- Department of Biology, Philipps-University Marburg, Marburg, Germany
Peroxisomes are eukaryotic organelles with critical functions in cellular energy and lipid metabolism. Depending on the organism, cell type, and developmental stage, they are involved in numerous other metabolic and regulatory pathways. Many peroxisomal functions require factors also relevant to other cellular compartments. Here, we review proteins shared by peroxisomes and at least one different site within the cell. We discuss the mechanisms to achieve dual targeting, their regulation, and functional consequences. Characterization of dual targeting is fundamental to understand how peroxisomes are integrated into the metabolic and regulatory circuits of eukaryotic cells.
Origin of peroxisomes
Peroxisomes are eukaryotic organelles containing enzymes for the breakdown of reactive oxygen species and fatty acids (Poirier et al., 2006; Wanders, 2014). Peroxisomes may have no direct endosymbiotic origin, are unlike mitochondria and plastids devoid of nucleic acids but show a certain degree of autonomy, and contain dedicated systems for protein import (Gabaldón, 2010; Smith and Aitchison, 2013). One scenario is their emergence from the endoplasmic reticulum (ER) to reduce the detrimental effects of reactive oxygen species on ER protein homeostasis (Gabaldón, 2014). This idea is supported by findings indicating that the import machinery for peroxisomal matrix proteins is similar to the ERAD system known to control the export of misfolded proteins from the ER (Gabaldón et al., 2006; Schlüter et al., 2006).
According to a different opinion, peroxisomes are derivatives of mitochondria, since both organelles are sites of fatty acid breakdown. Peroxisomes may have emerged during the evolution of ancient eukaryotic cells to reduce the ROS burden of their progenitor (Speijer, 2017). Phylogenetic analysis revealed a bacterial origin of three out of four β-oxidation enzymes (Bolte et al., 2015). This led the authors to propose that at least the present form of peroxisomes emerged after mitochondria since a major catabolic pathway of peroxisomes likely originated from this organelle (Bolte et al., 2015). Mitochondrial enzymes probably became retargeted to peroxisomes over time, and some of these still remained dually localized (Gabaldón, 2018). Indeed, recent work shows that the early diverged cryptophyte Guillardia theta contains peroxisomes, but enzymes for fatty acid oxidation seem to be only mitochondrial (Vasilev et al., 2022). Independent of the actual evolutionary scenario, both the ER and mitochondria likely contributed to the proteome of modern peroxisomes and still sustain their biogenesis.
Peroxisome functions
The biochemical functions of peroxisomes are versatile. We only provide an overview and mainly refer to review articles focused on a more detailed explanation of peroxisomes from different species. The coupled degradation of fatty acids and H2O2 is a prominent task, and peroxisomes owe their name to this process (De Duve and Baudhuin, 1966; Poirier et al., 2006). Remarkably, in several parasitic species including trypanosomes peroxisomes house the enzymes for glycolysis (Haanstra et al., 2016). This already highlights a fascinating feature of the organelle—it is highly adaptable to a specific lifestyle. Another example is the metabolism of methylotrophic yeasts that break down methanol and harbor this oxidative process inside of the peroxisome (van der Klei and Veenhuis, 2006b). Yeasts and filamentous fungi contain a large variety of biosynthetic pathways inside of peroxisomes including enzymes involved in the production of antibiotics, biotin, surface-active glycolipids, and siderophores (Meijer et al., 2010; Magliano et al., 2011; Tanabe et al., 2011; Gründlinger et al., 2013; Freitag et al., 2014; Stehlik et al., 2014). Furthermore, peroxisomes are important for virulence of several human- and plant-pathogenic fungi (Kretschmer et al., 2012a; Falter and Reumann, 2022).
Peroxisomes are essential for the regular development of humans, and mutations in peroxisomal proteins are associated with severe diseases including the Zellweger syndrome (Wanders, 2014). Besides their prominent function in fatty acid catabolism, mammalian peroxisomes are required for further processes such as the production of ether lipids and bile acids or metabolism of amino acids including D-amino acids (Wanders and Waterham, 2006; Wanders et al., 2016). Furthermore, peroxisomes play an important role in the development of the brain and their dysfunction may contribute to neurological pathologies including amyotrophic lateral sclerosis and Alzheimer’s disease (Berger et al., 2016). More recent evidence supports a major function of peroxisomes in regulating the response of the immune system to pathogenic attack in several animals (Odendall et al., 2014; Di Cara et al., 2017; Di Cara, 2020). Plant peroxisomes are critical for oil mobilization during early seedling development and play a role in the metabolism of the toxic by-product 2-phosphoglycolate derived from an O2-consuming side reaction of RuBisCO. 2-Phosphoglycolate needs to be removed from the plastid, where it can inhibit several enzymes and is recycled into 3-phosphoglycerate in a series of reactions involving enzymatic reactions in peroxisomes and mitochondria (Reumann and Weber, 2006; Hu et al., 2012; Dellero et al., 2016; Pan et al., 2020). Besides their metabolic tasks, peroxisomes are also emerging as cellular signaling platforms (Tripathi and Walker, 2016). In addition, they can act as proviral and antiviral organelles depending on the type of virus (Ferreira et al., 2022). Given this remarkable variability, it is likely that many more functions of peroxisomes remain elusive, which may often be specific to a particular organism.
Protein targeting to peroxisomes
To attach to a defined organelle, proteins usually possess targeting signals, which act as a molecular zip code (Blobel and Sabatini, 1971). Proteins designated for the peroxisomal matrix mostly contain C-terminal or N-terminal sequence motifs termed peroxisomal targeting signal type I (PTS1) or type II (PTS2) (Kunze et al., 2015; Francisco et al., 2017; Walter and Erdmann, 2019; Kunze, 2020; Bürgi et al., 2021). These signals are recognized and bound by soluble targeting factors in the cytosol. Cargo proteins can be imported in a folded state and even as oligomers (Francisco et al., 2017; Walter and Erdmann, 2019). Very large particles have been shown to be imported into the lumen of the organelle revealing a flexible import pore (Walton et al., 1995; Yang et al., 2018). Proteins without a defined signal can enter the peroxisome in complex with canonical cargo (Islinger et al., 2009; Schueren et al., 2014; Effelsberg et al., 2015; Al-Saryi et al., 2017a; Gabay-Maskit et al., 2020). This piggyback import may be a hallmark of import pathways, which accept folded or partially folded clients such as the nucleus and the peroxisome. Several proteins are known to contain elements that enable binding to the targeting factor Pex5 in the cytosol although they lack a classical PTS1 (van der Klei and Veenhuis, 2006a; Kempiński et al., 2020; Rosenthal et al., 2020; Yifrach et al., 2021). Peroxisomal membrane proteins (PMPs) can be directly inserted into the peroxisomal membrane aided by the chaperone Pex19 and the transmembrane protein Pex3, but some can also be sorted via the ER (for a review, see Kim and Hettema, 2015).
Mechanisms to achieve the dual localization of proteins
We have previously discussed mechanisms involved in dual targeting of peroxisomal proteins in greater detail (Ast et al., 2013). Therefore, they are only briefly discussed, and instead, the focus of this work is on more recent findings on the plethora of dually targeted proteins and their potential function. We provide single chapters focusing on proteins targeted to peroxisomes and at least one other organelle with a major focus on yeasts and filamentous fungi. In addition, we showcase parallels to other eukaryotes.
Dual targeting can be achieved through very different mechanisms. Gene duplication and subsequent development of isoforms with import signals for only one cellular compartment are often found in Saccharomyces cerevisiae, presumably due to the genome duplications, which happened during its evolution (Kellis et al., 2004; Yogev et al., 2011; Ast et al., 2013). Polypeptides with different targeting signals can also be generated from a single gene (Yogev et al., 2011; Ast et al., 2013), e.g., from alternative transcripts, alternative splicing (Natsoulis et al., 1986; Clausmeyer et al., 1999; Freitag et al., 2012; Strijbis et al., 2012), programmed readthrough of stop codons (for a review, see Schueren and Thoms, 2016), and noncanonical translation initiation (Monteuuis et al., 2019; Kremp et al., 2020). In addition, proteins can contain ambiguous targeting signals at their N-termini, which enable sorting into two cellular compartments. This is particularly prominent in plants for those proteins required in mitochondria and plastids—two organelles, which use a related protein import pathway involving N-terminal targeting signals and translocation of unfolded proteins (Carrie et al., 2009). We also discuss the dual targeting of proteins that contain N-terminal targeting signals, e.g., for mitochondria or the ER in combination with a C-terminal PTS1. It was suggested previously that the localization of these proteins might be dictated by the N-terminal signal since it can be bound by respective targeting factors during translation before the C-terminal PTS1 becomes accessible (Ast et al., 2013; Kunze and Berger, 2015). However, a recent study uncovered that proteins with competing N-terminal and C-terminal targeting signals localize in peroxisomes and mitochondria (Stehlik et al., 2020). Low efficiency or unusual peroxisomal targeting signals often provoke partial cytosolic retention due to low import rates or other reasons, e.g., the modification of the peroxisomal targeting machinery (Ast et al., 2013; Okumoto et al., 2020). Another focus of our review is on peroxisomal membrane proteins (PMPs) and membrane-associated proteins, which follow variable transit routes.
Typical enzymes with a role in multiple cellular compartments
One critical type of enzymes required inside of peroxisomes and in other compartments are NADH-dependent dehydrogenases. They can assist peroxisomal NAD+ regeneration, e.g., during β-oxidation. The reduced substrate can be translocated into the cytosol and exchanged with an oxidized molecule giving rise to a redox shuttle system (Visser et al., 2007). The existence of a peroxisomal redox shuttle was first demonstrated in S. cerevisiae—a peroxisomal isoenzyme of malate dehydrogenase (Mdh3p) containing a PTS1 was shown to be involved (Van Roermund et al., 1995). Malate can be generated from oxaloacetate enabling the reoxidation of NADH. The small molecules are thought to pass the peroxisomal membrane, albeit it is not fully understood if transporters or size-selective pore-forming proteins are involved (for a review, see Chornyi et al., 2021). Recently, it was shown that a second malate dehydrogenase Mdh2p from S. cerevisiae involved in glyoxylate metabolism binds Mdh3p and can enter peroxisomes via piggyback import resulting in dual localization (Gabay-Maskit et al., 2020). It is yet unclear how cells exactly benefit from having two MDH enzymes inside of peroxisomes, but a possible answer is their different activities (Steffan and McAlister-Henn, 1992; Gabay-Maskit et al., 2020). In addition to the described malate–oxaloacetate shuttle, S. cerevisiae contains a second shuttle system relying on the glycerol-3-phosphate dehydrogenase Gpd1p, which catalyzes the interconversion of glycerol-3-phosphate and dihydroxyacetone phosphate. Gpd1p is dually localized in the cytosol and in peroxisomes (Jung et al., 2010) (Figure 1). Mdh3p and Gpd1p exhibit redundant functions for NAD shuttling in conditions that require biosynthesis of lysine, the last step of which occurs inside of yeast peroxisomes (Al-Saryi et al., 2017a). Why cells employ multiple shuttle systems for NAD+/NADH and how this functionally connects peroxisomal metabolism to the metabolism of the entire cell requires further investigation. The presence of multiple systems, however, seems to be common. Many fungi contain a peroxisomal isoform of the NAD+-dependent glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) derived from alternative splicing or stop codon readthrough. Genetic data point to overlapping functions of peroxisomal GAPDH, GPD, and MDH presumably for the regulation of NAD+/NADH balance (Freitag et al., 2012).
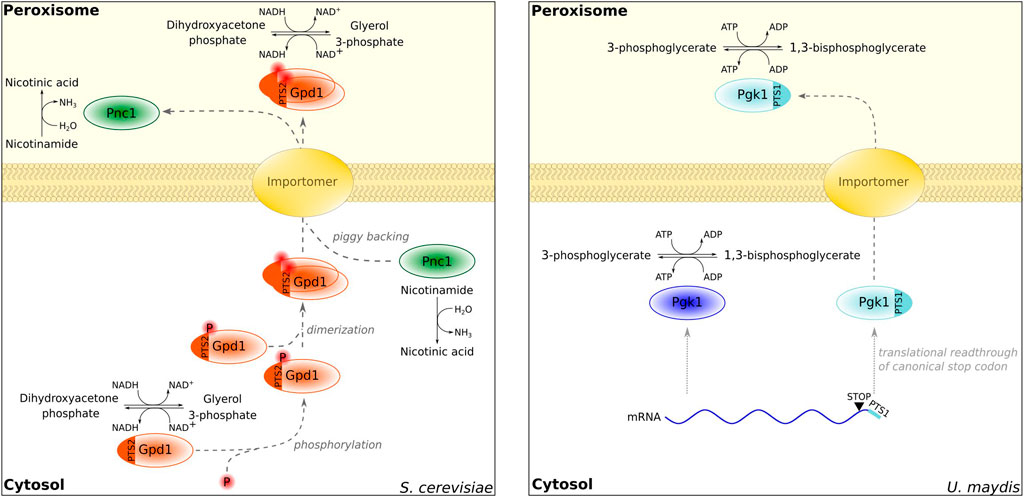
FIGURE 1. Examples of proteins localized in the cytosol and peroxisomes. Gpd1 distribution is controlled by phosphorylation [left; Jung et al. (2010)]. In addition, Gpd1 regulates the sorting of Pnc1 via piggybacking (Effelsberg et al. (2015), Kumar et al. (2016), Al-Saryi et al. (2017b)). An extended isoform of Pgk1 derived from translational readthrough is localized in peroxisomes in Ustilago maydis [right; Freitag et al. (2012)].
In mammalian cells, programmed stop codon readthrough triggers the synthesis of extended PTS1-containing isoforms of lactate dehydrogenase LDHB and malate dehydrogenase MDH1 (Schueren et al., 2014; Stiebler et al., 2014). This is in accordance with at least two pathways to control the NAD+/NADH ratio. Stop codon readthrough is a widespread mechanism to regulate the dual localization of central metabolic enzymes in fungi and animals; e.g., a peroxisomal isoform of the glycolytic enzyme phosphoglycerate kinase contains a readthrough-derived PTS1 (Freitag et al., 2012; Figure 1). Other prominent enzymes occurring in many cellular compartments are inorganic pyrophosphatases—several enzymes harbor putative PTS1-containing extensions, which can be activated by stop codon readthrough. These include the Caenorhabditis elegans Pyp-1 protein and PAP1 or PAP2 from different mammals (Stiebler et al., 2014). Inorganic pyrophosphatase is a central enzyme rendering energy-dependent reactions throughout the cell virtually irreversible (Kornberg, 1962) and may also speed up peroxisomal metabolism.
Why is stop codon readthrough such a prominent mechanism for the generation of peroxisomal isoforms? First, a trivial reason is the C-terminal position of PTS1—a simple prerequisite for this mechanism to activate a hidden PTS1. Furthermore, readthrough rates are low but seem to be sufficient to enable enzyme supply for the relatively small peroxisomal compartment in concentrations high enough to satisfy demands. Finally, the region behind the stop codon could be regarded as a playground for evolution and partial peroxisomal localization can be easily tested and rejected again. This may explain the previously observed patchy distribution of readthrough sites coupled to PTS1 motifs among different eukaryotic species (Freitag et al., 2012; Dunn et al., 2013; Stiebler et al., 2014; Hofhuis et al., 2016).
Peroxisomal NADPH turnover is involved in the oxidation of fatty acids with a cis double bond at an even position (Poirier et al., 2006). Early work in S. cerevisiae demonstrated a role of an isocitrate dehydrogenase as part of a peroxisomal NADPH shuttle (van Roermund et al., 1998). Isocitrate is oxidized to alpha-ketoglutarate and CO2 leading to the formation of NADPH. Idp1p is the mitochondrial isoform, Idp2p is the cytosolic isoform, and Idp3p is the peroxisomal isoform (Haselbeck and McAlister-Henn, 1991; Haselbeck and McAlister-Henn, 1993; van Roermund et al., 1998). In contrast, peroxisomal, mitochondrial, and cytosolic isocitrate dehydrogenases are derived from a single gene in the filamentous fungus Neurospora crassa (Szewczyk et al., 2001). In Arabidopsis thaliana, several enzymes of the pentose phosphate pathway including an isoform of the NADPH-dependent enzyme 6-phosphogluconate dehydrogenase (PGD) reside in peroxisomes and are critical for development (Corpas et al., 1998; Meyer et al., 2011; Hölscher et al., 2016). Dually targeted peroxisomal isoforms of PGD can be generated by alternative splicing or noncanonical translation initiation in different fungi (Strijbis et al., 2012; Kremp et al., 2020). Recently, in A. thaliana a transport protein for glucose-6-phosphate has been identified that reaches peroxisomes via the ER but is also found in plastids (Baune et al., 2020).
Additional carbohydrate-metabolizing enzymes contain cryptic or low-efficiency peroxisomal targeting signals in several fungi including not only glycolytic/gluconeogenetic enzymes but also enzymes of the non-oxidative part of the pentose phosphate pathway (Freitag et al., 2012; Freitag et al., 2018). This points to many molecular links of the peroxisomal metabolism to the central carbohydrate metabolism. In several fungal species, optimal growth on glucose medium requires intact peroxisomes—even on full medium in the logarithmic growth phase (Idnurm et al., 2007; Freitag et al., 2012; Camões et al., 2015; Ast et al., 2022). Whether these growth phenotypes are directly related to the dual targeting of glycolytic enzymes or pentose phosphate pathway enzymes is yet elusive.
Dually targeted proteins—the dark matter of the peroxisomal proteome
Minor destinations of proteins are very likely to be missed. A recent systematic study in S. cerevisiae uncovered many novel proteins residing in peroxisomes, as well as in other organelles or in the cytosol (Yifrach et al., 2021). Using systematic metabolomics analysis of mutants and overexpression strains, the authors suggest that several of these have potential peroxisomal functions. Among the newly identified dually localized proteins are direct substrates of Pex5, which lack a classical PTS1. This reveals a greater substrate repertoire of this targeting factor (Yifrach et al., 2021). Other work also points to the occurrence of many unusual variations of the PTS1 motif, which are recognized by Pex5 in several fungi (Freitag et al., 2012; Camões et al., 2015; Nötzel et al., 2016). An unexpected example is sequence motifs resembling a PTS1, which are located near the C-terminus of a protein rather than at its end. This type of signal was suggested to be responsible for peroxisomal localization of catalase from pumpkin and for human ataxia telangiectasia–mutated (ATM) kinase (Kamigaki et al., 2003; Zhang J. et al., 2015). It is still unclear how PTS1 motifs, which do not reside at the C-terminus, are recognized by Pex5 as this is thought to be a structural prerequisite for binding (Fodor et al., 2015).
An interesting example identified by Yifrach et al. (2021) is dually localized subunits of the GID (glucose-induced degradation-deficient) complex, which regulate the stability of fructose-1,6-bisphosphatase (FBP). The targeting of GID complex proteins to peroxisomes was found to increase FBP levels probably enhancing gluconeogenesis (Yifrach et al., 2021). These data indicate a novel function of peroxisomes in regulating carbohydrate mechanism upon glucose limitation. Similarly, in different yeasts nicotinamidase Pnc1 is targeted to peroxisomes more efficiently in response to certain stresses—this is achieved through hitchhiking on the PTS2 protein Gpd1 (Anderson et al., 2003; Effelsberg et al., 2015; Kumar et al., 2016; Al-Saryi et al., 2017b) (Figure 1). Sequestration of a protein inside of the peroxisomal lumen to remove it from its site of action anywhere in the cell may be a more widespread function of peroxisomes. They seem to be promiscuous sites among the cellular organelles and tolerate many different proteins and activities. Even the efficient production of toxic compounds was successfully engineered aided by the peroxisomal targeting of respective enzymes (Grewal et al., 2021). For each of the dually localized proteins, it will be critical to assess whether they fulfill a specific function inside of peroxisomes or whether their localization reflects a strategy to sequester them from a different cellular compartment.
Many functions of peroxisomes relying on dually targeted proteins are likely to remain established. Surprisingly, in Aspergillus nidulans the microtubule-organizing protein ApsB is partially localized to a subpopulation of peroxisomes, but its role at this location has not been understood (Zekert et al., 2010). Several proteins from A. thaliana show a dual localization to peroxisomes and to other compartments, often due to PTS1 motifs with low import efficiency (Reumann et al., 2007; Kataya and Reumann, 2010; Lingner et al., 2011). Interestingly, protein kinases were identified that often contain unusual but functional PTS1 motifs (Kataya et al., 2022). These findings are of particular interest as little is known about the phosphorylation-dependent regulation of enzymes inside of the peroxisomal lumen. More data are needed to assess which kinases play a crucial role in regulating the activity of the peroxisomal proteome. Particularly, the fact that some kinases contain transmembrane domains is puzzling with regard to the classical view of peroxisomal matrix protein import (Walter and Erdmann, 2019). Recent data, however, point to a function of the peroxisomal matrix protein import machinery in membrane protein translocation (Martenson et al., 2020). Together, all the mentioned studies emphasize a plethora of diverse peroxisomal proteins, for which a functional characterization is lacking so far. We expect more surprises concerning the proteome composition of peroxisomes in the future.
Peroxisomes—organelles in a twilight zone
Peroxisomes have been adapted for multiple purposes during evolution, e.g., as a seal for septal pores in fungal hyphae, as sites for methanol breakdown in different yeast, or as glycolytic organelles in trypanosomes (Jedd and Chua, 2000; van der Klei and Veenhuis, 2006b; Liu et al., 2011; Haanstra et al., 2016). Why do peroxisomes show this high degree of functional flexibility? For many species, cellular survival does not strictly depend on peroxisomal functions; hence, their repertoire of proteins could change without detrimental consequences. Furthermore, peroxisomes represent a cellular one-way road. They import cargo, grow, divide, and are degraded but probably do not fuse regularly in their mature form (Islinger et al., 2012; Kim and Hettema, 2015; Germain and Kim, 2020). This may explain their tolerance to a plethora of proteins and the coexistence of multiple peroxisome variants in a single cell as evident for the Woronin body biogenesis in ascomycetes.
Peroxisomes and mitochondria—striking similarities
Dually targeted soluble proteins
Mitochondria and peroxisomes cooperate in versatile metabolic processes, exchange many molecules, and also share proteins required for fission and quality control (Schrader et al., 2015; Wanders et al., 2016). This peroxisome–mitochondria connection has been excellently discussed before (Fransen et al., 2017; Costello et al., 2018), and we only focus on several examples. Both organelles can be cellular sites for β-oxidation of fatty acids—while in several fungi and plants, β-oxidation occurs exclusively inside of peroxisomes, other fungi and animals harbor full sets of β-oxidation enzymes inside of both organelles (Maggio-Hall and Keller, 2004; Poirier et al., 2006; Goepfert and Poirier, 2007; Kretschmer et al., 2012b; Camões et al., 2015). An important difference between the compartments is the mechanisms they use for the import of proteins: While mitochondrial proteins often rely on N-terminal targeting signals and can be imported in an unfolded or flexible state (Backes and Herrmann, 2017), import into the peroxisomal matrix is different.
To exchange intermediates of the β-oxidation pathway, substitution of a coenzyme A moiety with carnitine is one possibility to shuttle acetyl and acyl groups as acetyl- or acylcarnitine units (Antonenkov and Hiltunen, 2012). Acetylcarnitine transferases are dually localized enzymes, and isoforms with different targeting signals have been reported, which can be generated via alternative transcriptional and translational start sites or differential splicing (Corti et al., 1994; Elgersma et al., 1995; Ueda et al., 1998; Houten et al., 2020). Within mitochondria, carnitine is replaced by coenzyme A to enable further metabolization (Houten et al., 2020).
A second cellular pathway enables the exchange of C2 units between peroxisomes and mitochondria, albeit more indirectly. Citrate can be generated from oxaloacetate via citrate synthases, which are not only part of the Krebs cycle but also part of the glyoxylate cycle, partially located inside of peroxisomes (Kim et al., 1986; Kunze et al., 2006). The citrate synthase Cit2 has overlapping functions with acetylcarnitine transferase Cat2p in S. cerevisiae and is contained in peroxisomes (Van Roermund et al., 1995; van Roermund et al., 1999; Swiegers et al., 2001; Shai et al., 2018). Of interest, Cit2p can be targeted to mitochondria, as well as to peroxisomes, and can compensate for the absence of mitochondrial citrate synthase Cit1p (Kim et al., 1986; Hoppins et al., 2011; Lee et al., 2011; Morgenstern et al., 2017). Citrate can be shuttled via dedicated transport proteins linking the mitochondrial matrix to the cytosol (Kaplan et al., 1995; Palmieri, 2004).
How are proteins distributed that do not come in different isoforms but contain a mitochondrial targeting signal at the N-terminus and a PTS1 at the C-terminus? Previously, it was discussed that these usually end up in mitochondria, because the N-terminal signal will exit the ribosome first, can directly interact with the mitochondrial import machinery, and thus will dominate a PTS1—a hierarchy of targeting signals is likely to exist (Kunze and Berger, 2015). Indeed, the mitochondrial ribosomal protein Mrp7p contains a functional PTS1 in S. cerevisiae but does not localize in peroxisomes presumably because the N-terminal signal is recognized first (Neuberger et al., 2004). However, several bona fide peroxisomal proteins such as the thiolase Tes1p or the catalase Cta1p have been identified in highly purified mitochondria of S. cerevisiae (Morgenstern et al., 2017).
Another PTS1-containing protein—the protein phosphatase Ptc5p—has been recently shown to first target mitochondria where its N-terminus is proteolytically removed. Subsequently, the protein is translocated into the peroxisomal lumen via interaction with Pex5p (Stehlik et al., 2020) (Figure 2). Further proteins were also shown to be retargeted from mitochondria to the cytosol, e.g., S. cerevisiae fumarase (Stein et al., 1994; Yogev et al., 2010). The sorting of Ptc5p is different as the presence of Pex5p is critical for export from mitochondria, indicating an interaction between Ptc5p and Pex5p prior to full mitochondrial import. One target of Ptc5p in peroxisomes is Gpd1p (Stehlik et al., 2020), and dephosphorylation of Gpd1p increases its activity (Lee et al., 2012). In line with these data, a synthetic growth defect was observed for strains deleted for PTC5 and MDH3 (Costanzo et al., 2016; see the previous chapter on the redundancy of Gpd1p and Mdh3p). Together, these data suggest a network consisting of Ptc5p, Gpd1p, and Mdh3p to regulate peroxisomal NADH metabolism presumably in a manner adapting the different compartments to each other. In A. thaliana, the sorting of NAD(P)H dehydrogenases may resemble Ptc5p from S. cerevisiae as competing N- and C-terminal targeting signals for mitochondria and peroxisomes are involved (Carrie et al., 2008).
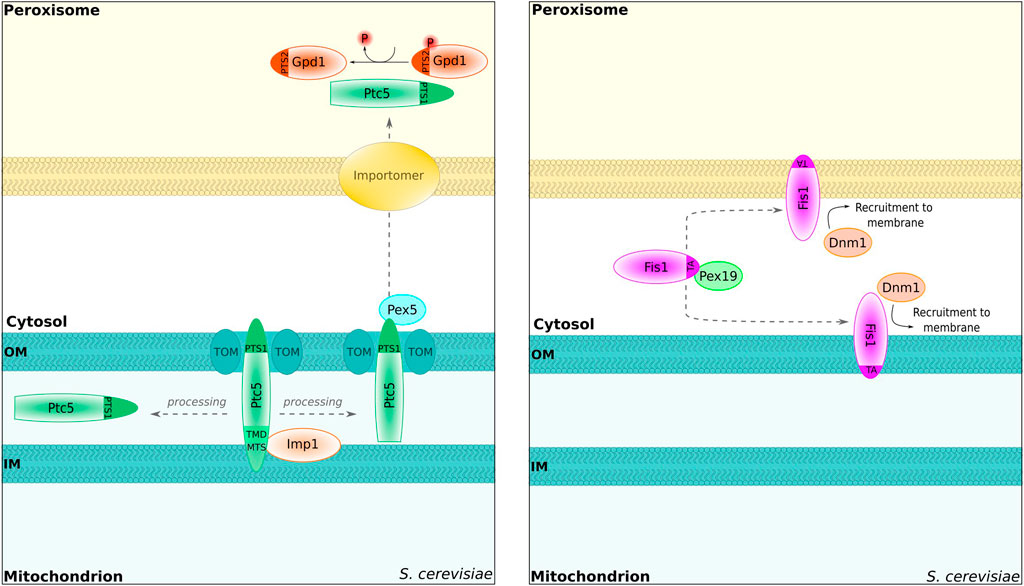
FIGURE 2. Proteins shared between peroxisomes and mitochondria. S. cerevisiae Ptc5p can reach the peroxisome via mitochondrial transit. The protein is proteolytically processed inside of the inner mitochondrial membrane by the peptidase Imp1 and subsequently translocated to peroxisomes in a Pex5-dependent manner [left; Stehlik et al. (2020)]. Inside of the peroxisome, Ptc5 dephosphorylates Gpd1 [left; Stehlik et al. (2020)]. TOM, translocator of the outer mitochondrial membrane. Importomer, complex for peroxisomal matrix protein import [Walter and Erdmann (2019)]. The tail-anchored protein Fis1 is involved in the fission process of peroxisomes and mitochondria [right; Kuravi et al. (2006)]. Targeting is regulated by Pex19 [Cichocki et al. (2018)]. Dnm1, dynamin-related GTPase for peroxisome fission Kuravi et al. (2006).
Targeting of proteins with competing targeting signals may represent a more generic approach to regulate the interaction and communication of compartments—the overexpression of several of these proteins increases the fraction of peroxisomes associated with mitochondria (Stehlik et al., 2020). In mammalian cells, the overexpression of the dual affinity protein ACBD2/ECI2 also enhances the proximity of peroxisomes and mitochondria (Fan et al., 2016). The protein Cnm1p with competing targeting signals at its termini can increase the attachment of mitochondria to the nuclear envelope in S. cerevisiae, suggesting a similar mechanism occurring between another pair of organelles (Eisenberg-Bord et al., 2021).
Dual targeting of mitochondrial membrane proteins
In addition to luminal proteins, peroxisomes and mitochondria share a number of membrane proteins or membrane-associated proteins with various tasks (also see Fransen et al., 2017; Costello et al., 2018). A key process controlled by overlapping factors is fission. Both in S. cerevisiae and in mammalian cells, the C-terminally tail-anchored protein Fis1p/FIS1, dynamin-related GTPases, and additional shared proteins such as the WD40 repeat-containing protein Caf4p belong to the factors involved in the fission process (Koch et al., 2003; Praefcke and McMahon, 2004; Koch et al., 2005; Kuravi et al., 2006; Motley et al., 2008; Motley et al., 2015; Castro et al., 2018). C-terminally tail-anchored membrane proteins such as FIS1 tend to be dually localized, and it was shown that the hydrophobicity of the transmembrane domain together with changes in the charge of the luminal tail determines targeting efficiency and hence the subcellular distribution (Costello et al., 2017a). Remarkably, the peroxisomal targeting factor Pex19p is required for the correct sorting of Fis1p to mitochondria and to peroxisomes in S. cerevisiae (Cichocki et al., 2018) (Figure 2). How the distribution of proteins such as Fis1p is adapted to the needs of both organelles is not known yet.
Further proteins operating at mitochondria and peroxisomes are ATPases termed Msp1p in S. cerevisiae or ATAD in mammals, which can extract superfluous or mistargeted tail-anchored proteins (Chen Y.-C. et al., 2014; Okreglak and Walter, 2014; Weir et al., 2017). Of interest, a variety of PMPs can be removed from mitochondria via Msp1p/ATAD, suggesting a broader range of substrates (Nuebel et al., 2021). Furthermore, these authors observed that many peroxisomal proteins are not degraded or downregulated in the absence of preexisting peroxisomes, but functional PMP-containing protein complexes assemble on mitochondria, which causes unfavorable consequences such as the import of PTS1 cargo into mitochondria (Nuebel et al., 2021). Mitochondrial dysfunction in the absence of functional peroxisomes is emerging as one cause of symptoms in patients lacking peroxisomes (Wanders, 2014; Lismont et al., 2019; Schrader et al., 2020; Nuebel et al., 2021). The ubiquitin ligase MARCH5—another protein for quality control—was shown to localize to peroxisomes in addition to its known location at mitochondria in mammals. At peroxisomes, MARCH5 is involved in controlling selective autophagy (Zheng et al., 2021).
A multi-subunit protein structure associated with both organelles in fungi is the ERMES (ER–mitochondria encounter structure) complex, which has a role in tethering of mitochondria to the ER and to peroxisomes to form putative three-way junctions (Kornmann et al., 2009; Cohen et al., 2014; Ušaj et al., 2015; Kundu and Pasrija, 2020). Moreover, the ERMES regulating GTPase Gem1p can be found in peroxisomes and mitochondria of S. cerevisiae (Kornmann et al., 2011; Cichocki et al., 2018). In mammalian cells, the Gem1p ortholog MIRO1 is dually localized as well and has a role in recruiting the lipid transfer protein VPS13D probably involved in sustaining organellar growth and lipid exchange (Castro et al., 2018; Baldwin et al., 2021; Guillén-Samander et al., 2021). Thus, mitochondria and peroxisomes use an overlapping set of proteins to connect to the ER.
A different mode of transport to accomplish the dual localization of selected proteins is vesicular trafficking from one organelle to the other. Dynamin-independent carriers were proposed to transport the ubiquitin ligase MAPL from mitochondria to peroxisomes (Neuspiel et al., 2008). More recently, it was suggested that de novo peroxisome formation involves mitochondria-derived vesicles (Sugiura et al., 2017).
Dual targeting and evolution of peroxisomes
All these data point to a close relationship of mitochondria and peroxisomes not only in terms of metabolism but also in terms of biogenesis, quality control, and turnover. This remarkable overlap might reflect peroxisome evolution as partially mitochondria-derived organelles (Bolte et al., 2015; Speijer, 2017). Are peroxisomes indeed outposts of mitochondria, and how can this be in line with the key role of the endoplasmic reticulum for peroxisome biogenesis (Joshi et al., 2017; see also in the next chapter)?
We speculate that peroxisomes may have emerged from an ancient ER-derived quality control compartment involved in clearing specific mitochondrial proteins, especially under conditions of oxidative stress. In this perspective, the ERAD-related peroxisomal import machinery (Schliebs et al., 2010) could be regarded as an extraction machine for mitochondrial proteins working in trans. Particularly, the oxidative enzymes of the β-oxidation pathway may be a major burden for mitochondrial metabolism, which may explain why this pathway has been completely transferred to peroxisomes of several species, e.g., S. cerevisiae (Hiltunen et al., 2003; Poirier et al., 2006; Speijer, 2017). The indirect targeting of the phosphatase Ptc5p via mitochondrial transit (Stehlik et al., 2020) could be a snapshot or remnant of this evolutionary scenario. In this effect, peroxisomes may represent ancient molecular vacuum cleaners of the mitochondrial surface (Figure 3), which became autonomous over time by hitchhiking the mitochondrial division and quality control machinery. Further shared proteins might still be obscure—one study indicated a role for the dually localized mitofusin ortholog Fzo1 from S. cerevisiae for tethering peroxisomes to mitochondria (Shai et al., 2018).
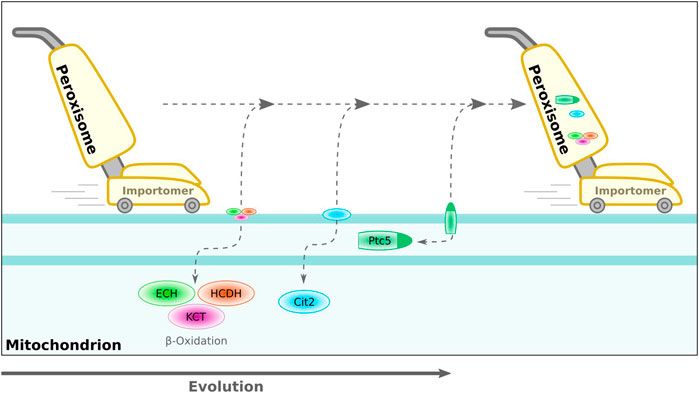
FIGURE 3. Hypothetical model for translocation of proteins from mitochondria to peroxisomes during evolution. Peroxisomes are indicated by a vacuum cleaner—this parallel is inferred from their unique import mode for folded proteins and the possible evolutionary origin of the importomer from an ER-derived quality control system (Gabaldón et al. (2006), Schlüter et al. (2006)). We speculate that over time, various mitochondrial enzymes developed into peroxisomal enzymes via a dually targeted intermediate. Sorting of the phosphatase Ptc5p from S. cerevisiae shows hallmarks for such an evolutionary scenario. ECH, enoyl-CoA hydratase; HCDH, 3-hydroxyacyl-CoA dehydrogenase; KCT, 3-ketoacyl-CoA-thiolase; Cit2, citrate synthase 2 (S. cerevisiae); Ptc5, PP2C-type phosphatase (S. cerevisiae).
Proteins localized to peroxisomes and the endoplasmic reticulum
The endoplasmic reticulum as a source for peroxisomal membrane
The endoplasmic reticulum (ER) is the major source of cellular lipids and has a key role in supporting the growth of the entire cell (Ferro-Novick et al., 2013). Early work already revealed intimate connections between this large-supply organelle and peroxisomes (Novikoff and Shin, 1964). Metabolites are exchanged between both compartments, e.g., during the biosynthesis of ether lipids (Wanders et al., 2016). In S. cerevisiae, the sorting of the multifunctional PMP Pex3—a key factor in regulating PMP import into peroxisomes—from the ER to peroxisomes was described (Hoepfner et al., 2005; Jansen and van der Klei, 2019). ER-derived vesicular carriers were reported to contain peroxisomal proteins, and machinery was uncovered that is relevant for the emergence of peroxisomal vesicles from the ER in yeasts (Titorenko et al., 2000; Lam et al., 2010; Agrawal et al., 2011; Van der Zand et al., 2012; Agrawal et al., 2016; Mast et al., 2018). Hence, vesicular trafficking from the ER represents one road for lipids and proteins to the peroxisome and also operates in mammalian cells (Kim et al., 2006; Aranovich et al., 2014; Sugiura et al., 2017). In S. cerevisiae and other yeasts, ER-resident microdomain-forming reticulon proteins play a critical role in tethering of preexisting peroxisomes (David et al., 2013; Mast et al., 2016; Wu et al., 2020; Ferreira and Carvalho, 2021). Interestingly, the same domains may be required for the budding of peroxisome precursors (Joshi et al., 2016; Joshi et al., 2018; Wang et al., 2018). As an alternative to vesicle formation, direct lipid transfer promotes the growth of peroxisomes and recruitment of lipid transfer proteins such as VPS13 is involved in peroxisome biogenesis both in yeasts and in mammals (Raychaudhuri and Prinz, 2008; Baldwin et al., 2021; Guillén-Samander et al., 2021; Yuan et al., 2022). How these mechanisms for lipid supply each contribute to peroxisome formation is an exciting question for future work. An intimate physical connection between peroxisomes and the ER is probably required for the correct activity of both mechanisms. This can, e.g., be supported through an interaction between the peroxisomal acyl-CoA binding protein ACBD5 and the ER membrane-associated protein VAPB in mammalian cells (Costello et al., 2017b; Hua et al., 2017).
Proteins found in the endoplasmic reticulum and peroxisomes
Many PMPs are synthesized in the vicinity of the peroxisome of S. cerevisiae (Zipor et al., 2009; Dahan et al., 2022), and direct insertion into the peroxisomal membrane was demonstrated for several of them (Sacksteder et al., 2000; Fang et al., 2004; Jones et al., 2004; Matsuzaki and Fujiki, 2008; Yagita et al., 2013; Chen Y. et al., 2014). Interestingly, other PMPs are synthesized proximal to the ER and may reach the peroxisome via ER transit (Jan et al., 2014). For Pex3, dual localization to peroxisomes and the ER of S. cerevisiae was shown by fractionation experiments (Mast et al., 2016). Hence, the ER is not only an intermediate but also a relevant steady-state location for certain PMPs, indicating functions beyond ER transit. Indeed, in mammalian cells ER-localized PEX3 is involved in the sorting of a protein designated for lipid droplets (Schrul and Kopito, 2016). In addition to PEX3, the soluble targeting factor PEX19 is associated with the ER and involved in targeting reticulon-homology proteins to this organelle besides its prominent function in peroxisome biogenesis (Yamamoto and Sakisaka, 2018; Zimmermann et al., 2021).
The coat protein I (COPI) complex localizes primarily along the organelles of the secretory pathway where it is involved in retrograde transport (Barlowe and Miller, 2013). In addition, this protein assembly can be associated with peroxisomes and also with mitochondria, but the molecular function of the localization at these organelles is not fully understood (Passreiter et al., 1998; Barlowe and Miller, 2013; David et al., 2013; Zabezhinsky et al., 2016). Remarkably, COPI-dependent sorting of a viral protein from peroxisomes to the ER was reported (McCartney et al., 2005). The small GTPase Arf1p—a major regulator of COPI—shows multiple subcellular localizations including peroxisomes and mitochondria in S. cerevisiae (Ackema et al., 2014; Yofe et al., 2017). So far, the role of COPI-dependent vesicular trafficking for peroxisome biogenesis is elusive although in S. cerevisiae, COPI components can be purified together with the subdomain-forming protein Pex30p, which is involved in peroxisome formation (David et al., 2013). Pex30p is also found at sites of lipid droplet formation in S. cerevisiae, and COPI was shown to be involved in formation of this organelle in Drosophila cells (Wilfling et al., 2014; Joshi et al., 2018; Wang et al., 2018; Ferreira and Carvalho, 2021). Further research is required to understand how key factors of the secretory pathway are dynamically distributed between the different organelles and how this coordinates organelle biogenesis at different sites. Nevertheless, the discussed examples emphasize that the ER and peroxisomes are possible alternative destinations of several factors required for the proper maintenance of each.
Another interesting example of dual targeting was described in A. thaliana—a purple acid phosphatase-containing competing targeting signals is localized to the ER and to peroxisomes (Kataya et al., 2016). Various other phosphatases with noncanonical PTS1 motifs were identified in this study that may reside inside of peroxisomes and in additional compartments including the nucleus (Kataya et al., 2016).
Dual targeting to the nucleus and peroxisomes
Different types of pathogenic attack trigger the nuclear localization of plant catalases (Inaba et al., 2011; Zhang M. et al., 2015). A recent study in A. thaliana uncovered that catalase is localized both in peroxisomes and in the nucleus even in the absence of any infection (Al-Hajaya et al., 2022). Regulation of dual targeting via metabolites such as H2O2 is likely to operate in plants similar to what was observed in mammals (Okumoto et al., 2020). Peroxisomes together with mitochondria have a key role in controlling ROS production, which is important for cellular signaling and needs to be maintained on a level acceptable for the integrity of cells, especially of the nucleus (Fransen et al., 2017; Sies, 2017; Li et al., 2021; Paradis et al., 2021).
Interestingly, the hypoxia-inducible transcription factor Hif1 and hydroxylases regulating its activity are found in peroxisomes and mitochondria (Khan et al., 2006; Li et al., 2019). Sequestration upon reoxygenation was suggested as a possible biological function of peroxisomal targeting (Khan et al., 2006).
Proteins shared by lipid droplets and peroxisomes
Lipid droplets are sites of cellular fat storage and mobilization. Their biosynthesis and function are both intricately linked to peroxisomes. Both organelles require overlapping proteins for biogenesis, e.g., the lipodystrophy protein seipin and other factors including Pex30p (Szymanski et al., 2007; Salo et al., 2016; Joshi et al., 2017; Joshi et al., 2018; Wang et al., 2018). The targeting factor PEX19 and ER-localized PEX3 facilitate the insertion of the membrane protein UBXD8 into a subdomain of the ER membrane, which turned out as a prerequisite for subsequent transfer to lipid droplets in mammalian cells (Schrul and Kopito, 2016). Upon depletion of PEX19, UBXD8 appeared predominantly in mitochondria. Of interest, a proper targeting of UBXD8 requires C-terminal farnesylation of PEX19 (Schrul and Kopito, 2016).
Peroxisomes and lipid droplets are physically connected, and membrane protrusions emerging from peroxisomes have been shown to attach to lipid droplets in S. cerevisiae (Binns et al., 2006). In A. thaliana, a dually localized lipase can be sorted from peroxisomes to lipid droplets via similar protrusions in the course of fat mobilization during seed germination (Thazar-Poulot et al., 2015). In a similar way, in fasting animals triglyceride lipases are translocated from peroxisomes to lipid droplets depending on the targeting factor PEX5. This process was proposed to involve sites of organelle contact (Kong et al., 2020) (Figure 4).
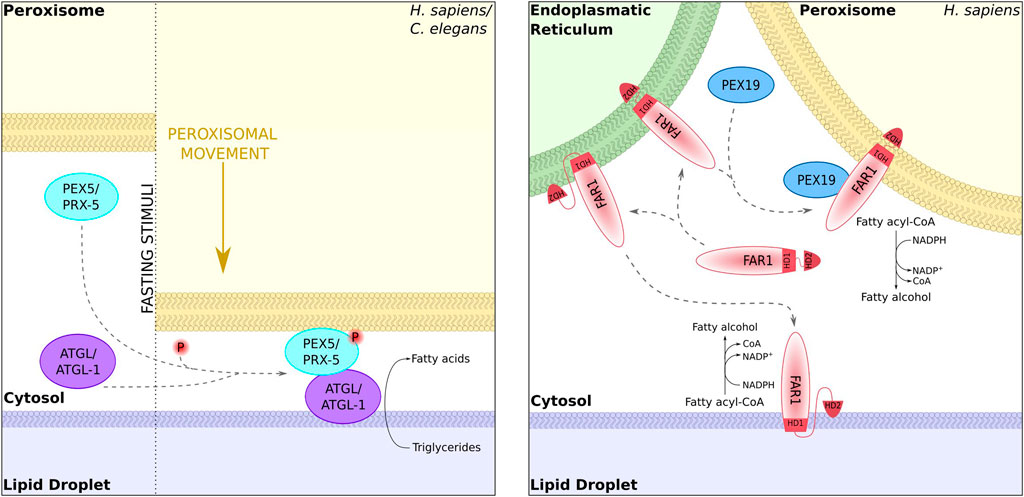
FIGURE 4. Dual targeting to peroxisomes and lipid droplets. The lipases ATGL-1 (Caenorhabditis elegans) and ATGL (mammalian cells) can target lipid droplets in a Pex5-dependent manner at sites of organelle contact [left; Kong et al. (2020)]. The fatty acid reductase Far1 is dually localized to peroxisomes and lipid droplets. Localization to one or the other organelle requires different topologies of the hydrophobic domains [right; Exner et al. (2019)].
Since peroxisomes are enclosed by a lipid bilayer and lipid droplets only by a monolayer, the dual localization of membrane proteins is likely to depend on dual topology. This was demonstrated for mammalian fatty acyl-CoA reductase 1, which is targeted to peroxisomes and lipid droplets and transits via the ER (Exner et al., 2019) (Figure 4).
What about the remaining cell?
Knowledge about proteins destined for peroxisomes and the plasma membrane or peroxisomes and endosomes is limited. It has been described that fungal peroxisomes hitchhike on motile endosomes to move through fungal hyphae (Guimaraes et al., 2015; Salogiannis et al., 2016). In A. nidulans, the association between endosomes and peroxisomes is mediated by the linker protein PxdA (Salogiannis et al., 2016). Proteins required for the maturation of endosomes such as members of the ESCRT (endosomal sorting complex required for transport) pathway (Henne et al., 2011) are involved in peroxisome biogenesis (Mast et al., 2018). Of interest, recent work revealed intraluminal vesicles inside of plant peroxisomes, which might also require the function of the ESCRT pathway, hinting at some parallels to multi-vesicular bodies (Wright and Bartel, 2020).
How to choose between the different destinations?
Insights into the molecular mechanisms that regulate the distribution of proteins among their different destinations are of interest to understand communication and homeostasis of cellular compartments. Studies on the dual localization of catalase in mammalian cells could represent a blueprint—it was shown that an elevated cytosolic concentration of H2O2 provokes cytosolic retention of this detoxifying enzyme (Walton et al., 2017). Recent studies showed phosphorylation of the peroxisomal membrane Pex14 in response to oxidative stress and during mitosis, which reduces the peroxisomal import of catalase to a greater extent than the import of other tested matrix proteins (Okumoto et al., 2020; Yamashita et al., 2020). Modification of the peroxisomal import machinery, hence, has an important role to adapt the peroxisomal and cytosolic protein composition to cellular demands. In fungi, the peroxisomal import of catalase is exceptional, and it relies on a noncanonical PTS1 and a second unusual peroxisomal targeting signal closer to the N-terminus (Kragler et al., 1993; Williams et al., 2012). Substitution of the C-terminus with a canonical targeting signal lowers catalase activity and leads to the aggregation of the enzyme (Williams et al., 2012). In S. cerevisiae, peroxisomal localization of the glyoxylate cycle enzyme citrate synthase 2 is reduced upon expression of a version of Pex14 mimicking a phosphorylated state (Schummer et al., 2020). Phosphorylation of the targeting factor Pex5 has been implicated in the regulation of pexophagy upon stress exposure in mammalian cells and occurs in different species (Zhang J. et al., 2015; Oeljeklaus et al., 2016). How phosphorylation of Pex5 modulates the import of specific cargo besides regulating peroxisome breakdown remains to be established. Redox regulation at an N-terminal cysteine residue is another way to control Pex5 activity (Ma et al., 2013).
Phosphorylation of dually localized cargo proteins is also involved in regulating their intracellular distribution (Figure 1). S. cerevisiae glycerol-3-phosphate dehydrogenase Gpd1 is phosphorylated in the vicinity of the PTS2 (Jung et al., 2010). While phosphomimetic variants of Gpd1 are efficiently targeted to the peroxisome, variants resembling non-phosphorylated Gpd1 are retained in the cytosol.
Many fungal species encode at least two versions of the PTS1 receptor Pex5 (Kiel et al., 2006). The expression of the Pex5 paralog Pex9 is induced upon incubation of S. cerevisiae cells in oleic acid medium and regulates the import of a subset of peroxisomal matrix proteins with specific targeting signals (Effelsberg et al., 2016; Yifrach et al., 2016; Yifrach et al., 2022). In the corn smut fungus Ustilago maydis, two Pex5 paralogs with different cargo selectivity are required for peroxisome function (Ast et al., 2022). The concentration or availability of different targeting factors at peroxisomes and at other organelles may emerge as an additional regulatory device to control the subcellular distribution of proteins.
Conclusion
Knowledge about proteins with various cellular destinations significantly increased in the last years. It probably will continue to grow rapidly as sophisticated approaches become more and more available, which allow the tracking of minor or transient destinations. Careful investigation to confirm whether dual or multiple localizations are of biological significance will be required. Targeted disruption of sorting signals leaving the overall protein function intact is one approach to reveal a function inside of a particular organelle (Kunze et al., 2002; Freitag et al., 2012).
The mechanisms to achieve dual or multiple targeting are highly variable, and how they are embedded in the regulatory circuits of the cell is not established in many of the cases. Proteins with different destinations may further emerge as critical factors regulating organellar interplay in terms of metabolism and molecule exchange, as well as in terms of biogenesis and proliferation. Thus, a better characterization of how the shared proteins are distributed and how this is regulated will improve our understanding of eukaryotic cells.
Author contributions
EB designed the figures. All authors contributed to literature searches and conceptualization. JF wrote the manuscript with inputs from EB and TS.
Funding
TS was supported by a fellowship from DAAD and by the Center for Synthetic Microbiology, Marburg. JF acknowledges funding from the DFG (Grant ID FR-3586/2-1). Open Access funding provided by the Open Acess Publication Fund of Philipps-Universität Marburg with support of the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation).
Acknowledgments
We thank Björn Sandrock for critical reading of the manuscript and Michael Bölker for support. We apologize to all scientists whose excellent work was not cited in this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ackema, K. B., Hench, J., Böckler, S., Wang, S. C., Sauder, U., Mergentaler, H., et al. (2014). The small GTPase Arf1 modulates mitochondrial morphology and function. EMBO J. 33, 2659–2675. doi:10.15252/embj.201489039
Agrawal, G., Fassas, S. N., Xia, Z.-J., and Subramani, S. (2016). Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. J. Cell Biol. 212, 335–348. doi:10.1083/jcb.201506141
Agrawal, G., Joshi, S., and Subramani, S. (2011). Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 108, 9113–9118. doi:10.1073/pnas.1018749108
Al-Hajaya, Y., Karpinska, B., Foyer, C. H., and Baker, A. (2022). Nuclear and peroxisomal targeting of catalase. Plant Cell Environ. 45, 1096–1108. doi:10.1111/pce.14262
Al-Saryi, N. A., Al-Hejjaj, M. Y., van Roermund, C. W. T., Hulmes, G. E., Ekal, L., Payton, C., et al. (2017a). Two NAD-linked redox shuttles maintain the peroxisomal redox balance in Saccharomyces cerevisiae. Sci. Rep. 7, 11868. doi:10.1038/s41598-017-11942-2
Al-Saryi, N. A., Hutchinson, J. D., Al-hejjaj, M. Y., Sedelnikova, S., Baker, P., and Hettema, E. H. (2017b). Pnc1 piggy-back import into peroxisomes relies on Gpd1 homodimerisation. Sci. Rep. 7, 42579. doi:10.1038/srep42579
Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., and Sinclair, D. A. (2003). Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423, 181–185. doi:10.1038/nature01578
Antonenkov, V. D., and Hiltunen, J. K. (2012). Transfer of metabolites across the peroxisomal membrane. Biochim. Biophys. Acta 1822, 1374–1386. doi:10.1016/j.bbadis.2011.12.011
Aranovich, A., Hua, R., Rutenberg, A. D., and Kim, P. K. (2014). PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J. Cell Sci. 127, 3675–3686. doi:10.1242/jcs.146282
Ast, J., Bäcker, N., Bittner, E., Martorana, D., Ahmad, H., Bölker, M., et al. (2022). Two Pex5 proteins with different cargo specificity are critical for peroxisome function in Ustilago maydis. Front. Cell Dev. Biol. 10, 858084. doi:10.3389/fcell.2022.858084
Ast, J., Stiebler, A. C., Freitag, J., and Bölker, M. (2013). Dual targeting of peroxisomal proteins. Front. Physiol. 4, 297. doi:10.3389/fphys.2013.00297
Backes, S., and Herrmann, J. M. (2017). Protein translocation into the intermembrane space and matrix of mitochondria: Mechanisms and driving forces. Front. Mol. Biosci. 4, 83. doi:10.3389/fmolb.2017.00083
Baldwin, H. A., Wang, C., Kanfer, G., Shah, H. V., Velayos-Baeza, A., Dulovic-Mahlow, M., et al. (2021). VPS13D promotes peroxisome biogenesis. J. Cell Biol. 220, e202001188. doi:10.1083/jcb.202001188
Barlowe, C. K., and Miller, E. A. (2013). Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 193, 383–410. doi:10.1534/genetics.112.142810
Baune, M.-C., Lansing, H., Fischer, K., Meyer, T., Charton, L., Linka, N., et al. (2020). The Arabidopsis plastidial glucose-6-phosphate transporter GPT1 is dually targeted to peroxisomes via the endoplasmic reticulum. Plant Cell 32, 1703–1726. doi:10.1105/tpc.19.00959
Berger, J., Dorninger, F., Forss-Petter, S., and Kunze, M. (2016). Peroxisomes in brain development and function. Biochim. Biophys. Acta 1863, 934–955. doi:10.1016/j.bbamcr.2015.12.005
Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., et al. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731. doi:10.1083/jcb.200511125
Blobel, G., and Sabatini, D. D. (1971). “Ribosome-membrane interaction in eukaryotic cells,” in Biomembranes. Editor L. A. Manson (Boston, MA: Springer), 193–195.
Bolte, K., Rensing, S. A., and Maier, U.-G. (2015). The evolution of eukaryotic cells from the perspective of peroxisomes: Phylogenetic analyses of peroxisomal beta-oxidation enzymes support mitochondria-first models of eukaryotic cell evolution. BioEssays 37, 195–203. doi:10.1002/bies.201400151
Bürgi, J., Ekal, L., and Wilmanns, M. (2021). Versatile allosteric properties in Pex5-like tetratricopeptide repeat proteins to induce diverse downstream function. Traffic 22 (5), 140–152. doi:10.1111/tra.12785
Camões, F., Islinger, M., Guimarães, S. C., Kilaru, S., Schuster, M., Godinho, L. F., et al. (2015). New insights into the peroxisomal protein inventory: Acyl-CoA oxidases and-dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophys. Acta 1853, 111–125. doi:10.1016/j.bbamcr.2014.10.005
Carrie, C., Giraud, E., and Whelan, J. (2009). Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 276, 1187–1195. doi:10.1111/j.1742-4658.2009.06876.x
Carrie, C., Murcha, M. W., Kuehn, K., Duncan, O., Barthet, M., Smith, P. M., et al. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079. doi:10.1016/j.febslet.2008.07.061
Castro, I. G., Richards, D. M., Metz, J., Costello, J. L., Passmore, J. B., Schrader, T. A., et al. (2018). A role for Mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic 19, 229–242. doi:10.1111/tra.12549
Chen, Y.-C., Umanah, G. K. E., Dephoure, N., Andrabi, S. A., Gygi, S. P., Dawson, T. M., et al. (2014). Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail-anchored proteins. EMBO J. 33, 1548–1564. doi:10.15252/embj.201487943
Chen, Y., Pieuchot, L., Loh, R. A., Yang, J., Kari, T. M. A., Wong, J. Y., et al. (2014). Hydrophobic handoff for direct delivery of peroxisome tail-anchored proteins. Nat. Commun. 5, 5790. doi:10.1038/ncomms6790
Chornyi, S., IJlst, L., van Roermund, C. W. T., Wanders, R. J. A., and Waterham, H. R. (2021). Peroxisomal metabolite and cofactor transport in humans. Front. Cell Dev. Biol. 8, 613892. doi:10.3389/fcell.2020.613892
Cichocki, B. A., Krumpe, K., Vitali, D. G., and Rapaport, D. (2018). Pex19 is involved in importing dually targeted tail-anchored proteins to both mitochondria and peroxisomes. Traffic 19, 770–785. doi:10.1111/tra.12604
Clausmeyer, S., Stürzebecher, R., and Peters, J. (1999). An alternative transcript of the rat renin gene can result in a truncated prorenin that is transported into adrenal mitochondria. Circ. Res. 84, 337–344. doi:10.1161/01.res.84.3.337
Cohen, Y., Klug, Y. A., Dimitrov, L., Erez, Z., Chuartzman, S. G., Elinger, D., et al. (2014). Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol. Biosyst. 10 (7), 1742–1748. doi:10.1039/c4mb00001c
Corpas, F., Barroso, J., Sandalio, L., Distefano, S., Palma, J., Luoiannez, J., et al. (1998). A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem. J. 330, 777–784. doi:10.1042/bj3300777
Corti, O., DiDonato, S., and Finocchiaro, G. (1994). Divergent sequences in the 5’region of cDNA suggest alternative splicing as a mechanism for the generation of carnitine acetyltransferases with different subcellular localizations. Biochem. J. 303, 37–41. doi:10.1042/bj3030037
Costanzo, M., VanderSluis, B., Koch, E. N., Baryshnikova, A., Pons, C., Tan, G., et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353 (6306), aaf1420. doi:10.1126/science.aaf1420
Costello, J. L., Castro, I. G., Camões, F., Schrader, T. A., McNeall, D., Yang, J., et al. (2017a). Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells. J. Cell Sci. 130, 1675–1687. doi:10.1242/jcs.200204
Costello, J. L., Castro, I. G., Hacker, C., Schrader, T. A., Metz, J., Zeuschner, D., et al. (2017b). ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 216, 331–342. doi:10.1083/jcb.201607055
Costello, J. L., Passmore, J. B., Islinger, M., and Schrader, M. (2018). Multi-localized proteins: The peroxisome-mitochondria connection. Subcell. Biochem. 89, 383–415. doi:10.1007/978-981-13-2233-4_17
Dahan, N., Bykov, Y. S., Boydston, E. A., Fadel, A., Gazi, Z., Hochberg-Laufer, H., et al. (2022). Peroxisome function relies on organelle-associated mRNA translation. Sci. Adv. 8, eabk2141. doi:10.1126/sciadv.abk2141
David, C., Koch, J., Oeljeklaus, S., Laernsack, A., Melchior, S., Wiese, S., et al. (2013). A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol. Cell. Proteomics 12, 2408–2425. doi:10.1074/mcp.M112.017830
De Duve, C., and Baudhuin, P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46, 323–357. doi:10.1152/physrev.1966.46.2.323
Dellero, Y., Jossier, M., Schmitz, J., Maurino, V. G., and Hodges, M. (2016). Photorespiratory glycolate-glyoxylate metabolism. J. Exp. Bot. 67 (10), 3041–3052. doi:10.1093/jxb/erw090
Di Cara, F. (2020). Peroxisomes in host defense. PLoS Pathog. 16, e1008636. doi:10.1371/journal.ppat.1008636
Di Cara, F., Sheshachalam, A., Braverman, N. E., Rachubinski, R. A., and Simmonds, A. J. (2017). Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity 47, 93–106. e7. doi:10.1016/j.immuni.2017.06.016
Dunn, J. G., Foo, C. K., Belletier, N. G., Gavis, E. R., and Weissman, J. S. (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2, e01179. doi:10.7554/eLife.01179
Effelsberg, D., Cruz-Zaragoza, L. D., Schliebs, W., and Erdmann, R. (2016). Pex9p is a new yeast peroxisomal import receptor for PTS1-containing proteins. J. Cell Sci. 129, 4057–4066. doi:10.1242/jcs.195271
Effelsberg, D., Cruz-Zaragoza, L. D., Tonillo, J., Schliebs, W., and Erdmann, R. (2015). Role of Pex21p for piggyback import of Gpd1p and Pnc1p into peroxisomes of Saccharomyces cerevisiae. J. Biol. Chem. 290, 25333–25342. doi:10.1074/jbc.M115.653451
Eisenberg-Bord, M., Zung, N., Collado, J., Drwesh, L., Fenech, E. J., Fadel, A., et al. (2021). Cnm1 mediates nucleus--mitochondria contact site formation in response to phospholipid levels. J. Cell Biol. 220, e202104100. doi:10.1083/jcb.202104100
Elgersma, Y., van Roermund, C. W., Wanders, R. J., and Tabak, H. F. (1995). Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 14, 3472–3479. doi:10.1002/j.1460-2075.1995.tb07353.x
Exner, T., Romero-Brey, I., Yifrach, E., Rivera-Monroy, J., Schrul, B., Zouboulis, C. C., et al. (2019). An alternative membrane topology permits lipid droplet localization of peroxisomal fatty acyl-CoA reductase 1. J. Cell Sci. 132, jcs223016. doi:10.1242/jcs.223016
Falter, C., and Reumann, S. (2022). The essential role of fungal peroxisomes in plant infection. Mol. Plant Pathol. 23 (6), 781–794. doi:10.1111/mpp.13180
Fan, J., Li, X., Issop, L., Culty, M., and Papadopoulos, V. (2016). ACBD2/ECI2-Mediated peroxisome-mitochondria interactions in leydig cell steroid biosynthesis. Mol. Endocrinol. 30, 763–782. doi:10.1210/me.2016-1008
Fang, Y., Morrell, J. C., Jones, J. M., and Gould, S. J. (2004). PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 164, 863–875. doi:10.1083/jcb.200311131
Ferreira, A. R., Marques, M., Ramos, B., Kagan, J. C., and Ribeiro, D. (2022). Emerging roles of peroxisomes in viral infections. Trends Cell Biol. 32, 124–139. doi:10.1016/j.tcb.2021.09.010
Ferreira, J. V., and Carvalho, P. (2021). Pex30-like proteins function as adaptors at distinct ER membrane contact sites. J. Cell Biol. 220, e202103176. doi:10.1083/jcb.202103176
Ferro-Novick, S., Rapoport, T. A., and Schekman, R. (2013). The endoplasmic reticulum. New York: Cold Spring Harbor Lab Press.
Fodor, K., Wolf, J., Reglinski, K., Passon, D. M., Lou, Y., Schliebs, W., et al. (2015). Ligand-induced compaction of the PEX5 receptor-binding cavity impacts protein import efficiency into peroxisomes. Traffic 16, 85–98. doi:10.1111/tra.12238
Francisco, T., Rodrigues, T. A., Dias, A. F., Barros-Barbosa, A., Bicho, D., and Azevedo, J. E. (2017). Protein transport into peroxisomes: Knowns and unknowns. BioEssays 39, 1700047. doi:10.1002/bies.201700047
Fransen, M., Lismont, C., and Walton, P. (2017). The peroxisome-mitochondria connection: How and why? Int. J. Mol. Sci. 18, 1126. doi:10.3390/ijms18061126
Freitag, J., Ast, J., and Bölker, M. (2012). Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525. doi:10.1038/nature11051
Freitag, J., Ast, J., Linne, U., Stehlik, T., Martorana, D., Bölker, M., et al. (2014). Peroxisomes contribute to biosynthesis of extracellular glycolipids in fungi. Mol. Microbiol. 93, 24–36. doi:10.1111/mmi.12642
Freitag, J., Stehlik, T., Stiebler, A. C., and Bölker, M. (2018). “The obvious and the hidden: Prediction and function of fungal peroxisomal matrix proteins,” in Proteomics of peroxisomes, Subcellular Biochemistry, Editor L. del Río, and M. Schrader (Singapore: Springer), 89. doi:10.1007/978-981-13-2233-4_6
Gabaldón, T. (2014). A metabolic scenario for the evolutionary origin of peroxisomes from the endomembranous system. Cell. Mol. Life Sci. 71, 2373–2376. doi:10.1007/s00018-013-1424-z
Gabaldón, T. (2018). “Evolution of the peroxisomal proteome BT - proteomics of peroxisomes: Identifying novel functions and regulatory networks,” in, eds. L. A. del Río, and M. Schrader (Singapore: Springer Singapore), 221–233.
Gabaldón, T. (2010). Peroxisome diversity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 765–773. doi:10.1098/rstb.2009.0240
Gabaldón, T., Snel, B., van Zimmeren, F., Hemrika, W., Tabak, H., and Huynen, M. A. (2006). Origin and evolution of the peroxisomal proteome. Biol. Direct 1, 8. doi:10.1186/1745-6150-1-8
Gabay-Maskit, S., Cruz-Zaragoza, L. D., Shai, N., Eisenstein, M., Bibi, C., Cohen, N., et al. (2020). A piggybacking mechanism enables peroxisomal localization of the glyoxylate cycle enzyme Mdh2 in yeast. J. Cell Sci. 133, jcs244376. doi:10.1242/jcs.244376
Germain, K., and Kim, P. K. (2020). Pexophagy: A model for selective autophagy. Int. J. Mol. Sci. 21, E578. doi:10.3390/ijms21020578
Goepfert, S., and Poirier, Y. (2007). Beta-oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 10, 245–251. doi:10.1016/j.pbi.2007.04.007
Grewal, P. S., Samson, J. A., Baker, J. J., Choi, B., and Dueber, J. E. (2021). Peroxisome compartmentalization of a toxic enzyme improves alkaloid production. Nat. Chem. Biol. 17, 96–103. doi:10.1038/s41589-020-00668-4
Gründlinger, M., Yasmin, S., Lechner, B. E., Geley, S., Schrettl, M., Hynes, M., et al. (2013). Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 88 (5), 862–875. doi:10.1111/mmi.12225
Guillén-Samander, A., Leonzino, M., Hanna IV, M. G., Tang, N., Shen, H., and De Camilli, P. (2021). VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J. Cell Biol. 220, e202010004. doi:10.1083/jcb.202010004
Guimaraes, S. C., Schuster, M., Bielska, E., Dagdas, G., Kilaru, S., Meadows, B. R. A., et al. (2015). Peroxisomes, lipid droplets, and endoplasmic reticulum “hitchhike” on motile early endosomes. J. Cell Biol. 211 (5), 945–954. doi:10.1083/jcb.201505086
Haanstra, J. R., González-Marcano, E. B., Gualdrón-López, M., and Michels, P. A. M. (2016). Biogenesis, maintenance and dynamics of glycosomes in trypanosomatid parasites. Biochim. Biophys. Acta 1863, 1038–1048. doi:10.1016/j.bbamcr.2015.09.015
Haselbeck, R. J., and McAlister-Henn, L. (1993). Function and expression of yeast mitochondrial NAD-and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 268, 12116–12122. doi:10.1016/s0021-9258(19)50315-5
Haselbeck, R. J., and McAlister-Henn, L. (1991). Isolation, nucleotide sequence, and disruption of the Saccharomyces cerevisiae gene encoding mitochondrial NADP(H)-specific isocitrate dehydrogenase. J. Biol. Chem. 266, 2339–2345. doi:10.1016/s0021-9258(18)52249-3
Henne, W. M., Buchkovich, N. J., and Emr, S. D. (2011). The ESCRT pathway. Dev. Cell 21, 77–91. doi:10.1016/j.devcel.2011.05.015
Hiltunen, J. K., Mursula, A. M., Rottensteiner, H., Wierenga, R. K., Kastaniotis, A. J., and Gurvitz, A. (2003). The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27, 35–64. doi:10.1016/S0168-6445(03)00017-2
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P., and Tabak, H. F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95. doi:10.1016/j.cell.2005.04.025
Hofhuis, J., Schueren, F., Nötzel, C., Lingner, T., Gärtner, J., Jahn, O., et al. (2016). The functional readthrough extension of malate dehydrogenase reveals a modification of the genetic code. Open Biol. 6, 160246. doi:10.1098/rsob.160246
Hölscher, C., Lutterbey, M.-C., Lansing, H., Meyer, T., Fischer, K., and von Schaewen, A. (2016). Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in Arabidopsis thaliana. Plant Physiol. 171, 192–205. doi:10.1104/pp.15.01301
Hoppins, S., Collins, S. R., Cassidy-Stone, A., Hummel, E., DeVay, R. M., Lackner, L. L., et al. (2011). A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340. doi:10.1083/jcb.201107053
Houten, S. M., Wanders, R. J. A., and Ranea-Robles, P. (2020). Metabolic interactions between peroxisomes and mitochondria with a special focus on acylcarnitine metabolism. Biochim. Biophys. Acta. Mol. Basis Dis. 1866, 165720. doi:10.1016/j.bbadis.2020.165720
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: Biogenesis and function. Plant Cell 24, 2279–2303. doi:10.1105/tpc.112.096586
Hua, R., Cheng, D., Coyaud, É., Freeman, S., Di Pietro, E., Wang, Y., et al. (2017). VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216, 367–377. doi:10.1083/jcb.201608128
Idnurm, A., Giles, S. S., Perfect, J. R., and Heitman, J. (2007). Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot. Cell 6, 60–72. doi:10.1128/EC.00214-06
Inaba, J., Kim, B. M., Shimura, H., and Masuta, C. (2011). Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol. 156, 2026–2036. doi:10.1104/pp.111.180042
Islinger, M., Grille, S., Fahimi, H. D., and Schrader, M. (2012). The peroxisome: An update on mysteries. Histochem. Cell Biol. 137, 547–574. doi:10.1007/s00418-012-0941-4
Islinger, M., Li, K. W., Seitz, J., Völkl, A., and Lüers, G. H. (2009). Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes--evidence for a natural piggyback import mechanism in mammals. Traffic 10, 1711–1721. doi:10.1111/j.1600-0854.2009.00966.x
Jan, C. H., Williams, C. C., and Weissman, J. S. (2014). Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521. doi:10.1126/science.1257521
Jansen, R. L. M., and van der Klei, I. J. (2019). The peroxisome biogenesis factors Pex3 and Pex19: Multitasking proteins with disputed functions. FEBS Lett. 593, 457–474. doi:10.1002/1873-3468.13340
Jedd, G., and Chua, N. H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231. doi:10.1038/35008652
Jones, J. M., Morrell, J. C., and Gould, S. J. (2004). PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164, 57–67. doi:10.1083/jcb.200304111
Joshi, A. S., Huang, X., Choudhary, V., Levine, T. P., Hu, J., and Prinz, W. A. (2016). A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J. Cell Biol. 215 (4), 515–529. doi:10.1083/jcb.201602064
Joshi, A. S., Nebenfuehr, B., Choudhary, V., Satpute-Krishnan, P., Levine, T. P., Golden, A., et al. (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat. Commun. 9, 2940. doi:10.1038/s41467-018-05277-3
Joshi, A. S., Zhang, H., and Prinz, W. A. (2017). Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 19, 876–882. doi:10.1038/ncb3579
Jung, S., Marelli, M., Rachubinski, R. A., Goodlett, D. R., and Aitchison, J. D. (2010). Dynamic changes in the subcellular distribution of Gpd1p in response to cell stress. J. Biol. Chem. 285, 6739–6749. doi:10.1074/jbc.M109.058552
Kamigaki, A., Mano, S., Terauchi, K., Nishi, Y., Tachibe-Kinoshita, Y., Nito, K., et al. (2003). Identification of peroxisomal targeting signal of pumpkin catalase and the binding analysis with PTS1 receptor. Plant J. 33, 161–175. doi:10.1046/j.0960-7412.2003.001605.x
Kaplan, R. S., Mayor, J. A., Gremse, D. A., and Wood, D. O. (1995). High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae (∗). J. Biol. Chem. 270, 4108–4114. doi:10.1074/jbc.270.8.4108
Kataya, A., Gautam, N., Jamshed, M., Muench, D. G., Samuel, M. A., Thelen, J. J., et al. (2022). Identification of Arabidopsis protein kinases that harbor functional type 1 peroxisomal targeting signals. Front. Cell Dev. Biol. 10, 745883. doi:10.3389/fcell.2022.745883
Kataya, A. R. A., and Reumann, S. (2010). Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal. Behav. 5, 171–175. doi:10.4161/psb.5.2.10527
Kataya, A., Schei, E., and Lillo, C. (2016). Towards understanding peroxisomal phosphoregulation in Arabidopsis thaliana. Planta 243, 699–717. doi:10.1007/s00425-015-2439-5
Kellis, M., Birren, B. W., and Lander, E. S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624. doi:10.1038/nature02424
Kempiński, B., Chełstowska, A., Poznański, J., Król, K., Rymer, Ł., Frydzińska, Z., et al. (2020). The peroxisomal targeting signal 3 (PTS3) of the budding yeast acyl-CoA oxidase is a signal patch. Front. Cell Dev. Biol. 8, 198. doi:10.3389/fcell.2020.00198
Khan, Z., Michalopoulos, G. K., and Stolz, D. B. (2006). Peroxisomal localization of hypoxia-inducible factors and hypoxia-inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia-reoxygenation. Am. J. Pathol. 169, 1251–1269. doi:10.2353/ajpath.2006.060360
Kiel, J. A. K. W., Veenhuis, M., and Van Der Klei, I. J. (2006). PEX genes in fungal genomes: Common, rare or redundant. Traffic 7, 1291–1303. doi:10.1111/j.1600-0854.2006.00479.x
Kim, K.-S., Rosenkrantz, M. S., and Guarente, L. (1986). Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol. Cell. Biol. 6, 1936–1942. doi:10.1128/mcb.6.6.1936
Kim, P. K., and Hettema, E. H. (2015). Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427, 1176–1190. doi:10.1016/j.jmb.2015.02.005
Kim, P. K., Mullen, R. T., Schumann, U., and Lippincott-Schwartz, J. (2006). The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 173, 521–532. doi:10.1083/jcb.200601036
Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M. A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. doi:10.1074/jbc.M211761200
Koch, A., Yoon, Y., Bonekamp, N. A., McNiven, M. A., and Schrader, M. (2005). A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 16 (11), 5077–5086. doi:10.1091/mbc.e05-02-0159
Kong, J., Ji, Y., Jeon, Y. G., Han, J. S., Han, K. H., Lee, J. H., et al. (2020). Spatiotemporal contact between peroxisomes and lipid droplets regulates fasting-induced lipolysis via PEX5. Nat. Commun. 11, 578. doi:10.1038/s41467-019-14176-0
Kornberg, A. (1962). “On the metabolic significance of phosphorolytic and pyrophosphorolytic reactions,” in Horizons in biochemistry. Editors H. Kasha,, and B. Pullman (New York, USA: Academic Press), 251–264.
Kornmann, B., Currie, E., Collins, S. R., Schuldiner, M., Nunnari, J., Weissman, J. S., et al. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. doi:10.1126/science.1175088
Kornmann, B., Osman, C., and Walter, P. (2011). The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. U. S. A. 108, 14151–14156. doi:10.1073/pnas.1111314108
Kragler, F., Langeder, A., Raupachova, J., Binder, M., and Hartig, A. (1993). Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J. Cell Biol. 120, 665–673. doi:10.1083/jcb.120.3.665
Kremp, M., Bittner, E., Martorana, D., Klingenberger, A., Stehlik, T., Bölker, M., et al. (2020). Non-AUG translation initiation generates peroxisomal isoforms of 6-phosphogluconate dehydrogenase in fungi. Front. Cell Dev. Biol. 8, 251. doi:10.3389/fcell.2020.00251
Kretschmer, M., Klose, J., and Kronstad, J. W. (2012a). Defects in mitochondrial and peroxisomal β-oxidation influence virulence in the maize pathogen Ustilago maydis. Eukaryot. Cell 11, 1055–1066. doi:10.1128/EC.00129-12
Kretschmer, M., Wang, J., and Kronstad, J. W. (2012b). Peroxisomal and mitochondrial β-oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 11, 1042–1054. doi:10.1128/EC.00128-12
Kumar, S., Singh, R., Williams, C. P., and van der Klei, I. J. (2016). Stress exposure results in increased peroxisomal levels of yeast Pnc1 and Gpd1, which are imported via a piggy-backing mechanism. Biochim. Biophys. Acta 1863, 148–156. doi:10.1016/j.bbamcr.2015.10.017
Kundu, D., and Pasrija, R. (2020). The ERMES (endoplasmic reticulum and mitochondria encounter structures) mediated functions in fungi. Mitochondrion 52, 89–99. doi:10.1016/j.mito.2020.02.010
Kunze, M., and Berger, J. (2015). The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 6, 259. doi:10.3389/fphys.2015.00259
Kunze, M., Kragler, F., Binder, M., Hartig, A., and Gurvitz, A. (2002). Targeting of malate synthase 1 to the peroxisomes of Saccharomyces cerevisiae cells depends on growth on oleic acid medium. Eur. J. Biochem. 269, 915–922. doi:10.1046/j.0014-2956.2001.02727.x
Kunze, M., Malkani, N., Maurer-Stroh, S., Wiesinger, C., Schmid, J. A., and Berger, J. (2015). Mechanistic insights into PTS2-mediated peroxisomal protein import: The co-receptor PEX5L drastically increases the interaction strength between the cargo protein and the receptor PEX7. J. Biol. Chem. 290 (8), 4928–4940. doi:10.1074/jbc.M114.601575
Kunze, M., Pracharoenwattana, I., Smith, S. M., and Hartig, A. (2006). A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim. Biophys. Acta 1763, 1441–1452. doi:10.1016/j.bbamcr.2006.09.009
Kunze, M. (2020). The type-2 peroxisomal targeting signal. Biochim. Biophys. Acta. Mol. Cell Res. 1867, 118609. doi:10.1016/j.bbamcr.2019.118609
Kuravi, K., Nagotu, S., Krikken, A. M., Sjollema, K., Deckers, M., Erdmann, R., et al. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119, 3994–4001. doi:10.1242/jcs.03166
Lam, S., Yoda, N., and Schekman, R. (2010). A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 107 (50), 21523–21528. doi:10.1073/pnas.1013397107
Lee, Y. J., Jang, J. W., Kim, K. J., and Maeng, P. J. (2011). TCA cycle-independent acetate metabolism via the glyoxylate cycle in Saccharomyces cerevisiae. Yeast 28, 153–166. doi:10.1002/yea.1828
Lee, Y. J., Jeschke, G. R., Roelants, F. M., Thorner, J., and Turk, B. E. (2012). Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol. Cell. Biol. 32, 4705–4717. doi:10.1128/MCB.00897-12
Li, H.-S., Zhou, Y.-N., Li, L., Li, S.-F., Long, D., Chen, X.-L., et al. (2019). HIF-1α protects against oxidative stress by directly targeting mitochondria. Redox Biol. 25, 101109. doi:10.1016/j.redox.2019.101109
Li, H., Lismont, C., Revenco, I., Hussein, M. A. F., Costa, C. F., and Fransen, M. (2021). The peroxisome-autophagy redox connection: A double-edged sword? Front. Cell Dev. Biol. 9, 814047. doi:10.3389/fcell.2021.814047
Lingner, T., Kataya, A. R., Antonicelli, G. E., Benichou, A., Nilssen, K., Chen, X.-Y., et al. (2011). Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23, 1556–1572. doi:10.1105/tpc.111.084095
Lismont, C., Revenco, I., and Fransen, M. (2019). Peroxisomal hydrogen peroxide metabolism and signaling in health and disease. Int. J. Mol. Sci. 20, 3673. doi:10.3390/ijms20153673
Liu, F., Lu, Y., Pieuchot, L., Dhavale, T., and Jedd, G. (2011). Import oligomers induce positive feedback to promote peroxisome differentiation and control organelle abundance. Dev. Cell 21 (3), 457–468. doi:10.1016/j.devcel.2011.08.004
Ma, C., Hagstrom, D., Polley, S. G., and Subramani, S. (2013). Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 288, 27220–27231. doi:10.1074/jbc.M113.492694
Maggio-Hall, L. A., and Keller, N. P. (2004). Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 54, 1173–1185. doi:10.1111/j.1365-2958.2004.04340.x
Magliano, P., Flipphi, M., Arpat, B. A., Delessert, S., and Poirier, Y. (2011). Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi. J. Biol. Chem. 286, 42133–42140. doi:10.1074/jbc.M111.279687
Martenson, J. S., Tam, H., McQuown, A. J., Reif, D., Zhou, J., and Denic, V. (2020). The importomer is a peroxisomal membrane protein translocase. bioRxiv. bioarchives preprints.
Mast, F. D., Herricks, T., Strehler, K. M., Miller, L. R., Saleem, R. A., Rachubinski, R. A., et al. (2018). ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217, 2087–2102. doi:10.1083/jcb.201706044
Mast, F. D., Jamakhandi, A., Saleem, R. A., Dilworth, D. J., Rogers, R. S., Rachubinski, R. A., et al. (2016). Peroxins Pex30 and Pex29 dynamically associate with reticulons to regulate peroxisome biogenesis from the endoplasmic reticulum. J. Biol. Chem. 291, 15408–15427. doi:10.1074/jbc.M116.728154
Matsuzaki, T., and Fujiki, Y. (2008). The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275–1286. doi:10.1083/jcb.200806062
McCartney, A. W., Greenwood, J. S., Fabian, M. R., White, K. A., and Mullen, R. T. (2005). Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17, 3513–3531. doi:10.1105/tpc.105.036350
Meijer, W. H., Gidijala, L., Fekken, S., Kiel, J. A. K. W., van den Berg, M. A., Lascaris, R., et al. (2010). Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 76, 5702–5709. doi:10.1128/AEM.02327-09
Meyer, T., Hölscher, C., Schwöppe, C., and von Schaewen, A. (2011). Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 66, 745–758. doi:10.1111/j.1365-313X.2011.04535.x
Monteuuis, G., Miścicka, A., Świrski Michałand Zenad, L., Niemitalo, O., Wrobel, L., Alam, J., et al. (2019). Non-canonical translation initiation in yeast generates a cryptic pool of mitochondrial proteins. Nucleic Acids Res. 47, 5777–5791. doi:10.1093/nar/gkz301
Morgenstern, M., Stiller, S. B., Lübbert, P., Peikert, C. D., Dannenmaier, S., Drepper, F., et al. (2017). Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852. doi:10.1016/j.celrep.2017.06.014
Motley, A. M., Galvin, P. C., Ekal, L., Nuttall, J. M., and Hettema, E. H. (2015). Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J. Cell Biol. 211, 1041–1056. doi:10.1083/jcb.201412066
Motley, A. M., Ward, G. P., and Hettema, E. H. (2008). Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121, 1633–1640. doi:10.1242/jcs.026344
Natsoulis, G., Hilger, F., and Fink, G. R. (1986). The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell 46 (2), 235–243. doi:10.1016/0092-8674(86)90740-3
Neuberger, G., Kunze, M., Eisenhaber, F., Berger, J., Hartig, A., and Brocard, C. (2004). Hidden localization motifs: Naturally occurring peroxisomal targeting signals in non-peroxisomal proteins. Genome Biol. 5, R97. doi:10.1186/gb-2004-5-12-r97
Neuspiel, M., Schauss, A. C., Braschi, E., Zunino, R., Rippstein, P., Rachubinski, R. A., et al. (2008). Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18, 102–108. doi:10.1016/j.cub.2007.12.038
Nötzel, C., Lingner, T., Klingenberg, H., and Thoms, S. (2016). Identification of new fungal peroxisomal matrix proteins and revision of the PTS1 consensus. Traffic 17, 1110–1124. doi:10.1111/tra.12426
Novikoff, A. B., and Shin, W.-Y. (1964). The endoplasmic reticulum in the Golgi zone and its relations to microbodies, Golgi apparatus and autophagic vacuoles in rat liver cells. J. Microsc. 3, 187–206.
Nuebel, E., Morgan, J. T., Fogarty, S., Winter, J. M., Lettlova, S., Berg, J. A., et al. (2021). The biochemical basis of mitochondrial dysfunction in Zellweger Spectrum Disorder. EMBO Rep. 22, e51991. doi:10.15252/embr.202051991
Odendall, C., Dixit, E., Stavru, F., Bierne, H., Franz, K. M., Durbin, A. F., et al. (2014). Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726. doi:10.1038/ni.2915
Oeljeklaus, S., Schummer, A., Mastalski, T., Platta, H. W., and Warscheid, B. (2016). Regulation of peroxisome dynamics by phosphorylation. Biochim. Biophys. Acta 1863, 1027–1037. doi:10.1016/j.bbamcr.2015.12.022
Okreglak, V., and Walter, P. (2014). The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. Proc. Natl. Acad. Sci. U. S. A. 111, 8019–8024. doi:10.1073/pnas.1405755111
Okumoto, K., El Shermely, M., Natsui, M., Kosako, H., Natsuyama, R., Marutani, T., et al. (2020). The peroxisome counteracts oxidative stresses by suppressing catalase import via Pex14 phosphorylation. Elife 9, e55896. doi:10.7554/eLife.55896
Palmieri, F. (2004). The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflügers Arch. 447, 689–709. doi:10.1007/s00424-003-1099-7
Pan, R., Liu, J., Wang, S., and Hu, J. (2020). Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 225, 1410–1427. doi:10.1111/nph.16134
Paradis, M., Kucharowski, N., Edwards, G., Palacios, S. M., Meyer, C., Stümpges, B., et al. (2021). The ER protein Creld regulates ER-mitochondria contact dynamics and respiratory complex 1 activity. ResearchSquare. preprint. doi:10.21203/rs.3.rs-911428/v1
Passreiter, M., Anton, M., Lay, D., Frank, R., Harter, C., Wieland, F. T., et al. (1998). Peroxisome biogenesis: Involvement of ARF and coatomer. J. Cell Biol. 141, 373–383. doi:10.1083/jcb.141.2.373
Poirier, Y., Antonenkov, V. D., Glumoff, T., and Hiltunen, J. K. (2006). Peroxisomal β-oxidation—A metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426. doi:10.1016/j.bbamcr.2006.08.034
Praefcke, G. J. K., and McMahon, H. T. (2004). The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, 133–147. doi:10.1038/nrm1313
Raychaudhuri, S., and Prinz, W. A. (2008). Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 105, 15785–15790. doi:10.1073/pnas.0808321105
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E., et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. doi:10.1105/tpc.107.050989
Reumann, S., and Weber, A. P. M. (2006). Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim. Biophys. Acta 1763, 1496–1510. doi:10.1016/j.bbamcr.2006.09.008
Rosenthal, M., Metzl-Raz, E., Bürgi, J., Yifrach, E., Drwesh, L., Fadel, A., et al. (2020). Uncovering targeting priority to yeast peroxisomes using an in-cell competition assay. Proc. Natl. Acad. Sci. U. S. A. 117, 21432–21440. doi:10.1073/pnas.1920078117
Sacksteder, K. A., Jones, J. M., South, S. T., Li, X., Liu, Y., and Gould, S. J. (2000). PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148, 931–944. doi:10.1083/jcb.148.5.931
Salo, V. T., Belevich, I., Li, S., Karhinen, L., Vihinen, H., Vigouroux, C., et al. (2016). Seipin regulates ER–lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716. doi:10.15252/embj.201695170
Salogiannis, J., Egan, M. J., and Reck-Peterson, S. L. (2016). Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J. Cell Biol. 212, 289–296. doi:10.1083/jcb.201512020
Schliebs, W., Girzalsky, W., and Erdmann, R. (2010). Peroxisomal protein import and ERAD: Variations on a common theme. Nat. Rev. Mol. Cell Biol. 11, 885–890. doi:10.1038/nrm3008
Schlüter, A., Fourcade, S., Ripp, R., Mandel, J. L., Poch, O., and Pujol, A. (2006). The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol. Biol. Evol. 23, 838–845. doi:10.1093/molbev/msj103
Schrader, M., Costello, J., Godinho, L. F., and Islinger, M. (2015). Peroxisome-mitochondria interplay and disease. J. Inherit. Metab. Dis. 38, 681–702. doi:10.1007/s10545-015-9819-7
Schrader, M., Kamoshita, M., and Islinger, M. (2020). Organelle interplay—Peroxisome interactions in health and disease. J. Inherit. Metab. Dis. 43, 71–89. doi:10.1002/jimd.12083
Schrul, B., and Kopito, R. R. (2016). Peroxin-dependent targeting of a lipid-droplet-destined membrane protein to ER subdomains. Nat. Cell Biol. 18, 740–751. doi:10.1038/ncb3373
Schueren, F., Lingner, T., George, R., Hofhuis, J., Dickel, C., Gärtner, J., et al. (2014). Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife 3, e03640. doi:10.7554/eLife.03640
Schueren, F., and Thoms, S. (2016). Functional translational readthrough: A systems biology perspective. PLoS Genet. 12, e1006196. doi:10.1371/journal.pgen.1006196
Schummer, A., Maier, R., Gabay-Maskit, S., Hansen, T., Mühlhäuser, W. W. D., Suppanz, I., et al. (2020). Pex14p phosphorylation modulates import of citrate synthase 2 into peroxisomes in Saccharomyces cerevisiae. Front. Cell Dev. Biol. 8, 549451. doi:10.3389/fcell.2020.549451
Shai, N., Yifrach, E., Roermund, C. W. T., Cohen, N., Bibi, C., IJlst, L., et al. (2018). Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9, 1761. doi:10.1038/s41467-018-03957-8
Sies, H. (2017). Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 11, 613–619. doi:10.1016/j.redox.2016.12.035
Smith, J. J., and Aitchison, J. D. (2013). Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14, 803–817. doi:10.1038/nrm3700
Speijer, D. (2017). Evolution of peroxisomes illustrates symbiogenesis. BioEssays 39, 1700050. doi:10.1002/bies.201700050
Steffan, J. S., and McAlister-Henn, L. (1992). Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase. J. Biol. Chem. 267, 24708–24715. doi:10.1016/s0021-9258(18)35822-8
Stehlik, T., Kremp, M., Kahnt, J., Bölker, M., and Freitag, J. (2020). Peroxisomal targeting of a protein phosphatase type 2C via mitochondrial transit. Nat. Commun. 11, 2355. doi:10.1038/s41467-020-16146-3
Stehlik, T., Sandrock, B., Ast, J., and Freitag, J. (2014). Fungal peroxisomes as biosynthetic organelles. Curr. Opin. Microbiol. 22, 8–14. doi:10.1016/j.mib.2014.09.011
Stein, I., Peleg, Y., Even-Ram, S., and Pines, O. (1994). The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 4770–4778. doi:10.1128/mcb.14.7.4770
Stiebler, A. C., Freitag, J., Schink, K. O., Stehlik, T., Tillmann, B. A. M., Ast, J., et al. (2014). Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in fungi and animals. PLoS Genet. 10, e1004685–9. doi:10.1371/journal.pgen.1004685
Strijbis, K., den Burg, J., F Visser, W., den Berg, M., and Distel, B. (2012). Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS Yeast Res. 12 (1), 61–68. doi:10.1111/j.1567-1364.2011.00761.x
Sugiura, A., Mattie, S., Prudent, J., and McBride, H. M. (2017). Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254. doi:10.1038/nature21375
Swiegers, J. H., Dippenaar, N., Pretorius, I. S., and Bauer, F. F. (2001). Carnitine-dependent metabolic activities in Saccharomyces cerevisiae: Three carnitine acetyltransferases are essential in a carnitine-dependent strain. Yeast 18, 585–595. doi:10.1002/yea.712
Szewczyk, E., Andrianopoulos, A., Davis, M. A., and Hynes, M. J. (2001). A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADP-dependent isocitrate dehydrogenase inAspergillus nidulans. J. Biol. Chem. 276, 37722–37729. doi:10.1074/jbc.M105645200
Szymanski, K. M., Binns, D., Bartz, R., Grishin, N. V., Li, W.-P., Agarwal, A. K., et al. (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A. 104, 20890–20895. doi:10.1073/pnas.0704154104
Tanabe, Y., Maruyama, J., Yamaoka, S., Yahagi, D., Matsuo, I., Tsutsumi, N., et al. (2011). Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J. Biol. Chem. 286, 30455–30461. doi:10.1074/jbc.M111.247338
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I., and Gaude, T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. U. S. A. 112 (13), 4158–4163. doi:10.1073/pnas.1403322112
Titorenko, V. I., Chan, H., and Rachubinski, R. A. (2000). Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of yarrowia lipolytica. J. Cell Biol. 148, 29–44. doi:10.1083/jcb.148.1.29
Tripathi, D. N., and Walker, C. L. (2016). The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol. 39, 109–112. doi:10.1016/j.ceb.2016.02.017
Ueda, M., Kawachi, H., Atomi, H., and Tanaka, A. (1998). Peroxisomal and mitochondrial carnitine acetyltransferase isozymes of the n-alkane-assimilating yeast, Candida tropicalis, occurred by alternative initiation of translation from the transcripts of a single gene. Biochim. Biophys. Acta 1397, 213–222. doi:10.1016/s0167-4781(98)00009-8
Ušaj, M. M., Brložnik, M., Kaferle, P., Žitnik, M., Wolinski, H., Leitner, F., et al. (2015). Genome-wide localization study of yeast Pex11 identifies peroxisome--mitochondria interactions through the ERMES complex. J. Mol. Biol. 427, 2072–2087. doi:10.1016/j.jmb.2015.03.004
van der Klei, I. J., and Veenhuis, M. (2006a). PTS1-independent sorting of peroxisomal matrix proteins by Pex5p. Biochim. Biophys. Acta 1763, 1794–1800. doi:10.1016/j.bbamcr.2006.08.013
van der Klei, I. J., and Veenhuis, M. (2006b). Yeast and filamentous fungi as model organisms in microbody research. Biochim. Biophys. Acta 1763, 1364–1373. doi:10.1016/j.bbamcr.2006.09.014
Van der Zand, A., Gent, J., Braakman, I., and Tabak, H. (2012). Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149, 397–409. doi:10.1016/j.cell.2012.01.054
Van Roermund, C. W., Elgersma, Y., Singh, N., Wanders, R. J., and Tabak, H. F. (1995). The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD (H) and acetyl-CoA under in vivo conditions. EMBO J. 14, 3480–3486. doi:10.1002/j.1460-2075.1995.tb07354.x
van Roermund, C. W. T., Hettema, E. H., Kal, A. J., van den Berg, M., Tabak, H. F., and Wanders, R. J. A. (1998). Peroxisomal β-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: Isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 17, 677–687. doi:10.1093/emboj/17.3.677
van Roermund, C. W. T., Hettema, E. H., van den Berg, M., Tabak, H. F., and Wanders, R. J. A. (1999). Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 18, 5843–5852. doi:10.1093/emboj/18.21.5843
Vasilev, J., Mix, A. K., Heimerl, T., Maier, U. G., and Moog, D. (2022). Inferred subcellular localization of peroxisomal matrix proteins of Guillardia theta suggests an important role of peroxisomes in cryptophytes. Front. Plant Sci. 13, 889662. doi:10.3389/fpls.2022.889662
Visser, W. F., Van Roermund, C. W. T., Ijlst, L., Waterham, H. R., and Wanders, R. J. A. (2007). Metabolite transport across the peroxisomal membrane. Biochem. J. 401, 365–375. doi:10.1042/BJ20061352
Walter, T., and Erdmann, R. (2019). Current advances in protein import into peroxisomes. Protein J. 38, 351–362. doi:10.1007/s10930-019-09835-6
Walton, P. A., Brees, C., Lismont, C., Apanasets, O., and Fransen, M. (2017). The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim. Biophys. Acta. Mol. Cell Res. 1864, 1833–1843. doi:10.1016/j.bbamcr.2017.07.013
Walton, P. A., Hill, P. E., and Subramani, S. (1995). Import of stably folded proteins into peroxisomes. Mol. Biol. Cell 6, 675–683. doi:10.1091/mbc.6.6.675
Wanders, R. J. A. (2014). Metabolic functions of peroxisomes in health and disease. Biochimie 98, 36–44. doi:10.1016/j.biochi.2013.08.022
Wanders, R. J. A., and Waterham, H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332. doi:10.1146/annurev.biochem.74.082803.133329
Wanders, R. J. A., Waterham, H. R., and Ferdinandusse, S. (2016). Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front. Cell Dev. Biol. 3, 83. doi:10.3389/fcell.2015.00083
Wang, S., Idrissi, F.-Z., Hermansson, M., Grippa, A., Ejsing, C. S., and Carvalho, P. (2018). Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat. Commun. 9, 2939. doi:10.1038/s41467-018-05278-2
Weir, N. R., Kamber, R. A., Martenson, J. S., and Denic, V. (2017). The AAA protein Msp1 mediates clearance of excess tail-anchored proteins from the peroxisomal membrane. Elife 6, e28507. doi:10.7554/eLife.28507
Wilfling, F., Thiam, A. R., Olarte, M.-J., Wang, J., Beck, R., Gould, T. J., et al. (2014). Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3, e01607. doi:10.7554/eLife.01607
Williams, C., Bener Aksam, E., Gunkel, K., Veenhuis, M., and van der Klei, I. J. (2012). The relevance of the non-canonical PTS1 of peroxisomal catalase. Biochim. Biophys. Acta 1823, 1133–1141. doi:10.1016/j.bbamcr.2012.04.006
Wright, Z. J., and Bartel, B. (2020). Peroxisomes form intralumenal vesicles with roles in fatty acid catabolism and protein compartmentalization in Arabidopsis. Nat. Commun. 11, 6221. doi:10.1038/s41467-020-20099-y
Wu, F., de Boer, R., Krikken, A. M., Akşit, A., Bordin, N., Devos, D. P., et al. (2020). Pex24 and Pex32 are required to tether peroxisomes to the ER for organelle biogenesis, positioning and segregation in yeast. J. Cell Sci. 133, jcs246983. doi:10.1242/jcs.246983
Yagita, Y., Hiromasa, T., and Fujiki, Y. (2013). Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200, 651–666. doi:10.1083/jcb.201211077
Yamamoto, Y., and Sakisaka, T. (2018). The peroxisome biogenesis factors posttranslationally target reticulon homology domain-containing proteins to the endoplasmic reticulum membrane. Sci. Rep. 8, 2322. doi:10.1038/s41598-018-20797-0
Yamashita, K., Tamura, S., Honsho, M., Yada, H., Yagita, Y., Kosako, H., et al. (2020). Mitotic phosphorylation of Pex14p regulates peroxisomal import machinery. J. Cell Biol. 219, e202001003. doi:10.1083/jcb.202001003
Yang, J., Pieuchot, L., and Jedd, G. (2018). Artificial import substrates reveal an omnivorous peroxisomal importomer. Traffic 19, 786–797. doi:10.1111/tra.12607
Yifrach, E., Chuartzman, S. G., Dahan, N., Maskit, S., Zada, L., Weill, U., et al. (2016). Characterization of proteome dynamics during growth in oleate reveals a new peroxisome-targeting receptor. J. Cell Sci. 129, 4067–4075. doi:10.1242/jcs.195255
Yifrach, E., Holbrook-Smith, D., Bürgi, J., Othman, A., Eisenstein, M., Van Roermund, C. W. T., et al. (2021). Systematic multi-level analysis of an organelle proteome reveals new peroxisomal functions. bioRxiv. bioarchives preprints.
Yifrach, E., Rudowitz, M., Cruz-Zaragoza, L. D., Tirosh, A., Gazi, Z., Peleg, Y., et al. (2022). Determining the targeting specificity of the selective peroxisomal targeting factor Pex9. bioRxiv. bioarchives preprints.
Yofe, I., Soliman, K., Chuartzman, S. G., Morgan, B., Weill, U., Yifrach, E., et al. (2017). Pex35 is a regulator of peroxisome abundance. J. Cell Sci. 130 (4), 791–804. doi:10.1242/jcs.187914
Yogev, O., Naamati, A., and Pines, O. (2011). Fumarase: A paradigm of dual targeting and dual localized functions. FEBS J. 278, 4230–4242. doi:10.1111/j.1742-4658.2011.08359.x
Yogev, O., Singer, E., Shaulian, E., Goldberg, M., Fox, T. D., Pines, O., et al. (2010). Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8, e1000328. doi:10.1371/journal.pbio.1000328
Yuan, W., Akşit, A., de Boer, R., Krikken, A. M., and van der Klei, I. J. (2022). Yeast Vps13 is crucial for peroxisome expansion in cells with reduced peroxisome-ER contact sites. Front. Cell Dev. Biol. 10, 842285. doi:10.3389/fcell.2022.842285
Zabezhinsky, D., Slobodin, B., Rapaport, D., and Gerst, J. E. (2016). An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep. 15, 540–549. doi:10.1016/j.celrep.2016.03.053
Zekert, N., Veith, D., and Fischer, R. (2010). Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot. Cell 9, 795–805. doi:10.1128/EC.00058-10
Zhang, J., Tripathi, D. N., Jing, J., Alexander, A., Kim, J., Powell, R. T., et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17, 1259–1269. doi:10.1038/ncb3230
Zhang, M., Li, Q., Liu, T., Liu, L., Shen, D., Zhu, Y., et al. (2015). Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. doi:10.1104/pp.114.252437
Zheng, J., Chen, X., Liu, Q., Zhong, G., and Zhuang, M. (2021). Ubiquitin ligase MARCH5 localizes to peroxisomes to regulate pexophagy. J. Cell Biol. 221, e202103156. doi:10.1083/jcb.202103156
Zimmermann, R., Lang, S., Lerner, M., Förster, F., Nguyen, D., Helms, V., et al. (2021). Quantitative proteomics and differential protein abundance analysis after the depletion of PEX3 from human cells identifies additional aspects of protein targeting to the ER. Int. J. Mol. Sci. 22, 13028. doi:10.3390/ijms222313028
Keywords: dual targeting, Endoplasmic reticulum, peroxisome, targeting signal, Mitochondria
Citation: Bittner E, Stehlik T and Freitag J (2022) Sharing the wealth: The versatility of proteins targeted to peroxisomes and other organelles. Front. Cell Dev. Biol. 10:934331. doi: 10.3389/fcell.2022.934331
Received: 02 May 2022; Accepted: 27 July 2022;
Published: 26 September 2022.
Edited by:
Michael Schrader, University of Exeter, United KingdomReviewed by:
María C. Romero-Puertas, Experimental Station of Zaidín (CSIC), SpainMarkus Kunze, Medical University of Vienna, Austria
Nicole Linka, Heinrich Heine University of Düsseldorf, Germany
Copyright © 2022 Bittner, Stehlik and Freitag. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Freitag, am9oYW5uZXNmcmVpdGFnQGdteC5uZXQ=
 Elena Bittner
Elena Bittner Thorsten Stehlik
Thorsten Stehlik Johannes Freitag
Johannes Freitag