
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 09 August 2022
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.967147
This article is part of the Research Topic iPS Cell Models of Human Single Gene Inheritance Diseases View all 4 articles
 Steven Paniagua1
Steven Paniagua1 Bilal Cakir1
Bilal Cakir1 Yue Hu2
Yue Hu2 Ferdi Ridvan Kiral1
Ferdi Ridvan Kiral1 Yoshiaki Tanaka1,3
Yoshiaki Tanaka1,3 Yangfei Xiang4
Yangfei Xiang4 Benjamin Patterson1
Benjamin Patterson1 Jeffrey R. Gruen5*
Jeffrey R. Gruen5* In-Hyun Park1*
In-Hyun Park1*Dyslexia, also known as reading disability, is defined as difficulty processing written language in individuals with normal intellectual capacity and educational opportunity. The prevalence of dyslexia is between 5 and 17%, and the heritability ranges from 44 to 75%. Genetic linkage analysis and association studies have identified several genes and regulatory elements linked to dyslexia and reading ability. However, their functions and molecular mechanisms are not well understood. Prominent among these is KIAA0319, encoded in the DYX2 locus of human chromosome 6p22. The association of KIAA0319 with reading performance has been replicated in independent studies and different languages. Rodent models suggest that kiaa0319 is involved in neuronal migration, but its role throughout the cortical development is largely unknown. In order to define the function of KIAA0319 in human cortical development, we applied the neural developmental model of a human embryonic stem cell. We knocked down KIAA0319 expression in hESCs and performed the cortical neuroectodermal differentiation. We found that neuroepithelial cell differentiation is one of the first stages of hESC differentiation that are affected by KIAA0319 knocked down could affect radial migration and thus differentiation into diverse neural populations at the cortical layers.
Reading is a complex trait influenced by both environmental and genetic factors. Dyslexia, also known as reading disability, is characterized by reading difficulty in the setting of normal intelligence and adequate education. Dyslexia is associated with long-term detrimental effects on educational achievement and socioeconomic status (Schatschneider and Torgesen, 2004). It is the most common learning disability, with a prevalence of 5–17% and a heritability of 0.34–0.76 (DeFries et al., 1987; Shaywitz and Shaywitz, 2005; Hawke et al., 2006). Genetic studies have identified nine dyslexia loci (DYX1-9), contingent on aspects of reading performance (Gayan et al., 1999; Fisher and DeFries, 2002). The most frequently replicated locus is DYX2 on chromosome 6p21.3, and the most frequently replicated dyslexia genes on DYX2 are KIAA0319 and DCDC2 (Deffenbacher et al., 2004; Francks et al., 2004; Meng et al., 2005; Brkanac et al., 2007; Zhao et al., 2016).
Animal models show that kiaa319 interacts with intracellular trafficking protein AP-2. Additionally, KIAA0319 undergoes N- and O- glycosylation, associated with the plasma membrane, suggesting that KIAA0319 plays a role in cell maintenance and cell-cell signaling in the endocytosis pathway (Velayos-Baeza et al., 2008; Levecque et al., 2009). Kiaa0319 is also associated with extracellular signaling pathways (Franquinho et al., 2017; Wu et al., 2020), via Smad2/3. Both endocytosis and extracellular signaling pathways are essential for regulating neurogenesis (Cope and Gould, 2019).
Several dyslexia genes, including kiaa0319 and dcdc2, affect neuronal radial migration during rat embryogenesis (Peschansky et al., 2010). Radial migration begins at the neuroepithelial cell stage of neurogenesis. Individual cortical layers are then populated by neuronal progenitor cells (Stockinger et al., 2011; Ji et al., 2017). Neuronal progenitor cells are differentiated into neurons, astrocytes, and oligodendrocytes, providing the diversity in neuronal subtypes essential for neural circuitry construction, maintenance, and function (Gotz and Huttner, 2005; Klausberger and Somogyi, 2008; Bergles and Richardson, 2015; Molofsky and Deneen, 2015; Durkee and Araque, 2019). Regulation of neuronal progenitor cell differentiation in the dentate gyrus of the hippocampus is associated with learning and memory and critical for propagating new neurons in the adult brain (Cameron and Glover, 2015).
While animal models provide insights into some of the cellular functions of dyslexia genes (Paracchini et al., 2006; Szalkowski et al., 2012), they do not account for the complexity of reading or the critical skills that subserve reading, such as decoding, phonological awareness, or orthographic coding. Modeling genetic variants associated with specific human neurons and tissues could provide a way to reference findings to brain regions critical to reading (Powers et al., 2016; Fitch, 2017; Kirby, 2017).
Gene editing approaches in pluripotent cells, such as human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), have been successfully used to model the effects of genetic variants on neuronal differentiation (Wang et al., 2017; Xiang et al., 2020). Pluripotent stem cells can be induced to differentiate into many cell types, including glutamatergic neurons, GABAergic neurons, and glial cells that populate the human brain (Damdimopoulou et al., 2016). Neuroectodermal differentiation of hESCs can reproduce developmental stages during embryonic brain development and gene expression patterns (Howard et al., 2008). Combined with gene editing tools, hESCs provide powerful experimental systems for dissecting the function of genes associated with neurodevelopmental or neuropsychiatric disorders (Kolagar et al., 2020).
The KIAA0319 variants associated with dyslexia are at the promoter region and thus the cell type and the temporal regulation of KIAA0319 expression is perhaps a major cause of the dyslexia. Here, as one of the first attempts to understand how KIAA0319 regulates the human cortical development, we set out to investigate the function of KIAA0319 in the context of the loss of function. We used a hESC model of cortical neural differentiation and probe the function of KIAA0319 during neurogenesis. We knocked down KIAA0319 in hESCs and performed neuroectodermal differentiation to examine how the neural differentiation is affected by loss of function of KIAA0319 throughout the neuronal development. We found that KIAA0319 is critical in neuroepithelial cell differentiation, which could affect radial migration and further differentiation into diverse populations of brain cells. These results suggest that one of the mechanisms whereby dysregulated expression of KIAA0319 could influence human cortical neurodevelopmental process is the neuroepithelial differentiation, which could underlie the complex traits such as reading.
To determine how KIAA0319 gene expression changes, we first examined transcriptome profiles of somatic tissues during human fetal development. KIAA0319 is strongly expressed throughout human cortical development and in all brain regions (Supplementary Figures S1A–D), (Pletikos et al., 2014; Cardoso-Moreira et al., 2019).
To study the potential effects of KIAA0319 on neurogenesis, we knocked down (KD) expression using CRISPRi/dCas9-KRAB in H7dCas9−KRAB-b6 hESCs (Figure 1A) targetting the transcription start site. Both qRT-PCR and immunofluorescence confirmed decreased expression of KIAA0319 in CRISPRi cells compared to controls during neuronal differentiation at days 7, 14, 21, and 28 (Figures 1B–D). We then assessed the expression of genes representative of transitional stages during neuronal differentiation by qRT-PCR and immunostaining on days 7, 14, 21, and 28 of cortical differentiation. We focused on neuronal stage lineage markers SOX10, PAX6, TBR2, TBR1, and MAP2 to quantitate the population of neuroepithelial, neuronal progenitor, immature neuron, and mature neurons, respectively. Compared to controls, SOX10 expression by qRT-PCR and immunofluorescence, a marker of neuroepithelial cells, was suppressed at day 21 in KIAA0319 KD cells (Figures 2A–C,E). Conversely, PAX6 expression by qRT-PCR and immunofluorescence, a marker of neuronal progenitor cells, increased in KIAA0319 KD cells on days 14 and 21 (Figures 2D–F). Together, these results suggest that KIAA0319 is critical for regulating the transition of SOX10 + neuroepithelial cells to PAX6+ neuronal progenitor cells.
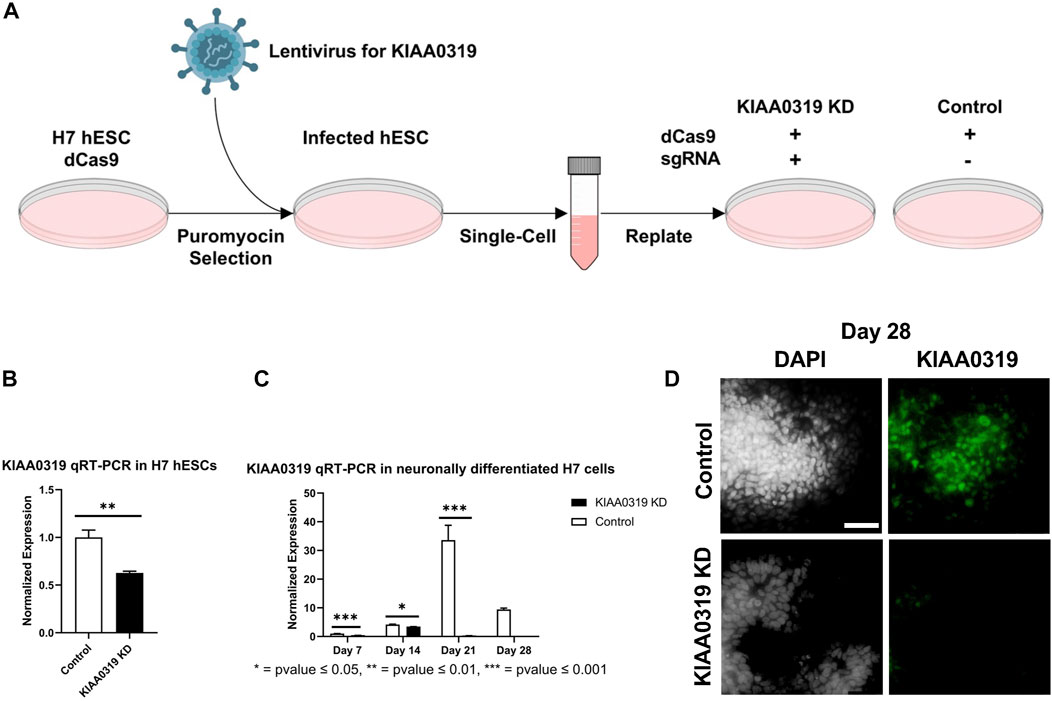
FIGURE 1. Depletion of KIAA0319 by CRISPR interference (CRISPRi) (A) Workflow for generating KIAA0319 KD using CRISPRi in hESCs. (B) Confirmation of KIAA0319 KD by qRT-PCR in H7 cells after transfection and puromycin selection. (C) Confirmation of KIAA0319 KD by qRT-PCR on days 7, 14, 21, and 28 of neuronal differentiation in H7 cells by qRT-PCR in neuronally differentiated H7 cells. (D) Immunofluorescence images of KIAA0319 KD and control on day 28. Green: KIAA0319, grey: DAPI. Scale bar: 50 µm.
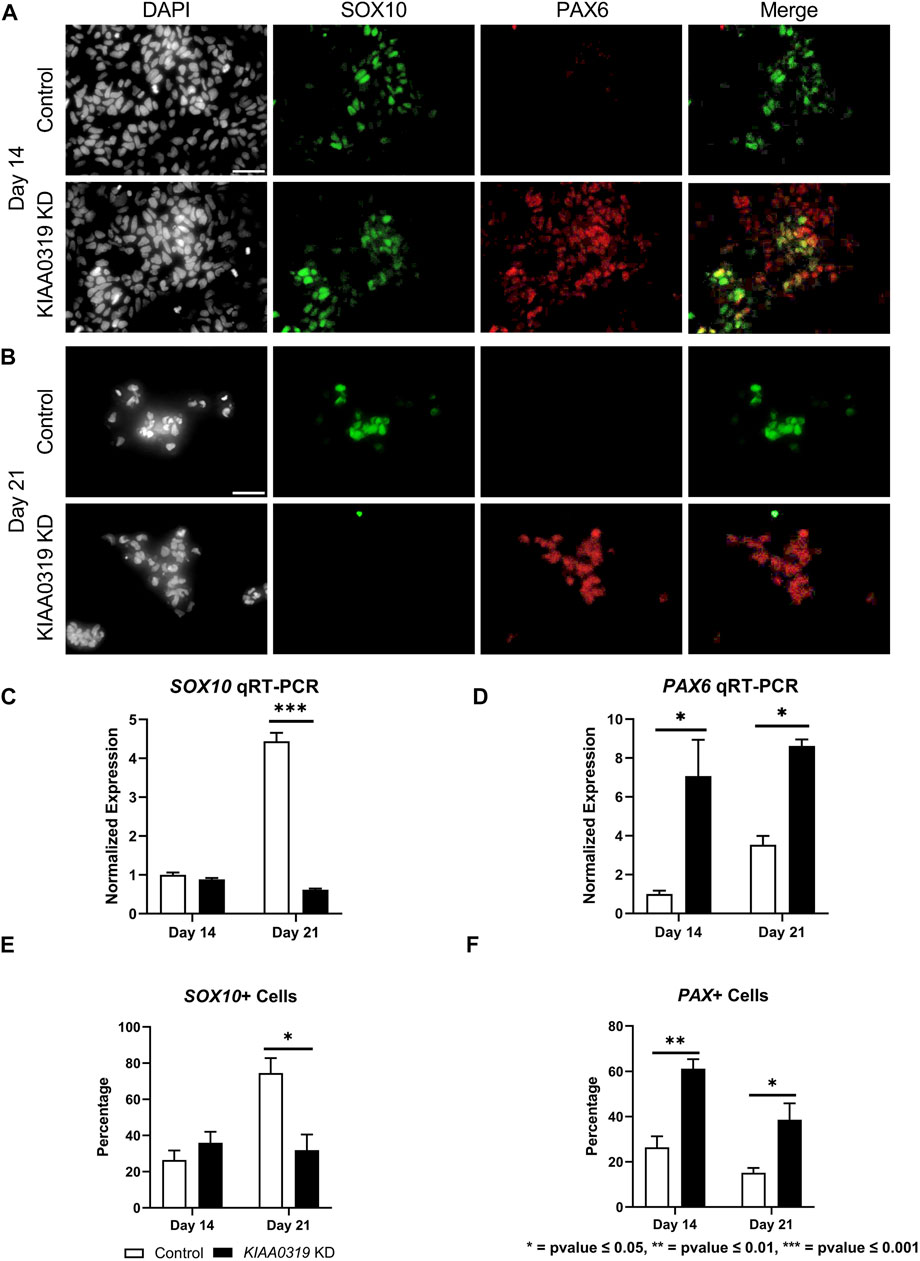
FIGURE 2. KIAA0319 KD decreases expression of neuronal epithelial marker SOX10 but increases neuronal progenitor marker PAX6. (A) After 14 days of neuronal differentiation, immunofluorescence was performed for SOX10 (green), PAX6 (red), and DAPI (grey). (B) After 21 days of neuronal differentiation, immunofluorescence was performed for SOX10 (green), PAX6 (red), and DAPI (grey). qRT-PCR for SOX10 (C) and PAX 6 (D) were performed on days 14 and 21 of KIAA0319 KD and Control H7 cells. The percentage of the population of SOX10+ (E) or PAX6+ (F) cells were calculated on day 14 and day 21 of differentiation for KIAA0319 KD and Control H7 cells.
Next, we characterized how KIAA0319 regulates PAX6+ neuronal progenitor cells at later stages of differentiation. We simultaneously measured KI67, a marker for proliferation, and PAX6 by qRT-PCR and immunofluorescence, and quantified the proliferation of neuronal progenitor cells at two later time points of differentiation (days 28 and 42) (Figures 3A,B). We found that the fraction of KI67 + cells remained relatively low in KIAA0319 KD cells compared to controls (Figure 3C), while the fraction of PAX6+ cells was relatively the same (Figure 3D). However, the fraction of KI67 + cells among PAX6+ cells decreased, suggesting an overall decrease in the population of proliferating neuronal progenitor cells (Figure 3E). Overall, these results suggest that KIAA0319 KD could be driving PAX6+ neuronal progenitor cells into cell cycle arrest, inducing apoptosis in the proliferating cells, or prematurely inducing transition into downstream stages of neuronal differentiation.
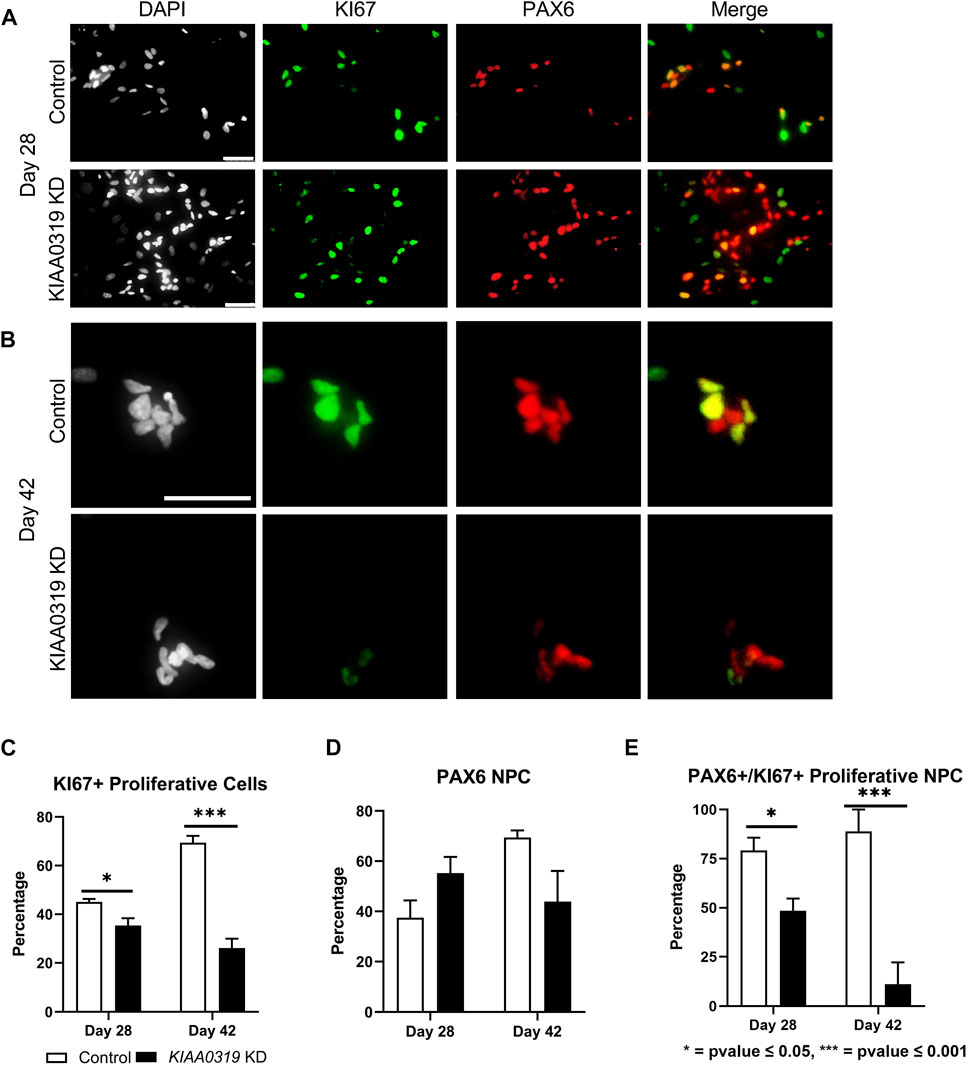
FIGURE 3. KIAA0319 KD decreases the percentage of proliferating neuronal progenitor cells. (A–B) Immunostaining of KIAA0319 KD and control cells for KI67 (green) and PAX6 (red) in Dapi (gray) on day 28 (A) and day 42 (B) of neuronal differentiation. Scale bar: 50 µm. Percentage of KI67 + cells (C) and PAX6+ population (D) among DAPI + cells at day 28 and day 42. Percentage of PAX6+ cells among KI67 + population at day 28 and day 42 (E).
To assess whether the decrease in KI67 + PAX6+ neuronal progenitor cells was due to apoptosis, we stained KD and control cells with cleaved caspase-3 antibody, a marker of cell death induction, at day 28 (Figure 4A). KIAA0319 KD did not induce apoptosis, suggesting that the decrease in KI67 + PAX6+ neuronal progenitor cells was not due to dying cells. Next, we examined markers at later stages of neuronal differentiation to quantify if KIAA0319 KD induces the premature transition from neuronal progenitor cells into downstream stages of neuronal differentiation. Compared to controls at day 28, KIAA0319 KD significantly decreased the expression of markers of intermediate progenitor cells (TBR2), immature neuronal cells (TBR1 and β-Tubulin), and mature neuronal cells (MAP2) (Figure 4B). The results of these experiments show that KIAA0319 KD arrests cells in a non-proliferative neuronal progenitor cell stage, and that KIAA0319 is important for transitioning neuronal epithelial cells into and through the neuronal progenitor cell stage and to later stages of neuronal differentiation.
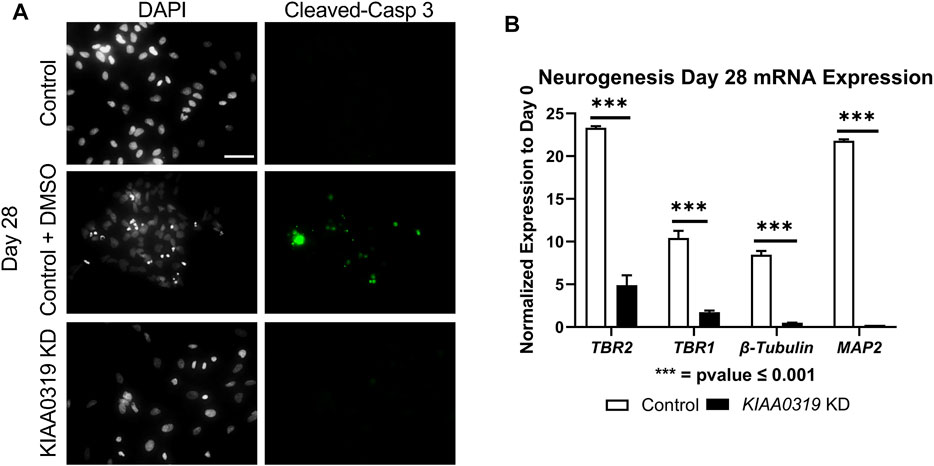
FIGURE 4. KIAA0319 KD cells do not continue differentiating. (A) Images of KIAA0319 KD, Control, Control + DMSO (Positive Apoptosis Control) at day 28 of neuronal differentiation stained with cleaved-caspase 3 (green) and DAPI (grey). (B) QRT-PCR at day 28 for neuronal lineage markers TBR1, TBR2, β-Tubulin, and MAP2.
To examine how KIAA0319 regulates proliferation in neuronal progenitor cells, we performed RNA-sequencing on the differentiating cells from day 7, 14, 21, and 28 (Figure 5A). Ingenuity Pathway Analysis (IPA) showed that endocytosis, exocytosis, lysosome, differentiation, proliferation, and metabolism are affected by the KIAA0319 KD (Figure 5B), which were previously implicated in cell-cycle arrest of neuronal progenitor cells (Kobayashi et al., 2019; Harada et al., 2021; Zhang et al., 2021). Individual gene analysis showed a consistent decrease in expression of genes associated with differentiation at neuroepithelial and neuronal progenitor cell stages (Figure 5C). The changes in expression of genes associated with extracellular matrix, metabolism, cell maintenance, and cell-cell signaling further support the findings that KIAA0319 KD drives neuronal progenitors into cell cycle arrest.
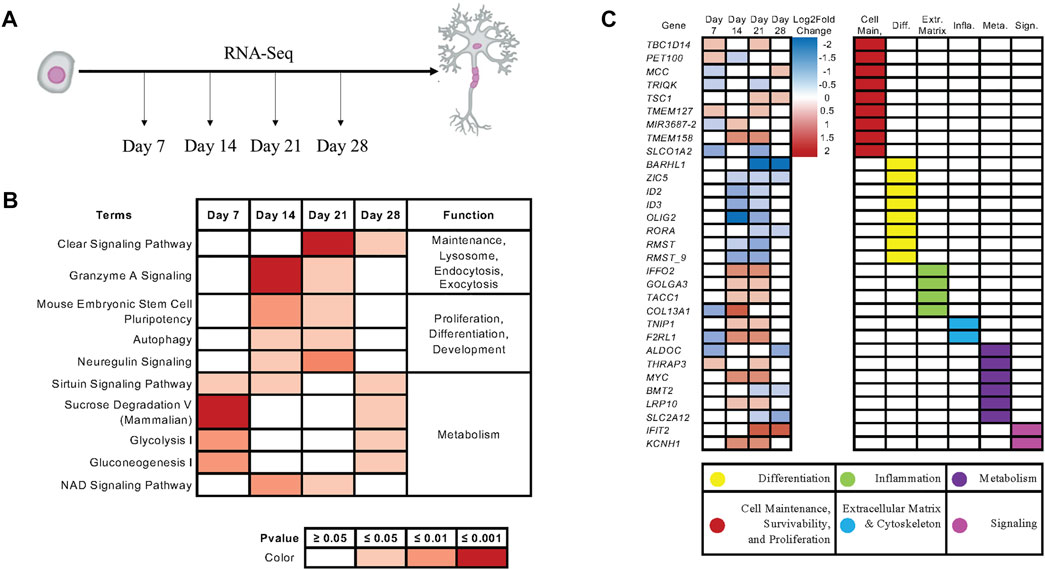
FIGURE 5. KIAA0319 KD RNA-Seq shows pathways that promote cell cycle arrest of neuronal progenitor cells. (A) RNA Sequencing performed at four distinct timepoints separated by 7-day intervals during cortical neuronal differentiation. (B) Ingenuity Pathway Analysis revealed enrichment of pathways appearing in multiple time points. (C) Ingenuity Pathway Analysis revealed enrichment of genes appearing in multiple time points.
Here, we tested our hypothesis that the clinical association of KIAA0319 with reading performance and dyslexia is a consequence of altered expression during early stages of neurogenesis. KIAA0319 KD showed a decrease in the SOX10 + neuroepithelial cells concurrent with an increase in the PAX6 neuronal progenitors and suppression of ensuing neurogenesis. These findings are consistent with a decreased proliferative state at the neuronal progenitor cells due to cell cycle arrest, highlighting the importance of tight regulation of KIAA0319 expression during a critical period of brain development.
Decreased expression of KIAA0319 appears to promote early exit from the neuroepithelial cell stage and entry into the neuronal progenitor cell stage. This suggests that KIAA0319 plays a critical role as an upstream gatekeeper for neurogenesis, slowing or enhancing downstream differentiation by arresting cells at the neuronal progenitor cell stage. Transcriptome analysis suggests that KIAA0319 could regulate cell proliferation through endocytosis-related pathways. Indeed, dysregulation of the endocytosis pathways has been shown to drive neuronal progenitor cells into a stage of cell cycle arrest (Zhang et al., 2021). Our results provide a new avenue for studying the regulation of neurogenesis in human neurons.
This is the first study to suggest that KIAA0319, which has been associated with reading performance in multiple independent studies, may act by regulating neurogenesis at a critical checkpoint between neuroepithelial cells and proliferating active neuronal progenitor cells. The population of mature adult neurons is tightly regulated by the proliferation of active neuronal progenitors. Our results suggest that KIAA0319 is a critical check on this transition by either actively driving neuronal progenitors into cell cycle arrest, or by blocking release from cell cycle arrest into proliferating active cells and development into downstream lineages.
The KIAA0319 genetic variants associated with reading and dyslexia in clinical studies are in the promoter region and regulate expression. Spatial and temporal regulation of KIAA0319 expression that are affected by the variants could affect an isolated high-order trait such as reading without global effects on brain development or cognition; children and adults with dyslexia typically have normal brain development and cognition. While this study has a limitation in its design to reveal the function of the KIAA0319 variants in human cortical neurogenesis, it is one of the first studies to show the role of KIAA0319 in early neurogenesis in human cells. Future studies will be needed to define the function of the specific KIAA0319 genetic variants associated with dyslexia in the genesis of specific cell lineages and brain development. Use of the 3D cell culture brain organoid models and single cell-based assay, like scRNA-seq, or scATAC-seq will be critical in reveaingl the cell type specific function of the variants.
HEK 293T cells were maintained in 6-well plates at 37°C with 5%CO2. Cells were tested for mycoplasma and passaged every 3 days using Gibco TrypLE™ Express with Phenol Red for dissociation. Culture media consisted of 88% v/v DMEM, 10% v/v FBS, 1% v/v Glutamax, and 1% v/v Penicillin/Streptomycin. H7dCas9−KRAB-b6 (WiCell, WA07, H7) human ESCs were maintained in feeder-free culture conditions using Matrigel-coated cell culture dishes and mTeSR1 media at 37°C and 5% CO2. Cells were tested for mycoplasma and passaged every 7 days following dissociation with Dispase (0.83 U/ml). All experiments involving hESCs were approved by the Yale Embryonic Stem Cell Research Oversight Committee (ESCRO).
Pre-differentiated cells were removed before hESC colonies, dissociated into single cells using Accutase, and plated on 24-well plates at 500,000 cells/well with 50 µM Y27632 and mTeSR1. Once plates reached 90% confluency, cells were grown for 10 days with neural induction media (47.5% v/v; DMEM/F12, 47.5% v/v Neurobasal media, 2% v/v B27 supplement, 1% v/v DMEM-NEAA, 1% v/v N2 supplement, 1% v/v Glutamax, 20 μg/ml Insulin, 100 nM LDN-193189, 50 µM β-Mercaptoethanol, 10 μM SB431542, 2 µM XAV939), changing media every day. Cells were dissociated using Accutase on day 11 and plated on new 24-well plates along with 50 µM Y27632 and progenitor expansion media (neural induction media without inhibitors: LDN-193189, SB431542, and XAV939), changing media every other day. On day 19, culture media was changed to maturation media (95% v/v Neurobasal media, 2% v/v B27 supplement, 1% v/v N2 Supplement, 1% v/v Glutamax, 1% v/v Penicillin/Streptomycin) with 25 ng/ml BDNF, changing media every 2 days for 2 weeks and every 4 days thereafter. Cells were dissociated using Accutase and replated on new 24-well plate on day 25 at 100,000 cells/well when confluency was reached.
Single Guide RNA (sgRNA) and reverse complement were designed using CRISPR PAM sites (crispr.mit.edu) targeting the transcription start site of KIAA0319 (KIAA0319 KD plasmid) and synthesized by Keck (sgRNA KIAA0319 KD and Control listed in Table 1). Oligos were annealed using 50% v/v NEB Annealing Buffer, 46% v/v ddH20, 2% v/v top oligo, and 2% v/v bottom oligo (Oligos listed in Table 1). Oligos were incubated at 95 °C and left to cool to room temperature. Plasmid pCRISPia-V2, encoding a Puromycin resistance gene, had a sgRNA insertion site opened using digestion enzymes BstXI and Blpl. Ligation reactions consisted of 70% v/v ddH2O, 10% v/v of 10X T4 ligase buffer, 10% v/v 1:20 diluted annealed oligos, 5% pCRISPia-V2 (totaling 100 ng of digested plasmid), and 5% v/v T4 Ligase at room temperature for 1 h.
Ligation reactions were transformed into One Shot™ TOP10 Chemically Competent cells following manufactures instructions and plated on 10 cm LB Agar + Ampicillin (100ug/ml) plates from Recombinant Technologies, LLC at 37°C for culture overnight. Surviving colonies were cultured overnight in Gibco LB Broth with Ampicillin at 100ug/ml in a 37°C shaker, followed by plasmid purification using QIAGEN Plasmid Mini Kit and sequence confirmation.
KIAA0319 KD and Control plasmids were individually transfected into HEK293T cells along with 2.5 ugs of packaging plasmids using X-treme GENE nine DNA transfection reagent following manufacturer’s instructions. Supernatants containing lentivirus were collected and concentrated 48 h after transfection and stored at −80°C. H7 cells were infected with KIAA0319 KD lentivirus and control lentivirus. Infected cultures were selected following puromycin selection for 5 days.
Four wells of a 24-well plate of H7 cells transfected with KIAA0319 KD or control samples were collected on days 7, 14, 21, and 28 of neuronal differentiation per replicate. Samples were homogenized using QIAGEN QIAshredder columns, and total RNA was isolated using the RNeasy Mini Kit following the manufacturer’s instructions. One ug of total RNA was used to synthesize cDNA using amfiRivert cDNA Synthesis Platinum Master Mix. Real-time quantitative PCR (RT-PCR) was performed using amfiSure qGreen Q-PCR Master Mix (2X), Low Rox on a CFX96 R-PCR System at amfiSure manufacture cycling conditions with primers listed in Table 2. Gene expression was quantified using the ∆∆CT method with GAPDH as a housekeeping gene control. t-test was performed for the statistical analysis. p values are given at each figure.
Twelve wells of a 24-well plate of KIAA0319 KD or Control cells were washed once in PBS before fixing in 4% formaldehyde/PBS at room temperature for 15 min and then washed in PBS (3 × for 15 min). In addition, samples were permeabilized by incubating with 0.1% Triton-100/PBS at room temperature for 1 hour and then washed in PBS (3 x for 15 min) before storing at 4°C in PBS.
Blocking was done with 3% BSA/0.1% Triton-100/PBS at 4°C for 2 hours before incubating with primary antibodies diluted to the manufacturer’s recommendation in 3% BSA/0.1% Triton-100/PBS at 4°C overnight. Samples were then washed in PBS (3 x for 15 min) before incubating with secondary antibody diluted in 3% BSA/0.1% Triton-100/PBS at room temperature for 1 h. Samples were washed in PBS (3 x for 15 min) before staining with DAPI to highlight nuclei for 5 minutes before final PBS wash. Immunofluorescence images were captured on a Leica TCS SP5 Spectral Confocal Microscope using Leica LAS AF software. Images were processed using ImageJ-Fiji.
Cell pellets from three biological replicates of KIAA0319 KD and controls for days 7, 14, 21, and 28 were collected. RNA were isolated from cell pellets, processed for library prep, and then subjected to 150bp paired-end sequencing on a NOVA-seq for 50 million reads per sample at the Yale Center for Genomic Analysis (YCGA).
FASTQ files were processed using an RNA-Sequencing pipeline developed at the YCGA. Raw RNA-Sequencing reads were trimmed and aligned using Hierarchical Indexing for Spliced Alignment of Transcripts 2 (HISAT2) (Kim et al., 2019). Aligned reads are then concatenated using StringTie/Ballgown, followed by quality control using Picard (Frazee et al., 2015; Pertea et al., 2015). R package DESeq2 was used to generate summary reports and heatmaps using default setting (Love et al., 2014) (Supplementary Figures S2–S5).
Ingenuity Pathway Analysis (QIAGEN IPA, QIAGEN Inc, content version: 70750971) (Krämer et al., 2014) was used for pathway bioinformatics analysis. Significant genes that expressed differently in KD and control groups were identified by DESeq2 using Benjamini–Hochberg correction to guarantee a false discovery rate (FDR) ≤ 0.05. IPA performed a core analysis based on these significant genes and identified the significant pathways for each time point using a right-tailed Fisher’s exact test with a p-value threshold of 0.05. Finally, the pathways and genes were grouped by GO Term.
The data presented in this study are deposited at the BioProject repository accession number PRJNA854587. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SP designed the experiments, collected data, performed data analysis, and wrote manuscript. BC, YH, FK, YT, YX, and BP collected data. JG and I-HP conceived the idea, and designed the experiments. All authors contributed to the article and approved the submitted version.
I-HP was partly supported by NIH (R01MH118344-01A1, R01MH118554-01A1), CSCRF (16-RMB-YALE-04), Kavli Foundation, Simons Foundation, and Nomis foundation. Support for JRG was provided by the Manton Foundation, and the National Institutes of Health (R15 HD087937, P50 HD027802, R01 HG010171).
We thank the Park lab and the Gruen lab members for the helpful discussion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.967147/full#supplementary-material
Bergles, D. E., and Richardson, W. D. (2015). Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 8 (2), a020453. doi:10.1101/cshperspect.a020453
Brkanac, Z., Chapman, N. H., Matsushita, M. M., Chun, L., Nielsen, K., Cochrane, E., et al. (2007). Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B (4), 556–560. doi:10.1002/ajmg.b.30471
Cameron, H. A., and Glover, L. R. (2015). Adult neurogenesis: Beyond learning and memory. Annu. Rev. Psychol. 66, 53–81. doi:10.1146/annurev-psych-010814-015006
Cardoso-Moreira, M., Halbert, J., Valloton, D., Velten, B., Chen, C., Shao, Y., et al. (2019). Gene expression across mammalian organ development. Nature 571 (7766), 505–509. doi:10.1038/s41586-019-1338-5
Cope, E. C., and Gould, E. (2019). Adult neurogenesis, glia, and the extracellular matrix. Cell Stem Cell 24 (5), 690–705. doi:10.1016/j.stem.2019.03.023
Damdimopoulou, P., Rodin, S., Stenfelt, S., Antonsson, L., Tryggvason, K., Hovatta, O., et al. (2016). Human embryonic stem cells. Best. Pract. Res. Clin. Obstet. Gynaecol. 31, 2–12. doi:10.1016/j.bpobgyn.2015.08.010
Deffenbacher, K. E., Kenyon, J. B., Hoover, D. M., Olson, R. K., Pennington, B. F., DeFries, J. C., et al. (2004). Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: Linkage and association analyses. Hum. Genet. 115 (2), 128–138. doi:10.1007/s00439-004-1126-6
DeFries, J. C., Fulker, D. W., and LaBuda, M. C. (1987). Evidence for a genetic aetiology in reading disability of twins. Nature 329 (6139), 537–539. doi:10.1038/329537a0
Durkee, C. A., and Araque, A. (2019). Diversity and specificity of astrocyte-neuron communication. Neuroscience 396, 73–78. doi:10.1016/j.neuroscience.2018.11.010
Fisher, S. E., and DeFries, J. C. (2002). Developmental dyslexia: Genetic dissection of a complex cognitive trait. Nat. Rev. Neurosci. 3 (10), 767–780. doi:10.1038/nrn936
Fitch, W. T. (2017). Empirical approaches to the study of language evolution. Psychon. Bull. Rev. 24 (1), 3–33. doi:10.3758/s13423-017-1236-5
Francks, C., Paracchini, S., Smith, S. D., Richardson, A. J., Scerri, T. S., Cardon, L. R., et al. (2004). A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am. J. Hum. Genet. 75 (6), 1046–1058. doi:10.1086/426404
Franquinho, F., Nogueira-Rodrigues, J., Duarte, J. M., Esteves, S. S., Carter-Su, C., Monaco, A. P., et al. (2017). The dyslexia-susceptibility protein KIAA0319 inhibits axon growth through Smad2 signaling. Cereb. Cortex 27 (3), 1732–1747. doi:10.1093/cercor/bhx023
Frazee, A. C., Pertea, G., Jaffe, A. E., Langmead, B., Salzberg, S. L., Leek, J. T., et al. (2015). Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 33 (3), 243–246. doi:10.1038/nbt.3172
Gayan, J., Smith, S. D., Cherny, S. S., Cardon, L. R., Fulker, D. W., Brower, A. M., et al. (1999). Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am. J. Hum. Genet. 64 (1), 157–164. doi:10.1086/302191
Gotz, M., and Huttner, W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6 (10), 777–788. doi:10.1038/nrm1739
Harada, Y., Yamada, M., Imayoshi, I., Kageyama, R., Suzuki, Y., Kuniya, T., et al. (2021). Cell cycle arrest determines adult neural stem cell ontogeny by an embryonic Notch-nonoscillatory Hey1 module. Nat. Commun. 12 (1), 6562. doi:10.1038/s41467-021-26605-0
Hawke, J. L., Wadsworth, S. J., and DeFries, J. C. (2006). Genetic influences on reading difficulties in boys and girls: The Colorado twin study. Dyslexia 12 (1), 21–29. doi:10.1002/dys.301
Howard, B. M., Zhicheng, M., Filipovic, R., Moore, A. R., Antic, S. D., Zecevic, N., et al. (2008). Radial glia cells in the developing human brain. Neuroscientist 14 (5), 459–473. doi:10.1177/1073858407313512
Ji, L., Bishayee, K., Sadra, A., Choi, S., Choi, W., Moon, S., et al. (2017). Defective neuronal migration and inhibition of bipolar to multipolar transition of migrating neural cells by Mesoderm-Specific Transcript, Mest, in the developing mouse neocortex. Neuroscience 355, 126–140. doi:10.1016/j.neuroscience.2017.05.003
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi:10.1038/s41587-019-0201-4
Kirby, S. (2017). Culture and biology in the origins of linguistic structure. Psychon. Bull. Rev. 24 (1), 118–137. doi:10.3758/s13423-016-1166-7
Klausberger, T., and Somogyi, P. (2008). Neuronal diversity and temporal dynamics: The unity of hippocampal circuit operations. Science 321 (5885), 53–57. doi:10.1126/science.1149381
Kobayashi, T., Piao, W., Takamura, T., Kori, H., Miyachi, H., Kitano, S., et al. (2019). Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat. Commun. 10 (1), 5446. doi:10.1038/s41467-019-13203-4
Kolagar, T. A., Farzaneh, M., Nikkar, N., and Khoshnam, S. E. (2020). Human pluripotent stem cells in neurodegenerative diseases: Potentials, advances and limitations. Curr. Stem Cell Res. Ther. 15 (2), 102–110. doi:10.2174/1574888X14666190823142911
Krämer, A., Green, J., Pollard, J., and Tugendreich, S. (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30 (4), 523–530. doi:10.1093/bioinformatics/btt703
Levecque, C., Velayos-Baeza, A., Holloway, Z. G., and Monaco, A. P. (2009). The dyslexia-associated protein KIAA0319 interacts with adaptor protein 2 and follows the classical clathrin-mediated endocytosis pathway. Am. J. Physiol. Cell Physiol. 297 (1), C160–C168. doi:10.1152/ajpcell.00630.2008
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Meng, H., Smith, S. D., Hager, K., Held, M., Liu, J., Olson, R. K., et al. (2005). DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl. Acad. Sci. U. S. A. 102 (47), 17053–17058. doi:10.1073/pnas.0508591102
Molofsky, A. V., and Deneen, B. (2015). Astrocyte development: A Guide for the perplexed. Glia 63 (8), 1320–1329. doi:10.1002/glia.22836
Paracchini, S., Thomas, A., Castro, S., Lai, C., Paramasivam, M., Wang, Y., et al. (2006). The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum. Mol. Genet. 15 (10), 1659–1666. doi:10.1093/hmg/ddl089
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L., et al. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi:10.1038/nbt.3122
Peschansky, V. J., Burbridge, T. J., Volz, A. J., Fiondella, C., Wissner-Gross, Z., Galaburda, A. M., et al. (2010). The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb. Cortex 20 (4), 884–897. doi:10.1093/cercor/bhp154
Pletikos, M., Sousa, A. M., Sedmak, G., Meyer, K. A., Zhu, Y., Cheng, F., et al. (2014). Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81 (2), 321–332. doi:10.1016/j.neuron.2013.11.018
Powers, N. R., Eicher, J. D., Miller, L. L., Kong, Y., Smith, S. D., Pennington, B. F., et al. (2016). The regulatory element READ1 epistatically influences reading and language, with both deleterious and protective alleles. J. Med. Genet. 53 (3), 163–171. doi:10.1136/jmedgenet-2015-103418
Schatschneider, C., and Torgesen, J. K. (2004). Using our current understanding of dyslexia to support early identification and intervention. J. Child. Neurol. 19 (10), 759–765. doi:10.1177/08830738040190100501
Shaywitz, S. E., and Shaywitz, B. A. (2005). Dyslexia (specific reading disability). Biol. Psychiatry 57 (11), 1301–1309. doi:10.1016/j.biopsych.2005.01.043
Stockinger, P., Maitre, J. L., and Heisenberg, C. P. (2011). Defective neuroepithelial cell cohesion affects tangential branchiomotor neuron migration in the zebrafish neural tube. Development 138 (21), 4673–4683. doi:10.1242/dev.071233
Szalkowski, C. E., Fiondella, C. G., Galaburda, A. M., Rosen, G. D., Loturco, J. J., Fitch, R. H., et al. (2012). Neocortical disruption and behavioral impairments in rats following in utero RNAi of candidate dyslexia risk gene Kiaa0319. Int. J. Dev. Neurosci. 30 (4), 293–302. doi:10.1016/j.ijdevneu.2012.01.009
Velayos-Baeza, A., Toma, C., Paracchini, S., and Monaco, A. P. (2008). The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum. Mol. Genet. 17 (6), 859–871. doi:10.1093/hmg/ddm358
Wang, P., Mokhtari, R., Pedrosa, E., Kirschenbaum, M., Bayrak, C., Zheng, D., et al. (2017). CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism 8, 11. doi:10.1186/s13229-017-0124-1
Wu, G. D., Li, Z. H., Li, X., Zheng, T., and Zhang, D. K. (2020). microRNA-592 blockade inhibits oxidative stress injury in Alzheimer's disease astrocytes via the KIAA0319-mediated Keap1/Nrf2/ARE signaling pathway. Exp. Neurol. 324, 113128. doi:10.1016/j.expneurol.2019.113128
Xiang, Y., Tanaka, Y., Patterson, B., Hwang, S. M., Hysolli, E., Cakir, B., et al. (2020). Dysregulation of BRD4 function underlies the functional abnormalities of MeCP2 mutant neurons. Mol. Cell 79 (1), 84–98. e89. doi:10.1016/j.molcel.2020.05.016
Zhang, J., Uchiyama, J., Imami, K., Ishihama, Y., Kageyama, R., Kobayashi, T., et al. (2021). Novel roles of small extracellular vesicles in regulating the quiescence and proliferation of neural stem cells. Front. Cell Dev. Biol. 9, 762293. doi:10.3389/fcell.2021.762293
Keywords: KIAA0319, cell cycle, hESCs, NPCs, differentiation
Citation: Paniagua S, Cakir B, Hu Y, Kiral FR, Tanaka Y, Xiang Y, Patterson B, Gruen JR and Park I-H (2022) Dyslexia associated gene KIAA0319 regulates cell cycle during human neuroepithelial cell development. Front. Cell Dev. Biol. 10:967147. doi: 10.3389/fcell.2022.967147
Received: 12 June 2022; Accepted: 07 July 2022;
Published: 09 August 2022.
Edited by:
Xujie Liu, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Yan Liu, Nanjing Medical University, ChinaCopyright © 2022 Paniagua, Cakir, Hu, Kiral, Tanaka, Xiang, Patterson, Gruen and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey R. Gruen, amVmZnJleS5ncnVlbkB5YWxlLmVkdQ==; In-Hyun Park, aW5oeXVuLnBhcmtAeWFsZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.