Corrigendum: Long non-coding RNAs as molecular biomarkers in cholangiocarcinoma
- 1Department of Gastroenterology, The Fourth School of Clinical Medicine, Zhejiang Chinese Medical University, Hangzhou, China
- 2Department of Gastroenterology, International Education College of Zhejiang Chinese Medical University, Hangzhou, China
- 3Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 4Key Laboratory of Integrated Traditional Chinese and Western Medicine for Biliary and Pancreatic Diseases of Zhejiang Province, Hangzhou, China
Cholangiocarcinoma (CCA) is a biliary system cancer that has the characteristics of strong invasiveness, poor prognosis, and few therapy choices. Furthermore, the absence of precise biomarkers for early identification and prognosis makes it hard to intervene in the early phase of initial diagnosis or recurring cholangiocarcinoma following surgery. Encouragingly, previous studies found that long non-coding RNA (lncRNA), a subgroup of RNA that is more than 200 nucleotides long, can affect cell proliferation, migration, apoptosis, and even drug resistance by altering numerous signaling pathways, thus reaching pro-cancer or anti-cancer outcomes. This review will take a retrospective view of the recent investigations on the work of lncRNAs in cholangiocarcinoma progression and the potential of lncRNAs serving as promising clinical biomarkers and therapeutic targets for CCA.
Introduction
Cholangiocarcinoma (CCA) is an extremely aggressive malignancy of the bile duct epithelium. Depending on the anatomical site of origin, CCA can be stratified into three categories: intrahepatic cholangiocarcinoma (iCCA), perihilar cholangiocarcinoma (pCCA), as well as distal cholangiocarcinoma (dCCA) (Rizvi and Gores, 2013). Cholangiocarcinoma is the 2nd most prevalent primary hepatobiliary cancer after HCC (hepatocellular carcinoma). It accounts for about 3% of all gastrointestinal malignant tumors (Banales et al., 2016). Cholangiocarcinoma, particularly intrahepatic cholangiocarcinoma, has become more common in the majority of countries around the world in recent decades (Florio et al., 2020; Van Dyke et al., 1999-2013; Saha et al., 2016). Even though extensive surgery is by far the most effective therapy, the 5-year rate of survival with CCA ranged between 20% and 40% following resection (van der Gaag et al., 2012; Nagino et al., 2013; Mazzaferro et al., 2020). Due to the lack of clinical symptoms, about 70% of patients have missed out on surgical resection at diagnosis time (Banales et al., 2020). Thus, it is necessary to find sensitive and specific biomarkers for CCA.
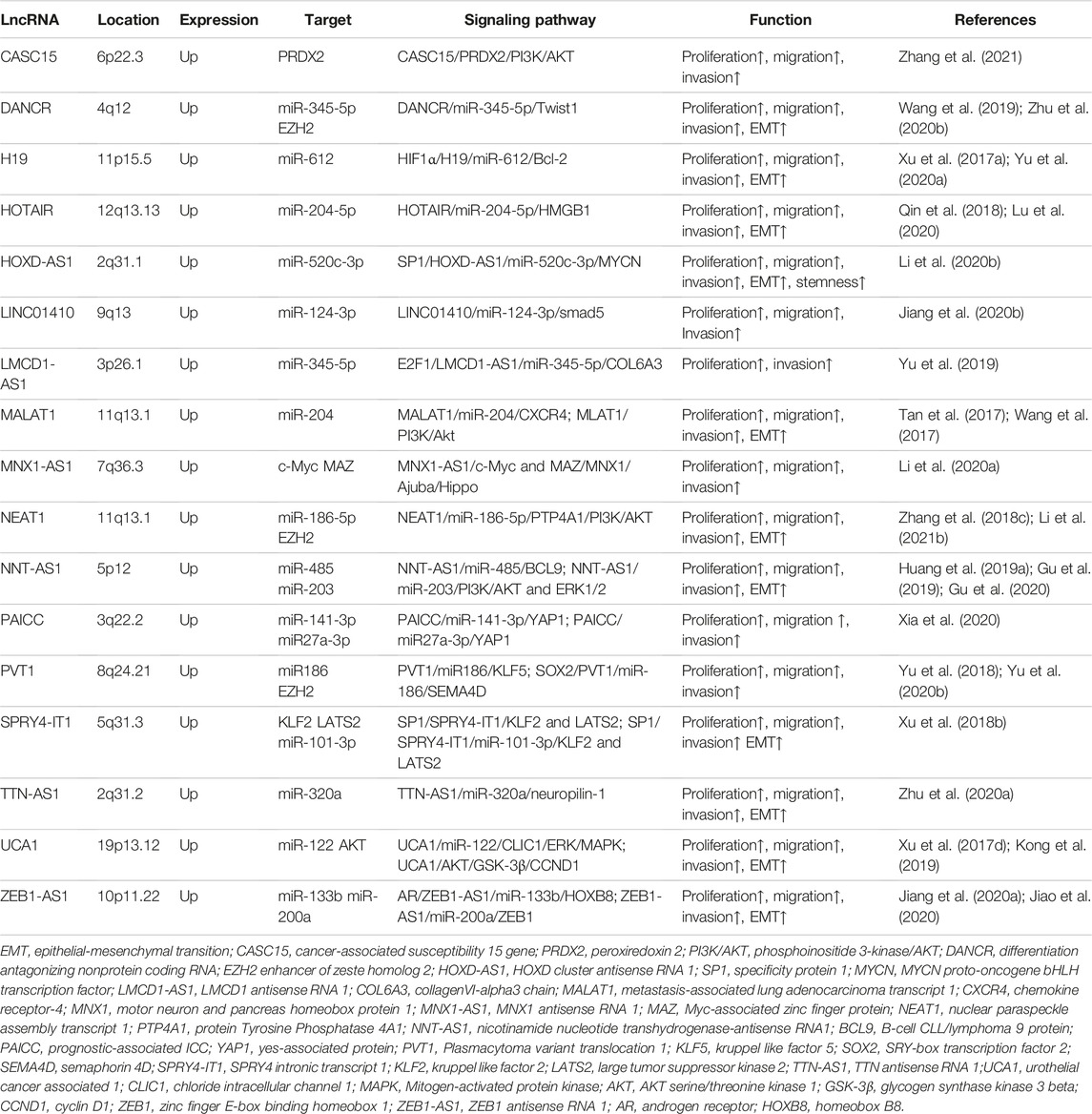
Long non-coding RNA (lncRNA) constitutes a non-coding RNA longer than 200 nucleotides and was previously thought to be translational noise since it lacks the ability to code for proteins (Kopp and Mendell, 2018). Nevertheless, mounting evidence indicates that lncRNAs are involved in carcinogenesis or tumor suppression (Rinn and Chang, 2020). LncRNAs are obtained by assembling factors and Pol II at promoters, followed by intron elongation and splicing, and finally by adding a methylated guanosine cap and polyadenylated tail (Guo et al., 2016). LncRNAs have extensive functions in different biological processes, involving chromatin modification, gene transcription, and translation (Quinn and Chang, 2016). LncRNAs serve extensive functions according to their cellular location. In the nucleus, the functions of lncRNAs mainly include chromatin interaction, RNA processing, and transcriptional regulation, whereas lncRNAs in the cytoplasm can interact with miRNAs, functioning as competitive endogenous RNAs (ceRNAs) to modulate mRNA and cellular signaling pathways (Schmitt and Chang, 2016). The dysregulation of lncRNAs has been widely observed to be remarkably linked to the development of cancers, consisting of liver cancer, gastrointestinal cancer, lung cancer, breast cancer, and hematological malignancies (Lv et al., 2017; Kim et al., 2018; Wu et al., 2019; Zhuo et al., 2019; Wang et al., 2020a; Pan et al., 2020). A lot of abnormally expressed lncRNAs have been discovered between CCA and adjacent normal tissues (Long et al., 2019). The purpose of this review is to summarize the role and underlying mechanism of lncRNAs in CCA progression (Figure 1; Table 1) and explore their potential as biomarkers and therapeutic targets.
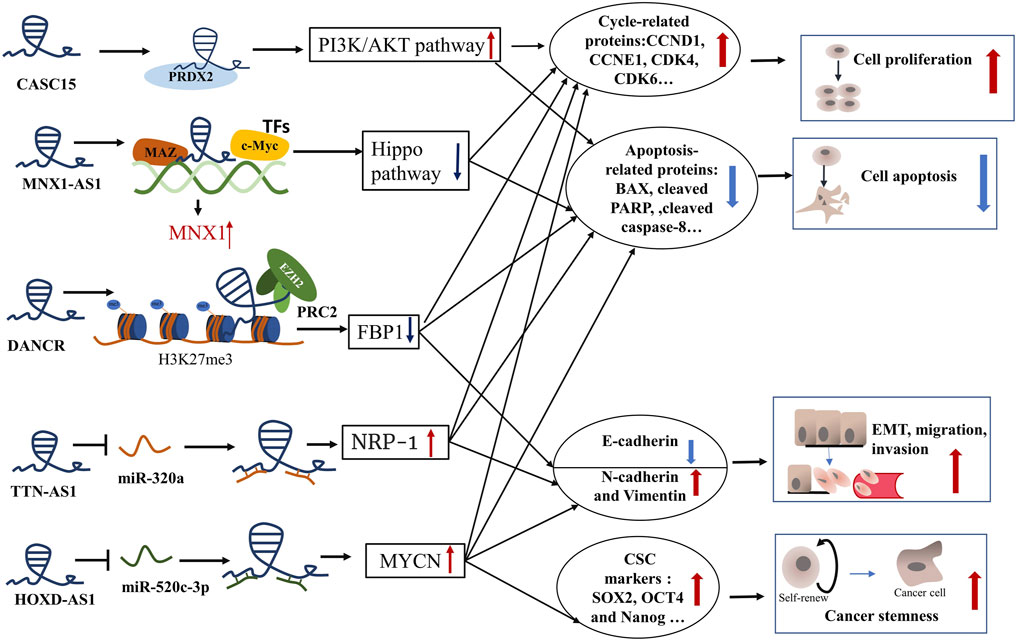
FIGURE 1. The role and mechanism of lncRNAs in CCA progression. CASC15, cancer-associated susceptibility 15 gene; PRDX2, peroxiredoxin 2; CCND1, cyclin D1; CCNE1, cyclin E1; CDK4, cyclin dependent kinase 4; CDK6, cyclin dependent kinase 6; MNX, motor neuron and pancreas homeobox protein 1; MNX1-AS1, MNX1 antisense RNA 1; MAZ, Myc-associated zinc finger protein; TFs, transcription factors; BAX, BCL2 associated X;PARP, poly ADP-ribose polymerase; DANCR, differentiation antagonizing nonprotein coding RNA; EZH2, enhancer of zeste homolog 2; H3K27me3, methylation of lysine 27 on histone 3; PRC2, polycomb repressive complex 2; FBP1, Fructose-1, 6-biphosphatase; TTN-AS1, titin-antisense RNA1; NRP-1, neuropilin-1; HOXD-AS1, HOXD cluster antisense RNA 1; MYCN, MYCN proto-oncogene, bHLH transcription factor; SOX2, SRY-box transcription factor 2; OCT4, organic cation/carnitine transporter4.
The Role of lncRNAs in Cholangiocarcinoma Progression
Proliferation and Anti-Apoptosis
The infinite proliferation of tumor cells is due to abnormal modulation of the cell cycle and anti-apoptosis. Knockdown of these upregulated oncogenic lncRNAs in CCA can induce cell cycle arrest and apoptosis by regulating cycle-related proteins and apoptosis-related proteins. Some key lncRNA-related signaling pathways were also involved in the pathogenesis of cholangiocarcinoma. For example, silencing lncRNA CASC15 hampered the expression of cell cycle regulators (CCND1, CCNE1, CDK4, and CDK6) and enhanced apoptosis-related proteins (BAX, cleaved PARP, and cleaved caspase-8) to suppress ICC cell cycle progression and promote cell apoptosis (Zhang et al., 2021). Further, the mechanism experiments showed that lncRNA CASC15 modulated the cell cycle by binding to the protein peroxiredoxin 2 (PRDX2) to active the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway (Zhang et al., 2021). In addition, lncRNAs can also recruit transcription factors to promote the tumorigenesis and progress of CCA. The Hippo pathway has a significant impact on mammalian organ size and participates in the development of various cancers (Ma et al., 2019). Li et al. (2020a) reported that lncRNA MNX1-AS1 bound to transcription factors (TFs) c-myc and MAZ to induce the expression of MNX1 in ICC cells. Then MNX1 prohibited the Hippo pathway by promoting the expression of Ajuba protein, which contributed to the cell proliferation of ICC (Li et al., 2020a). Besides, one of the most fundamental mechanisms of lncRNA tumorigenesis is chromatin remodeling in CCA. Research reported that lncRNA DANCR cooperated with the enhancer of zeste homolog 2 (EZH2) to inhibit the tumor suppressor gene FBP1 expression epigenetically through methylation of lysine 27 on histone 3 (H3K27me3), contributing to CCA growth in vitro and in vivo (Wang et al., 2019). Moreover, lncRNA can act as a ceRNA for miRNA and subsequently modulate proliferation-related and apoptosis-related mRNA in CCA, informing the lncRNA-miRNA-mRNA regulation network. The NF-κB pathway exerts an oncogenic role by boosting the expression of oncogenes involving considerable antiapoptotic genes (Sun, 2017). By inhibition of miR-140, lncRNA SNGH1 facilitated the expression of Toll-like receptor 4(TLR4) mRNA and protein and activate the NF-κB pathway to motivate CCA tumor growth (Li et al., 2019a). Collectively, lncRNAs can regulate CCA cell proliferation and apoptosis through binding to protein, recruiting TFs, chromatin remodeling, and sponging miRNA.
Migration and Invasion
Epithelial-mesenchymal transition (EMT) is a procedure in which tightly attached polarized epithelial cells obtain the phenotype of mesenchymal cells, allowing them to spread and colonize new territory (Gugnoni and Ciarrocchi, 2019). Therefore, EMT is a vital mechanism for tumor metastasis and invasion. Several lncRNAs, such as CCAT1, CCAT2, HOTAIR, TTN-AS1, ASAP1-IT1, DANCR, LINC00261, LINC00665, and SPRY4-IT1, have a close connection with EMT. These lncRNAs enhanced the migration along with the infiltration of CCA by increasing vimentin and N-cadherin expression but reducing E-cadherin expression (Zhang et al., 2017a; Xu et al., 2018a; Xu et al., 2018b; Guo et al., 2018; Qin et al., 2018; Zhu et al., 2020a; Zhu et al., 2020b; Gao et al., 2020; Lu et al., 2021). Most lncRNAs affect CCA cell migration, invasion, and EMT via the mechanism of ceRNA to target miRNAs, such as CCAT1, CCAT2, HOTAIR, TTN-AS1, DANCR, and SPRY4-IT1. The transforming growth factor β (TGF-β) pathway is critical for EMT (David et al., 2016). Zhu et al. (2020a) first reported that lncRNA TTN-AS1 upregulated neuropilin-1 (NRP-1) through sponging miR-320a. Further mechanistic studies manifested that neuropilin-1 as an important co-receptor activated the HGF/c-Met and TGF-β/TGF-βRI pathways contributing to progression, EMT, and angiogenesis in CCA cells (Zhu et al., 2020a). Guo et al. (2018) observed that the silence of lncRNA ASAP1-IT1 has the significant action of inhibiting CCA cell migration, proliferation, and reversing EMT through blocking the hedgehog signaling pathway. And rescue experiments demonstrated that the repressed EMT progress was motivated again by upregulating hedgehog-related Smo and Gli1 (Guo et al., 2018). However, the underlying mechanism that ASAP1-IT1 activates the hedgehog pathway remains unclear. CLIC1 can act as an essential motivator in cancer migration and invasion. It is reported that lncRNA-UCA1 enhanced the ability of CCA cell migration and invasion through inducing CLIC1 targeted by miR-12 and positively regulating the ERK/MAPK signaling pathway (Kong et al., 2019). On the other hand, some lncRNAs, such as CASC2, lnc-LFAR1, MIR22HG, and MEG3, act as protectors to restrain CCA growth and metastasis (Li et al., 2019b; Chen et al., 2019; Hu et al., 2019; Peng et al., 2020). For instance, highly expressed lncRNA CASC2 stimulated the expression of E-cadherin while decreasing the expression of N-cadherin and Vimentin by sponging miR-18a to induce SOCS5, thereby reversing the EMT process (Peng et al., 2020).
Cancer Stemness
Cancer stem cells (CSCs) have the capacity to self-renew and develop into a variety of different types of cancer cells, playing a key role in tumor chemosensitivity and recurrence. A small number of highly expressed lncRNAs, including lnc-PKD2-2-3, ZEB1-AS1, HOXD-AS1, and LINC00665, have participated in the regulation of CCA stemness (Qiu et al., 2019; Jiang et al., 2020a; Li et al., 2020b; Lu et al., 2021). Many studies report that cancer cell stemness has a strong effect on chemoresistance in various cancers (Smith and Macleod, 2019). For instance, the overexpression of lnc-PKD2-2-3 enhanced CSC markers (CD44, CD133, and OCT4) expression and sphere generation efficiency in CCA cell lines (Qiu et al., 2019). Moreover, lnc-PKD2-2-3 was observed to have been dramatically upregulated in CCA stem-like cells, which illustrated that lnc-PKD2-2-3 might act as a fresh CSC marker (Qiu et al., 2019). ZEB1-AS1, HOXD-AS1and LINC00665 enhanced the cancer cell stemness respectively through sponging miR-133b, miR-520c-3p, and miR-424-5p (Jiang et al., 2020a; Li et al., 2020b; Lu et al., 2021). So far, these lncRNAs function as ceRNA for miRNAs, which is the major mechanism is ceRNA to modulate tumor cell stemness in CCA cells. However, the mechanism and singling pathways of lncRNA on CCA cell stemness are complex and unclear, which need to be explored in depth.
Angiogenesis and Oxidative Stress
Recently, more attention has been paid to the vital function of the tumor microenvironment in tumor progression, which includes angiogenesis, oxidative stress, and immune escape (Whiteside, 2008). Vascular endothelial growth factor (VEGF) is considered to be an essential factor in regulating tumor angiogenesis (Siveen et al., 2017). The content of lncRNA DANCR was enriched in CCA cells and tissues. Downregulation of DANCR inhibited the levels of VEGF-C and VEGF-A in cholangiocarcinoma cell lines (Zhu et al., 2020b). It is also reported that lncRNA SNHG6 reinforced CCA proliferation and angiogenesis by modulating miR-101-3p-targeted E2F8 which is indispensable for tumor angiogenesis (Wang et al., 2020b). There is a common opinion that chronic inflammation and bile duct epithelial cell injury can facilitate CCA tumorigenesis and growth (Sirica et al., 2021). Oxidative stress-induced lncRNA HULC, along with H19, increased the level of inflammatory mediators IL-6 and CXCR4, which in turn activated the positive feedback pathway of inflammation, promoting tumorigenesis in CCA via working as a sponge of miR-372/miR-373 and let-7a/let-7b, respectively (Wang et al., 2016). In terms of the tumor microenvironment, research on the role and mechanism of lncRNA is still limited in CCA.
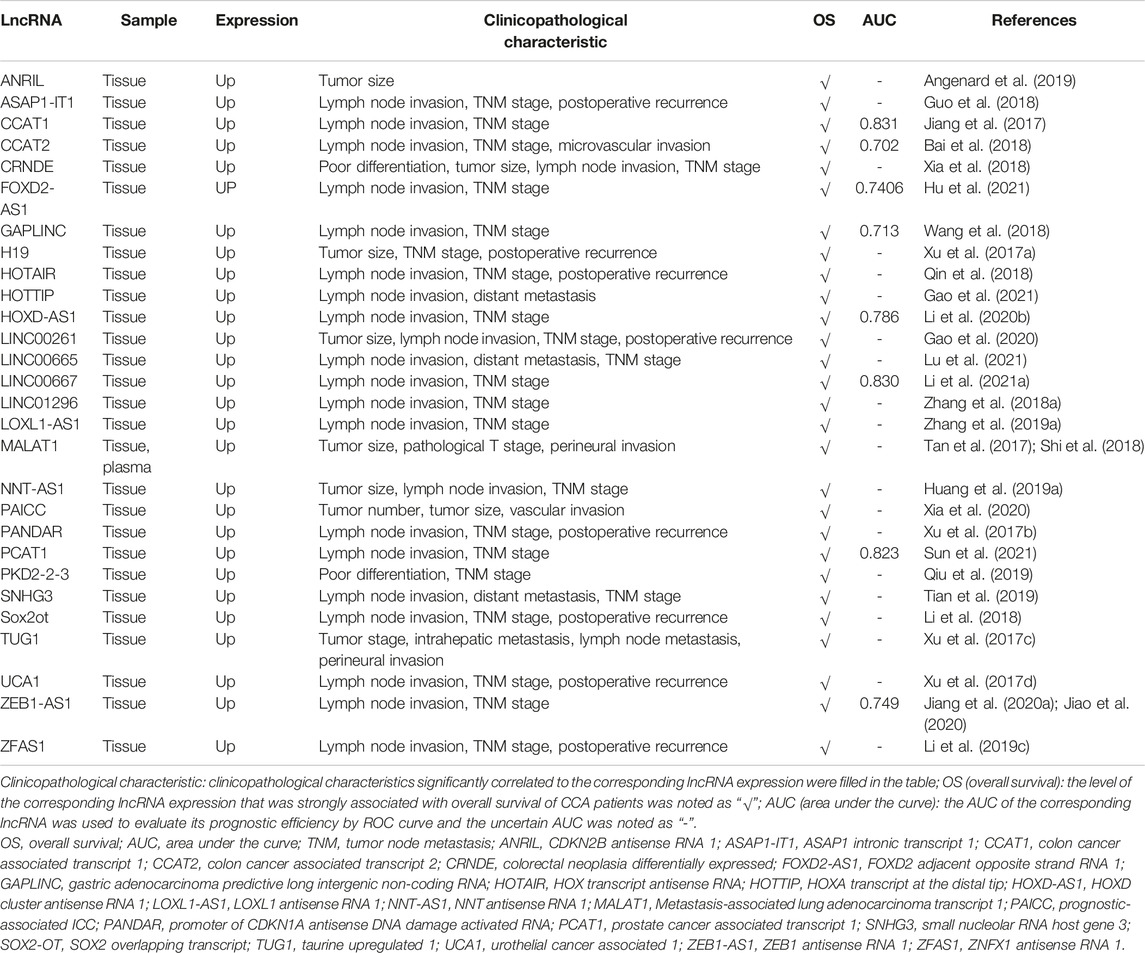
LncRNAs as Diagnostic and Prognostic Biomarkers in Cholangiocarcinoma
The value of tumor biomarkers lies in their ability to provide early diagnosis, predict disease prognosis, monitor disease recurrence, and offer guidance for disease treatment. Unfortunately, there is no effective and reliable biomarker for the diagnosis, as well as prognosis of cholangiocarcinoma. The most commonly used biomarkers are carbohydrate antigen 199 (CA199) and carcinoembryonic antigen (CEA) in the clinical diagnosis of CCA, whether used alone or in combination, but their value is limited due to the inconsistency in sensitivity (47.2%–98.2%) and specificity (89.7%–100%) (Tshering et al., 2018; Macias et al., 2019). Interestingly, many lncRNAs localized in exosomes can be secreted into bodily fluids (for instance, plasma, urine, and bile) as circulating RNA with high tissue specificity, becoming putative non-invasive biomarkers for many tumors (Di Meo et al., 2017; Xu et al., 2018c; Lapitz et al., 2020). However, there is little research about the diagnostic value of lncRNAs from exosomes or EVs in CCA. Ge et al. (2017) observed that exosomal lncRNAs of ENST0000588480.1 and ENST00000517758.1, which were isolated and identified from human bile samples, in contrast with normal individuals, showed significantly higher expression in CCA patients. The diagnostic value of these lncRNAs was examined in CCA, and the result showed that the combination of ENST00000588480.1 and ENST00000517758.1 harbored more excellent sensitivity versus CA199 (82.9% vs. 74.3%) (Ge et al., 2017). Notably, the level of both lncRNAs was negatively associated with survival time (Ge et al., 2017). In a recent study, a few lncRNAs were identified in serum and urine extracellular vesicles (EVs) from patients with CCA, primary sclerosing cholangitis (PSC), and healthy individuals to evaluate their prognostic value (Lapitz et al., 2020). In serum EVs, MALAT-1 and LOC100190986 possessed a high accuracy for the differentiation of CCA and PSC with the area under the curve (AUC) values of both 1.0 (Lapitz et al., 2020). In urine EVs, LOC100134868 displayed an excellent diagnostic value with an AUC of 0.896 (patients with CCA vs. healthy individuals) (Lapitz et al., 2020). Moreover, the diagnostic ability of some other lncRNAs in plasma or tissue, including PCAT1, CPS1-IT1, H19, PVT1, and NEF (Han et al., 2018; Shi et al., 2018; Liang et al., 2019), has also been verified in CCA.
In the last decade, it was found that many dysregulated lncRNAs in CCA tissues and cells were linked to the pathological characteristics and prognosis of patients with cholangiocarcinoma (Merdrignac et al., 2021). Xie et al. (2021) established a prediction model with five lncRNAs (HULC, AL359715.5, AC006504.8, AC090114.2, and AP00943.4) to distinguish the CCA patients with poor prognosis. In the validation cohort, the five-lncRNA module had a perfect predictive value with a AUC of 0.816 for prognosis in CCA. However, the cohort studies on the predictive effect of lncRNAs are still lacking. Most of the lncRNA-related research has focused on the mechanism and function of dysregulated lncRNAs in the tissues and cells of CCA. The following lncRNAs have been documented to be linked to the prognosis of cholangiocarcinoma (Table 2): ANRIL (Angenard et al., 2019), ASAP1-IT1 (Guo et al., 2018), CCAT1 (Zhang et al., 2017a), CCAT2 (Xu et al., 2018a), CRNDE (Xia et al., 2018), FOXD2-AS1 (Hu et al., 2022), GAPLINC (Wang et al., 2018), H19 (Xu et al., 2017a), HOTAIR (Qin et al., 2018), HOTTIP (Gao et al., 2021), HOXD-AS1 (Li et al., 2020b), LINC00261 (Gao et al., 2020), LINC00665 (Lu et al., 2021), LINC00667 (Li et al., 2021a), LINC01296 (Zhang et al., 2018a), LOXL1-AS1 (Zhang et al., 2019a), NNT-AS1 (Huang et al., 2019a), MALAT1 (Tan et al., 2017), PAICC (Xia et al., 2020), PANDAR (Xu et al., 2017b), PCAT1 (Sun et al., 2021), lnc-PKD2-2-3 (Qiu et al., 2019), SNHG3 (Tian et al., 2019), Sox2ot (Li et al., 2018), TUG1 (Xu et al., 2017c), UCA1 (Xu et al., 2017d), ZEB1-AS1 (Jiang et al., 2020a), ZFAS1 (Li et al., 2019c).
In this review, these lncRNAs for diagnosis and/or prognosis, including PCAT1, MALAT1, H19, NEF, CCAT1, CCAT2, ZEB1-AS1, will be described in detail.
Prostate Cancer-Associated Transcript 1
Prostate cancer-associated transcript 1(PCAT1) was initially identified as an overexpressed lncRNA in prostate cancer tissues (Prensner et al., 2011), which has also been proved to be abnormally expressed in various malignant tumors and to play a carcinogenic role (Wen et al., 2016; Bi et al., 2017; Xu et al., 2017e; Huang et al., 2018; Liu et al., 2019a; Shen et al., 2019). Additionally, dysfunctional lncRNA PCAT1, identified in body fluids, can be used as a prospective biomarker for esophageal squamous cell carcinoma, bladder carcinoma, and multiple myeloma (Shen et al., 2017; Huang et al., 2019b; Zhang et al., 2019b). Zhang et al. (2017b) discovered that PCAT1 was remarkably elevated in extrahepatic cholangiocarcinoma (ECC) tissues and cell lines, and could regulate ECC progression through the Wnt/β-catenin signaling pathway. An updated study uncovered that YY1-induced lncRNA PCAT1 promoted the progression of cholangiocarcinoma through the miR-216a-3p/BCL3 axis (Sun et al., 2021). Also, this study showed that the overexpressed PCAT1 was confirmed as the factor associated with the adverse outcome for CCA patients and made an effective prognostic prediction with an AUC of 0.823 (Sun et al., 2021). In terms of its diagnostic value, a study illustrated that the AUC of plasma PCAT1 was 0.784 by ROC (receiver operating characteristic curves), which indicated that circulating PCAT1 may act as an early diagnostic biomarker for HCC (Shi et al., 2018). In general, PCAT1 has the potential value in the diagnosis and prognosis of CCA, but clinical trials on a large scale are still needed to confirm it.
Metastasis-Associated Lung Adenocarcinoma Transcript 1
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA with a length of about 8.5 kb transcribed from human chromosome 11q13 in the nucleus (Ji et al., 2003; Eißmann et al., 2012). MALAT1, which was detected in body fluids, has been identified as a valuable biomarker either alone or in combination with other molecules in malignant tumors (Goyal et al., 2021), such as nasopharyngeal carcinoma (He et al., 2017), osteosarcoma (Huo et al., 2017), and bladder cancer (Zhan et al., 2018). In CCA, MALAT1 was established to be upregulated in tissues and cells, and over-expression of MALAT1 was remarkably correlated with adverse clinical features (Tan et al., 2017). Shi et al. (2018) showed that MALAT1 in plasma has excellent specificity and sensitivity as a diagnostic biomarker of hilar cholangiocarcinoma (AUC, 0.860; sensitivity, 81.1%; specificity, 90.9%). Further, when PCAT1, MALAT1, along with CPS1-IT1 were combined, the sensitivity and specificity increased to 85.5% and 93.2%, respectively (Shi et al., 2018).
H19
LncRNA H19 is a long-chain noncoding RNA transcribed by a paternally imprinted carcinoembryonic gene on chromosome 11p15.5. The content of H19 was elevated in fetal tissues and downregulated in various healthy tissues after birth (Gabory et al., 2010). A pooled analysis indicated that H19 may be a reliable biomarker to discriminate patients with tumors from normal individuals (Liu et al., 2019b). In cholangiocarcinoma, aberrant expression of H19 has been shown to promote proliferation and invasion through inflammatory and oxidative stress pathways in vitro (Wang et al., 2016; Xu et al., 2017a; Yu et al., 2020a). Besides, overexpression of H19 was a dismal predictor for CCA patients, which was significantly correlated with tumor size, TNM stage, as well as postoperative recurrence (Xu et al., 2017a). Remarkably, H19 was observed to have moderate sensitivity in distinguishing CCA tissues from normal ones with an AUC of 0.7422 (Han et al., 2018). Furthermore, a combination of AC005550.3, H19, C3P1, PVT1, along with LPAL2 showed more excellent sensitivity (93.75%) and specificity (81.25%) with an AUC of 0.8828 in differentiating CCA tissues from normal ones (Han et al., 2018). Thus, the establishment of an effective lncRNA-related identification panel plays an essential role in improving the detection rate of CCA.
Neighboring Enhancer of FOXA2
LncRNA neighboring enhancer of FOXA2 (NEF) was first discovered as a tumor suppressor in hepatocellular carcinoma by reversing the EMT process and inhibiting tumor metastasis (Liang et al., 2018). In intrahepatic cholangiocarcinoma, a study reported that lncRNA-NEF was dramatically downregulated in tumor tissues and upregulated lncRNA-NEF repressed cell migration and invasion by inhibition of runt-related transcription factor 1 (RUNX1) (Liang et al., 2019). Additionally, the study observed that the level of plasma lncRNA-NEF in patients (including patients at stage I or II) was significantly lower than in healthy controls (Liang et al., 2019). Plasma lncRNA-NEF exhibited excellent ability for early diagnosis with an AUC of 0.8642 (Liang et al., 2019). Therefore, plasma lncRNA-NEF may become the effective early diagnostic biomarker of intrahepatic cholangiocarcinoma.
Colon Cancer-Associated Transcript 1 and Colon Cancer-Associated Transcript 2
Colon cancer-associated transcript 1 (CCAT1) was initially identified as being overexpressed in colon cancer tissues, peripheral blood, and related lymph node tissues, but it was almost undetectable in normal human body tissues (Nissan et al., 2012). As a rising star among carcinogenic lncRNAs, CCAT1 has been closely related to many cancers’ occurrence, development, and drug resistance (Jin et al., 2020; Shan et al., 2020; Su et al., 2020; Wang et al., 2021). CCAT1 expression was dramatically increased in CCA tissues and was shown to be strongly linked to lymph node infiltration and late TNM stage (Zhang et al., 2017a; Jiang et al., 2017). Through multivariate analysis, it was established that increased CCAT1 expression is an independent prognostic factor and that it has a certain sensitivity (81.8%) and specificity (74.5%) for estimating overall survival via ROC analysis (Jiang et al., 2017). Xu et al. (2018a) and Bai et al. (2018) both documented that the level of CCAT2 expression was inversely linked with the overall survival rate of CCA patients. The study suggested that CCAT2 has practicalvaluein predicting the prognosis of patients with ICC, with an AUC of 0.702 and 0.715 for OS and PFS, respectively (Bai et al., 2018). Besides, a meta-analysis in 2019 showed that upregulated CCAT lncRNA families, especially CCAT2, can predict shorter overall survival (p < 0.001) (Dai et al., 2019). Therefore, CCAT1 and CCAT2 can be considered promising prognostic biomarkers of CCA.
ZEB1 Antisense RNA 1
ZEB1-AS1 is an antisense transcript originating from the promoter of Zinc finger E-box binding homeobox 1(ZEB1), a prominent factor in the EMT process of tumor epithelial cells (Li et al., 2016; Caramel et al., 2018). ZEB1-AS1 plays a carcinogenic factor in various digestive system tumors, for instance, hepatocellular carcinoma, esophageal cancer, colorectal cancer, and gastric cancer (Li et al., 2016; Zhang et al., 2018b; Zhao et al., 2019; Wu et al., 2020). Jiang et al. (2020a) proved that ZEB1-AS1 was overexpressed in CCA and promoted CCA growth along with metastasis in vivo, as well as in vitro experiments. Patients with high ZEB1-AS1 expression were clearly related to lymph node invasion, progressed TNM stage, along with the shorter survival time when compared to the low ZEB1-AS1 expression group (Jiang et al., 2020a). Regarding prognostic efficiency, the AUC of ZEB1-AS1 was 0.749 with 65.5% sensitivity and 80.0% specificity by ROC analysis (Jiang et al., 2020a). Hopefully, ZEB1-AS1 could become a sensitive, as well as specific prognostic biomarker for CCA.
The Role of lncRNAs in the Therapeutic Target of Cholangiocarcinoma
R0 radical surgical resection has remained the mainstay of potentially curative treatment (Valle et al., 2021). For individuals with advanced or unresectable cancer, systemic chemotherapy is the primary treatment option (Rizvi et al., 2018). Gemcitabine plus cisplatin constitutes the standard first-line chemotherapy for advanced cholangiocarcinoma (Valle et al., 2010). However, the median overall survival for this standard chemotherapy regimen is less than 1 year (Weigt and Malfertheiner, 2010). Recently, studies have identified lncRNAs as important players in CCA chemoresistance. So far, a few novel lncRNAs, including NEAT-1, LINC01714, HOXD-AS1, HOTTIP, LINC00665, and lnc-PKD2-2-3, have been revealed to modulate chemoresistance in CCA (Figure 2) (Lu et al., 2021; Qiu et al., 2019; Li et al., 2020b; Gao et al., 2021; Parasramka et al., 2017; Shen et al., 2020). These resistance-associated lncRNAs present new opportunities for the treatment of CCA. It was reported that both NEAT-1 and LINC01714 could positively regulate the sensitivity to gemcitabine in CCA cells. More specifically, Parasramka et al. (2017) noticed that gemcitabine cytotoxicity was strengthened in low BAP1 expressing CCA cells, and then NEAT1 was enriched in CCA cells after the knockout of BAP1. Further experiments revealed that BAP1-targeted lncRNA NEAT1 could enhance the responsivity of CCA cells to gemcitabine (Figure 2A) (Parasramka et al., 2017). Additionally, overexpressed LINC01714 facilitated the susceptibility of CCA cells to gemcitabine by reducing phosphorylated FOXO3-Ser318 (Figure 1B), which indicated that LINC01714 transcription promoter and gemcitabine combined with chemotherapy may have a synergistic effect on the treatment of advanced CCA patients (Shen et al., 2020). LncRNA HOTTIP and HOXD-AS1 were reported to regulate the sensitivity of CCA cells to gemcitabine and cisplatin through sponging miR-637 to increase LASP1 and sponging miR-520c-3p to increase MYCN, respectively (Figures 2C,D) (Li et al., 2020b; Gao et al., 2021). The acquisition of EMT and the existence of CSC are not only crucial mechanisms in the invasion and metastasis of CCA, but recent investigations have established that they may be closely linked to the chemoresistance of CCA (Huang et al., 2019c; Varamo et al., 2019). Lu et al. (2021) proved that LINC00665 can act as a ceRNA to modulate the miR-424-5p/BCL9L axis, thereby promoting EMT and cell stemness and finally enhancing gemcitabine resistance of CCA (Figure 2E). It is the first report that lnc-PKD2-2-3 enhanced drug resistance to 5-FU in CCA cells possibly by promoting CCA stemness (Figure 2F) (Qiu et al., 2019). Therefore, the above-mentioned upregulated lncRNAs may serve as biomarkers for chemotherapeutic drug resistance to guide clinical chemotherapy medication, and may also provide new therapeutic targets for cholangiocarcinoma. Moreover, Tu et al. (2020) first observed that the overexpression of PCAT6 can promote macrophage M2 polarization to repress the immune response of macrophages via sponging miR-326 and activating RhoA/ROCK pathways. In vitro, it was found that the tumor growth of PCAT6 knockout mice was significantly inhibited. Therefore, targeting PCAT6 may become potentially feasible immunotherapy for CCA treatment (Tu et al., 2020). Some lncRNAs may become underlying therapeutic targets by modulating previously mentioned signal pathways, including the PI3K/AKT, NF-κB, Hedgehog, Wnt/β-catenin, ERK/MAPK, TGFβ/Smad, Hippo, and RhoA/ROCK pathway (Figure 3). Although no drug has been developed to target lncRNA directly, several approaches have been successfully used to deplete specific oncogenic lncRNAs in cells, including RNA interference (RNAi) and antisense oligonucleotides (ASOs) (Lennox and Behlke, 2016). Of note, the prospect of lncRNA-targeted therapy is bright thanks to a deep dive into the mechanisms of lncRNAs and the development of emerging technologies.
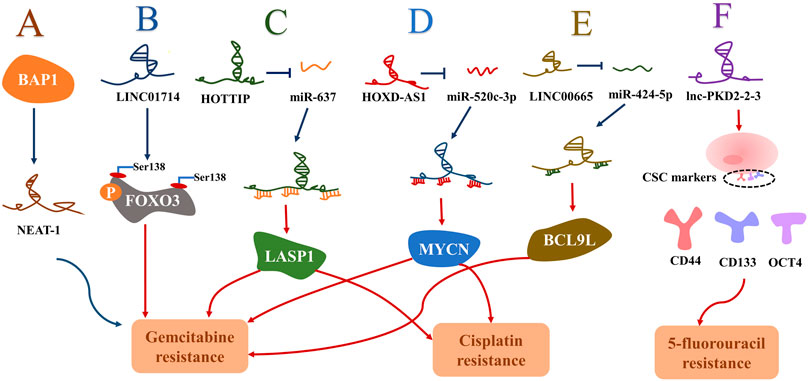
FIGURE 2. The chemo-resistance mechanism of lncRNAs in cholangiocarcinoma. Note: the blue arrow represents a negative correlation and the red arrow represents a positive correlation. (A): NAET-1 negatively regulated by BAP1 promoted the sensitivity of CCA cells to gemcitabine. (B): LINC01714 facilitated gemcitabine sensitivity by reducing phosphorylated FOXO3-Ser318. (C,D): HOTTIP and HOXD-AS1 promoted the gemcitabine and cisplatin resistance through sponging miR-637 to increase LASP1 and through sponging miR-520c-3p to increase MYCN, respectively. (E): LINC00665 enhanced BCL9L to promote gemcitabine resistance by sponging miR-424-5p. (F): lnc-PKD2-2-3 enhanced 5-FU resistance by facilitating cell stemness in CCA. Abbreviation: BAP1, BRCA-1 associated protein-1; NEAT-1, nuclear paraspeckle assembly transcript 1; FOXO3, Forkhead Box O3; HOTTIP, HOXA transcript at the distal tip; MYCN, MYCN proto-oncogene bHLH transcription factor; HOXD-AS1, HOXD cluster antisense RNA 1; LASP1, LIM and SH3 domain protein 1; BCL9L, B cell CLL/lymphoma 9-like; CSC, cancer stem cell; CD44, Cluster of differentiation-44; CD133, Cluster of differentiation-133; OCT4, octamer-binding transcription factor 4.
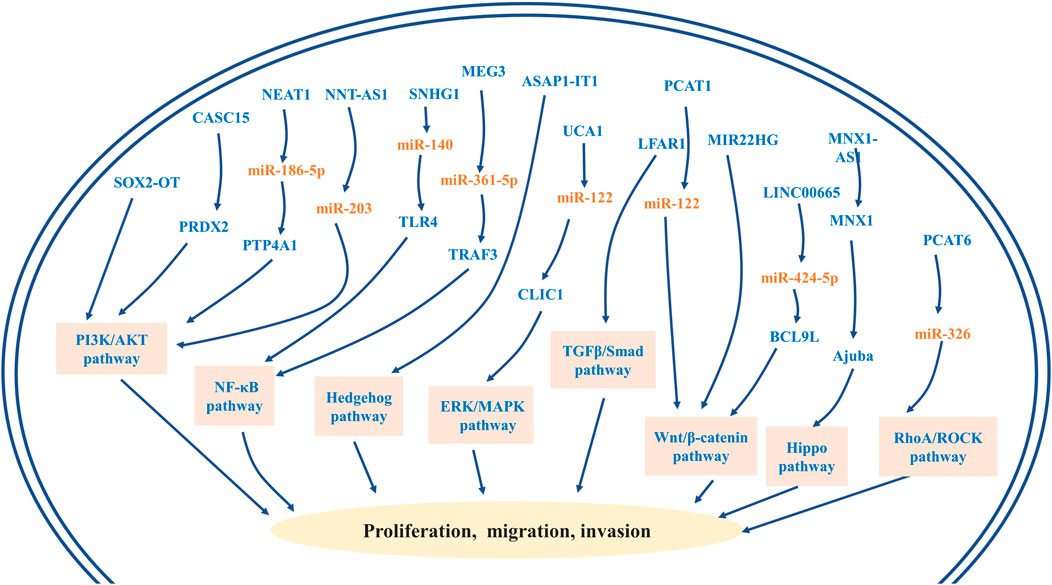
FIGURE 3. LncRNA-related oncogenic signaling pathways and their roles in the progression of cholangiocarcinoma, including PI3K/AKT, NF-κB, Hedgehog, Wnt/β-catenin, ERK/MAPK, TGFβ/Smad, Hippo, and RhoA/ROCK pathways.
Conclusion and Future Perspectives
Cholangiocarcinoma is a kind of malignant tumor with an adverse prognosis and growing prevalence. With the advancements in high-throughput sequencing and molecular gene technologies, it has been demonstrated that aberrantly expressed lncRNAs harbor diverse functions in the progression of malignancies, including CCA. However, the underlying molecular mechanism of most lncRNAs remains unclear in CCA. Thus, more basic biological research will be required. A rising number of improperly expressed lncRNAs have been detected in tissue, and serum and bile secretion of CCA patients. Due to their extensive distribution and relatively stable structure, lncRNAs harbor the potential to be extremely useful biomarkers for monitoring, diagnosis, along with prognosis. At present, aberrant expression of many lncRNAs in CCA tissues has been observed to be remarkably linked to adverse clinical features and OS, which also means that lncRNAs may be used as effective indicators for risk-stratification and predicting survival in CCA patients. Furthermore, the combination of several exosomal lncRNAs in bile and serum has shown excellent sensitivity and specificity for the diagnosis in CCA. Therefore, the exosomal lncRNA will become a rising star in CCA diagnosis and prognosis, and more studies should focus on exploring specific exosomal lncRNAs as reliable biomarkers. However, due to a lack of significant sample clinical cohort verification, the value of lncRNAs as clinical biomarkers is controversial in CCA. In addition, several lncRNAs, as biomarkers of chemotherapy resistance, can be used to guide clinical chemotherapy. Many lncRNAs are the critical molecules involved in activating multiple carcinogenic signaling pathways in CCA. Therefore, lncRNA is the novel promising therapeutic target for CCA.
Author Contributions
YW: Wrote the manuscript. KH: Edit and correct the paper. YH and YW: Performed literature analyses and prepared figures. JY: Provided article ideas and article review.
Funding
This study was supported by grants from the Zhejiang health committee (2021ZH003), Hangzhou science and technology commission (202004A14), and the Construction Fund of Medical Key Disciplines of Hangzhou (OO20190001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Angenard, G., Merdrignac, A., Louis, C., Edeline, J., and Coulouarn, C. (2019). Expression of Long Non-coding RNA ANRIL Predicts a Poor Prognosis in Intrahepatic Cholangiocarcinoma. Dig. Liver Dis. 51, 1337–1343. doi:10.1016/j.dld.2019.03.019
Bai, J. G., Tang, R. F., Shang, J. F., Qi, S., Yu, G. D., and Sun, C. (2018). Upregulation of Long Non-coding RNA CCAT2 I-ndicates a P-oor P-rognosis and P-romotes P-roliferation and M-etastasis in I-ntrahepatic C-holangiocarcinoma. Mol. Med. Rep. 17, 5328–5335. doi:10.3892/mmr.2018.8518
Banales, J. M., Cardinale, V., Carpino, G., Marzioni, M., Andersen, J. B., Invernizzi, P., et al. (2016). Cholangiocarcinoma: Current Knowledge and Future Perspectives Consensus Statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 13, 261–280. doi:10.1038/nrgastro.2016.51
Banales, J. M., Marin, J. J. G., Lamarca, A., Rodrigues, P. M., Khan, S. A., Roberts, L. R., et al. (2020). Cholangiocarcinoma 2020: the Next Horizon in Mechanisms and Management. Nat. Rev. Gastroenterol. Hepatol. 17, 557–588. doi:10.1038/s41575-020-0310-z
Bi, M., Yu, H., Huang, B., and Tang, C. (2017). Long Non-coding RNA PCAT-1 Over-expression Promotes Proliferation and Metastasis in Gastric Cancer Cells through Regulating CDKN1A. Gene 626, 337–343. doi:10.1016/j.gene.2017.05.049
Caramel, J., Ligier, M., and Puisieux, A. (2018). Pleiotropic Roles for ZEB1 in Cancer. Cancer Res. 78, 30–35. doi:10.1158/0008-5472.can-17-2476
Chen, C., Li, H., Wang, X., Wang, L., and Zeng, Q. (2019). Lnc-LFAR1 Affects Intrahepatic Cholangiocarcinoma Proliferation, Invasion, and EMT by Regulating the TGFβ/Smad Signaling Pathway. Int. J. Clin. Exp. Pathol. 12, 2455–2461.
Dai, K., Quan, J., Yan, F., Jin, X., Pan, X., Song, X., et al. (2019). lncRNAs as Potential Molecular Biomarkers in the Clinicopathology and Prognosis of Cholangiocarcinoma: a Systematic Review and Meta-Analysis. Ott 12, 1905–1915. doi:10.2147/ott.s188134
David, C. J., Huang, Y.-H., Chen, M., Su, J., Zou, Y., Bardeesy, N., et al. (2016). TGF-β Tumor Suppression through a Lethal EMT. Cell 164, 1015–1030. doi:10.1016/j.cell.2016.01.009
Di Meo, A., Bartlett, J., Cheng, Y., Pasic, M. D., and Yousef, G. M. (2017). Liquid Biopsy: a Step Forward towards Precision Medicine in Urologic Malignancies. Mol. Cancer 16, 80. doi:10.1186/s12943-017-0644-5
Eißmann, M., Gutschner, T., Hämmerle, M., Günther, S., Caudron-Herger, M., Groß, M., et al. (2012). Loss of the Abundant Nuclear Non-coding RNA MALAT1 Is Compatible with Life and Development. RNA Biol. 9, 1076–1087. doi:10.4161/rna.21089
Florio, A. A., Ferlay, J., Znaor, A., Ruggieri, D., Alvarez, C. S., Laversanne, M., et al. (2020). Global Trends in Intrahepatic and Extrahepatic Cholangiocarcinoma Incidence from 1993 to 2012. Cancer 126, 2666–2678. doi:10.1002/cncr.32803
Gabory, A., Jammes, H., and Dandolo, L. (2010). The H19 Locus: Role of an Imprinted Non-coding RNA in Growth and Development. Bioessays 32, 473–480. doi:10.1002/bies.200900170
Gao, J., Qin, W., Kang, P., Xu, Y., Leng, K., Li, Z., et al. (2020). Up-regulated LINC00261 Predicts a Poor Prognosis and Promotes a Metastasis by EMT Process in Cholangiocarcinoma. Pathol. - Res. Pract. 216, 152733. doi:10.1016/j.prp.2019.152733
Gao, K., Chen, S., and Yang, X. (2021). HOTTIP Enhances Gemcitabine and Cisplatin Resistance through Sponging miR-637 in Cholangiocarcinoma. Front. Oncol. 11, 664916. doi:10.3389/fonc.2021.664916
Ge, X., Wang, Y., Nie, J., Li, Q., Tang, L., Deng, X., et al. (2017). The Diagnostic/prognostic Potential and Molecular Functions of Long Non-coding RNAs in the Exosomes Derived from the Bile of Human Cholangiocarcinoma. Oncotarget 8, 69995–70005. doi:10.18632/oncotarget.19547
Goyal, B., Yadav, S. R. M., Awasthee, N., Gupta, S., Kunnumakkara, A. B., and Gupta, S. C. (2021). Diagnostic, Prognostic, and Therapeutic Significance of Long Non-coding RNA MALAT1 in Cancer. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1875, 188502. doi:10.1016/j.bbcan.2021.188502
Gu, Y., Li, C., Xiao, L., Li, J., Pei, H., Xu, D., et al. (2019). High Expression of Long Non-coding RNA NNT-AS1 Facilitates Progression of Cholangiocarcinoma through Promoting Epithelial-Mesenchymal Transition. Am. J. Transl Res. 11, 5438–5456.
Gu, Y., Zhu, Z., Pei, H., Xu, D., Jiang, Y., Zhang, L., et al. (2020). Long Non-coding RNA NNT-AS1 Promotes Cholangiocarcinoma Cells Proliferation and Epithelial-To-Mesenchymal Transition through Down-Regulating miR-203. Aging 12, 2333–2346. doi:10.18632/aging.102747
Gugnoni, M., and Ciarrocchi, A. (2019). Long Noncoding RNA and Epithelial Mesenchymal Transition in Cancer. Int. J. Mol. Sci. 20. doi:10.3390/ijms20081924
Guo, L., Zhou, Y., Chen, Y., Sun, H., Wang, Y., and Qu, Y. (2018). LncRNA ASAP1-IT1 Positively Modulates the Development of Cholangiocarcinoma via Hedgehog Signaling Pathway. Biomed. Pharmacother. 103, 167–173. doi:10.1016/j.biopha.2018.04.015
Guo, X., Gao, L., Wang, Y., Chiu, D. K. Y., Wang, T., and Deng, Y. (2016). Advances in Long Noncoding RNAs: Identification, Structure Prediction and Function Annotation. Brief. Funct. Genomics 15, 38–46. doi:10.1093/bfgp/elv022
Han, B.-W., Ye, H., Wei, P.-P., He, B., Han, C., Chen, Z.-H., et al. (2018). Global Identification and Characterization of lncRNAs that Control Inflammation in Malignant Cholangiocytes. BMC genomics 19, 735. doi:10.1186/s12864-018-5133-8
He, B., Zeng, J., Chao, W., Chen, X., Huang, Y., Deng, K., et al. (2017). Serum Long Non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma. Oncotarget 8, 41166–41177. doi:10.18632/oncotarget.17083
Hu, X., Tan, Z., Yang, Y., and Yang, P. (2019). Long Non-coding RNA MIR22HG Inhibits Cell Proliferation and Migration in Cholangiocarcinoma by Negatively Regulating the Wnt/β-catenin Signaling Pathway. J. Gene Med. 21, e3085. doi:10.1002/jgm.3085
Hu, Z., Huang, L., Wang, W., Guan, C., Zhao, Y., Liu, L., et al. (2021). Long Non-coding RNA FOXD2-AS1 Promotes Proliferation, Migration, and Invasion in Cholangiocarcinoma through Regulating miR-760/E2F3 Axis. Dig. Dis. Sci. 67, 546–558. doi:10.1007/s10620-021-06876-9
Hu, Z., Huang, L., Wang, W., Guan, C., Zhao, Y., Liu, L., et al. (2022). Long Non-coding RNA FOXD2-AS1 Promotes Proliferation, Migration, and Invasion in Cholangiocarcinoma through Regulating miR-760/E2F3 Axis. Dig. Dis. Sci. 67, 546–558. doi:10.1007/s10620-021-06876-9
Huang, J., Deng, G., Liu, T., Chen, W., and Zhou, Y. (2018). Long Noncoding RNA PCAT-1 Acts as an Oncogene in Osteosarcoma by Reducing P21 Levels. Biochem. biophysical Res. Commun. 495, 2622–2629. doi:10.1016/j.bbrc.2017.12.157
Huang, L., Cai, J., Guo, H., Gu, J., Tong, Y., Qiu, B., et al. (2019). ID3 Promotes Stem Cell Features and Predicts Chemotherapeutic Response of Intrahepatic Cholangiocarcinoma. Hepatology 69, 1995–2012. doi:10.1002/hep.30404
Huang, L., Jiang, X., Kang, P., Wang, Z., Leng, K., Ji, D., et al. (2019). Long Non-coding RNA NNT-AS1 Functions as an Oncogenic Gene through Modulating miR-485/BCL9 in Cholangiocarcinoma. Cmar 11, 7739–7749. doi:10.2147/cmar.s207801
Huang, L., Wang, Y., Chen, J., Wang, Y., Zhao, Y., Wang, Y., et al. (2019). Long Noncoding RNA PCAT1, a Novel Serum-Based Biomarker, Enhances Cell Growth by Sponging miR-326 in Oesophageal Squamous Cell Carcinoma. Cell Death Dis 10, 513. doi:10.1038/s41419-019-1745-4
Huo, Y., Li, Q., Wang, X., Jiao, X., Zheng, J., Li, Z., et al. (2017). MALAT1 Predicts Poor Survival in Osteosarcoma Patients and Promotes Cell Metastasis through Associating with EZH2. Oncotarget 8, 46993–47006. doi:10.18632/oncotarget.16551
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a Novel Noncoding RNA, and Thymosin β4 Predict Metastasis and Survival in Early-Stage Non-small Cell Lung Cancer. Oncogene 22, 8031–8041. doi:10.1038/sj.onc.1206928
Jiang, T., Wang, C., Zhu, Y., and Han, H. (2020). LINC01410 Promotes Cell Proliferation and Migration of Cholangiocarcinoma through Modulating miR-124-3p/SMAD5 axis. J. Gene Med. 22, e3162. doi:10.1002/jgm.3162
Jiang, X., Li, J., Wang, W., Hu, Z., Guan, C., Zhao, Y., et al. (2020). AR-induced ZEB1-AS1 Represents Poor Prognosis in Cholangiocarcinoma and Facilitates Tumor Stemness, Proliferation and Invasion through Mediating miR-133b/HOXB8. Aging 12, 1237–1255. doi:10.18632/aging.102680
Jiang, X. M., Li, Z. L., Li, J. L., Zheng, W. Y., Li, X. H., Cui, Y. F., et al. (2017). LncRNA CCAT1 as the Unfavorable Prognostic Biomarker for Cholangiocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 21, 1242–1247.
Jiao, M., Ning, S., Chen, J., Chen, L., Jiao, M., Cui, Z., et al. (2020). Long Non-coding RNA ZEB1-AS1 P-redicts a P-oor P-rognosis and P-romotes C-ancer P-rogression through the miR-200a/ZEB1 S-ignaling P-athway in I-ntrahepatic C-holangiocarcinoma. Int. J. Oncol. 56, 1455–1467. doi:10.3892/ijo.2020.5023
Jin, X., Liu, X., Zhang, Z., and Guan, Y. (2020). lncRNA CCAT1 Acts as a MicroRNA-218 Sponge to Increase Gefitinib Resistance in NSCLC by Targeting HOXA1. Mol. Ther. - Nucleic Acids 19, 1266–1275. doi:10.1016/j.omtn.2020.01.006
Kim, J., Piao, H.-L., Kim, B.-J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long Noncoding RNA MALAT1 Suppresses Breast Cancer Metastasis. Nat. Genet. 50, 1705–1715. doi:10.1038/s41588-018-0252-3
Kong, L., Wu, Q., Zhao, L., Ye, J., Li, N., and Yang, H. (2019). Upregulated lncRNA-UCA1 Contributes to Metastasis of Bile Duct Carcinoma through Regulation of miR-122/CLIC1and Activation of the ERK/MAPK Signaling Pathway. Cell Cycle 18, 1212–1228. doi:10.1080/15384101.2019.1593647
Kopp, F., and Mendell, J. T. (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Lapitz, A., Arbelaiz, A., O'Rourke, C. J., Lavin, J. L., Casta, A., Ibarra, C., et al. (2020). Patients with Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells 9. doi:10.3390/cells9030721
Lennox, K. A., and Behlke, M. A. (2016). Cellular Localization of Long Non-coding RNAs Affects Silencing by RNAi More Than by Antisense Oligonucleotides. Nucleic Acids Res. 44, 863–877. doi:10.1093/nar/gkv1206
Li, F., Chen, Q., Xue, H., Zhang, L., Wang, K., and Shen, F. (2020). LncRNA MNX1-AS1 Promotes Progression of Intrahepatic Cholangiocarcinoma through the MNX1/Hippo axis. Cel Death Dis 11, 894. doi:10.1038/s41419-020-03029-0
Li, J., Guan, C., Hu, Z., Liu, L., Su, Z., Kang, P., et al. (2021). Yin Yang 1-induced LINC00667 Up-Regulates Pyruvate Dehydrogenase Kinase 1 to Promote Proliferation, Migration and Invasion of Cholangiocarcinoma Cells by Sponging miR-200c-3p. Hum. Cel 34, 187–200. doi:10.1007/s13577-020-00448-1
Li, J., Jiang, X., Li, C., Liu, Y., Kang, P., Zhong, X., et al. (2019). LncRNA-MEG3 Inhibits Cell Proliferation and Invasion by Modulating Bmi1/RNF2 in Cholangiocarcinoma. J. Cell Physiol. 234, 22947–22959. doi:10.1002/jcp.28856
Li, J., Jiang, X., Li, Z., Huang, L., Ji, D., Yu, L., et al. (2020). SP1-induced HOXD-AS1 Promotes Malignant Progression of Cholangiocarcinoma by Regulating miR-520c-3p/MYCN. Aging 12, 16304–16325. doi:10.18632/aging.103660
Li, O., Jiang, B., Yi, W. M., Zhang, Y., Yang, P. Z., Guo, C., et al. (2021). LncRNA NEAT1 Promotes Cell Proliferation, Migration, and Invasion via the miR -186-5p/PTP4A1 axis in Cholangiocarcinoma. Kaohsiung J. Med. Sci. 37, 379–391. doi:10.1002/kjm2.12354
Li, T., Xie, J., Shen, C., Cheng, D., Shi, Y., Wu, Z., et al. (2016). Upregulation of Long Noncoding RNA ZEB1-AS1 Promotes Tumor Metastasis and Predicts Poor Prognosis in Hepatocellular Carcinoma. Oncogene 35, 1575–1584. doi:10.1038/onc.2015.223
Li, Z., Jiang, X., Huang, L., Li, J., Ji, D., Xu, Y., et al. (2019). Up-regulation of ZFAS1 Indicates Dismal Prognosis for Cholangiocarcinoma and Promotes Proliferation and Metastasis by Modulating USF1 via miR-296-5p. J. Cel Mol Med 23, 8258–8268. doi:10.1111/jcmm.14698
Li, Z., Li, J., Ji, D., Leng, K., Xu, Y., Huang, L., et al. (2018). Overexpressed Long Noncoding RNA Sox2ot Predicts Poor Prognosis for Cholangiocarcinoma and Promotes Cell Proliferation and Invasion. Gene 645, 131–136. doi:10.1016/j.gene.2017.12.017
Li, Z., Li, X., Du, X., Zhang, H., Wu, Z., Ren, K., et al. (2019). The Interaction between lncRNA SNHG1 and miR-140 in Regulating Growth and Tumorigenesis via the TLR4/NF-Κb Pathway in Cholangiocarcinoma. Oncol. Res. 27, 663–672. doi:10.3727/096504018x15420741307616
Liang, W.-C., Ren, J.-L., Wong, C.-W., Chan, S.-O., Waye, M. M.-Y., Fu, W.-M., et al. (2018). LncRNA-NEF Antagonized Epithelial to Mesenchymal Transition and Cancer Metastasis via Cis-Regulating FOXA2 and Inactivating Wnt/β-Catenin Signaling. Oncogene 37, 1445–1456. doi:10.1038/s41388-017-0041-y
Liang, Z., Zhu, B., Meng, D., Shen, X., Li, X., Wang, Z., et al. (2019). Down-regulation of lncRNA-NEF Indicates Poor Prognosis in Intrahepatic Cholangiocarcinoma. Biosci. Rep. 39. doi:10.1042/BSR20181573
Liu, H. P., Lv, D., Wang, J. Y., Zhang, Y., Chang, J. F., Liu, Z. T., et al. (2019). Long Noncoding RNA PCAT-1 Promoted Ovarian Cancer Cell Proliferation and Invasion by Suppressing KLF6. Eur. Rev. Med. Pharmacol. Sci. 23, 4650–4655. doi:10.26355/eurrev_201906_18044
Liu, Y., He, A., Liu, B., Huang, Z., and Mei, H. (2019). Potential Role of lncRNA H19 as a Cancer Biomarker in Human Cancers Detection and Diagnosis: A Pooled Analysis Based on 1585 Subjects. Biomed. Res. Int. 2019, 9056458. doi:10.1155/2019/9056458
Long, J., Xiong, J., Bai, Y., Mao, J., Lin, J., Xu, W., et al. (2019). Construction and Investigation of a lncRNA-Associated ceRNA Regulatory Network in Cholangiocarcinoma. Front. Oncol. 9, 649. doi:10.3389/fonc.2019.00649
Lu, M., Qin, X., Zhou, Y., Li, G., Liu, Z., Geng, X., et al. (2021). Long Non-coding RNA LINC00665 Promotes Gemcitabine Resistance of Cholangiocarcinoma Cells via Regulating EMT and Stemness Properties through miR-424-5p/BCL9L axis. Cel Death Dis 12, 72. doi:10.1038/s41419-020-03346-4
Lu, M., Qin, X., Zhou, Y., Li, G., Liu, Z., Yue, H., et al. (2020). LncRNA HOTAIR Suppresses Cell Apoptosis, Autophagy and Induces Cell Proliferation in Cholangiocarcinoma by Modulating the miR-204-5p/HMGB1 axis. Biomed. Pharmacother. 130, 110566. doi:10.1016/j.biopha.2020.110566
Lv, L., Wei, M., Lin, P., Chen, Z., Gong, P., Quan, Z., et al. (2017). Integrated mRNA and lncRNA Expression Profiling for Exploring Metastatic Biomarkers of Human Intrahepatic Cholangiocarcinoma. Am. J. Cancer Res. 7, 688–699.
Ma, S., Meng, Z., Chen, R., and Guan, K.-L. (2019). The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 88, 577–604. doi:10.1146/annurev-biochem-013118-111829
Macias, R. I. R., Kornek, M., Rodrigues, P. M., Paiva, N. A., Castro, R. E., Urban, S., et al. (2019). Diagnostic and Prognostic Biomarkers in Cholangiocarcinoma. Liver Int. 39 Suppl 1 (Suppl. 1), 108–122. doi:10.1111/liv.14090
Mazzaferro, V., Gorgen, A., Roayaie, S., Droz Dit Busset, M., and Sapisochin, G. (2020). Liver Resection and Transplantation for Intrahepatic Cholangiocarcinoma. J. Hepatol. 72, 364–377. doi:10.1016/j.jhep.2019.11.020
Merdrignac, A., Papoutsoglou, P., and Coulouarn, C. (2021). Long Noncoding RNAs in Cholangiocarcinoma. Hepatology 73, 1213–1226. doi:10.1002/hep.31534
Nagino, M., Ebata, T., Yokoyama, Y., Igami, T., Sugawara, G., Takahashi, Y., et al. (2013). Evolution of Surgical Treatment for Perihilar Cholangiocarcinoma. Ann. Surg. 258, 129–140. doi:10.1097/sla.0b013e3182708b57
Nissan, A., Stojadinovic, A., Mitrani-Rosenbaum, S., Halle, D., Grinbaum, R., Roistacher, M., et al. (2012). Colon Cancer Associated Transcript-1: a Novel RNA Expressed in Malignant and Pre-malignant Human Tissues. Int. J. Cancer 130, 1598–1606. doi:10.1002/ijc.26170
Pan, J., Fang, S., Tian, H., Zhou, C., Zhao, X., Tian, H., et al. (2020). lncRNA JPX/miR-33a-5p/Twist1 axis Regulates Tumorigenesis and Metastasis of Lung Cancer by Activating Wnt/β-Catenin Signaling. Mol. Cancer 19, 9. doi:10.1186/s12943-020-1133-9
Parasramka, M., Yan, I. K., Wang, X., Nguyen, P., Matsuda, A., Maji, S., et al. (2017). BAP1 Dependent Expression of Long Non-coding RNA NEAT-1 Contributes to Sensitivity to Gemcitabine in Cholangiocarcinoma. Mol. Cancer 16, 22. doi:10.1186/s12943-017-0587-x
Peng, L., Liu, Y. H., Nie, S., and Gao, M. (2020). LncRNA CASC2 Inhibits Cell Proliferation, Metastasis and EMT through miR-18a/SOCS5 axis in Cholangiocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 24, 8367–8376. doi:10.26355/eurrev_202008_22633
Prensner, J. R., Iyer, M. K., Balbin, O. A., Dhanasekaran, S. M., Cao, Q., Brenner, J. C., et al. (2011). Transcriptome Sequencing across a Prostate Cancer Cohort Identifies PCAT-1, an Unannotated lincRNA Implicated in Disease Progression. Nat. Biotechnol. 29, 742–749. doi:10.1038/nbt.1914
Qin, W., Kang, P., Xu, Y., Leng, K., Li, Z., Huang, L., et al. (2018). Long Non-coding RNA HOTAIR Promotes Tumorigenesis and Forecasts a Poor Prognosis in Cholangiocarcinoma. Sci. Rep. 8, 12176. doi:10.1038/s41598-018-29737-4
Qiu, G., Ma, D., Li, F., Sun, D., and Zeng, Z. (2019). lnc-PKD2-2-3, Identified by Long Non-coding RNA Expression Profiling, Is Associated with Pejorative Tumor Features and Poor Prognosis, Enhances Cancer Stemness and May Serve as Cancer Stem-Cell Marker in Cholangiocarcinoma. Int. J. Oncol. 55, 45–58. doi:10.3892/ijo.2019.4798
Quinn, J. J., and Chang, H. Y. (2016). Unique Features of Long Non-coding RNA Biogenesis and Function. Nat. Rev. Genet. 17, 47–62. doi:10.1038/nrg.2015.10
Rinn, J. L., and Chang, H. Y. (2020). Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 89, 283–308. doi:10.1146/annurev-biochem-062917-012708
Rizvi, S., and Gores, G. J. (2013). Pathogenesis, Diagnosis, and Management of Cholangiocarcinoma. Gastroenterology 145, 1215–1229. doi:10.1053/j.gastro.2013.10.013
Rizvi, S., Khan, S. A., Hallemeier, C. L., Kelley, R. K., and Gores, G. J. (2018). Cholangiocarcinoma - Evolving Concepts and Therapeutic Strategies. Nat. Rev. Clin. Oncol. 15, 95–111. doi:10.1038/nrclinonc.2017.157
Saha, S. K., Zhu, A. X., Fuchs, C. S., and Brooks, G. A. (2016). Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. The oncologist 21, 594–599. doi:10.1634/theoncologist.2015-0446
Schmitt, A. M., and Chang, H. Y. (2016). Long Noncoding RNAs in Cancer Pathways. Cancer cell 29, 452–463. doi:10.1016/j.ccell.2016.03.010
Shan, L., Liu, W., and Zhan, Y. (2020). Long Non-coding RNA CCAT1 Acts as an Oncogene and Promotes Sunitinib Resistance in Renal Cell Carcinoma. Front. Oncol. 10, 516552. doi:10.3389/fonc.2020.516552
Shen, S., Wang, J., Zheng, B., Tao, Y., Li, M., Wang, Y., et al. (2020). LINC01714 Enhances Gemcitabine Sensitivity by Modulating FOXO3 Phosphorylation in Cholangiocarcinoma. Mol. Ther. - Nucleic Acids 19, 446–457. doi:10.1016/j.omtn.2019.11.028
Shen, X., Shen, P., Yang, Q., Yin, Q., Wang, F., Cong, H., et al. (2019). Knockdown of Long Non-coding RNA PCAT-1 Inhibits Myeloma Cell Growth and Drug Resistance via P38 and JNK MAPK Pathways. J. Cancer 10, 6502–6510. doi:10.7150/jca.35098
Shen, X., Zhang, Y., Wu, X., Guo, Y., Shi, W., Qi, J., et al. (2017). Upregulated lncRNA-PCAT1 Is Closely Related to Clinical Diagnosis of Multiple Myeloma as a Predictive Biomarker in Serum. Cbm 18, 257–263. doi:10.3233/cbm-160158
Shi, J., Li, X., Zhang, F., Kong, L., Zhang, X., Cheng, Y., et al. (2018). The Plasma LncRNA Acting as Fingerprint in Hilar Cholangiocarcinoma. Cell Physiol Biochem 49, 1694–1702. doi:10.1159/000493613
Sirica, A. E., Strazzabosco, M., and Cadamuro, M. (2021). Intrahepatic Cholangiocarcinoma: Morpho-Molecular Pathology, Tumor Reactive Microenvironment, and Malignant Progression. Adv. Cancer Res. 149, 321–387. doi:10.1016/bs.acr.2020.10.005
Siveen, K. S., Prabhu, K., Krishnankutty, R., Kuttikrishnan, S., Tsakou, M., Alali, F. Q., et al. (2017). Vascular Endothelial Growth Factor (VEGF) Signaling in Tumour Vascularization: Potential and Challenges. Curr. Vasc. Pharmacol. 15, 339–351. doi:10.2174/1570161115666170105124038
Smith, A. G., and Macleod, K. F. (2019). Autophagy, Cancer Stem Cells and Drug Resistance. J. Pathol. 247, 708–718. doi:10.1002/path.5222
Su, T., Zhang, S. D., and Zhao, J. (2020). Long Non-coding RNA CCAT1 Regulates the Biological Behavior of Osteosarcoma Cells through the miR-454-3p/ZEB2 axis. Eur. Rev. Med. Pharmacol. Sci. 24, 11016–11025. doi:10.26355/eurrev_202011_23586
Sun, D., Zhao, Y., Wang, W., Guan, C., Hu, Z., Liu, L., et al. (2021). PCAT1 Induced by Transcription Factor YY1 Promotes Cholangiocarcinoma Proliferation, Migration and Invasion by Sponging miR-216a-3p to Up-Regulate Oncogene BCL3. Biol. Chem. 402, 207–219. doi:10.1515/hsz-2020-0276
Sun, S.-C. (2017). The Non-canonical NF-Κb Pathway in Immunity and Inflammation. Nat. Rev. Immunol. 17, 545–558. doi:10.1038/nri.2017.52
Tan, X., Huang, Z., and Li, X. (2017). Long Non-Coding RNA MALAT1 Interacts with miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J. Cel. Biochem. 118, 3643–3653. doi:10.1002/jcb.25862
Tian, D., Wei, X., Zhu, H., Zhu, L., Li, T., and Li, W. (2019). LncRNA-SNHG3 Is an Independent Prognostic Biomarker of Intrahepatic Cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 12, 2706–2712.
Tshering, G., Dorji, P. W., Chaijaroenkul, W., and Na-Bangchang, K. (2018). Biomarkers for the Diagnosis of Cholangiocarcinoma: A Systematic Review. Am. J. Trop. Med. Hyg. 98, 1788–1797. doi:10.4269/ajtmh.17-0879
Tu, J., Wu, F., Chen, L., Zheng, L., Yang, Y., Ying, X., et al. (2020). Long Non-coding RNA PCAT6 Induces M2 Polarization of Macrophages in Cholangiocarcinoma via Modulating miR-326 and RhoA-ROCK Signaling Pathway. Front. Oncol. 10, 605877.
Valle, J., Wasan, H., Palmer, D. H., Cunningham, D., Anthoney, A., Maraveyas, A., et al. (2010). Cisplatin Plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. N. Engl. J. Med. 362, 1273–1281. doi:10.1056/nejmoa0908721
Valle, J. W., Kelley, R. K., Nervi, B., Oh, D.-Y., and Zhu, A. X. (2021). Biliary Tract Cancer. The Lancet 397, 428–444. doi:10.1016/s0140-6736(21)00153-7
van der Gaag, N. A., Kloek, J. J., de Bakker, J. K., Musters, B., Geskus, R. B., Busch, O. R. C., et al. (2012). Survival Analysis and Prognostic Nomogram for Patients Undergoing Resection of Extrahepatic Cholangiocarcinoma. Ann. Oncol. 23, 2642–2649. doi:10.1093/annonc/mds077
Van Dyke, A. L., Shiels, M. S., Jones, G. S., Pfeiffer, R. M., Petrick, J. L., Beebe-Dimmer, J. L., et al. (1999). Biliary Tract Cancer Incidence and Trends in the United States by Demographic Group, 1999-2013. Cancer 125, 1489–1498. doi:10.1002/cncr.31942
Varamo, C., Peraldo-Neia, C., Ostano, P., Basiricò, M., Raggi, C., Bernabei, P., et al. (2019). Establishment and Characterization of a New Intrahepatic Cholangiocarcinoma Cell Line Resistant to Gemcitabine. Cancers (Basel) 11. doi:10.3390/cancers11040519
Wang, C., Mao, Z. p., Wang, L., Wu, G. h., Zhang, F. h., Wang, D. y., et al. (2017). Long Non-coding RNA MALAT1 Promotes Cholangiocarcinoma Cell Proliferation and Invasion by Activating PI3K/Akt Pathway. neo 64, 725–731. doi:10.4149/neo_2017_510
Wang, H., Wang, L., Tang, L., Luo, J., Ji, H., Zhang, W., et al. (2020). Long Noncoding RNA SNHG6 Promotes Proliferation and Angiogenesis of Cholangiocarcinoma Cells through Sponging miR-101-3p and Activation of E2F8. J. Cancer 11, 3002–3012. doi:10.7150/jca.40592
Wang, J., Sun, N., Han, W., Tong, L., Xu, T., and Li, G. (2021). Long Non-coding RNA CCAT1 Sponges miR-490 to Enhance Cell Proliferation and Migration of Non-small Cell Lung Cancer. Thorac. Cancer 12, 364–371. doi:10.1111/1759-7714.13758
Wang, N., Zhang, C., Wang, W., Liu, J., Yu, Y., Li, Y., et al. (2019). Long Noncoding RNA DANCR Regulates Proliferation and Migration by Epigenetically Silencing FBP1 in Tumorigenesis of Cholangiocarcinoma. Cel Death Dis 10, 585. doi:10.1038/s41419-019-1810-z
Wang, W.-T., Chen, T.-Q., Zeng, Z.-C., Pan, Q., Huang, W., Han, C., et al. (2020). The lncRNA LAMP5-AS1 Drives Leukemia Cell Stemness by Directly Modulating DOT1L Methyltransferase Activity in MLL Leukemia. J. Hematol. Oncol. 13, 78. doi:10.1186/s13045-020-00909-y
Wang, W.-T., Ye, H., Wei, P.-P., Han, B.-W., He, B., Chen, Z. H., et al. (2016). LncRNAs H19 and HULC, Activated by Oxidative Stress, Promote Cell Migration and Invasion in Cholangiocarcinoma through a ceRNA Manner. J. Hematol. Oncol. 9, 117. doi:10.1186/s13045-016-0348-0
Wang, X. P., Song, J., Liu, G. T., Wang, J. J., and Guo, H. F. (2018). Upregulation of Gastric Adenocarcinoma Predictive Long Intergenic Non-coding RNA Promotes Progression and Predicts Poor Prognosis in Perihilar Cholangiocarcinoma. Oncol. Lett. 16, 3964–3972. doi:10.3892/ol.2018.9137
Weigt, J., and Malfertheiner, P. (2010). Cisplatin Plus Gemcitabine versus Gemcitabine for Biliary Tract Cancer. Expert Rev. Gastroenterol. Hepatol. 4, 395–397. doi:10.1586/egh.10.45
Wen, J., Xu, J., Sun, Q., Xing, C., and Yin, W. (2016). Upregulation of Long Non Coding RNA PCAT-1 Contributes to Cell Proliferation, Migration and Apoptosis in Hepatocellular Carcinoma. Mol. Med. Rep. 13, 4481–4486. doi:10.3892/mmr.2016.5075
Whiteside, T. L. (2008). The Tumor Microenvironment and its Role in Promoting Tumor Growth. Oncogene 27, 5904–5912. doi:10.1038/onc.2008.271
Wu, G., Xue, M., Zhao, Y., Han, Y., Li, C., Zhang, S., et al. (2020). Long Noncoding RNA ZEB1-AS1 Acts as a Sponge of miR-141-3p to Inhibit Cell Proliferation in Colorectal Cancer. Int. J. Med. Sci. 17, 1589–1597. doi:10.7150/ijms.46698
Wu, Y., Yang, X., Chen, Z., Tian, L., Jiang, G., Chen, F., et al. (2019). m6A-induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1A-Induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1. Mol. Cancer 18, 87. doi:10.1186/s12943-019-1014-2
Xia, L., Chen, X., Yang, J., Zhu, S., Zhang, L., Yin, Q., et al. (2020). Long Non-coding RNA-PAICC Promotes the Tumorigenesis of Human Intrahepatic Cholangiocarcinoma by Increasing YAP1 Transcription. Front. Oncol. 10, 595533. doi:10.3389/fonc.2020.595533
Xia, X. L., Xue, D., Xiang, T. H., Xu, H. Y., Song, D. K., Cheng, P. G., et al. (2018). Overexpression of Long Non-coding RNA CRNDE Facilitates Epithelial-Mesenchymal Transition and Correlates with Poor Prognosis in Intrahepatic Cholangiocarcinoma. Oncol. Lett. 15, 4105–4112. doi:10.3892/ol.2018.7815
Xie, X., Wang, Y., Zhang, S., Li, J., Yu, Z., Ding, X., et al. (2021). A Novel Five-lncRNA Signature Panel Improves High-Risk Survival Prediction in Patients with Cholangiocarcinoma. Aging 13, 2959–2981. doi:10.18632/aging.202446
Xu, R., Rai, A., Chen, M., Suwakulsiri, W., Greening, D. W., and Simpson, R. J. (2018). Extracellular Vesicles in Cancer - Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol. 15, 617–638. doi:10.1038/s41571-018-0036-9
Xu, W., Chang, J., Du, X., and Hou, J. (2017). Long Non-coding RNA PCAT-1 Contributes to Tumorigenesis by Regulating FSCN1 via miR-145-5p in Prostate Cancer. Biomed. Pharmacother. 95, 1112–1118. doi:10.1016/j.biopha.2017.09.019
Xu, Y., Jiang, X., and Cui, Y. (2017). Upregulated Long Noncoding RNA PANDAR Predicts an Unfavorable Prognosis and Promotes Tumorigenesis in Cholangiocarcinoma. Ott 10, 2873–2883. doi:10.2147/ott.s137044
Xu, Y., Leng, K., Li, Z., Zhang, F., Zhong, X., Kang, P., et al. (2017). The Prognostic Potential and Carcinogenesis of Long Non-coding RNA TUG1 in Human Cholangiocarcinoma. Oncotarget 8, 65823–65835. doi:10.18632/oncotarget.19502
Xu, Y., Wang, Z., Jiang, X., and Cui, Y. (2017). Overexpression of Long Noncoding RNA H19 Indicates a Poor Prognosis for Cholangiocarcinoma and Promotes Cell Migration and Invasion by Affecting Epithelial-Mesenchymal Transition. Biomed. Pharmacother. 92, 17–23. doi:10.1016/j.biopha.2017.05.061
Xu, Y., Yao, Y., Jiang, X., Zhong, X., Wang, Z., Li, C., et al. (2018). SP1-induced Upregulation of lncRNA SPRY4-IT1 Exerts Oncogenic Properties by Scaffolding EZH2/LSD1/DNMT1 and Sponging miR-101-3p in Cholangiocarcinoma. J. Exp. Clin. Cancer Res. 37, 81. doi:10.1186/s13046-018-0747-x
Xu, Y., Yao, Y., Leng, K., Li, Z., Qin, W., Zhong, X., et al. (2017). Long Non-coding RNA UCA1 Indicates an Unfavorable Prognosis and Promotes Tumorigenesis via Regulating AKT/GSK-3β Signaling Pathway in Cholangiocarcinoma. Oncotarget 8, 96203–96214. doi:10.18632/oncotarget.21884
Xu, Y., Yao, Y., Qin, W., Zhong, X., Jiang, X., and Cui, Y. (2018). Long Non-coding RNA CCAT2 Promotes Cholangiocarcinoma Cells Migration and Invasion by Induction of Epithelial-To-Mesenchymal Transition. Biomed. Pharmacother. 99, 121–127. doi:10.1016/j.biopha.2018.01.061
Yu, A., Zhao, L., Kang, Q., Li, J., Chen, K., and Fu, H. (2020). SOX2 Knockdown Slows Cholangiocarcinoma Progression through Inhibition of Transcriptional Activation of lncRNA PVT1. Biochem. J. 477, 3527–3540. doi:10.1042/bcj20200219
Yu, A., Zhao, L., Kang, Q., Li, J., Chen, K., and Fu, H. (2020). Transcription Factor HIF1α Promotes Proliferation, Migration, and Invasion of Cholangiocarcinoma via Long Noncoding RNA H19/microRNA-612/Bcl-2 axis. Translational Res. 224, 26–39. doi:10.1016/j.trsl.2020.05.010
Yu, J., Zhang, B., Zhang, H., Qi, Y., Wang, Y., Wang, W., et al. (2019). E2F1-induced Upregulation of Long Non-coding RNA LMCD1-AS1 Facilitates Cholangiocarcinoma Cell Progression by Regulating miR-345-5p/COL6A3 Pathway. Biochem. biophysical Res. Commun. 512, 150–155. doi:10.1016/j.bbrc.2019.03.054
Yu, Y., Zhang, M., Liu, J., Xu, B., Yang, J., Wang, N., et al. (2018). Long Non-coding RNA PVT1 Promotes Cell Proliferation and Migration by Silencing ANGPTL4 Expression in Cholangiocarcinoma. Mol. Ther. - Nucleic Acids 13, 503–513. doi:10.1016/j.omtn.2018.10.001
Zhan, Y., Du, L., Wang, L., Jiang, X., Zhang, S., Li, J., et al. (2018). Expression Signatures of Exosomal Long Non-coding RNAs in Urine Serve as Novel Non-invasive Biomarkers for Diagnosis and Recurrence Prediction of Bladder Cancer. Mol. Cancer 17, 142. doi:10.1186/s12943-018-0893-y
Zhang, B., Zhou, M., Zou, L., Miao, J., Wang, Y., Li, Y., et al. (2019). Long Non-coding RNA LOXL1-AS1 Acts as a ceRNA for miR-324-3p to Contribute to Cholangiocarcinoma Progression via Modulation of ATP-Binding Cassette Transporter A1. Biochem. biophysical Res. Commun. 513, 827–833. doi:10.1016/j.bbrc.2019.04.089
Zhang, C., Li, J.-Y., Tian, F.-Z., Zhao, G., Hu, H., Ma, Y.-F., et al. (2018). Long Noncoding RNA NEAT1 Promotes Growth and Metastasis of Cholangiocarcinoma Cells. Oncol. Res. 26, 879–888. doi:10.3727/096504017x15024935181289
Zhang, D., Li, H., Xie, J., Jiang, D., Cao, L., Yang, X., et al. (2018). Long Noncoding RNA LINC01296 Promotes Tumor Growth and Progression by Sponging miR-5095 in Human Cholangiocarcinoma. Int. J. Oncol. 52, 1777–1786. doi:10.3892/ijo.2018.4362
Zhang, F., Wan, M., Xu, Y., Li, Z., Leng, K., Kang, P., et al. (2017). Long Noncoding RNA PCAT1 Regulates Extrahepatic Cholangiocarcinoma Progression via the Wnt/β-Catenin-Signaling Pathway. Biomed. Pharmacother. 94, 55–62. doi:10.1016/j.biopha.2017.07.025
Zhang, L.-l., Zhang, L.-f., Guo, X.-h., Zhang, D.-z., Yang, F., and Fan, Y.-y. (2018). Downregulation of miR-335-5p by Long Noncoding RNA ZEB1-AS1 in Gastric Cancer Promotes Tumor Proliferation and Invasion. DNA Cel. Biol. 37, 46–52. doi:10.1089/dna.2017.3926
Zhang, S., Du, L., Wang, L., Jiang, X., Zhan, Y., Li, J., et al. (2019). Evaluation of Serum Exosomal LncRNA-Based Biomarker Panel for Diagnosis and Recurrence Prediction of Bladder Cancer. J. Cel Mol Med 23, 1396–1405. doi:10.1111/jcmm.14042
Zhang, S., Xiao, J., Chai, Y., Du, Y. y., Liu, Z., Huang, K., et al. (2017). LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma through Suppressing miR-152. Dig. Dis. Sci. 62, 3050–3058. doi:10.1007/s10620-017-4759-8
Zhang, Y., Zhang, L., Lu, S., Xiang, Y., Zeng, C., He, T., et al. (2021). Long Non-coding RNA CASC15 Promotes Intrahepatic Cholangiocarcinoma Possibly through Inducing PRDX2/PI3K/AKT Axis. Cancer Res. Treat. 53, 184–198. doi:10.4143/crt.2020.192
Zhao, Y., Wang, N., Zhang, X., Liu, H., and Yang, S. (2019). LncRNA ZEB1-AS1 Down-regulation Suppresses the Proliferation and Invasion by Inhibiting ZEB1 Expression in Oesophageal Squamous Cell Carcinoma. J. Cel Mol Med 23, 8206–8218. doi:10.1111/jcmm.14692
Zhu, C. Y., Fan, C. R., Zhang, Y. L., Sun, Q. X., Yan, M. J., Wei, W., et al. (2020). LncRNA DANCR Affected Cell Growth, EMT and Angiogenesis by Sponging miR-345-5p through Modulating Twist1 in Cholangiocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 24, 2321–2334. doi:10.26355/eurrev_202003_20498
Zhu, H., Zhai, B., He, C., Li, Z., Gao, H., Niu, Z., et al. (2020). LncRNA TTN-AS1 Promotes the Progression of Cholangiocarcinoma via the miR-320a/neuropilin-1 axis. Cel Death Dis 11, 637. doi:10.1038/s41419-020-02896-x
Zhuo, W., Liu, Y., Li, S., Guo, D., Sun, Q., Jin, J., et al. (2019). Long Noncoding RNA GMAN, Up-Regulated in Gastric Cancer Tissues, Is Associated with Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology 156, 676–691. doi:10.1053/j.gastro.2018.10.054
Keywords: cholangiocarcinoma, lncRNA, biomarker, diagnosis, prognosis, therapeutic target
Citation: Wu Y, Hayat K, Hu Y and Yang J (2022) Long Non-Coding RNAs as Molecular Biomarkers in Cholangiocarcinoma. Front. Cell Dev. Biol. 10:890605. doi: 10.3389/fcell.2022.890605
Received: 06 March 2022; Accepted: 10 April 2022;
Published: 27 April 2022.
Edited by:
Ye Tian, Northwestern Polytechnical University, ChinaReviewed by:
Jingjia Li, Affiliated Hospital of North Sichuan Medical College, ChinaLulu Chen, Zigong First People’s Hospital, China
Ji Cao, Zhejiang University, China
Copyright © 2022 Wu, Hayat, Hu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Yang, yjf3303@zju.edu.cn
 Yanhua Wu
Yanhua Wu Khizar Hayat
Khizar Hayat Yufei Hu
Yufei Hu Jianfeng Yang
Jianfeng Yang