
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell Dev. Biol. , 05 September 2022
Sec. Cellular Biochemistry
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.916114
This article is part of the Research Topic Bioinorganic Chemistry of Metals in Cell Function and Disease View all 13 articles
A family of cytosolic copper (Cu) storage proteins (the Csps) bind large quantities of Cu(I) via their Cys-lined four-helix bundles, and the majority are cytosolic (Csp3s). The presence of Csp3s in many bacteria appears inconsistent with the current dogma that bacteria, unlike eukaryotes, have evolved not to maintain intracellular pools of Cu due to its potential toxicity. Sporulation in Bacillus subtilis has been used to investigate if a Csp3 binds Cu(I) in the cytosol for a target enzyme. The activity of the Cu-requiring endospore multi-Cu oxidase BsCotA (a laccase) increases under Cu-replete conditions in wild type B. subtilis. In the strain lacking BsCsp3 lower BsCotA activity is observed and is unaffected by Cu levels. BsCsp3 loaded with Cu(I) readily activates apo-BsCotA in vitro. Experiments with a high affinity Cu(I) chelator demonstrate that Cu(I) transfer from Cu(I)-BsCsp3 must occur via an associative mechanism. BsCsp3 and BsCotA are both upregulated during late sporulation. We hypothesise that BsCsp3 acquires cuprous ions in the cytosol of B. subtilis for BsCotA.
Copper (Cu) is essential for most organisms, but use of this metal ion is associated with significant risks due to its potential toxicity. The availability of Cu is regulated by the presence of high-affinity sites in both eukaryotes (Rae et al., 1999) and prokaryotes (Changela et al., 2003). Therefore, all intracellular Cu(I) is tightly bound to either proteins or small molecules, i.e. there is no ‘free’ Cu(I) (Rae et al., 1999; Changela et al., 2003; Festa and Thiele 2011). Import, cytosolic handling, trafficking to different locations, and storage of Cu have all been characterised in eukaryotic cells (Festa and Thiele 2011). In bacteria, some of these processes are either not thought to occur, or are not yet fully understood. For example, the plasma membrane protein CcoG, which reduces Cu(II) to the preferred intracellular oxidation state [Cu(I)] has only recently been identified in bacteria as a cytochrome oxidase (COX) assembly factor (Marckmann et al., 2019). The reduction of Cu(II) prior to import into eukaryotic cells has been known to happen for many years (Hassett and Kosman 1995; Festa and Thiele 2011). Excess Cu(I) is removed from the cytosol by probably the best-studied component of bacterial Cu homeostasis (homologues are present in eukaryotes); a Cu-transporting P-type ATPase (CopA), which can be assisted by the cytosolic Cu metallochaperone CopZ (Figure 1A) (Rensing et al., 2000; Solioz et al., 2010; Festa and Thiele 2011; Rensing and McDevitt 2013; Meydan et al., 2017). The toxicity of Cu can involve Cu(I) binding in place of the native metal in cytosolic iron-sulfur (Fe-S) cluster-containing proteins (Macomber and Imlay 2009), and Cu catalyses ROS formation (Solioz et al., 2010; Festa and Thiele 2011; Rensing and McDevitt 2013). The intracellular damage that Cu causes, and the current dearth of intracellular Cu-requiring enzymes (Ridge et al., 2008), has resulted in a prevailing view that bacteria have evolved not to use this metal ion in the cytosol (Ridge et al., 2008; Rensing and McDevitt 2013). However, there is no a priori reason why bacteria, like eukaryotes, cannot utilise Cu in this compartment if mechanisms are available to enable its safe handling, i.e., by ensuring tight chelation and specific delivery. The presence of cytosolic Cu storage proteins (Csps) that bind large quantities of Cu(I) with high affinity (Vita et al., 2015; Vita et al., 2016; Dennison et al., 2018; Lee and Dennison 2019) provide a possible route for intracellular Cu use in bacteria.
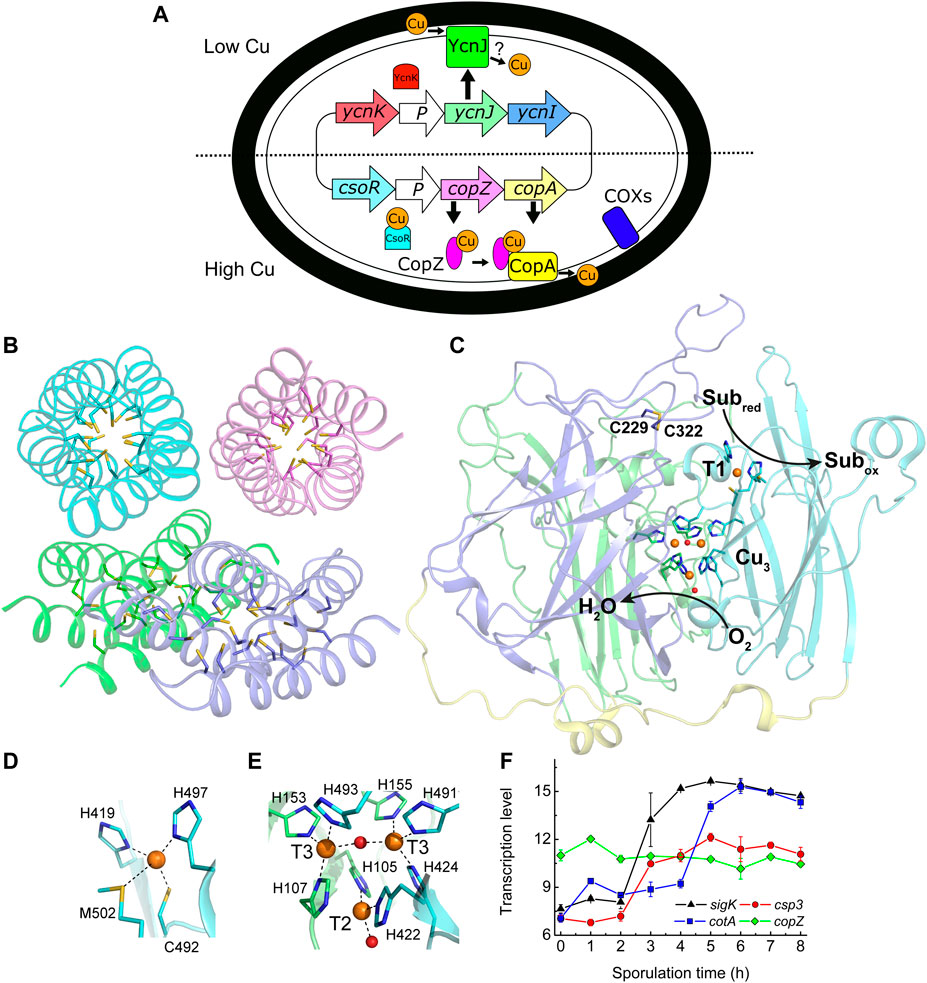
FIGURE 1. Copper handling, a cytosolic Cu(I) storage protein, a Cu-requiring enzyme, and their transcription during sporulation in B. subtilis. (A) An overview of Cu homeostasis in B. subtilis including Cu (orange circles, oxidation state undefined) export by CopA and CopZ (regulated by CsoR) (Smaldone and Helmann 2007), and import by YcnJ (regulated by YcnK) (Chillappagari et al., 2009; Hirooka et al., 2012). YcnI is membrane bound and binds Cu(II) in vitro, but its role in Cu homeostasis is unclear (Damle et al., 2021). The only currently known Cu-requiring enzymes in vegetative B. subtilis cells are two cytochrome oxidases (COXs) located on the plasma membrane (Lauraeus et al., 1991). (B) The crystal structure of Cu(I)-free BsCsp3 (PDB: 5FIG), a tetramer of four-helix bundles each with 19 Cys residues pointing into their cores enabling the binding of up to ∼20 Cu(I) ions per monomer (Vita et al., 2016). (C) The crystal structure of the endospore multi-Cu oxidase (a laccase) BsCotA (PDB: 1GSK, Enguita et al., 2003) with domains 1, 2, and 3 coloured green, slate and cyan, respectively (the linking regions are yellow). Substrates are oxidized (Subred to Subox) at the T1 Cu centre with electrons passed to the T2/T3 trinuclear (Cu3) cluster where oxygen is reduced to water. Also highlighted is the disulfide bond between Cys229 and Cys322. Detailed views of the T1 Cu site (D) and the Cu3 cluster (E) are shown. The side chains of coordinating residues are represented as sticks, Cu ions as orange spheres and the oxygen atoms of water (bound to the T2 Cu) and hydroxide (bridging the T3 Cu ions) ligands as red spheres in (C–E). (F) Transcription profiles (Nicolas et al., 2012) of the sigK (σK, which facilitates expression of outer and inner spore coat proteins, black triangles), csp3 (red circles), cotA (blue squares) and copZ (green diamonds) genes during sporulation.
The Csps were first identified in Gram-negative bacteria that oxidize methane (Vita et al., 2015). These methanotrophs can possess different Csp homologues, all having many Cys residues lining the cores of their four-helix bundles that enable the binding of a large number of Cu(I) ions (Vita et al., 2015; Vita et al., 2016; Dennison et al., 2018). A Csp exported from the cytosol (Csp1) stores up to 52 Cu(I) ions per tetramer for the particulate (membrane-bound) methane monooxygenase (pMMO) in the model methanotroph Methylosinus trichosporium OB3b (MtCsp1) (Vita et al., 2015). MtCsp1 is upregulated at the Cu concentrations required for methane oxidation by pMMO in switchover methanotrophs (Gu and Semrau 2017), which uses a soluble Fe MMO when Cu is limiting (DiSpirito et al., 2016). However, a cytosolic Csp homologue (MtCsp3) is not upregulated with pMMO in M. trichosporium OB3b (Gu and Semrau 2017).
The Gram-positive bacterium Bacillus subtilis is an ideal model system for investigating the role of a Csp3 as its Cu homeostasis system is well characterised (Figure 1A) (Radford et al., 2003; Smaldone and Helmann 2007; Chillappagari et al., 2009; Ma et al., 2009; Solioz et al., 2010; Hirooka et al., 2012; Damle et al., 2021). This includes the copZA operon (Cu efflux machinery, vide supra) and its Cu-sensing repressor CsoR (Radford et al., 2003; Smaldone and Helmann 2007; Ma et al., 2009; Solioz et al., 2010). The membrane protein YcnJ is upregulated under Cu-limiting conditions, controlled by the suggested repressor YcnK (Chillappagari et al., 2009; Hirooka et al., 2012), and has been proposed to play a role in Cu acquisition (Figure 1A). The gene for the membrane-anchored YcnI is part of the same (ycnKJI) operon and is also thought to be regulated by YcnK (Hirooka et al., 2012). The soluble domain of YcnI binds Cu(II) in vitro, and this protein has been suggested to function as a Cu metallochaperone (Damle et al., 2021. Cytosolic Cu(I) could be safely stored by the B. subtilis Csp3 homologue (YhjQ, herein BsCsp3) whose core is lined with 19 Cys residues (Figure 1B), enabling the binding of ∼80 Cu(I) ions per tetramer in vitro (Vita et al., 2016).
Only two families of Cu enzymes are currently known to be present in B. subtilis; two COXs (one without the CuA site in subunit II), located on the plasma membrane (Figure 1A) and the multi-Cu oxidase (MCO; a laccase) BsCotA (Lauraeus et al., 1991; Hullo et al., 2001; Martins et al., 2002; Enguita et al., 2003). Assembly of the COXs, including their acquisition of Cu has been extensively studied (for example von Wachenfeldt et al., 2021). BsCotA is an outer spore-coat (endospore) enzyme (McKenney et al., 2013) that possesses the typical type 1 (T1), 2 (T2) and 3 (T3) Cu sites of an MCO (Enguita et al., 2003), which are all involved in the catalytic cycle (see Figures 1C–E). BsCotA produces a melanin-like pigment thought to provide spores with protection against hydrogen peroxide and UV light (Hullo et al., 2001; McKenney et al., 2013). This enzyme is upregulated during the latter stages of sporulation, as is BsCsp3 (Figure 1F) (Nicolas et al., 2012).
Herein we demonstrate that, despite previous preliminary work from our laboratory (Vita et al., 2016), BsCsp3 does not provide resistance to toxicity caused by elevated Cu levels in B. subtilis. We have therefore tested the hypothesis that BsCsp3 binds Cu(I) ions in the cytosol for a Cu-requiring enzyme by investigating the effect of gene deletion on the activity of BsCotA in spores grown under Cu limiting and replete conditions. The data obtained indicate a role for BsCsp3 in ensuring maximum BsCotA activity. The ability of Cu(I)-BsCsp3 to activate apo-BsCotA has been confirmed in vitro. A model for how BsCotA is loaded with Cu during sporulation is proposed. This is the first example showing an enzyme acquiring Cu(I) in the cytosol of a bacterium, as well as identifying the protein from which the metal ion is obtained.
The presence of BsCsp3 with a high capacity for Cu(I) in the cytosol of B. subtilis (Vita et al., 2015; Vita et al., 2016; Dennison et al., 2018) would suggest a role in helping to prevent the issues associated with excess Cu (Macomber and Imlay 2009; Lee and Dennison 2019). The toxicity of Cu to bacteria is highlighted by how increasing Cu concentrations limited the growth of wild type (WT) B. subtilis in LB medium (Supplementary Figure S1). At ≥1.5 mM Cu cells started to grow more slowly, with a very small increase in the absorbance/OD observed only after more than 6 h at 2 mM added Cu, coinciding with elevated intracellular Cu concentrations (Supplementary Figure S2). Very similar growth and Cu accumulation results were obtained (Supplementary Figures S1, S2) for the B. subtilis strain (Δcsp3) lacking the csp3 gene (yhjQ). The growth studies reported herein demonstrate that BsCsp3 is not solely required in helping prevent the harmful effects of elevated Cu levels on B. subtilis (more details are provided in the legend to Supplementary Figure S1). Therefore, the protein apparently does not have a function like the eukaryotic cytosolic Cys-rich metallothioneins (Festa and Thiele 2011).
BsCotA, along with the two COXs (whose Cu acquisition is well-characterised, von Wachenfeldt et al., 2021), are the only known Cu-requiring enzymes present in spores. The csp3 and cotA genes are both upregulated (Nicolas et al., 2012) at similar stages during sporulation (Figure 1F). We have therefore studied whether BsCsp3 is involved in Cu(I) supply to BsCotA. This enzyme binds four Cu ions (Figures 1C–E), which are required to enable oxidation of the laccase substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) in vitro and in spores (Martins et al., 2002). We have used the oxidation of ABTS to assess the relative amounts of Cu-BsCotA in B. subtilis spores (Figure 2). For WT B. subtilis spores, the ability to oxidise ABTS increased approximately two-fold when 50 μM Cu is added to Difco sporulation medium (DSM) (Figures 2A,D, Supplementary Figures S3A,B). This indicated that unless supplemented, DSM does not contain sufficient Cu (the Cu concentration in DSM without any added Cu is ∼0.4 μM) to allow all of the BsCotA produced during sporulation to be active.
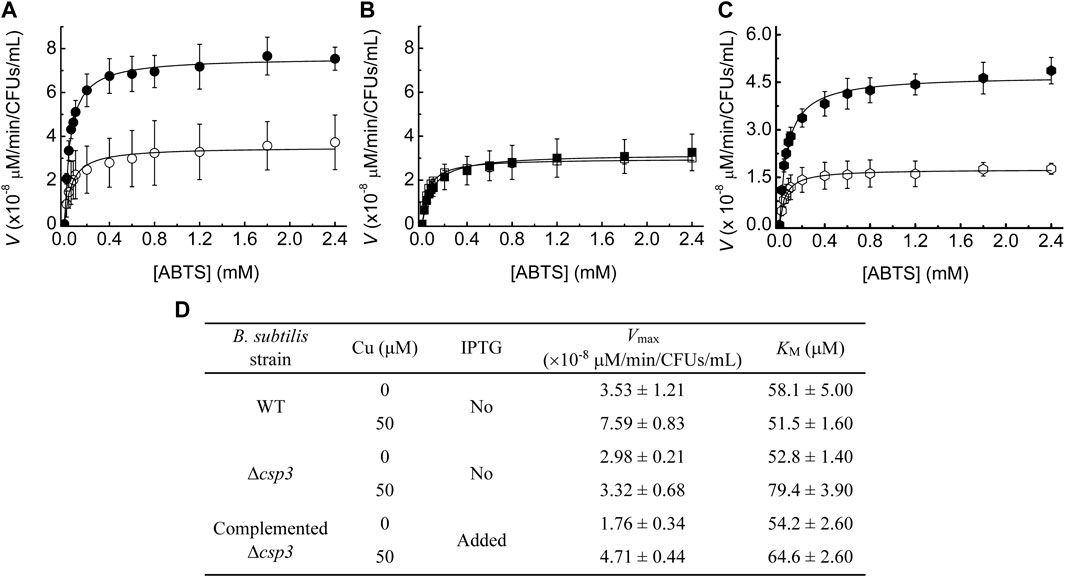
FIGURE 2. The influence of Cu levels and BsCsp3 on BsCotA activity in B. subtilis spores. Michaelis-Menten plots of BsCotA activity for heated purified spores from WT (A), Δcsp3 (B), and the complemented Δcsp3 (C) strains. Spores (A,B) were produced in DSM plus no (open symbols) and 50 μM (black filled symbols) added Cu(NO3)2. For the complemented Δcsp3 strain (C) sporulation was carried out in the presence of 1 mM IPTG with either no (open symbols) or 50 μM (black filled symbols) added Cu(NO3)2. Plots from which the initial rates for WT and Δcsp3 B. subtilis spores were obtained are shown in Supplementary Figure S3. Kinetics measurements were made in 100 mM citrate-phosphate buffer pH 4.0 at 37°C using three different sets of spores (averages and standard deviations are shown). (D) The Vmax and KM values obtained from the fits of the data in (A–C) to the Michaelis-Menten equation. The KM values for the oxidation of ABTS by BsCotA in spores are all in the range of those reported for purified enzyme (Martins et al., 2002; Durão et al., 2008; Fernandes et al., 2011).
The BsCotA activity of Δcsp3 B. subtilis spores grown in DSM without added Cu was similar to that for WT spores produced under the same conditions (Figures 2B,D, Supplementary Figures S3C,D). However, unlike for WT B. subtilis, supplementing DSM with Cu during sporulation had no effect on BsCotA activity for the Δcsp3 strain. These results indicate that BsCsp3 plays a role in Cu acquisition by BsCotA during sporulation, particularly under Cu-replete conditions. Some BsCotA activity remained for Δcsp3 B. subtilis spores, and an alternative mechanism of Cu acquisition by BsCotA must exist, which could also be responsible for the activity observed in the WT strain under Cu-limiting conditions.
To confirm that BsCsp3 is involved in Cu(I) supply to BsCotA, the Δcsp3 strain was complemented by introducing the csp3 gene at a different location (the amyE locus), which can be induced by isopropyl β-D-thiogalactopyranoside (IPTG). The highest BsCotA activity was obtained for spores of this strain grown in the presence of IPTG and Cu (Figures 2C,D). Under these conditions, activity was almost three-fold greater than without their addition, similar to the increase for WT B. subtilis spores under Cu-replete conditions (Figures 2A,D).
The above data support the hypothesis that BsCsp3 binds Cu(I) in the cytosol under Cu-replete conditions, which is used to metallate BsCotA. The ability of Cu(I)-BsCsp3 to activate apo (Cu-free)-BsCotA was therefore studied in vitro (Figures 3A–D). Apo-BsCotA is inactive (Figures 3A,C), whilst the addition of Cu produces enzyme that rapidly oxidises ABTS (for example see Figure 3A and Martins et al., 2002). Cu(I)-BsCsp3 readily activates apo-BsCotA (Figures 3B,C), giving similar reactivity to enzyme plus Cu(I) at 24 h (Figure 3B). This is consistent with Cu(I) transfer from Cu(I)-BsCsp3 to apo-BsCotA (apo-BsCsp3 does not activate apo-BsCotA, see Figures 3B,C), and >50% activation is achieved in 6 h (Figure 3C). The number of free thiols in this form of BsCotA, which possess four Cys residues, was routinely determined to be ∼2, consistent with the Cys229-Cys322 disulfide bond being present in the overexpressed enzyme purified from E. coli (Enguita et al., 2003). Reduction of the protein with dithiothreitol (DTT) resulted in ∼3.5 free thiols and thus cleavage of the Cys229-Cys322 disulfide. Reduced apo-BsCotA reacts much more rapidly with Cu(I)-BsCsp3 and >50% activation is achieved in just over 45 min (Figure 3D). Inactive apo-BsCotA was found to contain no detectable Cu (<0.2 equivalents) by atomic absorption spectrometry (AAS). After transfer, 4.11 ± 0.75 (n = 4) equivalents were bound, and when measured the absorbance values at 600 and 330 nm were consistent with full occupancy of the T1 and T3 sites with Cu(II), respectively (Durão et al., 2008). Another cytosolic Cu(I)-binding protein with a well-established role in Cu homeostasis (delivering Cu(I) to BsCopA, Radford et al., 2003) and a similar Cu(I) affinity (∼1017 M−1 at pH 7.5) to BsCsp3 (Badarau and Dennison 2011a; Vita et al., 2016) is BsCopZ (see Figure 1A). After incubation of apo-BsCotA with Cu(I)-BsCopZ for 24 h almost no activity is observed (Figure 3E), indicating Cu(I) transfer does not occur. A large excess of bathocuproine disulfonate (BCS) removes only ∼60% of Cu(I) from BsCsp3 in 24 h (Figure 3F; Supplementary Table S1). The slow kinetics for this reaction demonstrates that Cu(I) does not freely dissociate from BsCsp3, otherwise the [Cu(BCS)2]3- complex would rapidly form. We therefore assume associative mechanisms for the reactions of Cu(I)-BsCsp3 with BCS and also with apo-BsCotA, with the partners interacting prior to Cu(I) transfer.
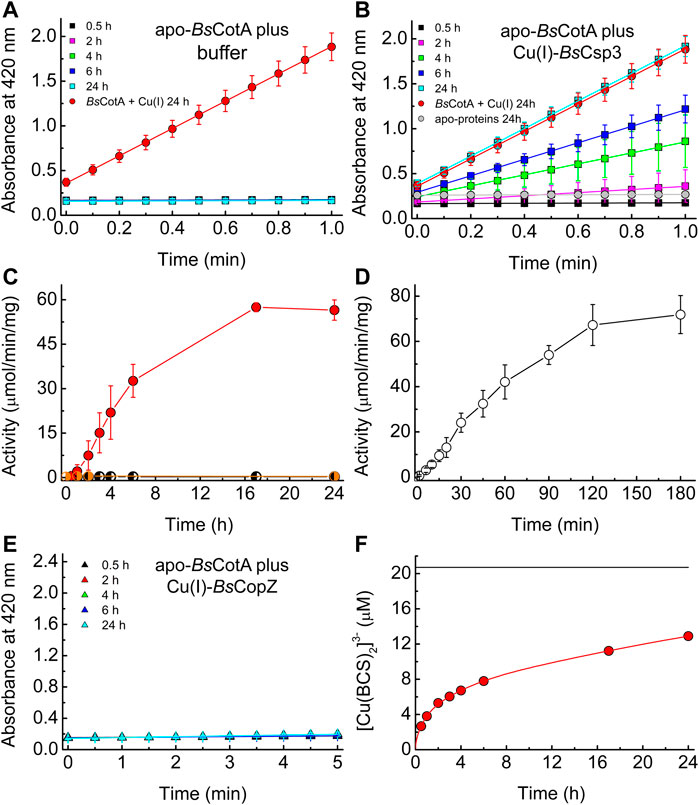
FIGURE 3. The activation of apo-BsCotA by Cu(I)-BsCsp3 and associative Cu(I) transfer. Plots of absorbance at 420 nm against time for the reaction with 2.4 mM ABTS (at 37°C) of mixtures of apo-BsCotA incubated with buffer (A) and Cu(I)-BsCsp3 (B) for up to 24 h. Also shown is the data obtained when apo-BsCotA was incubated with Cu(I) (red circles in A and B) and apo-BsCsp3 (grey circles in B) for 24 h. Mixtures were incubated under anaerobic conditions (apart from the reaction with apo-BsCsp3) in 20 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) pH 7.5 plus 200 mM NaCl (the buffer used in A), and BsCotA activity was measured in 100 mM citrate-phosphate buffer pH 4.0. (C) Plots of activity against incubation time of apo-BsCotA plus buffer alone (half-black circles), Cu(I)-BsCsp3 (red circles), and apo-BsCsp3 (half-grey circles) for up to 24 h (very similar values to those at 24 h were measured at 48 h for buffer and Cu(I)-BsCsp3). In (D) are activity data obtained when apo-BsCotA with the Cys229-Cys322 disulfide bond reduced, was mixed with Cu(I)-BsCsp3 for up to 180 min. (E) Plots of absorbance at 420 nm against time for the reaction with 2.4 mM ABTS (at 37°C) of apo-BsCotA incubated with Cu(I)-BsCopZ for up to 24 h (there was also no sign of activity after 48 h). All activity data are averages from three to six independent experiments (apart from apo-BsCotA plus apo-BsCsp3) with error bars showing standard deviations. (F) A plot of [Cu(BCS)2]3- concentration against time for BsCsp3 (1.08 µM) plus 18.0 equivalents of Cu(I) mixed with 2.5 mM BCS (red line) carried out in the same buffer as that used in (A–E) under anaerobic conditions. The data at 0.5, 1, 2, 3, 4, 6, 17, and 24 h, which correspond to times at which BsCotA activity was measured (C), are shown by red circles. The black line indicates the outcome of the same experiment but with 6.64 M guanidine-HCl present in the buffer. The Cu(I)-protein unfolds resulting in much faster removal of cuprous ions, giving the end point for the reaction (the value shown was obtained after 2 h). The average percentage removal compared to that for unfolded samples, and the standard deviations, from three independent experiments are listed in Supplementary Table S1.
In this study we have demonstrated that BsCsp3 binds cytosolic Cu(I) and plays a role in supplying Cu(I) to the Cu-requiring enzyme BsCotA during sporulation (Figure 2). This is not the only mechanism available to load BsCotA with Cu as some activity is observed in Δcsp3 B. subtilis. A possibility we considered was that the cytosolic Cu metallochaperone BsCopZ, as well as transferring Cu(I) to BsCopA (Figure 1A), may supply cuprous ions to BsCotA. The in vitro studies reported here show that despite the similar Cu(I) affinity to BsCsp3 (Badarau and Dennison 2011a; Vita et al., 2016), Cu(I)-BsCopZ cannot activate apo-BsCotA (Figure 3E), consistent with BsCopZ not being upregulated during sporulation (Figure 1F). Furthermore, BsCotA activity in Δcsp3 B. subtilis spores is unaffected when the Cu concentration is higher, conditions which would increase BsCopZ expression. A similar level of activity is determined for WT spores without supplementing DSM with Cu, conditions under which BsCopZ will not be upregulated. Collectively, these data exclude a potential role for BsCopZ in activating BsCotA. The source(s) of Cu(I) for activating BsCotA in the absence of BsCsp3, and also at lower intracellular concentrations of the metal ion, remain(s) to be established. Regardless, the lack of activation of apo-BsCotA by Cu(I)-BsCopZ in vitro highlights the specificity of activation by Cu(I)-BsCsp3. This is essential in a cell as it ensures Cu(I) is delivered to where it is needed, as observed for other Cu-homeostasis proteins (Pufahl et al., 1997; Rae et al., 1999; Schmidt et al., 1999; Lamb et al., 2001; Banci et al., 2006; Banci et al. 2010; Banci et al. 2011; Sala et al., 2019).
The high Cu(I) affinity (Vita et al., 2016) of BsCsp3 (1.5 × 1017 M−1), and the slow formation of [Cu(BCS)2]3- when BCS is added to protein fully loaded with Cu(I) (Figure 3F), indicates the transfer of cuprous ions from Cu(I)-BsCsp3 to apo-BsCotA has to occur via an associative mechanism (unassisted Cu(I) off-rates for BsCsp3 can be estimated to be ∼10–9 s−1, Dennison et al., 2018). This is consistent with the requirement for no intracellular ‘free’ Cu(I). For the acquisition of such tightly bound Cu(I) to be possible, metalation must take place once BsCotA has at least partially folded so the sites where Cu is required have formed. The T1 Cu site is closest to the surface, with its His497 ligand solvent exposed, and is ∼12.5–15.5 Å from the Cu3 cluster (Figure 1C). Therefore, BsCsp3 association at more than one location may be required to metalate all of the sites in folded BsCotA. Published Cu(I) affinities of T1 Cu sites (Badarau and Dennison 2011b; North and Wilcox 2019) are (2.1–4.0) × 1017 M−1, similar to the average Cu(I) affinity of BsCsp3 (Vita et al., 2016). Although Cu(I) affinities are not available for the Cu3 cluster, Cu(I) transfer from the storage protein to the enzyme should not be hindered thermodynamically (Banci et al., 2010; Badarau and Dennison 2011b). To facilitate access to the more buried Cu3 cluster (required for activity) the protein may need to be partially unfolded. The MCO CueO from E. coli undergoes a transition from an ‘open’ non-metallated folded form with accessible Cu sites, to a more ‘closed’ conformation after Cu has bound (Strolle et al., 2016). BsCotA has a disulfide bond between Cys229 and Cys322 (Figure 1C) that is ∼12–13 Å from the T1 Cu site and 17–23 Å from the Cu3 cluster (Enguita et al., 2003). Only two of the four Cys residues are reactive in BsCotA overexpressed and purified from E. coli (see Materials and Methods) and this form of the enzyme possesses the Cys229-Cys322 disulfide. In vitro activation is significantly faster in the absence of the disulfide (Figures 3C,D), and the formation of this bond in BsCotA may be linked to Cu(I) binding in vivo (vide infra). This is consistent with previous studies that found an increase in the rate of Cu(II) binding when this disulfide was removed (Fernandes et al., 2011), but with limited influence on overall structure and stability.
BsCsp3 and BsCotA expression are regulated by sigma factor K (SigK or σK), which is produced after the forespore has been engulfed by the mother cell (Figure 4). It appears the csp3 gene constitutes an operon with yhjR, an inner spore coat protein (Hosan et al., 2006) also regulated by SigK (Eichenberger et al., 2004) and co-expressed with Csp3 (Nicolas et al., 2012). Upregulation of the csp3 gene happens prior to cotA (Figure 1F), which would allow BsCsp3 to acquire Cu(I) before production of the enzyme requiring the metal. We propose that Cu(I) is transferred to BsCotA before it localizes to the spore coat. If Cu(I) acquisition occurs once BsCotA is part of the spore coat it is possible that YhjR plays a role in assisting this process. As discussed, BsCotA activation by Cu(I)-BsCsp3 in vitro is significantly faster in the absence of the Cys229-Cys322 disulfide, indicating that Cu(I) is acquired prior to the formation of this bond in vivo. Currently, there is only one known example of Cu acquisition by an enzyme from a partner protein in the cytosol. This is the eukaryotic Cu,Zn-superoxide dismutase (SOD1), which obtains Cu(I) from the Cu metallochaperone CCS (Rae et al., 1999; Schmidt et al., 1999; Lamb et al., 2001; Banci et al., 2010; Banci et al., 2011; Sala et al., 2019; Culotta et al., 1997; Wong et al., 2000). After many years of study, the activation of SOD1 by CCS is now fully understood and has been found to be linked to the formation of an essential disulfide bond in SOD1.
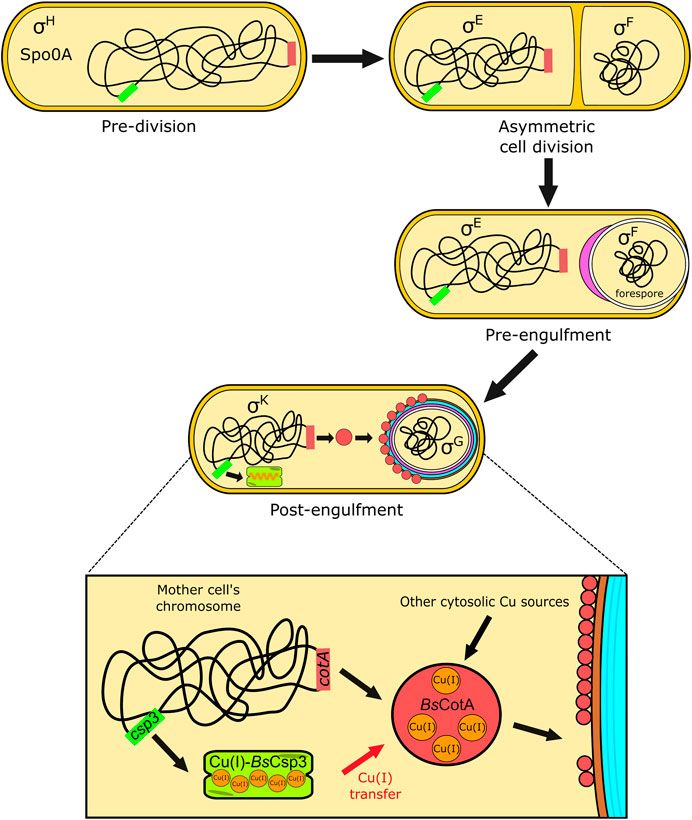
FIGURE 4. The proposed role of BsCsp3 in Cu(I) acquisition by BsCotA during sporulation in B. subtilis. The transcription factor Spo0A, along with σH, initiates sporulation. A septum asymmetrically divides the cell into the forespore and mother cell, with σE and σF, respectively, activated within these. The mother cell begins engulfment of the forespore and σE directs gene expression and initiation of spore coat (purple) formation. The expression of BsCsp3 and BsCotA now begins, promoted by σK (see Figure 1F) and coat assembly continues. We propose that BsCsp3 acquires Cu(I) during this stage of sporulation, which is transferred to BsCotA prior to insertion of the Cu-enzyme into the spore coat.
Added importance to understanding the correct metalation of BsCotA is provided by the observation that melanin production interferes with the phagocytosis of pathogenic yeast, and is required to allow survival in macrophages (Eisenman and Casadevall 2012). The related pigment made by Cu-loaded BsCotA is important for spore survival (Hullo et al., 2001; McKenney et al., 2013), and this may include within a host. Bacillus spores, and particularly those from B. cereus, cause food poisoning and are a common contaminant in a range of foods (Soni et al., 2016; Jessberger et al., 2020). The development of more effective inactivation approaches requires a better understanding of enzymes such as CotA that help protect spores. This includes establishing how they acquire essential cofactors including Cu ions.
WT B. subtilis 168 (genotype: trpC2) and the strain with the yhjQ gene deleted (genotype: trpC2 ΔyhjQ::erm, referred to herein as Δcsp3) strains were obtained from the Bacillus Genetic Stock Centre (BGSC) library (BGSCID 1A1 and BKE10600, respectively). These strains were checked by PCR (Supplementary Figures S4A,B, S5A; Supplementary Table S2). The disrupted csp3 gene was amplified by PCR using genomic DNA from the Δcsp3 strain with primers that hybridise ∼300 bp upstream and downstream of this region (Supplementary Figures S4B, S5A; Supplementary Table S2). The resulting fragment was sequenced with primers designed to hybridise ∼20 bp from the ends of the PCR product (Supplementary Table S2) and matches that of the erythromycin resistance gene.
To test the influence of Cu on WT and Δcsp3 strains, cultures were grown (agitation at 250 rpm) in LB medium at 37°C overnight, diluted 100-fold in LB and LB plus added Cu(NO3)2 (0.5–2.0 mM). The absorbance at 600 nm was measured at regular intervals for up to 12 h, and also after 24 h. Cells ( ∼ 35 ml) were collected and washed, including with buffer plus 10 mM ethylenediaminetetraacetic acid (EDTA) (Lee and Dennison 2019), and digested in 200 μl of 65% HNO3 (Ultrapur) for up to 3 days at room temperature. These mixtures were centrifuged at 12,000 g for 10 min, diluted in MilliQ water to give a final HNO3 concentration of 2% and analysed for Cu by AAS. The Cu concentration in the stock solution used for these studies was regularly determined by AAS, as described previously (Lee and Dennison 2019).
To insert the csp3 gene plus its ribosome binding site (RBS) into the amyE locus of the Δcsp3 strain, a region including an additional 28 bp at the 5′ end was amplified from WT B. subtilis 168 genomic DNA by PCR using primers; rbs_BsCsp3-F and rbs_BsCsp3-R (Supplementary Figures S4A, S5A; Supplementary Table S2). The product was cloned into pGEM-T (Promega) and the resulting rbs_csp3 fragment sub-cloned into pDR111, which possesses the IPTG-inducible Phyerspank promoter (Quisel et al., 2001; Britton at al. 2002), using HindIII and NheI to generate pDR111_rbs_csp3. To obtain a strain possessing an IPTG-inducible copy of the csp3 gene (trpC2 ΔyhjQ::erm amyE::Physpank-rbs_yhjQ, called complemented Δcsp3 herein), Δcsp3 B. subtilis was transformed with pDR111_rbs_csp3. Selection was achieved using spectinomycin (50 μg/ml) and successful integration into the chromosomal amyE (α-amylase) gene identified by growing on LB agar containing 1% starch and staining with iodine (Engman et al., 2012). Insertion of the csp3 gene was determined as described above (Supplementary Figures S4C, S5B; Supplementary Table S2), with the location and size of the fragment incorporated confirmed by PCR using the primers pDR111_int-F and pDR111_int-R (Supplementary Figures S4C, S5B; Supplementary Table S2).
WT, Δcsp3 and complemented Δcsp3 strains were grown overnight (agitation at 250 rpm) in 20 ml of DSM. Cultures were diluted 50-fold into 200 ml of DSM in a single 1 L Erlenmeyer flask and grown until the absorbance at 580 nm reached ∼0.5. This culture was split into four 50 ml cultures, each in a 250 ml Erlenmeyer flask, and 50 μM Cu(NO3)2 and 1 mM IPTG added when required. The cultures were grown (agitation at 250 rpm) for 48 h at 37°C and absorbance values at 580 nm measured at regular intervals. To purify spores (Tavares et al., 2013) cultures were centrifuged (4°C) for 10 min at 5,000 g, pellets re-suspended in 50 mM tris(hydroxymethyl)aminomethane (Tris) pH 7.2 plus 50 μg/ml lysozyme and incubated at 37°C for 1 h. After incubation and further centrifugation (4°C) for 10 min at 5,000 g, pellets were washed once in sterile MilliQ water and centrifuged. The pellets were re-suspended in 0.05% SDS by vortexing, centrifuged (4°C) for 10 min at 5,000 g and subsequently washed three times with sterile MilliQ water and stored at 4°C. The purity was checked by determining the colony-forming units (CFUs) of spore stocks that were unheated and those heated at 65°C for 1 h prior to growth on LB plates overnight at 37°C, and was typically >75%. As well as strains, spores used for kinetic experiments were verified by PCR (for example, Supplementary Figure S5) after germination in LB overnight at 37°C, using the primers listed in Supplementary Table S2.
For kinetic measurements of BsCotA activity, purified spores from the WT, Δcsp3 and complemented Δcsp3 strains were diluted with MilliQ water to give an absorbance at 580 nm of ∼1.2 (measured accurately), and heated at 65°C for 1 h prior to use. To determine the CFUs/ml for this suspension a 5 × 105-fold dilution in LB was plated (100 μl) onto LB agar, incubated at 37°C overnight and colonies counted. An aliquot of the heat-treated spore suspension (100 μl) was added to 900 μl of 100 mM citrate-phosphate buffer pH 4.0 plus 0.1–2.4 mM ABTS, and the absorbance at 420 nm (ε = 35,000 M−1cm−1) measured for 5 min at 37°C (Supplementary Figure S3). A control using 100 μl of buffer was also measured and showed no change in absorbance at 420 nm. The initial velocity (V0; typically reported in units of μM/min/CFUs/mL) was calculated, and plots of V0 against ABTS concentration (Figure 2) were fit to the Michaelis-Menten equation to determine Vmax (the maximum rate) and KM (the Michaelis constant). Comparing Vmax values calculated based on the absorbance at 580 nm of the heat-treated spore suspension, rather than using CFUs/mL, has no significant influence on the outcome of the study, but generally produces data with larger errors.
The cotA gene was amplified from B. subtilis genomic DNA using primers CotA_1F and CotA_1R listed in Supplementary Table S2, and cloned into pGEM-T. After removing the NdeI site in the gene by QuickChange site-directed mutagenesis (with primers CotA_2F and CotA_2R, Supplementary Table S2), the product was excised with NdeI and BamHI and re-cloned into pET11a. BsCotA was overexpressed in E. coli BL21 (DE3) (100 μM IPTG) grown at 20°C for 24 h. The protein was purified using a modified version of a published procedure (Martins et al., 2002). Cells from 0.5 to 2.0 L of culture were resuspended in 20 mM Tris pH 8.5, sonicated and centrifuged at 40,000 g for 30 min. The supernatant was diluted five-fold in 20 mM Tris pH 8.5 (sometimes plus 1 mM EDTA) and loaded onto a HiTrap Q HP column (1 or 5 ml) equilibrated in the same buffer. Proteins were eluted with a linear NaCl gradient (0–500 mM, total volume 50–200 ml) and fractions analysed using 18% SDS-PAGE. BsCotA-containing fractions were diluted with 20 mM Tris pH 7.6 (sometimes plus 1 mM EDTA) and loaded onto a HiTrap SP HP column (5 ml) and eluted with a linear NaCl gradient (0–500 mM, total volume, 200 ml). In some cases the BsCotA-containing fractions were heated at 70 °C for 30 min [BsCotA is a highly thermostable enzyme (Martins et al., 2002)], centrifuged at 40,000 g for 30 min, and the supernatant exchanged into 20 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) pH 7.5 plus 200 mM NaCl for further purification on a Superdex 75 10/300 GL gel-filtration column. Purified BsCotA had no detectable Cu (<0.2 equivalents) associated with it when analysed by AAS (Vita et al., 2015; Vita et al., 2016), and showed minimal ABTS oxidation activity (<0.3 μmol/min/mg).
BsCopZ was purified as described previously (Vita et al., 2016) and contains a small amount of bound Zn(II). Samples were therefore incubated with >10 equivalents of EDTA for 1 h and exchanged with 20 mM HEPES pH 7.5 plus 200 mM NaCl. The resulting protein had no Zn(II) associated with it and was reduced with DTT under anaerobic conditions and desalted as described previously (Vita et al., 2015; Vita et al., 2016).
BsCsp3 plus ∼18 equivalents of Cu(I) was prepared by adding the appropriate amount of a buffered solution of Cu(I) in an anaerobic chamber (Belle Technology, O2 << 2 ppm) to apo-protein in 20 mM HEPES pH 7.5 plus 200 mM NaCl, quantified using the 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB) assay carried out in the presence of ∼6.0 M guanidine hydrochloride (Vita et al., 2015; Vita et al., 2016). Apo-BsCotA was quantified using the absorbance at 280 nm (ε value of 84,739 M−1cm−1, Durão et al., 2008) and the number of free thiols determined with the DTNB assay (Vita et al., 2016). To reduce the Cys229-Cys322 disulfide (see Figure 1C), apo-BsCotA was incubated overnight in the anaerobic chamber with a 100-fold excess of DTT. The protein was desalted twice on a PD10 column and quantified from the absorbance at 280 nm, with thiols measured using the DTNB assay. Fully-reduced BsCopZ was also quantified using the DTNB assay and loaded with ∼0.8 equivalents of Cu(I) under anaerobic conditions. Cu(I)-BsCsp3 [∼3 μM binding ∼49–55 μM Cu(I)] was mixed with ∼1.1–1.3 μM of either as-isolated or reduced apo-BsCotA, requiring ∼4.4–5.2 μM Cu(I) to occupy all four Cu sites. Control experiments in which apo-BsCotA was incubated with a similar concentration (∼49–55 μM) of either Cu(I) or Cu(II) were also analyzed, as was a mixture of apo-BsCotA (1.1 μM) plus apo-BsCsp3 (3.1 μM). Cu(I)-BsCopZ [∼50–53 μM binding ∼40–42 μM Cu(I)] was separately added to ∼1 μM apo-BsCotA, requiring ∼4 μM Cu(I) to fill all Cu sites. Mixtures were incubated at room temperature in the anaerobic chamber (some experiments with Cu(II) were performed in air as was the reaction between the two apo-proteins) for up to 48 h. To measure activity, 10 μl of each mixture was added to 990 μl of aerated 100 mM citrate-phosphate buffer pH 4.0 plus 2.4 mM ABTS, and the absorbance at 420 nm measured for up to 5 min at 37°C (Figures 3A,B,E). A similar concentration of apo-BsCotA was also incubated anaerobically with just buffer (20 mM HEPES pH 7.5 plus 200 mM NaCl) and the lack of activity is clear (Figures 3A,D).
After some transfer experiments with reduced apo-BsCotA plus Cu(I)-BsCsp3 mixtures were loaded onto a 1 ml HiTrap SP HP column and eluted with a linear NaCl gradient (0–1 M, total volume 12 ml). Fractions containing BsCotA were combined, concentrated and a UV/Vis spectrum measured. This not only enabled protein quantification from the absorbance at 280 nm, but allowed Cu(II) occupancies of the T1 and T3 sites to be estimated from the absorbance at 600 (ε = 3,870 M−1cm−1) and 330 (ε = 3,639 M−1cm−1) nm, respectively (Durão et al., 2008). The total Cu content of BsCotA after transfer was measured using AAS (Vita et al., 2015; Vita et al., 2016).
The removal of Cu(I) by BCS (∼2.5 mM) was analysed for the Cu(I)-BsCsp3 [∼0.8–1.2 μM plus ∼18 equivalents of Cu(I)] samples used for activity experiments, both in the absence (folded BsCsp3) and presence (unfolding conditions) of guanidine-HCl (6.64 M) (Vita et al., 2015; Vita et al., 2016). The absorbance increase at 483 nm due to formation of [Cu(BCS)2]3- (ε = 12,500 M−1cm−1) (Badarau and Dennison 2011a) was measured over time at 22°C in 20 mM HEPES pH 7.5 plus 200 mM NaCl (Figure 3F).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
CD and JL conceived the project and designed the experiments. JL and RD performed the experiments and analysed data with help from CD. CD wrote the manuscript with help from JL and RD.
The Biotechnology and Biological Sciences Research Council (BBSRC) funded CD (grant BB/K008439/1) and RD (DTP grant BB/M011186/1). JL was partly supported by an Overseas Research Scholarship (ORS) award from Newcastle University.
We thank Prof. Heath Murray for help generating B. subtilis mutant strains, Dr. Mark Harrison for cloning the cotA gene and Dr. Gianpiero Landolfi for purifying BsCopZ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.916114/full#supplementary-material
Badarau, A., and Dennison, C. (2011a). Copper trafficking mechanism of CXXC-containing domains: Insight from the pH dependence of their Cu(I) affinities. J. Am. Chem. Soc. 133, 2983–2988. doi:10.1021/ja1091547
Badarau, A., and Dennison, C. (2011b). Thermodynamics of copper and zinc distribution in the cyanobacterium Synechocystis PCC 6803. Proc. Natl. Acad. Sci. U. S. A. 108, 13007–13012. doi:10.1073/pnas.1101448108
Banci, L., Bertini, I., Cantini, F., Felli, I. C., Gonnelli, L., Hadjiliadis, N., et al. (2006). The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat. Chem. Biol. 2, 367–368. doi:10.1038/nchembio797
Banci, L., Bertini, I., Cantini, F., Kozyreva, T., Massagni, C., Palumaa, P., et al. (2011). Human superoxide dismutase 1 (hSOD1) maturation through interaction with human copper chaperone for SOD1 (hCCS). Proc. Natl. Acad. Sci. U. S. A. 109, 13555–13560. doi:10.1073/pnas.1207493109
Banci, L., Bertini, I., Ciofi-Baffoni, S., Kozyreva, T., Zovo, K., and Palumaa, P. (2010). Affinity gradients drive copper to cellular destinations. Nature 465, 645–648. doi:10.1038/nature09018
Britton, R. A., Eichenberger, P., Gonzalez-Pastor, J. E., Fawcett, P., Monson, R., Losick, R., et al. (2002). Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184, 4881–4890. doi:10.1128/JB.184.17.4881-4890.2002
Changela, A., Chen, K., Xue, Y., Holschen, J., O’Halloran, T. V., and Mondragón, A. (2003). Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387. doi:10.1126/science.1085950
Chillappagari, S., Miethke, M., Trip, H., Kuipers, O. P., and Marahiel, M. M. (2009). Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 191, 2362–2370. doi:10.1128/JB.01616-08
Culotta, V. C., Klomp, L. W., Strain, J., Casareno, R. L., Krems, B., and Gitlin, J. D. (1997). The copper chaperone for superoxide dismutase. J. Biol. Chem. 272, 23469–23472. doi:10.1074/jbc.272.38.23469
Damle, M. S., Singh, A. N., Peters, S. C., Szalai, V. A., and Fisher, O. S. (2021). The YcnI protein from Bacillus subtilis contains a copper-binding domain. J. Biol. Chem. 297, 101078. doi:10.1016/j.jbc.2021.101078
Dennison, C., David, S., and Lee, J. (2018). Bacterial copper storage proteins. J. Biol. Chem. 293, 4616–4627. doi:10.1074/jbc.TM117.000180
DiSpirito, A. A., Semrau, J. D., Murrell, J. C., Gallagher, W. H., Dennison, C., and Vuilleumier, S. (2016). Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev. 80, 387–409. doi:10.1128/MMBR.00058-15
Durão, P., Chen, Z., Fernandes, A. T., Hilderbrandt, P., Murgida, D. H., Todorovic, S., et al. (2008). Copper incorporation into recombinant CotA laccase from Bacillus subtilis: Characterisation of fully copper loaded enzymes. J. Biol. Inorg. Chem. 13, 183–193. doi:10.1007/s00775-007-0312-0
Eichenberger, P., Fujita, M., Jensen, S. T., Conlon, E. M., Rudner, D. Z., Wang, S. T., et al. (2004). The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2, e328. doi:10.1371/journal.pbio.0020328
Eisenman, H. C., and Casadevall, A. (2012). Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940. doi:10.1007/s00253-011-3777-2
Engman, J., Rogstam, A., Frees, D., Ingmer, H., and von Wachenfeldt, C. (2012). The YjbH adaptor protein enhances proteolysis of the transcriptional regulator Spx in Staphylococcus aureus. J. Bacteriol. 194, 1186–1194. doi:10.1128/JB.06414-11
Enguita, F. J., Martins, L. O., Henriques, A. O., and Carrondo, M. A. (2003). Crystal structure of a bacterial endospore coat component. J. Biol. Chem. 278, 19416–19425. doi:10.1074/jbc.M301251200
Fernandes, A. T., Pereira, M. M., Silva, C. S., Lindley, P. F., Bento, I., Pinho Melo, E., et al. (2011). The removal of a disulfide bridge in CotA-lacces changes the slower motion dynamics involved in copper binding but has no effect on the thermodynamic stability. J. Biol. Inorg. Chem. 16, 641–651. doi:10.1007/s00775-011-0768-9
Festa, R. A., and Thiele, D. J. (2011). Copper: An essential metal in biology. Curr. Biol. 21, R877–R883. doi:10.1016/j.cub.2011.09.040
Gu, W., and Semrau, J. D. (2017). Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl. Microbiol. Biotechnol. 101, 8499–8516. doi:10.1007/s00253-017-8572-2
Hassett, R., and Kosman, D. J. (1995). Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J. Biol. Chem. 270, 128–134. doi:10.1074/jbc.270.1.128
Hirooka, K., Edahiro, T., Kimura, K., and Fujita, Y. (2012). Direct and indirect regulation of the ycnKJI operon involved in copper uptake through two transcriptional respresors, YcnK and CsoR, in Bacillus subtilis. J. Bacteriol. 194, 5675–5687. doi:10.1128/JB.00919-12
Hosan, K., Marlene, H., Paul, G., Derrell, C. M., Michele, M. O., Rong, W., et al. (2006). The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59, 487–502. doi:10.1111/j.1365-2958.2005.04968.x
Hullo, M. F., Moszer, I., Danchin, A., and Martin-Verstraete, I. (2001). CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183, 5426–5430. doi:10.1128/JB.183.18.5426-5430.2001
Jessberger, N., Dietrich, R., Granum, P. E., and Märtlbauer, E. (2020). The Bacillus cereus food infection as multifactorial process. Toxins 12, 701. doi:10.3390/toxins12110701
Lamb, A. L., Torres, A. S., O'Halloran, T. V., and Rosenzweig, A. C. (2001). Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8, 751–755. doi:10.1038/nsb0901-751
Lauraeus, M., Haltia, T., Saraste, M., and Wikström, M. (1991). Bacillus subtilis expresses two kinds of haem-A-containing terminal oxidases. Eur. J. Biochem. 197, 699–705. doi:10.1111/j.1432-1033.1991.tb15961.x
Lee, J., and Dennison, C. (2019). Cytosolic copper binding by a bacterial storage protein and interplay with copper efflux. Int. J. Mol. Sci. 20, 4144. doi:10.3390/ijms20174144
Ma, Z., Cowart, D. M., Scott, R. A., and Giedroc, D. P. (2009). Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry 48, 3325–3334. doi:10.1021/bi900115w
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106, 8344–8349. doi:10.1073/pnas.0812808106
Marckmann, D., Trasnea, P. I., Schimpf, J., Winterstein, C., Andrei, A., Schmoller, S., et al. (2019). The cbb3-type cytochrome oxidase assembly factor CcoG is a widely distributed cupric reductase. Proc. Natl. Acad. Sci. U. S. A. 116, 21166–21175. doi:10.1073/pnas.1913803116
Martins, L. O., Soares, C. M., Pereira, M. M., Teixeira, M., Costa, T., Jones, G. H., et al. (2002). Molecular and biochemical characterisation of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277, 18849–18859. doi:10.1074/jbc.M200827200
McKenney, P. T., Driks, A., and Eichenberger, P. (2013). The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11, 33–44. doi:10.1038/nrmicro2921
Meydan, S., Klepacki, D., Karthikeyan, S., Margus, T., Thomas, P., Jones, J. E., et al. (2017). Programmed ribosomal frameshifting generates a copper transporter and a copper chaperone from the same gene. Mol. Cell. 65, 207–219. doi:10.1016/j.molcel.2016.12.008
Nicolas, P., Mäder, U., Dervyn, E., Rochat, T., Leduc, A., Pigeonneau, N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. doi:10.1126/science.1206848
North, M. L., and Wilcox, D. E. (2019). Shift from entropic Cu2+ binding to enthaplic Cu1+ binding determines the reduction thermodynamics of blue copper proteins. J. Am. Chem. Soc. 141, 14329–14339. doi:10.1021/jacs.9b06836
Pufahl, R. A., Singer, C. P., Peariso, K. L., Lin, S. J., Schmidt, P. J., Fahrni, C. J., et al. (1997). Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278, 853–856. doi:10.1126/science.278.5339.853
Quisel, J. D., Burkholder, W. F., and Grossman, A. D. (2001). In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183, 6573–6578. doi:10.1128/JB.183.22.6573-6578.2001
Radford, D. S., Kihlken, M. A., Borrelly, G. P. M., Harwood, C. R., Le Brun, N. E., and Cavet, J. S. (2003). CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx-type ATPase CopA. FEMS Microbiol. Lett. 220, 105–112. doi:10.1016/S0378-1097(03)00095-8
Rae, T. D., Schmidt, P. J., Pufahl, R. A., Culotta, V. C., and O’Halloran, T. V. (1999). Undetectable intracellular free copper: The requirement for a copper chaperone for superoxide dismutase. Science 284, 805–808. doi:10.1126/science.284.5415.805
Rensing, C., Fan, B., Sharma, R., Mitra, B., and Rosen, B. P. (2000). CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97, 652–656. doi:10.1073/pnas.97.2.652(
Rensing, C., and McDevitt, S. F. (2013). The copper metallome in prokaryotic cells. Metall. Ions Life Sci. 12, 417–450. doi:10.1007/978-94-007-5561-1_12
Ridge, P. G., Zhang, Y., and Gladyshev, V. N. (2008). Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3, e1378. doi:10.1371/journal.pone.0001378
Sala, F. A., Wright, G. S. A., Antonyuk, S. V., Garratt, R. C., and Hasnain, S. S. (2019). Molecular recognition and maturation of SOD1 by its evolutionary destabilised cognate chaperone hCCS. PLoS Biol. 17, e3000141. doi:10.1371/journal.pbio.3000141
Schmidt, P. J., Rae, T. D., Pufahl, R. A., Hamma, T., Strain, J., O'Halloran, T. V., et al. (1999). Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 274, 23719–23725. doi:10.1074/jbc.274.34.23719
Smaldone, G. T., and Helmann, J. D. (2007). CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 153, 4123–4128. doi:10.1099/mic.0.2007/011742-0
Solioz, M., Abicht, H. K., Mermod, M., and Mancini, S. (2010). Response of Gram-positive bacteria to copper stress. J. Biol. Inorg. Chem. 15, 3–14. doi:10.1007/s00775-009-0588-3
Soni, A., Oey, I., Silcock, P., and Bremer, P. (2016). Bacillus spores in the food industry: A review on resistance and response to novel inactivation technologies. Comp. Rev. Food Sci. Food Saf. 15, 1139–1148. doi:10.1111/1541‒4337.12231
Strolle, P., Hou, B., and Brüser, T. (2016). The Tat substrate CueO is transported in an incomplete folding state. J. Biol. Chem. 291, 13520–13528. doi:10.1074/jbc.M116.729103
Tavares, M. B., Souza, R. D., Luiz, W. B., Cavalcante, R. C. M., Casaroli, C., Martins, E. G., et al. (2013). Bacillus subtilis endospores at high purity and recovery yields: Optimization of growth conditions and purification method. Curr. Microbiol. 66, 279–285. doi:10.1007/s00284-012-0269-2
Vita, N., Landolfi, G., Baslé, A., Platsaki, S., Lee, J., Waldron, K. J., et al. (2016). Bacterial cytosolic proteins with a high capacity for Cu(I) that protects against copper toxicity. Sci. Rep. 6, 39065. doi:10.1038/srep39065
Vita, N., Platsaki, S., Baslé, A., Allen, S. J., Paterson, N. G., Crombie, A. T., et al. (2015). A four-helix bundle stores copper for methane oxidation. Nature 525, 140–143. doi:10.1038/nature14854
von Wachenfeldt, C., Hallgren, J., and Hederstedt, L. (2021). YtkA (CtaK) and YozB (CtaM) function in the biogenesis of cytochrome c oxidase in Bacillus subtilis. Mol. Microbiol. 116, 184–199. doi:10.1111/mmi.14701
Keywords: copper, copper storage, bacterial copper homeostasis, bacterial laccases, sporulation, Bacillus subtilis
Citation: Lee J, Dalton RA and Dennison C (2022) Copper delivery to an endospore coat protein of Bacillus subtilis. Front. Cell Dev. Biol. 10:916114. doi: 10.3389/fcell.2022.916114
Received: 08 April 2022; Accepted: 26 July 2022;
Published: 05 September 2022.
Edited by:
Kourosh Honarmand Ebrahimi, King’s College London, United KingdomReviewed by:
Patrick Eichenberger, New York University, United StatesCopyright © 2022 Lee, Dalton and Dennison. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher Dennison, Y2hyaXN0b3BoZXIuZGVubmlzb25AbmNsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.