- 1School of Life Science, Beijing Institute of Technology, Beijing, China
- 2Key Laboratory of Molecular Medicine and Biological Diagnosis and Treatment, Beijing Institute of Technology, Ministry of Industry and Information Technology, Beijing, China
The repair of DNA damage is a complex process, which helps to maintain genome fidelity, and the ability of cancer cells to repair therapeutically DNA damage induced by clinical treatments will affect the therapeutic efficacy. In the past decade, great success has been achieved by targeting the DNA repair network in tumors. Recent studies suggest that DNA damage impacts cellular innate and adaptive immune responses through nucleic acid-sensing pathways, which play essential roles in the efficacy of DNA repair targeted therapy. In this review, we summarize the current understanding of the molecular mechanism of innate immune response triggered by DNA damage through nucleic acid-sensing pathways, including DNA sensing via the cyclic GMP-AMP synthase (cGAS), Toll-like receptor 9 (TLR9), absent in melanoma 2 (AIM2), DNA-dependent protein kinase (DNA-PK), and Mre11-Rad50-Nbs1 complex (MRN) complex, and RNA sensing via the TLR3/7/8 and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs). Furthermore, we will focus on the recent developments in the impacts of nucleic acid-sensing pathways on the DNA damage response (DDR). Elucidating the DDR-immune response interplay will be critical to harness immunomodulatory effects to improve the efficacy of antitumor immunity therapeutic strategies and build future therapeutic approaches.
1 Overview of DNA Damage and Repair Network
Up until now, chemotherapy and radiotherapy have remained the important treatment options for a variety of cancers (Hellmann et al., 2016; Chu et al., 2018; Qin et al., 2018; De Ruysscher et al., 2019; Grassberger et al., 2019; Galluzzi et al., 2020; Huang and Zhou, 2020; McLaughlin et al., 2020). The key mechanism of tumor cell death induced by standard chemotherapy and radiotherapy is DNA damage, leading to cell-cycle arrest and death directly or after S-phase DNA replication in the cell cycle. On the other hand, to deal with possible DNA lesions, tumors cells have evolved intricate repair mechanisms, and the ability to repair therapy-induced DNA damage would influence the therapeutic efficacy (Bouwman and Jonkers, 2012; Gavande et al., 2016). Here, we first overview the various types of DNA damage caused by radiotherapy and chemotherapy and the corresponding DNA damage repair pathways (see previous reviews for details) (Ciccia and Elledge, 2010; Giglia-Mari et al., 2011; Pinder et al., 2013; Chatterjee and Walker, 2017; Cussiol et al., 2019; Souliotis et al., 2020).
Some commonly used potent chemotherapy compounds (cyclophosphamide, dacarbazine, cisplatin, etc.) act by adding the alkyl groups to specific bases of DNA, yielding alkylated products such as O2-alkylthymine, O4-alkylthymine, O6-methylguanine, and O6-ethylguanine (Serrone et al., 2000; Emadi et al., 2009; Dasari and Tchounwou, 2014). Monofunctional alkylating agents have one active moiety and can only modify a single base, while bifunctional alkylating agents have two reaction sites, which can crosslink DNA to protein or another DNA base resulting in intra-strand crosslinks or inter-strand crosslinks. Radiotherapy by ionizing radiation (IR) can attack DNA directly by breaking the phosphodiester bond and the deoxyribose, and other indirect means have also been confirmed, for example, highly reactive oxygen species (ROS) produced from water radiolysis could result in the oxidization of the DNA desoxyribose moiety and the four nitrogenous bases (Henner et al., 1982; Desouky et al., 2015). Thereby, chemotherapy and radiotherapy can result in various kinds of DNA damage including base damage, single-strand breaks (SSBs), and double-strand breaks (DSBs). Among them, DSBs are thought to be the most harmful to cell survival and are the main mechanism to promote the therapeutic effect.
To remedy various DNA damage types, there has developed a complex DNA damage response (DDR) network (Giglia-Mari et al., 2011). The pathways involved in DNA damage repair mainly include direct reversal, base excision repair (BER), nucleotide excision repair (NER), non-homologous end joining (NHEJ), and homologous recombination (HR) pathway. Direct reversal, the simplest DNA repair pathway, depends primarily on a single protein and does not involve nucleotide removal, resynthesis, or ligation. For example, the O6-alkyl group of guanines can be removed by O6-methylguanine DNA methyltransferase (MGMT). BER and NER pathways take part in the DNA SSBs repairment. BER often participates in the repair of the small but highly mutagenic DNA lesions, which usually significantly undermine genomic fidelity and stability (Dianov and Hübscher, 2013). The BER pathway is initiated with the excision of the damaged base by any of 11 DNA glycosylases (Krokan et al., 1997), after which the exposed gap will be filled by a different set of proteins, among which the repair of single-base gaps require the short-patch pathway while polybasic gaps are in need of the long-patch pathway (Woodrick et al., 2017; Biau et al., 2019; Caldecott 2020). The NER pathway involves multiple steps requiring more than 30 proteins and is the main pathway used by mammals to remove bulky DNA lesions, including numerous chemical adducts, intra-strand crosslinking of DNA, and some forms of oxidative damage. In the NER pathway, the damaged bases are first recognized, then the DNA double-strand is unwound, and then the excision repair complex will remove the damaged bases followed by filling and ligating of the gap (Gillet and Schärer, 2006; Shuck et al., 2008).
DSB is a highly toxic gene damage that seriously threatens cellular homeostasis by affecting the transcription of genes, DNA replication, and chromosome segregation. Failure in repairing DSBs can lead to devastating chromosomal instabilities, and result in the dysregulation of gene expression and an increased hazard of carcinogenesis (Kryston et al., 2011). In human cells, two pathways, HDR and NHEJ, take part in DNA DSBs repair (Chang et al., 2017). HDR is generally considered to be participated in DSBs repair only during the S and G2 phases of the cell cycle given that it needs a homologous template to replace the damaged DNA segment in the genome, but it has been shown that centromeric DSBs in the G1 phase can activate the HDR pathway to maintain centromeric integrity recently (Yilmaz et al., 2021). The canonical HDR pathway is relatively slow but error-free, which needs numerous factors involved in homology search, Holliday junction formation, DNA synthesis, and the final DNA ligation (San Filippo et al., 2008; Krejci et al., 2012). Unlike the HR pathway, the NHEJ pathway does not need a DNA template and is active throughout the whole cell cycle, therefore it responds relatively quickly but is error-prone (Chang et al., 2017). In NHEJ pathway, four specific steps are involved including DNA termini recognition, bridging of the DNA ends, DNA end processing, and DNA ligation.
2 DNA Repair Targeted Therapy
Given that tumor cells could repair DNA damage induced by chemotherapy and radiotherapy in order to survive, the use of inhibitors of specific DNA repair pathways combined with DNA-damaging treatment can be efficacious. Some DNA repair inhibitors have been exploited as clinical agents targeting the proteins involved in sensing and conducting DNA damage signals as well as other proteins in DNA repair pathways (Mateo et al., 2019; Cheng et al., 2022).
DNA damage sensor proteins are key functional proteins to initiate repair and can sense multiple DNA damage signals, in which poly (ADP-ribosyl) polymerase-1 (PARP-1) is widely recognized to be the primary responder to SSBs while it could also bind and signal DSBs (Li and Yu 2013; Ceccaldi et al., 2015; Mateos-Gomez et al., 2015; Pandey and Black, 2021). After rapidly detecting the DNA damage, PARP-1 synthesizes of poly (ADP-ribose) (PAR) chains on itself and many different proteins near the damage site initiating recruitment of DNA repair complexes. The formation of the PAR chain can promote the release of PARP-1 from the position where it binds to the damaged DNA so that the other repair proteins could contact with the damaged site. Inhibition of PARP will reduce the synthesis of PAR chains, making PARP unable to dissociate from damaged DNA, thus preventing the recruitment of other repair proteins (Zandarashvili et al., 2020). The failure of PAR chain formation and the release of PARP from damaged DNA will lead to the enrichment of SSBs, which can be transformed into single-sided DSBs during DNA replication (Bixel and Hays, 2015). However, in the cells with the absence of intact DSBs repair pathways, such as in BRCA1 and BRCA2 mutated cells, the persistent DSBs are toxic and even deadly. Many small molecule PARP inhibitors (PARPi) targeting the catalytic activity of PARP-1 are now approved and clinically used in patients with breast, ovarian, prostate, and pancreatic cancers deficient in other DDR components, and the expanded utilities of small molecule PARP inhibitors in other cancer types are under consideration (Ramakrishnan Geethakumari et al., 2017; Zimmer et al., 2018; Hammel et al., 2020; Xie et al., 2020).
For DSBs, lupus Ku autoantigen protein (Ku) and the Mre11-Rad50-Nbs1 complex (MRN) play important roles. Ku is a protein heterodimer composed of Ku70/Ku80, which takes part in the NHEJ pathway and binds to DNA DSBs (Chen et al., 2021). Upon recognition and binding of DSBs, Ku recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) that assists in classical NHEJ repair. A class of compounds has been developed that abrogates the Ku-DNA end binding activity, inhibits cellular NHEJ, and enhances the cellular activity of radiomimetic agents and IR (Gavande et al., 2020). The MRN complex has nuclease activity and can bind DNA, so it can participate in the initial detection and processing of DSB (Rupnik et al., 2008), which is dependent on the nuclease activity of Mre11, the central factor of the MRN complex with endonuclease activity and 3′–5′ exonuclease activity (Stracker and Petrini, 2011). Upon bound to the damaged position, MRN recruits the DNA-damage signaling kinase ataxia-telangiectasia mutated (ATM), activates it, and triggers a series of signaling events that drive HR repair (Uziel et al., 2003). A class of inhibitors has been developed to selectively block the nuclease activity of Mre11 and prevent DNA damage repair (Dupré et al., 2008).
DNA damage signaling proteins trigger multiple post-translational modifications and the assembly of protein complexes, which amplify and diversify the DNA damage signals. Initially, one of three phosphatidylinositol-3 kinase-related kinases (PIKKs): DNA-PKcs, ATM, or ATM- and Rad3-Related (ATR) is activated by phosphorylation in response to DNA damage (Woods and Turchi, 2013). DNA-PKcs, forming a heterotrimeric complex with Ku, are required for proper DSBs repair by NHEJ. Using its activated kinase activity after being bound to the DNA terminus, it will phosphorylate itself and other target proteins to coordinates the NHEJ pathway (Falck et al., 2005; Uematsu et al., 2007). Following the appearance of DSB, the MRN complex activates ATM, which then phosphorylates histone H2AX as the main kinase (Burma et al., 2001). ATM could also phosphorylate checkpoint kinase 2 (Chk2) and p53 to impact cell cycle regulation and cytotoxicity (Cheng and Chen, 2010; Smith et al., 2010). ATR participates in HR, NER, long-patch BER, postreplication repair, interstrand cross-link repair, and replication fork restart after its activation by replication protein A (RPA)-coated ssDNA (Cimprich and Cortez, 2008). Several small molecules targeting three PIKKs such as VX-984 and CC-115 for DNA-PKcs, AZD0156 for ATM, VX-970, and AZD6738 for ATR are currently in different stages of clinical trials (Munster et al., 2016; Foote et al., 2018; Pike et al., 2018; Timme et al., 2018; Gorecki et al., 2020). In addition, checkpoint kinase 1 (Chk1) and Chk2, protein kinases that lie downstream of ATR and ATM, have also been utilized as therapeutic targets for drug development (Jobson et al., 2009; King et al., 2014).
3 Nucleic Acid-Sensing Pathways Connect DNA Damage to Innate Immunity
Cancer chemotherapy and radiotherapy aim to induce catastrophic DNA damage such as DSBs to cause cancer cell apoptosis, which can further aggravate the degree of DNA damage and promote the therapeutic effect when combined with DNA damage repair inhibitors. According to the severity of the DNA damage, some cancer cells directly initiate programmed cell death to clear the damaged genome beyond endurance. Besides, nucleic acid released from dying cells can activate the innate immune response of surrounding cells (Wang et al., 2021b). Even if DNA damage does not directly kill cells, increasing evidence indicates that the accumulation of nucleic acids in the cytoplasm caused by DNA damage can also trigger an inflammatory response within the cells (Nastasi et al., 2020). DNA damage-induced cytosolic nucleic acid shares common receptors (pattern recognition receptors, PRRs) and downstream effectors with those induced by viral or bacterial infections (Takeuchi and Akira, 2010; Taffoni et al., 2021), which are summarized below (Figure 1). And some agonists of these nucleic acid-sensing pathways are already in clinical trials (Table 1). Strikingly, recent research has indicated that the proteins that participated in DNA repair also play active roles in innate immune signaling.
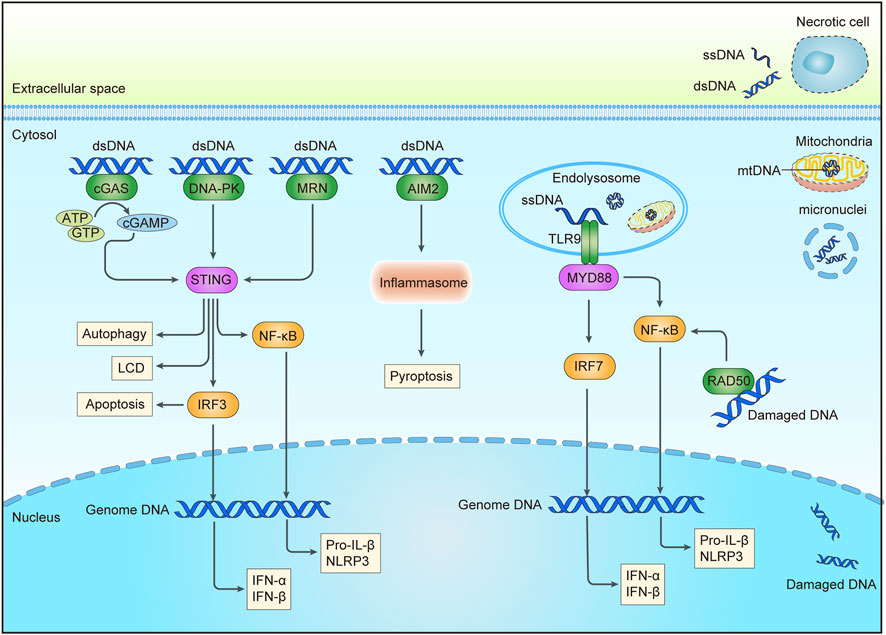
FIGURE 1. DNA sensing pathways triggered by DNA damage. An abnormal increase of intracellular DNA could come from the nucleus, micronuclei, or mitochondria after chemotherapy or radiation. DNA in endosomal may be from extracellular DNA of necrotic cells through endocytosis or cytoplasmic DNA through autophagy. Sensors for DNA are shown in green, including cGAS, DNA-PK, MRN, AIM2, and RAD50 in the cytoplasm, and TLR9 in the endolysosome. Adaptor molecules are shown in pink and downstream signaling molecules are shown in yellow. Activation of these pathways may result in the production of interferon (IFN) and other cytokines, apoptosis, pyroptosis, autophagy, etc., cGAS, cyclic GMP-AMP synthase; DNA-PK, DNA-dependent protein kinase complex; MRN, Mre11-Rad50-Nbs1 complex; AIM2, absent in melanoma 2; TLR9, Toll-like receptor 9; cGAMP, cyclic GMP-AMP; STING, stimulator of interferon genes; MYD88, myeloid differentiation primary response protein 88; IRF3/7, interferon regulatory factor 3/7; NF-κB, nuclear factor-κB; NLRP3, NOD-, LRR- and pyrin domain-containing 3; LCD, a lytic cell death program; dsDNA, double stranded DNA; ssDNA, single stranded DNA; mtDNA, mitochondrial DNA.
3.1 DNA Sensing Pathways
3.1.1 Cyclic GMP-AMP Synthase
Normally, DNA is trapped in the nucleus and mitochondria and rapidly degraded by nucleases in the cytoplasm and endolysosomes. DNA-sensing receptors could detect increased amounts of intracellular DNA. Currently, cyclic GMP-AMP synthase (cGAS), a nucleotidyltransferase (NTase), is the most widely accepted dsDNA sensor that acts followed by stimulator of interferon genes (STING) performing multiple functions (Kuchta et al., 2009; Sun et al., 2013; Wu et al., 2013). cGAS normally resides to be inactive. Initial studies considered cGAS as a cytoplasmic protein, in which cGAS could not interact with nuclear or mitochondrial DNA, but some recent works indicate that cGAS also resides in the nucleus constitutively (Gentili et al., 2019; Jiang et al., 2019; Volkman et al., 2019). cGAS can be activated not only by viral or bacterial infection-related DNA entering the cytoplasm but also by endogenous self-DNA, including cytosolic DNA from nucleus and mitochondria, DNA in cytoplasmic micronucleus, and chromatin in the nucleus (Li X.-D. et al., 2013; West et al., 2015; Dou et al., 2017; Glück et al., 2017; Harding et al., 2017; Mackenzie et al., 2017; Gratia et al., 2019). Upon binding to DNA, cGAS assembles into a dimer at an active state and converts ATP and GTP into the second messenger cyclic GMP-AMP (cGAMP) (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013; Zhang et al., 2013). The complete activation and stabilization of cGAS-DNA complexes require DNA lengths to exceed a certain threshold, allowing two or more cGAS molecules to bind to the same DNA to form oligomeric structures or condensates (Li X. et al., 2013; Zhang J.-Z. et al., 2014; Andreeva et al., 2017; Luecke et al., 2017; Du and Chen, 2018; Hooy and Sohn, 2018). The cyclic-dinucleotide sensor STING, an ∼40 kDa dimeric transmembrane protein located in the endoplasmic reticulum (ER), could detect and bind to cGAMP to make a conformational change, which results in its translocation from the ER to the Golgi apparatus and the activation of TANK-binding kinase 1 (TBK1) (Ishikawa and Barber, 2008; Burdette et al., 2011; Liu et al., 2015; Ergun et al., 2019; Shang et al., 2019; Zhang et al., 2019; Zhao et al., 2019). The activated TBK1 phosphorylates itself, STING, and the interferon regulatory factor 3 (IRF3), after which the active IRF3 dimer is transported to the nucleus to activate the expression of type I interferon genes. The cGAS-STING signaling can also lead to the transcription of pro-inflammatory cytokines-related genes via nuclear factor-κB (NF-κB) (de Oliveira Mann et al., 2019). In addition, STING is related to autophagy induction, however, its functional mechanism still remains to be elucidated (Watson et al., 2012). Growing studies suggest that the activation of STING can trigger cell death by various means. For example, STING could facilitate respective programmed cell death by inducing the production of many pro-apoptotic and pro-necroptotic molecules (Paludan et al., 2019). Furthermore, the accumulation of lysosomal STING could trigger lysosome membrane permeabilization, leading to the release of lysosomal hydrolases with cell death as a result (Gaidt et al., 2017). Besides, phosphorylated IRF3 downstream of STING can stimulate apoptosis by reducing the Bcl-xL-dependent suppression of the permeability of mitochondrial outer membrane in mitotic cells (Zierhut et al., 2019; Xie et al., 2021).
3.1.2 Toll-Like Receptors 9
Toll-like receptors (TLRs) are important components of innate immune responses induced by pathogenic microorganisms or tissue injury (Moresco et al., 2011). TLR, a highly conserved intracellular transmembrane protein, has emerged as a key PRR in the past 20 years. It exists in multiple types of cells, such as T cells, B cells, APC, epithelial cells, and endothelial cells (Chang 2010). All TLRs possess a leucine-rich-repeat (LRR) domain binding ligand extracellularly, a transmembrane domain, and a cytosolic Toll/IL-1 receptor (TIR) homology domain (Delneste et al., 2007). In all TLRs, TLR9 is found specifically in the endosomes and can be activated by single-stranded DNA containing unmethylated cytidine-phosphate-guanosine (CpG) dinucleotides escaping from the digestion of nucleases such as DNase II (Kumagai et al., 2008). TLR9 must undergo proteolytic processing in the endosomal compartments to complete ligand-mediated dimerization and activation (Majer et al., 2017). TLR9 not only triggers plasmacytoid dendritic cells to produce type I IFN and activate the polyclonal B cells through the myeloid differentiation primary response protein 88 (MYD88) and interferon-regulatory factor 7 (IRF7) signaling pathway, but also induces the production of inflammasome-related factors pro-interleukin-1β (pro-IL-1β) and NOD-, LRR- and pyrin domain-containing 3 (NLRP3) via NF-κB (Honda et al., 2005; Zhang X. et al., 2014). According to a recent study, TLR9 can sense mitochondrial DNA during mitophagy, and induce C-X-C motif chemokine ligand 10 (CXCL10) expression and CD8+ T cell recruitment, which reveals a novel role of chemotherapy in the innate immune response (Limagne et al., 2022).
3.1.3 Absent in Melanoma 2
The absent in melanoma 2 (AIM2), containing pyrin and HIN domains, takes an important part in inflammasome activation as the dsDNA-sensing receptor in the cytoplasm of cells (Fernandes-Alnemri et al., 2009; Hornung et al., 2009). The N-terminal pyrin domain (PYD) of AIM2 could interact with apoptosis-associated speck-like protein (ASC), which possesses a caspase recruitment domain (CARD) and a C-terminal HIN domain sensing cytoplasmic DNA (Hornung et al., 2009; Wang and Yin, 2017). The recruitment of ASC leads to the generation of fibrilar super-structures, to which Caspase-1 is bound by CARD-CARD interactions (Miao et al., 2011; Lugrin and Martinon, 2018). The oligomerization of ASC leads to the activation of some proteins such as the conversion of pro-IL-1β into the biologically active IL-1β. The inflammasome can also trigger pro-inflammatory pyroptosis (Liang et al., 2020). Gasdermins family proteins are the important factors mediating inflammatory cell death (Kovacs and Miao, 2017), in which Gasdermin D (GSDMD) can be cleaved by Caspase-1 to release the N-terminal of Gasdermin so that it can polymerize and cause perforation of the plasma membrane, allowing the intracellular substance to leak out, causing cell death (Lammert et al., 2020). It is well known that AIM2 can detect dsDNA in the cytosol, but recent studies have found that AIM2 can also sense DSBs directly within the nucleus to induce intestinal epithelial cells and bone marrow cells to initiate the caspase-1-dependent death (Hu et al., 2016).
3.1.4 DNA-PK Complex
Recent findings reveal that some proteins in DNA repair also take part in the sense of foreign DNA in the cytosol. DNA-dependent protein kinase (DNA-PK) complex assembled from Ku70/Ku80 heterodimer and the kinase subunit (DNA-PKcs) can sense DNA DSBs in response to repair the DNA damage in the NHEJ pathway, and Ku has also been reported to detect viral DNA in human cells as a PRR to induce type I and type III interferons or pro-inflammatory cytokines (Abe et al., 2019). DNA stimulates the Ku complex to translocate into the cytoplasm and bind to the dsDNA terminals using its middle domain independent of DNA sequence (Sui et al., 2021a; Sui et al., 2021b). Numerous studies have reported that STING is the downstream adaptor of Ku to induce type I/III interferons and inflammatory cytokines via phosphorylation of IRF3 (Sui et al., 2017). Ku complex is abundantly expressed in aged human and mouse CD4+ T cells, and it recognizes accumulating cytoplasmic DNA in the cytoplasm, which facilitates the recruitment of DNA-PKcs and phosphorylation of the kinase ZAK, ultimately promoting the proliferation and activation of CD4+ T cells (Wang et al., 2021a).
3.1.5 Mre11-Rad50-Nbs1 Complex
MRN complex consisting of two MRE11 subunits, two RAD50 units, and two NBS1 subunits, can sense and respond to DNA damage firstly in DDR to orchestrate DDR response in DSBs and replication fork collapse (Zhu et al., 2018; Tisi et al., 2020). The hetero-hexamers MRN complex could conduct ATP hydrolysis of RAD50, bind multiple DNA molecules, and link DNA molecules with exonuclease and endonuclease (Kondo et al., 2013). RAD50 could recognize cytosolic dsDNA and bind caspase-recruitment domain (CARD9), a pro-inflammatory signaling adaptor, to its zinc-hook region, which tends to recruit Bcl-10, leading to the activation of NF-κB and the generation of pro-inflammatory cytokine IL-1β (Roth et al., 2014).
3.2 RNA Sensing Pathways
3.2.1 Toll-Like Receptor 3/7/8
In mammalian cells, both single-stranded and double-stranded RNA can be recognized by PRR as pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) (Figure 2). RNA from necrotic cells may be internalized by cells via clathrin-dependent endocytosis, or it can enter cells after complexation with peptides or within immune complexes (Lövgren et al., 2004; Barrat et al., 2005; Itoh et al., 2008; Ganguly et al., 2009). The activation of TLR, a type I transmembrane protein, is required for inducing innate and adaptive immune responses, among which TLR3 can recognize double-stranded RNA and structured RNA containing a partial stem in secondary structures of single-stranded RNA, and TLR7/8 can recognize the fragments of single-stranded RNA (Alexopoulou et al., 2001; Karikó et al., 2004; Cavassani et al., 2008; Tatematsu et al., 2013). Depending on the intracellular compartments to recognize ligands, discriminate self-and non-self-derived nucleic acids, TLRs activate the downstream signaling pathways (Gay et al., 2014). Upon ligand binding, the dimers of TLRs will emerge after binding ligands to recruit the cytosolic adaptor proteins with TIR domain, such as MyD88 and Toll-like receptor adaptor molecule 1 (TRIF) (Yamamoto et al., 2003; Vyncke et al., 2016). The recruitment of MyD88 could activate NF-κB through type I interferon induction by IRF7 and tumor necrosis factor receptor-associated factor 6 (TRAF6). Intriguingly, among the TLRs, only TLR3 could signal to induce the generation of type I interferon and proinflammatory cytokines by the recruitment of TRIF rather than depending on MyD88 (Oshiumi et al., 2003).
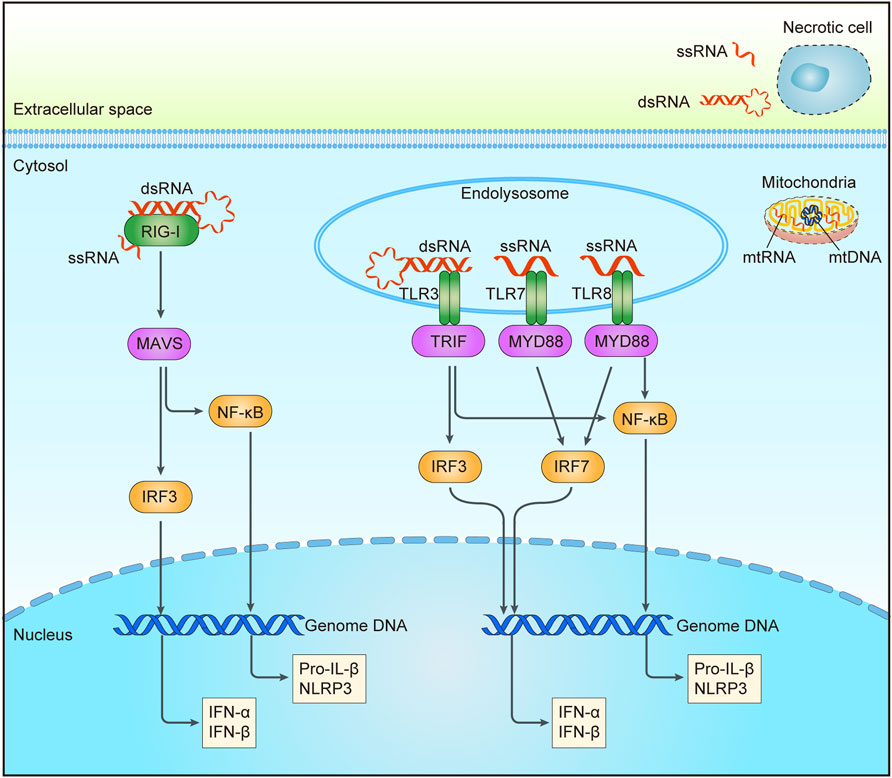
FIGURE 2. RNA sensing pathways triggered by DNA damage. Aberrant increase of intracellular RNA could be from mitochondria after chemotherapy or radiation. RNA in endosomal may be from extracellular RNA of necrotic cells through endocytosis or cytoplasmic RNA through autophagy. Sensors for RNA are shown in green, including RIG-1 in the cytoplasm, and TLR3/7/8 in the endolysosome. Adaptor molecules are shown in pink and downstream signaling molecules are shown in yellow. Activation of these pathways may result in the production of interferon (IFN) and other cytokines, etc. RIG-I, a retinoic acid-inducible gene I; TLR3/7/8, Toll-like receptor 3/7/8; MAVS, mitochondrial antiviral signaling protein; TRIF, Toll-like receptor adaptor molecule 1; MYD88, myeloid differentiation primary response protein 88; IRF3/7, interferon regulatory factor 3/7; NF-κB, nuclear factor-κB; dsRNA, double stranded RNA; ssRNA, single stranded RNA; mtRNA, mitochondrial RNA; mtDNA, mitochondrial DNA.
3.2.2 (RIG-I)-Like Receptors
RLRs are composed of retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology2 (LGP2), which can help the innate immune system sense cytosolic RNA (Kang et al., 2004; Yoneyama et al., 2004; Yoneyama et al., 2005; Ori et al., 2017). RIG-I and MDA5 contain two CARDs at the N-terminus, a DExD/H box RNA helicase domain in the central and a C-terminal domain (CTD), which mediate downstream signaling by sensing RNA. MDA5 preferentially recognizes dsRNA longer than 1 kb unlike RIG-I, which could bind relatively short dsRNA (Kato et al., 2008; Goubau et al., 2014). The CARD domains of RIG-I and MDA5 oligomerize after binding to dsRNA and tend to interact with the CARD of mitochondrial antiviral signaling protein (MAVS) (Kawai et al., 2005; Seth et al., 2005). Then the oligomerization of MAVS occurs after binding to RIG-I or MDA5 to form prion-like aggregates, upon which the downstream signaling pathways can be activated (Hou et al., 2011). Besides, MAVS could trigger the transcription of type I IFNs via IRF3 or IRF7 by activating TBK1, and the transcription of inflammatory cytokines via NF-κB by activating the IKK complex (IKKα, IKKβ, NEMO) (Goubau et al., 2013; Ori et al., 2017).
Recent research has found that the DNA fragments released from ionizing radiation-induced double-strand DNA breaks could activate both the cGAS/STING-dependent DNA-sensing pathway and the MAVS-dependent RNA sensing pathway (Feng et al., 2020). Interestingly, chemotherapeutic agents and ionizing radiation can also lead to mitochondrial DNA double-strand breaks (mtDSBs). After mtDNA breaks, BAX and BAK mediate herniation, then the mitochondrial RNA will be released into the cytoplasm, triggering the RIG-I-MAVS-dependent immune response (Tigano et al., 2021). These studies suggest that DNA damage can also activate innate immune responses through RNA sensing pathways.
4 The Impacts of Nucleic Acid-Sensing Pathways on DNA Repair
4.1 Cyclic GMP-AMP Synthase-Stimulator of Interferon Genes
As mentioned above, several DDR proteins, such as DNA-PK and MRN complex, can not only can participate in the DNA repair, but also take part in the onset of inflammatory responses. Conversely, PRRs, first discovered to sense immune-stimulatory nucleic acids, have also been found to take part in the regulation of DDR. Previously, cGAS-STING signaling was simply identified as a response pathway to cytosol dsDNA, however, they have recently been found to function in DNA repair, independent of interferon response. cGAS could inhibit the repair of DNA DSBs by HR without relying on STING and the catalytic activity of cGAS (Hopfner and Hornung, 2020). There are two mechanisms for cGAS-dependent HR inhibition. In one of them, because it has been found at the sites of chromosomal damage marked by PARP1 and γ-H2AX, cGAS could prevent the recruitment of proteins necessary for the HR process by interacting with γ-H2AX and PAR (Liu et al., 2018). On the other hand, subsequent work suggested that cGAS could prevent RAD51-DNA filaments to pair and the broken DNA strand from invading into the homologous strand by binding the homologous dsDNA template to form oligomeric clusters (Jiang et al., 2019). However, the detailed mechanism remains to be investigated further.
STING has been confirmed to promote DDR and enable cell survival without cGAS, though they are partners (Cheradame et al., 2021). Downregulation of STING increases cell death and makes breast cancer cells more sensitive to genotoxic treatment. Following chemotherapy regimens, some STINGs are found to be located at the inner nuclear membrane and bind the NHEJ proteins DNA-PKcs, Ku70, and Ku80, suggesting that STING may control NHEJ-mediated DNA repair by cooperating with DNA-PK (Ferguson et al., 2012; Morchikh et al., 2017; Sui et al., 2017; Cheradame et al., 2021). Regrettably, its specific mechanism remains unclear.
Recent research has proposed a novel mechanism that IR-induced DNA damages would trigger the phosphorylation and activation of phosphoribosyl pyrophosphate synthetases PRPS1/2 via ATM and cGAS/STING/TBK1, which could promote the synthesis of deoxyribonucleotide given that the PRPSs are the rate-limiting enzymes. Then the increased deoxyribonucleotide will help for DNA repair. Nevertheless, it remains to be clarified how the cells respond to DNA damage via the cGAS-STING pathway under different contexts (Kornberg et al., 1955; Hove-Jensen, 1988; Liu et al., 2021).
4.2 Toll-Like Receptors
TLRs are the key members of the innate immune system, functioning as the first line of defense against multiple injurious substances (Trinchieri and Sher, 2007; Barton and Kagan, 2009; Kawai and Akira, 2011). Recent literature has shown that activation of TLRs could also promote DNA repair by upregulating the expression of DNA repair genes, apart from upregulating cellular defense systems. Upon TLR9 stimulation, there is a significant increase of mRNAs to participate in adjusting cell cycles and DNA repair after CpG DNA is injected into the abdominal cavity (Zheng et al., 2008; Klaschik et al., 2010; Sommariva et al., 2011). And NER gene expression is increased by treating bone marrow-derived cell lines with the TLR7/8 agonist (Imiquimod) in vitro (Fishelevich et al., 2011). It has been deduced that TLR signaling pathways may result in transcriptional activation of DNA repair machinery through direct and indirect mechanisms. The promoter regions of many genes involved in DNA repair contain the binding sites of activator protein-1 (AP-1) (Xiao et al., 1993; Zhong et al., 2000). The transcriptional control of DNA repair might be linked to TLR agonist treatment through the transcriptional activation function of the AP-1. On the other hand, DNA repair can be promoted because the activated TLR could induce the generation of cytokines. The cytokines such as IL-12 may be sensed by their appropriate cytokine receptor, leading to the increased transcription levels of DNA repair genes (Majewski et al., 2010). However, increased DNA repair is detrimental for cancer treatment. The previous study has found that TLR9 agonist treatment upregulates the genes associated with DNA repair in immune cells but downregulates them in tumor cells, which is contributed to the death of cancer cells (Sommariva et al., 2011). Those conflicting data highlight the need for further research into how immune versus stromal cells, and normal versus cancerous cells respond to TLR agonists, as well as related DNA repair and cell survival.
4.3 AIM2-Like Receptors
AIM2-like receptors (ALRs) are a large family of structurally related proteins that are generally considered to act as intracellular DNA sensors alerting the innate immune system. Recent studies reveal that ALRs perform different functions outside the immune system. It has been demonstrated that DNA breaks are repaired more efficiently in mice and cells lacking ALRs (ALR−/− mice lack the entire ALR locus containing all 13 ALR genes on Chromosome 13), resulting in the stronger resistance to the genotoxic effects of irradiation and chemotherapy (Brunette et al., 2012; Gray et al., 2016; Jiang et al., 2021). Mechanistically, nuclear ALRs limit DNA repair machinery access to damaged sites by binding the chromatin, and self-oligomerization promoted chromatin compaction. These findings reveal that ALRs could be the possible target for new interventions against genotoxic tissue injury, but more research into the different members of the ALR family is needed.
5 Conclusion and Future Perspective
Targeting DDR factors in tumors has achieved outstanding success over the last decade, especially PARP inhibitors for cancer therapy. Recently, we have gained an improved understanding of the molecular mechanism of how DNA damage is interconnected to cellular innate immunity, which plays essential roles in the therapeutic efficacy of DNA repair targeted treatments (Pantelidou et al., 2019). Damaged nucleic acids also shape adaptive immune responses by activating innate immune cells. Accumulation of nucleic acids from necrotic cells can induce type I IFNs and other immune-regulatory cytokines production from bystander cells [e.g., dendritic cells (DCs)] to promote antitumor immunity through nucleic acid-sensing pathways. The DCs activated by the type I IFNs secretion will be transported to tumor-draining lymph nodes and cross-prime naïve CD8+ T lymphocytes (Ellermeier et al., 2013; Duewell et al., 2014; Klarquist et al., 2014; Woo et al., 2014). The cancer-induced host response and tumor rejection relies heavily on the immunological responses to danger DAMPs signals.
Immune checkpoint blockade (ICB), as a promising therapeutic strategy, has made tremendous strides in recent years providing an alternative to irradiation therapy or traditional chemotherapies. Unfortunately, the efficacy of ICB is limited to only a subgroup of cancer patients (depending on the type of the tumor), with an overall response rate of about 20% for all malignancies to date (Hargadon et al., 2018; Chowell et al., 2021). ICB is effective in “hot” tumors with T cell infiltration rather than in “cold” tumors lacking T cell infiltration (Petitprez et al., 2020). Novel strategies for activating innate immunity within the TME to promote the antitumor immune responses have emerged in recent years, with the goal of eradicating the disease in “cold” tumors (Iurescia et al., 2018). Agonists of the nucleic acid-sensing pathways (such as cGAMP, agonists of STING) have been applied to eradicate tumor mass and induce a durable anti-tumor immune response (Chin et al., 2020; Pan et al., 2020). However, since some PRRs are involved in DDR and tumorigenesis, the impact of nucleic acid-sensing pathways on DNA repair cannot be ignored in therapeutic strategies aiming at promoting the activation of nucleic acid-sensing pathways.
As DDR inhibitors can trigger innate immune responses, DDR inhibition could be effective in combination with ICBs. The DDR inhibitors that target PARP have been investigated the most in anticancer immunotherapies. PARP inhibition could increase CD8+ T-cell infiltration and IFN-γ generation in tumors, and promote the tumor regression when used accompanied with anti-PD-1 antibody (Pantelidou et al., 2019; Shen et al., 2019). Recently, many other regents inhibiting DDR components have been developed and used preclinically (Cleary et al., 2020). DDR inhibition strategies combined with other therapeutic strategies possess tremendous potential to improve the effectiveness of cancer treatment because of their immunomodulatory effect on radiation and chemotherapies and immune checkpoint blocking. Furthermore, in order to kill tumor cells more precisely, innovative approaches (such as nanoparticles, viral particles, and targeted deliveries) beneficial to the precise delivery of a chemotherapeutic drug, DDR inhibitors, or nucleic acid-sensing pathways agonists to tumors can be used to induce local specific antitumor immune responses, which could significantly expand the therapeutic window for their use in cancer immunotherapies (Gentili et al., 2015; Wilson et al., 2018; Wang et al., 2020; Xu et al., 2021).
Author Contributions
BX and AL drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32100554) to BX.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, T., Marutani, Y., and Shoji, I. (2019). Cytosolic DNA-Sensing Immune Response and Viral Infection. Microbiol. Immunol. 63, 51–64. doi:10.1111/1348-0421.12669
Ablasser, A., Goldeck, M., Cavlar, T., Deimling, T., Witte, G., Röhl, I., et al. (2013). cGAS Produces a 2′-5′-linked Cyclic Dinucleotide Second Messenger that Activates STING. Nature 498, 380–384. doi:10.1038/nature12306
Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001). Recognition of Double-Stranded RNA and Activation of NF-Κb by Toll-like Receptor 3. Nature 413, 732–738. doi:10.1038/35099560
Andreeva, L., Hiller, B., Kostrewa, D., Lässig, C., de Oliveira Mann, C. C., Jan Drexler, D., et al. (2017). cGAS Senses Long and HMGB/TFAM-bound U-Turn DNA by Forming Protein-DNA Ladders. Nature 549, 394–398. doi:10.1038/nature23890
Barrat, F. J., Meeker, T., Gregorio, J., Chan, J. H., Uematsu, S., Akira, S., et al. (2005). Nucleic Acids of Mammalian Origin Can Act as Endogenous Ligands for Toll-like Receptors and May Promote Systemic Lupus Erythematosus. J. Exp. Med. 202, 1131–1139. doi:10.1084/jem.20050914
Barton, G. M., and Kagan, J. C. (2009). A Cell Biological View of Toll-like Receptor Function: Regulation through Compartmentalization. Nat. Rev. Immunol. 9, 535–542. doi:10.1038/nri2587
Biau, J., Chautard, E., Verrelle, P., and Dutreix, M. (2019). Altering DNA Repair to Improve Radiation Therapy: Specific and Multiple Pathway Targeting. Front. Oncol. 9, 1009. Epub 2019/10/28. doi:10.3389/fonc.2019.01009
Bixel, K., and Hays, J. (2015). Olaparib in the Management of Ovarian Cancer. Pgpm 8, 127–135. Epub 2015/08/27. doi:10.2147/pgpm.s62809
Bouwman, P., and Jonkers, J. (2012). The Effects of Deregulated DNA Damage Signalling on Cancer Chemotherapy Response and Resistance. Nat. Rev. Cancer 12, 587–598. Epub 2012/08/25. doi:10.1038/nrc3342
Brunette, R. L., Young, J. M., Whitley, D. G., Brodsky, I. E., Malik, H. S., and Stetson, D. B. (2012). Extensive Evolutionary and Functional Diversity Among Mammalian AIM2-like Receptors. J. Exp. Med. 209, 1969–1983. doi:10.1084/jem.20121960
Burdette, D. L., Monroe, K. M., Sotelo-Troha, K., Iwig, J. S., Eckert, B., Hyodo, M., et al. (2011). STING Is a Direct Innate Immune Sensor of Cyclic Di-GMP. Nature 478, 515–518. doi:10.1038/nature10429
Burma, S., Chen, B. P., Murphy, M., Kurimasa, A., and Chen, D. J. (2001). ATM Phosphorylates Histone H2AX in Response to DNA Double-Strand Breaks. J. Biol. Chem. 276, 42462–42467. Epub 2001/09/26. doi:10.1074/jbc.c100466200
Caldecott, K. W. (2020). Mammalian DNA Base Excision Repair: Dancing in the Moonlight. DNA Repair 93, 102921. Epub 2020/10/23. doi:10.1016/j.dnarep.2020.102921
Cavassani, K. A., Ishii, M., Wen, H., Schaller, M. A., Lincoln, P. M., Lukacs, N. W., et al. (2008). TLR3 Is an Endogenous Sensor of Tissue Necrosis during Acute Inflammatory Events. J. Exp. Med. 205, 2609–2621. doi:10.1084/jem.20081370
Ceccaldi, R., Liu, J. C., Amunugama, R., Hajdu, I., Primack, B., O’Connor, M. I. R., et al. (2015). Homologous-recombination-deficient Tumours Are Dependent on Polθ-Mediated Repair. Nature 518, 258–262. doi:10.1038/nature14184
Chang, H. H. Y., Pannunzio, N. R., Adachi, N., and Lieber, M. R. (2017). Non-homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell Biol 18, 495–506. Epub 2017/05/18. doi:10.1038/nrm.2017.48
Chang, Z. L. (2010). Important Aspects of Toll-like Receptors, Ligands and Their Signaling Pathways. Inflamm. Res. 59, 791–808. doi:10.1007/s00011-010-0208-2
Chatterjee, N., and Walker, G. C. (2017). Mechanisms of DNA Damage, Repair, and Mutagenesis. Environ. Mol. Mutagen. 58, 235–263. Epub 2017/05/10. doi:10.1002/em.22087
Chen, X., Xu, X., Chen, Y., Cheung, J. C., Wang, H., Jiang, J., et al. (2021). Structure of an Activated DNA-PK and its Implications for NHEJ. Mol. Cell 81, 801–810. e803Epub 2021/01/02. doi:10.1016/j.molcel.2020.12.015
Cheng, B., Pan, W., Xing, Y., Xiao, Y., Chen, J., and Xu, Z. (2022). Recent Advances in DDR (DNA Damage Response) Inhibitors for Cancer Therapy. Eur. J. Med. Chem. 230, 114109. Epub 2022/01/21. doi:10.1016/j.ejmech.2022.114109
Cheng, Q., and Chen, J. (2010). Mechanism of P53 Stabilization by ATM after DNA Damage. Cell Cycle 9, 472–478. Feb 1Epub 2010/01/19. doi:10.4161/cc.9.3.10556
Cheradame, L., Guerrera, I. C., Gaston, J., Schmitt, A., Jung, V., Goudin, N., et al. (2021). STING Protects Breast Cancer Cells from Intrinsic and Genotoxic-Induced DNA Instability via a Non-canonical, Cell-Autonomous Pathway. Oncogene 40, 6627–6640. Epub 2021/10/10. doi:10.1038/s41388-021-02037-4
Chin, E. N., Yu, C., Vartabedian, V. F., Jia, Y., Kumar, M., Gamo, A. M., et al. (2020). Antitumor Activity of a Systemic STING-Activating Non-nucleotide cGAMP Mimetic. Science 369, 993–999. Epub 2020/08/21. doi:10.1126/science.abb4255
Chowell, D., Yoo, S.-K., Valero, C., Pastore, A., Krishna, C., Lee, M., et al. (2021). Improved Prediction of Immune Checkpoint Blockade Efficacy across Multiple Cancer Types. Nat. Biotechnol. 1, 1–8. Epub 2021/11/03. doi:10.1038/s41587-021-01070-8
Chu, W., Jin, W., Liu, D., Wang, J., Geng, C., Chen, L., et al. (2018). Diffusion-weighted Imaging in Identifying Breast Cancer Pathological Response to Neoadjuvant Chemotherapy: A Meta-Analysis. Oncotarget 9, 7088–7100. Epub 2018/02/23. doi:10.18632/oncotarget.23195
Ciccia, A., and Elledge, S. J. (2010). The DNA Damage Response: Making it Safe to Play with Knives. Mol. Cell 40, 179–204. Epub 2010/10/23. doi:10.1016/j.molcel.2010.09.019
Cimprich, K. A., and Cortez, D. (2008). ATR: an Essential Regulator of Genome Integrity. Nat. Rev. Mol. Cell Biol 9, 616–627. Epub 2008/07/03. doi:10.1038/nrm2450
Cleary, J. M., Aguirre, A. J., Shapiro, G. I., and D’Andrea, A. D. (2020). Biomarker-Guided Development of DNA Repair Inhibitors. Mol. Cell 78, 1070–1085. Jun 18Epub 2020/05/28. doi:10.1016/j.molcel.2020.04.035
Cussiol, J. R. R., Soares, B. L., and Oliveira, F. M. B. (2019). From Yeast to Humans: Understanding the Biology of DNA Damage Response (DDR) Kinases. Genet. Mol. Biol. 43, e20190071. Epub 2020/01/14. doi:10.1590/1678-4685-GMB-2019-0071
Dasari, S., and Tchounwou, P. B. (2014). Cisplatin in Cancer Therapy: Molecular Mechanisms of Action. Eur. J. Pharmacol. 740, 364–378. Epub 2014/07/25. doi:10.1016/j.ejphar.2014.07.025
de Oliveira Mann, C. C., Orzalli, M. H., King, D. S., Kagan, J. C., Lee, A. S. Y., and Kranzusch, P. J. (2019). Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-Κb Signaling Adaptation. Cell Rep. 27, 1165–1175. e1165. doi:10.1016/j.celrep.2019.03.098
De Ruysscher, D., Niedermann, G., Burnet, N. G., Siva, S., Lee, A. W. M., and Hegi-Johnson, F. (2019). Radiotherapy Toxicity. Nat. Rev. Dis. Primers 5, 13–20. doi:10.1038/s41572-019-0064-5
Delneste, Y., Beauvillain, C., and Jeannin, P. (2007). Immunité Naturelle. Med. Sci. (Paris) 23, 67–74. Epub 2007/01/11. doi:10.1051/medsci/200723167
Desouky, O., Ding, N., and Zhou, G. (2015). Targeted and Non-targeted Effects of Ionizing Radiation. J. Radiat. Res. Appl. Sci. 8, 247–254. doi:10.1016/j.jrras.2015.03.003
Dianov, G. L., and Hübscher, U. (2013). Mammalian Base Excision Repair: the Forgotten Archangel. Nucleic Acids Res. 41, 3483–3490. Apr 1Epub 2013/02/15. doi:10.1093/nar/gkt076
Diner, E. J., Burdette, D. L., Wilson, S. C., Monroe, K. M., Kellenberger, C. A., Hyodo, M., et al. (2013). The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 3, 1355–1361. doi:10.1016/j.celrep.2013.05.009
Dou, Z., Ghosh, K., Vizioli, M. G., Zhu, J., Sen, P., Wangensteen, K. J., et al. (2017). Cytoplasmic Chromatin Triggers Inflammation in Senescence and Cancer. Nature 550, 402–406. Epub 2017/10/05. doi:10.1038/nature24050
Du, M., and Chen, Z. J. (2018). DNA-induced Liquid Phase Condensation of cGAS Activates Innate Immune Signaling. Science 361, 704–709. Epub 2018/07/07. doi:10.1126/science.aat1022
Duewell, P., Steger, A., Lohr, H., Bourhis, H., Hoelz, H., Kirchleitner, S. V., et al. (2014). RIG-I-like Helicases Induce Immunogenic Cell Death of Pancreatic Cancer Cells and Sensitize Tumors toward Killing by CD8+ T Cells. Cell Death Differ 21, 1825–1837. Epub 2014/07/12. doi:10.1038/cdd.2014.96
Dupré, A., Boyer-Chatenet, L., Sattler, R. M., Modi, A. P., Lee, J. H., Nicolette, M. L., et al. (2008). A Forward Chemical Genetic Screen Reveals an Inhibitor of the Mre11-Rad50-Nbs1 Complex. Nat. Chem. Biol. 4, 119–125. doi:10.1038/nchembio.63
Ellermeier, J., Wei, J., Duewell, P., Hoves, S., Stieg, M. R., Adunka, T., et al. (2013). Therapeutic Efficacy of Bifunctional siRNA Combining TGF-Β1 Silencing with RIG-I Activation in Pancreatic Cancer. Cancer Res. 73, 1709–1720. doi:10.1158/0008-5472.can-11-3850
Emadi, A., Jones, R. J., and Brodsky, R. A. (2009). Cyclophosphamide and Cancer: golden Anniversary. Nat. Rev. Clin. Oncol. 6, 638–647. Epub 2009/09/30. doi:10.1038/nrclinonc.2009.146
Ergun, S. L., Fernandez, D., Weiss, T. M., and Li, L. (2019). STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 178, 290–301. Epub 2019/06/25. doi:10.1016/j.cell.2019.05.036
Falck, J., Coates, J., and Jackson, S. P. (2005). Conserved Modes of Recruitment of ATM, ATR and DNA-PKcs to Sites of DNA Damage. Nature 434, 605–611. Epub 2005/03/11. doi:10.1038/nature03442
Feng, X., Tubbs, A., Zhang, C., Tang, M., Sridharan, S., Wang, C., et al. (2020). ATR Inhibition Potentiates Ionizing Radiation-Induced Interferon Response via Cytosolic Nucleic Acid-Sensing Pathways. EMBO J. 39, e104036. doi:10.15252/embj.2019104036
Ferguson, B. J., Mansur, D. S., Peters, N. E., Ren, H., and Smith, G. L. (2012). DNA-PK Is a DNA Sensor for IRF-3-dependent Innate Immunity. elife 1, e00047. doi:10.7554/eLife.00047
Fernandes-Alnemri, T., Yu, J.-W., Datta, P., Wu, J., and Alnemri, E. S. (2009). AIM2 Activates the Inflammasome and Cell Death in Response to Cytoplasmic DNA. Nature 458, 509–513. Epub 2009/01/23. doi:10.1038/nature07710
Fishelevich, R., Zhao, Y., Tuchinda, P., Liu, H., Nakazono, A., Tammaro, A., et al. (2011). Imiquimod-induced TLR7 Signaling Enhances Repair of DNA Damage Induced by Ultraviolet Light in Bone Marrow-Derived Cells. J.I. 187, 1664–1673. Epub 2011/07/19. doi:10.4049/jimmunol.1100755
Foote, K. M., Nissink, J. W. M., McGuire, T., Turner, P., Guichard, S., Yates, J. W. T., et al. (2018). Discovery and Characterization of AZD6738, a Potent Inhibitor of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Kinase with Application as an Anticancer Agent. J. Med. Chem. 61, 9889–9907. doi:10.1021/acs.jmedchem.8b01187
Gaidt, M. M., Ebert, T. S., Chauhan, D., Ramshorn, K., Pinci, F., Zuber, S., et al. (2017). The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell 171, 1110–1124. e1118. doi:10.1016/j.cell.2017.09.039
Galluzzi, L., Humeau, J., Buqué, A., Zitvogel, L., and Kroemer, G. (2020). Immunostimulation with Chemotherapy in the Era of Immune Checkpoint Inhibitors. Nat. Rev. Clin. Oncol. 17, 725–741. Epub 2020/08/08. doi:10.1038/s41571-020-0413-z
Ganguly, D., Chamilos, G., Lande, R., Gregorio, J., Meller, S., Facchinetti, V., et al. (2009). Self-RNA-antimicrobial Peptide Complexes Activate Human Dendritic Cells through TLR7 and TLR8. J. Exp. Med. 206, 1983–1994. doi:10.1084/jem.20090480
Gao, P., Ascano, M., Wu, Y., Barchet, W., Gaffney, B. L., Zillinger, T., et al. (2013). Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell 153, 1094–1107. doi:10.1016/j.cell.2013.04.046
Gavande, N. S., VanderVere-Carozza, P. S., Hinshaw, H. D., Jalal, S. I., Sears, C. R., Pawelczak, K. S., et al. (2016). DNA Repair Targeted Therapy: The Past or Future of Cancer Treatment? Pharmacol. Ther. 160, 65–83. Epub 2016/02/21. doi:10.1016/j.pharmthera.2016.02.003
Gavande, N. S., VanderVere-Carozza, P. S., Pawelczak, K. S., Mendoza-Munoz, P., Vernon, T. L., Hanakahi, L. A., et al. (2020). Discovery and Development of Novel DNA-PK Inhibitors by Targeting the Unique Ku-DNA Interaction. Nucleic Acids Res. 48, 11536–11550. doi:10.1093/nar/gkaa934
Gay, N. J., Symmons, M. F., Gangloff, M., and Bryant, C. E. (2014). Assembly and Localization of Toll-like Receptor Signalling Complexes. Nat. Rev. Immunol. 14, 546–558. Epub 2014/07/26. doi:10.1038/nri3713
Gentili, M., Kowal, J., Tkach, M., Satoh, T., Lahaye, X., Conrad, C., et al. (2015). Transmission of Innate Immune Signaling by Packaging of cGAMP in Viral Particles. Science 349, 1232–1236. Epub 2015/08/01. doi:10.1126/science.aab3628
Gentili, M., Lahaye, X., Nadalin, F., Nader, G. P. F., Puig Lombardi, E., Herve, S., et al. (2019). The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Rep. 26, 2377–2393. e2313. doi:10.1016/j.celrep.2019.01.105
Giglia-Mari, G., Zotter, A., and Vermeulen, W. (2011). DNA Damage Response. Cold Spring Harbor Perspect. Biol. 3, a000745. Epub 2010/10/29. doi:10.1101/cshperspect.a000745
Gillet, L. C. J., and Schärer, O. D. (2006). Molecular Mechanisms of Mammalian Global Genome Nucleotide Excision Repair. Chem. Rev. 106, 253–276. Epub 2006/02/09. doi:10.1021/cr040483f
Glück, S., Guey, B., Gulen, M. F., Wolter, K., Kang, T. W., Schmacke, N. A., et al. (2017). Innate Immune Sensing of Cytosolic Chromatin Fragments through cGAS Promotes Senescence. Nat. Cell Biol 19, 1061–1070. doi:10.1038/ncb3586
Gorecki, L., Andrs, M., Rezacova, M., and Korabecny, J. (2020). Discovery of ATR Kinase Inhibitor Berzosertib (VX-970, M6620): Clinical Candidate for Cancer Therapy. Pharmacol. Ther. 210, 107518. Epub 2020/02/29. doi:10.1016/j.pharmthera.2020.107518
Goubau, D., Deddouche, S., and Reis e Sousa, C. (2013). Cytosolic Sensing of Viruses. Immunity 38, 855–869. Epub 2013/05/28. doi:10.1016/j.immuni.2013.05.007
Goubau, D., Schlee, M., Deddouche, S., Pruijssers, A. J., Zillinger, T., Goldeck, M., et al. (2014). Antiviral Immunity via RIG-I-Mediated Recognition of RNA Bearing 5′-diphosphates. Nature 514, 372–375. doi:10.1038/nature13590
Grassberger, C., Ellsworth, S. G., Wilks, M. Q., Keane, F. K., and Loeffler, J. S. (2019). Assessing the Interactions between Radiotherapy and Antitumour Immunity. Nat. Rev. Clin. Oncol. 16, 729–745. doi:10.1038/s41571-019-0238-9
Gratia, M., Rodero, M. P., Conrad, C., Bou Samra, E., Maurin, M., Rice, G. I., et al. (2019). Bloom Syndrome Protein Restrains Innate Immune Sensing of Micronuclei by cGAS. J. Exp. Med. 216, 1199–1213. Epub 2019/04/03. doi:10.1084/jem.20181329
Gray, E. E., Winship, D., Snyder, J. M., Child, S. J., Geballe, A. P., and Stetson, D. B. (2016). The AIM2-like Receptors Are Dispensable for the Interferon Response to Intracellular DNA. Immunity 45, 255–266. Aug 16Epub 2016/08/09. doi:10.1016/j.immuni.2016.06.015
Hammel, P., Zhang, C., Matile, J., Colle, E., Hadj-Naceur, I., Gagaille, M.-P., et al. (2020). PARP Inhibition in Treatment of Pancreatic Cancer. Expert Rev. Anticancer Ther. 20, 939–945. Epub 2020/09/17. doi:10.1080/14737140.2020.1820330
Harding, S. M., Benci, J. L., Irianto, J., Discher, D. E., Minn, A. J., and Greenberg, R. A. (2017). Mitotic Progression Following DNA Damage Enables Pattern Recognition within Micronuclei. Nature 548, 466–470. doi:10.1038/nature23470
Hargadon, K. M., Johnson, C. E., and Williams, C. J. (2018). Immune Checkpoint Blockade Therapy for Cancer: An Overview of FDA-Approved Immune Checkpoint Inhibitors. Int. Immunopharmacology 62, 29–39. Epub 2018/07/11. doi:10.1016/j.intimp.2018.06.001
Hellmann, M. D., Li, B. T., Chaft, J. E., and Kris, M. G. (2016). Chemotherapy Remains an Essential Element of Personalized Care for Persons with Lung Cancers. Ann. Oncol. 27, 1829–1835. Epub 2016/07/28. doi:10.1093/annonc/mdw271
Henner, W. D., Grunberg, S. M., and Haseltine, W. A. (1982). Sites and Structure of Gamma Radiation-Induced DNA Strand Breaks. J. Biol. Chem. 257, 11750–11754. Epub 1982/10/10. doi:10.1016/s0021-9258(18)33827-4
Honda, K., Ohba, Y., Yanai, H., Negishi, H., Mizutani, T., Takaoka, A., et al. (2005). Spatiotemporal Regulation of MyD88-IRF-7 Signalling for Robust Type-I Interferon Induction. Nature 434, 1035–1040. doi:10.1038/nature03547
Hooy, R. M., and Sohn, J. (2018). The Allosteric Activation of cGAS Underpins its Dynamic Signaling Landscape. Elife 7, e39984. Epub 2018/10/09. doi:10.7554/eLife.39984
Hopfner, K.-P., and Hornung, V. (2020). Molecular Mechanisms and Cellular Functions of cGAS-STING Signalling. Nat. Rev. Mol. Cell Biol 21, 501–521. doi:10.1038/s41580-020-0244-x
Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., et al. (2009). AIM2 Recognizes Cytosolic dsDNA and Forms a Caspase-1-Activating Inflammasome with ASC. Nature 458, 514–518. doi:10.1038/nature07725
Hou, F., Sun, L., Zheng, H., Skaug, B., Jiang, Q.-X., and Chen, Z. J. (2011). MAVS Forms Functional Prion-like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell 146, 448–461. Epub 2011/07/26. doi:10.1016/j.cell.2011.06.041
Hove-Jensen, B. (1988). Mutation in the Phosphoribosylpyrophosphate Synthetase Gene (Prs) that Results in Simultaneous Requirements for Purine and Pyrimidine Nucleosides, Nicotinamide Nucleotide, Histidine, and Tryptophan in Escherichia coli. J. Bacteriol. 170, 1148–1152. doi:10.1128/jb.170.3.1148-1152.1988
Hu, B., Jin, C., Li, H.-B., Tong, J., Ouyang, X., Cetinbas, N. M., et al. (2016). The DNA-Sensing AIM2 Inflammasome Controls Radiation-Induced Cell Death and Tissue Injury. Science 354, 765–768. Epub 2016/11/16. doi:10.1126/science.aaf7532
Huang, R.-X., and Zhou, P.-K. (2020). DNA Damage Response Signaling Pathways and Targets for Radiotherapy Sensitization in Cancer. Sig Transduct Target. Ther. 5, 60. Epub 2020/05/02. doi:10.1038/s41392-020-0150-x
Ishikawa, H., and Barber, G. N. (2008). STING Is an Endoplasmic Reticulum Adaptor that Facilitates Innate Immune Signalling. Nature 455, 674–678. doi:10.1038/nature07317
Itoh, K., Watanabe, A., Funami, K., Seya, T., and Matsumoto, M. (2008). The Clathrin-Mediated Endocytic Pathway Participates in dsRNA-Induced IFN-β Production. J. Immunol. 181, 5522–5529. doi:10.4049/jimmunol.181.8.5522
Iurescia, S., Fioretti, D., and Rinaldi, M. (2018). Targeting Cytosolic Nucleic Acid-Sensing Pathways for Cancer Immunotherapies. Front. Immunol. 9, 711. Epub 2018/04/25. doi:10.3389/fimmu.2018.00711
Jiang, H., Xue, X., Panda, S., Kawale, A., Hooy, R. M., Liang, F., et al. (2019). Chromatin-bound cGAS Is an Inhibitor of DNA Repair and Hence Accelerates Genome Destabilization and Cell Death. EMBO J. 38, e102718. doi:10.15252/embj.2019102718
Jiang, H., Swacha, P., and Gekara, N. O. (2021). Nuclear AIM2‐Like Receptors Drive Genotoxic Tissue Injury by Inhibiting DNA Repair. Adv. Sci. 8, 2102534. doi:10.1002/advs.202102534
Jobson, A. G., Lountos, G. T., Lorenzi, P. L., Llamas, J., Connelly, J., Cerna, D., et al. (2009). Cellular Inhibition of Checkpoint Kinase 2 (Chk2) and Potentiation of Camptothecins and Radiation by the Novel Chk2 Inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic Acid {4-[1-(guanidinohydrazone)-Ethyl]-Phenyl}-Amide]. J. Pharmacol. Exp. Ther. 331, 816–826. Epub 2009/09/11. doi:10.1124/jpet.109.154997
Kang, D.-c., Gopalkrishnan, R. V., Lin, L., Randolph, A., Valerie, K., Pestka, S., et al. (2004). Expression Analysis and Genomic Characterization of Human Melanoma Differentiation Associated Gene-5, Mda-5: a Novel Type I Interferon-Responsive Apoptosis-Inducing Gene. Oncogene 23, 1789–1800. Epub 2003/12/17. doi:10.1038/sj.onc.1207300
Karikó, K., Ni, H., Capodici, J., Lamphier, M., and Weissman, D. (2004). mRNA Is an Endogenous Ligand for Toll-like Receptor 3. J. Biol. Chem. 279, 12542–12550. doi:10.1074/jbc.M310175200
Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., et al. (2008). Length-dependent Recognition of Double-Stranded Ribonucleic Acids by Retinoic Acid-Inducible Gene-I and Melanoma Differentiation-Associated Gene 5. J. Exp. Med. 205, 1601–1610. doi:10.1084/jem.20080091
Kawai, T., and Akira, S. (2011). Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity 34, 637–650. doi:10.1016/j.immuni.2011.05.006
Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., et al. (2005). IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat. Immunol. 6, 981–988. doi:10.1038/ni1243
King, C., Diaz, H., Barnard, D., Barda, D., Clawson, D., Blosser, W., et al. (2014). Characterization and Preclinical Development of LY2603618: a Selective and Potent Chk1 Inhibitor. Invest. New Drugs 32, 213–226. Epub 2013/10/12. doi:10.1007/s10637-013-0036-7
Klarquist, J., Hennies, C. M., Lehn, M. A., Reboulet, R. A., Feau, S., and Janssen, E. M. (2014). STING-mediated DNA Sensing Promotes Antitumor and Autoimmune Responses to Dying Cells. J.I. 193, 6124–6134. Epub 2014/11/12. doi:10.4049/jimmunol.1401869
Klaschik, S., Tross, D., Shirota, H., and Klinman, D. M. (2010). Short- and Long-Term Changes in Gene Expression Mediated by the Activation of TLR9. Mol. Immunol. 47, 1317–1324. Epub 2009/12/17. doi:10.1016/j.molimm.2009.11.014
Kondo, T., Kobayashi, J., Saitoh, T., Maruyama, K., Ishii, K. J., Barber, G. N., et al. (2013). DNA Damage Sensor MRE11 Recognizes Cytosolic Double-Stranded DNA and Induces Type I Interferon by Regulating STING Trafficking. Proc. Natl. Acad. Sci. U.S.A. 110, 2969–2974. Epub 2013/02/08. doi:10.1073/pnas.1222694110
Kornberg, A., Lieberman, I., and Simms, E. S. (1955). Enzymatic Synthesis and Properties of 5-phosphoribosylpyrophosphate. J. Biol. Chem. 215, 389–402. Epub 1955/07/01. doi:10.1016/s0021-9258(18)66047-8
Kovacs, S. B., and Miao, E. A. (2017). Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 27, 673–684. Epub 2017/06/18. doi:10.1016/j.tcb.2017.05.005
Krejci, L., Altmannova, V., Spirek, M., and Zhao, X. (2012). Homologous Recombination and its Regulation. Nucleic Acids Res. 40, 5795–5818. Epub 2012/04/03. doi:10.1093/nar/gks270
Krokan, H. E., Standal, R., and Slupphaug, G. (1997). DNA Glycosylases in the Base Excision Repair of DNA. Biochem. J. 325 (Pt 1), 1–16. Epub 1997/07/01. doi:10.1042/bj3250001
Kryston, T. B., Georgiev, A. B., Pissis, P., and Georgakilas, A. G. (2011). Role of Oxidative Stress and DNA Damage in Human Carcinogenesis. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 711, 193–201. Epub 2011/01/11. doi:10.1016/j.mrfmmm.2010.12.016
Kuchta, K., Knizewski, L., Wyrwicz, L. S., Rychlewski, L., and Ginalski, K. (2009). Comprehensive Classification of Nucleotidyltransferase Fold Proteins: Identification of Novel Families and Their Representatives in Human. Nucleic Acids Res. 37, 7701–7714. doi:10.1093/nar/gkp854
Kumagai, Y., Takeuchi, O., and Akira, S. (2008). TLR9 as a Key Receptor for the Recognition of DNA☆. Adv. Drug Deliv. Rev. 60, 795–804. doi:10.1016/j.addr.2007.12.004
Lammert, C. R., Frost, E. L., Bellinger, C. E., Bolte, A. C., McKee, C. A., Hurt, M. E., et al. (2020). AIM2 Inflammasome Surveillance of DNA Damage Shapes Neurodevelopment. Nature 580, 647–652. Epub 2020/05/01. doi:10.1038/s41586-020-2174-3
Li, M., and Yu, X. (2013). Function of BRCA1 in the DNA Damage Response Is Mediated by ADP-Ribosylation. Cancer cell 23, 693–704. doi:10.1016/j.ccr.2013.03.025
Li, X.-D., Wu, J., Gao, D., Wang, H., Sun, L., and Chen, Z. J. (2013b). Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science 341, 1390–1394. Epub 2013/08/31. doi:10.1126/science.1244040
Li, X., Shu, C., Yi, G., Chaton, C. T., Shelton, C. L., Diao, J., et al. (2013a). Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity 39, 1019–1031. doi:10.1016/j.immuni.2013.10.019
Liang, F., Zhang, F., Zhang, L., and Wei, W. (2020). The Advances in Pyroptosis Initiated by Inflammasome in Inflammatory and Immune Diseases. Inflamm. Res. 69, 159–166. Epub 2020/01/15. doi:10.1007/s00011-020-01315-3
Limagne, E., Nuttin, L., Thibaudin, M., Jacquin, E., Aucagne, R., Bon, M., et al. (2022). MEK Inhibition Overcomes Chemoimmunotherapy Resistance by Inducing CXCL10 in Cancer Cells. Cancer cell 40, 136–152. Feb 14e112Epub 2022/01/21. doi:10.1016/j.ccell.2021.12.009
Liu, H., Zhang, H., Wu, X., Ma, D., Wu, J., Wang, L., et al. (2018). Nuclear cGAS Suppresses DNA Repair and Promotes Tumorigenesis. Nature 563, 131–136. Epub 2018/10/26. doi:10.1038/s41586-018-0629-6
Liu, R., Li, J., Shao, J., Lee, J.-H., Qiu, X., Xiao, Y., et al. (2021). Innate Immune Response Orchestrates Phosphoribosyl Pyrophosphate Synthetases to Support DNA Repair. Cell Metab. 33, 2076–2089. Epub 2021/08/04. doi:10.1016/j.cmet.2021.07.009
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of Innate Immune Adaptor Proteins MAVS, STING, and TRIF Induces IRF3 Activation. Science 347, aaa2630. Epub 2015/02/01. doi:10.1126/science.aaa2630
Lövgren, T., Eloranta, M. L., Båve, U., Alm, G. V., Rönnblom, L. J. A., and Rheumatology, R. O. (2004). Induction of Interferon‐α Production in Plasmacytoid Dendritic Cells by Immune Complexes Containing Nucleic Acid Released by Necrotic or Late Apoptotic Cells and Lupus IgG. Arthritis Rheum. 50, 1861–1872. doi:10.1002/art.20254
Luecke, S., Holleufer, A., Christensen, M. H., Jønsson, K. L., Boni, G. A., Sørensen, L. K., et al. (2017). cGAS Is Activated by DNA in a Length‐dependent Manner. EMBO Rep. 18, 1707–1715. doi:10.15252/embr.201744017
Lugrin, J., and Martinon, F. (2018). The AIM2 Inflammasome: Sensor of Pathogens and Cellular Perturbations. Immunol. Rev. 281, 99–114. doi:10.1111/imr.12618
Mackenzie, K. J., Carroll, P., Martin, C.-A., Murina, O., Fluteau, A., Simpson, D. J., et al. (2017). cGAS Surveillance of Micronuclei Links Genome Instability to Innate Immunity. Nature 548, 461–465. Epub 2017/07/25. doi:10.1038/nature23449
Majer, O., Liu, B., and Barton, G. M. (2017). Nucleic Acid-Sensing TLRs: Trafficking and Regulation. Curr. Opin. Immunol. 44, 26–33. Epub 2016/12/03. doi:10.1016/j.coi.2016.10.003
Majewski, S., Jantschitsch, C., Maeda, A., Schwarz, T., and Schwarz, A. (2010). IL-23 Antagonizes UVR-Induced Immunosuppression through Two Mechanisms: Reduction of UVR-Induced DNA Damage and Inhibition of UVR-Induced Regulatory T Cells. J. Invest. Dermatol. 130, 554–562. doi:10.1038/jid.2009.274
Mateo, J., Lord, C. J., Serra, V., Tutt, A., Balmaña, J., Castroviejo-Bermejo, M., et al. (2019). A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann. Oncol. 30, 1437–1447. Epub 2019/06/21. doi:10.1093/annonc/mdz192
Mateos-Gomez, P. A., Gong, F., Nair, N., Miller, K. M., Lazzerini-Denchi, E., and Sfeir, A. (2015). Mammalian Polymerase θ Promotes Alternative NHEJ and Suppresses Recombination. Nature 518, 254–257. doi:10.1038/nature14157
McLaughlin, M., Patin, E. C., Pedersen, M., Wilkins, A., Dillon, M. T., Melcher, A. A., et al. (2020). Inflammatory Microenvironment Remodelling by Tumour Cells after Radiotherapy. Nat. Rev. Cancer 20, 203–217. AprEpub 2020/03/13. doi:10.1038/s41568-020-0246-1
Miao, E. A., Rajan, J. V., and Aderem, A. (2011). Caspase-1-induced Pyroptotic Cell Death. Immunological Rev. 243, 206–214. doi:10.1111/j.1600-065x.2011.01044.x
Morchikh, M., Cribier, A., Raffel, R., Amraoui, S., Cau, J., Severac, D., et al. (2017). HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-Subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol. Cel. 67, 387–399. e385. doi:10.1016/j.molcel.2017.06.020
Moresco, E. M. Y., LaVine, D., and Beutler, B. (2011). Toll-like Receptors. Curr. Biol. 21, R488–R493. Epub 2011/07/12. doi:10.1016/j.cub.2011.05.039
Munster, P. N., Mahipal, A., Nemunaitis, J. J., Mita, M. M., Paz-Ares, L. G., Massard, C., et al. (2016). Phase I Trial of a Dual TOR Kinase and DNA-PK Inhibitor (CC-115) in Advanced Solid and Hematologic Cancers. J. Clin. Oncol. 34–2505. doi:10.1200/jco.2016.34.15_suppl.2505
Nastasi, C., Mannarino, L., and D’Incalci, M. (2020). DNA Damage Response and Immune Defense. Ijms 21, 7504. Epub 2020/10/16. doi:10.3390/ijms21207504
Ori, D., Murase, M., and Kawai, T. (2017). Cytosolic Nucleic Acid Sensors and Innate Immune Regulation. Int. Rev. Immunol. 36, 74–88. Epub 2017/03/24. doi:10.1080/08830185.2017.1298749
Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T., and Seya, T. (2003). TICAM-1, an Adaptor Molecule that Participates in Toll-like Receptor 3-mediated Interferon-β Induction. Nat. Immunol. 4, 161–167. doi:10.1038/ni886
Paludan, S. R., Reinert, L. S., and Hornung, V. (2019). DNA-stimulated Cell Death: Implications for Host Defence, Inflammatory Diseases and Cancer. Nat. Rev. Immunol. 19, 141–153. Epub 2019/01/16. doi:10.1038/s41577-018-0117-0
Pan, B. S., Perera, S. A., Piesvaux, J. A., Presland, J. P., Schroeder, G. K., Cumming, J. N., et al. (2020). An Orally Available Non-nucleotide STING Agonist with Antitumor Activity. Science 369, eaba6098. Epub 2020/08/21. doi:10.1126/science.aba6098
Pandey, N., and Black, B. E. (2021). Rapid Detection and Signaling of DNA Damage by PARP-1. Trends Biochem. Sci. 46, 744–757. Epub 2021/03/07. doi:10.1016/j.tibs.2021.01.014
Pantelidou, C., Sonzogni, O., De Oliveria Taveira, M., Mehta, A. K., Kothari, A., Wang, D., et al. (2019). PARP Inhibitor Efficacy Depends on CD8+ T-Cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer. Cancer Discov. 9, 722–737. Epub 2019/04/25. doi:10.1158/2159-8290.cd-18-1218
Petitprez, F., Meylan, M., de Reyniès, A., Sautès-Fridman, C., and Fridman, W. H. (2020). The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 11, 784. Epub 2020/05/28. doi:10.3389/fimmu.2020.00784
Pike, K. G., Barlaam, B., Cadogan, E., Campbell, A., Chen, Y., Colclough, N., et al. (2018). The Identification of Potent, Selective, and Orally Available Inhibitors of Ataxia Telangiectasia Mutated (ATM) Kinase: the Discovery of AZD0156 (8-{6-[3-(Dimethylamino) Propoxy] Pyridin-3-Yl}-3-Methyl-1-(tetrahydro-2 H-Pyran-4-Yl)-1, 3-dihydro-2 H-Imidazo [4, 5-c] Quinolin-2-One). J. Med. Chem. 61, 3823–3841. doi:10.1021/acs.jmedchem.7b01896
Pinder, J. B., Attwood, K. M., and Dellaire, G. (2013). Reading, Writing, and Repair: the Role of Ubiquitin and the Ubiquitin-like Proteins in DNA Damage Signaling and Repair. Front. Genet. 4, 45. Epub 2013/04/05. doi:10.3389/fgene.2013.00045
Qin, S.-Y., Cheng, Y.-J., Lei, Q., Zhang, A.-Q., and Zhang, X.-Z. (2018). Combinational Strategy for High-Performance Cancer Chemotherapy. Biomaterials 171, 178–197. Epub 2018/04/27. doi:10.1016/j.biomaterials.2018.04.027
Ramakrishnan Geethakumari, P., Schiewer, M. J., Knudsen, K. E., and Kelly, W. K. (2017). PARP Inhibitors in Prostate Cancer. Curr. Treat. Options. Oncol. 18, 37. Epub 2017/05/26. doi:10.1007/s11864-017-0480-2
Roth, S., Rottach, A., Lotz-Havla, A. S., Laux, V., Muschaweckh, A., Gersting, S. W., et al. (2014). Rad50-CARD9 Interactions Link Cytosolic DNA Sensing to IL-1β Production. Nat. Immunol. 15, 538–545. doi:10.1038/ni.2888
Rupnik, A., Grenon, M., and Lowndes, N. (2008). The MRN Complex. Curr. Biol. 18, R455–R457. Epub 2008/06/05. doi:10.1016/j.cub.2008.03.040
San Filippo, J., Sung, P., and Klein, H. (2008). Mechanism of Eukaryotic Homologous Recombination. Annu. Rev. Biochem. 77, 229–257. Epub 2008/02/16. doi:10.1146/annurev.biochem.77.061306.125255
Serrone, L., Zeuli, M., Sega, F. M., and Cognetti, F. (2000). Dacarbazine-based Chemotherapy for Metastatic Melanoma: Thirty-Year Experience Overview. J. Exp. Clin. Cancer Res. 19, 21–34. Epub 2000/06/07.
Seth, R. B., Sun, L., Ea, C.-K., and Chen, Z. J. (2005). Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-Κb and IRF3. Cell 122, 669–682. doi:10.1016/j.cell.2005.08.012
Shang, G., Zhang, C., Chen, Z. J., Bai, X.-c., and Zhang, X. (2019). Cryo-EM Structures of STING Reveal its Mechanism of Activation by Cyclic GMP-AMP. Nature 567, 389–393. doi:10.1038/s41586-019-0998-5
Shen, J., Zhao, W., Ju, Z., Wang, L., Peng, Y., Labrie, M., et al. (2019). PARPi Triggers the STING-dependent Immune Response and Enhances the Therapeutic Efficacy of Immune Checkpoint Blockade Independent of BRCAness. Cancer Res. 79, 311–319. Epub 2018/11/30. doi:10.1158/0008-5472.can-18-1003
Shuck, S. C., Short, E. A., and Turchi, J. J. (2008). Eukaryotic Nucleotide Excision Repair: from Understanding Mechanisms to Influencing Biology. Cell Res 18, 64–72. Epub 2008/01/02. doi:10.1038/cr.2008.2
Smith, J., Mun Tho, L., Xu, N., and Gillespie, D. A. (2010). The ATM-Chk2 and ATR-Chk1 Pathways in DNA Damage Signaling and Cancer. Adv. Cancer Res. 108, 73–112. doi:10.1016/b978-0-12-380888-2.00003-0
Sommariva, M., De Cecco, L., De Cesare, M., Sfondrini, L., Ménard, S., Melani, C., et al. (2011). TLR9 Agonists Oppositely Modulate DNA Repair Genes in Tumor versus Immune Cells and Enhance Chemotherapy Effects. Cancer Res. 71, 6382–6390. Epub 2011/09/01. doi:10.1158/0008-5472.can-11-1285
Souliotis, V. L., Vlachogiannis, N. I., Pappa, M., Argyriou, A., Ntouros, P. A., and Sfikakis, P. P. (2020). DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci. 21, 55. doi:10.3390/ijms21010055
Stracker, T. H., and Petrini, J. H. J. (2011). The MRE11 Complex: Starting from the Ends. Nat. Rev. Mol. Cell Biol 12, 90–103. Epub 2011/01/22. doi:10.1038/nrm3047
Sui, H., Zhou, M., Imamichi, H., Jiao, X., Sherman, B. T., Lane, H. C., et al. (2017). STING Is an Essential Mediator of the Ku70-Mediated Production of IFN-Λ1 in Response to Exogenous DNA. Sci. Signal. 10, eaah5054. doi:10.1126/scisignal.aah5054
Sui, H., Chen, Q., and Imamichi, T. (2021a). Cytoplasmic‐translocated Ku70 Senses Intracellular DNA and Mediates Interferon‐lambda1 Induction. Immunology 163, 323–337. doi:10.1111/imm.13318
Sui, H., Hao, M., Chang, W., and Imamichi, T. (2021b). The Role of Ku70 as a Cytosolic DNA Sensor in Innate Immunity and beyond. Front. Cel. Infect. Microbiol. 11, 761983. doi:10.3389/fcimb.2021.761983
Sun, L., Wu, J., Du, F., Chen, X., and Chen, Z. J. (2013). Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor that Activates the Type I Interferon Pathway. Science 339, 786–791. doi:10.1126/science.1232458
Taffoni, C., Steer, A., Marines, J., Chamma, H., Vila, I. K., and Laguette, N. (2021). Nucleic Acid Immunity and DNA Damage Response: New Friends and Old Foes. Front. Immunol. 12, 660560. Epub 2021/05/14. doi:10.3389/fimmu.2021.660560
Takeuchi, O., and Akira, S. (2010). Pattern Recognition Receptors and Inflammation. Cell. 140, 805–820. Epub 2010/03/23. doi:10.1016/j.cell.2010.01.022
Tatematsu, M., Nishikawa, F., Seya, T., and Matsumoto, M. (2013). Toll-like Receptor 3 Recognizes Incomplete Stem Structures in Single-Stranded Viral RNA. Nat. Commun. 4, 1833. Epub 2013/05/16. doi:10.1038/ncomms2857
Tigano, M., Vargas, D. C., Tremblay-Belzile, S., Fu, Y., and Sfeir, A. (2021). Nuclear Sensing of Breaks in Mitochondrial DNA Enhances Immune Surveillance. Nature 591, 477–481. Epub 2021/02/26. doi:10.1038/s41586-021-03269-w
Timme, C. R., Rath, B. H., O'Neill, J. W., Camphausen, K., and Tofilon, P. J. (2018). The DNA-PK Inhibitor VX-984 Enhances the Radiosensitivity of Glioblastoma Cells Grown In Vitro and as Orthotopic Xenografts. Mol. Cancer Ther. 17, 1207–1216. doi:10.1158/1535-7163.mct-17-1267
Tisi, R., Vertemara, J., Zampella, G., and Longhese, M. P. (2020). Functional and Structural Insights into the MRX/MRN Complex, a Key Player in Recognition and Repair of DNA Double-Strand Breaks. Comput. Struct. Biotechnol. J. 18, 1137–1152. doi:10.1016/j.csbj.2020.05.013
Trinchieri, G., and Sher, A. (2007). Cooperation of Toll-like Receptor Signals in Innate Immune Defence. Nat. Rev. Immunol. 7, 179–190. Epub 2007/02/24. doi:10.1038/nri2038
Uematsu, N., Weterings, E., Yano, K.-i., Morotomi-Yano, K., Jakob, B., Taucher-Scholz, G., et al. (2007). Autophosphorylation of DNA-PKCS Regulates its Dynamics at DNA Double-Strand Breaks. J. Cel. Biol. 177, 219–229. doi:10.1083/jcb.200608077
Uziel, T., Lerenthal, Y., Moyal, L., Andegeko, Y., Mittelman, L., and Shiloh, Y. (2003). Requirement of the MRN Complex for ATM Activation by DNA Damage. EMBO J. 22, 5612–5621. Epub 2003/10/09. doi:10.1093/emboj/cdg541
Volkman, H. E., Cambier, S., Gray, E. E., and Stetson, D. B. (2019). Tight Nuclear Tethering of cGAS Is Essential for Preventing Autoreactivity. Elife 8, e47491. doi:10.7554/eLife.47491
Vyncke, L., Bovijn, C., Pauwels, E., Van Acker, T., Ruyssinck, E., Burg, E., et al. (2016). Reconstructing the TIR Side of the Myddosome: a Paradigm for TIR-TIR Interactions. Structure 24, 437–447. Epub 2016/02/16. doi:10.1016/j.str.2015.12.018
Wang, B., and Yin, Q. (2017). AIM2 Inflammasome Activation and Regulation: A Structural Perspective. J. Struct. Biol. 200, 279–282. Epub 2017/08/17. doi:10.1016/j.jsb.2017.08.001
Wang, Q., Wang, Y., Ding, J., Wang, C., Zhou, X., Gao, W., et al. (2020). A Bioorthogonal System Reveals Antitumour Immune Function of Pyroptosis. Nature 579, 421–426. Epub 2020/03/20. doi:10.1038/s41586-020-2079-1
Wang, Y., Fu, Z., Li, X., Liang, Y., Pei, S., Hao, S., et al. (2021a). Cytoplasmic DNA Sensing by KU Complex in Aged CD4+ T Cell Potentiates T Cell Activation and Aging-Related Autoimmune Inflammation. Immunity 54, 632–647. Apr 13e639Epub 2021/03/06. doi:10.1016/j.immuni.2021.02.003
Wang, Y., Wang, M., Djekidel, M. N., Chen, H., Liu, D., Alt, F. W., et al. (2021b). eccDNAs Are Apoptotic Products with High Innate Immunostimulatory Activity. Nature 599, 308–314. Epub 2021/10/22. doi:10.1038/s41586-021-04009-w
Watson, R. O., Manzanillo, P. S., and Cox, J. S. (2012). Extracellular M. tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell 150, 803–815. Epub 2012/08/21. doi:10.1016/j.cell.2012.06.040
West, A. P., Khoury-Hanold, W., Staron, M., Tal, M. C., Pineda, C. M., Lang, S. M., et al. (2015). Mitochondrial DNA Stress Primes the Antiviral Innate Immune Response. Nature 520, 553–557. Epub 2015/02/03. doi:10.1038/nature14156
Wilson, D. R., Sen, R., Sunshine, J. C., Pardoll, D. M., Green, J. J., and Kim, Y. J. (2018). Biodegradable STING Agonist Nanoparticles for Enhanced Cancer Immunotherapy. Nanomedicine: Nanotechnology, Biol. Med. 14, 237–246. Epub 2017/11/12. doi:10.1016/j.nano.2017.10.013
Woo, S.-R., Fuertes, M. B., Corrales, L., Spranger, S., Furdyna, M. J., Leung, M. Y. K., et al. (2014). STING-dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity 41, 830–842. Nov 20Epub 2014/12/18. doi:10.1016/j.immuni.2014.10.017
Woodrick, J., Gupta, S., Camacho, S., Parvathaneni, S., Choudhury, S., Cheema, A., et al. (2017). A New Sub‐pathway of Long‐patch Base Excision Repair Involving 5′ gap Formation. Embo J. 36, 1605–1622. doi:10.15252/embj.201694920
Woods, D., and Turchi, J. J. (2013). Chemotherapy Induced DNA Damage Response. Cancer Biol. Ther. 14, 379–389. Epub 2013/02/06. doi:10.4161/cbt.23761
Wu, J., Sun, L., Chen, X., Du, F., Shi, H., Chen, C., et al. (2013). Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 339, 826–830. doi:10.1126/science.1229963
Xiao, W., Singh, K. K., Chen, B., and Samson, L. (1993). A Common Element Involved in Transcriptional Regulation of Two DNA Alkylation Repair Genes (MAG and MGT1) of Saccharomyces cerevisiae. Mol. Cell Biol 13, 7213–7221. Epub 1993/12/01. doi:10.1128/mcb.13.12.7213-7221.1993
Xie, B., Liang, X., Yue, W., Ma, J., Li, X., Zhang, N., et al. (2021). Targeting Cytokinesis Bridge Proteins to Kill High-CIN Type Tumors. Fundam. Res. 1, 752–766. doi:10.1016/j.fmre.2021.08.015
Xie, H., Wang, W., Xia, B., Jin, W., and Lou, G. (2020). Therapeutic Applications of PARP Inhibitors in Ovarian Cancer. Biomed. Pharmacother. 127, 110204. Epub 2020/05/19. doi:10.1016/j.biopha.2020.110204
Xu, S., Wang, L., and Liu, Z. (2021). Molecularly Imprinted Polymer Nanoparticles: An Emerging Versatile Platform for Cancer Therapy. Angew. Chem. Int. Ed. Engl. 19 (60), 3858–3869. Epub 2020/08/14. doi:10.1002/anie.202005309
Yamamoto, M., Sato, S., Hemmi, H., Hoshino, K., Kaisho, T., Sanjo, H., et al. (2003). Role of Adaptor TRIF in the MyD88-independent Toll-like Receptor Signaling Pathway. Science 301, 640–643. Epub 2003/07/12. doi:10.1126/science.1087262
Yilmaz, D., Furst, A., Meaburn, K., Lezaja, A., Wen, Y., Altmeyer, M., et al. (2021). Activation of Homologous Recombination in G1 Preserves Centromeric Integrity. Nature 600, 748–753. Epub 2021/12/03. doi:10.1038/s41586-021-04200-z
Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., et al. (2005). Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity. J. Immunol. 175, 2851–2858. Epub 2005/08/24. doi:10.4049/jimmunol.175.5.2851
Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., et al. (2004). The RNA Helicase RIG-I Has an Essential Function in Double-Stranded RNA-Induced Innate Antiviral Responses. Nat. Immunol. 5, 730–737. doi:10.1038/ni1087
Zandarashvili, L., Langelier, M. F., Velagapudi, U. K., Hancock, M. A., Steffen, J. D., Billur, R., et al. (2020). Structural Basis for Allosteric PARP-1 Retention on DNA Breaks. Science 368, eaax6367. Apr 3Epub 2020/04/04. doi:10.1126/science.aax6367
Zhang, C., Shang, G., Gui, X., Zhang, X., Bai, X.-c., and Chen, Z. J. (2019). Structural Basis of STING Binding with and Phosphorylation by TBK1. Nature 567, 394–398. Epub 2019/03/08. doi:10.1038/s41586-019-1000-2
Zhang, J.-Z., Liu, Z., Liu, J., Ren, J.-X., and Sun, T.-S. (2014a). Mitochondrial DNA Induces Inflammation and Increases TLR9/NF-Κb Expression in Lung Tissue. Int. J. Mol. Med. 33, 817–824. doi:10.3892/ijmm.2014.1650
Zhang, X., Shi, H., Wu, J., Zhang, X., Sun, L., Chen, C., et al. (2013). Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is an Endogenous High-Affinity Ligand for STING. Mol. Cel. 51, 226–235. doi:10.1016/j.molcel.2013.05.022
Zhang, X., Wu, J., Du, F., Xu, H., Sun, L., Chen, Z., et al. (2014b). The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Rep. 6, 421–430. Epub 2014/01/28. doi:10.1016/j.celrep.2014.01.003
Zhao, B., Du, F., Xu, P., Shu, C., Sankaran, B., Bell, S. L., et al. (2019). A Conserved PLPLRT/SD Motif of STING Mediates the Recruitment and Activation of TBK1. Nature 569, 718–722. Epub 2019/05/24. doi:10.1038/s41586-019-1228-x
Zheng, L., Asprodites, N., Keene, A. H., Rodriguez, P., Brown, K. D., and Davila, E. (2008). TLR9 Engagement on CD4 T Lymphocytes Represses γ-radiation-induced Apoptosis through Activation of Checkpoint Kinase Response Elements. Blood 111, 2704–2713. doi:10.1182/blood-2007-07-104141
Zhong, X., Thornton, K., and Reed, E. (2000). Computer Based Analyses of the 5'-flanking Regions of Selected Genes Involved in the Nucleotide Excision Repair Complex. Int. J. Oncol. 17, 375–380. Epub 2000/07/13. doi:10.3892/ijo.17.2.375
Zhu, M., Zhao, H., Limbo, O., and Russell, P. (2018). Mre11 Complex Links Sister Chromatids to Promote Repair of a Collapsed Replication fork. Proc. Natl. Acad. Sci. U S A. 115 (115), 8793–8798. Epub 2018/08/15. doi:10.1073/pnas.1808189115
Zierhut, C., Yamaguchi, N., Paredes, M., Luo, J.-D., Carroll, T., and Funabiki, H. (2019). The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell. 178, 302–315. e323Epub 2019/07/13. doi:10.1016/j.cell.2019.05.035
Keywords: DNA damage and repair, DDR inhibitors, nucleic acid-sensing pathways, innate immunity, immunotherapy
Citation: Xie B and Luo A (2022) Nucleic Acid Sensing Pathways in DNA Repair Targeted Cancer Therapy. Front. Cell Dev. Biol. 10:903781. doi: 10.3389/fcell.2022.903781
Received: 24 March 2022; Accepted: 08 April 2022;
Published: 26 April 2022.
Edited by:
Teng Ma, Capital Medical University, ChinaReviewed by:
Jianyu Wang, Chongqing Medical University, ChinaYang Pu, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2022 Xie and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingteng Xie, eGllYmluZ3RlbmdAYml0LmVkdS5jbg==; Aiqin Luo, YXFsdW9iaXRAMTYzLmNvbQ==
 Bingteng Xie
Bingteng Xie Aiqin Luo
Aiqin Luo