- 1Affiliated Stomatology Hospital of Guangzhou Medical University, Guangdong Engineering Research Center of Oral Restoration and Reconstruction, Guangzhou Key Laboratory of Basic and Applied Research of Oral Regenerative Medicine, Guangzhou, China
- 2Department of Basic Oral Medicine, School and Hospital of Stomatology, Guangzhou Medical University, Guangzhou, China
- 3Laboratory for Myology, Department of Human Movement Sciences, Faculty of Behavioural and Movement Sciences, Amsterdam Movement Sciences, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
Autologous bone marrow-derived mesenchymal stem cells (BMSCs) are more easily available and frequently used for bone regeneration in clinics. Osteogenic differentiation of BMSCs involves complex regulatory networks affecting bone formation phenomena. Non-coding RNAs (ncRNAs) refer to RNAs that do not encode proteins, mainly including microRNAs, long non-coding RNAs, circular RNAs, piwi-interacting RNAs, transfer RNA-derived small RNAs, etc. Recent in vitro and in vivo studies had revealed the regulatory role of ncRNAs in osteogenic differentiation of BMSCs. NcRNAs had both stimulatory and inhibitory effects on osteogenic differentiation of BMSCs. During the physiological condition, osteo-stimulatory ncRNAs are upregulated and osteo-inhibitory ncRNAs are downregulated. The opposite effects might occur during bone degenerative disease conditions. Intracellular ncRNAs and ncRNAs from neighboring cells delivered via exosomes participate in the regulatory process of osteogenic differentiation of BMSCs. In this review, we summarize the recent advances in the regulatory role of ncRNAs on osteogenic differentiation of BMSCs during physiological and pathological conditions. We also discuss the prospects of the application of modulation of ncRNAs function in BMSCs to promote bone tissue regeneration in clinics.
1 Introduction
The bone defect is mainly caused by trauma, severe infection, bone diseases, tumor resection, and various congenital malformations (Gaihre et al., 2017). The number of bone transplantation-related surgery is over two million all over the world (Li et al., 2018). Currently, autologous bone grafts are regarded as the gold standard for bone defect reconstruction (Nicot et al., 2020). The risks of autologous bone grafts such as limited source, infection, pain, loss of sensation, scars, and donor site morbidity limit the clinical applications (Younger and Chapman, 1989; Tessier et al., 2005). Allografts and synthetic bone grafts are used as alternatives to autologous bone grafts (Eppley et al., 2005). However, bone allografts may lead to complications such as fracture, nonunion, and infection (Delloye et al., 2014). While bone substitutes materials such as ceramics have osteoconductivity and weak osteoinductivity. The variable resorption rate and higher brittleness of biomaterial-based bone grafts lead to impaired graft osseointegration (Sohn and Oh, 2019). Stem cell-based approaches for bone tissue engineering have shown promising results in the clinic. The combination of precursor cells, bone grafts, and growth factors have the potential to replace auto-/allo-bone grafts (Steinhardt et al., 2008; El-Rashidy et al., 2017; Zhao et al., 2020). Studies have shown bone marrow-derived mesenchymal stem cells (BMSCs) as a promising source of seed cells for bone tissue engineering applications (Qi et al., 2017; Arthur and Gronthos, 2020; Chen et al., 2021; Jiang et al., 2021). Autologous or human leukocyte antigen matched allogeneic BMSCs are commonly used for bone regeneration in clinics.
The osteogenic differentiation of BMSCs is a complex process, which is regulated by multiple signaling pathways. Various non-coding RNAs (ncRNAs) had been reported to regulate the osteogenic differentiation of BMSCs. NcRNAs are transcribed from the genome, do not directly translate into proteins, but participate in the protein translation process of coding mRNAs (Guttman et al., 2013). MicroRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), ribosomal RNAs (rRNAs), transfert RNAs (tRNAs), tRNA-derived small RNAs (tsRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and PIWI-interacting RNAs (piRNAs) are key ncRNAs that regulate basic cellular function such as cell metabolism (Sun et al., 2020a), proliferation (Song et al., 2016), autophagy, apoptosis (Li et al., 2018) as well as various diseases (Peng et al., 2021; Li et al., 2020) (Figure 1). MiRNAs promote mRNAs degradation and regulate mRNAs translation, and participate in various cellular processes (Zealy et al., 2017). LncRNAs have many biological functions, including genes imprinting, chromatin modification, cell cycle, apoptosis, mRNA decay, and protein translation regulation (Zhu et al., 2013). CircRNAs may be by-products of precursor mRNAs. It demonstrated that circRNAs act as a sponge to regulate the function of miRNAs, participate in the splicing of target genes, translate genes into proteins, and interact with RNA-binding proteins (RBPs) (Zang et al., 2020). The report showed that rRNAs are an important part of ribosomes, which are widely involved in cell translation (Sloan et al., 2017). The main function of tRNAs is to carry amino acids and enter ribosomes for protein synthesis with the participation of mRNAs (Liu R. et al., 2021). TsRNAs are produced by tRNAs cleavage, which participate in the processes such as RNAs silencing, ribosome biogenesis, retrotransposition, epigenetics, and regulate translation. tsRNAs also indirectly regulate gene expression by binding RBPs (Chen et al., 2021; Liu et al., 2021a). SnRNAs are the main component of RNA spliceosomes in the post-transcriptional processing of eukaryotes and participate in the processing of precursor RNAs (pre-mRNAs) (Karijolich and Yu, 2010). SnoRNAs modify snRNAs and rRNAs, and participate in the processing of rRNAs during the maturation of ribosomal subunits (Xing and Chen, 2018). PiRNAs maintain the structure of the genome and mRNAs stability, and regulate protein synthesis by binding with members of PIWI protein family (Xu et al., 2020). The piRNA complex formed by the combination of piRNA and PIWI further regulates the function of germ and stem cells by silencing the process of gene transcription (Xu et al., 2020). NcRNAs also mediate osteogenic differentiation of mesenchymal stem cells (MSCs) via regulating various signaling pathways (Yang et al., 2018; Mazziotta et al., 2021). It had been demonstrated that Runt-related transcription factor 2 (RUNX2) is the main regulator responsible for the differentiation of MSCs into preosteoblasts (Bruderer et al., 2014). RUNX2 expression is regulated by several signaling pathways, especially bone morphogenetic protein (BMP) and Wnt (Narayanan et al., 2019). As an important factor in Wnt/β-catenin signaling pathway, β-catenin also regulates genes related to osteogenic differentiation (Zhang and Wang, 2020). Furthermore, Osterix (OSX) is an osteoblast-specific transcription factor, which activates a repertoire of genes during preosteoblasts differentiation into mature osteoblasts (Sinha and Zhou, 2013). Reports from the literature had shown ncRNAs in MSCs as possible targets to induce osteogenic differentiation and bone regeneration (Liu et al., 2018; Peng et al., 20182018; Yang et al., 2019; Hu et al., 2020; Chen et al., 2021). This review focuses on the regulatory role of ncRNAs in the osteogenic differentiation of BMSCs to provide detailed information for the application of ncRNAs in BMSCs-based bone tissue engineering. We also summarize the recent advances, challenges, and prospects of targeting ncRNAs in BMSCs for bone tissue engineering applications in the clinic.
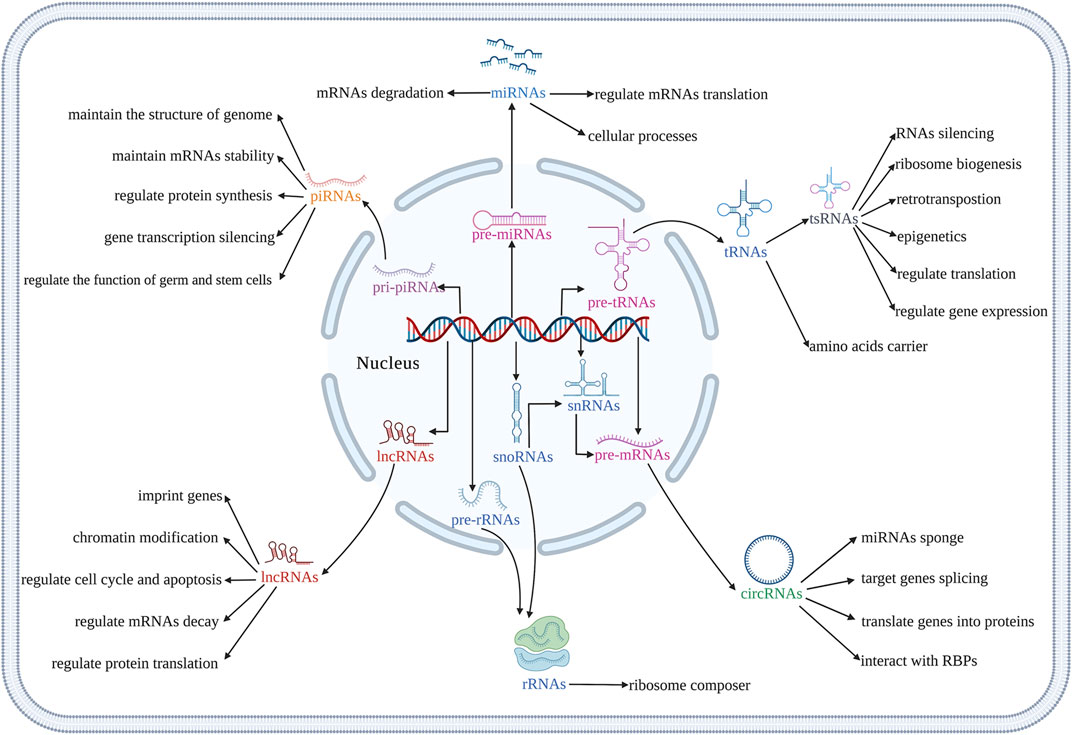
FIGURE 1. Illustration of biological functions of different kinds of ncRNAs. (Created with BioRender.com).
2 MiRNAs Involved in the Osteogenic Differentiation of BMSCs
2.1 The Biogenesis and Function of miRNAs
MiRNAs are a broad family consisting of single-stranded ncRNAs, ranging in size from 19 to 25 nucleotides (Lu and Rothenberg, 2018). MiRNAs were first discovered in Caenorhabditis elegans controlling gene expression in 1993 (Lee et al., 1993). The classical production of miRNAs is a multi-step process that requires the participation of multiple enzymes. The gene encoding miRNA is mainly transcribed by RNA polymerase II in the nucleus to produce a primary miRNA (pri-miRNA). The pri-miRNA is processed into a pre-miRNA by a microprocessor containing the RNase III enzyme Drosha. Exportin-5 is a cytoplasmic transport protein that transports pre-miRNA from the nucleus into the cytoplasm with the assistance of Ran-mediated guanosine triphosphate. Subsequently, the pre-miRNA is further processed by the RNase III enzyme Dicer, and finally, the mature miRNA is released (Lin and Gregory, 2015). Intriguingly, the maturity of some miRNAs bypass one or more steps in the classical pathway. These nonclassical miRNAs are similar to classical miRNAs in structure and function (Divisato et al., 2021). The maturation process of nonclassical miRNAs, derived from introns, snoRNAs, endogenous short hairpin RNAs, and tRNAs, does not depend on the processing of Drosha/Dgcr8, but only Dicer (Abdelfattah et al., 2014). Dicer is almost indispensable in the production of both standard and non-standard miRNA. But surprisingly, several miRNAs can also be produced in the absence of Dicer, such as miR-451 (Abdelfattah et al., 2014).
Usually, the gene silencing mechanism is determined by the degree and nature of complementarity between the miRNA binding sites and the 3′ untranslated region (3′UTR) of its target genes. The target gene undergoes degradation when the miRNAs and target genes are fully complementary (Huntzinger and Izaurralde, 2011). However, miRNAs inhibit the translation of the target genes while the binding is not complementary (Huntzinger and Izaurralde, 2011). MiRNAs are involved in various cellular processes, such as proliferation, differentiation, apoptosis, etc., (Morgado et al., 2016; Wang et al., 2019; Ding et al., 2019). Drosha and Dicer are endonucleases involved in miRNA synthesis, which are closely related to the osteogenic differentiation of BMSCs (Macfarlane and Murphy, 2010; Feng et al., 2020). Knockout of Dicer or Drosha inhibits the osteogenic differentiation of BMSCs (Oskowitz et al., 2008). Furthermore, miRNAs could directly regulate the osteogenic differentiation of BMSCs through complex mechanisms (Mazziotta et al., 2021).
2.2 Mechanisms Involved in miRNAs-Induced Osteogenic Differentiation of BMSCs
2.2.1 The Regulation in Physiological Conditions
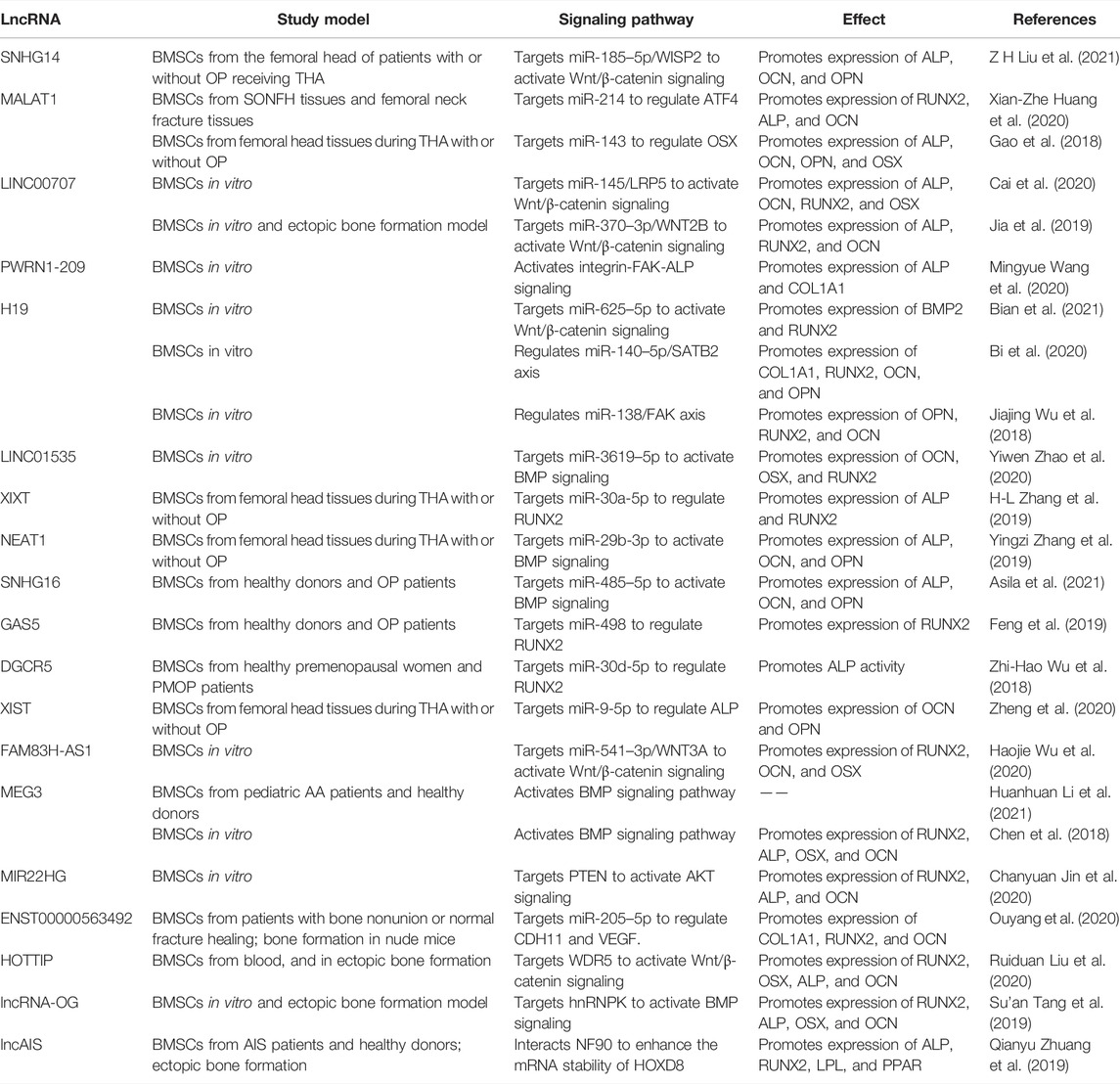
A range of miRNAs has the potential to promote osteogenic differentiation of BMSCs. BMP3 is the most abundant member of BMP family, accounting for about 65% of the total content (Bahamonde and Lyons, 2001). MiR-34a promotes the osteogenic differentiation of BMSCs by directly targeting BMP3 (Zeng et al., 2021). MiR-19b significantly promotes the osteogenic differentiation of BMSCs by targeting WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) and Samd ubiquitin regulatory factor 2 (SMURF2) through the kruppel like factor (KLF) 5/β-catenin signaling pathway (Huang et al., 2021) (Figure 2 and Table 1). Short-term or intermittent hypoxia is an inducer of osteogenic differentiation of BMSCs (Ciapetti et al., 2016; Sha et al., 2017). Epigallocatechin gallate promotes osteogenic differentiation of BMSCs under hypoxia, in which miR-210 is upregulated and targets to inhibit ephrin-A3 (EFNA3) (Qiu et al., 2016). Overexpression of miR-27b and miR-130a promotes the osteogenic differentiation of BMSCs by directly targeting peroxisome proliferator-activated receptor γ (PPARγ) to increase RUNX2 expression (Seenprachawong et al., 2018). Wang et al. found that miR-28 upregulation inhibits signal transducer and activator of transcription 1 (STAT1) expression, thus promoting the osteogenic differentiation of BMSCs (Wang et al., 2022). The expression level of miR-34c-5p is increased during the osteogenic differentiation of BMSCs (Liu et al., 2021b). B-cell lymphoma 2 (BCL2) is an anti-apoptotic protein (Ebrahim et al., 2016), and miR-34c-5p promotes the osteogenic differentiation of BMSCs via inhibition of BCL2 expression and upregulation of RUNX2 and osteocalcin (OCN) (Liu et al., 2021b). MiR-99a-5p boosts osteogenic differentiation of BMSCs, while downregulation of miR-99a-5p expression inhibits the differentiation, but the regulatory mechanism is unclear (Xu et al., 2018).
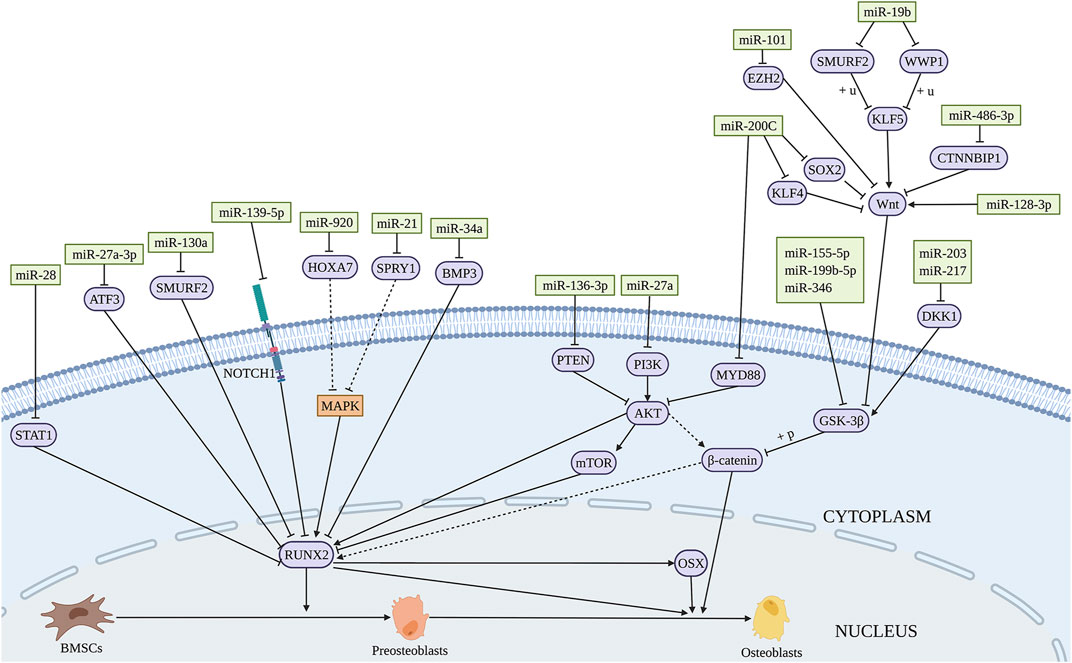
FIGURE 2. Illustration of the role and regulatory mechanism of miRNAs-induced osteogenic differentiation of BMSCs. Several miRNAs promote osteogenic differentiation of BMSCs by regulating the expression of target genes and related signaling pathways. (Created with BioRender.com).
2.2.2 The Regulation of miRNAs in Pathological Conditions
MiRNAs participate in the BMSCs differentiation in several diseases such as osteoporosis (OP), osteonecrosis, etc. OP is one common disease in the elderly and menopausal women (Tella and Gallagher, 2014). Postmenopausal osteoporosis (PMOP) is a common type of OP caused by estrogen deficiency. The osteogenic differentiation potential of BMSCs is compromised in OP patients (Zeng et al., 2021). The expression of miR-486-3p is significantly downregulated in the bone marrow of OP patients. Catenin beta interacting protein 1 (CTNNBIP1) is an inhibitor of Wnt/β-catenin signaling and mechanistically, miR-486-3p promotes the osteogenic differentiation of BMSCs by targeting CTNNBIP1 to active the Wnt/β-catenin pathway (Zhang et al., 2021). MiR-27a-3p shows lower serum level in OP patients compared with the control group. Overexpression of miR-27a-3p promotes the osteogenic differentiation of BMSCs by directly targeting activating transcription factor (ATF) 3 (Fu et al., 2019). MiR-27a is significantly decreased in the serum of PMOP patients. And miR-27a promotes the expression of osteogenesis-related markers such as alkaline phosphatase (ALP), RUNX2, and OCN by targeting myocyte enhancer factor 2C (MEF2C) (You et al., 2016). Similarly, miR-203 which is downregulated in the serum of OP patients increases the levels of osteogenic genes by targeting dickkopf 1 (DKK1) (Qiao et al., 2018). DKK1 is an important molecule in the development of embryo and adult bone, and is involved in the occurrence of OP (Glinka et al., 1998). Sprouty 1 (SPRY1) is a negative regulator of fibroblast growth factor (FGF) and extracellular signal-regulated kinase-mitogen-activated protein kinase (ERK-MAPK) signaling pathways, which is considered to be related to promoting MSCs osteogenesis (Ge et al., 2007; Ng et al., 2008). Yang et al. (2013) found that miR-21 is downregulated in BMSCs from estrogen deficiency-induced OP and promotes the osteogenic differentiation of BMSCs by targeting SPRY1.
Exogenous usage of glucocorticoids is the main risk factor for nontraumatic osteonecrosis of the femoral head (ONFH), which is termed as glucocorticoids associated ONFH and belongs to one type of steroid-associated osteonecrosis of the femoral head (SONFH). It had been demonstrated that miR-155-5p promotes osteogenic differentiation of BMSCs from SONFH by targeting glycogen synthetase kinase 3 beta (GSK-3β) and activating β-catenin signaling (Wu et al., 2021). Dai et al. (2019) reported that the expression level of miR-217 in BMSCs from patients with SONFH is decreased significantly, and miR-217 promotes the osteogenic differentiation of BMSCs by targeting DKK1. While miR-27a is downregulated in BMSCs from patients with SONFH. MiR-27a impairs the activation of phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway by targeting PI3K, thereby reversing the inhibitory effect of glucocorticoids on osteogenic differentiation of BMSCs (Tang et al., 2021).
2.2.3 The Regulation of miRNAs in Bone Regeneration and Bone Tissue Engineering
Si(OH)4 inhibits nuclear factor kappa B (NF-κB) by inducing the expression of miR-146a that activates RUNX2 expression to promote osteogenic differentiation of BMSCs (Zhou et al., 2016), suggesting miR-146a upregulation as a possible approach to promote the bone regenerative potential of BMSCs. Overexpression of miR-200c activates the AKT/β-catenin signaling pathway by targeting myeloid differentiation factor 88 (MYD88), which promotes the osteogenic differentiation of BMSCs (Xia et al., 2019). Similarly, miR-200c was found to promote the osteogenic differentiation of BMSCs in vitro by targeting sex-determining region Y-box 2 (SOX2)-mediated Wnt signaling and KLF4 (Akkouch et al., 2019). SOX2 is a major transcription factor affecting stem cell differentiation (Ma et al., 2014). Moreover, Hong et al. (2016) showed that polyethylenimine nanoparticle-based delivery of miR-200c improves the osteogenic differentiation of BMSCs and promotes bone regeneration. MiR-21 delivered by chitosan/hyaluronic acid nanoparticles promotes the osteogenesis of BMSC sheets (Zhao et al., 2016). These findings indicate the possible applications of nanomaterial-based exogenous miRNAs delivery for bone regeneration. Thus, the increased expression of osteogenesis promoting miRNAs has the potential application in bone repair. The regulatory function and mechanism of miRNAs-induced osteogenesis in BMSCs are summarized in Figure 2 and Table 1.
2.3 Mechanisms Involved in miRNAs-Inhibited Osteogenic Differentiation of BMSCs
2.3.1 The Regulation of miRNAs in Physiological Conditions
Reports from literature had shown the inhibitory role of various miRNAs in the osteogenic differentiation of BMSCs. Low-density lipoprotein receptor-related protein 5 (LRP5) is an important Wnt receptor and plays an important role in Wnt/β-catenin signaling pathway. Li et al. found that miR-23a decreases the osteogenic differentiation of BMSCs by targeting LRP5 (Wang et al., 2016) (Figure 3 and Table 2). MiR-98 inhibits the osteogenic differentiation of BMSCs by targeting BMP2 (Zhang et al., 2017). Furthermore, miR-145 inhibits the osteogenic differentiation of BMSCs by targeting semaphorin 3A (SEMA3A) (Jin et al., 2020). Retinol (vitamin A) is a micronutrient essential for cell proliferation and differentiation. Its metabolite, retinoic acid, can promote osteoblast differentiation together with BMP2 (Skillington et al., 2002). MiR-223 regulates retinol metabolism by directly inhibiting the expression of retinoic acid-inducible dehydrogenase reductase 3 (DHRS3), reducing the osteogenic differentiation of BMSCs (Zhang et al., 2018). MiRNAs mediate the osteogenic differentiation processes of drugs, factors, etc. Overexpression of miR-625-5p reverses the promoting effect of quercetin on osteogenic differentiation of BMSCs (Bian et al., 2021). Wang et al. (2016) showed that miR-150-3p targets β-catenin and inhibits tumor necrosis factor-α (TNF-α) induced osteogenic differentiation of BMSCs, which inhibits the inflammation response during bone formation. Lin et al. showed that interleukin-1β (IL−1β) inhibits osteogenic differentiation of BMSCs via miR-496-mediated inhibition of β-catenin signaling. This study claimed miR-496 as a possible target to treat inflammation-related bone loss (Huang and Chen, 2017). MiR-143-3p is involved in cadmium suppression of the Wnt/β-catenin pathway and inhibits osteogenic differentiation of BMSCs by targeting adenosine diphosphate-ribosylation factor-like protein 6 (ARL6) (Wu et al., 2020). Therefore, miR-143-3p could be targeted to treat cadmium poisoning-related bone loss. MiR-153 is a mechanosensitive miRNA that inhibits the osteogenic differentiation of BMSCs by directly targeting BMP receptor (BMPR) 2 (Cao et al., 2015).
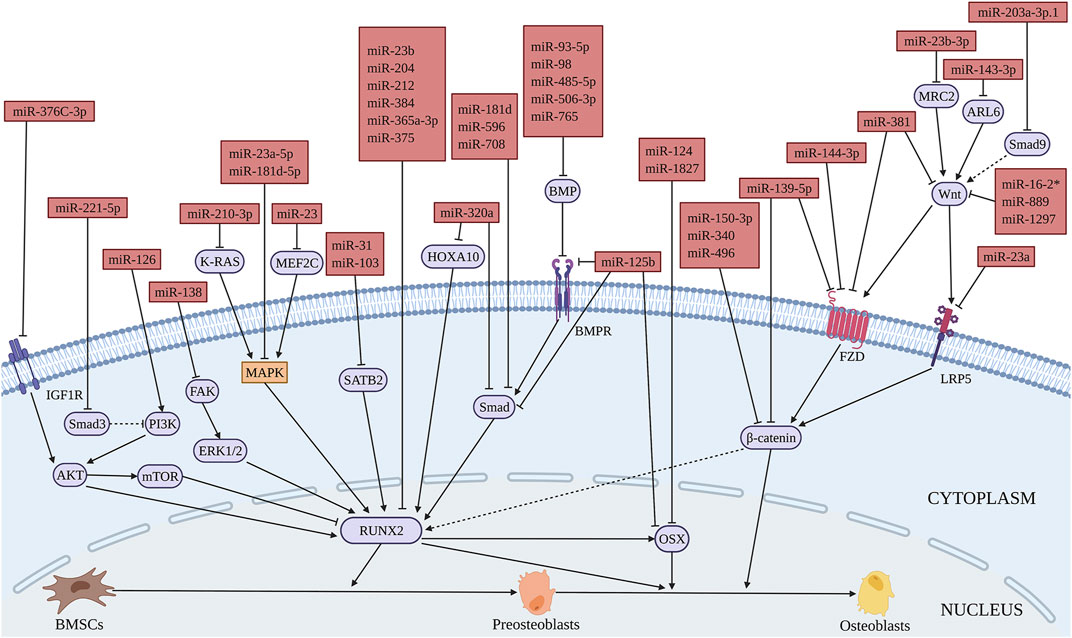
FIGURE 3. Illustration of the role and regulatory mechanism of miRNAs-inhibited osteogenic differentiation of BMSCs. (Created with BioRender.com).
2.3.2 The Regulation of miRNAs in Pathological Conditions
MiR-23, miR-16-2*, miR-210-3p, miR-889 were found to be upregulated in bone tissues or BMSCs from OP patients. MiR-23 overexpression significantly inhibits the osteogenic differentiation of BMSCs by targeting MEF2C through the MEF2C/MAPK signaling pathway, thus accelerating OP development (Jiang et al., 2020). MiR-16-2* could interfere with Wnt signal transduction by targeting WNT5A to inhibit osteogenic differentiation of BMSCs (Duan et al., 2018). Furthermore, miR-210-3p inhibits the osteogenic differentiation of BMSCs by targeting Kirsten rat sarcoma viral oncogene (K-RAS) and the downstream MAPK signal (Hu et al., 2021). MiR-889 reduces the osteogenic capability of BMSCs by targeting WNT7A through the Wnt/β-catenin signaling pathway (Xu et al., 2019). In BMSCs of age-associated OP, miR-29b-1-5p significantly downregulates the expression of stromal cell-derived factor 1 (CXCL12)/C-X-C chemokine receptor type 4 (SDF-1(CXCL12)/CXCR4) axis as well as BMP2 and RUNX2, thus negatively regulating the osteogenic differentiation of BMSCs (Eisa et al., 2021). The levels of miR-200a-3p, miR-365a-3p, miR-579-3p, and miR-1297 are increased significantly in the serum of OP patients (Cheng et al., 2019; Luo et al., 2019; Lv et al., 2019; Wang et al., 2019). MiR-365a-3p decreases the osteogenic differentiation of BMSCs by targeting RUNX2 and promotes the progress of OP (Cheng et al., 2019). Sirtuin1 (SIRT1) is an important regulator of Wnt signaling pathway, which promotes the expression of downstream differentiation related factors by de-acetylating β-catenin, thus regulating the differentiation of MSCs (Simic et al., 2013). MiR-579-3p inhibits the osteogenic differentiation of BMSCs by targeting SIRT1 (Luo et al., 2019). MiR-1297 overexpression interferes with the regulation of the Wnt signaling pathway by targeting WNT5A, thereby inhibiting the osteogenic differentiation of BMSCs (Wang et al., 2019). MiR-375 was shown to be increased in the serum of OP patients. Polypeptide drug teriparatide promotes osteogenic differentiation of BMSCs through decreasing miR-375, while the increased expression of miR-375 reverses this process (Lei et al., 2019). Forkhead box (FOX) O1 belongs to the forkhead family and is a key transcription factor regulating cell physiological function, including osteoblasts (Kitamura et al., 2005; Kim et al., 2012). Yang et al. (2021). reported a higher expression of miR-1271-5p, which is higher in osteoporotic trabecular bone tissues, and inhibits the osteogenic differentiation of BMSCs by downregulating its target FOXO1 as well as the expression of RUNX2, ALP, and OCN. In addition, miR-133 expression is significantly enhanced in BMSCs from PMOP patients. Solute carrier family 39 member one (SLC39A1) encodes zinc transporter 1, which plays an important role in the initiation of MSCs osteogenic lineage (Tang et al., 2006). MiR-133 regulates the osteogenic differentiation of BMSCs by inhibiting SLC39A1 expression (106). It has been demonstrated that diabetes increases the risk of OP (Chau et al., 2003). MiR-337 negatively regulates osteogenic differentiation of BMSCs by targeting ras-related protein 1A (RAP1A) under hyperglycemic conditions (Liu et al., 2021). Dead-box helicase 17 (DDX17) regulates the RUNX2 expression in osteoblast differentiation (Fuller-Pace and Ali, 2008), and miR-9-5p knockout promotes the osteogenic differentiation of BMSCs through targeting DDX17 under hyperglycemic conditions (He et al., 2021).
The role of miRNAs in ONFH-related diseases is also reported. Zhang et al. (2017) found that miR-93-5p is upregulated in the peripheral blood of trauma-induced ONFH patients, which inhibits osteogenic differentiation of BMSCs by targeting BMP2. MiR-181d and miR-596 are upregulated in the bone marrow of SONFH patients, while miR-708 is increased in BMSCs. These miRNAs inhibit the osteogenic differentiation of BMSCs by targeting Smad3, thereby promoting the progression of SONFH (Hao et al., 2016; Xie et al., 2018; Fu et al., 2020). Furthermore, miR-144-3p was found to be downregulated in BMSCs from patients with SONFH, and inhibit the osteogenic differentiation of BMSCs by targeting frizzled (FZD) 4 (Sun et al., 2020b). Special AT-rich sequence-binding protein 2 (SATB2) is a key regulator involved in gene expression and chromatin remodeling. SATB2 overexpression can induce the differentiation of pluripotent stem cells in vitro and significantly enhance bone regeneration and bone repair in vivo (Zhou. et al., 2016). In BMSCs from ethanol-induced osteonecrosis, the expression of miR-31 is increased. MiR-31 inhibits the osteogenesis of BMSCs by targeting SATB2 (Yu et al., 2019). Thus, the expression of these miRNAs could be inhibited in BMSCs to treat ONFH.
In addition, miRNAs play an important role in hematologic diseases. The expression of miR-203a-3p.1 is significantly decreased in BMSCs from patients with multiple myeloma (MM). MiR-203a-3p.1 inhibits the osteogenic differentiation of BMSCs by directly targeting Smad9 through the WNT3A/β-catenin signaling pathway (Fan et al., 2019a). MiR-135b is abnormally upregulated in BMSCs from MM patients. Mechanistically, miR-135b directly targets Smad5 and negatively regulates its expression, finally inhibiting the osteogenic differentiation of BMSCs (Xu et al., 2013). Myeloma bone disease (MBD) is one of the clinical features of MM. Aggressive osteolysis and low bone mass phenotype are frequently observed in MBD patients. Fan et al. (2019b) showed that miR-221-5p inhibition significantly promotes the osteogenic differentiation of BMSCs from MBD patients by targeting Smad3 and activating the PI3K/AKT/mTOR signaling pathway. Inhibition of these miRNAs in bone marrow might prevent MBD-induced bone loss. Ten-eleven translocation (TET) family is an important epigenetic modifier, which can demethylate DNA and play a key role in stem cell differentiation (Dawlaty et al., 2014; Su et al., 2019). MiR-144-3p inhibits osteogenic differentiation of BMSCs of patients with aplastic anemia (AA) by inhibiting TET2 (Li et al., 2020). Furthermore, miR-204 inhibits the osteogenic differentiation of BMSCs from AA by directly inhibiting RUNX2 (Zhao et al., 2014).
2.3.3 The Regulation of miRNAs in Bone Regeneration and Bone Tissue Engineering
BMSCs infected with these miRNAs sponges may be used in regenerative medicine. Titanium surface modification can change the shape and activity of MSCs, promote the differentiation of these cells into osteoblast lineage and upregulate osteogenic genes. MiR-23a inhibits the osteogenic differentiation of BMSCs on the surface of titanium nanotubes by targeting CXCL12 (Zhuang et al., 2019). Furthermore, the micro-arc oxidation surface of titanium implant promotes osteogenic differentiation by activating ERK1/2-miR-1827-OSX, while the overexpression of miR-1827 significantly inhibits the osteogenic differentiation of BMSCs (Liu et al., 2020). MiR-181d-5p regulates the implants’ surface roughness-induced osteogenesis. Inhibition of miR-181d-5p enhances osteogenic differentiation of BMSCs by targeting MAPK1 (Liu et al., 2021). Furthermore, the addition of miR-23a and miR-1827 inhibitors in BMSCs with titanium may increase bone integration. MiR-138 inhibits osteogenic differentiation of BMSCs by targeting focal adhesion kinase (FAK) signaling, thus reducing the ectopic bone formation of BMSCs in vivo by the combination of hydroxyapatite/tricalcium phosphate (HA/TCP) scaffolds (Eskildsen et al., 2011). MiR-125b inhibits osteogenic differentiation of BMSCs by targeting BMPR1b. The application of demineralized bone matrix with BMSCs treated with miR-125b inhibitor could be used to repair bone defects in vivo (Wang et al., 2017). BMSC transfected with miR-124 combined with HA/TCP scaffolds were subcutaneously transplanted into nude mice, demonstrating the inhibitory effect of miR-124 on the formation of ectopic bone in vivo (Qadir et al., 2015). Thus, sponges of these inhibitory miRNAs on osteogenic differentiation may be used in bone regeneration and bone repair. The regulatory function and mechanism of miRNAs that inhibit osteogenesis in BMSCs are summarized in Figure 3 and Table 2.
3 LncRNAs and Osteogenic Differentiation of BMSCs
3.1 The Biogenesis and Function of lncRNAs
LncRNAs are a group of ncRNAs with a length >200 nucleotides. According to their gene structure and the position relationship with protein-coding genes, lncRNAs are categorized into five groups: (Gaihre et al., 2017): long intergenic ncRNAs, which are located between coding genes, (Li et al., 2018), intronic lncRNAs, which originate from the intronic region of coding genes, (Nicot et al., 2020), antisense lncRNAs, which share same sequences with coding mRNA on the non-coding strand genes, (Tessier et al., 2005), bidirectional RNA, which possess the same transcription start sites with coding genes, and (Younger and Chapman, 1989) sense RNAs, which overlap with coding mRNAs on the coding strand of genes (Ponting et al., 2009; McCabe and Rasmussen, 2021). Also, lncRNAs could be divided depending on their functions and regulatory mechanisms as decoy lncRNAs, guide lncRNAs, scaffold lncRNAs, stabilizing lncRNAs, and competitive endogenous lncRNAs (ceRNAs) (McCabe and Rasmussen, 2021). LncRNAs can derive from diverse sequences of genes, including their own sequences and other promoter sequences, as well as the enhancer sequences. The biogenesis of lncRNAs differs with cell type and cell stage (Jiang and Zhang, 2021). Sharing a similar biogenesis process to mRNAs, lncRNAs are transcribed by RNA polymerase II and then capped at the 5′ region, polyadenylated at the 3′ region, and spliced (Goff and Rinn, 2015). In addition, they are expressed in a specific spatial and temporal manner influencing their functions. Recent studies demonstrate the existence of a small open reading frame in lncRNAs, which indicates their potential in various cellular processes (Ji et al., 2015).
3.2 LncRNAs Promote the Osteogenic Differentiation of BMSCs
LncRNAs are now known to exert influence on diverse biological processes, such as cell cycle (Guiducci and Stojic, 2021), proliferation (Liu et al., 2021), metastasis (Hong et al., 2021), and differentiation (Li. et al., 2021), as well as several diseases (Li et al., 2021; Liu et al., 2021; Xin and Liu, 2021). Moreover, emerging evidence shows that lncRNAs participate in the osteogenic differentiation of BMSCs. During osteogenic differentiation, lncRNAs may play their biological functions via four major ways including serving as miRNAs sponges or precursors (Huang et al., 2015), modulating epigenetic modification (Huo et al., 2017), and mediating other regulatory mechanisms.
3.2.1 The Regulation of lncRNAs in Physiological Conditions
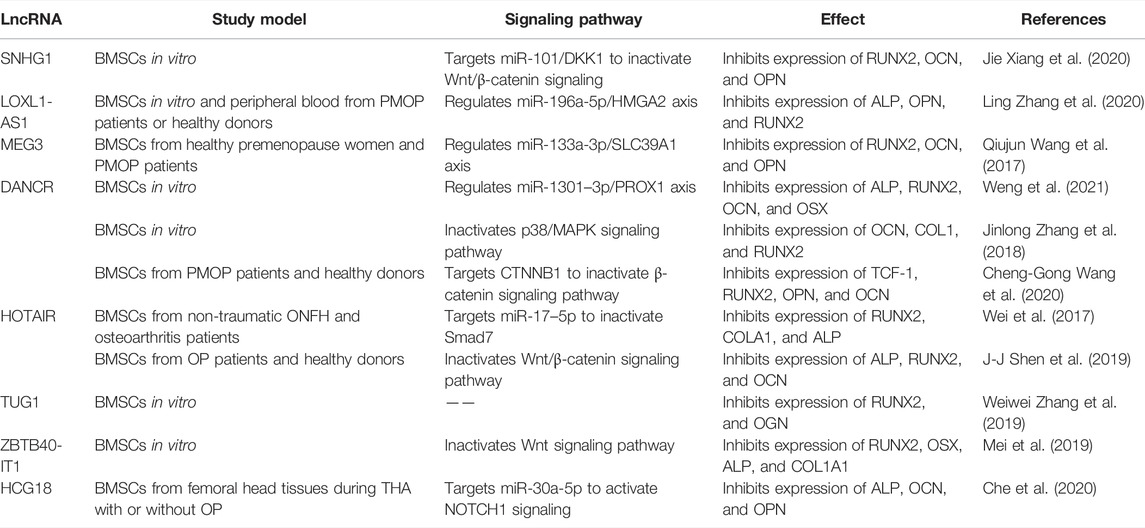
Several lncRNAs are involved in promoting osteogenesis through direct interaction with miRNAs. H19 is one of the most studied lncRNA in osteogenic differentiation. The increased expression of H19 during fetal development indicates its highly conserved characteristic throughout evolution. H19 not only influences various biological processes such as RNA progression, and cellular proliferation but also implicates in multiple human disorders (Shermane Lim et al., 2021; Wang and Qi, 2021). Quercetin has been proved to affect osteogenesis and osteoclastgenesis by regulation of a number of mechanisms, including mediating the expression of osteoprotegerin, and MAPK signaling (Wong et al., 2020). Quercetin also plays a significant role in accelerating osteogenesis via interaction with H19 by sponging miR-625-5p, and ultimately activates Wnt/β-catenin pathway (Bian et al., 2021) (Figure 4 and Table 3). Bi et al. (2020) found that H19 expression is increased in a time-dependent manner during osteogenesis. Further studies elucidated that H19 promotes osteogenic differentiation via miR-140-5p/SATB2 axis in BMSCs. Besides, H19 binds to miR-138, an miRNA targeting the gene encoding FAK called PTK2, thus upregulates downstream FAK expression, playing an important role in mechanical tension-induced osteogenic differentiation of BMSCs (Wu et al., 2018). Cai et al. (2020) found that LINC00707 is increased during osteogenic differentiation. Further studies demonstrated that LINC00707 modulates LRP5 expression by sponging miR-145, which activates the Wnt/β-catenin pathway and promotes the osteogenic differentiation of BMSCs. LINC01535 contributes to the osteogenic process via acting as a sponge of miR-3619-5p to alter BMP2 expression (Zhao et al., 2020). Similarly, lncRNA NEAT1 binds with miR-29b-3p which targets BMP1 to accelerate the osteogenic process (Zhang et al., 2019).
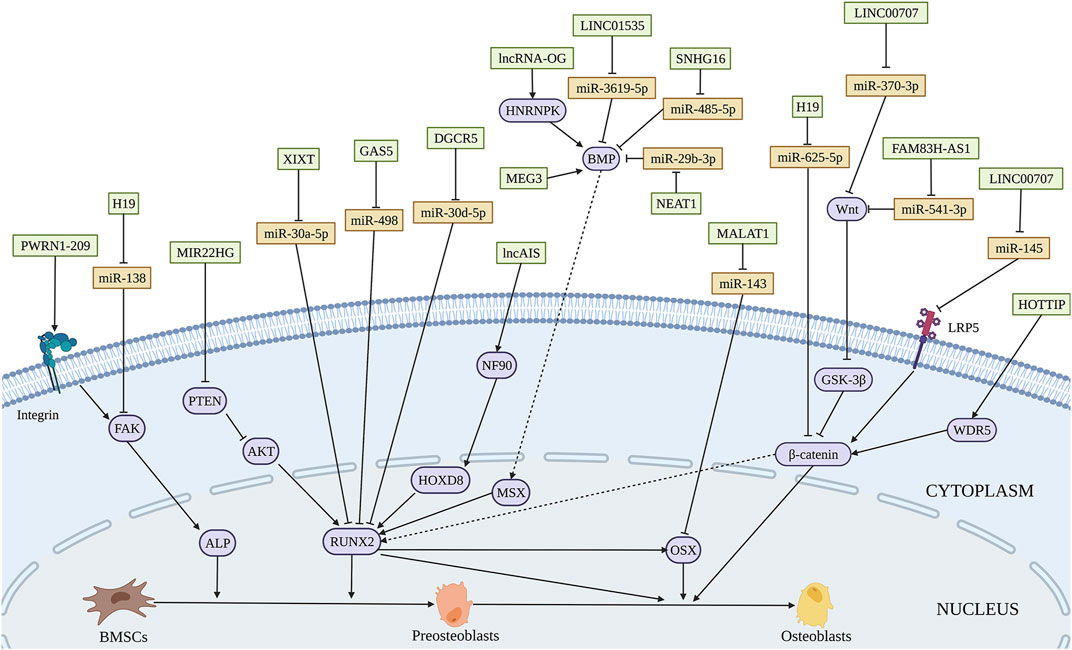
FIGURE 4. Illustration of the role and regulatory mechanism of lncRNAs-induced osteogenic differentiation of BMSCs. Various lncRNAs promote BMSCs osteogenesis through sponging miRNA (orange), activating epigenetic regulation, and mediating other regulatory mechanisms. (Created with BioRender.com).
LncRNAs could influence osteogenic differentiation of BMSCs by epigenetic regulation. WD Repeat-Containing Protein 5 (WDR5) is a transcription factor binding with the promoter of β-catenin. Upregulation of lncRNA HOX transcript at the distal tip (HOTTIP) promotes ectopic bone formation in vivo. The interaction of HOTTIP and WDR5 facilitates WDR5 translocation into the nucleus and β-catenin transcription, thus increasing osteogenic differentiation (Liu et al., 2020). Tang et al. (2019) used customized microarrays to reveal a novel lncRNA, osteogenesis-associated lncRNA (lncRNA-OG), which was upregulated by almost 12-fold during BMSCs osteogenesis. LncRNA-OG overexpression induces osteogenic differentiation of BMSCs in vitro and ectopic bone formation in nude mice. Mechanically, lncRNA-OG regulates BMP signaling pathway through direct interaction with heterogeneous nuclear ribonucleoprotein K (hnRNPK). Moreover, hnRNPK is associated with lncRNA-OG transcriptional activity by involving in the H3K27 acetylation of the lncRNA-OG promoter.
3.2.2 The Regulation of lncRNAs in Pathological Conditions
LncRNAs dysregulation is widely associated with bone-related diseases. The dysfunction of osteogenesis plays a key role in SONFH. A decline in lncRNA MALAT1 expression was found in SONFH tissues (Huang et al., 2020). ATF4, a vital regulator in bone formation, transactivates numerous osteogenic genes like RUNX2, BSP, OSX (Chan et al., 2021). MALAT1 influences ATF4 expression through sponging miR-214, ultimately increasing osteogenesis (Huang et al., 2020). What’s more, the expression of MALAT1 is significantly lower in BMSCs from discarded femoral head tissues under THA with OP than that without OP. MALAT1 could elevate an essential osteogenesis-related gene OSX expression through miR-143, affecting the osteogenic process and the development of OP (Gao et al., 2018). LncRNAs are related to the development and therapy of OP. LncRNA XIXT is downregulated, while miRNA-30a-5p is upregulated in the serum of OP patients. Mechanistically, lncRNA XIXT promotes osteogenesis by serving as a sponge of miR-30a-5p to upregulate RUNX2 (Zhang et al., 2019). Aberrant expression of small nucleolar RNA host gene 16 (SNHG16) had been reported in BMSCs from OP patients. And the promoting effect of SNHG16 on the osteogenic differentiation of BMSCs is modulated by SNHG16/miR-485-5p/BMP7 axis (Asila et al., 2021). Feng et al. (2019) demonstrated that lncRNA GAS5 is downregulated in BMSCs isolated from OP patients. Osteoblastic differentiation is promoted by the regulatory effect of GAS5 on miR-498, leading to increased RUNX2 expression and alleviating the development of OP. DEP domain-containing mTOR interacting protein (DEPTOR) is the endogenous inhibitor of mTOR, which is crucial to osteogenic differentiation and involved in OP. DEPTOR binds with the promoter of lncRNA maternally expressed 3 (nonprotein coding) (MEG3) to inhibit its transcription, consequently inactivating BMP4 signaling to restrain the osteogenic differentiation of BMSCs. Further study showed that downregulation of DEPTOR contributes to bone formation in vivo (Chen et al., 2018). Downregulated in BMSCs from osteoporosis patients, lncRNA X inactivate-specific transcript (XIST) promotes osteoblast differentiation and represses OP by regulating miR-9-5p and increasing the expression of its target ALP (Zheng et al., 2020). Sharing similar expression pattern in BMSCs from patients with PMOP, lncRNA DGCR5 upregulates RUNX2 to induce osteogenic differentiation, by sponging miR-30d-5p, thus, beneficial to delaying PMOP development (Wu et al., 2018). It has proved that osteomyelitis impedes the differentiation of BMSCs. A decline in the expression of lncRNA FAM83H-AS1 was identified in BMSCs during staphylococcal protein A-induced osteomyelitis. Mechanically, FAM83H-AS1 improves osteogenic differentiation of BMSCs by serving as a ceRNA of miR-541-3p, which brings augmentation in the expression of WNT3A, a critical member of the Wu et al. (2020)signaling pathway. Besides, AA, a common hematological disease, is characterized by inhibition of osteoblastic differentiation. Lower expression of MEG3 was detected in BMSCs of AA patients. DNA cytosine-5-methyltransferase 1 is correlated with the hypermethylation of the MEG3 promoter. MEG3 increases the transcriptional activity of BMP4 and positively affects osteoblastic differentiation of BMSCs (Li et al., 2021). Decreased osteogenic capability of BMSCs exhibits in adolescent idiopathic scoliosis (AIS) patients. Zhuang et al. (2019) reported downregulation of novel lncAIS in BMSCs from AIS patients. The interplay between lncAIS and NF90 promotes HOXD8 mRNA stability and eventually promotes the osteogenesis in normal BMSCs in vitro and in vivo.
3.2.3 The Regulation of lncRNAs in Bone Regeneration and Bone Tissue Engineering
LncRNAs, which have a promoting effect on osteogenic differentiation, are overexpressed in BMSCs. The modified BMSCs with beneficial lncRNAs loaded on biomaterials maybe used to repair bone defect in vivo. A novel lncRNA Prader-willi region ncRNAs 1–209 (PWRN1-209) was proved to enhance osteoblast differentiation on microtopography titanium surfaces possibly through integrin/FAK/ALP signaling (Wang et al., 2020). Acting as a ceRNA for miR-370-3p, LINC00707 influences osteogenic differentiation via the Wnt/β-catenin pathway in vitro. And LINC00707 modified BMSCs loaded on HA/TCP promote ectopic bone formation in NOD/SCID mice (Jia et al., 2019). Various studies have revealed the indispensable role of the PTEN/AKT pathway in bone formation (Nielsen-Preiss et al., 2003). Targeting PTEN/AKT pathway, lncRNA MIR22HG serves as a positive regulator of osteogenic differentiation in BMSCs in vitro and Bio-Oss mediated ectopic bone formation in vivo (Jin et al., 2020). Reports demonstrated that the poor osteogenic potential of BMSCs typifies bone nonunion (Tawonsawatruk et al., 2014). LncRNA ENST00000563492 functions as a sponge of miR-205-5p to elevate Cadherin-11 (CDH11) and vascular endothelial growth factor (VEGF) expression, enhancing osteogenesis of BMSCs in vitro and bone formation by a combination of matrigel in vivo. ENST00000563492 was considered a new therapeutic target for bone nonunion (Ouyang et al., 2020). The osteogenesis promoting lncRNAs may be further evaluated.
3.3 LncRNAs That Inhibit Osteogenic Differentiation in BMSCs
Likewise, multiple lncRNAs are involved in the suppression of osteogenic differentiation of BMSCs by sponging pro-osteogenic miRNAs. The expression of SNHG1 is decreased in a time-dependent manner during osteogenic differentiation. LncRNA SNHG1 inhibits osteogenesis via the miR-101/DKK1 axis and modulation of the Wnt/β-catenin signaling pathway by acting as a ceRNAs of miR-101 (Xiang et al., 2020) (Figure 5 and Table 4). LncRNA differentiation antagonizing non-protein coding RNA (DANCR) is downregulated during osteogenesis. DANCR inhibits osteogenesis of BMSCs as a sponge of miR-1301-3p which modulates prospero homeobox 1 (PROX1) expression (Weng et al., 2021). DANCR can also mediate cell proliferation and osteoblastic differentiation through the inactivation of the p38/MAPK pathway (Zhang et al., 2018). Inhibitory lnRNAs are related to bone diseases. Abnormal expression of lncRNA LOXL1 antisense RNA 1 (LOXL1-AS1) was found in peripheral blood from PMOP patients. LOXL1-AS1 suppresses osteogenic differentiation by mediating HMGA2 expression and subsequent C/EBPβ-mediated PPARγ expression by binding with miR-196a-5p in BMSCs from PMOP (Zhang et al., 2020). The expression of lncRNA MEG3 is upregulated in BMSCs of patients with PMOP. MEG3 regulates miR-133a-3p, accompanied by decreased SLC39A1 expression, to repress the osteogenic differentiation of BMSCs (Wang et al., 2017). The inhibitory effect of MEG3 on the osteogenesis of BMSCs is different from the previous reports (Chen et al., 2018; Li et al., 2021), which may be resulted from the BMSCs isolated from patients with different diseases. Increased in BMSCs from OP patients, lncRNA HCG18, which regulates miR-30a-5p/notch receptor 1 (NOTCH1) axis, suppresses osteogenic differentiation of BMSCs in OP patients and mice (Che et al., 2020). Wang et al. identified the aberrant expression of lncRNA DANCR, miR-320a, and CTNNB1 in BMSCs derived from OP patients. Furthermore, during the osteogenesis in BMSCs, DANCR and miR-320a regulate the Wnt/β-catenin signaling pathway through CTNNB1 inhibition, ultimately inhibiting the process (Wang et al., 2020). It has been suggested that the abnormal osteoblast differentiation of BMSCs is responsible for the pathogenesis of nontraumatic ONFH. In BMSCs of patients with nontraumatic ONFH, lncRNA HOX transcript antisense RNA (HOTAIR) expression was remarkably higher than normal. The sponging effect of HOTAIR on miR-17-5p leads to the decreased expression of Smad7, thus suppressing osteoblast differentiation of BMSCs in ONFH (Wei et al., 2017).
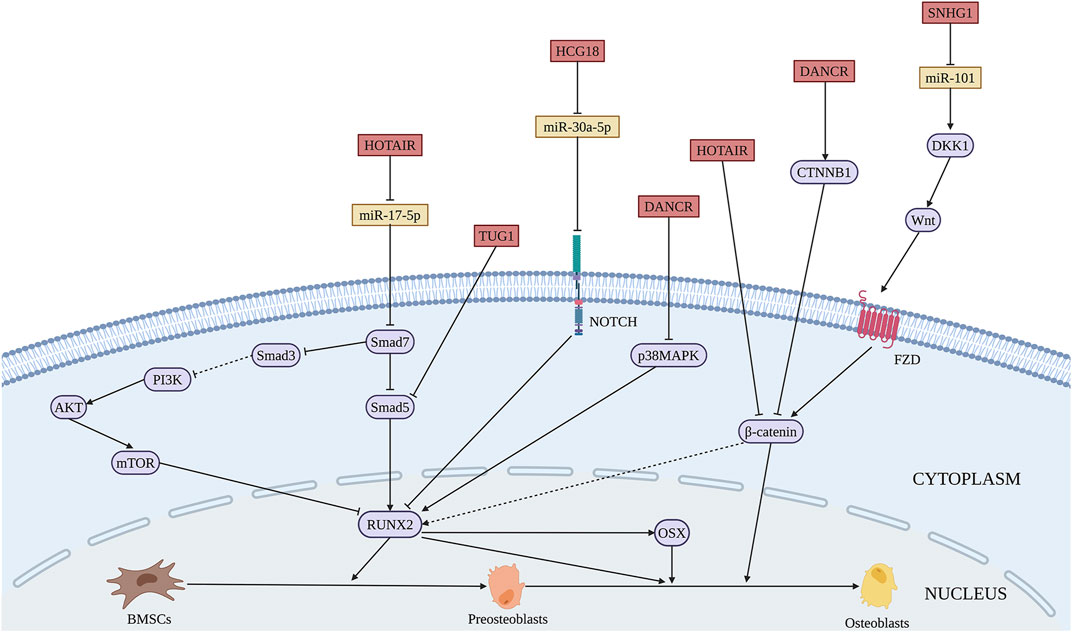
FIGURE 5. Illustration of the role and regulatory mechanism of lncRNAs-inhibited osteogenic differentiation of BMSCs. Some lncRNAs inhibit BMSCs osteogenesis through sponging pro-osteogenic miRNAs (orange). (Created with BioRender.com).
Additionally, lncRNAs inhibit the osteogenic process via epigenetic regulation. LncRNA ZBTB40-IT1 exerts an adverse effect on osteogenic differentiation in the manner of modulating WNT4, a crucial gene of the Wnt signaling pathway, while ZBTB40 has the opposite function (Mei et al., 2019). HOTAIR is significantly upregulated in OP patients both in serum and BMSCs levels. It suppresses the differentiation of BMSCs into osteoblasts through the Wnt/β-catenin signaling pathway (Shen et al., 2019). Taurine Upregulated Gene 1 (TUG1), a notably increased lncRNA after irradiation, abolishes the Smad5 signaling using the reciprocal action with the 50–90 amino acid region of Smad5 and blocking the nuclear translocation of p-Smad5 that serves as a negative regulator of osteogenic differentiation (Zhang et al., 2019). Thus, silencing the expression of inhibitory lncRNAs may increase the application potential in bone regeneration.
4 CircRNAs and the Osteogenic Differentiation of BMSCs
4.1 The Biogenesis and Function of circRNAs
CircRNAs are a kind of covalently closed ncRNAs (Kristensen et al., 2019). Unlike linear RNAs, circRNAs are more stable due to the lack of 5′ to 3′ polarity and polyadenylated tail. CircRNAs were first discovered in eukaryotic cells and were found in almost all organisms. They are abundant and evolutionarily conservative in eukaryotic cells (Hsu and Coca-Prados, 1979). Since then, thousands of circRNAs have been found in animals ranging from Drosophila melanogaster to Homo sapiens (Huang et al., 2017). CircRNAs are mostly produced from exons and have a wide variety of species, such as exon circRNAs, exon-intron circRNAs, intron circRNAs, antisense circRNAs, intergenic circRNAs, and sensory-overlap circRNAs (Guarnerio et al., 2019). The production of circRNAs mainly depends on two mechanisms. RBPs bind to introns with long inverted repeats at two ends of linear RNA and promote the binding of the two ends of linear RNA together to allow circRNAs formation. Some RBPs have been found to promote the formation of some circRNAs including the splicing factor muscleblind (Ashwal-Fluss et al., 2014), Quaking (Conn et al., 2015), RNA-binding motif protein 20 (Khan et al., 2016), and the RBP FUS (Errichelli et al., 2017), Muscleblind (Ashwal-Fluss et al., 2014), and so on. Furthermore, the RNA pairing of the complementary sequences at two ends of linear RNAs leads to circRNA formation (Patop et al., 2019).
Functionally, circRNAs play an important role in regulating gene expression in various ways, such as modulating transcription, alternative splicing, RNA processing reactions, being translated into polypeptides, interacting with RBPs, and sequestrating of miRNAs or proteins (Kristensen et al., 2019). Several studies have revealed that circRNAs are involved in the physiological and pathological processes, such as OP (Shen et al., 2020), osteosarcoma (Liu et al., 2017), Alzheimer disease, diabetes mellitus, malignant tumors (Li et al., 2018), and osteoarthritis (Ouyang et al., 2017; Shen et al., 2019). Also, circRNAs are implicated in neuronal function, innate immune responses, cell proliferation, and pluripotency (Li et al., 2019; Shi et al., 2020; Li and Chen, 2021). CircRNAs participate in the osteogenic differentiation of several kinds of MSCs including BMSCs by sponging miRNAs (Gu et al., 2017; Ouyang et al., 2019; Peng et al., 2019).
4.2 CircRNAs That Promote Osteogenic Differentiation in BMSCs
Circ_0113689 originated from gene DAB1 binds miR-1270 and miR-944 to enhance the osteogenic differentiation of BMSCs, finally exerted promoting role in chondrogenesis through NOTCH/RBPJ pathway (Chia et al., 2020) (Figure 6 and Table 5). During NOTCH/RBPJ signaling pathway, the Notch intracellular domain translocates to the nucleus and binds with RBPJ and co-activators, forming a complex that induces the transcription of downstream gene DAB1 (Luo et al., 2019). CircRNA AFF4 activates the expression of fibronectin type III domain-containing protein 5 (FNDC5)/Irisin through Smad1/5 pathway via sponging miR-135a-5 p, which induces the osteogenic differentiation of BMSCs in vitro and ectopic bone formation in vivo (Liu et al., 2021). Moreover, circ_AFF4 was reported to promote osteoblastic proliferation by acting as a miR-7223-5p sponge (Mi et al., 2019). During the bone-related diseases progression, the expressions of circ_0076906 is greatly decreased both in the bone tissue and serum of OP patients. Circ_0076906 promotes osteogenic differentiation of BMSCs through regulating to miR-1305 and its target osteoglycin (OGN), finally alleviates the OP progression (Wen et al., 2020). Circ_0006393 was decreased in the bone tissue of patients with glucocorticoid-induced OP. Further study demonstrated that circ_0006393 overexpression increases bone metabolism through miR-145-5p-FOXO1 pathway (Wang et al., 2019). CircFOXP1 regulates PTEN gene expression, thereby promoting the osteogenic differentiation of BMSCs through PI3K/AKT pathway, which may be used as the therapeutic targets in bone-related diseases such as ONFH (Xin et al., 2021). In addition, The expressions of circ_0000219 and circ_0005936 are significantly decreased in the bone marrow tissue of ONFH patients, which may be related to the proliferation and osteogenic capacity of BMSCs from ONFH patients (Xiang et al., 2020).
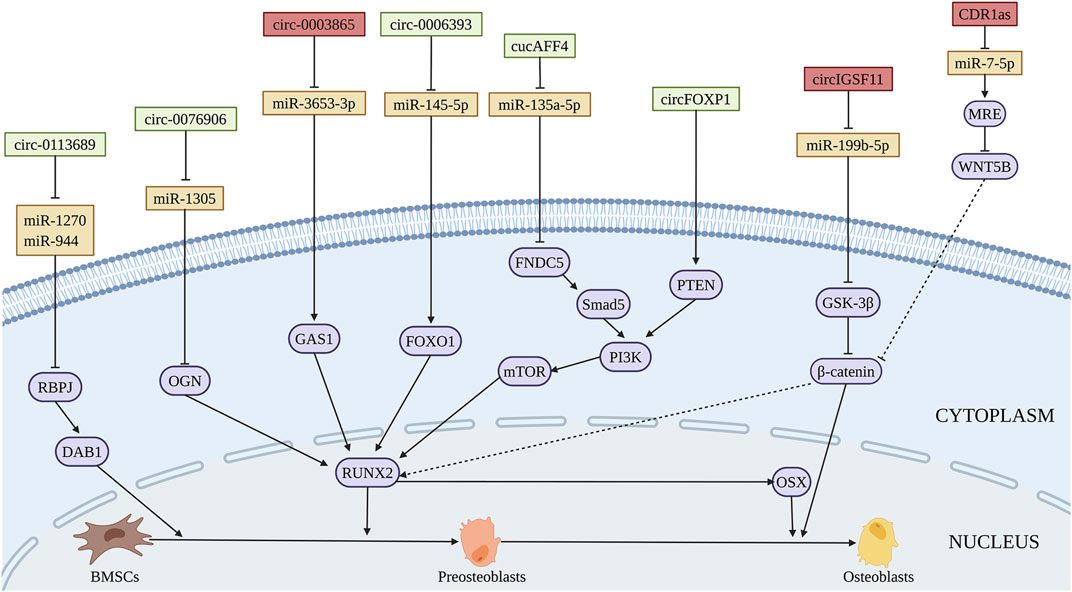
FIGURE 6. Illustration of the role and regulatory mechanism of circRNAs-promoted (green) and inhibited (red) osteogenic differentiation of BMSCs. Some circRNAs inhibit BMSCs osteogenesis through sponging pro-osteogenic miRNAs (orange). (Created with BioRender.com).
4.3 CircRNAs That Inhibit Osteogenic Differentiation in BMSCs
CircIGSF11 is downregulated during osteogenic differentiation of BMSCs. Silencing of circIGSF11 may promote osteogenesis through regulating miR-199b-5p of BMSCs (Ouyang et al., 2019). Chen et al. (2020) determined that circRNA CDR1as suppresses the expression of CDR1as and WNT5B via sponging miR-7-5p, which inhibits the osteogenic differentiation of BMSCs from patients with SONFH. Furthermore, circ_0003865 sponges miR-3653-3p to regulate growth arrest-specific gene 1 (GAS1) gene expression through NF-κB pathways, thereby inhibiting the osteogenic differentiation of BMSCs in the bone marrow tissue of OP patients (Wang et al., 2021). The expression of hsa-circ-0000885 was upregulated in peripheral blood mononuclear cells of OP patients. Circ-0000885 silencing has the potential to promote cell proliferation, osteogenic differentiation, and inhibit apoptosis of BMSCs (Zhao et al., 2021). The regulatory function and mechanism of circRNAs on osteogenic differentiation of BMSCs are summarized in Figure 6 and Table 5.
5 PiRNAs and Osteogenic Differentiation of BMSCs
In addition, piRNAs also participate in the osteogenic differentiation of BMSCs. PiRNAs are a kind of linear ncRNAs with a length of 26–31 nucleotides, which are to perform their biological functions by binding with PIWI protein (PIWIL) proteins (Iwasaki et al., 2015). The piRNA biogenesis pathways are complex and conserved, including de novo piRNA production, the ping-pong cycle, and self-amplification mechanisms, resulting in mature piRNAs (Zhang et al., 2022). Some studies have suggested that piRNA plays an essential role in maintaining the functionality of stem cells, formatting, and differentiating germ cells and somatic cells (Vagin et al., 2006; Lin et al., 2020; Li et al., 2021). In the Drosophila germline, the binding of piRNA and repeat-associated small interfering RNA ensure genomic stability by silencing transposable elements and participate in the whole process of spermatogonial generation, development, and differentiation (Vagin et al., 2006). The change of chromatin state during cell differentiation creates a circumstance in which specific transposons can be expressed, the binding of piRNA and PIWIL SMEDWI-2 participates in the regulation of somatic differentiation by specifically silencing these transposons in different cell types (Li et al., 2021). Reports demonstrated that piRNAs are involved in the osteogenic differentiation of BMSCs. RNA sequencing confirmed that 8 piRNAs are upregulated and 46 piRNAs are downregulated in the early osteogenic differentiation of BMSCs, but it is not clear whether these piRNAs are involved in the osteogenic differentiation of BMSCs (Della Bella et al., 2020). According to these dysregulated piRNAs, Liu et al. further confirmed that the binding of piR-36741 and PIWIL4 protein suppresses methyltransferase like 3-mediated BMP2 m6A level and promotes BMP2 expression, thereby increasing the osteogenic differentiation in BMSCs (Liu et al., 2021). The functional regulation and mechanism of piRNAs on osteogenic differentiation in BMSCs should be revealed. And the potential in bone regeneration of piRNAs and whether piRNAs are involved in bone-related diseases should be further evaluated.
6 Exsosomal ncRNAs and Osteogenesis
Exosomes are a kind of extracellular vesicles with a diameter of 40–100 nm (Raposo and Stoorvogel, 2013). Exosomes exist in human body fluids such as saliva, blood, and breast milk (Admyre et al., 2007; Michael et al., 2010), and can be secreted by various cells including MSCs (Raposo and Stahl, 2019). Exosomes are encapsulated by lipid bilayers, which could protect their contents from degradation. According to the different source cells, the components of exosome contents include miRNAs, lncRNAs, proteins, lipids, amino acids, etc., (Kalluri and LeBleu, 2020). Exosomes deliver these small molecules to recipient cells, thus participating in bone regeneration and other processes (Huang et al., 2020).
The expression of miR-199b, miR-218a, miR-148a, miR-135b, miR-203, miR-219, miR-299-5p, and miR-302b were significantly increased during the osteogenic differentiation of BMSCs, while the expression of miR-221, miR-155, miR-885-5p, miR-181a and miR-320c is decreased (Xu et al., 2014). Li et al. (2021) found that exosomal miR-101 derived from BMSCs promotes the osteogenic differentiation of BMSCs by targeting F-box and WD repeat domain containing 7 (FBXW7), as well as modulating FBXW7-mediated hypoxia-inducible factor-1α (HIF1α)/FOXP3 axis (Figure 7). In addition, overexpression of exosomal miR-375-5p derived from human adipose mesenchymal stem cells (AMSCs) promotes the osteogenic differentiation of BMSCs by targeting insulin-like growth factor binding protein 3 (IGFBP3) (Chen et al., 2019). Some exosomal miRNAs inhibit the osteogenic differentiation of BMSCs. Jiang et al. (2018) found an increased level of miR-21 in exosomes extracted from BMSCs in OP patients that inhibits the osteogenic differentiation of BMSCs by targeting Smad7. Furthermore, exosomal miR-23a secreted by human gingival fibroblasts inhibits the osteogenic capacity of BMSCs by targeting CXCL12 (Zhuang and Zhou, 2020). Exosomal miR-100-5p inhibits the osteogenic differentiation of BMSCs by targeting BMPR2 through BMPR2/Smad1/5/9 signaling pathway (Yang et al., 2021). Intriguingly, exosomal miR-1260a, which is also derived from BMSCs treated with Fe3O4 and a static magnetic field, promotes osteogenic differentiation of BMSCs by targeting, which provides the potential for bone regeneration of tissue engineering (Wu et al., 2021).
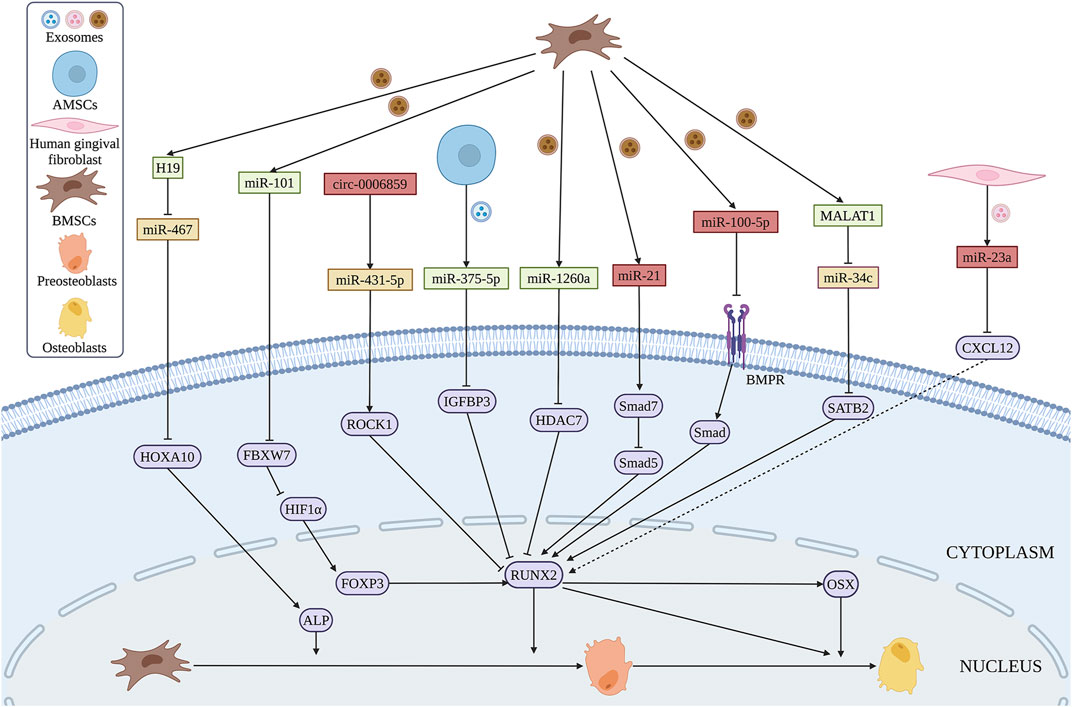
FIGURE 7. Exosomal ncRNAs derived from BMSCs (brown), AMSCs (blue) and human gingival fibroblasts (pink) can promote (green) or inhibit (red) osteogenic differentiation of BMSCs via various regulatory mechanisms. (Created with BioRender.com).
Similar to miRNAs, several lncRNAs from exosomes of BMSCs exert great functions in osteogenic differentiation. SATB2 was proved to promote osteogenic differentiation of BMSCs in patients with osteonecrosis (Yang et al., 2019). Exosomes containing MALAT1 promote osteogenic differentiation of BMSCs through the interaction between MALAT1 and miR-34c/SATB2 axis in vitro (Yang et al., 2019). Moreover, exosomal H19 derived from BMSCs reverses poor the osteogenic differentiation of BMSCs during obesity-induced fracture healing via miR-467/HOXA10 axis (Zhi et al., 2021).
CircRNAs are abundant in exosomes, and more than one thousand exosomal circRNAs have been identified in human serum (Fanale et al., 2018). The expression of circ_0006859 in exosomes is upregulated in OP patients. Exosomal circ_0006859 suppresses osteogenesis of BMSCs by sponging miR-431-5p and then elevated Rho-associated coiled-coil containing protein kinase 1 (ROCK1) expression (Zhi et al., 2021).
TsRNAs, which are classified into tRNA-derived stress-induced RNAs and tRNA-derived fragments are small fragments of RNAs generated from tRNAs by specific ribonucleases, such as dicer and angiogenin (Zong et al., 2021). Several reports showed that tsRNAs are involved in the physical and diseases processes (Kim et al., 2017; Zhu et al., 2019). Furthermore, tsRNAs are found to be dysregulated in the exomes during osteogenic differentiation of BMSCs. Yan et al. demonstrated that several tsRNAs including Ser-ACT, Ser-GCT, Sup-TTA, Phe-GAA, Ile-AAT, Lys-TTT, Leu-TAG, and Thr-CGT are significantly upregulated, while the expression of Gly-CCC, Gly-GCC, and His-GTG is downregulated in the exosomes during the osteogenic differentiation in BMSCs. However, the specific function of these tsRNAs should be further revealed. The regulatory function and mechanism, and potential application in bone regeneration of more ncRNAs should be further explored. Exosomal ncRNAs in the regulating of osteogenic differential in BMSCs are shown in Figure 7.
7 Challenges and Perspectives
BMSCs are an important source of stem cells in bone tissue engineering, which have a good application prospect in the fields of bone tissue engineering and bone regeneration. BMSCs also have great potential in the therapy of bone-related diseases such as OP and OFNH (Qi et al., 2017). The osteogenic differentiation of BMSCs is a complex physiological process. There is increasing evidence that this process is regulated by different epigenetic factors, including ncRNAs. This review focused on the function and regulatory mechanism of ncRNAs in the osteogenic differentiation of BMSCs. It has made great progress on the osteogenic differentiation of BMSCs regulated by ncRNAs. However, there are still much more to know about the functions and mechanisms of ncRNAs in regulating the osteogenic differentiation of BMSCs.
Firstly, more ncRNAs should be explored by sequencing. As we reviewed above, exosomes derived ncRNAs are not fully elucidated. Currently, only a few piRNAs and tsRNAs are identified during the osteogenic differentiation of BMSCs. And the function and regulation of these new piRNAs and tsRNAs are not clear. To our knowledge, no reports show that other ncRNAs such as rRNAs, snRNAs, snoRNAs are involved in influencing the osteogenic capability of BMSCs, which needs much more attention. Mostly, the regulatory mechanism of lncRNAs and circRNAs are involved in sponging miRNAs during the osteogenic processes of BMSCs. PiRNAs may also have the binding potential to lncRNAs and circRNAs. Thus, the regulatory network needs much deeper mining through bioinformatic analysis. Furthermore, the epigenetic regulation of lncRNAs such as methylation, and binding of transcription factors during the osteogenesis of BMSCs should be paid more attention.
Secondly, lncRNA and circRNA-encoded small peptides were identified by the computational and analytical methods used to forecast prospective ncRNAs encoding oligopeptides (Wu et al., 2020). These peptides have specific biological functions such as tumor development and inflammatory responses (Huang. et al., 2017; Niu et al., 2020; Wu et al., 2020; Gao et al., 2021). However, few reports show that lncRNA and circRNA-encoded peptides participate in the osteogenic differentiation of MSCs.
Finally, more efforts should be made to increase the clinical application for bone regeneration and bone-related diseases in the future. These ncRNAs influenced the osteogenic differentiation of BMSCs has the potential as targets by overexpression or inhibition. The efficient delivery system into BMSCs is rather important. These ncRNAs could be incorporated into exosomes and then delivered into the BMSCs. These modified BMSCs alone or in combination with biomaterials can be directly injected into bone defect sites.
8 Conclusion
In summary, research on the function of ncRNAs in the regulation of osteogenic differentiation of BMSCs has made great processes. The ncRNAs could be biomarkers of bone-related diseases. MiRNAs, circRNAs, and lncRNAs are the most extensively investigated ncRNAs for their regulatory role in the osteogenic differentiation of BMSCs and the underlying mechanisms. The osteogenic differentiation regulation potentials of piRNAs, tsRNAs, rRNAs, snRNAs, and snoRNAs are still unclear. The current understanding of the regulatory role of different miRNAs, circRNAs, and lncRNAs in osteogenic differentiation of BMSCs could be applied for bone tissue regeneration, such uses of exosomes or nanoparticles carrying osteo-stimulatory ncRNAs. However, the clinical applications of ncRNAs in bone tissue engineering are hardly reported. Therefore, future preclinical studies are mandatory to evaluate the efficacy and safety of ncRNAs-mediated BMSCs-based bone tissue engineering.
Author Contributions
ZZ, JP, and WH conceived the project and revised the manuscript; XC, WX, MZ, YS, SX, and HC wrote the first draft and drew the figures.
Funding
This project was supported by the National Undergraduate Training Program for Innovation and Entrepreneurship (202110570027), the General Guiding Project of Guangzhou (20201A011105), and the Medical Scientific Research Foundation of Guangdong Province (B2020027).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelfattah, A. M., Park, C., and Choi, M. Y. (2014). Update on Non-canonical microRNAs. Biomol. Concepts 5 (4), 275–287. doi:10.1515/bmc-2014-0012
Admyre, C., Johansson, S. M., Qazi, K. R., Filén, J.-J., Lahesmaa, R., Norman, M., et al. (2007). Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 179 (3), 1969–1978. doi:10.4049/jimmunol.179.3.1969
Akkouch, A., Eliason, S., Sweat, M. E., Romero-Bustillos, M., Zhu, M., Qian, F., et al. (2019). Enhancement of MicroRNA-200c on Osteogenic Differentiation and Bone Regeneration by Targeting Sox2-Mediated Wnt Signaling and Klf4. Hum. Gene Ther. 30 (11), 1405–1418. doi:10.1089/hum.2019.019
Arthur, A., and Gronthos, S. (2020). Clinical Application of Bone Marrow Mesenchymal Stem/Stromal Cells to Repair Skeletal Tissue. Ijms 21 (24), 9759. doi:10.3390/ijms21249759
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell. 56 (1), 55–66. doi:10.1016/j.molcel.2014.08.019
Asila, A., Yang, X., Kaisaer, Y., and Ma, L. (2021). SNHG16/miR‐485‐5p/BMP7 axis Modulates Osteogenic Differentiation of Human Bone Marrow‐derived Mesenchymal Stem Cells. J. Gene Med. 23 (3), e3296. doi:10.1002/jgm.3296
Bahamonde, M. E., and Lyons, K. M. (2001). BMP3: to Be or Not to Be a BMP. J. Bone Jt. Surgery-American Volume 83 (Pt 1), S1–S56. doi:10.2106/00004623-200100001-00008
Bi, H., Wang, D., Liu, X., Wang, G., and Wu, X. (2020). Long Non-coding RNA H19 Promotes Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Regulating microRNA-140-5p/SATB2 axis. J. Biosci. 45, 56. doi:10.1007/s12038-020-0024-y
Bian, W., Xiao, S., Yang, L., Chen, J., and Deng, S. (2021). Quercetin Promotes Bone Marrow Mesenchymal Stem Cell Proliferation and Osteogenic Differentiation through the H19/miR-625-5p axis to Activate the Wnt/β-Catenin Pathway. BMC Complement. Med. Ther. 21 (1), 243. doi:10.1186/s12906-021-03418-8
Bruderer, M., Richards, R. G., Alini, M., and Stoddart, M. (2014). Role and Regulation of RUNX2 in Osteogenesis. eCM 28, 269–286. doi:10.22203/ecm.v028a19
Cai, W. L., Zeng, W., Liu, H. H., Zhu, B. Y., Liu, J. L., and Liu, Y. (2020). LncRNA LINC00707 Promotes Osteogenic Differentiation of hBMSCs through the Wnt/β-Catenin Pathway Activated by LINC00707/miR-145/LRP5 axis. Eur. Rev. Med. Pharmacol. Sci. 24 (1), 18–28. doi:10.26355/eurrev_202001_19891
Camp, E., Pribadi, C., Anderson, P. J., Zannettino, A. C. W., and Gronthos, S. (2018). miRNA-376c-3p Mediates TWIST-1 Inhibition of Bone Marrow-Derived Stromal Cell Osteogenesis and Can Reduce Aberrant Bone Formation of TWIST-1 Haploinsufficient Calvarial Cells. Stem Cells Dev. 27 (23), 1621–1633. doi:10.1089/scd.2018.0083
Cao, Y., Lv, Q., and Lv, C. (2015). MicroRNA-153 Suppresses the Osteogenic Differentiation of Human Mesenchymal Stem Cells by Targeting Bone Morphogenetic Protein Receptor Type II. Int. J. Mol. Med. 36 (3), 760–766. doi:10.3892/ijmm.2015.2275
Chan, W. C. W., Tan, Z., To, M. K. T., and Chan, D. (2021). Regulation and Role of Transcription Factors in Osteogenesis. Ijms 22 (11), 5445. doi:10.3390/ijms22115445
Chau, D. L., Edelman, S. V., and Chandran, M. (2003). Osteoporosis and Diabetes. Curr. Diab Rep. 3 (1), 37–42. doi:10.1007/s11892-003-0051-8
Che, M., Gong, W., Zhao, Y., and Liu, M. (2020). Long Noncoding RNA HCG18 Inhibits the Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells in Osteoporosis by Targeting miR-30a-5p/NOTCH1 axis. Mol. Med. 26 (1), 106. doi:10.1186/s10020-020-00219-6
Chen, G., Wang, Q., Li, Z., Yang, Q., Liu, Y., Du, Z., et al. (2020). Circular RNA CDR1as Promotes Adipogenic and Suppresses Osteogenic Differentiation of BMSCs in Steroid-Induced Osteonecrosis of the Femoral Head. Bone 133, 115258. doi:10.1016/j.bone.2020.115258
Chen, L., Luo, W., Wang, Y., Song, X., Li, S., Wu, J., et al. (2021). Directional Homing of Glycosylation-Modified Bone Marrow Mesenchymal Stem Cells for Bone Defect Repair. J. Nanobiotechnol 19 (1), 228. doi:10.1186/s12951-021-00969-3
Chen, Q., Zhang, X., Shi, J., Yan, M., and Zhou, T. (2021). Origins and Evolving Functionalities of tRNA-Derived Small RNAs. Trends Biochem. Sci. 46 (10), 790–804. doi:10.1016/j.tibs.2021.05.001
Chen, S., Jia, L., Zhang, S., Zheng, Y., and Zhou, Y. (2018). DEPTOR Regulates Osteogenic Differentiation via Inhibiting MEG3-Mediated Activation of BMP4 Signaling and Is Involved in Osteoporosis. Stem Cell. Res. Ther. 9 (1), 185. doi:10.1186/s13287-018-0935-9
Chen, S., Tang, Y., Liu, Y., Zhang, P., Lv, L., Zhang, X., et al. (2019). Exosomes Derived from miR‐375‐overexpressing Human Adipose Mesenchymal Stem Cells Promote Bone Regeneration. Cell. Prolif. 52 (5), e12669. doi:10.1111/cpr.12669
Chen, S., Yang, L., Jie, Q., Lin, Y.-S., Meng, G.-L., Fan, J.-Z., et al. (2014). MicroRNA-125b Suppresses the Proliferation and Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Med. Rep. 9 (5), 1820–1826. doi:10.3892/mmr.2014.2024
Chen, Y., Yang, Y.-R., Fan, X.-L., Lin, P., Yang, H., Chen, X.-Z., et al. (2019). miR-206 Inhibits Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells by Targetting Glutaminase. Biosci. Rep. 39 (3), BSR20181108. doi:10.1042/bsr20181108
Chen, Y., Yu, H., Zhu, D., Liu, P., Yin, J., Liu, D., et al. (2020). miR‐136‐3p Targets PTEN to Regulate Vascularization and Bone Formation and Ameliorates Alcohol‐induced Osteopenia. FASEB J. 34 (4), 5348–5362. doi:10.1096/fj.201902463RR
Cheng, F., Yang, M. M., and Yang, R. H. (2019). MiRNA-365a-3p Promotes the Progression of Osteoporosis by Inhibiting Osteogenic Differentiation via Targeting RUNX2. Eur. Rev. Med. Pharmacol. Sci. 23 (18), 7766–7774. doi:10.26355/eurrev_201909_18986
Chia, W., Liu, J., Huang, Y.-G., and Zhang, C. (2020). A Circular RNA Derived from DAB1 Promotes Cell Proliferation and Osteogenic Differentiation of BMSCs via RBPJ/DAB1 axis. Cell. Death Dis. 11 (5), 372. doi:10.1038/s41419-020-2572-3
Ciapetti, G., Granchi, D., Fotia, C., Savarino, L., Dallari, D., Del Piccolo, N., et al. (2016). Effects of Hypoxia on Osteogenic Differentiation of Mesenchymal Stromal Cells Used as a Cell Therapy for Avascular Necrosis of the Femoral Head. Cytotherapy 18 (9), 1087–1099. doi:10.1016/j.jcyt.2016.06.005
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 160 (6), 1125–1134. doi:10.1016/j.cell.2015.02.014
Dai, Z., Jin, Y., Zheng, J., Liu, K., Zhao, J., Zhang, S., et al. (2019). MiR-217 Promotes Cell Proliferation and Osteogenic Differentiation of BMSCs by Targeting DKK1 in Steroid-Associated Osteonecrosis. Biomed. Pharmacother. 109, 1112–1119. doi:10.1016/j.biopha.2018.10.166
Dawlaty, M. M., Breiling, A., Le, T., Barrasa, M. I., Raddatz, G., Gao, Q., et al. (2014). Loss of Tet Enzymes Compromises Proper Differentiation of Embryonic Stem Cells. Dev. Cell. 29 (1), 102–111. doi:10.1016/j.devcel.2014.03.003
Della Bella, E., Menzel, U., Basoli, V., Tourbier, C., Alini, M., and Stoddart, M. J. (2020). Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells 9 (2), 398. doi:10.3390/cells9020398
Delloye, C., van Cauter, M., Dufrane, D., Francq, B. G., Docquier, P. L., and Cornu, O. (2014). Local Complications of Massive Bone Allografts: an Appraisal of Their Prevalence in 128 Patients. Acta Orthop. Belg 80 (2), 196–204.
Deng, L., Hu, G., Jin, L., Wang, C., and Niu, H. (2018). Involvement of microRNA-23b in TNF-α-Reduced BMSC Osteogenic Differentiation via Targeting Runx2. J. Bone Min. Metab. 36 (6), 648–660. doi:10.1007/s00774-017-0886-8
Ding, Y., Wang, L., Zhao, Q., Wu, Z., and Kong, L. (2019). MicroRNA-93 I-nhibits C-hondrocyte A-poptosis and I-nflammation in O-steoarthritis by T-argeting the TLR4/NF-κB S-ignaling P-athway. Int. J. Mol. Med. 43 (2), 779–790. doi:10.3892/ijmm.2018.4033
Divisato, G., Piscitelli, S., Elia, M., Cascone, E., and Parisi, S. (2021). MicroRNAs and Stem-like Properties: The Complex Regulation Underlying Stemness Maintenance and Cancer Development. Biomolecules 11 (8), 1074. doi:10.3390/biom11081074
Du, K., Li, Z., Fang, X., Cao, T., and Xu, Y. (2017). Ferulic Acid Promotes Osteogenesis of Bone Marrow-Derived Mesenchymal Stem Cells by Inhibiting microRNA-340 to Induce β-catenin Expression through Hypoxia. Eur. J. Cell. Biol. 96 (6), 496–503. doi:10.1016/j.ejcb.2017.07.002
Duan, L., Zhao, H., Xiong, Y., Tang, X., Yang, Y., Hu, Z., et al. (2018). miR-16-2* Interferes with WNT5A to Regulate Osteogenesis of Mesenchymal Stem Cells. Cell. Physiol. Biochem. 51 (3), 1087–1102. doi:10.1159/000495489
Ebrahim, A. S., Sabbagh, H., Liddane, A., Raufi, A., Kandouz, M., and Al-Katib, A. (2016). Hematologic Malignancies: Newer Strategies to Counter the BCL-2 Protein. J. Cancer Res. Clin. Oncol. 142 (9), 2013–2022. doi:10.1007/s00432-016-2144-1
Eisa, N. H., Sudharsan, P. T., Herrero, S. M., Herberg, S. A., Volkman, B. F., Aguilar-Pérez, A., et al. (2021). Age-associated Changes in microRNAs Affect the Differentiation Potential of Human Mesenchymal Stem Cells: Novel Role of miR-29b-1-5p Expression. Bone 153, 116154. doi:10.1016/j.bone.2021.116154
El-Rashidy, A. A., Roether, J. A., Harhaus, L., Kneser, U., and Boccaccini, A. R. (2017). Regenerating Bone with Bioactive Glass Scaffolds: A Review of In Vivo Studies in Bone Defect Models. Acta Biomater. 62, 1–28. doi:10.1016/j.actbio.2017.08.030
Eppley, B. L., Pietrzak, W. S., and Blanton, M. W. (2005). Allograft and Alloplastic Bone Substitutes: a Review of Science and Technology for the Craniomaxillofacial Surgeon. J. Craniofac Surg. 16 (6), 981–989. doi:10.1097/01.scs.0000179662.38172.dd
Errichelli, L., Dini Modigliani, S., Laneve, P., Colantoni, A., Legnini, I., Capauto, D., et al. (2017). FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat. Commun. 8, 14741. doi:10.1038/ncomms14741
Eskildsen, T., Taipaleenmäki, H., Stenvang, J., Abdallah, B. M., Ditzel, N., Nossent, A. Y., et al. (2011). MicroRNA-138 Regulates Osteogenic Differentiation of Human Stromal (Mesenchymal) Stem Cells In Vivo. Proc. Natl. Acad. Sci. U.S.A. 108 (15), 6139–6144. doi:10.1073/pnas.1016758108
Fan, F. Y., Deng, R., Qiu, L., Wen, Q., Zeng, Y., Gao, L., et al. (2019a). miR-203a-3p.1 is Involved in the Regulation of Osteogenic Differentiation by Directly Targeting Smad9 in MM-MSCs. Oncol. Lett. 18 (6), 6339–6346. doi:10.3892/ol.2019.10994
Fan, F. Y., Deng, R., Lai, S. H., Wen, Q., Zeng, Y., Gao, L., et al. (2019b). Inhibition of MicroRNA-221-5p Induces Osteogenic Differentiation by Directly Targeting Smad3 in Myeloma Bone Disease Mesenchymal Stem Cell. Oncol. Lett. 18 (6), 6536–6544. doi:10.3892/ol.2019.10992
Fanale, D., Taverna, S., Russo, A., and Bazan, V. (2018). Circular RNA in Exosomes. Adv. Exp. Med. Biol. 1087, 109–117. doi:10.1007/978-981-13-1426-1_9
Feng, J., Wang, J. X., and Li, C. H. (2019). LncRNA GAS5 Overexpression Alleviates the Development of Osteoporosis through Promoting Osteogenic Differentiation of MSCs via Targeting microRNA-498 to Regulate RUNX2. Eur. Rev. Med. Pharmacol. Sci. 23 (18), 7757–7765. doi:10.26355/eurrev_201909_18985
Feng, L., Zhang, J.-f., Shi, L., Yang, Z.-m., Wu, T.-y., Wang, H.-x., et al. (2020). MicroRNA-378 Suppressed Osteogenesis of MSCs and Impaired Bone Formation via Inactivating Wnt/β-Catenin Signaling. Mol. Ther. - Nucleic Acids 21, 1017–1028. doi:10.1016/j.omtn.2020.07.018
Fu, L., Liu, H., and Lei, W. (2020). MiR-596 Inhibits Osteoblastic Differentiation and Cell Proliferation by Targeting Smad3 in Steroid-Induced Osteonecrosis of Femoral Head. J. Orthop. Surg. Res. 15 (1), 173. doi:10.1186/s13018-020-01688-5
Fu, Y. C., Zhao, S. R., Zhu, B. H., Guo, S. S., and Wang, X. X. (2019). MiRNA-27a-3p Promotes Osteogenic Differentiation of Human Mesenchymal Stem Cells through Targeting ATF3. Eur. Rev. Med. Pharmacol. Sci. 23 (3 Suppl. l), 73–80. doi:10.26355/eurrev_201908_18632
Fuller-Pace, F. V., and Ali, S. (2008). The DEAD Box RNA Helicases P68 (Ddx5) and P72 (Ddx17): Novel Transcriptional Co-regulators. Biochem. Soc. Trans. 36 (Pt 4), 609–612. doi:10.1042/bst0360609
Gaihre, B., Uswatta, S., and Jayasuriya, A. (2017). Reconstruction of Craniomaxillofacial Bone Defects Using Tissue-Engineering Strategies with Injectable and Non-injectable Scaffolds. Jfb 8 (4), 49. doi:10.3390/jfb8040049
Gao, Y., Xiao, F., Wang, C., Wang, C., Cui, P., Zhang, X., et al. (2018). Long Noncoding RNA MALAT1 Promotes Osterix Expression to Regulate Osteogenic Differentiation by Targeting miRNA‐143 in Human Bone Marrow‐derived Mesenchymal Stem Cells. J. Cell. Biochem. 119 (8), 6986–6996. doi:10.1002/jcb.26907
Ge, C., Xiao, G., Jiang, D., and Franceschi, R. T. (2007). Critical Role of the Extracellular Signal-Regulated Kinase-MAPK Pathway in Osteoblast Differentiation and Skeletal Development. J. Cell. Biol. 176 (5), 709–718. doi:10.1083/jcb.200610046
Glinka, A., Wu, W., Delius, H., Monaghan, A. P., Blumenstock, C., and Niehrs, C. (1998). Dickkopf-1 Is a Member of a New Family of Secreted Proteins and Functions in Head Induction. Nature 391 (6665), 357–362. doi:10.1038/34848
Goff, L. A., and Rinn, J. L. (2015). Linking RNA Biology to lncRNAs. Genome Res. 25 (10), 1456–1465. doi:10.1101/gr.191122.115
Gu, X., Li, M., Jin, Y., Liu, D., and Wei, F. (2017). Identification and Integrated Analysis of Differentially Expressed lncRNAs and circRNAs Reveal the Potential ceRNA Networks during PDLSC Osteogenic Differentiation. BMC Genet. 18 (1), 100. doi:10.1186/s12863-017-0569-4
Guarnerio, J., Zhang, Y., Cheloni, G., Panella, R., Mae Katon, J., Simpson, M., et al. (2019). Intragenic Antagonistic Roles of Protein and circRNA in Tumorigenesis. Cell. Res. 29 (8), 628–640. doi:10.1038/s41422-019-0192-1
Guiducci, G., and Stojic, L. (2021). Long Noncoding RNAs at the Crossroads of Cell Cycle and Genome Integrity. Trends Genet. 37 (6), 528–546. doi:10.1016/j.tig.2021.01.006
Guttman, M., Russell, P., Ingolia, N. T., Weissman, J. S., and Lander, E. S. (2013). Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 154 (1), 240–251. doi:10.1016/j.cell.2013.06.009
Hao, C., Yang, S., Xu, W., Shen, J. K., Ye, S., Liu, X., et al. (2016). MiR-708 Promotes Steroid-Induced Osteonecrosis of Femoral Head, Suppresses Osteogenic Differentiation by Targeting SMAD3. Sci. Rep. 6, 22599. doi:10.1038/srep22599
He, C., Liu, M., Ding, Q., Yang, F., and Xu, T. (2021). Upregulated miR-9-5p Inhibits Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells under High Glucose Treatment. J. Bone Min. Metab. 40 (2), 208–219. doi:10.1007/s00774-021-01280-9
Hong, J., Guo, F., Lu, S.-Y., Shen, C., Ma, D., Zhang, X., et al. (2021). F. Nucleatum Targets lncRNA ENO1-IT1 to Promote Glycolysis and Oncogenesis in Colorectal Cancer. Gut 70 (11), 2123–2137. doi:10.1136/gutjnl-2020-322780
Hong, L., Sharp, T., Khorsand, B., Fischer, C., Eliason, S., Salem, A., et al. (2016). MicroRNA-200c Represses IL-6, IL-8, and CCL-5 Expression and Enhances Osteogenic Differentiation. PLoS One 11 (8), e0160915. doi:10.1371/journal.pone.0160915
Hsu, M.-T., and Coca-Prados, M. (1979). Electron Microscopic Evidence for the Circular Form of RNA in the Cytoplasm of Eukaryotic Cells. Nature 280 (5720), 339–340. doi:10.1038/280339a0
Hu, M., Zhu, X., Yuan, H., Li, H., Liao, H., and Chen, S. (2021). The Function and Mechanism of the miR‐210‐3p/KRAS axis in Bone Marrow‐derived Mesenchymal Stem Cell from Patients with Osteoporosis. J. Tissue Eng. Regen. Med. 15 (8), 699–711. doi:10.1002/term.3215
Hu, Z., Zhang, L., Wang, H., Wang, Y., Tan, Y., Dang, L., et al. (2020). Targeted Silencing of miRNA-132-3p Expression Rescues Disuse Osteopenia by Promoting Mesenchymal Stem Cell Osteogenic Differentiation and Osteogenesis in Mice. Stem Cell. Res. Ther. 11 (1), 58. doi:10.1186/s13287-020-1581-6
Huang, C.-C., Kang, M., Lu, Y., Shirazi, S., Diaz, J. I., Cooper, L. F., et al. (2020). Functionally Engineered Extracellular Vesicles Improve Bone Regeneration. Acta Biomater. 109, 182–194. doi:10.1016/j.actbio.2020.04.017
Huang, J.-Z., Chen, M., Chen, D., Gao, X.-C., Zhu, S., Huang, H., et al. (2017). A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell. 68 (1), 171–184. doi:10.1016/j.molcel.2017.09.015
Huang, J., and Chen, L. (2017). IL-1β Inhibits Osteogenesis of Human Bone Marrow-Derived Mesenchymal Stem Cells by Activating FoxD3/microRNA-496 to Repress Wnt Signaling. Genesis 55 (7), e23040. doi:10.1002/dvg.23040
Huang, J., Meng, Y., Liu, Y., Chen, Y., Yang, H., Chen, D., et al. (2016). MicroRNA-320a Regulates the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Targeting HOXA10. Cell. Physiol. Biochem. 38 (1), 40–48. doi:10.1159/000438607
Huang, S., Yang, B., Chen, B. J., Bliim, N., Ueberham, U., Arendt, T., et al. (2017). The Emerging Role of Circular RNAs in Transcriptome Regulation. Genomics 109 (5-6), 401–407. doi:10.1016/j.ygeno.2017.06.005
Huang, X.-Z., Huang, J., Li, W.-Z., Wang, J.-J., Song, D.-Y., and Ni, J.-D. (2020). LncRNA-MALAT1 Promotes Osteogenic Differentiation through Regulating ATF4 by Sponging miR-214: Implication of Steroid-Induced Avascular Necrosis of the Femoral Head. Steroids 154, 108533. doi:10.1016/j.steroids.2019.108533
Huang, Y., Xu, Y., Feng, S., He, P., Sheng, B., and Ni, J. (2021). miR-19b Enhances Osteogenic Differentiation of Mesenchymal Stem Cells and Promotes Fracture Healing through the WWP1/Smurf2-Mediated KLF5/β-Catenin Signaling Pathway. Exp. Mol. Med. 53 (5), 973–985. doi:10.1038/s12276-021-00631-w
Huang, Y., Zheng, Y., Jia, L., and Li, W. (2015). Long Noncoding RNA H19 Promotes Osteoblast Differentiation via TGF-β1/Smad3/HDAC Signaling Pathway by Deriving miR-675. STEM CELLS 33 (12), 3481–3492. doi:10.1002/stem.2225
Huang, Z., Cai, Z., Qian, J., Wang, J., and Hu, N. (2020). Effect of Micro RNA-335-5p Regulating Bone Morphogenetic Protein 2 on Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 34 (6), 781–786. doi:10.7507/1002-1892.201910097
Huntzinger, E., and Izaurralde, E. (2011). Gene Silencing by microRNAs: Contributions of Translational Repression and mRNA Decay. Nat. Rev. Genet. 12 (2), 99–110. doi:10.1038/nrg2936
Huo, S., Zhou, Y., He, X., Wan, M., Du, W., Xu, X., et al. (2017). Insight into the Role of Long Non-coding RNAs during Osteogenesis in Mesenchymal Stem Cells. Cscr 13 (1), 52–59. doi:10.2174/1574888x12666171115124112
Iwasaki, Y. W., Siomi, M. C., and Siomi, H. (2015). PIWI-interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 84, 405–433. doi:10.1146/annurev-biochem-060614-034258
Ji, Z., Song, R., Regev, A., and Struhl, K. (2015). Many lncRNAs, 5'UTRs, and Pseudogenes Are Translated and Some Are Likely to Express Functional Proteins. Elife 4, e08890. doi:10.7554/eLife.08890
Jia, B., Wang, Z., Sun, X., Chen, J., Zhao, J., and Qiu, X. (2019). Long Noncoding RNA LINC00707 Sponges miR-370-3p to Promote Osteogenesis of Human Bone Marrow-Derived Mesenchymal Stem Cells through Upregulating WNT2B. Stem Cell. Res. Ther. 10 (1), 67. doi:10.1186/s13287-019-1161-9
Jiang, K., Teng, G. D., and Chen, Y. Q. (2020). MicroRNA‐23 Suppresses Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells by Targeting the MEF2C‐mediated MAPK Signaling Pathway. J. Gene Med. 22 (10), e3216. doi:10.1002/jgm.3216
Jiang, L. B., Tian, L., and Zhang, C. G. (2018). Bone Marrow Stem Cells-Derived Exosomes Extracted from Osteoporosis Patients Inhibit Osteogenesis via microRNA-21/SMAD7. Eur. Rev. Med. Pharmacol. Sci. 22 (19), 6221–6229. doi:10.26355/eurrev_201810_16028
Jiang, Y., Zhang, P., Zhang, X., Lv, L., and Zhou, Y. (2021). Advances in Mesenchymal Stem Cell Transplantation for the Treatment of Osteoporosis. Cell. Prolif. 54 (1), e12956. doi:10.1111/cpr.12956
Jiang, Z.-F., and Zhang, L. (2021). LncRNA: A Potential Research Direction in Intestinal Barrier Function. Dig. Dis. Sci. 66 (5), 1400–1408. doi:10.1007/s10620-020-06417-w
Jin, C., Jia, L., Tang, Z., and Zheng, Y. (2020). Long Non-coding RNA MIR22HG Promotes Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells via PTEN/AKT Pathway. Cell. Death Dis. 11 (7), 601. doi:10.1038/s41419-020-02813-2
Jin, Y., Hong, F., Bao, Q., Xu, Q., Duan, R., Zhu, Z., et al. (2020). MicroRNA-145 Suppresses Osteogenic Differentiation of Human Jaw Bone Marrow Mesenchymal Stem Cells Partially via Targeting Semaphorin 3A. Connect. Tissue Res. 61 (6), 577–585. doi:10.1080/03008207.2019.1643334
Kalluri, R., and LeBleu, V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Karijolich, J., and Yu, Y.-T. (2010). Spliceosomal snRNA Modifications and Their Function. RNA Biol. 7 (2), 192–204. doi:10.4161/rna.7.2.11207
Khan, M. A. F., Reckman, Y. J., Aufiero, S., van den Hoogenhof, M. M. G., van der Made, I., Beqqali, A., et al. (2016). RBM20 Regulates Circular RNA Production from the Titin Gene. Circ. Res. 119 (9), 996–1003. doi:10.1161/circresaha.116.309568
Kim, H. K., Fuchs, G., Wang, S., Wei, W., Zhang, Y., Park, H., et al. (2017). A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature 552 (7683), 57–62. doi:10.1038/nature25005
Kim, K. M., Park, S. J., Jung, S.-H., Kim, E. J., Jogeswar, G., Ajita, J., et al. (2012). miR-182 Is a Negative Regulator of Osteoblast Proliferation, Differentiation, and Skeletogenesis through Targeting FoxO1. J. Bone Min. Res. 27 (8), 1669–1679. doi:10.1002/jbmr.1604
Kitamura, Y. I., Kitamura, T., Kruse, J.-P., Raum, J. C., Stein, R., Gu, W., et al. (2005). FoxO1 Protects against Pancreatic β Cell Failure through NeuroD and MafA Induction. Cell. Metab. 2 (3), 153–163. doi:10.1016/j.cmet.2005.08.004
Kong, R., Gao, J., Ji, L., and Zhao, D. (2020). MicroRNA-126 Promotes Proliferation, Migration, Invasion and Endothelial Differentiation while Inhibits Apoptosis and Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells. Cell. Cycle 19 (17), 2119–2138. doi:10.1080/15384101.2020.1788258
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell. 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lei, N. B., Liang, X., Wang, P., Liu, Q., and Wang, W. G. (2019). Teriparatide Alleviates Osteoporosis by Promoting Osteogenic Differentiation of hMSCs via miR-375/RUNX2 axis. Eur. Rev. Med. Pharmacol. Sci. 23 (24), 11043–11050. doi:10.26355/eurrev_201912_19812
Li, D., Taylor, D. H., and van Wolfswinkel, J. C. (2021). PIWI-mediated Control of Tissue-specific Transposons Is Essential for Somatic Cell Differentiation. Cell. Rep. 37 (1), 109776. doi:10.1016/j.celrep.2021.109776
Li, H., Xu, J.-D., Fang, X.-H., Zhu, J.-N., Yang, J., Pan, R., et al. (2020). Circular RNA circRNA_000203 Aggravates Cardiac Hypertrophy via Suppressing miR-26b-5p and miR-140-3p Binding to Gata4. Cardiovasc Res. 116 (7), 1323–1334. doi:10.1093/cvr/cvz215
Li, H., Xu, X., Wang, D., Zhang, Y., Chen, J., Li, B., et al. (2021). Hypermethylation-mediated Downregulation of Long Non-coding RNA MEG3 Inhibits Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells and Promotes Pediatric Aplastic Anemia. Int. Immunopharmacol. 93, 107292. doi:10.1016/j.intimp.2020.107292
Li, I., and Chen, Y. G. (2021). Emerging Roles of Circular RNAs in Innate Immunity. Curr. Opin. Immunol. 68, 107–115. doi:10.1016/j.coi.2020.10.010
Li, J. J., Ebied, M., Xu, J., and Zreiqat, H. (2018). Current Approaches to Bone Tissue Engineering: The Interface between Biology and Engineering. Adv. Healthc. Mater 7 (6), e1701061. ARTN 1701061. doi:10.1002/adhm.201701061
Li, J., Shi, Q., Wang, Q., Tan, X., Pang, K., Liu, X., et al. (2019). Profiling Circular RNA in Methamphetamine-Treated Primary Cortical Neurons Identified Novel circRNAs Related to Methamphetamine Addiction. Neurosci. Lett. 701, 146–153. doi:10.1016/j.neulet.2019.02.032
Li, J., Wu, X., Shi, Y., and Zhao, H. (2021). FGD5-AS1 Facilitates the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells via Targeting the miR-506-3p/BMP7 axis. J. Orthop. Surg. Res. 16 (1), 665. doi:10.1186/s13018-021-02694-x
Li, N., Liu, L., Liu, Y., Luo, S., Song, Y., and Fang, B. (2020). miR-144-3p Suppresses Osteogenic Differentiation of BMSCs from Patients with Aplastic Anemia through Repression of TET2. Mol. Ther. - Nucleic Acids 19, 619–626. doi:10.1016/j.omtn.2019.12.017
Li, R., Ruan, Q., Yin, F., and Zhao, K. (2021). MiR-23b-3p Promotes Postmenopausal Osteoporosis by Targeting MRC2 and Regulating the Wnt/β-Catenin Signaling Pathway. J. Pharmacol. Sci. 145 (1), 69–78. doi:10.1016/j.jphs.2020.11.004
Li, T., Li, H., Li, T., Fan, J., Zhao, R. C., and Weng, X. (2014). MicroRNA Expression Profile of Dexamethasone-Induced Human Bone Marrow-Derived Mesenchymal Stem Cells during Osteogenic Differentiation. J. Cell. Biochem. 115 (10), 1683–1691. doi:10.1002/jcb.24831
Li, X., Yang, L., and Chen, L.-L. (2018). The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell. 71 (3), 428–442. doi:10.1016/j.molcel.2018.06.034
Li, Y., Jiang, J., Liu, W., Wang, H., Zhao, L., Liu, S., et al. (2018). microRNA-378 Promotes Autophagy and Inhibits Apoptosis in Skeletal Muscle. Proc. Natl. Acad. Sci. U.S.A. 115 (46), E10849–e10858. doi:10.1073/pnas.1803377115
Li, Y., Wang, J., Ma, Y., Du, W., Feng, K., and Wang, S. (2021). miR‐101‐loaded Exosomes Secreted by Bone Marrow Mesenchymal Stem Cells Requires the FBXW7/HIF1α/FOXP3 axis, Facilitating Osteogenic Differentiation. J. Cell. Physiol. 236 (6), 4258–4272. doi:10.1002/jcp.30027
Lin, K.-Y., Wang, W.-D., Lin, C.-H., Rastegari, E., Su, Y.-H., Chang, Y.-T., et al. (2020). Piwi Reduction in the Aged Niche Eliminates Germline Stem Cells via Toll-GSK3 Signaling. Nat. Commun. 11 (1), 3147. doi:10.1038/s41467-020-16858-6
Lin, S., and Gregory, R. I. (2015). MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 15 (6), 321–333. doi:10.1038/nrc3932
Lin, Y. P., Liao, L. M., Liu, Q. H., Ni, Y., Zhong, Y., and Yu, S. (2021). MiRNA-128-3p Induces Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells via Activating the Wnt3a Signaling. Eur. Rev. Med. Pharmacol. Sci. 25 (3), 1225–1232. doi:10.26355/eurrev_202102_24826
Lin, Z., He, H., Wang, M., and Liang, J. (2019). MicroRNA‐130a Controls Bone Marrow Mesenchymal Stem Cell Differentiation towards the Osteoblastic and Adipogenic Fate. Cell. Prolif. 52 (6), e12688. doi:10.1111/cpr.12688
Liu, B., Cao, J., Wang, X., Guo, C., Liu, Y., and Wang, T. (2021a). Deciphering the tRNA-Derived Small RNAs: Origin, Development, and Future. Cell. Death Dis. 13 (1), 24. doi:10.1038/s41419-021-04472-3
Liu, B., Gan, W., Jin, Z., Wang, M., Cui, G., Zhang, H., et al. (2021b). The Role of miR-34c-5p in Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Ijsc 14 (3), 286–297. doi:10.15283/ijsc20188
Liu, C., Liu, A.-S., Zhong, D., Wang, C.-G., Yu, M., Zhang, H.-W., et al. (2021). Circular RNA AFF4 Modulates Osteogenic Differentiation in BM-MSCs by Activating SMAD1/5 Pathway through miR-135a-5p/FNDC5/Irisin axis. Cell. Death Dis. 12 (7), 631. doi:10.1038/s41419-021-03877-4
Liu, H., Zhong, L., Yuan, T., Chen, S., Zhou, Y., An, L., et al. (2018). MicroRNA-155 Inhibits the Osteogenic Differentiation of Mesenchymal Stem Cells Induced by BMP9 via Downregulation of BMP Signaling Pathway. Int. J. Mol. Med. 41 (6), 3379–3393. doi:10.3892/ijmm.2018.3526
Liu, J., Chen, M., Ma, L., Dang, X., and Du, G. (2021). piRNA-36741 Regulates BMP2-Mediated Osteoblast Differentiation via METTL3 Controlled m6A Modification. Aging 13 (19), 23361–23375. doi:10.18632/aging.203630
Liu, L., Zeng, D., Chen, Y., Zhou, J., Liao, Y., and Shi, B. (2020). Microarc Oxidation Surface of Titanium Implants Promote Osteogenic Differentiation by Activating ERK1/2-miR-1827-Osterix. Vitro Cell.Dev.Biol.-Animal 56 (4), 296–306. doi:10.1007/s11626-020-00444-7
Liu, N., Kataoka, M., Wang, Y., Pu, L., Dong, X., Fu, X., et al. (2021). LncRNA LncHrt Preserves Cardiac Metabolic Homeostasis and Heart Function by Modulating the LKB1-AMPK Signaling Pathway. Basic Res. Cardiol. 116 (1), 48. doi:10.1007/s00395-021-00887-3
Liu, R., Li, Z., Song, E., Hu, P., Yang, Q., Hu, Y., et al. (2020). LncRNA HOTTIP Enhances Human Osteogenic BMSCs Differentiation via Interaction with WDR5 and Activation of Wnt/β-Catenin Signalling Pathway. Biochem. Biophysical Res. Commun. 524 (4), 1037–1043. doi:10.1016/j.bbrc.2020.02.034
Liu, S., Yang, X., Zhong, X., Li, L., and Zhang, X. (2021). Involvement of miR-337 in High Glucose-Suppressed Osteogenic Differentiation in Bone Marrow Mesenchymal Stem Cells via Negative Regulation of Rap1A. Vitro Cell.Dev.Biol.-Animal 57 (3), 350–358. doi:10.1007/s11626-021-00553-x
Liu, W., Zhang, J., Zou, C., Xie, X., Wang, Y., Wang, B., et al. (2017). Microarray Expression Profile and Functional Analysis of Circular RNAs in Osteosarcoma. Cell. Physiol. Biochem. 43 (3), 969–985. doi:10.1159/000481650
Liu, Y., Wang, Y., Cheng, X., Zheng, Y., Lyu, M., Di, P., et al. (2021). MiR-181d-5p Regulates Implant Surface Roughness-Induced Osteogenic Differentiation of Bone Marrow Stem Cells. Mater. Sci. Eng. C 121, 111801. doi:10.1016/j.msec.2020.111801
Liu, Z. H., Qi, D. D., Li, X., Zhang, S. Q., Zhao, Y., Fu, L. X., et al. (2021). LncRNA SNHG14 Promotes Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells via Regulating miR-185-5p/WISP2 axis. J. Biol. Regul. Homeost. Agents 35 (2), 605–615. doi:10.23812/20-391-a
Long, C., Cen, S., Zhong, Z., Zhou, C., and Zhong, G. (2021). FOXO3 Is Targeted by miR-223-3p and Promotes Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells by Enhancing Autophagy. Hum. Cell. 34 (1), 14–27. doi:10.1007/s13577-020-00421-y
Long, H., Sun, B., Cheng, L., Zhao, S., Zhu, Y., Zhao, R., et al. (2017). miR-139-5p Represses BMSC Osteogenesis via Targeting Wnt/β-Catenin Signaling Pathway. DNA Cell. Biol. 36 (8), 715–724. doi:10.1089/dna.2017.3657
Long, H., Zhu, Y., Lin, Z., Wan, J., Cheng, L., Zeng, M., et al. (2019). miR-381 Modulates Human Bone Mesenchymal Stromal Cells (BMSCs) Osteogenesis via Suppressing Wnt Signaling Pathway during Atrophic Nonunion Development. Cell. Death Dis. 10 (7), 470. doi:10.1038/s41419-019-1693-z
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141 (4), 1202–1207. doi:10.1016/j.jaci.2017.08.034
Lu, X., Deng, M., He, H., Zeng, D., and Zhang, W. (2013). [miR-125b Regulates Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells by Targeting Smad4]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 38 (4), 341–346. doi:10.3969/j.issn.1672-7347.2013.04.002
Luo, B., Yang, J. F., Wang, Y. H., Qu, G. B., Hao, P. D., Zeng, Z. J., et al. (2019). MicroRNA-579-3p Promotes the Progression of Osteoporosis by Inhibiting Osteogenic Differentiation of Mesenchymal Stem Cells through Regulating Sirt1. Eur. Rev. Med. Pharmacol. Sci. 23 (16), 6791–6799. doi:10.26355/eurrev_201908_18717
Luo, Z., Shang, X., Zhang, H., Wang, G., Massey, P. A., Barton, S. R., et al. (2019). Notch Signaling in Osteogenesis, Osteoclastogenesis, and Angiogenesis. Am. J. Pathology 189 (8), 1495–1500. doi:10.1016/j.ajpath.2019.05.005
Lv, H., Yang, H., and Wang, Y. (2020). Effects of miR-103 by Negatively Regulating SATB2 on Proliferation and Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. PLoS One 15 (5), e0232695. doi:10.1371/journal.pone.0232695
Lv, R., Pan, X., Song, L., Sun, Q., Guo, C., Zou, S., et al. (2019). MicroRNA-200a-3p Accelerates the Progression of Osteoporosis by Targeting Glutaminase to Inhibit Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Biomed. Pharmacother. 116, 108960. doi:10.1016/j.biopha.2019.108960
Ma, K., Song, G., An, X., Fan, A., Tan, W., Tang, B., et al. (2014). miRNAs Promote Generation of Porcine-Induced Pluripotent Stem Cells. Mol. Cell. Biochem. 389 (1-2), 209–218. doi:10.1007/s11010-013-1942-x
Macfarlane, L.-A., and R. Murphy, P. (2010). MicroRNA: Biogenesis, Function and Role in Cancer. Cg 11 (7), 537–561. doi:10.2174/138920210793175895
Mazziotta, C., Lanzillotti, C., Iaquinta, M. R., Taraballi, F., Torreggiani, E., Rotondo, J. C., et al. (2021). MicroRNAs Modulate Signaling Pathways in Osteogenic Differentiation of Mesenchymal Stem Cells. Ijms 22 (5), 2362. doi:10.3390/ijms22052362
McCabe, E. M., and Rasmussen, T. P. (2021). lncRNA Involvement in Cancer Stem Cell Function and Epithelial-Mesenchymal Transitions. Seminars Cancer Biol. 75, 38–48. doi:10.1016/j.semcancer.2020.12.012
Mei, B., Wang, Y., Ye, W., Huang, H., Zhou, Q., Chen, Y., et al. (2019). LncRNA ZBTB40-IT1 Modulated by Osteoporosis GWAS Risk SNPs Suppresses Osteogenesis. Hum. Genet. 138 (2), 151–166. doi:10.1007/s00439-019-01969-y
Mi, B., Xiong, Y., Chen, L., Yan, C., Endo, Y., Liu, Y., et al. (2019). CircRNA AFF4 Promotes Osteoblast Cells Proliferation and Inhibits Apoptosis via the Mir-7223-5p/PIK3R1 axis. Aging 11 (24), 11988–12001. doi:10.18632/aging.102524
Michael, A., Bajracharya, S., Yuen, P., Zhou, H., Star, R., Illei, G., et al. (2010). Exosomes from Human Saliva as a Source of microRNA Biomarkers. Oral Dis. 16 (1), 34–38. doi:10.1111/j.1601-0825.2009.01604.x
Mollazadeh, S., Fazly Bazzaz, B. S., Neshati, V., de Vries, A. A. F., Naderi-Meshkin, H., Mojarad, M., et al. (2019). Overexpression of MicroRNA-148b-3p Stimulates Osteogenesis of Human Bone Marrow-Derived Mesenchymal Stem Cells: the Role of MicroRNA-148b-3p in Osteogenesis. BMC Med. Genet. 20 (1), 117. doi:10.1186/s12881-019-0854-3
Morgado, A. L., Rodrigues, C. M. P., and Solá, S. (2016). MicroRNA-145 Regulates Neural Stem Cell Differentiation through the Sox2-Lin28/let-7 Signaling Pathway. Stem Cells 34 (5), 1386–1395. doi:10.1002/stem.2309
Narayanan, A., Srinaath, N., Rohini, M., and Selvamurugan, N. (2019). Regulation of Runx2 by MicroRNAs in Osteoblast Differentiation. Life Sci. 232, 116676. doi:10.1016/j.lfs.2019.116676
Ng, F., Boucher, S., Koh, S., Sastry, K. S. R., Chase, L., Lakshmipathy, U., et al. (2008). PDGF, TGF-β, and FGF Signaling Is Important for Differentiation and Growth of Mesenchymal Stem Cells (MSCs): Transcriptional Profiling Can Identify Markers and Signaling Pathways Important in Differentiation of MSCs into Adipogenic, Chondrogenic, and Osteogenic Lineages. Blood 112 (2), 295–307. doi:10.1182/blood-2007-07-103697
Nicot, R., Schlund, M., Touzet-Roumazeille, S., Ferri, J., and Raoul, G. (2020). Unicortical Calvarial Autologous Bone Graft Harvest. Plast. Reconstr. Surg. Glob. Open 8 (11), e3241. doi:10.1097/gox.0000000000003241
Nielsen-Preiss, S. M., Silva, S. R., and Gillette, J. M. (2003). Role of PTEN and Akt in the Regulation of Growth and Apoptosis in Human Osteoblastic Cells. J. Cell. Biochem. 90 (5), 964–975. doi:10.1002/jcb.10709
Niu, L., Lou, F., Sun, Y., Sun, L., Cai, X., Liu, Z., et al. (2020). A Micropeptide Encoded by lncRNA MIR155HG Suppresses Autoimmune Inflammation via Modulating Antigen Presentation. Sci. Adv. 6 (21), eaaz2059. doi:10.1126/sciadv.aaz2059
Oskowitz, A. Z., Lu, J., Penfornis, P., Ylostalo, J., McBride, J., Flemington, E. K., et al. (2008). Human Multipotent Stromal Cells from Bone Marrow and microRNA: Regulation of Differentiation and Leukemia Inhibitory Factor Expression. Proc. Natl. Acad. Sci. U.S.A. 105 (47), 18372–18377. doi:10.1073/pnas.0809807105
Ouyang, Q., Wu, J., Jiang, Z., Zhao, J., Wang, R., Lou, A., et al. (2017). Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients. Cell. Physiol. Biochem. 42 (2), 651–659. doi:10.1159/000477883
Ouyang, Z., Tan, T., Zhang, X., Wan, J., Zhou, Y., Jiang, G., et al. (2019). CircRNA Hsa_circ_0074834 Promotes the Osteogenesis-Angiogenesis Coupling Process in Bone Mesenchymal Stem Cells (BMSCs) by Acting as a ceRNA for miR-942-5p. Cell. Death Dis. 10 (12), 932. doi:10.1038/s41419-019-2161-5
Ouyang, Z., Tan, T., Zhang, X., Wan, J., Zhou, Y., Jiang, G., et al. (2020). LncRNA ENST00000563492 Promoting the Osteogenesis-Angiogenesis Coupling Process in Bone Mesenchymal Stem Cells (BMSCs) by Functions as a ceRNA for miR-205-5p. Cell. Death Dis. 11 (6), 486. doi:10.1038/s41419-020-2689-4
Patop, I. L., Wüst, S., and Kadener, S. (2019). Past, Present, and Future of Circ RNA S. Embo J. 38 (16), e100836. doi:10.15252/embj.2018100836
Peng, F., Gong, W., Li, S., Yin, B., Zhao, C., Liu, W., et al. (2021). circRNA_010383 Acts as a Sponge for miR-135a, and its Downregulated Expression Contributes to Renal Fibrosis in Diabetic Nephropathy. Diabetes 70 (2), 603–615. doi:10.2337/db20-0203
Peng, S., Cao, L., He, S., Zhong, Y., Ma, H., Zhang, Y., et al. (20182018). An Overview of Long Noncoding RNAs Involved in Bone Regeneration from Mesenchymal Stem Cells. Stem Cells Int. 2018, 1–11. doi:10.1155/2018/8273648
Peng, W., Zhu, S., Chen, J., Wang, J., Rong, Q., and Chen, S. (2019). Hsa_circRNA_33287 Promotes the Osteogenic Differentiation of Maxillary Sinus Membrane Stem Cells via miR-214-3p/Runx3. Biomed. Pharmacother. 109, 1709–1717. doi:10.1016/j.biopha.2018.10.159
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and Functions of Long Noncoding RNAs. Cell. 136 (4), 629–641. doi:10.1016/j.cell.2009.02.006
Qadir, A. S., Um, S., Lee, H., Baek, K., Seo, B. M., Lee, G., et al. (2015). miR-124 Negatively Regulates Osteogenic Differentiation and In Vivo Bone Formation of Mesenchymal Stem Cells. J. Cell. Biochem. 116 (5), 730–742. doi:10.1002/jcb.25026
Qi, M., Zhang, L., Ma, Y., Shuai, Y., Li, L., Luo, K., et al. (2017). Autophagy Maintains the Function of Bone Marrow Mesenchymal Stem Cells to Prevent Estrogen Deficiency-Induced Osteoporosis. Theranostics 7 (18), 4498–4516. doi:10.7150/thno.17949
Qiao, L., Liu, D., Li, C. G., and Wang, Y. J. (2018). MiR-203 Is Essential for the Shift from Osteogenic Differentiation to Adipogenic Differentiation of Mesenchymal Stem Cells in Postmenopausal Osteoporosis. Eur. Rev. Med. Pharmacol. Sci. 22 (18), 5804–5814. doi:10.26355/eurrev_201809_15906
Qiu, Y., Chen, Y., Zeng, T., Guo, W., Zhou, W., and Yang, X. (2016). EGCG Ameliorates the Hypoxia-Induced Apoptosis and Osteogenic Differentiation Reduction of Mesenchymal Stem Cells via Upregulating miR-210. Mol. Biol. Rep. 43 (3), 183–193. doi:10.1007/s11033-015-3936-0
Raposo, G., and Stahl, P. D. (2019). Extracellular Vesicles: a New Communication Paradigm? Nat. Rev. Mol. Cell. Biol. 20 (9), 509–510. doi:10.1038/s41580-019-0158-7
Raposo, G., and Stoorvogel, W. (2013). Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell. Biol. 200 (4), 373–383. doi:10.1083/jcb.201211138
Ren, G., Sun, J., Li, M. M., Zhang, Y. D., Li, R. H., and Li, Y. M. (2018). MicroRNA-23a-5p Regulates Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Targeting Mitogen-Activated Protein Kinase-13. Mol. Med. Rep. 17 (3), 4554–4560. doi:10.3892/mmr.2018.8452
Seenprachawong, K., Tawornsawutruk, T., Nantasenamat, C., Nuchnoi, P., Hongeng, S., and Supokawej, A. (2018). miR-130a and miR-27b Enhance Osteogenesis in Human Bone Marrow Mesenchymal Stem Cells via Specific Down-Regulation of Peroxisome Proliferator-Activated Receptor γ. Front. Genet. 9, 543. doi:10.3389/fgene.2018.00543
Sha, Y., Lv, Y., Xu, Z., Yang, L., Hao, X., and Afandi, R. (2017). MGF E Peptide Pretreatment Improves the Proliferation and Osteogenic Differentiation of BMSCs via MEK-Erk1/2 and PI3K-Akt Pathway under Severe Hypoxia. Life Sci. 189, 52–62. doi:10.1016/j.lfs.2017.09.017
Shen, J. J., Zhang, C. H., Chen, Z. W., Wang, Z. X., Yang, D. C., Zhang, F. L., et al. (2019). LncRNA HOTAIR Inhibited Osteogenic Differentiation of BMSCs by Regulating Wnt/β-Catenin Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (17), 7232–7246. doi:10.26355/eurrev_201909_18826
Shen, S., Wu, Y., Chen, J., Xie, Z., Huang, K., Wang, G., et al. (2019). CircSERPINE2 Protects against Osteoarthritis by Targeting miR-1271 and ETS-Related Gene. Ann. Rheum. Dis. 78 (6), 826–836. doi:10.1136/annrheumdis-2018-214786
Shen, W., Sun, B., Zhou, C., Ming, W., Zhang, S., and Wu, X. (2020). CircFOXP1/FOXP1 Promotes Osteogenic Differentiation in Adipose‐derived Mesenchymal Stem Cells and Bone Regeneration in Osteoporosis via miR‐33a‐5p. J. Cell. Mol. Med. 24 (21), 12513–12524. doi:10.1111/jcmm.15792
Shermane Lim, Y. W., Xiang, X., Garg, M., Le, M. T., Li-Ann Wong, A., Wang, L., et al. (2021). The Double-Edged Sword of H19 lncRNA: Insights into Cancer Therapy. Cancer Lett. 500, 253–262. doi:10.1016/j.canlet.2020.11.006
Shi, Y., Fang, N., Li, Y., Guo, Z., Jiang, W., He, Y., et al. (2020). Circular RNA LPAR3 Sponges microRNA‐198 to Facilitate Esophageal Cancer Migration, Invasion, and Metastasis. Cancer Sci. 111 (8), 2824–2836. doi:10.1111/cas.14511
Simic, P., Zainabadi, K., Bell, E., Sykes, D. B., Saez, B., Lotinun, S., et al. (2013). SIRT1 Regulates Differentiation of Mesenchymal Stem Cells by Deacetylating β‐catenin. EMBO Mol. Med. 5 (3), 430–440. doi:10.1002/emmm.201201606
Sinha, K. M., and Zhou, X. (2013). Genetic and Molecular Control of Osterix in Skeletal Formation. J. Cell. Biochem. 114 (5), 975–984. doi:10.1002/jcb.24439
Skillington, J., Choy, L., and Derynck, R. (2002). Bone Morphogenetic Protein and Retinoic Acid Signaling Cooperate to Induce Osteoblast Differentiation of Preadipocytes. J. Cell. Biol. 159 (1), 135–146. doi:10.1083/jcb.200204060
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K.-D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the Ribosome: The Influence of rRNA Modification on Eukaryotic Ribosome Biogenesis and Function. RNA Biol. 14 (9), 1138–1152. doi:10.1080/15476286.2016.1259781
Sohn, H.-S., and Oh, J.-K. (2019). Review of Bone Graft and Bone Substitutes with an Emphasis on Fracture Surgeries. Biomater. Res. 23, 9. doi:10.1186/s40824-019-0157-y
Song, G., Shen, Y., Ruan, Z., Li, X., Chen, Y., Yuan, W., et al. (2016). LncRNA-uc.167 Influences Cell Proliferation, Apoptosis and Differentiation of P19 Cells by Regulating Mef2c. Gene 590 (1), 97–108. doi:10.1016/j.gene.2016.06.006
Steinhardt, Y., Aslan, H., Regev, E., Zilberman, Y., Kallai, I., Gazit, D., et al. (2008). Maxillofacial-derived Stem Cells Regenerate Critical Mandibular Bone Defect. Tissue Eng. Part A 14 (11), 1763–1773. doi:10.1089/ten.tea.2008.0007
Su, P.-H., Hsu, Y.-W., Huang, R.-L., Chen, L.-Y., Chao, T.-K., Liao, C.-C., et al. (2019). TET1 Promotes 5hmC-dependent Stemness, and Inhibits a 5hmC-independent Epithelial-Mesenchymal Transition, in Cervical Precancerous Lesions. Cancer Lett. 450, 53–62. doi:10.1016/j.canlet.2019.01.033
Sun, J., Wang, Y., Li, Y., and Zhao, G. (2014). Downregulation of PPARγ by miR-548d-5p Suppresses the Adipogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells and Enhances Their Osteogenic Potential. J. Transl. Med. 12, 168. doi:10.1186/1479-5876-12-168
Sun, Z., Zhang, Q., Yuan, W., Li, X., Chen, C., Guo, Y., et al. (2020a). MiR-103a-3p Promotes Tumour Glycolysis in Colorectal Cancer via hippo/YAP1/HIF1A axis. J. Exp. Clin. Cancer Res. 39 (1), 250. doi:10.1186/s13046-020-01705-9
Sun, Z., Wu, F., Yang, Y., Liu, F., Mo, F., Chen, J., et al. (2020b). MiR-144-3p Inhibits BMSC Proliferation and Osteogenic Differentiation via Targeting FZD4 in Steroid-Associated Osteonecrosis. Cpd 25 (45), 4806–4812. doi:10.2174/1381612825666190930094019
Tang, J., Yu, H., Wang, Y., Duan, G., Wang, B., Li, W., et al. (2021). miR-27a Promotes Osteogenic Differentiation in Glucocorticoid-Treated Human Bone Marrow Mesenchymal Stem Cells by Targeting PI3K. J. Mol. Histol. 52 (2), 279–288. doi:10.1007/s10735-020-09947-9
Tang, J. Z., Lin, X., Zhong, J. Y., Xu, F., Wu, F., Liao, X. B., et al. (2019). miR-124 Regulates the Osteogenic Differentiation of Bone Marrow-derived Mesenchymal Stem Cells by Targeting Sp7. Mol. Med. Rep. 19 (5), 3807–3814. doi:10.3892/mmr.2019.10054
Tang, S. a., Xie, Z., Wang, P., Li, J., Wang, S., Liu, W., et al. (2019). LncRNA-OG Promotes the Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells under the Regulation of hnRNPK. Stem Cells 37 (2), 270–283. doi:10.1002/stem.2937
Tang, Z., Sahu, S. N., Khadeer, M. A., Bai, G., Franklin, R. B., and Gupta, A. (2006). Overexpression of the ZIP1 Zinc Transporter Induces an Osteogenic Phenotype in Mesenchymal Stem Cells. Bone 38 (2), 181–198. doi:10.1016/j.bone.2005.08.010
Tawonsawatruk, T., Kelly, M., and Simpson, H. (2014). Evaluation of Native Mesenchymal Stem Cells from Bone Marrow and Local Tissue in an Atrophic Nonunion Model. Tissue Eng. Part C. Methods 20 (6), 524–532. doi:10.1089/ten.TEC.2013.0465
Tella, S. H., and Gallagher, J. C. (2014). Prevention and Treatment of Postmenopausal Osteoporosis. J. Steroid Biochem. Mol. Biol. 142, 155–170. doi:10.1016/j.jsbmb.2013.09.008
Tessier, P., Kawamoto, H., Posnick, J., Raulo, Y., Tulasne, J. F., and Wolfe, S. A. (2005). Complications of Harvesting Autogenous Bone Grafts: a Group Experience of 20,000 Cases. Plastic Reconstr. Surg. 116 (5 Suppl. l), 72s–73s. doi:10.1097/01.prs.0000173841.59063.7e
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A Distinct Small RNA Pathway Silences Selfish Genetic Elements in the Germline. Science 313 (5785), 320–324. doi:10.1126/science.1129333
Wang, C.-G., Hu, Y.-H., Su, S.-L., and Zhong, D. (2020). LncRNA DANCR and miR-320a Suppressed Osteogenic Differentiation in Osteoporosis by Directly Inhibiting the Wnt/β-Catenin Signaling Pathway. Exp. Mol. Med. 52 (8), 1310–1325. doi:10.1038/s12276-020-0475-0
Wang, H., Xie, Z., Hou, T., Li, Z., Huang, K., Gong, J., et al. (2017). MiR-125b Regulates the Osteogenic Differentiation of Human Mesenchymal Stem Cells by Targeting BMPR1b. Cell. Physiol. Biochem. 41 (2), 530–542. doi:10.1159/000457013
Wang, H., Meng, Y., Cui, Q., Qin, F., Yang, H., Chen, Y., et al. (2016). MiR-101 Targets the EZH2/Wnt/β-Catenin the Pathway to Promote the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. Sci. Rep. 6, 36988. doi:10.1038/srep36988
Wang, H., Tan, Z., Hu, H., Liu, H., Wu, T., Zheng, C., et al. (2019). microRNA-21 Promotes Breast Cancer Proliferation and Metastasis by Targeting LZTFL1. BMC Cancer 19 (1), 738. doi:10.1186/s12885-019-5951-3
Wang, J. L., Wei, X., Wang, A. G., Bai, Y., and Wu, X. J. (2020). KCNQ1OT1 Regulates Osteogenic Differentiation of hBMSC by miR-320a/Smad5 axis. Eur. Rev. Med. Pharmacol. Sci. 24 (6), 2843–2854. doi:10.26355/eurrev_202003_20648
Wang, L., and Qi, L. (2021). The Role and Mechanism of Long Non-coding RNA H19 in Stem Cell Osteogenic Differentiation. Mol. Med. 27 (1), 86. doi:10.1186/s10020-021-00350-y
Wang, M., Dai, T., Meng, Q., Wang, W., and Li, S. (2022). Regulatory Effects of miR-28 on Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Bioengineered 13 (1), 684–696. doi:10.1080/21655979.2021.2012618
Wang, M., Ge, X., Zheng, Y., Wang, C., Zhang, Y., and Lin, Y. (2020). Microarray Analysis Reveals that lncRNA PWRN1 ‐209 Promotes Human Bone Marrow Mesenchymal Stem Cell Osteogenic Differentiation on Microtopography Titanium Surface In Vitro. J. Biomed. Mater Res. 108 (7), 2889–2902. doi:10.1002/jbm.b.34620
Wang, N., Zhou, Z., Wu, T., Liu, W., Yin, P., Pan, C., et al. (2016). TNF-α-induced NF-Κb Activation Upregulates microRNA-150-3p and Inhibits Osteogenesis of Mesenchymal Stem Cells by Targeting β-catenin. Open Biol. 6 (3), 150258. doi:10.1098/rsob.150258
Wang, Q., Wang, C. H., and Meng, Y. (2019). microRNA-1297 Promotes the Progression of Osteoporosis through Regulation of Osteogenesis of Bone Marrow Mesenchymal Stem Cells by Targeting WNT5A. Eur. Rev. Med. Pharmacol. Sci. 23 (11), 4541–4550. doi:10.26355/eurrev_201906_18029
Wang, Q., Cai, J., Cai, X.-h., and Chen, L. (2013). miR-346 Regulates Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Targeting the Wnt/β-Catenin Pathway. PLoS One 8 (9), e72266. doi:10.1371/journal.pone.0072266
Wang, Q., Li, Y., Zhang, Y., Ma, L., Lin, L., Meng, J., et al. (2017). LncRNA MEG3 Inhibited Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells from Postmenopausal Osteoporosis by Targeting miR-133a-3p. Biomed. Pharmacother. 89, 1178–1186. doi:10.1016/j.biopha.2017.02.090
Wang, X. B., Li, P. B., Guo, S. F., Yang, Q. S., Chen, Z. X., Wang, D., et al. (2019). circRNA_0006393 Promotes Osteogenesis In glucocorticoid-induced O-steoporosis by S-ponging miR-145-5p and U-pregulating FOXO1. Mol. Med. Rep. 20 (3), 2851–2858. doi:10.3892/mmr.2019.10497
Wang, X., Chen, T., Deng, Z., Gao, W., Liang, T., Qiu, X., et al. (2021). Melatonin Promotes Bone Marrow Mesenchymal Stem Cell Osteogenic Differentiation and Prevents Osteoporosis Development through Modulating Circ_0003865 that Sponges miR-3653-3p. Stem Cell. Res. Ther. 12 (1), 150. doi:10.1186/s13287-021-02224-w
Wei, B., Wei, W., Zhao, B., Guo, X., and Liu, S. (2017). Long Non-coding RNA HOTAIR Inhibits miR-17-5p to Regulate Osteogenic Differentiation and Proliferation in Non-traumatic Osteonecrosis of Femoral Head. PLoS One 12 (2), e0169097. doi:10.1371/journal.pone.0169097
Wen, J., Guan, Z., Yu, B., Guo, J., Shi, Y., and Hu, L. (2020). Circular RNA Hsa_circ_0076906 Competes with OGN for miR-1305 Biding Site to Alleviate the Progression of Osteoporosis. Int. J. Biochem. Cell. Biol. 122, 105719. doi:10.1016/j.biocel.2020.105719
Weng, W., Di, S., Xing, S., Sun, Z., Shen, Z., Dou, X., et al. (2021). Long Non-coding RNA DANCR Modulates Osteogenic Differentiation by Regulating the miR-1301-3p/PROX1 axis. Mol. Cell. Biochem. 476 (6), 2503–2512. doi:10.1007/s11010-021-04074-9
Wong, S. K., Chin, K.-Y., and Ima-Nirwana, S. (2020). Quercetin as an Agent for Protecting the Bone: A Review of the Current Evidence. Ijms 21 (17), 6448. doi:10.3390/ijms21176448
Wu, D., Chang, X., Tian, J., Kang, L., Wu, Y., Liu, J., et al. (2021). Bone Mesenchymal Stem Cells Stimulation by Magnetic Nanoparticles and a Static Magnetic Field: Release of Exosomal miR-1260a Improves Osteogenesis and Angiogenesis. J. Nanobiotechnol 19 (1), 209. doi:10.1186/s12951-021-00958-6
Wu, F., Huang, W., Yang, Y., Liu, F., Chen, J., Wang, G., et al. (2021). miR‐155‐5p Regulates Mesenchymal Stem Cell Osteogenesis and Proliferation by Targeting GSK3B in Steroid‐associated Osteonecrosis. Cell. Biol. Int. 45 (1), 83–91. doi:10.1002/cbin.11470
Wu, H., Cao, F., Zhou, W., Wang, G., Liu, G., Xia, T., et al. (2020). Long Noncoding RNA FAM83H-AS1 Modulates SpA-Inhibited Osteogenic Differentiation in Human Bone Mesenchymal Stem Cells. Mol. Cell. Biol. 40 (5), e00362–19. doi:10.1128/mcb.00362-19
Wu, J., Zhao, J., Sun, L., Pan, Y., Wang, H., and Zhang, W.-B. (2018). Long Non-coding RNA H19 Mediates Mechanical Tension-Induced Osteogenesis of Bone Marrow Mesenchymal Stem Cells via FAK by Sponging miR-138. Bone 108, 62–70. doi:10.1016/j.bone.2017.12.013
Wu, L., Song, J., Xue, J., Xiao, T., Wei, Q., Zhang, Z., et al. (2020). MircoRNA-143-3p Regulating ARL6 Is Involved in the Cadmium-Induced Inhibition of Osteogenic Differentiation in Human Bone Marrow Mesenchymal Stem Cells. Toxicol. Lett. 331, 159–166. doi:10.1016/j.toxlet.2020.06.001
Wu, P., Mo, Y., Peng, M., Tang, T., Zhong, Y., Deng, X., et al. (2020). Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and circRNA. Mol. Cancer 19 (1), 22. doi:10.1186/s12943-020-1147-3
Wu, Z.-h., Huang, K.-h., Liu, K., Wang, G.-t., and Sun, Q. (2018). DGCR5 Induces Osteogenic Differentiation by Up-Regulating Runx2 through miR-30d-5p. Biochem. Biophysical Res. Commun. 505 (2), 426–431. doi:10.1016/j.bbrc.2018.09.033
Gao, X., Xia, X., Li, F., Zhang, M., Zhou, H., Wu, X., et al. Circular RNA-Encoded Oncogenic E-Cadherin Variant Promotes Glioblastoma Tumorigenicity through Activation of EGFR-STAT3 Signalling. Nat. Cell. Biol., 23(3), 278, 291.+ (2021) doi:10.1038/s41556-021-00639-4
Xia, P., Gu, R., Zhang, W., Shao, L., Li, F., Wu, C., et al. (2019). MicroRNA‐200c Promotes Osteogenic Differentiation of Human Bone Mesenchymal Stem Cells through Activating the AKT/β‐Catenin Signaling Pathway via Downregulating Myd88. J. Cell. Physiology 234 (12), 22675–22686. doi:10.1002/jcp.28834
Xiang, J., Fu, H. Q., Xu, Z., Fan, W. J., Liu, F., and Chen, B. (2020). lncRNA SNHG1 Attenuates Osteogenic Differentiation via the miR-101/DKK1 axis in Bone Marrow Mesenchymal Stem Cells. Mol. Med. Rep. 22 (5), 3715–3722. doi:10.3892/mmr.2020.11489
Xiang, S., Li, Z., and Weng, X. (2020). Changed Cellular Functions and Aberrantly Expressed miRNAs and circRNAs in Bone Marrow Stem cells in Osteonecrosis of the Femoral Head. Int. J. Mol. Med. 45 (3), 805–815. doi:10.3892/ijmm.2020.4455
Xie, Y., Hu, J. Z., and Shi, Z. Y. (2018). MiR-181d Promotes Steroid-Induced Osteonecrosis of the Femoral Head by Targeting SMAD3 to Inhibit Osteogenic Differentiation of hBMSCs. Eur. Rev. Med. Pharmacol. Sci. 22 (13), 4053–4062. doi:10.26355/eurrev_201807_15393
Xin, C., and Liu, J. (2021). Long Non-coding RNAs in Parkinson's Disease. Neurochem. Res. 46 (5), 1031–1042. doi:10.1007/s11064-021-03230-3
Xin, W., Yuan, S., Wang, B., Qian, Q., and Chen, Y. (2021). Hsa_circ_0066523 Promotes the Proliferation and Osteogenic Differentiation of Bone Mesenchymal Stem Cells by Repressing PTEN. Bone & Jt. Res. 10 (8), 526–535. doi:10.1302/2046-3758.108.Bjr-2020-0127.R2
Xing, Y.-H., and Chen, L.-L. (2018). Processing and Roles of snoRNA-Ended Long Noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 53 (6), 596–606. doi:10.1080/10409238.2018.1508411
Xu, G., Ding, Z., and Shi, H.-f. (2019). The Mechanism of miR-889 Regulates Osteogenesis in Human Bone Marrow Mesenchymal Stem Cells. J. Orthop. Surg. Res. 14 (1), 366. doi:10.1186/s13018-019-1399-z
Xu, H., Wang, J., Ye, X., Ming, J., Li, S. J., Wang, X., et al. (2018). Effect of MiR-99a-5p on the Differential Ability of Human Bone Marrow Mesenchymal Stem Cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi 26 (2), 563–568. doi:10.7534/j.issn.1009-2137.2018.02.043
Xu, J.-F., Yang, G.-h., Pan, X.-H., Zhang, S.-J., Zhao, C., Qiu, B.-S., et al. (2014). Altered microRNA Expression Profile in Exosomes during Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. PLoS One 9 (12), e114627. doi:10.1371/journal.pone.0114627
Xu, J., Yang, X., Zhou, Q., Zhuang, J., and Han, S. (2020). Biological Significance of piRNA in Liver Cancer: a Review. Biomarkers 25 (6), 436–440. doi:10.1080/1354750x.2020.1794041
Xu, S., Cecilia Santini, G., De Veirman, K., Vande Broek, I., Leleu, X., De Becker, A., et al. (2013). Upregulation of miR-135b Is Involved in the Impaired Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Multiple Myeloma Patients. PLoS One 8 (11), e79752. doi:10.1371/journal.pone.0079752
Yang, C., Liu, X., Zhao, K., Zhu, Y., Hu, B., Zhou, Y., et al. (2019). miRNA-21 Promotes Osteogenesis via the PTEN/PI3K/Akt/HIF-1α Pathway and Enhances Bone Regeneration in Critical Size Defects. Stem Cell. Res. Ther. 10 (1), 65. doi:10.1186/s13287-019-1168-2
Yang, N., Wang, G., Hu, C., Shi, Y., Liao, L., Shi, S., et al. (2013). Tumor Necrosis Factor α Suppresses the Mesenchymal Stem Cell Osteogenesis Promoter miR-21 in Estrogen Deficiency-Induced Osteoporosis. J. Bone Min. Res. 28 (3), 559–573. doi:10.1002/jbmr.1798
Yang, Q., Jia, L., Li, X., Guo, R., Huang, Y., Zheng, Y., et al. (2018). Long Noncoding RNAs: New Players in the Osteogenic Differentiation of Bone Marrow- and Adipose-Derived Mesenchymal Stem Cells. Stem Cell. Rev Rep 14 (3), 297–308. doi:10.1007/s12015-018-9801-5
Yang, Q., Zhou, Y., Wang, T., Cai, P., Fu, W., Wang, J., et al. (2021). MiRNA‐1271‐5p Regulates Osteogenic Differentiation of Human Bone Marrow‐derived Mesenchymal Stem Cells by Targeting Forkhead Box O1 (FOXO1). Cell. Biol. Int. 45 (7), 1468–1476. doi:10.1002/cbin.11585
Yang, W., Zhu, W., Yang, Y., Guo, M., Qian, H., Jiang, W., et al. (2021). Exosomal miR-100-5p Inhibits Osteogenesis of hBMSCs and Angiogenesis of HUVECs by Suppressing the BMPR2/Smad1/5/9 Signalling Pathway. Stem Cell. Res. Ther. 12 (1), 390. doi:10.1186/s13287-021-02438-y
Yang, X., Yang, J., Lei, P., and Wen, T. (2019). LncRNA MALAT1 Shuttled by Bone Marrow-Derived Mesenchymal Stem Cells-Secreted Exosomes Alleviates Osteoporosis through Mediating microRNA-34c/SATB2 axis. Aging 11 (20), 8777–8791. doi:10.18632/aging.102264
You, L., Pan, L., Chen, L., Gu, W., and Chen, J. (2016). MiR-27a Is Essential for the Shift from Osteogenic Differentiation to Adipogenic Differentiation of Mesenchymal Stem Cells in Postmenopausal Osteoporosis. Cell. Physiol. Biochem. 39 (1), 253–265. doi:10.1159/000445621
Younger, E. M., and Chapman, M. W. (1989). Morbidity at Bone Graft Donor Sites. J. Orthop. Trauma 3 (3), 192–195. doi:10.1097/00005131-198909000-00002
Yu, L., Xu, Y., Qu, H., Yu, Y., Li, W., Zhao, Y., et al. (2019). Decrease of MiR‐31 Induced by TNF‐α Inhibitor Activates SATB2/RUNX2 Pathway and Promotes Osteogenic Differentiation in Ethanol‐induced Osteonecrosis. J. Cell. Physiology 234 (4), 4314–4326. doi:10.1002/jcp.27210
Zang, J., Lu, D., and Xu, A. (2020). The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J. Neurosci. Res. 98 (1), 87–97. doi:10.1002/jnr.24356
Zealy, R. W., Wrenn, S. P., Davila, S., Min, K.-W., and Yoon, J.-H. (2017). microRNA-Binding Proteins: Specificity and Function. WIREs RNA 8 (5), e1414. doi:10.1002/wrna.1414
Zeng, H.-B., Dong, L.-Q., Huang, Y.-L., Xu, C., Zhao, X.-H., and Wu, L.-G. (2021). USF2 Reduces BMP3 Expression via Transcriptional Activation of miR-34a, Thus Promoting Osteogenic Differentiation of BMSCs. J. Bone Min. Metab. 39 (6), 997–1008. doi:10.1007/s00774-021-01254-x
Zha, J.-p., Wang, X.-q., and Di, J. (2020). MiR-920 Promotes Osteogenic Differentiation of Human Bone Mesenchymal Stem Cells by Targeting HOXA7. J. Orthop. Surg. Res. 15 (1), 254. doi:10.1186/s13018-020-01775-7
Zhang, G. P., Zhang, J., Zhu, C. H., Lin, L., Wang, J., Zhang, H. J., et al. (2017). Micro RNA ‐98 Regulates Osteogenic Differentiation of Human Bone Mesenchymal Stromal Cells by Targeting BMP 2. J. Cell. Mol. Med. 21 (2), 254–264. doi:10.1111/jcmm.12961
Zhang, H. L., Du, X. Y., and Dong, Q. R. (2019). LncRNA XIXT Promotes Osteogenic Differentiation of Bone Mesenchymal Stem Cells and Alleviates Osteoporosis Progression by Targeting miRNA-30a-5p. Eur. Rev. Med. Pharmacol. Sci. 23 (20), 8721–8729. doi:10.26355/eurrev_201910_19266
Zhang, J., Chen, S., and Liu, K. (2022). Structural Insights into piRNA Biogenesis. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1865 (2), 194799. doi:10.1016/j.bbagrm.2022.194799
Zhang, J., Tao, Z., and Wang, Y. (2018). Long Non-coding RNA DANCR R-egulates the P-roliferation and O-steogenic D-ifferentiation of H-uman B-one-D-erived M-arrow M-esenchymal S-tem C-ells via the p38�MAPK P-athway. Int. J. Mol. Med. 41 (1), 213–219. doi:10.3892/ijmm.2017.3215
Zhang, L., Liu, Y., Feng, B., Liu, L.-G., Zhou, Y.-C., and Tang, H. (2021). MiR-138-5p Knockdown Promotes Osteogenic Differentiation through FOXC1 Up-Regulation in Human Bone Mesenchymal Stem Cells. Biochem. Cell. Biol. 99 (3), 296–303. doi:10.1139/bcb-2020-0163
Zhang, S., Liu, Y., Zheng, Z., Zeng, X., Liu, D., Wang, C., et al. (2018). MicroRNA-223 Suppresses Osteoblast Differentiation by Inhibiting DHRS3. Cell. Physiol. Biochem. 47 (2), 667–679. doi:10.1159/000490021
Zhang, W., Chen, L., Wu, J., Li, J., Zhang, X., Xiang, Y., et al. (2019). Long Noncoding RNA TUG1 Inhibits Osteogenesis of Bone Marrow Mesenchymal Stem Cells via Smad5 after Irradiation. Theranostics 9 (8), 2198–2208. doi:10.7150/thno.30798
Zhang, Y., Chen, B., Li, D., Zhou, X., and Chen, Z. (2019). LncRNA NEAT1/miR-29b-3p/BMP1 axis Promotes Osteogenic Differentiation in Human Bone Marrow-Derived Mesenchymal Stem Cells. Pathology - Res. Pract. 215 (3), 525–531. doi:10.1016/j.prp.2018.12.034
Zhang, Y., Jiang, Y., Luo, Y., and Zeng, Y. (2020). Interference of miR-212 and miR-384 Promotes Osteogenic Differentiation via Targeting RUNX2 in Osteoporosis. Exp. Mol. Pathology 113, 104366. doi:10.1016/j.yexmp.2019.104366
Zhang, L., Xie, H., and Li, S. (2020). LncRNA LOXL1-AS1 Controls Osteogenic and Adipocytic Differentiation of Bone Marrow Mesenchymal Stem Cells in Postmenopausal Osteoporosis through Regulating the miR-196a-5p/Hmga2 axis. J. Bone Min. Metab. 38 (6), 794–805. doi:10.1007/s00774-020-01123-z
Zhang, Y., Liu, Y., Wu, M., Wang, H., Wu, L., Xu, B., et al. (2020a). MicroRNA-664a-5p Promotes Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells by Directly Downregulating HMGA2. Biochem. Biophysical Res. Commun. 521 (1), 9–14. doi:10.1016/j.bbrc.2019.09.122
Zhang, Y., Zhou, L., Zhang, Z., Ren, F., Chen, L., and Lan, Z. (2020b). miR-10a-5p I-nhibits O-steogenic D-ifferentiation of B-one M-arrow-derived M-esenchymal S-tem C-ells. Mol. Med. Rep. 22 (1), 135–144. doi:10.3892/mmr.2020.11110
Zhang, Y., Sun, Y., Liu, J., Han, Y., and Yan, J. (2020). MicroRNA-346-5p Regulates Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells by Inhibiting Transmembrane Protein 9. BioMed Res. Int. 2020, 1–7. doi:10.1155/2020/8822232
Zhang, Y., Sun, Y., and Zhang, Y. (2015). MiR-133 Is Involved in Estrogen Deficiency-Induced Osteoporosis through Modulating Osteogenic Differentiation of Mesenchymal Stem Cells. Med. Sci. Monit. 21, 1527–1534. doi:10.12659/msm.894323
Zhang, Y., and Wang, X. (2020). Targeting the Wnt/β-Catenin Signaling Pathway in Cancer. J. Hematol. Oncol. 13 (1), 165. doi:10.1186/s13045-020-00990-3
Zhang, Y., Wei, Q.-S., Ding, W.-B., Zhang, L.-L., Wang, H.-C., Zhu, Y.-J., et al. (2017). Increased microRNA-93-5p Inhibits Osteogenic Differentiation by Targeting Bone Morphogenetic Protein-2. PLoS One 12 (8), e0182678. doi:10.1371/journal.pone.0182678
Zhang, Z., Jiang, W., Hu, M., Gao, R., and Zhou, X. (2021). MiR-486-3p Promotes Osteogenic Differentiation of BMSC by Targeting CTNNBIP1 and Activating the Wnt/β-Catenin Pathway. Biochem. Biophysical Res. Commun. 566, 59–66. doi:10.1016/j.bbrc.2021.05.098
Zhao, J., Wang, C., Song, Y., and Fang, B. (2014). Arsenic Trioxide and microRNA-204 Display Contrary Effects on Regulating Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells in Aplastic Anemia. Acta Biochim. Biophys. Sin. (Shanghai) 46 (10), 885–893. doi:10.1093/abbs/gmu082
Zhao, L., Zhao, J., Yu, J.-J., and Zhang, C. (2020). Irregular Bone Defect Repair Using Tissue-Engineered Periosteum in a Rabbit Model. Tissue Eng. Regen. Med. 17 (5), 717–727. doi:10.1007/s13770-020-00282-4
Zhao, R., Li, Y., Lin, Z., Wan, J., Xu, C., Zeng, Y., et al. (2016). miR-199b-5p Modulates BMSC Osteogenesis via Suppressing GSK-3β/β-Catenin Signaling Pathway. Biochem. Biophysical Res. Commun. 477 (4), 749–754. doi:10.1016/j.bbrc.2016.06.130
Zhao, Y., Chen, Y., Hu, X., Zhang, N., and Wang, F. (2020). lncRNA LINC01535 Upregulates BMP2 Expression Levels to Promote Osteogenic Differentiation via Sponging miR-3619-5p. Mol. Med. Rep. 22 (6), 5428–5435. doi:10.3892/mmr.2020.11635
Zhao, Y. S., Lin, P., Tu, Y. C., An, T., Wu, Y. P., and Li, X. F. (2021). Lentivirus Mediated siRNA Hsa-Circ-0000885 Transfection of BMSCs and Osteoclast Co-culture System on Cell Differentiation, Proliferation and Apoptosis. Zhongguo Gu Shang 34 (10), 978–984. doi:10.12200/j.issn.1003-0034.2021.10.017
Zhao, Y., Wang, Z., Wu, G., Wei, M., Liu, Q., Zhou, J., et al. (2016). Improving the Osteogenesis of Human Bone Marrow Mesenchymal Stem Cell Sheets by microRNA-21-Loaded Chitosan/hyaluronic Acid Nanoparticles via Reverse Transfection. Ijn 11, 2091–2105. doi:10.2147/ijn.S104851
Zheng, C., Bai, C., Sun, Q., Zhang, F., Yu, Q., Zhao, X., et al. (2020). Long Noncoding RNA XIST Regulates Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells by Targeting miR-9-5p. Mech. Dev. 162, 103612. doi:10.1016/j.mod.2020.103612
Zhi, F., Ding, Y., Wang, R., Yang, Y., Luo, K., and Hua, F. (2021). Exosomal Hsa_circ_0006859 Is a Potential Biomarker for Postmenopausal Osteoporosis and Enhances Adipogenic versus Osteogenic Differentiation in Human Bone Marrow Mesenchymal Stem Cells by Sponging miR-431-5p. Stem Cell. Res. Ther. 12 (1), 157. doi:10.1186/s13287-021-02214-y
Zhou, P., Wu, G., Zhang, P., Xu, R., Ge, J., Fu, Y., et al. (2016). SATB2-Nanog axis Links Age-Related Intrinsic Changes of Mesenchymal Stem Cells from Craniofacial Bone. Aging 8 (9), 2006–2022. doi:10.18632/aging.101041
Zhou, X., Moussa, F. M., Mankoci, S., Ustriyana, P., Zhang, N., Abdelmagid, S., et al. (2016). Orthosilicic Acid, Si(OH)4, Stimulates Osteoblast Differentiation In Vitro by Upregulating miR-146a to Antagonize NF-Κb Activation. Acta Biomater. 39, 192–202. doi:10.1016/j.actbio.2016.05.007
Zhu, J., Fu, H., Wu, Y., and Zheng, X. (2013). Function of lncRNAs and Approaches to lncRNA-Protein Interactions. Sci. China Life Sci. 56 (10), 876–885. doi:10.1007/s11427-013-4553-6
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., et al. (2019). Exosomal tRNA-Derived Small RNA as a Promising Biomarker for Cancer Diagnosis. Mol. Cancer 18 (1), 74. ARTN 74. doi:10.1186/s12943-019-1000-8
Zhuang, Q., Ye, B., Hui, S., Du, Y., Zhao, R. C., Li, J., et al. (2019). Long Noncoding RNA lncAIS Downregulation in Mesenchymal Stem Cells Is Implicated in the Pathogenesis of Adolescent Idiopathic Scoliosis. Cell. Death Differ. 26 (9), 1700–1715. doi:10.1038/s41418-018-0240-2
Zhuang, X.-m., and Zhou, B. (2020). Exosome Secreted by Human Gingival Fibroblasts in Radiation Therapy Inhibits Osteogenic Differentiation of Bone Mesenchymal Stem Cells by Transferring miR-23a. Biomed. Pharmacother. 131, 110672. doi:10.1016/j.biopha.2020.110672
Zhuang, X.-M., Zhou, B., and Yuan, K.-F. (2019). Role of P53 Mediated miR-23a/CXCL12 Pathway in Osteogenic Differentiation of Bone Mesenchymal Stem Cells on Nanostructured Titanium Surfaces. Biomed. Pharmacother. 112, 108649. doi:10.1016/j.biopha.2019.108649
Zong, T., Yang, Y., Zhao, H., Li, L., Liu, M., Fu, X., et al. (2021). tsRNAs: Novel Small Molecules from Cell Function and Regulatory Mechanism to Therapeutic Targets. Cell. Prolif. 54 (3), e12977. doi:10.1111/cpr.12977
Glossary
AA aplastic anemia
AIS adolescent idiopathic scoliosis
AKT protein kinase B
ALP alkaline phosphatase
AMSC adipose mesenchymal stem cells
ARL6 adenosine diphosphate-ribosylation factor-like protein 6
ATF activating transcription factor
BCL2 B-cell lymphoma 2
BMP bone morphogenetic protein
BMPR BMP receptor
BMSC bone marrow derived mesenchymal stem cell
CDH11 cadherin-11
ceRNA competitive endogenous lncRNAs
circRNA circular RNA
CNNB1 Targets catenin beta 1
COL1 collagen type I
COL1A1 collagen type I α1
CTNNBIP1 catenin beta interacting protein 1
CXCR4 C-X-C chemokine receptor type 4
DANCR differentiation antagonizing non-protein coding RNA
DDX17 dead-box helicase 17
DEPTOR DEP domain-containing mTOR interacting protein
DHRS3 retinoic acid-inducible dehydrogenase reductase 3
DKK1 dickkopf 1
DLX distal-less
EFNA3 ephrin-A3
ERK extracellular signal-regulated kinase
EZH2 enhancer of zeste homolog 2
FAK focal adhesion kinase
FBXW7 F-box and WD repeat domain containing 7
FGF fibroblast growth factor
FNDC5 fibronectin type III domain-containing protein 5
FOX forkhead box
FZD frizzled
GAS1 growth arrest-specific gene 1
GLS glutaminase
GSK-3β glycogen synthetase kinase 3 beta
H3K27 trimethylation of lysine 27 on histone H3
HA/TCP hydroxyapatite/tricalcium phosphate
HIF1α hypoxia-inducible factor-1α
HMGA2 high-mobility group A2
hnRNPK heterogeneous nuclear ribonucleoprotein K
HOTAIR HOX transcript antisense RNA
HOTTIP HOX transcript at the distal tip
HOX homeobox
IGF1R insulin growth factor 1 receptor
IGFBP3 insulin-like growth factor binding protein 3
IL-1β interleukin-1β
KLF kruppel like factor
K-RAS kirsten rat sarcoma viral oncogene
lncRNA long non-coding RNA
lncRNA-OG osteogenesis-associated lncRNA
LOXL1-AS1 LOXL1 antisense RNA 1
LRP5 low-density lipoprotein receptor-related protein 5
MAPK mitogen-activated protein kinase
MEF2C myocyte enhancer factor 2 C
MEG3 maternally expressed 3 (nonprotein coding)
miRNA microRNA
MRC2 type 2 mannose receptor C
MSC mesenchymal stem cell
mTOR mammalian target of rapamycin
MYD88 myeloid differentiation factor 88
ncRNA non-coding RNA
NF-κB nuclear factor kappa B
NOTCH1 notch recepetor 1
OCN osteocalcin
OGN osteoglycin
ONFH osteonecrosis of the femoral head
OP osteoporosis
OPN osteopontin
OSX osterix
PI3K phosphoinositide 3-kinase
piRNA PIWI-interacting RNA
PIWIL PIWI protein
PMOP postmenopausal osteoporosis
PPARγ peroxisome proliferator-activated receptor γ
pre-miRNA precursor miRNA
pri-miRNA primary miRNA
PROX1 prospero homeobox 1
PTEN phosphatase and tensin homolog deleted on chromosome ten
PWRN1-209 Prader-willi region ncRNAs 1–-209
RANKL receptor or activator of NF-κB ligand
RAP1A ras-related protein 1 AA
RBP RNA-binding protein
ROCK1 Rho associated coiled-coil containing protein kinase 1
rRNA ribosomal RNA
RUNX2 runt-related transcription factor 2
SATB2 special AT-rich binding protein homeobox 2
SDF-1 stromal cell-derived factor 1
SEMA3A semaphorin 3 AA
SIRT1 sirtuin1
SLC39A1 solute carrier family 39 member 1
SMURF2 Smad ubiquitin regulatory factor 2
SNHG16 small nucleolar RNA host gene 16
snoRNA small nucleolar RNA
snRNA small nuclear RNA
SONFH steroid-associated osteonecrosis of the femoral head
SOX2 sex determining region Y-box 2
SPRY1 Sprouty 1
STAT1 signal transducer and activator of transcription 1
TCF-1 T-cell factor 1
TET2 ten-eleven translocation 2
TGF-β transforming growth factor-β
THA total hip arthroplasty
TNF-α tumor necrosis factor-α
tRNA transfert RNA
tsRNA tRNA-derived small RNA
TUG1 taurine upregulated gene 1
VEGF vascular endothelial growth factor
WDR5 WD Repeat-Containing Protein 5
WISP2 WNT1 inducible signaling pathway protein 2
WNT3A Wnt family member 3 AA
WWP1 WW domain-containing E3 ubiquitin protein ligase 1
XIST X inactivate-specific transcript
Keywords: BMSCs, ncRNAs, osteogenic differentiation, bone regeneration, bone tissue engineering
Citation: Chen X, Xie W, Zhang M, Shi Y, Xu S, Cheng H, Wu L, Pathak JL and Zheng Z (2022) The Emerging Role of Non-Coding RNAs in Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Front. Cell Dev. Biol. 10:903278. doi: 10.3389/fcell.2022.903278
Received: 24 March 2022; Accepted: 25 April 2022;
Published: 16 May 2022.
Edited by:
Zhong Wang, University of Michigan, United StatesReviewed by:
Giuseppina Divisato, University of Naples Federico II, ItalySean Shishu Huang Sichuan University, China
Copyright © 2022 Chen, Xie, Zhang, Shi, Xu, Cheng, Wu, Pathak and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhichao Zheng, emhpY2hhb3poZW5nQGFsaXl1bi5jb20=; Janak L. Pathak, ai5wYXRoYWtAZ3pobXUuZWR1LmNu; Lihong Wu, d2NhbmhvbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xiaoying Chen
Xiaoying Chen Wei Xie
Wei Xie Ming Zhang
Ming Zhang Yuhan Shi
Yuhan Shi Shaofen Xu
Shaofen Xu Haoyu Cheng
Haoyu Cheng Lihong Wu
Lihong Wu Janak L. Pathak
Janak L. Pathak Zhichao Zheng
Zhichao Zheng