
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol. , 21 October 2022
Sec. Morphogenesis and Patterning
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.929808
This article is part of the Research Topic Creating Asymmetry and Keeping Symmetry: the Underlying Genetic and Signaling Mechanisms View all 12 articles
The epithalamus of zebrafish shows morphological and molecular left-right (L-R) asymmetry, but such asymmetry is not apparent in tetrapods. To provide further insight into the evolutionary diversity of brain L-R asymmetry, we have now examined the developing brains of reptile embryos for expression of Nodal, Lefty, and Pitx2. Two turtle species, the Chinese softshell turtle and the red-eared slider turtle, showed left-sided expression of these three genes in the developing forebrain, with this expression occurring after Nodal expression at the lateral plate and the L-R organizer has disappeared. Nodal activity, as revealed by the detection of phosphorylated Smad2/3, was also apparent in the neural epithelium on the left side in both turtle species. In the Chinese softshell turtle, the habenula did not show apparent asymmetry in size and the parapineal organ was absent, but the expression of Kctd12 in the habenula showed a small yet reproducible asymmetry. In contrast to the turtles, L-R asymmetric expression of Nodal, Lefty, Pitx2, or Kctd12 was not detected in the developing brain of the Madagascar ground gecko. The transcriptional enhancer (ASE) responsible for the asymmetric expression of Nodal, Lefty, and Pitx2 was conserved among reptiles, including the Chinese softshell turtle and Madagascar ground gecko. Our findings suggest that Nodal, Lefty, and Pitx2 have the potential to be asymmetrically expressed in the developing brain of vertebrates, but that their expression varies even among reptiles.
Left-right (L-R) asymmetry of the brain is evident at anatomic and functional levels. However, the evolution of brain asymmetry and its functional importance remain largely unknown. Most of the progress in the characterization of such mechanisms has been made with the zebrafish epithalamus, a forebrain region that includes the centrally located pineal organ, the left-sided parapineal organ, and the bilateral habenular nuclei (Concha et al., 2000; Liang et al., 2000; Bianco and Wilson, 2009; Aizawa et al., 2011; Roberson and Halpern, 2018). Nodal, Lefty, and Pitx2 genes are expressed simultaneously and asymmetrically on the left side of the developing diencephalon, with such expression determining the direction of epithalamus laterality (Roussigne et al., 2009; Signore et al., 2016). For example, whereas the parapineal is located on the left side of the pineal organ in wild-type zebrafish, the position of the parapineal organ is randomized in Nodal mutants (Concha et al., 2000; Liang et al., 2000). The Nodal-Lefty-Pitx2 cassette is also expressed asymmetrically in the diencephalon of basal vertebrates such as lamprey and catshark (Lagadec et al., 2015).
The morphological and molecular asymmetry of the epithalamus varies among vertebrates (Concha and Wilson, 2001). Morphological asymmetry is apparent in cyclostomes (such as lamprey), chondrichthyans (such as catshark), and actinopterygians (such as zebrafish), but not in tetrapods including mammals. Asymmetric Nodal expression has been detected in teleosts, lamprey, and catshark but not in tetrapods, suggesting that it is an ancient feature of vertebrates but was lost during evolution. To clarify the evolutionary conservation of molecular brain asymmetry among vertebrates, we have now examined Nodal, Lefty, and Pitx2 expression in the developing brain of reptiles, including two turtles and a gecko. Unexpectedly, we found that Nodal, Lefty, and Pitx2 are expressed asymmetrically in the forebrain of the two turtles but not in that of the gecko.
Fertilized eggs of the Chinese softshell turtle (Pelodiscus sinensis) and red-eared slider turtle (Trachemys scripta elegans) were obtained from Daiwa Farm (Taku, Saga, Japan) and Himeji City Aquarium, respectively. At the time of oviposition, turtle embryos were at the late gastrulation stage, which is slightly too early for the study of L-R patterning. The eggs were therefore incubated for 3–38 days (depending on the type of analysis) at room temperature (for early-stage embryos of Chinese softshell turtle), followed by setting at a temperature of 28° C (for late-stage embryos of Chinese softshell turtle), or at 28°C (red-eared slider turtle embryos) before recovery of embryos for analysis. Madagascar ground geckos (Paroedura picta) were maintained as described previously (Noro et al., 2009; Yoshida et al., 2016) by the Laboratory for Animal Resources and Genetic Engineering at the RIKEN Center for Biosystems Dynamics Research. At the time of oviposition, gecko embryos are at the 11- to 16-somite stage, when L-R asymmetric gene expression in the lateral plate has disappeared (Kajikawa et al., 2020). For examination of gene expression in the developing brain, gecko embryos were recovered from the oviduct before oviposition (9–10 days after the previous oviposition) or at 0–1 day postoviposition (dpo). Both turtle and gecko embryos were staged as described previously (Yntema, 1968; Hubert et al., 1985; Yoshida et al., 2016).
Whole-mount in situ hybridization for turtle and gecko embryos was performed as described previously (Yoshida et al., 2016). The probes for Nodal, Lefty, and Otx5 mRNAs were also performed as described previously (Yoshida et al., 2016). In the case of probes for Pitx2, Not2, and Kctd12, corresponding cDNA fragments were cloned from the embryos of turtle or gecko, and their nucleotide sequences were verified by Sanger sequencing. Sequences of the primers used for cloning and amino acid sequence homology of encoded genes are listed in Supplementary Table S1.
Nissl staining of the developing brain was performed with cresyl violet. To estimate the volume of the habenula, the area (mm2) of the whole habenula (or the lateral habenula and medial habenula individually) was measured with ImageJ for each Nissl section. The volume (mm3) of the habenula was estimated by multiplying the habenula area (mm2) in each section and the thickness of sections (10 μm). The area of the habenula region positive for Kctd12 expression was similarly measured with ImageJ for each section and was expressed as an arbitrary unit.
Turtle embryos were subjected to immunostaining with a monoclonal antibody to phosphorylated Smad2/3 (Cell Signaling Technology, catalog no. 3108) as described previously (Kawasumi et al., 2011; Kajikawa et al., 2020). Immunostained embryos were sectioned at a thickness of 8 µm and were observed with an FV1000 confocal microscope (Olympus).
Nodal, Lefty, and Pitx2 genes of reptiles were identified in the NCBI gene database (https://www.ncbi.nlm.nih.gov/gene). Their nucleotide sequences were obtained from the NCBI gene database or Reptiliomix (https://transcriptome.riken.jp/reptiliomix). The FoxH1-binding sequences (ASE-like sequences) in each gene were manually identified. Amino acid sequence homology of an encoded protein was analyzed by CLUSTALW (https://www.genome.jp//tools-bin/clustalw).
When we previously examined Nodal expression in Chinese softshell turtle (P. sinensis) embryos (Kajikawa et al., 2020), we noticed that those at later developmental stages, after asymmetric expression of Nodal in the lateral plate mesoderm (LPM) had disappeared, showed left-sided Nodal expression in the forebrain (Figure 1A). Our present study revealed that Lefty (Figure 1B) and Pitx2 (Figure 1C) are also expressed on the left side of the forebrain at similar stages. Frontal sections of these embryos confirmed left-sided expression of Nodal, Lefty, and Pitx2 in the neural tube of the forebrain. The level of phosphorylated Smad2/3 has been shown to reflect the level of Nodal activity in mouse embryos (Kawasumi et al., 2011). Immunostaining with an antibody to phosphorylated Smad2/3 showed that Nodal activity is increased on the left side of the neural tube in P. sinensis (Figure 1D).
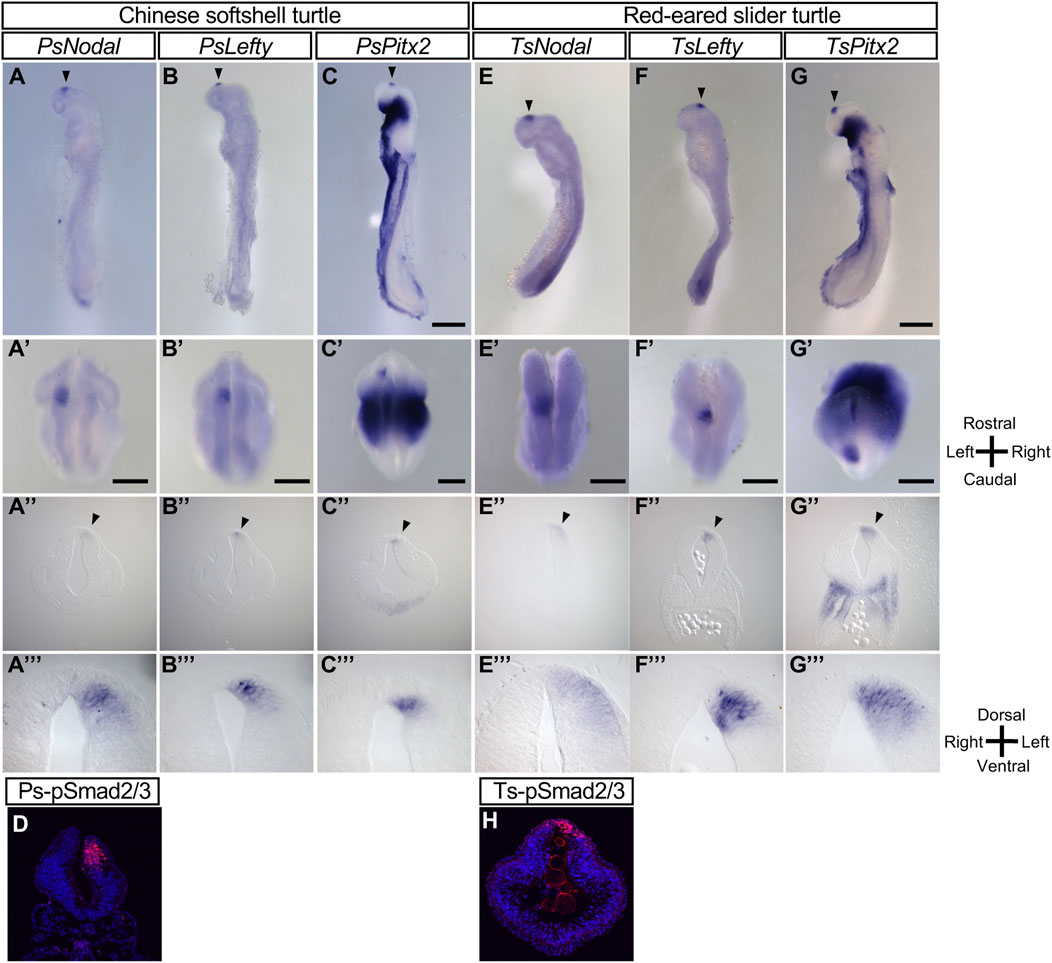
FIGURE 1. L-R asymmetric expression of Nodal, Lefty, and Pitx2 in the developing brain of the Chinese softshell turtle and red-eared slider turtle. (A–C), (E–G) Whole-mount in situ hybridization analysis of the expression domains of Nodal (A,E), Lefty (B,F), and Pitx2 (C,G) in the developing brain of Chinese softshell turtle embryos at 3 dpo (A–C) and of red-eared slider turtle embryos at 4 dpo (E–G). Top, middle, and bottom panels show left lateral views, dorsal views, and frontal sections, respectively. Domains manifesting L-R asymmetric expression are indicated by closed arrowheads. Rostral-caudal, dorsal-ventral, and L-R axes are indicated. Scale bars, 0.5 mm (top panels) and 0.2 mm (middle panels). Dorsal views, frontal sections, and higher magnification images of frontal sections of embryo in (A–C), (E–G) are shown in (A–C), (E–G), (A–C), (E–G), and (A–C), (E–G), respectively. (D,H) Nodal activity in Chinese softshell turtle (D) and red-eared slider turtle (H) embryos at 3 and 4 dpo, respectively, as determined by immunofluorescence staining of phosphorylated (p) Smad2/3 (red fluorescence). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue fluorescence). Nodal activity is asymmetric (L > R) in the diencephalon.
Given the marked asymmetric gene expression apparent in the forebrain of the Chinese softshell turtle, we examined another turtle species, the red-eared slider turtle (T. scripta elegans). Nodal, Lefty, and Pitx2 all showed left-sided expression in the developing forebrain of this species (Figures 1E–G). Nodal signaling (as reflected by phosphorylated Smad2/3) was also activated on the left side of the forebrain (Figure 1H).
The L-R asymmetry of the pineal complex (composed of the pineal organ and parapineal organ) and the habenula varies among the vertebrate taxa. The pineal complex is derived from the dorsal region of the diencephalon. In zebrafish embryos, left-sided expression of Nodal, Lefty, and Pitx2 occurs in the dorsal diencephalon and is followed by asymmetric expression of downstream genes such as Nrp1 (neuropilin 1) and Kctd12 (leftover) in the dorsal habenula (Gamse et al., 2003; Kuan et al., 2007). In the Chinese softshell turtle, asymmetric expression of Pitx2 persists longer than that of Nodal and was apparent in the diencephalon even at 7 dpo (Figure 2A-2A‴). We obtained cDNA clones for Not2 (a likely ortholog of Flh in zebrafish) and Kctd12 from the Chinese softshell turtle, and examined the expression of these genes in the developing brain. In zebrafish, Flh expression marks the pineal organ at the midline and the parapineal located on the left side (Concha et al., 2003). In the Chinese softshell turtle, Not2 was expressed at the dorsal side of the forebrain at 7 dpo (Figure 2B-2B‴), with this expression domain likely representing the future pineal organ in this species. In Nissl-stained sections of the Chinese softshell turtle at 38 dpo, the pineal organ was apparent, whereas the parapineal organ was not detected (Figure 2K). Unlike zebrafish, the habenula of the turtle consists of the lateral habenula and medial habenula on both sides (Figure 2D’), and there was no apparent L-R asymmetry in the morphology of the habenula (Figures 2C–F, Supplementary Figure S1). The volume of the habenula on the right and left sides was estimated from Nissl-stained sections (Supplementary Figure S1), which suggested no notable L-R difference (1.308 × 10−2mm3 for the left habenula vs 1.361 × 10−2mm3 for the right habenula). However, in some sections of the habenula (such as Figure 2D, 2D’), the lateral habenula on the left side looked larger than that on the right side.
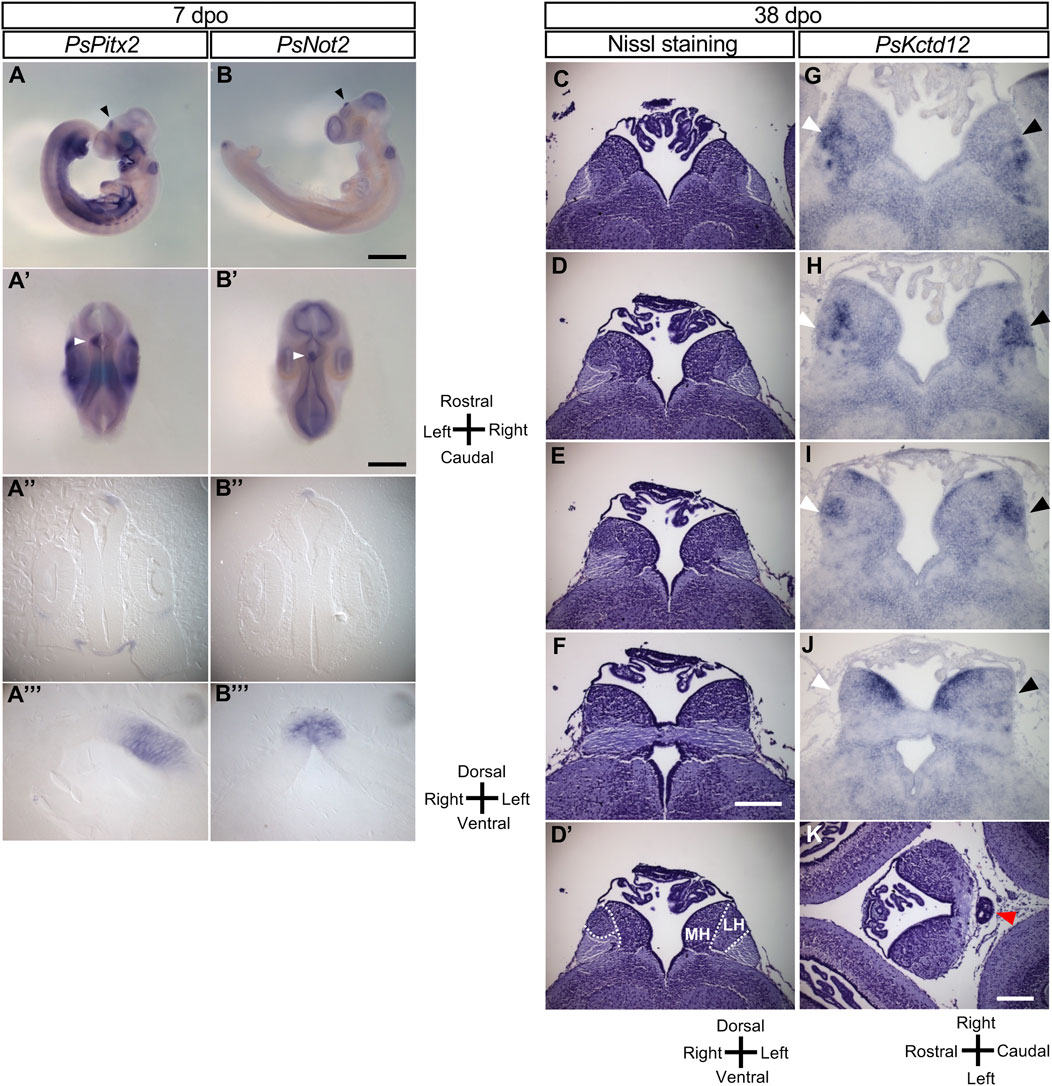
FIGURE 2. Molecular and morphological L-R asymmetry of the diencephalon in Chinese softshell turtle embryos at later stages. (A,B) Whole-mount in situ hybridization analysis of Pitx2 (A) and Not2 (B) in the developing brain of embryos at 7 dpo. Left lateral views (A,B), dorsal views (A,B), frontal sections (A,B), and higher magnification views of the frontal sections (A,B) are shown for each embryo. Closed arrowheads indicate the regions showing L-R asymmetric expression in the developing brain. Scale bars, 1.0 mm (top panels) and 0.5 mm (second from the top panels). (C–F) Nissl staining of frontal sections of embryos at 38 dpo. Sections at the level of the habenula are shown from the rostral side (C) to the caudal side (F). There was no apparent L-R asymmetry in morphology although a subtle asymmetry may exist in (D). Dotted lines in 2(D) indicate subdivision of the habenula shown in (D). MH and LH denote the medial habenula and lateral habenula, respectively. Scale bar, 0.2 mm. (G–J) Kctd12 expression in the habenula as examined by in situ hybridization with frontal sections of embryos at 38 dpo. Frontal sectional levels in (G–J) are similar to those in (C–F), respectively. Regions positive for Kctd12 expression on the right and left sides are indicated by white and black arrowheads, respectively. (K) Nissl-stained section of a Chinese softshell turtle at 38 dpo. The pineal organ is marked by the red arrowhead.
In zebrafish, Kctd12 is expressed in the dorsal habenula on the left side (Gamse et al., 2003; Kuan et al., 2007). In the Chinese softshell turtle, Kctd12 expression was mainly found in the lateral habenula and showed a small but reproducible asymmetry at 38 dpo (Figures 2G–J; Supplementary Figure 2D–F). To quantify Kctd12 expression, the area clearly positive for Kctd12 expression in the habenula was measured for the left and right sides of a series of sections obtained from three turtle embryos. The total positive area was 116,883 (arbitrary units) for the left habenula and 95,188 for the right habenula in the embryo shown in Figure 2. In the remaining two embryos, the L-R ratio was 32,618 (left) vs 25,227 (right) or 31,603 (left) vs 28,425 (right), showing only a subtle difference. When sections of the habenula were carefully examined, however, there was a small but significant asymmetry depending on the level along the rostral–caudal axis. At the most rostral level, there was no obvious L-R difference in the location, shape, or size of positive areas (Figure 2G). At caudal levels (Figure 2H), the Kctd12-positive area was located in the lateral region of the lateral habenula on the left side, whereas it was found in the more medial region on the right side. At more caudal levels (Figure 2I), the location was symmetric but the expression level seemed higher on the left side. At the most caudal level (Figure 2J), bilateral Kctd12 expression was only detected in the dorsal-medial habenula. A similar pattern of Kctd12 expression was observed in the habenula of two other embryos (Supplementary Figure S2).
We next examined whether the L-R asymmetric expression of the Nodal-Lefty-Pitx2 module in the developing brain is conserved among reptiles. With the use of cDNA clones for Nodal, Lefty, and Pitx2 obtained from the Madagascar ground gecko (P. picta), we examined the expression of these genes in embryos of this species. In turtles, asymmetric gene expression in the developing brain was detected after that in the LPM had disappeared (Figure 1). Given that Nodal expression in the LPM of gecko embryos was detected before oviposition (Kajikawa et al., 2020), we recovered gecko embryos at various stages both before oviposition (9–10 days after the previous oviposition) and at 0 to 1 dpo. However, expression of neither Nodal, Lefty, nor Pitx2 was detected in the forebrain at any stage examined (Figures 3A–J). Whereas asymmetric expression of a longer time, with this difference, is due to different enhancer sequences (Shiratori et al., 2001). However, Pitx2 expression was not detected in the forebrain at any stage examined between the 5- and 14-somite stages (Figures 3C–J). These observations thus suggested that the Nodal-Lefty-Pitx2 module is not asymmetrically expressed in the developing brain of the gecko.
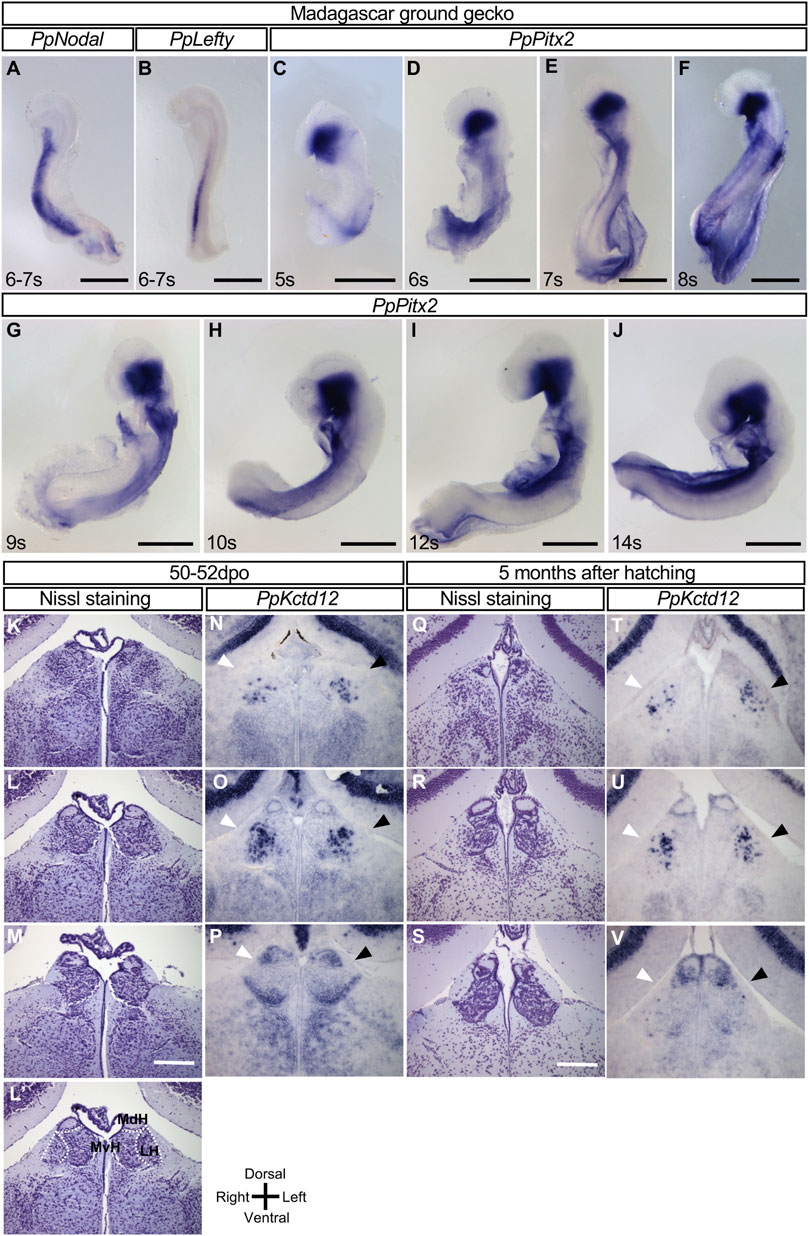
FIGURE 3. Lack of L-R asymmetric expression of Nodal, Lefty, and Pitx2 in the developing brain of the Madagascar ground gecko. (A–J) Whole-mount in situ hybridization analysis of Nodal (A), Lefty (B), and Pitx2 (C–J) expression in the developing brain of gecko embryos. These genes are not expressed in the developing brain, whereas their expression is apparent in other regions. The embryos were obtained before oviposition (9–10 days after the previous oviposition) (A–F) or at 0 to 1 dpo (G–J). The number of somites (s) is indicated for each embryo. Scale bars, 0.5 mm. (K–P) The habenula of gecko at the 50–52 dpo. Nissl staining of frontal sections at the level of the habenula is shown from the rostral side (K) to the caudal side (M). Dotted lines in (L′’) indicate the subdivision of the habenula shown in section (L). MdH, MvH, and LH denote the medial dorsal habenula, medial ventral habenula, and lateral habenula, respectively. Kctd12 expression in the habenula as examined by in situ hybridization with frontal sections are shown in (N–P). Frontal sectional levels in (N) to (p) are similar to those in (K) to (M), respectively. Regions positive for Kctd12 expression on the right and left sides are indicated by white and black arrowheads, respectively. (Q–V) The habenula of gecko at 5 months after hatching. Nissl staining of frontal sections at the level of the habenula are shown from the rostral side (Q) to the caudal side (S). Kctd12 expression in the habenula as examined by in situ hybridization with frontal sections are shown in (T–V). Frontal sectional levels in (T–V) are similar to those in (Q–S), respectively. Regions positive for Kctd12 expression on the right and left sides are indicated by white and black arrowheads, respectively.
We also examined the habenula of gecko embryos. The medial habenula was subdivided into dorsal-medial and ventral-medial habenula (Figure 3L’), but there was no L-R difference in the morphology or size at 50–52 dpo or 5 months after hatching (Figure 3K-M, 3Q-S, Supplementary Figure S3, S4). We have also examined Kctd12 expression in the gecko habenula. Kctd12 expression was detected in the habenula (mainly, the lateral habenula in the rostral region and the dorsal-medial habenula in the caudal region), but there was no notable asymmetry in its expression pattern at the two different stages (Figures 3N–P,T–V).
Asymmetric expression of Nodal, Lefty, and Pitx2 in the LPM is highly conserved among vertebrates and is induced by Nodal signaling via a FoxH1-dependent enhancer (ASE). This, thus, appears to be the case for the LPM of fish, frog, and chick embryos. Simultaneous expression of Nodal, Lefty, and Pitx2 in the developing forebrain of turtle embryos suggests that expression of these three genes is induced by the Nodal signaling. Indeed, a cluster of FoxH1-binding sequences resembling those of ASE is apparent in Nodal, Lefty, and Pitx2 of the Chinese softshell turtle at conserved positions: in intron 1 for Nodal, in the upstream region for Lefty, and in intron 2 for Pitx2 (Figure 4). ASE-like sequences are also present in the corresponding regions of Nodal, Lefty, and Pitx2 of the Madagascar ground gecko (Figure 4).
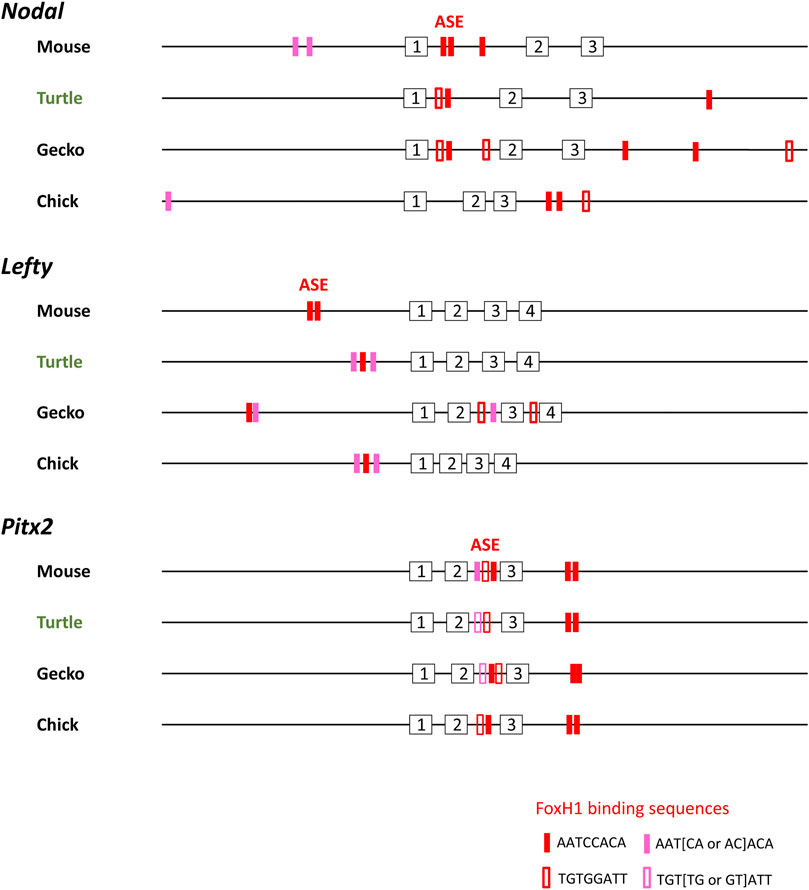
FIGURE 4. Transcriptional enhancer (ASE) sequences of Nodal, Lefty, and Pitx2 genes of amniotes. FoxH1-binding sequences (red and pink boxes) present within a 30-kbp region of Nodal, Lefty, and Pitx2 genes of mouse (Mus musculus), reptiles, and chicken (Gallus) are shown. Analysis of Nodal genes was previously performed (Kajikawa et al., 2020). For mouse, which has two Lefty genes, Lefty2 is shown. Red closed and open boxes correspond to AATCCACA and TGTGGATT, respectively. Pink closed and open boxes denote ATT [AC or CA]ACA and TGT [GT or TG]ATT, respectively. Numbered boxes indicate exons. Turtle and gecko refer to the Chinese softshell turtle (Pelodiscus sinensis) and Madagascar ground gecko (Paroedura picta), respectively. Nodal, Lefty, and Pitx2 genes of other turtles (green sea turtle and painted turtle) show similar configurations.
The epithalamus shows L-R asymmetry of morphology, gene expression, and connectivity in many of the vertebrates studied, but its laterality varies among species. For instance, the habenula manifests varied L-R asymmetry in size, with the left side being larger or smaller than or equal in size to the right, depending on the species (Concha and Wilson, 2001). In the Chinese softshell turtle, the habenula showed no apparent asymmetry of size, whereas it did show a small but reproducible asymmetry in the subdivision pattern and Kctd12 expression. The parapineal organ is also variable among vertebrate taxa, being present in some species but absent in others. Lizards possess a parietal eye, an organ equivalent to the parapineal organ of teleosts (Engbretson et al., 1981). In the Chinese softshell turtle, whereas the pineal organ was clearly evident, the parapineal was not detected. As previously suggested (Concha and Wilson, 2001), the connection between the parapineal organ and the left habenula does not necessarily correlate with asymmetries in size and organization of the habenula, and the parapineal organ may have been lost several times independently during evolution.
Our data suggest that asymmetric expression of the Nodal-Lefty-Pitx2 module in the developing turtle forebrain is induced by Nodal signaling. If this is the case, then what might be the origin of the Nodal protein that initiates Nodal expression in the turtle brain? Nodal protein produced at the L-R organizer (blastopore) of turtle embryos may travel to the diencephalon, either by passive diffusion or by active transport, where it then activates the expression of the Nodal gene itself. A similar mechanism is thought to operate in mouse embryos, in which Nodal protein produced at the node is activated and then transported to the left side of the LPM, where it activates the Nodal expression (Oki et al., 2007). Alternatively, an unknown mechanism may initiate expression of Nodal on the left side of the turtle diencephalon, possibly similar to that by which Nodal expression is induced at the left side of the L-R organizer in chick and reptile embryos (Levin et al., 1995; Kajikawa et al., 2020). The distance between the L-R organizer and the forebrain is relatively large, however, and Nodal expression in the forebrain takes place about 1 day after that at the L-R organizer has disappeared. These spatial and temporal constraints between Nodal expression at the L-R organizer and that in the developing brain may leave the latter scenario also possible.
Why is the Nodal-Lefty-Pitx2 module not expressed in the developing brain of other reptiles such as geckos and in that of mammals? If L-R asymmetric expression of Nodal in the brain, unlike that in the LPM, is indeed not induced by the Nodal derived from the L-R organizer, then the unknown signal that activates Nodal expression in the diencephalon of turtle embryos may be absent in other reptiles and in mammals.
The left-sided expression of Nodal in the LPM is highly conserved among vertebrates, without a single exception having been identified to date. In contrast, the expression pattern of the Nodal-Lefty-Pitx2 module in the developing brain appears to be highly varied among vertebrates. Even within reptiles, turtles are positive, whereas the gecko is negative for the L-R asymmetric expression of this module. A phylogenetic tree of reptiles based on the genomic sequences of these genes and overall genomic sequences shows that the turtle is far distant from the gecko (Hara et al., 2018). In this regard, it will be of interest to see whether the alligator, a reptile positioned close to the turtle, also shows asymmetric gene expression in the developing brain.
The raw data supporting the conclusion of this article will be made available by the authors on requests /Supplementary Material.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the RIKEN Kobe Branch.
EK performed all experiments and analyzed data. TM and MT prepared eggs of red-eared slider turtle. HK prepared gecko eggs. HH wrote the manuscript.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (no. 17H01435 and 20H00472).
We thank Mari Taniguchi and Tatsuya Hirasawa for their suggestions on the red-eared slider turtle and for introducing Himeji City Aquarium.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.929808/full#supplementary-material.
Aizawa, H., Amo, R., and Okamoto, H. (2011). Phylogeny and ontogeny of the habenular structure. Front. Neurosci. 5, 138. doi:10.3389/fnins.2011.00138
Bianco, I. H., and Wilson, S. W. (2009). The habenular nuclei: A conserved asymmetric relay station in the vertebrate brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 1005–1020. doi:10.1098/rstb.2008.0213
Concha, M. L., Burdine, R. D., Russell, C., Schier, A. F., and Wilson, S. W. (2000). A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28, 399–409. doi:10.1016/s0896-6273(00)00120-3
Concha, M. L., Russell, C., Regan, J. C., Tawk, M., Sidi, S., Gilmour, D. T., et al. (2003). Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron 39, 423–438. doi:10.1016/s0896-6273(03)00437-9
Concha, M. L., and Wilson, S. W. (2001). Asymmetry in the epithalamus of vertebrates. J. Anat. 199, 63–84. doi:10.1046/j.1469-7580.2001.19910063.x
Engbretson, G. A., Reiner, A., and Brecha, N. (1981). Habenular asymmetry and the central connections of the parietal eye of the lizard. J. Comp. Neurol. 198, 155–165. doi:10.1002/cne.901980113
Gamse, J. T., Thisse, C., Thisse, B., and Halpern, M. E. (2003). The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development 130, 1059–1068. doi:10.1242/dev.00270
Hara, Y., Takeuchi, M., Kageyama, Y., Tatsumi, K., Hibi, M., Kiyonari, H., et al. (2018). Madagascar ground gecko genome analysis characterizes asymmetric fates of duplicated genes. BMC Biol. 16, 40. doi:10.1186/s12915-018-0509-4
Hubert, J., Seve, A. P., Bouvier, D., Masson, C., Bouteille, M., and Monsigny, M. (1985). In situ ultrastructural localization of sugar-binding sites in lizard granulosa cell nuclei. Biol. Cell 55, 15–20. doi:10.1111/j.1768-322x.1985.tb00404.x
Kajikawa, E., Horo, U., Ide, T., Mizuno, K., Minegishi, K., Hara, Y., et al. (2020). Nodal paralogues underlie distinct mechanisms for visceral left-right asymmetry in reptiles and mammals. Nat. Ecol. Evol. 4, 261–269. doi:10.1038/s41559-019-1072-2
Kawasumi, A., Nakamura, T., Iwai, N., Yashiro, K., Saijoh, Y., Belo, J. A., et al. (2011). Left-right asymmetry in the level of active Nodal protein produced in the node is translated into left-right asymmetry in the lateral plate of mouse embryos. Dev. Biol. 353, 321–330. doi:10.1016/j.ydbio.2011.03.009
Kuan, Y. S., Yu, H. H., Moens, C. B., and Halpern, M. E. (2007). Neuropilin asymmetry mediates a left-right difference in habenular connectivity. Development 134, 857–865. doi:10.1242/dev.02791
Lagadec, R., Laguerre, L., Menuet, A., Amara, A., Rocancourt, C., Pericard, P., et al. (2015). The ancestral role of nodal signalling in breaking L/R symmetry in the vertebrate forebrain. Nat. Commun. 6, 6686. doi:10.1038/ncomms7686
Levin, M., Johnson, R. L., Stern, C. D., Kuehn, M., and Tabin, C. (1995). A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 82, 803–814. doi:10.1016/0092-8674(95)90477-8
Liang, J. O., Etheridge, A., Hantsoo, L., Rubinstein, A. L., Nowak, S. J., Izpisua Belmonte, J. C., et al. (2000). Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development 127, 5101–5112. doi:10.1242/dev.127.23.5101
Noro, M., Uejima, A., Abe, G., Manabe, M., and Tamura, K. (2009). Normal developmental stages of the Madagascar ground gecko Paroedura pictus with special reference to limb morphogenesis. Dev. Dyn. 238, 100–109. doi:10.1002/dvdy.21828
Oki, S., Hashimoto, R., Okui, Y., Shen, M. M., Mekada, E., Otani, H., et al. (2007). Sulfated glycosaminoglycans are necessary for Nodal signal transmission from the node to the left lateral plate in the mouse embryo. Development 134, 3893–3904. doi:10.1242/dev.009464
Roberson, S., and Halpern, M. E. (2018). Development and connectivity of the habenular nuclei. Semin. Cell Dev. Biol. 78, 107–115. doi:10.1016/j.semcdb.2017.10.007
Roussigne, M., Bianco, I. H., Wilson, S. W., and Blader, P. (2009). Nodal signalling imposes left-right asymmetry upon neurogenesis in the habenular nuclei. Development 136, 1549–1557. doi:10.1242/dev.034793
Shiratori, H., Sakuma, R., Watanabe, M., Hashiguchi, H., Mochida, K., Sakai, Y., et al. (2001). Two-step regulation of left-right asymmetric expression of Pitx2: Initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7, 137–149. doi:10.1016/s1097-2765(01)00162-9
Signore, I. A., Palma, K., and Concha, M. L. (2016). Nodal signalling and asymmetry of the nervous system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150401. doi:10.1098/rstb.2015.0401
Yntema, C. L. (1968). A series of stages in the embryonic development of Chelydra serpentina. J. Morphol. 125, 219–251. doi:10.1002/jmor.1051250207
Keywords: brain, left-right asymmetry, Nodal, reptile, turtle
Citation: Kajikawa E, Miki T, Takeda M, Kiyonari H and Hamada H (2022) Left-right asymmetric expression of the Nodal-Lefty-Pitx2 module in developing turtle forebrain. Front. Cell Dev. Biol. 10:929808. doi: 10.3389/fcell.2022.929808
Received: 27 April 2022; Accepted: 03 October 2022;
Published: 21 October 2022.
Edited by:
Daniel Grimes, University of Oregon, United StatesReviewed by:
Miguel Luis Concha, University of Chile, ChileCopyright © 2022 Kajikawa, Miki, Takeda, Kiyonari and Hamada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eriko Kajikawa, ZXJpa28ua2FqaWthd2FAcmlrZW4uanA=; Hiroshi Hamada, aGlyb3NoaS5oYW1hZGFAcmlrZW4uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.