- Genómica Funcional y Ciencia de Datos, Instituto de Investigaciones en Medicina Traslacional (IIMT), CONICET-Universidad Austral, Buenos Aires, Argentina
Ubiquitination and ubiquitin-like post-translational modifications control the activity and stability of different tumor suppressors and oncoproteins. Hence, regulation of this enzymatic cascade offers an appealing scenario for novel antineoplastic targets discovery. Among the different families of enzymes that participate in the conjugation of Ubiquitin, deubiquitinating enzymes (DUBs), responsible for removing ubiquitin or ubiquitin-like peptides from substrate proteins, have attracted increasing attention. In this regard, increasing evidence is accumulating suggesting that the modulation of the catalytic activity of DUBs represents an attractive point of therapeutic intervention in cancer treatment. In particular, different lines of research indicate that USP19, a member of the DUBs, plays a role in the control of tumorigenesis and cancer dissemination. This review aims at summarizing the current knowledge of USP19 wide association with the control of several cellular processes in different neoplasms, which highlights the emerging role of USP19 as a previously unrecognized prognosis factor that possesses both positive and negative regulation activities in tumor biology. These observations indicate that USP19 might represent a novel putative pharmacologic target in oncology and underscores the potential of identifying specific modulators to test in clinical settings.
Introduction
Following translation, proteins can undergo several posttranslational modifications (PTMs) to modulate their activity, such as phosphorylation, methylation, glycosylation, acetylation, sumoylation and ubiquitination. These modifications represent a very important component in the physiological regulation of different pathways, including protein degradation, DNA repair activity, gene regulation and signal transduction, among others (Millar et al., 2019). Since growth regulatory proteins that drive tumorigenesis are modified by PTMs (Krueger and Srivastava, 2006), understating the mechanisms by which these modifications regulate oncogenic, or tumor suppressive pathways is of great relevance to restrain their effects upon pathological scenarios (Konstantinopoulos et al., 2007).
Moreover, the alteration in the levels and functionality of the components comprising the pathways responsible for the different PTMs, is related to different pathologies, including cancer (Xu et al., 2018; Sharma et al., 2019; Chen et al., 2020; Vellosillo and Minguez, 2021). In particular, ubiquitin-related PTMs are under active study as their dysregulation has been linked with the onset and progression of different oncological disorders (Reinstein and Ciechanover, 2006; Shi and Grossman, 2010).
Ubiquitination
Ubiquitination is the covalent attachment of ubiquitin (an 8-kDa 76 amino-acid molecule) to target proteins, and it plays crucial roles in the regulation of target proteins activity, stability, subcellular localization and trafficking, and interaction with other proteins (Damgaard, 2021). Therefore, this modification affects a great number of biological processes (Mevissen and Komander, 2017).
Protein ubiquitination is a tightly regulated process which involves the activity of two groups of enzymes, namely, E1/E2/E3 ligases and deubiquitinating enzymes (DUBs) (Figure 1).
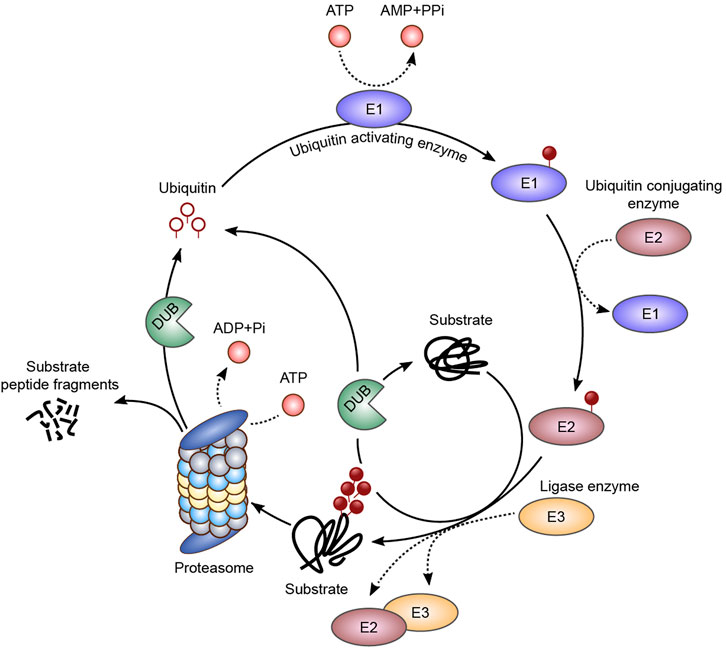
FIGURE 1. Ubiquitination pathway. The ubiquitin molecule is activated by an E1 ubiquitin activating enzyme, in an ATP-dependent step, and a thioester intermediate is formed (E1-S-ubiquitin). The ubiquitin molecule is then transferred to an E2 conjugating enzyme (E2-S-ubiquitin), and then to the final substrate by an E3 ligase. Ubiquitin bound as monomers or polymers with different topologies are associated with different biological outputs, such as regulation of enzymatic activity, localization, protein-protein interactions, among others. Sequential ubiquitin conjugations form a polyubiquitin chain on the substrate, which can be recognized and degraded by the 26S proteasome. The deubiquitinating enzymes (DUBs) are responsible for the ubiquitin molecules recycling and chain editing.
The attachment of ubiquitination moieties to target proteins is catalyzed by the sequential action of a ubiquitin ATP-dependent activating enzyme (E1), which transfers the ubiquitin molecule to a ubiquitin conjugating enzyme (E2) by trans-thiolation, and by a ubiquitin ligase (E3), which provide substrate specificity to ubiquitin conjugation (Ciechanover, 1994; Hershko and Ciechanover, 1998; Komander and Rape, 2012).
This modification can occur as ubiquitin monomers or polymer chains, and since the ubiquitin molecule contains eight ubiquitination sites (seven internal lysine residues -Lys 6, 11, 27, 29, 33, 48 and 63- and a primary amine at the N-terminus), various types of ubiquitin chains with different length and shape might form (Akutsu et al., 2016; Yau and Rape, 2016; Dwane et al., 2017; Kwon and Ciechanover, 2017; Ohtake and Tsuchiya, 2017).
Furthermore, the ubiquitin molecule is subject to other PTMs such as phosphorylation, acetylation (Ohtake et al., 2015; Wauer et al., 2015; Huguenin-Dezot et al., 2016), and modification with ubiquitin-like proteins such as interferon (IFN)-stimulated gene 15 (ISG15) (Fan et al., 2015) and small ubiquitin-related modifier (SUMO) (Lamoliatte et al., 2013). Therefore, these modifications broaden the ubiquitin code versatility, as they affect not only ubiquitin interactions but also the formation and topology of the polyubiquitin chain.
The nature of the ubiquitin chain determines the outcome of the substrate protein, and different molecular signals are induced in the cell (Ikeda and Dikic, 2008; Sadowski and Sarcevic, 2010), affecting biological processes such as protein stability through proteasome degradation, DNA repair and replication, signal transduction, gene regulation, molecule trafficking and endocytosis, etc. (Hershko and Ciechanover, 1998; Haglund and Dikic, 2005; Komander and Rape, 2012; Yau and Rape, 2016).
The deubiquitinating enzymes are proteases that reverse the modification of proteins by a single ubiquitin or ubiquitin-like protein, and remodel polyubiquitin/ubiquitin-like chains on target proteins. They hydrolyze the isopeptide bond between the ubiquitin and the substrate residue of either the target protein or another ubiquitin molecule (Komander et al., 2009; Komander and Rape, 2012). The human genome encodes nearly 100 DUBs, each with distinct substrate specificities and catalytic properties, which confer high precision upon ubiquitin chains processing (Komander et al., 2009; Mevissen and Komander, 2017). Consequently, individual DUBs likely confer specific actions (Komander et al., 2009; Huang and Dixit, 2016) and pharmacological modulation of their catalytic activity should lead to desired outcomes upon physiological or pharmacological scenarios.
Based on sequence and structural similarities, DUBs have been classified into seven families: Ubiquitin-specific proteases (USPs), Ubiquitin C-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph (Josephin) domain (MJD) proteases, Jab1/MPN domain-associated metallo-iso-peptidases (JAMM/MPM+), Zinc finger UB-specific proteases (ZUP/ZUFSP), and monocyte chemotactic protein-induced proteins (MCPIP). Except for the JAMMs, which are zinc-dependent metalloproteases, the remaining families are cysteine proteases (Reyes-Turcu et al., 2009; Hanpude et al., 2015; Mevissen and Komander, 2017; Kwasna et al., 2018).
General Properties of USP19
Human ubiquitin-specific protease 19 (USP19) is a modular deubiquitinating enzyme that belongs to the largest family of DUBs, the USPs (Nijman et al., 2005; Reyes-Turcu et al., 2009). This family is characterized by the presence of a highly conserved USP catalytic domain fold (Hu et al., 2002; Hu et al., 2005; Avvakumov et al., 2006; Renatus et al., 2006; Komander et al., 2008), which holds two well-conserved motifs (Cys and His boxes), each containing the critical residues for the enzymatic activity. Moreover, USP19 contains two CHORD-SGT1/P23 domains (namely CS1 and CS2) at its N-terminus, which are relevant for the interaction with other proteins, as well as for the intra-molecular inhibition and regulation of the catalytic core (Xue et al., 2020) (Figure 2).
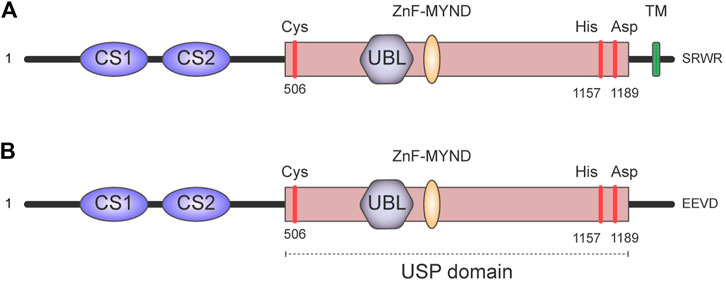
FIGURE 2. Domain architecture of USP19. It contains two CHORD-SGT1 domains (namely CS1 and CS2) at its N-terminus and a large USP domain with a ubiquitin-like domain (UBL) and myeloid translocation protein 8, Nervy protein, Deaf-1 zinc finger (MYND Zn-finger). The positions of the amino acids Cys, His, and Asp in the catalytic triad are indicated in red. There are multiple USP19 isoforms generated by alternative splicing. In particular, alternative splicing of the last exon generates isoforms with a cytoplasmic localization or isoforms anchored to the endoplasmic reticulum. This schematic depicts: (A) the USP19 isoform that contains a transmembrane (TM) domain which anchors USP19 to the endoplasmic reticulum. (B) The soluble USP19 isoform has a relatively hydrophilic region and an EEVD motif in the C-terminus instead.
USP19 presents different isoforms generated by alternative splicing, and the most distinctive feature—structurally and functionally–is that some of them have a cytoplasmic localization, while others have a transmembrane domain that serves as anchorage to the endoplasmic reticulum (Hassink et al., 2009) (Figure 2).
Like other DUBs, USP19 is covalently modified by PTMs such as phosphorylation and ubiquitination, which affect its activity and half-life, respectively (Matsuoka et al., 2007; Velasco et al., 2013).
Functionally, USP19 has mainly been associated with protein quality control and cellular homeostasis (Hassink et al., 2009; Lee et al., 2014; Wiles et al., 2015; He et al., 2016; He et al., 2017), muscle development (Combaret et al., 2005; Sundaram et al., 2009; Wiles et al., 2015), and it has been shown that it controls the half-life of several proteins such as HIF1-α (Altun et al., 2012), BECN1 (Cui et al., 2016), TGFßRI (Zhang et al., 2012), TRAF3 (Gu et al., 2017), HRD1 (Harada et al., 2016), TAK1 (Lei et al., 2019), KPC1 (Lu et al., 2009), c-IAPs one and 2 (Mei et al., 2011), HDAC1/2 (Wu et al., 2017), COROA2 (Lim et al., 2016), LRP6 (Perrody et al., 2016) and MARCH6 (Nakamura et al., 2014), therefore affecting cellular processes relevant in tumorigenesis such as DNA damage repair, apoptosis, the TGF-β Pathway, hypoxia and angiogenesis, immunity, proliferation, ERAD and autophagy.
The Role of USP19 in Cancer Malignancy
Disrupted regulation of protein ubiquitination is a trigger of cancer, among other diseases. Not surprisingly, alterations in the levels of the ubiquitination cascade components -including the DUBs-have been associated with multiple neoplasms (Shi and Grossman, 2010; Deng et al., 2020; Sun et al., 2020).
In the last couple of years, increasing evidence has begun to demonstrate that USP19 is associated with tumor progression and that it represents a novel prognostic factor for the outcome of several malignant diseases. In particular, it has been shown that USP19 plays both positive and negative roles in the onset and development of diverse neoplasms, in a tissue-specific manner. Consequently, in the following paragraphs, results denoting USP19 relevance in different signaling pathways regulating cell proliferation and cell-cycle progression, as well as tumor growth and metastasis will be presented, therefore unveiling the importance of conducting extensive studies to further the study of USP19’s dual role in tumorigenesis under different molecular scenarios, and to establish its significance as a potential new target for the clinical treatment of cancer.
USP19 as a DUB Negatively Regulating Tumorigenesis
A couple of recent papers presented results indicating that USP19 negatively affected proliferation and migration in clear cell renal cell (Hu et al., 2020) and serous ovarian carcinomas (Kang et al., 2021).
Hu and others utilized clear cells renal cancer (ccRCC) cell lines in vitro and demonstrated that overexpression of USP19 levels negatively affected migration and proliferation, and the opposite occurred upon USP19 silencing. They validated their results using in vivo models and observed that USP19 downregulation promoted tumor growth in a xenograft model. Moreover, they conducted in silico analyses and observed that USP19 mRNA levels were significantly lower in ccRCC than normal tissues, and that low USP19 expression was associated with disease progression and poor prognostic outcomes in a The Cancer Genome Atlas (TCGA) cohort of patients (Hu et al., 2020). These results were consistent with a previous work by Liu and collaborators, who performed an in silico analysis and observed that isoform uc003cvz.3, which is mainly localized in the cytoplasm, serves as an indicator of poor outcome in patients with advanced stage ccRCC (Liu et al., 2013).
Similarly, Kang et al. applied a machine learning model on RNA-sequencing data from 51 patients who received conventional therapies for high-grade serous ovarian carcinoma (HGSC) and identified USP19 and RPL23 as candidate prognostic markers. Specifically, they showed that patients with lower USP19 or higher RPL23 mRNA levels had worse prognoses and they validated their model using publicly available data from the TCGA (Kang et al., 2021). They also observed that USP19 levels positively correlated with TOP3B and XRN2, which regulate genome instability (Kang et al., 2021). Based on this observation, and considering that USP19 interacts with and deubiquitinates HDAC1/2 in order to regulate DNA damage repair and chromosomal stability (Wu et al., 2017) and that both ccRCC and HGSC are characterized by high genomic instability, it is plausible that USP19-mediated deubiquitination of key regulators associated with DSB repair or genome instability might be responsible for the worse prognosis observed in ccRCC and HGSC patients with low USP19 levels (Kang et al., 2021).
In addition, Shahriyari L and collaborators (Shahriyari et al., 2019) described the existence of a correlation between the expression of USP19, RBM15B and the tumor suppressor gene BAP1 (BRCA1 associated protein-1) in different type of cancers. All three genes are in proximity of the 3p21 tumor suppressor region, which is commonly altered in many cancers, suggesting that USP19 could play a functional role in BAP1 molecular mechanism of action or its alteration could be a byproduct of chromosomal rearrangement affecting other genes. Although further characterization is required, this observation highlights the potential of USP19 as a putative prognostic biomarker in different cancers.
USP19 Positively Regulates Tumor Growth and Metastasis
Opposite to the role of USP19 as a tumor suppressor, recent work has also established that antagonism of USP19 expression conferred a prominent antiproliferative and antitumorigenic response in diverse neoplasms: Ewing sarcoma, gastric, breast and colorectal cancers (Gierisch et al., 2019; Dong et al., 2020; Rossi et al., 2021; Zhu et al., 2021), suggesting pro-tumorigenic roles in these tissues.
Ewing sarcoma is the second most common pediatric bone and soft tissue tumor, which is characterized by the presence of a chimeric oncoprotein, EWS-FLI1, due to a genetic translocation between chromosomes 22 and 11 (Desmaze et al., 1997). Gierisch and collaborators demonstrated that this protein, which maintains tumor cells survival, is regulated by USP19 in a post translational manner, and dependent on its catalytic activity (Gierisch et al., 2019). Downregulation of USP19 levels resulted in a reduction of EWS-FLI1 levels, hence decreasing tumor cells growth and colony formation capability, whereas the opposite occurred upon USP19 (TM isoform) overexpression. Using in vivo experiments, the authors demonstrated that tumor growth was delayed when USP19 levels were reduced.
On the other hand, Dong and others analyzed USP19 relevance in gastric cancer (Dong et al., 2020). Their results revealed that USP19 TM isoform overexpression enhanced cell proliferation and exhibited anti-apoptotic properties, as well as it increased cells migration and spreading capabilities in vitro; the opposite was observed upon USP19 silencing (multiple isoforms). Furthermore, they showed that increased USP19 (TM isoform) levels enhanced MMP2/MMP9 protein expression and enzyme activity, and that genetic alteration of USP19 levels affected tumorigenesis using in vivo models. Finally, using a cohort of 212 gastric cancer patients, the authors observed that USP19 expression was significantly increased in gastric cancer tissues, compared to normal gastric tissues, and the high level of USP19 expression was positively correlated with a poorer prognosis.
Similarly, our group analyzed USP19 clinical significance in breast cancer (Rossi et al., 2021). We demonstrated that USP19 positively regulates breast tumor cells migration and invasion in vitro, and that genetic silencing reduces cells motility, whereas its overexpression increases migratory and invasive capabilities—dependent on USP19’s catalytic activity and ER localization. Our results also indicated that USP19 does not affect breast cancer cells proliferation in two dimensions, in concordance with Lu and collaborators (Lu et al., 2011), but significantly modulates proliferation and invasion if cells are grown embedded in extracellular matrix proteins and basement membrane proteins. In vivo experiments showed that USP19 silencing reduces tumorigenicity and delays tumor onset and growth, and the opposite was observed upon wild type USP19 overexpression (but not when overexpressing a catalytically dead mutant, or a cytoplasmic version of USP19). Using experimental metastasis assays, we verified that USP19 silencing reduces cells’ ability to engraft in secondary tissues, and using in silico approaches and TCGA data, we demonstrated that the Wnt pathway is activated in patient samples expressing high levels of USP19. In concordance with these results, we observed a positive correlation between USP19 and LRP6 levels (a Wnt pathway coreceptor). Functional analysis on USP19 overexpressing cells indicated that LRP6 silencing reverted migratory and invasive phenotypes, possibly as a downstream USP19 effector. Finally, we conducted a retrospective analysis on early breast cancer patients which revealed that USP19 expression levels correlated with poor outcome and reduced distant metastasis free survival, hence serving as a prognostic factor in early breast cancer patients.
Lastly, a very recent publication by Zhu and collaborators studied USP19 pertinence in colorectal carcinogenesis (Zhu et al., 2021). Their work showed that ERK2 signaling is responsible for lipid synthesis mediated by cytoplasmic-localized malic enzyme 1 (ME1) phosphorylation, which is overexpressed in a variety of cancers (including colorectal cancer). USP19-mediated ME1 stabilization is enhanced by phosphorylation, generating oncogenic phenotypes, and either USP19 deletion or a point mutation in ME1 protein that prevents ubiquitination, represses colorectal carcinogenesis. Of note, USP19 catalytic activity is necessary to ensure ME1 stabilization. Finally, the authors showed that the USP19-ME1 signaling axis is dysregulated in human colorectal cancer samples, and that USP19 is upregulated during colorectal carcinogenesis pathogenesis and spontaneous tumor development.
Supplementary Table S1 summarizes USP19 relevance in different cancers, and whether is catalytic activity or specific isoform is important in each type of neoplasm.
Concluding Remarks
Various studies have linked USP19 to different cancers, and either its overexpression or silencing may dysregulate the function of several proteins with oncogenic or tumor-suppressive properties, which in the long run may impact on the onset and development of tumors. Since USP19 has different isoforms, and divergent effects have been observed in different cancers, it is plausible to assume that this difference could be explained by the effect these isoforms exert on differing substrates. Moreover, USP19 is a fundamental deubiquitinase with pivotal roles in several cellular processes related to tumorigenesis, including DNA damage repair (Wu et al., 2017), apoptosis (Mei et al., 2011), the TGF-β Pathway (Zhang et al., 2012), hypoxia and angiogenesis (Altun et al., 2012; Boscaro et al., 2020), immunity (Cui et al., 2016; Jin et al., 2016; Gu et al., 2017; Lei et al., 2019; Wu et al., 2019; Liu et al., 2021), proliferation (Lu et al., 2009), ERAD (Hassink et al., 2009) and autophagy (Cui et al., 2016). Given its versatility, USP19’s role on tumorigenesis and metastasis might also be determined by a combinatorial effect on diverse signaling pathways rather than a specific substrate. In this respect, more studies should be performed to analyze the association of USP19 with cancer-related signaling pathways and putative targets, regulatory mechanisms affecting its expression and to search for molecular alterations shared by tumors across different tissues and new targets to better understand how USP19 is affecting cell survival and cellular homeostasis.
Taken together, the findings described here implicate USP19 as a previously unrecognized target for the development of novel therapeutic alternatives for cancer treatments.
Author Contributions
FR and MR designed the article, and acquired, analyzed, and interpreted data for writing the article.
Funding
This work was supported by the grants UA2020 from Universidad Austral awarded to FR and MR. MR was funded by grant PICT 2018-03688 from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.889166/full#supplementary-material
References
Akutsu, M., Dikic, I., and Bremm, A. (2016). Ubiquitin Chain Diversity at a Glance. J. Cell Sci. 129, 875–880. doi:10.1242/jcs.183954
Altun, M., Zhao, B., Velasco, K., Liu, H., Hassink, G., Paschke, J., et al. (2012). Ubiquitin-specific Protease 19 (USP19) Regulates Hypoxia-Inducible Factor 1α (HIF-1α) during Hypoxia. J. Biol. Chem. 287, 1962–1969. doi:10.1074/jbc.m111.305615
Avvakumov, G. V., Walker, J. R., Xue, S., Finerty, P. J., Mackenzie, F., Newman, E. M., et al. (2006). Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8). J. Biol. Chem. 281, 38061.doi:10.1074/jbc.m606704200
Boscaro, C., Carotti, M., Albiero, M., Trenti, A., Fadini, G. P., Trevisi, L., et al. (2020). Non‐genomic Mechanisms in the Estrogen Regulation of Glycolytic Protein Levels in Endothelial Cells. FASEB J. 34, 12768. doi:10.1096/fj.202001130r
Chen, L., Liu, S., and Tao, Y. (2020). Regulating Tumor Suppressor Genes: Post-translational Modifications. Sig Transduct. Target Ther. 5, 90. doi:10.1038/s41392-020-0196-9
Ciechanover, A. (1994). The Ubiquitin-Proteasome Proteolytic Pathway. Cell 79, 13–21. doi:10.1016/0092-8674(94)90396-4
Combaret, L., Adegoke, O. A. J., Bedard, N., Baracos, V., Attaix, D., and Wing, S. S. (2005). USP19 Is a Ubiquitin-specific Protease Regulated in Rat Skeletal Muscle during Catabolic States. Am. J. Physiology-Endocrinology Metabolism 288, E693–E700. doi:10.1152/ajpendo.00281.2004
Cui, J., Jin, S., and Wang, R.-F. (2016). The BECN1-USP19 axis Plays a Role in the Crosstalk between Autophagy and Antiviral Immune Responses. Autophagy 12, 1210–1211. doi:10.1080/15548627.2016.1173801
Damgaard, R. B. (2021). The Ubiquitin System: from Cell Signalling to Disease Biology and New Therapeutic Opportunities. Cell Death Differ. 28, 423–426. doi:10.1038/s41418-020-00703-w
Deng, L., Meng, T., Chen, L., Wei, W., and Wang, P. (2020). The Role of Ubiquitination in Tumorigenesis and Targeted Drug Discovery. Sig Transduct. Target Ther. 5, 11. doi:10.1038/s41392-020-0107-0
Desmaze, C., Brizard, F., Turc-Carel, C., Melot, T., Delattre, O., Thomas, G., et al. (1997). Multiple Chromosomal Mechanisms Generate an EWS/FLI1 or an EWS/ERG Fusion Gene in Ewing Tumors. Cancer Genet. Cytogenet. 97, 12–19. doi:10.1016/s0165-4608(96)00326-3
Dong, Z., Guo, S., Wang, Y., Zhang, J., Luo, H., Zheng, G., et al. (2020). USP19 Enhances MMP2/MMP9-Mediated Tumorigenesis in Gastric Cancer. Onco Targets Ther. 13, 8495–8510. doi:10.2147/ott.s240543
Dwane, L., Gallagher, W. M., Ní Chonghaile, T., and O'Connor, D. P. (2017). The Emerging Role of Non-traditional Ubiquitination in Oncogenic Pathways. J. Biol. Chem. 292, 3543–3551. doi:10.1074/jbc.r116.755694
Fan, J.-B., Arimoto, K.-l., Motamedchaboki, K., Yan, M., Wolf, D. A., and Zhang, D.-E. (2015). Identification and Characterization of a Novel ISG15-Ubiquitin Mixed Chain and its Role in Regulating Protein Homeostasis. Sci. Rep. 5, 12704. doi:10.1038/srep12704
Gierisch, M. E., Pedot, G., Walser, F., Lopez-Garcia, L. A., Jaaks, P., Niggli, F. K., et al. (2019). USP19 Deubiquitinates EWS-FLI1 to Regulate Ewing Sarcoma Growth. Sci. Rep. 9, 951. doi:10.1038/s41598-018-37264-5
Gu, Z., Shi, W., Zhang, L., Hu, Z., and Xu, C. (2017). USP19 Suppresses Cellular Type I Interferon Signaling by Targeting TRAF3 for Deubiquitination. Future Microbiol. 12, 767–779. doi:10.2217/fmb-2017-0006
Haglund, K., and Dikic, I. (2005). Ubiquitylation and Cell Signaling. EMBO J. 24, 3353–3359. doi:10.1038/sj.emboj.7600808
Hanpude, P., Bhattacharya, S., Dey, A. K., and Maiti, T. K. (2015). Deubiquitinating Enzymes in Cellular Signaling and Disease Regulation. IUBMB Life 67, 544–555. doi:10.1002/iub.1402
Harada, K., Kato, M., and Nakamura, N. (2016). USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase. Int. J. Mol. Sci. 17. doi:10.3390/ijms17111829
Hassink, G. C., Zhao, B., Sompallae, R., Altun, M., Gastaldello, S., Zinin, N. V., et al. (2009). The ER‐resident Ubiquitin‐specific Protease 19 Participates in the UPR and Rescues ERAD Substrates. EMBO Rep. 10, 755–761. doi:10.1038/embor.2009.69
He, W.-T., Xue, W., Gao, Y.-G., Hong, J.-Y., Yue, H.-W., Jiang, L.-L., et al. (2017). HSP90 Recognizes the N-Terminus of Huntingtin Involved in Regulation of Huntingtin Aggregation by USP19. Sci. Rep. 7, 14797. doi:10.1038/s41598-017-13711-7
He, W.-T., Zheng, X.-M., Zhang, Y.-H., Gao, Y.-G., Song, A.-X., van der Goot, F. G., et al. (2016). Cytoplasmic Ubiquitin-specific Protease 19 (USP19) Modulates Aggregation of Polyglutamine-Expanded Ataxin-3 and Huntingtin through the HSP90 Chaperone. PLoS One 11, e0147515. doi:10.1371/journal.pone.0147515
Hershko, A., and Ciechanover, A. (1998). The Ubiquitin System. Annu. Rev. Biochem. 67, 425–479. doi:10.1146/annurev.biochem.67.1.425
Hu, M., Li, P., Li, M., Li, W., Yao, T., Wu, J., et al. (2002). Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex with Ubiquitin Aldehyde. Cell 111, 1041–1054. doi:10.1016/s0092-8674(02)01199-6
Hu, M., Li, P., Song, L., Jeffrey, P. D., Chenova, T. A., Wilkinson, K. D., et al. (2005). Structure and Mechanisms of the Proteasome-Associated Deubiquitinating Enzyme USP14. EMBO J. 24, 3747–3756. doi:10.1038/sj.emboj.7600832
Hu, W., Su, Y., Fei, X., Wang, X., Zhang, G., Su, C., et al. (2020). Ubiquitin Specific Peptidase 19 Is a Prognostic Biomarker and Affect the Proliferation and Migration of Clear Cell Renal Cell Carcinoma. Oncol. Rep. 43, 1964–1974. doi:10.3892/or.2020.7565
Huang, X., and Dixit, V. M. (2016). Drugging the Undruggables: Exploring the Ubiquitin System for Drug Development. Cell Res. 26, 484–498. doi:10.1038/cr.2016.31
Huguenin-Dezot, N., De Cesare, V., Peltier, J., Knebel, A., Kristaryianto, Y. A., Rogerson, D. T., et al. (2016). Synthesis of Isomeric Phosphoubiquitin Chains Reveals that Phosphorylation Controls Deubiquitinase Activity and Specificity. Cell Rep. 16, 1180–1193. doi:10.1016/j.celrep.2016.06.064
Ikeda, F., and Dikic, I. (2008). Atypical Ubiquitin Chains: New Molecular Signals. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 9, 536–542. doi:10.1038/embor.2008.93
Jin, S., Tian, S., Chen, Y., Zhang, C., Xie, W., Xia, X., et al. (2016). USP 19 Modulates Autophagy and Antiviral Immune Responses by Deubiquitinating Beclin‐1. EMBO J. 35, 866–880. doi:10.15252/embj.201593596
Kang, H., Choi, M. C., Kim, S., Jeong, J. Y., Kwon, A. Y., Kim, T. H., et al. (2021). USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma. Cancers (Basel) 13, 976. doi:10.3390/cancers13163976
Komander, D., Clague, M. J., and Urbé, S. (2009). Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563. doi:10.1038/nrm2731
Komander, D., Lord, C. J., Scheel, H., Swift, S., Hofmann, K., Ashworth, A., et al. (2008). The Structure of the CYLD USP Domain Explains its Specificity for Lys63-Linked Polyubiquitin and Reveals a B Box Module. Mol. Cell 29, 451–464. doi:10.1016/j.molcel.2007.12.018
Komander, D., and Rape, M. (2012). The Ubiquitin Code. Annu. Rev. Biochem. 81, 203–229. doi:10.1146/annurev-biochem-060310-170328
Konstantinopoulos, P. A., Karamouzis, M. V., and Papavassiliou, A. G. (2007). Post-translational Modifications and Regulation of the RAS Superfamily of GTPases as Anticancer Targets. Nat. Rev. Drug Discov. 6, 541–555. doi:10.1038/nrd2221
Krueger, K. E., and Srivastava, S. (2006). Posttranslational Protein Modifications: Current Implications for Cancer Detection, Prevention, and Therapeutics. Mol. Cell. Proteomics 5, 1799–1810. doi:10.1074/mcp.r600009-mcp200
Kwasna, D., Abdul Rehman, S. A., Natarajan, J., Matthews, S., Madden, R., De Cesare, V., et al. (2018). Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell 70, 150–164. doi:10.1016/j.molcel.2018.02.023
Kwon, Y. T., and Ciechanover, A. (2017). The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 42, 873–886. doi:10.1016/j.tibs.2017.09.002
Lamoliatte, F., Bonneil, E., Durette, C., Caron-Lizotte, O., Wildemann, D., Zerweck, J., et al. (2013). Targeted Identification of SUMOylation Sites in Human Proteins Using Affinity Enrichment and Paralog-specific Reporter Ions. Mol. Cell. Proteomics 12, 2536–2550. doi:10.1074/mcp.m112.025569
Lee, J.-G., Kim, W., Gygi, S., and Ye, Y. (2014). Characterization of the Deubiquitinating Activity of USP19 and its Role in Endoplasmic Reticulum-Associated Degradation. J. Biol. Chem. 289, 3510–3517. doi:10.1074/jbc.m113.538934
Lei, C.-Q., Wu, X., Zhong, X., Jiang, L., Zhong, B., and Shu, H.-B. (2019). USP19 Inhibits TNF-α- and IL-1β-Triggered NF-Κb Activation by Deubiquitinating TAK1. J. Immunol. 203, 259–268. doi:10.4049/jimmunol.1900083
Lim, K.-H., Choi, J.-H., Park, J.-H., Cho, H.-J., Park, J.-J., Lee, E.-J., et al. (2016). Ubiquitin Specific Protease 19 Involved in Transcriptional Repression of Retinoic Acid Receptor by Stabilizing CORO2A. Oncotarget 7, 34759. doi:10.18632/oncotarget.8976
Liu, Q., Zhao, S., Su, P. F., and Yu, S. (2013). Gene and Isoform Expression Signatures Associated with Tumor Stage in Kidney Renal Clear Cell Carcinoma. BMC Syst. Biol. 7 (Suppl. 5), S7–S11. doi:10.1186/1752-0509-7-S5-S7
Liu, T., Wang, L., Liang, P., Wang, X., Liu, Y., Cai, J., et al. (2021). USP19 Suppresses Inflammation and Promotes M2-like Macrophage Polarization by Manipulating NLRP3 Function via Autophagy. Cell Mol. Immunol. 18, 2431–2442. doi:10.1038/s41423-020-00567-7
Lu, Y., Adegoke, O. A. J., Nepveu, A., Nakayama, K. I., Bedard, N., Cheng, D., et al. (2009). USP19 Deubiquitinating Enzyme Supports Cell Proliferation by Stabilizing KPC1, a Ubiquitin Ligase for P27 Kip1. Mol. Cell Biol. 29, 547–558. doi:10.1128/mcb.00329-08
Lu, Y., Bedard, N., Chevalier, S., and Wing, S. S. (2011). Identification of Distinctive Patterns of USP19-Mediated Growth Regulation in Normal and Malignant Cells. PLoS One 6, e15936. doi:10.1371/journal.pone.0015936
Matsuoka, S., Ballif, B. A., Smogorzewska, A., McDonald, E. R., Hurov, K. E., Luo, J., et al. (2007). ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 316, 1160–1166. doi:10.1126/science.1140321
Mei, Y., Hahn, A. A., Hu, S., and Yang, X. (2011). The USP19 Deubiquitinase Regulates the Stability of C-IAP1 and C-IAP2. J. Biol. Chem. 286, 35380. doi:10.1074/jbc.m111.282020
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 86, 159–192. doi:10.1146/annurev-biochem-061516-044916
Millar, A. H., Heazlewood, J. L., Giglione, C., Holdsworth, M. J., Bachmair, A., and Schulze, W. X. (2019). The Scope, Functions, and Dynamics of Posttranslational Protein Modifications. Annu. Rev. Plant Biol. 70, 119–151. doi:10.1146/annurev-arplant-050718-100211
Nakamura, N., Harada, K., Kato, M., and Hirose, S. (2014). Ubiquitin-specific Protease 19 Regulates the Stability of the E3 Ubiquitin Ligase MARCH6. Exp. Cell Res. 328, 207–216. doi:10.1016/j.yexcr.2014.07.025
Nijman, S. M. B., Luna-Vargas, M. P. A., Velds, A., Brummelkamp, T. R., Dirac, A. M. G., Sixma, T. K., et al. (2005). A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell 123, 773–786. doi:10.1016/j.cell.2005.11.007
Ohtake, F., Saeki, Y., Sakamoto, K., Ohtake, K., Nishikawa, H., Tsuchiya, H., et al. (2015). Ubiquitin Acetylation Inhibits Polyubiquitin Chain Elongation. EMBO Rep. 16, 192–201. doi:10.15252/embr.201439152
Ohtake, F., and Tsuchiya, H. (2017). The Emerging Complexity of Ubiquitin Architecture. J. Biochem. 161, 125–133. doi:10.1093/jb/mvw088
Perrody, E., Abrami, L., Feldman, M., Kunz, B., Urbé, S., and van der Goot, F. G. (2016). Ubiquitin-dependent Folding of the Wnt Signaling Coreceptor LRP6. eLife 5, 19083. doi:10.7554/eLife.19083
Reinstein, E., and Ciechanover, A. (2006). Narrative Review: Protein Degradation and Human Diseases: the Ubiquitin Connection. Ann. Intern Med. 145, 676–684. doi:10.7326/0003-4819-145-9-200611070-00010
Renatus, M., Parrado, S. G., D'Arcy, A., Eidhoff, U., Gerhartz, B., Hassiepen, U., et al. (2006). Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2. Structure 14, 1293–1302. doi:10.1016/j.str.2006.06.012
Reyes-Turcu, F. E., Ventii, K. H., and Wilkinson, K. D. (2009). Regulation and Cellular Roles of Ubiquitin-specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 78, 363–397. doi:10.1146/annurev.biochem.78.082307.091526
Rossi, F. A., Enriqué Steinberg, J. H., Calvo Roitberg, E. H., Joshi, M. U., Pandey, A., Abba, M. C., et al. (2021). USP19 Modulates Cancer Cell Migration and Invasion and Acts as a Novel Prognostic Marker in Patients with Early Breast Cancer. Oncogenesis 10, 28. doi:10.1038/s41389-021-00318-x
Sadowski, M., and Sarcevic, B. (2010). Mechanisms of Mono- and Poly-Ubiquitination: Ubiquitination Specificity Depends on Compatibility between the E2 Catalytic Core and Amino Acid Residues Proximal to the Lysine. Cell Div. 5, 19. doi:10.1186/1747-1028-5-19
Shahriyari, L., Abdel-Rahman, M., and Cebulla, C. (2019). BAP1 Expression Is Prognostic in Breast and Uveal Melanoma but Not Colon Cancer and is Highly Positively Correlated with RBM15B and USP19. PLoS One 14, e0211507. doi:10.1371/journal.pone.0211507
Sharma, B. S., Prabhakaran, V., Desai, A. P., Bajpai, J., Verma, R. J., and Swain, P. K. (2019). Post-Translational Modifications (PTMs), From a Cancer Perspective: An Overview. Oncogen 2, 1–12. doi:10.35702/onc.10012
Shi, D., and Grossman, S. R. (2010). Ubiquitin Becomes Ubiquitous in Cancer: Emerging Roles of Ubiquitin Ligases and Deubiquitinases in Tumorigenesis and as Therapeutic Targets. Cancer Biol. Ther. 10, 737–747. doi:10.4161/cbt.10.8.13417
Sun, T., Liu, Z., and Yang, Q. (2020). The Role of Ubiquitination and Deubiquitination in Cancer Metabolism. Mol. Cancer 19, 146. doi:10.1186/s12943-020-01262-x
Sundaram, P., Pang, Z., Miao, M., Yu, L., and Wing, S. S. (2009). USP19-deubiquitinating Enzyme Regulates Levels of Major Myofibrillar Proteins in L6 Muscle Cells. Am. J. Physiology-Endocrinology Metabolism 297, E1283. doi:10.1152/ajpendo.00409.2009
Velasco, K., Zhao, B., Callegari, S., Altun, M., Liu, H., Hassink, G., et al. (2013). An N-Terminal SIAH-Interacting Motif Regulates the Stability of the Ubiquitin Specific Protease (USP)-19. Biochem. Biophysical Res. Commun. 433, 390–395. doi:10.1016/j.bbrc.2013.02.094
Vellosillo, P., and Minguez, P. (2021). A Global Map of Associations between Types of Protein Posttranslational Modifications and Human Genetic Diseases. iScience 24, 102917. doi:10.1016/j.isci.2021.102917
Wauer, T., Swatek, K. N., Wagstaff, J. L., Gladkova, C., Pruneda, J. N., Michel, M. A., et al. (2015). Ubiquitin Ser65 Phosphorylation Affects Ubiquitin Structure, Chain Assembly and Hydrolysis. EMBO J. 34, 307–325. doi:10.15252/embj.201489847
Wiles, B., Miao, M., Coyne, E., Larose, L., Cybulsky, A. V., and Wing, S. S. (2015). USP19 Deubiquitinating Enzyme Inhibits Muscle Cell Differentiation by Suppressing Unfolded-Protein Response Signaling. Mol. Biol. Cell 26, 913–923. doi:10.1091/mbc.e14-06-1129
Wu, M., Tu, H.-Q., Chang, Y., Tan, B., Wang, G., Zhou, J., et al. (2017). USP19 Deubiquitinates HDAC1/2 to Regulate DNA Damage Repair and Control Chromosomal Stability. Oncotarget 8, 2197–2208. doi:10.18632/oncotarget.11116
Wu, X., Lei, C., Xia, T., Zhong, X., Yang, Q., and Shu, H.-B. (2019). Regulation of TRIF-Mediated Innate Immune Response by K27-Linked Polyubiquitination and Deubiquitination. Nat. Commun. 10, 4115. doi:10.1038/s41467-019-12145-1
Xu, H., Wang, Y., Lin, S., Deng, W., Peng, D., Cui, Q., et al. (2018). PTMD: A Database of Human Disease-Associated Post-translational Modifications. Genomics, Proteomics Bioinforma. 16, 244–251. doi:10.1016/j.gpb.2018.06.004
Xue, W., Zhang, S.-X., He, W.-T., Hong, J.-Y., Jiang, L.-L., and Hu, H.-Y. (2020). Domain Interactions Reveal Auto-Inhibition of the Deubiquitinating Enzyme USP19 and its Activation by HSP90 in the Modulation of Huntingtin Aggregation. Biochem. J. 477, 4295–4312. doi:10.1042/bcj20200536
Yau, R., and Rape, M. (2016). The Increasing Complexity of the Ubiquitin Code. Nat. Cell Biol. 18, 579–586. doi:10.1038/ncb3358
Zhang, L., Zhou, F., Drabsch, Y., Gao, R., Snaar-Jagalska, B. E., Mickanin, C., et al. (2012). USP4 Is Regulated by AKT Phosphorylation and Directly Deubiquitylates TGF-β Type I Receptor. Nat. Cell Biol. 14, 717–726. doi:10.1038/ncb2522
Keywords: PTMs, ubiquitination, DUBs, USP19, tumorigenesis, metastasis
Citation: Rossi FA and Rossi M (2022) Emerging Role of Ubiquitin-Specific Protease 19 in Oncogenesis and Cancer Development. Front. Cell Dev. Biol. 10:889166. doi: 10.3389/fcell.2022.889166
Received: 03 March 2022; Accepted: 26 April 2022;
Published: 12 May 2022.
Edited by:
Long Zhang, Zhejiang University, ChinaReviewed by:
Simon Sipen Wing, McGill University, CanadaCopyright © 2022 Rossi and Rossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Rossi, bXJvc3NpLWNvbmljZXRAYXVzdHJhbC5lZHUuYXI=
 Fabiana Alejandra Rossi
Fabiana Alejandra Rossi Mario Rossi
Mario Rossi