- 1Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Life Sciences, Hebei Agricultural University, Baoding, Hebei, China
- 3National Germplasm Center of Domestic Animal Resources, Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 4State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 5Department of Human Genetics David Geffen School of Medicine University of California Los Angeles, Los Angeles, CA, United States
Correct reprogramming of the DLK1-DIO3 imprinted region is critical for the development of cloned animals. However, in pigs, the imprinting and regulation of the DLK1-DIO3 region has not been systematically analyzed. The objective of this study was to investigate the imprinting status and methylation regulation of the DLK1-DIO3 region in wild-type and cloned neonatal pigs. We mapped the imprinting control region, IG-DMR, by homologous alignment and validated it in sperm, oocytes, fibroblasts, and parthenogenetic embryos. Subsequently, single nucleotide polymorphism-based sequencing and bisulfite sequencing polymerase chain reaction were conducted to analyze imprinting and methylation in different types of fibroblasts, as well as wild-type and cloned neonatal pigs. The results showed that Somatic cell nuclear transfer (SCNT) resulted in hypermethylation of the IG-DMR and aberrant gene expression in the DLK1-DIO3 region. Similar to wild-type pigs, imprinted expression and methylation were observed in the surviving cloned pigs, whereas in dead cloned pigs, the IG-DMR was hypermethylated and the expression of GTL2 was nearly undetectable. Our study reveals that abnormal imprinting of the DLK1-DIO3 region occurs in cloned pigs, which provides a theoretical basis for improving the cloning efficiency by gene editing to correct abnormal imprinting.
Introduction
Somatic cell nuclear transfer (SCNT) is an assisted reproduction technology applied in the production of genetically modified (transgenic) animals, multiplication of elite animals, and protection of endangered species. Although the SCNT technology has been well developed in most domesticated and laboratory animals, the efficiency remains low, which limits its widespread application. Cloned animals show high rates of abortion during the perinatal period and reduced neonatal viability due to obesity, immunodeficiency, and respiratory defects (Ogura et al., 2013; Loi et al., 2016). A growing number of studies have demonstrated that cloned animals often undergo epigenetic modification errors, with gene imprinting being a major cause of developmental disorders in cloned animals (Matoba and Zhang 2018). The erasure and reconstruction of gene imprinting during somatic cell nuclear transfer cannot fully mimic normal gamete fertilization and embryo development, resulting in the loss of gene imprinting, which in turn affects the development of cloned embryos.
Imprinted genes are usually present in clusters, and the delta-like homolog one gene and the type III iodothyronine deiodinase gene (DLK1-DIO3) imprinted domains are located on chromosomes 12 (Schmidt et al., 2000) and 14 (Miyoshi et al., 2000) in mouse and human, respectively. The DLK1-DIO3 imprinting domain spans a region of 825 kb in the mouse and contains multiple coding and non-coding transcripts (da Rocha et al., 2008). The main genes are the paternally expressed imprinted genes DLK1, RTL1, and DIO3, and the maternally expressed imprinted gene GTL2. The genetic spacer region DMR (intergenic DMR, IG-DMR) located between GTL2 and DLK1 has been demonstrated to regulate the expression of the entire DLK1-DIO3 region. The GTL2 gene has been shown to repress the expression of the maternally expressed gene DLK1 in cis-regulation by recruiting polycomb repressive complex II (PRC2) in mice (Zhao et al., 2010; Kaneko et al., 2014; Das et al., 2015; Sanli et al., 2018). Previous studies have reported that the control of GTL2 expression by the IG-DMR is essential for the maintenance of imprinting in the DLK1-DIO3 domain (Lin et al., 2003; Kota et al., 2014; Das et al., 2015; Luo et al., 2016).
Loss of imprinting of DLK1-DIO3 is associated with severe developmental defects and malignant tumorigenesis (da Rocha et al., 2008; Jelinic and Shaw 2007; Khoury et al., 2010; Manodoro et al., 2014). The DLK1-DIO3 imprinted domain plays an important role in the derivation and culture of induced pluripotent stem cells and embryonic stem cells in mice (Liu et al., 2010; Carey et al., 2011; Stadtfeld et al., 2012; Mo et al., 2015). A recent study successfully generated bimaternal and bipaternal mice by knocking out three maternal imprinting regions (including the IG-DMR) and seven paternal imprinting regions in parthenogenetic haploid stem cells and parthenogenetic haploid stem cells, respectively. The bimaternal mice survived to adulthood, while the bipaternal mice survived more than 48 h (Li et al., 2018). These results suggest that proper maintenance of imprinting in the DLK1-DIO3 region is critical for embryonic development.
The expression of imprinting genes is controlled by DNA methylation marks that are established in germ cells in a sex-specific manner by cis-regulated differentially methylated regions (DMRs) called imprinting control regions (ICRs). Differentially methylated regions within imprinted loci undergo binding of specific transcription factors and modification by chromatin modifiers, ultimately establishing single allele expression patterns (Ferguson-Smith and Bourc’his 2018). Countless reports in the human and mouse have shown that the IG-DMR is the ICR of the DLK1-DIO3 imprinting region. (Rocha et al., 2008; Saito et al., 2017; Tucci et al., 2019), but there are no reports in large mammals such as the pig.
Previous studies have demonstrated that the DLK1-DIO3 region is silenced in cloned pig embryos, and the survival rate of cloned pigs is significantly improved by dosage compensation of the RTL1 gene, indicating that the imprinting of the DLK1-DIO3 region is abnormal in cloned pigs (Yu et al., 2018). Currently, there are few studies on the DLK1-DIO3 region. In addition, the location of the IG-DMR imprinting regulatory region in the DLK1-DIO3 region and the imprinting state of the DLK1-DIO3 region in pigs remain unknown. Therefore, a detailed and systematic study of the porcine DLK1-DIO3 imprinting region is necessary to provide a theoretical basis for improving the efficiency of pig cloning.
In the present study, the conserved sequences of the IG-DMRs in the mouse, human, and sheep were used for a comparative analysis of the porcine genome, and the location of the IG-DMR in the pig was localized. Methylation analysis of the IG-DMR showed that the methylation of cloned fetal fibroblasts and neonatal cloned dead pigs was higher than that of corresponding donor cells and wild-type neonates. In addition, the expression of the DLK1-DIO3 region was aberrant, and the expression of GTL2 was nearly completely lost in neonatal cloned dead pigs, indicating abnormal imprinting in the DLK1-DIO3 imprinted domain.
Materials and methods
Primary cell isolation and culture
Porcine fetal fibroblasts (PEFs) were isolated from 20 to 30-day-old embryos of forward and backward crosses of laboratory minipigs, Duroc and Rongchang. Porcine fetal fibroblasts were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum, 1% non-essential amino acids (Invitrogen), and 1% penicillin–streptomycin (Gibco) in a constant temperature and humidified incubator at 37.5 °C and 5% CO2.
Collection and in vitro maturation of porcine oocytes
The bilateral ovaries of slaughterhouse sows were harvested within 30 min of slaughtering and dried with sterilized gauzes. The ovaries were placed in wide-mouth thermos flasks filled in sterilized saline (containing both antibiotics) at a temperature of 30–35°C and transported back to the laboratory within 2 h. The follicular fluid from follicles of 3–6 mm in diameter was collected with an 18-gauge needle connected to a filter pump in a 50-ml centrifuge tube and undisturbed for 45–60 min. The supernatant was discarded, and Poly (vinyl alcohol)-Phosphate Buffered Saline (PVA-PBS) solution was added to the precipitate, followed by resuspension in a 60-mm cell culture dish. The intact cumulus and oocytes (COCs) were collected with a mouth pipette under a body view microscope and cultured in an incubator at 38.5°C with 5% CO2 for approximately 40 h. The matured COCs were transferred into T2 solution containing l mg/ml hyaluronidase pre-warmed to 38.5°C, followed by gentle pipetting. The oocytes with clear perivitelline gaps and homogenized cytoplasm were selected under the body view microscope and placed in T2 solution pre-warmed to 38.5°C.
Nuclear transfer
The mature oocytes were removed by micro-manipulation. Next, the individual donor cells were injected into the perivitelline space, and fusion was completed with two 1.2 kV/cm DC pulses (1-s interval) of 30 μs in fusion medium [0.3 M mannitol, 1.0 mM CaCl2, 0.1 mM MgCl2, 0.5 mM HEPES (pH 7.0–7.4)] using a BTX electronic cell manipulator. The oocytes were then incubated for 30 min in porcine zygote medium-3 (PZM-3), and the fusion percentage was calculated under a stereomicroscope. Fifty fused embryos were placed into a four-well dish (Nunc) containing 500 μl of PZM-3 pre-warmed to 38.5°C and incubated at 5% CO2 with maximum humidity.
Embryo transfer
The day-1 NT zygotes were surgically transferred into surrogate mothers (250–300 zygotes per surrogate). Approximately 25 days later, the pregnancy status of each surrogate was determined by ultrasonography.
Sample collection
The pregnant sows were executed on gestational day 114, and the skin, fat, muscle, and peritoneum were incised sequentially in the lower abdomen. The uterine body and uterine horns were ligated with hemostats, and the uterus was removed for dissection. An incision in the uterus was made to detach the placenta containing the fetus from the uterus. Finally, the fetus was removed for anatomical sampling. These tissues are subsequently used to extract RNA or DNA.
RNA and DNA extraction
Fetuses and placentae were frozen in liquid N2 and grounded in a mortar with a pestle. Half of each tissue was used for RNA extraction, and the remaining half was used for DNA extraction. RNA was extracted using the RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol. To eliminate contamination of RNA with DNA, genomic DNA was removed by treatment with DNase I. The RNA was eluted in RNase free water and stored at −80°C. The DNA samples were extracted using the QIAamp DNA Mini Kit (Qiagen), according to the manufacturer’s protocol. The DNA was eluted in sterile RNase-free water and stored at -20 °C until use.
RNA reverse transcription and quantitative polymerase chain reaction analysis
One microgram of total RNA was converted to a final volume of 20 μl of cDNA using the HiScript®III First Strand cDNA Synthesis Kit. The universal SYBR qPCR Master Mix was used for quantitative polymerase chain reactions. The procedure included 40 cycles of pre-denaturation (95°C, 30 s) and amplification (95°C, 10 s; 60°C, 30 s), followed by melting curve analysis (95°C, 15 s; 60°C, 1 min; 95°C, 15 s). The expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. The primer information is provided in Supplementary Table S1. Statistical analyses were performed using Excel and graphs were prepared using GraphPad Prism 5, and the statistical significance was set at 0.05.
Bisulfite genomic sequencing analysis
Genomic DNA was extracted from tissue samples using the TIANamp Genomic DNA Kit. For bisulfite genomic sequencing, 1 μg of gDNA was subjected to bisulfite treatment using the EpiTect Bisulfite Kit (Qiagen, Germany), according to the manufacturer’s protocol. Bisulfite sequencing PCR primers were designed using a web-based methylation primer tool (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The PCR products were cloned into the pMD19-T vector. At least ten randomly selected clones were sequenced. The sequences were aligned using a web-based quantification tool for methylation analysis (QUMA; http://quma.cdb.riken.jp/).
Examination of the allelic expression status of DLK1, GTL2, and DIO3
All heterozygous individuals corresponding to each SNP were used to analyze the allelic expression of DLK1, GTL2, and DIO3 by quantitative PCR. The cDNA templates were reverse transcribed from fetal and placental samples by random hexamer priming. The primers DK-F and DK-R were used for DLK1 expression analysis, the primers GT-F and GT-R were used for GTL2 expression analysis, and the primers DO-F and DO-R were used for DIO3 expression analysis. The cDNA templates were amplified for 35 cycles. The PCR products were extracted from agarose gels using the Gel Recovery Kit (Sangon, Shanghai, China) and subjected to direct sequencing.
Results
Identification of the porcine IG-DMR
A previous study has reported that conserved repeats exist in the IG-DMRs of the human, mouse, and sheep (Paulsen et al., 2001). To investigate whether such repeats exist in the porcine IG-DMR, the conserved repeats from the human, mouse, and sheep were aligned with the porcine DLK1-GTL2 region. As a result, conserved repeats were observed 5 kb upstream of GTL2 in the pig (Figure 1A). The repeat sequence of the pig included 13 repeats, and the conserved sequence was GTTGCCCGCGGTCCGCCA.
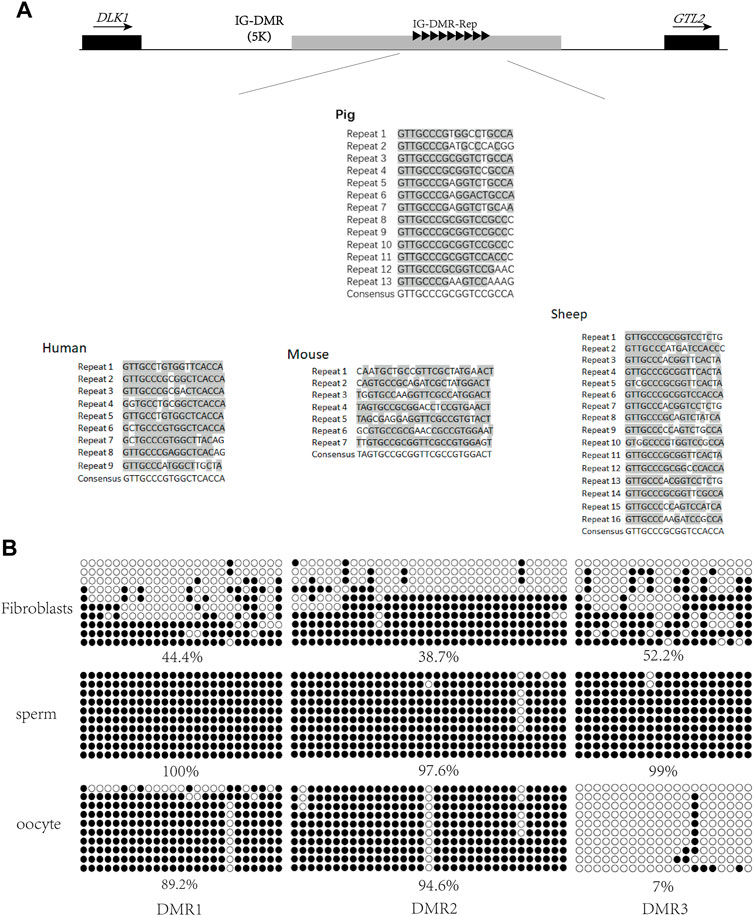
FIGURE 1. Screening of porcine IG-DMR. (A) Analysis of the IG-DMR tandem repeat sequences in pigs by interspecies conserved sequences. The black squares represent the positions of DLK1 and GTL2, respectively. The arrow direction is the gene transcription direction. The gray rectangle represents the IG-DMR candidate region (5 kb). The black triangle represents the conserved interspecies tandem repeat sequence, and the gray areas of the tandem repeat sequences in human, mouse, sheep, and pig are the conserved sequence dinucleotides. (B) Analysis of methylation in porcine fibroblasts, sperm, and oocytes Each circle represents a CpG dinucleotide. The degree of methylation (%) is based on methylated CpGs/all CpGs; open circles indicate unmethylated CpGs and filled circles indicate methylated CpGs.
To map the porcine IG-DMR, we used a website tool (MethPrimer) to predict the three DMRs around the repeats and designed methylation primers to analyze this region. The methylation detection of the three DMRs was carried out in sperm, oocytes, and fibroblasts. The results showed that DMR3 was hypomethylated in oocytes, hypermethylated in sperm, and half hypermethylated and half hypomethylated in fibroblasts, suggesting DMR3 is the IG-DMR regulating imprinting in the DLK1-GTL2 region (Figure 1B).
Conserved imprinting of the porcine DLK1-DIO3 region
To examine the imprinting status of the DLK1-DIO3 region (Figure 2A), we performed allele-specific expression analysis in 20-days (20-d) Rongchang (mother) and Duroc (father) crossbred pigs. To distinguish the expression between paternal and maternal alleles, we searched for SNPs in the DLK1-DIO3 region between the two pig breeds using the UCSC database (UCSC Genome Browser Home). Three SNPs (rs81211138, rs325797437, and rs343094622) were identified in DLK1, GTL2, and RTL1, respectively, and verified in the genomes of the female parent Rongchang pig and the male parent Duroc pig (Table 1; Figure 2B). The SNP rs81211138 in DLK1 was C in Rongchang and T in Duroc. The SNP rs325797437 in GTL2 was T in Rongchang and C in Duroc. The SNP rs343094622 in DIO3 was C in Rongchang and A in Duroc (Figure 2C). Quantitative polymerase chain reactions were performed to detect the expression of the three genes in fetuses and placentae of 20-d crossbred pigs. The PCR products were sequenced and showed monoallelic expression of DLK1, GTL2, and DIO3 in the three crossbred pigs.
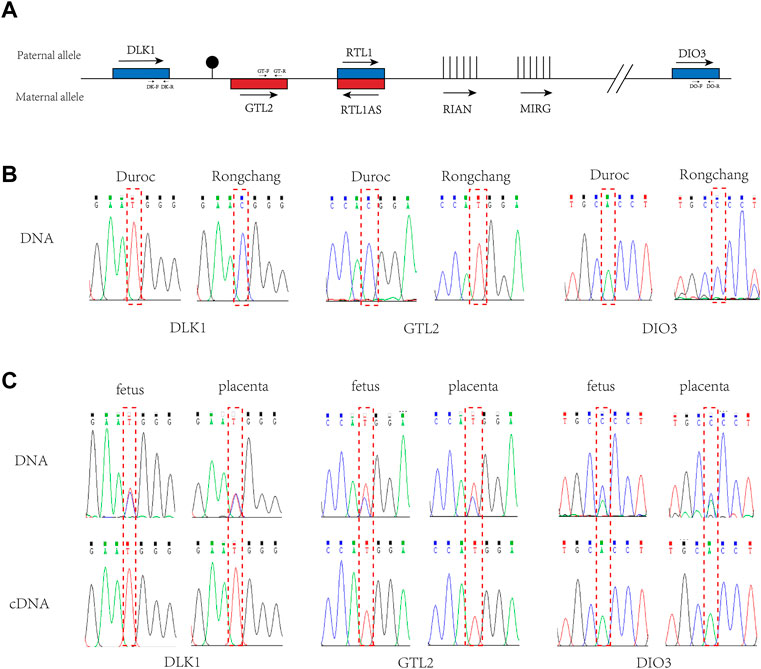
FIGURE 2. Imprinted expression of DLK1, GTL2, and DIO3 in the DLK1-DIO3 region. (A). Structure diagram of the DLK1-DIO3 imprinted domain. Blue squares represent paternal expression imprinted genes, and red squares represent maternal expression imprinted bases. The arrow direction is the transcription direction, and vertical lines represent non-coding RNA. Primer positions and directions are marked on the corresponding genes. (B). Validation of SNPs in Rongchang and Duroc pig genomes. (C). Allele-specific expression analysis of DLK1, GTL2, and DIO3 in the fetus and placenta of the three crossbred pigs. The above sequencing results used the genome as a template for PCR, and the following sequencing results used cDNA as a template for PCR. The red-dashed boxes indicate SNP loci.
Combined with SNP analysis, DLK1 and DIO3 were determined to be expressed by the paternal chromosome and GTL2 was determined to be expressed by the maternal chromosome. The results indicated that DLK1 and DIO3 in the porcine DLK1-DIO3 imprinted domain were maternally imprinted genes and GTL2 was paternally imprinted, consistent with the imprinting status of the mouse DLK1-DIO3 imprinted domain (Edwards et al., 2008; Li et al., 2008; Coster et al., 2012). These results reveal that imprinting of the DLK1-DIO3 domain is conserved in mammals (Coster et al., 2012; Kalish et al., 2014).
IG-DMR is hypomethylated in porcine parthenogenetic embryos
The genome of parthenogenetic embryos is derived from oocytes. Methylation analysis of the IG-DMR was conducted on parthenogenetic embryos at different developmental stages (Figure 3). The results showed that the IG-DMR in parthenogenetic embryos remained hypomethylated from the two-cell stage to the blastocyst stage, consistent with the hypomethylation of the IG-DMR in the female parent.
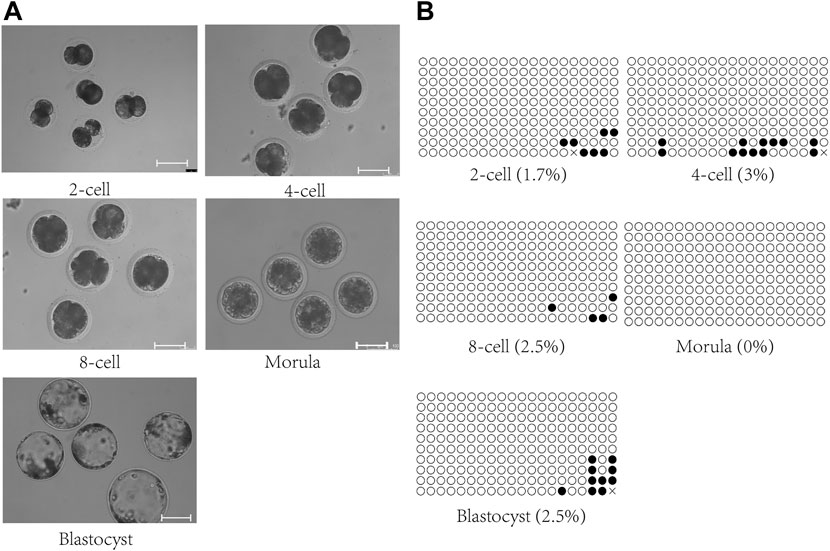
FIGURE 3. Methylation analysis of the IG-DMR in parthenogenetic embryos. (A). Parthenogenetic embryos at different developmental stages. Scale bars, 100 μm. (B). Methylation analysis of the IG-DMR in parthenogenetic embryos. Each circle represents a CpG dinucleotide. The degree of methylation (%) is based on methylated CPGs/all CPGs; open circles indicate unmethylated CpGs, and filled circles indicate methylated CpGs.
SCNT alters imprinting and methylation of the DLK1-DIO3 region
The data from mice and some pigs showed that the methylation of the IG-DMR and the gene expression of the DLK1-DIO3 imprinted domain were abnormal in unisexual embryos and cloned animals (Wei et al., 2010; Aronson et al., 2021; Wei et al., 2022). Therefore, we initially performed methylation analysis of the IG-DMR and examined gene imprinting expression of the DLK1-DIO3 region in porcine fetal fibroblasts (pEF-1, pEF-2) (Figure 4A), fetal fibroblasts derived from porcine somatic cell nuclear transfer embryos (pEF-1-NT, pEF-2-NT) (Figure 4B), and fetal fibroblasts derived from porcine parthenogenetic embryos (PApEF-1, PApEF-2, PApEF-3) (Figure 4C). PEF-1 and PEF-2 were donor cells for nuclear transfer embryos of PEF-1-NT and PEF-2-NT, respectively. The IG-DMR methylation levels of PEF-1-NT and PEF-2-NT were higher than those of PEF-1 and PEF-2. The IG-DMRs of PApEF-1, PApEF-2, and PApEF-3 were hypomethylated, consistent with previous results in early parthenogenetic embryos (Sato et al., 2011). Correspondingly, the GTL2 expression level was decreased in pEF-1-NT and pEF-2-NT, whereas it was increased in PApEF-1, PApEF-2, and PApEF-3, indicating the IG-DMR regulates the expression of GTL2. The expression levels of maternally imprinted genes DLK1, RTL1, and DIO3 were not significantly different among the three types of cells (Figure 4D). These results reveal that the methylation status of the IG-DMR is abnormal in cloned and parthenogenetic embryos, which affects the expression of the paternally imprinted gene GTL2.
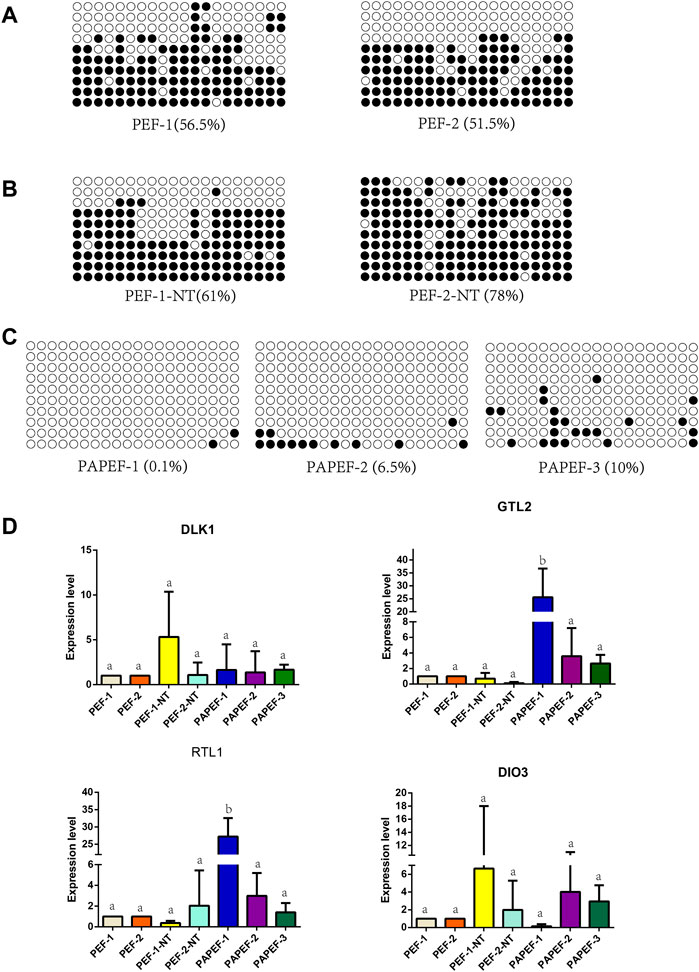
FIGURE 4. Methylation analysis of IG-DMR gene imprinting expression of the DLK1-DIO3 region in pEF, NTpEF, and PApEF. (A) Analysis of IG-DMR methylation levels in pEF, (B) NTpEF, and (C) PApEF. Each circle represents one CpG dinucleotide. The degree of methylation (%) is based on methylated CPGs/all CPGs; open circles indicate unmethylated CpGs, and filled circles indicate methylated CpGs. (D) Analysis of DLK1, GTL2, RTL1, and DIO3 gene expression in pEF, NTpEF, and PApEF. Horizontal coordinates are samples, and vertical coordinates are gene expression levels. Different letters (A,B) indicate significant differences (p < 0.05).
IG-DMR is abnormally methylated in cloned pigs
To investigate in detail the methylation status of the IG-DMR in cloned pigs, we performed methylation assays on various tissues from wild-type neonatal and cloned neonatal pigs. Wild-type pigs are born naturally at term. The newborn cloned pigs were obtained by caesarean section at term, of which #7 died during birth, while #2 and #28503 survived at birth. IG-DMR methylation analysis was performed on various tissues (heart, liver, spleen, lung, kidney, muscle, and brain) from these pigs. The methylation level in cloned pig #2 was not significantly different from that in wild-type pigs (Figures 5A,B; Supplementary Figure S1A), and the methylation degree of each tissue was approximately 50%, whereas the IG-DMR in cloned pig #7 was abnormally hypermethylated, and the methylation level of each tissue was higher than 88% (Figure 5C). Therefore, we hypothesize that reprogramming errors occur in the methylation of the IG-DMR during somatic reprogramming.

FIGURE 5. Analysis of IG-DMR methylation in wild-type neonatal pigs and cloned neonatal pigs. (A) Analysis of IG-DMR methylation in wild-type neonatal pigs. (B) Analysis of methylation in the surviving cloned piglet #2. (C) Analysis of methylation in dead cloned piglet #7. Each circle represents a CpG dinucleotide. The degree of methylation (%) is based on methylated CpGs/all CpGs; open circles indicate unmethylated CpGs, and filled circles indicate methylated CpGs.
Abnormal imprinted expression in cloned pigs
We analyzed the gene expression of the DLK1-DIO3 imprinted domain in wild-type neonatal pigs and cloned neonatal pigs. The results of quantitative PCR showed that DLK1 was highly expressed in the muscle (Figures 6A; Supplementary Figure S1B), DIO3 was highly expressed in the lungs (Figure 6D), and GTL2 and RTL1 were highly expressed in the brain (Figures 6B,C). The expression level of GTL2 in cloned pig #2 was higher than that in wild-type pigs, with no significant differences in the expression levels of maternally imprinted genes in most tissues. However, GTL2 expression was nearly undetectable in cloned pig #7, and DLK1 expression was elevated in most tissues. The hypermethylation of the IG-DMR in cloned pig #7 inhibited the expression of GTL2, indicating aberrant methylation of the IG-DMR was an important cause of death of cloned pig #7.
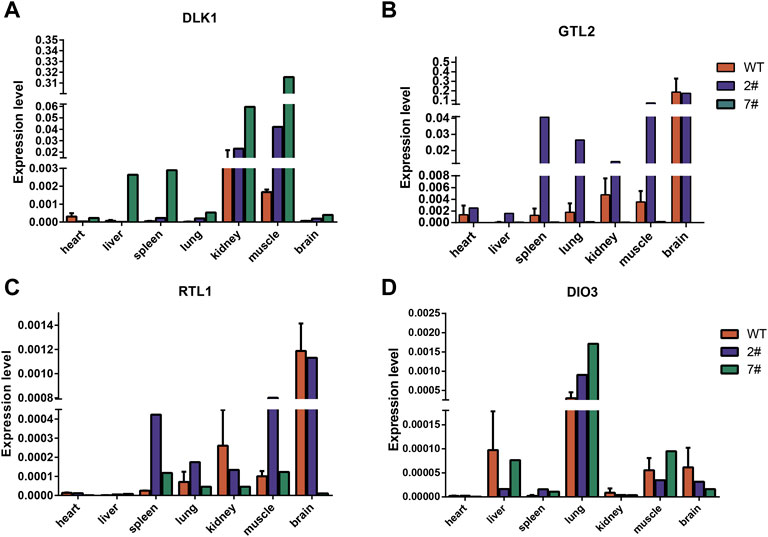
FIGURE 6. Analysis of DLK1-DIO3 imprinted domain gene expression in wild-type neonatal pigs and cloned neonatal pigs. (A) The expression of DLK1 in wild-type neonatal pigs and cloned neonatal pigs. (B) The expression of GTL2 in wild-type neonatal pigs and cloned neonatal pigs. (C) The expression of RTL1 in wild-type neonatal pigs and cloned neonatal pigs. (D) The expression of DIO3 in wild-type neonatal pigs and cloned neonatal pigs. The quantitative results are presented for tissue samples at horizontal coordinates and gene expression levels at vertical coordinates.
Discussion
Genomic imprinting is critical for several life processes. The DLK1-DIO3 imprinted domain is a cluster of genes essential for mammalian development. Numerous studies performed in the human and mouse have demonstrated that the IG-DMR is a key regulatory element of DLK1-DIO3 and that it can also control maternally imprinted gene expression by regulating GTL2 expression (Lin et al., 2003; Nowak et al., 2011; Kota et al., 2014; Luo et al., 2016; Wang et al., 2017; Saito et al., 2018). However, the imprinting of the DLK1-DIO3 region in the pig and the location of the IG-DMR remain unknown. We examined the expression of DLK1, GTL2, and DIO3 in Rongchang and Duroc crossbred pigs. The expression of male and female alleles was analyzed by SNPs in both pig breeds. The results showed that DLK1 and DIO3 were paternally expressed in the fetus and placenta, while GTL2 was maternally expressed, consistent with the results observed in the human and mouse. Thus, the imprinting of the DLK1-DIO3 region is well conserved in mammals.
Furthermore, we identified the tandem repeats in the pig based on the intermediate conserved repeat sequences in the human, mouse, and sheep IG-DMR and located it in the pig IG-DMR (Engemann et al., 2000; Okamura et al., 2000; Onyango et al., 2000; Paulsen et al., 2000). These tandem repeats are highly conserved, thus verifying the reliability of the pig IG-DMR locus. Nine tandem repeats were found in the human, seven conserved tandem repeats in the mouse, 16 conserved tandem repeats in the sheep, and 13 conserved tandem repeats, which spanned approximately 2.5 kb, in the pig.
The analysis of the methylation of the IG-DMR and the expression of each gene in the imprinted domain of DLK1-DIO3 in cloned pigs showed abnormal hypermethylation of the IG-DMR in dead cloned pigs and the loss of GTL2 expression in various tissues. GTL2 is widely involved in various cell processes, including transcriptional repression and RNA interference-mediated mRNA degradation (Charlier et al., 2001) GTL2 is expressed in the embryo and placenta, as well as in the adult, and the brain is the main site of its expression (Rocha et al., 2008). Maternal deletion of GTL2 and its DMR in the mouse leads to death due to alveolar hypoplasia and hepatocyte necrosis. We speculate that the loss of GTL2 expression in various tissues may affect the growth and development of cloned pigs.
Previous studies in the mouse have indicated that Gtl2 suppresses the expression of the parental allele (Rocha et al., 2008). In our study, the expression levels of the maternal imprinting genes DLK1 and DIO3 were higher in the tissues of cloned surviving and dead pigs than those in wild-type pigs, whereas the DLK1 expression level was significantly higher in the muscle and kidneys and that of DIO3 was significantly higher in the lungs of wild-type pigs and cloned surviving pigs. However, the RTL1 expression level was unchanged. RTL1 is mainly expressed in embryonic and placental tissues, where it is essential for normal placental development, and both paternal and maternal RTL1 deletion can lead to growth retardation and prenatal death (Yu et al., 2018). The antisense strand of the RTL1 gene encodes an RNA transcript and two maternally expressed microRNAs that are complementary to RTL1 (Seitz et al., 2003). We speculate that RTL1 was not elevated like the other maternally imprinted genes in cloned dead pigs because RTL1as, which is expressed by the antisense strand, also participates in the regulation of RTL1 expression.
In conclusion, we have localized the porcine IG-DMR and demonstrated that the porcine DLK1-DIO3 region was imprinted. In addition, abnormal DLK1-DIO3 region imprinting was observed in cloned dead pigs by methylation and quantitative PCR analyses. Our study provides a theoretical basis for the future correction of aberrant methylation in the IG-DMR by epigenetic editing and provides new directions for improving the cloning efficiency in pigs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by The Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China IAS 2021-240.
Author contributions
HYZ, DY, HBZ, and SL designed the research; JL, DY, JW, CL, QL, JW, WD, SZ, YP, HH, and XZ performed the experiments and analyzed the data. JL, HYZ, DY, JW, HBZ, and SL wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Agricultural Science and Technology Innovation Program (ASTIP-IAS04-1, ASTIP-IAS06) and the Hebei Province Natural Science Foundation of China (C2020204004).
Acknowledgments
We would like to thank all those who helped us during the study and the writing of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.964045/full#supplementary-material
References
Aronson, B. E., Scourzic, L., Shah, V., Swanzey, E., Kloetgen, A., Polyzos, A., et al. (2021). A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus. Dev. Cell 56 (22), 3052–3065.e5. doi:10.1016/j.devcel.2021.10.004.e3055
Carey, B. W., Markoulaki, S., Hanna, J. H., Faddah, D. A., Buganim, Y., Kim, J., et al. (2011). Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9 (6), 588–598. doi:10.1016/j.stem.2011.11.003
Charlier, C., Segers, K., Wagenaar, D., Karim, L., Berghmans, S., Jaillon, O., et al. (2001). Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11 (5), 850–862. doi:10.1101/gr.172701
Coster, A., Madsen, O., Heuven, H. C., Dibbits, B., Groenen, M. A., van Arendonk, J. A., et al. (2012). The imprinted gene DIO3 is a candidate gene for litter size in pigs. PLoS One 7 (2), e31825. doi:10.1371/journal.pone.0031825
da Rocha, S. T., Edwards, C. A., Ito, M., Ogata, T., and Ferguson-Smith, A. C. (2008). Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24 (6), 306–316. doi:10.1016/j.tig.2008.03.011
Das, P. P., Hendrix, D. A., Apostolou, E., Buchner, A. H., Canver, M. C., Beyaz, S., et al. (2015). PRC2 is required to maintain expression of the maternal gtl2-rian-mirg locus by preventing de novo DNA methylation in mouse embryonic stem cells. Cell Rep. 12 (9), 1456–1470. doi:10.1016/j.celrep.2015.07.053
Edwards, C. A., Mungall, A. J., Matthews, L., Ryder, E., Gray, D. J., Pask, A. J., et al. (2008). The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6 (6), e135. doi:10.1371/journal.pbio.0060135
Engemann, S., Strödicke, M., Paulsen, M., Franck, O., Reinhardt, R., Lane, N., et al. (2000). Sequence and functional comparison in the beckwith-wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9 (18), 2691–2706. doi:10.1093/hmg/9.18.2691
Ferguson-Smith, A. C., and Bourc'his, D. (2018). The discovery and importance of genomic imprinting. Elife 7, e42368. doi:10.7554/eLife.42368
Jelinic, P., and Shaw, P. (2007). Loss of imprinting and cancer. J. Pathol. 211 (3), 261–268. doi:10.1002/path.2116
Kalish, J. M., Jiang, C., and Bartolomei, M. S. (2014). Epigenetics and imprinting in human disease. Int. J. Dev. Biol. 58 (2-4), 291–298. doi:10.1387/ijdb.140077mb
Kaneko, S., Son, J., Bonasio, R., Shen, S. S., and Reinberg, D. (2014). Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 28 (18), 1983–1988. doi:10.1101/gad.247940.114
Khoury, H., Suarez-Saiz, F., Wu, S., and Minden, M. D. (2010). An upstream insulator regulates DLK1 imprinting in AML. Blood 115 (11), 2260–2263. doi:10.1182/blood-2009-03-212746
Kota, S. K., Llères, D., Bouschet, T., Hirasawa, R., Marchand, A., Begon-Pescia, C., et al. (2014). ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev. Cell 31 (1), 19–33. doi:10.1016/j.devcel.2014.08.009
Li, X. P., Do, K. T., Kim, J. J., Huang, J., Zhao, S. H., Lee, Y., et al. (2008). Molecular characteristics of the porcine DLK1 and MEG3 genes. Anim. Genet. 39 (2), 189–192. doi:10.1111/j.1365-2052.2007.01693.x
Li, Z. K., Wang, L. Y., Wang, L. B., Feng, G. H., Yuan, X. W., Liu, C., et al. (2018). Generation of bimaternal and bipaternal mice from hypomethylated haploid ESCs with imprinting region deletions. Cell Stem Cell 23 (5), 665–676. doi:10.1016/j.stem.2018.09.004
Lin, S.-P., Youngson, N., Takada, S., Seitz, H., Reik, W., Paulsen, M., et al. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35 (1), 97–102. doi:10.1038/ng1233
Liu, L., Luo, G. Z., Yang, W., Zhao, X., Zheng, Q., Lv, Z., et al. (2010). Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 285 (25), 19483–19490. doi:10.1074/jbc.M110.131995
Loi, P., Iuso, D., Czernik, M., and Ogura, A. (2016). A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol. 34 (10), 791–797. doi:10.1016/j.tibtech.2016.03.008
Luo, Z., Lin, C., Woodfin, A. R., Bartom, E. T., Gao, X., Smith, E. R., et al. (2016). Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 30 (1), 92–101. doi:10.1101/gad.270413.115
Manodoro, F., Marzec, J., Chaplin, T., Miraki-Moud, F., Moravcsik, E., Jovanovic, J. V., et al. (2014). Loss of imprinting at the 14q32 domain is associated with microRNA overexpression in acute promyelocytic leukemia. Blood 123 (13), 2066–2074. doi:10.1182/blood-2012-12-469833
Matoba, S., and Zhang, Y. (2018). Somatic cell nuclear transfer reprogramming: Mechanisms and applications. Cell Stem Cell 23 (4), 471–485. doi:10.1016/j.stem.2018.06.018
Miyoshi, N., Wagatsuma, H., Wakana, S., Shiroishi, T., Nomura, M., Aisaka, K., et al. (2000). Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes cells. 5 (3), 211–220. doi:10.1046/j.1365-2443.2000.00320.x
Mo, C. F., Wu, F. C., Tai, K. Y., Chang, W. C., Chang, K. W., Kuo, H. C., et al. (2015). Loss of non-coding RNA expression from the DLK1-DIO3 imprinted locus correlates with reduced neural differentiation potential in human embryonic stem cell lines. Stem Cell Res. Ther. 6 (1), 1. doi:10.1186/scrt535
Nowak, K., Stein, G., Powell, E., He, L. M., Naik, S., Morris, J., et al. (2011). Establishment of paternal allele-specific DNA methylation at the imprinted mouse Gtl2 locus. Epigenetics 6 (8), 1012–1020. doi:10.4161/epi.6.8.16075
Ogura, A., Inoue, K., and Wakayama, T. (2013). Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368 (1609), 20110329. doi:10.1098/rstb.2011.0329
Okamura, K., Hagiwara-Takeuchi, Y., Li, T., Vu, T. H., Hirai, M., Hattori, M., et al. (2000). Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: Toward the structural basis for species-specific imprinting. Genome Res. 10 (12), 1878–1889. doi:10.1101/gr.139200
Onyango, P., Miller, W., Lehoczky, J., Leung, C. T., Birren, B., Wheelan, S., et al. (2000). Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10 (11), 1697–1710. doi:10.1101/gr.161800
Paulsen, M., El-Maarri, O., Engemann, S., Strödicke, M., Franck, O., Davies, K., et al. (2000). Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 9 (12), 1829–1841. doi:10.1093/hmg/9.12.1829
Paulsen, M., Takada, S., Youngson, N. A., Benchaib, M., Charlier, C., Segers, K., et al. (2001). Comparative sequence analysis of the imprinted dlk1–gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2–H19 region. Genome Res. 11 (12), 2085–2094. doi:10.1101/gr.206901
Rocha, S. T. d., Edwards, C. A., Ito, M., Ogata, T., and Ferguson-Smith, A. C. (2008). Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24 (6), 306–316. doi:10.1016/j.tig.2008.03.011
Saito, T., Hara, S., Kato, T., Tamano, M., Muramatsu, A., Asahara, H., et al. (2018). A tandem repeat array in IG-DMR is essential for imprinting of paternal allele at the Dlk1-Dio3 domain during embryonic development. Hum. Mol. Genet. 27 (18), 3283–3292. doi:10.1093/hmg/ddy235
Saito, T., Hara, S., Tamano, M., Asahara, H., and Takada, S. (2017). Deletion of conserved sequences in IG-DMR at Dlk1-Gtl2 locus suggests their involvement in expression of paternally expressed genes in mice. J. Reprod. Dev. 63 (1), 101–109. doi:10.1262/jrd.2016-135
Sanli, I., Lalevée, S., Cammisa, M., Perrin, A., Rage, F., Llères, D., et al. (2018). Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis. Cell Rep. 23 (2), 337–348. doi:10.1016/j.celrep.2018.03.044
Sato, S., Yoshida, W., Soejima, H., Nakabayashi, K., and Hata, K. (2011). Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development. Genomics 98 (2), 120–127. doi:10.1016/j.ygeno.2011.05.003
Schmidt, J. V., Matteson, P. G., Jones, B. K., Guan, X. J., and Tilghman, S. M. (2000). The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 14 (16), 1997–2002. doi:10.1101/gad.14.16.1997
Seitz, H., Youngson, N., Lin, S. P., Dalbert, S., Paulsen, M., Bachellerie, J. P., et al. (2003). Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34 (3), 261–262. doi:10.1038/ng1171
Stadtfeld, M., Apostolou, E., Ferrari, F., Choi, J., Walsh, R. M., Chen, T., et al. (2012). Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 44 (4), 398–392. doi:10.1038/ng.1110
Tucci, V., Isles, A. R., Kelsey, G., and Ferguson-Smith, A. C. (2019). Genomic imprinting and physiological processes in mammals. Cell 176 (5), 952–965. doi:10.1016/j.cell.2019.01.043
Wang, Y., Shen, Y., Dai, Q., Yang, Q., Zhang, Y., Wang, X., et al. (2017). A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 45 (3), 1177–1185. doi:10.1093/nar/gkw1051
Wei, Y., Yang, C. R., and Zhao, Z. A. (2022). Viable offspring derived from single unfertilized mammalian oocytes. Proc. Natl. Acad. Sci. U. S. A. 119 (12), e2115248119. doi:10.1073/pnas.2115248119
Wei, Y., Zhu, J., Huan, Y., Liu, Z., Yang, C., Zhang, X., et al. (2010). Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell. Reprogr. 12 (2), 213–222. doi:10.1089/cell.2009.0090
Yu, D., Wang, J., Zou, H., Feng, T., Chen, L., Li, J., et al. (2018). Silencing of retrotransposon-derived imprinted gene RTL1 is the main cause for postimplantational failures in mammalian cloning. Proc. Natl. Acad. Sci. U. S. A. 115 (47), E11071–E11080. doi:10.1073/pnas.1814514115
Keywords: pig, IG-DMR, DNA methylation, genomic imprinting, SCNT
Citation: Li J, Yu D, Wang J, Li C, Wang Q, Wang J, Du W, Zhao S, Pang Y, Hao H, Zhao X, Zhu H, Li S and Zou H (2022) Identification of the porcine IG-DMR and abnormal imprinting of DLK1-DIO3 in cloned pigs. Front. Cell Dev. Biol. 10:964045. doi: 10.3389/fcell.2022.964045
Received: 08 June 2022; Accepted: 12 July 2022;
Published: 10 August 2022.
Edited by:
Lin Yuan, China Medical University, ChinaReviewed by:
Shoulong Deng, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaXiangpeng Dai, Jilin University, China
Copyright © 2022 Li, Yu, Wang, Li, Wang, Wang, Du, Zhao, Pang, Hao, Zhao, Zhu, Li and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Yu, eWR3MDIzQDE2My5jb20=; Huabin Zhu, emh1aHVhYmluQGNhYXMuY24=; Shijie Li, bGlzaGlqaWUyMDAwNUAxNjMuY29t; Huiying Zou, em91aHVpeWluZ0BjYWFzLmNu
†These authors have contributed equally to this work
 Junliang Li
Junliang Li Dawei Yu
Dawei Yu Jing Wang4†
Jing Wang4† Chongyang Li
Chongyang Li Jing Wang
Jing Wang Weihua Du
Weihua Du Shanjiang Zhao
Shanjiang Zhao Yunwei Pang
Yunwei Pang Xueming Zhao
Xueming Zhao Huabin Zhu
Huabin Zhu Huiying Zou
Huiying Zou