- 1Clinical Medical Laboratory Center, Jining First People’s Hospital, Jining Medical University, Jining, China
- 2Cheeloo College of Medicine, Shandong University, Jinan, China
- 3Department of Thyroid and Breast Surgery, Jining First People’s Hospital, Jining Medical University, Jining, China
Ubiquitination is a critical type of protein post-translational modification playing an essential role in many cellular processes. To date, more than eight types of ubiquitination exist, all of which are involved in distinct cellular processes based on their structural differences. Studies have indicated that activation of the ubiquitination pathway is tightly connected with inflammation-related diseases as well as cancer, especially in the non-proteolytic canonical pathway, highlighting the vital roles of ubiquitination in metabolic programming. Studies relating degradable ubiquitination through lys48 or lys11-linked pathways to cellular signaling have been well-characterized. However, emerging evidence shows that non-degradable ubiquitination (linked to lys6, lys27, lys29, lys33, lys63, and Met1) remains to be defined. In this review, we summarize the non-proteolytic ubiquitination involved in tumorigenesis and related signaling pathways, with the aim of providing a reference for future exploration of ubiquitination and the potential targets for cancer therapies.
1 Introduction
1.1 The Ubiquitin-Proteasome System
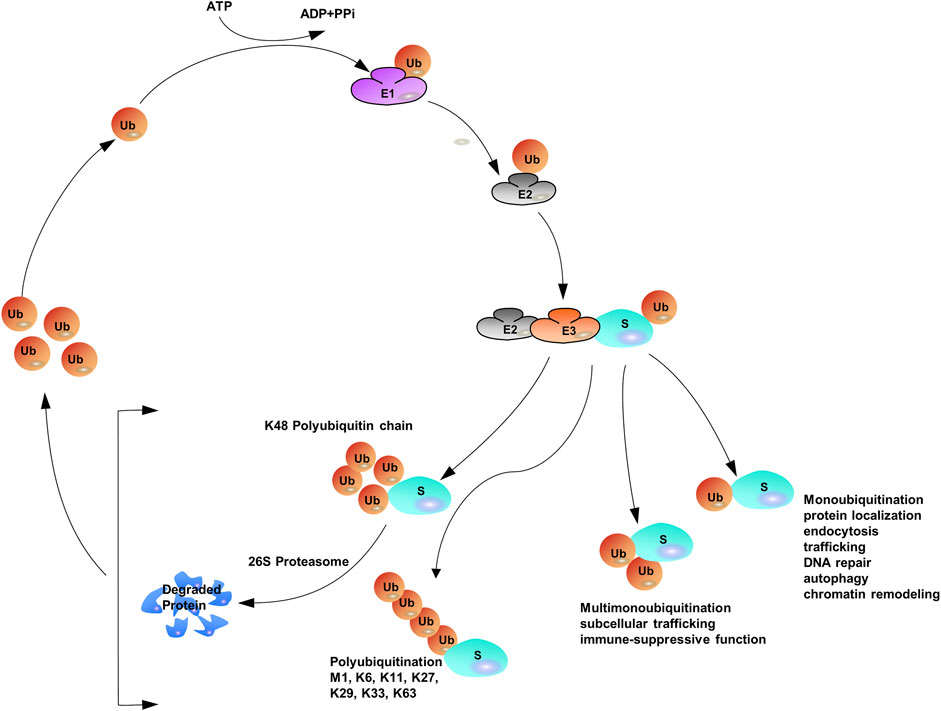
Ubiquitination, also known as ubiquitylation, refers to the process by which ubiquitin (Ub, a small and highly conserved protein), with the help of a series of special enzymes, classifies proteins in cells, selects target proteins, and modifies those proteins (McDowell and Philpott, 2016; Seeler and Dejean, 2017; Rape, 2018). Ubiquitination plays fundamental roles in many cellular events such as cell proliferation (Kwon and Ciechanover, 2017; Werner et al., 2017; Senft et al., 2018; Song and Luo, 2019), cell cycle (Teixeira and Reed, 2013; Darling et al., 2017; Gilberto and Peter, 2017), DNA repair (Alpi and Patel, 2009; Abbas and Dutta, 2011), immune response (Heaton et al., 2016; Rudnicka and Yamauchi, 2016; Manthiram et al., 2017), transcription (Zhou et al., 2018; Imam et al., 2019; Sun et al., 2019), angiogenesis (Zhang et al., 2022), metastasis (Rossi and Rossi, 2022), and apoptosis (Xu et al., 2017; Zhou et al., 2017).
Protein ubiquitination requires three different enzymes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases (Heap et al., 2017; O'Connor and Huibregtse, 2017). E1 enzymes activate the ubiquitin polypeptide in an ATP-dependent manner, and the activated forms are then conjugated to E2 enzymes through the formation of thioester bonds (Wang et al., 2018). Finally, E3 ubiquitin ligases recognize both E2 enzymes and specific target substrates that confer specificity to the system, and then the Ub can be transferred from E2 enzymes to the target substrates to complete the ubiquitination process (Rittinger and Ikeda, 2017; Yau et al., 2017). The coordination of E1, E2, and E3 enzymes earmarks target proteins with a wide variety of ubiquitin modifications such that distinct ubiquitin modifications transmit different cellular signals (Khaminets et al., 2016; Venuto and Merla, 2019). Specific ubiquitin-binding domains (UBDs) can identify ubiquitylated substrate proteins (Kristariyanto et al., 2015; Kung et al., 2019; Polykratis et al., 2019; Spiliotopoulos et al., 2019). UBDs utilize diverse mechanisms to interact with various surface patches on ubiquitin molecules or different ubiquitin linkages (Hicke et al., 2005; Dikic et al., 2009; Komander and Rape, 2012). The hydrophobic surfaces of ubiquitin molecules, such as isoleucine 36 (Ile36) and isoleucine 44 (Ile44), are the structural basis for the recognition of ubiquitination signals. Different ubiquitin chains have various spatial structures and thus expose different hydrophobic surfaces, which can be recognized by specific UBDs. Proteins containing UBDs recognize and transmit the functional signals represented by ubiquitin chains (Buetow and Huang, 2016). Similar to many other protein post-translational modifications (PTMs), ubiquitination can be preserved through cleavage by deubiquitylating enzymes (DUBs) (Herhaus and Sapkota, 2014; Nishi et al., 2014; Clague et al., 2019) (Figure 1).
The versatility of ubiquitination is determined by the complex assembly pattern of ubiquitin molecules on the target protein. Ubiquitins can be attached to one or multiple lysine residues with either a single ubiquitin molecule (mono- and multi-mono-ubiquitination, respectively) or ubiquitin polymers (poly-ubiquitination) (Eger et al., 2010; Akutsu et al., 2016; Chitale and Richly, 2017; van der Heden van Noort et al., 2017). Poly-ubiquitin chains comprising only one single linkage are often assumed to be homotypic (Jeusset and McManus, 2017; Leestemaker and Ovaa, 2017), whereas heterotypic chains adopt multiple linkages within the same polymer (branched or non-branched) (Akturk et al., 2018). In a poly-ubiquitin chain, ubiquitin moieties can be linked through any of the seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) or N-terminal methionine (Met1) (Dittmar and Winklhofer, 2019; Spit et al., 2019; Liao et al., 2022), resulting in an almost unlimited number of poly-ubiquitin chain topologies (Varadan et al., 2004; Sadowski and Sarcevic, 2010; Dondelinger et al., 2016; O'Connor and Huibregtse, 2017; Padala et al., 2017). Further complexity is added to ubiquitination when the ubiquitin polypeptide is modified by phosphorylation (Dong et al., 2017) or acetylation (Choudhary et al., 2009; Ohtake et al., 2015). Given the sophisticated assembly of protein ubiquitination, it has been often referred to as the “ubiquitin code” (Komander and Rape, 2012). Proteomics studies have shown that Lys48-linked chains are predominant in cells (>50% of all linkages), and Lys63-linked chains rank the second abundant chain form, however, researchers have begun to characterize the remain chain types, which were considered to be “atypical” ubiquitin modifications (linked through Lys6, Lys11, Lys27, Lys29, Lys33 and Met1) (Xu et al., 2009; Dammer et al., 2011; Kim et al., 2011; Wagner et al., 2011; Ziv et al., 2011).
1.2 Structural Features of Poly-Ubiquitin Chains
Poly-ubiquitin chains occur when a single ubiquitin molecule is repeatedly connected in series with another ubiquitin lysine residue. Substrate proteins can be distinguished by poly-ubiquitin chains by attaching between different types of deubiquition formations, single mono-ubiquitination events, multiple mono-ubiquitinations events, homotypic ubiquitination events and heterotypic ubiquitination events (branched and non-branched ubiquitination) (Sokratous et al., 2014; Yau and Rape, 2016; Zhao et al., 2017). This also led to the formation of eight different homotypic chains. The key distinguishing feature of how this can be achieved is the specific combination of E2/E3 enzymes, thereby triggering distinct cellular fates of substrate proteins. However, some of the reported E2/E3 enzyme combinations were not 100% specific for targeted linkage. It has been reported that two E1 enzymes were selected for ubiquitin in humans: UBA1 and UBA6 (Barghout and Schimmer, 2021). Humans also encoded 40 E2 conjugation enzymes cooperate with approximately 600 E3 ligase enzymes (Hodson et al., 2014). The E3s were categorized into three groups: RING/U-box, HECT, and RING between RING (RBR) (Wang et al., 2020).
With the assistance of E1, E2, and E3, mono-ubiquitination occurs when a single ubiquitin is attached to its target proteins, then Ub molecules are added to the model in linear ways one by one (Torres et al., 2009; Tang et al., 2011). The sequential addition model of Ub on the substrate contributes to the elongation of the Ub lines. When secondary Ub molecules are connected to specific lysine residues, they are called homotypic chains. If any of the attached adjacent Ub molecules are linked to each other by different lysine residues (mixed or branched model), a heteropic structure is formed. For homotypic chains, reports have found that different chain types are closely related to the confirmation of the structure, either “compact” or “open”. Generally, non-proteolytic ubiquitination Lys63 linkages and Met1 linkages, adopt “open” ones. Contrast to the aforementioned linkages, internal structural molecules interact with each other among degradable linkages, like Lys6, Lys11 and Lys48 linkages, and those linkages display “open” conformation. Furthermore, all ubiquitin moieties can be modified by acetylation or phosphorylation to add additional layers of complexity (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014; Ordureau et al., 2014; Ohtake et al., 2015; Swaney et al., 2015) (Figure 1).
Numerous studies have found that ubiquitin acetylation inhibits poly-ubiquitination elongation, and phosphoubiquitin leads to mitophagy. Any PTM chain can be changed to ubiquitin chains, which may prevent or facilitate ubiquitin interactions. Protein phosphorylation is linked to ubiquitination for proteasomal degradation. Reports have shown that the phosphorylation of ULK1 by MAPK1/3 kinase interacts with BTRC, which leads to subsequent proteasomal degradation and attenuates breast cancer bone metastasis (Deng et al., 2020). However, the stability of some proteins is also regulated by phosphorylation. Reports have also shown that Aurora B-mediated phosphorylation of ubiquitin specific protease 13 (USP13) at Serine 114 promoted the stability of Aurora B.
1.3 Encoding and Decoding the Ubiquitin Code
Encoding and decoding the ubiquitin code is performed by factors that recognize Ub chains and connect the substrate proteins to the downstream response (Ji and Kwon, 2017; Kwon and Ciechanover, 2017). Recognition of chains occurs through discrete domains and affinity binders with specificity for a particular Ub substrate and chain type (Fu et al., 2012; Suryadinata et al., 2014; Kniss et al., 2018; Michel et al., 2018). This complex system consists of the conjugation of diverse mono, multi-mono and polymeric chains (Rösner et al., 2015; Ji and Kwon, 2017). The interpretation of how, when, and why the ubiquitin codes are written, read, and erased emerged to be characterized. Ubiquitination is a powerful decoration process of proteins and is typically actualized by “ubiquitinase” (Zientara-Rytter and Subramani, 2019). Ubiquitin can be successfully linked to one of the seven lysine residues, all of which can be characterized as poly-ubiquitin chains (Laplantine et al., 2009; Regev et al., 2015; Bax et al., 2019). Poly-ubiquitin chains with different topologies depended upon the lysine residues (which were chosen to be attached) and the substantial chain length, determine the lucky chance of the target proteins and regulate diverse cellular processes, known as “ubiquitin code” (Dittmar and Selbach, 2017; Chatr-Aryamontri et al., 2018; Fottner et al., 2019; O'Donnell, 2019; Song and Luo, 2019). Recent discoveries have deepened our understanding of the whole picture of the ubiquitin code, the interplay between “writers” (E1/E2/E3s), “erasers” (DUBs) and “readers” (ubiquitin binding domain containing proteins) (Tanno and Komada, 2013; Heride et al., 2014; Rogerson et al., 2015; Di Lello and Hymowitz, 2016; Smeenk and Mailand, 2016). Studies have highlighted that mono-ubiquitination can be catalyzed by different E2 and E3 enzymes, acting either individually or together to determine specific substrates. Notably, the linkage specificity of E3s containing RING or U-box domains is likely dictated by E2. As for the HECT E3s class of enzymes, HECT domain swaps can activate the acceptor lysine and are sufficient to determine the linkage specificity. RBR E3s, however, are somewhat complex. RBR E3s display linkage specificity in multiple chains, Met1-, Lys63-, Lys48-, and Lys27-linked chains, as well as mono-ubiquitination, while cooperating with E2 to synthesize Lys-linked or Met1-linked chains. The Ub tag attached to a certain substrate, which is preferably achieved through the cooperation of specific E2/E3 enzyme pairs, represents a complex yet specific message encoded by the cell (Kim and Huibregtse, 2009; David et al., 2011; Turek et al., 2018). Interestingly, this is achieved by Ub receptors which are equipped with one or more UBDs (Mattern et al., 2019). Ubiquitin recognition achieved by UBDs can translate written code into specific outcomes. There are more than 20 families of UBDs that bind to different patches on Ub surrounding hydrophobic patches, either lle44 or lle36 (Dikic et al., 2009; Hendriks et al., 2018). Moreover, UBDs are able to sense unique 3D conformations of distinct chain types, which are associated with diverse biological activities, meaning that this small motif-containing protein can help recognize versatile signals and affect the desired effect (Lee et al., 2006; Penengo et al., 2006; Reyes-Turcu et al., 2006; Bomar et al., 2010). In addition, ubiquitin-interacting proteins which served as “decoders” of the ubiquitin message, participated in the downstream regulation of the ubiquitinated substrate (Heideker and Wertz, 2015; Leznicki and Kulathu, 2017; Mevissen and Komander, 2017). These “decoders” may specifically reverse the ubiquitination process or function as receptors for the transfer of the targeted substrate toward downstream signaling components and/or subcellular compartments (Hu et al., 2002; Lin et al., 2008; Komander et al., 2009). To date, 55 ubiquitin specific proteases (USPs), 14 ovarian tumor DUBs (OTUs), 10 JAMM family DUBs, 4 ubiquitin C-terminal hydrolases (UCHs) and 4 Josephin domain DUBs have been identified. Encoding and decoding ubiquitin codes are responsible for all levels of epigenetic changes, and by changing substrate protein activities, they can also activate and repress effects on gene transcription depending on their target proteins and the ubiquitin chain types, all of which are connectively related to the process of tumor proliferation (Kim and Baek, 2006; Zheng et al., 2008; Fradet-Turcotte et al., 2013; Yeh et al., 2018). Ubiquitin code signaling is frequently dysregulated in numerous cancer types and can function as a tumor suppressor or tumor promoter, suggesting a potential target for cancer therapy (Loch and Strickler, 2012; Choudhry et al., 2018; Emanuelli et al., 2019).
1.4 Physiological Functions of Non-proteolytic Poly-Ubiquitin Chains
The simplest version of ubiquitination or mono-ubiquitination confers non-degradative activities including protein localization (Yang et al., 2017), endocytosis (Shih et al., 2002; Windheim et al., 2008), trafficking, DNA repair (Xie et al., 2014; Whiteaker et al., 2018), autophagy (Chen S. et al., 2017; Zheng et al., 2020; Leng et al., 2021) and chromatin remodeling (Cole et al., 2021). When mono-ubiquitination is further modified, multiple lysine residues of the substrate are yielded to induce multi-mono-ubiquitination. Emerging investigations have found that this process connects ubiquitin with subcellular trafficking (Yin et al., 2010; Cooray et al., 2011) and immune-suppressive functions (Zhu et al., 2018). As for poly-ubiquitination, Lys48-linked poly-ubiquitination leads to the degradation of substrates (Komander and Rape, 2012). In contrast, Lys63-linked poly-ubiquitination exerts critical signaling functions in regulating protein stability, including nuclear factor κB (NF-κB) signaling (Syed et al., 2006; Wu Z. et al., 2014; Gallo et al., 2014), endocytosis (Galan and Haguenauer-Tsapis, 1997), DNA damage responses, and immune responses (Wu and Karin, 2015; Hrdinka et al., 2016; Liu et al., 2017; Paul and Wang, 2017). Emerging experiments have shown that incorrect regulation of cellular processes (either tumour inhibitors or promoters) contributes to cancer pathogenesis and progression (Shirane et al., 1999). Additionally, the cellular functions of atypical ubiquitin linkages (except Lys11-linked ubiquitin) are supposed to be non-degradable (Kulathu and Komander, 2012; Iwai, 2014, 2015; Meza Gutierrez et al., 2018) and in most cases, activities involved in Lys63-linked poly-ubiquitin chains are also considered to be non-degradable (Liu et al., 2015).
Of note, other chains also regulate specific physiological functions: Lys6-linked poly-ubiquitin was supposed to be indirectly linked to DNA damage response with the help of heterodimeric ubiquitin E3 ligase BRCA1–BARD1 (Wu-Baer et al., 2003; Wu-Baer et al., 2010). Linear (M1) chains can regulate NF-κB activation (Behrends and Harper, 2011; Chen et al., 2015; Borghi et al., 2018), whereas Lys11 linkage acts as a powerful degradation signal in heterotypic ubiquitin conjugates (Locke et al., 2014; Mevissen et al., 2016) and lys27 linkage can prompt mitochondrial depolarization and mediate translocation of the E3 ligase Parkin, which accumulates Lys27-linked linkages on mitochondrial protein voltage-dependent anion-selective channel protein1 (VDAC1). This exact Lys27-linked translocation leads to Parkinson’s disease in the presence of Parkin (Geisler et al., 2010; Glauser et al., 2011). Lys27 linkage has also been demonstrated to be related with the DNA damage response and innate immune response (Xue et al., 2018). The propagation of Wnt/β-catenin signaling through Lys29-or Lys11-linked ubiquitin chains is closely associated with cancer pathogenesis and is involved in protein ubiquitination at multiple levels (Hay-Koren et al., 2011). Several studies have reported the functions of both Lys29-and Lys33-linked chains in the regulation of AMPK-related protein kinases (Al-Hakim et al., 2008), and the Lys33 linkage is negatively regulated by T-cell antigen receptors (TCRs), which indirectly affect cellular activities in tumors (Huang et al., 2010). In this review, we will focus on recent progresses in non-degradable ubiquitin chains in tumorigenesis and the essential proteins involved in non-proteolytic ubiquitination.
2 The Roles of Non-Proteolytic Ubiquitination in Tumorigenesis
Non-proteolytic ubiquitination, including both mono-ubiquitination and poly-ubiquitination (mainly K6-, K27-, K29-, K33-, K63-, and M1-linked poly-ubiquitination), has become key regulators in a variety of cancers. How they function as signaling entities in the pathogenesis and progression of cancer will be beneficial to gain an in-depth understanding of ubiquitination (Figure 2).
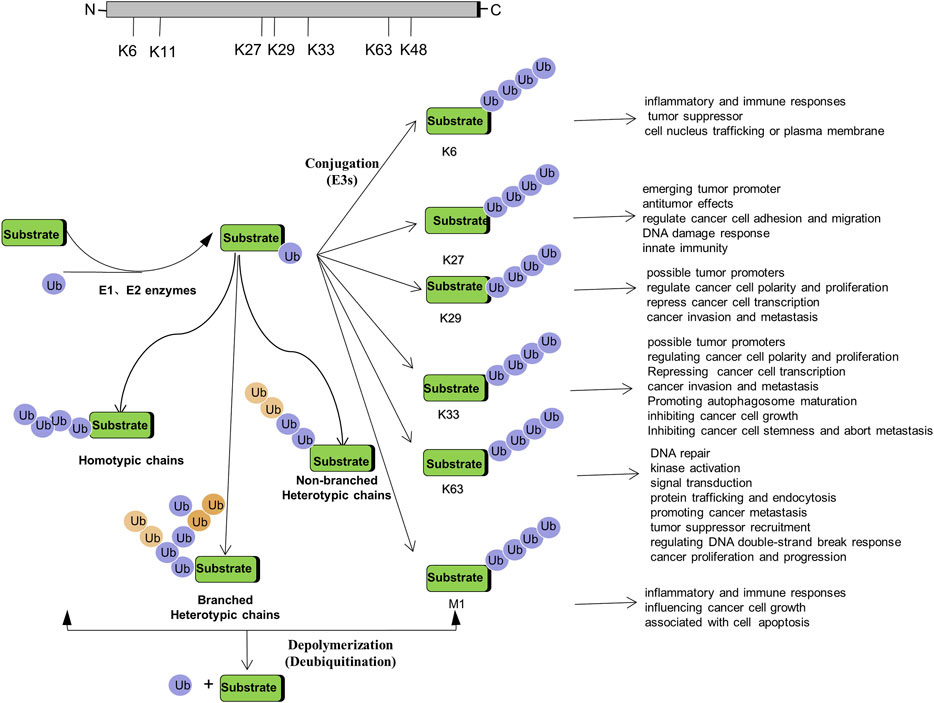
FIGURE 2. Regulation of Tumorigenesis by Non-proteolytic Ubiquitination: Linked through Lys6, Lys27, Lys 29, Lys 33, Lys 63, M1 poly-ubiquitination.
2.1 Mono-Ubiquitination
Mono-ubiquitination has been suggested to be even more dynamic than previously thought, and its functions have been deciphered by various ubiquitin-binding proteins. It has been demonstrated that the cellular functionality of ubiquitin is mediated by mono-ubiquitin and/or poly-ubiquitin. In fact, mono-ubiquitination is associated with tumorigenesis, not limited to membrane transportation, endocytosis, receptor internalization, degradation in lysosomes, and protein reprocessing (Haglund et al., 2003). BMI1 interacts with histone H2A through mono-ubiquitination, repressing multiple genes, such as INK4A/ARF, which function in the pRb and p53 pathways, thereby facilitating cancer progression (Lin et al., 2015). Interestingly, in the presence of UbE2E1, PRC1 catalyzes the mono-ubiquitination of H2A, contributing to cancer cell proliferation (Wheaton et al., 2017). Mono-ubiquitination is also involved in the process of USP22 regulating histone H2B and exhibits both oncogenic and tumor-suppresser roles in cancer development (Jeusset and McManus, 2017). In addition, reversible mono-ubiquitination activity plays an essential role in balancing TGF-β/SMAD signaling, which is involved in cancer initiation and progression (Xie et al., 2014). Mono-ubiquitination also regulates forkhead box O (FOXO) proteins, which control specific gene expression programs that are vital for slowing the onset of cancer in aging individuals (Greer and Brunet, 2008). Strikingly, FANCL cooperates with, UBE2T and catalyzes mono-ubiquitination, which participated in the regulation of Fanconi Anemia pathway, leading to chromosome instability and promoting tumorigenesis (Machida et al., 2006; Hodson et al., 2014; Miles et al., 2015; Sun et al., 2020; Wang S. et al., 2021) (Table 1, 2).
2.2 Linear (M1) Linkage
2.2.1 The LUBAC Complex Encodes the M1 Linkage
Emerging evidences connect Met1-linked ubiquitin chains to NF-κB signaling, which enables physiological regulation of inflammation and immune responses (Emmerich et al., 2011; Borghi et al., 2018). Indeed, mutations and deficiencies involved in the formation and dissolution of Met1-linked poly-ubiquitin chains have been extensively illustrated in immune-related disorders (Tokunaga and Iwai, 2012; Fiil et al., 2013; Fiil and Gyrd-Hansen, 2014). Until now, linear ubiquitin chain assembly complex (LUBAC) is the only known E3 ubiquitin ligase assembling this type of chain (Kirisako et al., 2006; Walczak et al., 2012). The multi-subunit E3 ligase comprises catalytically active hybrid organic-inorganic perovskite (HOIP, also known as RNF31) and two adaptor proteins, HOIP1L (also known as RBCK1) and SHANK-associated RH domain-interacting protein (SHARPIN) (Tokunaga et al., 2011). Both HoIL-1 and SHARPIN can co-operate with HOIP, which might be a central part of the LUBAC complex (Ikeda et al., 2011; Elton et al., 2015). The HOIP orthologue, linear ubiquitin E3 ligase (LUBEL), modifies Kenny with M1-linked linear ubiquitin chains in Drosophila and is indispensable for inflammatory responses by activating Imd pathway (Aalto et al., 2019). Additionally, HOIP, with the help of cIAP1, is recruited to the linear ubiquitination of the TNFR2 signaling complex and activates canonical NF-κB, thereby facilitating cancer progression (Borghi et al., 2018). Multiple investigations found that HOIP, presumably through M1-linked ubiquitination, was proved to be connected with sorts of malignancies, including breast and prostate cancer (Guo et al., 2015; Zhu et al., 2016). Furthermore, previous experiments highlight that HOIL-1 interacts with HOIP, which adds a Mi-linked poly-ubiquitin chain to specific NF-κB signaling proteins, suggesting its link to a diversity of immune disorders, antiviral signaling (Elton et al., 2015), iron and xenobiotic metabolism (Elton et al., 2015), apoptosis (Emmerich et al., 2011; Lewis et al., 2015; Sasaki and Iwai, 2015), and cancer (Queisser et al., 2014; Taminiau et al., 2016). Another member of the LUBAC family, SHARPIN, is a novel component of the LUBAC complex. Spontaneous mutation of this tiny gene led to dysregulation of the NF-κB signaling pathway through linear ubiquitinate of NEMO (the key modulator of NF-κB). This systematic linear ubiquitination is obvious and can induce immune system disorders in SHARPIN-deficient mice (Tokunaga et al., 2011). Interestingly, SHARPIN-containing complexes can also interact with NEMO to activate NF-κB pathway (Ikeda et al., 2011). Moreover, NEMO was identified to be modified by LUBAC, generating M1-linked chains that were recognized by the UBAN domain of NEMO, causing conformational changes in the intertwined helices of NEMO dimers (Haas et al., 2009; Belgnaoui et al., 2012; Tokunaga and Iwai, 2012; Noad et al., 2017). NF-κB signaling regulates human cellular activities in different ways and is balanced by ubiquitination and deubiquitination (Adhikari et al., 2007; Lork et al., 2017) (Table 2).
2.2.2 OTULIN Dissembles M1-Linked Poly-Ubiquitin Chains
OTULIN, a methionine 1 (M1)-specific deubiquitinase (DUB), is a rare member of the OTU family of DUBs. Its proximal ubiquitin moiety cannot break down isopeptide linkages of ubiquitin chains, but can efficiently cleave peptide bonds present in the linear chains (Keusekotten et al., 2013; Rivkin et al., 2013). OTULIN presents negative regulation in the cellular process of immune homeostasis and inflammation (Damgaard et al., 2019). Depletion of OTULIN resulted in an increase in the formation of linear Ub chains and demonstrated proteasome dysregulation as the cause of NF-κB positive activation, which in turn restricts bacterial proliferation (Takiuchi et al., 2014; van Wijk et al., 2017). Moreover, the deficiency of OTULIN led to the inability to remove M1-linked poly-ubiquitin signals, which are typically conjugated by the LUBAC, resulting in LUBAC degradation and dysregulation of TNF signaling and cell death (Tokunaga, 2013; Damgaard et al., 2016). Notably, the function of LUBAC is controlled by cylindromatosis (CYLD), which interacts with the LUBAC core subunit HOIP to generate Met1-linked ubiquitin. However, this interaction can be weakened by the Met1-Ub-specific deubiquitinase OTULIN. In addition, through the deubiquitinase function of OTULIN, LUBAC can regulate Met1-Ub to ensure an advisable response to innate immune activity (Elliott et al., 2016; Hrdinka et al., 2016). The CYLD/TRAF3 complex has also been reported to regulate NF-κB-mediated inflammation and interferon signaling, which defines a subset of head and neck cancers that harbor human papillomavirus (Chen T. et al., 2017). Notably, accumulating evidence has manifested that linear ubiquitin chains play essential roles in ensuring appropriate activity of inflammatory responses and innate immune signaling (Tokunaga and Iwai, 2012; Tokunaga, 2013; Jing et al., 2017). The linear ubiquitination of cFLIP is directly induced by RNF31, a catalytic subunit of LUBAC, at Lys-351 and Lys-353, contributing to TNFα-induced apoptosis, thereby protecting cells from apoptosis (Tang et al., 2018) (Table 3).
2.2.3 Linear Ubiquitination in Tumorigenesis
M1-linked ubiquitination, specifically N-terminal Met1-linked ubiquitination, is able to form eight different inter-ubiquitin linkages via its N-terminal methionine (M1). It can be specifically catalyzed by LUBAC. For instance, EGFR recruits PKP2 to the plasma membrane and cooperates with LUBAC (HOIP), activating linear ubiquitination of NEMO, which is critical for tumor cell proliferation (Hua et al., 2021). Also, Epsins 1/2 promotes NEMO linear ubiquitination via LUBAC, driving breast cancer development (Song et al., 2021). Moreover, LUBAC (SHARPIN) regulated β-catenin activity through linear ubiquitination, promoting gastric tumorigenesis (Zhang et al., 2021). Interestingly, OTULIN exclusively cleaves M1-linked ubiquitination and exhibits a high affinity for linear ubiquitination. LUBAC and linear ubiquitination have been found to be relevant to TNF signaling. Interestingly, OTULIN was shown to remove linear ubiquitination from LUBAC-modified proteins, which is critical for various cellular activities (Draber et al., 2015). Moreover, the deubiquitinating enzyme, CYLD, was also identified for disassembly Met1-linked-Ub (mostly the immune system). A previous report demonstrated that modification of proliferating cell nuclear antigen (PCNA) induces apoptosis and inhibits tumor growth through the linear ubiquitin chain (Qin et al., 2018).
2.3 Lys63 Linkage
2.3.1 The Writer Enzymes for the Lys63 Linkage
It has been well established that Lys63 chains (ubiquitin chains topology lysine 63 poly-ubiquitin linkages) regulate and trigger distinct cellular signaling, including kinase activation, signal transduction, protein trafficking, endocytosis and DNA repair (Spence et al., 1995; Hofmann and Pickart, 1999; Komander and Rape, 2012; Wu and Karin, 2015; Hrdinka et al., 2016). The E2 conjugating enzyme complex Ubc13/Uev1A preferentially assembles the K63-pUb chain (Smith et al., 2013; Zhang et al., 2018). Furthermore, Tax can be recruited to K63-pUb by E3 ligase RNF8 and Ubc13/Uev1A, which allows the activation of TGFβ-activating kinase 1 (TAK1), followed by multiple downstream signaling pathways such as the IKK and JNK pathways. These ultimately lead to DNA damage repair, cytokinesis, and the genomic instability in ATL cells (Ho et al., 2015; Lee et al., 2017). Similarly, tumor necrosis factor receptor associated factor 6 (TRAF6) can interact with the E2 conjunction enzyme Ubc13/Uev1A in a RING-dependent manner, catalyzing Lys63-linked TRAF6 auto-ubiquitination. This activates IKK and NF-κB, thereby promoting TAK1 and IKK to trigger spontaneous osteoclast differentiation (Lamothe et al., 2007a; Lenoir et al., 2021). Another report also showed that TRAF6, in a RING-dependent fashion, catalyzed auto-ubiquitination by conjugating with ubc13/Uev1A, activating the AKT pathway, and promoting cell migration in breast cancer (Lamothe et al., 2007b; Niu et al., 2021).
The Ubc13/Uev1A complex has been shown to conjugate Lys63-linked poly-ubiquitination of substrate proteins, which contribute to breast cancer metastasis via NF-кB signaling regulation (Wu Z. et al., 2014; 2017). It is also reported that Ubc13 can catalyze K63-linked protein poly-ubiquitination, which is indispensable for the activation of non-SMAD signaling by TAK1 and p38, whose activity controls breast cancer metastatic spread and lung colonization (Wu X. et al., 2014). Interacting with Ubc13, RNF213 mediates autoubiquitination and controls inflammatory responses and angiogenic activities (Habu and Harada, 2021). RNF8 was demonstrated to activate Ubc13 and recruit K63-linked poly-ubiquitin conjugation to histones H2A/H2AX, thus contributing to breast cancer predisposition (Vuorela et al., 2011). Moreover, RNF8 utilizes the RING domain, mediating Lys63-linked Ub chains, which is required for DNA double-strand break (DSB) signaling and the downstream BRCA1 tumor suppressor recruitment or lung cancer cell proliferation (Hodge et al., 2016; Xu et al., 2021). Inhibiting the Ubc13/Uev1A complex specifically, which is critical for Lys63-linked ubiquitination, promotes ubiquitin conjugation at the Lys147 site, thereby upregulating NF-кB signaling in multiple myeloma and other cancers (Gallo et al., 2014). UbcH6 and NEDD4 regulate angiogenesis and tumor growth (Sun et al., 2021). Ube2w accompanies the E3 ligase RNF4 function in distinct DNA repair pathways through Lys63-linked chains and BRIC6 (also named BRUCE) acting on K63-linked ubiquitinylate in unstimulated cells, which regulates the DNA double-strand break response through bridging USP8 and BRCT-repeat inhibitor of hTERT expression (BRIT1) in a deubiquitination manner (Ge et al., 2015; Maure et al., 2016). Poly-ubiquitination of histone H1 depends on Ubc13 and RNF8, which prolong pre-existing ubiquitin modifications to K63-linked chains, thereby stimulating RNF8-Ubc13 mediated DNA damage response (Mandemaker et al., 2017). SKP2 triggers non-proteolytic K63-linked ubiquitination, which is crucial for cancer initiation and progression by positively regulating cancer cell survival and glycolysis. The depletion of SKP2 restricts cancer stem cell proliferation and survival (Chan et al., 2013). However, the non-proteolytic K63-linked ubiquitination triggered by SKP2 can be reversed and modulated by debiquitinase ovarian tumor domain-containing protein 1 (OTUD1) (Yao et al., 2018) and USP10 (Liao et al., 2019). USP10 has been identified as a novel deubiquitinase of SKP2 that modulates and stabilizes SKP2. Indeed, USP10 can recognize and remove Lys63-linked ubiquitin chains from Bcr-Abl, leading to positive activities in chronic myeloid leukemia cells. OTUD1 interacts with p53 and is essential for constant stabilization of p53. Its overexpression dramatically induces the cell cycle and apoptosis (Mevissen et al., 2013; Piao et al., 2017). In addition, several more DUBs have been defined as having linkage specificity for Lys63-linked ubiquitination. CYLD, a tumor suppressor, inhibits NF-κB signaling by cleaving K63-linked ubiquitination of NEMO/IKKγ, thus reducing its stability and averting the IKK complex from phosphorylation of IκB (Chen T. et al., 2017). Furthermore, CYLD was associated with the catalytic LUBAC subunit HOIP to counteract Lys63-Ub and Met1-Ub conjugation to receptor-interacting protein kinase 2 (RIPK2), leading to restriction of innate immune signaling and cytokine production (Hrdinka et al., 2016). TRAF-binding protein domain (TRABID) was demonstrated to bind and cleave Lys63-linked ubiquitin moieties on APC tumor suppressor substrates, which led to the disruption of APC and activation of Wnt signaling in colorectal cancer cell lines (Tran et al., 2008). The USP17/TRAF2/TRAF3 complex acts to stabilize its client proteins, enhancing inflammatory responses and stemness in lung cancer cells (Lu et al., 2018). USP20 deubiquitinates TRAF6 and Tax, and may function as a key regulator of adult T cell leukemia (ATL) leukemogenesis through suppressing NF-κB activation (Yasunaga et al., 2011). In terms of actively removing Lys63-linked poly-ubiquitin chains on GβL, OTUD7B appears to be the primary regulator in governing mTOR2 integrity, which is essential for NF-κB activation in cancer biology (Wang B. et al., 2017). Further detailed analysis of Lys63-linked ubiquitination is required to better understand how ubiquitin chains function in numerous cellular events. However, identification of this linkage will provide an ideal point to understand the potential mechanism in the cellular regulation of the incoming ubiquitin research (Table 1, 2).
2.3.2 Disassembly of Lys63 Poly-Ubiquitin Chains by DUBs
Although a number of DUBs have been identified, the disassembly ability of the substrate and its physiological role, especially for Lys63 poly-ubiquitin chains, is poorly defined. Previous studies have shown that K63-specific DUBs, A20, and CYLD present the anti-tumor effects by regulating NF-κB signaling (Heyninck and Beyaert, 2005; Sun, 2010; Meng et al., 2021; Zhu et al., 2021). USP10 reduces K63-linked ubiquitination chains and functions as a tumor suppressor (He et al., 2021). Stimulated by the pro-inflammatory cytokine TNF, A20 exerts two major functions: sequential de-ubiquitination and catalyzing receptor-interacting protein (RIP), resulting in downregulation of NF-κB signaling. Absence of A20 led to hyper-accumulation of Lys63 poly-ubiquitin in NF-κB signaling. Like A20, CYLD disassembles the Lys63 poly-ubiquitin recruiting signal and degrades IKKκ. In this way, USP4 and OTUD1 cleaves K63-linked ubiquitination chains, and regulates downstream NF-κB signaling (Fan et al., 2011; Liang et al., 2013; Wu et al., 2022) (Table 3).
2.3.3 The Multifaceted Functions of Lys63 Poly-Ubiquitin Chains in Cancer
Lys63-linked chains have been described as non-degradative signals and are the core of inflammatory system and the NF-κB pathway, which is involved in either mono-ubiquitination or poly-ubiquitination. TGFβ type I receptor (TBRI) was shown to interact with TRAF6 through Lys63-dependent poly-ubiquitination in order to promote tumor invasion (Mu et al., 2011). It has also been reported that polo-like kinase 1 (PLK1) phosphorylates kruppel-like factor 4 (KLF4), leading to the recruitment of TRAF6, resulting in Lys63-linked ubiquitination and promoting nasopharyngeal tumorigenesis (Mai et al., 2019). Moreover, TRAF6 functions in hepatocarcinogenesis by regulating TRAF6/HDAC3/c-Myc signal axis. Mechanistically, TRAF6 interacts with histone deacetylase 3 (HDAC3) via Lys63-linked ubiquitin chains, which enhances c-Myc stability, and its overexpression has been proposed to facilitate HCC progression (Wu et al., 2020). Similarly, HUWE1 promotes K63-linked ubiquitinatination of c-Myc, thereby promoting retinoblastoma cell proliferation (Dai et al., 2021). It was also demonstrated that ASK1-dependent phosphorylation and lys63-linked poly-ubiquitination of LKB1 is critical for its activation. Tankyrase and RNF146 act as upstream regulators of the LKB1/AMPK pathway to promote Lys63-linked ubiquitination resulting in AMPK activation and tumor suppression (Li et al., 2019). Lys63-linked non-proteolytic ubiquitination of Hexokinase 2 by HectH9 effectively disrupts glycolysis activation and facilitates apoptosis in prostate cancer cells (Lee H. J. et al., 2019). SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) methylates Akt and functions as a scaffold to recruit JMJD2A, which then binds TRAF6 and Skp2-SCF to the Akt complex, thereby promoting tumor development in lung cancer (Wang et al., 2019). Notably, numbers of E3 ligase have been discovered to catalyze K63-linked ubiquitination chains, which are involved in activating NF-κB or Akt signaling pathway, acting as tumor promoters and facilitating tumor progression (Ji et al., 2021; Xu et al., 2021; Yu et al., 2021; Zhou et al., 2021; Zhu et al., 2021; He et al., 2022). Lys63 poly-ubiquitin chains were also found to be involved in driving the cell cycle and promoting cancer progression (Kolapalli et al., 2021). In contrast, tripartite motif-containing 31 (TRIM31) directly ubiquitinates p53 via K63-linked ubiquitination, resulting in tumor-suppressing effects (Guo et al., 2021) (Table 2).
2.4 Lys6 Linkages–Tumor Suppressors
Lys6-linked ubiquitin chains are less abundant in resting cells and their functional implications are unclear (Durcan et al., 2014; Michel et al., 2017). It has been reported that BRCA1 can be autoubiquitinated and then bound by UBX domain containing protein 1 (UBXN1) through K6-linked poly-ubiquitin chains. Interestingly, UBXN1 regulates BRCA1 expression upon ubiquitination. Autoubiquitinated forms of BRCA1 act as tumor suppressors and inhibit their enzymatic function (Wu-Baer et al., 2010). Moreover, BRCA1 auto-ubiquitination occurs in a way that the BRCA1/BARD1 complex conducts polymerization by conjugation with K6-linked polymers, which imparts cellular properties to its natural enzymatic substrates (Wu-Baer et al., 2003), further linking BRCA1 auto-ubiquitination to the tumor suppressor. C terminus HSC70-interacting protein (CHIP) was reported to bind to fas-associated protein with death domain (FADD) to induce K6-linked poly-ubiquitination of FADD, which was demonstrated to be essential for the prevention of cell death (Seo et al., 2018).
It has also been revealed that community-based learning collaborative (CBLC) assembles K6- and K11-linked poly-ubiquitin on EGFR and positively regulates its stability. The sustained activation of EGFR is largely dependent on CBLC-mediated ubiquitination, and its dysregulation is preferentially destined for membrane recycling, which plays an important role in non-small-cell lung carcinoma (NSCLC) progression (Hong et al., 2018).
Structural findings indicated that USP30 efficiently cleaves Lys6-linked Ub chains, which abrogates parkin-mediated Ub-chain formation in mitochondria. Dysfunction of this novel distal Ub-recognition mechanism is associated with physiological disorders such as hepatocellular carcinoma (Sato et al., 2017; Wang et al., 2022). Although an extraordinary progress has been made over the last 2 decades, the detailed functional consequences of Lys6 modifications require further investigation (Tables 2, 3).
2.5 Lys27 linkages—emerging Tumor Promoter
Emerging investigations have demonstrated that K27-linked poly-ubiquitination is crucial for promoting cancer development, such as facilitating cell proliferation, invasion, and metastasis (Peng et al., 2011; Yin et al., 2019). ITCH, an E3 ubiquitin ligase, was shown to generate Lys27-linked poly-ubiquitination of the transcription factor TIEG1, which inhibits TIEG1 nuclear translocation and subsequent Treg development (Peng et al., 2011). Additionally, TGF-β, in the presence of cytokine IL-6, can efficiently promote mono- and poly-ubiquitination of TIEG1 and modulate Treg/Th17 differentiation (Peng et al., 2011). Yet, it seems that tumor immunity in TIEG1−/− mice were apparently enhanced by hampering Treg development and increasing Th17 response, suggesting its pro-tumor effects (Peng et al., 2011). In the presence of proinflammatory cytokines, ITCH can catalyze BRAF to disrupt 14–3–3-mediated inhibition of BRAF kinase activity, resulting in ΜEK/ERK signaling activation. This ubiquitin function plays an essential role in supporting the proliferation and invasion abilities of melanoma cells (Yin et al., 2019). Another report connected Lys27-linked ubiquitination to melanoma cell invasive properties, in which HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 (HACE1) decorated fibronection with Lys27 Ub moieties (El-Hachem et al., 2018). As mentioned in this study, upregulation of fibronectin in turn modulates the transcription levels of integrin subunit alpha V (ITGAV) and integrin β1 (ITGB1), leading to an increased invasive power of melanoma cells (El-Hachem et al., 2018). TRAF4 was found to increase TrkA kinase activity through K27- and K29-linked ubiquitination upon nerve growth factor (NGF) stimulation, followed by the recruitment of downstream adaptor proteins and increased metastasis in prostate cancer (Singh et al., 2018).
Strikingly, reports have also revealed that Lys27-linked ubiquitination is implicated in the DNA damage response (DDR) and innate immunity. A previous study demonstrated that RNF168 can target the N-terminal tail of histones H2A/H2A.X, generating Lys27-linked ubiquitin chains. In response to DNA damage, K27 ubiquitination is the major source of PTMs that mark chromatin. Meanwhile, the activition of DDR can be inhibited by mutation of K27, which hinders the localization of 53BP1 and BRCA1 to DDR foci (Gatti et al., 2015). DDR is responsible for the recognition, signal transduction and repair of DNA damage. The inactivation of DDR will result in the accumulation of cell mutations and the increase of genomic instability, which play an essential role in cancer initiation (Klinakis et al., 2020). It is also indicated that K27 and the related DDR mediators may be the potential targets for the development of anti-tumor drugs. cGAS is subjected to K27 poly-ubiquitination by RNF185, facilitating cGAS-mediated innate immune response. TRIM-mediated ubiquitination by binding to residue K27 activates TBK1 recruitment to mitochondrial antiviral signaling (MAVS) and promotes innate immunity (Wang Q. et al., 2017). TRIM31 and TRIM40 can also mediate K27-linked ubiquitination, thereby regulating innate and adaptive immunity (Wang X. et al., 2021; Shen et al., 2021). Specifically, TRIM40 interacts with Riok3, resulting in RIG-I and MDA5 degradation through K27-linked ubiquitination, and negatively regulates innate immunity (Shen et al., 2021). Nevertheless, TRIM31 catalyzes K27-linked ubiquitination of SYK, facilitating antifungal immunity (Wang X. et al., 2021). Additionally, auto-ubiquitination of TRIM23 through K27-linked ubiquitination was found to mediate autophagy via activation of TBK1 (Sparrer et al., 2017). Moreover, E3 ubiquitin ligase Hectd3 decorates Stata3 with non-degradative K27-linked poly-ubiquitin chains and Malt1 with K27/K29-linked poly-ubiquitin chains, leading to signaling-related ubiquitination in neuroinflammation (Cho et al., 2019) (Table 3). In fact, the innate immune system plays a key role in the formation of tumor. The response of it is generally affected by a variety of immune cells and cytokines in tumor microenvironment (Wenbo and Wang, 2017). In this way, it is beneficial to clarify the regulatory effects of K27 on tumor innate immunity for understanding the mechanisms of cancer.
2.6 Lys29 and Lys33 linkage—possible Tumor Promoters
Recent proteomic data have identified the role of Lys29-and Lys33-linked ubiquitin chains in various biological processes, including the control of AMPK-mediated mitochondrial function and Wnt-induced transcription signaling (Jiang et al., 2015; Nusse and Clevers, 2017). AMPK-related kinases, AMPK-related kinase 5 (ARK5, also known as NUAK1) and microtubule-affinity-regulating kinase 4 (MARK4), which are mediated by unconventional Lys29/Lys33 linkage, are involved in cell polarity and proliferation (Al-Hakim et al., 2008). One of the underlying mechanisms is that USP9X specifically identifies Lys29/Lys33-conjugated ubiquitin chains on NUAK1 and MARK4 (Al-Hakim et al., 2008). However, it should be noted that the ubiquitination of NUAK1 and MARK4 suppresses their phosphorylations rather than restores their stability and facilitates LKB1 activation. Interestingly, the TRABID core domain N-terminal Npl4-like zinc finger (NAZF1) preferentially hydrolyzes K29/K33-linked diUb, and this novel AnkUBD displays TRABID linkage specificity (Licchesi et al., 2011). Additionally, TRABID interacts with APC tumor suppressor protein, recruits TCF target genes, and activates their transcription in colorectal cancer cells (Tran et al., 2008). The OUT family DUB TRABID preferentially abolishes Smad ubiquitination regulatory factor 1-induced K29/K33-linked poly-ubiquitin chains from UV radiation resistance associated gene (UVRAG), thereby promoting autophagosome maturation and inhibiting cell proliferation in hepatocellular cancer (Feng et al., 2019). Considering its linkage specificity, it would be insightful to explore the potential positive regulation of TRABID in cancer tumorigenesis via these two pathways.
K29-linked ubiquitin chains play important roles in driving cancer invasion and metastasis, as well as in the positive regulation of immunity (Singh et al., 2018; Cho et al., 2019; Gao et al., 2021). Several studies have also manifested that, with the assistance of Cbl-b and ITCH, T cell receptor-zeta (TCR-zeta) was decorated with a K33 linage, accompanied by positively activated T cells and immune responses (Huang et al., 2010). In addition, OTUD1 directly deubiquitinates the inhibitor SMAD7 of TGF-β pathway and aborates Lys33-linked poly-ubiquitin chains, which inhibits cell stemness and suppresses metastasis (Zhang et al., 2017). Acetylation of GLDC inhibits its enzymatic activity and facilitates K33-linked ubiquitination by NF-X1, thereby suppressing glioma tumor growth (Liu et al., 2021). Ultimately, Lys29-and Lys33-linked ubiquitin chains appear to have more complicated cellular functions that remain to be characterized (Table 2, 3).
2.7 Mixed Linkage and Branched Poly-Ubiquitination
Mixed linkage chains send mixed signaling messages that can be identified by different linkage-specific receptors. Our understanding of the mixed linkages and branched poly-ubiquitin is limited. Previous reports have elucidated that Tax in combination with UbcH2, Uhc5c, or UbcH7, can catalyze the construction of free mixed-linkage poly-ubiquitin chains, which are responsible for IKK-NF-κB activation and induction of T cell transformation (Wang et al., 2016). Furthermore, Brcc 36 isopeptidase complex (BRISC), the JAMM/MPN + family of DUBs, preferentially cleaves K63 linkages within mixed-linkage chains. In addition, RING1B was found to generate atypical mixed poly-ubiquitin chains and mediate mono-ubiquitination of H2A (Ben-Saadon et al., 2006). A recent study showed that SPOP triggered mixed-linkage ubiquitination of MyD88 in human lymphoma cells and mouse HSCs, suggesting that the SPOP-MyD88 pathway plays a critical role in hematopoietic neoplasms (Jin et al., 2020).
3 Conclusion and Furture Perspectives
Non-proteolytic ubiquitination, the molecular switch in cell fate regulation, plays a crucial role in post-translational protein modifications. Diverse ubiquitination enzymes are essential for ubiquitination linkages that are necessary for normal metabolism and physiological functions. Meanwhile, it is also the root cause of physiological disorders, such as cancer. The aberrant regulation of the UPS is typically achieved by ubiquitination enzymes, DUBs, 20S proteasome catalytic core particles and 19S proteasome regulatory particles (Rape, 2018). In general, specific ubiquitination enzymes determine ubiquitin linking with one of the seven lysine residues to form distinctive styles of poly-ubiquitin chains, deciding the fate of substrate proteins. It is noteworthy that the ubiquitin proteasome system functions as a theoretical target for drug screening, and the study of ubiquitination will provide more insights into the development of anti-tumor drugs. Recently, the quantities of E3s inhibitors have already been processed in preclinical models of cancer immunotherapy. It has been shown that E3 enzyme Smac mimetics (SMs) were the promising immune modulators for cancer therapy as the antagonists targeting E3 ligases IAPs (Cossu et al., 2019). The small molecule inhibitor AMG-232, targeting another E3-ubiquitin ligase oncogenic mouse double minute 2 homolog (MDM2), was shown to strengthen T cell killing of cancer cells, especially when combined with an anti-PD-1 monoclonal antibody (Sahin et al., 2020). Several other proteasome inhibitors have been confirmed to be clinically effective against malignancies as well. The neddylation (NAE) inhibitor pevonedistat has already been tested in multiple clinical trials and has shown positive effects in patients with AML or advanced solid tumors (Barghout and Schimmer, 2021). Small molecule inhibitors based on deubiquitinase have been widely used in experimental anti-tumor therapy, most of which are still in the preclinical research stage. Due to dose-limiting toxicity, the first deubiquitinase inhibitor VLX1570 was terminated in the clinical trial phase. No other deubiquitinase inhibitors have been approved for clinical studies since then. Also, the discovered deubiquitinase inhibitors related to tumor treatment are mainly concentrated in the USP family, and the relationship between the inhibitors of non-USP family members and the treatment of malignant tumors needs to be further studied (Zhang et al., 2022). Interestingly, the innovative approaches of proteolysis targeting chimeras (PROTACs) and molecular glues might facilitate clinical cancer therapy (Cruz Walma et al., 2022; Kung and Weber, 2022; Zhou and Xu, 2022). Consequently, it is essential further to investigate the role of ubiquitination enzymes in tumorigenesis. Meanwhile, targeting different ubiquitination, including K6-, K27-, K29-, K33-, K63-, and M1-linked poly-ubiquitination, may also be one of the directions for cancer drug discovery. Although great progresses have been achieved with the development of anti-cancer drugs aimed at the UPS, numerous challenges still stand in the way. Some candidate inhibitors have emerged as drug resistant or have limited efficacy in patients. Despite these questions, the discovery of new drugs targeting single or multiple segments of the UPS is still worthy of future research.
Author Contributions
Conceptualization, SJ and JH; writing original draft preparation, XY; writing review and editing, QL, FL, SJ and JH; visualization: XT and TY; supervision: SJ and JH; funding acquisition: SJ and XY. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (grant nos. 81873249 and 82074360) and the Young Taishan Scholars Program of Shandong Province (grant no. tsqn201909200) and Shandong Province Medicine and Health Project (grant no2019WS091).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aalto, A. L., Mohan, A. K., Schwintzer, L., Kupka, S., Kietz, C., and Walczak, H. (2019). M1-linked Ubiquitination by LUBEL Is Required for Inflammatory Responses to Oral Infection in Drosophila. Cell Death Differ. 26 (5), 860–876. doi:10.1038/s41418-018-0164-x
Abbas, T., and Dutta, A. (2011). CRL4Cdt2: Master Coordinator of Cell Cycle Progression and Genome Stability. Cell Cycle 10 (2), 241–249. doi:10.4161/cc.10.2.14530
Adhikari, A., Xu, M., and Chen, Z. J. (2007). Ubiquitin-mediated Activation of TAK1 and IKK. Oncogene 26 (22), 3214–3226. doi:10.1038/sj.onc.1210413
Adhikary, S., Marinoni, F., Hock, A., Hulleman, E., Popov, N., Beier, R., et al. (2005). The Ubiquitin Ligase HectH9 Regulates Transcriptional Activation by Myc and Is Essential for Tumor Cell Proliferation. Cell 123 (3), 409–421. doi:10.1016/j.cell.2005.08.016
Ahmed, N., Zeng, M., Sinha, I., Polin, L., Wei, W. Z., Rathinam, C., et al. (2011). The E3 Ligase Itch and Deubiquitinase Cyld Act Together to Regulate Tak1 and Inflammation. Nat. Immunol. 12 (12), 1176–1183. doi:10.1038/ni.2157
Akturk, A., Wasilko, D. J., Wu, X., Liu, Y., Zhang, Y., Qiu, J., et al. (2018). Mechanism of Phosphoribosyl-Ubiquitination Mediated by a Single Legionella Effector. Nature 557 (7707), 729–733. doi:10.1038/s41586-018-0147-6
Akutsu, M., Dikic, I., and Bremm, A. (2016). Ubiquitin Chain Diversity at a Glance. J. Cell Sci. 129 (5), 875–880. doi:10.1242/jcs.183954
Al-Hakim, A. K., Zagorska, A., Chapman, L., Deak, M., Peggie, M., and Alessi, D. R. (2008). Control of AMPK-Related Kinases by USP9X and Atypical Lys(29)/Lys(33)-Linked Polyubiquitin Chains. Biochem. J. 411 (2), 249–260. doi:10.1042/bj20080067
Ali, A. M., Pradhan, A., Singh, T. R., Du, C., Li, J., Wahengbam, K., et al. (2012). FAAP20: a Novel Ubiquitin-Binding FA Nuclear Core-Complex Protein Required for Functional Integrity of the FA-BRCA DNA Repair Pathway. Blood 119 (14), 3285–3294. doi:10.1182/blood-2011-10-385963
Alpi, A. F., and Patel, K. J. (2009). Monoubiquitylation in the Fanconi Anemia DNA Damage Response Pathway. DNA Repair (Amst) 8 (4), 430–435. doi:10.1016/j.dnarep.2009.01.019
An, J., Mo, D., Liu, H., Veena, M. S., Srivatsan, E. S., Massoumi, R., et al. (2008). Inactivation of the CYLD Deubiquitinase by HPV E6 Mediates Hypoxia-Induced NF-kappaB Activation. Cancer Cell 14 (5), 394–407. doi:10.1016/j.ccr.2008.10.007
Barghout, S. H., and Schimmer, A. D. (2021). E1 Enzymes as Therapeutic Targets in Cancer. Pharmacol. Rev. 73 (1), 1–58. doi:10.1124/pharmrev.120.000053
Bax, M., McKenna, J., Do-Ha, D., Stevens, C. H., Higginbottom, S., Balez, R., et al. (2019). The Ubiquitin Proteasome System Is a Key Regulator of Pluripotent Stem Cell Survival and Motor Neuron Differentiation. Cells 8 (6). doi:10.3390/cells8060581
Behrends, C., and Harper, J. W. (2011). Constructing and Decoding Unconventional Ubiquitin Chains. Nat. Struct. Mol. Biol. 18 (5), 520–528. doi:10.1038/nsmb.2066
Belgnaoui, S. M., Paz, S., Samuel, S., Goulet, M. L., Sun, Q., Kikkert, M., et al. (2012). Linear Ubiquitination of NEMO Negatively Regulates the Interferon Antiviral Response through Disruption of the MAVS-TRAF3 Complex. Cell Host Microbe 12 (2), 211–222. doi:10.1016/j.chom.2012.06.009
Ben-Saadon, R., Zaaroor, D., Ziv, T., and Ciechanover, A. (2006). The Polycomb Protein Ring1B Generates Self Atypical Mixed Ubiquitin Chains Required for its In Vitro Histone H2A Ligase Activity. Mol. Cell 24 (5), 701–711. doi:10.1016/j.molcel.2006.10.022
Bhatnagar, S., Gazin, C., Chamberlain, L., Ou, J., Zhu, X., Tushir, J. S., et al. (2014). TRIM37 Is a New Histone H2A Ubiquitin Ligase and Breast Cancer Oncoprotein. Nature 516 (7529), 116–120. doi:10.1038/nature13955
Bomar, M. G., D'Souza, S., Bienko, M., Dikic, I., Walker, G. C., and Zhou, P. (2010). Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases Iota and Rev1. Mol. Cell 37 (3), 408–417. doi:10.1016/j.molcel.2009.12.038
Borghi, A., Haegman, M., Fischer, R., Carpentier, I., Bertrand, M. J. M., Libert, C., et al. (2018). The E3 Ubiquitin Ligases HOIP and cIAP1 Are Recruited to the TNFR2 Signaling Complex and Mediate TNFR2-Induced Canonical NF-Κb Signaling. Biochem. Pharmacol. 153, 292–298. doi:10.1016/j.bcp.2018.01.039
Brickner, J. R., Soll, J. M., Lombardi, P. M., Vagbo, C. B., Mudge, M. C., Oyeniran, C., et al. (2017). A Ubiquitin-dependent Signalling axis Specific for ALKBH-Mediated DNA Dealkylation Repair. Nature 551 (7680), 389–393. doi:10.1038/nature24484
Buetow, L., and Huang, D. T. (2016). Structural Insights into the Catalysis and Regulation of E3 Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 17 (10), 626–642. doi:10.1038/nrm.2016.91
Cai, F., Chen, P., Chen, L., Biskup, E., Liu, Y., Chen, P. C., et al. (2014). Human RAD6 Promotes G1-S Transition and Cell Proliferation through Upregulation of Cyclin D1 Expression. PLoS One 9 (11), e113727. doi:10.1371/journal.pone.0113727
Chakraborty, A., Diefenbacher, M. E., Mylona, A., Kassel, O., and Behrens, A. (2015). The E3 Ubiquitin Ligase Trim7 Mediates C-Jun/AP-1 Activation by Ras Signalling. Nat. Commun. 6, 6782. doi:10.1038/ncomms7782
Chan, C. H., Morrow, J. K., Li, C. F., Gao, Y., Jin, G., Moten, A., et al. (2013). Pharmacological Inactivation of Skp2 SCF Ubiquitin Ligase Restricts Cancer Stem Cell Traits and Cancer Progression. Cell 154 (3), 556–568. doi:10.1016/j.cell.2013.06.048
Chang, J. H., Xiao, Y., Hu, H., Jin, J., Yu, J., Zhou, X., et al. (2012). Ubc13 Maintains the Suppressive Function of Regulatory T Cells and Prevents Their Conversion into Effector-like T Cells. Nat. Immunol. 13 (5), 481–490. doi:10.1038/ni.2267
Charlaftis, N., Suddason, T., Wu, X., Anwar, S., Karin, M., and Gallagher, E. (2014). The MEKK1 PHD Ubiquitinates TAB1 to Activate MAPKs in Response to Cytokines. EMBO J. 33 (21), 2581–2596. doi:10.15252/embj.201488351
Chatr-Aryamontri, A., van der Sloot, A., and Tyers, M. (2018). At Long Last, a C-Terminal Bookend for the Ubiquitin Code. Mol. Cell 70 (4), 568–571. doi:10.1016/j.molcel.2018.05.006
Chen, M., Zhao, Z., Meng, Q., Liang, P., Su, Z., Wu, Y., et al. (2020). TRIM14 Promotes Noncanonical NF-kappaB Activation by Modulating P100/p52 Stability via Selective Autophagy. Adv. Sci. (Weinh) 7 (1), 1901261. doi:10.1002/advs.201901261
Chen, S., Jing, Y., Kang, X., Yang, L., Wang, D. L., Zhang, W., et al. (2017a). Histone H2B Monoubiquitination Is a Critical Epigenetic Switch for the Regulation of Autophagy. Nucleic Acids Res. 45 (3), 1144–1158. doi:10.1093/nar/gkw1025
Chen, T., Zhang, J., Chen, Z., and Van Waes, C. (2017b). Genetic Alterations in TRAF3 and CYLD that Regulate Nuclear Factor kappaB and Interferon Signaling Define Head and Neck Cancer Subsets Harboring Human Papillomavirus. Cancer 123 (10), 1695–1698. doi:10.1002/cncr.30659
Chen, Y., He, L., Peng, Y., Shi, X., Chen, J., Zhong, J., et al. (2015). The Hepatitis C Virus Protein NS3 Suppresses TNF-α-Stimulated Activation of NF-Κb by Targeting LUBAC. Sci. Signal 8 (403), ra118. doi:10.1126/scisignal.aab2159
Chitale, S., and Richly, H. (2017). Timing of DNA Lesion Recognition: Ubiquitin Signaling in the NER Pathway. Cell Cycle 16 (2), 163–171. doi:10.1080/15384101.2016.1261227
Cho, J. J., Xu, Z., Parthasarathy, U., Drashansky, T. T., Helm, E. Y., Zuniga, A. N., et al. (2019). Hectd3 Promotes Pathogenic Th17 Lineage through Stat3 Activation and Malt1 Signaling in Neuroinflammation. Nat. Commun. 10 (1), 701. doi:10.1038/s41467-019-08605-3
Choudhary, C., Kumar, C., Gnad, F., Nielsen, M. L., Rehman, M., Walther, T. C., et al. (2009). Lysine Acetylation Targets Protein Complexes and Co-regulates Major Cellular Functions. Science 325 (5942), 834–840. doi:10.1126/science.1175371
Choudhry, H., Zamzami, M. A., Omran, Z., Wu, W., Mousli, M., Bronner, C., et al. (2018). Targeting microRNA/UHRF1 Pathways as a Novel Strategy for Cancer Therapy. Oncol. Lett. 15 (1), 3–10. doi:10.3892/ol.2017.7290
Clague, M. J., Urbé, S., and Komander, D. (2019). Breaking the Chains: Deubiquitylating Enzyme Specificity Begets Function. Nat. Rev. Mol. Cell Biol. 20 (6), 338–352. doi:10.1038/s41580-019-0099-1
Cole, A. J., Dickson, K. A., Liddle, C., Stirzaker, C., Shah, J. S., Clifton-Bligh, R., et al. (2021). Ubiquitin Chromatin Remodelling after DNA Damage Is Associated with the Expression of Key Cancer Genes and Pathways. Cell Mol. Life Sci. 78 (3), 1011–1027. doi:10.1007/s00018-020-03552-5
Cooray, S. N., Guasti, L., and Clark, A. J. (2011). The E3 Ubiquitin Ligase Mahogunin Ubiquitinates the Melanocortin 2 Receptor. Endocrinology 152 (11), 4224–4231. doi:10.1210/en.2011-0147
Cossu, F., Milani, M., Mastrangelo, E., and Lecis, D. (2019). Targeting the BIR Domains of Inhibitor of Apoptosis (IAP) Proteins in Cancer Treatment. Comput. Struct. Biotechnol. J. 17, 142–150. doi:10.1016/j.csbj.2019.01.009
Cruz Walma, D. A., Chen, Z., Bullock, A. N., and Yamada, K. M. (2022). Ubiquitin Ligases: Guardians of Mammalian Development. Nat. Rev. Mol. Cell Biol. 23 (5), 350–367. doi:10.1038/s41580-021-00448-5
Dai, H., Zeng, W., Zeng, W., Yan, M., Jiang, P., Li, Y., et al. (2021). HELZ2 Promotes K63-Linked Polyubiquitination of C-Myc to Induce Retinoblastoma Tumorigenesis. Med. Oncol. 39 (1), 11. doi:10.1007/s12032-021-01603-w
Damgaard, R. B., Elliott, P. R., Swatek, K. N., Maher, E. R., Stepensky, P., Elpeleg, O., et al. (2019). OTULIN Deficiency in ORAS Causes Cell Type-specific LUBAC Degradation, Dysregulated TNF Signalling and Cell Death. EMBO Mol. Med. 11 (3). doi:10.15252/emmm.201809324
Damgaard, R. B., Walker, J. A., Marco-Casanova, P., Morgan, N. V., Titheradge, H. L., Elliott, P. R., et al. (2016). The Deubiquitinase OTULIN Is an Essential Negative Regulator of Inflammation and Autoimmunity. Cell 166 (5), 1215–1230. doi:10.1016/j.cell.2016.07.019
Dammer, E. B., Na, C. H., Xu, P., Seyfried, N. T., Duong, D. M., Cheng, D., et al. (2011). Polyubiquitin Linkage Profiles in Three Models of Proteolytic Stress Suggest the Etiology of Alzheimer Disease. J. Biol. Chem. 286 (12), 10457–10465. doi:10.1074/jbc.M110.149633
Darling, S., Fielding, A. B., Sabat-Pośpiech, D., Prior, I. A., and Coulson, J. M. (2017). Regulation of the Cell Cycle and Centrosome Biology by Deubiquitylases. Biochem. Soc. Trans. 45 (5), 1125–1136. doi:10.1042/bst20170087
David, Y., Ternette, N., Edelmann, M. J., Ziv, T., Gayer, B., Sertchook, R., et al. (2011). E3 Ligases Determine Ubiquitination Site and Conjugate Type by Enforcing Specificity on E2 Enzymes. J. Biol. Chem. 286 (51), 44104–44115. doi:10.1074/jbc.M111.234559
Deng, R., Zhang, H. L., Huang, J. H., Cai, R. Z., Wang, Y., Chen, Y. H., et al. (2020). MAPK1/3 Kinase-dependent ULK1 Degradation Attenuates Mitophagy and Promotes Breast Cancer Bone Metastasis. Autophagy, 1–19. doi:10.1080/15548627.2020.1850609
Di Lello, P., and Hymowitz, S. G. (2016). Unveiling the Structural and Dynamic Nature of the Ubiquitin Code. Structure 24 (4), 498–499. doi:10.1016/j.str.2016.03.013
Dikic, I., Wakatsuki, S., and Walters, K. J. (2009). Ubiquitin-binding Domains - from Structures to Functions. Nat. Rev. Mol. Cell Biol. 10 (10), 659–671. doi:10.1038/nrm2767
Dittmar, G., and Selbach, M. (2017). Deciphering the Ubiquitin Code. Mol. Cell 65 (5), 779–780. doi:10.1016/j.molcel.2017.02.011
Dittmar, G., and Winklhofer, K. F. (2019). Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification. Front. Chem. 7, 915. doi:10.3389/fchem.2019.00915
Dondelinger, Y., Darding, M., Bertrand, M. J., and Walczak, H. (2016). Poly-ubiquitination in TNFR1-Mediated Necroptosis. Cell Mol. Life Sci. 73 (11-12), 2165–2176. doi:10.1007/s00018-016-2191-4
Dong, X., Gong, Z., Lu, Y. B., Liu, K., Qin, L. Y., Ran, M. L., et al. (2017). Ubiquitin S65 Phosphorylation Engenders a pH-Sensitive Conformational Switch. Proc. Natl. Acad. Sci. U. S. A. 114 (26), 6770–6775. doi:10.1073/pnas.1705718114
Draber, P., Kupka, S., Reichert, M., Draberova, H., Lafont, E., de Miguel, D., et al. (2015). LUBAC-recruited CYLD and A20 Regulate Gene Activation and Cell Death by Exerting Opposing Effects on Linear Ubiquitin in Signaling Complexes. Cell Rep. 13 (10), 2258–2272. doi:10.1016/j.celrep.2015.11.009
Durcan, T. M., Tang, M. Y., Pérusse, J. R., Dashti, E. A., Aguileta, M. A., McLelland, G. L., et al. (2014). USP8 Regulates Mitophagy by Removing K6-Linked Ubiquitin Conjugates from Parkin. Embo J. 33 (21), 2473–2491. doi:10.15252/embj.201489729
Eger, S., Scheffner, M., Marx, A., and Rubini, M. (2010). Synthesis of Defined Ubiquitin Dimers. J. Am. Chem. Soc. 132 (46), 16337–16339. doi:10.1021/ja1072838
El-Hachem, N., Habel, N., Naiken, T., Bzioueche, H., Cheli, Y., Beranger, G. E., et al. (2018). Uncovering and Deciphering the Pro-invasive Role of HACE1 in Melanoma Cells. Cell Death Differ. 25 (11), 2010–2022. doi:10.1038/s41418-018-0090-y
Elliott, P. R., Leske, D., Hrdinka, M., Bagola, K., Fiil, B. K., McLaughlin, S. H., et al. (2016). SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling. Mol. Cell 63 (6), 990–1005. doi:10.1016/j.molcel.2016.08.001
Elton, L., Carpentier, I., Verhelst, K., Staal, J., and Beyaert, R. (2015). The Multifaceted Role of the E3 Ubiquitin Ligase HOIL-1: beyond Linear Ubiquitination. Immunol. Rev. 266 (1), 208–221. doi:10.1111/imr.12307
Emanuelli, A., Manikoth Ayyathan, D., Koganti, P., Shah, P. A., Apel-Sarid, L., Paolini, B., et al. (2019). Altered Expression and Localization of Tumor Suppressive E3 Ubiquitin Ligase SMURF2 in Human Prostate and Breast Cancer. Cancers (Basel) 11 (4). doi:10.3390/cancers11040556
Emmerich, C. H., Schmukle, A. C., and Walczak, H. (2011). The Emerging Role of Linear Ubiquitination in Cell Signaling. Sci. Signal 4 (204), re5. doi:10.1126/scisignal.2002187
Fan, L., Xu, S., Zhang, F., Cui, X., Fazli, L., Gleave, M., et al. (2020). Histone Demethylase JMJD1A Promotes Expression of DNA Repair Factors and Radio-Resistance of Prostate Cancer Cells. Cell Death Dis. 11 (4), 214. doi:10.1038/s41419-020-2405-4
Fan, Y. H., Yu, Y., Mao, R. F., Tan, X. J., Xu, G. F., Zhang, H., et al. (2011). USP4 Targets TAK1 to Downregulate TNFalpha-Induced NF-kappaB Activation. Cell Death Differ. 18 (10), 1547–1560. doi:10.1038/cdd.2011.11
Feng, X., Jia, Y., Zhang, Y., Ma, F., Zhu, Y., Hong, X., et al. (2019). Ubiquitination of UVRAG by SMURF1 Promotes Autophagosome Maturation and Inhibits Hepatocellular Carcinoma Growth. Autophagy 15 (7), 1130–1149. doi:10.1080/15548627.2019.1570063
Fiil, B. K., Damgaard, R. B., Wagner, S. A., Keusekotten, K., Fritsch, M., Bekker-Jensen, S., et al. (2013). OTULIN Restricts Met1-Linked Ubiquitination to Control Innate Immune Signaling. Mol. Cell 50 (6), 818–830. doi:10.1016/j.molcel.2013.06.004
Fiil, B. K., and Gyrd-Hansen, M. (2014). Met1-linked Ubiquitination in Immune Signalling. Febs J. 281 (19), 4337–4350. doi:10.1111/febs.12944
Fottner, M., Brunner, A. D., Bittl, V., Horn-Ghetko, D., Jussupow, A., Kaila, V. R. I., et al. (2019). Site-specific Ubiquitylation and SUMOylation Using Genetic-Code Expansion and Sortase. Nat. Chem. Biol. 15 (3), 276–284. doi:10.1038/s41589-019-0227-4
Fradet-Turcotte, A., Canny, M. D., Escribano-Díaz, C., Orthwein, A., Leung, C. C., Huang, H., et al. (2013). 53BP1 Is a Reader of the DNA-Damage-Induced H2A Lys 15 Ubiquitin Mark. Nature 499 (7456), 50–54. doi:10.1038/nature12318
Fu, Q. S., Song, A. X., and Hu, H. Y. (2012). Structural Aspects of Ubiquitin Binding Specificities. Curr. Protein Pept. Sci. 13 (5), 482–489. doi:10.2174/138920312802430581
Galan, J. M., and Haguenauer-Tsapis, R. (1997). Ubiquitin Lys63 Is Involved in Ubiquitination of a Yeast Plasma Membrane Protein. Embo J. 16 (19), 5847–5854. doi:10.1093/emboj/16.19.5847
Gallo, L. H., Meyer, A. N., Motamedchaboki, K., Nelson, K. N., Haas, M., and Donoghue, D. J. (2014). Novel Lys63-Linked Ubiquitination of IKKβ Induces STAT3 Signaling. Cell Cycle 13 (24), 3964–3976. doi:10.4161/15384101.2014.988026
Gao, P., Ma, X., Yuan, M., Yi, Y., Liu, G., Wen, M., et al. (2021). E3 Ligase Nedd4l Promotes Antiviral Innate Immunity by Catalyzing K29-Linked Cysteine Ubiquitination of TRAF3. Nat. Commun. 12 (1), 1194. doi:10.1038/s41467-021-21456-1
Gatti, M., Pinato, S., Maiolica, A., Rocchio, F., Prato, M. G., Aebersold, R., et al. (2015). RNF168 Promotes Noncanonical K27 Ubiquitination to Signal DNA Damage. Cell Rep. 10 (2), 226–238. doi:10.1016/j.celrep.2014.12.021
Ge, C., Che, L., Ren, J., Pandita, R. K., Lu, J., Li, K., et al. (2015). BRUCE Regulates DNA Double-Strand Break Response by Promoting USP8 Deubiquitination of BRIT1. Proc. Natl. Acad. Sci. U. S. A. 112 (11), E1210–E1219. doi:10.1073/pnas.1418335112
Geisler, S., Holmström, K. M., Skujat, D., Fiesel, F. C., Rothfuss, O. C., Kahle, P. J., et al. (2010). PINK1/Parkin-mediated Mitophagy Is Dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12 (2), 119–131. doi:10.1038/ncb2012
Gilberto, S., and Peter, M. (2017). Dynamic Ubiquitin Signaling in Cell Cycle Regulation. J. Cell Biol. 216 (8), 2259–2271. doi:10.1083/jcb.201703170
Glauser, L., Sonnay, S., Stafa, K., and Moore, D. J. (2011). Parkin Promotes the Ubiquitination and Degradation of the Mitochondrial Fusion Factor Mitofusin 1. J. Neurochem. 118 (4), 636–645. doi:10.1111/j.1471-4159.2011.07318.x
Greer, E. L., and Brunet, A. (2008). FOXO Transcription Factors in Ageing and Cancer. Acta Physiol. (Oxf) 192 (1), 19–28. doi:10.1111/j.1748-1716.2007.01780.x
Guo, J., Liu, X., and Wang, M. (2015). miR-503 Suppresses Tumor Cell Proliferation and Metastasis by Directly Targeting RNF31 in Prostate Cancer. Biochem. Biophys. Res. Commun. 464 (4), 1302–1308. doi:10.1016/j.bbrc.2015.07.127
Guo, Y., Li, Q., Zhao, G., Zhang, J., Yuan, H., Feng, T., et al. (2021). Loss of TRIM31 Promotes Breast Cancer Progression through Regulating K48- and K63-Linked Ubiquitination of P53. Cell Death Dis. 12 (10), 945. doi:10.1038/s41419-021-04208-3
Ha, K., Ma, C., Lin, H., Tang, L., Lian, Z., Zhao, F., et al. (2017). The Anaphase Promoting Complex Impacts Repair Choice by Protecting Ubiquitin Signalling at DNA Damage Sites. Nat. Commun. 8, 15751. doi:10.1038/ncomms15751
Haas, T. L., Emmerich, C. H., Gerlach, B., Schmukle, A. C., Cordier, S. M., Rieser, E., et al. (2009). Recruitment of the Linear Ubiquitin Chain Assembly Complex Stabilizes the TNF-R1 Signaling Complex and Is Required for TNF-Mediated Gene Induction. Mol. Cell 36 (5), 831–844. doi:10.1016/j.molcel.2009.10.013
Habu, T., and Harada, K. H. (2021). UBC13 Is an RNF213-Associated E2 Ubiquitin-Conjugating Enzyme, and Lysine 63-linked Ubiquitination by the RNF213-UBC13 axis Is Responsible for Angiogenic Activity. FASEB Bioadv 3 (4), 243–258. doi:10.1096/fba.2019-00092
Haglund, K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P. P., and Dikic, I. (2003). Multiple Monoubiquitination of RTKs Is Sufficient for Their Endocytosis and Degradation. Nat. Cell Biol. 5 (5), 461–466. doi:10.1038/ncb983
Hay-Koren, A., Caspi, M., Zilberberg, A., and Rosin-Arbesfeld, R. (2011). The EDD E3 Ubiquitin Ligase Ubiquitinates and Up-Regulates Beta-Catenin. Mol. Biol. Cell 22 (3), 399–411. doi:10.1091/mbc.E10-05-0440
He, M., Chaurushiya, M. S., Webster, J. D., Kummerfeld, S., Reja, R., Chaudhuri, S., et al. (2019). Intrinsic Apoptosis Shapes the Tumor Spectrum Linked to Inactivation of the Deubiquitinase BAP1. Science 364 (6437), 283–285. doi:10.1126/science.aav4902
He, Y., Jiang, S., Mao, C., Zheng, H., Cao, B., Zhang, Z., et al. (2021). The Deubiquitinase USP10 Restores PTEN Activity and Inhibits Non-small Cell Lung Cancer Cell Proliferation. J. Biol. Chem. 297 (3), 101088. doi:10.1016/j.jbc.2021.101088
He, Y. M., Zhou, X. M., Jiang, S. Y., Zhang, Z. B., Cao, B. Y., Liu, J. B., et al. (2022). TRIM25 Activates AKT/mTOR by Inhibiting PTEN via K63-Linked Polyubiquitination in Non-small Cell Lung Cancer. Acta Pharmacol. Sin. 43 (3), 681–691. doi:10.1038/s41401-021-00662-z
Heap, R. E., Gant, M. S., Lamoliatte, F., Peltier, J., and Trost, M. (2017). Mass Spectrometry Techniques for Studying the Ubiquitin System. Biochem. Soc. Trans. 45 (5), 1137–1148. doi:10.1042/bst20170091
Heaton, S. M., Borg, N. A., and Dixit, V. M. (2016). Ubiquitin in the Activation and Attenuation of Innate Antiviral Immunity. J. Exp. Med. 213 (1), 1–13. doi:10.1084/jem.20151531
Heideker, J., and Wertz, I. E. (2015). DUBs, the Regulation of Cell Identity and Disease. Biochem. J. 465 (1), 1–26. doi:10.1042/bj20140496
Hendriks, I. A., Lyon, D., Su, D., Skotte, N. H., Daniel, J. A., Jensen, L. J., et al. (2018). Site-specific Characterization of Endogenous SUMOylation across Species and Organs. Nat. Commun. 9 (1), 2456. doi:10.1038/s41467-018-04957-4
Herhaus, L., and Sapkota, G. P. (2014). The Emerging Roles of Deubiquitylating Enzymes (DUBs) in the TGFβ and BMP Pathways. Cell Signal 26 (10), 2186–2192. doi:10.1016/j.cellsig.2014.06.012
Heride, C., Urbé, S., and Clague, M. J. (2014). Ubiquitin Code Assembly and Disassembly. Curr. Biol. 24 (6), R215–R220. doi:10.1016/j.cub.2014.02.002
Heyninck, K., and Beyaert, R. (2005). A20 Inhibits NF-kappaB Activation by Dual Ubiquitin-Editing Functions. Trends Biochem. Sci. 30 (1), 1–4. doi:10.1016/j.tibs.2004.11.001
Hicke, L., Schubert, H. L., and Hill, C. P. (2005). Ubiquitin-binding Domains. Nat. Rev. Mol. Cell Biol. 6 (8), 610–621. doi:10.1038/nrm1701
Ho, Y. K., Zhi, H., Bowlin, T., Dorjbal, B., Philip, S., Zahoor, M. A., et al. (2015). HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation. PLoS Pathog. 11 (8), e1005102. doi:10.1371/journal.ppat.1005102
Hodge, C. D., Ismail, I. H., Edwards, R. A., Hura, G. L., Xiao, A. T., Tainer, J. A., et al. (2016). RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity that Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment. J. Biol. Chem. 291 (18), 9396–9410. doi:10.1074/jbc.M116.715698
Hodson, C., Purkiss, A., Miles, J. A., and Walden, H. (2014). Structure of the Human FANCL RING-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection. Structure 22 (2), 337–344. doi:10.1016/j.str.2013.12.004
Hofmann, R. M., and Pickart, C. M. (1999). Noncanonical MMS2-Encoded Ubiquitin-Conjugating Enzyme Functions in Assembly of Novel Polyubiquitin Chains for DNA Repair. Cell 96 (5), 645–653. doi:10.1016/s0092-8674(00)80575-9
Hong, S. Y., Kao, Y. R., Lee, T. C., and Wu, C. W. (2018). Upregulation of E3 Ubiquitin Ligase CBLC Enhances EGFR Dysregulation and Signaling in Lung Adenocarcinoma. Cancer Res. 78 (17), 4984–4996. doi:10.1158/0008-5472.Can-17-3858
Hrdinka, M., Fiil, B. K., Zucca, M., Leske, D., Bagola, K., Yabal, M., et al. (2016). CYLD Limits Lys63- and Met1-Linked Ubiquitin at Receptor Complexes to Regulate Innate Immune Signaling. Cell Rep. 14 (12), 2846–2858. doi:10.1016/j.celrep.2016.02.062
Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J. W., et al. (2002). Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex with Ubiquitin Aldehyde. Cell 111 (7), 1041–1054. doi:10.1016/s0092-8674(02)01199-6
Hua, F., Hao, W., Wang, L., and Li, S. (2021). Linear Ubiquitination Mediates EGFR-Induced NF-kappaB Pathway and Tumor Development. Int. J. Mol. Sci. 22 (21). doi:10.3390/ijms222111875
Huang, H., Jeon, M. S., Liao, L., Yang, C., Elly, C., Yates, J. R., et al. (2010). K33-linked Polyubiquitination of T Cell Receptor-Zeta Regulates Proteolysis-independent T Cell Signaling. Immunity 33 (1), 60–70. doi:10.1016/j.immuni.2010.07.002
Ikeda, F., Deribe, Y. L., Skånland, S. S., Stieglitz, B., Grabbe, C., Franz-Wachtel, M., et al. (2011). SHARPIN Forms a Linear Ubiquitin Ligase Complex Regulating NF-Κb Activity and Apoptosis. Nature 471 (7340), 637–641. doi:10.1038/nature09814
Imam, S., Kömürlü, S., Mattick, J., Selyutina, A., Talley, S., Eddins, A., et al. (2019). K63-Linked Ubiquitin Is Required for Restriction of HIV-1 Reverse Transcription and Capsid Destabilization by Rhesus TRIM5α. J. Virol. 93 (14). doi:10.1128/jvi.00558-19
Infante, P., Faedda, R., Bernardi, F., Bufalieri, F., Lospinoso Severini, L., Alfonsi, R., et al. (2018). Itch/beta-arrestin2-dependent Non-proteolytic Ubiquitylation of SuFu Controls Hedgehog Signalling and Medulloblastoma Tumorigenesis. Nat. Commun. 9 (1), 976. doi:10.1038/s41467-018-03339-0
Iwai, K. (2014). Diverse Roles of the Ubiquitin System in NF-Κb Activation. Biochim. Biophys. Acta 1843 (1), 129–136. doi:10.1016/j.bbamcr.2013.03.011
Iwai, K. (2015). “Linear Polyubiquitination: A Crucial Regulator of NF-Κb Activation,” in Innovative Medicine: Basic Research and Development. Editors K. Nakao, N. Minato, and S. Uemoto (Tokyo: Springer Copyright), 51–59. doi:10.1007/978-4-431-55651-0_4
Jachimowicz, R. D., Beleggia, F., Isensee, J., Velpula, B. B., Goergens, J., Bustos, M. A., et al. (2019). UBQLN4 Represses Homologous Recombination and Is Overexpressed in Aggressive Tumors. Cell 176 (3), 505–519. e522. doi:10.1016/j.cell.2018.11.024
Jeusset, L. M., and McManus, K. J. (2017). Ubiquitin Specific Peptidase 22 Regulates Histone H2B Mono-Ubiquitination and Exhibits Both Oncogenic and Tumor Suppressor Roles in Cancer. Cancers (Basel) 9 (12). doi:10.3390/cancers9120167
Ji, C. H., and Kwon, Y. T. (2017). Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells 40 (7), 441–449. doi:10.14348/molcells.2017.0115
Ji, J., Ding, K., Luo, T., Zhang, X., Chen, A., Zhang, D., et al. (2021). TRIM22 Activates NF-kappaB Signaling in Glioblastoma by Accelerating the Degradation of IkappaBalpha. Cell Death Differ. 28 (1), 367–381. doi:10.1038/s41418-020-00606-w
Jiang, S., Park, D. W., Gao, Y., Ravi, S., Darley-Usmar, V., Abraham, E., et al. (2015). Participation of Proteasome-Ubiquitin Protein Degradation in Autophagy and the Activation of AMP-Activated Protein Kinase. Cell Signal 27 (6), 1186–1197. doi:10.1016/j.cellsig.2015.02.024
Jin, X., Shi, Q., Li, Q., Zhou, L., Wang, J., Jiang, L., et al. (2020). CRL3-SPOP Ubiquitin Ligase Complex Suppresses the Growth of Diffuse Large B-Cell Lymphoma by Negatively Regulating the MyD88/NF-kappaB Signaling. Leukemia 34 (5), 1305–1314. doi:10.1038/s41375-019-0661-z
Jing, H., Fang, L., Ding, Z., Wang, D., Hao, W., Gao, L., et al. (2017). Porcine Reproductive and Respiratory Syndrome Virus Nsp1α Inhibits NF-Κb Activation by Targeting the Linear Ubiquitin Chain Assembly Complex. J. Virol. 91 (3). doi:10.1128/jvi.01911-16
Kane, L. A., Lazarou, M., Fogel, A. I., Li, Y., Yamano, K., Sarraf, S. A., et al. (2014). PINK1 Phosphorylates Ubiquitin to Activate Parkin E3 Ubiquitin Ligase Activity. J. Cell Biol. 205 (2), 143–153. doi:10.1083/jcb.201402104
Kazlauskaite, A., Kondapalli, C., Gourlay, R., Campbell, D. G., Ritorto, M. S., Hofmann, K., et al. (2014). Parkin Is Activated by PINK1-dependent Phosphorylation of Ubiquitin at Ser65. Biochem. J. 460 (1), 127–139. doi:10.1042/bj20140334
Keusekotten, K., Elliott, P. R., Glockner, L., Fiil, B. K., Damgaard, R. B., Kulathu, Y., et al. (2013). OTULIN Antagonizes LUBAC Signaling by Specifically Hydrolyzing Met1-Linked Polyubiquitin. Cell 153 (6), 1312–1326. doi:10.1016/j.cell.2013.05.014
Khaminets, A., Behl, C., and Dikic, I. (2016). Ubiquitin-Dependent and Independent Signals in Selective Autophagy. Trends Cell Biol. 26 (1), 6–16. doi:10.1016/j.tcb.2015.08.010
Kim, H. C., and Huibregtse, J. M. (2009). Polyubiquitination by HECT E3s and the Determinants of Chain Type Specificity. Mol. Cell Biol. 29 (12), 3307–3318. doi:10.1128/mcb.00240-09
Kim, K. I., and Baek, S. H. (2006). SUMOylation Code in Cancer Development and Metastasis. Mol. Cells 22 (3), 247–253.
Kim, W., Bennett, E. J., Huttlin, E. L., Guo, A., Li, J., Possemato, A., et al. (2011). Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 44 (2), 325–340. doi:10.1016/j.molcel.2011.08.025
Kirisako, T., Kamei, K., Murata, S., Kato, M., Fukumoto, H., Kanie, M., et al. (2006). A Ubiquitin Ligase Complex Assembles Linear Polyubiquitin Chains. Embo J. 25 (20), 4877–4887. doi:10.1038/sj.emboj.7601360
Klinakis, A., Karagiannis, D., and Rampias, T. (2020). Targeting DNA Repair in Cancer: Current State and Novel Approaches. Cell Mol. Life Sci. 77 (4), 677–703. doi:10.1007/s00018-019-03299-8
Kniss, A., Schuetz, D., Kazemi, S., Pluska, L., Spindler, P. E., Rogov, V. V., et al. (2018). Chain Assembly and Disassembly Processes Differently Affect the Conformational Space of Ubiquitin Chains. Structure 26 (2), 249–258. e244. doi:10.1016/j.str.2017.12.011
Kolapalli, S. P., Sahu, R., Chauhan, N. R., Jena, K. K., Mehto, S., Das, S. K., et al. (2021). RNA-binding RING E3-Ligase DZIP3/hRUL138 Stabilizes Cyclin D1 to Drive Cell-Cycle and Cancer Progression. Cancer Res. 81 (2), 315–331. doi:10.1158/0008-5472.CAN-20-1871
Komander, D., Clague, M. J., and Urbé, S. (2009). Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 10 (8), 550–563. doi:10.1038/nrm2731
Komander, D., and Rape, M. (2012). The Ubiquitin Code. Annu. Rev. Biochem. 81, 203–229. doi:10.1146/annurev-biochem-060310-170328
Koyano, F., Okatsu, K., Kosako, H., Tamura, Y., Go, E., Kimura, M., et al. (2014). Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature 510 (7503), 162–166. doi:10.1038/nature13392
Kristariyanto, Y. A., Abdul Rehman, S. A., Campbell, D. G., Morrice, N. A., Johnson, C., Toth, R., et al. (2015). K29-selective Ubiquitin Binding Domain Reveals Structural Basis of Specificity and Heterotypic Nature of K29 Polyubiquitin. Mol. Cell 58 (1), 83–94. doi:10.1016/j.molcel.2015.01.041
Kulathu, Y., and Komander, D. (2012). Atypical Ubiquitylation - the Unexplored World of Polyubiquitin beyond Lys48 and Lys63 Linkages. Nat. Rev. Mol. Cell Biol. 13 (8), 508–523. doi:10.1038/nrm3394
Kung, C. P., and Weber, J. D. (2022). It's Getting Complicated-A Fresh Look at P53-MDM2-ARF Triangle in Tumorigenesis and Cancer Therapy. Front. Cell Dev. Biol. 10, 818744. doi:10.3389/fcell.2022.818744
Kung, W. W., Ramachandran, S., Makukhin, N., Bruno, E., and Ciulli, A. (2019). Structural Insights into Substrate Recognition by the SOCS2 E3 Ubiquitin Ligase. Nat. Commun. 10 (1), 2534. doi:10.1038/s41467-019-10190-4
Kwon, Y. T., and Ciechanover, A. (2017). The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 42 (11), 873–886. doi:10.1016/j.tibs.2017.09.002
Lamothe, B., Besse, A., Campos, A. D., Webster, W. K., Wu, H., and Darnay, B. G. (2007a). Site-specific Lys-63-Linked Tumor Necrosis Factor Receptor-Associated Factor 6 Auto-Ubiquitination Is a Critical Determinant of I Kappa B Kinase Activation. J. Biol. Chem. 282 (6), 4102–4112. doi:10.1074/jbc.M609503200
Lamothe, B., Campos, A. D., Webster, W. K., Gopinathan, A., Hur, L., and Darnay, B. G. (2008). The RING Domain and First Zinc Finger of TRAF6 Coordinate Signaling by Interleukin-1, Lipopolysaccharide, and RANKL. J. Biol. Chem. 283 (36), 24871–24880. doi:10.1074/jbc.M802749200
Lamothe, B., Webster, W. K., Gopinathan, A., Besse, A., Campos, A. D., and Darnay, B. G. (2007b). TRAF6 Ubiquitin Ligase Is Essential for RANKL Signaling and Osteoclast Differentiation. Biochem. Biophys. Res. Commun. 359 (4), 1044–1049. doi:10.1016/j.bbrc.2007.06.017
Laplantine, E., Fontan, E., Chiaravalli, J., Lopez, T., Lakisic, G., Véron, M., et al. (2009). NEMO Specifically Recognizes K63-Linked Poly-Ubiquitin Chains through a New Bipartite Ubiquitin-Binding Domain. Embo J. 28 (19), 2885–2895. doi:10.1038/emboj.2009.241
Lee, B. L., Singh, A., Mark Glover, J. N., Hendzel, M. J., and Spyracopoulos, L. (2017). Molecular Basis for K63-Linked Ubiquitination Processes in Double-Strand DNA Break Repair: A Focus on Kinetics and Dynamics. J. Mol. Biol. 429 (22), 3409–3429. doi:10.1016/j.jmb.2017.05.029
Lee, H. J., Li, C. F., Ruan, D., He, J., Montal, E. D., Lorenz, S., et al. (2019a). Non-proteolytic Ubiquitination of Hexokinase 2 by HectH9 Controls Tumor Metabolism and Cancer Stem Cell Expansion. Nat. Commun. 10 (1), 2625. doi:10.1038/s41467-019-10374-y
Lee, H. J., Li, C. F., Ruan, D., Powers, S., Thompson, P. A., Frohman, M. A., et al. (2016). The DNA Damage Transducer RNF8 Facilitates Cancer Chemoresistance and Progression through Twist Activation. Mol. Cell 63 (6), 1021–1033. doi:10.1016/j.molcel.2016.08.009
Lee, J. T., Shan, J., Zhong, J., Li, M., Zhou, B., Zhou, A., et al. (2013). RFP-Mediated Ubiquitination of PTEN Modulates its Effect on AKT Activation. Cell Res. 23 (4), 552–564. doi:10.1038/cr.2013.27
Lee, S. B., Kim, J. J., Han, S. A., Fan, Y., Guo, L. S., Aziz, K., et al. (2019b). The AMPK-Parkin axis Negatively Regulates Necroptosis and Tumorigenesis by Inhibiting the Necrosome. Nat. Cell Biol. 21 (8), 940–951. doi:10.1038/s41556-019-0356-8
Lee, S., Tsai, Y. C., Mattera, R., Smith, W. J., Kostelansky, M. S., Weissman, A. M., et al. (2006). Structural Basis for Ubiquitin Recognition and Autoubiquitination by Rabex-5. Nat. Struct. Mol. Biol. 13 (3), 264–271. doi:10.1038/nsmb1064
Lee, S. W., Li, C. F., Jin, G., Cai, Z., Han, F., Chan, C. H., et al. (2015). Skp2-dependent Ubiquitination and Activation of LKB1 Is Essential for Cancer Cell Survival under Energy Stress. Mol. Cell 57 (6), 1022–1033. doi:10.1016/j.molcel.2015.01.015
Lee, Y. R., Chen, M., Lee, J. D., Zhang, J., Lin, S. Y., Fu, T. M., et al. (2019c). Reactivation of PTEN Tumor Suppressor for Cancer Treatment through Inhibition of a MYC-WWP1 Inhibitory Pathway. Science 364 (6441). doi:10.1126/science.aau0159
Leestemaker, Y., and Ovaa, H. (2017). Tools to Investigate the Ubiquitin Proteasome System. Drug Discov. Today Technol. 26, 25–31. doi:10.1016/j.ddtec.2017.11.006
Leng, S., Huang, W., Chen, Y., Yang, Y., Feng, D., Liu, W., et al. (2021). SIRT1 Coordinates with the CRL4B Complex to Regulate Pancreatic Cancer Stem Cells to Promote Tumorigenesis. Cell Death Differ. 28 (12), 3329–3343. doi:10.1038/s41418-021-00821-z
Lenoir, J. J., Parisien, J. P., and Horvath, C. M. (2021). Immune Regulator LGP2 Targets Ubc13/UBE2N to Mediate Widespread Interference with K63 Polyubiquitination and NF-kappaB Activation. Cell Rep. 37 (13), 110175. doi:10.1016/j.celrep.2021.110175
Lewis, M. J., Vyse, S., Shields, A. M., Boeltz, S., Gordon, P. A., Spector, T. D., et al. (2015). UBE2L3 Polymorphism Amplifies NF-Κb Activation and Promotes Plasma Cell Development, Linking Linear Ubiquitination to Multiple Autoimmune Diseases. Am. J. Hum. Genet. 96 (2), 221–234. doi:10.1016/j.ajhg.2014.12.024
Leznicki, P., and Kulathu, Y. (2017). Mechanisms of Regulation and Diversification of Deubiquitylating Enzyme Function. J. Cell Sci. 130 (12), 1997–2006. doi:10.1242/jcs.201855
Li, N., Wang, Y., Neri, S., Zhen, Y., Fong, L. W. R., Qiao, Y., et al. (2019). Tankyrase Disrupts Metabolic Homeostasis and Promotes Tumorigenesis by Inhibiting LKB1-AMPK Signalling. Nat. Commun. 10 (1), 4363. doi:10.1038/s41467-019-12377-1
Li, Z., Younger, K., Gartenhaus, R., Joseph, A. M., Hu, F., Baer, M. R., et al. (2015). Inhibition of IRAK1/4 Sensitizes T Cell Acute Lymphoblastic Leukemia to Chemotherapies. J. Clin. Invest 125 (3), 1081–1097. doi:10.1172/JCI75821
Liang, L., Fan, Y., Cheng, J., Cheng, D., Zhao, Y., Cao, B., et al. (2013). TAK1 Ubiquitination Regulates Doxorubicin-Induced NF-kappaB Activation. Cell Signal 25 (1), 247–254. doi:10.1016/j.cellsig.2012.09.003
Liao, Y., Liu, N., Xia, X., Guo, Z., Li, Y., Jiang, L., et al. (2019). USP10 Modulates the SKP2/Bcr-Abl axis via Stabilizing SKP2 in Chronic Myeloid Leukemia. Cell Discov. 5, 24. doi:10.1038/s41421-019-0092-z
Liao, Y., Sumara, I., and Pangou, E. (2022). Non-proteolytic Ubiquitylation in Cellular Signaling and Human Disease. Commun. Biol. 5 (1), 114. doi:10.1038/s42003-022-03060-1
Licchesi, J. D., Mieszczanek, J., Mevissen, T. E., Rutherford, T. J., Akutsu, M., Virdee, S., et al. (2011). An Ankyrin-Repeat Ubiquitin-Binding Domain Determines TRABID's Specificity for Atypical Ubiquitin Chains. Nat. Struct. Mol. Biol. 19 (1), 62–71. doi:10.1038/nsmb.2169
Lin, S. C., Chung, J. Y., Lamothe, B., Rajashankar, K., Lu, M., Lo, Y. C., et al. (2008). Molecular Basis for the Unique Deubiquitinating Activity of the NF-kappaB Inhibitor A20. J. Mol. Biol. 376 (2), 526–540. doi:10.1016/j.jmb.2007.11.092
Lin, X., Ojo, D., Wei, F., Wong, N., Gu, Y., and Tang, D. (2015). A Novel Aspect of Tumorigenesis-BMI1 Functions in Regulating DNA Damage Response. Biomolecules 5 (4), 3396–3415. doi:10.3390/biom5043396
Liu, B., Zhang, M., Chu, H., Zhang, H., Wu, H., Song, G., et al. (2017). The Ubiquitin E3 Ligase TRIM31 Promotes Aggregation and Activation of the Signaling Adaptor MAVS through Lys63-Linked Polyubiquitination. Nat. Immunol. 18 (2), 214–224. doi:10.1038/ni.3641
Liu, R., Zeng, L. W., Gong, R., Yuan, F., Shu, H. B., and Li, S. (2021). mTORC1 Activity Regulates Post-translational Modifications of glycine Decarboxylase to Modulate glycine Metabolism and Tumorigenesis. Nat. Commun. 12 (1), 4227. doi:10.1038/s41467-021-24321-3
Liu, Z., Gong, Z., Jiang, W. X., Yang, J., Zhu, W. K., Guo, D. C., et al. (2015). Lys63-linked Ubiquitin Chain Adopts Multiple Conformational States for Specific Target Recognition. Elife 4. doi:10.7554/eLife.05767
Loch, C. M., and Strickler, J. E. (2012). A Microarray of Ubiquitylated Proteins for Profiling Deubiquitylase Activity Reveals the Critical Roles of Both Chain and Substrate. Biochim. Biophys. Acta 1823 (11), 2069–2078. doi:10.1016/j.bbamcr.2012.05.006
Locke, M., Toth, J. I., and Petroski, M. D. (2014). Lys11- and Lys48-Linked Ubiquitin Chains Interact with P97 during Endoplasmic-Reticulum-Associated Degradation. Biochem. J. 459 (1), 205–216. doi:10.1042/bj20120662
Lork, M., Verhelst, K., and Beyaert, R. (2017). CYLD, A20 and OTULIN Deubiquitinases in NF-Κb Signaling and Cell Death: So Similar, yet So Different. Cell Death Differ. 24 (7), 1172–1183. doi:10.1038/cdd.2017.46
Lu, C. H., Yeh, D. W., Lai, C. Y., Liu, Y. L., Huang, L. R., Lee, A. Y., et al. (2018). USP17 Mediates Macrophage-Promoted Inflammation and Stemness in Lung Cancer Cells by Regulating TRAF2/TRAF3 Complex Formation. Oncogene 37 (49), 6327–6340. doi:10.1038/s41388-018-0411-0
Ma, J., Shi, Q., Cui, G., Sheng, H., Botuyan, M. V., Zhou, Y., et al. (2021). SPOP Mutation Induces Replication Over-firing by Impairing Geminin Ubiquitination and Triggers Replication Catastrophe upon ATR Inhibition. Nat. Commun. 12 (1), 5779. doi:10.1038/s41467-021-26049-6
Machida, Y. J., Machida, Y., Chen, Y., Gurtan, A. M., Kupfer, G. M., D'Andrea, A. D., et al. (2006). UBE2T Is the E2 in the Fanconi Anemia Pathway and Undergoes Negative Autoregulation. Mol. Cell 23 (4), 589–596. doi:10.1016/j.molcel.2006.06.024
Mai, J., Zhong, Z. Y., Guo, G. F., Chen, X. X., Xiang, Y. Q., Li, X., et al. (2019). Polo-like Kinase 1 Phosphorylates and Stabilizes KLF4 to Promote Tumorigenesis in Nasopharyngeal Carcinoma. Theranostics 9 (12), 3541–3554. doi:10.7150/thno.32908
Mandemaker, I. K., van Cuijk, L., Janssens, R. C., Lans, H., Bezstarosti, K., Hoeijmakers, J. H., et al. (2017). DNA Damage-Induced Histone H1 Ubiquitylation Is Mediated by HUWE1 and Stimulates the RNF8-Rnf168 Pathway. Sci. Rep. 7 (1), 15353. doi:10.1038/s41598-017-15194-y
Manthiram, K., Zhou, Q., Aksentijevich, I., and Kastner, D. L. (2017). The Monogenic Autoinflammatory Diseases Define New Pathways in Human Innate Immunity and Inflammation. Nat. Immunol. 18 (8), 832–842. doi:10.1038/ni.3777
Massoumi, R., Chmielarska, K., Hennecke, K., Pfeifer, A., and Fassler, R. (2006). Cyld Inhibits Tumor Cell Proliferation by Blocking Bcl-3-dependent NF-kappaB Signaling. Cell 125 (4), 665–677. doi:10.1016/j.cell.2006.03.041
Mattern, M., Sutherland, J., Kadimisetty, K., Barrio, R., and Rodriguez, M. S. (2019). Using Ubiquitin Binders to Decipher the Ubiquitin Code. Trends Biochem. Sci. 44 (7), 599–615. doi:10.1016/j.tibs.2019.01.011
Maure, J. F., Moser, S. C., Jaffray, E. G., F Alpi, A., and Hay, R. T. (2016). Loss of Ubiquitin E2 Ube2w Rescues Hypersensitivity of Rnf4 Mutant Cells to DNA Damage. Sci. Rep. 6, 26178. doi:10.1038/srep26178
McDowell, G. S., and Philpott, A. (2016). New Insights into the Role of Ubiquitylation of Proteins. Int. Rev. Cell Mol. Biol. 325, 35–88. doi:10.1016/bs.ircmb.2016.02.002
Meng, Y., Liu, C., Shen, L., Zhou, M., Liu, W., Kowolik, C., et al. (2019). TRAF6 Mediates Human DNA2 Polyubiquitination and Nuclear Localization to Maintain Nuclear Genome Integrity. Nucleic Acids Res. 47 (14), 7564–7579. doi:10.1093/nar/gkz537
Meng, Z., Xu, R., Xie, L., Wu, Y., He, Q., Gao, P., et al. (2021). A20/Nrdp1 Interaction Alters the Inflammatory Signaling Profile by Mediating K48- and K63-Linked Polyubiquitination of Effectors MyD88 and TBK1. J. Biol. Chem. 297 (1), 100811. doi:10.1016/j.jbc.2021.100811
Mevissen, T. E., Hospenthal, M. K., Geurink, P. P., Elliott, P. R., Akutsu, M., Arnaudo, N., et al. (2013). OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell 154 (1), 169–184. doi:10.1016/j.cell.2013.05.046
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 86, 159–192. doi:10.1146/annurev-biochem-061516-044916
Mevissen, T. E. T., Kulathu, Y., Mulder, M. P. C., Geurink, P. P., Maslen, S. L., Gersch, M., et al. (2016). Molecular Basis of Lys11-Polyubiquitin Specificity in the Deubiquitinase Cezanne. Nature 538 (7625), 402–405. doi:10.1038/nature19836
Meza Gutierrez, F., Simsek, D., Mizrak, A., Deutschbauer, A., Braberg, H., Johnson, J., et al. (2018). Genetic Analysis Reveals Functions of Atypical Polyubiquitin Chains. Elife 7. doi:10.7554/eLife.42955
Michel, M. A., Komander, D., and Elliott, P. R. (2018). Enzymatic Assembly of Ubiquitin Chains. Methods Mol. Biol. 1844, 73–84. doi:10.1007/978-1-4939-8706-1_6
Michel, M. A., Swatek, K. N., Hospenthal, M. K., and Komander, D. (2017). Ubiquitin Linkage-specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling. Mol. Cell 68 (1), 233–246. e235. doi:10.1016/j.molcel.2017.08.020
Miles, J. A., Frost, M. G., Carroll, E., Rowe, M. L., Howard, M. J., Sidhu, A., et al. (2015). The Fanconi Anemia DNA Repair Pathway Is Regulated by an Interaction between Ubiquitin and the E2-like Fold Domain of FANCL. J. Biol. Chem. 290 (34), 20995–21006. doi:10.1074/jbc.M115.675835
Min, Y., Kim, M. J., Lee, S., Chun, E., and Lee, K. Y. (2018). Inhibition of TRAF6 Ubiquitin-Ligase Activity by PRDX1 Leads to Inhibition of NFKB Activation and Autophagy Activation. Autophagy 14 (8), 1347–1358. doi:10.1080/15548627.2018.1474995
Morris, J. R., and Solomon, E. (2004). BRCA1 : BARD1 Induces the Formation of Conjugated Ubiquitin Structures, Dependent on K6 of Ubiquitin, in Cells during DNA Replication and Repair. Hum. Mol. Genet. 13 (8), 807–817. doi:10.1093/hmg/ddh095
Mu, Y., Sundar, R., Thakur, N., Ekman, M., Gudey, S. K., Yakymovych, M., et al. (2011). TRAF6 Ubiquitinates TGFbeta Type I Receptor to Promote its Cleavage and Nuclear Translocation in Cancer. Nat. Commun. 2, 330. doi:10.1038/ncomms1332
Myant, K. B., Cammareri, P., Hodder, M. C., Wills, J., Von Kriegsheim, A., Gyorffy, B., et al. (2017). HUWE1 Is a Critical Colonic Tumour Suppressor Gene that Prevents MYC Signalling, DNA Damage Accumulation and Tumour Initiation. EMBO Mol. Med. 9 (2), 181–197. doi:10.15252/emmm.201606684
Ni, X., Kou, W., Gu, J., Wei, P., Wu, X., Peng, H., et al. (2019). TRAF6 Directs FOXP3 Localization and Facilitates Regulatory T-Cell Function through K63-Linked Ubiquitination. EMBO J. 38 (9). doi:10.15252/embj.201899766
Nishi, R., Wijnhoven, P., le Sage, C., Tjeertes, J., Galanty, Y., Forment, J. V., et al. (2014). Systematic Characterization of Deubiquitylating Enzymes for Roles in Maintaining Genome Integrity. Nat. Cell Biol. 16 (10), 10161011–10261018. doi:10.1038/ncb3028
Nishikawa, H., Ooka, S., Sato, K., Arima, K., Okamoto, J., Klevit, R. E., et al. (2004). Mass Spectrometric and Mutational Analyses Reveal Lys-6-Linked Polyubiquitin Chains Catalyzed by BRCA1-BARD1 Ubiquitin Ligase. J. Biol. Chem. 279 (6), 3916–3924. doi:10.1074/jbc.M308540200
Niu, J., Shi, Y., Iwai, K., and Wu, Z. H. (2011). LUBAC Regulates NF-kappaB Activation upon Genotoxic Stress by Promoting Linear Ubiquitination of NEMO. EMBO J. 30 (18), 3741–3753. doi:10.1038/emboj.2011.264
Niu, T., Wu, Z., and Xiao, W. (2021). Uev1A Promotes Breast Cancer Cell Migration by Up-Regulating CT45A Expression via the AKT Pathway. BMC Cancer 21 (1), 1012. doi:10.1186/s12885-021-08750-3
Noad, J., von der Malsburg, A., Pathe, C., Michel, M. A., Komander, D., and Randow, F. (2017). LUBAC-Synthesized Linear Ubiquitin Chains Restrict Cytosol-Invading Bacteria by Activating Autophagy and NF-Κb. Nat. Microbiol. 2, 17063. doi:10.1038/nmicrobiol.2017.63
Nowsheen, S., Aziz, K., Aziz, A., Deng, M., Qin, B., Luo, K., et al. (2018). L3MBTL2 Orchestrates Ubiquitin Signalling by Dictating the Sequential Recruitment of RNF8 and RNF168 after DNA Damage. Nat. Cell Biol. 20 (4), 455–464. doi:10.1038/s41556-018-0071-x
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169 (6), 985–999. doi:10.1016/j.cell.2017.05.016
O'Connor, H. F., and Huibregtse, J. M. (2017). Enzyme-substrate Relationships in the Ubiquitin System: Approaches for Identifying Substrates of Ubiquitin Ligases. Cell Mol. Life Sci. 74 (18), 3363–3375. doi:10.1007/s00018-017-2529-6
O'Donnell, M. A. (2019). Gustavo Silva: Translating the Ubiquitin Code. J. Cell Biol. 218 (1), 3–4. doi:10.1083/jcb.201812083
Ohtake, F., Saeki, Y., Sakamoto, K., Ohtake, K., Nishikawa, H., Tsuchiya, H., et al. (2015). Ubiquitin Acetylation Inhibits Polyubiquitin Chain Elongation. EMBO Rep. 16 (2), 192–201. doi:10.15252/embr.201439152
Ordureau, A., Sarraf, S. A., Duda, D. M., Heo, J. M., Jedrychowski, M. P., Sviderskiy, V. O., et al. (2014). Quantitative Proteomics Reveal a Feedforward Mechanism for Mitochondrial PARKIN Translocation and Ubiquitin Chain Synthesis. Mol. Cell 56 (3), 360–375. doi:10.1016/j.molcel.2014.09.007
Padala, P., Soudah, N., Giladi, M., Haitin, Y., Isupov, M. N., and Wiener, R. (2017). The Crystal Structure and Conformations of an Unbranched Mixed Tri-ubiquitin Chain Containing K48 and K63 Linkages. J. Mol. Biol. 429 (24), 3801–3813. doi:10.1016/j.jmb.2017.10.027
Park, H. Y., Go, H., Song, H. R., Kim, S., Ha, G. H., Jeon, Y. K., et al. (2014). Pellino 1 Promotes Lymphomagenesis by Deregulating BCL6 Polyubiquitination. J. Clin. Invest 124 (11), 4976–4988. doi:10.1172/JCI75667
Paul, A., and Wang, B. (2017). RNF8- and Ube2S-dependent Ubiquitin Lysine 11-Linkage Modification in Response to DNA Damage. Mol. Cell 66 (4), 458–472. e455. doi:10.1016/j.molcel.2017.04.013
Penengo, L., Mapelli, M., Murachelli, A. G., Confalonieri, S., Magri, L., Musacchio, A., et al. (2006). Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin. Cell 124 (6), 1183–1195. doi:10.1016/j.cell.2006.02.020
Peng, D. J., Zeng, M., Muromoto, R., Matsuda, T., Shimoda, K., Subramaniam, M., et al. (2011). Noncanonical K27-Linked Polyubiquitination of TIEG1 Regulates Foxp3 Expression and Tumor Growth. J. Immunol. 186 (10), 5638–5647. doi:10.4049/jimmunol.1003801
Piao, S., Pei, H. Z., Huang, B., and Baek, S. H. (2017). Ovarian Tumor Domain-Containing Protein 1 Deubiquitinates and Stabilizes P53. Cell Signal 33, 22–29. doi:10.1016/j.cellsig.2017.02.011
Polykratis, A., Martens, A., Eren, R. O., Shirasaki, Y., Yamagishi, M., Yamaguchi, Y., et al. (2019). A20 Prevents Inflammasome-dependent Arthritis by Inhibiting Macrophage Necroptosis through its ZnF7 Ubiquitin-Binding Domain. Nat. Cell Biol. 21 (6), 731–742. doi:10.1038/s41556-019-0324-3
Qin, Z., Jiang, W., Wang, G., Sun, Y., and Xiao, W. (2018). Linear Ubiquitin Chain Induces Apoptosis and Inhibits Tumor Growth. Apoptosis 23 (1), 16–26. doi:10.1007/s10495-017-1433-8
Queisser, M. A., Dada, L. A., Deiss-Yehiely, N., Angulo, M., Zhou, G., Kouri, F. M., et al. (2014). HOIL-1L Functions as the PKCζ Ubiquitin Ligase to Promote Lung Tumor Growth. Am. J. Respir. Crit. Care Med. 190 (6), 688–698. doi:10.1164/rccm.201403-0463OC
Raimondi, M., Cesselli, D., Di Loreto, C., La Marra, F., Schneider, C., and Demarchi, F. (2019). USP1 (Ubiquitin Specific Peptidase 1) Targets ULK1 and Regulates its Cellular Compartmentalization and Autophagy. Autophagy 15 (4), 613–630. doi:10.1080/15548627.2018.1535291
Rape, M. (2018). Ubiquitylation at the Crossroads of Development and Disease. Nat. Rev. Mol. Cell Biol. 19 (1), 59–70. doi:10.1038/nrm.2017.83
Regev, O., Roth, Z., Korman, M., Khalaila, I., and Gur, E. (2015). A Kinetic Model for the Prevalence of Mono- over Poly-Pupylation. Febs J. 282 (21), 4176–4186. doi:10.1111/febs.13413
Ren, Y., Xu, X., Mao, C. Y., Han, K. K., Xu, Y. J., Cao, B. Y., et al. (2020). RNF6 Promotes Myeloma Cell Proliferation and Survival by Inducing Glucocorticoid Receptor Polyubiquitination. Acta Pharmacol. Sin. 41 (3), 394–403. doi:10.1038/s41401-019-0309-6
Reyes-Turcu, F. E., Horton, J. R., Mullally, J. E., Heroux, A., Cheng, X., and Wilkinson, K. D. (2006). The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 124 (6), 1197–1208. doi:10.1016/j.cell.2006.02.038
Rittinger, K., and Ikeda, F. (2017). Linear Ubiquitin Chains: Enzymes, Mechanisms and Biology. Open Biol. 7 (4). doi:10.1098/rsob.170026
Rivkin, E., Almeida, S. M., Ceccarelli, D. F., Juang, Y. C., MacLean, T. A., Srikumar, T., et al. (2013). The Linear Ubiquitin-specific Deubiquitinase Gumby Regulates Angiogenesis. Nature 498 (7454), 318–324. doi:10.1038/nature12296
Rogerson, D. T., Sachdeva, A., Wang, K., Haq, T., Kazlauskaite, A., Hancock, S. M., et al. (2015). Efficient Genetic Encoding of Phosphoserine and its Nonhydrolyzable Analog. Nat. Chem. Biol. 11 (7), 496–503. doi:10.1038/nchembio.1823
Rösner, D., Schneider, T., Schneider, D., Scheffner, M., and Marx, A. (2015). Click Chemistry for Targeted Protein Ubiquitylation and Ubiquitin Chain Formation. Nat. Protoc. 10 (10), 1594–1611. doi:10.1038/nprot.2015.106
Rossi, F. A., and Rossi, M. (2022). Emerging Role of Ubiquitin-specific Protease 19 in Oncogenesis and Cancer Development. Front. Cell Dev. Biol. 10, 889166. doi:10.3389/fcell.2022.889166
Rudnicka, A., and Yamauchi, Y. (2016). Ubiquitin in Influenza Virus Entry and Innate Immunity. Viruses 8 (10). doi:10.3390/v8100293
Sadowski, M., and Sarcevic, B. (2010). Mechanisms of Mono- and Poly-Ubiquitination: Ubiquitination Specificity Depends on Compatibility between the E2 Catalytic Core and Amino Acid Residues Proximal to the Lysine. Cell Div. 5, 19. doi:10.1186/1747-1028-5-19
Sahin, I., Zhang, S., Navaraj, A., Zhou, L., Dizon, D., Safran, H., et al. (2020). AMG-232 Sensitizes High MDM2-Expressing Tumor Cells to T-Cell-Mediated Killing. Cell Death Discov. 6, 57. doi:10.1038/s41420-020-0292-1
Sahu, S. K., Tiwari, N., Pataskar, A., Zhuang, Y., Borisova, M., Diken, M., et al. (2017). FBXO32 Promotes Microenvironment Underlying Epithelial-Mesenchymal Transition via CtBP1 during Tumour Metastasis and Brain Development. Nat. Commun. 8 (1), 1523. doi:10.1038/s41467-017-01366-x
Sasaki, K., and Iwai, K. (2015). Roles of Linear Ubiquitinylation, a Crucial Regulator of NF-Κb and Cell Death, in the Immune System. Immunol. Rev. 266 (1), 175–189. doi:10.1111/imr.12308
Sato, Y., Okatsu, K., Saeki, Y., Yamano, K., Matsuda, N., Kaiho, A., et al. (2017). Structural Basis for Specific Cleavage of Lys6-Linked Polyubiquitin Chains by USP30. Nat. Struct. Mol. Biol. 24 (11), 911–919. doi:10.1038/nsmb.3469
Seeler, J. S., and Dejean, A. (2017). SUMO and the Robustness of Cancer. Nat. Rev. Cancer 17 (3), 184–197. doi:10.1038/nrc.2016.143
Senft, D., Qi, J., and Ronai, Z. A. (2018). Ubiquitin Ligases in Oncogenic Transformation and Cancer Therapy. Nat. Rev. Cancer 18 (2), 69–88. doi:10.1038/nrc.2017.105
Seo, J., Lee, E. W., Shin, J., Seong, D., Nam, Y. W., Jeong, M., et al. (2018). K6 Linked Polyubiquitylation of FADD by CHIP Prevents Death Inducing Signaling Complex Formation Suppressing Cell Death. Oncogene 37 (36), 4994–5006. doi:10.1038/s41388-018-0323-z
Shanbhag, N. M., Rafalska-Metcalf, I. U., Balane-Bolivar, C., Janicki, S. M., and Greenberg, R. A. (2010). ATM-Dependent Chromatin Changes Silence Transcription in Cis to DNA Double-Strand Breaks. Cell 141 (6), 970–981. doi:10.1016/j.cell.2010.04.038
Shekhar, M. P., Gerard, B., Pauley, R. J., Williams, B. O., and Tait, L. (2008). Rad6B Is a Positive Regulator of Beta-Catenin Stabilization. Cancer Res. 68 (6), 1741–1750. doi:10.1158/0008-5472.CAN-07-2111
Shen, Y., Tang, K., Chen, D., Hong, M., Sun, F., Wang, S., et al. (2021). Riok3 Inhibits the Antiviral Immune Response by Facilitating TRIM40-Mediated RIG-I and MDA5 Degradation. Cell Rep. 35 (12), 109272. doi:10.1016/j.celrep.2021.109272
Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D., and Hicke, L. (2002). Epsins and Vps27p/Hrs Contain Ubiquitin-Binding Domains that Function in Receptor Endocytosis. Nat. Cell Biol. 4 (5), 389–393. doi:10.1038/ncb790
Shirane, M., Hatakeyama, S., Hattori, K., Nakayama, K., and Nakayama, K. (1999). Common Pathway for the Ubiquitination of IkappaBalpha, IkappaBbeta, and IkappaBepsilon Mediated by the F-Box Protein FWD1. J. Biol. Chem. 274 (40), 28169–28174. doi:10.1074/jbc.274.40.28169
Singh, A. N., Oehler, J., Torrecilla, I., Kilgas, S., Li, S., Vaz, B., et al. (2019). The P97-Ataxin 3 Complex Regulates Homeostasis of the DNA Damage Response E3 Ubiquitin Ligase RNF8. EMBO J. 38 (21), e102361. doi:10.15252/embj.2019102361
Singh, R., Karri, D., Shen, H., Shao, J., Dasgupta, S., Huang, S., et al. (2018). TRAF4-mediated Ubiquitination of NGF Receptor TrkA Regulates Prostate Cancer Metastasis. J. Clin. Invest 128 (7), 3129–3143. doi:10.1172/JCI96060
Smeenk, G., and Mailand, N. (2016). Writers, Readers, and Erasers of Histone Ubiquitylation in DNA Double-Strand Break Repair. Front. Genet. 7, 122. doi:10.3389/fgene.2016.00122
Smith, C. J., Berry, D. M., and McGlade, C. J. (2013). The E3 Ubiquitin Ligases RNF126 and Rabring7 Regulate Endosomal Sorting of the Epidermal Growth Factor Receptor. J. Cell Sci. 126 (Pt 6), 1366–1380. doi:10.1242/jcs.116129
Sokratous, K., Hadjisavvas, A., Diamandis, E. P., and Kyriacou, K. (2014). The Role of Ubiquitin-Binding Domains in Human Pathophysiology. Crit. Rev. Clin. Lab. Sci. 51 (5), 280–290. doi:10.3109/10408363.2014.915287
Somasagara, R. R., Spencer, S. M., Tripathi, K., Clark, D. W., Mani, C., Madeira da Silva, L., et al. (2017). RAD6 Promotes DNA Repair and Stem Cell Signaling in Ovarian Cancer and Is a Promising Therapeutic Target to Prevent and Treat Acquired Chemoresistance. Oncogene 36 (48), 6680–6690. doi:10.1038/onc.2017.279
Song, K., Cai, X., Dong, Y., Wu, H., Wei, Y., Shankavaram, U. T., et al. (2021). Epsins 1 and 2 Promote NEMO Linear Ubiquitination via LUBAC to Drive Breast Cancer Development. J. Clin. Invest 131 (1). doi:10.1172/JCI129374
Song, L., and Luo, Z. Q. (2019). Post-Translational Regulation of Ubiquitin Signaling. J. Cell Biol. 218 (6), 1776–1786. doi:10.1083/jcb.201902074
Sparrer, K. M. J., Gableske, S., Zurenski, M. A., Parker, Z. M., Full, F., Baumgart, G. J., et al. (2017). TRIM23 Mediates Virus-Induced Autophagy via Activation of TBK1. Nat. Microbiol. 2 (11), 1543–1557. doi:10.1038/s41564-017-0017-2
Spence, J., Sadis, S., Haas, A. L., and Finley, D. (1995). A Ubiquitin Mutant with Specific Defects in DNA Repair and Multiubiquitination. Mol. Cell Biol. 15 (3), 1265–1273. doi:10.1128/mcb.15.3.1265
Spiliotopoulos, A., Blokpoel Ferreras, L., Densham, R. M., Caulton, S. G., Maddison, B. C., Morris, J. R., et al. (2019). Discovery of Peptide Ligands Targeting a Specific Ubiquitin-like Domain-Binding Site in the Deubiquitinase USP11. J. Biol. Chem. 294 (2), 424–436. doi:10.1074/jbc.RA118.004469
Spit, M., Rieser, E., and Walczak, H. (2019). Linear Ubiquitination at a Glance. J. Cell Sci. 132 (2). doi:10.1242/jcs.208512
Sun, J., Zhu, Z., Li, W., Shen, M., Cao, C., Sun, Q., et al. (2020). UBE2T-regulated H2AX Monoubiquitination Induces Hepatocellular Carcinoma Radioresistance by Facilitating CHK1 Activation. J. Exp. Clin. Cancer Res. 39 (1), 222. doi:10.1186/s13046-020-01734-4
Sun, L., Amraei, R., and Rahimi, N. (2021). NEDD4 Regulates Ubiquitination and Stability of the Cell Adhesion Molecule IGPR-1 via Lysosomal Pathway. J. Biomed. Sci. 28 (1), 35. doi:10.1186/s12929-021-00731-9
Sun, S. C. (2010). CYLD: a Tumor Suppressor Deubiquitinase Regulating NF-kappaB Activation and Diverse Biological Processes. Cell Death Differ. 17 (1), 25–34. doi:10.1038/cdd.2009.43
Sun, X., Ding, Y., Zhan, M., Li, Y., Gao, D., Wang, G., et al. (2019). Usp7 Regulates Hippo Pathway through Deubiquitinating the Transcriptional Coactivator Yorkie. Nat. Commun. 10 (1), 411. doi:10.1038/s41467-019-08334-7
Suryadinata, R., Roesley, S. N., Yang, G., and Sarčević, B. (2014). Mechanisms of Generating Polyubiquitin Chains of Different Topology. Cells 3 (3), 674–689. doi:10.3390/cells3030674
Swaney, D. L., Rodríguez-Mias, R. A., and Villén, J. (2015). Phosphorylation of Ubiquitin at Ser65 Affects its Polymerization, Targets, and Proteome-wide Turnover. EMBO Rep. 16 (9), 1131–1144. doi:10.15252/embr.201540298
Sy, S. M., Jiang, J., O, W. S., Deng, Y., and Huen, M. S. (2013). The Ubiquitin Specific Protease USP34 Promotes Ubiquitin Signaling at DNA Double-Strand Breaks. Nucleic Acids Res. 41 (18), 8572–8580. doi:10.1093/nar/gkt622
Syed, N. A., Andersen, P. L., Warrington, R. C., and Xiao, W. (2006). Uev1A, a Ubiquitin Conjugating Enzyme Variant, Inhibits Stress-Induced Apoptosis through NF-kappaB Activation. Apoptosis 11 (12), 2147–2157. doi:10.1007/s10495-006-0197-3
Takiuchi, T., Nakagawa, T., Tamiya, H., Fujita, H., Sasaki, Y., Saeki, Y., et al. (2014). Suppression of LUBAC-Mediated Linear Ubiquitination by a Specific Interaction between LUBAC and the Deubiquitinases CYLD and OTULIN. Genes cells.. 19 (3), 254–272. doi:10.1111/gtc.12128
Taminiau, A., Draime, A., Tys, J., Lambert, B., Vandeputte, J., Nguyen, N., et al. (2016). HOXA1 Binds RBCK1/HOIL-1 and TRAF2 and Modulates the TNF/NF-κB Pathway in a Transcription-independent Manner. Nucleic Acids Res. 44 (15), 7331–7349. doi:10.1093/nar/gkw606
Tang, J., Luo, Y., Tian, Z., Liao, X., Cui, Q., Yang, Q., et al. (2020). TRIM11 Promotes Breast Cancer Cell Proliferation by Stabilizing Estrogen Receptor Alpha. Neoplasia 22 (9), 343–351. doi:10.1016/j.neo.2020.06.003
Tang, L. Y., Yamashita, M., Coussens, N. P., Tang, Y., Wang, X., Li, C., et al. (2011). Ablation of Smurf2 Reveals an Inhibition in TGF-β Signalling through Multiple Mono-Ubiquitination of Smad3. Embo J. 30 (23), 4777–4789. doi:10.1038/emboj.2011.393
Tang, Y., Joo, D., Liu, G., Tu, H., You, J., Jin, J., et al. (2018). Linear Ubiquitination of cFLIP Induced by LUBAC Contributes to TNFα-Induced Apoptosis. J. Biol. Chem. 293 (52), 20062–20072. doi:10.1074/jbc.RA118.005449
Tanno, H., and Komada, M. (2013). The Ubiquitin Code and its Decoding Machinery in the Endocytic Pathway. J. Biochem. 153 (6), 497–504. doi:10.1093/jb/mvt028
Teixeira, L. K., and Reed, S. I. (2013). Ubiquitin Ligases and Cell Cycle Control. Annu. Rev. Biochem. 82, 387–414. doi:10.1146/annurev-biochem-060410-105307
Tikoo, S., Madhavan, V., Hussain, M., Miller, E. S., Arora, P., Zlatanou, A., et al. (2013). Ubiquitin-dependent Recruitment of the Bloom Syndrome Helicase upon Replication Stress Is Required to Suppress Homologous Recombination. EMBO J. 32 (12), 1778–1792. doi:10.1038/emboj.2013.117
Tokunaga, F., and Iwai, K. (2012). Linear Ubiquitination: a Novel NF-Κb Regulatory Mechanism for Inflammatory and Immune Responses by the LUBAC Ubiquitin Ligase Complex. Endocr. J. 59 (8), 641–652. doi:10.1507/endocrj.ej12-0148
Tokunaga, F. (2013). Linear Ubiquitination-Mediated NF-Κb Regulation and its Related Disorders. J. Biochem. 154 (4), 313–323. doi:10.1093/jb/mvt079
Tokunaga, F., Nakagawa, T., Nakahara, M., Saeki, Y., Taniguchi, M., Sakata, S., et al. (2011). SHARPIN Is a Component of the NF-kappaB-Activating Linear Ubiquitin Chain Assembly Complex. Nature 471 (7340), 633–636. doi:10.1038/nature09815
Torres, M. P., Lee, M. J., Ding, F., Purbeck, C., Kuhlman, B., Dokholyan, N. V., et al. (2009). G Protein Mono-Ubiquitination by the Rsp5 Ubiquitin Ligase. J. Biol. Chem. 284 (13), 8940–8950. doi:10.1074/jbc.M809058200
Tran, H., Hamada, F., Schwarz-Romond, T., and Bienz, M. (2008). Trabid, a New Positive Regulator of Wnt-Induced Transcription with Preference for Binding and Cleaving K63-Linked Ubiquitin Chains. Genes Dev. 22 (4), 528–542. doi:10.1101/gad.463208
Turek, I., Tischer, N., Lassig, R., and Trujillo, M. (2018). Multi-tiered Pairing Selectivity between E2 Ubiquitin-Conjugating Enzymes and E3 Ligases. J. Biol. Chem. 293 (42), 16324–16336. doi:10.1074/jbc.RA118.004226
van der Heden van Noort, G. J., Kooij, R., Elliott, P. R., Komander, D., and Ovaa, H. (2017). Synthesis of Poly-Ubiquitin Chains Using a Bifunctional Ubiquitin Monomer. Org. Lett. 19 (24), 6490–6493. doi:10.1021/acs.orglett.7b03085
van Wijk, S. J. L., Fricke, F., Herhaus, L., Gupta, J., Hotte, K., Pampaloni, F., et al. (2017). Linear Ubiquitination of Cytosolic Salmonella Typhimurium Activates NF-kappaB and Restricts Bacterial Proliferation. Nat. Microbiol. 2, 17066. doi:10.1038/nmicrobiol.2017.66
Varadan, R., Assfalg, M., Haririnia, A., Raasi, S., Pickart, C., and Fushman, D. (2004). Solution Conformation of Lys63-Linked Di-ubiquitin Chain Provides Clues to Functional Diversity of Polyubiquitin Signaling. J. Biol. Chem. 279 (8), 7055–7063. doi:10.1074/jbc.M309184200
Venuto, S., and Merla, G. (2019). E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 8 (5). doi:10.3390/cells8050510
Vuorela, M., Pylkas, K., and Winqvist, R. (2011). Mutation Screening of the RNF8, UBC13 and MMS2 Genes in Northern Finnish Breast Cancer Families. BMC Med. Genet. 12, 98. doi:10.1186/1471-2350-12-98
Wagner, S. A., Beli, P., Weinert, B. T., Nielsen, M. L., Cox, J., Mann, M., et al. (2011). A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell Proteomics 10 (10), M111–M013284. doi:10.1074/mcp.M111.013284
Walczak, H., Iwai, K., and Dikic, I. (2012). Generation and Physiological Roles of Linear Ubiquitin Chains. BMC Biol. 10, 23. doi:10.1186/1741-7007-10-23
Wang, B., Jie, Z., Joo, D., Ordureau, A., Liu, P., Gan, W., et al. (2017a). TRAF2 and OTUD7B Govern a Ubiquitin-dependent Switch that Regulates mTORC2 Signalling. Nature 545 (7654), 365–369. doi:10.1038/nature22344
Wang, B., Ma, A., Zhang, L., Jin, W. L., Qian, Y., Xu, G., et al. (2015). POH1 Deubiquitylates and Stabilizes E2F1 to Promote Tumour Formation. Nat. Commun. 6, 8704. doi:10.1038/ncomms9704
Wang, C., Long, W., Peng, C., Hu, L., Zhang, Q., Wu, A., et al. (2016). HTLV-1 Tax Functions as a Ubiquitin E3 Ligase for Direct IKK Activation via Synthesis of Mixed-Linkage Polyubiquitin Chains. PLoS Pathog. 12 (4), e1005584. doi:10.1371/journal.ppat.1005584
Wang, F., Fu, X., Chen, P., Wu, P., Fan, X., Li, N., et al. (2017b). SPSB1-mediated HnRNP A1 Ubiquitylation Regulates Alternative Splicing and Cell Migration in EGF Signaling. Cell Res. 27 (4), 540–558. doi:10.1038/cr.2017.7
Wang, F., Gao, Y., Zhou, L., Chen, J., Xie, Z., Ye, Z., et al. (2022). USP30: Structure, Emerging Physiological Role, and Target Inhibition. Front. Pharmacol. 13, 851654. doi:10.3389/fphar.2022.851654
Wang, G., Long, J., Gao, Y., Zhang, W., Han, F., Xu, C., et al. (2019). SETDB1-mediated Methylation of Akt Promotes its K63-Linked Ubiquitination and Activation Leading to Tumorigenesis. Nat. Cell Biol. 21 (2), 214–225. doi:10.1038/s41556-018-0266-1
Wang, P., Dai, X., Jiang, W., Li, Y., and Wei, W. (2020). RBR E3 Ubiquitin Ligases in Tumorigenesis. Semin. Cancer Biol. 67 (Pt 2), 131–144. doi:10.1016/j.semcancer.2020.05.002
Wang, Q., Huang, L., Hong, Z., Lv, Z., Mao, Z., Tang, Y., et al. (2017c). The E3 Ubiquitin Ligase RNF185 Facilitates the cGAS-Mediated Innate Immune Response. PLoS Pathog. 13 (3), e1006264. doi:10.1371/journal.ppat.1006264
Wang, S., Wang, R., Peralta, C., Yaseen, A., and Pavletich, N. P. (2021a). Structure of the FA Core Ubiquitin Ligase Closing the ID Clamp on DNA. Nat. Struct. Mol. Biol. 28 (3), 300–309. doi:10.1038/s41594-021-00568-8
Wang, X., Zhang, H., Shao, Z., Zhuang, W., Sui, C., Liu, F., et al. (2021b). TRIM31 Facilitates K27-Linked Polyubiquitination of SYK to Regulate Antifungal Immunity. Signal Transduct. Target Ther. 6 (1), 298. doi:10.1038/s41392-021-00711-3
Wang, Y., Shi, M., Feng, H., Zhu, Y., Liu, S., Gao, A., et al. (2018). Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 173 (5), 1231–1243. e1216. doi:10.1016/j.cell.2018.04.023
Wenbo, L., and Wang, J. (2017). Uncovering the Underlying Mechanism of Cancer Tumorigenesis and Development under an Immune Microenvironment from Global Quantification of the Landscape. J. R. Soc. Interface 14 (131). doi:10.1098/rsif.2017.0105
Werner, A., Manford, A. G., and Rape, M. (2017). Ubiquitin-Dependent Regulation of Stem Cell Biology. Trends Cell Biol. 27 (8), 568–579. doi:10.1016/j.tcb.2017.04.002
Wertz, I. E., O'Rourke, K. M., Zhou, H., Eby, M., Aravind, L., Seshagiri, S., et al. (2004). De-ubiquitination and Ubiquitin Ligase Domains of A20 Downregulate NF-kappaB Signalling. Nature 430 (7000), 694–699. doi:10.1038/nature02794
Wheaton, K., Sarkari, F., Stanly Johns, B., Davarinejad, H., Egorova, O., Kaustov, L., et al. (2017). UbE2E1/UBCH6 Is a Critical In Vivo E2 for the PRC1-Catalyzed Ubiquitination of H2A at Lys-119. J. Biol. Chem. 292 (7), 2893–2902. doi:10.1074/jbc.M116.749564
Whiteaker, J. R., Zhao, L., Ivey, R. G., Sanchez-Bonilla, M., Moore, H. D., Schoenherr, R. M., et al. (2018). Targeted Mass Spectrometry Enables Robust Quantification of FANCD2 Mono-Ubiquitination in Response to DNA Damage. DNA Repair (Amst) 65, 47–53. doi:10.1016/j.dnarep.2018.03.003
Windheim, M., Peggie, M., and Cohen, P. (2008). Two Different Classes of E2 Ubiquitin-Conjugating Enzymes Are Required for the Mono-Ubiquitination of Proteins and Elongation by Polyubiquitin Chains with a Specific Topology. Biochem. J. 409 (3), 723–729. doi:10.1042/bj20071338
Wu, B., Qiang, L., Zhang, Y., Fu, Y., Zhao, M., Lei, Z., et al. (2022). The Deubiquitinase OTUD1 Inhibits Colonic Inflammation by Suppressing RIPK1-Mediated NF-kappaB Signaling. Cell Mol. Immunol. 19 (2), 276–289. doi:10.1038/s41423-021-00810-9
Wu, H. T., Kuo, Y. C., Hung, J. J., Huang, C. H., Chen, W. Y., Chou, T. Y., et al. (2016). K63-polyubiquitinated HAUSP Deubiquitinates HIF-1alpha and Dictates H3K56 Acetylation Promoting Hypoxia-Induced Tumour Progression. Nat. Commun. 7, 13644. doi:10.1038/ncomms13644
Wu, H., Yang, T. Y., Li, Y., Ye, W. L., Liu, F., He, X. S., et al. (2020). Tumor Necrosis Factor Receptor-Associated Factor 6 Promotes Hepatocarcinogenesis by Interacting with Histone Deacetylase 3 to Enhance C-Myc Gene Expression and Protein Stability. Hepatology 71 (1), 148–163. doi:10.1002/hep.30801
Wu, X., and Karin, M. (2015). Emerging Roles of Lys63-Linked Polyubiquitylation in Immune Responses. Immunol. Rev. 266 (1), 161–174. doi:10.1111/imr.12310
Wu, X., Zhang, W., Font-Burgada, J., Palmer, T., Hamil, A. S., Biswas, S. K., et al. (2014a). Ubiquitin-conjugating Enzyme Ubc13 Controls Breast Cancer Metastasis through a TAK1-P38 MAP Kinase Cascade. Proc. Natl. Acad. Sci. U. S. A. 111 (38), 13870–13875. doi:10.1073/pnas.1414358111
Wu, Z., Shen, S., Zhang, Z., Zhang, W., and Xiao, W. (2017). Erratum to: Ubiquitin-Conjugating Enzyme Complex Uev1A-Ubc13 Promotes Breast Cancer Metastasis through Nuclear Factor-Кb Mediated Matrix Metalloproteinase-1 Gene Regulation. Breast Cancer Res. 19 (1), 41. doi:10.1186/s13058-017-0833-6
Wu, Z., Shen, S., Zhang, Z., Zhang, W., and Xiao, W. (2014b). Ubiquitin-conjugating Enzyme Complex Uev1A-Ubc13 Promotes Breast Cancer Metastasis through Nuclear Factor-Small Ka, CyrillicB Mediated Matrix Metalloproteinase-1 Gene Regulation. Breast Cancer Res. 16 (4), R75. doi:10.1186/bcr3692
Wu-Baer, F., Lagrazon, K., Yuan, W., and Baer, R. (2003). The BRCA1/BARD1 Heterodimer Assembles Polyubiquitin Chains through an Unconventional Linkage Involving Lysine Residue K6 of Ubiquitin. J. Biol. Chem. 278 (37), 34743–34746. doi:10.1074/jbc.C300249200
Wu-Baer, F., Ludwig, T., and Baer, R. (2010). The UBXN1 Protein Associates with Autoubiquitinated Forms of the BRCA1 Tumor Suppressor and Inhibits its Enzymatic Function. Mol. Cell Biol. 30 (11), 2787–2798. doi:10.1128/MCB.01056-09
Xiao, N., Li, H., Luo, J., Wang, R., Chen, H., Chen, J., et al. (2012). Ubiquitin-specific Protease 4 (USP4) Targets TRAF2 and TRAF6 for Deubiquitination and Inhibits TNFalpha-Induced Cancer Cell Migration. Biochem. J. 441 (3), 979–986. doi:10.1042/BJ20111358
Xie, F., Zhang, Z., van Dam, H., Zhang, L., and Zhou, F. (2014). Regulation of TGF-Beta Superfamily Signaling by SMAD Mono-Ubiquitination. Cells 3 (4), 981–993. doi:10.3390/cells3040981
Xing, L., Tang, X., Wu, K., Huang, X., Yi, Y., and Huan, J. (2020). TRIM27 Functions as a Novel Oncogene in Non-triple-negative Breast Cancer by Blocking Cellular Senescence through P21 Ubiquitination. Mol. Ther. Nucleic Acids 22, 910–923. doi:10.1016/j.omtn.2020.10.012
Xu, L., Jia, Y., Yang, X. H., Han, F., Zheng, Y., Ni, Y., et al. (2017). MicroRNA-130b Transcriptionally Regulated by Histone H3 Deacetylation Renders Akt Ubiquitination and Apoptosis Resistance to 6-OHDA. Biochim. Biophys. Acta Mol. Basis Dis. 1863 (6), 1678–1689. doi:10.1016/j.bbadis.2017.04.012
Xu, P., Duong, D. M., Seyfried, N. T., Cheng, D., Xie, Y., Robert, J., et al. (2009). Quantitative Proteomics Reveals the Function of Unconventional Ubiquitin Chains in Proteasomal Degradation. Cell 137 (1), 133–145. doi:10.1016/j.cell.2009.01.041
Xu, X., Kalac, M., Markson, M., Chan, M., Brody, J. D., Bhagat, G., et al. (2020). Reversal of CYLD Phosphorylation as a Novel Therapeutic Approach for Adult T-Cell Leukemia/lymphoma (ATLL). Cell Death Dis. 11 (2), 94. doi:10.1038/s41419-020-2294-6
Xu, Y., Hu, Y., Xu, T., Yan, K., Zhang, T., Li, Q., et al. (2021). RNF8-mediated Regulation of Akt Promotes Lung Cancer Cell Survival and Resistance to DNA Damage. Cell Rep. 37 (3), 109854. doi:10.1016/j.celrep.2021.109854
Xue, B., Li, H., Guo, M., Wang, J., Xu, Y., Zou, X., et al. (2018). TRIM21 Promotes Innate Immune Response to RNA Viral Infection through Lys27-Linked Polyubiquitination of MAVS. J. Virol. 92 (14). doi:10.1128/jvi.00321-18
Yang, J. M., Schiapparelli, P., Nguyen, H. N., Igarashi, A., Zhang, Q., Abbadi, S., et al. (2017). Characterization of PTEN Mutations in Brain Cancer Reveals that Pten Mono-Ubiquitination Promotes Protein Stability and Nuclear Localization. Oncogene 36 (26), 3673–3685. doi:10.1038/onc.2016.493
Yang, Y., Yang, C., Li, T., Yu, S., Gan, T., Hu, J., et al. (2020). The Deubiquitinase USP38 Promotes NHEJ Repair through Regulation of HDAC1 Activity and Regulates Cancer Cell Response to Genotoxic Insults. Cancer Res. 80 (4), 719–731. doi:10.1158/0008-5472.CAN-19-2149
Yao, F., Zhou, Z., Kim, J., Hang, Q., Xiao, Z., Ton, B. N., et al. (2018). SKP2- and OTUD1-Regulated Non-proteolytic Ubiquitination of YAP Promotes YAP Nuclear Localization and Activity. Nat. Commun. 9 (1), 2269. doi:10.1038/s41467-018-04620-y
Yasunaga, J., Lin, F. C., Lu, X., and Jeang, K. T. (2011). Ubiquitin-specific Peptidase 20 Targets TRAF6 and Human T Cell Leukemia Virus Type 1 Tax to Negatively Regulate NF-kappaB Signaling. J. Virol. 85 (13), 6212–6219. doi:10.1128/JVI.00079-11
Yau, R. G., Doerner, K., Castellanos, E. R., Haakonsen, D. L., Werner, A., Wang, N., et al. (2017). Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 171 (4), 918–933. doi:10.1016/j.cell.2017.09.040
Yau, R., and Rape, M. (2016). The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 18 (6), 579–586. doi:10.1038/ncb3358
Yeh, C. H., Bellon, M., and Nicot, C. (2018). FBXW7: a Critical Tumor Suppressor of Human Cancers. Mol. Cancer 17 (1), 115. doi:10.1186/s12943-018-0857-2
Yin, H., Gui, Y., Du, G., Frohman, M. A., and Zheng, X. L. (2010). Dependence of Phospholipase D1 Multi-Monoubiquitination on its Enzymatic Activity and Palmitoylation. J. Biol. Chem. 285 (18), 13580–13588. doi:10.1074/jbc.M109.046359
Yin, Q., Han, T., Fang, B., Zhang, G., Zhang, C., Roberts, E. R., et al. (2019). K27-linked Ubiquitination of BRAF by ITCH Engages Cytokine Response to Maintain MEK-ERK Signaling. Nat. Commun. 10 (1), 1870. doi:10.1038/s41467-019-09844-0
Yu, X., Li, W., Deng, Q., Liu, H., Wang, X., Hu, H., et al. (2020a). MYD88 L265P Elicits Mutation-specific Ubiquitination to Drive NF-kappaB Activation and Lymphomagenesis. Blood. doi:10.1182/blood.2020004918
Yu, X., Li, W., Liu, H., Deng, Q., Wang, X., Hu, H., et al. (2020b). Ubiquitination of the DNA-Damage Checkpoint Kinase CHK1 by TRAF4 Is Required for CHK1 Activation. J. Hematol. Oncol. 13 (1), 40. doi:10.1186/s13045-020-00869-3
Yu, Z., Li, X., Yang, M., Huang, J., Fang, Q., Jia, J., et al. (2021). TRIM41 Is Required to Innate Antiviral Response by Polyubiquitinating BCL10 and Recruiting NEMO. Signal Transduct. Target Ther. 6 (1), 90. doi:10.1038/s41392-021-00477-8
Zhang, H., Hu, H., Greeley, N., Jin, J., Matthews, A. J., Ohashi, E., et al. (2014). STAT3 Restrains RANK- and TLR4-Mediated Signalling by Suppressing Expression of the E2 Ubiquitin-Conjugating Enzyme Ubc13. Nat. Commun. 5, 5798. doi:10.1038/ncomms6798
Zhang, L., Liu, Q., Liu, K. W., Qin, Z. Y., Zhu, G. X., Shen, L. T., et al. (2021). SHARPIN Stabilizes Beta-Catenin through a Linear Ubiquitination-independent Manner to Support Gastric Tumorigenesis. Gastric Cancer 24 (2), 402–416. doi:10.1007/s10120-020-01138-5
Zhang, L., Zhou, F., Garcia de Vinuesa, A., de Kruijf, E. M., Mesker, W. E., Hui, L., et al. (2013a). TRAF4 Promotes TGF-Beta Receptor Signaling and Drives Breast Cancer Metastasis. Mol. Cell 51 (5), 559–572. doi:10.1016/j.molcel.2013.07.014
Zhang, Q., Karnak, D., Tan, M., Lawrence, T. S., Morgan, M. A., and Sun, Y. (2016). FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol. Cell 61 (3), 419–433. doi:10.1016/j.molcel.2015.12.010
Zhang, X., Meng, T., Cui, S., Liu, D., Pang, Q., and Wang, P. (2022). Roles of Ubiquitination in the Crosstalk between Tumors and the Tumor Microenvironment (Review). Int. J. Oncol. 61 (1). doi:10.3892/ijo.2022.5374
Zhang, X., Zhang, J., Zhang, L., van Dam, H., and ten Dijke, P. (2013b). UBE2O Negatively Regulates TRAF6-Mediated NF-kappaB Activation by Inhibiting TRAF6 Polyubiquitination. Cell Res. 23 (3), 366–377. doi:10.1038/cr.2013.21
Zhang, Y., Li, Y., Yang, X., Wang, J., Wang, R., Qian, X., et al. (2018). Uev1A-Ubc13 Catalyzes K63-Linked Ubiquitination of RHBDF2 to Promote TACE Maturation. Cell Signal 42, 155–164. doi:10.1016/j.cellsig.2017.10.013
Zhang, Z., Fan, Y., Xie, F., Zhou, H., Jin, K., Shao, L., et al. (2017). Breast Cancer Metastasis Suppressor OTUD1 Deubiquitinates SMAD7. Nat. Commun. 8 (1), 2116. doi:10.1038/s41467-017-02029-7
Zhao, X., Lutz, J., Höllmüller, E., Scheffner, M., Marx, A., and Stengel, F. (2017). Identification of Proteins Interacting with Ubiquitin Chains. Angew. Chem. Int. Ed. Engl. 56 (49), 15764–15768. doi:10.1002/anie.201705898
Zheng, J., Wang, B., Zheng, R., Zhang, J., Huang, C., Zheng, R., et al. (2020). Linc-RA1 Inhibits Autophagy and Promotes Radioresistance by Preventing H2Bub1/USP44 Combination in Glioma Cells. Cell Death Dis. 11 (9), 758. doi:10.1038/s41419-020-02977-x
Zheng, Y. G., Wu, J., Chen, Z., and Goodman, M. (2008). Chemical Regulation of Epigenetic Modifications: Opportunities for New Cancer Therapy. Med. Res. Rev. 28 (5), 645–687. doi:10.1002/med.20120
Zhi, H., Guo, X., Ho, Y. K., Pasupala, N., Engstrom, H. A. A., Semmes, O. J., et al. (2020). RNF8 Dysregulation and Down-Regulation during HTLV-1 Infection Promote Genomic Instability in Adult T-Cell Leukemia. PLoS Pathog. 16 (5), e1008618. doi:10.1371/journal.ppat.1008618
Zhou, H., Liu, Y., Zhu, R., Ding, F., Wan, Y., Li, Y., et al. (2017). FBXO32 Suppresses Breast Cancer Tumorigenesis through Targeting KLF4 to Proteasomal Degradation. Oncogene 36 (23), 3312–3321. doi:10.1038/onc.2016.479
Zhou, H., Wertz, I., O'Rourke, K., Ultsch, M., Seshagiri, S., Eby, M., et al. (2004). Bcl10 Activates the NF-kappaB Pathway through Ubiquitination of NEMO. Nature 427 (6970), 167–171. doi:10.1038/nature02273
Zhou, L., Jiang, H., Du, J., Li, L., Li, R., Lu, J., et al. (2018). USP15 Inhibits Multiple Myeloma Cell Apoptosis through Activating a Feedback Loop with the Transcription Factor NF-κBp65. Exp. Mol. Med. 50 (11), 1–12. doi:10.1038/s12276-018-0180-4
Zhou, L., and Xu, G. (2022). The Ubiquitination-dependent and -Independent Functions of Cereblon in Cancer and Neurological Diseases. J. Mol. Biol. 434 (5), 167457. doi:10.1016/j.jmb.2022.167457
Zhou, W., Zhong, Z., Lin, D., Liu, Z., Zhang, Q., Xia, H., et al. (2021). Hypothermic Oxygenated Perfusion Inhibits HECTD3-Mediated TRAF3 Polyubiquitination to Alleviate DCD Liver Ischemia-Reperfusion Injury. Cell Death Dis. 12 (2), 211. doi:10.1038/s41419-021-03493-2
Zhu, F., Yi, G., Liu, X., Zhu, F., Zhao, A., Wang, A., et al. (2018). Ring Finger Protein 31-mediated Atypical Ubiquitination Stabilizes Forkhead Box P3 and Thereby Stimulates Regulatory T-Cell Function. J. Biol. Chem. 293 (52), 20099–20111. doi:10.1074/jbc.RA118.005802
Zhu, G., Herlyn, M., and Yang, X. (2021). TRIM15 and CYLD Regulate ERK Activation via Lysine-63-Linked Polyubiquitination. Nat. Cell Biol. 23 (9), 978–991. doi:10.1038/s41556-021-00732-8
Zhu, J., Li, X., Su, P., Xue, M., Zang, Y., and Ding, Y. (2020). The Ubiquitin Ligase RNF181 Stabilizes ERalpha and Modulates Breast Cancer Progression. Oncogene 39 (44), 6776–6788. doi:10.1038/s41388-020-01464-z
Zhu, J., Zhuang, T., Yang, H., Li, X., Liu, H., and Wang, H. (2016). Atypical Ubiquitin Ligase RNF31: the Nuclear Factor Modulator in Breast Cancer Progression. BMC Cancer 16, 538. doi:10.1186/s12885-016-2575-8
Zientara-Rytter, K., and Subramani, S. (2019). The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy. Cells 8 (1). doi:10.3390/cells8010040
Keywords: ubiquitin, atypical ubiquitination, ubiquitin E2 conjugating enzyme, ubiquitin E3 ligase, tumorigenesis, ubiquitin-proteasome system
Citation: Yin X, Liu Q, Liu F, Tian X, Yan T, Han J and Jiang S (2022) Emerging Roles of Non-proteolytic Ubiquitination in Tumorigenesis. Front. Cell Dev. Biol. 10:944460. doi: 10.3389/fcell.2022.944460
Received: 15 May 2022; Accepted: 15 June 2022;
Published: 06 July 2022.
Edited by:
Haitao Wang, Center for Cancer Research (NIH), United StatesReviewed by:
Jianping Guo, Sun Yat-sen University, ChinaPetro Starokadomskyy, University of Texas Southwestern Medical Center, United States
Jianlin Liu, Dana–Farber Cancer Institute, United States
Shivani Dixit, National Cancer Institute (NIH), United States
Copyright © 2022 Yin, Liu, Liu, Tian, Yan, Han and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Han, aGFuajM2NjlAMTYzLmNvbQ==; Shulong Jiang, am5zbGppYW5nQDE2My5jb20=
 Xiu Yin
Xiu Yin Qingbin Liu
Qingbin Liu Fen Liu
Fen Liu Xinchen Tian
Xinchen Tian Tinghao Yan
Tinghao Yan Jie Han
Jie Han Shulong Jiang
Shulong Jiang