors = 5, min.dist = 0.001. To assign cell identities to clusters FindAllMarkers was called in Seurat using default parameters. For consistency with the original source publication of the 6 h.p.f. scRNA-seq data we cross-referenced the marker genes for each cluster in the present study with the clusters defined by Wagner et al. (2018). Significant positive markers in each of the 14 clusters defined by our analyses in Seurat were overlapped with the top 20 markers for each identity defined by Wagner et al. (2018). Cell identities were then assigned based on maximum concordance with markers defined by Wagner et al. (2018).
Results
Eomes isoforms, conservation and expression
In mouse the three annotated Eomes transcripts give rise to three structurally distinct isoforms including the full length (FL) product, a splice variant having an alternative splice acceptor site within exon 6 leading to loss of a 19 amino acid variant region (∆VR), and a transcript with an alternative mRNA 3’ cleavage site leading to loss of exon 6 and its encoded CTD (∆CTD) (Figure 1A). The highly conserved VR sequence is known to be phosphorylated at three amino acid residues in mouse spleen and kidney (Figure 1A) (Huttlin et al., 2010). The internal exon 6 splicing event emerged in the tetrapod lineage through a synonymous single nucleotide change in an arginine codon (CG>AG) introducing a splice acceptor sequence.
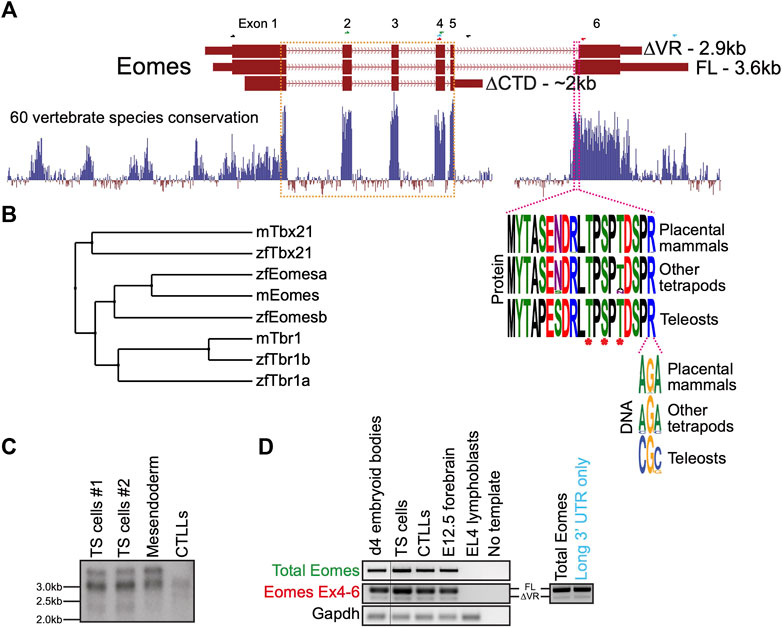
FIGURE 1. Mouse Eomes has multiple isoforms, including a mammalian-specific alternative splicing event. (A) Gene model with conservation track and sequence logos for variant region. All transcripts are Ensembl version 107 annotations - ∆VR transcript is ENSMUST00000111763; FL transcript is ENSMUST00000035020; ∆CTD transcript is ENSMUST00000150633. Annotated transcript sizes are indicated, as well as amino acid conservation of the VR between placental mammals, other tetrapods and teleosts, and the variation within the terminal VR arginine codon. The VR is defined by the amino acids present in ENSMUSP00000035020 (encoded by ENSMUST00000035020) that are absent from ENSMUSP00000107393 (encoded by ENSMUST00000111763). The T-box is outlined in orange and the VR in pink. Asterisks indicate known phosphorylated amino acid residues. RT-PCR primer pairs are indicated as half arrows and colour-coded as follows: black–to establish connectivity between the annotated start codon and ∆CTD isoform 3′ UTR; green–to assess total Eomes through amplification of exon 2–4; blue–to amplify Eomes cDNA between exon 4 and the distal 3′ UTR; red–to assess alternate splicing at exon 6. (B) BLOSUM62 average distance evolutionary tree of the Tbr1 subfamily showing relationships between mouse and zebrafish genes. (C) Northern blot showing Eomes transcripts in different cell types using a probe against the T-box. Data for two independent trophoblast stem (TS) cell lines are shown. Mesendoderm is P19Cl6 cells after 4 days of DMSO induced differentiation. CTLLs are IL-2-dependent T-cell lymphocytes derived from ATCC TIB-214. (D) RT-PCR showing relative levels of FL and ∆VR isoforms (left), and nested PCR showing FL/∆VR ratio for long 3′ UTR transcripts (right). Day 4 differentiated embryoid bodies contain cells mimicking embryonic endoderm. CTLLs are IL-2-dependent T-cell lymphocytes derived from ATCC TIB-214. EL4 cells are a negative control for Eomes expression. Gapdh is a loading control. Locations of primer pairs used for RT-PCR are shown in panel A and the text colour-coded accordingly. Nested PCR to analyse exon 6 splicing in transcripts containing the long 3′UTR was performed using the blue primer pair in panel A, followed by the red primer pair.
Because the ∆CTD transcript annotation has an incomplete 5′ end, it remains unclear whether it encodes the entire NTD. However, our RT-PCR analysis using primers located in the ∆CTD 3’ UTR and at the FL/∆VR start codon suggests that exon 1 coding information is intact (not shown). The CTD encoded by exon 6 has been shown to function in transcriptional activation (Picozzi et al., 2009), suggesting that the ∆CTD isoform is likely to be functionally compromised in comparison to FL and ∆VR isoforms. Consistent with this, the CTD is more highly conserved than the NTD (Figure 1A). Functional differences between FL and ∆VR isoforms, however, are yet to be examined. Both eomesa transcripts identified in zebrafish encode the same protein (Bruce et al., 2003). Relatively little is known about the single annotated zebrafish eomesb transcript, which appears to be more divergent from the ancestral gene (Figure 1B).
Murine Eomes is expressed in numerous cell types including trophoblast stem cells (TSCs), mesendoderm, and T lymphocytes. To identify Eomes transcripts we initially performed Northern blot analysis (Figure 1C). Transcript sizes corresponding to all three annotated isoforms were detectable but the ∆CTD transcript was underrepresented. The FL and ∆VR annotations display different 3′ UTR lengths. To test whether the two distinct upper bands detected by Northern blot correspond to alternative exon 6 splicing events or merely different UTR lengths we next performed nested PCR (Figure 1D). We found that the long 3′ UTR is associated with both the FL and ∆VR coding isoforms. Strikingly, the ratio of FL/∆VR is similar for both total Eomes and the long 3’ UTR transcripts. The abundance of the different coding transcripts therefore appears to be independent of UTR length. Further analysis through cloning Eomes intron5/exon6 to replace intron2/exon3 of the human HBB gene in an expression construct followed by transfection into HeLa cells revealed that the ratio of FL/∆VR splicing is consistent with the wild type Eomes gene, suggesting that the low levels of the ∆VR isoform are due to weaker splicing consensus sequences, thus favouring the FL isoform (Supplementary Figure S1). However, analysis of Eomes proteins by Western blot indicates that various N-terminal truncations occur which cannot be accounted for by the annotated coding transcripts (Supplementary Figure S2). We conclude that FL is clearly the most abundant of the three annotated coding isoforms. Importantly, this predominant isoform expressed by mouse cells corresponds to the single eomesa isoform in zebrafish.
Both mouse full length and ∆VR isoforms are functionally equivalent to zebrafish eomesa in early development
Zebrafish eomesa loss of function causes less severe phenotypes compared with the dramatic defects observed in Eomes mutant mouse embryos. To further explore mouse and zebrafish Eomes functional capabilities we overexpressed either mouse Eomes FL or ∆VR mRNAs in zebrafish embryos for comparison with those overexpressing zebrafish eomesa.
Zebrafish Eomesa represses ectoderm genes such as vgll4l and zic3 during blastula stages and activates mesendoderm genes including organizer markers noto and chrd at the onset of gastrulation (Bruce et al., 2003; Nelson et al., 2014). Injecting equimolar quantities of zebrafish eomesa mRNA, or mouse FL or ∆VR Eomes isoforms at the one-cell stage, we found that each was equally able to repress vgll4l and zic3 in mid/late blastulas (4 h post-fertilisation - h.p.f.) and induce noto and chrd in early gastrulas (6 h.p.f.; Figures 2A–D).
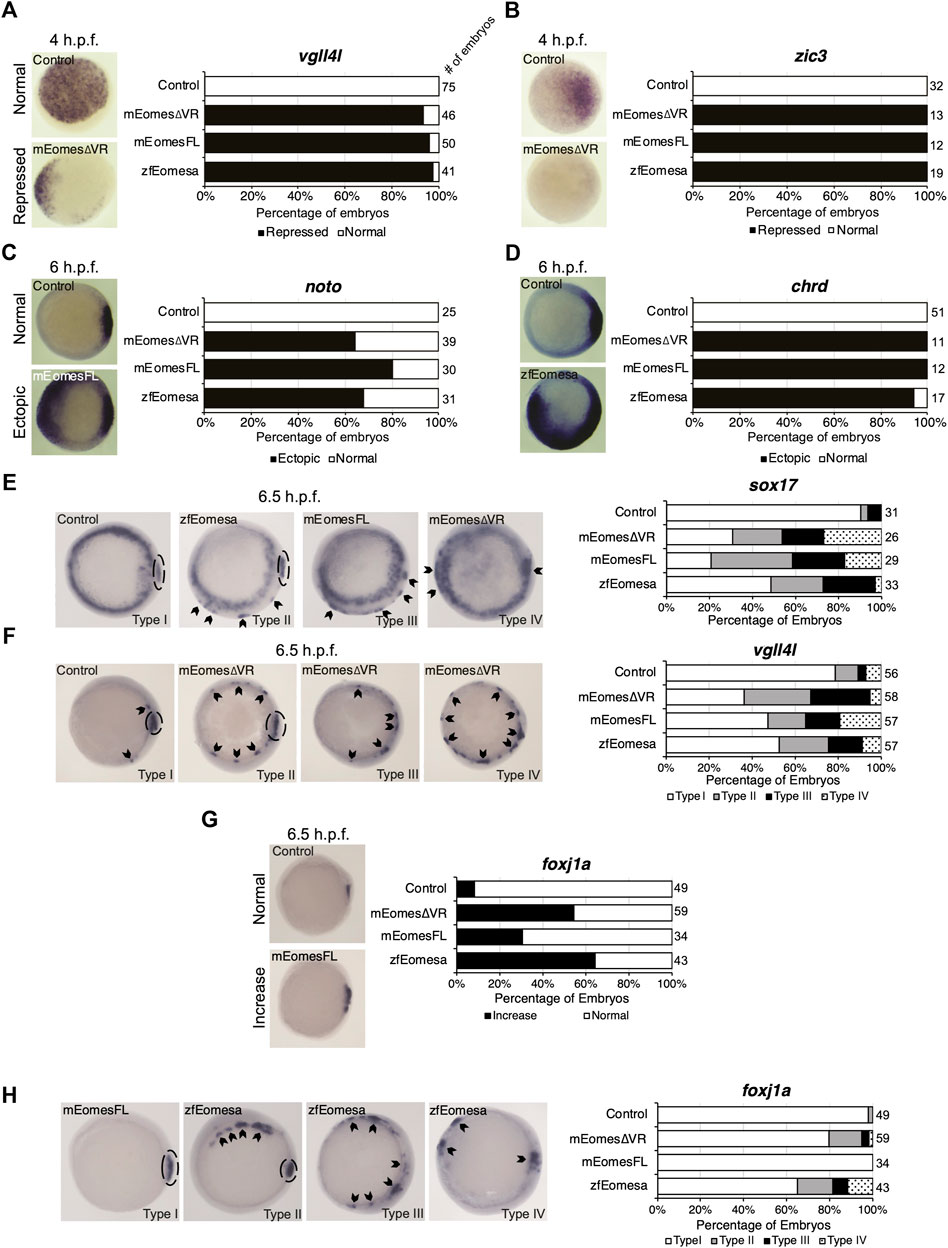
FIGURE 2. Both FL and ∆VR isoforms of mouse Eomes are functionally equivalent to zebrafish Eomesa in the early embryo. WISH analysis of ectoderm markers vgll4l and zic3 in mid/late blastulas (4 h.p.f.) embryos (A,B), organiser markers noto and chrd (C,D) in early gastrulas (6 h.p.f.), or DFC markers sox17, vgll4l and foxj1a (E–H) in early/mid gastrulas (6.5 h.p.f.). Embryos have been injected at the 1 cell stage to overexpress either mouse EomesFL, Eomes∆VR or zebrafish Eomesa. N = 2. Total numbers of embryos scored per condition are indicated. Representative images of expression patterns per gene per category are shown. (A–D) Animal views; dorsal to the right. (E) Animal views; dorsal to the right. (F–H) Vegetal views; dorsal to the right. Panel G indicates the percentage of embryos with greater intensity of dorsal foxj1a WISH staining, while panel H indicates percentages of embryos with ectopic foxj1a staining. Type I–wild type expression; Type II–ectopic dorsolateral expression with clear primary dorsal DFC cluster; Type III–dorsolateral expression with no defined primary cluster; Type IV–ectopic expression in the ventral margin. Dotted ovals indicate primary DFC clusters. Arrowheads indicate ectopic DFC marker expression.
Zebrafish Eomesa is suggested to induce DFCs, based on observation of ectopic sox17 expression in cells of the outer margin in early gastrulas (7 h.p.f.) on eomesa overexpression (Bjornson et al., 2005). We sought to determine whether these ectopic sox17 + cells express a broader range of DFC markers (sox17, vgll4l and foxj1a), and also whether they can be similarly induced by mouse Eomes. Notably, though Eomesa represses expression of vgll4l during blastula stages, during gastrulation vgll4l is expressed in DFCs, and has recently been identified as a key regulator of DFC proliferation, survival and function (Fillatre et al., 2019). Repression of vgll4l at later stages in DFCs would consequently be inconsistent with promoting DFC formation.
We found that eomesa, and Eomes FL or ∆VR isoforms were all similarly able to upregulate both sox17 and vgll4l in the outer margin of gastrulas. We further note that there was a diversity of phenotypes beyond wild type expression (type I) wherein ectopic dorsolateral expression was markedly observed in individual cells/small clusters outside the primary dorsal DFC cluster (type II), where there was dorsolateral expression with no defined primary cluster (type III), and where ectopic expression was also observed in the ventral margin (type IV) (Figures 2E,F). Conversely, for foxj1a we note that while eomesa, Eomes FL and ∆VR can induce a greater intensity of dorsal staining, and expansion of the dorsal DFC cluster, ectopic expression in the ventrolateral margin is rare compared to other DFC markers (Figures 2G,H).
We conclude that both FL and ∆VR mouse Eomes isoforms are functionally highly similar to zebrafish Eomesa in these contexts. Moreover, Eomesa regulation of vgll4l appears to be context-specific, repressing its expression during blastula stages while inducing its expression in DFCs during gastrulation. We further conclude that additional factors likely to be predominantly dorsally localised are required for robust upregulation of foxj1a compared to sox17 and vgll4l.
Eomesa and non-mammalian T-box factor Tbx16 redundantly regulate mixl1 expression at the initiation of zebrafish endoderm formation
Since results above strongly suggest mouse Eomes is functionally similar to zebrafish Eomesa, do eomesa mutants have comparatively mild defects due to functional redundancy with other T-box factors? Eomesb is not appreciably expressed during early zebrafish development (Figure 3A), and is not upregulated in eomesa mutants (Nelson et al., 2014) thus it seems unlikely that it compensates for loss of Eomesa. We recently identified a key role for the non-placental mammal T-box factor, Tbx16 in endoderm formation, with Brachyury homologue Tbxta having a more minor role (Nelson et al., 2017). Both of these factors show zygotic upregulation concomitant with declining eomesa mRNA levels and prior to expression of key markers of presumptive endoderm (e.g., gata5 and mixl1), and endoderm (e.g., gata5, sox32 and sox17; Figure 3A), thus may compensate for the early loss of expression of such markers in MZeomesa mutants (Du et al., 2012).
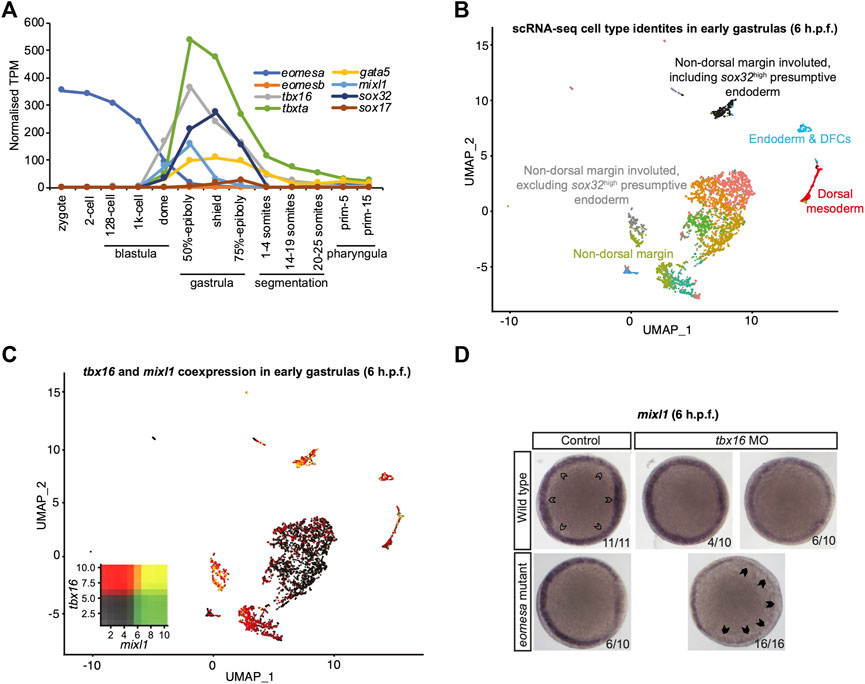
FIGURE 3. Eomesa and Tbx16 are redundantly required for mixl1 expression in the presumptive endoderm. (A) Timing of expression of T-box factors (eomesa, eomesb, tbxta, tbx16) under study and presumptive endoderm (mixl1, gata5, sox32) and endoderm markers (gata5, sox32, sox17) indicated by bulk RNA-seq data from (White et al., 2017). Gene expression is shown as transcripts per million (TPM). Stages are as defined by (Kimmel et al., 1995). (B) UMAP clustering analysis of single-cell RNA-seq data for early gastrulas (6 h.p.f.) zebrafish embryos (Wagner et al., 2018) indicating colour-coded cell type identities. Cell types relevant to the present study are labelled. The identities of all cell types are indicated in Supplementary Figure S3. (C) UMAP clustering analysis of single-cell RNA-seq data for early gastrulas (6 h.p.f.) zebrafish embryos indicating co-expression of tbx16 and mixl1 (Wagner et al., 2018). Heatmap insets indicate overall expression levels per gene and co-expression. Overlapping expression is shown in yellow. (D) WISH analysis of mixl1 in early gastrulas (5.7–6 h.p.f.) zebrafish embryos in wild type or eomesa mutant embryos (see Methods for information on genotype) with and without Tbx16 morpholino knockdown. Total numbers of embryos and fractions as categorised are indicated. Animal views; dorsal to the right. Open arrowheads indicate normal mixl1 expression at the blastoderm margin. Closed arrowheads indicate profound loss of mixl1 expression on tbx16 knockdown in eomesa mutants.
Moreover, single-cell RNA-seq data (Wagner et al., 2018) demonstrate that tbx16 is robustly co-expressed with the critical endodermal regulator mixl1 in the presumptive endoderm at early gastrulation stages (6 h.p.f.), suggesting the potential for Tbx16 to upregulate mixl1 expression to initiate endoderm specification, as our previous published analyses suggest (Nelson et al., 2017) (Figures 3B,C). Additional specific detail on cluster identities in Figures 3B,C is provided in Supplementary Figure S3.
To test whether Tbx16 functions redundantly with Eomesa during endoderm formation next we performed antisense morpholino knockdown of Tbx16 in wild type and eomesa mutant embryos. We found that while mixl1 expression is reduced on loss of Eomesa or Tbx16 alone, loss of both TFs leads to more striking loss of mixl1 (Figure 3D). Eomesa and Tbx16 therefore collaboratively activate mixl1 expression, strongly suggesting that Tbx16 relieves the requirement for Eomesa in zebrafish endoderm formation.
Tbx16 and eomesa overexpression do not equivalently induce endoderm fate in concert with mixl1 and gata5
Eomesa and Mixl1 bind upstream of endoderm master regulator sox32 to positively regulate its expression prior to endoderm specification (Nelson et al., 2014; Nelson et al., 2017). Eomesa, Mixl1 and Gata5 can physically interact and their combined overexpression has been shown to induce ectopic endoderm gene expression in late blastulas and early gastrulas (Bjornson et al., 2005). Combined expression of Tbxta with Mixl1 and Gata5, however, is insufficient to induce sox32 expression (Bjornson et al., 2005). This is consistent with limited co-expression between tbxta and sox32 around the time of endoderm specification (Figure 4A). However, tbx16 and sox32 are substantially co-expressed in the endoderm at the onset of gastrulation, (Figures 3B, 4B). Tbx16 is also critical for endoderm progenitor induction (Nelson et al., 2017). We therefore extended our study to test whether Tbx16 can induce ectopic endoderm marker expression in cells at the animal pole (i.e. in cells where Tbx16, Mixl1 and Gata5 are absent in wild type embryos) through co-overexpression with Mixl1 and Gata5, as Eomesa can.
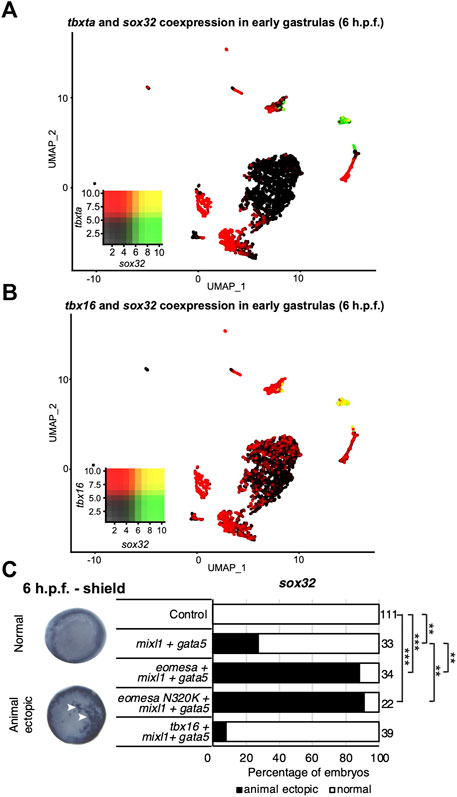
FIGURE 4. Tbx16 is substantially co-expressed with sox32 but cannot induce it in combination with mixl1 and gata5. (A,B) UMAP clustering plots of whole embryo single-cell RNA-seq data in early gastrulas (6 h.p.f.) indicating co-expression of sox32 with tbxta and tbx16. Heatmap insets indicate overall expression levels per gene and co-expression. Overlapping expression is shown in yellow. (C) WISH analysis of the ability of eomesa, eomesaN320K and tbx16 in combination with gata5 and mixl to induce sox32 expression at the animal pole of early gastrulas (6 h.p.f.). N = 2. Total numbers of embryos scored per condition are indicated. Representative images of expression patterns per gene per category are shown. Animal views; dorsal to the right. Arrowheads indicate ectopic expression. Fisher’s Exact two-tailed probability test p values: **P ≤ 5 × 10−6; ***P ≤ 5 × 10−12.
As expected, combined overexpression of eomesa, mixl1 and gata5 induces ectopic sox32 expression at the animal pole (Figure 4C). However, tbx16 did not synergise with mixl1 and gata5 to upregulate sox32 in the animal pole. This may suggest that Tbx16 and Eomesa are not equally capable of forming a complex with Mixl1 and Gata5 to induce endoderm and/or DFC fate, or alternatively that there are other key components of the complex which are capable of interaction with Eomesa/Mixl1/Gata5, but not Tbx16/Mixl1/Gata5. We conclude that Eomesa and Tbx16 perform similar functions in overlapping processes in the developing zebrafish embryo, but appear to do so via distinct molecular mechanisms.
T-box factors co-expressed with eomesa do not share its potent abilities to upregulate dorsal marker genes
Tbx16 and Eomesa lack significant sequence homology, especially outside the T-box domain (Supplementary Figure S4). However, Xenopus Eomes and its Tbx16 orthologue VegT have been suggested to display similar specificity in part due to a single shared asparagine residue within the T-box, rendering them functionally distinct from the Tbxta orthologue Xbra, which has a lysine in the equivalent position (Conlon et al., 2001) (Supplementary Figure S4). We therefore sought to address the following questions: 1) Are Tbx16, Tbxta and/or Eomesb capable of inducing Eomesa target genes in early gastrulas; 2) Is the T-box asparagine residue critical for Eomesa function; 3) Are key Eomesa functions dependent on the highly conserved CTD or relatively poorly conserved NTD.
We injected equimolar quantities of mRNA corresponding to each wild type T-box factor, or Eomesa ∆NTD, ∆CTD or N320K mutants and assessed the effect on dorsal mesoderm marker noto and DFC/endoderm marker sox32, and DFC marker vgll4l. Deletion of the NTD or CTD ablated Eomesa ability to induce noto expression, while N320K mutation had no significant effect (Figure 5A). All other T-box factors failed to produce a consistent or compelling induction of ectopic noto (Figure 5A). Consistent with previous results, Eomesa overexpression led to ectopic sox32 expression in the outer marginal cells indicative of increased numbers of DFC-like cells but not endoderm (Bjornson et al., 2005) (Figure 5B). NTD and especially CTD deletions markedly reduced ectopic sox32 induction, however, N320K mutation had little effect (Figure 5B). We further note that the N320K mutation had no discernible effect on the ability of Eomesa to synergise with Mixl1 and Gata5 to induce sox32 expression at the animal pole, suggesting that this mutation within the T-box does not interfere with the known T-box interactions with Mixl1 and Gata5 (Bjornson et al., 2005) (Figure 4C). Overexpression of tbx16, tbxta and eomesb did not lead to ectopic sox32 induction highlighting functional distinctions with eomesa (Figure 5B). Vgll4l expression showed similar induction to sox32 at the margin by both wild type Eomesa and ∆NTD and N320K Eomesa (Figure 5C). We have previously shown by ChIP-seq that at sphere stage Eomesa binds in the first intron of vgll4l (Nelson et al., 2014). Analysis of Tbx16 ChIP-seq data in mid/late gastrulas (8–8.5 h.p.f.) (Nelson et al., 2017) also shows Tbx16 binding at close matches to the known T-box consensus sequence within vgll4l intron 1, suggesting Tbx16 does have the potential to directly participate in regulation of vgll4l during gastrulation (Figure 5D). However, tbx16 overexpression suggests that it is not individually sufficient to strongly drive ectopic vgll4l expression (Figure 5C).
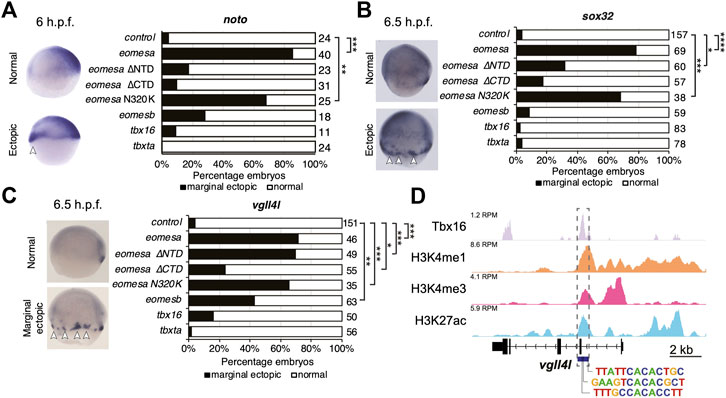
FIGURE 5. Eomesa is a more potent inducer of endoderm, organiser and dorsal forerunner cell markers than other T-box factors. (A–C) WISH analysis of dorsal mesoderm marker noto (A) and dorsal forerunner cell markers sox32 (B) and vgll4l (C) on overexpression of various wild type and mutant T-box factors. mRNAs injected at the 1 cell stage; WISH performed at stages as indicated. N = 2. Total numbers of embryos scored per condition are indicated. Representative images of expression patterns per gene per category are shown. Lateral views; dorsal to the right. Arrowheads indicate ectopic expression. Fisher’s Exact two-tailed probability test p values: *P ≤ 5 × 10−4; **P ≤ 5 × 10−6; ***P ≤ 5 × 10−12; ****P ≤ 5 × 10−30. (D) ChIP-seq data in mid/late gastrulas (8–8.5 h.p.f.) indicating Tbx16 binding within the vgll4l promoter (Bogdanovic et al., 2012; Nelson et al., 2017). Scale is reads per million reads (RPM). Putative T-box binding sites identified using JASPAR are indicated (Fornes et al., 2020).
We next tested whether Tbx16 and Tbxta can act combinatorially to induce dorsal marker gene expression. We found that on Tbx16/Tbxta combinatorial overexpression DFC markers vgll4l and sox17 were expressed over a broader region of the dorsolateral margin but showed concomitant reduction in the primary DFC cluster (Supplementary Figure S5A). Conversely, foxj1a expression remained localised to the dorsal margin, again suggesting that foxj1a expression is somewhat dependent on additional dorsally localised regulators (Supplementary Figure S5A). Furthermore, while tbx16 and tbxta are coexpressed with noto in early gastrulas (Supplementary Figures S5B,C), their combinatorial overexpression does not lead to expanded noto expression (Supplementary Figure S5D). We therefore conclude that combined activities of Tbx16 and Tbxta are not sufficient to induce dorsal fates, unlike Eomesa.
Overall this suggests that Eomesb, Tbx16 and Tbxta do not individually have similar abilities to Eomesa in inducing dorsal mesoderm and DFC gene expression, that the previously reported N/K amino acid difference between Eomesa/Tbx16 and Tbxta does not appreciably influence specificity and function in this context, and that deletion of the relatively poorly conserved Eomesa NTD does not result in complete loss of function.
Eomesa regulates vgll4l expression and dorsal forerunner cell formation through interlocking feedforward loops via sox32
Results above suggest that Eomesa overexpression induces DFC fate based on ectopic expression of markers including sox32 and vgll4l (Figures 2F, 5B,C). To resolve the conflict between the observed Eomesa-mediated repression of vgll4l expression at mid/late blastula stages (4 h.p.f.) but induction by early gastrulation (6 h.p.f.) we explored the role of Eomesa target Sox32 in vgll4l induction. We found that sox32 overexpression through one-cell stage mRNA injection led to a dramatic upregulation of vgll4l expression in early gastrulas (6 h.p.f.; Figure 6A), suggesting localised Eomesa-mediated upregulation of vgll4l at the margin occurs through Sox32 rather than a switch in Eomesa function directly at the vgll4l locus. To test whether Sox32 is required for induction of vgll4l expression we performed knockdown using a previously validated and widely used antisense morpholino (Dickmeis et al., 2001). This knockdown clearly resulted in loss of DFC vgll4l expression (Figure 6B). Furthermore, vgll4l induction in locations outside the dorsal margin, caused by combinatorial eomesa/mixl1/gata5 overexpression was also profoundly abrogated by Sox32 knockdown. Thus, during gastrula stages induction of vgll4l is via Sox32 rather than direct Eomesa activities.
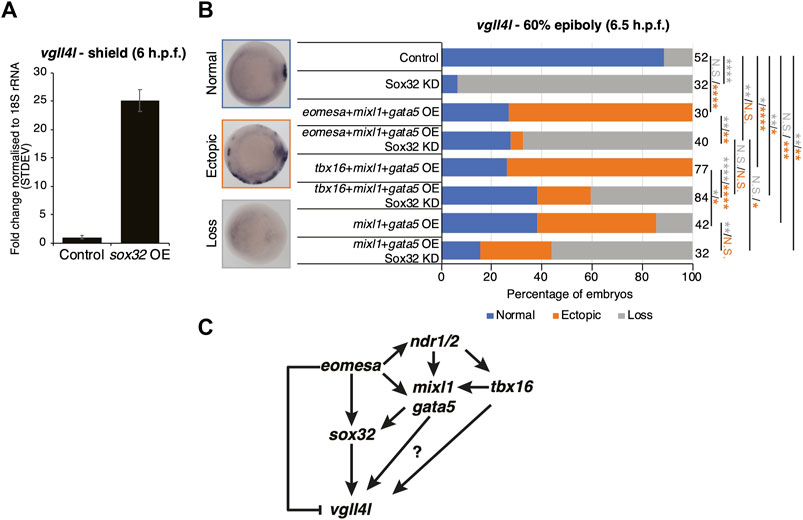
FIGURE 6. Eomesa activation of vgll4l is through feedforward loops via sox32 and its upstream activators. (A) qRT-PCR analysis of vgll4l expression in early gastrulas (6 h.p.f.) in control embryos and those injected with sox32 mRNA at the 1 cell stage. Expression is represented as fold change relative to control normalised to 18S rRNA. (B) WISH analysis of the ability of vgll4l expression in early gastrulas (6 h.p.f.) in embryos injected with mRNAs at the one-cell stage as indicated, with and without Sox32 morpholino knockdown. N = 2. Total numbers of embryos scored per condition are indicated. Representative images of expression patterns per gene per category are shown. Animal views; dorsal to the right. Fisher’s Exact two-tailed probability test p values: *P ≤ 5 × 10−2; **P ≤ 5 × 10−4; ***P ≤ 5 × 10−8; ****P ≤ 5 × 10−12; N.S. = not significant. Orange asterisks indicate significant differences in fractions of embryos exhibiting ectopic expression vs. other categories. Grey asterisks indicate significant differences in fractions of embryos exhibiting loss of expression vs. other categories. (C) Model for Eomesa regulation of vgll4l expression in dorsal forerunner cells indicating a type 3 incoherent feedforward loop on the left as Eomesa represses vgll4l directly while activating via Sox32, and potential type 1 coherent feedforward loops on the right as Eomesa activates positive regulators of sox32 and potentially also vgll4l.
Vgll4l expression was similarly upregulated outside the dorsal margin by combinatorial mixl1/gata5 overexpression, and partially blocked by Sox32 KD (Figure 6B). However, vgll4l expression in mixl1/gata5 overexpressing embryos was not as profoundly reduced on Sox32 KD as was the case for eomesa/mixl1/gata5 overexpression. It is not completely clear whether this is because mixl1/gata5 can activate vgll4l independent of Sox32 function, or due to the absence of Eomesa-mediated repression of vgll4l. Addition of tbx16 to mixl1/gata5 overexpression led to a significant increase in the fraction of embryos exhibiting ectopic vgll4l expression compared to mixl1/gata5 alone (Figure 6B). We note that overexpression of tbx16 alone was insufficient to enhance vgll4l expression in a significant fraction of embryos, and showed no ability to induce sox32 expression (Figures 5B,C). It seems likely that while tbx16 alone does not exert a strong influence at the vgll4l locus, mixl1 and gata5 can provide a context for tbx16 to enhance vgll4l expression. This likely involves Tbx16 binding to vgll4l intron 1 (Figure 5D) rather than occurring via sox32, since Tbx16 cannot induce sox32 expression, either individually or in combination with Mixl1 and Gata5 (Figures 4C, 5B).
Overall the present results combined with previous published reports from ourselves and others suggest a model wherein Eomesa acts within interlocking incoherent type 3 and coherent type 1 feedforward loops (Mangan and Alon, 2003) to repress vgll4l while combining with Nodal downstream effectors Mixl1 and Gata5 to activate sox32, which in turn activates vgll4l around the time of DFC specification. In addition to this, our analyses indicate that both mouse Eomes FL and ∆VR isoforms are functionally equivalent to Eomesa, suggesting phenotypic differences between zebrafish and mouse Eomes loss-of-function mutants are not likely to be driven by functional divergence, but rather redundancy with co-expressed factors in zebrafish such as Tbx16.
Discussion
Phenotypic differences between mouse and zebrafish Eomes loss-of-function mutants are not due to molecular divergence
T-box transcription factors are an ancient family of genes with many key roles in embryogenesis and disease. Lineage-specific differences that occurred in the family during vertebrate evolution have resulted in altered gene complements and diversity of splice isoforms in distinct evolutionary lineages (DeBenedittis and Jiao, 2011; Papaioannou, 2014). While AS events in specific evolutionary lineages have led to functional diversification of certain T-box factors, we have shown that Eomes loss-of-function phenotypic differences between mouse and zebrafish are unlikely to be due to evolutionary differences in Eomes protein function, but rather a degree of compensation by Tbx16 which is present in zebrafish but not placental mammals.
Though AS is an evolutionary means of increasing functional diversity within the proteome, our data suggests that Eomes exon 6 AS is not functionally important in the context of early development. In the case of the ∆VR splicing event a synonymous mutation created an alternative splice acceptor, however, our data suggests it is hardly used leading to majority production of the FL isoform. That the ∆VR isoform arose and is maintained in the tetrapod lineage may be due to substantial functional similarlity of FL and ∆VR isoforms, leading to a lack of selective pressure.
In overexpression studies the ∆VR isoform has the ability to induce trophoblast markers in embryonic stem cells (Niwa et al., 2005), and cardiac mesoderm markers in embryonal carcinoma cells (Costello et al., 2011). Moreover, both FL and ∆VR isoforms can induce organizer and DFC markers while repressing ectoderm markers on overexpression in zebrafish. These observations provide further evidence of their functional equivalence.
The present data demonstrate that the ∆VR and ∆CTD isoforms are only weakly expressed compared with FL Eomes. While we find no evidence for the functional importance of the VR it is intriguing that it is both highly conserved and known to be phosphorylated (Huttlin et al., 2010). It is possible that these isoforms potentially make substantial contributions in contexts we have not explored. The present evidence, however, suggests that in mice, as in zebrafish the FL isoform is the more important molecule.
Given the complexities of mouse Eomes mutant phenotypes it would be interesting to explore isoform-specific functions in mouse. This could be achieved through either modifying the endogenous Eomes locus in embryonic stem cells (ESCs) such that only specific isoforms could be expressed, or using isoform-specific inducible transgenes in Eomes null mutant ESCs. This could be combined with directed differentiation procedures to determine whether there are detectable isoform-specific functions in relevant cell types. Whether zebrafish Eomesa, or other T-box factors can functionally substitute for mouse Eomes could also be tested in a similar system. Alternatively, the genetically modified ESCs could be used to make mice in an attempt to study isoform and orthologue functions in vivo.
Functional similarities and differences of Eomesa and Tbx16
We previously demonstrated that Eomesa and Tbx16 display overlapping genomic binding profiles in early zebrafish embryos (Nelson et al., 2017). Whether they are functionally redundant, however, had not been explored. The present experiments strongly suggest that Eomesa and Tbx16 redundantly regulate the homeodomain transcription factor mixl1, which has key conserved functions in endoderm formation in zebrafish and mouse (Kikuchi et al., 2000; Hart et al., 2002). Mixl1 mutants suffer profound loss of endoderm (Kikuchi et al., 2000). Reduced expression of mixl1 in the margin of late blastulas/early gastrulas is coupled with reduced numbers of endoderm progenitors in late gastrulation in both MZeomesa mutants, and on tbx16 knockdown (Du et al., 2012; Nelson et al., 2017). It therefore seems likely that the enhanced reduction of mixl1 expression on tbx16 knockdown in eomesa mutants in the present study would lead to increased loss of endoderm progenitors. Our previous RNA-seq analyses indicate that expression of tbx16 is not significantly different in MZeomesa mutants (Nelson et al., 2014). It therefore seems likely that Tbx16 partially compensates for loss of Eomesa during zebrafish endoderm formation. Our study therefore highlights a consistent requirement for T-box function in vertebrate endoderm formation. Interestingly, while multiple orthologous T-box factors have similar expression domains in early zebrafish and mouse embryogenesis, those domains are typically expanded in zebrafish (Wardle and Papaioannou, 2008). Coupled with its rapid rate of development and the greater number of T-box factors in zebrafish, there is likely to be a higher degree of T-box factor co-expression, enhancing the probability of redundancy.
While Eomesa and Tbx16 share some redundant functions we also identified key differences. It was previously shown that Eomesa can combine with Mixl1 and Gata5 to drive expression of sox32 at the animal pole (Bjornson et al., 2005). However, Tbx16 does not appear to have the same ability as Eomesa to drive sox32 expression either individually or in combination with Mixl1 and Gata5, even though sox32 expressing cells appear to exhibit tbx16 expression in single-cell RNA-seq data. This is consistent with previous observations that Meomesa and MZeomesa mutants have reduced expression of sox32 during gastrulation without complete loss (Du et al., 2012; Xing et al., 2022). It therefore seems likely that Tbx16 is sufficient to rescue certain Eomesa functions but cannot completely compensate for its loss. It is further notable that Tbx16 does not seem individually able to induce the dorsal mesoderm marker noto as Eomesa can. However, given that Eomesa acts upstream of Nodal (Xu et al., 2014; Xing et al., 2022) it seems likely that major differences in outcome between eomesa and tbx16 overexpression stem from enhanced Nodal signalling on eomesa overexpression. It will be interesting to learn more about the common and unique functional activities of Eomes and Tbx16 that drive target gene expression.
Eomesa and Tbx16 are only distantly related within the T-box family (25.3% of Tbx16 amino acid identity), with the majority of conserved amino acids occurring within the T-box domain. Whether they are likely to act in similar protein complexes to regulate their target genes is therefore unclear. A key study in Xenopus, however, suggested the specificity of target gene induction is primarily mediated by the T-box itself, rather than NTDs and CTDs (Conlon et al., 2001). The same study demonstrated a single asparagine to lysine substitution in Xenopus Eomes and VegT T-box domains, alter their inductive properties to mimic Brachyury (Conlon et al., 2001). Importantly, both Eomesa and Tbx16 (which has been proposed as the zebrafish orthologue of Xenopus VegT (Griffin et al., 1998)) share the same critical asparagine. Our data suggest that the N320K mutation has little effect on induction of Eomesa target genes explored here, is unlikely to prevent T-box interaction with co-factors Mixl1 and Gata5, or substantially account for differences with Tbxta in endoderm and DFC formation. In fact, analysis of single-cell RNA-seq data suggests that the greater importance of Eomesa and Tbx16 in endoderm formation is more likely to be attributable to lesser Tbxta expression in the endoderm. It is therefore possible that Eomesa and Tbx16 also have overlapping roles in endoderm formation downstream of driving mixl1 expression in presumptive endoderm that are yet to be elucidated.
Interestingly, participation of zebrafish Tbx16 in processes controlled by Eomes in mice is not limited to endoderm formation. For example, while Eomes acts upstream of basic helix-loop-helix transcription factor gene Mesp1 to specify cardiac mesoderm in mice (Costello et al., 2011), Tbx16 regulates the orthologous gene mespaa in zebrafish (Garnett et al., 2009). Eomes loss-of-function leads to aberrant mesoderm cell migration during mouse gastrulation, while tbx16 zebrafish mutants also exhibit cell-autonomous defects in mesoderm migration (Ho and Kane, 1990; Arnold et al., 2008a). Remarkably, potentially interesting parallels continue to emerge, such as the requirements for zebrafish Tbx16 and mouse Eomes in blood progenitors (Rohde et al., 2004; Harland et al., 2021). It is therefore possible that the presence of Tbx16 in teleost fish has led to a reduced requirement for Eomes in multiple developmental contexts.
The present study focuses on early embryonic development, however, Eomes is known to have later roles in neurological development, as well as in the immune system. Importantly, Eomes is an key regulator of neurogenesis in the subventricular zone, and loss leads to microcephaly and severe behavioural defects (Arnold et al., 2008b). Though eomesa is equivalently expressed in the telencephalon of developing zebrafish larvae, whether null mutants have an equivalent phenotype is unknown (Mione et al., 2001; Du et al., 2012). If they do not, however, it is unlikely to be due to redundancy with tbx16, which is absent from the developing brain. Similarly, it is unclear whether eomesa mutants exhibit defects in the immune system, such as in T cell differentiation and NK cell development and function as in mammals (Simonetta et al., 2016; D'Cruz et al., 2009). Both eomesa and eomesb are co-expressed in lymphocytes in fish, however, suggesting they may be redundant in the immune system (Takizawa et al., 2007; Takizawa et al., 2014).
While T-box factor redundancy during development is not a novel concept e.g. (Amacher et al., 2002; Garnett et al., 2009; Jahangiri et al., 2012; Gentsch et al., 2013; Nelson et al., 2017), the molecular basis for this redundancy (or indeed T-box factor molecular interactions in general) is not well understood. In future it will be interesting to study whether redundant T-box factors recruit similar co-factors to regulate gene expression, and whether this occurs through conserved or divergent amino acid sequences and structural motifs.
On the roles of Eomesa, Tbx16 and Tbxta in dorsal mesoderm
While Eomesa is capable of inducing dorsal mesoderm markers such as noto and chrd in zebrafish embryos, it is notable that their expression is normal in the absence of Eomesa (Bruce et al., 2003; Du et al., 2012). However, it is also notable that tbxta and tbx16 do not show altered expression in eomesa mutants in published WISH and RNA-seq datasets (Du et al., 2012; Nelson et al., 2014). It is therefore possible that Tbx16 and Tbxta are amongst factors compensating for the loss of Eomesa. In support of this, Tbxta has been shown to directly activate noto expression, and tbxta mutants fail to maintain noto expression in mid/late gastrulation stages, leading to loss of notochord (Melby et al., 1997; Morley et al., 2009). Similarly, Tbx16 is required to maintain chrd expression in axial structures at mid/late gastrulation stages (Miller-Bertoglio et al., 1997; Warga et al., 2013). The reduced expression of both noto in tbxta mutants and chrd in tbx16 mutants follows the decline in eomesa mRNA expression levels, suggesting that Tbxta and Tbx16 maintain the expression of dorsal mesoderm markers in the absence of Eomesa. Nevertheless, our results indicate that of these T-box factors, only Eomesa is sufficient to induce ectopic dorsal mesoderm marker expression. This suggests key differences in the molecular functions of these T-box factors. It is possible that co-factors required for dorsal mesoderm induction by Eomesa are localised throughout the margin while those required by Tbxta and Tbx16 are restricted to the dorsal margin. Alternatively, given Eomesa regulates expression of Nodal pathway ligands which are required for dorsal mesoderm fates, it is possible that Eomesa but not Tbxta/Tbx16 is capable of expanding dorsal mesoderm through upregulation of Nodal signalling (Du et al., 2012; Xu et al., 2014; Xing et al., 2022).
Eomesa, Tbx16, Mixl1 and Gata5 activities during dorsal forerunner cell formation
Loss of Eomesa leads to upregulation of vgll4l during blastula stages whereas overexpression of eomesa causes repression of vgll4l (Nelson et al., 2014). The present experiments suggests that Eomesa acts within feedforward loops to repress vgll4l expression until activators including Sox32 accumulate to drive vgll4l in DFCs at the onset of gastrulation. Given that Eomesa is maternally contributed and not spatially restricted in early development (Du et al., 2012), while accumulation of vgll4l activators is principally driven by Nodal at the dorsal margin, this suggests a model wherein Eomesa controls the specificity and timing of vgll4l induction. Eomesa mRNA steadily declines during blastula stages as expression of mixl1, gata5, tbx16 and sox32 increase, and is virtually undetectable at the onset of gastrulation, (Figure 3A and Bruce et al., 2003; Figiel et al., 2021). While Eomesa protein does persist through gastrulation (Du et al., 2012) it seems likely that temporal and spatial changes in abundance of vgll4l activators and repressors acting within these feedforward loops cooperatively regulate the specificity of vgll4l expression during DFC specification.
Genetic data, however, suggest that our model is likely to be incomplete. While Sox32 is required for correct DFC formation (Alexander et al., 1999; Essner et al., 2005), upstream regulators mixl1 and gata5 are not required for DFC formation individually or in combination. Rather mixl1 and gata5 seem to be strictly required upstream of sox32 for correct endoderm formation (Reiter et al., 1999; Kikuchi et al., 2000; Reiter et al., 2001). While we cannot discount the possibility of mixl1 and gata5 expression in precursors of DFCs, present evidence suggests that there are either alternative upstream regulators of sox32 in DFCs vs. endoderm, or additional redundant factors in DFCs rescuing the requirement for mixl1 and gata5. However, given the apparent requirement for Nodal signalling in DFC formation (Alexander and Stainier, 1999), it seems likely that whatever the upstream regulators of sox32 expression in DFCs they will be Nodal-dependent. Overall this highlights a lack of understanding of the gene regulatory networks that direct DFC vs. endoderm formation, which will be a key focus of our future work.
Recent evidence suggests Vgll4l is required for tbx16 expression during DFC formation (Fillatre et al., 2019). That Tbx16 binds the vgll4l promoter during gastrulation could suggest that complex regulatory loops control DFC formation and maintenance. The ability of Eomesa to induce ectopic DFCs during early gastrulation combined with expression of mouse Eomes in progenitors of the node and requirement for correct node formation (Arnold et al., 2008a; Costello et al., 2011) suggests the potential for a conserved role in establishment of left-right asymmetry with some degree of redundancy with Tbx16 in zebrafish. However, a role for the vgll4l mammalian homologue Vgll4 in left-right asymmetry has yet to be determined. Further study of the mechanistic parallels in T-box factor mediated formation of zebrafish DFCs and mouse node would be beneficial to gain a more detailed evo-devo understanding of this process.
The diversity of DFC marker gene induction observed in this study was particularly striking, and points to dorsally localised determinants of DFC identity and function that are less readily induced by Eomesa and Eomes. We found that between them Eomesa and mouse Eomes isoforms were able to induce sox32, sox17, vgll4l and foxj1a. Sox32 is required for maintenance of DFC identify and formation of the left-right organiser (Alexander et al., 1999; Essner et al., 2005) while its downstream target sox17 is required for correct left-right organiser function (Aamar and Dawid, 2010). Vgll4l is a key mediator of Hippo signalling and regulates epigenetic programming of DFC by controlling the expression of writers and readers of DNA methylation, influencing DFC proliferation, apoptosis and ciliagenesis (Fillatre et al., 2019). Foxj1a is the master regulator if motile cilia formation (Yu et al., 2008). That the Eomes-mediated induction of foxj1a was more restricted to the dorsal margin than that of other markers suggests that Vgll4l and the Sox and T-box factors known to be involved in DFC formation are not sufficient to fully induce DFC identity. Other localised cues (physical, mechanical, signalling or cell intrinsic factors) are therefore likely to be involved in foxj1a induction.
Overall we conclude that enhanced AS in mammals has not significantly altered Eomes function in early embryogenesis. Rather we conclude that the different degrees of T-box factor co-expression and the presence/absence of additional factors including Tbx16 has modulated the severity of the Eomes null mutant phenotype in the embryo proper between mouse and zebrafish. Furthermore, we conclude that in zebrafish Eomesa participates in DFC formation through directing feedforward loops via sox32 to control vgll4l expression. Our results therefore provide novel insights into evolutionary differences in vertebrate endoderm formation, and the gene regulatory networks involved in controlling the zebrafish left-right organiser formation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The single-cell RNA-seq data used in the study is from a published paper, and is available in NCBI GEO (accession GSM3067190).
Ethics statement
The animal study was reviewed and approved by Animal Welfare and Ethical Review Body, University of Warwick Animal Welfare and Ethical Review Body, King’s College London University of Toronto Animal Care Committee.
Author contributions
Conceived and designed the experiments: ACN and CDT. Performed the experiments: CDT, ACN, AEEB, SJC, FCW, RE, and EV. Analysed the data: ACN, CDT, and MDW. Wrote the paper: ACN.
Funding
This research was funded in whole or in part by the BBSRC Midlands Integrative Biosciences Training Partnership (BB/M01116X/1). For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising from this submission. This research was also funded in part, by the Wellcome Trust through a Wellcome Seed Award in Science to ACN (210177/Z/18/Z), and a Wellcome Trust Programme grant (102811) to Elizabeth J. Robertson. The research was also funded by Horizon 2020 MSCA-ITN project ZENCODE-ITN (GA no: 643062) to FCW. The research was also funded by a Natural Sciences and Engineering Research Council grant to Ashley Bruce (458019). CDT has a PhD studentship funded by the BBSRC Midlands Integrative Biosciences Training Partnership (BB/M01116X/1). RE was funded by the MRC Doctoral Training Partnership in Interdisciplinary Biomedical Research (MR/N014294/1).
Acknowledgments
We thank Elizabeth Bikoff and Elizabeth Robertson for reagents, valuable conversations and critical review of the manuscript. We also thank Karuna Sampath for valuable discussions and generous access to equipment, and Ita Costello, Claire Simon and Daniel Hebenstreit for valuable discussions, the Warwick zebrafish facility for zebrafish care and acknowledge the contributions of Henrietta Lacks and her family to this research. We also thank Jeffrey Amack and H. Joseph Yost for reagents.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.982477/full#supplementary-material
References
Ashe, H. L., Monks, J., Wijgerde, M., Fraser, P., and Proudfoot, N. J. (1997). Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11, 2494–2509. doi:10.1101/gad.11.19.2494
Aamar, E., and Dawid, I. B. (2010). Sox17 and chordin are required for formation of Kupffer's vesicle and left-right asymmetry determination in zebrafish. Dev. Dyn. 239, 2980–2988. doi:10.1002/dvdy.22431
Adams, S. E., Johnson, I. D., Braddock, M., Kingsman, A. J., Kingsman, S. M., and Edwards, R. M. (1988). Synthesis of a gene for the HIV transactivator protein TAT by a novel single stranded approach involving in vivo gap repair. Nucleic Acids Res. 16, 4287–4298. doi:10.1093/nar/16.10.4287
Ahn, D., You, K. H., and Kim, C. H. (2012). Evolution of the tbx6/16 subfamily genes in vertebrates: Insights from zebrafish. Mol. Biol. Evol. 29, 3959–3983. doi:10.1093/molbev/mss199
Alexander, J., Rothenberg, M., Henry, G. L., and Stainier, D. Y. (1999). Casanova plays an early and essential role in endoderm formation in zebrafish. Dev. Biol. 215, 343–357. doi:10.1006/dbio.1999.9441
Alexander, J., and Stainier, D. Y. (1999). A molecular pathway leading to endoderm formation in zebrafish. Curr. Biol. 9, 1147–1157. doi:10.1016/S0960-9822(00)80016-0
Amacher, S. L., Draper, B. W., Summers, B. R., and Kimmel, C. B. (2002). The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development 129, 3311–3323. doi:10.1242/dev.129.14.3311
Arnold, S. J., Hofmann, U. K., Bikoff, E. K., and Robertson, E. J. (2008a). Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501–511. doi:10.1242/dev.014357
Arnold, S. J., Huang, G. J., Cheung, A. F., Era, T., Nishikawa, S., Bikoff, E. K., et al. (2008b). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479–2484. doi:10.1101/gad.475408
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–D995. doi:10.1093/nar/gks1193
Bisgrove, B. W., Snarr, B. S., Emrazian, A., and Yost, H. J. (2005). Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer's vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 287, 274–288. doi:10.1016/j.ydbio.2005.08.047
Bjornson, C. R., Griffin, K. J., Farr, G. H., 3R. D., Terashima, A., Himeda, C., Kikuchi, Y., et al. (2005). Eomesodermin is a localized maternal determinant required for endoderm induction in zebrafish. Dev. Cell 9, 523–533. doi:10.1016/j.devcel.2005.08.010
Bogdanovic, O., Fernandez-Minan, A., Tena, J. J., De La Calle-Mustienes, E., Hidalgo, C., Van Kruysbergen, I., et al. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043–2053. doi:10.1101/gr.134833.111
Bruce, A. E., Howley, C., Zhou, Y., Vickers, S. L., Silver, L. M., King, M. L., et al. (2003). The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development 130, 5503–5517. doi:10.1242/dev.00763
Conlon, F. L., Fairclough, L., Price, B. M., Casey, E. S., and Smith, J. C. (2001). Determinants of T box protein specificity. Development 128, 3749–3758. doi:10.1242/dev.128.19.3749
Costello, I., Pimeisl, I. M., Drager, S., Bikoff, E. K., Robertson, E. J., and Arnold, S. J. (2011). The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 13, 1084–1091. doi:10.1038/ncb2304
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. doi:10.1101/gr.849004
D'Cruz, L. M., Rubinstein, M. P., and Goldrath, A. W. (2009). Surviving the crash: Transitioning from effector to memory CD8+ T cell. Semin. Immunol. 21, 92–98. doi:10.1016/j.smim.2009.02.002
Debenedittis, P., and Jiao, K. (2011). Alternative splicing of T-box transcription factor genes. Biochem. Biophys. Res. Commun. 412, 513–517. doi:10.1016/j.bbrc.2011.08.010
Dickmeis, T., Mourrain, P., Saint-Etienne, L., Fischer, N., Aanstad, P., Clark, M., et al. (2001). A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 15, 1487–1492. doi:10.1101/gad.196901
Du, S., Draper, B. W., Mione, M., Moens, C. B., and Bruce, A. (2012). Differential regulation of epiboly initiation and progression by zebrafish Eomesodermin A. Dev. Biol. 362, 11–23. doi:10.1016/j.ydbio.2011.10.036
Eggermont, J., and Proudfoot, N. J. (1993). Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 12, 2539–2548. doi:10.1002/j.1460-2075.1993.tb05909.x
Essner, J. J., Amack, J. D., Nyholm, M. K., Harris, E. B., and Yost, H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247–1260. doi:10.1242/dev.01663
Faial, T., Bernardo, A. S., Mendjan, S., Diamanti, E., Ortmann, D., Gentsch, G. E., et al. (2015). Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development 142, 2121–2135. doi:10.1242/dev.117838
Figiel, D. M., Elsayed, R., and Nelson, A. C. (2021). Investigating the molecular guts of endoderm formation using zebrafish. Brief. Funct. Genomics 20, 394–406. doi:10.1093/bfgp/elab013
Fillatre, J., Fauny, J. D., Fels, J. A., Li, C., Goll, M., Thisse, C., et al. (2019). TEADs, Yap, Taz, Vgll4s transcription factors control the establishment of Left-Right asymmetry in zebrafish. Elife 8, e45241. doi:10.7554/eLife.45241
Fornes, O., Castro-Mondragon, J. A., Khan, A., Van Der Lee, R., Zhang, X., Richmond, P. A., et al. (2020). Jaspar 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92. doi:10.1093/nar/gkz1001
Garnett, A. T., Han, T. M., Gilchrist, M. J., Smith, J. C., Eisen, M. B., Wardle, F. C., et al. (2009). Identification of direct T-box target genes in the developing zebrafish mesoderm. Development 136, 749–760. doi:10.1242/dev.024703
Gentsch, G. E., Monteiro, R. S., and Smith, J. C. (2017). Cooperation between T-box factors regulates the continuous segregation of germ layers during vertebrate embryogenesis. Curr. Top. Dev. Biol. 122, 117–159. doi:10.1016/bs.ctdb.2016.07.012
Gentsch, G. E., Owens, N. D., Martin, S. R., Piccinelli, P., Faial, T., Trotter, M. W., et al. (2013). In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep. 4, 1185–1196. doi:10.1016/j.celrep.2013.08.012
Glasauer, S. M. K., and Neuhauss, S. C. F. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289, 1045–1060. doi:10.1007/s00438-014-0889-2
Goujon, M., Mcwilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. doi:10.1093/nar/gkq313
Griffin, K. J., Amacher, S. L., Kimmel, C. B., and Kimelman, D. (1998). Molecular identification of spadetail: Regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125, 3379–3388. doi:10.1242/dev.125.17.3379
Grinblat, Y., and Sive, H. (2001). Zic Gene expression marks anteroposterior pattern in the presumptive neurectoderm of the zebrafish gastrula. Dev. Dyn. 222, 688–693. doi:10.1002/dvdy.1221
Hafemeister, C., and Satija, R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. doi:10.1186/s13059-019-1874-1
Harland, L. T. G., Simon, C. S., Senft, A. D., Costello, I., Greder, L., Imaz-Rosshandler, I., et al. (2021). The T-box transcription factor Eomesodermin governs haemogenic competence of yolk sac mesodermal progenitors. Nat. Cell Biol. 23, 61–74. doi:10.1038/s41556-020-00611-8
Hart, A. H., Hartley, L., Sourris, K., Stadler, E. S., Li, R., Stanley, E. G., et al. (2002). Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development 129, 3597–3608. doi:10.1242/dev.129.15.3597
Ho, R. K., and Kane, D. A. (1990). Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature 348, 728–730. doi:10.1038/348728a0
Huttlin, E. L., Jedrychowski, M. P., Elias, J. E., Goswami, T., Rad, R., Beausoleil, S. A., et al. (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. doi:10.1016/j.cell.2010.12.001
Jahangiri, L., Nelson, A. C., and Wardle, F. C. (2012). A cis-regulatory module upstream of deltaC regulated by Ntla and Tbx16 drives expression in the tailbud, presomitic mesoderm and somites. Dev. Biol. 371, 110–120. doi:10.1016/j.ydbio.2012.07.002
Jowett, T., and Lettice, L. (1994). Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 10, 73–74. doi:10.1016/0168-9525(94)90220-8
Karolchik, D., Hinrichs, A. S., Furey, T. S., Roskin, K. M., Sugnet, C. W., Haussler, D., et al. (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. doi:10.1093/nar/gkh103
Keren, H., Lev-Maor, G., and Ast, G. (2010). Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 11, 345–355. doi:10.1038/nrg2776
Kikuchi, Y., Trinh, L. A., Reiter, J. F., Alexander, J., Yelon, D., and Stainier, D. Y. (2000). The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 14, 1279–1289. doi:10.1101/gad.14.10.1279
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi:10.1002/aja.1002030302
Lolas, M., Valenzuela, P. D., Tjian, R., and Liu, Z. (2014). Charting Brachyury-mediated developmental pathways during early mouse embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 111, 4478–4483. doi:10.1073/pnas.1402612111
Mangan, S., and Alon, U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U. S. A. 100, 11980–11985. doi:10.1073/pnas.2133841100
Marcellini, S., Technau, U., Smith, J. C., and Lemaire, P. (2003). Evolution of brachyury proteins: Identification of a novel regulatory domain conserved within bilateria. Dev. Biol. 260, 352–361. doi:10.1016/s0012-1606(03)00244-6
Mcwilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600. doi:10.1093/nar/gkt376
Melby, A. E., Kimelman, D., and Kimmel, C. B. (1997). Spatial regulation of floating head expression in the developing notochord. Dev. Dyn. 209, 156–165. doi:10.1002/(SICI)1097-0177(199706)209:2<156::AID-AJA2>3.0.CO;2-H
Melville, M., Lun, A., Djekidel, M. N., and Hao, Y. (2020). uwot: The uniform manifold approximation and projection (UMAP) method for dimensionality reduction. R package version 0.1.11. Available at: https://cran.r-project.org/web/packages/uwot/index.html.
Miller-bertoglio, V. E., Fisher, S., Sanchez, A., Mullins, M. C., and Halpern, M. E. (1997). Differential regulation of chordin expression domains in mutant zebrafish. Dev. Biol. 192, 537–550. doi:10.1006/dbio.1997.8788
Mione, M., Shanmugalingam, S., Kimelman, D., and Griffin, K. (2001). Overlapping expression of zebrafish T-brain-1 and eomesodermin during forebrain development. Mech. Dev. 100, 93–97. doi:10.1016/s0925-4773(00)00501-3
Morley, R. H., Lachani, K., Keefe, D., Gilchrist, M. J., Flicek, P., Smith, J. C., et al. (2009). A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc. Natl. Acad. Sci. U. S. A. 106, 3829–3834. doi:10.1073/pnas.0808382106
Nelson, A. C., Cutty, S. J., Gasiunas, S. N., Deplae, I., Stemple, D. L., and Wardle, F. C. (2017). In vivo regulation of the zebrafish endoderm progenitor niche by T-box transcription factors. Cell Rep. 19, 2782–2795. doi:10.1016/j.celrep.2017.06.011
Nelson, A. C., Cutty, S. J., Niini, M., Stemple, D. L., Flicek, P., Houart, C., et al. (2014). Global identification of Smad2 and Eomesodermin targets in zebrafish identifies a conserved transcriptional network in mesendoderm and a novel role for Eomesodermin in repression of ectodermal gene expression. BMC Biol. 12, 81. doi:10.1186/s12915-014-0081-5
Nelson, A. C., Pillay, N., Henderson, S., Presneau, N., Tirabosco, R., Halai, D., et al. (2012). An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J. Pathol. 228, 274–285. doi:10.1002/path.4082
Niwa, H., Toyooka, Y., Shimosato, D., Strumpf, D., Takahashi, K., Yagi, R., et al. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929. doi:10.1016/j.cell.2005.08.040
Nowotschin, S., Costello, I., Piliszek, A., Kwon, G. S., Mao, C. A., Klein, W. H., et al. (2013). The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo. Genes Dev. 27, 997–1002. doi:10.1101/gad.215152.113
Papaioannou, V. E. (2014). The T-box gene family: Emerging roles in development, stem cells and cancer. Development 141, 3819–3833. doi:10.1242/dev.104471
Pearce, E. L., Mullen, A. C., Martins, G. A., Krawczyk, C. M., Hutchins, A. S., Zediak, V. P., et al. (2003). Control of effector CD8(+) T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043. doi:10.1126/science.1090148
Picozzi, P., Wang, F., Cronk, K., and Ryan, K. (2009). Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes. J. Biol. Chem. 284, 2397–2408. doi:10.1074/jbc.M808704200
Probst, S., and Arnold, S. J. (2017). Eomesodermin-at dawn of cell fate decisions during early embryogenesis. Curr. Top. Dev. Biol. 122, 93–115. doi:10.1016/bs.ctdb.2016.09.001
Ramani, A. K., Calarco, J. A., Pan, Q., Mavandadi, S., Wang, Y., Nelson, A. C., et al. (2011). Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 21, 342–348. doi:10.1101/gr.114645.110
Reiter, J. F., Alexander, J., Rodaway, A., Yelon, D., Patient, R., Holder, N., et al. (1999). Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 13, 2983–2995. doi:10.1101/gad.13.22.2983
Reiter, J. F., Kikuchi, Y., and Stainier, D. Y. (2001). Multiple roles for Gata5 in zebrafish endoderm formation. Development 128, 125–135. doi:10.1242/dev.128.1.125
Rohde, L. A., Oates, A. C., and Ho, R. K. (2004). A crucial interaction between embryonic red blood cell progenitors and paraxial mesoderm revealed in spadetail embryos. Dev. Cell 7, 251–262. doi:10.1016/j.devcel.2004.07.010
Russ, A. P., Wattler, S., Colledge, W. H., Aparicio, S. A., Carlton, M. B., Pearce, J. J., et al. (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99. doi:10.1038/35003601
Sebe-pedros, A., Ariza-Cosano, A., Weirauch, M. T., Leininger, S., Yang, A., Torruella, G., et al. (2013). Early evolution of the T-box transcription factor family. Proc. Natl. Acad. Sci. U. S. A. 110, 16050–16055. doi:10.1073/pnas.1309748110
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi:10.1038/msb.2011.75
Simonetta, F., Pradier, A., and Roosnek, E. (2016). T-Bet and eomesodermin in NK cell development, maturation, and function. Front. Immunol. 7, 241. doi:10.3389/fimmu.2016.00241
Speir, M. L., Zweig, A. S., Rosenbloom, K. R., Raney, B. J., Paten, B., Nejad, P., et al. (2016). The UCSC genome browser database: 2016 update. Nucleic Acids Res. 44, D717–D725. doi:10.1093/nar/gkv1275
Strumpf, D., Mao, C. A., Yamanaka, Y., Ralston, A., Chawengsaksophak, K., Beck, F., et al. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102. doi:10.1242/dev.01801
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., et al. (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902. doi:10.1016/j.cell.2019.05.031
Takizawa, F., Araki, K., Ito, K., Moritomo, T., and Nakanishi, T. (2007). Expression analysis of two Eomesodermin homologues in zebrafish lymphoid tissues and cells. Mol. Immunol. 44, 2324–2331. doi:10.1016/j.molimm.2006.11.018
Takizawa, F., Araki, K., Ohtani, M., Toda, H., Saito, Y., Lampe, V. S., et al. (2014). Transcription analysis of two Eomesodermin genes in lymphocyte subsets of two teleost species. Fish. Shellfish Immunol. 36, 215–222. doi:10.1016/j.fsi.2013.11.004
Talbot, W. S., Trevarrow, B., Halpern, M. E., Melby, A. E., Farr, G., Postlethwait, J. H., et al. (1995). A homeobox gene essential for zebrafish notochord development. Nature 378, 150–157. doi:10.1038/378150a0
Teo, A. K., Arnold, S. J., Trotter, M. W., Brown, S., Ang, L. T., Chng, Z., et al. (2011). Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238–250. doi:10.1101/gad.607311
Tsankov, A. M., Gu, H., Akopian, V., Ziller, M. J., Donaghey, J., Amit, I., et al. (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349. doi:10.1038/nature14233
Vesterlund, L., Jiao, H., Unneberg, P., Hovatta, O., and Kere, J. (2011). The zebrafish transcriptome during early development. BMC Dev. Biol. 11, 30. doi:10.1186/1471-213X-11-30
Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G., and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. doi:10.1126/science.aar4362
Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. doi:10.1038/nature07509
Wardle, F. C., and Papaioannou, V. E. (2008). Teasing out T-box targets in early mesoderm. Curr. Opin. Genet. Dev. 18, 418–425. doi:10.1016/j.gde.2008.07.017
Warga, R. M., Mueller, R. L., Ho, R. K., and Kane, D. A. (2013). Zebrafish Tbx16 regulates intermediate mesoderm cell fate by attenuating Fgf activity. Dev. Biol. 383, 75–89. doi:10.1016/j.ydbio.2013.08.018
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi:10.1093/bioinformatics/btp033
Westerfield, M. (2000). The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th Ed. Eugene: University of Oregon Press.
White, R. J., Collins, J. E., Sealy, I. M., Wali, N., Dooley, C. M., Digby, Z., et al. (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6, e30860. doi:10.7554/eLife.30860
Windner, S. E., Doris, R. A., Ferguson, C. M., Nelson, A. C., Valentin, G., Tan, H., et al. (2015). Tbx6, Mesp-b and Ripply1 regulate the onset of skeletal myogenesis in zebrafish. Development 142, 1159–1168. doi:10.1242/dev.113431
Xing, C., Shen, W., Gong, B., Li, Y., Yan, L., and Meng, A. (2022). Maternal factors and nodal autoregulation orchestrate nodal gene expression for embryonic mesendoderm induction in the zebrafish. Front. Cell Dev. Biol. 10, 887987. doi:10.3389/fcell.2022.887987
Xu, P., Zhu, G., Wang, Y., Sun, J., Liu, X., Chen, Y. G., et al. (2014). Maternal Eomesodermin regulates zygotic nodal gene expression for mesendoderm induction in zebrafish embryos. J. Mol. Cell Biol. 6, 272–285. doi:10.1093/jmcb/mju028
Yates, A., Akanni, W., Amode, M. R., Barrell, D., Billis, K., Carvalho-Silva, D., et al. (2016). Ensembl 2016. Nucleic Acids Res. 44, D710–D716. doi:10.1093/nar/gkv1157
Keywords: Eomes, T-box, vgll4l, left-right organiser, zebrafish
Citation: Talbot CD, Walsh MD, Cutty SJ, Elsayed R, Vlachaki E, Bruce AEE, Wardle FC and Nelson AC (2022) Eomes function is conserved between zebrafish and mouse and controls left-right organiser progenitor gene expression via interlocking feedforward loops. Front. Cell Dev. Biol. 10:982477. doi: 10.3389/fcell.2022.982477
Received: 30 June 2022; Accepted: 28 July 2022;
Published: 25 August 2022.
Edited by:
Silvia L. López, CONICET Instituto de Biología Celular y Neurociencias, ArgentinaReviewed by:
Hiroshi Hamada, RIKEN Center for Biosystems Dynamics Research, JapanSimone Probst, University of Freiburg, Germany
Copyright © 2022 Talbot, Walsh, Cutty, Elsayed, Vlachaki, Bruce, Wardle and Nelson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew C. Nelson, A.Nelson.1@warwick.ac.uk
†Present address: Stephen J. Cutty, Institute of Clinical Sciences, Imperial College London, Hammersmith Hospital Campus, London, United Kingdom
 Conor D. Talbot
Conor D. Talbot