- College of Agriculture, Ningxia University, Yinchuan, China
Functional cells in embryonic myogenesis and postnatal muscle development undergo multiple stages of proliferation and differentiation, which are strict procedural regulation processes. N6-methyladenosine (m6A) is the most abundant RNA modification that regulates gene expression in specific cell types in eukaryotes and regulates various biological activities, such as RNA processing and metabolism. Recent studies have shown that m6A modification-mediated transcriptional and post-transcriptional regulation plays an essential role in myogenesis. This review outlines embryonic and postnatal myogenic differentiation and summarizes the important roles played by functional cells in each developmental period. Furthermore, the key roles of m6A modifications and their regulators in myogenesis were highlighted, and the synergistic regulation of m6A modifications with myogenic transcription factors was emphasized to characterize the cascade of transcriptional and post-transcriptional regulation during myogenesis. This review also discusses the crosstalk between m6A modifications and non-coding RNAs, proposing a novel mechanism for post-transcriptional regulation during skeletal muscle development. In summary, the transcriptional and post-transcriptional regulatory mechanisms mediated by m6A and their regulators may help develop new strategies to maintain muscle homeostasis, which are expected to become targets for animal muscle-specific trait breeding and treatment of muscle metabolic diseases.
1 Introduction
The skeletal muscle is mainly composed of a large number of muscle fibers, a small amount of adipose tissue, and connective tissue, which are highly heterogeneous striated muscles that contain muscle cells, immune cells, and nerves. Accounting for approximately 40% of an animal’s body weight, the skeletal muscle is the organism’s largest motor and metabolic organ and plays a vital role in metabolism and energy balance (Mayeuf-Louchart et al., 2015; Cong et al., 2020). Embryonic myogenesis and postnatal muscle development are precisely regulated by multiple mechanisms that involve myoblast progenitor cell proliferation, differentiation, and fusion to form muscle fibers (Zhu et al., 2021). Skeletal muscle development involves multiple stages of proliferation and differentiation, and research on its formation process and molecular regulation mechanisms has always been a hot topic in the field of molecular genetics. Currently, research on skeletal muscle growth and development mainly focuses on the functional identification of key genes and the post-transcriptional regulatory mechanisms mediated by non-coding RNAs (such as lncRNA, circRNA, and microRNA) (Zhao et al., 2011). In addition to being regulated by a series of specific transcription factors and signaling pathways, epigenetic modifications are also involved in a variety of biological processes in muscle development (McKinnell et al., 2008; Yang et al., 2021). As the most common methylation modification in RNA (Deng et al., 2018), N6-methyladenosine (m6A) represents a new type of post-transcriptional gene regulation, which is tissue specific and spatio-temporally specific.
To date, more than 150 different RNA modifications have been identified (Yang et al., 2018), which play an important role in tissue development and homeostasis by controlling the cell state transition (Frye et al., 2018). The m6A modification is the most abundant methylation modification in eukaryotic cells, and it is widely distributed in mRNA and non-coding RNA and can regulate gene expression through “epitranscriptomics” without changing the sequence of RNA molecules (Roundtree et al., 2017a; Helm and Motorin, 2017; Fazi and Fatica, 2019). m6A modifications in eukaryotic RNA are reversible (Jia et al., 2012) and can be deposited, removed, and recognized by a series of methyltransferase complexes (writers), demethylases (erasers), and m6A-binding proteins (readers) (Lu et al., 2019). They are involved in the regulation of biological processes, such as disease occurrence (Han et al., 2019; Wang et al., 2020), embryonic development (Mendel et al., 2018), tissue development, and cell proliferation and differentiation (Zhang et al., 2017; Lee et al., 2019) by regulating RNA metabolic activities such as precursor RNA splicing (Haussmann et al., 2016), mRNA translocation (Fustin et al., 2013), stability (Geula et al., 2015), and translation (Coots et al., 2017). However, the post-transcriptional mechanisms of m6A modifications in the regulation of muscle development remain largely unknown.
Recent studies have shown that m6A methylation plays an important role in muscle stem cell maintenance, myocyte proliferation, and cell differentiation (Kudou et al., 2017; Wang et al., 2017; Dorn et al., 2019; Mathiyalagan et al., 2019; Gheller et al., 2020; Lin et al., 2020; Zhang et al., 2020). This review aims to provide an overview of the mechanisms of myogenesis under transcriptional and m6A methylation-mediated post-transcriptional regulation and emphasizes the important role of m6A modification and its regulators in coordinating functional signaling factors at different stages of skeletal muscle development. Based on the latest research in epigenetic modifications, this study also discusses the interaction between m6A modification and non-coding RNA and explores different levels of co-regulation in myogenesis.
2 Biological characteristics of skeletal muscle development
Both embryonic myogenesis and postnatal muscle development undergo a series of cell proliferation and differentiation processes from the embryonic stage to early growth and development, and mesoderm cells undergo repeated mitosis and massive proliferation to form mononuclear myoblasts. Furthermore, myoblasts proliferate and fuse into multinucleated myotubes (Velleman et al., 2013). During myogenesis, mononuclear myoblasts withdraw from the cell cycle, lose their ability to divide, and migrate from the cell center to the cell membrane to form myofibers (Picard et al., 2010). Single myoblasts that cannot be fused are isolated between the myofiber basement membrane and muscle cell membrane and finally form muscle satellite cells (MuSCs) with stem cell characteristics. The proliferative capacity of MuSCs decreases with age during animal growth and development. However, MuSCs can proliferate and undergo myogenic differentiation when myofibers are damaged, participating in their repair and regeneration (Zhang et al., 2018), thus ensuring normal growth and development of animals.
2.1 Embryonic myogenesis
The genesis and development of the skeletal muscle in the animal embryo is a complex physiological and biochemical process that primarily includes muscle cell genesis, myofiber formation and maturation, and the accumulation of myofiber number (Figure 1). The skeletal muscles of vertebrates originate from the paraxial mesoderm during embryonic development (Buckingham and Vincent, 2009). The paraxial mesoderm differentiates to produce somites, which act as the “helmsman of fate” and control the direction of myogenesis and osteogenesis (Biressi et al., 2007). The somite matures and divides again during embryo development. Driven by the sonic hedgehog (Shh) signal secreted by the neural tube and notochord, a part of the epithelial cells of the dorsal develops into a dermomyotome (Buckingham, 2001). Pluripotent stem cells located in the somite differentiate into muscle progenitor cells (MPCs) under the conditions of various signal molecules and embryonic environments. With the proliferation of dermomyotome cells, the number of MPC increases continuously, and the MPC in the middle of the dermomyotome continues to migrate downward to form the first skeletal muscle tissue: the myotome. MPCs are further exfoliated from the dermomyotome and fused with the myotome to form skeletal muscle, and some MPCs migrate to the extremities to form the limb skeletal muscle (Parker et al., 2003; Jensen et al., 2010).
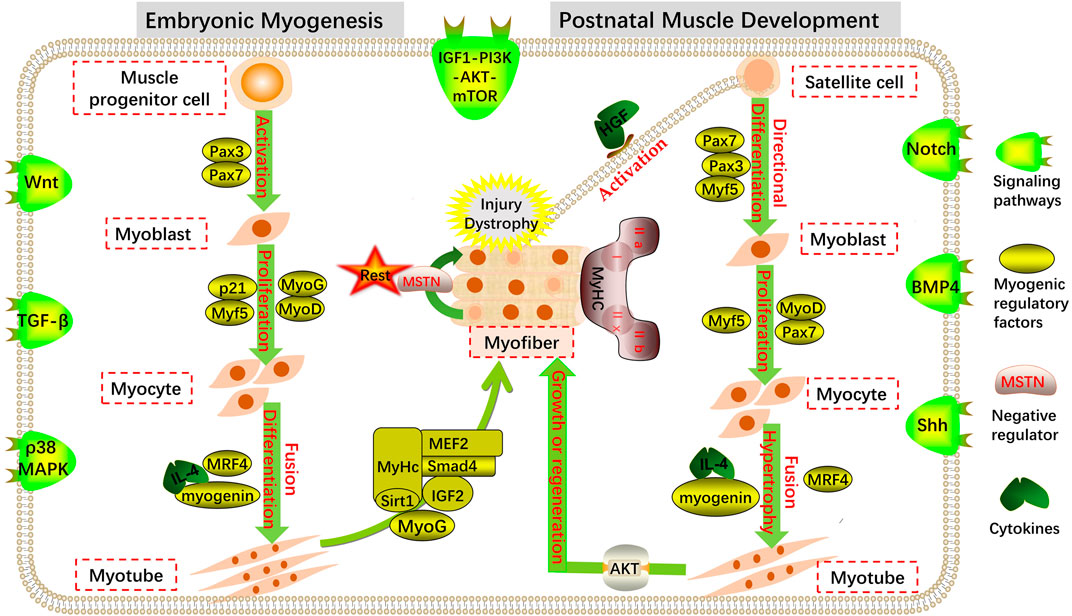
FIGURE 1. Regulation of myogenesis. Coordinated expression of multiple transcription factors and signaling pathways at different stages of embryonic myogenesis and postnatal muscle development maintain the dynamic balance of muscle cell proliferation and differentiation.
During primary muscle development, MPC-expressing Pax3, Pax7, and low Myf5 genes in the myotome stratify from somites and migrate to more distant muscle tissues to form mononuclear myoblast precursors, which are then induced by myogenic determinants to differentiate further into myoblasts (Kassar-Duchossoy et al., 2005; Bajard et al., 2006). During the development of the secondary muscle in the later stage of the embryo, myoblasts proliferate, and the expression of the muscle differentiation factor, myogenin, and other genes increases (Tajbakhsh and Buckingham, 2000). After a series of proliferation, myoblasts withdraw from the proliferation cycle and are irreversible. Then, they undergo terminal differentiation by expressing muscle-specific proteins and different types of cell adhesion factors and fuse to form fusiform multinucleated myotubes containing non-striated myofibrils (Walsh and Perlman, 1997; Sabourin and Rudnicki, 2000). The multinucleated cells already contain myofibrils composed of myosin and actin. When myofibril filaments are arranged in rows to produce transverse striations, the myotubes further develop into mature myofiber-forming skeletal muscles with a relatively perfect structure and function (Buckingham et al., 2003; Buckingham, 2006; Buckingham and Relaix, 2007). The embryonic stage is the main period of myofiber quantity fixation and differentiation in most mammals.
2.2 Postnatal skeletal muscle development
The number of skeletal myofibers after birth is fixed, and muscle growth is mainly derived from increased cell volume (Sassoon, 1993). In this process, the length of myofibers increases with the length and number of sarcomeres, and at the same time, myofibrils increase the diameter of myofibers through multiple divisions. Notably, MuSCs are essential for the developmental growth of myofibers, which can replenish the nuclei of the postnatal myocyte pool, thus contributing to the increase in myonuclei during the early postnatal stage (Pallafacchina et al., 2013; Fukada and Ito, 2021). When some MuSCs divide and proliferate, their nuclei fuse with myofibers, maintaining the relative balance between myocyte nuclei and cytoplasm and promoting myocyte enlargement, thus causing the skeletal muscle to exhibit a growth state (Pallafacchina et al., 2013; Fukada and Ito, 2021). MuSCs are pre-eminent stem cells that maintain the regeneration of postnatal myofibers and are generally in a quiescent state of mitosis (Partridge, 2004; Dhawan and Rando, 2005). However, when mature myofibers are damaged, MuSCs can be activated and re-enter the myogenic pathway, differentiate into myoblasts, and fuse to produce new myofibers after a series of proliferation and differentiation processes (Collins et al., 2005; Dhawan and Rando, 2005; Le Grand and Rudnicki, 2007; Tedesco et al., 2010) (Figure 1).
The growth and development of the postnatal skeletal muscle are accompanied by the transformation and maturation of myofiber types. According to the different expression activities of ATP enzymes in myofibers, myofibers are divided into slow oxidation, fast oxidation, fast glycolysis, and intermediate oxidation types (Schiaffino and Reggiani, 1996). It has been discovered that in slow myofiber activity, unlike in fast myofibers, protein synthesis and degradation rates are higher (Li and Goldberg, 1976). Furthermore, muscle activity is mainly reflected in protein metabolism, and the regulation of muscle mass and myofiber size mainly depends on the balance between protein synthesis and degradation in myofibers (Goll et al., 2008; Yin et al., 2021). When myofibers are stimulated by load or synthetic metabolic hormones after development and maturity, the total protein synthesis rate of the skeletal muscle is greater than the degradation rate, and the size of myofibers increases to promote muscle growth. When an organism is starved, diseased, or stimulated by catabolic hormones, the synthesis rate of skeletal muscle protein is reduced, resulting in the loss of the balanced state of synthesis and degradation and therefore decreased skeletal muscle mass and muscle atrophy (Burd et al., 2010; Goodman, 2014; Mckendry et al., 2021; Sartori et al., 2021).
2.3 The critical role played by muscle cells
Totipotent muscle cells (myocytes) exist throughout skeletal muscle growth and development during the embryonic and postnatal stages of vertebrates, and the myocytes in the somites begin to be active in the early stages of myogenesis. During various stages of animal life, myocytes promote the formation and development of muscle tissue through a series of proliferation and differentiation processes (Figure 1).
2.3.1 Myoblasts
Myoblasts are derived from muscle progenitor cells, which are the basic materials for myofiber formation. The proliferation and differentiation of myoblasts play an important role in muscle development. After muscle injury, stationary MuSCs activate and commit to myoblasts and further proliferate, fuse, and eventually mature into myofibers and restore the contraction ability of injured muscles (Zhang et al., 2018). Therefore, myoblasts are the driving force of skeletal muscle development and an effective tool for treating many diseases with poor prognoses, such as clinical muscular atrophy.
During secondary muscle development, myoblast proliferation and fusion are regulated by a variety of molecular networks. Myoblasts play a decisive role in muscle development under the direct or indirect action of a series of regulatory factors. Owing to the fundamental constitutive role of myoblasts in muscle development, maintenance, and adaptation, the study of regulatory mechanisms of myoblast differentiation has become a key direction in skeletal muscle growth and development, and as an in vitro model, it is widely used in muscle growth, differentiation, migration, apoptosis, and other related studies (Sharples and Stewart, 2011; Cai et al., 2020).
2.3.2 Satellite cells
MuSCs are myoblast precursor cells with the ability to proliferate and self-renew in the later stages of embryonic development and play an essential role in the growth and regeneration of newborn muscle (Chargé and Rudnicki, 2004; Endo, 2007; Relaix and Zammit, 2012; Yin et al., 2013). MuSCs are in the G0 phase and typically remain quiescent. When the skeletal muscle is stimulated externally, the basement membrane secretes the hepatocyte growth factor (HGF), which binds to receptors on the surface of MuSCs and activates quiescent-stage MuSCs (Tatsumi et al., 1998). Furthermore, under the induction of HGF and other related factors, some activated MuSCs re-enter the cell cycle and migrate along the myofiber to the damaged site (Bischoff, 1997). MuSCs proliferate massively after migrating to the injury site and generate sufficient myoblasts, which subsequently proliferate and fuse under the regulation of a series of muscle-specific factors to further develop into mature myofibers for skeletal muscle growth, maintenance, and regeneration (Halevy et al., 2004; Zammit et al., 2004; Rizzi et al., 2012). Inactive MuSCs re-enter the quiescent state and become reserve cells for the next cell cycle (Zammit et al., 2004). The self-renewal and myogenic differentiation of MuSCs maintain a dynamic balance during muscle development, which is essential for the normal function of MuSCs and the maintenance of homeostasis in the internal environment (De Luca et al., 2013).
In summary, the ability of MuSCs to remain quiescent plays a crucial role in the long-term maintenance of a functional stem cell pool during skeletal muscle development and regeneration. Myoblasts and MuSCs are the most important functional cells involved in muscle growth and development. The dynamic balance between their proliferation and differentiation is critical for maintaining the fate of skeletal muscle cells, thus ensuring normal growth and development of the organism.
3 RNA m6A-modified enzyme system
m6A is the most characteristic methylation modification in eukaryotic RNA, in which three key proteins—writers, erasers, and readers—are involved in maintaining the dynamic balance of its modification.
3.1 m6A methyltransferase (writers)
m6A methyltransferase is composed of the m6A–METTL core complex (MAC) and m6A–METTL-associated complex (MACOM) (Lence et al., 2019), and multiple subunits of the complex are co-transcribed and bound to RNA to catalyze methylation. The m6A–METTL complex includes methyltransferase 3 (METTL3) and methyltransferase 14 (METTL14), which can form stable heterodimers in vitro. METTL3 is highly conserved, including the SAM-binding domain and methyltransferase active domain, which catalyzes the formation of m6A (Wang et al., 2016a). METTL14 lacks the catalytic active domain of the enzyme but can promote the binding of METTL3 and RNA. When MELLT14 and METTL3 bind, the methylation catalytic ability of the complex is significantly enhanced (Wang et al., 2016b).
Further studies have shown that some m6A–METTL-associated complexes play an important role in guiding the methylation of specific target sites. Wilms’ tumor 1-associated protein (WTAP) lacks methylase activity, but it can promote m6A deposition by recruiting the METTL3–METTL14 complex to localize in nuclear plaques (Ping et al., 2014). Vir-like m6A methyltransferase associated (VIRMA) can promote the specific deposition of m6A in the 3′UTR (Yue et al., 2018). KIAA1429 also has the catalytic activity of m6A methylation, which affects splicing by regulating the level of m6A (Schwartz et al., 2014). RNA-binding protein 15 (RBM15) and its analog RBM15B contain three RNA recognition motif (RRM) domains that interact with the WTAP–METTL3 complex at specific sites to promote m6A methylation (Yang et al., 2018). CCCH type 13 zinc finger protein (ZC3H13) regulates the nuclear localization of the WTAP–Virilizer–Hakai complex and the self-renewal of mouse embryonic stem cells (ESCs) by promoting m6A methylation (Wen et al., 2018). Although the m6A modification mechanism of the key complex has been defined, future studies may identify additional subunits of the methyltransferase complex that promote or inhibit the occurrence of m6A by identifying specific loci and thereby accurately regulating gene expression.
3.2 m6A demethylase (erasers)
The m6A modification is a dynamic and reversible regulatory process whose activity can be counteracted by demethylases, fat mass and obesity-associated (FTO) gene, and ALKBH5 in gene regulation. The FTO gene is the first protease discovered to have m6A demethylation activity and belongs to the Fe (II) and α-KG (ketoglutaric acid)-dependent ALKB dioxygenase family. FTO exhibits catalytic activity both in vitro and in vivo. During demethylation, m6A is catalyzed to form N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (fm6A), which are extremely unstable and eventually decompose into adenine (A) (Jia et al., 2012; Fu et al., 2013). In the intracranial glioma model of nude mice, mice bearing FTO shRNA-1-infected U251 cells had significantly shorter survival times, and when mice were co-infected with FTO-Mut, they survived longer. This suggests that FTO inhibits the in vivo progression of gliomas (Tao et al., 2020). An ovariectomized mouse model demonstrated that FTO can promote osteoporosis by demethylating the osteogenic marker, Runx2 mRNA (Wang et al., 2021). In C57BL/6N mice, FTO deficiency results in weight loss, marked reduction in white adipose tissue, and promotion of the conversion of white adipocytes to brown or beige adipocytes (Ronkainen et al., 2015). Moreover, FTO-mediated mRNA m6A demethylation can affect preadipocyte differentiation and lipid deposition by regulating the expression of fat-related genes, such as C/EBPβ, PPARγ, and ANGPTL4, and plays an important regulatory role in lipid metabolism and lipid disorders (Yang et al., 2022b). It has been shown that inhibiting the expression of FTO can considerably increase the total m6A level of polyadenylated RNA (Jia et al., 2012). Additionally, FTO also uses N6,2′-O-dimethyladenosine (m6Am) on single-stranded RNA as a substrate and has higher demethylase activity (Mauer et al., 2017). ALKBH5, also derived from the ALKB protein family, is the second m6A demethylase discovered, and its catalytic activity is similar to that of FTO; however, it can directly demethylate m6A to A through one reaction. ALKBH5 plays a role in specific sequences, showing a preference for m6A in common sequences and can completely remove m6A methylation modifications from single-stranded RNA (Zheng et al., 2013).
3.3 m6A-binding protein (readers)
Following m6A modification in eukaryotes, respective downstream biological functions require specific recognition by reader proteins to proceed normally. Currently known m6A-binding proteins include the YT521-B homology (YTH) family, IGF2BP, and HNRNP.
The YTH domain family members mainly include YTHDF1/2/3 and YTHDC1/2, which contain a YTH domain that can selectively bind to the m6A site on RNA (Theler et al., 2014; Hsu et al., 2017). YTHDF2 has a strong binding ability, which can identify m6A sites and regulate the degradation of modified genes (Wang et al., 2014a). Studies have shown that YTHDF2 can enter the nucleus during heat stress stimulation, prevent FTO from de-modifying m6A in the 5′UTR, and promote translation in a non-hat-dependent manner (Zhou et al., 2015). YTHDF1 can interact with the translation initiation factor, eIF, to enhance the translation efficiency of m6A modified genes by promoting ribosome enrichment of methylated genes (Wang et al., 2015). YTHDF3 is the first reader protein that binds to nuclear-exported m6A-modified RNA, assisting YTHDF1 and YTHDF2 in regulating the degradation or translation of target genes, respectively (Li et al., 2017; Shi et al., 2017). However, recent studies have found that YTHDF1, YTHDF2, and YTHDF3 directly affect mRNA degradation in an m6A-dependent manner but do not participate in the translation regulation of mRNA (Lasman et al., 2020).
YTHDC1, located in the nucleus, is highly conserved, which can selectively recruit the pre-mRNA splicing factor, SRSF3, and promote its binding to m6A-modified mRNA. Furthermore, it inhibits the binding of SRSF10 and mRNA, promotes the retention of exons modified by m6A, and regulates mRNA splicing (Xiao et al., 2016). Additionally, YTHDC1 interacts with SRSF3 and nuclear RNA output factor 1 to promote the nuclear output of m6A-modified mRNA, which plays an important role in the metabolic regulation of mRNA (Roundtree et al., 2017b). YTHDC2 preferentially binds to m6A in a common motif to improve translation efficiency and reduce the abundance of its target mRNA (Yang et al., 2018). The insulin-like growth factor-2 mRNA-binding protein (IGF2BP) family consists of three homologous coding genes, IGF2BP1, IGF2BP2, and IGF2BP3, which contain an RNA recognition domain and a ribonucleoprotein K domain. IGF2BPs promote the stability and storage of target genes in an m6A-dependent manner through the ribonucleoprotein K domain (Huang et al., 2018). Finally, a family of HNRNP proteins mediates the “m6A-switch” mechanism, and its member, HNRNPA2B1, can directly bind to m6A and cooperate with METTL3 to regulate alternative splicing events and primary microRNA processing. The other two members, HNRNPC11 and HNRNPG, do not directly bind to m6A but regulate the processing of RNA transcripts containing m6A (Yang et al., 2018).
4 Regulation of skeletal muscle growth and development by RNA m6A modifications
m6A modification regulates various biological activities, such as RNA processing and metabolism in eukaryotes. Its dynamic and reversible mode of action may affect gene expression and cell fate by regulating various RNA-related cellular signaling pathways. Given the role of m6A modification in gene expression and the regulation of functional cellular mechanisms, there is emerging evidence that m6A modification and its regulators play an essential role in the growth and development of the skeletal muscle (Kudou et al., 2017; Wang et al., 2017; Gheller et al., 2020; Zhang et al., 2020; Deng et al., 2021a; Li et al., 2021; Petrosino et al., 2022) (Figure 2). Therefore, this review focused on the cascade regulation of m6A modification during embryonic and postnatal skeletal muscle development to provide evidence for exploring a new mechanism of m6A regulation of myogenesis.
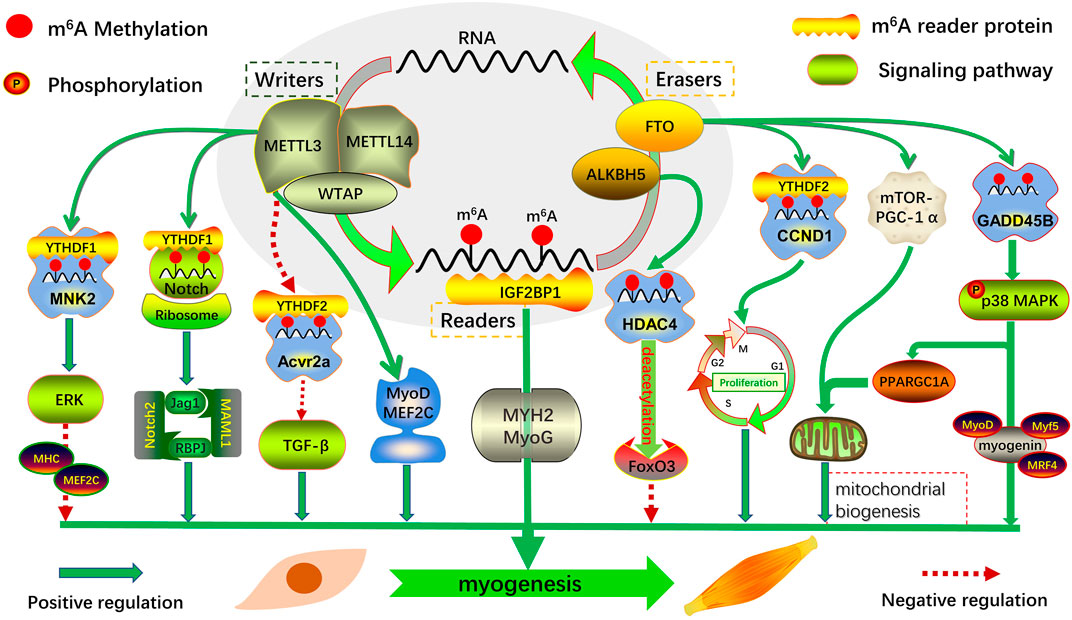
FIGURE 2. Regulatory role of m6A modification and its regulators in skeletal muscle myogenesis. Myogenic genes that mediate myogenesis are regulated transcriptionally and post-transcriptionally by m6A-related proteins in multiple ways: regulating the transcription of key factors in an m6A-dependent manner; directly regulating the expression of related transcription factors; and coordinating with other transcription factors or post-transcriptional modifications.
4.1 Embryonic m6A methylation
In recent years, many studies have shown that m6A plays an important role in regulating the embryonic development of eukaryotes. During early embryonic development in zebrafish, m6A modification promotes maternal–zygote transition through YTHDF2-dependent maternal mRNA clearance (Zhao et al., 2017). In addition, m6A participates in the metabolism of mRNA in embryonic stem cells, maintains cell self-renewal, and regulates the state transition and pluripotency of embryonic cells as a determining factor of cell fate at the transcriptional level (Batista et al., 2014).
The regulatory mechanism of m6A in embryonic cell fate, which exists in a tissue-specific form at different stages of embryonic development, has been continuously explored. It maintains the dynamic balance between biological processes by regulating functional gene transcription and post-transcriptional expression. Determining the number of skeletal muscle fibers in animal embryos involves complex regulatory mechanisms, and the expression of key muscle-specific transcription factors is precisely regulated by m6A methylation (Figure 2). Therefore, in-depth exploration of the potential mechanism of m6A in embryonic skeletal muscle development is of key interest in developmental muscle biology.
Analysis of two key stages of pectoral muscle development in Dingan goose embryos revealed a negative correlation between m6A methylation and gene expression. Moreover, most m6A-modified differentially methylated genes were significantly enriched in muscle-related pathways, such as the Wnt, mTOR, and FoxO signaling pathways (Xu et al., 2021). Combined with miRNA-seq, potential m6A-miRNA-PDK3 was screened, which revealed the key role of m6A-modified miRNA in muscle growth and development of Dingan goose embryos (Xu et al., 2021). Analysis of m6A distribution in goat muscles at two key developmental stages revealed that the m6A peak in the longissimus at embryonic day 75 was significantly higher than that in the newborn stage, and m6A-modified genes were mainly enriched in actin binding, myotubular differentiation, MAPK, Wnt, and other signaling pathways related to skeletal muscle development (Deng et al., 2021a). During the differentiation of goat primary myoblasts, FTO expression was negatively correlated with global m6A levels. Following FTO knockdown in myoblasts, m6A levels of GADD45B mRNA were increased, whereas its protein expression and phosphorylation levels of p38 MAPK were significantly decreased, and myotube formation was attenuated. This demonstrated that the FTO-mediated m6A demethylates GADD45B and activates the p38 MAPK pathway, which in turn promotes myogenic differentiation of goat skeletal muscle (Deng et al., 2021a).
The expression of IGF2BP1 is continuously downregulated in the six stages of skeletal muscle growth and development in the embryonic pig stage. Combined with RIP-seq, m6A-modified myogenic marker genes, MYH2 and MyoG, were identified as target genes of IGF2BP1 (Zhang et al., 2020). Loss-of-function experiments were performed in myoblasts, and it was found that knocking down IGF2BP1 significantly downregulated MYH2 and MyoG mRNA expression and significantly inhibited myotube formation. The same phenotypic changes were observed with METTL14 knockdown (Zhang et al., 2020), demonstrating that m6A is a key epigenetic factor in embryonic myogenesis. These results suggest that dynamic changes in m6A modification levels during embryonic skeletal muscle development play an essential role in regulating myogenesis.
4.2 Postnatal m6A methylation
Studies have found that m6A methylation has commonality and discrepancy in regulatory functions in different developmental stages of the organism, and it also plays an essential role in postnatal skeletal muscle development and muscle regeneration (Figure 2). Through whole-transcriptome m6A methylation map analysis of the muscle tissue of wild boar and Landrace and Rongchang pigs, it was found that most of the nuclear-related genes containing m6A encode transcription factors, indicating that m6A modification is involved in transcriptional regulation, and the two coordinately regulate gene expression (Tao et al., 2017). This is consistent with the results of a recent study on m6A profiles in the longissimus dorsi of Landrace and Jinhua pigs (Jiang et al., 2019), revealing the potential biological role of m6A modification in regulating muscle growth and development.
4.2.1 The critical role of m6A modulators in myogenic differentiation
A previous study found that after METTL3 knockdown in proliferating C2C12 cells, the overall levels of m6A modification decreased, resulting in premature differentiation of myoblasts, thus demonstrating that METTL3-mediated m6A methylation is an important regulator of myoblast state transition (Gheller et al., 2020). Both the mRNA and protein levels of METTL3/14 and WTAP were significantly downregulated during C2C12 cell differentiation and were negatively correlated with the expression of MHC and MEF2C. Consistent with in vitro findings, the expression of METTL3/14 is significantly downregulated in mouse embryonic hindlimb muscles during skeletal muscle growth and development (Xie et al., 2021a). Through genome-wide expression and gain/loss of function analysis, METTL3/14-mediated m6A methylation was found to inhibit myogenic differentiation by enhancing MNK2-ERK signaling (Xie et al., 2021a). This was consistent with the findings of Liang et al. (2021) and Gheller et al. (2020), suggesting that METTL3/14-mediated m6A methylation has a common inhibitory effect on myogenesis. However, METTL3 knockdown in proliferative C2C12 cells significantly downregulated MyoD mRNA expression and inhibited myoblast differentiation, and its m6A-mediated modification stabilized MyoD mRNA levels by promoting mRNA processing, thereby maintaining myoblast myogenic potential (Kudou et al., 2017). This is consistent with the findings of Zhang et al. (2020), who showed that METTL14 knockdown downregulates MyoD mRNA expression and significantly inhibits myotube formation.
Investigating METTL3/14 promotion or inhibition of myoblast differentiation, we found that m6A modification may mediate the expression of myogenic transcription factors by regulating specific signaling axes, thereby inducing myogenic differentiation. In contrast, the upstream of m6A-modified myogenic transcription factors may be regulated by other transcriptional elements in multiple ways, leading to the opposite result of myogenic differentiation. It has been suggested that m6A modification in myogenesis has multiple functions, but the current study only analyzed the potential mechanism of m6A modification-mediated differentiation induced by myogenic transcription factors. Future research needs to explore the key upstream/downstream factors co-regulated by RNA m6A methylation and myogenic transcription factors to explain the specific functional mechanism of m6A modification in regulating myogenic differentiation.
FTO-mediated m6A plays an important role in fat mass and lipogenesis (Song et al., 2020). Considering that the skeletal muscle participates in the body’s metabolic regulation and is similar to fat-related functions, it can be considered that FTO has an important regulatory effect on its myogenic differentiation. FTO expression is increased during the differentiation of mouse myoblasts into myotubes, whereas FTO silencing inhibits differentiation and impairs skeletal muscle development in endogenous FTO-null mice (Wang et al., 2017). Further exploration revealed that FTO-mediated m6A modification promoted myogenic differentiation through the mTOR-PGC-1α–mitochondrial axis. Interestingly, FTO overexpression, in vitro, does not significantly promote myoblast differentiation, presumably because of the high abundance of endogenous FTO expression, which is sufficient to support muscle differentiation (Wang et al., 2017). This is similar to the results of Church et al. (2010), who found that FTO overexpression did not increase lean mass in male mice.
Furthermore, the potential role of m6A modification in goat skeletal muscle was explored, and it was found that FTO-mediated m6A demethylation activity upregulates CCND1 expression in a YTHDF2-dependent manner and that silencing of FTO can also induce autophagy during myogenic differentiation (Deng et al., 2021b). Moreover, the global m6A modification in goat embryonic skeletal muscle was significantly higher than that in newborn fetuses. Functional studies have shown that FTO-mediated m6A modification can promote the expression of GADD45B and myogenic differentiation by activating the p38 MAPK pathway (Deng et al., 2021a).
These results provide a new insight that FTO promotes myogenesis by regulating the expression of related genes and may serve as a new target for myogenic differentiation.
4.2.2 m6A methylation regulates skeletal muscle homeostasis and regeneration
MuSCs are required for maintaining skeletal muscle homeostasis and regeneration after injury. This study found that the proliferative activity of MuSCs was reduced after METTL3 knockdown, resulting in a significant decrease in both m6A modification and protein expression levels of key genes of the Notch signaling pathway. Furthermore, YTHDF1 is positively correlated with the mRNA translation efficiency of Notch signaling pathway components, revealing a novel post-transcriptional mechanism whereby the METTL3-m6A-YTHDF1 axis further controls MuSC fate and muscle regeneration by regulating the Notch signaling pathway (Liang et al., 2021). As a key factor in the MAPK signaling pathway, the protein kinase, MNK2, can target the phosphorylation-activated ERK signaling pathway and maintain muscle homeostasis (Hu et al., 2012; Maimon et al., 2014).
By establishing a mouse muscle injury regeneration model, it was found that METTL3/14-MNK2 could activate MuSCs and promote their proliferation in the early stages of muscle regeneration, thereby controlling ERK signaling (Xie et al., 2021a). These results suggest that the METTL3/14-m6A-MNK2-ERK signaling axis is required to regulate early muscle regeneration after acute injury. In the establishment of a BaCl2-induced mouse skeletal muscle injury model, the global m6A levels in muscle tissue were significantly increased 3 days after injury. In addition, m6A also plays an important regulatory function during the state transition of MuSCs (Gheller et al., 2020), indicating that m6A is a key epigenetic modifier of skeletal muscle regeneration.
4.2.3 m6A methylation regulates skeletal muscle hypertrophic response
Maintaining skeletal muscle mass is critical for an organism’s health. Evaluation of the m6A modification signature of skeletal muscle hypertrophic growth in mice with mechanical overload revealed that global m6A levels and METTL3 expression were significantly increased in overloaded muscles (Petrosino et al., 2022). The myofiber-specific METTL3 mouse model was constructed using gain-of-function and loss-of-function experiments, and further validation revealed that m6A content and the myofiber cross-sectional area increased in muscles overexpressing METTL3. Moreover, METTL3 can regulate the post-transcriptional process of the myostatin pathway, and its mediated m6A methylation affects TGF-β superfamily signaling by inhibiting the translation of the activin receptor, Acvr2a mRNA, thereby promoting hypertrophic growth of the skeletal muscle (Petrosino et al., 2022). These results reveal a novel post-transcriptional mechanism to regulate muscle gene-specific expression, namely, that m6A modification regulates muscle growth through the translation of activin receptors and is required to maintain muscle mass and function in vivo.
4.2.4 m6A may be an effective modulator for the treatment of muscle-related diseases
MuSC transition from quiescent to activated and proliferative to differentiated states after skeletal muscle injury and multiple m6A-modifying genes are associated with MuSC function during the differentiation process. When METTL3 is knocked down in primary murine MuSCs, the proliferation of MuSCs is slowed and the engraftment ability of their primary transplantation is enhanced, but their serial transplantation ability is lacking (Gheller et al., 2020).
In addition to FTO, ALKBH5 may also be involved in regulating mRNA processing and metabolism related to skeletal muscle growth and development. ALKBH5-mediated m6A modification plays an important role in FoxO3-dependent neurogenic muscle atrophy (Liu et al., 2022). To verify the specific mechanism, m6A-seq and Co-IP combined with loss-of-function experiments were performed, and it was found that ALKBH5 activates FoxO3 signaling in an m6A-HDAC4-dependent manner in denervated muscles, resulting in loss of skeletal muscle mass and denervation muscle atrophy (Liu et al., 2022). These results suggest that ALKBH5 may be a potential therapeutic target for the treatment of neurogenic muscle atrophy.
In conclusion, m6A is an essential epigenetic regulator of MuSC function; further work is needed to determine the fate of specific m6A-modified proteins based on their binding activity to muscle mRNA, which is expected to be better applied to treat muscle atrophy and other related diseases.
5 Transcriptional and post-transcriptional regulation of myogenesis
Transcriptional and post-transcriptional regulation at the RNA level can rapidly respond to the regulation of the corresponding mechanisms when biological functions occur. In eukaryotic cells, mRNA efficiently initiates protein synthesis by adding a 5′ 7-methyl-guanosine cap and a 3′ poly(A) tail. However, mRNA-directed protein synthesis is blocked by sequence-specific RNA-binding proteins (Sonenberg and Hinnebusch, 2009; Nwokoye et al., 2021). This way of promotion or repression reveals the importance of RNA transcriptional regulation. Early studies have found that one of the main functions of m6A in mammalian cells is to mediate mRNA degradation, suggesting a possible negative correlation between m6A methylation and mRNA stability and transcription levels (Wang et al., 2014a; Wang et al., 2014b; Liu et al., 2014; Ping et al., 2014). Like DNA methylation and histone modification, RNA methylation, an important post-transcriptional epigenetic modification, plays an important role in regulating gene expression in specific cell types (Heck and Wilusz, 2019).
Therefore, future studies are needed to elucidate the synergistic regulation of m6A and myogenic transcription factors to explain the cascade response of transcriptional and post-transcriptional regulation during myogenesis.
5.1 Interactions between m6A-related proteins and myogenic transcription factors
Studies have reported a critical role of MyoD methylation modification during myogenic differentiation. During C2C12 cell proliferation, high m6A levels in the 5′ UTR can promote the efficient processing of MyoD mRNA and actively maintain its stability. Furthermore, MyoD mRNA levels and myotube formation are significantly inhibited when METTL3 is knocked down (Kudou et al., 2017). Through the analysis of m6A at six prenatal stages in pigs, it was identified that the m6A reader, IGF2BP1, is continuously downregulated, which can regulate mRNA stability and translation (Huang et al., 2018). The same phenotypic changes were observed after knockdown of METTL14 and IGF2BP1 in C2C12 cells, inhibiting myoblast differentiation and significantly downregulating MyHC, MyoD, and MyoG expression (Zhang et al., 2020). These results suggest that the m6A modification-mediated key transcription factor, MyoD, is a positive regulator of skeletal muscle differentiation.
The Notch signaling pathway plays an important role in regulating MuSCs and muscle regeneration and is necessary for MuSC maintenance, activation, proliferation, and differentiation (Brack et al., 2008; Buas and Kadesch, 2010; Bjornson et al., 2012; Gerli et al., 2019). It was found that the mRNA molecules of the receptor (Notch2), transcription factor (RBPJ), and activator (MAML1) of the Notch signaling pathway in myoblasts are also regulated by m6A modification (Liang et al., 2021). Through in vivo and in vitro validation of m6A MeRIP-seq and cellular functions, it was found that METTL3-mediated m6A modification significantly inhibited the translation efficiency of Notch signaling pathway components, thereby promoting MuSC and muscle regeneration (Liang et al., 2021). These results revealed a novel post-transcriptional mechanism for m6A regulation of mRNA methylation of key transcription factors in MuSC fate and muscle regeneration.
MEF2C, a member of the myocyte enhancer factor 2 (MEF2) family, induces the expression of muscle-specific genes, mainly by binding to basic helix–loop–helix proteins in myogenic regulatory factors. It can also bind to the promoter and enhancer regions of transcription factors that assist in regulating myoblast differentiation during muscle development (Molkentin et al., 1995; Black and Olson, 1998; Kim et al., 2008). Recent studies have revealed that m6A levels of MEF2C mRNA are significantly increased during bovine myoblast differentiation, and its expression is post-transcriptionally regulated by m6A modifications. Through gain-of-function and loss-of-function analyses, METTL3 was found to regulate myogenic differentiation by promoting the translation of MEF2C mRNA in an m6A-YTHDF1-dependent manner (Yang et al., 2022a). Furthermore, it is worth noting that both the mRNA and protein levels of METTL3 were significantly increased in MEF2C-overexpressing myoblasts. Genomic analysis and ChIP-qPCR demonstrated that MEF2C binds directly to the METTL3 promoter as a transcription factor to promote its expression (Yang et al., 2022a). This positive feedback loop during myogenic differentiation explains the transcriptional and post-transcriptional cascade regulatory mechanisms.
Therefore, an in-depth study of the novel mechanisms of transcription factors in coordinating gene transcription and RNA m6A modification is required to shed more light on the functional mechanisms of skeletal muscle growth and development.
5.2 Mechanism of action of m6A-related proteins and non-coding RNAs during myogenesis
Myogenesis is a highly coordinated process involving multiple mechanisms, the programmed occurrence of which is controlled by specific genes and epigenetic modifications. An increasing number of studies have shown that non-coding RNAs (ncRNAs) or m6A-mediated transcriptional/post-transcriptional regulation play an important role in gene expression in skeletal muscle development. Moreover, m6A has been identified as a key regulatory factor that affects the function of ncRNAs, thus participating in body growth and development. Therefore, we have summarized recent studies to elucidate the molecular mechanisms of m6A-regulated miRNAs and long non-coding RNAs (lncRNAs) in myogenesis.
5.2.1 Methyltransferase 3 regulates muscle-specific miRNAs through transcriptional and post-transcriptional regulation
miRNAs are a class of highly conserved small RNA molecules that participate in biological processes by degrading target genes or inhibiting post-transcriptional translation. During miRNA biogenesis, METTL3-mediated m6A methylation markers can promote the binding and processing of primary miRNA to DGCR8 and promote the maturation of miRNA in a global and non-cell-type-specific manner (Alarcon et al., 2015b). In addition, the m6A-labeled nuclear reader and effector, HNRNPA2B1, can interact with the DGCR8 protein to promote the processing of primary miRNAs (Alarcon et al., 2015a). These results are consistent with those of Alarcon et al. (2015b). This study found that METTL3 overexpression significantly downregulated the expression of muscle-specific miR-1a, miR-133a, miR-133b, and miR-206 in differentiated C2C12 cells and a mouse model of muscle injury regeneration. Combined with the immunoprecipitation results, it was demonstrated that METTL3 inhibited muscle-specific miRNAs through m6A modification of primary miRNAs during skeletal muscle differentiation (Diao et al., 2021). These results are contrary to the previous findings of Alarcon et al. (2015b).
Therefore, Diao et al. (2021) further explored the complex mechanism by which METTL3 inhibits muscle-specific miRNAs. They found that in differentiated C2C12 cells, METTL3 overexpression significantly suppressed the expression of myogenic transcription factors, MEF2A/C and SRF, whereas the expression of epigenetic regulators, HDAC1, HDAC4, and HDAC8, was significantly upregulated. The METTL3-overexpressing C2C12 cells were then subjected to MEF2C overexpression and HDAC inhibitor TSA treatment, and it was noted that the expression of miR-1a, miR-133a, miR-133b, and miR-206 was significantly upregulated (Diao et al., 2021). These results suggest that METTL3 represses muscle-specific miRNA expression by repressing MEF2C and promoting HDAC family epigenetic regulators. Taken together, METTL3 can repress muscle-specific miRNAs at the transcriptional and post-transcriptional levels and plays an important role in muscle function maintenance and anti-differentiation.
5.2.2 The critical role of methyltransferase 3-mediated lncRNA in myogenesis
LncRNAs are mainly involved in regulating gene expression at the transcriptional and post-transcriptional levels. It has been reported that lncRNA can directly induce the binding of chromatin remodeling proteins to target genes and change histone modification or DNA methylation status according to specific genome sites, thus regulating the expression of functional genes (Chen and Xue, 2016). In addition to the aforementioned epigenetic modification, m6A-methylated lncRNAs have been well-characterized. For example, METTL3 binds to RBM15 and RBM15B in a WTAP-dependent manner and promotes lncRNA XIST-mediated gene silencing through m6A-YTHDC1 (Patil et al., 2016). Furthermore, lncRNAs are involved in a variety of biological processes, and their post-transcriptional regulation plays an essential role in myogenesis. Recent studies have found that during myogenesis, m6A methylation levels of lncRNA are positively correlated with the transcriptional abundance of lncRNA, and m6A methylation modifies lncRNA to positively or negatively regulate its adjacent mRNA. Functional verification showed that METTL3-mediated m6A-lncRNA Brip1os is involved in muscle differentiation by negatively regulating the expression of Tbx2 mRNA (Xie et al., 2021b), revealing a novel mechanism of post-transcriptional regulation during skeletal muscle development.
6 Summary and outlook
The discovery of RNA m6A modifications has greatly broadened our understanding of the mechanisms underlying gene expression regulation. In recent years, many studies have confirmed that m6A modifications are the important components of myogenic regulatory networks. Based on this, we summarized current research on m6A modifications in skeletal muscle growth and development and highlighted the mechanisms of transcriptional and post-transcriptional regulation of m6A modifications with myogenic transcription factors, which may help to better explore the synergistic regulation at different levels during myogenesis. In addition, other myogenic transcription factors are expected to be modified by m6A or regulate the expression of m6A-related proteins in the form of transcription factors, and the interaction of multiple factors uncovers new transcriptional regulatory mechanisms in myogenesis.
The state transition of MuSCs and myoblasts is accompanied by dramatic changes in transcriptional regulation. Transcriptional changes affecting the muscle state are influenced by various aspects of ncRNAs (miRNA, lncRNA, and circRNA), DNA methylation, histone modifications, and chromatin remodeling. The current study found that miRNAs, lncRNAs, and circRNAs have m6A modifications and that m6A exerts facilitative and inhibitory effects by altering the expression of targeted ncRNAs. However, it remains unclear if ncRNAs can participate in myogenic differentiation by regulating the stability of m6A-related enzymes, and the biological mechanisms by which the two crosstalk during skeletal muscle development are still unknown. Multiomics should be integrated to explore the interactions between m6A and other epigenetic modifications in skeletal muscle development and elucidate the functional signals, regulating skeletal muscle development.
The role of m6A-regulated RNA processing and metabolism in myogenesis requires further investigation. In summary, METTL3/14 was found to promote or inhibit myoblast differentiation, although the cause of this heterogeneity has not been identified. Therefore, future studies should exclude analytical errors due to tissue and cell heterogeneity, apply single-cell transcriptome technology to functional studies of myoblasts, and comprehensively explore the specific functional mechanisms of the synergistic regulation of RNA m6A methylation and myogenic transcription factors.
Moreover, we should focus on the special mechanism between m6A and functional cellular markers and look for “classic” drugs targeting new-type m6A-related enzymes to provide new targets and ideas for the early prevention of skeletal muscle aging and treatment of muscle metabolic diseases.
Author contributions
JZ and YG conceived and managed the project. BY and JL wrote the manuscript. TM, XF, and RM revised the manuscript. All the authors approved the final version of the manuscript.
Funding
This study was supported by the Ningxia Hui Autonomous Region Key R and D Program (2022BBF02034).
Acknowledgments
We appreciate the help and support from all participants who took part in the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., Tavazoie, S. F., et al. (2015a). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Bajard, L., Relaix, F., Lagha, M., Rocancourt, D., Daubas, P., Buckingham, M. E., et al. (2006). A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes. Dev. 20 (17), 2450–2464. doi:10.1101/gad.382806
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell. Stem Cell. 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Biressi, S., Molinaro, M., and Cossu, G. (2007). Cellular heterogeneity during vertebrate skeletal muscle development. Dev. Biol. 308 (2), 281–293. doi:10.1016/j.ydbio.2007.06.006
Bischoff, R. (1997). Chemotaxis of skeletal muscle satellite cells, Dev. Dyn. 208. Saint Louis, MO: COM, 5052–5115. doi:10.1002/(SICI)1097-0177(199704)
Bjornson, C. R., Cheung, T. H., Liu, L., Tripathi, P. V., Steeper, K. M., Rando, T. A., et al. (2012). Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30 (2), 232–242. doi:10.1002/stem.773
Black, B. L., and Olson, E. N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol. 14, 167–196. doi:10.1146/annurev.cellbio.14.1.167
Brack, A. S., Conboy, I. M., Conboy, M. J., Shen, J., and Rando, T. A. (2008). A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell. Stem Cell. 2 (1), 50–59. doi:10.1016/j.stem.2007.10.006
Buas, M. F., and Kadesch, T. (2010). Regulation of skeletal myogenesis by Notch. Exp. Cell. Res. 316 (18), 3028–3033. doi:10.1016/j.yexcr.2010.05.002
Buckingham, M., Bajard, L., Chang, T., Daubas, P., Hadchouel, J., Meilhac, S., et al. (2003). the formation of skeletal muscle: From somite to limb. J. Anat. 202, 59–68. doi:10.1046/j.1469-7580.2003.00139.x
Buckingham, M. (2006). Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 16 (5), 525–532. doi:10.1016/j.gde.2006.08.008
Buckingham, M., and Relaix, F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell. Dev. Biol. 23, 645–673. doi:10.1146/annurev.cellbio.23.090506.123438
Buckingham, M. (2001). Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11, 440–448. doi:10.1016/s0959-437x(00)00215-x
Buckingham, M., and Vincent, S. D. (2009). Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 19 (5), 444–453. doi:10.1016/j.gde.2009.08.001
Burd, N. A., Holwerda, A. M., Selby, K. C., West, D. W., Staples, A. W., Cain, N. E., et al. (2010). Resistance exercise volume affects myofibrillar protein synthesis and anabolic signalling molecule phosphorylation in young men. J. Physiol. 588 (Pt 16), 3119–3130. doi:10.1113/jphysiol.2010.192856
Cai, S., Zhu, Q., Guo, C., Yuan, R., Zhang, X., Nie, Y., et al. (2020). MLL1 promotes myogenesis by epigenetically regulating Myf5. Cell. Prolif. 53 (2), e12744. doi:10.1111/cpr.12744
Chargé, S. B., and Rudnicki, M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238. doi:10.1152/physrev.00019.2003
Chen, J., and Xue, Y. (2016). Emerging roles of non-coding RNAs in epigenetic regulation. Sci. China. Life Sci. 59 (3), 227–235. doi:10.1007/s11427-016-5010-0
Church, C., Moir, L., McMurray, F., Girard, C., Banks, G. T., Teboul, L., et al. (2010). Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42 (12), 1086–1092. doi:10.1038/ng.713
Collins, C. A., Olsen, I., Zammit, P. S., Heslop, L., Petrie, A., Partridge, T. A., et al. (2005). Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 122 (2), 289–301. doi:10.1016/j.cell.2005.05.010
Cong, X. X., Gao, X. K., Rao, X. S., Wen, J., Liu, X. C., Shi, Y. P., et al. (2020). Rab5a activates IRS1 to coordinate IGF-AKT-mTOR signaling and myoblast differentiation during muscle regeneration. Cell. Death Differ. 27 (8), 2344–2362. doi:10.1038/s41418-020-0508-1
Coots, R. A., Liu, X. M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). M(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell. 68 (3), 504–514. e507. doi:10.1016/j.molcel.2017.10.002
De Luca, G., Ferretti, R., Bruschi, M., Mezzaroma, E., and Caruso, M. (2013). Cyclin D3 critically regulates the balance between self-renewal and differentiation in skeletal muscle stem cells. Stem Cells 31 (11), 2478–2491. doi:10.1002/stem.1487
Deng, K., Fan, Y., Liang, Y., Cai, Y., Zhang, G., Deng, M., et al. (2021a). FTO-mediated demethylation of GADD45B promotes myogenesis through the activation of p38 MAPK pathway. Mol. Ther. Nucleic Acids 26, 34–48. doi:10.1016/j.omtn.2021.06.013
Deng, K., Zhang, Z., Ren, C., Liang, Y., Gao, X., Fan, Y., et al. (2021b). FTO regulates myoblast proliferation by controlling CCND1 expression in an m(6)A-YTHDF2-dependent manner. Exp. Cell. Res. 401 (2), 112524. doi:10.1016/j.yexcr.2021.112524
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., Chen, J., et al. (2018). RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell. Res. 28 (5), 507–517. doi:10.1038/s41422-018-0034-6
Dhawan, J., and Rando, T. A. (2005). Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell. Biol. 15 (12), 666–673. doi:10.1016/j.tcb.2005.10.007
Diao, L. T., Xie, S. J., Lei, H., Qiu, X. S., Huang, M. C., Tao, S., et al. (2021). METTL3 regulates skeletal muscle specific miRNAs at both transcriptional and post-transcriptional levels. Biochem. Biophys. Res. Commun. 552, 52–58. doi:10.1016/j.bbrc.2021.03.035
Dorn, L. E., Lasman, L., Chen, J., Xu, X. Y., Hund, T. J., Medvedovic, M., et al. (2019). The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation 139, 533–545. doi:10.1161/CIRCULATIONAHA.118.036146
Endo, T. (2007). Stem cells and plasticity of skeletal muscle cell differentiation: Potential application to cell therapy for degenerative muscular diseases. Regen. Med. 2, 243–256. doi:10.2217/17460751.2.3.243
Fazi, F., and Fatica, A. (2019). Interplay between N(6)-methyladenosine (m(6)A) and non-coding RNAs in cell development and cancer. Front. Cell. Dev. Biol. 7, 116. doi:10.3389/fcell.2019.00116
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Science 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4, 1798. doi:10.1038/ncomms2822
Fukada, S. I., and Ito, N. (2021). Regulation of muscle hypertrophy: Involvement of the Akt-independent pathway and satellite cells in muscle hypertrophy. Exp. Cell. Res. 409 (2), 112907. doi:10.1016/j.yexcr.2021.112907
Fustin, J. M., Doi, M., Yamaguchi, Y., Hida, H., Nishimura, S., Yoshida, M., et al. (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 155 (4), 793–806. doi:10.1016/j.cell.2013.10.026
Gerli, M. F. M., Moyle, L. A., Benedetti, S., Ferrari, G., Ucuncu, E., Ragazzi, M., et al. (2019). Combined Notch and PDGF signaling enhances migration and expression of stem cell markers while inducing perivascular cell features in muscle satellite cells. Stem Cell. Rep. 12 (3), 461–473. doi:10.1016/j.stemcr.2019.01.007
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. doi:10.1126/science.1261417
Gheller, B. J., Blum, J. E., Fong, E. H. H., Malysheva, O. V., Cosgrove, B. D., Thalacker-Mercer, A. E., et al. (2020). A defined N6-methyladenosine (m(6)A) profile conferred by METTL3 regulates muscle stem cell/myoblast state transitions. Cell. Death Discov. 6 (1), 95. doi:10.1038/s41420-020-00328-5
Goll, D. E., Neti, G., Mares, S. W., and Thompson, V. F. (2008). Myofibrillar protein turnover: The proteasome and the calpains. J. Anim. Sci. 86 (14 Suppl. l), E19–E35. doi:10.2527/jas.2007-0395
Goodman, C. A. (2014). The role of mTORC1 in regulating protein synthesis and skeletal muscle mass in response to various mechanical stimuli. Rev. Physiol. Biochem. Pharmacol. 166, 43–95. doi:10.1007/112_2013_17
Halevy, O., Piestun, Y., Allouh, M. Z., Rosser, B. W., Rinkevich, Y., Reshef, R., et al. (2004). Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev. Dyn. 231 (3), 489–502. doi:10.1002/dvdy.20151
Han, J., Wang, J. Z., Yang, X., Yu, H., Zhou, R., Lu, H. C., et al. (2019). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18 (1), 110. doi:10.1186/s12943-019-1036-9
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540 (7632), 301–304. doi:10.1038/nature20577
Heck, A. M., and Wilusz, C. J. (2019). Small changes, big implications: The impact of m(6)A RNA methylation on gene expression in pluripotency and development. Biochim. Biophys. Acta. Gene Regul. Mech. 1862 (9), 194402. doi:10.1016/j.bbagrm.2019.07.003
Helm, M., and Motorin, Y. (2017). Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 18 (5), 275–291. doi:10.1038/nrg.2016.169
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell. Res. 27 (9), 1115–1127. doi:10.1038/cr.2017.99
Hu, S. I., Katz, M., Chin, S., Qi, X., Cruz, J., Ibebunjo, C., et al. (2012). MNK2 inhibits eIF4G activation through a pathway involving serine-arginine-rich protein kinase in skeletal muscle. Sci. Signal. 5, ra14. doi:10.1126/scisignal.2002466
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell. Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Jensen, P. B., Pedersen, L., Krishna, S., and Jensen, M. H. (2010). A Wnt oscillator model for somitogenesis. Biophys. J. 98 (6), 943–950. doi:10.1016/j.bpj.2009.11.039
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2012). Erratum: Corrigendum: N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 8 (12), 1008. doi:10.1038/nchembio1212-1008a
Jiang, Q., Sun, B., Liu, Q., Cai, M., Wu, R., Wang, F., et al. (2019). MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m(6)A-YTHDF1-dependent mechanism. FASEB J. 33 (2), 2971–2981. doi:10.1096/fj.201801393RRR
Kassar-Duchossoy, L., Giacone, E., Gayraud-Morel, B., Jory, A., Gomes, D., Tajbakhsh, S., et al. (2005). Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes. Dev. 19 (12), 1426–1431. doi:10.1101/gad.345505
Kim, M. S., Fielitz, J., McAnally, J., Shelton, J. M., Lemon, D. D., McKinsey, T. A., et al. (2008). Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol. Cell.. Biol. 28 (11), 3600–3609. doi:10.1128/MCB.00189-08
Kudou, K., Komatsu, T., Nogami, J., Maehara, K., Harada, A., Saeki, H., et al. (2017). The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 7 (9), 170119. doi:10.1098/rsob.170119
Lasman, L., Krupalnik, V., Viukov, S., Mor, N., Aguilera-Castrejon, A., Schneir, D., et al. (2020). Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes. Dev. 34 (19-20), 1373–1391. doi:10.1101/gad.340695.120
Le Grand, F., and Rudnicki, M. A. (2007). Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell. Biol. 19 (6), 628–633. doi:10.1016/j.ceb.2007.09.012
Lee, H., Bao, S., Qian, Y., Geula, S., Leslie, J., Zhang, C., et al. (2019). Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell. Biol. 21 (6), 700–709. doi:10.1038/s41556-019-0318-1
Lence, T., Paolantoni, C., Worpenberg, L., and Roignant, J. Y. (2019). Mechanistic insights into m(6)A RNA enzymes. Biochim. Biophys. Acta. Gene Regul. Mech. 1862 (3), 222–229. doi:10.1016/j.bbagrm.2018.10.014
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell. Res. 27 (3), 444–447. doi:10.1038/cr.2017.10
Li, J. B., and Goldberg, A. L. (1976). Effects of food deprivation on protein synthesis and degradation in rat skeletal muscles. Am. J. Physiol. 231, 441–448. doi:10.1152/ajplegacy.1976.231.2.441
Li, J., Pei, Y., Zhou, R., Tang, Z., and Yang, Y. (2021). Regulation of RNA N(6)-methyladenosine modification and its emerging roles in skeletal muscle development. Int. J. Biol. Sci. 17 (7), 1682–1692. doi:10.7150/ijbs.56251
Liang, Y., Han, H., Xiong, Q., Yang, C., Wang, L., Ma, J., et al. (2021). METTL3-mediated m(6)A methylation regulates muscle stem cells and muscle regeneration by Notch signaling pathway. Stem Cells Int. 2021, 9955691. doi:10.1155/2021/9955691
Lin, J., Zhu, Q., Huang, J., Cai, R., and Kuang, Y. (2020). Hypoxia promotes vascular smooth muscle cell (VSMC) differentiation of adipose-derived stem cell (ADSC) by regulating mettl3 and paracrine factors. Stem Cells Int. 2020, 2830565. doi:10.1155/2020/2830565
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, Y., Zhou, T., Wang, Q., Fu, R., Zhang, Z., Chen, N., et al. (2022). m(6) A demethylase ALKBH5 drives denervation-induced muscle atrophy by targeting HDAC4 to activate FoxO3 signalling. J. Cachexia Sarcopenia Muscle 13 (2), 1210–1223. doi:10.1002/jcsm.12929
Lu, Z., Zhang, L., Li, Q., Liu, E., Du, L. X., Chu, M., et al. (2019). N6-methyladenosine modification in mRNA and its research advance in animals. Acta Veterinaria Zootechnica Sinica 50, 1–13.
Maimon, A., Mogilevsky, M., Shilo, A., Golan-Gerstl, R., Obiedat, A., Ben-Hur, V., et al. (2014). Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell. Rep. 7 (2), 501–513. doi:10.1016/j.celrep.2014.03.041
Mathiyalagan, P., Adamiak, M., Mayourian, J., Sassi, Y., Liang, Y., Agarwal, N., et al. (2019). FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation 139, 518–532. doi:10.1161/CIRCULATIONAHA.118.033794
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
Mayeuf-Louchart, A., Staels, B., and Duez, H. (2015). Skeletal muscle functions around the clock. Diabetes Obes. Metab. 17, 39–46. doi:10.1111/dom.12517
Mckendry, J., Stokes, T., Mcleod, J. C., and Phillips, S. M. J. C. P. (2021). Resistance exercise, aging, disuse, and muscle protein metabolism. Compr. Physiol. 11 (3), 2249–2278. doi:10.1002/cphy.c200029
McKinnell, I. W., Ishibashi, J., Le Grand, F., Punch, V. G., Addicks, G. C., Greenblatt, J. F., et al. (2008). Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell. Biol. 10 (1), 77–84. doi:10.1038/ncb1671
Mendel, M., Chen, K. M., Homolka, D., Gos, P., Pandey, R. R., McCarthy, A. A., et al. (2018). Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 71 (6), 986–1000. doi:10.1016/j.molcel.2018.08.004
Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995). Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 83, 1125–1136. doi:10.1016/0092-8674(95)90139-6
Nwokoye, E. C., AlNaseem, E., Crawford, R. A., Castelli, L. M., Jennings, M. D., Kershaw, C. J., et al. (2021). Overlapping regions of Caf20 mediate its interactions with the mRNA-5'cap-binding protein eIF4E and with ribosomes. Sci. Rep. 11 (1), 13467. doi:10.1038/s41598-021-92931-4
Pallafacchina, G., Blaauw, B., and Schiaffino, S. (2013). Role of satellite cells in muscle growth and maintenance of muscle mass. Nutr. Metab. Cardiovasc. Dis. 23 (Suppl. 1), S12–S18. doi:10.1016/j.numecd.2012.02.002
Parker, M. H., Seale, P., and Rudnicki, M. A. (2003). Looking back to the embryo: Defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4 (7), 497–507. doi:10.1038/nrg1109
Partridge, T. (2004). Reenthronement of the muscle satellite cell. Cell. 119 (4), 447–448. doi:10.1016/j.cell.2004.10.022
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Petrosino, J. M., Hinger, S. A., Golubeva, V. A., Barajas, J. M., Dorn, L. E., Iyer, C. C., et al. (2022). The m(6)A methyltransferase METTL3 regulates muscle maintenance and growth in mice. Nat. Commun. 13 (1), 168. doi:10.1038/s41467-021-27848-7
Picard, B., Berri, C., Lefaucheur, L., Molette, C., Sayd, T., Terlouw, C., et al. (2010). Skeletal muscle proteomics in livestock production. Brief. Funct. Genomics 9 (3), 259–278. doi:10.1093/bfgp/elq005
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell. Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Relaix, F., and Zammit, P. S. (2012). Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 139 (16), 2845–2856. doi:10.1242/dev.069088
Rizzi, R., Bearzi, C., Mauretti, A., Bernardini, S., Cannata, S., Gargioli, C., et al. (2012). Tissue engineering for skeletal muscle regeneration. Muscles Ligaments Tendons J. 2, 230–234.
Ronkainen, J., Huusko, T. J., Soininen, R., Mondini, E., Cinti, F., Makela, K. A., et al. (2015). Fat mass- and obesity-associated gene Fto affects the dietary response in mouse white adipose tissue. Sci. Rep. 5, 9233. doi:10.1038/srep09233
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell. 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. doi:10.7554/eLife.31311
Sabourin, L. A., and Rudnicki, M. A. (2000). The molecular regulation of myogenesis. Clin. Genet. 57, 16–25. doi:10.1034/j.1399-0004.2000.570103.x
Sartori, R., Romanello, V., and Sandri, M. (2021). Mechanisms of muscle atrophy and hypertrophy: Implications in health and disease. Nat. Commun. 12 (1), 330. doi:10.1038/s41467-020-20123-1
Sassoon, D. A. (1993). Myogenic regulatory factors: Dissecting their role and regulation during vertebrate embryogenesis. Dev. Biol. 156, 11–23. doi:10.1006/dbio.1993.1055
Schiaffino, S., and Reggiani, C. (1996). Molecular diversity of myofibrillar proteins: Gene regulation and functional significance. Physiol. Rev. 76 (2), 371–423. doi:10.1152/physrev.1996.76.2.371
Schwartz, S., Mumbach, M. R., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G. G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell. Rep. 8 (1), 284–296. doi:10.1016/j.celrep.2014.05.048
Sharples, A. P., and Stewart, C. E. (2011). Myoblast models of skeletal muscle hypertrophy and atrophy. Curr. Opin. Clin. Nutr. Metab. Care 14 (3), 230–236. doi:10.1097/MCO.0b013e3283457ade
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell. Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 136 (4), 731–745. doi:10.1016/j.cell.2009.01.042
Song, T., Yang, Y., Jiang, S., and Peng, J. (2020). Novel insights into adipogenesis from the perspective of transcriptional and RNA N6-methyladenosine-mediated post-transcriptional regulation. Adv. Sci. 7 (21), 2001563. doi:10.1002/advs.202001563
Tajbakhsh, S., and Buckingham, M. (2000). The birth of muscle progenitor cells in the mouse: Spatiotemporal considerations. Curr. Top. Dev. Biol. 48, 225–268. doi:10.1016/s0070-2153(08)60758-9
Tao, B., Huang, X., Shi, J., Liu, J., Li, S., Xu, C., et al. (2020). FTO interacts with FOXO3a to enhance its transcriptional activity and inhibits aggression in gliomas. Signal Transduct. Target. Ther. 5 (1), 130. doi:10.1038/s41392-020-00234-3
Tao, X., Chen, J., Jiang, Y., Wei, Y., Chen, Y., Xu, H., et al. (2017). Transcriptome-wide N (6) -methyladenosine methylome profiling of porcine muscle and adipose tissues reveals a potential mechanism for transcriptional regulation and differential methylation pattern. BMC Genomics 18 (1), 336. doi:10.1186/s12864-017-3719-1
Tatsumi, R., Anderson, J. E., Nevoret, C. J., Halevy, O., and Allen, R. E. (1998). HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev. Biol. 194 (1), 114–128. doi:10.1006/dbio.1997.8803
Tedesco, F. S., Dellavalle, A., Diaz-Manera, J., Messina, G., and Cossu, G. (2010). Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J. Clin. Investig.. 120 (1), 11–19. doi:10.1172/JCI40373
Theler, D., Dominguez, C., Blatter, M., Boudet, J., and Allain, F. H. (2014). Solution structure of the YTH domain in complex with N6-methyladenosine RNA: A reader of methylated RNA. Nucleic Acids Res. 42 (22), 13911–13919. doi:10.1093/nar/gku1116
Velleman, S. G., Song, Y., Shin, J., and McFarland, D. C. (2013). Modulation of Turkey myogenic satellite cell differentiation through the shedding of glypican-1. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 164 (1), 36–43. doi:10.1016/j.cbpa.2012.10.007
Walsh, K., and Perlman, H. (1997). Cell cycle exit upon myogenic differentiation. Curr. Opin. Genet. Dev. 7, 597–602. doi:10.1016/s0959-437x(97)80005-6
Wang, J., Fu, Q., Yang, J., Liu, J. L., Hou, S. M., Huang, X., et al. (2021). RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating Runx2 mRNA and inhibiting osteogenic differentiation. Aging (Albany NY) 13 (17), 21134–21141. doi:10.18632/aging.203377
Wang, P., Doxtader, K. A., and Nam, Y. (2016a). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, Q., Chen, C., Ding, Q., Zhao, Y., Wang, Z., Chen, J., et al. (2020). METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69 (7), 1193–1205. doi:10.1136/gutjnl-2019-319639
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016b). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi:10.1038/nature18298
Wang, X., Huang, N., Yang, M., Wei, D., Tai, H., Han, X., et al. (2017). FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis.. Cell. Death Dis. 8 (3), e2702. doi:10.1038/cddis.2017.122
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., Zhao, J. C., et al. (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell. Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 69 (6), 1028–1038. e1026. doi:10.1016/j.molcel.2018.02.015
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xie, S. J., Lei, H., Yang, B., Diao, L. T., Liao, J. Y., He, J. H., et al. (2021a). Dynamic m(6)A mRNA methylation reveals the role of METTL3/14-m(6)A-MNK2-ERK signaling axis in skeletal muscle differentiation and regeneration. Front. Cell. Dev. Biol. 9, 744171. doi:10.3389/fcell.2021.744171
Xie, S. J., Tao, S., Diao, L. T., Li, P. L., Chen, W. C., Zhou, Z. G., et al. (2021b). Characterization of long non-coding RNAs modified by m(6)A RNA methylation in skeletal myogenesis. Front. Cell. Dev. Biol. 9, 762669. doi:10.3389/fcell.2021.762669
Xu, T., Xu, Z., Lu, L., Zeng, T., Gu, L., Huang, Y., et al. (2021). Transcriptome-wide study revealed m6A regulation of embryonic muscle development in Dingan goose (Anser cygnoides orientalis). BMC Genomics 22 (1), 270. doi:10.1186/s12864-021-07556-8
Yang, X., Ning, Y., Abbas Raza, S. H., Mei, C., and Zan, L. (2022a). MEF2C expression is regulated by the post-transcriptional activation of the METTL3-m(6)A-YTHDF1 axis in myoblast differentiation. Front. Vet. Sci. 9, 900924. doi:10.3389/fvets.2022.900924
Yang, Y., Fan, X., Yan, J., Chen, M., Zhu, M., Tang, Y., et al. (2021). A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 49 (3), 1313–1329. doi:10.1093/nar/gkaa1203
Yang, Y., Hsu, P. J., Chen, Y. S., and Yang, Y. G. (2018). Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell. Res. 28 (6), 616–624. doi:10.1038/s41422-018-0040-8
Yang, Z., Yu, G. L., Zhu, X., Peng, T. H., and Lv, Y. C. (2022b). Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes. Dis. 9 (1), 51–61. doi:10.1016/j.gendis.2021.01.005
Yin, H., Price, F., and Rudnicki, M. A. (2013). Satellite cells and the muscle stem cell niche. Physiol. Rev. 93 (1), 23–67. doi:10.1152/physrev.00043.2011
Yin, L., Li, N., Jia, W., Wang, N., Liang, M., Yang, X., et al. (2021). Skeletal muscle atrophy: From mechanisms to treatments. Pharmacol. Res. 172, 105807. doi:10.1016/j.phrs.2021.105807
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell. Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zammit, P. S., Golding, J. P., Nagata, Y., Hudon, V., Partridge, T. A., Beauchamp, J. R., et al. (2004). Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J. Cell. Biol. 166 (3), 347–357. doi:10.1083/jcb.200312007
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 31 (4), 591–606. e596. doi:10.1016/j.ccell.2017.02.013
Zhang, W., Xu, Y., Zhang, L., Wang, S., Yin, B., Zhao, S., et al. (2018). Synergistic effects of TGFβ2, WNT9a, and FGFR4 signals attenuate satellite cell differentiation during skeletal muscle development. Aging Cell. 17, e12788. doi:10.1111/acel.12788
Zhang, X., Yao, Y., Han, J., Yang, Y., Chen, Y., Tang, Z., et al. (2020). Longitudinal epitranscriptome profiling reveals the crucial role of N(6)-methyladenosine methylation in porcine prenatal skeletal muscle development. J. Genet. Genomics 47 (8), 466–476. doi:10.1016/j.jgg.2020.07.003
Zhao, B. S., Wang, X., Beadell, A. V., Lu, Z., Shi, H., Kuuspalu, A., et al. (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542 (7642), 475–478. doi:10.1038/nature21355
Zhao, X., Mo, D., Li, A., Gong, W., Xiao, S., Zhang, Y., et al. (2011). Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS One 6 (5), e19774. doi:10.1371/journal.pone.0019774
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., Qian, S. B., et al. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 (7574), 591–594. doi:10.1038/nature15377
Keywords: N6-methyladenosine (m6A) modification, myogenesis, skeletal muscle development, transcriptional regulation, epigenetic
Citation: Yu B, Liu J, Zhang J, Mu T, Feng X, Ma R and Gu Y (2022) Regulatory role of RNA N6-methyladenosine modifications during skeletal muscle development. Front. Cell Dev. Biol. 10:929183. doi: 10.3389/fcell.2022.929183
Received: 27 April 2022; Accepted: 28 June 2022;
Published: 05 August 2022.
Edited by:
De-Li Shi, Sorbonne University, FranceReviewed by:
Brandon J. Gheller, Cornell University, United StatesJianjun Jin, Northwest A and F University, China
Yalan Yang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Yu, Liu, Zhang, Mu, Feng, Ma and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Zhang, emhhbmdqa2F0aHlAMTI2LmNvbQ==
 Baojun Yu
Baojun Yu Jiamin Liu
Jiamin Liu Tong Mu
Tong Mu Xiaofang Feng
Xiaofang Feng Yaling Gu
Yaling Gu