- 1Department of Health Sciences, Università Degli Studi di Milano, Milan, Italy
- 2“Aldo Ravelli” Center for Neurotechnology and Experimental Brain Therapeutics, Università Degli Studi di Milano, Milan, Italy
Chromatinopathies are defined as genetic disorders caused by mutations in genes coding for protein involved in the chromatin state balance. So far 82 human conditions have been described belonging to this group of congenital disorders, sharing some molecular features and clinical signs. For almost all of these conditions, no specific treatment is available. For better understanding the molecular cascade caused by chromatin imbalance and for envisaging possible therapeutic strategies it is fundamental to combine clinical and basic research studies. To this end, animal modelling systems represent an invaluable tool to study chromatinopathies. In this review, we focused on available data in the literature of animal models mimicking the human genetic conditions. Importantly, affected organs and abnormalities are shared in the different animal models and most of these abnormalities are reported as clinical manifestation, underlying the parallelism between clinics and translational research.
Introduction
Rare diseases are defined as conditions having a prevalence lower than 1:2000 and, nevertheless, they are estimated to be over 6,000, deeply impacting patients, families, caregivers, and health systems (Eurordis 2005). Among rare diseases, some have recently been ascribed to the so-called chromatinopathies, a heterogeneous group of Mendelian disorders affecting the epigenetic machinery (Fahrner and Bjornsson, 2014). By 2019, 70 epigenetic machinery genes have been identified, when mutated these genes are responsible for 82 human conditions. These genes were further expanded to 295 by Bjornsson and collegues (Boukas et al., 2019), and a review on monogenetic neurodevelopmental disorders (Ciptasari and van Bokhoven, 2020) is available. In this review we will focus on the 70 firstly described and better characterized causative genes. Such genes code for different epigenetic components presenting protein domains exerting writer, eraser, reader, and remodeler activities or a combination of these functions (e.g., a protein could include both a writer and a reader domain) (Fahrner and Bjornsson, 2019). Patients affected by chromatinopathies display shared clinical features, such as intellectual disability (ID) and abnormal growth, and shared etiology, which mainly relies on disruption of dosage-sensitive genes leading to haploinsufficiency (Bjornsson, 2015; Fahrner and Bjornsson, 2019).
Considering the advent of next generation sequencing (NGS) technologies, which led to the identification of a growing number of candidate variants, validation of pathogenicity can be difficult. Considering that many of the epigenetic machinery disorders are rare and/or “ultrarare”, human-based studies are often intrinsically challenging (Shen et al., 2015).
Among in vivo models, mice (Mus musculus) are the most commonly used for human genetic diseases mainly for their homology to human genome, their size, their strains that are highly inbred giving uniformed experimental conditions and reproducibility, their lifespan and for the possibility to perform genetic manipulation to obtain models of monogenic diseases, through available transgenic technologies (Dow and Lowe, 2012). Despite these advantages, mice have high husbandry costs which make them suboptimal for candidate variants assessment and drug screening. For these purposes fruit flies (Drosophila melanogaster) represent another valid option, considering the rapid generation time, relatively low-cost housing, and high experimental manipulability, even if evolutionary more distant from humans compared to mammals (Moulton and Letsou, 2016). Another animal commonly used for high-throughput studies is zebrafish (Danio rerio), which is a vertebrate with a high homology in the genome. It can be exploited for a deep characterization of disorders involving embryogenesis, due to its transparent embryos and larvae, and it represents a valid model for studying organs and structures shared with humans (Santoriello and Zon, 2012). For example, this model has been used to evaluate 3D genome organization of the epigenetic machinery (Labudina and Horsfield, 2021).
In this work we report on animal models for chromatinopathies, focusing on how these models recapitulate genotype-phenotype correlation and analyzing affected functions; moreover, we highlight the importance of an integrative approach for epigenetic machinery disorders.
Animal models for chromatinopathies
In the last 30 years many animal models have been used to study chromatinopathies. The possibility to exploit animal models for studying the molecular mechanisms underlying a disorder, is a pivotal step for confirming etiology and pathogenic variants validation, and a valuable tool for preclinical analysis of possible therapeutic approaches. To date, the majority of translational research in this field mainly focuses on mouse (Mus musculus), zebrafish (Danio rerio) and invertebrates as fruit fly (Drosophila melanogaster). We also included studies on other modeling systems such as Caenorhabditis elegans, medaka fish (Oryzias latipes), Xenopus laevis, rat (Rattus norvegicus), chicken (Gallus gallus domesticus), rabbit (Oryctolagus cuniculus) and monkey (Macaca fascicularis). In Supplementary Table S1 animal models discussed in this review are detailed.
Genetic heterogenous syndromes
Some chromatinopathies are known to be caused by mutations in different components of the epigenetic machinery, such as Rubinstein-Taybi (RSTS1, OMIM #180849; RSTS2, OMIM #613684), Kleefstra (KLEFS1, OMIM #610253; KLEFS2, OMIM #617768) and Kabuki (KS1, OMIM #147920; KS2 OMIM #300867) syndromes. In this cases, “canonical causative genes” exert the same function on the open/close chromatin equilibrium (Figure 1). Patients affected by these syndromes show a recognizable phenotype, which can vary in severity depending on the mutated gene. Therefore, it is interesting to investigate animal models-related phenotypes recapitulating syndromes with these molecular/clinical features (below 3 examples are discussed in details) for understanding whether the human genotype-phenotype correlations are well translated in vivo (Supplementary Table S1).
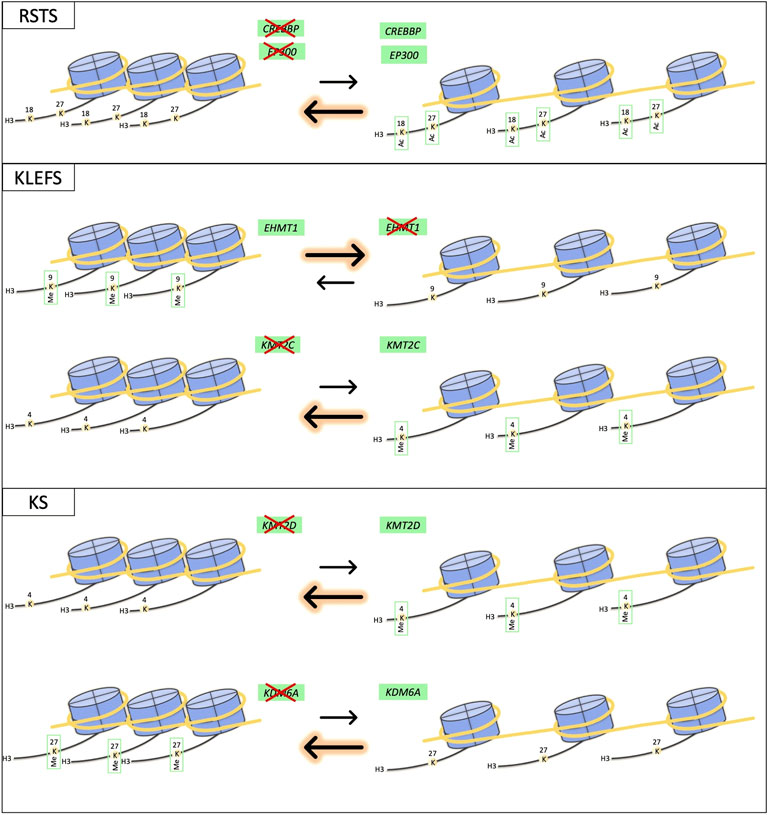
FIGURE 1. Schematic representation of effects on chromatin equilibrium for RSTS, KLEFS and KS syndromes. The drawing shows the impact of abnormal function of proteins coded by different genes having common effects on chromatin state equilibrium.
Rubinstein-Taybi syndrome
RSTS is a rare neurodevelopmental multisystem malformation syndrome characterized by developmental delay and intellectual disability, growth retardation, skeletal anomalies including broad/short thumbs and/or big toes and distinctive facial features. RSTS causative genes CREBBP and EP300 code for two writers of the epigenetic machinery, CBP and p300, both histone acetyltransferases (HAT) which alterations are responsible respectively for the 60% (RSTS1) and 10% (RSTS2) of cases; p300 disruption is associated with milder phenotypes in humans (Cohen et al., 2020). Homozygous mice for Cbp or p300 (ortholog genes of human CREBBP and EP300, respectively) show embryonic lethality (Tanaka et al., 1997) and, interestingly, it has been observed also in p300 heterozygous mutants by Yao and colleagues (Yao et al., 1998). Cbp heterozygous mice are viable although displaying skeletal and cardiac abnormalities, growth retardation and memory deficits (Oike et al., 1999). In addition, craniofacial aspects and developmental delay associated with RSTS have been reported in p300 mutant mice (Viosca et al., 2010). Therefore, loss of function of Cbp and p300 leads to these similar defects in mouse models (Tanaka et al., 1997; Yao et al., 1998; Oike et al., 1999; Viosca et al., 2010) together with multilineage defects in hematopoiesis (Kung et al., 2000; Kasper et al., 2002), which in Cbp ± mice can increase the incidence of hematologic malignancies, as observed in RSTS patients (Kung et al., 2000; Boot et al., 2018). Mice lacking normal Cbp functions have been well characterized for memory and behavior capabilities, displaying synaptic plasticity deficits (Bourtchouladze et al., 2003; Alarcón et al., 2004; Wood et al., 2005), long-term memory (Tanaka et al., 1997; Oike et al., 1999; Korzus et al., 2004; Wood et al., 2006) and neuroadaptation impairment (Lopez-Atalaya et al., 2011), ASD-relevant repetitive behaviors, hyperactivity, and social interaction deficits (Zheng et al., 2016). Furthermore, Cbp seems to play a role in energy homeostasis, with mice showing increased insulin sensitivity and glucose tolerance (Yamauchi et al., 2002). In zebrafish, specific inhibition of cbp/p300 leads to a muscular dystrophy-like phenotype (Fauquier et al., 2018), while ep300 knockdown causes skeletal, cardiac and neural abnormalities (Babu et al., 2018) modelling defects present in patients. Conversely, a Drosophila model for EP300-related RSTS phenotype does not exist, leaving the study of dCBP mutant flies, named nejire (nejP/+), as the only option for Drosophila studies of RSTS. Hemizygous nej are embryonic lethal (Akimaru et al., 1997; Di Fede et al., 2021), while nejire mutants affect the eye specification and cell fate determination (Kumar et al., 2004). Similarly to what happens in mouse, knockdown of dCBP causes behavioral alterations (Boyles et al., 2010; Sethi et al., 2019), affects nervous system development (Kirilly et al., 2011) and learning, due to altered development of mushroom bodies, associative center in invertebrate brains (Li et al., 2018).
Kleefstra syndrome
Another chromatinopathy caused by variants in two known causative genes coding for proteins members of the epigenetic machinery is Kleefstra syndrome (KLEFS) characterized by a variable phenotype including severe intellectual disability, hypotonia, brachy (micro)cephaly, seizures, heart defects, and typical facies. Patients affected from this disorder present pathogenic variants in EHMT1 or KMT2C/MLL3, coding for two methyltransferases and epigenetic writers (respectively EHMT1 and KMT2C/MLL3), which give rise to clinically overlapping phenotypes in human (KLEFS1 and KLEFS2) (Koemans et al., 2017). However, this does not seem to be reflected in mouse models, observing on the one hand severe growth retardation and embryonic lethality in Ehmt1 knockout mice (Tachibana et al., 2005), on the other hand only partial embryonic lethality and features such as stunted growth, lower fertility, very little white fat, unusual hyperproliferation, hydronephrosis, kidney abnormalities and even ureter epithelial tumors, upon Mll3 inactivation in mice (Lee et al., 2009). Despite a mouse model heterozygous for Kmt2c (Kmt2c+/-) has never been described, Ehmt1 ± mice recapitulate closely KLEFS phenotype, displaying autistic-like features (Balemans et al., 2010), learning deficits and synaptic dysfunction (Balemans et al., 2013), delayed postnatal development and increased expression of bone developmental genes (Balemans et al., 2014), increased adult cell proliferation in the hippocampus and enhanced pattern separation ability (Benevento et al., 2017), impaired cognitive abilities and hypoactive behavior (Iacono et al., 2018). Loss of Drosophila EHMT1 and KMT2C orthologs, G9a and trr respectively, appears to be rather convergent in flies, leading to neurodevelopmental impairment, with defects of peripheral dendrite development, larval locomotor behavior, non-associative learning, and courtship memory observed in Ehmt1 mutants (Kramer et al., 2011), short-term memory impairment caused by trr knockdown in mushroom bodies (Koemans et al., 2017) and developmental phenotypes in trr catalytic mutant alleles after environmental stress (Rickels et al., 2017).
Kabuki syndrome
Kabuki syndrome (KS) is a genetic condition characterized by growth deficiency, intellectual disability, minor skeletal anomalies and distinctive facial features caused by mutations in KMT2D (60% of cases) or in KDM6A (6% of cases), coding for two different components of the epigenetic machinery, a writer and an eraser respectively. Although the affected proteins are a histone methyltransferase and a histone demethylase, they lead to indistinguishable conditions (KS1 and KS2). This could be explained by the role of these two proteins: KMT2D methylates lysine 4 of histone 3 (H3K4), while KDM6A demethylates lysine 27 of histone 3 (H3K27), both enzymes operating two epigenetic modifications associated to a common downstream effect on chromatin state (open) (Fahrner and Bjornsson, 2014). Between KS1 and KS2 only slight phenotypic differences were found in a large cohort of patients, with a major incidence of hypotonia for KMT2D patients and postnatal growth retardation for KDM6A mutation group (Miyake et al., 2013). Also, mice models for KS1 and KS2 show similarities recapitulating some aspects of the human disorder. Homozygous mice deficient for the orthologs of human KMT2D and KDM6A, Kmt2d/Mll2 and Utx respectively, show both embryonic lethality and developmental retardation (Glaser et al., 2006; Lee et al., 2012, 2013; Welstead et al., 2012; Thieme et al., 2013). In addition to these features, loss of Utx leads also to heart malformations and defective cardiovascular development (Lee et al., 2012; Shpargel et al., 2012; Welstead et al., 2012; Thieme et al., 2013), neural tube closure defects (Welstead et al., 2012) and cranioschisis (Thieme et al., 2013). Conditional mouse knockouts show that both Kmt2d and Utx are involved in myogenesis (Lee et al., 2013; Faralli et al., 2016) and in neural development since their loss can lead to hippocampal memory defects (Bjornsson, 2015) and post-migratory embryonic neural crest deficiencies (Shpargel et al., 2017). Furthermore, Utx depletion in adult females leads to myelodysplasia (Thieme et al., 2013) and in females neural crest induces severe craniofacial abnormalities (Shpargel et al., 2017), annexing tumorigenesis and dysmorphisms to the features shared with human phenotypes. Particularly, one of the most interesting mouse models is the N-ethyl-N-nitrosourea (ENU)-induced mutant mouse named bate palmas (bapa) which presents a missense mutation in Kmt2d and features such as psychomotor and behavior impairments (i.e., hypotonia), fine motor coordination and hyperactivity, closely modelling brain-associated aspects of KS1 (Yamamoto et al., 2019). To date, only Drosophila model for KDM6A (dUTX) loss has been studied, which results in semi-lethality and Trithorax-like phenotype (Herz et al., 2010), while both kmt2d and kdm6a zebrafish morphants have been generated, displaying skeletal and craniofacial abnormalities (Bögershausen et al., 2015; Van Laarhoven et al., 2015; Tsai et al., 2018), impairment of cardiac and angiogenic development (Van Laarhoven et al., 2015; De Los Angeles Serrano et al., 2019) and neurodevelopmental defects (Van Laarhoven et al., 2015; Tsai et al., 2018), resembling the ones observed in KS patients.
The three chromatinopathies cited above clearly represent a good example of how alterations in different genes can result in the same phenotype and, by extension, in the same disorder. This is possible in case of either common protein functions, as for RSTS and KLEFS, or different epigenetic actors, as for KS, with shared effect on chromatin balance.
Genetic convergence for divergent phenotypes
Another important feature of chromatinopathies is the blurred limit among syndromes: in some cases molecular diagnosis does not coincide with the initial clinical one, complicating genotype-phenotype correlation for these disorders. Below three examples of overlapping molecular/clinical syndromes (i.e. genes coding for proteins of the epigenetic machinery “canonical” for one chromatinopathy, found altered in other chromatinopathies) are discussed in details (Figure 2A). For these cases, animal models could be an extremely relevant tool for elucidating pathogenetic mechanisms of these human disorders. ⁃
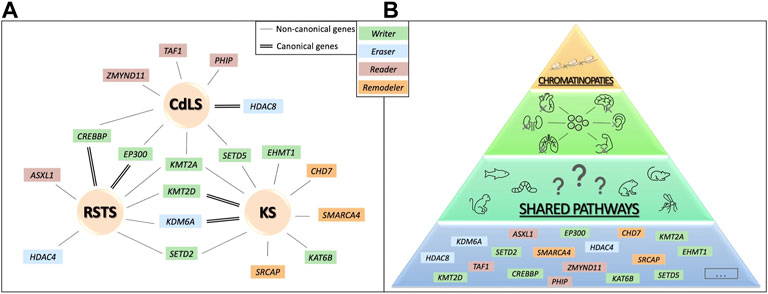
FIGURE 2. Genetic convergence for different human phenotype. In (A), canonical and non-canonical causative genes are shown for RSTS, CdLS and KS. In (B) a schematic representation of the hypothesis of possible shared molecular pathways upon chromatin balance disruption resulting in organ abnormal development that could be studied combining animal models and human data.
Rubinstein-Taybi syndrome
Indeed, clinically diagnosed RSTS patients found negative for CREBPP/EP300 mutations resulted in presenting pathogenic variants in genes causative of other chromatinopathies such as Bohring-Opitz syndrome (BOPS, OMIM #605039) (pts #80 and #173 in (Negri et al., 2019)), KS (pt #95 in (Negri et al., 2019)), Wiedemann-Steiner syndrome (WDSTS, OMIM #605130) (pt #103 in (Negri et al., 2019) and six patients in (Di Fede et al., 2020)), Brachydactyly-Mental Retardation syndrome (BDMR, OMIM #600430) (pt #GDB1427 in (Squeo et al., 2020)) and Luscan-Lumish syndrome (LLS, OMIM #616831) (pt #18–2,798 in (Squeo et al., 2020)). Causative genes of these syndromes are not all writers of the epigenetic machinery as those responsible for RSTS (CREBBP and EP300). Indeed, only KS (KMT2D), WDSTS (KMT2A) and LLS (SETD2) genes share their epigenetic function with CREBBP and EP300, while the other KS causative gene KDM6A, BDMR gene HDAC4 and BOPS gene ASXL1 are two erasers and a reader, respectively. Despite molecular differences some patients were found to have common clinical signs and interestingly similarities can be observed in animals modelling these human disorders (Supplementary Table S1, Table 1). In vivo systems found in literature for all these syndromes show that if mutated, those genes impact on viability and growth (Sinclair et al., 1992; Yu et al., 1995; Akimaru et al., 1997; Tanaka et al., 1997; Sinclair et al., 1998; Yao et al., 1998; Katsani et al., 2001; Kumar et al., 2004; Vega et al., 2004; Glaser et al., 2006; Herz et al., 2010; Hu et al., 2010; Viosca et al., 2010; Lee et al., 2012; Abdel-Wahab et al., 2013; Thieme et al., 2013; Wang et al., 2014; Hsu et al., 2017; Tsai et al., 2018; Gjini et al., 2019; Liu et al., 2020), neural and brain development (Tanaka et al., 1997; Sinclair et al., 1998; Yao et al., 1998; Oike et al., 1999; Bourtchouladze et al., 2003; Wood et al., 2006; Boyles et al., 2010; Gupta et al., 2010; Hu et al., 2010; Kirilly et al., 2011; Lopez-Atalaya et al., 2011; Welstead et al., 2012; Huang et al., 2015; Van Laarhoven et al., 2015; Zheng et al., 2016; Babu et al., 2018; Li et al., 2018; Tsai et al., 2018; Yamamoto et al., 2019), hematopoiesis (Kung et al., 2000; Kasper et al., 2002; Fisher et al., 2010; Wan et al., 2011; Thieme et al., 2013; Gjini et al., 2019), and lead to skeletal and craniofacial abnormalities (Sinclair et al., 1992; Yu et al., 1995; Tanaka et al., 1997; Sinclair et al., 1998; Vega et al., 2004; Hu et al., 2010; Viosca et al., 2010; Welstead et al., 2012; Abdel-Wahab et al., 2013; Bögershausen et al., 2015; Van Laarhoven et al., 2015; Babu et al., 2018; Tsai et al., 2018), resembling human phenotypes.

TABLE 1. Correlation between function/organ involvement and genes belonging to the writers-erasers-readers-remodelers groups.
Kabuki syndrome
As described for RSTS patients, initial clinical diagnosis for Kabuki syndrome (KS) was not always confirmed at molecular level. KS patients negative for mutations in KMT2D or KMD6A were found carriers of pathogenic variants in genes related to WDSTS, KLEFS, Mental Retardation Autosomal Dominant 23 (MRD23, OMIM #615761), Say-Barber-Biesecker-Young-Simpson or Ohdo syndrome (SBBYSS, OMIM #603736), Coffin-Siris syndrome-4 (CSS4, OMIM #614609), Floating Harbor syndrome (FLHS, OMIM #136140), CHARGE syndrome (CHARGE, OMIM #214800) and LLS (e.g. pts GDB1054, GDB1405, GDB1400, #18–2,261, GDB1128, GDB1185, GDB1311, GDB1404, GDB1422, GDB1154, GDB1406, GDB1433 in (Squeo et al., 2020)). As KMT2D, most of the causative genes of the aforementioned syndromes are writers of the epigenetic machinery (KMT2A, EHMT1, SETD5, KAT6B and SETD2), except for the remodelers SMARCA4 (CSS4), SRCAP (FLHS) and CHD7 (CHARGE). Animal models mimicking these disorders (Supplementary Table S1, Table 1) do not reach adulthood when ortholog genes are depleted, as observed in mouse (Yu et al., 1995; Bultman et al., 2000; Bosman et al., 2005; Tachibana et al., 2005; Glaser et al., 2006; Hu et al., 2010; Lee et al., 2012; Thieme et al., 2013; Osipovich et al., 2016), zebrafish (Thieme et al., 2013; Tsai et al., 2018) and fruit fly (Braun et al., 1997; Ruhf et al., 2001; Herz et al., 2010), with the only exception of KAT6B mutant mice getting through the postnatal period (Thomas et al., 2000; Merson et al., 2006). Interestingly, despite the variety of mutants used for studying these disorders, they display anomalies impacting similar neural aspects: neural tube closure in mouse (Hu et al., 2010; Welstead et al., 2012; Osipovich et al., 2016); neuronal differentiation in zebrafish (Van Laarhoven et al., 2015) and Xenopus (Seo et al., 2005); neural development and neurogenesis in rodents (Merson et al., 2006; Layman et al., 2009; Clayton-Smith et al., 2011; Moore et al., 2019; Sessa et al., 2019), zebrafish (Huang et al., 2015; Van Laarhoven et al., 2015; Tsai et al., 2018) and fruit flies (Braun et al., 1997; Melicharek et al., 2010; Kramer et al., 2011); cognitive ability and synaptic functions in mice (Layman et al., 2009; Gupta et al., 2010; Balemans et al., 2013; Iacono et al., 2018; Sessa et al., 2019) and Drosophila (Melicharek et al., 2010; Kramer et al., 2011); psychomotor functions and autistic-like behavior in mice (Balemans et al., 2010; Deliu et al., 2018; Moore et al., 2019; Sessa et al., 2019; Yamamoto et al., 2019) and Drosophila (Melicharek et al., 2010; Kramer et al., 2011). In addition, patients affected from syndromes such as KS, MRD32, CSS4 and CHARGE often display heart defects and cardiovascular anomalies (Kosho et al., 2014; Koemans et al., 2017; van Ravenswaaij-Arts and Martin, 2017; Powis et al., 2018) as well as animals modelling these disorders (Bosman et al., 2005; Takeuchi et al., 2011; Lee et al., 2012; Shpargel et al., 2012; Welstead et al., 2012; Thieme et al., 2013; Van Laarhoven et al., 2015; Osipovich et al., 2016; De Los Angeles Serrano et al., 2019). Although similar defects have been observed also in WDSTS, KLEFS1 and SBBYSS patients (Lemire et al., 2012; Willemsen et al., 2012; Baer et al., 2018), in vivo systems for these syndromes do not show anomalies in this organ.
Cornelia de lange syndrome
CdLS is a rare heterogenous developmental disorder characterized by multiorgan abnormalities leading to severe growth delay, distinctive facial feature, psychomotor deficit, intellectual disability, behavioral problems, and limb malformations. Patients firstly diagnosed by geneticists for having CdLS (CdLS1, OMIM #122470; CdLS2, OMIM #300590; CdLS3, OMIM #610759; CdLS4, OMIM #614701; CdLS5, OMIM #300882), not always carried mutations in one of the seven known genes causative of this syndrome. Instead, the genetic alteration can be found in a gene associated with another syndrome even though patients display typical characteristics described in the CdLS consensus (Kline et al., 2018). As a matter of fact, CdLS is now considered a “clinical spectrum” rather than an isolated syndrome with unique features (Selicorni et al., 2021). Genes found mutated in patients with an initial diagnosis of CdLS are: EP300 ((Aoi et al., 2019) - Patient 6; (Cucco et al., 2020); - Patient A; (Squeo et al., 2020); - GDB1418; (Woods et al., 2014); - Case Report)), KMT2A ((Yuan et al., 2015) - CdLS-3 (BAB4964); (Parenti et al., 2017); - Patient 12; (Aoi et al., 2019); - Patient 27; (Demir et al., 2021); - Case Report)); TAF1 ((O’Rawe et al., 2015) - Individual 4A; (Cheng et al., 2020); - Individual 13) causative of Mental Retardation X-Linked Syndromic 33 (MRXS33, OMIM #300966)); ZMYND11((Aoi et al., 2019) - Patient 53) causative of Mental Retardation Autosomal Dominant 30 (MRD30, OMIM #616083)); PHIP ((Aoi et al., 2019) - Patient 56) associated to Developmental Delay Intellectual Disability Obesity and Dysmorphism or Chung-Jansen syndrome (CHUJANS, OMIM #617991)), CREBBP ((Tang et al., 2019) - Patient 3)); SETD5 ((Squeo et al., 2020) - GDB1400) causative of MRD23). These genes, involved in chromatin regulation, are associated with syndromes different from CdLS, but studies conducted exploiting animal models highlighted some similar characteristics that can be found in the human patients affected with CdLS. Almost all animals mutated in the above mentioned genes display impaired viability or overall defects in growth (HDAC8, EP300, KMT2A, TAF1, CREBBP, SETD5) (Yu et al., 1995; Akimaru et al., 1997; Tanaka et al., 1997; Yao et al., 1998; Wassarman et al., 2000; Katsani et al., 2001; Kumar et al., 2004; Haberland et al., 2009; Viosca et al., 2010; Osipovich et al., 2016; Deliu et al., 2018; Gudmundsson et al., 2019), usually with mild to severe problems affecting the embryonic neurodevelopment (HDAC8, EP300, KMT2A, TAF1, CREBBP, SETD5) (Tanaka et al., 1997; Yao et al., 1998; Oike et al., 1999; Bourtchouladze et al., 2003; Wood et al., 2006; Haberland et al., 2009; Boyles et al., 2010; Gupta et al., 2010; Kirilly et al., 2011; Lopez-Atalaya et al., 2011; Huang et al., 2015; O’Rawe et al., 2015; Osipovich et al., 2016; Zheng et al., 2016; Babu et al., 2018; Deliu et al., 2018; Li et al., 2018; Gudmundsson et al., 2019; Sessa et al., 2019) and skeletal abnormalities often found in the craniofacial district (Yu et al., 1995; Tanaka et al., 1997; Haberland et al., 2009; Viosca et al., 2010; Babu et al., 2018; Sun et al., 2018; Sessa et al., 2019) (HDAC8, EP300, KMT2A, ZMYND11, CREBBP, SETD5) (Supplementary Table S1, Table 1).
Lessons from the models
In vivo systems representative of depletion or alteration of chromatin regulators, listed in Supplementary Table S1, show many parallelisms with patients affected by chromatinopathies. As shown in Figure 3 genes causative of human chromatinopathies impact common organ function/development also in different animal models. As portrayed in Figure 3A and Table 1, the most compromised functions in most animal models we found reported for all the players of the epigenetic machinery (writers, erasers, readers, and remodelers) resulted to be viability, growth, and neural and skeletal development. These features can be identified in patients who often present main clinical signs such as growth delay, intellectual disability (ID), facial dysmorphisms and skeletal anomalies (Fahrner and Bjornsson, 2014). In addition, most individuals affected by chromatinopathies have loss of function mutations leading to haploinsufficiency, due to the fundamental role of causative genes in embryonic development whose complete loss would be often at odds with life (Bjornsson, 2015). Of note, the vascular system together with organs such as heart and eyes seem particularly affected in writers models (Figure 3A), memory in writers and remodelers models (Figure 3A), while fertility resulted mostly altered in readers models (Figure 3B). Furthermore, about 20% of these in vivo systems display defects in hematopoiesis and altered mechanisms leading to tumorigenesis (Figure 3 and Table 1). Interestingly, although somatic mutations in genes of the epigenetic apparatus have been found in different types of tumors, cancer predisposition due to germline mutations is a feature common to some chromatinopathies. For instance, a peculiar aspect of RSTS is the increased incidence of benign and malignant tumors (Boot et al., 2018), which can be observed in the Cbp ± mouse model for this syndrome (Kung et al., 2000). Tumors susceptibility has been studied also for BOPS and was found that, although isolated reports on BOPS children seem to suggest a greater risk for Wilms tumor, sporadic malignancies in absence of other BOPS findings more frequently harbor somatic ASXL1 pathogenetic variants (Russell et al., 2018) which increase the risk of myelodysplastic syndrome as shown in mice (Abdel-Wahab et al., 2013; Wang et al., 2014). A tumor predisposition was also found in patients positive for germline mutation in MSH6 and SMARCA4 who developed respectively Hereditary nonpolyposis colorectal cancer type 5 and rhabdoid tumors (Biegel et al., 2014; Lepore Signorile et al., 2021) and this increased risk of developing malignancies was observed also in their corresponding animal models (Edelmann et al., 1997; De Wind et al., 1999; Bultman et al., 2000; Feitsma et al., 2008; Peled et al., 2010). Conversely, for Immunodeficiency-centromeric instability-facial anomalies syndrome 1 (ICF1, OMIM #242860), Borjeson-Forssmann-Lehmann syndrome (BFLS, OMIM #301900) and KS a cancer association has only been hypothesized so far, as patients who developed Hodgkin lymphoma, adrenocortical adenoma, acute lymphoblastic leukemia, Burkitt’s lymphoma and solid tumors (Ijichi et al., 1996; Scherer et al., 2003; Ehrlich et al., 2008; Chao et al., 2010a,b; Wang et al., 2019), and animal models presenting hematopoietic tumors (Shah et al., 2010; Thieme et al., 2013; Hsu et al., 2019) have been reported for these syndromes.
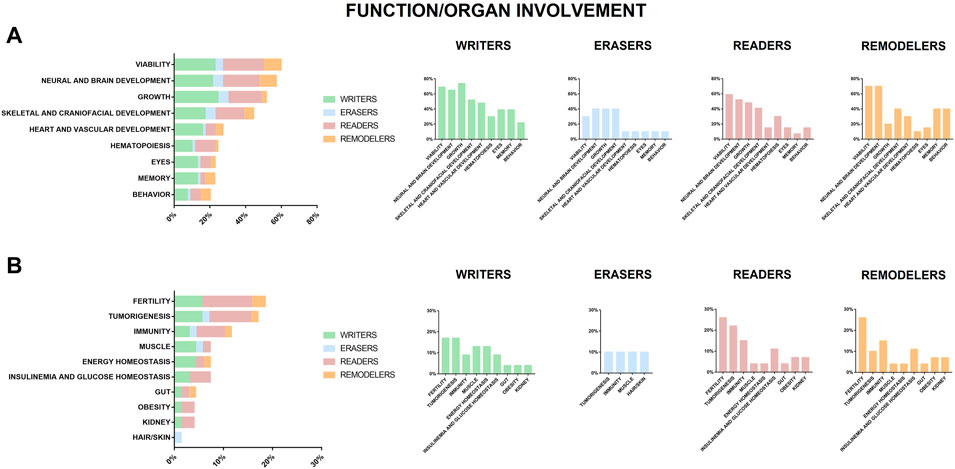
FIGURE 3. Function or organ found impaired or altered in animal models of chromatinopathies. Genes coding for writers (green), erasers (blue), readers (pink) and remodelers (orange), for which haploinsufficiency results in human abnormalities. (A) reports functions/organs in animal models altered in more than 20% of analyzed genes and (B) those in <20%.
Conclusion
In vivo models require genetic manipulation and shared homology with the human genome, leading to the possibility of mimicking the human genetic disease for studying for possibly shared molecular mechanisms responsible for clinical phenotypes and examine physically or temporarily inaccessible tissues (Figure 2B). Animal models for chromatinopathies have proved to be a valuable tool for dissecting mechanisms underlying these disorders and altered functions due to mutations in genes of the epigenetic apparatus. More variants in genes that can be grouped in the chromatinopathies are increasingly reported as well as animal models for their study. Hence, we propose a table with details of the first 70 well characterized genes, with the possibility of expanding such collection in an open science format (https://www.shorturl.at/nuV78). Importantly, affected organs and abnormalities are shared in the different animal models (listed in Supplementary Table S1), generated for a better understanding for the effects of loss or alteration of epigenetic genes (represented in Figure 3 and Table 1). Most of these abnormalities can also be found in patients affected by chromatinopathies, pointing out once again the parallelism between clinics and translational research. Importantly, for better dissecting each organ/function abnormalities in these rare conditions many studies are undergoing exploiting also stem cells and organoids for combining human data and animal model information. Interestingly, Boukas and colleagues (Boukas et al., 2019) recently demonstrated that a large subset of genes belonging to the epigenetic machinery are highly co-expressed, intolerant to variation and independently enriched for genes affecting neurological function. This suggests a link between these properties, highlighting once again the interconnection between epigenetic regulators. This aspect can be observed in applied research, where modelling disorders leads to phenotypes resembling not only the human disease but also shared features among chromatinopathies. Thus, to ensure the understanding of molecular mechanisms characterizing these disorders an integrative approach should be supported.
Author contributions
EDF and PG literature review; EDF, PG, CG, and VM manuscript writing; AL, CP, SC, ET, EAC, SA, and AP critical revision.
Funding
The authors are grateful to the following funding sources: “Aldo Ravelli” Center for Neurotechnology and Experimental Brain Therapeutics - Università degli Studi di Milano (to AL, AP, CG and VM), Intramural funding - Dipartimento DISS, Linea 2, Università degli Studi di Milano (to CG and VM).
Acknowledgments
Translational Medicine Ph.D. scholarship - Università degli Studi di Milano (to ED, PG and CP) and Fondazione Umberto Veronesi fellowship (to AL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.979512/full#supplementary-material
References
Abdel-Wahab, O., Gao, J., Adli, M., Dey, A., Trimarchi, T., Chung, Y. R., et al. (2013). Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210, 2641–2659. doi:10.1084/jem.20131141
Akimaru, H., Chen, Y., Dai, P., Hou, D. X., Nonaka, M., Smolik, S. M., et al. (1997). Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386, 735–738. doi:10.1038/386735a0
Alarcón, J. M., Malleret, G., Touzani, K., Vronskaya, S., Ishii, S., Kandel, E. R., et al. (2004). Chromatin acetylation, memory, and ltp are impaired in CBP+/- mice: A model for the cognitive deficit in rubinstein-taybi syndrome and its amelioration. Neuron 42, 947–959. doi:10.1016/j.neuron.2004.05.021
Aoi, S., Ohashi, T., Bamba, R., Fujiki, S., Tamura, D., Funato, T., et al. (2019). Neuromusculoskeletal model that walks and runs across a speed range with a few motor control parameter changes based on the muscle synergy hypothesis. Sci. Rep. 9, 369. doi:10.1038/s41598-018-37460-3
Babu, A., Kamaraj, M., Basu, M., Mukherjee, D., Kapoor, S., Ranjan, S., et al. (2018). Chemical and genetic rescue of an ep300 knockdown model for Rubinstein Taybi Syndrome in zebrafish. Biochim. Biophys. Acta. Mol. Basis Dis. 1864, 1203–1215. doi:10.1016/j.bbadis.2018.01.029
Baer, S., Afenjar, A., Smol, T., Piton, A., Gérard, B., Alembik, Y., et al. (2018). Wiedemann-steiner syndrome as a major cause of syndromic intellectual disability: A study of 33 French cases. Clin. Genet. 94, 141–152. doi:10.1111/cge.13254
Balemans, M. C. M., Ansar, M., Oudakker, A. R., van Caam, A. P. M., Bakker, B., Vitters, E. L., et al. (2014). Reduced Euchromatin histone methyltransferase 1 causes developmental delay, hypotonia, and cranial abnormalities associated with increased bone gene expression in Kleefstra syndrome mice. Dev. Biol. 386, 395–407. doi:10.1016/j.ydbio.2013.12.016
Balemans, M. C. M., Huibers, M. M. H., Eikelenboom, N. W. D., Kuipers, A. J., van Summeren, R. C. J., Pijpers, M. M. C. A., et al. (2010). Reduced exploration, increased anxiety, and altered social behavior: Autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav. Brain Res. 208, 47–55. doi:10.1016/j.bbr.2009.11.008
Balemans, M. C. M., Nadif kasri, N., Kopanitsa, M. V., Afinowi, N. O., Ramakers, G., Peters, T. A., et al. (2013). Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Hum. Mol. Genet. 22, 852–866. doi:10.1093/hmg/dds490
Benevento, M., Oomen, C. A., Horner, A. E., Amiri, H., Jacobs, T., Pauwels, C., et al. (2017). Haploinsufficiency of EHMT1 improves pattern separation and increases hippocampal cell proliferation. Sci. Rep. 7, 40284. doi:10.1038/srep40284
Biegel, J. A., Busse, T. M., and Weissman, B. E. (2014). SWI/SNF chromatin remodeling complexes and cancer. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 350–366. doi:10.1002/AJMG.C.31410
Bjornsson, H. T. (2015). The Mendelian disorders of the epigenetic machinery. Genome Res. 25, 1473–1481. doi:10.1101/gr.190629.115
Bögershausen, N., Tsai, I. C., Pohl, E., Kiper, P. O. S., Beleggia, F., Ferda Percin, E., et al. (2015). RAP1-mediated MEK/ERK pathway defects in Kabuki syndrome. J. Clin. Invest. 125, 3585–3599. doi:10.1172/JCI80102
Boot, M. V., van Belzen, M. J., Overbeek, L. I., Hijmering, N., Mendeville, M., Waisfisz, Q., et al. (2018). Benign and malignant tumors in Rubinstein–Taybi syndrome. Am. J. Med. Genet. A 176, 597–608. doi:10.1002/ajmg.a.38603
Bosman, E. A., Penn, A. C., Ambrose, J. C., Kettleborough, R., Stemple, D. L., and Steel, K. P. (2005). Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 14, 3463–3476. doi:10.1093/hmg/ddi375
Boukas, L., Havrilla, J. M., Hickey, P. F., Quinlan, A. R., Bjornsson, H. T., and Hansen, K. D. (2019). Coexpression patterns define epigenetic regulators associated with neurological dysfunction. Genome Res. 29, 532–542. doi:10.1101/gr.239442.118
Bourtchouladze, R., Lidge, R., Catapano, R., Stanley, J., Gossweiler, S., Romashko, D., et al. (2003). A mouse model of Rubinstein-Taybi syndrome: Defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc. Natl. Acad. Sci. U. S. A. 100, 10518–10522. doi:10.1073/pnas.1834280100
Boyles, R. S., Lantz, K. M., Poertner, S., Georges, S. J., and Andres, A. J. (2010). Presenilin controls CBP levels in the adult Drosophila central nervous system. PLoS One 5, e14332. doi:10.1371/journal.pone.0014332
Braun, A., Lemaitre, B., Lanot, R., Zachary, D., and Meister, M. (1997). Drosophila immunity: Analysis of larval hemocytes by P-element-mediated enhancer trap. Genetics 147, 623–634. doi:10.1093/genetics/147.2.623
Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., et al. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 6, 1287–1295. doi:10.1016/S1097-2765(00)00127-1
Chao, H. T., Chen, H., Samaco, R. C., Xue, M., Chahrour, M., Yoo, J., et al. (2010a). Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 468, 263–269. doi:10.1038/nature09582
Chao, M. M., Todd, M. A., Kontny, U., Neas, K., Sullivan, M. J., Hunter, A. G., et al. (2010b). T-cell acute lymphoblastic leukemia in association with Börjeson-Forssman-Lehmann syndrome due to a mutation in PHF6. Pediatr. Blood Cancer 55, 722–724. doi:10.1002/PBC.22574
Cheng, C., Deng, P. Y., Ikeuchi, Y., Yuede, C., Li, D., Rensing, N., et al. (2018). Characterization of a mouse model of börjeson-forssman-lehmann syndrome. Cell. Rep. 25, 1404–1414. e6. doi:10.1016/j.celrep.2018.10.043
Cheng, H., Capponi, S., Wakeling, E., Marchi, E., Li, Q., Zhao, M., et al. (2020). Missense variants in TAF1 and developmental phenotypes: Challenges of determining pathogenicity. Hum. Mutat. 41, 449–464. doi:10.1002/HUMU.23936
Ciptasari, U., and van Bokhoven, H. (2020). The phenomenal epigenome in neurodevelopmental disorders. Hum. Mol. Genet. 29, R42–R50. doi:10.1093/HMG/DDAA175
Clayton-Smith, J., O’Sullivan, J., Daly, S., Bhaskar, S., Day, R., Anderson, B., et al. (2011). Whole-exome-sequencing identifies mutations in histone acetyltransferase gene KAT6B in individuals with the say-barber-biesecker variant of Ohdo syndrome. Am. J. Hum. Genet. 89, 675–681. doi:10.1016/j.ajhg.2011.10.008
Cohen, J. L., Schrier Vergano, S. A., Mazzola, S., Strong, A., Keena, B., McDougall, C., et al. (2020). EP300-related Rubinstein–Taybi syndrome: Highlighted rare phenotypic findings and a genotype–phenotype meta-analysis of 74 patients. Am. J. Med. Genet. A 182, 2926–2938. doi:10.1002/ajmg.a.61883
Cucco, F., Sarogni, P., Rossato, S., Alpa, M., Patimo, A., Latorre, A., et al. (2020). Pathogenic variants in EP300 and ANKRD11 in patients with phenotypes overlapping Cornelia de Lange syndrome. Am. J. Med. Genet. A 182, 1690–1696. doi:10.1002/AJMG.A.61611
De Los Angeles Serrano, M., Demarest, B. L., Tone-Pah-Hote, T., Tristani-Firouzi, M., and Joseph Yost, H. (2019). Inhibition of notch signaling rescues cardiovascular development in Kabuki Syndrome. PLoS Biol. 17, e3000087. doi:10.1371/journal.pbio.3000087
De Wind, N., Dekker, M., Claij, N., Jansen, L., Van Klink, Y., Radman, M., et al. (1999). HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 23, 359–362. doi:10.1038/15544
Deliu, E., Arecco, N., Morandell, J., Dotter, C. P., Contreras, X., Girardot, C., et al. (2018). Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat. Neurosci. 21, 1717–1727. doi:10.1038/s41593-018-0266-2
Demir, S., Gürkan, H., Öz, V., Yalçlntepe, S., Atll, E., and Atll, E. (2021). Wiedemann-steiner syndrome as a differential diagnosis of cornelia de Lange syndrome using targeted next-generation sequencing: A case report. Mol. Syndromol. 12, 46–51. doi:10.1159/000511971
Di Fede, E., Massa, V., Augello, B., Squeo, G., Scarano, E., Perri, A. M., et al. (2020). Expanding the phenotype associated to KMT2A variants: Overlapping clinical signs between wiedemann–steiner and rubinstein–taybi syndromes. Eur. J. Hum. Genet. 29, 88–98. doi:10.1038/s41431-020-0679-8
Di Fede, E., Ottaviano, E., Grazioli, P., Ceccarani, C., Galeone, A., Parodi, C., et al. (2021). Insights into the role of the microbiota and of short-chain fatty acids in rubinstein-taybi syndrome. Int. J. Mol. Sci. 22, 3621. doi:10.3390/IJMS22073621
Dow, L. E., and Lowe, S. W. (2012). Life in the fast lane: Mammalian disease models in the genomics era. Cell. 148, 1099–1109. doi:10.1016/j.cell.2012.02.023
Edelmann, W., Yang, K., Umar, A., Heyer, J., Lau, K., Fan, K., et al. (1997). Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 91, 467–477. doi:10.1016/S0092-8674(00)80433-X
Ehrlich, M., Sanchez, C., Shao, C., Nishiyama, R., Kehrl, J., Kuick, R., et al. (2008). ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity 41, 253–271. doi:10.1080/08916930802024202
Eurordis, November (2005). Rare Diseases : Understanding this Public Health Priority” – www.eurordis.org.
Fahrner, J. A., and Bjornsson, H. T. (2019). Mendelian disorders of the epigenetic machinery: Postnatal malleability and therapeutic prospects. Hum. Mol. Genet. 28, R254–R264. doi:10.1093/HMG/DDZ174
Fahrner, J. A., and Bjornsson, H. T. (2014). Mendelian disorders of the epigenetic machinery: Tipping the balance of chromatin States. Annu. Rev. Genomics Hum. Genet. 15, 269–293. doi:10.1146/annurev-genom-090613-094245
Faralli, H., Wang, C., Nakka, K., Benyoucef, A., Sebastian, S., Zhuang, L., et al. (2016). UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 126, 1555–1565. doi:10.1172/JCI83239
Fauquier, L., Azzag, K., Parra, M. A. M., Quillien, A., Boulet, M., Diouf, S., et al. (2018). CBP and P300 regulate distinct gene networks required for human primary myoblast differentiation and muscle integrity. Sci. Rep. 8, 12629. doi:10.1038/s41598-018-31102-4
Feitsma, H., Kuiper, R. V., Korving, J., Nijman, I. J., and Cuppen, E. (2008). Zebrafish with mutations in mismatch repair genes develop neurofibromas and other tumors. Cancer Res. 68, 5059–5066. doi:10.1158/0008-5472.CAN-08-0019
Fisher, C. L., Pineault, N., Brookes, C., Helgason, C. D., Ohta, H., Bodner, C., et al. (2010). Loss-of-function additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood 115, 38–46. doi:10.1182/blood-2009-07-230698
Gjini, E., Jing, C. B., Nguyen, A. T., Reyon, D., Gans, E., Kesarsing, M., et al. (2019). Disruption of asxl1 results in myeloproliferative neoplasms in zebrafish. Dis. Model. Mech. 12, dmm035790. doi:10.1242/dmm.035790
Glaser, S., Schaft, J., Lubitz, S., Vintersten, K., van der Hoeven, F., Tuftteland, K. R., et al. (2006). Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133, 1423–1432. doi:10.1242/dev.02302
Gudmundsson, S., Wilbe, M., Filipek-Górniok, B., Molin, A. M., Ekvall, S., Johansson, J., et al. (2019). TAF1, associated with intellectual disability in humans, is essential for embryogenesis and regulates neurodevelopmental processes in zebrafish. Sci. Rep. 9, 10730. doi:10.1038/s41598-019-46632-8
Gupta, S., Kim, S. Y., Artis, S., Molfese, D. L., Schumacher, A., Sweatt, J. D., et al. (2010). Histone methylation regulates memory formation. J. Neurosci. 30, 3589–3599. doi:10.1523/JNEUROSCI.3732-09.2010
Haberland, M., Mokalled, M. H., Montgomery, R. L., and Olson, E. N. (2009). Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes. Dev. 23, 1625–1630. doi:10.1101/gad.1809209
Herz, H.-M., Madden, L. D., Chen, Z., Bolduc, C., Buff, E., Gupta, R., et al. (2010). The H3K27me3 demethylase dUTX is a suppressor of notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 30, 2485–2497. doi:10.1128/mcb.01633-09
Hsu, Y. C., Chen, T. C., Lin, C. C., Yuan, C. T., Hsu, C. L., Hou, H. A., et al. (2019). Phf6-null hematopoietic stem cells have enhanced self-renewal capacity and oncogenic potentials. Blood Adv. 3, 2355–2367. doi:10.1182/bloodadvances.2019000391
Hsu, Y. C., Chiu, Y. C., Lin, C. C., Kuo, Y. Y., Hou, H. A., Tzeng, Y. S., et al. (2017). The distinct biological implications of Asxl1 mutation and its roles in leukemogenesis revealed by a knock-in mouse model. J. Hematol. Oncol. 10, 139. doi:10.1186/s13045-017-0508-x
Hu, M., Sun, X. J., Zhang, Y. L., Kuang, Y., Hu, C. Q., Wu, W. L., et al. (2010). Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc. Natl. Acad. Sci. U. S. A. 107, 2956–2961. doi:10.1073/PNAS.0915033107
Huang, Y. C., Shih, H. Y., Lin, S. J., Chiu, C. C., Ma, T. L., Yeh, T. H., et al. (2015). The epigenetic factor Kmt2a/Mll1 regulates neural progenitor proliferation and neuronal and glial differentiation. Dev. Neurobiol. 75, 452–462. doi:10.1002/dneu.22235
Iacono, G., Dubos, A., Méziane, H., Benevento, M., Habibi, E., Mandoli, A., et al. (2018). Increased H3K9 methylation and impaired expression of Protocadherins are associated with the cognitive dysfunctions of the Kleefstra syndrome. Nucleic Acids Res. 46, 4950–4965. doi:10.1093/nar/gky196
Ijichi, O., Kawakami, K., Matsuda, Y., Ikarimoto, N., Miyata, K., Takamatsu, H., et al. (1996). A case of Kabuki make-up syndrome with EBV+Burkitt’s lymphoma. Acta Paediatr. Jpn. 38, 66–68. doi:10.1111/J.1442-200X.1996.TB03439.X
Kasper, L. H., Boussouar, F., Ney, P. A., Jackson, C. W., Rehg, J., Van Deursen, J. M., et al. (2002). A transcription-factor-binding surface of coactivator p300 is required for haematopoiesis. Nature 419, 738–743. doi:10.1038/nature01062
Katsani, K. R., Arredondo, J. J., Kal, A. J., and Verrijzer, C. P. (2001). A homeotic mutation in the trithorax SET domain impedes histone binding. Genes. Dev. 15, 2197–2202. doi:10.1101/gad.201901
Kirilly, D., Wong, J. J. L., Lim, E. K. H., Wang, Y., Zhang, H., Wang, C., et al. (2011). Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron 72, 86–100. doi:10.1016/j.neuron.2011.08.003
Kline, A. D., Moss, J. F., Selicorni, A., Bisgaard, A. M., Deardorff, M. A., Gillett, P. M., et al. (2018). Diagnosis and management of cornelia de Lange syndrome: First international consensus statement. Nat. Rev. Genet. 19, 649–666. doi:10.1038/s41576-018-0031-0
Koemans, T. S., Kleefstra, T., Chubak, M. C., Stone, M. H., Reijnders, M. R. F., de Munnik, S., et al. (2017). Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 13, e1006864. doi:10.1371/journal.pgen.1006864
Korzus, E., Rosenfeld, M. G., and Mayford, M. (2004). CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972. doi:10.1016/j.neuron.2004.06.002
Kosho, T., Okamoto, N., Imai, Y., Ohashi, H., van Eerde, A. M., Chrzanowska, K., et al. (2014). Genotype-phenotype correlation of coffin-siris syndrome caused by mutations in SMARCB1, SMARCA4, SMARCE1, and ARID1A. Am. J. Med. Genet. C Semin. Med. Genet. 166, 262–275. doi:10.1002/ajmg.c.31407
Kramer, J. M., Kochinke, K., Oortveld, M. A. W., Marks, H., Kramer, D., de Jong, E. K., et al. (2011). Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 9, e1000569. doi:10.1371/journal.pbio.1000569
Kumar, J. P., Jamal, T., Doetsch, A., Turner, F. R., and Duffy, J. B. (2004). CREB binding protein functions during successive stages of eye development in Drosophila. Genetics 168, 877–893. doi:10.1534/genetics.104.029850
Kung, A. L., Rebel, V. I., Bronson, R. T., Ch’ng, L. E., Sieff, C. A., Livingston, D. M., et al. (2000). Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes. Dev. 14, 272–277. doi:10.1101/gad.14.3.272
Labudina, A., and Horsfield, J. A. (2021). The three-dimensional genome in zebrafish development. Brief. Funct. Genomics 20, elab008–377. doi:10.1093/BFGP/ELAB008
Layman, W. S., McEwen, D. P., Beyer, L. A., Lalani, S. R., Fernbach, S. D., Oh, E., et al. (2009). Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum. Mol. Genet. 18, 1909–1923. doi:10.1093/hmg/ddp112
Lee, J. E., Wang, C., Xu, S., Cho, Y. W., Wang, L., Feng, X., et al. (2013). H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, e01503. doi:10.7554/eLife.01503
Lee, S., Kim, D. H., Goo, Y. H., Lee, Y. C., Lee, S. K., and Lee, J. W. (2009). Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol. Endocrinol. 23, 610–619. doi:10.1210/me.2008-0455
Lee, S., Lee, J. W., and Lee, S. K. (2012). UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell. 22, 25–37. doi:10.1016/j.devcel.2011.11.009
Lemire, G., Campeau, P. M., and Lee, B. H. (2012). KAT6B disorders. Available at: https://www.ncbi.nlm.nih.gov/books/NBK114806/(Accessed December 28, 2020).
Lepore Signorile, M., Disciglio, V., Di Carlo, G., Pisani, A., Simone, C., and Ingravallo, G. (2021). From genetics to histomolecular characterization: An insight into colorectal carcinogenesis in lynch syndrome. Int. J. Mol. Sci. 22, 6767. doi:10.3390/IJMS22136767
Li, K. L., Zhang, L., Yang, X. M., Fang, Q., Yin, X. F., Wei, H. M., et al. (2018). Histone acetyltransferase CBP-related H3K23 acetylation contributes to courtship learning in Drosophila. BMC Dev. Biol. 18, 20. doi:10.1186/s12861-018-0179-z
Liu, D. J., Zhang, F., Chen, Y., Jin, Y., Zhang, Y. L., Chen, S. B., et al. (2020). setd2 knockout zebrafish is viable and fertile: differential and developmental stress-related requirements for Setd2 and histone H3K36 trimethylation in different vertebrate animals. Cell. Discov. 61 6, 72–15. doi:10.1038/s41421-020-00203-8
Lopez-Atalaya, J. P., Ciccarelli, A., Viosca, J., Valor, L. M., Jimenez-Minchan, M., Canals, S., et al. (2011). CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J. 30, 4287–4298. doi:10.1038/emboj.2011.299
Melicharek, D. J., Ramirez, L. C., Singh, S., Thompson, R., and Marenda, D. R. (2010). Kismet/CHD7 regulates axon morphology, memory and locomotion in a Drosophila model of CHARGE syndrome. Hum. Mol. Genet. 19, 4253–4264. doi:10.1093/hmg/ddq348
Merson, T. D., Dixon, M. P., Collin, C., Rietze, R. L., Bartlett, P. F., Thomas, T., et al. (2006). The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 26, 11359–11370. doi:10.1523/JNEUROSCI.2247-06.2006
Miyake, N., Mizuno, S., Okamoto, N., Ohashi, H., Shiina, M., Ogata, K., et al. (2013). KDM6A point mutations cause Kabuki syndrome. Hum. Mutat. 34, 108–110. doi:10.1002/humu.22229
Moore, S. M., Seidman, J. S., Ellegood, J., Gao, R., Savchenko, A., Troutman, T. D., et al. (2019). Setd5 haploinsufficiency alters neuronal network connectivity and leads to autistic-like behaviors in mice. Transl. Psychiatry 9, 24. doi:10.1038/s41398-018-0344-y
Moulton, M. J., and Letsou, A. (2016). Modeling congenital disease and inborn errors of development in Drosophila melanogaster. Dis. Model. Mech. 9, 253–269. doi:10.1242/dmm.023564
Negri, G., Magini, P., Milani, D., Crippa, M., Biamino, E., Piccione, M., et al. (2019). Exploring by whole exome sequencing patients with initial diagnosis of rubinstein–taybi syndrome: The interconnections of epigenetic machinery disorders. Hum. Genet. 138, 257–269. doi:10.1007/s00439-019-01985-y
Oike, Y., Hata, A., Mamiya, T., Kaname, T., Noda, Y., Suzuki, M., et al. (1999). Truncated CBP protein leads to classical rubinstein-taybi syndrome phenotypes in mice: Implications for a dominant-negative mechanism. Hum. Mol. Genet. 8, 387–396. doi:10.1093/HMG/8.3.387
O’Rawe, J. A., Wu, Y., Dörfel, M. J., Rope, A. F., Au, P. Y. B., Parboosingh, J. S., et al. (2015). TAF1 variants are associated with dysmorphic features, intellectual disability, and neurological manifestations. Am. J. Hum. Genet. 97, 922–932. doi:10.1016/j.ajhg.2015.11.005
Osipovich, A. B., Gangula, R., Vianna, P. G., and Magnuson, M. A. (2016). Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development 143, 4595–4607. doi:10.1242/dev.141465
Parenti, I., Teresa-Rodrigo, M. E., Pozojevic, J., Ruiz Gil, S., Bader, I., Braunholz, D., et al. (2017). Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum. Genet. 136, 307–320. doi:10.1007/S00439-017-1758-Y
Peled, J. U., Sellers, R. S., Iglesias-Ussel, M. D., Shin, D. M., Montagna, C., Zhao, C., et al. (2010). Msh6 protects mature B cells from lymphoma by preserving genomic stability. Am. J. Pathol. 177, 2597–2608. doi:10.2353/ajpath.2010.100234
Powis, Z., Farwell Hagman, K. D., Mroske, C., McWalter, K., Cohen, J. S., Colombo, R., et al. (2018). Expansion and further delineation of the SETD5 phenotype leading to global developmental delay, variable dysmorphic features, and reduced penetrance. Clin. Genet. 93, 752–761. doi:10.1111/cge.13132
Rickels, R., Herz, H. M., Sze, C. C., Cao, K., Morgan, M. A., Collings, C. K., et al. (2017). Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653. doi:10.1038/ng.3965
Ruhf, M. L., Braun, A., Papoulas, O., Tamkun, J. W., Randsholt, N., and Meister, M. (2001). The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development 128, 1429–1441. doi:10.1242/dev.128.8.1429
Russell, B., Tan, W.-H., and Graham, J. M. (2018). “Bohring-opitz syndrome,” in GeneReviews (qeios). doi:10.32388/uq2hhr
Santoriello, C., and Zon, L. I. (2012). Hooked! modeling human disease in zebrafish. J. Clin. Invest. 122, 2337–2343. doi:10.1172/JCI60434
Scherer, S., Theile, U., Beyer, V., Ferrari, R., Kreck, C., and Rister, M. (2003). Patient with Kabuki syndrome and acute leukemia. Am. J. Med. Genet. A 122A, 76–79. doi:10.1002/AJMG.A.20261
Selicorni, A., Mariani, M., Lettieri, A., and Massa, V. (2021). Cornelia de Lange syndrome: From a disease to a broader spectrum. Genes. 12, 1075. doi:10.3390/GENES12071075
Seo, S., Richardson, G. A., and Kroll, K. L. (2005). The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 132, 105–115. doi:10.1242/dev.01548
Sessa, A., Fagnocchi, L., Mastrototaro, G., Massimino, L., Zaghi, M., Indrigo, M., et al. (2019). SETD5 regulates chromatin methylation state and preserves global transcriptional fidelity during brain development and neuronal wiring. Neuron 104, 271–289. e13. doi:10.1016/j.neuron.2019.07.013
Sethi, S., Lin, H. H., Shepherd, A. K., Volkan, P. C., Su, C. Y., and Wang, J. W. (2019). Social context enhances hormonal modulation of pheromone detection in Drosophila. Curr. Biol. 29, 3887–3898. e4. doi:10.1016/j.cub.2019.09.045
Shah, M. Y., Vasanthakumar, A., Barnes, N. Y., Figueroa, M. E., Kamp, A., Hendrick, C., et al. (2010). DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 70, 5840–5850. doi:10.1158/0008-5472.CAN-10-0847
Shen, T., Lee, A., Shen, C., and Lin, C. J. (2015). The long tail and rare disease research: The impact of next-generation sequencing for rare Mendelian disorders. Genet. Res. 97, e15. doi:10.1017/S0016672315000166
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964. doi:10.1371/journal.pgen.1002964
Shpargel, K. B., Starmer, J., Wang, C., Ge, K., and Magnuson, T. (2017). UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc. Natl. Acad. Sci. U. S. A. 114, E9046–E9055. doi:10.1073/pnas.1705011114
Sinclair, D. A., Campbell, R. B., Nicholls, F., Slade, E., and Brock, H. W. (1992). Genetic analysis of the additional sex combs locus of Drosophila melanogaster. Genetics 130, 817–825. doi:10.1093/genetics/130.4.817
Sinclair, D. A., Milne, T. A., Hodgson, J. W., Shellard, J., Salinas, C. A., Kyba, M., et al. (1998). The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development 125, 1207–1216. doi:10.1242/dev.125.7.1207
Squeo, G. M., Augello, B., Massa, V., Milani, D., Colombo, E. A., Mazza, T., et al. (2020). Customised next-generation sequencing multigene panel to screen a large cohort of individuals with chromatin-related disorder. J. Med. Genet. 57, 760–768. doi:10.1136/jmedgenet-2019-106724
Sun, X., Chen, J., Zhang, Y., Munisha, M., Dougan, S., and Sun, Y. (2018). Mga modulates Bmpr1a activity by antagonizing Bs69 in zebrafish. Front. Cell. Dev. Biol. 6, 126. doi:10.3389/fcell.2018.00126
Tachibana, M., Ueda, J., Fukuda, M., Takeda, N., Ohta, T., Iwanari, H., et al. (2005). Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes. Dev. 19, 815–826. doi:10.1101/gad.1284005
Takeuchi, J. K., Lou, X., Alexander, J. M., Sugizaki, H., Delgado-OlguÃ- N, P., Holloway, A. K., et al. (2011). Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2, 187. doi:10.1038/ncomms1187
Tanaka, Y., Naruse, I., Maekawa, T., Masuya, H., Shiroishi, T., and Ishii, S. (1997). Abnormal skeletal patterning in embryos lacking a single cbp allele: A partial similarity with rubinstein-taybi syndrome. Proc. Natl. Acad. Sci. U. S. A. 94, 10215–10220. doi:10.1073/pnas.94.19.10215
Tang, H., Guo, J., Linpeng, S., and Wu, L. (2019). Next generation sequencing identified two novel mutations in NIPBL and a frame shift mutation in CREBBP in three Chinese children. Orphanet J. Rare Dis. 14, 45. doi:10.1186/S13023-019-1022-8
Thieme, S., Gyárfás, T., Richter, C., Özhan, G., Fu, J., Alexopoulou, D., et al. (2013). The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 121, 2462–2473. doi:10.1182/blood-2012-08-452003
Thomas, T., Voss, A. K., Chowdhury, K., and Gruss, P. (2000). Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 127, 2537–2548. doi:10.1242/dev.127.12.2537
Tsai, I. C., McKnight, K., McKinstry, S. U., Maynard, A. T., Tan, P. L., Golzio, C., et al. (2018). Small molecule inhibition of RAS/MAPK signaling ameliorates developmental pathologies of Kabuki Syndrome. Sci. Rep. 8, 10779. doi:10.1038/s41598-018-28709-y
Van Laarhoven, P. M., Neitzel, L. R., Quintana, A. M., Geiger, E. A., Zackai, E. H., Clouthier, D. E., et al. (2015). Kabuki syndrome genes KMT2D and KDM6A: Functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24, 4443–4453. doi:10.1093/hmg/ddv180
van Ravenswaaij-Arts, C., and Martin, D. M. (2017). New insights and advances in CHARGE syndrome: Diagnosis, etiologies, treatments, and research discoveries. Am. J. Med. Genet. C Semin. Med. Genet. 175, 397–406. doi:10.1002/AJMG.C.31592
Vega, R. B., Matsuda, K., Oh, J., Barbosa, A. C., Yang, X., Meadows, E., et al. (2004). Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 119, 555–566. doi:10.1016/j.cell.2004.10.024
Viosca, J., Lopez-Atalaya, J. P., Olivares, R., Eckner, R., and Barco, A. (2010). Syndromic features and mild cognitive impairment in mice with genetic reduction on p300 activity: Differential contribution of p300 and CBP to Rubinstein-Taybi syndrome etiology. Neurobiol. Dis. 37, 186–194. doi:10.1016/j.nbd.2009.10.001
Wan, X., Hu, B., Liu, J. X., Feng, X., and Xiao, W. (2011). Zebrafish mll gene is essential for hematopoiesis. J. Biol. Chem. 286, 33345–33357. doi:10.1074/JBC.M111.253252
Wang, J., Li, Z., He, Y., Pan, F., Chen, S., Rhodes, S., et al. (2014). Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood 123, 541–553. doi:10.1182/blood-2013-05-500272
Wang, Y. R., Xu, N. X., Wang, J., and Wang, X. M. (2019). Kabuki syndrome: Review of the clinical features, diagnosis and epigenetic mechanisms. World J. Pediatr. 15, 528–535. doi:10.1007/S12519-019-00309-4
Wassarman, D. A., Aoyagi, N., Pile, L. A., and Schlag, E. M. (2000). TAF250 is required for multiple developmental events in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 97, 1154–1159. doi:10.1073/pnas.97.3.1154
Welstead, G. G., Creyghton, M. P., Bilodeau, S., Cheng, A. W., Markoulaki, S., Young, R. A., et al. (2012). X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. U. S. A. 109, 13004–13009. doi:10.1073/pnas.1210787109
Willemsen, M. H., Vulto-Van Silfhout, A. T., Nillesen, W. M., Wissink-Lindhout, W. M., Van Bokhoven, H., Philip, N., et al. (2012). Update on Kleefstra syndrome. Mol. Syndromol. 2, 202–212. doi:10.1159/000335648
Wood, M. A., Attner, M. A., Oliveira, A. M. M., Brindle, P. K., and Abel, T. (2006). A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn. Mem. 13, 609–617. doi:10.1101/lm.213906
Wood, M. A., Kaplan, M. P., Park, A., Blanchard, E. J., Oliveira, A. M. M., Lombardi, T. L., et al. (2005). Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 12, 111–119. doi:10.1101/lm.86605
Woods, S. A., Robinson, H. B., Kohler, L. J., Agamanolis, D., Sterbenz, G., and Khalifa, M. (2014). Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am. J. Med. Genet. A 164A, 251–258. doi:10.1002/AJMG.A.36237
Yamamoto, P. K., de Souza, T. A., Antiorio, A. T. F. B., Zanatto, D. A., Garcia-Gomes, M. de S. A., Alexandre-Ribeiro, S. R., et al. (2019). Genetic and behavioral characterization of a Kmt2d mouse mutant, a new model for Kabuki Syndrome. Genes., Brain Behav. 18, e12568. doi:10.1111/gbb.12568
Yamauchi, T., Oike, Y., Kamon, J., Waki, H., Komeda, K., Tsuchida, A., et al. (2002). Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat. Genet. 30, 221–226. doi:10.1038/ng829
Yao, T. P., Oh, S. P., Fuchs, M., Zhou, N. D., Ch’ng, L. E., Newsome, D., et al. (1998). Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 93, 361–372. doi:10.1016/S0092-8674(00)81165-4
Yu, B. D., Hess, J. L., Horning, S. E., Brown, G. A. J., and Korsmeyer, S. J. (1995). Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508. doi:10.1038/378505A0
Yuan, B., Pehlivan, D., Karaca, E., Patel, N., Charng, W. L., Gambin, T., et al. (2015). Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Invest. 125, 636–651. doi:10.1172/JCI77435
Keywords: chromatinopathies, animal models, rare diseases, mus musculus, drosophila melanogaster, Danio rerio
Citation: Di Fede E, Grazioli P, Lettieri A, Parodi C, Castiglioni S, Taci E, Colombo EA, Ancona S, Priori A, Gervasini C and Massa V (2022) Epigenetic disorders: Lessons from the animals–animal models in chromatinopathies. Front. Cell Dev. Biol. 10:979512. doi: 10.3389/fcell.2022.979512
Received: 27 June 2022; Accepted: 05 September 2022;
Published: 26 September 2022.
Edited by:
Ronja Hollstein, University Hospital Bonn, GermanyReviewed by:
Binnaz Yalcin, Institut National de la Santé et de la Recherche Médicale, FranceVanja Nagy, Ludwig Boltzmann Institute for Rare and Undiagnosed Diseases, Austria
Copyright © 2022 Di Fede, Grazioli, Lettieri, Parodi, Castiglioni, Taci, Colombo, Ancona, Priori, Gervasini and Massa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valentina Massa, dmFsZW50aW5hLm1hc3NhQHVuaW1pLml0
§Present address: Chiara Parodi, EHS Department, Columbia University, New York, NY, United States
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Elisabetta Di Fede
Elisabetta Di Fede Paolo Grazioli
Paolo Grazioli Antonella Lettieri
Antonella Lettieri Chiara Parodi
Chiara Parodi Silvia Castiglioni
Silvia Castiglioni Esi Taci
Esi Taci Elisa Adele Colombo
Elisa Adele Colombo Silvia Ancona
Silvia Ancona Alberto Priori
Alberto Priori Cristina Gervasini
Cristina Gervasini Valentina Massa
Valentina Massa