
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 13 April 2022
Sec. Epigenomics and Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.881255
This article is part of the Research Topic Regulatory Mechanisms of Gene Expression, Chromatin Structure and Nuclear Dynamics in Gametogenesis View all 6 articles
 Yasuhiko Kato*
Yasuhiko Kato* Hajime Watanabe
Hajime WatanabeThe cladoceran crustacean Daphnia produces only females by parthenogenesis in a healthy population. However, in response to environmental declines such as crowding and lack of foods, it produces eggs destined to become males that are genetically identical to females. During the development of the sexually committed eggs, DM domain-containing transcription factor Doublesex1 (Dsx1) orchestrates male trait formation globally both in somatic and gonadal tissues. Recent studies have revealed that Dsx1 expression is tightly controlled at transcriptional, post-transcriptional, and epigenetic levels to avoid sexual ambiguity. In this review, together with basic information on Dsx1 structure and expression, we introduce the multi-layered Dsx1 regulation and discuss how each regulation is interconnected for controlling male development in environmental sex-determining Daphnia.
Sex determination is a fundamental biological process that regulates differentiation not only of somatic tissues but also of gonads, resulting in the change of physiology and behavior in animals. It is broadly divided into two categories: genetic and environmental. Genetic sex determination is more spread in animals from invertebrates to vertebrates and relies on the difference of genetic elements between males and females (Bachtrog et al., 2014). In environmental sex determination, environmental cues trigger expression of the sex-determining gene and lead to any of male and female developmental programs. An environmental sex-determination system is also found in diverse taxa such as rotifers, nematodes, crustaceans, insects, fishes, and reptiles (Korpelainen, 1990). Exactly how the transient environmental cue initiates the sex-determining cascade and changes the developmental program has been of interest, but the regulatory mechanisms have remained unknown because the animals using the environmental sex-determination system have been poor genetic models.
The cladoceran crustacean is one of the models for studying environmental sex determination. In a healthy population, it produces females parthenogenetically. In contrast, when environmental quality declines, it also produces males that are genetically identical to females. The male then mates with a sexual female that also appears with environmental stressors, resulting in the generation of resting eggs that can withstand environmental hardships such as desiccation (Hebert, 1978). The commitment of sex to males occurs in the oocyte of the mother (Banta and Brown, 1929), meaning that the adult female senses the decline of the surrounding environment and transmits its environmental information to the next generation during oogenesis. Exposure of the crustacean sesquiterpenoid, methyl farnesoate, or its analog such as Fenoxycarb, also stimulates the production of male offspring (Olmstead and Leblanc, 2002; Tatarazako et al., 2003). This sesquiterpenoid-dependent commitment occurs at the late stages of oogenesis (Figure 1) (Olmstead and Leblanc, 2002; Kato et al., 2010). Taken together, in the cladoceran crustacean, the adult female senses the environmental cue and possibly converts it into sesquiterpenoid by the neuroendocrine system. Thereafter, this hormonal signaling induces sexual commitment of the oocytes to males, which in turn leads to the switching of the developmental program from the female-type to the male-type for male trait development.
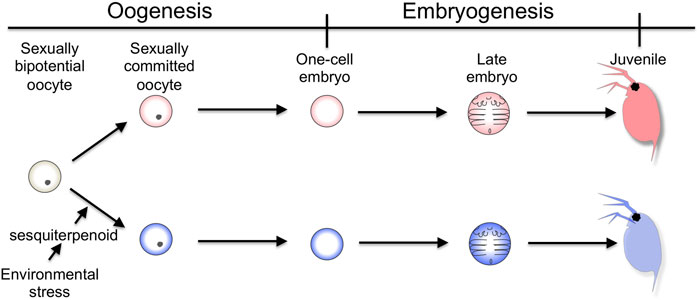
FIGURE 1. Timing of sexual commitment in environmental sex determination of D. magna. Under the healthy condition, the sexually bipotential oocyte is committed to be developed into a female. In contrast, by the mother, the information of environmental decline is detected and converted into sesquiterpenoid signaling, which commits the sexual fate of the oocyte to male.
Doublesex (Dsx) gene codes for the transcription factor harboring the DM domain and oligomerization domain and was firstly identified in Drosophila melanogaster as a sex determination gene (Hildreth, 1965). The DM domain is a DNA-binding domain that consists of an intertwined zinc-binding module followed by an alpha-helical recognition domain (Erdman and Burtis, 1993; Erdman et al., 1996). The oligomerization domain is known as the ubiquitin-associated domain and is required for dimerization that enhances the binding to the target sequence. Dsx gene is subjected to the sex-specific splicing, resulting in male- and female-specific isoforms (Baker and Wolfner, 1988; Burtis and Baker, 1989; Nagoshi et al., 1988). Each isoform shares the same DM domain but differs in the C-terminal oligomerization domain, which changes the interacting proteins and contributes to sex-specific gene regulation (Bayrer et al., 2005). In Caenorhabditis elegans, the male regulator mab-3 gene was found to have the DM domain (Shen and Hodgkin, 1988). The orthologous genes have been further identified in vertebrates as “Doublesex- and mab-3 regulated transcription factor (DMRT)” and have been proved to function as a sexual regulator (Raymond et al., 2000; Matsuda et al., 2002; Ge et al., 2017; Smith et al., 2009). In contrast to deep conservation of this gene family for sex-related development, the regulators that act upstream of the DMRT genes in the sex-determining cascade are diverse in animals (Wilkins, 1995; Herpin and Schartl, 2015).
Here we review the characterization and regulation of Dsx ortholog named Doublesex1 (Dsx1) in the cladoceran crustacean Daphnia magna. We will introduce the functional conservation of Dsx1 in sexual development and describe how this ancestral sexual regulator is controlled by environmental cues and during embryogenesis in D. magna.
D. magna has two Dsx orthologs, Dsx1 and Dsx2 (Kato et al., 2011). Because Dsx2 seems to have lost sex-related function (Kato et al., 2011), its characterization and potential function is not focused in this review. D. magna Dsx1 has the DM domain and oligomerization domain at the N-terminus and C-terminus respectively like Drosophila and the other insect Dsx proteins (Kato et al., 2011) (Figure 2A). The amino acid sequence of Dsx1 DM domain is well-conserved including zinc-chelating amino acids (Figure 2B). The oligomerization domain shows a more divergent sequence, but three non-polar amino acids involved in the formation of the interface are conserved (Figure 2C), suggesting that modes of transcriptional regulation of the D. magna Dsx1 are similar to those of Drosophila Dsx.
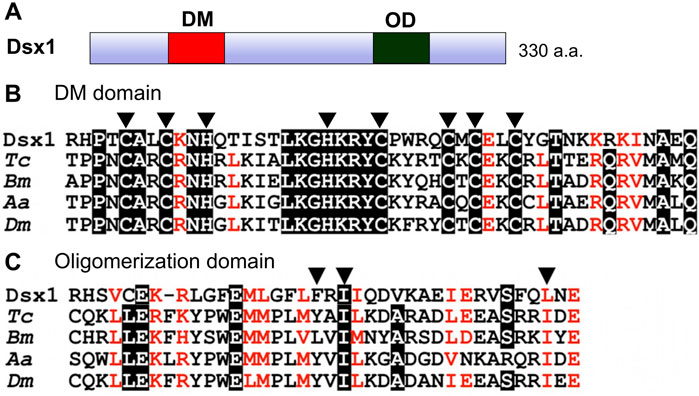
FIGURE 2. Domain organization of Dsx1. (A) Schematic representation of Dsx1. DM: DM domain, OD: Oligomerization domain. (B) Amino acid sequence alignment of DM domains from arthropod Dsx orthologs. Arrowheads indicate zinc-chelating amino acids. (C) Amino acid sequence alignment of oligomerization domains of arthropod Dsx orthologs. Arrowheads indicate three non-polar amino acids involved in the formation of the interface. Dsx1, D. magna Dsx1; Tc, Tribolium castaneum; Bm, Bombyx mori; Aa, Aedes aegypti; Dm, Drosophila melanogaster. Adapted and modified from Kato et al. (2011).
The D. magna Dsx1 gene is approximately 20 kb in length and consists of four exons (Kato et al., 2011) (Figure 3). The first and second introns are approximately 9 and 8 kb, each of which is considerably longer than the average size of introns (392 bp) on the D. magna genome, which may imply that important regulatory sequences are in the introns. Two transcripts, Dsx1α and Dsx1β, are produced from the Dsx1 locus by alternative promoter usage: Dsx1α utilizes exon 3 and exon 4, and Dsx1β is encoded in exons 1, 2, and 4. The Dsx1 ORF is included in exon 4 and is not spliced in a sex-specific manner, demonstrating that, unlike Drosophila and other insects’ Dsx, the same polypeptide is produced in both sexes. The 3′ UTR region contains four alternative polyadenylation sites, resulting in the synthesis of mRNAs with different 3′ UTR lengths (Kato et al., 2011). Even though Dsx1 gene is not regulated by sex-specific splicing, these sequence characteristics suggest that this gene is intricately regulated at transcriptional and post-transcriptional levels.
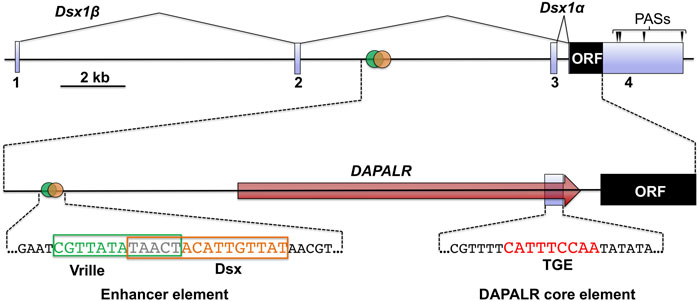
FIGURE 3. Genomic structure of Dsx1 gene. Exons of Dsx1 are indicated with light blue boxes. The red arrow shows the region and direction of DAPALR. Polyadenylation sites are indicated with arrowheads. The enhancer element that includes the potential Vrille and Dsx binding sites is shown with its sequence. The position of the DAPALR core element is shown with the sequence that is similar to the TRA-2/GLI element (TGE). Adapted and modified from Kato et al. (2018).
Temporal Dsx1 expression is divided into three phases: initiation, upregulation, and maintenance (Figure 4) (Mohamad Ishak et al., 2017). Before 6-hour post ovulation (hpo), Dsx1α mRNA is synthesized both in males and females (initiation phase). Around the timing of gastrulation, from 6 to 9 hpo, its expression is upregulated only in males (upregulation phase). In contrast to Dsx1α mRNA, Dsx1β mRNA is deposited as a maternal RNA in both sexes. After consumption of maternal RNAs, zygotic Dsx1β expression begins in both sexes and its male-specific upregulation occurs at 3 h later than the timing of Dsx1α upregulation. This difference of timing for male-specific upregulation of each isoform suggests that maternal sesquiterpenoid signaling first influences the Dsx1α promoter Thereafter, in males, expression of both isoforms is steadily maintained. Dsx1α is further upregulated at the instar 5 when the secondary sex characteristics appear. In females, the Dsx1 locus is persistently silenced except for synthesis of the maternal Dsx1β mRNA (Nong et al., 2017).
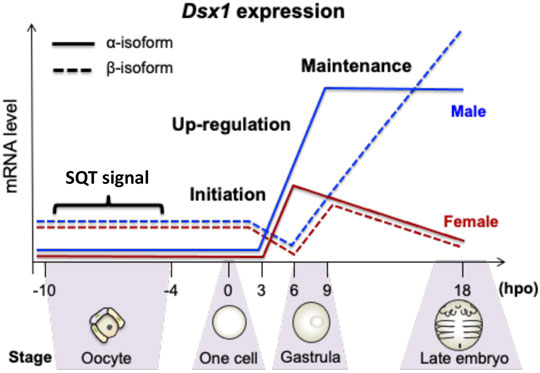
FIGURE 4. Temporal expression patterns of Dsx1 isoforms during late oogenesis and early embryogenesis. hpo, hour post ovulation; SQT, sesquiterpenoid. Adapted from Mohamad Ishak et al. (2017).
To investigate where the Dsx1 activation occurs throughout development, the transgenic D. magna for visualizing the Dsx1 expression was generated (Nong et al., 2017). TALENs that introduce double-strand break at the start codon of Dsx1 in its exon 4 was constructed and used for non-homologous end-joining mediated knock-in of the donor plasmid harboring the mCherry ORF and Dsx1 3′ UTR (Figure 5). Coinjection of the TALEN mRNAs with the donor plasmid led to the establishment of two transgenic lines (Line A and Line B) showing mCherry fluorescence. Both lines have a single copy of the mCherry reporter on one of the Dsx1 alleles. In Line A, another Dsx1 allele is mutated with a 6-bp deletion at the start codon, which suggests that this line does not produce any intact Dsx1 protein (Nong et al., 2020). In contrast, Line B has a wild-type Dsx1 gene on another allele. This genotype enables Line B males to develop sex-specific morphology such as elongation of first antennae and to have the reproductive ability for producing sexual eggs by copulating with females, even though those male-specific traits are slightly feminized (Nong et al., 2017). Localization of mCherry fluorescence was consistent with Dsx1 mRNA localization that was analyzed by in situ hybridization (Kato et al., 2011). Therefore, Line B has been used as a Dsx1 reporter line that recapitulates Dsx1 expression (Mohamad Ishak et al., 2017; Kato et al., 2018; Perez et al., 2021).
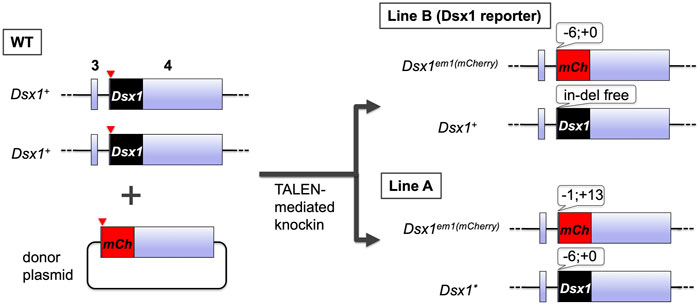
FIGURE 5. Generation and genotypes of the transgenic D. magna harboring the mCherry coding sequence at the Dsx1 locus. The TALANs induced a double-strand break at the start codon of Dsx1 ORF on exon 4. Concomitantly, it also cleaved near the start codon of mCherry ORF fused with Dsx1 3′ UTR on the donor plasmid, resulting in the generation of two transgenic lines, Line A and Line B.
Live imaging of mCherry fluorescence in Line B male embryos revealed the dynamic change of spatial expression patterns in early embryos (Nong et al., 2020) (Figure 6). During the initiation phase of Dsx1, no clear mCherry signal was detected probably due to low Dsx1 expression levels (Figure 6, stage i and ii). In the upregulation phase, two mCherry signals were detected at 11 hpo. One was the cell cumulus appearing around the invagination, and the other was around the blastopore (Figure 6, stage iii). The mCherry-expressing cumulus moved to the vicinity of the blastopore, and the signal appeared to weaken gradually (Figure 6, stage iv). In parallel, another mCherry-expressing population migrated from the blastopore to the posterior region (Figure 6, stage iv). When naupliar segmentation occurred, the population of cells showing strong mCherry fluorescence converged in the posterior growth zone although it is not clear whether the two mCherry-expressing cell populations merged (Figure 6, stage v). In other animals, cell populations showing similar migration behavior are known as a primary organizer that induces differentiation in the adjacent area (Bénazéraf and Pourquié, 2013; Akiyama-Oda and Oda, 2003). This suggests that the migrating cell population may induce sexual differentiation in their surrounding cells. The posterior growth zone is known to supply the progenitor cells for the formation of thoracic appendages (Martin and Kimelman, 2009), suggesting that, in male embryos of D. magna, this region distributes masculinized progenitor cells for the development of the male-specific structure of thoracic appendages. Strong mCherry fluorescence was also observed in the buds of the first antennae, and in parallel, strong mCherry fluorescence was detected at the boundary between the head and thoracic segments (Figure 6, stage v). Later, at 18 h, more intense mCherry fluorescence was detected in the first thoracic appendage (Figure 6, stage vi), where hooks for female capture are formed at the later juvenile stage. During these earlier embryonic stages, rather broader mCherry fluorescence patterns were observed, indicating that the embryos are still sexually immature. Localization of mCherry fluorescence of female embryos was similar but its intensity was weaker compared to male embryos until the naupliar segmentation stage. Thereafter, there was no mCherry fluorescence in female embryos (Figure 6).
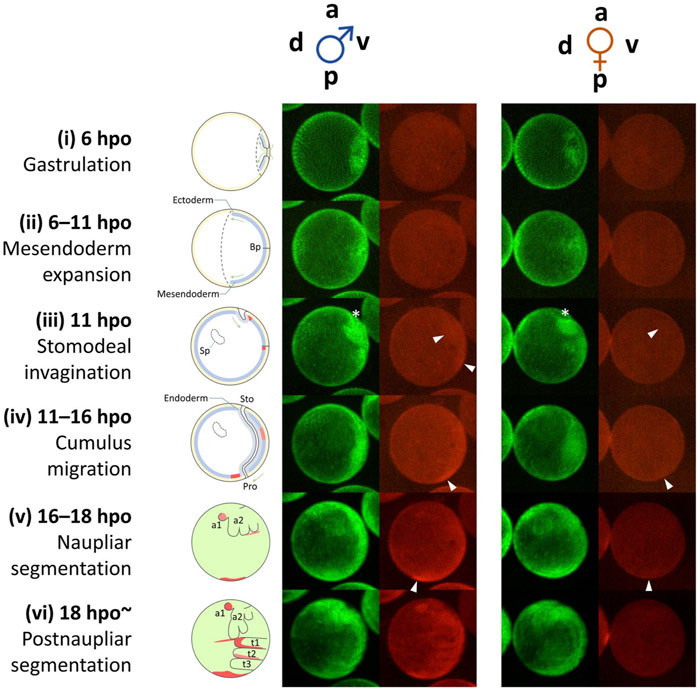
FIGURE 6. Dynamic progression of Dsx1-expressing cells during early embryogenesis from gastrulation until thoracic segmentation. Schematic diagrams on the left illustrate the simple anatomy of the embryos at the corresponding time points. The diagrams from stages (i) to (iv) show images of embryonic sections. The diagrams (v) and (vi) show superficial views of embryos. a, anterior; p, posterior; v, ventral; d, dorsal; Bp, blastopore; Sp, Scheitelplatten; Sto, stomodeum; Pro, proctodeum; t1–3, thoracic segments; a1, first antenna; a2, second antenna; hpo, hours post ovulation. White arrowheads indicate mCherry-expressing cell clusters that migrate in an anterior-to-posterior direction. An asterisk indicates the site of invagination. Adapted from Nong et al. (2017).
As embryonic development progressed toward the first instar stage that corresponded to 72 hpo, mCherry signal was localized to organs characteristic of males, first antennae, first thoracic leg, external genitalia, and testis (Figure 7, instar 1). The expression of Dsx1 in these male-specific traits stably continued until the fourth instar stage (Figure 7, instar 4). After males showed secondary sex characteristics at the instar 5, mCherry fluorescence expanded to the other several male-specific structures, including the carapace edge below the helmet, the tip of the penis, and the skeletal muscle (Figure 7, instar 7) (Nong et al., 2020). These imaging results suggest that Dsx1 is tightly regulated by time and place for controlling male development.
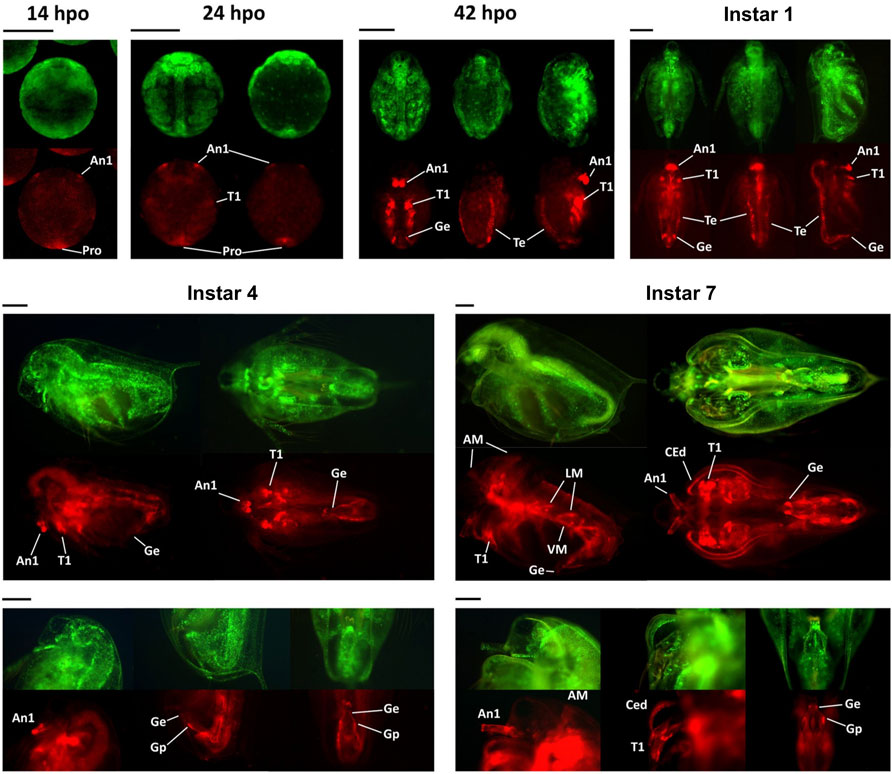
FIGURE 7. Spatially and temporally specific expressions of mCherry in the Dsx1 reporter strain. All embryos and daphniids shown in this figure are males of the Line B. hpo, hours post-ovulation; dpo, days post-ovulation. For the 14 hpo panel: ventral view. For the 24 hpo panel: ventral view (left) and dorsal view (right). For the 42 hpo and instar 1 panels: ventral view (left), dorsal view (middle), and side view with the ventral side facing right (right). A1, first antenna; T1, first thoracic leg; Pro, proctodeum; Ge, genital (i.e., penis in this case); Te, testis; AM, antennal muscle; LM, lateral muscle; VM, ventral muscle; CEd, carapace edge; Gp, gonopore. Scale bars = 200 μm. Adapted from Nong et al. (2017).
A previous study reported that knockdown of Dsx1 led to feminization in males (Kato et al., 2011). Long dsRNA targeting Dsx1 mRNA was injected into eggs that were destined to be developed into males. By Dsx1 RNAi, the first antennae of males became short like those of females (Figure 8A) and a copulatory hook for capturing females was lost from the first thoracic leg (Figure 8B). In addition to the feminization of somatic tissues, the gonads also showed sex reversal from testes to ovaries (Figure 8C). When the chemically modified Dsx1-targeting siRNA was injected, the effects of Dsx1 silencing in adults became severe enough to allow the knockdown males for laying eggs (Kato et al., 2018), showing that Dsx1 is the key sex determination factor in D. magna.
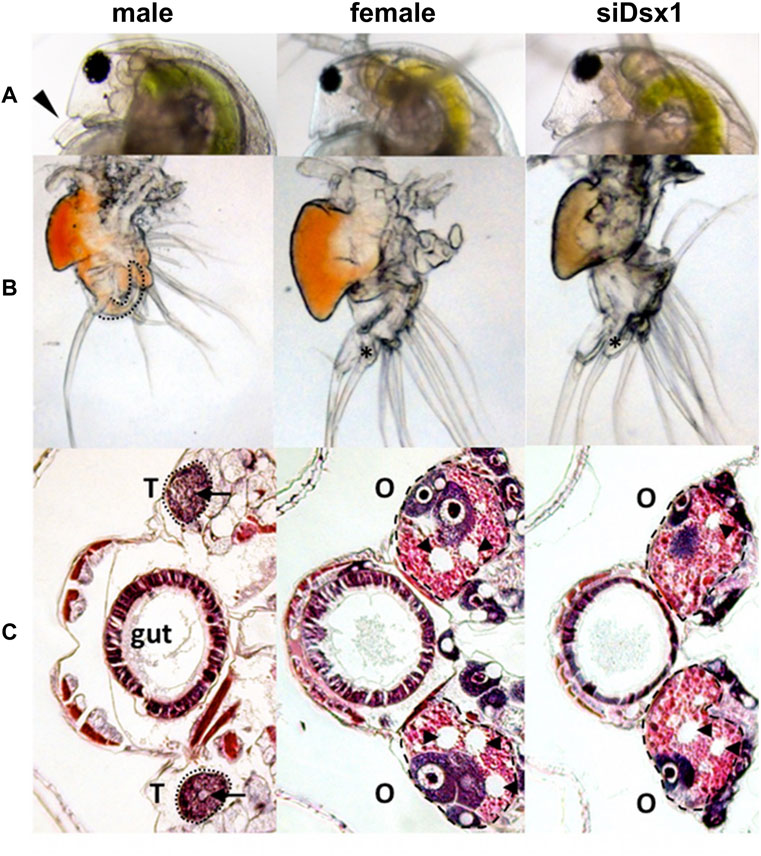
FIGURE 8. Dimorphic development of D. magna. Eggs induced to become males were obtained from D. magna. After injection of the synthesized dsRNA, sexually dimorphic phenotypes were examined at the fifth or sixth instar except for the first antennae (third instar). The first two columns represent normal male and female phenotypes, respectively. The third and fourth columns represent phenotypes of individuals injected with dsRNAs of dsx1 and dsx2, respectively. (A) Lateral view of the head. Arrowheads indicate the first antennae. (B) First thoracic limb. The dotted line shows the outline of the stout chitinized hook. A female-type long filament corresponding to the hook is labeled with an asterisk. (C): Gonad. Daphnids were embedded in paraffin and sectioned, followed by standard hematoxylin and eosin staining. Dorsal is left, ventral is right. Dotted circled lines show gonads at both sides of a gut. T and O indicate testis and ovary, respectively. Arrowheads indicate large lipid droplets lying among the eosinophilic yolk granules. Arrows indicate lumens into which the mature spermatozoa are released. Adapted from Kato et al. (2011).
Phenotyping of the Dsx1 mutants Line A and Line B aforementioned also showed Dsx1 dependency of male trait development (Figure 4) (Nong et al., 2017; Nong et al., 2020). Compared to Line B males, Line A males showed more severe feminization throughout development, which were obvious in the first antennae and carapace (Figure 9A). The first antennae of Line A were as short as those of wild-type females in younger juveniles in contrast to Line B males with rather elongated first antennae. At instar 5 when the secondary sex characteristics appeared, the difference in the degree of feminization between the two mutants became clearer. Line B male showed the transformation of first antennae and carapace into the mature adult-like structures at this instar whereas Line A gradually transformed morphology of those two male traits across the following instar stages. Feminization of Line A males was also observed throughout the body such as larger body size (Figures 9B,C), the modest opening of the ventral carapace (Figure 9C), bulky head (Figure 9D), female-like genital and anus (Figure 9E). In addition, Line A male developed ovary-like darkened gonads but did not produce any eggs (Figures 9B,C). Male-specific traits are developed exclusively dependent on Dsx1 activity in D. magna.
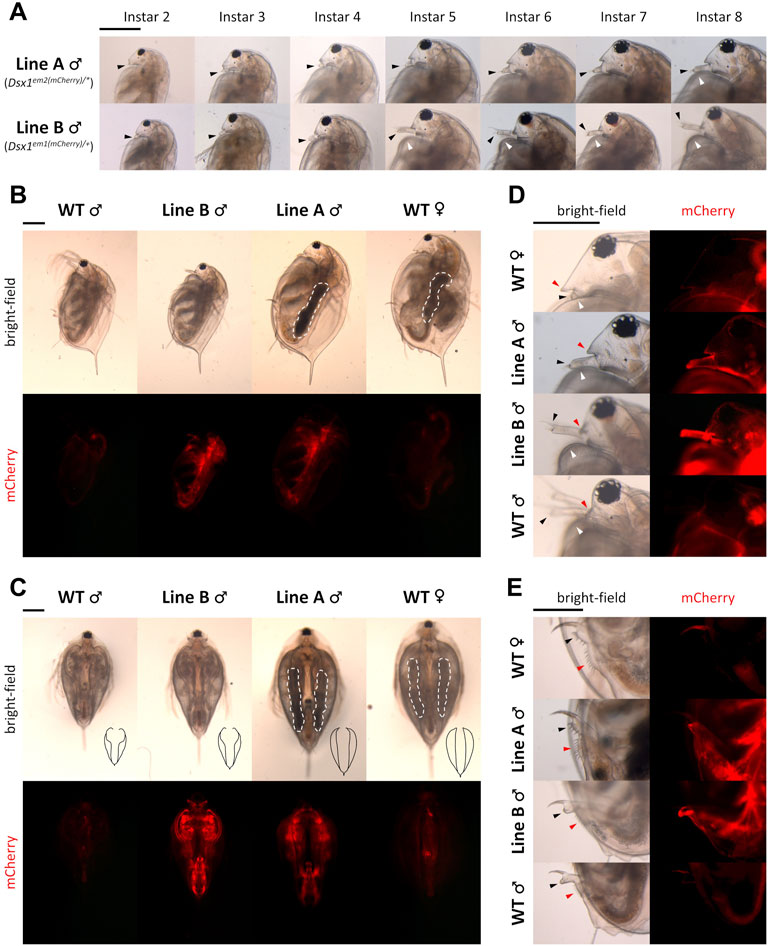
FIGURE 9. Feminization in the phenotype of Dsx1 mutants. Head region with a focus on the first pair of antennae of Line A males and Line B males of various ages. Until instar 4, juvenile stages of daphniids continue. Transition to fully mature adults usually occurs when the animal reaches instar 5, which is approximately 1 week after birth. Black-arrowheads: first antennae. White arrowheads: carapace curve that reveals copulation hooks in males. (B) Full body, side view of instar 8 daphniids. White dotted lines indicate ovary-like gonads, which appeared darker because of the accumulation of yolk protein. (C) Full body, ventral view of instar 8 daphniids. Black drawings at the bottom right corner of each bright-field photo are tracing lines scaled down to a 1:2.26 ratio depicting the outline of the carapace edge. White dotted lines have the same meaning as in panel (B). (D) Head region, side view of instar 8 daphniids. Black and white arrowheads have the same meaning as in panel (A). Red arrowheads indicate the rostrum. (E) Genital and anal region, side view of instar 8 daphniids. Black arrowheads: genital. Red arrowheads: anus. Each panel from (A–E) has one scale bar indicating 0.5 mm, which is shared among all photos of the same panel. Adapted from Nong et al. (2020).
On the D. magna genome, upstream of the Dsx1α transcription start site, there is an element that resembles the consensus sequence of the binding site for the bZip transcription factor Vrille. This element overlaps a potential Dsx binding site (Figure 3) (Mohamad Ishak et al., 2017). A similar element harboring both the bZip transcription factor and Dsx binding sites had been characterized well as an enhancer of yolk protein gene in Drosophila (An and Wensink, 1995). Importantly, in D. magna, male-specific expression of Vrille occurred at 6 hpo before upregulation of Dsx1 begins (Figure 10A) even though Vrille did not show any sexually dimorphic expression in the later stages of embryos (Figure 10B). Vrille knockdown reduced Dsx1 expression in male embryos (Figures 10C,D) whereas its overexpression in females activated Dsx1 expression and in turn led to the development of male trait, elongation of first antennae (Mohamad Ishak et al., 2017). Disruption of the Vrille binding site on the Dsx1α promoter region inhibited Dsx1 activation in male embryos, demonstrating that Vrille is required for Dsx1 activation during the upregulation phase. Interestingly, there was a potential binding site of the sesquiterpenoid receptor Methoprene-tolerant (Met) in the Vrille promoter, suggesting the possibility that the Met may directly regulate Vrille expression (Mohamad Ishak et al., 2017). In Drosophila, Vrille functions as a sex regulatory factor downstream of Dsx (Lebo et al., 2009). Vrille might be repeatedly co-opted in the sex-determining pathways with the different hierarchical position in animals as reported in the other sex-determining genes (Herpin and Schartl, 2015).
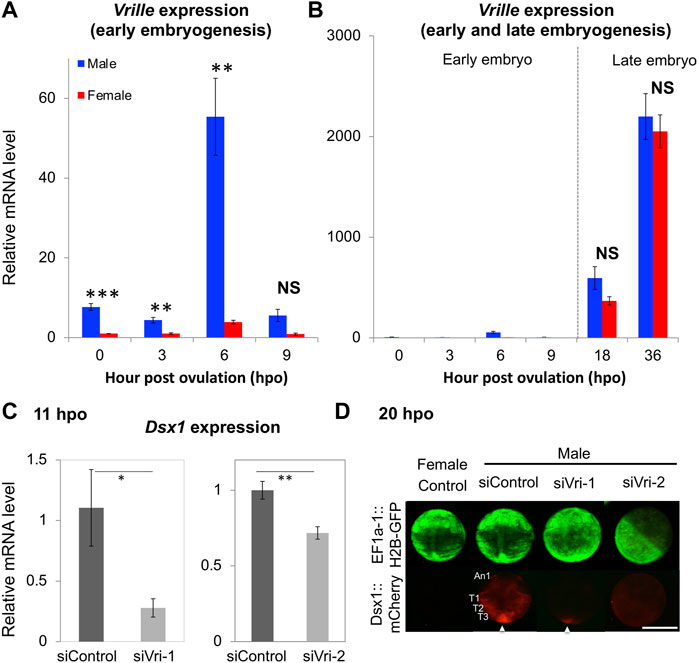
FIGURE 10. Vrille (Vri) expression in wild-type and knockdown embryos of D. magna. (A) Sexual dimorphism of Vri expression at early embryonic stages (0, 3, 6, and 9-hpo) of wild-type. (B) Non-sex-specific Vri expression in the late embryos (18 and 36-hpo) of wild-type. The Vri expression levels were normalized to the reference gene expression levels (ribosomal protein L32). The normalized Vri expression in female embryos at 0-hpo was set to one. (C) The Dsx1 expression level at 11 h after siRNA injection was measured by qRT-PCR. siControl, siVri-1 and siVri-2 indicate control-, Vri_siRNA_1, and Vri_siRNA_2 injected males, respectively. Each experiment was performed in biological triplicate. For each replicate, nine or ten embryos were pooled in one tube, subjected to total RNA extraction, and used for quantitation. (D) Spatial expression pattern of mCherry as a reporter of Dsx1 gene expression in Vrille knockdown embryos at 20-hpo. An1, first antennae; T1, thoracic appendage 1; T2, thoracic appendage 2; T3, thoracic appendage 3; An arrowhead indicates the posterior growth zone; Scale bars: 200 μm. (Student’s t-test; *p < 0.05; **p < 0.01; ***p < 0.001; NS, No significant different). Adapted from Mohamad Ishak et al. (2017).
A comprehensive epigenomic analysis of Daphnia pulex revealed that, on the Dsx1 locus, there were sex differences of active and repressive histone marks, histone H3 trimethylation at lysine 4 (H3K4me3), and histone H3 trimethylation at lysine 27 (H3K27me3) in addition DNA methylation status (Kvist et al., 2020). The H3K4me3 level was around 300-fold higher but the H3K27me3 level was around 5,000-fold lower in males compared to those in females. CpG methylation level was higher in females. These epigenetic marks suggest opened and closed chromatin states in males and females respectively. Although epigenetic marks on D. magna Dsx1 remain unknown, a previous study found that Dsx1 enhancer element-targeting Crispr/Cas9 could not induce any mutation on the Dsx1 enhancer element in female embryos but resulted in indel formation in males (Mohamad Ishak et al., 2017). CRISPR/Cas9 is known to have weaker activity against the closed chromatin (Chen et al., 2016; Daer et al., 2016; Jensen et al., 2017), supporting the sex difference of the epigenetic state on the Dsx1 locus.
The RNA binding protein Alan shepard (Shep) ortholog was proved as a post-transcriptional regulator for Dsx1α mRNA in D. magna (Perez et al., 2021). The relationship between Shep and Dsx1α mRNA was found unexpectedly based on phenotypic analysis of female embryos injected with DsRed2 reporter RNA harboring the Dsx1α or Dsx1β 5′ UTR (Kato et al., 2018). Despite lacking the Dsx1 ORF, the chimeric DsRed2 RNA including the Dsx1α 5′ UTR induced development of the male trait and activation of Dsx1 in females. The RNA that coded for only Dsx1α 5′ UTR also led to Dsx1 activation. To investigate the molecular mechanism underlying the unexpected function of Dsx1α 5′ UTR for Dsx1 regulation, the proteins interacting with Dsx1α 5′ UTR were examined by the MS analysis with D. magna lysates and one of the identified proteins was Shep. D. magna Shep gene is expressed both in males and females during embryogenesis. Silencing of Shep in the Dsx1 reporter line increased mCherry expression in male and female embryos (Figures 11A,C). In contrast, overexpression of Shep reduced the mCherry expression in the Dsx1 reporter line (Figures 11B,F). However, both loss- and gain-of-function did not change Dsx1 transcript levels (Figures 11D–F), suggesting that Shep represses Dsx1 expression at a post-transcriptional level. Dsx1α 5′ UTR has a sequence like “tra-2 and GLI element (TGE)” (Figure 3), which has been identified as a binding site of Sup-26, an ortholog of Shep in C. elegans. In vitro reporter assay demonstrated that Shep represses translation of Dsx1α mRNA via TGE (Figure 13A) (Perez et al., 2021).
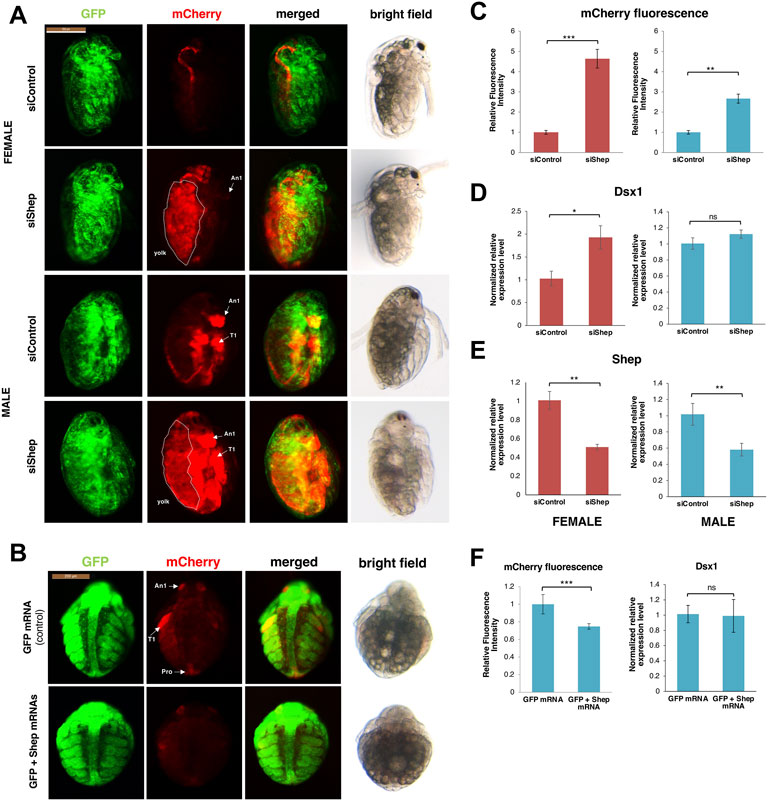
FIGURE 11. Shep loss of function analysis. (A) Lateral view of female and male embryos of Dsx1 reporter strain injected with control siRNA and Shep siRNA and observed at 48 h after injection. mCherry fluorescence allowed visualization of Dsx1 expression while GFP fluorescence in the nucleus enabled observation of body structures. The merged images of mCherry and GFP and the bright field images were used to understand the localization pattern of mCherry expression. An1: first antennae, T1: first thoracic leg, dotted lines: yolk area. (B) Ventral view of male embryos of Dsx1 reporter strain injected with GFP mRNA as control and GFP plus Shep mRNA observed at 30 h after injection. mCherry fluorescence allowed visualization of Dsx1 expression while GFP fluorescence in the nucleus enabled observation of body structures. The merged images of mCherry and GFP and the bright field images were used to understand the localization pattern of mCherry expression. An1: first antennae, T1: first thoracic legs, Ge: genital. (C) Relative mCherry fluorescence intensity calculated between Shep siRNA- and control siRNA-injected female (red) and male (blue) embryos. Error bars indicate the standard error of the mean (n = 5). (D) Gene expression profile of Dsx1 in control siRNA- and Shep siRNA-injected female (red) and male (blue) embryos. (E) Gene expression profile of Shep in control siRNA- and Shep siRNA-injected female (red) and male (blue) embryos. (F) Relative mCherry fluorescence intensity calculated between GFP mRNA- and GFP plus Shep mRNA-injected male embryos (left figure). Error bars indicate the standard error of the mean (n = 5). Gene expression profile of Dsx1 in GFP mRNA- and GFP plus Shep mRNA-injected male embryos (right figure). RT-qPCR results are shown as expression levels normalized with housekeeping genes L32, L8, and Cyclophilin and relatively compared to the control. Error bars indicate the standard error of the mean (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant (Student’s t-test). Adapted from Perez et al. (2021).
Dsx1 gene transactivation function of the Dsx1α 5′ UTR is attributed to the inclusion of this 5′ UTR sequence within long noncoding RNA DAPALR (Dsx1 alpha promoter associated long RNA) (Figure 12A). This lncRNA is transcribed from upstream of the transcription start site of Dsx1α mRNA in sense orientation and overlaps Dsx1α 5′ UTR (Figure 3) (Kato et al., 2018). DAPALR is a capped and non-polyadenylated RNA. This lncRNA expression is male-specific as well as Dsx1 but its expression level is around 10 times lower than that of Dsx1. DAPALR is activated by Vrille, which may suggest co-regulation of DAPALR with Dsx1. RNAi-mediated knockdown of DAPALR in males led to their feminization both in somatic tissues and germline, which resulted in the production of offspring (Figure 12B). This feminized phenotype was caused by a reduction of the Dsx1 transcript level (Figure 12C). In contrast, overexpression of DAPALR in females induced upregulation of the Dsx1 gene, resulting in the development of the male trait, elongation of the first antennae (Figures 12D–F). DAPALR activates Dsx1 in trans and Dsx1α 5′ UTR functions as a transactivation element of DAPALR (Kato et al., 2018).
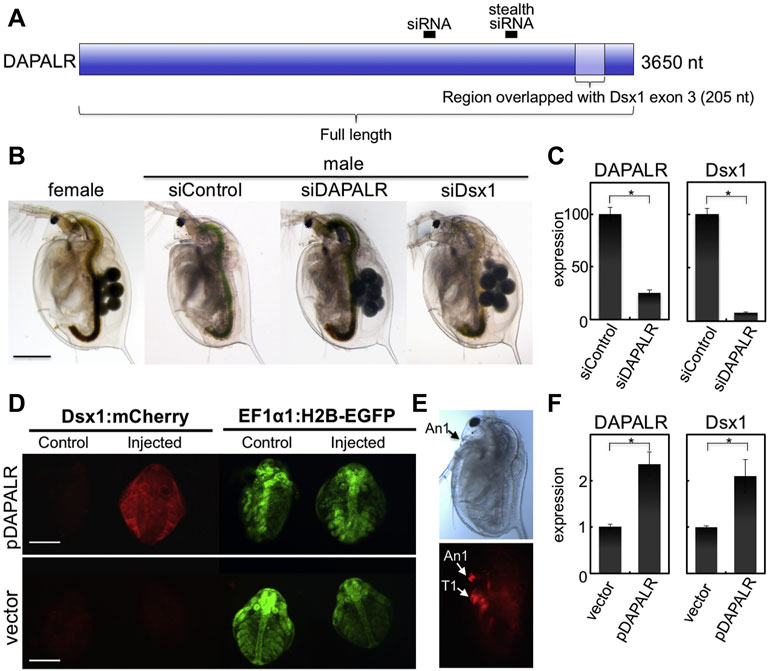
FIGURE 12. Knockdown and overexpression of DAPALR. (A) Position of small interference RNAs (siRNAs) and RNAs used for knockdown and overexpression of DAPALR. The siRNAs were designed in the DAPALR-specific region. The full-length of DAPALR or its region overlapped with Dsx1 exon 3 (Dsx1α-isoform specific region) and was ectopically expressed in females. (B) Lateral view of D. magna injected with stealth siRNAs. DAPALR- and Dsx1-stealth siRNA-injected daphniids produced eggs in their brood chamber located in the dorsal region, in contrast to control-stealth siRNA-injected embryos. Scale bar, 500 μm. (C) Gene expression profile of DAPALR and Dsx1 in embryos injected with stealth siRNAs of DAPALR (left panel) and Dsx1 (right panel). Error bars indicate the standard error of the mean (n = 3). ∗p < 0.05 (Student’s t test). (D) Ventral view of female embryos of the Dsx1-reporter strain injected with DAPALR-expression plasmid (pDAPALR) and control plasmid (vector). (E) Lateral view of a female juvenile overexpressing DAPALR. An1, first antennae; T1, thoracic appendage 1. Scale bar, 200 μm. (F) Gene expression profile of DAPALR and Dsx1 in embryos injected with pDAPALR (left panel) and vector (right panel). Error bars indicate the standard error of the mean (n = 3). ∗p < 0.05 (Student’s t test). Adapted from Kato et al. (2018).
In in vitro translation system including the reporter mRNA harboring Dsx1α 5′ UTR, DAPALR addition canceled the Shep-dependent translational suppression (Perez et al., 2021). Similarly, Dsx1α 5′ UTR had the ability for canceling the Shep function (Figure 13A). Thus, Dsx1α 5′ UTR was also designated as a “DAPALR core element” (Figure 3). Importantly, DAPALR and its core element inhibited the function of Shep as a translational repressor in a dose-dependent manner (Figure 13B). DAPALR functions as a decoy of Shep through its core element that corresponds to Dsx1α 5′ UTR, which in turn increases Dsx1 expression.
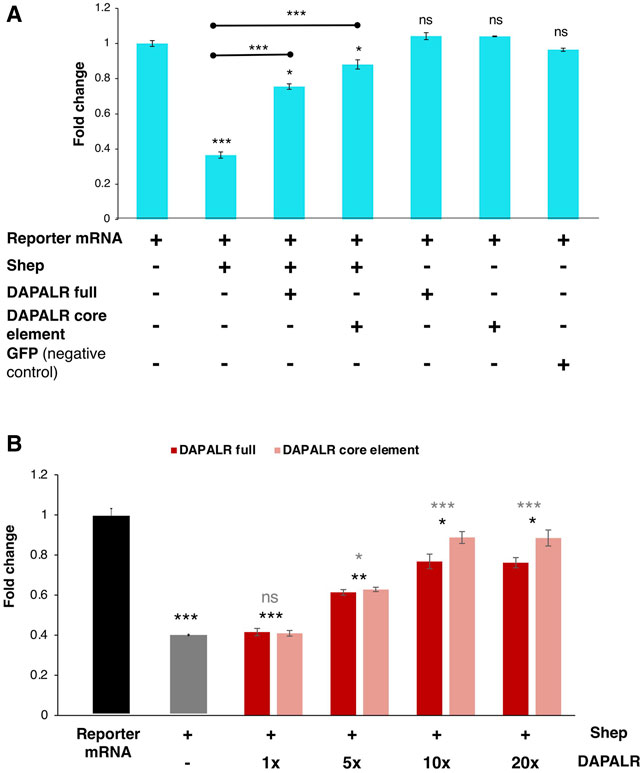
FIGURE 13. Dsx1 post-transcription regulation by DAPALR and Shep in vitro. (A) Relative luciferase activity after in vitro translation assay of Dsx1 5′ UTR-Luc reporter mRNA with intact TGE upon addition of Shep mRNA, DAPALR full RNA, DAPALR core element, and GFP mRNA (negative control). Samples were compared against the expression of the Dsx1 5′ UTR-Luc reporter mRNA without the addition of any other mRNAs. The endpoints of the line above the bars show which samples were additionally compared statistically. (B) Relative luciferase activity after in vitro translation assay of Dsx1 5′ UTR-Luc reporter mRNA with Shep and different concentrations of full region of DAPALR and its core element. Error bars indicate the standard error of the mean, n = 3. Black asterisks show significant statistics compared with the expression of the Dsx1 5′ UTR-Luc reporter mRNA. Gray asterisks show significant statistics compared with the Reporter mRNA with Shep. Error bars indicate the standard error of the mean, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant (Student’s t-test). Adapted from Perez et al. (2021).
Figure 14 shows how each regulation for Dsx1 expression is potentially interconnected. Adult parthenogenetic females detect environmental cues and convert them to sesquiterpenoid signaling that leads to the sexual commitment of oocytes from females to males. This hormone also leads to the activation of the Vrille gene at the gastrulation stage. Vrille may bind to the Dsx1 enhancer element and triggers activation of Dsx1, a master regulator of male development. Importantly, the active Dsx1 locus also produces lncRNA DAPALR that overlaps with Dsx1α 5′ UTR in the same orientation. With a potential Dsx binding site in the Dsx1 intron 2, Dsx1 possibly activates its own expression via a positive feedback loop, which may contribute to the maintenance of Dsx1 expression throughout development. Loss- and gain-of-function analyses of Dsx1 and its regulators such as Vrille, Shep, and DAPALR provided us evidence that ectopic Dsx1 expression in females resulted in intersex phenotypes. However, the sexual ambiguity of Daphnia is rare in nature, suggesting robust regulation of Dsx1 expression. To achieve stringent suppression of Dsx1 expression in females, Shep might be used for translational repression of unintentional Dsx1α mRNA by promoter leakage that may cause the autoregulation of Dsx1. In contrast, in males, DAPALR evicts Shep and unlocks Dsx1 translation. This post-transcriptional regulation may function as a fail-safe system by which Dsx1 expression is guaranteed and sexual ambiguity is avoided (Perez et al., 2021). It is also possible that the epigenetic state of the Dsx1 locus may be changed from closed to opened chromatin as a memory of the environmental information that the mother has experienced.
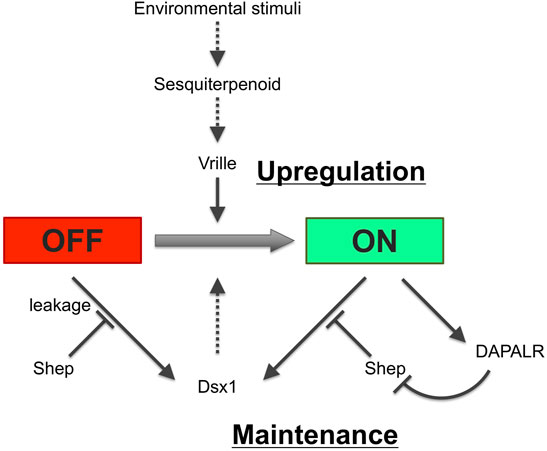
FIGURE 14. Potential regulatory network for Dsx1 expression. Solid and dotted lines indicate validated and non-validated relationships by experiments. Figure inspiration was drawn from Huang et al. (2015).
The mechanistic bases of the unique Dsx1 control system in the upregulation and maintenance phases during development would be an important future direction. Understanding the molecular pathway between the maternal sesquiterpenoid signaling and Dsx1 upregulation could not only provide us evolutionary insights into the diversity of upstream components in the sex determination cascades of animals but also lead to unraveling mechanisms of transgenerational regulation of development in response to environmental stimuli. As described in this review, the masculinized cells that migrate the ventral region in early embryos might be a key cell population that lets its surrounding cells know the sexual fate. For understanding how the male-specific Dsx1 expression is maintained, it would be important to clarify the positive-feedback regulation of Dsx1 and the factors involved in the epigenetic regulations. To prove the decoy model of DAPALR and Shep, the stoichiometry of the post-transcriptional regulators and the target Dsx1 mRNA must be investigated. For these future objectives and challenges, in vivo functional analysis of Dsx1 regulators in Daphnia must be moved from the individual level to tissue and cellular levels. By using CRISPR/Cas or TALEN-mediated knock-in (Nakanishi et al., 2015; Nakanishi et al., 2016; Kumagai et al., 2017), specific cell population needs to be fluorescently labeled, isolated, and subjected to single-cell analysis. This approach will allow us to move away from current analyses using heterogeneous cell populations and to identify Dsx1 regulators that control developmental stage-, tissue- and cell-specific traits at high resolution, which provides us a comprehensive understanding of Dsx1 regulation for environmental sex determination in D. magna.
YK wrote the initial manuscript draft and generated the figures and tables. HW revised the manuscript. Both authors approved the final article.
This work was supported by JSPS KAKENHI Grant Numbers 21K19298 (YK), 20H04923 (YK), 19H05423 (YK), 21H03602 (HW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all the members of the HW laboratory for their support.
Akiyama-Oda, Y., and Oda, H. (2003). Early Patterning of the Spider Embryo: a Cluster of Mesenchymal Cells at the Cumulus Produces Dpp Signals Received by Germ Disc Epithelial Cells. Development 130, 1735–1747. doi:10.1242/dev.00390
An, W., and Wensink, P. C. (1995). Integrating Sex- and Tissue-specific Regulation within a Single Drosophila Enhancer. Genes Dev. 9, 256–266. doi:10.1101/gad.9.2.256
Bachtrog, D., Mank, J. E., Peichel, C. L., Kirkpatrick, M., Otto, S. P., Ashman, T.-L., et al. (2014). Sex Determination: Why So many Ways of Doing it? Plos Biol. 12, e1001899–13. doi:10.1371/journal.pbio.1001899
Baker, B. S., and Wolfner, M. F. (1988). A Molecular Analysis of Doublesex, a Bifunctional Gene that Controls Both Male and Female Sexual Differentiation in D. melanogaster. Genes Dev. 2, 477–489. doi:10.1101/gad.2.4.477
Banta, A. M., and Brown, L. A. (1929). Control of Sex in Cladocera. III. Localization of the Critical Period for Control of Sex. Proc. Natl. Acad. Sci. U.S.A. 15, 71–81. doi:10.1073/pnas.15.2.71
Bayrer, J. R., Zhang, W., and Weiss, M. A. (2005). Dimerization of Doublesex Is Mediated by a Cryptic Ubiquitin-Associated Domain Fold. J. Biol. Chem. 280, 32989–32996. doi:10.1074/jbc.m507990200
Bénazéraf, B., and Pourquié, O. (2013). Formation and Segmentation of the Vertebrate Body axis. Annu. Rev. Cel Dev. Biol. 29, 1–26. doi:10.1146/annurev-cellbio-101011-155703
Burtis, K. C., and Baker, B. S. (1989). Drosophila Doublesex Gene Controls Somatic Sexual Differentiation by Producing Alternatively Spliced mRNAs Encoding Related Sex-specific Polypeptides. Cell 56, 997–1010. doi:10.1016/0092-8674(89)90633-8
Chen, X., Rinsma, M., Janssen, J. M., Liu, J., Maggio, I., and Gonçalves, M. A. F. V. (2016). Probing the Impact of Chromatin Conformation on Genome Editing Tools. Nucleic Acids Res. 44, 6482–6492. doi:10.1093/NAR/GKW524
Daer, R. M., Cutts, J. P., Brafman, D. A., and Haynes, K. A. (2016). The Impact of Chromatin Dynamics on Cas9-Mediated Genome Editing in Human Cells. ACS Synth. Biol. 6, 428–438. doi:10.1021/ACSSYNBIO.5B00299
Erdman, S. E., and Burtis, K. C. (1993). The Drosophila Doublesex Proteins Share a Novel Zinc finger Related DNA Binding Domain. EMBO J. 12, 527–535. doi:10.1002/j.1460-2075.1993.tb05684.x
Erdman, S. E., Chen, H.-J., and Burtis, K. C. (1996). Functional and Genetic Characterization of the Oligomerization and DNA Binding Properties of the Drosophila Doublesex Proteins. Genetics 144, 1639–1652. doi:10.1093/genetics/144.4.1639
Ge, C., Ye, J., Zhang, H., Zhang, Y., Sun, W., Sang, Y., et al. (2017). Dmrt1 Induces the Male Pathway in a Turtle with Temperature-dependent Sex Determination. Development 144, 2222–2233. doi:10.1242/DEV.152033
Hebert, P. D. N. (1978). The Population Bilogy of Daphnia (Crustacea, Daphnidae). Biol. Rev. 53, 387–426. doi:10.1111/j.1469-185x.1978.tb00860.x
Herpin, A., and Schartl, M. (2015). Plasticity of Gene‐regulatory Networks Controlling Sex Determination: of Masters, Slaves, Usual Suspects, Newcomers, and Usurpators. EMBO Rep. 16, 1260–1274. doi:10.15252/EMBR.201540667
Hildreth, P. E. (1965). Doublesex, a Recessive Gene that Tranforms Both Males and Females of Drosophila into Intersexes. Genetics 51, 659–678. doi:10.1093/GENETICS/51.4.659
Huang, L., Yuan, Z., Liu, P., and Zhou, T. (2015). Effects of Promoter Leakage on Dynamics of Gene Expression. BMC Syst. Biol. 9, 16. doi:10.1186/S12918-015-0157-Z
Jensen, K. T., Fløe, L., Petersen, T. S., Huang, J., Xu, F., Bolund, L., et al. (2017). Chromatin Accessibility and Guide Sequence Secondary Structure Affect CRISPR-Cas9 Gene Editing Efficiency. FEBS Lett. 591, 1892–1901. doi:10.1002/1873-3468.12707
Kato, Y., Kobayashi, K., Oda, S., Tatarazako, N., Watanabe, H., and Iguchi, T. (2010). Sequence Divergence and Expression of a Transformer Gene in the Branchiopod Crustacean, Daphnia magna. Genomics 95, 160–165. doi:10.1016/j.ygeno.2009.12.00510.1016/j.ygeno.2009.12.005
Kato, Y., Kobayashi, K., Watanabe, H., and Iguchi, T. (2011). Environmental Sex Determination in the Branchiopod Crustacean Daphnia magna: Deep Conservation of a Doublesex Gene in the Sex-Determining Pathway. Plos Genet. 7, e1001345. doi:10.1371/journal.pgen.1001345
Kato, Y., Perez, C. A. G., Mohamad Ishak, N. S., Nong, Q. D., Sudo, Y., Matsuura, T., et al. (2018). A 5′ UTR-Overlapping LncRNA Activates the Male-Determining Gene Doublesex1 in the Crustacean Daphnia magna. Curr. Biol. 28, 1811–1817. doi:10.1016/j.cub.2018.04.029
Korpelainen, H. (1990). Sex Ratios and Conditions Required for Environmental Sex Determination in Animals. Biol. Rev. 65, 147–184. doi:10.1111/j.1469-185x.1990.tb01187.x
Kumagai, H., Nakanishi, T., Matsuura, T., Kato, Y., and Watanabe, H. (2017). CRISPR/Cas-mediated Knock-In via Non-homologous End-Joining in the Crustacean Daphnia magna. PLoS One 12, e0186112. doi:10.1371/journal.pone.0186112
Kvist, J., Athanàsio, C. G., Pfrender, M. E., Brown, J. B., Colbourne, J. K., and Mirbahai, L. (2020). A Comprehensive Epigenomic Analysis of Phenotypically Distinguishable, Genetically Identical Female and Male Daphnia pulex. BMC Genomics 21, 1–17. doi:10.1186/S12864-019-6415-5
Lebo, M. S., Sanders, L. E., Sun, F., and Arbeitman, M. N. (2009). Somatic, Germline and Sex Hierarchy Regulated Gene Expression during Drosophila Metamorphosis. BMC Genomics 10, 80. doi:10.1186/1471-2164-10-80
Martin, B. L., and Kimelman, D. (2009). Wnt Signaling and the Evolution of Embryonic Posterior Development. Curr. Biol. 19, R215–R219. doi:10.1016/j.cub.2009.01.052
Matsuda, M., Nagahama, Y., Shinomiya, A., Sato, T., Matsuda, C., Kobayashi, T., et al. (2002). DMY Is a Y-specific DM-Domain Gene Required for Male Development in the Medaka Fish. Nature 417, 559–563. doi:10.1038/NATURE751
Mohamad Ishak, N. S., Nong, Q. D., Matsuura, T., Kato, Y., and Watanabe, H. (2017). Co-option of the bZIP Transcription Factor Vrille as the Activator of Doublesex1 in Environmental Sex Determination of the Crustacean Daphnia magna. Plos Genet. 13, e1006953. doi:10.1371/journal.pgen.1006953
Nagoshi, R. N., McKeown, M., Burtis, K. C., Belote, J. M., and Baker, B. S. (1988). The Control of Alternative Splicing at Genes Regulating Sexual Differentiation in D. melanogaster. Cell 53, 229–236. doi:10.1016/0092-8674(88)90384-4
Nakanishi, T., Kato, Y., Matsuura, T., and Watanabe, H. (2015). TALEN-mediated Homologous Recombination in Daphnia magna. Sci. Rep. 5, 18312. doi:10.1038/srep18312
Nakanishi, T., Kato, Y., Matsuura, T., and Watanabe, H. (2016). TALEN-mediated Knock-In via Non-homologous End Joining in the Crustacean Daphnia magna. Sci. Rep. 6, 36252. doi:10.1038/srep36252
Nong, Q. D., Matsuura, T., Kato, Y., and Watanabe, H. (2020). Two Doublesex1 Mutants Revealed a Tunable Gene Network Underlying Intersexuality in Daphnia magna. PLoS One 15, e0238256. doi:10.1371/JOURNAL.PONE.0238256
Nong, Q. D., Mohamad Ishak, N. S., Matsuura, T., Kato, Y., and Watanabe, H. (2017). Mapping the Expression of the Sex Determining Factor Doublesex1 in Daphnia magna Using a Knock-In Reporter. Sci. Rep. 7, 13521. doi:10.1038/s41598-017-13730-4
Olmstead, A. W., and Leblanc, G. A. (2002). Juvenoid Hormone Methyl Farnesoate Is a Sex Determinant in the crustaceanDaphnia magna. J. Exp. Zool. 293, 736–739. doi:10.1002/jez.10162
Perez, C. A. G., Adachi, S., Nong, Q. D., Adhitama, N., Matsuura, T., Natsume, T., et al. (2021). Sense-overlapping lncRNA as a Decoy of Translational Repressor Protein for Dimorphic Gene Expression. Plos Genet. 17, e1009683. doi:10.1371/journal.pgen.1009683
Raymond, C. S., Murphy, M. W., O'Sullivan, M. G., Bardwell, V. J., and Zarkower, D. (2000). Dmrt1, a Gene Related to Worm and Fly Sexual Regulators, Is Required for Mammalian Testis Differentiation. Genes Dev. 14, 2587–2595. doi:10.1101/gad.834100
Shen, M. M., and Hodgkin, J. (1988). mab-3, a Gene Required for Sex-specific Yolk Protein Expression and a Male-specific Lineage in C. elegans. Cell 54, 1019–1031. doi:10.1016/0092-8674(88)90117-1
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The Avian Z-Linked Gene DMRT1 Is Required for Male Sex Determination in the Chicken. Nature 461, 267–271. doi:10.1038/nature08298
Tatarazako, N., Oda, S., Watanabe, H., Morita, M., and Iguchi, T. (2003). Juvenile Hormone Agonists Affect the Occurrence of Male Daphnia. Chemosphere 53, 827–833. doi:10.1016/S0045-6535(03)00761-6
Keywords: Doublesex1, environmental sex determination, Daphnia magna, transcriptional regulation, post-transcriptional regulation, epigenetic regulation, male development
Citation: Kato Y and Watanabe H (2022) Regulation of Doublesex1 Expression for Environmental Sex Determination in the Cladoceran Crustacean Daphnia. Front. Cell Dev. Biol. 10:881255. doi: 10.3389/fcell.2022.881255
Received: 22 February 2022; Accepted: 22 March 2022;
Published: 13 April 2022.
Edited by:
Soichiro Yamanaka, The University of Tokyo, JapanReviewed by:
Masataka Gakyo Suzuki, The University of Tokyo, JapanCopyright © 2022 Kato and Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuhiko Kato, a2F0b195YXN1aGlrb0BiaW8uZW5nLm9zYWthLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.