
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 30 June 2022
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.946219
This article is part of the Research TopicNeurocyte-endotheliocyte Crosstalk-mediated Tissue RegenerationView all 4 articles
 Ganggang Zhai1
Ganggang Zhai1 Likang Xiao1
Likang Xiao1 Chenyang Jiang1
Chenyang Jiang1 Songkai Yue1
Songkai Yue1 Meng Zhang1
Meng Zhang1 Jia zheng1
Jia zheng1 Zeming Liu2*
Zeming Liu2* Yonghui Dong1,3*
Yonghui Dong1,3*Osteoarthritis (OA) is the most common joint disease, usually occurring in middle-aged and elderly people. However, current treatment for OA in its early stages is ineffective, and drug therapy is often ineffective in slowing the progression of the disease. In fact, a deeper understanding of the underlying molecular mechanisms of OA could help us to better develop effective therapeutic measures. N6-methyladenosine (m6A) is a methylation that occurs at the adenosine N6-position, which is the most common internal modification on eukaryotic mRNAs. The role and mechanisms of m6A in mammalian gene regulation have been extensively studied. The “Writer”, “eraser”, and “reader” proteins are key proteins involved in the dynamic regulation of m6A modifications. Recent studies on post-transcriptional regulation alone have shown that m6a modification has an important role in the development of OA. This paper summarizes the specific regulatory processes of M6A in disease and reviews the role of m6A in OA, describing its pathophysiological role and molecular mechanisms, as well as its future research trends and potential clinical applications in OA.
Osteoarthritis (OA) is a chronic, degenerative joint disease and is one of the most common clinical joint disorders (Stonestrom, 2018). With the increasing aging of the global population, the incidence of OA is increasing year by year. The pathology is characterized by degeneration or destruction of the articular cartilage, secondary hyperplasia of the surrounding bone, and degeneration of the synovium and meniscus. It is often characterized by pain, limitation of movement, and joint deformity (Langworthy et al., 2010). The pathogenesis of OA is still not fully understood, but numerous studies have shown that the excessive inflammatory response and apoptosis during OA progression leads to the destruction of chondrocytes, which ultimately causes the disease to progress (Chen et al., 2017). Few drugs are clinically effective in relieving OA, and there is a lack of more satisfactory treatments for patients with advanced disease other than joint replacement surgery. Therefore, great efforts are needed to further investigate the biology of osteoarthritis in order to develop new therapeutic options (Richmond et al., 2010).
Post-transcriptional modifications are important regulators of a variety of physiological processes and disease progression, and thus have received increasing attention in bioscience research (Harvey et al., 2018). Among the numerous RNA modifications, N6-methyladenosine (m6A) is the most abundant mRNA modification (Jonkhout et al., 2017). m6A RNA methylation is the addition of a methyl group to the nitrogenous base position at position six of the adenine residue of RNA (Ma et al., 2019). m6A modifications affect almost all aspects of mRNA metabolism, including maintenance of RNA stability, RNA selective splicing and translation, thereby affecting apoptosis, autophagy and immune response (Dominissini et al., 2012). With the rapid development of molecular biology and sequencing, m6A modifications have been reported to be associated with almost all cellular functions and multiple diseases. In recent years, it has been shown that m6A modification promotes the inflammatory response, chondrocyte apoptosis and extracellular matrix imbalance in OA, which ultimately leads to the exacerbation of OA. In addition to this, previous work has found that the progression of OA can be inhibited by treatment with demethylation in a mouse model (Li et al., 2021). In this paper, we briefly describe the mechanism of m6A modification and its important role in RNA synthesis, transport and translation. Next, we summarize the role of m6A modification in osteoarthritic diseases and discuss future research directions.
M6A-related enzymes determine whether m6A is methylated or demethylated. In other words, the type of m6A-related enzymes reveals the function and significance of m6A. M6A-related enzymes include “writers”- Methylesterase, “erasers”-Demethylase and “readers”-m6A Reading Protein. The process of M6A modified RNA includes writer addition, reader recognition and eraser removal. (Wu et al., 2016). Methyltransferase like protein3 (METTL3), Methyltransferase like protein14 (METTL14), Wilms’ tumor 1-associated protein (WTAP) and other proteins can form writers with various binding proteins to regulate the methylation of RNA. On the contrary, m6A erasers protein can catalyze RNA demethylation, which is a part of m6A modification in reversible dynamic process. Readers include YTH domain family and heterogeneous nuclear ribonucleoprotein (hnRNP) family, whose role is to recognize m6A-mediated physiological behavior and influence RNA function (Zaccara et al., 2019). The dynamic process of m6A regulation is shown in Figure 1.
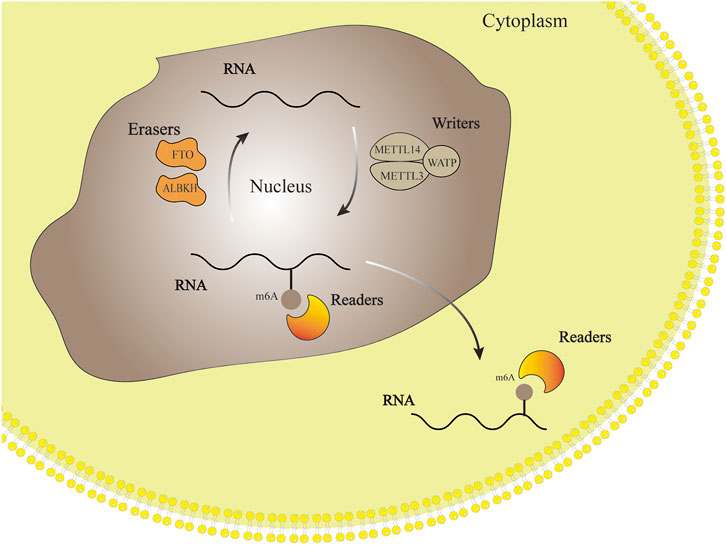
FIGURE 1. RNA methylation modification process. Writers, erasers and readers add, remove and identify methyl groups respectively. This process makes the m6A modification flexible and reversible.
Writer is the methyltransferase complex which is composed of the catalytic subunit METTL3 and other accessory subunits including METTL14, WTAP, VIRMA, RBM15, and ZC3H13 (Wang et al., 2016). The most important components are METTL14, WTAP, and METTL3. METTL3 can combine with methyl alone to catalyze m6A formation and promote protein translation in cytoplasm. Although METTL14 can’t catalyze the formation of m6A methyl modification independently, it can form heterodimers with METL3, and the methylation activity of this complex is higher than that of METL3 alone. Therefore, it can be seen that these proteins are not isolated in organisms, but form a complex to perform the catalytic function jointly (Liu et al., 2014). WTAP has the functions of stabilizing heterodimers, helping to locate nuclear spots, promoting RNA degradation and regulating cell differentiation and proliferation (Chen et al., 2019). RBM15 binds to the m6A complex and recruits it to specific RNA binding sites. VIRMA and ZC3H13 play a regulatory role in the process of methylation respectively (He et al., 2019).
Erasers, which are demethylases, are essential for the m6A modification to remain a dynamic and reversible process (Wang et al., 2014). Eraser can demethylate the base modified by M6A by demethylase. Two types of Erasers are known, FTO and ALKBH5. FTO is the first demethylase identified, first thought to be a gene associated with obesity, enriched in the brain, especially in neurons, and plays an important regulatory role in the central nervous system (Zhang et al., 2019). The FTO-mediated demethylation process is stepwise, m6A is first converted to N6-hydroxymethyladenosine (hm6A), then to N6-formyladenosine (f6A), and finally reduced to the original adenosine acid (Wu et al., 2015). AKKB homolog 5 (ALKBH5) is the second demethylase identified after FTO, which is highly expressed in the testis and is essential for spermatogenesis (Zhou et al., 2017). Its mediated demethylation process has no intermediate products. The differences about the m6A metabolism mechanism between FTO and ALKBH5 result in distinct biochemical outcomes and different biological functions.
The main function of Readers is to recognize bases modified by m6A and regulate RNA processing, transport, translation and stability (Wang et al., 2020). Up to now, the m6A reader proteins that have been identified include the YTH family (YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2), the hnRNP family (HNRNPA2B1, HNRNPC, HNRNPG) and the IGF2BP family (IGF2BP1, IGF2BP2, IGF2BP3). Among them, the YTH domain-containing protein was the first reader to be discovered (Huang et al., 2018). Human YTHDC1 has been proved to be involved in RNA splicing. In addition, it has been proved that YTHDF2 can reduce the stability of its targeted methylated mRNA transcript and control its span, while YTHDF1 can ensure the efficient expression of protein from its shared region. These studies have proved that YTHDF family proteins have potential synergistic effect in regulating gene expression (Sun et al., 2020).
Although our current understanding of the precise mechanism of m6A RNA modification is limited, various studies have examined the effects of m6A modification on the regulation of RNA transcripts.
The maintenance of normal tissue and anatomical structure of bone is the result of the complementary and balanced functions of osteoblasts and osteoclasts in bone (Nakashima, 2016). Imbalance between osteoblasts and osteoclasts often leads to lesions of bone. Found that the expression of PIWI-interacting RNA-36741 (pri-36741) and METTL3 were upregulated during the induction of osteoblast differentiation using primary human bone marrow mesenchymal stem cells (BMSCs). After human silencing the expression of Pri-36741 and METTL3, the osteoblast markers decreased, indicating that METL3 plays a regulatory role in osteoblast differentiation. Ultimately, it was found that in terms of specific regulatory mechanisms, the pri-36741-PIWIL4 complex binds directly to METTL3, inhibits METTL3 methylation regulation of bone morphogenetic protein-2 (BMP2), and promotes BMP2 mRNA and protein expression, thereby promoting osteoblast differentiation Liu et al. (2021). In addition to this, found that the level of m6A in bone marrow mesenchymal stem cells (BMSCs) was decreased in paraffin sections of femurs from mice knocked out of Mettl3, and found that the trabeculae of the distal femoral epiphysis of mice exhibited a pathological phenotype similar to that of osteoporosis by μCT. After the study, it was found that knockdown Mettl3 mice BMSCs had poor osteogenic differentiation ability, which led to a decrease in osteoblasts and osteocytes, bone loss, and even the development of osteoporosis. In terms of specific regulatory mechanisms, mettl3-deficient BMSCs are responsible for osteoporosis through the regulation of the PTH/Pth1r signaling axis to inhibit osteoblast differentiation Wu et al. (2018). In recently, Wang et al. found that RNA methylase METTL3 acted on the 1956bp m6A functional site in circ_0008542, which promoted the competitive binding between circ_0008542 and miRNA-185-5p, resulting in the increase of target gene RANK and the initiation of osteoclast bone resorption (Wang et al., 2021). This indicates that m6A is fully involved in the regulation of bone homeostasis. If bone homeostasis is destroyed, many bone metabolic diseases such as osteosarcoma, osteoporosis and osteoarthritis may occur. The specific role of m6A modification regulatory mechanism in bone development is shown in Figure 2.
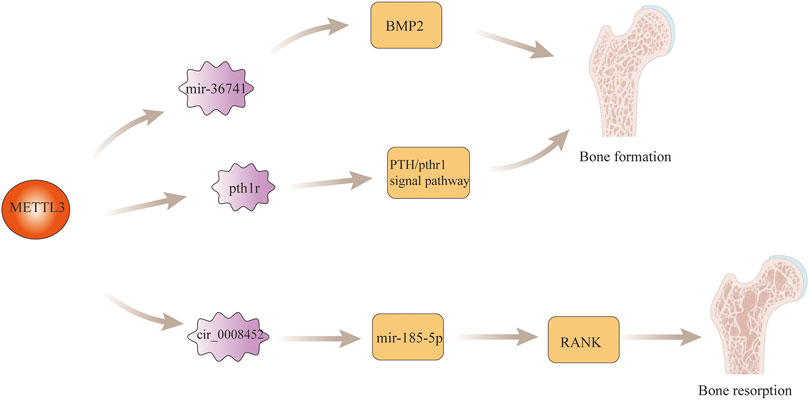
FIGURE 2. m6A modification in bone development. M6a modification regulates osteoblast differentiation by regulating MBP2 gene expression and the PTH/Pth1r signaling axis. In addition, m6a modification also promotes the process of bone resorption by regulating the expression of RANK gene.
Many findings have shown that mettle3 is aberrantly expressed in cartilage in OA patients or OA mouse models. Simulated in vitro inflammatory state using IL-6-treated chondroprogenitor cells ATDC5 and found that METTL3 mRNA levels, percentage of m6A methylation in total mRNA and apoptosis rate of chondrocytes were increased compared to the chondrocyte group not given IL-6 treatment. Then silencing METTL3 expression revealed reduced chondrocyte apoptosis, suppressed inflammatory response, suppressed cellular inflammatory response signal NF-kB and reduced extracellular matrix (ECM) synthesis. Furthermore, in an in vivo experiment in a mouse model of OA, injection of methylation inhibitors affected METTL3 mRNA expression but significantly inhibited total m6A levels, inflammatory conditions, and ECM degradation Liu et al. (2019). Additionally, Chen et al. found that cellular senescence markers and METTL3 mRNA expression were upregulated and autophagy markers were downregulated in synovial tissues from OA patients and in vitro OA models. And in vitro cartilage experiments revealed that senescent synovial tissues were able to cause cartilage dysfunction and aging. To assess whether autophagy mediates FLS senescence, it was finally found that metttl3-mediated ATG7 m6A modification could inhibit cellular autophagy, and the next inhibition of autophagy was through the senescence regulator (GATA4) to regulate the process of cellular senescence, thus affecting the progression of OA (Chen et al., 2022a). However, found that METTL3 expression was reduced in knee cartilage of OA patients compared to cartilage of femoral neck fracture patients, and this was also demonstrated in an OA model constructed using IL-1β-treated SW1353 cells. Next, it was found that overexpression of METTL3 increased the level of inflammatory factors, promoted the expression of p-65 protein and p-ERK, activated the NF-κB signaling pathway and thus aggravated OA progression, and overexpression of METTL3 also regulated the balance between TIMPs and MMPs, thus affecting the degradation of ECM. The contradictory results may result from the limited potential of SW1353 cells as a cell line to mimic primary human chondrocytes and the differences in femoral cartilage compared to knee cartilage Sang et al. (2021).
The FTO gene, encoding a 2-oxoglutarate-dependent nucleic acid demethylase (Gerken et al., 2007), has been demonstrated in related studies to be an obesity-associated gene. Some studies have suggested that the FTO gene mediates the progression of OA through obesity. However, some argue that the causal relationship between obesity and OA is difficult to establish (Pang et al., 2013), thus rejecting this theory. For example, compared patients with temporomandibular joint osteoarthritis (TMJOA)with the general population and concluded that the frequency of the FTO polymorphism allele was significantly different between the two groups, while the differences in weight, age, and sex were not statistically significant Takaoka et al. (2022). Yang et al. found that lncRNA AC008 was highly expressed in human OA cartilage samples, and found that aberrant expression of AC008 affected OA progression by regulating chondrocyte viability, chondrocyte apoptosis and ECM degradation. To investigate the specific regulatory mechanism of AC008, miR-328-3p was suggested to be the target of AC008 by bioinformatics analysis and dual luciferase reporter gene assay; while AQP1 and ANKH were the targets of miR-328-3p. In addition, the study verified the reversal of chondrocyte apoptosis by decreasing m6A levels of AC008 after FTO overexpression in chondrocytes, and finally concluded that FTO-dependent m6A demethylation-mediated upregulation of AC008 promotes osteoarthritis progression through the miR-328-3p-AQP1/ANKH axis (Yang et al., 2021).
The main mechanism by which M6A acts is through the recruitment of m6A binding proteins (Meyer and Jaffrey, 2017). In fact, studies on m6A readers in OA are currently lacking. Recently, in a mouse cartilage OA model, demonstrated that Mettl3 was highly expressed in cartilage and caused apoptosis and autophagy in chondrocytes by directly interacting with Bcl2 mRNA and increasing Bcl2 mRNA methylation levels. It was also confirmed by protein-RNA interaction predictor (PRIdictor) analysis and mRNA stability test that Bcl2 mRNA methylation was mediated by YTHDF1 to produce a regulatory effect on OA He et al. (2022). As to whether the hnRNP family is related to the molecular mechanism of OA, no relevant studies have been found and further exploration is worthwhile. The regulatory mechanism of m6A is shown in Table 1 and Figure 3.
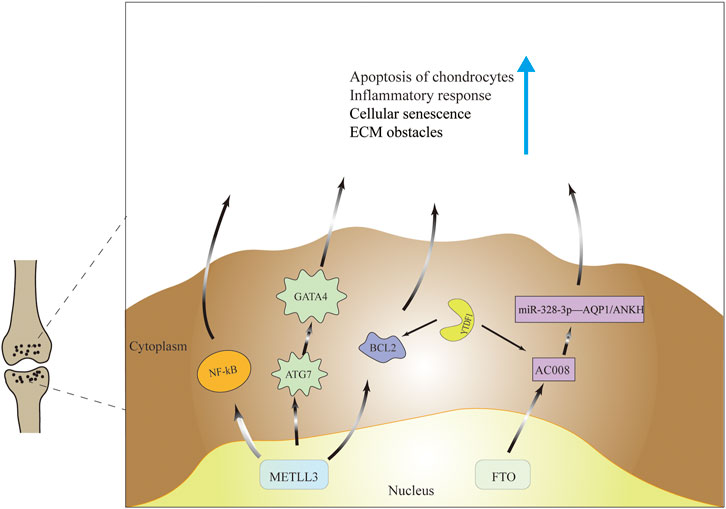
FIGURE 3. Molecular mechanisms of m6a modifications in the pathogenesis of OA. M6A modification ultimately cause cartilage apoptosis, cellular senescence, increased inflammatory response and extracellular matrix imbalance through regulation of gene expression and signaling pathways.
As the most common RNA modification in mammals, m6A modifications are also common in other skeletal system diseases. In spinal degenerative diseases, many studies have suggested a regulatory role for m6A modifications. Demonstrated that ALKBH5-mediated mRNA demethylation accelerated intervertebral disc (IVD) aging through ex vivo experiments. AlKBH5 caused disc aging and nucleus pulposus degeneration by demethylating DNMT3B transcripts Li et al. (2022). Found that in the nucleus pulposus of patients with disc degeneration, the mRNA content of MAT2A was lower than normal, while the mRNA of the demethylase metttl16 was higher. Following this, in vivo and in vitro experiments were constructed to conclusively demonstrate that m6A modification of MAT2A pre-mRNA by METTL16 may cause increased apoptosis in the nucleus pulposus and intervertebral disc degeneration under oxidative stress Chen et al. (2022b). In addition, many studies have confirmed that m6A modification has an important regulatory role in osteosarcoma (Patil et al., 2016). Identified METTL3 as an oncogene in osteosarcoma. Knockdown of METTL3 inhibited the proliferation, migration and invasion of human osteosarcoma cell lines SAOS-2 and MG63 by suppressing m6A methylation levels and the expression of ATPase family AAA structural domain protein 2 (ATAD2) Zhou et al. (2020). Recently, Yuan et al. showed that the demethylase ALKBH5 can silence the pre-miR-181b-1/YAP signaling axis, thereby inhibiting tumor progression in osteosarcoma. In addition, upregulation of ALKBH5 expression also contributes to chemoresistance in osteosarcoma patients and predicts worsening of their metastasis-free survival (WANG et al., 2019).
Given that abnormal m6A modifications are closely related to cartilage degeneration, cartilage apoptosis, and inflammatory response in the pathogenesis of OA (Chen et al., 2021). Therefore, m6A modification may bring new breakthroughs for the early diagnosis as well as the treatment of OA. On the one hand, many studies have pointed to the presence of m6A hypermethylation levels and dysregulation of m6A-related enzymes in cartilage or synovial samples from OA patients (Zhang et al., 2020). However, because cartilage and synovial samples are difficult to obtain, there is a lack of studies to confirm the presence of abnormal expression of m6A modification-related markers in the peripheral blood of OA patients, so exploring whether such differences exist could be the next research focus, which is important for the early diagnosis or screening of OA. On the other hand, the current treatment modalities for OA in the world are still mainly conservative and surgical; conservative treatment includes lifestyle changes and medication, and surgical treatment includes arthroscopic debridement and joint replacement surgery (Katz et al., 2021). But the current mainstream medications have no significant effect on stopping the progression of OA, while surgical treatment is more applicable to patients with advanced OA who have lost joint function. Therefore, it is necessary to develop new therapeutic strategies for OA. In a mouse model of OA, m6A-related inhibitors have been shown to be effective in improving the inflammatory status and degradation of ECM and cellular aging in OA, thereby slowing down the progression of the disease. Unfortunately, however, the development of m6A-related drugs is only at the experimental stage, and relevant studies on clinical patients are lacking. Further studies are needed to investigate whether m6A inhibitors can effectively inhibit the progression of OA patients.
M6A modification has emerged as an important factor in the development and progression of OA. It can accelerate or inhibit OA progression in different ways. There is no doubt that the emergence of m6A regulation provides new insights into the molecular mechanisms of OA and may contribute to the development of new and more effective therapeutic approaches. However, the full understanding of the mechanisms of m6A modifications in OA is still in its infancy, and several knowledge gaps remain. Firstly, the role of m6A-related regulatory factors in OA, other than METTL3, has been less studied. Secondly researchers have noted the potential of m6A as a therapeutic target for OA, but few studies have focused on the clinical application of effective and specific drugs targeting the m6A enzyme. All, it is necessary to accelerate its translation to the clinic.
GZ and YD conceived and designed the study. JZ and ZL supervised the study. GZ, SY, MZ, and LX prepared the manuscript. CJ and GZ prepared the figures. All authors approved the final manuscript.
This study was supported by the Henan Provincial Natural Science Foundation of China (212300410242), Henan Province medical science and technology tackling key problems provincial and ministerial joint construction Youth Project (SBGJ202103009) and Henan Young and Middle-aged Health Science and Technology Innovation Excellent Youth Talent Training Project of China (YXKC2021047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer QD declared a shared affiliation with the author ZL to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Chen, D., Shen, J., Zhao, W., Wang, T., Han, L., Hamilton, J. L., et al. (2017). Osteoarthritis: toward a Comprehensive Understanding of Pathological Mechanism. Bone Res. 5, 16044. doi:10.1038/boneres.2016.44
Chen, J., Tian, Y., Zhang, Q., Ren, D., Zhang, Q., Yan, X., et al. (2021). Novel Insights into the Role of N6-Methyladenosine RNA Modification in Bone Pathophysiology. Stem Cells Dev. 30 (1), 17–28. doi:10.1089/scd.2020.0157
Chen, P. B., Shi, G. X., Liu, T., Li, B., Jiang, S. D., Zheng, X. F., et al. (2022a). Oxidative Stress Aggravates Apoptosis of Nucleus Pulposus Cells through m6A Modification of MAT2A Pre-mRNA by METTL16. Oxid. Med. Cell Longev. 2022, 4036274. doi:10.1155/2022/4036274
Chen, X., Gong, W., Shao, X., Shi, T., Zhang, L., Dong, J., et al. (2022b). METTL3-mediated m6A Modification of ATG7 Regulates Autophagy-GATA4 axis to Promote Cellular Senescence and Osteoarthritis Progression. Ann. Rheum. Dis. 81 (1), 87–99. doi:10.1136/annrheumdis-2021-221091
Chen, Y., Peng, C., Chen, J., Chen, D., Yang, B., He, B., et al. (2019). WTAP Facilitates Progression of Hepatocellular Carcinoma via m6A-HuR-dependent Epigenetic Silencing of ETS1. Mol. Cancer 18 (1), 127. doi:10.1186/s12943-019-1053-8
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Gerken, T., Girard, C. A., Tung, Y.-C. L., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The Obesity-Associated FTO Gene Encodes a 2-oxoglutarate-dependent Nucleic Acid Demethylase. Science 318 (5855), 1469–1472. doi:10.1126/science.1151710
Harvey, Z. H., Chen, Y., and Jarosz, D. F. (2018). Protein-Based Inheritance: Epigenetics beyond the Chromosome. Mol. Cell 69 (2), 195–202. doi:10.1016/j.molcel.2017.10.030
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-Methyladenosine and its Role in Cancer. Mol. Cancer 18 (1), 176. doi:10.1186/s12943-019-1109-9
He, Y., Wang, W., Xu, X., Yang, B., Yu, X., Wu, Y., et al. (2022). Mettl3 Inhibits the Apoptosis and Autophagy of Chondrocytes in Inflammation through Mediating Bcl2 Stability via Ythdf1-Mediated m6A Modification. Bone 154, 116182. doi:10.1016/j.bone.2021.116182
Huang, X., Zhang, H., Guo, X., Zhu, Z., Cai, H., and Kong, X. (2018). Insulin-like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1) in Cancer. J. Hematol. Oncol. 11 (1), 88. doi:10.1186/s13045-018-0628-y
Jonkhout, N., Tran, J., Smith, M. A., Schonrock, N., Mattick, J. S., and Novoa, E. M. (2017). The RNA Modification Landscape in Human Disease. Rna 23 (12), 1754–1769. doi:10.1261/rna.063503.117
Katz, J. N., Arant, K. R., and Loeser, R. F. (2021). Diagnosis and Treatment of Hip and Knee Osteoarthritis. Jama 325 (6), 568–578. doi:10.1001/jama.2020.22171
Langworthy, M. J., Saad, A., and Langworthy, N. M. (2010). Conservative Treatment Modalities and Outcomes for Osteoarthritis: The Concomitant Pyramid of Treatment. Physician Sportsmed. 38 (2), 133–145. doi:10.3810/psm.2010.06.1792
Li, G., Luo, R., Zhang, W., He, S., Wang, B., Liang, H., et al. (2022). m6A Hypomethylation of DNMT3B Regulated by ALKBH5 Promotes Intervertebral Disc Degeneration via E4F1 Deficiency. Clin. Transl. Med. 12 (3), e765. doi:10.1002/ctm2.765
Li, H., Xiao, W., He, Y., Wen, Z., Cheng, S., Zhang, Y., et al. (2021). Novel Insights into the Multifaceted Functions of RNA N6-Methyladenosine Modification in Degenerative Musculoskeletal Diseases. Front. Cell Dev. Biol. 9 (null), 766020. doi:10.3389/fcell.2021.766020
Liu, J., Chen, M., Ma, L., Dang, X., and Du, G. (2021). piRNA-36741 Regulates BMP2-Mediated Osteoblast Differentiation via METTL3 Controlled m6A Modification. Aging 13 (19), 23361–23375. doi:10.18632/aging.203630
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, Q., Li, M., Jiang, L., Jiang, R., and Fu, B. (2019). METTL3 Promotes Experimental Osteoarthritis Development by Regulating Inflammatory Response and Apoptosis in Chondrocyte. Biochem. Biophysical Res. Commun. 516 (1), 22–27. doi:10.1016/j.bbrc.2019.05.168
Ma, C.-J., Ding, J.-H., Ye, T.-T., Yuan, B.-F., and Feng, Y.-Q. (2019). AlkB Homologue 1 Demethylates N3-Methylcytidine in mRNA of Mammals. ACS Chem. Biol. 14 (7), 1418–1425. doi:10.1021/acschembio.8b01001
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi:10.1146/annurev-cellbio-100616-060758
Pang, H., Luo, F., Dai, F., Wu, X.-H., and Xu, J.-Z. (2013). Genome-wide Association Study for Osteoarthritis. Lancet 381 (9864), 372–373. doi:10.1016/s0140-6736(13)60167-1
Patil, D. P., Chen, C.-K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional repressionA RNA Methylation Promotes XIST-Mediated Transcriptional Repression [J]. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Richmond, J., Hunter, D., Irrgang, J., Jones, M. H., Snyder-Mackler, L., Van Durme, D., et al. (2010). American Academy of Orthopaedic Surgeons Clinical Practice Guideline on the Treatment of Osteoarthritis (OA) of the Knee. J. Bone Jt. Surgery-American Volume 92 (4), 990–993. doi:10.2106/jbjs.i.00982
Sang, W., Xue, S., Jiang, Y., Lu, H., Zhu, L., Wang, C., et al. (2021). METTL3 Involves the Progression of Osteoarthritis Probably by Affecting ECM Degradation and Regulating the Inflammatory Response. Life Sci. 278, 119528. doi:10.1016/j.lfs.2021.119528
Stonestrom, A. J. (2018). The Key Role of Epigenetics in Human Disease. N. Engl. J. Med. 379 (4), 400. doi:10.1056/NEJMc1805989
Sun, J., Bie, X. M., Wang, N., Zhang, X. S., and Gao, X.-Q. (2020). Genome-wide Identification and Expression Analysis of YTH Domain-Containing RNA-Binding Protein Family in Common Wheat. BMC Plant Biol. 20 (1), 351. doi:10.1186/s12870-020-02505-1
TakaokA, R., Kuyama, K., Yatani, H., Ishigaki, S., Kayashima, H., Koishi, Y., et al. (2022). Involvement of an FTO Gene Polymorphism in the Temporomandibular Joint Osteoarthritis. Clin. Oral Invest 26 (3), 2965–2973. doi:10.1007/s00784-021-04278-9
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, T., Kong, S., Tao, M., and Ju, S. (2020). The Potential Role of RNA N6-Methyladenosine in Cancer Progression. Mol. Cancer 19 (1), 88. doi:10.1186/s12943-020-01204-7
Wang, W., Qiao, S.-C., Wu, X.-B., Sun, B., Yang, J.-G., Li, X., et al. (2021). Circ_0008542 in Osteoblast Exosomes Promotes Osteoclast-Induced Bone Resorption through m6A Methylation. Cell Death Dis. 12 (7), 628. doi:10.1038/s41419-021-03915-1
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent Regulation of Messenger RNA Stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, Y., Zeng, L., Liang, C., Zan, R., Ji, W., Zhang, Z., et al. (2019). Integrated Analysis of Transcriptome-wide m6A Methylome of Osteosarcoma Stem Cells Enriched by Chemotherapy. Epigenomics 11 (15), 1693–1715. doi:10.2217/epi-2019-0262
Wu, J., Xiao, H., Wang, T., Hong, T., Fu, B., Bai, D., et al. (2015). N6-Hydroperoxymethyladenosine: a New Intermediate of Chemical Oxidation of N6-Methyladenosine Mediated by Bicarbonate-Activated Hydrogen Peroxide. Chem. Sci. 6 (5), 3013–3017. doi:10.1039/c5sc00484e
Wu, R., Jiang, D., Wang, Y., and Wang, X. (2016). N 6-Methyladenosine (m6A) Methylation in mRNA with A Dynamic and Reversible Epigenetic Modification. Mol. Biotechnol. 58 (7), 450–459. doi:10.1007/s12033-016-9947-9
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m6A RNA Methylation Regulates the Fate of Bone Marrow Mesenchymal Stem Cells and Osteoporosis. Nat. Commun. 9 (1), 4772. doi:10.1038/s41467-018-06898-4
Yang, J., Zhang, M., Yang, D., Ma, Y., Tang, Y., Xing, M., et al. (2021). m6A-mediated Upregulation of AC008 Promotes Osteoarthritis Progression through the miR-328-3p‒AQP1/ANKH axis. Exp. Mol. Med. 53 (11), 1723–1734. doi:10.1038/s12276-021-00696-7
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cell Biol. 20 (10), 608–624. doi:10.1038/s41580-019-0168-5
Zhang, W., He, L., Liu, Z., Ren, X., Qi, L., Wan, L., et al. (2020). Multifaceted Functions and Novel Insight into the Regulatory Role of RNA N6-Methyladenosine Modification in Musculoskeletal Disorders. Front. Cell Dev. Biol. 8, 870. doi:10.3389/fcell.2020.00870
Zhang, X., Wei, L.-H., Wang, Y., Xiao, Y., Liu, J., Zhang, W., et al. (2019). Structural Insights into FTO's Catalytic Mechanism for the Demethylation of Multiple RNA Substrates. Proc. Natl. Acad. Sci. U.S.A. 116 (8), 2919–2924. doi:10.1073/pnas.1820574116
Zhou, C., Molinie, B., Daneshvar, K., Pondick, J. V., Wang, J., Van Wittenberghe, N., et al. (2017). Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-type-specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 20 (9), 2262–2276. doi:10.1016/j.celrep.2017.08.027
Keywords: N6-methyladenosine, molecular mechanisms, osteoarthritis, biomarker, therapeutic target
Citation: Zhai G, Xiao L, Jiang C, Yue S, Zhang M, zheng J, Liu Z and Dong Y (2022) Regulatory Role of N6-Methyladenosine (m6A) Modification in Osteoarthritis. Front. Cell Dev. Biol. 10:946219. doi: 10.3389/fcell.2022.946219
Received: 17 May 2022; Accepted: 10 June 2022;
Published: 30 June 2022.
Edited by:
Yuan Xiong, Huazhong University of Science and Technology, ChinaReviewed by:
Wei Xiang, Renmin Hospital of Wuhan University, ChinaCopyright © 2022 Zhai, Xiao, Jiang, Yue, Zhang, zheng, Liu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghui Dong, ZG9uZ3loQHp6dS5lZHUuY24=; Zeming Liu, Nm15dEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.