- 1Department of Biological Sciences, Indian Institute of Science Education and Research, Berhampur, Transit campus (Govt. ITI Building), Berhampur, Odisha, India
- 2EMBL, Rome, Italy
- 3Department of Biology, Indian Institute of Science Education and Research, Pune, Maharashtra, India
The molecular mechanisms that regulate stem cell pluripotency and differentiation has shown the crucial role that methylation plays in this process. DNA methylation has been shown to be important in the context of developmental pathways, and the role of histone methylation in establishment of the bivalent state of genes is equally important. Recent studies have shed light on the role of RNA methylation changes in stem cell biology. The dynamicity of these methylation changes not only regulates the effective maintenance of pluripotency or differentiation, but also provides an amenable platform for perturbation by cellular stress pathways that are inherent in immune responses such as inflammation or oncogenic programs involving cancer stem cells. We summarize the recent research on the role of methylation dynamics and how it is reset during differentiation and de-differentiation.
Introduction
The field of ‘Epigenetics’ has led to a “paradigm shift” in several domains of biomedical research (Deichmann, 2016). Waddington proposed the “epigenetic landscape” (EL) model in 1940, depicting a series of developmental options that a differentiating cell in the embryo could choose from. Epigenetics is now defined as “mitotically and/or meiotically heritable alterations in gene function that cannot be explained by changes in the DNA sequence.” The pluripotency of the undifferentiated cell and the eventual development of specific cell types is heavily reliant on the coordinated action of hundreds of transcription factors that bind to particular DNA regions to activate or repress cell lineage specific gene transcription (Srivastava and DeWitt, 2016). This establishment phase most closely reflects what is regarded as Conrad Waddington’s description of epigenetics, namely the study of the mechanisms by which the genotype gives the developmental phenotype. The maintenance phase usually involves a plethora of non-DNA sequence-specific chromatin cofactors that accumulate and maintain chromatin states through multiple cell divisions and for extended periods of time—sometimes even in the absence of the initial transcription factors (Schuettengruber et al., 2017).
Methylation dynamics in stem cells
The stem cells have been excellent cellular models to understand the molecular mechanisms of epigenetics. Stem cells are capable of self-renewal and differentiation to all three lineages, and can be classified as follows: a) Naïve stem cells (derived from the zygote of the mammalian embryo, capable of self-renewal and unrestricted differentiation potential), b) Primed stem cells/Epiblast stem cells (EpiSCs) (that originate from the zygotic stage immediately after maternal redetermination post implantation, capable of self-renewal but have a more lineage restricted differentiation potential, c) Embryonic stem cells (ESCs) (derived from the inner cell mass of the blastocyst, capable of self-renewal and multi-lineage differentiation potential, d) Adult stem cells (ASCs), found in adult tissues and organs within their respective niche responsible for maintaining tissue homeostasis, repair and regeneration. These stem cells remain in a quiescent state till activation by a signal like cell damage, and capable of self-renewal and multi-lineage differentiation potential, e) Cancer stem cells (CSCs) that are derived from the dedifferentiation of cancer cells or from the malignant transformation of normal stem cells. These cells like any other stem cells have self-renewal abilities and multi-lineage differentiation potential and play a major role in the prognosis of the disease (Zhou and Zhang, 2008; Harikumar and Meshorer et al., 2015; Morena et al., 2018).
These unique characteristics of a stem cell are regulated by molecular mechanisms that involve transcription factors, signalling pathways, epigenetics and epitranscriptomics. Transcription factors such as Oct3/4, Sox2, c-Myc and Nanog bind to their target genes and regulate their expression (Harikumar and Meshorer et al., 2015). Many signalling pathways such as the JAK/STAT, PI(3)K, MAPK, Wnt, Notch, Smad and FGF pathways play major roles in regulating stemness. The epigenome dynamics contributes to the regulation of stemness which includes biochemical modification of DNA, RNA, histone proteins, and chromatin. These modifications are carried out by specific enzymes where the “writer” and “eraser” proteins catalyse the addition and removal of the modifications respectively, while other proteins called “reader” proteins specifically recognize these modifications (Figure 1).
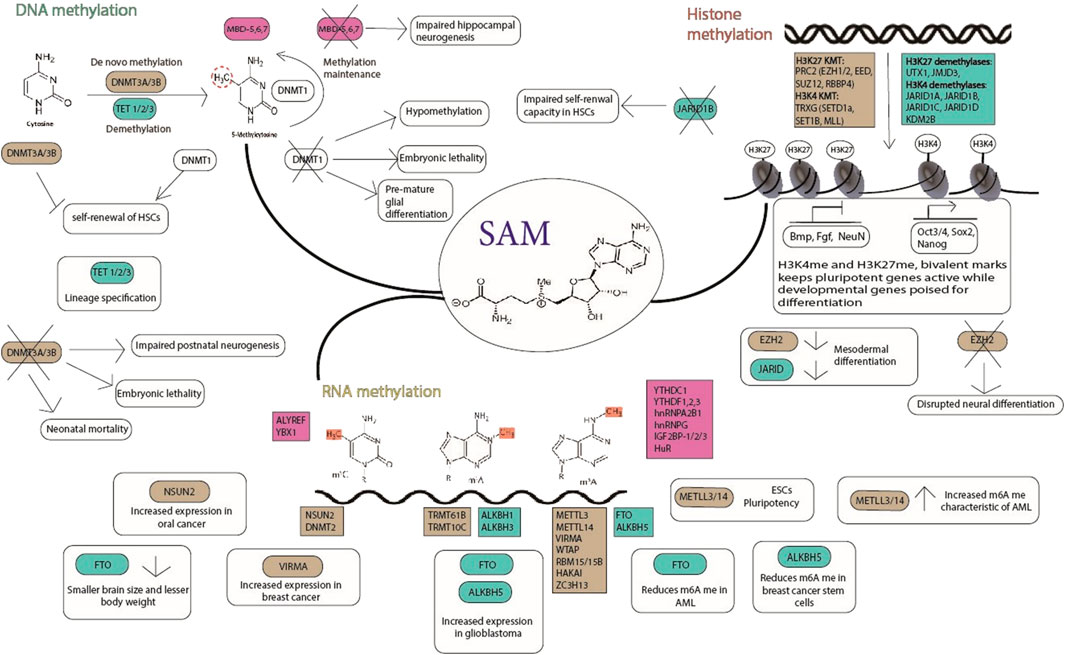
FIGURE 1. Methylation dynamics in stem cells; the readers (pink), writers (brown), and erasers (green) are indicated. SAM (S-adenosyl methionine) is the common methyl donor for histone, DNA and RNA methylation. The writers, erasers and readers play important roles in maintaining pluripotency and lineage commitment of stem cells. Their role in physiology and pathophysiology is depicted in the figure.
DNA Methylation: A family of DNA methyltransferases (DNMTs), catalyzes the Methylation of cytosine’s fifth carbon position, leading to 5-methylcytosine (5 mC) formation. DNMT1 copies existing methylation patterns for inheritance during DNA replication, while DNMT3A and DNMT3B act as de novo methyltransferases to create new methylation patterns (Bestor, 2000). A group of methyl-CpG-binding proteins acts as readers, interpreting the 5 mC signal and mediating its role. While DNA methylation can be ‘passively diluted’ by cell division, mechanisms for enzymatic DNA methylation removal have been recently discovered. The ten eleven translocation 1 (TET1) enzymes, catalyzes the conversion of 5mC to 5hydroxymethylcytosine (5hmC) (Tahiliani et al., 2009). Following that, three TET family proteins were discovered to be able to oxidize 5hmC to 5formylcytosine (5 fC) and then to 5carboxylcytosine (5caC) (Ito et al., 2010; He et al., 2011; Ito et al., 2011). In addition, the deaminases activation induced cytidine deaminase (AID; also known as AICDA) and apolipoprotein B mRNA editing enzyme catalytic polypeptides (APOBECs) can convert 5hmC to 5-hydroxymethyluracil (5hmU). To complete the active DNA demethylation process, thymine DNA glycosylase excises all of these derivatives and replaces them with an unmodified cytosine through the base-excision repair (BER) pathway (Wu and Zhang, 2014).
Histone methylation and demethylation: Histone methylation is a dynamic process that plays important functions in differentiation and development (Eissenberg and Shilatifard, 2010). Basic residues like lysine and arginine undergo methylation and can have several methylations on their side chains (Greer and Shi, 2012). H3K4me3 and H3K27me3 are two histone modifications that have been linked to active and repressive transcription, respectively. A variety of lysine methyltransferases (KMTs) as writers and lysine demethylases as erasers can mediate dynamic methylation of lysine residues. Many proteins, including the well-known PcG repressive complex (PRC) and Trithorax active complex (TRXG), have KMT properties (Schwartz and Pirrotta, 2007; Greer and Shi, 2012). Methylation of H3K4, H3K36, and H3K79 is associated with transcriptional activation, and methylation of H3K9, H4K20, and H3K27 is related with transcriptional repression. Notably, “bivalent domains” which are thought to be crucial for maintaining pluripotency by silencing developmental genes in embryonic stem cells (ESCs) and keeping them ready for activation during developmental stage—are formed when large regions of H3K27 methylation co-occur with smaller regions of H3K4 methylation marks (Bernstein et al., 2006; Hu et al., 2013).
RNA Methylation: More than 100 post-transcriptionally modified ribonucleosides have been found in various forms of RNA (Jia et al., 2013). N6-methyladenosine (m6A) is a conserved modification found in most eukaryotic nuclear RNAs, as well as some viral RNAs replicating in the host nuclei (Carroll et al., 1990). m6A was discovered as an abundant nucleotide modification in eukaryotic messenger RNA in 1970 (Desrosiers et al., 1974). In global cellular RNAs, m6A is found in 0.1–0.4% of all adenosines and accounts for almost half of all methylated ribonucleotides. m6A modification is enriched in long internal exons, upstream of stop codons, and the 3′-UTR of mRNA, suggesting roles in translational regulation, affecting RNA binding protein affinities, or distinctive m6A derived transcriptome topology (Dominissini et al., 2012; Meyer et al., 2012; Batista et al., 2014). The discovery of proteins involved in m6A regulation, as well as their roles as “writers” (m6A methyltransferases), “erasers” (m6A demethylases), and “readers” (effectors recognizing m6A), has been one of the most significant achievements in this field of study (Lee et al., 2014), together facilitate various functional outcomes, including nuclear RNA export, splicing, mRNA stability, circRNA translation, miRNA biogenesis, and lncRNA metabolism (Roignant and Soller, 2017; Yang et al., 2017) thus regulating physiological and pathological events such as Yeast meiosis, plant development, immunoregulation obesity, and carcinogenesis (Wang et al., 2017; Wei et al., 2017).
The epigenome in embryonic stem cells
Nucleosomes of stem cells show a higher level of modifications marks that are involved in active gene expression such as histone H3 lysine four trimethylation (H3K4me3), histone H4 lysine 9 and 14 acetylation (H3K9ac, H3K14ac). The two methyl modifications on H3K4 and H3K27 form a bivalent chromatin mark which is seen in the chromatin of stem cells. In stem cells, the highly conserved non-coding elements (HNCE) were found to be enriched with bivalent histone modifications, an active chromatin mark, H3K4me3 and a repressive chromatin mark, H3K27me3 (Bernstein, et al., 2006; Harikumar and Meshorer, 2015). These modifications are also abundant at promoter regions of genes that code for other factors required during development (Lessard and Crabtree, 2010). It is proposed that this bivalent chromatin mark resolves and there is activation of a few genes to regulate stemness while keeping other genes required for development poised for activation during development and cell differentiation (Bernstein et al., 2006; Lessard and Crabtree, 2010; Harikumar and Meshorer, 2015; Paranjpe and Veenstra, 2015). Recent studies have shown that many lineage-commitment genes have the bivalent mark and RNA polymerase II may be stalled at the promoters of these genes. During differentiation, the chromatin modifications are resolved into either an active or repressed state depending on the lineage commitment and these modifications can be newly established or maintained in differentiating cells (De Gobbi et al., 2011). Many early genes involved in the determination of the mesodermal lineage including various members of the GATA and Tbx families, Mixl1, and Brachyury, have bivalent domains in ES cells, supporting the notion that they are important early contributors (Pan et al., 2007). Histone arginine methylation has been shown to be important for pluripotency maintenance as well as lineage specification (Torres-Padilla et al., 2007; Selvi et al., 2015; Cui et al., 2017). Recent studies have shown that the RNA modifications have an important role in stem cell maintenance. The writer proteins are involved in controlling the expression of critical transcripts that are essential for stem cell self-renewal. m6A is shown to regulate molecular switches for differentiation and generation of EpiSCs, as well as in adult stem cells, like myeloid differentiation of hematopoietic stem cells (HSCs) (Morena et al., 2018).
The epigenome during differentiation
The embryonic stem cells undergo multiple rounds of differentiation, resulting in multipotent or unipotent adult stem cell progenitors. Extrinsic differentiation signals and intrinsic pathways interact and tightly regulate how stem cells differentiate. The formation of neurons and other ectodermal lineage cell types, has been one of the most well studied differentiation pathways. The perturbation of DNA methylation, histone methylation or RNA methylation leads to defects in neurogenesis. In mice, a mutation in any of the three main Dnmt genes causes significant developmental defects and embryonic or early postnatal death (Li et al., 1992; Okano et al., 1999). Methyl-CpG binding domain protein 1 (MBD1) binds to hypermethylated CpG islands in gene promoter regions preferentially, and its absence impairs adult hippocampal neurogenesis and genomic stability in vitro (Zhao et al., 2003). PcG proteins and TRXG have also been linked to neurogenesis regulation. In embryonic cortical NPCs, deletion of Enhancer of zeste homologue two in PRC2 (Ezh2) causes a global loss of H3K27me3, derepression of a large number of neuronal genes, and disrupted neuronal differentiation (Pereira et al., 2010). The RNA demethylase FTO has been shown to be expressed in adult neural stem cells and neurons and exhibits dynamic expression during postnatal neurodevelopment.
The role of the epigenome on differentiation has also been well studied in the hematopoietic stem cells (HSC). Hox genes, critical for maintaining the balance between self-renewal and differentiation of HSC and progenitor cells are associated with bivalent domains in undifferentiated ESCs and its sequential expression during differentiation is regulated by PcG and TRXG proteins (Deng et al., 2013). Hematopoietic specific genes such as CD45, CD34 among others exhibited repressive DNA methylation marks prior to differentiation of the ESC which are subsequently lost upon differentiation correlating with gene expression (Suelves et al., 2016). DNMT3a and DNMT3b act to repress self-renewal genes in HSCs and their combined loss enhances self-renewal by activating β-catenin signalling (Sharma and Gurudutta, 2016). DNMT1 aids in efficient hematopoietic differentiation and is crucial for the progression of cells to multipotent progenitors to lineage-restricted myeloid and lymphoid progenitor cells. DNMT3b is responsible for the de novo methylation of hematopoietic genes during early embryogenesis (Suelves et al., 2016). Deletion of the histone demethylase JARID1b compromises the self-renewal capability of the HSCs (Sharma & Gurudutta, 2016). The RNA m6A modification writer METTL, has also been shown to be essential for the symmetric division of HSCs (Cheng et al., 2019).
The epigenome in CSCs, during dedifferentiation
Cancer Stem Cells (CSCs) are a small subpopulation of cells within tumors, which are capable of self-renewal, differentiation, and tumorigenicity when transplanted into an animal host. CSCs can be distinguished from other cells within the tumor by differences in their cell division and gene expression (Rosen and Jordan, 2009). The first evidence for the presence of CSCs was shown in a study where leukemia initiating cell population from AML patients was identified based on the expression of (CD34+/CD38−) cell surface markers, by transplantation into severe combined immune-deficient (SCID) mice (Lapidot et al., 1994). The existence of Glioma stem cells (GSC) was first hypothesized in 2002, when it was considered to have migrated from the sub-ventricular niche. (Ignatova et al., 2002). It has now been shown that the genome-wide distribution of epigenetic signatures is associated with the differential programming of GSC and Neuronal Stem Cells (NSC) (Valor Luis and Hervás-Corpión 2020). CSCs are resistant to conventional chemotherapy or radiation treatment and can contribute to metastasis through the dedifferentiation process (Meirelles et al., 2012). High methylation can contribute to the self-renewing ability of CSCs during tumor progression (Muñoz et al., 2012). The epigenome modifications of CSCs play a major role in recurrence, metastasis, and therapeutic failure.
Resetting the epigenome through methylation dynamics
The dynamicity of the methylation mark on DNA, histones or RNA serves as an important biochemical rheostat for regulating stem cell pluripotency and lineage commitment along with other regulatory factors (Berdasco and Esteller, 2011; Völker-Albert et al., 2020; Sun et al., 2021). The reversible nature of these modifications provide an easy and efficient modulatory node that is used by cancer stem cells (Vincent et al., 2019; Carvalho, 2020). The expression of transcription factors, signalling pathways and other regulatory proteins in stem cell biology are under the control of this reversible modification.
A meta analysis of the available datasets was done to assess the changes that occur during these stages, as shown in Figure 2A. The transcription factors Oct3/4, Sox2, and Nanog expression are upregulated in ESCs because they are the core transcription factors in maintaining the pluripotency of the embryonic stem cells (Boyer et al., 2005). At the same time, Elf5, Gata4, Wt1, Stat6, Klf2, Tbx3, Cdx2, etc., are downregulated in ESCs. In Neural Stem Cells (NSCs) (NSCs), almost all of the TFs have average expression levels (Figure 2A, Panel I), with Sox2 at the highest level of expression. The cancer stem cells (CSCs) in gliomas, that would have undergone a dedifferentiation, show a very different expression level compared to the NSCs. Sox2, Sox9, and HIF1A show increased expression, whereas Cdx2, Esrrb, Wt1, etc., show decreased expression in CSCs. These expression levels could be the markers of cancer stem cells (Zhao et al., 2017). On comparing the three germ layers (Ectoderm, Endoderm, and Mesoderm), the expression level of TFs changes significantly, especially in the mesodermal lineage. The cells or tissues showing the elevated expression of the Eomes, Hif1a, Gata6, Gata4, Sox17, Otx2, etc., can be identified as an endodermal lineage. In addition to this, there is an expression of pluripotency factors such as Oct3/4 and Nanog. In ectodermal cells, we see the upregulated expression of Hif1a, Twist1, Sox2. Interestingly, the expression profile of ectodermal cells is somewhat similar to the CSCs.
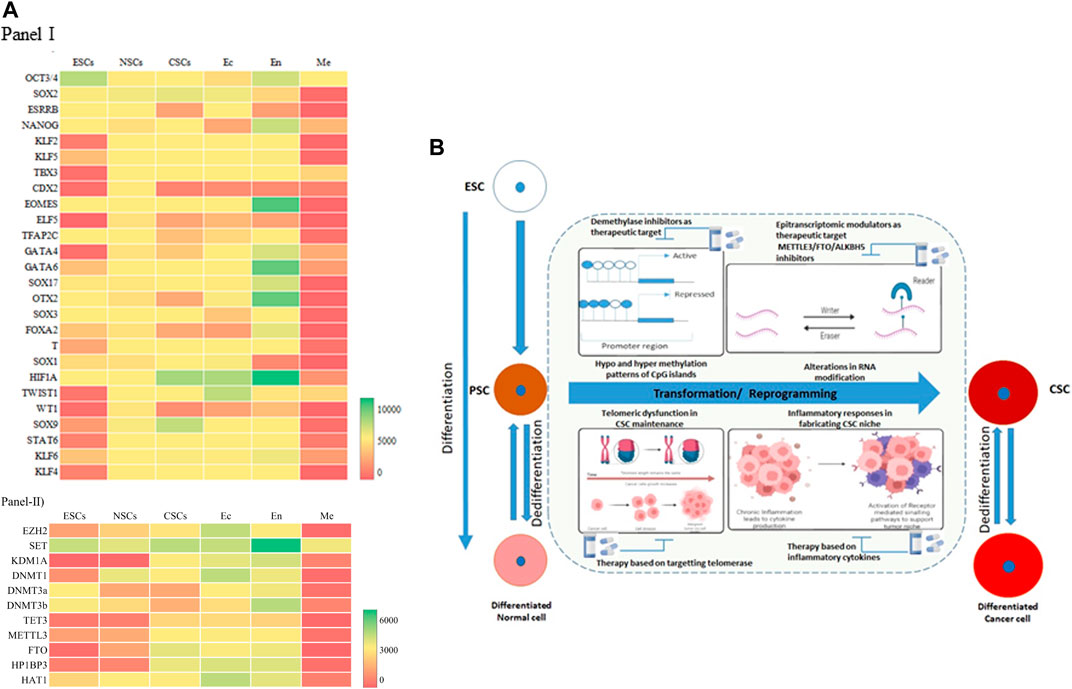
FIGURE 2. Resetting the Epigenome Dynamics in Differentiation and Dedifferentiation. (A) Heat map of Panel I, Transcription Factors (26) expression and Panel II modifiers (11) such as Histone methyltransferase/demethylase, DNMT/demethylase, RNA methyltransferase/demethylase in different cell types- embryonic stem cells (ESCs), neural stem cells (NSCs), cancer stem cells (CSCs), ectoderm (Ec), endoderm (En), mesoderm (Me). The red color indicates a lower expression level, and the green indicates a high expression level. Yellow represents intermediary expression levels. The intensity of the color indicates the expression level. Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) was used to gather the data for the six types of cells/tissues EMBRYONIC STEM CELLS (Datasets- GSE220881 and GSE775182), NEURAL STEM CELLS (Datasets- GSE380453 and GSE353904), CANCER STEM CELLS (Datasets- GSE433785 and GSE42906), ECTODERM (Datasets- GSE339037 and GSE1442418), ENDODERM (Datasets- GSE1080479, GSE5528310, and GSE2413511), MESODERM (Datasets- GSE18216112 and GSE11477613). Excel software was used to plot the heatmap. The selected data is converted into the heat map using conditional formatting (color scales). (B) A combination of various oncogenic events triggers transition of pluripotent stem cells and differentiated cells to cancer stem cells. Differential methylation at the CpG islands triggers oncogenes and transcription factors leading to emergence of tumor heterogeneity and CSCs. Alterations in global epitranscriptomic profile also regulate the reprogramming or dedifferentiation events. Cytokine and Interleukins acts in paracrine manner leading to cancer inflammation and crafts a niche for emergence of stem like cells by activating downstream signalling pathways. Telomere associated quiescence supports the stem like cellsin the tumor micro environment elevating their self-renewal capacity.
The epigenome modifiers such as Histone methyltransferases/demethylases, DNMTs/demethylases, RNA methyltransferases/demethylases also have dynamic expressions in the different cell types (Rwigemera et al., 2021). In ESCs, most of the transcription factors have moderate expression. SET has a higher expression level as opposed to the KDM1A, TET3, and FTO (Chung and Sidhu, 2008). NSCs also follow the same trends as ESCs (Figure 2A, Panel II). Ectodermal cells have higher expression levels of epigenome modifiers. Most epigenome modifiers have lower expression in the mesodermal cells except SET, KDM1A, DNMT3b, and HAT1. In CSCs, all the modifiers express moderately, except SET, FTO, and HP1BP3. This suggests an intermediary state of gene expression in the CSCs, where additional environmental factors can then come into play and facilitate tumour manifestation. It has been shown that Glioma stem cells (GSC), once formed, are also regulated by various signalling pathways, coordinated by epigenetic reprogramming. GSCs are reported to overexpress histone demethylase KDM4C, which removes H3K9me3 from Wnt target genes, promoting Wnt/Signalling Pathway and thereby stem cell maintenance (Chen et al., 2020; Kumar et al., 2022). Epigenetic regulators maintain tumoral hierarchy through two mechanisms, either through inhibition of self-renewal property of cancer cells thereby maintaining heterogeneity, or by facilitating CSCs in evading differentiation and maintenance of stem cell phenotype (Wainwright and Scaffidi, 2017; Valor Luis and Hervás-Corpión, 2020; Tao et al., 2022). RNA Methyltransferase, METTL3-mediated RNA stabilization positively regulates major signalling pathways such as Notch, NFκB, Wnt, c-Myc, TGFβ, involved in cancer stem cell maintenance and proliferation in several cancers including Glioma and Leukemia maintenance and tumorigenesis implying its oncogenic role (Visvanathan et al., 2018).
In this context, the inflammatory pathway has been shown to be recognized as a major component of tumorigenesis in various cancers. Solid tumors are also associated with Tumor Associated Macrophages (TAM) which constitute various immune infiltrating cells. These TAMs and stromal cells secrete cytokines such as Interleukin 1(IL1), IL6 and TNFα acts in paracrine fashion for sustenance and reprogramming of CSCs, by altering epigenetic mechanisms and thereby regulating transduction pathways such as NFκB, STAT3 and SMADs. (Biswas et al., 2013). These inflammatory pathways interconnect to form molecular regulatory circuits in resetting the networks for maintaining CSCs (Liu et al., 2021). Chronic inflammation can initiate DNA damage response in preneoplastic lesions, leading to telomere loss (Shay and Wright 2010). This triggers segregational defects, activation of telomerase and setting in of genomic instability, one of the major hallmarks of cancer. Patient derived CSCs in glioma have demonstrated shortened telomeres along with telomerase expression indicating the fact that GSCs are not quiescent and have the capacity for aberrant self-renewal properties (Koeneman et al., 1998). A summary of the alterations in reprogramming/transformation and de-differentiation is represented in Figure 2B. Inflammation regulates the acquisition and maintenance of the cancer stem cell phenotype by stimulating epithelial mesenchymal transitions. Many inflammatory factors like IL-1β, TGF-β, IL-6 can regulate the DNA methylation patterns that induce cancer initiation and progression in cancers such as gastric cancer, ovarian cancer, and liver cancer (Liu et al., 2021). The exact mechanisms of how the epigenome dynamics facilitates this process warrants further investigation which will provide useful therapeutic intervention prospects.
Author contributions
AR, SSP, IK, SN, HS, and RSB wrote the manuscript. IK and HS created the Figure 1, SN created Figure 2A, SP created Figure 2B.
Funding
Work in the authors’ laboratory is supported by research funding from Indian Institute of Science Education and Research Berhampur, Department of Science and Technology-SERB. IK is DST INSPIRE fellow. RSB is a DBT Ramalingaswami Fellow. Department of Science and Technology-SERB-SRG 2020.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Batista, P., Molinie, B., Wang, J., Qu, K., Zhang, J., and Li, L. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi:10.1016/j.stem.2014.09.019
Berdasco, M., and Esteller, M. (2011). DNA methylation in stem cell renewal and multipotency - stem Cell Research & Therapy. Biomed. Cent. 2 (5), 42. doi:10.1186/scrt83
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental gene in embryonic stem cells. Cell 125, 315–326. doi:10.1016/j.cell.2006.02.041
Bestor, T. H. (2000). The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402. doi:10.1093/hmg/9.16.2395
Biswas, S. K., Allavena, P., and Mantovani, A. (2013). Tumor-associated macrophages: Functional diversity, clinical significance, and open questions. Semin. Immunopathol. 35, 585–600. doi:10.1007/s00281-013-0367-7
Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucke, P. J., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. doi:10.1016/j.cell.2005.08.020
Carroll, S. M., Narayan, P., and Rottman, F. M. (1990). N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol. Cell Biol. 10, 4456–4465. doi:10.1128/mcb.10.9.445610.1128/mcb.10.9.4456-4465.1990
Carvalho, J. (2020). Cell reversal from a differentiated to a stem-like state at cancer initiation. Front. Oncol. 10, 541. doi:10.3389/fonc.2020.00541
Chen, Y., Fang, R., Yue, C., Chang, G., and Li, P. Q. (2020). Wnt-induced stabilization of KDM4C is required for wnt/β-catenin target gene expression and glioblastoma tumorigenesis. Cancer Res. 80, 1049–1063. doi:10.1158/0008-5472.CAN-19-1229
Cheng, Y., Luo, H., Izzo, F., Pickering, B. F., Nguyen, D., Myers, R., et al. (2019). m6A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 28, 1703–1716. doi:10.1016/j.celrep.2019.07.032
Chung, H., and Sidhu, K. S. (2008). Epigenetic modifications of embryonic stem cells: Current trends and relevance in developing regenerative medicine. Stem Cells Cloning 17, 11–21. doi:10.2147/sccaa.s3566
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634. doi:10.1016/j.celrep.2017.02.059
De Gobbi, M., Garrick, D., Lynch, M., Vermimmen, D., and Huges, J. R. (2011). Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin 4, 9. doi:10.1186/1756-8935-4-9
Deichmann, U. (2016). Epigenetics: The origins and evolution of a fashionable topic. Dev. Biol. 416, 249–254. doi:10.1016/j.ydbio.2016.06.005
Deng, C., Li, Y., Liang, S., Cui, K., Salz, T., Yang, H., et al. (2013). USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet. 9 (6), e1003524. doi:10.1371/journal.pgen.1003524
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc. Nat. Acad. Sci. 71, 3971–3975. doi:10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi:10.1038/nature11112
Eissenberg, J. C., and Shilatifard, A. (2010). Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249. doi:10.1016/j.ydbio.2009.08.017
Greer, E. L., and Shi, Y. (2012). Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13 (5), 343–357. doi:10.1038/nrg3173
Harikumar, A., and Meshorer, E. (2015). Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 16, 1609–1619. doi:10.15252/embr.201541011
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307. doi:10.1126/science.1210944
Hu, D., Garruss, A. S., Gao, X., Morgan, M. A., Cook, M., Smith, E. R., et al. (2013). The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1093–1097. doi:10.1038/nsmb.2653
Ignatova, T. N., Kukekov, V. G., Laywell, E. D., Suslov, O. N., Vrionis, F. D., and Steindler, D,A. (2002). Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39, 193–206. doi:10.1002/glia.10094
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi:10.1038/nature09303
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi:10.1126/science.1210597
Jia, G., Fu, Y., and He, C. (2013). Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115. doi:10.1016/j.tig.2012.11.003
Koeneman, K. S., Pan, C. X., Jin, J. K., Pyle, J. M., Flanigan, R. C., Shankey, T. V., et al. (1998). Telomerase activity, telomere length, and DNA ploidy in prostatic intraepithelial neoplasia (PIN). J. Urol. 160, 1533–1539. doi:10.1016/s0022-5347(01)62608-9
Kumar, Easwar V., Nambiar, R., De Souza, C., Nguyen, A., Chien, J., and Lam, K. S. (2022). Targeting epigenetic modifiers of tumor plasticity and cancer stem cell behavior. Cells 11, 1–23. doi:10.3390/cells11091403
Lapidot, T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., et al. (1994). A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. doi:10.1038/367645a0
Lee, M., Kim, B., and Kim, V. (2014). Emerging roles of RNA modification: m6A and U-tail. Cell 158, 980–987. doi:10.1016/j.cell.2014.08.005
Lessard, J. A., and Crabtree, G. R. (2010). Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 26, 503–532. doi:10.1146/annurev-cellbio-051809-102012
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi:10.1016/0092-8674(92)90611-f
Liu, Z., Ren, Y., Meng, L., Li, L., Beatson, R., Deng, J., et al. (2021). Epigenetic signaling of cancer stem cells during inflammation. Front. Cell Dev. Biol. 9, 772211. doi:10.3389/fcell.2021.772211
Meirelles, K., Benedict, L. A., Dombkowski, D., Pepin, D., Preffer, F. I., Teixeira, J., et al. (2012). Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc. Natl. Acad. Sci. U. S. A. 109, 2358–2363. doi:10.1073/pnas.1120733109
Meyer, K., Saletore, Y., Zumbo, P., Elemento, O., Mason, C., and Jaffrey, S. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Morena, F., Argentati, C., Bazzucchi, M., Emiliani, C., and Martino, S. (2018). Above the epitranscriptome: RNA modifications and stem cell identity. Genes 9, 329. doi:10.3390/genes9070329
Muñoz, P., Iliou, M. S., and Esteller, M. (2012). Epigenetic alterations involved in cancer stem cell reprogramming. Mol. Oncol. 6, 620–636. doi:10.1016/j.molonc.2012.10.006
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi:10.1016/s0092-8674(00)81656-6
Pan, G., Tian, S., Nie, J., Yang, C., Ruotti, V., Wei, H., et al. (2007). Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1, 299–312. doi:10.1016/j.stem.2007.08.003
Paranjpe, S., and Veenstra, J. G. C. (2015). Establishing pluripotency in early development. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1849, 626–636. doi:10.1016/j.bbagrm.2015.03.006
Pereira, J. D., Sansom, S. N., Smith, J., Dobenecker, M. W., Tarakhovsky, A., and Livesey, F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. 107, 15957–15962. doi:10.1073/pnas.1002530107
Roignant, J. Y., and Soller, M. (2017). m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 33, 380–390. doi:10.1016/j.tig.2017.04.003
Rosen, J. M., and Jordan, C. T. (2009). The increasing complexity of the cancer stem cell paradigm. Science 324, 1670–1673. doi:10.1126/science.1171837
Rwigemera, A., omri-Charai, R. E., Lecante, L. L., and Delbes, G. (2021). Dynamics in the expression of epigenetic modifiers and histone modifications in perinatal rat germ cells during de novo DNA methylation†. Biol. Reproduction 104, 361–373. doi:10.1093/biolre/ioaa206
Schuettengruber, B., Bourbon, H. M., Di Croce, L., and Cavalli, G. (2017). Genome regulation by polycomb and Trithorax: 70 Years and counting. Cell 171, 34–57. doi:10.1016/j.cell.2017.08.002
Schwartz, Y. B., and Pirrotta, V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22. doi:10.1038/nrg1981
Selvi, B. R., Swaminathan, A., Maheshwari, U., Nagabhushana, A., Mishra, R. K., and Kundu, T. K. (2015). CARM1 regulates astroglial lineage through transcriptional regulation of Nanog and posttranscriptional regulation by miR92a. Mol. Biol. Cell 26, 316–326. doi:10.1091/mbc.E14-01-0019
Sharma, S., and Gurudutta, G. (2016). Epigenetic regulation of hematopoietic stem cells. Int. J. Stem Cells 9, 36–43. doi:10.15283/ijsc.2016.9.1.36
Shay, J. W., and Wright, W. E. (2010). Telomeres and telomerase in normal and cancer stem cells. FEBS Lett. 584, 3819–3825. doi:10.1016/j.febslet.2010.05.026
Srivastava, D., and DeWitt, N. (2016). In vivo cellular reprogramming: The next generation. Cell 166, 1386–1396. doi:10.1016/j.cell.2016.08.055
Suelves, M., Carrió, E., Núñez-Álvarez, Y., and Peinado, M. A. (2016). DNA methylation dynamics in cellular commitment and differentiation. Briefings Funct. Genomics 15, 443–453. doi:10.1093/bfgp/elw017
Sun, W., Zhang, B., Bie, Q., Ma, N., Liu, N., and Shao, Z. (2021). The role of RNA methylation in regulating stem cell fate and function-focus on m6A. Stem Cells Int. 21, 8874360. doi:10.1155/2021/8874360
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi:10.1126/science.1170116
Tao, M., Li, X., He, L., Rong, X., Wang, H., Pan, J., et al. (2022). Decreased RNA m6A methylation enhances the process of the epithelial mesenchymal transition and vasculogenic mimicry in glioblastoma. Am. J. Cancer Res. 15, 893–906.
Torres-Padilla, M. E., Parfitt, D. E., Kouzarides, T., and Zernicka-Goetz, M. (2007). Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218. doi:10.1038/nature05458
Valor Luis, M., and Hervás-Corpión, I. (2020). The epigenetics of glioma stem cells: A brief overview. Front. Oncol. 10, 1–8. doi:10.3389/fonc.2020.602378
Vincent, A., Ouelkdite-Oumouchal, A., Souidi, M., Leclerc, J., Neve, B., and Seuningen, I. V. (2019). Colon cancer stemness as a reversible epigenetic state: Implications for anticancer therapies - PMC. PubMed Cent. (PMC) 11, 920–936. doi:10.4252/wjsc.v11.i11.920
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi:10.1038/onc.2017.351
Völker-Albert, M., Bronkhorst, A., Holdenrieder, S., and Imhof, A. (2020). Histone modifications in stem cell development and their clinical implications. Stem Cell Rep. 15, 1196–1205. doi:10.1016/j.stemcr.2020.11.002
Wainwright, E. N., and Scaffidi, P. (2017). Epigenetics and cancer stem cells: Unleashing, hijacking, and restricting cellular plasticity. Trends Cancer 3, 372–386. doi:10.1016/j.trecan.2017.04.004
Wang, S., Sun, C., Li, J., Zhang, E., Ma, Z., Xu, W., et al. (2017). Roles of RNA methylation by means of N6-76 methyladenosine (m6A) in human cancers. Cancer Lett. 408, 112–120. doi:10.1016/j.canlet.2017.08.030
Wei, W., Ji, X., Guo, X., and Ji, S. (2017). Regulatory role of N6-methyladenosine (m6A) methylation in RNA processing and human diseases. J. Cell Biochem. 118, 2534–2543. doi:10.1002/jcb.25967
Wu, H., and Zhang, Y. (2014). Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 156, 45–68. doi:10.1016/j.cell.2013.12.019
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641. doi:10.1038/cr.2017.31
Zhao, W., Li, Y., and Zhang, X. (2017). Stemness-related markers in cancer. Cancer Transl. Med. 3, 87–95. doi:10.4103/ctm.ctm_69_16
Zhao, X., Ueba, T., Christie, B. R., Barkho, B., McConnell, M. J., Nakashima, K., et al. (2003). Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. 100, 6777–6782. doi:10.1073/pnas.1131928100
Keywords: chromatin, epigenetics, epitranscriptomic modification, cancer stem cells, RNA methylation, readers
Citation: Roy A, Padhi SS, Khyriem I, Nikose S, Sankar S. H H and Bharathavikru RS (2022) Resetting the epigenome: Methylation dynamics in cancer stem cells. Front. Cell Dev. Biol. 10:909424. doi: 10.3389/fcell.2022.909424
Received: 01 April 2022; Accepted: 01 September 2022;
Published: 26 September 2022.
Edited by:
Chandrima Das, Saha Institute of Nuclear Physics (SINP), IndiaReviewed by:
Simone Sidoli, Albert Einstein College of Medicine, United StatesRob Illingworth, The University of Edinburgh; CRM, United Kingdom
Copyright © 2022 Roy, Padhi, Khyriem, Nikose, Sankar S. H and Bharathavikru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruthrotha Selvi Bharathavikru, YnJzZWx2aUBpaXNlcmJwci5hYy5pbg==
 Aiendrila Roy1,2
Aiendrila Roy1,2 Saket Nikose
Saket Nikose Ruthrotha Selvi Bharathavikru
Ruthrotha Selvi Bharathavikru