- 1Małopolska Centre of Biotechnology, Jagiellonian University, Krakow, Poland
- 2Department of Material Chemistry, Graduate School of Engineering, Kyoto University, Kyoto, Japan
- 3Faculty of Biology-Oriented Science and Technology, Kindai University, Wakayama, Japan
- 4Department of Cell Biology, National Institute for Basic Biology, Okazaki, Japan
- 5Department of Biology, Faculty of Science and Engineering, Konan University, Kobe, Japan
- 6Department of Basic Biology, School of Life Science, SOKENDAI (The Graduate University for Advanced Studies), Okazaki, Japan
Peroxisomes are present in eukaryotic cells and have essential roles in various biological processes. Plant peroxisomes proliferate by de novo biosynthesis or division of pre-existing peroxisomes, degrade, or replace metabolic enzymes, in response to developmental stages, environmental changes, or external stimuli. Defects of peroxisome functions and biogenesis alter a variety of biological processes and cause aberrant plant growth. Traditionally, peroxisomal function-based screening has been employed to isolate Arabidopsis thaliana mutants that are defective in peroxisomal metabolism, such as lipid degradation and photorespiration. These analyses have revealed that the number, subcellular localization, and activity of peroxisomes are closely related to their efficient function, and the molecular mechanisms underlying peroxisome dynamics including organelle biogenesis, protein transport, and organelle interactions must be understood. Various approaches have been adopted to identify factors involved in peroxisome dynamics. With the development of imaging techniques and fluorescent proteins, peroxisome research has been accelerated. Image-based analyses provide intriguing results concerning the movement, morphology, and number of peroxisomes that were hard to obtain by other approaches. This review addresses image-based analysis of peroxisome dynamics in plants, especially A. thaliana and Marchantia polymorpha.
1 Introduction
Peroxisomes are present in eukaryotic cells and have important roles in various biological processes. In plants, peroxisomes are responsible for photorespiration, which is required to salvage byproducts of photosynthesis, and biosynthesis of plant hormones such as jasmonic acid and auxin, in addition to metabolism of fatty acids and detoxification of reactive oxygen species (ROS), which are common functions of peroxisomes in plant, mammalian, and yeast cells (Kamada et al., 2003). Peroxisomes are multiplied by division of pre-existing peroxisomes and degraded in response to developmental stages, environmental changes, and external stimuli. All peroxisomal proteins are encoded by the nuclear genome, and matrix proteins are transported to peroxisomes after translation in the cytosol. Many factors involved in the biosynthesis and functions of peroxisomes are conserved among various organisms. The factors responsible for biosynthesis of peroxisomes are collectively called PEROXINs (PEXs). More than 30 PEXs and their isoforms have been reported (Hu et al., 2012; Baker et al., 2016; Yuan et al., 2016; Fujiki et al., 2020; Jansen et al., 2021). However, some PEXs are unique to an organism. For example, the intraperoxisomal protein PEX8, PEX17, which is part of the docking complex on the peroxisomal membrane, and the PTS2 co-receptor PEX20 are reportedly involved in peroxisomal protein transport in fungi (Purdue et al., 1998; Agne et al., 2003; Montilla-Martinez et al., 2015; Jansen et al., 2021), but have not been identified in plants or animals. Fatty acid degradation via the β-oxidation pathway is a common type of metabolism in peroxisomes among various organisms. Although β-oxidation proceeds both in peroxisomes and mitochondria in mammalian cells, it occurs only in peroxisomes in plants and fungi (Poirier et al., 2006). Plant peroxisomes are also closely connected to photosynthesis, a unique plant system. The absolute byproduct glycolate-2-phosphate produced by RubisCO during photosynthesis is recycled to glycerate via photorespiration in peroxisomes and mitochondria to increase the photosynthetic efficiency (Peterhansel et al., 2010). In addition, peroxisomes are closely associated with chloroplasts when photosynthesis is active. Therefore, it is not sufficient to use information from yeast and animals to understand the molecular regulation that controls the morphology and dynamics of plant peroxisomes, and peroxisomal proteins in plants must be identified.
Peroxisome research has been accelerated by the application of imaging techniques such as the use of fluorescent proteins. In 2002, three groups visualized peroxisomes with GFP (Figure 1A; Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002). Visualization of peroxisomes was simple and did not affect their functions or dynamics. It only required expression of the fusion gene encoding peroxisome targeting signal (PTS) 1 or PTS2 added to the C- or N-terminus of GFP, respectively. Additional reagents and treatments were not required to observe GFP-labeled peroxisomes. Observation of GFP-labeled peroxisomes under a fluorescence microscope provided important information about peroxisome dynamics such as their morphology, number, size, intracellular distribution, movement, and interactions with other subcellular components, which was hard to obtain by traditional approaches. In particular, live imaging is a powerful technique in the plant peroxisome research field and provides useful information such as the velocity, direction of movement, and morphological changes of peroxisomes (Supplementary Movie S1; Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002). In those days, electron microscopic analysis was the only way to observe peroxisome dynamics, especially their shape and size. This is because, unlike mitochondria and other organelles, there are no dyes to specifically stain peroxisomes and, unlike chloroplasts, peroxisomes do not emit autofluorescence, which is occasionally used to monitor chloroplast dynamics in living cells. Electron microscopic analysis has been a powerful tool to investigate ultrafine structures of peroxisomes (Figure 1B). However, electron micrographs are static images and therefore do not provide temporal information. Meanwhile, although the resolution of fluorescence images is inferior to that of electron micrographs, researchers can obtain spatiotemporal information from observations under a fluorescence microscope. A confocal laser scanning microscope can generate 3D images containing information about the distribution of peroxisomes in the whole cell.
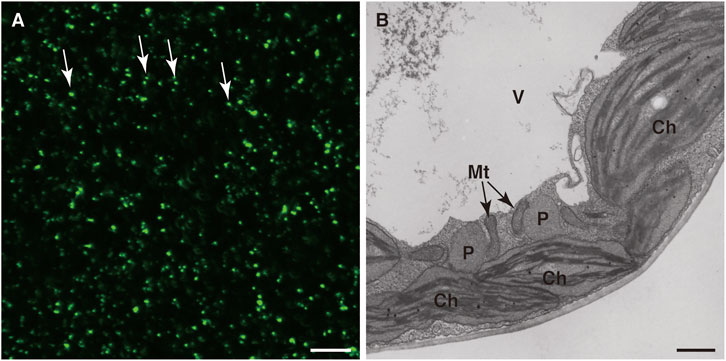
FIGURE 1. Detection of peroxisomes in leaf cells. Fluorescence microscopic analysis of GFP (A) and electron microscopic analysis (B) were performed of transgenic A. thaliana (GFP-PTS1) expressing the fusion gene of GFP with PTS1 under the regulation of the constitutive promoter. (A) A lot of peroxisomes were visualized as spherical structures (Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002). Some representative peroxisomes are indicated by arrows. Bar, 20 µm. (B) Transmission electron microscopic observation of GFP-PTS1 plants (Mano et al., 2002). P, peroxisome; Mt, mitochondrion; Ch, chloroplast; V, vacuole. Bar, 1 µm.
Thus, it has become easier for researchers to obtain information about the dynamics of peroxisomes in plant cells using imaging analysis. Peroxisomes are maintained by sophisticated machinery that regulates their biogenesis and functions, such as their elongation, division, and protein transport. Disturbance of these regulatory mechanisms can cause peroxisome anomalies in cells. This is expected to result in abnormal peroxisomes, and the introduction of fluorescent peroxisome markers allows direct observation of such aberrations. For example, if the mechanisms controlling peroxisome proliferation, such as their elongation and division, were defective, peroxisomes with an abnormal size and morphology would be observed. If the efficiency of peroxisomal protein transport was decreased, GFP fluorescence would be observed in the cytosol as well as in peroxisomes. Based on these insights, Arabidopsis thaliana aberrant peroxisome morphology (apem) mutants were isolated and analyzed, which provided useful information about peroxisome dynamics (Mano et al., 2004; Mano et al., 2006; Goto et al., 2011; Goto-Yamada et al., 2014a). These studies are described in Section 2.2 in more detail. Among various important peroxisome functions, photorespiration is a metabolic system involving reactions in chloroplasts, mitochondria, and peroxisomes, and interactions among these three organelles support efficient photorespiratory activity (Oikawa et al., 2015). A. thaliana peroxisome unusual positioning (peup) mutants were screened based on an abnormal pattern of peroxisome positioning in cells (Shibata et al., 2013). In this screening, green and red fluorescence, which was derived from GFP-labeled peroxisomes and autofluorescence emitted by chloroplasts, was used to analyze the intracellular distributions of both organelles. The detailed studies of peup mutants are described in Section 2.3.
As described in Section 3, the liverwort Marchantia polymorpha has become a useful model plant for plant life science research due to several advantages, such as the availability of well-developed genetic resources and methods to introduce exogeneous genes for visualizing subcellular compartments and to perform genome editing with the CRISPR/Cas9 system (Bowman, 2016; Bowman et al., 2017; Iwasaki et al., 2021; Kohchi et al., 2021). Like in A. thaliana, peroxisomes are easily visualized with fluorescent proteins in M. polymorpha, and therefore M. polymorpha is becoming a useful material in the plant peroxisome research field (Ogasawara et al., 2013; Kimura and Kodama, 2016; Mano et al., 2018). By comparing the molecular mechanisms regulating peroxisome dynamics in A. thaliana and M. polymorpha, we can determine whether mechanisms related to plant peroxisomes are conserved among all plant species or are specific to particular plant species.
This review comprehensively addresses image-based analysis of peroxisomes. In particular, we describe the identification and characterization of factors involved in peroxisome dynamics based on analyses of mutants with peroxisome defects in A. thaliana, and a combination of imaging and bioinformatics analyses in M. polymorpha.
2 Imaging Analysis of A. thaliana Mutants With Peroxisome Defects
2.1 Introduction of Image-Based Screening to Identify New Mutants With Peroxisome Defects
The forward genetics approach to identify novel mutants that display an abnormality of peroxisomes followed by characterization of the gene products responsible is a powerful method to improve our knowledge of peroxisome dynamics, metabolism, and biosynthesis. Efficient isolation to obtain promising mutants is key for successful research. The model flowering plant A. thaliana has been used to screen mutants with peroxisome defects because genetic resources and information are abundant (Koornneef and Meinke, 2010). Various approaches have been adopted to identify mutants with peroxisome defects. Traditionally, peroxisomal function-based screening has been performed to identify a number of mutants that are defective in lipid metabolism and photorespiration, contributing to the identification of several peroxisome-related genes, such as those encoding enzymes involved in metabolism and PEXs (Somerville and Ogren, 1980; Somerville and Ogren, 1981; Hayashi et al., 1998; Hayashi et al., 2000; Zolman et al., 2000; Zolman et al., 2001; Hayashi et al., 2002; Zolman and Bartel, 2004). Screening relied on morphological differences from wild-type (WT) plants, such as dwarfism and short roots, as a result of indirect effects. To obtain novel mutants with peroxisome defects, including peroxisome dynamics-deficient mutants, another screening approach is employed: visualized peroxisome-based mutant screening. The first set of mutants, called the apem mutant series, was isolated by focusing on plant peroxisome dynamics, i.e., their morphology, movement, number, and subcellular localization (Table 1). As a supplementary note, the abbreviation apm was initially used, but has been replaced with apem to avoid confusion with other A. thaliana mutants. The mutants were screened from the pool of ethyl methanesulfonate (EMS)-mutagenized A. thaliana (accession Columbia) plants, which expressed the peroxisome marker GFP-PTS1, based on a GFP fluorescence pattern that differed from that in WT plants (Mano et al., 2002). Approximately 37,000 M2 plants were examined under a fluorescence microscope, and 82 mutants were isolated. These mutants were classified into four groups: 1) elongated peroxisomes, 2) enlarged peroxisomes, 3) mislocalization of GFP-PTS1 protein to the cytosol, and 4) other distributions of GFP (Mano et al., 2004; Mano et al., 2006; Goto et al., 2011; Goto-Yamada et al., 2014a). In addition, the same mutagenized seed pool was screened for differences in the pattern of interactions between peroxisomes and chloroplasts. In these mutants, designated peup, the size and morphology of peroxisomes were almost identical to those in the parent plants, but the intracellular distributions of peroxisomes and chloroplasts were dramatically altered (Shibata et al., 2013; Goto-Yamada et al., 2019). Apart from apem and peup mutants, screenings based on visualized peroxisomes were also reported by other groups (Zhang and Hu, 2009; Rinaldi et al., 2016). In addition, Lingard et al. (2009) used GFP fused with ISOCITRATE LYASE (ICL), which encodes a glyoxylate cycle enzyme in peroxisomes, under the regulation of the ICL promoter to investigate peroxisome-associated protein degradation (Lingard et al., 2009; Burkhart et al., 2013). In this section, we introduce imaging analysis-based peroxisome research. We first outline various apem and peup mutants, and then describe reports in A. thaliana in comparison with other organisms.
2.2 Analysis of Peroxisome Biogenesis, Proliferation, and Quality Control With apem Mutants
2.2.1 apem1/drp3a
The apem1 (previously known as apm1) mutant exhibits elongated and a reduced number of peroxisomes in a variety of cells throughout the plant (Figure 2). Mitochondria are also elongated, but other organelles such as chloroplasts, nuclei, the Golgi apparatus, and the endoplasmic reticulum (ER) are not. The APEM1 gene encodes DYNAMIN-RELATED PROTEIN 3A (DRP3A), a member of the dynamin superfamily that has a pivotal role in vesicle division and organelle fission and fusion (Mano et al., 2004; Praefcke and McMahon, 2004). In addition to DRP3A, its closest homolog, DRP3B, is also involved in peroxisome and mitochondria fission, and plant- and alga-specific DRP5B affects peroxisome, chloroplast, and mitochondria fission (Fujimoto et al., 2009; Aung and Hu, 2012). Interestingly, forward genetic screening isolated a number of independent lines possessing mutations at the DRP3A locus (Mano et al., 2004; Praefcke and McMahon, 2004), but not the DRP3B or DRP5B locus (Aung and Hu, 2012). Various experimental data indicate that DRP3A is the primary protein responsible for peroxisome fission (Fujimoto et al., 2009; Zhang and Hu, 2009; Aung and Hu, 2012).
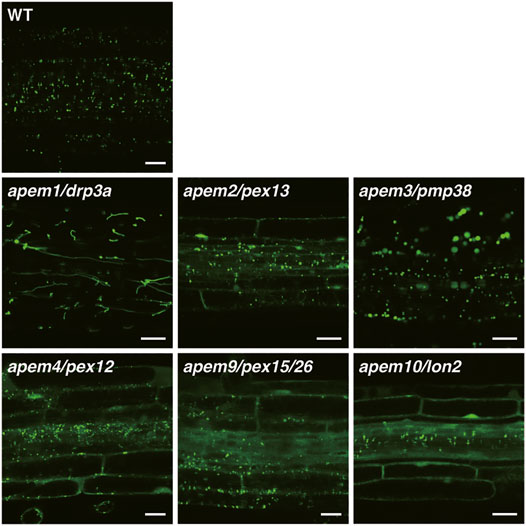
FIGURE 2. GFP fluorescence in root tissue of the WT plant and apem mutants expressing the peroxisome marker GFP-PTS1 (Mano et al., 2004; Mano et al., 2006; Goto et al., 2011; Goto-Yamada et al., 2014a). Bars, 20 µm.
2.2.2 apem2/pex13
In the apem2 (previously known as apm2) mutant, GFP-PTS1 protein is located in the cytosol as well as in peroxisomes (Figure 2). The APEM2 gene encodes the integral peroxisomal membrane protein PEX13 (Mano et al., 2006). Peroxisomal matrix proteins are transported to peroxisomes by their receptor PEX5 or PEX7, and translocate into the peroxisomal matrix through the pore formed by the receptor and the docking complex consisting of PEX14 and PEX13 (and PEX17 in fungi) on the peroxisomal membrane. The apem2 mutation introduces a stop codon at position 263 instead of glutamine in the C-terminal region. The C-terminus of PEX13 interacts with PEX14 and PEX5 via the Src homology 3 (SH3) domain in fungi (Bottger et al., 2000; Douangamath et al., 2002). However, A. thaliana PEX13 lacks an obvious SH3 domain, and a yeast two-hybrid assay showed that PEX13 interacts with PEX7, but not PEX14 or PEX5 (Mano et al., 2006; Boisson-Dernier et al., 2008). Another group reported a different pex13 mutation in which Glu is substituted by Lys only 20 amino acids upstream of the apem2 mutation (Woodward et al., 2014), supporting the importance of the C-terminal region of plant PEX13. Boisson-Dernier et al. (2008) isolated the A. thaliana abstinence by mutual consent (amc) mutant, which disrupts male-female gametophyte recognition (Boisson-Dernier et al., 2008). amc is a PEX13 loss-of-function mutant, showing that peroxisomes play a role in the reproductive process (Boisson-Dernier et al., 2008; Goto-Yamada et al., 2014b). These results indicate that PEX13 and other peroxisomal biogenesis factors evolved differently in plants in comparison with other organisms.
2.2.3 apem3/pmp38/pxn
The apem3 mutant exhibits enlarged peroxisomes, and the diameter of some can reach more than 10 µm (Figure 2; Mano et al., 2011). Introduction of the apem1/drp3a mutation, which impairs peroxisome division, into apem3 does not affect the enlarged peroxisome phenotype, and DRP3A protein is detected on apem3 enlarged peroxisomes. In addition, division-arrested apem1/drp3a peroxisomes do not show the same level of enlargement as those in the apem3 mutant (Mano et al., 2011). These results suggest that the enlarged peroxisomes observed in apem3 do not arise due to perturbation of peroxisome division. The APEM3 gene encodes PEROXISOMAL MEMBRANE PROTEIN 38 (PMP38), which is also known as PEROXISOMAL NAD+ CARRIER (PXN) because it can transport NAD+ into the peroxisomal matrix (Fukao et al., 2001; Eubel et al., 2008; Bernhardt et al., 2012). Blockade of NAD+ supply reduces the efficiency of lipid metabolism in peroxisomes and induces the accumulation of long-chain fatty acids (Bernhardt et al., 2012). Other mutants defective in fatty acid β-oxidation also contain enlarged peroxisomes (Hayashi et al., 2001). In addition, the enlargement of peroxisomes in the pmp38/pxn mutant is suppressed by disruption of PEROXISOMAL ABC TRANSPORTER1 (PXA1), which transports fatty acids into peroxisomes (Rinaldi et al., 2016). These results demonstrate that the enlargement of peroxisomes in apem3/pmp38/pxn mutants is due to the accumulation of fatty acids, which may produce hydrogen peroxide and damage peroxisomes (Rinaldi et al., 2016).
2.2.4 apem4/pex12
In the apem4 (previously known as apm4) mutant, GFP-PTS1 protein is located in the cytosol as well as in peroxisomes (Figure 2). The APEM4 gene encodes PEX12 (Mano et al., 2006), which is one of the RING-finger domain-containing peroxins (PEX2, PEX10, and PEX12) involved in peroxisomal protein transport. In fungi, mono-ubiquitination of the peroxisomal protein receptor Pex5 is mediated by Pex4 and Pex12, which are E2 and E3 ligases, respectively (Platta et al., 2009), and this is required for recycling of Pex5 from the peroxisomal membrane to the cytosol. Three A. thaliana RING peroxins, PEX2, PEX10, and PEX12, exhibit E3 ubiquitin ligase activity in vitro (Kaur et al., 2013). Indeed, the apem4/pex12 mutant displayed accumulation of PEX5 protein in the peroxisomal membrane fraction, while the pex12-1 mutant exhibited elevated PEX5 and PEX7 levels (Mano et al., 2006; Kao et al., 2016). The apem4 mutant, as well as another pex12 mutant, exhibits suppression of not only PTS1- but also PTS2-directed protein transport. These results indicate that a PEX12 defect impairs PEX5 and PEX7 recycling. A. thaliana PEX12 can bind to PEX7 (Singh et al., 2009). Therefore, a defect of PEX12 also can lead to a decrease in the efficiency of PEX7-dependent PTS2 transport. Interestingly, the apem4 mutation, which replaces Arg170 with Lys, is adjacent to the pex12-1 mutation, which replaces Glu171 with Lys (Mano et al., 2006; Kao et al., 2016). However, these amino acid residues are not conserved among organisms, and the function of the region containing these two amino acid residues has not been clarified. The findings that mutations at two positions affect peroxisome transport imply the specific function of this region of PEX12.
2.2.5 apem9/pex15/pex26
Like apem2 and apem4, the apem9 mutant was isolated on the basis of a phenotype in which peroxisomal proteins accumulate in the cytosol and exhibits defects in both PTS1- and PTS2-dependent transport (Figure 2; Goto et al., 2011). APEM9 encodes a protein of unknown function, homologs of which are found only in plant genomes. Hydropathy profile analysis of APEM9 suggested that it is similar to yeast PEX15, which can recruit the PEX1/PEX6 complex from the cytosol to peroxisomal membranes (Goto et al., 2011). PEX15 is a tail-anchored peroxisomal membrane protein that is involved in recycling of PEX5 in fungi (Birschmann et al., 2003), and PEX26 was reported to be a PEX15 ortholog in mammalian cells (Matsumoto et al., 2003). The secondary structure of APEM9 appears to be more similar to that of PEX26 than PEX15, although sequence similarity is quite low (Goto et al., 2011). The AAA+ ATPases PEX1 and PEX6 form a heterooligomer and can function as an unfoldase to extract PEX5 from the membrane (Ciniawsky et al., 2015; Gardner et al., 2018; Pedrosa et al., 2018). The apem9 mutation substitutes Gly278 with Glu in the transmembrane domain, which affects the peroxisomal localization of APEM9 and the PEX1/PEX6 complex (Goto et al., 2011). DAYU (a synonym of APEM9) binds to PEX13 and PEX16 (Li et al., 2014). As described above, PEX13 is a component of the PEX5 docking complex, and bridging the docking complex closer to the recycling machinery may make export of PEX5 efficient. Unlike apem2/pex13, mutants defective in APEM9/PEX15/PEX26 or PEX6 displayed a decreased amount of PEX5, and the PEX5 level was increased by treatment with the proteasome inhibitor MG132, suggesting that PEX5 undergoes proteasomal degradation when recycling machinery does not extract it properly (Gonzalez et al., 2017).
2.2.6 apem10/lon2
The apem10 mutant exhibits a decreased level of punctate peroxisomal GFP signals and accumulation of GFP fluorescence in the cytosol (Figure 2). The apem10 mutation replaces Gln144 with a stop codon in the peroxisomal LON PROTEASE 2 (LON2) protein. Immunostaining of the peroxisomal membrane protein ASCORBATE PEROXIDASE (APX) showed that the number of peroxisomes is reduced in the apem10 mutant (Goto-Yamada et al., 2014a). This indicates that peroxisomes are degraded and that matrix proteins, such as GFP-PTS1, accumulate in the cytosol (Lingard and Bartel, 2009; Goto-Yamada et al., 2014a). The apem10/lon2 phenotype is accentuated with age. Accumulation of GFP-PTS1 in the cytosol is not observed in newly emerging young leaves, but is prominent in well-developed mature leaves in which peroxisomes are enlarged occasionally (Farmer et al., 2013; Goto-Yamada et al., 2014a). In addition, the phenotype of enlarged peroxisomes in apem10 mutants was suppressed under high CO2 conditions, in which the photorespiratory pathway was not required, indicating that the apem10 phenotype correlates with peroxisomal activity (Goto-Yamada et al., 2014a). Induction of autophagy deficiency rescued the apem10/lon2 phenotype (Farmer et al., 2013; Goto-Yamada et al., 2014a). Peroxisomal metabolic systems contain a variety of oxidases and produce hydrogen peroxide, which is a threat to peroxisomal proteins and membranes (Nishimura et al., 1983; Corpas et al., 2020). Studies of plant LON2 and autophagy revealed the existence of two independent peroxisome maintenance processes: 1) LON2 degrades abnormal and/or obsolete matrix proteins inside peroxisomes and 2) when peroxisomes are not sufficiently restored by LON2, autophagy degrades abnormal peroxisomes (Farmer et al., 2013; Shibata et al., 2013; Goto-Yamada et al., 2014a). A lack of LON2 induces peroxisome degradation via autophagy (called pexophagy). Plant peroxisomes alter their metabolic systems in response to their environment and developmental changes. The molecular mechanisms to replace enzymes responsible for each type of metabolism have long been discussed. The quality control system of peroxisomes, which involves the two aforementioned coordinated degradation processes, explains the mechanism underlying peroxisomal functional transition and a new model was proposed (Goto-Yamada et al., 2015). Interestingly, the protease activity of the C-terminal serine peptidase domain seems to contribute to degradation of peroxisomal proteins, but not to inhibition of pexophagy, which is dependent on the N-terminal chaperone domain (Goto-Yamada et al., 2014a). The mechanisms underlying inhibition and induction of pexophagy remain to be investigated.
2.3 Analysis of Organelle-Organelle Interactions
2.3.1 Physical Interactions of Peroxisomes With Other Organelles
Leaf peroxisomes function in many metabolic pathways, some of which also involve other organelles such as mitochondria and chloroplasts (Mano and Nishimura, 2005; Hayashi and Nishimura, 2006; Nyathi and Baker, 2006; Hu et al., 2012; Kao et al., 2018; Oikawa et al., 2019). Therefore, it is thought that the close localization of peroxisomes, mitochondria, and chloroplasts contributes to efficient metabolite flow. In fact, electron micrographs showed these three organelles in close contact with each other (Frederick and Newcomb, 1969; Tolbert, 1982; Nishimura et al., 1986; Oikawa et al., 2019; Baillie et al., 2020). As described above, visualization of peroxisomes using fluorescent proteins enables analysis of their positioning in living cells (Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002). Peroxisomes actively move on actin filaments using myosin motors and interact with other organelles such as chloroplasts and mitochondria (Jedd and Chua, 2002; Mano et al., 2002; Mathur et al., 2002; Goto-Yamada et al., 2015; Oikawa et al., 2015; Oikawa et al., 2019; Baillie et al., 2020; Mathur, 2021). Peroxisomes in dark-adapted cells change their shape from spherical to elliptical in order to strengthen their interactions with chloroplasts under the photosynthetic condition (Oikawa et al., 2015; Oikawa et al., 2019). The strength of interactions between peroxisomes and chloroplasts in the dark and light was measured using a femtosecond laser to evaluate adhesion strength directly in living leaf cells. This revealed that light has a strong positive effect on adhesion (Oikawa et al., 2015; Hosokawa et al., 2016). An optical tweezer was used to measure the interaction strength between a peroxisome and a chloroplast in vitro (Gao et al., 2016). These studies revealed the existence of a physical interaction between peroxisomes and chloroplasts, and suggest that this interaction has physiological significance for plant cellular function.
The tethering factor(s) that connects a peroxisome and a chloroplast remains unclear, but PEX10, a C3HC4 zing RING-finger peroxisomal membrane protein, is one candidate (Schumann et al., 2007). Expression of dominant-negative PEX10 disturbed the interaction of peroxisomes with chloroplasts and photorespiration. Further studies are required to clarify whether PEX10 functions as a tethering factor between a peroxisome and a chloroplast directly and whether other PEXs are involved in this interaction.
It was recently reported that a large complex of glycolysis enzymes, a phosphoglycerate mutase-enolase metabolon, plays a role in the interaction between mitochondria and chloroplasts (Zhang et al., 2020). A direct interaction between mitochondria and chloroplasts has been clearly shown by analyzing mitochondrial movement (Oikawa et al., 2021). It is interesting to investigate whether enzymes in the metabolite pathway participate in the interaction between peroxisomes and chloroplasts similar to the interaction between mitochondria and chloroplasts. Determination of the mechanism underlying the peroxisome-chloroplast interaction will help to elucidate the role of organelle interactions in plants.
Glyoxysomes, one of the peroxisomes, engage in the degradation of reserve oil stored in the oil body via β-oxidation and the glyoxylate cycle. A. thaliana peroxisome defective 1 (ped1) was defective in fatty acid β-oxidation (Hayashi et al., 1998). Detailed electron microscopic analysis revealed that the glyoxysomes in etiolated cotyledons of the ped1 mutant appeared abnormal, having tubular structures that are derived from invagination of the glyoxysomal membrane. (Hayashi et al., 2001). These invagination sites were always in contact with oil bodies, proposing that direct interaction between glyoxysomes and lipid bodies is involved in the process of fatty acid metabolism (Hayashi et al., 2001). A. thaliana sugar dependent 1 (sdp1) mutant was identified from the pool of ethyl methanesulfonate (EMS)-mutagenized A. thaliana, which expressed the fusion gene encoding OLEOSIN, one of oil body membrane proteins, with GFP, as having larger and more oil body aggregates compared with the wild-type plant (Cui et al., 2016). SDP1 is a triacylglycerol (TAG) lipase that resides on the oil body membrane, and hydrolyzes TAG to produce fatty acids. From the analyses using the sdp1 mutant, Cui et al. (2016) showed that sucrose is a key factor for peroxisome-oil body interaction dependent on actin filaments, and that PEROXISOME DEFFECTIVE 3 (PED3), a peroxisomal ATP binding cassette transporter, is the potential anchor protein to the membranes of these organelles (Cui et al., 2016).
The analysis of mutants accumulating excess peroxisomes described below and several other reports indicate that autophagic peroxisomal degradation, or pexophagy, is one of the major peroxisomal quality control mechanisms, along with maintenance by the chaperone-proteinase LON2/APEM10 (Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013; Goto-Yamada et al., 2014a). Mutants with defective autophagy fail to form autophagosomes and subsequently degrade peroxisomes. In these mutants, the cisterna-like membrane structure associated with peroxisomes and the ATG8 protein, one of the autophagosome components, were detected on autophagosome membrane structures by immunoelectron microscopy (Yoshimoto et al., 2014). Reduction-oxidation sensitive green fluorescent protein (roGFP) analysis revealed that the peroxisomes of autophagy-deficient mutants are highly oxidized, and that mCherry-ATG8a proteins selectively assemble on the oxidized peroxisomes (Shibata et al., 2013). Peroxisomes are oxidized by hydrogen peroxide produced in the process of peroxisome function, and such damaged peroxisomes are selectively recognized and eliminated by autophagy.
2.3.2 peup Mutants
It is crucial to study mutants in order to understand the biological significance of peroxisome movement and positioning (interactions with other organelles) for cellular function. A. thaliana peup mutants were isolated from the EMS-mutagenized seed pools that were used to obtain apem mutants by the following method (Table 1; Shibata et al., 2013; Goto-Yamada et al., 2019). Leaves of the mutant lines were put on an agar plate under 100 μmol m−2 s−1 light to distribute chloroplasts perpendicular in leaf mesophyll cells (Kagawa et al., 2001; Oikawa et al., 2008; Wada and Kong, 2018). In WT cells, peroxisomes reside in a similar location as chloroplasts at the cell periphery because they mostly interact with chloroplasts (Oikawa et al., 2015). It was expected that if the mutants were defective in the peroxisome motility system that regulates peroxisome localization or in tethering factors that connect a peroxisome with a chloroplast, peroxisomes would exhibit abnormal positioning or remain in the cytosol distant from the chloroplast. About 10,000 plants were screened under a fluorescence microscope, and more than 50 peup mutants, which displayed peroxisome aggregation and diffuse localization in the cytosol due to a defect in interactions with chloroplasts, were obtained. Of them, peup1, peup2, and peup4 exhibited remarkable peroxisome aggregation and an increased number of peroxisomes (Figure 3; Shibata et al., 2013). Furthermore, the mutants displayed earlier senescence than WT plants in normal air conditions (Shibata et al., 2013; Yoshimoto et al., 2014). PEUP1, PEUP2, and PUEP4 encode autophagy-related (ATG) 2, ATG18a, and ATG7 proteins, respectively (Table 1; Shibata et al., 2013). These mutants accumulated undegraded peroxisomes containing inactive catalase aggregates, which were observed as high-density regions in peroxisomes in electron micrographs (Shibata et al., 2013). The undegraded peroxisomes are defective in interactions with chloroplasts and movement in the cytosol (Yoshimoto et al., 2014).
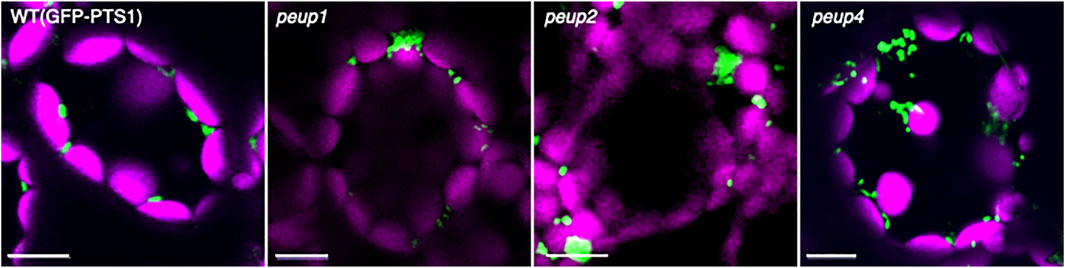
FIGURE 3. Peroxisome aggregation in peup mutants. Representative images of peroxisomes (green) and chloroplasts (magenta) in leaf mesophyll cells of the WT plant and peup mutants (Shibata et al., 2013; Goto-Yamada et al., 2019). Peroxisomes associate with chloroplasts in the WT plant, whereas peroxisomes partially form aggregates in peup mutants. Bars, 10 µm.
Other PEUP genes, PEUP17 and PEUP22, were recently revealed to encode ATG5 and ATG7, respectively (Table 1). Analysis of peup17 and peup22 demonstrated that sucrose starvation induces a type of microautophagy in root tip cells and that ATG genes are involved in this process (Goto-Yamada et al., 2019). Peroxisomes in other peup mutants are spherical with reduced motility or form small aggregates with aberrant motility. These mutants are expected to have defects in gene products that regulate the interaction between a peroxisome and a chloroplast or peroxisome mobility, such as tethering factors or receptors of motor proteins. They will be useful materials to study peroxisome quality control via autophagy and organelle interactions.
2.4 Conclusion of Analyses of apem and peup Mutants
The apem and peup mutant series were isolated based on the imaging technique in our laboratory and are summarized in Figure 4. APEM1/DRP3A was identified as a major component of the peroxisome division machinery. APEM2/PEX13, APEM4/PEX12, and APEM9/PEX15/PEX26 were identified as a group of peroxisome biogenesis factors. Like in animals, many plant PEX mutants with T-DNA insertions causing complete protein dysfunction display lethality, as reported in studies of PEX2, PEX10, PEX12, PEX13, APEM9/PEX15/PEX26, and PEX16 (Lin et al., 1999; Hu et al., 2002; Sparkes et al., 2003; Fan et al., 2005; Boisson-Dernier et al., 2008; Goto et al., 2011). Therefore, partial loss of function of each PEX, rather than complete abolition of PEX function, is desirable to study plant peroxisomes. EMS-induced mutagenesis causes single nucleotide substitutions and is therefore expected to induce a milder loss of function than null mutations. The functions of the regions of PEX13 and PEX12 that are affected by the apem2 and apem4 mutations, respectively, are unknown, and further analysis is required to understand how these regions contribute to the functions of the proteins and their interactions with other proteins. APME9 is functionally equivalent to PEX15/PEX26 found in fungi and mammals. It has no detectable sequence similarity to PEX15 or PEX26, which emphasizes the major advantage of the forward genetic approach, i.e., the discovery of novel factors. Analysis of APEM10/LON2 revealed that its chaperone and protease functions, as well as autophagy acting in concert with these functions, are required for peroxisome quality control. APEM3/PMP38/PXN is a membrane transporter that supplies NAD+ to the peroxisomal matrix. Depletion of NAD+ induces accumulation of fatty acids, and their toxicity may result in enlargement of peroxisomes in apem3. The study of PEUP1/ATG2, PEUP2/ATG18A, and PEUP4/ATG7 provided evidence that damaged peroxisomes accumulate a massive amount of inactivated catalase, and abnormal oxidative conditions induce pexophagy. In addition, the study of PEUP17/ATG5 and PEUP22/ATG7 has shed light on a new type of microautophagy induced by starvation.
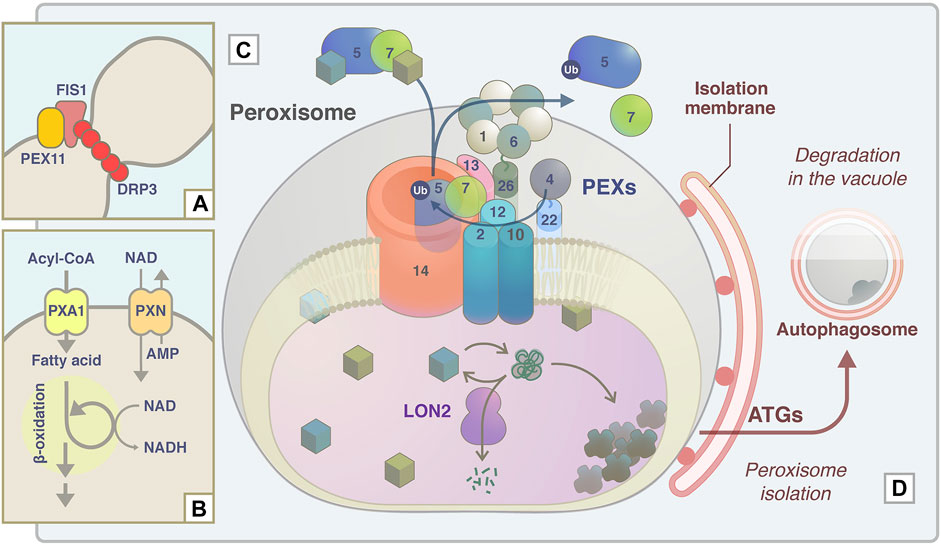
FIGURE 4. Schematic model of APEM protein functions in peroxisome proliferation, lipid metabolism, protein transport machinery, and quality control. (A) During peroxisome fission, DRP3A/APEM1 is recruited to the peroxisome division site together with DRP3B in a PEX11- and FIS1-dependent manner (Kao et al., 2018). DRP proteins are polymerized and constrict to divide peroxisomes. (B) PXN/APEM3 import NAD into the peroxisomal matrix and this is required for optimal fatty acid β-oxidation. (C) Peroxisomal matrix proteins are captured by the receptor PEX5 or PEX7. The PEX5-PEX7-cargo complex translocates to peroxisomes by binding to the docking complex consisting of PEX14 and PEX13/APEM2. The E2 ubiquitin ligase PEX4 and the E3 ligase PEX2/PEX10/PEX12 supposedly ubiquitinate PEX5 to export it from the peroxisomal membrane with/without the force generated by the APEM9/PEX15/PEX26-tethered AAA-ATPase PEX1-PEX6 complex. Experimental data support the interactions between PEX13 and PEX7 (Mano et al., 2006), PEX13 and PEX15/PEX26 (Li et al., 2014), and PEX7 and PEX12 (Singh et al., 2009). (D) Damaged and/or unwanted peroxisomal proteins are supposedly maintained or degraded by the chaperone/protease activity of LON2/APEM10 protein. Excess damaged proteins accumulate inside peroxisomes. Peroxisomes become oxidative upon catalase inactivation and aggregation, and these peroxisomes are targeted for pexophagy to be degraded in the vacuole (Shibata et al., 2013). ATG proteins, including ATG2/PEUP1, ATG18A/PEUP2, ATG7/PEUP4/PEUP22, and ATG5/PEUP17, are involved in this process.
2.5 Other Imaging Analyses of Peroxisomes
2.5.1 Imaging-Based Mutant Screening Other Than That of apem and peup Mutants
The screening strategy, e.g., the parental strain to be mutagenized and the criteria for isolation of mutants, depends on the experiments. The most reported imaging-based approach is mutagenesis and screening of strains with visualized peroxisomes as described in the above section. Zhang and Hu. (2009) reported the screening and identification of A. thaliana peroxisome division/proliferation deficient (pdd) mutants to identify factors involved in peroxisome division and proliferation pathways. pdd mutants were isolated from parental EMS-mutagenized A. thaliana, which expressed 35S promoter-driven YFP-PTS1. They identified pdd1 and pdd2 as DRP3A alleles (Aung and Hu, 2009; Zhang and Hu, 2009). Rinaldi et al. (2016) reported a massive number of mutants, with 34 novel alleles of 15 genes involved in oil body mobilization, fatty acid β-oxidation, the glyoxylate cycle, peroxisome fission, and pexophagy (Rinaldi et al., 2016). These mutants were isolated from EMS-mutagenized GFP-PTS1-expressing plants, and the mutations were determined by a combination of map-based cloning and direct or whole genome sequencing. Although it had been reported that deficiencies in enzymes involved in peroxisomal β-oxidation led to swollen peroxisomes, this had not been proven. One of the main achievements of this report is genetically proving that accumulation of fatty acids inside peroxisomes leads to enlargement of peroxisomes using a number of isolated mutants defective in peroxisomal metabolism and transporters (Rinaldi et al., 2016). The visualization of peroxisomes is also effective in mammalian cells to isolate peroxisome-deficient mutants. Ghaedi et al. (1999) generated Chinese Hamster Ovary cells stably transformed with GFP-PTS1 or PTS2-GFP, and mutagenized these cells with N-methyl-N’-nitro-N-nitrosoguanidine. The mutant phenotypes were investigated by observation under a fluorescence microscope (Ghaedi et al., 1999). Another group employed a similar technique, and these studies identified several genes related to peroxisome biogenesis (Ito et al., 2000; Ghaedi and Fujiki, 2008). Comprehensive, imaging-based, large-scale screening has been achieved in yeast. Yeast is a very useful tool for functional analysis of proteins because of the ease of genetic analyses and the large number of established analytical tools. A collection of gene deletions covering 96% of yeast open reading frames, called a modified synthetic genetic array (SGA), is available, and automated screening is also possible (Giaever et al., 2002). Wolinski et al. (2009) established an experimental platform that can be connected to a SGA, enabling qualitative, quantitative, and automated large-scale analysis of GFP-labeled peroxisomes in yeast cells (Wolinski et al., 2009). The technique identified two novel genes that had not been previously linked to peroxisome biogenesis as well as all known factors required for PTS1-dependent protein transport. Cohen et al. (2014) employed dual reporters to visualize peroxisomes. Cherry fused with PTS1 (Cherry-PTS1) and GFP-tagged peroxisomal membrane protein Ant1 (GFP-Ant1) allow assessment of the efficiency of not only matrix protein sorting but also membrane protein transport and peroxisome formation (Cohen et al., 2014). Use of a combination of the SGA approach and multiple reporters identified a P-type ATPase and ion transporter in the ER membrane (Spf1), which is required for delivery of peroxisomal membrane proteins to peroxisomes, and revealed that peroxisomes localize in close contact with mitochondria and ER (Cohen et al., 2014).
In addition to mutant screening using strains with visualized peroxisomes, mutant screenings focusing on specific peroxisomal factors or phenomena have also been reported. Burkhart et al. (2013) focused on degradation of peroxisomal enzymes in glyoxysomes, which are a type of peroxisomes in which the glyoxylate cycle occurs and are found in cotyledons during early germination of seedlings (Burkhart et al., 2013). ISOCITRATE LYASE (ICL), a glyoxysomal enzyme, is required for lipid conversion to sucrose during post-germinative growth and becomes unnecessary once photosynthesis starts in seedlings (Nishimura et al., 1982; Titus and Becker, 1985). To identify components required for degradation of unwanted peroxisomal matrix proteins, a transgenic plant expressing GFP-ICL was mutagenized with EMS. Mutants that retained GFP-ICL longer than the WT, named persistent GFP-ICL fluorescence (pfl) mutants, were isolated (Burkhart et al., 2013). In the screening, proteins involved in the docking complex (PEX14) and recycling complex (PEX2, PEX6, and PEX10) of the matrix protein transport machinery and a β-oxidation enzyme (PED1/KAT2) were identified from the pfl mutants (Burkhart et al., 2013; Burkhart et al., 2014). From these analyses, the authors concluded that efficient degradation of peroxisomal matrix proteins requires proteins to be sorted inside peroxisomes and also seems to require an appropriate metabolic level of peroxisomes and the entire peroxisomal protein transport system (Burkhart et al., 2013). A unique and large-scale screening was performed in yeast to study the priority of peroxisomal protein targeting. Peroxisomal proteins containing the PTS1 targeting signal are captured by the receptor PEX5 and transported to peroxisomes. If the level of cargo becomes high, the occupancy of PEX5 increases and only proteins with a high targeting priority will localize to peroxisomes. Rosenthal et al. (2020) generated yeast strains that express varying levels of PTS1 fused to mCherry (mCherry-SKL) by changing the copy number of mCherry-SKL in the construct (Rosenthal et al., 2020). Around 90 strains expressing peroxisomal proteins tagged with GFP were transformed with low or high levels of mCherry-SKL, and the localization of GFP to peroxisomes was measured in each strain using an automated microscopy platform.
2.5.2 Visualization of Peroxisomes Using Other Imaging Technical Methods
In the correlative light and electron microscopy (CLEM) method, fluorescence and dyes in a sample are observed with an optical microscope, and then the same area is observed with an electron microscope (Razi and Tooze, 2009; Jahn et al., 2012). Although various CLEM methods have been developed and reported, they have mainly used cultured animal cells, and there are few reports on methods suitable for plant tissues and cells. Toyooka (2016) developed a new CLEM method to accurately capture the localization of fluorescently labeled biomolecules in plant tissues and cells at high resolution, and applied the method to A. thaliana with GFP-labeled peroxisomes (Toyooka, 2016). In yeast Hansenula polymorpha, the peroxisome-vacuole contact site was visualized using the CLEM method, and Pex3 is shown to be involved in the formation of peroxisome-vacuole contact sites (Wu et al., 2019). Bykov et al. (2019) developed a new methodology, MultiCLEM, to allow systematic, parallel, high-throughput screening for traits using the CLEM with computer image analysis (Bykov et al., 2019). By applying MultiCLEM to different yeast strains with GFP-labelled peroxisomes, they successfully identified peroxisomes in both fluorescence and electron microscopic images (Bykov et al., 2019). Since this methodology apparently can be scaled up to higher throughputs, not limited to yeast, it is expected to enable electron microscopy a powerful screening method.
Three-dimensional ultrastructural images with quantitative information can be reconstructed from image data obtained by transmission electron microscopy or focused ion beam scanning electron microscopy (FIB-SEM). Recently, Zechmann et al. (2021) reported that quantitative changes of the volumes of viral inclusion bodies, chloroplast fine structures, mitochondria, and peroxisomes using reconstituted 3D image data (Zechmann et al., 2021). They reconstituted 3D images during the process of Tobacco mosaic virus and Zucchini yellow mosaic virus infection in tobacco and pumpkin plants from serial sections obtained by transmission electron microscopy and extracted quantitative information on the size and number of peroxisomes and other organelles (Zechmann et al., 2021). In mouse liver hepatocytes, the wrappER, a curved wrapping type of rough ER accumulates fatty acid and fatty acid-binding proteins of the lipocalin family and regulates intracellular and systemic lipid flux by establishing extensive contact with almost all mitochondria. Ilacqua et al. (2022) showed that the wrappER contacts with peroxisomes in addition to mitochondria by analyzing a large portion of the cell volume of the hepatocytes by serial section electron tomography coupled to 3D reconstruction. Xu et al. (2017) reported an extended FIB-SEM system for high volume 3D imaging suitable for connectomics (Xu et al., 2017). Using this new system, the authors have successfully imaged large, complex samples of mammalian neural tissue, Drosophila brain, and Chlamydomonas reinhardtii in entirety with sufficient detail to allow high-quality reconstruction of connections. The introduction of these new imaging techniques is expected to make it possible to analyze peroxisome dynamics at higher resolution, more easily, and with a larger volume of data.
3 Evolution of Peroxisome Dynamics in Land Plants
3.1 The Liverwort M. polymorpha as a Model
Our current understanding of the biogenesis and function of peroxisomes in land plants is largely based on the studies using A. thaliana as described above. To obtain more insights into the evolution of peroxisome dynamics in land plants, yet another model plant that is divergent from A. thaliana is needed: the liverwort M. polymorpha. This bryophyte species is an early diverging land plant and thus retains features of ancestral land plants. Its main form during its gametophyte-dominant life cycle is a complex thalloid structure with cupules containing gemmae for asexual propagation and rhizoids on the ventral and dorsal surfaces, respectively (Figures 5A,B; Shimamura, 2016). Like many other bryophyte species, M. polymorpha is dioicous and has heteromorphic sex chromosomes: U with the sex determining gene for female and V for male (Haupt, 1932; Bowman et al., 2017; Montgomery et al., 2020; Iwasaki et al., 2021). Under long-day conditions enriched with far-red light, M. polymorpha initiates transition from the vegetative to reproductive phase, generating sexual organs (Figures 5C,D; Chiyoda et al., 2008; Inoue et al., 2019). Motile sperm are released from a male reproductive organ, the antheridiophore, and navigate to a female reproductive organ, the archegoniophore. Sperm can be readily collected from male plants and applied to female plants, meaning genetic crosses of M. polymorpha are easily performed. After fertilization, a zygote continues mitotic division to form a diploid multicellular sporangium. Meiotic division of spore mother cells in each sporangium produces as many as 300,000 haploid spores (O'Hanlon, 1926), which is advantageous for forward genetics by mutagenesis. The genome of M. polymorpha has a set of regulatory systems comparable with that in angiosperms but in a remarkably less redundant form, presumably representing the situation in ancestral land plants (Bowman et al., 2017). Its low genetic redundancy, together with the molecular and genetic tools described below, makes M. polymorpha a model plant of choice for both forward and reverse genetics to elucidate the molecular machineries that operate in land plants (Ishizaki et al., 2016; Sauret-Güeto et al., 2020; Kohchi et al., 2021).
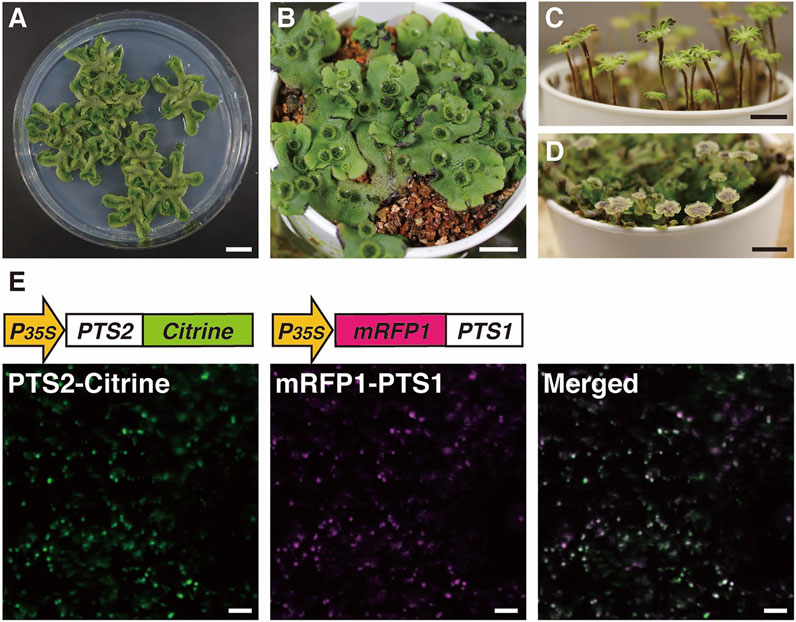
FIGURE 5. Images of M. polymorpha and visualization of peroxisomes using Citrine- and mRFP1-fused proteins. Vegetative haploid life form (thallus) on agar plate (A) and vermiculite (B) (Shimamura, 2016). Female (C) and male (D) sexual organs from the haploid thallus of a female plant or a male plant, respectively (Chiyoda et al., 2008; Inoue et al., 2019). Bars, 1 cm. (E) Fluorescence in peroxisomes was observed in thallus epidermal cells expressing both pro35S:PTS2-Citrine and pro35S:mRFP1-PTS1 genes (Mano et al., 2018). Bars, 10 μm.
Genetic transformation of M. polymorpha has been well-established (Chiyoda et al., 2008; Ishizaki et al., 2008; Kubota et al., 2013) and continuously improved (Tsuboyama-Tanaka and Kodama, 2015; Tsuboyama and Kodama, 2018; Iwakawa et al., 2021; Seo et al., 2021). A wide range of gateway vectors for Agrobacterium-mediated transformation have been developed and made available for simple gene transfer, conditional gene expression and deletion (Nishihama et al., 2016), reporter assays (Ishizaki et al., 2015; Mano et al., 2018), and CRISPR/Cas9-mediated genome editing (Sugano et al., 2018; Sugano and Nishihama, 2018). Gene targeting including knock-in by homologous recombination is also feasible (Ishizaki et al., 2013; Yamaoka et al., 2018; Yasui et al., 2019; Kato et al., 2020).
There are web-based databases where genomic and related resources can be accessed, including MarpolBase (https://marchantia.info, Bowman et al., 2017; Montgomery et al., 2020), Phytozome (https://phytozome-next.jgi.doe.gov/info/Mpolymorpha_v3_1), and Ensembl Plants (https://plants.ensembl.org/Marchantia_polymorpha/Info/Index). MarpolBase is an up-to-date comprehensive site dedicated to M. polymorpha studies, where researchers can browse, search, and retrieve sequences and genes, design CRISPR/Cas9 target sites, and browse lists of M. polymorpha-related literature. The guideline for M. polymorpha gene nomenclature is also available at this site (Bowman, 2016).
3.2 Visualization of Peroxisomes in M. polymorpha
As described in Section 2, visualization of peroxisomes with fluorescent proteins in A. thaliana greatly helped to unveil the molecular dynamics of plant peroxisomes. To investigate whether the regulatory mechanisms of peroxisome dynamics that were clarified using A. thaliana are conserved among plant species or species-specific, transgenic M. polymorpha plants expressing Citrine-PTS1, mRFP1-PTS1, and PTS2-Citrine have been generated (Figure 5E; Ogasawara et al., 2013; Kimura and Kodama, 2016; Mano et al., 2018). Research using these transgenic plants revealed that the morphology, size, and movement of peroxisomes in M. polymorpha resemble those in A. thaliana (Mano et al., 2018). Moreover, peroxisomes relocated from the periclinal cell wall to the anticlinal cell wall after cold treatment (Ogasawara et al., 2013) and this relocation was mediated via actin filaments (Kimura and Kodama, 2016), suggesting that similar mechanisms mediate subcellular positioning of peroxisomes in response to environmental stimuli in M. polymorpha and A. thaliana (Oikawa et al., 2015). To generate transgenic M. polymorpha with visualized peroxisomes, PTS1 or PTS2 was fused to fluorescent proteins, meaning that both PTS1- and PTS2-dependent protein transport pathways could be analyzed. Genes encoding proteins with high similarities to PEX5 and PEX7, which are receptors for PTS1 and PTS2, respectively, are present in the M. polymorpha genome (Table 2). This indicates that both pathways were required from the beginning of evolution of land plants.
3.3 Bioinformatics Analysis to Identify Peroxisomal Genes
Peroxisome biogenesis requires a set of specialized proteins, peroxins, encoded by PEX genes. A. thaliana has 22 PEX genes (Table 2; Cross et al., 2016), while M. polymorpha has 18. Most PEX genes, except for PEX3, PEX11c/d/e, and PEX19, in A. thaliana have a single counterpart in M. polymorpha. Duplication and triplication of PEX3/PEX19 and PEX11, respectively, in A. thaliana explains why there are more PEX genes than in M. polymorpha. Phylogenetic analysis revealed that duplication of PEX11a and PEX11b predates the divergence of Zygnematales and Embryophytes, and the divergence of PEX11a/b and PEX11c/d/e likely occurred even earlier (Figure 6), suggesting that PEX11 should be further categorized into three subclasses, PEX11a, PEX11b, and others, increasing the total number of PEX subclasses in land plants to 18. It should be noted that M. polymorpha has the complete set of 18 PEX genes without duplication, which makes it suitable for functional and evolutionary analyses. The set of PEX genes in M. polymorpha appears to have been already established in the common ancestor of Zygnematales and Embryophytes, although there are a few missing genes in Mesotaenium endlicherianum and Klebsormidium nitens, which could be explained by secondary loss in these lineages and/or the presence of sequence gaps (Table 2).
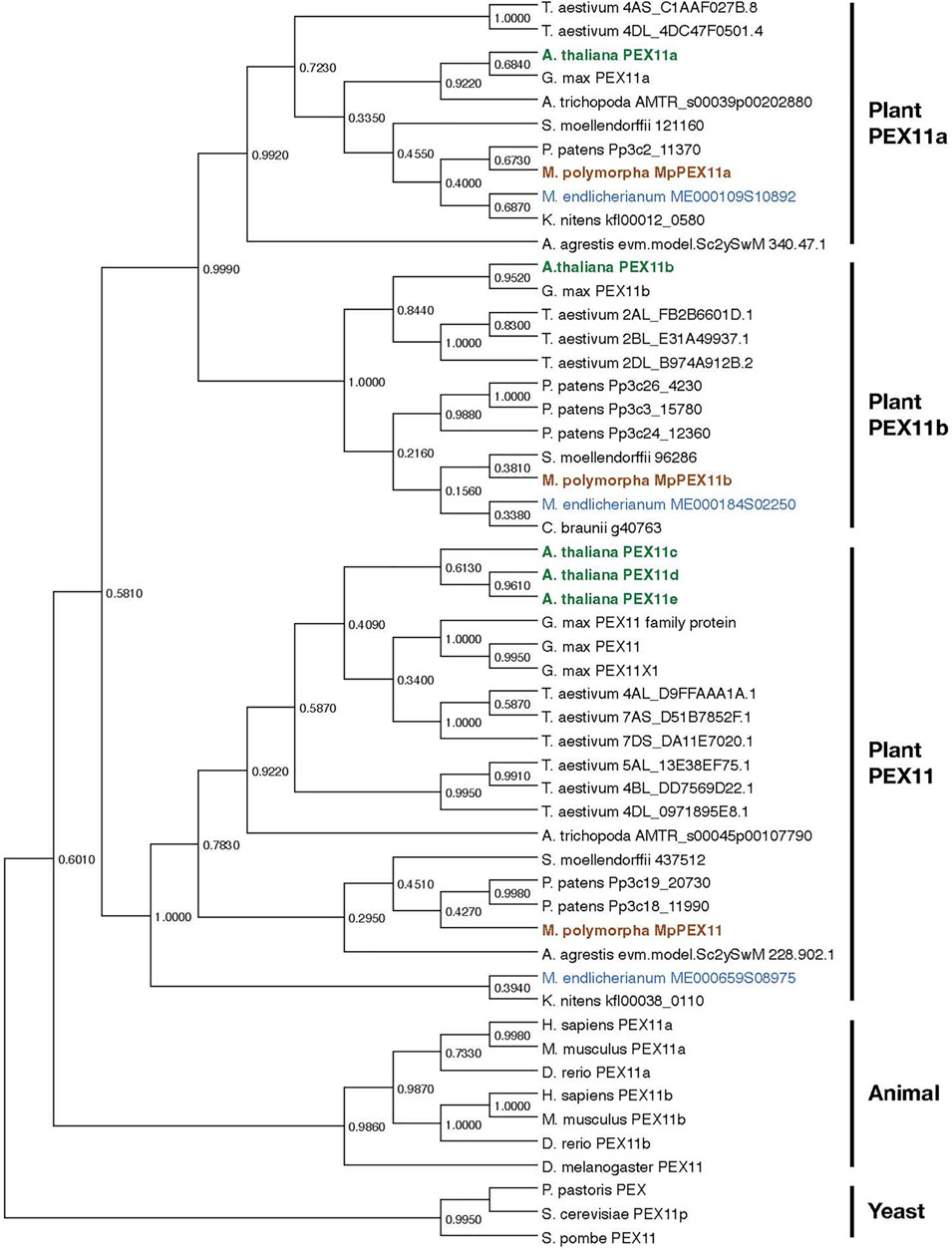
FIGURE 6. Phylogenetic relationships of PEX11 subfamilies. The numbers are the proportion of trees in which the associated sequences cluster together. Sequences of A. thaliana, M. polymorpha, and M. endlicherianum are colored as representatives from among angiosperms, bryophytes, and algae, respectively. The phylogenetic tree for PEX11 homologs was inferred using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992) with MEGA11 (Stecher et al., 2020; Tamura et al., 2021). All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). Orthologue sequences in plants were obtained from the datasets for C. braunii (Nishiyama et al., 2018), M. endlicherianum (Cheng et al., 2019), K. nitens (Hori et al., 2014), M. polymorpha (Montgomery et al., 2020), P. patens (Lang et al., 2018), A. agrestis (Li et al., 2020), S. moellendorffii (Banks et al., 2011), A. trichopoda (Amborella Genome Project, 2013), A. thaliana (Berardini et al., 2015), G. max (Schmutz et al., 2010), and T. aestivum (International Wheat Genome Sequencing Consortium, 2014). Other PEX11 species used in this analysis are P. pastoris (ANZ76138.1), S. cerevisiae (AJT75217.1), S. pombe (NP_595177.1), D. melanogaster (NP_611071.1), D. rerio (NP_001096590.1 and NP_001039319.1), H. sapiens (NP_003838.1 and NP_003837.1), and M. musculus (NP_035198.1 and NP_001155859.1).
3.4 Genome Editing to Analyze Peroxisome Dynamics in M. polymorpha
Genome editing is a powerful tool for functional analysis of gene products and is applied in various organisms. CRISPR/Cas9-based vectors with high efficiency have been established and used in M. polymorpha (Sugano et al., 2018; Sugano and Nishihama, 2018). As described above, bioinformatics analysis of peroxisomal genes in A. thaliana revealed the presence of orthologous genes in the M. polymorpha genome. For example, Mp6g18570 shows high similarity to At3g19190, which is the responsible gene in the peup1/atg2 mutant (Shibata et al., 2013). Norizuki et al. (2019) performed CRISPR/Cas9-based genome editing of several M. polymorpha ATG genes including Mp6g18570 (Norizuki et al., 2019). Mp6g18570-edited M. polymorpha exhibited earlier senescence than the WT plant (Norizuki et al., 2019), consistent with the phenotype of the A. thaliana peup1/atg2 mutant. PEUP1/ATG2 has a role in autophagy (Shibata et al., 2013), demonstrating the existence of a similar degradation system in M. polymorpha. As described above, some A. thaliana peroxisomal genes, such as PEX3, PEX11, and PEX19, constitute a gene family, and they have single counterparts in M. polymorpha. This is true of other genes encoding metabolic enzymes that function inside peroxisomes; the number of genes constituting the family is decreased in M. polymorpha. Therefore, M. polymorpha is a good material to investigate the functions of gene products in peroxisome research because the generation of mutants with knockout and/or knockdown of gene products requires manipulation of fewer genes and thus is easier. This approach will uncover the mechanisms underlying peroxisome dynamics and diversification of peroxisomes during the evolution of plants, accelerating peroxisome research.
4 Conclusion
A. thaliana mutants with peroxisomes defects that were obtained using transgenic A. thaliana with visualized peroxisomes as a parent material have greatly helped to identify essential components for regulation of peroxisome dynamics. A similar approach in which plants with visualized peroxisomes, including M. polymorpha, as a parent material are randomly mutagenized can be used to obtain valuable mutants based on imaging analysis. Together with plants with visualized peroxisomes, genome editing of M. polymorpha target genes identified by bioinformatic analysis can be performed to investigate the dynamics and diversity of peroxisomes in land plants.
Screening and phenotyping of a large number of mutants take an enormous amount of time. Automated screening, as in the yeast example described above, could dramatically reduce the experimental time. Li et al. (2021) recently established Deep Learning of the Morphology of Organelles (DeepLearnMOR), which can categorize mutant phenotypes and identify an abnormal morphology with 97% accuracy (Li et al., 2021), and can be used to accurately and quantitatively analyze phenotypes. It is hoped that this new technology will accelerate and advance identification of mutants in peroxisome biology in the near future.
5 Future Prospects for Plant Peroxisome Research
As described in this review, imaging-based mutant screening has identified various factors involved in peroxisome dynamics and have elucidated their molecular mechanisms. However, there are still unresolved issues in plant peroxisome research that remain to be addressed. For example, autophagosomes have been reported to access damaged peroxisomes, but how autophagosomes detect internal peroxisomal abnormalities and recognize only abnormal peroxisomes in plants is not understood. Peroxisomes interact with other organelles, such as endoplasmic reticulum, chloroplasts, and mitochondria, at membrane contact sites (MCS) between organelles to exchange metabolites and signals, thereby playing a role in maintaining cellular homeostasis (Prinz, 2014; Perico and Sparkes, 2018). However, the mechanism of MCS formation between peroxisomes and other organelles is not yet fully understood. The morphology and movement of plant peroxisomes are influenced by ROS induced by environmental stresses such as high light and high temperature. ROS alter the organelle membrane structure, resulting in the formation of peroxules from peroxisomes (Mathur, 2021). However, little is known about the dynamics of peroxisomal membrane lipids. Recent studies have also revealed that peroxisomes have essential roles in reproductive processes, such as pollen fertility, male-female recognition, and embryo development after fertilization (Sparkes et al., 2003; Fan et al., 2005; Boisson-Dernier et al., 2008; Goto et al., 2011; Goto-Yamada et al., 2014b). However, the roles of peroxisomes and molecular mechanisms underlying their functions in the reproductive process are less well understood than in the analysis of roots and leaves.
To address the above issues, it is necessary to introduce new imaging techniques such as super-resolution microscopy analysis using the stimulated release depletion method and structured illumination microscopy combined with chemical approaches (Ovečka et al., 2022). In addition, quantitative methods using fluorescent probes to visualize ROS (de Torres Zabala et al., 2015), redox state (Exposito-Rodriguez et al., 2017), ATP (Voon et al., 2018), and NADH/NAD+ (Lim et al., 2020), split fluorescent proteins, and FRET techniques (Shai et al., 2018; Vallese et al., 2020) are also expected to be available for isolation of interesting peroxisome mutants. Furthermore, combining femtosecond laser and optical tweezers (Oikawa et al., 2015; Gao et al., 2016) with biochemical approaches such as proteome (or lipidome) analyses by immunoprecipitation and mass spectrometry is expected to be useful in the search for proteins that mediate peroxisome tethering with other cellular structures. Further advances in imaging technology are expected to elucidate various peroxisome-mediated biological phenomena and the molecular mechanisms that control them, such as the interaction between peroxisomes and other organelles.
Author Contributions
SG-Y and SM conceived and coordinated the study. KTY performed the bioinformatics analysis. SG-Y, KO, KH, and SM performed the other experiments. SG-Y, KO, KTY, and SM wrote the paper. MK and MN reviewed and edited the paper. All authors analyzed the results and approved the final version of the manuscript.
Funding
This work was supported in part by JSPS KAKENHI (Grant Numbers 20059035, 2057004, 22112523, 17K07457, and 20K06711 awarded to SM) and the National Science Centre, Poland (UMO-2019/34/E/NZ3/00299 awarded to SG-Y).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the staff at the Model Plant Facilities at the NIBB Bioresource Center, the Functional Genomics Facility, and the Spectrography and Bioimaging Facility at the NIBB Core Research Facilities for technical support. We are also grateful to Chihiro Nakamori, Masami Araki, and Azusa Matsuda for supporting the experiments and taking care of the plants as technical staff.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.883491/full#supplementary-material
Supplementary Movie S1 | Movement of peroxisomes in root cells. Time-lapse images were collected every 2 s for 1 min. Most peroxisomes show non-directional and random motion. The movie is shown at double speed.
References
Agne, B., Meindl, N. M., Niederhoff, K., Einwächter, H., Rehling, P., and Sickmann, A. (2003). Pex8p: An Intraperoxisomal Organizer of the Peroxisomal Import Machinery. Mol. Cel 11, 635–646.doi:10.1016/s1097-2765(03)00062-5
Amborella Genome Project (2013). The Amborella Genome and the Evolution of Flowering Plants. Science 342 (6165), 1241089. doi:10.1126/science.1241089
Aung, K., and Hu, J. (2012). Differential Roles of Arabidopsis Dynamin-Related Proteins DRP3A, DRP3B, and DRP5B in Organelle Division. J. Integr. Plant Biol. 54 (11), 921–931. doi:10.1111/j.1744-7909.2012.01174.x
Aung, K., and Hu, J. (2009). The Arabidopsis Peroxisome Division Mutant Pdd2 Is Defective in the DYNAMIN-RELATED PROTEIN3A (DRP3A) Gene. Plant Signal. Behav. 4 (6), 542–544. doi:10.4161/psb.4.6.8699
Baillie, A. L., Falz, A. L., Müller-Schüssele, S. J., and Sparkes, I. (2020). It Started with a Kiss: Monitoring Organelle Interactions and Identifying Membrane Contact Site Components in Plants. Front. Plant Sci. 11, 517. doi:10.3389/fpls.2020.00517
Baker, A., Lanyon-Hogg, T., and Warriner, S. L. (2016). Peroxisome Protein Import: a Complex Journey. Biochem. Soc. Trans. 44 (3), 783–789. doi:10.1042/bst20160036
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., dePamphilis, C., et al. (2011). The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 332 (6032), 960–963. doi:10.1126/science.1203810
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis Information Resource: Making and Mining the "gold Standard" Annotated Reference Plant Genome. Genesis 53 (8), 474–485. doi:10.1002/dvg.22877
Bernhardt, K., Wilkinson, S., Weber, A. P. M., and Linka, N. (2012). A Peroxisomal Carrier Delivers NAD+ and Contributes to Optimal Fatty Acid Degradation during Storage Oil Mobilization. Plant J. 69, 1–13. doi:10.1111/j.1365-313x.2011.04775.x
Birschmann, I., Stroobants, A. K., van den Berg, M., Schäfer, A., Rosenkranz, K., Kunau, W. H., et al. (2003). Pex15p of Saccharomyces cerevisiae Provides a Molecular Basis for Recruitment of the AAA Peroxin Pex6p to Peroxisomal Membranes. Mol. Biol. Cel 14 (6), 2226–2236. doi:10.1091/mbc.e02-11-0752
Boisson-Dernier, A., Frietsch, S., Kim, T.-H., Dizon, M. B., and Schroeder, J. I. (2008). The Peroxin Loss-Of-Function Mutation Abstinence by Mutual Consent Disrupts Male-Female Gametophyte Recognition. Curr. Biol. 18, 63–68. doi:10.1016/j.cub.2007.11.067
Bottger, G., Barnett, P., Klein, A. T., Kragt, A., Tabak, H. F., and Distel, B. (2000). Saccharomyces cerevisiae PTS1 Receptor Pex5p Interacts with the SH3 Domain of the Peroxisomal Membrane Protein Pex13p in an Unconventional, Non-PXXP-related Manner. Mol. Biol. Cel 11 (11), 3963–3976. doi:10.1091/mbc.11.11.3963
Bowman, J. L. (2016). A Brief History of Marchantia from greece to Genomics. Plant Cel Physiol 57 (2), 210–229. doi:10.1093/pcp/pcv044
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into Land Plant Evolution Garnered from the Marchantia Polymorpha Genome. Cell, 287–304. doi:10.1016/j.cell.2017.09.030
Burkhart, S. E., Kao, Y. T., and Bartel, B. (2014). Peroxisomal Ubiquitin-Protein Ligases Peroxin2 and Peroxin10 Have Distinct but Synergistic Roles in Matrix Protein Import and Peroxin5 Retrotranslocation in Arabidopsis. Plant Physiol. 166 (3), 1329–1344. doi:10.1104/pp.114.247148
Burkhart, S. E., Lingard, M. J., and Bartel, B. (2013). Genetic Dissection of Peroxisome-Associated Matrix Protein Degradation in Arabidopsis thaliana. Genetics 193, 125–141. doi:10.1534/genetics.112.146100
Bykov, Y. S., Cohen, N., Gabrielli, N., Manenschijn, H., Welsch, S., Chlanda, P., et al. (2019). High-throughput Ultrastructure Screening Using Electron Microscopy and Fluorescent Barcoding. J. Cel Biol. 218 (8), 2797–2811. doi:10.1083/jcb.201812081
Cheng, S., Xian, W., Fu, Y., Marin, B., Keller, J., Wu, T., et al. (2019). Genomes of Subaerial Zygnematophyceae Provide Insights into Land Plant Evolution. Cell 179 (5), 1057–1067. doi:10.1016/j.cell.2019.10.019
Chiyoda, S., Ishizaki, K., Kataoka, H., Yamato, K. T., and Kohchi, T. (2008). Direct Transformation of the Liverwort Marchantia Polymorpha L. By Particle Bombardment Using Immature Thalli Developing from Spores. Plant Cel Rep 27, 1467–1473. doi:10.1007/s00299-008-0570-5
Ciniawsky, S., Grimm, I., Saffian, D., Girzalsky, W., Erdmann, R., and Wendler, P. (2015). Molecular Snapshots of the Pex1/6 AAA+ Complex in Action. Nat. Commun. 6, 7331. doi:10.1038/ncomms8331
Cohen, Y., Klug, Y. A., Dimitrov, L., Erez, Z., Chuartzman, S. G., Elinger, D., et al. (2014). Peroxisomes Are Juxtaposed to Strategic Sites on Mitochondria. Mol. Biosyst. 10 (7), 1742–1748. doi:10.1039/c4mb00001c
Corpas, F. J., González-Gordo, S., and Palma, J. M. (2020). Plant Peroxisomes: A Factory of Reactive Species. Front. Plant Sci. 11, 853. doi:10.3389/fpls.2020.00853
Cross, L. L., Ebeed, H. T., and Baker, A. (2016). Peroxisome Biogenesis, Protein Targeting Mechanisms and PEX Gene Functions in Plants. Biochim. Biophys. Acta 1863 (5), 850–862. doi:10.1016/j.bbamcr.2015.09.027
Cui, S., Hayashi, Y., Otomo, M., Mano, S., Oikawa, K., Hayashi, M., et al. (2016). Sucrose Production Mediated by Lipid Metabolism Suppresses the Physical Interaction of Peroxisomes and Oil Bodies during Germination of Arabidopsis thaliana. J. Biol. Chem. 291, 19734–19745. doi:10.1074/jbc.m116.748814
de Torres Zabala, M., Littlejohn, G., Jayaraman, S., Studholme, D., Bailey, T., Lawson, T., et al. (2015). Chloroplasts Play a central Role in Plant Defence and Are Targeted by Pathogen Effectors. Nat. Plants 1, 15074. doi:10.1038/nplants.2015.74
Douangamath, A., Filipp, F. V., Klein, A. T. J., Barnett, P., Zou, P., Voorn-Brouwer, T., et al. (2002). Topology for Independent Binding of α-helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain. Mol. Cel 10, 1007–1017. doi:10.1016/s1097-2765(02)00749-9
Eubel, H., Meyer, E. H., Taylor, N. L., Bussell, J. D., O'Toole, N., Heazlewood, J. L., et al. (2008). Novel Proteins, Putative Membrane Transporters, and an Integrated Metabolic Network Are Revealed by Quantitative Proteomic Analysis of Arabidopsis Cell Culture Peroxisomes. Plant Physiol. 148, 1809–1829. doi:10.1104/pp.108.129999
Exposito-Rodriguez, M., Laissue, P. P., Yvon-Durocher, G., Smirnoff, N., and Mullineaux, P. M. (2017). Photosynthesis-dependent H2O2 Transfer from Chloroplasts to Nuclei Provides a High-Light Signalling Mechanism. Nat. Commun. 8 (1), 49. doi:10.1038/s41467-017-00074-w
Fan, J., Quan, S., Orth, T., Awai, C., Chory, J., and Hu, J. (2005). The Arabidopsis PEX12 Gene Is Required for Peroxisome Biogenesis and Is Essential for Development. Plant Physiol. 139, 231–239. doi:10.1104/pp.105.066811
Farmer, L. M., Rinaldi, M. A., Young, P. G., Danan, C. H., Burkhart, S. E., and Bartel, B. (2013). Disrupting Autophagy Restores Peroxisome Function to an Arabidopsis Lon2 Mutant and Reveals a Role for the LON2 Protease in Peroxisomal Matrix Protein Degradation. Plant Cell 25 (10), 4085–4100. doi:10.1105/tpc.113.113407
Frederick, S. E., and Newcomb, E. H. (1969). Cytochemical Localization of Catalase in Leaf Microbodies (Peroxisomes). J. Cel Biol. 43 (2), 343–353. doi:10.1083/jcb.43.2.343
Fujiki, Y., Abe, Y., Imoto, Y., Tanaka, A. J., Okumoto, K., Honsho, M., et al. (2020). Recent Insights into Peroxisome Biogenesis and Associated Diseases. J. Cel Sci. 133 (9). doi:10.1242/jcs.236943
Fujimoto, M., Arimura, S.-i., Mano, S., Kondo, M., Saito, C., Ueda, T., et al. (2009). Arabidopsis Dynamin-Related Proteins DRP3A and DRP3B Are Functionally Redundant in Mitochondrial Fission, but Have Distinct Roles in Peroxisomal Fission. Plant J. 58, 388–400. doi:10.1111/j.1365-313x.2009.03786.x
Fukao, Y., Hayashi, Y., Mano, S., Hayashi, M., and Nishimura, M. (2001). Developmental Analysis of a Putative ATP/ADP Carrier Protein Localized on Glyoxysomal Membranes during the Peroxisome Transition in Pumpkin Cotyledons. Plant Cel Physiol 42, 835–841. doi:10.1093/pcp/pce108
Gao, H., Metz, J., Teanby, N. A., Ward, A. D., Botchway, S. W., Coles, B., et al. (2016). In Vivo quantification of Peroxisome Tethering to Chloroplasts in Tobacco Epidermal Cells Using Optical Tweezers. Plant Physiol. 170 (1), 263–272. doi:10.1104/pp.15.01529
Gardner, B. M., Castanzo, D. T., Chowdhury, S., Stjepanovic, G., Stefely, M. S., Hurley, J. H., et al. (2018). The Peroxisomal AAA-ATPase Pex1/Pex6 Unfolds Substrates by Processive Threading. Nat. Commun. 9 (1), 135. doi:10.1038/s41467-017-02474-4
Ghaedi, K., and Fujiki, Y. (2008). Isolation and Characterization of Novel Phenotype CHO Cell Mutants Defective in Peroxisome Assembly, Using ICR191 as a Potent Mutagenic Agent. Cell Biochem. Funct. 26 (6), 684–691. doi:10.1002/cbf.1493
Ghaedi, K., Kawai, A., Okumoto, K., Tamura, S., Shimozawa, N., Suzuki, Y., et al. (1999). Isolation and Characterization of Novel Peroxisome Biogenesis-Defective Chinese Hamster Ovary Cell Mutants Using green Fluorescent Protein. Exp. Cel Res. 248 (2), 489–497. doi:10.1006/excr.1999.4413
Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., et al. (2002). Functional Profiling of the Saccharomyces cerevisiae Genome. Nature, 387–391. doi:10.1038/nature00935
Gonzalez, K. L., Fleming, W. A., Kao, Y. T., Wright, Z. J., Venkova, S. V., Ventura, M. J., et al. (2017). Disparate Peroxisome-Related Defects in Arabidopsis Pex6 and Pex26 Mutants Link Peroxisomal Retrotranslocation and Oil Body Utilization. Plant J. 92 (1), 110–128. doi:10.1111/tpj.13641
Goto, S., Mano, S., Nakamori, C., and Nishimura, M. (2011). Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 Is a Peroxin that Recruits the PEX1-PEX6 Complex to Peroxisomes. Plant Cell 23, 1573–1587. doi:10.1105/tpc.110.080770
Goto-Yamada, S., Mano, S., Nakamori, C., Kondo, M., Yamawaki, R., Kato, A., et al. (2014a). Chaperone and Protease Functions of LON Protease 2 Modulate the Peroxisomal Transition and Degradation with Autophagy. Plant Cel Physiol 55 (3), 482–496. doi:10.1093/pcp/pcu017
Goto-Yamada, S., Mano, S., and Nishimura, M. (2014b). in The Role of Peroxisomes in Plant Reproductive Processes” in Sexual Reproduction in Animals and Plants. Editors H. Sawada, N. Inoue, and M. Iwano (Tokyo, Japan: Springer), 419–429. doi:10.1007/978-4-431-54589-7_35
Goto-Yamada, S., Mano, S., Yamada, K., Oikawa, K., Hosokawa, Y., Hara-Nishimura, I., et al. (2015). Dynamics of the Light-dependent Transition of Plant Peroxisomes. Plant Cel Physiol 56 (7), 1264–1271. doi:10.1093/pcp/pcv081
Goto-Yamada, S., Oikawa, K., Bizan, J., Shigenobu, S., Yamaguchi, K., Mano, S., et al. (2019). Sucrose Starvation Induces Microautophagy in Plant Root Cells. Front. Plant Sci. 10, 1604. doi:10.3389/fpls.2019.01604
Haupt, G. (1932). Beitra ̈ge zur Zytologie der Gattung Marchantia (L.). Z. Induktstamm. Vererbungsl Abstamm. Vererbungsl. 62, 367–428. doi:10.1007/bf01948611
Hayashi, M., and Nishimura, M. (2006). Arabidopsis thaliana - A Model Organism to Study Plant Peroxisomes. Biochim. Biophys. Acta 1763, 1382–1391. doi:10.1016/j.bbamcr.2006.08.014
Hayashi, M., Nito, K., Takei-Hoshi, R., Yagi, M., Kondo, M., Suenaga, A., et al. (2002). Ped3p Is a Peroxisomal ATP-Binding Cassette Transporter that Might Supply Substrates for Fatty Acid β-oxidation. Plant Cel Physiol 43, 1–11. doi:10.1093/pcp/pcf023
Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T., and Nishimura, M. (2000). AtPex14p Maintains Peroxisomal Functions by Determining Protein Targeting to Three Kinds of Plant Peroxisomes. EMBO J. 19, 5701–5710. doi:10.1093/emboj/19.21.5701
Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric Acid-Resistant Mutants of Arabidopsis Have Defects in Glyoxysomal Fatty Acid β-oxidation. Plant Cell 10 (2), 183–195. doi:10.1105/tpc.10.2.183
Hayashi, Y., Hayashi, M., Hayashi, H., Hara-Nishimura, I., and Nishimua, M. (2001). Direct Interaction between Glyoxysomes and Lipid Bodies in Cotyledons of the Arabidopsis thaliana Ped1 Mutant. Protoplasma 218 (1/2), 83–94. doi:10.1007/BF01288364
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium Flaccidum Genome Reveals Primary Factors for Plant Terrestrial Adaptation. Nat. Commun. 5, 3978. doi:10.1038/ncomms4978
Hosokawa, Y., Iino, T., Oikawa, K., Mano, S., Yamada, K., and Nishimura, M. (2016). Quantification of the Adhesion Strength between Peroxisomes and Chloroplasts by Femtosecond Laser Technology. Bio-Protocol 6, e1834. doi:10.21769/bioprotoc.1834
Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J., and Chory, J. (2002). A Role for Peroxisomes in Photomorphoenesis and Development of Arabidopsis. Science 297, 405–409. doi:10.1126/science.1073633
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant Peroxisomes: Biogenesis and Function. Plant Cell 24, 2279–2303. doi:10.1105/tpc.112.096586
Ilacqua, N., Anastasia, I., Raimondi, A., Lemieux, P., de Aguiar Vallim, T. Q., Toth, K., et al. (2022). A Three-Organelle Complex Made by wrappER Contacts with Peroxisomes and Mitochondria Responds to Liver Lipid Flux Changes. J. Cel Sci. 135 (5). doi:10.1242/jcs.259091
Inoue, K., Nishihama, R., Araki, T., and Kohchi, T. (2019). Reproductive Induction Is a Far-Red High Irradiance Response that Is Mediated by Phytochrome and PHYTOCHROME INTERACTING FACTOR in Marchantia Polymorpha. Plant Cel Physiol 60 (5), 1136–1145. doi:10.1093/pcp/pcz029
International Wheat Genome Sequencing Consortium (2014). A Chromosome-Based Draft Sequence of the Hexaploid Bread Wheat (Triticum aestivum) Genome. Science 345 (6194), 1251788. doi:10.1126/science.1251788
Ishizaki, K., Chiyoda, S., Yamato, K. T., and Kohchi, T. (2008). Agrobacterium-mediated Transformation of the Haploid Liverwort Marchantiapolymorpha L., an Emerging Model for Plant Biology. Plant Cel Physiol 49 (7), 1084–1091. doi:10.1093/pcp/pcn085
Ishizaki, K., Johzuka-Hisatomi, Y., Ishida, S., Iida, S., and Kohchi, T. (2013). Homologous Recombination-Mediated Gene Targeting in the Liverwort Marchantia Polymorpha L. Sci. Rep. 3, 1532. doi:10.1038/srep01532
Ishizaki, K., Nishihama, R., Ueda, M., Inoue, K., Ishida, S., Nishimura, Y., et al. (2015). Development of Gateway Binary Vector Series with Four Different Selection Markers for the Liverwort Marchantia Polymorpha. PLoS One 10 (9), e0138876. doi:10.1371/journal.pone.0138876
Ishizaki, K., Nishihama, R., Yamato, K. T., and Kohchi, T. (2016). Molecular Genetic Tools and Techniques for Marchantia Polymorpha Research. Plant Cel Physiol 57 (2), 262–270. doi:10.1093/pcp/pcv097
Ito, M., Ito, R., Huang, Y., Miura, S., Imamura, A., Suzuki, Y., et al. (2000). Rapid Isolation and Characterization of CHO Mutants Deficient in Peroxisome Biogenesis Using the Peroxisomal Forms of Fluorescent Proteins. Biochim. Biophys. Acta 1496 (2-3), 232–242. doi:10.1016/s0167-4889(00)00019-7
Iwakawa, H., Melkonian, K., Schlüter, T., Jeon, H. W., Nishihama, R., Motose, H., et al. (2021). Agrobacterium-mediated Transient Transformation of Marchantia Liverworts. Plant Cel Physiol 62 (11), 1718–1727. doi:10.1093/pcp/pcab126
Iwasaki, M., Kajiwara, T., Yasui, Y., Yoshitake, Y., Miyazaki, M., Kawamura, S., et al. (2021). Identification of the Sex-Determining Factor in the Liverwort Marchantia Polymorpha Reveals Unique Evolution of Sex Chromosomes in a Haploid System. Curr. Biol. 31 (24), 5522–5532. doi:10.1016/j.cub.2021.10.023
Jahn, K. A., Barton, D. A., Kobayashi, K., Ratinac, K. R., Overall, R. L., and Braet, F. (2012). Correlative Microscopy: Providing New Understanding in the Biomedical and Plant Sciences. Micron 43 (5), 565–582. doi:10.1016/j.micron.2011.12.004
Jansen, R. L. M., Santana-Molina, C., van den Noort, M., Devos, D. P., and van der Klei, I. J. (2021). Comparative Genomics of Peroxisome Biogenesis Proteins: Making Sense of the PEX Proteins. Front. Cel Dev. Biol. 9, 654163. doi:10.3389/fcell.2021.654163
Jedd, G., and Chua, N.-H. (2002). Visualization of Peroxisomes in Living Plant Cells Reveals Acto-myosin-dependent Cytoplasmic Streaming and Peroxisome Budding. Plant Cel Physiol 43, 384–392. doi:10.1093/pcp/pcf045
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput. Appl. Biosci. 8 (3), 275–282. doi:10.1093/bioinformatics/8.3.275
Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., et al. (2001). Arabidopsis NPL1: a Phototropin Homolog Controlling the Chloroplast High-Light Avoidance Response. Science 291 (5511), 2138–2141. doi:10.1126/science.291.5511.2138
Kamada, T., Nito, K., Hayashi, H., Mano, S., Hayashi, M., and Nishimura, M. (2003). Functional Differentiation of Peroxisomes Revealed by Expression Profiles of Peroxisomal Genes in Arabidopsis thaliana. Plant Cel Physiol 44, 1275–1289. doi:10.1093/pcp/pcg173
Kao, Y.-T., Gonzalez, K. L., and Bartel, B. (2018). Peroxisome Function, Biogenesis, and Dynamics in Plants. Plant Physiol. 176, 162–177. doi:10.1104/pp.17.01050
Kao, Y. T., Fleming, W. A., Ventura, M. J., and Bartel, B. (2016). Genetic Interactions between PEROXIN12 and Other Peroxisome-Associated Ubiquitination Components. Plant Physiol. 172 (3), 1643–1656. doi:10.1104/pp.16.01211
Kato, H., Mutte, S. K., Suzuki, H., Crespo, I., Das, S., Radoeva, T., et al. (2020). Design Principles of a Minimal Auxin Response System. Nat. Plants 6 (5), 473–482. doi:10.1038/s41477-020-0662-y
Kaur, N., Zhao, Q., Xie, Q., and Hu, J. (2013). Arabidopsis RING Peroxins Are E3 Ubiquitin Ligases that Interact with Two Homologous Ubiquitin Receptor Proteins. J. Integr. Plant Biol. 55 (1), 108–120. doi:10.1111/jipb.12014
Kim, J., Lee, H., Lee, H. N., Kim, S. H., Shin, K. D., and Chung, T. (2013). Autophagy-related Proteins Are Required for Degradation of Peroxisomes in Arabidopsis Hypocotyls during Seedling Growth. Plant Cell 25 (12), 4956–4966. doi:10.1105/tpc.113.117960
Kimura, S., and Kodama, Y. (2016). Actin-dependence of the Chloroplast Cold Positioning Response in the Liverwort Marchantia Polymorpha L. PeerJ 4, e2513. doi:10.7717/peerj.2513
Kohchi, T., Yamato, K. T., Ishizaki, K., Yamaoka, S., and Nishihama, R. (2021). Development and Molecular Genetics of Marchantia Polymorpha. Annu. Rev. Plant Biol. 72, 677–702. doi:10.1146/annurev-arplant-082520-094256
Koornneef, M., and Meinke, D. (2010). The Development of Arabidopsis as a Model Plant. Plant J. 61 (6), 909–921. doi:10.1111/j.1365-313X.2009.04086.x
Kubota, A., Ishizaki, K., Hosaka, M., and Kohchi, T. (2013). Efficient Agrobacterium-Mediated Transformation of the Liverwort Marchantia Polymorpha Using Regenerating Thalli. Biosci. Biotechnol. Biochem. 77 (1), 167–172. doi:10.1271/bbb.120700
Lang, D., Ullrich, K. K., Murat, F., Fuchs, J., Jenkins, J., Haas, F. B., et al. (2018). The Physcomitrella Patens Chromosome-Scale Assembly Reveals moss Genome Structure and Evolution. Plant J. 93 (3), 515–533. doi:10.1111/tpj.13801
Li, F. W., Nishiyama, T., Waller, M., Frangedakis, E., Keller, J., Li, Z., et al. (2020). Anthoceros Genomes Illuminate the Origin of Land Plants and the Unique Biology of Hornworts. Nat. Plants 6 (3), 259–272. doi:10.1038/s41477-020-0618-2
Li, J., Peng, J., Jiang, X., Rea, A. C., Peng, J., and Hu, J. (2021). DeepLearnMOR: a Deep-Learning Framework for Fluorescence Image-Based Classification of Organelle Morphology. Plant Physiol. 186, 1786–1799. doi:10.1093/plphys/kiab223
Li, X.-R., Li, H.-J., Yuan, L., Liu, M., Shi, D.-Q., Liu, J., et al. (2014). Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 Is a Key Regulator of Peroxisome Biogenesis and Plays Critical Roles during Pollen Maturation and Germination in Planta. Plant Cell 26 (2), 619–635. doi:10.1105/tpc.113.121087
Lim, S. L., Voon, C. P., Guan, X., Yang, Y., Gardeström, P., and Lim, B. L. (2020). In Planta Study of Photosynthesis and Photorespiration Using NADPH and NADH/NAD+ Fluorescent Protein Sensors. Nat. Commun. 11 (1), 3238. doi:10.1038/s41467-020-17056-0
Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A., and Goodman, H. M. (1999). The Pex16p Homolog SSE1 and Storage Organelle Formation in Arabidopsis Seeds. Science 284, 328–330. doi:10.1126/science.284.5412.328
Lingard, M. J., and Bartel, B. (2009). Arabidopsis LON2 Is Necessary for Peroxisomal Function and Sustained Matrix Protein Import. Plant Physiol. 151, 1354–1365. doi:10.1104/pp.109.142505
Lingard, M. J., Monroe-Augustus, M., and Bartel, B. (2009). Peroxisome-associated Matrix Protein Degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 4561–4566. doi:10.1073/pnas.0811329106
Mano, S., Nakamori, C., Fukao, Y., Araki, M., Matsuda, A., Kondo, M., et al. (2011). A Defect of Peroxisomal Membrane Protein 38 Causes Enlargement of Peroxisomes. Plant Cel Physiol 52 (12), 2157–2172. doi:10.1093/pcp/pcr147
Mano, S., Nakamori, C., Hayashi, M., Kato, A., Kondo, M., and Nishimura, M. (2002). Distribution and Characterization of Peroxisomes in Arabidopsis by Visualization with GFP: Dynamic Morphology and Actin-dependent Movement. Plant Cel Physiol 43, 331–341. doi:10.1093/pcp/pcf037
Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimua, M. (2004). An Arabidopsis Dynamin-Related Protein, DRP3A, Controls Both Peroxisomal and Mitochondrial Division. Plant J. 38, 487–498. doi:10.1111/j.1365-313x.2004.02063.x
Mano, S., Nakamori, C., Nito, K., Kondo, M., and Nishimura, M. (2006). The Arabidopsis Pex12 and Pex13 Mutants Are Defective in Both PTS1- and PTS2-dependent Protein Transport to Peroxisomes. Plant J. 47, 604–618. doi:10.1111/j.1365-313x.2006.02809.x
Mano, S., Nishihama, R., Ishida, S., Hikino, K., Kondo, M., Nishimura, M., et al. (2018). Novel Gateway Binary Vectors for Rapid Tripartite DNA Assembly and Promoter Analysis with Various Reporters and Tags in the Liverwort Marchantia Polymorpha. PLoS One 13 (10), e0204964. doi:10.1371/journal.pone.0204964
Mano, S., and Nishimura, M. (2005). Plant Peroxisomes. Vit. Hor. 72, 111–154. doi:10.1016/s0083-6729(05)72004-5
Mathur, J., Mathur, N., and Hülskamp, M. (2002). Simultaneous Visualization of Peroxisomes and Cytoskeletal Elements Reveals Actin and Not Microtubule-Based Peroxisome Motility in Plants. Plant Physiol. 128, 1031–1045. doi:10.1104/pp.011018
Mathur, J. (2021). Organelle Extensions in Plant Cells. Plant Physiol. 185 (3), 593–607. doi:10.1093/plphys/kiaa055
Matsumoto, N., Tamura, S., and Fujiki, Y. (2003). The Pathogenic Peroxin Pex26p Recruits the Pex1p-Pex6p AAA ATPase Complexes to Peroxisomes. Nat. Cel Biol. 5, 454–460. doi:10.1038/ncb982
Montgomery, S. A., Tanizawa, Y., Galik, B., Wang, N., Ito, T., Mochizuki, T., et al. (2020). Chromatin Organization in Early Land Plants Reveals an Ancestral Association between H3K27me3, Transposons, and Constitutive Heterochromatin. Curr. Biol. 30 (4), 573–588. e577. doi:10.1016/j.cub.2019.12.015
Montilla-Martinez, M., Beck, S., Klümper, J., Meinecke, M., Schliebs, W., Wagner, R., et al. (2015). Distinct Pores for Peroxisomal Import of PTS1 and PTS2 Proteins. Cell Rep 13 (10), 2126–2134. doi:10.1016/j.celrep.2015.11.016
Nishihama, R., Ishida, S., Urawa, H., Kamei, Y., and Kohchi, T. (2016). Conditional Gene Expression/deletion Systems for Marchantia Polymorpha Using its Own Heat-Shock Promoter and Cre/loxP-Mediated Site-specific Recombination. Plant Cel Physiol 57 (2), 271–280. doi:10.1093/pcp/pcv102
Nishimura, M., Akhmedov, Y. D., Strzalka, K., and Akazawa, T. (1983). Purification and Characterization of Glycolate Oxidase from Pumpkin Cotyledons. Arch. Biochem. Biophys. 222, 397–402. doi:10.1016/0003-9861(83)90536-2
Nishimura, M., Bhusawang, P., Strzalka, K., and Akazawa, T. (1982). Developmental Formation of Glutamine Synthetase in Greening Pumpkin Cotyledons and its Subcellular Localization. Plant Physiol. 70, 353–356. doi:10.1104/pp.70.2.353
Nishimura, M., Yamaguchi, J., Mori, H., Akazawa, T., and Yokota, S. (1986). Immunocytochemical Analysis Shows that Glyoxysomes Are Directly Transformed to Leaf Peroxisomes during Greening of Pumpkin Cotyledon. Plant Physiol. 80, 313–316. doi:10.1104/pp.81.1.313
Nishiyama, T., Sakayama, H., de Vries, J., Buschmann, H., Saint-Marcoux, D., Ullrich, K. K., et al. (2018). The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 174 (2), 448–464. e424. doi:10.1016/j.cell.2018.06.033
Norizuki, T., Kanazawa, T., Minamino, N., Tsukaya, H., and Ueda, T. (2019). Marchantia Polymorpha, a New Model Plant for Autophagy Studies. Front. Plant Sci. 10, 935. doi:10.3389/fpls.2019.00935
Nyathi, Y., and Baker, A. (2006). Plant Peroxisomes as a Source of Signalling Molecules. Biochim. Biophys. Acta 1763 (12), 1478–1495. doi:10.1016/j.bbamcr.2006.08.031
O'Hanlon, M. E. (1926). Germination of Spores and Early Stages in Development of Gametophyte of Marchantia Polymorpha. Bot. Gaz. 82, 215–222. doi:10.1086/333650
Ogasawara, Y., Ishizaki, K., Kohchi, T., and Kodama, Y. (2013). Cold-induced Organelle Relocation in the Liverwort Marchantia Polymorpha L. Plant Cell Envrion 36, 1520–1528. doi:10.1111/pce.12085
Oikawa, K., Hayashi, M., Hayashi, Y., and Nishimura, M. (2019). Re-evaluation of Physical Interaction between Plant Peroxisomes and Other Organelles Using Live-Cell Imaging Techniques. J. Integr. Plant Biol. 61, 836–852. doi:10.1111/jipb.12805
Oikawa, K., Imai, T., Thagun, C., Toyooka, K., Yoshizumi, T., Ishikawa, K., et al. (2021). Mitochondrial Movement during its Association with Chloroplasts in Arabidopsis thaliana. Commun. Biol. 4 (1), 292. doi:10.1038/s42003-021-01833-8
Oikawa, K., Matsunaga, S., Mano, S., Kondo, M., Yamada, K., Hayashi, M., et al. (2015). Physical Interaction between Peroxisomes and Chloroplasts Elucidated by In Situ Laser Analysis. Nat. Plants 1 (4), 15035. doi:10.1038/nplants.2015.35
Oikawa, K., Yamasato, A., Kong, S. G., Kasahara, M., Nakai, M., Takahashi, F., et al. (2008). Chloroplast Outer Envelope Protein CHUP1 Is Essential for Chloroplast anchorage to the Plasma Membrane and Chloroplast Movement. Plant Physiol. 148 (2), 829–842. doi:10.1104/pp.108.123075
Ovečka, M., Sojka, J., Tichá, M., Komis, G., Basheer, J., Marchetti, C., et al. (2022). Imaging Plant Cells and Organs with Light-Sheet and Super-resolution Microscopy. Plant Physiol. 188 (2), 683–702. doi:10.1093/plphys/kiab349
Pedrosa, A. G., Francisco, T., Bicho, D., Dias, A. F., Barros-Barbosa, A., Hagmann, V., et al. (2018). Peroxisomal Monoubiquitinated PEX5 Interacts with the AAA ATPases PEX1 and PEX6 and Is Unfolded during its Dislocation into the Cytosol. J. Biol. Chem. 293 (29), 11553–11563. doi:10.1074/jbc.RA118.003669
Perico, C., and Sparkes, I. (2018). Plant Organelle Dynamics: Cytoskeletal Control and Membrane Contact Sites. New Phytol. 220 (2), 381–394. doi:10.1111/nph.15365
Peterhansel, C., Horst, I., Niessen, M., Blume, C., Kebeish, R., Kürkcüoglu, S., et al. (2010). Photorespiration. Arabidopsis Book 8, e0130. doi:10.1199/tab.0130
Platta, H. W., Magraoui, F. E., Bäumer, B. E., Schlee, D., Girzalsky, W., and Erdmann, R. (2009). Pex2 and Pex12 Function as Protein-Ubiquitin Ligases in Peroxisomal Protein Import. Mol. Cel. Biol. 29, 5505–5516. doi:10.1128/mcb.00388-09
Poirier, Y., Antonenkov, V. D., Glumoff, T., and Hiltunen, J. K. (2006). Peroxisomal β-oxidation--α Metabolic Pathway with Multiple Functions. Biochim. Biophys. Acta 1763 (12), 1413–1426. doi:10.1016/j.bbamcr.2006.08.034
Praefcke, G. J., and McMahon, H. T. (2004). The Dynamin Superfamily: Universal Membrane Tubulation and Fission Molecules? Nat. Rev. Mol. Cel Biol. 5 (2), 133–147. doi:10.1038/nrm1313
Prinz, W. A. (2014). Bridging the gap: Membrane Contact Sites in Signaling, Metabolism, and Organelle Dynamics. J. Cel Biol. 205 (6), 759–769. doi:10.1083/jcb.201401126
Purdue, P. E., Yang, X., and Lazarow, P. B. (1998). Pex18p and Pex21p, a Novel Pair of Related Peroxins Essential for Peroxisomal Targeting by the PTS2 Pathway. J. Cel Biol. 143 (7), 1859–1869. doi:10.1083/jcb.143.7.1859
Razi, M., and Tooze, S. A. (2009). Correlative Light and Electron Microscopy. Methods Enzymol. 452, 261–275. doi:10.1016/s0076-6879(08)03617-3
Rinaldi, M. A., Patel, A. B., Park, J., Lee, K., Strader, L. C., and Bartel, B. (2016). The Roles of β-oxidation and Cofactor Homeostasis in Peroxisome Distribution and Function in Arabidopsis thaliana. Genetics 204 (3), 1089–1115. doi:10.1534/genetics.116.193169
Rosenthal, M., Metzl-Raz, E., Bürgi, J., Yifrach, E., Drwesh, L., Fadel, A., et al. (2020). Uncovering Targeting Priority to Yeast Peroxisomes Using an In-Cell Competition Assay. Proc. Natl. Acad. Sci. USA 117 (35), 21432–21440. doi:10.1073/pnas.1920078117
Sauret-Güeto, S., Frangedakis, E., Silvestri, L., Rebmann, M., Tomaselli, M., Markel, K., et al. (2020). Systematic Tools for Reprogramming Plant Gene Expression in a Simple Model, Marchantia Polymorpha. ACS Synth. Biol. 9 (4), 864–882. doi:10.1021/acssynbio.9b00511
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome Sequence of the Palaeopolyploid Soybean. Nature 463 (7278), 178–183. doi:10.1038/nature08670
Schumann, U., Prestele, J., O'Geen, H., Brueggeman, R., Wanner, G., and Gietl, C. (2007). Requirement of the C3HC4 Zinc RING finger of the Arabidopsis PEX10 for Photorespiration and Leaf Peroxisome Contact with Chloroplasts. Proc. Natl. Acad. Sci. USA 104, 1069–1074. doi:10.1073/pnas.0610402104
Seo, D. H., Kim, S., Seo, H. J., Lee, S. E., Lim, C. J., Yang, H. W., et al. (2021). A Simple Protocol for Thallus Culture-Based Genetic Transformation of the Liverwort Marchantia Polymorpha. J. Plant Biol. doi:10.1007/s12374-021-09339-w
Shai, N., Yifrach, E., van Roermund, C. W. T., Cohen, N., Bibi, C., Bibi, L. I. J., et al. (2018). Systematic Mapping of Contact Sites Reveals Tethers and a Function for the Peroxisome-Mitochondria Contact. Nat. Commun. 9 (1), 1761. doi:10.1038/s41467-018-03957-8
Shibata, M., Oikawa, K., Yoshimo, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly Oxidized Peroxisomes Are Selectively Degraded via Autophagy in Arabidopsis. Plant Cell 25, 4967–7983. doi:10.1105/tpc.113.116947
Shimamura, M. (2016). Marchantia Polymorpha: Taxonomy, Phylogeny and Morphology of a Model System. Plant Cel Physiol 57 (2), 230–256. doi:10.1093/pcp/pcv192
Singh, T., Hayashi, M., Mano, S., Arai, Y., Goto, S., and Nishimura, M. (2009). Molecular Components Required for the Targeting of PEX7 to Peroxisomes in Arabidopsis thaliana. Plant J. 60, 488–498. doi:10.1111/j.1365-313x.2009.03970.x
Somerville, C. R., and Ogren, W. L. (1980). Photorespiration Mutants of Arabidopsis thaliana Deficient in Serine-Glyoxylate Aminotransferase Activity. Proc. Natl. Acad. Sci. USA 77, 2684–2687. doi:10.1073/pnas.77.5.2684
Somerville, C. R., and Ogren, W. L. (1981). Photorespiration-deficient Mutants of Arabidopsis thaliana Lacking Mitochondrial Serine Transhydroxymethylase Activity. Plant Physiol. 67, 666–671. doi:10.1104/pp.67.4.666
Sparkes, I. A., Brandizzi, F., Slocombe, S. P., El-Shami, M., Hawes, C., and Baker, A. (2003). An Arabidopsis Pex10 Null Mutant Is Embryo Lethal, Implicating Peroxisomes in an Essential Role during Plant Embryogenesis. Plant Physiol. 133, 1809–1819. doi:10.1104/pp.103.031252
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 37 (4), 1237–1239. doi:10.1093/molbev/msz312
Sugano, S. S., and Nishihama, R. (2018). CRISPR/Cas9-based Genome Editing of Transcription Factor Genes in Marchantia Polymorpha. Methods Mol. Biol. 1830, 109–126. doi:10.1007/978-1-4939-8657-6_7
Sugano, S. S., Nishihama, R., Shirakawa, M., Takagi, J., Matsuda, Y., Ishida, S., et al. (2018). Efficient CRISPR/Cas9-based Genome Editing and its Application to Conditional Genetic Analysis in Marchantia Polymorpha. PLoS One 13 (10), e0205117. doi:10.1371/journal.pone.0205117
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Titus, D. E., and Becker, W. M. (1985). Investigation of the Glyoxysome-Peroxisome Transition in Germinating Cucumber Cotyledons Using Double-Label Immunoelectron Microscopy. J. Cel Biol. 101, 1288–1299. doi:10.1083/jcb.101.4.1288
Tolbert, N. E. (1982). Leaf Peroxismes. Annal. N Y Acad. Sci. 386 (1), 254–268. doi:10.1111/j.1749-6632.1982.tb21421.x
Toyooka, K. (2016). Development of Correlative Light and Electron Microscopy to Observe green Fluorescent Protein-Labeled Organelles Embedded in Resin Using Field-Emission Electron Scanning Microscope. Plant Morphol. 28, 15–21. doi:10.5685/plmorphol.28.15
Tsuboyama, S., and Kodama, Y. (2018). AgarTrap Protocols on Your Benchtop: Simple Methods for Agrobacterium-Mediated Genetic Transformation of the Liverwort Marchantia Polymorpha. Plant Biotechnol. 35 (2), 93–99. doi:10.5511/plantbiotechnology.18.0312b
Tsuboyama-Tanaka, S., and Kodama, Y. (2015). AgarTrap‐mediated Genetic Transformation Using Intact Gemmae/gemmalings of the Liverwort Marchantia Polymorpha L. J. Plant Res. 128, 337–344. doi:10.1007/s10265-014-0695-2
Vallese, F., Catoni, C., Cieri, D., Barazzuol, L., Ramirez, O., Calore, V., et al. (2020). An Expanded Palette of Improved SPLICS Reporters Detects Multiple Organelle Contacts In Vitro and In Vivo. Nat. Commun. 11 (1), 6069. doi:10.1038/s41467-020-19892-6
Voon, C. P., Guan, X., Sun, Y., Sahu, A., Chan, M. N., Gardeström, P., et al. (2018). ATP Compartmentation in Plastids and Cytosol of Arabidopsis thaliana Revealed by Fluorescent Protein Sensing. Proc. Natl. Acad. Sci. USA 115 (45), E10778–e10787. doi:10.1073/pnas.1711497115
Wada, M., and Kong, S. G. (2018). Actin-mediated Movement of Chloroplasts. J. Cel Sci. 131 (2). doi:10.1242/jcs.210310
Wolinski, H., Petrovic, U., Mattiazzi, M., Petschnigg, J., Heise, B., Natter, K., et al. (2009). Imaging-based Live Cell Yeast Screen Identifies Novel Factors Involved in Peroxisome Assembly. J. Proteome Res. 8 (1), 20–27. doi:10.1021/pr800782n
Woodward, A. W., Fleming, W. A., Burkhart, S. E., Ratzel, S. E., Bjornson, M., and Bartel, B. (2014). A Viable Arabidopsis Pex13 Missense Allele Confers Severe Peroxisomal Defects and Decreases PEX5 Association with Peroxisomes. Plant Mol. Biol. 86 (1-2), 201–214. doi:10.1007/s11103-014-0223-8
Wu, H., de Boer, R., Krikken, A. M., Akşit, A., Yuan, W., and van der Klei, I. J. (2019). Peroxisome Development in Yeast Is Associated with the Formation of Pex3-dependent Peroxisome-Vacuole Contact Sites. Biochim. Biophys. Acta Mol. Cel Res. 1866 (3), 349–359. doi:10.1016/j.bbamcr.2018.08.021
Xu, C. S., Hayworth, K. J., Lu, Z., Grob, P., Hassan, A. M., García-Cerdán, J. G., et al. (2017). Enhanced FIB-SEM Systems for Large-Volume 3D Imaging. elife 6. doi:10.7554/eLife.25916
Yamaoka, S., Nishihama, R., Yoshitake, Y., Ishida, S., Inoue, K., Saito, M., et al. (2018). Generative Cell Specification Requires Transcription Factors Evolutionarily Conserved in Land Plants. Curr. Biol. 28 (3), 479–486. doi:10.1016/j.cub.2017.12.053
Yasui, Y., Tsukamoto, S., Sugaya, T., Nishihama, R., Wang, Q., Kato, H., et al. (2019). GEMMA CUP-ASSOCIATED MYB1, an Ortholog of Axillary Meristem Regulators, Is Essential in Vegetative Reproduction in Marchantia Polymorpha. Curr. Biol. 29 (23), 3987–3995. doi:10.1016/j.cub.2019.10.004
Yoshimoto, K., Shibata, M., Kondo, M., Oikawa, K., Sato, M., Toyooka, K., et al. (2014). Organ-specific Quality Control of Plant Peroxisomes Is Mediated by Autophagy. J. Cel Sci. 127 (Pt 6), 1161–1168. doi:10.1242/jcs.139709
Yuan, W., Veenhuis, M., and van der Klei, I. J. (2016). The Birth of Yeast Peroxisomes. Biochim. Biophys. Acta 1863 (5), 902–910. doi:10.1016/j.bbamcr.2015.09.008
Zechmann, B., Müller, M., Möstl, S., and Zellnig, G. (2021). Three-dimensional Quantitative Imaging of Tobacco Mosaic Virus and Zucchini Yellow Mosaic Virus Induced Ultrastructural Changes. Protoplasma 258 (6), 1201–1211. doi:10.1007/s00709-021-01626-0
Zhang, X., and Hu, J. (2009). Two Small Protein Families, DYNAMIN-RELATED PROTEIN3 and FISSION1, Are Required for Peroxisome Fission in Arabidopsis. Plant J. 57 (1), 146–159. doi:10.1111/j.1365-313X.2008.03677.x
Zhang, Y., Sampathkumar, A., Kerber, S. M., Swart, C., Hille, C., Seerangan, K., et al. (2020). A Moonlighting Role for Enzymes of Glycolysis in the Co-localization of Mitochondria and Chloroplasts. Nat. Commun. 11 (1), 4509. doi:10.1038/s41467-020-18234-w
Zolman, B. K., and Bartel, B. (2004). An Arabidopsis Indole-3-Butyric Acid-Response Mutant Defective in PEROXIN6, an Apparent ATPase Implicated in Peroxisomal Function. Proc. Natl. Acad. Sci. USA 101, 1786–1791. doi:10.1073/pnas.0304368101
Zolman, B. K., Silva, I. D., and Bartel, B. (2001). The Arabidopsis Pxa1 Mutant Is Defective in an ATP-Binding Cassette Transporter-like Protein Required for Peroxisomal Fatty Acid β-oxidation. Plant Physiol. 127, 1266–1278. doi:10.1104/pp.010550
Keywords: apem mutant, Arabidopsis thaliana, imaging, Marchantia polymorpha, peup mutant, peroxisome
Citation: Goto-Yamada S, Oikawa K, Yamato KT, Kanai M, Hikino K, Nishimura M and Mano S (2022) Image-Based Analysis Revealing the Molecular Mechanism of Peroxisome Dynamics in Plants. Front. Cell Dev. Biol. 10:883491. doi: 10.3389/fcell.2022.883491
Received: 25 February 2022; Accepted: 15 April 2022;
Published: 03 May 2022.
Edited by:
Yajin Ye, Nanjing Forestry University, ChinaReviewed by:
Jinbo Shen, Zhejiang Agriculture and Forestry University, ChinaShuh-Ichi Nishikawa, Niigata University, Japan
Copyright © 2022 Goto-Yamada, Oikawa, Yamato, Kanai, Hikino, Nishimura and Mano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoji ManobWFub0BuaWJiLmFjLmpw
 Shino Goto-Yamada
Shino Goto-Yamada Kazusato Oikawa
Kazusato Oikawa Katsuyuki T. Yamato
Katsuyuki T. Yamato Masatake Kanai
Masatake Kanai Kazumi Hikino4
Kazumi Hikino4 Shoji Mano
Shoji Mano