
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell Dev. Biol. , 11 July 2022
Sec. Epigenomics and Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.940197
This article is part of the Research Topic Manipulating the epigenome to improve cloning efficiency View all 5 articles
Over the last 25 years, cloned animals have been produced by transferring somatic cell nuclei into enucleated oocytes (SCNT) in more than 20 mammalian species. Among domestic animals, pigs are likely the leading species in the number of clones produced by SCNT. The greater interest in pig cloning has two main reasons, its relevance for food production and as its use as a suitable model in biomedical applications. Recognized progress in animal cloning has been attained over time, but the overall efficiency of SCNT in pigs remains very low, based on the rate of healthy, live born piglets following embryo transfer. Accumulating evidence from studies in mice and other species indicate that new strategies for promoting chromatin and epigenetic reprogramming may represent the beginning of a new era for pig cloning.
The transfer of somatic cell nuclei to enucleated oocytes has proved that differentiated cells can be reverted to a totipotent state (Wilmut et al., 1997), and has helped to attain a better understanding of cell differentiation and reprogramming through chromatin and epigenetic mechanisms (Matoba and Zhang, 2018; de Macedo et al., 2021). The SCNT technology has also been applied to clone animals for different purposes, including the preservation of endangered species, replication of companion and production animals, and creation of transgenic and gene-edited animals for agricultural and biomedical purposes (Bordignon, 2019). Pigs are important for both food production and biomedical research (Gutierrez et al., 2015). Thus, pigs with unique traits and genomes of agricultural and biomedical importance have been created by SCNT, nonetheless, with low and variable efficiency. In addition of ameliorating technical aspects (e.g., oocyte maturation, enucleation, activation, embryo culture, cell fusion, cell cycle coordination), researchers have identified some roadblock mechanisms for cell reprogramming and tested new approaches to facilitate chromatin and epigenetic remodeling in SCNT embryos (Figure 1). These approaches may not only facilitate the erasure of repressive epigenetic marks that are incompatible with cell reprogramming but may also enhance the acquisition or reestablishment of specific marks required for normal embryo development and success of SCNT. This article highlights the recent progress in understanding and improving SCNT efficiency in pigs.
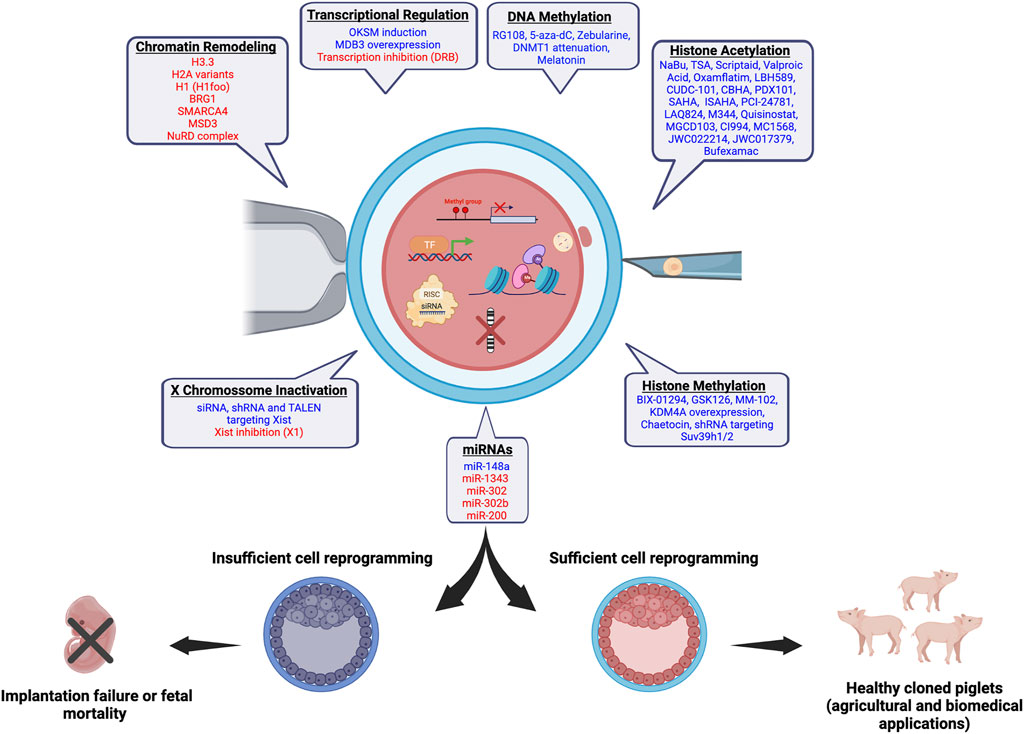
FIGURE 1. Chromatin and epigenetic reprogramming impacting SCNT efficiency in pigs. In blue, chromatin/epigenetic modifiers that have been previously shown to play a role on development of pig SCNT embryos and cloning efficiency are listed. In red, modifiers that are believed to have a beneficial effect on the development of pig SCNT embryos based on studies in other species are listed. The figure was created with BioRender.com.
Successful development of SCNT embryos depends on the reestablishment of cell totipotency, which relies on changes in chromatin structure, epigenetic marks, and transcriptional profile. Nucleosomes’ positioning and occupancy affect chromatin function by either facilitating or restricting DNA access to transcription factors (TFs) and other regulators, which impacts both the reacquisition of cell totipotency following SCNT and cloning efficiency (Matoba and Zhang, 2018). Nucleosome occupancy seems to be rapidly, but likely insufficiently, reprogrammed in SCNT embryos. In pig SCNT embryos, nucleosome occupancy 10 h after nuclear transfer was lower in promoter sequences but higher in coding sequences, compared to nuclear donor cells, indicating a reprogramming process (Tao et al., 2017). Chromatin accessibility in mouse SCNT embryos was reprogrammed to a similar state of fertilized zygotes within 12 h, except for reprogramming resistant regions enriched for histone three lysine nine trimethylation (H3K9me3) (Djekidel et al., 2018).
The incorporation of maternally-derived histones facilitates chromatin remodeling in SCNT embryos. The histone H3 is replaced by the maternally-derived H3.3 in bovine (Wang et al., 2020) and mouse (Wen et al., 2014a) SCNT embryos. This switch is involved in the activation of pluripotency genes such as the Octamer-binding transcript factor (Oct-4 or Pou5f1) and Nanog homeobox (Nanog) (Wen et al., 2014b), and reduction of epigenetic barriers for cell reprogramming, such as H3K27me3 and H3K9me3 (Wen et al., 2014b; Wang et al., 2020). In murine SCNT embryos, H2A and H2A.Z were replaced by the H2A.X variant (Nashun et al., 2011). H2A.X participates in the DNA damage response, and has been negatively correlated with embryo quality (Bohrer et al., 2013). The macro H2A variant, which is involved in the repression of pluripotency genes and considered a barrier for cell reprogramming (Gaspar-Maia et al., 2013), was also stripped from the chromatin after SCNT in mice (Chang et al., 2010). In addition, somatic variants of the linker histone H1 are reprogrammed after nuclear transfer (Bordignon et al., 1999), and are replaced by the oocyte-specific variant H1foo in SCNT embryos (Teranishi et al., 2004; Yun et al., 2012). Although H1foo′s role in chromatin reprogramming in SCNT embryos remains poorly understood, induction of H1foo expression along with the pluripotency factors Oct-4, SRY-Box Transcription Factor 2 (Sox2) and Kruppel Like Factor 4 (Klf4) improved the efficiency of mouse induced pluripotent cells (iPSCs) production (Kunitomi et al., 2016).
Chromatin relaxation, which is induced by remodeling complexes such as the transcription activator Brahma-related gene 1 (BRG1 or SMARCA4), a member of the SWItch/sucrose nonfermentable (SWI/SNF) complex, represents another important feature for attaining cell totipotency and normal embryo development (Hansis et al., 2004; Egli and Eggan, 2010; Glanzner et al., 2017). The methyl-CpG-binding domain protein 3 (MBD3), a core component of the nucleosome remodeling and deacetylation (NuRD) complex, is also essential for embryo development (Hendrich et al., 2001). The NuRD complex regulates nucleosome occupancy and recruitment of TFs (Bornelov et al., 2018). Lower MBD3 expression was detected in pig SCNT embryos compared to control embryos, and its overexpression in SCNT embryos improved blastocyst formation and total cell number (Wang et al., 2019) (Table 1).
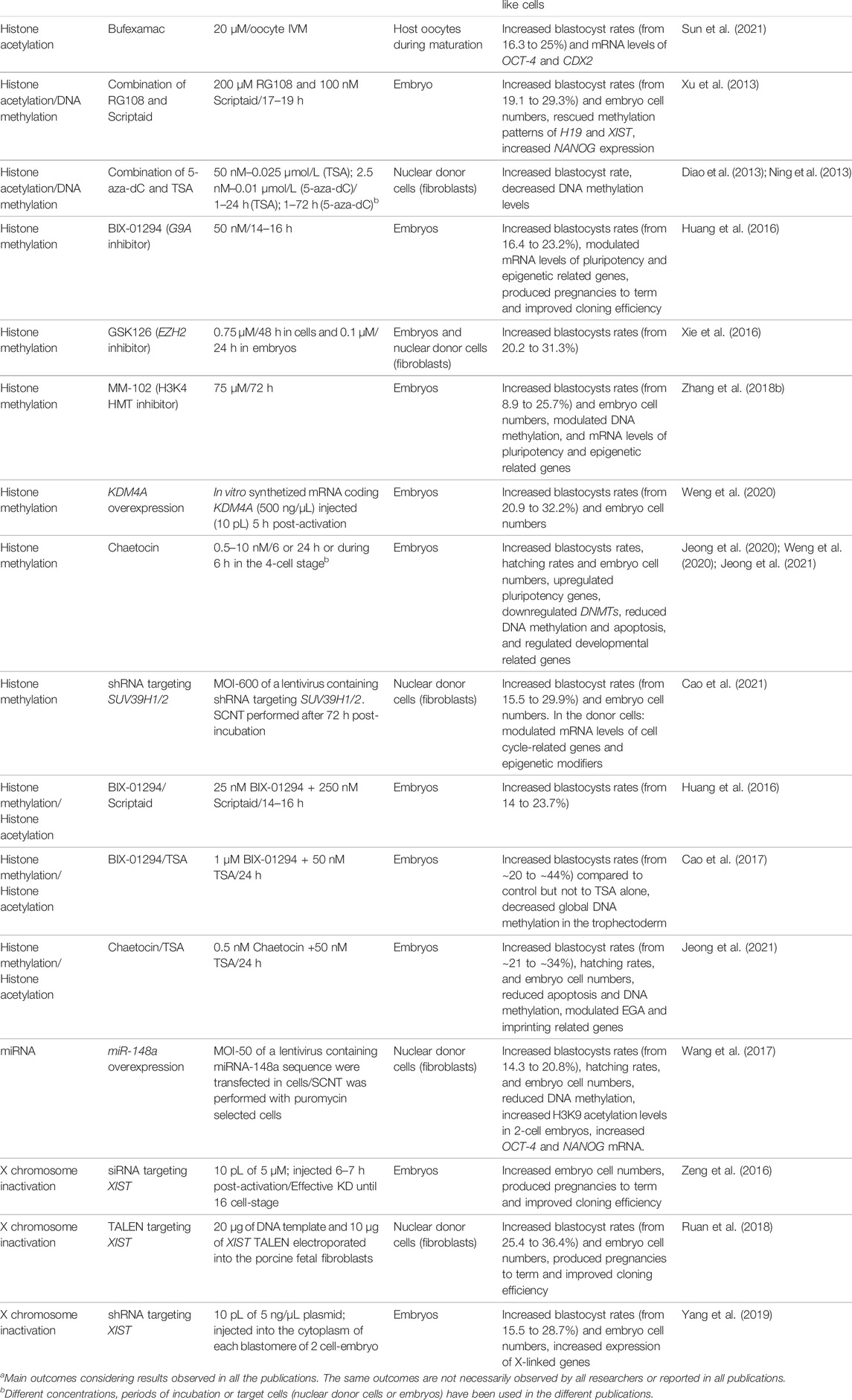
TABLE 1. Impact of different modifiers of chromatin or epigenetic marks on development of SCNT embryos and pig cloning efficiency.
Proper regulation of TFs is crucial for reprograming cell totipotency and successful development of SCNT embryos (Table 1). Overexpression of the iPSC inducing factors OCT-4, SOX2, KLF4 and c-MYC (OSKM) in porcine cells prior to nuclear transfer improved blastocyst rates and quality of SCNT embryos (Song Z. et al., 2014; Kim et al., 2019). The co-expression of OKSM and the estrogen-related receptor B (ESRRB), which is abundantly expressed in pig embryos (Yu et al., 2021), improved iPSCs production by regulating pluripotency factors (Shi et al., 2020). In addition, pig iPSCs overexpressing ESRRB showed higher potential for trophectoderm differentiation when injected into 8-cell stage embryos (Yu et al., 2021).
The impact of OCT-4 on development of SCNT embryos has been investigated in multiple different species. In pigs, expression of OCT-4 and OCT-4 related genes, such as Calcium binding and coiled-coil domain 2 (NDP52l1), and Developmental pluripotency associated 2, three and 5 (DPPA2,3,5), was lower in SCNT than control embryos (Lee et al., 2006). The Double homeobox (Dux) transcription factor and its upstream regulators Dppa2 and four have been identified as major regulators of embryonic genome activation (EGA) (Eckersley-Maslin et al., 2019). Overexpression of Dux in mouse SCNT embryos improved development and normalized EGA transcripts, comparable to levels of control embryos produced by fertilization (Yang et al., 2020). Despite of these relevant findings in other species, the impact of manipulating EGA regulators has not been studied in porcine SCNT embryos.
Proper reprogramming to totipotency in SCNT embryos can be hampered by both lack of gene activation and failure to inactivate the transcriptional memory of somatic nuclei following nuclear transfer (Ng and Gurdon, 2005; Matoba et al., 2014). The transcriptional memory of somatic cells was more efficiently reprogrammed in SCNT embryos that cleaved early and produced higher blastocyst rates than late cleaving embryos (Liu et al., 2020). In addition, attenuation of the transcriptional memory for 15 h after nuclear transfer, using the inhibitor of RNA polymerase II, 5,6-Dichlorobenzimidazole 1-β-d-ribofuranoside (DRB), improved gene expression in bovine SCNT embryos and increased blastocyst cell numbers (Rissi et al., 2018). However, more studies are needed to determine the impact of transcriptional inhibition following SCNT on cloning efficiency in pigs.
DNA methylation is usually associated with transcriptional silencing. Substantial DNA demethylation occurs during early embryo development but remethylation happens at later stages (Reik et al., 2001; Ivanova et al., 2020). In SCNT embryos, timely DNA demethylation and remethylation should occur to enable proper gene expression and cell reprogramming. However, donor cells are usually highly methylated and demethylation seems to be incomplete in SCNT embryos (Cao et al., 2020). In mice, several genes important for early development, including Dppa2/4, Oocyte-specific homeobox 6 (Obox6) and TEA domain transcription factor 4 (Tead4), failed to activate in SCNT embryos due to abnormal DNA methylation (Cao et al., 2020). In pigs, 4-cell SCNT embryos presented higher DNA methylation levels and expression of the DNA methyl transferase 1 (DNMT1) than control embryos (Deshmukh et al., 2011; Song et al., 2017). Attenuation of DNMT1 in nuclear donor cells 36 h prior to nuclear transfer decreased DNA methylation levels of OCT-4, NANOG and SOX2, increased their expression at EGA and blastocyst stages, and improved blastocyst rates of pig SCNT embryos (Song et al., 2017). A similar effect was observed by attenuating DNMT1 expression after nuclear transfer, which normalized the methylation status of OCT-4 and the Thy-1 cell surface antigen (THY1), a fibroblast marker, promoting OCT-4 activation and THY1 silencing in pig SCNT embryos (Huan et al., 2015b). Overexpression of MBD3 in pig SCNT embryos corrected methylation levels of NANOG, OCT-4 and long interspersed nuclear elements (LINEs), and increased blastocyst rates and quality (Wang et al., 2019).
Treatment of nuclear donor cells with 5-aza-2′-deoxycytidine (5-aza-dC), which incorporates onto DNA during replication leading to hypomethylation by inhibiting Dnmt1 action (Stresemann and Lyko, 2008), has been another strategy tested to improve SCNT efficiency. This approach increased the development and quality of pig SCNT embryos (Diao et al., 2013; Kumar et al., 2013), and enhanced transcript levels of DNMT1, two and 3 (Kumar et al., 2013; Huan et al., 2015b). Treatment of pig SCNT embryos with 5-aza-dC for 24 h after nuclear transfer increased embryo development, which was associated with a decrease in methylation levels of NANOG and an increase in the expression of NANOG and SOX2 (Huan et al., 2015a). Treatment with other inhibitors of DNA methyltransferases, including Zeburaline and RG108, also increased development of pig SCNT embryos (Zhai et al., 2018; Taweechaipaisankul et al., 2019b). A combined treatment of RG108 with Scriptaid, a histone deacetylase inhibitor, rescued defective methylation patters in the imprinted gene H19/insulin-like growth factor 2 (IGF2), and increased development of pig SCNT embryos (Xu et al., 2013). Moreover, melatonin was shown to favor DNA methylation reprogramming, and expression of imprinted, pluripotency and EGA related genes in pig SCNT embryos (Qu et al., 2020) (Table 1). Overexpression of the Ten-eleven translocation 3 (TET3) enhanced SCNT efficiency in bovine (Zhang et al., 2020), and goats (Han et al., 2018), however, this has not been investigated in pigs yet.
Pig oocytes have high levels of acetylated histones H3 and H4, especially at the germinal vesicle (GV) stage (Endo et al., 2005). Histone acetylation levels are controlled by acetyltransferases (HAT) and deacetylases (HDAC) enzymes (Endo et al., 2011). Pig oocytes express several HDACs, which regulate histone acetylation levels during oocyte maturation (Endo et al., 2008). The acetylation status of the lysine nine in the histone 3 (H3K9ac) has been proposed as a biomarker for embryo outcome, since it can be altered by culture conditions and it affects EGA and embryo development (Rollo et al., 2017). Normal maturation of pig oocytes is affected by HDACs inhibition (Jin Y.-X. et al., 2014; Huang et al., 2021).
There is consensus, based on studies conducted in different species, that development of SCNT embryos is improved by treatment with HDAC inhibitors (HDACi). Several HDACi molecules were shown to enhance pig SCNT efficiency and cell reprogramming. This includes PDX101 (Belinosat), LBH589 (Panobinostat), CI994, CUDC-101, MGCD0103, MC1568, JWC022214 (HDACi-14), JWC017379 (HDACi-79), PCI-24781, m-carboxycin- namic acid bishydroxamide (CBHA), Suberoylanilide hydroxamic acid (SAHA) 4-iodo-SAHA (ISAHA), Quisinostat, Bufexamac, M344, LAQ824, Oxamflatin, Sodium butyrate (NaBu), Valproic acid, and the more commonly used Scriptaid, and Trichostatin A (TSA) (Table 1). HDACi have also been associated with other molecules, including DNA methylation modifying agents (Table 1).
Although HDACi treatment has become an important component of SCNT protocols, the mechanism by which it promotes cell reprogramming has not been fully elucidated. In addition of enabling a more permissive chromatin state and improving gene expression by increasing acetylation levels, HDCAi treatment may also facilitate DNA damage repair in embryos (Bohrer et al., 2014; Wang et al., 2015). There is evidence that the effect of HDACi treatment in SCNT embryos is influenced by the differentiation state of the nuclear donor cell (Kishigami et al., 2006; Martinez-Diaz et al., 2010), and also by cell cycle interactions between the host cytoplast and the nuclear donor cell at the time of nuclear transfer (Rissi et al., 2016).
Histone methylation is crucial for cell reprograming and normal embryo development by controlling important embryo features such as EGA, cell differentiation and DNA damage response (Dahl et al., 2016; Qin et al., 2016; Glanzner et al., 2017; Glanzner et al., 2018; Jambhekar et al., 2019; Rissi et al., 2019; Glanzner et al., 2020). In the context of cell reprogramming in SCNT embryos, interest in histone methylation has gained more emphasis after an increase of 3.4-fold in blastocyst rates (88 vs. 26%) and 8.7-fold development to term (8.7 vs. 1%) was obtained by expressing the demethylase Kdm4d in mouse SCNT embryos (Matoba et al., 2014). In addition, Kdm4d expression in human SCNT embryos improved the establishment of SCNT-derived embryonic stem cell cultures (Chung et al., 2015). The impact of manipulating the expression of specific demethylases on development of SCNT embryos has been studied in several species including sheep (Zhang Y. et al., 2018), cattle (Liu et al., 2018; Zhou et al., 2019), and swine (Rissi et al., 2019; Glanzner et al., 2020). Among the histone methylation markers studied, H3K9me3 and H3K27me3 have been identified as the most important barriers of SCNT success (Matoba et al., 2014; Xie et al., 2016). In pigs, SCNT efficiency increased by attenuating H3K9me3 levels, either by suppressing specific methyltransferases (SUV39H1/2, G9A) (Huang et al., 2016; Jeong et al., 2020; Weng et al., 2020; Cao et al., 2021), or by expressing the demethylase KDM4A (Weng et al., 2020). Similarly, the inhibition of the H3K27 methyltransferase EZH2 increased pig SCNT efficiency, while the inhibition of the H3K27 demethylase UTX (also known as KDM6A) decreased the efficiency (Xie et al., 2016). There is also evidence supporting an important role for H3K4 methylation, which is normally associated with an accessible chromatin state and transcriptional activity, in the regulation of SCNT embryos (Hormanseder et al., 2017). In pigs, depletion of H3K4 methylation increased blastocyst rates and improved gene expression patterns in SCNT embryos (Zhang Z. et al., 2018).
The association of different epigenetic modifiers has been tested to improve pig SCNT efficiency. For example, increasing histone acetylation along with decreasing H3K9me3 levels (Cao et al., 2017; Jeong et al., 2021) or decreasing DNA methylation (Huang et al., 2016; Cao et al., 2017), improved gene expression patterns in SCNT embryos and pig cloning efficiency (Table 1).
Micro RNAs (miRNAs) have important roles on cell reprogramming and regulation of normal embryo development (Blakaj and Lin, 2008; Pauli et al., 2011; O'Brien et al., 2018). miRNAs are small RNA sequences (∼22/23 nucleotides) that regulate gene functions by pairing to target mRNAs and inducing their degradation or repressing translation (Bartel, 2004; O'Brien et al., 2018). There is evidence that miRNAs are required for normal oocyte growth and maturation, and early embryogenesis in several species, including pigs (Prather et al., 2009; Kaczmarek et al., 2020). For example, deletion of the RNase III endonuclease DICER, which is important for the biogenesis of miRNAs, impaired mouse embryo development due to a decrease in mature miRNAs (Tang et al., 2007). During early embryo development, miRNAs regulate the silencing of transcripts that are no longer necessary for development, modulate chromatin rearrangements, and promote cell pluripotency (Tang et al., 2007; Prather et al., 2009; Pauli et al., 2011). They also contribute to the regulation of embryo implantation and embryo-maternal communication (Kaczmarek et al., 2020). Examples of miRNAs identified to have important roles on embryo gene silencing, cell pluripotency and cell differentiation include miR-430, miR-125, miR-145 and let-7 (Blakaj and Lin, 2008; Bartel, 2009; Pauli et al., 2011).
Studies in pigs revealed that miRNAs, including miR-1343, miR-302, miR-302b and miR-200, are involved in the acquisition and maintenance of cell pluripotency by regulating the expression of TFs, such as SOX2 and OCT-4 (Ma et al., 2014; Qiao et al., 2019; Xie et al., 2019). However, the impact of miRNAs during cell reprogramming and development of pig SCNT embryos has not been extensively explored. In mice, miR-125b was identified as a crucial factor for cell reprogramming in SCNT embryos by regulating the expression of methyltransferases (e.g., Suv39h1) that control H3K9me levels and chromatin accessibility (Zhang J. et al., 2017). In pigs, overexpression of miR-148a in nuclear donor cells downregulated DNMT1 expression and increased blastocyst rates and total cell numbers in SCNT embryos (Wang et al., 2017) (Table 1). Treatment of pig SCNT embryos with the HDACi Scriptaid attenuated DNMT1 expression and H3K9me3 levels, as well as increased miR-152 expression, suggesting a link between miRNAs, DNMT1, and histone methylation and acetylation (Liang et al., 2015). However, more studies are required to dissect the regulation and cross talk between miRNAs and epigenetic regulation in pig SCNT embryos.
Long non-coding RNAs (lncRNAs) are known to regulate several biological processes such as chromatin function, signaling pathways, and mRNA stability (Statello et al., 2021). The X-inactive specific transcript gene produces a lncRNA (Xist), which is a major regulator of X chromosome inactivation (XCI) (Penny et al., 1996). During normal embryo development, XCI is required for compensation of X-linked genes between males and females. In mouse embryos, inactivation of the paternally-derived X chromosome starts at the early cleavage stages of development (Okamoto et al., 2011). In pig embryos, XCI is random and only observed in the late epiblast stage, as evidenced by the reduction of biallelic expression of X-linked genes and increase in H3K27me3 levels (Ramos-Ibeas et al., 2019).
Proper reprogramming of the X chromosome seems a critical component of SCNT efficiency (Table 1). Downregulation of X-linked genes due to the ectopic expression of Xist was detected in mouse SCNT embryos (Matoba et al., 2011). Similarly, XIST and X-linked genes were aberrantly expressed in pig SCNT embryos (Park C.-H. et al., 2012; Mao et al., 2015), and SCNT fetuses having abnormal development (Yuan et al., 2014; Ruan et al., 2018). Inactivation of XIST in nuclear donor cells or its attenuation in SCNT embryos improved blastocyst rates and cloning efficiency in mice and pigs (Matoba et al., 2011; Zeng et al., 2016; Ruan et al., 2018; Yang et al., 2019). A recent study demonstrated that the small molecule X1 can target a specific motif of Xist and blocks initiation of XCI (Aguilar et al., 2022). This may represent a new alternative for preventing early inaction of X chromosome in SCNT embryos, however, more studies are needed to evaluate efficiency and toxicity in pig embryos.
Recent progress in xenotransplantation of pig organs, along with the evolution of technologies for editing the pig genome has expanded interest in the production of cloned pigs by SCNT. Since it was proven that SCNT can make somatic cells regain totipotency, researchers in the field have been attempting to increase the efficiency of this reproductive method, but the progress has been modest. While in vitro development to the blastocyst stage of SCNT embryos is often similar to fertilized embryos, development to term and rate of alive cloned piglets remain low, confirming that not all chromatin functions regulating development have been reset properly. Cumulating evidence, mainly from mouse studies, pointed out that epigenetic marks, such as DNA and histone methylation, as well as histone acetylation, transcription factors, and non-coding RNAs, can all affect cell reprogramming and SCNT efficiency. Moreover, efforts to modulate these factors in SCNT embryos to mimic fertilized embryos by using molecules, or either attenuating or overexpressing genes, has shown encouraging results that improved not only the blastocyst rate and quality, but also development to term of cloned animals (Figure 1). In addition, some attempts taken to modulate multiple factors have further ameliorated mouse cloning efficiency, suggesting this route could be explored to improve SCNT protocols in other species, including pigs. For example, inhibition of HDAC and transcription along with either attenuation or expression of specific modulators of histones or DNA methylation may likely improve porcine SCNT efficiency. It is worth highlighting however, that there are fundamental differences between species in the regulation of early development, including the timing of EGA and first cell lineage specification, which should be taken in consideration when translating findings from one species to another.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Conceptualization, WGG, MPM, KG, and VB; writing: original draft preparation and figures, WGG, MPM, and KG; review and editing, WGG and VB. All authors have read and agreed to the content of the manuscript.
Research in our laboratory is funded by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canada Foundation for Innovation (CFI), le Consortium de Recherche et Innovations en Bioprocédés Industriels au Québec (CRIBIQ), and Mitacs. KG was supported by a scholarship from the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES). MPM is supported by a scholarship from the Fonds de recherche du Québec—Nature et technologie (FRQNT). KG and MPM received financial support from the NSERC—Collaborative Research and Training Experience Program—Genome Editing for Food Security and Environmental Sustainability (GEFSES).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilar, R., Spencer, K. B., Kesner, B., Rizvi, N. F., Badmalia, M. D., Mrozowich, T., et al. (2022). Targeting Xist with Compounds that Disrupt RNA Structure and X Inactivation. Nature 604, 160–166. doi:10.1038/s41586-022-04537-z
Bartel, D. P. (2009). MicroRNAs: Target Recognition and Regulatory Functions. Cell. 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Blakaj, A., and Lin, H. (2008). Piecing Together the Mosaic of Early Mammalian Development through microRNAs. J. Biol. Chem. 283 (15), 9505–9508. doi:10.1074/jbc.R800002200
Bohrer, R. C., Che, L., Gonçalves, P. B. D., Duggavathi, R., and Bordignon, V. (2013). Phosphorylated Histone H2A.X in Porcine Embryos Produced by IVF and Somatic Cell Nuclear Transfer. Reproduction 146 (4), 325–333. doi:10.1530/rep-13-0271
Bohrer, R. C., Duggavathi, R., and Bordignon, V. (2014). Inhibition of Histone Deacetylases Enhances DNA Damage Repair in SCNT Embryos. Cell. Cycle 13 (13), 2138–2148. doi:10.4161/cc.29215
Bordignon, V. (2019). “Animal Cloning - State of the Art and Applications,” in Comprehensive Biotechnology. Editor M. Moo-Young. 3th edn (Netherlands: Elsevier), 450–465. doi:10.1016/b978-0-444-64046-8.00266-4
Bordignon, V., Clarke, H. J., and Smith, L. C. (1999). Developmentally Regulated Loss and Reappearance of Immunoreactive Somatic Histone H1 on Chromatin of Bovine Morula-Stage Nuclei Following Transplantation into Oocytes1. Biol. Reprod. 61 (1), 22–30. doi:10.1095/biolreprod61.1.22
Bornelöv, S., Reynolds, N., Xenophontos, M., Gharbi, S., Johnstone, E., Floyd, R., et al. (2018). The Nucleosome Remodeling and Deacetylation Complex Modulates Chromatin Structure at Sites of Active Transcription to Fine-Tune Gene Expression. Mol. Cell. 71 (1), 56–72. e54. doi:10.1016/j.molcel.2018.06.003
Cao, L., Dai, X., Huang, S., Shen, K., Shi, D., and Li, X. (2021). Inhibition of Suv39h1/2 Expression Improves the Early Development of Debao Porcine Somatic Cell Nuclear Transfer Embryos. Reprod. Dom. Anim. 56 (7), 992–1003. doi:10.1111/rda.13942
Cao, P., Li, H., Zuo, Y., and Nashun, B. (2020). Characterization of DNA Methylation Patterns and Mining of Epigenetic Markers during Genomic Reprogramming in SCNT Embryos. Front. Cell. Dev. Biol. 8, 570107. doi:10.3389/fcell.2020.570107
Cao, Z., Hong, R., Ding, B., Zuo, X., Li, H., Ding, J., et al. (2017). TSA and BIX-01294 Induced Normal DNA and Histone Methylation and Increased Protein Expression in Porcine Somatic Cell Nuclear Transfer Embryos. PLoS One 12 (1), e0169092. doi:10.1371/journal.pone.0169092
Cervera, R. P., Martí-Gutiérrez, N., Escorihuela, E., Moreno, R., and Stojkovic, M. (2009). Trichostatin A Affects Histone Acetylation and Gene Expression in Porcine Somatic Cell Nucleus Transfer Embryos. Theriogenology 72 (8), 1097–1110. doi:10.1016/j.theriogenology.2009.06.030
Chang, C.-C., Gao, S., Sung, L.-Y., Corry, G. N., Ma, Y., Nagy, Z. P., et al. (2010). Rapid Elimination of the Histone Variant MacroH2A from Somatic Cell Heterochromatin after Nuclear Transfer. Cell. Reprogr. 12 (1), 43–53. doi:10.1089/cell.2009.0043
Chawalit, S., Nguyen, N. T., Tseng, J.-K., Lo, N.-W., Tu, C.-F., and Ju, J.-C. (2012). Trichostatin A and Ascorbic Acid Assist in the Development of Porcine Handmade Cloned Embryos via Different Physiologic Pathways. Reprod. Sci. 19 (9), 976–986. doi:10.1177/1933719112440049
Chung, Y. G., Matoba, S., Liu, Y., Eum, J. H., Lu, F., Jiang, W., et al. (2015). Histone Demethylase Expression Enhances Human Somatic Cell Nuclear Transfer Efficiency and Promotes Derivation of Pluripotent Stem Cells. Cell. Stem Cell. 17 (6), 758–766. doi:10.1016/j.stem.2015.10.001
Cong, P., Zhu, K., Ji, Q., Zhao, H., and Chen, Y. (2013). Effects of Trichostatin A on Histone Acetylation and Methylation Characteristics in Early Porcine Embryos after Somatic Cell Nuclear Transfer. Animal Sci. J. 84 (9), 639–649. doi:10.1111/asj.12059
Dahl, J. A., Jung, I., Aanes, H., Greggains, G. D., Manaf, A., Lerdrup, M., et al. (2016). Broad Histone H3K4me3 Domains in Mouse Oocytes Modulate Maternal-To-Zygotic Transition. Nature 537 (7621), 548–552. doi:10.1038/nature19360
Das, Z. C., Gupta, M. K., Uhm, S. J., and Lee, H. T. (2010). Increasing Histone Acetylation of Cloned Embryos, but Not Donor Cells, by Sodium Butyrate Improves TheirIn VitroDevelopment in Pigs. Cell. Reprogr. 12 (1), 95–104. doi:10.1089/cell.2009.0068
de Macedo, M. P., Glanzner, W. G., Gutierrez, K., and Bordignon, V. (2021). Chromatin Role in Early Programming of Embryos. Anim. Front. 11 (6), 57–65. doi:10.1093/af/vfab054
Deshmukh, R. S., Østrup, O., Østrup, E., Vejlsted, M., Niemann, H., Lucas-Hahn, A., et al. (2011). DNA Methylation in Porcine Preimplantation Embryos Developed In Vivo and Produced by In Vitro Fertilization, Parthenogenetic Activation and Somatic Cell Nuclear Transfer. Epigenetics 6 (2), 177–187. doi:10.4161/epi.6.2.13519
Diao, Y.-F., Naruse, K.-J., Han, R.-X., Li, X.-X., Oqani, R. K., Lin, T., et al. (2013). Treatment of Fetal Fibroblasts with DNA Methylation Inhibitors And/or Histone Deacetylase Inhibitors Improves the Development of Porcine Nuclear Transfer-Derived Embryos. Animal Reproduction Sci. 141 (3-4), 164–171. doi:10.1016/j.anireprosci.2013.08.008
Djekidel, M. N., Inoue, A., Matoba, S., Suzuki, T., Zhang, C., Lu, F., et al. (2018). Reprogramming of Chromatin Accessibility in Somatic Cell Nuclear Transfer Is DNA Replication Independent. Cell. Rep. 23 (7), 1939–1947. doi:10.1016/j.celrep.2018.04.036
Eckersley-Maslin, M., Alda-Catalinas, C., Blotenburg, M., Kreibich, E., Krueger, C., and Reik, W. (2019). Dppa2 and Dppa4 Directly Regulate the Dux-Driven Zygotic Transcriptional Program. Genes. Dev. 33 (3-4), 194–208. doi:10.1101/gad.321174.118
Egli, D., and Eggan, K. (2010). Recipient Cell Nuclear Factors Are Required for Reprogramming by Nuclear Transfer. Development 137 (12), 1953–1963. doi:10.1242/dev.046151
Endo, T., Imai, A., Shimaoka, T., Kano, K., and Naito, K. (2011). Histone Exchange Activity and its Correlation with Histone Acetylation Status in Porcine Oocytes. Reproduction 141 (4), 397–405. doi:10.1530/REP-10-0164
Endo, T., Kano, K., and Naito, K. (2008). Nuclear Histone Deacetylases Are Not Required for Global Histone Deacetylation during Meiotic Maturation in Porcine Oocytes1. Biol. Reprod. 78 (6), 1073–1080. doi:10.1095/biolreprod.107.067397
Endo, T., Naito, K., Aoki, F., Kume, S., and Tojo, H. (2005). Changes in Histone Modifications during In Vitro Maturation of Porcine Oocytes. Mol. Reprod. Dev. 71 (1), 123–128. doi:10.1002/mrd.20288
Gaspar-Maia, A., Qadeer, Z. A., Hasson, D., Ratnakumar, K., Adrian Leu, N., Leroy, G., et al. (2013). MacroH2A Histone Variants Act as a Barrier upon Reprogramming towards Pluripotency. Nat. Commun. 4, 1565. doi:10.1038/ncomms2582
Glanzner, W. G., Gutierrez, K., Rissi, V. B., de Macedo, M. P., Lopez, R., Currin, L., et al. (2020). Histone Lysine Demethylases KDM5B and KDM5C Modulate Genome Activation and Stability in Porcine Embryos. Front. Cell. Dev. Biol. 8, 151. doi:10.3389/fcell.2020.00151
Glanzner, W. G., Rissi, V. B., de Macedo, M. P., Mujica, L. K. S., Gutierrez, K., Bridi, A., et al. (2018). Histone 3 Lysine 4, 9, and 27 Demethylases Expression Profile in Fertilized and Cloned Bovine and Porcine Embryos†. Biol. Reprod. 98 (6), 742–751. doi:10.1093/biolre/ioy054
Glanzner, W. G., Wachter, A., Coutinho, A. R. S., Albornoz, M. S., Duggavathi, R., GonÇAlves, P. B. D., et al. (2017). Altered Expression of BRG1 and Histone Demethylases, and Aberrant H3K4 Methylation in Less Developmentally Competent Embryos at the Time of Embryonic Genome Activation. Mol. Reprod. Dev. 84 (1), 19–29. doi:10.1002/mrd.22762
Gutierrez, K., Dicks, N., Glanzner, W. G., Agellon, L. B., and Bordignon, V. (2015). Efficacy of the Porcine Species in Biomedical Research. Front. Genet. 6, 293. doi:10.3389/fgene.2015.00293
Han, C., Deng, R., Mao, T., Luo, Y., Wei, B., Meng, P., et al. (2018). Overexpression of Tet3 in Donor Cells Enhances Goat Somatic Cell Nuclear Transfer Efficiency. FEBS J. 285 (14), 2708–2723. doi:10.1111/febs.14515
Hansis, C., Barreto, G., Maltry, N., and Niehrs, C. (2004). Nuclear Reprogramming of Human Somatic Cells by xenopus Egg Extract Requires BRG1. Curr. Biol. 14 (16), 1475–1480. doi:10.1016/j.cub.2004.08.031
Hendrich, B., Guy, J., Ramsahoye, B., Wilson, V. A., and Bird, A. (2001). Closely Related Proteins MBD2 and MBD3 Play Distinctive but Interacting Roles in Mouse Development. Genes. Dev. 15 (6), 710–723. doi:10.1101/gad.194101
Hörmanseder, E., Simeone, A., Allen, G. E., Bradshaw, C. R., Figlmüller, M., Gurdon, J., et al. (2017). H3K4 Methylation-dependent Memory of Somatic Cell Identity Inhibits Reprogramming and Development of Nuclear Transfer Embryos. Cell. Stem Cell. 21 (1), 135–143. e136. doi:10.1016/j.stem.2017.03.003
Hou, L., Ma, F., Yang, J., Riaz, H., Wang, Y., Wu, W., et al. (2014). Effects of Histone Deacetylase Inhibitor Oxamflatin onIn VitroPorcine Somatic Cell Nuclear Transfer Embryos. Cell. Reprogr. 16 (4), 253–265. doi:10.1089/cell.2013.0058
Huan, Y. J., Zhu, J., Xie, B. T., Wang, J. Y., Liu, S. C., Zhou, Y., et al. (2013). Treating Cloned Embryos, but Not Donor Cells, with 5-Aza-2'-Deoxycytidine Enhances the Developmental Competence of Porcine Cloned Embryos. J. Reproduction Dev. 59 (5), 442–449. doi:10.1262/jrd.2013-026
Huan, Y., Wang, H., Wu, Z., Zhang, J., Zhu, J., Liu, Z., et al. (2015a). Epigenetic Modification of Cloned Embryos ImprovesNanogReprogramming in Pigs. Cell. Reprogr. 17 (3), 191–198. doi:10.1089/cell.2014.0103
Huan, Y., Wu, Z., Zhang, J., Zhu, J., Liu, Z., and Song, X. (2015b). Epigenetic Modification Agents Improve Gene-specific Methylation Reprogramming in Porcine Cloned Embryos. PLoS One 10 (6), e0129803. doi:10.1371/journal.pone.0129803
Huang, J., Zhang, H., Yao, J., Qin, G., Wang, F., Wang, X., et al. (2016). BIX-01294 Increases Pig Cloning Efficiency by Improving Epigenetic Reprogramming of Somatic Cell Nuclei. Reproduction 151 (1), 39–49. doi:10.1530/REP-15-0460
Huang, R., Sui, L., Fu, C., Zhai, Y., Dai, X., Zhang, S., et al. (2021). HDAC11 Inhibition Disrupts Porcine Oocyte Meiosis via Regulating α-tubulin Acetylation and Histone Modifications. Aging 13 (6), 8849–8864. doi:10.18632/aging.202697
Huang, Y., Tang, X., Xie, W., Zhou, Y., Li, D., Yao, C., et al. (2011). Histone Deacetylase Inhibitor Significantly Improved the Cloning Efficiency of Porcine Somatic Cell Nuclear Transfer Embryos. Cell. Reprogr. 13 (6), 513–520. doi:10.1089/cell.2011.0032
Ivanova, E., Canovas, S., Garcia-Martínez, S., Romar, R., Lopes, J. S., Rizos, D., et al. (2020). DNA Methylation Changes during Preimplantation Development Reveal Inter-species Differences and Reprogramming Events at Imprinted Genes. Clin. Epigenet 12 (1), 64. doi:10.1186/s13148-020-00857-x
Jambhekar, A., Dhall, A., and Shi, Y. (2019). Roles and Regulation of Histone Methylation in Animal Development. Nat. Rev. Mol. Cell. Biol. 20 (10), 625–641. doi:10.1038/s41580-019-0151-1
Jeong, P.-S., Sim, B.-W., Park, S.-H., Kim, M. J., Kang, H.-G., Nanjidsuren, T., et al. (2020). Chaetocin Improves Pig Cloning Efficiency by Enhancing Epigenetic Reprogramming and Autophagic Activity. Ijms 21 (14), 4836. doi:10.3390/ijms21144836
Jeong, P.-S., Yang, H.-J., Park, S.-H., Gwon, M. A., Joo, Y. E., Kim, M. J., et al. (2021). Combined Chaetocin/Trichostatin A Treatment Improves the Epigenetic Modification and Developmental Competence of Porcine Somatic Cell Nuclear Transfer Embryos. Front. Cell. Dev. Biol. 9, 709574. doi:10.3389/fcell.2021.709574
Jin, J.-X., Kang, J.-D., Li, S., Jin, L., Zhu, H.-Y., Guo, Q., et al. (2015). PXD101 Significantly Improves Nuclear Reprogramming and the In Vitro Developmental Competence of Porcine SCNT Embryos. Biochem. Biophysical Res. Commun. 456 (1), 156–161. doi:10.1016/j.bbrc.2014.11.051
Jin, J.-X., Lee, S., Taweechaipaisankul, A., Kim, G. A., and Lee, B. C. (2017a). The HDAC Inhibitor LAQ824 Enhances Epigenetic Reprogramming and In Vitro Development of Porcine SCNT Embryos. Cell. Physiol. Biochem. 41 (3), 1255–1266. doi:10.1159/000464389
Jin, J.-X., Li, S., Gao, Q.-S., Hong, Y., Jin, L., Zhu, H.-Y., et al. (2013). Significant Improvement of Pig Cloning Efficiency by Treatment with LBH589 after Somatic Cell Nuclear Transfer. Theriogenology 80 (6), 630–635. doi:10.1016/j.theriogenology.2013.06.006
Jin, J.-X., Li, S., Hong, Y., Jin, L., Zhu, H.-Y., Guo, Q., et al. (2014a). CUDC-101, a Histone Deacetylase Inhibitor, Improves the In Vitro and In Vivo Developmental Competence of Somatic Cell Nuclear Transfer Pig Embryos. Theriogenology 81 (4), 572–578. doi:10.1016/j.theriogenology.2013.11.011
Jin, L., Guo, Q., Zhang, G.-L., Xing, X.-X., Xuan, M.-F., Luo, Q.-R., et al. (2018). The Histone Deacetylase Inhibitor, CI994, Improves Nuclear Reprogramming and In Vitro Developmental Potential of Cloned Pig Embryos. Cell. Reprogr. 20 (3), 205–213. doi:10.1089/cell.2018.0001
Jin, L., Guo, Q., Zhu, H.-Y., Xing, X.-X., Zhang, G.-L., Xuan, M.-F., et al. (2017b). Histone Deacetylase Inhibitor M344 Significantly Improves Nuclear Reprogramming, Blastocyst Quality, and In Vitro Developmental Capacity of Cloned Pig Embryos. J. Anim. Sci. 95 (3), 1388–1395. doi:10.2527/jas.2016.1240
Jin, L., Guo, Q., Zhu, H.-Y., Xing, X.-X., Zhang, G.-L., Xuan, M.-F., et al. (2017c). Quisinostat Treatment Improves Histone Acetylation and Developmental Competence of Porcine Somatic Cell Nuclear Transfer Embryos. Mol. Reprod. Dev. 84 (4), 340–346. doi:10.1002/mrd.22787
Jin, L., Zhu, H.-Y., Guo, Q., Li, X.-C., Zhang, Y.-C., Cui, C.-D., et al. (2017d). Effect of Histone Acetylation Modification with MGCD0103, a Histone Deacetylase Inhibitor, on Nuclear Reprogramming and the Developmental Competence of Porcine Somatic Cell Nuclear Transfer Embryos. Theriogenology 87, 298–305. doi:10.1016/j.theriogenology.2016.09.011
Jin, L., Zhu, H.-Y., Guo, Q., Li, X.-C., Zhang, Y.-C., Zhang, G.-L., et al. (2016). PCI-24781 Can Improve In Vitro and In Vivo Developmental Capacity of Pig Somatic Cell Nuclear Transfer Embryos. Biotechnol. Lett. 38 (9), 1433–1441. doi:10.1007/s10529-016-2141-0
Jin, Y.-X., Zhao, M.-H., Zheng, Z., Kwon, J.-S., Lee, S.-K., Cui, X.-S., et al. (2014b). Histone Deacetylase Inhibitor Trichostatin A Affects Porcine Oocyte Maturation In Vitro. Reprod. Fertil. Dev. 26 (6), 806–816. doi:10.1071/RD13013
Kaczmarek, M. M., Najmula, J., Guzewska, M. M., and Przygrodzka, E. (2020). MiRNAs in the Peri-Implantation Period: Contribution to Embryo-Maternal Communication in Pigs. Ijms 21 (6), 2229. doi:10.3390/ijms21062229
Kim, M. J., Oh, H. J., Choi, Y. B., Lee, S., Setyawan, E. M. N., Lee, S. H., et al. (2018). Suberoylanilide Hydroxamic Acid during In Vitro Culture Improves Development of Dog-Pig Interspecies Cloned Embryos but Not Dog Cloned Embryos. J. Reproduction Dev. 64 (3), 277–282. doi:10.1262/jrd.2017-112
Kim, S.-J., Kwon, H.-S., Kwon, D.-k., Koo, O.-J., Moon, J.-H., Park, E.-J., et al. (2019). Production of Transgenic Porcine Embryos Reconstructed with Induced Pluripotent Stem-like Cells Derived from Porcine Endogenous Factors Using piggyBac System. Cell. Reprogr. 21 (1), 26–36. doi:10.1089/cell.2018.0036
Kim, Y. J., Ahn, K. S., Kim, M., and Shim, H. (2011). Comparison of Potency between Histone Deacetylase Inhibitors Trichostatin A and Valproic Acid on Enhancing In Vitro Development of Porcine Somatic Cell Nuclear Transfer Embryos. Vitro Cell. Dev.Biol.-Animal 47 (4), 283–289. doi:10.1007/s11626-011-9394-7
Kishigami, S., Mizutani, E., Ohta, H., Hikichi, T., Thuan, N. V., Wakayama, S., et al. (2006). Significant Improvement of Mouse Cloning Technique by Treatment with Trichostatin A after Somatic Nuclear Transfer. Biochem. Biophysical Res. Commun. 340 (1), 183–189. doi:10.1016/j.bbrc.2005.11.164
Kumar, B. M., Maeng, G.-H., Lee, Y.-M., Lee, J.-H., Jeon, B.-G., Ock, S.-A., et al. (2013). Epigenetic Modification of Fetal Fibroblasts Improves Developmental Competency and Gene Expression in Porcine Cloned Embryos. Vet. Res. Commun. 37 (1), 19–28. doi:10.1007/s11259-012-9542-x
Kunitomi, A., Yuasa, S., Sugiyama, F., Saito, Y., Seki, T., Kusumoto, D., et al. (2016). H1foo Has a Pivotal Role in Qualifying Induced Pluripotent Stem Cells. Stem Cell. Rep. 6 (6), 825–833. doi:10.1016/j.stemcr.2016.04.015
Lee, D.-K., Park, C.-H., Choi, K.-H., Jeong, Y.-i., Uh, K.-J., Hwang, J. Y., et al. (2016). Aggregation of Cloned Embryos in Empty Zona Pellucida Improves Derivation Efficiency of Pig ES-like Cells. Zygote 24 (6), 909–917. doi:10.1017/S0967199416000241
Lee, E., Lee, S. H., Kim, S., Jeong, Y. W., Kim, J. H., Koo, O. J., et al. (2006). Analysis of Nuclear Reprogramming in Cloned Miniature Pig Embryos by Expression of Oct-4 and Oct-4 Related Genes. Biochem. Biophysical Res. Commun. 348 (4), 1419–1428. doi:10.1016/j.bbrc.2006.08.004
Li, J., Svarcova, O., Villemoes, K., Kragh, P. M., Schmidt, M., Bøgh, I. B., et al. (2008). High In Vitro Development after Somatic Cell Nuclear Transfer and Trichostatin A Treatment of Reconstructed Porcine Embryos. Theriogenology 70 (5), 800–808. doi:10.1016/j.theriogenology.2008.05.046
Liang, S., Zhao, M.-H., Choi, J.-w., Kim, N.-H., and Cui, X.-S. (2015). Scriptaid Treatment Decreases DNA Methyltransferase 1 Expression by Induction of MicroRNA-152 Expression in Porcine Somatic Cell Nuclear Transfer Embryos. PLoS One 10 (8), e0134567. doi:10.1371/journal.pone.0134567
Liu, L., Liu, Y., Gao, F., Song, G., Wen, J., Guan, J., et al. (2012). Embryonic Development and Gene Expression of Porcine SCNTEmbryos Treated with Sodium Butyrate. J. Exp. Zool. 318 (3), 224–234. doi:10.1002/jez.b.22440
Liu, X., Wang, Y., Gao, Y., Su, J., Zhang, J., Xing, X., et al. (2018). H3K9 Demethylase KDM4E Is an Epigenetic Regulator for Bovine Embryonic Development and a Defective Factor for Nuclear Reprogramming. Development 145 (4), dev158261. doi:10.1242/dev.158261
Liu, Z., Xiang, G., Xu, K., Che, J., Xu, C., Li, K., et al. (2020). Transcriptome Analyses Reveal Differential Transcriptional Profiles in Early- and Late-Dividing Porcine Somatic Cell Nuclear Transfer Embryos. Genes. 11 (12), 1499. doi:10.3390/genes11121499
Luo, B., Ju, S., Muneri, C. W., and Rui, R. (2015). Effects of Histone Acetylation Status on the Early Development ofIn VitroPorcine Transgenic Cloned Embryos. Cell. Reprogr. 17 (1), 41–48. doi:10.1089/cell.2014.0041
Ma, K., Song, G., An, X., Fan, A., Tan, W., Tang, B., et al. (2014). miRNAs Promote Generation of Porcine-Induced Pluripotent Stem Cells. Mol. Cell. Biochem. 389 (1-2), 209–218. doi:10.1007/s11010-013-1942-x
Mao, J., Zhao, M.-T., Whitworth, K. M., Spate, L. D., Walters, E. M., O'Gorman, C., et al. (2015). Oxamflatin Treatment Enhances Cloned Porcine Embryo Development and Nuclear Reprogramming. Cell. Reprogr. 17 (1), 28–40. doi:10.1089/cell.2014.0075
Martinez-Diaz, M. A., Che, L., Albornoz, M., Seneda, M. M., Collis, D., Coutinho, A. R. S., et al. (2010). Pre- and Postimplantation Development of Swine-Cloned Embryos Derived from Fibroblasts and Bone Marrow Cells after Inhibition of Histone Deacetylases. Cell. Reprogr. 12 (1), 85–94. doi:10.1089/cell.2009.0047
Matoba, S., Inoue, K., Kohda, T., Sugimoto, M., Mizutani, E., Ogonuki, N., et al. (2011). RNAi-Mediated Knockdown of Xist Can Rescue the Impaired Postimplantation Development of Cloned Mouse Embryos. Proc. Natl. Acad. Sci. U.S.A. 108 (51), 20621–20626. doi:10.1073/pnas.1112664108
Matoba, S., Liu, Y., Lu, F., Iwabuchi, K. A., Shen, L., Inoue, A., et al. (2014). Embryonic Development Following Somatic Cell Nuclear Transfer Impeded by Persisting Histone Methylation. Cell. 159 (4), 884–895. doi:10.1016/j.cell.2014.09.055
Matoba, S., and Zhang, Y. (2018). Somatic Cell Nuclear Transfer Reprogramming: Mechanisms and Applications. Cell. Stem Cell. 23 (4), 471–485. doi:10.1016/j.stem.2018.06.018
Miyoshi, K., Kawaguchi, H., Maeda, K., Sato, M., Akioka, K., Noguchi, M., et al. (2016). Birth of Cloned Microminipigs Derived from Somatic Cell Nuclear Transfer Embryos that Have Been Transiently Treated with Valproic Acid. Cell. Reprogr. 18 (6), 390–400. doi:10.1089/cell.2016.0025
Miyoshi, K., Mori, H., Mizobe, Y., Akasaka, E., Ozawa, A., Yoshida, M., et al. (2010). Valproic Acid EnhancesIn VitroDevelopment and Oct-3/4 Expression of Miniature Pig Somatic Cell Nuclear Transfer Embryos. Cell. Reprogr. 12 (1), 67–74. doi:10.1089/cell.2009.0032
Mohana Kumar, B., Song, H.-J., Cho, S.-K., Balasubramanian, S., Choe, S.-Y., and Rho, G.-J. (2007). Effect of Histone Acetylation Modification with Sodium Butyrate, a Histone Deacetylase Inhibitor, on Cell Cycle, Apoptosis, Ploidy and Gene Expression in Porcine Fetal Fibroblasts. J. Reproduction Dev. 53 (4), 903–913. doi:10.1262/jrd.18180
Nashun, B., Akiyama, T., Suzuki, M. G., and Aoki, F. (2011). Dramatic Replacement of Histone Variants during Genome Remodeling in Nuclear-Transferred Embryos. Epigenetics 6 (12), 1489–1497. doi:10.4161/epi.6.12.18206
Ng, R. K., and Gurdon, J. B. (2005). Epigenetic Memory of Active Gene Transcription Is Inherited through Somatic Cell Nuclear Transfer. Proc. Natl. Acad. Sci. U.S.A. 102 (6), 1957–1962. doi:10.1073/pnas.0409813102
Ning, S.-F., Li, Q.-Y., Liang, M.-M., Yang, X.-G., Xu, H.-Y., Lu, Y.-Q., et al. (2013). Methylation Characteristics and Developmental Potential of Guangxi Bama Minipig (Sus scrofa Domestica) Cloned Embryos from Donor Cells Treated with Trichostatin A and 5-Aza-2′-Deoxycytidine. Zygote 21 (2), 178–186. doi:10.1017/S0967199411000797
No, J.-G., Hur, T.-Y., Zhao, M., Lee, S., Choi, M.-K., Nam, Y.-S., et al. (2018). Scriptaid Improves the Reprogramming of Donor Cells and Enhances Canine-Porcine Interspecies Embryo Development. Reprod. Biol. 18 (1), 18–26. doi:10.1016/j.repbio.2017.11.001
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Okamoto, I., Patrat, C., Thépot, D., Peynot, N., Fauque, P., Daniel, N., et al. (2011). Eutherian Mammals Use Diverse Strategies to Initiate X-Chromosome Inactivation during Development. Nature 472 (7343), 370–374. doi:10.1038/nature09872
Ongaratto, F. L., Rodriguez-Villamil, P., Bertolini, M., and Carlson, D. F. (2020). Influence of Oocyte Selection, Activation with a Zinc Chelator and Inhibition of Histone Deacetylases on Cloned Porcine Embryo and Chemically Activated Oocytes Development. Zygote 28 (4), 286–290. doi:10.1017/S0967199419000856
Park, C.-H., Jeong, Y. H., Jeong, Y.-I., Lee, S.-Y., Jeong, Y.-W., Shin, T., et al. (2012a). X-Linked Gene Transcription Patterns in Female and Male In Vivo, In Vitro and Cloned Porcine Individual Blastocysts. PLoS One 7 (12), e51398. doi:10.1371/journal.pone.0051398
Park, S.-J., Park, H.-J., Koo, O.-J., Choi, W.-J., Moon, J.-h., Kwon, D.-K., et al. (2012b). Oxamflatin Improves Developmental Competence of Porcine Somatic Cell Nuclear Transfer Embryos. Cell. Reprogr. 14 (5), 398–406. doi:10.1089/cell.2012.0007
Pauli, A., Rinn, J. L., and Schier, A. F. (2011). Non-coding RNAs as Regulators of Embryogenesis. Nat. Rev. Genet. 12 (2), 136–149. doi:10.1038/nrg2904
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X Chromosome Inactivation. Nature 379 (6561), 131–137. doi:10.1038/379131a0
Prather, R. S., Ross, J. W., Isom, S. C., and Green, J. A. (2009). Transcriptional, Post-transcriptional and Epigenetic Control of Porcine Oocyte Maturation and Embryogenesis. Soc. Reprod. Fertil. Suppl. 66, 165–176.
Qiao, S., Deng, Y., Li, S., Yang, X., Shi, D., and Li, X. (2019). Partially Reprogrammed Induced Pluripotent Stem Cells Using MicroRNA Cluster miR-302s in Guangxi Bama Minipig Fibroblasts. Cell. Reprogr. 21 (5), 229–237. doi:10.1089/cell.2019.0035
Qin, H., Zhao, A., Zhang, C., and Fu, X. (2016). Epigenetic Control of Reprogramming and Transdifferentiation by Histone Modifications. Stem Cell. Rev Rep 12 (6), 708–720. doi:10.1007/s12015-016-9682-4
Qu, J., Sun, M., Wang, X., Song, X., He, H., and Huan, Y. (2020). Melatonin Enhances the Development of Porcine Cloned Embryos by Improving DNA Methylation Reprogramming. Cell. Reprogr. 22 (3), 156–166. doi:10.1089/cell.2019.0103
Ramos-Ibeas, P., Sang, F., Zhu, Q., Tang, W. W. C., Withey, S., Klisch, D., et al. (2019). Pluripotency and X Chromosome Dynamics Revealed in Pig Pre-gastrulating Embryos by Single Cell Analysis. Nat. Commun. 10 (1), 500. doi:10.1038/s41467-019-08387-8
Reik, W., Dean, W., and Walter, J. (2001). Epigenetic Reprogramming in Mammalian Development. Science 293 (5532), 1089–1093. doi:10.1126/science.1063443
Rissi, V. B., Glanzner, W. G., de Macedo, M. P., Gutierrez, K., Baldassarre, H., Gonçalves, P. B. D., et al. (2019). The Histone Lysine Demethylase KDM7A Is Required for Normal Development and First Cell Lineage Specification in Porcine Embryos. Epigenetics 14 (11), 1088–1101. doi:10.1080/15592294.2019.1633864
Rissi, V. B., Glanzner, W. G., de Macedo, M. P., Mujica, L. K. S., Campagnolo, K., Gutierrez, K., et al. (2019). Inhibition of RNA Synthesis during Scriptaid Exposure Enhances Gene Reprogramming in SCNT Embryos. Reproduction 157 (2), 123–133. doi:10.1530/REP-18-0366
Rissi, V. B., Glanzner, W. G., Mujica, L. K. S., Antoniazzi, A. Q., Gonçalves, P. B. D., and Bordignon, V. (2016). Effect of Cell Cycle Interactions and Inhibition of Histone Deacetylases on Development of Porcine Embryos Produced by Nuclear Transfer. Cell. Reprogr. 18 (2), 8–16. doi:10.1089/cell.2015.0052
Rollo, C., Li, Y., Jin, X. L., and O’Neill, C. (2017). Histone 3 Lysine 9 Acetylation Is a Biomarker of the Effects of Culture on Zygotes. Reproduction 154 (4), 375–385. doi:10.1530/REP-17-0112
Ruan, D., Peng, J., Wang, X., Ouyang, Z., Zou, Q., Yang, Y., et al. (2018). XIST Derepression in Active X Chromosome Hinders Pig Somatic Cell Nuclear Transfer. Stem Cell. Rep. 10 (2), 494–508. doi:10.1016/j.stemcr.2017.12.015
Shi, B., Gao, D., Zhong, L., Zhi, M., Weng, X., Xu, J., et al. (2020). IRF-1 Expressed in the Inner Cell Mass of the Porcine Early Blastocyst Enhances the Pluripotency of Induced Pluripotent Stem Cells. Stem Cell. Res. Ther. 11 (1), 505. doi:10.1186/s13287-020-01983-2
Siriboon, C., Li, T.-S., Yu, C.-W., Chern, J.-W., and Ju, J.-C. (2018). Novel Histone Deacetylase Inhibitors and Embryo Aggregation Enhance Cloned Embryo Development and ES Cell Derivation in Pigs. PLoS One 13 (9), e0204588. doi:10.1371/journal.pone.0204588
Song, X., Liu, Z., He, H., Wang, J., Li, H., Li, J., et al. (2017). Dnmt1s in Donor Cells Is a Barrier to SCNT-Mediated DNA Methylation Reprogramming in Pigs. Oncotarget 8 (21), 34980–34991. doi:10.18632/oncotarget.16507
Song, Y., Hai, T., Wang, Y., Guo, R., Li, W., Wang, L., et al. (2014a). Epigenetic Reprogramming, Gene Expression and In Vitro Development of Porcine SCNT Embryos Are Significantly Improved by a Histone Deacetylase Inhibitor-M-Carboxycinnamic Acid Bishydroxamide (CBHA). Protein Cell. 5 (5), 382–393. doi:10.1007/s13238-014-0034-3
Song, Z., Ji, Q., Zhao, H., Nie, Y., He, Z., Chen, Y., et al. (2014b). Ectopic Expression of Reprogramming Factors Enhances the Development of Cloned Porcine Embryos. Biotechnol. Lett. 36 (10), 1953–1961. doi:10.1007/s10529-014-1580-8
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell. Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Stresemann, C., and Lyko, F. (2008). Modes of Action of the DNA Methyltransferase Inhibitors Azacytidine and Decitabine. Int. J. Cancer 123 (1), 8–13. doi:10.1002/ijc.23607
Sun, J., Cui, K., Li, Z., Gao, B., Jiang, J., Liu, Q., et al. (2020). Histone Hyperacetylation May Improve the Preimplantation Development and Epigenetic Status of Cloned Embryos. Reprod. Biol. 20 (2), 237–246. doi:10.1016/j.repbio.2020.02.005
Sun, J., Cui, K., Li, Z., Lu, X., Xu, Z., Liu, Q., et al. (2017). Suberoylanilide Hydroxamic Acid, a Novel Histone Deacetylase Inhibitor, Improves the Development and Acetylation Level of Miniature Porcine Handmade Cloning Embryos. Reprod. Dom. Anim. 52 (5), 763–774. doi:10.1111/rda.12977
Sun, J., Liu, Q., Lv, L., Sun, R., Li, Z. P., Huang, B., et al. (2021). HDAC6 Is Involved in the Histone Deacetylation of In Vitro Maturation Oocytes and the Reprogramming of Nuclear Transplantation in Pig. Reprod. Sci. 28 (9), 2630–2640. doi:10.1007/s43032-021-00533-2
Tang, F., Kaneda, M., O’Carroll, D., Hajkova, P., Barton, S. C., Sun, Y. A., et al. (2007). Maternal microRNAs Are Essential for Mouse Zygotic Development. Genes. Dev. 21 (6), 644–648. doi:10.1101/gad.418707
Tao, C., Li, J., Zhang, X., Chen, B., Chi, D., Zeng, Y., et al. (2017). Dynamic Reorganization of Nucleosome Positioning in Somatic Cells after Transfer into Porcine Enucleated Oocytes. Stem Cell. Rep. 9 (2), 642–653. doi:10.1016/j.stemcr.2017.06.004
Taweechaipaisankul, A., Jin, J.-X., Lee, S., Kim, G. A., Suh, Y. H., Ahn, M. S., et al. (2019a). Improved Early Development of Porcine Cloned Embryos by Treatment with Quisinostat, a Potent Histone Deacetylase Inhibitor. J. Reproduction Dev. 65 (2), 103–112. doi:10.1262/jrd.2018-098
Taweechaipaisankul, A., Kim, G. A., Jin, J. X., Lee, S., Qasim, M., Kim, E. H., et al. (2019b). Enhancement of Epigenetic Reprogramming Status of Porcine Cloned Embryos with Zebularine, a DNA Methyltransferase Inhibitor. Mol. Reprod. Dev. 86 (8), 1013–1022. doi:10.1002/mrd.23178
Teranishi, T., Tanaka, M., Kimoto, S., Ono, Y., Miyakoshi, K., Kono, T., et al. (2004). Rapid Replacement of Somatic Linker Histones with the Oocyte-specific Linker Histone H1foo in Nuclear Transfer. Dev. Biol. 266 (1), 76–86. doi:10.1016/j.ydbio.2003.10.004
Wang, H., Cui, W., Meng, C., Zhang, J., Li, Y., Qian, Y., et al. (2018). MC1568 Enhances Histone Acetylation during Oocyte Meiosis and Improves Development of Somatic Cell Nuclear Transfer Embryos in Pig. Cell. Reprogr. 20 (1), 55–65. doi:10.1089/cell.2017.0023
Wang, H., Luo, Y., Lin, Z., Lee, I.-W., Kwon, J., Cui, X.-S., et al. (2015). Effect of ATM and HDAC Inhibition on Etoposide-Induced DNA Damage in Porcine Early Preimplantation Embryos. PLoS One 10 (11), e0142561. doi:10.1371/journal.pone.0142561
Wang, P., Li, X., Cao, L., Huang, S., Li, H., Zhang, Y., et al. (2017). MicroRNA-148a Overexpression Improves the Early Development of Porcine Somatic Cell Nuclear Transfer Embryos. PLoS One 12 (6), e0180535. doi:10.1371/journal.pone.0180535
Wang, X., Shi, J., Cai, G., Zheng, E., Liu, D., Wu, Z., et al. (2019). Overexpression of MBD3 Improves Reprogramming of Cloned Pig Embryos. Cell. Reprogr. 21 (5), 221–228. doi:10.1089/cell.2019.0008
Wang, Y., Li, Y., Luan, D., Kang, J., He, R., Zhang, Y., et al. (2020). Dynamic Replacement of H3.3 Affects Nuclear Reprogramming in Early Bovine SCNT Embryos. Theriogenology 154, 43–52. doi:10.1016/j.theriogenology.2020.05.031
Wen, D., Banaszynski, L. A., Liu, Y., Geng, F., Noh, K.-M., Xiang, J., et al. (2014a). Histone Variant H3.3 Is an Essential Maternal Factor for Oocyte Reprogramming. Proc. Natl. Acad. Sci. U.S.A. 111 (20), 7325–7330. doi:10.1073/pnas.1406389111
Wen, D., Banaszynski, L. A., Rosenwaks, Z., Allis, C. D., and Rafii, S. (2014b). H3.3 Replacement Facilitates Epigenetic Reprogramming of Donor Nuclei in Somatic Cell Nuclear Transfer Embryos. Nucleus 5 (5), 369–375. doi:10.4161/nucl.36231
Weng, X.-g., Cai, M.-m., Zhang, Y.-t., Liu, Y., Liu, C., and Liu, Z.-h. (2020). Improvement in the In Vitro Development of Cloned Pig Embryos after Kdm4a Overexpression and an H3K9me3 Methyltransferase Inhibitor Treatment. Theriogenology 146, 162–170. doi:10.1016/j.theriogenology.2019.11.027
Whitworth, K. M., Mao, J., Lee, K., Spollen, W. G., Samuel, M. S., Walters, E. M., et al. (2015). Transcriptome Analysis of PigIn Vivo,In Vitro-Fertilized, and Nuclear Transfer Blastocyst-Stage Embryos Treated with Histone Deacetylase Inhibitors Postfusion and Activation Reveals Changes in the Lysosomal Pathway. Cell. Reprogr. 17 (4), 243–258. doi:10.1089/cell.2015.0022
Wilmut, I., Schnieke, A. E., McWhir, J., Kind, A. J., and Campbell, K. H. S. (1997). Viable Offspring Derived from Fetal and Adult Mammalian Cells. Nature 385 (6619), 810–813. doi:10.1038/385810a0
Xie, B., Zhang, H., Wei, R., Li, Q., Weng, X., Kong, Q., et al. (2016). Histone H3 Lysine 27 Trimethylation Acts as an Epigenetic Barrier in Porcine Nuclear Reprogramming. Reproduction 151 (1), 9–16. doi:10.1530/REP-15-0338
Xie, Y., Cao, H., Zhang, Z., Zhang, S., and Wang, H. (2019). Molecular Network of miR-1343 Regulates the Pluripotency of Porcine Pluripotent Stem Cells via Repressing OTX2 Expression. RNA Biol. 16 (1), 82–92. doi:10.1080/15476286.2018.1559688
Xu, W., Li, Z., Yu, B., He, X., Shi, J., Zhou, R., et al. (2013). Effects of DNMT1 and HDAC Inhibitors on Gene-specific Methylation Reprogramming during Porcine Somatic Cell Nuclear Transfer. PLoS One 8 (5), e64705. doi:10.1371/journal.pone.0064705
Yamanaka, K.-i., Sugimura, S., Wakai, T., Kawahara, M., and Sato, E. (2009). Acetylation Level of Histone H3 in Early Embryonic Stages Affects Subsequent Development of Miniature Pig Somatic Cell Nuclear Transfer Embryos. J. Reproduction Dev. 55 (6), 638–644. doi:10.1262/jrd.20245
Yang, L., Liu, X., Song, L., Di, A., Su, G., Bai, C., et al. (2020). Transient Dux Expression Facilitates Nuclear Transfer and Induced Pluripotent Stem Cell Reprogramming. EMBO Rep. 21 (9), e50054. doi:10.15252/embr.202050054
Yang, X., Wu, X., Yang, Y., Gu, T., Hong, L., Zheng, E., et al. (2019). Improvement of Developmental Competence of Cloned Male Pig Embryos by Short Hairpin Ribonucleic Acid (shRNA) Vector-Based but Not Small Interfering RNA (siRNA)-Mediated RNA Interference (RNAi) of Xist Expression. J. Reproduction Dev. 65 (6), 533–539. doi:10.1262/jrd.2019-070
Yu, S., Zhang, R., Shen, Q., Zhu, Z., Zhang, J., Wu, X., et al. (2021). ESRRB Facilitates the Conversion of Trophoblast-like Stem Cells from Induced Pluripotent Stem Cells by Directly Regulating CDX2. Front. Cell. Dev. Biol. 9, 712224. doi:10.3389/fcell.2021.712224
Yuan, L., Wang, A., Yao, C., Huang, Y., Duan, F., Lv, Q., et al. (2014). Aberrant Expression of Xist in Aborted Porcine Fetuses Derived from Somatic Cell Nuclear Transfer Embryos. Ijms 15 (12), 21631–21643. doi:10.3390/ijms151221631
Yun, Y., Zhao, G.-m., Wu, S.-j., Li, W., and Lei, A.-m. (2012). Replacement of H1 Linker Histone during Bovine Somatic Cell Nuclear Transfer. Theriogenology 78 (6), 1371–1380. doi:10.1016/j.theriogenology.2012.06.004
Zeng, F., Huang, Z., Yuan, Y., Shi, J., Cai, G., Liu, D., et al. (2016). Effects of RNAi-Mediated Knockdown of Xist on the Developmental Efficiency of Cloned Male Porcine Embryos. J. Reproduction Dev. 62 (6), 591–597. doi:10.1262/jrd.2016-095
Zhai, Y., Zhang, Z., Yu, H., Su, L., Yao, G., Ma, X., et al. (2018). Dynamic Methylation Changes of DNA and H3K4 by RG108 Improve Epigenetic Reprogramming of Somatic Cell Nuclear Transfer Embryos in Pigs. Cell. Physiol. Biochem. 50 (4), 1376–1397. doi:10.1159/000494598
Zhang, J., Hao, L., Wei, Q., Zhang, S., Cheng, H., Zhai, Y., et al. (2020). TET3 Overexpression Facilitates DNA Reprogramming and Early Development of Bovine SCNT Embryos. Reproduction 160 (3), 379–391. doi:10.1530/REP-20-0021
Zhang, J., Qu, P., Zhou, C., Liu, X., Ma, X., Wang, M., et al. (2017a). MicroRNA-125b Is a Key Epigenetic Regulatory Factor that Promotes Nuclear Transfer Reprogramming. J. Biol. Chem. 292 (38), 15916–15926. doi:10.1074/jbc.M117.796771
Zhang, L., Huang, Y., Wu, Y., Si, J., Huang, Y., Jiang, Q., et al. (2017b). Scriptaid Upregulates Expression of Development-Related Genes, Inhibits Apoptosis, and Improves the Development of Somatic Cell Nuclear Transfer Mini-Pig Embryos. Cell. Reprogr. 19 (1), 19–26. doi:10.1089/cell.2016.0033
Zhang, Y., Li, J., Villemoes, K., Pedersen, A. M., Purup, S., and Vajta, G. (2007). An Epigenetic Modifier Results in Improved In Vitro Blastocyst Production after Somatic Cell Nuclear Transfer. Cloning Stem Cells 9 (3), 357–363. doi:10.1089/clo.2006.0090
Zhang, Y., Wang, Q., Liu, K., Gao, E., Guan, H., and Hou, J. (2018a). Treatment of Donor Cells with Recombinant KDM4D Protein Improves Preimplantation Development of Cloned Ovine Embryos. Cytotechnology 70 (5), 1469–1477. doi:10.1007/s10616-018-0224-6
Zhang, Z., Zhai, Y., Ma, X., Zhang, S., An, X., Yu, H., et al. (2018b). Down-Regulation of H3K4me3 by MM-102 Facilitates Epigenetic Reprogramming of Porcine Somatic Cell Nuclear Transfer Embryos. Cell. Physiol. Biochem. 45 (4), 1529–1540. doi:10.1159/000487579
Zhao, J., Hao, Y., Ross, J. W., Spate, L. D., Walters, E. M., Samuel, M. S., et al. (2010). Histone Deacetylase Inhibitors ImproveIn VitroandIn VivoDevelopmental Competence of Somatic Cell Nuclear Transfer Porcine Embryos. Cell. Reprogr. 12 (1), 75–83. doi:10.1089/cell.2009.0038
Zhao, J., Ross, J. W., Hao, Y., Spate, L. D., Walters, E. M., Samuel, M. S., et al. (2009). Significant Improvement in Cloning Efficiency of an Inbred Miniature Pig by Histone Deacetylase Inhibitor Treatment after Somatic Cell Nuclear Transfer1. Biol. Reprod. 81 (3), 525–530. doi:10.1095/biolreprod.109.077016
Keywords: pig, cloning, SCNT, embryo development, chromatin, epigenetics, histone acetylation, histone methylation
Citation: Glanzner WG, de Macedo MP, Gutierrez K and Bordignon V (2022) Enhancement of Chromatin and Epigenetic Reprogramming in Porcine SCNT Embryos—Progresses and Perspectives. Front. Cell Dev. Biol. 10:940197. doi: 10.3389/fcell.2022.940197
Received: 10 May 2022; Accepted: 20 June 2022;
Published: 11 July 2022.
Edited by:
Sadie L. Marjani, Central Connecticut State University, United StatesReviewed by:
Bojiang Li, Shenyang Agricultural University, ChinaCopyright © 2022 Glanzner, de Macedo, Gutierrez and Bordignon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vilceu Bordignon, dmlsY2V1LmJvcmRpZ25vbkBtY2dpbGwuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.