- 1Environmental and Life Sciences Graduate Program, Trent University, Peterborough, ON, Canada
- 2Department of Biology, Trent University, Peterborough, ON, Canada
MFSD8 is a transmembrane protein that has been reported to transport chloride ions across the lysosomal membrane. Mutations in MFSD8 are associated with a subtype of Batten disease called CLN7 disease. Batten disease encompasses a family of 13 inherited neurodegenerative lysosomal storage diseases collectively referred to as the neuronal ceroid lipofuscinoses (NCLs). Previous work identified an ortholog of human MFSD8 in the social amoeba D. discoideum (gene: mfsd8, protein: Mfsd8), reported its localization to endocytic compartments, and demonstrated its involvement in protein secretion. In this study, we further characterized the effects of mfsd8 loss during D. discoideum growth and early stages of multicellular development. During growth, mfsd8− cells displayed increased rates of proliferation, pinocytosis, and expansion on bacterial lawns. Loss of mfsd8 also increased cell size, inhibited cytokinesis, affected the intracellular and extracellular levels of the quorum-sensing protein autocrine proliferation repressor A, and altered lysosomal enzyme activity. During the early stages of development, loss of mfsd8 delayed aggregation, which we determined was at least partly due to impaired cell-substrate adhesion, defects in protein secretion, and alterations in lysosomal enzyme activity. Overall, these results show that Mfsd8 plays an important role in modulating a variety of processes during the growth and early development of D. discoideum.
Introduction
Batten disease, clinically known as the neuronal ceroid lipofuscinoses (NCLs), is a family of neurodegenerative disorders that affect people of all ages and ethnicities (Mole and Cotman, 2015). Clinical symptoms of the disease include vision loss leading to blindness, seizures, deterioration in motor and cognitive function, and premature death (Schulz et al., 2013). There are 13 different subtypes of NCL, each one resulting from a mutation in a distinct ceroid lipofuscinosis neuronal (CLN) gene (CLN1-8, CLN10-14) (Mole and Cotman, 2015). CLN genes encode soluble lysosomal proteins (PPT1/CLN1, TPP1/CLN2, CLN5, CTSD/CLN10 and CTSF/CLN13), lysosomal membrane proteins (CLN3, MFSD8/CLN7, and ATP13A2/CLN12), membrane proteins localizing to the endoplasmic reticulum (CLN6 and CLN8), cytoplasmic proteins (GRN/CLN11 and KCTD7/CLN14), and a protein that localizes to synaptic vesicles (DNAJC5/CLN4) (Cárcel-Trullols et al., 2015). While these proteins have a wide range of localizations and functions, it has been suggested that they function in shared or convergent cellular pathways (Persaud-Sawin et al., 2007; Huber, 2020).
Mutations in major facilitator superfamily domain-containing 8 (MFSD8) cause a late-infantile form of NCL called CLN7 disease (Aiello et al., 2009). In mammalian cells, MFSD8 is proteolytically cleaved and has been reported to transport chloride ions across the lysosomal membrane (Siintola et al., 2007; Sharifi et al., 2010; Steenhuis et al., 2010; Steenhuis et al., 2012; Wang et al., 2021). However, how MFSD8 influences cellular processes related to NCL pathology is not fully understood. Previous work using Mfsd8-deficient mouse embryonic fibroblasts (MEFs) showed that loss of Mfsd8 alters the amounts of several soluble lysosomal proteins (Danyukova et al., 2018). In addition, loss of Mfsd8 in mice affects autophagy, neuronal cell survival, and the size and trafficking of lysosomes (Brandenstein et al., 2016; von Kleist et al., 2019; Wang et al., 2021). Further work in human MFSD8 knockout HEK293T cells revealed the role of MFSD8 in regulating lysosomal chloride conductance, luminal calcium content, lysosomal membrane potential, and lysosomal pH (Wang et al., 2021).
The social amoeba D. discoideum is a eukaryotic microbe that is used as a biomedical model organism for studying a variety of human diseases, including the NCLs (Huber, 2021; Kirolos et al., 2021; Mathavarajah et al., 2021; Pain et al., 2021). During the growth phase of the life cycle, haploid amoebae feed on nutrients and divide by mitosis (Mathavarajah et al., 2017). Removal or depletion of the food source triggers a 24-h multicellular developmental program that begins with the chemotactic aggregation of cells into multicellular mounds and ends with the formation of fruiting bodies that are composed of terminally differentiated spores held atop slender stalks of terminally differentiated stalk cells (Mathavarajah et al., 2017). As a result, D. discoideum can be used to study a variety of fundamental cellular and developmental processes. The D. discoideum genome encodes homologs of 11 of the 13 CLN proteins (Huber, 2016). Recent work on the D. discoideum homologs of human tripeptidyl peptidase 1 (TPP1)/CLN2 (Tpp1), CLN3 (Cln3), and CLN5 (Cln5) has provided valuable new insight into the localizations and functions of these proteins in human cells (Huber and Mathavarajah, 2019; McLaren et al., 2019; Smith et al., 2019; Huber, 2020; Huber et al., 2020a; Huber, 2021; McLaren et al., 2021). The D. discoideum homolog of human MFSD8 (gene: mfsd8, protein: Mfsd8) is expressed throughout the life cycle and a proteomic analysis revealed that Mfsd8 is present in the macropinocytic pathway (Rot et al., 2009; Journet et al., 2012). Recent work in D. discoideum showed that Mfsd8 localizes to endocytic compartments, including acidic intracellular vesicles and late endosomes, influences the secretion of two other CLN protein homologs, Cln5 and cathepsin D (CtsD), and interacts with a diversity of proteins during growth and the early stages of multicellular development (Huber et al., 2020b).
In this study, we further characterized the function of Mfsd8 in D. discoideum by assessing the effects of mfsd8 loss on growth and the early stages of multicellular development. Results presented here support a function for Mfsd8 in cell proliferation, pinocytosis, cytokinesis, protein secretion, lysosomal enzyme activity, aggregation, and cell-substrate adhesion. We then integrated these findings into an emerging model summarizing the known roles of Mfsd8 in D. discoideum.
Materials and Methods
Cell Lines, Antibodies, and Chemicals
Cell lines were grown and maintained on SM/2 agar with Klebsiella aerogenes at 21°C (Fey et al., 2007). Cells were also grown axenically in HL5 medium at 21°C and 150 rpm. For all experiments, cells were harvested in the mid-log phase of growth (1–5 × 106 cells/ml). Cultures were supplemented with 100 μg/ml ampicillin and 300 μg/ml streptomycin sulfate to prevent bacterial growth. AX4, hereafter referred to as WT, was the parental cell line for mfsd8−, which was purchased from the Genome Wide Dictyostelium Insertion (GWDI) bank via the Dicty Stock Center (https://remi-seq.org) (Fey et al., 2019; Gruenheit et al., 2021). Blasticidin S hydrochloride (10 μg/ml) was used to select mfsd8− cells. HL5 and low-fluorescence HL5 were purchased from Formedium (Hunstanton, Norfolk, United Kingdom). 2-N-morpholinoethanesulfonic acid (MES) was purchased from Fisher Scientific Company (Ottawa, ON, Canada). Rabbit polyclonal antibodies against autocrine proliferation repressor (AprA) and countin (CtnA) were provided as gifts by Dr. Richard Gomer (Brock and Gomer, 1999; Brock and Gomer, 2005). Rabbit polyclonal antibody against calcium dependent cell adhesion protein (CadA) was generated and validated in a previous study (McLaren et al., 2021). Mouse monoclonal anti-α-actinin (47-18-9) and mouse monoclonal anti-discoidin (DscA) (80-52-13) were purchased from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA, United States) (Stadler et al., 1984). Mouse monoclonal anti-β-actin was purchased from Santa Cruz Biotechnology Incorporated (Dallas, TX, United States). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from New England Biolabs (Whitby, ON, Canada). p-Nitrophenyl-β-D-glucopyranoside (487507) (substrate for β-glucosidase), p-Nitrophenyl-a-D-glucopyranoside (487506) (substrate for α-glucosidase), 4-Nitrophenyl-α-D-mannopyranoside (N2127) (substrate for α-mannosidase), o-Nitrophenyl-β-D-galactopyranoside (48712-M) (substrate for β-galactosidase), 4-Nitrophenyl-α-D-galactopyranoside (N0877) (substrate for α-galactosidase), 4-Nitrophenyl N-acetyl-β-D-glucosaminide (N9376) (substrate for N-acetylglucosaminidase, NAG), Ala-Ala-Phe-7-amido-4-methylcoumarin (A3401) (substrate for TPP1), Fluorogenic Cathepsin B Substrate III (219392) (substrate for cathepsin B, CTSB), and β-glucosidase (49290) were purchased from Sigma Aldrich Canada (Oakville, ON, Canada). 4-Methylumbelliferyl 6-thio-Palmitate-β-D-Glucopyranoside (19524) (substrate for palmitoyl-protein thioesterase 1, PPT1) and Fluorogenic Cathepsin F Substrate (80350-BP) (substrate for cathepsin F, CTSF) were purchased from Cedarlane Laboratories (Burlington, ON, Canada). The Cathepsin D Activity Assay Kit (10013-596) was purchased from VWR International (Mississauga, ON, Canada).
Cell Proliferation, Pinocytosis, Cell Size, and Cytokinesis Assays
To assess cell proliferation, cells in the mid-log phase of growth were washed thrice with fresh HL5. Cells were then diluted to 1–2 × 105 cells/ml in HL5 and incubated at 21°C and 150 rpm. Cell concentrations were measured every 24 h over a 96-h growth period using a hemocytometer. The pinocytosis assay was conducted using a method described elsewhere (Rivero and Maniak, 2006; Huber et al., 2014). Briefly, cells in the mid-log phase of growth were placed in 5 ml of HL5 at a density of 5 × 106 cells/ml. 100 μl of a 20 mg/ml fluorescein isothiocyanate (FITC)-dextran (70,000 Mr) stock solution was added to the 5 ml suspension, which was then incubated for 120 min at 21°C and 150 rpm. 500 μl were collected every 15 min, washed twice with ice-cold Sorenson’s buffer (2 mM Na2HPO4, 14.6 mM KH2PO4, pH 6.0) and lysed with 1 ml of buffer containing 50 mM Na2HPO4 (pH 9.3) and 0.2% Triton-X. Lysates were added to separate wells of black bottom 96-well plates and fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (460/40 nm for excitation, 528/20 nm for emission) (BioTek Instruments Incorporated, Winooski, VT, United States). To measure cell area, cells in the mid-log phase of growth were deposited onto a hemocytometer. Cells were imaged with a Nikon Ts2R-FL inverted microscope equipped with a Nikon Digital Sight Qi2 monochrome camera (Nikon Canada Incorporated Instruments Division, Mississauga, ON, Canada). The area of each individual cell was quantified using Fiji/ImageJ (Schindelin et al., 2012). To assess cytokinesis, cells (5 × 105 total) in the mid-log phase of growth were collected and deposited onto coverslips placed inside separate wells of a 12-well dish. Coverslips were then submerged in low-fluorescence HL5 and incubated overnight at 21°C. The following day, cells were fixed in −80°C methanol for 45 min and mounted onto slides with Prolong Gold Anti-Fade Reagent containing DAPI (Fisher Scientific Company, Ottawa, ON, Canada). Cells were then imaged using a Nikon Ts2R-FL inverted microscope equipped with a Nikon Digital Sight Qi2 monochrome camera. For each independent experiment, the number of nuclei within each cell (at least 100) was scored and expressed as a percentage of the total number of cells analyzed.
Plaque Expansion on Bacterial Lawns
Three full inoculation loops of K. aerogenes were collected and resuspended in KK2 buffer (0.7 g/L K2HPO4 and 2.2 g/L KH2PO4, pH 6.5). 25 μl of the suspension were then deposited onto SM/2 agar and incubated at 21°C. Two days later, D. discoideum cells in the mid-log phase of growth were harvested, washed thrice with KK2 buffer, and resuspended in KK2 buffer to obtain a final concentration of 0.4 × 106 cells/ml. Cells (1 × 102 total) were deposited onto the center of the K. aerogenes lawns. Plaques were captured at the indicated time points using a Leica EZ4W stereomicroscope equipped with an internal 5MP CMOS camera (Leica Microsystems Incorporated, Concord, ON, Canada). Plaque diameters were quantified using Fiji/ImageJ.
Autocrine Proliferation Repressor Protein Levels During Growth
Cells from the proliferation assay described above were harvested after 24, 48, and 72 h of growth (Huber et al., 2014). Cells were lysed with buffer containing 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.5% NP40, and a protease inhibitor tablet (PIA32965) (Fisher Scientific Company, Ottawa, ON, Canada). Samples of conditioned media (CM) were collected and standardized based on cell number (1 × 106 total). Whole cell (WC) lysates and equal volumes of CM were separated by SDS-PAGE and analyzed by western blotting. The following primary and secondary antibodies were used: anti-AprA (1∶1000), anti-β-actin (1∶1000), and HRP-conjugated secondary antibodies (1:2000). Protein bands were imaged using the ChemiDoc Imaging System (Bio-Rad Laboratories Limited, Mississauga, ON, Canada) and quantified using Fiji/ImageJ.
Aggregation Assay
Aggregation was examined using a method described previously with minor modifications (Huber, 2017). Briefly, cells (6 × 106 total) harvested from the mid-log phase of growth were deposited into separate wells of a 6-well dish. Cells were allowed to adhere to the surface of the dish for 1 h after which time they were washed two times with KK2 buffer, and then starved in 1 ml of KK2 buffer. Cells were imaged at the indicated times with a Nikon Ts2R-FL inverted microscope equipped with a Nikon 10 Digital Sight Qi2 monochrome camera. A conditioned buffer (CB) swap experiment was conducted to test the effect of proteins secreted by WT cells on the aggregation of mfsd8− cells (Huber et al., 2017). Briefly, the CB from WT cells starved for 2 h in 1.5 ml of KK2 buffer was collected and spun down to remove any cells present in the buffer. mfsd8− cells were submerged in either 1 ml of KK2 buffer (control) or CB collected from starving WT cells. Cells were imaged at the indicated times with a Nikon Ts2R-FL inverted microscope equipped with a Nikon 10 Digital Sight Qi2 monochrome camera. Aggregation was also examined on 0.5% agar/KK2 (Huber and Mathavarajah, 2018). Briefly, cells in the mid-log phase of growth were harvested from HL5, washed two times with KK2 buffer, and plated (1.5 × 108 cells/ml) in 0.5 μl volumes on 0.5% agar/KK2. Cell spots were imaged at 0 and 5 h using a Nikon Ts2R-FL inverted microscope equipped with a Nikon Digital Sight Qi2 monochrome camera. Images were viewed using NIS Elements Basic Research and analyzed using Fiji/ImageJ. The area after 5 h was expressed as a percentage of the area at 0 h to provide a measure of the amount of aggregation.
Protein Secretion During Aggregation
Cells (8 × 106 total) in the mid-log phase of growth were deposited into 60 mm × 15 mm Petri dishes and allowed to adhere for 1 h, after which time the HL5 was removed. Adherent cells were washed two times with KK2 buffer and then submerged in 4 ml of KK2 buffer for 4 h, after which time the CB was collected and concentrated using an Amicon Ultra-4 centrifugal filter unit (UFC801024) (Fisher Scientific Company, Ottawa, ON, Canada) according to the manufacturer’s instructions. Adherent cells were lysed with NP40 lysis buffer (recipe noted above). SDS-PAGE and western blotting were then used to determine the effect of mfsd8 loss on the intracellular and extracellular amounts of CtnA, CadA, and DscA. The following primary and secondary antibodies were used: anti-CtnA (1∶1000), anti-CadA (1∶1000), anti-DscA (1:1000), anti-β-actin (1∶1000), anti-α-actinin (1∶1000), and HRP-conjugated secondary antibodies (1:2000). Protein bands were imaged using the ChemiDoc Imaging System (Bio-Rad Laboratories Limited, Mississauga, ON, Canada) and quantified using Fiji/ImageJ.
Cell-Substrate Adhesion Assay
Cell-substrate adhesion was assessed using a previously described method with minor modifications (Huber et al., 2017; Huber and Mathavarajah, 2018). Briefly, cells (6 × 106 total) were deposited into separate wells of a 6-well dish and allowed to adhere for 1 h after which time they were washed twice with KK2 buffer and starved in 1.5 ml of KK2 buffer for 4 h. After 4 h, cells were shaken at 150 rpm for 30 min. Samples of CB were collected and cells in CB were counted using a hemocytometer to determine cell dissociation. Cells remaining on the dish were also lysed with NP40 lysis buffer (recipe noted above) and protein concentrations of the lysates were quantified using the Qubit Protein Assay Kit (Q33211) and a Qubit 2.0 Fluorometer (Fisher Scientific Company, Ottawa, ON, Canada).
Enzyme Activity Assays
To assess various enzyme activities, cells were grown in HL5 overnight to confluency in 60 mm × 15 mm Petri dishes (8 × 106 total). Growth-phase cells and cells starved for 4 h in KK2 buffer were then lysed with buffer containing 50 mM MES pH 6.53 and 0.1% NP40, unless stated otherwise (Phillips and Gomer, 2015). Protein concentrations of lysates were quantified using the Qubit Protein Assay Kit and a Qubit 2.0 Fluorometer, and 150 μg of protein was used for the enzyme assays described below. For all enzyme assays, the absorbance and fluorescence values for the experimental samples were corrected using a lysis buffer control.
Lysosomal hydrolases: To assay α-galactosidase activity, lysates were added to 18 μl of 2 mM of 4-Nitrophenyl-α-D-galactopyranoside in sodium citrate/phosphate buffer (pH 4.5) (Kilpatrick and Stirling, 1976). Samples were then incubated for 45 min at 37°C and quenched with equal volume of 1 M sodium glycinate buffer (pH 10.4). The reactions were then deposited into 96-well black clear bottom plates and the absorbance (405 nm) was measured using a BioTek Synergy HTX plate reader. To assay β-galactosidase activity, lysates were added to 36 μl of 25 mM of o-Nitrophenyl-β-D-galactopyranoside in 100 mM citrate buffer (pH 4.0) (Maruhn, 1976). The reaction mixtures were incubated for 45 min at 37°C, after which time an equal volume of 2-amino-2-methyl-1-propnanol/HCl solution was added. The reactions were then deposited into 96-well black clear bottom plates and the absorbance (405 nm) was measured using a BioTek Synergy HTX plate reader. To assay α-glucosidase activity, WC lysates were incubated in 2 mM p-Nitrophenyl-a-D-glucopyranoside within 0.1 M sodium succinate (pH 6.0) in a total volume of 150 μl (Wimmer et al., 1997). The reaction solution was incubated for 1 h at 65°C followed by the addition of 300 μl of 1 M sodium carbonate to quench the reaction. The quenched reactions were then deposited into 96-well black clear bottom plates and the absorbance (395 nm) was measured using a BioTek Synergy HTX plate reader. To assay β-glucosidase activity, lysates were added to 18 μl of 10 mM of p-Nitrophenyl-β-D-glucopyranoside in 50 mM acetate buffer (pH 5.0) (Coston and Loomis, 1969). The reactions were incubated at 35°C for 45 min. The reactions were then quenched with an equal volume of 1 M sodium carbonate and deposited into 96-well black clear bottom plates and the absorbance (405 nm) was measured using a BioTek Synergy HTX plate reader. To assay α-mannosidase activity, lysates were added to 18 μl of 5 mM of 4-Nitrophenyl-α-D-mannopyranoside in 5 mM acetate buffer (pH 5.0) (Loomis, 1970). Samples were incubated for 45 min at 35°C after which time an equal volume of 1 M Na2CO3 was added to quench the reactions. The quenched reactions were then deposited into 96-well black clear bottom plates and the absorbance (405 nm) was measured using a BioTek Synergy HTX plate reader. To assay NAG activity, WC lysates were added to 75 µl of 7 mM of 4-Nitrophenyl N-acetyl-β-D-glucosaminide in 100 mM acetate buffer (pH 5.0). Samples were then incubated for 5 min at 35°C, after which time an equal volume of 1 M Na2CO3 was added to quench the reactions (Loomis, 1969; Huber and Mathavarajah, 2018). The quenched reactions were then deposited into 96-well black clear bottom plates and the absorbance (405 nm) was measured using a BioTek Synergy HTX plate reader.
Ppt1 and Tpp1: To assay Ppt1 activity, lysates were added to 5 µl of 9 mM of 4-Methylumbelliferyl 6-thio-Palmitate-β-D-Glucopyranoside dissolved in McIlvaine phosphate/citric-acid buffer (pH 4) supplemented with 15 mM dithiothreitol and 0.375% Triton X-100. The reactions were incubated at 37°C for 1 h after which time they were boiled for 3 min at 95°C (van Diggelen et al., 1999; Brand et al., 2018). Once the reactions were cooled, 2.75 µl of 2.5 M NaOH and 4 µl of 0.025 U/μl β-glucosidase were added to the reactions and incubated for another hour at 37°C. The reaction was quenched by adding 0.5 M sodium carbonate-bicarbonate buffer containing 0.025% Triton X-100 (pH 10.7). The quenched reactions were then deposited into 96-well opaque black bottom plates and the fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (360/40 nm for excitation, 460/40 nm for emission). To assay Tpp1 activity, cells were lysed with buffer containing 50 mM sodium phosphate (pH 6.5) and 0.5% NP40, and then added to 80 µl of 200 μM Ala-Ala-Phe-7-amido-4-methylcoumarin dissolved in reaction buffer (150 mM NaCl, 100 mM sodium acetate, pH 4.5, 0.1% Triton X-100) (Stumpf et al., 2017). Reactions were incubated in the dark at 37°C for 1 h and then quenched by adding stop solution (150 mM NaCl, 100 mM sodium acetate, pH 4.3). The quenched reactions were then deposited into 96-well opaque black bottom plates and the fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (360/40 nm for excitation, 460/40 nm for emission).
Cathepsins: To assay CtsB activity, methods were adapted with minor revisions (Barrett, 1980). Briefly, WC lysates were incubated in reaction buffer (352 mM KH2PO4, 48 mM Na2HPO4, 4 mM EDTA, pH 6.0) with 8 mM cysteine in a total volume of 150 μl and incubated at 40°C for 5 min. A final concentration of 5 μM of substrate was added to the reaction solution, which was then incubated at 40°C for 30 min. The reaction solutions were quenched with 200 μl of 100 mM sodium chloroacetate (30 mM NaC2H3O2, 70 mM HC2H3O2, pH 4.3). The quenched reactions were then deposited into 96-well opaque black bottom plates and the fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (360/40 nm for excitation, 460/40 nm for emission). CtsD activity was measured following the manufacturer’s instructions specified in the Cathepsin D Activity Assay Kit. Briefly, cells were lysed with 100 µl of chilled cell lysis buffer (provided in kit) and incubated on ice for 10 min. The solution was then centrifuged for 5 min. Following centrifugation, 27.5 µg of clear cell lysate was added to 100 µl of reaction buffer (provided in kit) mixed with 2 µl of substrate. Reactions were incubated at 37°C for 1 h and 30 min. The reactions were then deposited into 96-well opaque black bottom plates and the fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (360/40 nm for excitation, 460/40 nm for emission). To assay CtsF activity, WC lysates were added to 0.5 µl of 25% HCl and 1 µl of 250 μg/ml pepsin. 50 µl reactions were incubated for 1 h at 37°C after which time 0.24 µl of 0.5 mM Fluorogenic Cathepsin F Substrate followed by 5.76 µl dimethyl sulfoxide and 70 µl of 0.1 M sodium phosphate buffer containing 1 mM EDTA and 0.1% (v/v) PEG 6000 (pH 6.5) were added to the reaction mixture (total volume of 120 µl). Reactions were incubated at 27°C for 1 h (Fonovic et al., 2004). The reactions were then deposited into 96-well opaque black bottom plates and the fluorescence was measured using a BioTek Synergy HTX plate reader and the following filters (360/40 nm for excitation, 460/40 nm for emission).
Statistical Analyses
Numeric data are reported as the means ± SEM. Statistical analyses for all experiments were performed using GraphPad Prism 8 (GraphPad Software Incorporated, La Jolla, CA, United States). A one-sample t-test was used for evaluating data where the raw data between biological replicates was inherently variable (e.g., quantifying pixel intensity from a western blot). A p-value < 0.05 was considered significant for all analyses and n represents the number of independent experiments that were performed. Details on the specific statistical analyses performed are found in the figure captions.
Results
Expression Profile of mfsd8 During the D. discoideum Life Cycle
A previous study in D. discoideum showed that Mfsd8 localizes to endocytic compartments, linked the function of Mfsd8 to protein secretion, and revealed the Mfsd8 interactome during growth and starvation (Huber et al., 2020b). To gain further insight into the function of Mfsd8 in D. discoideum, here, we performed an in-depth characterization of an mfsd8− cell line. The expression of mfsd8 increases during the early stages of development, which involves the chemotactic aggregation of cells into multicellular mounds (Rot et al., 2009; Mathavarajah et al., 2017). Following mound formation, mfsd8 expression decreases dramatically reaching its lowest level just prior to terminal differentiation of pre-spore and pre-stalk cells. Expression then rises slightly during fruiting body formation. This expression profile suggested to us that Mfsd8 primarily functions during growth and the early stages of development. As a result, we focused our analysis on the effects of mfsd8-deficiency on processes that occur during these stages of the life cycle.
mfsd8− Cells Display Increased Proliferation and Accumulation of Fluorescein Isothiocyanate-Dextran During Growth
When cultured in liquid growth medium, mfsd8− cells proliferated at a significantly increased rate compared to WT cells (Figure 1A). Since D. discoideum cells ingest extracellular liquid nutrients through macropinocytosis, we assessed whether the rate of pinocytosis was affected by mfsd8-deficiency (Hacker et al., 1997). Cells were incubated in liquid growth medium containing FITC-dextran for a 120-min period and the amount of intracellular fluorescence was measured every 15 min. Like proliferation, the rate of FITC-dextran accumulation was significantly increased in mfsd8− cells relative to WT cells (Figure 1B).
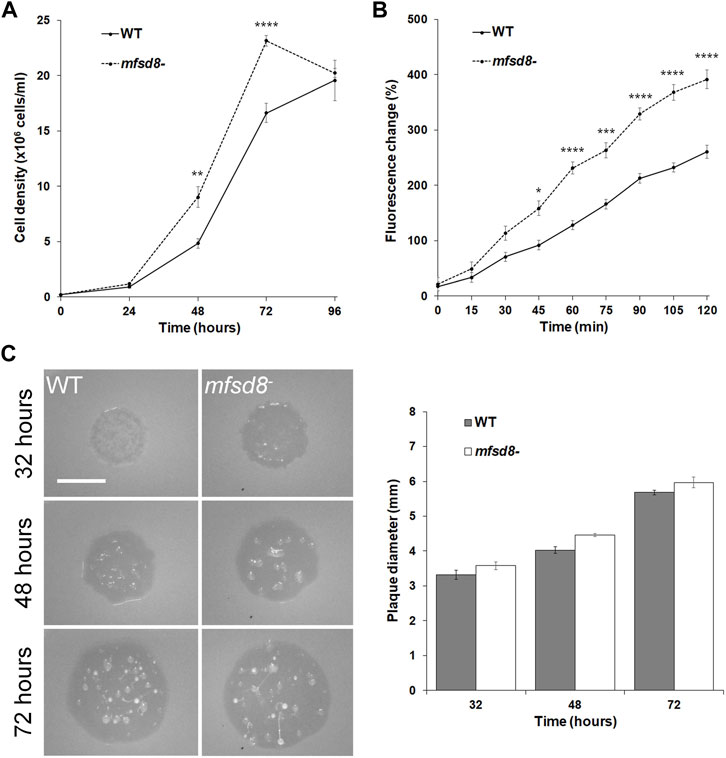
FIGURE 1. Effect of mfsd8-deficiency on cell proliferation, pinocytosis, and plaque expansion. (A) Effect of mfsd8-deficiency on cell proliferation. WT and mfsd8− cells were grown axenically in HL5 medium. Cell densities were measured every 24 h over a 96-h period. Data presented as mean concentration (×106 cells/ml) ± SEM (n = 5). Statistical significance was assessed using two-way ANOVA followed by Bonferroni post-hoc analysis. **p < 0.05 and ****p < 0.0001 vs. WT at the indicated time points. (B) Effect of mfsd8-deficiency on pinocytosis. WT and mfsd8− cells were incubated in HL5 containing FITC-dextran for 120 min. Cells were collected, washed, and lysed every 15 min. The fluorescence of the lysates was then measured using a plate reader. The data were corrected for background signals and expressed as the mean fluorescence change (%) relative to the 0-min time point. Data presented as mean fluorescence change (%) ± SEM (n = 3). Statistical significance was assessed using two-way ANOVA followed by Bonferroni post-hoc analysis. *p < 0.05, ***p < 0.001, and ****p < 0.0001 vs. WT at the indicated time points. (C) Effect of mfsd8-deficiency on plaque expansion on bacterial lawns. WT and mfsd8− cells grown axenically in HL5 were harvested, washed with KK2 buffer, and deposited onto lawns of K. aerogenes. Plaques were imaged at the indicated time points and their diameters were quantified using Fiji/ImageJ. Scale bar = 20 μm. Data presented as mean plaque diameter (mm) ± SEM (n = 4).
Since loss of mfsd8 impacted cell proliferation and the accumulation of FITC-dextran in liquid growth medium, we tested whether mfsd8− cells would display the same phenotype when grown on bacteria lawns. Cell lines were plated on lawns of K. aerogenes and plaque formation was monitored. In general, we observed that mfsd8− cells formed plaques earlier than WT cells and the plaques were larger than WT plaques at all time points examined (Figure 1C). Of note, 32 h after depositing cells onto lawns, mfsd8− plaques were translucent, while WT plaques were semi-translucent, indicating that mfsd8− cells cleared bacterial lawns earlier than WT cells. When feeding on bacterial lawns, D. discoideum amoebae chemotactically respond to folic acid that is secreted by bacteria (Pan et al., 1972). However, we observed no significant effect of mfsd8-deficiency on folic acid-mediated chemotaxis (Supplementary Figure S1). Together, these results suggest that Mfsd8 regulates cell proliferation, pinocytosis, and growth on bacterial lawns in D. discoideum.
Loss of mfsd8 Increases Cell Size and Inhibits Cytokinesis During Growth
When examining the proliferation of mfsd8− cells, we observed that mfsd8− cells appeared larger than WT cells. Indeed, when we quantified the area of cells cultured in liquid growth medium, we found that mfsd8− cultures contained a significantly higher proportion of cells > 200 μm2 compared to WT cultures, and a correlated lower proportion of cells 100–200 μm2 (Figure 2A).
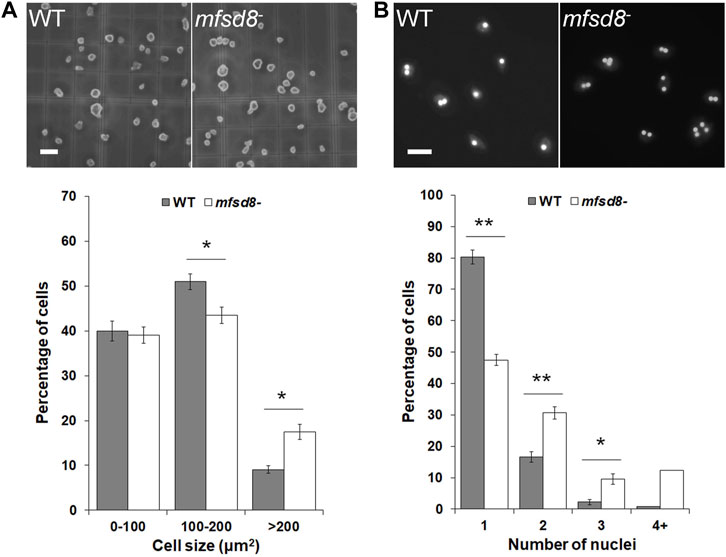
FIGURE 2. Effect of mfsd8-deficiency on cell size and cytokinesis. (A) Effect of mfsd8-deficiency on cell size. WT and mfsd8− cells grown axenically in HL5 were deposited onto a hemocytometer and imaged. Scale bar = 20 μm. The area of each individual cell was quantified using Fiji/ImageJ. Data was placed into different cell area bins and expressed as a mean percentage of total cells analyzed ± SEM (n = 5). Statistical significance was assessed using two-way ANOVA followed by Bonferroni post-hoc analysis. *p < 0.05 vs. WT. (B) Effect of mfsd8-deficiency on cytokinesis. WT and mfsd8− cells grown axenically in HL5 were collected and deposited onto coverslips placed inside separate wells of a 12-well dish. Cells on coverslips were then submerged in low-fluorescence HL5 overnight at 21°C. The following day, cells were fixed in −80°C methanol mounted onto slides with Prolong Gold Anti-Fade Reagent containing DAPI. Scale bar = 20 μm. The number of nuclei within each cell was scored. For each experiment, 10 random images, each containing at least 10 cells, were obtained from each coverslip. Data presented as mean percentage of total cells analyzed ±SEM (n = 7). Statistical significance was assessed using two-way ANOVA followed by Bonferroni post-hoc analysis. *p < 0.05 and **p < 0.01 vs. WT.
When cultured in liquid growth medium, the majority of D. discoideum cells are mononucleated. However, it is well established that axenically grown cultures of D. discoideum can also contain polynucleated cells, which form due to defects in cytokinesis and tend to be larger than mononucleated cells (Waddell et al., 1987). Based on these findings, we examined the proportions of mononucleated and polynucleated cells in mfsd8− cultures. We observed that loss of mfsd8 significantly decreased the proportion of mononucleated cells in culture and increased the proportion of polynucleated cells (Figure 2B). Together, these findings showed that mfsd8-deficiency increases cell size and reduces cytokinesis during the growth phase of the D. discoideum life cycle.
Loss of mfsd8 Alters the Levels of Autocrine Proliferation Repressor During Growth
To gain insight into the possible mechanisms underlying the increased proliferation of mfsd8− cells, the intracellular and extracellular levels of AprA were examined. AprA functions extracellularly to repress cell proliferation in D. discoideum and has previously been shown to be aberrantly secreted by cln3− cells, which like mfsd8− cells, also display increased rates of proliferation (Brock and Gomer, 2005; Huber et al., 2014). WC lysates and CM from WT and mfsd8− cells were collected after 24, 48, and 72 h of axenic growth (Figure 1A). When analyzed by western blotting, anti-AprA detected two protein bands; one at 60 kDa and the other at 55 kDa, which is consistent with the banding pattern reported in previous studies (Brock and Gomer, 2005; Huber et al., 2014) (Figure 3). Intracellularly, the amounts of 60 kDa AprA remained constant during all stages of axenic growth for both WT and mfsd8− cultures (Figure 3A). However, the amounts of 60 kDa AprA in mfsd8− cells were lower at all time points analyzed compared to WT cells. The amounts of intracellular 55 kDa AprA decreased in WT and mfsd8− cells during axenic growth but there were no differences between cell lines. In samples of CM, loss of mfsd8 reduced the extracellular amount of 60 kDa AprA after 24 and 48 h of axenic growth, compared to WT cells, but there was no effect at the 72-h time point (Figure 3B). In addition, CM collected from mfsd8− cells after 72 h of axenic growth contained more 55 kDa AprA than CM collected from WT cells. Blots containing samples of CM were also probed with anti-β-actin to validate that CM did not contain intracellular proteins due to cell lysis (data not shown). Since AprA functions as a proliferation repressor, these findings suggest that altered levels of intracellular and extracellular AprA may have contributed to the increased rate of proliferation of mfsd8− cells.
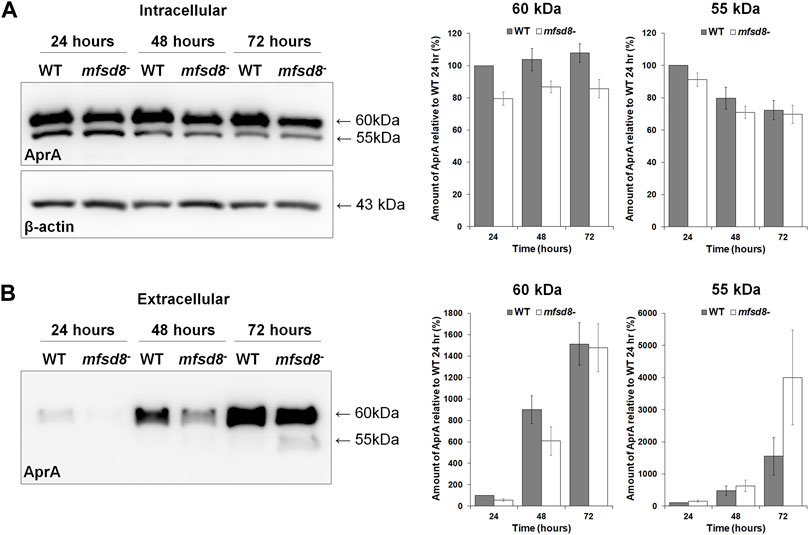
FIGURE 3. Effect of mfsd8-deficiency on the intracellular and extracellular levels of AprA during growth. WT and mfsd8− cells were grown in HL5, harvested after 24, 48, and 72 h of growth, and lysed. Conditioned media (CM) was also harvested at each time point. (A) Whole cell lysates (15 µg) were separated by SDS-PAGE and analyzed by western blotting with anti-AprA and anti-β-actin (loading control). Molecular weight markers (in kDa) are shown to the right of each blot. Protein bands were quantified using Fiji/ImageJ. The amounts of AprA were normalized against the amounts of β-actin. Data presented as mean amount of AprA relative to WT 24 h (%) ± SEM (n = 7). (B) Samples of CM were harvested at each time point and standardized against cell number. Equal volumes of CM (15 μl) were separated by SDS-PAGE and analyzed by western blotting with anti-AprA. Molecular weight markers (in kDa) are shown to the right of each blot. Protein bands were quantified using Fiji/ImageJ. Data presented as mean amount of AprA relative to WT 24 h (%) ± SEM (n = 7).
mfsd8-Deficiency Increases the Intracellular Activities of Several Lysosomal Enzymes During Growth
Lysosomal enzymes play an essential role in degrading material internalized by amoebae during the growth phase of the D. discoideum life cycle to provide cells with nutrients (Ashworth and Quance, 1972). Interestingly, in mice, loss of Mfsd8 has been shown to affect the levels of several lysosomal enzymes (Danyukova et al., 2018). Thus, we were interested in assessing lysosomal enzyme activity in mfsd8− cells during growth. Loss of mfsd8 significantly increased the intracellular activities of α-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, and N-acetylglucosaminidase (Figure 4). The activity of β-galactosidase was also slightly elevated but not statistically significant (p = 0.08). mfsd8-deficiency also elevated the activities of Ppt1 and CtsF. In humans, mutations in PPT1 and CTSF cause the CLN1 and CLN13 subtypes of NCL, respectively (Mole and Cotman, 2015). Finally, there was no effect of mfsd8 loss on the activities of Tpp1, CtsB, or CtsD. In humans, mutations in TPP1 and CTSD cause the CLN2 and CLN10 subtypes of NCL, respectively, and CTSB has been identified as a potential biomarker for CLN6 disease (Huber, 2021). Together, these data suggest that mfsd8− cells increase the activities of some, but not all, lysosomal enzymes to support their increased rates of proliferation and pinocytosis.
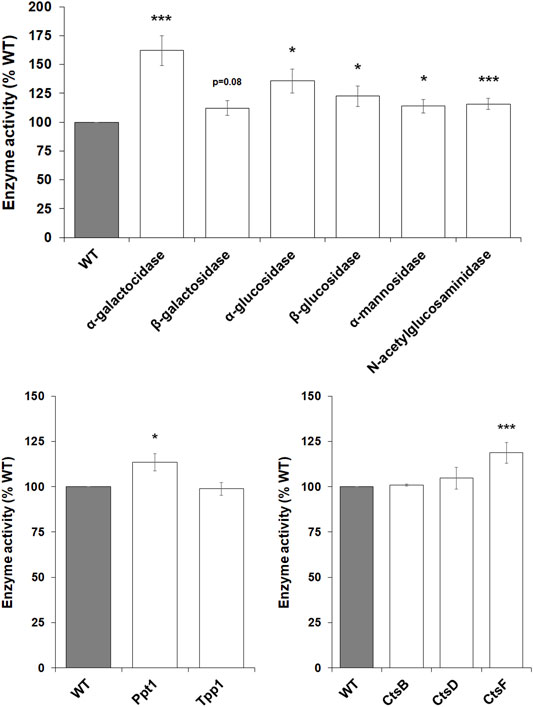
FIGURE 4. Effect of mfsd8-deficiency on enzyme activity during growth. WT and mfsd8− cells grown axenically in HL5 were collected and lysed. The activities of α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, N-acetylglucosaminidase, palmitoyl-protein thioesterase 1 (Ppt1), tripeptidyl peptidase 1 (Tpp1), cathepsin B (CtsB), cathepsin D (CtsD), and cathepsin F (CtsF) were assessed as described in the Materials and Methods. Data presented as mean enzyme activity (% WT) ± SEM (n > 4). Statistical significance was determined using the one-sample t-test (mean, 100; two-tailed) vs. WT. *p < 0.05 and ***p < 0.001 vs. WT.
Loss of mfsd8 Delays Aggregation During the Early Stages of Multicellular Development
Based on the increased expression of mfsd8 during the early stages of development (Rot et al., 2009), we suspected that loss of mfsd8 would affect cellular processes during this stage of the life cycle. Therefore, we performed two assays to examine the impact of mfsd8-deficiency on aggregation. In the first assay, cells adhered to Petri dishes were submerged in KK2 buffer to initiate the developmental program. After 8 h, there was a noticeable delay in the aggregation of mfsd8− cells compared to WT cells (Figure 5A). A delay in mfsd8− aggregation was also observed when cells were deposited on KK2-buffered agar (Figure 5B). In D. discoideum, mound formation occurs through the chemotactic aggregation of cells towards 3′,5′-cyclic adenosine monophosphate (cAMP) (Konijn et al., 1967). However, we did not observe a significant effect of mfsd8-deficiency on cAMP-mediated chemotaxis (Supplementary Figure S1).
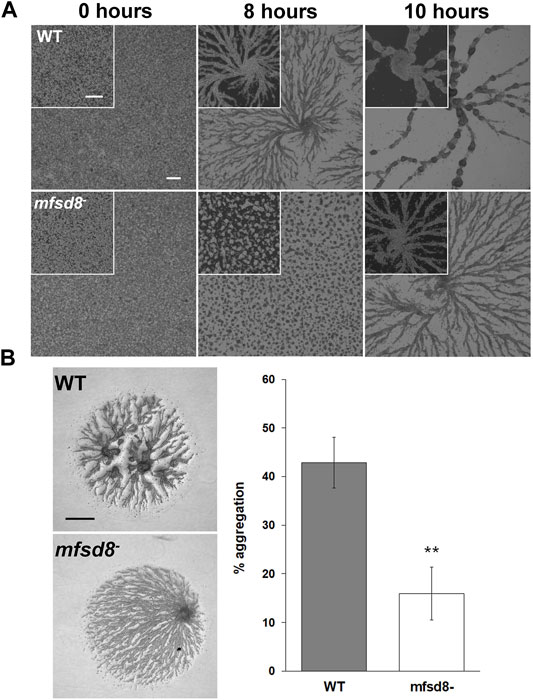
FIGURE 5. Effect of mfsd8-deficiency on aggregation. (A) WT and mfsd8− cells grown axenically in HL5 were submerged in KK2 buffer and imaged at the indicated time points. Scale bars = 250 μm. Images are representative of six independent experiments. (B) WT and mfsd8− cells grown axenically in HL5 were harvested, washed with KK2 buffer, and deposited onto 0.5% agar/KK2. Images were taken once cells were deposited and after 5 h. Scale bar = 250 μm. The amount of aggregation of each cell line was quantified by measuring the area of the cell spot that remained after 5 h of starvation, subtracting it from the initial area, and then expressing the difference as a percentage of the area of the cell spot at 0 h. Data presented as mean % aggregation ± SEM (n = 9). Statistical significance was assessed using the Kruskal-Wallis test followed by the Dunn’s multiple comparisons test. **p < 0.01.
A previous study showed that mfsd8-deficiency alters the secretion of two other CLN protein homologs in D. discoideum, Cln5 and CtsD, during the early stages of development (Huber et al., 2020b). Thus, we performed a CB swap experiment to examine the possible role of altered secretion in the delayed aggregation of mfsd8− cells. For this assay, we starved WT cells for 2 h and then collected the CB. In nine independent experiments, we observed that incubating mfsd8− cells in CB collected from WT cells partially restored the delayed aggregation of mfsd8− cells relative to WT cells (Figure 6). We also examined the effect of mfsd8-deficiency on cell-substrate adhesion, which plays an essential role in aggregation (Tarantola et al., 2014). During the early stages of development, loss of mfsd8 caused more cells to de-adhere from Petri dishes relative to WT cells (Figure 7). Combined, these data suggest that reduced cell-substrate adhesion along with aberrant protein secretion likely contribute to the delayed aggregation of mfsd8− cells during the early stages of D. discoideum development.
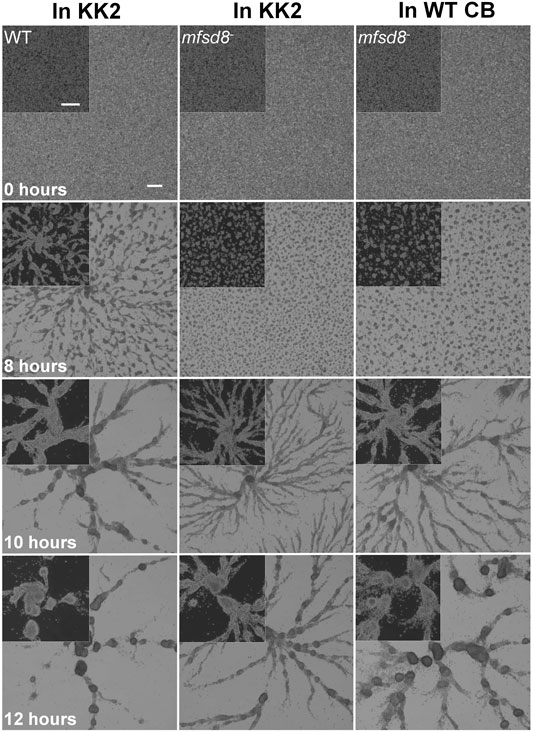
FIGURE 6. Effect of conditioned buffer on the aggregation of mfsd8− cells. WT cells were submerged in KK2 buffer for 2 h, after which time, the conditioned buffer (CB) of starving WT cells was collected and added to mfsd8− cells at the onset of starvation. Images are representative of nine independent experiments. Scale bar = 250 μm.
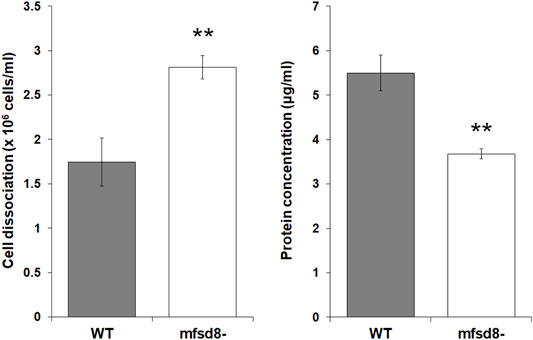
FIGURE 7. Effect of mfsd8-deficiency on cell-substrate adhesion. WT and mfsd8− cells were submerged in KK2 buffer for 4 h, after which time they were shaken at 150 rpm for 30 min. Samples of conditioned buffer were collected to measure cell dissociation, while cells remaining on the dish were lysed to assess protein concentration. Data presented as mean cell dissociation (×106 cells/ml) and protein concentration (μg/ml) ± SEM (n = 9). Statistical significance was assessed using one-way ANOVA followed by Bonferroni post-hoc analysis. **p < 0.01 vs. WT.
mfsd8-Deficiency Affects the Intracellular and Extracellular Amounts of Proteins Involved in Aggregation and Cell Adhesion
To gain further insight into the cellular mechanisms affected by mfsd8-deficiency, we starved WT and mfsd8− cells for 4 h and analyzed the intracellular and extracellular amounts of CtnA, CadA, and DscA. CtnA is a component of a secreted 450 kDa protein complex that regulates group size during aggregation by repressing cell-cell adhesion (Roisin-Bouffay et al., 2000). CadA and DscA, which were previously shown to interact with Mfsd8 (Huber et al., 2020b), both play important roles in regulating cell adhesion during the early stages of development (Cano and Pestaña, 1984; Springer et al., 1984; Knecht et al., 1987; Brar and Siu, 1993). In addition, loss of dscA impairs cell-substrate adhesion during aggregation (Springer et al., 1984; Bastounis et al., 2016). In this study, loss of mfsd8 significantly reduced the intracellular level of CtnA but increased the amount of the protein in CB, suggesting that secretion of CtnA was increased due to mfsd8-deficiency (Figure 8). Loss of mfsd8 had no effect on the amount of CadA inside cells but significantly increased its level extracellularly. Finally, loss of mfsd8 reduced the intracellular and extracellular levels of DscA. Combined, these data indicate that mfsd8-deficiency affects the intracellular and extracellular amounts of proteins involved in aggregation and cell adhesion, which could explain the delayed aggregation and reduced adhesion of mfsd8− cells during the early stages of development.
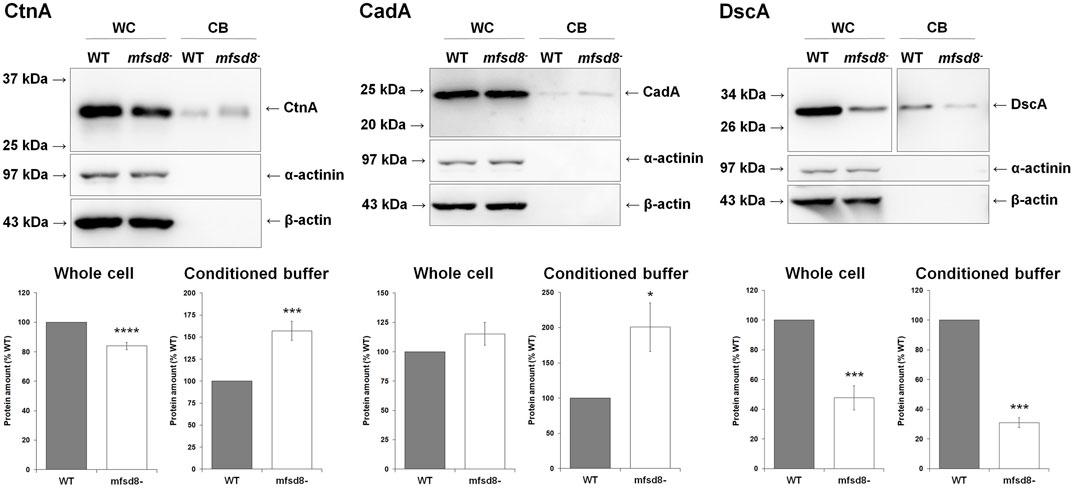
FIGURE 8. Effect of mfsd8-deficiency on the intracellular and extracellular levels of CtnA, CadA, and DscA. Cells grown axenically in HL5 were starved for 4 h in KK2 buffer, after which time whole cell (WC) lysates and conditioned buffer (CB) were collected. WC lysates (15 μg) and concentrated CB (0.25 μg) were separated by SDS-PAGE and analyzed by western blotting with anti-CtnA, anti-CadA, anti-DscA, anti-α-actinin (loading and fractionation control), and anti-β-actin (loading and fractionation control). Protein bands from WC lysates were standardized against the levels of α-actinin and β-actin. Data presented as mean protein amount (% WT) ± SEM (n = 8). Statistical significance was assessed using the one sample t-test. *p < 0.05, ***p < 0.001, and ****p < 0.0001 vs. WT.
Loss of mfsd8− Alters Lysosomal Enzyme Activities During the Early Stages of Development
During multicellular development, D. discoideum amoebae rely on autophagy and the actions of several lysosomal enzymes to provide cells with energy and building blocks required for fueling aggregation and fruiting body formation (Loomis, 1969; Loomis, 1970; Dimond et al., 1973; Kilpatrick and Stirling, 1976; Knecht et al., 1985; Otto et al., 2003; Kiel, 2010). As noted above, previous work in mice showed that loss of Mfsd8 affects the levels of several lysosomal enzymes (Danyukova et al., 2018). Thus, we assessed the effect of mfsd8-deficiency on lysosomal enzyme activity during the early stages of D. discoideum development. After 4 h of starvation, we observed increased intracellular activity of α-mannosidase and decreased activity of Ppt1 and CtsF (Figure 9). There was no significant effect of mfsd8-deficiency on the activities of α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetylglucosaminidase, Tpp1, CtsB, or CtsD. Combined, these findings show that loss of mfsd8 alters the activities of some, but not all, lysosomal enzymes during aggregation.
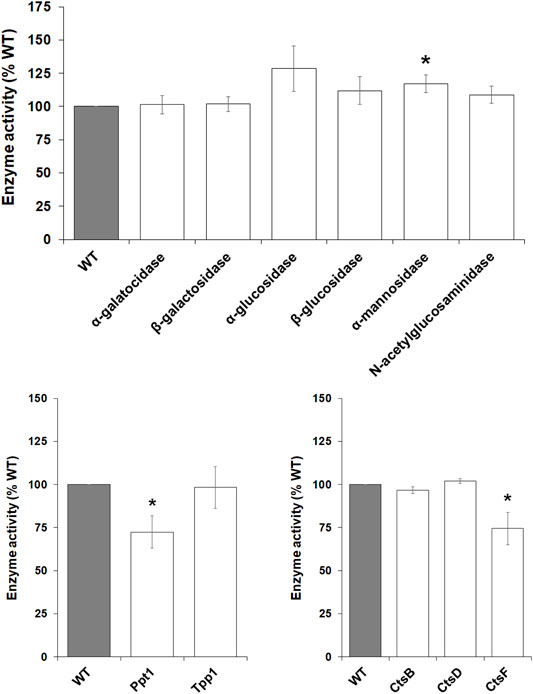
FIGURE 9. Effect of mfsd8-deficiency on enzyme activity during starvation. WT and mfsd8− cells grown axenically in HL5 were starved in KK2 buffer for 4 h, after which time cells were lysed. The activities of α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, N-acetylglucosaminidase, palmitoyl-protein thioesterase 1 (Ppt1), tripeptidyl peptidase 1 (Tpp1), cathepsin B (CtsB), cathepsin D (CtsD), and cathepsin F (CtsF) were assessed as described in the Materials and Methods. Data presented as mean enzyme activity (% WT) ± SEM (n > 4). Statistical significance was determined using the one-sample t-test (mean, 100; two-tailed) vs. WT. *p < 0.05 and ***p < 0.001 vs. WT.
Discussion
In this study, we examined the cellular roles of Mfsd8 during the growth and early development of D. discoideum (Figure 10). During growth, mfsd8-deficiency enhanced cell proliferation, FITC-dextran accumulation, and growth on bacterial lawns. The increased proliferation of mfsd8− cells correlated with altered levels of the proliferation repressor AprA and increased activity of several lysosomal enzymes. We also showed that Mfsd8 functions during the early stages of development to regulate cell adhesion, protein secretion, lysosomal enzyme activity, and aggregation. Together, this study provides new insights into the multifaceted roles of MFSD8 in the eukaryotic cell.
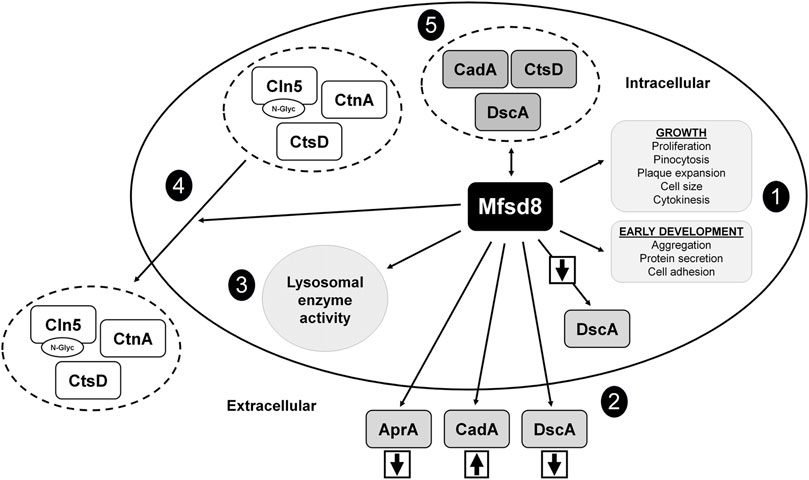
FIGURE 10. Current model depicting the functions of Mfsd8 in D. discoideum. (1) Loss of mfsd8 affects cell proliferation, pinocytosis, plaque expansion, cell size, and cytokinesis during growth, and aggregation, protein secretion, and cell adhesion during the early stages of development. (2) Loss of mfsd8 reduces the intracellular and extracellular amounts of DscA during the early stages of development and increases the extracellular amount of CadA. Loss of mfsd8 decreases the extracellular amount of AprA during growth. (3) Loss of mfsd8 affects lysosomal enzyme activity during growth and the early stages of development. (4) Mfsd8 regulates the secretion of Cln5, CtsD, and CtnA during the early stages of development. (5) Mfsd8 interacts with CadA, CtsD, and DscA during growth and the early stages of development.
The enhanced proliferation and accumulation of FITC-dextran observed in mfsd8− cells suggests that the increased proliferation was at least partly due to increased nutrient uptake. When grown on bacterial lawns, mfsd8− cells form plaques earlier than WT cells, further supporting their increased rate of growth. However, we showed that loss of mfsd8 has no effect on folic acid-mediated chemotaxis, which drives bacterial acquisition during feeding (Pan et al., 1972), suggesting that Mfsd8 influences mechanisms other than folic acid signalling during bacterial uptake.
Mfsd8 localizes to the macropinocytosis pathway in D. discoideum (Journet et al., 2012). In addition, previous work has shown that Mfsd8 interacts with proteins that negatively regulate endocytosis such as the Ras GTPase RapA and the nucleoside diphosphate kinase NdkC-1 (Seastone et al., 1999; Annesley et al., 2011). As a result, the loss of mfsd8 could have impacted the ability of these proteins to limit the rate of endocytosis in mfsd8− cells. Mfsd8-interactors also include transport proteins as well as proteins that localize to endocytic vesicles and the cytoskeleton (e.g., actin-10, myosin-2 heavy chain, tubulin alpha chain, and V-ATPase subunit B) (Huber et al., 2020b). Since endocytic processes are highly dependent on cytoskeletal elements for membrane invagination and vesicle transport (Kumari et al., 2010; Mooren et al., 2012), these observations provide insight into the mechanisms underlying the role of Mfsd8 in pinocytosis.
Loss of mfsd8 increases cell size, which like proliferation, could be explained by increased liquid nutrient uptake. However, we also observed that mfsd8-deficiency reduces cytokinesis, which is consistent with previous work in D. discoideum that correlated reduced cytokinesis and enhanced pinocytosis to increased cell size (Adachi, 2001; Winckler et al., 2001; Lim et al., 2005). In addition, enhanced proliferation and reduced cytokinesis have been observed in other D. discoideum mutants (Brock and Gomer, 2005; Bakthavatsalam et al., 2008; Huber et al., 2014; Mathavarajah et al., 2018). In this study, we observed reduced cytokinesis of mfsd8− cells submerged in growth medium on dishes. We suspect the cytokinesis defect could be due to aberrant adhesion, which is consistent with our observations of reduced adhesion of mfsd8− cells and previous work in D. discoideum linking aberrant cytokinesis to defects in adhesion (Nagasaki et al., 2009; Tsujioka et al., 2012). Finally, Mfsd8 was previously shown to interact with several proteins involved in cytokinesis, including actin-10, RapA, myosin-2 heavy chain, and Ras-like protein RasG (Tuxworth et al., 1997; Huber et al., 2020b). Together, these findings support a role for Mfsd8 in cytokinesis.
In D. discoideum, AprA is a secreted factor that represses cell proliferation and shares structural and functional similarity with human dipeptidyl peptidase 4 (Brock and Gomer, 2005; Herlihy et al., 2013; Herlihy et al., 2017). In addition to repressing cell proliferation, AprA also coordinates cytokinesis following mitosis and helps to reduce the formation of multinucleated cells (Brock and Gomer, 2005). In this study, extracellular AprA accumulated in parallel with cell density, which aligns with observations in previous studies (Brock and Gomer, 2005; Huber et al., 2014). However, loss of mfsd8 reduced the intracellular and extracellular levels of 60 kDa AprA suggesting that Mfsd8 may regulate proliferation by modulating both the synthesis and secretion of AprA. Contrary to the levels of 60 kDa AprA, loss of mfsd8 dramatically increased the extracellular levels of 55 kDa AprA as cells approached the stationary phase of axenic growth. While the identity of the 55 kDa band is not known, it has been proposed to be a AprA cleavage product (Huber et al., 2014). Therefore, its increased presence in mfsd8− CM during the later stages of axenic growth could be due to AprA degradation once cells reach the stationary phase of growth, which is consistent with our observation of mfsd8− cells reaching stationary phase earlier than WT cells. Combined, these observations suggest that loss of mfsd8 affects the synthesis and secretion of AprA, which may have played a role in the aberrant proliferation and cytokinesis observed in mfsd8− cells.
Lysosomes play an essential role in degrading endocytosed material to simple metabolites (Tjelle et al., 1996; Pillay et al., 2002). A recent study using MEFs derived from a Mfsd8−/− mouse showed that loss of Mfsd8 alters the amounts of soluble lysosomal proteins (Danyukova et al., 2018). In addition, our previous work showed that 61% of Mfsd8-interactors during growth have catalytic activity (Huber et al., 2020b). In this study, we observed that loss of mfsd8 increases the intracellular activities of several lysosomal enzymes, including α-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, N-acetylglucosaminidase, Ppt1, and CtsF. While these observations suggest that Mfsd8 plays a role in regulating lysosomal enzyme activity, the increased activities of these and potentially other lysosomal enzymes could also be explained by mfsd8− cells ingesting material at an increased rate. The increased lysosomal enzyme activity would allow material to be digested at an enhanced rate to prevent its accumulation and/or provide macromolecules required to fuel the increased rate of proliferation. Although one could argue that the increased intracellular fluorescence in mfsd8− cells could be due to reduced degradation of the internalized material, this seems unlikely since the increased lysosomal enzyme activity suggests that degradation of internalized material is normal in mfsd8− cells. Finally, while loss of mfsd8 affected CtsF activity, there are several proteins in D. discoideum that share sequence similarity with human CTSF (Huber et al., 2020a). Therefore, it is currently not known which CTSF-like protein(s) in D. discoideum is/are affected by mfsd8-deficiency.
Here, we showed that loss of mfsd8 delays aggregation but has no effect on cAMP-mediated chemotaxis. In addition, submerging mfsd8− cells in CB harvested from starving WT cells partially restored the timing of aggregation to WT levels. These findings support a regulatory role for Mfsd8 in processes involved in aggregation and suggest that Mfsd8 may regulate protein secretion. In a previous study, we showed that mfsd8-deficiency alters the secretion of Cln5 and CtsD (Huber et al., 2020b). Here, we showed that loss of mfsd8 also modulates the secretion of CtnA. However, we did not observe any obvious effects of mfsd8-deficiency on global protein secretion during growth or starvation (data not shown). Thus, it seems that Mfsd8 may regulate the secretion of selected proteins via an undetermined pathway to regulate aggregation.
mfsd8-deficiency also affected cell adhesion during the early stages of development. Consistent with this phenotype, we observed increased extracellular amounts of CadA in mfsd8− CB and reduced intracellular and extracellular amounts of DscA. Thus, it appears that altered levels of these two cell adhesion proteins likely contributed to the reduced adhesion of mfsd8− cells. This is further supported by work that has reported impaired cell-substrate adhesion for cells lacking dscA (Springer et al., 1984; Bastounis et al., 2016). Intriguingly, both CadA and DscA were identified in the Mfsd8-interactome (Huber et al., 2020b). In addition, the increased secretion of CtnA also likely contributed to the reduced adhesion of mfsd8− cells, since increased extracellular CtnA has been shown to reduce adhesion (Roisin-Bouffay et al., 2000).
During multicellular development, D. discoideum amoebae utilize autophagy and lysosomal enzymes to provide cells with energy required for fruiting body formation (Loomis, 1969; Loomis, 1970; Dimond et al., 1973; Kilpatrick and Stirling, 1976; Knecht et al., 1985; Otto et al., 2003; Kiel, 2010). In this study, we observed increased activity of α-mannosidase in mfsd8− cells and reduced activity of Ppt1 and CtsF. Intriguingly, α-mannosidase activity is also increased in Mfsd8−/− MEFs suggesting that the regulation of α-mannosidase may be an evolutionarily conserved function of MFSD8 (Danyukova et al., 2018). However, this effect may be cell type-dependent since Damme et al. (2014) reported unaltered activity of α-mannosidase in protein extracts generated from the cerebral cortex and liver of aged Mfsd8-depleted mice. In addition, a reduced amount of intracellular PPT1 was reported in human MFSD8 knockout HAP1 cells (Danyukova et al., 2018). Finally, Danyokova et al. (2018) reported reduced amounts of intracellular CLN5 and CTSD, which is supported by our findings in D. discoideum of reduced intracellular levels of Cln5 and CtsD in mfsd8− cells due to increased secretion (Danyukova et al., 2018; Huber et al., 2020b). Combined, these findings support a role for MFSD8 in regulating lysosomal enzyme activity. However, at present, we are unable to determine if the altered activities of α-mannosidase, Ppt1, and CtsF in mfsd8− cells contributed to the delayed aggregation or if the delayed aggregation altered the activities of those enzymes.
Collectively, our data shows that Mfsd8 plays a pleiotropic role in regulating D. discoideum growth and the early stages of multicellular development. While the molecular function of Mfsd8 in D. discoideum is not known, several of the phenotypes we uncovered could be explained by Mfsd8 functioning as a chloride channel, as has been reported in mammalian models (Wang et al., 2021). Future research to resolve the molecular function of Mfsd8 in D. discoideum should provide clarity on how loss of mfsd8 affects the diversity of processes we uncovered in this study. Intriguingly, aberrant phenotypes observed in cells lacking mfsd8 are also seen in other D. discoideum NCL models, further suggesting that CLN proteins function in shared or convergent biological pathways (Journet et al., 1999; Huber et al., 2014; Huber, 2017; Huber et al., 2017; Huber and Mathavarajah, 2018; Mathavarajah et al., 2018; Smith et al., 2019; Huber, 2020; McLaren et al., 2021). Overall, the findings of this study have provided novel insight into the roles of Mfsd8 in D. discoideum, which could be used to inform research in mammalian models of CLN7 disease.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
SQY data acquisition and analysis, writing-original draft, review and editing; WK: data acquisition and analysis, writing-review and editing; RH: conceptualization, data acquisition and analysis, writing–review and editing, funding acquisition, supervision. All authors read and approved the final draft of the manuscript.
Funding
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04855 to RH). SQY was supported by an Ontario Graduate Scholarship. WK was supported by a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and a Queen Elizabeth II Graduate Scholarship in Science and Technology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Sabateeshan Mathavarajah for providing technical assistance during the early stages of this work and Dr. Richard Gomer for providing antibodies against AprA and CtnA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.930235/full#supplementary-material
Abbreviations
AprA, autocrine proliferation repressor A; CadA, calcium-dependent cell adhesion protein A; cAMP, 3′,5′-cyclic adenosine monophosphate; CB, conditioned starvation buffer; CLN, ceroid lipofuscinosis neuronal; CM, conditioned growth media; CtnA, countin; CTSB, cathepsin B; CTSD, cathepsin D; CTSF, cathepsin F; DscA, discoidin A; FITC, fluoroscein isothiocyanate; HRP, horseradish peroxidase; MEF, mouse embryonic fibroblast; MES, 2-N-morpholinoethanesulfonic acid; MFSD8, major facilitator superfamily domain-containing 8; NAG, N-acetylglucosaminidase; NCL, neuronal ceroid lipofuscinosis; PPT1, palmitoyl-protein thioesterase 1; TPP1, tripeptidyl peptidase 1; WC, whole cell; WT, wild type.
References
Adachi, H. (2001). Identification of Proteins Involved in Cytokinesis of Dictyostelium. Cell Struct. Funct. 26, 571–575. doi:10.1247/csf.26.571
Aiello, C., Terracciano, A., Simonati, A., Discepoli, G., Cannelli, N., Claps, D., et al. (2009). Mutations in MFSD8/CLN7 are a Frequent Cause of Variant-Late Infantile Neuronal Ceroid Lipofuscinosis. Hum. Mutat. 30, E530–E540. doi:10.1002/humu.20975
Annesley, S. J., Bago, R., Bosnar, M. H., Filic, V., Marinović, M., Weber, I., et al. (2011). Dictyostelium discoideum Nucleoside Diphosphate Kinase C Plays a Negative Regulatory Role in Phagocytosis, Macropinocytosis and Exocytosis. PLoS One 6, e26024. doi:10.1371/journal.pone.0026024
Ashworth, J. M., and Quance, J. (1972). Enzyme Synthesis in Myxamoebae of the Cellular Slime Mould Dictyostelium discoideum during Growth in Axenic Culture. Biochem. J. 126, 601–608. doi:10.1042/bj1260601
Bakthavatsalam, D., Brock, D. A., Nikravan, N. N., Houston, K. D., Hatton, R. D., and Gomer, R. H. (2008). The Secreted Dictyostelium Protein CfaD is a Chalone. J. Cell Sci. 121, 2473–2480. doi:10.1242/jcs.026682
Barrett, A. J. (1980). Fluorimetric Assays for Cathepsin B and Cathepsin H with Methylcoumarylamide Substrates. Biochem. J. 187, 909–912. doi:10.1042/bj1870909
Bastounis, E., Álvarez-González, B., del Álamo, J. C., Lasheras, J. C., and Firtel, R. A. (2016). Cooperative Cell Motility during Tandem Locomotion of Amoeboid Cells. MBoC 27, 1262–1271. doi:10.1091/mbc.e15-12-0836
Brand, S., Roy, S., Schröder, P., Rathmer, B., Roos, J., Kapoor, S., et al. (2018). Combined Proteomic and In Silico Target Identification Reveal a Role for 5-Lipoxygenase in Developmental Signaling Pathways. Cell Chem. Biol. 25, 1095–1106. doi:10.1016/j.chembiol.2018.05.016
Brandenstein, L., Schweizer, M., Sedlacik, J., Fiehler, J., and Storch, S. (2016). Lysosomal Dysfunction and Impaired Autophagy in a Novel Mouse Model Deficient for the Lysosomal Membrane Protein Cln7. Hum. Mol. Genet. 25, 777–791. doi:10.1093/hmg/ddv615
Brar, S. K., and Siu, C. H. (1993). Characterization of the Cell Adhesion Molecule Gp24 in Dictyostelium discoideum. Mediation of Cell-Cell Adhesion via a Ca(2+)-dependent Mechanism. J. Biol. Chem. 268, 24902–24909. doi:10.1016/s0021-9258(19)74550-5
Brock, D. A., and Gomer, R. H. (1999). A Cell-Counting Factor Regulating Structure Size in Dictyostelium. Genes & Dev. 13, 1960–1969. doi:10.1101/gad.13.15.1960
Brock, D. A., and Gomer, R. H. (2005). A Secreted Factor Represses Cell Proliferation in Dictyostelium. Development 132, 4553–4562. doi:10.1242/dev.02032
Cárcel-Trullols, J., Kovács, A. D., and Pearce, D. A. (2015). Cell Biology of the NCL Proteins: What They Do and Don't Do. Biochim. Biophys. Acta 1852, 2242–2255. doi:10.1016/j.bbadis.2015.04.027
Cano, A., and Pestaña, A. (1984). The Role of Membrane Lectins in Dictyostelium discoideum Aggregation as Ascertained by Specific Univalent Antibodies against Discoidin I. J. Cell. Biochem. 25, 31–43. doi:10.1002/jcb.240250104
Coston, M. B., and Loomis, W. F. (1969). Isozymes of β-Glucosidase in Dictyostelium discoideum. J. Bacteriol. 100, 1208–1217. doi:10.1128/jb.100.3.1208-1217.1969
Damme, M., Brandenstein, L., Fehr, S., Jankowiak, W., Bartsch, U., Schweizer, M., et al. (2014). Gene Disruption of Mfsd8 in Mice Provides the First Animal Model for CLN7 Disease. Neurobiol. Dis. 65, 12–24. doi:10.1016/j.nbd.2014.01.003
Danyukova, T., Ariunbat, K., Thelen, M., Brocke-Ahmadinejad, N., Mole, S. E., and Storch, S. (2018). Loss of CLN7 Results in Depletion of Soluble Lysosomal Proteins and Impaired mTOR Reactivation. Hum. Mol. Genet. 27, 1711–1722. doi:10.1093/hmg/ddy076
Dimond, R. L., Brenner, M., and Loomis, W. F. (1973). Mutations Affecting N -Acetylglucosaminidase in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 70, 3356–3360. doi:10.1073/pnas.70.12.3356
Fey, P., Kowal, A. S., Gaudet, P., Pilcher, K. E., and Chisholm, R. L. (2007). Protocols for Growth and Development of Dictyostelium discoideum. Nat. Protoc. 2, 1307–1316. doi:10.1038/nprot.2007.178
Fey, P., Dodson, R. J., Basu, S., Hartline, E. C., and Chisholm, R. L. (2019). dictyBase and the Dicty Stock Center (Version 2.0) - A Progress Report. Int. J. Dev. Biol. 63, 563–572. doi:10.1387/ijdb.190226pf
Fonovic, M., Brömme, D., Turk, V., and Turk, B. (2004). Human Cathepsin F: Expression in Baculovirus System, Characterization and Inhibition by Protein Inhibitors. Biol. Chem. 385, 505–509. doi:10.1515/BC.2004.059
Gruenheit, N., Baldwin, A., Stewart, B., Jaques, S., Keller, T., Parkinson, K., et al. (2021). Mutant Resources for Functional Genomics in Dictyostelium discoideum Using REMI-Seq Technology. BMC Biol. 19, 172. doi:10.1186/s12915-021-01108-y
Hacker, U., Albrecht, R., and Maniak, M. (1997). Fluid-phase Uptake by Macropinocytosis in Dictyostelium. J. Cell Sci. 110, 105–112. doi:10.1242/jcs.110.2.105
Herlihy, S. E., Pilling, D., Maharjan, A. S., and Gomer, R. H. (2013). Dipeptidyl Peptidase IV is a Human and Murine Neutrophil Chemorepellent. J. Immunol. 190, 6468–6477. doi:10.4049/jimmunol.1202583
Herlihy, S. E., Tang, Y., Phillips, J. E., and Gomer, R. H. (2017). Functional Similarities between the dictyostelium Protein AprA and the Human Protein Dipeptidyl-Peptidase IV. Protein Sci. 26, 578–585. doi:10.1002/pro.3107
Huber, R. J., and Mathavarajah, S. (2018). Secretion and Function of Cln5 during the Early Stages of Dictyostelium Development. Biochim. Biophys. Acta Mol. Cell Res. 1865, 1437–1450. doi:10.1016/j.bbamcr.2018.07.017
Huber, R. J., and Mathavarajah, S. (2019). Comparative Transcriptomics Reveals Mechanisms Underlying Cln3-Deficiency Phenotypes in Dictyostelium. Cell. Signal. 58, 79–90. doi:10.1016/j.cellsig.2019.02.004
Huber, R. J., Myre, M. A., and Cotman, S. L. (2014). Loss of Cln3 Function in the Social Amoeba Dictyostelium discoideum Causes Pleiotropic Effects that are Rescued by Human CLN3. PLoS One 9, e110544. doi:10.1371/journal.pone.0110544
Huber, R. J., Myre, M. A., and Cotman, S. L. (2017). Aberrant Adhesion Impacts Early Development in a Dictyostelium Model for Juvenile Neuronal Ceroid Lipofuscinosis. Cell Adhesion Migr. 11, 399–418. doi:10.1080/19336918.2016.1236179
Huber, R. J., Hughes, S. M., Liu, W., Morgan, A., Tuxworth, R. I., and Russell, C. (2020a). The Contribution of Multicellular Model Organisms to Neuronal Ceroid Lipofuscinosis Research. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165614. doi:10.1016/j.bbadis.2019.165614
Huber, R. J., Mathavarajah, S., and Yap, S. Q. (2020b). Mfsd8 Localizes to Endocytic Compartments and Influences the Secretion of Cln5 and Cathepsin D in Dictyostelium. Cell. Signal. 70, 109572. doi:10.1016/j.cellsig.2020.109572
Huber, R. J. (2016). Using the Social Amoeba Dictyostelium to Study the Functions of Proteins Linked to Neuronal Ceroid Lipofuscinosis. J. Biomed. Sci. 23, 83. doi:10.1186/s12929-016-0301-0
Huber, R. J. (2017). Loss of Cln3 Impacts Protein Secretion in the Social Amoeba Dictyostelium. Cell. Signal. 35, 61–72. doi:10.1016/j.cellsig.2017.03.022
Huber, R. J. (2020). Molecular Networking in the Neuronal Ceroid Lipofuscinoses: Insights from Mammalian Models and the Social Amoeba Dictyostelium discoideum. J. Biomed. Sci. 27, 64. doi:10.1186/s12929-020-00653-y
Huber, R. J. (2021). Altered Protein Secretion in Batten Disease. Dis. Model Mech. 14, dmm049152. doi:10.1242/dmm.049152
Journet, A., Chapel, A., Jehan, S., Adessi, C., Freeze, H., Klein, G., et al. (1999). Characterization of Dictyostelium discoideum Cathepsin D. J. Cell Sci. 112, 3833–3843. doi:10.1242/jcs.112.21.3833
Journet, A., Klein, G., Brugière, S., Vandenbrouck, Y., Chapel, A., Kieffer, S., et al. (2012). Investigating the Macropinocytic Proteome of Dictyostelium Amoebae by High-Resolution Mass Spectrometry. Proteomics 12, 241–245. doi:10.1002/pmic.201100313
Kiel, J. A. K. W. (2010). Autophagy in Unicellular Eukaryotes. Phil. Trans. R. Soc. B 365, 819–830. doi:10.1098/rstb.2009.0237
Kilpatrick, D. C., and Stirling, J. L. (1976). Properties and Developmental Regulation of an α-d-galactosidase from Dictyostelium discoideum. Biochem. J. 158, 409–417. doi:10.1042/bj1580409
Kirolos, S. A., Rijal, R., Consalvo, K. M., and Gomer, R. H. (2021). Using Dictyostelium to Develop Therapeutics for Acute Respiratory Distress Syndrome. Front. Cell Dev. Biol. 9, 710005. doi:10.3389/fcell.2021.710005
Knecht, D. A., Green, E. D., Loomis, W. F., and Dimond, R. L. (1985). Developmental Changes in the Modification of Lysosomal Enzymes in Dictyostelium discoideum. Dev. Biol. 107, 490–502. doi:10.1016/0012-1606(85)90330-6
Knecht, D. A., Fuller, D. L., and Loomis, W. F. (1987). Surface Glycoprotein, Gp24, Involved in Early Adhesion of Dictyostelium discoideum. Dev. Biol. 121, 277–283. doi:10.1016/0012-1606(87)90160-6
Konijn, T. M., Van De Meene, J. G., Bonner, J. T., and Barkley, D. S. (1967). The Acrasin Activity of Adenosine-3',5'-Cyclic Phosphate. Proc. Natl. Acad. Sci. U.S.A. 58, 1152–1154. doi:10.1073/pnas.58.3.1152
Kumari, S., Mg, S., and Mayor, S. (2010). Endocytosis Unplugged: Multiple Ways to Enter the Cell. Cell Res. 20, 256–275. doi:10.1038/cr.2010.19
Lim, C. J., Zawadzki, K. A., Khosla, M., Secko, D. M., Spiegelman, G. B., and Weeks, G. (2005). Loss of the Dictyostelium RasC Protein Alters Vegetative Cell Size, Motility and Endocytosis. Exp. Cell Res. 306, 47–55. doi:10.1016/j.yexcr.2005.02.002
Loomis, W. F. (1969). Acetylglucosaminidase, an Early Enzyme in the Development of Dictyostelium discoideum. J. Bacteriol. 97, 1149–1154. doi:10.1128/jb.97.3.1149-1154.1969
Loomis, W. F. (1970). Developmental Regulation of α-Mannosidase in Dictyostelium discoideum. J. Bacteriol. 103, 375–381. doi:10.1128/jb.103.2.375-381.1970
Maruhn, D. (1976). Rapid Colorimetric Assay of β-galactosidase and N-Acetyl-β-Glucosaminidase in Human Urine. Clin. Chim. Acta 73, 453–461. doi:10.1016/0009-8981(76)90147-9
Mathavarajah, S., Flores, A., and Huber, R. J. (2017). Dictyostelium discoideum: A Model System for Cell and Developmental Biology. Curr. Protoc. Essent. Lab. Tech. 15, 14.1.1–14.1.19. doi:10.1002/cpet.15
Mathavarajah, S., McLaren, M. D., and Huber, R. J. (2018). Cln3 Function Is Linked to Osmoregulation in a Dictyostelium Model of Batten Disease. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3559–3573. doi:10.1016/j.bbadis.2018.08.013
Mathavarajah, S., VanIderstine, C., Dellaire, G., and Huber, R. J. (2021). Cancer and the Breakdown of Multicellularity: What Dictyostelium discoideum, a Social Amoeba, Can Teach Us. Bioessays 43, e2000156. doi:10.1002/bies.202000156
McLaren, M., Mathavarajah, S., and Huber, R. (2019). Recent Insights into NCL Protein Function Using the Model Organism Dictyostelium discoideum. Cells 8, 115. doi:10.3390/cells8020115
McLaren, M. D., Mathavarajah, S., Kim, W. D., Yap, S. Q., and Huber, R. J. (2021). Aberrant Autophagy Impacts Growth and Multicellular Development in a Dictyostelium Knockout Model of CLN5 Disease. Front. Cell Dev. Biol. 9, 657406. doi:10.3389/fcell.2021.657406
Mole, S. E., and Cotman, S. L. (2015). Genetics of the Neuronal Ceroid Lipofuscinoses (Batten Disease). Biochim. Biophys. Acta Mol. Basis Dis. 1852, 2237–2241. doi:10.1016/j.bbadis.2015.05.011
Mooren, O. L., Galletta, B. J., and Cooper, J. A. (2012). Roles for Actin Assembly in Endocytosis. Annu. Rev. Biochem. 81, 661–686. doi:10.1146/annurev-biochem-060910-094416
Nagasaki, A., Kanada, M., and Uyeda, T. Q. (2009). Cell Adhesion Molecules Regulate Contractile Ring-independent Cytokinesis in Dictyostelium discoideum. Cell Res. 19, 236–246. doi:10.1038/cr.2008.318
Otto, G. P., Wu, M. Y., Kazgan, N., Anderson, O. R., and Kessin, R. H. (2003). Macroautophagy is Required for Multicellular Development of the Social Amoeba Dictyostelium discoideum. J. Biol. Chem. 278, 17636–17645. doi:10.1074/jbc.m212467200
Pain, E., Shinhmar, S., and Williams, R. S. B. (2021). Using Dictyostelium to Advance Our Understanding of the Role of Medium Chain Fatty Acids in Health and Disease. Front. Cell Dev. Biol. 9, 722066. doi:10.3389/fcell.2021.722066
Pan, P., Hall, E. M., and Bonner, J. T. (1972). Folic Acid as Second Chemotactic Substance in the Cellular Slime Moulds. Nat. New Biol. 237, 181–182. doi:10.1038/newbio237181a0
Persaud-Sawin, D.-A., Mousallem, T., Wang, C., Zucker, A., Kominami, E., and Boustany, R.-M. N. (2007). Neuronal Ceroid Lipofuscinosis: A Common Pathway? Pediatr. Res. 61, 146–152. doi:10.1203/pdr.0b013e31802d8a4a
Phillips, J. E., and Gomer, R. H. (2015). Partial Genetic Suppression of a Loss-Of-Function Mutant of the Neuronal Ceroid Lipofuscinosis-Associated Protease TPP1 in Dictyostelium discoideum. Dis. Model Mech. 8, 147–156. doi:10.1242/dmm.018820
Pillay, C. S., Elliott, E., and Dennison, C. (2002). Endolysosomal Proteolysis and its Regulation. Biochem. J. 363, 417–429. doi:10.1042/bj3630417
Rivero, F., and Maniak, M. (2006). Quantitative and Microscopic Methods for Studying the Endocytic Pathway. Methods Mol. Biol. 346, 423–438. doi:10.1385/1-59745-144-4:423
Roisin-Bouffay, C., Jang, W., Caprette, D. R., and Gomer, R. H. (2000). A Precise Group Size in Dictyostelium is Generated by a Cell-Counting Factor Modulating Cell-Cell Adhesion. Mol. Cell 6, 953–959. doi:10.1016/s1097-2765(05)00082-1
Rot, G., Parikh, A., Curk, T., Kuspa, A., Shaulsky, G., and Zupan, B. (2009). Dictyexpress: A Dictyostelium discoideum Gene Expression Database with an Explorative Data Analysis Web-Based Interface. BMC Bioinform. 10, 265. doi:10.1186/1471-2105-10-265
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Schulz, A., Kohlschütter, A., Mink, J., Simonati, A., and Williams, R. (2013). NCL Diseases - Clinical Perspectives. Biochim. Biophys. Acta Mol. Basis Dis. 1832, 1801–1806. doi:10.1016/j.bbadis.2013.04.008
Seastone, D. J., Zhang, L., Buczynski, G., Rebstein, P., Weeks, G., Spiegelman, G., et al. (1999). The Small MrRas-like GTPase Rap1 and the Phospholipase C Pathway Act to Regulate Phagocytosis inDictyostelium Discoideum. MBoC 10, 393–406. doi:10.1091/mbc.10.2.393
Sharifi, A., Kousi, M., Sagné, C., Bellenchi, G. C., Morel, L., Darmon, M., et al. (2010). Expression and Lysosomal Targeting of CLN7, a Major Facilitator Superfamily Transporter Associated with Variant Late-Infantile Neuronal Ceroid Lipofuscinosis. Hum. Mol. Genet. 19, 4497–4514. doi:10.1093/hmg/ddq381
Siintola, E., Topcu, M., Aula, N., Lohi, H., Minassian, B. A., Paterson, A. D., et al. (2007). The Novel Neuronal Ceroid Lipofuscinosis Gene MFSD8 Encodes a Putative Lysosomal Transporter. Am. J. Hum. Genet. 81, 136–146. doi:10.1086/518902
Smith, P. K., Sen, M. G., Fisher, P. R., and Annesley, S. J. (2019). Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests that Cytopathological Outcomes Result from Altered TOR Signalling. Cells 8, 469. doi:10.3390/cells8050469
Springer, W. R., Cooper, D. N. W., and Barondes, S. H. (1984). Discoidin I is Implicated in Cell-Substratum Attachment and Ordered Cell Migration of Dictyostelium discoideum and Resembles Fibronectin. Cell 39, 557–564. doi:10.1016/0092-8674(84)90462-8
Stadler, J., Bauer, G., Westphal, M., and Gerisch, G. (1984). Monoclonal Antibody against Cytoplasmic Lectins of Dictyostelium discoideum: Cross-Reactivity with a Membrane Glycoprotein, Contact Site A, and with E. coli β-galactosidase and Lac Repressor. Hoppe Seylers Z Physiol. Chem. 365, 283–288. doi:10.1515/bchm2.1984.365.1.283
Steenhuis, P., Herder, S., Gelis, S., Braulke, T., and Storch, S. (2010). Lysosomal Targeting of the CLN7 Membrane Glycoprotein and Transport via the Plasma Membrane Require a Dileucine Motif. Traffic 11, 987–1000. doi:10.1111/j.1600-0854.2010.01073.x
Steenhuis, P., Froemming, J., Reinheckel, T., and Storch, S. (2012). Proteolytic Cleavage of the Disease-Related Lysosomal Membrane Glycoprotein CLN7. Biochim. Biophys. Acta Mol. Basis Dis. 1822, 1617–1628. doi:10.1016/j.bbadis.2012.05.015
Stumpf, M., Müller, R., Gaßen, B., Wehrstedt, R., Fey, P., Karow, M. A., et al. (2017). A Tripeptidyl Peptidase 1 is a Binding Partner of the Golgi pH Regulator (GPHR) in Dictyostelium. Dis. Model Mech. 10, 897–907. doi:10.1242/dmm.029280
Tarantola, M., Bae, A., Fuller, D., Bodenschatz, E., Rappel, W.-J., and Loomis, W. F. (2014). Cell Substratum Adhesion during Early Development of Dictyostelium discoideum. PloS One 9, e106574. doi:10.1371/journal.pone.0106574
Tjelle, T. E., Brech, A., Juvet, L. K., Griffiths, G., and Berg, T. (1996). Isolation and Characterization of Early Endosomes, Late Endosomes and Terminal Lysosomes: Their Role in Protein Degradation. J. Cell Sci. 109, 2905–2914. doi:10.1242/jcs.109.12.2905
Tsujioka, M., Yumura, S., Inouye, K., Patel, H., Ueda, M., and Yonemura, S. (2012). Talin Couples the Actomyosin Cortex to the Plasma Membrane during Rear Retraction and Cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 109, 12992–12997. doi:10.1073/pnas.1208296109
Tuxworth, R. I., Cheetham, J. L., Machesky, L. M., Spiegelmann, G. B., Weeks, G., and Insall, R. H. (1997). Dictyostelium RasG is Required for Normal Motility and Cytokinesis, but Not Growth. J. Cell Biol. 138, 605–614. doi:10.1083/jcb.138.3.605
van Diggelen, O. P., Keulemans, J. L. M., Winchester, B., Hofman, I. L., Vanhanen, S. L., Santavuori, P., et al. (1999). A Rapid Fluorogenic Palmitoyl-Protein Thioesterase Assay: Pre- and Postnatal Diagnosis of INCL. Mol. Genet. Metabolism 66, 240–244. doi:10.1006/mgme.1999.2809
von Kleist, L., Ariunbat, K., Braren, I., Stauber, T., Storch, S., and Danyukova, T. (2019). A Newly Generated Neuronal Cell Model of CLN7 Disease Reveals Aberrant Lysosome Motility and Impaired Cell Survival. Mol. Genet. Metabolism 126, 196–205. doi:10.1016/j.ymgme.2018.09.009
Waddell, D. R., Duffy, K., and Vogel, G. (1987). Cytokinesis is Defective in Dictyostelium Mutants with Altered Phagocytic Recognition, Adhesion, and Vegetative Cell Cohesion Properties. J. Cell Biol. 105, 2293–2300. doi:10.1083/jcb.105.5.2293
Wang, Y., Zeng, W., Lin, B., Yao, Y., Li, C., Hu, W., et al. (2021). CLN7 is an Organellar Chloride Channel Regulating Lysosomal Function. Sci. Adv. 7, eabj9608. doi:10.1126/sciadv.abj9608
Wimmer, B., Lottspeich, F., Ritter, J., and Bronnenmeier, K. (1997). A Novel Type of Thermostable α-D-glucosidase from Thermoanaerobacter Thermohydrosulfuricus Exhibiting Maltodextrinohydrolase Activity. Biochem. J. 328, 581–586. doi:10.1042/bj3280581
Keywords: Batten disease, CLN7, Dictyostelium discoideum, growth, development, lysosome, MFSD8, neuronal ceroid lipofuscinoses
Citation: Yap SQ, Kim WD and Huber RJ (2022) Mfsd8 Modulates Growth and the Early Stages of Multicellular Development in Dictyostelium discoideum. Front. Cell Dev. Biol. 10:930235. doi: 10.3389/fcell.2022.930235
Received: 27 April 2022; Accepted: 23 May 2022;
Published: 09 June 2022.
Edited by:
Paola Gavazzo, National Research Council (CNR), ItalyReviewed by:
Satoshi Sawai, The University of Tokyo, JapanRitva Tikkanen, University of Giessen, Germany
Copyright © 2022 Yap, Kim and Huber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Huber, cm9iZXJ0aHViZXJAdHJlbnR1LmNh
 Shyong Quan Yap
Shyong Quan Yap William D. Kim
William D. Kim Robert J. Huber
Robert J. Huber